Abstract
While almost all mycobacterial species are saprophytic environmental organisms, a few, such as Mycobacterium tuberculosis, have evolved to cause transmissible human infection. By analysing the recent emergence and spread of the environmental organism Mycobacterium abscessus through the global Cystic Fibrosis population, we have defined key, generalisable steps involved in the pathogenic evolution of mycobacteria. We show that epigenetic modifiers, acquired through horizontal gene transfer, cause saltational increases in pathogenic potential of specific environmental clones. Allopatric parallel evolution during chronic lung infection then promotes rapid increases in virulence, through mutations in a discrete gene network that enhance growth within macrophages but impair fomite survival. As a consequence, we observe constrained pathogenic evolution while person-to-person transmission remains indirect, but postulate accelerated pathogenic adaptation once direct transmission is possible, as observed for M. tuberculosis. Our findings indicate how key interventions, such as early treatment and cross-infection control, might restrict existing, and prevent new, emergent mycobacterial pathogens.
Mycobacterium abscessus, a rapidly growing multidrug-resistant species of nontuberculous mycobacteria (NTM), has recently emerged as a major threat to individuals with Cystic Fibrosis (CF) and other chronic lung conditions [1–5]. In CF, M. abscessus causes accelerated inflammatory lung damage [6], is frequently impossible to treat [2–4], and prevents safe lung transplantation [5, 7]. Infection rates are increasing globally [2–4], driven in part by indirect person-to-person transmission of M. abscessus [8–11], probably through the generation of long-lived infectious aerosols and via fomite spread [9].
Currently over 70% of infections in CF patients are caused by genetically clustered (and thus transmitted) isolates, of which the majority are from three dominant circulating clones (DCCs) that have emerged within the past 50 years and have spread globally [9] (Figure 1A). Clustered isolates are more virulent than non-clustered isolates (when tested in vitro and in vivo) and result in worse clinical outcomes [9], suggesting that they are evolving from environmental saprophytes into obligate lung pathogens (potentially in a similar way to the ancestral Mycobacterium tuberculosis over 6000 years ago [12–14]) and thus provide a unique opportunity to define the critical (and generalisable) steps involved in pathogenic evolution of mycobacteria.
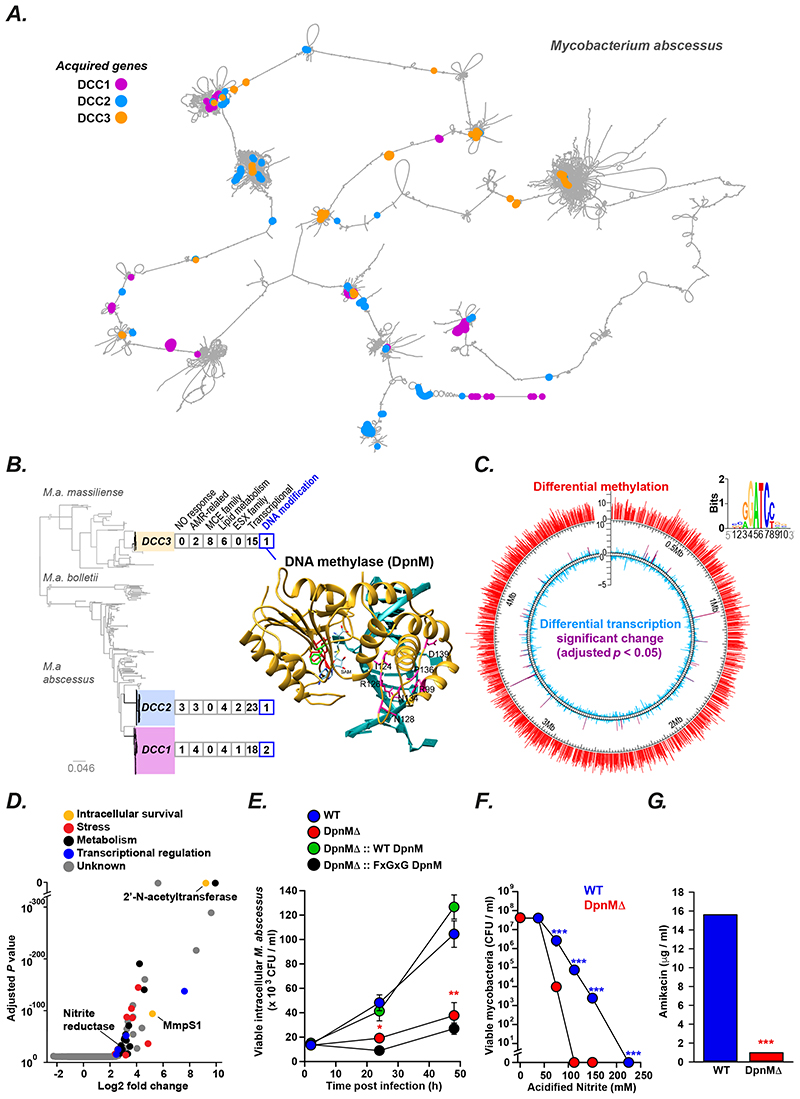
Figure 1. Saltational evolution of M. abscessus dominant circulating clones.
(A) Pangenome graph of M. abscessus (constructed using Panaroo [15]), where nodes represent clusters of orthologous genes and two nodes are connected by an edge if they are adjacent on a contig in any sample from the population, defines gene gain events associated with the emergence of DCC1 (purple), DCC2 (blue), and DCC3 (orange). For illustration purposes, the graph has been ordered against M. abscessus ATCC19997, and any long-range edges cut [34]. (B) Pangenome analysis of the three dominant circulating clones of M. abscessus (DCC1-3) revealed horizontal acquisition of potential virulence genes (complete gene list in Supplementary Table 1). All DCCs have independently acquired genes involved in DNA modification including a putative DNA methylase (DpnM) found in DCC3 (modelled structure shown: DPPY motif red; F42 green; bound DNA blue; predicted DNA-recognition residues magenta). (C) Genome-wide differential methylation (detected by SMRT sequencing; red) and transcription (monitored through RNAseq; global blue, significant differences purple) between wild type (WT) and DpnM knockout (DpnMΔ) M. abscessus, with predicted methylation motif shown as weblogo below. (D) Volcano plot of differentially expressed genes (log2 fold change greater than 2 or less than -2 with a corrected p-value less than 0.05) between WT and ΔDpnM M. abscessus, annotated by predicted function. (E) Survival in primary human macrophages was impaired in DpnMΔ (red) compared to wild type (blue), complemented by expression of wild type DpnM (green) but not DpnM mutant unable to bind substrate (black). All experiments were performed at least in triplicate on at least three separate occasions and data represented as the mean ± s.e with statistical significance determined using Student t-test. (F, G) DpnMΔ bacteria (red) are more susceptible to acidified nitrite (F) and amikacin (G) than wild type controls (blue). Experiments were performed in triplicate on at least three separate occasions. Results from representative experiements shown as mean ± s.e with statistical significance determined using two-tailed Student t-test (* p < 0.05, ** p <0.01. *** p < 0.001).
Horizontal gene transfer drives saltational evolution
We initially sought to understand how the DCCs may have emerged. We first examined whether mutational variation (through single nucleotide polymorphisms; SNPs) might explain the enhanced fitness of DCCs. However, analysis of the ratio of non-synonymous to synonymous SNPs (dN/dS) failed to provide evidence for positive selection on proteincoding genes, as the phylogenetic branches leading to the last common ancestors for each DCC demonstrated strong purifying selection (dN/dS < 1), at similar levels to that seen for non-DCC branches (Supplementary Figure 1).
We therefore examined whether gene acquisition through horizontal transfer might explain the increased fitness of the DCCs. To accurately analyse the accessory genome, we generated a pangenome graph (Figure 1A), with nodes as clusters of orthologous genes and two nodes linked by an edge if they are adjacent in any contig [15], and found that each DCC had acquired a functionally similar repertoire of accessory genes (Figure 1B; Supplementary Figure 2, Supplementary Table 1). We observed a significant enrichment of genes involved in transcriptional regulation or DNA modification in the DCCs compared to non DCC isolates (Fishers exact test p < 0.001), acquisition of which would be expected to lead to large phenotypic differences and thus saltational changes in evolutionary fitness. To explore this process experimentally, we focused on a putative DNA methyltransferase (DpnM; Figure 1B), that was acquired by DCC3, a clone of particular interest as it was responsible for several large outbreaks in CF centres in the UK [8] and US [9].
Comparing wild-type and knockout strains, we found that dpnM was responsible for N6-adenine DNA methylation (with RGATCY as the dominant motif) and that a dpnM knock-out caused global changes in gene expression (Figure 1B). We identified 52 differentially expressed genes, many of which are implicated in mycobacterial stress responses, metabolic re-wiring, transcriptional regulation, and intracellular survival (Figure 1C; Supplementary Table 2) including: an efflux pump (mmpS1; MAB_2649), reported to enhance growth within macrophages [16]; a nitrite reductase (MAB_3521c), potentially contributing to nitric oxide tolerance [17]; and a 2’-N-acetyltransferase (MAB_4395), orthologs of which have been shown to inactivate aminoglycoside antibiotics [18] and promote macrophage infection [19].
As predicted from these transcriptional changes, the DpnM knockout showed: impaired survival in primary human macrophages (Figure 1D), which could be functionally complemented by the wild type methylase (but not a mutant enzyme unable to bind S– adenosyl methionine); reduced tolerance to nitric oxide (Figure 1E); and increased susceptibility to amikacin (Figure 1F). It is therefore likely that these phenotypes were enhanced on initial acquisition of dpnM by DCC3.
Our results therefore indicate that horizontal gene transfer, particularly of global transcriptional regulators, can provide an important mechanism for creating large phenotypic variance in environmental M. abscessus isolates, and consequently enabling saltational evolution towards enhanced human infectivity. Importantly, we believe this process may be generalisable across mycobacterial species. Graphical pangenome analysis indicates that gene gain/loss events are associated with the pathogenic evolution of several virulent clones including: Cluster 1a within Mycobacterium avium [20]; Clone A within Mycobacterium canettii [21], and the monophyletic M. tuberculosis complex (MTBC) from an M. canettii-like ancestor (Supplementary Figures 2 & 3, Supplementary Table 3 & 4)
Chronic lung infection leads to allopatric evolution
We next examined whether ongoing adaptation of infecting M. abscessus clones could further promote pathogenicity. We leveraged longitudinal sample collections from 18 chronically infected individuals (Supplementary Figure 4, Supplementary Table 5) to explore the development of, and fluctuations in, within-host population diversity, using deep sequencing of sweeps of colonies from each sputum to capture both consensus SNPs and minority variants.
