Abstract
Bacteria and archaea are frequently attacked by viruses and other mobile genetic elements and rely on dedicated antiviral defense systems, such as restriction endonucleases and CRISPR, to survive. The enormous diversity of viruses suggests that more types of defense systems exist than are currently known. By systematic defense gene prediction and heterologous reconstitution, here we discover 29 widespread antiviral gene cassettes, collectively present in 32% of all sequenced bacterial and archaeal genomes, that mediate protection against specific bacteriophages. These systems incorporate enzymatic activities not previously implicated in antiviral defense, including RNA editing and retron satellite DNA synthesis. In addition, we computationally predict a diverse set of other putative defense genes that remain to be characterized. These results highlight an immense array of molecular functions that microbes use against viruses.
Introduction
Bacterial and archaeal viruses are the most abundant, and possibly the most diverse, biological entities on earth (1, 2). To resist frequent and varied attacks by viruses, prokaryotes possess multiple antiviral defense systems. These include the adaptive immune system CRISPR-Cas, which provides immunity by memorizing past infection events (3), and a variety of innate immune systems, such as restriction-modification (RM) systems that target specific, pre-defined sequences within the viral DNA; abortive infection (Abi) systems that induce cell dormancy or death upon viral infection; and additional systems with mechanisms that have not yet been elucidated (4). Antiviral defense systems range in complexity from a single small protein (e.g., certain types of Abi systems) to 10 or more proteins acting in concert (e.g., type I and type III CRISPR-Cas systems). Conversely, viruses have evolved strategies to counteract many of these defense systems, including anti-CRISPR and anti-restriction proteins (5, 6). Given the vast diversity of viruses and their complex patterns of co-evolution with defense systems (7-9), more types of defense systems with diverse mechanisms can be expected to exist than are currently known.
Domain-independent prediction of uncharacterized defense systems
Many antiviral defense genes in bacterial and archaeal genomes show a distinctive tendency to cluster together within defense ‘islands’ (7, 10). As a consequence, an uncharacterized gene whose homologs consistently occur next to, for instance, restriction-modification genes has an increased likelihood of being involved in defense (11, 12). Using this principle, a recent analysis (4) identified and validated 10 new defense systems, based on the requirement that each (putative) system contain at least one annotated protein domain that is enriched within defense islands.
We hypothesized that additional, unknown defense systems exist which either lack annotated domains, or only contain domains that are not typically associated with defense but have been co-opted in specific instances to perform defense functions. To test this hypothesis, we developed an expanded computational approach in which putative novel defense systems are predicted independent of domain annotations (Fig. 1A). We analyzed all bacterial and archaeal genomes available in GenBank as of November 2018, collectively encoding 620 million proteins. To identify candidate novel defense genes, we first compiled a list of all genes within 10 kb or 10 open reading frames away from known defense systems (see Methods). This initial list (n = 8.7 x 106) which evidently contained both novel defense genes and non-defense ones, was clustered to yield 6 x 105 representative sequences (‘seeds’). To distinguish between defense and non-defense seeds, we identified all homologs of each seed present in GenBank and analyzed their gene neighborhoods. The seed was predicted to be a defense gene if these neighborhoods resembled those of known defense gene—in particular, if a high percentage of homologs were located in proximity to known defense genes and displayed context diversity (Fig. 1B, fig. S1, and Methods). All clustering and homolog detection steps were performed based on amino acid sequences, without invoking existing domain annotations and thus allowing the prediction of novel types of defense genes.
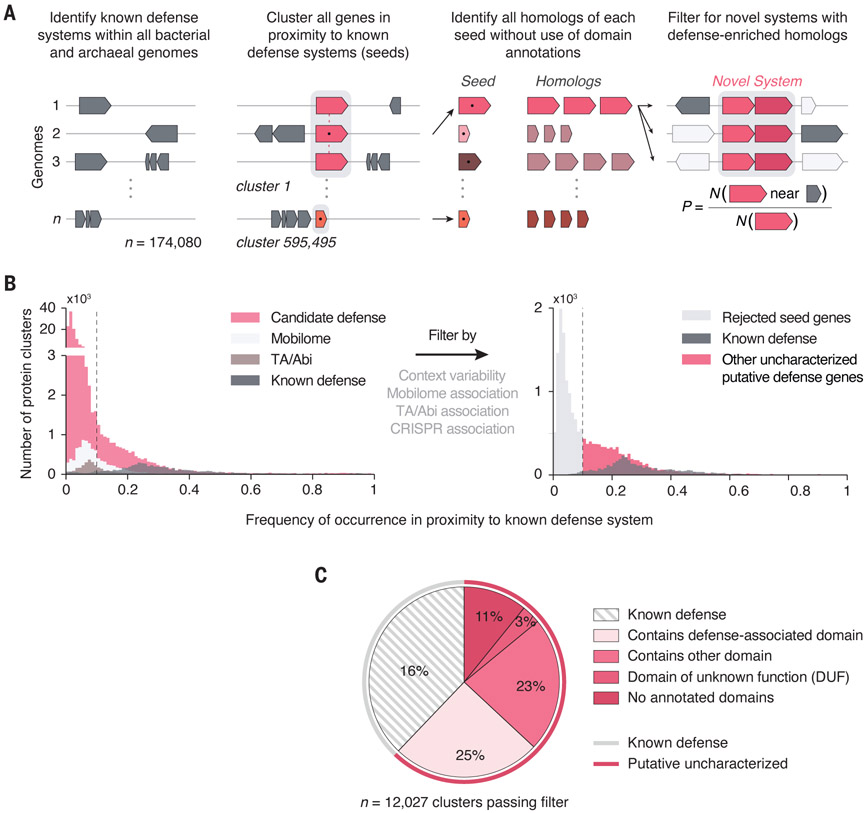
Fig. 1:
Domain-independent prediction of putative antiviral defense systems. (A) Computational pipeline to identify uncharacterized putative defense systems across all sequenced bacterial and archaeal genomes. Defense systems were predicted on the basis of analysis of amino acid sequences, independent of domain annotations. (B) Histograms of defense association frequencies before filtering and after neighborhood context–based filtering (minimum 50 homologs). Seeds to the right of the dashed line (0.1) were selected for further analysis. TA, toxin-antitoxin. (C) Pie chart of the domain diversity among predicted defense genes, based on additional analysis using HHpred against pfam domains.
