SUMMARY
Ductal carcinoma in situ (DCIS) is a pre-invasive lesion that is thought to be a precursor to invasive breast cancer (IBC). To understand the changes in the tumor microenvironment (TME) accompanying transition to IBC, we used multiplexed ion beam imaging by time of flight (MIBI-TOF) and a 37-plex antibody staining panel to interrogate 79 clinically annotated surgical resections using machine learning tools for cell segmentation, pixel-based clustering, and object morphometrics. Comparison of normal breast with patient-matched DCIS and IBC revealed coordinated transitions between four TME states that were delineated based on the location and function of myoepithelium, fibroblasts, and immune cells. Surprisingly, myoepithelial disruption was more advanced in DCIS patients that did not develop IBC, suggesting this process could be protective against recurrence. Taken together, this HTAN Breast PreCancer Atlas study offers insight into drivers of IBC relapse and emphasizes the importance of the TME in regulating these processes.
In brief
A spatial imaging atlas of patient-matched ductal carcinoma in situ and invasive breast cancer depicts coordinated changes in the tumor microenviroment associated with invasive relapse, suggesting a potential protective role of myoepithelial disruption against invasive progression.
Graphical Abstract

INTRODUCTION
Ductal carcinoma in situ (DCIS) is a pre-invasive lesion of tumor cells within the breast duct that are isolated from the surrounding stroma by a near-continuous layer of myoepithelium and basement membrane proteins. This histologic property is the primary feature that distinguishes DCIS from invasive breast cancer (IBC), where this barrier is absent and tumor cells are in direct contact with the stroma (Figure 1A). DCIS comprises 20% of new breast cancer diagnoses, but unlike IBC, is not a life-threatening disease in itself. However, if left untreated, up to half of patients with DCIS develop IBC within 10 years (Betsill et al., 1978; Erbas et al., 2006; Eusebi et al., 1994; Page et al., 1982; Ryser et al., 2019), leading to the current practice of surgical intervention for all DCIS patients.
Figure 1. A longitudinal cohort of DCIS patients with or without subsequent invasive relapse.
(A) Schematic of the tumor stages and patient sample numbers profiled in this study, including normal breast tissue, primary DCIS, and ipsilateral IBC relapses; 9/12 IBC samples were paired with primary DCIS samples.
(B) Primary DCIS samples consisted of two outcome groups: progressors, who recurred with ipsilateral invasive disease with a median of 9.1 years, and non-progressors, who never recurred within a median follow-up of 11.4 years.
Sequencing-based approaches have been used extensively over the last decade to identify molecular mechanisms that could explain the connection between DCIS and IBC. Genomic profiling has identified recurrent copy number variants that are more prevalent in high-grade DCIS lesions (Afghahi et al., 2015; Buerger et al., 1999; Fujii et al., 1996). Comparison of DCIS and IBC lesions from the same patient has provided clues into the clonal evolution from in situ to invasive disease by revealing genomic alterations that are acquired during this transition (Casasent et al., 2018; Kim et al., 2015; Newburger et al., 2013). To date, however, these findings have not consistently explained this transition. Similarly, the utility of tumor phenotyping by single-plex immunohistochemical tissue staining has been limited as well.
In light of this uncertainty, clinical management has favored treating all patients presumptively as progressors to IBC with surgery, radiation therapy, and pharmacological interventions, all of which carry risks for adverse events. Consequently, this approach is likely to be overly aggressive for patients who do not progress (non-progressors). Thus, understanding what drives DCIS to transition to IBC is a critical unmet need and opportunity for prevention. Surprisingly, despite all the information now known about the genetic and functional state of tumor cells in DCIS, histopathology remains the only reliable way to diagnose it. Thus, DCIS is an intrinsically structured entity for which the spatial orientation of tumor, myoepithelial, and stromal cells are defining characteristics.
To understand how DCIS structure and single-cell function are interrelated, we used tools previously developed by our lab for highly multiplexed subcellular imaging to analyze a large cohort of human archival tissue samples covering the spectrum of breast cancer progression, from in situ to invasive disease, in a spatially resolved manner (Keren et al., 2019; McCaffrey et al., 2020). In previous work, we used multiplexed ion beam imaging by time of flight (MIBI-TOF) to identify rule sets governing the tumor microenvironment (TME) structure in triple-negative breast cancer that were highly predictive of the composition of immune infiltrates, the expression of immune checkpoint drug targets, and 10-year overall survival (Keren et al., 2018). This effort provided a framework for how TME structure and composition could be used more generally as a surrogate readout to understand the functional response to neoplasia. With this in mind, we sought to determine to the extent to which similar themes involving myoepithelial, stromal, and immune cells in the DCIS TME might play pivotal roles in breast cancer progression. These cell types have been implicated previously in promoting local invasion (Gil Del Alcazar et al., 2017; Barsky and Karlin, 2005; Ibrahim et al., 2020), metastasis (Pelon et al., 2020; Shani et al., 2020), and correlation with clinical progression (Yang et al., 2018; Zhou et al., 2018).
