Abstract
Reactive oxygen species (ROS) are essential for life and involved in the regulation of almost all biological processes. ROS production is critical for plant development, response to abiotic stresses, and immune responses. ROS can also cause direct oxidative damage and organisms must therefore balance ROS production and quenching. Here, we focus on recent discoveries in ROS biology emphasizing abiotic and biotic stress responses. Recent advancements have resulted in the identification of one of the first sensors for extracellular ROS and highlighted waves of ROS production during stress signaling in Arabidopsis. Enzymes that produce ROS, including NADPH oxidases, exhibit precise regulation through diverse posttranslational modifications. Discoveries highlight the importance of both N- and C-terminal regulation of NADPH oxidases through protein phosphorylation and cysteine oxidation. Here, we discuss advancements in ROS compartmentalization, systemic ROS waves, ROS sensing, and posttranslational modification of ROS producing enzymes as well as identify areas where foundational gaps remain.
Introduction
Generation of reactive oxygen species (ROS) is a necessary process in all living organisms1. Production of ROS including singlet oxygen (1O2), superoxide (O2●−), hydroxyl radicals (OH●), hydrogen peroxide (H2O2), as well as reactive nitrogen species such as nitric oxide (NO) are required for basic biological processes and stress responses2. However, ROS and NO can also cause direct oxidative damage to DNA, lipids, and proteins. All living organisms must therefore balance ROS production and ROS quenching. Like other eukaryotes, plants utilize ROS as signaling molecules in multiple biological processes including abiotic and biotic stress responses3,4,5.
Multiple environmental factors induce abiotic stress in plants, including extreme temperatures, drought, salinity, osmotic stress, excess light, ozone, and heavy metals 6, 7. ROS contribute to stress acclimation, metabolic changes, and induction of programmed cell death 8. Biotic signals also lead to enhanced ROS production. However, ROS are not only important during the response to external cues but basal production is critical for cell expansion and localized production is important for numerous events including pollen tube elongation, root hair growth, cell polarity and Casparian strip formation9, 10,11,12,13. ROS production in response to pathogen infection and abiotic stimuli, including salt stress or wounding, has been linked to local and systemic signal transduction and is integrated with calcium signaling and with the deposition of callose at plasmodesmata14. Importantly, both abiotic and biotic stresses lead to ROS production in several different subcellular compartments and these events must be tightly coordinated and regulated 2,15,16,17.
Although ROS production is critical for growth, signaling, and development, their reactivity are a double-edged sword. To regulate ROS toxicity, plants compartmentalize ROS production, utilize ROS scavengers, and tightly regulate ROS production temporally and spatially. Previous reviews have focused on ROS production during metabolism, biotic, and abiotic stress6,18,19. Here, we focus on recent discoveries in ROS biology including compartmentalization, systemic ROS propagation, and sensing of extracellular ROS in plants. We also highlight recent insight into the regulation of NADPH oxidase (NOX) activity through diverse post-translational modifications.
Compartmentalized ROS production
Specificity of oxidative signaling is partially achieved by ROS production in different subcellular compartments, including the apoplast and intracellular organelles (Fig. 1)20. ROS compartmentalization can also reduce the deleterious nature of highly reactive species. Intracellular ROS accumulation is in large part due to metabolic byproducts produced by chloroplasts and mitochondria17. Plastid produced ROS are also important signaling messengers within cells. Eukaryotic cells have evolved a complex network of signals that allow plastids to regulate gene expression in the nucleus, a process known as retrograde signaling. In many cases, ROS reactivity and permeability allows these molecules to relay signaling to the nucleus21. H2O2 originating from chloroplasts and mitochondria operates as a retrograde signal by interacting with transcription factors (TFs) affecting nuclear gene expression. The Arabidopsis NAC DOMAIN-CONTAINING PROTEIN17 (ANAC017) TF is localized to the ER and released upon increased levels of H2O2, where it alters nuclear gene expression22. RADIDAL-INDUCED CELL DEATH1 (RCD1) also interacts with ANAC017 and is proposed to inhibit ANAC017 activity, potentially in a ROS dependent manner23. The chloroplast can also act as an environmental sensor, regulating cellular communication and gene expression. Most chloroplast ROS-dependent retrograde signaling is associated with singlet oxygen, which is primarily generated by photosystem II as a photosynthesis by-product and functions near its site of generation24. Singlet oxygen can trigger chloroplast-to-nucleus retrograde signaling, resulting in alteration of nuclear gene expression, stress response and programmed cell death25.
Figure 1. Biphasic ROS production and cell-to-cell communication.
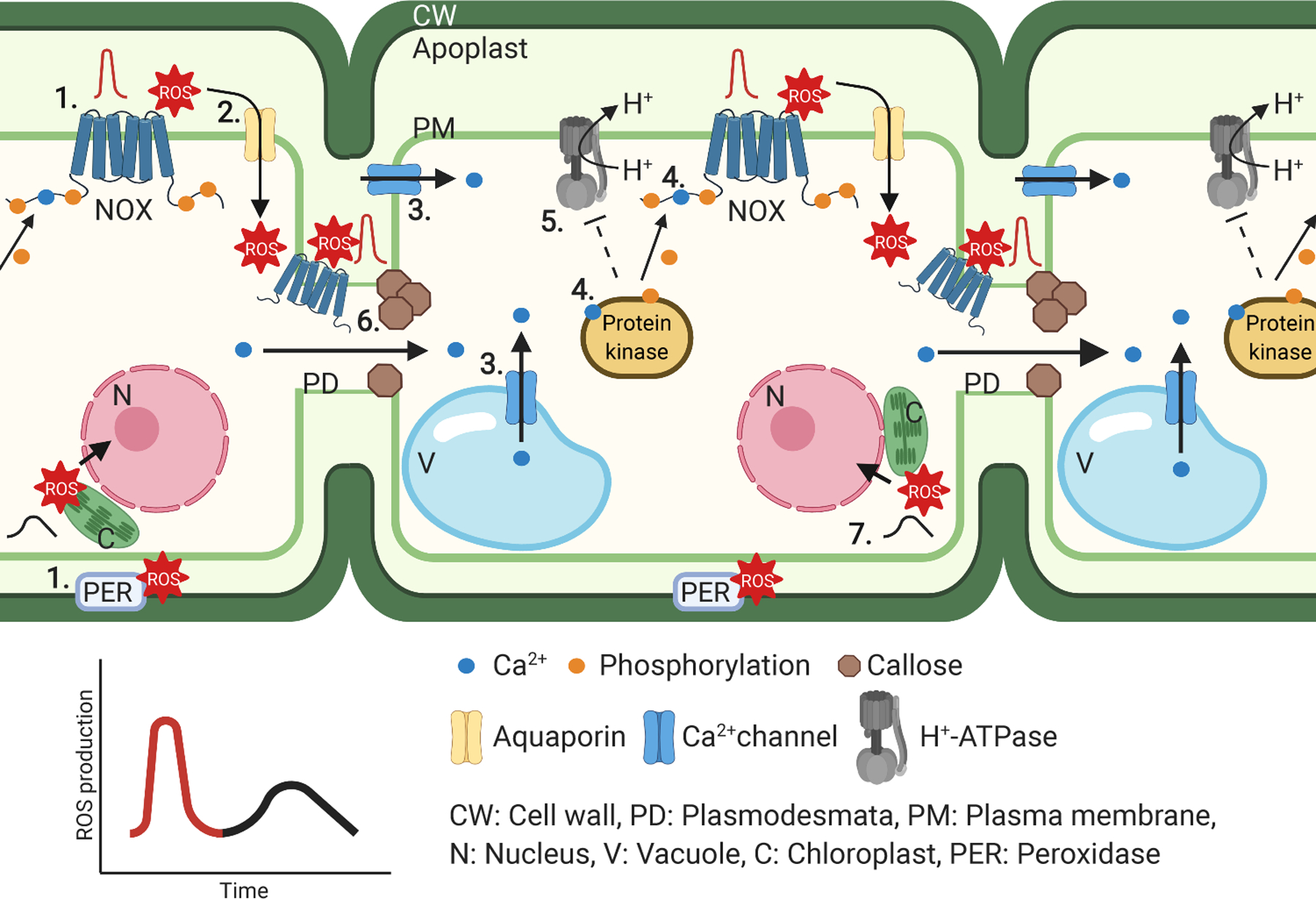
1. Stimuli induce rapid initial apoplastic ROS production by NADPH oxidases (NOXs) and peroxidases (PER). 2. Apoplastic ROS can enter into the cytosol via aquaporins. 3. ROS then activates Ca2+ influx into the cytosol from the apoplast and vacuole. 4. Ca2+ binds to NOXs or protein kinases, which amplifies NOX activation. 5. Apoplastic ROS accumulation frequently coincides with extracellular alkalization by inhibition of H+-ATPases. 6. NOX-dependent ROS can restrict symplastic signaling by modulating callose deposition at plasmodesma. 7. After initial rapid systemic signaling, a second ROS burst with contributions from the chloroplast may affect nuclear gene expression.
