Abstract
In diseased organs, stress-activated signaling cascades alter chromatin, triggering maladaptive cell state transitions. Fibroblast activation is a common tissue stress response that worsens lung, liver, kidney and heart disease, yet its mechanistic basis remains obscure1,2. Pharmacologic BET inhibition alleviates cardiac dysfunction3-7, providing a tool to interrogate and modulate cardiac cell states as a potential therapeutic approach. Here, we leverage single-cell epigenomic interrogation of hearts dynamically exposed to BET inhibitors to reveal a reversible transcriptional switch underlying fibroblast activation. Resident cardiac fibroblasts demonstrated robust toggling between the quiescent and activated state in a manner directly correlating with BET inhibitor exposure and cardiac function. Single-cell chromatin accessibility revealed novel DNA elements whose accessibility dynamically correlated with cardiac performance. Among the most dynamic elements was an enhancer regulating the transcription factor MEOX1, which was specifically expressed in activated fibroblasts, occupied putative regulatory elements of a broad fibrotic gene program, and was required for TGFβ-induced fibroblast activation. Selective CRISPR inhibition of the single most dynamic cis-element within the enhancer blocked TGFβ-induced Meox1 activation. We identify MEOX1 as a central regulator of fibroblast activation associated with cardiac dysfunction, and also demonstrate its upregulation upon activation of human lung, liver and kidney fibroblasts. The plasticity and specificity of BET-dependent regulation of MEOX1 in tissue fibroblasts provide new trans- and cis- targets for treating fibrotic disease.
In many human diseases, dynamic changes in gene expression fuel progressive organ dysfunction. As such, targeting gene transcription has emerged as a new therapeutic strategy in a variety of chronic diseases, including heart failure, a common and lethal condition afflicting 24 million people worldwide8. Among strategies to therapeutically target the gene regulatory apparatus, small molecule inhibitors of bromodomain and extra-terminal domain (BET) proteins (BRD2, BRD3, BRD4 and BRDT) have emerged as potent tools to reversibly interdict enhancer-to-promoter signaling in vivo9. BET proteins are a highly conserved family of ubiquitously expressed acetyl-lysine reader proteins that co-activate transcription, and systemic administration of BET bromodomain inhibitors can ameliorate heart failure in mouse models3. Because the cell types most affected by BET inhibition in these models are not known and systemic administration of potent BET bromodomain inhibitors is likely too broad to be therapeutically tractable for chronic cardiovascular indications, we leveraged single cell transcriptomic and epigenomic interrogation of heart tissue in the setting of intermittent BET bromodomain inhibitor exposure to reveal cell states and cis- and trans- targets critical for disease pathogenesis and therapeutic efficacy.
Transcriptional plasticity of fibroblasts
One month of treatment with a small molecule BET bromodomain inhibitor, JQ19, commenced 18 days after heart failure induction by left ventricle (LV) pressure overload via transvers aortic constriction (TAC), significantly improved LV systolic function in mice (Fig.1a). Withdrawal of JQ1 for 2 weeks led to regression of LV systolic function, demonstrating therapeutic reversibility of BET inhibition. We isolated adult cardiomyocytes (CMs) and found that the effects of JQ1 on their transcriptome were modest compared to the previously published transcriptomic signature of whole LV tissue (<3% overlap, Extended Data Fig. 1a)5. To determine cardiac cell types affected by JQ1, we performed single cell RNA sequencing (scRNAseq) of >35,000 non-CMs from four experimental groups using the 10X Genomics platform: Sham, TAC vehicle-treated (TAC), TAC JQ1-treated (TAC JQ1), and TAC JQ1-treated followed by JQ1 withdrawal (TAC JQ1 withdrawn) (Fig. 1b). Unsupervised clustering of the scRNAseq identified cardiac cell subpopulations, including fibroblasts, endothelial cells, myeloid cells and epicardial cells (Fig. 1c and Extended Data Fig. 1b,c). Among fibroblasts, TAC caused a large shift in cell state, and JQ1 treatment resulted in reversion to a state that closely approached the Sham state (Fig. 1d). Withdrawal of JQ1 shifted the fibroblast population back to a TAC-like stressed state (Fig. 1d). Dynamic JQ1-dependent transcriptomic shifts also occurred in the endothelial and myeloid compartments, but cell states elicited by JQ1 were less like Sham than those elicited in fibroblasts (Fig. 1d and Extended Data Fig. 1d,e).
Figure 1: Heart failure reversibility with BET inhibition correlates with myofibroblast cell state.
a. Left ventricle (LV) ejection fraction in indicated groups with treatment and withdrawal of JQ1 (50mg/kg/d). ****p-val=1.46x10−7 for TAC JQ1 vs. TAC JQ1 withdrawn at day 62 (one-way ANOVA with Tukey post hoc test). Sham (n=4), TAC (n=6), TAC JQ1 (n=10), TAC JQ1 withdrawn (n=14). b. scRNA- and ATACseq library generation workflow. c, d. UMAP plot of non-CM cells captured from hearts in (b) colored by cluster (c) and sample identity (d), n=35,551. e. UMAP plot of fibroblasts colored by sample identity, n=13,937. f. Fibroblast Periostin (Postn) expression shown as UMAP feature and violin plots. g. Normalized expression score of the 260 genes increased in TAC vs. Sham across fibroblast samples and associated GO terms (Fisher exact test). h. LV fibrosis in TAC JQ1 (n=4) and TAC JQ1 withdrawn (n=8) mice by picrosirius red staining and quantification. (*p=0.0081, Mann-Whitney with Tukey post hoc test). Bar=250um
a,h Data are shown as means ± SEM.