Here, we report the first systematic, high-dimensional analysis of breast cancer progression using the Washington University Resource Archival Human Breast Tissue (RAHBT) cohort, a clinically annotated set of archived tissue from patients diagnosed with DCIS and IBC. Because the DCIS patient population is complicated by differences in age, parity status, tumor subtype, and treatment course, a well-conceived cohort design is crucial for identifying meaningful features amidst these confounding variables. The RAHBT cohort was therefore composed of primary DCIS tumors from women who later progressed to IBC that were matched by age and year of diagnosis with DCIS from women who did not have a subsequent ipsilateral breast event. We used MIBI-TOF and a 37-plex antibody staining panel to comprehensively define the cellular composition and structural characteristics in normal breast tissue, DCIS, and IBC relapses. These findings were corroborated by transcriptomic data acquired from adjacent co-registered tissue regions isolated by laser capture microdissection. We used the 433 parameters quantified in these analyses to build a random forest classifier for predicting which DCIS patients would later progress to IBC based on the original resection specimen. This classifier was heavily weighted for spatially informed parameters quantifying breast cancer TME structure, particularly those relating to ductal myoepithelium. Surprisingly, myoepithelial loss was more pronounced in samples from DCIS patients that did not recur and was typically associated with a more reactive stroma. Taken together, the studies reported here provide insight into potential etiologies of DCIS progression that will guide development of future diagnostics and serve as a template for how to conduct similar analyses of pre-invasive cancers.
RESULTS
A longitudinal cohort of DCIS patients with or without subsequent invasive relapse
The goal of this study was to explore two central questions of breast cancer progression. First, how does the structure, composition, and function of breast tissue change with progression from DCIS to IBC? Second, what distinguishes DCIS lesions in patients that later develop IBC (progressors) from those that do not (non-progressors)? To examine these questions, we mapped the phenotype, structure, and spatial distribution of tumor, myoepithelium, stroma, and immune cells of 79 archival formalin-fixed paraffin-embedded patient tissues from the RAHBT cohort (Figure 1A; Table S1).
Patient samples included normal breast tissue (N = 9, reduction mammoplasty), primary DCIS (N = 58), and IBC (N = 12). Of the 58 primary DCIS samples, 44 were from non-progressors (median follow-up = 11.4 years), while the remaining 14 were from progressors (median time to subsequent breast event = 9.1 years; Figure 1B). Importantly, all IBC tissues were ipsilateral breast events from patients with a prior diagnosis of DCIS, 9/12 of which were longitudinal samples that were matched to a progressor DCIS sample.
A single-cell phenotypic atlas of DCIS epithelium and its microenvironment
As part of the Human Tumor Atlas Network (HTAN) PreCancer Atlas, we created a multiomic atlas of breast cancer progression using co-registered adjacent serial sections cut from each RAHBT tissue microarray (TMA) block. For this study, these tissues were used for hematoxylin and eosin (H&E) histochemical staining, RNA transcriptome laser-capture microdissection (LCM-Smart-3SEQ), and highly multiplexed imaging (MIBI-TOF; Figure 2A). The location of DCIS-containing ducts in H&E sections were manually demarcated by a breast pathologist. This information was then used to guide spatial co-registration of LCM-Smart-3SEQ and MIBI-TOF analyses to ensure that the same ductal and stromal regions were sampled with each technique (Foley et al., 2019).
Figure 2. A single-cell phenotypic atlas of DCIS epithelium and its microenvironment.
(A) Depiction of the parallel tissue analysis methods used in this study, including H&E staining, laser-capture microdissection (LCM) of stroma and epithelium with RNA-seq, and MIBI-TOF with an overview of the MIBI-TOF workflow.
(B) Markers used in the MIBI-TOF panel, grouped by target cell type or protein class.
(C) Cell lineage assignments based on normalized expression of lineage markers (heatmap columns). Rows are ordered by absolute abundance (bar plot, left), while columns are hierarchically clustered (euclidean distance, average linkage). Myoep, myoepithelial cell; Mono, monocyte; Endo, endothelial cell; APC, antigen-presenting cell; Macs, macrophages; ImmOther, immune other; MonoDC, monocyte-derived dendritic cell; dnT, double-negative T cell; DC, dendritic cell.
(D) Representative MIBI image of a DCIS tumor with a nine-color overlay of major cell lineage markers. Inset showing the corresponding H&E image; scale bar: 100 μm. Pt., patient.
(E) A cell phenotype map (CPM) showing cell identity by color, as defined in C, overlaid onto the cell segmentation mask; scale bar: 100 μm.
(F) Region masks marking stroma (pink), myoepithelial (cyan), and ductal (blue) tissue regions; scale bar: 100 μm.
(G) Heatmap of normalized marker expression for four tumor cell subsets including luminal (CK7/PanCK/ECAD+), CK5/7-low (PanCK+, ECAD+ only), Basal (CK5/PanCK/ECAD+), and EMT (VIM/PanCK/ECAD+), with an accompanying bar graph of cell subset prominence.
(H) Images of DCIS tumors with diversity in tumor cell subsets including basal/luminal heterogeneity (left) and EMT tumor cells (right); scale bar, 100 μm.
(I) Heatmap of normalized marker expression for four fibroblast cell subsets including resting fibroblasts (VIM+ only, Resting), myofibroblasts (SMA/VIM+, Myo), cancer-associated fibroblasts (FAP/VIM+, CAFs), and normal fibroblasts (CD36/VIM+, Normal).