Apoplastic ROS production is primarily produced by cell wall peroxidases and plasma membrane-localized flavin-containing NAPDH oxidases (NOXs), referred to as RESPIRATORY BURST OXIDASE HOMOLOGS (RBOHs) in plants6,26. Plants possess 2–10 NOX isoforms, which produce superoxide. Extracellular ROS can modify cell wall components, extracellular proteins, and plasma membrane proteins18. Plant NOXs play distinct roles in developmental processes such as pollen tube growth, root hair formation, leaf cell expansion and Casparian strip formation as well as defense responses10,11,18,27,28,29. During pathogen invasion, plants use pattern recognition receptors (PRRs) that recognize conserved molecular features, so-called microbe-associated molecular patterns (MAMPs), resulting in pattern-triggered immunity (PTI); plant immune receptors can also recognize pathogen effectors delivered inside plant cells resulting in effector-triggered immunity (ETI)30. PRR activation leads to apoplastic ROS production through NOX and peroxidase activity18,26,31. RBOHD is the highest expressed NOX in Arabidopsis and its ability to produce ROS has been linked to callose deposition, cell wall lignification, stomatal closure, and systemic acquired resistance (SAR)17, 18, 32. Homologs of RBOHD in monocots and dicots play similar roles33,34.
Of the many produced species, H2O2 is considered to be the key signaling molecule, due to its relatively long half-life, its ability to oxidize proteins and move across the plasma membrane via aquaporins35, 36, and its similar molecular properties to H2O (Fig. 1). The Arabidopsis PLASMA MEMBRANE INTRINSIC PROTEIN (PIP) 1;4 and PIP2;1 aquaporins are implicated in the influx of apoplastic H2O2 to the cytosol during plant immune signaling and ABA-dependent stomatal closure, respectively37,35. Interestingly, not all aquaporins are permeable for H2O2, but the biochemical basis for this selectivity remains elusive36,38. Once inside the cell, ROS produced in the extracellular space can react with intracellular proteins or can be detoxified via intracellular scavenging systems. Notably, in animal erythrocytes, superoxide can pass the cellular membrane through anion exchange 1/solute carrier 4A1 (SLC4A1)39. Plant homologs of SLC4A11 mediate borate transport40, but so far there is no evidence for superoxide transport by anion channels in plants. Rather, NOX activation leads to production of superoxide which is thought to either act in the extracellular space or to be dismutated to H2O2, which can enter the cytosol through aquaporins 17, 18, 35, 36.
Not only is ROS compartmentalized by subcellular localization, but there are accumulation hotspots within a compartment, akin to previous reports of hotspots of Ca2+ ions41,42,43,44. Localized ROS hotspots likely provide signaling specificity within the cytosol and chloroplast. However, the ability to visualize and measure distinct ROS with high spatial, temporal, and quantitative resolution remains a major challenge in biological research. Current methods for measuring ROS production include the use of histochemical stains, chemiluminescent and fluorescent probes, electrochemical detection, and the detection of protein oxidation via mass spectrometry 45, 46, 47, 48, 49. Currently, no single technique combines all the desired aspects of specificity, sensitivity and quantification. Future development of new and innovative detection platforms, including direct detection, will provide a deeper understanding of ROS production and compartmentalization.
ROS Waves: Mediators of plant systemic signaling
Over the past decade, multiple studies have demonstrated the presence of rapid long distance auto-propagating ROS signals, known as the ROS wave (Fig. 1)50. The ROS wave can be induced by biotic and abiotic stimuli and is followed by gene expression changes in distal tissue. The function of the ROS wave has been viewed as (1) an essential signal that alerts cells and tissues to an impending stress, (2) a link between abiotic and biotic signals, and (3) a coordinator of the whole-plant systemic stomatal response to different stresses50.
The ROS wave is dependent on NOX activity and is integrated with systemic Ca2+ and pH signaling51. Apoplastic H2O2 activates the ROS sensor HYDROGEN-PEROXIDE-INDUCED CA2+ INCREASES 1 (HPCA1) and induces Ca2+ influx into cytosol52. Ca2+-binding to CALCIUM-DEPENDENT PROTEIN KINASES (CPKs) and NOXs leads to amplification of apoplastic ROS production which may result in the propagation of a systemic signal. Due to the high cytosolic ROS scavenging capacity it is unlikely that intracellular ROS contribute directly to symplastic cell-to-cell communication via plasmodesmata. Nevertheless, NOXs can localize to plasmodesmata and apoplastic ROS production participate in the regulation of symplastic signaling by inducing callose deposition at plasmodesmata, which results in a restriction of the plasmodesmal aperture, thereby limiting cell-to-cell communication53 (Fig. 1).
A contribution of the ROS wave in systemic acquired acclimation (SAA) has been previously suggested54 and recent work proposes that vascular bundles play a major role in transmitting systemic ROS signals55. ROS production is also required for systemic acquired resistance (SAR), but the relationship between the ROS wave and SAR is not obvious. An avirulent pathogen causing local programmed cell death (PCD) can induce SAR through generation of the mobile metabolite N-hydroxypipecolic acid (NHP) and subsequent accumulation of the phytohormone salicylic acid (SA) in distal tissues56,57. SA regulates SAR via the redox-sensitive transcriptional co-regulator NONEXPRESSER OF PR GENES 1 (NPR1; see ROS perception section). Activation of SAR requires the aquaporin AtPIP1;4 for the cytoplasmic import of apoplastic H2O235. Local inoculation with the avirulent bacterial pathogen Pseudomonas syringae pv. tomato avrRpt2 triggered ROS accumulation in distal Arabidopsis leaves, which was reduced in rbohD and rbohF single mutants8, 58. In rbohD and rbohF single mutants, the levels of the NHP precursor pipecolic acid in the distal tissue are significantly lower than in wild type plants32. These results suggest a function of ROS as an inducer or mediator of SAR. While the ROS wave is a very early response and starts within minutes of the perception of a local stimulus59, the production of mobile NHP occurs hours after pathogen infection in the primary leaves, which may represent the time required for effector delivery.
Two peaks of ROS production have been described during PTI60 (Fig. 1). The first is a rapid ROS burst largely mediated by NOXs60 that participates in the ROS wave (Fig. 1). Recently it has been reported that ETI enhances the second PTI-induced ROS burst, which begins several hours after pathogen perception and is suppressed by diphenyleneiodonium chloride (DPI), an inhibitor of flavin-containing proteins60. Oxidation of apoplastic proteins is also temporarily enhanced during ETI61. Moreover, ETI induces an accumulation of components associated with PTI such as receptor like cytoplasmic kinases (RLCKs), CPKs, Ca2+-permeable channels and NOXs, which are also involved in the propagation of the ROS wave62. These data suggest that NOX activity is required for the second ROS burst which could propagate systemically as a “second ROS wave”. Research on the ROS wave has so far mostly focused on rapid signal propagation (maximum 30 min). Hence, it would be highly interesting to investigate whether avirulent pathogens induce not only a biphasic ROS burst but also a biphasic ROS wave in the future. In addition to NOXs, a strong chloroplastic contribution to the second ROS burst has been described in response to microbial features and ozone. Chloroplastic ROS also play a role in ETI-induced cell death in response to pathogen attack63,64,65. For example, the chloroplast-localized redox-sensitive THIMET OLIGOPEPTIDASE 1 (TOP1) binds to SA and regulates ETI-induced PCD66,61, 67. Thus, redox-sensitive TOP might play dual roles in induction of cell death and SA-dependent SAR. These data demonstrate tight integration of systemic signaling during ETI over multiple time scales. Compared to the biphasic ROS burst in response to biotic stimuli, the presence of distinct ROS peaks in response to abiotic stress and wounding is less clear. However, in response to stress, apoplastic ROS accumulation also lead to changes in cytosolic Ca2+ concentration ([Ca2+]cyto) and amplifies stress responses. Thus, multiple mechanisms collectively contribute to the initial, systemic, and sustained ROS production in response to biotic and abiotic stimuli.
ROS perception
For a signaling molecule to function, it must be sensed. Unlike peptides or other non-reactive molecules, it is unlikely that ROS are perceived in a receptor-ligand manner. Instead, ROS are likely perceived through redox modifications, resulting in altered protein structure, localization, activity, and protein-protein interactions. While apoplastic ROS could be perceived in the extracellular space, its diffusion across the plasma membrane through aquaporin and anion channels38 can also facilitate perception in the cytosol (Figs. 1 and 2). Intriguingly, these extra- and intracellular ROS sensing mechanisms are not mutually exclusive and could enhance sensitivity or the temporal resolution of ROS as a signaling molecule 17, 68. A main mechanism for ROS sensing is the oxidative modification of cysteines, which has been extensively investigated and reviewed69, 70. The availability of different oxidation states results in a diverse range of cysteine oxidative posttranslational modifications (PTMs). In addition to cysteine, oxidation of methionine and tyrosine can affect protein structure and function71, 72.
Figure 2. ROS perception by the HPCA1 receptor.