After all filtering and curation steps, we identified a total of 7,472 seeds (table S1) that represented putative defense genes, along with 4,555 seeds for known defense genes under the same analysis parameters (Fig. 1C and table S2). These seeds were analyzed with additional, more sensitive analysis of their domain content (table S3). Of the uncharacterized genes, 1,687 (23%) had either no annotated domains or contained only domains of unknown function (DUFs), and an additional 2,756 (37%) contained only domains that are different from the characteristic domains of known defense genes. These results suggest the existence of a diverse set of defense genes with mechanisms that remain to be investigated.
Candidate defense systems exhibit antiviral activity in a heterologous system
To characterize the functional diversity among the predicted defense genes, we selected 48 candidate systems to test experimentally for defense activity. Candidate systems were prioritized based on the presence of predicted molecular functions not previously implicated in defense; broad phylogenetic distribution; the presence of at least one protein larger than 300 amino acids (to increase the likelihood of the presence of enzymes); and, for multi-gene systems, conservation of the component genes. Because wild-type bacterial strains are likely to harbor multiple active defense systems, thereby maintaining phage resistance even if one of the systems were knocked out (13), we elected to assay activity by heterologous reconstitution. For each system, 1-4 homologs were selected, cloned from the source organism into the low-copy vector pACYC and transformed into Escherichia coli (Fig. 2A), comprising a total of 395 kb of exogenous DNA (see tables S4-11 for sequence, accession, and source organism information). Three previously identified defense systems, BREX type I (13, 14), Druantia type I (4), and the abortive infection reverse transcriptase RT-Abi-P2 (15) were included as positive controls. Each system was then challenged with a diverse panel of coliphages with dsDNA, ssDNA, or ssRNA genomes, and phage sensitivity of the bacteria was compared to that observed with the empty vector control (Fig. 2B-C).
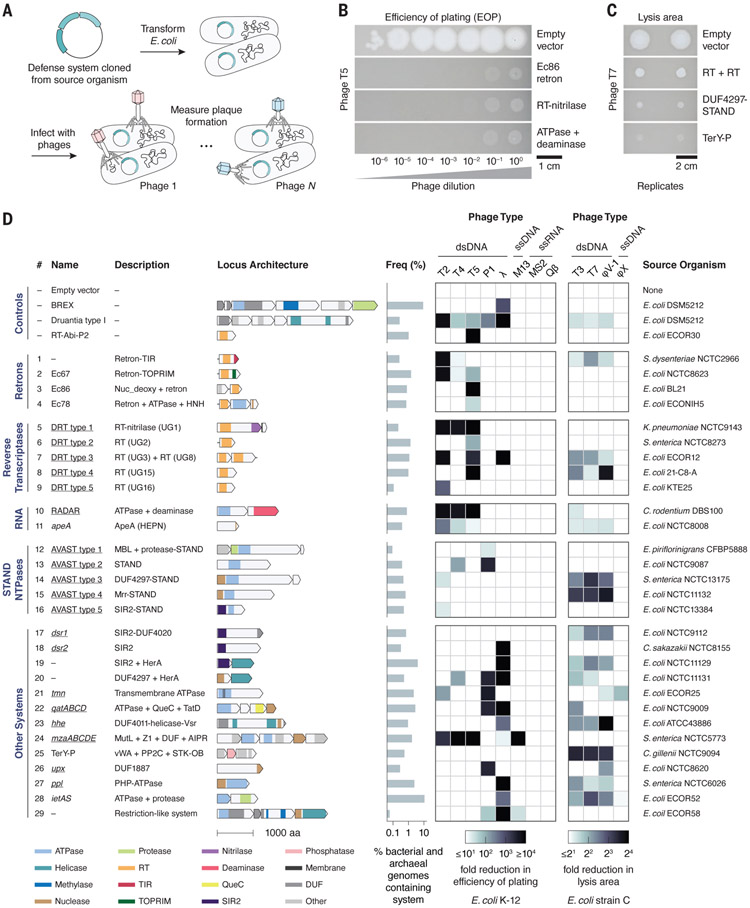
Fig. 2:
Candidate defense systems exhibit antiviral activity in a heterologous system. (A) Experimental validation pipeline using phage plaque assays on E. coli heterologously expressing a cloned candidate defense system. (B) Example plaques and (C) zones of lysis for six candidate defense systems. (D) Anti-phage activity across a panel of 12 coliphages with dsDNA, ssDNA, or ssRNA genomes (mean of two replicates). The bar graph shows the abundance of each system in sequenced bacterial and archaeal genomes. Domains: RT, reverse transcriptase; TIR, Toll/interleukin-1 receptor homology domain; TOPRIM, topoisomerase-primase domain; QueC, 7-cyano-7-deazaguanine synthase-like domain; SIR2, sirtuin; membrane, transmembrane helix; DUF, domain of unknown function. Proposed gene names: DRT, defense-associated reverse transcriptase; RADAR, phage restriction by an adenosine deaminase acting on RNA; AVAST, antiviral ATPase/NTPase of the STAND superfamily; dsr, defense-associated sirtuin; tmn, transmembrane NTPase; qat, QueC-like associated with ATPase and TatD DNase; hhe, HEPN, helicase, and Vsr endonuclease; mza, MutL, Z1, and AIPR; upx, uncharacterized (P)D-(D/E)-XK defense protein; ppl, polymerase/histidinol phosphatase-like; HerA, helicase; MBL, metallo β-lactamase.
