Graphical abstract

Keywords: fMRI, Independent component analysis, Brain disorders, Reproducible and comparable biomarkers, NeuroMark
Highlights
-
•
Propose a new pipeline to link brain changes among different datasets, studies, and disorders.
-
•
Identify reproducible biomarkers in schizophrenia using independent data.
-
•
Find both common and unique brain impairments in schizophrenia and autism.
-
•
Reveal gradual changes from healthy controls to mild cognitive impairment to Alzheimer’s disease.
-
•
Obtain high classification accuracy (~90%) between bipolar disorder and major depressive disorder.
Abstract
Many mental illnesses share overlapping or similar clinical symptoms, confounding the diagnosis. It is important to systematically characterize the degree to which unique and similar changing patterns are reflective of brain disorders. Increasing sharing initiatives on neuroimaging data have provided unprecedented opportunities to study brain disorders. However, it is still an open question on replicating and translating findings across studies. Standardized approaches for capturing reproducible and comparable imaging markers are greatly needed. Here, we propose a pipeline based on the priori-driven independent component analysis, NeuroMark, which is capable of estimating brain functional network measures from functional magnetic resonance imaging (fMRI) data that can be used to link brain network abnormalities among different datasets, studies, and disorders. NeuroMark automatically estimates features adaptable to each individual subject and comparable across datasets/studies/disorders by taking advantage of the reliable brain network templates extracted from 1828 healthy controls as guidance. Four studies including 2442 subjects were conducted spanning six brain disorders (schizophrenia, autism spectrum disorder, mild cognitive impairment, Alzheimer’s disease, bipolar disorder, and major depressive disorder) to evaluate validity of the proposed pipeline from different perspectives (replication of brain abnormalities, cross-study comparison, identification of subtle brain changes, and multi-disorder classification using identified biomarkers). Our results highlight that NeuroMark effectively identified replicated brain network abnormalities of schizophrenia across different datasets; revealed interesting neural clues on the overlap and specificity between autism and schizophrenia; demonstrated brain functional impairments present to varying degrees in mild cognitive impairments and Alzheimer's disease; and captured biomarkers that achieved good performance in classifying bipolar disorder and major depressive disorder.
1. Introduction
In the neuroscience field, increasing data-sharing initiatives have accelerated the use of neuroimaging data to study brain disorders in the clinic (Poline et al., 2012, Poldrack and Gorgolewski, 2014, Woo et al., 2017). Access to multi-site datasets affords unprecedented opportunities to perform large-scale analysis across disorders. Brain functional connectivity and other related measures derived from functional magnetic resonance imaging (fMRI) data are powerful for characterizing brain organization and its abnormalities in brain disorders. Mounting evidence has supported that functional connectivity is a reliable indicator of brain disorders for early diagnosis and treatments (Liu et al., 2008, Whitfield-Gabrieli et al., 2009, Li et al., 2020). Many approaches have been utilized to capture neuroimaging features informative of functional connectivity, including region of interest (ROI) or seed derived connectivity analysis (Tzourio-Mazoyer et al., 2002, Dosenbach et al., 2010), decomposition-based independent component analysis (ICA) (Calhoun et al., 2001, McKeown et al., 2003, Calhoun and Adali, 2012), self-activation detection method such as amplitude of low frequency fluctuation and regional homogeneity (Zang et al., 2004), as well as clustering techniques to group brain voxels (Du et al., 2012). ROI analysis and ICA are two commonly used strategies for studying functional connectivity. While ROI-based methods typically require fixed brain regions according to prior experience or knowledge (e.g. atlas), ICA, a data-driven method, is capable of capturing functional networks while retaining more single-subject variability (Yu et al., 2017). ICA leverages the hidden spatial–temporal information to extract spatially independent components (ICs), each of which includes brain voxels sharing co-varying patterns. In addition, ICA performs component extraction and noise component removal simultaneously (Du et al., 2016), which might provide better signal-to-noise ratio (SNR) level for the functional connectivity estimation. ICA also enables separation of overlapping but distinct functional activity (Xu et al., 2015), which cannot be achieved by using atlas-based analysis. To overcome the limitation of ICA that components from different subjects might not have spatial correspondence, group ICA methods were developed (Calhoun et al., 2001, Beckmann et al., 2009, Du and Fan, 2013, Calhoun and de Lacy, 2017), which perform ICA on the group data to estimate the group-level components, and then utilize a back-reconstruction strategy to extract individual-level functional networks and associated time-courses.
However, there is still a big challenge on group ICA that the identified components might vary across different group ICA runs due to the difference of the data properties, affecting its ability to replicate and compare findings across different studies. For example, Allen et al. (Allen et al., 2014) identified 50 intrinsic connectivity networks (ICNs) arranged into seven functional domains, while Marusak, Calhoun et al. (2017) characterized 52 ICNs sorted to three domains, despite they implemented the same model order in the group ICA. Such discrepancy hinders the direct comparisons across the results. Another potential problem of group ICA is that conventional ICA-based classification studies typically performed group ICA on all subjects’ fMRI data to make the resulting features consistent across training and testing sets (Demirci et al., 2008, Rashid et al., 2016). However, this operation can be biased, as the feature extraction should be independent from the testing data. Accordingly, it is essential to adopt an ICA approach to estimate brain network measures in an unbiased way that fully persist individual property while also being able to be compared across subjects from various datasets/studies/disorders.
From a clinical perspective, it is very important to evaluate sensitivity and specificity as well as similarity and overlap between disorders that share clinically-overlapping symptoms using neuroimaging data. For example, both schizoaffective and bipolar disorders experience hallucinations and delusions that are typical features of schizophrenia (SZ) (Cosgrove and Suppes, 2013), which can make their clinical differentiation difficult. As such, it would be beneficial to study the shared and unique brain impairments among them for better diagnosis and accurate treatments. Unfortunately, there is a paucity of studies that perform a direct comparison of symptom-related disorders and the validation of brain changes using large-sample datasets, probably due to the limited ability of analytic methods to characterize individual variability (Zuo et al., 2014) and reliability (Noble et al., 2019). Furthermore, since the traditional diagnosis relying on symptom assessment could be imperfect, it may be promising to develop new biologically-based types across the psychotic illnesses by combining the use of neuroimage-derived features (Marquand et al., 2016). Taking SZ and autism spectrum disorder (ASD) into account, although they are conceptualized as distinct illnesses, they have also been revisited in recent years due to their shared phenotypic and genotypic expression (Hommer and Swedo, 2015). There have been studies that explore subtypes for mental disorders such as schizophrenia by performing clustering techniques on neuroimage-derived features (Yang et al., 2014, Sun et al., 2015, Dwyer et al., 2018, Honnorat et al., 2019, Chand et al., 2020). Hence, effective methods that can accurately capture biologically meaningful subject-specific features that are still comparable across different individuals will benefit the refinement of current disorder categories. As more neuroimaging data are now available than ever before, ICA based method has a great potential to study the shared and unique brain network abnormalities across brain disorders. But the traditional ICA methods cannot be implemented directly on multiple datasets due to the inconsistency of IC estimations and arrangements across different ICA runs, which greatly influence the ability for comparing and validating the brain changes among disorders. Therefore, there is a need for advanced ICA techniques that can accurately estimate subject-specific network features representing interpretable biomarkers and achieve corresponding network features to accelerate the evaluation of biomarkers in terms of their generalizability, reproducibility and relationship to other data.
In this study, we propose a pipeline called NeuroMark, which leverages an adaptive-ICA technique such as group information guided ICA (GIG-ICA) (Du and Fan, 2013) or spatially constrained ICA (Lin et al., 2010) to fully automate the estimation and labeling of individual-subject connectivity features, by incorporating an additional input of spatial network priors derived from independent large samples. We implemented the NeuroMark pipeline to four studies involving six brain disorders and >2400 subject samples, in order to validate its performance from different aspects. We expected that replicable brain changes in patients with certain mental disorder can be found on independent datasets. We were interested in the cross-study comparison to link related mental disorders using NeuroMark. We also hoped that NeuroMark is able to extract subtle group difference across progressively developing disorders. Another concern was its ability in capturing effective biomarkers for the classification on challengeable brain disorders.
2. Materials and methods
In this section, we first introduce the NeuroMark, and then apply it to four example studies to assess its capacity.
2.1. NeuroMark
The flowchart of NeuroMark is displayed in Fig. 1. First, ICs were estimated using different datasets of large-sample healthy controls (HCs). Second, reproducible ICNs were identified by matching and inspecting the spatial maps of ICs from different datasets. Next, using the highly replicated ICNs as the network templates, an adaptive-ICA method (Du and Fan, 2013, Du et al., 2016a, Salman et al., 2019) was applied to automatically estimate subject-specific functional networks and associated time-courses (TCs). Finally, different functional connectivity features such as static or dynamic functional network connectivity (FNC) were computed and then evaluated. The following describes the details.
Fig. 1.
Schematic flowchart of the NeuroMark pipeline. Step 1: Calculate group-level independent components (ICs) from two independent datasets, human connectome project (HCP) and genomics superstruct project (GSP) datasets, respectively. Step 2: Match ICs using correlations between their spatial maps and then identify highly replicated intrinsic connectivity networks (ICNs) as the network templates. Step 3: Calculate the individual-level ICNs and their related time courses (TCs) by taking the network templates as prior information in adaptive-ICA. Functional connectivity features such as static and dynamic functional network connectivity (FNC) can be obtained and then compared across datasets, studies, and disorders.
2.1.1. Identifying reliable functional network templates
The spatial network priors (i.e., the ICN templates) were obtained using two independent HC resting-state fMRI datasets from the human connectome project (HCP, http://www.humanconnectomeproject.org/data/) and genomics superstruct project (GSP, https://www.nitrc.org/projects/gspdata). GSP dataset was preprocessed using statistical parametric mapping (SPM12, http://www.fil.ion.ucl.ac.uk/spm/). Rigid body motion correction was performed to correct subject head motion, followed by the slice-timing correction to account for timing difference in slice acquisition. The fMRI data were subsequently warped into the standard Montreal Neurological Institute (MNI) space using an echo planar imaging (EPI) template and were resampled to 3 × 3 × 3 mm3 isotropic voxels. The resampled fMRI images were further smoothed using a Gaussian kernel with a full width at half maximum (FWHM) = 6 mm. For the HCP dataset, we downloaded the preprocessed data from online and resliced them to the same spatial resolution (3 × 3 × 3 mm3) with the preprocessed GSP data using SPM12. More details in terms of the preprocessing on HCP data can be found online (http://www.humanconnectomeproject.org/data/).
We performed quality control (QC) for all the preprocessed data. The detailed QC procedure can be found in the supplementary materials. In total, 1005 individuals from the GSP dataset and 823 individuals from the HCP dataset were chosen after QC for further analysis. Please see Table S1 for the age, gender and motion information. For the GSP data, all imaging data were captured on Siemens 3T MAGNETOM Tim Trio MRI systems using the vendor-supplied 12-channel phase-array head coil. Although five different scanners were used to acquire data, each scanner used the exact same sequences, parameters, and instructions. The HCP subjects were scanned on a customized Siemens 3T “Connectome Skyra”, using a standard 32-channel Siemens receive head coil and a “body” transmission coil designed by Siemens specifically for the smaller space available using the special gradients of the Connectome scanners. The GSP and HCP datasets have different temporal resolution (TR for GSP data = 3 s and TR for HCP data = 0.72 s). Among the 1005 selected subjects in the GSP dataset, 935 subjects are right handedness, 61 subjects are left handedness, and 9 subjects use both hands. The mean education year of selected 1005 subjects is 14.45 years, with the standard deviation as 1.91. 612 subjects are white and not Hispanic, and 393 subjects are other race/ethnicities. Among the selected 823 subjects in the HCP dataset, 747 subjects are right handedness, 74 subjects are left handedness, and 2 are both handedness. The mean education year of 823 subjects is 14.88 years, with the standard deviation as 1.82. 549 subjects are white and and not Hispanic, and 274 are other race/ethnicities. We used the two datasets with different scanning information, preprocessing procedures, and demographic distributions, as we wanted to capture the functional networks that are reproducible across different conditions so that the selected network templates can be reliable for various independent data.