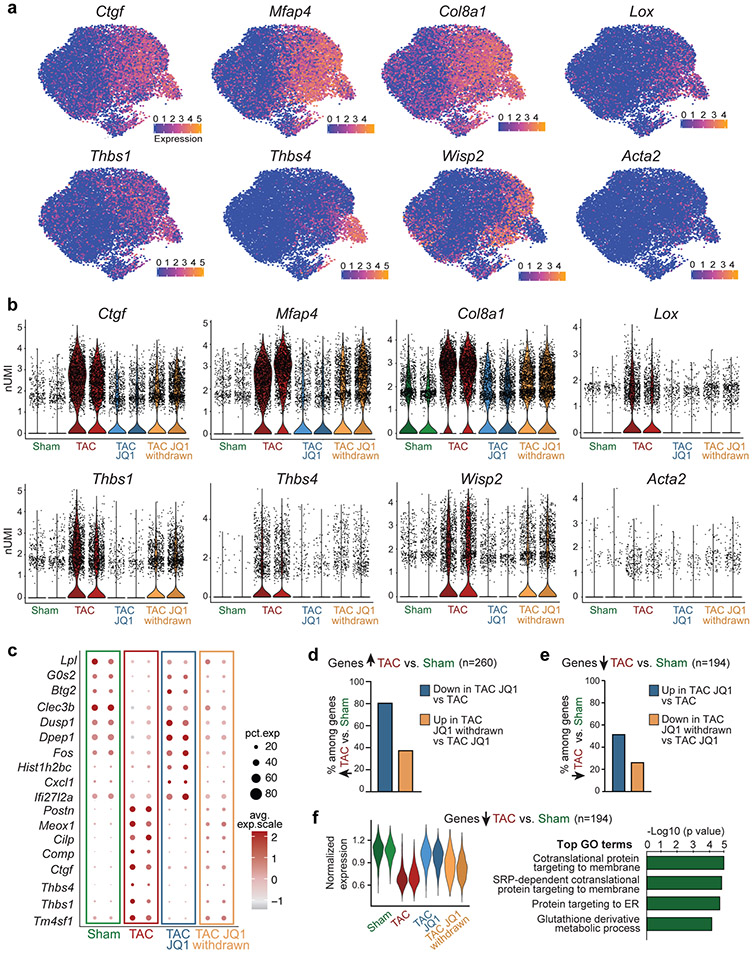
Subsetting the 13,937 individual fibroblast transcriptomes better illustrated the response to JQ1 in the TAC state (Fig.1e). Cardiac stress is known to trigger the transition of resident fibroblasts into a contractile and synthetic state called the myofibroblast10. The myofibroblast-enriched gene Postn10, together with many other markers of fibroblast activation, were upregulated in TAC, downregulated with JQ1 treatment, and re-emerged upon JQ1 withdrawal (Fig. 1f and Extended Data Fig. 2a-c). We used a cumulative score to plot expression of 260 fibroblast genes upregulated by stress across all samples and found this gene set was highly sensitive to JQ1 exposure (Fig.1g) and enriched for extracellular matrix (ECM), TGFβ signaling, and proliferative gene ontology (GO) terms (Fig.1g). Differential expression (DE) analysis between TAC:TAC-JQ1 or TAC-JQ1:TAC-JQ1-withdrawn among these 260 genes demonstrated that 81% were significantly downregulated by JQ1, and 38% of those were upregulated again when JQ1 was withdrawn, highlighting the transcriptional reversibility of the stress-induced signature with BET inhibition (Extended Data Fig. 2d). Consistent with transcriptomic data, JQ1 withdrawal led to increased LV fibrosis (Fig. 1h), which may reflect shifts in fibroblast state that influence or are affected by other cell types through heterotypic interactions or paracrine factors. JQ1 exposure also restored expression of a fraction of downregulated genes between Sham and TAC in fibroblasts (Extended Data Fig. 2e,f), suggesting a dual effect of JQ1 in suppressing stress-induced gene programs and restoring basal gene programs. Among the 9 fibroblast subclusters, JQ1 treated fibroblasts (cluster 1) were closest to Sham (cluster 0, 4) by hierarchical analysis (Extended Data Fig. 3a-c). While most of the JQ1-withdrawn cells clustered together with TAC cells in the three subclusters associated with greatest ECM upregulation (clusters 2,3,5, Extended Data Fig. 3d-f), some clustered with Sham suggesting they achieved stable restoration of gene expression, at least for two weeks after drug withdrawal (Extended Data Fig. 3a-c).
Enhancers during fibroblast activation
To probe changes in chromatin accessibility and enhancer activation modulated by JQ1 in non-CMs during heart failure pathogenesis, we integrated scRNAseq with single cell Assay for Transposase-Accessible Chromatin sequencing (scATACseq) data from the same hearts (Fig. 1b and Extended Data 4a)11,12. We identified 490,020 accessible sites distributed among 31,766 individual cells and assigned cellular identity based on chromatin signature (Extended Data Fig. 4b,c). To focus on distal regulatory elements, we excluded accessible sites in promoters and gene bodies and defined a catalog of fibroblast-, myeloid- or endothelial-enriched distal elements that were used for all subsequent analyses (Extended Data Fig. 4d). Fibroblasts had increased chromatin accessibility after TAC that was reversibly attenuated with JQ1 treatment, a feature less evident in myeloid and endothelial cells (Extended Data Fig. 5a). To dissect dynamic and reversible changes in chromatin activation, we defined open and closed distal elements across the 4 samples and excluded regions constitutively open across all conditions (Extended Data Fig. 5b). We discovered a cluster of highly dynamic fibroblast distal elements that were closed in Sham, opened in TAC, closed by JQ1, and robustly re-accessible following JQ1 withdrawal (Cluster 2, Extended Data Fig. 5b). GO analysis showed that these regions were proximal to genes controlling heart growth and ECM organization, two hallmark features of adverse cardiac remodeling and fibrosis. We also identified a large cluster of fibroblast regions that opened from Sham to TAC that were insensitive to JQ1, highlighting a signature of stress-responsive chromatin activation that is BET-independent (Cluster 9, Extended Data Fig. 5b). In fibroblasts, transcription factor (TF) binding motifs for CEBPB, JUN and MEOX1 were enriched in accessible regions in the Sham-to-TAC transition with subsequent loss of enrichment upon BET inhibition that was then re-acquired with JQ1 withdrawal (Extended Data Fig. 5c).
To identify functionally relevant enhancers among the scATACseq peaks, we performed precision nuclear run-on sequencing (PROseq)13 on cultured fibroblasts in vitro to map genome-wide RNA polymerase II nascent transcription, which robustly indicates enhancer activity14 and gene transcription. Since PROseq requires large quantities of cells, we generated an immortalized cell line derived from primary adult mouse cardiac fibroblasts and treated it with TGFβ, a canonical stimulus for eliciting fibroblast activation in vitro (Extended Data Fig. 6a). Induction of Postn, Ctgf and several other markers validated that this cell line transitioned to the activated state upon TGFβ exposure (Extended Data Fig. 6b). PROseq identified a set of distal and gene elements that were significantly more transcribed after TGFβ stimulation and were associated with fibroblast activation (Fig. 2a and Extended Data Fig. 6c-f). Using our scATACseq data, we identified distal elements that were either opening or closing between Sham and TAC in vivo, and assessed the PROseq signal in the cultured fibroblasts at these same regions. We found that TGFβ-stimulated transcription at distal elements in vitro correlated with regions of chromatin that open in fibroblasts in vivo during heart failure pathogenesis, whereas nascent transcription at chromatin regions that closed in vivo was not notably altered (Fig. 2b and Extended Data Fig. 6g for protein coding genes). Visualization of the Postn locus illustrates this dynamic regulation (Fig. 2c), in which there is chromatin opening in vivo after TAC with dynamic sensitivity to JQ1 exposure. scATACseq co-accessibility analysis between the Postn promoter and a TGFβ-dependent region within this area (Peak10/11) showed low co-accessibility in the Sham state, robust increase with TAC, and modulation with JQ1 exposure (Fig. 2c and Extended Data Fig. 6h). We used CRISPR interference (CRISPRi) comprising catalytically inactive Cas9 (dCas9) fused to the KRAB repressor protein15 to specifically inhibit individual regulatory elements within this large Postn enhancer (Fig. 2d and Extended Data Fig. 6i,j). The Peak10/11 cis-element was uniquely required for Postn transactivation upon TGFβ stimulation (Fig. 2d). CRISPRi-mediated deposition of the repressive histone mark H3K9me3 was specific to the Peak10/11 region and did not affect the Postn promoter (Extended Data Fig. 6k).
