Abstract
Over the past 20 years, the zebrafish has emerged as a powerful model organism for studying cardiac development. Its ability to survive without an active circulation and amenability to forward genetics has led to the identification of numerous mutants whose study has helped elucidate new mechanisms in cardiac development. Furthermore, its transparent, externally developing embryos have allowed detailed cellular analyses of heart development. In this review, we discuss the molecular and cellular processes involved in zebrafish heart development from progenitor specification to development of the valve and the conduction system. We focus on imaging studies that have uncovered the cellular bases of heart development and on zebrafish mutants with cardiac abnormalities whose study has revealed novel molecular pathways in cardiac cell specification and tissue morphogenesis.
Keywords: zebrafish, heart, genetic screen, mutant, imaging
INTRODUCTION
During development, the embryonic heart undergoes a series of complex morphogenetic and differentiation processes to form the mature cardiac structures (7, 10, 23, 111). Problems with these processes can lead to congenital heart defects. Understanding the mechanisms that form the heart is a difficult challenge, as early heart defects cause lethality in most vertebrates. One notable exception is the zebrafish, which develops a functional heart within 24 hours post-fertilization (hpf) but can survive through the first week of development without a functioning cardiovascular system. Taking advantage of this property, along with the external development of the zebrafish embryo, relatively fast generation time, and high number of offspring, a number of forward genetic screens have been performed. These screens identified dozens of mutants with defects in cardiac structure or function (2, 12, 18, 22, 115). Complementary to forward genetic approaches, a number of tools and techniques allow disruption of specific genes of interest. Morpholino antisense oligonucleotides allow for rapid disruption of genes of interest (27) but often cause nonspecific effects, particularly in the cardiovascular system. Emerging technologies such as zinc finger nucleases (28), TAL effector nucleases (TALENs) (45, 98), and TILLING (74) enable generation or identification of mutations in genes of interest and do not suffer from the same off-target effects as morpholinos do. Recently, powerful transgenic tools have allowed spatial and temporal manipulation of gene expression, and advances in microscopy have utilized the transparency of zebrafish embryos and larvae to monitor complex developmental processes in vivo (31, 76, 99, 102). Furthermore, many small molecules enter zebrafish after addition to the water, which, along with the ability to raise embryos and larvae in multi-well plates, allows for rapid in vivo screening of chemical libraries (8). Automated screening technologies continue to improve, and platforms capable of performing high-throughput confocal imaging with cellular resolution have been recently described (81).
Remarkably, the zebrafish heart also shows high regenerative capacity and can reform a fully functional heart even after substantial damage (16, 36, 48, 55, 86, 87, 100, 129). Because the regenerative process seems to reactivate developmental pathways (55), understanding zebrafish heart development also helps us understand the unusual cardiac regenerative potential of this model organism.
Despite being a two-chambered organ, the zebrafish heart exhibits many similarities to the amniote heart, and cellular and molecular studies clearly illustrate the common evolutionary origin of these structures. For this reason, many scientists who have traditionally used the mouse or chick to study heart development have now fully embraced the zebrafish model system. Here, we present an overview of the processes involved in cell differentiation, morphogenesis, and physiology during zebrafish heart development, with particular emphasis on findings made possible by the unique properties of this powerful model system.
EARLY CARDIAC PROGENITORS
Cardiac progenitors can first be tagged at the blastula stage. Fate mapping places myocardial progenitors close to the embryonic margin in lateral fields on both side of the embryo (53, 113). Even at this early stage, atrial and ventricular progenitors are segregated, with ventricular progenitors occupying a position closer to the margin and more dorsal than their atrial counterparts (53). Meanwhile, endocardial progenitors occupy a similar area of the margin (63), but do not appear to be organized by chamber (53). Myocardial progenitors retain their relative positions through gastrulation, and by the 6–9 somite stages are found within the posterior half of the anterior lateral plate mesoderm (ALPM) (Figure 1a). This territory expresses the transcription factor genes hand2 and gata4 (101, 134). Ventricular progenitors are enriched in the medial aspect of this region, which expresses the transcription factor gene nkx2.5, whereas atrial progenitors are enriched laterally. Progenitors are commonly visualized by the expression of nkx2.5 and the cardiac regulatory myosin light chain gene myl7 (previously cmlc2) (35). Ventricular progenitors remain medial to the atrial progenitors, and these two populations can be distinguished by their expression of ventricular myosin heavy chain (vmhc) and atrial myosin heavy chain (amhc/myh6), respectively (133).
Figure 1.
Overview of zebrafish heart development. (a–c) Heart development before looping. Embryos are shown in a dorsal view. (a) At 12 hours post-fertilization (hpf) (15 somites), myocardial progenitors are found in the anterior lateral plate mesoderm (ALPM), with ventricular progenitors more medial than atrial progenitors. Endocardial progenitors lie anterior in the ALPM. (b) Myocardial and endocardial progenitors migrate to the midline and fuse by 19 hpf (20 somites) to create the cardiac cone. (c) Through a process of involution, the myocardium rearranges to surround the endocardium and form the linear heart tube by 24 hpf. (d–e) Later heart development. Heart sections are shown from a ventral view. (d ) By 48 hpf, the heart has looped to form a right-sided ventricle and left-sided atrium. Distinct outer and inner curvatures can be distinguished, and the outer curvature (OC) of the ventricle has started to balloon. The atrioventricular (AV) myocardium has functionally differentiated to introduce a conduction delay, and pacemaker activity has become restricted to the sinoatrial (SA) ring. The proepicardial organ has coalesced near the AV canal. (e) By 120 hpf, the OC of the ventricle has more fully ballooned, and trabecular ridges have started to appear. The AV valve leaflets are well formed and prevent retrograde flow. The epicardium has expanded over the entire exterior surface of the myocardium. The SA nodal tissue is most often restricted to a few cells on the right side of the atrium several cell diameters away from the SA ring.
Several signaling pathways have been shown to regulate the number of myocardial progenitors, including retinoic acid (RA) (52, 112, 130), Wnt (124), Hedgehog (Hh) (127), fibroblast growth factor (Fgf) (68), bone morphogenic protein (Bmp) (69, 89), and Nodal (53, 89). Although Hh signaling seems to be important for establishing progenitors for both chambers (118), Bmp inhibition greatly decreases atrial progenitor specification (69). In contrast, decreasing Nodal or Fgf signaling suppresses both atrial and ventricular specification but with a greater effect on ventricular progenitors (53, 68, 89). Meanwhile, although inhibition of RA signaling increases both atrial and ventricular progenitor populations to the same degree, it appears to accomplish this effect through different mechanisms (130). Whereas ventricular populations increase because of an increased number of ventricular progenitors within the blastula, atrial populations seem to increase because of an increase in the number of progeny per early progenitor. Other studies have revealed interesting temporal roles for these signaling pathways. For example, activation of Wnt signaling at pregastrulation stages increases progenitor numbers, whereas activation an hour after the onset of gastrulation significantly decreases their numbers, possibly through induction of programmed cell death in cardiac progenitors (26, 124). Though the heat-shock inducible transgenic animals used in these studies allowed for temporal analysis of Wnt signaling in cardiac specification, genetic loss-of-function studies are needed to dissect the role of this pathway in this process. Looking forward, although many pathways have been identified to modulate cardiac progenitor specification, how these different pathways interact to produce the correct number and distribution of progenitors remains an open question, as does the elucidation of the gene regulatory networks downstream of these signaling pathways. Importantly, it is very likely that the role of these pathways and their effectors in cardiac cell specification is conserved throughout evolution, and thus that the zebrafish model will continue to contribute significantly to these studies.
Once progenitors are specified, they migrate to the correct location within the ALPM, where they further differentiate. Two studies have implicated G-protein coupled receptors (GPCRs) in this migration (103, 136). The grinch mutant has greatly diminished heart fields and, occasionally, completely lacks a heart (103). This mutation affects agtrl1b, which encodes a GPCR for the signaling peptide Apelin. Cell transplantation data using morpholino knockdown of agtr1b suggested that the receptor works cell-autonomously to limit both cardiac potential and cell movement during gastrulation. Surprisingly, transgenic overexpression of apelin before gastrulation, but not during, also inhibits heart formation. Hence, these data support a model where localized Apelin/Agtr1b signaling directs the movement of myocardial progenitors to the correct location within the ALPM, where they can receive myocardial differentiation signals.
Once in the ALPM, interactions with other tissues influence cardiac cell differentiation. Within the ALPM, vascular and hematopoietic precursors neighbor the heart field anteriorly and pectoral fin precursors posteriorly. The absence of vascular and hematopoietic precursors in the cloche (clo) mutant (114) likely leads to the observed expansion of hand2 expression and ectopic cardiomyocyte differentiation in the anterior ALPM, which causes an increase in atrial cell number (101). Conversely, expansion of vascular and hematopoietic progenitors by injection of scl and etsrp RNA reduces the size of the myocardial domain. Interestingly, knocking down etsrp in an etsrp:GFP bacterial artificial chromosome (BAC) transgenic line in which the vascular and hematopoietic progenitors are marked revealed that etsrp:GFP positive cells became cardiomyocytes, indicating that Etsrp functions in vascular progenitors at least in part to prevent cardiomyocyte differentiation (79). On the posterior side, RA signaling is required cell-autonomously in the forelimb field for its specification (130). RA-signaling inhibition leads to a posterior expansion of Fgf signaling, which represses forelimb specification (110). Suppression of specification or differentiation of the forelimb field allows the heart field to expand posteriorly. The RA downstream target gene hoxb5b is expressed in the forelimb field territory, and upon morpholino knockdown of this gene, atrial progenitors expand into the forelimb field, indicating that hoxb5b regulates an unknown signal that limits atrial progenitor expansion (130).
A network of transcription factors controls cardiomyocyte differentiation. As in amniotes, a combination of the GATA transcription factors Gata4, Gata5, and Gata6 regulates heart formation (40, 84, 88, 89, 122). faust mutants, which carry a mutation in gata5, show a variable decrease in cardiac progenitors, and gata5 overexpression is sufficient to produce ectopic beating tissue (88, 89). The variability of the faust cardiac phenotype may suggest that other GATA factors are compensating for the loss of Gata5 function. In support of this hypothesis, combinatorial morpholino-mediated knockdown of gata5 and gata6 results in a complete absence of nkx2.5 positive cardiac progenitors (40, 84). Analysis of null alleles of these genes is needed to confirm this result. Recent work suggests that GATA factors work with the Brg1 associated factor (BAF) chromatin remodeling complex to promote cardiac specification (65, 116), and overexpression of gata5 and the BAF component smarcd3b dramatically increases cardiac progenitor specification cell-autonomously (65). Intriguingly, these two factors also cell-autonomously drive overexpressing cells to migrate to the cardiogenic regions of the ALPM, even from neurogenic regions, and dramatically increase cardiac incorporation in transplant experiments. This finding opens up a number of interesting questions about the identity and mode of action of the downstream factors that drive this induced migratory behavior and what they can tell us about heart development.
