Abstract
Background:
The role of the lipidome as a biomarker for Parkinson’s disease (PD) is a relatively new field that currently only focuses on PD diagnosis.
Objective:
To identify a relevant lipidome signature for PD severity markers.
Methods:
Disease severity of 149 PD patients was assessed by the Unified Parkinson’s Disease Rating Scale (UPDRS) and the Montreal Cognitive Assessment (MoCA). The lipid composition of whole blood samples was analyzed, consisting of 517 lipid species from 37 classes; these included all major classes of glycerophospholipids, sphingolipids, glycerolipids, and sterols. To handle the high number of lipids, the selection of lipid species and classes was consolidated via analysis of interrelations between lipidomics and disease severity prediction using the random forest machine-learning algorithm aided by conventional statistical methods.
Results:
Specific lipid classes dihydrosphingomyelin (dhSM), plasmalogen phosphatidylethanolamine (PEp), glucosylceramide (GlcCer), dihydro globotriaosylceramide (dhGB3), and to a lesser degree dihydro GM3 ganglioside (dhGM3), as well as species dhSM(20:0), PEp(38:6), PEp(42:7), GlcCer(16:0), GlcCer(24:1), dhGM3(22:0), dhGM3(16:0), and dhGB3(16:0) contribute to PD severity prediction of UPDRS III score. These, together with age, age at onset, and disease duration, also contribute to prediction of UPDRS total score. We demonstrate that certain lipid classes and species interrelate differently with the degree of severity of motor symptoms between men and women, and that predicting intermediate disease stages is more accurate than predicting less or more severe stages.
Conclusions:
Using machine-learning algorithms and methodologies, we identified lipid signatures that enable prediction of motor severity in PD. Future studies should focus on identifying the biological mechanisms linking GlcCer, dhGB3, dhSM, and PEp with PD severity.
Keywords: Parkinson’s disease (PD), PD severity, Lipidomics, Lipidome, UPDRS, Machine learning
Introduction
The role of lipids, specifically glycosphingolipids, in Parkinson’s disease (PD) pathogenesis has been highlighted by recent discoveries [1]. The association between PD and lysosomal lipid hydrolases, specifically glucocerebrosidase (GBA) and potentially others (e.g., SMPD1), further supports the need to examine the role of lipids as biomarkers in PD [2]. GBA plays an important role in the glycosphingolipid metabolic pathway, encoding the lysosomal enzyme β-glucosidase (GCase), which hydrolyzes glucosylceramide and glucosylsphingosine to ceramide and sphingosine, respectively. In GCase deficient cells, α-synuclein aggregation has also been linked to reduced ceramide production [3].
In a previous study, we tested the association between plasma lipid concentration and PD diagnosis [4]. Using univariate logistic regression, two lipid classes, monosialodihexosylgangliosides (GM3) and triglycerides (TG), were significantly different between PD and healthy controls. A link between GM3 and PD pathology had been previously demonstrated through research on the high affinity of α-synuclein via its ganglioside-binding domain to GM3, and by a study demonstrating that saturating membranes with GM3 accelerates the aggregation of α-synuclein [5, 6]. However, the analysis was limited to univariate regression models, which assumed variable independencies and did not explore interactions across lipids or the potential of joint impact on the probability of diagnosis. In this study, neither GM3 nor TG were found significant to PD severity prediction of the Unified Parkinson’s Disease Rating Scale (UPDRS) score.
Recently, machine learning (ML) has been used in biomedical studies for better predictions and richer insights in PD [7–20] and other neurodegenerative diseases [21]. For example, neural networks and regression trees examined the ability to diagnose PD based on biomedical voice measurements [9], and other regression trees such as a random forest (RF) were used to estimate the UPDRS score based on a speech test [16]. Similarly, ML algorithms predicted PD severity using non-motor PD symptoms [7] and a voice data set [15].
Here, we extend our previous investigation of linear connections between lipids and PD diagnosis [4] to a prediction of disease severity using the multivariate non-linear RF algorithm. We also identify interrelations among a lipidome of 517 species from 37 classes and PD severity measured by the UPDRS and the Montreal Cognitive Assessment (MoCA) scores. To our knowledge, this research is the first to study the interrelations between the lipidome and PD severity using a combination of a relatively large cohort and multivariate non-linear ML algorithms.
Material and Methods
Participants and clinical evaluation
The cohort analyzed was as described in an earlier study [4]. In brief, participants in the ‘Spot’ study [22, 23] were recruited between 2010–2016 from the Center for Parkinson’s Disease at Columbia University Irving Medical Center in New York, NY. Among the participants, we randomly selected PD patients (n=150). All participants were non-carriers of SNCA, LRRK2, and GBA mutations (with the exception of one PD case who was a carrier of the GBA variant E326K, n=1). Evaluation of all participants included the MoCA [24], and the UPDRS in the “on” state. Medical history, current medications, family history of PD, and demographics were also collected. The original study also included 100 controls, which were not analyzed here, as our primary aim was to test the link between lipids and disease severity. Data from these controls are presented only in Figure 2 for illustrative purposes. Plasma collection for lipidomics was as previously described [4]. In brief, blood samples were collected in single EDTA tubes (10cc in volume) and centrifuged, after which 1cc plasma aliquots were extracted and stored in a −80°C freezer within one hour of collection. Table 1 includes all the lipids measured in the study and lipid class membership. Informed consent was signed by all participants and the Columbia University IRB approved all study procedures. Since the UPDRS III and UPDRS total score were missing for a single patient, in this analysis, we used the data of 149 participants.
FIG. 2.
(A) Radar plot representing UPDRS III scrores and average values of the most contributing lipid classes (Table 3), age, AAO, and gender for three PD stages: early (low severity; yellow), intermediate (red), and late (high severity; blue) in comparison to control (green). The controls are 100 subjects genetically unrelated frequency-matched by gender and age (Avg.=66.11 years, std.=9.4) with average and std. BMI values of 25.7 and 4.4, respectively. AAO for the control was set to the value of the low severity. Numbers in parentheses indicate numbers of observations (patients). As we move along an axis from the center/origin of the outer polygon to any of its vertices, the corresponding variable increases its value, e.g., towards higher age, AAO, and lipid class concentrations (and for higher percentages of men than women for the gender variable). The figure demonstrates differences between PD patient groups in three tertiles of UPDRS III scores of 0–11 (lower tertile), 12–20 (middle tertile), and 21–48 (upper tertile) in comparison to control. Among the patients, early stage patients (lowest tertile), mostly women, are recognized by the lowest values of GlcCer (and, of course, of age and AAO values), with extreme values of dhSM, PEp, and PE; the latter value is extremely larger than those of the intermediate and late stage patients (and similar to that of control). Patients with intermediate UPDRS III scores (middle tertile) have dhSM and PEp values similar to those of early stage patients, but also the highest levels of GlcCer and lowest levels of dhGB3. The patients with the highest UPDRS III scores (upper tertile), as expected, are older and have higher AAO values, highest levels of dhGB3, intermediate levels of GlcCer, and PE, and the lowest values of PEp and dhSM. When considering also the control group, we notice two results: a) a perfect order of values from control to low, intermediate, and up to high severities for dhSM and almost perfect order for GlcCer and PE (the two highest severities swapped their order although are very similar in values), and b) for most lipid classes (except PEp and dhGB3), lipid levels of controls are at the extreme ends, i.e., having either the lowest or highest level. (B) Similar to (A) for a combination of average (over the patients) values of dhGM3 and its most contributing species. The figure shows that patients in the lower UPDRS III score tertile have the highest values of dhGM3, dhGM3(22:0), and dhGM3(22:1) but the lowest values of dhGM3(16:0). The case is nearly opposite for patients in the highest UPDRS III score tertile, i.e., the lowest values of dhGM3, dhGM3(22:0), and dhGM3(22:1), and relatively high values of dhGM3(16:0). Interestingly, while the intermediate disease severity is characterized by intermediate values of dhGM3, dhGM3(22:0), and dhGM3(22:1), it is also characterized by the highest values of dhGM3(16:0). (C) Similar to (A) for a combination of average (over the patients) values of GlcCer and its most contributing species together with age and AAO. The figure shows patients in the higher tertile of UPDRS III scores were older with later AAOs, but also had the highest values of GlcCer(16:0) and relatively high values of GlcCer and all of its other species. On the other hand, patients with the lowest UPDRS III scores are the youngest with the lowest AAO on average and have the lowest values of GlcCer and its species, except for GlcCer(24:1), where these patients have the highest plasma levels. (D) The same as (C) including the control group for a reference (the small values of the control subjects changed the scale drastically so previous differences among PD severities were diminished), showing that none of the species of the GlcCer class is expressed in the control subjects, and thereby emphasizing the role of this lipid class in PD (C).
