The terpene synthase gene family contributes to variations in cannabis metabolite profiles.
Abstract
Cannabis (Cannabis sativa) resin is the foundation of a multibillion dollar medicinal and recreational plant bioproducts industry. Major components of the cannabis resin are the cannabinoids and terpenes. Variations of cannabis terpene profiles contribute much to the different flavor and fragrance phenotypes that affect consumer preferences. A major problem in the cannabis industry is the lack of proper metabolic characterization of many of the existing cultivars, combined with sometimes incorrect cultivar labeling. We characterized foliar terpene profiles of plants grown from 32 seed sources and found large variation both within and between sets of plants labeled as the same cultivar. We selected five plants representing different cultivars with contrasting terpene profiles for clonal propagation, floral metabolite profiling, and trichome-specific transcriptome sequencing. Sequence analysis of these five cultivars and the reference genome of cv Purple Kush revealed a total of 33 different cannabis terpene synthase (CsTPS) genes, as well as variations of the CsTPS gene family and differential expression of terpenoid and cannabinoid pathway genes between cultivars. Our annotation of the cv Purple Kush reference genome identified 19 complete CsTPS gene models, and tandem arrays of isoprenoid and cannabinoid biosynthetic genes. An updated phylogeny of the CsTPS gene family showed three cannabis-specific clades, including a clade of sesquiterpene synthases within the TPS-b subfamily that typically contains mostly monoterpene synthases. The CsTPSs described and functionally characterized here include 13 that had not been previously characterized and that collectively explain a diverse range of cannabis terpenes.
The pistillate flowers of cannabis (Cannabis sativa) are densely covered with glandular trichomes that produce and accumulate a resin that is rich in cannabinoids as well as monoterpenes and sesquiterpenes (Turner et al., 1978; Brenneisen and elSohly, 1988; Livingston et al., 2020). Cannabinoids are responsible for the various medicinal and psychoactive properties of cannabis. The terpenes of cannabis resin, which include more than a dozen different monoterpenes and over a hundred different sesquiterpenes, account for much of the diverse organoleptic impressions of cannabis products (Fig. 1; Fischedick et al., 2010; Casano et al., 2011; Booth and Bohlmann, 2019). Cannabis is broadly categorized into three major chemotypes based on the ratio of Δ9-tetrahydrocannabinolic acid (THCA) to cannabidiolic acid (CBDA). Type I has high amounts of THCA; type II has approximately equal amounts of THCA and CBDA, and type III is CBDA-dominant (de Meijer et al., 2003). Across these three major chemotypes, terpene profiles show much variation between different cultivars, with myrcene, limonene, α-pinene, α-terpinene, or β-caryophyllene as major variable components (Fischedick et al., 2010; Fischedick, 2017; Richins et al., 2018; Reimann-Philipp et al., 2019).
Figure 1.
Terpene and cannabinoid biosynthetic pathways. Precursors and intermediates are shown in black, final product classes in green, and enzyme names in purple. Cannabinoid pathway: FAD, Fatty acid desaturase; LOX, lipoxygenase; HPL, hydroperoxide lyase; AAE, acyl activating enzyme. MEP pathway: CMK, 4-Diphosphocytidyl-2-C-methyl-d-erythritol kinase; MDS, 2-C-methyl-d-erythritol 2,4-cyclodiphosphate synthase; MEV pathway: HMGS, 3-HMG-CoA synthase; HMGR, HMG-CoA reductase; MK, MEV kinase; PMK, MEV-3-phosphate kinase; MPDC, MEV-5-pyrophosphate decarboxylase; IDI, isopentenyl diphosphate isomerase; FPPS, farnesyl diphosphate synthase.
Terpene synthases (TPSs), which are encoded in large TPS gene families with several subfamilies, produce the diversity of cyclic and acyclic terpene core structures found in plants (Chen et al., 2011). In angiosperms, the TPS-a subfamily generally contains sesquiterpene synthases (sesqui-TPSs), and the TPS-b subfamily contains primarily monoterpene synthases (mono-TPSs) and hemiterpene synthases. Acyclic monoterpenes are also produced by members of the TPS-g subfamily. The TPS gene family has undergone lineage-specific expansions, leading to blooms of related TPS enzymes, as shown for example in grapevine (Vitis vinifera; Martin et al., 2010), eucalyptus (Eucalyptus grandis; Külheim et al., 2015), and tomato (Solanum lycopersicum; Falara et al., 2011). Terpenes and cannabinoids share common isoprenoid precursors (Fig. 1). The most abundant cannabinoids in different cannabis cultivars are THCA and CBDA, which are produced by cannabinoid synthases from cannabigerolic acid (CBGA; Sirikantaramas et al., 2004; Taura et al., 2007). CBGA is formed by condensation of the monoterpene precursor geranyl diphosphate (GPP) with the aromatic polyketide olivetolic acid (OA; Fellermeier and Zenk, 1998).
At least 55 different CsTPS gene models have previously been reported (Supplemental Table S1), but only 14 have been functionally characterized, including eight mono-TPSs and six sesqui-TPSs (Gunnewich et al., 2007; Booth et al., 2017; Allen et al., 2019; Zager et al., 2019; Livingston et al., 2020). The 14 functionally characterized CsTPSs account for some of the major terpenes in cannabis (e.g. α-pinene, limonene, myrcene, β-caryophyllene) as well as some of the rare compounds (e.g. terpinene, hedycaryol, and alloaromadendrene). However, much of the terpene variation in cannabis remains to be explored. While this article was in preparation, Zager et al. (2019) reported gene networks associated with terpenoid biosynthesis in seven different cannabis cultivars, revealing relationships between gene expression and terpenoid accumulation.
Cv Purple Kush (PK) has been established as a reference for genomic research in cannabis (van Bakel et al., 2011; Booth et al., 2017; Laverty et al., 2019). Here, we report the terpene profile of cv PK and its genome annotation for CsTPS genes and other genes of terpenoid and cannabinoid biosynthesis. We investigated variations in terpene profiles in flowers (Fig. 2) of six different cannabis cultivars, including cv PK, based on metabolite analysis, trichome-specific RNA-sequencing (RNA-seq) transcriptome analysis, and functional characterization of CsTPSs.
Figure 2.
Stages of floral maturation. Drawing showing four stages of floral maturation within the inflorescence. Representative photographs are of a cv PK inflorescence at four different stages, from youngest (1) to oldest (4). Different stages are characterized as follows: (1) very pale pistils and few to no stalked trichomes; (2) no browned pistils and ∼50% stalked trichomes; (3) pistils beginning to brown, with entirely stalked trichomes; (4) entirely browned pistils, with brown or amber trichome heads. For this study, metabolite analyses were performed at stages 1 and 3.
RESULTS
Annotation of the cv PK Reference Genome
As a foundation for our study of CsTPS genes and their role in terpenoid variation in different cannabis cultivars, we annotated CsTPS genes and other genes involved in isoprenoid and cannabinoid biosynthesis in the cv PK reference genome. We identified 19 complete CsTPS gene models in cv PK (Fig. 3), including four clusters of two to five genes that are more similar in sequence to one another than they are to those of any other gene model. Sequences for CsTPS6 were identified at two adjacent loci. In addition, five partial CsTPS genes were found in the cv PK genome, likely representing pseudogenes. We also located gene models for all known steps in isoprenoid and cannabinoid biosynthesis, including the plastidial methylerythritol phosphate (MEP) pathway leading to cannabinoids and monoterpenes. Many of the isoprenoid pathway genes, notably 1-deoxy-d-xylulose-5-phosphate synthase (DXS) and 1-deoxy-d-xylulose-5-phosphate reductase (DXR), have multiple copies. Similar to the CsTPSs, several other isoprenoid and cannabinoid pathway genes are arranged in multicopy clusters, namely genes encoding DXS, DXR, two copies of the polyketide synthase (PKS) responsible for producing OA (Taura et al., 2009), OA cyclase (OAC), aromatic prenyltransferases involved in cannabinoid and cannflavin biosynthesis (aPTs; Page and Boubakir, 2011; Luo et al., 2019; Rea et al., 2019), and cannabinoid synthases THCAS and CBDAS. None of the CsTPS gene models clustered with any other genes known to be related to terpenoid or cannabinoid biosynthesis. While there are no obvious biosynthetic clusters, CBDAS, GPP synthase (GPPS) small subunit, and CsTPS9 are positioned within a 10-megabase region of the cv PK genome.
Figure 3.
Genome locations of genes related to terpenoid and cannabinoid biosynthesis. Scaffolds are from Laverty et al. (2019). TPSs are shown in pink, UbiA family prenyltransferases in blue, MEP pathway genes in green, and cannabinoid biosynthetic genes in black. Loci with identical labels represent duplicated genes.
Variation of Foliar Terpene Profiles within and between Cultivars
To explore variation of terpene biosynthesis in cultivars with different terpene profiles, we initially grew plants from 32 seeds, which according to the supplier’s information, represented eight different cultivars: cv Lemon Skunk (LS), cv CBD Skunk Haze (CSH), cv Blue Cheese (BC), cv Afghan Kush (AK), cv Chocolope (Choc), cv Blueberry, cv Vanilla Kush, and cv Jack Herer. The initial metabolite analysis was done with leaf samples to enable subsequent selection of individual plants for clonal propagation. Once plants have reached the flowering stage, propagation from cuttings becomes inefficient.
In total, we detected 48 different terpene peaks in the gas chromatography-mass spectrometry (GC/MS) analysis of foliar extracts across all 32 individuals, of which 11 were annotated as monoterpenes and 37 as sesquiterpenes (Supplemental Fig. S1). Of these, only three monoterpenes, namely myrcene, α-pinene, and limonene, and two sesquiterpenes, β-caryophyllene and α-humulene, were present in every individual. To select plants representing the most contrasting terpene profiles for further study, we performed a principal component analysis (PCA). Principal components (PCs) 1 and 2 account for 26.4% and 19.7%, respectively, of the terpene variation among the 32 plants (Fig. 4A). Most plants cluster toward the lower end of PC2. All plants labeled as cv CSH clustered together. Only one cv Jack Herer seed germinated, so variability and clustering could not be assessed for this cultivar. For the other plants, there was as much variation among plants with the same cultivar label as between plants labeled as different cultivars. Five individual plants, one from each quadrant and one from near the center of the PCA plot, were selected for clonal propagation and detailed characterization including terpene and cannabinoid analysis of flowers, floral trichome transcriptome sequencing, transcript expression analysis, and CsTPS discovery and characterization. The selected individuals represent plants identified as belonging to the cultivars cv AK, cv BC, cv Choc, cv CSH, and cv LS.
Figure 4.