Another major player in cardiac specification is the transcription factor Hand2. It is expressed in all myocardial progenitors, more highly in ventricular progenitors, and functions to increase their number (101, 134). Hand2 is also necessary for maintenance of the expression of tbx5, a T-box transcription factor gene mutated in Holt-Oram syndrome and necessary for heart and pectoral fin development in fish (33, 134). nkx2.5 expression is not abolished in hand2 mutants, but rather a population of nkx2.5-positive, myl7-negative cells can be found adjacent to the population of nkx2.5-positive, myl7-positive cells, suggesting that hand2 is also necessary for the differentiation of nkx2.5-expressing cells (101). Morpholino knockdown experiments suggest that Nkx2.5 itself is not critical for progenitor specification within the ALPM but that instead it plays a redundant role with the closely related transcription factor Nkx2.7 to promote heart tube extension (117, 123). Nkx2.5 and Nkx2.7 seem to regulate the size of the two chambers, as combinatorial knockdown of both genes increases the number of atrial cardiomyocytes during heart tube stages while diminishing the number of ventricular cells produced by 52 hpf. Genetic analysis of these and other transcription factors as well as their downstream targets will be necessary to understand how the underlying gene regulatory network drives cardiomyocyte specification and differentiation.
MIDLINE MIGRATION
As they differentiate, the bilateral populations of myocardial precursors, sandwiched between the overlying anterior endoderm and underlying yolk syncytial layer (YSL), migrate toward the embryonic midline. The populations make contact with each other by 18 hpf, first posteriorly, then anteriorly. They then merge, forming a shallow cone with ventricular precursors at its dorsal apex (Figure 1b) (113). A number of mutations have been identified that perturb this process, leading to cardia bifida, where two hearts form in lateral positions. Mutants with defects in endoderm formation display cardia bifida, revealing that the endoderm plays a critical role in cardiac migration (1, 35, 56, 88). Although this role for the endoderm was shown more than a decade ago, the underlying mechanisms remain unknown. Another series of mutants have revealed a role for sphingolipid signaling in midline migration. The miles apart (mil ) mutant displays cardia bifida and is caused by mutations in the sphingosine-1-phosphate (S1P) receptor gene s1p2 (60). mil/s1p2 appears to act cell-nonautonomously in midline migration of the cardiac precursors and probably acts within the endoderm, as complete replacement of the anterior endoderm of mil mutants with wild-type endoderm can rescue the phenotype (78). Meanwhile, mutations in spinster homolog 2 (spns2), a gene of previously unknown function, cause phenotypes similar to those in mil/s1p2 mutants (51, 78). Biochemical studies revealed that Spns2 can function as an S1P transporter (51). Targeted injection of spns2 morpholinos into the YSL causes cardia bifida (78), and injection of wild-type spns2 RNA within the YSL rescues the mutant phenotype, suggesting that spns2 functions in the YSL and most likely provides a source of S1P during midline migration (51). Despite these advances, the precise role of S1P in midline migration remains elusive.
One way the endoderm and YSL may be regulating midline migration is through the extracellular matrix (ECM). Fibronectin, an important ECM protein also required for mouse heart development (34), surrounds myocardial precursors during midline migration, and the natter (nat) mutation in fibronectin causes cardia bifida (121). Recently, both the YSL and the myocardial precursors themselves have been implicated in regulating fibronectin deposition. Disruption of the transcription factor Mtx1 specifically in the YSL using targeted morpholino injections leads to cardia bifida accompanied by a profound reduction in fibronectin expression (5, 97). Additionally, disruption of the proteoglycan gene syndecan 2 (sdc2) in the YSL causes cardia bifida associated with normal expression of fibronectin, but undetectable fibronectin fibrillogenesis (5). Interestingly, this phenotype can be rescued by injection of sdc2 RNA into the YSL but not into the whole embryo, suggesting that sdc2 in the YSL is necessary to provide a substrate for fibronectin fibrillogenesis in the embryo. Finally, the myocardial precursors themselves were recently found to regulate fibronectin deposition. hand2 mutants have reduced numbers of myocardial precursors and a defect in midline migration (134). Transplant experiments show that the defect in precursor production is cell-autonomous, but that the defect in midline migration is cell nonautonomous (32). Interestingly, hand2 mutants show increased levels of fibronectin, and the hand2 midline migration defect can be ameliorated by heterozygosity of the nat/fn1tl43c allele. These data suggest that expression of hand2 within cells of the ALPM, possibly within the myocardial precursors themselves, limits excessive fibronectin deposition that itself can prevent midline migration. These studies suggest that a precise level of ECM components is necessary for midline migration but do not reveal precisely how fibronectin influences cell behavior, a fundamental question zebrafish studies should help resolve.
Close examination of fibronectin mutants suggest that ECM components may be acting through regulation of myocardial epithelial organization. During their migration to the midline, myocardial precursors maintain a polarized epithelial organization (121). fibronectin mutants exhibit defects in the organization and polarization of this epithelium, with some cells even losing coherence with the rest of the monolayer and migrating separately. Although this phenotype suggests that epithelial organization is necessary for midline migration, cardiomyocytes in embryos homozygous for mutations in genes that regulate epithelial polarity eventually do migrate to the midline (42, 47, 75, 77, 85, 131). The possibility remains that maternal contribution of these epithelial polarity proteins is sufficient to support midline migration. The analysis of maternal zygotic (MZ) epithelial polarity mutants would be necessary to resolve whether maternal polarity factors are required for midline migration, as one might expect.
ENDOCARDIAL MIGRATION
In order to form a functional heart, the endocardium must also incorporate into the developing organ. While the myocardium is migrating towards the midline, endocardial precursors, located more anteriorly in the ALPM (Figure 1a) (101), stream medially and posteriorly, where they proceed to coat the interior of the forming heart cone (Figure 1b) (15). This process requires the basic helix-loop-helix (bHLH) transcription factor Scl/Tal1. A truncating mutation removing the bHLH domain leads to the defective posterior migration of the endocardial precursors, which in turn delays the anterior merging of the bilateral myocardial populations, resulting in a delay in cardiac cone formation and the clustering of endocardial cells at the arterial pole, leading to a nonfunctional heart. Recent work has also shown that endocardial migration requires Vegf signaling, which is regulated in a complex, dose-dependent manner by Slit2 signaling through the Robo1 receptor (29). The level of Slit2/Robo1 signaling is in turn tuned by the microRNA mir-218, which lies within an intron of slit2 and targets Robo receptors. Much work remains to be done to determine how these and other as yet unidentified factors direct endocardial midline migration as well as to understand how this process occurs in amniotes.
Recent data suggest that myocardial factors also play a role in endocardial midline migration. MZ mutants of tmem2, which encodes a transmembrane protein of unknown function, exhibit midline migration defects of both the endocardium and myocardium (120). Interestingly, transgenic reexpression of tmem2 in the myocardium of these animals rescues both myocardial and endocardial midline migration, whereas reexpression in the endocardium alone rescues neither, illustrating how myocardial factors are necessary for endocardial midline migration.
Reciprocally, the endocardium may also play a role in fine-tuning the migration of myocardial precursors. Myocardial precursor movements during midline migration can be separated into two types: a linear midline migration and a curved movement that closes the cardiac cone (41). In the absence of endocardium in clo mutants, this second movement does not occur, leading to a dysmorphic cardiac cone. Interestingly, in mil/s1p2 cardia bifida mutants, this second curving movement still occurs in the absence of midline migration. Removing the endocardium from this background by creating mil;clo double mutants abrogates this curving movement, further suggesting that the endocardium regulates this process. The signals that direct this curving movement remain to be elucidated. In addition, the isolation and further analysis of the clo gene will be necessary to confirm this model. Myocardial-endocardial interactions are a constant theme during heart formation and function and will remain a rich avenue of investigation.
HEART-TUBE ASSEMBLY
After endocardial and myocardial precursors come together to form the cardiac cone, they migrate asymmetrically to form the linear heart tube (Figure 1c). Although cardiac precursors migrate to the midline in epithelial polarity mutants, has/prkci mutant embryos fail to assemble a linear heart tube (42, 85), and detailed analysis of embryos injected with morpholinos targeting either has/prkci or nok/mpp5a showed that knockdown of these genes causes defects in the formation and tilting of the cardiac cone and heart-tube elongation (91). Time-lapse imaging has shown that myocardial cells undergo an asymmetric involution to transition from a cone to a linear tube, whereby cells on the right side of the cone move ventrally and medially (92, 107). The direction of this involution is controlled by global left/right signaling pathways. In order to accomplish this rearrangement, the cardiomyocytes must migrate as a cohesive epithelial sheet, and disruption of this epithelium in has and nok morphants leads to uncoordinated, nondirected migration and failure to create the heart tube (92). This migration also requires the action of hyaluronan synthase, which is expressed in the migrating myocardium and produces the ECM molecule hyaluronic acid (107). Thus, epithelial coherence and polarity, as well as the ECM, are necessary to create the linear heart tube, a complex process which undoubtedly requires many more regulators.
LOOPING AND BALLOONING
After its initial formation at 24 hpf, the heart tube gradually extends and undergoes a series of morphogenetic changes leading to the emergence of an anterior, right-sided ventricle and a more posterior, left-sided atrium separated by a constricted region called the atrioventricular canal (AVC). In order to form this asymmetrically positioned heart, the cardiomyocytes undergo a number of asymmetric movements, eventually culminating in the rightward looping of the heart tube. Possible cellular mechanisms underlying these asymmetric movements have been reviewed elsewhere (9, 10). As the heart loops, the chambers form as bulges in the heart tube in a process called ballooning. This bulging creates two distinct surfaces within the chambers: the concave inner curvature (IC) and the convex outer curvature (OC) (Figure 1d). In the ventricle, this morphogenetic change is accomplished by differential shape changes within the IC and the OC (6, 25). Measurements of individual cell shapes reveal that, although the surface area of both IC and OC cells increase relative to cells in the linear heart tube, OC cells, but not IC cells, increase in length, measured by a decrease in circularity. Failure of these cells to elongate leads to a smaller, rounder ventricle.
These shape changes seem to depend on cardiac function. In the weak atrium (wea/myh6) mutant, which lacks atrial contraction because of defects in atrial sarcomere formation, ventricular cells fail to increase their surface area and fail to elongate (6). Transplantation of wea mutant cells into wild-type embryos revealed that this effect is cell nonautonomous, as wea mutant ventricular cardiomyocytes within otherwise wild-type hearts elongate normally. In contrast, half-hearted (haf/vmhc) mutants, which fail to form sarcomeres in the ventricle, develop enlarged ventricles, with individual cells increasing their surface area and elongating more than in wild type. This effect is eliminated in wea;haf double mutants, as well as in silent heart (sih) mutants, which lack sarcomeres in both chambers because of a mutation in cardiac troponin T2a (tnnt2a) (6, 106). Taken together, these data suggest that sensing fluid flow and/or myocardial stretch increases myocardial surface area and causes OC-cell elongation, and ventricular contractility modulates the extent of these shape changes.
