Abstract
The outbreak of existing public health distress is threatening the entire world with emergence and rapid spread of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). The novel coronavirus disease 2019 (COVID-19) is mild in most people. However, in some elderly people with co-morbid conditions, it may progress to pneumonia, acute respiratory distress syndrome (ARDS) and multi organ dysfunction leading to death. COVID-19 has caused global panic in the healthcare sector and has become one of the biggest threats to the global economy. Drug discovery researchers are expected to contribute rapidly than ever before. The complete genome sequence of coronavirus had been reported barely a month after the identification of first patient. Potential drug targets to combat and treat the coronavirus infection have also been explored. The iterative structure-based drug design (SBDD) approach could significantly contribute towards the discovery of new drug like molecules for the treatment of COVID-19. The existing antivirals and experiences gained from SARS and MERS outbreaks may pave way for identification of potential drug molecules using the approach. SBDD has gained momentum as the essential tool for faster and costeffective lead discovery of antivirals in the past. The discovery of FDA approved human immunodeficiency virus type 1 (HIV-1) inhibitors represent the foremost success of SBDD. This systematic review provides an overview of the novel coronavirus, its pathology of replication, role of structure based drug design, available drug targets and recent advances in in-silico drug discovery for the prevention of COVID-19. SARSCoV- 2 main protease, RNA dependent RNA polymerase (RdRp) and spike (S) protein are the potential targets, which are currently explored for the drug development.
Keywords: Coronavirus 2, Structure based drug design, Proteases, RNA dependent RNA polymerase, Spike (S) protein
Highlights
-
•
Review opens up about the contribution, and significance of SBDD towards discovery of novel therapeutics for the treatment of COVID-19.
-
•
Potential targets/proteins of COVID-19 for development of therapeutic agents are discussed.
-
•
An up-to-date review of all the various, available COVID-19 treatments and their efficacy.
1. Introduction
In early December 2019, an outbreak of novel coronavirus disease 2019 (COVID-19) occurred in Wuhan City, China caused by a novel severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). Ever since its emergence in China, the infectious disease has progressed into a serious threat, wreaking havoc around the world. On March 11, 2020, the World Health Organization (WHO) declared COVID-19 outbreak a global pandemic. It has now reached almost every country in the world, closing borders, shutting down institutions, work places, ceasing economic activity and forcing entire continents to act on emergency response protocols. The most rapid spread of the virus is now in United states of America, India and Brazil (Tuttle, 2020; Cevik et al., 2020; Sohrabi et al., 2020). As per WHO COVID-19 dashboard, on April 6th, 2021, there had been 131,309,792 confirmed cases of infections and 2,854,276 deaths reported and these numbers keep increasing every day. The global scientific community is in action and researchers have learned a lot about the virus. Still, there are many questions that need answering viz. Why do people respond so differently to the same infection? people show strikingly different responses, some remain asymptomatic whereas, other seemingly healthy people develop severe and at times, fatal pneumonias (McCoy et al., 2020). Multiple teams of researchers are also investigating the possibility of a genetic component (Murray et al., 2020). A genome-wide study in Spain and Italy, involving 1980 COVID-19 patients, identified two genetic variants locus on chromosomes found at 9q34 and 3p21.31 (McCoy et al., 2020; Ovsyannikova et al., 2020). The later one i.e. 3p21.31 has been repeatedly associated with patients in hospitalization (GenOMICC Investigators IICOVID-19 Human Genetics Initiative23andMe InvestigatorsBRACOVID InvestigatorsGen-COVID Investigators %J Nature, 2020). Further, new genome-wide associations on chromosomes may also help in the identification of mechanistic targets for the development of therapeutics. Several genetic variants were found recently, including loss-of-function variants in TLR7 with impairment in type I and II IFN responses (Fallerini et al., 2021; Zhang et al., 2020a). Studies have also showed striking variations between social and ethnic groups. These disparities are likely linked to lower socio-economic equality as well as access to treatment (McCoy et al., 2020; Ovsyannikova et al., 2020). The vaccinations have already been started on war footings. Immunologists are working efficiently to determine the development of immunity, its impact and duration of protection of the vaccines. Variations (both genetic and epigenetic) in the ACE2, TMPRSS2 and FURIN genes, pertaining to the cellular entry mechanism of the SARS-CoV-2, as well as the molecules involved in the modulation of their expression viz., CALM, ADAM-17, AR and ESR could contribute to the discrepancies in individual disease-risk score (Ragia and Manolopoulos, 2020). Although, most researchers agree that the SARS-CoV-2 originated in horseshoe bats, its transmission from bats to humans has not yet been established and so has the intermediary animal through which it passed to humans (Helmy et al., 2020; Bai et al., 2020; Latinne et al., 2020). The discovery of RaTG13 bat coronavirus and molecular and serological evidence of SARS-CoV-2 related coronavirus (SC2r-CoVs) actively circulating in bats in Southern Asia suggested a high probability of its bat origin (Wacharapluesadee et al., 2021).
The entire human race is waiting for any sign of hope in countering the COVID-19 pandemic. Immunologists, pharmacologists and drug discovery researchers around the globe are working continuously to develop variety of potential COVID-19 treatments (Holmes et al., 2020; Fauci et al., 2020). Each of the treatment approach is facing challenges in conducting clinical trials during the COVID-19 health emergency. Currently, most of the treatment regimens focus on the management of symptoms, excluding few antiviral medications (Ahmed et al., 2020). The potential symptoms of COVID-19 infection are enormously close to that of severe acute respiratory syndrome (SARS) and middle east respiratory syndrome (MERS) viral infections. The quest for SARS-CoV-2 vaccines commenced in January 2020 with deciphering of the SARS-CoV-2 genome. Since then, several vaccine candidates have been undergoing trials in humans, many of which have been approved for usage (Dong et al., 2020a; Krammer, 2020; Mulligan et al., 2020; Xia et al., 2020; Wang et al., 2020a; Gao et al., 2020a).
The modern computational drug discovery techniques i.e. virtual screening and molecular docking will play pivotal role in discovery of novel bioactive compounds in context to COVID-19 (van Hilten et al., 2019; Pillaiyar et al., 2020). Recently developed protein crystal structures from parts of the virus/host and homology models are already being used as early state approaches. More refined techniques that account for the electronic description of the biological processes such as molecular dynamics and quantum mechanics, could also be utilized in the development process (Liu et al., 2020; Al-Khafaji et al., 2020; Mittal et al., 2020).
Structure based drug design (SBDD) is a highly validated and essential design approach in the modern drug discovery process (Anderson, 2012; Mandal and Mandal, 2009). It directs the discovery of a lead compound which could show potent target inhibitory activity through a faster and cost-efficient approach. In order to rapidly achieve new drug compounds for clinical use, a combination of SBDD, virtual drug screening, and high-throughput screening approaches could be employed for anti-COVID-19 drug discovery (Kim et al., 2020; Choudhury, 2020). In light of the remarkable computational approaches that are being used for the discovery of novel drug like compounds in the context of COVID-19, we foresee that potent inhibitor(s) could be discovered through structure-based drug design in the coming days. The current review opens up the contributions and significance of SBDD towards discovery of novel therapeutic approaches for the treatment of COVID-19.
2. Structure based drug design (SBDD)
SBDD is one of the inherent tools of the principal industrial drug discovery programs. The tool helps in overcoming the limitations of conventional drug discovery process by accelerating it. Comparatively, lower amount of time, cost, and labour is spent in the computational drug design and can have a huge impact in the discovery of the new drug molecules. In drug discovery process, the identification of molecules that show significant interactions with the therapeutic targets is of the paramount importance (Batool et al., 2019; Anderson, 2003). An effective workflow of SBDD is depicted in Fig. 1. In the discovery and optimization of the lead, SBDD is a more specific, efficient and rapid process. The knowledge of 3D structures of the biological targets is a prerequisite for SBDD. Due to the advances in bioinformatics and Human Genome Project completion, vast number of 3D structures of the target proteins are now available. SBDD methods analyse the information of 3-dimensional macromolecular targets, mostly of proteins or RNA, to identify key sites and interactions that are essential for their respective biological function. It is mainly used for binding energy analysis, interaction of ligand-protein and evaluation of the conformational changes during the process of docking (Bohacek et al., 1996; Wilson and Lill, 2011; Buchanan, 2002; Singh et al., 2006; KJJocmedicine, 2007). The development of protease inhibitor, Saquinavir (Fig. 2 (Tuttle, 2020)) by Roche, has made HIV-AIDS a manageable and treatable disease. There are other drugs also (Fig. 2; Indinavir (Cevik et al., 2020), Nelfinavir (Sohrabi et al., 2020), Amprenavir (McCoy et al., 2020), Fosamprenavir (Murray et al., 2020), Tipranavir (Ovsyannikova et al., 2020), and Atazanavir (GenOMICC Investigators IICOVID-19 Human Genetics Initiative23andMe InvestigatorsBRACOVID InvestigatorsGen-COVID Investigators %J Nature, 2020)) discovered by SBDD from the same class that are approved by the FDA and are available in the market (Agnello et al., 2019). SBDD is an iterative process, which undergoes multiple cycles of iteration until optimization of the lead molecule is achieved. The drug discovery process is composed of different phases including, the discovery phase, development phase, clinical trial phase, and registry phase. In the discovery phase, extraction, purification and determination of target protein structure are achieved (Batool et al., 2019). SBDD tools can be used to dock the commercially available small molecule databases into the binding cavities of the specific target. The 3D structure of the target molecule provides knowledge of electrostatic properties of the binding sites, including the presence of different cavities, clefts, and allosteric pockets. The compounds are ranked on the basis of steric and electrostatic interactions with the binding site of the target. In the next stage of cycle, the top ranked hits are synthesized, optimized and tested in vitro for biochemical assays. The selected compounds are evaluated for their efficacy, affinity and potency experimentally. In the initial phase, the 3D structure of target protein complexed with the potential lead is obtained. The 3D structure provides the details of intermolecular features that help in the molecular recognition process and ligand binding. The structure of ligand-protein complex helps in the analysis of different binding confirmations, binding pocket identification and interaction of ligand-protein. It also helps in the elucidation of conformational changes due to binding of ligand and mechanistic studies (Kalyaanamoorthy and Chen, 2011; Ferreira et al., 2015; Fang, 2012). De novo drug design offers an attractive opportunity to produce novel molecular structures from scratch with desired pharmacological properties. When the 3D structure of the biomolecule of interest is available, a structure-based de novo drug discovery approach can be used as a valuable starting point (Fischer et al., 2019; Bung et al., 2021).
Fig. 1.
Workflow of structure-based drug design (SBDD) in the drug discovery process.
Fig. 2.
Drugs developed through structure-based drug design.
