Abstract
In the mammalian lung, an apparently homogenous mesh of capillary vessels surrounds each alveolus, forming the vast respiratory surface across which oxygen transfers to blood1. Here, we use single cell analysis to elucidate the cell types, development, renewal, and evolution of the alveolar capillary endothelium. We show that alveolar capillaries are mosaics; like the epithelium lining the alveolus, the alveolar endothelium is made up of two intermingled cell types, with complex ‘swiss cheese’-like morphologies and distinct functions. One cell type we term the ‘aerocyte’ is specialized for gas exchange and leukocyte trafficking, and unique to the lung. The other, termed gCap (general capillary) is specialized to regulate vasomotor tone and functions as a stem/progenitor cell in capillary homeostasis and repair. The two cell types develop from bipotent progenitors, they mature gradually, and are affected differently in disease and aging. This cell type specialization is conserved between mouse and human but is not found in alligator or turtle lungs, suggesting that specialized capillary cells arose during evolution of the mammalian lung. The discovery of capillary cell type specialization transforms our understanding of the structure, function, regulation, and maintenance of the air-blood barrier and gas exchange in health, disease, and evolution.
Lungs have evolved complex and diverse architectures that combine a large surface with an exquisitely thin barrier for efficient exchange of oxygen and carbon dioxide between air and blood. In mammalian lungs, gas exchange occurs in tightly packed alveoli, the terminal airspaces of the bronchial tree, surrounded by walls containing a dense capillary network (Fig. 1a). The discovery of alveoli and their associated capillaries by Malpighi inaugurated what became the study of the structural basis of gas exchange, providing the foundation for modern respiratory physiology and pulmonary medicine2,3. Efforts to understand the cellular structure of the barrier have focused mainly on epithelial cells, beginning with the recognition that alveoli are lined by a continuous epithelium composed of intermixed alveolar type 1 (AT1) and AT2 cell types4. AT1 cells are large, thin and highly extended cells that comprise 95% of the respiratory surface across which diffusion occurs, whereas cuboidal AT2 cells secrete surfactant that prevents alveolar collapse1. There has been much recent progress in understanding the development, maintenance, and repair of the alveolar epithelium5–7. The cells of the alveolar endothelium – the other side of the air-blood barrier – have received less attention.
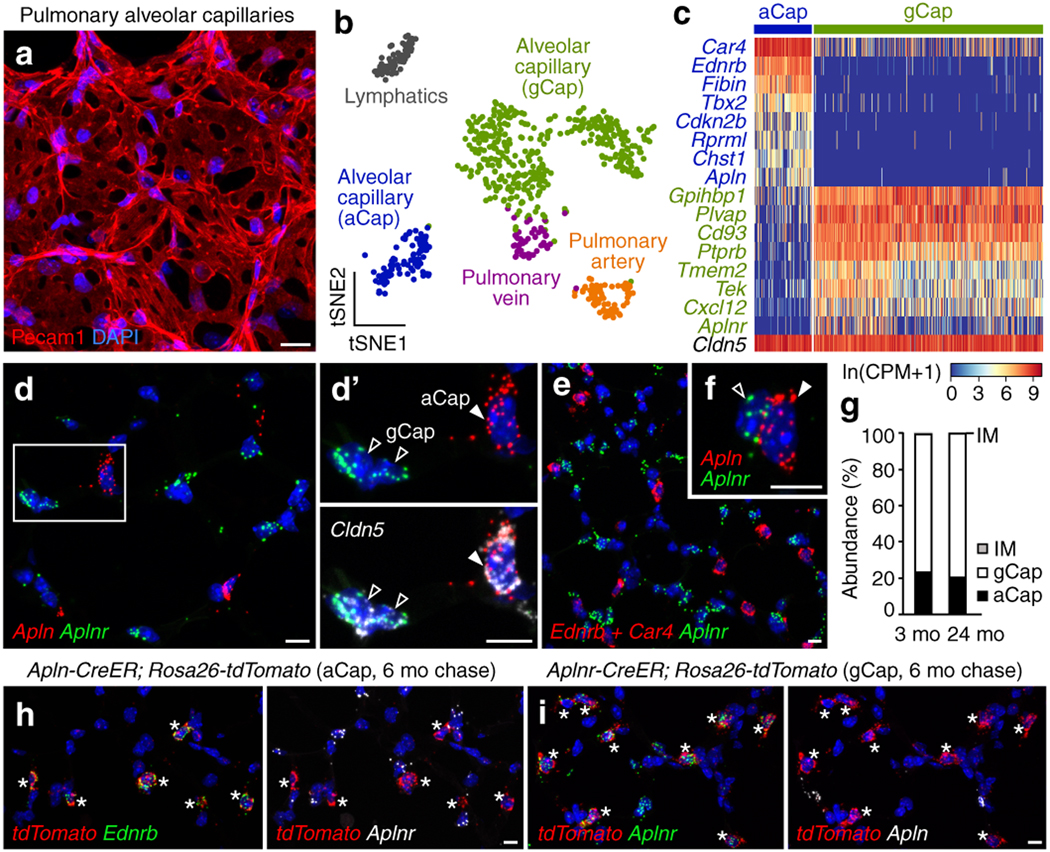
Figure 1. Two stable, intermingled alveolar capillary cell types.
a, Alveolar capillaries in adult mouse lung immunostained for Pecam1. b, tSNE plot of endothelial cell populations annotated in scRNAseq data for adult mouse lung13. c, Heatmap of expression of capillary subset markers and general endothelial marker Cldn5 in individual capillary cells. d-f, smFISH for capillary subset markers Apln, or Ednrb and Car4 (aCap) and Aplnr (gCap) in adult mouse lung. d’, f, Individual aCap and gCap cells. g, Relative abundance of aCap, gCap, and cells that co-express aCap and gCap markers (IM) in lungs from 3- and 24-month old mice (data as mean; n=500 cells scored per mouse; 2 mice per group). h, i, Co-expression of tdTomato lineage label (asterisks) and aCap marker Ednrb but not gCap marker Aplnr (h) or gCap marker Aplnr but not aCap marker Apln (i) in lungs harvested six months after mature aCap (h) or gCap (i) cells were lineage labeled. Blue, DAPI. Scale bars, 10 μm.
Intermingled alveolar capillary cell types
We systematically defined the cellular diversity of the pulmonary endothelium by single-cell RNA-sequencing (scRNAseq) and mapping in adult mouse lung, and identified two molecularly distinct capillary cell populations in the alveolus (Figs. 1b-d, ED1a-g, Supplementary Tables 1,2). The two subsets are interspersed throughout the gas exchange region, creating apparently random diversity within the alveolar capillary network, and their relative abundance does not change significantly with age (Figs. 1e-g, ED1h-j). To test whether the capillary populations are stable cell types or interconverting cell states, we used complementary genetic strategies to permanently label each subset (using either Apln-CreER or Aplnr-CreER) and analyzed expression of subset markers in labeled cells after 48 hours, one, or six months to determine if the labeled population continues to express the markers, or if cells turn them off and start to express markers of the other population. We found minimal (0.2% at 6 months) interconversion (Figs. 1h,i, ED1k-n), indicating the populations are not transient cell states. We conclude that the alveolar capillary network is composed of two intermingled, stable cell types, which we call gCap (‘general’ capillary cells) and aCap (‘aerocytes’, see below).
Aerocytes are specialized for gas exchange
We used sparse cell labeling and deep imaging to visualize individual capillary cells in three dimensions. aCap cells are remarkably complex, large cells (spanning >100μm; 21×103 μm3 mean volume) with ramified extensions that surround pores (mean 6/cell, range 2–9), giving cells the appearance of ‘swiss cheese’ (Figs. 2a,c, ED2f-h, Supplementary Video 1). The cells have a variety of sizes and shapes, and a single cell frequently spans multiple alveoli. Such surprising morphologic complexity has also been described for AT1 cells1.
Figure 2. Specialized alveolar capillary cell types in gas exchange and capillary renewal.
a, b, Single aerocyte (a) or gCap (b) cells in adult Apln-CreER; Rosa26-Confetti (a) or Aplnr-CreER; Rosa26-Confetti (b) lungs. c, Quantification of individual cell volumes. Bar indicates mean (19 aCap and 17 gCap cells scored from n=2 mice). d, aCap and gCap cells in adult Cdh5-CreER; Rosa26-Confetti lung form multicellular tubes (asterisks) within capillaries surrounding a single alveolus (dotted outline). Blue, elastin fibers (a, b, d). e, Schematic of alveolar capillary network. Asterisks indicate multicellular tubes. RBC, red blood cell. f-i, Transmission electron micrographs of adult mouse alveolar walls. f, f’ Thick and thin regions of the air-blood barrier. g-i, Apln-CreER; Rosa26-tdTomato (g, h) or Aplnr-CreER; Rosa26-tdTomato (i) lungs immunostained for tdTomato (heavy black stain). Labeled aerocytes (g, h) but not gCap cells (i) are associated with thin regions (dashed lines). j, Quantification of percent of each labeled cell type associated with thick or thin regions (n=2 mice of each genotype, 21 labeled aCap cells and 24 labeled gCap cells scored). k, Schematic representation of air-blood barrier. AT1, alveolar type 1 epithelial cell. l, m, Analysis of proliferation of lineage labeled aCap (l) or gCap cells (m) during adult homeostasis, detected by cumulative EdU incorporation for 6 weeks. 0.1% (mean) of labeled aCap cells incorporated EdU (l) compared to 7.7% of labeled gCap cells (m) (n=400–4,000 cells scored per lung in n=2 mice of each genotype). n, Quantification of fraction of EdU+ lineage labeled aCap or gCap cells during indicated intervals after elastase (El) or mock injury with saline as control (Ctrl; data as mean; n=200–1,600 cells scored per lung in 2–4 mice of each genotype per time point and treatment group; see Methods for exact sample sizes). o, o’ smFISH for lineage label (tdTomato), aCap (Ednrb) and gCap (Aplnr) markers in gCap lineage labeled lung six weeks after elastase injury. Blue, DAPI (l, m, o). p, Quantification of fraction of gCap (left) or aCap (right) lineage labeled cells expressing aCap (Ednrb), gCap (Aplnr or Ptprb), or both markers (intermediate (IM) cells) six weeks after elastase injury in injured and uninjured (control) regions (n=500–1000 cells scored per region; 3 injured and control regions scored in n=2 mice of each genotype). q, Relative abundance of capillary cell types in injured and uninjured (control) regions, six weeks after elastase administration (n=800–5600 cells scored per region; 3 injured and control regions; n=3 mice). Scale bars, 10 μm (a, b, d, l, m, o, o’), 2 μm (f-i).
gCap cells have a related but less extreme morphology. They are smaller (<40μm; 4×103 μm3 mean volume), have fewer pores (mean 3/cell, range 1–6), and are less extensively branched, rarely spanning multiple alveoli (Figs. 2b,c, ED2e,g,h, Supplementary Video 2). The two cell types fit together to form multicellular tubes (Fig. 2d,e, Supplementary Video 3). The mean surface area of aCap cells is four to five times greater than that of gCap cells (Fig. ED2g), but they are four-fold less abundant (Fig. 1g), hence each contributes about half the capillary surface area. The morphologies of both types, especially aCap cells, are strikingly different from capillary cell morphologies elsewhere in the lung, within the bronchial circulation, and in other organs (Fig. ED3a-g), likely reflecting the unique design and function of the pulmonary circulation. Comparison of their molecular diversity (Fig. ED3h-l) suggests capillary cells in other organs are more similar to gCap cells (supporting the name ‘general’ capillary) whereas aCap cells are unique to lung.
Capillaries are asymmetrically positioned within alveolar walls such that only some of the endothelium is tightly apposed to squamous AT1 cells to form thin regions of the gas exchange surface where the barrier to diffusion is minimized, while other (‘thick’) regions are separated from epithelium by stromal cells and connective tissue (Fig. 2f)3. To look for differences in localization of the cell types in these structurally distinct regions, we performed immuno-electron microscopy (EM) on lungs in which aCap and gCap cells were separately labeled. We found thin regions are composed entirely of aCap cells, whereas gCap cells are positioned in contact with stromal cells in thick regions (Figs. 2g-j, ED4a-c). Because of their close association with AT1 cells within thin regions of the respiratory surface (Fig. 2k) and their expansive morphology, reflecting a specialized role in gas exchange analogous to AT1 cells, we term aCap cells ‘aerocytes’.
gCap cells are capillary stem cells
Little is known about how alveolar capillaries are maintained throughout life and repaired upon alveolar damage7. To examine behaviors of capillary cell types in alveolar homeostasis, we first analyzed proliferation by cumulative labeling with ethynyldeoxyuridine (EdU) for six weeks in mice in which either gCap cells or aerocytes were genetically labeled. Capillary cell turnover was slow8, but surprisingly, proliferation was almost entirely restricted to gCap cells (7.7% EdU+ gCap cells; Fig. 2l,m). We detected extremely rare EdU+ solitary aCap cells (2 of 4401 cells), whereas EdU+ gCap cells were present as clusters of up to 10 cells, indicative of focal proliferation.