We observed anti-phage activity for 29 of the 48 tested candidates (60%) (fig. S2). Systems from source organisms outside the Enterobacteriaceae family, which consists of Escherichia and closely-related genera including Salmonella and Klebsiella, had little to no activity, suggesting the importance of host compatibility. The most active representative in each of these 29 systems (representing 4% of the uncharacterized defense seeds) was further tested with an expanded panel of phages in two E. coli strains (Fig. 2D and fig. S3). All 29 systems were active against at least one dsDNA phage, and four were active against ssDNA phages (M13 or φX174). Phage specificity was typically narrow and varied widely across systems. The abundance of these defense systems among the sequenced bacterial and archaeal genomes spans two orders of magnitude, ranging from ~0.1% to ~10% of the genomes (Fig. 2D). Overall, 32% of all sequenced bacterial and archaeal genomes contain at least one of these defense systems, which are broadly distributed across bacterial and archaeal phyla (fig. S4).
RADAR contains a divergent adenosine deaminase that edits RNA in response to phage infection
We identified a two-gene cassette consisting of an adenosine triphosphatase (ATPase) (~900 residues) and a divergent adenosine deaminase (~900 residues) that was active against dsDNA phages T2, T3, T4, and T5. Because deaminase activity had not been previously implicated in antiviral defense, we focused on this system for further investigation. The system appears in diverse defense contexts and forms three subtypes (Fig. 3A and fig. S5A). In most cases, it consists of the ATPase and deaminase only, but some variants also include a small membrane protein, either a SLATT domain (16) or the type VI-B CRISPR ancillary protein Csx27 (17). Mutations in the ATPase Walker B motif or in the putative divalent metal cation-binding HxH motif of the deaminase abolished defense activity, whereas the SLATT domain membrane protein was required for resistance against phage T5 but not against phage T2 (Fig. 3B).
Fig. 3:
RADAR mediates RNA editing in response to phage infection. (A) Examples of genomic loci containing three subtypes of RADAR (standalone, Csx27-associated, and SLATT-associated). (B) Essentiality of the core RADAR genes rdrAB and the accessory gene rdrD against phages T2 and T5. D215A, Asp215→Ala; H168A, His168→Ala; H170A, His170→Ala; WT, wild type. (C) Representative RNA sequencing (RNAseq) reads from E. coli expressing either RADAR or an empty vector control. (D) Expression of phage T2 RNA relative to total host RNA in E. coli containing RADAR. Each dot represents a phage gene. Cells were infected at a MOI of 2. The p value was determined by a Wilcoxon signed-rank test. (E) Representative editing sites in the host and phage transcriptomes, with corresponding predicted RNA secondary structures. (F) Growth kinetics of RADAR-containing E. coli in comparison with an empty vector control under varying MOI by phage T2.
Given the large size of the deaminase compared to typical metabolic adenosine deaminases and its sequence divergence due to large insertions within the deaminase domain (fig. S5B), we hypothesized that it acts on nucleic acids rather than on free nucleosides or nucleotides. To test this hypothesis, we performed whole-transcriptome sequencing and found an enrichment of A to G substitutions in sequencing reads at specific sites in the presence of phage, whereas C, G, or U bases were not affected (Fig. 3C and fig. S6A), consistent with RNA editing of adenosine to inosine. Furthermore, the overall expression of phage genes, including early genes, was reduced by ~100-fold even at a multiplicity of infection (MOI) of 2 (Fig. 3D). Since most of the cells in the culture were expected to be infected, this suggested that defense activity occurs early in the infection cycle, which was not evident from efficiency of plating (EOP) alone.
