Abstract
We showed that severe acute respiratory syndrome coronavirus 2 is probably a novel recombinant virus. Its genome is closest to that of severe acute respiratory syndrome–related coronaviruses from horseshoe bats, and its receptor-binding domain is closest to that of pangolin viruses. Its origin and direct ancestral viruses have not been identified.
Keywords: severe acute respiratory syndrome coronavirus 2, SARS-CoV-2, viruses, genomic analysis, recombinant virus, recombination, coronavirus disease, COVID-19, bats, respiratory infections, zoonoses
Seventeen years after the severe acute respiratory syndrome (SARS) epidemic, an outbreak of pneumonia, now called coronavirus disease (COVID-19), was reported in Wuhan, China. Some of the early case-patients had a history of visiting the Huanan Seafood Wholesale Market, where wildlife mammals are sold, suggesting a zoonotic origin. The causative agent was rapidly isolated from patients and identified to be a coronavirus, now designated as severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) by the International Committee on Taxonomy of Viruses (1). SARS-CoV-2 has spread rapidly to other places; 113,702 cases and 4,012 deaths had been reported in 110 countries/areas as of March 10, 2020 (2). In Hong Kong, 130 cases and 3 deaths had been reported.
SARS-CoV-2 is a member of subgenus Sarbecovirus (previously lineage b) in the family Coronaviridae, genus Betacoronavirus, and is closely related to SARS-CoV, which caused the SARS epidemic during 2003, and to SARS-related-CoVs (SARSr-CoVs) in horseshoe bats discovered in Hong Kong and mainland China (3–5). Whereas SARS-CoV and Middle East respiratory syndrome coronavirus were rapidly traced to their immediate animal sources (civet and dromedaries, respectively), the origin of SARS-CoV remains obscure.
SARS-CoV-2 showed high genome sequence identities (87.6%–87.8%) to SARSr-Rp-BatCoV-ZXC21/ZC45, detected in Rhinolophus pusillus bats from Zhoushan, China, during 2015 (6). A closer-related strain, SARSr-Ra-BatCoV-RaTG13 (96.1% genome identity with SARS-CoV-2), was recently reported in Rhinolophus affinis bats captured in Pu’er, China, during 2013 (7). Subsequently, Pangolin-SARSr-CoV/P4L/Guangxi/2017 (85.3% genome identities to SARS-CoV-2) and related viruses were also detected in smuggled pangolins captured in Nanning, China, during 2017 (8) and Guangzhou, China, during 2019 (9). To elucidate the evolutionary origin and pathway of SARS-CoV-2, we performed an in-depth genomic, phylogenetic, and recombination analysis in relation to SARSr-CoVs from humans, civets, bats, and pangolins (10).
The Study
We downloaded 4 SARS-CoV-2, 16 human/civet-SARSr-CoV, 63 bat-SARSr-CoV and 2 pangolin-SARSr-CoV genomes from GenBank and GISAID (https://www.gisaid.org). We also sequenced the complete genome of SARS-CoV-2 strain HK20 (GenBank accession no. MT186683) from a patient with COVID-19 in Hong Kong. We performed genome, phylogenetic, and recombination analysis as described (11).
The 5 SARS-CoV-2 genomes had overall 99.8%–100% nt identities with each other. These genomes showed 96.1% genome identities with SARSr-Ra-BatCoV-RaTG13, 87.8% with SARSr-Rp-BatCoV-ZC45, 87.6% with SARSr-Rp-BatCoV-ZXC21, 85.3% with pangolin-SARSr-CoV/P4L/Guangxi/2017, and 73.8%–78.6% with other SARSr-CoVs, including human/civet-SARSr-CoVs (Table 1).
Table 1. Comparison of pairwise percentage amino acid identities between SARS-CoV-2 HK20, 4 other strains of SARS-CoV-2, and other SARSr-CoVs with complete genomes available*.
| Virus | nsp1 | nsp2 | nsp3 | nsp4 | nsp5 | nsp6 | nsp7 | nsp8 | nsp9 | nsp10 | nsp12 | nsp13 | nsp14 | nsp15 | nsp16 | Spike | NTD | CTD | S2 | ORF3 | ORF8 | E | M | N | Genome |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SARS-CoV-2 HKU-SZ-005b_2020/Guangdong/2020 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 99.3 | 100 | 100 | 100 | 100 | 99.2 | 100 | 100 | 100 | 100 |