3. Tools used for SBDD
SBDD approach includes important stages of drug discovery such as ‘hit identification’ and ‘hit-to-lead’ optimization. The initial phase comprises the identification of a number of chemical molecules, known as ‘hits’, that ideally exhibit some range of potential effect along with specificity against the particular biological target (Kalyaanamoorthy and Chen, 2011). Whereas, the latter phase consists of evaluation of the early identified hits to identify the potential lead molecules before entering into a large-scale lead optimization. In the course of the past decades, there has been a sharp escalation in the innovative software packages (Table 1), which contribute immensely in carrying out the different iterative phases of SBDD effectively. Even though these software resources have much to offer, it has eventually become an exacting to pick successful strategies and tools for effective discovery of lead compound (Halperin et al., 2002; Salemme et al., 1997). The iterative process of SBDD is depicted in Fig. 1. The initial phase of SBDD is the selection of potential target, followed by the ligand identification. The target may be a macromolecular protein or enzyme involved in the biosynthetic pathways. After selection of the target, extraction, purification and determination of the structure of the protein are the crucial processes. The 3D structure of the protein is developed by using sophisticated techniques viz. X-ray crystallography (XRD), nuclear magnetic resonance (NMR) spectroscopy and cryo-electron microscopy (cryo-EM) etc. Once the structure of the target is identified, it is necessary to investigate the electrostatic properties of the binding site such as presence of cavities, clefts, amino acid sequences and allosteric pockets (R Laurie et al., 2006; Grinter and Zou, 2014; Koutsoukas et al., 2011). By using the various available computer algorithms, drug like molecules from the large databases of small molecules or some of the fragments of compounds into the binding cavities of the target protein can be identified (Table 1) (Wishart et al., 2006). Furthermore, the top scoring molecules with high affinity towards target protein are synthesized and tested in in-vitro biochemical assays. The ligands with desired therapeutic activities are further evaluated for efficacy, affinity and potency studies. Simultaneously, the availability of the 3D structure of the target protein in complexation with the promising ligand is identified, which offers detailed evidence of intermolecular features aiding in the molecular identification process and binding pattern of the ligand. Structural insights into the complex of ligand–protein assist the analysis of variety of binding conformations and interactions of ligand–protein (Kapetanovic, 2008; Wallace et al., 1995; Leach et al., 2006; Gohlke et al., 2000). Various essential computational tools and databases which help experimental biologists with predictive insights, accelerate the ongoing research efforts to find therapeutics against the COVID-19 Infection (Kangabam et al., 2020). For the immediate use by the scientific community, more than 800 SARS-CoV-2 proteins structures have been deposited with the Protein Data Bank. Most of the structures deposited are the two proteases of virus and spike glycoprotein (Papageorgiou and Mohsin, 2020). The available databases with structural features of SARS-CoV-2 provide insights into the viral transmission and reveal novel therapeutic targets. The molecular docking and molecular dynamics simulation approach have been employed for potential protein targets of SARS-CoV-2 (Table 2) with various compounds to identify potential drugs for SARS-CoV-2.
Table 1.
Drug discovery software tools and packages.
| Sl No. | Molecular Docking software | Description | Source |
|---|---|---|---|
| 1. | Autodock | Flexible ligand and protein side chain. It predicts binding of small molecules to the receptor of known 3D structure. | http://autodock.scripps.edu/ |
| 2. | DOCK | Flexible ligand. DOCK algorithm performed rigid body docking with geometric matching algorithm. | http://dock.compbio.ucsf.edu/ |
| 3. | GOLD | Flexible ligand and partial flexible of protein. Docking program based on Genetic algorithm | https://www.ccdc.cam.ac.uk/ |
| 4. | Glide | Docking Program based on Exhaustive search. It exists in Standard precision, extra precision and virtual high throughput screening models. | https://www.schrodinger.com/glide/ |
| 5. | GlamDock | GlamDock work on the basis of Monte-Carlo with minimization (basin hopping) search in a hybrid interaction matching/internal coordinate search space. | http://www.chil2.de/Glamdock.html |
| 6. | FRED | Fast Exhaustive docking performs a systematic and non-stochastic examination of all possible poses of protein-ligand complex. | https://www.eyesopen.com/oedocking |
| 7. | GEMDOCK | It is a program for computing ligand confirmation and orientation. It's a Generic Evolutionary method for molecular docking. | http://gemdock.life.nctu.edu.tw/dock/ |
| 8. | HomDock | It is similarity-based docking program. It is a combination of two different tool such as GMA molecular alignment and GlamDock | http://www.chil2.de/HomDock.html |
| 9. | iGEMDOCK | It is a Graphical Environment for the reorganization of pharmacological interactions as well as virtual screening. | http://gemdock.life.nctu.edu.tw/dock/igemdock.php |
| 10. | ICM | Ligand and protein flexible. It useful for ligand-protein docking, protein-protein docking and peptide-protein docking. | http://www.molsoft.com/docking.html |
| 11. | FlexX, Flex-Ensemble (FlexE) | Flexible ligand in FlexX, flexible protein and flexible ligand in FlexE. It is an Incremental build-based docking program | https://www.biosolveit.de/flexx/index.html?ct=1 |
| 12. | vLifeDock | It provides three methods, Grid based docking, GA docking and GRIP docking. | https://www.vlifesciences.com/products/VLifeMDS/VLifeDock.php |
| 13. | FITTED | Flexibility Induced Through Targeted Evolutionary Description. Flexible ligand and protein. high accuracy programs with unique, customizable features | http://fitted.ca/ |
| 14. | DAIM-SEED-FFLD | It allows Decomposition and Identification of Molecules, Solvation Energy for Exhaustive Docking, Fragment-based Flexible Ligand Docking | http://www.biochem-caflisch.uzh.ch/download/ |
| 15. | Molegro Virtual Docker | Handle all aspects of docking process from molecule preparation to binding site determination. | https://molegrovirtualdocker.weebly.com/ |
| 16. | Autodock Vina | Flexible ligand and protein side chain. It improves the accuracy of the binding mode prediction as compared to Autodock | http://vina.scripps.edu/ |
| 17. | VinaMPI | It is open-source parallelization of Autodock vina. It reduces time of virtual screening. | https://github.com/mokarrom/mpi-vina |
| 18. | FlipDock | Flexible ligand and protein. It uses two FlexTree data structures to show complex of protein-ligand. | http://flipdock.scripps.edu/what-is-flipdock |
| 19. | POSIT | Ligand guided pose prediction (POSIT) uses the information from bound ligand for the improvement of pose prediction. | https://www.eyesopen.com/oedocking |
| 20. | idock | It's a tool of multithreaded virtual screening for docking of flexible ligand. | https://github.com/HongjianLi/idock |
| 21. | Rosetta Ligand | Included with de novo design, ligand docking, enzyme design and prediction of biological macromolecule structure. | https://www.rosettacommons.org/software |
| 22. | rDock | It is one of the fast and versatile open-source docking program. Suitable for campaigns of High throughput virtual screening and Binding mode prediction | http://rdock.sourceforge.net/ |
| 23. | Lead Finder | It introduces about 3 scoring functions in virtual screening experiment | http://moltech.ru/ |
| 24. | ADAM | Flexible ligand. Best suited solution for rational molecular design in case of unknown 3D structure of the target protein. | http://www.immd.co.jp/en/product_2.html |
| 25. | DockoMatic | It is intended to ease and automate the job of Auto Dock for the high throughput screening. | https://sourceforge.net/projects/dockomatic/ |
Table 2.
Structure and function of potential SARS-CoV-2 proteins.
| Sr. No. | Name of the Protein | Structure model | Function | Experimental Structure |
|---|---|---|---|---|
| Host translation inhibitor nsp1 GenBank: QHD43415_1 |
 |
It inhibits host translation by interacting with host 40 S subunit in ribosomal complexes, including the 43 S pre-initiation complex and the non-translating 80 S ribosome. | PDB: 7k3n, 7k7p |
|
| 1. | Non-structural protein 2 (nsp2) GenBank: QHD43415_2 |
 |
May play an important role in the modulation of host cell survival signaling pathway by interacting with host PHB and PHB2. | Only remote homologues were identified, no models have been built. |
| 2. | Papain-like proteinase (PLpro) GenBank: QHD43415_3 |
 |
Responsible for the cleavages located at the N-terminus of the replicase polyprotein. | PDB: 7KAG(range:1–111); 6W6Y(range:207–379); 6W9C(range:748–1060); |
| 3. | Non-structural protein 4 (nsp4) GenBank: QHD43415_4 |
 |
It Participates in the assembly of virally-induced cytoplasmic double-membrane vesicles necessary for viral replication. | NA (range:NA) |
| Main proteinase 3CL-PRO GenBank: QHD43415_5 |
 |
It cleaves the C-terminus of replicase polyprotein at 11 sites. It recognizes substrates containing the core sequence. | PDB: 6LU7(range:1–306) |
|
| 4. | RNA-directed RNA polymerase (RdRp) GenBank: QHD43415_11 |
 |
It Responsible for replication and transcription of the viral RNA genome. | PDB: 6M71(range:1–932); |
| 5. | Helicase (Hel). GenBank: QHD43415_12 |
 |
It's a Multi-functional protein with a zinc-binding domain in N-terminus displaying RNA and DNA duplex-unwinding activities with 5′ to 3′ polarity. | PDB: 5RL9(range:1–601); |
| 6. | Surface glycoprotein (Spike) GenBank: QHD43416 |
 |
S1 (13–685): attaches the virion to the cell membrane by interacting with host receptor, initiating the infection. S2 (686–1273): mediates fusion of the virion and cellular membranes by acting as a class I viral fusion protein. | PDB: 6VYB (open state) (range:1–1273); 6VXX (closed state) (range:1–1273); 6LXT(range:912–988,1164-1202); |
| 7. | Envelope Small Membrane Protein GenBank: QHD43418 |
 |
It Plays a central role in virus morphogenesis and assembly. It also plays a role in the induction of apoptosis | PDB: 7K3G(range:8–39) |
| 8. | Membrane Glycoprotein GenBank: QHD43419 |
 |
It's a component of the viral envelope that plays a central role in virus morphogenesis and assembly via its interactions with other viral proteins. | NA(range:NA) |
| 9. | ORF7a protein (ORF7a) GenBank: QHD43421 |
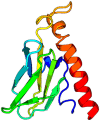 |
It Plays a role as antagonist of host tetherin (BST2), disrupting its antiviral effect. | PDB: 6w37, 7ci3 |
4. Computational drug repurposing
Drug repurposing (also called repositioning, reprofiling, redirecting, or rediscovering) is a strategy for identifying new uses and indications for existing or failed drugs. The scope of drug repurposing has increased in recent years as pharmaceutical companies seek potential inexpensive alternatives to compensate for the high costs along with the disappointing rate of success associated with the drug discovery pipeline (Baker et al., 2018). This strategy offers various advantages over the conventional drug development path such as, lower risk of failure, reduction in the time frame of drug development, and lower investment (Pushpakom et al., 2019). A plethora of computational methodologies have offered an avenue to repurpose drugs on small as well as large scales with the availability of high throughput data (Sadeghi and Keyvanpour, 2019). Computational drug repurposing approaches applied to COVID19 can be classified as network-based models, structure-based approaches and machine/deep learning approaches (Dotolo et al., 2020). Multiple drugs could be interrogated against particular targets involved in disease using structure-based drug design strategies. In the context of COVID19, significant efforts have been put into the computational studies for the repurposing of previous FDA-approved drugs (Wang and Guan, 2020).