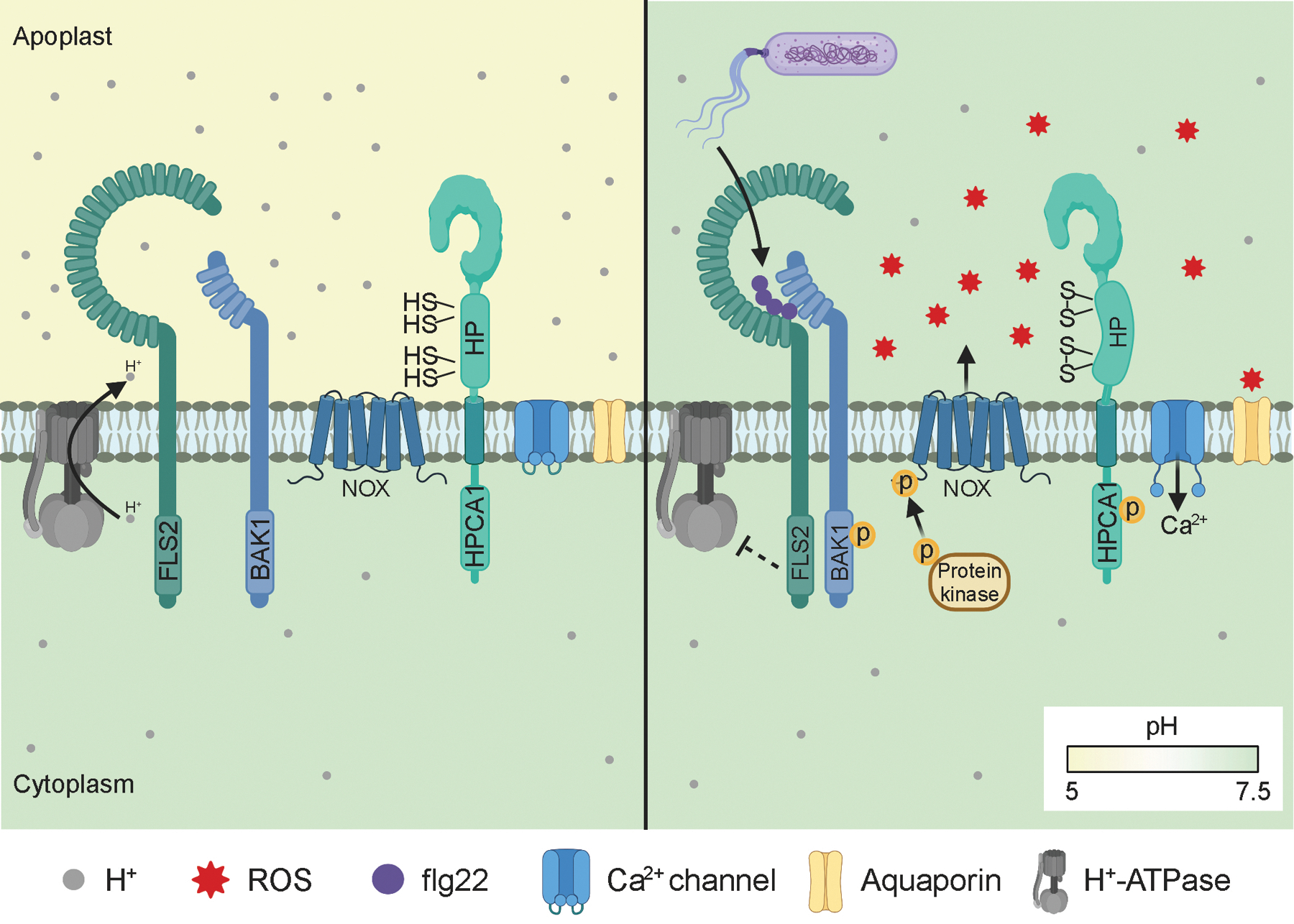
Left: The flagellin receptor complex and ROS receptor HPCA1 in the absence of pathogen perception. Right: Perception of the immunogenic flagellin epitope flg22 results in FLS2-BAK1 complex formation, inhibition of the H+-ATPase resulting in alkalization of the apoplast, and activation of NOX-induced ROS production. The increase in extracellular pH enables cysteine modification of HPCA1’s hydrogen peroxidase (HP) domain, increasing HPCA1’s kinase activity and downstream ROS signalling.
Within ROS-sensitive proteins, the reactivity of cysteines with ROS is strongly correlated with the local electrostatic environment that affects the pKa of cysteine residues. Cysteines are prone to oxidation at pH levels higher than their pKa. The resting pH of the cytosol and apoplast is considered to be approximately 7.5 and 5, respectively73. This suggests that cysteines in the apoplast could be less readily oxidized compared to those in the cytosol. However, biotic and abiotic stimuli trigger a transient alkalization of the apoplast to pH 6 −774, increasing the reactivity of cysteines with ROS. Inhibition of plasma membrane (PM)-H+-ATPase activity, by phosphorylation and by accumulation of cations in the apoplast, is responsible for apoplastic alkalization74 (Fig. 2). Interestingly, the duration and magnitude of apoplastic alkalization differs between salinity, drought, and attack by powdery mildew74. Thus, stimulus-dependent apoplastic alkalization may be an important component of ROS signaling specificity. More localized modulation of pH is also possible, where distinct pH microenvironments impact oxidation of cysteine thiols75.
The first identified ROS sensors are the prokaryotic OxyR and SoxR transcription factors that are directly oxidized by ROS76, 77. The functional homolog of OxyR in yeast, Yap, is not directly oxidized by ROS but is targeted by the thiol-peroxidase Oxidant receptor peroxidase (Orp1). Oxidized Orp1 oxidizes Yap1, potentially masking its nuclear export signal, resulting in nuclear localization of Yap1 and upregulation of antioxidant gene expression78. This thiol-peroxidase based redox relay has also been implicated in ABA-induced stomatal closure. It has been proposed that Arabidopsis GLUTATHIONE PEROXIDASE 3 (GPX3) is oxidized by H2O2 and relays oxidizing equivalents to ABA-INSENSITIVE 2 (ABI2) in the cytosol to inhibit phosphatase activity79. NPR1 is a SA receptor whose localization and self-association is another intriguing example of ROS-mediated protein regulation80, 81. NPR1 participates in global reprogramming towards defense and re-localizes to the nucleus in a redox dependent manner. At a resting state, NPR1 forms oligomers in the cytoplasm82. However, after the ROS burst triggered by pathogen perception, SA induces a reduction of the cytoplasm82 leading to activation of the thioredoxin TRX-5h, which in turn catalyzes the release of NPR1 monomers via reduction of inter-molecular disulfide bonds83. The nuclear localization signal is unmasked in monomeric NPR1, enabling nuclear localization, transcription factor binding, and global defense gene expression84.
Cysteine oxidation can also regulate the activity of metabolic enzymes, such as GLYCERALDEHYDE-3-PHOSPHATE DEHYDROGENASE (GAPDH), a conserved glycolytic enzyme catalyzing the conversion of glyceraldehyde-3-phosphate (G3P) and NAD into NADH and 1,3-bisphosphoglycerate (1,3-BPG)85. Oxidation of the catalytic cysteine in the active site of GAPDH blocks its catalytic activity, alters its subcellular localization and affects protein-protein interactions85, 86. Reducing conditions in the cytoplasm increase the affinity of cytosolic GAPDH isoforms (GAPCs) for the mitochondrial voltage-dependent anion-selective channel (VDCA) leading to localization of the GAPCs to the outer mitochondrial membrane. Oxidizing conditions, on the other hand, induce GAPC translocation to the nucleus by cysteine oxidation87. Under oxidizing conditions GAPCs can also activate plasma membrane-localized PHOSPHOLIPASE D δ (PLDδ), which plays a role in ABA- and H2O2-induced stomatal closure88.
The perception of apoplastic ROS in plants has remained enigmatic until recently. CYSTEINE-RICH RECEPTOR-LIKE KINASES (CRKs) have been proposed as candidate ROS sensors based on their extracellular cysteine-rich regions as well as their transcriptional induction in response to stress89, 90. Structural analyses of the cysteine-rich extracellular region of plasmodesmata localized proteins, close relatives of CRKs, suggest that the cysteines stabilize the ectodomain structure and may not be accessible for efficient ROS perception91. Recently, another receptor-like kinase (RLK) with extracellular leucine-rich repeat (LRR) domain, HPCA1, has recently been identified to be involved in direct ROS sensing52 (Fig. 2). HPCA1 possesses a unique extracellular domain, the hydrogen peroxidase (HP) domain, which contains four cysteines. Extracellular H2O2 modifies cysteine residues in HPCA1’s HP domain, resulting in increased kinase activity. Subsequently, active HPCA1 triggers activation of guard cell Ca2+ channels and stomatal closure. RLK-mediated MAMP perception also induces extracellular ROS production and activates a calmodulin-gated calcium channel leading to an increase in [Ca2+]cyto92. However, while HPCA1 is required for the extracellular H2O2-induced increase in [Ca2+]cyto, HPCA1 is not involved in the increase in [Ca2+]cyto in response to perception of the MAMPs flg22 or elf2652. Thus, it is likely that other extracellular ROS sensors exist which mediate extracellular ROS perception in response to MAMPs and pathogen infection. Protein function can be exquisitely modulated by PTM cross-talk and the role of ROS in these processes is an emerging field of interest in both plants and animals. For example, the activity of the human serine/threonine kinase AURORA A is regulated by oxidation of cysteine 290 which inhibits autophosphorylation of threonine 288 in the conserved activation loop (T-loop)93. Comparative evolutionary analysis across kingdoms showed that ~11.5% of all protein kinases contain an analogous Cys residue to Cys290 in Aurora A93. Phosphorylation can also trigger a switch of protein conformation resulting in the exposure and consequent oxidation of cryptic redox-sensitive cysteines94. In plants, the direct interplay between phosphorylation and cysteine oxidation at a molecular level has not yet been described. However, multiple kinases are redox-regulated. For example, H2O2 activates the Brassica napus MAP kinase MPK4, which in turn positively regulates ROS production95. Oxidation of redox-sensitive cysteine residues is crucial for activation of MPK3 and MPK6 in both rice and Arabidopsis96, 97. Moreover, cysteine oxidation inactivates the calcium-dependent kinase AtCPK2198 . Future research will likely reveal extensive cross-talk between phosphorylation and cysteine oxidation in plants.