Alveolar capillary cell proliferation can be induced by acute lung injury8,9. To investigate the role of the cell types in capillary repair, we used a mouse model of emphysema10 (Fig. ED5a,b) in which elastase-induced alveolar damage is accompanied, we found, by robust capillary cell proliferation. Lineage labeled gCap cells proliferated as early as day 3 after elastase instillation (17% EdU+), with almost all (93%) gCap cells in injured regions EdU+ at 6 weeks (Figs. 2n, ED5c-f). Aerocytes rarely proliferated, even after injury (0.2% EdU+ at 3 days, 1.7% at 6 weeks).
We examined the fate of lineage labeled gCap cells after injury and found labeled aerocytes as well as gCap cells, demonstrating aerocytes are generated from gCap cells during repair (Fig. 2o,p). We also detected rare, lineage labeled aerocytes in the absence of injury, after extended chases (3.4% lineage labeled aCap cells at 14 months; Fig. ED1n), indicating intermittent generation of aerocytes from gCap cells during homeostasis. We conclude gCap cells replenish the alveolar capillary endothelium during maintenance and repair, functioning as specialized stem/progenitor cells.
Even after 6 weeks of recovery from injury, the cellular composition of the alveolar capillary network is altered (Fig. 2q), suggesting repair is abnormal or incomplete at this stage. Aberrant or insufficient repair may underlie vascular changes in lung diseases, such as emphysema and interstitial lung disease, as well as respiratory distress syndromes accompanying severe injury or virus-induced alveolar damage as in Covid-1911,12. Understanding the behavior of gCap cells, and what signals activate their proliferation and reprogramming, may offer a strategy to restore the normal pattern.
Molecular functions of capillary cell types
We used scRNAseq profiles13 to discover common and additional type-specific capillary cell functions. We found only a small number of genes (Scn7a, Mapt) expressed by all/most, and only, alveolar capillary cells, suggesting that few if any molecular functions are carried out by both capillary cell types but not other lung endothelial cells (Fig. ED1a,b). In contrast, we identified many genes with roles in physiology, immune interactions, and signaling whose expression differed between the cell types, revealing further specialization (Figs. 3a, ED4d, Supplementary Table 2).
Figure 3. Molecular functions of alveolar capillary cell types.
a, Dot plots showing expression in aerocytes and gCap cells for selected differentially expressed genes. b-f, Diagrams of proposed specialized alveolar capillary cell functions. Aerocyte genes, blue; gCap genes, green; aerocyte/gCap genes, yellow; pericyte genes16, purple. d, gCap cells express Edn1, eNOS (encoded by Nos3) and prostaglandin I2 synthase (encoded by Ptgis), making them a unique source of vasomodulators. Edn1 can signal to Ednra expressed on pericytes or to Ednrb expressed on aerocytes, which may feed back to gCap cells (dashed arrow) to regulate vasodilator production. f, Lipoprotein lipase (encoded by Lpl), anchored to the lumen of aerocytes by Gpihbp1, converts circulating lipoprotein triglycerides to monoglycerides, which are broken down to free fatty acids by monoacylglycerol lipase (encoded by Mgll) expressed by gCap cells.
Some functions appear unique to one cell type. Aerocytes are likely the site of leukocyte trafficking, primarily a capillary function in the lung14, as they specifically express adhesion and leukocyte sequestration genes (Fig. 3a,b). gCap cells, in contrast, express MHC class II components, suggesting they present antigens (Fig. 3a,c). gCap cells may also play a specialized role in vasomotor control (see below; Fig. 3a,d). Other functions appear to be distributed across the cell types. They produce distinct pro- and anticoagulants suggesting each plays a different role in hemostasis (Fig. 3a,e), and may cooperate in lipid metabolism, forming an ‘assembly line’ producing fatty acids (Fig. 3a,f).
Our analysis also revealed the cell types can signal one another. Aerocytes are a source of ligands (Apln, Kitl) that signal through cognate receptors (Aplnr, Kit) displayed by gCap cells; conversely gCap cells produce ligands (Edn1, Vegfa) with cognate receptors (Ednrb, Kdr) on aerocytes (Figs. 3a, ED4d,e). Such bidirectional signaling indicates the cell types can regulate each other.
We also identified specialized signaling interactions with other alveolar cell types. Aerocytes express Ednrb and Kdr, suggesting they interact with AT1 cells (Figs. 3a, ED4d,e). gCap cells produce a vasoconstrictor (Edn1) that can signal to Ednra on pericytes; they also express eNOS and prostaglandin I2 synthase making them a source of vasodilators (Figs. 3a,d, ED4d,e). This suggests gCap cells regulate vasomotor tone through interactions with pericytes. These specialized signaling relationships reflect distinct associations of the capillary cell types with surrounding cells, revealing functional compartmentalization within the alveolus (Fig. ED4f).
Capillary cell type development and aging
To determine when the cell types emerge during development, we investigated their origin in embryonic lung where a dense vascular plexus surrounds branching airways (Fig. 4a). Using lineage tracing, we found that this plexus, composed of small (2×103 μm3 mean volume), simple, proliferating endothelial cells, gives rise to both alveolar capillary subsets (Figs. 4b-d, ED6a-c, Supplementary Data 1,2). The near complete labeling of the capillary network suggests plexus is the major or sole source of aerocytes and gCap cells. To determine whether individual plexus cells can give rise to both capillary cell types, we performed a clonal analysis. Clones contained both aerocytes and gCap cells, demonstrating that plexus cells are bipotent (Figs. 4e,f, ED6d-f, Supplementary Video 4).
Figure 4. Development and evolution of specialized alveolar capillary cell types.
a, Plexus surrounding airways in immunostained E13.5 mouse lung. b, Sparsely labeled plexus cells in E12.5 Aplnr-CreER; Rosa26-Confetti lung immunostained for RFP. c, Near complete labeling of alveolar capillaries in Aplnr-CreER; Rosa26-tdTomato lung administered tamoxifen at E12.5. Endomucin, green; tdTomato, red. d, smFISH to detect tdTomato (white) in aCap (Apln, red) and gCap (Aplnr, green) cells in P60 Aplnr-CreER; Rosa26-tdTomato lung lineage labeled at E12.5. e, Clone in Aplnr-CreER; Rosa26-Confetti lung composed of aCap and gCap cells derived from a single YFP-expressing plexus cell labeled at E14.5. f, Schematics depicting the origin of both alveolar capillary cell types from single bipotent cells in the embryonic plexus. g, Individual aerocytes in Apln-CreER; Rosa26-Confetti lungs at indicated stages. Dots, pores. h, Quantification of individual cell volumes at indicated times (n>10 cells scored for each cell type at each time point from n=2 mice; see Methods for exact cell number; adult aCap and gCap cells from Fig. 2c). Bar, mean value. i, Quantification of Vwf-expressing capillary cells in lungs from 3- and 24-month old mice (mean± s.d.; n>500 cells scored per mouse; 3 mice per age group). j, smFISH for capillary cell type markers in alveolar capillary cells in adult human lung (75 year old male). k, Heatmap of expression of cell type markers in individual capillary cells16. l, Human adenocarcinoma vessel containing cells co-expressing EDNRB (aCap marker) and PTPRB (gCap marker). See also Fig. ED9a. m-o, Dot plots showing expression in aCap and gCap cells for selected conserved genes (Type 0, leukocyte trafficking; m), human- and mouse-specialized genes (Type 1, hemostasis; n), or genes (Type 2, antigen presentation; o) which switch cell type between species (mouse13, human16). See Fig. ED10c-e. p, Schematic illustrating alligator lung faveolus. q, Faveolar capillary network in alligator lung immunostained for CLDN5. r, r’ Co-expression (asterisks) of mammalian alveolar capillary cell type markers EDNRB (aCap) and APLNR (gCap) in faveolar capillary cells (CLDN5, white) in alligator lung. Blue, DAPI (d, j, l, r) or elastin fibers (e, g). Scale bars, 10 μm.
Aerocytes first emerge at E17.5 and begin to acquire their ‘swiss cheese’ morphology during embryonic development (Figs. 4g,h, ED2, ED6g,h). This is consistent with a recent study that identified emerging aerocytes and characterized their gene expression and morphology15. Capillary cell type emergence is gradual and asynchronous, and both cell types continue to mature molecularly and morphologically after birth (Figs. 4g,h, ED2, ED6h-j, ED7, Supplementary Data 3,4). These results uncover a remarkable transformation of plexus into the alveolar capillary network beginning in embryonic lung.
Endothelial cell phenotypes change with age, and may contribute to age-related disease. We detected widespread induction of von Willebrand Factor (Vwf), considered a marker of endothelial dysfunction, in lungs of aged mice. Vwf is induced specifically in gCap cells, but not aerocytes (Figs. 4i, ED6k-m), indicating differential regulation of the cell types during aging.
Human alveolar capillary cell types
We identified both cell types intermingled within human alveolar capillary networks (Figs. 4j,k, ED8a-i, Supplementary Video 5), demonstrating they have been conserved in mammalian evolution16. Similar to mice, aerocytes emerge during embryonic development (Fig. ED8j).
The mosaic capillary cell pattern, however, is lost or altered in lung tumors. We observed abundant intermediate cells co-expressing markers of both cell types in human adenocarcinoma vessels (Figs. 4l, ED9a). Cell composition is also altered in mouse adenomas; tumor vessels are composed of gCaps and intermediates, with few/no aerocytes (Fig. ED9b-f).
As in mouse, we identified genes with key roles in physiology, immune interactions, and signaling with specialized expression in the human cell types (Fig. ED10a,b, Supplementary Table 3). Many genes with cell type specificity in mice are also expressed by the corresponding human cell type (Figs. 4m; ED10c)16. However, we also identified mouse-human differences. Some genes show specialized expression in only one species, including genes involved in functions distributed between the cell types (Figs. 4n, ED10d). For other genes the cell type with specialized expression switches between mouse and human, presumably reflecting species-specific functional differences (Figs. 4o, ED10e).
Our analysis suggests some cell type-specific functions are conserved between mouse and human. For example, specialized leukocyte trafficking genes are restricted to aerocytes, and gCap cells may regulate vasomotor tone in both species (Figs. 4m, ED10a-c). But capillary cell types can also gain (or lose) functions: in mouse gCap cells preferentially express MHC class II genes, whereas in human these genes are expressed by aerocytes (Figs. 4o, ED10a,b,e).
Evolution of capillary cell specialization
To investigate evolutionary origins of the cell types, we examined capillary cell diversity in lungs from American alligator (Alligator mississippiensis) and Western painted turtle (Chrysemys picta bellii), reptiles from distinct phylogenetic groups (Fig. ED11a). Alligator and turtle respiratory systems are, in many ways, representative of the ancestral amniote condition17 and gas exchange occurs across a thick air-blood barrier in faveoli, which are surrounded by capillary nets strikingly resembling those of mouse and human alveoli (Figs. 4p,q, ED11b-d,g-k,m, Supplementary Video 6). We found that in each species lung capillary cells express markers of both mammalian cell types (Figs. 4r, ED11e,f,l). However, in contrast to alveolar capillary cells, alligator and turtle faveolar capillary cells co-express these cell type markers, suggesting they may lack the cell specialization of mammalian lungs.
Discussion
Here we show that the alveolar capillary endothelium, like the alveolar epithelium, is composed of two intermingled cell types. Such cell type specialization may have evolved to optimize gas exchange within the complex environment of the alveolus, balancing and integrating structure and function. Aerocytes and AT1 cells are both large, complex cells tightly apposed in the thinnest regions of the gas exchange surface – separated only by a shared, compositionally unique basement membrane18 – facilitating diffusion. This specialized interface may be particularly important in lung injury. In pulmonary edema, seen early in ARDS, fluid accumulates in thick regions, initially protecting thin regions and preserving gas exchange19,20. The alveolar capillary cell types arise, like the epithelium, from bipotent progenitors, through distinct maturation programs6, with aerocytes first emerging as AT1 differentiation begins, highlighting coordination between the cell types critical for gas exchange15. During adult life, the alveolar endothelium is maintained and repaired by gCap cells, which like AT2 cells are ‘bifunctional’ stem/progenitor cells5,6 that also serve physiologic functions. Separating the progenitor function from aerocytes and AT1 cells may be another mechanism for preserving gas exchange surface. Capillary changes may underlie pathology in the lung (Figs. 4i,l, ED9) and other organs, making it essential to now explore and map the full heterogeneity of capillary cell types, states, and specializations in health, aging, and disease; to identify changes in capillary composition and which cell types change, how they change, and the consequences for organ function.
Methods
Mice.
