Abstract
In this work, we propose a free-breathing magnetic resonance fingerprinting (MRF) method that can be used to obtain B1+-robust quantitative T1 maps of the abdomen in a clinically acceptable time. A three-dimensional MRF sequence with a radial stack-of-stars trajectory was implemented, and its k-space acquisition ordering was adjusted to improve motion-robustness in the context of MRF. The flip angle pattern was optimized using the Cramér–Rao Lower Bound, and the encoding efficiency of sequences with 300, 600, 900 and 1800 flip angles was evaluated. To validate the sequence, a movable multicompartment phantom was developed. Reference multiparametric maps were acquired under stationary conditions using a previously validated MRF method. Periodic motion of the phantom was used to investigate the motion-robustness of the proposed sequence. The best performing sequence length (600 flip angles) was used to image the abdomen during a free-breathing volunteer scan. When using a series of 600 or more flip angles, the estimated T1 values in the stationary phantom showed good agreement with the reference scan. Phantom experiments revealed that motion-related artifacts can appear in the quantitative maps and confirmed that a motion-robust k-space ordering is essential. The in vivo scan demonstrated that the proposed sequence can produce clean parameter maps while the subject breathes freely. Using this sequence, it is possible to generate B1+-robust quantitative maps of T1 and B1+ next to M0-weighted images under free-breathing conditions at a clinically usable resolution within 5 min.
Keywords: abdominal imaging, Cramér–Rao lower bound, magnetic resonance fingerprinting, quantitative imaging, respiratory motion
1 ∣. INTRODUCTION
Most routine clinical magnetic resonance imaging (MRI) measurements depict a relative contrast between tissues. In addition to the tissue properties, the measured signal intensities depend on experimental factors, such as RF excitation and receive sensitivities.1 Consequently, the obtained qualitative contrast depends on the specific sequence parameters and scanner hardware used to acquire the data. Quantitative MRI, on the other hand, strives to directly measure physical or chemical properties of tissues, most notably the longitudinal (T1) and transverse (T2) relaxation times. Compared with qualitative images, such quantitative maps enable more straightforward comparisons between scans from different patients, different time points, or using different hardware.2,3
Quantification of MR parameters can be performed using several techniques. Historically, relaxometry measurements were performed by fitting exponentials to a series of inversion times or echo times to quantify T1 or T2 relaxation times, respectively.1 However, such measurements are too slow for routine clinical usage. Over the years many techniques have been developed to provide a better balance between accuracy and acquisition speed, such as the Look-Locker method4 or DESPOT1 and DESPOT2,5 among others. Recently, MR fingerprinting (MRF) has been proposed for the fast and simultaneous quantification of multiple parameters.6 This method uses a variable flip angle train that produces unique signal evolutions, so-called “fingerprints”, for different tissue parameter combinations. By comparing the measured fingerprints with the entries in a precomputed dictionary, the underlying tissue parameters, such as T1, T2 and equilibrium magnetization (M0), can be estimated.
Several challenges arise when attempting quantitative imaging of the abdomen. Intestinal gas in the abdomen can cause susceptibility artifacts, especially for field strengths of 3 T and higher.7 Furthermore, the relatively short wavelength of the RF can lead to interferences in the B1+ field, resulting in an inhomogeneous excitation.7,8 Moreover, respiratory, cardiac, gastrointestinal and voluntary motion can cause other image artifacts9 and can corrupt the MRF-based parameter maps.10 Although breathholds can be used to eliminate respiratory motion during an MRF experiment,11 it places restrictive limits on scan time and thus on spatial resolution or volumetric coverage. Besides, not all patients can hold their breath (e.g. pediatric patients).12,13 Therefore, it is preferable that the sequence is inherently robust against motion, such that the subject can breathe freely during the scan.14
In this work, we present a free-breathing 3D MRF sequence to generate quantitative T1 and B1+ maps, as well as M0-weighted images. The k-space ordering was adjusted to improve robustness against motion, while B1+-related artifacts were addressed by including B1+ in the parameter estimation. The flip angles were optimized to improve the efficiency of the sequence and to decrease the scan time. The method was validated using a movable phantom and subsequently evaluated in vivo during free-breathing abdominal measurements.
2 ∣. METHODS
2.1 ∣. Sequence design
Building on prior work,15 an MRF sequence was designed to generate a distinct signal pattern for every combination of T1 and B1+. By including the B1+ value in the parameter estimation, the effect of an inhomogeneous excitation field can be dealt with.16,17 Since most B0-robust T2-encoding solutions are particularly motion sensitive,10 only fast low angle shot (FLASH) segments with a repetition time (TR) of 5 ms were used. These segments were gradient- and RF-spoiled to avoid T2 dependence.