We performed ICA on the GSP and HCP datasets separately to yield reliable ICs. First, principal component analysis (PCA) was performed on each individual subject to reduce fMRI data to 110 principal components (PCs), which preserved >95% variance of the original data. Then, the individual-level PCs of each subject were concatenated across different subjects (1005 subjects for GSP or 823 subjects for HCP) and reduced into 100 PCs via another PCA at the group level. Next, the Infomax algorithm (Bell and Sejnowski, 1995) was applied to decompose the 100 PCs into 100 ICs. This procedure was repeated 100 times using the ICASSO technique (Himberg and Hyvarinen, 2003), in which the best ICA run was selected to generate 100 reliable ICs for each dataset (Ma et al., 2011). In our study, we computed the skewness of each estimated IC and flipped the IC if its skewness was negative.
Two groups of ICs were matched using a greedy spatial correlation analysis and then inspected to find reproducible ICNs as the network templates. A similarity matrix C (size: 100 × 100) was obtained by computing the absolute value of Pearson correlation coefficients between the spatial maps of ICs from GSP and that from HCP. Based on the matrix C, the pair of ICs with the maximum correlation value were selected and considered as the first-matched IC pair. If their original correlation value was negative, one of the ICs was sign-flipped. After identifying a matched IC pair, the correlation values related to them in the matrix C were set to zero, resulting in a new similarity matrix Cnew. As such, the matching procedure was repeated continually on the updated correlation matrices until the final matched IC pair was found. IC pairs were considered to be reproducible if they showed a higher spatial correlation than a given threshold 0.4, a more strict threshold than previous work (Smith et al., 2009). Next, we characterized a subset of these reproducible ICs as ICNs if they exhibited peak values in gray matter, had low spatial overlap with known vascular, ventricular, motion and other artifacts, and exhibited dominant low-frequency fluctuations in their TCs. Five fMRI experts carefully inspected those matched ICs, then labeled meaningful ICNs and assigned them to different functional domains. ICs with more than three votes were identified as meaningful ICNs. This resulted in two groups of highly similar ICNs from the HCP and GSP datasets. Finally, the ICNs captured from the GSP dataset were taken as the network templates as their spatial maps were smoother with less noises. Hereinafter, we use to denote the number of network templates.
2.1.2. Estimating subject-specific brain functional networks and relevant connectivity features
Based on the fMRI data of each subject, the subject-specific ICNs were computed by adaptive-ICA, an approach that automatically and adaptively estimates individual-level independent components using the prior network templates as guidance. Two ICA algorithms (Lin et al., 2010, Du and Fan, 2013) available in the group ICA of fMRI toolbox (GIFT) (http://trendscenter.org/software/) can be used for adaptive-ICA. In this paper, we extended GIG-ICA (Du and Fan, 2013) for the adaptive-ICA due to its superiority, by taking the obtained network templates and each subject's fMRI data as input. Basically, there are two objective functions in GIG-ICA, one of which is to optimize the independence of networks, while the other is to optimize the comparability between one subject-specific network and its related network template. Using simulations, our previous studies (Du and Fan, 2013, Du et al., 2016a) demonstrated that subject-specific independent components can be obtained with higher accuracy in GIG-ICA compared to other group ICA methods. Using test–retest fMRI data, GIG-ICA yielded higher intra class coefficients (ICCs) in the estimated networks than the independent vector analysis (IVA) (Du et al., 2017). Our another work (Salman et al., 2019) supported that GIG-ICA can result in higher classification accuracy than the dual regression method in distinguishing schizophrenia patients from healthy controls. In the original GIG-ICA algorithm, group-level components used as guidance are computed from its own group data (Du and Fan, 2013, Du et al., 2016a). In this paper, we used the labeled and ordered network templates validated from two independent datasets as the spatial priors for guidance in estimating subject-specific networks.
The multiple-objective optimization denoted by (1) represents how one subject-specific network can be estimated using a network template as guidance.
| (1) |
In (1), denotes the network template, and represents the estimated corresponding network of the subject, where is the whitened representing fMRI data matrix of the subject. Here, is the unmixing column vector, which is to be solved in the optimization functions. The first function is for optimizing the independence measure of , which is reflected using , i.e., the negentropy of . Here, is a Gaussian variable with zero mean and unit variance; is a nonquadratic function. The second function is used to measure the correspondence between and . denotes the expectation of variable. A linear weighted sum method is applied to combine the two objective functions (Du and Fan, 2013), with the weights as 0.5, in order to solve the optimization problem. Our method can automatically result in Z-scored networks. It is also worth pointing out that the resulting components (networks) will be very stable across different runs using our method. Thus, for each subject, all subject-specific networks corresponding to the network templates and their relevant TCs are estimated from the data. In summary, using NeuroMark, all subject-specific functional networks will not only be comparable across different datasets/studies/disorders as well as between previously analyzed data and new coming impendent data, but also show subject-unique characteristics.
Using NeuroMark, multiple network features can be obtained, including spatial functional networks, functional connectivity between networks, graph measures of functional organization, and frequency information of networks’ fluctuations, from both static and dynamic perspectives. Taking functional network connectivity (FNC) as an example, static FNC (sFNC) can be obtained by computing the Pearson correlations between TCs of ICNs to yield a sFNC matrix reflecting the interaction between any two networks. While the spatial map of each ICN reflects intra-connectivity within brain functional network, sFNC matrix represents inter-connectivity strengths between different ICNs. Dynamic FNC (dFNC) can also be investigated through a sliding time window approach (Hutchison et al., 2013, Allen et al., 2014), in which a tapered window obtained by convolving a rectangle with a Gaussian is often used to segment the entire TC of each ICN into several short TCs. For each window, the connectivity matrix is computed using the windowed TCs of ICNs to measure the functional connectivity between ICNs within the window. Thus, for each subject, the connectivity matrix of each window can be concatenated to form an array (size: , here is the number of ICNs and is the number of windows), representing the temporal changes of FNC along time.
2.2. Studies for validating NeuroMark
In the paper, we performed four different studies to fully evaluate our proposed NeuroMark pipeline from different angles. Totally, we included resting-state fMRI data from 2442 scans after the QC processing for the experiments. Table 1 summarizes the age, gender, and motion information for each of the four studies. Study 1 focused on the investigation of the reproducibility of brain functional connectivity abnormalities using independent SZ data from the Function Biomedical Informatics Research Network (FBIRN) and the Maryland Psychiatric Research Center (MPRC). We hoped that reproducible brain changes can be found between the two datasets. In study 2, we identified the functional changes in ASD using the release 1 of Autism Brain Imaging Data Exchange (ABIDEI) data, and then linked the results to study 1, aiming to show the ability of NeuroMark in the cross-disorder comparison. We were interested in similar and unique changes between SZ and ASD. In study 3, we worked on the dynamic connectivity analysis on progressively developing disorders, using data of Alzheimer's disease (AD) patients, mild cognitive impairment (MCI) patients, and HCs from Alzheimer’s Disease Neuroimaging Initiative (ADNI) dataset, in order to show the ability of NeuroMark in exploring subtle group differences. NeuroMark was expected to be able to reveal a progressive change from HC to MCI to AD. In study 4, our goal was to verify the classification capability in distinguishing challengeable brain disorders ( bipolar disorder (BD) and major depressive disorder (MDD)) using network features estimated from NeuroMark.
Table 1.
The demographic and motion information of the datasets used in studies 1–4.
 |
Note: Two-sample t-tests and analysis of variance (ANOVA) were used to examine the group differences in the age, motion translations and rotations. Chi-square test was performed to examine the gender difference.
In the following studies, NeuroMark was applied to each subject's fMRI data to extract the subject-specific ICNs and their related TCs. Since the network templates were used to guide the estimation of the subject-specific ICNs, we investigated if the correspondence between the network templates and the subject-specific ICNs as well as the correspondence between the ICNs from different subjects can be well maintained, and going further if the unique characteristics of each subject's ICNs still were preserved. To measure the correspondence between the network templates and the subject-specific ICNs, for each subject we first computed the spatial similarity between each network template and the estimated ICN using Pearson correlation coefficients, and then averaged the correlation coefficients across all ICNs to reflect its template-ICN similarity. We investigated if the template-ICN similarity shows reliability across different studies and groups by summarizing the template-ICN similarity for all subjects in each dataset (e.g. both HC and SZ subjects in FBIRN) and for the subjects in each group (e.g. HC or SZ subjects in FBIRN). In addition to the template-ICN similarity, we also assessed the similarity across the subject-specific ICNs to verify that the network patterns can be subject-unique meanwhile they are still corresponding (comparable) across subjects. For the ICNs guided by one network template, we computed Pearson correlation coefficients across those subject-specific ICNs, and then averaged the coefficients to reflect the inter-subject similarity of the ICN. The mean of the inter-subject similarity across all ICNs was used to summarize the inter-subject ICN similarity. We also demonstrate the inter-subject ICN similarity for all subjects in each dataset and for the subjects in each group to reflect the data variation.
2.2.1. Study 1: Investigating static functional network connectivity (sFNC) abnormalities in schizophrenia: A replication study
In the first study, we assessed the ability of NeuroMark to identify reproducible disease abnormalities across different datasets. NeuroMark was implemented to two independent SZ datasets. The discovery dataset was from the FBIRN including 210 SZ patients and 195 HCs. The replication dataset was collected at the MPRC, including 251 SZ patients and 327 HCs. Both datasets was preprocessed using the same preprocessing pipeline used for the GSP dataset. Subject inclusion criteria was also similar, which required participants with head motion <= 3° and <= 3 mm and with functional data providing near full brain successful normalization. We retained 137 SZ patients and 144 HCs in the FBIRN dataset, and 150 SZ patients and 238 HCs in the MPRC dataset for the further analysis.
We applied NeuroMark to each dataset to extract subject-specific ICNs and their related TCs. The following steps were done to remove noise sources of TCs before the sFNC computation, including 1) detrending linear, quadratic, and cubic trends; 2) conducting multiple regressions of the six realignment parameters and their temporal derivatives; 3) de-spiking to detect and remove outliers; and 4) band-pass filtering with [0.01–0.15] Hz. Then, a sFNC matrix was obtained for each subject via computing Pearson correlation coefficients between the post-processed TCs. The strength of each sFNC (i.e. each element in the sFNC matrix) was transformed to Fisher’s Z-score. After that, for each sFNC, we investigated difference in the connectivity strength between HCs and SZ patients by performing a two-tailed two-sample t-test (p < 0.05 with Bonferroni correction) for the FBIRN and MPRC datasets separately, after controlling the age, gender and site effects (see the supplementary materials). Finally, we compared the significant sFNC difference identified from the two datasets to highlight reproducible functional connectivity abnormalities in SZ.
2.2.2. Study 2: Investigating the common and unique static functional network connectivity (sFNC) alterations in autism spectrum disorder (ASD) and SZ: multi-study comparison
Since we obtained the brain functional network templates using data from large-sample population independent from the data being analyzed, it is feasible to link multiple independent studies using NeuroMark. In this study, we investigated sFNC changes in ASD compared to HCs, and then compared the results with those captured in study 1 for searching the common and unique impairments between SZ and ASD. The ASD data was from ABIDEI, provided by the National Institute of Mental Health. The ABIDEI dataset includes 539 individuals with ASD and 573 HCs. We performed the same preprocessing and subject selection as introduced in the study 1. Consequently, 398 ASD individuals and 471 age-matched HCs in the ABIDEI dataset were remained.
Similar to study 1, Neuromark was applied to the ABIDEI dataset to estimate ICNs and corresponding TCs for each subject. Then, we investigated the HC vs. ASD differences on sFNC measures using two-tailed two-sample t-tests (p < 0.05 with Bonferroni correction). With the help of NeuroMark, we compared the symptom-related disorders (i.e. SZ and ASD) in terms of their overlapping and unique sFNC alterations based on the results from study 1 and the results obtained from the above analyses. The age, gender, and site effect were regressed out before the statistical analyses (see the supplementary materials). In addition, for the commonly changed FNCs between SZ and ASD, we also computed the correlation between FNC measures and clinical symptoms. For SZ, the symptoms scores included the positive and negative syndrome scale (PANSS) positive score and PANSS negative score. Symptoms of ASD included autism diagnostic observation schedule (ADOS) total score and social responsiveness scale (SRS). The significance level was set to p < 0.05 for the correlation analyses.