The following mouse strains were used: C57BL/6 (C57BL/6NCrl, Charles River Laboratories, strain code: 027) was the wild-type strain. Apln-CreER (Aplntm1.1(cre/ERT2)Bzsh)21 (provided by Bin Zhou), Aplnr-CreER (Tg(Aplnr-cre/ERT2)#Krh)22 (provided by Kristy Red-Horse), Cdh5-CreER (Tg(Cdh5-cre/ERT2)1Rha)23 (provided by Ralf Adams), and Sftpc-CreER (Sftpctm1(cre/ERT2,rtTA)Hap)24 (provided by Harold Chapman) were used for conditional expression of Cre recombinase. Rosa26-tdTomato (Gt(ROSA)26Sortm14(CAG-tdTomato)Hze)25 (The Jackson Laboratory, strain #007914), which expresses cytoplasmic tdTomato following recombination, and Rosa26-Confetti (Gt(ROSA)26Sortm1(CAG-Brainbow2.1)Cle)26 (The Jackson Laboratory, strain #017492), which expresses either membrane targeted Cerulean CFP, nuclear GFP, cytoplasmic EYFP, or cytoplasmic RFP following recombination, were used as Cre reporters. KrasLSL-G12D (Krastm4Tyj/J)27 (The Jackson Laboratory, strain # 008179) was used to express a constitutively active form of KRAS from the endogenous locus following Cre-mediated recombination. All experimental animals and embryos were heterozygous (or hemizygous) for indicated alleles. Only female animals/embryos were used for experiments with Apelin-CreER, since Apelin is X-linked21. Noon of the day a vaginal plug was detected was considered as E0.5. The day a litter was born was considered as P0. For induction of Cre recombinase activity, tamoxifen (Sigma, T5648) was dissolved in corn oil and administered by intraperitoneal (i.p.) injection unless otherwise noted. Adult lungs were perfused, inflated with 2% low melting point agarose (Invitrogen), and collected as previously described6. Heart, brain, small intestine, thyroid, and kidney were collected following perfusion of the left heart with Ca2+- and Mg2+-free phosphate-buffered saline, pH 7.4 (PBS; Gibco). Postnatal (P7) retinas28 and embryonic lungs29 were collected and prepared as previously described.
Tamoxifen dose, administration and tissue collection schedules for individual experiments (unless noted elsewhere) were:
For cell type stability pulse/chase experiments (presented in Fig. 1h, i, Extended Data Fig. 1k-n): 4 mg tamoxifen to adults, harvested after 48 hours, 1, 6, or 14 months.
For cell morphology (sparse labeling) experiments: 2 mg (Fig. 2a, b, Extended Data Fig. 2e, f), 0.5 mg (Extended Data Fig. 3a), 1 mg (Fig. 2d, Extended Data Fig. 3b-f) tamoxifen to adults, harvested after 5–7 days. 2 mg (Fig. 4b) or 0.5 mg (Extended Data Fig. 2a) tamoxifen to pregnant dams at E11.5, harvested at E12.5, 2 mg tamoxifen at E17.5, harvested at E18.5 (Fig. 4g, Extended Data Fig. 2b), 0.5 mg tamoxifen at E18.5, harvested at P0 (Extended Data Fig. 2c), or 3 mg tamoxifen at E18.5, harvested at P0 (Extended Data Fig. 2d). 0.2 mg tamoxifen at P5 by intragastric injection, harvested at P7 (Extended Data Fig. 3g), or 0.05 mg tamoxifen by intragastric injection at P5, harvested at P7 or P14 (Fig. 4g).
For lineage tracing experiments: 4 mg tamoxifen administered to pregnant dams; lungs harvested from progeny at P21 or P60 (Fig. 4c, d, Extended Data Fig. 6c, Supplementary Data 1). 0.5 mg tamoxifen administered at P7, harvested at P21 (Extended Data Fig. 6f).
For maximal labeling experiments: two or three 4 mg tamoxifen doses (administered 48 hours apart) to Apln-CreER; Rosa26-tdTomato (Fig. 2g, h, Extended Data Fig. 4a, b) or Aplnr-CreER; Rosa26-tdTomato (Fig. 2i, Extended Data Fig. 4a, c) adult animals, harvested 5–14 days after the first dose.
To induce adenoma formation: 4 mg tamoxifen to Sftpc-CreER; KrasLSL-G12D/+ adult animals (Extended Data Fig. 9b, c, f), harvested 3 weeks later.
Mice were housed and bred in the animal facility at Stanford University in accordance with Institutional Animal Care and Use Committee guidance, and were maintained on a 12 hour light/dark cycle with food and water provided ad libitum. Adult mice were 2–6 months old, unless otherwise noted. All mouse experiments were approved by the Stanford University Institutional Animal Care and Use Committee.
Human tissue.
De-identified normal human adult lung tissue from 69 and 75 year old males and a 66 year old female was obtained from the Stanford Tissue Bank. De-identified aborted human fetal lung tissue (17 and 23 weeks) was obtained in collaboration with the Stanford Family Planning Research Team, Department of Obstetrics & Gynecology, Division of Family Planning Services and Research, Stanford University School of Medicine. De-identified human tissue representing well-differentiated invasive lung adenocarcinoma (confirmed by pathological evaluation by S.Y.T.) from a 41 year old female was obtained from archival diagnostic material in collaboration with the Stanford Department of Pathology, Stanford University School of Medicine. Tissue collection and use in research were approved by the Stanford Institutional Review Board.
Alligators.
Lungs were collected from juvenile (body mass, 1.3 kg) and adult (body mass, 14 kg) American alligators (Alligator mississippiensis, Daudin; male), acquired from the Rockefeller Wildlife Refuge (Grand Chenier, Louisiana). Lungs were inflated with sterile PBS or 10% formalin for single-molecule fluorescent in situ hybridization (smFISH), or 4% paraformaldehyde (PFA; Electron Microscopy Sciences [EMS])/PBS for immunostaining. Experiments were approved by the University of Utah Animal Care and Use Committee.
Turtles.
Lungs were collected from two adult (body mass, 262g and 281g) Western painted turtles (Chrysemys picta bellii; male; The Turtle Source). Lungs were inflated with 10% formalin for smFISH, or 4% PFA/PBS for immunostaining. Experiments were approved by the University of Utah Institutional Animal Care and Use Committee.
Immunostaining.
Immunostaining was performed using previously published protocols6,29 with modifications for adult mouse, human, alligator, and turtle tissues as described below. Adult mouse, alligator, and turtle lungs and human lung tissue pieces were fixed in 4% PFA/PBS at 4°C for 2–3 hours, then dehydrated through a PBS/methanol series into 100% methanol and stored at −20°C. Immediately prior to sectioning, tissue was rehydrated through a methanol/PBT (PBT: 0.1% Tween-20/PBS) series into PBT. 350 μm sections were cut from adult mouse lung lobes on a vibratome (Leica Biosystems). Alligator and turtle lungs, and human lung pieces, were manually cut with a platinum coated double edge razor blade (EMS) into rough sections 0.5 mm to 3 mm thick. Sections were incubated with primary antibody for 3 nights and secondary antibody for 2 nights. Sections stained using peroxidase-conjugated secondary antibodies were incubated in tyramide reagents (Perkin Elmer; 1:100) for 45 minutes. Stained sections were post-fixed in 4% PFA/PBS at 4°C for 1 hour, dehydrated into methanol, and cleared in either benzyl alcohol:benzyl benzoate (1:2; BABB) or Vectashield (Vector Laboratories) for confocal imaging.
Immunostaining of the human adenocarcinoma sample was performed on a formalin fixed paraffin embedded tumor section using the BOND automated staining system with ER2 epitope retrieval solution and the BOND Polymer Refine Detection system (Leica Biosystems), which includes a haematoxylin counterstain. Adjacent sections were used for immunostaining and smFISH.
Primary antibodies used, at indicated concentrations: CD34 (BD Biosciences, 347660; 1:160); Claudin5 (Abcam, ab53765; 1:300); E-Cadherin (BD Biosciences, 610181; 1:100); Endomucin (Invitrogen, eBioV.7C7, 14–5851−82; 1:300); Integrin alpha8 (R&D, AF4076; reconstituted to 1 mg/ml, used at 1:500); Pecam1 (rat anti-mouse; BD Biosciences, 553370; 1:5000 for staining embryonic lung, 1:500 for staining adult lung); Pecam1 (mouse anti-human; R&D, BBA7; reconstituted to 0.5 mg/ml in PBS, used at 1:200); tdTomato (Rockland, 600–401−379; 1:300); VE-Cadherin (R&D, AF938; reconstituted to 0.5 mg/ml in PBS, used at 1:300).
Secondary antibodies used, at indicated concentrations: Donkey anti-goat IgG, Alexa Fluor 568 conjugated (Invitrogen, A11057; 1:250); Horse anti-mouse IgG, peroxidase conjugated (Vector Laboratories, PI-2000; 1:150); Goat anti-rabbit IgG, peroxidase conjugated (Vector Laboratories, PI-1000; 1:125–1:250); Goat anti-rabbit IgG, Alexa 568 conjugated (Invitrogen, A11036; 1:250); Goat anti-rat IgG, Alexa 488 conjugated (Invitrogen, A11006; 1:250), for embryonic lung; Donkey, anti-rat IgG, Alexa 647 conjugated (Jackson Immunoresearch, 712–605-153; 1:250); Goat anti-rat IgG, biotin conjugated (Vector Laboratories, BA-9401; 1:250), for embryonic lung; Goat anti-rat IgG, peroxidase conjugated (Vector Laboratories, PI-9401; 1:250), for adult lung. 4’,6-Diamidino-2-Phenylindole, Dihydrochloride (DAPI; Invitrogen, D1306; reconstituted in PBS, used at 2 μg/ml) to stain nuclei and/or Alexa Fluor 350 hydrazide (Invitrogen, A10439; reconstituted to 0.5 mg/ml in PBS, used at 1:100) or Alexa Fluor 633 hydrazide (Invitrogen, A30634; reconstituted to 0.5 mg/ml in PBS, used at 1:500–1:1000) to visualize elastin fibers, were added along with secondary antibody.
Single-molecule fluorescent in situ hybridization (smFISH).
Mouse, alligator, and turtle lungs, inflated as described above, human lung, or adult mouse kidney tissue were fixed in 10% neutral buffered formalin (Fisher Scientific) for 24 hours at room temperature and transferred to 70% ethanol (made up in PBS) following 3 brief washes in PBS for embedding in paraffin. Sections were cut at 6 μm. smFISH was performed using a proprietary high sensitivity RNA amplification and detection technology (RNAscope, Advanced Cell Diagnostics), according to the manufacturer’s instructions using the indicated proprietary probes, the RNAscope Multiplex Fluorescent Reagent Kit (v2), and TSA Plus reagents (Perkin Elmer; 1:500 dilution for Cy3 and Cy5 dyes, 1:250 dilution for FITC) or Opal dyes (Akoya Biosciences, 1:500 dilution for Opal 570 and 620 dyes, 1:250 dilution for Opal 520 and 690 dyes). Following smFISH, sections were incubated in DAPI (used at 2 μg/ml in PBS) for 5 minutes to counterstain nuclei and mounted in Prolong Gold antifade reagent (Invitrogen). Proprietary (Advanced Cell Diagnostics) probes used were: Mouse, Mm-Aplnr (436171), Mm-Vwf (499111), Mm-Ednrb (473801, 473801-C2, 473801-C3), Mm-Apln (415371-C2), Mm-Ptprb (481391-C2), Mm-H2-Ab1 (414731-C2), Mm-Car4 (468421-C3), Mm-Gpihbp1 (540631-C3), Mm-Cldn5 (491611-C3), Mm-Pecam1 (316721-C3), tdTomato (317041-C3); Human, Hs-EDN1 (459381), Hs-PTPRB (588141), Hs-CA4 (438561), Hs-EDNRB (528301, 528301-C2), Hs-CLDN5 (517141-C2, 517141-C3), Hs-VWF (560461-C3), Hs-APLN (449971-C3); Alligator, Ami-APLNR (576071), Ami-PTPRB (828711), Ami-EDNRB (576081-C2), Ami-CA4 (828621-C2), Ami-CLDN5 (576091-C3); Western Painted Turtle, Cpi-APLNR (828481), Cpi-EDNRB (828471-C2), Cpi-CLDN5 (828461-C3).
For quantification of capillary cell type abundance, aCap and gCap cells were detected using probes for Ednrb or Ptprb, respectively, and the pan-endothelial probe Cldn5. Cldn5-expressing alveolar cells with 2 or more Ednrb puncta and 0–1 Ptprb puncta were classified ‘aCap’, cells with 2 or more Ptprb puncta and 0–1 Ednrb puncta were classified as ‘gCap’, and cells with 2 or more Ednrb and 2 or more Ptprb puncta were classified as capillary intermediate (IM) cells. 500 Cldn5-expressing alveolar cells were scored per lung in 5–10 random fields of view taken with a Plan-Apochromat 25× objective (Carl Zeiss Microscopy), using Volocity software (Quorum Technologies). For quantification of capillary cell type distribution, capillary cells were scored in the last generation of alveoli immediately adjacent to the pleura and in intra-acinar regions of left and right cranial lobes from 3 month old mice. For quantification of capillary cell type abundance in mouse adenomas, capillary cells were scored in sections from tumors with intra-acinar (rather than pleural) location and round, compact morphologies with clear boundaries between tumor and surrounding alveolar tissue. For quantification of Vwf induction with age, Ptprb+ Cldn5+ alveolar cells were classified as ‘gCap’ and Ptprb- Cldn5+ alveolar cells were classified as ‘aCap’. Cells with 3 or more Vwf puncta were scored as positive. For quantification of aerocyte emergence in the fetal human lung, CA4/APLN/EDNRB triple positive cells with 5 or more puncta for each transcript were classified as ‘emerging aerocytes’. For quantification of capillary fate conversion upon elastase injury, aCap and gCap cells were detected using probes for Ednrb (aCap) and Ptprb or Aplnr (gCap) in injured areas, identified as regions with enlarged airspaces and remodeled elastin fibers, in 3–4 month old Apln-CreER; Rosa26-tdTomato and Aplnr-CreER; Rosa26-tdTomato lungs harvested six weeks after elastase instillation. For quantification of capillary cell type abundance in the human lung, CLDN5+ cells with 2 or more EDNRB puncta and 0–1 PTPRB (or EDN1) puncta were classified as ‘aCap’, CLDN5+ cells with 2 or more PTPRB (or EDN1) puncta and 0–1 EDNRB puncta were classified as ‘gCap’, and CLDN5+ cells with 2 or more EDNRB and 2 or more PTPRB (or EDN1) puncta were classified as ‘IM’.