Figure 2: Reversibility of fibroblast chromatin states reveals DNA elements that correlate with heart function.
a. PROseq heatmap of differentially transcribed distal regions (Wald test with Benjamini/Hochberg correction) in unstimulated (Unstim) vs. TGFβ-treated fibroblasts and associated top GO terms (binomial test). Mean signal for 2 replicates is shown. b. PROseq coverage in Unstim and TGFβ-treated fibroblasts in vitro of scATACseq peaks opening (2,553) or closing (567) between Sham and TAC in fibroblasts in vivo. c. scATACseq from fibroblasts in vivo and PROseq at the Postn locus. A highly transcribed region (Peak10/11) is highlighted in pink with co-accessibility (CoA) between the Postn promoter and Peak10/11. d. Postn expression by qPCR in Unstim or TGFβ-treated fibroblasts in CRISPRi lines targeting either Peak8 (***p-val=0.0001), Peak10/11 or Peak19 (***p-val=0.0001). One-way ANOVA followed by Tukey post hoc test. Values normalized to CRISPRi control line under unstim conditions (shown in Extended Data Fig. 6i). Data are means ± SEM. e. Schematic of correlation analysis between LV ejection fraction and chromatin accessibility highlighting a negative or positive correlation. f. Volcano plot showing correlation coefficients (from analysis in Fig. 2e) and corresponding p-val of 470 super-enhancers in fibroblasts.
Chromatin state and cardiac function
To unveil distal elements involved in progression and reversal of heart failure, we assembled a catalog of cell population–enriched large enhancers (also known as super-enhancers)16 using scATACseq data in the diseased heart (TAC) (Extended Data Fig. 7a) and correlated the degree of accessibility of these enhancers in fibroblast, myeloid and endothelial cells with LV ejection fraction in each treatment setting described. This correlation analysis between a measure of enhancer chromatin accessibility and a physiological trait (LV ejection fraction) is summarized schematically in Figure 2e. Enhancer elements were defined as having a negative correlation if their accessibility was anti-correlated with heart function (i.e., opening from Sham to TAC) or a positive correlation if closing from Sham to TAC. Of the 470 fibroblast super-enhancers identified, 48 showed strong negative correlation while 22 showed strong positive correlation (volcano plot in Fig. 2f and Extended Data Fig. 7b,c). To determine if the super-enhancers responded similarly in the TGF-β-responsive fibroblast cell line, we performed ChIPseq with an antibody to H3K27ac, an active enhancer mark, with or without TGFβ-treatment (Extended Data Fig. 7d). Enhancers identified in vivo characterized by negative correlation with heart function showed increased H3K27ac deposition following TGFβ treatment, while those with positive correlation showed decreased H3K27ac (Extended Data Fig. 7e).
One of the most negatively correlated elements in fibroblasts in vivo was a large enhancer downstream of Meox1 (Fig. 2f and Extended Data Fig. 8a), a homeodomain-containing TF required for sclerotome development17. Meox1 was highly upregulated in myofibroblasts following TAC18 (Fig. 3a and Extended Data Fig. 8b). JQ1 exposure abolished Meox1 expression while JQ1 withdrawal led to its robust re-induction (Fig. 3a).
Figure 3: Chromatin accessibility and nascent transcription identify a cis-element controlling Meox1 expression.
a. UMAP plot of fibroblasts colored by sample identity or Meox1 expression with associated violin plots. b. Mouse Meox1 locus showing from top to bottom: scATACseq from fibroblasts in vivo; ChIPseq for BRD4 (GSE46668), H3K27ac and CTCF (ENCSR000CDF and ENCSR000CBI) in the adult heart; coverage of PROseq in Unstim and TGFβ-treated fibroblasts in vitro; and co-accessibility measures between Meox1 promoter and Peak9 region in fibroblasts. A highly transcribed region (Peak9/10) is highlighted in pink. c. Schematic showing CRISPRi targeting of 3 regions within the Meox1 enhancer (top). Meox1 expression by qPCR in Unstim or TGFβ-treated fibroblasts upon CRISPRi targeting either Peak5 (****p-val=1.82e−10), Peak9/10 or Peak13 (****p-val=1.64e−8). Values normalized to CRISPRi control line under unstim conditions (Extended Data Fig. 10e). d. Meox1 expression by qPCR in Unstim or TGFβ conditions in WT (****p-val=1.99e−8) or Peak9/10 deleted cells (****p-val=1.99e−8). e. Peak9/10 eRNA (top, **** p-val=1.76e−8 and ****P=5.29e−6) and Meox1 (bottom, **** p-val=9.80e−9 and **** p-val=1.29e−8) expression by qPCR in Unstim and TGFβ conditions with control or Smad3-targeting siRNAs.
c-e Data are means ± SEM. One-way ANOVA followed by Tukey post hoc test.
Stress-induced cis-regulation of Meox1
The enhancer downstream of Meox1 was extremely sensitive to stress and JQ1 exposure in fibroblasts, but not in myeloid and endothelial cells (Fig. 3b and Extended Data Fig. 8c). From Sham to TAC conditions, 10 peaks became accessible in fibroblasts; many of these closed with JQ1 treatment back to a Sham level, and re-opened when JQ1 was withdrawn (Fig. 3b and Extended Data Fig. 8c-e). Publicly available BRD4 and H3K27ac ChIPseq data from adult mouse LV tissue corroborated these active enhancer marks at the Meox1 locus, suggesting this enhancer might regulate Meox1 (Fig. 3b). Indeed, Meox1 mRNA expression was induced in cultured fibroblasts treated with TGFβ (Extended Data Fig. 9a)19; this activation was suppressed by JQ1, and, among BET proteins, was most sensitive to BRD4 dosage (Extended Data Fig. 9a-c).