2.2.3. Study 3: Investigating the dynamic functional network connectivity (dFNC) abnormalities in Alzheimer's disease (AD) and mild cognitive impairment (MCI)
In this study, we aimed to show that NeuroMark can effectively capture subtle differences in dynamic functional network connectivity (dFNC) features among progressively developing brain disorders. The dFNC changes were compared among Alzheimer's disease (AD) patients, mild cognitive impairment (MCI) patients, and HCs. We used the publicly available ADNI dataset. Using the same preprocessing and subject selection procedures, we had a total of 838 scans (104 scans of AD patients, 470 scans of MCI patients, and 264 scans of HCs) for analysis.
For each scan, we estimated dFNC using a sliding window approach (Allen et al., 2014). Since the fMRI data have different temporal resolutions, we performed interpolation on the TCs with longer repetition time (TR) to construct new TCs with the same temporal resolution as those data with smallest TR and the same length of data. This procedure helped to control the potential impacts on the dynamic analysis caused by the different temporal resolutions. In this paper, the tapered window was obtained by convolving a rectangle (window size = 40 TRs = 24.3 s) with a Gaussian (σ = 3) function. This window was slid in steps of 1 TR, resulting in total = 468 windows for yielding dFNC matrices.
A K-means clustering analysis (Allen et al., 2014) was implemented on the time-varying connectivity patterns to capture occurred connectivity states in time and across subjects. L1 norm was used as the distance function with the upper triangular () values in the dFNC matrices as features. The optimal number of clusters was determined as five by the elbow criterion, which was within a reasonable range (4 ~ 7) and consistent with the previous dFNC studies on different brain disorders (Rashid et al., 2014, Abrol et al., 2017, Du et al., 2018, Fu et al., 2018a, Fu et al., 2018b, Fu et al., 2019).
Regarding each connectivity state, we computed its fraction rate of occurrence for each subject by computing the percentage of the number of time windows assigned to the state in the number of total windows. To investigate group differences in the fraction rate of each state, analysis of variance (ANOVA) was performed after regressing out age and gender. If the ANOVA resulted in a significant diagnosis effects, a generalized linear model (GLM) including age and gender was conducted to examine the group difference between any paired groups.
2.2.4. Study 4: Classification between bipolar disorder (BD) and major depressive disorder (MDD)
In study 4, our goal was to test the ability of NeuroMark for capturing functional network markers that can be used for the classification of symptom-related disorders. We focused on classifying BD and MDD, both of which can exhibit strong depressive symptoms and are difficult to distinguish in clinical diagnosis. Resting-state fMRI data including 32 patients with BD Type I and 34 patients with MDD were used for the two-group classification. More details of the subject information can be found in a previous study (Osuch et al., 2018).
Using NeuroMark, the subject-specific ICNs and TCs were estimated for each subject. Due to cerebellum being partially missing in the scanned data, the cerebellar ICNs were not estimated in this study. In order to evaluate if the spatial maps of ICNs captured by the NeuroMark pipeline can be powerful biomarkers to classify BD and MDD patients, we used an unbiased 10-fold cross-validation framework, in which nine of ten folds were used as the training data and the remaining fold was used as the testing data successively. Consistent to the previous work (Osuch et al., 2018), we applied support vector machine with sigmoid kernel for classification. The feature selection and model training were performed only based on the training data.
Feature selection plays a key role in classification, especially for the high-dimensional network measures. In this work, we extracted the most discriminative ICN from each functional domain, and then combined the discriminative ICNs from all functional domains as features. In order to find the most discriminative ICN for each domain, we used an inner 10 times of 10-fold cross-validation procedure within the training set based on a forward ICN-selection technique. Basically, the ICNs were added one by one based on the classification accuracy on the inner testing data, evaluated using the model built using the inner training data. Then, for each run in the inner 10-fold procedure, the optimal ICN combination corresponding to the highest classification accuracy can be found. The ICN with the highest occurring frequency in the optimal ICN combination sets (across different repeats) was validated as the most discriminative ICN for that domain. After that, the combined discriminative ICNs from different domains were used as features to train a model using the outer training data. While the previous study (Osuch et al., 2018) used group information from its own data, the subject-specific ICN features computed using NeuroMark were more unbiased. Fig. 2 shows our classification method.
Fig. 2.
The pipeline of classifying BD and MDD patients using brain functional networks (i.e. ICNs) as features, in which an unbiased 10-fold cross-validation procedure was applied.
To quantify the classification results, we evaluated multiple measures including the individual class accuracy, individual class precision, overall accuracy, balanced accuracy, and balanced precision (Cuadros-Rodriguez et al., 2016) based on the predicted and diagnosis labels. Different measures reflect the results from different angles. The individual class accuracy reported the ratio of correctly classified subjects of a particular class to the total number of subjects in the class. The individual class precision was defined as the number of correctly classified subjects of a particular class divided by the total number of subjects predicted as the class. The overall accuracy was computed as the ratio of correctly classified subjects of all classes to the total number of subjects of all classes. Additionally, we also computed the mean of individual class accuracies, called as the balanced accuracy. The individual class precision values were also averaged to represent the balanced precision. For each measure, we show the results from different repeats using both boxplot and violinplot, respectively.
3. Results
3.1. Reliable network templates
Fig. 3 displays the 53 ICNs from the GSP dataset that were set as the network templates in the NeuroMark pipeline. The correlation matrix computed using the 53 matched ICNs from the two datasets is shown in Fig. 4, indicating that these ICNs are highly reproducible between the GSP and HCP datasets. Regarding the original 100 group-level independent components, there were 81 components matched between the two datasets with correlation > 0.4, 58 components with correlation > 0.6, and 18 components with correlation > 0.8, as shown in Fig. 5(A). If only considering the meaningful ICNs, there were 53 ICNs with correlation > 0.4, 44 ICNs with correlation > 0.6, and 16 ICNs with correlation > 0.8, as shown in Fig. 5(B). It is worth pointing out that the spatial correlation reflecting the similarity between the matched two groups of functional networks or components in Figs. 4 and 5(A)–(B) were obtained by using the whole-brain voxels (i.e. 60,358 voxels), since those voxels were taken as input for NeuroMark. If the correlation was calculated only using the important voxels with positive Z-scores in each ICN, the correspondence (represented by correlation) between the two groups of ICNs would be higher (see Fig. 5(C)). The positive Z-scores in each ICN represented the voxels that mostly contributed to the ICN, because that the skewness of each ICN was changed to be positive, as mentioned in the section 2.1.1.
Fig. 3.
Visualization of the identified network templates, which were divided into seven functional domains based on their anatomical and functional properties. In each subfigure, one color in the composite maps corresponds to an ICN.
Fig. 4.
The spatial correlation matrix between the matched two groups of functional networks. It is seen that the diagonal values are high, indicating the selected network templates are common and reproducible between the GSP and HCP datasets.
Fig. 5.
Correspondence between the HCP and GSP datasets. (A) The number of matched components with the correlation > 0.4, >0.5, >0.6, >0.7, >0.8, and > 0.9, respectively. (B) The number of matched meaningful networks with the correlation > 0.4, >0.5, >0.6, >0.7, >0.8, and > 0.9, respectively, in which the correlation was computed using the whole-brain voxels. (C) The number of matched meaningful networks with the correlation > 0.4, >0.5, >0.6, >0.7, >0.8, and > 0.9, respectively, in which the correlation was computed only using the important voxels with positive Z-scores. Note: corr means correlation.
Those ICNs were arranged into seven functional domains according to their functional and anatomical roles (Allen et al., 2014), including the subcortical (SC: 5 ICNs), auditory (AU: 2 ICNs), sensorimotor (SM: 9 ICNs), visual (VI: 9 ICNs), cognitive control (CC: 17 ICNs), default mode (DM: 7 ICNs) and cerebellar (CB: 4 ICNs) domains. The detailed component labels and peak coordinates are provided in Table 2.
Table 2.
Information of the extracted network templates. For each template, its functional domain, primary brain region and peak coordinate are included. Here, each network template is represented by one independent component (IC). IC ID is shown along with the brain region name.
| Primary regions in ICNs (IC ID) | X | Y | Z | Primary regions in ICNs (IC ID) | X | Y | Z |
|---|---|---|---|---|---|---|---|
| Sub-cortical domain (SC) | Cognitive-control domain (CC) | ||||||
| Caudate (IC 69) | 6.5 | 10.5 | 5.5 | Inferior parietal lobule ([IPL], IC 68) | 45.5 | −61.5 | 43.5 |
| Subthalamus/hypothalamus (IC 53) | −2.5 | −13.5 | −1.5 | Insula (IC 33) | −30.5 | 22.5 | −3.5 |
| Putamen (IC 98) | −26.5 | 1.5 | −0.5 | Superior medial frontal gyrus ([SMFG], IC 43) | −0.5 | 50.5 | 29.5 |
| Caudate (IC 99) | 21.5 | 10.5 | −3.5 | Inferior frontal gyrus ([IFG], IC 70) | −48.5 | 34.5 | −0.5 |
| Thalamus (IC 45) | −12.5 | −18.5 | 11.5 | Right inferior frontal gyrus ([R IFG], IC 61) | 53.5 | 22.5 | 13.5 |
| Auditory domain (AU) | Middle frontal gyrus ([MiFG], IC 55) | −41.5 | 19.5 | 26.5 | |||
| Superior temporal gyrus ([STG], IC 21) | 62.5 | –22.5 | 7.5 | Inferior parietal lobule ([IPL], IC 63) | −53.5 | −49.5 | 43.5 |
| Middle temporal gyrus ([MTG], IC 56) | −42.5 | −6.5 | 10.5 | Left inferior parietal lobue ([R IPL], IC 79) | 44.5 | −34.5 | 46.5 |
| Sensorimotor domain (SM) | Supplementary motor area ([SMA], IC 84) | −6.5 | 13.5 | 64.5 | |||
| Postcentral gyrus ([PoCG], IC 3) | 56.5 | −4.5 | 28.5 | Superior frontal gyrus ([SFG], IC 96) | −24.5 | 26.5 | 49.5 |
| Left postcentral gyrus ([L PoCG], IC 9) | −38.5 | –22.5 | 56.5 | Middle frontal gyrus ([MiFG], IC 88) | 30.5 | 41.5 | 28.5 |
| Paracentral lobule ([ParaCL], IC 2) | 0.5 | –22.5 | 65.5 | Hippocampus ([HiPP], IC 48) | 23.5 | −9.5 | −16.5 |
| Right postcentral gyrus ([R PoCG], IC 11) | 38.5 | −19.5 | 55.5 | Left inferior parietal lobue ([L IPL], IC 81) | 45.5 | −61.5 | 43.5 |
| Superior parietal lobule ([SPL], IC 27) | −18.5 | −43.5 | 65.5 | Middle cingulate cortex ([MCC], IC 37) | −15.5 | 20.5 | 37.5 |
| Paracentral lobule ([ParaCL], IC 54) | −18.5 | −9.5 | 56.5 | Inferior frontal gyrus ([IFG], IC 67) | 39.5 | 44.5 | −0.5 |
| Precentral gyrus ([PreCG], IC 66) | −42.5 | −7.5 | 46.5 | Middle frontal gyrus ([MiFG], IC 38) | −26.5 | 47.5 | 5.5 |
| Superior parietal lobule ([SPL], IC 80) | 20.5 | −63.5 | 58.5 | Hippocampus ([HiPP], IC 83) | −24.5 | −36.5 | 1.5 |
| Postcentral gyrus ([PoCG], IC 72) | −47.5 | −27.5 | 43.5 | Default-mode domain (DM) | |||
| Visual domain (VI) | Precuneus (IC 32) | −8.5 | −66.5 | 35.5 | |||
| Calcarine gyrus ([CalcarineG], IC 16) | −12.5 | −66.5 | 8.5 | Precuneus (IC 40) | −12.5 | −54.5 | 14.5 |
| Middle occipital gyrus ([MOG], IC 5) | –23.5 | −93.5 | −0.5 | Anterior cingulate cortex ([ACC], IC 23) | −2.5 | 35.5 | 2.5 |
| Middle temporal gyrus ([MTG], IC 62) | 48.5 | −60.5 | 10.5 | Posterior cingulate cortex ([PCC], IC 71) | −5.5 | −28.5 | 26.5 |
| Cuneus (IC 15) | 15.5 | −91.5 | 22.5 | Anterior cingulate cortex ([ACC], IC 17) | −9.5 | 46.5 | −10.5 |
| Right middle occipital gyrus ([R MOG], IC 12) | 38.5 | −73.5 | 6.5 | Precuneus (IC 51) | −0.5 | −48.5 | 49.5 |
| Fusiform gyrus (IC 93) | 29.5 | −42.5 | −12.5 | Posterior cingulate cortex ([PCC], IC 94) | −2.5 | 54.5 | 31.5 |
| Inferior occipital gyrus ([IOG], IC 20) | −36.5 | −76.5 | −4.5 | Cerebellar domain (CB) | |||
| Lingual gyrus ([LingualG], IC 8) | −8.5 | −81.5 | −4.5 | Cerebellum ([CB], IC 13) | −30.5 | −54.5 | −42.5 |
| Middle temporal gyrus ([MTG], IC 77) | −44.5 | −57.5 | −7.5 | Cerebellum ([CB], IC 18) | –32.5 | −79.5 | −37.5 |
| Cerebellum ([CB], IC 4) | 20.5 | −48.5 | −40.5 | ||||
| Cerebellum ([CB], IC 7) | 30.5 | −63.5 | −40.5 | ||||
3.2. Subject-specific ICNs with both correspondence and uniqueness
As mentioned in the section 2.2, we assessed the correspondence and uniqueness of the subject-specific ICNs. Fig. 6(A) and (B) show the similarity between the network templates and the subject-specific ICNs for all subjects in each dataset (e.g. FBIRN) and the subjects in each group of dataset (e.g. FBIRN-SZ), using boxplots. It is observed from Fig. 6(A)–(B) that the mean template-ICN similarity across different subjects was close to 0.5, which met our expectation designed in the objective function optimization. Our results support that the template-ICN correspondence was well maintained. Furthermore, the correspondence measure was relatively reliable across different datasets and groups, indicating that our method is effective. Fig. 6(C) and (D) demonstrate the inter-subject ICN similarity for all subjects in each dataset and the subjects in each group, respectively. The mean of the inter-subject similarity across different ICNs is shown using a bar. The results suggest that the subject-specific ICNs presented unique patterns while they were still comparable across subjects.