TABLE 1.
Lipids measured in the study and their class membership
| Type | Abbrev. | Full name |
|---|---|---|
| Neutral Lipids | FC | Free Cholesterol |
| CE | Cholesterol Ester | |
| MG | Monoacylglycerol | |
| DG | Diacylglycerol | |
| TG | Triacylglycerol | |
| Sphingolipids | Cer | Ceramide |
| dhCer | Dihydroceramide | |
| SM | Sphingomyelin | |
| dhSM | Dihydrosphingomyelin | |
| GalCer | Galactosylceramide | |
| Sulf | Sulfatide | |
| dhSulf | DihydroSulfatide | |
| GlcCer | Glucosylceramide | |
| LacCer | Lactosylceramide | |
| dhLacCer | Dihydrolactosylceramide | |
| GM3 | Monosialodihexosylganglioside | |
| dhGM3 | DihydroMonosialodihexosylganglioside | |
| GB3 | Globotriaosylceramide | |
| dhGB3 | DihydroGlobotriaosylceramide | |
| Phospholipids | PA | Phosphatidic acid |
| PC | Phosphatidylcholine | |
| PCe | Ether phosphatidylcholine | |
| PE | Phosphatidylethanolamine | |
| PEp | Plasmalogen phosphatidylethanolamine | |
| PG | Phosphatidylglycerol | |
| PI | Phosphatidylinositol | |
| PS | Phosphatidylserine | |
| LPC | Lysophosphatidylcholine | |
| LPCe | lysophosphatidylcholine ether | |
| LPE | Lysophosphatidylethanolamine | |
| LPEp | lysophosphatidylethanolamine plasmalogen | |
| Phospholipids | LPI | Lysophosphatidylinositol |
| BMP | Bis(monoacylglycero)phosphate | |
| APG | Acyl Phosphatidylglycerol | |
| NAPE | N-Acylphosphatidylethanolamines | |
| NAPEp | N-Acylphosphatidylethanolamine plasmalogen | |
| NAPS | N-Acyl Phosphatidylserine |
Analysis of lipid classes and PD severity
The first phase of the study focused on identifying the most influential lipid classes that predicted PD severity as determined by the UPDRS and MoCA scores. We predicted both the UPDRS III and UPDRS total scores, although the latter composes of UPDRS I, II, and III and thus depends on the former. The 37 lipid classes, two clinical variables [age at onset (AAO) and disease duration], demographic variables (i.e., age, gender, height, BMI, marital status), and education were the predicting variables.
The root mean square error (RMSE) [25], which is the most common measure to evaluate prediction performance, was calculated as an average over a Monte Carlo cross validation, where in each of 200 datasets, 80% of the (149) observations were sampled randomly (without replacement) for training the algorithm, and the remaining 20% used for testing it. This resampling method was chosen to reinforce the significance of the research results when experimenting with the cohort.
In this phase, we wanted first to validate our ML algorithm, the RF, in predicting PD severity. The RF is a popular accurate classifier, which was shown in many previous studies to be efficient and beneficial in various clinical [21, 26–28] and non-clinical [29, 30] domains. The RF classifier [31, 32] holds no assumptions about the data distribution, can cope with very complex problems with minimum overfitting [31, 33–35], and ranks variables by their contribution to accurate (or informative) prediction [36–38]. In this phase, we selected variables that were ranked in the top 30% as contributory by RF. For further explanation about the RF, see the Supplementary material.
In our case, we expected the RF to help interpret the prediction results by indicating interrelations among lipid classes and species with different stages in the disease. Since the RF has never been used in predicting PD severity, we first validated it through conventional statistical methods. Each statistical method is limited by the assumptions it makes, hence we employed three of them. Each statistical method explored the data from a different angle and selected contributing variables, as specified below. The RF together with the statistical methods create an ensemble of classifiers (though not independent) that is used here as “a panel of experts”.
The statistical models that were used to validate and then to enforce the RF algorithm are: (1) Univariate linear regression (UR), where a variable is selected if considered statistically significant with p<0.05; (2) Multivariate linear regression (MR), implemented by considering variables using sequential backward selection, sequential forward selection, or sequential floating forward selection (i.e., stepwise), where the model that achieves the lowest Akaike information criterion (AIC) [39] is chosen, and all variables that are statistically significant (p<0.05) are selected; (3) Lasso regression [40], which is based on ordinary least squares with a penalty factor for the number of variables, forces some of the coefficients to equal to zero, and thereby selects the variables with coefficients greater than zero.
We let each of the four methods identify the most contributing variables among the predictors and kept only variables that at least three methods indicated as contributing highly to prediction of the PD severity.
Analysis of lipid species and PD severity
The second phase was done only for the UPDRS since the first phase results for the MoCA showed poor relation to the lipidome. The same methodology of the first phase was implemented here. However, in this phase, we faced another challenge since the number of variables exceeded the number of examples (patients); that is, the 517 lipid species and eight demographical and clinical variables were much greater than the 149 patients, which made variable selection even more challenging than in the first phase. To accommodate this, we selected only lipid classes that had at least two supporting methods (three methods were employed in the first phase). Then for the lipid species of these classes, we added the eight demographical and clinical variables and let the RF rank these variables as in the first phase. Finally, we sequentially removed lower-ranked variables as long as it did not worsen the RMSE, while keeping at least 10% of the variables to allow for retention of a sufficient number of them.
First in our research plan, we identified and ranked the most contributing lipid classes and species for UPDRS total and UPDRS III scores in a two-stage approach (first classes and then species of the most contributing classes) using the RF classifier that is validated using statistical methods. Second, we examined values of plasma concentrations of the most contributing lipid classes and species in different PD severity levels and considered their sensitivity to gender and age. Third, we identified lipid signatures (combinations of classes, species, and demographics) for different severity levels, and evaluated their interrelations with age, AAO, and disease duration and the impact of the combined signatures on the prediction model accuracy.