In addition to contraction forces, electrical conduction also appears to be required for OC morphogenesis. The dococ (dco) mutant exhibits uncoordinated conduction and contraction within the ventricle, caused by a mutation in the connexin gene gap junction protein alpha 3 (gja3) (20). This mutation also causes defects in heart morphogenesis, namely a lack of constriction of the AVC and a smaller, rounder ventricle. These defects are not secondary to disrupted flow, as sih;dco double mutant OC cells are significantly less elongated than sih mutant OC cells even though the beat is completely disrupted in both settings. A similar flow-independent defect in elongation was also observed in sih;dco double mutant AV myocardial cells. This study provides the first example of conduction affecting cardiomyocyte morphogenesis independent of contractility. Although still in its early stages, zebrafish research on structure/function relationships in the ballooning myocardium offers a powerful and highly relevant in vivo view of how forces and electrical signals shape cardiac morphogenesis.
PROLIFERATION AND GROWTH
In addition to undergoing morphogenetic movements, the developing heart must also increase its myocardial mass. This increase can happen through addition of differentiating cells from outside the forming heart or by proliferation of existing cardiomyocytes. During heart-tube stages, the main mode of cardiac growth is through differentiation rather than proliferation. Populations of cells at the arterial and venous poles contribute to myocardial growth by late addition of cells at both ends (24, 37, 62, 139). The population contributing to the arterial pole shares many characteristics with the secondary heart field (SHF) seen in amniotes. These cells are marked by the expression of latent TGF-β binding protein 3 (ltbp3), and contribute to myocardial and endothelial cells as well as smooth muscle cells of the outflow tract. The process of cardiac expansion by these cells requires Fgf (24), TGF-β (139), Bmp, and Hh signaling (37). Interestingly, although islet-1, a transcription factor gene important in the SHF in mouse, can be detected in some cells that contribute to the arterial pole up through 48 hpf, its mutation does not disrupt late addition to the arterial pole but does disrupt late addition to the venous pole. SHF development is a very active area of investigation in amniotes, and it will be interesting to see whether zebrafish studies will contribute novel concepts to this field.
After 48 hpf, cardiomyocyte proliferation progressively increases (64). Mutant studies have revealed some of the molecular determinants of cardiomyocyte proliferation in zebrafish. A mutation in a gene encoding a subunit of the cardiac L-type calcium channel results in a silent ventricle with greatly reduced numbers of cardiomyocytes at 72 hpf despite normal expression patterns of myofibrillar components and cardiac transcription factor genes up to 48 hpf (93). This effect implicates calcium signaling and/or cardiac function in the control of late increases in cardiomyocyte number. Embryos harboring a mutation in erbb2, which encodes a tyrosine kinase receptor of the epidermal growth factor receptor (EGFR) family, exhibit a complete lack of myocardial proliferation between 3–5 days post-fertilization (dpf) even though heart function appears unaffected (64). In contrast, an activating mutation in the gene encoding the DNA-stimulated ATPase Reptin in the liebeskummer (lik) mutant results in a dramatic cell-autonomous increase in ventricular cardiomyocyte proliferation between 48 and 72 hpf, and this increase can be pheno-copied by morpholino-mediated knockdown of Pontin, another DNA-stimulated ATPase that is known to be a Reptin antagonist in other contexts (95). The ability to manipulate and visualize single cardiomyocytes should make the zebrafish particularly well suited to gaining a better understanding of the mechanisms that control cardiomyocyte proliferation in vivo.
A complex of endocardial proteins plays a role in regulating the overall size of the heart, seemingly independent of proliferation (66, 67). The zebrafish mutants heart of glass (heg), santa (san), and valentine (vtn) share a similar phenotype, namely a lack of circulation at 28–30 hpf and an enlarged, thin-walled heart by 48 hpf. san and vtn encode Krit (Ccm1) and Ccm2, respectively, proteins previously associated with familial cerebral cavernous malformations (CCMs) in humans (61), and heg encodes a previously unidentified large transmembrane protein expressed in the endocardium (67). Combinatorial injection of low doses of morpholinos targeting san, vtn, and heg recapitulate the enlarged heart phenotype, suggesting that heg acts within the same pathway as san and vtn (66), and recently Ccm1, Ccm2, and Heg were shown to form a complex in vitro, further suggesting that Heg serves as a novel member of the Ccm pathway (57). A third member of this complex, Ccm3, has been reported to cause a similar cardiac phenotype as in san, val, and heg mutants (127, 138), but more recent work has challenged those findings, indicating that morpholinos targeting Ccm3 disrupt cardiac morphology but do not result in grossly dilated hearts (135). Genetic loss-of-function studies are needed to resolve this discrepancy. Importantly, comparison of sih single mutants to heg;sih double mutants reveals that heg;sih hearts are enlarged relative to sih hearts, indicating that the enlarged heart phenotype is not secondary to the lack of flow or dependent on contractility, at least in heg mutants. The number of endocardial and myocardial cells appeared to be unaffected in heg mutants at 72 hpf and in 48 hpf embryos injected with morpholinos targeting san/krit1 or vtn/ccm2. Thus, some other mechanism must explain the enlarged hearts. It was initially suggested that wild-type hearts underwent concentric growth to create a multilayered myocardium, and that these mutants arranged the same number of cells in a monolayer, resulting in an enlarged heart. In support of this proposal, fixed sections through wild-type hearts at 72 hpf appeared to have a multilayered myocardium, and mutant hearts appeared to remain single layered. However, embryonic heart morphology is not well maintained in fixed tissues, and recent confocal studies on live embryos have revealed that the wild-type myocardium is single layered until the start of trabeculation around 60 hpf, well after the large-heart phenotype emerges (4, 6, 83). Many questions remain regarding the etiology of CCMs, and continued studies in zebrafish should help address them.
FORMATION OF INTERNAL CARDIAC STRUCTURES
In order to function properly, the heart must form a number of internal structures. In the next section, we focus on two of these structures: the AV valve and the electrical conduction system. For the purposes of this review, two other important structures, the epicardium and coronary vessels, are not discussed, as they have been recently reviewed elsewhere (9).
Atrioventricular Canal and Valve Development
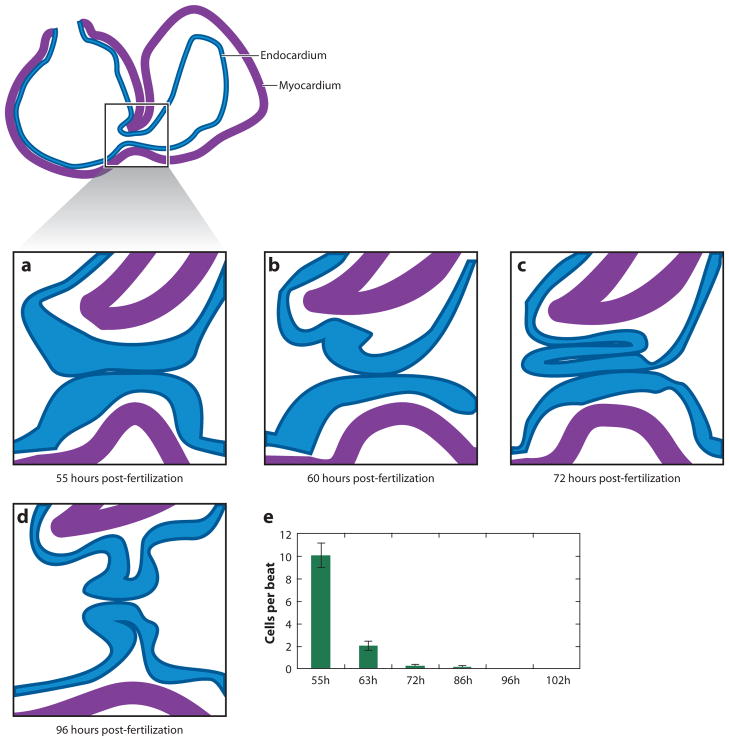
The valve structures in the AVC are crucial for maintaining unidirectional blood flow in the heart. The first sign of AVC differentiation occurs at 37 hpf, when a constriction between the forming atrium and ventricle forms, and bmp4 and versican, previously expressed in the whole heart tube, become restricted to the forming AVC myocardium (12, 128). Similarly, notch1b, expressed in the entire ventricular endocardium at 36 hpf, becomes restricted to the AVC endocardium by 45 hpf. By 48 hpf the endocardial ring, a noticeable thickening of the AVC endocardium, has formed and the AVC prevents retrograde blood flow to some extent. The AVC becomes progressively more efficient at blocking retrograde flow over developmental time, completely blocking retrograde flow by 72 hpf (99). Analysis of valve formation has been complicated by dramatic changes to the morphology of the valve upon fixation. Care must be taken in analyzing cellular architecture in fixed preparations. On a cellular level, the first noticeable event is the emergence at the AV boundary of cuboidal endocardial cells that express the cell adhesion molecule Alcama. By 55 hpf, a single layer of polarized cuboidal endocardial cells that stain strongly for Alcama has emerged at both the superior and inferior portions of the AVC. By 60 hpf, endocardial cells in the superior AVC send projections into the ECM between the AVC endocardium and myocardium. Cells in the inferior aspect do the same at 80 hpf. By 76 hpf, live imaging reveals a primitive valve leaflet two cells thick on the superior aspect of the AVC, which becomes readily apparent by 85 hpf (99). The inferior valve leaflet becomes apparent by 102 hpf. The cells in the two layers of the valve leaflets assume different morphologies, with those in the layer closest to the AVC remaining cuboidal and those in the layer closest to the myocardium assuming a rounded shape. Importantly, live imaging suggests that, unlike in amniotes, this leaflet appears to form without a complete endothelial-mesenchymal transition (EMT), but rather through a direct invagination of AVC endocardial cells (Figure 2). By 7 dpf, the two leaflets are clearly formed and are connected to the myocardial trabeculae.
Figure 2.
Zebrafish valve development. (a) By 55 hpf, the atrioventricular (AV) endocardium forms a thick monolayer. (b) By 60 hpf, the endocardial layer shows signs of involution. (c) By 72 hpf, a superior leaflet has formed through the involution of a few distinct AV endocardial cells. (d ) By 96 hpf, both superior and inferior leaflets have formed. (e) Retrograde flow of blood cells observed over developmental time. By 72 hpf, retrograde flow has essentially ceased. Modified from 107.
A number of signaling pathways have been implicated in AVC development in zebrafish, many of which show parallel functions in amniotes. Detailed analyses have revealed the function of many of these pathways both at a cellular and molecular level. Notch signaling, known to be important for promoting EMT in mouse endocardium (119), seems to play multiple roles in zebrafish AVC development. First, inhibition of Notch signaling starting at 24 hpf results in the appearance at 60 hpf of ectopic cuboidal, Alcama-positive endocardial cells in the ventricle, and constitutive activation of Notch signaling in the endocardium eliminates the differentiation of cuboidal endocardial cells in the valve, suggesting that Notch activity restricts AVC differentiation in the ventricle (12). Second, inhibition of Notch signaling starting at 36 hpf results in a disorganized AVC endocardium with reduced Alcama expression, suggesting a later role in maintaining AVC identity. Calcineurin signaling has also been implicated in two stages in mouse AVC development, first for endocardial EMT and then for remodeling (17). In fish, inhibition of calcineurin from the one-cell stage blocks AVC differentiation, and inhibition starting at 48 hpf causes a disorganized AVC endocardium (12). Combining inhibitor treatments with live imaging approaches allows for the identification of how different signaling pathways contribute to valve function. Inhibition of ErbB/Neuregulin, TGF-β, or prostaglandin signaling results in different degrees of retrograde flow at 72 hpf, occurring through three different mechanisms (99). ErbB inhibition interferes with valve geometry, leading to an open valve during atrial relaxation, TGF-β inhibition creates incompressible valve cells that do not seal the AVC, and inhibition of prostaglandin signaling changes myocardial structures, shifting the superior valve surface and preventing it from maintaining a seal. These functional readouts provide a new level of detail on how different signaling pathways contribute to valve formation.
