Abstract
Background
Gastric cancer (GC) is one of the most common solid malignant tumors worldwide with a high-recurrence-rate. Identifying the molecular signatures and specific biomarkers of GC might provide novel clues for GC prognosis and targeted therapy.
Methods
Gene expression profiles were obtained from the ArrayExpress and Gene Expression Omnibus database. Differentially expressed genes (DEGs) were picked out by R software. The hub genes were screened by cytohubba plugin. Their prognostic values were assessed by Kaplan–Meier survival analyses and the gene expression profiling interactive analysis (GEPIA). Finally, qRT-PCR in GC tissue samples was established to validate these DEGs.
Results
Total of 295 DEGs were identified between GC and their corresponding normal adjacent tissue samples in E-MTAB-1440, GSE79973, GSE19826, GSE13911, GSE27342, GSE33335 and GSE56807 datasets, including 117 up-regulated and 178 down-regulated genes. Among them, 7 vital upregulated genes (HMMR, SPP1, FN1, CCNB1, CXCL8, MAD2L1 and CCNA2) were selected. Most of them had a significantly worse prognosis except SPP1. Using qRT-PCR, we validated that their transcriptions in our GC tumor tissue were upregulated except SPP1 and FN1, which correlated with tumor relapse and predicts poorer prognosis in GC patients.
Conclusions
We have identified 5 upregulated DEGs (HMMR, CCNB1, CXCL8, MAD2L1, and CCNA2) in GC patients with poor prognosis using integrated bioinformatical methods, which could be potential biomarkers and therapeutic targets for GC treatment.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12885-021-08358-7.
Keywords: Gastric cancer, Bioinformatics analysis, Differentially expressed genes
Background
Gastric cancer (GC), the fifth most frequently diagnosed cancer and the third leading cause of cancer-related death [1], has become a major global health challenge. About 934,000 new GC cases and 700,000 mortalities occurred annually [2]. Despite improvement in diagnosis and treatment, the prognosis of GC patients remains poor, which has become an active topic of clinical and basic research. Genetic mutations, epigenetic alterations and aberrant molecular signaling pathways are involved in the processes of gastric carcinogenesis, spread and metastasis [3]. In particular, the new molecular characteristics can be applied in early risk assessment, the identification of better specific biomarkers, and the improvement of clinic treatment and survival.
In recent decades, microarray and high-throughput sequencing have been considered as reliable techniques to quickly detect differentially expressed genes (DEGs) [4] that are able to make various slice data be produced and stored in public databases. Consequently, many valuable clues could be explored for new research on the base of these data. However, with the data getting updated, a large amount of genetic information uploaded to public databases was not used effectively.
In this study, we downloaded related mRNA expression datasets from ArrayExpress and Gene Expression Omnibus. A set of DEGs in these datasets were extracted by comparing gene expression profiles of carcinoma specimen and adjacent normal tissues. By analyzing the GO and Kyoto Encyclopedia of Gene and Genome (KEGG) pathway enrichment [5, 6], along with the construction of protein–protein interaction (PPI) network [7], we selected vital genes. After evaluating the clinical prognosis of these genes and their transcriptional factor (TF) regulatory network, we further validated these genes by quantitative real-time PCR (qRT-PCR) in GC tissue samples.
Methods
Gastric cancer microarray data information
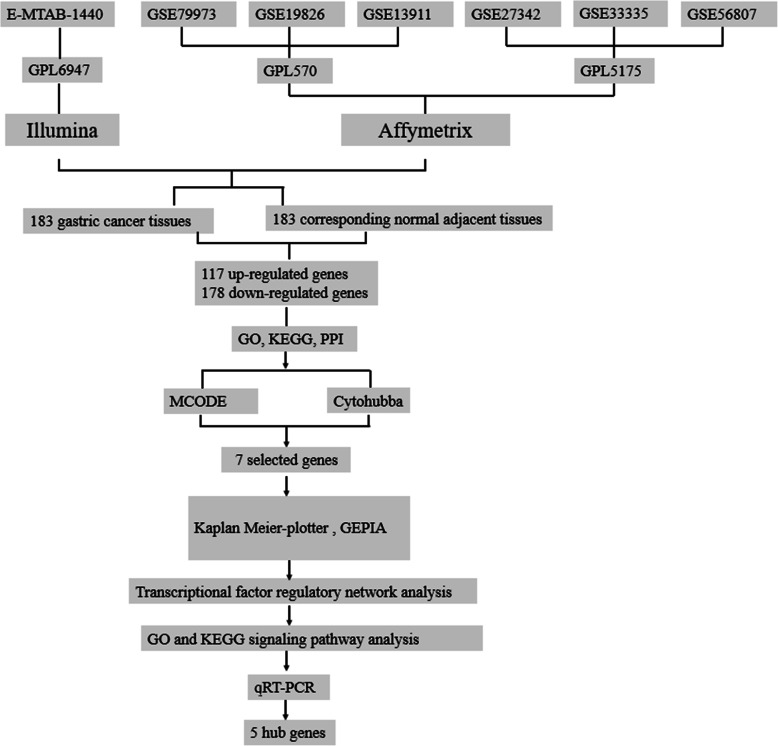
Microarray data information of GC and adjacent gastric tissues were obtained from Arrayexpress (https://www.ebi.ac.uk/arrayexpress/) and NCBI-GEO (https://www.ncbi.nlm.nih.gov/geo). When “gastric cancer” was used as a keyword to perform queries, we selected the original studies of RNA assay and array assay in Homo sapiens which samples with available clinical information for analysis. The expression microarray datasets E-MTAB-1440, GSE79973, GSE19826, GSE13911, GSE27342, GSE33335 and GSE56807 were downloaded. Overall, 183 patients with gastric cancer enrolled in this study. The workflow chart is shown in Fig.1.
Fig. 1.
Flowchart of the multistep screening strategy used in this study on bioinformatics data in Arrayexpress and GEO database
Gene expression profile data
Microarray data of 7 databases were on account of three platforms. E-MTAB-1440 genome-wide gene expression profile data were generated from the Illumina platform GPL6947 (A-MEXP-1171-Illumina Human HT-12 v3.0 Expression BeadChip). GSE19826, GSE13911 and GSE27342 microarray data from the Affymetrix platform GPL570 (HG-U133_Plus_2 Affymetrix Human Genome U133 Plus 2.0 Arrays) and GSE27342, GSE33335 and GSE56807 microarray data from the Affymetrix platform GPL5175(HuEx-1_0-st-v1 Affymetrix Gene Chip Human Exon 1.0 ST Array version 1). Detailly, GPL6947 dataset consisted of 20 GC tissues and 20 adjacent normal gastric samples. GPL570 and GPL5175 respectively include 53 and 110 GC tissues as well as same number of matched normal specimen.
Data processing of DEGs
Significant DEGs between GC specimen and normal gastric tissues specimen were analyzed via software and packages from Bioconductor (http://www.bioconductor.org/) in R (version 3.6.0). The microarray data were first preprocessed using the RMA (robust multi-array average) which contains background adjustment, normalization with the quantile method, and expression calculations. The probes were removed when they were not able to be matched to a specific gene symbol, and the average value was taken as the expression value for each gene when different probes matched to the same gene symbol. Then the statistically significant DEGs was selected by Moderated T statistic approach with “limma [8]” and “oligo [9]” package of Bioconductor. After preprocessing, SVA batch difference processing of combat was used to consolidate these 7 datasets to obtain the final dataset (GC tissues: corresponding normal adjacent tissues =183:183). Finally, DEGs were annotated through annotation table downloaded from the GEO website. The resulting P values were adjusted by the default Benjamini & Hochberg (BH) false discovery rate method. The adj. P value < 0.05, P value < 0.05 and |log fold change (FC)| > 0.58 were considered as significantly different for DEGs.
Protein–protein interactions (PPI) network and module analysis
Information of DEGs’ protein experimental interactions and prediction was obtained by Search Tool for the Retrieval of Interacting Genes (STRING, Version 11.0, http://www.string-db.org/) [10] with the parameters set to species = Homo sapiens, and PPI score ≥ 0.4 (medium confidence) [11]. Subsequently, a specific PPI network of DEGs was constructed by cytoscape (version 3.7.2, http://www.cytoscape.org/) [12] based on the interactions retrieved from STRING. The gene-interaction relationship was represented by nodes and edges graphically for better visualization, which included phosphorylation, dephosphorylation, inhibition and activation. In the signaling network, the size of the cycle was considered as the frequency of the gene interaction. The most prominent central genes in the network indicated the genes with the highest frequency. In addition, the molecular complex detection (MCODE) analysis (Version 3.7.2, http://apps.cytoscape.org/apps/MCODE) [13] in cytoscape was used to identify the significant modules of the PPI network with degree cut-off 2, max depth 100, k-core 2, and node score cutoff 0.2. To screen the hub genes that may be involved in GC, we applied the cytohubba plug-in, using various parameters such as degree, betweenness centrality, and closeness. The DEGs from cytohubba were then subjected to VEEN analysis using the online tool (http://bioinformatics.psb.ugent.be/webtools/Venn/), and overlapping genes were considered selected genes.
Evaluation of prognostic value of selected genes
Expression and prognostic values of the hub genes were analyzed using two online datasets, Kaplan Meier-plotter dataset (http://kmplot.com/analysis/) and Gene Expression Profiling Interactive Analysis (GEPIA, http://gepia.cancer-pku.cn) [14]. The hazard ratio (HR) with 95% confidence intervals and log rank p value were calculated and displayed on the plot. GEPIA was established for customized genomic analysis based on the Cancer Genome Atlas (TCGA) database, which was used to compare poor prognosis related hub genes expression between GC patients and healthy people.