Fig. 6.
The correspondence and uniqueness of the subject-specific ICNs. Similarity between the network templates and subject-specific ICNs is shown in (A) for all subjects in each dataset (e.g. FBIRN) and (B) for the subjects in each group of dataset (e.g. FBIRN-SZ), using boxplots. In (A) and (B), each sample in the boxplots denotes the template-ICN similarity of one subject. Similarity across the subject-specific ICNs is shown in (C) for all subjects in each dataset and (D) for the subjects in each group of dataset, using bars. In (C) and (D), the inter-subject ICN similarity is shown using a bar. Note: the correlations were computed based on the whole-brain voxels.
3.3. Validity of NeuroMark
3.3.1. Study 1: Reproducible static functional network connectivity (sFNC) alterations in schizophrenia
Fig. 7(A) and (D) show the mean sFNC matrices across subjects for the FBIRN and MPRC datasets, respectively. The sFNC matrix showed a similar pattern between the two datasets, indicating the comparability of connectivity measures computed by NeuroMark. Regarding the FBIRN dataset, Fig. 7(B) and (C) display the T-values obtained from two-sample t-tests and the significant group differences after the multiple comparisons correction, respectively. Similar group differences were found using the MPRC dataset, as shown in Fig. 7(E) and (F), indicating reproducible SZ-related functional connectivity abnormalities captured by the NeuroMark framework.
Fig. 7.
Results of study 1, which shows that there are reproducible sFNC alterations of SZ between the FBIRN and MPRC datasets. (A) and (D): Mean sFNC matrices across all subjects for the FBIRN and MPRC datasets, respectively. (B) and (E): T-values of all sFNCs obtained from two-sample t-tests for FBIRN and MPRC, respectively. (C) and (F): T-values for the sFNCs passing the multiple comparisons correction (p < 0.05 with Bonferroni correction) for FBIRN and MPRC, respectively. “BFN” denotes Bonferroni correction. (G): Mean sFNC strength across subjects for the HC and SZ groups in the common impairments between FBIRN and MPRC data. For each commonly changed sFNC, the averaged values in SZ patients of FBIRN dataset, SZ patients of MPRC dataset, and HCs of the two datasets are shown, respectively.
Regarding both datasets, the brain functional abnormalities in SZ were mainly located in the connectivity between the SC and CB domains, between the SC and AU domains, as well as between the SC and SM domains. Furthermore, the common significantly changed functional connectivity was evaluated based on Fig. 7(C) and (F). Compared to HC, SZ showed decreased connectivity strength between thalamus and cerebellum, caudate and cerebellum, subthalamus and cerebellum, but increased strengths between thalamus and postcentral gyrus, thalamus and superior temporal gyrus, caudate and postcentral gyrus, caudate and superior temporal gyrus, as well as subthalamus and superior temporal gyrus. Fig. 7(G) demonstrates that although the FBIRN dataset showed slightly higher mean functional connectivity strengths than the MPRC dataset, their common impaired connectivity in SZ patients was highly close. Taken together, our results suggest that NeuroMark can help to identify functional connectivity abnormalities that are validated across datasets.
3.3.2. Study 2: Autism and schizophrenia show common and also unique static functional network connectivity (sFNC) alterations
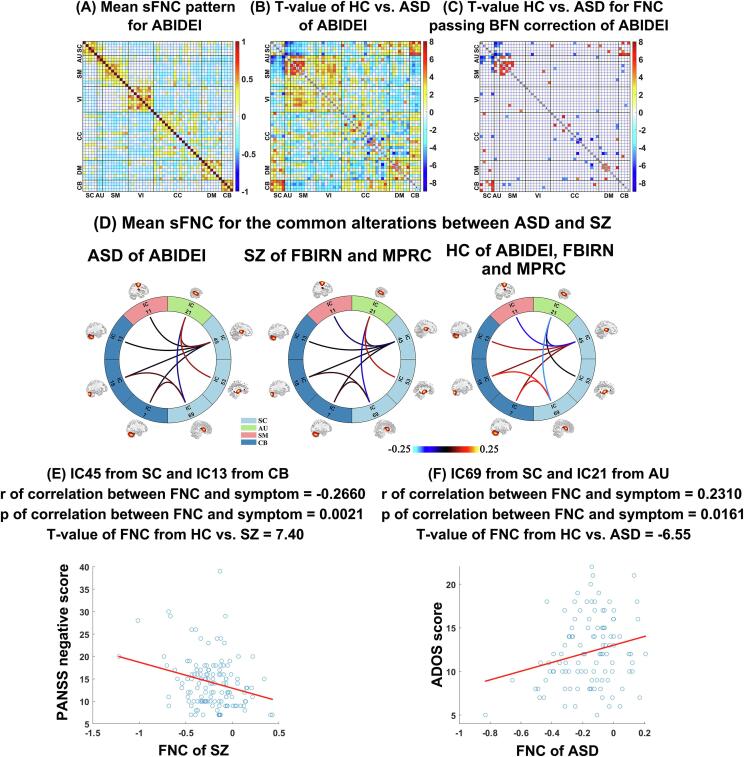
For ABIDEI data, the mean sFNC matrix computed by averaging the sFNC matrices across different subjects (Fig. 8(A)) showed a similar connectivity pattern as that in study 1, again demonstrating that the network features computed using NeuroMark are corresponding and comparable. Statistical analysis showed significant group differences between HC and ASD primarily involving the SC, CB, AU and SM domains, as shown in Fig. 8(B) and (C).
Fig. 8.
Results of study 2, which supports that SZ and ASD show common alterations in sFNC. (A): Mean sFNC pattern across all subjects for ABIDEI. (B) and (C): T-values of all sFNCs and T-values of the sFNCs passing the multiple comparisons correction (p < 0.05 with Bonferroni correction), obtained from two-sample t-tests of HC vs. ASD for ABIDEI. “BFN” denotes Bonferroni correction. (D): Mean sFNC strength of each group (ASD, SZ and HC) in the common impairments between SZ and ASD. For each commonly impaired sFNC, the averaged connectivity values in ASD patients of ABIDEI, SZ patients of FBIRN and MPRC, and HCs of the three datasets are shown, respectively. (E) and (F): The significant correlations (r and p values) between FNC measures and clinical symptoms, with p < 0.05. The T-value from testing group difference between HC and disorder by two-sample t-test is also included in each subfigure. Taking (F) for an instance, it shows the correlation between FNC measure (corresponding to IC 69 and IC 21) and ADOS score in ASD patients. The T-value of FNC measure from two-sample t-test between HC and ASD is also shown in the title part.
While our findings in terms of ASD alterations could stand on its own as a result, we were interested in learning additional information by linking study 1 and the above-mentioned analysis. By comparing their results, we found that SZ and ASD had significant overlapping brain abnormalities between SC domain and AU/SM/CB domains. Specifically, we identified nine atypical sFNC overlapping between SZ and ASD. Among the nine overlaps between SZ and ASD impairments, four functional connectivity showed decrease between cerebellum and thalamus (or caudate); four functional connectivity showed increase between superior temporal gyrus and subcortical regions; and the remaining one showed increase between postcentral gyrus and thalamus (Fig. 8(D)). Two of the nine commonly changed FNCs showed significant correlations (p < 0.05) with the symptoms in one of the two disorders (Fig. 8(E)–(F)), while they also showed a similar correlation pattern in the other disorder. Furthermore, the correlations accorded with the group differences. Taking the connectivity between IC 69 from sub-cortical domain and IC 21 from auditory domain for an instance, the correlation between the FNC measures and ADOS scores of AD patients was positive and the T-value of HC vs. ASD was <0, supporting greater FNC strengths in this connectivity could be associated with severer ASD condition. SZ and ASD also had their unique brain abnormalities in sFNC. For example, SZ showed atypical low functional connectivity within the VI domain and the ASD showed changed functional connectivity within CC domain and decreased functional connectivity within DM domain.
In summary, our NeuroMark framework can identify functional connectivity abnormalities that are comparable across different brain disorders, which would provide great convenience for searching the unique and common brain changes in multiple brain disorders and thus further advance our understanding of the underlying interrelationships between them.
3.3.3. Study 3: Mild cognitive impairment (MCI) demonstrates intermediate dynamic functional network connectivity (FNC) changes between healthy controls (HCs) and Alzheimer's disease (AD)
The identified reoccurring brain states with obviously distinct connectivity patterns were displayed in Fig. 9. Specifically, the state 2 which accounts for > 50% of all windows resembles the sFNC patterns; the state 1 shows negative connectivity strengths between SM and VI; and the state 3 in contrast shows a strong positive connection between SM and VI. The statistical analysis revealed that compared to HCs, AD patients had a significantly increased occurrence in those weakly-connected dFNC states (i.e. state 2 and state 5) but decreased occurrence in those states with strong connectivity pattern (i.e. state 1 and state 3, which showed strong correlated and anticorrelated connectivity strengths). Although there was no significant group difference in the state occurrences between MCI and HC/AD, MCI showed a similar changing trend as AD but with a relatively weaker degree (Fig. 9, upper panel). When dividing the MCI group into early MCI (EMCI) and late MCI (LMCI) group, three of the four states still show similar gradually changing patterns on their occurrence (increase or decrease, from HC to EMCI to LMCI to AD) (Fig. 9, bottom panel).
Fig. 9.