(J) Images of DCIS tumors with distinct stroma makeup of fibroblast subsets including normal fibroblast enriched (left) and CAF enriched (right); scale bar: 100 μm.
(K) Area plots of the frequency of tumor subsets (top), fibroblast subsets (middle), and immune lineages (bottom) in all DCIS, IBC, and normal patient samples profiled in this study. Tissue and PAM50 subtype are denoted by color in the top row.
MIBI-TOF imaging was performed on each RAHBT TMA using a 37-plex metal-conjugated antibody staining panel (Figure 2B; Figure S1; Table S2), acquiring one 500 × 500 μm region of interest per core. A deep learning pipeline (Mesmer) was subsequently used to annotate single cells in each image (mean = 875 cells per image, standard deviation = 316 cells; Figure S2; STAR Methods “low-level image processing” and “single-cell segmentation”) (Greenwald et al., 2021a; Keren et al., 2018; McCaffrey et al., 2020; Moen et al., 2019; Van Valen et al., 2016). We then used FlowSOM to identify tumor cells, fibroblasts, myoepithelium, endothelium, and 12 types of immune cells (Figure 2C; Figures S2C–S2H) (Van Gassen et al., 2015). Overall, we assigned 95% of segmented cells (n = 69,151 single cells) to one of these 16 cell classes that had an aggregate frequency range of 0.7%–58.3%; a robustness analysis of FlowSOM assignments can be found in Figures S3A–S3D comparing these cluster-based measurements to phenotypic assignments by manual gating. To examine how cell type and function varied with respect to tissue structure (Figure 2D), these data were combined to generate cell phenotype maps (Figure 2E) and tissue compartment masks (Figure 2F) demarcating the epithelium, stroma, and myoepithelium.
DCIS epithelial and stromal tissue compartments were predominantly composed of epithelial cells and fibroblasts, respectively, which were each composed of four major phenotypic subsets. Epithelial cells consisted of luminal (56.9% ± 33.7), basal (4.4% ± 6.6), epithelial-to-mesenchymal (EMT, 2.3% ± 2.8), and CK5/7-low (36.2% ± 33.5) subsets defined by variable expression of vimentin, CK7, and CK5 (Figure 2G, H, Figure S2G). Fibroblasts consisted of normal fibroblasts (12.1% ± 15), myofibroblasts (23.5% ± 16), resting fibroblasts (47% ± 20.3), and cancer-associated fibroblasts (CAFs; 17.4% ± 18.2 of fibroblasts) that were defined by variable coexpression of CD36, fibroblast activation protein (FAP), and smooth muscle actin (SMA) (Figures 2I and 2J; Figure S2H). Per-patient interrogation of epithelial, fibroblast, and immune cell subsets across DCIS, IBC, and normal breast revealed that all phenotypic subsets were observed in all tissue types, including ER−, HER2−, and AR-defined functional subsets, with primary DCIS tumors showing high interpatient heterogeneity in cellular and PAM50 subtype makeup (Figure 2K; Figures S4A–S4C). These data indicate that beyond the presence of myoepithelial cells, DCIS tumors have a diverse epithelial, stromal, and immune makeup that cannot be differentiated from IBC solely based on the presence of discrete cell types.
Transition to DCIS and IBC is marked by coordinated changes in the TME
In the previous section, we defined normal, DCIS, and IBC samples in terms of bulk cellular composition in a manner that was agnostic to the spatial location of each cell population. Next, to interrogate potential spatial differentiators of disease state, and to understand how tissue composition, cellular organization, and structure are interrelated, we augmented these compositional data with a description of the spatial distribution of each cell subset within the TME. First, to determine the proportion of each cell population residing within ductal or stromal regions, we used regional masks demarcating the epithelium and stroma to quantify the frequency of each cell type in these regions (“tissue compartment enrichment,” Figure 3A; Figure S5A; STAR Methods “single-cell phenotyping and composition”; note: due to loss of myoepithelium in IBC, this compartment was not analyzed in these samples). Next, we used two cell-cell proximity metrics—pairwise cell distances and cell neighborhoods—to capture preferential spatial interactions between discrete cell types (“cellular spatial enrichment analysis,” Figure 3A; Figure S5B; STAR Methods “region masking”). In addition to this more general cell-centric approach, we also developed custom tools for capturing specific morphologic and phenotypic attributes of the thin monolayer of myoepithelium-encapsulating ductal epithelial cells and the structure of stromal collagen (“TME morphometrics,” Figure 3A; Figures S5D–S5G; STAR Methods “myoepithelial morphology analysis, “myoepithelial pixel clustering analysis,” and “collagen morphometrics”). Taken together, this analysis yielded a digitized TME profile consisting of 433 parameters quantifying both the cellular composition and spatial structure of each patient sample.
Figure 3. Transition to DCIS and IBC is marked by coordinated changes in the TME.
(A) Schematic of the classes of spatial features quantified in all samples, including the measurement of cell type prevalence in specific tissue regions (1: Tissue compartment enrichment), the calculation of paired cell-cell spatial enrichment or spatially enriched cell neighborhoods (2: cell-cell proximity), and morphometric features of the myoepithelial layer and collagen fibers (3: morphometrics).
(B) Area plot of the distribution of each feature class in the features that significantly differ between normal breast tissue, DCIS, and IBC states by Kruskal-Wallis H test (p < 0.05).