Transcriptional factor (TF) regulatory network construction
NetworkAnalyst (http://www.networkanalyst.ca/faces/home.xhtml) is used to explore TF-gene interactions for the input genes and assess the effect of the TF on the expression and functional pathways of the hub gene. In this study, the TFs of the hub genes were predicted from this database and a transcriptional regulatory network was constructed and visualized by the cytoscape software.
Analysis of significant functions and pathway enrichment
After computing hub genes and evaluating prognosis, the database for annotation, visualization and integrated discovery (DAVID 6.8, http://david.abcc.ncifcrf.gov/) [6] was applied to re- analyze the KEGG pathway and Gene Ontology annotations for selected hub genes. P-value < 0.05, and count ≥2 were considered to indicate significance.
Validation of selected DEGs’ transcription in fresh GC tissue specimens using quantitative real-time PCR
We analyzed samples from 10 GC patients who underwent tumor resection at the Department of Pathology, Shanxi Cancer Hospital (Shanxi, China). The detailed clinicopathological information for all the enrolled patients was available. GC and their corresponding normal adjacent tissue samples were immediately frozen in liquid nitrogen and stored at − 80 °C until further processing. Every specimen was anonymously handled based on ethical standards. All patients provided written informed consent and our study was approved by the hospital’s Ethical Review Committee.
The total RNA was extracted using Trizol reagent and reverse-transcribed into complementary DNA (cDNA) for quantitative real-time polymerase chain reaction (qRT-PCR) following the manufacturer’s instructions. GAPDH gene served as an endogenous control. The primer sequences of selected genes (HMMR, SPP1, FN1, CCNB1, CXCL8, MAD2L1 and CCNA2) used in the experiment are illustrated in Table 1. Each sample was tested in triplicates, and each sample underwent a melting curve analysis to check for the specificity of amplification. The relative expression level was determined as a ratio between the hub genes and the internal control GAPDH in the same mRNA sample, and calculated by the comparative CT method. Levels of hub genes’ expression were calculated by the 2−ΔΔCt method [15, 16].
Table 1.
Primer sequences of PCR
| gene | Forward primer(5–3) | Reverse primer(5–3) |
|---|---|---|
| HMMR | GCTAAGCAAGAAGGCATGGA | CCACTTGATCTGAAGCACAAC |
| SPP1 | GCCGAGGTGATAGTGTGGTT | AACGGGGATGGCCTTGTATG |
| FN1 | AATAGATGCAACGATCAGGACA | GCAGGTTTCCTCGATTATCCTT |
| CCNB1 | GACTTTGCTTTTGTGACTGACA | CCCAGACCAAAGTTTAAAGCTC |
| CXCL8 | AACTGAGAGTGATTGAGAGTGG | ATGAATTCTCAGCCCTCTTCAA |
| MAD2L1 | ACGGTGACATTTCTGCCACT | TGGTCCCGACTCTTCCCATT |
| CCNA2 | AGAAACAGCCAGACATCACTAA | TTCAAACTTTGAGGCTAACAGC |
| GAPDH | ACAGTCAGCCGCATCTTCTT | ACGACCAAATCCGTTGACTC |
Statistical analysis
Demographic and clinical data were analyzed using Chi-squared test, student’s t-test or paired t-test to evaluate group balance of variables. All statistical analyses were performed using SPSS 26.0, the GraphPad Prism V8.0 and R 3.6.0. Two-tailed P < 0.05 were considered statistically significant.
Results
Identification of DEGs in GC
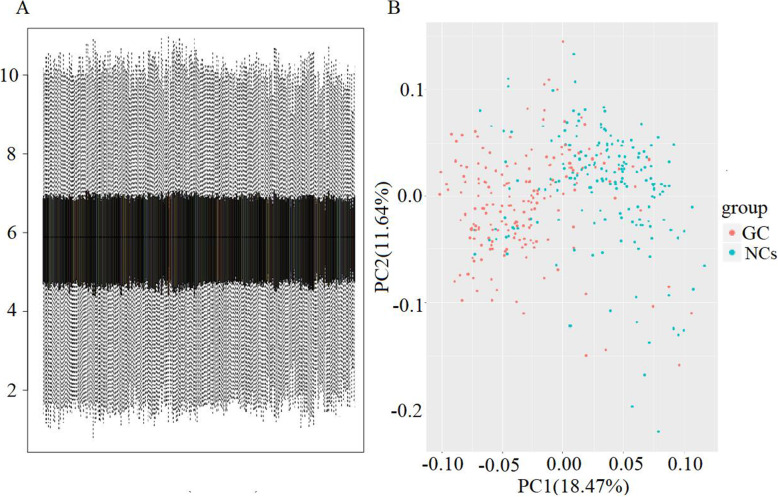
A total of 366 samples were included in the present study: 183GC and 183 adjacent normal tissues used as normal controls (NCs). Via R software, a total of 3224 DEGs (gastric cancer tissues vs. NCs), including 117 up-regulated and 178 down-regulated genes were selected. The statistical metrics for key DEGs was shown in Supplemental Table 1. The data distributions were neat after background adjustment and normalization with the RMA method, and values with an unchanged position in the boxplot were used for subsequent analysis.
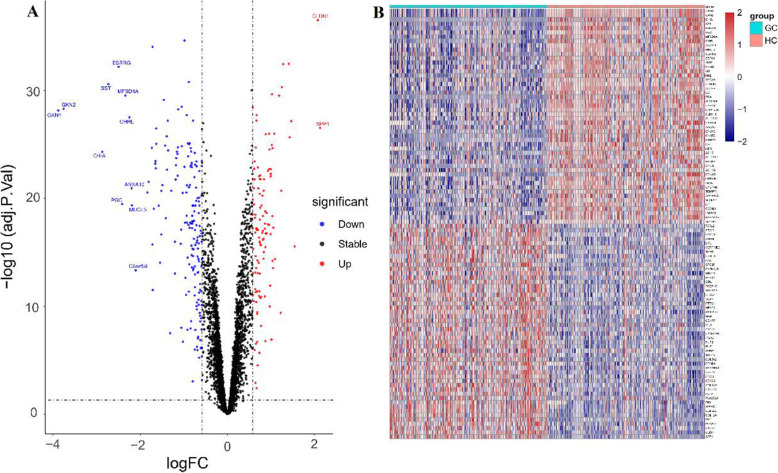
(Figure 2A). Principal component analysis (PCA) was conducted to obtain better insights into the data. The DEGs of GC and normal tissues were relatively well separated in 2D score plot PCA. (Fig. 2B) The volcano plots of DEGs were shown in Fig.3A. DEGs expression heatmaps of the top 50 significant up-regulated genes and top 50 significant down-regulated genes were depicted in Fig. 3B, and hierarchical clustering analysis revealed that DEGs can be easily distinguished from GC tissues and normal gastric tissues.
Fig. 2.
Data normalization and the distribution of differentially expressed genes (DEGs). (A) Box plots illustrating data normalization: the data distributions were neat after background adjustment and normalization. (B) Principal component analysis (PCA): each point in the PCA diagram represents a sample, and the distance between samples reflects the difference. After batch correction, individuals with similar genetic background were clustered together, and obvious stratification was observed between GC and adjacent normal gastric tissue samples. Gastric cancer, GC. normal controls, NCs
Fig. 3.
Volcano plot and heatmap of DEGs. (A) Volcanic map of DEGs: each colored dot represents a DEG based on the criteria of P < 0.05 and |log FC| > 0.58; red: up-regulation, blue: downregulation, black: normally expressed mRNAs. (B) Heatmap of top 50 significant up-regulated and down-regulated DEGs expressed in mRNAs microarrays. The horizontal axis shows clusters of DEGs, and the right vertical axis represents each sample. Gene expression levels were indicated by colors: red: high expression level and blue: low expression level
PPI and modular analysis
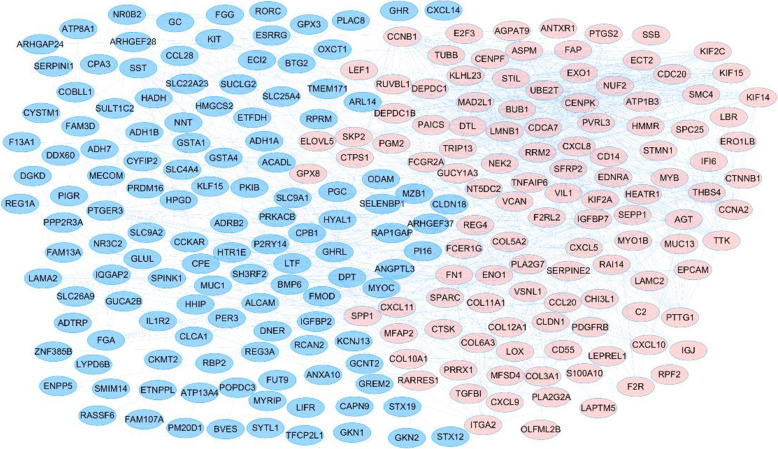
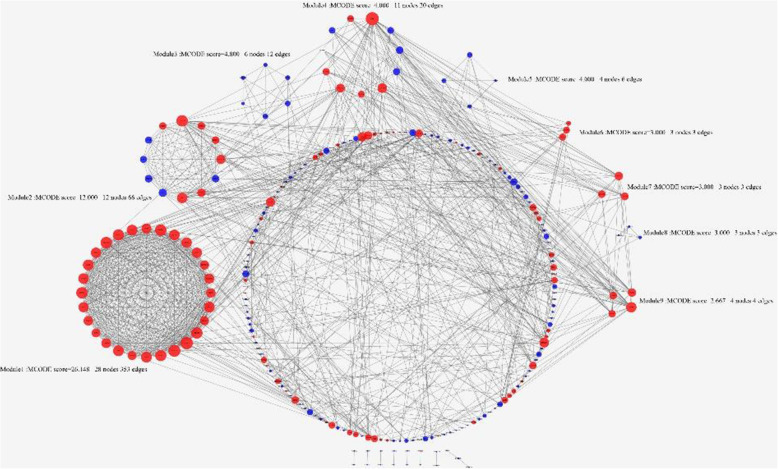
Based on the STRING online database, a total of 295 DEGs were imported into the DEG PPI network complex which included 291 nodes and 1016 edges. All the parameters were set as defaults [10]. The average node degree of PPI network was 6.98 and the local clustering coefficient was 0.446. To further investigate the PPI, the PPI network was visualized by cytoscape. (Fig. 4) Nine modules were exhibited after analyzing the entire PPI network by MCODE plug-in (Fig. 5& Supplemental Table 2).