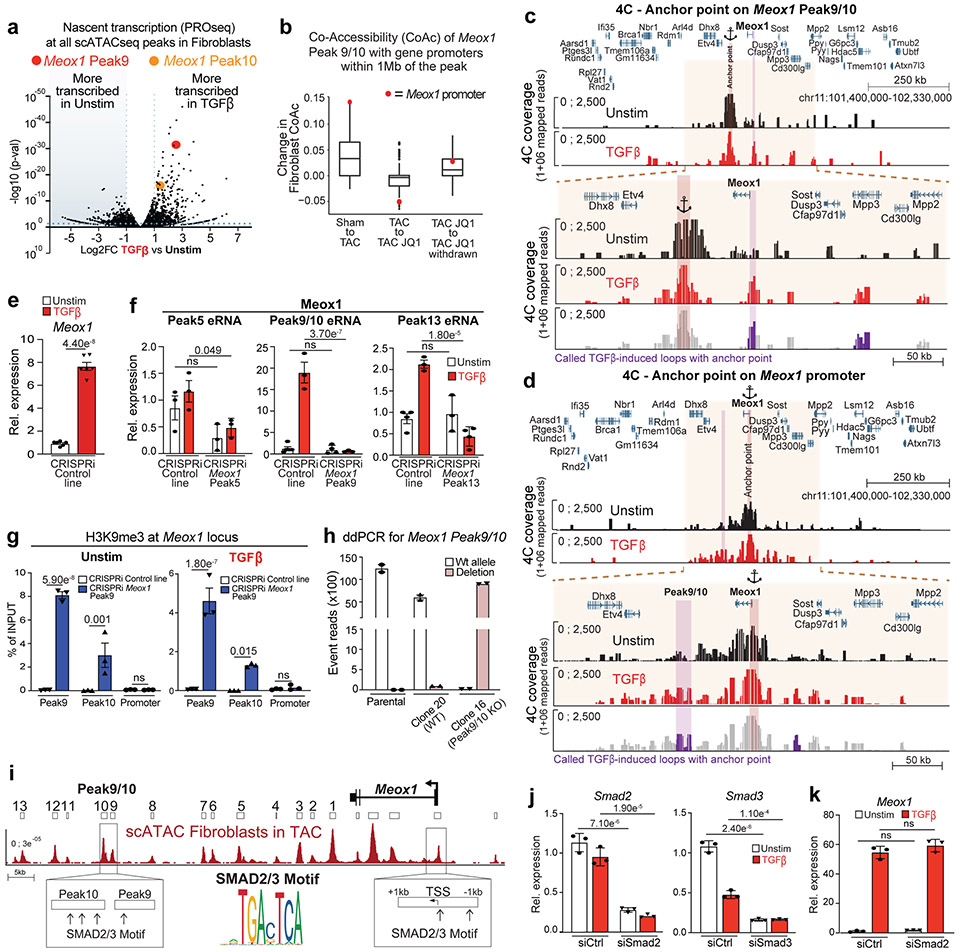
Among the scATACseq peaks in this locus that showed increased accessibility with TAC in vivo, PROseq identified a region located 62-65 kilobases (kb) downstream of the Meox1 promoter (Peak 9/10) that featured a striking increase in nascent transcription following TGFβ stimulation (Fig. 3b). This DNA element showed more TGFβ-stimulated transcription than the Meox1 gene body itself and was one of the most differentially transcribed regions across the whole genome in response to TGFβ (Fig. 3b and Extended Data Fig. 10a), suggesting a functionally relevant enhancer14. The Meox1 promoter and the Peak 9/10 region showed low co-accessibility in the Sham state, a strong increase in co-accessibility with TAC, and modulation of co-accessibility with BET inhibition (Fig. 3b and Extended Data Fig. 10b). Chromosome conformation capture analysis of this locus in fibroblasts revealed a robust increase in contact between the Peak9/10 region and the Meox1 promoter in response to TGFβ (Extended Data Fig. 10c,d). CRISPRi revealed that the Peak 9/10 cis-element was specifically required for Meox1 transactivation upon TGFβ stimulation, while other accessible regions identified in vivo were not (Fig. 3c and Extended Data Fig. 10e,f). Guide RNAs targeting Peak9/10 led to localized deposition of the repressive H3K9me3 mark in the Peak9/10 element but not on the Meox1 promoter, supporting specificity of Peak 9/10 silencing (Extended Data Fig. 10g). CRISPR-Cas9 based excision revealed that deletion of Peak 9/10 abolished Meox1 transactivation upon TGFβ stimulation (Fig. 3d and Extended Data Fig. 10h). Multiple motifs of SMAD2/3, transcriptional mediators of TGFβ signaling, were present in both Peak 9/10 and the Meox1 promoter (Extended Data Fig. 10i), and knockdown of Smad3, but not Smad2, led to a significant reduction of Peak9/10 transcription and Meox1 expression (Fig. 3e and Extended Data Fig. 10j,k). Thus, Peak9/10 represents a stress-activated enhancer for Meox1 during the activated myofibroblast transition.
MEOX1 regulates pro-fibrotic function
Meox1 knockdown reduced formation of αSMA-positive stress fibers and αSMA/Acta2 expression in TGFβ treated fibroblasts (Extended Data Fig. 11a-d). Moreover, Meox1 depletion attenuated TGFβ-stimulated collagen-gel contraction and EdU incorporation, two functional hallmarks of myofibroblasts in disease pathogenesis (Fig. 4a and Extended Data Fig. 11e). MEOX1 occupancy by ChIPseq at active chromatin (marked by H3K27ac) revealed increased recruitment of MEOX1 at stress-responsive distal elements upon TGFβ treatment (Fig. 4b, Extended Data Fig. 11f-h). Upon Meox1 knockdown, transcription of the stress-responsive distal elements bound by MEOX1 in TGFβ-treated fibroblasts, detected by PROseq, was diminished (Fig. 4c and Extended Data Fig. 11i-k). These distal chromatin loci were near genes linked to tissue morphogenesis, regulation of growth, and positive regulation of cell migration and motility (Fig. 4c). Correspondingly, Meox1 knockdown in the setting of TGFβ stimulation resulted in downregulation of over 400 genes compared to TGFβ alone, while 863 were more highly transcribed (Fig. 4d and Extended Data Fig. 11l-o). GO analysis of the Meox1-dependent genes again revealed enrichment for processes associated with fibroblast activation, such as regulation of cell migration, motility and proliferation (Fig. 4d). Among these genes were classical markers of cardiac myofibroblast activation, including Ctgf and Postn, which showed MEOX1 enrichment at their promoters and proximal regulatory elements (including the Postn Peak10/11 enhancer described in Figure 2c,d). These elements also featured decreased transcription following Meox1 depletion (Fig. 4e and Extended Data Fig. 11p). Thus, MEOX1 functions as an essential transcriptional mediator of the fibroblast to myofibroblast switch associated with fibrotic disease.