Histology.
Haematoxylin and eosin (H&E) staining was performed using standard protocols on formalin fixed, paraffin embedded alligator, turtle, and fetal human lung tissue, processed as described above for smFISH. Adjacent sections were used for H&E staining and smFISH.
Electron microscopy and ultrastructural analysis.
To visualize capillaries within alveolar walls, perfused and inflated adult mouse lungs were fixed in 2% glutaraldehyde in PBS for 1 hour at room temperature. Tissue was manually cut with a platinum coated double edge razor blade (EMS) into rough sections. Samples were post-fixed in Karnovsky’s fixative (2% Glutaraldehyde [EMS] and 4% paraformaldehyde [EMS] in 0.1M sodium cacodylate [EMS] pH 7.4) for 1 hour, and incubated in cold aqueous 1% osmium tetroxide (EMS), washed, stained in 1% uranyl acetate for 2 hours, dehydrated into 100% ethanol, infiltrated with Embed 812 resin (EMS) and cured at 65°C overnight. Sections (75–90 nm) were collected on formvar/carbon-coated slot copper grids, observed in the JEM-1400 transmission electron microscope (JEOL) with a 120 kV beam and imaged with an Orius SC1000 (Gatan) digital camera.
For immuno-EM, adult perfused and inflated Apln-CreER; Rosa26-tdTomato and Aplnr-CreER; Rosa26-tdTomato lungs, were fixed in 4% PFA/0.1% glutaraldehyde (EMS) in PBS for 3 hours at 4 degrees. 200 μm sections were cut on a vibratome and then immunostained for tdTomato as described above. 3,3’-diaminobenzidine (DAB)/nickel (Vector Laboratories, SK-4100) was used as a substrate for the peroxidase conjugated to the secondary antibody. Sections were incubated in DAB/nickel working solution (prepared following the manufacturer’s instructions) for 6–15 minutes at room temperature. After washing, samples were processed for electron microscopy as described, omitting uranyl acetate staining. To determine the percent of aerocytes or gCap cells associated with thin or thick regions of the air-blood barrier, capillaries with complete lumens and containing immunolabeled endothelial cells were identified on sections viewed by electron microscopy. Labeled endothelial cells (n=2 mice of each genotype, 21 labeled aCap cells and 24 labeled gCap cells) were scored as being associated either with the thin region (defined as regions where the endothelial cell is tightly apposed to the epithelium) or thick region (defined as regions where the endothelial cell is clearly separated from the epithelium by stromal cells or connective tissue fibers). Samples were observed by electron microscopy at multiple magnifications in order to confirm endothelial cell labeling, AT1 cell identity, and separation between endothelium and epithelium. As a control, we also scored the association of unlabeled capillary cells. Importantly, we found that in capillaries containing labeled gCap cells, all endothelial cells associated with thin regions (n=12 scored) were unlabeled, consistent with the conclusion that only aCap cells are associated with thin regions. Some sections were scored by an investigator blinded to the genotype of the sample, and similar results were obtained. Representative electron micrographs (Fig. 2g-i) were pseudocolored in Adobe Illustrator (Extended Data Fig. 4a).
Pulse/chase labeling experiments.
To determine the stability of the two alveolar capillary cell populations in the adult mouse lung, aCap cells were labeled using the Apln-CreER knock-in allele combined with the Rosa26-tdTomato Cre reporter and gCap cells were labeled with the Aplnr-CreER BAC transgenic allele combined with the Rosa26-tdTomato Cre reporter. Co-expression of tdTomato and a marker for the respective capillary populations (Apln or Ednrb for aCap, Aplnr or Ptprb for gCap) was detected after a 1, 6, or 14 month chase by smFISH. 500–2000 lineage labeled cells were scored per lung in at least 3 random fields of view taken with a Plan-Apochromat 25x oil objective (Carl Zeiss Microscopy), using Volocity software (Quorum Technologies).
The fidelity of Apln-CreER was confirmed by dosing an Apln-CreER; Rosa26-tdTomato mouse with 4 mg tamoxifen followed by a 48 hour chase. Tamoxifen-dependent Cre recombination was observed only in Ednrb-expressing cells (n=604 scored cells; 598 cells co-expressed tdTomato and Ednrb, but not Aplnr; 6 cells co-expressed tdTomato, Ednrb, and Aplnr. Because the Apln-CreER knock-in allele is a loss of function allele, we used Ednrb rather than Apln as an aCap marker).
The fidelity of Aplnr-CreER was established by dosing an Aplnr-CreER; Rosa26-tdTomato mouse with 4 mg tamoxifen followed by a 48 hour chase. Tamoxifen-dependent Cre recombination was observed only in Aplnr-expressing cells (n=1879 scored cells; 1860 cells co-expressed tdTomato and Aplnr, but not Apln; 19 cells co-expressed tdTomato, Aplnr, and Apln).
Sparse labeling of endothelial cells and analysis of cell morphology.
To visualize single endothelial cells in the lung and other organs, mice carrying inducible CreER alleles were administered limiting doses of tamoxifen (see ‘Mice‘ for details). Organs were harvested as described, fixed in 2% PFA/PBS for 5 hours at 4°C, and cut into 200–250 μm sections on a vibratome. To preserve endogenous fluorescence for imaging, tissue was not dehydrated into methanol. For some experiments, lung sections were stained with Alexa Fluor 350 hydrazide (Invitrogen, A10439; 1:100) or Alexa Fluor 633 hydrazide (Invitrogen, A30634; 1:1000–1:5000) to visualize elastin fibers30. To visualize the vasculature, tamoxifen-dosed Cdh5-CreER; Rosa26-Confetti mice were injected with 0.2 ml DyLight 649 labeled Lycopersicon Esculentum (Tomato) Lectin (Vector Laboratories, DL-1178; 1 mg/ml) and humanely sacrificed after 5 minutes. Sections were cleared and mounted in CUBIC131 for confocal imaging. Sparse labeling, with individual fluorescent cells well separated from other cells, was verified. Volume and surface area of individual plexus (n=18 cells at E12.5 from n=2 mice), aCap (n=23 cells at P0; n=14 (volume and surface area); n=16 (pores) at P7; n=19 cells at 4 months from n=2 mice), and gCap cells (n=12 cells at P0; n=11 cells at P7; n=17 cells at 4 months from n=2 mice) were automatically calculated from computed 3D surfaces using Imaris software (Bitplane). Pore number was determined by counting using the original confocal stack viewed in 3D in Imaris.
Clonal analysis.
Lungs from P25 Aplnr-CreER; Rosa26-Confetti mice (n=2) whose dams were administered limiting doses of tamoxifen (0.06 mg administered by i.p. injection) at E14.5 were harvested as described in Methods above, fixed in 4% PFA/PBS for 2 hours at 4°C, and cut into 300 μm sections on a vibratome. Sections were stained with Alexa Fluor 633 hydrazide to visualize elastin fibers as above. To preserve endogenous fluorescence for imaging, sections were cleared and mounted in CUBIC131. The tamoxifen dose was chosen to yield small numbers of well-separated clones for each fluorescent reporter, such that some sections did not contain any clones. Only YFP- and RFP-expressing clones were analyzed, because cell number and type could not be identified in nuclear GFP- and membrane targeted Cerulean CFP-expressing clones.
Following confocal imaging, clones were visualized in 3D in Imaris. Cell type composition was scored on the basis of cell morphology. For some clones, cell type identity could not be definitively assigned to all cells in the clone, especially where cell boundaries could not be determined. In these cases, the number of cells of a given type that could be identified is given as a lower limit in Extended Data Fig. 6e. (For example, in clone 28, which contains 40 cells, there are at least 4 aCap and 30 gCap cells.)
Detection of capillary cell proliferation by EdU labeling.
The synthetic deoxyribonucleoside analog 5-Ethynyl-2’-Deoxyuridine (EdU, Carbosynth, NE08701) was administered to adult Apln-CreER; Rosa26-tdTomato and Aplnr-CreER; Rosa26-tdTomato mice in drinking water at 0.2 mg/ml 3 weeks after tamoxifen injection (two doses of 4 mg administered by i.p. injection 48 hours apart) for 6 weeks. Lungs were harvested as described, fixed in 4% PFA/PBS for 5 hours at 4°C, and cut into 300 μm sections on a vibratome. EdU was detected using click chemistry to covalently attach Alexa Fluor 647 azide to EdU alkyne incorporated into DNA during S phase of the cell cycle (Invitrogen, C10340; Click-iT EdU Alexa Fluor 647 Imaging Kit), by incubating vibratome sections in Click-iT reaction cocktail for 4 hours at room temperature.
Alveolar injury with elastase.
Elastase solution (0.8 μg/μl) was prepared by dissolving elastase (Worthington, LS002292) in PBS and stored at −20°C. A single dose of elastase (40 μg) or saline was delivered by intratracheal instillation into the lungs of avertin (1.2% in saline) anesthetized adult Apln-CreER; Rosa26-tdTomato and Aplnr-CreER; Rosa26-tdTomato mice 3 weeks after tamoxifen injection (two doses of 4 mg administered by i.p. injection 48 hours apart) to allow tamoxifen clearance. EdU was administered as described above starting the night before elastase injury for 3 days, 1 week, or 6 weeks. Lungs were harvested as described, fixed in 4% PFA/PBS for 5 hours at 4°C, and cut into 300 μm sections on a vibratome. EdU was detected using click chemistry as described above. Sections were incubated in Alexa Fluor 488 hydrazide (Invitrogen, A10436; reconstituted to 0.5 mg/ml in PBS, used at 1:100) and DAPI (2 μg/ml) in 0.1% Triton-X-100/PBS overnight at 4°C. After washing, sections were cleared and imaged in CUBIC131. EdU incorporation was analyzed in lineage labeled cells in Apln-CreER; Rosa26-tdTomato lungs (3 days, control: n=2 mice, 296 and 297 cells counted; 1 week, control: n=2 mice, 404 and 295 cells counted; 6 weeks, control: n=2 mice, 424 and 3977 cells counted; 3 days, elastase: n=2 mice, 767 and 1018 cells counted; 1 week, elastase: n=4 mice, 431, 877, 253, and 223 cells counted; 6 weeks, elastase: n=3 mice, 1252, 420, and 1054 cells counted) and in Aplnr-CreER; Rosa26-tdTomato lungs (3 days, control: n=2 mice, 1416 and 1569 cells counted; 1 week, control: n=2 mice, 1286 and 1129 cells counted; 6 weeks, control: n=2 mice, 1172 and 3390 cells counted; 3 days, elastase: n=3 mice, 1286, 810, and 1267 cells counted; 1 week, elastase: n=3 mice, 844, 623, and 707 cells counted; 6 weeks, elastase: n=2 mice, 967 and 1246 cells counted).
Acquisition and processing of images.
The image of the immunostained embryonic lung in Fig. 4a was captured using a Zeiss Axioskop microscope and the MRC-1000 Laser Scanning Confocal Imaging System (Bio-Rad) and processed in ImageJ (NIH). All other fluorescent samples were imaged on an LSM 780 (Carl Zeiss Microscopy) or LSM 880 equipped with Airyscan (Carl Zeiss Microscopy) confocal microscope, and images were processed in Zen (Carl Zeiss Microscopy) or Imaris (Bitplane) software. 2D confocal images presented are maximum intensity projections of z-stacks. H&E stains were imaged on a slide scanning system (Philips), and images were processed using QuPath software32.
Analysis of single-cell RNA-sequencing (scRNAseq) data.
Processed scRNAseq Smart-Seq2 data for adult (3 month old) mouse lung were obtained from the Tabula Muris resource (https://tabula-muris.ds.czbiohub.org)13 as gene count tables with de-multiplexed and aligned reads. Cells with fewer than 500 detected genes or 50,000 reads were excluded. Data were log-transformed: ln(counts per million+1) [ln(CPM+1)]. Expression profiles of cells were clustered using the R software package Seurat33 (version 2.3). Highly variable genes were selected using the ‘FindVariableGenes’ function (dispersion (mean/variance) z-score > 0.5) for linear dimensionality reduction using principal component analysis. The number of principal components was selected by inspection of the plot of variance explained. Cells were clustered by constructing a shared nearest neighbor graph and clusters were visualized by t-distributed stochastic neighbor embedding (tSNE). Lung endothelial cells (n=693 cells, Fig. 1b, c, Fig. 3, Extended Data Fig. 1a, b, Extended Data Fig. 3h, Extended Data Fig. 4d, e, Extended Data Fig. 10a) were identified by Cldn5, Pecam1, and Cdh5 expression and subclustered. Artery (n=76 cells), vein (n=54 cells), and lymphatics (n=68 cells) clusters were annotated using canonical markers (Gja5 and Bmx for arteries; Nr2f2 for veins; Vwf for arteries and veins; Pdpn and Prox1 for lymphatics). For annotation of the remaining clusters (aCap, n=101 cells; gCap, n=394 cells), cluster markers were identified by differential expression analysis using the Wilcoxon rank sum test with Bonferroni correction (p < 0.01) as implemented in the ‘FindMarkers’ function in Seurat, and specific cluster markers were selected for localization of the cells by smFISH (Apln or Ednrb for aCap, Aplnr or Ptprb for gCap). The apparent separation of the cluster we annotated as ‘gCap’ into two populations (see tSNE plot in Fig. 1b) is likely due to batch effects, based on the correlation of gene expression differences with technical differences, and was not analyzed further.
