In recent years, the worldwide spread of the so-called high-risk clones of multidrug-resistant or extensively drug-resistant (MDR/XDR) Pseudomonas aeruginosa has become a public health threat. This article reviews their mechanisms of resistance, epidemiology, and clinical impact and current and upcoming therapeutic options. In vitro and in vivo treatment studies and pharmacokinetic and pharmacodynamic (PK/PD) models are discussed.
KEYWORDS: Pseudomonas aeruginosa
SUMMARY
In recent years, the worldwide spread of the so-called high-risk clones of multidrug-resistant or extensively drug-resistant (MDR/XDR) Pseudomonas aeruginosa has become a public health threat. This article reviews their mechanisms of resistance, epidemiology, and clinical impact and current and upcoming therapeutic options. In vitro and in vivo treatment studies and pharmacokinetic and pharmacodynamic (PK/PD) models are discussed. Polymyxins are reviewed as an important therapeutic option, outlining dosage, pharmacokinetics and pharmacodynamics, and their clinical efficacy against MDR/XDR P. aeruginosa infections. Their narrow therapeutic window and potential for combination therapy are also discussed. Other “old” antimicrobials, such as certain β-lactams, aminoglycosides, and fosfomycin, are reviewed here. New antipseudomonals, as well as those in the pipeline, are also reviewed. Ceftolozane-tazobactam has clinical activity against a significant percentage of MDR/XDR P. aeruginosa strains, and its microbiological and clinical data, as well as recommendations for improving its use against these bacteria, are described, as are those for ceftazidime-avibactam, which has better activity against MDR/XDR P. aeruginosa, especially strains with certain specific mechanisms of resistance. A section is devoted to reviewing upcoming active drugs such as imipenem-relebactam, cefepime-zidebactam, cefiderocol, and murepavadin. Finally, other therapeutic strategies, such as use of vaccines, antibodies, bacteriocins, anti-quorum sensing, and bacteriophages, are described as future options.
INTRODUCTION
There are several major reasons why the emergence and dissemination of multidrug-resistant (MDR) and extensively drug-resistant (XDR) Pseudomonas aeruginosa strains have recently become issues of public health concern. First, P. aeruginosa causes severe infections, particularly in health care settings and in immunocompromised patients. Second, it has an outstanding capacity for being selected and for spreading antimicrobial resistance in vivo (1, 2). Third, the successful worldwide spread of the so-called “high-risk” clones of P. aeruginosa poses a threat to global public health that needs to be studied and managed with urgency and determination (3).
The lack of therapeutic alternatives means that infections caused by these antibiotic-resistant bacteria pose a considerable threat regarding morbidity and mortality worldwide. The impact of inadequate therapy in these infections is significant; indeed, the World Health Organization reported in 2017 that carbapenem-resistant P. aeruginosa was listed in the “critical” group for which new antibiotics were urgently required (4).
Recent years have witnessed an increasing prevalence of MDR and XDR P. aeruginosa strains, with rates of between 15% and 30% in some geographical areas (5–7). Most countries in Europe report rates of resistance of more than 10% for all antimicrobial groups under surveillance (8). Combined resistance is also common in P. aeruginosa. In 2015, the European Centers for Disease Prevention and Control stated that 13.7% of P. aeruginosa isolates were resistant to at least three antimicrobial groups and 5.5% to all five antimicrobial groups under surveillance (EARS-Net) (8). According to data from the United States, MDR P. aeruginosa is the cause of 13% of severe health care-associated infections (9).
The solutions to this crisis are to allocate more resources to basic and clinical research and to infection control and antimicrobial stewardship, to develop new antimicrobials, and to optimize the use of those that are currently available. This article reviews the current definitions and mechanisms of multidrug resistance in P. aeruginosa and the epidemiology of high-risk clones disseminated worldwide. Based on the information available, current and upcoming therapeutic options are reviewed, including clinical studies and, where these are lacking, in vitro and animal studies. It should be noted that most clinical studies have methodological limitations and that interpretation of the evidence is difficult.
OVERVIEW OF P. AERUGINOSA RESISTANCE MECHANISMS
Intrinsic P. aeruginosa Resistance (Intrinsic Resistome)
P. aeruginosa has a remarkable array of mechanisms of antibiotic resistance in its arsenal, including multiple chromosomal determinants as well as the complex regulatory pathways involved in intrinsic and adaptive resistance (1, 2, 10–13). The mechanisms thought to have the greatest effect on the lower natural susceptibility of P. aeruginosa compared to other Gram-negative microorganisms are inducible AmpC cephalosporinase expression, constitutive (MexAB-OprM) and inducible (MexXY) efflux pump production, and low outer membrane permeability. Since the aminopenicillins and a number of cephalosporins (cefoxitin, in particular) are strong inducers of expression and are also efficiently hydrolyzed by AmpC, inducible β-lactamase production has a key role in the natural resistance of P. aeruginosa to these agents. Inducible AmpC expression plays a decisive role in the natural reduced susceptibility of P. aeruginosa to imipenem, since the hydrolytic stability of this antibiotic is to some degree affected by its high inducer potency. Two other chromosomal β-lactamases, the OXA enzyme OXA-50/PoxB (14, 15) and the more recently described imipenemase (PA5542) (16), may also have an impact on intrinsic β-lactam susceptibility levels, although their role in intrinsic and/or acquired resistance requires further elucidation. Constitutive expression of the MexAB-OprM efflux pump plays a major role in lower basal levels of susceptibility to the vast majority of β-lactams (except for imipenem) and fluoroquinolones, whereas inducible production of MexXY has a major effect on the intrinsic low-level resistance to aminoglycosides (17). In addition to these well-known resistance determinants, an analysis of mutant libraries resulting from whole-genome screening has revealed a large set of genes, referred to collectively as the intrinsic resistome, which have an effect on antibiotic susceptibility (2, 16, 18, 19).
Acquisition of Resistance through Chromosomal Gene Mutations (Mutational Resistome)
Apart from its vast intrinsic resistome, P. aeruginosa shows an outstanding ability to develop further antimicrobial resistance to all available antibiotics via the acquisition of chromosomal mutations. Table 1 provides a summary of the main genes known to increase resistance levels and thus shape the P. aeruginosa mutational resistome (3).
TABLE 1.
Main genes known to be involved in P. aeruginosa mutational antibiotic resistance
| Gene(s) | Resistance mechanism | Antibiotics affected |
|---|---|---|
| gyrA | Quinolone target modification (DNA gyrase) | Fluoroquinolones |
| gyrB | Quinolone target modification (DNA gyrase) | Fluoroquinolones |
| parC | Quinolone target modification (DNA topoisomerase IV) | Fluoroquinolones |
| parE | Quinolone target modification (DNA topoisomerase IV) | Fluoroquinolones |
| pmrA, pmrB, phoQ, cprS, colR, colS | Lipopolysaccharide modification (addition of the 4-amino-4-deoxy-l-arabinose moiety to the lipid A portion) | Polymyxins |
| parR | Lipopolysaccharide modification (addition of the 4-amino-4-deoxy-l-arabinose moiety to the lipid A portion) | Polymyxins |
| OprD downregulation | Imipenem, meropenem | |
| MexEF-OprN hyperproduction | Fluoroquinolones | |
| MexXY hyperproduction | Fluoroquinolones, aminoglycosides, cefepime | |
| parS | Lipopolysaccharide modification (addition of the 4-amino-4-deoxy-l-arabinose moiety to the lipid A portion) | Polymyxins |
| OprD downregulation | Imipenem, meropenem | |
| MexEF-OprN hyperproduction | Fluoroquinolones | |
| MexXY hyperproduction | Fluoroquinolones, aminoglycosides, cefepime | |
| mexR, nalC, nalD | MexAB-OprM hyperproduction | Fluoroquinolones, ceftazidime, cefepime, piperacillin-tazobactam, meropenem, ceftazidime-avibactam |
| nfxB | MexCD-OprJ hyperproduction | Fluoroquinolones, cefepime |
| mexS | MexEF-OprN hyperproduction | Fluoroquinolones |
| OprD downregulation | Imipenem, meropenem | |
| mexT | MexEF-OprN hyperproduction | Fluoroquinolones |
| OprD downregulation | Imipenem, meropenem | |
| cmrA, mvaT, PA3271 | MexEF-OprN hyperproduction | Fluoroquinolones |
| mexZ, PA5471.1, amgS | MexXY hyperproduction | Fluoroquinolones, aminoglycosides, cefepime |
| oprD | OprD porin inactivation | Imipenem, meropenem |
| ampC | AmpC structural modification | Ceftolozane-tazobactam, ceftazidime-avibactam |
| ampD, ampDh2, ampDh3, ampR, dacB, mpl | AmpC hyperproduction | Ceftazidime, cefepime, piperacillin-tazobactam |
| ftsI | β-Lactam target modification (PBP3) | Ceftazidime, cefepime, piperacillin-tazobactam, ceftolozane-tazobactam, ceftazidime-avibactam, meropenem |
| fusA1 | Aminoglycoside target modification (elongation factor G) | Aminoglycosides |
| glpT | Inactivation of transporter protein GlpT | Fosfomycin |
| rpoB | Rifampin target modification, RNA polymerase β-chain | Rifampin |
Overproduction of chromosomal AmpC cephalosporinase, involving a broad range of genes belonging to the complex regulatory cell wall recycling pathways, is probably the most common mutation-driven β-lactam resistance mechanism. It has been detected in over 20% of P. aeruginosa clinical isolates (13, 20, 21). Mutational inactivations of dacB (which encodes PBP4) and ampD (which encodes an N-acetylmuramyl-l-alanine amidase) are known to be the most common mechanisms of ampC hyperproduction and β-lactam resistance (21, 22). Inactivation of PBP4 has also been demonstrated to activate the CreBC/BlrAB two-component system, increasing resistance levels further (21). Specific mutations leading to modification of the conformation of the transcriptional regulator AmpR, which regulates ampC overexpression and β-lactam resistance, have also been detected in clinical strains. These mutations include D135N, documented in species other than P. aeruginosa, and the R154H mutation, associated with the epidemic MDR/XDR ST175 high-risk clone (13). Mutations in various other genes have been found to upregulate ampC, including those encoding other amidases (AmpDh2/AmpDh3), other penicillin-binding proteins (PBP5 or PBP7), lytic transglycosylases (MltB and SltB1), MPL (UDP-N-acetylmuramate:l-alanyl-γ-d-glutamyl-meso-diaminopimelate ligase), and NuoN (NADH dehydrogenase I chain N). Nevertheless, further analysis of their effect on β-lactam resistance in natural strains is still required (13).
Apart from AmpC hyperproduction, recent studies have highlighted the fact that mutations leading to the structural modification of AmpC may be the cause of resistance to β-lactams, including the novel β-lactam–β-lactamase inhibitor combinations ceftolozane-tazobactam and ceftazidime-avibactam (23–26). Another study detected several amino acid variants in AmpC in a small proportion (approximately 1%) of P. aeruginosa clinical isolates that were linked to ceftolozane-tazobactam and ceftazidime-avibactam resistance (27). To date, over 300 Pseudomonas-derived cephalosporinase (PDC) variants have been reported, some of which confer increased ceftolozane-tazobactam and ceftazidime-avibactam resistance. An updated database of PDC variants is available free from Antonio Oliver’s laboratory at https://arpbigidisba.com. In addition to β-lactamases, there is growing evidence of the role of PBP modification in β-lactam resistance, especially mutations in PBP3 (encoded by ftsI). Recent data from cystic fibrosis (CF) patients (28, 29), epidemic strains (30, 31), and in vitro studies (32, 33) have shown that specific mutations in PBP3 play a role in the emergence of β-lactam resistance. Those most frequently reported are R504C/R504H and F533L, located in domains involved in the stabilization of the β-lactam–PBP3 inactivation complex (34).
Loss of the carbapenem-specific porin OprD may be the result either of inactivating mutations/insertion sequences in the oprD gene or of remote mutations that upregulate efflux system MexEF-OprN or CzcCBA with concomitant downregulation of oprD expression. Mutational inactivation or downregulation of the OprD porin (along with inducible AmpC production) drives imipenem resistance and decreased meropenem susceptibility. The prevalence of imipenem resistance is frequently above 20%, and most of the isolates involved are OprD deficient (20, 35). OprD inactivation frequently acts synergistically with AmpC overexpression to drive resistance to all the classic antipseudomonal β-lactams (36). Mutational overexpression of one of the four major efflux pumps of P. aeruginosa also plays a major role in mutation-driven resistance (17, 20, 37, 38). Overexpression of MexAB-OprM and MexXY is common (10% to 30%) among clinical isolates, whereas the prevalence of MexCD-OprJ and MexEF-OprN overexpression is considerably lower (<5%). MexAB-OprM has the widest substrate spectrum, and mutation-driven overexpression of this efflux pump results in reduced susceptibility to fluoroquinolones and all β-lactams (except imipenem). The combination of MexAB-OprM overexpression and OprD inactivation is one of the major causes of resistance to meropenem among clinical strains (35). Apart from its role in intrinsic aminoglycoside resistance, mutation-driven hyperproduction of MexXY is a common driver of resistance to cefepime in clinical strains (39). MexCD-OprJ or MexEF-OprN hyperproduction is less prevalent and mainly affects fluoroquinolones, although the mutations (mexT/mexS) that drive MexEF-OprN hyperproduction also determine resistance to imipenem due to the repression of oprD (40). Overexpression of MexCD-OprJ, which is particularly prevalent in chronic infections, also drives increased cefepime MICs, despite determining increased susceptibility to several β-lactams and aminoglycosides (41).
Apart from efflux pump overexpression, P. aeruginosa fluoroquinolone resistance frequently arises from mutations in DNA gyrases (GyrA and GyrB) and type IV topoisomerases (ParC and ParE) (42). The prevalence of fluoroquinolone resistance varies according to geography but is over 30 to 40% in multiple countries. Studies have recently shown that, in addition to MexXY overexpression and horizontally acquired mechanisms (see below), aminoglycoside resistance may result from mutations in fusA1, encoding elongation factor G, and indeed, specific FusA1 mutations have been shown to confer aminoglycoside resistance in vitro (43, 44) and in clinical strains, particularly among CF patients (29, 44, 45). The role of specific fusA1 mutations in resistance has also been demonstrated using site-directed mutagenesis (46).
Finally, while the prevalence of colistin resistance remains relatively low (<5%), it has grown recently, possibly because of increased use of colistin as a last-resort agent for the treatment of infections caused by MDR/XDR strains. Colistin resistance frequently results from the modification of the lipid A moiety of lipopolysaccharide (LPS) following the addition of 4-amino-4-deoxy-l-arabinose (47). The mutations involved are frequently linked to the two-component regulatory systems PmrAB and PhoPQ, which lead to activation of the arnBCADTEF operon. More recently, it has been shown that mutations in the ParRS two-component regulator not only drive colistin resistance by activating the arnBCADTEF operon but also lead to an MDR profile through overexpression of MexXY and downregulation of OprD (12). Two other two-component regulators (ColRS and CprRS) are also known to be involved in polymyxin resistance (48).
Horizontally Acquired Resistance Mechanisms (Horizontally Acquired Resistome)
In addition to mutational resistance, which is relatively frequent, transferable resistance in P. aeruginosa is another area of increasing concern. Indeed, there is a growing prevalence worldwide of the most troublesome of transferable β-lactamases, the extended-spectrum β-lactamases (ESBLs) and carbapenemases (especially class B carbapenemases, or metallo-β-lactamases [MBLs]), although the distribution is not uniform and ranges from below 1% to nearly 50%, depending on the hospital and geographic area (49). Furthermore, the challenge of detecting transferable β-lactamases in P. aeruginosa may mean that their prevalence has been underestimated in several areas (50). The genes encoding ESBLs and carbapenemases are generally found in class 1 integrons along with determinants of aminoglycoside resistance. These integrons are often inserted into transposable elements located on the bacterial chromosome, although the involvement of conjugative elements is increasingly reported (51–54). Transferable β-lactamases detected so far in P. aeruginosa were recently reviewed by Potron et al. (55). The most frequently reported ESBLs in P. aeruginosa include those in class D (such as OXA-2 or OXA-10 variants) and class A (PER, VEB, GES, BEL, and PME). Class A ESBLs typically documented in the order Enterobacteriales (such as TEM, SHV, or CTX-M β-lactamases) are infrequently documented in P. aeruginosa. With respect to the carbapenemases, MBLs are by far the most prevalent in P. aeruginosa, with the VIM and IMP types being the most frequent and the most geographically widespread. The SPM MBL is prevalent in Brazil, and NDM, GIM, and FIM are detected only occasionally. Finally, the worldwide prevalence of class A carbapenemases in P. aeruginosa is low, although GES and KPC enzymes have been detected in several countries (54).
Transferable aminoglycoside resistance is most frequently driven by aminoglycoside-modifying enzymes encoded in class 1 integrons. Those most commonly described in P. aeruginosa are acetyltransferases from the AAC(3′) (gentamicin) and AAC(6′) (tobramycin including amikacin or not) groups and nucleotidyltransferase ANT(2′)-I (gentamicin and tobramycin) (1). Nevertheless 16S rRNA methyltransferases (such as Rmt or Arm), which confer resistance to all aminoglycosides on the market, including the novel plazomicin, also represent major emerging threats (55). Transferable fluoroquinolone resistance driven mainly by Qnr determinants such as QnrVC1 has occasionally been detected (56). A very recent study has also reported the occurrence of plasmid-mediated quinolone resistance apparently driven by a novel phosphotransferase, CrpP (57).
The novel combinations ceftolozane-tazobactam and ceftazidime-avibactam are known to be relatively stable against AmpC hydrolysis (58, 59), relying on the stability of ceftolozane against hydrolysis by AmpC in the case of ceftolozane-tazobactam and on the inhibitory activity of avibactam against AmpC in the case of ceftazidime-avibactam. However, recent in vitro and in vivo data indicate that the development of resistance to both agents may be the result of a combination of mutations leading to hyperproduction and the structural modification of AmpC (23, 25–27). Available in vitro and in vivo data also suggest that specific PBP3 mutations may reduce susceptibility to both combinations. On the other hand, overexpression of different efflux pumps seems to affect ceftazidime-avibactam susceptibility more than that of ceftolozane-tazobactam (27, 60).
With respect to acquired β-lactamases, neither ceftolozane-tazobactam nor ceftazidime-avibactam shows activity against MBL-producing strains. However, ceftazidime-avibactam, but not ceftolozane-tazobactam, may show activity against isolates producing class A carbapenemases such as GES enzymes (61). Likewise, the activity of ceftolozane-tazobactam and ceftazidime-avibactam against ESBL-producing P. aeruginosa isolates is variable, but it is generally favorable in the case of ceftazidime-avibactam. Finally, extended-spectrum mutations in horizontally acquired OXA-type β-lactamases may lead to the emergence of resistance to both agents (25, 62, 63).
EPIDEMIOLOGY OF MULTIDRUG-RESISTANT P. AERUGINOSA: DEFINITIONS AND PREVALENCE
Over the last decades, various definitions of MDR P. aeruginosa profiles have been used, although the consensus definition that is probably most widely used at present is the one published by Magiorakos et al. (64) in 2012. Multidrug resistance (MDR) was defined as nonsusceptibility (intermediate plus resistant [I+R]) to at least one agent in at least 3 antibiotic classes, extensive drug resistance (XDR) as nonsusceptibility to at least one agent in all but 1 or 2 antibiotic classes, and pan-drug resistance (PDR) as nonsusceptibility to all agents in all classes. The following classes and antibiotics were recommended for testing: antipseudomonal cephalosporins (ceftazidime and cefepime), antipseudomonal penicillins plus β-lactamase inhibitors (ticarcillin-clavulanate and piperacillin-tazobactam), monobactams (aztreonam), antipseudomonal carbapenems (imipenem, meropenem, and doripenem), aminoglycosides (gentamicin, tobramycin, amikacin, and netilmicin), fluoroquinolones (ciprofloxacin and levofloxacin), phosphonic acids (fosfomycin), and polymyxins (colistin and polymyxin B). While this proposal was certainly useful for the harmonization of definitions of P. aeruginosa resistance profiles, several other aspects remain to be considered. First, even if a single definition is used, the result will vary depending on whether EUCAST or CLSI breakpoints are used. Second, the comprehensive application of the proposed definition is limited by the lack of clinical breakpoints (both CLSI and EUCAST) for one of the agents (fosfomycin). Similarly, until 2019, EUCAST breakpoints for aztreonam considered P. aeruginosa intrinsically nonsusceptible to this agent and therefore not applicable to MDR/XDR/PDR definitions based on acquired resistance. Finally, the current definition does not consider recently introduced antipseudomonal agents such as ceftazidime-avibactam or ceftolozane-tazobactam.
Regardless of the question of definitions mentioned above and the mechanisms involved, the prevalence of MDR P. aeruginosa is probably increasing worldwide, although with major geographical differences. The prevalence of MDR P. aeruginosa has increased over the last few decades and is now within the 15 to 30% range in multiple areas (5–7, 56). Furthermore, a significant proportion of MDR strains also meet the criteria for classification as XDR, which further restricts the treatment options available. As an example, a recent (2017) large-scale (51 hospitals) multicenter study of P. aeruginosa infections performed in Spain showed that 26% of isolates were MDR and 65% of those (17% of all isolates) met the criteria for XDR, and most were susceptible only to colistin including amikacin or not (56). Indeed, in many hospitals worldwide, colistin-only-sensitive (COS) profiles are not uncommon and pan-drug resistance has already been documented (65, 66). However, resistance to the novel antipseudomonal agents ceftolozane-tazobactam and ceftazidime-avibactam was not considered in most of these studies. While the overall prevalence of resistance to these new therapeutic options is below 10%, there is considerable geographical variation depending on the prevalence of acquired β-lactamases such as ESBLs or carbapenemases (31, 66–70).
Epidemic High-Risk Clones
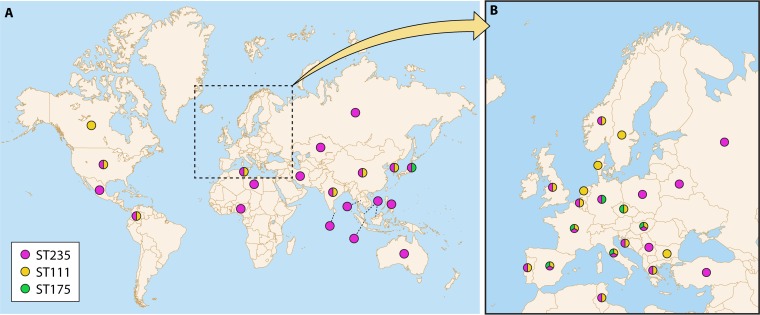
An analysis of the molecular epidemiology of P. aeruginosa isolates obtained from hospital-acquired infections, CF patients, or the environment typically reveals high clonal diversity, with most isolates being linked to unique genotypes, However, a closer look shows that this is true for antibiotic-susceptible isolates but not for those showing MDR/XDR phenotypes. Indeed, there have been multiple epidemic outbreak reports and alerts of MDR/XDR strains in the hospital environment for decades. More recent studies have provided further evidence of the MDR/XDR global clones, referred to as “high-risk” clones, disseminated in several hospitals worldwide (71). P. aeruginosa high-risk clones were recently reviewed (3). A map of the worldwide distribution of the most prevalent high-risk clones is provided in Fig. 1 and a summary of their characteristics in Table 2.
FIG 1.
World distribution (A) and European distribution (B) of ST235, ST111, and ST175 based on published data. (Reproduced from reference 3 with permission from Elsevier.)
TABLE 2.
Characteristics of the three major global P. aeruginosa high-risk clones
| Characteristic | ST111 | ST175 | ST235 |
|---|---|---|---|
| O-antigen serotype | O12 | O4 | O11 |
| Type III secretion system | ExoS | ExoS | ExoU |
| Virulencea | ++ | + | +++ |
| Worldwide distribution | ++ | + | +++ |
| Transferable resistance | ++ | + | +++ |
| Mutational resistance | ++ | +++ | ++ |
With regard to the prevalence and impact of high-risk clones, a 2008–2009 multicenter study of P. aeruginosa bacteremia carried out in Spain revealed that the vast majority of susceptible isolates were represented by single genotypes but that clonal diversity was much lower among MDR and, especially, XDR strains (72, 73). Seventy-three of 81 (90%) XDR isolates in fact were found to belong to just 3 clones, which were those of the major international MDR/XDR high-risk clones: ST175 (62), ST111 (9), and ST235 (5, 10). In a multicenter study performed 7 years later, ST175 continued to be the most common high-risk clone (68% of XDR isolates) (31). The same pattern was found in several studies worldwide, with most of the MDR/XDR isolates being linked to these and a few other clones (74–78). Of the three major high-risk clones, ST235, associated with serotype O11, is without doubt the most widespread, being found in many countries across all five continents (3, 79). ST111 (serotype O12) also has a worldwide distribution and has so far been documented on every continent except Oceania. Finally, ST175 (serotype O4) is widely distributed in several European countries. Interestingly, even though susceptible isolates have probably been studied less than MDR isolates, the available information suggests that these clones are infrequent among susceptible isolates. Apart from the 3 major high-risk clones, ST277 is of particular significance, being widely disseminated in Brazil (79). ST244 is also frequently detected in several countries but is not always linked to MDR/XDR profiles (72, 80). Other recently reported emerging high-risk clones include ST308 and ST395 (81, 82).
The link between high-risk clones and horizontally acquired resistance mechanisms is overwhelming, and most ESBL- or MBL-producing P. aeruginosa isolates belong to a few clones, with ST235 being the most frequent, followed by ST111 (3). A recent genomic analysis suggested that the specific presence in ST235 of DprA, a determinant involved in homologous recombination present in transformable species, likely increases the ability of this high-risk clone to acquire and maintain foreign resistance elements at a higher rate than other P. aeruginosa clones (79). A significant relationship between high-risk clones and mutation-driven resistance mechanisms has also been reported. For example, the mechanisms responsible for the XDR phenotype of the ST175 clone, which is widespread in Spanish and French hospitals, are combinations of specific mutations in AmpR (G154R), OprD (Q142X), and MexZ (G195E) and 3 quinolone-resistance determining region (QRDR) mutations (GyrA T83I and D87N and ParC S87W) (30). An analysis of the resistomes of large worldwide collections of P. aeruginosa strains also showed that mutation-driven mechanisms were frequent among ST111 and ST235 clones; most of them had QRDR mutations (frequently GyrA T83I and ParC S87L) and often showed a mutated oprD (31, 83, 84).
The pathogenicity of epidemic high-risk clones is another major issue that should be taken into account (85). Current evidence suggests that virulence among the different high-risk clones is variable. We considered virulence as the capacity to produce more severe infections and/or higher mortality in acute infections according to results with animal models and clinical experience (5, 61, 66, 86, 87). These studies specifically show that the ExoU+ ST235 high-risk clone is highly virulent and associated with very high mortality, whereas the virulence of ST175 appears to be particularly low. Apart from virulence, determining which factors drive the success of the high-risk clones is another major issue that certainly needs clarification. A recent study evaluated a panel of eight biological characteristics potentially associated with the success of these clones (73). Surprisingly, the three major high-risk clones (ST111, ST175, and ST235) were found to be defective in the three types of motility and pigment (pyoverdine and pyocyanin) production and also showed reduced fitness in vitro. On the other hand, high-risk clones displayed increased spontaneous mutant frequencies and biofilm growth. Other recent studies have demonstrated enhanced biofilm formation and decreased motility in high-risk clones (88). Hence, there are similarities between these biological markers defined for P. aeruginosa high-risk clones and those typically resulting from adaptation to chronic infection (73). Nevertheless, further analysis, including information from whole-genome sequencing (WGS) (30, 31, 73, 84, 89, 90), is needed for a more complete understanding of the factors driving the success of high-risk clones.
CLINICAL IMPACT OF MULTIDRUG RESISTANCE IN P. AERUGINOSA
One of the main consequences of multidrug resistance is the difficulty of selecting an appropriate empirical antibiotic treatment. Patients with infections due to MDR/XDR pathogens are at an increased risk of receiving inadequate initial antimicrobial therapy (91, 92). Delays in receipt of effective antibiotic therapy are associated with worse outcomes and higher mortality rates in patients with P. aeruginosa bloodstream infections (93–96), and MDR/XDR patterns are also associated with a greater likelihood of inadequate empirical treatment in these infections (97–100). Furthermore, directed therapies used for MDR/XDR infections are usually second- or third-line antimicrobial agents and are thus less effective than those used to treat infections caused by susceptible strains (85). Nevertheless, the direct relationship between multidrug resistance and clinical outcome remains unclear (97, 101, 102). Although it is generally assumed that infections caused by MDR bacteria are associated with poor outcomes (94, 97, 103–105), infection outcomes depend not only on delays in receiving adequate antimicrobial therapy or use of suboptimal directed therapy but also on factors associated with the host or the pathogen (5, 85, 98, 106–108). With respect to the host, MDR/XDR P. aeruginosa colonization and infection usually occur in patients with multiple underlying diseases, which may explain the worse outcome (109, 110). Consequently, mortality in these patients may be due to severe preexisting comorbidities (111, 112). With respect to the pathogen, the biological implications of antibiotic resistance for virulence in P. aeruginosa is currently a hot topic (85, 86). It is generally assumed that acquisition of resistance mechanisms is associated with fitness costs that lead to decreased virulence in MDR/XDR strains (86, 113–117). Nevertheless, it has also been reported that some resistance mutations are not associated with fitness costs (115, 118), and other reports have claimed that MDR strains would be able to develop compensatory or suppressor mutations that allow them to recover their initial fitness so that, in the end, they do not lose virulence (113–115, 119). As mentioned previously, P. aeruginosa has a large number of virulence factors (5, 85, 108, 120). One of the most important virulence determinants is the type III secretion system (TTSS) (5, 120–122), which injects effector cytotoxins (ExoS, ExoT, ExoU, and/or ExoT) into the host cells (5, 120, 122). ExoU is the most potent of the four effector exotoxins identified, and its expression correlates with a poor prognosis (5, 85, 120–123). A recent clinical study conducted in patients with P. aeruginosa bacteremia demonstrated that the exoU+ genotype was associated with increased early mortality and suggested that it would be a useful prognostic biomarker in P. aeruginosa infections (5). Apart from the TTSS, other virulence determinants of P. aeruginosa have recently been described, such as toxin ExlA, which induces plasma membrane disruption of host cells, conferring increased virulence to the bacterium (108, 124). The HigB/HigA toxin/antitoxin system may influence some virulence factors of P. aeruginosa, such as pyocyanin, swarming, and biofilm formation (125). With respect to the impact of multidrug resistance on the virulence of P. aeruginosa, Peña et al. (5) found an association between some TTSS genotypes and antibiotic resistance patterns, with the exoU+ genotype being less frequent in MDR strains (5). Other studies also suggest an association between the TTSS and certain resistance profiles (115, 122, 123). The exoU+ genotype is present in only one of the three most prevalent high-risk clones worldwide, ST235 (5, 86). As previously noted, this high-risk clone is more virulent than the other prevalent clones, ST175 and ST111 (85, 86) (Table 2). Several experimental and clinical studies, however, suggest potentially reduced virulence in MDR/XDR P. aeruginosa (5, 73, 86, 117, 126–129). Experimental in vitro studies have shown that MDR strains have a lower growth rate and are defective in virulence determinants such as bacterial motility or pigment production (73, 86, 117). Experimental in vivo animal models have demonstrated that MDR/XDR P. aeruginosa strains are less able to produce infection, an inflammatory response, and mortality than susceptible strains (86, 126, 127, 129). Clinical studies also support the impaired virulence of MDR P. aeruginosa strains (5, 100, 107, 111, 130), and some of them showed that infections caused by MDR/XDR strains were not associated with higher mortality, even though they were more frequently managed with delayed adequate therapy (5, 98–100, 111, 114, 131). Taking these studies into account, we conclude that XDR strains may be associated with fitness costs and reduced virulence, but the data should be interpreted with caution, because, as mentioned above, at least one of the international XDR high-risk clone strains maintains high virulence regardless of its resistance profile. More studies are needed to clarify this.