Results of study 3. The results revealed gradually changing patterns from healthy controls (HCs) to early mild cognitive impairment (EMCI) to late MCI (LMCI) to Alzheimer’s disease (AD), measured by dynamic functional network connectivity (dFNC) measures. Upper: Group differences in the fraction rate of occurrences of dFNC states among HC, MCI, and AD. Middle: The discriminating dFNC states, along with the count of subjects that have at least one window clustered into the state. Bottom: Group differences in the fraction rate of occurrences of dFNC states among HC, EMCI, LMCI, and AD. Regarding the fraction rate of occurrences in each state, bar and error bar represent the mean and the standard error of mean, respectively. Significant group differences (false discovery rate corrected, q = 0.05) are indicated by asterisks.
3.3.4. Study 4: Functional network features captured by the NeuroMark serve as reliable biomarkers for classifying bipolar disorder (BD) and major depressive disorder (MDD)
Our results show that the functional networks (i.e. ICNs) estimated by NeuroMark can serve as reliable biomarkers for separating BD and MDD. As shown in Fig. 10, the mean overall classification accuracy is 91.3%, the mean balanced accuracy is 91.2%, and the mean balanced precision is 91.5%. The individual-class accuracy for the BD group is 88.7% (precision is 93.8%), while the individual-class accuracy for MDD is 93.3% (precision is 90.0%). Importantly, we observed that some functional networks represented by IC 56, IC 33, IC 40, IC 98, IC 80, and IC 20 were frequently selected as features in all cross-validation runs (Fig. 11), which might suggest that they play a key role in separating BD and MDD. These functional networks engaged middle temporal gyrus, insula, precuneus, putamen, superior parietal lobule, and inferior occipital gyrus.
Fig. 10.
Results of study 4. The evaluated measures included individual-class accuracy of bipolar disorder (BD) and major depressive disorder (MDD) (BD_acc and MDD_acc), individual-class precision (BD_prec and MDD_prec), overall accuracy (Overall_acc), balanced accuracy (Bala_acc), and balanced precision (Bala_prec). For each measure, we show the values from 100 classification runs using both boxplot and violinplot, respectively.
Fig. 11.
Spatial maps of six most discriminative ICNs, each of which was selected from one functional domain (i.e., SC, AU, SM, VI, CC, and DM).
4. Discussion
The current diagnosis of brain disorders overwhelmingly relies on the patterns of clinical symptoms. Neuroimaging measures may hold more objective, biology-based quantification of brain abnormalities and consequently serve as potential biomarkers to guide diagnosis and treatment. However, the human brain is highly complex and the neuroimaging signals are confounded by various noises. This requires the neuroscience community to analyze big-data samples that might be assembled from multi-site studies to achieve enough statistical power for capturing more reliable findings. Studying the brain alterations in multiple brain disorders via using neuroimaging data collected from different studies would advance our understanding of their underlying mechanisms and relationships, which might help to redefine the disorder categories or develop new subtypes (Marquand et al., 2016, Du et al., 2018).
Functional connectivity, which evaluates the interaction of spatially distributed brain regions, has been suggested to be associated with cognition and many mental activities (Bressler and Menon, 2010). Studies of functional connectivity also improve the understanding of functional alterations caused by brain disorders (Sheline et al., 2009, Öngür et al., 2010, Xia et al., 2019). ICA is a promising method that can be used to extract functional connectivity measures (Calhoun et al., 2001, Allen et al., 2014), like intra-network connectivity (spatial maps of ICNs) and inter-network connectivity (functional network connectivity). Compared with ROI-based analysis, ICA is capable of extracting functional connectivity features that retain more individual-level variability (Yu et al., 2017), which might provide more statistical power benefiting the analysis of cross-disorders brain abnormalities. However, ICA is a data-driven approach which might result in different components identified across data. Such inconsistency of identified components and their arrangements may hinder finding replication and cross-study comparison.
To address this issue, we proposed the NeuroMark pipeline, a priori-driven ICA informed by reliable network templates to achieve linked analyses among different datasets, studies, and disorders. In this pipeline, the network templates were first generated using two large-sample HC populations (including 1828 subjects) in order to automatically maintain inter-subject correspondence of networks. The use of such prior information can greatly reduce the search space and improve the likelihood of detecting useful biomarkers (Cohen et al., 2017). Then, subject-specific ICNs and TCs were estimated based on the network templates, and this procedure would not only maintain the correspondence of extracted networks but also achieve more individual variances by optimizing the subject-specific network independence (Lin et al., 2010, Du and Fan, 2013).
To assess the efficacy of NeuroMark, we performed four studies including six different brain disorders. The performance of NeuroMark was evaluated from different angles. The overall results clearly support that NeuroMark is capable of capturing functional connectivity abnormalities that can be replicated across datasets and can be compared across brain disorders. NeuroMark also shows promising ability in characterizing atypical dynamic functional connectivity features across progressively developing disorders. More importantly, the functional connectivity features extracted by NeuroMark can serve as reliable biomarkers that can be used for the classification of brain disorders.
In study 1, by using the NeuroMark pipeline, we explored the whole-brain sFNC abnormalities in two independent SZ datasets. Similar abnormalities in sFNC were observed across datasets, mainly involving the subcortical, sensory, and cerebellar networks. Specifically, SZ shows increased sFNC between subcortical networks (including thalamus and caudate) and sensory networks. Being validated across datasets, these findings are consistent with and significantly extend previous reports about hyperconnectivity between subcortical and sensory regions using both atlas-based and ICA approaches (Woodward et al., 2012, Anticevic et al., 2014, Damaraju et al., 2014a, Damaraju et al., 2014b, Fu et al., 2018a, Fu et al., 2018b). For example, Woodward et al. have shown that SZ exhibits increased connectivity between thalamus and somatosensory regions by evaluating the connectivity between thalamic region and the other cortical regions (Woodward et al., 2012). Chen et al. (2019) revealed that thalamic hyperconnectivity with sensorimotor areas is related to the severity of cognitive deficits and clinical symptoms in SZ. Studies using ICA also reveal that increased strengths of FNC between subcortical and sensory networks in SZ patients from both static and dynamic perspectives (Damaraju et al., 2014a, Damaraju et al., 2014b). The subcortical regions are important sensory gates that receive and deliver information to the cortical regions, supporting the basic brain functions and behaviors. The thalamus is known for its important roles in visual, auditory, motor activity, emotion, memory, and sensorimotor association functions, and the caudate nucleus integrates spatial information with motor behavior formulation. Considering the negative functional connectivity between subcortical and sensory regions in our results, our observed increased sFNC in SZ might indicate dysfunctional connectivity between those regions. Such disconnection between subcortical and sensory regions would influence the information flow in the SZ brain, which might further result in cognitive deficits and other clinical symptoms in SZ. We observed replicated decreased sFNC between subcortical networks and cerebellum, which are also in line with previous findings (Andreasen et al., 1998, Anticevic et al., 2014). Using a large sample of SZ patients, Anticevic et al. found hypoconnectivity between the thalamus and cerebellar regions in patients (Anticevic et al., 2014). Although cerebellum has been previously viewed as a key node of motor coordination, more and more documents show that it would participate in a broad range of cognitive functions in humans (Buckner, 2013). Recent studies using large-scale neuroimaging data have documented the potential associations between the structure and function of the cerebellum and schizophrenia (Cao and Cannon, 2019). Our results provide additional evidence supporting that the cerebellar dysfunction, especially the dysfunction in cortical-subcortical-cerebellar circuitry involved in the pathogenesis of schizophrenia (Cao and Cannon, 2019). Overall, the results of study 1 highlight the potential of NeuroMark as a tool for capturing reproducible connectivity changes.
In study 2, the NeuroMark pipeline was applied to an open source data for studying the sFNC abnormalities in ASD. With the help of NeuroMark, the extracted ICNs not only retained subject-specific variability but also had the same order and arrangement as that in the study 1, making it feasible to compare the results from two independent studies. Combining their results, we found that subcortical networks were significantly affected by both SZ and ASD. Similar to SZ, decreased sFNC between cerebellum and thalamus/caudate and increased sFNC between thalamus and superior temporal gyrus/postcentral gyrus were identified in ASD. One previous study (Cerliani et al., 2015) using ICA-based fMRI analysis also showed increased connectivity between networks encompassing thalamus with the sensorimotor networks in ASD. Indeed, the disruption of subcortical-cortical brain connectivity, especially between thalamic and sensory regions has been widely documented in literature (Minshew and Keller, 2010, Fu et al., 2019). The broken connection between thalamus and the cortical regions is suggested to a potential cause of the atypical sensory processing in ASD (Fu et al., 2019). Our results indicate that atypical thalamic-to-cortical connectivity in SZ and ASD might underlie the same impairments of cerebello-thalamo-cortical pathways in both diseases (Andreasen et al., 1998, Bailey et al., 1998). While our findings are consistent with previous work, this paper directly shows, for the first time, the overlap between SZ and ASD with respect to these changes. Besides those common sFNC abnormalities, we found that ASD also showed unique changes on sFNC involving the cognitive-control and default mode domains. Our results are in line with the abnormal functional connectivity which is observed both within the DMN and between the DMN and networks involved in higher cognitive processing (de Lacy et al., 2017). Another dynamic functional connectivity study found that children with ASD diagnoses spend less time in the dynamic state with strong DMN connectivity, which might result in overall decreased DMN connectivity, aligned with our findings (Rashid et al., 2018). SZ and ASD are currently conceptualized as distinct disorders, however there is overlap in symptoms such as social withdrawal and communication impairment (Ford et al., 2017). Historically, schizophrenia and autism were even once considered to be the same disorder expressed at different developmental periods. The exploration of common and unique brain changes in SZ and ASD using a standardized pipeline like NeuroMark would help to advance our understanding of the neuronal relationship between these clinically-overlapping diseases and provide biological evidence hinting at their possible underlying mechanisms.
The human brain is highly dynamic system that spontaneous brain activity and connectivity is rich with dynamic properties (Leonardi and Van De Ville, 2015). Increasing evidence has shown that the research into dynamic functional connectivity would provide more information on brain functions and organization that cannot be probed by its static counterpart (Chang and Glover, 2010). The study of dynamic functional connectivity also improves the understanding of impaired brain and the functional alterations caused by brain disorders (Du et al., 2016). In study 3, we tested the performance of NeuroMark on capturing dynamic functional connectivity features from large scale imaging data. Via the NeuroMark framework, the atypical dFNCs were studied on brain disorders which manifest symptom severity along a continuous spectrum (MCI to AD). Five states with highly variable connectivity patterns were identified, demonstrating the flexibility in the functional coordination between brain networks during the resting state. Compared to HCs, AD patients exhibited more occurrence in weakly-connected states but fewer occurrence in strongly-connected states, consistent with our previous findings observed from another independent data (Fu et al., 2019). We speculate that the increased occurrence in the weakly-connected states but decreased occurrence in the strongly-connected states would result in disrupted inter-region communication which influences the maintenance of the basic brain functions in dementia brain. Similar observations were found in other brain disorders, including bipolar disorder (Rashid et al., 2014), schizophrenia (Damaraju et al., 2014a, Damaraju et al., 2014b, Du et al., 2016b), and autism (Fu et al., 2018a, Fu et al., 2018b). Interestingly, we observed that MCI showed similar changing trends as AD, with a weaker degree than AD. MCI is suggested to be an intermediate stage between HC and AD and our results demonstrate that the biological changes in MCI also show an intermediate condition between HC and AD. We further divided the MCI group into EMCI and LMCI, such phenomenon can also be observed in the dynamic features from HC to EMCI to LMCI to AD, suggesting a continual change in neurodegenerative diseases. The converging results of study 3 show that NeuroMark is an effective framework that can capture subtle dFNC differences that help to characterize progression in cognitive impairment in dementia.