5. SARS-CoV-2 genomics
SARS-CoV-2, a positive-stranded RNA virus belonging to orthocoronavirinae subfamily of coronaviridae family, is classified into five sub-genera including Alphacoronavirus, Betacoronavirus, Deltacoronavirus, and Gammacoronavirus (Cascella et al., 2020). Human CoV infections are mainly caused by the alpha- and beta-CoVs. The members of β-coronaviruses are SARS coronavirus (SARS-CoV) and MERS coronavirus (MERS-CoV), which can cause severe and potentially fatal respiratory tract infections (Jin et al., 2020a; Guo et al., 2020). The newly identified SARS-CoV-2 was found to share a genome sequence identity of 79.5% and 50% with SARS-CoV and MERS-CoV, respectively (Jin et al., 2020a). SARS-CoV-2 genome is approximately 30,000 nucleotides long. Constant mutations are making some of the single-nucleotide variations (SNVs) more resilient to the environment, with increased transmission efficiency. Apart from the 5′ leading and 3′ terminal sequences, the genome includes 11 coding regions for encoding spike glycoprotein (S), envelop protein (E), transmembrane glycoprotein (M), nucleocapsid protein (N), along with numerous open reading frames (ORFs) (ORF1ab, ORF3a, ORF6, ORF7a, ORF7b, ORF8, and ORF10) of different lengths wherein, ORF1ab includes ORF1a (containing nonstructural proteins genes nsp1 to nsp11) and ORF1b (containing nsp12 to nsp16) (Lung et al., 2020).
6. SARS-CoV-2 pathogenesis and molecular mechanism studies
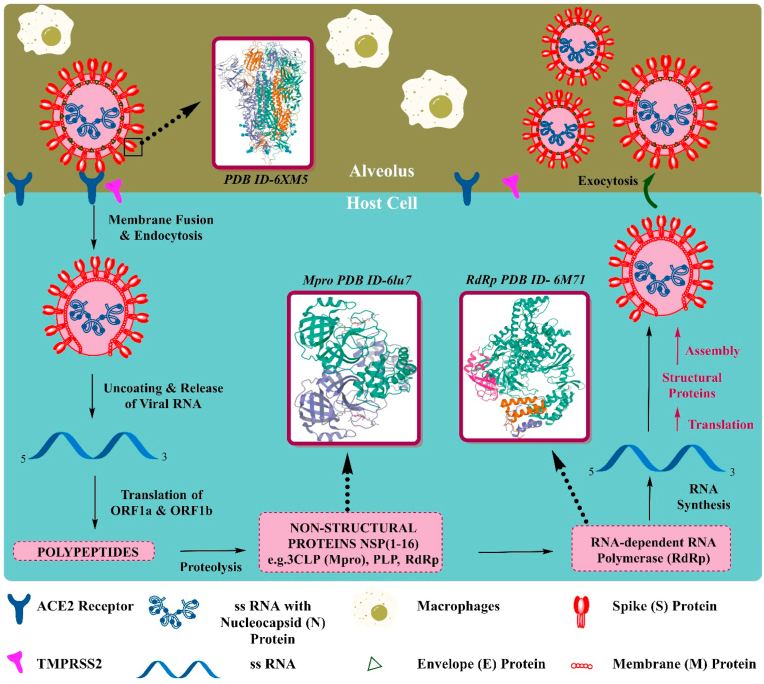
The virus undergoes various crucial steps after coming in contact with the host (Fig. 3): i) Attachment to the host cells (ii) Penetration into host the cells through endocytosis (iii) Biosynthesis of viral protein by using viral mRNA and (iv) Maturation and release of viral particles (Yuki et al., 2020). The functional and viral structure of the protein is mainly regulated by the four important structural proteins including the membrane protein(M), the envelope protein (E), the spike protein (S) and the nucleo-capsid protein(N) (Vellingiri et al., 2020). The entry of virus into the host cell is mediated through virion S-glycoprotein present on the surface of coronavirus, which can attach to ACE2 receptor in the lower respiratory tract in humans (Guo et al., 2020). The cell surface-associated transmembrane protease serine 2 (TMPRSS2) facilitates cell entry of the virus through the S-protein (Sanders et al., 2020). Once it enters the cells, the viral genome RNA is released into the cytoplasm of the cell. The uncoated RNA translates into two different polyproteins (pp1a and pp1ab), which encodes for non-structural protein and then synthesizes structural proteins after that viral genome starts to replicate (Guo et al., 2020; Li et al., 2020a). The newly formed envelope glycoproteins, genomic RNA, and nucleocapsid get assembled and form buds of the viral particle, mediated by endoplasmic reticulum (Guo et al., 2020). Finally, particle of virus containing vesicles fuses with the plasma membrane and the virus is released (Li et al., 2020a).
Fig. 3.
Life cycle of severe acute respiratory syndrome coronavirus (SARS-Covid-19) and its potential targets.
As the scientific research is progressing in COVID19, the molecular mechanism studies and structural informations are coming into the picture. Along with pathogenesis, other key biological processes involved in SARS-Cov-2 infection include the effect of disharmonic inflammatory signature (Parackova et al., 2020), the hallmark of circulating platelet-derived extracellular vesicles (Cappellano et al., 2021), and alteration of infectivity of the virus in the presence of osteopontin and furin (Adu-Agyeiwaah et al., 2020). Structural information at the atomic level can further improve understanding of the interactions between SARS-CoV-2 and its variants with infection susceptible cells. It will also help to identify robust target regions for antibody neutralization. The assistance of structure-based drug design may be vital in the fight against COVID 19. Several studies have shown that proteins of different coronaviruses including SARS-CoV-2, SARS-CoV and MERS-CoV display variations in the sequence, resulting in differences in the three-dimensional (3D) structures and thus also in the shape of their binding contacts (Hatmal et al., 2020). The SARS-CoV-2 entry into the host cells is an essential determinant of viral infectivity and pathogenesis and is also an essential target for host immune surveillance and human intervention strategies (Shang et al., 2020). The S glycoprotein, a major structural protein of the virus responsible for its entry by binding to the Angiotensin-Converting Enzyme 2 (ACE2) on the human host cells, attracted significant attention as target for the drug and vaccine development against SARS-CoV-2. The development of neutralizing antibodies, small molecules, and mini proteins aims to disrupt ACE2 binding to the receptor-binding-domain (RBD) (Papageorgiou and Mohsin, 2020). Another well-known SARS-CoV-2's main protease (Mpro) is mainly responsible for viral maturation and is the most attractive antiviral drug target. Mpro responsible for the cleavage, is necessary for the maturation of 12 nonstructural proteins (Nsp4-Nsp16) and critical proteins i.e., RNA-dependent RNA polymerase (RdRp, Nsp12) and helicase. The inhibition of Mpro is reported to prevent viral replication in multiple studies and is now the important target in drug discovery (Huynh et al., 2020). The understanding of various functional proteins of SARS-CoV-2 involved in the viral RNA replication machinery has also significantly improved our ability to design therapeutics (Romano et al., 2020).
7. Potential targets/proteins of COVID-19 for development of therapeutic agents
The drugs or therapies acting on coronavirus can be categorized on the basis of i) Specific pathways including inhibition of functional proteins involved in RNA synthesis ii) Blockage of the structural protein(s) Responsible for binding to human cell receptors iii) Alteration of factors that restore host innate immunity and acts on host's receptor or enzyme to prevent entry of virus into the cell (Wu et al., 2020). The novel coronavirus encodes both structural and non-structural proteins. The structural proteins include spike (S) protein, membrane protein(M), envelope protein(E), and the nucleocapsid protein(N) (Ahmed et al., 2020). Several protein targets are included in the database, encoded by both virus and human genomes. Identifying potential drug targets and proteins (both viral and human) to reduce the false-negative and enhance the success rate are the major challenges for the search of potential drug molecule (Shi et al., 2020). The potential and promising targets for COVID-19 are included in the following sections.
7.1. SARS-CoV-2 main protease
SARS-CoV-2 is related to the other beta coronaviruses, including SARS-CoV. Further, 96.08% of identity has been observed between Mpro of SARS-CoV-2 and SARS CoV on sequence comparisons (Kandeel and Al-Nazawi, 2020). The main protease (Mpro) target received significant attention as compared to the other corona viral targets studied in the past, particularly in the first SARS-CoV (Ullrich et al., 2020). The main protease (Mpro) is one of the conserved and attractive drug targets for the discovery of an anti-coronavirus drug, due to its important role in post-translational processing of polyproteins (Zhang et al., 2020b; Havranek and Islam, 2020; Kumar et al., 2020). The replicase gene encodes with two overlapping polyproteins i.e. pp1a and pp1ab, that are essential for the transcription and viral replication (Jin et al., 2020b). Mpro breaks the polyproteins by proteolytic processing and releases the functional polypeptide essential for replicating new viruses (Havranek and Islam, 2020; Jin et al., 2020b). Polyprotein 1 ab (pp1ab) and Mpro affect at least 11 cleavage sites and viral replication can be prevented by inhibiting the enzyme (Zhang et al., 2020b). Further, the inhibitors are unlikely to be toxic due to the non-homologous sequence of 2019-nCOV Mpro to human host-pathogen (Naik et al., 2020). Different crystal structures of the main protease (Mpro) of novel COVID-19 are deposited in the Protein data bank PDB (Berman et al., 2000) to identify potential compounds. The crystal structure of main protease with PDB ID's 6LU7 was consistently used in the in silico virtual screening for identification of potential inhibitors. Further, the crystal structure of SARS-CoV-2 Mpro complexed with N3 is determined in resolution of 2.1 Å. It contains about 1–306 residues and each protomer is composed of three different domains. The inhibitor, N3 forms multiple hydrogen bondings with the main chain of the residues present in the substrate-binding pocket to lock the inhibitor (Jin et al., 2020c) (Fig. 5). The hydrogen bond and van der waals interactions between the inhibitor and residue in the substrate-binding pockets of Mpro in the crystallographic electron density maps of N3, is suitable to guide the designing of improved compounds (Arafet et al., 2021).