Processed scRNAseq MARS-Seq data for embryonic (E12.5-E19.5) and postnatal (P0, P1, P7, and 2 month old) mouse lung by Cohen et al.34 were obtained from GEO (accession number GSE119228). Cells with fewer than 500 unique molecular identifiers were discarded. Cells from adult lung processed differently than tissue from embryonic and postnatal time points were also eliminated. Data were log-transformed: ln(unique molecular identifiers per ten thousand+1) [ln(UP10K+1)] and analyzed with Seurat as described above. Cells were initially clustered separately at each developmental stage and endothelial cells were identified by Cldn5, Pecam1, and Cdh5 expression. Annotated endothelial cells were combined from all time points (n=4378 cells) and clustered again. Contaminant hematopoietic cells were eliminated by filtering out Ptprc-expressing cells (log-transformed expression levels > 0.5). Artery, vein, and lymphatics clusters were identified using canonical markers (Gja5 and Bmx for arteries; Nr2f2 for veins; Vwf for arteries and veins; Pdpn and Prox1 for lymphatics) following correction for cell cycle as described by Su et al.35 and eliminated from the analysis, resulting in a dataset of 3094 plexus and capillary cells (64 cells at E12.5, 404 cells at E16.5, 296 cells at E18.5, 117 cells at E19.5, 1016 cells at P0, 426 cells at P1, 438 cells at P7, and 333 cells at 2 months (adult); see Extended Data Fig. 6h-j, Extended Data Fig. 7, Supplementary Data 2, 3, and 4).
Single cell trajectories were constructed for plexus and capillary cells (n=3094) using Monocle236. Mature aCap and gCap markers, identified by differential gene expression analysis in Seurat, were used for ordering of the cells, which produced a tree-shaped branched developmental trajectory (Extended Data Fig. 6h), with plexus cells located along the stem before the branchpoint, mature aCap cells at the tip of one branch, and mature gCap cells at the tip of the second branch. Genes with branch dependent expression were identified by branched expression analysis modeling (BEAM) (n=1119 genes; q-value < 0.05), as implemented in the ‘BEAM’ function in Monocle236. Genes that vary as a function of pseudotime were identified by differential expression analysis (n=3734 genes; q-value < 0.05), as implemented in the ‘differentialGeneTest’ function in Monocle2. The genes identified by the two approaches (n=4129) were clustered hierarchically and plotted as a branched heatmap (Extended Data Fig. 7b, Supplementary Data 4) to visualize groups of genes that co-vary across pseudotime.
Processed scRNAseq Smart-Seq2 data for adult (3 month old) mouse heart and brain were obtained from the Tabula Muris resource13 as Seurat objects with annotated clusters. Brain and heart endothelial cells were identified by Cldn5, Pecam1, Esam, and Cdh5 expression and subclustered. Artery, vein, and lymphatics clusters were identified using canonical markers (Gja5, Bmx, and Vegfc for arteries; Vwf for arteries and veins; Il1r1 for veins; Pdpn and Prox1 for lymphatics)37 and excluded from the analysis. The remaining cells were annotated as capillaries based on expression of Car438,39.
Processed scRNAseq droplet (10X) data for adult (1, 3, 18, 21, and 30 month old) mouse lung, kidney, and mammary gland were obtained from the Tabula Muris Senis resource (https://tabula-muris-senis.ds.czbiohub.org)40. Scanpy single-cell objects were imported in R and analyzed with Seurat as described above. Cells with fewer than 500 detected genes or 1,000 unique molecular identifiers were discarded. Kidney, mammary gland, and lung endothelial cells were identified by Pecam1, Cdh5, and Esam (for kidney and mammary gland) or Cldn5 (for lung) expression and subclustered. Contaminant hematopoietic cells were eliminated by filtering out Ptprc-expressing cells (log-transformed expression levels > 0.5). Contaminant stromal and epithelial cells were eliminated in the mammary gland dataset by filtering out cells that express Col1a1, Pdgfrb, or Epcam. Artery, vein, and lymphatics clusters were identified using canonical markers as above and excluded. Lung capillary cells were annotated using aerocyte (Apln, Car4, Ednrb, Tbx2) and gCap markers (Aplnr, Gpihbp1, Lpl). Clusters with Ehd3, Sost, and Meg3 expression were annotated as glomerular capillaries in the kidney dataset. Clusters with expression of Gpihbp1, Rbp7, and Car4, and little/low expression of Vwf, were annotated as capillaries in the mammary gland dataset.
Alveolar aCap and gCap signature scores (defined as the sum of normalized and scaled gene expression values for the significant alveolar aCap or gCap marker genes: Bonferroni corrected p-value < 0.01; average normalized expression fold change > 1; % expression > 40) were calculated for annotated lung (n=495), heart (n=753) and brain (n=441) capillary endothelial cells in the Tabula Muris Smart-Seq2 data13 (Extended Data Fig. 3h), and for annotated lung (n=2050), glomerular (n=126), and mammary gland (n=173) capillary endothelial cells in the Tabula Muris Senis droplet data40 (Extended Data Fig. 3i).
Smart-Seq2 and droplet (10X) data for adult human lung (patient 1, 75 year old male)16 were analyzed as described above. Endothelial cells were identified by CLDN5 expression, subclustered, and annotated using markers (GJA5 for arteries, ACKR1 for vein, PDPN for lymphatics, EDNRB for aerocytes, EDN1 for gCap cells, COL15A1 for bronchial vessels; droplet: n=1497 cells; 211 artery cells, 154 vein cells, 33 lymphatic cells, 230 bronchial endothelial cells, 315 aerocytes, 554 gCap cells; Fig. 4k, m-o, Extended Data Fig. 8b, c, Extended Data Fig. 10; Smart-Seq2, n=599 cells, Extended Data Fig. 10).
To identify differences in cell type-specific expression between mouse and human, we first identified genes that are differentially expressed (p < 0.01, Wilcoxon rank sum test with Bonferroni correction) between the alveolar capillary cell types in each species using scRNAseq Smart-Seq2 data for adult mouse lung endothelial cells obtained from the Tabula Muris resource13 and Smart-Seq2 data for adult human lung endothelial cells (patient 1, 75 year old male)16. Lists of differentially expressed genes (see Supplementary Tables 2 and 3) were then compared in order to identify genes specifically expressed in the same cell type in the two species (Type 0), genes specifically expressed in one cell type in one species but not the other (Type 1), or genes specifically expressed in different cell types in the two species (Type 2). Selected genes of each type are shown in Fig. 4m-o and Extended Data Fig. 10c-e. A complete analysis of the evolutionary changes between alveolar capillary cell types in mouse vs. human lungs is presented in the accompanying manuscript (see Supplementary Table 7 in ref. 16).
Statistics and reproducibility.
Data analysis and statistical tests were performed using R software (version 3.5.1). Data are represented as mean ± standard deviation (s.d.) for sample sizes larger than two. For comparison of two groups, a two-sided Wilcoxon rank sum test was conducted at 5% significance level. For Fig. 1a, the image is representative of n=10 mice; for Fig. 1d, d’, smFISH was performed on samples from n=5 mice; for Fig. 1e, smFISH was performed on samples from n=2 mice; for Fig. 1f, smFISH was performed on samples from n=5 mice; for Fig. 1h, smFISH was performed on samples from two lobes of n=1 mouse; for Fig. 1i, smFISH was performed on samples from two lobes of n=1 mouse; for Fig. 2a, the image is representative of n=5 mice; for Fig. 2b, the image is representative of n=5 mice; for Fig. 2d, the image is representative of n=2 mice; for Fig. 2f-i, each image is representative of n=2 mice; for Fig. 2l, images are representative of n=3 mice; for Fig. 2m, images are representative of n=2 mice; for Fig. 2o, o’, smFISH was performed on samples from n=2 mice; for Fig. 4a, the image is representative of n=10 embryos; for Fig. 4b, the image is representative of n=2 embryos; for Fig. 4c, images are representative of n=2 mice; for Fig. 4d, smFISH was performed on samples from n=2 mice; for Fig. 4e, clones were examined in n=2 mice, see clone table in Extended Data Fig. 6e; for Fig. 4g, images are representative of n=2 mice at each timepoint; for Fig. 4j, smFISH was performed on samples from n=3 humans; for Fig. 4l, smFISH was performed on samples from a single adenocarcinoma; for Fig. 4q, the image is representative of multiple lung regions from a single alligator; for Fig. 4r, r’, smFISH was performed on samples from n=2 alligators (1 juvenile; 1 adult); for Extended Data Fig. 1c, smFISH was performed on samples from n=5 mice; for Extended Data Fig. 1d, smFISH was performed on samples from n=2 mice; for Extended Data Fig. 1e, e’, smFISH was performed on samples from n=2 mice; for Extended Data Fig. 1f, g, smFISH was performed on samples from n=1 mouse; for Extended Data Fig. 1j, smFISH was performed on samples from n=2 mice at each age; for Extended Data Fig. 1k, l, smFISH was performed on samples from two lobes of n=1 mouse of each genotype; for Extended Data Fig. 2a-d; images are representative of n=2 embryos or mouse pups at each timepoint; for Extended Data Fig. 2e, f, images are representative of n=5 mice; for Extended Data Fig. 3a, the image is representative of n=2 mice; for Extended Data Fig. 3b-f, images are representative of n=4 mice; for Extended Data Fig. 3g, the image is representative of n=2 mice; for Extended Data Fig. 3j-l, smFISH was performed on samples from n=2 mice; for Extended Data Fig. 4a-c, images are representative of n=2 mice of each genotype; for Extended Data Fig. 5a, the images are representative of n=2 mice at each timepoint; for Extended Data Fig. 5c, the images are representative of n=2 mice of each genotype; for Extended Data Fig. 5d, the images are representative of n=2 Apln-CreER; Rosa26-tdTomato mice and n=3 Aplnr-CreER; Rosa26-tdTomato mice; for Extended Data Fig. 5e, the images are representative of n=4 Apln-CreER; Rosa26-tdTomato mice and n=3 Aplnr-CreER; Rosa26-tdTomato mice; for Extended Data Fig. 5f, the images are representative of n=3 Apln-CreER; Rosa26-tdTomato mice and n=2 Aplnr-CreER; Rosa26-tdTomato mice; for Extended Data Fig. 6a, b, smFISH was performed on samples from n=2 embryos; for Extended Data Fig. 6c, smFISH was performed on samples from two lobes of n=1 mouse; for Extended Data Fig. 6d, e, clones were examined in n=2 mice; for Extended Data Fig. 6f, f’, f’’, smFISH was performed on samples from two lobes of n=1 mouse; for Extended Data Fig. 6g, smFISH was performed on samples from n=3 embryos; for Extended Data Fig. 6k, l, smFISH was performed on samples from n=3 mice at each age; for Extended Data Fig. 8a, the images are representative of n=5 human lungs; for Extended Data Fig. 8d-h, smFISH was performed on samples from n=3 humans; for Extended Data Fig. 8j, j’, histology and smFISH were performed on samples from a single human fetal lung from each time point (17 weeks and 23 weeks; see figure legend); for Extended Data Fig. 9a, a’, a’’, immunostaining and smFISH were performed on samples from a single adenocarcinoma; for Extended Data Fig. 9b, b’, c, c’, f, smFISH was performed on samples from n=2 mice; for Extended Data Fig. 11c, d, images are representative of multiple regions from a single alligator lung; for Extended Data Fig. 11e, f, smFISH was performed on samples from n=2 alligators (1 juvenile, 1 adult); for Extended Data Fig. 11g, the image is representative of multiple regions from a single alligator lung; for Extended Data Fig. 11i, the image is representative of multiple regions from a single turtle lung; for Extended Data Fig. 11j, k, images are representative of n=2 turtles; for Extended Data Fig. 11l, l’, smFISH was performed on samples from n=2 turtles; for Extended Data Fig. 11m, the image is representative of n=2 turtles; for Supplementary Data 1, images are representative of n=2 mice. For all graphs, the number of biologically independent samples is reported in the Figure Legends, or in the ‘Alveolar injury with elastase’ (for Fig. 2n) or ‘Sparse labeling of endothelial cells and analysis of cell morphology’ (for Fig. 2c, 4h, and Extended Data Fig. 2g, h) sections of the Methods. Sample size calculations were not performed. Mice of the appropriate genotype or age were allocated into experimental groups (control vs elastase injury) at random. The investigators were not blinded to sample allocation.
Data availability.
The scRNAseq datasets analyzed are available in the GEO repository under the accession numbers GSE109774 (Tabula Muris13, https://tabula-muris.ds.czbiohub.org), GSE132042 (Tabula Muris Senis40, https://tabula-muris-senis.ds.czbiohub.org), and GSE119228 (Cohen et al.34) or on Synapse under the accession number syn21041850 (Human Lung Cell Atlas16, https://hlca.ds.czbiohub.org). Source data used in all graphs are provided in the Source Data files.