| SARS-CoV-2 Wuhan-Hu-1/Hubei/2019 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 99.9 |
| SARS-CoV-2 WIV02/Hubei/2019 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 99.7 | 99.3 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 99.8 |
| SARS-CoV-2 HKU-SZ-002a_2020/Guangdong/2020 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 99.3 | 100 | 100 | 100 | 100 | 99.2 | 100 | 100 | 100 | 99.8 |
| SARSr-Ra-BatCoV RaTG13/R.affinis/Yunnan/2013 | 96.7 | 98.3 | 96.7 | 99.6 | 99.3 | 99.7 | 100 | 100 | 100 | 100 | 99.6 | 99.7 | 99.2 | 97.7 | 100 | 96.7 | 98.6 | 89.3 | 99.1 | 97.8 | 95.0 | 100 | 99.5 | 99.0 | 96.1 |
| SARSr-Rp-BatCoV bat-SL-CoVZC45/R.pusillus/Zhejiang/2015 | 95.6 | 95.3 | 93.9 | 96.8 | 99.0 | 97.9 | 100 | 97.5 | 97.3 | 97.1 | 95.9 | 99.2 | 94.5 | 89.0 | 98.0 | 80.2 | 66.3 | 65.5 | 90.8 | 90.9 | 94.2 | 100 | 98.6 | 94.3 | 87.8 |
| SARSr-Rp-BatCoV bat-SL-CoVZXC21/R.pusillus/Zhejiang/2015 | 95.6 | 96.4 | 93.0 | 96.2 | 99.0 | 97.6 | 98.8 | 96.0 | 96.5 | 97.8 | 95.6 | 98.7 | 94.7 | 88.2 | 98.0 | 79.6 | 65.2 | 64.5 | 90.5 | 92.0 | 94.2 | 100 | 98.6 | 94.3 | 87.6 |
| Pangolin-SARSr-CoV Guangxi/P4L/2017 | 90.6 | 88.4 | 85.5 | 90.2 | 97.1 | 96.2 | 100 | 97.5 | 97.3 | 97.1 | 97.9 | 98.2 | 97.0 | 94.5 | 97.7 | 91.7 | 88.0 | 86.8 | 95.6 | 89.5 | 87.6 | 100 | 98.2 | 93.8 | 85.3 |
| SARSr-Rm-BatCoV Longquan-140/R.monoceros/Zhejiang/2012 | 83.9 | 82.8 | 75.9 | 81.2 | 95.8 | 86.6 | 100 | 97.5 | 97.3 | 94.3 | 95.7 | 99.0 | 94.9 | 89.0 | 94.3 | 75.8 | 49.3 | 66.0 | 90.5 | 61.5 | – | 93.4 | 90.5 | 89.5 | 80.1 |
| SARSr-Rs-BatCoV Rs4231/R.sinicus/Yunnan/2013 | 85.0 | 67.9 | 76.3 | 81.2 | 96.1 | 87.2 | 100 | 97.0 | 97.3 | 97.8 | 96.4 | 99.3 | 95.6 | 89.0 | 93.3 | 76.3 | 52.2 | 77.2 | 87.5 | 73.8 | 57.9 | 93.4 | 90.1 | 90.8 | 79.5 |
| SARSr-Rs-BatCoV Rs3367/R.sinicus/Yunnan/2012 | 85.0 | 67.4 | 76.1 | 80.4 | 96.1 | 86.9 | 100 | 97.5 | 97.3 | 97.8 | 96.4 | 99.3 | 95.4 | 88.7 | 93.0 | 76.8 | 53.7 | 76.6 | 87.8 | 74.2 | 57.9 | 94.7 | 90.1 | 90.5 | 79.5 |
| SARSr-Rs-BatCoV Rs9401/R.sinicus/Yunnan/2015 | 84.4 | 68.3 | 76.3 | 80.8 | 96.1 | 87.2 | 100 | 97.5 | 97.3 | 97.8 | 96.1 | 99.3 | 95.6 | 89.0 | 93.0 | 76.6 | 53.7 | 76.1 | 87.7 | 74.2 | 57.9 | 93.4 | 89.6 | 90.5 | 79.5 |
| SARSr-Rs-BatCoV RsSHC014/R.sinicus/Yunnan/2011 | 85.0 | 67.9 | 75.9 | 81.0 | 95.8 | 87.2 | 100 | 97.5 | 97.3 | 97.8 | 96.4 | 99.3 | 95.1 | 89.0 | 92.6 | 77.0 | 53.7 | 77.7 | 87.9 | 74.2 | 58.7 | 94.7 | 90.1 | 90.3 | 79.4 |
| SARSr-Ra-BatCoV LYRa11/R.affinis/Yunnan/2011 | 82.2 | 68.0 | 76.1 | 80.8 | 95.8 | 86.6 | 100 | 97.5 | 96.5 | 96.4 | 96.1 | 98.8 | 95.1 | 89.0 | 93.0 | 76.0 | 49.3 | 75.1 | 88.3 | 76.0 | 57.9 | 93.4 | 89.6 | 91.0 | 79.4 |
| SARSr-Rs-BatCoV Rs4237/R.sinicus/Yunnan/2013 | 83.9 | 67.7 | 76.1 | 81.0 | 96.1 | 87.6 | 98.8 | 97.5 | 97.3 | 97.8 | 96.7 | 99.5 | 95.4 | 89.0 | 94.6 | 75.0 | 50.0 | 65.0 | 89.1 | 72.0 | 57.9 | 94.7 | 90.5 | 90.8 | 79.4 |
| SARSr-Rs-BatCoV Rs4247/R.sinicus/Yunnan/2013 | 83.9 | 67.7 | 76.3 | 81.0 | 96.1 | 87.6 | 98.8 | 97.5 | 97.3 | 97.8 | 96.6 | 99.3 | 95.6 | 89.0 | 94.6 | 75.2 | 49.7 | 65.5 | 89.3 | 72.4 | 57.0 | 93.4 | 90.1 | 90.5 | 79.4 |
| Human SARS-CoV TOR2/Toronto/Mar2003 | 84.4 | 68.3 | 76.0 | 80.0 | 96.1 | 87.2 | 98.8 | 97.5 | 97.3 | 97.1 | 96.4 | 99.7 | 95.1 | 88.7 | 93.3 | 75.4 | 51.9 | 74.1 | 87.1 | 72.4 | – | 94.7 | 90.5 | 90.5 | 79.4 |
| Human SARS-CoV GZ02/Guangdong/Feb2003 | 83.9 | 68.3 | 76.1 | 79.8 | 96.1 | 87.2 | 98.8 | 97.5 | 97.3 | 97.1 | 96.4 | 99.3 | 95.1 | 88.7 | 93.6 | 75.7 | 52.6 | 74.6 | 87.3 | 73.1 | 29.7 | 94.7 | 90.5 | 90.5 | 79.4 |
| SARSr-Rs-BatCoV Rs4081/R.sinicus/Yunnan/2012 | 85.0 | 67.9 | 76.3 | 81.0 | 96.1 | 87.6 | 100 | 97.5 | 97.3 | 97.8 | 96.4 | 99.2 | 95.8 | 89.0 | 94.3 | 74.4 | 48.1 | 64.0 | 89.0 | 72.0 | 58.7 | 94.7 | 90.1 | 90.3 | 79.3 |
