Abstract
Introduction:
Clinical trials directed at mechanistic target of rapamycin (mTOR) inhibition have yielded disappointing results in glioblastoma (GBM). A major mechanism of resistance involves the activation of a salvage pathway stimulating internal ribosome entry site (IRES)-mediated protein synthesis. PRMT5 activity has been implicated in the enhancement of IRES activity.
Methods:
We analyzed the expression and activity of PRMT5 in response to mTOR inhibition in GBM cell lines and short-term patient cultures. To determine whether PRMT5 conferred resistance we used genetic and pharmacological approaches to ablate PRMT5 activity and assessed the effects on in vitro and in vivo sensitivity. Mutational analyses of the requisite IRES-trans-acting factor (ITAF), hnRNP A1 determined whether PRMT5-mediated methylation was necessary for ITAF RNA binding and IRES activity.
Results:
PRMT5 activity is stimulated in response to mTOR inhibitors. Knockdown or treatment with a PRMT5 inhibitor blocked IRES activity and sensitizes GBM cells. Ectopic expression of non-methylatable hnRNP A1 mutants demonstrated that methylation of either arginine residues 218 or 225 was sufficient to maintain IRES binding and hnRNP A1-dependent cyclin D1 or c-MYC IRES activity, however a double R218K/R225K mutant was unable to do so. The PRMT5 inhibitor EPZ015666 displayed synergistic anti-GBM effects in vitro and in a xenograft mouse model in combination with PP242.
Conclusions:
These results demonstrate that PRMT5 activity is stimulated upon mTOR inhibition in GBM. Our data further support a signaling cascade in which PRMT5-mediated methylation of hnRNP A1 promotes IRES RNA binding and activation of IRES-mediated protein synthesis and resultant mTOR inhibitor resistance.
Keywords: PRMT5, mTOR, PP242, rapamycin, EPZ015666, drug resistance, glioblastoma
Introduction
Glioblastoma is one of the most prevalent central nervous system tumors with a dismal median survival of typically only twelve months [1]. The deadliness of this tumor is due to the recalcitrant nature of the tumors to surgical resection and the inevitable development of drug resistance [2]. The mutational landscape of GBMs includes EGFR amplification or constitutively activating mutations, PTEN loss and hyperactivation of the phosphatidylinositol 3-kinase (PI3K) signaling cascade [3]. These molecular alterations are seen in approximately ninety percent of GBMs and converge on the downstream effector mTOR, resulting in its hyperactivation [4]. mTOR is a major regulator of protein synthesis, metabolism and autophagy in the cell and as a result controls tumor cell proliferation, survival and drug resistance [5, 6]. mTOR exists in at least two complexes, mTOR complex 1 and 2 (mTORC1 and 2) and each contain unique regulatory subunits dictating substrate specificities [7].
The prototypic mTOR inhibitor rapamycin, and other first generation allosteric inhibitors have failed in the clinic for GBM as a result of loss of feedback regulation promoting AKT activation [8]. The second generation direct mTOR kinase inhibitors (TORKIs) which block both mTORC1 and mTORC2 activity await complete evaluation in GBM. Recently, an inhibitor targeted to mTORC1, Rapalink-1 drove the regression of intracranial tumors improving survival compared with earlier-generation inhibitors however this agent also requires further evaluation [9]. Taken together, these studies emphasize the potential role of mTOR kinase inhibitors in the treatment of GBM.
The signaling relationships between mTORCs suggest the possibility of several mechanisms of mTOR inhibitor resistance [1, 10]. Our studies have demonstrated that both allosteric and direct mTOR kinase inhibitors are able to activate a transcript-specific protein synthesis salvage pathway capable of maintaining the translation of critical cell cycle mRNAs resulting in resistance to mTOR therapies [11, 12]. Transcript-specific enhancement of translation upon activation of the salvage pathway is mediated via IRES-dependent protein synthesis and requires the ITAF, hnRNP A1 [13]. Recently, we have described the identification of a small molecule inhibitor of hnRNP A1 which blocks cyclin D1 and c-MYC IRES activity and synergizes with mTOR inhibitors to achieve strong anti-GBM activities [14].
PRMT5 is a type II methyltransferase which is essential for viability and normal development and whose expression is increased in GBM and elevated expression is negatively correlated with patient survival [15, 16]. Additionally, PRMT5 regulates the expression and stability of critical transcription factors and signaling receptors that control normal stem cell and oligodendrocyte progenitor cell function [17]. Further evidence in support of the importance of PRMT5 in gliomagenesis is the observation that GBM cells with depleted PRMT5 fail to grow as intracranial xenografts [15, 18]. Data also now show that small molecule inhibitors of PRMT5 demonstrate antitumor efficacy in GBM [19]. PRMT5 was recently demonstrated to activate IRES-mediated translation upon symmetrical di-methylation of arginine (SDMA) residues in hnRNP A1 [20]. PRMT5 was shown to di-methylate hnRNP A1 on two residues, R218 and R225, which facilitated the interaction of hnRNP A1 with IRES RNAs to promote IRES-mediated translation. In this report we examined whether PRMT5 is capable of regulating GBM tumor responses to mTOR inhibitors. We demonstrate that mTOR inhibition results in stimulation of PRMT5 activity in GBM and patient-derived cell lines. PRMT5 knockdown sensitizes GBM cells to mTOR inhibition and suppresses mTOR inhibitor-induced IRES activity. It is shown that PRMT5-mediated SDMA of hnRNP A1 is required for its binding to IRES RNAs and stimulation of IRES activity in response to mTOR inhibition. We also examine the anti-tumor effects of co-therapy utilizing specific PRMT5 and TORKI inhibitors in vitro and in xenografted mice.
Materials and methods
Details regarding cell cultures, reagents, in vitro and in vivo protocols and data analyses are described in Online Resource 1 Supplemental Materials and Methods.