Recent advances in understanding redox modifications and ROS perception in plants highlight additional outstanding biological questions. What is the importance of localized ROS production and pH for perception and redox modification of proteins? Can cysteine oxidation directly regulate NOXs or modulate the activity of the kinases involved in NOX activation? What are the identities of additional ROS receptors beyond HPCA1 and is there specificity in perception in different tissue types? Finally, how is ROS perception by apoplastic sensors or sensor domains integrated with ROS influx through aquaporins?
Regulation of NOX Activity
Not only can ROS directly regulate diverse biological processes and protein activity, but ROS producing enzymes are also subject to diverse PTMs. PTMs enable precise and rapid regulation of protein activity, stability, and interaction profiles. Additional layers of regulation can be achieved through PTM interplay, or cross-talk, where different modifications work in concert to achieve signaling specificity. Here, we highlight diverse PTMs that regulate NOX activity, both positively and negatively, in plants.
NOX and superoxide production
Plant NOXs are not only central enzymes for stress response, but also play a role in development18. RBOHD is the most highly expressed NOX in Arabidopsis and is required for ROS production upon immune perception and abiotic cues. RBOHF is also involved in regulating stomatal closure, the responses to abiotic signals and Casparian strip formation18. Other plant NOXs are involved in various developmental processes99.
NOXs produce superoxide, which is highly reactive and unstable making it difficult to accurately measure19. Superoxide can be dismutated spontaneously or more rapidly by the action of superoxide dismutase (SOD) to H2O217. The existence of an extracellular SODs in plants has been proposed100, 101. However, the high dismutation rate of superoxide even in the absence of a SOD might offer sufficient protection its toxic effects102. H2O2 is frequently considered to be the key signaling molecule following NOX activation and NOX activity is often assessed by measuring H2O2 levels after dismutation. In plants, many studies focus on the activity of H2O2 (Fig. 1). Gene expression studies suggest a certain amount of specificity between different types of ROS19 and the importance of extracellular superoxide as a signaling molecule is an underexplored area. In animals, superoxide can react with tyrosyl radicals resulting in the generation of tyrosine hydroperoxide103. These findings raise multiple outstanding questions. Does NOX-produced superoxide carry biological activity on its own? What is the relative importance of the balance between extracellular superoxide and H2O2 levels? Are apoplastic SODs involved in rapid conversion of extracellular superoxide?
N-terminal NOX regulation
Plant NOXs are composed of six trans-membrane domains, which contain an active ferric oxidoreductase domain, as well as N- and C-terminal extensions which are localized in the cytoplasm and are highly similar to mammalian NOXs. The N-terminus of NOXs contain Ca2+ binding EF-hands and PA might contribute to the regulation of some plant NOXs104. In contrast, the C-terminus contains FAD and NADPH binding domains. Tissue-specific expression patterns (e.g. pollen specific AtRBOHH and J) and stress stimulus-inducible expression profiles of NOX genes suggest spatio-temporal transcriptional regulation105, 106. In addition, NOX enzyme activity is also regulated at the post-translational level. Phosphorylation of the N-terminal region at conserved residues through distinct kinases has been demonstrated to affect NOX enzymatic activity. In Arabidopsis RBOHD, specific N-terminal residues are phosphorylated by distinct kinases (Fig. S1a). Examples include the RLK DOESN’T RESPOND TO NUCLEOTIDES 1 (DORN1) which phosphorylates S22 and T24, BOTRYTIS INDUCED KINASE 1 (BIK1) which phosphorylates S39 and S343, and CALCIUM DEPENDENT PROTEIN KINASES (CPKs) which phosphorylate S133, S148, and S163107, 108, 109, 110, 111.
The N-terminus of RBOHs functions as a hub for a multitude of kinases which induce ROS production. S339 is phosphorylated by BIK1 and the MAP4K SERINE/THREONINE KINASE 1 (SIK1), while S347 is phosphorylated by BIK1, CPKs and SIK1108, 109, 110, 111, 112. Phosphorylation of those residues (with the exception of S148) is linked to NOX activation, in particular phosphorylation of S343 and S347 during immune responses113. Intriguingly, phosphorylation of S163 contributed to negative regulation of the ROS producing activity and a S163A variant of RBOHD displayed enhanced ROS production following chitin treatment14, 113 (Fig. S1c). The interaction between multiple kinases with specific and convergent sites allows for flexibility in activation of RBOHD in response to independent stimuli and developmental stages. SCHENGEN1 (SGN1) phosphorylates unidentified residues in the N-terminus of both RBOHD and RBOHF in vitro27. OPEN STOMATA 1 (OST1), CALCINEURIN B-LIKE INTERACTING PROTEIN KINASE 26 (CIPK26) and SCHENGEN 1 (SGN1) were also found to modulate ROS-production of RBOHD and RBOHF in HEK293T cells. In particular, CIPK26-induced phosphorylation of RBOHF S13/S130/S132 and S174 residues was found to enhance and reduce ROS production, respectively (Fig. S1d). However, the phosphorylation status of RBOHF residues has not been elucidated in planta15,27.
Different kinases might target key residues of RBOHs in a tissue- or stimulus dependent manner. For example, BIK1, CPKs, SIK1, DORN1 and OST1 stimulate NOX-dependent ROS production in response to MAMPs, ATP and ABA, while SGN1 regulates the Casparian strip formation in the roots27, 37, 107, 108, 109, 110, 111, 112. The RLCK MARIS has been implicated in activation of RBOHH/J in pollen tube growth114. However, the preference of distinct kinases for specific sites and the importance of kinase specificity in different tissues awaits future investigation.
C-terminal NOX regulation
Since excessive ROS accumulation is detrimental, a coordinated balance between positive and negative regulation of ROS production is required. Some of the most pressing questions in ROS biology focus on how responses are appropriately regulated to ensure robust and efficient outputs pre- and post-activation. Recent studies highlight the importance of diverse C-terminal PTMs for fine-tuning NOX activity.
S-nitrosation is the addition of a nitrosonium ion (NO+) to the reactive thiol of a cysteine residue thereby forming S-nitrosothiol (SNO)115. S-nitrosothiols can be also produced by oxidation of thiol to thiyl radical followed by addition of NO (S-nitrosylation)116. While NO production in plants is still poorly understood, the best characterized way includes the enzymatic and non/enzymatic reduction of nitrite to NO117. In general, the predominant mechanism for S-nitrosothiol formation is S-nitrosation118. In plants, the production of nitric oxide increases upon pathogen perception and induces protein S-nitrosation with important implications in the regulation of immune responses119. RBOHD is S-nitrosated on C890 in vitro by exposure to S-nitrosoglutathione (GSNO) and Cys-NO and interestingly also in vivo upon infection with a bacterial pathogen120. Transgenic expression of a C890A variant of RBOHD (RBOHDC890A) in the Arabidopsis rbohD mutant background leads to increased ROS production compared to rbohD plants complemented with wild type RBOHD upon pathogen infection120. S-nitrosation of C890 destabilizes FAD binding, thereby reducing NOX activity120. Together, this suggests that pathogen-induced NO production provides a negative feedback loop limiting ROS production by RBOHD. Intriguingly, C890 is evolutionarily conserved across plants and humans, corresponding to C537121 and C694122 in human NOX2 and NOX5, respectively, and S-nitrosation also leads to decreased enzymatic activity (Fig. 3c,d; Fig. S1b). This conservation highlights the importance of cysteine S-nitrosation for downregulating NOX activity. Another PTM that affects NOX activity is persulfidation, the covalent addition of a thiol group to cysteine123. Cysteine persulfidation modulates protein structure, localization, intermolecular interactions and enzyme activities123. In plants, a major driver of persulfidation is hydrogen sulfide (H2S), a gaseus molecule that is produced in the cytosol by cysteine-degrading enzymes123. ABA triggers persulfidation of RBOHD. Persulfidation of RBOHD is abolished in plants expressing RBOHDC825A/C890A and those plants also exhibit reduced ROS production following ABA treatment123. Cysteine persulfidation of RBOHD likely results in an increased negative charge, which enhances FAD binding and subsequently enzymatic activity. Cysteine residues might be important regulation sites for NOXs as Cysteine residues in both N and C terminal regions are conserved (Fig S1b).
Figure 3. Conservation of the NOX C-terminus across plants and conservation of critical residues in RBOHD and human NOX5.