Fig. 5.
Molecular interactions of compound N3 with SARS-CoV-2 Mpro (Protein Data Bank ID: 6LU7).
7.2. SARS-CoV-2 RNA dependent RNA polymerase (RdRp)
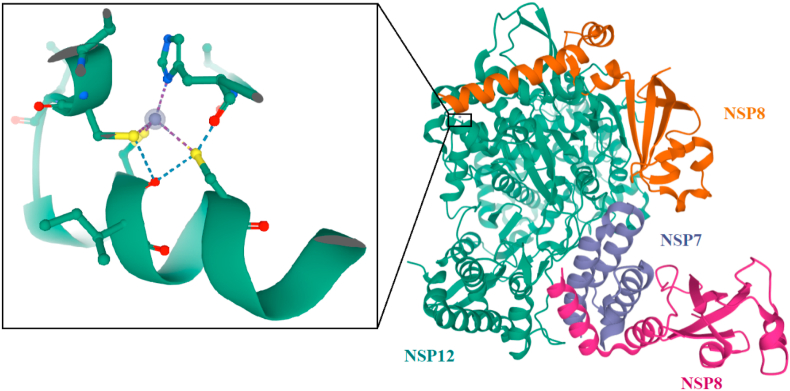
The enzyme, RNA-dependent RNA polymerase (RdRp), also named as nsp12, plays an important role in replicating and transcribing the life cycle of the COVID 19 virus by catalyzing the synthesis of viral RNA with the help of co-factors, nsp7 and nsp8 (Gao et al., 2020b). The nsp12 polymerase was predicted to contain about 932 amino acids located in the polyprotein (Mirza and Froeyen, 2020). On comparison, the amino acid sequences of RdRp in both severe acute respiratory syndrome coronavirus (SARS-CoV) and SARS-CoV-2 were found to be remarkably similar (Lung et al., 2020). It has a deep groove as an active site for RNA polymerization and variations in the residue are distal to the active site (Lung et al., 2020). In the process of replicating RNA, nsp12subunit is required to bind with NSP7 and NSP8 co-factors to improve its capability (Ruan et al., 2020). The compounds that disrupt the binding of nsp7 or nsp8 to nsp12 could inhibit the activity of RdRpnsp12 (Ruan et al., 2020). Therefore, nsp12 is considered as the primary target to identify potential compounds for the treatment of COVID-19 viral infection (Gao et al., 2020b).
The cryo-EM structure of PDB ID 6NUR showed the nsp12 polymerase bound with the co-factor NSP7-NSP8 with a second subunit of NSP8 (Kirchdoerfer and Ward, 2019) (Fig. 6). The crystal structure contained two metal binding sites, to which zinc atoms were assigned (Farag et al., 2020).
Fig. 6.
The crystal structure of NSP12 polymerase bound with co-factor NSP7-NSP8 with a second subunit of NSP8 in complex with zinc atom (Protein Data Bank ID: 6NUR).
7.3. SARS-CoV-2 spike (S) protein
The entry mechanism of SARS-CoV-2 into human cells begins with the binding of SARS-CoV-2 spike (S) protein to the human angiotensin-converting enzyme 2 (ACE2) receptor through the receptor-binding domain (Senathilake et al., 2020). S protein is trimeric, cleaved into S1 and S2 subunits and mediates receptor recognition and membrane fusion function. S1 subunit contains the receptor-binding domain (RBD), which binds to the peptide domain of the ACE2 and S2 is responsible for membrane fusion (Yan et al., 2020). The receptor-binding domain is the key functional component of S1 subunit and is required for binding of SARS-SoV-2 to ACE2 (Shi, Wang, Zhang, Wang). The variation in attachment and the entry receptor observed among different coronaviruses are due to the use of distinct domains within the S1 subunit (Walls et al., 2020). It was reported that S protein mediates activation of host immune response against the virus (Narkhede et al., 2020). S protein, an important target of neutralizing antibodies upon infection, can be used for designing therapeutics and vaccines (Walls et al., 2020).
7.4. Miscellaneous proteins
The other target proteins of SARS-Co-2 include E protein, TMPRSSS2, ORF7a, ACE2, helicase, C-terminal RNA binding domain (CRBD), N-terminal RNA binding domain (NRBD), Nsp3 (Nsp3b, Nsp3c, PLpro, and Nsp3e), Nsp1, Nsp7_Nsp8 complex, Nsp9-Nsp10, and Nsp14-Nsp16 and have also been considered for the virtual screening (Wu et al., 2020).
8. Clinically available drugs for COVID-19 evaluated through SBDD
8.1. Remdesivir
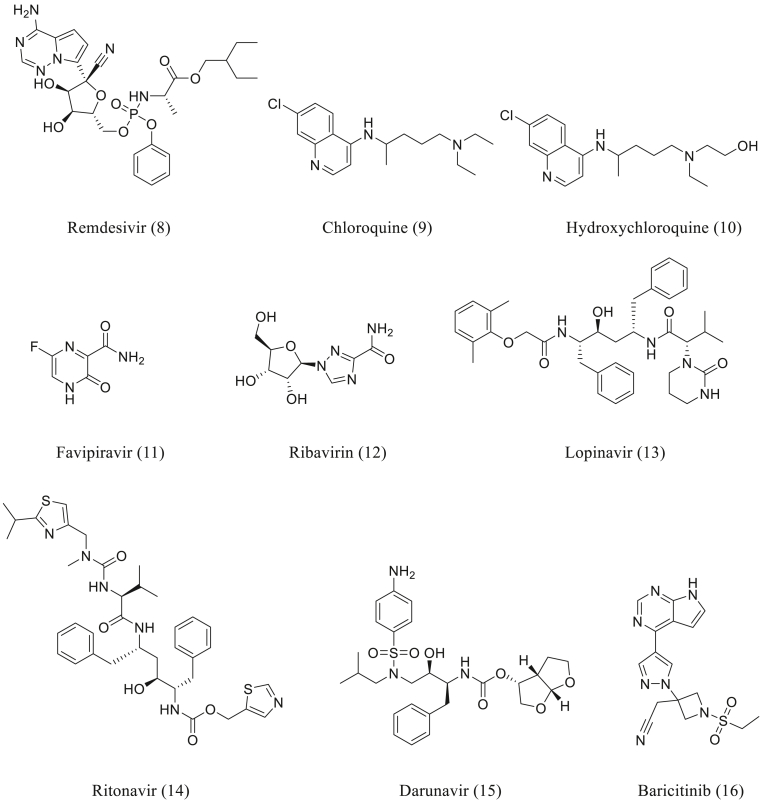
Remdesivir (Fig. 4 (Fallerini et al., 2021)) was emerged from the joint collaborative efforts of Gilead Sciences, U.S. Centers for Disease Control and Prevention (CDC) and the U.S. Army Medical Research Institute of Infectious Diseases (USAMRIID). The journey of the discovery of Remdesivir started with the compilation of nucleoside analogues library based on the prior knowledge of effective nucleoside based antiviral compounds (Eastman et al., 2020). It is a prodrug, which gets converted into an active metabolite of C-adenosine nucleoside triphosphate analog (Sanders et al., 2020). Remdesivir shows broad-spectrum antiviral activity against RNA viruses viz. filoviruses, paramyxoviruses, and coronaviruses (Rosa and Santos, 2020). The drug has been found to be potent, in in vitro, against SARS-CoV-2 with EC50 and EC90 of 0.77 μM and 1.76 μM, respectively (Sanders et al., 2020). One of the clinical trials (NCT04292899, NCT04292730, NCT04257656, NCT04252664, NCT04280705) of Remdesivir, on the patients with COVID 19, produced faster clinical improvement than that of placebo (Wang et al., 2020b). Although, questions did arise with respect to the efficacy of the drug. A more recent double-blind, randomized, placebo-controlled ACTT-1 trial in hospitalized patients with COVID-19 established a shortened recovery time compared to placebo and fewer cases of progression to more severe respiratory disease (Beigel et al., 2020). The molecular docking studies of the drug was carried out by using different protein structures of SARS-CoV-2. The active metabolite of Remdesivir (RemTP) has a relatively substantial binding free energy of −8.28 ± 0.65 kcal/mol, against SARS-CoV-2 nsp12 RNA-dependent RNA polymerase (RdRp), as compared to the natural substrate ATP (- 4.14 ± 0.89 kcal/mol), which is essential for the polymerization (Zhang and Zhou, 2020). Remdesivir has good binding affinity for 2019-nCoV Mpro with N3 peptide (PDB ID: 6LU7) with docking score of −8.2 kcal/mol and it forms a strong hydrogen bond with Mpro residues such as Cys145, and His164 (Naik et al., 2020).
Fig. 4.
Drugs being repurposed for COVID-19.
8.2. Chloroquine/hydroxychloroquine
Chloroquine (Fig. 4 (Zhang et al., 2020a)) is the first natural antimalarial agent derived from the bark of the cinchona tree and the synthetic route of its analogues originated from the works of Paul Ehrlich's group. The study of its structural variations led to the discovery of the Hydroxychloroquine with 3 folds less toxicity (Al-Bari, 2015). Both the drugs, Chloroquine and Hydroxychloroquine (Fig. 4 (9 and 10)) are widely used for the prevention and treatment of malaria. Hydroxychloroquine is already under clinical trials for the treatment of acquired immune deficiency syndrome (AIDS) (Rosa and Santos, 2020). It is reported to have antiviral as well as immune-modulating properties (Vellingiri et al., 2020). The antiviral activity of Chloroquine and Hydroxychloroquine's is considered to be by blocking the viral entry into cells due to inhibition of glycosylation of host receptors, proteolytic processing, and endosomal acidification (Sanders et al., 2020).
Hydroxychloroquine (EC50 = 0.72 μM) was found to be more potent than chloroquine (EC50 = 5.47 μM) in in vitro evaluation, when tested using SARS-CoV-2 infected Vero cells. (Yao et al., 2020). However, the combinatorial therapy involving Chloroquine/Hydroxychloroquine with Azithromycin showed a prolongation of the corrected QTc interval as an important adverse effect. Most of the observational studies involving hospitalized COVID-19 patients, Hydroxychloroquine did not show greatly lowered or an increased risk of death/intubation (Geleris et al., 2020; Li et al., 2020b). It was followed by randomized, double-blind, placebo-controlled trials involving non-hospitalized adults with COVID-19, which showed a lack of difference in the symptom severity score over 14 days for the drug versus placebo (Skipper et al., 2020). Hydroxychloroquine showed considerable binding efficacy and inhibited various drug targets of SARS-CoV-2 including Spike glycoprotein, RNA dependent RNA polymerase, Chimeric RBD, Main protease, Non-structural Protein 3, Non-structural Protein 9, and ADP-ribose-1 monophosphatase. The Structural and molecular modeling studies of Chloroquine and Hydroxychloroquine against SARS-CoV-2 protein revealed that chloroquine (or active derivative, hydroxychloroquine) binds to sialic acids and gangliosides of the host cell with high affinity, which ultimately cease binding of viral S protein to gangliosides (Fantini et al., 2020).