Results
mTOR inhibitors enhance PRMT5 activity
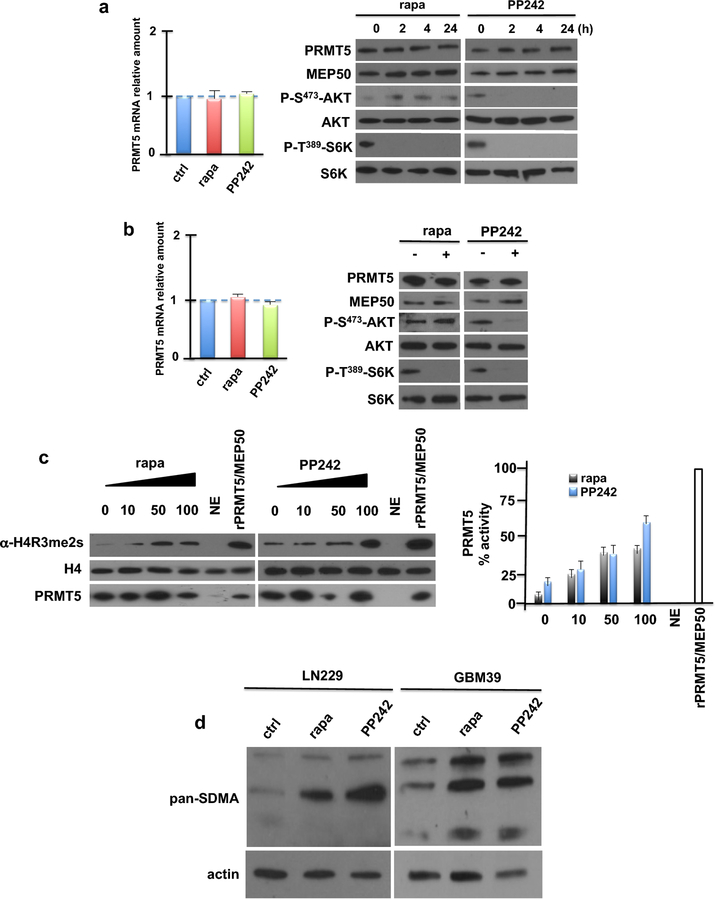
To investigate if PRMT5 played a role in the response of GBM cells to mTOR inhibition we examined whether PRMT5 transcript or protein levels were altered in response to rapamycin or PP242 exposure in LN229 cells. Neither PRMT5 mRNA (left panel) nor protein (right panel) was affected by exposure (Fig. 1a). Expression of the PRMT5 cofactor, MEP50, was also unaffected by mTOR inhibition. Similarly, PRMT5 mRNA or protein levels did not change in GBM patient-derived cells (GBM39) upon mTOR inhibitor exposure (Fig. 1b). However, upon exposure to either rapamycin or PP242, PRMT5 activity was markedly stimulated as determined by an in vitro methylation assay of immunoprecipitated PRMT5 from LN229 cells utilizing histone H4 as a substrate (Fig. 1c, left panel). A dose-dependent increase in PRMT5 activity was observed in response to rapamycin or PP242 (Fig. 1c, right panel). PRMT5 activity was also stimulated by PP242 in U87 cells expressing a constitutively active EGFRvIII allele, one of the most common genetic events driving GBM [21] (Online Resource 2 Suppl. Fig. S2). Immunoblotting of lysates obtained from LN229 or GBM39 cells treated with rapamycin or PP242 and probed with antibody recognizing symmetric di-methyl arginine motifs also displayed enhanced levels of methylated species in response to mTOR inhibition (Fig. 1d). These data demonstrate that PRMT5 activity is stimulated by mTOR inhibition.
Fig. 1.
PRMT5 expression and activity in response to mTOR inhibition. a Expression of PRMT5 mRNA (left panel) and protein (right panel) in LN229 cells following treatment with rapamycin (rapa) or PP242 (50 nM) for the indicated time points. b Effects of mTOR inhibitors on PRMT5 mRNA (left panel) and protein (right panel) in short-term patient derived GBM39 cells. Cells were treated for 24 h at 50 nM with each inhibitor. c LN229 cells expressing Flag-PRMT5 were treated with the indicated concentrations of either rapa or PP242 (0–100 nM) for 4 h and subsequently PRMT5/MEP50 complexes were immunoprecipitated using Flag antibodies. In vitro methylation activity of the complexes was then determined utilizing H4 as a substrate and activity detected by immunoblotting reactions with symmetric dimethylarginine (R3me2s)-specific antibodies. Reactions were also probed for H4 and PRMT5. NE, no extract used in reactions (negative control); recombinant PRMT5/MEP50 was added to positive control reactions (left panel). In vitro PRMT5 activity of LN229 cells treated with mTOR inhibitors displayed graphically (right panel). d LN229 or GBM39 cells were treated with mTOR inhibitors (50 nM, 24 h) and extracts immunoblotted with pan-SDMA motif and actin antibodies.
Inhibition of PRMT5 sensitizes GBM cells to mTOR inhibitors
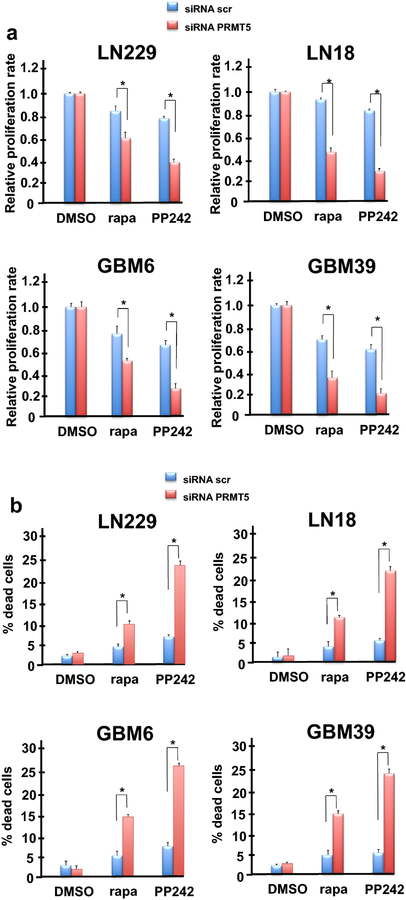
To determine whether a specific role for PRMT5 exists in preventing mTOR inhibitor mediated cell death we blocked PRMT5 expression via small interfering RNAs (siRNA) in established GBM lines and patient-derived cell lines and assessed its effects on mTOR inhibitor sensitivity. Knockdown of PRMT5 significantly sensitized LN229, LN18, GBM6 and GBM39 cells to rapamycin and PP242 treatment inhibiting growth as compared to controls (Fig. 2a). SiRNA targeting PRMT5 effectively reduced expression by greater then 95% in patient-derived cells and below detectable levels in established GBM lines as determined via immunoblot while a nontargeting scrambled control siRNA had no appreciable effect on expression (Online Resource 3 Suppl. Fig. S3). Additionally, cytotoxicity as determined by trypan blue exclusion assays, demonstrated a significant increase in rapamycin or PP242-mediated cell death following knockdown of PRMT5 (Fig. 2b). We noted that rapamycin induced markedly less cell death, even in the context of PRMT5 knockdown as compared to PP242. As rapamycin does not significantly inhibit mTORC2 activity relative to PP242, this suggested that sustained mTORC2 activity might play a role in tumor cell survival. These data demonstrate that PRMT5 contributes to mTOR inhibitor resistance by suppressing cell death and can be reversed by genetic inhibition of PRMT5 in GBM cells.
Fig. 2.