Results
Data description
Table 2 presents descriptive statistics for the demographical and clinical data of the 149 PD patients’ cohort. The AAO is platykurtic (kurtosis=−0.52, skewness =0.029), the disease duration is leptokurtic and positively skewed (kurtosis=1.99, skewness =1.35), with 82% of the patients with disease durations shorter than 10 years. Gender, by design, is evenly distributed )75 males and 74 females). The medication dosage distribution for different UPDRS III scores is presented in Supplementary Figure S1, showing, as expected, that the dosage increases with the severity.
TABLE 2.
Descriptive statistics of demographical and clinical data of the 149 PD patients’ cohort
| Variable | Mean (Min, Max) | Std. |
|---|---|---|
| Age (years) | 66.5 (43, 87) | 9.2 |
| Height (inches) | 66.4 (55, 76) | 3.9 |
| BMI | 25.8 (17, 50) | 4.8 |
| Age at onset (years) | 60.8 (43, 81) | 9.0 |
| Disease duration (years) | 5.7 (0, 27) | 5.2 |
| Levodopa equivalent dose (mg) | 465.8 (0, 1500) | 396.6 |
| UPDRS III score | 16.3 (0, 48) | 9.3 |
| UPDRS total score | 25.8 (3, 72) | 13.8 |
| MoCA | 25.7 (12, 30) | 3.2 |
UPDRS: Unified Parkinson’s Disease Rating Scale; MoCA: Montreal Cogntive Assessment
Lipid classes for prediction of disease severity
Table 3 shows the most contributing variables (as defined above and based on at least three out of four algorithms) from all lipid classes and the demographical and clinical variables for the UPDRS total and UPDRS III scores. As expected, age and disease duration were found among the most influential variables especially for UPDRS total score. Our data shows that dihydroglobotriaosylceramide (dhGB3), dihydrosphingomyelin )dhSM), glucosylceramide (GlcCer), and plasmalogen phosphatidylethanolamine (PEp) are the most contributory lipid classes in prediction of both UPDRS total and UPDRS III scores. While the UR and MR sometimes gave conflicting results regarding the significance of a variable, RF and Lasso were more consistent. Similar to age and disease duration, higher GlcCer, dhGB3, and globotriaosylceramide (GB3) values were associated with higher scores (the first two classes for both UPDRS total and III scores). In contrast, dhSM and PEp (for both UPDRS total and III scores) as well as monoacylglycerol (MG), phosphatidylethanolamine (PE), and dihydro GM3 (dhGM3) (only for UPDRS III score) affected UPDRS negatively. In addition, lipid classes sulfatide (Sulf), free cholesterol (FC), acyl phosphatidylglycerol (APG), lysophosphatidylcholine (LPC), phosphatidylglycerol (PG), and dhGM3, as well as lipid classes triacylglycerol (TG), dihydrolactosylceramide (dhLacCer), lysophosphatidylinositol (LPI), APG, and Sulf had only two supporting methods for UPDRS total and UPDRS III scores, respectively, and thus are not presented in Table 3. Additionally, BMI was not found to be a contributing variable.
TABLE 3.
Most contributing lipid classes for UPDRS scores prediction
|
UPDRS total
| ||||||
| No. of methods | Variable | Direction | UR (p-value) | MR (p-value) | Lasso | RF |
|
| ||||||
| 4 | Age | Positive | 3.91e-06 | 1.81e-06 | + | + |
| 3 | Duration | Positive | 1.35e-05 | + | + | |
| 3 | dhSM | Negative | 0.0024 | + | + | |
| 3 | PEp | Negative | 0.0035 | + | + | |
| 3 | AAO | Positive | 0.0333 | 0.0022 | + | |
| 3 | GlcCer | Positive | 0.0001 | + | + | |
| 3 | dhGB3 | Positive | 0.0127 | + | + | |
| 3 | GB3 | Positive | 0.048 | + | + | |
|
UPDRS III | ||||||
| No. of Methods | Variable | Direction | UR (p-value) | MR (p-value) | Lasso | RF |
|
| ||||||
| 4 | Age | Positive | 0.0002 | 1.42 e-06 | + | + |
| 4 | MG | Negative | 0.0177 | 0.0037 | + | + |
| 4 | dhGB3 | Positive | 0.0207 | 0.0001 | + | + |
| 3 | dhSM | Negative | 0.0064 | + | + | |
| 3 | PEp | Negative | 0.0231 | + | + | |
| 3 | Duration | Positive | 0.0286 | + | + | |
| 3 | PE | Negative | 0.0304 | 7.57 e-05 | + | |
| 3 | dhGM3 | Negative | 0.0415 | 0.0337 | + | |
| 3 | GlcCer | Positive | 0.0038 | + | + | |
Lipid acronyms: Dihydrosphingomyelin (dhSM), plasmalogen phosphatidylethanolamine (PEp), glucosylceramide (GlcCer), globotriaosylceramide (GB3), dihydro globotriaosyl ceramide(dhGB3), monoacylglycerol (MG), phosphatidylethanolamine (PE), dihydro GM3 ganglioside (dhGM3)
Univariate linear regression (UR)
Multivariate linear regression (MR)
Lipid classes and clinical and demographic variables identified as contributing/statistically significant to severity prediction [as calculated by UPDRS total (upper section) and UPDRS III (lower section)] scores by at least three methods. Also shown is the variable impact/direction (as positive/negative) based on the Lasso regression (since the Lasso coefficient for AAO was zero, we used Ridge regression to get the direction of impact for this variable). The p-value is presented for significant variables only. Variables supported by four or three methods are sorted according first to the UR and then MR p-values, and those common to UPDRS total and UPDRS III scores are in bold.
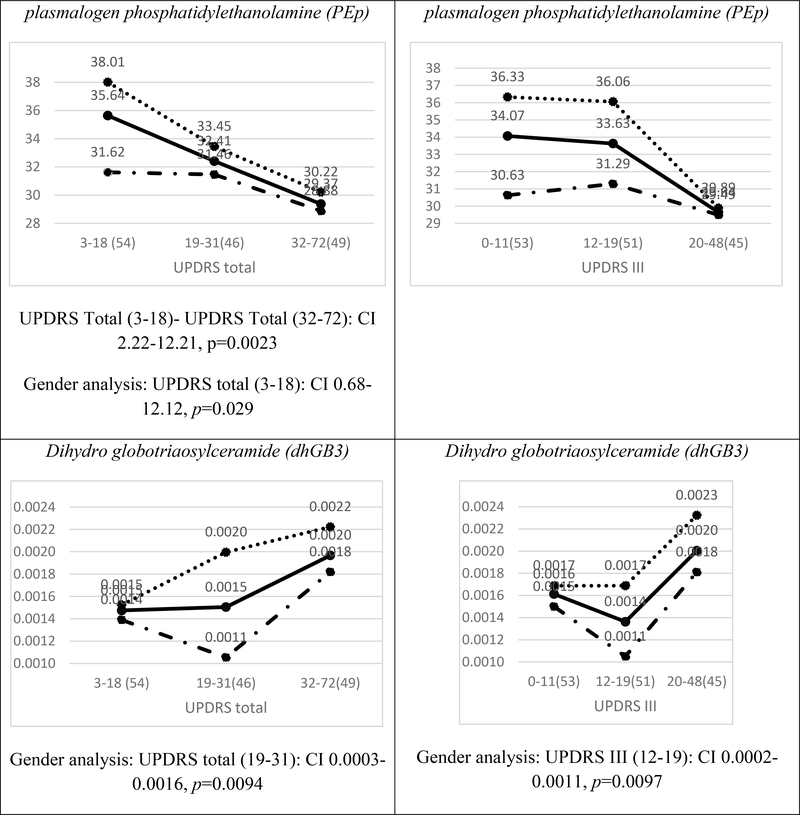
Average plasma concentrations divided by tertiles of the UPDRS III and UPDRS total scores for the cohort are presented in Figure 1 for dhSM, GlcCer, PEp, and dhGB3. Figure 1 shows that in most of the cases (e.g., dhSM and PEp), a clear trend can be observed (either increasing or decreasing monotonicity) between the scores in the lower and higher cohorts for both UPDRS total and UPDRS III scores. The figure also demonstrates that while female patients have higher concentrations than male patients in the lower score ranges UPDRS (both UPDRS total and UPDRS III scores), the difference almost always becomes insignificant in the higher severity level.