| SARS-As-BatCoV As6526/Aselliscus stoliczkanus/Yunnan/2014 | 84.4 | 67.9 | 76.3 | 81.0 | 95.8 | 87.2 | 100 | 97.5 | 97.3 | 97.8 | 96.1 | 99.3 | 95.6 | 89.0 | 94.6 | 75.1 | 49.7 | 66.0 | 88.9 | 71.6 | 58.7 | 94.7 | 91.0 | 90.0 | 79.3 |
| Human SARS-CoV BJ01/Beijing | 84.4 | 68.3 | 76.0 | 80.4 | 96.1 | 87.2 | 98.8 | 97.5 | 97.3 | 97.1 | 96.4 | 99.5 | 95.1 | 88.7 | 93.3 | 75.6 | 52.2 | 74.1 | 87.3 | 72.4 | – | 94.7 | 90.5 | 90.5 | 79.3 |
| Civet SARS-CoV SZ3/Shenzhen/2013 | 84.4 | 68.5 | 76.1 | 80.6 | 96.1 | 87.2 | 98.8 | 97.5 | 97.3 | 97.1 | 96.4 | 99.5 | 95.1 | 88.7 | 93.6 | 75.7 | 51.9 | 75.1 | 87.3 | 72.4 | 29.7 | 94.7 | 90.1 | 90.5 | 79.3 |
| Civet SARS-CoV SZ16/Shenzhen/2013 | 84.4 | 68.5 | 76.0 | 80.6 | 96.1 | 87.2 | 98.8 | 97.5 | 97.3 | 97.1 | 96.4 | 99.5 | 95.1 | 88.7 | 93.6 | 75.7 | 51.9 | 75.1 | 87.3 | 72.4 | 29.7 | 94.7 | 90.1 | 90.5 | 79.3 |
| SARSr-Ra-BatCoV YN2018A/R.affinis/Yunnan?/2016 | 84.4 | 67.9 | 76.3 | 80.8 | 96.1 | 86.9 | 100 | 97.5 | 97.3 | 97.8 | 96.5 | 99.5 | 95.4 | 89.0 | 64.1 | 75.2 | 49.7 | 65.5 | 89.1 | 71.3 | 57.9 | 94.7 | 89.6 | 90.3 | 79.2 |
| SARSr-Rs-BatCoV Rs4255/R.sinicus/Yunnan/2013 | 85.0 | 67.9 | 76.1 | 80.8 | 96.1 | 87.2 | 100 | 97.5 | 97.3 | 97.8 | 96.1 | 99.3 | 95.6 | 88.7 | 94.3 | 74.5 | 48.1 | 65.0 | 89.0 | 72.0 | 58.7 | 94.7 | 89.2 | 90.3 | 79.2 |
| SARSr-Rs-BatCoV Rp3/R.sinicus/Guangxi/2004 | 82.8 | 67.7 | 75.4 | 80.6 | 95.8 | 87.2 | 100 | 96.0 | 96.5 | 97.8 | 96.2 | 99.2 | 95.4 | 88.4 | 93.3 | 75.0 | 49.3 | 65.5 | 89.3 | 74.5 | 56.2 | 94.7 | 90.1 | 90.3 | 79.2 |
| SARSr-Rs-BatCoV Rs4084/R.sinicus/Yunnan/2012 | 84.4 | 68.3 | 76.3 | 80.4 | 95.8 | 87.2 | 100 | 97.5 | 97.3 | 97.8 | 96.2 | 98.5 | 95.1 | 88.7 | 92.6 | 76.9 | 53.7 | 77.2 | 87.9 | 74.2 | – | 94.7 | 90.1 | 90.0 | 79.2 |
| Civet SARS-CoV PC4–227/Guangdong/2004 | 84.4 | 68.2 | 76.0 | 80.6 | 96.1 | 86.9 | 98.8 | 97.0 | 96.5 | 97.1 | 96.2 | 99.3 | 94.9 | 88.7 | 93.0 | 75.4 | 52.2 | 75.1 | 86.7 | 72.4 | – | 94.7 | 90.1 | 90.0 | 79.2 |
| SARSr-Rs-BatCoV HuB2013/R.sinicus/Hubei/2013 | 83.9 | 69.3 | 76.0 | 81.0 | 96.1 | 86.6 | 100 | 97.5 | 97.3 | 97.8 | 96.1 | 99.0 | 95.6 | 88.2 | 94.0 | 74.9 | 47.8 | 66.0 | 89.4 | 75.3 | 49.6 | 93.4 | 91.0 | 91.0 | 79.1 |
| SARSr-Rm-BatCoV Rm1/R.macrotis/Hubei/2004 | 80.0 | 69.1 | 74.8 | 80.6 | 95.8 | 86.6 | 100 | 97.0 | 96.5 | 97.1 | 95.9 | 98.8 | 94.9 | 87.0 | 93.0 | 74.4 | 47.6 | 65.0 | 89.0 | 74.5 | 57.0 | 93.4 | 91.0 | 90.0 | 79.1 |
| SARSr-Rs-BatCoV Anlong-103/Guizhou/R.sinicus/2013 | 84.4 | 68.2 | 75.8 | 80.8 | 96.1 | 86.9 | 98.8 | 97.5 | 97.3 | 97.1 | 96.5 | 99.3 | 95.6 | 89.0 | 93.6 | 75.1 | 50.2 | 64.5 | 89.1 | 72.0 | 29.7 | 94.7 | 89.6 | 89.6 | 79.1 |
| Civet SARS-CoV civet020/Guangdong/2004 | 84.4 | 68.0 | 75.9 | 80.6 | 96.1 | 87.2 | 97.6 | 97.0 | 97.3 | 97.1 | 96.4 | 99.3 | 94.7 | 88.7 | 93.3 | 75.4 | 52.2 | 75.1 | 86.7 | 72.4 | 29.7 | 94.7 | 90.1 | 90.0 | 79.1 |
| SARSr-Rs-BatCoV HKU3-12/R.sinicus/Hong Kong/2007 | 83.9 | 68.8 | 75.3 | 81.8 | 95.8 | 86.6 | 100 | 97.5 | 96.5 | 97.8 | 95.9 | 98.7 | 94.7 | 88.2 | 93.3 | 75.8 | 49.0 | 66.5 | 90.5 | 73.5 | 57.0 | 94.7 | 91.0 | 89.1 | 79.0 |
| SARSr-Rs-BatCoV HKU3-7/R.sinicus/Guangdong/2006 | 83.9 | 68.5 | 75.5 | 81.4 | 95.8 | 87.2 | 100 | 96.5 | 97.3 | 97.8 | 95.8 | 99.2 | 94.5 | 87.9 | 94.0 | 75.7 | 49.0 | 66.5 | 90.3 | 73.5 | 57.0 | 93.4 | 90.1 | 90.3 | 79.0 |
| SARSr-Rs-BatCoV HKU3–8/R.sinicus/Guangdong/2006 | 83.9 | 68.5 | 75.6 | 81.4 | 95.4 | 87.2 | 100 | 96.5 | 97.3 | 97.8 | 95.8 | 99.2 | 94.5 | 87.9 | 94.0 | 75.9 | 49.3 | 66.5 | 90.5 | 73.5 | – | 92.1 | 89.6 | 90.3 | 79.0 |
| SARSr-Rs-BatCoV HKU3–1/R.sinicus/Hong Kong/2005 | 83.9 | 69.0 | 75.2 | 81.4 | 95.4 | 87.2 | 100 | 96.5 | 97.3 | 97.8 | 95.6 | 98.7 | 94.7 | 88.2 | 93.3 | 75.7 | 49.0 | 66.5 | 90.3 | 73.1 | 57.0 | 94.7 | 91.0 | 89.5 | 79.0 |
| SARSr-Rs-BatCoV HKU3–3/R.sinicus/Hong Kong/2005 | 83.9 | 69.0 | 75.2 | 81.4 | 95.4 | 87.2 | 100 | 96.5 | 97.3 | 97.8 | 95.6 | 98.7 | 94.7 | 88.2 | 93.3 | 75.7 | 49.0 | 66.5 | 90.3 | 73.1 | 57.0 | 94.7 | 91.0 | 89.5 | 79.0 |