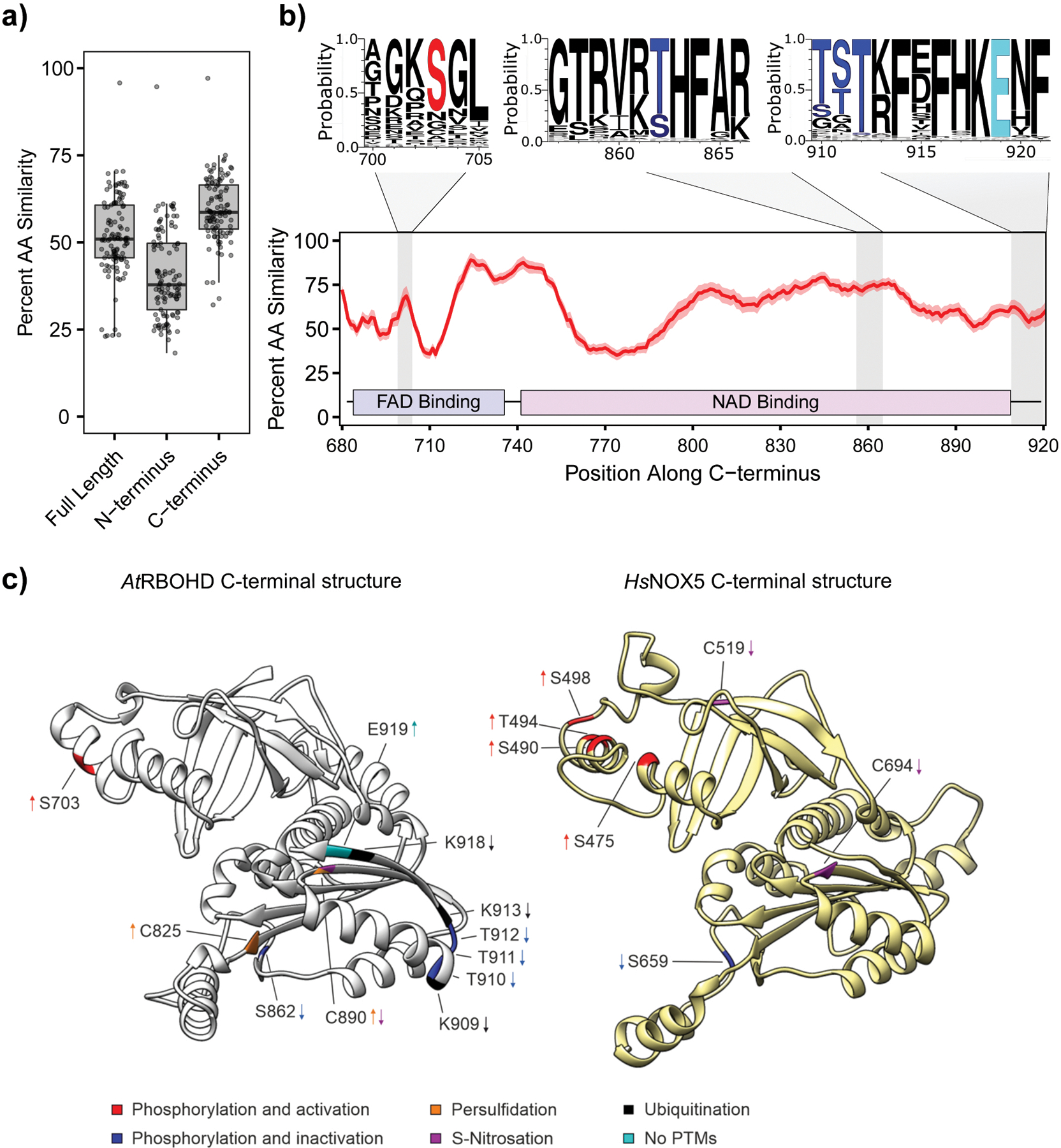
(a) Conservation of plant NOX homologs when comparing full length, N-terminal (residues 1–376) or C-terminal regions (residues 680–920). Percent amino acid (AA) similarity was compared using AtRBOHD as a reference across 112 plant NOXs. (b) Sliding window of average amino acid similarity displayed in red with a 95% confidence interval in light red along the C-termini for plant NOX homologs. AtRBOHD was used as a reference with a 7 amino acid sliding window. Regions known to be regulated via posttranslational modifications (PTMs) are highlighted in grey and displayed by weblogos with modified residues differentially colored. (c) Structural models of (left) AtRBOHD C-terminus (residues 613–920) and (right) HsNOX5 β-isoform C-terminus (residues 401–719). Residues are labelled with different colours, based on the type of post-translational modifications (PTMs) underlying their regulatory role. Positive and negative regulatory outputs of the PTMs are specified by up- and down-arrows, respectively. Red label indicates positive regulation by phosphorylation, blue indicates negative regulation by phosphorylation, black specify ubiquitination, purple specify S-nitrosylation and orange indicates persulfidation. Residues highlighted with light blue were not found to be post-translationally modified, but were experimentally shown to modulate ROS producing activity.
Broadly, across 112 NOXs presently known in the plant kingdom, the C-terminus is more conserved in amino acid sequence than full length proteins when compared to Arabidopsis RBOHD (Fig. 3a). Additionally, residues known to be post-translationally modified are conserved, especially in functionality, along the C-terminus of NOX homologs (Fig. 3b). While the N-terminus functions as a hub for activation, PTMs on C-terminal residues can have opposing effects on ROS production. As described above, S-nitrosation inhibits while persulfidation enhances RBOHD activity. Interestingly, phosphorylation of the C-terminal region is also critical for regulation of human NOXs. Ataxia telangiectasia mutated (ATM)-kinase-dependent phosphorylation of NOX2 S486 leads to reduced NOX2 activity121. Human NOX5 can be activated by phosphorylation of S475, S490, T494, and S498 by PROTEIN KINASE C (PKC) and CALCIUM/CALMODULIN-DEPENDENT KINASE II (CAMKII; Fig. 3b, c). However, CAMKII also participates in negative regulation of NOX5 activity by phosphorylation of S659100 (Fig. 3c). Recent work has identified kinases that interact with residues in the C-terminus of RBOHD. CRK2 phosphorylates RBOHD S703 and enhances production of ROS during recognition of flg2268 (Fig. 3b,c). The kinase AvrPphB SUSCEPTIBLE 1-LIKE 13 (PBL13) also interacts with the C-terminal region of RBOHD, however phosphorylation of RBOHD T912 by PBL13 dampens ROS production124 (Fig. 3b,c). These results suggest that at the NOX C-terminus multiple kinases converge to achieve robust, precise, and selective control of NOX activity.
C-terminal PTMs play a key role in calibrating the enzymatic activity of RBOHD and also regulate ROS by fine-tuning the abundance of RBOHD. Ubiquitination, which is characterized by the addition of one or more ubiquitin molecules to lysine (K) residues, is involved in nearly all aspects of eukaryotic biology125, 126 and plays a crucial role in regulating immune signalling127. For example, ubiquitination negatively regulates crucial molecular players of immune responses in Arabidopsis, including FLAGELLIN-SENSING 2 (FLS2)128, BIK1129, and the chitin receptor LYSIN-MOTIF RECEPTOR LIKE KINASE 5 (LYK5)130. Recent work demonstrates interplay between phosphorylation and ubiquitination to negatively regulate RBOHD124 (Fig. 3c). PBL13 can phosphorylate RBOHD T912 in vitro, and the abundance of RBOHDT912D is reduced compared to wild-type RBOHD in planta124. PBL13 interacts with the PBL13 interacting RING domain E3 (PIRE) ligase, a RING domain E3 ubiquitin ligase124. Strikingly, RBOHD ubiquitination was enhanced in complementation lines expressing the phosphomimic RBOHDT912D, while the phosphonull RBOHDT912A decreased ubiquitination levels. Importantly, deletions of pbl13 and pire exhibited enhanced RBOHD protein accumulation and enhanced ROS production triggered by flg22. Furthermore, ubiquitination results in endocytosis and vacuolar-mediated degradation124. Both T912 and C-terminal lysine residues are conserved in other plant RBOHs, indicating that phosphorylation and ubiquitination may be a conserved mechanism for RBOH regulation (Fig. 3). These results suggest posttranslational control of NOX protein abundance through cross talk between phosphorylation and ubiquitination.
The C-terminus of RBOHs act as a conserved hub to modulate production of ROS and NOX abundance. Phosphomimetic mutants of S862 and T912 lead to decreased ROS production and abundance of RBOHD, respectively, and these residues are highly conserved across plant NOXs (Fig. 3b). A recent Arabidopsis screen led to the identification of a mutant defective in LPS-triggered ROS burst131. The mutation responsible for the decreased RBOHD activity was mapped to residue E919, which is conserved in all Arabidopsis RBOHs6 (Fig. 3). Intriguingly, mutation of this residue to a lysine (E919L) had no effect on protein accumulation, localization, or association with BIK1. These finding suggest a role for E919 as an “off switch” for RBOHD. An important focus of future research will be to determine how multiple kinases dynamically regulate ROS production by interacting with conserved residues in N- and C-terminal regions. Given the conservation of C-terminal residues across diverse NOXs, it is possible that different organisms enlist specific modifying enzymes to fine-tune NOX abundance at similar residues to regulate the amplitude and duration of ROS production.
Conclusions
ROS are essential for life, but must be appropriately regulated in response to development as well as diverse stimuli. Recent scientific advances highlight the complexity of ROS effect on proteins, enzymatic activity, and cellular processes. Future advances in the field of ROS will shed light on a fundamental component of all cellular life, with significant implications for crop improvement and protection. We hope the ideas presented in this article serve as an invitation for the scientific community to push the field forward.