IN VITRO AND IN VIVO TREATMENT MODELS: ANTIMICROBIAL COMBINATION OPTIONS
In Vitro Models
Pharmacodynamic interactions between drugs and bacteria have been studied in several in vitro models. Static systems can be used for rapid determination of time-kill behavior (132). Dynamic models such as the one-compartment in vitro model (IVM) and the two-compartment hollow-fiber infection model (HFIM) provide information that allows the development of dosing regimens that improve therapeutic results (133). Studies of dose fractionation, the suppression of resistant mutants, combination therapy, and the magnitude of the index required to obtain a specific amount of bacterial kill can be performed with both systems (132). Compared with the one-compartmental model, the hollow-fiber model allows the bacterial load to remain constant, biohazardous organisms to be safely contained, and absorption and elimination curves and rapid half-lives (t1/2s) to be modeled (133).
In vitro studies have been conducted to find “optimal treatment options” against MDR or XDR P. aeruginosa. Combination antibiotic therapy for MDR/XDR P. aeruginosa is generating interest because of the potential severity of these infections and the high risk of resistance selection with monotherapy. The possibilities of expanding the spectrum of coverage, achieving additive or synergistic antibacterial effects, and suppressing emerging resistance are all factors that favor the use of combination therapy (7).
Several studies have examined in vitro interactions between various antipseudomonal antibiotics (e.g., carbapenems, colistin and polymyxin B, fosfomycin, aminoglycosides, and quinolones). A number of methods of detecting synergy have been employed, including the microdilution checkerboard technique, gradient diffusion (Etest), time-kill curve assays (134, 135), and dynamic models. Reported synergistic drug combinations against MDR/XDR P. aeruginosa include colistin-ceftazidime (136), colistin-rifampin (137), cefepime-tobramycin (138), ceftazidime-avibactam-amikacin (139), colistin-doripenem, imipenem, and meropenem (140–142), meropenem-levofloxacin (143), imipenem-levofloxacin and colistin-levofloxacin (144), meropenem-ciprofloxacin (145), polymyxin B-enrofloxacin (146), fosfomycin-amikacin (147), and even some double-β-lactam combinations (148). Table 3 provides a list of studies of different drug combinations against MDR/XDR P. aeruginosa, showing the in vitro study model used, type of drug interactions, and whether or not suppression of resistance was achieved. Nevertheless, these studies have not led to clear recommendations for clinical practice, and there is a lack of consensus about which antibiotic combinations should be used against these difficult-to-treat infections to improve the therapeutic response and reduce selection of resistant mutants (135).
TABLE 3.
In vitro and in vivo models in which combination therapy options against MDR/XDR P. aeruginosa have been studied
| Drug combination (yr, reference) | P. aeruginosa type | Study modela | Type of drug interaction | Suppression of resistanceb |
|---|---|---|---|---|
| Cefepime-aztreonam (1998, 148) | Strain 164, including wild-type, partially derepressed, and fully derepressed phenotypes | HFIM | Synergistic | — |
| Meropenem-levofloxacin (2010, 143) | Wild-type strain PAO1 and isogenic MexAB-OprM-overexpressing strain | HFIM | Synergistic | Yes |
| Colistin-doripenem (2011, 140) | Colistin-heteroresistant reference strain (ATCC 27853) and colistin-resistant MDR clinical isolate | IVM | Additive or synergistic | Emergence of colistin-resistant subpopulations of ATCC 27853 reduced and delayed with combination therapy |
| Cefepime-tobramycin (2012, 138) | Wild-type strain PAO1 and its isogenic AmpC stably derepressed mutant | HFIM | Additive | Yes |
| Colistin-doripenem (2014, 361) | MDR | HFIM | Synergistic | Yes |
| Imipenem-levofloxacin, colistin-levofloxacin (2015, 144) | MDR | IVM | Synergistic | — |
| Colistin-meropenem (2016, 142) | Wild-type strain (ATCC 27853) and a meropenem-resistant strain (ARU552) | PK/PD model | Synergistic | Yes |
| Fosfomycin-amikacin (2017, 147) | Strain PA SAT 290 | HFIM | Synergistic | Yes |
| Ceftolozane-tazobactam- meropenem (2018, 157) | Strain ST175 | HFIM | Synergistic | Yes |
| Ceftolozane-tazobactam-colistin or -amikacin (2018, 154) | MDR | IVM | Synergistic | Yes |
| Ceftolozane-tazobactam-amikacin (2018, 155) | 5 strains, 3 with OprD mutation and AmpC overexpression | IVM | Additive | Yes |
| Enrofloxacin-polymyxin B (2018, 16) | Strain 12196 | IVM and HFIM | Synergistic | Yes |
| Meropenem-tobramycin (2018, 362) | PAO1 wild-type strain and its isogenic hypermutable PAO ΔmutS strain | HFIM | Synergistic | — |
| Ceftazidime-avibactam-amikacin (2018, 139) | 3 carbapenem-resistant P. aeruginosa isolates with ceftazidime-avibactam MICs of 4/4 to 8/4 μg/ml and AMK-I MICs of 8 to 64 μg/ml | Chemostat model | Quicker killing with the combination | — |
| Meropenem-ciprofloxacin (2018, 145) | Hypermutable | HFIM | Synergistic at high doses | Yes |
| Ceftolozane-tazobactam- meropenem and ceftazidime, alone and in combination with colistin (continuous infusion) (2019, 151) | MDR-HUB1 (ceftolozane-tazobactam and meropenem-susceptible), XDR-HUB2, (ceftolozane-tazobactam susceptible and meropenem resistant), MDR-HUB3 (ceftolozane-tazobactam resistant and meropenem-susceptible) | Pharmacodynamic in vitro model of biofilm | Combinations of colistin plus ceftolozane-tazobactam and meropenem were the most appropriate treatments for biofilm-related infections caused by XDR and MDR P. aeruginosa strains, respectively | Yes |
| Colistin-rifampin (2009, 158) | MDR | AM | Synergistic | Yes |
| Colistin-aztreonam (2017, 159) | MDR | AM | Synergistic | Yes |
| Imipenem-tobramycin (2017, 238) | MDR | AM | Synergistic | Yes |
IVM, in vitro one-compartment model; HFIM, hollow-fiber infection model; AM, animal model.
—, not evaluated.
The lack of antipseudomonal agents in the pipeline adds a further complication to this situation (149), although in recent years some progress has been made with the development of new molecules and new β-lactamase inhibitor combinations (150, 151). The new cephalosporin ceftolozane (formerly known as CXA-101) (58) in combination with tazobactam has shown promising characteristics for the treatment of P. aeruginosa infection (152). Since 2010, many in vitro studies of the role of ceftolozane against MDR and XDR P. aeruginosa have been carried out. VanScoy et al. (153) studied the effect of ceftolozane-tazobactam against two isolates of P. aeruginosa: an ATCC strain and a clinical isolate. Against the wild-type isolate (MIC of 0.5 mg/liter), resistance was not selected by any dose; against the clinical P. aeruginosa isolate (MIC of 4 mg/liter), however, although resistance was suppressed by a ceftolozane-tazobactam dose of 2 g/1 g every 8 h, resistance selection was observed with intermediate dosing regimens (125/62.5 through 1,000/500 mg) (153). For this reason, combination therapy is also starting to be studied for some MDR/XDR infections. Some recent studies have shown synergistic effects of ceftolozane-tazobactam with colistin and amikacin (154–156). Interestingly, in a hollow-fiber infection model, the combination therapy of ceftolozane-tazobactam plus meropenem had a synergistic effect on cell killing and also prevented resistance selection against XDR P. aeruginosa strains belonging to the ST175 clone (157).
Clinical trials are needed to confirm the results of these models, which are nonetheless very useful for deciding which trials should be developed.
In Vivo Models
There are few in vivo studies related to different antibiotic options and combinations against MDR/XDR P. aeruginosa. In a mouse model of pneumonia, intranasal colistin combined with rifampin was beneficial for synergistic antibacterial activity (158). High-dose colistin showed a 1.5-log10 CFU reduction against MDR P. aeruginosa infections in a neutropenic mouse thigh model (159). In the same study, the combination of high-dose colistin with aztreonam was even better, showing a 2.5-log10 CFU reduction. Yadav et al. recently demonstrated substantially enhanced killing in vivo against an MDR P. aeruginosa clinical isolate with an optimized imipenem-plus-tobramycin combination regimen (160).
CURRENTLY AVAILABLE ANTIMICROBIALS FOR THE TREATMENT OF MDR AND XDR P. AERUGINOSA INFECTIONS
Polymyxins
Increased bacterial resistance to antibiotics in conjunction with the lack of new drugs in the pipeline has become a major clinical and public health concern worldwide, which is especially worrisome in the case of MDR, XDR and PDR P. aeruginosa (3, 4). Although novel agents such as ceftolozane-tazobactam and ceftazidime-avibactam have expanded the therapeutic arsenal (67, 157, 161–165), polymyxins continue to represent the only therapeutic option in some cases.
Two polymyxins are available for clinical use: colistin (polymyxin E) and polymyxin B. They were released in the 1950s and were not subjected to the same drug development procedures and regulatory scrutiny that are needed for modern drugs, so our underlying pharmacological knowledge of these two polymyxins has until relatively recently been less than reliable (166–168). In recent years, however, a significant amount of preclinical and clinical data about these “old drugs” has emerged (169–173). The chemistry of polymyxins is very important for their antibacterial activity. Polymyxins are positively charged, enabling them to interact with phosphate groups in lipid A of the lipopolysaccharide (LPS) that are negatively charged (168). Polymyxins also have hydrophobic regions that can interact with the LPS (174). The result of these interactions is disruption of the bacterial cell membrane (174–176), which is the first step in the mechanism of action. Nevertheless, the final mechanism involved in bacterial cell death remains unknown (168). Recent studies performed on P. aeruginosa have argued against the traditional idea that colistin exerts its bactericidal effect by creating holes in the cytoplasmic membrane (177–179). New studies should explore other hypotheses, such as that bacterial killing is due to phospholipid exchange between the outer and cytoplasmic membranes, inhibition of respiratory enzymes, and/or formation of reactive oxygen species (179).
Since colistin and polymyxin B differ by only a single amino acid in the peptide ring (174), it is not surprising that they have similar antibacterial spectra, mainly against Gram-negative bacilli (174). In spite of their similar chemical structures, however, they are used in different forms when administered to patients parenterally. Polymyxin B is administered directly as an active antibiotic, whereas colistin is administered as an inactive prodrug, colistin methanesulfonate (CMS) (168, 176), which must be converted into colistin after administration (180). The use of one or the other polymyxin varies according to geographical area. In Europe and Australia, the only available form is colistin (in the form of CMS), whereas in the United States, Brazil, Malaysia, and Singapore, clinicians can use either colistin or the polymyxin B parenteral formulation (168).
Intravenous colistin dosing is controversial. Initially, low doses of CMS were used in clinics, based on the manufacturer’s instructions (181–183), but thanks to more recent pharmacokinetic and pharmacodynamic (PK/PD) data from population studies, it is now possible to provide an update of recommended dosages (171, 172, 184, 185). Some clinical studies evaluated the efficacy of parenteral colistin at higher doses (4.5 IU administered every 12 h), following a loading dose of 9 million IU (186, 187), although there are no clinical data available for the outcomes for patients receiving doses based on the equation proposed by Garonzik et al. (170) and updated by Nation et al. (171, 172) in 2016. In an attempt to translate PK/PD knowledge to clinical practice, Sorlí et al. studied the impact of colistin plasma concentrations on clinical outcome in 91 patients with infections caused by MDR/XDR P. aeruginosa (188). The mean colistin plasma concentrations in this cohort of patients were 1.67 ± 1.42 mg/liter, which is lower than those proposed in other studies and in the recent polymyxin use guidelines (170–172, 185). Nevertheless, 79.9% of patients achieved clinical cure, and colistin plasma concentrations were not observed to be statistically related to clinical cure (188). The same group demonstrated that a high plasma concentration of colistin was an independent risk factor for nephrotoxicity (183, 189). In conclusion, although PK/PD studies have concluded that higher doses of colistin should be used, there is a lack of clinical studies on the outcomes for patients treated according to more recent recommendations (171, 172, 185). In the case of urinary tract infections, colistin is a good option because concentrations of formed colistin in urine are high (185). Moreover, and because of this, in urinary tract infections the colistin dose could be lower than in other invasive infections (190). However, no clinical data are available to confirm this option.
There are several published clinical studies focused on colistin for treating MDR/XDR P. aeruginosa infections. The majority are single-center retrospective series with low numbers of patients, with two exceptions comprising more than 100 patients (182, 183) and with very different patient profiles (intensive care unit [ICU], cancer, hematologic, pneumological, etc.). The most frequent infectious source was low respiratory tract infection. Colistin doses were variable and adjusted for renal function. Combination therapy was administered to 51 to 100% of patients in these series. The clinical response at different time points varied between 52% and 79% and was higher than 70% in half of the studies. Mortality (at different time points) was between 11% and 61%. No information about resistance selection was given in these studies (181, 182, 188, 191–201) (Table 4).
TABLE 4.
Clinical studies providing outcome information for infections due to MDR/XDR P. aeruginosa treated with systemic antimicrobial therapya
Abbreviations: AB, antibiotic; AKI, acute kidney injury; BAL, bronchoalveolar lavage; BAT, best available therapy; BJI, bone and joint infection; BSI, bloodstream infection; CAZ-AVI, ceftazidime-avibactam; cIAI, complicated intra-abdominal infection; COL, colistin; Cr, plasma creatinine concentration; CLCR, creatinine clearance; cUTI, complicated urinary tract infection; CLABSI, central line-associated bloodstream infection; DOR, doripenem; EOT, end of treatment; GRF, estimated glomerular filtration rate; IE, infective endocarditis; FDA, Food and Drug Administration; FOS, fosfomycin; HCT, hematopoietic cell transplant; IAI, intra-abdominal infection; ID, infectious diseases; LAVD, left ventricular assist device; MDR, multidrug resistant; MU, millions of international units; NA , not applicable; PIP-TAZ, piperacillin-tazobactam; PJI, prosthetic joint infection; PNEU, pneumonia; SSI, surgical site infection; RIFLE, risk, injury, failure, loss, and end-stage kidney disease; RTI, respiratory tract infection (other than pneumonia or not specified); SAPS-II, simplified acute physiology score; SSTI, skin and soft tissue infection; TOL/TZ, ceftolozane-tazobactam; UTI, urinary tract infection; VAP, ventilator-associated pneumonia; XDR, extensively drug-resistant.
++, high quality; +, acceptable; −, low quality. Adapted from the Scottish Intercollegiate Guidelines Network (SIGN) (https://www.sign.ac.uk/methodology.html).
Study including patients with MDR Acinetobacter spp., MDR P. aeruginosa, and/or other MDR GNB infections; only specific information available on P. aeruginosa infections is reported.
High dose, TOL/TZ dose of 3 g/8 h (2 g ceftolozane + 1 g tazobactam every 8 h) or the equivalent after adjusting for renal function.
The study includes infections due to ceftazidime-resistant Gram-negative pathogens and not specifically multidrug-resistant Gram-negative pathogens.
The question of whether combination therapy might improve patient outcomes is another major issue to be considered for the use of polymyxins in the treatment of MDR/XDR P. aeruginosa infections. Data from PK studies confirm that colistin plasma concentrations following the dosing suggestions of the European Medicines Agency (EMA) and FDA are low and inadequate for the treatment of MDR/XDR P. aeruginosa infections (165–167, 178, 182, 183). These findings highlight the importance of considering colistin combination therapy for MDR/XDR P. aeruginosa infections. Zusman et al. (202) recently published a systematic review about polymyxins in combination or as monotherapy against carbapenem-resistant Gram-negative bacteria (GNB) and showed that polymyxin combined with carbapenems or tigecycline and/or aminoglycosides had an unadjusted association with survival, but when biased studies were excluded from the analysis, there was no association between combination therapy and survival. The majority of the studies did not include P. aeruginosa infections (202). In a cohort of patients with pneumonia due to XDR P. aeruginosa, Khawcharoenporn et al. showed that combination therapy with 2 active drugs was associated with better survival than active monotherapy, including colistin in the majority of cases (Table 4) (203). Interestingly, a recent prospective clinical series of bone and joint infections due to MDR/XDR P. aeruginosa also showed better clinical outcomes with colistin in combination therapy, in comparison with β-lactam or colistin as monotherapy (204) (Table 4). Larger clinical series and randomized clinical trials with invasive MDR/XDR P. aeruginosa infections are needed to confirm these data. Until then, the recent expert-panel guidelines for optimal use of polymyxins recommend that for the treatment of MDR/XDR P. aeruginosa infections, polymyxin should be used in combination with one or more additional agents to which the pathogen displays a susceptible MIC (185).
With respect to polymyxin B, there is limited clinical experience with MDR/XDR P. aeruginosa infections (166, 205–209). The studies are retrospective series with low numbers of patients, except for one with 126 cases (208). Bacteremia and pneumonia were the predominant indications. Clinical response is insufficiently studied, and mortality rates are worryingly high (Table 4).
Nephrotoxicity is a common adverse effect of systemically administered polymyxins (210, 211). This adverse effect is dose-limiting for both polymyxins (colistin and polymyxin B), although polymyxin B seems to be less nephrotoxic (205). In the case of colistin, plasma concentrations associated with renal damage overlap those required for a bacterial effect (212). Colistin plasma concentrations have been demonstrated to be the most important risk factor for the development of acute kidney injury (AKI). An average steady-state plasma colistin concentration of greater than ∼2 mg/liter is considered to be an independent risk factor for colistin-associated nephrotoxicity (183, 189, 213). These data highlight the narrow therapeutic window of colistin. In this scenario, therapeutic drug monitoring could be a useful clinical tool to maximize clinical goals while minimizing potential nephrotoxicity (185).
Both polymyxin B and CMS have been administered as inhalation therapy for the treatment of pneumonia, bronchiectasis, and chronic P. aeruginosa infection and for pulmonary exacerbations in patients with cystic fibrosis. Once again, most of the studies of inhaled administration were performed with CMS. A recent meta-analysis focused on the use of inhaled colistin monotherapy for respiratory infections in non-CF patients (214). The analysis included 10 studies of patients diagnosed with pneumonia and 2 studies of those with ventilator-associated tracheobronchitis. Overall all-cause mortality was 33.8% and the clinical success rate was 70.4% (214). The authors of this meta-analysis concluded that the outcomes for patients receiving therapy with inhaled CMS as monotherapy were encouraging and deserved further consideration for the treatment of respiratory tract infections caused by MDR GNB. Another recent meta-analysis analyzed the combination of inhaled and intravenous (i.v.) colistin. The studies used low-quality data, which suggested that the combination does not lower mortality in patients with MDR Gram-negative infections except when a low i.v. colistin dose is administered. The results for MDR/XDR P. aeruginosa infections were not specifically extracted (215).
Regarding studies performed in patients with infections caused by P. aeruginosa, Athanassa et al. performed a pharmacokinetic study of inhaled CMS in mechanically ventilated critically ill patients (216). The study included 8 patients with P. aeruginosa infection receiving 80 mg of CMS every 8 h. Colistin concentrations in epithelial lining fluid (ELF) were 5-fold higher than those achieved in serum, although ELF concentrations at 4 and 8 h were below the EUCAST breakpoints. Based on these data, the authors concluded that inhaled colistin can achieve high drug concentrations in the lungs, although a dose of 80 mg every 8 h may not be suitable for the treatment of infections caused by MDR GNB (216). Lu et al. (217) compared the clinical outcomes for 122 patients with ventilator-associated pneumonia (VAP) caused by P. aeruginosa and Acinetobacter baumannii strains susceptible to β-lactams, aminoglycosides, or quinolones and treated with i.v. antibiotics for 14 days with those for patients with VAP caused by MDR P. aeruginosa or A. baumannii treated with nebulized colistin (5 million IU every 8 h) either in monotherapy (n = 28) or in combination with i.v. aminoglycosides. With several methodological limitations, they concluded that nebulized CMS was noninferior to intravenous β-lactams associated with aminoglycosides or quinolones (217). With respect to the use of CMS in patients with ventilator-associated tracheobronchitis, Maskin et al. demonstrated in a study of 17 patients infected with MDR P. aeruginosa that inhaled CMS was able to reduce the volume of tracheal secretions, purulence, and bacterial load (218).
There is little clinical information about the use of nebulized polymyxin B. A recent study of inhaled polymyxin B against P. aeruginosa in a mouse lung infection model highlighted the advantage of pulmonary delivery of polymyxin B over intravenous administration for achieving high levels of drug exposure in ELF (219). A clinical study performed by Pereira et al. that focused on the use of nebulized polymyxin B as salvage therapy for pneumonia and initial treatment of tracheobronchitis caused by MDR GNB (220) concluded that inhaled polymyxin B was useful as salvage therapy for hospital-acquired pneumonia caused by MDR GNB that failed i.v. treatment and also when used alone in the treatment of P. aeruginosa tracheobronchitis. Taking all these results into account, we consider that inhaled polymyxins should be considered for the treatment of lower respiratory tract infections caused by MDR/XDR P. aeruginosa. The evidence is not strong enough to consider inhaled therapy alone for pneumonia, where a combination of intravenous and inhaled polymyxins would be a good option. In the case of tracheobronchitis, inhaled therapy alone could be used, although more dosage studies and clinical series are needed.
Another scenario in which polymyxins can play an important role is in the treatment of central nervous system infections (CNS) due to MDR/XDR bacteria. When MDR organisms are the cause of infection, CNS mortality has been reported to be as high as 71% (221). This is partly due to the fact that only a proportion of the intravenous antibiotic dose reaches the site of infection in these difficult-to-treat infections (222–224). Hence, high intravenous doses are required to achieve bacterial killing. Peripheral administration of colistin, however, is neither effective nor safe for CNS infection, due to extensive renal reabsorption and the risk of colistin-associated nephrotoxicity (225). To overcome this problem, intrathecal or intraventricular delivery of polymyxins has generally been used in clinical practice and has become the only therapeutic option for the treatment of MDR GNB CNS infections that are resistant to all other antibiotics. Although most clinical experience with this administration route has been reported for infections caused by Acinetobacter baumannii, there are some reports of infections caused by MDR/XDR P. aeruginosa (226–233) that have had good clinical outcomes. Even though the intrathecal route in this setting is mandatory, intrathecal polymyxin therapy has never been optimized according to PK/PD indices (225). The current IDSA guidelines suggest an intrathecal dose of 10 mg of CMS or 5 mg of polymyxin B once daily (234). The recent international consensus guidelines on the use of polymyxins recommend an intraventricular or intrathecal dose of 125,000 IU CMS (∼4.1 mg colistin base activity) or 5 mg (50,000 IU) polymyxin B per day (185).
In clinical practice and even in the guidelines, both the dose and the duration of treatment of CNS infections are chosen empirically, since no PK/PD targets have so far been established. For this reason, and as Nation et al. pointed out, it is important in the near future to define optimal targets for the optimization of dosage regimens for the administration of polymyxins by the intrathecal route (169).
Carbapenems
Like all β-lactam antibiotics, carbapenems exhibit time-dependent antibacterial activity. Different in vitro and in vivo studies have identified the PK/PD parameter most predictive of efficacy as the percentage of the dosing interval that unbound or free serum drug concentrations exceed the MIC for the pathogen (fT > MIC). Old in vitro and in vivo PK/PD studies initially defined an fT > MIC of ≥40% as the optimal value for the bactericidal activity of carbapenems (235). A similar value was identified in a murine thigh model infected with P. aeruginosa strains overexpressing MexA-MexB-OprM efflux pumps at both standard and high inocula (236).
With the currently approved antibiotic doses and short-term infusion regimens, the probability of achieving optimal PK/PD target exposures across all patient populations and susceptible pathogens is greater than 80% (http://www.eucast.org/documents/rd/; accessed 25 October 2018). This probability is considerably reduced, however, in the case of infections caused by less susceptible or even resistant pathogens, such as MDR or PDR P. aeruginosa. New strategies aimed at achieving the desired targets are therefore required. In this scenario, numerous studies have assessed new dosing strategies, such as increasing the dose or using prolonged infusion administration.
Various studies aimed at defining the optimal carbapenem dose for these difficult-to-treat P. aeruginosa infections in different special populations show that high doses may be needed.
One pharmacodynamic study used Monte Carlo simulation to evaluate different dosage regimens of meropenem administered in intermittent or extended (3-h) infusions against populations of Enterobacteriaceae, P. aeruginosa, and Acinetobacter species with different susceptibilities. MIC data and distributions were derived from the Meropenem Yearly Susceptibility Test Information Collection (MYSTIC), a multicenter, longitudinal surveillance program in 14 American centers. A total of 276 isolates of P. aeruginosa were included, 22.1% of them with MIC values of >4 mg/liter. A meropenem dosage of 1 g/8 h in extended infusion, or 2 g/8 h in intermittent/extended infusion, was required for exposure of 50% fT > MIC against all susceptible P. aeruginosa isolates (MIC values of ≤4 mg/liter). However, for organisms considered intermediate-resistant to meropenem (MIC = 8 mg/liter), only the higher-dose regimen of 2 g/8 h in extended infusion achieved adequate bactericidal exposure. The authors suggested the highest dose of meropenem (2 g/8 h) administered by extended infusion for treatment of intermediate or resistant P. aeruginosa (237).
An in vitro infection model also using Monte Carlo simulation evaluated the optimal dosage of imipenem combined with tobramycin against carbapenem- and aminoglycoside-resistant P. aeruginosa clinical isolates. The simulated doses that obtained the best antibacterial activity were imipenem at 4 or 5 g/day in continuous infusion combined with tobramycin (238). The same group confirmed the adequacy of this dosage regimen in a neutropenic mouse thigh model of XDR P. aeruginosa infection (160). In one study of bloodstream infections that included 237 isolates of P. aeruginosa with reduced susceptibility to carbapenems, different carbapenem dosing regimens were tested: imipenem at 0.5 to 1 g/6 h by 0.5- and 3-h infusion, meropenem at 1 to 2 g/8 h by 0.5- and 3-h infusion, and doripenem at 0.5 to 2 g/8 h by 1- and 4-h infusion. A T > MIC of 40% was considered to be the optimal PK/PD ratio. The results showed that meropenem at 2 g/8 h infused over 3 h and doripenem at 1 g/8 h infused over 4 h showed the best efficacy against P. aeruginosa with reduced susceptibility to carbapenems (239). Interestingly, in a case report of a critically ill double-lung transplant patient with pneumonia due to MDR P. aeruginosa with a meropenem MIC of 32 mg/liter who received meropenem in continuous infusion (8 g meropenem/24 h), a clinical cure was achieved (240).
Other Classical Antipseudomonal β-Lactams
There are very few data regarding the role of some classical antipseudomonal β-lactams such as cefepime, ceftazidime, piperacillin-tazobactam, and aztreonam in monotherapy against MDR/XDR P. aeruginosa infections.
Aztreonam could be a possible option for the treatment of Ambler class B MBL-producing Gram-negative bacteria, including P. aeruginosa. One series assessed its clinical efficacy in MBL-producing P. aeruginosa infections. In that study, the mortality rate was 30%, but most cases involved combination therapy, and the sample size was too small to be able to draw definitive conclusions (241). Another series included nine patients with MBL-producing Pseudomonas infections receiving i.v. colistin combined with aztreonam or piperacillin-tazobactam, and seven (77.8%) of these patients had favorable outcomes and survived (242).
A case report described an immunocompromised patient with an MDR P. aeruginosa wound infection who was successfully treated with high-dose aztreonam administered in continuous infusion (8.4 g/day) (243). Another case report described a patient undergoing hemodialysis who developed MDR P. aeruginosa bacteremia with a cefepime MIC of 16 mg/liter and was successfully treated with an extended-infusion regimen (3 h) of this antibiotic (244).
A severely immunodepressed patient with MDR P. aeruginosa bacteremia with a ceftazidime MIC of 64 mg/liter was treated with high-dose ceftazidime administered in continuous infusion (6.5 to 9.6 g/day) with clinical success (243).
As previously mentioned, some in vitro combination assays, such as those with cefepime-tobramycin (138) and cefepime-aztreonam (148), have shown additive or synergistic effects against MDR P. aeruginosa.
Based on the type of strain and resistance phenotype and genotype, a possible strategy in individual cases could be to use one of these drugs at high doses administered in prolonged infusion in a combination therapeutic regimen.
Aminoglycosides
Some aminoglycosides remain active against several MDR/XDR P. aeruginosa strains (245, 246). Although they can be used in monotherapy in urinary tract infections (247), aminoglycosides could be used in combination with other antimicrobials for the treatment of more severe infections caused by MDR/XDR P. aeruginosa.
With respect to their pharmacodynamics, numerous in vitro and in vivo studies have demonstrated that aminoglycosides have concentration-dependent antibacterial activity and that a peak concentration (maximum concentration [Cmax]/MIC) of ≥8 to 10 is the best PK/PD predictor of efficacy (248). This value should be reached during the first 24 to 48 h of treatment. This PK/PD index was associated with better clinical cure rates in a retrospective clinical study performed in patients with P. aeruginosa bacteremia that was not specifically caused by MDR or XDR P. aeruginosa strains (249).
A few studies in recent years have set out to optimize dosing regimens to combat MDR GNB such as P. aeruginosa. One PK model, cited above, evaluated the optimal dose of tobramycin and imipenem against carbapenem- and aminoglycoside-resistant P. aeruginosa clinical isolates (238). The authors concluded that a 7-mg/kg dose of tobramycin every 24 h, given in 0.5-h infusions, combined with imipenem was needed to achieve adequate bacterial killing and prevent regrowth at 48 h.
One strategy used to treat infections caused by XDR P. aeruginosa was to administer very high doses of aminoglycosides combined with continuous renal clearance techniques to prevent renal toxicity. The results showed high survival rates, although the number of included patients was limited (250, 251).
In the case of severe or deep infections such as pneumonia or meningitis due to MDR or XDR P. aeruginosa, other routes of administration can be used for aminoglycosides. For the treatment of pneumonia, inhaled amikacin allows high drug concentrations to be achieved at the site of infection (e.g., ELF) and prevents high systemic exposures that can potentially cause systemic toxicity. The use of inhaled antibiotics (polymyxins or aminoglycosides), however, is currently recommended only as adjunctive therapy for infections caused by Gram-negative bacilli susceptible only to aminoglycosides or polymyxins, and in combination with other systemically administered agents (252).
Meningitis is another difficult-to-treat infection. The efficacy of intravenous aminoglycosides is limited due to poor penetration into the central nervous system, which leads to low and inadequate concentrations at the site of infection. In cases of this kind, administration of intraventricular aminoglycosides may be needed. A recent case of postsurgical meningitis caused by PDR P. aeruginosa was successfully treated with a combination of intravenous cefepime administered by continuous infusion and combined with intravenous and intraventricular amikacin (253). Although the strain had an MIC for amikacin of 32 mg/liter, the achievement of concentrations of 200 mg/liter in the central nervous system was sufficient for resolution of infection.
Fosfomycin
Because of its excellent in vitro bactericidal activity against a wide spectrum of organisms, including MDR P. aeruginosa, intravenous fosfomycin in combination with other antimicrobials has reemerged for the treatment of infections caused by MDR bacteria (254, 255). One proposed therapeutic option is to use fosfomycin with carbapenems, a combination that has shown good synergistic activity against different P. aeruginosa isolates. This combination has also demonstrated better clinical outcomes, especially when the carbapenem is administered in extended infusion (256–258).