| SARSr-Rs-BatCoV HKU3–9/R.sinicus/Hong Kong/2006 | 83.9 | 69.0 | 75.2 | 81.4 | 95.1 | 87.2 | 100 | 96.5 | 97.3 | 97.8 | 95.6 | 98.8 | 94.7 | 87.6 | 93.3 | 75.7 | 49.0 | 66.5 | 90.2 | 72.7 | 57.0 | 94.7 | 91.0 | 89.3 | 79.0 |
| SARSr-Rf-BatCoV Rf4092/R.ferrumequinum/Yunnan/2012 | 84.4 | 67.7 | 76.1 | 81.0 | 96.1 | 87.2 | 98.8 | 96.5 | 97.3 | 97.8 | 96.5 | 99.5 | 95.1 | 89.0 | 93.0 | 72.6 | 39.9 | 66.0 | 88.6 | 71.3 | 28.9 | 94.7 | 89.6 | 90.5 | 79.0 |
| SARSr-Rs-BatCoV Anlong-112/Guizhou/R.sinicus/2013 | 83.9 | 68.2 | 76.0 | 80.8 | 96.1 | 87.2 | 98.8 | 97.5 | 97.3 | 97.8 | 96.5 | 99.3 | 95.6 | 89.0 | 93.6 | 75.1 | 50.2 | 64.5 | 89.1 | 72.0 | 29.7 | 94.7 | 89.6 | 89.6 | 79.0 |
| SARSr-Rs-BatCoV HKU3–2/R.sinicus/Hong Kong/2005 | 83.9 | 69.0 | 75.3 | 81.4 | 95.4 | 87.2 | 100 | 96.5 | 97.3 | 97.8 | 95.6 | 98.8 | 94.7 | 87.6 | 93.3 | 75.7 | 49.0 | 66.5 | 90.3 | 73.1 | 57.0 | 94.7 | 91.0 | 89.8 | 78.9 |
| SARSr-Rs-BatCoV HKU3–13/R.sinicus/Hong Kong/2007 | 83.9 | 69.0 | 75.1 | 81.4 | 95.4 | 87.2 | 100 | 96.5 | 97.3 | 97.8 | 95.6 | 98.8 | 94.5 | 87.6 | 93.3 | 75.7 | 49.0 | 66.5 | 90.2 | 72.7 | 57.0 | 94.7 | 91.0 | 89.3 | 78.9 |
| UNVERIFIED: SARSr-Rp-BatCoV F46/R.pusillus/Yunnan/2012 | 81.7 | 68.3 | 75.8 | 80.8 | 96.1 | 86.2 | 98.8 | 97.5 | 96.5 | 97.1 | 96.4 | 99.3 | 95.6 | 89.6 | 93.6 | 72.6 | 39.9 | 66.0 | 88.9 | 70.9 | 26.6 | 94.7 | 89.6 | 90.5 | 78.9 |
| Human SARS-CoV GZ0401/Guangdong/Dec2003 | 84.4 | 68.2 | 45.9 | 80.6 | 96.1 | 87.2 | 97.6 | 97.0 | 97.3 | 97.1 | 96.2 | 99.3 | 94.9 | 88.7 | 93.3 | 75.3 | 51.9 | 75.1 | 86.7 | 64.4 | 29.7 | 94.7 | 84.7 | 90.0 | 78.9 |
| SARSr-Rl-BatCoV SC2018/Rhinolophus sp./2016 | 83.3 | 69.3 | 74.9 | 81.2 | 95.8 | 86.9 | 100 | 97.5 | 97.3 | 97.8 | 96.1 | 98.8 | 95.4 | 86.7 | 92.3 | 74.4 | 47.4 | 66.0 | 88.9 | 70.9 | 55.4 | 94.7 | 90.1 | 90.7 | 78.8 |
| SARSr-Rf-BatCoV YNLF_31C/R.ferrumequinum/Yunnan/2013 | 78.9 | 69.0 | 75.8 | 80.8 | 95.8 | 86.6 | 97.6 | 97.0 | 97.3 | 97.1 | 96.2 | 99.2 | 95.6 | 88.7 | 93.0 | 73.4 | 49.3 | 65.5 | 86.2 | 70.9 | 29.7 | 94.7 | 89.6 | 90.0 | 78.7 |
| Civet SARS-CoV B039/Guangdong/2004 | 84.4 | 68.2 | 75.9 | 80.6 | 96.1 | 87.2 | 98.8 | 97.0 | 97.3 | 97.1 | 96.4 | 99.5 | 94.7 | 88.7 | 93.3 | 75.4 | 52.2 | 75.1 | 86.7 | 72.4 | 29.7 | 94.7 | 90.1 | 90.0 | 78.6 |
| Civet SARS-CoV civet007/Guangdong/2004 | 84.4 | 68.0 | 75.9 | 80.6 | 96.1 | 87.2 | 98.8 | 97.0 | 97.3 | 97.1 | 96.4 | 99.5 | 94.9 | 88.7 | 93.0 | 75.3 | 51.9 | 75.1 | 86.6 | 72.0 | 29.7 | 94.7 | 90.1 | 90.0 | 78.6 |
| Civet SARS-CoV A022/Guangdong/2004 | 84.4 | 68.2 | 76.0 | 80.6 | 96.1 | 87.2 | 98.8 | 97.0 | 97.3 | 97.1 | 96.4 | 99.5 | 94.9 | 88.7 | 92.6 | 75.3 | 51.9 | 75.1 | 86.6 | 72.0 | 29.7 | 94.7 | 90.1 | 89.8 | 78.5 |
| SARSr-Rf-BatCoV Rf1/R.ferrumequinum/Hubei/2004 | 78.9 | 68.8 | 75.3 | 81.6 | 96.1 | 86.6 | 100 | 97.0 | 97.3 | 97.1 | 96.2 | 99.0 | 94.9 | 88.2 | 91.9 | 73.6 | 50.0 | 66.0 | 86.2 | 68.7 | 31.3 | 93.4 | 89.6 | 89.1 | 78.4 |
| SARSr-Rp-BatCoV Rp/Shaanxi2011/R.pusillus/Shaanxi/2011 | 84.4 | 69.3 | 75.8 | 81.2 | 96.1 | 86.6 | 100 | 97.5 | 97.3 | 97.8 | 96.1 | 99.2 | 96.0 | 88.4 | 93.6 | 74.7 | 48.0 | 66.0 | 89.1 | 72.4 | 58.7 | 93.4 | 90.5 | 91.0 | 78.4 |
| SARSr-Rf-BatCoV Jiyuan-84/R.ferrumequinum/ Henan/2012 | 77.8 | 68.8 | 75.3 | 81.4 | 95.4 | 86.9 | 100 | 97.0 | 97.3 | 97.1 | 96.2 | 99.0 | 95.8 | 89.0 | 91.9 | 73.3 | 49.7 | 66.0 | 85.8 | 70.2 | 28.9 | 93.4 | 89.6 | 89.8 | 78.3 |
| SARSr-Cp-BatCoV Cp/Yunnan2011/C. plicata/Yunnan/2011 | 81.7 | 68.0 | 76.5 | 81.0 | 96.1 | 86.6 | 100 | 97.0 | 96.5 | 97.8 | 96.0 | 98.0 | 94.7 | 89.3 | 93.0 | 74.5 | 49.0 | 65.5 | 88.7 | 72.0 | 57.9 | 94.7 | 91.4 | 90.3 | 78.2 |
| SARSr-Ra-BatCoV YN2018B/R.affinis/Yunnan?/2016 | 85.0 | 68.2 | 76.3 | 80.6 | 96.1 | 87.2 | 100 | 97.5 | 97.3 | 97.8 | 96.4 | 99.3 | 95.6 | 89.0 | 93.0 | 76.6 | 53.4 | 76.1 | 87.8 | 73.8 | 57.9 | 94.7 | 90.1 | 90.8 | 78.1 |