FIG. 1.
Average plasma concentrations (pmol/μl) for different ranges of the UPDRS total score (left) and UPDRS III score (right) for four contributory lipid classes common to UPDRS total and III scores (Table 3). Numbers in parentheses indicate numbers of observations (patients) in different ranges of severities. The solid, dotted, and broken lines represent all patients, female patients, and male patients, respectively. The distributions of females and males over UPDRS III and UPDRS total scores ranges are given at the top of the figure. The confidence interval (CI) results for multi comparisons between the severities are based on a post-hoc Tukey HSD test and those between females and males on a two-sample t-test [both CIs are presented below each plot only for statistically significant (p<0.05) differences].
Figure 2A demonstrates differences between PD patients in three tertiles of their UPDRS III scores with respect to values of their lipid classes and demographics, showing distinct demographic-lipidomic signatures of patients with different disease severities in comparison with controls.
Finally, the results for the MoCA indicate four variables that had three or four supporting methods: age, education, disease duration, and PE. Age and disease duration were negatively correlated with the MoCA score, the older the patient or the longer the disease duration, the lower the score. Education was positively correlated with MoCA score; the more educated the patient, the higher the score. Note that the MoCA analysis is the sole place where education came up as contributable. PE has a strong monotonic negative impact on the MoCA score (data not presented).
Lipid species for prediction of disease severity
We started this analysis with 517 species and 8 demographical and clinical variables and then minimized this list using the feature selection ML methodology (described in the Material and Methods section), identifying the species that contributed the most for PD severity. This was implemented for UPDRS III and UPDSR total scores, as presented in Tables 4 and 5, respectively. As in the lipid class analysis, age and disease duration are highly ranked in predicting PD severity in the lipid species analysis of UPDRS total score (Table 4). Predictors of the UPDRS total score included 18 species from 11 classes, but the majority of the influential species (n=15; 83%, Table 4) are from five classes: PEp (5), GB3 (3), dhSM (3), GlcCer (2), and dhGM3 )2). In addition to age and AAO that are influential on both UPDRS total and UPDRS III scores, half of the species (9/18) are influential on both scores (with the same directionality). Most of the classes (9/12) identified for UPDRS III in the first phase have representative species in Table 5. There are three species, highlighted in dark grey, whose directions are not corresponding with those of their class directions. Two of them, PEp (42:7) and dhGM3(16:0), also appear influential on the UPDRS total score with no corresponding directions with their classes (Table 4).
TABLE 4.
Most contributing lipid species for UPDRS total score prediction
| Rank | Variable (species) | Direction | Class |
UR (p-value) | |
|---|---|---|---|---|---|
| Name | Direction | ||||
| 1 | Age | Positive | Age | Positive | 3.91e-06 |
| 2 | Duration | Positive | Duration | Positive | 1.35e -05 |
| 3 | PEp(38:6) | Negative | PEp | Negative | 0.0002 |
| 4 | GB3(24:1) | Negative | GB3 | Positive | |
| 5 | GlcCer(16:0) | Positive | GlcCer | Positive | |
| 6 | dhSM(22:0) | Negative | dhSM | Negative | 0.0009 |
| 7 | dhGB3(16:0) | Positive | dhGB3 | Positive | |
| 8 | PEp(40:6) | Negative | PEp | Negative | 0.0006 |
| 9 | GB3(18:0) | Positive | GB3 | Positive | |
| 10 | PEp(42:7) | Positive | PEp | Negative | |
| 11 | GB3(16:0) | Positive | GB3 | Positive | |
| 12 | dhSM(20:0) | Negative | dhSM | Negative | 0.00012 |
| 13 | GlcCer(24:1) | Positive | GlcCer | Positive | |
| 14 | AAO | Positive | AAO | Positive | 0.0333 |
| 15 | APG(34:2) | Negative | APG | Negative | |
| 16 | PEp(40:7) | Negative | PEp | Negative | 0.0021 |
| 17 | dhGM3(22:0) | Negative | dhGM3 | Negative | 0.0230 |
| 18 | PEp(36:2) | Negative | PEp | Negative | |
| 19 | dhGM3(16:0) | Positive | dhGM3 | Negative | |
| 20 | PG(32:0) | Negative | PG | Negative | 2.43 e-05 |
| 21 | dhSM(18:1) | Negative | dhSM | Negative | |
Lipid acronyms: Plasmalogen phosphatidylethanolamine (PEp), globotriaosylceramide (GB3), glucosylceramide, (GlcCer), dihydrosphingomyelin )dhSM), dihydro globotriaosylceramide (dhGB3), acyl phosphatidylglycerol (APG), dihydro GM3 ganglioside (dhGM3), phosphatidylglycerol (PG)
Univariate linear regression (UR)
Most contributing variables/species for prediction of UPDRS total score sorted in descending order of importance according to the RF. Light gray indicates that a species and its class have the same directionality based on the Lasso regression (unless the Lasso coefficient is zero; in these cases, we used Ridge regression to get the direction of impact), and dark gray indicates they have different directionalities. Bold indicates the variable is also contributing to UPDRS III score (Table 5). UR p-values were computed in a post-processing phase and are presented only for significant variables.
TABLE 5.