(C) Column plot comparing the prevalence of each feature class in features that differ between tissue states, and total measured features.
(D) Heatmap of the distinguishing feature prevalence in normal breast tissue, DCIS, and recurrent IBC samples. K-means clustering separated features into four groups of distinct feature-enrichment patterns in the tissues states, including those highest in normal tissue and low in IBC (TME1: normal enriched), those highest in DCIS (TME2: DCIS enriched), and those highest in IBC and low in normal (TME3: IBC enriched). Features are organized by descending false-discovery rate Q value within each TME. Color indicates mean over tissue state, Z scored per feature across tissue states.
(E) Area plot of the distribution of the cellular compartment of the distinguishing features in each TME cluster.
We then compared these profiles for normal, DCIS, and IBC tissues to address our first question: how do the composition and structure of the TME change with progression to IBC? We applied the Kruskal-Wallis H test to discern which aspects of tissue composition and structure were significantly distinctive of each clinical group (p < 0.05; Table S3; STAR Methods “distinguishing feature analysis”). This analysis identified 137 parameters that were preferentially enriched or depleted in normal, DCIS, or IBC tissue, with spatially agnostic (cell type, cell state) and spatially informed metrics accounting for 39% and 61% of differentially expressed parameters, respectively (Figure 3B; Figure S5H; Table S3). Notably, all three categories of spatially informed parameters were overrepresented. For example, morphometrics were 3-fold enriched, accounting for 16% of distinguishing parameters but only 5% of all parameters (Figure 3C).
To organize distinguishing features into interpretable TME signatures, we performed k-means clustering to yield four clusters defining the breast tissue states: TME1, TME2, and TME3 uniquely distinguished normal, DCIS, and IBC samples, respectively, and TME4 consisted of features that were specifically depleted in DCIS samples (Figure S5I; Table S3). Not surprisingly given its enrichment in normal breast, TME1 was typified by myoepithelium with high cellularity, thickness, and continuity (Figure 3D; Table S3) (Ding et al., 2019). Additionally, this robust myoepithelial layer in TME1 was paired with elevated CD36 expression in endothelium and immune cells (Figure 3D; TME1 “CD36+ immune and endothelial cells”), consistent with normative lipid metabolism in homeostatic breast tissue. TME2 was specifically enriched in DCIS tumors and was typified by increased myoepithelial proliferation (%Ki67+), stromal mast cells, and CD4 T cells. Notably, TME2 contained the highest proportion of tumor and myoepithelial parameters (Figure 3D; TME3 “pS6+, CK5+, Ki67+ myoepithelium”), suggesting that the transition to in situ disease involves a coordinated shift in the function of these two lineages (Figure 3E). IBC-enriched TME3 was stroma-predominant (50%) and had surprisingly few distinctive tumor parameters (4%; Figure 3E; Table S3).
Along these lines, we noted when comparing TME2 and TME3 that—aside from the pathognomonic loss of ductal myoepithelium—the most distinctive property delineating DCIS from IBC samples was an increase in stromal desmoplasia (collagen deposition, CAF frequency, and proliferation). To further evaluate whether these trends reflected changes specific to the interval between a new DCIS diagnosis and ipsilateral invasive relapse, we compared these parameters in a subset of sample pairs in which both DCIS and IBC tissue had been procured longitudinally from the same patient (N = 9). We found that the degree of statistical significance in this lesser-powered pairwise analysis and the larger unpaired analysis were linearly correlated (R2 = 0.58, p = 3E-15) and that the salient trends reflected in TME2 and TME3 occurred at the patient level (Figure 4A; Table S3). These significant longitudinal changes included a reduction in mast cells, resting fibroblasts, and normal fibroblasts in the stroma between paired patient samples (Figure 4B), reflecting a transition where normal fibroblasts in primary DCIS samples (Figure 4C, green arrows) were supplanted by CAFs (Figure 4C, pink arrows) in patients’ subsequent later invasive breast events (Figure S6A).
Figure 4. Increased desmoplasia and ECM remodeling distinguish primary DCIS from their IBC recurrence.
(A) Paired vertical scatterplot of the stromal density of mast cells in the primary DCIS diagnosis and subsequent IBC recurrence in individual patients; paired Mann-Whitney test.
(B) The stromal density of normal fibroblasts is compared in longitudinal samples from single patients as in (A).
(C) Representative MIBI image overlays showing the primary DCIS diagnosis (left) and invasive recurrence (right) from patient 1023. Green arrows, normal fibroblasts, orange arrows, CAFs; scale bar: 100 μm.
(D) Example of dense MIBI collagen signal, collagen fiber object segmentation, and subsequent fiber area and orientation measurement, with fiber-fiber alignment denoted by fiber color.
(E) Scatterplot comparing summed stromal density of CAFs and myofibroblasts versus collagen fiber density.
(F) Volcano plot of ECM-related gene expression for the top and bottom CAF-enriched DCIS tumors.