Figure 4: MEOX1 regulates fibroblast plasticity and profibrotic function.
a. Images and quantification (n=4 per condition) of fibroblasts seeded on compressible collagen gel matrices in Unstim or TGFβ conditions with a control or a Meox1-targeting siRNA. Two-way ANOVA followed by Bonferroni correction for TGFβ siCtrl vs. TGFβ siMeox1. **p-val=0.0082 (24h), **p-val =0.0015 (48h) and ****p-val =2.21e−5 (72h). b. MEOX1 ChIPseq coverage in Unstim and TGFβ-treated cells at distal elements identified by high enrichment of H3K27ac in Unstim (left, 2194 regions) or TGFβ-treated (right, 2898 regions) cells. c. PROseq coverage under indicated conditions at distal elements with high H3K27ac enrichment in TGFβ vs. Unstim bound by MEOX1 (427 regions) and associated top GO terms (binomial test). d. PROseq coverage of differentially transcribed genes (Wald test with Benjamini/Hochberg correction) in TGFβ-treated fibroblasts with Ctrl or Meox1 siRNA and associated top GO terms (Fisher exact test). Mean signal for 2 replicates shown. e. Coverage of indicated ChIPseq or PROseq with or without TGFβ in cardiac fibroblasts at the Ctgf locus with Ctrl or Meox1 siRNA. f. UMAP plot of adult human cardiac fibroblasts colored by cluster identity (left) and POSTN or MEOX1 expression (right). g. MEOX1 expression by qPCR in primary human fibroblasts from indicated tissues. One-way ANOVA followed by Tukey post hoc test for Unstim vs. TGFβ (****p-val=1.29e−5 lung, **** p-val=1.49e−6 liver, ****p-val =5.83e−5 kidney) and TGFβ vs. TGFβ+JQ1 (****p-val=3.91e−6 lung, **** p-val=1.06e−6 liver, **** p-val=4.54e−5 kidney). h. Model of stress-induced activation of fibroblasts regulated by Meox1.
a,g Data are shown as means ± SEM.
MEOX1 is upregulated in human fibrosis
Single cell data from the human adult heart20 showed that MEOX1 was expressed in activated fibroblasts and, together with POSTN, was one of the top genes determining the cluster of activated fibroblasts (Fig. 4f and Extended Data Fig. 12a). An atlas of chromatin accessibility from the human fetal heart21 indicated that the syntenic region of Peak9/10 was characterized by the strongest signal of accessible chromatin in the MEOX1 distal element in fibroblasts (Extended Data Fig. 12b). Similar to our findings in the heart, MEOX1 expression was induced by TGFβ and suppressed by JQ1 in fibroblasts from human lung, liver and kidney, three other organs that often develop pathological fibrosis in the setting of chronic organ dysfunction (Fig. 4g). Furthermore, MEOX1 expression was significantly up-regulated in heart tissue from patients with cardiomyopathy and in lung tissue from patients with idiopathic pulmonary fibrosis, two human diseases that prominently feature pathological fibrosis (Extended Data Fig. 12c,d)22.
Conclusion
This study uncovers an essential MEOX1-dependent transcriptional switch governing cellular plasticity in the fibroblast compartment during chronic heart failure pathogenesis and demonstrates that this maladaptive cell state transition is a druggable feature of disease (Fig. 4h). Our work highlights that single-cell based interrogation of cell states in a diseased tissue, coupled with temporally-controlled perturbation of transcription signaling, can be leveraged to discover the plasticity of cell states and molecular mechanisms critical for progression and reversal of fibrotic diseases. In contrast to the broad effects of systemic BET inhibition, mechanistic refinement converging on cell-type specific enhancers in the context of a complex tissue offers the opportunity to develop therapeutic approaches that are tailored to targeted gene regulation in defined cell compartments. These findings may inform new therapeutic strategies for a wide variety of chronic disorders that feature maladaptive remodeling of cell state and tissue architecture.
Extended Data
Extended Data Figure 1: Single cell transcriptional landscape of non-cardiomyocytes in heart failure during intermittent exposure to BET bromodomain inhibition.
a. Venn diagram showing overlap of TAC-induced and JQ1-suppressed genes (Log2FC>0.5; adj. p value<0.05: FDR, Benjamini-Hochberg) between bulk RNA-Seq from undissociated LV tissue5 and extracted CMs. b. Heatmap showing the top 5 markers per cluster in the scRNAseq. Total cells n=35,551 in 9 clusters. c. UMAP plots showing cluster identity of all cells (n=35,551) and expression of Dcn, Postn,Ctgf, Lyz2 and Fabp4. d,e UMAP plot colored by sample identity of myeloid (d, n=7,986) and endothelial (e, n=10,672) cells.
Extended Data Figure 2: Reversible effect of JQ1 exposure on the transcriptional signature of baseline and stressed fibroblasts.
a. Expression of known fibroblast stress-related genes shown as UMAP feature plot in fibroblasts (total cells n = 13,937). b. Expression by sample of known fibroblast stress-related genes shown as violin plot in fibroblasts (y axis is normalized UMI levels). c. Dot plot showing expression (avg.exp.scale) and cell percentage of top differentially expressed (DE) marker genes between samples. d. Percentages of the 260 genes upregulated in fibroblasts in TAC vs. Sham that are significantly downregulated in TAC JQ1 vs. TAC (blue bar) and upregulated in TAC JQ1 withdrawn vs. TAC JQ1 (yellow bar). e. Percentages of the 194 genes significantly downregulated in TAC vs. Sham that are significantly upregulated in TAC JQ1 vs. TAC (blue bar) and downregulated in TAC JQ1 withdrawn vs TAC JQ1 (yellow bar). f. Violin plot showing normalized expression score across fibroblast samples of the 194 genes significantly downregulated in TAC vs. Sham and associated TOP GO terms (Fisher exact test).
Extended Data Figure 3: Fibroblast subclusters associated to stress-related gene programs are depleted in Sham and TAC JQ1 cells.
a. UMAP plot of fibroblasts colored by sample identity, n = 13,937. b. UMAP plot of fibroblast subclusters colored by cluster identity. Tree diagram showing cluster relationship is shown to the right. Total cells n = 13,937. c. Histograms showing percentage of each sample in each fibroblast cluster. d. Heatmap showing the top 10 markers per fibroblast cluster in the scRNAseq. Total cells n = 13,937 in 9 clusters. e. Periostin (Postn) expression in fibroblasts by clusters as violin plot (y axis is normalized UMI levels). f. Comparative GO term analysis (Fisher exact test) between fibroblast clusters for fibroblast stress-related biological processes (top GO terms when analyzing the genes driving fibroblast cluster 2,3 and 5).
Extended Data Figure 4: Defining a catalog of cell population–enriched distal elements in fibroblasts, myeloid and endothelial cells using scATACseq.
a. Schematic highlighting the approach to integrate scRNAseq with scATACseq12. See extended methods for details. b. Total scATACseq proximal and distal peaks identified in all cells. c. scATACseq tSNE plot showing clusters and cell number of fibroblasts, myeloid and endothelial cells after integration with scRNAseq. d. Venn diagrams showing sample replicate convergence of cell-enriched distal elements found with scATACseq.