We developed, and then experimentally validated (Supplementary Figure 4), a new method (MV-trees) to reconstruct the evolutionary trajectories of individual M. abscessus subclones within each patient. We observed that many within-host variants co-occurred at near-identical frequencies over time, implying linkage on the same genetic background as a single haplotype (Figure 2A). We then inferred an ancestor-descendent relationship between related pairs of haplotypes if the first was consistently found at an equal or higher frequency than the second over time. The resultant directed graphs are acyclic, demonstrate conditional independence, and can therefore be pruned through transitive reduction to provide direct evolutionary relationships (Figure 2B), thus revealing both the phylogenetic history (Figure 2C) and temporal frequencies (Figure 2D) of subclones within an individual.
Figure 2. Within-host allopatric evolution of M. abscessus.
(A-D) M. abscessus subclone evolution during chronic infection, shown for one representative individual (P6), illustrating (for a subset of mutations) (A) changes in allele (top) and haplotype (bottom) frequencies over time, (B) inferred ancestor-descendent relationship between related pairs of haplotypes (top), pruned through transitive reduction to provide direct evolutionary relationships (bottom), and (C) resultant phylogenetic reconstruction. (D) Fishplot [33] visualisation of the evolution of all inferred subclones from P6 over time. (E) Relationship (pairwise comparisons) of subclone repertoire within (alpha diversity) and between (beta diversity) sputum samples from 18 patients chronically infected with M. abscessus. (F) Detection of communities of subclones (based on co-occurrence frequency analysis) in 18 CF patients chronically infected with M. abscessus, including with UNG and Nth hypermutator clones (red boxes). Edge thickness represents co-occurrence frequency within (black) and between (red) communities. (G) Subclone communities of M. abscessus within P6 permitting (H) deconvolution of the fishplot (shown in D). Subclones coloured consistently across A-D, G, and H.
In all but two of the patients, we found that the total number (gamma diversity) and the genetic divergence of subclones were linearly correlated with the duration of infection, suggesting a clock-like evolutionary process (Supplementary Figure 5). In two individuals however, we observed much larger than expected subclone repertoires (and genetic differences) due to the acquisition of hypermutator phenotypes, suggesting that rapid and unpredictable expansions of within-host diversity can occur, potentially accelerating ongoing M. abscessus evolution.
We assumed that hypermutation was driven (in Patient 13) by a premature stop codon in uracil DNA glycoylase (UDG), an enzyme responsible for removing mis-incorporated uracil from DNA, and (in Patient 14) by a frameshift mutation in Nth endonuclease III, an enzyme that repairs damaged cytosines [9] (Supplementary Figure 6).
When we explored the temporal changes in subclone frequencies in individuals, we found rapid fluctuations in apparent population composition over time (Figure 2D) and a greater difference in subclone repertoire between (beta diversity) than within (alpha diversity) sputum samples (Figure 2E), suggesting selective sampling (by each collected sputum) of a small subset of multiple infecting populations from presumably distinct anatomical areas of the lung.
Using density-based clustering, we found that there were repeated patterns in the subclone repertoire present in individual sputa, not explicable by the timing of sample collection (Supplementary Figure 7), and indicating the presence (in 17 of 18 patients studied) of multiple discrete communities of subclones (Figure 2F). These communities were genetically closely related (Supplementary Figure 8) and most likely represent the outcome of distinct, spatially segregated (allopatric) adaptive evolution, made possible by lobar anatomy of the lung. By examining each community separately, we were able to capture the temporal dynamics in subclone frequencies within their specific niche (Figure 2G, H).
Convergent evolution of M. abscessus within and between infected individuals
As expected from allopatric evolution, we saw frequent examples of non-synonymous mutations occurring in the same gene in independent lineages within individuals (Figure 3A). Of the 18 patients with reconstructed subclone histories, we detected within-host parallel evolution in 13 individuals, with 30 different genes accumulating more non-synonymous SNPs than would be expected by chance. Eight loci were found repeatedly mutated in multiple patients (suggesting critical host adaptations), comprising genes associated with smooth-to-rough morphotype transition (GPL locus), macrolide resistance (23S rRNA), cell wall biosynthesis (ubiA), and global regulation (phoR, crp/fnr, engA, a tetR family member, and ideR; Figure 3B).
Figure 3. Parallel evolution of M. abscessus within and between patients.
(A) Example of within-host parallel evolution in P4 where several subclones have independently acquired a non-synonymous mutation in phoR (red), engA (yellow), and embC (pale blue). (B) Within-patient parallel evolution in 18 patients (each patient represented by a concentric circle) of 7 genes (chromosomal position relative to reference (ATCC19997) strain shown). Size of circles represent the number of non-synonymous SNPs (or any mutation in the case of 23S) present in each patient. (C) Manhattan plot identifying genes with more non-synonymous mutations than would be expected by chance across 201 patients (size of circle indicates the number of patients mutations were identified in used a one tailed binomial test. The p-values were corrected for multiple testing using Benjamini Hochberg method, with nonsignificant values (>0.01) shown in the shaded grey area. (D) Network analysis (using String) suggests that many of the genes undergoing parallel evolution may be functionally related. Edge thickness represents strength of evidence for direct interaction. (E) Impact of inducible CRISPRi knockdown of selected genes on M. abscessus survival in primary human macrophages at 2h (grey) and 24h (black) post infection (24h/2h ratios shown above), bacterial viability assessed by colony forming units (CFU). (F) Intracellular survival within primary human macrophages (at 2h (grey) and 24h (black) of wild type (WT) M. abscessus, PhoPR knockout (PhoPRD) mutants alone, or expressing empty vector (EV), wild type PhoPR (PhoPRwt) or PhoPR containing a patient-derived PhoR mutation (PhoPRmut). Experiments were performed in at least triplicate on at least three separate occasions. Results from representative experiments shown as mean ± s.e with statistical significance determined using two-tailed Student t-test (* p < 0.05, ** p <0.01. *** p < 0.001). (G,H) Infection of bENaC-tg mice with wild type (WT, black), PhoPRΔ (white), PhoPRΔ::PhoPRwt (blue) or PhoPRD::PhoPRmut (red) M. abscessus showing (G) bacterial burden in the lungs and (H) representative histology (arrows denote granuloma (left) and mycobacteria (right). Statistical significance determined using two-tailed Student t-test (* p < 0.05, ** p <0.01. *** p < 0.001).
As an orthogonal approach, we also identified genes accumulating an excess of non-synonymous consensus SNPs during M. abscessus infection, by examining isolates from 201 CF patients with longitudinal samples [9] (Figure 3C). Many of the genes identified by these within- and between-patient analyses form part of a single functional network (Figure 3D) and are implicated in the control of macrophage invasion by mycobacteria [22–26]. Five genes (phoR, ubiA, ideR, engA and crp/fnr) are likely to be under very strong evolutionary pressure, since they were identified in both analyses, and appear to be specifically important for lung adaptation. Examining genomic data from laparoscopy-associated M. abscessus wound infections [27], we found that non-synonymous mutations in these genes occurred at significantly lower rates than we observe during pulmonary disease (χ2 test: p = 0.02).
To explore the functional impact of deleterious mutations in these genes, we created isogenic inducible knockdown mutants (using a modified CRISPR-dCas9 system, Supplementary Methods) and screened them for phenotypic differences during macrophage infection. We found that all five gene hypomorphs were both less readily phagocytosed and showed enhanced intracellular survival (Figure 3E), implying that within-host evolutionary pressures may be focused on avoiding macrophage killing.
We found that mutations in these genes were non-randomly distributed, with significant enrichment in specific domains (Supplementary Figure 9), suggesting that the SNPs might be functionally equivalent. Most noticeably, within phoR, the most common gene to acquire non-synonymous mutations during lung infection, almost 70% of all within-host mutations occurred in the sensor loop (Supplementary Figure 9; Fisher’s exact test p < 0.001). PhoR, a histidine kinase response regulator, is part of the PhoPR two-component system that in M. tuberculosis is required for macrophage survival and in vivo virulence [25].
Deletion of phoPR in M. abscessus, however, resulted in decreased macrophage uptake and increased intracellular survival (Figure 3F; Supplementary Figure 10), phenocopying the behaviour of the phoR hypomorph (Figure 3E), and suggesting a very different role for PhoPR in M. abscessus compared to M. tuberculosis. While we were able to complement the phenotype by merodiploid expression of wild type phoR, expression of the patient-adapted phoR (containing a T140K sensor loop mutation) resulted in even greater intracellular survival (Figure 3F).
Similarly, infection of βENaC-tg mice, which phenocopy CF lung disease [28], also revealed that the phoPR knockout mutant was more pathogenic than wild type M. abscessus and could be complemented by wildtype (but not patient-adapted) phoR (Figure 3G & H), confirming that the mutations acquired during lung infection in CF patients result in increased virulence through loss of protein function.
Thus, our results indicate that within-host allopatric evolution drives pathogenic adaptation of specific lineages of M. abscessus and is influenced by the chronicity of infection, as well as presumably total bacterial community size, mutation rate, and the stringency of various selection pressures. The impact of these adaptive changes on lineage virulence will, however, depend on the presence of person-to-person transmission allowing multiple rounds of within-host evolution for that lineage.
Indirect transmission constrains pathogenic evolution
We therefore measured the transmissibility of the most frequently occurring adaptive mutations between patients (through analysis of a collection of clinical isolates of M. abscessus from 532 individuals with CF from around the World [9]), by determining the proportion of mutations that are unique to a single patient, or have been shared amongst multiple clustered patients.
We found that within-host adaptive mutations had impacts on transmission fitness that were variously mild (for example the 16S and 23S rRNA mutations causing aminoglycoside and macrolide antibiotic resistance); moderate (phoR sensor loop mutations); or severe (such as mutations in the GPL locus causing smooth-to-rough morphotype transitions), while non-adaptive mutations, such as those in non-sensor regions of phoR, were transmitted freely (Figure 4A).
Figure 4. Constrained evolution of M. abscessus.