Most contributing lipid species for UPDRS III score prediction
| Rank | Variables (species) | Direction | Class |
UR (p-value) | |
|---|---|---|---|---|---|
| Name | Direction | ||||
| 1 | dhGB3(16:0) | Positive | dhGB3 | Positive | 0.021 |
| 2 | Age | Positive | Age | Positive | 0.0002 |
| 3 | AAO | Positive | AAO | Positive | 0.0138 |
| 4 | LPI(18:0) | Negative | LPI | Negative | |
| 5 | PE(36:1) | Negative | PE | Negative | 0.0031 |
| 6 | dhGM3(22:0) | Negative | dhGM3 | Negative | 0.0146 |
| 7 | PEp(38:5) | Negative | PEp | Negative | 0.0172 |
| 8 | GlcCer(24:1) | Positive | GlcCer | Positive | |
| 9 | APG(38:1) | Negative | APG | Negative | 0.0161 |
| 10 | GlcCer(16:0) | Positive | GlcCer | Positive | |
| 11 | PEp(38:6) | Negative | PEp | Negative | 0.0056 |
| 12 | APG(34:2) | Negative | APG | Negative | |
| 13 | dhGM3(22:1) | Negative | dhGM3 | Negative | |
| 14 | PE(40:6) | Negative | PE | Negative | 0.0183 |
| 15 | LPI(18:1) | Positive | LPI | Negative | |
| 16 | dhSM(20:0) | Negative | dhSM | Negative | 0.0071 |
| 17 | MG(16:0) | Negative | MG | Negative | 0.0042 |
| 18 | GlcCer(22:0) | Positive | GlcCer | Positive | |
| 19 | PEp(42:7) | Positive | PEp | Negative | |
| 20 | dhGM3(16:0) | Positive | dhGM3 | Negative | |
Lipid acronyms: Dihydro globotriaosylceramide (dhGB3), lysophosphatidylinositol (LPI), phosphatidylethanolamine (PE), dihydro GM3 ganglioside (dhGM3), Plasmalogen phosphatidylethanolamine (PEp), glucosylceramide, (GlcCer), acyl phosphatidylglycerol (APG), dihydrosphingomyelin )dhSM), monoacylglycerol (MG)
Univariate linear regression (UR)
Most contributing variables/species for prediction of UPDRS III score sorted in descending order of importance according to the RF. Light gray indicates a species and its class have the same directionality based on the Lasso regression (unless the Lasso coefficient is zero; in these cases we used Ridge regression to get the direction of impact), and dark gray indicates they have different directionalities. Bold indicates the variable is also contributing to the total UPDRS score (Table 4). UR p-values were computed in a post-processing phase and are presented only for significant variables.
Average plasma concentrations of the cohort divided by tertiles based on the UPDRS III score are presented in Figure 3 for most of the contributing lipid species in Table 5. In most of the cases [e.g., GlcCer(16:0), dhSM(20:0), dhGM3(22:0), and dhGM3(22:1)], the cohort concentration increases/decreases linearly with the disease severity, whereas in other cases [e.g., GlcCer(24:1) and dhGM3(16:0)], a point in an intermediate severity shows a change in the trend of the concentration. We believe this change point is responsible for the mismatch in directionality of impact between a species and its class, as was observed before [e.g., dhGM3(16:0) in Tables 4 and 5]. We also see a similar pattern to that of lipid classes (Figure 1), where female patients usually have higher concentrations in the low and medium severities, but the difference to male patients usually vanishes for the highest UPDRS III scores. Similar to the lipid class analysis, we show in Figures 2B-2D radar plots presenting distinct demographic-lipidomic signatures of patients with different disease severities in comparison with controls.
FIG. 3.
Average plasma concentrations (pmol/μl) for different ranges of the UPDRS III score for species that were identified among the eighteenth important ones (Table 4). The solid line represents all the PD patients. Numbers in parentheses indicate numbers of observations (patients) in different ranges of severities. The dotted line represents female patients and the broken line the male. The distributions of females and males over UPDRS score ranges are given at the top of the figure. The CI results for the comparisons between the severities are based on a post hoc Tukey HSD test and those between females and males on a two-sample [both Cis t-test, are presented below each plot only for statistically significant (p<0.05) differences.]. Note that dhGB3(16:0) is the only species in the dhGB3 class, what explains the identity between the corresponding figures here and in Figure 1.
Prediction model accuracy for UPDRS III and UPDRS total scores as measured by the RMSE is presented in Figure 4, with an average of 6.81 and 9.42, respectively. We see that our models are most accurate in predicting UPDRS III and UPDRS total scores in those 50% of the patients whose scores range between 9 and 22, with RMSE values of 2.85–5.72 and 4.48–7.56, respectively, and less accurate in those extreme scores (with UPDRS III score under 5 or over 27). As Supplementary Figure S2 shows, this difference in accuracy is because of few extreme prediction errors due to outliers and high variability in the patient data in the extreme severity ranges.
FIG. 4.
Average prediction RMSE values for UPDRS III and UDPRS total scores and relatively equally populated UPDRS III score ranges. , where n is the number of observations (patients), yj is the actual UPDRS total and UPDRS III scores for the jth patient, and is the corresponding predicted value. Numbers in parentheses indicate numbers of patients for different severity ranges. The figure presents the RMSE of the RF trained using the most contributing lipid species (Tables 4 and 5) in eight ranges of UPDRS III score severities. While the average RMSE for UPDRS III and UPDRS total scores for all 149 patients is 6.81 and 9.42, respectively (not shown in the figure), the figure shows that for 50% (75) of the patients, those with UPDRS III score between 9 to 22 (the four middle severity ranges), the average RMSE in predicting UPDRS III score is between 2.85 and 5.72, and that in predicting UPDRS total score is between 4.48 and 7.56. These RMSE values are much smaller than the 9.3 and 13.8 standard deviations of UPDRS III and UPDRS total scores in our data, respectively (Table 2). For another 25% (38) of the patients, in severity ranges of 6–8 and 23–26, the average error is between 7.07 and 7.57 for UPDRS III score and between 9.79 and 10.11 for UPDRS total score, which are also smaller compared with the corresponding 9.3/13.8 standard deviations. However, for extreme (low and high) UPDRS III score, 24% (36) of the patients have average error higher than the standard deviation, which is attributed to the high variance of, and outliers in, RMSE in these severity ranges (Supplementary Figure S2). Also note from comparing the graphs for UPDRS total and UPDRS III scores, that errors due to UPDRS I and UPDRS II scores (the difference between UPDRS total and III scores) are greater for extreme severity values.
Supplementary Figures S3A and S3B show the result of excluding age, AAO, and disease duration (both the variables and their interactions with the lipids) from the models, relying only on the prediction capability of the contributing lipid species (Tables 4 and 5). While excluding age and AAO (duration was not found contributory to prediction in Table 5 probably because it is a derivation of the former two) did not affect UPDRS III score (Supplementary Figure S3A; RMSE increase of only 3%−4% for all severities), our data show that the absence of these two variables together with duration increased the prediction error for UPDRS total score for most of the severities (Supplementary Figure S3B) between 26% and 198%. We believe that age, AAO, and disease duration mostly affect UPSRS II score (Activities of Daily Living) and especially UPDRS I score (Mentation, Behavior, and Mood) and to a lesser degree the motor examination in the test (UPDRS III score). Supplementary Table S1 shows ranked by RF values of variable importance (ordered by the Gini index) for UPDRS total and UPDRS III scores, reinforcing that age and duration (much more than AAO) contribute to more accurate prediction of UPDRS total score, whereas age and AAO are less influential compared to lipid species (and duration is not even ranked) for accurate prediction of UPDRS III score (compare Gini index values of these variables between UPDRS total and UPDRS III scores). However, the lowest (under 10) and highest (above 36) severities for UPDRS total score (Supplementary Figure S3B) show the opposite picture. The improvement in the average error while excluding age, AAO, and duration is explained by a skewed disease duration in the lowest range (the 25th and 48th percentiles are almost identical) and high variability of this variable in the highest range that together undermine the accuracy of the model that uses all variables.