To reduce the sensitivity of the sequence to motion, a radial readout sampling pattern was chosen.14 This trajectory samples the center of k-space, which encodes the global image contrast with every readout. Consequently, the motion during acquisition is averaged out in the resulting image. Furthermore, undersampling artifacts are expressed as streaks in the image,18 instead of the less desirable ghosts that occur with a Cartesian sampling pattern. These streaks lead to incoherent, noise-like artifacts in the reconstructed images, which can be filtered out by the dictionary-matching process. The extension to 3D can be made by stacking multiple partitions on top of each other, where each partition consists of several radial readouts, to form a stack-of-stars trajectory.14
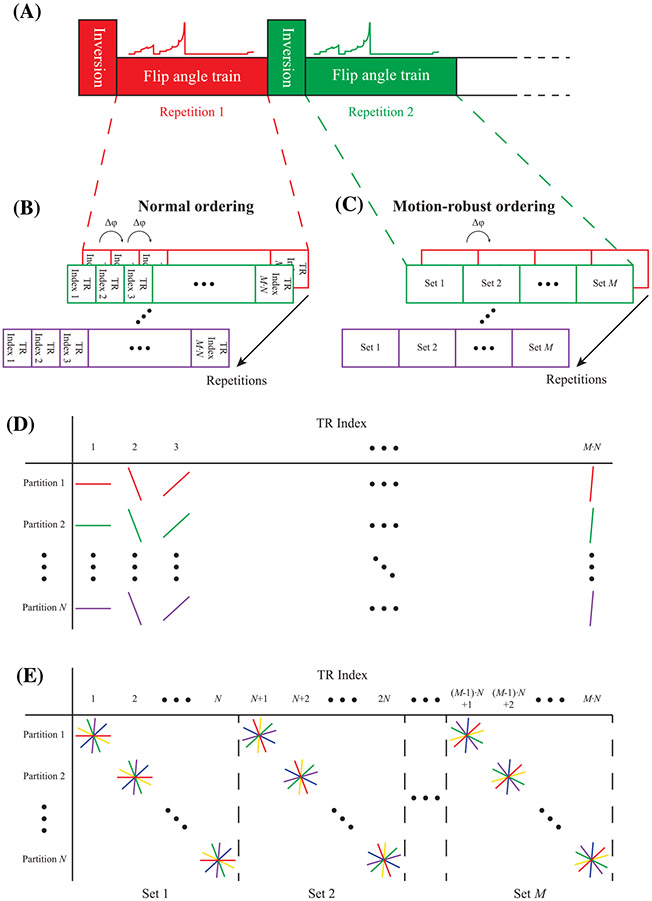
The simplest stack-of-stars implementation, hereafter referred to as normal ordering, keeps the partition index fixed during the flip angle sequence (Figure 1). The readout angle is changed by 111.25 degrees (the golden angle) every readout.19 The flip angle sequence is then repeated once for every partition, after which one shot has been acquired. To increase the sampling density, multiple shots can be acquired. Using this ordering, lines in adjacent partitions are acquired using the same flip angle, and thus have the same contrast weighting. However, this ordering scheme results in a time interval of several seconds between adjacent lines in the partition direction, and any motion during this time results in motion artifacts in the fingerprints.
FIGURE 1.
Proposed motion-robust 3D MRF sequence. (A), The data are acquired using a repeated flip angle sequence, each generating a fingerprint encoding T1 and B1+. Every repetition is preceded by a nonselective adiabatic inversion pulse. Each repetition is indicated with a separate color. (B), In the normal ordering, each fingerprint sequence consists of M·N TRs. Each TR index generates a separate k-space. (C), In the motion-robust ordering, each fingerprint sequence consists of M sets, containing N TRs each. All readouts acquired during the TR indices corresponding to one set are grouped together to form a single k-space. (D), Readout lines acquired for the normal ordering. Every TR, the readout angle is incremented with the golden angle (Δφ = 111.25 degrees), while the partition index stays the same. This results in a single undersampled k-space for every TR index. All lines of the same color are acquired sequentially during the same repetition. (E), Readout lines acquired for the motion-robust ordering. Every TR, the readout angle stays the same, but the partition index is increased. Note how readouts in adjacent partitions with the same angle are acquired successively in the motion-robust ordering, while in the normal ordering, the next partition is sampled during the next repetition of the sequence
To obtain motion robustness while using a stack-of-stars sampling pattern, all partitions are typically acquired in quick succession before changing the readout angle.14 However, during an MRF experiment, the MR signal is in a transient state because the flip angle changes for every TR index. Hence, it is not possible to sample all partitions with the same contrast weighting in quick succession. Therefore, to retain motion robustness, we propose to increase the TR index and the partition index simultaneously (hereafter referred to as motion-robust ordering; see Figure 1). For every readout, the next partition is sampled, until the last partition is reached, after which the first partition is sampled again. To remain consistent with the normal ordering, one shot is defined to be completed after the flip angle sequence has been repeated as many times as the number of partitions. For each repetition, the sequence of partition indices remains the same, but all readout angles are incremented with the golden angle.
To allow reconstruction of the data with the motion-robust ordering, the measured signals are divided into sets, with each set containing the data from as many consecutive TR indices as the number of partitions. Note that this will introduce some contrast mixing during the Fourier transform along the partition direction. When the flip angles change smoothly, it is our hypothesis that the differences in signal intensities along the partition direction will remain acceptable.
For both ordering schemes, data acquisition was started from the second repetition onwards to ensure that the initial magnetization at the start of each repetition was identical.20
2.2 ∣. Dictionary construction
The Bloch equations21 were used to create a precomputed dictionary consisting of 17,600 simulated fingerprints, each with a unique combination of T1 and B1+ values. The T1 values ranged from 50 to 3764 ms with relative increments of 2.5%, and B1+ effects were implemented by multiplying the desired flip angle with a relative weighting factor ranging from 0.02 to 2.0 in steps of 0.02. The complete sequence consisted of gradient echoes with both gradient- and RF-spoiling, which prevented the formation of stimulated echoes and thus eliminated any T2 contrast.16 In addition, the echo time (TE) was held fixed, resulting in a constant scaling of the fingerprints caused by T2* effects. However, since this effect is indistinguishable from M0 contrast, T2* effects were not simulated in the dictionary.
For every combination of T1 and B1+ in the dictionary, the whole sequence was simulated for two repetitions. The first repetition was used to obtain the initial magnetization during a continuous scan, while the second repetition was used to obtain the fingerprint. Next, for the motion-robust ordering, the fingerprint signals from Npartitions consecutive readouts were grouped into sets (as in Figure 1), and the signals within each set were averaged. The dictionary was compressed by projecting the fingerprints onto the first five singular vectors of the dictionary.22 This rank-5 approximation retained more than 99.5% of the energy in all dictionaries, where the energy is defined as the sum of the squared singular values. Finally, all simulated and averaged fingerprints were normalized to have unit Euclidean norm.
2.3 ∣. Sequence optimization
Besides making the sequence motion robust, the flip angle pattern was optimized to bring the acquisition time down to under 5 min. Four different patterns were designed, corresponding to a sequence of 300, 600, 900 and 1800 flip angles. Consecutive flip angles were placed 5 ms apart, with a nonselective adiabatic inversion pulse23 at the start of each sequence. Note, however, that the optimization algorithm can set the flip angle to 0 degrees, thus effectively creating a delay.