| SARSr-Rs-BatCoV Rs7327/R.sinicus/Yunnan/2014 | 84.4 | 67.9 | 76.2 | 80.8 | 96.1 | 87.2 | 100 | 97.5 | 97.3 | 97.8 | 96.2 | 99.2 | 95.6 | 89.0 | 93.0 | 76.7 | 53.7 | 76.1 | 87.8 | 73.8 | 57.0 | 94.7 | 90.1 | 90.5 | 78.1 |
| SARSr-Rs-BatCoV Rs4874/R.sinicus/Yunnan/2013 | 85.0 | 67.9 | 76.3 | 81.0 | 96.1 | 87.6 | 100 | 97.5 | 97.3 | 97.8 | 96.2 | 98.7 | 95.3 | 88.7 | 93.3 | 76.5 | 52.2 | 76.6 | 87.9 | 74.2 | 57.9 | 94.7 | 89.6 | 90.3 | 78.1 |
| SARSr-Rs-BatCoV WIV16/R.sinicus/Yunnan/2013 | 85.0 | 67.9 | 76.2 | 81.0 | 96.1 | 87.6 | 100 | 97.5 | 97.3 | 97.8 | 96.1 | 99.3 | 94.7 | 88.7 | 93.3 | 76.5 | 52.2 | 76.6 | 87.9 | 74.2 | 57.9 | 94.7 | 90.1 | 90.3 | 78.1 |
| SARSr-Ra-BatCoV YN2018D/R.affinis/Yunnan?/2016 | 85.0 | 68.2 | 76.3 | 81.0 | 96.1 | 87.2 | 100 | 97.5 | 97.3 | 97.8 | 96.5 | 99.3 | 95.4 | 89.0 | 94.0 | 74.4 | 48.1 | 64.0 | 89.1 | 72.7 | 57.9 | 94.7 | 76.1 | 90.5 | 77.9 |
| SARSr-Rf-BatCoV SX2013/R.ferrumequinum/Shanxi/2013 | 78.3 | 69.1 | 75.3 | 81.2 | 95.8 | 86.6 | 100 | 96.5 | 97.3 | 97.1 | 95.9 | 99.0 | 95.8 | 89.0 | 91.9 | 73.3 | 49.7 | 66.0 | 85.8 | 70.9 | 30.5 | 92.1 | 89.6 | 89.8 | 77.9 |
| SARSr-Rf-BatCoV HeB2013/R.ferrumequinum/Hebei/2013 | 78.9 | 69.1 | 75.1 | 81.6 | 95.8 | 86.6 | 100 | 97.0 | 97.3 | 97.1 | 96.1 | 99.0 | 95.8 | 88.7 | 91.9 | 73.4 | 49.7 | 66.0 | 85.9 | 70.5 | 30.5 | 93.4 | 89.6 | 89.8 | 77.7 |
| SARSr-Rs-BatCoV YN2013/R.sinicus/Yunnan/2013 | 83.9 | 67.9 | 76.1 | 80.6 | 96.1 | 87.2 | 100 | 97.5 | 97.3 | 97.8 | 96.4 | 99.3 | 95.8 | 88.4 | 93.3 | 75.1 | 50.9 | 64.0 | 89.0 | 71.3 | 29.7 | 94.7 | 91.0 | 90.8 | 77.5 |
| SARSr- Rs-BatCoV Rs672/2006/R.sinicus/Guizhou/2006 | 85.0 | 67.9 | 68.3 | 80.4 | 95.4 | 87.2 | 100 | 97.5 | 97.3 | 97.8 | 96.5 | 98.8 | 95.6 | 88.2 | 94.3 | 74.2 | 47.8 | 64.0 | 89.0 | 71.6 | 58.7 | 94.7 | 89.6 | 90.5 | 77.5 |
| SARSr-Rs-BatCoV GX2013/R.sinicus/Guangxi/2013 | 83.3 | 68.3 | 75.9 | 80.2 | 95.8 | 87.2 | 100 | 97.5 | 96.5 | 97.1 | 96.5 | 99.3 | 95.4 | 89.0 | 94.0 | 75.1 | 49.7 | 66.5 | 89.0 | 72.7 | 29.7 | 94.7 | 91.4 | 89.6 | 77.3 |
| SARSr-Rf-BatCoV 16BO133/R.ferrumequinum/Korea/2016 | 78.9 | 69.1 | 67.6 | 81.2 | 95.1 | 85.9 | 100 | 96.5 | 97.3 | 97.1 | 96.1 | 98.5 | 94.9 | 88.2 | 92.6 | 71.2 | 39.0 | 65.0 | 86.6 | 68.0 | – | 93.4 | 89.6 | 88.8 | 76.9 |
| SARSr-Rf-BatCoV JL2012/R.ferrumequinum/Jilin/2012 | 78.9 | 69.0 | 74.8 | 81.2 | 94.8 | 85.9 | 98.8 | 96.5 | 97.3 | 97.1 | 96.2 | 98.3 | 95.1 | 87.9 | 92.6 | 71.2 | 39.5 | 65.0 | 86.6 | 68.0 | – | 92.1 | 89.6 | 89.3 | 76.9 |
| SARSr-Rf-BatCoV JTMC15/R.ferrumequinum/Jilin/2013 | 78.9 | 69.0 | 67.2 | 81.2 | 94.8 | 85.9 | 98.8 | 96.5 | 97.3 | 97.1 | 96.2 | 98.3 | 94.9 | 88.2 | 92.6 | 71.2 | 39.3 | 65.0 | 86.6 | 68.0 | – | 90.8 | 90.1 | 89.3 | 76.4 |
| SARSr-Rs-BatCoV BtKY72/Rhinolophus sp./Kenya/2007 | 82.2 | 63.2 | 71.6 | 82.2 | 95.4 | 82.1 | 94.0 | 95.5 | 97.3 | 95.0 | 95.0 | 96.7 | 92.8 | 87.3 | 90.6 | 71.6 | 46.6 | 72.7 | 82.5 | 63.6 | 19.1 | 94.7 | 86.9 | 86.2 | 75.5 |
| SARSr-Rb-BatCoV BM48–31/BGR/2008/R.blasii/Bulgaria/2008 | 81.7 | 62.4 | 71.3 | 81.0 | 94.1 | 83.8 | 95.2 | 96.5 | 98.2 | 95.0 | 95.2 | 97.7 | 93.5 | 89.9 | 88.6 | 71.3 | 44.1 | 69.7 | 83.4 | 62.5 | – | 92.1 | 85.9 | 87.6 | 75.2 |
*SARS-CoV-2 strains are highlighted in tan, and SARSr-CoV strains that had the highest end identity for each gene with SARS-CoV-2 are highlighted in red. SARSr-CoV strains were listed in descending order according to complete genome identities to SARS-CoV-2 HK20/Hong Kong/2020.. Pairwise amino acid identities could not be calculated for some strains because of completely or partially truncated ORF8. CTD, C-terminal domain; E, envelope; M, matrix; N, nucleocapsid; nsp, nonstructural protein; NTD, N-terminal domain; ORF, open reading frame; S, spike; SARS-CoV- 2, severe acute respiratory syndrome coronavirus 2; SARSr-CoV, severe acute respiratory syndrome–related coronavirus; –, not applicable..