Discussion
In this current age where large databases (e.g., of omics data) are available for analysis, traditional statistical tools may not be sufficient to gain deep clinical understanding of interrelations within the data. Lipidomics data are a classical example of such an instance. First, lipids are inherently related to one another and assumptions of independence often made by statistical models may not be accurate. Second, as is the case of this study, the amount of data collected on each participant exceeds the number of participants (525 lipid species together with clinic and demographic variables vs. 149 participants), which further requires a careful variable selection procedure as we have applied here. Our contribution in this study is in: 1) providing an RF-based ensemble for PD severity prediction and statistically supported lipidome feature selection that is both accurate and informative; and 2) identifying severity-related lipid class and species signatures that can be used as age-gender-specific PD severity markers.
Our analysis showed several influential lipid classes, and, within each class, several lipid species that contribute to high PD severity prediction accuracy, especially in intermediate severities. This analysis also demonstrated interesting interrelations between lipid classes/species and different UPDRS score ranges representing different stages of the disease. We showed that different lipid classes/species are expressed in different disease stages. Although this result needs further validation, it may open new avenues of research in investigating the roles of the lipids in the progression of PD. A key finding is that plasma levels of four specific lipid classes may predict UPDRS performance of people with PD. Higher levels of GlcCer and dhGB3 are associated with worse performance, and higher levels of dhSM and PEp are associated with better (lower scores) UPDRS performance. Our models did not identify key associations with cognitive performance as measured by the MoCA.
Glucosylceramide is a main substrate of glucocerebrosidase (GCase), which is encoded by GBA. Carriers of GBA mutations with PD have faster motor and cognitive progression [41]. Studies have shown faster alpha synuclein aggregation in the presence of glucosylceramide [42]. Of note, our study participants were non-carriers of GBA mutations (with the exception of one), highlighting the potential role of glucosylceramide levels even among non-carriers. The association between higher glucosylceramide levels and more severe PD phenotype has been previously reported in a cohort of 52 PD cases (26 with normal cognition and 28 with impaired cognition) [43]. However, a similar association was not reported with motor functioning [43]. In summary, our findings of higher glucosylceramide levels predicting more severe motor phenotype are consistent with the literature and support the consideration of this lipid class as a drug target.
GB3 is a globoside that contains glucosylceramide as its base cerebroside. This glycosphingolipid and its analogs [44] are the primary lipids accumulated in Fabry disease, a lysosomal storage disease. In Fabry disease, the accumulation of GB3 and its analogs is due to the deficiency of the enzyme α-galactosidase A [45]. We have previously shown that reduced α-galactosidase A is associated with PD status,[46] and that there may be a higher incidence of PD among Fabry disease patients [47]. The cause of the elevations in these specific dehydrosphingolipids is unknown. However, we note that increases in this class of lipids could be the result of reduced activity of Ceramide synthases 5/6 (DEGS) due to hypoxic stress as the lipid desaturases are known to be modulated by oxygen concentrations.
dhSM is one of the few phospholipids not synthetized from glycerol. In our data, dhSM levels are higher among those with lower UPDRS scores. This finding correlates with observations in Alzheimer’s disease (AD) where it is believed that high levels of SM and dhSM are protective, and lower levels of SM and dhSM are observed in AD patients [48]. The mechanism behind lower dhSM levels and AD or PD progression is unknown. Reduced levels may be the result of an increased rate of ganglioside biosynthesis and a reduction in the pool dihydro ceramides for the synthesis of dhSM. dhCer is the precursor of dhSM (as Cer is for SM). All four sphingolipids are enriched in the brain and are major components of neuronal membranes. Alterations of these sphingolipids have been reported in AD and PD [48–54]. In particular, decrease of SM has been observed in samples from PD patients [55–57].
PE and PEp are also phospholipids. PEp is a plasmalogen phospholipid almost identical to PE, other than the vinyl-ethyl bond in place of the ether bond. The presence of vinyl-ether bonds confers plasmalogens with specific biophysical properties that differ from non-plasmalogen phospholipids [59] such as an increase in the saturation of cellular membranes.
Plasmalogens are highly abundant in the nervous system. The physiological relevance of alterations in plasmalogen synthesis is underscored by disorders such as Zellweger syndrome or chondrodysplasia [60]. While the link between elevation in PEp and better performance on motor examination is unknown, we note that elevations in the synthesis and levels of plasmalogens have been associated with reductions in the biosynthesis of non-plasmalogen glycerophospholipids. Two independent studies in different models linked lower PE levels with PD, supporting this hypothesis. In the first study, cellular and animal models of familial PD that carried pathogenic mutations in SNCA showed significant reduction in PE synthesis [61]. Likewise, Chan et al., 2017 showed reduced levels of PE in plasma samples from male idiopathic Parkinson’s disease (iPD) patients compared to controls [4]. Additionally, Riekkinen et al., 1975 found significant reductions in the levels of PE in the substantia nigra [62]. The relatively high abundance of PEp makes this phospholipid an ideal biomarker candidate since it is easy to detect by regular and consistent lipidomics analysis.
Strengths and limitations
One strength of our study is the novel machine-learning technologies applied to the data. These offer a solid prediction framework around the RF by: 1) designing a careful, statistically-supported training-validation-test ML methodology; 2) initially applying the classifier to select lipid classes (and demographics) that contributed to the smallest prediction error; 3) repeating this application with only highly ranked classes (and demographics) to select the most contributing lipid species; 4) validating and reinforcing this selection using statistical methods; and 5) profiling gender-age lipid signatures in relation to PD severity, establishing lipid-based markers of disease progression.
A second strength of the study is the extensive unbiased lipidomic analysis we applied using our ML methodology to validate our findings. The cohort is carefully phenotyped, and SNCA, GBA (with the exception of one), and LRRK2 mutation carriers were not included. The main limitation of the study is that the participants did not fast at time of the blood extraction. Fasting status could affect the levels of blood sphingolipids [63]. Therefore, ideally, our findings should be confirmed in larger studies, ideally including fasting participants with longitudinal data points, like in the Parkinson’s Progression Markers Initiative (PPMI) study.
In conclusion, we demonstrated lipid signatures for motor disease severity in PD. The links between PD status, disease severity, and lipidomics require further investigation, but we believe ML adds an extra tool that improves the level of interpretation for large and interconnected lipidomics data.
Supplementary Material
Acknowledgement including sources of support
Hila Avisar was supported by the Ben-Gurion University High-tech, Bio-tech, Chem-tech–STEM Fellowship. The lipidomic analysis was funded by the Parkinson’s Foundation and the NIH (K0NS080915).
Financial disclosure: The lipidomic analysis was funded by the Parkinson’s Foundation and the NIH (K02NS080915). RNA is funded by the NIH, DoD, the Parkinson’s Foundation, and the Michael. J. Fox Foundation. He received consultation fees from Sanofi and Janssen. HA, CGL, EAG, MS, AKC, and BL have nothing to disclose.
Footnotes
Conflict of interest
The authors have no conflict of interest to report.