Extended Data Figure 5: scATAC defines chromatin accessibility in heart failure during intermittent exposure to BET bromodomain inhibition.
a. Chromatin accessibility at distal elements between samples in fibroblasts, myeloid and endothelial cells. Boxplots show 25th, 50th, and 75th percentile, with whiskers extending to the furthest value no further than 1.5 times the interquartile range (IQR). The 10% most extreme points were trimmed for better visualization (these never included points within the whiskers of the boxplot). Numbers above boxplots show significant p values, statistical significance (Two-sided Wilcoxon rank-sum test) is shown for: Sham vs. TAC; Sham vs. TAC JQ1; TAC vs. TAC JQ1 and TAC JQ1 vs. TAC JQ1 withdrawn. b. Dynamic accessibility (mean with 95% confidence interval) of distal elements in fibroblasts (n=4394), myeloid (n=1325) and endothelial (n=1626) cells clustered by trend across samples. Accessibility trend and top GO terms (binomial test) associated to clusters 2,6,9,11,12 and 14 are shown for fibroblasts. For myeloid and endothelial cells, only top GO terms (binomial test) associated to cluster 2 are shown. c. Enrichment scores for TF motif accessibility in distal elements between samples for the ten most expressed TFs in TAC in fibroblasts, myeloid and endothelial cells.
Extended Data Figure 6: Nascent transcription in TGFβ-treated cells identifies stress-responsive distal and gene elements.
a. Schematic of isolation and immortalization of mouse adult cardiac fibroblasts. b. Expression by qPCR of canonical markers of activated fibroblasts in Unstim and TGFβ-treated cells. Unpaired t-test (Two-tailed). c. Pearson correlation of the two independent biological replicates of PROseq in Unstim and TGFβ-treated cells. d,e. Heatmap of PROSeq coverage of differentially transcribed distal regions (d) and protein coding genes (e) between Unstim and TGFβ-treated fibroblasts. Wald test with Benjamini/Hochberg correction. Signal for replicate 1 & 2 is shown. Top associated GO terms are shown for panel e (Fisher exact test). f. PROseq tag density (± 5kb gene body) in Unstim and TGFβ-treated cells in the genes differentially transcribed in Unstim vs. TGFβ. Top panel: genes upregulated with TGFβ; Bottom panel: genes downregulated with TGFβ. g. PROseq tag density (± 5kb gene body) in Unstim and TGFβ-treated cells in the set of genes upregulated (left, n=260) or downregulated (right, n=194) in TAC vs. Sham in fibroblasts in vivo. h. Co-Accessibility (CoAc) change in fibroblasts of Postn peak 10/11 elements with the promoters of genes within 1Mb of the peak. Change in CoAc with the Postn promoter is in red. N = 27 genes within 1mb. Boxplots show 25th, 50th, and 75th percentile, with whiskers extending to the furthest value no further than 1.5 times the interquartile range (IQR). i. Postn expression measured by qPCR in Unstim and TGFβ-treated in the CRISPRi control line. Unpaired t-test (Two-tailed). j. Postn Peak8, 10/11 and 19 eRNA expression measured by qPCR in Unstim and TGFβ-treated fibroblasts in a CRISPRi control line and lines targeting either Peak8, Peak10/11 or Peak19. Values are normalized to CRISPRi control line in the Unstim condition. One-way ANOVA followed by Sidak’s correction, statistical significance is shown between Unstim samples and TGFβ-treated samples. k. ChIP qPCR data showing enrichment over chromatin input of H3K9me3 in Control and Postn Peak10/11 CRISPRi lines. Unstim condition (left) and TGFβ-treated condition (right). Regions amplifying Peak10, Peak11 and Postn promoters are shown. One-way ANOVA followed by Sidak’s correction, statistical significance is shown between Control and Postn Peak10/11 CRISPRi lines.
b,i-k, Numbers above histograms show significant p-val. Data are shown as means ± SEM.
Extended Data Figure 7: Characterization of a catalog of super-enhancers in fibroblasts, myeloid and endothelial cells.
a. Distribution of accessibility in fibroblast, myeloid and endothelial cells in TAC state identifies a class of distal regions (super-enhancers, SE) where the accessibility falls over the inflection point of the curve. b,c. Volcano plots showing correlation coefficients and corresponding p-val (refer to analysis depicted in Fig. 2e) of 239 SEs in myeloid (b) and 267 SEs endothelial (c) cells. d. Distribution of H3K27ac in Unstim and TGFβ-treated fibroblasts identifies a class of distal regions (super-enhancers, SE) where the accessibility falls over the inflection point of the curve. e. Fraction of H3K27ac in Unstim and TGFβ-treated fibroblasts for the enhancers identified in vivo having a negative (left, =264) or positive (right, n=206) correlation with heart function (based on analysis depicted in Fig. 2e). Boxplots show 25th, 50th, and 75th percentile, with whiskers extending to the furthest value no further than 1.5 times the interquartile range (IQR).
Extended Data Figure 8: Dynamic changes in chromatin accessibility at the Meox1 super-enhancer.
a. Comparison of LV ejection fraction with chromatin accessibility at the Meox1 super-enhancer in fibroblasts, myeloid and endothelial cells. b. UMAP plot of Meox1 expression in all non-CM cells (n=35,551). c. Chromatin accessibility at the Meox1 super-enhancer between samples in fibroblasts, myeloid and endothelial cells. Boxplots show 25th, 50th, and 75th percentile, with whiskers extending to the furthest value no further than 1.5 times the interquartile range (IQR). Sample size is: Fibroblast (n=676, 979, 1,906, 1,654), Endothelial (n=731, 1,666, 1,030, 851), Myeloid (n=631, 1,080, 1,021, 712). Numbers above boxplots show significant p values, statistical significance (Two-sided Wilcoxon rank-sum test) is shown for: Sham vs. TAC; Sham vs. TAC JQ1; TAC vs. TAC JQ1 and TAC JQ1 vs. TAC JQ1 withdrawn. d. scATACseq average signal across cells in fibroblast samples at the Meox1 super enhancer identifies multiple dynamic peaks in heart failure with pulsatile exposure to BET inhibition. e. Chromatin accessibility trend between samples (mean with 95% confidence interval) in all identified Meox1 super enhancer peaks.
Extended Data Figure 9: Brd4-dependent regulation of Meox1 expression.
a. Meox1 expression measured by qPCR in Unstim and TGFβ-treated fibroblasts, with or without JQ1. b. Expression measured by qPCR of individual BET genes in Unstim or TGFβ-treated fibroblasts with siRNA targeting either Ctrl, Brd2, Brd3 or Brd4. Statistical significance is shown between Unstim samples and TGFβ-treated samples. c. Meox1 expression measured by qPCR in Unstim or TGFβ-treated fibroblasts with siRNA targeting either Ctrl, Brd2, Brd3 or Brd4. Statistical significance is shown between the TGFβ siCtrl sample and the other TGFβ-treated samples.
a-c, All analyzed samples were biological replicates. Numbers above bar graphs show significant p-val (One-way ANOVA followed by Tukey post hoc test). Data are shown as means ± SEM.