As a measure of optimality, the Cramér–Rao Lower Bound (CRLB)24 was used. This measure expresses the minimum variance of a set of estimated parameters, in this case T1 and B1+, obtained using an unbiased estimator, here the MRF reconstruction. The CRLB has previously been used to find optimal parameters for an MRI experiment, as well as for MRF in particular.25-28
The measurements are assumed to be subject to white Gaussian noise:
| (1) |
where sn is the measured signal intensity in the nth set, Mn is the normalized and averaged signal intensity of the nth set as calculated by the same simulator used for the dictionary, is the vector of all estimated parameters (T1 and B1+ in this case), is the vector containing all flip angles in the sequence, and is normally distributed noise with standard deviation σ. Since the fingerprints are normalized, the standard deviation of the measurement noise should be scaled identically to obtain the right σ. The Fisher information matrix (FIM), which can be used to obtain the CRLB,24 can be written as:
| (2) |
with the FIM, σ the standard deviation scaled according to the normalization of the fingerprint, and the Jacobian of the signal Mn with respect to the estimated parameters θ. Taking the inverse of the FIM gives a matrix, whose diagonal elements indicate the CRLBs of the estimated parameters given the flip angle sequence24:
| (3) |
These variances were normalized by the square of the true T1 and B1+ values, respectively, to give the coefficient of variation (COV) for both parameters. To make the MRF sequence sensitive to both T1 and B1+, the sum of these two relative CRLBs (the trace of the covariance matrix) was minimized.
The CRLB of a parameter gives an estimate of the sensitivity around a specific parameter value. Since we want the MRF experiment to be sensitive to a range of parameter values, the relative CRLBs for Nl different combinations of T1 and B1+ values were calculated and averaged. Furthermore, all flip angles were limited to 60 degrees, corresponding to the peak transmit voltage for an average subject when using an apodized sinc-shaped RF-pulse of 1.5 ms with a time-bandwidth factor of 8. The final optimization problem is given by Equation 4:
| (4) |
Here, is the lth parameter combination, is a diagonal matrix with weighting factors for the parameters, tr(·) denotes the trace of a matrix, αn is the nth flip angle in the sequence, and q is the total number of flip angles. We used , which normalized the variances of both parameters.
During optimization, the number of flip angles was fixed. To investigate the influence of different sequence lengths, the sequence was optimized for 300, 600, 900 and 1800 flip angles, using T1 values between 500 and 1500 ms in steps of 250 ms, and relative B1+ values of 0.75, 1.0 and 1.25, giving Nl = 15. The sequences were initialized with smooth random flip angle patterns with values between 0 and 10 degrees. The optimization was repeated for 20 different initializations. The optimized sequence with the lowest objective value after optimization was selected. There was no explicit delay time between subsequent repetitions of the sequence. However, note that the optimization algorithm was able to set the flip angles to negligible low values, thus effectively creating a delay that allowed the magnetization to relax towards equilibrium.
The optimization problem given in Equation 4 was implemented in MATLAB (MathWorks, Natick, MA, USA). Automatic differentiation,27,29 implemented using the CasADI toolbox,30 was used to calculate the Jacobian in Equation 2, as well as the gradient and Hessian-times-vector product of the objective function in Equation 4. The minimization problem was solved with the MATLAB function fmincon. The code used to optimize the flip angles is available on our bitbucket: https://bitbucket.org/MaxvRiel/free-breathing-mr-fingerprinting/.
2.4 ∣. Imaging experiments
All experiments were performed on a clinical 3-T MRI scanner (Prisma, Siemens Healthineers, Erlangen, Germany). A phantom containing seven glass tubes with different T1 values was placed on a cart made from LEGO bricks (The Lego Group, Billund, Denmark). This cart was placed on a slope of approximately 7 degrees and connected to a motor just outside the scanner room, which was controlled using the RWTH Mindstorms NXT Toolbox for MATLAB (RWTH Aachen University, Aachen, Germany). An 18-channel body coil (Siemens) was placed over the phantom and the cart (Figure 2). The phantom was scanned while stationary and while moving back and forth with frequencies of 0.22, 0.24 and 0.30 Hz, which are within the range of normal breathing frequencies in adults.31 The peak-to-peak amplitude of the motion was 32 mm. All scans were performed with both ordering schemes. A previously validated 2D MRF implementation with slice profile correction15 was used to obtain reference T1 values in the absence of motion. In addition, to validate the quantitative B1+ values, a gold standard B1+ map was obtained using the saturation prepared turbo-FLASH method.32
FIGURE 2.
Experimental setup. (A), The phantom placed on a movable cart. This cart is placed on a ramp. Under the influence of gravity, the cart moves to the right, while a rope connected to a motor can pull the cart to the left. (B), The 18-channel body coil that was used to acquire the data was placed over the movable phantom
In order to keep the same acquisition time of 4 min 30 s for the different flip angle trains, more shots were used for shorter sequences. This resulted in six, three, two and one shots for the sequences with 300, 600, 900 and 1800 TRs, respectively.
Abdominal scans were performed on six healthy volunteers after obtaining written informed consent. The study was approved by the institutional review board at the New York University School of Medicine. The same scanner and body coil were used as during the phantom scan. The volunteers were instructed to breathe normally, but to avoid taking deep breaths (by sighing or yawning). The parameters used for both experiments are summarized in Table 1. In addition, a DESPOT1 scan5 was performed for one volunteer to compare the resulting T1 maps. Furthermore, for one volunteer we acquired data at a higher resolution of 1.0 mm × 1.0 mm × 3.0 mm and with five shots.
TABLE 1.
Image acquisition parameters used in the phantom and in vivo experiments
| Parameter | Value (phantom) | Value (in vivo) |
|---|---|---|
| Repetition time (TR) | 5.0 ms | |
| Echo time (TE) | 2.4 ms | |
| Inversion time (TI) | 10 ms | |
| Number of partitions | 30 | |
| Acquisition time | 4 min 30 s | |
| Resolution | 1.0 mm × 1.0 mm × 6.0 mm | 1.3 mm × 1.3 mm × 3.0 mm |
| Field of view | 256 mm × 256 mm × 180 mm | 420 mm × 420 mm × 90 mm |
2.5 ∣. Image reconstruction
After acquiring all the data, the receive channels were compressed to five (phantom) or 10 (in vivo) virtual coils for every slice independently, retaining more than 98.5% of the energy in all datasets. A fast Fourier transform (FFT) was performed in the partition direction. A rank-5 approximation of the k-space data was made using the same five singular vectors computed for the dictionary compression.22 Next, every partition was reconstructed separately using nonuniform FFT (NUFFT),33 and the virtual coil channels were combined using a matched-filter reconstruction.34
For every voxel, the dot product between the fingerprint and the dictionary was maximized to identify the T1 and B1+ values associated with that voxel. The Euclidean norm corresponding to the (unnormalized) dictionary entry was used to estimate M0. The image reconstruction, dictionary construction and dictionary matching were all implemented in MATLAB. The code used to reconstruct the parameter maps is available at https://bitbucket.org/MaxvRiel/free-breathing-mr-fingerprinting/.