Supplementary Material
Acknowledgements
BC and GC are supported by a grant from the National Institutes of Health (NIH 1R35GM136402). BC is partially supported by the UC Davis Dean’s Distinguished Graduate Fellowship. MW acknowledges funding from the Academy of Finland (Decision 323917). SK is supported by JSPS KAKENHI (grant 20K05831).
Footnotes
Competing interests
The authors declare no competing interests. No figures contain third party material.
Data availability statement
Data used to generate Fig. 3 a, b and Fig. S1 a, b as well as detailed methodology and scripts are available in the GitHub repository (https://github.com/DanielleMStevens/ROS_production_review). The NOX C-terminal alignments are available in wasabi (http://was.bi?id=gedS1F).
References
- 1.Mhamdi A, Van Breusegem F. Reactive oxygen species in plant development. Development 145, (2018). [DOI] [PubMed] [Google Scholar]
- 2.Mittler R ROS Are Good. Trends Plant Sci 22, 11–19 (2017). [DOI] [PubMed] [Google Scholar]
- 3.Dietz KJ, Turkan I, Krieger-Liszkay A. Redox- and Reactive Oxygen Species-Dependent Signaling into and out of the Photosynthesizing Chloroplast. Plant Physiol 171, 1541–1550 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Huang S, Van Aken O, Schwarzländer M, Belt K, Millar AH. The Roles of Mitochondrial Reactive Oxygen Species in Cellular Signaling and Stress Response in Plants. Plant Physiol 171, 1551–1559 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sandalio LM, Romero-Puertas MC. Peroxisomes sense and respond to environmental cues by regulating ROS and RNS signalling networks. Ann Bot 116, 475–485 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Huang H, Ullah F, Zhou DX, Yi M, Zhao Y. Mechanisms of ROS Regulation of Plant Development and Stress Responses. Front Plant Sci 10, 800 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhao C, Zhang H, Song C, Zhu J-K, Shabala S. Mechanisms of Plant Responses and Adaptation to Soil Salinity. The Innovation 1, 100017 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.El-Shetehy M, Wang C, Shine MB, Yu K, Kachroo A, Kachroo P. Nitric oxide and reactive oxygen species are required for systemic acquired resistance in plants. Plant Signal Behav 10, e998544 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schmidt R, Kunkowska AB, Schippers JH. Role of Reactive Oxygen Species during Cell Expansion in Leaves. Plant Physiol 172, 2098–2106 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kaya H, et al. Ca2+-activated reactive oxygen species production by Arabidopsis RbohH and RbohJ is essential for proper pollen tube tip growth. Plant Cell 26, 1069–1080 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Takeda S, Gapper C, Kaya H, Bell E, Kuchitsu K, Dolan L. Local positive feedback regulation determines cell shape in root hair cells. Science 319, 1241–1244 (2008). [DOI] [PubMed] [Google Scholar]
- 12.Mangano S, et al. Molecular link between auxin and ROS-mediated polar growth. Proc Natl Acad Sci U S A 114, 5289–5294 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee Y, Rubio MC, Alassimone J, Geldner N. A mechanism for localized lignin deposition in the endodermis. Cell 153, 402–412 (2013). [DOI] [PubMed] [Google Scholar]
- 14.Cheval C, et al. Chitin perception in plasmodesmata characterizes submembrane immune-signaling specificity in plants. Proc Natl Acad Sci U S A 117, 9621–9629 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Han JP, et al. Fine-tuning of RBOHF activity is achieved by differential phosphorylation and Ca. New Phytol 221, 1935–1949 (2019). [DOI] [PubMed] [Google Scholar]
- 16.Smirnoff N, Arnaud D. Hydrogen peroxide metabolism and functions in plants. New Phytol 221, 1197–1214 (2019). [DOI] [PubMed] [Google Scholar]
- 17.Waszczak C, Carmody M, Kangasjärvi J. Reactive Oxygen Species in Plant Signaling. Annu Rev Plant Biol 69, 209–236 (2018). [DOI] [PubMed] [Google Scholar]
- 18.Kadota Y, Shirasu K, Zipfel C. Regulation of the NADPH Oxidase RBOHD During Plant Immunity. Plant Cell Physiol 56, 1472–1480 (2015). [DOI] [PubMed] [Google Scholar]
- 19.Vaahtera L, Brosché M, Wrzaczek M, Kangasjärvi J. Specificity in ROS signaling and transcript signatures. Antioxid Redox Signal 21, 1422–1441 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Willems P, et al. The ROS Wheel: Refining ROS Transcriptional Footprints. Plant Physiol 171, 1720–1733 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mielecki J, Gawroński P, Karpiński S. Retrograde Signaling: Understanding the Communication between Organelles. Int J Mol Sci 21, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ng S, et al. A membrane-bound NAC transcription factor, ANAC017, mediates mitochondrial retrograde signaling in Arabidopsis. Plant Cell 25, 3450–3471 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shapiguzov A, et al. Arabidopsis RCD1 coordinates chloroplast and mitochondrial functions through interaction with ANAC transcription factors. Elife 8, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chan KX, Phua SY, Crisp P, McQuinn R, Pogson BJ. Learning the Languages of the Chloroplast: Retrograde Signaling and Beyond. Annu Rev Plant Biol 67, 25–53 (2016). [DOI] [PubMed] [Google Scholar]
- 25.op den Camp RG, et al. Rapid induction of distinct stress responses after the release of singlet oxygen in Arabidopsis. Plant Cell 15, 2320–2332 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Daudi A, et al. The apoplastic oxidative burst peroxidase in Arabidopsis is a major component of pattern-triggered immunity. Plant Cell 24, 275–287 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fujita S, et al. SCHENGEN receptor module drives localized ROS production and lignification in plant roots. EMBO J 39, e103894 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lassig R, Gutermuth T, Bey TD, Konrad KR, Romeis T. Pollen tube NAD(P)H oxidases act as a speed control to dampen growth rate oscillations during polarized cell growth. Plant J 78, 94–106 (2014). [DOI] [PubMed] [Google Scholar]
- 29.Müller K, Carstens AC, Linkies A, Torres MA, Leubner-Metzger G. The NADPH-oxidase AtrbohB plays a role in Arabidopsis seed after-ripening. New Phytol 184, 885–897 (2009). [DOI] [PubMed] [Google Scholar]
- 30.Lolle S, Stevens D, Coaker G. Plant NLR-triggered immunity: from receptor activation to downstream signaling. Curr Opin Immunol 62, 99–105 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.O’Brien JA, et al. A peroxidase-dependent apoplastic oxidative burst in cultured Arabidopsis cells functions in MAMP-elicited defense. Plant Physiol 158, 2013–2027 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang C, et al. Pipecolic acid confers systemic immunity by regulating free radicals. Sci Adv 4, eaar4509–eaar4509 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li X, et al. Tomato SlRbohB, a member of the NADPH oxidase family, is required for disease resistance against Botrytis cinerea and tolerance to drought stress. Front Plant Sci 6, 463 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wong HL, et al. Regulation of rice NADPH oxidase by binding of Rac GTPase to its N-terminal extension. Plant Cell 19, 4022–4034 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tian S, et al. Plant Aquaporin AtPIP1;4 Links Apoplastic H2O2 Induction to Disease Immunity Pathways. Plant Physiol 171, 1635–1650 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bienert GP, Chaumont F. Aquaporin-facilitated transmembrane diffusion of hydrogen peroxide. Biochim Biophys Acta 1840, 1596–1604 (2014). [DOI] [PubMed] [Google Scholar]
- 37.Grondin A, Rodrigues O, Verdoucq L, Merlot S, Leonhardt N, Maurel C. Aquaporins Contribute to ABA-Triggered Stomatal Closure through OST1-Mediated Phosphorylation. Plant Cell 27, 1945–1954 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Möller MN, Cuevasanta E, Orrico F, Lopez AC, Thomson L, Denicola A. Diffusion and Transport of Reactive Species Across Cell Membranes. Adv Exp Med Biol 1127, 3–19 (2019). [DOI] [PubMed] [Google Scholar]
- 39.Lynch RE, Fridovich I. Permeation of the erythrocyte stroma by superoxide radical. J Biol Chem 253, 4697–4699 (1978). [PubMed] [Google Scholar]
- 40.Reithmeier RA, Casey JR, Kalli AC, Sansom MS, Alguel Y, Iwata S. Band 3, the human red cell chloride/bicarbonate anion exchanger (AE1, SLC4A1), in a structural context. Biochim Biophys Acta 1858, 1507–1532 (2016). [DOI] [PubMed] [Google Scholar]
- 41.de Rezende FF, et al. Integrin α7β1 is a redox-regulated target of hydrogen peroxide in vascular smooth muscle cell adhesion. Free Radic Biol Med 53, 521–531 (2012). [DOI] [PubMed] [Google Scholar]
- 42.Noctor G, Foyer CH. Intracellular Redox Compartmentation and ROS-Related Communication in Regulation and Signaling. Plant Physiol 171, 1581–1592 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bononi A, et al. Mitochondria-associated membranes (MAMs) as hotspot Ca(2+) signaling units. Adv Exp Med Biol 740, 411–437 (2012). [DOI] [PubMed] [Google Scholar]
- 44.Hancock JT. Considerations of the importance of redox state for reactive nitrogen species action. J Exp Bot 70, 4323–4331 (2019). [DOI] [PubMed] [Google Scholar]
- 45.Thordal-Christensen H, Zhang Z, Wei Y, Collinge DB. Subcellular localization of H2O2 in plants. H2O2 accumulation in papillae and hypersensitive response during the barley—powdery mildew interaction.) (1997).