Other experiments have assessed the use of fosfomycin in combination with β-lactams, aminoglycosides, or colistin (259). In one of these, fosfomycin was administered to 5 patients undergoing orthotopic liver transplantation, 3 of whom had infections due to XDR P. aeruginosa with a MIC for fosfomycin of <16 mg/liter and another due to XDR Klebsiella pneumoniae and P. aeruginosa with MICs of 32 mg/liter (259). In two of the patients, the infection was eradicated, but in the other three, treatment failed (in two the clinical response was poor, and the third developed a superinfection).
This so-called “old” antibiotic has also been associated with new antimicrobials, such as ceftazidime-avibactam or ceftolozane-tazobactam (260, 261). A patient with XDR P. aeruginosa meningitis was successfully treated with a 3-g/8-h dose of ceftolozane-tazobactam associated with a 4-g/6-h dose of fosfomycin (261). Nevertheless, the doses of fosfomycin used in these cases varied considerably, which provides evidence that the optimal dose of this antibiotic for the treatment of difficult-to-treat infections is yet to be defined.
In a systematic review of the clinical and microbiological effectiveness of fosfomycin for the treatment of MDR, XDR, or PDR nonfermenting Gram-negative bacterial infections, the fosfomycin dose for P. aeruginosa infections ranged from 2 g/12 h to 5 g/8 h in combination with other antimicrobials (254).
Several studies have evaluated different dosage regimens of fosfomycin in combination with carbapenems for the treatment of non-MDR and MDR P. aeruginosa clinical isolates based on PK/PD target attainment. In one of these, Monte Carlo simulation was used to calculate the probability of target attainment for different fosfomycin and carbapenem doses and infusion times (262). In the case of non-MDR P. aeruginosa isolates, prolonged infusion of a carbapenem combined with fosfomycin in continuous infusion at 16 to 24 g/day obtained the best PK/PD ratios. However, for the MDR P. aeruginosa isolates, none of the fosfomycin and carbapenem combinations achieved the PK/PD targets. It should be borne in mind that the clinical isolates tested in this study, which was carried out in Thailand, had very high fosfomycin MIC values, and the results cannot be extrapolated to other settings (262).
More clinical series and trials are needed to define the future role of fosfomycin in these infections, including the optimal dose and possible combinations.
NEW ANTIMICROBIALS AGAINST MDR AND XDR P. AERUGINOSA
Although a clear distinction has often been made between old and new antipseudomonal antibiotics (263, 264), two antibiotics resulting from the combination of old and new drugs have been released in recent years (265, 266).
Ceftolozane-Tazobactam
Ceftolozane-tazobactam is an effective combination against several MDR Gram-negative bacilli, especially MDR/XDR P. aeruginosa. Ceftolozane is one of the most active antipseudomonals. Its activity against P. aeruginosa exceeds that of the rest of the antipseudomonal β-lactams by between 20% and 25% (267). Ceftolozane inhibits PBPs and non-ESBL TEM and SHV variants and AmpC enzymes, while tazobactam targets class A serine β-lactamases and ESBLs. Ceftolozane also acts against non-ESBL class D oxacillinases, but it lacks activity against carbapenemases (268).
In a number of studies, MDR/XDR P. aeruginosa susceptibility to ceftolozane-tazobactam has been shown to be variable, with rates varying between 55% and 96.6% depending on the series and countries (31, 67, 150, 245, 246, 269–274). The data from these studies are shown in Table 5.
TABLE 5.
Ceftolozane-tazobactam activity against resistant strains of P. aeruginosa
| Authors (journal, yr, reference)a | Total no. of centers (country[ies]) | Total no. of P. aeruginosa strains | Susceptibilityb
|
Comments | ||
|---|---|---|---|---|---|---|
| Type of strain | No. | % | ||||
| Giani et al. (JAC, 2018, 68) | 20 (Italy) | 935 | Resistant to rest of β-lactams | 183 | 59.6 | Ceftolozane-tazobactam retained activity against 64 (46.8%) of the strains that were resistant to all other agents except colistin |
| Humphries et al. (AAC, 2017, 271) | 3 (USA) | 309 | Resistant to rest of β-lactams | 105 | 55 | 55 (29%) strains analyzed were susceptible to ceftazidime-avibactam |
| Pfaller et al. (JAC, 2017, 272) | 17 (European countries) | 603 | 4 resistance phenotypes (ceftazidime, cefepime, meropenem, piperacillin-tazobactam) | CFZ NS,139; CP NS, 124; MER NS, 126; TZP NS, 162 | CFZ NS, 65.5; CP NS, 61.3; MER NS, 65.9; TZP NS, 70.4 | 91.7% of the strains analyzed were globally susceptible |
| Del Barrio-Tofiño et al. (AAC, 2017, 31) | 9 (Spain) | 150 | XDR | 101 | 68.7 | MIC90, >64 mg/liter |
| Walkty et al. (JAC, 2018, 246) | 10–15 (Canada) | 3,229 | MDR | 462 | 90.5 | MIC90, 4 mg/liter |
| XDR | 84 | 78.6 | MIC90, 16 mg/liter | |||
| Pfaller et al. (IJAA, 2017, 245) | 14 (Asia-Pacific, minus China, Australia, and New Zealand) | 489 | MDR | 134 | 67.2 | MIC90, >32 mg/liter |
| Sader et al. (JAC, 2014, 269) | 31 (13 European countries) | 2,191 | MDR | 698 | 57.4 | MIC90, ≤8 mg/liter |
| XDR | 538 | 46.3 | ||||
| Buehrle et al. (AAC, 2016, 150) | 1 (USA) | 38 | Meropenem resistant | 38 | 92 | Median MIC90 (range): ceftolozane-tazobactam, 1 (0.25–64) mg/liter; ceftazidime-avibactam, 4 (2–>32) mg/liter |
| Grupper et al. (AAC, 2017, 273) | 34 (USA) | 290 | Meropenem, aztreonam, cefepime, piperacillin-tazobactam NS | 103 | 91 | |
| Livermore et al. (JAC, 2018, 67) | Not specified (UK) | 1,384 | AmpC derepressed | 147 | 96.6 | 94.6% of the strains analyzed were susceptible to ceftazidime-avibactam |
| Tato et al. (IJAA, 2015, 274) | 10 (Spain) | 500 | MDR | 34 | 64.7 | MIC90, >64 mg/liter |
JAC, Journal of Antimicrobial Chemotherapy; AAC, Antimicrobial Agents and Chemotherapy; IJAA, International Journal of Antimicrobial Agents.
CFZ, ceftazidime; CP, ciprofloxacin; MER, meropenem; TZP, piperacillin-tazobactam; NS, nonsusceptible.
With respect to its PK/PD indices, the bactericidal efficacy of ceftolozane-tazobactam, as with other cephalosporins, is correlated with the percentage of time the plasma drug concentration is above the MIC for the target organism (%T > MIC) (151). Monte Carlo simulations have been performed to study ceftolozane-tazobactam dosing regimens and to define the optimal dose of this drug against infections caused by MDR P. aeruginosa with MIC values of between 4 and 32 mg/liter, testing different doses, infusion times, and renal function statuses (275). The multiple scenarios simulated identified the current ceftolozane-tazobactam dose of 1/0.5 g as optimal for MICs of ≤32 mg/liter (creatinine clearance [CLCR], 15 to 50 ml/min), ≤16 mg/liter (CLCR, 51 to 120 ml/min), and ≤8 mg/liter (CLCR, 121 to 180 ml/min). In simulations of augmented renal clearance across infections with MICs of 4 to 32 mg/liter, extended infusions of 4 to 5 h had a higher probability of target attainment (PTA) than shorter and continuous infusions (275). Another study simulated four ceftolozane-tazobactam doses ranging from 250/125 mg to 2/1 g every 8 h, with infusion durations of 1 to 7 h and continuous infusions. The PTA target was defined as 40% fT> MIC (276). The results showed that the current dose of 1/0.5 g was optimal for MICs of ≤32 mg/liter and different renal function values. In patients with augmented renal clearance, however, extended infusions of 4 to 5 h provided higher PTAs than intermittent infusions. On the other hand, another population PK study in patients with pneumonia, which included kinetics in the ELF, also simulated different dosage regimens and concluded that a dose of 2 g/1g was necessary to achieve a >90% PTA (actual, 98%) in ELF against pathogens with MICs of ≤8 mg/liter (277). As with other β-lactam antibiotics, administration of the drug over a prolonged period by extended or continuous infusion is a potential strategy for improving the probability of attaining the PK/PD target. Until recently, however, very little evidence of evaluations of extended or continuous infusion of ceftolozane-tazobactam has been available. One case report described a patient with urinary tract infection caused by MDR P. aeruginosa who was successfully treated with no adverse events in an outpatient setting with a 4/0.5-g dose of ceftolozane-tazobactam every 24 h given as continuous infusion (278). Another case report evaluated the pharmacokinetics of this antibiotic in a critically ill patient with an MDR P. aeruginosa prosthetic hip joint infection receiving continuous venovenous hemofiltration who was treated with a 1/0.5-g dose of ceftolozane-tazobactam every 8 h administered as extended infusion over 4 h (279). An outpatient with a lung abscess caused by carbapenem-resistant P. aeruginosa with a ceftolozane-tazobactam MIC90 of 2 mg/liter obtained favorable clinical results after 3 g/1.5 g of ceftolozane-tazobactam administered in continuous infusion (280). In these last two cases, the serum concentration analysis confirmed that these dosing regimens were adequate for the achievement of the desired PK/PD target (279, 280).
Some series of clinical experience with ceftolozane-tazobactam in MDR/XDR P. aeruginosa infections have been published (Table 4). These studies are mainly retrospective, with short series of patients, except for a study of 205 patients (281). The main indication in these series was respiratory tract infection, and different doses of ceftolozane-tazobactam were used, sometimes in combination therapy. Cure rates were close to 70%, with emergence of resistance of 4 to 14% in some studies and mortality rates, measured at different times, ranging from 0% (in small series of cases) to 27% (26, 62, 165, 281–288).
In summary, ceftolozane-tazobactam might be a good option for the treatment of MDR/XDR P. aeruginosa infections that are susceptible to this drug, but it should be used with caution and with optimization of dosing and, probably, infusion times. Combination therapy could be considered for high-inoculum infections in order to prevent selection of resistance in vivo (Table 6). However, more clinical studies are needed to fully confirm these statements. Specifically, observational studies on the use of this drug and on the possible selection of resistant mutants in the real world, clinical trials comparing monotherapy with combination therapy, and larger series of MDR/XDR P. aeruginosa infections would be useful in the near future.
TABLE 6.
Suggested antimicrobial therapy options for the most prevalent resistance profiles of MDR/XDR P. aeruginosaa
| Resistance profile | Resistance mechanism(s) | High-risk clones where they are more frequent | Treatment optionsb |
|---|---|---|---|
| PTZ R, CAZ R, ATM R, MER R, TOL/TZ S, CAZ/AVI S, AMK S COL S | AmpC overexpression + OprD deficiency | ST175 | COL, POLY-B, TOL/TZ, CAZ/AVI, AMK |
| PTZ R, CAZ R, ATM S, MER R, TOL/TZ R, CAZ/AVI R, AMK S COL S | MBL production | ST235, ST111 (ST175) | COL, POLY-B, ATM, AMK |
| PTZ R, CAZ R, ATM R, MER R, TOL/TZ R, CAZ/AVI S, AMK S COL S | Class A carbapenemase (such as GES enzymes) or combinations of certain ESBLs with OprD deficiency | ST235 | COL, POLY-B, CAZ/AVI |
Abbreviations: PTZ, piperacillin-tazobactam; CAZ, ceftazidime; ATM, aztreonam; MER, meropenem; TOL/TZ, ceftolozane-tazobactam; CAZ/AVI, ceftazidime-avibactam; AMK, amikacin; COL, colistin; POLY-B, polymyxin B.
Administration of β-lactams in extended or continuous infusion and/or combination with intravenous colistin, polymyxin B, or amikacin should be considered in severe infections. Amikacin or colistin in monotherapy is acceptable for urinary tract infection. Therapeutic drug monitoring of colistin or amikacin is recommended. Nebulized colistin (2 to 5 MU/8 h) as adjunctive therapy in lower respiratory tract infections should be considered.
Ceftazidime-Avibactam
Avibactam contains a diazabicyclooctane nucleus and acts as a broad-spectrum inhibitor that is effective against enzymes with a nucleophilic serine residue (289). It has no activity against MBL-producing strains (290). The addition of avibactam to ceftazidime protects the cephalosporin from enzymatic degradation caused by P. aeruginosa strains (mainly due to Amp-C enzymes but also due to ESBLs and class A carbapenemases such as GES enzymes) and leads to decreased MICs of ceftazidime, which is more marked when combined with higher doses of avibactam (291).
Several in vitro studies have demonstrated that ceftazidime-avibactam displays good activity against large collections of MDR/XDR P. aeruginosa strains collected in different parts of the world and at different times, with inhibition rates varying between 66.1% and 86.5% (162, 292–295) (Table 7). In another series including 5,716 strains of P. aeruginosa collected in the INFORM study, ceftazidime-avibactam showed 92.4% activity against all the strains tested (296). Although its activity was low against MBL-positive strains, it was the second most active agent after colistin. Likewise, in another in vitro study, several P. aeruginosa strains with different levels of resistance were exposed to β-lactam antibiotics and 74.1% of pan-β-lactam-resistant isolates were susceptible to ceftazidime-avibactam with an MIC90 of 16 mg/liter (59). Similarly, activity against strains of P. aeruginosa from patients with urinary tract infections in U.S. hospitals showed a MIC90 of 32 mg/liter for strains resistant to ceftazidime, meropenem, or piperacillin-tazobactam (297). The INFORM 2012–2014 study analyzed the activity of ceftazidime-avibactam against 7,062 strains of P. aeruginosa and found that 563 (8%) of them showed resistance to this antibiotic. Half of these were explained as due to possession of genes encoding MBLs (298). Another study assessed ceftazidime-avibactam activity against clinical isolates, 41 of them P. aeruginosa, in a phase III trial of complicated urinary tract infections. The range of MICs for these strains was 4 to 16 mg/liter (299).
TABLE 7.
Ceftazidime-avibactam activity against MDR and XDR P. aeruginosa
| Authors (journal, yr, reference)a | Total no. of centers (country or continents)b | Total no. of strains | MDR strains |
XDR strains |
Comments | ||||
|---|---|---|---|---|---|---|---|---|---|
| No. | % | MIC90 (mg/liter) | No. | % | MIC90 (mg/liter) | ||||
| Sader et al. (IJAA, 2015 292) | 71 (USA) | 3,082 | 436 | 80.7 | ≤8 | 247 | 74.5 | ≤8 | Susceptibility rates for ceftazidime, piperacillin-tazobactam and meropenem, 8.5%–22.9% (MDR strains) and 2.0%–13.4% (XDR strains) |
| Sader et al. (AAC, 2015, 293) | 75 (USA) | 3,902 | 338 | 81.0 | 16 | 338 | 73.7 | 32 | Colistin efficacy against MDR and XDR strains (EUCAST), 99.7% |
| Stone et al. (JAC, 2018, 162) | ? (Europe, North and South America, Asia, and Africa) | 565 | 56 | 66.1 | 64 | Data from adult phase III clinical trials | |||
| Sader et al. (AAC, 2017, 295) | INFORM study (USA) | 7.868 | 1,562 | 86.5 | 16 | 717 | 75.9 | 32 | Amikacin efficacy, 87.1% (MDR strains) and 80.8% (XDR strains); colistin efficacy, >99% (both types of strains) |
| Sader et al. (AAC, 2017, 294) | ? (USA) | 3,402 | 613 | 82.7 | 16 | 365 | 76.2 | 32 | Colistin was the most active antibiotic (99.6% susceptibility) |
JAC, Journal of Antimicrobial Chemotherapy; AAC, Antimicrobial Agents and Chemotherapy; IJAA, International Journal of Antimicrobial Agents.
A question mark indicates that the total number of centers was not given in the indicated article.
In order to analyze the emergence of P. aeruginosa resistance to ceftazidime-avibactam, a study was developed to assess the evolution of this microorganism after exposure to the antibiotic. Interestingly, the studied strains developed mutants resistant to ceftazidime-avibactam, mainly through the efflux pumps PA14_45890 and PA14_45910 (60).
Regarding PK/PD parameters, after multiple doses of ceftazidime-avibactam at 2 g/0.5g, the Cmax was 113.0/15.0 mg/ml and the area under the curve (AUC) was 348.2/42.2 mg · h/liter. With respect to the PK/PD properties of ceftazidime-avibactam, a new ratio has been proposed that can be calculated in vitro or in vivo, defined as %fT > CT, with CT being the “concentration threshold” for avibactam (300). In an HFIM, several CT values were tested to determine which one best correlated with efficacy against ceftazidime-resistant P. aeruginosa strains (MICs of 32 to 128 mg/liter for ceftazidime and 2 to 16 mg/liter for ceftazidime-avibactam) (301).
A PK/PD study was designed to evaluate the predictive performance of the susceptibility cutoff points established by the regulatory agencies for ceftazidime-avibactam against different bacteria (161). The results were consistent in the case of susceptible P. aeruginosa strains, but the cutoff points were challenged when strains resistant to several antibiotics included in clinical trials were considered. The model was considered unreliable for the analysis of ceftazidime-avibactam activity against resistant P. aeruginosa, probably because these strains had mechanisms of resistance that could not be reversed by adding avibactam to ceftazidime.
Various in vitro and in vivo PK studies have evaluated the PK/PD parameters of ceftazidime-avibactam against different Gram-negative microorganisms. An fT > MIC for at least 50% of the dosing interval has been shown to achieve the maximum bacterial kill (302). For avibactam, which does not have antibacterial activity at clinically relevant concentrations, a minimum free avibactam concentration (threshold concentration [fCT]) needed to achieve sufficient β-lactamase inhibition to restore the activity of ceftazidime was defined. The estimated critical concentration threshold (CT) was ≤0.15 mg/liter (303). In a neutropenic mouse lung and thigh infection model of ceftazidime-resistant P. aeruginosa expressing AmpC and/or TEM-24 β-lactamase, achievement of a free T > CT of 40% to 50% and an fCT of 1 mg/liter for avibactam exceeded the exposures associated with stasis, 1-log10 kill, and 2-log10 kill of P. aeruginosa (300). Another in vitro study evaluated the bactericidal activity of ceftazidime-avibactam against 18 P. aeruginosa isolates and 15 Enterobacteriaceae isolates, including wild-type isolates and ESBL, KPC, and/or AmpC producers (304). At 6 h, the authors observed time-dependent and bactericidal activity against all Enterobacteriaceae isolates and a lower degree of initial killing against all P. aeruginosa isolates. At 24 h, ceftazidime-avibactam did not have any bactericidal activity, and bacterial regrowth was detected in both species.
Based on this PK/PD target, the optimal dose for its achievement has been evaluated in population PK models, and a dose of 2/0.5 g ceftazidime-avibactam every 8 h administered intravenously over 2 h has been recommended for patients with normal renal function. This selected dose allows the PK/PD target to be achieved against Enterobacteriaceae and P. aeruginosa isolates using the ceftazidime-avibactam breakpoints of ≤8/4 mg/liter (305). These population PK models of ceftazidime-avibactam were built using PK data from five phase III trials in patients with complicated intra-abdominal infections, complicated urinary tract infections, and nosocomial (including ventilator-associated) pneumonia (306). This clinical dose was further validated in an HFIM and in neutropenic and immunocompetent mouse thigh infection models against different P. aeruginosa isolates with ceftazidime-avibactam MICs of 4 to 16 mg/liter (307).
Clinical studies with ceftazidime-avibactam in MDR/XDR P. aeruginosa infections are scarce and contain low numbers of patients. Doses used were 2/0.5 g/8 h, sometimes prescribed in combination. The cure rates were close to 80%, and most failures occurred in respiratory tract infections. There is limited information on mortality, microbiological eradication, recurrence, or the emergence of resistance (163, 308). Related to this, and rather worryingly, ceftazidime-avibactam-resistant P. aeruginosa isolates were identified in 9/355 (2.5%) of microbiologically evaluable patients in a phase III clinical trial that compared ceftazidime-avibactam with meropenem in nosocomial pneumonia (309).
In summary, in vitro studies have shown that ceftazidime-avibactam might be a good option for the treatment of MDR/XDR P. aeruginosa infections, but clinical experience is currently limited. Depending on the underlying mechanisms of resistance, ceftazidime-avibactam could be the best option for some MDR/XDR P. aeruginosa strains, such as those harboring Class A carbapenemases (such as GES enzymes) or combinations of certain ESBLs with OprD deficiency (Table 6). Larger series of MDR/XDR P. aeruginosa infections treated with ceftazidime-avibactam are needed.
CRITICAL EVALUATION OF CLINICAL STUDIES PROVIDING INFORMATION ON OUTCOMES OF INFECTIONS DUE TO MDR/XDR P. AERUGINOSA
Table 4 provides a summary of clinical studies including 5 or more patients that have analyzed the outcomes for patients with MDR/XDR P. aeruginosa infections treated with different systemic antibiotic regimens. Although some studies addressed only MDR/XDR P. aeruginosa infections, many others considered them jointly with other MDR GNB infections, including those caused by A. baumannii and/or Enterobacteriaceae. Among these, only those that specifically detail any outcome for patients with MDR/XDR P. aeruginosa infections are included here. The design and quality of the studies were evaluated according to the Scottish Intercollegiate Guidelines Network (SIGN) method (https://www.sign.ac.uk/methodology.html).
Most publications have analyzed patients treated with colistin (180–188, 299, 300) and, more recently, with ceftolozane-tazobactam (26, 62, 165, 281–287). There are some articles about patients treated with polymyxin B (207–209), ceftazidime-avibactam (163, 308), and aminoglycosides (250) or with different combinations of antimicrobials (203, 204, 206, 258). Overall, the number of studies is limited. Most are retrospective studies of case series or cohorts and have all the limitations inherent to this type of design, as well as small sample sizes. At the same time, most studies have other significant limitations. First, they include patients with heterogeneous baseline characteristics (age, comorbidities, and immunocompetence), infection sites, percentages of polymicrobial infections (often not provided), and severity at presentation, differences in pathogen susceptibility (MDR and XDR P. aeruginosa) and MICs of the antimicrobials (frequently not provided), use of different antimicrobial doses or dose adjustments according to the patient’s degree of renal dysfunction (especially in the case of colistin), delays in time to effective therapy, use of different antimicrobial combinations in an unplanned way, use of the antibiotic being studied after failure of initial (empirical and/or directed) antimicrobial therapy, a variety of treatment durations, and different amounts of information, or none at all, about source control. Despite this heterogeneity, outcomes are usually presented in aggregated form, making them difficult to interpret. Furthermore, in the few studies comparing different antibiotics, doses, or antimicrobial combinations, the invariably small sample sizes make it difficult to adjust for all other variables affecting the outcomes. Second, the included studies do not use a unanimous definition of multidrug-resistant P. aeruginosa. Third, the outcomes considered in different studies (clinical and microbiological responses and mortality) are frequently defined in different ways and/or are evaluated at different time points during clinical evolution. In the particular case of colistin and ceftolozane-tazobactam, an additional problem is that different studies have used different doses, with a tendency to increase them over time, which makes it difficult to compare results between publications.
As previously mentioned, until very recently, colistin was the only alternative for many cases of MDR P. aeruginosa infection. The use of this drug is complicated due to its narrow therapeutic window and frequent nephrotoxicity and by the fact that an adequate dosage has not yet been properly determined. More recently, the availability of ceftolozane-tazobactam and ceftazidime-avibactam represents a major step forward, mainly because they are active against several MDR/XDR P. aeruginosa strains, with limited side effects. However, it is difficult to use published data as a basis for comparing outcomes with these antimicrobials against those with colistin. There is a lack of clinical studies with ceftazidime-avibactam. In the case of ceftolozane-tazobactam, some clinical studies allow for some comparisons. With respect to 28- to 30-day all-cause mortality, a 32% to 47% rate was reported for colistin in 3 studies (181, 183, 192) and a 10% to 28% rate for ceftolozane-tazobactam in 4 studies (26, 62, 281, 282). As a result of the limitations of the studies that have been mentioned already, these apparent differences should be interpreted with caution. The emergence of resistant P. aeruginosa mutants during treatment with ceftolozane-tazobactam is of particular concern. This fact supports the use of ceftolozane-tazobactam at high doses, preferably in extended or continuous infusion, and also raises the question of the possible advantages of combining antibiotics in difficult-to-treat and high-inoculum infections, at least in the first days of treatment. Several studies have addressed the potential advantages of combination therapy (203, 204, 206, 258) and suggested possible alternatives: the use of β-lactams in extended or continuous infusion (in the case of MICs classified as intermediate) combined with colistin, as well as different dual therapies of combinations including doripenem (with intermediate MICs) in extended infusion, fosfomycin, or colistin. In 3 studies, the combinations showed better results than monotherapy (203, 204, 206). Nevertheless, the limitations of these studies prevent definitive conclusions from being drawn. Only one well-designed clinical trial has addressed this issue, with a comparison of the colistin-plus-meropenem combination versus colistin alone (187). Unfortunately, the study lacked enough power to reach conclusions in the case of MDR P. aeruginosa.
Another aspect that has led to several publications is the possible usefulness or advantage of administration of inhaled versus intravenous colistin or of a combination of both of them in the case of respiratory infections. Two recent meta-analyses addressed this question in patients with MDR GNB (214, 215), although the studies do not provide specific results for MDR/XDR P. aeruginosa infections. Only studies that explicitly detail the outcomes for patients with MDR P. aeruginosa respiratory infections are included here (199, 217). No definitive conclusions can be reached from these studies. This is another of the many questions in the treatment of these complex infections that remain unresolved. However, based on data on other MDR GNB infections, nebulized colistin seems acceptable as adjuvant therapy when treating MDR/XDR P. aeruginosa respiratory infections.
Considering all of the reviewed studies and data, we have defined some general therapeutic recommendations based on the most prevalent resistance profiles of MDR/XDR P. aeruginosa (Table 6).
INVESTIGATIONAL AGENTS WITH ACTIVITY AGAINST MDR/XDR P. AERUGINOSA
Imipenem-Relebactam
Relebactam is an active β-lactamase inhibitor of class A and class C β-lactamases (310), and in combination with imipenem (plus cilastatin), it can restore imipenem activity against resistant strains, including AmpC-producing P. aeruginosa (311). One study set out to assess the in vitro activity of imipenem-relebactam against 3,143 clinical strains of non-Proteus Enterobacteriaceae and P. aeruginosa collected at 21 U.S. hospitals participating in the SMART program (312). Of all P. aeruginosa strains tested, 94.4% (846/896) were susceptible to this antibiotic, compared with 74.7% (669/896) that were susceptible to imipenem. The in vitro activities of the imipenem-relebactam combination were 78% against imipenem-resistant P. aeruginosa strains and 82.2% against MDR P. aeruginosa strains. Only colistin and amikacin showed activity similar or superior to that of imipenem-relebactam. Another study evaluated its in vitro activity against strains in the ESKAPE (Enterococcus faecium, Staphylococcus aureus, Klebsiella pneumoniae, Acinetobacter baumannii, Pseudomonas aeruginosa, and Enterobacter species) group, which includes MDR P. aeruginosa (313). Imipenem-relebactam showed activity against 94% (796/845) of P. aeruginosa strains, in contrast to 70.3% (594/845) in the case of imipenem. Additionally, 80.5% of the strains of these imipenem-resistant microorganisms (MIC90, 16 μg/ml) were susceptible to imipenem-relebactam with a MIC90 of 2 μg/ml. Only amikacin showed activity comparable to that of imipenem-relebactam, with 95.6% susceptibility.
A double-blind, phase I clinical trial evaluated the pharmacokinetics and safety of relebactam, administered alone or in combination with imipenem-cilastatin (314). Administration of a single dose of relebactam over a dose range of 25 to 1,150 mg showed a terminal half-life (t1/2) of between 1.3 and 1.8 h, with pharmacokinetics similar to those of imipenem, which supports combination with this antibiotic given with the same frequency of infusion. The AUC and Cmax increased exponentially with the dose. The AUC target of 13.1 mg · h/liter was obtained with a 125-mg dose of relebactam administered with imipenem, but the AUC reached higher values when relebactam was administered as the sole drug. The pharmacokinetics of imipenem-cilastatin were similar when administered as standard combination therapy or together with relebactam. Administration of a single 125-mg dose of relebactam with 500 mg imipenem showed AUC values for relebactam that were 22% and 25% higher in adult and elderly women, respectively, than in men in each age group. Both elderly men and women had 41% and 45% higher mean plasma concentrations than the corresponding group of adult men. Relebactam was excreted almost completely in the urine, with the percentages ranging between 94.7% and 100% in the 24 h after administration of a single dose. The PK parameters for imipenem were comparable across all the groups studied. Tolerability was good, with drowsiness being the most frequently observed adverse effect. In study 2, 7-day administration of multiple doses of 125 mg of relebactam with 500 mg of imipenem showed similar t1/2 values on days 1 and 7, and accumulation of relebactam did not occur. The AUC target of 13.1 mg · h/liter was again reached with the same doses of relebactam. Again, when the compounds were administered at the described doses, the drug was generally well tolerated.
A phase II clinical trial evaluated different doses of relebactam combined with imipenem-cilastatin in patients with complicated intra-abdominal infections (315). Both the 125-mg and 250-mg doses of relebactam combined with 500 mg of imipenem showed 100% microbiological eradication in patients with P. aeruginosa infections. Of a total of 5 strains of imipenem-resistant P. aeruginosa, 3 showed susceptibility to the combination with relebactam. To date, no clinical experiences with imipenem-relebactam and MDR/XDR P. aeruginosa infections have been published. However, considering its characteristics and antimicrobial activity, this drug could play an important role in the near future in therapy against MDR/XDR P. aeruginosa.
Cefiderocol
Cefiderocol is a siderophore cephalosporin with activity against multiple Gram-negative organisms, including strains that are resistant to other antibiotics (316). Cefiderocol acts by binding to ferric iron, which enables it to use bacterial iron transporters to penetrate the external bacterial membrane. It also has high stability against β-lactamases, including serine-dependent β-lactamases and MBLs (317, 318). The ability of cefiderocol to neutralize AmpC overproduction, its stability against these enzymes, and its ability to induce AmpC in Enterobacter cloacae and P. aeruginosa have been studied (318). While the MICs of ceftazidime and cefepime for the P. aeruginosa PAO1 strain increased 4- to 16-fold due to the inactivation of AmpD and DacB, cefiderocol MICs were only slightly affected. The effect of AmpC inactivation on the MICs of these antibiotics was very limited. Similar results were observed when the effect on AmpC overproduction was studied. Hence, cefiderocol was found to be a very stable cephalosporin against these enzymes.
Another in vitro experiment evaluated the activity of cefiderocol against 1,873 clinical isolates of Gram-negative organisms from 52 countries (319). A total of 262 strains of MDR P. aeruginosa were exposed to various antibiotics; the MIC90 values of colistin and cefiderocol were 1 mg/liter, versus >8 mg/liter for ciprofloxacin and >64 mg/liter for meropenem, cefepime, ceftazidime-avibactam, and ceftolozane-tazobactam. The same finding was also observed when the activities of several antibiotics against 100 strains of imipenem-resistant P. aeruginosa were studied (320). The MIC90 of cefiderocol was 1 mg/liter, and this drug was the most active of all the antipseudomonal antibiotics studied.
A mouse model of infection was used to determine the PK/PD characteristics of this antibiotic against different strains of P. aeruginosa with cefiderocol MICs in the range of 0.063 to 0.5 mg/liter (321). When the %fT> MIC was calculated for the different strains, it was observed that the probability of achieving the therapeutic target was 100% against all strains tested at a dose of 166 mg/kg/8 h. While the results of ongoing clinical trials and other clinical studies are awaited, this drug holds great promise for the treatment of MDR/XDR P. aeruginosa infections.