2.6 ∣. Image analysis
In order to analyze the parameter maps of the moving phantom, a region of interest (ROI) was drawn in each of the seven tubes. The mean and standard deviation of all estimated parameters were calculated for each tube. The estimated T1 values were validated against those from the reference scan.
Three ROIs were drawn in each in vivo dataset, where care was taken to avoid any vessels. The mean and standard deviation of T1 values were determined from these ROIs, and these values were compared with values from the literature.
3 ∣. RESULTS
3.1 ∣. Flip angle patterns
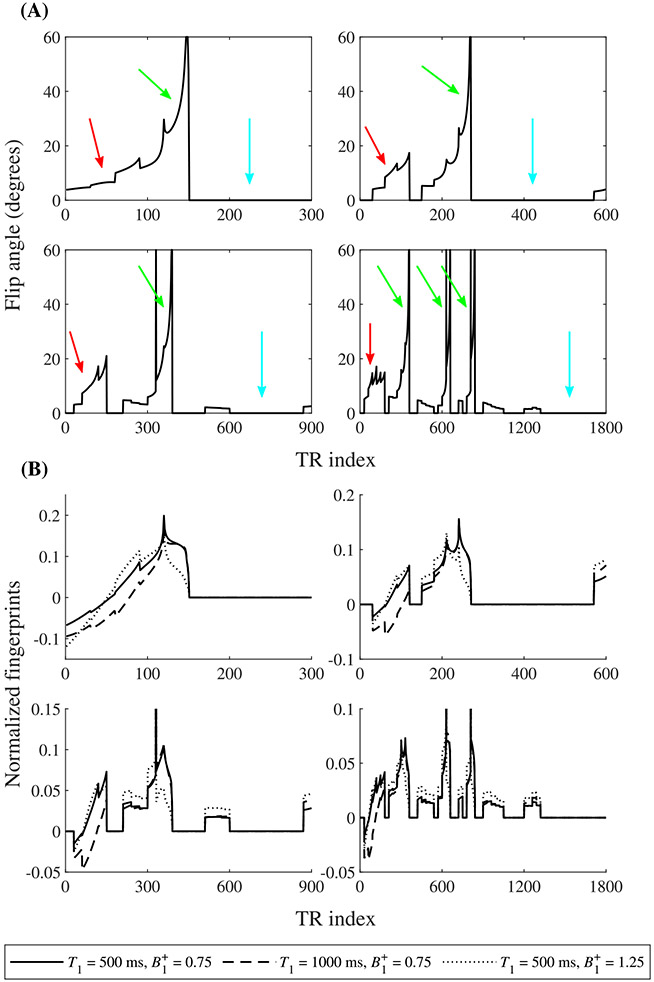
The flip angles optimized for different train lengths (Figure 3) showed similar patterns. Within each set of 30 consecutive TRs (the number of partitions), the flip angles generally increase smoothly, which results in smoothly varying signal intensities within each set. The flip angles show large jumps only between sets.
FIGURE 3.
Results of the flip angle optimization. (A), Optimized flip angle patterns for 300, 600, 900 and 1800 TRs (left to right, top to bottom). Note the smoothly varying flip angles within each set. Indicated are the main T1-encoding part (red arrows), the main B1+-encoding part (green arrows) and the delay (light blue arrows). (B), Normalized fingerprints for different combinations of T1 and B1+ values for the same four flip angle patterns
Furthermore, the flip angles of the first few sets in all sequences follow a similar pattern, which occurs around the zero-crossing of the inverted magnetization. Hence, it is mostly dependent on T1 and mainly serves to encode T1 in the fingerprint (solid and dashed lines in Figure 3B).
Next, there is a recurring pattern that consists of a single high flip angle, followed by a set of increasing flip angles. This pattern is particularly visible in the sequences with 900 and 1800 TRs but is also present in the sequences with 300 and 600 TRs. It mainly serves to provide B1+-sensitive signals (solid and dotted lines in Figure 3B). The single high flip angle partly saturates the signal, with the amount of saturation depending on the B1+ value. The succeeding excitations then provide the B1+-dependent signal for the fingerprint. Note that this pattern is repeated several times in the sequence with 1800 TRs.
Since the magnetization is saturated after this B1+-encoding pattern, any subsequent excitations would not generate much signal. The optimization resulted in multiple segments with very small flip angles. These segments do not provide much useful information. Instead, they decrease patient discomfort as the scanner produces more noise. Therefore, we decided to set all flip angles smaller than 1.5 degrees to zero. During these segments, the magnetization can recover before the next repetition of the sequence.
3.2 ∣. Phantom results
The T1 maps from the phantom scan for all four optimized sequences can be seen in Figure 4. For M0, T1, and B1+ maps, acquired using all tested motion speeds, see Figures S1-S8. The sequence with 300 flip angles and six shots slightly underestimated higher T1 values, while the longest flip angle train (1800 flip angles, one shot) showed large variability in the T1 estimates. This is also visible when looking at the correlation between the estimated and the reference T1 values (Figures 5 and S9). The other two sequences showed a better compromise between the number of shots and encoding capability.
FIGURE 4.
T1 maps of the phantom scan without (left) and with (right) motion, for both ordering schemes, and for all four sequence lengths for which the flip angles were optimized. For the maps with motion, the motion speeds with the most severe artifacts were selected for each sequence separately. Note the severe motion-related artifacts visible when using the normal ordering, which are greatly reduced by using the motion-robust ordering
FIGURE 5.