Fig. 4.
PPI network of the DEGs in GC. The PPI network of DEGs was constructed using Cytoscape. The nodes meant proteins; the edges meant the interaction of proteins. Upregulated genes are marked in light red; downregulated genes are marked in blue. PPI: protein–protein interaction; DEG: differentially expressed gene; GC: gastric cancer
Fig. 5.
The 9 modules identified in the PPI network. Module analysis via Cytoscape software (degree cutoff = 2, node score cutoff = 0.2, k-core = 2, and max. Depth = 100). Node size represents the degree score; lines represent interactions
Identification of the selected genes
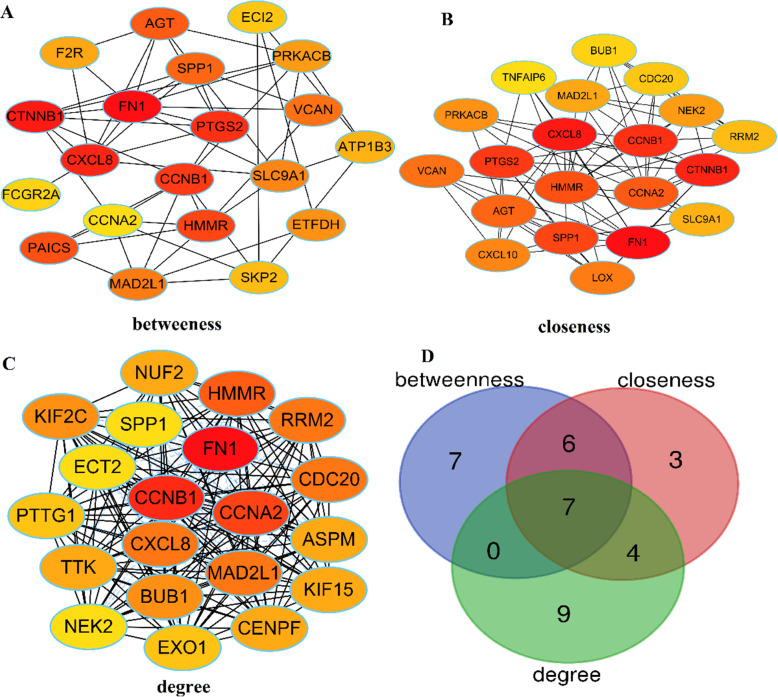
The vital genes were determined from the PPI network by cytohubba plug-in. All the gene code and edge were calculated (Fig.6A.B.C& Supplemental Table 3). Three groups of DEGs calculated from degree, betweenness centrality and closeness were subjected to VEEN analysis (Fig.6D& Supplemental Table 3 & Supplemental Table 4). The overlapping genes were sequentially listed as follows: HMMR (hyaluronan mediated motility receptor), SPP1 (secreted phosphoprotein 1), FN1(fibronectin 1), CCNB1 (cyclin B1), CXCL8 (C-X-C motif chemokine ligand 8), MAD2L1 (mitotic arrest deficient 2 like 1), CCNA2 (cyclin A2). (Table 2) Besides, the selected genes also showed significant enrichment in modules by MCODE analysis (Fig. 5). Some of these genes exhibited potential prognostic values for patients with GC.
Fig. 6.
Seven hub genes selected from PPI network. (A) Hub genes screened by betweenness centrality according to cytoHubba plug-in. (B) Hub genes screened by closeness according to cytoHubba plug-in. (C) Hub genes screened by degree according to cytoHubba plug-in. (D) Venn diagram of DEGs. Hub genes were HMMR, SPP1, FN1, CCNB1, CXCL8, MAD2L1, and CCNA2; PPI, protein–protein interaction
Table 2.
Details of seven selected genes
| N | Gene symbol | Full name | Function |
|---|---|---|---|
| 1 | HMMR | hyaluronan mediated motility receptor | the driver of tumor progression, plays a important role in the modulation of motor activities and the maintenance of genome stability. |
| 2 | SPP1 | secreted phosphoprotein 1 | The protein encoded by this gene is involved in the attachment of osteoclasts to the mineralized bone matrix |
| 3 | FN1 | fibronectin 1 | Fibronectin 1 (FN1) is involved in cell adhesion and migration processes including embryogenesis, wound healing, blood coagulation, host defense, metastasis, and implicated in various biochemical processes. |
| 4 | CCNB1 | cyclin B1 | an important regulator in cell cycle machinery, is a monitoring protein in mitosis and expressed primarily in G2/M phase which is critical for controlling the cell cycle at the G2/M (mitosis) transition |
| 5 | CXCL8 | C-X-C motif chemokine ligand 8 | a chemokine that acts as an important multifunctional cytokine to modulate tumour proliferation, invasion and migration in an autocrine or paracrine manner. |
| 6 | MAD2L1 | mitotic arrest deficient 2 like 1 | a component of the mitotic spindle assembly checkpoint that prevents the onset of anaphase until all chromosomes are properly aligned at the metaphase plate |
| 7 | CCNA2 | cyclin A2 | The protein encoded by this gene belongs to the highly conserved cyclin family, whose members function as regulators of the cell cycle. |
Survival analysis of selected genes by the Kaplan Meier plotter and GEPIA
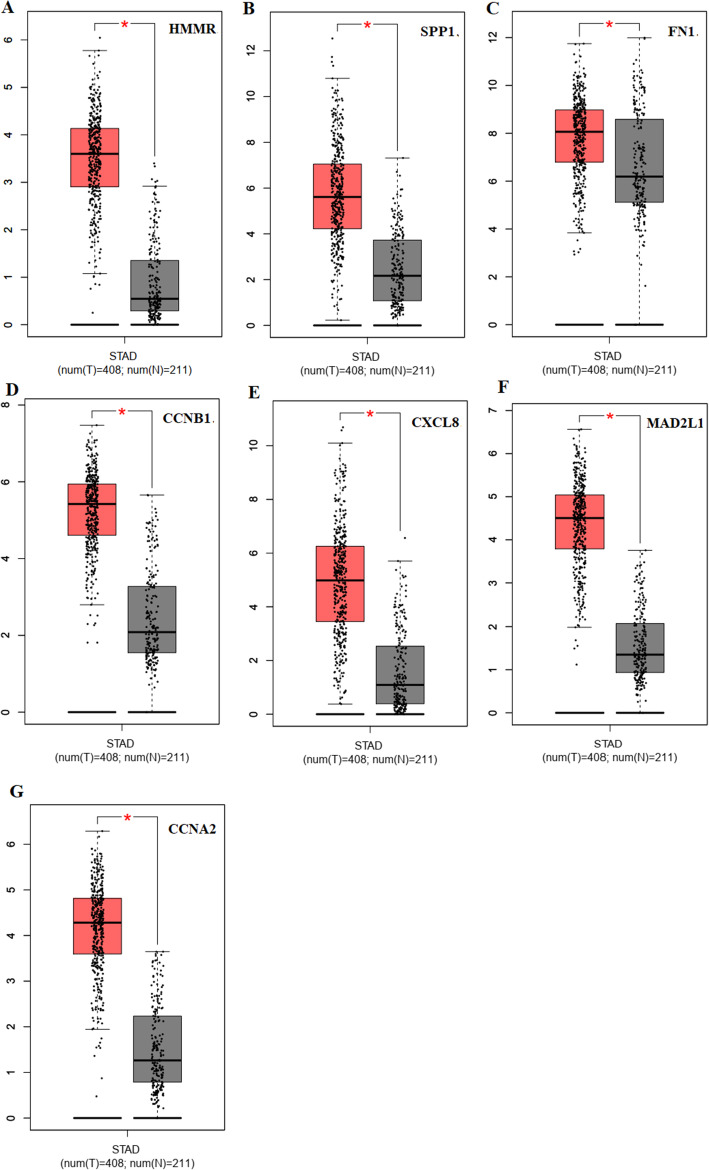
To further analyze the prognostic value of the selected genes, the overall survivals (OS) with selected genes were analyzed for 875 patients with GC by using the Kaplan-Meier plotter. It was found that most of the genes had a significantly worse survival (P < 0.05, Fig.7). High expression of HMMR (P = 5.0e-9), FN1 (P = 1.0e-6), CCNB1(P = 9.5e-7), CXCL8(P = 1.5e-5), MAD2L1(P = 2.4e-8), CCNA2(P = 9.9e-8) were correlated with significantly worse OS in GC patients, while SPP1 expression was not relevant to survival (P = 0.2713). Then, we used GEPIA to dig up the expression levels of selected genes in GC patients and healthy controls. Results reflected that, contrasted to normal samples, all the selected genes reflected high expressed in GC samples (P < 0.05, Fig. 8).