In study 4, we tested whether the functional connectivity features captured by NeuroMark can serve as reliable biomarkers that help to distinguish different disease groups. The spatial maps of the ICNs extracted via NeuroMark were used as the input in the classification of BD and MDD patients, who have highly overlapping depressive symptoms and usually are difficult to be separated in clinical practice. We achieved >90% overall accuracy and balance accuracy, which are comparable with previous work that tried to distinguish BD and MDD (Jie et al., 2015, Osuch et al., 2018). However, Osuch et al. performed component decomposition using all the MDD and BD data, which makes them difficult to extend their findings on the new coming subjects or on another independent data. Our results suggest that although the use of functional network features and sophisticated classification algorithms is not practical in routine clinical due to the difficulty to implement the approaches to the new coming data, the NeuroMark framework might help to overcome this. Our results also indicate that the most important regions that help to discriminate BD and MDD are middle temporal gyrus, insula, precuneus, putamen, superior parietal gyrus, and inferior occipital gyrus. The abnormalities in these regions have been widely reported in MDD (Wang et al., 2012, Peng et al., 2015, Schreiner et al., 2019) or BD (Favre et al., 2014) literature. Our results argue that although these regions shows atypical patterns in both MDD and BD, their abnormalities may pertain to the different pathophysiology of mood disorders and may be particularly helpful for separating patients with challenging mood diagnoses.
Although NeuroMark shows great promise on extracting comparable and reproducible functional connectivity abnormalities that can serve as valuable markers across brain disorders, one limitation is that the present network templates were obtained only based on two independent HC datasets. The templates can be progressively improved and refined as more datasets collected under different conditions are included, hopefully to generate functional network templates with greater reproducibility. Since matching group-level networks across multiple groups is difficult, an efficient matching technique will be needed in order to find the highly reproducible (matched) networks as templates. In addition, the network templates were estimated using a higher model-order (the number of ICs = 100). In future, we will explore network estimation at different parcellation levels. Considering the ability to link different datasets, studies, and disorders in our method, we also plan to provide a cloud computation platform that implements this approach. Our hope is that by using NeuroMark, functional connectivity features can be widely studied and compared among numerous brain disorders. In this paper, we did not compare the NeuroMark with other pipelines (such as other group ICA and ROI methods) in the four studies, because we are more interested in the application of the proposed pipeline in a broad range. However, our previous studies (Du and Fan, 2013, Du et al., 2016a, Salman et al., 2019) have shown the superiority of the core of NeuroMark (i.e. GIG-ICA) in its powerful capability of estimating individual networks. In this work, we validated that the unique property of network features can be captured in NeuroMark while the correspondence between the network templates and individual ICNs is well maintained. We also found that the template-ICN similarity is relatively stable across different datasets and populations (as shown in the section 3.2). Furthermore, we also investigated the correlation between the network templates and the subject-specific ICNs across groups with different age and head motion parameters. As shown in the supplementary section S4, our results indicate that age and head motion did not greatly influence the correspondence measure. We wanted to point out that in this work we carefully regressed out the age, gender, and site effects before the statistical analyses (see the section S3) and handled the motion in the preprocessing and FNC computation steps, which minimized the contamination of these effects on the group differences.
We also wanted to mention that in this paper we identified the group differences under the guidance of diagnosis labels in studies 1–3 and classifying different subjects using models and features obtained by believing in the class labels in study 4. Our work accords with most of previous studies in the neuroscience field (Du et al., 2018), however, it is obvious that the diagnosis labels could be inaccurate. Refining mental disorder categories with the help of neuroimaging technique has attracted a lot of interests (Insel et al., 2010, Cuthbert and Insel, 2013, Fusar‐Poli et al., 2019), and unsupervised or semi-supervised clustering is the mostly popular method to solve the regrouping problem (Marquand et al., 2016). The biggest difficulty is to select good measures which can reflect subject-unique characteristics while still be corresponding across subjects for being used as features in clustering, under the situation of lack of group label information. We believe that NeuroMark helps to facilitate this issue, as our studies support that subtle data variation can be captured and linking of data can be easily achieved by NeuroMark.
Although the current NeuroMark framework was only applied for the analysis of fMRI datasets, it can also be expanded to other modalities. Taking structural MRI for example, source-based morphometry (SBM) (Xu et al., 2009, Bergsland et al., 2018), a multivariate version of voxel-based morphometry (VBM), applies ICA to gray matter maps to detect common covariation among subjects and subject-associated weights. It is apparent that results of SBM vary across different datasets and runs. Using our method taking reliable priors as guidance, the covariation patterns can be linked, thus resulting in comparable weights as features across different data (see regression based example here (Silva et al., 2014)). Group ICA is also useful for analyzing electroencephalography (EEG) data. Previous studies (Huster et al., 2015, Huster and Raud, 2018) extracted EEG sources by concatenating the data across the spatial dimension (see also the EEGIFT software: http://trendscenter.org/software). Generating a priori sources to guide the individual source computation in different modalities will be an ongoing effort.
In summary, we proposed an ICA-based framework to generalize and standardize the calculation of possible functional connectivity features that leverages the benefits of a data-driven approach and also provides comparability across multiple analyses. Via four different example studies, we highlight the validity of this framework. We hope this will be a useful stepping stone towards eventual application of such approaches in the clinic.
Author contributions
Yuhui Du proposed the whole analysis method and conducted the analyses on studies 1 and 2. Yuhui Du and Zening Fu drafted the manuscript and prepared the figures. Zening Fu worked on study 3 and revised the paper. Jing Sui, Shuang Gao, Yuhui Du, and Ying Xing worked on study 4. Yuhui Du, Zening Fu, Dongdong Lin, Mustafa Salman, Anees Abrol, and Md Abdur Rahaman worked on the data preprocessing, network template extraction, and network measure computation. L Elliot Hong and Peter Kochunov collected the MPRC data and revised the paper. Jiayu Chen revised the paper. Vince Calhoun supervised the work and edited the paper. All authors have given final approval of this version of the article.
CRediT authorship contribution statement
Yuhui Du: Methodology, Investigation, Writing - review & editing, Funding acquisition. Zening Fu: Writing - review & editing, Investigation. Jing Sui: Writing - review & editing. Shuang Gao: Writing - review & editing, Investigation. Ying Xing: Writing - review & editing, Investigation. Dongdong Lin: Writing - review & editing. Mustafa Salman: Writing - review & editing. Anees Abrol: Writing - review & editing. Md Abdur Rahaman: Writing - review & editing. Jiayu Chen: Writing - review & editing. L. Elliot Hong: Writing - review & editing, Data collection. Peter Kochunov: Writing - review & editing, Data collection. Elizabeth A. Osuch: Writing - review & editing, Data collection. Vince D. Calhoun: Writing - review & editing, Funding acquisition.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
This work was supported by National Natural Science Foundation of China (Grant No. 61703253 to YHD, 61773380 to JS), National Institutes of Health grants 5P20RR021938/P20GM103472 & R01EB020407 and National Science Foundation grant 1539067 (to VDC), and the 1331 Engineering Project of Shanxi Province, China.
Data collection and sharing for Study 3 was funded by the Alzheimer's Disease Neuroimaging Initiative (ADNI) (National Institutes of Health Grant U01 AG024904) and DOD ADNI (Department of Defense award number W81XWH-12-2-0012). ADNI is funded by the National Institute on Aging, the National Institute of Biomedical Imaging and Bioengineering, and through generous contributions from the following: AbbVie, Alzheimer’s Association; Alzheimer’s Drug Discovery Foundation; Araclon Biotech; BioClinica, Inc.; Biogen; Bristol-Myers Squibb Company; CereSpir, Inc.; Cogstate; Eisai Inc.; Elan Pharmaceuticals, Inc.; Eli Lilly and Company; EuroImmun; F. Hoffmann-La Roche Ltd and its affiliated company Genentech, Inc.; Fujirebio; GE Healthcare; IXICO Ltd.; Janssen Alzheimer Immunotherapy Research & Development, LLC.; Johnson & Johnson Pharmaceutical Research & Development LLC.; Lumosity; Lundbeck; Merck & Co., Inc.; Meso Scale Diagnostics, LLC.; NeuroRx Research; Neurotrack Technologies; Novartis Pharmaceuticals Corporation; Pfizer Inc.; Piramal Imaging; Servier; Takeda Pharmaceutical Company; and Transition Therapeutics. The Canadian Institutes of Health Research is providing funds to support ADNI clinical sites in Canada. Private sector contributions are facilitated by the Foundation for the National Institutes of Health (www.fnih.org). The grantee organization is the Northern California Institute for Research and Education, and the study is coordinated by the Alzheimer’s Therapeutic Research Institute at the University of Southern California. ADNI data are disseminated by the Laboratory for Neuro Imaging at the University of Southern California.
Data availability statement
The code and network templates used in this paper are available online (www.yuhuidu.com and http://trendscenter.org/software).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.nicl.2020.102375.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- Abrol A., Damaraju E., Miller R.L., Stephen J.M., Claus E.D., Mayer A.R., Calhoun V.D. Replicability of time-varying connectivity patterns in large resting state fMRI samples. Neuroimage. 2017;163:160–176. doi: 10.1016/j.neuroimage.2017.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen, E.A., Damaraju, E., Plis, S.M., Erhardt, E.B., Eichele, T., Calhoun, V.D., 2014. Tracking whole-brain connectivity dynamics in the resting state. Cereb. Cortex 24(3), 663–676. [DOI] [PMC free article] [PubMed]