References
- [1].Ryckman AE, Brockhausen I, Walia JS (2020) Metabolism of Glycosphingolipids and Their Role in the Pathophysiology of Lysosomal Storage Disorders. Int J Mol Sci 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Murphy KE, Halliday GM (2014) Glucocerebrosidase deficits in sporadic Parkinson disease. Autophagy 10, 1350–1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Kim MJ, Jeon S, Burbulla LF, Krainc D (2018) Acid ceramidase inhibition ameliorates alpha-synuclein accumulation upon loss of GBA1 function. Hum Mol Genet 27, 1972–1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Chan RB, Perotte AJ, Zhou B, Liong C, Shorr EJ, Marder KS, Kang UJ, Waters CH, Levy OA, Xu Y, Shim HB, Pe’er I, Di Paolo G, Alcalay RN (2017) Elevated GM3 plasma concentration in idiopathic Parkinson’s disease: A lipidomic analysis. PLoS One 12, e0172348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Di Pasquale E, Fantini J, Chahinian H, Maresca M, Taieb N, Yahi N (2010) Altered ion channel formation by the Parkinson’s-disease-linked E46K mutant of alpha-synuclein is corrected by GM3 but not by GM1 gangliosides. J Mol Biol 397, 202–218. [DOI] [PubMed] [Google Scholar]
- [6].Grey M, Dunning CJ, Gaspar R, Grey C, Brundin P, Sparr E, Linse S (2015) Acceleration of alpha-synuclein aggregation by exosomes. J Biol Chem 290, 2969–2982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Armananzas R, Bielza C, Chaudhuri KR, Martinez-Martin P, Larranaga P (2013) Unveiling relevant non-motor Parkinson’s disease severity symptoms using a machine learning approach. Artif Intell Med 58, 195–202. [DOI] [PubMed] [Google Scholar]
- [8].Chen H-L, Huang C-C, Yu X-G, Xu X, Sun X, Wang G, Wang S-J (2013) An efficient diagnosis system for detection of Parkinson’s disease using fuzzy k-nearest neighbor approach. Expert Systems with Applications 40, 263–271. [Google Scholar]
- [9].Das R (2010) A comparison of multiple classification methods for diagnosis of Parkinson disease. Expert Systems with Applications 37, 1568–1572. [Google Scholar]
- [10].Ma C, Ouyang J, Chen HL, Zhao XH (2014) An efficient diagnosis system for Parkinson’s disease using kernel-based extreme learning machine with subtractive clustering features weighting approach. Comput Math Methods Med 2014, 985789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Ozcift A, Gulten A (2011) Classifier ensemble construction with rotation forest to improve medical diagnosis performance of machine learning algorithms. Computer Methods and Programs in Biomedicine 104, 443–451. [DOI] [PubMed] [Google Scholar]
- [12].Prashanth R, Roy SD, Mandal PK, Ghosh S (2014) Parkinson’s disease detection using olfactory loss and REM sleep disorder features. Conf Proc IEEE Eng Med Biol Soc 2014, 5764–5767. [DOI] [PubMed] [Google Scholar]
- [13].Ramani (2011) Parkinson disease classification using data mining algorithms. International journal of computer applications 32. [Google Scholar]
- [14].Shahbakhi M (2014) Speech Analysis for Diagnosis of Parkinson’s Disease Using Genetic Algorithm and Support Vector Machine. Journal of biomedical science and engineering 07, 147–156. [Google Scholar]
- [15].Sriram T, Rao M, Narayana G, Vital T, Dowluru KS (2013) Intelligent Parkinson Disease Prediction Using Machine Learning Algorithms. IJEIT 3, 212–215. [Google Scholar]
- [16].Tsanas A, Little MA, McSharry PE, Ramig LO (2011) Nonlinear speech analysis algorithms mapped to a standard metric achieve clinically useful quantification of average Parkinson’s disease symptom severity. J R Soc Interface 8, 842–855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Tsanas A, Little MA, McSharry PE, Spielman J, Ramig LO (2012) Novel speech signal processing algorithms for high-accuracy classification of Parkinson’s disease. IEEE Trans Biomed Eng 59, 1264–1271. [DOI] [PubMed] [Google Scholar]
- [18].Bind STA, Sahani AK (2015) A Survey of Machine Learning Based Approaches for Parkinson Disease Prediction International Journal of Computer Science and Information Technologies 6, 1648–1655. [Google Scholar]
- [19].Gil DJM (2009) Diagnosing Parkinson by using artificial neural networks and support vector machines. Global Journal of Computer Science and Technology 9, 63–71. [Google Scholar]
- [20].Tahir NMH (2012) Parkinson Disease Gait Classification based on Machine Learning Approach. Journal of Applied Sciences 12, 180–185. [Google Scholar]
- [21].Gordon J, Lerner B (2019) Insights into Amyotrophic Lateral Sclerosis from a Machine Learning Perspective. J Clin Med 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Sakanaka K, Waters CH, Levy OA, Louis ED, Chung WK, Marder KS, Alcalay RN (2014) Knowledge of and interest in genetic results among Parkinson disease patients and caregivers. J Genet Couns 23, 114–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Swanson CR, Berlyand Y, Xie SX, Alcalay RN, Chahine LM, Chen-Plotkin AS (2015) Plasma apolipoprotein A1 associates with age at onset and motor severity in early Parkinson’s disease patients. Mov Disord 30, 1648–1656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Nasreddine ZS, Phillips NA, Bedirian V, Charbonneau S, Whitehead V, Collin I, Cummings JL, Chertkow H (2005) The Montreal Cognitive Assessment, MoCA: a brief screening tool for mild cognitive impairment. J Am Geriatr Soc 53, 695–699. [DOI] [PubMed] [Google Scholar]
- [25].Ferri C, Hernández-Orallo J, Modroiu R (2009) An experimental comparison of performance measures for classification. Pattern Recognition Letters 30, 27–38. [Google Scholar]
- [26].Hever G, Cohen L, O’Connor MF, Matot I, Lerner B, Bitan Y (2020) Machine learning applied to multi-sensor information to reduce false alarm rate in the ICU. J Clin Monit Comput 34, 339–352. [DOI] [PubMed] [Google Scholar]
- [27].Sarica A, Cerasa A, Quattrone A (2017) Random Forest Algorithm for the Classification of Neuroimaging Data in Alzheimer’s Disease: A Systematic Review. Front Aging Neurosci 9, 329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Kashi S, Feingold-Polak R, Lerner B, Rokach L, Levy-Tzedek S (2020) A machine-learning model for automatic detection of movement compensations in stroke patients. IEEE Transactions on Emerging Topics in Computing, 1–1. [Google Scholar]
- [29].Roguet A, Eren AM, Newton RJ, McLellan SL (2018) Fecal source identification using random forest. Microbiome 6, 185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Svetnik V, Liaw A, Tong C, Culberson JC, Sheridan RP, Feuston BP (2003) Random forest: a classification and regression tool for compound classification and QSAR modeling. J Chem Inf Comput Sci 43, 1947–1958. [DOI] [PubMed] [Google Scholar]
- [31].Breiman L (2001) Random Forests. Machine Learning 45, 5–32. [Google Scholar]
- [32].Liaw A, Wiener M (2001) Classification and Regression by RandomForest. Forest 23. [Google Scholar]