Several mutants have revealed other signaling pathways involved in valvulogenesis. In wild-type animals, Wnt activity can be strongly detected in the developing valve at 72 hpf (46). This activation is associated with proliferation of the valvular endocardium. Constitutive activation of the Wnt pathway by mutations in apc leads to increased proliferation throughout the endocardium and diffuse expression of AVC markers, such as notch1b, bmp4, versican, and has2. Interestingly, global inhibition of Wnt signaling by dkk1 RNA injection can also cause a failure of AVC differentiation with regurgitation of blood from ventricle to atrium, suggesting that precise spatiotemporal control of Wnt signaling is necessary for valve formation. bungee mutants fail to form a valve and show a redistribution of notch1b expression throughout the ventricular endocardium as well as a lack of spp1 staining, which normally marks endocardial cells undergoing EMT (49). The mutation was mapped to the protein kinase D2 ( pkd2) locus and inhibits kinase activity. Unlike the wild-type version, this mutant Pkd2 cannot phosphorylate and prevent the nuclear localization of histone deacetylase 5 (HDAC5) in vitro, and morpholinos against Hdac5 partially rescued the bungee valve phenotype. Interestingly, HDAC5 is known to repress klf2a, which has previously been shown to be important for zebrafish valve development (125), suggesting that a Pkd2/Hdac5/Klf signaling axis may be important for zebrafish valve development. It remains to be determined what pathways activate Pkd2 in the valvular endocardium.
Another series of mutants recently revealed a role for the transmembrane protein Tmem2 in valvulogenesis (109, 120). Embryos harboring mutations in this gene exhibit expanded AV marker expression into the ventricular endocardium. Rescue experiments suggest both cell-autonomous and cell nonautonomous roles for Tmem2 in the endocardium, as aberrant Alcama expression can be rescued by reexpression of Tmem2 in endocardial cells (120) and by morpholino knockdown of bmp4, which is expressed in the myocardium (109). As this protein is biochemically uncharacterized, it represents an exciting avenue for future research.
ECM proteins also play a role in AVC differentiation. As mentioned previously, two of the best markers for AVC differentiation are the ECM genes cspg2 (versican) and hyaluronan synthase 2 (has2), which become restricted to the AVC endocardium by 48 hpf (46, 128). One of the earliest zebrafish heart mutants cloned was jekyll, which shows toggling of blood between the two heart chambers. The jekyll mutation affects the UDP-glucose dehydrogenase (ugdh) gene, which encodes a critical enzyme in the synthesis of hyaluronic acid, heparan sulfate, and chondroitin sulfate glycosaminoglycans. Additionally, a recent morpholino study implicated nephronectin, an ECM protein previously uncharacterized in heart development, as acting through Bmp signaling and has2 production to ensure proper AVC differentiation (82). It is currently unknown exactly how these ECM proteins influence valve formation, but it will be interesting to study how they interact with and influence molecular and mechanical signals necessary for valve development.
One of the most interesting observations in AVC development has been that fluid forces play a role in valve formation. Mutants defective in cardiac contraction exhibit impaired valve formation: For example, AVC endocardial cells in sih/tnnt2a mutants fail to undergo cuboidal morphogenesis and do not express Alcama (11, 12). Furthermore, chemical inhibition of contraction results in a dose-dependent decrease in endocardial ring formation. This failure of differentiation is most likely due to lack of fluid flow, as physical occlusion of either the inflow or outflow tracts with 50 μm glass beads inserted at 37 hpf resulted in a failure of valve formation, along with failure of looping and multiple other cardiac defects (43). Recently, high-speed observations of blood flow have allowed for measurement of wall shear stress (WSS) in live 48 hpf zebrafish embryos over the course of each heart beat (125). Manipulations that changed flow patterns, ranging from genetic manipulation of cardiac contractility to morpholino-based manipulation of blood viscosity, revealed the importance of retrograde flow at early stages of valve formation for the completion of this process. Increases in valve defects correlated with a significant reduction in the expression of the known oscillatory flow-sensitive gene klf2a and knockdown of klf2a, although not changing retrograde flow, led to failed valve formation and a decrease in valve marker expression. Although separating the role of oscillatory flow from other flow parameters is difficult, these data suggest that Klf2a activity downstream of oscillatory flow plays a role in valve formation. More studies are necessary to identify the molecular pathway upstream of klf2a responsible for sensing fluid forces, as well as the effector genes downstream of Klf2a. Because it can survive dramatic changes in blood flow, the zebrafish constitutes a powerful system for understanding the interactions between these myriad players in valve development. Furthermore, although zebrafish cardiac valves are structurally smaller and simpler than those in amniotes, it has become clear that the mechanisms underlying their formation are highly conserved. Thus, the zebrafish should continue to provide a fertile ground of research to address the formation of the cardiac valves, which are often affected in congenital heart disease.
Conduction System
In order to generate a coordinated beat, the heart relies on a specialized network of cardiomyocytes termed the cardiac conduction system (CCS). In vertebrates, the electrical signal initiates in the sinoatrial (SA) node and travels through the atrium/atria until it reaches specialized tissue called the AV node, where it slows dramatically. It then enters the ventricular fast conduction system, which rapidly conducts the signal to the apex of the heart and then spreads from the apex to the rest of the ventricle (30). As a result, this conduction pattern allows the apex-to-base contraction necessary to eject blood from the ventricle. Such a conduction path has been observed in both larval and adult zebrafish (22, 44, 105). Interestingly, many of the electrical properties of the zebrafish heart resemble those of the human heart (59, 71, 105, 137). Recently, the physiological development of the CCS in zebrafish has been traced using calcium-sensitive dyes, as well as a transgenic line that expresses a genetically encoded calcium-sensitive fluorescent protein (22, 71). Coordinated, regular conduction develops as soon as the heart starts beating at 24 hpf, indicating the presence of pacemaker tissue. However, AV conduction delay between the forming chambers is not observed until 40 hpf (71). By 48 hpf, both calcium imaging and patch clamp experiments confirm that atrial, ventricular, and AV myocardial cells have developed distinct electrical properties. Additionally, functional experiments using optogenetics have revealed that pacemaker activity starts out diffusely localized to the venous pole but becomes restricted to a small area on the dorsal right quadrant of the SA ring by 3 dpf (4). This gradual right-sided restriction is consistent with previous mapping in developing chick hearts (50).
Maturation of ventricular conduction patterns begins at 72 hpf, when high-speed imaging of membrane potentials reveals that the conduction velocity in the outer curvature becomes markedly faster than that seen in the inner curvature (80). Also around this time, myocardial sheets known as trabeculae begin to form in the developing ventricle (64, 83). In other species, the fast conduction system is thought to form initially within these trabeculae (90, 104). In the zebrafish, by 96 hpf conduction waves clearly pass from the AVC to the trabeculae before entering the rest of the ventricular myocardium (22). By 21 dpf, apex-to-base conduction is evident. In the adult, the electrical impulse passes from the AVC into the trabecular network at two clearly defined points, propagates to the apex, and then travels to the base of the heart (105). Severing the two connection points results in complete AV block. Thus, the trabeculae appear to serve as the fast conduction system in embryonic and adult zebrafish.
Only recently have studies begun to provide insight into the molecular mechanisms underlying the development of the different components of the CCS. The initial difference in conduction velocity observed between the inner and outer curvatures appears to be dependent on Wnt11 signaling, which may act at least in part by inhibiting L-type calcium channels, thereby decreasing calcium conductance (80). The conduction properties of the AV canal, meanwhile, are dependent on a network of transcription factors. Foxn4 in AVC myocytes directly activates tbx2b in cooperation with Tbx5, resulting in AVC differentiation (21). Additionally, in a recent forward genetic screen, a mutant in tcf2 was identified that lacks AV delay (22). Signals from the endocardium are also likely important for AV conduction tissue development, as clo mutants fail to develop an AV conduction delay (71). In support of this idea, neuregulin and notch1b are both expressed in the AV endocardium, and morpholino knockdown of either of these genes inhibits AV delay. Trabecular formation is also dependent on the presence of the endocardium and requires signaling through the neuregulin coreceptor ErbB2 cell-autonomously within the myocardium, demonstrating that some ventricular conduction components rely on the same signals as the AVC myocardium (64, 83). Interestingly, different components of the conduction system respond differently to cardiac function. sih/tnnt2a mutants completely lack trabeculae and amhc mutants show a variable decrease in trabecular formation (22, 83), whereas neither AV conduction (22, 71) nor the OC/IC conduction ratio (80) are affected in contractile mutants. Although a clear demonstration of Purkinje-like cells in the zebrafish heart remains a major quest, the data so far point to the existence of a specialized conduction system. Given the importance of this tissue in heart function and our limited understanding of its formation, one should approach it using a variety of model systems.
CARDIAC PHYSIOLOGY
Establishment and Maintenance of Cardiac Contractility
In order to pump blood effectively, the heart must not only form the correct morphology, but must also form contractile sarcomeres and regulate their contractility. Because the zebrafish can develop in the first week without a functional heart, it is an excellent in vivo model for studying proteins involved in sarcomerogenesis and cardiac contractility. Large-scale genetic screens have identified many mutations that cause defects in sarcomere assembly and contractility. These have included some mutations that completely disrupt sarcomere assembly in one or both chambers, including mutations in sarcomeric genes (titin, myl7, tnnt2, vmhc, amhc) (6, 14, 96, 106, 132). These screens have also identified mutants with impaired contractility but intact sarcomeres, which have illuminated new mechanisms for maintaining cardiac contractility. A truncating mutation in the essential cardiac myosin light chain gene (cmlc1) surprisingly does not affect sarcomeres (70), which is in stark contrast to the absence of sarcomeres observed in a morpholino knockdown of the same gene (19, 70). Examination of the carboxyl terminus of Cmlc1 deleted in the mutant led to the identification of a highly conserved serine residue that might constitute a previously unknown posttranslational modification site. Myocyte-specific expression of wild-type cmlc1 rescues myocyte contractility in the mutant, whereas a version in which this serine amino acid is mutated to an alanine cannot. Although the precise role of this serine is unknown, this mutant reveals a novel sarcomeric modification site that contributes to contractility.