Foliar terpene profiles differentiate cannabis plants grown from seeds. A, First two dimensions (Dim) of a PCA of foliar terpene profiles from 32 cannabis plants. Dim1 accounts for 26.35% of the variance between individuals and Dim2 accounts for 19.73%. Colors indicate the names under which seeds were obtained. Boxed points are individuals that were chosen for clonal propagation and further characterization. B, Unsupervised hierarchical cluster analysis of 46 terpenoid peaks (x axis) in 32 cannabis seedlings. Ward’s minimum variance was used as the clustering method. Seven clusters, indicated by the colored boxes, were determined by inertia gain.
Hierarchical cluster analysis of foliar terpenes from the 32 plants was used to determine which compounds account for most of the differences between individuals, and to identify compounds that co-occur. Of the 48 total terpene peaks identified, 23 were found to account for the significant variation in seven groups. Bisabolol contributed the most to differentiation between cultivars, followed by (E)-β-farnesene (Fig. 4B). Two guaiane-type sesquiterpenes clustered together and apart from other compounds. A guaiane- and an eremophilane-type sesquiterpene also clustered together and apart from other compounds. The two sesquiterpenes β-caryophyllene and α-humulene, which are produced by the same CsTPS (Booth et al., 2017), formed a unique cluster. Myrcene was a member of this clade, but did not cluster with any other compounds. The remaining 39 compounds grouped into a larger cluster. The monoterpenes camphene and α-pinene clustered with a guaiane-type sesquiterpene and three cadinane-type compounds. The same group also included β-bisabolene and eudesma-3,7(11)-diene, which were closely related to the largest cluster consisting of another bisabolane-type sesquiterpene, an unidentified sesquiterpene, terpinolene, linalool, limonene, and a himachalane-type sesquiterpene. The remaining compounds did not account for a significant proportion of the variation between terpene profiles.
Flower and Foliar Metabolite \Pprofiles from Clonal Plants of Six Cultivars
Three clonal replicates were made from each of the selected five plants and grown in a hydroponic growth chamber, and flowering was induced after 5 weeks of vegetative growth. Terpenes and cannabinoids were analyzed in samples from flowers and foliage of all 15 plants. For all five cultivars, terpene profiles were qualitatively similar in foliage and flower samples, but quantities of terpenes were much higher in flowers (Fig. 5; Supplemental Table S2). The five cultivars included four that are THCA-dominant and one with approximately equal amounts of THCA and CBDA (Supplemental Table S3). Foliar terpenes were dominated by sesquiterpenes (Fig. 5; Supplemental Fig. S1), with a total terpene content between 0.5 and 1.1 mg g−1 dry weight (DW). In contrast, terpene levels were between 4.9 and 7.3 mg g−1 DW in juvenile flowers (Fig. 2) and between 9.3 and 13.6 mg g−1 DW in mature flowers at 15 d post floral initiation (DPI; Fig. 5; Supplemental Fig. S1). The proportion of monoterpene increased as flowers developed from juvenile to mature. In total, 15 different monoterpenes and 27 different sesquiterpenes were separated by GC/MS and quantified in mature flowers of the five cultivars (Table 1; Supplemental Fig. S2). In addition, several other terpenes were below the limit of quantification. Myrcene was the most abundant terpene in cv CHS, cv AK, and cv BC. In cv LS and cv Choc, the most abundant monoterpenes were (+)-α-pinene and (−)-limonene, respectively. In four of the five cultivars, β-caryophyllene was the dominant sesquiterpene. In cv AK, germacrene B was the dominant sesquiterpene. (E)-β-farnesene was present in all samples and was a major component of cv Choc and cv AK.
Figure 5.
Terpene content in leaves and flowers of five different cannabis cultivars. Fan leaves were taken from flowering plants at ∼14 DPI. Juvenile flowers of stage 1 (Fig. 2) were sampled in triplicate, at the same time as the leaves, from three clones of each cultivar. Mature flowers of stage 3 (Fig. 2) were sampled in triplicate from three clones of each cultivar between 51 and 60 DPI. Error bars represent the mean ± se across nine samples.
Table 1. Amounts of terpenes in mature flowers of different cannabis cultivars.
Each value is the mean of three replicates from three clones of each cultivar, with the sd of nine samples in parentheses. Metabolites marked with an asterisk have been verified using authentic standards; others were identified based on MS and RI. Compounds below the limit of quantification across all samples are omitted. RI was calculated on a DB-Wax GC column. Note that cv PK data were obtained from plants grown for a separate study, and data may not be directly comparable with those of the other five cultivars. n.d., Not detected; tr, trace (<1 μg).
| RI | Compound Identifier | Mean (sd) [µg g−1 DW] | |||||
|---|---|---|---|---|---|---|---|
| cv LS | cv Choc | cv AK | cv CSH | cv BC | cv PK | ||
| 934 | (+)-α-pinene* | 1,849(768) | 199 (134) | n.d. | 1,409 (2) | 1,024 (206) | 86 (25) |
| 934 | (−)-α-pinene* | 139 (58) | 98 (61) | 94 (74) | n.d. | 10 (10) | 10 (3) |
| 1,028 | (+)-Camphene* | 37 (16) | n.d. | n.d. | n.d. | 21 (13) | n.d. |
| 1,028 | (−)-Camphene* | 59 (25) | 92 (107) | 18 (3) | 48 (4) | 30 (5) | 44 (14) |
| 1,067 | (+)-β-pinene* | 174 (88) | 83 (13) | n.d. | n.d. | 57 (12) | n.d. |
| 1,067 | (−)-β-pinene* | 522 (219) | 312 (65) | 615 (166) | 563 (50) | 517 (108) | 1,337 (104) |
| 1,123 | Myrcene* | 1,387 (286) | 1,369 (1273) | 6,222 (1674) | 7,680 (1065) | 4,268 (275) | 5,683 (1,583) |
| 1,158 | (−)-Limonene* | 951 (118) | 2,644 (380) | 525 (151) | 627 (77) | 314 (42) | 2,405 (726) |
| 1,167 | β-phellandrene* | 60 (11) | 162 (21) | 51 (15) | 97 (9) | 46 (26) | n.d. |
| 1,206 | (E)-β-ocimene* | 214 (38) | 1,382 (1776) | 316 (103) | 191 (201) | n.d. | n.d. |
| 1,237 | Terpinolene* | 31 (7) | 300 (123) | 33 (24) | 35 (5) | 12 (1) | 35 (11) |
| 1,416 | Monoterpene alcohol | 8 (3) | n.d. | tr | 5 (0.8) | tr | n.d. |
| 1,434 | δ-elemene | tr | tr | tr | 5 (1) | tr | 392 (115) |
| 1,488 | (+)-linalool* | 215 (58) | 382 (444) | 149 (42) | 55 (21) | 149 (23) | 503 (128) |
| 1,498 | 2-pinanol | 87 (26) | 160 (195) | 44 (9) | 47 (3) | 36 (4) | 99 (31) |
| 1,515 | Sesquiterpene 1 | 5 (6) | 15 (16) | 19 (2) | tr | 4 (1) | 10 (10) |
| 1,523 | Fenchol* | 53 (26) | 87 (105) | 14 (2) | 28 (3) | 9 (2) | n.d. |
| 1,529 | Sesquiterpene 2 | 5 (9) | 42 (44) | 38 (12) | n.d. | n.d. | n.d. |
| 1,533 | Sesquiterpene 3 | tr | 3 (4) | 13 (4) | n.d. | n.d. | n.d. |
| 1,541 | α-bergamotene | 24 (25) | 224 (204) | 268 (34) | 18 (7) | 66 (3) | 320 (103) |
| 1,546 | Guiane 1 | 116 (45) | 41 (34) | tr | 13 (3) | 7 (7) | n.d. |
| 1,554 | β-caryophyllene* | 271 (167) | 314 (211) | 743 (129) | 156 (23) | 242 (51) | 348 (69) |
| 1,595 | γ-elemene | 154 (68) | 104 (94) | 265 (52) | 68 (3) | 142 (34) | 317 (71) |
| 1,617 | (E)-β-farnesene* | 19 (14) | 357 (410) | 295 (111) | 8 (6) | 54 (45) | 367 (104) |
| 1,620 | Sesquiterpene 4 | 4 (7) | tr | 13 (2) | n.d. | 2 (1) | n.d. |
| 1,623 | α-humulene* | 169 (96) | 362 (163) | 820 (163) | 140 (28) | 220 (45) | 108 (21) |
| 1,636 | Borneol | 16 (13) | 32 (39) | 12 (9) | 12 (2) | n.d. | n.d. |
| 1,641 | (+)-α-terpineol* | 210 (87) | 384 (459) | 139 (27) | 113 (10) | 83 (2) | 235 (72) |
| 1,664 | Guiane 2 | 196 (131) | 2 (4) | 38 (4) | 4 (4) | 17 (7) | n.d. |
| 1,671 | Eudesmane 1 | 552 (442) | 665 (502) | 677 (213) | 40 (23) | 330 (160) | 564 (85) |
| 1,677 | α-selinene | 121 (87) | 40 (9) | 131 (22) | 94 (26) | 40 (10) | 57 (33) |
| 1,682 | Eudesma-3,7(11)-diene | 92 (71) | 24 (25) | 204 (33) | 50 (15) | 89 (12) | 334 (96) |
| 1,696 | α-farnesene | n.d. | n.d. | n.d. | 2 (3) | 23 (11) | 58 (43) |
| 1,700 | Sesquiterpene 5 | 489 (321) | 190 (114) | 645 (81) | 475 (207) | 281 (49) | 91 (64) |
| 1,713 | Valencene | n.d. | n.d. | n.d. | n.d. | 1 (0.2) | 37 (11) |
| 1,714 | δ-selinene | 137 (95) | 110 (102) | 359 (43) | 90 (16) | 154 (37) | 101 (29) |
| 1,721 | Cyclounatriene | 130 (149) | 20 (6) | 42 (10) | 86 (30) | 29 (13) | n.d. |
| 1,724 | Sesquiterpene 6 | 45 (35) | 24 (25) | 155 (35) | 42 (14) | 31 (11) | n.d. |
| 1,729 | Sesquiterpene 7 | 489 (321) | 190 (114) | 645 (81) | 475 (207) | 281 (49) | 73 (29) |
| 1,773 | Germacrene B* | 764 (315) | 489 (422) | 1268 (275) | 344 (24) | 711 (179) | 247 (71) |
| 1,931 | Caryophyllene oxide | 2 (3) | n.d. | 3 (2) | n.d. | n.d. | n.d. |
| 2,067 | Guiaol | n.d. | 492 (116) | n.d. | 506 (219) | n.d. | n.d. |
| 2,136 | γ-eudesmol | 77 (51) | 383 (225) | 59 (9) | 412 (219) | 101 (59) | 43 (32) |
| 2,158 | α-bisabolol* | 235 (178) | n.d. | 24 (35) | 327 (18) | 10 (11) | 93 (26) |
| 2,108 | Bulnesol | n.d. | 354 (372) | n.d. | 312 (151) | n.d. | n.d. |
A terpene profile for cv PK (Table 1) was produced with three clones of the cv PK plant that was sequenced for the reference genome; however, these plants were grown under different conditions. The terpene content of floral trichomes of cv PK, induced to flower after 4 weeks of vegetative growth, peaked at 21 mg g−1 DW. In cv PK flowers, we detected 49 different terpenes, including 15 monoterpenes and 34 sesquiterpenes. Monoterpenes were dominated by myrcene, (−)-limonene, and (+)-linalool, with lesser amounts of (+)-β-pinene, α-terpineol, (−)-α-pinene, (−)-camphene, and (Z)-β-ocimene. The most abundant sesquiterpene was β-caryophyllene, followed by γ-elemene and a eudesmane-type olefin.