- 46.Dikalov SI, Harrison DG. Methods for detection of mitochondrial and cellular reactive oxygen species. Antioxid Redox Signal 20, 372–382 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kauffman ME, et al. MitoSOX-Based Flow Cytometry for Detecting Mitochondrial ROS. React Oxyg Species (Apex) 2, 361–370 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fichman Y, Miller G, Mittler R. Whole-Plant Live Imaging of Reactive Oxygen Species. Molecular Plant 12, 1203–1210 (2019). [DOI] [PubMed] [Google Scholar]
- 49.Nietzel T, et al. The fluorescent protein sensor roGFP2-Orp1 monitors in vivo H. New Phytol 221, 1649–1664 (2019). [DOI] [PubMed] [Google Scholar]
- 50.Fichman Y, Mittler R. Rapid systemic signaling during abiotic and biotic stresses: is the ROS wave master of all trades? Plant J 102, 887–896 (2020). [DOI] [PubMed] [Google Scholar]
- 51.Gilroy S, et al. ROS, Calcium, and Electric Signals: Key Mediators of Rapid Systemic Signaling in Plants. Plant Physiol 171, 1606–1615 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wu F, et al. Hydrogen peroxide sensor HPCA1 is an LRR receptor kinase in Arabidopsis. Nature 578, 577–581 (2020). [DOI] [PubMed] [Google Scholar]
- 53.Cheval C, Faulkner C. Plasmodesmal regulation during plant-pathogen interactions. New Phytol 217, 62–67 (2018). [DOI] [PubMed] [Google Scholar]
- 54.Mittler R, Blumwald E. The roles of ROS and ABA in systemic acquired acclimation. Plant Cell 27, 64–70 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zandalinas SI, Fichman Y, Mittler R. Vascular Bundles Mediate Systemic Reactive Oxygen Signaling during Light Stress. The Plant Cell 32, 3425–3435 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chen YC, et al. -hydroxy-pipecolic acid is a mobile metabolite that induces systemic disease resistance in. Proc Natl Acad Sci U S A 115, E4920–E4929 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Návarová H, Bernsdorff F, Döring AC, Zeier J. Pipecolic acid, an endogenous mediator of defense amplification and priming, is a critical regulator of inducible plant immunity. Plant Cell 24, 5123–5141 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wu J, et al. Deficient plastidic fatty acid synthesis triggers cell death by modulating mitochondrial reactive oxygen species. Cell Research 25, 621–633 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gilroy S, et al. A tidal wave of signals: calcium and ROS at the forefront of rapid systemic signaling. Trends Plant Sci 19, 623–630 (2014). [DOI] [PubMed] [Google Scholar]
- 60.Yuan M, et al. Pattern-recognition receptors are required for NLR-mediated plant immunity. bioRxiv, 2020.2004.2010.031294 (2020). [DOI] [PMC free article] [PubMed]
- 61.McConnell EW, et al. Proteome-Wide Analysis of Cysteine Reactivity during Effector-Triggered Immunity. Plant Physiology 179, 1248–1264 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ngou BPM, Ahn H-K, Ding P, Jones JDG. Mutual Potentiation of Plant Immunity by Cell-surface and Intracellular Receptors. bioRxiv, 2020.2004.2010.034173 (2020). [DOI] [PubMed]
- 63.Liu Y, Ren D, Pike S, Pallardy S, Gassmann W, Zhang S. Chloroplast-generated reactive oxygen species are involved in hypersensitive response-like cell death mediated by a mitogen-activated protein kinase cascade. Plant J 51, 941–954 (2007). [DOI] [PubMed] [Google Scholar]
- 64.Rossi FR, Krapp AR, Bisaro F, Maiale SJ, Pieckenstain FL, Carrillo N. Reactive oxygen species generated in chloroplasts contribute to tobacco leaf infection by the necrotrophic fungus Botrytis cinerea. Plant J 92, 761–773 (2017). [DOI] [PubMed] [Google Scholar]
- 65.Zurbriggen MD, et al. Chloroplast-generated reactive oxygen species play a major role in localized cell death during the non-host interaction between tobacco and Xanthomonas campestris pv. vesicatoria. Plant J 60, 962–973 (2009). [DOI] [PubMed] [Google Scholar]
- 66.Moreau M, et al. The Arabidopsis oligopeptidases TOP1 and TOP2 are salicylic acid targets that modulate SA-mediated signaling and the immune response. Plant J 76, 603–614 (2013). [DOI] [PubMed] [Google Scholar]
- 67.Westlake TJ, Ricci WA, Popescu GV, Popescu SC. Dimerization and thiol sensitivity of the salicylic acid binding thimet oligopeptidases TOP1 and TOP2 define their functions in redox-sensitive cellular pathways. Frontiers in Plant Sciences 6, 327 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kimura S, Waszczak C, Hunter K, Wrzaczek M. Bound by Fate: The Role of Reactive Oxygen Species in Receptor-Like Kinase Signaling. Plant Cell 29, 638–654 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Akter S, et al. Cysteines under ROS attack in plants: a proteomics view. J Exp Bot 66, 2935–2944 (2015). [DOI] [PubMed] [Google Scholar]
- 70.Waszczak C, Akter S, Jacques S, Huang J, Messens J, Van Breusegem F. Oxidative post-translational modifications of cysteine residues in plant signal transduction. J Exp Bot 66, 2923–2934 (2015). [DOI] [PubMed] [Google Scholar]
- 71.Jacques S, et al. Protein Methionine Sulfoxide Dynamics in Arabidopsis thaliana under Oxidative Stress. Mol Cell Proteomics 14, 1217–1229 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Jacques S, Ghesquière B, Van Breusegem F, Gevaert K. Plant proteins under oxidative attack. Proteomics 13, 932–940 (2013). [DOI] [PubMed] [Google Scholar]
- 73.Felle HH. pH: Signal and Messenger in Plant Cells. Plant Biology 3, 577–591 (2001). [Google Scholar]
- 74.Geilfus CM. The pH of the Apoplast: Dynamic Factor with Functional Impact Under Stress. Mol Plant 10, 1371–1386 (2017). [DOI] [PubMed] [Google Scholar]
- 75.Wojtkowiak JW, Verduzco D, Schramm KJ, Gillies RJ. Drug resistance and cellular adaptation to tumor acidic pH microenvironment. Mol Pharm 8, 2032–2038 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Toledano MB, Kullik I, Trinh F, Baird PT, Schneider TD, Storz G. Redox-dependent shift of OxyR-DNA contacts along an extended DNA-binding site: a mechanism for differential promoter selection. Cell 78, 897–909 (1994). [DOI] [PubMed] [Google Scholar]
- 77.Gaudu P, Moon N, Weiss B. Regulation of the soxRS oxidative stress regulon reversible oxidation of the Fe-S centers of SoxR in vivo. Journal of Biological Chemistry 272, 5082–5086 (1997). [DOI] [PubMed] [Google Scholar]
- 78.Delaunay A, Pflieger D, Barrault MB, Vinh J, Toledano MB. A thiol peroxidase is an H2O2 receptor and redox-transducer in gene activation. Cell 111, 471–481 (2002). [DOI] [PubMed] [Google Scholar]
- 79.Miao Y, et al. An Arabidopsis glutathione peroxidase functions as both a redox transducer and a scavenger in abscisic acid and drought stress responses. Plant Cell 18, 2749–2766 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Ding Y, Sun T, Ao K, Peng Y, Zhang Y, Li X. Opposite Roles of Salicylic Acid Receptors NPR1 and NPR3/NPR4 in Transcriptional Regulation of Plant Immunity. Cell 173, 1454–1467.e1415 (2018). [DOI] [PubMed] [Google Scholar]
- 81.Dong X NPR1, all things considered. Curr Opin Plant Biol 7, 547–552 (2004). [DOI] [PubMed] [Google Scholar]
- 82.Mou Z, Fan W, Dong X. Inducers of plant systemic acquired resistance regulate NPR1 function through redox changes. Cell 113, 935–944 (2003). [DOI] [PubMed] [Google Scholar]
- 83.Tada Y, et al. Plant immunity requires conformational changes of NPR1 via S-nitrosylation and thioredoxins. Science 321, 952–956 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Kinkema M, Fan W, Dong X. Nuclear localization of NPR1 is required for activation of PR gene expression. Plant Cell 12, 2339–2350 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Zaffagnini M, Fermani S, Costa A, Lemaire SD, Trost P. Plant cytoplasmic GAPDH: redox post-translational modifications and moonlighting properties. Front Plant Sci 4, 450 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Hancock JT, et al. Proteomic identification of glyceraldehyde 3-phosphate dehydrogenase as an inhibitory target of hydrogen peroxide in Arabidopsis. Plant Physiol Biochem 43, 828–835 (2005). [DOI] [PubMed] [Google Scholar]
- 87.Schneider M, Knuesting J, Birkholz O, Heinisch JJ, Scheibe R. Cytosolic GAPDH as a redox-dependent regulator of energy metabolism. BMC Plant Biol 18, 184 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Uraji M, et al. Cooperative function of PLDδ and PLDα1 in abscisic acid-induced stomatal closure in Arabidopsis. Plant Physiol 159, 450–460 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Bourdais G, et al. Large-Scale Phenomics Identifies Primary and Fine-Tuning Roles for CRKs in Responses Related to Oxidative Stress. PLoS Genet 11, e1005373 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Kimura S, et al. CRK2 and C-terminal Phosphorylation of NADPH Oxidase RBOHD Regulate Reactive Oxygen Species Production in Arabidopsis. Plant Cell 32, 1063–1080 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Vaattovaara A, et al. Mechanistic insights into the evolution of DUF26-containing proteins in land plants. Commun Biol 2, 56 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Tian W, et al. A calmodulin-gated calcium channel links pathogen patterns to plant immunity. Nature 572, 131–135 (2019). [DOI] [PubMed] [Google Scholar]