RNA editing occurred only when both the defense system and the phage were present; expression of the defense system without the phage resulted in a near-baseline level of editing, and no editing was detected in the absence of the system. Mutations in the ATPase or deaminase active sites abolished editing, and no DNA editing was detected (fig. S6B). Editing sites were broadly distributed throughout the E. coli transcriptome (Fig. 3E, fig. S6A, fig. S7, and table S12), and editing could also be induced by co-expressing specific phage proteins with the system (fig. S8 and table S13). RNA secondary structure predictions indicated a characteristic stem-loop structure at strong editing sites; specific adenosines in loops were edited with up to ~90% frequency, whereas adenosines within the stem were not edited within the limit of detection (Fig. 3E and fig. S7). Finally, some of the editing sites are likely to be deleterious to the host cell, resulting in nonsynonymous mutations such as at the UAA stop codon of the transfer messenger RNA (tmRNA) (fig. S8B), which rescues ribosomes stalled during translation (18).
Based on these results, we named this system phage restriction by an adenosine deaminase acting on RNA (RADAR). Growth kinetics at varying phage multiplicity of infection (MOI) revealed a threshold MOI above which RADAR-expressing cells had a lower optical density at 600 nm (OD600) compared to the empty vector control, suggestive of RADAR-mediated growth arrest (Fig. 3F). Together with the abundance and broad distribution of editing sites in the host transcriptome (fig. S6-7), these results are consistent with an editing-dependent abortive infection mechanism that is activated by phage.
A widespread family of defense systems containing reverse transcriptases
We discovered that a family of uncharacterized reverse transcriptases (RTs) are active defense systems. Although most RTs in prokaryotes are components of mobile retroelements, distinct clades of RTs that lack the hallmarks of mobility also exist, including 16 ‘unknown groups’ (UGs) (19-22). We independently identified many of these uncharacterized RTs via our pipeline, suggesting that they might be defense genes (Fig. 4A). Indeed, six of these candidates (UG1, UG2, UG3, UG8, UG15, and UG16) provided robust protection against dsDNA phages. In all cases, mutations in the RT active site [(Y/F)xDD to (Y/F)xAA, where x is any amino acid] abolished activity (Fig. 4B and fig. S9A-B). We named these genes defense-associated RTs (DRTs).
Fig. 4:
Diverse families of RTs mediate antiviral defense. (A) Examples of genomic loci containing two RT-based defense systems (DRT type 1 and type 3), with two representative subtypes shown for each system. (B) Essential components of non-retron RTs (left panel) and retrons (right panel). TM, transmembrane; ncRNA, noncoding RNA; msr/msd: genes encoding msRNA and msDNA, respectively; a2, retron 5’ inverted repeat. (C) Effect of defense RTs on the expression of phage T2 genes in E. coli infected at an MOI of 2. (D) RNAseq reads mapping to the DRT type 3 system. (E) Predicted secondary structure of the highly expressed noncoding RNA identified in (D).
Each of these RT systems displayed a distinct pattern of phage resistance (Fig. 2D). Moreover, while UG2 (drt2), UG15 (drt4), and UG16 (drt5) act as individual genes, the UG3 (drt3a) and UG8 (drt3b) RTs are components of the same defense system (DRT type 3), with both RTs required for defense activity. Like RADAR, some subtypes of the UG1 (DRT type 1) and DRT type 3 systems are also associated with small membrane proteins (Fig. 4A). Moreover, DRT type 1 encompasses a much larger protein (~1200 residues) than the other five RTs and also contains a C-terminal nitrilase domain. Mutation of the catalytic cysteine of the nitrilase to alanine (C1119A) abolished activity (Fig. 4B). Nitrilases typically function in processes unrelated to defense, such as nucleotide metabolism and small molecule biosynthesis (23). Thus, DRT type 1, which is divergent from typical nitrilases and forms a distinct clade in the phylogenetic tree of the nitrilase family (fig. S10), exemplifies a non-defense domain that was apparently co-opted for a defense function.
To further characterize these RTs, we performed whole transcriptome sequencing of RT-expressing E. coli during phage infection. These experiments revealed substantial differences in phage gene expression across the different RTs (Fig. 4C). For instance, DRT type 1 strongly suppressed the expression of phage late genes, such as capsid proteins, whereas early and middle genes were not substantially affected, suggesting that it is active prior to the late stage of infection but does not prevent the injection of phage DNA into the host cell. In contrast, DRT type 3 did not strongly suppress expression of any of the phage genes, despite growing at a rate similar to DRT type 1 during phage infection (fig. S11A). Transcriptome sequencing also identified a highly expressed, structured non-coding RNA at the 3’ end of the DRT type 3 system that is required for activity (Fig. 4B, D-E).