Two mutants in combination helped to shed light on how cardiomyocytes sense stretch. First, a nonsense mutation in phospholipase C γ1 ( plcγ1) leads to progressive loss of ventricular contractility starting at 48 hpf, leading to a silent ventricle by 60 hpf (94). Sarcomeric structure remains normal in these mutants. Further analysis indicates that PLCγ1 functions downstream of Vegf signaling through the receptor Flt-1 to increase calcium transients in cardiomyocytes, leading to increased contractility. Second, main squeeze (msq) mutants, which carry a mutation in the integrin-linked kinase (ilk) gene, exhibit a progressive loss of ventricular contractility with seemingly normal sarcomeric structure (13). Integrin-linked kinase (ILK) colocalizes with α-actinin at Z disks, but also binds β-parvin, which in turn binds integrins, providing a link between the ECM and the sarcomere. In keeping with this model, embryos carrying one copy of a separately isolated nonsense allele of ilk, termed lost contact (loc), are sensitized to morpholino knockdown of the ECM component laminin α 4, suggesting that ILK function may depend on integrin-mediated interactions with laminins (58). The msq mutation decreases kinase activity and ILK’s affinity for β-parvin, and although expression of wild-type Ilk can rescue msq mutant embryos, mutants which lack either kinase activity or the ability to bind to β-parvin cannot. Additionally, msq mutants can be rescued by injection of RNA encoding activated, membrane-associated protein kinase B (PKB) or Vegf. Overall, analysis of these two mutants suggests a stretch response pathway in which ILK serves as a link between the costamere and the ECM through β-parvin and integrins. In this model, stretch along the costamere/ECM axis leads to ILK activation. Kinase activation in turn activates PKB, leading to expression of VEGF, which activates PLCγ1 downstream of Flt-1, increasing calcium transients, and thus increasing contractility upon increased stretch. Although this pathway needs further confirmation in other systems, it is an attractive model for how force sensing may connect to increased contractility and possibly contraction-dependent differentiation, as seen in the ventricular trabeculae.
HUMAN DISEASE
Insights into Human Disease
The zebrafish has emerged as a unique model for studying human disease. The combination of forward and reverse genetic techniques available in the zebrafish, coupled with the possibility of performing in vivo drug screens (8), allows a number of different approaches for dissecting human diseases. The similarity between the electrical properties of zebrafish and human hearts (105, 137) makes the fish particularly useful for modeling conduction defects. Mutations in the zebrafish homolog of the hERG potassium channel gene have led to models of short and long QT syndromes, which had been difficult to generate in murine systems, probably because of inherent differences in the channels used for conduction in mice and humans (3, 39). Meanwhile, morpholino technology has been used to model Barth syndrome (54), and dilated cardiomyopathy (126). The ease of manipulation of the zebrafish embryo can also facilitate identification of the function of human mutations. One study identified mutant forms of the BMP receptor ALK2 in patients with AV septal (AVS) defects. Injection of RNA for the mutant receptors into zebrafish embryos led to the discovery of a dominant-negative form of the receptor associated with AVS defects (108). The zebrafish can also be used to validate new human heart disease genes. A search through publicly available microarray data identified nexilin as a gene of unknown function enriched in the heart (38). Nexilin was found to be a component of sarcomeric Z-disks, and morpholino-based inhibition led to enlarged ventricles and decreased fractional shortening reminiscent of dilated cardiomyopathy (DCM). Given the DCM phenotype of the nexilin knockdown, a cohort of DCM patients was sequenced, and three heterozygous mutations from nine patients were identified. RNA injection of the mutant forms of nexilin into zebrafish embryos confirmed that these mutations led to dominant negative forms of the protein that disrupted heart function, providing support for a causative role of these mutations in DCM. Finally, using the forward genetic potential of zebrafish can identify new genetic modifiers of heart disease. One study used an intriguing approach to screen for modulators of cardiac repolarization. Treating embryos with inhibitors of zERG causes a 2:1 heart block. Treating embryos from a collection of mutant fish with zERG inhibitors and screening for enhancement or repression of the heart block phenotype led to the identification of a number of modifiers of repolarization (73). Cross-referencing the identified genes with human genome-wide association studies looking at altered cardiac repolarization revealed that one gene, GINS3, was in a region highly associated with human QT interval defects. These studies highlight the multiple advantages of the zebrafish system for examining human heart disease and pave the way for continued contributions from this model system.
SUMMARY POINTS.
Large-scale screening approaches available in zebrafish make it a powerful platform for uncovering novel mechanisms of human development and disease. With a large number of cardiac mutants already identified and analyzed, the zebrafish provides a unique genetic model for studying heart development.
Advances in imaging and transgenesis combined with the transparency of the zebrafish embryo and larvae allow for analysis of cellular behaviors during heart formation. These tools have already allowed for thorough analyses of cell migration during heart tube formation. Further advances promise to elucidate cellular behaviors within the beating heart.
Mechanical forces and electrical signals contribute substantially to heart development and morphogenesis. These factors influence the formation of the trabeculae and the atrioventricular valve as well as other key cardiac structures essential for a fully functional heart. Because zebrafish embryos and larvae do not require a heartbeat for their survival, they serve as a powerful model for studying the contribution of mechanical and electrical influences on cardiac development.
Acknowledgments
We thank D. Yelon, T. Mikawa, J. Huisken, I. Scott, N. Chi, D. Beis, D. Hassel, R. Arnaout, K. Mellman, and M. Bussen for helpful comments. Our work on cardiac development and function was supported in part by NIH grant RO1 HL54737 and the Packard Foundation.
Footnotes
DISCLOSURE STATEMENT
The authors are not aware of any affiliations, memberships, funding, or financial holdings that might be perceived as affecting the objectivity of this review.
LITERATURE CITED
- 1.Alexander J, Rothenberg M, Henry GL, Stainier DY. Casanova plays an early and essential role in endoderm formation in zebrafish. Dev Biol. 1999;215(2):343–57. doi: 10.1006/dbio.1999.9441. [DOI] [PubMed] [Google Scholar]
- 2.Alexander J, Stainier DY, Yelon D. Screening mosaic F1 females for mutations affecting zebrafish heart induction and patterning. Dev Genet. 1998;22(3):288–99. doi: 10.1002/(SICI)1520-6408(1998)22:3<288::AID-DVG10>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 3.Arnaout R, Ferrer T, Huisken J, Spitzer K, Stainier DYR, et al. Zebrafish model for human long QT syndrome. Proc Natl Acad Sci USA. 2007;104(27):11316–21. doi: 10.1073/pnas.0702724104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Arrenberg AB, Stainier DYR, Baier H, Huisken J. Optogenetic control of cardiac function. Science. 2010;330(6006):971–74. doi: 10.1126/science.1195929. [DOI] [PubMed] [Google Scholar]
- 5.Arrington CB, Yost HJ. Extra-embryonic syndecan 2 regulates organ primordia migration and fibrillogenesis throughout the zebrafish embryo. Development. 2009;136(18):3143–52. doi: 10.1242/dev.031492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Auman HJ, Coleman H, Riley HE, Olale F, Tsai HJ, Yelon D. Functional modulation of cardiac form through regionally confined cell shape changes. PLoS Biol. 2007;5(3):e53. doi: 10.1371/journal.pbio.0050053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Auman HJ, Yelon D. Vertebrate organogenesis: getting the heart into shape. Curr Biol. 2004;14(4):R152–53. [PubMed] [Google Scholar]
- 8.Baker M. Screening: the age of fishes. Nat Meth. 2011;8(1):47–51. doi: 10.1038/nmeth0111-47. [DOI] [PubMed] [Google Scholar]
- 9.Bakkers J. Zebrafish as a model to study cardiac development and human cardiac disease. Cardiovasc Res. 2011;91(2):279–88. doi: 10.1093/cvr/cvr098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bakkers J, Verhoeven MC, Abdelilah-Seyfried S. Shaping the zebrafish heart: from left-right axis specification to epithelial tissue morphogenesis. Dev Biol. 2009;330(2):213–20. doi: 10.1016/j.ydbio.2009.04.011. [DOI] [PubMed] [Google Scholar]
- 11.Bartman T, Walsh EC, Wen KK, McKane M, Ren J, et al. Early myocardial function affects endocardial cushion development in zebrafish. PLoS Biol. 2004;2(5):673–81. doi: 10.1371/journal.pbio.0020129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Beis D, Bartman T, Jin S-W, Scott IC, D’Amico LA, et al. Genetic and cellular analyses of zebrafish atrioventricular cushion and valve development. Development. 2005;132(18):4193–204. doi: 10.1242/dev.01970. [DOI] [PubMed] [Google Scholar]
- 13.Bendig G, Grimmler M, Huttner IG, Wessels G, Dahme T, et al. Integrin-linked kinase, a novel component of the cardiac mechanical stretch sensor, controls contractility in the zebrafish heart. Genes Dev. 2006;20(17):2361–72. doi: 10.1101/gad.1448306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Berdougo E, Coleman H, Lee DH, Stainier DYR, Yelon D. Mutation of weak atrium/atrial myosin heavy chain disrupts atrial function and influences ventricular morphogenesis in zebrafish. Development. 2003;130(24):6121–29. doi: 10.1242/dev.00838. [DOI] [PubMed] [Google Scholar]