Extended Data Figure 10: The Peak9/10 Meox1 enhancer is strongly transcribed following TGFβ stimulation.
a. Volcano plot showing log2FC of PROseq signal of all identified distal scATACseq peaks in fibroblasts (n=9211) between Unstim and TGFβ-treated fibroblasts. Meox1 peak 9 (red) and 10 (orange) are highlighted. b. Co-Accessibility (CoAc) change in fibroblasts of Meox1 peak 9/10 elements with the promoters of genes within 1Mb of the peak. Change in CoAc with the Meox1 promoter is in red. N=115 genes within 1mb. Boxplots show 25th, 50th, and 75th percentile, with whiskers extending to the furthest value no further than 1.5 times the interquartile range (IQR). c,d. Chromosome conformation capture (4C) using Meox1 Peak9/10 region (c) or Meox1 promoter (d) as anchor point. 4C coverage in Unstim and TGFβ-treated fibroblasts are shown in a 922kb (top) and 328kb (bottom) genomic regions. Last track represents the called TGFβ-induced loops with the anchor point (colored in purple). e. Meox1 expression measured by qPCR in Unstim and TGFβ-treated fibroblasts in the CRISPRi control line. Unpaired t-test (Two-tailed). f. Meox1 Peak5, 9/10 and 13 eRNA expression measured by qPCR in Unstim and TGFβ-treated fibroblasts in a CRISPRi control line and lines targeting either Peak5, Peak9/10 or Peak13. Values are normalized to CRISPRi control line in the Unstim condition. One-way ANOVA followed by Sidak’s correction, statistical significance is shown between Unstim samples and TGFβ-treated samples. g. ChIP qPCR data showing enrichment over chromatin input of H3K9me3 in Control and Meox1 Peak9/10 CRISPRi lines. Unstim condition (left) and TGFβ-treated condition (right). Regions amplifying Peak9, Peak10 and Meox1 promoters are shown. One-way ANOVA followed by Sidak’s correction, statistical significance is shown between CRISPRi control and targeted lines. h. Droplet digital (dd) PCR amplifying a WT or mutated region of Meox1 Peak9/10 DNA. Parental fibroblast cell line, WT (clone 20 – isogenic control exposed to CRISPR Cas9 and gRNAs) and Peak9/10 KO (clone 16) cell lines are shown. i. Schematic showing the Meox1 locus with the scATACseq average signal across fibroblasts in TAC. SMAD2/3 motifs (Jaspar - MA1622.1) in the Peak9/10 region and in the Meox1 promoter (±1 kb from TSS) are highlighted. j. Expression measured by qPCR of Smad2 (left) and Smad3 (right) in Unstim or TGFβ-treated fibroblasts with siRNA targeting Ctrl and either Smad2 (left) or Smad3 (right). One-way ANOVA followed by Tukey post hoc test. k. Meox1 expression measured by qPCR in Unstim or TGFβ-treated fibroblasts with siRNA targeting either Ctrl or Smad2. One-way ANOVA followed by Tukey post hoc test.
e-g and j,k Numbers above histograms show significant p-val. Data are shown as means ± SEM.
Extended Data Figure 11: MEOX1 is a regulator of fibroblast activation.
a,b. Meox1 expression measured by qPCR in mouse primary cardiac fibroblasts (a) and immortalized cardiac fibroblasts (b) in Unstim condition, TGFβ siCtrl and TGFβ siMeox1. One-way ANOVA followed by Tukey post hoc test. c. Immunofluorescence staining of αSMA in Unstim, and TGFβ cells with a control or a Meox1-targeting siRNA. Nuclei are marked by Hoechst; scale bar =100μm. Quantification of αSMA staining is shown to the right (two independent experiments). Fold change intensity is normalized to cell number. One-way ANOVA followed by Tukey post hoc test. d. αSMA expression measured by qPCR in Unstim condition, TGFβ siCtrl and TGFβ siMeox1. One-way ANOVA followed by Tukey post hoc test. e. Representative images of Edu incorporation in Unstim condition, TGFβ siCtrl and TGFβ siMeox1. DAPI (blue), Edu (red) and CellMask (green). Quantification (two independent experiments) on the right. One-way ANOVA followed by Tukey post hoc test. f. Meox1 expression measured by qPCR in WT and Meox1 over-expression (o/e) mouse immortalized cardiac fibroblasts. Unpaired t-test (Two-tailed). g. Pearson correlation of the three replicates of MEOX1 anti-HA ChIPseq in Unstim and TGFβ-treated cells. h. MEOX1 anti-HA ChIPseq coverage in all protein coding genes (± 2kb gene body) in Unstim and TGFβ-treated fibroblasts. i. Pearson correlation of the two independent biological replicates of PROseq in TGFβ siCtrl and TGFβ siMeox1. j. PROseq coverage in Unstim, TGFβ siCtrl and TGFβ siMeox1 at the distal elements defined as more transcribed in TGFβ vs. Unstim (2101 sites, see Fig. 2a) that are either bound by MEOX1 (496 regions – top panel) or not (1,605 regions – bottom panel). k. PROseq coverage in Unstim, TGFβ siCtrl and TGFβ siMeox1 at the distal elements with high H3K27ac enrichment in Unstim bound by MEOX1 (379 regions). l. PROseq coverage of differentially transcribed genes (Wald test followed by Benjamini/Hochberg) in TGFβ-treated fibroblasts with Ctrl or Meox1 siRNA and associated top GO terms. Signal for replicate 1 & 2 is shown. m. PROseq tag density (± 5kb gene body) in TGFβ siCtrl and TGFβ siMeox1 in genes upregulated in TGFβ siCtrl vs. TGFβ siMeox1 (left); and genes upregulated in TGFβ siMeox1 vs. TGFβ siCtrl (right). n. Violin plot showing normalized expression score of genes upregulated in TGFβ siCtrl vs. TGFβ siMeox1 in PROseq that were captured in the scRNAseq. Expression of Sham and TAC fibroblast samples is depicted. o. Number of MEOX1-bound genes in MEOX1 ChIPseq (in TGFβ-treated cells) in ±2kb gene body, ±100kb gene body or ±500kb gene body in genes differentially transcribed in PROseq: upregulated in TGFβ vs. Unstim (left); downregulated in TGFβ vs. Unstim (center left); upregulated in TGFβ siCtrl vs. TGFβ siMeox1 (center right); upregulated in TGFβ siMeox1 vs. TGFβ siCtrl (right). p. Coverage of MEOX1 ChIP (TGFβ-treated cells), H3K27ac ChIPseq (Unstim and TGFβ-treated cells) and PROseq (Unstim condition, TGFβ siCtrl and TGFβ siMeox1) at the Postn locus. The Postn Peak10/11 region is highlighted in red.
a-f, Numbers above bar graphs show significant p-val. Data are shown as means ± SEM.