Retrons mediate anti-phage defense
We also found that retrons, a distinct class of RTs that produce extrachromosomal satellite DNA (multi-copy ssDNA, msDNA), are active anti-phage defense systems. The retron msDNA is produced from the 5’ UTR of its own mRNA and is covalently linked to an internal guanosine of the RNA via a 2’-5’ phosphodiester bond (24). First identified over 30 years ago, retrons have been harnessed for bacterial genome engineering (25), but their native biological function has remained unknown. We found that the original E. coli retrons Ec67 (26) and Ec86 (27), as well as a homolog of the Ec78 retron (28) and a novel TIR (Toll/interleukin 1 receptor) domain-associated retron, mediate defense against dsDNA phages. Of note, the Ec86 retron is natively present in the widely-used laboratory E. coli strain BL21. Mutations in the (Y/F)xDD active site motif of the RT, as well as at the branching guanosine, abolished activity, indicating that the defense function depends on msDNA synthesis (Fig. 4B and fig. S9C). Furthermore, perturbations to the msDNA also abolished activity (fig. S11), suggesting that its structure, and not simply formation, is essential for the defense activity. Indeed, a single nucleotide mismatch in the msDNA hairpin reduced activity by 100-1000 fold, but introducing a second mutation on the complementary strand to restore the structure of the msDNA also restored wild-type activity (fig. S11). Notably, these retrons are associated with other domains, including TOPRIM (topoisomerase-primase) (29), TIR (30), a nucleoside deoxyribosyltransferase-like enzyme, and the Septu defense system (4), all of which are required for activity (Fig. 4B).
Additional molecular functions of defense systems
We investigated several additional systems with diverse components (Fig. 5 and fig. S12). These include a three-gene system containing a von Willebrand factor A (vWA) metal ion binding protein, a serine/threonine protein phosphatase, and a serine/threonine protein kinase that provided strong protection against T7-like phages (T3, T7, and φV-1). This system, dubbed the TerY-phosphorylation triad (TerY-P), has been previously analyzed computationally in the context of tellurite resistance-associated stress response and might operate as a phosphorylation switch that couples the activities of the kinase and the phosphatase (31).
Fig. 5:
Domain architectures and mutational analysis of additional defense systems. Graphics show domains identified by using HHpred, and stars indicate locations of active site mutations. Bar graphs (n = 4 replicates per bar) show either log10 fold change of efficiency of plating (for phages T2, P1, and λ) or fold change in the area of the zone of lysis (for phages T7 and φV-1) relative to the empty vector control. MBL, metallo β-lactamase; SIR2, sirtuin; HerA, helicase; QueC, 7-cyano-7-deazaguanine synthase-like domain; Vsr, very short patch repair endonuclease; TatD, DNase; vWA, von Willebrand factor type A; Prot phos, serine/threonine protein phosphatase; PHP, polymerase/histidinol phosphatase; MTase, methyltransferase; PLD, phospholipase D; DUF, domain of unknown function.
Additional systems include proteins containing a SIR2 (sirtuin) deacetylase domain that is also present in the recently-discovered Thoeris system (4) and has also been detected in the same neighborhoods with prokaryotic Argonaute proteins (32); ApeA, a predicted HEPN-family abortive infection protein (33) and a putative ancestor of the type VI CRISPR effector Cas13; a ~1300 residue P-loop ATPase containing an unusual insertion of two transmembrane helices into the ATPase domain, similar to the KAP ATPases (34); and a four-gene cassette containing a 7-cyano-7-deazaguanine synthase-like protein (QueC), suggestive of small molecule biosynthesis. All of these components are essential for defense activity (Fig. 5). Further investigation is required to understand the mechanisms by which these systems sense and respond to phage infection.
Finally, we also demonstrate defense functions for several predicted NTPases of the STAND (signal transduction ATPases with numerous associated domains) superfamily (Fig. 5). This expansive superfamily consists of multidomain proteins that include eukaryotic ATPases and GTPases involved in programmed cell death and various forms of signal transduction (35, 36). Typically, STAND NTPases contain a C-terminal helical sensor domain that, upon target recognition, induces oligomerization via ATP or GTP hydrolysis, leading to activation of the N-terminal effector domain. The role of the STAND NTPases in prokaryotes has long remained enigmatic (35, 37); the few for which experimental data are available contain a helix-turn-helix domain and have been shown to regulate transcription (36). Several STAND NTPases were active against dsDNA phages (Fig. 2D); these proteins contained different putative effector domains, including DUF4297 (a putative PD-(D/E)xK-family nuclease), an Mrr-like nuclease, SIR2, a trypsin-like serine protease, and an uncharacterized helical domain. We named these systems antiviral ATPases/NTPases of the STAND superfamily (AVAST). As homologs of essential eukaryotic programmed cell death effectors, AVAST systems are likely to function via an abortive infection mechanism, i.e. by causing growth arrest or programmed cell death in infected hosts.
Discussion
These findings substantially expand the space of protein domains, molecular functions, and interactions that are employed by bacteria and archaea in antiviral defense. Some of these functions, including RNA editing, have not been previously implicated in defense mechanisms. The high success rate of defense system prediction based on the evolutionary conservation of their proximity to previously identified defense genes supports the defense island concept (4, 7, 10) and demonstrates its growing utility at the time of rapid expansion of sequence databases. Furthermore, the computational approach implemented in this work provided for a substantial expansion of the range of the identified putative defense systems. Many of these previously unknown systems contain enzymatic activities as well as predicted sensor components that potentially could be engineered for novel biotechnology applications.