(A) Transmission rates of mutations are compared between all non-unique SNPs (grey), non-synonymous non-sensor loop mutations in PhoR (pink), mutations conferring aminoglycoside or macrolide resistance in 16S and 23S rRNA respectively (green), non-synonymous PhoR sensor loop mutations (red), and non-synonymous mutations affecting GPL production (blue). Corresponding sizes clades with shared mutations (represented as number of patients per outbreak cluster) are also shown (right). (B, C) Phylogenetic tree of isolates from patients within transmission chains, showing examples of (B) transmission of adaptive sensor-loop SNPs and (C) preferential cross-infection by subclones with un-evolved phoR. (D) Impaired survival on fomites of (i) phoR knockout mutants (red) and (ii) rough isolates with GPL mutations (blue) compared to isogenic controls (black). Experiments were performed in at least triplicate on at least three separate occasions. Results from representative experiments shown as mean ± s.e with statistical significance determined using two-tailed Student t-test (* p < 0.05, ** p <0.01. *** p < 0.001).
Our findings imply that pathogenic evolution of M. abscessus may be constrained by the competing requirement to maintain transmission fitness, and suggest that long-term within-patient maintenance of less host-adapted ancestral subclones (Supplementary Figure 10) may be important for successful spread to other patients.
For example, while we could identify transmission of adaptive sensor-loop SNPs between some patients (Figure 4B), we also found instances where there appeared to be preferential cross-infection by subclones with un-evolved phoR, despite the parallel evolution of multiple adaptive phoR mutations in that individual (Figure 4C).
Since we have previously shown that transmission of M. abscessus between CF patients is indirect [8], probably through the generation of long-lived infectious aerosols or via fomites [9], we wondered whether mutations that maximally increase virulence might concomitantly impair environmental survival, and thereby explain their transmission fitness cost. In support of this model, we found that both phoR knockout mutants and rough isolates with GPL mutations (which also show increased virulence in vitro and in vivo [26, 29]) demonstrate impaired survival on fomites compared to isogenic controls (Figure 4D), and represent a clear barrier to optimal pathogenic evolution.
Discussion
Our results point to what may be a generalisable model for mycobacterial pathogenic evolution (Supplementary Figure 12). Initially, horizontal gene acquisition (particularly of genes with global transcriptional effects) by environmental clones drives saltational evolution across fitness landscapes, increasing virulence of particular strains, and giving rise to the ancestors of the dominant circulating clones of M. abscessus, as well as to virulent clones in other mycobacterial species including monophyletic MTBC (Supplementary Figure 2 & 3). We highlight the importance of changes in DNA modification, particularly methylation, in driving pathogenic evolution of clones, which has occurred through gene acquisition in M. abscessus and M. avium clones, and gene loss in M. canettii. We note that lineage specific differences in DNA methylation have also been suggested to alter M. tuberculosis behaviour [30].
Next, allopatric within-host adaptation during chronic infection drives increased intracellular survival within macrophages and inflammatory lung damage. Pathogenic evolution is however constrained while transmission is via environmental intermediaries, since the most highly adapted strains lose transmission fitness through reduced fomite survival.
Ultimately, we predict that opportunities for direct transmission of emergent mycobacteria (potentially through increases in population density and/or host susceptibility) will permit unconstrained, accelerated evolution into an obligate pathogen (accompanied by permanent loss of the smooth morphotype); as occurred in M. tuberculosis an estimated 4,000-6,000 years ago [12].
Our findings thus define the key steps involved in the evolution of mycobacteria but also have immediate implications for the clinical management of M. abscessus. They highlight: the importance of minimising within-host adaptation, potentially through immediate treatment of infected individuals rather than waiting (as international guidelines currently recommend [1,2,5]) for radiological and symptom changes; the necessity (given allopatric evolution within the lungs) of testing several colonies from multiple samples to define the behaviour and antibiotic resistance of infecting strains; and the critical need to prevent person-to-person transmission (through enhanced infection control measures [31,32]) in order to block multiple rounds of pathogenic evolution.
Material and methods
Whole genome dataset
We utilized a previously described (9) collection of whole genome data for 1173 clinical M. abscessus samples collected from 526 patients, obtained from UK CF clinics and their associated regional reference laboratories, as well as CF Centres in the US (UNC Chapel Hill), the Republic of Ireland (Dublin), mainland Europe (Denmark, Sweden, The Netherlands), and Australia (Queensland). All sequence data associated with this study is deposited in the European Nucleotide Archive under project accession ERP001039.
Evolutionary analyses of DCCs
Raw reads from a dataset consisting of one isolate per patient (n=526) were mapped to the M. a. abscessus ATCC19977(35) reference genome using BWA-MEM (v. 0.7.12)(36) using default parameters. Variants were called using Samtools (v.1.2.1) and Bcftools (v.1.2.1) (37). A maximum likelihood phylogenetic tree was inferred from these sites using RAxML (v.v.8.2.8)(38). In order to analyze the genetic changes occurring on the branches leading to the last common ancestors (LCA) of the DCCs, the SNPs were mapped back onto the phylogeny using the ACCTRAN parsimony algorithm (custom script written by Simon Harris). To identify whether a change in selection pressure had occurred on the branches leading to the LCA of each of the DCCs, which could be indicative of adaptation to a novel environment, the ratio of nonsynonymous SNPs per nonsynonymous site to synonymous SNPs per synonymous sites (dN/dS) was calculated for each branch of the phylogeny using the Nei-Gojobori method(39).
Pangenome analyses
Datasets for pangenome analyses were obtained from previous publications for M. abscessus (40–44), M. avium (20), M. tuberculosis complex (45–58) and M. canettii (21, (59, 60). Samples obtained as Illumina sequencing reads were assembled de novo as previously described (61). Samples from M. tuberculosis lineages 1-6 were obtained as PacBio sequencing reads and assembled using SMRT v2.2.0 (https://github.com/sanger-pathogens). A summary of the samples used in pangenome reconstruction for each species is provided in Supplementary Table 3.
All sample assemblies were annotated using the run_prokka function in Panaroo version 1.2.2 (15). This function annotates each sample using Prokka(62) with the same gene model for each sample. We ran the run_prokka function on each species independently. Samples that were outliers based on their number of genes and contigs were removed, as inferred using the panaroo-qc scripts (https://github.com/gtonkinhill/panaroo). Pangenomes were reconstructed for each species independently using Panaroo v1.2.2 (15) with clean-mode set to moderate.
Clades of interest were DCC1, DCC2 and DCC3 in M. abscessus, cluster Ia in M. avium, the M. tuberculosis complex and clone 1A in M. canettii. Genes that were gained or lost leading to a clade were identified through phylogenetic reconstruction. We mapped samples from each species or subspecies against their corresponding reference genome (ATCC 19977 for M. a. abscessus, CIP_108297 for M. a. massiliense, TH135 for M. avium, H37Rv for M. tuberculosis and M. canettii) using the multiple_mappings_to_bam pipeline (https://github.com/sanger-pathogens/bact-gen-scripts) with BWA-MEM as the aligner. A phylogenetic tree was reconstructed on the variable positions for each alignment using RAxML version 8.2.9 (63) with the GTR model of nucleotide substitution and gamma rate heterogeneity with four gamma classes. Each accessory gene was reconstructed onto the respective phylogenetic tree using PastML (64). Genes gained leading to a clade of interest were included if they were present in ≥70% of samples in the clade and ≤30% of remaining samples in the species. Genes lost leading to a clade of interest were included if they were present in ≤30% of samples from the clade and ≥70% of remaining samples in the species. The lists of genes identified can be found in Supplementary Tables 1 and 4. To supplement our analysis of M. abscessus DCCs, any gene gain or loss events annotated as “hypothetical” underwent further functional analysis: MetaCyc (65), which is a non-redundant database of metabolic pathways, enzymes and reactions, was interrogated. Distantly related homologs were identified using simultaneous BLAST(66) and HMMER(67) searches were done using the following databases: PFAM(68), SCOP(69), NCBI(70), Uniprot(71), ESTHER(72) and on the Transporter Classification Database(73).
For illustration purposes the pangenome graphs were simplified so that they were ordered against an appropriate reference genome, and any long-range edges were cut. Briefly, pairs of genes that were more than 100 genes apart in the reference genome and had a path connecting the two genes using only non-reference genes in the graph were identified. For each long-range connection, a minimal set of edges was identified and removed from the graph to cut that path. This was performed iteratively until there were no more long-range edges (34).
The final graphs were visualised using Cytoscape v3.7.1 (74), and arranged using the organic graph layout from the yFiles plugin.
Structural model of DpnM
The homology model for M. abscessus DpnM was built using the SAM-bound DNA adenine methyltransferase from Streptococcus pneumoniae as template (PDB ID: 2DPM(75), percent identity of 35% and similarity of 66%) using Modeller (v9.21)(76). The best model was selected using the DOPE (Discrete Optimised Potential Energy) score(77). The residue of interest (F42) was mapped onto the SAM bound modelled structure using UCSF Chimera(78). The model for the mutant F42S was obtained using the swapaa command in Chimera. DNA-bound form was modelled using the E. coli DNA adenyl methyltransferase structure (PDB ID: 2G1P).
Construction of dpnM knockout strain and complementation
A representative of DCC3 (BIR1049, accession GCF_900137475.1) was chosen as a strain for knockout construction. Deletion of the inserted mobile element in BIR1049 was carried out using a modified protocol of mutagenesis by recombineering for M. abscessus(79). Briefly, primers were designed to amplify 1000bp flanking regions upstream and downstream of the mobile element containing dpnM. A streptomycin cassette (obtained from pHP45W) was cloned between the upstream and downstream fragments of the target gene to create an allelic exchange substrate (AES). A modified version of pJV53 containing the xylE gene (pJV53-xylE) was used to create a recombineering strain of M. a. massiliense isolate BIR1049 (BIR1049-pJV53-xylE). BIR1049-pJV53-xylE was grown to OD=0.5, induced for 4h with 0.2% acetamide and electroporated with the AES. Transformants were plated on 7H11 agar supplemented with ADC containing selective antibiotic (200 μg/mL streptomycin). Clones were selected and checked for the AES by PCR. In order to remove pJV53-xylE, selected clones were grown in liquid broth under streptomycin selection only for two weeks, plated and checked by 1% catechol to confirm they lost the pJV53-xylE plasmid. Thereafter, mutant colonies are kanamycin-sensitive streptomycin-resistant.