Transcriptomes of Floral Trichomes Are Enriched for Terpene and Cannabinoid Biosynthesis
We produced 15 separate trichome-specific transcriptomes from three plants for each of the five cultivars. Trichome heads were isolated from mature flowers (Fig. 2; Supplemental Fig. S3) from individual clonal plants prior to signs of floral senescence. Mature flowers are characterized by apparent lack of unstalked glandular trichomes, as glandular trichomes have matured to the stalked stage (Livingston et al., 2020), and with >80% of pistils turning from white/green to brown. Total RNA was extracted from isolated trichome heads and used for RNA-seq. We initially assembled sequences from all five cultivars into a single pooled transcriptome. The normalization of a pooled transcriptome allows quantitative comparison among cultivars. The pooled assembly contained 599,285 nonredundant contigs with an average length of 511 bp (Supplemental Table S4). The trichome transcriptome raw sequence data are deposited in the National Center for Biotechnology Information (NCBI) Sequence Read Archive (accession no. PRJNA599437).
To ensure that the time point of floral and glandular trichome development selected for RNA isolation represented active terpene and cannabinoid biosynthesis, we examined the transcriptome for genes of these pathways. In general, the pooled transcriptome assembly included at least one full-length transcript corresponding to each known step in the cannabinoid and terpene biosynthetic pathways. The 200 most highly expressed genes in the trichome transcriptome included isoprenoid biosynthesis enzymes [(E)-4-hydroxy-3-methyl-but-2-enyl pyrophosphate (HMB-PP) synthase (HDS), HMB-PP reductase (HDR), isopentenyl diphosphate isomerase, and GPPS], six cannabinoid biosynthetic enzymes (CBDAS, PKS, OAC, THCAS, and two CBGA synthases [aPT1 and aPT4]), and seven CsTPSs (Supplemental Table S5). Three contigs annotated as fatty acid desaturase, which may be involved in biosynthesis of cannabinoid fatty acid precursors or cell membranes, were also highly expressed. Additionally, several contigs annotated as lipid transfer proteins or ABCG transporters were highly abundant.
PCA was done on the complete set of 15 trichome transcriptomes. The three replicates of each cultivar clustered together, and cultivars were well differentiated (Fig. 6A). cv LS and cv Choc were the two cultivars most similar to each other, while cv AK had the most distance from the other cultivars. We used unsupervised cluster analysis to test for patterns of expression of contigs annotated as terpene or cannabinoid biosynthesis. Contigs were selected by mutual best tBLASTn hit against known sequences involved in isoprenoid and cannabinoid biosynthesis (Fig. 6B). The 26 contigs identified as putatively involved in resin biosynthesis clustered into four groups. Contigs associated with the core MEP pathway (DXS, DXR, HDS, and GPPS) clustered with cannabinoid biosynthetic genes acyl activating enzyme, aPT1, aPT4, and THCAS. Mevalonate (MEV) pathway genes grouped into two clusters, which also included the MEP pathway gene methylerythritol phosphate cytidyltransferase, CBDAS, and an aPT4 contig. A second cluster of MEV contigs were much less highly expressed on average, and also included isopentenyl phosphate kinase (IPK) and CBDAS. The final cluster had the highest average expression levels, and included cannabinoid biosynthetic genes PKS and OAC, as well as the MEP pathway gene HDR and a version of IPK.
Figure 6.
Gene expression in floral trichomes of five cannabis cultivars. A, Whole-transcriptome PCA of the first two dimensions (Dim). B, Heatmap and expression of contigs representing genes annotated as terpene or cannabinoid biosynthesis. Colors indicate row-wise Z-score, or standard deviations from the mean. Gray bars at right show the average log2 CPM across 24 samples for eight individuals with three technical replicates. For the bar diagram, cv Choc 3 was treated as an outlier and not included in the log-mean expression results. C, Volcano plots showing differentially expressed contigs for four cultivars compared to cv BC. The P-values were determined using a modified Student’s t test with the R package “limma” (Ritchie et al., 2015). Significance categories were not significant (NS; gray), significant at a log2 fold change of 2 (Log2 FC; green); significant at an adjusted P-value of 0.05 (P; blue); and significant by both fold change and adjusted P-value (P & Log2 FC; red). Contigs labeled with names are those shown with yellow diamonds, representing transcripts that may be associated with resin biosynthesis. Green numbers indicate the number of transcript contigs in each cultivar with abundance significantly higher compared to cv BC, and red numbers the number of transcript contigs significantly lower compared to cv BC. AAE1, Acyl activating enzyme; PKS, polyketide synthase; OAC, olivetolic acid cyclase; 1-deoxy-d-xylulose-5-phosphate reductase: MCT, 2-C-methyl-d-erythritol 4-phosphate cytidylyltransferase; HDS, HMB-PP synthase; HDR, HMB-PP reductase; GPPS lsu, GPPS large subunit; GPPS ssu: GPPS small subunit; HMGS, 3-HMG-CoA synthase; HMGR, HMG-CoA reductase; MK, MEV kinase; PMK, MEV-3-phosphate kinase; FPPS, farnesyl diphosphate synthase.
Next, we performed a differential gene expression analysis across the five cultivars with all contigs that had expression levels of at least 100 counts per million (CPM). cv BC was used as the reference, because it placed near the center of the PCA of foliar terpene variation (Fig. 4A), and the other four cultivars were compared against cv BC. Differential gene expression analysis was performed with an adjusted P-value cutoff of 0.05 and a log2 fold change cutoff of 3. In total, across the cultivars, 25,218 contigs were differentially expressed relative to cv BC; 19,987 were upregulated and 19,263 were downregulated. Contigs were identified with at least 95% identity to known enzymes involved in cannabinoid and terpene biosynthesis, but most were not significantly differentially expressed in any cultivar compared to cv BC (Fig. 6C). Most notably, CBDAS was highly upregulated in cv CSH, the only cultivar to produce CBD as a major cannabinoid. CsPT1 was downregulated in cv CSH, and CsPT4 was downregulated in cv LS. HMGR, a component of the MEV pathway, was upregulated in cv LS. OAC was downregulated in cv Choc.
CsTPS Gene Discovery
For discovery and quantification of CsTPS transcripts, separate transcriptomes were assembled for each cultivar (Supplemental Table S4). While separate transcriptomes do not permit quantitative comparison between cultivars, they eliminate the risk of quantitation errors from mapping kmers from similar transcripts between cultivars. We used the RNA-Bloom assembler (Nip et al., 2019), designed for single-cell RNA-seq libraries, to capture the diversity of sequences across the five cultivars while reducing the possibility of chimeric contigs. Contigs with >98% predicted amino acid sequence identity were collapsed under the longest representative sequence. These five single-cultivar transcriptomes and the previously published cv PK trichome transcriptome (van Bakel et al., 2011) were searched with BLASTX to identify known (Gunnewich et al., 2007; Booth et al., 2017; Zager et al., 2019; Livingston et al., 2020) as well as new CsTPS sequences. Sequences representing all but three of the previously functionally characterized and unique CsTPSs (a total of 18; Supplemental Table S1) were present in the transcriptomes of at least one of the five cultivars (Booth et al., 2017; Allen et al., 2019; Zager et al., 2019; Livingston et al., 2020). The three missing CsTPSs were CsTPS13, CsTPS14, and CsTPS33. When we screened the transcriptomes of the six different cultivars, we found a total of 33 unique and apparently full-length CsTPS sequences, including two that we annotated as copalyl diphosphate synthase (CsTPS65) and ent-kaurene synthase (CsTPS66) of gibberellin biosynthesis.
A phylogeny of the predicted amino acid sequences of the 33 CsTPSs together with TPSs from other plant species placed CsTPSs into the subfamilies TPS-a, TPS-b, TPS-c, TPS-e/f, and TPS-g (Fig. 7; Supplemental Table S6). Within the TPS-a subfamily, all CsTPSs fall into one cluster with TPSs from hops (Humulus lupulus) as the nearest noncannabis members. Within TPS-b, the CsTPSs fall into two clades, which we named the CsTPS-b1 and the CsTPS-b2 clades, with two hop mono-TPSs as the nearest relatives. Three CsTPSs fall into TPS-g, but do not cluster together. TPS-c and TPS-e/f each contain one CsTPS.
Figure 7.
Maximum-likelihood phylogeny of CsTPS relative to other plant TPSs. CsTPSs are in bold. The size of purple dots represents the size of bootstrap values from 100 bootstrap replicates. TPS subfamilies are color coded as follows: TPS-a (purple), TPS-b (orange), TPS-d (brown), TPS-c (black), TPS-e/f (red), and TPS-g (green). Colored lines outside the tree show the location of CsTPSs within the corresponding subfamilies. The tree scale, 0.5, represents 50% sequence difference.
CsTPS Gene Expression in Five Different Cultivars
We used the separate trichome transcriptome assemblies to determine for each cultivar expression of CsTPSs and to correlate CsTPS gene expression and terpene profiles in each of the five different cultivars. The analysis was limited to predicted CsTPS sequences of 400 amino acids or longer to reduce quantification ambiguity, which allowed expression analysis for 18 different CsTPSs (Fig. 8). Transcript abundance was calculated within each cultivar relative to mean transcripts per million (tpm) values of all contigs across three clonal replicates. Each of the 18 different CsTPSs was highly expressed in at least one cultivar (Fig. 8). CsTPS18, CsTPS29, and CsTPS35, which belong to the TPS-g subfamily, were the only CsTPSs with above-mean transcript abundance in all five cultivars.
Figure 8.

Transcript abundance of CsTPS genes in floral trichomes of five different Cannabis cultivars. Values are log2 fold change compared to average CPM for each cultivar. Colored “X” symbols indicate individual data points and black box plots show quartiles and outliers.