Correlation plots between the estimated T1 values (vertical axis) and the corresponding T1 values from the reference scan (horizontal axis) for each map in Figure 4. Each point indicates the mean T1 values of both scans within one single tube, with the standard deviation depicted as error bars. The dashed line is the identity line. Note the increased deviation from the identity line for the normal ordering compared with the motion-robust ordering
The sequence with 600 flip angles was selected to be investigated further, as this sequence showed the highest accuracy for the T1 estimation when using the motion-robust ordering. Phantom scans without motion also indicated that the obtained quantitative B1+ values correlate well with the saturation prepared turbo-FLASH method (Figure S10).
In Figure 6, the estimated parameter maps for this sequence with both ordering schemes, and both with and without motion, are shown. Without motion, both ordering schemes showed good agreement with the reference scan. In the presence of motion, however, the normal ordering showed notable motion artifacts in all parameter maps, and a large deviation from the identity line in the T1 correlation plots. The normalized root mean square error, comparing the estimated T1 values of the tubes measured in the absence of motion with the measurements including motion, was also reduced from 0.17 for the normal ordering to 0.05 for the motion-robust ordering.
FIGURE 6.
M0-weighted (top row), quantitative T1 (middle row) and quantitative B1+ (bottom row) maps of the phantom, both without (left) and with (right) motion, and with both ordering schemes, using the optimized sequence with 600 flip angles and three shots. Note the motion-related artifacts in the parameter maps acquired with the normal ordering
The severity of the observed motion artifacts depends on the frequency of the motion. Presented here are the results obtained with the speeds that caused the most severe artifacts when using the normal ordering. Nevertheless, it should be noted that the motion-robust ordering performed well independent of the speed of the motion (Figure S4).
3.3 ∣. In vivo results
Since the standard deviation of the T1 values obtained using the sequence with 600 flip angles was the lowest, this sequence was evaluated in vivo (Figure 7). The estimated T1 values inside the liver (Table 2) were comparable with values found in the literature.11,35 Note the variations of the excitation field strength in the B1+ maps, which are simultaneously quantified using our method.
FIGURE 7.
In vivo M0-weighted (top row), quantitative T1 (middle row) and quantitative B1+ (bottom row) maps of all six volunteers for the normal ordering and the motion-robust ordering, using the optimized sequence with 600 flip angles and three shots. The motion-robust ordering reveals more details in the T1 map and removes the motion-related artifacts visible in the maps of all three parameters. Areas with strong artifacts are highlighted with yellow ROIs
TABLE 2.
Distribution (mean ± standard deviation) of in vivo T1 values in the liver
| T1 – Normal ordering | T1 – Motion-robust ordering | |
|---|---|---|
| Volunteer 1 | 689 ± 58 ms | 725 ± 65 ms |
| Volunteer 2 | 596 ± 64 ms | 587 ± 53 ms |
| Volunteer 3 | 731 ± 58 ms | 724 ± 60 ms |
| Volunteer 4 | 657 ± 67 ms | 657 ± 67 ms |
| Volunteer 5 | 756 ± 58 ms | 760 ± 66 ms |
| Volunteer 6 | 646 ± 69 ms | 632 ± 59 ms |
| Literature | 745 ± 65 ms11 767 ± 82 ms35 |
The motion-robust ordering resulted in a sharper T1 map, where the vessels in the liver are much more visible, when compared with the normal ordering. Furthermore, there are severe motion artifacts visible in the maps acquired using the normal ordering, as highlighted by the yellow ROIs in Figure 7. These artifacts are no longer visible when using the motion-robust ordering.
To investigate the effect of including B1+ in the dictionary, we reconstructed one dataset with the motion-robust ordering while constraining the relative B1+ value to 1.0. The results can be seen in Figure 8. The estimated T1 maps are very similar. This indicates that B1+ can be estimated without decreasing the quality of the T1 estimates. Moreover, the T1 difference map resembles the pattern seen in the B1+ map, indicating that omitting the estimation of B1+ influences the estimation of the T1 values. The estimated T1 values as determined by DESPOT1 can be seen in Figure 8 as well. Here, some very clear B1+-related artifacts are visible, which lead to reduced T1 values.
FIGURE 8.
In vivo M0-weighted, (top row) quantitative T1 (middle row) and quantitative B1+ (bottom row) maps of volunteer 4, as estimated by four different reconstruction methods. The first two columns correspond to the 3D MRF sequence with the normal ordering and the motion-robust ordering, respectively, as in Figure 7. For the third column, the motion-robust ordering was used, but the relative B1+ value in the dictionary was fixed to 1. The top figure in the fourth column shows the estimated T1 map from the DESPOT1 sequence. Notice the severe B1+ artifacts when using this last method. The bottom figure in the fourth column shows the difference map between the T1 maps when using the motion-robust ordering, with and without fixing B1+ during matching. Notice that the difference map resembles the B1+ pattern
Figure 9 shows the estimated parameter maps for volunteer 6 at a higher resolution. Since the number of shots was increased from three to five to keep the signal-to-noise ratio (SNR) sufficiently high, the acquisition time increased as well. However, the higher resolution reveals the differences between the two orderings schemes.
FIGURE 9.
In vivo quantitative M0 (top row), T1 (middle row) and B1+ (bottom row) maps of volunteer 6, at a resolution of 1.3 mm × 1.3 mm × 3.0 mm (left two columns) and 1.0 mm × 1.0 mm × 3.0 mm (right two columns). Each image consists of an axial, a coronal and a sagittal view. Notice the motion artifacts when using the normal ordering at both resolutions
4 ∣. DISCUSSION
In this work, we have demonstrated a 3D MRF sequence for free-breathing abdominal T1 mapping. In addition, small effects of excitation field inhomogeneities were accounted for by including B1+ in the parameter estimation. Note that receive sensitivities and T2* effects are included in the M0 maps besides the proton density, as these quantities cannot be estimated separately using our method. Therefore, we refer to them as M0-weighted images, instead of quantitative M0 maps.
Both phantom and in vivo scans showed that motion artifacts are reduced when using the motion-robust ordering. Because the MR signal in an MRF experiment is not in steady state, the signal intensity within each set is not constant in the partition direction when using the motion-robust ordering scheme. An additional constraint could be added to the optimization problem to enforce signal smoothness. However, the absence of such a constraint did not result in significant artifacts in the parameter maps when using the motion-robust ordering.