To generate a complementation of dpnM the gene was PCR amplified, digested with EcoRI and HindIII and ligated to pMV306H-hsp60 cut with the same enzymes. The plasmid was electroporated into BIR1049DdpnM and transformants were selected on 7H11 ADC plates with Hygromycin 1mg/ml and confirmed by PCR.
BIR1049ΔdpnM and complementation strains were validated by whole genome sequencing.
SMRT sequencing of BIR1049ΔdpnM
BIR1049 and BIR1049ΔdpnM were grown for in 10 mL of 7H9 broth supplemented with ADC and glycerol at 37°C. Culture tubes were spun in a centrifuge at 1,900 xg for 5 minutes and the pellet was resuspended in 250 μL TE buffer. The suspension was transferred into a tube containing 500 μL of 0.1 mm silica beads and subjected to three 30-second pulses with 30 seconds rest between pulses using a mini bead beater (BioSpec, USA). DNA extraction was performed using the QIAmp DNA mini kit (QIAGEN, UK) and elution performed using 100 μL of MilliQ water.
The Pacific Biosciences RSII instrument was used to perform SMRT sequencing on eight isolates at the Wellcome Trust Sanger Institute (accession ERP010248). One SMRT-cell was used per isolate. Post sequencing analysis was performed using the SMRT-analysis.2.3.0 pipeline available via the SMRT-portal. The sequencing reads were assembled using HGAP v3(80). This involves three steps. Firstly, pre-assembly which aims to produce long and accurate sequences. This is followed by the assembly of these high quality sequences into a draft genome (GCF_900137475.1) and finally, the correction of the draft assembly by the PacBio RS_Resequencing protocol and Quiver (v1). The approximate genome size parameter was set to 5Mbp (approximately the size of the M. a. abscessus genome) and the target coverage was set to 25. RS_Modification_and_Motif_Analysis.1 was run using the SMRT analysis software v2.3.0 embedded in the SMRT-portal. Briefly, this protocol uses SFilter to remove short reads and sequencing adapters. The filtered reads are then mapped to the assembly produced by HGAP using BlasR v1(81). Kinetic analysis is then applied to the alignment of the reads to the reference enabling the identification of the modified bases by detecting bases where the interpulse duration ratio (IPDR) was significantly different from that of the in silico control(82). The modified motifs recognized by the methylases present in the genome were then identified using Motif Finder v1, with a minimum modification quality (MODQV) threshold of 30.
RNA extraction
Mycobacterial RNA was extracted from BIR1049 and BIR1049DdpnM using a combination of bead beating and RNAeasy mini kit (QIAGEN, UK). Bacterial cultures were grown in DifcoTM Middlebrook 7H9 broth supplemented with ADC, Tween 80, and glycerol until culture saturation at 37°C with 100 xg. One hundred microliters of saturated cultures were used to inoculate 10 ml of 7H9 broth supplemented with ADC and glycerol and cultures were grown at 37°C with 100 xg until mid-logarithmic stage. Culture tubes were spun in a centrifuge at 1,900 xg for 10 minutes and the pellet stored immediately at -80°C until extraction. At the time of extraction, the pellets were removed from -80°C and placed in ice. The frozen pellets were resuspended in 700 μL of RLT buffer containing 1% β-mercaptoethanol, and transferred to a tube containing 500 μL of 0.1 mm silica beads and subjected to two 2-minute pulses with 1 minute rest in between using a mini bead beater (BioSpec, USA). Samples were spun in a centrifuge for 2 minutes at 7,200 xg and 700 μL of the supernatant were transferred into a gDNA eliminator column. Additional 500 μL of RLT buffer containing 1% β-mercaptoethanol were added, the samples spun in a centrifuge at 7,200 xg and 300 μL of the supernatant were added to the same gDNA eliminator column. The columns were spun in a centrifuge at 8,600 xg for 1 minute and the flow-through collected. One volume of 70% ethanol was added to the flow-through, transferred onto a RNA mini column, and spun in a centrifuge at 8,600 xg for 1 minute, discarding the flow-through. The columns were then washed with 700 μL of RW1 buffer, followed by two washes with 500 μL of RPE buffer. RNA was eluted using 30 μL of RNase-free water.
RNAseq of BIR1049ΔdpnM
RNAseq was performed on BIR1049 and BIR1049DdpnM at the Wellcome Trust Sanger Institute using the Illumina HiSeq 2500 platform (accession ERP016362). Gene expression values were computed from the read alignments to the coding sequencing to generate the number of reads mapping and reads per kilobase per million (RPKM). Only reads with a mapping quality score of 10 were included in the count. Genes differentially expressed in the presence and absence of the methyltransferase were determined using DESeq2 (v.1.20)(83). P-values were corrected for multiple testing using the Benjamini Hochberg method. Significantly differentially expressed genes were identified as those with a log2 fold change greater than 2 or less than -2 with a corrected p-value less than 0.05. Experiments were performed in triplicate biological replicates for each condition.
Inference of subclonal population structure (Minority Variant-trees)
Whole genome sequencing (as described above) was carried out on longitudinal plate sweep samples (Figure 5A). Reads were mapped to the appropriate subspecies reference genome (M. a.. abscessus (NC_010394.1)(35), M. a. massiliense or M. a. bolletii de novo assemblies as described previously (84)) using SMALT (85). Samples with less than 20x average read depth, or an excess of heterozygous positions (indicating contamination) were excluded from analysis.
Figure 5. Method for inferring subclone population structure.
(A) Bacterial colony sweeps were taken from culture of longitudinal samples for each patient and DNA extracted. (B) DNA was deep sequenced and minority variants called (see Methods for details). (C, D) We tracked allele frequencies over time and noticed that many variants followed the same frequency trajectory, suggesting that they form a haplotype or subclone. (E) We then used MV-trees to infer the underlying subclone population structure and subsequently pruned the resultant trees, given their acyclic directed graph relationship, using transitive reduction (see Methods for details).
Consensus variants that passed stringent quality filters were called as described previously (8), and variable positions between same-patient isolates identified. In order to detect minority variants (Figure 5B), where all reads do not agree on a consensus sequence, stringent filters were applied. As a first step, stringent mapping was used where in addition to the default SMALT (85) parameters, a minimum nucleotide identity of 0.98 was applied, which avoided the mapping of reads with more than one mis-match which could be considered poor quality. To distinguish sequencing errors from true variants, a minority variant had to be supported by at least 5 reads, where at least 2 reads were mapped to each strand, and a strand bias P-value cutoff of 0.05 (calculated by bcftools(37)) was applied. To avoid heterozygous positions which may arise due to mis-mapping, the base positions had to have a depth of coverage within a normal range (+/- 50% of the average). The alternative (non-reference) allele frequency for the all within-patient variant positions was extracted.
We noticed that many variants followed the same allele frequency trajectory (Figure 5C), suggesting that they formed the same haplotype or subclone (Figure 5D). We developed MV-trees (https://github.com/JosieMB/MV_trees.git) to exploit these patterns to infer the underlying subclonal population structure. In order to identify variants which had the same allele frequency trajectory over time, density-based clustering was performed using DBSCAN (86) in R (version 3.4.2) using allele frequency as input and minPts as 2. The eps value was selected by generating a k-nearest neighbor distance plot (kNNdistplot function) with k as 2. Variants whose allele frequency trajectory clustered into the same group were considered a single subclone. Variants that could not be clustered (referred to as “noise” by the algorithm) were considered singleton variant subclones. Subclonal frequencies could then be tracked over time using the mean allele frequency of the assigned variants for each time point (Figure 5D).
Quality filtering was performed to remove clusters where over 20% of the variants were within a read length (150 nucleotides) of one another, indicating variation due to mis-mapping or recombination. Similarly, singleton variants that were located within 150 nucleotides of other singleton variants were also removed.
The resultant allele frequency trajectories can be used to infer ancestral relationships between the subclones. For example, if mean allele frequency trajectory of subclone A follows a similar path to subclone B but is sometimes higher, then subclone A is the ancestor to B. However if subclones have allele frequency trajectories that are dissimilar, then they are considered unlinked and don’t have an ancestor-descendent relationship.
The ancestor-descendent relationships between the subclones were inferred in a pairwise fashion (here referred to as subclone A and subclone B) if the following requirements were satisfied (allowing ±0.05 to allow random deviation due to variation in sequencing depth):
-
for all time points the mean AF (allele frequency) of subclone A > subclone B
atleast once the mean AF of subclone A > subclone B
atleast once the mean AF of subclone A + subclone B > 1
If all three conditions are not met but that the mean AF of subclone A + subclone B is always ≤ 1 then they are considered sister taxa (unlinked).
If, atleast once, the mean AF of subclone A + subclone B > 1 but that at different time points subclone A < subclone B or subclone A> subclone B then this relationship is considered evolutionary incompatible. This means subclone A or B is problematic which could be caused by homoplasy or recombination. In order to identify problematic clones or singleton variants, the node with the most number of problematic relationships is removed and the ancestordescendent relationship inference is re-run. This is repeated iteratively until no more problematic relationships remain.
This process infers all ancestor-descendent relationships which then need to be reduced down to direct parent-child relationships. Assuming that the evolutionary relationships can be represented by a single tree structure (acyclic directed graph) this is achieved through transitive reduction (Figure 5E) as implemented in the ‘nem’ R package (87). The resultant tree is drawn using igraph (88). We found that common drug resistance mutations were often filtered from the analysis due to being homoplasious and associated with multiple clones. In order to include these variants their likely position on the tree was assessed manually using the AF data and added to the final trees.
Validation of subclonal relationship inference
In order to validate the inferred subclonal trees generated from sweep data, we whole genome sequenced single colony purified samples from the same collection. Three samples were selected from three patients, where the sample was predicted to contain multiple subclones from the sweep data. Thirty single colonies were randomly picked from each for whole genome sequencing (as previously). Consensus variants were called as previously, and a maximum likelihood phylogenetic tree (RAxML) was constructed for each patient. The resultant tree was compared with the subclonal tree inferred from the sweep data. In two of the cases the maximum-likelihood tree formed a subtree of the total subclonal tree with identical internal tree structure (Supplementary Figure 4). Additional variants were identified on the tips of the single-colony trees not identified in the subclonal tree, which were unique to single colonies so occurred at a low frequency and were filtered from the analysis. In patient 6 an additional drug resistance mutation was identified as an internal node which was likely excluded from the sweep analysis as it occurred at the same position as another drug resistance mutation. Currently the method assumes variants are biallelic.