Fig. 7.
Genes associated with patient’s survival outcomes by applying the K-M method. Prognostic curves of most selected genes showed a significantly worse survival rate (P < 0.05). The red lines represent patients with high gene expression, and black lines with a low gene expression. HR: hazard ratio.
Fig. 8.
Differentially expressed genes related with poor prognosis were analyzed using GEPIA website. These genes had significantly upregulated expression in gastric cancers compared to normal specimen (*P < 0.05). The red and gray boxes represent cancer and normal tissues, respectively. STAD: Stomach adenocarcinoma
Transcriptional factor regulatory network analysis of selected genes
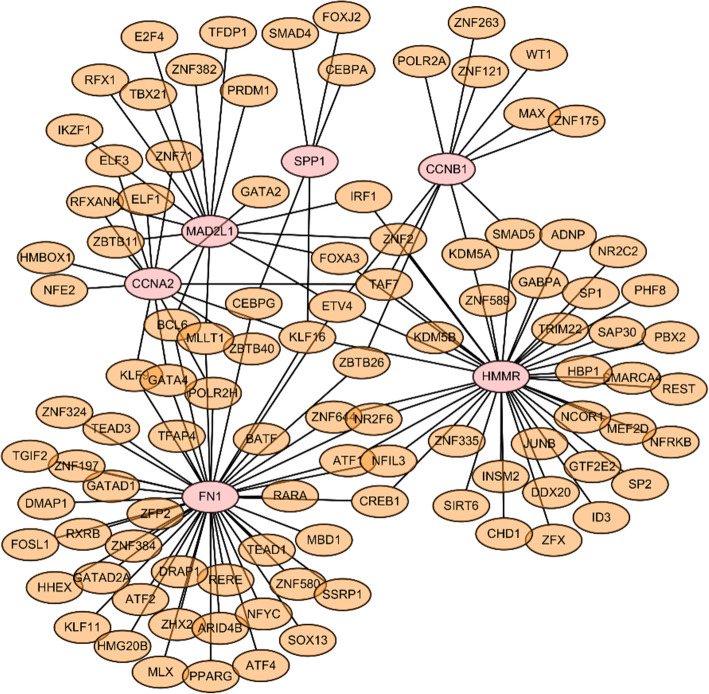
For the genes we identified, a gene-TF regulatory network was constructed including 129 interaction pairs among the selected genes and 102 TFs (Fig. 9 & Supplemental Table 5). While HMMR was found to be regulated by 39 TFs, SPP1 by 5 TFs, FN1 by 45 TFs, CCNB1 by 11 TFs, and CCNA2 by 14 TFs. In addition, various TFs were found to regulate more than one hub gene, and twenty TFs were identified with a connectivity degree ≥ 2 in the gene-TF regulatory network, which means that these TFs have close interactions with these hub DEGs. For example, zinc finger protein 2 (ZNF2) was predicted to regulate HMMR, and MAD2L1; ETS variant transcription factor 4 (ETV4) was found to regulate HMMR, FN1, MAD2L1, and CCNB1; Kruppel like factor 16 (KLF16) was found to regulate HMMR, SPP1, FN1, and CCNA2.
Fig. 9.
The hub gene-transcription factor (TF) regulatory network. Pink circle stands for the hub gene and orange node stands for the transcription factor
Analysis of 7 selected genes via gene ontology and pathway enrichment
To understand the possible pathway of these 7 selected DEGs, KEGG pathway enrichment was re-analyzed via DAVID (P < 0.05). GO analysis revealed 7 selected genes that are involved in a number of biological processes (BP), including positive regulation of fibroblast proliferation, cell division, and negative regulation of ubiquitin-protein ligase activity involved in mitotic cell cycle. In terms of cellular components, 7 selected genes were mostly enriched in spindle pole, extracellular space, and extracellular region. The 7 selected genes were mainly associated with protein binding in terms of molecular functions. With regards to the KEGG pathway analysis of the 7 selected genes, ten pathways were enriched: ‘ECM-receptor interaction’, ‘Progesterone-mediated oocyte maturation’, ‘Cell cycle’, ‘Amoebiasis’, ‘Toll-like receptor signaling pathway’, and ‘Oocyte meiosis’. Detailed results are displayed in Table 3.These results suggested that Toll-like receptor signaling pathway and Cell cycle played extremely important roles in progesterone resistance and should be further studied.
Table 3.
Analysis of 7 selected genes by GO and KEGG pathway enrichment. FDR, false discovery rate. GO, gene ontology; BP, biological process, CC, cellular component; MF, molecular function; KEGG, Kyoto Encyclopedia of Genes and Genomes; DEMs, differentially expressed miRNAs
| Category | Term | Count | % | PValue | Genes | FDR |
|---|---|---|---|---|---|---|
| GOTERM_BP_DIRECT | GO:0048146 ~ positive regulation of fibroblast proliferation | 3 | 0.263 | 1.51E-04 | CCNB1, CCNA2, FN1 | 0.174735 |
| GOTERM_BP_DIRECT | GO:0051301 ~ cell division | 3 | 0.263 | 0.006147477 | CCNB1, MAD2L1, CCNA2 | 6.892135 |
| GOTERM_BP_DIRECT | GO:0051436 ~ negative regulation of ubiquitin-protein ligase activity involved in mitotic cell cycle | 2 | 0.175 | 0.025106263 | CCNB1, MAD2L1 | 25.50655 |
| GOTERM_BP_DIRECT | GO:0051437 ~ positive regulation of ubiquitin-protein ligase activity involved in regulation of mitotic cell cycle transition | 2 | 0.175 | 0.026854324 | CCNB1, MAD2L1 | 27.03882 |
| GOTERM_BP_DIRECT | GO:0022617 ~ extracellular matrix disassembly | 2 | 0.175 | 0.026854324 | SPP1, FN1 | 27.03882 |
| GOTERM_BP_DIRECT | GO:0031145 ~ anaphase-promoting complex-dependent catabolic process | 2 | 0.175 | 0.027901906 | CCNB1, MAD2L1 | 27.94322 |
| GOTERM_BP_DIRECT | GO:0071456 ~ cellular response to hypoxia | 2 | 0.175 | 0.033820471 | CCNB1, CCNA2 | 32.86331 |
| GOTERM_BP_DIRECT | GO:0000086 ~ G2/M transition of mitotic cell cycle | 2 | 0.175 | 0.047971221 | CCNB1, HMMR | 43.40814 |
| GOTERM_BP_DIRECT | GO:0042787 ~ protein ubiquitination involved in ubiquitin-dependent protein catabolic process | 2 | 0.175 | 0.053446406 | CCNB1, MAD2L1 | 47.06463 |
| GOTERM_BP_DIRECT | GO:0030198 ~ extracellular matrix organization | 2 | 0.175 | 0.068031098 | SPP1, FN1 | 55.77698 |
| GOTERM_BP_DIRECT | GO:0001525 ~ angiogenesis | 2 | 0.175 | 0.077092848 | CXCL8, FN1 | 60.50819 |
| GOTERM_CC_DIRECT | GO:0000922 ~ spindle pole | 2 | 0.175 | 0.035359174 | CCNB1, MAD2L1 | 26.96429 |
| GOTERM_CC_DIRECT | GO:0005615 ~ extracellular space | 3 | 0.263 | 0.067058677 | CXCL8, SPP1, FN1 | 45.44016 |
| GOTERM_CC_DIRECT | GO:0005576 ~ extracellular region | 3 | 0.263 | 0.092078284 | CXCL8, SPP1, FN1 | 56.9647 |
| GOTERM_MF_DIRECT | GO:0005515 ~ protein binding | 7 | 0.613 | 0.019847501 | CCNB1, MAD2L1, CXCL8, CCNA2, SPP1, FN1, HMMR | 13.51086 |
| KEGG_PATHWAY | hsa04512:ECM-receptor interaction | 3 | 0.263 | 0.002294922 | SPP1, FN1, HMMR | 2.054976 |
| KEGG_PATHWAY | hsa04914:Progesterone-mediated oocyte maturation | 3 | 0.263 | 0.002294922 | CCNB1, MAD2L1, CCNA2 | 2.054976 |
| KEGG_PATHWAY | hsa04110:Cell cycle | 3 | 0.263 | 0.004611117 | CCNB1, MAD2L1, CCNA2 | 4.090835 |
| KEGG_PATHWAY | hsa05146:Amoebiasis | 2 | 0.175 | 0.088997082 | CXCL8, FN1 | 56.93087 |
| KEGG_PATHWAY | hsa04620:Toll-like receptor signaling pathway | 2 | 0.175 | 0.088997082 | CXCL8, SPP1 | 56.93087 |
| KEGG_PATHWAY | hsa04114:Oocyte meiosis | 2 | 0.175 | 0.093026279 | CCNB1, MAD2L1 | 58.62208 |
The transcription levels of selected genes were verified within GC tissues
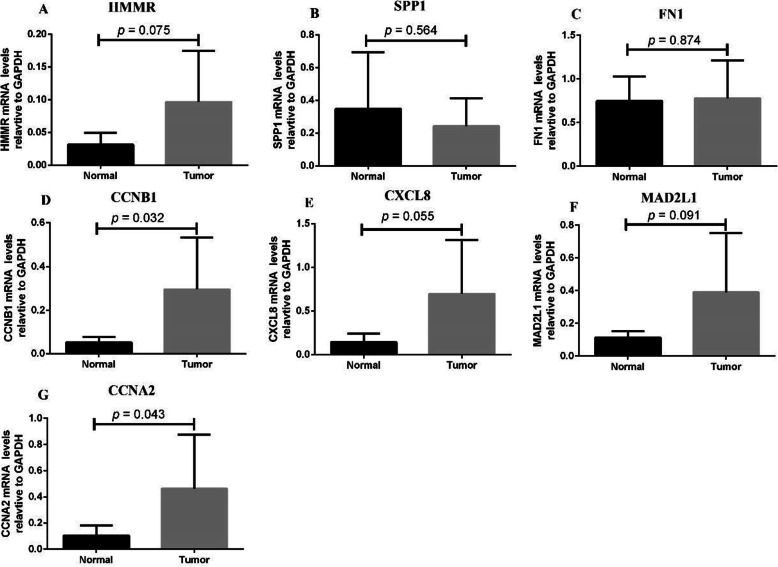
To further verify the results of bioinformatics analysis, we applied qRT-PCR to validate the mRNA levels of HMMR, SPP1, FN1, CCNB1, CXCL8, MAD2L1 and CCNA2 in 10 paired tumor and adjacent normal tissues with qRT-PCR. Among the genes we validated, HMMR, CCNB1, CXCL8, MAD2L1, and CCNA2 showed increasing expression levels in GC. As illustrated in Fig. 10, high expression of CCNB1 and CCNA2 significantly correlates with tumor relapse and predicts poorer prognosis in GC patients (P < 0.05). The expression of HMMR, CXCL8 and MAD2L1 shows an increasing trend in GC, whereas COL1A2 and SPP1 expression levels might not affect the prognosis of patients with GC. We identified 5 hub genes including HMMR, CCNB1, CXCL8, MAD2L1, and CCNA2 with poor prognosis in GC on the basis of integrated bioinformatical methods, which could be potential biomarkers and therapeutic targets for GC treatment.