Extended Data Figure 12: MEOX1 is expressed in human activated fibroblasts.
a. POSTN (left) and MEOX1 (right) expression in human adult fibroblast clusters (y axis is normalized UMI levels). b. Track showing scATACseq average signal across fibroblasts in the human fetal heart21 in the MEOX1 locus. The syntenic region of Peak9/10 is highlighted in red. c. Bulk RNAseq data of human MEOX1 expression (FPKM) in heart tissue in controls (Ctrl) and individuals with dilated cardiomyopathy (DCM) or hypertrophic cardiomyopathy (HCM) (GSE141910). Unpaired t-test with Benjamini/Hochberg correction. d. Bulk RNAseq data of human MEOX1 expression (raw counts) in lung tissue between controls (Ctrl) and individuals with idiopathic pulmonary fibrosis (GSE134692)22. Unpaired t-test (Two-tailed). c,d, Numbers above figure show significant p-val.
Supplementary Material
Acknowledgments
We thank the Srivastava laboratory, Joke van Bemmel, Mauro Costa, Nathan Palpant, Woo Jun Shim, Phillip Grote, Elphège P. Nora, Arjun A. Rao, Alexis J. Combes, Paola Benaglio, and Kathryn N. Ivey and Jin Yang from Tenaya Therapeutics for critical discussion and feedback; Jun Qi, Deyao Li and Logan Sigua for providing JQ1; we acknowledge the UCSF Center for Advanced Technology (CAT), Gladstone Genomics Core and Flow Cytometry Core for their technical support and the Gladstone Animal Facility for support with mouse colonies; and Ana Catarina Silva (ana@anasilvaillustrations.com) for help with figure editing and design. M.A. is supported by the Swiss National Science Foundation (P400PM_186704). P.F.P and K.S.P. are supported by NIH P01 HL098707, HL098179, Gladstone Institutes, and the San Simeon Fund. A.P. is supported by the Tobacco-Related Disease Research Program (578649), A.P. Giannini Foundation (P0527061), Michael Antonov Charitable Foundation Inc., and Sarnoff Cardiovascular Research foundation. J.G.T. is supported by NIH F32 HL147463. B.G.T. is supported by the American Heart Association (18POST34080175). R.J. is supported by the Burroughs Wellcome Fund and funds from the Allen Foundation and American Heart Association. T.A.M. is supported by NIH R01 HL116848, NIH R01 HL147558, NIH R01 DK119594 and NIH R01 HL150225. T.A.M. and J.G.T. were supported by the American Heart Association (16SFRN31400013). S.M.H. was supported by NIH R01 HL127240. D.S. is supported by NIH P01 HL146366, NIH R01 HL057181, NIH R01 HL015100, and by the Roddenberry Foundation, the L.K. Whittier Foundation, the Sattui Family, and the Younger Family Fund. D.S. and T.A.M. are supported by NIH R01 HL127240. This work was also supported by NIH/NCRR grant C06 RR018928 to the Gladstone Institutes.
Footnotes
Competing interests
D.S. is scientific co-founder, shareholder and director of Tenaya Therapeutics. S.M.H. is an executive, officer, and shareholder of Amgen, Inc., a scientific co-founder and shareholder of Tenaya Therapeutics, and serves on the scientific advisory board of DZHK (German Centre for Cardiovascular Research). T.A.M. received funding from Italfarmaco for an unrelated project.
Code availability
All scRNAseq analyses were performed using standard protocols with the Seurat R package (v2.3.4). Custom codes relevant to the scATACseq analysis are available at: https://github.com/PFPrzytycki/FibroSwitch
Data availability
All source data, including sequencing reads and single-cell expression matrices have been deposited in NCBI’s Gene Expression Omnibus and are accessible through GEO series accession number GSE155882.
Bibliography
- 1.Rockey DC, Bell PD & Hill JA Fibrosis--A Common Pathway to Organ Injury and Failure. N. Engl. J. Med 373, 96 (2015). [DOI] [PubMed] [Google Scholar]
- 2.Jun J-I & Lau LF Resolution of organ fibrosis. J. Clin. Invest 128, 97–107 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Anand P et al. BET bromodomains mediate transcriptional pause release in heart failure. Cell 154, 569–582 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Spiltoir JI et al. BET acetyl-lysine binding proteins control pathological cardiac hypertrophy. J. Mol. Cell Cardiol 63, 175–179 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Duan Q et al. BET bromodomain inhibition suppresses innate inflammatory and profibrotic transcriptional networks in heart failure. Sci. Transl. Med 9, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stratton MS et al. Dynamic chromatin targeting of BRD4 stimulates cardiac fibroblast activation. Circ. Res 125, 662–677 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Antolic A et al. BET bromodomain proteins regulate transcriptional reprogramming in genetic dilated cardiomyopathy. JCI Insight 5, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Virani SS et al. Heart Disease and Stroke Statistics-2020 Update: A Report From the American Heart Association. Circulation 141, e139–e596 (2020). [DOI] [PubMed] [Google Scholar]
- 9.Filippakopoulos P et al. Selective inhibition of BET bromodomains. Nature 468, 1067–1073 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kanisicak O et al. Genetic lineage tracing defines myofibroblast origin and function in the injured heart. Nat. Commun 7, 12260 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Buenrostro JD, Wu B, Chang HY & Greenleaf WJ ATAC-seq: A Method for Assaying Chromatin Accessibility Genome-Wide. Curr. Protoc. Mol. Biol 109, 21.29.1–21.29.9 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Przytycki PF & Pollard KS CellWalker integrates single-cell and bulk data to resolve regulatory elements across cell types in complex tissues. Genome Biol. 22, 61 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mahat DB et al. Base-pair-resolution genome-wide mapping of active RNA polymerases using precision nuclear run-on (PRO-seq). Nat. Protoc 11, 1455–1476 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.De Santa F et al. A large fraction of extragenic RNA pol II transcription sites overlap enhancers. PLoS Biol. 8, e1000384 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gilbert LA et al. CRISPR-mediated modular RNA-guided regulation of transcription in eukaryotes. Cell 154, 442–451 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Whyte WA et al. Master transcription factors and mediator establish super-enhancers at key cell identity genes. Cell 153, 307–319 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Skuntz S et al. Lack of the mesodermal homeodomain protein MEOX1 disrupts sclerotome polarity and leads to a remodeling of the cranio-cervical joints of the axial skeleton. Dev. Biol 332, 383–395 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lu D et al. Meox1 accelerates myocardial hypertrophic decompensation through Gata4. Cardiovasc. Res 114, 300–311 (2018). [DOI] [PubMed] [Google Scholar]
- 19.Wei Z et al. Molecular Mechanism of Mesenchyme Homeobox 1 in Transforming Growth Factor β1-Induced P311 Gene Transcription in Fibrosis. Front. Mol. Biosci 7, 59 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Litviňuková M et al. Cells of the adult human heart. Nature 588, 466–472 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Domcke S et al. A human cell atlas of fetal chromatin accessibility. Science 370, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sivakumar P et al. RNA sequencing of transplant-stage idiopathic pulmonary fibrosis lung reveals unique pathway regulation. ERJ Open Research 5, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All source data, including sequencing reads and single-cell expression matrices have been deposited in NCBI’s Gene Expression Omnibus and are accessible through GEO series accession number GSE155882.