- 15.Bussmann J, Bakkers J, Schulte-Merker S. Early endocardial morphogenesis requires Scl/Tal1. PLoS Genet. 2007;3(8):e140. doi: 10.1371/journal.pgen.0030140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chablais F, Veit J, Rainer G, JaŸwińska A. The zebrafish heart regenerates after cryoinjury-induced myocardial infarction. BMC Dev Biol. 2011;11:21. doi: 10.1186/1471-213X-11-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chang C-P, Neilson JR, Bayle JH, Gestwicki JE, Kuo A, et al. A field of myocardial-endocardial NFAT signaling underlies heart valve morphogenesis. Cell. 2004;118(5):649–63. doi: 10.1016/j.cell.2004.08.010. [DOI] [PubMed] [Google Scholar]
- 18.Chen JN, Haffter P, Odenthal J, Vogelsang E, Brand M, et al. Mutations affecting the cardiovascular system and other internal organs in zebrafish. Development. 1996;123:293–302. doi: 10.1242/dev.123.1.293. [DOI] [PubMed] [Google Scholar]
- 19.Chen Z, Huang W, Dahme T, Rottbauer W, Ackerman MJ, Xu X. Depletion of zebrafish essential and regulatory myosin light chains reduces cardiac function through distinct mechanisms. Cardiovasc Res. 2008;79(1):97–108. doi: 10.1093/cvr/cvn073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chi NC, Bussen M, Brand-Arzamendi K, Ding C, Olgin JE, et al. Cardiac conduction is required to preserve cardiac chamber morphology. Proc Natl Acad Sci USA. 2010;107(33):14662–67. doi: 10.1073/pnas.0909432107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chi NC, Shaw RM, De Val S, Kang G, Jan LY, et al. Foxn4 directly regulates tbx2b expression and atrioventricular canal formation. Genes Dev. 2008;22(6):734–39. doi: 10.1101/gad.1629408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chi NC, Shaw RM, Jungblut B, Huisken J, Ferrer T, et al. Genetic and physiologic dissection of the vertebrate cardiac conduction system. PLoS Biol. 2008;6(5):e109. doi: 10.1371/journal.pbio.0060109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Christoffels VM, Habets PEMH, Franco D, Campione M, de Jong F, et al. Chamber formation and morphogenesis in the developing mammalian heart. Dev Biol. 2000;223(2):266–78. doi: 10.1006/dbio.2000.9753. [DOI] [PubMed] [Google Scholar]
- 24.de Pater E, Clijsters L, Marques SR, Lin Y-F, Garavito-Aguilar ZV, et al. Distinct phases of cardiomyocyte differentiation regulate growth of the zebrafish heart. Development. 2009;136(10):1633–41. doi: 10.1242/dev.030924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Deacon DC, Nevis KR, Cashman TJ, Zhou Y, Zhao L, et al. The miR-143-adducin3 pathway is essential for cardiac chamber morphogenesis. Development. 2010;137(11):1887–96. doi: 10.1242/dev.050526. [DOI] [PubMed] [Google Scholar]
- 26.Dohn TE, Waxman JS. Distinct phases of Wnt/β-catenin signaling direct cardiomyocyte formation in zebrafish. Dev Biol. 2012;361(2):364–76. doi: 10.1016/j.ydbio.2011.10.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Eisen JS, Smith JC. Controlling morpholino experiments: don’t stop making antisense. Development. 2008;135(10):1735–43. doi: 10.1242/dev.001115. [DOI] [PubMed] [Google Scholar]
- 28.Ekker SC. Zinc finger-based knockout punches for zebrafish genes. Zebrafish. 2008;5(2):121–23. doi: 10.1089/zeb.2008.9988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fish JE, Wythe JD, Xiao T, Bruneau BG, Stainier DYR, et al. A Slit/miR-218/Robo regulatory loop is required during heart tube formation in zebrafish. Development. 2011;138(7):1409–19. doi: 10.1242/dev.060046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fishman GI. Understanding conduction system development: a hop, skip and jump away? Circ Res. 2005;96(8):809–11. doi: 10.1161/01.RES.0000165653.83279.20. [DOI] [PubMed] [Google Scholar]
- 31.Forouhar AS, Liebling M, Hickerson A, Nasiraei-Moghaddam A, Tsai H-J, et al. The embryonic vertebrate heart tube is a dynamic suction pump. Science. 2006;312(5774):751–53. doi: 10.1126/science.1123775. [DOI] [PubMed] [Google Scholar]
- 32.Garavito-Aguilar ZV, Riley HE, Yelon D. Hand2 ensures an appropriate environment for cardiac fusion by limiting fibronectin function. Development. 2010;137(19):3215–20. doi: 10.1242/dev.052225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Garrity DM, Childs S, Fishman MC. The heartstrings mutation in zebrafish causes heart/fin Tbx5 deficiency syndrome. Development. 2002;129(19):4635–45. doi: 10.1242/dev.129.19.4635. [DOI] [PubMed] [Google Scholar]
- 34.George EL, Georges-Labouesse EN, Patel-King RS, Rayburn H, Hynes RO. Defects in mesoderm, neural tube and vascular development in mouse embryos lacking fibronectin. Development. 1993;119(4):1079–91. doi: 10.1242/dev.119.4.1079. [DOI] [PubMed] [Google Scholar]
- 35.Glickman NS, Yelon D. Cardiac development in zebrafish: coordination of form and function. Semin Cell Dev Biol. 2002;13(6):507–13. doi: 10.1016/s1084952102001040. [DOI] [PubMed] [Google Scholar]
- 36.González-Rosa JM, Martín V, Peralta M, Torres M, Mercader N. Extensive scar formation and regression during heart regeneration after cryoinjury in zebrafish. Development. 2011;138(9):1663–74. doi: 10.1242/dev.060897. [DOI] [PubMed] [Google Scholar]
- 37.Hami D, Grimes AC, Tsai H-J, Kirby ML. Zebrafish cardiac development requires a conserved secondary heart field. Development. 2011;138(11):2389–98. doi: 10.1242/dev.061473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hassel D, Dahme T, Erdmann J, Meder B, Huge A, et al. Nexilin mutations destabilize cardiac Z-disks and lead to dilated cardiomyopathy. Nat Med. 2009;15(11):1281–88. doi: 10.1038/nm.2037. [DOI] [PubMed] [Google Scholar]
- 39.Hassel D, Scholz EP, Trano N, Friedrich O, Just S, et al. Deficient zebrafish ether-à-go-go-related gene channel gating causes short-QT syndrome in zebrafish reggae mutants. Circulation. 2008;117(7):866–75. doi: 10.1161/CIRCULATIONAHA.107.752220. [DOI] [PubMed] [Google Scholar]
- 40.Holtzinger A, Evans T. Gata5 and Gata6 are functionally redundant in zebrafish for specification of cardiomyocytes. Dev Biol. 2007;312(2):613–22. doi: 10.1016/j.ydbio.2007.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Holtzman NG, Schoenebeck JJ, Tsai H-J, Yelon D. Endocardium is necessary for cardiomyocyte movement during heart tube assembly. Development. 2007;134(12):2379–86. doi: 10.1242/dev.02857. [DOI] [PubMed] [Google Scholar]
- 42.Horne-Badovinac S, Lin D, Waldron S, Schwarz M, Mbamalu G, et al. Positional cloning of heart and soul reveals multiple roles for PKC lambda in zebrafish organogenesis. Curr Biol. 2001;11(19):1492–502. doi: 10.1016/s0960-9822(01)00458-4. [DOI] [PubMed] [Google Scholar]
- 43.Hove JR, Koster RW, Forouhar AS, Acevedo-Bolton G, Fraser SE, Gharib M. Intracardiac fluid forces are an essential epigenetic factor for embryonic cardiogenesis. Nature. 2003;421(6919):172–77. doi: 10.1038/nature01282. [DOI] [PubMed] [Google Scholar]
- 44.Hu N, Sedmera D, Yost HJ, Clark EB. Structure and function of the developing zebrafish heart. Anat Rec. 2000;260(2):148–57. doi: 10.1002/1097-0185(20001001)260:2<148::AID-AR50>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- 45.Huang P, Xiao A, Zhou M, Zhu Z, Lin S, Zhang B. Heritable gene targeting in zebrafish using customized TALENs. Nat Biotech. 2011;29(8):699–700. doi: 10.1038/nbt.1939. [DOI] [PubMed] [Google Scholar]
- 46.Hurlstone AFL, Haramis A-PG, Wienholds E, Begthel H, Korving J, et al. The Wnt/β-catenin pathway regulates cardiac valve formation. Nature. 2003;425(6958):633–37. doi: 10.1038/nature02028. [DOI] [PubMed] [Google Scholar]
- 47.Jensen AM, Westerfield M. Zebrafish mosaic eyes is a novel FERM protein required for retinal lamination and retinal pigmented epithelial tight junction formation. Curr Biol. 2004;14(8):711–17. doi: 10.1016/j.cub.2004.04.006. [DOI] [PubMed] [Google Scholar]
- 48.Jopling C, Sleep E, Raya M, Martí M, Raya A, Belmonte JCI. Zebrafish heart regeneration occurs by cardiomyocyte dedifferentiation and proliferation. Nature. 2010;464(7288):606–9. doi: 10.1038/nature08899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Just S, Berger IM, Meder B, Backs J, Keller A, et al. Protein kinase D2 controls cardiac valve formation in zebrafish by regulating histone deacetylase 5 activity. Circulation. 2011;124(3):324–34. doi: 10.1161/CIRCULATIONAHA.110.003301. [DOI] [PubMed] [Google Scholar]
- 50.Kamino K, Hirota A, Fujii S. Localization of pacemaking activity in early embryonic heart monitored using voltage-sensitive dye. Nature. 1981;290(5807):595–97. doi: 10.1038/290595a0. [DOI] [PubMed] [Google Scholar]
- 51.Kawahara A, Nishi T, Hisano Y, Fukui H, Yamaguchi A, Mochizuki N. The sphingolipid transporter spns2 functions in migration of zebrafish myocardial precursors. Science. 2009;323(5913):524–27. doi: 10.1126/science.1167449. [DOI] [PubMed] [Google Scholar]
- 52.Keegan BR, Feldman JL, Begemann G, Ingham PW, Yelon D. Retinoic acid signaling restricts the cardiac progenitor pool. Science. 2005;307(5707):247–49. doi: 10.1126/science.1101573. [DOI] [PubMed] [Google Scholar]
- 53.Keegan BR, Meyer D, Yelon D. Organization of cardiac chamber progenitors in the zebrafish blastula. Development. 2004;131(13):3081–91. doi: 10.1242/dev.01185. [DOI] [PubMed] [Google Scholar]
- 54.Khuchua Z, Yue Z, Batts L, Strauss AW. A zebrafish model of human barth syndrome reveals the essential role of tafazzin in cardiac development and function. Circ Res. 2006;99(2):201–8. doi: 10.1161/01.RES.0000233378.95325.ce. [DOI] [PubMed] [Google Scholar]
- 55.Kikuchi K, Holdway JE, Werdich AA, Anderson RM, Fang Y, et al. Primary contribution to zebrafish heart regeneration by gata4+ cardiomyocytes. Nature. 2010;464(7288):601–5. doi: 10.1038/nature08804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kikuchi Y, Trinh LA, Reiter JF, Alexander J, Yelon D, Stainier DY. The zebrafish bonnie and clyde gene encodes a Mix family homeodomain protein that regulates the generation of endodermal precursors. Genes Dev. 2000;14(10):1279. [PMC free article] [PubMed] [Google Scholar]
- 57.Kleaveland B, Zheng X, Liu JJ, Blum Y, Tung JJ, et al. Regulation of cardiovascular development and integrity by the heart of glass-cerebral cavernous malformation protein pathway. Nat Med. 2009;15(2):169–76. doi: 10.1038/nm.1918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Knöll R, Postel R, Wang J, Krätzner R, Hennecke G, et al. Laminin-α4 and integrin-linked kinase mutations cause human cardiomyopathy via simultaneous defects in cardiomyocytes and endothelial cells. Circulation. 2007;116(5):515–25. doi: 10.1161/CIRCULATIONAHA.107.689984. [DOI] [PubMed] [Google Scholar]