Fig. 10.
Validation of 7 selected gene expression in gastric cancer samples was performed by qRT-PCR analysis. (A) HMMR; (B) SPP1; (C) FN1; (D) CCNB1; (E) CXCL8; (F) MAD2L1; (G) CCNA2. Expression of these DEGs was normalized against GAPDH expression. The statistical significance of differences was calculated by the Student’s t-test
Discussion
GC is a gastroenterological malignancy with high rates of prevalence and mortality [1, 2, 17, 18]. Therefore, sensitive and specific biomarkers of GC are urgently needed to be detected. In the present study, bioinformatic methods are promising methods to analyze the critical genes and pathways, which might provide novel clues for diagnosis, therapy, and prognosis of GC. We integrated seven gene expression profile datasets from different groups and used R software and bioinformatics to deeply analyze these datasets. DEGs PPI network was successfully constructed via the STRING online database and cytoscape software. Seven vital regulated genes including HMMR, SPP1, FN1, CCNB1, CXCL8, MAD2L1, and CCNA2 were screened from the PPI network complex by cytohubba plug-in of cytoscape.
Through Kaplan Meier plotter analysis, we found that most of the selected genes were associated with a significantly worse survival, except SPP1. The expression of the genes was higher in GC samples than normal samples by GEPIA analysis. Importantly, using qRT-PCR, we could validate the higher mRNA expression of the selected genes based our bioinformatics analysis; most selected genes, except SPP1 and FN1, were upregulated in tumor tissue. They showed the same trend in expression as predicted by bioinformatics verifying the accuracy of our method. In the light of important roles in cells, the selected hub genes in GC (HMMR, CCNB1, CXCL8, MAD2L1, and CCNA2) may represent potential prognostic biomarkers and/or therapeutic targets for GC.
For a more in-depth understanding of these DEGs, we analyzed the selected genes for GO and KEGG enrichment analyses and found that ‘Cell cycle’ signaling pathways was significant enriched. HMMR, CCNB1, MAD2L1 and CCNA2 play important roles in cell cycle. HMMR, a cell surface hyaluronan receptor and mitotic spindle protein and the driver of tumor progression [19] [20] [21], plays an important role in the modulation of motor activities and the maintenance of genome stability [22, 23]. High expression of HMMR significantly correlates with tumor relapse [24, 25] and predicts poorer prognosis in GC patients. Furthermore, HMMR has been identified as a promising target for antibody therapy to block the extracellular function of HMMR on the surface of tumor cells [26], which might be a potential prognostic marker or therapeutic target against the disease. The protein encoded by CCNB1 gene is an important monitoring protein in mitosis, which is necessary for proper controlling the cell cycle at the G2/M transition phase [27]. Previous studies have reported that the CCNB1–Cdk1 complex is a key regulator of mitotic entry [28]. Recently, increasing evidence demonstrated that CCNB1 was over-expressed in considerable cancers with poor prognosis, including hepatocellular carcinoma [29, 30], breast cancer [31, 32], and pancreatic cancer [33, 34]. The expression of CCNB1 is often used to estimate prognosis after treatment with anticancer drugs [29, 35]. Studies had shown that CCNB1 were associated with gastric cancer [36, 37]. HnRNPR-CCNB1/CENPF axis may be a potential therapeutic target for GC treatment [38].
The function of MAD2L1 is to maintain the separation state of chromosomes during the dissociation of mitotic chromosomes and spindle, and to play a role in the checkpoint during mitosis [39, 40]. Abnormal regulation of MAD2L1 is associated with chromosomal instability and a large number of aneuploidy, which can lead to tumorigenesis [41]. Studies have found that MAD2L1 is overexpressed in lung adenocarcinoma tissues, and the overexpression of MAD2L1 may indicate poor prognosis and increased risk of tumor recurrence in patients, which can be used as a prognostic marker for lung adenocarcinoma [39]. Our bioinformatics analysis showed that MAD2L1 was highly expressed in tumor tissues compared with normal tissues. MAD2L1 is a pro-oncogene which is upregulated in GC [42, 43], and we need to further study its specific mechanism. The protein encoded by CCNA2 belongs to the highly conserved cyclin family, whose members function as regulators of the cell cycle at the G1/S and the G2/M transitions [44]. CCNA2 is overexpressed in several human cancers and closely related to tumor progression and shorter survival in lung, breast, and colorectal cancer [45–49]. Poor prognosis in GC patients related with high expressions of cyclins [50].CCNA2 is a novel predictive biomarker of sensitivity to PLK1 inhibitors for the treatment of advanced gastric cancer [51], whose overexpression was an indicator of poor prognosis. Limited by few studies about evaluating the expression and prognostic role of CCNA2 in GC patients, more efforts are necessary to confirm expression pattern and prognostic role of CCNA2 in GC patients.
CXCL8 is a member of the CXC chemokine family that acts as an important multifunctional cytokine to modulate tumor proliferation, invasion and migration in an autocrine or paracrine manner. Neovascularization, which provides a basis for fostering tumor growth and metastasis, is now recognized as a critical function of CXCL8 in the tumor microenvironment [52]. CXCL8 signaling axis also plays an indispensable role in colorectal carcinoma [53, 54], renal cell carcinoma, pancreatic cancer, thyroid tumors, gastric cancer [55, 56], and lymphomas [57]. Aberrant activation of CXCL8 in cancer-associated fibroblasts is correlated with poorer survival in gastric cancer patients [58]. Microarray analysis revealed that protein tyrosine phosphatase receptor delta-inactivation-induced CXCL8 promotes angiogenesis and metastasis in gastric cancer [59]. Interruption of the related signaling pathways may thus provide promising therapeutic avenues for tumors. Studies have found that CXCL8 is predominantly secreted by macrophages and contributes to the immunosuppressive microenvironment by inducing PD-L1+ macrophages in GC [60]. CXCL8 could be an early detection marker for perineural invasion-related GC, with a potential to be utilized as individual therapy targets [61]. CXCL8 inhibitors may drive antitumor response, providing potential therapeutic effects for patients with gastric cancer.
To further screen the TFs in hub genes, we constructed a gene-TF regulatory network and found IRF1, ETV4, KLFs, and SMAD5 that were meaningful in GC. It was reported that MTMR2 mediated epithelial-mesenchymal transition through the IFNγ/STAT1/IRF1 pathway to promote GC invasion and metastasis [62]. KIF2A expression is a potential target for GC therapy, which can be upregulated by transcription factor ETV4 [63]. Krüppel-like factors (KLFs) have been extensively investigated in multi-cancers, which plays a significant role in GC progression and could be a new therapeutic target for GC patients. Interestingly, SMAD5 was frequently altered in human GC [64]. The intricate interaction between TFs and other hub genes made great contribution to the development of cancer.