PRMT5 knockdown sensitizes GBM and patient-derived lines to mTOR-targeted therapies. a Cell proliferation was determined following treatment of lines with rapa or PP242 (100 nM, 48 h) as indicated in the context of PRMT5 knockdown via siRNAs. non-targeting siRNAs (siRNA scr) were utilized as a control. Data are means +S.D., n=3. * P < 0.05. b Effects of PRMT5 knockdown and mTOR inhibitors on LN229, LN18, GBM6 and GBM39 cell line death as determined by trypan blue exclusion assays. Data are means +S.D., n=3. * P < 0.05.
Knockdown of PRMT5 blocks IRES activity
As the induction of IRES activity has been demonstrated to be a major mode of drug resistance to mTOR inhibitors, we assessed whether cyclin D1 or c-MYC IRES activity was affected by PRMT5 downregulation in GBM lines and patient-derived GBM cells. IRES reporter assays were conducted utilizing dicistronic mRNA reporters containing the indicated IRES sequences within the intercistronic region between the Renilla and firefly open reading frames. Thus, Renilla luciferase activity is a readout of cap-dependent translation while firefly translation is driven by the respective IRES. Reporter plasmids were transiently transfected into LN229, LN18, GBM6 or GBM39 cells in which PRMT5 was knocked down via siRNAs. Cyclin D1 and c-MYC IRES activities were markedly induced by PP242 relative to control or scrambled siRNA transfected groups (Online Resource 4 Suppl. Fig. S4). However, in cells treated with siRNAs targeting PRMT5 both cyclin D1 and c-MYC IRES activities were curtailed following PP242 exposure. These data demonstrate that PRMT5 is required for cyclin D1 and c-MYC IRES activity in response to mTOR inhibition.
R218/R225 methylation of the ITAF hnRNP A1 is required for IRES activity in response to mTOR inhibition
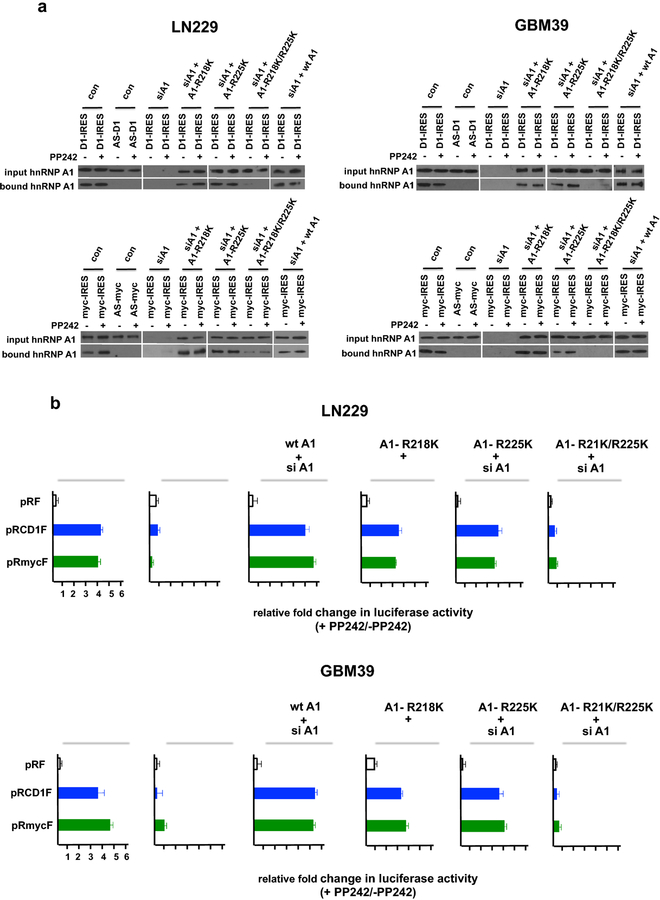
Our previous data demonstrated that hnRNP A1 constitutively binds to the cyclin D1 and c-MYC IRESs and is required for mTOR inhibitor-induced IRES activity [13]. As symmetrical di-methylation of arginines 218 and 225 by PRMT5 has recently been shown to activate cyclin D1 and c-MYC IRES activity [20] we sought to determine whether these methylation events were involved in mTOR inhibitor resistance in GBM. We utilized an RNA pull-down assay to evaluate whether cyclin D1 or c-MYC IRES sequences would bind native hnRNP A1, as well as hnRNP A1 mutants containing lysine substitutions at amino acids 218, 225 or both (R218K, R225K, R218K/R225K). HnRNP A1 was precipitated from LN229 or GBM39 lysates extracted from cells treated without or with PP242 by either of the IRES RNAs (Fig. 3a; LN229; cyclin D1 IRES, upper left panel; c-MYC IRES, lower left panel; GBM39; cyclin D1 IRES, upper right panel; c-MYC IRES, lower right panel) in the sense orientation, but no hnRNP A1 was detected in precipitates of the IRES RNAs in the antisense orientation. Furthermore, this data, consistent with our previous experiments, demonstrate that hnRNP A1 is constitutively bound to either the cyclin D1 or c-MYC IRESs independent of PP242 exposure. However, in LN229 or GBM39 cells in which constructs expressing the hnRNP A1 R218K, R225K or R218K/R225K mutants were transfected 24 h post hnRNP A1 siRNA transfection, we observed that either of the single nonmethylatable (R218K or R225K) hnRNP A1 mutants were effectively pulled-down by cyclin D1 or c-MYC IRESs, while the double (R218K/R225K) hnRNP A1 displayed significantly reduced binding irrespective of PP242 treatment. We also determined the effects of these mutant hnRNP A1 alleles on cyclin D1 and c-MYC IRES reporter activity in the absence or presence of PP242. LN229 or GBM39 cells transfected with the single R218K or R225K hnRNP A1 mutants and either the cyclin D1 or c-MYC IRES reporters 24 h post hnRNP A1 siRNA transfection displayed a marked induction of PP242 induced IRES activity (Fig. 3b). However, consistent with the IRES binding data, LN229 or GBM39 cells transfected with the double (R218K/R225K) mutant did not exhibit significant PP242-induced IRES activity. These data suggest that PRMT5 directed methylation of either arginine residues 218 or 225 is necessary for hnRNP A1 cyclin D1 or c-MYC IRES binding and mTOR inhibitor-induced IRES activity.
Fig. 3.