8.3. Favipiravir
Favipiravir (T-705) (Fig. 4 (Helmy et al., 2020) was discovered and synthesized by Toyoma Chemical Co., Ltd. through phenotypic screening against influenza virus (Furuta et al., 2013). Favipiravir (11), a prodrug of purine nucleotide, is converted into an active phosphoribosylated form of Favipiravir ribofuranosyl-5′-triphosphate in the cells, that inhibits RNA polymerase activity (Sanders et al., 2020). It mainly shows anti-influenza virus activity but also has been demonstrated to block the replication of flavi-, filo-, alpha-, bunya-, noro-, arena-, and other RNA viruses (Dong et al., 2020b). Favipiravir inhibited SARS-CoV-2 in vitro with the EC50 of 61.88 μM/L in Vero E6 cells (Sanders et al., 2020). The clinical trials for the treatment of COVID 19 were initiated by the Clinical Medical Research Center of the National Infectious Diseases and the Third People's Hospital of Shenzhen. Preliminary results showed potent antiviral action (Dong et al., 2020b). The in silico, activity of Favipiravir, against COVID 19, were evaluated with both protease (6LU7) and polymerase (6NUR) macromolecules. The molecular docking simulation results showed interaction of different tautomers of Favipiravir with targets (Harismah and Mirzaei, 2020).The structural analysis of Favipiravir bound to the replicating polymerase complex of SARSCoV-2 in the pre-catalytic state revealed that, it inhibited viral replication mainly by inducing mutations in progeny RNAs which impaired the elongation of RNA products (Harismah and Mirzaei, 2020).
8.4. Ribavirin
Ribavirin (Fig. 4 (12)) was first synthesized in 1972 by Sidewell and colleagues as a broad-spectrum antiviral acting against various virus families, mainly the negative RNA strand virus and respiratory syncytial virus (Brillanti et al., 2011). It is a guanine analog with broad-spectrum antiviral activity, inhibiting viral RNA-dependent RNA polymerase (Sanders et al., 2020). This drug is generally used for the treatment of hepatitis C as a combination with interferon α (IFN) (Vellingiri et al., 2020). Ribavirin has shown in vitro activity towards WIV04 strain of 2019-nCoV and is undergoing clinical trials (NCT04276688, NCT04306497) for the treatment of COVID-19 (Khalili et al., 2020). Further, promising results were obtained in molecular docking studies of ribavirin performed against both the protein structure, SARS-CoV-2 RdRp and COVID-19 main protease in complexation with an inhibitor N3(PDB ID 6LU7). The results with main protease produced a binding affinity of −5.6 (kcal/mol) in the pocket of chain A, owing to the formation of four hydrogen bonds (Narkhede et al., 2020).However, docking of Ribavirin against SARS-CoV-2 RdRp (PDB ID: 6NUR, chain A) revealed interactions established by 13H-bonds with W508, Y510, K512, C513, D514, N582, D651 (Sohrabi et al., 2020), A653 (Sohrabi et al., 2020), and W691 of the SARS-CoV-2 RdRp with the binding energy of −7.8 kcal/mol (Elfiky, 2020).
8.5. Lopinavir-ritonavir
The ritonavir (ABT-538) (Fig. 4 (Wacharapluesadee et al., 2021), a symmetry-based HIV protease inhibitor was identified with the lower rate of metabolism and high oral bioavailability (Kempf et al., 1998). The combination of Lopinavir (Fig. 4 (13)) and Ritonavir (Fig. 4 (14)) were approved by the USFDA for the treatment of HIV (Sanders et al., 2020). Both the drugs act through inhibition of HIV protease (Rosa and Santos, 2020). Ritonavir also inhibits another enzymes, cytochrome P450 3A4, responsible for the metabolism of Lopinavir, that indirectly prolongs bioavailability of Lopinavir (Choy et al., 2020). The combination of Lopinavir/Ritonavir was first recommended by the National Health Commission (NHC) of the People's Republic of China for the experimental treatment of COVID-19 (Dong et al., 2020b). Lopinavir and ritonavir showed in vitro antiviral activity against SARS-CoV-2 in Vero E6 cells with an EC50 of 26.1 μM.16 and EC50 of >100 μM respectively (Maciorowski et al., 2020).
The antiviral agents reported, numbering 61, were docked against COVID-19 main protease co-crystallized structure (PDB IDs: 5R7Y, 5R7Z, 5R80, 5R81, 5R82). Among all the agents, only Lopinavir showed binding with all the protein structures with docking scores of less than −6.5 kcal/mol. Further, Lopinavir and Ritonavir were effective in in silico, in vitro and animal models but were less effective in the clinical studies (Shah et al., 2020).
8.6. Darunavir
Darunavir (Fig. 4 (15)) was design based on the ‘Backbone binding concept’ which has emerged as a useful approach to combat drug resistance. It is a multi-drug resistant HIV-1 virus inhibitor and received US FDA approval in 2006 for the treatment of patients harboring highly drug-resistant viruses, further approval was extended to all HIV/AIDS patients in 2008 (Ghosh and Chapsal, 2013). The structure based drug design strategies targeting HIV-1 protease backbone resulted Darunavir, as one of the exceedingly potent nonpeptidyl Inhibitors (Ghosh et al., 2007). It was found to show in vitro inhibition of SARS-CoV-2 infection and potential efficacy in the treatment of COVID-19 (Dong et al., 2020b). Cell based experiments indicated that Darunavir significantly inhibited viral replication at a concentration of 300 μM in in vitro and its inhibition efficiency was 280-folds in comparison to the untreated group (Dong et al., 2020b). Darunavir in combination with cobicistat is undergoing clinical trials in patients with COVID 19 and is approved by the US FDA for the treatment of AIDS (Rosa and Santos, 2020). The model, papain-like viral protease (PLVP) of orf1ab polyprotein of SARS-CoV-2 has been developed and docking of drugs is being carried out. The inhibitory effect of Darunavir on SARS-CoV-2 observed in the study may be due to its inhibitory effect on PLVP (Lin et al., 2020). The binding free energy of −11.96 ± 1.99 kcal/mol by Darunavir suggested that it may potentially inhibit SARS-CoV-2 Mpro with high affinities (Ngo et al., 2020).
8.7. Baricitinib
Baricitinib (Fig. 4 (16)) was approved by FDA in 2017 for the treatment of rheumatoid arthritis (RA) (Mullard, 2018). It is a Janus kinase (JAK) inhibitor mainly used in the management of rheumatoid arthritis (Santos et al., 2020). The inhibition of JAK interrupts the signaling of multiple cytokines pathways, that are likely to be effective in patients with COVID-19 (Stebbing et al., 2020). In a study by machine learning tool (BenevolentAI), it was evident that Baricitinib reduced the ability of virus to infect the lung cells (Richardson et al., 2020). It is reported to bind with the adaptor-associated kinase-1 (AAK1), that plays an important role in the clathrin-mediated viral endocytosis (Santos et al., 2020). The drug may have antiviral activity due to its high affinity towards AAK1-binding, which disrupts the viral entry and intracellular assembly of virus particles (Richardson et al., 2020). The docking simulations studies between Baricitinib and SARS-CoV-2 main protease (PDB ID: 6LU7) were also performed. The receptor-ligand complex showed ten interaction sites with the amino acid residues of domain III of receptor and binding energy of −6.5 (kcal/mol) (Marinho et al., 2020).
9. Conclusions
SBDD is a faster, highly cost effective and efficient tool for the lead drug discovery in comparison to the traditional approaches. It has rapidly gained the acceptance of researchers to design and optimize ligands and can greatly influence the discovery of new drug molecules for the treatment of COVID-19. Over past two decades, technological advancements in experimental approaches including computational, X-ray crystallography and NMR etc. have facilitated SBDD approach to its vital position in drug discovery. Owing to the substantial increase in the availability of huge number of 3D structure of targets and the rapid progression in computational chemistry, SBDD is now a fundamental strategy for both lead generation and optimization. Multiple drugs can be interrogated with the help of SBDD strategies against particular target(s) involved in the disease process. Remdesivir (Fig. 4 (Fallerini et al., 2021)), analyzed using SBDD approach, is a potential ‘molecule of hope’ for the treatment of SARS-CoV-2. It is used in restricted emergency to patients with moderate to severe COVID-19 infection. This triphosphate nucleotide analog inhibits the viral RNA synthesis by a specific mechanism of delayed chain termination in SARS-CoV-2. The key interactions of Remdesivir with crucial Mpro residues, Cys145, and His164 also suggest that the drug molecules are breakthrough analogs for the treatment of COVID-19. SBDD approach has been widely realized and accepted as a crucial part of COVID-19 drug research and it serves the drug discovery researchers in making an effort to expedite and an added level of confidence.
10. Future perspectives
The approaches and methodologies used in the designing of drugs have undergone transformation over a period of time. At present, drug design approaches accomplish and drive new technological advances to solve the varied, new, difficult setbacks, which occur along the path to drug discovery. As the advances in structural genomics, bioinformatics, and computational power continue to expand, further successes in structure-based drug design are likely to follow. The global population is going to face radical health emergencies in the future, which could be overcome with the help of technological and knowledge-based advancements. SBDD would be more beneficial not just for the discovery of lead compounds but also in the entire process of optimization and development. Allosteric drug design is one of the novel approaches in drug discovery, where the identification of allosteric sites and allosteric modulation of protein function are mainly carried out. The functional activity of protein can be regulated through alteration of its confirmation intrinsically on the binding of a ligand at an allosteric site, that is topographically distinct from the orthosteric site (Lu et al., 2014). The SBDD methods can be used to discover and optimize allosteric modulator properties, which are responsible for the alteration of ligand binding, signaling, function of the protein and represent a massive opportunity for the development of novel therapeutics (Congreve et al., 2017). The investigation of the allosteric nature of the S-ACE2 complex will help to identify and quantify the extent to which the S protein binds to ACE2. It also allows the design and test of allosteric drugs with enhanced bioactivity (Di Paola et al., 2020). The Gaussian accelerated molecular dynamics are also used to elucidate cryptic and allosteric pockets present within the SARS-CoV-2 protease (Sztain et al., 2020). The allosteric drug discovery approach is developing rapidly and may play a crucial role in identifying therapeutics for SARS-CoV-2 (Di Paola et al., 2020). Every year, new targets are being identified and the structures of targets are being determined at an astonishing rate. Our capability to capture a quantitative picture of the interactions between macromolecules and ligands is also advancing. The progresses made in SBDD technology holds an important role in the identification of new ‘lead’ compounds to treat future, unexpected pandemics.