Most predicted proteins of SARS-CoV-2 showed high amino acid sequence identities with that of SARSr-Ra-BatCoV RaTG13, except the receptor-binding domain (RBD) region. SARS-CoV-2 possessed an intact open reading frame 8 without the 29-nt deletion found in most human SARS-CoVs. The concatenated conserved replicase domains for coronavirus species demarcation by the International Committee on Taxonomy of Viruses showed >92.9% aa identities (threshold >90% for same species) between SARS-CoV-2 and other SARSr-CoVs, supporting their classification under the same coronavirus species (Table 2) (1).
Table 2. Percentage amino acid identity between 7 conserved domains of the replicase polyprotein for species demarcation in SARS-CoV-2 and selected members of the subgenus Sarbecovirus*.
| Virus | % Amino acid identity compared with that for SARS-CoV-2 |
|||||||
|---|---|---|---|---|---|---|---|---|
| ADRP | nsp5 | nsp12 | nsp13 | nsp14 | nsp15 | nsp16 | Seven concatenated domains | |
| Human SARS-CoV TOR2/Toronto/Mar2003 | 79.3 | 96.1 | 96.4 | 99.8 | 95.1 | 88.7 | 93.3 | 95.0 |
| Civet SARS-CoV SZ3/Shenzhen/2013 | 79.3 | 96.1 | 96.4 | 99.7 | 95.1 | 88.7 | 93.6 | 95.0 |
| Civet SARS-CoV PC4–136/Guangdong/2004 | 78.4 | 96.1 | 96.4 | 99.3 | 94.7 | 88.7 | 93.3 | 94.8 |
| Human SARS-CoV GZ0402 | 78.4 | 95.8 | 96.2 | 99.5 | 94.9 | 51.7 | 93.3 | 94.8 |
| SARSr-Rs-BatCoV HKU3–1/R.sinicus/Hong Kong/2005 | 79.3 | 95.4 | 95.6 | 98.8 | 94.7 | 88.2 | 93.3 | 94.5 |
| SARSr-Rp-BatCoV Rp/Shaanxi2011/R.pusillus/Shaanxi/2011 | 78.4 | 96.1 | 96.1 | 99.3 | 96.0 | 88.4 | 93.6 | 95.0 |
| SARSr-Rs-BatCoV Rs672/2006/R.sinicus/Guizhou/2006 | 80.2 | 95.4 | 96.5 | 99.0 | 95.6 | 88.2 | 94.3 | 95.0 |
| SARSr-Rs-BatCoV WIV1 | 80.2 | 95.8 | 96.2 | 99.5 | 95.4 | 89.0 | 93.0 | 95.0 |
| SARSr-Rf-BatCoV YNLF_31C/R.ferrumequinum/Yunnan/2013 | 80.2 | 95.8 | 96.2 | 99.3 | 95.6 | 88.7 | 93.0 | 95.0 |
| SARSr-Rf-BatCoV 16BO133/R.ferrumequinum/Korea/2016 | 79.3 | 95.1 | 96.1 | 98.7 | 94.9 | 88.2 | 92.6 | 94.6 |
| SARSr-Rf-BatCoV JTMC15/R.ferrumequinum/Jilin/2013 | 79.3 | 94.8 | 96.2 | 98.5 | 94.9 | 88.2 | 92.6 | 94.5 |
| SARSr-Rs-BatCoV BtKY72/Rhinolophus sp./Kenya/2007 | 74.8 | 95.4 | 95.0 | 96.8 | 92.8 | 87.3 | 90.6 | 92.9 |
| SARSr-Rm-BatCoV Longquan-140/R.monoceros/Zhejiang/2012 | 79.3 | 95.8 | 95.7 | 99.2 | 94.9 | 89.0 | 94.3 | 94.8 |
| SARSr-Rb-BatCoV BM48–31/BGR/2008/R.blasii/Bulgaria/2008 | 78.4 | 94.1 | 95.2 | 97.8 | 93.5 | 89.9 | 88.6 | 93.5 |
| SARSr-Rp-BatCoV ZXC21/R.pusillus/Zhejiang/2015 | 88.3 | 99.0 | 95.6 | 98.8 | 94.7 | 88.2 | 98.0 | 95.4 |
| SARSr-Rp-BatCoV ZC45/R.pusillus/Zhejiang/2017 | 92.8 | 99.0 | 95.9 | 99.3 | 94.5 | 89.0 | 98.0 | 95.8 |
| Pangolin-SARSr-CoV Guangxi/P4L/2017 | 85.5 | 97.1 | 97.9 | 98.2 | 97.0 | 94.5 | 97.7 | 96.9 |
| SARSr-Ra-BatCoV RaTG13/R.affinis/Yunnan/2013 | 96.7 | 99.3 | 99.6 | 99.7 | 99.2 | 97.7 | 100 | 99.2 |
*ADRP, ADP-ribose-1′′ phosphatase; nsp, nonstuctural protein; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2; SARSr-CoV, severe acute respiratory syndrome–related coronavirus.
Unlike other members of the subgenus Sarbecovirus, SARS-CoV-2 has a spike protein that contains a unique insertion that results in a potential cleavage site at the S1/S2 junction, which might enable proteolytic processing that enhances cell–cell fusion. SARS-CoV-2 was demonstrated to use the same receptor, human angiotensin-converting enzyme 2 (hACE2), as does SARS-CoV (7). The predicted RBD region of SARS-CoV-2 spike protein, corresponding to aa residues 318–513 of SARS-CoV (12), showed the highest (97% aa) identities with pangolin-SARSr-CoV/MP789/Guangdong and 74.1%–77.7% identities with human/civet/bat-SARSr-CoVs known to use hACE2 (Table 1). Moreover, similar to the human/civet/bat-SARSr-CoV hACE2-using viruses, the 2 deletions (5 aa and 12 aa) found in all other SARSr-BatCoVs (10) were absent in SARS-CoV-2 RBD (Appendix Figure 1). Of the 5 critical residues needed for RBD-hACE2 interaction in SARSr-CoVs (13), 3 (F472, N487, and Y491) were present in SARS-CoV-2 RBD and pangolin SARSr-CoV/MP789/2019-RBD.
Phylogenetic analysis showed that the RNA-dependent RNA polymerase gene of SARS-CoV-2 is most closely related to that of SARSr-Ra-BatCoV RaTG13, whereas its predicted RBD is closest to that of pangolin-SARSr-CoVs (Figure 1). This finding suggests a distinct evolutionary origin for SARS-CoV-2 RBD, possibly as a result of recombination. Moreover, the SARS-CoV-2 RBD was also closely related to SARSr-Ra-BatCoV RaTG13 and the hACE2-using cluster containing human/civet-SARSr-CoVs and Yunnan SARSr-BatCoVs previously successfully cultured in VeroE6 cells (4,5).
Figure 1.