- Andreasen N.C., Paradiso S., O'Leary D.S. Cognitive dysmetria“ as an integrative theory of schizophrenia: a dysfunction in cortical-subcortical-cerebellar circuitry? Schizophr. Bull. 1998;24(2):203–218. doi: 10.1093/oxfordjournals.schbul.a033321. [DOI] [PubMed] [Google Scholar]
- Anticevic A., Cole M.W., Repovs G., Murray J.D., Brumbaugh M.S., Winkler A.M., Savic A., Krystal J.H., Pearlson G.D., Glahn D.C. Characterizing thalamo-cortical disturbances in schizophrenia and bipolar illness. Cerebral Cortex. 2014;24(12):3116–3130. doi: 10.1093/cercor/bht165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey A., Luthert P., Dean A., Harding B., Janota I., Montgomery M., Rutter M., Lantos P. A clinicopathological study of autism. Brain. 1998;121(Pt 5):889–905. doi: 10.1093/brain/121.5.889. [DOI] [PubMed] [Google Scholar]
- Beckmann C., Mackay C., Filippini N., Smith S. Group comparison of resting-state FMRI data using multi-subject ICA and dual regression. Neuroimage. 2009;47(Supplement 1):S148. [Google Scholar]
- Bell A.J., Sejnowski T.J. An information-maximization approach to blind separation and blind deconvolution. Neural Comput. 1995;7(6):1129–1159. doi: 10.1162/neco.1995.7.6.1129. [DOI] [PubMed] [Google Scholar]
- Bergsland N., Horakova D., Dwyer M.G., Uher T., Vaneckova M., Tyblova M., Seidl Z., Krasensky J., Havrdova E., Zivadinov R. Gray matter atrophy patterns in multiple sclerosis: a 10-year source-based morphometry study. Neuroimage Clin. 2018;17:444–451. doi: 10.1016/j.nicl.2017.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bressler S.L., Menon V. Large-scale brain networks in cognition: emerging methods and principles. Trends Cogn. Sci. 2010;14(6):277–290. doi: 10.1016/j.tics.2010.04.004. [DOI] [PubMed] [Google Scholar]
- Buckner R. The cerebellum and cognitive function: 25 years of insight from anatomy and neuroimaging. Neuron. 2013;80(3):807–815. doi: 10.1016/j.neuron.2013.10.044. [DOI] [PubMed] [Google Scholar]
- Calhoun V.D., Adali T. Multisubject independent component analysis of fMRI: a decade of intrinsic networks, default mode, and neurodiagnostic discovery. IEEE Rev. Biomed. Eng. 2012;5:60–73. doi: 10.1109/RBME.2012.2211076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calhoun V.D., Adali T., Pearlson G.D., Pekar J.J. A method for making group inferences from functional MRI data using independent component analysis. Hum. Brain Mapp. 2001;14(3):140–151. doi: 10.1002/hbm.1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calhoun V.D., de Lacy N. Ten key observations on the analysis of resting-state functional MR imaging data using independent component analysis. Neuroimaging Clin. North Am. 2017;27(4):561–579. doi: 10.1016/j.nic.2017.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao H., Cannon T.D. Cerebellar dysfunction and schizophrenia: from “Cognitive Dysmetria” to a potential therapeutic target. Am. J. Psychiatry. 2019;176(7):498–500. doi: 10.1176/appi.ajp.2019.19050480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerliani L., Mennes M., Thomas R.M., Di Martino A., Thioux M., Keysers C. Increased functional connectivity between subcortical and cortical resting-state networks in autism spectrum disorder. JAMA Psychiatry. 2015;72(8):767. doi: 10.1001/jamapsychiatry.2015.0101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chand, G.B., Dwyer, D.B., Erus, G., Sotiras, A., Varol, E., Srinivasan, D., Doshi, J., Pomponio, R., Pigoni, A., Dazzan, P., Kahn, R.S., Schnack, H.G., Zanetti, M.V., Meisenzahl, E., Busatto, G.F., Crespo-Facorro, B., Pantelis, C., Wood, S.J., Zhuo, C., Shinohara, R.T., Shou, H., Fan, Y., Gur, R.C., Gur, R.E., Satterthwaite, T.D., Koutsouleris, N., Wolf, D.H., Davatzikos, C., 2020. Two distinct neuroanatomical subtypes of schizophrenia revealed using machine learning. Brain. [DOI] [PMC free article] [PubMed]
- Chang C., Glover G.H. Time–frequency dynamics of resting-state brain connectivity measured with fMRI. NeuroImage. 2010;50(1):81–98. doi: 10.1016/j.neuroimage.2009.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen P., Ye E., Jin X., Zhu Y., Wang L. Association between thalamocortical functional connectivity abnormalities and cognitive deficits in schizophrenia. Sci. Rep. 2019;9(1) doi: 10.1038/s41598-019-39367-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen J.D., Daw N., Engelhardt B., Hasson U., Li K., Niv Y., Norman K.A., Pillow J., Ramadge P.J., Turk-Browne N.B., Willke T.L. Computational approaches to fMRI analysis. Nat. Neurosci. 2017;20(3):304–313. doi: 10.1038/nn.4499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosgrove V.E., Suppes T. Informing DSM-5: biological boundaries between bipolar I disorder, schizoaffective disorder, and schizophrenia. BMC Med. 2013;11:127. doi: 10.1186/1741-7015-11-127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuadros-Rodriguez L., Perez-Castano E., Ruiz-Samblas C. Quality performance metrics in multivariate classification methods for qualitative analysis. Trac-Trends Anal. Chem. 2016;80:612–624. [Google Scholar]
- Cuthbert B.N., Insel T.R. Toward the future of psychiatric diagnosis: the seven pillars of RDoC. BMC Med. 2013;11:126. doi: 10.1186/1741-7015-11-126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damaraju E., Allen E., Belger A., Ford J., McEwen S., Mathalon D., Mueller B., Pearlson G., Potkin S., Preda A. Dynamic functional connectivity analysis reveals transient states of dysconnectivity in schizophrenia. NeuroImage: Clin. 2014;5:298–308. doi: 10.1016/j.nicl.2014.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damaraju E., Allen E.A., Belger A., Ford J.M., McEwen S., Mathalon D.H., Mueller B.A., Pearlson G.D., Potkin S.G., Preda A., Turner J.A., Vaidya J.G., van Erp T.G., Calhoun V.D. Dynamic functional connectivity analysis reveals transient states of dysconnectivity in schizophrenia. Neuroimage Clin. 2014;5:298–308. doi: 10.1016/j.nicl.2014.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Lacy N., Doherty D., King B.H., Rachakonda S., Calhoun V.D. Disruption to control network function correlates with altered dynamic connectivity in the wider autism spectrum. Neuroimage Clin. 2017;15:513–524. doi: 10.1016/j.nicl.2017.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demirci O., Clark V.P., Calhoun V.D. A projection pursuit algorithm to classify individuals using fMRI data: application to schizophrenia. NeuroImage. 2008;39(4):1774–1782. doi: 10.1016/j.neuroimage.2007.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dosenbach N.U., Nardos B., Cohen A.L., Fair D.A., Power J.D., Church J.A., Nelson S.M., Wig G.S., Vogel A.C., Lessov-Schlaggar C.N., Barnes K.A., Dubis J.W., Feczko E., Coalson R.S., Pruett J.R., Jr., Barch D.M., Petersen S.E., Schlaggar B.L. Prediction of individual brain maturity using fMRI. Science. 2010;329(5997):1358–1361. doi: 10.1126/science.1194144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Y., Fryer S.L., Fu Z., Lin D., Sui J., Chen J., Damaraju E., Mennigen E., Stuart B., Loewy R.L., Mathalon D.H., Calhoun V.D. Dynamic functional connectivity impairments in early schizophrenia and clinical high-risk for psychosis. Neuroimage. 2018;180(Pt B):632–645. doi: 10.1016/j.neuroimage.2017.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Y., Allen E.A., He H., Sui J., Wu L., Calhoun V.D. Artifact removal in the context of group ICA: a comparison of single-subject and group approaches: artifact removal in the context of group ICA. Hum. Brain Mapp. 2016;37(3):1005–1025. doi: 10.1002/hbm.23086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Y., Fan Y. Group information guided ICA for fMRI data analysis. Neuroimage. 2013;69:157–197. doi: 10.1016/j.neuroimage.2012.11.008. [DOI] [PubMed] [Google Scholar]
- Du, Y., Fu, Z.N., Calhoun, V.D., 2018. Classification and prediction of brain disorders using functional connectivity: promising but challenging, Front. Neurosci. 12. [DOI] [PMC free article] [PubMed]
- Du, Y., Li, H.M., Wu, H., Fan, Y., 2012. Identification of subject specific and functional consistent ROIs using semi-supervised learning. Proc. SPIE, Medical Imag. 2012: Imag. Proc. 8314.
- Du Y., Lin D.D., Yu Q.B., Sui J., Chen J.Y., Rachakonda S., Adali T., Calhoun V.D. Comparison of IVA and GIG-ICA in brain functional network estimation using fMRI data. Front. Neurosci. 2017;11:267. doi: 10.3389/fnins.2017.00267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Y., Pearlson G.D., Yu Q., He H., Lin D., Sui J., Wu L., Calhoun V.D. Interaction among subsystems within default mode network diminished in schizophrenia patients: a dynamic connectivity approach. Schizophrenia Res. 2016;170(1):55–65. doi: 10.1016/j.schres.2015.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dwyer, D.B., Cabral, C., Kambeitz-Ilankovic, L., Sanfelici, R., Kambeitz, J., Calhoun, V.D., Falkai, P., Pantelis, C., Meisenzahl, E., Koutsouleris, N., 2018. Brain subtyping enhances the neuroanatomical discrimination of schizophrenia, Schizophr. Bull. 44(5) 1060–1069. [DOI] [PMC free article] [PubMed]
- Favre P., Baciu M., Pichat C., Bougerol T., Polosan M. fMRI evidence for abnormal resting-state functional connectivity in euthymic bipolar patients. J. Affect Disord. 2014;165:182–189. doi: 10.1016/j.jad.2014.04.054. [DOI] [PubMed] [Google Scholar]
- Ford T.C., Apputhurai P., Meyer D., Crewther D.P. Confirmatory factor analysis of autism and schizophrenia spectrum traits. Personality Individual Diff. 2017;110:80–84. [Google Scholar]
- Fu Z., Caprihan A., Chen J., Du Y., Adair J.C., Sui J., Rosenberg G.A., Calhoun V.D. Altered static and dynamic functional network connectivity in Alzheimer's disease and subcortical ischemic vascular disease: shared and specific brain connectivity abnormalities. Hum. Brain Mapp. 2019;40(11):3203–3221. doi: 10.1002/hbm.24591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Z., Tu Y., Di X., Du Y., Pearlson G.D., Turner J.A., Biswal B.B., Zhang Z., Calhoun V.D. Characterizing dynamic amplitude of low-frequency fluctuation and its relationship with dynamic functional connectivity: an application to schizophrenia. Neuroimage. 2018;180(Pt B):619–631. doi: 10.1016/j.neuroimage.2017.09.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Z., Tu Y., Di X., Du Y., Sui J., Biswal B.B., Zhang Z., de Lacy N., Calhoun V.D. Transient increased thalamic-sensory connectivity and decreased whole-brain dynamism in autism. NeuroImage. 2018 doi: 10.1016/j.neuroimage.2018.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Z., Tu Y., Di X., Du Y., Sui J., Biswal B.B., Zhang Z., de Lacy N., Calhoun V.D. Transient increased thalamic-sensory connectivity and decreased whole-brain dynamism in autism. Neuroimage. 2019;190:191–204. doi: 10.1016/j.neuroimage.2018.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fusar‐Poli P., Solmi M., Brondino N., Davies C., Chae C., Politi P., Borgwardt S., Lawrie S.M., Parnas J., McGuire P. Transdiagnostic psychiatry: a systematic review. World Psychiatry. 2019;18(2):192–207. doi: 10.1002/wps.20631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Himberg, J., Hyvarinen, A., 2003. ICASSO: software for investigating the reliability of ICA estimates by clustering and visualization. In: 2003 IEEE XIII Workshop on Neural Networks for Signal Processing – Nnsp'03, pp. 259–268.