Extended Data
Extended Data Figure 1. Molecular characterization and mapping of the two alveolar capillary cell types.
a, Feature plots showing log-transformed expression of marker genes used to annotate five molecularly distinct clusters (see tSNE plot in Fig. 1b) derived by unsupervised clustering of lung endothelial cells in the Tabula Muris scRNAseq Smart-seq2 data13 for adult mouse lung. We identified two endothelial cell clusters as capillaries based on expression of carbonic anhydrases (Car4, Car14) that catalyze the conversion of bicarbonate to carbon dioxide41, Gpihbp1, a lipoprotein binding protein that localizes to the luminal membrane of alveolar capillaries42, and lipoprotein lipase (Lpl), which is transported to the capillary lumen by Gpihbp143. b, Heatmap showing log-transformed expression of selected cluster markers in individual pulmonary artery, vein, lymphatics, and alveolar capillary (aCap, gCap) endothelial cells identified in the Tabula Muris Smart-Seq2 data13. c, Co-expression of alveolar capillary cell type markers Apln (aCap) and Aplnr (gCap) in a single alveolar capillary intermediate (IM) endothelial cell (arrow; marked by Cldn5) in adult mouse lung, as detected by smFISH. d, Co-expression of aCap markers (Apln and Ednrb) in a subset (aCap cells) of alveolar endothelial cells (marked by Cldn5) in adult mouse lung. e, Co-expression of aCap markers Ednrb and Car4 but not Aplnr, a gCap marker, in a subset (aCap cells) of adult mouse lung alveolar endothelial cells. e’, Boxed region from e at higher magnification. Filled arrowhead indicates aCap cell expressing high levels of Car4. Open arrowhead points to gCap cell expressing low levels of Car4. f, g, Co-expression of gCap markers Gpihbp1 (f) or H2-Ab1 (g), a major histocompatibility complex (MHC) class II gene, and Aplnr in a subset (gCap cells) of adult mouse lung alveolar endothelial cells. h, i, Quantification of the relative abundance (in % of capillary endothelial cells) of the two alveolar capillary populations (aCap, gCap) and rare cells (IM) that co-express aCap and gCap markers at the pleura and in intra-acinar regions (h), or in different lobes (i, left vs. right cranial) in lungs from 3-month old mice. Data shown as mean ± s.d.; n=500 cells scored per mouse; n=3 mice; p-values comparing cell type abundance by two-sided Wilcoxon rank sum test: aCap (h, 0.2; i, 0.2), gCap (h, 0.4; i, 0.2), IM (h, 0.7; i, 0.8). n.s., not significant. j, aCap (marked by Ednrb) and gCap (marked by Ptprb) cells in lungs from 3- and 24-month old mice. Endothelial cells are marked by Cldn5. k, l, Co-expression of tdTomato lineage label (asterisks) and either aCap marker Ednrb but not gCap marker Aplnr in an Apln-CreER; Rosa26-tdTomato lung (k) or gCap marker Aplnr but not aCap marker Apln in an Aplnr-CreER; Rosa26-tdTomato lung (l) harvested one month after mature aCap (k) or gCap (l) cells were lineage labeled. Lineage labeled cells (asterisks) continue to express the marker (Ednrb, aCap; Aplnr, gCap) of the labeled population. m, n, Quantification of percent of lineage labeled cells in Apln-CreER; Rosa26-tdTomato (m) or Aplnr-CreER; Rosa26-tdTomato (n) lungs that continue to express the aCap marker, gCap marker, or markers of both cell types (IM) after 48 hours, 1, 6, or 14 months (500–2000 cells scored at each time point from multiple regions in each of two separate lobes from one lung). Blue (c-g, j-l), DAPI. Scale bars, 10 μm.
Extended Data Figure 2. Alveolar capillary cell morphologies.
a-d, Additional examples of single cytoplasmic YFP- or RFP-expressing plexus cells at E12.5 (a), aerocytes at E18.5 (b), gCap cells at P0 (c), and aerocytes at P0 (d). Plexus and gCap cells were labeled in Aplnr-CreER; Rosa26-Confetti lungs; aerocytes were labeled in Apln-CreER; Rosa26-Confetti lungs. Pores, present in some aerocytes by E18.5, but not plexus or P0 gCap cells, are marked by dots. e, f, Additional examples of single gCap cells (e) or aerocytes (f) expressing cytoplasmic RFP or YFP in adult Aplnr-CreER; Rosa26-Confetti (e) or Apln-CreER; Rosa26-Confetti (f) lungs. Dotted circles outline two alveoli spanned by a single aerocyte (f). Note that gCap cells are less morphologically diverse than aerocytes. g, h, Quantification of individual plexus, aerocyte (aCap), or gCap cell surface area (g) and pore number (h) at indicated times. n>10 cells scored for each cell type at each time point from n=2 lungs; see Methods for exact cell number. Blue, elastin fibers highlighting the alveolar entrance. Grey bar, mean value. Scale bars, 10 μm.
Extended Data Figure 3. Capillary cells in the lung and other organs.
a, Bronchial (systemic) capillary cell immunostained for tdTomato (red) in adult Aplnr-CreER; Rosa26-tdTomato lung. Airway epithelium is immunostained for E-cadherin (blue). b-f, Single capillary cells labeled with membrane targeted CFP (b) or cytoplasmic YFP (c-f) in brain (b), heart (c), small intestine (d), kidney glomerulus (e), or thyroid (f), where capillaries are arranged in baskets similar to pulmonary alveoli, from an adult Cdh5-CreER; Rosa26-Confetti mouse. Capillaries labeled with tomato lectin (blue, b-f). e’, Cell in boxed region in e at higher magnification, along with other examples of labeled glomerular endothelial cells. g, Single endothelial tip cell expressing cytoplasmic YFP in developing (P7) Apln-CreER; Rosa26-Confetti mouse retina. Filled arrowheads, filopodia. h, i, Scatter plots of alveolar aCap and gCap signature scores assigned to annotated lung, heart, and brain capillary endothelial cells identified in the Tabula Muris scRNAseq Smart-seq2 data13 (h) or annotated lung, kidney glomerular, and mammary gland capillary endothelial cells identified in the Tabula Muris Senis droplet data for 1, 3, 18, 21, and 30 month old mice40 (i). Lung capillary cells segregate into two clusters, with annotated gCap cells having high gCap and low aCap signature scores, and annotated aCap cells having low gCap and high aCap signature scores. Heart (h), brain (h), glomerular (i), and mammary gland (i) capillary cells have high gCap and low aCap signature scores and each form a single cluster near gCap but not aCap cells, suggesting that they are more similar to gCap cells and any heterogeneity within these other capillary cell populations is different from that in the lung. j-l, Some aCap and gCap markers (aCap, Ednrb (j, k); gCap, Ptprb, j, or Gpihbp1, k) are broadly co-expressed in glomerular endothelial cells (Pecam1) in adult mouse kidney whereas expression of others (l: Apln, aCap or Aplnr, gCap) is detected only in small numbers of cells or not at all. Dotted lines outline glomeruli. j’-l’, Boxed regions from j-l at higher magnification. Arrows (j’, k’) point to cells co-expressing the markers. Blue (j-l), DAPI. Scale bars, 10 μm.
Extended Data Figure 4. Functional compartmentalization of the alveolus.
a, Transmission electron micrographs of alveolar walls from adult mouse lungs, pseudocolored to highlight cells and connective tissue (extracellular matrix (ECM) or fibers). Apln-CreER; Rosa26-tdTomato (to label aerocytes) or Aplnr-CreER; Rosa26-tdTomato (to label gCap cells) lungs were immunostained for tdTomato (detected with DAB and NiCl2, heavy black stain). Labeled aerocytes but not gCap cells are associated with thin regions of the air-blood barrier (delimited by dashed lines), where the epithelium is tightly apposed to the endothelium. Some thin regions do not contain labeled aerocytes since aerocyte labeling using Apelin-CreER is inefficient. Thick regions, where the epithelium is separated from the endothelium by fibers or other cells, can be associated with either aerocytes or gCap cells. AT1 cell, alveolar type 1 epithelial cell; AT2 cell, alveolar type 2 epithelial cell. Micrographs in top left, middle, and bottom panels are shown without pseudocoloring in Fig. 2g-i. b, c, gCap cells associate with fibroblasts. Alveoli with labeled aerocytes (aCap) in Apln-CreER; Rosa26-tdTomato (b) or gCap cells in Aplnr-CreER; Rosa26-tdTomato (c) lungs immunostained for integrin α8 to show alveolar fibroblasts16. Asterisks mark aerocytes that do not associate with fibroblasts (in b) and gCap cells that do (in c). Example regions of alveoli not covered by fibroblasts are dotted. Note that the dotted region in b is occupied by an aerocyte not overlaid by fibroblasts, whereas fibroblasts overlay gCap cells in c. d, Selected genes with known functions, which are differentially expressed between the two alveolar capillary cell types in adult mouse lung13. Pro, procoagulants; anti, anticoagulants44,45. e, Summary of signaling interactions between capillary cell types and surrounding cells in the mouse alveolus. Arrows indicate direction of signaling. f, Schematic representation of an alveolus highlighting proposed specialized functions and signaling interactions of alveolar capillary cell types. AT1, alveolar type 1 epithelial cell; AT2, alveolar type 2 epithelial cell. Scale bars, 2 μm (a), 10 μm (b, c).
Extended Data Figure 5. Proliferation of gCap cells following elastase injury.
a, Elastin fibers (black), labeled with fluorescent hydrazide, in lungs of wild-type mice treated with elastase or mock-treated with saline as control. Loss of elastin fibers in injured areas (dashed outlines) is apparent one day after intratracheal instillation of elastase. At one month after injury, regions containing abnormally thickened elastin fibers and enlarged airspaces are evident. Boxed areas are shown at higher magnification. b, Scheme for detecting proliferation of alveolar capillary cells following elastase injury. aCap and gCap cells were lineage labeled in adult Apln-CreER; Rosa26-tdTomato (for aCap cells) or Aplnr-CreER; Rosa26-tdTomato (for gCap cells) mice 3 weeks prior to elastase administration, then EdU was administered in drinking water and capillary cells were analyzed at the indicated times after injury. c-f, Proliferation analyzed by cumulative EdU incorporation in lineage labeled (red) aCap cells (top panels in c-f) and gCap cells (bottom panels in c-f) at 3 days (c, d), 1 week (e), or 6 weeks (f) after instillation of saline (as control; c) or elastase (d-f). Boxed areas are shown at higher magnification, with cell bodies of individual capillary cells outlined. Following treatment with elastase, gCap cells proliferated in injured areas. White, elastin fibers. Scale bars (c-f), 10 μm.
Extended Data Figure 6. Development, maturation, and aging of specialized alveolar capillary cell types.
a, Expression of Aplnr throughout plexus (labeled by Pecam1) surrounding a developing airway at E12.5, detected by smFISH. b, Expression of mature aerocyte markers Apln and Ednrb in plexus (labeled by Pecam1) surrounding a developing airway at E12.5. Plexus cells expressing Apln but not Ednrb are marked by asterisks. Ednrb is expressed at high levels in Pecam1-negative (stromal) cells, but rarely and only at low levels in Pecam1-positive endothelial cells (arrow) at this stage. c, The subpopulation of Apln-expressing cells within the Aplnr+ plexus gives rise to both capillary cell types, as shown by expression of tdTomato transcripts in both aerocyte (filled arrowheads, marked by Ednrb) and gCap (open arrowheads, marked by Aplnr) cells of P21 Apln-CreER; Rosa26-tdTomato lung lineage labeled at E12.5, demonstrating that even plexus cells that express an aerocyte marker are uncommitted. c’, Lineage labeled aerocyte (filled arrowhead) and gCap (open arrowheads) cells shown at higher magnification. d, Alveolar capillary clone in a P25 Aplnr-CreER; Rosa26-Confetti lung composed of both aCap and gCap cells (higher magnification at right) derived from a single YFP-expressing plexus cell labeled at E14.5. e, Composition of Aplnr-CreER; Rosa26-Confetti clones induced at E14.5 and analyzed at P25, ordered by clone size. All analyzed clones, with the exception of clone number 1, contained both aCap and gCap cells. Some clones also contained cells located in larger vessels (LV). Both coherent clones (in which all cells are touching) and dispersed clones were observed, suggesting that there can be cell movement during capillary development. The number of cells of each type was scored from 3D renderings of confocal z-stacks, as described in the Methods. Clone 4 is shown in Fig. 4e; clone 28 is shown in Extended Data Fig. 6d and Supplementary Video 4. RCr, right cranial lobe; RAc, right accessory lobe, RCd, right caudal lobe; L, left lobe. f, The Aplnr+ population remains uncommitted even after birth. The population labeled at P7 in an Aplnr-CreER; Rosa26-tdTomato lung gives rise to both aCap and gCap cells, as shown by co-expression of tdTomato lineage label and either aCap marker Apln or gCap marker Aplnr at P21, detected by smFISH. f’, f’’, Lineage labeled aerocyte and gCap cells shown at higher magnification. g, Expression of aerocyte markers Car4, Ednrb, and Apln in developing mouse lung at indicated stages. Aerocytes (asterisks), identified by marker co-expression, begin to emerge at E17.5. Insets, cells in boxed regions at higher magnification. h, Plexus and capillary cells identified in scRNAseq data for developing mouse lung34 arranged as tree-shaped branched developmental trajectory inferred using Monocle236. Cells collected at the indicated stages are colored to show plexus located along the stem before the branchpoint, maturing and mature aerocytes along the bottom branch, and maturing and mature gCap cells along the top branch. Note that cells from a single late embryonic or early postnatal time point are found at multiple positions along the trajectory. This analysis is consistent with that presented in ref. 15. Each cell type undergoes distinct and asynchronous molecular and morphological maturation (see Extended Data Fig. 2), suggesting that the two capillary cell specialization programs are likely under separate genetic control. i, Heatmap showing log-transformed expression levels in individual plexus and capillary cells for selected genes differentially expressed during aerocyte emergence and maturation. Many aerocyte markers are expressed in emerging aerocytes; others are only expressed in mature aerocytes. j, Developmental trajectory plot (top) and feature plots showing log-transformed expression for selected gCap markers expressed only in mature gCap cells (upper branch tip). k, l, Expression of Vwf in the gas exchange region at 3 (k) and 24 months (l), as detected by smFISH. k’, l’, Boxed regions from k and l shown at higher magnification. Endothelial cells are marked by Cldn5 expression. At 24 months, Vwf is induced in gCap cells (marked by Ptprb, open arrowheads), but not aerocytes (filled arrowhead). m, Schematic representation of Vwf induction in gCap cells with aging. At 24 months, Vwf is more broadly expressed in alveolar capillaries than at 3 months but only in gCap cells (marked by Ptprb). Blue, DAPI (a-c, f, g, k’, l’). Scale bars, 10 μm.