CRediT authorship contribution statement
Nilesh Gajanan Bajad: Conceptualization, Methodology, Writing – original draft. Swetha Rayala: Conceptualization, Methodology. Gopichand Gutti: Conceptualization, Methodology, Writing – review & editing. Anjali Sharma: Writing – review & editing. Meenakshi Singh: Supervision. Ashok Kumar: Supervision. Sushil Kumar Singh: Supervision.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper. NGB, SR, GG and AS are thankful to Indian Institute of Technology ( Banaras Hindu University) Varanasi for providing teaching assistantships to them.
References
- Adu-Agyeiwaah Y., Grant M.B., Obukhov A.G.J.C. The potential role of osteopontin and furin in worsening disease outcomes in COVID-19 patients with pre-existing diabetes. 2020;9(11):2528. doi: 10.3390/cells9112528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agnello S., Brand M., Chellat M.F., Gazzola S., Riedl RJACIE A structural view on medicinal chemistry strategies against drug resistance. 2019;58(11):3300–3345. doi: 10.1002/anie.201802416. [DOI] [PubMed] [Google Scholar]
- Ahmed S.F., Quadeer A.A., McKay MRJV Preliminary identification of potential vaccine targets for the COVID-19 coronavirus (SARS-CoV-2) based on SARS-CoV immunological studies. 2020;12(3):254. doi: 10.3390/v12030254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Bari MAAJJoAC Chloroquine analogues in drug discovery: new directions of uses, mechanisms of actions and toxic manifestations from malaria to multifarious diseases. 2015;70(6):1608–1621. doi: 10.1093/jac/dkv018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Khafaji K., AL-DuhaidahawiL D., Taskin Tok TJJoBS . 2020. Dynamics. Using integrated computational approaches to identify safe and rapid treatment for SARS-CoV-2; pp. 1–11. (just-accepted) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson A.C.J.C. biology. The process of structure-based drug design. 2003;10(9):787–797. doi: 10.1016/j.chembiol.2003.09.002. [DOI] [PubMed] [Google Scholar]
- Anderson A.C. Springer; 2012. Structure-based Functional Design of Drugs: from Target to Lead Compound. Molecular Profiling; pp. 359–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arafet K., Serrano-Aparicio N., Lodola A., Mulholland A.J., González F.V., Świderek K., et al. Mechanism of inhibition of SARS-CoV-2 M pro by N3 peptidyl Michael acceptor explained by QM/MM simulations and design of new derivatives with tunable chemical reactivity. 2021;12(4):1433–1444. doi: 10.1039/d0sc06195f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai Y., Yao L., Wei T., Tian F., Jin D.-Y., Chen L., et al. Presumed asymptomatic carrier transmission of COVID-19. 2020;323(14):1406–1407. doi: 10.1001/jama.2020.2565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker N.C., Ekins S., Williams A.J., Tropsha AJDdt. A bibliometric review of drug repurposing. 2018;23(3):661–672. doi: 10.1016/j.drudis.2018.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batool M., Ahmad B., Choi SJIjoms. A structure-based drug discovery paradigm. 2019;20(11):2783. doi: 10.3390/ijms20112783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beigel J.H., Tomashek K.M., Dodd L.E., Mehta A.K., Zingman B.S., Kalil A.C., et al. 2020. Remdesivir for the Treatment of Covid-19. [DOI] [PubMed] [Google Scholar]
- Berman H.M., Westbrook J., Feng Z., Gilliland G., Bhat T.N., Weissig H., et al. vol. 28. 2000. pp. 235–242. (The Protein Data Bank). 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohacek R.S., McMartin C., Guida WCJMrr The art and practice of structure-based drug design: a molecular modeling perspective. 1996;16(1):3–50. doi: 10.1002/(SICI)1098-1128(199601)16:1<3::AID-MED1>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- Brillanti S., Mazzella G., Roda E.J.D., Disease L. Ribavirin for chronic hepatitis C: and the mystery goes on. 2011;43(6):425–430. doi: 10.1016/j.dld.2010.10.007. [DOI] [PubMed] [Google Scholar]
- Buchanan SGJCOiDD Development. Structural genomics: bridging functional genomics and structure-based drug design. 2002;5(3):367–381. [PubMed] [Google Scholar]
- Bung N., Krishnan S.R., Bulusu G., Roy AJFmc. De novo design of new chemical entities for SARS-CoV-2 using artificial intelligence. 2021;13:575–585. doi: 10.4155/fmc-2020-0262. 06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cappellano G., Raineri D., Rolla R., Giordano M., Puricelli C., Vilardo B., et al. Circulating platelet-derived extracellular vesicles are a hallmark of sars-cov-2 infection. 2021;10(1):85. doi: 10.3390/cells10010085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cascella M., Rajnik M., Cuomo A., Dulebohn S.C., Di Napoli R. StatPearls Publishing; 2020. Features, Evaluation and Treatment Coronavirus (COVID-19) Statpearls [internet] [PubMed] [Google Scholar]
- Cevik M., Bamford C., Ho A.J.C.M., Infection . 2020. COVID-19 Pandemic–A Focused Review for Clinicians. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choudhury CJJoBS . 2020. Dynamics. Fragment tailoring strategy to design novel chemical entities as potential binders of novel corona virus main protease; pp. 1–15. (just-accepted) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choy K.-T., Wong A.Y.-L., Kaewpreedee P., Sia S.-F., Chen D., Hui K.P.Y., et al. 2020. Remdesivir, Lopinavir, Emetine, and Homoharringtonine Inhibit SARS-CoV-2 Replication in Vitro; p. 104786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Congreve M., Oswald C., Marshall FHJTips Applying structure-based drug design approaches to allosteric modulators of GPCRs. 2017;38(9):837–847. doi: 10.1016/j.tips.2017.05.010. [DOI] [PubMed] [Google Scholar]
- Di Paola L., Hadi-Alijanvand H., Song X., Hu G., Giuliani AJJopr. The discovery of a putative allosteric site in the SARS-CoV-2 spike protein using an integrated structural/dynamic approach. 2020;19(11):4576–4586. doi: 10.1021/acs.jproteome.0c00273. [DOI] [PubMed] [Google Scholar]
- Dong Y., Dai T., Wei Y., Zhang L., Zheng M., Zhou FJSt, et al. A systematic review of SARS-CoV-2 vaccine candidates. 2020;5(1):1–14. doi: 10.1038/s41392-020-00352-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong L., Hu S., Gao JJDd. therapeutics. Discovering drugs to treat coronavirus disease 2019 (COVID-19) 2020;14(1):58–60. doi: 10.5582/ddt.2020.01012. [DOI] [PubMed] [Google Scholar]
- Dotolo S., Marabotti A., Facchiano A., Tagliaferri RJBiB. 2020. A Review on Drug Repurposing Applicable to COVID-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eastman R.T., Roth J.S., Brimacombe K.R., Simeonov A., Shen M., Patnaik S., et al. 2020. Remdesivir: A Review of its Discovery and Development Leading to Emergency Use Authorization for Treatment of COVID-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elfiky AAJLs. 2020. Ribavirin, Remdesivir, Sofosbuvir, Galidesivir, and Tenofovir against SARS-CoV-2 RNA Dependent RNA Polymerase (RdRp): A Molecular Docking Study; p. 117592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fallerini C., Daga S., Mantovani S., Benetti E., Picchiotti N., Francisci D., et al. vol. 10. 2021. (Association of Toll-like Receptor 7 Variants with Life-Threatening COVID-19 Disease in Males: Findings from a Nested Case-Control Study). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang YJEoodd. Ligand–receptor interaction platforms and their applications for drug discovery. 2012;7(10):969–988. doi: 10.1517/17460441.2012.715631. [DOI] [PubMed] [Google Scholar]
- Fantini J., Di Scala C., Chahinian H., Yahi NJIjoaa. 2020. Structural and Molecular Modeling Studies Reveal a New Mechanism of Action of Chloroquine and Hydroxychloroquine against SARS-CoV-2 Infection; p. 105960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farag A., Wang P., Ahmed M., Sadek H. 2020. Identification of FDA Approved Drugs Targeting COVID-19 Virus by Structure-Based Drug Repositioning. [Google Scholar]
- Fauci A.S., Lane H.C., Redfield R.R. Mass Medical Soc; 2020. Covid-19—navigating the Uncharted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira L.G., Dos Santos R.N., Oliva G., Andricopulo A.D.J.M. Molecular docking and structure-based drug design strategies. 2015;20(7):13384–13421. doi: 10.3390/molecules200713384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer T., Gazzola S., RJEoodd Riedl. Approaching target selectivity by de novo drug design. 2019;14(8):791–803. doi: 10.1080/17460441.2019.1615435. [DOI] [PubMed] [Google Scholar]
- Furuta Y., Gowen B.B., Takahashi K., Shiraki K., Smee D.F., Barnard DLJAr. Favipiravir (T-705), a novel viral RNA polymerase inhibitor. 2013;100(2):446–454. doi: 10.1016/j.antiviral.2013.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Q., Bao L., Mao H., Wang L., Xu K., Yang M., et al. 2020. Development of an Inactivated Vaccine Candidate for SARS-CoV-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Y., Yan L., Huang Y., Liu F., Zhao Y., Cao L., et al. Structure of the RNA-dependent RNA polymerase from COVID-19 virus. 2020;368(6492):779–782. doi: 10.1126/science.abb7498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geleris J., Sun Y., Platt J., Zucker J., Baldwin M., Hripcsak G., et al. 2020. Observational Study of Hydroxychloroquine in Hospitalized Patients with Covid-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GenOMICC Investigators II, Covid-19 Human Genetics Initiative, 23andMe Investigators, Bracovid Investigators, Gen-Covid Investigators %J Nature . 2020. Genetic Mechanisms of Critical Illness in COVID-19. [Google Scholar]
- Ghosh A.K., Chapsal B.D. Elsevier; 2013. Design of the Anti-HIV Protease Inhibitor Darunavir. Introduction to Biological and Small Molecule Drug Research and Development; pp. 355–384. [Google Scholar]
- Ghosh A.K., Dawson Z.L., Mitsuya H.J.B. Chemistry m. Darunavir, a conceptually new HIV-1 protease inhibitor for the treatment of drug-resistant HIV. 2007;15(24):7576–7580. doi: 10.1016/j.bmc.2007.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gohlke H., Hendlich M., GJJomb Klebe. Knowledge-based scoring function to predict protein-ligand interactions. 2000;295(2):337–356. doi: 10.1006/jmbi.1999.3371. [DOI] [PubMed] [Google Scholar]