In cv AK trichomes, CsTPS5, CsTPS16, CsTPS18, and CsTPS35 showed the highest expression, while CsTPS2 and CsTPS25 transcripts were barely detected. Expression levels of CsTPS3, CsTPS4, CsTPS9, CsTPS17, CsTPS20, CsTPS21, CsTPS22, CsTPS23, CsTPS29, CsTPS32, and CsTPS36 were similar to the mean trichome transcript abundance, defined as within 4-fold log2 CPM of the mean. In cv BC, CsTPS5, CsTPS36, CsTPS18, and CsTPS9 were highly expressed, CsTPS25 transcripts were barely detected, and CsTPS23 and CsTPS20 transcript levels were low relative to mean trichome transcript abundance. The other 11 CsTPSs were expressed at levels similar to the mean transcript abundance. In cv Choc trichomes, CsTPS35 was the most highly expressed CsTPS. CsTPS22, CsTPS25, and CsTPS32 transcripts were detected at low levels. The remaining 14 CsTPSs were expressed at levels similar to the mean trichome transcript abundance. In cv CSH, the most highly expressed CsTPS transcripts were CsTPS4 and CsTPS32. CsTPS25 was detected at low levels, with the remaining 15 CsTPSs expressed at levels similar to the mean trichome transcript abundance. cv LS was the only cultivar with above-mean expression of CsTPS25 and the only one with below-mean expression of CsTPS1. The most highly expressed CsTPS in cv LS was CsTPS21. Aside from CsTPS21, all CsTPSs in cv LS were similar to the mean transcript abundance.
Functions of CsTPSs
The CsTPS phylogeny suggests that all but two, CsTPS65 and CsTPS66, encode mono-TPS or sesqui-TPS enzymes (Fig. 7). For functional characterization, CsTPS enzymes were produced in and purified from Escherichia coli, assayed with GPP and farnesyl diphosphate (FPP) as substrates, and their products identified by GC/MS (Fig. 9; Supplemental Figs. S4 and S5). We identified 11 CsTPS members of the TPS-a subfamily. Of these, six were previously characterized (Booth et al., 2017; Zager et al., 2019). Zager and colleagues reported the identities of CsTPS16, a germacrene B synthase, and CsTPS20, a hedycaryol synthase. We were able to confirm the activities of both of these enzymes as germacrene B and hedycaryol synthases, respectively, using CsTPS16 and CsTPS20 cloned from cv PK [CsTPS16(PK) and CsTPS20(PK), respectively; Supplemental Fig. S6]. CsTPS28 is closely related to CsTPS20 (Fig. 7). With FPP as a substrate, products detected for CsTPS28(PK) included a eudesmane-type compound (25%) with a retention index (RI) of 1,505, α-selinene (23%), and a cadinane-type sesquiterpene (17%) with a RI of 1,598. An additional product was initially annotated as β-elemene (28%); however, since β-elemene may result from thermal rearrangement of germacrene A, we re-examined this product using cold GC injection. Under these conditions the distinct β-elemene peak was replaced by a broader peak, indicative of product rearrangement during the separation or detection process (Supplemental Fig. S4). The most diverse product profile in the TPS-a subfamily was detected with CsTPS22(PK), which produces 13 different sesquiterpenes and himachalane (20%) as the major product. Other products were a eudesmane-type sesquiterpene (10%) with a RI of 1,505, an unidentified sesquiterpene (10%) with a RI of 1,528 and base peak 121, a cadinane type sesquiterpene (7%) with a RI of 1,498, eudesma-3,7(11)-diene (7%), cubenol (6%), a eudesmane-type compound (6%) with a RI of 1,557, a sesquiterpene alcohol (6%) with a RI of 1,716 and base peak 206.4, γ-eudesmol (4%), an unidentified compound (4%) with a RI of 1,552 and base peak 161, α-humulene (3%), nerolidol (3%), and β-elemene (2%). CsTPS25(LS) produced (E)-β-farnesene (56%) as its major product, as well as a cadinane-type compound (22%) with a RI of 1,494, (Z,E)-α-farnesene (15%), and nerolidol (7%). CsTPS24 clusters together with CsTPS25 and CsTPS16. No products were found in enzyme assays with CsTPS24 with either GPP or FPP as substrate.
Figure 9.
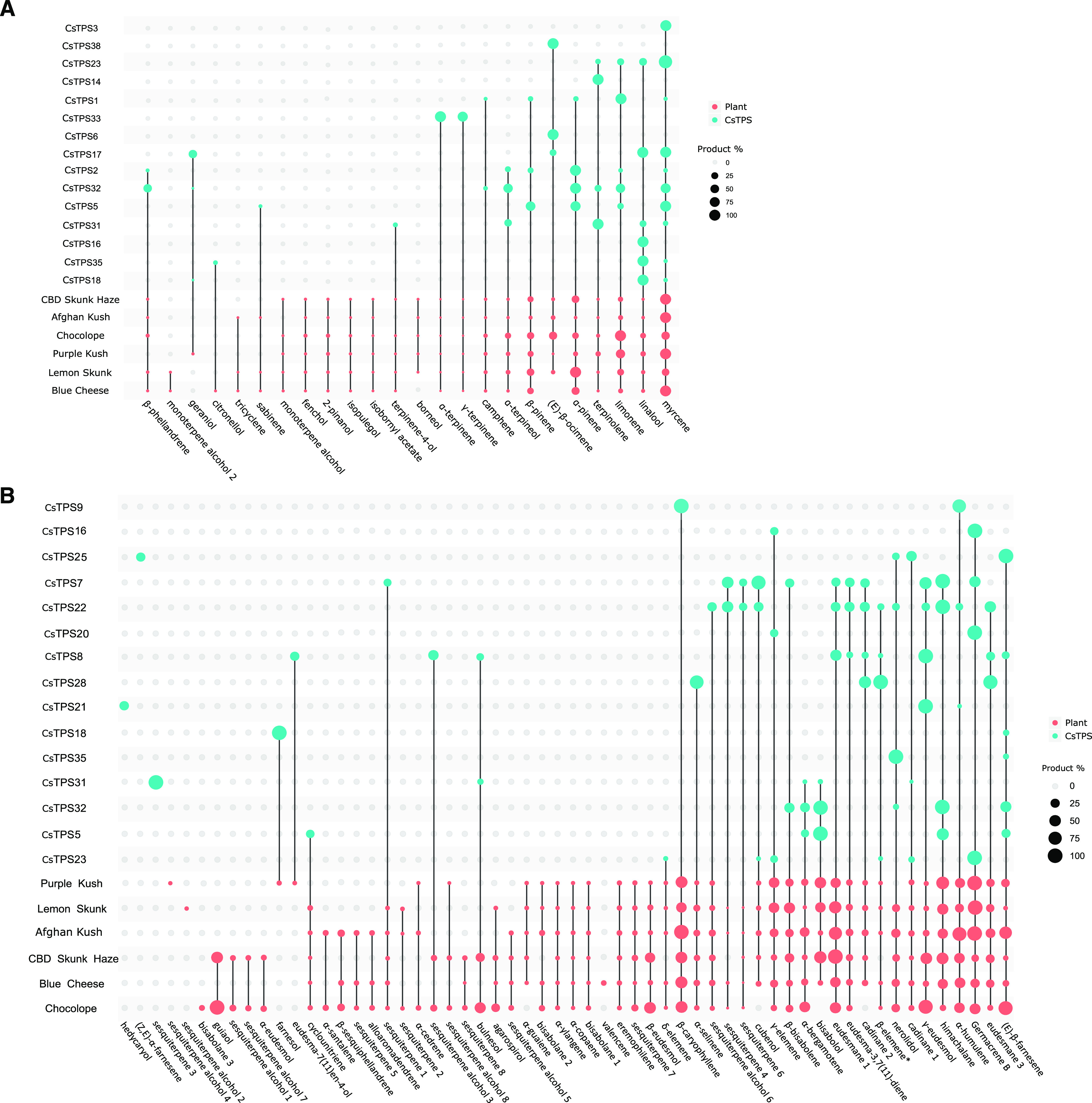
Products of functionally characterized CsTPSs and their representation in cannabis floral trichome terpene profiles of different cultivars. A, Monoterpenes. B, Sesquiterpenes. CsTPS gene identification and cultivar names are shown on the y axis and compounds on the x axis. Dot size corresponds to the percentage of each compound compared to the most abundant product of a given CsTPS (blue dots) or floral metabolite (pink dots). β-elemene is marked with an asterisk because it may be a degradation product of germacrene A.
Within the TPS-b subfamily, the CsTPS-b1 clade contains three members that had not been previously described, specifically CsTPS17, CsTPS23, and CsTPS36. CsTPS17(BC) was functionally characterized as a mono-TPS that produced myrcene (34%) and linalool (34%) as equal major products along with minor products geraniol (16%), (E)-β-ocimene (8%), and α-terpineol (9%). CsTPS23(LS) also is a mono-TPS that produced myrcene (53%) as its major product and the minor products linaloo (20%), limonene (15%), and terpinolene (12%). We were unable to obtain a function for CsTPS36. The CsTPS-b2 clade contains four members, CsTPS5, CsTPS30, CsTPS31 and CsTPS32. Unlike clade b1 members, these CsTPSs do not possess the predicted plastidial target peptides that are typical of plant mono-TPSs. CsTPS5 (cv Finola [FN]) and CsTPS30(PK) were previously characterized as myrcene synthases (Booth et al., 2017), while CsTPS31 and CsTPS32 had not been previously characterized. Given the lack of target peptides, CsTPS5, CsTPS31, and CsTPS32 were assayed here with both GPP and FPP. CsTPS30 was previously characterized as a myrcene synthase from cv PK (Booth et al., 2017). In assays with GPP, the major product of CsTPS5(PK) [96% amino acid identity to CsTPS5(FN)] was α-pinene (33%), with less abundant products myrcene (18%), α-terpineol (18%), limonene (17%), and β-pinene (14%). When assayed with FPP, CsTPS5(PK) produced mainly α-bisabolol (46%), as well as himachalane (27%), (E)-β-farnesene (11%), α-bergamotene (7%), and a compound tentatively identified as a cyclounitriene (9%). CsTPS31(PK) produced terpinolene (57%) as a major product with GPP, as well as α-terpineol (19%), linalool (14%), β-pinene (6%) and terpinen-4-ol (4%). Using FPP as a substrate, the major product (91%) of CsTPS31(PK) was an unknown sesquiterpene with a RI of 1,916 and base peak 93. It also produced 6% bulnesol, 2% α-bisabolol, and trace amounts of α-bergamotene and a cadinane-type sesquiterpene with a RI of 1,494. CsTPS32(PK) produced eight different monoterpenes from GPP: geraniol (23%), α-pinene (20%), myrcene (16%), limonene (13%), β-phellandrene (10%), terpinolene (5%), α-terpineol (13%), and camphene (1%). With FPP, CsTPS32(PK) produced himachalane (32%), α-bisabolol (31%), (E)-β-farnesene (14%), β-bisabolene (12%), α-bergamotene (10%), and nerolidol (2%).
CsTPS18 and CsTPS19, members of the TPS-g subfamily that differ by one amino acid, were recently reported by Zager et al. (2019) while this work was in preparation. Here, we refer to homologs of these genes (>95% amino acid identity) as CsTPS18. We confirmed CsTPS18/1 as a linalool/nerolidol synthase, with CsTPS18(Choc) producing exclusively (−)-linalool (Supplemental Fig. S5). We functionally characterized TPS-g subfamily members CsTPS35 and CsTPS29. CsTPS35(LS) produced acyclic terpenes with both GPP and FPP. With GPP, it produced mostly linalool (93%), with minor amounts of citronellol (5%) and myrcene (2%). Using FPP, CsTPS35(LS) produced nerolidol (95%) and (E)-β-farnesene (5%). CsTPS29(BC) produced exclusively linalool from GPP, and no products were detected when CsTPS29 was assayed with FPP.