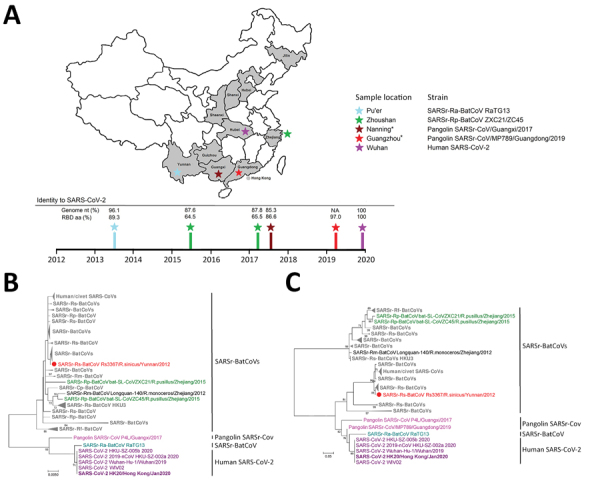
Geographic and phylogenetic comparisons of SARS-CoV-2 isolates with closely related viruses. A) Locations in China where SARS-CoV-2 first emerged (Wuhan), and where closely related viruses were found, including SARSr-Ra-BatCoV RaTG13 (Pu’er), Pangolin-SARSr-CoVs (Guangzhou and Nanning), and SARSr-Rp-BatCoV ZC45 (Zhoushan). Time of sampling and percentage genome identities to SARS-CoV-2 are shown. *Guangzhou and Nanning. The geographic origin of smuggled pangolins remains unknown. B, C) Phylogenetic analyses of RdRp (B) and RBD (C) domains of SARSr-CoVs. Trees were constructed by using maximum-likelihood methods with Jones-Taylor-Thornton plus gamma plus invariant sites (RdRp) and Whelan and Goldman plus gamma (RBD) substitution models. A total of 745 aa residues for RdRp and 177 aa residues for RBD were included in the analyses. Numbers at nodes represent bootstrap values, which were calculated from 1,000 trees. Only bootstrap values >70% are shown. Purple indicates SARS-CoV-2 (strain HK20 in bold); teal indicates SARSr-Ra-BatCoV RaTG13; pink indicates pangolin SARSr-CoVs; green indicates SARSr-Rp-BatCoVs ZXC21 and ZC45; red indicates SARSr-Rs-BatCoV Rs3367; black indicates SARSr-Rs-BatCoV Longquan-140; gray indicates remaining SARSr-BatCoVs. Dots indicate SARSr-BatCoVs reported to use angiotensin-converting enzyme 2 as receptor. Scale bars indicate estimated number of amino acid substitutions per 200 aa residues for RdRp and per 20 aa residues for RBD. SARS-CoV-2, severe acute respiratory syndrome coronavirus 2; SARSr-CoV, severe acute respiratory syndrome–related coronavirus; NA, not available; RBD, receptor-binding domain; RdRp, RNA-dependent RNA polymerase.
To identify putative recombination events, we performed sliding window analysis using SARS-CoV-2-HK20 as query and SARSr-Ra-BatCoV RaTG13, pangolin-SARSr-CoV/P4L/Guangxi/2017, SARSr-Rp-BatCoV ZC45, SARSr-Rs-BatCoV Rs3367, and SARSr-Rs-BatCoV Longquan-140 as potential parents (Figure 2; Appendix Figures 2). A similarity plot showed that SARS-CoV-2 is most closely related to SARSr-Ra-BatCoV RaTG13 in the entire genome, except for its RBD, which is closest to pangolin-SARSr-CoV/MP789/Guangdong, and shows potential recombination breakpoints. Moreover, different regions of SARS-CoV-2 genome showed different similarities to pangolin-SARSr-CoV/P4L/Guangxi/2017, SARSr-Rp-BatCoV ZC45, SARSr-Rs-BatCoV Rs3367, and SARSr-Rs-BatCoV Longquan-140, as supported by phylogenetic analysis (Appendix Figures 2, 3).
Figure 2.
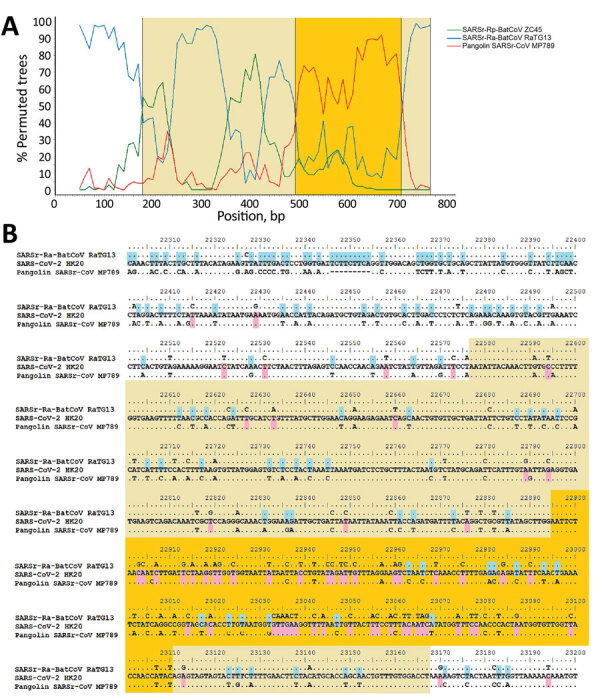
Bootscan analysis and nucleotide sequence alignment for SARS-CoV-2 isolates and closely related viruses. A) Boot scan analysis using the partial spike gene (positions 22397–23167) of SARS-CoV-2 strain HK20 as query sequence. Bootscanning was conducted with Simplot version 3.5.1 (https://sray.med.som) (F84 model; window size, 100 bp; step, 10 bp) on nucleotide alignment, generated with ClustalX (http://www.clustal.org). B) Multiple alignment of nucleotide sequences from genome positions 22300 to 23700. Yellow indicates receptor binding domain; orange indicates receptor binding motif; pink indicates bases conserved between SARS-CoV-2 HK20 and Pangolin-SARSr-CoV/MP789/Guangdong/2019; and blue indicates bases conserved between SARS-CoV-2 HK20 and SARSr-Ra-BatCoVs RaTG13.
Sequence alignment around the RBD supported potential recombination between SARSr-Ra-BatCoV RaTG13 and pangolin-SARSr-CoV/MP789/Guangdong/2019 and the receptor-binding motif region showing exceptionally high sequence similarity to that of pangolin-SARSr-CoV/MP789/Guangdong/2019. This finding suggested that SARS-CoV-2 might be a recombinant virus between viruses closely related to SARSr-Ra-BatCoV RaTG13 and pangolin-SARSr-CoV/MP789/Guangdong/2019.
Conclusions
Despite the close relatedness of SARS-CoV-2 to bat and pangolin viruses, none of the existing SARSr-CoVs represents its immediate ancestor. Most of the genome region of SARS-CoV-2 is closest to SARSr-Ra-BatCoV-RaTG13 from an intermediate horseshoe bat in Yunnan, whereas its RBD is closest to that of pangolin-SARSr-CoV/MP789/Guangdong/2019 from smuggled pangolins in Guangzhou. Potential recombination sites were identified around the RBD region, suggesting that SARS-CoV-2 might be a recombinant virus, with its genome backbone evolved from Yunnan bat virus–like SARSr-CoVs and its RBD region acquired from pangolin virus–like SARSr-CoVs.