- [33].Breiman L (1996) Bagging Predictors. Machine Learning 24, 123–140. [Google Scholar]
- [34].Bauer E, Kohavi R (1999) An Empirical Comparison of Voting Classification Algorithms: Bagging, Boosting, and Variants. Machine Learning 36, 105–139. [Google Scholar]
- [35].Jensen DD, Cohen PR (2000) Multiple Comparisons in Induction Algorithms. Machine Learning 38, 309–338. [Google Scholar]
- [36].Calle ML, Urrea V (2011) Letter to the editor: Stability of Random Forest importance measures. Brief Bioinform 12, 86–89. [DOI] [PubMed] [Google Scholar]
- [37].Genuer R, Poggi J-M, Tuleau-Malot C (2010) Variable selection using random forests. Pattern Recognition Letters 31, 2225–2236. [Google Scholar]
- [38].Strobl C, Boulesteix AL, Kneib T, Augustin T, Zeileis A (2008) Conditional variable importance for random forests. BMC Bioinformatics 9, 307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Burnham KP Multimodel Inference Understanding AIC and BIC in Model Selection. Sociological methods & research 33, 261–304. [Google Scholar]
- [40].Tibshirani R (1996) Regression Shrinkage and Selection Via the Lasso. Journal of the Royal Statistical Society: Series B (Methodological) 58, 267–288. [Google Scholar]
- [41].Gan-Or Z, Liong C, Alcalay RN (2018) GBA-Associated Parkinson’s Disease and Other Synucleinopathies. Curr Neurol Neurosci Rep 18, 44. [DOI] [PubMed] [Google Scholar]
- [42].Mazzulli JR, Xu YH, Sun Y, Knight AL, McLean PJ, Caldwell GA, Sidransky E, Grabowski GA, Krainc D (2011) Gaucher disease glucocerebrosidase and alpha-synuclein form a bidirectional pathogenic loop in synucleinopathies. Cell 146, 37–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Mielke MM, Maetzler W, Haughey NJ, Bandaru VV, Savica R, Deuschle C, Gasser T, Hauser AK, Graber-Sultan S, Schleicher E, Berg D, Liepelt-Scarfone I (2013) Plasma ceramide and glucosylceramide metabolism is altered in sporadic Parkinson’s disease and associated with cognitive impairment: a pilot study. PLoS One 8, e73094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Manwaring V, Boutin M, Auray-Blais C (2013) A metabolomic study to identify new globotriaosylceramide-related biomarkers in the plasma of Fabry disease patients. Anal Chem 85, 9039–9048. [DOI] [PubMed] [Google Scholar]
- [45].Abe A, Gregory S, Lee L, Killen PD, Brady RO, Kulkarni A, Shayman JA (2000) Reduction of globotriaosylceramide in Fabry disease mice by substrate deprivation. J Clin Invest 105, 1563–1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Alcalay RN, Wolf P, Levy OA, Kang UJ, Waters C, Fahn S, Ford B, Kuo SH, Vanegas N, Shah H, Liong C, Narayan S, Pauciulo MW, Nichols WC, Gan-Or Z, Rouleau GA, Chung WK, Oliva P, Keutzer J, Marder K, Zhang XK (2018) Alpha galactosidase A activity in Parkinson’s disease. Neurobiol Dis 112, 85–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Wise AH, Yang A, Naik H, Stauffer C, Zeid N, Liong C, Balwani M, Desnick RJ, Alcalay RN (2018) Parkinson’s disease prevalence in Fabry disease: A survey study. Mol Genet Metab Rep 14, 27–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Mielke MM, Haughey NJ, Bandaru VV, Weinberg DD, Darby E, Zaidi N, Pavlik V, Doody RS, Lyketsos CG (2011) Plasma sphingomyelins are associated with cognitive progression in Alzheimer’s disease. J Alzheimers Dis 27, 259–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Cutler RG, Kelly J, Storie K, Pedersen WA, Tammara A, Hatanpaa K, Troncoso JC, Mattson MP (2004) Involvement of oxidative stress-induced abnormalities in ceramide and cholesterol metabolism in brain aging and Alzheimer’s disease. Proc Natl Acad Sci U S A 101, 2070–2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Grimm MO, Grimm HS, Patzold AJ, Zinser EG, Halonen R, Duering M, Tschape JA, De Strooper B, Muller U, Shen J, Hartmann T (2005) Regulation of cholesterol and sphingomyelin metabolism by amyloid-beta and presenilin. Nat Cell Biol 7, 1118–1123. [DOI] [PubMed] [Google Scholar]
- [51].Jana A, Pahan K (2004) Fibrillar amyloid-beta peptides kill human primary neurons via NADPH oxidase-mediated activation of neutral sphingomyelinase. Implications for Alzheimer’s disease. J Biol Chem 279, 51451–51459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Kalvodova L, Kahya N, Schwille P, Ehehalt R, Verkade P, Drechsel D, Simons K (2005) Lipids as modulators of proteolytic activity of BACE: involvement of cholesterol, glycosphingolipids, and anionic phospholipids in vitro. J Biol Chem 280, 36815–36823. [DOI] [PubMed] [Google Scholar]
- [53].Mattson MP, Cutler RG, Jo DG (2005) Alzheimer peptides perturb lipid-regulating enzymes. Nat Cell Biol 7, 1045–1047. [DOI] [PubMed] [Google Scholar]
- [54].Puglielli L, Ellis BC, Saunders AJ, Kovacs DM (2003) Ceramide stabilizes beta-site amyloid precursor protein-cleaving enzyme 1 and promotes amyloid beta-peptide biogenesis. J Biol Chem 278, 19777–19783. [DOI] [PubMed] [Google Scholar]
- [55].Bras J, Singleton A, Cookson MR, Hardy J (2008) Emerging pathways in genetic Parkinson’s disease: Potential role of ceramide metabolism in Lewy body disease. FEBS J 275, 5767–5773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Fabelo N, Martin V, Santpere G, Marin R, Torrent L, Ferrer I, Diaz M (2011) Severe alterations in lipid composition of frontal cortex lipid rafts from Parkinson’s disease and incidental Parkinson’s disease. Mol Med 17, 1107–1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Haughey NJ (2010) Sphingolipids in neurodegeneration. Neuromolecular Med 12, 301–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Yasuda T, Al Sazzad MA, Jantti NZ, Pentikainen OT, Slotte JP (2016) The Influence of Hydrogen Bonding on Sphingomyelin/Colipid Interactions in Bilayer Membranes. Biophys J 110, 431–440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Braverman NE, Moser AB (2012) Functions of plasmalogen lipids in health and disease. Biochim Biophys Acta 1822, 1442–1452. [DOI] [PubMed] [Google Scholar]
- [60].Mani SK, Decker GL, Glasser SR (1991) Hormonal responsiveness by immature rabbit uterine epithelial cells polarized in vitro. Endocrinology 128, 1563–1573. [DOI] [PubMed] [Google Scholar]
- [61].Guardia-Laguarta C, Area-Gomez E, Rub C, Liu Y, Magrane J, Becker D, Voos W, Schon EA, Przedborski S (2014) alpha-Synuclein is localized to mitochondria-associated ER membranes. J Neurosci 34, 249–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Riekkinen P, Rinne UK, Pelliniemi TT, Sonninen V (1975) Interaction between dopamine and phospholipids. Studies of the substantia nigra in Parkinson disease patients. Arch Neurol 32, 25–27. [DOI] [PubMed] [Google Scholar]
- [63].Hammad SM, Pierce JS, Soodavar F, Smith KJ, Al Gadban MM, Rembiesa B, Klein RL, Hannun YA, Bielawski J, Bielawska A (2010) Blood sphingolipidomics in healthy humans: impact of sample collection methodology. J Lipid Res 51, 3074–3087. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.