DISCUSSION
The CsTPS Gene Family
Previous estimates of the size of the CsTPS family varied from ∼30 to 50 different genes (Booth et al., 2017; Allen et al., 2019). The present analysis of the cv PK reference genomes identified 19 complete and five partial CsTPS genes. The transcriptomes reported here and in two recent studies (Booth et al., 2017; Zager et al., 2019) cover 11 different cannabis cultivars. Screening of these cultivar-specific transcriptomes for CsTPS genes revealed variations of the CsTPS gene family, variations of CsTPS transcript expression, and variations of CsTPS enzyme functions with respect to their mono- and sesquiterpene products. Among the different cultivars, some CsTPS genes were more variable in their transcriptome representation across cultivars than others. For example, the CsTPS9 gene, which encodes a β-caryophyllene/α-humulene synthase, was expressed in the transcriptomes of all cultivars reported to date, including this study. The same is the case for CsTPS5, which encodes an enzyme that uses both GPP and FPP and produces multiple monoterpenes and sesquiterpenes, respectively. By contrast, the CsTPS2 gene, which encodes an α-pinene synthase, was not found in the cv PK genome or in the cv PK and cv AK transcriptomes, but was present in the transcriptomes of other cultivars. CsTPS8, which encodes a multiproduct sesquiterpene synthase, was only detected in transcriptomes of cv FN, cv Choc, and cv CSH. Considering these variations, which are based on the analysis of 11 different cultivars, we expect that the full suite of CsTPS genes that differ by sequence, expression, and function, and which contribute to different terpene profiles, will be substantially larger across the many cannabis cultivars that exist around the world. In this study, we used a conservative cutoff of 95% amino acid identity to assign sequences to the same CsTPS identifier to avoid separating minor variants. However, it should be noted that even at this cutoff, minor sequence variation may result in variation of enzyme function. Assigning unique gene identifiers to transcript sequences based on 100% identity would result in a larger number of apparently different CsTPSs (Allen et al., 2019; Zager et al., 2019).
Within the plant TPS phylogeny, CsTPSs of the TPS-a and TPS-b subfamilies cluster with TPS sequences of its close relative hops (Fig. 7). In both subfamilies, we found cannabis-specific expansions, suggesting that the diversity of CsTPSs described here for mono- and sesquiterpene biosynthesis may have resulted from progressive and relatively recent multiplications of a few ancestral CsTPSs. The TPS-b subfamily has two distinct CsTPS blooms, identified here as CsTPS-b1 and CsTPS-b2. The CsTPS-b2 group includes four members, of which all but one (CsTPS30) lack a predicted plastid target peptide. Two of these TPSs produce α-bisabolol, a sesquiterpene found in many cannabis cultivars, but which is not a product of any of the functionally characterized CsTPS-a group enzymes. These results suggested that indeed some CsTPS-b members contribute to sesquiterpene production in cannabis trichomes, although the TPS-b group has been previously described as including mostly mono-TPSs in other plant species (Chen et al., 2011). In sandalwood (Santalum spp.), a member of TPS-b also functions as a sesquiterpene synthase (Jones et al., 2011). It is striking that in both cannabis and sandalwood, these TPS-b members produce bisabolane-type sesquiterpenes, which may be due to similar routes of active site evolution from their respective monoterpene synthase ancestors (Gao et al., 2012).
Relatedness of CsTPS Functions
The expansion of CsTPSs provided an opportunity to assess whether different products of closely related CsTPSs may arise through similar cyclization cascades. We tested this hypothesis with a focus on sesqui-TPSs because of their usually complex cyclization cascades. The sesquiterpenes identified in the different cannabis cultivars of this study, including cv PK, belong to 11 sesquiterpene parent skeletons that may originate from six central carbocationic intermediates (Fig. 10A; Degenhardt et al., 2009). The farnesane, elemane, and germacrene sesquiterpenes of the cannabis resin may be formed by CsTPSs via either a farnesyl or nerolidyl cation. The eudesmane and humulane sesquiterpenes, which were abundant in the leaf and flower metabolite profiles, most likely arise from (E,E)-germacranedienyl and (E,E)-humulyl cations, respectively, which are formed by 10,1 or 11,1 closure of the farnesyl cation. Four different types of cannabis sesquiterpenes may be formed via the (Z,E)-germacranedienyl cation, the elemane and germacrene compounds, and the cadinane and guaiane skeletons. The nerolidyl cation can also cyclize into the (Z,E)-humulyl cation via 11,1 closure, leading to formation of the himachalane and aromadendrane sesquiterpenes. While the latter compounds are generally present in cannabis terpene profiles, they were not abundant in the cultivars of this study. The bisabolane sesquiterpenes are likely formed from the bisabolane carbocation, which is generally the result of 6,1 closure of the nerolidyl cation.
Figure 10.
Proposed routes of sesquiterpene formation by CsTPS and correlation with CsTPS sequence relatedness. A, Schematic of carbocation intermediates and sesquiterpene classes (according to Degenhardt et al. [2009]) for sesquiterpenes identified in Cannabis floral trichomes. B, Intermediates and major and minor products of CsTPSs described in this article. Intermediates include all major proposed cationic intermediates, and “major product” is the class of the most abundant sesquiterpene product of each enzyme. 1, (E,E)-farnesyl diphosphate; 2, (E,E)-farnesyl cation; 3, farnesane skeleton; 4, nerolidyl cation; 5, bisabolyl cation; 6, (E,E)-germacranedienyl cation; 7, (E,E)-humulyl cation; 8, (Z,E)-germacranedienyl cation; 9, (Z,E)-humulyl cation; 10, bisabolane skeleton; 11, elemane skeleton; 12: eudesmane skeleton; 13, humulane skeleton; 14, cadinane skeleton; 15, germacrane skeleton; 16, guaiane skeleton; 17, aromadendrane skeleton; 18, himachalane skeleton.
We attempted to correlate CsTPS positions in the TPS phylogeny (Fig. 7) with their assumed cyclization reactions (Fig. 10B). CsTPS18 and CsTPS35 are related enzymes that each produce acyclic farnesane compounds. CsTPS5, CsTPS31, and CsTPS32 are related enzymes in the CsTPS-b1 group and share many of the same products and likely the same intermediates, bisabolyl and (Z,E)-humulyl cations. The more closely related CsTPS5 and CsTPS32 share three of four of the same product skeletons and are likely to share the same four potential intermediates. CsTPS8(FN), CsTPS28(PK), and CsTPS21(PK) share only four products between them, but their major and secondary products could all be formed from the (E,E)-germacranedienyl cation. Similarly, CsTPS7(FN) and CsTPS22(PK), which are closely related, may also share three of four intermediate carbocations. CsTPS25(LS), which groups with CsTPSs that have mostly cyclic primary products, produces predominantly acyclic sesquiterpenes. Its secondary product, however, is a cadinane sesquiterpene, which may be a result of its recent evolution from a cyclic-product sesqui-TPS. Overall, we found that similar proposed cyclization routes are more commonly shared between closely related CsTPSs than between more distantly related CsTPSs.
Assessing CsTPS Expression and CsTPS Products to Explain Cannabis Metabolite Profiles
Of the total 61 apparently unique CsTPSs, only 14 had been functionally characterized prior to this work (Supplemental Table S1; Gunnewich et al., 2007; Booth et al., 2017; Zager et al., 2019; Livingston et al., 2020). Here we describe the functional characterization of 13 additional CsTPSs and validation of the functions of several others. The CsTPSs described here, together with those previously reported, account for most of the terpenes identified in the cannabis cultivars of this study. One of the objectives of this work was to explore to what extent information on CsTPS expression and CsTPS function can be used to predict terpene profiles in cannabis trichome extracts. We found that with current knowledge, metabolite profiles can only be partially predicted, and substantially more information is required about the CsTPS proteome, enzyme kinetics, and substrate availability. Across the different cultivars, CsTPSs and other genes for terpene biosynthesis, as well as cannabinoid biosynthesis genes, were highly expressed in floral trichomes (Fig. 6B). This observation is in agreement with previous reports on the cannabis MEP and MEV pathways and selected CsTPS genes previously reported by Booth et al., (2017), Braich et al. (2019), and Livingston et al. (2020).
A single-time point transcript assessment is likely to be insufficient to explain the accumulation of terpene profiles, which occurs over longer periods of time. However, at a qualitative level, we found some general agreement between the presence of terpene products of CsTPSs expressed in a given cultivar (Fig. 8) and the metabolites that accumulate in the trichomes of that cultivar (Fig. 9). For example, cv AK and cv PK, which had no detectable transcript expression of the α-pinene synthase CsTPS2, also had the lowest proportion of α-pinene compared to the other cultivars (Figs. 8 and 9). Similarly, cv AK has a high proportion of nerolidol and relatively high expression of the linalool/nerolidol synthase CsTPS35. An example of a case where current knowledge of CsTPS expression and CsTPS function could not quantitatively explain metabolite profiles is the high proportion of (E)-β-farnesene in the metabolite profile of cv Choc. This cultivar did not reveal high levels of transcripts of any of the three CsTPSs known to encode enzymes that produce (E)-β-farnesene as a major product (CsTPS5, CsTPS25, and CsTPS32). Some possible explanations are that one or more of these CsTPSs may be a highly efficient enzyme, this protein may be highly stable, or additional (E)-β-farnesene synthases may exist to account for the level of (E)-β-farnesene in cv Choc. Similarly, the β-caryophyllene/α-humulene synthase CsTPS9 did not show particularly high transcript levels in any of the cultivars, although these two sesquiterpenes are commonly among the most abundant in cannabis. There are also several terpenes in the metabolite profiles that cannot yet be accounted for by products of known CsTPS functions. These compounds may be the products of CsTPSs that remain to be characterized or minor products of CsTPSs that were below the detection level under assay conditions but accumulate to detectable levels in trichomes over the course of flower development. We also observed the opposite, where a CsTPS product is not found in the metabolite profile despite high transcript levels. Notably, the hedycaryol synthase CsTPS20 was highly expressed across several cultivars, but hedycaryol was not observed in the metabolite profile in any of the cultivars. The labile hedycaryol may be subject to modification (Hattan et al., 2016).