Because bats are the major reservoir of SARSr-CoVs and the pangolins harboring SARSr-CoVs were captured from the smuggling center, it is possible that pangolin SARSr-CoVs originated from bat viruses as a result of animal mixing, and there might be an unidentified bat virus containing an RBD nearly identical to that of SARS-CoV-2 and pangolin SARSr-CoV. Similar to SARS-CoV, SARS-CoV-2 is most likely a recombinant virus originated from bats.
The ability of SARS-CoV-2 to emerge and infect humans is likely explained by its hACE2-using RBD region, which is genetically similar to that of culturable Yunnan SARSr-BatCoVs and human/civet-SARSr-CoVs. Most SARSr-BatCoVs have not been successfully cultured in vitro, except for some Yunnan strains that had human/civet SARS-like RBDs and were shown to use hACE2 (4,5). For example, SARSr-Rp-BatCoV ZC45, which has an RBD that is more divergent from that of human/civet-SARSr-CoVs, did not propagate in VeroE6 cells (6). Factors that determine hACE2 use among SARSr-CoVs remain to be elucidated.
Although the Wuhan market was initially suspected to be the epicenter of the epidemic, the immediate source remains elusive. The close relatedness among SARS-CoV-2 strains suggested that the Wuhan outbreak probably originated from a point source with subsequent human-to-human transmission, in contrast to the polyphyletic origin of Middle East respiratory syndrome coronavirus (14). If the Wuhan market was the source, a possibility is that bats carrying the parental SARSr-BatCoVs were mixed in the market, enabling virus recombination. However, no animal samples from the market were reported to be positive. Moreover, the first identified case-patient and other early case-patients had not visited the market (15), suggesting the possibility of an alternative source.
Because the RBD is considered a hot spot for construction of recombinant CoVs for receptor and viral replication studies, the evolutionarily distinct SARS-CoV-2 RBD and the unique insertion of S1/S2 cleavage site among Sarbecovirus species have raised the suspicion of an artificial recombinant virus. However, there is currently no evidence showing that SARS-CoV-2 is an artificial recombinant, which theoretically might not carry signature sequences. Further surveillance studies in bats are needed to identify the possible source and evolutionary path of SARS-CoV-2.
Additional information on possible bat origin of severe acute respiratory syndrome coronavirus virus 2.
Acknowledgments
This study was partly supported by the theme-based research scheme (project no. T11-707/15-R) of the University Grant Committee; Health and Medical Research Fund of the Food and Health Bureau of HKSAR; Consultancy Service for Enhancing Laboratory Surveillance of Emerging Infectious Disease for the HKSAR Department of Health and the University Development Fund of the University of Hong Kong.
Biography
Dr. Lau is a professor and head of the Department of Microbiology at The University of Hong Kong, Hong Kong, China. Her primary research interest is using microbial genomics for studying emerging infectious diseases, including coronaviruses.
Footnotes
Suggested citation for this article: Lau SKP, Luk HKH, Wong ACP, Li KSM, Zhu L, He Z, et al. Possible bat origin of severe acute respiratory syndrome coronavirus 2. Emerg Infect Dis. 2020 Jul [date cited]. https://doi.org/10.3201/eid2607.200092
These authors contributed equally to this article.
References
- 1.Coronaviridae Study Group of the International Committee on Taxonomy of Viruses. The species Severe acute respiratory syndrome-related coronavirus: classifying 2019-nCoV and naming it SARS-CoV-2. Nat Microbiol. 2020;5:536–44. 10.1038/s41564-020-0695-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.World Health Organization. Coronavirus disease 2019 (COVID-19) situation report 50, March 10, 2020. [cited 2020 Apr 11]. https://www.who.int/docs/default-source/coronaviruse/situation-reports/20200310-sitrep-50-covid-19.pdf
- 3.Lau SK, Woo PC, Li KS, Huang Y, Tsoi HW, Wong BH, et al. Severe acute respiratory syndrome coronavirus-like virus in Chinese horseshoe bats. Proc Natl Acad Sci U S A. 2005;102:14040–5. 10.1073/pnas.0506735102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ge XY, Li JL, Yang XL, Chmura AA, Zhu G, Epstein JH, et al. Isolation and characterization of a bat SARS-like coronavirus that uses the ACE2 receptor. Nature. 2013;503:535–8. 10.1038/nature12711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hu B, Zeng LP, Yang XL, Ge XY, Zhang W, Li B, et al. Discovery of a rich gene pool of bat SARS-related coronaviruses provides new insights into the origin of SARS coronavirus. PLoS Pathog. 2017;13:e1006698. 10.1371/journal.ppat.1006698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hu D, Zhu C, Ai L, He T, Wang Y, Ye F, et al. Genomic characterization and infectivity of a novel SARS-like coronavirus in Chinese bats. Emerg Microbes Infect. 2018;7:154. 10.1038/s41426-018-0155-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhou P, Yang XL, Wang XG, Hu B, Zhang L, Zhang W, et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–3. 10.1038/s41586-020-2012-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu P, Chen W, Chen JP. Viral metagenomics revealed sendai virus and coronavirus infection of Malayan pangolins (Manis javanica). Viruses. 2019;11:E979. 10.3390/v11110979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lam TT, Shum MH, Zhu HC, Tong YG, Ni XB, Liao YS, et al. Identifying SARS-CoV-2 related coronaviruses in Malayan pangolins. Nature. 2020; Epub ahead of print. 10.1038/s41586-020-2169-0 [DOI] [PubMed] [Google Scholar]
- 10.Luk HKH, Li X, Fung J, Lau SKP, Woo PCY. Molecular epidemiology, evolution and phylogeny of SARS coronavirus. Infect Genet Evol. 2019;71:21–30. 10.1016/j.meegid.2019.03.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lau SKP, Li KS, Huang Y, Shek CT, Tse H, Wang M, et al. Ecoepidemiology and complete genome comparison of different strains of severe acute respiratory syndrome-related Rhinolophus bat coronavirus in China reveal bats as a reservoir for acute, self-limiting infection that allows recombination events. J Virol. 2010;84:2808–19. 10.1128/JVI.02219-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wong SK, Li W, Moore MJ, Choe H, Farzan M. A 193-amino acid fragment of the SARS coronavirus S protein efficiently binds angiotensin-converting enzyme 2. J Biol Chem. 2004;279:3197–201. 10.1074/jbc.C300520200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li W, Zhang C, Sui J, Kuhn JH, Moore MJ, Luo S, et al. Receptor and viral determinants of SARS-coronavirus adaptation to human ACE2. EMBO J. 2005;24:1634–43. 10.1038/sj.emboj.7600640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lau SK, Wernery R, Wong EY, Joseph S, Tsang AK, Patteril NA, et al. Polyphyletic origin of MERS coronaviruses and isolation of a novel clade A strain from dromedary camels in the United Arab Emirates. Emerg Microbes Infect. 2016;5:e128. 10.1038/emi.2016.129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huang C, Wang Y, Li X, Ren L, Zhao J, Hu Y, et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497–506. 10.1016/S0140-6736(20)30183-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional information on possible bat origin of severe acute respiratory syndrome coronavirus virus 2.