All optimized flip angle trains included a segment without any data acquisition. Other MRF implementations used a delay time between flip angle segments6,16 to allow the magnetization to recover. In this work, the delay time was not set explicitly. Instead, the total time of data acquisition was set by determining the number of TRs. The optimization algorithm could thus find the optimal compromise between delay time and data acquisition. In particular, when using more TRs, the optimization resulted in longer delays.
When optimizing the three shortest sequences (300, 600 and 900 TRs), the same optimal objective value was reached for the majority of the 20 different random initializations. By contrast, the longest sequence (1800 TRs) was more affected by local minima in the optimization. This might indicate that this sequence could be optimized even further by increasing the number of initializations. However, it is important to note that the goal of the sequence optimization was not to find the most optimal sequence, but instead to get more time-efficient sequences, which is achieved even when the optimization reaches a local minimum.
One important thing to note is that the phantom was moved at different speeds, but not all speeds showed the motion artifacts when using the normal ordering (Figures S1-S8). This was probably caused by interference between the motion pattern and the timing of the sequence, which repeats the flip angle train every few seconds (the exact duration depends on the number of flip angles in the sequence). In other words, if the phantom happens to be in the same place at the start of each flip angle train, the motion artifacts are minimal. However, human subjects have a wide range of breathing frequencies. Moreover, these breathing rhythms can be irregular. Consequently, as demonstrated in the in vivo scan, the proposed motion-robust ordering is important to prevent motion artifacts.
Different sequence lengths were compared using the moving phantom. Shorter sequences are less flexible in their encoding but are faster to acquire per repetition. To compare different sequence lengths, we used more shots (and thus more repetitions) for shorter sequences to keep the total acquisition time constant. For the shortest sequence, the parameter estimations were less accurate, especially for the tubes with longer T1 values. Most likely, the duration of one repetition was too short to observe the slow dynamics of the long T1 samples. By increasing the sequence length to 600 and 900 TRs, the estimated quantitative values were improved when using the motion-robust ordering. For the longest sequence with 1800 TRs, however, the accuracy decreased again, presumably because the undersampling artifacts were more pronounced in the reconstructed images due to the lower number of shots. We found an optimal compromise between encoding capability and acquisition speed around 600 TRs, corresponding to a 3000-ms interval between two inversion pulses. Note that our optimization routine sets some of these flip angles to zero to create delays.
Besides the change in acquisition ordering, there is an additional difference between the normal ordering and the motion-robust ordering. In order to be able to perform a Fourier transform in the partition direction, the data need to be grouped together into sets for the motion-robust ordering. To investigate what effect this has on the parameter maps, we reconstructed the normal-ordering dataset while also grouping together sets of 30 successive TR indices (Figure S11). The differences in the resulting parameter maps were only minor, suggesting that most of the differences between the two ordering schemes are caused by the change in k-space ordering, and not by taking together the data of several readouts during the Fourier transform. In addition, we performed a simulation experiment to investigate the difference in matching error for small T1 values (Figure S12). This analysis showed that there is no noticeable difference in the resulting error, whether the fingerprints were averaged or not.
The volunteers in this study were instructed to breathe normally. We acquired one additional dataset for one volunteer, who we asked to breathe deeply (Figure S13). Although the quality of the resulting maps is still acceptable, we do expect that our method will break down when the subject has irregular breathing or has unexpected motion such as hiccups.
Although considerable effort went into optimizing the sequence, there is still room for further improvements. It is possible that the insertion of inversion or saturation pulses in the sequence could improve the encoding capability. Currently, the optimization algorithm is unable to do this, since the peak flip angle is limited to 60 degrees to account for the peak power the system can provide. Additionally, it could be investigated whether regularization could improve the optimization algorithm. For example, the signal differences within one set could be minimized by adding a term in Equation 4. These signal variations could act as a filter applied to the images,36 and by making the fingerprint signals more smooth, the effect of this filter is reduced.26,27 Furthermore, in this work only spoiled gradient echoes were used. The inclusion of fast imaging with steady-state precession (FISP) segments, as used before in MRF,16,37 could add valuable clinical information. However, this would increase the complexity for both the dictionary construction and the optimization algorithm. Moreover, FISP segments are notoriously sensitive to motion.10 Finally, it is important to keep in mind that the CRLB assumes only white Gaussian noise on the data, while the data is also subject to radial undersampling artifacts. These artifacts are non-Gaussian and depend on both the parameter maps and the sequence parameters. Taking them into account during the sequence optimization could result in better sequences.38,39
Besides the sequence itself, the reconstruction process could be improved as well. Low-rank methods40 or parallel imaging techniques41 could be used to reduce the undersampling artifacts and improve the quantitative maps. These methods could also be used to increase the number of partitions, thereby increasing the field of view.
Changing the acquisition ordering is not the only method that can be used to reduce motion artifacts.42 The acquisition can be prospectively triggered to only acquire data at the same respiratory position. Alternatively, the data can be retrospectively binned into several respiratory phases. However, these methods will only address respiratory motion, and come at the cost of longer acquisition times or less available data, respectively. Another approach would be to actively correct the motion in the data, as has been done for rigid motion in brain MRF.43-46 However, motion in the abdomen is nonrigid. Therefore, further studies will be needed to explore if such a motion-correction step is feasible in the abdomen.
The goal of this work was to introduce a new free-breathing MRF sequence. Additional studies will have to be performed to show the repeatability and reproducibility of our method, especially regarding the B1+ maps, because it has not yet been compared quantitatively with other motion-robust mapping strategies.47,48 Finally, to investigate the performance of the sequence with different pathologies, several clinical scans with patients will have to be performed as well.
5 ∣. CONCLUSION
A free-breathing MRF sequence was demonstrated for B1+-robust quantitative abdominal T1 mapping. Four different flip angle patterns were optimized. A movable phantom was used to validate the sequences. The flip angle train with 600 TRs provided the best trade-off between T1-encoding power and sampling density. In vivo measurements confirmed the advantage of the motion-robust k-space ordering. With this free-breathing MRF implementation it is possible to collect crisp M0-weighted images and clean T1 maps of the abdomen at a clinically usable resolution within 5 min.