TPS Gene Family Variation and Variation of Terpene Metabolite Profiles
The CsTPS gene family appears to be of a size similar to that reported for other plant species with extensive terpene diversity (Chen et al., 2011). Variation of the composition of the TPS gene family, or variation of TPS gene expression, within a given plant species has been linked to variation of terpene profiles in a number of different systems. This includes both cultivated and noncultivated plants, as well as angiosperms and gymnosperms. For example, in grapevine, members of a large VvTPS gene family are differentially expressed between tissues, developmental stages, and cultivars, leading to differences in terpene profiles depending on the specific combination of TPS genes that are expressed during flowering and fruit ripening (Martin et al., 2009, 2010; Drew et al., 2016; Smit et al., 2019). In rice (Oryza spp.), lineage-specific blooms of similar TPS genes contributed to variation of terpene defenses between different rice species (Chen et al., 2020). Similarly, in corn (Zea mays), variation of expression of ZmTPS genes encoding β-caryophyllene synthase is central to the variation of terpene-mediated indirect defense against corn borer (Ostrinia nubilalis; Köllner et al., 2008). In Sitka spruce (Picea sitchensis), a gymnosperm, copy number variation and variation of expression of PsTPS genes encoding (+)-3-carene synthase caused variation of monoterpene composition associated with insect resistance (Hall et al., 2011; Roach et al., 2014).
The variation of terpene metabolite profiles and CsTPS gene family expression described here for different cannabis cultivars highlights the apparently unlimited opportunity for humans to expand, design, and shape interesting terpene profiles in cannabis. While some of this potential has already been realized through traditional selection and propagation of cannabis strains over past decades and centuries, knowledge of the CsTPS gene family and its contribution to terpene variation will allow cannabis breeders to substantially increase the symphonic diversity of terpene compositions.
CONCLUSION
By identifying suites of CsTPS genes in six cannabis cultivars, we demonstrated variations of expression and function that contribute to the different terpene profiles in cannabis cultivars. The enzymes described here, together with other recent studies on terpene biosynthesis in cannabis (Booth et al., 2017; Livingston et al., 2020; Zager et al., 2019), bring the number of characterized cannabis CsTPSs to 30 across 14 cultivars.
MATERIALS AND METHODS
Plant Material
Cannabis (Cannabis sativa) seeds were provided by Anandia Labs, a subsidiary of Aurora Cannabis (www.auroramj.com) under a Health Canada research license, and plants were grown at their laboratory. Seeds were surface sterilized in 5% (w/v) Plant Preservative Mixture (www.plantcelltechnology.com) and placed in petri dishes between filter paper soaked with 0.5% (w/v) of this mixture. Germination occurred within 2 to 10 d. Germinated seeds were planted in soil (Sunshine Mix 4, Sun Gro Horticulture; www.sungro.com) supplemented with Florikote 14-14-14 controlled-release fertilizer (www.americanhort.com). During the vegetative growth stage, plants were kept under an 18 h/6 h light/dark cycle under T5 HO light bulbs. Plants were fertilized twice weekly with Peter’s Excel 15-15-15 water-soluble fertilizer (pH 5.6–5.8; www.domyown.com). After ∼2 weeks of growth under the 18 h/6 h light/dark cycle, plants were moved to a surface under high-pressure sodium light bulbs and a 12 h/12 h light/dark cycle to induce flowering. During the flowering stage, plants were fertilized twice weekly with MaxiBloom 15-15-14 water-soluble fertilizer (pH 5.6–5.8; www.generallyhydroponics.ca). Between fertilizations, plants were continuously watered with tap water in hydroponic chambers.
For clonal propagation of plants of five different cultivars, cv LS, cv CSH, cv BC, cv AK, and cv Choc, cuttings were taken from well-established stock plants in the vegetative stage and surface sterilized with 5% (v/v) bleach. Cut ends were dipped in 0.4% (w/v) indole-3-butyric acid rooting hormone (www.valleyindoor.com), placed in rockwool cubes soaked for 1 h in pH 6 water, and kept in trays under a clear plastic dome to maintain humidity and promote rooting. Rooted cuttings in rockwool cubes were moved into hydroponic chambers. Female cv PK plants were clonally propagated and grown as described above, but cuttings in rockwool were transferred directly into soil. All plants were grown in growth chambers (BC Northern Lights) under LED lights (3000K 80 CRI spectrum, 1200 W equivalent, BC Northern Lights). The plants were subjected to vegetative growth for 2 to 3 weeks using an 18 h/6 h light/dark cycle and watered with Peter’s Excel (15-5-15). To induce flower development, the light cycle was switched to 12 h/12 h, and plants were watered with Maxibloom (5-15-14).
Harvesting of Leaf and Flower Samples
Leaves (three per plant) were removed from plants 4 weeks post germination with scissors and placed into 50-mL Falcon tubes. Flowers were harvested for trichome isolation by removal of entire inflorescences of plants at two stages, 1 week post induction of flowering and at midstage maturity, between 51 and 60 d post induction of flowering. The time of midstage maturity harvest was based on three criteria: (1) all glandular trichomes had matured to have a stalk; (2) 50% of pistils had begun to brown; and (3) trichome heads were translucent and had not changed color to appear amber or brown (Fig. 2). Flowers were taken from several nodes along the stem. For metabolite analysis, individual florets were removed using scissors and forceps and placed in a 1.5-mL Eppendorf tube. Fresh weight of harvested plant material was recorded and plant material was kept on ice for up to 60 min prior to extractions. After extraction, plant material was dried at 60°C for 16 h and DW was determined.
Terpene Extraction and Analysis
Intact plant material was extracted with three washes using 0.5 mL pentane per 100 mg fresh weight. For the first extraction, plant material was vortexed for 30 s in pentane to disrupt trichomes and then shaken at room temperature for 4 h. For the second and third extractions, the same plant material was shaken in pentane at room temperature for 1 h. The three pentane extracts were combined, centrifuged at 4,300g for 10 min, filtered through a 0.45-μm nylon membrane (Gelman Sciences/Thermo Fisher Scientific) to remove precipitated waxes and starch, and used for terpene analysis.
For the initial screening of terpene profiles in foliage harvested from plants in vegetative growth at 4 weeks post germination, each extract was analyzed by GC/MS on an Agilent 7890A GC coupled with an Agilent 7000A triple-quad MS. An Agilent HP-5 column (5% [w/w] phenyl methylpolysiloxane; 30-m length, 0.25-mm i.d., and 0.25-μm film thickness; 19091S-433HP-5MS, Agilent) was used. The injector was operated in pulsed-splitless mode at 250°C. He gas was used as the carrier with a flow rate of 1 mL min−1 and 30-s pulse at 25 psi. The oven program was 50°C for 3 min, increased by 10°C min−1 to 90°C, then by 20°C min−1 to 120°C, by 10°C min−1 to 150°C, and by 15°C min−1 to 320°C, then held at 320°C for 5 min, giving a total run time of 27.8 min. The mass spectrometer was operated in electron ionization mode at 70 eV and data acquisition was made in full-scan mode with a mass range of 40 to 500 atomic mass units.
Analysis of terpenes in extracts from flowers and TPS assay products was done on an Agilent 6890 GC coupled with an Agilent 5973 mass selective detector. An Agilent DB-Wax column (60-m length, 0.25-mm i.d., and 0.25-μm film thickness; 122-7062, Agilent) was used. The injector was operated in pulsed-splitless mode at 250°C; except for cold injection of the CsTPS28 products, the injector temperature was set to 50°C. He gas was used as the carrier with a flow rate of 1 mL min−1 and 30-s pulses at 25 psi. The initial oven temperature was 40°C, which was increased by 10°C min−1 to 100°C, by 3°C min−1 to 130°C, and by 30°C min−1 to 250°C, then held for 12 min. The mass spectrometer was operated in electron ionization mode at 70 eV and data acquisition was made in full-scan mode with a mass range of 40 to 500 atomic mass units.
Chiral analysis was done using a Cyclodex-B column (30-m length, 250-μm internal diameter, 0.25-μm film thickness; 122-2532E, Agilent). The injector was operated in pulsed-splitless mode at 240°C. He gas was used as the carrier with a flow rate of 1 mL min−1 and 30-s pulses at 25 psi. The initial oven temperature was 40°C for 1 min, increased by 3°C min−1 to 80°C, then increased by 25°C min−1 to 240°C, then held for 5 min. The mass spectrometer was operated in electron ionization mode at 70 eV.
Terpenes were identified by comparison of RIs and mass spectra using authentic standards and Wiley09 and NIST08 mass spectral libraries (http://chemdata.nist.gov/). RIs of terpenes were calculated by the retention time of a standard mixture of n-alkanes (C8–C20). Compounds were compared to authentic standards for the following metabolites: alloaromadendrene (Fluka), α-bisabolol (Fluka), bisabolene (mix of isomers; Bedoukian Research), cadinene (native; Penta MFC), camphene (Sigma-Aldrich), β-caryophyllene (Sigma-Aldrich), (1,8)-cineole (Sigma-Aldrich), citronellol (Bedoukian Research), farnesene (mix of enantiomers; Bedoukian Research), geraniol (Fluka), germacrene D (FL-Treatt), α-humulene (Sigma), limonene (Sigma-Aldrich), linalool (Sigma-Aldrich), myrcene (Sigma-Aldrich), nerolidol (Sigma-Aldrich), ocimene (mix of enantiomers, Sigma), β-phellandrene (Fluka), α-pinene (Sigma-Aldrich), β-pinene (Sigma-Aldrich), terpinen-4-ol (Fluka), α-terpinene (Sigma-Aldrich), γ-terpinene (Sigma-Aldrich), α-terpineol (SAFC), terpinolene (Fluka), and valencene (Sigma-Aldrich). Identifications of bergamotene, δ-selinene, selinane-type, and guaiane-type sesquiterpenes were supported by comparison to Citrus bergamia (Bergamot), Guiaicum officinale (guaiac wood), and Pimenta racemose (Bay) essential oils (www.lgbotanicals.com). Quantification was determined relative to a standard curve of authentic standards. Where no quantitative standard was available, compounds were quantified using the curve of a compound of the same terpene parent skeleton.
Trichome Isolation
Flowers were collected at midstage maturity from all branches of three clonal plants for each cultivar and incubated in water containing 5 mm aurintricarboxylic acid and 1 mm thiourea for 1 to 4 h on ice. After incubation, tissue abrasion to remove trichomes was achieved using a BeadBeater with 30 to 60 g of tissue with 100 g of 1-mm-diameter zirconia/silica beads and 20 g of XAD-4 in enough trichome RNA purification buffer (TRPB; 25 mm HEPES [pH 7.3], 200 mm sorbitol, 10 mm Suc, 5 mm dithiothreitol, 5 mm aurintricarboxylic acid, 1 mm thiourea, 0.6% [w/v] methyl cellulose, and 1% [w/v] polyvinylpyrrolidone 40,000) to fill the BeadBeater chamber completely (total volume 350 mL). Floral tissue was abraded 3× for 15 s with a 30-s rest on ice between abrasions.