Sputum repertoire and community analysis
Sputum samples from the same patient with similar subclonal composition were clustered by subclonal repertoire using DBSCAN(86) in R (version 3.4.2), using frequency of subclone as input.
Community analysis was used to determine if the inferred subclones co-occurred with other subclones in a non-random fashion. This was achieved by constructing networks based on frequency of co-occurrence between subclones. The number of times subclones co-occurred in the same sputum was counted and then corrected for overall prevalence of the subclone. Pairwise frequencies of co-occurrence between each subclone in a patient was then used to create an undirected weighted graph implemented by “graph.adjacency” in the igraph package (version 1.2.4.1)(89). Hypothetical communities were identified as nodes that were densely connected to themselves but sparsely connected to others, as implemented by the “edge.betweenness.community” function in the igraph package(89).
Intra- and inter-patient parallel evolution
Genes were identified as having evidence of intra-host parallel evolution when they accumulated more than one non-synonymous variant on independent branches on the inferred subclonal trees. Genes with inter-patient parallel evolution were those that acquired non-synonymous variants in multiple patients at a rate higher than considered by chance using a ‘burden of mutation’ approach(90). This method estimates the ρSN (synonymous mutation rate) by dividing the observed number of synonymous SNPs by the number of coding sequence bases in the reference genome. The expected nonsynonymous mutation rate (ρNS) is then estimated using the following equation: ρNS = ρSN X R. R represents the ratio of nonsynonymous sites to synonymous sites and is determined by permuting every base of every codon in silico and identifying whether it resulted in a synonymous or nonsynonymous change. This was done on a per gene level, with the number of synonymous SNPs accumulated per gene used to estimate the value of ρSN per site per gene. If no synonymous SNPs were observed in a gene the synonymous mutation rate estimated for the whole genome was used. Finally, to obtain the expected number of nonsynonymous SNPs per gene, ρNS was multiplied by the gene length.
To determine if the observed number of nonsynonymous SNPs was significantly greater than the expected, a one tailed binomial test was used. The p-values were corrected for multiple testing using Benjamini Hochberg method, and we applied a significance threshold of 0.01.
STRING-based analysis of protein-protein interactions
M. abscessus proteins that were frequently mutated within patients were mapped to their M. tuberculosis orthologues using the SYNERGY orthogroup resource [91]. The STRING database (v11.0) was queried with those genes with clear orthologues to infer a protein network based on protein-protein interactions allowing up to 30 additional proteins directly interacting with the input set of proteins requiring a minimum confidence of 0.4 [92].
Measurement of mutation transmissibility
Mutations were identified across the global collection of M. abscessus isolates [9] that occurred in genes of interest:
Frameshift and nonsense mutations in the GPL loci MAB_4098 and MAB_4099)
Nonsynonymous mutations in phoR (sensor loop and non sensor loop region as described in figure 3F)
Known antibiotic resistance mutations in 23s rRNA for macrolides (A2058 A2059) and aminoglycosides (A1408 C1409).
Mutations on the deep branches leading to the three subspecies were excluded. Mutations were considered transmitted (shared by more than one patient in a monophyletic clade) or non-transmitted (unique to one patient). The statistical significance of the observed differences in transmissibility was calculated through comparison to all mutations using a Fishers exact test.
Construction of M. abscessus knockout and complemented strains
ΔdpnM: A representative of DCC3 (BIR1049, accession GCF_900137475.1) was chosen as a strain for knockout construction. Deletion of the inserted mobile element in BIR1049 was carried out using a modified protocol of mutagenesis by recombineering for M. abscessus [93]. Briefly, primers were designed to amplify 1000bp flanking regions upstream and downstream of the mobile element containing dpnM. A streptomycin cassette (obtained from pHP45Ω) was cloned between the upstream and downstream fragments of the target gene to create an allelic exchange substrate (AES). A modified version of pJV53 containing the xylE gene (pJV53-xylE) was used to create a recombineering strain of M. a. massiliense isolate BIR1049 (BIR1049-pJV53-xylE). BIR1049-pJV53-xylE was grown to OD = 0.5, induced for 4h with 0.2% acetamide and electroporated with the AES. Transformants were plated on 7H11 agar supplemented with ADC containing selective antibiotic (200 μg/mL streptomycin). Clones were selected and checked for the AES by PCR. In order to remove pJV53-xylE, selected clones were grown in liquid broth under streptomycin selection only for two weeks, plated and checked by 1% catechol to confirm loss of the pJV53-xylE plasmid. Mutant colonies were this characterised as kanamycin-sensitive and streptomycin-resistant. To generate a complementation of dpnM the gene was PCR amplified, digested with EcoRI and HindIII and ligated to pMV306H-hsp60 cut with the same enzymes. The plasmid was electroporated into BIR1049DdpnM and transformants were selected on 7H11 ADC plates with Hygromycin 1mg/ml and confirmed by PCR. BIR1049DdpnM and complementation strains were validated by whole genome sequencing.
ΔphoPR: A knockout strain of the phoPR locus was generated in M. a massiliense CIP108297, using the same method as described above for dpnM. For complementation of phoPR, the operon plus a 130bp upstream region containing the promoter was PCR amplified from M. massiliense CIP108297 or from clinical strains containing mutations in PhoR, digested with XbaI and HindIII and ligated to pMV306-xylE cut with the same enzymes. The construct was then electroporated into CIP108297DphoPR and transformants were selected in 7H11 ADC plates with Kanamycin 200μg/ml and confirmed by PCR.
CRISPR interference (CRISPRi) using dCas9 in M. abscessus
We optimised a previously described tetracycline inducible CRISPr Interference system [94] for M. abscessus ATCC19977 utilizing a dCas9 encoding plasmid (pTetInt-dcas9-Hyg) and a second vector (pGRNAz) containing a custom designed small-guide RNA (sgRNA) cassette. The dCas9-expressing strains were cultivated in Middlebrook 7H9 broth supplemented with 1 × ADC, 0.05% Tween-80 and 0.8% glycerol, hygromycin 1 mg/mL, Zeocin 300 μg/mL. Induction of gene silencing was achieved by adding anhydrotetracycline (ATc) 100 ng/mL. Three 20 nucleotide guides were designed per gene of interest and annealed and cloned between sphI and aclI of the pGRNAz. The dCas9-expressing strains were then transformed with the sgRNA encoding vectors.
SMRT sequencing of BIR1049 WT and ΔdpnM
BIR1049 and BIR1049DdpnM were grown for in 10 mL of 7H9 broth supplemented with ADC and glycerol at 37 °C. Culture tubes were spun in a centrifuge at 1,900 x g for 5 minutes, the pellet resuspended in 250 μL TE buffer, and then transferred into a tube containing 500 μL of 0.1 mm silica beads and subjected to three 30-second pulses with 30 seconds rest between pulses using a mini bead beater (BioSpec, USA). DNA extraction was performed using the QIAmp DNA mini kit (QIAGEN, UK) and elution performed using 100 μL of MilliQ water.
SMRT sequencing on eight isolates was performed at the Wellcome Sanger Institute (using a Pacific Biosciences RSII instrument). One SMRT-cell was used per isolate. Post sequencing analysis was performed using the SMRT-analysis.2.3.0 pipeline available via the SMRT-portal. The sequencing reads were assembled using HGAP v3 [95]. This involves three steps. Firstly, pre-assembly which aims to produce long and accurate sequences. This is followed by the assembly of these high-quality sequences into a draft genome (GCF_900137475.1) and finally, the correction of the draft assembly by the PacBio RS_Resequencing protocol and Quiver (v1). The approximate genome size parameter was set to 5Mbp (approximately the size of the M. a. abscessus genome) and the target coverage was set to 25. RS_Modification_and_Motif_Analysis.1 was run using the SMRT analysis software v2.3.0 embedded in the SMRT-portal. Briefly, this protocol uses SFilter to remove short reads and sequencing adapters. The filtered reads are then mapped to the assembly produced by HGAP using BlasR v1 [96]. Kinetic analysis is then applied to the alignment of the reads to the reference enabling the identification of the modified bases by detecting bases where the interpulse duration ratio (IPDR) was significantly different from that of the in silico control [97] The modified motifs recognized by the methylases present in the genome were then identified using Motif Finder v1, with a minimum modification quality (MODQV) threshold of 30.
RNAseq of BIR1049 WT and ΔdpnM
RNA extraction: Mycobacterial RNA was extracted from BIR1049 and BIR1049DdpnM using a combination of bead beating and RNAeasy mini kit (QIAGEN, UK). Bacterial cultures were grown in DifcoTM Middlebrook 7H9 broth supplemented with ADC, Tween 80, and glycerol until culture saturation at 37 °C with 100 x g. One hundred microliters of saturated cultures were used to inoculate 10 ml of 7H9 broth supplemented with ADC and glycerol and cultures were grown at 37 °C with 100 x g until mid-logarithmic stage. Culture tubes were spun in a centrifuge at 1,900 x g for 10 minutes and the pellet stored immediately at -80 °C until extraction. At the time of extraction, the pellets were removed from -80 °C and placed in ice. The frozen pellets were resuspended in 700 μL of RLT buffer containing 1% β-mercaptoethanol, and transferred to a tube containing 500 μL of 0.1 mm silica beads and subjected to two 2-minute pulses with 1 minute rest in between using a mini bead beater (BioSpec, USA). Samples were spun in a centrifuge for 2 minutes at 7,200 x g and 700 μL of the supernatant were transferred into a gDNA eliminator column. Additional 500 μL of RLT buffer containing 1% β-mercaptoethanol were added, the samples spun in a centrifuge at 7,200 x g and 300 μL of the supernatant were added to the same gDNA eliminator column. The columns were spun in a centrifuge at 8,600 x g for 1 minute and the flow-through collected. One volume of 70% ethanol was added to the flow-through, transferred onto a RNA mini column, and spun in a centrifuge at 8,600 x g for 1 minute, discarding the flow-through. The columns were then washed with 700 μL of RW1 buffer, followed by two washes with 500 μL of RPE buffer. RNA was eluted using 30 μL of RNase-free water.
RNAseq: RNAseq was performed on BIR1049 and BIR1049DdpnM at the Wellcome Trust Sanger Institute using the Illumina HiSeq 2500 platform on at least three biological replicates per condition. Gene expression values were computed from the read alignments to the coding sequencing to generate the number of reads mapping and reads per kilobase per million (RPKM). Only reads with a mapping quality score of 10 were included in the count. Genes differentially expressed in the presence and absence of the methyltransferase were determined using DESeq2 (v.1.20) [98]. P-values were corrected for multiple testing using the Benjamini-Hochberg method. Significantly differentially expressed genes were identified as those with a log2 fold change greater than 2 or less than -2 with a corrected p-value less than 0.05.