Supplementary Material
ACKNOWLEDGEMENTS
The research reported in this publication was supported by the NIH/NIBIB grant R01 EB026456, NIH/NIAMS grant R01 AR070297, and was performed under the rubric of the Center for Advanced Imaging Innovation and Research, an NIBIB Biomedical Technology Resource Center (P41 EB017183). Furthermore, we would like to express our gratitude towards the Holland Scholarship from the Dutch Ministry of Education, Culture and Science, as well as the Professor Huson award and the BMT Bachelorbeurs from the Eindhoven University of Technology for supporting this project.
Abbreviations used:
- B1+
excitation field strength
- COV
coefficient of variation
- CRLB
Cramér–Rao Lower Bound
- DESPOT1
driven equilibrium single pulse observation of T1
- DESPOT2
driven equilibrium single pulse observation of T2
- FFT
fast Fourier transform
- FIM
Fisher information matrix
- FISP
fast imaging with steady-state precession
- FLASH
fast low angle shot
- M0
equilibrium magnetization
- MRF
magnetic resonance fingerprinting
- MRI
magnetic resonance imaging
- NUFFT
nonuniform fast Fourier transform
- ROI
region of interest
- SNR
signal-to-noise ratio
- T1
longitudinal relaxation time
- T2
transverse relaxation time
- TE
echo time
- TR
repetition time
Footnotes
SUPPORTING INFORMATION
Additional supporting information may be found online in the Supporting Information section at the end of this article.
REFERENCES
- 1.Brown RW, Cheng Y-CN, Haacke EM, Thompson MR, Venkatesan R. Magnetic Resonance Imaging: Physical Principles and Sequence Design. 2nd ed. Hoboken, NJ: John Wiley & Sons; 2014. [Google Scholar]
- 2.Cheng H-LM, Stikov N, Ghugre NR, Wright GA. Practical medical applications of quantitative MR relaxometry. J Magn Reson Imaging. 2012; 36(4):805–824. [DOI] [PubMed] [Google Scholar]
- 3.Deoni SCL. Quantitative relaxometry of the brain. Top Magn Reson Imaging. 2010;21(2):101–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Look DC, Locker DR. Time saving in measurement of NMR and EPR relaxation times. Rev Sci Instrum. 1970;41(2):250–251. [Google Scholar]
- 5.Deoni SCL, Rutt BK, Peters TM. Rapid combined T1 and T2 mapping using gradient recalled acquisition in the steady state. Magn Reson Med. 2003; 49(3):515–526. [DOI] [PubMed] [Google Scholar]
- 6.Ma D, Gulani V, Seiberlich N, et al. Magnetic resonance fingerprinting. Nature. 2013;495(7440):187–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Merkle EM, Dale BM. Abdominal MRI at 3.0 T: The basic revisited. Am J Roentgenol. 2006;186(6):1524–1532. [DOI] [PubMed] [Google Scholar]
- 8.Collins CM, Liu W, Schreiber W, Yang QX, Smith MB. Central brightening due to constructive interference with, without, and despite dielectric resonance. J Magn Reson Imaging. 2005;21(2):192–196. [DOI] [PubMed] [Google Scholar]
- 9.Yang RK, Roth CG, Ward RJ, DeJesus JO, Mitchell DG. Optimizing abdominal MR imaging: approaches to common problems. Radiographics. 2010; 30(1):185–199. [DOI] [PubMed] [Google Scholar]
- 10.Yu Z, Zhao T, Assländer J, Lattanzi R, Sodickson DK, Cloos MA. Exploring the sensitivity of magnetic resonance fingerprinting to motion. Magn Reson Imaging. 2018;54:241–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen Y, Jian Y, Pahwa S, et al. MR fingerprinting for rapid quantitative abdominal imaging. Radiology. 2016;279(1):278–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bernstein MA, King KF, Zhou XJ. Basics of physiologic gating, triggering, and monitoring. Handb MRI Pulse Seq. 2004;443–490. [Google Scholar]
- 13.Masaracchia MM, Tsapakos MJ, McNulty NJ, Beach ML. Changing the paradigm for diagnostic MRI in pediatrics: Don't hold your breath. Paediatr Anesth. 2017;27(9):880–884. [DOI] [PubMed] [Google Scholar]
- 14.Block KT, Chandarana H, Milla S, et al. Towards routine clinical use of radial stack-of-stars 3D gradient-echo sequences for reducing motion sensitivity. J Korean Soc Magn Reson Med. 2014;18(2):87–106. [Google Scholar]
- 15.Cloos MA, Assländer J, Abbas B, et al. Rapid radial T 1 and T 2 mapping of the hip articular cartilage with magnetic resonance fingerprinting. J Magn Reson Imaging. 2019;50(3):810–815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cloos MA, Knoll F, Zhao T, et al. Multiparametric imaging with heterogeneous radiofrequency fields. Nat Commun. 2016;7(1):1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Buonincontri G, Schulte RF, Cosottini M, Tosetti M. Spiral MR fingerprinting at 7 T with simultaneous B1 estimation. Magn Reson Imaging. 2017; 41:1–6. [DOI] [PubMed] [Google Scholar]
- 18.Scheffler K, Hennig J. Reduced circular field-of-view imaging. Magn Reson Med. 1998;40(3):474–480. [DOI] [PubMed] [Google Scholar]
- 19.Winkelmann S, Schaeffter T, Koehler T, Eggers H, Doessel O. An optimal radial profile order based on the golden ratio for time-resolved MRI. IEEE Trans Med Imaging. 2007;26(1):68–76. [DOI] [PubMed] [Google Scholar]
- 20.Amthor T, Doneva M, Koken P, Sommer K, Meineke J, Börnert P. Magnetic resonance fingerprinting with short relaxation intervals. Magn Reson Imaging. 2017;41:22–28. [DOI] [PubMed] [Google Scholar]
- 21.Bloch F Nuclear induction. Phys Ther Rev. 1946;70(7-8):460–474. [Google Scholar]
- 22.McGivney DF, Pierre E, Ma D, et al. SVD compression for magnetic resonance fingerprinting in the time domain. IEEE Trans Med Imaging. 2014;33(12): 2311–2322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ordidge RJ, Wylenzinska M, Hugg JW, Butterworth E, Fraconi F. Frequency offset corrected inversion (FOCI) pulses for use in localized spectroscopy. Magn Reson Med. 1996;36(4):562–566. [DOI] [PubMed] [Google Scholar]