Studied have proved that Toll-like receptor (TLR) signaling pathways play important roles in development of GC. TLR signaling pathways are involved in innate and adaptive immunity responses [65] and activation of both inflammatory and carcinogenic processes [66]. Thus, the pattern of the host’s immune response beyond genetic and environmental factors is also essential for understanding the pathology of GC [67]. TLRs, a class of transmembrane receptors [68], play an important role in defense against Helicobacter pylori (H. pylori) widely known as a class I carcinogen in GC [68]. Therefore, the abnormal expression of TLRs is closely related to tumorigenesis and cancer progression and a better understanding of TLRs will provide new diagnostic or predictive markers for the diagnosis of GC.
We failed to validate SPP1 as a DEG in our fresh GC samples, which may be as a result of the small sample size and inter-sample variation. The protein encoded by SPP1 plays an important role in tumorigenesis, invasion and metastasis [10, 69]. Overexpressed SPP1 expression had been confirmed in various types of cancers [70–73]. A study based on gastric cancer cell lines indicated that the elevated expression of SPP1 is a critical determinant of poor prognosis [74]. In addition, in a recent study, SPP1 rs4754 polymorphism was observed to be associated with the risk of gastric cancer and has an important effect in gastric carcinogenesis [75]. However, it has been reported that SPP1 might not affect the prognosis of patients with GC [73], which needs more study in the future.
All above, we found that high expression of 5 validated hub genes should promote the progress of GC patients, suggesting that their antagonism may improve the prognosis of GC. Although some of these genes were found before, our study could validate and explain the expression status of these genes and their impact on prognosis in GC again. These findings provide a set of useful driving genes and key pathways of cancers, which are worth future investigating for novel therapeutic targets, a prognostic evaluation index, and the detailed pathogenesis of them in GCs.
However, there were several limitations of the present study. Firstly, validation with qRT-PCR study need more tumor and adjacent normal tissues samples. Second, more experiments, such as immunohistochemistry and Western blot, should be conducted to confirm the protein levels in GC.
Conclusion
Taken above, our bioinformatics analysis identified 295 DEGs of GC. Among them, HMMR, CCNB1, CXCL8, MAD2L1 and CCNA2 were verified and considered as Hub genes were associated with disease prognosis, which could be predictive and therapeutic targets.
Supplementary Information
Additional file 1 Table S1. All 295 commonly DEGs were detected from seven profile datasets, including 178 down-regulated genes and 117 up-regulated genes in the GC tissues compared to normal gastric tissues. Table S2. Significant models were obtained from the PPI network based on the MCODE analysis in Cytoscape. Table S3. The determined selected genes by using the cytoHubba plugin such as degree, betweenness centrality, and closeness. Table S4. The determined selected genes of Venn diagram. Table S5. The gene-TF regulatory network was constructed including 129 interaction pairs among 7 genes and 102 TFs.
Acknowledgments
We acknowledge and appreciate our colleagues for valuable efforts and comments on this paper.
Abbreviations
- GC
Gastric cancer
- DEGs
Differentially expressed genes
- GEPIA
Gene Expression Profiling Interactive analysis
- KEGG
Kyoto Encyclopedia of Gene and Genome
- PPI
protein–protein interaction
- TF
Transcriptional Factor
- qRT-PCR
quantitative real-time PCR
- RMA
Robust Multi-array Average
- BH
Benjamini & Hochberg
- MCODE
Molecular complex detection
- DAVID
Database for annotation, visualization and integrated discovery
- cDNA
complementary DNA
- NCs
Normal Controls
- PCA
Principal component analysis
- HMMR
Hyaluronan Mediated Motility Receptor
- SPP1
Secreted phosphoprotein 1
- FN1
Fibronectin 1
- CCNB1
Cyclin B1
- CXCL8
C-X-C motif chemokine ligand 8
- MAD2L1
Mitotic arrest deficient 2 like 1
- CCNA2
cyclin A2
- OS
Overall Survivals
- ZNF2
Zinc finger protein 2
- KLF16
Kruppel like factor 16
- BP
Biological Processes
- KLFs
Krüppel-like factors
- TLR
Toll-like receptor
- H. pylori
Helicobacter pylori
Authors’ contributions
Study design and manuscript writing: JZ and XL1. Data collection, quality assessment, analysis and interpretation of data: JZ, JQ, MQ, XL2 and XC. Manuscript revising: SZ, CG and HZ. All authors proofed and approved the submitted version of the article.
Funding
This project was supported by Graduate Students Outstanding Innovation Project Foundation of Shanxi Province (2C592020079). The funding bodies played no role in the design of the study and collection, analysis, and interpretation of data and in writing the manuscript.
Availability of data and materials
All data generated or analyzed during this study are included in this published article.
Declarations
Ethics approval and consent to participate
This trial is approved by the Ethics Committee for Clinical Research of Shanxi Cancer Hospital (ethics number: 2018-KY-0184).
Consent for publication
Not Applicable.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Xiao-Qing Lu, Jia-Qian Zhang are equally and should be regarded as co-first authors.
References
- 1.Bray F, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA Cancer J Clin. 2016;66(1):7–30. doi: 10.3322/caac.21332. [DOI] [PubMed] [Google Scholar]
- 3.Nagini S. Carcinoma of the stomach: a review of epidemiology, pathogenesis, molecular genetics and chemoprevention. World J Gastrointest Oncol. 2012;4(7):156–169. doi: 10.4251/wjgo.v4.i7.156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vogelstein B, Papadopoulos N, Velculescu VE, Zhou S, Diaz LA, Kinzler KW. Cancer genome landscapes. Science. 2013;339(6127):1546–1558. doi: 10.1126/science.1235122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Thomas PD. The gene ontology and the meaning of biological function. Methods Mol Biol. 2017;1446:15–24. doi: 10.1007/978-1-4939-3743-1_2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Da H, Sherman WBT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4(1):44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 7.Khunlertgit N, Yoon BJ. Incorporating topological information for predicting robust cancer subnetwork markers in human protein-protein interaction network. BMC Bioinformatics. 2016;17(Suppl 13):351. doi: 10.1186/s12859-016-1224-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ritchie ME, et al. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43(7):e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Carvalho BS, Irizarry RA. A framework for oligonucleotide microarray preprocessing. Bioinformatics. 2010;26(19):2363–2367. doi: 10.1093/bioinformatics/btq431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Szklarczyk D, Morris JH, Cook H, Kuhn M, Wyder S, Simonovic M, Santos A, Doncheva NT, Roth A, Bork P, Jensen LJ, von Mering C. The STRING database in 2017: quality-controlled protein-protein association networks, made broadly accessible. Nucleic Acids Res. 2017;45(D1):D362–d368. doi: 10.1093/nar/gkw937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Szklarczyk D, Franceschini A, Wyder S, Forslund K, Heller D, Huerta-Cepas J, Simonovic M, Roth A, Santos A, Tsafou KP, Kuhn M, Bork P, Jensen LJ, von Mering C. STRING v10: protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 2015;43(Database issue):D447–D452. doi: 10.1093/nar/gku1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13(11):2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bader GD, Hogue CW. An automated method for finding molecular complexes in large protein interaction networks. BMC Bioinformatics. 2003;4(1):2. doi: 10.1186/1471-2105-4-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Szasz AM, et al. Cross-validation of survival associated biomarkers in gastric cancer using transcriptomic data of 1,065 patients. Oncotarget. 2016;7(31):49322–49333. doi: 10.18632/oncotarget.10337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ferguson B, Bokka NR, Maddipati KR, Ayilavarapu S, Weltman R, Zhu L, Chen W, Zheng WJ, Angelov N, van Dyke TE, Lee CT. Distinct profiles of specialized pro-resolving lipid mediators and corresponding receptor gene expression in periodontal inflammation. Front Immunol. 2020;11:1307. doi: 10.3389/fimmu.2020.01307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xu Y, Liang C, Cai X, Zhang M, Yu W, Shao Q. High centromere protein-a (CENP-A) expression correlates with progression and prognosis in gastric Cancer. Onco Targets Ther. 2020;13:13237–13246. doi: 10.2147/OTT.S263512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Van Cutsem E, et al. Gastric cancer. Lancet. 2016;388(10060):2654–2664. doi: 10.1016/S0140-6736(16)30354-3. [DOI] [PubMed] [Google Scholar]