Mouse infection experiments
Specific-pathogen-free βENaC transgenic mice, were purchased from the Jackson Laboratories, Bar Harbor, ME (Stock No: 006438-B6.Cg-Tg(Scgb1a1-Scnn1b)6608Bouc/J). Mice were rederived and maintained at Colorado State University (CSU) and were given sterile water, mouse chow and enrichment for the course of experiment. All experimental protocols were approved by the Animal Care and Use Committee of Colorado State University (Approvals: IRB #14-032B, IACUC#1020). Mycobacteria were grown at 30 °C in Middlebrook 7H9 broth supplemented with 10% (v/v) oleic acid/albumin/dextrose/catalase (OADC) enrichment and 0.05% Tween 80 or on 7H10 agar containing 10% (vol/vol) OADC with appropriate antibiotics.
Experimental infections: Mice (6 weeks old, females) were challenged with wild type (WT) M. abscessus, phoPR knockout mutant (PhoPRΔ), PhoPRΔ::PhoPRwt (blue) or PhoPRΔ::PhoPRmut. using an intratracheal infection calibrated to deliver 1 x 109 bacilli per animal. At days 1, 10, and 20 following infection, bacterial loads in the lungs, spleen, and liver were determined and lung histology examined. Bacterial counts were determined by plating serial dilutions of whole organ homogenates on 7H11-OADC agar and counting colonyforming units after 5-10 days incubation at 30 °C. At least five animals were infected for each condition at each time point with data presented as the mean ± s.e. using Student t-test to determine statistical significance. For histological analysis, the whole lung from each mouse was fixed with 10% formalin in phosphate buffered saline (PBS). Tissue sections were stained using haematoxylin and eosin and acid-fast stain as previously reported [9].
Colorado State University’s (CSU) animal care program follows the recommendations of the NRC Guide for the Care and Use of Laboratory Animals (National Research Council, 2010), the requirements of the Public Health Service (PHS) Grants Administration Manual, and The Animal Welfare Act as amended. CSU files assurances with the DHHS Office of Extramural Research, Office of Laboratory Animal Welfare (OLAW), the Public Health Service, and adheres to NIH standards and practices for grantees. CSU’s Animal Welfare Assurance Number is A3572-01. CSU animal research facilities have been accredited by the Association for Assessment and Accreditation of Laboratory Animal Care International (AAALAC). All care and use of animals is overseen by the Institutional Animal Care and Use Committee (IACUC). The CSU Laboratory Animal Resources has a fully trained staff that includes multiple laboratory animal staff veterinarians as well as an AAALAC accredited Laboratory Animal Veterinarian residency training program that currently has multiple trainees in various stages of post-DVM clinical and research training.
Primary Macrophage and THP1 cell infection assays with M. abscessus
Primary human macrophages were generated from peripheral blood samples from consented healthy volunteers (males and females aged 24-54) as previously described [99] under Regional ethics approval REC 12/WA/0148.
Briefly, Peripheral Blood Mononuclear Cells (PBMCs) were isolated from citrated samples by density gradient separation (Lympholyte; Cedarlane Labs), and subjected to CD14+ positive selection (MACS Miltenyi Biotec). CD14+ cells were counted, added to 24 well sterile tissue culture plates at 0.2 x 106 cells / well, and then differentiated in macrophages with either (i) recombinant human macrophage colony-stimulating factor (M-CSF; 200 ng/ml) or (ii) granulocyte-macrophage colony-stimulating factor (GM-CSF; 200 ng/ml) followed by interferon-γ (IFN-γ 50 ng/ml) during culture in DMEM media containing 10 % fetal calf serum, 100 U/ml penicillin, and 100 μg/ml streptomycin (Sigma), with antibiotics removed before infection experiments. Cells were maintained at 37°C 5% CO2
At Day 7, macrophages were washed twice with sterile phosphate buffered saline (PBS), incubated with M. abscessus at an MOI of 3:1 (for phoPR experiments) or 5:1 (for dpnM and CRISPRi experiments) for 2h (in culture media), washed twice in PBS, and then incubated for indicated times in culture media. Viable intracellular M. abscessus was assessed at specified time points by washing macrophages twice with sterile PBS and then lysing the cells (in sterile water) and plating on 7H11 or Columbia Blood Agar plates to enumerate colony forming units (as previously [9]).
THP1 cells (obtained directly from ATCC; ATCC-TIB-202) were differentiated using 12ng/ml PMA (Sigma) as previously [1], infected for 2 hours with M. abscessus at specified MOIs, and then washed and incubated and treated as above.
All experiments were performed at least in triplicate biological replicates on at least three separate occasions and data represented as the mean ± s.e with statistical significance determined using Student t-test.
Fomite experiments
Cultures were grown in 7H9+ADC liquid media, centrifuged, resuspended in sterile PBS, and colony forming units determined by serial dilution on 7H11 agar plates (to give the input bacterial number. 20 μl of culture was added to sterile glass cover slips in wells of a sterile 24 well tissue culture plate, which were air dried, and stored in the open in a CL2 microbiological safety cabinet. At 24h, 48h, 7 days and 14 days, 100μl of sterile PBS was added to the cover slips in the well and incubated for 10 minutes at RT. The PBS was mixed in the well by pipetting and then plated in serial dilution onto 7H11 plates and colony forming units measured after incubation at 37°C 5% CO2.
All experiments were performed at least in triplicate biological replicates on at least three separate occasions and data represented as the mean ± s.e with statistical significance determined using Student t-test.
Resistance to amikacin of M. abscessus
BIR1049 WT and ΔdpnM strains was determined by MIC determination using broth microdilution (in 7H9 + ADC liquid media) according to CLSI standard protocols (Clinical and Laboratory Standards Institute M07 11th edition; clsi.org).
All experiments were performed at least in triplicate biological replicates on at least three separate occasions.
Mycobacterial resistance to nitric oxide
To evaluate resistance to nitric oxide, we exposed M. abscessus to a NO-producing solution of sodium nitrite and citric acid [70]. M. abscessus BIR1049 WT and DdpnM (grown in liquid culture) were resuspended in 7H9 with ADC in 100 μl aliquots. Sodium Nitrite (at final concentrations as indicated in Figure 1F) and citric acid (at final concentration of 0.1M) were added to the aliquots which were then mixed by gentle inversion, and incubated for 24 hours at 37 °C. Samples were then plated onto Columbia Blood Agar plates and colony forming units enumerated at Day 5. Experiments were performed in at least triplicate biological replicates and on three separate occasions (with representative experiment shown in Figure 1F).
Supplementary Material
One sentence summary.
The evolutionary steps involved for environmental mycobacteria to become lung pathogens
Acknowledgements
We would like to thank Simon Harris for help with the ACCTRAN parsimony algorithm and all contributors to the global M. abscessus isolate collection.
Funding
This work was supported by The Wellcome Trust (107032AIA (RAF, DMG, SB), 10224/Z/15/Z JMB, 098051(JP)); The UK Cystic Fibrosis Trust (Innovation Hub grant 001 (RAF, TLB, JP, SB, IEE), SRC 002 & 010 (IE, DR-R, TLB, DO, JB, DV, MJ, DMG, DR-R, IE, JP, RAF); NIHR Cambridge Biomedical Research Centre (RAF, KPB); and The Botnar Foundation (6063; RAF, AW, TLB, SM, JP).
Footnotes
Author contributions: JMB and RAF conceived the project. JMB, JP and RAF designed the experiments and wrote the manuscript. JMB developed the MV trees method for Inference of subclonal population structure. JMB, IE, CR, AW, BB performed the bioinformatic analyses. SM and TLB performed the computational structural modelling. JB, MJ, and DR-R generated knockout and complemented bacterial strains. SB developed the M. abscessus CRISPRi method. KPB, SB, JB, DR-R, IEE, DA, CP performed the in vitro experiments. CMP, DV, and DJO performed the mouse infection experiments. DMG, KT, GM, KJ, AJF analysed clinical metadata. JP and RAF provided supervisory support.
Competing interests: none.
Ethical approvals: Ethical approval for the study was obtained nationally for centres in England and Wales from the National Research Ethics Service (NRES; REC reference: 12/EE/0158) and the National Information Governance Board (NIGB; ECC 3-03 (f)/2012), for Scottish centres from NHS Scotland Multiple Board Caldicott Guardian Approval (NHS Tayside AR/SW), and locally for other centres. Primary cell in vitro experiments were authorised by Regional ethics approval REC 12/WA/0148.
Data and Materials availability
All sequencing data associated with this study is deposited in the European Nucleotide Archive under the following project accession numbers: shortread DNA sequence data, ERP001039; RNA-seq data, ERP016362; SMRT-seq data, ERP010248. The code for MV-trees can be found at: https://github.com/JosieMB/MV_trees.git (Zenodo DOI: 10.5281/zenodo.4569470). All phylogenetic trees associated with this study are deposited in TreeBase (http://purl.org/phylo/treebase/phylows/study/TB2:S27748).