Despite similarities in domain architectures among some of the identified defense systems, their phage specificities differ significantly, emphasizing the importance of multiple defense mechanisms for the survival of prokaryotes in the arms race against viruses. These observations are compatible with the concept of distributed microbial immunity, according to which defense systems encoded in different genomes collectively protect microbial communities from the diverse viromes they confront (38). Additionally, several of the identified defense systems incorporate molecular functions from typically non-defense sources, highlighting the versatility of activities that are recruited for antiviral defense. These include the RADAR deaminase, nitrilases, RTs, and retrons. The demonstration of defense functions for multiple RTs, which are generally associated with mobile genetic elements (MGEs), is consistent with the ‘guns for hire’ paradigm whereby enzymes are shuttled between MGEs and defense systems during microbial evolution (8). Finally, most of these defense systems do not appear to be substantially enriched within prophages, suggesting that they are dedicated host defense genes, rather than virus superinfection exclusion modules (fig. S13 and Methods).
The overall patchy pattern of phage specificity observed for the different defense systems was unexpected. In some cases, the same system exhibited widely varying levels of protection against similar phages; for instance, retron Ec67 and DRT type 3 offered full protection against phage T2 but poor protection against phage T4, which is ~82% identical to T2. We hypothesize that phage-encoded inhibitors or anti-defense genes that have yet to be discovered may play a significant role in determining the specificity of defense systems. Indeed, many phage-encoded proteins have no known function and remain to be investigated for anti-defense activity.
With the exception of RADAR, we do not yet know the mechanisms of most of the identified defense systems. The range of domains contained within these systems indicates that they employ diverse biochemical activities. Additional experimental characterization is required to elucidate the effector functions for these systems and the molecular basis of anti-phage action and specificity. The identification of these defense systems, as well as others we have predicted computationally, provides a foundation for further mechanistic investigation.
The results described here have broad implications for understanding antiviral resistance and host-virus interactions in natural populations of microbes, as well as for technological applications such as the development of anti-bacterial therapeutics, nucleic acid editing, molecular detection, and targeted cell destruction.
Supplementary Material
Acknowledgements:
We thank Alim Ladha, Jonathan Strecker, Soumya Kannan, Joe Kreitz, and Guilhem Faure for valuable discussions and experimental assistance; Rhiannon Macrae for a critical reading of the manuscript; Susan Richards and Rich Belliveau for assistance with ordering bacterial strains; and the entire Zhang lab for support and advice.
Funding: K.S.M. and E.V.K. are supported by intramural funds of the U.S. Department of Health and Human Services (to National Library of Medicine). F.Z. is supported by the National Institutes of Health (grants 1R01-HG009761, 1R01-MH110049, and 1DP1-HL141201); the Howard Hughes Medical Institute; the Open Philanthropy Project, the Harold G. and Leila Mathers and Edward Mallinckrodt, Jr. Foundations; the Poitras Center for Psychiatric Disorders Research at MIT; the Hock E. Tan and K. Lisa Yang Center for Autism Research at MIT; and by the Phillips family and J. and P. Poitras.
Footnotes
Competing interests: F.Z. is a scientific advisor and cofounder of Editas Medicine, Beam Therapeutics, Pairwise Plants, Arbor Biotechnologies, and Sherlock Biosciences. L.G, J.L.S.-B., and F.Z. are co-inventors on US provisional application no. 62/928,269, which includes bacterial defense systems described in this manuscript.
Data and materials availability: Expression plasmids are available from Addgene (nos. 157879 to 157912) under the Uniform Biological Material Transfer Agreement. The genome assembly of E. coli phage φV-1 has been deposited in GenBank (accession number MT542512). All other data are available in the manuscript or the supplementary materials.