- 18.Chen W, Zheng R, Baade PD, Zhang S, Zeng H, Bray F, Jemal A, Yu XQ, He J. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66(2):115–132. doi: 10.3322/caac.21338. [DOI] [PubMed] [Google Scholar]
- 19.Telmer PG, Tolg C, McCarthy JB, Turley EA. How does a protein with dual mitotic spindle and extracellular matrix receptor functions affect tumor susceptibility and progression? Commun Integr Biol. 2011;4(2):182–185. doi: 10.4161/cib.4.2.14270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dunsch AK, Hammond D, Lloyd J, Schermelleh L, Gruneberg U, Barr FA. Dynein light chain 1 and a spindle-associated adaptor promote dynein asymmetry and spindle orientation. J Cell Biol. 2012;198(6):1039–1054. doi: 10.1083/jcb.201202112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Maxwell CA, Keats JJ, Crainie M, Sun X, Yen T, Shibuya E, Hendzel M, Chan G, Pilarski LM. RHAMM is a centrosomal protein that interacts with dynein and maintains spindle pole stability. Mol Biol Cell. 2003;14(6):2262–2276. doi: 10.1091/mbc.e02-07-0377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen H, Connell M, Mei L, Reid GSD, Maxwell CA. The nonmotor adaptor HMMR dampens Eg5-mediated forces to preserve the kinetics and integrity of chromosome segregation. Mol Biol Cell. 2018;29(7):786–796. doi: 10.1091/mbc.E17-08-0531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Manning AL, Compton DA. SnapShot: nonmotor proteins in spindle assembly. Cell. 2008;134(4):694–694.e1. doi: 10.1016/j.cell.2008.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yang D, Ma Y, Zhao P, Ma J, He C. Systematic screening of protein-coding gene expression identified HMMR as a potential independent indicator of unfavorable survival in patients with papillary muscle-invasive bladder cancer. Biomed Pharmacother. 2019;120:109433. doi: 10.1016/j.biopha.2019.109433. [DOI] [PubMed] [Google Scholar]
- 25.Zhang H, Ren L, Ding Y, Li F, Chen X, Ouyang Y, Zhang Y, Zhang D. Hyaluronan-mediated motility receptor confers resistance to chemotherapy via TGFbeta/Smad2-induced epithelial-mesenchymal transition in gastric cancer. FASEB J. 2019;33(5):6365–6377. doi: 10.1096/fj.201802186R. [DOI] [PubMed] [Google Scholar]
- 26.Hamilton SR, Fard SF, Paiwand FF, Tolg C, Veiseh M, Wang C, McCarthy JB, Bissell MJ, Koropatnick J, Turley EA. The hyaluronan receptors CD44 and Rhamm (CD168) form complexes with ERK1,2 that sustain high basal motility in breast cancer cells. J Biol Chem. 2007;282(22):16667–16680. doi: 10.1074/jbc.M702078200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Strauss B, Harrison A, Coelho PA, Yata K, Zernicka-Goetz M, Pines J. Cyclin B1 is essential for mitosis in mouse embryos, and its nuclear export sets the time for mitosis. J Cell Biol. 2018;217(1):179–193. doi: 10.1083/jcb.201612147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nakayama Y, Yamaguchi N. Role of cyclin B1 levels in DNA damage and DNA damage-induced senescence. Int Rev Cell Mol Biol. 2013;305:303–337. doi: 10.1016/B978-0-12-407695-2.00007-X. [DOI] [PubMed] [Google Scholar]
- 29.Chai N, Xie HH, Yin JP, Sa KD, Guo Y, Wang M, Liu J, Zhang XF, Zhang X, Yin H, Nie YZ, Wu KC, Yang AG, Zhang R. FOXM1 promotes proliferation in human hepatocellular carcinoma cells by transcriptional activation of CCNB1. Biochem Biophys Res Commun. 2018;500(4):924–929. doi: 10.1016/j.bbrc.2018.04.201. [DOI] [PubMed] [Google Scholar]
- 30.Zhuang L, Yang Z, Meng Z. Upregulation of BUB1B, CCNB1, CDC7, CDC20, and MCM3 in tumor tissues predicted worse overall survival and disease-free survival in hepatocellular carcinoma patients. Biomed Res Int. 2018;2018:7897346. doi: 10.1155/2018/7897346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kongsema M, Wongkhieo S, Khongkow M, Lam EW, Boonnoy P, Vongsangnak W, Wong-Ekkabut J. Molecular mechanism of Forkhead box M1 inhibition by thiostrepton in breast cancer cells. Oncol Rep. 2019;42(3):953–962. doi: 10.3892/or.2019.7225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu B, Liu Y, Wang Y, Xie C, Gan M, Han T, Cao J, Wang J. CyclinB1 deubiquitination by USP14 regulates cell cycle progression in breast cancer. Pathol Res Pract. 2019;215(10):152592. doi: 10.1016/j.prp.2019.152592. [DOI] [PubMed] [Google Scholar]
- 33.Zhang H, Zhang X, Li X, Meng WB, Bai ZT, Rui SZ, Wang ZF, Zhou WC, Jin XD. Effect of CCNB1 silencing on cell cycle, senescence, and apoptosis through the p53 signaling pathway in pancreatic cancer. J Cell Physiol. 2018;234(1):619–631. doi: 10.1002/jcp.26816. [DOI] [PubMed] [Google Scholar]
- 34.Zhou L, Li J, Zhao YP, Cui QC, Zhou WX, Guo JC, You L, Wu WM, Zhang TP. The prognostic value of cyclin B1 in pancreatic cancer. Med Oncol. 2014;31(9):107. doi: 10.1007/s12032-014-0107-4. [DOI] [PubMed] [Google Scholar]
- 35.Gu J, Liu X, Li J, He Y. MicroRNA-144 inhibits cell proliferation, migration and invasion in human hepatocellular carcinoma by targeting CCNB1. Cancer Cell Int. 2019;19(1):15. doi: 10.1186/s12935-019-0729-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu P, Wang X, Hu CH, Hu TH. Bioinformatics analysis with graph-based clustering to detect gastric cancer-related pathways. Genet Mol Res. 2012;11(3):3497–3504. doi: 10.4238/2012.September.26.5. [DOI] [PubMed] [Google Scholar]
- 37.Shi Q, Wang W, Jia Z, Chen P, Ma K, Zhou C. ISL1, a novel regulator of CCNB1, CCNB2 and c-MYC genes, promotes gastric cancer cell proliferation and tumor growth. Oncotarget. 2016;7(24):36489–36500. doi: 10.18632/oncotarget.9269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chen EB, Qin X, Peng K, Li Q, Tang C, Wei YC, Yu S, Gan L, Liu TS. HnRNPR-CCNB1/CENPF axis contributes to gastric cancer proliferation and metastasis. Aging (Albany NY) 2019;11(18):7473–7491. doi: 10.18632/aging.102254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cheng Y, Li K, Diao D, Zhu K, Shi L, Zhang H, Yuan D, Guo Q, Wu X, Liu D, Dang C. Expression of KIAA0101 protein is associated with poor survival of esophageal cancer patients and resistance to cisplatin treatment in vitro. Lab Investig. 2013;93(12):1276–1287. doi: 10.1038/labinvest.2013.124. [DOI] [PubMed] [Google Scholar]
- 40.Liu L, Chen X, Xie S, Zhang C, Qiu Z, Zhu F. Variant 1 of KIAA0101, overexpressed in hepatocellular carcinoma, prevents doxorubicin-induced apoptosis by inhibiting p53 activation. Hepatology. 2012;56(5):1760–1769. doi: 10.1002/hep.25834. [DOI] [PubMed] [Google Scholar]
- 41.Kato T, Daigo Y, Aragaki M, Ishikawa K, Sato M, Kaji M. Overexpression of KIAA0101 predicts poor prognosis in primary lung cancer patients. Lung Cancer. 2012;75(1):110–118. doi: 10.1016/j.lungcan.2011.05.024. [DOI] [PubMed] [Google Scholar]
- 42.Wang Y, Wang F, He J, du J, Zhang H, Shi H, Chen Y, Wei Y, Xue W, Yan J, Feng Y, Gao Y, Li D, Han J, Zhang J. miR-30a-3p targets MAD2L1 and regulates proliferation of gastric Cancer cells. Onco Targets Ther. 2019;12:11313–11324. doi: 10.2147/OTT.S222854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kim Y, Choi JW, Lee JH, Kim YS. Spindle assembly checkpoint MAD2 and CDC20 overexpressions and cell-in-cell formation in gastric cancer and its precursor lesions. Hum Pathol. 2019;85:174–183. doi: 10.1016/j.humpath.2018.10.029. [DOI] [PubMed] [Google Scholar]
- 44.Arsic N, Bendris N, Peter M, Begon-Pescia C, Rebouissou C, Gadéa G, Bouquier N, Bibeau F, Lemmers B, Blanchard JM. A novel function for cyclin A2: control of cell invasion via RhoA signaling. J Cell Biol. 2012;196(1):147–162. doi: 10.1083/jcb.201102085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ko E, Kim Y, Cho EY, Han J, Shim YM, Park J, Kim DH. Synergistic effect of Bcl-2 and cyclin A2 on adverse recurrence-free survival in stage I non-small cell lung cancer. Ann Surg Oncol. 2013;20(3):1005–1012. doi: 10.1245/s10434-012-2727-2. [DOI] [PubMed] [Google Scholar]
- 46.Gao T, Han Y, Yu L, Ao S, Li Z, Ji J. CCNA2 is a prognostic biomarker for ER+ breast cancer and tamoxifen resistance. PLoS One. 2014;9(3):e91771. doi: 10.1371/journal.pone.0091771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gopinathan L, Tan SLW, Padmakumar VC, Coppola V, Tessarollo L, Kaldis P. Loss of Cdk2 and cyclin A2 impairs cell proliferation and tumorigenesis. Cancer Res. 2014;74(14):3870–3879. doi: 10.1158/0008-5472.CAN-13-3440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bukholm IR, Bukholm G, Nesland JM. Over-expression of cyclin a is highly associated with early relapse and reduced survival in patients with primary breast carcinomas. Int J Cancer. 2001;93(2):283–287. doi: 10.1002/ijc.1311. [DOI] [PubMed] [Google Scholar]