To quantify how this shift in fibroblast phenotype relates to the extent of stromal desmoplasia, we compared the shape, length, and density of individual collagen fibers with CAF location, frequency, and phenotype (Figure 4D, Figures S6B–S6D; STAR Methods “collagen morphometrics”). Collagen fiber density was linearly correlated with the presence of stromal CAFs and myofibroblasts (R2 = 0.4; Figure 4E), suggesting a direct relationship between CAF activation and the extent of collagen fibrillization. Finally, to identify changes in the proportion of collagen isoforms accompanying CAF activation, we compared transcript levels in stroma of CAF high- and low-density tumors using LCM RNA sequencing (RNA-seq). The majority of collagen species were upregulated in CAF-high tumors with COL5A2, COL3A1, and COL1A1 (p < 0.01; Figure 4F; Table S3). In addition, CAF-high tumors showed increased deposition of fibronectin (FN1; p < 0.05), SPARC (p < 0.01), and periostin (POSTN; p < 0.01), which have been shown to promote a pro-invasive stromal niche (Barth et al., 2005; Malanchi et al., 2011).
Identifying DCIS features correlated with risk of invasive progression
We next leveraged both spatially informed and agnostic parameters to examine our second central question: what distinguishes DCIS lesions that later progress to IBC from those that do not? We compared tissue procured at the time of diagnosis in two sets of patients with primary DCIS. The first set, referred to as “progressor,” consisted of 14 patients who had a subsequent ipsilateral invasive recurrence following a diagnosis of pure DCIS (median time to recurrence = 9.1 years). The second set, referred to as “non-progressor,” consisted of 44 patients with pure DCIS that did not have a breast event following tumor resection (median time of follow = 11.4 years).
To identify predictive features of the TME, we trained a random forest classifier to predict which patients would relapse with invasive disease based on cell-type prevalence, tissue compartment enrichment, cell-cell proximity, and morphometrics for each sample (Figure 5A; Table S1). Although sample size precluded us from being able to eliminate patient demographics and differences in clinical therapy as confounders in this analysis, treatment regimens known to affect recurrence rates (mastectomy, radiation, tamoxifen) were well distributed between the progressor and non-progressor patients (Figure S6E). Likewise, no significant differences in classifier predictions were identified with respect to these variables (Figure S6F).
Figure 5. Identifying DCIS features correlated with risk of invasive progression.
(A) Schematic of the outcome groups of primary DCIS: “progressors,” who recurred with ipsilateral IBC, and “non-progressors,” who showed no recurrence within 11 years of follow-up. MIBI features (N = 433) of numerous feature classes were used to train a random forest classifier to differentiate progressor and non-progressor samples. Classifier specificity was then tested on a withheld set of 20% of patients in a test group.
(B) AUC plot of classifier sensitivity and specificity.
(C) Classifier accuracy is compared for 10 runs with known progressor/non-progressor labels and 10 runs with randomly permuted progressor/non-progressor labels. p = 0.02, Wilcoxon signed rank test.
(D) Bar plot of features with top classifier importance ranked by average Gini importance across the unpermuted 10 runs. Orange, enriched for progressors; green, enriched for non-progressors. The parent feature class for each feature is shown and whether that class leveraged spatial information.
(E) Column plot of the sum of Gini importance of features separated by their corresponding cellular compartment.
After removing sparse and overly correlated parameters, we randomly split the patient population 80/20 into training and test sets, respectively (Figure S6G). We evaluated classifier accuracy in the withheld test set, where the model achieved an area under the curve (AUC) of 0.74 (Figure 5B). To control for variation due to the random partitioning of training and test sets, we repeated this approach with 10 different seeds, resulting in 10 different training test partitions, and maintained a median AUC of 0.74 (Figure 5C). For additional rigor, we trained classifiers on randomly permuted patient group labels for each seed and compared the distribution of resultant AUCs to the unpermuted models. Pairwise comparison of these replicates demonstrated significantly superior accuracy when using unpermuted data (median AUC of 0.74 [red] versus 0.48 [blue], p = 0.02), demonstrating that the model’s predictive power is predicated on the distinct biological features of progressors and non-progressors.
To understand the biology being leveraged by the model to accurately discriminate pre-invasive from indolent DCIS tumors, we ranked the top 20 features based on Gini importance (Table S4). These features primarily consisted of metrics related to the phenotype of myoepithelium and the spatial distribution of multiple immune cell subsets (Figures 5D and 5E). Notably, spatially informed metrics describing cell densities, cell neighborhoods, pairwise cell distances, collagen structure, and multiplexed subcellular features were overrepresented and accounted for 15 of the top 20 Gini-ranked metrics in the model, while representing less than half of total measured features (Figures S6H and S6I; Table S4.