Murepavadin
The need to find antibiotics with new mechanisms of action has given rise to certain agents that are able to interact with external bacterial membranes composed of phospholipids and lipopolysaccharides (322). Murepavadin is able to interact with the membrane protein LptD of P. aeruginosa as a peptidomimetic antibiotic with specific activity against this microorganism. It would therefore be the first microorganism-specific antimicrobial molecule (323). The activity of murepavadin against 785 strains of XDR P. aeruginosa has been studied and compared with those of other antibiotics, such as colistin, ceftolozane-tazobactam, and tobramycin (324). The activity of this antibiotic was excellent, since it inhibited 97.8% of isolates studied at concentrations of <2 mg/liter and showed 8 times higher activity than colistin.
A phase I study aimed to assess the PK behavior of murepavadin in healthy volunteers after a single dose ranging from 0.05 mg/kg to 4.5 mg/kg and after multiple doses ranging from 1 mg/kg to 5 mg/kg every 12 h (325). The AUCs ranged between 12,500 ng · h/ml for the lowest multiple dose and 74,500 ng · h/ml for the highest, with a mean t1/2 of between 6.17 h and 7.15 h, respectively. A further study compared the PKs of this antibiotic in patients with different degrees of renal function (326). The mean values ranged between 71.13 and 27.52 ng · h/ml for AUC, between 2.4 and 7 liters/h for clearance, between 1.4 and 7.8 h for t1/2, and between 80.9 and 76.3 liters for the volume of distribution in patients with the worst renal function (mean creatinine clearance of 25.5 ml/min) versus healthy volunteers, respectively. In healthy volunteers, the ELF/plasma ratio for this antibiotic was practically 1.
In a phase II study in patients with ventilator-associated pneumonia, murepavadin showed a clinical cure in 10 of the 12 patients with confirmed P. aeruginosa infection (327). Likewise, the mortality rate was 8%, a value that should be interpreted with caution due to the low number of patients included in the study. Murepavadin, the first microorganism-specific antipseudomonal molecule, is a promising therapeutic alternative due to its excellent antimicrobial activity and PK data.
Cefepime-Zidebactam
Zidebactam, like avibactam, is a non-β-lactam drug belonging to the diazabicyclooctane group and has a high affinity for the PBP2 locus of Gram-negative microorganisms as well as a high capacity for β-lactamase inhibition (328). A study assessed the activity of zidebactam alone or combined with cefepime against several species of Gram-negative microorganisms, including 50 strains of P. aeruginosa (329). Zidebactam showed activity against two strains of NDM-positive P. aeruginosa at concentrations of 8 and 32 mg/liter. An analysis of the in vitro behavior of this drug against 1,291 strains of P. aeruginosa was performed, which included 43 strains of MDR P. aeruginosa: 10 strains that are susceptible to cefepime, 21 strains with overexpression of AmpC or efflux pumps, and 12 MBL-producing strains (330). The MIC90 of the combination against cefepime-susceptible strains was 2 mg/liter, versus 4 mg/liter for cefepime. For strains with overexpression of AmpC or efflux pumps, the 1:1 combination had a cefepime-zidebactam MIC of 8 mg/liter, versus 16 mg/liter for the 2:1 combination and 64 mg/liter and 32 mg/liter for cefepime and zidebactam alone, respectively. Finally, for the MBL-producing strains, the MICs of the 1:1 and 2:1 combinations were 8 mg/liter and 16 mg/liter, respectively. These values show the very high in vitro activity of this new antibiotic against MDR/XDR P. aeruginosa.
A study evaluated the plasma and intrapulmonary pharmacokinetics of multiple doses of 2 g cefepime and 1 g zidebactam administered to healthy volunteers and also carried out a safety analysis (331). After the seventh dose, the mean plasma PK values of cefepime were a Cmax of 139.5 μg/ml and an AUC of 327.0 mg · h/liter, while those of zidebactam were a Cmax of 60.0 mg/liter and an AUC of 139.5 mg · h/liter. The mean concentrations of cefepime and zidebactam in ELF were reached at 1.25 h after administration, with values of 35.24 mg/liter and 14.61 mg/liter, respectively, and a plasma/ELF ratio of 2.41. In the case of alveolar macrophages, the highest mean concentrations of cefepime and zidebactam were 16.99 mg/liter at 8 h and 2.06 mg/liter at 6 h, respectively. One volunteer presented a moderate hypersensitivity reaction that was drug related. These data, together with the very high in vitro activity of this new antibiotic against MDR/XDR P. aeruginosa, define it as an excellent future option for these infections.
Bacteriophages
Bacteriophages were developed more than a century ago but were superseded by antibiotics, largely because phage activity is frequently limited to particular strains; now, however, they are being reinvestigated due to their activity against difficult-to-treat strains (332). Phage therapy has a special place in Eastern Europe, notably Russia, Georgia, and Poland. There is limited experience regarding the effectiveness of phage use. They have mostly been used in phage mixtures with activity against different resistant strains of P. aeruginosa for the treatment of infections and have shown promising results, some of them pending publication.
A review showed that the number of studies with phages for the treatment or prevention of P. aeruginosa infection is limited, and most or all have been developed in patients with cystic fibrosis and used as inhaled therapy (333). Given that some infections in this population are produced by mucoid strains of MDR P. aeruginosa, the phages used should have activity against both biofilm-producing and non-biofilm-producing strains. Combination therapy with active antibiotics and phages could be a possible option that should be evaluated in future studies. One study used an in vitro model and two mouse models to determine the activity of a combination of 6 phages against biofilm-producing MDR P. aeruginosa strains in patients with cystic fibrosis (334). The results with the bacteriophage cocktail used were encouraging and showed good capacity of the phages in reducing bacterial load and biofilm formation. This could be a useful strategy for local treatment of deep infections, such as bone and joint infections caused by XDR P. aeruginosa (335). Another study was based on the isolation of different P. aeruginosa phages from hospital sewage samples, specifically SL1, SL2, and SL4 (336). The 5 strains of MDR P. aeruginosa were affected by at least one of the phages studied, and no bacterial regrowth was observed. Phage SL2 showed the highest activity against planktonic strains, while SL4 was the best against the biofilm model. The highest survival rate was achieved with SL1. However, the activity of the phage cocktail was not better than that of the most active phage used individually.
The use of inhaled antibiotics is currently spreading, and one study set out to evaluate three different types of nebulizer for administering bacteriophage PEV44, which is active against P. aeruginosa (337). The authors concluded that the Omron NE22 nebulizer best maintained phage viability. The efficacy and safety of inhaled administration of P. aeruginosa phage therapy were evaluated in a mouse model of pulmonary infection caused by MDR P. aeruginosa (338). The results demonstrated that intratracheal administration of dry phage powder significantly reduced the bacterial load of MDR P. aeruginosa in the lungs of the mice and resulted in minimal damage to the lung tissue.
Another strategy that has been studied for the treatment of infections caused by resistant microorganisms is the use of phages combined with antibiotics (339). Phage-drug combination therapy has been shown to be superior to the activity of each agent separately. Phage-antibiotic synergy (PAS) was studied in an in vitro model in which phages of the family Myoviridae, genus Pbunavirus, showed synergy with multiple antibiotics, and in the case of the phage active against P. aeruginosa, with piperacillin and ceftazidime (340).
Bacteriocins
Bacteriocins are substances with antimicrobial activity that are produced by some bacteria. It has been proposed that they could be used clinically for the treatment for infections caused by multidrug-resistant microorganisms (341). One study assessed the activity of three R-type pyocins produced by strains of P. aeruginosa against clinical isolates of this microorganism in patients with cystic fibrosis (342). These substances showed potential therapeutic activity that should be considered in future clinical studies.
Anti-Quorum-Sensing Strategies
Quorum-sensing molecules are regulators of virulence mechanisms that are present in diverse microorganisms, including MDR/XDR P. aeruginosa (343, 344). These regulators are also involved in the formation of biofilm and in the regulation of gene expression that underlies collective behaviors in cellular populations. Interfering with these molecules has been proposed as an alternative or complementary tool against MDR bacteria by inhibiting their pathogenicity and biofilm formation (345). The strategies to interfere with quorum sensing are directed against the biosynthesis, accumulation, and detection of the signals derived from small molecules that act as self-inductors of the propagation of quorum sensing (346). Among the strategies to inhibit quorum sensing are the interference with transcriptional factors related to DNA transcription and signal interference of the quorum sensing once it has been detected, as has been applied successfully with P. aeruginosa (347), as well as the inhibition of bacterial enzymes related to this process in different bacteria, including P. aeruginosa (348).
Vaccines and Monoclonal Antibodies
One of the strategies for combating infections caused by MDR/XDR microorganisms involves the use of vaccines or monoclonal antibodies, which are emerging as novel tools for preventing the acquisition of MDR P. aeruginosa infections in high-risk patients (349). Some vaccines, namely IC43, KB001-A, and KBPA-101, have already been tested in critically ill patients (350). A study performed in ventilated critically ill patients confirmed the immunogenicity of IC43 vaccination in this population (351).
With respect to antibodies, new active targets aimed at neutralizing virulence factors such as the P. aeruginosa type III secretion system (TTSS) are under development as monoclonal antibodies and vaccines (352). PcrV is an essential protein for TTSS activity and the one that has been used mostly as a target. The aim of one study was to develop monoclonal antibodies that neutralize the virulence of the TTSS by acting on the PcrV protein (353). In a clinical study, KB001-A, a pegylated antibody that inhibits the function of the PcrV protein, was administered to cystic fibrosis patients infected with P. aeruginosa. Although the endpoint of clinical efficacy was not achieved, possible benefits in the regulation of infection and inflammation in these patients were observed (354). KBPA-101 is another human monoclonal antibody obtained from healthy volunteers who were given a polysaccharide conjugate vaccine with P. aeruginosa toxin A (355). This antibody showed linear pharmacokinetics in healthy volunteers and no adverse effects. Consequently, it has been proposed for future use as an alternative for the prevention of P. aeruginosa infections (356). Another monoclonal antibody (V2L2MD) showed very good prophylactic protection in several mouse models of P. aeruginosa infection. In a rabbit model of pneumonia, MEDI3902, a selective monoclonal antibody against PcrV proteins, and Psl exopolysaccharide showed a highly protective effect against a highly virulent P. aeruginosa strain (357). This protective activity was observed in another experiment that demonstrated the high specificity of this monoclonal antibody against the PcrV epitopes of most of the P. aeruginosa strains studied, as well as maintenance of its protective effect (358). These results led to a phase I study in healthy volunteers (359) that demonstrated the efficacy and tolerability of this drug. At present, this antibody has been included in a phase II proof-of-concept trial for evaluating the prevention of nosocomial pneumonia in patients colonized by P. aeruginosa (360).
ACKNOWLEDGMENTS
Juan P. Horcajada is supported by Plan Nacional de I + D+i 2013–2016 and Instituto de Salud Carlos III, Subdirección General de Redes y Centros de Investigación Cooperativa, Ministerio de Economía, Industria y Competitividad, Spanish Network for Research in Infectious Diseases (REIPI RD16/0016), by grant PI17/00251 cofinanced by European Development Regional Fund ERDF “A Way To Achieve Europe,” Operative Program Intelligent Growth 2014–2020, and by the European Union through the 11th call of the Innovative Medicines Initiative (grant COMBACTE). Milagro Montero is supported by Plan Nacional de I + D+i 2013–2016 and Instituto de Salud Carlos III, Subdirección General de Redes y Centros de Investigación Cooperativa, Ministerio de Economía, Industria y Competitividad grant PI16/00669 cofinanced by European Development Regional Fund ERDF “A Way To Achieve Europe,” Operative Program Intelligent Growth 2014–2020. Antonio Oliver is supported by Plan Nacional de I + D+i 2013–2016 and Instituto de Salud Carlos III, Subdirección General de Redes y Centros de Investigación Cooperativa, Ministerio de Economía, Industria y Competitividad, Spanish Network for Research in Infectious Diseases (REIPI RD16/0016), and by grants PI15/00088 and PI18/00076 cofinanced by European Development Regional Fund ERDF “A Way To Achieve Europe,” Operative Program Intelligent Growth 2014–2020. He is also supported by the European Union through the 11th call of the Innovative Medicines Initiative (grant COMBACTE-MAGNET).
Juan P. Horcajada has received fees as a speaker and participant in advisory board meetings from Angelini, Pfizer, MSD, and Zanbom and a research grant from MSD. Antonio Oliver has received research grants and fees as a speaker and participant in advisory board meetings from MSD and Pfizer. Natividad Benito has received fees (for lectures, speaker bureaus, and/or expert testimony) and/or nonfinancial support from MSD, Pfizer, Angellini, Novartis, Gilead, and AstraZeneca. Santiago Grau has received fees as a speaker from Pfizer, Angellini, Kern, and MSD and research grants from Astellas Pharma and Pfizer. Sonia Luque is supported by a travel research grant from the Spanish Society of Infectious Diseases and Clinical Microbiology (SEIMC). The other authors have no potential conflicts of interest.
We thank Janet Dawson for English editing.
Biographies

Juan P. Horcajada, M.D., Ph.D., is Head of the Department of Infectious Diseases at Hospital del Mar, Barcelona, Spain, and Aggregate Professor in the Department of Medicine, Faculty of Medicine, at Universitat Autònoma de Barcelona (UAB) (Barcelona, Spain). He is also coordinator of the Infectious Pathology and Antimicrobials Research Group (IPAR) at the Institut Hospital del Mar d’Investigations Mèdiques (IMIM) of Barcelona, Spain. He is a member of the Executive Committee of the Spanish Network for Research in Infectious Diseases (REIPI), advisor of the Spanish national Bacterial Resistance Control Program (PRAN), Spanish Medicines Agency, Ministry of Health of Spain, Spanish and Catalan cocoordinator of national and regional Antimicrobial Stewardship Programs (PROA), and Spanish delegate of the European Committee for Infection Control (EUCIC) of the European Society of Clinical Microbiology and Infectious Diseases (ESCMID). His main research interests include the clinical investigation of antimicrobial-resistant infections, with a particular focus in Pseudomonas aeruginosa infections, and the study of the impact of pharmacokinetics and pharmacodynamics in the clinical management of multidrug-resistant and complex infections. Dr. Horcajada has published over 200 articles in peer-reviewed journals.

Milagro Montero, M.D., Ph.D., is an internal medicine specialist and a young researcher who is focused on the study of XDR Pseudomonas aeruginosa, as evidenced by her doctoral thesis and several papers published over the last 5 years. She has also been a research collaborator on a significant number of competitive grant-receiving projects, many of them on the treatment of XDR P. aeruginosa. She did a 1-year postdoctoral stay thanks to a grant for young researchers awarded by the Institute Hospital del Mar Barcelona Medical Research (IMIM). This training was at the Institute for Clinical and Pharmacodynamics (ICPD), Latham, New York, one of the worldwide reference centers for the study of pharmacokinetics and pharmacodynamics of antibiotics. Currently she is involved in different PK/PD research projects investigating treatment of infectious diseases.

Antonio Oliver is a Doctor in Pharmacy (Complutense University of Madrid, 2002) and Specialist in Clinical Microbiology (Hospital Ramón y Cajal, Madrid, 2001). Since 2002 he has worked as a clinical microbiologist in the Department of Microbiology of Hospital Son Espases (formerly Hospital Son Dureta) in Palma de Mallorca. Since 2003 Dr. Oliver has been an associate professor of microbiology in the University of the Balearic Islands and is the principal investigator of the Antimicrobial Resistance and Pathogenicity of Bacterial Infections Group of the Health Research Institute of the Balearic Islands (IdISBa). Dr. Oliver is a member of the Scientific Committee of the Spanish Network for Research in Infectious Diseases (REIPI) and an ESCMID fellow. His main research interest is the investigation of the mechanisms of adaptation and antimicrobial resistance in bacterial pathogens, with a particular focus on Pseudomonas aeruginosa infections and the development of novel strategies to combat them. Within this research line, Dr. Oliver has published over 200 articles in peer-reviewed journals.

Luisa Sorlí, M.D., Ph.D., obtained her medical degree from the University of Valencia and her Ph.D. from the Universitat Autònoma de Barcelona. She is a specialist in internal medicine and a faculty member of the Infectious Diseases Department at the Hospital del Mar in Barcelona, Spain. She is a member of the Infectious Pathology and Antimicrobials Research Group (IPAR), Institut Hospital del Mar d’Investigacions Mèdiques (IMIM), and of the Spanish Network for Research in Infectious Diseases (REIPI). Her research interests have focused on the treatment of patients with multidrug-resistant Gram-negative bacterial infections and on the pharmacokinetics of colistin in the clinics.

Sònia Luque, Pharm D., Ph.D., obtained her Pharm D. degree at the University of Barcelona and her Ph.D. from the University Autònoma of Barcelona. She is a faculty member of the Pharmacy Department at the Hospital del Mar, Barcelona, Spain. She has a master's degree in application and control of antimicrobial therapy from the Universitat Autònoma of Barcelona. She has been working on the study of the pharmacokinetic and pharmacodynamics of antimicrobials and is currently involved in various national and international research projects. She is a member of the Infectious Pathology and Antimicrobials Research Group (IPAR), Institut Hospital del Mar d’Investigacions Mèdiques (IMIM), of the Spanish Network for Research in Infectious Diseases (REIPI), and of the Pharmacokinetics/Pharmacodynamics (PK/PD) group of the Anti-Infectives Study Group (EPASG) of the European Society of Clinical Microbiology and Infectious Diseases (ESCMID). She was awarded a postdoctoral fellowship at the Center for Pharmacodynamics of the University of Liverpool. She has published over 30 articles in the field of the infectious diseases in different international journals.

Silvia Gómez-Zorrilla, M.D., Ph.D., is faculty member of the Infectious Diseases Service at the Hospital del Mar and an associate professor at Pompeu Fabra University (Barcelona, Spain). She completed her medical degree at Complutense University of Madrid and her specialty in internal medicine and her predoctoral studies at Hospital Universitari de Bellvitge (Barcelona, Spain). She obtained her Ph.D. from the University of Barcelona. She has a postgraduate degree in research in clinical science. Dr. Gómez-Zorrilla’s main research interests include multidrug-resistant Gram-negative infections, mainly P. aeruginosa, and the prevention and control of multidrug-resistant organism. Her Ph.D. thesis was titled “Impact of multidrug resistance on the pathogenicity of Pseudomonas aeruginosa: an epidemiological, clinical and experimental perspective.” She is also involved in various international clinical research projects investigating new antimicrobial agents.

Natividad Benito, M.D., Ph.D., is a specialist in internal medicine. She is Consultant in the Infectious Diseases Unit at the Hospital de la Santa Creu i Sant Pau and Associate Professor in the Department of Medicine of the Universitat Autònoma de Barcelona (UAB) (Barcelona, Spain). She has a master’s degree in epidemiology and clinical research from UAB. Currently, she is President of the Study Group for Bone and Joint Infections (GEIO) of the Spanish Society of Infectious Diseases and Clinical Microbiology (SEIMC), as well as a member of the Spanish Antibiogram Committee (COESANT). Her research interests have focused on different aspects of the prevention, diagnosis, and treatment of complex infections (bone and joint, endovascular, and central nervous system infections, health care-associated and multidrug-resistant infections, infections in immunocompromised patients).

Santiago Grau is Head of Department of Pharmacy at the Hospital del Mar, Barcelona, and Aggregate Professor in the Department of Pharmacology, Faculty of Medicine, in the Universitat Autònoma de Barcelona (Barcelona, Spain). He also holds the following positions: Master in Infectious Diseases, Faculty of Medicine, Universitat Autònoma of Barcelona; member of the Infectious Pathology and Antimicrobials Research Group (IPAR) in the Hospital del Mar Medical Research Institute (IMIM); Coordinator of Master in Use and Control of Antimicrobial Therapy, Faculty of Medicine, Universitat Autònoma of Barcelona; advisor of the Bacterial Resistance Control Program (PRAN), Spanish Medicines Agency, Ministry of Health of Spain; member of the Scientific Committee for the Antimicrobial Stewardship Program of the European Society of Clinical Microbiology and Infectious Diseases (ESCMID), and member of the Technical Committee of the Nosocomial Infections Surveillance in Catalonia, Spain.
REFERENCES
- 1.Poole K. 2011. Pseudomonas aeruginosa: resistance to the max. Front Microbiol 2:65. doi: 10.3389/fmicb.2011.00065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Breidenstein EBM, de la Fuente-Núñez C, Hancock R. 2011. Pseudomonas aeruginosa: all roads lead to resistance. Trends Microbiol 19:419–426. doi: 10.1016/j.tim.2011.04.005. [DOI] [PubMed] [Google Scholar]
- 3.Oliver A, Mulet X, López-Causapé C, Juan C. 2015. The increasing threat of Pseudomonas aeruginosa high-risk clones. Drug Resist Updat 21–22:41–59. doi: 10.1016/j.drup.2015.08.002. [DOI] [PubMed] [Google Scholar]
- 4.Tacconelli E, Carrara E, Savoldi A, Harbarth S, Mendelson M, Monnet DL, Pulcini C, Kahlmeter G, Kluytmans J, Carmeli Y, Ouellette M, Outterson K, Patel J, Cavaleri M, Cox EM, Houchens CR, Grayson ML, Hansen P, Singh N, Theuretzbacher U, Magrini N, Aboderin AO, Al-Abri SS, Awang Jalil N, Benzonana N, Bhattacharya S, Brink AJ, Burkert FR, Cars O, Cornaglia G, Dyar OJ, Friedrich AW, Gales AC, Gandra S, Giske CG, Goff DA, Goossens H, Gottlieb T, Guzman Blanco M, Hryniewicz W, Kattula D, Jinks T, Kanj SS, Kerr L, Kieny M-P, Kim YS, Kozlov RS, Labarca J, Laxminarayan R, Leder K, Leibovici L, Levy-Hara G, Littman J, Malhotra-Kumar S, Manchanda V, Moja L, Ndoye B, Pan A, Paterson DL, Paul M, Qiu H, Ramon-Pardo P, Rodríguez-Baño J, Sanguinetti M, Sengupta S, Sharland M, Si-Mehand M, Silver LL, Song W, Steinbakk M, Thomsen J, Thwaites GE, van der Meer JW, Van Kinh N, Vega S, Villegas MV, Wechsler-Fördös A, Wertheim HFL, Wesangula E, Woodford N, Yilmaz FO, Zorzet A, WHO Pathogens Priority List Working Group. 2018. Discovery, research, and development of new antibiotics: the WHO priority list of antibiotic-resistant bacteria and tuberculosis. Lancet Infect Dis 18:318–327. doi: 10.1016/S1473-3099(17)30753-3. [DOI] [PubMed] [Google Scholar]
- 5.Pena C, Cabot G, Gomez-Zorrilla S, Zamorano L, Ocampo-Sosa A, Murillas J, Almirante B, Pomar V, Aguilar M, Granados A, Calbo E, Rodriguez-Bano J, Rodriguez-Lopez F, Tubau F, Martinez-Martinez L, Oliver A, Gurgui M, Sorde R, Larrosa N, Martin C, Fontanals D, de Cueto M, Navarro MD, Torre-Cisneros J, Casal M, Lara R, Natera C, Rivero A, Peña C, Cabot G, Gómez-Zorrilla S, Zamorano L, Ocampo-Sosa A, Murillas J, Almirante B, Pomar V, Aguilar M, Granados A, Calbo E, Rodríguez-Baño J, Rodríguez-López F, Tubau F, Martínez-Martínez L, Oliver A, Spanish Network for Research in Infectious Diseases (REIPI). 2015. Influence of virulence genotype and resistance profile in the mortality of Pseudomonas aeruginosa bloodstream infections. Clin Infect Dis 60:539–548. doi: 10.1093/cid/ciu866. [DOI] [PubMed] [Google Scholar]
- 6.Walkty A, Lagace-Wiens P, Adam H, Baxter M, Karlowsky J, Mulvey MR, McCracken M, Zhanel GG. 2017. Antimicrobial susceptibility of 2906 Pseudomonas aeruginosa clinical isolates obtained from patients in Canadian hospitals over a period of 8 years: Results of the Canadian Ward surveillance study (CANWARD), 2008–2015. Diagn Microbiol Infect Dis 87:60–63. doi: 10.1016/j.diagmicrobio.2016.10.003. [DOI] [PubMed] [Google Scholar]
- 7.Sader HS, Castanheira M, Duncan LR, Flamm RK. 2018. Antimicrobial susceptibility of Enterobacteriaceae and Pseudomonas aeruginosa isolates from United States medical centers stratified by infection type: results from the International Network for Optimal Resistance Monitoring (INFORM) surveillance program. Diagn Microbiol Infect Dis 92:69–74. doi: 10.1016/j.diagmicrobio.2018.04.012. [DOI] [PubMed] [Google Scholar]
- 8.ECDC. 2015. Antimicrobial resistance surveillance in Europe 2015. Annual report of the European Antimicrobial Resistance Surveillance Network (EARS-Net). European Centre for Disease Prevention and Control. [Google Scholar]
- 9.CDC. 2013. Antibiotic resistance threats in the United States, 2013. CDC, Atlanta, GA. [Google Scholar]
- 10.Lister PD, Wolter DJ, Hanson ND. 2009. Antibacterial-resistant Pseudomonas aeruginosa: clinical impact and complex regulation of chromosomally encoded resistance mechanisms. Clin Microbiol Rev 22:582–610. doi: 10.1128/CMR.00040-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Skiada A, Markogiannakis A, Plachouras D, Daikos GL. 2011. Adaptive resistance to cationic compounds in Pseudomonas aeruginosa. Int J Antimicrob Agents 37:187–193. doi: 10.1016/j.ijantimicag.2010.11.019. [DOI] [PubMed] [Google Scholar]
- 12.Muller C, Plésiat P, Jeannot K. 2011. A two-component regulatory system interconnects resistance to polymyxins, aminoglycosides, fluoroquinolones, and β-lactams in Pseudomonas aeruginosa. Antimicrob Agents Chemother 55:1211–1221. doi: 10.1128/AAC.01252-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Juan C, Torrens G, González-Nicolau M, Oliver A. 2017. Diversity and regulation of intrinsic β-lactamases from non-fermenting and other Gram-negative opportunistic pathogens. FEMS Microbiol Rev 41:781–815. doi: 10.1093/femsre/fux043. [DOI] [PubMed] [Google Scholar]
- 14.Girlich D, Naas T, Nordmann P. 2004. Biochemical characterization of the naturally occurring oxacillinase OXA-50 of Pseudomonas aeruginosa. Antimicrob Agents Chemother 48:2043–2048. doi: 10.1128/AAC.48.6.2043-2048.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kong K-F, Jayawardena SR, Del Puerto A, Wiehlmann L, Laabs U, Tümmler B, Mathee K. 2005. Characterization of poxB, a chromosomal-encoded Pseudomonas aeruginosa oxacillinase. Gene 358:82–92. doi: 10.1016/j.gene.2005.05.027. [DOI] [PubMed] [Google Scholar]
- 16.Fajardo A, Hernando-Amado S, Oliver A, Ball G, Filloux A, Martinez JL. 2014. Characterization of a novel Zn2+-dependent intrinsic imipenemase from Pseudomonas aeruginosa. J Antimicrob Chemother 69:2972–2978. doi: 10.1093/jac/dku267. [DOI] [PubMed] [Google Scholar]