- 59.Kopp R, Schwerte T, Pelster B. Cardiac performance in the zebrafish breakdance mutant. J Exp Biol. 2005;208(11):2123–34. doi: 10.1242/jeb.01620. [DOI] [PubMed] [Google Scholar]
- 60.Kupperman E, An S, Osborne N, Waldron S, Stainier DY. A sphingosine-1-phosphate receptor regulates cell migration during vertebrate heart development. Nature. 2000;406(6792):192–95. doi: 10.1038/35018092. [DOI] [PubMed] [Google Scholar]
- 61.Labauge P, Denier C, Bergametti F, Tournier-Lasserve E. Genetics of cavernous angiomas. Lancet Neurol. 2007;6(3):237–44. doi: 10.1016/S1474-4422(07)70053-4. [DOI] [PubMed] [Google Scholar]
- 62.Lazic S, Scott IC. Mef2cb regulates late myocardial cell addition from a second heart field–like population of progenitors in zebrafish. Dev Biol. 2011;354(1):123–33. doi: 10.1016/j.ydbio.2011.03.028. [DOI] [PubMed] [Google Scholar]
- 63.Lee RK, Stainier DY, Weinstein BM, Fishman MC. Cardiovascular development in the zebrafish. II Endocardial progenitors are sequestered within the heart field. Development. 1994;120(12):3361–66. doi: 10.1242/dev.120.12.3361. [DOI] [PubMed] [Google Scholar]
- 64.Liu J, Bressan M, Hassel D, Huisken J, Staudt D, et al. A dual role for ErbB2 signaling in cardiac trabeculation. Development. 2010;137(22):3867–75. doi: 10.1242/dev.053736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lou X, Deshwar AR, Crump JG, Scott IC. Smarcd3b and Gata5 promote a cardiac progenitor fate in the zebrafish embryo. Development. 2011;138(15):3113–23. doi: 10.1242/dev.064279. [DOI] [PubMed] [Google Scholar]
- 66.Mably JD, Chuang LP, Serluca FC, Mohideen M-APK, Chen J-N, Fishman MC. santa and valentine pattern concentric growth of cardiac myocardium in the zebrafish. Development. 2006;133(16):3139–46. doi: 10.1242/dev.02469. [DOI] [PubMed] [Google Scholar]
- 67.Mably JD, Mohideen M-AP, Burns CG, Chen J-N, Fishman MC. Heart of glass regulates the concentric growth of the heart in zebrafish. Curr Biol. 2003;13(24):2138–47. doi: 10.1016/j.cub.2003.11.055. [DOI] [PubMed] [Google Scholar]
- 68.Marques SR, Lee Y, Poss KD, Yelon D. Reiterative roles for FGF signaling in the establishment of size and proportion of the zebrafish heart. Dev Biol. 2008;321(2):397–406. doi: 10.1016/j.ydbio.2008.06.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Marques SR, Yelon D. Differential requirement for BMP signaling in atrial and ventricular lineages establishes cardiac chamber proportionality. Dev Biol. 2009;328(2):472–82. doi: 10.1016/j.ydbio.2009.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Meder B, Laufer C, Hassel D, Just S, Marquart S, et al. A Single Serine in the Carboxyl Terminus of Cardiac Essential Myosin Light Chain-1 Controls Cardiomyocyte Contractility In Vivo. Circ Res. 2009;104(5):650–59. doi: 10.1161/CIRCRESAHA.108.186676. [DOI] [PubMed] [Google Scholar]
- 71.Milan DJ, Giokas AC, Serluca FC, Peterson RT, MacRae CA. Notch1b and neuregulin are required for specification of central cardiac conduction tissue. Development. 2006;133(6):1125–32. doi: 10.1242/dev.02279. [DOI] [PubMed] [Google Scholar]
- 72.Milan DJ, Jones IL, Ellinor PT, MacRae CA. In vivo recording of adult zebrafish electrocardiogram and assessment of drug-induced QT prolongation. Am J Physiol Heart Circ Physiol. 2006;291(1):H269–73. doi: 10.1152/ajpheart.00960.2005. [DOI] [PubMed] [Google Scholar]
- 73.Milan DJ, Kim AM, Winterfield JR, Jones IL, Pfeufer A, et al. Drug-sensitized zebrafish screen identifies multiple genes, including GINS3, as regulators of myocardial repolarization. Circulation. 2009;120(7):553–59. doi: 10.1161/CIRCULATIONAHA.108.821082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Moens CB, Donn TM, Wolf-Saxon ER, Ma TP. Reverse genetics in zebrafish by TILLING. Brief Funct Genomic Proteomic. 2008;7(6):454–59. doi: 10.1093/bfgp/eln046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Munson C, Huisken J, Bit-Avragim N, Kuo T, Dong PD, et al. Regulation of neurocoel morphogenesis by Pard6γb. Dev Biol. 2008;324(1):41–54. doi: 10.1016/j.ydbio.2008.08.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ohn J, Tsai H-J, Liebling M. Joint dynamic imaging of morphogenesis and function in the developing heart. Organogenesis. 2009;5(4):166–73. doi: 10.4161/org.5.4.10568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Omori Y, Malicki J. Oko meduzy and related crumbs genes are determinants of apical cell features in the vertebrate embryo. Curr Biol. 2006;16(10):945–57. doi: 10.1016/j.cub.2006.03.058. [DOI] [PubMed] [Google Scholar]
- 78.Osborne N, Brand-Arzamendi K, Ober EA, Jin S-W, Verkade H, et al. The spinster homolog, two of hearts, is required for sphingosine 1-phosphate signaling in zebrafish. Curr Biol. 2008;18(23):1882–88. doi: 10.1016/j.cub.2008.10.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Palencia-Desai S, Kohli V, Kang J, Chi NC, Black BL, Sumanas S. Vascular endothelial and endocardial progenitors differentiate as cardiomyocytes in the absence of Etsrp/Etv2 function. Development. 2011;138(21):4721–32. doi: 10.1242/dev.064998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Panáková D, Werdich AA, Macrae CA. Wnt11 patterns a myocardial electrical gradient through regulation of the L-type Ca(2+) channel. Nature. 2010;466(7308):874–78. doi: 10.1038/nature09249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Pardo-Martin C, Chang T-Y, Koo BK, Gilleland CL, Wasserman SC, Yanik MF. High-throughput in vivo vertebrate screening. Nat Meth. 2010;7(8):634–36. doi: 10.1038/nmeth.1481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Patra C, Diehl F, Ferrazzi F, van Amerongen MJ, Novoyatleva T, et al. Nephronectin regulates atrioventricular canal differentiation via Bmp4-Has2 signaling in zebrafish. Development. 2011;138(20):4499–509. doi: 10.1242/dev.067454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Peshkovsky C, Totong R, Yelon D. Dependence of cardiac trabeculation on neuregulin signaling and blood flow in zebrafish. Dev Dyn. 2011;240(2):446–56. doi: 10.1002/dvdy.22526. [DOI] [PubMed] [Google Scholar]
- 84.Peterkin T, Gibson A, Patient R. Redundancy and evolution of GATA factor requirements in development of the myocardium. Dev Biol. 2007;311(2):623–35. doi: 10.1016/j.ydbio.2007.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Peterson RT, Mably JD, Chen JN, Fishman MC. Convergence of distinct pathways to heart patterning revealed by the small molecule concentramide and the mutation heart-and-soul. Curr Biol. 2001;11(19):1481–91. doi: 10.1016/s0960-9822(01)00482-1. [DOI] [PubMed] [Google Scholar]
- 86.Poss KD. Getting to the heart of regeneration in zebrafish. Semin Cell Dev Biol. 2007;18(1):36–45. doi: 10.1016/j.semcdb.2006.11.009. [DOI] [PubMed] [Google Scholar]
- 87.Poss KD, Wilson LG, Keating MT. Heart regeneration in zebrafish. Science. 2002;298(5601):2188–90. doi: 10.1126/science.1077857. [DOI] [PubMed] [Google Scholar]
- 88.Reiter JF, Alexander J, Rodaway A, Yelon D, Patient R, et al. Gata5 is required for the development of the heart and endoderm in zebrafish. Genes Dev. 1999;13(22):2983–95. doi: 10.1101/gad.13.22.2983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Reiter JF, Verkade H, Stainier DY. Bmp2b and Oep promote early myocardial differentiation through their regulation of gata5. Dev Biol. 2001;234(2):330–38. doi: 10.1006/dbio.2001.0259. [DOI] [PubMed] [Google Scholar]
- 90.Rentschler S, Vaidya DM, Tamaddon H, Degenhardt K, Sassoon D, et al. Visualization and functional characterization of the developing murine cardiac conduction system. Development. 2001;128(10):1785–92. doi: 10.1242/dev.128.10.1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Rohr S. Heart and soul/PRKCi and nagie oko/Mpp5 regulate myocardial coherence and remodeling during cardiac morphogenesis. Development. 2006;133(1):107–15. doi: 10.1242/dev.02182. [DOI] [PubMed] [Google Scholar]
- 92.Rohr S, Otten C, Abdelilah-Seyfried S. Asymmetric involution of the myocardial field drives heart tube formation in zebrafish. Circ Res. 2008;102(2):e12–19. doi: 10.1161/CIRCRESAHA.107.165241. [DOI] [PubMed] [Google Scholar]
- 93.Rottbauer W, Baker K, Wo ZG, Mohideen MA, Cantiello HF, Fishman MC. Growth and function of the embryonic heart depend upon the cardiac-specific L-type calcium channel α1 subunit. Dev Cell. 2001;1(2):265–75. doi: 10.1016/s1534-5807(01)00023-5. [DOI] [PubMed] [Google Scholar]
- 94.Rottbauer W, Just S, Wessels G, Trano N, Most P, et al. VEGF-PLCγ1 pathway controls cardiac contractility in the embryonic heart. Genes Dev. 2005;19(13):1624–34. doi: 10.1101/gad.1319405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Rottbauer W, Saurin AJ, Lickert H, Shen X, Burns CG, et al. Reptin and pontin antagonistically regulate heart growth in zebrafish embryos. Cell. 2002;111(5):661–72. doi: 10.1016/s0092-8674(02)01112-1. [DOI] [PubMed] [Google Scholar]
- 96.Rottbauer W, Wessels G, Dahme T, Just S, Trano N, et al. Cardiac myosin light chain-2: a novel essential component of thick-myofilament assembly and contractility of the heart. Circ Res. 2006;99(3):323–31. doi: 10.1161/01.RES.0000234807.16034.fe. [DOI] [PubMed] [Google Scholar]
- 97.Sakaguchi T, Kikuchi Y, Kuroiwa A, Takeda H, Stainier DYR. The yolk syncytial layer regulates myocardial migration by influencing extracellular matrix assembly in zebrafish. Development. 2006;133(20):4063–72. doi: 10.1242/dev.02581. [DOI] [PubMed] [Google Scholar]
- 98.Sander JD, Cade L, Khayter C, Reyon D, Peterson RT, et al. Targeted gene disruption in somatic zebrafish cells using engineered TALENs. Nat Biotech. 2011;29(8):697–98. doi: 10.1038/nbt.1934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Scherz PJ, Huisken J, Sahai-Hernandez P, Stainier DYR. High-speed imaging of developing heart valves reveals interplay of morphogenesis and function. Development. 2008;135(6):1179–87. doi: 10.1242/dev.010694. [DOI] [PubMed] [Google Scholar]