Myoepithelial breakdown and phenotypic change between progressors and non-progressors
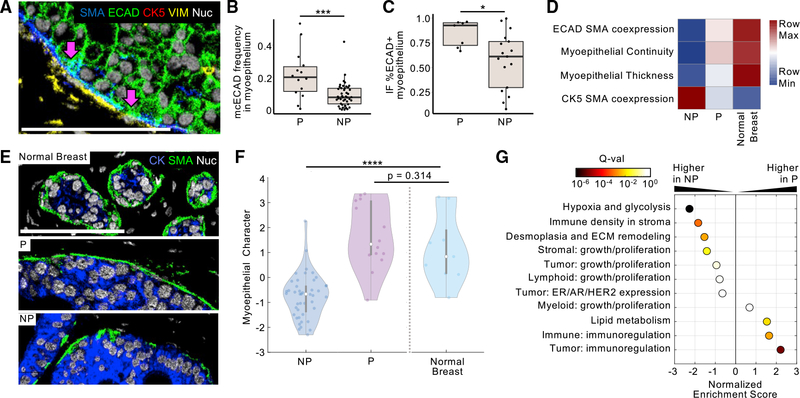
In the above analysis, myoepithelial structure and phenotype were overrepresented among the top Gini-ranked classifier features (Figure 5D), with myoepithelial expression of E-cadherin (ECAD) being the most discriminative feature. This parameter quantifies ECAD coexpression at the pixel level exclusively in periductal SMA-positive pixels (Figure 6A, pink arrows) and was significantly elevated in progressor samples (p = 0.001; Figure 6B; Figures S7A–S7C). We validated this finding using multi-color immunofluorescence for ECAD and SMA. Pixel-level coexpression in immunofluorescence measurements was higher in progressors than non-progressors (p = 0.034) and was well correlated with patient-matched values attained by MIBI (Figure 6C; Figures S7D and S7E).
Figure 6. Myoepithelial breakdown and phenotypic change between progressors and non-progressors.
(A) Representative MIBI image overlay of a DCIS progressor tumor with ECAD coexpression in the SMA+ myoepithelium; scale bar: 100 μm.
(B) Boxplot comparing the frequency of ECAD+/SMA+ myoepithelial coexpression cluster in progressor (P) and non-progressor (NP) tumors. ***p < 0.001, *p < 0.05, Mann-Whitney test.
(C) Boxplot comparing the frequency of the ECAD+ myoepithelium in immunofluorescence analysis between P and NP tumors.
(D) Heatmap of select myoepithelial feature prominence in NP tumors, P tumors, and normal breast tissue.
(E) Representative images of myoepithelial integrity in normal breast tissue, a P DCIS tumor, and a NP tumor.
(F) Violin plot of the distribution of linear discriminate analysis-derived “myoepithelial character” values in NP and P tumors as well as normal breast tissue; Kruskal-Wallis test.
(G) Gene set enrichment analysis of all measured features was used to determine which tissue feature ontologies were enriched in tumors with high or low myoepithelial character scores. Normalized enrichment score is given for each feature ontology; points are colored by significance (false discovery rate Q value).
In our analyses comparing normal tissue, DCIS, and IBC, we observed the highest myoepithelial ECAD expression in normal breast tissue (Figure 3; Table S3). To our surprise, on comparing normal samples with respect to DCIS clinical subgroups, we found that ECAD expression in normal ductal myoepithelium was more similar to progressor samples than non-progressor samples (Figure 6D). A similar trend was observed with other morphologic and phenotypic properties: progressor DCIS samples more closely resembled normal samples than non-progressor samples. For example, myoepithelium in non-progressors was thinner and less continuous than in progressor and normal samples (Figures 6D and 6E). To examine this difference more comprehensively, we trained a linear discriminant analysis model to differentiate progressors and non-progressors using all myoepithelial parameters exclusively, with only DCIS samples in the training set (STAR Methods “myoepithelial features LDA”). Composite scores (myoepithelial character) for DCIS samples calculated with the resultant model proficiently separated progressors from non-progressors (progressor mean = 1.65 ± 1.32, non-progressor mean = −0.75 ± 0.88; Figure 6F, left). We then used the trained model to quantify the myoepithelial character of normal samples. In line with Figure 6D, normal breast samples diverged significantly from non-progressor samples (p = 2.64E-4) but were statistically indistinguishable from progressor samples (p = 0.314; Table S5).
These data suggest that the loss of normal-like features, reflected in myoepithelial character composite scores, serves a protective function in non-progressors in preventing IBC relapse. To understand how this loss might influence recurrence outcomes, we used a method derived from gene set enrichment analysis to identify ontologies that were correlated with high or low myoepithelial character (Table S5; STAR Methods “feature ontology enrichment analysis”). Low scores typical of non-progressors were enriched for parameter ontologies relating to hypoxia, glycolysis, stromal immune density, and desmoplasia/remodeling of the extracellular matrix (ECM; Figure 6G; Table S5). Conversely, high myoepithelial character scores typically seen in progressors were enriched for immunoregulatory marker expression (PDL1, IDO1, COX2, PD1) in tumor and immune cells (Figure 6G; Table S5). Taken together, these results suggest that myoepithelial loss serves a protective, tumor-sensing function that favors fibroblast and immune-cell activation in the surrounding stroma.
DISCUSSION
Here, we report the first spatial atlas of breast cancer progression. The central focus of this study was to characterize features in primary DCIS that are associated with risk of invasive relapse, where tumor cells have breached the duct and invaded the surrounding stroma. Previous work examining breast cancer progression has attributed this transition either to tumor-intrinsic factors (Bartova et al., 2014; Fujii et al., 1996; Perez et al., 2015; Rakovitch et al., 2012) or to specific features of stromal cells in the surrounding TME (Aguiar et al., 2015; Gil Del Alcazar et al., 2017; Aponte-López et al., 2018; Ding et al., 2019; Sprague et al., 2021). By simultaneously mapping both of these entities in intact human tissue, we sought to treat the DCIS TME as a single ecosystem in which progression to invasive disease depends on an evolving spatial distribution and function of multiple cell types, rather than on any single cell subset.