- 17.Li X-Z, Plésiat P, Nikaido H. 2015. The challenge of efflux-mediated antibiotic resistance in Gram-negative bacteria. Clin Microbiol Rev 28:337–418. doi: 10.1128/CMR.00117-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Alvarez-Ortega C, Wiegand I, Olivares J, Hancock REW, Martinez JL. 2010. Genetic determinants involved in the susceptibility of Pseudomonas aeruginosa to beta-lactam antibiotics. Antimicrob Agents Chemother 54:4159–4167. doi: 10.1128/AAC.00257-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dötsch A, Becker T, Pommerenke C, Magnowska Z, Jänsch L, Häussler S. 2009. Genomewide identification of genetic determinants of antimicrobial drug resistance in Pseudomonas aeruginosa. Antimicrob Agents Chemother 53:2522–2531. doi: 10.1128/AAC.00035-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cabot G, Ocampo-Sosa AA, Tubau F, Macia MD, Rodríguez C, Moya B, Zamorano L, Suárez C, Peña C, Martínez-Martínez L, Oliver A, Spanish Network for Research in Infectious Diseases (REIPI). 2011. Overexpression of AmpC and efflux pumps in Pseudomonas aeruginosa isolates from bloodstream infections: prevalence and impact on resistance in a Spanish multicenter study. Antimicrob Agents Chemother 55:1906–1911. doi: 10.1128/AAC.01645-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Moya B, Dötsch A, Juan C, Blázquez J, Zamorano L, Haussler S, Oliver A. 2009. Beta-lactam resistance response triggered by inactivation of a nonessential penicillin-binding protein. PLoS Pathog 5:e1000353. doi: 10.1371/journal.ppat.1000353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Juan C, Maciá MD, Gutiérrez O, Vidal C, Pérez JL, Oliver A. 2005. Molecular mechanisms of beta-lactam resistance mediated by AmpC hyperproduction in Pseudomonas aeruginosa clinical strains. Antimicrob Agents Chemother 49:4733–4738. doi: 10.1128/AAC.49.11.4733-4738.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cabot G, Bruchmann S, Mulet X, Zamorano L, Moyà B, Juan C, Haussler S, Oliver A. 2014. Pseudomonas aeruginosa ceftolozane-tazobactam resistance development requires multiple mutations leading to overexpression and structural modification of AmpC. Antimicrob Agents Chemother 58:3091–3099. doi: 10.1128/AAC.02462-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lahiri SD, Alm RA. 2016. Identification of novel VEB β-lactamase enzymes and their impact on avibactam inhibition. Antimicrob Agents Chemother 60:3183–3186. doi: 10.1128/AAC.00047-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fraile-Ribot PA, Cabot G, Mulet X, Periañez L, Martín-Pena ML, Juan C, Pérez JL, Oliver A. 2018. Mechanisms leading to in vivo ceftolozane/tazobactam resistance development during the treatment of infections caused by MDR Pseudomonas aeruginosa. J Antimicrob Chemother 73:658–663. doi: 10.1093/jac/dkx424. [DOI] [PubMed] [Google Scholar]
- 26.Haidar G, Philips NJ, Shields RK, Snyder D, Cheng S, Potoski BA, Doi Y, Hao B, Press EG, Cooper VS, Clancy CJ, Nguyen MH. 2017. Ceftolozane-tazobactam for the treatment of multidrug-resistant Pseudomonas aeruginosa infections: clinical effectiveness and evolution of resistance. Clin Infect Dis 65:110–120. doi: 10.1093/cid/cix182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Berrazeg M, Jeannot K, Ntsogo Enguéné VY, Broutin I, Loeffert S, Fournier D, Plésiat P. 2015. Mutations in β-lactamase AmpC increase resistance of Pseudomonas aeruginosa isolates to antipseudomonal cephalosporins. Antimicrob Agents Chemother 59:6248–6255. doi: 10.1128/AAC.00825-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Diaz Caballero J, Clark ST, Coburn B, Zhang Y, Wang PW, Donaldson SL, Tullis DE, Yau YCW, Waters VJ, Hwang DM, Guttman DS. 2015. Selective sweeps and parallel pathoadaptation drive Pseudomonas aeruginosa evolution in the cystic fibrosis lung. mBio 6:e00981-15. doi: 10.1128/mBio.00981-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.López-Causapé C, Sommer LM, Cabot G, Rubio R, Ocampo-Sosa AA, Johansen HK, Figuerola J, Cantón R, Kidd TJ, Molin S, Oliver A. 2017. Evolution of the Pseudomonas aeruginosa mutational resistome in an international cystic fibrosis clone. Sci Rep 7:5555. doi: 10.1038/s41598-017-05621-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cabot G, López-Causapé C, Ocampo-Sosa AA, Sommer LM, Domínguez MÁ, Zamorano L, Juan C, Tubau F, Rodríguez C, Moyà B, Peña C, Martínez-Martínez L, Plesiat P, Oliver A. 2016. Deciphering the resistome of the widespread Pseudomonas aeruginosa sequence type 175 international high-risk clone through whole-genome sequencing. Antimicrob Agents Chemother 60:7415–7423. doi: 10.1128/AAC.01720-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.del Barrio-Tofiño E, López-Causapé C, Cabot G, Rivera A, Benito N, Segura C, Montero MM, Sorlí L, Tubau F, Gómez-Zorrilla S, Tormo N, Durá-Navarro R, Viedma E, Resino-Foz E, Fernández-Martínez M, González-Rico C, Alejo-Cancho I, Martínez JA, Labayru-Echverria C, Dueñas C, Ayestarán I, Zamorano L, Martinez-Martinez L, Horcajada JP, Oliver A. 2017. Genomics and susceptibility profiles of extensively drug-resistant Pseudomonas aeruginosa isolates from Spain. Antimicrob Agents Chemother 61:e01589-17. doi: 10.1128/AAC.02352-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cabot G, Zamorano L, Moyà B, Juan C, Navas A, Blázquez J, Oliver A. 2016. Evolution of Pseudomonas aeruginosa antimicrobial resistance and fitness under low and high mutation rates. Antimicrob Agents Chemother 60:1767–1778. doi: 10.1128/AAC.02676-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cabot G, Florit-Mendoza L, Sánchez-Diener I, Zamorano L, Oliver A. 2018. Deciphering β-lactamase-independent β-lactam resistance evolution trajectories in Pseudomonas aeruginosa. J Antimicrob Chemother 73:3322–3331. doi: 10.1093/jac/dky364. [DOI] [PubMed] [Google Scholar]
- 34.Han S, Zaniewski RP, Marr ES, Lacey BM, Tomaras AP, Evdokimov A, Miller JR, Shanmugasundaram V. 2010. Structural basis for effectiveness of siderophore-conjugated monocarbams against clinically relevant strains of Pseudomonas aeruginosa. Proc Natl Acad Sci U S A 107:22002–22007. doi: 10.1073/pnas.1013092107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Riera E, Cabot G, Mulet X, García-Castillo M, del Campo R, Juan C, Cantón R, Oliver A. 2011. Pseudomonas aeruginosa carbapenem resistance mechanisms in Spain: impact on the activity of imipenem, meropenem and doripenem. J Antimicrob Chemother 66:2022–2027. doi: 10.1093/jac/dkr232. [DOI] [PubMed] [Google Scholar]
- 36.Moyá B, Beceiro A, Cabot G, Juan C, Zamorano L, Alberti S, Oliver A. 2012. Pan-β-lactam resistance development in Pseudomonas aeruginosa clinical strains: molecular mechanisms, penicillin-binding protein profiles, and binding affinities. Antimicrob Agents Chemother 56:4771–4778. doi: 10.1128/AAC.00680-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hocquet D, Berthelot P, Roussel-Delvallez M, Favre R, Jeannot K, Bajolet O, Marty N, Grattard F, Mariani-Kurkdjian P, Bingen E, Husson M-O, Couetdic G, Plésiat P. 2007. Pseudomonas aeruginosa may accumulate drug resistance mechanisms without losing its ability to cause bloodstream infections. Antimicrob Agents Chemother 51:3531–3536. doi: 10.1128/AAC.00503-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Solé M, Fàbrega A, Cobos-Trigueros N, Zamorano L, Ferrer-Navarro M, Ballesté-Delpierre C, Reustle A, Castro P, Nicolás JM, Oliver A, Martínez JA, Vila J. 2015. In vivo evolution of resistance of Pseudomonas aeruginosa strains isolated from patients admitted to an intensive care unit: mechanisms of resistance and antimicrobial exposure. J Antimicrob Chemother 70:3004–3013. doi: 10.1093/jac/dkv228. [DOI] [PubMed] [Google Scholar]
- 39.Guénard S, Muller C, Monlezun L, Benas P, Broutin I, Jeannot K, Plésiat P. 2014. Multiple mutations lead to MexXY-OprM-dependent aminoglycoside resistance in clinical strains of Pseudomonas aeruginosa. Antimicrob Agents Chemother 58:221–228. doi: 10.1128/AAC.01252-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Köhler T, Epp SF, Curty LK, Pechère JC. 1999. Characterization of MexT, the regulator of the MexE-MexF-OprN multidrug efflux system of Pseudomonas aeruginosa. J Bacteriol 181:6300–6305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mulet X, Moyá B, Juan C, Macià MD, Pérez JL, Blázquez J, Oliver A. 2011. Antagonistic interactions of Pseudomonas aeruginosa antibiotic resistance mechanisms in planktonic but not biofilm growth. Antimicrob Agents Chemother 55:4560–4568. doi: 10.1128/AAC.00519-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bruchmann S, Dötsch A, Nouri B, Chaberny IF, Häussler S. 2013. Quantitative contributions of target alteration and decreased drug accumulation to Pseudomonas aeruginosa fluoroquinolone resistance. Antimicrob Agents Chemother 57:1361–1368. doi: 10.1128/AAC.01581-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Feng Y, Jonker MJ, Moustakas I, Brul S, ter Kuile BH. 2016. Dynamics of mutations during development of resistance by Pseudomonas aeruginosa against five antibiotics. Antimicrob Agents Chemother 60:4229–4236. doi: 10.1128/AAC.00434-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.López-Causapé C, Cabot G, Del Barrio-Tofiño E, Oliver A. 2018. The versatile mutational resistome of Pseudomonas aeruginosa. Front Microbiol 9:685. doi: 10.3389/fmicb.2018.00685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Greipel L, Fischer S, Klockgether J, Dorda M, Mielke S, Wiehlmann L, Cramer N, Tümmler B. 2016. Molecular epidemiology of mutations in antimicrobial resistance loci of Pseudomonas aeruginosa isolates from airways of cystic fibrosis patients. Antimicrob Agents Chemother 60:6726–6734. doi: 10.1128/AAC.00724-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bolard A, Plésiat P, Jeannot K. 2017. Mutations in Gene fusA1 as a novel mechanism of aminoglycoside resistance in clinical strains of Pseudomonas aeruginosa. Antimicrob Agents Chemother 62:e01835-17. doi: 10.1128/AAC.01835-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Olaitan AO, Morand S, Rolain J-M. 2014. Mechanisms of polymyxin resistance: acquired and intrinsic resistance in bacteria. Front Microbiol 5:643. doi: 10.3389/fmicb.2014.00643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gutu AD, Sgambati N, Strasbourger P, Brannon MK, Jacobs MA, Haugen E, Kaul RK, Johansen HK, Høiby N, Moskowitz SM. 2013. Polymyxin resistance of Pseudomonas aeruginosa phoQ mutants is dependent on additional two-component regulatory systems. Antimicrob Agents Chemother 57:2204–2215. doi: 10.1128/AAC.02353-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Patel G, Bonomo RA. 2011. Status report on carbapenemases: challenges and prospects. Expert Rev Anti Infect Ther 9:555–570. doi: 10.1586/eri.11.28. [DOI] [PubMed] [Google Scholar]
- 50.Juan C, Conejo MC, Tormo N, Gimeno C, Pascual Á, Oliver A. 2013. Challenges for accurate susceptibility testing, detection and interpretation of β-lactam resistance phenotypes in Pseudomonas aeruginosa: results from a Spanish multicentre study. J Antimicrob Chemother 68:619–630. doi: 10.1093/jac/dks439. [DOI] [PubMed] [Google Scholar]
- 51.Diene SMM, Rolain J-M. 2014. Carbapenemase genes and genetic platforms in Gram-negative bacilli: Enterobacteriaceae, Pseudomonas and Acinetobacter species. Clin Microbiol Infect 20:831–838. doi: 10.1111/1469-0691.12655. [DOI] [PubMed] [Google Scholar]
- 52.van der Zee A, Kraak WB, Burggraaf A, Goessens WHF, Pirovano W, Ossewaarde JM, Tommassen J. 2018. Spread of carbapenem resistance by transposition and conjugation among Pseudomonas aeruginosa. Front Microbiol 9:2057. doi: 10.3389/fmicb.2018.02057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Botelho J, Grosso F, Quinteira S, Mabrouk A, Peixe L. 2017. The complete nucleotide sequence of an IncP-2 megaplasmid unveils a mosaic architecture comprising a putative novel blaVIM-2-harbouring transposon in Pseudomonas aeruginosa. J Antimicrob Chemother 72:2225–2229. doi: 10.1093/jac/dkx143. [DOI] [PubMed] [Google Scholar]
- 54.Botelho J, Grosso F, Peixe L. 2018. Unravelling the genome of a Pseudomonas aeruginosa isolate belonging to the high-risk clone ST235 reveals an integrative conjugative element housing a blaGES-6 carbapenemase. J Antimicrob Chemother 73:77–83. doi: 10.1093/jac/dkx337. [DOI] [PubMed] [Google Scholar]
- 55.Potron A, Poirel L, Nordmann P. 2015. Emerging broad-spectrum resistance in Pseudomonas aeruginosa and Acinetobacter baumannii: mechanisms and epidemiology. Int J Antimicrob Agents 45:568–585. doi: 10.1016/j.ijantimicag.2015.03.001. [DOI] [PubMed] [Google Scholar]
- 56.del Barrio-Tofiño E, Zamorano L, Cortes-Lara S, López-Causapé C, Sánchez-Diener I, Cabot G, Bou G, Martínez-Martínez L, Oliver A, Galán F, Gracia I, Rodríguez MA, Martín L, Sánchez JM, Viñuela L, García MV, Lepe JA, Aznar J, López-Hernández I, Seral C, Javier Castillo-García F, López-Calleja AI, Aspiroz C, de la Iglesia P, Ramón S, Riera E, Cruz Pérez M, Gallegos C, Calvo J, Dolores Quesada M, Marco F, Hoyos Y, Pablo Horcajada J, Larrosa N, González JJ, Tubau F, Capilla S, Pérez-Moreno MO, Centelles MJ, Padilla E, Rivera A, Mirelis B, Elisa Rodríguez-Tarazona R, Arenal-Andrés N, del Pilar Ortega M, Megías G, García I, Colmenarejo C, González JC, Martínez NM, Gomila B, Giner S, Tormo N, Garduño E, Agulla JA, Seoane A, Pita J, Vidal IP, Guzmán DM, García M, Pérez del Molino ML, Barbeito G, Artiles F, Azcona-Gutiérrez JM, Sáenz Y, Antonio Oteo J, González A, Villa J, Chaves F, Cercenado E, Alarcón T, Zurita ND, Merino I, Morosini MI, Cantón R, Isabel Sánchez M, Moreno L, Yagüe G, Leiva J, Luis Barrios J, Canut A, Oteo J. 2019. Spanish nationwide survey on Pseudomonas aeruginosa antimicrobial resistance mechanisms and epidemiology. J Antimicrob Chemother 74:1825–1835. doi: 10.1093/jac/dkz147. [DOI] [PubMed] [Google Scholar]
- 57.Chávez-Jacobo VM, Hernández-Ramírez KC, Romo-Rodríguez P, Pérez-Gallardo RV, Campos-García J, Gutiérrez-Corona JF, García-Merinos JP, Meza-Carmen V, Silva-Sánchez J, Ramírez-Díaz MI. 2018. CrpP is a novel ciprofloxacin-modifying enzyme encoded by the Pseudomonas aeruginosa pUM505 plasmid. Antimicrob Agents Chemother 62:e02629-17. doi: 10.1128/AAC.02629-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Moya B, Zamorano L, Juan C, Perez JL, Ge Y, Oliver A. 2010. Activity of a new cephalosporin, CXA-101 (FR264205), against β-lactam-resistant Pseudomonas aeruginosa mutants selected in vitro and after antipseudomonal treatment of intensive care unit patients. Antimicrob Agents Chemother 54:1213–1217. doi: 10.1128/AAC.01104-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Torrens G, Cabot G, Ocampo-Sosa AA, Conejo MC, Zamorano L, Navarro F, Pascual Á, Martínez-Martínez L, Oliver A. 2016. Activity of ceftazidime-avibactam against clinical and isogenic laboratory Pseudomonas aeruginosa isolates expressing combinations of most relevant β-lactam resistance mechanisms. Antimicrob Agents Chemother 60:6407–6410. doi: 10.1128/AAC.01282-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sanz-García F, Hernando-Amado S, Martínez JL. 2018. Mutation-driven evolution of Pseudomonas aeruginosa in the presence of either ceftazidime or ceftazidime-avibactam. Antimicrob Agents Chemother 62:e01379-18. doi: 10.1128/AAC.01379-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Recio R, Villa J, Viedma E, Orellana MÁ, Lora-Tamayo J, Chaves F. 2018. Bacteraemia due to extensively drug-resistant Pseudomonas aeruginosa sequence type 235 high-risk clone: facing the perfect storm. Int J Antimicrob Agents 52:172–179. doi: 10.1016/j.ijantimicag.2018.03.018. [DOI] [PubMed] [Google Scholar]
- 62.Díaz-Cañestro M, Periañez L, Mulet X, Martin-Pena ML, Fraile-Ribot PA, Ayestarán I, Colomar A, Nuñez B, Maciá M, Novo A, Torres V, Asensio J, López-Causapé C, Delgado O, Pérez JL, Murillas J, Riera M, Oliver A. 2018. Ceftolozane/tazobactam for the treatment of multidrug resistant Pseudomonas aeruginosa: experience from the Balearic Islands. Eur J Clin Microbiol Infect Dis 37:2191–2200. doi: 10.1007/s10096-018-3361-0. [DOI] [PubMed] [Google Scholar]
- 63.Fraile-Ribot PA, Mulet X, Cabot G, del Barrio-Tofiño E, Juan C, Pérez JL, Oliver A. 2017. In vivo emergence of resistance to novel cephalosporin–β-lactamase inhibitor combinations through the duplication of amino acid D149 from OXA-2 β-lactamase (OXA-539) in sequence type 235 Pseudomonas aeruginosa. Antimicrob Agents Chemother 61:e01117-17. doi: 10.1128/AAC.01117-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Magiorakos A-P, Srinivasan A, Carey RB, Carmeli Y, Falagas ME, Giske CG, Harbarth S, Hindler JF, Kahlmeter G, Olsson-Liljequist B, Paterson DL, Rice LB, Stelling J, Struelens MJ, Vatopoulos A, Weber JT, Monnet DL. 2012. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin Microbiol Infect 18:268–281. doi: 10.1111/j.1469-0691.2011.03570.x. [DOI] [PubMed] [Google Scholar]
- 65.Souli M, Galani I, Giamarellou H. 2008. Emergence of extensively drug-resistant and pandrug-resistant Gram-negative bacilli in Europe. Euro Surveill 13:19045. doi: 10.2807/ese.13.47.19045-en. [DOI] [PubMed] [Google Scholar]
- 66.Viedma E, Juan C, Acosta J, Zamorano L, Otero JR, Sanz F, Chaves F, Oliver A. 2009. Nosocomial spread of colistin-only-sensitive sequence type 235 Pseudomonas aeruginosa isolates producing the extended-spectrum beta-lactamases GES-1 and GES-5 in Spain. Antimicrob Agents Chemother 53:4930–4933. doi: 10.1128/AAC.00900-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Livermore DM, Meunier D, Hopkins KL, Doumith M, Hill R, Pike R, Staves P, Woodford N. 2018. Activity of ceftazidime/avibactam against problem Enterobacteriaceae and Pseudomonas aeruginosa in the UK, 2015–16. J Antimicrob Chemother 73:648–657. doi: 10.1093/jac/dkx438. [DOI] [PubMed] [Google Scholar]
- 68.Giani T, Arena F, Pollini S, Di Pilato V, D’Andrea MM, Henrici De Angelis L, Bassetti M, Rossolini GM, Vismara C, Luzzaro F, Cavallo R, Dusi PA, Pagani E, Sarti M, Farina C, Rigoli R, Scarparo C, Pecile P, Cusi MG, Mencacci A, Manso E, Spanu T, Labonia M, Tassi V, Amato G, Stefani S, Giraldi C, Rassu M. 2018. Italian nationwide survey on Pseudomonas aeruginosa from invasive infections: activity of ceftolozane/tazobactam and comparators, and molecular epidemiology of carbapenemase producers. J Antimicrob Chemother 73:664–671. doi: 10.1093/jac/dkx453. [DOI] [PubMed] [Google Scholar]
- 69.Flamm RK, Farrell DJ, Sader HS, Jones RN. 2014. Ceftazidime/avibactam activity tested against Gram-negative bacteria isolated from bloodstream, pneumonia, intra-abdominal and urinary tract infections in US medical centres (2012). J Antimicrob Chemother 69:1589–1598. doi: 10.1093/jac/dku025. [DOI] [PubMed] [Google Scholar]
- 70.Evans SR, Tran TTT, Hujer AM, Hill CB, Hujer KM, Mediavilla JR, Manca C, Domitrovic TN, Perez F, Farmer M, Pitzer KM, Wilson BM, Kreiswirth BN, Patel R, Jacobs MR, Chen L, Fowler VG, Chambers HF, Bonomo RA, Antibacterial Resistance Leadership Group (ARLG). 2018. Rapid molecular diagnostics to inform empiric use of ceftazidime/avibactam and ceftolozane/tazobactam against Pseudomonas aeruginosa: PRIMERS IV. Clin Infect Dis doi: 10.1093/cid/ciy801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Woodford N, Turton JF, Livermore DM. 2011. Multiresistant Gram-negative bacteria: the role of high-risk clones in the dissemination of antibiotic resistance. FEMS Microbiol Rev 35:736–755. doi: 10.1111/j.1574-6976.2011.00268.x. [DOI] [PubMed] [Google Scholar]
- 72.Cabot G, Ocampo-Sosa AA, Domínguez MA, Gago JF, Juan C, Tubau F, Rodríguez C, Moyà B, Peña C, Martínez-Martínez L, Oliver A, Spanish Network for Research in Infectious Diseases (REIPI). 2012. Genetic markers of widespread extensively drug-resistant Pseudomonas aeruginosa high-risk clones. Antimicrob Agents Chemother 56:6349–6357. doi: 10.1128/AAC.01388-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Mulet X, Cabot G, Ocampo-Sosa AA, Domínguez MA, Zamorano L, Juan C, Tubau F, Rodríguez C, Moyà B, Peña C, Martínez-Martínez L, Oliver A, Spanish Network for Research in Infectious Diseases (REIPI). 2013. Biological markers of Pseudomonas aeruginosa epidemic high-risk clones. Antimicrob Agents Chemother 57:5527–5535. doi: 10.1128/AAC.01481-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Croughs PD, Klaassen CHW, van Rosmalen J, Maghdid DM, Boers SA, Hays JP, Goessens WHF, Dutch Antibiotic Resistance Surveillance Group. 2018. Unexpected mechanisms of resistance in Dutch Pseudomonas aeruginosa isolates collected during 14 years of surveillance. Int J Antimicrob Agents 52:407–410. doi: 10.1016/j.ijantimicag.2018.05.009. [DOI] [PubMed] [Google Scholar]
- 75.Castanheira M, Deshpande LM, Costello A, Davies TA, Jones RN. 2014. Epidemiology and carbapenem resistance mechanisms of carbapenem-non-susceptible Pseudomonas aeruginosa collected during 2009-11 in 14 European and Mediterranean countries. J Antimicrob Chemother 69:1804–1814. doi: 10.1093/jac/dku048. [DOI] [PubMed] [Google Scholar]
- 76.Cholley P, Thouverez M, Hocquet D, van der Mee-Marquet N, Talon D, Bertrand X. 2011. Most multidrug-resistant Pseudomonas aeruginosa isolates from hospitals in eastern France belong to a few clonal types. J Clin Microbiol 49:2578–2583. doi: 10.1128/JCM.00102-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Guzvinec M, Izdebski R, Butic I, Jelic M, Abram M, Koscak I, Baraniak A, Hryniewicz W, Gniadkowski M, Tambic Andrasevic A. 2014. Sequence types 235, 111, and 132 predominate among multidrug-resistant pseudomonas aeruginosa clinical isolates in Croatia. Antimicrob Agents Chemother 58:6277–6283. doi: 10.1128/AAC.03116-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Mano Y, Saga T, Ishii Y, Yoshizumi A, Bonomo RA, Yamaguchi K, Tateda K. 2015. Molecular analysis of the integrons of metallo-β-lactamase-producing Pseudomonas aeruginosa isolates collected by nationwide surveillance programs across Japan. BMC Microbiol 15:41. doi: 10.1186/s12866-015-0378-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Treepong P, Kos VN, Guyeux C, Blanc DS, Bertrand X, Valot B, Hocquet D. 2018. Global emergence of the widespread Pseudomonas aeruginosa ST235 clone. Clin Microbiol Infect 24:258–266. doi: 10.1016/j.cmi.2017.06.018. [DOI] [PubMed] [Google Scholar]
- 80.Chen Y, Sun M, Wang M, Lu Y, Yan Z. 2014. Dissemination of IMP-6-producing Pseudomonas aeruginosa ST244 in multiple cities in China. Eur J Clin Microbiol Infect Dis 33:1181–1187. doi: 10.1007/s10096-014-2063-5. [DOI] [PubMed] [Google Scholar]
- 81.Petitjean M, Martak D, Silvant A, Bertrand X, Valot B, Hocquet D. 2017. Genomic characterization of a local epidemic Pseudomonas aeruginosa reveals specific features of the widespread clone ST395. Microb Genomics 3:e000129. doi: 10.1099/mgen.0.000129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Abdouchakour F, Aujoulat F, Licznar-Fajardo P, Marchandin H, Toubiana M, Parer S, Lotthé A, Jumas-Bilak E. 2018. Intraclonal variations of resistance and phenotype in Pseudomonas aeruginosa epidemic high-risk clone ST308: a key to success within a hospital? Int J Med Microbiol 308:279–289. doi: 10.1016/j.ijmm.2017.11.008. [DOI] [PubMed] [Google Scholar]
- 83.Kos VN, Déraspe M, McLaughlin RE, Whiteaker JD, Roy PH, Alm RA, Corbeil J, Gardner H. 2015. The resistome of Pseudomonas aeruginosa in relationship to phenotypic susceptibility. Antimicrob Agents Chemother 59:427–436. doi: 10.1128/AAC.03954-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Jaillard M, van Belkum A, Cady KC, Creely D, Shortridge D, Blanc B, Barbu EM, Dunne WM, Zambardi G, Enright M, Mugnier N, Le Priol C, Schicklin S, Guigon G, Veyrieras J-B. 2017. Correlation between phenotypic antibiotic susceptibility and the resistome in Pseudomonas aeruginosa. Int J Antimicrob Agents 50:210–218. doi: 10.1016/j.ijantimicag.2017.02.026. [DOI] [PubMed] [Google Scholar]
- 85.Juan C, Peña C, Oliver A. 2017. Host and pathogen biomarkers for severe Pseudomonas aeruginosa infections. J Infect Dis 215:S44–S51. doi: 10.1093/infdis/jiw299. [DOI] [PubMed] [Google Scholar]
- 86.Gómez-Zorrilla S, Juan C, Cabot G, Camoez M, Tubau F, Oliver A, Dominguez MA, Ariza J, Peña C. 2016. Impact of multidrug resistance on the pathogenicity of Pseudomonas aeruginosa: in vitro and in vivo studies. Int J Antimicrob Agents 47:368–374. doi: 10.1016/j.ijantimicag.2016.02.010. [DOI] [PubMed] [Google Scholar]
- 87.Sánchez-Diener I, Zamorano L, López-Causapé C, Cabot G, Mulet X, Peña C, del Campo R, Cantón R, Doménech-Sánchez A, Martínez-Martínez L, Arcos SC, Navas A, Oliver A. 2017. Interplay among resistance profiles, high-risk clones, and virulence in the Caenorhabditis elegans Pseudomonas aeruginosa infection model. Antimicrob Agents Chemother 61:e01586-17. doi: 10.1128/AAC.01586-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Lee J-Y, Peck KR, Ko KS. 2013. Selective advantages of two major clones of carbapenem-resistant Pseudomonas aeruginosa isolates (CC235 and CC641) from Korea: antimicrobial resistance, virulence and biofilm-forming activity. J Med Microbiol 62:1015–1024. doi: 10.1099/jmm.0.055426-0. [DOI] [PubMed] [Google Scholar]
- 89.Turton JF, Wright L, Underwood A, Witney AA, Chan Y-T, Al-Shahib A, Arnold C, Doumith M, Patel B, Planche TD, Green J, Holliman R, Woodford N. 2015. High-resolution analysis by whole-genome sequencing of an international lineage (sequence type 111) of Pseudomonas aeruginosa associated with metallo-carbapenemases in the United Kingdom. J Clin Microbiol 53:2622–2631. doi: 10.1128/JCM.00505-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Witney AA, Gould KA, Pope CF, Bolt F, Stoker NG, Cubbon MD, Bradley CR, Fraise A, Breathnach AS, Butcher PD, Planche TD, Hinds J. 2014. Genome sequencing and characterization of an extensively drug-resistant sequence type 111 serotype O12 hospital outbreak strain of Pseudomonas aeruginosa. Clin Microbiol Infect 20:O609–O618. doi: 10.1111/1469-0691.12528. [DOI] [PubMed] [Google Scholar]
- 91.Paterson DL, Rice LB. 2003. Empirical antibiotic choice for the seriously ill patient: are minimization of selection of resistant organisms and maximization of individual outcome mutually exclusive?. Clin Infect Dis 36:1006–1012. doi: 10.1086/374243. [DOI] [PubMed] [Google Scholar]
- 92.Guillamet CV, Vazquez R, Noe J, Micek ST, Kollef MH. 2016. A cohort study of bacteremic pneumonia. Medicine (Baltimore, MD) 95:e4708. doi: 10.1097/MD.0000000000004708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Kang C, Kim S, Kim H, Park S, Choe Y, Oh M, Kim E, Choe K. 2003. Pseudomonas aeruginosa bacteremia: risk factors for mortality and influence of delayed receipt of effective antimicrobial therapy on clinical outcome. Clin Infect Dis 37:745–751. doi: 10.1086/377200. [DOI] [PubMed] [Google Scholar]
- 94.Micek ST, Lloyd AE, Ritchie DJ, Reichley RM, Fraser VJ, Kollef MH. 2005. Pseudomonas aeruginosa bloodstream infection: importance of appropriate initial antimicrobial treatment. Antimicrob Agents Chemother 49:1306–1311. doi: 10.1128/AAC.49.4.1306-1311.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Lodise TP, Patel N, Kwa A, Graves J, Furuno JP, Graffunder E, Lomaestro B, McGregor JC. 2007. Predictors of 30-day mortality among patients with Pseudomonas aeruginosa bloodstream infections: impact of delayed appropriate antibiotic selection. Antimicrob Agents Chemother 51:3510–3515. doi: 10.1128/AAC.00338-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Park S-Y, Park HJ, Moon SM, Park K-H, Chong YP, Kim M-N, Kim S-H, Lee S-O, Kim YS, Woo JH, Choi S-H. 2012. Impact of adequate empirical combination therapy on mortality from bacteremic Pseudomonas aeruginosa pneumonia. BMC Infect Dis 12:308. doi: 10.1186/1471-2334-12-308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Hirsch EB, Tam VH. 2010. Impact of multidrug-resistant Pseudomonas aeruginosa infection on patient outcomes. Expert Rev Pharmacoecon Outcomes Res 10:441–451. doi: 10.1586/erp.10.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Gómez-Zorrilla S, Morandeira F, Castro MJ, Tubau F, Periche E, Cañizares R, Dominguez MA, Ariza J, Peña C. 2017. Acute inflammatory response of patients with Pseudomonas aeruginosa infections: a prospective study. Microb Drug Resist 23:523–530. doi: 10.1089/mdr.2016.0144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Planquette B, Timsit JF, Misset BY, Schwebel C, Azoulay E, Adrie C, Vesin A, Jamali S, Zahar JR, Allaouchiche B, Souweine B, Darmon M, Dumenil AS, Goldgran-Toledano D, Mourvillier BH, Bédos JP. 2013. Pseudomonas aeruginosa ventilator-associated pneumonia: predictive factors of treatment failure. Am J Respir Crit Care Med 188:69–76. doi: 10.1164/rccm.201210-1897OC. [DOI] [PubMed] [Google Scholar]