- Grinter S.Z., Zou X.J.M. Challenges, applications, and recent advances of protein-ligand docking in structure-based drug design. 2014;19(7):10150–10176. doi: 10.3390/molecules190710150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y.-R., Cao Q.-D., Hong Z.-S., Tan Y.-Y., Chen S.-D., Jin H.-J., et al. The origin, transmission and clinical therapies on coronavirus disease 2019 (COVID-19) outbreak–an update on the status. 2020;7(1):1–10. doi: 10.1186/s40779-020-00240-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halperin I., Ma B., Wolfson H., Nussinov R.J.P.S. Function, Bioinformatics. Principles of docking: an overview of search algorithms and a guide to scoring functions. 2002;47(4):409–443. doi: 10.1002/prot.10115. [DOI] [PubMed] [Google Scholar]
- Harismah K., Mirzaei MJAJoC Section B: natural products, chemistry M. Favipiravir: structural analysis and activity against COVID-19. 2020;2(2):55–60. [Google Scholar]
- Hatmal MmM., Alshaer W., Al-Hatamleh M.A., Hatmal M., Smadi O., Taha M.O., et al. Comprehensive structural and molecular comparison of spike proteins of SARS-CoV-2, SARS-CoV and MERS-CoV, and their interactions with ACE2. 2020;9(12):2638. doi: 10.3390/cells9122638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Havranek B., Islam SMJJoBS . 2020. Dynamics. An in silico approach for identification of novel inhibitors as potential therapeutics targeting COVID-19 main protease; pp. 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helmy Y.A., Fawzy M., Elaswad A., Sobieh A., Kenney S.P., Shehata AAJJoCM The COVID-19 pandemic: a comprehensive review of taxonomy, genetics, epidemiology, diagnosis, treatment, and control. 2020;9(4):1225. doi: 10.3390/jcm9041225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes E.A., O'Connor R.C., Perry V.H., Tracey I., Wessely S., Arseneault L., et al. 2020. Multidisciplinary Research Priorities for the COVID-19 Pandemic: a Call for Action for Mental Health Science. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huynh T., Wang H., Luan BJTjopcl. In silico exploration of the molecular mechanism of clinically oriented drugs for possibly inhibiting SARS-CoV-2’s main protease. 2020;11(11):4413–4420. doi: 10.1021/acs.jpclett.0c00994. [DOI] [PubMed] [Google Scholar]
- Jin Y., Yang H., Ji W., Wu W., Chen S., Zhang W., et al. Virology, epidemiology, pathogenesis, and control of COVID-19. 2020;12(4):372. doi: 10.3390/v12040372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin Z., Du X., Xu Y., Deng Y., Liu M., Zhao Y., et al. 2020. Structure of M Pro from SARS-CoV-2 and Discovery of its Inhibitors; pp. 1–5. [DOI] [PubMed] [Google Scholar]
- Jin Z., Du X., Xu Y., Deng Y., Liu M., Zhao Y., et al. Structure of M pro from SARS-CoV-2 and discovery of its inhibitors. 2020;582(7811):289–293. doi: 10.1038/s41586-020-2223-y. [DOI] [PubMed] [Google Scholar]
- Kalyaanamoorthy S., Chen Y-PPJDdt Structure-based drug design to augment hit discovery. 2011;16(17–18):831–839. doi: 10.1016/j.drudis.2011.07.006. [DOI] [PubMed] [Google Scholar]
- Kandeel M., Al-Nazawi MJLs. 2020. Virtual Screening and Repurposing of FDA Approved Drugs against COVID-19 Main Protease; p. 117627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kangabam R., Sahoo S., Ghosh A., Roy R., Silla Y., Misra N., et al. 2020. Next-generation Computational Tools and Resources for Coronavirus Research: from Detection to Vaccine Discovery; p. 104158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapetanovic IJC-bi. Computer-aided drug discovery and development (CADDD): in silico-chemico-biological approach. 2008;171(2):165–176. doi: 10.1016/j.cbi.2006.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kempf D.J., Sham H.L., Marsh K.C., Flentge C.A., Betebenner D., Green B.E., et al. Discovery of ritonavir, a potent inhibitor of HIV protease with high oral bioavailability and clinical efficacy. 1998;41(4):602–617. doi: 10.1021/jm970636+. [DOI] [PubMed] [Google Scholar]
- Khalili J.S., Zhu H., Mak N.S.A., Yan Y., Zhu YJJomv. 2020. Novel Coronavirus Treatment with Ribavirin: Groundwork for an Evaluation Concerning COVID-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J., Zhang J., Cha Y., Kolitz S., Funt J., Chong R.E., et al. Advanced bioinformatics rapidly identifies existing therapeutics for patients with coronavirus disease-2019 (COVID-19) 2020;18(1):1–9. doi: 10.1186/s12967-020-02430-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirchdoerfer R.N., Ward ABJNc. Structure of the SARS-CoV nsp12 polymerase bound to nsp7 and nsp8 co-factors. 2019;10(1):1–9. doi: 10.1038/s41467-019-10280-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KJJoc Lundstrom, medicine m. Structural genomics and drug discovery. 2007;11(2):224–238. doi: 10.1111/j.1582-4934.2007.00028.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koutsoukas A., Simms B., Kirchmair J., Bond P.J., Whitmore A.V., Zimmer S., et al. From in silico target prediction to multi-target drug design: current databases, methods and applications. 2011;74(12):2554–2574. doi: 10.1016/j.jprot.2011.05.011. [DOI] [PubMed] [Google Scholar]
- Krammer F.J.N. SARS-CoV-2 vaccines in development. 2020;586(7830):516–527. doi: 10.1038/s41586-020-2798-3. [DOI] [PubMed] [Google Scholar]
- Kumar D., Chandel V., Raj S., Rathi B.J.C.B.L. In silico identification of potent FDA approved drugs against Coronavirus COVID-19 main protease: a drug repurposing approach. 2020;7(3):166–175. [Google Scholar]
- Latinne A., Hu B., Olival K.J., Zhu G., Zhang L., Li H., et al. 2020. Origin and Cross-Species Transmission of Bat Coronaviruses in China. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Leach A.R., Shoichet B.K., CEJJomc Peishoff. Prediction of protein− ligand interactions. Docking and scoring: successes and gaps. 2006;49(20):5851–5855. doi: 10.1021/jm060999m. [DOI] [PubMed] [Google Scholar]
- Li X., Geng M., Peng Y., Meng L., Lu SJJoPA . 2020. Molecular Immune Pathogenesis and Diagnosis of COVID-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Wang Y., Agostinis P., Rabson A., Melino G., Carafoli E., et al. Is hydroxychloroquine beneficial for COVID-19 patients? 2020;11(7):1–6. doi: 10.1038/s41419-020-2721-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin S., Shen R., He J., Li X., Guo XJb. 2020. Molecular Modeling Evaluation of the Binding Effect of Ritonavir, Lopinavir and Darunavir to Severe Acute Respiratory Syndrome Coronavirus 2 Proteases. [Google Scholar]
- Liu C., Zhou Q., Li Y., Garner L.V., Watkins S.P., Carter L.J., et al. ACS Publications; 2020. Research and Development on Therapeutic Agents and Vaccines for COVID-19 and Related Human Coronavirus Diseases. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu S., Huang W., Zhang JJDdt. Recent computational advances in the identification of allosteric sites in proteins. 2014;19(10):1595–1600. doi: 10.1016/j.drudis.2014.07.012. [DOI] [PubMed] [Google Scholar]
- Lung J., Lin Y.S., Yang Y.H., Chou Y.L., Shu L.H., Cheng Y.C., et al. The potential chemical structure of anti-SARS-CoV-2 RNA-dependent RNA polymerase. 2020;92(6):693–697. doi: 10.1002/jmv.25761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maciorowski D., Idrissi S.Z.E., Gupta Y., Medernach B.J., Burns M.B., Becker D.P., et al. A review of the preclinical and clinical efficacy of remdesivir, hydroxychloroquine, and lopinavir-ritonavir treatments against COVID-19. 2020;25(10):1108–1122. doi: 10.1177/2472555220958385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandal S., Mandal SKJEjop Rational drug design. 2009;625(1–3):90–100. doi: 10.1016/j.ejphar.2009.06.065. [DOI] [PubMed] [Google Scholar]
- Marinho E.M., de Andrade Neto J.B., Silva J., da Silva C.R., Cavalcanti B.C., Marinho E.S., et al. 2020. Virtual Screening Based on Molecular Docking of Possible Inhibitors of Covid-19 Main Protease; p. 104365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCoy J., Wambier C.G., Vano-Galvan S., Shapiro J., Sinclair R., Müller Ramos P., et al. Racial variations in COVID-19 deaths may Be due to androgen receptor genetic variants associated with prostate cancer and androgenetic alopecia. Are Anti-Androgens a Potential Treatment for COVID-19? 2020 doi: 10.1111/jocd.13455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirza M.U., Froeyen MJJoPA . 2020. Structural elucidation of SARS-CoV-2 vital proteins: computational methods reveal potential drug candidates against main protease, Nsp12 polymerase and Nsp13 helicase. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittal L., Kumari A., Srivastava M., Singh M., Asthana SJJoBS . 2020. Dynamics. Identification of potential molecules against COVID-19 main protease through structure-guided virtual screening approach; pp. 1–26. (just-accepted) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullard AJNRDD FDA approves Eli Lilly's baricitinib. 2018;17(7):460. doi: 10.1038/nrd.2018.112. [DOI] [PubMed] [Google Scholar]
- Mulligan M.J., Lyke K.E., Kitchin N., Absalon J., Gurtman A., Lockhart S., et al. Phase I/II study of COVID-19 RNA vaccine BNT162b1 in adults. 2020;586(7830):589–593. doi: 10.1038/s41586-020-2639-4. [DOI] [PubMed] [Google Scholar]
- Murray M.F., Kenny E.E., Ritchie M.D., Rader D.J., Bale A.E., Giovanni M.A., et al. 2020. COVID-19 Outcomes and the Human Genome; pp. 1–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naik V.R., Munikumar M., Ramakrishna U., Srujana M., Goudar G., Naresh P., et al. 2020. Remdesivir (GS-5734) as a Therapeutic Option of 2019-nCOV Main Protease–In Silico Approach; pp. 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narkhede R.R., Cheke R.S., Ambhore J.P., Shinde SDJs. The molecular docking study of potential drug candidates showing anti-COVID-19 activity by exploring of therapeutic targets of SARS-CoV-2. 2020;5:8. [Google Scholar]
- Ngo S.T., Quynh Anh Pham N., Thi Le L., Pham D.-H., Vu VVJJoci . 2020. Modeling. Computational determination of potential inhibitors of SARS-CoV-2 main protease. [DOI] [PubMed] [Google Scholar]