- 49.Handa K, Yamakawa M, Takeda H, Kimura S, Takahashi T. Expression of cell cycle markers in colorectal carcinoma: superiority of cyclin a as an indicator of poor prognosis. Int J Cancer. 1999;84(3):225–233. doi: 10.1002/(SICI)1097-0215(19990621)84:3<225::AID-IJC5>3.0.CO;2-A. [DOI] [PubMed] [Google Scholar]
- 50.Zhang HP, Li SY, Wang JP, Jun L. Clinical significance and biological roles of cyclins in gastric cancer. Onco Targets Ther. 2018;11:6673–6685. doi: 10.2147/OTT.S171716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lee Y, et al. Pharmacogenomic Analysis Reveals CCNA2 as a Predictive Biomarker of Sensitivity to Polo-Like Kinase I Inhibitor in Gastric Cancer. Cancers (Basel). 2020:12(6). [DOI] [PMC free article] [PubMed]
- 52.Liu Q, Li A, Tian Y, Wu JD, Liu Y, Li T, Chen Y, Han X, Wu K. The CXCL8-CXCR1/2 pathways in cancer. Cytokine Growth Factor Rev. 2016;31:61–71. doi: 10.1016/j.cytogfr.2016.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Brew R, Erikson JS, West DC, Kinsella AR, Slavin J, Christmas SE. Interleukin-8 as an autocrine growth factor for human colon carcinoma cells in vitro. Cytokine. 2000;12(1):78–85. doi: 10.1006/cyto.1999.0518. [DOI] [PubMed] [Google Scholar]
- 54.Xiao YC, Yang ZB, Cheng XS, Fang XB, Shen T, Xia CF, Liu P, Qian HH, Sun B, Yin ZF, Li YF. CXCL8, overexpressed in colorectal cancer, enhances the resistance of colorectal cancer cells to anoikis. Cancer Lett. 2015;361(1):22–32. doi: 10.1016/j.canlet.2015.02.021. [DOI] [PubMed] [Google Scholar]
- 55.Yasumoto K, Okamoto S, Mukaida N, Murakami S, Mai M, Matsushima K. Tumor necrosis factor alpha and interferon gamma synergistically induce interleukin 8 production in a human gastric cancer cell line through acting concurrently on AP-1 and NF-kB-like binding sites of the interleukin 8 gene. J Biol Chem. 1992;267(31):22506–22511. doi: 10.1016/S0021-9258(18)41701-2. [DOI] [PubMed] [Google Scholar]
- 56.Kitadai Y, Takahashi Y, Haruma K, Naka K, Sumii K, Yokozaki H, Yasui W, Mukaida N, Ohmoto Y, Kajiyama G, Fidler IJ, Tahara E. Transfection of interleukin-8 increases angiogenesis and tumorigenesis of human gastric carcinoma cells in nude mice. Br J Cancer. 1999;81(4):647–653. doi: 10.1038/sj.bjc.6690742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Isaza-Correa JM, Liang Z, van den Berg A, Diepstra A, Visser L. Toll-like receptors in the pathogenesis of human B cell malignancies. J Hematol Oncol. 2014;7(1):57. doi: 10.1186/s13045-014-0057-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Naito Y, Yamamoto Y, Sakamoto N, Shimomura I, Kogure A, Kumazaki M, Yokoi A, Yashiro M, Kiyono T, Yanagihara K, Takahashi RU, Hirakawa K, Yasui W, Ochiya T. Cancer extracellular vesicles contribute to stromal heterogeneity by inducing chemokines in cancer-associated fibroblasts. Oncogene. 2019;38(28):5566–5579. doi: 10.1038/s41388-019-0832-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bae WJ, Ahn JM, Byeon HE, Kim S, Lee D. PTPRD-inactivation-induced CXCL8 promotes angiogenesis and metastasis in gastric cancer and is inhibited by metformin. J Exp Clin Cancer Res. 2019;38(1):484. doi: 10.1186/s13046-019-1469-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lin C, He H, Liu H, Li R, Chen Y, Qi Y, Jiang Q, Chen L, Zhang P, Zhang H, Li H, Zhang W, Sun Y, Xu J. Tumour-associated macrophages-derived CXCL8 determines immune evasion through autonomous PD-L1 expression in gastric cancer. Gut. 2019;68(10):1764–1773. doi: 10.1136/gutjnl-2018-316324. [DOI] [PubMed] [Google Scholar]
- 61.Jia X, Lu M, Rui C, Xiao Y. Consensus-expressed CXCL8 and MMP9 identified by meta-analyzed Perineural invasion gene signature in gastric Cancer microarray data. Front Genet. 2019;10:851. doi: 10.3389/fgene.2019.00851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Jing JJ, Wang ZY, Li H, Sun LP, Yuan Y. Key elements involved in Epstein-Barr virus-associated gastric cancer and their network regulation. Cancer Cell Int. 2018;18(1):146. doi: 10.1186/s12935-018-0637-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhang X, Wang Y, Liu X, Zhao A, Yang Z, Kong F, Sun L, Yu Y, Jiang L. KIF2A promotes the progression via AKT signaling pathway and is upregulated by transcription factor ETV4 in human gastric cancer. Biomed Pharmacother. 2020;125:109840. doi: 10.1016/j.biopha.2020.109840. [DOI] [PubMed] [Google Scholar]
- 64.Jiang L, Liu JY, Shi Y, Tang B, He T, Liu JJ, Fan JY, Wu B, Xu XH, Zhao YL, Qian F, Cui YH, Yu PW. MTMR2 promotes invasion and metastasis of gastric cancer via inactivating IFNgamma/STAT1 signaling. J Exp Clin Cancer Res. 2019;38(1):206. doi: 10.1186/s13046-019-1186-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.De Re V, et al. Polymorphism in Toll-Like Receptors and Helicobacter Pylori Motility in Autoimmune Atrophic Gastritis and Gastric Cancer. Cancers (Basel) 2019;11:5. doi: 10.3390/cancers11050648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Susi MD, Lourenço CM, Rasmussen LT, Payão SLM, Rossi AFT, Silva AE, Oliveira-Cucolo JG. Toll-like receptor 9 polymorphisms and helicobacter pylori influence gene expression and risk of gastric carcinogenesis in the Brazilian population. World J Gastrointest Oncol. 2019;11(11):998–1010. doi: 10.4251/wjgo.v11.i11.0000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Song M, Rabkin CS, Camargo MC. Gastric Cancer: an evolving disease. Curr Treat Options Gastroenterol. 2018;16(4):561–569. doi: 10.1007/s11938-018-0203-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Varga MG, Peek RM. DNA transfer and toll-like receptor modulation by helicobacter pylori. Curr Top Microbiol Immunol. 2017;400:169–193. doi: 10.1007/978-3-319-50520-6_8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Endo H, Ikeda K, Urano T, Horie-Inoue K, Inoue S. Terf/TRIM17 stimulates degradation of kinetochore protein ZWINT and regulates cell proliferation. J Biochem. 2012;151(2):139–144. doi: 10.1093/jb/mvr128. [DOI] [PubMed] [Google Scholar]
- 70.Likui W, Hong W, Shuwen Z. Clinical significance of the upregulated osteopontin mRNA expression in human colorectal cancer. J Gastrointest Surg. 2010;14(1):74–81. doi: 10.1007/s11605-009-1035-z. [DOI] [PubMed] [Google Scholar]
- 71.Chen X, Xiong D, Ye L, Yang H, Mei S, Wu J, Chen S, Mi R. SPP1 inhibition improves the cisplatin chemo-sensitivity of cervical cancer cell lines. Cancer Chemother Pharmacol. 2019;83(4):603–613. doi: 10.1007/s00280-018-3759-5. [DOI] [PubMed] [Google Scholar]
- 72.Xu C, Sun L, Jiang C, Zhou H, Gu L, Liu Y, Xu Q. SPP1, analyzed by bioinformatics methods, promotes the metastasis in colorectal cancer by activating EMT pathway. Biomed Pharmacother. 2017;91:1167–1177. doi: 10.1016/j.biopha.2017.05.056. [DOI] [PubMed] [Google Scholar]
- 73.Zhuo C, Li X, Zhuang H, Tian S, Cui H, Jiang R, Liu C, Tao R, Lin X. Elevated THBS2, COL1A2, and SPP1 expression levels as predictors of gastric Cancer prognosis. Cell Physiol Biochem. 2016;40(6):1316–1324. doi: 10.1159/000453184. [DOI] [PubMed] [Google Scholar]
- 74.Higashiyama M, Ito T, Tanaka E, Shimada Y. Prognostic significance of osteopontin expression in human gastric carcinoma. Ann Surg Oncol. 2007;14(12):3419–3427. doi: 10.1245/s10434-007-9564-8. [DOI] [PubMed] [Google Scholar]
- 75.Chen LZ, He CY, Su X, Peng JL, Chen DL, Ye Z, Yang DD, Wang ZX, Wang F, Shao JY, Xu RH. SPP1 rs4754 and its epistatic interactions with SPARC polymorphisms in gastric cancer susceptibility. Gene. 2018;640:43–50. doi: 10.1016/j.gene.2017.09.053. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1 Table S1. All 295 commonly DEGs were detected from seven profile datasets, including 178 down-regulated genes and 117 up-regulated genes in the GC tissues compared to normal gastric tissues. Table S2. Significant models were obtained from the PPI network based on the MCODE analysis in Cytoscape. Table S3. The determined selected genes by using the cytoHubba plugin such as degree, betweenness centrality, and closeness. Table S4. The determined selected genes of Venn diagram. Table S5. The gene-TF regulatory network was constructed including 129 interaction pairs among 7 genes and 102 TFs.
Data Availability Statement
All data generated or analyzed during this study are included in this published article.