- 100.Kaminski C, Timsit JF, Dubois Y, Zahar JR, Garrouste-Orgeas M, Vesin A, Azoulay E, Feger C, Dumenil AS, Adrie C, Cohen Y, Allaouchiche B. 2011. Impact of ureido/carboxypenicillin resistance on the prognosis of ventilator-associated pneumonia due to Pseudomonas aeruginosa. Crit Care 15:R112. doi: 10.1186/cc10136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Arvanitis M, Anagnostou T, Kourkoumpetis TK, Ziakas PD, Desalermos A, Mylonakis E. 2014. The impact of antimicrobial resistance and aging in VAP outcomes: experience from a large tertiary care center. PLoS One 9:e89984-17. doi: 10.1371/journal.pone.0089984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Blot S, Vandewoude K, De Bacquer D, Colardyn F. 2002. Nosocomial bacteremia caused by antibiotic-resistant gram-negative bacteria in critically ill patients: clinical outcome and length of hospitalization. Clin Infect Dis 34:1600–1606. doi: 10.1086/340616. [DOI] [PubMed] [Google Scholar]
- 103.Klevens RM, Edwards JR, Gaynes RP. 2008. The impact of antimicrobial‐resistant, health care–associated infections on mortality in the United States. Clin Infect Dis 47:927–930. doi: 10.1086/591698. [DOI] [PubMed] [Google Scholar]
- 104.Vardakas KZ, Rafailidis PI, Konstantelias AA, Falagas ME. 2013. Predictors of mortality in patients with infections due to multi-drug resistant Gram negative bacteria: the study, the patient, the bug or the drug? J Infect 66:401–414. doi: 10.1016/j.jinf.2012.10.028. [DOI] [PubMed] [Google Scholar]
- 105.Neidell MJ, Cohen B, Furuya Y, Hill J, Jeon CY, Glied S, Larson EL. 2012. Costs of healthcare-and community-associated infections with antimicrobial-resistant versus antimicrobial-susceptible organisms. Clin Infect Dis 55:807–815. doi: 10.1093/cid/cis552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Cosgrove SE. 2006. The relationship between antimicrobial resistance and patient outcomes: mortality, length of hospital stay, and health care costs. Clin Infect Dis 42:S82–S89. doi: 10.1086/499406. [DOI] [PubMed] [Google Scholar]
- 107.Peña C, Gómez-Zorrilla S, Oriol I, Tubau F, Dominguez MA, Pujol M, Ariza J. 2013. Impact of multidrug resistance on Pseudomonas aeruginosa ventilator-associated pneumonia outcome: predictors of early and crude mortality. Eur J Clin Microbiol Infect Dis 32:413–420. doi: 10.1007/s10096-012-1758-8. [DOI] [PubMed] [Google Scholar]
- 108.McCarthy KL, Wailan AM, Jennison AV, Kidd TJ, Paterson DL. 2018. P. aeruginosa blood stream infection isolates: a “full house” of virulence genes in isolates associated with rapid patient death and patient survival. Microb Pathog 119:81–85. doi: 10.1016/j.micpath.2018.03.062. [DOI] [PubMed] [Google Scholar]
- 109.Gómez-Zorrilla S, Camoez M, Tubau F, Periche E, Cañizares R, Dominguez MA, Ariza J, Peña C. 2014. Antibiotic pressure is a major risk factor for rectal colonization by multidrug-resistant Pseudomonas aeruginosa in critically ill patients. Antimicrob Agents Chemother 58:5863–5870. doi: 10.1128/AAC.03419-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Peña C, Gómez-Zorrilla S, Suarez C, Dominguez MA, Tubau F, Arch O, Oliver A, Pujol M, Ariza J. 2012. Extensively drug-resistant Pseudomonas aeruginosa: risk of bloodstream infection in hospitalized patients. Eur J Clin Microbiol Infect Dis 31:2791–2797. doi: 10.1007/s10096-012-1629-3. [DOI] [PubMed] [Google Scholar]
- 111.Peña C, Suarez C, Gozalo M, Murillas J, Almirante B, Pomar V, Aguilar M, Granados A, Calbo E, Rodríguez-Baño J, Rodríguez F, Tubau F, Martínez-Martínez L, Oliver A, Spanish Network for Research in Infectious Diseases (REIPI). 2012. Prospective multicenter study of the impact of carbapenem resistance on mortality in Pseudomonas aeruginosa bloodstream infections. Antimicrob Agents Chemother 56:1265–1272. doi: 10.1128/AAC.05991-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.McCarthy KL, Paterson DL. 2017. Long-term mortality following Pseudomonas aeruginosa bloodstream infection. J Hosp Infect 95:292–299. doi: 10.1016/j.jhin.2016.11.014. [DOI] [PubMed] [Google Scholar]
- 113.Andersson DI. 2006. The biological cost of mutational antibiotic resistance: any practical conclusions? Curr Opin Microbiol 9:461–465. doi: 10.1016/j.mib.2006.07.002. [DOI] [PubMed] [Google Scholar]
- 114.Andersson DI, Hughes D. 2010. Antibiotic resistance and its cost: is it possible to reverse resistance? Nat Rev Microbiol 8:260–271. doi: 10.1038/nrmicro2319. [DOI] [PubMed] [Google Scholar]
- 115.Olivares Pacheco J, Alvarez-Ortega C, Alcalde Rico M, Martínez JL. 2017. Metabolic compensation of fitness costs is a general outcome for antibiotic-resistant Pseudomonas aeruginosa mutants overexpressing efflux pumps. mBio 8:e00500-17. doi: 10.1128/mBio.00500-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Sun Z, Jiao X, Peng Q, Jiang F, Huang Y, Zhang J, Yao F. 2013. Antibiotic resistance in Pseudomonas aeruginosa is associated with decreased fitness. Cell Physiol Biochem 31:347–354. doi: 10.1159/000343372. [DOI] [PubMed] [Google Scholar]
- 117.Deptuła A, Gospodarek E. 2010. Reduced expression of virulence factors in multidrug-resistant Pseudomonas aeruginosa strains. Arch Microbiol 192:79–84. doi: 10.1007/s00203-009-0528-1. [DOI] [PubMed] [Google Scholar]
- 118.Skurnik D, Roux D, Cattoir V, Danilchanka O, Lu X, Yoder-Himes DR, Han K, Guillard T, Jiang D, Gaultier C, Guerin F, Aschard H, Leclercq R, Mekalanos JJ, Lory S, Pier GB. 2013. Enhanced in vivo fitness of carbapenem-resistant oprD mutants of Pseudomonas aeruginosa revealed through high-throughput sequencing. Proc Natl Acad Sci U S A 110:20747–20752. doi: 10.1073/pnas.1221552110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Suárez C, Peña C, Gavaldà L, Tubau F, Manzur A, Dominguez MA, Pujol M, Gudiol F, Ariza J. 2010. Influence of carbapenem resistance on mortality and the dynamics of mortality in Pseudomonas aeruginosa bloodstream infection. Int J Infect Dis 14(Suppl 3):e73–e78. doi: 10.1016/j.ijid.2009.11.019. [DOI] [PubMed] [Google Scholar]
- 120.Agnello M, Finkel SE, Wong-Beringer A. 2016. Fitness cost of fluoroquinolone resistance in clinical isolates of Pseudomonas aeruginosa differs by type III secretion genotype. Front Microbiol 7:907–912. doi: 10.3389/Ffmicb.2016.01591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Engel J, Balachandran P. 2009. Role of Pseudomonas aeruginosa type III effectors in disease. Curr Opin Microbiol 12:61–66. doi: 10.1016/j.mib.2008.12.007. [DOI] [PubMed] [Google Scholar]
- 122.El-Solh AA, Hattemer A, Hauser AR, Alhajhusain A, Vora H. 2012. Clinical outcomes of type III Pseudomonas aeruginosa bacteremia. Crit Care Med 40:1157–1163. doi: 10.1097/CCM.0b013e3182377906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Hauser AR. 2014. Pseudomonas aeruginosa virulence and antimicrobial resistance: two sides of the same coin? Crit Care Med 42:201–202. doi: 10.1097/CCM.0b013e3182a120cd. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Reboud E, Elsen S, Bouillot S, Golovkine G, Basso P, Jeannot K, Attrée I, Huber P. 2016. Phenotype and toxicity of the recently discovered exlA-positive Pseudomonas aeruginosa strains collected worldwide. Environ Microbiol 18:3425–3439. doi: 10.1111/1462-2920.13262. [DOI] [PubMed] [Google Scholar]
- 125.Wood TL, Wood TK. 2016. The HigB/HigA toxin/antitoxin system of Pseudomonas aeruginosa influences the virulence factors pyochelin, pyocyanin, and biofilm formation. Microbiologyopen 5:499–511. doi: 10.1002/mbo3.346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Giamarellos-Bourboulis EJ, Koussoulas V, Panagou C, Adamis T, Baziaka F, Skiadas I, Perrea D, Dionyssiou-Asteriou A, Giamarellou H. 2004. Experimental sepsis using Pseudomonas aeruginosa: the significance of multi-drug resistance. Int J Antimicrob Agents 24:357–361. doi: 10.1016/j.ijantimicag.2004.03.025. [DOI] [PubMed] [Google Scholar]
- 127.Giamarellos-Bourboulis EJ, Tzepi I, Tsovolou I, Spyridaki A, Tsaganos T, Vaki I, Kotsaki A, Polychronopoulos V. 2011. Impact of multidrug resistance on experimental empyema by Pseudomonas aeruginosa. Respiration 82:46–53. doi: 10.1159/000326893. [DOI] [PubMed] [Google Scholar]
- 128.Giamarellos-Bourboulis EJ, Plachouras D, Tzivra A, Kousoulas V, Bolanos N, Raftogiannis M, Galani I, Dontas I, Dionyssiou-Asteriou A, Giamarellou H. 2004. Stimulation of innate immunity by susceptible and multidrug-resistant Pseudomonas aeruginosa: an in vitro and in vivo study. Clin Exp Immunol 135:240–246. doi: 10.1111/j.1365-2249.2003.02365.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Abdelraouf K, Kabbara S, Ledesma KR, Poole K, Tam VH. 2011. Effect of multidrug resistance-conferring mutations on the fitness and virulence of Pseudomonas aeruginosa. J Antimicrob Chemother 66:1311–1317. doi: 10.1093/jac/dkr105. [DOI] [PubMed] [Google Scholar]
- 130.Gómez-Zorrilla S, Camoez M, Tubau F, Cañizares R, Periche E, Dominguez MA, Ariza J, Peña C. 2015. Prospective observational study of prior rectal colonization status as a predictor for subsequent development of Pseudomonas aeruginosa clinical infections. Antimicrob Agents Chemother 59:5213–5219. doi: 10.1128/AAC.04636-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Di Pasquale M, Ferrer M, Esperatti M, Crisafulli E, Giunta V, Li Bassi G, Rinaudo M, Blasi F, Niederman M, Torres A. 2014. Assessment of severity of ICU-acquired pneumonia and association with etiology. Crit Care Med 42:303–312. doi: 10.1097/CCM.0b013e3182a272a2. [DOI] [PubMed] [Google Scholar]
- 132.Drusano GL, Louie A, MacGowan A, Hope W. 2015. Suppression of emergence of resistance in pathogenic bacteria: keeping our powder dry, part 1. Antimicrob Agents Chemother 60:1183–1193. doi: 10.1128/AAC.02177-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Drusano GL. 2017. Pre-clinical in vitro infection models. Curr Opin Pharmacol 36:100–106. doi: 10.1016/j.coph.2017.09.011. [DOI] [PubMed] [Google Scholar]
- 134.Zusman O, Avni T, Leibovici L, Adler A, Friberg L, Stergiopoulou T, Carmeli Y, Paul M. 2013. Systematic review and meta-analysis of in vitro synergy of polymyxins and carbapenems. Antimicrob Agents Chemother 57:5104–5111. doi: 10.1128/AAC.01230-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Petrosillo N, Ioannidou E, Falagas ME. 2008. Colistin monotherapy vs. combination therapy: evidence from microbiological, animal and clinical studies. Clin Microbiol Infect 14:816–827. doi: 10.1111/j.1469-0691.2008.02061.x. [DOI] [PubMed] [Google Scholar]
- 136.Gunderson BW, Ibrahim KH, Hovde LB, Fromm TL, Reed MD, Rotschafer JC. 2003. Synergistic activity of colistin and ceftazidime against multiantibiotic-resistant Pseudomonas aeruginosa in an in vitro pharmacodynamic model. Antimicrob Agents Chemother 47:905–909. doi: 10.1128/AAC.47.3.905-909.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Timurkaynak F, Can F, Azap OK, Demirbilek M, Arslan H, Karaman SO. 2006. In vitro activities of non-traditional antimicrobials alone or in combination against multidrug-resistant strains of Pseudomonas aeruginosa and Acinetobacter baumannii isolated from intensive care units. Int J Antimicrob Agents 27:224–228. doi: 10.1016/j.ijantimicag.2005.10.012. [DOI] [PubMed] [Google Scholar]
- 138.Drusano GL, Bonomo RA, Bahniuk N, Bulitta JB, VanScoy B, DeFiglio H, Fikes S, Brown D, Drawz SM, Kulawy R, Louie A. 2012. Resistance emergence mechanism and mechanism of resistance suppression by tobramycin for cefepime for Pseudomonas aeruginosa. Antimicrob Agents Chemother 56:231–242. doi: 10.1128/AAC.05252-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Almarzoky Abuhussain SS, Kuti JL, Nicolau DP. 2018. Antibacterial activity of human simulated epithelial lining fluid concentrations of ceftazidime-avibactam alone or in combination with amikacin inhale (BAY41-6551) against carbapenem-resistant Pseudomonas aeruginosa and Klebsiella pneumoniae. Antimicrob Agents Chemother 62:e00113-18. doi: 10.1128/AAC.00113-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Bergen PJ, Tsuji BT, Bulitta JB, Forrest A, Jacob J, Sidjabat HE, Paterson DL, Nation RL, Li J. 2011. Synergistic killing of multidrug-resistant Pseudomonas aeruginosa at multiple inocula by colistin combined with doripenem in an in vitro pharmacokinetic/pharmacodynamic model. Antimicrob Agents Chemother 55:5685–5695. doi: 10.1128/AAC.05298-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Bergen PJ, Forrest A, Bulitta JB, Tsuji BT, Sidjabat HE, Paterson DL, Li J, Nation RL. 2011. Clinically relevant plasma concentrations of colistin in combination with imipenem enhance pharmacodynamic activity against multidrug-resistant Pseudomonas aeruginosa at multiple inocula. Antimicrob Agents Chemother 55:5134–5142. doi: 10.1128/AAC.05028-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Mohamed AF, Kristoffersson AN, Karvanen M, Nielsen EI, Cars O, Friberg LE. 2016. Dynamic interaction of colistin and meropenem on a WT and a resistant strain of Pseudomonas aeruginosa as quantified in a PK/PD model. J Antimicrob Chemother 71:1279–1290. doi: 10.1093/jac/dkv488. [DOI] [PubMed] [Google Scholar]
- 143.Louie A, Grasso C, Bahniuk N, Van Scoy B, Brown DL, Kulawy R, Drusano GL. 2010. The combination of meropenem and levofloxacin is synergistic with respect to both Pseudomonas aeruginosa kill rate and resistance suppression. Antimicrob Agents Chemother 54:2646–2654. doi: 10.1128/AAC.00065-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Safarika A, Galani I, Pistiki A, Giamarellos-Bourboulis EJ. 2015. Time–kill effect of levofloxacin on multidrug-resistant Pseudomonas aeruginosa and Acinetobacter baumannii: synergism with imipenem and colistin. Eur J Clin Microbiol Infect Dis 34:317–323. doi: 10.1007/s10096-014-2231-7. [DOI] [PubMed] [Google Scholar]
- 145.Rees VE, Yadav R, Rogers KE, Bulitta JB, Wirth V, Oliver A, Boyce JD, Peleg AY, Nation RL, Landersdorfer CB. 2018. Meropenem combined with ciprofloxacin combats hypermutable Pseudomonas aeruginosa from respiratory infections of cystic fibrosis patients. Antimicrob Agents Chemother 62:e01150-18. doi: 10.1128/AAC.01150-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Lin Y-W, Yu HH, Zhao J, Han M-L, Zhu Y, Akter J, Wickremasinghe H, Walpola H, Wirth V, Rao GG, Forrest A, Velkov T, Li J. 2018. Polymyxin B in combination with enrofloxacin exerts synergistic killing against extensively drug-resistant Pseudomonas aeruginosa. Antimicrob Agents Chemother 62:e00028-18. doi: 10.1128/AAC.00028-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Sime FB, Johnson A, Whalley S, Santoyo-Castelazo A, Montgomery AB, Walters KA, Lipman J, Hope WW, Roberts JA. 2017. Pharmacodynamics of aerosolized fosfomycin and amikacin against resistant clinical isolates of Pseudomonas aeruginosa and Klebsiella pneumoniae in a hollow-fiber infection model: experimental basis for combination therapy. Antimicrob Agents Chemother 61:e01763-16. doi: 10.1128/AAC.01763-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Lister PD, Sanders WE, Sanders CC. 1998. Cefepime-aztreonam: a unique double beta-lactam combination for Pseudomonas aeruginosa. Antimicrob Agents Chemother 42:1610–1619. doi: 10.1128/AAC.42.7.1610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Boucher HW, Talbot GH, Benjamin DK, Bradley J, Guidos RJ, Jones RN, Murray BE, Bonomo RA, Gilbert D. 2013. 10 x ’20 progress—development of new drugs active against Gram-negative bacilli: an update from the Infectious Diseases Society of America. Clin Infect Dis 56:1685–1694. doi: 10.1093/cid/cit152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Buehrle DJ, Shields RK, Chen L, Hao B, Press EG, Alkrouk A, Potoski BA, Kreiswirth BN, Clancy CJ, Nguyen MH. 2016. Evaluation of the in vitro activity of ceftazidime-avibactam and ceftolozane-tazobactam against meropenem-resistant Pseudomonas aeruginosa isolates. Antimicrob Agents Chemother 60:3227–3231. doi: 10.1128/AAC.02969-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Craig WA, Andes DR. 2013. In vivo activities of ceftolozane, a new cephalosporin, with and without tazobactam against Pseudomonas aeruginosa and Enterobacteriaceae, including strains with extended-spectrum β-lactamases, in the thighs of neutropenic mice. Antimicrob Agents Chemother 57:1577–1582. doi: 10.1128/AAC.01590-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Farrell DJ, Flamm RK, Sader HS, Jones RN. 2013. Antimicrobial activity of ceftolozane-tazobactam tested against Enterobacteriaceae and Pseudomonas aeruginosa with various resistance patterns isolated in U.S. hospitals (2011–2012). Antimicrob Agents Chemother 57:6305–6310. doi: 10.1128/AAC.01802-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.VanScoy BD, Mendes RE, Castanheira M, McCauley J, Bhavnani SM, Jones RN, Friedrich LV, Steenbergen JN, Ambrose PG. 2014. Relationship between ceftolozane-tazobactam exposure and selection for Pseudomonas aeruginosa resistance in a hollow-fiber infection model. Antimicrob Agents Chemother 58:6024–6031. doi: 10.1128/AAC.02310-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Rico Caballero V, Almarzoky Abuhussain S, Kuti JL, Nicolau DP. 2018. Efficacy of human-simulated exposures of ceftolozane-tazobactam alone and in combination with amikacin or colistin against multidrug-resistant Pseudomonas aeruginosa in an in vitro pharmacodynamic model. Antimicrob Agents Chemother 62:e02384-17. doi: 10.1128/AAC.02384-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Noel AR, Bowker KE, Attwood M, MacGowan AP. 2018. Antibacterial effect of ceftolozane/tazobactam in combination with amikacin against aerobic Gram-negative bacilli studied in an in vitro pharmacokinetic model of infection. J Antimicrob Chemother 73:2411–2417. doi: 10.1093/jac/dky225. [DOI] [PubMed] [Google Scholar]
- 156.Gomez-Junyent J, Benavent E, Sierra Y, El Haj C, Soldevila L, Torrejon B, Rigo-Bonnin R, Tubau F, Ariza J, Murillo O. 2019. Efficacy of ceftolozane/tazobactam, alone and in combination with colistin, against multidrug-resistant Pseudomonas aeruginosa in an in vitro biofilm pharmacodynamic model. Int J Antimicrob Agents 53:612–619. doi: 10.1016/j.ijantimicag.2019.01.010. [DOI] [PubMed] [Google Scholar]
- 157.Montero M, VanScoy BD, López-Causapé C, Conde H, Adams J, Segura C, Zamorano L, Oliver A, Horcajada JP, Ambrose PG. 2018. Evaluation of ceftolozane-tazobactam in combination with meropenem against Pseudomonas aeruginosa sequence type 175 in a hollow-fiber infection model. Antimicrob Agents Chemother 62:e00026-18. doi: 10.1128/AAC.00026-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Aoki N, Tateda K, Kikuchi Y, Kimura S, Miyazaki C, Ishii Y, Tanabe Y, Gejyo F, Yamaguchi K. 2009. Efficacy of colistin combination therapy in a mouse model of pneumonia caused by multidrug-resistant Pseudomonas aeruginosa. J Antimicrob Chemother 63:534–542. doi: 10.1093/jac/dkn530. [DOI] [PubMed] [Google Scholar]
- 159.Yamagishi Y, Hagihara M, Kato H, Hirai J, Nishiyama N, Koizumi Y, Sakanashi D, Suematsu H, Nakai H, Mikamo H. 2017. In vitro and in vivo pharmacodynamics of colistin and aztreonam alone and in combination against multidrug-resistant Pseudomonas aeruginosa. Chemotherapy 62:105–110. doi: 10.1159/000449367. [DOI] [PubMed] [Google Scholar]
- 160.Yadav R, Bulitta JB, Wang J, Nation RL, Landersdorfer CB. 2017. Evaluation of pharmacokinetic/pharmacodynamic model-based optimized combination regimens against multidrug-resistant Pseudomonas aeruginosa in a murine thigh infection model by using humanized dosing schemes. Antimicrob Agents Chemother 61:e01268-17. doi: 10.1128/AAC.01268-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Nichols WW, Stone GG, Newell P, Broadhurst H, Wardman A, MacPherson M, Yates K, Riccobene T, Critchley IA, Das S. 2018. Ceftazidime-avibactam susceptibility breakpoints against Enterobacteriaceae and Pseudomonas aeruginosa. Antimicrob Agents Chemother 62:e02590-17. doi: 10.1128/AAC.02590-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Stone GG, Newell P, Gasink LB, Broadhurst H, Wardman A, Yates K, Chen Z, Song J, Chow JW. 2018. Clinical activity of ceftazidime/avibactam against MDR Enterobacteriaceae and Pseudomonas aeruginosa: pooled data from the ceftazidime/avibactam phase III clinical trial programme. J Antimicrob Chemother 73:2519–2523. doi: 10.1093/jac/dky204. [DOI] [PubMed] [Google Scholar]
- 163.Rodríguez-Núñez O, Ripa M, Morata L, de la Calle C, Cardozo C, Fehér C, Pellicé M, Valcárcel A, Puerta-Alcalde P, Marco F, García-Vidal C, Del Río A, Soriano A, Martínez-Martínez JA. 2018. Evaluation of ceftazidime/avibactam for serious infections due to multidrug-resistant and extensively drug-resistant Pseudomonas aeruginosa. J Glob Antimicrob Resist 15:136–139. doi: 10.1016/j.jgar.2018.07.010. [DOI] [PubMed] [Google Scholar]
- 164.Monogue ML, Nicolau DP. 2018. Antibacterial activity of ceftolozane/tazobactam alone and in combination with other antimicrobial agents against MDR Pseudomonas aeruginosa. J Antimicrob Chemother 73:942–952. doi: 10.1093/jac/dkx483. [DOI] [PubMed] [Google Scholar]
- 165.Xipell M, Paredes S, Fresco L, Bodro M, Marco F, Martínez JA, Soriano A. 2018. Clinical experience with ceftolozane/tazobactam in patients with serious infections due to resistant Pseudomonas aeruginosa. J Glob Antimicrob Resist 13:165–170. doi: 10.1016/j.jgar.2018.01.010. [DOI] [PubMed] [Google Scholar]
- 166.Zavascki AP, Goldani LZ, Li J, Nation RL. 2007. Polymyxin B for the treatment of multidrug-resistant pathogens: a critical review. J Antimicrob Chemother 60:1206–1215. doi: 10.1093/jac/dkm357. [DOI] [PubMed] [Google Scholar]
- 167.Li J, Nation RL, Turnidge JD, Milne RW, Coulthard K, Rayner CR, Paterson DL. 2006. Colistin: the re-emerging antibiotic for multidrug-resistant Gram-negative bacterial infections. Lancet Infect Dis 6:589–601. doi: 10.1016/S1473-3099(06)70580-1. [DOI] [PubMed] [Google Scholar]
- 168.Nation RL, Velkov T, Li J. 2014. Colistin and polymyxin B: peas in a pod, or chalk and cheese? Clin Infect Dis 59:88–94. doi: 10.1093/cid/ciu213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Nation RL, Li J, Cars O, Couet W, Dudley MN, Kaye KS, Mouton JW, Paterson DL, Tam VH, Theuretzbacher U, Tsuji BT, Turnidge JD. 2015. Framework for optimisation of the clinical use of colistin and polymyxin B: the Prato polymyxin consensus. Lancet Infect Dis 15:225–234. doi: 10.1016/S1473-3099(14)70850-3. [DOI] [PubMed] [Google Scholar]
- 170.Garonzik SM, Li J, Thamlikitkul V, Paterson DL, Shoham S, Jacob J, Silveira FP, Forrest A, Nation RL. 2011. Population pharmacokinetics of colistin methanesulfonate and formed colistin in critically ill patients from a multicenter study provide dosing suggestions for various categories of patients. Antimicrob Agents Chemother 55:3284–3294. doi: 10.1128/AAC.01733-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Nation RL, Garonzik SM, Thamlikitkul V, Giamarellos-Bourboulis EJ, Forrest A, Paterson DL, Li J, Silveira FP. 2016. Dosing guidance for intravenous colistin in critically-ill patients. Clin Infect Dis 64:ciw839. doi: 10.1093/cid/ciw839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Nation RL, Garonzik SM, Li J, Thamlikitkul V, Giamarellos-Bourboulis EJ, Paterson DL, Turnidge JD, Forrest A, Silveira FP. 2016. Updated US and European dose recommendations for intravenous colistin: how do they perform? Clin Infect Dis 62:552–558. doi: 10.1093/cid/civ964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Couet W, Grégoire N, Marchand S, Mimoz O. 2012. Colistin pharmacokinetics: the fog is lifting. Clin Microbiol Infect 18:30–39. doi: 10.1111/j.1469-0691.2011.03667.x. [DOI] [PubMed] [Google Scholar]
- 174.Velkov T, Roberts KD, Nation RL, Thompson PE, Li J. 2013. Pharmacology of polymyxins: new insights into an ‘old’ class of antibiotics. Future Microbiol 8:711–724. doi: 10.2217/fmb.13.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Velkov T, Thompson PE, Nation RL, Li J. 2010. Structure–activity relationships of polymyxin antibiotics. J Med Chem 53:1898–1916. doi: 10.1021/jm900999h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Poirel L, Jayol A, Nordmann P. 2017. Polymyxins: antibacterial activity, susceptibility testing, and resistance mechanisms encoded by plasmids or chromosomes. Clin Microbiol Rev 30:557–596. doi: 10.1128/CMR.00064-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Zhang L, Dhillon P, Yan H, Farmer S, Hancock RE. 2000. Interactions of bacterial cationic peptide antibiotics with outer and cytoplasmic membranes of Pseudomonas aeruginosa. Antimicrob Agents Chemother 44:3317–3321. doi: 10.1128/aac.44.12.3317-3321.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Alhanout K, Malesinki S, Vidal N, Peyrot V, Rolain JM, Brunel JM. 2010. New insights into the antibacterial mechanism of action of squalamine. J Antimicrob Chemother 65:1688–1693. doi: 10.1093/jac/dkq213. [DOI] [PubMed] [Google Scholar]
- 179.O’Driscoll NH, Cushnie T, Matthews KHH, Lamb AJJ, O’Driscoll NHH, Cushnie T, Matthews KHH, Lamb AJJ, O’Driscoll NH, Cushnie T, Matthews KHH, Lamb AJJ, O’Driscoll NHH, Cushnie T, Matthews KHH, Lamb A. 2018. Colistin causes profound morphological alteration but minimal cytoplasmic membrane perforation in populations of Escherichia coli and Pseudomonas aeruginosa. Arch Microbiol 200:793–802. doi: 10.1007/s00203-018-1485-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Bergen PJ, Li J, Rayner CR, Nation RL. 2006. Colistin methanesulfonate is an inactive prodrug of colistin against Pseudomonas aeruginosa. Antimicrob Agents Chemother 50:1953–1958. doi: 10.1128/AAC.00035-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Benattar YD, Omar M, Zusman O, Yahav D, Zak-Doron Y, Altunin S, Elbaz M, Daitch V, Granot M, Leibovici L, Paul M. 2016. The effectiveness and safety of high-dose colistin: prospective cohort study. Clin Infect Dis 63:1605–1612. doi: 10.1093/cid/ciw684. [DOI] [PubMed] [Google Scholar]
- 182.Montero M, Horcajada JP, Sorlí L, Alvarez-Lerma F, Grau S, Riu M, Sala M, Knobel H. 2009. Effectiveness and safety of colistin for the treatment of multidrug-resistant Pseudomonas aeruginosa infections. Infection 37:461–465. doi: 10.1007/s15010-009-8342-x. [DOI] [PubMed] [Google Scholar]
- 183.Sorlí L, Luque S, Grau S, Berenguer N, Segura C, Montero MM, Álvarez-Lerma F, Knobel H, Benito N, Horcajada JP. 2013. Trough colistin plasma level is an independent risk factor for nephrotoxicity: a prospective observational cohort study. BMC Infect Dis 13:380. doi: 10.1186/1471-2334-13-380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Plachouras D, Karvanen M, Friberg LE, Papadomichelakis E, Antoniadou A, Tsangaris I, Karaiskos I, Poulakou G, Kontopidou F, Armaganidis A, Cars O, Giamarellou H. 2009. Population pharmacokinetic analysis of colistin methanesulfonate and colistin after intravenous administration in critically ill patients with infections caused by gram-negative bacteria. Antimicrob Agents Chemother 53:3430–3436. doi: 10.1128/AAC.01361-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Tsuji BT, Pogue JM, Zavascki AP, Paul M, Daikos GL, Forrest A, Giacobbe DR, Viscoli C, Giamarellou H, Karaiskos I, Kaye D, Mouton JW, Tam VH, Thamlikitkul V, Wunderink RG, Li J, Nation RL, Kaye KS. 2019. International consensus guidelines for the optimal use of the polymyxins: endorsed by the American College of Clinical Pharmacy (ACCP), European Society of Clinical Microbiology and Infectious Diseases (ESCMID), Infectious Diseases Society of America (IDS). Pharmacotherapy 39:10–39. doi: 10.1002/phar.2209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Dalfino L, Puntillo F, Mosca A, Monno R, Spada ML, Coppolecchia S, Miragliotta G, Bruno F, Brienza N. 2012. High-dose, extended-interval colistin administration in critically ill patients: is this the right dosing strategy? A preliminary study. Clin Infect Dis 54:1720–1726. doi: 10.1093/cid/cis286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Paul M, Daikos GL, Durante-Mangoni E, Yahav D, Carmeli Y, Benattar YD, Skiada A, Andini R, Eliakim-Raz N, Nutman A, Zusman O, Antoniadou A, Pafundi PC, Adler A, Dickstein Y, Pavleas I, Zampino R, Daitch V, Bitterman R, Zayyad H, Koppel F, Levi I, Babich T, Friberg LE, Mouton JW, Theuretzbacher U, Leibovici L. 2018. Colistin alone versus colistin plus meropenem for treatment of severe infections caused by carbapenem-resistant Gram-negative bacteria: an open-label, randomised controlled trial. Lancet Infect Dis 18:391–400. doi: 10.1016/S1473-3099(18)30099-9. [DOI] [PubMed] [Google Scholar]