- Hommer R.E., Swedo S.E. Schizophrenia and autism--related disorders. Schizophr. Bull. 2015;41(2):313–314. doi: 10.1093/schbul/sbu188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honnorat N., Dong A., Meisenzahl-Lechner E., Koutsouleris N., Davatzikos C. Neuroanatomical heterogeneity of schizophrenia revealed by semi-supervised machine learning methods. Schizophr. Res. 2019;214:43–50. doi: 10.1016/j.schres.2017.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huster R.J., Plis S.M., Calhoun V.D. Group-level component analyses of EEG: validation and evaluation. Front. Neurosci. 2015;9:254. doi: 10.3389/fnins.2015.00254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huster R.J., Raud L. A tutorial review on multi-subject decomposition of EEG. Brain Topogr. 2018;31(1):3–16. doi: 10.1007/s10548-017-0603-x. [DOI] [PubMed] [Google Scholar]
- Hutchison R.M., Womelsdorf T., Gati J.S., Everling S., Menon R.S. Resting-state networks show dynamic functional connectivity in awake humans and anesthetized macaques: Dynamic Functional Connectivity. Hum. Brain Mapp. 2013;34(9):2154–2177. doi: 10.1002/hbm.22058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Insel T., Cuthbert B., Garvey M., Heinssen R., Pine D.S., Quinn K., Sanislow C., Wang P. Research Domain Criteria (RDoC): toward a new classification framework for research on mental disorders. AJP. 2010;167(7):748–751. doi: 10.1176/appi.ajp.2010.09091379. [DOI] [PubMed] [Google Scholar]
- Jie N.F., Zhu M.H., Ma X.Y., Osuch E.A., Wammes M., Theberge J., Li H.D., Zhang Y., Jiang T.Z., Sui J., Calhoun V.D. Discriminating bipolar disorder from major depression based on SVM-FoBa: efficient feature selection with multimodal brain imaging data. IEEE Trans. Auton. Mental Dev. 2015;7(4):320–331. doi: 10.1109/TAMD.2015.2440298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonardi N., Van De Ville D. On spurious and real fluctuations of dynamic functional connectivity during rest. Neuroimage. 2015;104:430–436. doi: 10.1016/j.neuroimage.2014.09.007. [DOI] [PubMed] [Google Scholar]
- Li A., Zalesky A., Yue W., Howes O., Yan H., Liu Y., Fan L., Whitaker K.J., Xu K., Rao G., Li J., Liu S., Wang M., Sun Y., Song M., Li P., Chen J., Chen Y., Wang H., Liu W., Li Z., Yang Y., Guo H., Wan P., Lv L., Lu L., Yan J., Song Y., Wang H., Zhang H., Wu H., Ning Y., Du Y., Cheng Y., Xu J., Xu X., Zhang D., Wang X., Jiang T., Liu B. A neuroimaging biomarker for striatal dysfunction in schizophrenia. Nat. Med. 2020;26(4):558–565. doi: 10.1038/s41591-020-0793-8. [DOI] [PubMed] [Google Scholar]
- Lin Q.H., Liu J., Zheng Y.R., Liang H., Calhoun V.D. Semiblind spatial ICA of fMRI using spatial constraints. Hum. Brain Mapp. 2010;31(7):1076–1088. doi: 10.1002/hbm.20919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Liang M., Zhou Y., He Y., Hao Y., Song M., Yu C., Liu H., Liu Z., Jiang T. Disrupted small-world networks in schizophrenia. Brain. 2008;131(Pt 4):945–961. doi: 10.1093/brain/awn018. [DOI] [PubMed] [Google Scholar]
- Ma S., Correa N.M., Li X.L., Eichele T., Calhoun V.D., Adali T. Automatic identification of functional clusters in FMRI data using spatial dependence. IEEE Trans. Biomed. Eng. 2011;58(12):3406–3417. doi: 10.1109/TBME.2011.2167149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marquand A.F., Wolfers T., Mennes M., Buitelaar J., Beckmann C.F. Beyond lumping and splitting: a review of computational approaches for stratifying psychiatric disorders. Biol. Psychiatry Cogn. Neurosci. Neuroimag. 2016;1(5):433–447. doi: 10.1016/j.bpsc.2016.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marusak H.A., Calhoun V.D., Brown S., Crespo L.M., Sala‐Hamrick K., Gotlib I.H., Thomason M.E. Dynamic functional connectivity of neurocognitive networks in children. Hum. Brain Mapp. 2017;38(1):97–108. doi: 10.1002/hbm.23346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKeown M.J., Hansen L.K., Sejnowsk T.J. Independent component analysis of functional MRI: what is signal and what is noise? Curr. Opin. Neurobiol. 2003;13(5):620–629. doi: 10.1016/j.conb.2003.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minshew N.J., Keller T.A. The nature of brain dysfunction in autism: functional brain imaging studies. Curr. Opin. Neurol. 2010;23(2):124–130. doi: 10.1097/WCO.0b013e32833782d4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noble S., Scheinost D., Constable R.T. A decade of test-retest reliability of functional connectivity: a systematic review and meta-analysis. Neuroimage. 2019;116157 doi: 10.1016/j.neuroimage.2019.116157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Öngür D., Lundy M., Greenhouse I., Shinn A.K., Menon V., Cohen B.M., Renshaw P.F. Default mode network abnormalities in bipolar disorder and schizophrenia. Psychiatry Res. Neuroimag. 2010;183(1):59–68. doi: 10.1016/j.pscychresns.2010.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osuch E., Gao S., Wammes M., Théberge J., Williamson P., Neufeld R.J., Du Y., Sui J., Calhoun V.D. Complexity in mood disorder diagnosis: fMRI connectivity networks predicted medication-class of response in complex patients. Acta Psychiatr. Scand. 2018;138(5):472–482. doi: 10.1111/acps.12945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng D., Liddle E.B., Iwabuchi S.J., Zhang C., Wu Z., Liu J., Jiang K., Xu L., Liddle P.F., Palaniyappan L., Fang Y. Dissociated large-scale functional connectivity networks of the precuneus in medication-naive first-episode depression. Psychiatry Res. 2015;232(3):250–256. doi: 10.1016/j.pscychresns.2015.03.003. [DOI] [PubMed] [Google Scholar]
- Poldrack R.A., Gorgolewski K.J. Making big data open: data sharing in neuroimaging. Nat. Neurosci. 2014;17(11):1510–1517. doi: 10.1038/nn.3818. [DOI] [PubMed] [Google Scholar]
- Poline J.B., Breeze J.L., Ghosh S., Gorgolewski K., Halchenko Y.O., Hanke M., Haselgrove C., Helmer K.G., Keator D.B., Marcus D.S., Poldrack R.A., Schwartz Y., Ashburner J., Kennedy D.N. Data sharing in neuroimaging research. Front. Neuroinform. 2012;6:9. doi: 10.3389/fninf.2012.00009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rashid B., Arbabshirani M.R., Damaraju E., Cetin M.S., Miller R., Pearlson G.D., Calhoun V.D. Classification of schizophrenia and bipolar patients using static and dynamic resting-state fMRI brain connectivity. Neuroimage. 2016;134:645–657. doi: 10.1016/j.neuroimage.2016.04.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rashid B., Blanken L.M.E., Muetzel R.L., Miller R., Damaraju E., Arbabshirani M.R., Erhardt E.B., Verhulst F.C., van der Lugt A., Jaddoe V.W.V., Tiemeier H., White T., Calhoun V.D. Connectivity dynamics in typical development and its relationship to autistic traits and autism spectrum disorder. Hum. Brain. Mapp. 2018;39(8):3127–3142. doi: 10.1002/hbm.24064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rashid B., Damaraju E., Pearlson G.D., Calhoun V.D. Dynamic connectivity states estimated from resting fMRI Identify differences among Schizophrenia, bipolar disorder, and healthy control subjects. Front. Hum. Neurosci. 2014;8:897. doi: 10.3389/fnhum.2014.00897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salman M.S., Du Y., Lin D., Fu Z., Fedorov A., Damaraju E., Sui J., Chen J., Mayer A., Rosse S., Mathalon D.H., Ford J.M., Erp T.V., Calhoun V.D. Group ICA for identifying biomarkers in Schizophrenia: 'Adaptive' networks via spatially constrained ICA show more sensitivity to group differences than spatio-temporal regression. Neuroimage Clin. 2019;22:101747. doi: 10.1016/j.nicl.2019.101747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schreiner M.W., Klimes-Dougan B., Cullen K.R. Neural correlates of suicidality in adolescents with major depression: resting-state functional connectivity of the precuneus and posterior cingulate cortex. Suicide Life Threat Behav. 2019;49(3):899–913. doi: 10.1111/sltb.12471. [DOI] [PubMed] [Google Scholar]
- Sheline Y.I., Barch D.M., Price J.L., Rundle M.M., Vaishnavi S.N., Snyder A.Z., Mintun M.A., Wang S., Coalson R.S., Raichle M.E. The default mode network and self-referential processes in depression. PNAS. 2009;106(6):1942–1947. doi: 10.1073/pnas.0812686106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva, R.F., Castro, E., Gupta, C.N., C. M., Arbabshirani, M., Potluru, V.K., Plis, S.M., Calhoun, V.D., 2014. The tenth annual MLSP competition: schizophrenia classification challenge. In: 2014 IEEE International Workshop on Machine Learning for Signal Processing (MLSP), pp. 1–6.
- Smith S.M., Fox P.T., Miller K.L., Glahn D.C., Fox P.M., Mackay C.E., Filippini N., Watkins K.E., Toro R., Laird A.R., Beckmann C.F. Correspondence of the brain's functional architecture during activation and rest. Proc. Natl. Acad. Sci. 2009;106(31):13040–13045. doi: 10.1073/pnas.0905267106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun, H., Lui, S., Yao, L., Deng, W., Xiao, Y., Zhang, W., Huang, X., Hu, J., Bi, F., Li, T., Sweeney, J.A., Gong, Q., 2015. Two patterns of white matter abnormalities in medication-naive patients with first-episode schizophrenia revealed by diffusion tensor imaging and cluster analysis. JAMA Psychiatry 72(7), 678–686. [DOI] [PubMed]
- Tzourio-Mazoyer N., Landeau B., Papathanassiou D., Crivello F., Etard O., Delcroix N., Mazoyer B., Joliot M. Automated anatomical labeling of activations in SPM using a macroscopic anatomical parcellation of the MNI MRI single-subject brain. NeuroImage. 2002;15(1):273–289. doi: 10.1006/nimg.2001.0978. [DOI] [PubMed] [Google Scholar]
- Wang L., Hermens D.F., Hickie I.B., Lagopoulos J. A systematic review of resting-state functional-MRI studies in major depression. J. Affect. Disord. 2012;142(1–3):6–12. doi: 10.1016/j.jad.2012.04.013. [DOI] [PubMed] [Google Scholar]
- Whitfield-Gabrieli S., Thermenos H.W., Milanovic S., Tsuang M.T., Faraone S.V., McCarley R.W., Shenton M.E., Green A.I., Nieto-Castanon A., LaViolette P., Wojcik J., Gabrieli J.D.E., Seidman L.J. Hyperactivity and hyperconnectivity of the default network in schizophrenia and in first-degree relatives of persons with schizophrenia. Proc. Natl. Acad. Sci. 2009;106(4):1279–1284. doi: 10.1073/pnas.0809141106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo C.W., Chang L.J., Lindquist M.A., Wager T.D. Building better biomarkers: brain models in translational neuroimaging. Nat. Neurosci. 2017;20(3):365–377. doi: 10.1038/nn.4478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodward N.D., Karbasforoushan H., Heckers S. Thalamocortical dysconnectivity in schizophrenia. AJP. 2012;169(10):1092–1099. doi: 10.1176/appi.ajp.2012.12010056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia, M., Womer, F.Y., Chang, M., Zhu, Y., Zhou, Q., Edmiston, E.K., Jiang, X., Wei, S., Duan, J., Xu, K., Tang, Y., He Y., Wang, F., 2019. Shared and distinct functional architectures of brain networks across psychiatric disorders. Schizophr. Bull. 45(2) 450–463. [DOI] [PMC free article] [PubMed]
- Xu J., Calhoun V.D., Worhunsky P.D., Xiang H., Li J., Wall J.T., Pearlson G.D., Potenza M.N. Functional network overlap as revealed by fMRI using sICA and its potential relationships with functional heterogeneity, balanced excitation and inhibition, and sparseness of neuron activity. PLoS One. 2015;10(2):e0117029. doi: 10.1371/journal.pone.0117029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu L., Groth K.M., Pearlson G., Schretlen D.J., Calhoun V.D. Source-based morphometry: the use of independent component analysis to identify gray matter differences with application to schizophrenia. Hum. Brain Mapp. 2009;30(3):711–724. doi: 10.1002/hbm.20540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z., Xu Y., Xu T., Hoy C.W., Handwerker D.A., Chen G., Northoff G., Zuo X.N., Bandettini P.A. Brain network informed subject community detection in early-onset schizophrenia. Sci. Rep. 2014;4:5549. doi: 10.1038/srep05549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Q.B., Du Y., Chen J.Y., He H., Sui J., Pearlson G., Calhoun V.D. Comparing brain graphs in which nodes are regions of interest or independent components: a simulation study. J. Neurosci. Methods. 2017;291:61–68. doi: 10.1016/j.jneumeth.2017.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zang Y., Jiang T., Lu Y., He Y., Tian L. Regional homogeneity approach to fMRI data analysis. NeuroImage. 2004;22(1):394–400. doi: 10.1016/j.neuroimage.2003.12.030. [DOI] [PubMed] [Google Scholar]
- Zuo X.N., Anderson J.S., Bellec P., Birn R.M., Biswal B.B., Blautzik J., Breitner J.C., Buckner R.L., Calhoun V.D., Castellanos F.X., Chen A., Chen B., Chen J., Chen X., Colcombe S.J., Courtney W., Craddock R.C., Di Martino A., Dong H.M., Fu X., Gong Q., Gorgolewski K.J., Han Y., He Y., He Y., Ho E., Holmes A., Hou X.H., Huckins J., Jiang T., Jiang Y., Kelley W., Kelly C., King M., LaConte S.M., Lainhart J.E., Lei X., Li H.J., Li K., Li K., Lin Q., Liu D., Liu J., Liu X., Liu Y., Lu G., Lu J., Luna B., Luo J., Lurie D., Mao Y., Margulies D.S., Mayer A.R., Meindl T., Meyerand M.E., Nan W., Nielsen J.A., O'Connor D., Paulsen D., Prabhakaran V., Qi Z., Qiu J., Shao C., Shehzad Z., Tang W., Villringer A., Wang H., Wang K., Wei D., Wei G.X., Weng X.C., Wu X., Xu T., Yang N., Yang Z., Zang Y.F., Zhang L., Zhang Q., Zhang Z., Zhang Z., Zhao K., Zhen Z., Zhou Y., Zhu X.T., Milham M.P. An open science resource for establishing reliability and reproducibility in functional connectomics. Sci Data. 2014;1:140049. doi: 10.1038/sdata.2014.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The code and network templates used in this paper are available online (www.yuhuidu.com and http://trendscenter.org/software).