- 24.Kay SM. Fundamentals of Statistical Signal Processing: Estimation Theory. Vol. 1. Upper Saddle River, NJ: Prentice Hall PTR; 1993. [Google Scholar]
- 25.Teixeira RPAG, Malik SJ, Hajnal JV. Joint system relaxometry (JSR) and Cramér-Rao Lower Bound optimization of sequence parameters: A framework for enhanced precision of DESPOT T1 and T2 estimation. Magn Reson Med. 2018;79(1):234–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhao B, Haldar JP, Liao C, et al. Optimal experiment design for magnetic resonance fingerprinting: Cramér-Rao Bound meets spin dynamics. IEEE Trans Med Imaging. 2019;38(3):844–861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee PK, Watkins LE, Anderson TI, Buonincontri G, Hargreaves BA. Flexible and efficient optimization of quantitative sequences using automatic differentiation of Bloch simulations. Magn Reson Med. 2019;82(4):1438–1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Assländer J, Lattanzi R, Sodickson DK, Cloos MA. Optimized quantification of spin relaxation times in the hybrid state. Magn Reson Med. 2019;82(4): 1385–1397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Baydin GA, Pearlmutter BA, Radul AA, Siskind JM. Automatic differentiation in machine learning: a survey. J Mach Learn Res. 2018;18:1–43. [Google Scholar]
- 30.Andersson JAE, Gillis J, Horn G, Rawlings JB, Diehl M. CasADi – A software framework for nonlinear optimization and optimal control. Math Program Comput. 2019;11(1):1–36. [Google Scholar]
- 31.Lindh WQ, Pooler MS, Tamparo CD, Dahl BM. Delmar's Comprehensive Medical Assisting: Administrative and Clinical Competencies. 4thed. Clifton Park, NY: Delmar; 2010. [Google Scholar]
- 32.Chung S, Kim D, Breton E, Axel L. Rapid B1+ mapping using a preconditioning RF pulse with turboFLASH readout. Magn Reson Med. 2010;64(2): 439–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fessler JA, Sutton BP. Nonuniform Fast Fourier Transforms using min-max interpolation. IEEE Trans Signal Process. 2003;51(2):560–574. [Google Scholar]
- 34.Roemer PB, Edelstein WA, Hayes CE, Souza SP, Mueller OM. The NMR phased array. Magn Reson Med. 1990;16:192–225. [DOI] [PubMed] [Google Scholar]
- 35.Obmann VC, Mertineit N, Marx C, et al. Liver MR relaxometry at 3T – segmental normal T1 and T2* values in patients without focal or diffuse liver disease and in patients with increased liver fat and elevated liver stiffness. Sci Rep. 2019;9(1):1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Deichmann R, Good CD, Josephs O, Ashburner J, Turner R. Optimization of 3-D MP-RAGE sequences for structural brain imaging. Neuroimage. 2000; 12(1):112–127. [DOI] [PubMed] [Google Scholar]
- 37.Jiang Y, Ma D, Seiberlich N, Gulani V, Griswold MA. MR fingerprinting using fast imaging with steady state precession (FISP) with spiral readout. Magn Reson Med. 2015;74:1621–1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kara D, Fan M, Hamilton J, Griswold M, Seiberlich N, Brown R. Parameter map error due to normal noise and aliasing artifacts in MR fingerprinting. Magn Reson Med. 2019;81(5):3108–3123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stolk CC, Sbrizzi A. Understanding the combined effect of k-space undersampling and transient states excitation in MR fingerprinting reconstructions. IEEE Trans Med Imaging. 2018;38(10):2445–2455. [DOI] [PubMed] [Google Scholar]
- 40.Assländer J, Cloos MA, Knoll F, Sodickson DK, Hennig J, Lattanzi R. Low rank alternating direction method of multipliers reconstruction for MR fingerprinting. Magn Reson Med. 2018;79(1):83–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liao C, Bilgic B, Manhard MK, et al. 3D MR fingerprinting with accelerated stack-of-spirals and hybrid sliding-window and GRAPPA reconstruction. Neuroimage. 2017;162:13–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zaitsev M, Maclaren J, Herbst M. Motion artifacts in MRI: A complex problem with many partial solutions. J Magn Reson Imaging. 2015;42(4):887–901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mehta BB, Ma D, Pierre EY, Jiang Y, Coppo S, Griswold MA. Image reconstruction algorithm for motion insensitive MR Fingerprinting (MRF): MORF. Magn Reson Med. 2018;80(6):2485–2500. [DOI] [PubMed] [Google Scholar]
- 44.Xu Z, Ye H, Lyu M, et al. Rigid motion correction for magnetic resonance fingerprinting with sliding-window reconstruction and image registration. Magn Reson Imaging. 2019;57:303–312. [DOI] [PubMed] [Google Scholar]
- 45.Cruz G, Jaubert O, Schneider T, Botnar RM, Prieto C. Rigid motion-corrected magnetic resonance fingerprinting. Magn Reson Med. 2019;81(2): 947–961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kurzawski JW, Cencini M, Peretti L, et al. Retrospective rigid motion correction of three-dimensional magnetic resonance fingerprinting of the human brain. Magn Reson Med. 2020;84(5):1–10. [DOI] [PubMed] [Google Scholar]
- 47.Weingärtner S, Zimmer F, Metzger GJ, Uğurbil K, Van de Moortele P-F, Akçakaya M. Motion-robust cardiac B1+ mapping at 3T using interleaved Bloch-Siegert shifts. Magn Reson Med. 2017;78(2):670–677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dietrich S, Kolbitsch C, Schaeffter T, Schmitter S. 3D Radial Phase Encoded Flip Angle Imaging at Ultra-High Field Strength. ISMRM 27 Annual Meeting, 2019;#4560. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.