- 188.Sorlí L, Luque S, Segura C, Campillo N, Montero M, Esteve E, Herrera S, Benito N, Alvarez-Lerma F, Grau S, Horcajada JP. 2017. Impact of colistin plasma levels on the clinical outcome of patients with infections caused by extremely drug-resistant Pseudomonas aeruginosa. BMC Infect Dis 17:11. doi: 10.1186/s12879-016-2117-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Horcajada JP, Sorlí L, Luque S, Benito N, Segura C, Campillo N, Montero M, Esteve E, Mirelis B, Pomar V, Cuquet J, Martí C, Garro P, Grau S. 2016. Validation of a colistin plasma concentration breakpoint as a predictor of nephrotoxicity in patients treated with colistin methanesulfonate. Int J Antimicrob Agents 48:725–727. doi: 10.1016/j.ijantimicag.2016.08.020. [DOI] [PubMed] [Google Scholar]
- 190.Luque S, Escaño C, Sorli L, Li J, Campillo N, Horcajada JP, Salas E, Grau S. 2017. Urinary concentrations of colistimethate and formed colistin after intravenous administration in patients with multidrug-resistant Gram-negative bacterial infections. Antimicrob Agents Chemother 61:e02595-16. doi: 10.1128/AAC.02595-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Hachem RY, Chemaly RF, Ahmar CA, Jiang Y, Boktour MR, Rjaili GA, Bodey GP, Raad II. 2007. Colistin is effective in treatment of infections caused by multidrug-resistant Pseudomonas aeruginosa in cancer patients. Antimicrob Agents Chemother 51:1905–1911. doi: 10.1128/AAC.01015-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Markou N, Apostolakos H, Koumoudiou C, Athanasiou M, Koutsoukou A, Alamanos I, Gregorakos L. 2003. Intravenous colistin in the treatment of sepsis from multiresistant Gram-negative bacilli in critically ill patients. Crit Care 7:R78. doi: 10.1186/cc2358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Linden PK, Kusne S, Coley K, Fontes P, Kramer DJ, Paterson D. 2003. Use of parenteral colistin for the treatment of serious infection due to antimicrobial-resistant Pseudomonas aeruginosa. Clin Infect Dis 37:e154–e160. doi: 10.1086/379611. [DOI] [PubMed] [Google Scholar]
- 194.Reina R, Estenssoro E, Sáenz G, Canales HS, Gonzalvo R, Vidal G, Martins G, Das Neves A, Santander O, Ramos C. 2005. Safety and efficacy of colistin in Acinetobacter and Pseudomonas infections: a prospective cohort study. Intensive Care Med 31:1058–1065. doi: 10.1007/s00134-005-2691-4. [DOI] [PubMed] [Google Scholar]
- 195.Michalopoulos AS, Tsiodras S, Rellos K, Mentzelopoulos S, Falagas ME. 2005. Colistin treatment in patients with ICU-acquired infections caused by multiresistant Gram-negative bacteria: the renaissance of an old antibiotic. Clin Microbiol Infect 11:115–121. doi: 10.1111/j.1469-0691.2004.01043.x. [DOI] [PubMed] [Google Scholar]
- 196.Cheng C-Y, Sheng W-H, Wang J-T, Chen Y-C, Chang S-C. 2010. Safety and efficacy of intravenous colistin (colistin methanesulphonate) for severe multidrug-resistant Gram-negative bacterial infections. Int J Antimicrob Agents 35:297–300. doi: 10.1016/j.ijantimicag.2009.11.016. [DOI] [PubMed] [Google Scholar]
- 197.Falagas ME, Rafailidis PI, Ioannidou E, Alexiou VG, Matthaiou DK, Karageorgopoulos DE, Kapaskelis A, Nikita D, Michalopoulos A. 2010. Colistin therapy for microbiologically documented multidrug-resistant Gram-negative bacterial infections: a retrospective cohort study of 258 patients. Int J Antimicrob Agents 35:194–199. doi: 10.1016/j.ijantimicag.2009.10.005. [DOI] [PubMed] [Google Scholar]
- 198.Durakovic N, Radojcic V, Boban A, Mrsic M, Sertic D, Serventi-Seiwerth R, Nemet D, Labar B. 2011. Efficacy and safety of colistin in the treatment of infections caused by multidrug-resistant Pseudomonas aeruginosa in patients with hematologic malignancy: a matched pair analysis. Intern Med 50:1009–1013. doi: 10.2169/internalmedicine.50.4270. [DOI] [PubMed] [Google Scholar]
- 199.Naesens R, Vlieghe E, Verbrugghe W, Jorens P, Ieven M. 2011. A retrospective observational study on the efficacy of colistin by inhalation as compared to parenteral administration for the treatment of nosocomial pneumonia associated with multidrug-resistant Pseudomonas aeruginosa. BMC Infect Dis 11:317. doi: 10.1186/1471-2334-11-317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Vicari G, Bauer SR, Neuner EA, Lam SW. 2013. Association between colistin dose and microbiologic outcomes in patients with multidrug-resistant Gram-negative bacteremia. Clin Infect Dis 56:398–404. doi: 10.1093/cid/cis909. [DOI] [PubMed] [Google Scholar]
- 201.Samonis G, Vardakas KZ, Kofteridis DP, Dimopoulou D, Andrianaki AM, Chatzinikolaou I, Katsanevaki E, Maraki S, Falagas ME. 2014. Characteristics, risk factors and outcomes of adult cancer patients with extensively drug-resistant Pseudomonas aeruginosa infections. Infection 42:721–728. doi: 10.1007/s15010-014-0635-z. [DOI] [PubMed] [Google Scholar]
- 202.Zusman O, Altunin S, Koppel F, Dishon Benattar Y, Gedik H, Paul M. 2017. Polymyxin monotherapy or in combination against carbapenem-resistant bacteria: systematic review and meta-analysis. J Antimicrob Chemother 72:29–39. doi: 10.1093/jac/dkw377. [DOI] [PubMed] [Google Scholar]
- 203.Khawcharoenporn T, Chuncharunee A, Maluangnon C, Taweesakulvashra T, Tiamsak P. 2018. Active monotherapy and combination therapy for extensively drug-resistant Pseudomonas aeruginosa pneumonia. Int J Antimicrob Agents 52:828–834. doi: 10.1016/j.ijantimicag.2018.09.008. [DOI] [PubMed] [Google Scholar]
- 204.Ribera A, Benavent E, Lora-Tamayo J, Tubau F, Pedrero S, Cabo X, Ariza J, Murillo O. 2015. Osteoarticular infection caused by MDR Pseudomonas aeruginosa: the benefits of combination therapy with colistin plus β-lactams. J Antimicrob Chemother 70:3357–3365. doi: 10.1093/jac/dkv281. [DOI] [PubMed] [Google Scholar]
- 205.Rigatto MH, Oliveira MS, Perdigão-Neto L V., Levin AS, Carrilho CM, Tanita MT, Tuon FF, Cardoso DE, Lopes NT, Falci DR, Zavascki AP. 2016. Multicenter prospective cohort study of renal failure in patients treated with colistin versus polymyxin B. Antimicrob Agents Chemother 60:2443–2449. doi: 10.1128/AAC.02634-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Rigatto MH, Vieira FJ, Antochevis LC, Behle TF, Lopes NT, Zavascki AP. 2015. Polymyxin B in combination with antimicrobials lacking in vitro activity versus polymyxin B in monotherapy in critically ill patients with Acinetobacter baumannii or Pseudomonas aeruginosa infections. Antimicrob Agents Chemother 59:6575–6580. doi: 10.1128/AAC.00494-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Nelson BC, Eiras DP, Gomez-Simmonds A, Loo AS, Satlin MJ, Jenkins SG, Whittier S, Calfee DP, Furuya EY, Kubin CJ. 2015. Clinical outcomes associated with polymyxin B dose in patients with bloodstream infections due to carbapenem-resistant Gram-negative rods. Antimicrob Agents Chemother 59:7000–7006. doi: 10.1128/AAC.00844-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Elias LS, Konzen D, Krebs JM, Zavascki AP. 2010. The impact of polymyxin B dosage on in-hospital mortality of patients treated with this antibiotic. J Antimicrob Chemother 65:2231–2237. doi: 10.1093/jac/dkq285. [DOI] [PubMed] [Google Scholar]
- 209.Furtado GHC, d’Azevedo PA, Santos AF, Gales AC, Pignatari ACC, Medeiros EAS. 2007. Intravenous polymyxin B for the treatment of nosocomial pneumonia caused by multidrug-resistant Pseudomonas aeruginosa. Int J Antimicrob Agents 30:315–319. doi: 10.1016/j.ijantimicag.2007.05.017. [DOI] [PubMed] [Google Scholar]
- 210.Dai C, Li J, Tang S, Li J, Xiao X. 2014. Colistin-induced nephrotoxicity in mice involves the mitochondrial, death receptor, and endoplasmic reticulum pathways. Antimicrob Agents Chemother 58:4075–4085. doi: 10.1128/AAC.00070-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Azad MAK, Finnin BA, Poudyal A, Davis K, Li J, Hill PA, Nation RL, Velkov T, Li J. 2013. Polymyxin B induces apoptosis in kidney proximal tubular cells. Antimicrob Agents Chemother 57:4329–4335. doi: 10.1128/AAC.02587-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Zavascki AP, Nation RL. 2017. Nephrotoxicity of polymyxins: is there any difference between colistimethate and polymyxin B? Antimicrob Agents Chemother 61:e02319-16. doi: 10.1128/AAC.02319-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Forrest A, Garonzik SM, Thamlikitkul V, Giamarellos-Bourboulis EJ, Paterson DL, Li J, Silveira FP, Nation RL. 2017. Pharmacokinetic/toxicodynamic analysis of colistin-associated acute kidney injury in critically ill patients. Antimicrob Agents Chemother 61:e01367-17. doi: 10.1128/AAC.01367-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Vardakas KZ, Voulgaris GL, Samonis G, Falagas ME. 2018. Inhaled colistin monotherapy for respiratory tract infections in adults without cystic fibrosis: a systematic review and meta-analysis. Int J Antimicrob Agents 51:1–9. doi: 10.1016/j.ijantimicag.2017.05.016. [DOI] [PubMed] [Google Scholar]
- 215.Vardakas KZ, Mavroudis AD, Georgiou M, Falagas ME. 2018. Intravenous plus inhaled versus intravenous colistin monotherapy for lower respiratory tract infections: a systematic review and meta-analysis. J Infect 76:321–327. doi: 10.1016/j.jinf.2018.02.002. [DOI] [PubMed] [Google Scholar]
- 216.Athanassa ZE, Markantonis SL, Fousteri M-ZF, Myrianthefs PM, Boutzouka EG, Tsakris A, Baltopoulos GJ. 2012. Pharmacokinetics of inhaled colistimethate sodium (CMS) in mechanically ventilated critically ill patients. Intensive Care Med 38:1779–1786. doi: 10.1007/s00134-012-2628-7. [DOI] [PubMed] [Google Scholar]
- 217.Lu Q, Luo R, Bodin L, Yang J, Zahr N, Aubry A, Golmard J-L, Rouby J-J, Nebulized Antibiotics Study Group. 2012. Efficacy of high-dose nebulized colistin in ventilator-associated pneumonia caused by multidrug-resistant Pseudomonas aeruginosa and Acinetobacter baumannii. Anesthesiology 117:1335–1347. doi: 10.1097/ALN.0b013e31827515de. [DOI] [PubMed] [Google Scholar]
- 218.Maskin LP, Setten M, Rodríguez PO, Bonelli I, Attie S, Stryjewski ME, Valentini R. 2015. Inhaled colistimethate sodium in ventilator-associated tracheobronchitis due to multidrug-resistant Gram-negative bacteria. Int J Antimicrob Agents 45:199–200. doi: 10.1016/j.ijantimicag.2014.09.010. [DOI] [PubMed] [Google Scholar]
- 219.Lin Y-W, Zhou Q, Onufrak NJ, Wirth V, Chen K, Wang J, Forrest A, Chan H-K, Li J. 2017. Aerosolized polymyxin B for treatment of respiratory tract infections: determination of pharmacokinetic-pharmacodynamic indices for aerosolized polymyxin B against Pseudomonas aeruginosa in a mouse lung infection model. Antimicrob Agents Chemother 61:e00211-17. doi: 10.1128/AAC.00211-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Pereira GH, Muller PR, Levin AS. 2007. Salvage treatment of pneumonia and initial treatment of tracheobronchitis caused by multidrug-resistant Gram-negative bacilli with inhaled polymyxin B. Diagn Microbiol Infect Dis 58:235–240. doi: 10.1016/j.diagmicrobio.2007.01.008. [DOI] [PubMed] [Google Scholar]
- 221.Kim B-N, Peleg AY, Lodise TP, Lipman J, Li J, Nation R, Paterson DL. 2009. Management of meningitis due to antibiotic-resistant Acinetobacter species. Lancet Infect Dis 9:245–245. doi: 10.1016/S1473-3099(09)70055-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Antachopoulos C, Karvanen M, Iosifidis E, Jansson B, Plachouras D, Cars O, Roilides E. 2010. Serum and cerebrospinal fluid levels of colistin in pediatric patients. Antimicrob Agents Chemother 54:3985–3987. doi: 10.1128/AAC.01799-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Ziaka M, Markantonis SL, Fousteri M, Zygoulis P, Panidis D, Karvouniaris M, Makris D, Zakynthinos E. 2013. Combined intravenous and intraventricular administration of colistin methanesulfonate in critically ill patients with central nervous system infection. Antimicrob Agents Chemother 57:1938–1940. doi: 10.1128/AAC.01461-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Markantonis SL, Markou N, Fousteri M, Sakellaridis N, Karatzas S, Alamanos I, Dimopoulou E, Baltopoulos G. 2009. Penetration of colistin into cerebrospinal fluid. Antimicrob Agents Chemother 53:4907–4910. doi: 10.1128/AAC.00345-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Velkov T, Dai C, Ciccotosto GD, Cappai R, Hoyer D, Li J. 2018. Polymyxins for CNS infections: pharmacology and neurotoxicity. Pharmacol Ther 181:85–90. doi: 10.1016/j.pharmthera.2017.07.012. [DOI] [PubMed] [Google Scholar]
- 226.Berlana D, Llop JM, Fort E, Badia MB, Jódar R. 2005. Use of colistin in the treatment of multiple-drug-resistant gram-negative infections. Am J Health Syst Pharm 62:39–47. doi: 10.1093/ajhp/62.1.39. [DOI] [PubMed] [Google Scholar]
- 227.Gump WC, Walsh JW. 2005. Intrathecal colistin for treatment of highly resistant Pseudomonas ventriculitis. Case report and review of the literature. J Neurosurg 102:915–917. doi: 10.3171/jns.2005.102.5.0915. [DOI] [PubMed] [Google Scholar]
- 228.Quinn AL, Parada JP, Belmares J, O’Keefe JP. 2005. Intrathecal colistin and sterilization of resistant Pseudomonas aeruginosa shunt infection. Ann Pharmacother 39:949–952. doi: 10.1345/aph.1E485. [DOI] [PubMed] [Google Scholar]
- 229.Schina M, Spyridi E, Daoudakis M, Mertzanos E, Korfias S. 2006. Successful treatment of multidrug-resistant Pseudomonas aeruginosa meningitis with intravenous and intrathecal colistin. Int J Infect Dis 10:178–179. doi: 10.1016/j.ijid.2005.03.005. [DOI] [PubMed] [Google Scholar]
- 230.Tsolaki V, Karvouniaris M, Manoulakas E, Kotlia P, Karadontas V, Fotakopoulos G, Zakynthinos E, Makris D. 2018. Intraventricular CNS treatment with colistin-tigecycline combination: a case series. J Crit Care 47:338–341. doi: 10.1016/j.jcrc.2018.07.025. [DOI] [PubMed] [Google Scholar]
- 231.Imberti R, Iotti GA, Regazzi M. 2014. Intraventricular or intrathecal colistin for the treatment of central nervous system infections caused by multidrug-resistant Gram-negative bacteria. Expert Rev Anti Infect Ther 12:471–478. doi: 10.1586/14787210.2014.896740. [DOI] [PubMed] [Google Scholar]
- 232.Baiocchi M, Catena V, Zago S, Badolati L, Baccarin M. 2010. Intrathecal colistin for treatment of multidrug resistant (MDR) Pseudomonas aeruginosa after neurosurgical ventriculitis. Infez Med 18:182–186. [PubMed] [Google Scholar]
- 233.Gilbert B, Morrison C. 2017. Evaluation of intraventricular colistin utilization: a case series. J Crit Care 40:161–163. doi: 10.1016/j.jcrc.2017.04.010. [DOI] [PubMed] [Google Scholar]
- 234.Tunkel AR, Hasbun R, Bhimraj A, Byers K, Kaplan SL, Scheld WM, van de Beek D, Bleck TP, Garton HJL, Zunt JR. 2017. 2017 Infectious Diseases Society of America’s clinical practice guidelines for healthcare-associated ventriculitis and meningitis. Clin Infect Dis 64:e34–e65. doi: 10.1093/cid/ciw861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Craig WA. 1998. Pharmacokinetic/pharmacodynamic parameters: rationale for antibacterial dosing of mice and men. Clin Infect Dis 26:1–10. (Quiz, 26:11–12.) doi: 10.1086/516284. [DOI] [PubMed] [Google Scholar]
- 236.Ong CT, Tessier PR, Li C, Nightingale CH, Nicolau DP. 2007. Comparative in vivo efficacy of meropenem, imipenem, and cefepime against Pseudomonas aeruginosa expressing MexA-MexB-OprM efflux pumps. Diagn Microbiol Infect Dis 57:153–161. doi: 10.1016/j.diagmicrobio.2006.06.014. [DOI] [PubMed] [Google Scholar]
- 237.Kuti JL, Dandekar PK, Nightingale CH, Nicolau DP. 2003. Use of Monte Carlo simulation to design an optimized pharmacodynamic dosing strategy for meropenem. J Clin Pharmacol 43:1116–1123. doi: 10.1177/0091270003257225. [DOI] [PubMed] [Google Scholar]
- 238.Yadav R, Bulitta JB, Nation RL, Landersdorfer CB. 2017. Optimization of synergistic combination regimens against carbapenem- and aminoglycoside-resistant clinical Pseudomonas aeruginosa isolates via mechanism-based pharmacokinetic/pharmacodynamic modeling. Antimicrob Agents Chemother 61:e01011-16. doi: 10.1128/AAC.01011-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Lim T-P, Wang R, Poh GQ, Koh T-H, Tan T-Y, Lee W, Teo JQ-M, Cai Y, Tan T-T, Ee PLR, Kwa AL. 2018. Integrated pharmacokinetic-pharmacodynamic modeling to evaluate empiric carbapenem therapy in bloodstream infections. Infect Drug Resist 11:1591–1596. doi: 10.2147/IDR.S168191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Domenig C, Traunmüller F, Kozek S, Wisser W, Klepetko W, Steininger R, Spiss C, Thalhammer F. 2001. Continuous beta-lactam antibiotic therapy in a double-lung transplanted patient with a multidrug-resistant Pseudomonas aeruginosa infection. Transplantation 71:744–745. doi: 10.1097/00007890-200103270-00009. [DOI] [PubMed] [Google Scholar]
- 241.Zavascki AP, Barth AL, Goldani LZ. 2008. Nosocomial bloodstream infections due to metallo-β-lactamase-producing Pseudomonas aeruginosa. J Antimicrob Chemother 61:1183–1185. doi: 10.1093/jac/dkn082. [DOI] [PubMed] [Google Scholar]
- 242.Sabuda DM, Laupland K, Pitout J, Dalton B, Rabin H, Louie T, Conly J. 2008. Utilization of colistin for treatment of multidrug-resistant Pseudomonas aeruginosa. Can J Infect Dis Med Microbiol 19:413–418. doi: 10.1155/2008/743197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Moriyama B, Henning SA, Childs R, Holland SM, Anderson VL, Morris JC, Wilson WH, Drusano GL, Walsh TJ. 2010. High-dose continuous infusion β-lactam antibiotics for the treatment of resistant Pseudomonas aeruginosa infections in immunocompromised patients. Ann Pharmacother 44:929–935. doi: 10.1345/aph.1M717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Heil EL, Lowery A V., Thom KA, Nicolau DP. 2015. Treatment of multidrug-resistant Pseudomonas aeruginosa using extended-infusion antimicrobial regimens. Pharmacother J Hum Pharmacol Drug Ther 35:54–58. doi: 10.1002/phar.1514. [DOI] [PubMed] [Google Scholar]
- 245.Pfaller MA, Shortridge D, Sader HS, Castanheira M, Flamm RK. 2018. Ceftolozane/tazobactam activity against drug-resistant Enterobacteriaceae and Pseudomonas aeruginosa causing healthcare-associated infections in the Asia-Pacific region (minus China, Australia and New Zealand): report from an Antimicrobial Surveillance Programme (2013-2015). Int J Antimicrob Agents 51:181–189. doi: 10.1016/j.ijantimicag.2017.09.016. [DOI] [PubMed] [Google Scholar]
- 246.Walkty A, Adam H, Baxter M, Lagacé-Wiens P, Karlowsky JA, Hoban DJ, Zhanel GG. 2018. In vitro activity of ceftolozane/tazobactam versus antimicrobial non-susceptible Pseudomonas aeruginosa clinical isolates including MDR and XDR isolates obtained from across Canada as part of the CANWARD study, 2008–16. J Antimicrob Chemother 73:703–708. doi: 10.1093/jac/dkx468. [DOI] [PubMed] [Google Scholar]
- 247.Britt NS, Ritchie DJ, Kollef MH, Burnham C-AD, Durkin MJ, Hampton NB, Micek ST. 2018. Importance of site of infection and antibiotic selection in the treatment of carbapenem-resistant Pseudomonas aeruginosa sepsis. Antimicrob Agents Chemother 62:e02400-17. doi: 10.1128/AAC.02400-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Craig WA, Redington J, Ebert SC. 1991. Pharmacodynamics of amikacin in vitro and in mouse thigh and lung infections. J Antimicrob Chemother 27(Suppl C):29–40. doi: 10.1093/jac/27.suppl_c.29. [DOI] [PubMed] [Google Scholar]
- 249.Zelenitsky SA, Harding GKM, Sun S, Ubhi K, Ariano RE. 2003. Treatment and outcome of Pseudomonas aeruginosa bacteraemia: an antibiotic pharmacodynamic analysis. J Antimicrob Chemother 52:668–674. doi: 10.1093/jac/dkg403. [DOI] [PubMed] [Google Scholar]
- 250.Brasseur A, Hites M, Roisin S, Cotton F, Vincent J-L, De Backer D, Jacobs F, Taccone FS. 2016. A high-dose aminoglycoside regimen combined with renal replacement therapy for the treatment of MDR pathogens: a proof-of-concept study. J Antimicrob Chemother 71:1386–1394. doi: 10.1093/jac/dkv491. [DOI] [PubMed] [Google Scholar]
- 251.Layeux B, Taccone FS, Fagnoul D, Vincent JL, Jacobs F. 2010. Amikacin monotherapy for sepsis caused by panresistant Pseudomonas aeruginosa. Antimicrob Agents Chemother 54:4939–4941. doi: 10.1128/AAC.00441-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Kalil AC, Metersky ML, Klompas M, Muscedere J, Sweeney DA, Palmer LB, Napolitano LM, O’Grady NP, Bartlett JG, Carratalà J, El Solh AA, Ewig S, Fey PD, File TM, Restrepo MI, Roberts JA, Waterer GW, Cruse P, Knight SL, Brozek JL. 2016. Management of adults with hospital-acquired and ventilator-associated pneumonia: 2016 clinical practice guidelines by the Infectious Diseases Society of America and the American Thoracic Society. Clin Infect Dis 63:e61–e111. doi: 10.1093/cid/ciw353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Molinaro M, Morelli P, De Gregori M, De Gregori S, Giardini I, Tordato F, Monzillo V, Pocaterra D, Casari E. 2018. Efficacy of intraventricular amikacin treatment in pan-resistant Pseudomonas aeruginosa postsurgical meningitis. Infect Drug Resist 11:1369–1372. doi: 10.2147/IDR.S169271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Falagas ME, Kastoris AC, Karageorgopoulos DE, Rafailidis PI. 2009. Fosfomycin for the treatment of infections caused by multidrug-resistant non-fermenting Gram-negative bacilli: a systematic review of microbiological, animal and clinical studies. Int J Antimicrob Agents 34:111–120. doi: 10.1016/j.ijantimicag.2009.03.009. [DOI] [PubMed] [Google Scholar]
- 255.Michalopoulos AS, Livaditis IG, Gougoutas V. 2011. The revival of fosfomycin. Int J Infect Dis 15:e732–e739. doi: 10.1016/j.ijid.2011.07.007. [DOI] [PubMed] [Google Scholar]
- 256.Samonis G, Maraki S, Karageorgopoulos DE, Vouloumanou EK, Falagas ME. 2012. Synergy of fosfomycin with carbapenems, colistin, netilmicin, and tigecycline against multidrug-resistant Klebsiella pneumoniae, Escherichia coli, and Pseudomonas aeruginosa clinical isolates. Eur J Clin Microbiol Infect Dis 31:695–701. doi: 10.1007/s10096-011-1360-5. [DOI] [PubMed] [Google Scholar]
- 257.Apisarnthanarak A, Mundy LM. 2010. Use of high-dose 4-hour infusion of doripenem, in combination with fosfomycin, for treatment of carbapenem-resistant Pseudomonas aeruginosa pneumonia. Clin Infect Dis 51:1352–1354. doi: 10.1086/657249. [DOI] [PubMed] [Google Scholar]
- 258.Apisarnthanarak A, Mundy LM. 2012. Carbapenem-resistant Pseudomonas aeruginosa pneumonia with intermediate minimum inhibitory concentrations to doripenem: combination therapy with high-dose, 4-h infusion of doripenem plus fosfomycin versus intravenous colistin plus fosfomycin. Int J Antimicrob Agents 39:271–272. doi: 10.1016/j.ijantimicag.2011.11.012. [DOI] [PubMed] [Google Scholar]
- 259.Pyrpasopoulou A, Pitsava G, Iosifidis E, Imvrios G, Massa E, Mouloudi E, Goulis I, Chatzidrosou E, Antachopoulos C, Fouzas I, Roilides E. 2018. Intravenous fosfomycin in patients with liver disease for extensively drug-resistant Gram-negative bacteria. J Infect 77:448–454. doi: 10.1016/j.jinf.2018.07.013. [DOI] [PubMed] [Google Scholar]
- 260.Winkler ML, Papp-Wallace KM, Hujer AM, Domitrovic TN, Hujer KM, Hurless KN, Tuohy M, Hall G, Bonomo RA. 2015. Unexpected challenges in treating multidrug-resistant Gram-negative bacteria: resistance to ceftazidime-avibactam in archived isolates of Pseudomonas aeruginosa. Antimicrob Agents Chemother 59:1020–1029. doi: 10.1128/AAC.04238-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261.Frattari A, Savini V, Polilli E, Cibelli D, Talamazzi S, Bosco D, Consorte A, Fazii P, Parruti G. 2018. Ceftolozane-tazobactam and fosfomycin for rescue treatment of otogenous meningitis caused by XDR Pseudomonas aeruginosa: case report and review of the literature. IDCases 14:e00451. doi: 10.1016/j.idcr.2018.e00451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Asuphon O, Montakantikul P, Houngsaitong J, Kiratisin P, Sonthisombat P. 2016. Optimizing intravenous fosfomycin dosing in combination with carbapenems for treatment of Pseudomonas aeruginosa infections in critically ill patients based on pharmacokinetic/pharmacodynamic (PK/PD) simulation. Int J Infect Dis 50:23–29. doi: 10.1016/j.ijid.2016.06.017. [DOI] [PubMed] [Google Scholar]
- 263.Maraolo AE, Cascella M, Corcione S, Cuomo A, Nappa S, Borgia G, De Rosa FG, Gentile I. 2017. Management of multidrug-resistant Pseudomonas aeruginosa in the intensive care unit: state of the art. Expert Rev Anti Infect Ther 15:861–871. doi: 10.1080/14787210.2017.1367666. [DOI] [PubMed] [Google Scholar]
- 264.Wright H, Bonomo RA, Paterson DL. 2017. New agents for the treatment of infections with Gram-negative bacteria: restoring the miracle or false dawn? Clin Microbiol Infect 23:704–712. doi: 10.1016/j.cmi.2017.09.001. [DOI] [PubMed] [Google Scholar]
- 265.Burrows LL. 2018. The therapeutic pipeline for Pseudomonas aeruginosa infections. ACS Infect Dis 4:1041–1047. doi: 10.1021/acsinfecdis.8b00112. [DOI] [PubMed] [Google Scholar]
- 266.Koulenti D, Song A, Ellingboe A, Abdul-Aziz MH, Harris P, Gavey E, Lipman J. 2018. Infections by multidrug-resistant Gram-negative bacteria: what’s new in our arsenal and what’s in the pipeline? Int J Antimicrob Agents 53:211–224. doi: 10.1016/j.ijantimicag.2018.10.011. [DOI] [PubMed] [Google Scholar]
- 267.Goodlet KJ, Nicolau DP, Nailor MD. 2017. In vitro comparison of ceftolozane-tazobactam to traditional beta-lactams and ceftolozane-tazobactam as an alternative to combination antimicrobial therapy for Pseudomonas aeruginosa. Antimicrob Agents Chemother 61:e01350-17. doi: 10.1128/AAC.01350-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268.Papp-Wallace KM, Bonomo RA. 2016. New β-lactamase inhibitors in the clinic. Infect Dis Clin North Am 30:441–464. doi: 10.1016/j.idc.2016.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269.Sader HS, Farrell DJ, Castanheira M, Flamm RK, Jones RN. 2014. Antimicrobial activity of ceftolozane/tazobactam tested against Pseudomonas aeruginosa and Enterobacteriaceae with various resistance patterns isolated in European hospitals (2011-12). J Antimicrob Chemother 69:2713–2722. doi: 10.1093/jac/dku184. [DOI] [PubMed] [Google Scholar]
- 270.Wasmuth E V, Lima CD. 2017. The Rrp6 C-terminal domain binds RNA and activates the nuclear RNA exosome. Nucleic Acids Res 45:846–860. doi: 10.1093/nar/gkw1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271.Humphries RM, Hindler JA, Wong-Beringer A, Miller SA. 2017. Activity of ceftolozane-tazobactam and ceftazidime-avibactam against beta-lactam-resistant Pseudomonas aeruginosa isolates. Antimicrob Agents Chemother 61:e01858-17. doi: 10.1128/AAC.01858-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 272.Pfaller MA, Bassetti M, Duncan LR, Castanheira M. 2017. Ceftolozane/tazobactam activity against drug-resistant Enterobacteriaceae and Pseudomonas aeruginosa causing urinary tract and intraabdominal infections in Europe: report from an antimicrobial surveillance programme (2012–15). J Antimicrob Chemother 72:1386–1395. doi: 10.1093/jac/dkx009. [DOI] [PubMed] [Google Scholar]
- 273.Grupper M, Sutherland C, Nicolau DP. 2017. Multicenter evaluation of ceftazidime-avibactam and ceftolozane-tazobactam inhibitory activity against meropenem-nonsusceptible Pseudomonas aeruginosa from blood, respiratory tract, and wounds. Antimicrob Agents Chemother 61:e00875-17. doi: 10.1128/AAC.00875-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 274.Tato M, García-Castillo M, Bofarull AM, Cantón R. 2015. In vitro activity of ceftolozane/tazobactam against clinical isolates of Pseudomonas aeruginosa and Enterobacteriaceae recovered in Spanish medical centres: results of the CENIT study. Int J Antimicrob Agents 46:502–510. doi: 10.1016/j.ijantimicag.2015.07.004. [DOI] [PubMed] [Google Scholar]
- 275.Natesan S, Pai MP, Lodise TP. 2017. Determination of alternative ceftolozane/tazobactam dosing regimens for patients with infections due to Pseudomonas aeruginosa with MIC values between 4 and 32 mg/L. J Antimicrob Chemother 72:2813–2816. doi: 10.1093/jac/dkx221. [DOI] [PubMed] [Google Scholar]
- 276.Xiao AJ, Miller BW, Huntington JA, Nicolau DP. 2016. Ceftolozane/tazobactam pharmacokinetic/pharmacodynamic-derived dose justification for phase 3 studies in patients with nosocomial pneumonia. J Clin Pharmacol 56:56–66. doi: 10.1002/jcph.566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 277.Melchers MJ, Mavridou E, Seyedmousavi S, van Mil AC, Lagarde C, Mouton JW. 2015. Plasma and epithelial lining fluid pharmacokinetics of ceftolozane and tazobactam alone and in combination in mice. Antimicrob Agents Chemother 59:3373–3376. doi: 10.1128/AAC.04402-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 278.Jones BM, Smith B, Bland CM. 2017. Use of continuous-infusion ceftolozane/tazobactam in a multidrug-resistant Pseudomonas aeruginosa urinary tract infection in the outpatient setting. Ann Pharmacother 51:715–716. doi: 10.1177/1060028017701938. [DOI] [PubMed] [Google Scholar]