Supplementary Materials
References
- 1.Suttle CA, Viruses: unlocking the greatest biodiversity on Earth. Genome 56, 542–544 (2013). [DOI] [PubMed] [Google Scholar]
- 2.Cobián Güemes AG et al. , Viruses as Winners in the Game of Life. Annu Rev Virol 3, 197–214 (2016). [DOI] [PubMed] [Google Scholar]
- 3.Hille F et al. , The Biology of CRISPR-Cas: Backward and Forward. Cell 172, 1239–1259 (2018). [DOI] [PubMed] [Google Scholar]
- 4.Doron S et al. , Systematic discovery of antiphage defense systems in the microbial pangenome. Science 359, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Samson JE, Magadán AH, Sabri M, Moineau S, Revenge of the phages: defeating bacterial defences. Nat Rev Microbiol 11, 675–687 (2013). [DOI] [PubMed] [Google Scholar]
- 6.Bondy-Denomy J, Pawluk A, Maxwell KL, Davidson AR, Bacteriophage genes that inactivate the CRISPR/Cas bacterial immune system. Nature 493, 429–432 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Makarova KS, Wolf YI, Koonin EV, Comparative genomics of defense systems in archaea and bacteria. Nucleic Acids Res 41, 4360–4377 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Koonin EV, Makarova KS, Wolf YI, Krupovic M, Evolutionary entanglement of mobile genetic elements and host defence systems: guns for hire. Nat Rev Genet, (2019). [DOI] [PubMed] [Google Scholar]
- 9.Faure G et al. , CRISPR-Cas in mobile genetic elements: counter-defence and beyond. Nat Rev Microbiol 17, 513–525 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Makarova KS, Wolf YI, Snir S, Koonin EV, Defense islands in bacterial and archaeal genomes and prediction of novel defense systems. J Bacteriol 193, 6039–6056 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shmakov SA, Makarova KS, Wolf YI, Severinov KV, Koonin EV, Systematic prediction of genes functionally linked to CRISPR-Cas systems by gene neighborhood analysis. Proc Natl Acad Sci U S A 115, E5307–E5316 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shmakov SA et al. , Systematic prediction of functionally linked genes in bacterial and archaeal genomes. Nat Protoc 14, 3013–3031 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gordeeva J et al. , BREX system of Escherichia coli distinguishes self from non-self by methylation of a specific DNA site. Nucleic Acids Res 47, 253–265 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Goldfarb T et al. , BREX is a novel phage resistance system widespread in microbial genomes. EMBO J 34, 169–183 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Odegrip R, Nilsson AS, Haggård-Ljungquist E, Identification of a gene encoding a functional reverse transcriptase within a highly variable locus in the P2-like coliphages. J Bacteriol 188, 1643–1647 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Burroughs AM, Zhang D, Schäffer DE, Iyer LM, Aravind L, Comparative genomic analyses reveal a vast, novel network of nucleotide-centric systems in biological conflicts, immunity and signaling. Nucleic Acids Res 43, 10633–10654 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Makarova KS, Gao L, Zhang F, Koonin EV, Unexpected connections between type VI-B CRISPR-Cas systems, bacterial natural competence, ubiquitin signaling network and DNA modification through a distinct family of membrane proteins. FEMS Microbiol Lett 366, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rae CD, Gordiyenko Y, Ramakrishnan V, How a circularized tmRNA moves through the ribosome. Science 363, 740–744 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zimmerly S, Wu L, An Unexplored Diversity of Reverse Transcriptases in Bacteria. Microbiol Spectr 3, MDNA3-0058-2014 (2015). [DOI] [PubMed] [Google Scholar]
- 20.Toro N, Nisa-Martínez R, Comprehensive phylogenetic analysis of bacterial reverse transcriptases. PLoS One 9, e114083 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kojima KK, Kanehisa M, Systematic survey for novel types of prokaryotic retroelements based on gene neighborhood and protein architecture. Mol Biol Evol 25, 1395–1404 (2008). [DOI] [PubMed] [Google Scholar]
- 22.Simon DM, Zimmerly S, A diversity of uncharacterized reverse transcriptases in bacteria. Nucleic Acids Res 36, 7219–7229 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pace HC, Brenner C, The nitrilase superfamily: classification, structure and function. Genome Biol 2, REVIEWS0001 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Simon AJ, Ellington AD, Finkelstein IJ, Retrons and their applications in genome engineering. Nucleic Acids Res 47, 11007–11019 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Farzadfard F, Lu TK, Synthetic biology. Genomically encoded analog memory with precise in vivo DNA writing in living cell populations. Science 346, 1256272 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lampson BC et al. , Reverse transcriptase in a clinical strain of Escherichia coli: production of branched RNA-linked msDNA. Science 243, 1033–1038 (1989). [DOI] [PubMed] [Google Scholar]
- 27.Lim D, Maas WK, Reverse transcriptase-dependent synthesis of a covalently linked, branched DNA-RNA compound in E. coli B. Cell 56, 891–904 (1989). [DOI] [PubMed] [Google Scholar]
- 28.Lima TM, Lim D, A novel retron that produces RNA-less msDNA in Escherichia coli using reverse transcriptase. Plasmid 38, 25–33 (1997). [DOI] [PubMed] [Google Scholar]
- 29.Aravind L, Leipe DD, Koonin EV, Toprim--a conserved catalytic domain in type IA and II topoisomerases, DnaG-type primases, OLD family nucleases and RecR proteins. Nucleic Acids Res 26, 4205–4213 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Horsefield S et al. , NAD. Science 365, 793–799 (2019). [DOI] [PubMed] [Google Scholar]