Methylation of hnRNP A1 is required for mTOR-inhibitor induced IRES-binding and activity. a RNA-pull down assays utilizing biotinylated cyclin D1 and c-MYC IRES RNAs. Cytoplasmic extracts of LN229 or GBM39 cells in which hnRNP A1 has been knocked down via siRNA and expressing the indicated wt hnRNP A1 or non-methylatable hnRNP A1 mutant (A1-R218K, A1-R225K or A1-R218K/R225K) constructs in the absence or presence of PP242 (50 nM, 24 h) were incubated with biotinylated cyclin D1 (top panels) or c-MYC (bottom panels) IRES RNAs and precipitated with streptavidin-Sepharose beads. AS, antisense RNA sequence. Input and bound fractions were analyzed by immunoblotting using hnRNP A1 antibodies. b LN229 or GBM39 cells were transfected with control siRNA or siRNA targeting hnRNP A1 and 24 h later either co-transfected with the control pRF, cyclin D1 or c-MYC IRES reporters and the indicated non-methylatable hnRNP A1 mutant (A1-R218K, A1-R225K or A1-R218K/R225K) in the absence or presence of PP242 (50 nM, 24 h). Relative Renilla and firefly luciferase activities were subsequently determined. Data are means + S.D., n = 3.
Pharmacological inhibition of PRMT5 curtails PP242-induced cyclin D1 and c-MYC IRES activity
We also determined whether a pharmacologic approach to inhibiting PRMT5 via the inhibitor EPZ015666 would have effects on PP242-induced IRES activity. We conducted co-treatment experiments in several GBM lines and patient derived cells, and monitored cyclin D1 and c-MYC IRES activity. PP242 induced significant cyclin D1 and c-MYC IRES activities in LN229 and LN18 cells which was markedly suppressed by combining EPZ015666 treatment (Online Resource 5 Suppl. Fig. S5). Similarly, in two patient-derived cell lines (GBM6 & GBM39), PP242 also markedly induced cyclin D1 and c-MYC IRES activity that was blocked by EPZ015666. These data show that hnRNP A1-dependent PP242-induced cyclin D1 and c-MYC IRES activity requires PRMT5 function in GBM.
Synergistic antiproliferative effects of PRMT5 and mTORC1/2 inhibitors
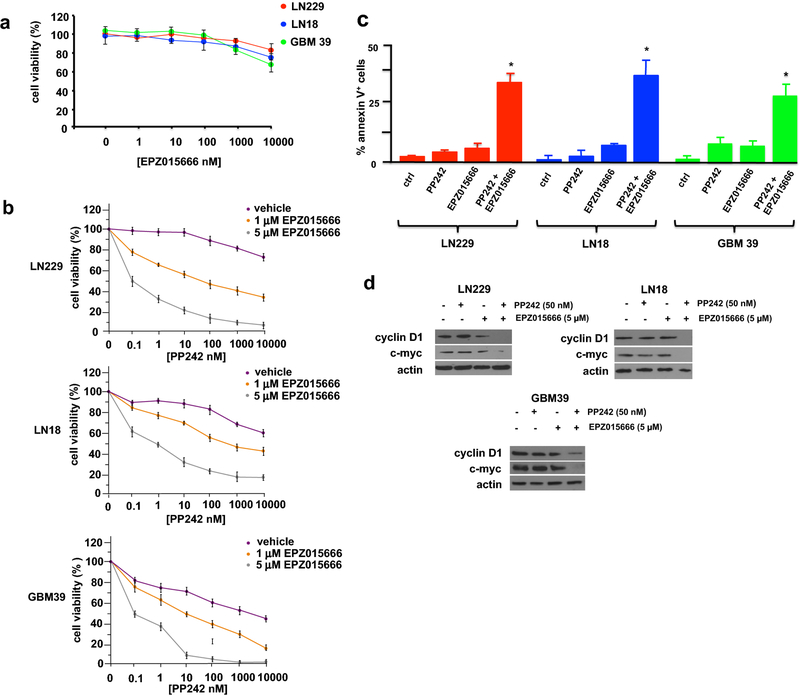
To determine whether the combination of PRMT5 and PP242 inhibitors would have effects on in vitro growth we initially determined if GBM lines were sensitive to growth inhibition following exposure to EPZ015666 alone. No significant inhibition of cell growth was observed at concentrations up to 10 µM in GBM cell lines or patient-derived cells (Fig. 4a). We subsequently tested the effects EPZ015666 on LN229 and LN18 GBM lines and short-term patient derived GBM39 cells following PP242 exposure. Treatment of these lines and the short-term patient derived GBM39 cells with EPZ015666 at 1 and 5 µM concentrations resulted in synergistic inhibition of cell growth over a wide range of concentrations of PP242 (Fig. 4b; CI = 0.45 at ED50 ratio of 1:100). We additionally determined whether combination treatment with EPZ015666 and PP242 would induce apoptosis. PP242 or EPZ015666 alone resulted in minimal induction of apoptosis in the lines tested, however the combination of the two inhibitors displayed a marked increase in apoptotic cell numbers (Fig. 4c). Cyclin D1 and c-MYC expression was also significantly reduced in GBM lines and the short-term patient derived GBM39 cells cotreated with PP242 and EPZ015666 (Fig. 4d).
Fig. 4.
Synergistic anti-GBM effects of mTORC1/2 and PRMT5 inhibitors in vitro. a Viability of LN229, LN18 and GBM39 cells following 48 h in culture in EPZ015666 at the indicated concentrations. Data represent ±S.D. of 3 independent experiments. b Combination analysis of PP242 and EPZ015666 in LN229, LN18 and GBM39 cells. Cells were treated with the indicated doses PP242 alone or in combination with EPZ015666 for 48 h and percent cell viability was determined relative to control cultures. Control cells were treated with DMSO vehicle. Data are means ±S.D., n = 3. c Percent apoptotic cells as determined via annexin V-FITC staining in LN229, LN18 and GBM39 cells treated with the indicated inhibitors for 48 h (PP242, 50 nM; EPZ015666, 5 µM; PP242, 50 nM + EPZ015666, 5 µM). Data are means +S.D., n = 3. d Protein levels of cyclin D1, c-MYC and actin in LN229, LN18 and GBM39 cells following the indicated treatments with PP242, EPZ015666 or both compounds at 24 h.