- 93.Byrne DP, et al. Aurora A regulation by reversible cysteine oxidation reveals evolutionarily conserved redox control of Ser/Thr protein kinase activity. Sci Signal 13, (2020). [DOI] [PubMed] [Google Scholar]
- 94.Behring JB, et al. Spatial and temporal alterations in protein structure by EGF regulate cryptic cysteine oxidation. Science Signaling 13, eaay7315 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Zhang T, Zhu M, Song WY, Harmon AC, Chen S. Oxidation and phosphorylation of MAP kinase 4 cause protein aggregation. Biochim Biophys Acta 1854, 156–165 (2015). [DOI] [PubMed] [Google Scholar]
- 96.Kovtun Y, Chiu WL, Tena G, Sheen J. Functional analysis of oxidative stress-activated mitogen-activated protein kinase cascade in plants. Proc Natl Acad Sci U S A 97, 2940–2945 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Xie G, Sasaki K, Imai R, Xie D. A redox-sensitive cysteine residue regulates the kinase activities of OsMPK3 and OsMPK6 in vitro. Plant Sci 227, 69–75 (2014). [DOI] [PubMed] [Google Scholar]
- 98.Ueoka-Nakanishi H, Sazuka T, Nakanishi Y, Maeshima M, Mori H, Hisabori T. Thioredoxin h regulates calcium dependent protein kinases in plasma membranes. FEBS J 280, 3220–3231 (2013). [DOI] [PubMed] [Google Scholar]
- 99.Kaya H, et al. Comparative analysis of the reactive oxygen species-producing enzymatic activity of Arabidopsis NADPH oxidases. The Plant Journal 98, 291–300 (2019). [DOI] [PubMed] [Google Scholar]
- 100.Pandey D, Gratton JP, Rafikov R, Black SM, Fulton DJ. Calcium/calmodulin-dependent kinase II mediates the phosphorylation and activation of NADPH oxidase 5. Mol Pharmacol 80, 407–415 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Streller S, Krömer S, Wingsle G. Isolation and purification of mitochondrial Mn-superoxide dismutase from the gymnosperm Pinus sylvestris L. Plant Cell Physiol 35, 859–867 (1994). [PubMed] [Google Scholar]
- 102.Alscher RG, Erturk N, Heath LS. Role of superoxide dismutases (SODs) in controlling oxidative stress in plants. Journal of Experimental Botany 53, 1331–1341 (2002). [PubMed] [Google Scholar]
- 103.Winterbourn CC, Parsons-Mair HN, Gebicki S, Gebicki JM, Davies MJ. Requirements for superoxide-dependent tyrosine hydroperoxide formation in peptides. Biochem J 381, 241–248 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Zhang Y, et al. Phospholipase dalpha1 and phosphatidic acid regulate NADPH oxidase activity and production of reactive oxygen species in ABA-mediated stomatal closure in Arabidopsis. Plant Cell 21, 2357–2377 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Hu CH, et al. NADPH Oxidases: The Vital Performers and Center Hubs during Plant Growth and Signaling. Cells 9, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Morales J, Kadota Y, Zipfel C, Molina A, Torres MA. The Arabidopsis NADPH oxidases RbohD and RbohF display differential expression patterns and contributions during plant immunity. J Exp Bot 67, 1663–1676 (2016). [DOI] [PubMed] [Google Scholar]
- 107.Chen D, et al. Extracellular ATP elicits DORN1-mediated RBOHD phosphorylation to regulate stomatal aperture. Nat Commun 8, 2265 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Kadota Y, et al. Direct regulation of the NADPH oxidase RBOHD by the PRR-associated kinase BIK1 during plant immunity. Mol Cell 54, 43–55 (2014). [DOI] [PubMed] [Google Scholar]
- 109.Li L, et al. The FLS2-associated kinase BIK1 directly phosphorylates the NADPH oxidase RbohD to control plant immunity. Cell Host Microbe 15, 329–338 (2014). [DOI] [PubMed] [Google Scholar]
- 110.Dubiella U, et al. Calcium-dependent protein kinase/NADPH oxidase activation circuit is required for rapid defense signal propagation. Proceedings of the National Academy of Sciences 110, 8744–8749 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Gao X, et al. Bifurcation of Arabidopsis NLR immune signaling via Ca 2+-dependent protein kinases. PLoS Pathog 9, e1003127 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Zhang M, et al. The MAP4 Kinase SIK1 Ensures Robust Extracellular ROS Burst and Antibacterial Immunity in Plants. Cell Host Microbe 24, 379–391.e375 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Kadota Y, et al. Quantitative phosphoproteomic analysis reveals common regulatory mechanisms between effector- and PAMP-triggered immunity in plants. New Phytol 221, 2160–2175 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Boisson-Dernier A, Franck CM, Lituiev DS, Grossniklaus U. Receptor-like cytoplasmic kinase MARIS functions downstream of CrRLK1L-dependent signaling during tip growth. Proc Natl Acad Sci U S A 112, 12211–12216 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Fernando V, Zheng X, Walia Y, Sharma V, Letson J, Furuta S. S-Nitrosylation: An Emerging Paradigm of Redox Signaling. Antioxidants (Basel) 8, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Heinrich TA, da Silva RS, Miranda KM, Switzer CH, Wink DA, Fukuto JM. Biological nitric oxide signalling: chemistry and terminology. Br J Pharmacol 169, 1417–1429 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Astier J, Gross I, Durner J. Nitric oxide production in plants: an update. Journal of Experimental Botany 69, 3401–3411 (2017). [DOI] [PubMed] [Google Scholar]
- 118.Gupta KJ, et al. Recommendations on terminology and experimental best practice associated with plant nitric oxide research. New Phytol 225, 1828–1834 (2020). [DOI] [PubMed] [Google Scholar]
- 119.Lindermayr C, Durner J. S-Nitrosylation in plants: pattern and function. J Proteomics 73, 1–9 (2009). [DOI] [PubMed] [Google Scholar]
- 120.Yun BW, et al. S-nitrosylation of NADPH oxidase regulates cell death in plant immunity. Nature 478, 264–268 (2011). [DOI] [PubMed] [Google Scholar]
- 121.Beaumel S, et al. Down-regulation of NOX2 activity in phagocytes mediated by ATM-kinase dependent phosphorylation. Free Radic Biol Med 113, 1–15 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Qian J, et al. Nitric oxide reduces NADPH oxidase 5 (Nox5) activity by reversible S-nitrosylation. Free Radic Biol Med 52, 1806–1819 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Shen J, et al. Persulfidation-based Modification of Cysteine Desulfhydrase and the NADPH Oxidase RBOHD Controls Guard Cell Abscisic Acid Signaling. Plant Cell 32, 1000–1017 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Lee D, et al. Regulation of reactive oxygen species during plant immunity through phosphorylation and ubiquitination of RBOHD. Nat Commun 11, 1838 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Swatek KN, Komander D. Ubiquitin modifications. Cell Res 26, 399–422 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Hershko A, Ciechanover A. The ubiquitin system. Annu Rev Biochem 67, 425–479 (1998). [DOI] [PubMed] [Google Scholar]
- 127.Mithoe SC, Menke FL. Regulation of pattern recognition receptor signalling by phosphorylation and ubiquitination. Curr Opin Plant Biol 45, 162–170 (2018). [DOI] [PubMed] [Google Scholar]
- 128.Lu D, et al. Direct ubiquitination of pattern recognition receptor FLS2 attenuates plant innate immunity. Science 332, 1439–1442 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Ma X, et al. Ligand-induced monoubiquitination of BIK1 regulates plant immunity. Nature 581, 199–203 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Liao D, et al. Arabidopsis E3 ubiquitin ligase PLANT U-BOX13 (PUB13) regulates chitin receptor LYSIN MOTIF RECEPTOR KINASE5 (LYK5) protein abundance. New Phytol 214, 1646–1656 (2017). [DOI] [PubMed] [Google Scholar]
- 131.Li QY, Li P, Myint Phyu Sin Htwe N, Shangguan KK, Liang Y. Antepenultimate residue at the C-terminus of NADPH oxidase RBOHD is critical for its function in the production of reactive oxygen species in Arabidopsis. J Zhejiang Univ Sci B 20, 713–727 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data used to generate Fig. 3 a, b and Fig. S1 a, b as well as detailed methodology and scripts are available in the GitHub repository (https://github.com/DanielleMStevens/ROS_production_review). The NOX C-terminal alignments are available in wasabi (http://was.bi?id=gedS1F).