- 100.Schnabel K, Wu C-C, Kurth T, Weidinger G. Regeneration of cryoinjury induced necrotic heart lesions in zebrafish is associated with epicardial activation and cardiomyocyte proliferation. PLoS ONE. 2011;6(4):e18503. doi: 10.1371/journal.pone.0018503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Schoenebeck JJ, Keegan BR, Yelon D. Vessel and blood specification override cardiac potential in anterior mesoderm. Dev Cell. 2007;13(2):254–67. doi: 10.1016/j.devcel.2007.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Schoenebeck JJ, Yelon D. Illuminating cardiac development: advances in imaging add new dimensions to the utility of zebrafish genetics. Semin Cell Dev Biol. 2007;18:27–35. doi: 10.1016/j.semcdb.2006.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Scott IC, Masri B, D’Amico LA, Jin S-W, Jungblut B, et al. The G protein-coupled receptor Agtrl1b regulates early development of myocardial progenitors. Dev Cell. 2007;12(3):403–13. doi: 10.1016/j.devcel.2007.01.012. [DOI] [PubMed] [Google Scholar]
- 104.Sedmera D, Reckova M, Bigelow MR, deAlmeida A, Stanley CP, et al. Developmental transitions in electrical activation patterns in chick embryonic heart. Anat Rec Part A Discov Mol Cell Evol Biol. 2004;280(2):1001–9. doi: 10.1002/ar.a.20107. [DOI] [PubMed] [Google Scholar]
- 105.Sedmera D, Reckova M, deAlmeida A, Sedmerova M, Biermann M, et al. Functional and morphological evidence for a ventricular conduction system in zebrafish and Xenopus hearts. Am J Physiol Heart Circ Physiol. 2003;284(4):H1152–60. doi: 10.1152/ajpheart.00870.2002. [DOI] [PubMed] [Google Scholar]
- 106.Sehnert AJ, Huq A, Weinstein BM, Walker C, Fishman M, Stainier DYR. Cardiac troponin T is essential in sarcomere assembly and cardiac contractility. Nat Genet. 2002;31(1):106–10. doi: 10.1038/ng875. [DOI] [PubMed] [Google Scholar]
- 107.Smith KA, Chocron S, von der Hardt S, de Pater E, Soufan A, et al. Rotation and asymmetric development of the zebrafish heart requires directed migration of cardiac progenitor cells. Dev Cell. 2008;14(2):287–97. doi: 10.1016/j.devcel.2007.11.015. [DOI] [PubMed] [Google Scholar]
- 108.Smith KA, Joziasse IC, Chocron S, van Dinther M, Guryev V, et al. Dominant-negative ALK2 allele associates with congenital heart defects. Circulation. 2009;119(24):3062–69. doi: 10.1161/CIRCULATIONAHA.108.843714. [DOI] [PubMed] [Google Scholar]
- 109.Smith KA, Lagendijk AK, Courtney AD, Chen H, Paterson S, et al. Transmembrane protein 2 (Tmem2) is required to regionally restrict atrioventricular canal boundary and endocardial cushion development. Development. 2011;138(19):4193–98. doi: 10.1242/dev.065375. [DOI] [PubMed] [Google Scholar]
- 110.Sorrell MRJ, Waxman JS. Restraint of Fgf8 signaling by retinoic acid signaling is required for proper heart and forelimb formation. Dev Biol. 2011;358(1):44–55. doi: 10.1016/j.ydbio.2011.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Srivastava D. Making or breaking the heart: from lineage determination to morphogenesis. Cell. 2006;126(6):1037–48. doi: 10.1016/j.cell.2006.09.003. [DOI] [PubMed] [Google Scholar]
- 112.Stainier DY, Fishman MC. Patterning the zebrafish heart tube: acquisition of anteroposterior polarity. Dev Biol. 1992;153(1):91–101. doi: 10.1016/0012-1606(92)90094-w. [DOI] [PubMed] [Google Scholar]
- 113.Stainier DY, Lee RK, Fishman MC. Cardiovascular development in the zebrafish. I Myocardial fate map and heart tube formation. Development. 1993;119(1):31–40. doi: 10.1242/dev.119.1.31. [DOI] [PubMed] [Google Scholar]
- 114.Stainier DY, Weinstein BM, Detrich HW, Zon LI, Fishman MC. Cloche, an early acting zebrafish gene, is required by both the endothelial and hematopoietic lineages. Development. 1995;121(10):3141–50. doi: 10.1242/dev.121.10.3141. [DOI] [PubMed] [Google Scholar]
- 115.Stainier D, Fouquet B, Chen J, Warren K, Weinstein B, et al. Mutations affecting the formation and function of the cardiovascular system in the zebrafish embryo. Development. 1996;123(1):285–92. doi: 10.1242/dev.123.1.285. [DOI] [PubMed] [Google Scholar]
- 116.Takeuchi JK, Lou X, Alexander JM, Sugizaki H, Delgado-Olguin P, et al. Chromatin remodelling complex dosage modulates transcription factor function in heart development. Nat Commun. 2011;2:187. doi: 10.1038/ncomms1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Targoff KL, Schell T, Yelon D. Nkx genes regulate heart tube extension and exert differential effects on ventricular and atrial cell number. Dev Biol. 2008;322(2):314–21. doi: 10.1016/j.ydbio.2008.07.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Thomas NA, Koudijs M, van Eeden FJM, Joyner AL, Yelon D. Hedgehog signaling plays a cell-autonomous role in maximizing cardiac developmental potential. Development. 2008;135(22):3789–99. doi: 10.1242/dev.024083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Timmerman LA, Grego-Bessa J, Raya A, Bertrán E, Pérez-Pomares JM, et al. Notch promotes epithelial-mesenchymal transition during cardiac development and oncogenic transformation. Genes Dev. 2004;18(1):99–115. doi: 10.1101/gad.276304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Totong R, Schell T, Lescroart F, Ryckebüsch L, Lin Y-F, et al. The novel transmembrane protein Tmem2 is essential for coordination of myocardial and endocardial morphogenesis. Development. 2011;138(19):4199–205. doi: 10.1242/dev.064261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Trinh LA, Stainier DY. Fibronectin regulates epithelial organization during myocardial migration in zebrafish. Dev Cell. 2004;6(3):371–82. doi: 10.1016/s1534-5807(04)00063-2. [DOI] [PubMed] [Google Scholar]
- 122.Trinh LA, Yelon D, Stainier DYR. Hand2 regulates epithelial formation during myocardial differentiation. Curr Biol. 2005;15(5):441–46. doi: 10.1016/j.cub.2004.12.083. [DOI] [PubMed] [Google Scholar]
- 123.Tu C-T, Yang T-C, Tsai H-J. Nkx2.7 and Nkx2.5 function redundantly and are required for cardiac morphogenesis of zebrafish embryos. PLoS ONE. 2009;4(1):e4249. doi: 10.1371/journal.pone.0004249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Ueno S, Weidinger G, Osugi T, Kohn AD, Golob JL, et al. Biphasic role for Wnt/β-catenin signaling in cardiac specification in zebrafish and embryonic stem cells. Proc Natl Acad Sci USA. 2007;104(23):9685–90. doi: 10.1073/pnas.0702859104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Vermot J, Forouhar AS, Liebling M, Wu D, Plummer D, et al. Reversing blood flows act through klf2a to ensure normal valvulogenesis in the developing heart. PLoS Biol. 2009;7(11):e1000246. doi: 10.1371/journal.pbio.1000246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Vogel B, Meder B, Just S, Laufer C, Berger I, et al. In-vivo characterization of human dilated cardiomyopathy genes in zebrafish. Biochem Biophys Res Commun. 2009;390(3):516–22. doi: 10.1016/j.bbrc.2009.09.129. [DOI] [PubMed] [Google Scholar]
- 127.Voss K, Stahl S, Hogan BM, Reinders J, Schleider E, et al. Functional analyses of human and zebrafish 18-amino acid in-frame deletion pave the way for domain mapping of the cerebral cavernous malformation 3 protein. Hum Mutat. 2009;30(6):1003–11. doi: 10.1002/humu.20996. [DOI] [PubMed] [Google Scholar]
- 128.Walsh EC, Stainier DY. UDP-glucose dehydrogenase required for cardiac valve formation in zebrafish. Science. 2001;293(5535):1670–73. doi: 10.1126/science.293.5535.1670. [DOI] [PubMed] [Google Scholar]
- 129.Wang J, Panáková D, Kikuchi K, Holdway JE, Gemberling M, et al. The regenerative capacity of zebrafish reverses cardiac failure caused by genetic cardiomyocyte depletion. Development. 2011;138(16):3421–30. doi: 10.1242/dev.068601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Waxman JS, Keegan BR, Roberts RW, Poss KD, Yelon D. Hoxb5b acts downstream of retinoic acid signaling in the forelimb field to restrict heart field potential in zebrafish. Dev Cell. 2008;15(6):923–34. doi: 10.1016/j.devcel.2008.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Wei X, Malicki J. nagie oko, encoding a MAGUK-family protein, is essential for cellular patterning of the retina. Nat Genet. 2002;31(2):150–57. doi: 10.1038/ng883. [DOI] [PubMed] [Google Scholar]
- 132.Xu X, Meiler SE, Zhong TP, Mohideen M, Crossley DA, et al. Cardiomyopathy in zebrafish due to mutation in an alternatively spliced exon of titin. Nat Genet. 2002;30(2):205–9. doi: 10.1038/ng816. [DOI] [PubMed] [Google Scholar]
- 133.Yelon D, Horne SA, Stainier DY. Restricted expression of cardiac myosin genes reveals regulated aspects of heart tube assembly in zebrafish. Dev Biol. 1999;214(1):23–37. doi: 10.1006/dbio.1999.9406. [DOI] [PubMed] [Google Scholar]
- 134.Yelon D, Ticho B, Halpern ME, Ruvinsky I, Ho RK, et al. The bHLH transcription factor hand2 plays parallel roles in zebrafish heart and pectoral fin development. Development. 2000;127(12):2573–82. doi: 10.1242/dev.127.12.2573. [DOI] [PubMed] [Google Scholar]
- 135.Yoruk B, Gillers BS, Chi NC, Scott IC. Ccm3 functions in a manner distinct from Ccm1 and Ccm2 in a zebrafish model of CCM vascular disease. Dev Biol. 2011;362(2):121–31. doi: 10.1016/j.ydbio.2011.12.006. [DOI] [PubMed] [Google Scholar]
- 136.Zeng X-XI, Wilm TP, Sepich DS, Solnica-Krezel L. Apelin and its receptor control heart field formation during zebrafish gastrulation. Dev Cell. 2007;12(3):391–402. doi: 10.1016/j.devcel.2007.01.011. [DOI] [PubMed] [Google Scholar]
- 137.Zhang P-C, Llach A, Sheng XY, Hove-Madsen L, Tibbits GF. Calcium handling in zebrafish ventricular myocytes. Am J Physiol Regul Integr Comp Physiol. 2010;300(1):R56–66. doi: 10.1152/ajpregu.00377.2010. [DOI] [PubMed] [Google Scholar]
- 138.Zheng X, Xu C, Di Lorenzo A, Kleaveland B, Zou Z, et al. CCM3 signaling through sterile 20-like kinases plays an essential role during zebrafish cardiovascular development and cerebral cavernous malformations. J Clin Invest. 2010;120(8):2795–804. doi: 10.1172/JCI39679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Zhou Y, Cashman TJ, Nevis KR, Obregon P, Carney SA, et al. Latent TGF-[bgr] binding protein 3 identifies a second heart field in zebrafish. Nature. 2011;474(7353):645–48. doi: 10.1038/nature10094. [DOI] [PMC free article] [PubMed] [Google Scholar]