Tissue was filtered through 350- and 105-μm nylon mesh, and filtrate was collected on 40-μm mesh. Purified trichome heads were then collected in a 15-mL Falcon tube and rinsed 3× with TRPB without methyl cellulose and polyvinylpyrrolidone 40,000. Purity of the trichome head preparation was determined by light microscopy (Supplemental Fig. S3). Trichome heads were pelleted by centrifugation at 200 g for 1 min. Pellets were weighed and then flash-frozen in liquid nitrogen and stored at −80°C.
RNA Isolation, Transcriptome Sequencing, and Assembly
Trichome pellets (200 μg) were used for RNA isolation. RNA was isolated using PureLink Plant RNA Reagent (Thermo Fisher Scientific) according to the manufacturer’s protocol. RNA concentration, purity, and integrity were determined using an Agilent 2100 Bioanalyzer microchip. Three replicates of trichome RNA from each clone were used for RNA-seq. Total RNA in a volume of 15 μL at 100 ng μL−1 was used for each sample. Sequencing was performed by the McGill University and Génome Québec Innovation Centre (Montreal, Canada), who performed strand-specific library preparation without heating the samples. Sequencing was performed on an Illumina HiSeq2000 platform using 100 bp paired-end sequencing. All samples were pooled and sequenced on four lanes, generating ∼1.5 billion paired-end reads in total. Quality of the sequences was assessed with FastQC (www.bioinformatics.babraham.ac.uk/projects/fastqc/). Reads that mapped to cannabis ribosomal RNA sequences, downloaded from NCBI, using Bowtie2 were removed. Adapters were trimmed with BBDuk from the BBTools software suite (www.sourceforge.net/projects/bbmap/). To improve the contiguity of the assembly, overlapping paired-end reads were joined by BBMerge to generate longer single-end reads. All merged and unmerged reads were pooled and first assembled with Trinity (version 2.6.5) to generate 599,285 nonredundant contigs with an average length of 511 bp. To gain insight into cultivar-specific sequences, we reassembled all of the unmerged sequences using RNA-Bloom (version 0.9.8; Nip et al., 2019) to generate five separate assemblies, one per cultivar, with an average of 260,000 nonredundant contigs and average length of 1,400 bp.
TransDecoder (version 5.5.0) predicted on average 170,000 open reading frames (ORFs) for each assembly. Predicted peptides translated from the ORFs were clustered at 95% amino acid identity, using CD-HIT (version 4.8.1; Fu et al., 2012) to collapse possible allelic variants. Predicted peptides from each assembly were then pooled together and clustered again at 98% amino acid identity to further reduce variations between cultivars to a total of 55,550 sequences. Salmon (version 0.14; Patro et al., 2017) was used to quantify the level of expression on the corresponding ORFs for downstream differential expression analysis.
CsTPS Gene Identification and Genome Annotation
CsTPS candidate genes were identified using the transcriptome assemblies described above as the subject of a tBLASTn search using 100 previously characterized TPS genes from cannabis and other plant species. The completeness of the CsTPS predictions was confirmed by a hmmscan domain search. Gene and splice site prediction on the cv PK reference genome was performed using the Exonerate algorithm (Curwen et al., 2004) from a list of all characterized CsTPS sequences. N-terminal transit peptides were predicted using the TargetP and LOCALIZER tools (Emanuelsson et al., 2007; Sperschneider et al., 2017).
CsTPS cDNA Cloning and Functional Characterization
Complementary DNA (cDNA) was made from trichome RNA using the Maxima First Strand cDNA synthesis kit (Thermo Fisher Scientific). cDNA was amplified using gene-specific primer, and ligated into a pJET vector (Clontech). Sequences were verified by Sanger sequencing, and full-length or N-terminally truncated sequences were subcloned into expression vectors pET28b+ (EMD Millipore) or pASK-IBA37 (IBA Lifesciences), which both carry an N-terminal 6-HIS tag. Full-length CsTPS36BC synthesis was done by IDT (www.idtdna.com). Plasmids were transformed into Escherichia coli strain BL21DE3 for heterologous protein expression, as previously described (Roach et al., 2014). Heterologous protein production was induced using 200 μm isopropylthio-β-galactoside (pET28) or 200 ng mL−1 anhydrotetracycline in methanol (IBA37), and protein was expressed at 18°C overnight. Cells were harvested by centrifugation and lysed by freeze-thaw cycles, warming the pellet to 4°C then freezing in liquid N2. Recombinant protein was purified using the GE healthcare HIS SpinTrap kit (www.gehealthcare.com). The binding buffer for purification was 20 mm HEPES (pH 7.5), 500 mm NaCl, 25 mm imidazole, and 5% (v/v) glycerol. Cells were lysed in binding buffer supplemented with Roche complete protease inhibitor tablets and 0.1 mg mL−1 lysozyme. The elution buffer was 20 mm HEPES (pH 7.5), 500 mm NaCl, 500 mm imidazole, and 5% (v/v) glycerol. Purified protein was desalted through Sephadex into TPS assay buffer: 25 mm HEPES (pH 7.3), 100 mm KCl, 10 mm MgCl2, 5% (v/v) glycerol, and 5 mm dithiothreitol. Protein purity was determined by western blotting using mouse monoclonal anti-polyHis antibody from Sigma-Aldrich (www.sigmaaldrich.com). In vitro assays were performed using 50 to 100 μL of freshly purified protein and TPS assay buffer to a final volume of 500 μL. Isoprenoid diphosphate substrates (www.isoprenoids.com) were dissolved in 50% (v/v) methanol and added to assays at a final concentration of 16 μm GPP or 13 μm FPP. Assays were overlaid with 500 μL pentane with 1.25 μm isobutyl benzene as internal standard. Assays were shaken at 40 rpm at 30°C for 4 h. Reactions were stopped, and products were extracted by vigorous vortexing of the assay vial for 30 s and then centrifuged at 4,300g for 15 min to separate phases. Assay products were determined using the same GC/MS equipment, program, and identification method as for floral terpene extracts described above (see “Terpene Extraction and Analysis”).
Phylogenetic Analysis
ClustalW alignment of translated CsTPS and TPS sequences from other plants and maximum-likelihood phylogeny construction were done in CLC Main Workbench 7. Phylogeny construction used the neighbor-joining method, with 100 bootstrap replicates. Tree visualization and labeling were performed on iTOL (Letunic and Bork, 2019).
Hierarchical Clustering, PCA, Heatmaps, and Differential Expression Analysis
Hierarchical clustering and PCA of the initial 32 seedlings used peak area for each compound normalized to tissue DW and internal standard isobutyl benzene. Clustering was performed using the R function hclust (Kaufman and Rousseeuw, 1990), with Pearson’s correlation as a distance measure for metabolites (rows) and Spearman correlation for individual plants (columns). Dendrogram clusters were determined with the number of clusters set to the maximum, where inertia gain is >1. PCA and visualization used the R package “FactoMineR” (Lê et al., 2008) with default settings. Heatmaps were generated using the R package gplots (https://cran.r-project.org/web/packages/gplots/), with scale = “row” and z-scores used to normalize rows. Transcript abundance was calculated as the mean normalized counts per million of three replicates for each clone. Read counts estimated with Sailfish were normalized using DESeq2 R package version 1.6.1 for differential expression analysis (Love et al., 2014). Transcripts with a normalized CPM < 100 in three or more samples were discarded. The false discovery rate was set at 5%. Differentially expressed genes were defined by an adjusted log2 fold-change >2 and a normalized P-value <0.05.
Accession Numbers
Raw sequence read data associated with the trichome transcriptome sequencing is deposited in the NCBI Sequence Read Archive under accession number PRJNA599437. TPS sequences are deposited under the following accession numbers: MN967481 [CsTPS5(PK)]; MN967478 (CsTPS16); MN967470 (CsTPS17); MN967473 (CsTPS18); MN967469 (CsTPS20); MN967483 (CsTPS21); MN967477 (CsTPS22); MN967480 (CsTPS23); MN967472 (CsTPS25); MN967479 (CsTPS26); MN967482 (CsTPS28); MN967468 (CsTPS29); MN967474 (CsTPS31); MN967484 (CsTPS32); MN967476 (CsTPS34); MN967475 (CsTPS35); MN967471 (CsTPS36); MT295506 (CsTPS65); and MT295505 (CsTPS66).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Representative extracted ion chromatograms of foliar terpene extracts from five cannabis plants.
Supplemental Figure S2. Representative mass spectra for all compounds identified in floral terpene extracts.
Supplemental Figure S3. Trichome head isolates from five cultivars.
Supplemental Figure S4. Representative mass spectra for all CsTPS products.
Supplemental Figure S5. Extracted ion chromatograms showing stereochemical determination of monoterpenes in cannabis juvenile floral terpene extracts and CsTPS enzyme assays.
Supplemental Figure S6. Total ion chromatograms and mass spectra for CsTPS18, CsTPS16, 2 and CsTPS20.
Supplemental Table S1. Cannabis TPSs (CsTPS) previously published or reported here.
Supplemental Table S2. Identification and amounts of foliar terpene in 32 different cannabis seedlings.
Supplemental Table S3. Foliar cannabinoid content in 32 cannabis seedlings.
Supplemental Table S4. Assembly statistics for six transcriptome assemblies used.
Supplemental Table S5. Highly expressed contigs in five cannabis cultivars.
Supplemental Table S6. Accession numbers of TPS sequences used to construct phylogeny.
Acknowledgments
We thank Dr. Carol Ritland and Angela Chiang for administrative and technical support. We thank Erin Gilchrist, Jose Celedon, Samantha Mishos, Eva Chou, and members of the Anandia team for assistance with plant growth, access to data, and discussion.
Footnotes
This work was supported by the Natural Science and Engineering Research Council of Canada (to J.B.) and by Genome British Columbia (a Sector Innovation Project grant to J.B.).
Articles can be viewed without a subscription.