Effects of EPZ015666 and PP242 combination treatment in GBM xenografts
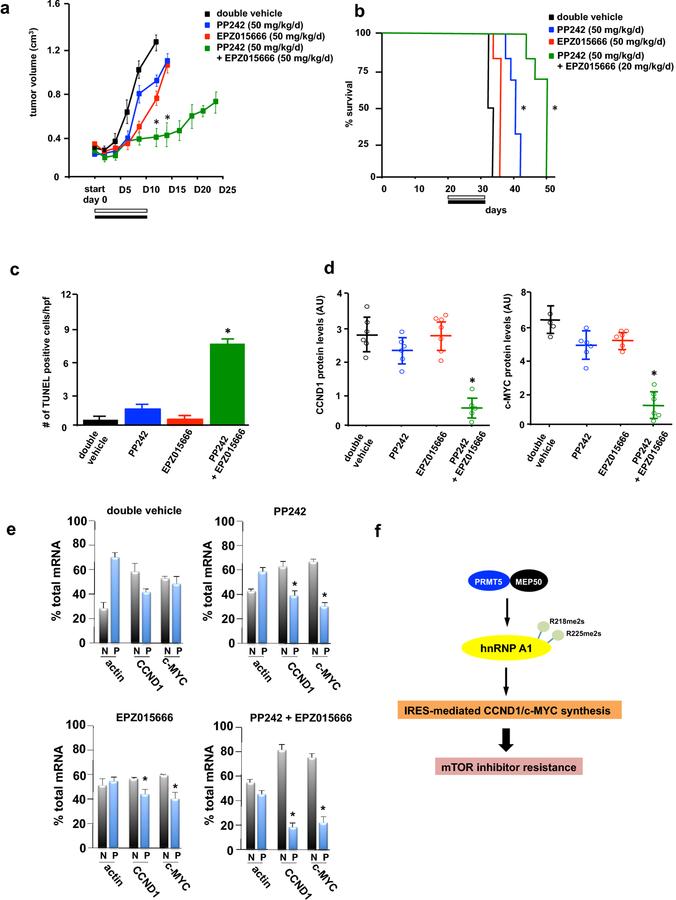
To examine whether PRMT5 and mTOR inhibition would demonstrate anti-GBM effects in vivo, we conducted xenograft studies utilizing LN229 cells in mice. Mice were subcutaneously implanted with cells and once palpable tumors formed (~200 mm3), mice were randomized into treatment groups and treated with double vehicle, PP242 (50 mg/kg/d), EPZ015666 (50 mg/kg/d) and the combination PP242 (50 mg/kg/d) plus EPZ015666 (50 mg/kg/d). Xenografts receiving monotherapy with PP242 displayed a significant inhibition of tumor growth (29% inhibition at end of regimen; tumor growth delay 5.5 days) (Fig. 5a). Tumor growth following treatment with EPZ015666 alone was reduced (30% inhibition at end of regimen; tumor growth delay 5.5 days). However, the combination of PP242 and EPZ015666 was markedly more effective as compared to either of the monotherapies alone (75% inhibition at end of regimen; tumor growth delay, 16.5 days). The overall survival of mice receiving monotherapy with PP242 was significantly extended relative to double vehicle (Fig. 5b). However, co-therapy with PRMT5 and mTORC1/2 inhibitors significantly extended survival relative to either of the monotherapies, consistent with the effects on xenograft growth. No overt indications of toxicity of this combination of inhibitors were observed as the mice did not display weight loss and tolerated this dosing regimen well. A significant induction of apoptosis was also observed in the combination treated mice as monitored by TUNEL staining of tumor sections from harvested tumors at autopsy (Fig. 5c, see also Online Resource 6 Suppl. Fig. S6). These data were in agreement with the effects of combination treatment that were observed in vitro (see Fig. 4c). We examined these harvested tumors for cyclin D1 and c-MYC protein levels via Western blot analyses and mice receiving combination therapy displayed a marked reduction in expression (Fig. 5d, left and right panels). To determine whether the reductions seen in cyclin D1 and c-MYC protein expression were the result of actual changes in mRNA translational efficiencies, we performed polysome analysis on freshly harvested tumors following the final day of inhibitor dosing. In tumors from double vehicle treated mice 42% of cyclin D1 and 50% c-MYC mRNAs were associated with highly translated polysomes (Fig. 5e). Tumors from mice receiving PP242 therapy exhibited reduced cyclin D1 and c-MYC mRNA translational states (35% and 30% polysomal, respectively). Effective inhibition of eIF-4F-mediated translation was evident by the marked redistribution of actin mRNA from polysomes to monosomal/non-ribosomal material in PP242 treated tumors. Mice that received EPZ015666 monotherapy also displayed a modest yet significant reduction in cyclin D1 and c-MYC mRNA which was associated with polysomes. EPZ015666 treatment resulted in a redistribution of cyclin D1 and c-MYC transcripts to 45% and 42% present within polysomal fractions, respectively. However, tumors from mice treated with the combination of inhibitors displayed a marked redistribution of cyclin D1 and c-MYC mRNAs to monosomal/non-ribosomal fractions with only 18% and 20% of mRNAs associated with polysomes, respectively.
Fig. 5.
Co-therapy of GBM xenografts with mTOR and PRMT5 inhibitors. a Xenograft tumor volume in SCID mice implanted with LN229 cells and treated with the indicated schedules for 10 consecutive days and tumor growth assessed every two days following the initiation of treatment (start, day 0). *, P < 0.05 (significantly different then double vehicle) n=6 mice per treatment group. b Overall survival of mice with subcutaneous implanted LN229 tumors receiving the indicated treatment schedules. Treatments were initiated upon tumors reaching ~ 100 mm3 in size. *, P < 0.05, n=6 mice per treatment group. c Apoptotic cells were identified by TUNEL assays of sections prepared from harvested tumors at day 12 following initiation of treatment regimens. Data are expressed as the number of positive apoptotic bodies divided by high power field (hpf; 10–12 hpf/tumor). Values are means +S.D., * P < 0.05. d Cyclin D1(left panel) and c-MYC (right panel) protein levels in tumors. Values are means ±S.D. *, P < 0.05 (significantly different from vehicle). Protein levels were quantified by Western analyses of harvested tumors from mice with the corresponding treatments as indicated. Band intensities were quantified by densitometry analyses via ImageJ software. e Polysome distributions of cyclin D1, c-MYC and actin transcripts from tumors harvested from mice receiving the indicated treatments. Tumor cell extracts were separated via sucrose density gradient centrifugation, fractionated and pooled into nonribosomal, monosomal fraction (N, white bars) and polysomal fraction (P, black bars). Purified mRNAs were subsequently used in qrt-PCR analyses to determine distributions of the indicated mRNAs across the gradients. Means and +S.D. values are shown for quadruplicate measurements. *, P < 0.05. f mTOR inhibition leads to activation of PRMT5 and symmetric dimethylation of hnRNP A1 at arginine residues 218 and 225. Either of these methylation events is sufficient to promote hnRNP A1 binding to cyclin D1 and c-MYC IRESs resulting in enhanced protein synthesis of these determinants and mTOR inhibitor resistance in GBM.