- 31.Anantharaman V, Iyer LM, Aravind L, Ter-dependent stress response systems: novel pathways related to metal sensing, production of a nucleoside-like metabolite, and DNA-processing. Mol Biosyst 8, 3142–3165 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Makarova KS, Wolf YI, van der Oost J, Koonin EV, Prokaryotic homologs of Argonaute proteins are predicted to function as key components of a novel system of defense against mobile genetic elements. Biol Direct 4, 29 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Anantharaman V, Makarova KS, Burroughs AM, Koonin EV, Aravind L, Comprehensive analysis of the HEPN superfamily: identification of novel roles in intra-genomic conflicts, defense, pathogenesis and RNA processing. Biol Direct 8, 15 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Aravind L, Iyer LM, Leipe DD, Koonin EV, A novel family of P-loop NTPases with an unusual phyletic distribution and transmembrane segments inserted within the NTPase domain. Genome Biol 5, R30 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Leipe DD, Koonin EV, Aravind L, STAND, a class of P-loop NTPases including animal and plant regulators of programmed cell death: multiple, complex domain architectures, unusual phyletic patterns, and evolution by horizontal gene transfer. J Mol Biol 343, 1–28 (2004). [DOI] [PubMed] [Google Scholar]
- 36.Danot O, Marquenet E, Vidal-Ingigliardi D, Richet E, Wheel of Life, Wheel of Death: A Mechanistic Insight into Signaling by STAND Proteins. Structure 17, 172–182 (2009). [DOI] [PubMed] [Google Scholar]
- 37.Koonin EV, Aravind L, Origin and evolution of eukaryotic apoptosis: the bacterial connection. Cell Death Differ 9, 394–404 (2002). [DOI] [PubMed] [Google Scholar]
- 38.Bernheim A, Sorek R, The pan-immune system of bacteria: antiviral defence as a community resource. Nat Rev Microbiol 18, 113–119 (2020). [DOI] [PubMed] [Google Scholar]
- 39.Hyatt D et al. , Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics 11, 119 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Punta M et al. , The Pfam protein families database. Nucleic Acids Res 40, D290–301 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Marchler-Bauer A et al. , CDD/SPARCLE: functional classification of proteins via subfamily domain architectures. Nucleic Acids Res 45, D200–D203 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Steinegger M, Söding J, MMseqs2 enables sensitive protein sequence searching for the analysis of massive data sets. Nat Biotechnol 35, 1026–1028 (2017). [DOI] [PubMed] [Google Scholar]
- 43.Steinegger M, Söding J, Clustering huge protein sequence sets in linear time. Nat Commun 9, 2542 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Roberts RJ, Vincze T, Posfai J, Macelis D, REBASE--a database for DNA restriction and modification: enzymes, genes and genomes. Nucleic Acids Res 43, D298–299 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cohen D et al. , Cyclic GMP-AMP signalling protects bacteria against viral infection. Nature, (2019). [DOI] [PubMed] [Google Scholar]
- 46.Ofir G et al. , DISARM is a widespread bacterial defence system with broad anti-phage activities. Nat Microbiol 3, 90–98 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Katoh K, Misawa K, Kuma K, Miyata T, MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res 30, 3059–3066 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zimmermann L et al. , A Completely Reimplemented MPI Bioinformatics Toolkit with a New HHpred Server at its Core. J Mol Biol 430, 2237–2243 (2018). [DOI] [PubMed] [Google Scholar]
- 49.Petricciani JC, Chu FC, Johnson JB, Meyer HM, Bacteriophages in live virus vaccines. Proc Soc Exp Biol Med 144, 789–792 (1973). [DOI] [PubMed] [Google Scholar]
- 50.Milstien JB, Walker JR, Petricciani JC, Bacteriophages in live virus vaccines: lack of evidence for effects on the genome of rhesus monkeys. Science 197, 469–470 (1977). [DOI] [PubMed] [Google Scholar]
- 51.Xu B, Ma X, Xiong H, Li Y, Complete genome sequence of 285P, a novel T7-like polyvalent E. coli bacteriophage. Virus Genes 48, 528–533 (2014). [DOI] [PubMed] [Google Scholar]
- 52.Picelli S et al. , Tn5 transposase and tagmentation procedures for massively scaled sequencing projects. Genome Res 24, 2033–2040 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Miller ES et al. , Bacteriophage T4 genome. Microbiol Mol Biol Rev 67, 86–156 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Turner DH, Mathews DH, NNDB: the nearest neighbor parameter database for predicting stability of nucleic acid secondary structure. Nucleic Acids Res 38, D280–282 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhou Y, Liang Y, Lynch KH, Dennis JJ, Wishart DS, PHAST: a fast phage search tool. Nucleic Acids Res 39, W347–352 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Arndt D et al. , PHASTER: a better, faster version of the PHAST phage search tool. Nucleic Acids Res 44, W16–21 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Strecker J et al. , RNA-guided DNA insertion with CRISPR-associated transposases. Science 365, 48–53 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Klompe SE, Vo PLH, Halpin-Healy TS, Sternberg SH, Transposon-encoded CRISPR-Cas systems direct RNA-guided DNA integration. Nature 571, 219–225 (2019). [DOI] [PubMed] [Google Scholar]
- 59.Koonin EV, Makarova KS, Wolf YI, Evolutionary Genomics of Defense Systems in Archaea and Bacteria. Annu Rev Microbiol 71, 233–261 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yamamoto S, Kiyokawa K, Tanaka K, Moriguchi K, Suzuki K, Novel toxin-antitoxin system composed of serine protease and AAA-ATPase homologues determines the high level of stability and incompatibility of the tumor-inducing plasmid pTiC58. J Bacteriol 191, 4656–4666 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.