Meeting this goal required first assembling a large, well-annotated, and diversified pool of human breast cancer tissue: the RAHBT cohort. This effort was motivated in part by the success of similar works investigating invasive disease (Molecular Taxonomy of Breast Cancer International Consortium, The Cancer Genome Atlas) that have provided deep insights into breast tumor composition and have served as authoritative resources in breast cancer research (Cancer Genome Atlas Network, 2012; Curtis et al., 2012). The Breast PreCancer Atlas constructed a unique set of archival human surgical resections that captured the full spectrum of breast cancer progression, from normal tissue, to primary DCIS, and onto patient-paired ipsilateral IBC recurrences. Here, assembling all these cases into TMAs has enabled a one-of-a-kind workflow for multiomics analyses in which genomic, transcriptomic, and proteomic techniques are performed not only on the same samples but on co-registered serial sections of the same local region of tissue.
Here, we analyzed these TMAs using MIBI-TOF and a 37-marker staining panel to map breast cancer progression and to understand why some patients with DCIS relapse with invasive disease while others do not. Our results suggest that coordinated transformation of ductal myoepithelium and surrounding stroma plays a central role in determining clinical outcome by establishing a tumor-permissive niche that favors local invasion. Relative to normal tissue, the thin myoepithelial layer in DCIS samples was less phenotypically diverse and more proliferative (Figure 3D). Curiously, these changes were accompanied by an influx of stromal CD4 T cells and mast cells that subsequently declined in IBC. Aside from the canonical loss of myoepithelium, stromal desmoplasia in IBC was the most consistent, distinctive aspect of invasive progression and was marked by higher numbers of proliferating CAFs and densely aligned fibrillar collagen (Figure 4) (Conklin et al., 2011; Esbona et al., 2018; Friedman et al., 2020).
Typified changes in TME structure and function were not only discriminative of DCIS and IBC but also separated DCIS progressors from non-progressors. Using 433 spatial and compositional parameters drawn exclusively from original primary DCIS samples, we built a random forest classifier model to predict which patients would relapse with an ipsilateral invasive tumor following initial DCIS diagnosis (AUC = 0.74, p = 0.02). On examining the relative weighting given to each parameter in the model, two compelling and overarching insights emerged. First, spatially informed metrics relating cell function to structure and morphology were significantly over-represented relative to non-spatial metrics. Second, the most influential features were primarily related to myoepithelium and stroma rather than to the tumor cells themselves.
Given its loss in IBC, ductal myoepithelium has long been thought to act as a barrier that deters local invasion by partitioning in situ carcinoma cells away from the surrounding stroma (Barsky and Karlin, 2005; Jones et al., 2003; Sirka et al., 2018). Initially, we hypothesized that a more intact and robust myoepithelial barrier resembling normal breast tissue would be protective against invasive progression. Surprisingly, however, our data seem to suggest the opposite: DCIS samples with more continuous myoepithelium and high ECAD expression were at higher risk of ipsilateral invasive recurrence following primary DCIS surgical excision. Retention of these normal-like myoepithelial traits correlated with fewer stromal immune cells and CAFs (Figure 6G). Conversely, the thin, discontinuous, low-ECAD myoepithelium present in non-progressor tumors was correlated with a more reactive desmoplastic stroma with more immune cells, CAFs, and collagen remodeling. Given the relationships uncovered here between myoepithelial integrity and reactive stromal, our observations are consistent with a model in which a compromised myoepithelial barrier promotes stromal sensing of tumor, which provides protection against future invasive relapse.
Taken together, the analyses reported here deliver a comprehensive, multi-compartmental atlas of preinvasive breast cancer that illustrates the full continuum of tissue structure and function starting from a homeostatic state in normal breast through in situ and invasive disease, including matched longitudinal samples. Combining this comprehensive dataset with extensive patient follow-up has enabled identification of tumor features that are associated with risk of invasive relapse in DCIS patients and offers a framework for exciting follow-on efforts.
Limitations of study
Limitations with respect to the patient cohort, imaging methodology, data analysis, and results interpretations should be noted in framing the significance of this work. First, while our long-term goal is to determine risk factors that promote DCIS progression in the absence of surgical intervention, all tissue analyzed here was procured from women who underwent surgical intervention for their disease. Additionally, the sample size used in this study, particularly when examining longitudinal changes in DCIS patients that progressed to IBC, are relatively modest and were procured from a single medical center. Consequently, the possibility that statistically significant features attributed here to inherent differences in DCIS and IBC are instead driven by sampling bias cannot be definitively ruled out. Thus, follow-up on multi-center studies examining these factors in patients undergoing or forgoing surgical intervention will be needed to test this hypothesis and assess its clinical utility for prospective risk stratification.
With respect to cell identification in tissue sections, accurate cell enumeration in two dimensional images can be confounded by partial overlap of cellular features that are difficult to discern without spatial information in the z-dimension. Cell classification and clustering introduce another potential source of variance as well. Unsupervised approaches where the number of cell clusters is arbitrarily specified by the user can be prone to overclustering and misclassification of low-frequency cell populations. In this work, we attempted to mitigate these issues using a hybrid approach where iterative rounds of FlowSOM were used to hierarchically stratify cell populations based on well-vetted multiparameter phenotypes. To assess robustness, we have included in the supplemental information a comparison of these estimates with those attained by manual gating, which did not reveal material differences between the two approaches. To enable further interrogation of this work as new tools for computational image analysis become available, we have made all images, cell masks, tissue masks, and data tables publicly available.