References
- 1.Griffith DE, et al. An official ATS/IDSA statement: diagnosis, treatment, and prevention of nontuberculous mycobacterial diseases. Am J Respir Crit Care Med. 2007;175:367–416. doi: 10.1164/rccm.200604-571ST. [DOI] [PubMed] [Google Scholar]
- 2.Floto RA, et al. Cystic Fibrosis Foundation and European Cystic Fibrosis Society Consensus Recommendations for the Management of Nontuberculous Mycobacteria in Individuals with Cystic Fibrosis. Thorax. 2016;71(Suppl 1):i1–i22. doi: 10.1136/thoraxjnl-2015-207360. (2015) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Richards CJ, Olivier KN. Nontuberculous Mycobacteria in Cystic Fibrosis. Semin Respir Crit Care Med. 2019;40:737–750. doi: 10.1055/s-0039-1693706. [DOI] [PubMed] [Google Scholar]
- 4.Martiniano SL, Nick JA, Daley CL. Nontuberculous Mycobacterial Infections in Cystic Fibrosis. Thorac Surg Clin. 2019;29:95–108. doi: 10.1016/j.thorsurg.2018.09.008. [DOI] [PubMed] [Google Scholar]
- 5.Haworth CS, et al. British Thoracic society guidelines for the management of non-tuberculous mycobacterial pulmonary disease (NTM-PD) Thorax. 2017;72:ii1–ii64. doi: 10.1136/thoraxjnl-2017-210927. [DOI] [PubMed] [Google Scholar]
- 6.Qvist T, et al. Comparing the harmful effects of nontuberculous mycobacteria and gram negative bacteria on lung function in patients with cystic fibrosis. J Cyst Fibros. 2016;15:380–385. doi: 10.1016/j.jcf.2015.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Taylor JL, Palmer SM. Mycobacterium abscessus chest wall and pulmonary infection in a cystic fibrosis lung transplant recipient. J Heart Lung Transplant. 2006;25:985–8. doi: 10.1016/j.healun.2006.04.003. [DOI] [PubMed] [Google Scholar]
- 8.Bryant JM, et al. Whole-genome sequencing to identify transmission of Mycobacterium abscessus between patients with cystic fibrosis: a retrospective cohort study. Lancet. 2013;381(9877):1551–60. doi: 10.1016/S0140-6736(13)60632-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bryant JM, et al. Emergence and spread of a human-transmissible multidrugresistant nontuberculous mycobacterium. Science. 2016;354(6313):751–757. doi: 10.1126/science.aaf8156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Aitken ML, et al. Respiratory outbreak of Mycobacterium abscessus subspecies massiliense in a lung transplant and cystic fibrosis center. Am J Respir Crit Care Med. 2012;185(2):231–2. doi: 10.1164/ajrccm.185.2.231. [DOI] [PubMed] [Google Scholar]
- 11.Yan J, et al. Investigating transmission of Mycobacterium abscessus amongst children in an Australian cystic fibrosis centre. J Cyst Fibros. 2019:S1569. doi: 10.1016/j.jcf.2019.02.011. [DOI] [PubMed] [Google Scholar]
- 12.Gagneux S. Ecology and evolution ofMycobacterium tuberculosis . Nature Reviews Microbiology. 2018;16:202–213. doi: 10.1038/nrmicro.2018.8. [DOI] [PubMed] [Google Scholar]
- 13.Orgeur M, Brosch R. Evolution of virulence in the Mycobacterium tuberculosis complex. Current Opinions in Microbiology. 2018;41:68–75. doi: 10.1016/j.mib.2017.11.021. [DOI] [PubMed] [Google Scholar]
- 14.Bos KI, et al. Pre-Columbian mycobacterial genomes reveal seals as a source of New World human tuberculosis. Nature. 2014;514(7523):494–7. doi: 10.1038/nature13591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tonkin-Hill G, et al. Producing Polished Prokaryotic Pangenomes with the Panaroo Pipeline. bioRxiv. 2020:2020.01.28.922989. doi: 10.1186/s13059-020-02090-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dubois V, et al. Mycobacterium abscessus virulence traits unraveled by transcriptomic profiling in amoeba and macrophages. PLoS Pathog. 2019;15:e1008069. doi: 10.1371/journal.ppat.1008069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vázquez-Torres A, Bäumler A. Nitrate, nitrite and nitric oxide reductases: from the last universal common ancestor to modern bacterial pathogens. Curr Opin Microbiol. 2016;29:1–8. doi: 10.1016/j.mib.2015.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sanz-García F, et al. Mycobacterial Aminoglycoside Acetyltransferases: A Little of Drug Resistance, and a Lot of Other Roles. Front Microbiol. 2019;10 doi: 10.3389/fmicb.2019.00046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shin DM, et al. Mycobacterium tuberculosis eis regulates autophagy, inflammation, and cell death through redox-dependent signalling. PLoS Pathog. 2010;6:e1001230. doi: 10.1371/journal.ppat.1001230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Uchiya K-i, et al. Comparative genome analyses of Mycobacterium avium reveal genomic features of its subspecies and strains that cause progression of pulmonary disease. Scientific Reports. 2017;7:39750. doi: 10.1038/srep39750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Blouin Y, et al. Progenitor “Mycobacterium canettii” Clone Responsible for Lymph Node Tuberculosis Epidemic, Djibouti. Emerging Infectious Diseases. 2014;20:21–28. doi: 10.3201/eid2001.130652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rickman L, et al. A member of the cAMP receptor protein family of transcription regulators in Mycobacteriium tuberculosis is required for virulence in mice and controls transcription of the rpfA gene coding for a resuscitation promoting factor. Mol Microbiol. 2005;56:1274–1286. doi: 10.1111/j.1365-2958.2005.04609.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kudhair BK, et al. Structure of a Wbl protein and implications for NO sensing by M.tuberculosis . Nature Comm. 2017;8:2280. doi: 10.1038/s41467-017-02418-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Marcela Rodriguez G, et al. IdeR an essential gene in Mycobacterium tuberculosis: Role of IdeR in Iron-dependent gene expression, iron metabolism, and oxidative stress response. Infection and Immunity. 2002;70:3371–3381. doi: 10.1128/IAI.70.7.3371-3381.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ryndak M, Wang S, Smith I. PhoP, a key player in Mycobacterium tuberculosis virulence. Trends in Microbiology. 2008;16:528–534. doi: 10.1016/j.tim.2008.08.006. [DOI] [PubMed] [Google Scholar]
- 26.Johnansen MD, Herrmann J-L, Kremer L. Non-tuberculous mycobacteria and the rise ofMycobacterium abscessus . Nature Reviews Microbiology. 2020 doi: 10.1038/s41579-020-0331-1. [DOI] [PubMed] [Google Scholar]
- 27.Everall I, et al. Genomic epidemiology of a national outbreak of post-surgical Mycobacterium abscessus wound infections in Brazil. Microb Genom. 2017;3(5):e000111. doi: 10.1099/mgen.0.000111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhou Z, et al. The ENaC-overexpressing mouse as a model of cystic fibrosis lung disease. J Cyst Fibros. 2011;10(Suppl 2):S172–82. doi: 10.1016/S1569-1993(11)60021-0. [DOI] [PubMed] [Google Scholar]
- 29.Byrd TF, Lyons CR. Preliminary characterization of a Mycobacterium abscessus mutant in human and murine models of infection. Infection and Immunity. 1999;67:4700–4707. doi: 10.1128/iai.67.9.4700-4707.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shell SS, et al. DNA methylation impacts gene expression and ensures hypoxic survival ofMycobacterium tuberculosis . PLoS Pathog. 2013;9:e1003419. doi: 10.1371/journal.ppat.1003419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cystic Fibrosis Trust. Mycobacterium abscessus Recommendations for infection prevention and control. 2017 http://www.cysticfibrosis.org.uk/media/care/ntm-guidelines-mar-2018.
- 32.Kapnadak SG, et al. Infection control strategies that successfully controlled an outbreak of Mycobacterium absxcessus at a cystic fibrosis center. American Journal of Infection Control. 2016;44:154–159. doi: 10.1016/j.ajic.2015.08.023. [DOI] [PubMed] [Google Scholar]
- 33.Miller CA, et al. Visualizing tumor evolution with the fishplot package for R. BMC Genomics. 2016 doi: 10.1186/s12864-016-3195-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Weimann A. Using a reference genome to produce less convoluted layouts. 2020 Available from: https://gtonkinhill.github.io/panaroo/-/vis/layout.
- 35.Ripoll F, et al. Non mycobacterial virulence genes in the genome of the emerging pathogen Mycobacterium abscessus. PloS one. 2009;4:e5660. doi: 10.1371/journal.pone.0005660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li H, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stamatakis A. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 2006;22:2688–2690. doi: 10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]
- 39.Nei M, Gojobori T. Simple methods for estimating the numbers of synonymous and nonsynonymous nucleotide substitutions. Mol Biol Evol. 1986;3:418–426. doi: 10.1093/oxfordjournals.molbev.a040410. [DOI] [PubMed] [Google Scholar]
- 40.Bryant JM, et al. Emergence and spread of a human-transmissible multidrugresistant nontuberculous mycobacterium. Science. 2016;354:751–757. doi: 10.1126/science.aaf8156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yan J, et al. Investigating transmission of Mycobacterium abscessus amongst children in an Australian cystic fibrosis centre. J Cyst Fibros. 2019 doi: 10.1016/j.jcf.2019.02.011. [DOI] [PubMed] [Google Scholar]
- 42.Li B, et al. Relationship between Antibiotic Susceptibility and Genotype in Mycobacterium abscessus Clinical Isolates. Frontiers in Microbiology. 2017;8 doi: 10.3389/fmicb.2017.01739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hasan NA, et al. Population Genomics of Nontuberculous Mycobacteria Recovered from United States Cystic Fibrosis Patients. bioRxiv. 2019 [Google Scholar]
- 44.Doyle RM, et al. Cross-transmission is not the source of new Mycobacterium abscessus infections in a multi-centre cohort of cystic fibrosis patients. Clinical Infectious Diseases. 2019 doi: 10.1093/cid/ciz526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chiner-Oms Á, et al. Genome-wide mutational biases fuel transcriptional diversity in the Mycobacterium tuberculosis complex. Nature Communications. 2019;10:3994. doi: 10.1038/s41467-019-11948-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nebenzahl-Guimaraes H, et al. Genomic characterization of Mycobacterium tuberculosis lineage 7 and a proposed name: ‘Aethiops vetus’. Microbial Genomics. 2016;2:e000063. doi: 10.1099/mgen.0.000063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ngabonziza JCS, et al. A sister lineage of the Mycobacterium tuberculosis complex discovered in the African Great Lakes region. Nature Communications. 2020;11:2917. doi: 10.1038/s41467-020-16626-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Coscolla M, et al. Novel Mycobacterium tuberculosis Complex Isolate from a Wild Chimpanzee. Emerging Infectious Diseases journal-CDC. 2013 Jun;19(6) doi: 10.3201/eid1906.121012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dippenaar A, et al. Whole genome sequence analysis of Mycobacterium suricattae. Tuberculosis. 2015;95:682–688. doi: 10.1016/j.tube.2015.10.001. [DOI] [PubMed] [Google Scholar]