Extended Data Figure 7. Emergence and maturation of the alveolar capillary cell types in developing mouse lung.
a, tSNE plots with plexus and capillary endothelial cells from embryonic, postnatal, and adult stages (n=3094 cells) identified in scRNAseq MARS-Seq data for developing mouse lung34. Cells from each indicated stage are colored black, except emerging/maturing aerocytes (aCap), first evident at E18.5 in this dataset, light blue; mature aerocytes, dark blue; mature gCap cells, dark green. See also feature plots in Supplementary Data 3. b, Branched heatmap showing gene expression changes during the transformation of plexus into mature alveolar capillary cell types. Normalized expression values (z-scores) are plotted for genes with branch-dependent expression, identified by branched expression analysis modeling (BEAM) (n=1119 genes at q-value < 0.05), and genes that vary as a function of pseudotime, identified by differential expression analysis as implemented in Monocle2 (n=3734 genes at q-value < 0.05), in plexus and capillary cells. Cells are ordered by ascending pseudotime values with plexus in the middle, mature aerocytes (aCap) on the left, and mature gCap cells on the right. Genes (n=4129) are grouped by expression pattern and selected genes are indicated on the heatmap. Branchpoint genes change their expression where the stem splits into aCap and gCap branches. Capillary genes are expressed in both aCap and gCap cells, but not plexus. An uncropped version of the heatmap is shown in Supplementary Data 4. c, Developmental trajectory plot (top) and feature plots (below) showing log-transformed expression for selected genes in plexus (cells located along the stem before the branchpoint), aerocytes (bottom branch), and gCap cells (top branch).
Extended Data Figure 8. Specialized alveolar capillary cell types in the human lung.
a, Alveolar capillary network in adult human lung immunostained for PECAM1 and VE-Cadherin. Boxed region shown at higher magnification. b, tSNE plot of annotated artery, vein, lymphatics, bronchial endothelial, aerocyte (aCap) and gCap cell clusters identified in scRNAseq droplet data for adult human lung16 (75 year old male). c, Heatmap showing log-transformed expression of selected cluster markers in individual endothelial cells. L., lymphatics. d-g, Expression of aerocyte (EDNRB, d, or APLN, f) and gCap (EDN1, e, g or VWF, g) markers in alveolar endothelial cells (marked by CLDN5, d-f), detected by smFISH in adult human lung. d’-g’, d’’-g’’, Aerocytes and gCap cells shown at higher magnification. h, As in the mouse lung (see Extended Data Fig. 1c), occasional human alveolar capillary cells co-express aerocyte (APLN) and gCap (EDN1) markers. h’, Boxed region shown at higher magnification with clustered capillary intermediate cells (arrows) near a large vessel (dotted line in h) and a nearby single cell. i, Quantification of the relative abundance (in % of capillary endothelial cells) of the two alveolar capillary populations (aCap, gCap) and intermediate (IM) cells that co-express aCap and gCap markers in adult human lung (n=2 individuals; 69 and 75 year old males; 500–600 capillary cells scored per lung; data as mean). j, j’ Co-expression of aerocyte markers EDNRB, APLN, and CA4 in emerging aerocytes (dotted outlines) but not plexus cells (asterisks) in fetal human lung (23 weeks gestational age, corresponding to E16.5–17.5 in mouse46). At 23 weeks, 6% of CA4-expressing cells also express APLN and EDNRB at high levels (emerging aerocytes; 5 or more puncta per cell), compared to 0% at 17 weeks (500 CA4+ cells scored at each time point from one lung). Inset, adjacent haematoxylin and eosin (H&E)-stained section. j’, Boxed region from j at higher magnification. Blue (a, d-h, j), DAPI. Scale bars, 10 μm.
Extended Data Figure 9. Altered capillary cell patterns in mouse and human lung tumors.
a, Human lung adenocarcinoma with tumor vessels immunostained for CD34 (brown), an endothelial marker expressed by both aCap and gCap cells16. Blue, haematoxylin counterstain. a’, Co-expression of alveolar capillary cell type markers EDNRB (aCap) and PTPRB (gCap) in vessels surrounding tumor acinus (boxed in a), as detected in adjacent section by smFISH. a’’, Boxed region from a’ showing two endothelial cells (marked by CLDN5) co-expressing aerocyte and gCap markers (dotted outlines) at higher magnification. Merged image also shown in Fig. 4l. b-c’, Co-expression (asterisks) of alveolar capillary cell type markers (Ednrb, b, b’, or Apln, c, c’ (aCap), and Ptprb, b, b’, or Aplnr, c, c’ (gCap)) in a subset of endothelial cells (marked by Cldn5, b’) of mouse lung adenomas induced by conditional expression of an activating Kras mutation in AT2 cells6. Open arrowheads point to gCap cells. b’, c’, Boxed regions in b and c shown at higher magnification. d, Quantification of the relative abundance (in % of capillary endothelial cells) of the two alveolar capillary populations (aCap, gCap) and intermediate cells (IM) that co-express aCap and gCap markers in adenomas (n=1,332 cells scored in 21 tumor sections from n=2 mice) and alveoli (n=11,219 cells scored in n=5 mice at 3 months). e, Scatter plot of gCap:IM ratios in individual adenomas (n=21). Grey bar, mean value. The fraction of intermediate cells varies from tumor to tumor even within the same lung, perhaps reflecting different stages of tumor development. f, f’, Expression of MHC class II gene H2-Ab1 (normally expressed in gCap cells; see Extended Data Fig. 1g) is lost in gCap cells (open arrowhead) in mouse adenomas, suggesting that they may lose their antigen presentation function. H2-Ab1 is also not expressed by the abundant tumor capillary cells that co-express Aplnr and Ednrb (asterisk). f’, Boxed region in f shown at higher magnification. Scale bars, a, 200 μm; a’-c’, f, f’, 10 μm.
Extended Data Figure 10. Conserved and species-specific alveolar capillary cell specialization.
a, Dot plots showing log-transformed average expression levels and percent expression in aerocytes (aCap) and gCap cells for selected genes with known physiologic and immune functions, ligands and receptors/co-receptors, and transcription factors, which are differentially expressed by the two capillary cell types in adult mouse lung (Tabula Muris scRNAseq Smart-Seq2 data13) or human lung (Human Lung Cell Atlas16; droplet (10X) or Smart-Seq2 data; 75 year old male). b, Selected differentially expressed genes in the human lung with known functions. c-e, Diagrams of selected proposed specialized alveolar capillary functions. Selected genes involved in leukocyte trafficking and vasomotor control47 (see legend to Fig. 3d) show the same specialized expression pattern in mouse and human (c; Type 0), whereas some genes involved in hemostasis show specialized expression only in the human lung (d; Type 1), and some antigen presenting genes48 show specialized expression that switches cell type between mouse and human (e; Type 2). Blue, genes expressed in aerocytes; green, genes expressed in gCap cells; yellow, genes expressed in both aerocytes and gCap cells; purple, genes expressed in pericytes16.
Extended Data Figure 11. Faveolar capillary cell types in alligator and turtle.
a, Cladogram illustrating phylogenetic relationships between amniote taxa with different lung structures. The relationship between testudines and other diapsids is unresolved (dotted line). Ma, million years.b, h, Internal anatomy of alligator (Alligator mississippiensis; b) and turtle (Emys orbicularis; h) lungs, reproduced from ref. 49. Alligator and turtle lungs have multiple separate airspaces (‘chambers’), smooth muscle, a branched vasculature, and terminal airspaces known as faveoli where gas exchange occurs across a thick air-blood barrier50,51. c, i, Gas exchange regions of American alligator (Alligator mississippiensis; c) and Western painted turtle (Chrysemys picta bellii, i) lungs, stained with haematoxylin and eosin. d, Faveolar capillary network in American alligator lung immunostained for Claudin 5 (red; endothelium). Green, elastin fibers. e, Co-expression (asterisks) of alveolar capillary cell type markers CA4 (aCap) and PTPRB (gCap) in faveolar endothelial cells (marked by CLDN5) in juvenile American alligator lung as detected by smFISH. e’, Faveolar capillary cell in boxed region from e shown at higher magnification. Note that faveolar capillary cells – reminiscent of the intermediate cells found in adult mouse and human lungs – appear to differ in gene expression from the developmental precursors identified in the mouse vascular plexus in which aCap and gCap markers are only rarely co-expressed (Supplementary Data 2). f, Co-expression (asterisks) of alveolar capillary cell type markers EDNRB (aCap) and PTPRB (gCap) in faveolar endothelial cells (marked by CLDN5) in adult American alligator lung. f’, Faveolar capillary cell in boxed region from f shown at higher magnification. g, Alligator faveolar septum immunostained for Claudin 5 (red; endothelium) and E-cadherin (white; epithelium). Dotted line, capillary lumen. j, k, Faveolar capillary network in Western painted turtle lung immunostained for Claudin 5 (red) to label endothelial cells. Neighboring faveoli are separated by septa with a capillary layer on each side of the septal wall (j). A capillary lumen is outlined (j, dotted line). l, Co-expression (asterisks) of alveolar capillary cell type markers (EDNRB, aCap and APLNR, gCap) in faveolar endothelial cells (marked by CLDN5) in adult turtle lung. l’, Two faveolar capillary cells in boxed region from l shown at higher magnification. m, Faveolar septum in turtle lung immunostained for Claudin 5 (red) to label endothelial cells in the capillary layer on each side of septal wall and E-cadherin (white) to label respiratory epithelium. A capillary lumen is outlined (dotted line). Blue, DAPI. Scale bars, 200 μm (c, i), 10 μm (d-g, j-m).
Supplementary Material
Acknowledgements.
We thank Steve Quake and the Chan Zuckerberg Biohub for our collaborative single-cell RNA-sequencing studies of the mouse and human lung, Ralf Adams, Harold Chapman, and Kristy Red-Horse for sharing mouse lines, Patrick Bogard, Youcef Ouadah, and SoRi Jang for advice and discussion, Ondine Cleaver for an antibody recommendation, David Cornfield and the Stanford Center for Excellence in Pulmonary Biology for providing resources and space, the Department of Obstetrics & Gynecology for access to fetal lung tissue, Ruth Elsey and the Rockefeller Wildlife Refuge for providing the alligators, F. Hernán Espinoza for sharing the expression pattern of Aplnr in the developing plexus, John Perrino and Ibanri Phanwar of the Cell Sciences Imaging Facility for advice, sample preparation, and assistance with EM analysis, Larry Taylor, the Department of Comparative Medicine Animal Histology Services, the Department of Pathology Immunohistochemistry lab, and the Human Pathology/Histology Service Center for technical assistance, Maya Kumar, Edda Spiekerkoetter, and Youcef Ouadah for critical reading of the manuscript, and Ewald Weibel (1929–2019) for his inspiring, careful observations of the cellular structure of the alveolus. Silhouettes in Extended Data Fig. 11 are from PhyloPic (phylopic.org). This work was supported by the Vera Moulton Wall Center for Pulmonary Vascular Disease at Stanford and by grants from the Austrian Science Fund (J-3373) and the American Heart Association (16POST27250261) to A.G., the National Science Foundation (IOS 1055080) to C.G.F., and the National Heart, Lung, and Blood Institute (K99 HL135258) to M.G. K.J.T. was supported by a Paul and Mildred Berg Stanford Graduate Fellowship. M.A.K. is an investigator of the Howard Hughes Medical Institute. Funding for the instrumentation used in the EM experiments was provided by an ARRA Award (1S10RR026780–01) from the National Center for Research Resources.
Footnotes
Competing interest declaration. The authors declare no competing interests.