Discussion
In this report we describe the induction of PRMT5 activity in response to mTOR inhibitors. Blockade of PRMT5 via RNAi led to heightened sensitivity of GBM tumor cells to mTOR inhibition and markedly inhibited IRES activity in response to PP242. An analysis of non-methylatable alleles of hnRNP A1 suggested that either arginine 218 or 225 methylation via PRMT5 is sufficient to mediate hnRNP A1 binding to IRES RNAs and to stimulate IRES-mediated translation of cyclin D1 and c-MYC-based reporters following PP242 exposure. Pharmacologic inhibition of PRMT5 utilizing EPZ015666 in combination with PP242 induced synergistic anti-GBM effects in vitro and in xenografts. Taken together, these data suggest that PRMT5 mediated methylation of hnRNP A1 stimulates IRES activity resulting in GBM cell resistance to mTOR inhibition.
PRMT5 is a type II methyltransferase and participates in cellular signaling and gene regulation by methylating histones and nonhistone proteins [22–26]. Several proteins are regulated by PRMT5 at the post-transcriptional level via IRES-dependent translation and include cyclin D1, c-MYC, HIF1α and ESR1 [20]. The ability of PRMTs to extensively methylate hnRNPs [27–29] has been appreciated for some time, however, the recent identification of the SDMA modifications on hnRNP A1 which regulate IRES activity extend the known functions of PRMT5 [20, 22]. These methylation events appear to modulate the ability of hnRNP A1 to bind RNA and promote IRES activity. We have previously demonstrated that AKT-mediated phosphorylation of serine 199 on hnRNP A1 suppresses IRES activity [11] and that this ITAF is constitutively bound to the cyclin D1 and c-MYC IRESs irrespective of mTOR inhibitor exposure [11, 13]. HnRNP A1 is known to facilitate RNA strand annealing activity [30, 31] and a critical function of this property may be to facilitate the formation of a productive IRES conformation which is able to recruit the small ribosomal subunit [32, 33]. Our studies suggested that serine 199 phosphorylation of hnRNP A1 abolished its ITAF function by inhibiting annealing activity [11]. Our data (see Fig. 3a) also support the ability of PRMT5-mediated arginine 218 or 225 di-methylation to regulate hnRNP A1 binding to IRES RNAs, while serine 199 phosphorylation appears to regulate IRES activity via effects on hnRNP A1-mediated RNA annealing properties. It will be interesting to determine if other post-translational modifications participate in further crosstalk mechanisms regulating the overall ITAF activity of hnRNP A1 and mTOR inhibitor resistance.
mTOR has become recognized as a validated target as a result of its persistent hyperactivation in GBM due to EGFR, PDGFRα or c-MET amplification, EGFRvIII mutation or PTEN loss [4]. Sensitivity to EGFR-targeted therapies has been demonstrated to be dependent on mTOR inhibition [34, 35] and this suggests a potential mechanism by which EGFR tyrosine kinase inhibitors may also significantly upregulate PRMT5 activity leading to drug resistance. As mTOR inhibitors typically induce a cytostatic response in GBM, with minimal clinical benefit, the ability of a combination therapeutic strategy, via pharmacologic inhibition of PRMT5 and mTOR, to induce a cytotoxic response may represent an advancement in therapeutic options. Our data demonstrating a synergistic induction of cell death in response to both drugs further supports this notion (see Figs. 4c & 5c). However, poor blood-brain-barrier (BBB) penetration may limit the efficacy of these inhibitors as preclinical data suggests that EPZ015666 may only be able to access late-stage tumors with a compromised BBB [36]. The BBB-penetrant mTOR kinase inhibitor AZD2014 may find utility in this regard [37].
The mechanism by which mTOR inhibition results in activation of PRMT5 remains to be elucidated. Curiously, MEP50 has been reported to be a cyclin D1/Cdk4 substrate and phosphorylation on Thr-5 markedly increases PRMT5/MEP50 activity [38]. It is tempting to speculate that mTOR inhibitor-induced cyclin D1 IRES-mediated protein synthesis may promote downstream cyclin D1/CDK4 complex formation and subsequent phosphorylation of MEP50 to activate PRMT5 methyltransferase activity. This feed-forward mechanism may result in continued PRMT5/hnRNP A1 signaling, however it is unlikely to be the initiating signaling event leading to activation of PRMT5 in response to mTOR inhibitor exposure. An increased understanding of the upstream regulators of PRMT5 is required.
Collectively, our current work supports a model wherein mTOR inhibition activates PRMT5 leading to dimethylation of hnRNP A1 stimulating its binding to cyclin D1 and c-MYC IRES RNAs and promoting mRNA translation resulting in drug resistance in GBM (Fig. 5f). Our functional analysis revealed that PRMT5 is necessary for cyclin D1 and c-MYC IRES activity and mTOR inhibitor resistance. Future studies directed towards addressing the mechanism(s) of PRMT5 activation by mTOR inhibitors will be of significant importance.
Supplementary Material
Acknowledgements
We thank Mark Schroeder and Drs. Jacob Fleischmann, Norimoto Yanagawa, Jann Sarkaria and Paul Mischel for cell lines and reagents. We also thank Dr. Alan Lichtenstein for comments on the manuscript.
Funding
This work was supported, in whole or in part, by Merit Review Award Number I01 BX002665 from the United States (U.S.) Department of Veterans Affairs Biomedical Laboratory R&D (BLRD) Service and NIH R01CA217820 grants.
Footnotes
Publisher's Disclaimer: This Author Accepted Manuscript is a PDF file of an unedited peer-reviewed manuscript that has been accepted for publication but has not been copyedited or corrected. The official version of record that is published in the journal is kept up to date and so may therefore differ from this version.
Compliance with ethical standards
All animal experiments were performed under an approved Institutional Animal Care and Use Committee protocol and conformed to the guidelines established by the Association for the Assessment and Accreditation of Laboratory Animal Care.
The authors declare no competing financial interests.