- 279.Oliver WD, Heil EL, Gonzales JP, Mehrotra S, Robinett K, Saleeb P, Nicolau DP. 2016. Ceftolozane-tazobactam pharmacokinetics in a critically ill patient on continuous venovenous hemofiltration. Antimicrob Agents Chemother 60:1899–1901. doi: 10.1128/AAC.02608-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 280.Stewart A, Roberts JA, Wallis SC, Allworth AM, Legg A, McCarthy KL. 2018. Evidence of clinical response and stability of ceftolozane/tazobactam used to treat a carbapenem-resistant Pseudomonas aeruginosa lung abscess on an outpatient antimicrobial program. Int J Antimicrob Agents 51:941–942. doi: 10.1016/j.ijantimicag.2018.02.008. [DOI] [PubMed] [Google Scholar]
- 281.Gallagher JC, Satlin MJ, Elabor A, Saraiya N, McCreary EK, Molnar E, El-Beyrouty C, Jones BM, Dixit D, Heil EL, Claeys KC, Hiles J, Vyas NM, Bland CM, Suh J, Biason K, McCoy D, King MA, Richards L, Harrington N, Guo Y, Chaudhry S, Lu X, Yu D. 2018. Ceftolozane-tazobactam for the treatment of multidrug-resistant Pseudomonas aeruginosa infections: a multicenter study. Open Forum Infect Dis 5:ofy280. doi: 10.1093/ofid/ofy280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 282.Castón JJ, De la Torre Á, Ruiz-Camps I, Sorlí ML, Torres V, Torre-Cisneros J. 2017. Salvage therapy with ceftolozane-tazobactam for multidrug-resistant Pseudomonas aeruginosa infections. Antimicrob Agents Chemother 61:e02136-16. doi: 10.1128/AAC.02136-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 283.Munita JM, Aitken SL, Miller WR, Perez F, Rosa R, Shimose LA, Lichtenberger PN, Abbo LM, Jain R, Nigo M, Wanger A, Araos R, Tran TT, Adachi J, Rakita R, Shelburne S, Bonomo RA, Arias CA. 2017. Multicenter evaluation of ceftolozane/tazobactam for serious infections caused by carbapenem-resistant Pseudomonas aeruginosa. Clin Infect Dis 65:158–161. doi: 10.1093/cid/cix014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 284.Hakki M, Lewis JS. 2018. Ceftolozane-tazobactam therapy for multidrug-resistant Pseudomonas aeruginosa infections in patients with hematologic malignancies and hematopoietic-cell transplant recipients. Infection 46:431–434. doi: 10.1007/s15010-018-1125-5. [DOI] [PubMed] [Google Scholar]
- 285.Dinh A, Wyplosz B, Kernéis S, Lebeaux D, Bouchand F, Duran C, Béraud G, Lazaro P, Davido B, Hénard S, Canouï E, Ferry T, Wolff M. 2017. Use of ceftolozane/tazobactam as salvage therapy for infections due to extensively drug-resistant Pseudomonas aeruginosa. Int J Antimicrob Agents 49:782–783. doi: 10.1016/j.ijantimicag.2017.04.001. [DOI] [PubMed] [Google Scholar]
- 286.Escolà-Vergé L, Pigrau C, Los-Arcos I, Arévalo Á, Viñado B, Campany D, Larrosa N, Nuvials X, Ferrer R, Len O, Almirante B. 2018. Ceftolozane/tazobactam for the treatment of XDR Pseudomonas aeruginosa infections. Infection 46:461–468. doi: 10.1007/s15010-018-1133-5. [DOI] [PubMed] [Google Scholar]
- 287.Dietl B, Sánchez I, Arcenillas P, Cuchi E, Gómez L, González de Molina FJ, Boix-Palop L, Nicolás J, Calbo E. 2018. Ceftolozane/tazobactam in the treatment of osteomyelitis and skin and soft-tissue infections due to extensively drug-resistant Pseudomonas aeruginosa: clinical and microbiological outcomes. Int J Antimicrob Agents 51:498–502. doi: 10.1016/j.ijantimicag.2017.11.003. [DOI] [PubMed] [Google Scholar]
- 288.Bassetti M, Castaldo N, Cattelan A, Mussini C, Righi E, Tascini C, Menichetti F, Mastroianni CM, Tumbarello M, Grossi P, Artioli S, Carrannante N, Cipriani L, Coletto D, Russo A, Digaetano M, Losito R, Peghin M, Capone A, Nicolè S, Vena A. 2018. Ceftolozane/tazobactam for the treatment of serious P. aeruginosa infections: a multicenter nationwide clinical experience. Int J Antimicrob Agents 53:408–415. doi: 10.1016/j.ijantimicag.2018.11.001. [DOI] [PubMed] [Google Scholar]
- 289.Coleman K. 2011. Diazabicyclooctanes (DBOs): a potent new class of non-beta-lactam beta-lactamase inhibitors. Curr Opin Microbiol 14:550–555. doi: 10.1016/j.mib.2011.07.026. [DOI] [PubMed] [Google Scholar]
- 290.Abboud MI, Damblon C, Brem J, Smargiasso N, Mercuri P, Gilbert B, Rydzik AM, Claridge TDW, Schofield CJ, Frère JM. 2016. Interaction of avibactam with class B metallo-β-lactamases. Antimicrob Agents Chemother 60:5655–5662. doi: 10.1128/AAC.00897-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 291.Sy SKB, Zhuang L, Beaudoin ME, Kircher P, Tabosa MAM, Cavalcanti NCT, Grunwitz C, Pieper S, Schuck VJ, Nichols WW, Derendorf H. 2017. Potentiation of ceftazidime by avibactam against β-lactam-resistant Pseudomonas aeruginosa in an in vitro infection model. J Antimicrob Chemother 72:1109–1117. [DOI] [PubMed] [Google Scholar]
- 292.Sader HS, Castanheira M, Flamm RK, Mendes RE, Farrell DJ, Jones RN. 2015. Ceftazidime/avibactam tested against Gram-negative bacteria from intensive care unit (ICU) and non-ICU patients, including those with ventilator-associated pneumonia. Int J Antimicrob Agents 46:53–59. doi: 10.1016/j.ijantimicag.2015.02.022. [DOI] [PubMed] [Google Scholar]
- 293.Sader HS, Castanheira M, Mendes RE, Flamm RK, Farrell DJ, Jones RN. 2015. Ceftazidime-avibactam activity against multidrug-resistant Pseudomonas aeruginosa isolated in U.S. medical centers in 2012 and 2013. Antimicrob Agents Chemother 59:3656–3659. doi: 10.1128/AAC.05024-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 294.Sader HS, Castanheira M, Flamm RK. 2017. Antimicrobial activity of ceftazidime-avibactam against Gram-negative bacteria isolated from patients hospitalized with pneumonia in U.S. medical centers, 2011 to 2015. Antimicrob Agents Chemother 61:e02083-16. doi: 10.1128/AAC.02083-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 295.Sader HS, Castanheira M, Shortridge D, Mendes RE, Flamm RK. 2017. Antimicrobial activity of ceftazidime-avibactam tested against multidrug-resistant Enterobacteriaceae and Pseudomonas aeruginosa isolates from U.S. medical centers, 2013 to 2016. Antimicrob Agents Chemother 61:e01045-17. doi: 10.1128/AAC.01045-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 296.Kazmierczak KM, de Jonge BLM, Stone GG, Sahm DF. 2018. In vitro activity of ceftazidime/avibactam against isolates of Pseudomonas aeruginosa collected in European countries: INFORM global surveillance 2012–15. J Antimicrob Chemother 73:2777–2781. doi: 10.1093/jac/dky267. [DOI] [PubMed] [Google Scholar]
- 297.Sader HS, Castanheira M, Flamm RK, Jones RN. 2016. Antimicrobial activities of ceftazidime-avibactam and comparator agents against Gram-negative organisms isolated from patients with urinary tract infections in U.S. medical centers, 2012 to 2014. Antimicrob Agents Chemother 60:4355–4360. doi: 10.1128/AAC.00405-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 298.Nichols WW, de Jonge BLMM, Kazmierczak KM, Karlowsky JA, Sahm DF. 2016. In vitro susceptibility of global surveillance isolates of Pseudomonas aeruginosa to ceftazidime-avibactam (INFORM 2012 to 2014). Antimicrob Agents Chemother 60:4743–4749. doi: 10.1128/AAC.00220-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 299.Stone GG, Bradford PA, Yates K, Newell P. 2017. In vitro activity of ceftazidime/avibactam against urinary isolates from patients in a phase 3 clinical trial programme for the treatment of complicated urinary tract infections. J Antimicrob Chemother 72:1396–1399. doi: 10.1093/jac/dkw561. [DOI] [PubMed] [Google Scholar]
- 300.Berkhout J, Melchers MJ, van Mil AC, Seyedmousavi S, Lagarde CM, Schuck VJ, Nichols WW, Mouton JW. 2016. Pharmacodynamics of ceftazidime and avibactam in neutropenic mice with thigh or lung infection. Antimicrob Agents Chemother 60:368–375. doi: 10.1128/AAC.01269-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 301.Coleman K, Levasseur P, Girard A-M, Borgonovi M, Miossec C, Merdjan H, Drusano G, Shlaes D, Nichols WW. 2014. Activities of ceftazidime and avibactam against β-lactamase-producing Enterobacteriaceae in a hollow-fiber pharmacodynamic model. Antimicrob Agents Chemother 58:3366–3372. doi: 10.1128/AAC.00080-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 302.Andes D, Craig WA. 2002. Animal model pharmacokinetics and pharmacodynamics: a critical review. Int J Antimicrob Agents 19:261–268. doi: 10.1016/S0924-8579(02)00022-5. [DOI] [PubMed] [Google Scholar]
- 303.Nichols WW, Newell P, Critchley IA, Riccobene T, Das S. 2018. Avibactam pharmacokinetic/pharmacodynamic targets. Antimicrob Agents Chemother 62:e02446-17. doi: 10.1128/AAC.02446-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 304.Keepers TR, Gomez M, Celeri C, Nichols WW, Krause KM. 2014. Bactericidal activity, absence of serum effect, and time-kill kinetics of ceftazidime-avibactam against β-lactamase-producing Enterobacteriaceae and Pseudomonas aeruginosa. Antimicrob Agents Chemother 58:5297–5305. doi: 10.1128/AAC.02894-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 305.Sy SKB, Zhuang L, Sy S, Derendorf H. 2018. Clinical pharmacokinetics and pharmacodynamics of ceftazidime–avibactam combination: a model-informed strategy for its clinical development. Clin Pharmacokinet 58:545. doi: 10.1007/s40262-018-0705-y. [DOI] [PubMed] [Google Scholar]
- 306.Li J, Lovern M, Green ML, Chiu J, Zhou D, Comisar C, Xiong Y, Hing J, MacPherson M, Wright JG, Riccobene T, Carrothers TJ, Das S. 2018. Ceftazidime-avibactam population pharmacokinetic modeling and pharmacodynamic target attainment across adult indications and patient subgroups. Clin Transl Sci 12:151–163. doi: 10.1111/cts.12585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 307.Crandon JL, Schuck VJ, Banevicius MA, Beaudoin ME, Nichols WW, Tanudra MA, Nicolau DP. 2012. Comparative in vitro and in vivo efficacies of human simulated doses of ceftazidime and ceftazidime-avibactam against Pseudomonas aeruginosa. Antimicrob Agents Chemother 56:6137–6146. doi: 10.1128/AAC.00851-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 308.Carmeli Y, Armstrong J, Laud PJ, Newell P, Stone G, Wardman A, Gasink LB. 2016. Ceftazidime-avibactam or best available therapy in patients with ceftazidime-resistant Enterobacteriaceae and Pseudomonas aeruginosa complicated urinary tract infections or complicated intra-abdominal infections (REPRISE): a randomised, pathogen-directed. Lancet Infect Dis 16:661–673. doi: 10.1016/S1473-3099(16)30004-4. [DOI] [PubMed] [Google Scholar]
- 309.Torres A, Zhong N, Pachl J, Timsit J, Kollef M, Chen Z, Song J, Taylor D, Laud PJ, Stone GG, Chow JW. 2018. Ceftazidime-avibactam versus meropenem in nosocomial pneumonia, including ventilator-associated pneumonia (REPROVE): a randomised, double-blind, phase 3 non-inferiority trial. Lancet Infect Dis 18:285–295. doi: 10.1016/S1473-3099(17)30747-8. [DOI] [PubMed] [Google Scholar]
- 310.Olsen I. 2015. New promising β-lactamase inhibitors for clinical use. Eur J Clin Microbiol Infect Dis 34:1303–1308. doi: 10.1007/s10096-015-2375-0. [DOI] [PubMed] [Google Scholar]
- 311.Livermore DM, Warner M, Mushtaq S. 2013. Activity of MK-7655 combined with imipenem against Enterobacteriaceae and Pseudomonas aeruginosa. J Antimicrob Chemother 68:2286–2290. doi: 10.1093/jac/dkt178. [DOI] [PubMed] [Google Scholar]
- 312.Karlowsky JA, Lob SH, Kazmierczak KM, Young K, Motyl MR, Sahm DF. 2018. In vitro activity of imipenem-relebactam against clinical isolates of Gram-negative bacilli isolated in hospital laboratories in the United States as part of the SMART 2016 program. Antimicrob Agents Chemother 62:e00169-18. doi: 10.1128/AAC.00169-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 313.Lob SH, Hackel MA, Kazmierczak KM, Young K, Motyl MR, Karlowsky JA, Sahm DF, Sahm F, Sahm DF. 2017. In vitro activity of imipenem-relebactam against Gram-negative ESKAPE pathogens isolated by clinical laboratories in the United States in 2015 (results from the SMART global surveillance program). Antimicrob Agents Chemother 61:e02209-16. doi: 10.1128/AAC.02209-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 314.Rhee EG, Rizk ML, Calder N, Nefliu M, Warrington SJ, Schwartz MS, Mangin E, Boundy K, Bhagunde P, Colon-Gonzalez F, Jumes P, Liu Y, Butterton JR. 2018. Pharmacokinetics, safety, and tolerability of single and multiple doses of relebactam, a β-lactamase inhibitor, in combination with imipenem and cilastatin in healthy participants. Antimicrob Agents Chemother 62:e00280-18. doi: 10.1128/AAC.00280-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 315.Lucasti C, Vasile L, Sandesc D, Venskutonis D, McLeroth P, Lala M, Rizk ML, Brown ML, Losada MC, Pedley A, Kartsonis NA, Paschke A. 2016. Phase 2, dose-ranging study of relebactam with imipenem-cilastatin in subjects with complicated intra-abdominal infection. Antimicrob Agents Chemother 60:6234–6243. doi: 10.1128/AAC.00633-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 316.Ito-Horiyama T, Ishii Y, Ito A, Sato T, Nakamura R, Fukuhara N, Tsuji M, Yamano Y, Yamaguchi K, Tateda K. 2016. Stability of novel siderophore cephalosporin S-649266 against clinically relevant carbapenemases. Antimicrob Agents Chemother 60:4384–4386. doi: 10.1128/AAC.03098-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 317.Matsumoto S, Singley CM, Hoover J, Nakamura R, Echols R, Rittenhouse S, Tsuji M, Yamano Y. 2017. Efficacy of cefiderocol against carbapenem-resistant Gram-negative bacilli in immunocompetent-rat respiratory tract infection models recreating human plasma pharmacokinetics. Antimicrob Agents Chemother 61:e00700-17. doi: 10.1128/AAC.00700-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 318.Ito A, Nishikawa T, Ota M, Ito-Horiyama T, Ishibashi N, Sato T, Tsuji M, Yamano Y. 2018. Stability and low induction propensity of cefiderocol against chromosomal AmpC β-lactamases of Pseudomonas aeruginosa and Enterobacter cloacae. J Antimicrob Chemother 73:3049–3052. doi: 10.1093/jac/dky317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 319.Hackel MA, Tsuji M, Yamano Y, Echols R, Karlowsky JA, Sahm DF. 2018. In vitro activity of the siderophore cephalosporin, cefiderocol, against carbapenem-nonsusceptible and multidrug-resistant isolates of gram-negative bacilli collected worldwide in 2014 to 2016. Antimicrob Agents Chemother 62:e01968-17. doi: 10.1128/AAC.01968-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 320.Hsueh S-C, Lee Y-J, Huang Y-T, Liao C-H, Tsuji M, Hsueh P-R. 2018. In vitro activities of cefiderocol, ceftolozane/tazobactam, ceftazidime/avibactam and other comparative drugs against imipenem-resistant Pseudomonas aeruginosa and Acinetobacter baumannii, and Stenotrophomonas maltophilia, all associated with bloodstream infections in Taiwan. J Antimicrob Chemother 74:380–386. doi: 10.1093/jac/dky425. [DOI] [PubMed] [Google Scholar]
- 321.Ghazi IM, Monogue ML, Tsuji M, Nicolau DP. 2018. Pharmacodynamics of cefiderocol, a novel siderophore cephalosporin, in a Pseudomonas aeruginosa neutropenic murine thigh model. Int J Antimicrob Agents 51:206–212. doi: 10.1016/j.ijantimicag.2017.10.008. [DOI] [PubMed] [Google Scholar]
- 322.Henderson JC, Zimmerman SM, Crofts AA, Boll JM, Kuhns LG, Herrera CM, Trent MS. 2016. The power of asymmetry: architecture and assembly of the Gram-negative outer membrane lipid bilayer. Annu Rev Microbiol 70:255–278. doi: 10.1146/annurev-micro-102215-095308. [DOI] [PubMed] [Google Scholar]
- 323.Andolina G, Bencze LC, Zerbe K, Müller M, Steinmann J, Kocherla H, Mondal M, Sobek J, Moehle K, Malojčić G, Wollscheid B, Robinson JA. 2018. A peptidomimetic antibiotic interacts with the periplasmic domain of LptD from Pseudomonas aeruginosa. ACS Chem Biol 13:666–675. doi: 10.1021/acschembio.7b00822. [DOI] [PubMed] [Google Scholar]
- 324.Sader HS, Flamm RK, Dale GE, Rhomberg PR, Castanheira M. 2018. Murepavadin activity tested against contemporary (2016–17) clinical isolates of XDR Pseudomonas aeruginosa. J Antimicrob Chemother 73:2400–2404. doi: 10.1093/jac/dky227. [DOI] [PubMed] [Google Scholar]
- 325.Wach A, Dembowsky K, Dale GE. 2018. Pharmacokinetics and safety of intravenous murepavadin infusion in healthy adult subjects administered single and multiple ascending doses. Antimicrob Agents Chemother 62:e02355-17. doi: 10.1128/AAC.02355-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 326.Dale GE, Halabi A, Petersen-Sylla M, Wach A, Zwingelstein C. 2018. Pharmacokinetics, tolerability, and safety of murepavadin, a novel antipseudomonal antibiotic, in subjects with mild, moderate, or severe renal function impairment. Antimicrob Agents Chemother 62:e00490-18. doi: 10.1128/AAC.00490-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 327.Armaganidis A, Zakynthinos S, Mandragos C. 2017. Efficacy of murepavadin coadministered with standard of care in a phase 2 study in patients with ventilator-associated pneumonia due to suspected or documented Pseudomonas aeruginosa infections, poster A459. In Abstr 37th Int Symp Intensive Care Emergency Med. ISICEM, Brussels, Belgium. [Google Scholar]
- 328.Sader HS, Rhomberg PR, Flamm RK, Jones RN, Castanheira M. 2017. WCK 5222 (cefepime/zidebactam) antimicrobial activity tested against Gram-negative organisms producing clinically relevant β-lactamases. J Antimicrob Chemother 72:1696–1703. doi: 10.1093/jac/dkx050. [DOI] [PubMed] [Google Scholar]
- 329.Livermore DM, Mushtaq S, Warner M, Vickers A, Woodford N. 2017. In vitro activity of cefepime/zidebactam (WCK 5222) against Gram-negative bacteria. J Antimicrob Chemother 72:1373–1385. doi: 10.1093/jac/dkw593. [DOI] [PubMed] [Google Scholar]
- 330.Sader HS, Castanheira M, Huband M, Jones RN, Flamm RK. 2017. WCK 5222 (cefepime-zidebactam) antimicrobial activity against clinical isolates of gram-negative bacteria collected worldwide in 2015. Antimicrob Agents Chemother 61:e00072-17. doi: 10.1128/AAC.00072-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 331.Rodvold KA, Gotfried MH, Chugh R, Gupta M, Patel A, Chavan R, Yeole R, Friedland HD, Bhatia A. 2018. Plasma and intrapulmonary concentrations of cefepime and zidebactam following intravenous administration of WCK 5222 to healthy adult subjects. Antimicrob Agents Chemother 62:e00682-18. doi: 10.1128/AAC.00682-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 332.Hill C, Mills S, Ross RP. 2018. Phages & antibiotic resistance: are the most abundant entities on earth ready for a comeback?. Future Microbiol 13:711–726. doi: 10.2217/fmb-2017-0261. [DOI] [PubMed] [Google Scholar]
- 333.Rossitto M, Fiscarelli EV, Rosati P. 2018. Challenges and promises for planning future clinical research into bacteriophage therapy against Pseudomonas aeruginosa in cystic fibrosis. An argumentative review. Front Microbiol 9:775. doi: 10.3389/fmicb.2018.00775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 334.Forti F, Roach DR, Cafora M, Pasini ME, Horner DS, Fiscarelli EV, Rossitto M, Cariani L, Briani F, Debarbieux L, Ghisotti D. 2018. Design of a broad-range bacteriophage cocktail that reduces Pseudomonas aeruginosa biofilms and treats acute infections in two animal models. Antimicrob Agents Chemother 62:e02573-17. doi: 10.1128/AAC.02573-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 335.Ferry T, Boucher F, Fevre C, Perpoint T, Chateau J, Petitjean C, Josse J, Chidiac C, L’hostis G, Leboucher G, Laurent F, Ferry T, Valour F, Perpoint T, Boibieux A, Biron F, Miailhes P, Ader F, Becker A, Roux S, Triffault-Fillit C, Conrad A, Bosch A, Daoud F, Lippman J, Braun E, Chidiac C, Lustig S, Servien E, Gaillard R, Schneider A, Gunst S, Batailler C, Fessy M-H, Herry Y, Viste A, Chaudier P, Courtin C, Louboutin L, Martres S, Trouillet F, Barrey C, Jouanneau E, Jacquesson T, Mojallal A, Braye F, Boucher F, Shipkov H, Chateau J, Gleizal A, Aubrun F, Dziadzko M, Macabéo C, Laurent F, Rasigade J-P, Dupieux C, Craighero F, Boussel L, Pialat J-B, Morelec I, Janier M, Giammarile F, Tod M, Gagnieu M-C, Goutelle S, Mabrut E. 2018. Innovations for the treatment of a complex bone and joint infection due to XDR Pseudomonas aeruginosa including local application of a selected cocktail of bacteriophages. J Antimicrob Chemother 73:2901–2903. doi: 10.1093/jac/dky263. [DOI] [PubMed] [Google Scholar]
- 336.Latz S, Krüttgen A, Häfner H, Buhl EM, Ritter K, Horz HP. 2017. Differential effect of newly isolated phages belonging to PB1-like, phiKZ-like and LUZ24-like viruses against multi-drug resistant Pseudomonas aeruginosa under varying growth conditions. Viruses 9:315–319. doi: 10.3390/v9110315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 337.Astudillo A, Leung SSY, Kutter E, Morales S, Chan HK. 2018. Nebulization effects on structural stability of bacteriophage PEV 44. Eur J Pharm Biopharm 125:124–130. doi: 10.1016/j.ejpb.2018.01.010. [DOI] [PubMed] [Google Scholar]
- 338.Chang RYK, Chen K, Wang J, Wallin M, Britton W, Morales S, Kutter E, Li J, Chan H-K. 2018. Proof-of-principle study in a murine lung infection model of antipseudomonal activity of phage PEV20 in a dry-powder formulation. Antimicrob Agents Chemother 62:e01714-17. doi: 10.1128/AAC.01714-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 339.Comeau AM, Tétart F, Trojet SN, Prère MF, Krisch HM. 2007. Phage-antibiotic synergy (PAS): β-lactam and quinolone antibiotics stimulate virulent phage growth. PLoS One 2:e799. doi: 10.1371/journal.pone.0000799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 340.Uchiyama J, Shigehisa R, Nasukawa T, Mizukami K, Takemura-Uchiyama I, Ujihara T, Murakami H, Imanishi I, Nishifuji K, Sakaguchi M, Matsuzaki S. 2018. Piperacillin and ceftazidime produce the strongest synergistic phage–antibiotic effect in Pseudomonas aeruginosa. Arch Virol 163:1941–1948. doi: 10.1007/s00705-018-3811-0. [DOI] [PubMed] [Google Scholar]
- 341.Karpiński TM, Szkaradkiewicz AK. 2013. Characteristic of bacteriocines and their application. Pol J Microbiol 62:223–235. [PubMed] [Google Scholar]
- 342.Redero M, López-Causapé C, Aznar J, Oliver A, Blázquez J, Prieto AI. 2018. Susceptibility to R-pyocins of Pseudomonas aeruginosa clinical isolates from cystic fibrosis patients. J Antimicrob Chemother 73:2770–2776. doi: 10.1093/jac/dky261. [DOI] [PubMed] [Google Scholar]
- 343.Dickey SW, Cheung GYC, Otto M. 2017. Different drugs for bad bugs: antivirulence strategies in the age of antibiotic resistance. Nat Rev Drug Discov 16:457–471. doi: 10.1038/nrd.2017.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 344.Heras B, Scanlon MJ, Martin JL. 2015. Targeting virulence not viability in the search for future antibacterials. Br J Clin Pharmacol 79:208–215. doi: 10.1111/bcp.12356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 345.Papenfort K, Bassler BL. 2016. Quorum sensing signal–response systems in Gram-negative bacteria. Nat Rev Microbiol 14:576–588. doi: 10.1038/nrmicro.2016.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 346.Köhler T, Perron GG, Buckling A, van Delden C. 2010. Quorum sensing inhibition selects for virulence and cooperation in Pseudomonas aeruginosa. PLoS Pathog 6:e1000883. doi: 10.1371/journal.ppat.1000883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 347.Eibergen NR, Moore JD, Mattmann ME, Blackwell HE. 2015. Potent and selective modulation of the RhlR quorum sensing receptor by using non-native ligands: an emerging target for virulence control in Pseudomonas aeruginosa. Chembiochem 16:2348–2356. doi: 10.1002/cbic.201500357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 348.Fleitas Martínez O, Rigueiras PO, Pires ÁDS, Porto WF, Silva ON, de la Fuente-Nunez C, Franco OL. 2018. Interference with quorum-sensing signal biosynthesis as a promising therapeutic strategy against multidrug-resistant pathogens. Front Cell Infect Microbiol 8:444. doi: 10.3389/fcimb.2018.00444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 349.Wagner EK, Maynard JA. 2018. Engineering therapeutic antibodies to combat infectious diseases. Curr Opin Chem Eng 19:131–141. doi: 10.1016/j.coche.2018.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 350.Vincent J-L. 2014. Vaccine development and passive immunization for Pseudomonas aeruginosa in critically ill patients: a clinical update. Future Microbiol 9:457–463. doi: 10.2217/fmb.14.10. [DOI] [PubMed] [Google Scholar]
- 351.Rello J, Krenn C-G, Locker G, Pilger E, Madl C, Balica L, Dugernier T, Laterre P-F, Spapen H, Depuydt P, Vincent J-L, Bogár L, Szabó Z, Völgyes B, Máñez R, Cakar N, Ramazanoglu A, Topeli A, Mastruzzo MA, Jasovich A, Remolif CG, del Carmen Soria L, Andresen Hernandez MA, Ruiz Balart C, Krémer I, Molnár Z, von Sonnenburg F, Lyons A, Joannidis M, Burgmann H, Welte T, Klingler A, Hochreiter R, Westritschnig K. 2017. A randomized placebo-controlled phase II study of a Pseudomonas vaccine in ventilated ICU patients. Crit Care 21:22. doi: 10.1186/s13054-017-1601-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 352.Baer M, Sawa T, Flynn P, Luehrsen K, Martinez D, Wiener-Kronish JP, Yarranton G, Bebbington C. 2009. An engineered human antibody fab fragment specific for Pseudomonas aeruginosa PcrV antigen has potent antibacterial activity. Infect Immun 77:1083–1090. doi: 10.1128/IAI.00815-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 353.Warrener P, Varkey R, Bonnell JC, DiGiandomenico A, Camara M, Cook K, Peng L, Zha J, Chowdury P, Sellman B, Stover CK. 2014. A novel anti-PcrV antibody providing enhanced protection against Pseudomonas aeruginosa in multiple animal infection models. Antimicrob Agents Chemother 58:4384–4391. doi: 10.1128/AAC.02643-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 354.Jain R, Beckett VV, Konstan MW, Accurso FJ, Burns JL, Mayer-Hamblett N, Milla C, VanDevanter DR, Chmiel JF. 2018. KB001-A, a novel anti-inflammatory, found to be safe and well-tolerated in cystic fibrosis patients infected with Pseudomonas aeruginosa. J Cyst Fibros 17:484–491. doi: 10.1016/j.jcf.2017.12.006. [DOI] [PubMed] [Google Scholar]
- 355.Lang AB, Rudeberg A, Schoni MH, Que JU, Furer E, Schaad UB. 2004. Vaccination of cystic fibrosis patients against Pseudomonas aeruginosa reduces the proportion of patients infected and delays time to infection. Pediatr Infect Dis J 23:504–510. doi: 10.1097/01.inf.0000129688.50588.ac. [DOI] [PubMed] [Google Scholar]
- 356.Lazar H, Horn MP, Zuercher AW, Imboden MA, Durrer P, Seiberling M, Pokorny R, Hammer C, Lang AB. 2009. Pharmacokinetics and safety profile of the human anti-Pseudomonas aeruginosa serotype O11 immunoglobulin M monoclonal antibody KBPA-101 in healthy volunteers. Antimicrob Agents Chemother 53:3442–3446. doi: 10.1128/AAC.01699-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 357.Le HN, Quetz JS, Tran VG, Le VTM, Aguiar-Alves F, Pinheiro MG, Cheng L, Yu L, Sellman BR, Stover CK, DiGiandomenico A, Diep BA. 2018. MEDI3902 correlates of protection against severe Pseudomonas aeruginosa pneumonia in a rabbit acute pneumonia model. Antimicrob Agents Chemother 62:e02565-17. doi: 10.1128/AAC.02565-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 358.Tabor DE, Oganesyan V, Keller AE, Yu L, McLaughlin RE, Song E, Warrener P, Rosenthal K, Esser M, Qi Y, Ruzin A, Stover CK, DiGiandomenico A. 2018. Pseudomonas aeruginosa PcrV and Psl, the molecular targets of bispecific antibody MEDI3902, are conserved among diverse global clinical isolates. J Infect Dis 218:1983–1994. doi: 10.1093/infdis/jiy438. [DOI] [PubMed] [Google Scholar]
- 359.Ali SO, Yu XQ, Robbie GJ, Wu Y, Shoemaker K, Yu L, DiGiandomenico A, Keller AE, Anude C, Hernandez-Illas M, Bellamy T, Falloon J, Dubovsky F, Jafri HS. 2018. Phase 1 study of MEDI3902, an investigational anti-Pseudomonas aeruginosa PcrV and Psl bispecific human monoclonal antibody, in healthy adults. Clin Microbiol Infect 25:629.e1–629.e6. doi: 10.1016/j.cmi.2018.08.004. [DOI] [PubMed] [Google Scholar]
- 360.François B, Chastre J, Eggiman P, Laterre PF, Torres A, Sanchez M, Esser MT, Bishop B, Bonten M, Goosens H, Jafri HS. 2016. The SAATELLITE and EVADE clinical studies within the COMBACTE consortium: a public-private collaborative effort in designing and performing clinical trials for novel antibacterial drugs to prevent nosocomial pneumonia. Clin Infect Dis 63:S46–S51. doi: 10.1093/cid/ciw245. [DOI] [PubMed] [Google Scholar]
- 361.Ly NS, Bulitta JB, Rao GG, Landersdorfer CB, Holden PN, Forrest A, Bergen PJ, Nation RL, Li J, Tsuji BT. 2015. Colistin and doripenem combinations against Pseudomonas aeruginosa: profiling the time course of synergistic killing and prevention of resistance. J Antimicrob Chemother 70:1434–1442. doi: 10.1093/jac/dku567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 362.Landersdorfer CB, Rees VE, Yadav R, Rogers KE, Kim TH, Bergen PJ, Cheah S-E, Boyce JD, Peleg AY, Oliver A, Shin BS, Nation RL, Bulitta JB. 2018. Optimization of a meropenem-tobramycin combination dosage regimen against hypermutable and nonhypermutable Pseudomonas aeruginosa via mechanism-based modeling and the hollow-fiber infection model. Antimicrob Agents Chemother 62:e02055-17. doi: 10.1128/AAC.02055-17. [DOI] [PMC free article] [PubMed] [Google Scholar]