References
- 1.Hsia CC, Hyde DM & Weibel ER Lung structure and the intrinsic challenges of gas exchange. Compr Physiol 6, 827–895 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Malpighi M “Dissertationes Epistolicæ de Pulmonibus” in Opera Omnia, 320–332 (Pieter van der Aa, 1687). https://www.biodiversitylibrary.org/bibliography/566#. [Google Scholar]
- 3.Weibel ER Morphological basis of alveolar-capillary gas exchange. Physiol Rev 53, 419–495 (1973). [DOI] [PubMed] [Google Scholar]
- 4.Bertalanffy FD & Leblond CP Structure of respiratory tissue. Lancet 269, 1365–1368 (1955). [DOI] [PubMed] [Google Scholar]
- 5.Barkauskas CE, Cronce MJ, Rackley CR, Bowie EJ, Keene DR, Stripp BR, Randell SH, Noble PW & Hogan BL Type 2 alveolar cells are stem cells in adult lung. J Clin Invest 123, 3025–3036 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Desai TJ, Brownfield DG & Krasnow MA Alveolar progenitor and stem cells in lung development, renewal and cancer. Nature 507, 190–194 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Basil MC, Katzen J, Engler AE, Guo M, Herriges MJ, Kathiriya JJ, Windmueller R, Ysasi AB, Zacharias WJ, Chapman HA, Kotton DN, Rock JR, Snoeck HW, Vunjak-Novakovic G, Whitsett JA & Morrisey EE The cellular and physiological basis for lung repair and regeneration: Past, present, and future. Cell Stem Cell 26, 482–502 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu Q, Huang X, Zhang H, Tian X, He L, Yang R, Yan Y, Wang QD, Gillich A & Zhou B c-kit(+) cells adopt vascular endothelial but not epithelial cell fates during lung maintenance and repair. Nat Med 21, 866–868 (2015). [DOI] [PubMed] [Google Scholar]
- 9.Niethamer TK, Stabler CT, Leach JP, Zepp JA, Morley MP, Babu A, Zhou S & Morrisey EE Defining the role of pulmonary endothelial cell heterogeneity in the response to acute lung injury. Elife 9 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hamakawa H, Bartolak-Suki E, Parameswaran H, Majumdar A, Lutchen KR & Suki B Structure-function relations in an elastase-induced mouse model of emphysema. Am J Respir Cell Mol Biol 45, 517–524 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Golden A & Bronk TT Diffuse interstitial fibrosis of lungs; a form of diffuse interstitial angiosis and reticulosis of the lungs. AMA Arch Intern Med 92, 106–114 (1953). [PubMed] [Google Scholar]
- 12.Ackermann M, Verleden SE, Kuehnel M, Haverich A, Welte T, Laenger F, Vanstapel A, Werlein C, Stark H, Tzankov A, Li WW, Li VW, Mentzer SJ & Jonigk D Pulmonary vascular endothelialitis, thrombosis, and angiogenesis in Covid-19. N Engl J Med (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tabula Muris Consortium. Single-cell transcriptomics of 20 mouse organs creates a Tabula Muris. Nature 562, 367–372 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lien DC, Wagner WW Jr., Capen RL, Haslett C, Hanson WL, Hofmeister SE, Henson PM & Worthen GS Physiological neutrophil sequestration in the lung: Visual evidence for localization in capillaries. J Appl Physiol 62, 1236–1243 (1987). [DOI] [PubMed] [Google Scholar]
- 15.Vila Ellis L, Cain MP, Hutchison V, Flodby P, Crandall ED, Borok Z, Zhou B, Ostrin EJ, Wythe JD & Chen J Epithelial Vegfa specifies a distinct endothelial population in the mouse lung. Dev Cell (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Travaglini KJ, Nabhan AN, Lolita P, Sinha R, Gillich A, Sit RV, Chang S, Conley SD, Mori Y, Seita J, Berry GJ, Shrager JB, Metzger RJ, Kuo CS, Neff N, Weissman IL, Quake SR & Krasnow MA A molecular cell atlas of the human lung from single cell RNA sequencing. Nature (Accompanying Manuscript). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lambertz M, Grommes K, Kohlsdorf T & Perry SF Lungs of the first amniotes: Why simple if they can be complex? Biol Lett 11, 20140848 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vaccaro CA & Brody JS Structural features of alveolar wall basement membrane in the adult rat lung. J Cell Biol 91, 427–437 (1981). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bachofen M & Weibel ER Structural alterations of lung parenchyma in the adult respiratory distress syndrome. Clin Chest Med 3, 35–56 (1982). [PubMed] [Google Scholar]
- 20.Szidon JP, Pietra GG & Fishman AP The alveolar-capillary membrane and pulmonary edema. N Engl J Med 286, 1200–1204 (1972). [DOI] [PubMed] [Google Scholar]
Online-only References
- 21.Tian X, Hu T, Zhang H, He L, Huang X, Liu Q, Yu W, He L, Yang Z, Zhang Z, Zhong TP, Yang X, Yang Z, Yan Y, Baldini A, Sun Y, Lu J, Schwartz RJ, Evans SM, Gittenberger-de Groot AC, Red-Horse K & Zhou B Subepicardial endothelial cells invade the embryonic ventricle wall to form coronary arteries. Cell Res 23, 1075–1090 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen HI, Sharma B, Akerberg BN, Numi HJ, Kivela R, Saharinen P, Aghajanian H, McKay AS, Bogard PE, Chang AH, Jacobs AH, Epstein JA, Stankunas K, Alitalo K & Red-Horse K The sinus venosus contributes to coronary vasculature through VEGFC-stimulated angiogenesis. Development 141, 4500–4512 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang Y, Nakayama M, Pitulescu ME, Schmidt TS, Bochenek ML, Sakakibara A, Adams S, Davy A, Deutsch U, Luthi U, Barberis A, Benjamin LE, Makinen T, Nobes CD & Adams RH Ephrin-B2 controls Vegf-induced angiogenesis and lymphangiogenesis. Nature 465, 483–486 (2010). [DOI] [PubMed] [Google Scholar]
- 24.Chapman HA, Li X, Alexander JP, Brumwell A, Lorizio W, Tan K, Sonnenberg A, Wei Y & Vu TH Integrin alpha6beta4 identifies an adult distal lung epithelial population with regenerative potential in mice. J Clin Invest 121, 2855–2862 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Madisen L, Zwingman TA, Sunkin SM, Oh SW, Zariwala HA, Gu H, Ng LL, Palmiter RD, Hawrylycz MJ, Jones AR, Lein ES & Zeng H A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat Neurosci 13, 133–140 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Snippert HJ, van der Flier LG, Sato T, van Es JH, van den Born M, Kroon-Veenboer C, Barker N, Klein AM, van Rheenen J, Simons BD & Clevers H Intestinal crypt homeostasis results from neutral competition between symmetrically dividing Lgr5 stem cells. Cell 143, 134–144 (2010). [DOI] [PubMed] [Google Scholar]
- 27.Jackson EL, Willis N, Mercer K, Bronson RT, Crowley D, Montoya R, Jacks T & Tuveson DA Analysis of lung tumor initiation and progression using conditional expression of oncogenic K-ras. Genes Dev 15, 3243–3248 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pitulescu ME, Schmidt I, Benedito R & Adams RH Inducible gene targeting in the neonatal vasculature and analysis of retinal angiogenesis in mice. Nat Protoc 5, 1518–1534 (2010). [DOI] [PubMed] [Google Scholar]
- 29.Metzger RJ, Klein OD, Martin GR & Krasnow MA The branching programme of mouse lung development. Nature 453, 745–750 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shen Z, Lu Z, Chhatbar PY, O’Herron P & Kara P An artery-specific fluorescent dye for studying neurovascular coupling. Nat Methods 9, 273–276 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Susaki EA, Tainaka K, Perrin D, Yukinaga H, Kuno A & Ueda HR Advanced CUBIC protocols for whole-brain and whole-body clearing and imaging. Nat Protoc 10, 1709–1727 (2015). [DOI] [PubMed] [Google Scholar]
- 32.Bankhead P, Loughrey MB, Fernandez JA, Dombrowski Y, McArt DG, Dunne PD, McQuaid S, Gray RT, Murray LJ, Coleman HG, James JA, Salto-Tellez M & Hamilton PW QuPath: Open source software for digital pathology image analysis. Sci Rep 7, 16878 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Butler A, Hoffman P, Smibert P, Papalexi E & Satija R Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat Biotechnol 36, 411–420 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cohen M, Giladi A, Gorki AD, Solodkin DG, Zada M, Hladik A, Miklosi A, Salame TM, Halpern KB, David E, Itzkovitz S, Harkany T, Knapp S & Amit I Lung single-cell signaling interaction map reveals basophil role in macrophage imprinting. Cell 175, 1031–1044 (2018). [DOI] [PubMed] [Google Scholar]
- 35.Su T, Stanley G, Sinha R, D’Amato G, Das S, Rhee S, Chang AH, Poduri A, Raftrey B, Dinh TT, Roper WA, Li G, Quinn KE, Caron KM, Wu S, Miquerol L, Butcher EC, Weissman I, Quake S & Red-Horse K Single-cell analysis of early progenitor cells that build coronary arteries. Nature 559, 356–362 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Qiu X, Mao Q, Tang Y, Wang L, Chawla R, Pliner HA & Trapnell C Reversed graph embedding resolves complex single-cell trajectories. Nat Methods 14, 979–982 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chen MB, Yang AC, Yousef H, Lee D, Chen W, Schaum N, Lehallier B, Quake SR & Wyss-Coray T Brain endothelial cells are exquisite sensors of age-related circulatory cues. Cell Rep 30, 4418–4432 e4414 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ghandour MS, Langley OK, Zhu XL, Waheed A & Sly WS Carbonic anhydrase IV on brain capillary endothelial cells: A marker associated with the blood-brain barrier. Proc Natl Acad Sci U S A 89, 6823–6827 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sender S, Decker B, Fenske CD, Sly WS, Carter ND & Gros G Localization of carbonic anhydrase IV in rat and human heart muscle. J Histochem Cytochem 46, 855–861 (1998). [DOI] [PubMed] [Google Scholar]
- 40.Tabula Muris Consortium, Pisco AO, Schaum N, McGeever A, Karkanias J, Neff NF, Darmanis S, Wyss-Coray T & Quake SR A single cell transcriptomic atlas characterizes aging tissues in the mouse. Nature, in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fleming RE, Crouch EC, Ruzicka CA & Sly WS Pulmonary carbonic anhydrase IV: Developmental regulation and cell-specific expression in the capillary endothelium. Am J Physiol 265, L627–635 (1993). [DOI] [PubMed] [Google Scholar]
- 42.Beigneux AP, Davies BS, Gin P, Weinstein MM, Farber E, Qiao X, Peale F, Bunting S, Walzem RL, Wong JS, Blaner WS, Ding ZM, Melford K, Wongsiriroj N, Shu X, de Sauvage F, Ryan RO, Fong LG, Bensadoun A & Young SG Glycosylphosphatidylinositol-anchored high-density lipoprotein-binding protein 1 plays a critical role in the lipolytic processing of chylomicrons. Cell Metab 5, 279–291 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Davies BS, Beigneux AP, Barnes RH 2nd, Tu Y, Gin P, Weinstein MM, Nobumori C, Nyren R, Goldberg I, Olivecrona G, Bensadoun A, Young SG & Fong LG GPIHBP1 is responsible for the entry of lipoprotein lipase into capillaries. Cell Metab 12, 42–52 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Esnouf MP Biochemistry of blood coagulation. Br Med Bull 33, 213–218 (1977). [DOI] [PubMed] [Google Scholar]
- 45.Davie EW, Fujikawa K & Kisiel W The coagulation cascade: Initiation, maintenance, and regulation. Biochemistry 30, 10363–10370 (1991). [DOI] [PubMed] [Google Scholar]
- 46.Nikolic MZ, Sun D & Rawlins EL Human lung development: Recent progress and new challenges. Development 145 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Goldenberg NM & Kuebler WM Endothelial cell regulation of pulmonary vascular tone, inflammation, and coagulation. Compr Physiol 5, 531–559 (2015). [DOI] [PubMed] [Google Scholar]
- 48.Kreisel D, Richardson SB, Li W, Lin X, Kornfeld CG, Sugimoto S, Hsieh C-S, Gelman AE & Krupnick AS MHC class II expression by pulmonary nonhematopoietic cells plays a critical role in controlling local inflammatory responses. The Journal of Immunology 185, 3809 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Milani A Beiträge zur Kenntnis der Reptilienlunge. II. Zoologische Jahrbücher. Abteilung für Anatomie und Ontogenie der Tiere 10, 93–156 (1897). [Google Scholar]
- 50.Sanders RK & Farmer CG The pulmonary anatomy of Alligator mississippiensis and its similarity to the avian respiratory system. Anatomical Record 295, 699–714 (2012). [DOI] [PubMed] [Google Scholar]
- 51.Maina JN & West JB Thin and strong! The bioengineering dilemma in the structural and functional design of the blood-gas barrier. Physiol Rev 85, 811–844 (2005). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The scRNAseq datasets analyzed are available in the GEO repository under the accession numbers GSE109774 (Tabula Muris13, https://tabula-muris.ds.czbiohub.org), GSE132042 (Tabula Muris Senis40, https://tabula-muris-senis.ds.czbiohub.org), and GSE119228 (Cohen et al.34) or on Synapse under the accession number syn21041850 (Human Lung Cell Atlas16, https://hlca.ds.czbiohub.org). Source data used in all graphs are provided in the Source Data files.