References
- 1.Cloughesy TF, Cavenee WK, Mischel PS (2014) Glioblastoma: from molecular pathology to targeted treatment. Annu Rev Pathol 9: 1–25 doi: 10.1146/annurev-pathol-011110-130324 [DOI] [PubMed] [Google Scholar]
- 2.Wen PY, Kesari S (2008) Malignant gliomas in adults. N Engl J Med 359: 492–507 doi: 10.1056/NEJMra0708126 [DOI] [PubMed] [Google Scholar]
- 3.Brennan CW, Verhaak RG, McKenna A, Campos B, Noushmehr H, Salama SR, Zheng S, Chakravarty D, Sanborn JZ, Berman SH, Beroukhim R, Bernard B, Wu CJ, Genovese G, Shmulevich I, Barnholtz-Sloan J, Zou L, Vegesna R, Shukla SA, Ciriello G, Yung WK, Zhang W, Sougnez C, Mikkelsen T, Aldape K, Bigner DD, Van Meir EG, Prados M, Sloan A, Black KL, Eschbacher J, Finocchiaro G, Friedman W, Andrews DW, Guha A, Iacocca M, O’Neill BP, Foltz G, Myers J, Weisenberger DJ, Penny R, Kucherlapati R, Perou CM, Hayes DN, Gibbs R, Marra M, Mills GB, Lander E, Spellman P, Wilson R, Sander C, Weinstein J, Meyerson M, Gabriel S, Laird PW, Haussler D, Getz G, Chin L, Network TR (2013) The somatic genomic landscape of glioblastoma. Cell 155: 462–477 doi: 10.1016/j.cell.2013.09.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cancer Genome Atlas Research N (2008) Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature 455: 1061–1068 doi: 10.1038/nature07385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cornu M, Albert V, Hall MN (2013) mTOR in aging, metabolism, and cancer. Curr Opin Genet Dev 23: 53–62 doi: 10.1016/j.gde.2012.12.005 [DOI] [PubMed] [Google Scholar]
- 6.Saxton RA, Sabatini DM (2017) mTOR Signaling in Growth, Metabolism, and Disease. Cell 169: 361–371 doi: 10.1016/j.cell.2017.03.035 [DOI] [PubMed] [Google Scholar]
- 7.Kim LC, Cook RS, Chen J (2017) mTORC1 and mTORC2 in cancer and the tumor microenvironment. Oncogene 36: 2191–2201 doi: 10.1038/onc.2016.363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cloughesy TF, Yoshimoto K, Nghiemphu P, Brown K, Dang J, Zhu S, Hsueh T, Chen Y, Wang W, Youngkin D, Liau L, Martin N, Becker D, Bergsneider M, Lai A, Green R, Oglesby T, Koleto M, Trent J, Horvath S, Mischel PS, Mellinghoff IK, Sawyers CL (2008) Antitumor activity of rapamycin in a Phase I trial for patients with recurrent PTEN-deficient glioblastoma. PLoS Med 5: e8 doi: 10.1371/journal.pmed.0050008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fan Q, Aksoy O, Wong RA, Ilkhanizadeh S, Novotny CJ, Gustafson WC, Truong AY, Cayanan G, Simonds EF, Haas-Kogan D, Phillips JJ, Nicolaides T, Okaniwa M, Shokat KM, Weiss WA (2017) A Kinase Inhibitor Targeted to mTORC1 Drives Regression in Glioblastoma. Cancer Cell 31: 424–435 doi: 10.1016/j.ccell.2017.01.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Iwanami A, Gini B, Zanca C, Matsutani T, Assuncao A, Nael A, Dang J, Yang H, Zhu S, Kohyama J, Kitabayashi I, Cavenee WK, Cloughesy TF, Furnari FB, Nakamura M, Toyama Y, Okano H, Mischel PS (2013) PML mediates glioblastoma resistance to mammalian target of rapamycin (mTOR)-targeted therapies. Proc Natl Acad Sci U S A 110: 4339–4344 doi: 10.1073/pnas.1217602110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Martin J, Masri J, Cloninger C, Holmes B, Artinian N, Funk A, Ruegg T, Anderson L, Bashir T, Bernath A, Lichtenstein A, Gera J (2011) Phosphomimetic substitution of heterogeneous nuclear ribonucleoprotein A1 at serine 199 abolishes AKT-dependent internal ribosome entry site-transacting factor (ITAF) function via effects on strand annealing and results in mammalian target of rapamycin complex 1 (mTORC1) inhibitor sensitivity. J Biol Chem 286: 16402–16413 doi: 10.1074/jbc.M110.205096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shi Y, Sharma A, Wu H, Lichtenstein A, Gera J (2005) Cyclin D1 and c-myc internal ribosome entry site (IRES)-dependent translation is regulated by AKT activity and enhanced by rapamycin through a p38 MAPK- and ERK-dependent pathway. J Biol Chem 280: 10964–10973 doi: 10.1074/jbc.M407874200 [DOI] [PubMed] [Google Scholar]
- 13.Jo OD, Martin J, Bernath A, Masri J, Lichtenstein A, Gera J (2008) Heterogeneous nuclear ribonucleoprotein A1 regulates cyclin D1 and c-myc internal ribosome entry site function through Akt signaling. J Biol Chem 283: 23274–23287 doi: 10.1074/jbc.M801185200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Holmes B, Lee J, Landon KA, Benavides-Serrato A, Bashir T, Jung ME, Lichtenstein A, Gera J (2016) Mechanistic Target of Rapamycin (mTOR) Inhibition Synergizes with Reduced Internal Ribosome Entry Site (IRES)-mediated Translation of Cyclin D1 and c-MYC mRNAs to Treat Glioblastoma. J Biol Chem 291: 14146–14159 doi: 10.1074/jbc.M116.726927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yan F, Alinari L, Lustberg ME, Martin LK, Cordero-Nieves HM, Banasavadi-Siddegowda Y, Virk S, Barnholtz-Sloan J, Bell EH, Wojton J, Jacob NK, Chakravarti A, Nowicki MO, Wu X, Lapalombella R, Datta J, Yu B, Gordon K, Haseley A, Patton JT, Smith PL, Ryu J, Zhang X, Mo X, Marcucci G, Nuovo G, Kwon CH, Byrd JC, Chiocca EA, Li C, Sif S, Jacob S, Lawler S, Kaur B, Baiocchi RA (2014) Genetic validation of the protein arginine methyltransferase PRMT5 as a candidate therapeutic target in glioblastoma. Cancer Res 74: 1752–1765 doi: 10.1158/0008-5472.CAN-13-0884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Han X, Li R, Zhang W, Yang X, Wheeler CG, Friedman GK, Province P, Ding Q, You Z, Fathallah-Shaykh HM, Gillespie GY, Zhao X, King PH, Nabors LB (2014) Expression of PRMT5 correlates with malignant grade in gliomas and plays a pivotal role in tumor growth in vitro. J Neurooncol 118: 61–72 doi: 10.1007/s11060-014-1419-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shailesh H, Zakaria ZZ, Baiocchi R, Sif S (2018) Protein arginine methyltransferase 5 (PRMT5) dysregulation in cancer. Oncotarget 9: 36705–36718 doi: 10.18632/oncotarget.26404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Banasavadi-Siddegowda YK, Russell L, Frair E, Karkhanis VA, Relation T, Yoo JY, Zhang J, Sif S, Imitola J, Baiocchi R, Kaur B (2017) PRMT5-PTEN