- 50.Cousins DV, Peet RL, Gaynor WT, Williams SN, Gow BL. Tuberculosis in imported hyrax (Procavia capensis) caused by an ususual variant belonging to the Mycobacterium tuberculosis complex. Veterinary Microbiology. 1994;42:135–145. doi: 10.1016/0378-1135(94)90013-2. [DOI] [PubMed] [Google Scholar]
- 51.Grandjean L, et al. Convergent evolution and topologically disruptive polymorphisms among multidrug-resistant tuberculosis in Peru. PLOS ONE. 2017;12:e0189838. doi: 10.1371/journal.pone.0189838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Broeckl S, et al. Investigation of intra-herd spread of Mycobacterium caprae in cattle by generation and use of a whole-genome sequence. Veterinary Research Communications. 2017;41:113–128. doi: 10.1007/s11259-017-9679-8. [DOI] [PubMed] [Google Scholar]
- 53.Boniotti MB, et al. Detection and Molecular Characterization of Mycobacterium microti Isolates in Wild Boar from Northern Italy. Journal of Clinical Microbiology. 2014;52:2834–2843. doi: 10.1128/JCM.00440-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Alexander KA, Larsen MH, Robbe-Austerman S, Stuber TP, Camp PM. Draft Genome Sequence of the Mycobacterium tuberculosis Complex Pathogen m mungi, Identified in a Banded Mongoose (Mungos mungo) in Northern Botswana. Genome Announcements. 2016;4 doi: 10.1128/genomeA.00471-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Walker TM, et al. Whole-genome sequencing for prediction of Mycobacterium tuberculosis drug susceptibility and resistance: a retrospective cohort study. The Lancet Infectious Diseases. 2015;15:1193–1202. doi: 10.1016/S1473-3099(15)00062-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Silva-Pereira TT, et al. Genome sequencing of Mycobacterium pinnipedii strains: genetic characterization and evidence of superinfection in a South American sea lion (Otaria flavescens) BMC Genomics. 2019;20:1030. doi: 10.1186/s12864-019-6407-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Crispell J, et al. Using whole genome sequencing to investigate transmission in a multi-host system: bovine tuberculosis in New Zealand. BMC Genomics. 2017;18:180. doi: 10.1186/s12864-017-3569-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Coscolla M, et al. Phylogenomics of Mycobacterium africanum reveals a new lineage and a complex evolutionary history. bioRxiv. 2020:2020.2006.2010.141788. doi: 10.1099/mgen.0.000477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Supply P, et al. Genomic analysis of smooth tubercle bacilli provides insights into ancestry and pathoadaptation of Mycobacterium tuberculosis. Nat Genet. 2013;45:172–179. doi: 10.1038/ng.2517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Blouin Y, et al. Progenitor “Mycobacterium canettii” Clone Responsible for Lymph Node Tuberculosis Epidemic, Djibouti. Emerging Infectious Diseases journal-CDC. 2014 Jan;20(1) doi: 10.3201/eid2001.130652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Page AJ, et al. Robust high-throughput prokaryote de novo assembly and improvement pipeline for Illumina data. Microbial Genomics. 2016;2:e000083. doi: 10.1099/mgen.0.000083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Seemann T. Prokka: rapid prokaryotic genome annotation. Bioinformatics. 2014;30:2068–2069. doi: 10.1093/bioinformatics/btu153. [DOI] [PubMed] [Google Scholar]
- 63.Stamatakis A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;30:1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ishikawa SA, Zhukova A, Iwasaki W, Gascuel O. A Fast Likelihood Method to Reconstruct and Visualize Ancestral Scenarios. Molecular Biology and Evolution. 2019;36:2069–2085. doi: 10.1093/molbev/msz131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Karp PD, Riley M, Paley SM, Pellegrini-Toole A. The MetaCyc Database. Nucleic Acids Res. 2002;30:59–61. doi: 10.1093/nar/30.1.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 67.Finn RD, Clements J, Eddy SR. HMMER web server: interactive sequence similarity searching. Nucleic Acids Res. 2011;39:W29–37. doi: 10.1093/nar/gkr367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Finn RD, et al. Pfam: the protein families database. Nucleic Acids Res. 2014;42:D222–230. doi: 10.1093/nar/gkt1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Andreeva A, Howorth D, Chothia C, Kulesha E, Murzin AG. SCOP2 prototype: a new approach to protein structure mining. Nucleic Acids Res. 2014;42:D310–314. doi: 10.1093/nar/gkt1242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Coordinators NR. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2018;46:D8–D13. doi: 10.1093/nar/gkx1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Consortium U. UniProt: a worldwide hub of protein knowledge. Nucleic Acids Res. 2019;47:D506–D515. doi: 10.1093/nar/gky1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lenfant N, et al. ESTHER, the database of the α/β-hydrolase fold superfamily of proteins: tools to explore diversity of functions. Nucleic Acids Res. 2013;41:D423–429. doi: 10.1093/nar/gks1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Saier MH, et al. The Transporter Classification Database (TCDB): recent advances. Nucleic Acids Res. 2016;44:D372–379. doi: 10.1093/nar/gkv1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Shannon P, et al. Cytoscape: A Software Environment for Integrated Models of Biomolecular Interaction Networks. Genome Research. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Tran PH, Korszun ZR, Cerritelli S, Springhorn SS, Lacks SA. Crystal structure of the DpnM DNA adenine methyltransferase from the DpnII restriction system of streptococcus pneumoniae bound to S-adenosylmethionine. Structure. 1998;6:1563–1575. doi: 10.1016/s0969-2126(98)00154-3. [DOI] [PubMed] [Google Scholar]
- 76.Sali A, Blundell TL. Comparative protein modelling by satisfaction of spatial restraints. J Mol Biol. 1993;234:779–815. doi: 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]
- 77.Shen MY, Sali A. Statistical potential for assessment and prediction of protein structures. Protein Sci. 2006;15:2507–2524. doi: 10.1110/ps.062416606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Pettersen EF, et al. UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 79.Medjahed H, Singh AK. Genetic manipulation of Mycobacterium abscessus. Curr Protoc Microbiol. 2010;Chapter10 doi: 10.1002/9780471729259.mc10d02s18. Unit 10D.12. [DOI] [PubMed] [Google Scholar]
- 80.Chin CS, et al. Nonhybrid, finished microbial genome assemblies from long-read SMRT sequencing data. Nat Methods. 2013;10:563–569. doi: 10.1038/nmeth.2474. [DOI] [PubMed] [Google Scholar]
- 81.Chaisson MJ, Tesler G. Mapping single molecule sequencing reads using basic local alignment with successive refinement (BLASR): application and theory. BMC Bioinformatics. 2012;13:238. doi: 10.1186/1471-2105-13-238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Flusberg BA, et al. Direct detection of DNA methylation during single-molecule, real-time sequencing. Nat Methods. 2010;7:461–465. doi: 10.1038/nmeth.1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Bryant JM, et al. Whole-genome sequencing to identify transmission of Mycobacterium abscessus between patients with cystic fibrosis: a retrospective cohort study. Lancet. 2013;381:1551–1560. doi: 10.1016/S0140-6736(13)60632-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Ponstingl H. SMALT v0.5.8. Vol. 2012 Wellcome Trust Sanger Institute; Hinxton: 2011. [Google Scholar]
- 86.Hahsler M. R package version 0.9-5. 2015. [Google Scholar]
- 87.Fröhlich H, et al. Analyzing gene perturbation screens with nested effects models in R and bioconductor. Bioinformatics. 2008;24:2549–2550. doi: 10.1093/bioinformatics/btn446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Csardi G, Nepusz T. The igraph software package for complex network research. InterJournal Complex Systems. 2006:1695. [Google Scholar]
- 89.Csardi G, Nepusz T. The igraph software package for complex network research. Inter JournalComplex Systems. 2006;1695 [Google Scholar]
- 90.Ding L, et al. Somatic mutations affect key pathways in lung adenocarcinoma. Nature. 2008;455:1069–1075. doi: 10.1038/nature07423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.McGuire AM, et al. Comparative analysis of mycobacterium and related actinomycetes yields insight into the evolution of Mycobacterium tuberculosis pathogenesis. BMC Genomics. 2012;13:120. doi: 10.1186/1471-2164-13-120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Szklarczyk D, et al. The STRING database in 2017: quality-controlled protein-protein association networks, made broadly accessible. Nucleic Acids Res. 2017;45(D1):D362–D368. doi: 10.1093/nar/gkw937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Medjahed H, Singh AK. Genetic manipulation of Mycobacterium abscessus. Curr Protoc Microbiol. 2010;Chapter 10 doi: 10.1002/9780471729259.mc10d02s18. Unit 10D.12. [DOI] [PubMed] [Google Scholar]
- 94.Choudhary E, Thakur P, Pareek M, Agarwal N. Gene silencing by CRISPR interference in mycobacteria. Nat Commun. 2015;6:6267. doi: 10.1038/ncomms7267. [DOI] [PubMed] [Google Scholar]
- 95.Chin CS, et al. Nonhybrid, finished microbial genome assemblies from long-read SMRT sequencing data. Nat Methods. 2013;10:563–569. doi: 10.1038/nmeth.2474. [DOI] [PubMed] [Google Scholar]
- 96.Chaisson MJ, Tesler G. Mapping single molecule sequencing reads using basic local alignment with successive refinement (BLASR): application and theory. BMC Bioinformatics. 2012;13:238. doi: 10.1186/1471-2105-13-238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Flusberg BA, et al. Direct detection of DNA methylation during single-molecule, realtime sequencing. Nat Methods. 2010;7:461–465. doi: 10.1038/nmeth.1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Hepburn L, et al. A Spaetzle-like role for Nerve Growth Factor b in vertebrate immunity to Staphylococcus aureus. Science. 2014;346:641–646. doi: 10.1126/science.1258705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Weller R, Finnen MJ. The effects of topical treatment with acidified nitrite on wound healing in normal and diabetic mice. Nitric Oxide. 2006;15:395–399. doi: 10.1016/j.niox.2006.04.002. [DOI] [PubMed] [Google Scholar]
- 101.Chiner-Oms Á, et al. Genome-wide mutational biases fuel transcriptional diversity in the Mycobacterium tuberculosis complex. Nature Communications. 2019;10(1):3994. doi: 10.1038/s41467-019-11948-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All sequencing data associated with this study is deposited in the European Nucleotide Archive under the following project accession numbers: shortread DNA sequence data, ERP001039; RNA-seq data, ERP016362; SMRT-seq data, ERP010248. The code for MV-trees can be found at: https://github.com/JosieMB/MV_trees.git (Zenodo DOI: 10.5281/zenodo.4569470). All phylogenetic trees associated with this study are deposited in TreeBase (http://purl.org/phylo/treebase/phylows/study/TB2:S27748).