- Ovsyannikova I.G., Haralambieva I.H., Crooke S.N., Poland G.A., Kennedy R.B.J.I.R. The role of host genetics in the immune response to SARS-CoV-2 and COVID-19 susceptibility and severity. 2020;296(1):205–219. doi: 10.1111/imr.12897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papageorgiou A.C., Mohsin I.J.C. The SARS-CoV-2 spike glycoprotein as a drug and vaccine target: structural insights into its complexes with ACE2 and antibodies. 2020;9(11):2343. doi: 10.3390/cells9112343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parackova Z., Zentsova I., Bloomfield M., Vrabcova P., Smetanova J., Klocperk A., et al. Disharmonic inflammatory signatures in COVID-19: augmented neutrophils' but impaired monocytes' and dendritic cells' responsiveness. 2020;9(10):2206. doi: 10.3390/cells9102206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pillaiyar T., Meenakshisundaram S., Manickam MJDdt. Recent discovery and development of inhibitors targeting coronaviruses. 2020;25(4):668–688. doi: 10.1016/j.drudis.2020.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pushpakom S., Iorio F., Eyers P.A., Escott K.J., Hopper S., Wells A., et al. Drug repurposing: progress, challenges and recommendations. 2019;18(1):41–58. doi: 10.1038/nrd.2018.168. [DOI] [PubMed] [Google Scholar]
- R Laurie A.T., Jackson R.M.J.C.P., Science P. Methods for the prediction of protein-ligand binding sites for structure-based drug design and virtual ligand screening. 2006;7(5):395–406. doi: 10.2174/138920306778559386. [DOI] [PubMed] [Google Scholar]
- Ragia G., Manolopoulos V.G.J.P. 2020. Assessing COVID-19 Susceptibility through Analysis of the Genetic and Epigenetic Diversity of ACE2 Mediated SARS-CoV-2 Entry. 0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson P., Griffin I., Tucker C., Smith D., Oechsle O., Phelan A., et al. Baricitinib as potential treatment for 2019-nCoV acute respiratory disease. 2020;395(10223) doi: 10.1016/S0140-6736(20)30304-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romano M., Ruggiero A., Squeglia F., Maga G., Berisio R.J.C. A structural view of SARS-CoV-2 RNA replication machinery: RNA synthesis, proofreading and final capping. 2020;9(5):1267. doi: 10.3390/cells9051267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosa S.G.V., Santos WCJRPdSP Clinical trials on drug repositioning for COVID-19 treatment. 2020;44:e40. doi: 10.26633/RPSP.2020.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruan Z., Liu C., Guo Y., He Z., Huang X., Jia X., et al. NSP12; 2020. SARS-CoV-2 and SARS-CoV: Virtual Screening of Potential Inhibitors Targeting RNA-dependent RNA Polymerase Activity. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadeghi S.S., Keyvanpour MRJIAToCB . 2019. Bioinformatics. An analytical review of computational drug repurposing. [DOI] [PubMed] [Google Scholar]
- Salemme F.R., Spurlino J., Bone R.J.S. Serendipity meets precision: the integration of structure-based drug design and combinatorial chemistry for efficient drug discovery. 1997;5(3):319–324. doi: 10.1016/s0969-2126(97)00189-5. [DOI] [PubMed] [Google Scholar]
- Sanders J.M., Monogue M.L., Jodlowski T.Z., Cutrell J.B.J.J. Pharmacologic treatments for coronavirus disease 2019 (COVID-19): a review. 2020;323(18):1824–1836. doi: 10.1001/jama.2020.6019. [DOI] [PubMed] [Google Scholar]
- Santos J., Brierley S., Gandhi M.J., Cohen M.A., Moschella P.C., Declan A.B.J.V. Repurposing therapeutics for potential treatment of SARS-CoV-2: a review. 2020;12(7):705. doi: 10.3390/v12070705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senathilake K., Samarakoon S., Tennekoon K.J.P. 2020. Virtual Screening of Inhibitors against Spike Glycoprotein of SARS-CoV-2: A Drug Repurposing Approach. [Google Scholar]
- Shah B., Modi P., Sagar S.R.J.L.S. 2020. In silico studies on therapeutic agents for COVID-19: drug repurposing approach; p. 117652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang J., Wan Y., Luo C., Ye G., Geng Q., Auerbach A., et al. Cell entry mechanisms of SARS-CoV-2. 2020;117(21):11727–11734. doi: 10.1073/pnas.2003138117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y., Zhang X., Mu K., Peng C., Zhu Z., Wang X., et al. 2020. D3Targets-2019-nCoV: a Webserver for Predicting Drug Targets and for Multi-Target and Multi-Site Based Virtual Screening against COVID-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi X, Wang Q, Zhang L, Wang X. Jun Lan1, 4, Jiwan Ge1, 4, Jinfang Yu1, 4, Sisi Shan2, 4, Huan Zhou3, Shilong Fan1, Qi Zhang2.
- Singh S., Malik B.K., Sharma D.K.J.B. Molecular drug targets and structure based drug design. A holistic approach. 2006;1(8):314. doi: 10.6026/97320630001314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skipper C.P., Pastick K.A., Engen N.W., Bangdiwala A.S., Abassi M., Lofgren S.M., et al. Hydroxychloroquine in nonhospitalized adults with early COVID-19: a randomized trial. 2020;173(8):623–631. doi: 10.7326/M20-4207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sohrabi C., Alsafi Z., O'Neill N., Khan M., Kerwan A., Al-Jabir A., et al. 2020. World Health Organization Declares Global Emergency: A Review of the 2019 Novel Coronavirus (COVID-19) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stebbing J., Phelan A., Griffin I., Tucker C., Oechsle O., Smith D., et al. COVID-19: combining antiviral and anti-inflammatory treatments. 2020;20(4):400–402. doi: 10.1016/S1473-3099(20)30132-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sztain T., Amaro R., McCammon J.A.J.B. 2020. Elucidation of Cryptic and Allosteric Pockets within the SARS-CoV-2 Protease. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuttle KRJNRN . 2020. Impact of the COVID-19 Pandemic on Clinical Research; pp. 1–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ullrich S., Nitsche C.J.B., Letters M.C. 2020. The SARS-CoV-2 Main Protease as Drug Target; p. 127377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Hilten N., Chevillard F., Kolb PJJoci. modeling. Virtual compound libraries in computer-assisted drug discovery. 2019;59(2):644–651. doi: 10.1021/acs.jcim.8b00737. [DOI] [PubMed] [Google Scholar]
- Vellingiri B., Jayaramayya K., Iyer M., Narayanasamy A., Govindasamy V., Giridharan B., et al. 2020. COVID-19: A Promising Cure for the Global Panic; p. 138277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wacharapluesadee S., Tan C.W., Maneeorn P., Duengkae P., Zhu F., Joyjinda Y., et al. Evidence for SARS-CoV-2 related coronaviruses circulating in bats and pangolins in Southeast Asia. 2021;12(1):1–9. doi: 10.1038/s41467-021-21240-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace A.C., Laskowski R.A., Thornton JMJPe. design, selection. LIGPLOT: a program to generate schematic diagrams of protein-ligand interactions. 1995;8(2):127–134. doi: 10.1093/protein/8.2.127. [DOI] [PubMed] [Google Scholar]
- Walls A.C., Park Y.-J., Tortorici M.A., Wall A., McGuire A.T., Veesler D.J.C. 2020. Structure, Function, and Antigenicity of the SARS-CoV-2 Spike Glycoprotein. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Guan Y.J.M.R.R. 2020. COVID-19 Drug Repurposing: A Review of Computational Screening Methods, Clinical Trials, and Protein Interaction Assays. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Zhang Y., Huang B., Deng W., Quan Y., Wang W., et al. Development of an inactivated vaccine candidate, BBIBP-CorV, with potent protection against SARS-CoV-2. 2020;182(3):713–721. doi: 10.1016/j.cell.2020.06.008. e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Zhang D., Du G., Du R., Zhao J., Jin Y., et al. 2020. Remdesivir in Adults with Severe COVID-19: a Randomised, Double-Blind, Placebo-Controlled, Multicentre Trial. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson G.L., Lill MAJFmc Integrating structure-based and ligand-based approaches for computational drug design. 2011;3(6):735–750. doi: 10.4155/fmc.11.18. [DOI] [PubMed] [Google Scholar]
- Wishart D.S., Knox C., Guo A.C., Shrivastava S., Hassanali M., Stothard P., et al. DrugBank: a comprehensive resource for in silico drug discovery and exploration. 2006;34(Suppl. l_1):D668–D672. doi: 10.1093/nar/gkj067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C., Liu Y., Yang Y., Zhang P., Zhong W., Wang Y., et al. 2020. Analysis of Therapeutic Targets for SARS-CoV-2 and Discovery of Potential Drugs by Computational Methods. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia S., Duan K., Zhang Y., Zhao D., Zhang H., Xie Z., et al. Effect of an inactivated vaccine against SARS-CoV-2 on safety and immunogenicity outcomes: interim analysis of 2 randomized clinical trials. 2020;324(10):951–960. doi: 10.1001/jama.2020.15543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan R., Zhang Y., Li Y., Xia L., Guo Y., Zhou Q.J.S. Structural basis for the recognition of SARS-CoV-2 by full-length human ACE2. 2020;367(6485):1444–1448. doi: 10.1126/science.abb2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao X., Ye F., Zhang M., Cui C., Huang B., Niu P., et al. In vitro antiviral activity and projection of optimized dosing design of hydroxychloroquine for the treatment of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) 2020;71(15):732–739. doi: 10.1093/cid/ciaa237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuki K., Fujiogi M., Koutsogiannaki SJCi. 2020. COVID-19 pathophysiology: a review. 108427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., Zhou R. 2020. Binding Mechanism of Remdesivir to SARS-CoV-2 RNA Dependent RNA Polymerase. [DOI] [PubMed] [Google Scholar]
- Zhang Q., Bastard P., Liu Z., Le Pen J., Moncada-Velez M., Chen J., et al. Inborn errors of type I IFN immunity in patients with life-threatening COVID-19. 2020;370(6515) doi: 10.1126/science.abd4570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., Lin D., Sun X., Curth U., Drosten C., Sauerhering L., et al. Crystal structure of SARS-CoV-2 main protease provides a basis for design of improved α-ketoamide inhibitors. 2020;368(6489):409–412. doi: 10.1126/science.abb3405. [DOI] [PMC free article] [PubMed] [Google Scholar]