molecular pathway regulates senescence and self-renewal of primary glioblastoma neurosphere cells. Oncogene 36: 263–274 doi: 10.1038/onc.2016.199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Banasavadi-Siddegowda YK, Welker AM, An M, Yang X, Zhou W, Shi G, Imitola J, Li C, Hsu S, Wang J, Phelps M, Zhang J, Beattie CE, Baiocchi R, Kaur B (2018) PRMT5 as a druggable target for glioblastoma therapy. Neuro Oncol 20: 753–763 doi: 10.1093/neuonc/nox206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gao G, Dhar S, Bedford MT (2017) PRMT5 regulates IRES-dependent translation via methylation of hnRNP A1. Nucleic Acids Res 45: 4359–4369 doi: 10.1093/nar/gkw1367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mellinghoff IK, Wang MY, Vivanco I, Haas-Kogan DA, Zhu S, Dia EQ, Lu KV, Yoshimoto K, Huang JH, Chute DJ, Riggs BL, Horvath S, Liau LM, Cavenee WK, Rao PN, Beroukhim R, Peck TC, Lee JC, Sellers WR, Stokoe D, Prados M, Cloughesy TF, Sawyers CL, Mischel PS (2005) Molecular determinants of the response of glioblastomas to EGFR kinase inhibitors. N Engl J Med 353: 2012–2024 doi: 10.1056/NEJMoa051918 [DOI] [PubMed] [Google Scholar]
- 22.Stopa N, Krebs JE, Shechter D (2015) The PRMT5 arginine methyltransferase: many roles in development, cancer and beyond. Cell Mol Life Sci 72: 2041–2059 doi: 10.1007/s00018-015-1847-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pal S, Vishwanath SN, Erdjument-Bromage H, Tempst P, Sif S (2004) Human SWI/SNF-associated PRMT5 methylates histone H3 arginine 8 and negatively regulates expression of ST7 and NM23 tumor suppressor genes. Mol Cell Biol 24: 9630–9645 doi: 10.1128/MCB.24.21.9630-9645.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pollack BP, Kotenko SV, He W, Izotova LS, Barnoski BL, Pestka S (1999) The human homologue of the yeast proteins Skb1 and Hsl7p interacts with Jak kinases and contains protein methyltransferase activity. J Biol Chem 274: 31531–31542 [DOI] [PubMed] [Google Scholar]
- 25.Chen H, Lorton B, Gupta V, Shechter D (2017) A TGFbeta-PRMT5-MEP50 axis regulates cancer cell invasion through histone H3 and H4 arginine methylation coupled transcriptional activation and repression. Oncogene 36: 373–386 doi: 10.1038/onc.2016.205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tarighat SS, Santhanam R, Frankhouser D, Radomska HS, Lai H, Anghelina M, Wang H, Huang X, Alinari L, Walker A, Caligiuri MA, Croce CM, Li L, Garzon R, Li C, Baiocchi RA, Marcucci G (2016) The dual epigenetic role of PRMT5 in acute myeloid leukemia: gene activation and repression via histone arginine methylation. Leukemia 30: 789–799 doi: 10.1038/leu.2015.308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rajpurohit R, Paik WK, Kim S (1992) Enzymatic methylation of heterogeneous nuclear ribonucleoprotein in isolated liver nuclei. Biochim Biophys Acta 1122: 183–188 [DOI] [PubMed] [Google Scholar]
- 28.Dreyfuss G, Kim VN, Kataoka N (2002) Messenger-RNA-binding proteins and the messages they carry. Nat Rev Mol Cell Biol 3: 195–205 doi: 10.1038/nrm760 [DOI] [PubMed] [Google Scholar]
- 29.Jean-Philippe J, Paz S, Caputi M (2013) hnRNP A1: the Swiss army knife of gene expression. Int J Mol Sci 14: 18999–19024 doi: 10.3390/ijms140918999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Idriss H, Kumar A, Casas-Finet JR, Guo H, Damuni Z, Wilson SH (1994) Regulation of in vitro nucleic acid strand annealing activity of heterogeneous nuclear ribonucleoprotein protein A1 by reversible phosphorylation. Biochemistry 33: 11382–11390 [DOI] [PubMed] [Google Scholar]
- 31.Kumar A, Wilson SH (1990) Studies of the strand-annealing activity of mammalian hnRNP complex protein A1. Biochemistry 29: 10717–10722 [DOI] [PubMed] [Google Scholar]
- 32.Mitchell SA, Spriggs KA, Coldwell MJ, Jackson RJ, Willis AE (2003) The Apaf-1 internal ribosome entry segment attains the correct structural conformation for function via interactions with PTB and unr. Mol Cell 11: 757–771 [DOI] [PubMed] [Google Scholar]
- 33.Pickering BM, Mitchell SA, Spriggs KA, Stoneley M, Willis AE (2004) Bag-1 internal ribosome entry segment activity is promoted by structural changes mediated by poly(rC) binding protein 1 and recruitment of polypyrimidine tract binding protein 1. Mol Cell Biol 24: 5595–5605 doi: 10.1128/MCB.24.12.5595-5605.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu F, Mischel PS (2018) Targeting epidermal growth factor receptor co-dependent signaling pathways in glioblastoma. Wiley Interdiscip Rev Syst Biol Med 10 doi: 10.1002/wsbm.1398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang MY, Lu KV, Zhu S, Dia EQ, Vivanco I, Shackleford GM, Cavenee WK, Mellinghoff IK, Cloughesy TF, Sawyers CL, Mischel PS (2006) Mammalian target of rapamycin inhibition promotes response to epidermal growth factor receptor kinase inhibitors in PTEN-deficient and PTEN-intact glioblastoma cells. Cancer Res 66: 7864–7869 doi: 10.1158/0008-5472.CAN-04-4392 [DOI] [PubMed] [Google Scholar]
- 36.Braun CJ, Stanciu M, Boutz PL, Patterson JC, Calligaris D, Higuchi F, Neupane R, Fenoglio S, Cahill DP, Wakimoto H, Agar NYR, Yaffe MB, Sharp PA, Hemann MT, Lees JA (2017) Coordinated Splicing of Regulatory Detained Introns within Oncogenic Transcripts Creates an Exploitable Vulnerability in Malignant Glioma. Cancer Cell 32: 411–426 e411 doi: 10.1016/j.ccell.2017.08.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kahn J, Hayman TJ, Jamal M, Rath BH, Kramp T, Camphausen K, Tofilon PJ (2014) The mTORC1/mTORC2 inhibitor AZD2014 enhances the radiosensitivity of glioblastoma stem-like cells. Neuro Oncol 16: 29–37 doi: 10.1093/neuonc/not139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Aggarwal P, Vaites LP, Kim JK, Mellert H, Gurung B, Nakagawa H, Herlyn M, Hua X, Rustgi AK, McMahon SB, Diehl JA (2010) Nuclear cyclin D1/CDK4 kinase regulates CUL4 expression and triggers neoplastic growth via activation of the PRMT5 methyltransferase. Cancer Cell 18: 329–340 doi: 10.1016/j.ccr.2010.08.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.