References
- Allen KD, McKernan K, Pauli C, Roe J, Torres A, Gaudino R(2019) Genomic characterization of the complete terpene synthase gene family from Cannabis sativa. PLoS One 14: e0222363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Bakel H, Stout JM, Cote AG, Tallon CM, Sharpe AG, Hughes TR, Page JE(2011) The draft genome and transcriptome of Cannabis sativa. Genome Biol 12: R102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Booth JK, Bohlmann J(2019) Terpenes in Cannabis sativa—From plant genome to humans. Plant Sci 284: 67–72 [DOI] [PubMed] [Google Scholar]
- Booth JK, Page JE, Bohlmann J(2017) Terpene synthases from Cannabis sativa. PLoS One 12: e0173911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braich S, Baillie RC, Jewell LS, Spangenberg GC, Cogan NOI(2019) Generation of a comprehensive transcriptome atlas and transcriptome dynamics in medicinal cannabis. Sci Rep 9: 16583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenneisen R, elSohly MA(1988) Chromatographic and spectroscopic profiles of Cannabis of different origins: Part I. J Forensic Sci 33: 1385–1404 [PubMed] [Google Scholar]
- Casano S, Grassi G, Martini V, Michelozzi M(2011) Variations in terpene profiles of different strains of Cannabis sativa L. Acta Hortic (925): 115–121 [Google Scholar]
- Chen F, Tholl D, Bohlmann J, Pichersky E(2011) The family of terpene synthases in plants: A mid-size family of genes for specialized metabolism that is highly diversified throughout the kingdom. Plant J 66: 212–229 [DOI] [PubMed] [Google Scholar]
- Chen H, Köllner TG, Li G, Wei G, Chen X, Zeng D, Qian Q, Chen F(2020) Combinatorial evolution of a terpene synthase gene cluster explains terpene variations in Oryza. Plant Physiol 182: 480–492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curwen V, Eyras E, Andrews TD, Clarke L, Mongin E, Searle SMJ, Clamp M(2004) The Ensembl automatic gene annotation system. Genome Res 14: 942–950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Degenhardt J, Köllner TG, Gershenzon J(2009) Monoterpene and sesquiterpene synthases and the origin of terpene skeletal diversity in plants. Phytochemistry 70: 1621–1637 [DOI] [PubMed] [Google Scholar]
- Drew DP, Andersen TB, Sweetman C, Møller BL, Ford C, Simonsen HT(2016) Two key polymorphisms in a newly discovered allele of the Vitis vinifera TPS24 gene are responsible for the production of the rotundone precursor α-guaiene. J Exp Bot 67: 799–808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emanuelsson O, Brunak S, von Heijne G, Nielsen H(2007) Locating proteins in the cell using TargetP, SignalP and related tools. Nat Protoc 2: 953–971 [DOI] [PubMed] [Google Scholar]
- Falara V, Akhtar TA, Nguyen TTH, Spyropoulou EA, Bleeker PM, Schauvinhold I, Matsuba Y, Bonini ME, Schilmiller AL, Last RL, et al. (2011) The tomato terpene synthase gene family. Plant Physiol 157: 770–789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fellermeier M, Zenk MH(1998) Prenylation of olivetolate by a hemp transferase yields cannabigerolic acid, the precursor of tetrahydrocannabinol. FEBS Lett 427: 283–285 [DOI] [PubMed] [Google Scholar]
- Fischedick JT.(2017) Identification of terpenoid chemotypes among high (−)-trans-Δ9-tetrahydrocannabinol-producing Cannabis sativa L. cultivars. Cannabis Cannabinoid Res 2: 34–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischedick JT, Hazekamp A, Erkelens T, Choi YH, Verpoorte R(2010) Metabolic fingerprinting of Cannabis sativa L., cannabinoids and terpenoids for chemotaxonomic and drug standardization purposes. Phytochemistry 71: 2058–2073 [DOI] [PubMed] [Google Scholar]
- Fu L, Niu B, Zhu Z, Wu S, Li W(2012) CD-HIT: Accelerated for clustering the next-generation sequencing data. Bioinformatics 28: 3150–3152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Y, Honzatko RB, Peters RJ(2012) Terpenoid synthase structures: A so far incomplete view of complex catalysis. Nat Prod Rep 29: 1153–1175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunnewich N, Page JE, Koellner T, Degenhardt J, Kutchan TM(2007) Functional expression and characterization of trichome-specific (−)-limonene synthase and (+)-α-pinene synthase from Cannabis sativa. Nat Prod Commun 2: 223–232 [Google Scholar]
- Hall DE, Robert JA, Keeling CI, Domanski D, Quesada AL, Jancsik S, Kuzyk MA, Hamberger B, Borchers CH, Bohlmann J(2011) An integrated genomic, proteomic and biochemical analysis of (+)-3-carene biosynthesis in Sitka spruce (Picea sitchensis) genotypes that are resistant or susceptible to white pine weevil. Plant J 65: 936–948 [DOI] [PubMed] [Google Scholar]
- Hattan J, Shindo K, Ito T, Shibuya Y, Watanabe A, Tagaki C, Ohno F, Sasaki T, Ishii J, Kondo A, et al. (2016) Identification of a novel hedycaryol synthase gene isolated from Camellia brevistyla flowers and floral scent of Camellia cultivars. Planta 243: 959–972 [DOI] [PubMed] [Google Scholar]
- Jones CG, Moniodis J, Zulak KG, Scaffidi A, Plummer JA, Ghisalberti EL, Barbour EL, Bohlmann J(2011) Sandalwood fragrance biosynthesis involves sesquiterpene synthases of both the terpene synthase (TPS)-a and TPS-b subfamilies, including santalene synthases. J Biol Chem 286: 17445–17454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufman L, Rousseeuw PJ(1990) Finding Groups in Data : An Introduction to Cluster Analysis. Wiley-Interscience, Hoboken, NJ [Google Scholar]
- Köllner TG, Held M, Lenk C, Hiltpold I, Turlings TCJ, Gershenzon J, Degenhardt J(2008) A maize (E)-β-caryophyllene synthase implicated in indirect defense responses against herbivores is not expressed in most American maize varieties. Plant Cell 20: 482–494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Külheim C, Padovan A, Hefer C, Krause ST, Köllner TG, Myburg AA, Degenhardt J, Foley WJ(2015) The Eucalyptus terpene synthase gene family. BMC Genomics 16: 450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laverty KU, Stout JM, Sullivan MJ, Shah H, Gill N, Holbrook L, Deikus G, Sebra R, Hughes TR, Page JE, et al. (2019) A physical and genetic map of Cannabis sativa identifies extensive rearrangements at the THC/CBD acid synthase loci. Genome Res 29: 146–156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lê S, Josse J, Husson F(2008) FactoMineR: An R Package for multivariate analysis. J Stat Softw 25: 1–18 [Google Scholar]
- Letunic I, Bork P(2019) Interactive Tree Of Life (iTOL) v4: Recent updates and new developments. Nucleic Acids Res 47(W1): W256–W259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livingston SJ, Quilichini TD, Booth JK, Wong DCJ, Rensing KH, Laflamme-Yonkman J, Castellarin SD, Bohlmann J, Page JE, Samuels AL(2020) Cannabis glandular trichomes alter morphology and metabolite content during flower maturation. Plant J 101: 37–56 [DOI] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S(2014) Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15: 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo X, Reiter MA, d’Espaux L, Wong J, Denby CM, Lechner A, Zhang Y, Grzybowski AT, Harth S, Lin W, et al. (2019) Complete biosynthesis of cannabinoids and their unnatural analogues in yeast. Nature 567: 123–126 [DOI] [PubMed] [Google Scholar]
- Martin DM, Aubourg S, Schouwey MB, Daviet L, Schalk M, Toub O, Lund ST, Bohlmann J(2010) Functional annotation, genome organization and phylogeny of the grapevine (Vitis vinifera) terpene synthase gene family based on genome assembly, FLcDNA cloning, and enzyme assays. BMC Plant Biol 10: 226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin DM, Toub O, Chiang A, Lo BC, Ohse S, Lund ST, Bohlmann J(2009) The bouquet of grapevine (Vitis vinifera L. cv. Cabernet Sauvignon) flowers arises from the biosynthesis of sesquiterpene volatiles in pollen grains. Proc Natl Acad Sci USA 106: 7245–7250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Meijer EPM, Bagatta M, Carboni A, Crucitti P, Moliterni VMC, Ranalli P, Mandolino G(2003) The inheritance of chemical phenotype in Cannabis sativa L. Genetics 163: 335–346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nip KM, Chiu R, Yang C, Chu J, Mohamadi H, Warren RL, Birol I(2019) RNA-Bloom provides lightweight reference-free transcriptome assembly for single cells. bioRxiv 701607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page JE, Boubakir Z, inventors (November 11, 2011) Aromatic prenyltransferase from Cannabis. United States Patent Application No. 8,884,100
- Patro R, Duggal G, Love MI, Irizarry RA, Kingsford C(2017) Salmon provides fast and bias-aware quantification of transcript expression. Nat Methods 14: 417–419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rea KA, Casaretto JA, Al-Abdul-Wahid MS, Sukumaran A, Geddes-McAlister J, Rothstein SJ, Akhtar TA(2019) Biosynthesis of cannflavins A and B from Cannabis sativa L. Phytochemistry 164: 162–171 [DOI] [PubMed] [Google Scholar]
- Reimann-Philipp U, Speck M, Orser C, Johnson S, Hilyard A, Turner H, Stokes AJ, Small-Howard AL(2019) Cannabis chemovar nomenclature misrepresents chemical and genetic diversity; survey of variations in chemical profiles and genetic markers in nevada medical cannabis samples. Cannabis Cannabinoid Res doi:10.1089/can.2018.0063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richins RD, Rodriguez-Uribe L, Lowe K, Ferral R, O’Connell MA(2018) Accumulation of bioactive metabolites in cultivated medical Cannabis. PLoS One 13: e0201119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, Smyth GK(2015) limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res 43: e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roach CR, Hall DE, Zerbe P, Bohlmann J(2014) Plasticity and evolution of (+)-3-carene synthase and (−)-sabinene synthase functions of a sitka spruce monoterpene synthase gene family associated with weevil resistance. J Biol Chem 289: 23859–23869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sirikantaramas S, Morimoto S, Shoyama Y, Ishikawa Y, Wada Y, Shoyama Y, Taura F(2004) The gene controlling marijuana psychoactivity: Molecular cloning and heterologous expression of Δ1-tetrahydrocannabinolic acid synthase from Cannabis sativa L. J Biol Chem 279: 39767–39774 [DOI] [PubMed] [Google Scholar]
- Smit SJ, Vivier MA, Young PR(2019) Linking terpene synthases to sesquiterpene metabolism in grapevine flowers. Front Plant Sci 10: 177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sperschneider J, Catanzariti A-M, DeBoer K, Petre B, Gardiner DM, Singh KB, Dodds PN, Taylor JM(2017) LOCALIZER: Subcellular localization prediction of both plant and effector proteins in the plant cell. Sci Rep 7: 44598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taura F, Sirikantaramas S, Shoyama Y, Yoshikai K, Shoyama Y, Morimoto S(2007) Cannabidiolic-acid synthase, the chemotype-determining enzyme in the fiber-type Cannabis sativa. FEBS Lett 581: 2929–2934 [DOI] [PubMed] [Google Scholar]
- Taura F, Tanaka S, Taguchi C, Fukamizu T, Tanaka H, Shoyama Y, Morimoto S(2009) Characterization of olivetol synthase, a polyketide synthase putatively involved in cannabinoid biosynthetic pathway. FEBS Lett 583: 2061–2066 [DOI] [PubMed] [Google Scholar]
- Turner JC, Hemphill JK, Mahlberg PG(1978) Quantitative determination of cannabinoids in individual glandular trichomes of Cannabis sativa L. (Cannabaceae). Am J Bot 65: 1103–1106 [Google Scholar]
- Zager JJ, Lange I, Srividya N, Smith A, Lange BM(2019) Gene networks underlying cannabinoid and terpenoid accumulation in Cannabis. Plant Physiol 180: 1877–1897 [DOI] [PMC free article] [PubMed] [Google Scholar]