Finally, key questions remain of how myoepithelial integrity is causally related to disease recurrence in progressors. While we find correlations between low myoepithelial integrity and increased stromal immune infiltration, collagen deposition, and CAFs, due to the observational nature of this study we cannot determine if myoepithelial breakdown triggers this shift in the tumor microenvironment or vice versa. In future studies we plan to confirm this relationship in a larger independent cohort of DCIS patients who progressed to invasive disease and to probe its association with stromal desmoplasia in functional models.
STAR★METHODS
RESOURCE AVAILABILITY
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by corresponding author, and lead contact, Michael Angelo (mangelo0@stanford.edu).
Materials availability
This study did not generate new unique reagents.
Data and code availability
All single channel images and area masks are present as single Tiffs in this public Mendeley Data repository. The DOI is listed in the key resources table. Accession numbers are listed in the key resources table.
All original code has been deposited at Mendeley and is publicly available as of the date of publication. DOIs are listed in the key resources table.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
KEY RESOURCES TABLE.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
|
| ||
| Antibodies | ||
|
| ||
| A full list of antibodies is provided in Table S2 | N/A | N/A |
|
| ||
| Biological samples | ||
|
| ||
| The Resource of Archival Breast Tissue (RAHBT) cohort TMAs were compiled at Washington University in St. Louis, all patient information is included in Table S1 | N/A | N/A |
|
| ||
| Chemicals, peptides, and recombinant proteins | ||
|
| ||
| TBS IHC Wash Buffer with Tween 20 | Cell Marque | Cat#935B-09 |
| PBS IHC Wash Buffer with Tween 20 | Cell Marque | Cat#934B-09 |
| Target Retrieval Solution, pH 9, (3:1) | Agilent (Dako) | Cat#S2375 |
| Avidin/Biotin Blocking Kit | Biolegend | Cat#927301 |
| Gelatin (cold water fish skin) | Sigma-Aldrich | Cat#G7765–250 |
| Xylene Histological grade | Sigma-Aldrich | Cat#534056–500 |
| Glutaraldehyde 8% Aqueous Solution EM Grade | EMS | Cat#16020 |
| Normal Donkey serum | Sigma-Aldrich | Cat#D9663–10ML |
| Bovine Albumin (BSA) | Fisher | Cat#BP1600–100 |
| Centrifugal filters (0.1 μm) | Millipore | Cat#UFC30VV00 |
|
| ||
| Critical commercial assays | ||
|
| ||
| MIBItag Conjugation Kit | IONpath | Cat#600XXX |
| ImmPRESS UNIVERSAL (Anti-Mouse/Anti-Rabbit) IgG KIT (HRP) | Vector Laboratories | Cat#MP-7500–15 |
| ImmPACT DAB (For HRP Substrate) | Vector Laboratories | Cat#SK-4105 |
|
| ||
| Deposited data | ||
|
| ||
| All image data, single cell data, and tissue feature data, is located in a public Mendeley data repository: https://data.mendeley.com/datasets/d87vg86zd8 | Mendeley Data | https://data.mendeley.com/datasets/d87vg86zd8 |
|
| ||
| Software and algorithms | ||
|
| ||
| Data analysis was done using MATLAB 2016 | Mathworks | N/A |
| Data analysis was done using R 3.6.1 | R | N/A |
| Data analysis was done using Python | Python | N/A |
| Analysis code for MATLAB, R, and Python is available at the Mendeley data and code repository: https://data.mendeley.com/datasets/d87vg86zd8 | Mendeley Data | https://data.mendeley.com/datasets/d87vg86zd8 |
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Patient cohort
We utilized a retrospective study cohort of patients from the Washington University Resource of Archival Tissue (RAHBT). The study was approved by the Washington University in St. Louis Institutional Review Board (IRB ID #: 201707090).
The RAHBT cohort includes women ages 18 and older with documented cases of premalignant breast disease (DCIS) and contained two outcome groups: non-progressors, which was composed of patients with DCIS who had no new breast event following resection (median follow-up = 11.4 years), and progressors, which was composed of patients with DCIS who had a new ipsilateral invasive breast cancer event following primary DCIS resection (median time to new event = 9.1 years). For each progressor, we matched two non-progressors who remained free from recurrent lesions, based on age at diagnosis (±5 years) and type of definitive surgery (mastectomy or lumpectomy).
Table S1 summarizes the data for the patients in the cohort. Patients with cancer diagnosis prior to qualifying premalignant lesions were excluded from the study. After exclusion, the study included samples from 70 patients, with a median age at diagnosis of 54 years, diagnosed between 1986 to 2017. Median time to recurrence was 9.1 years for invasive lesions and 5.3 years for pre-malignant lesions. For women in the cohort with no recurrence, follow-up extended to 132 months, on average. Treatment of initial DCIS ranged from lumpectomy with radiation (approximately half of cases), lumpectomy with no radiation (20%), and mastectomy with no radiation (30%). The RAHBT cohort is composed of African American women (26%) and white women (74%). Patient ages and additional clinical data are provided in Table S1.
METHOD DETAILS
TMA construction
For each DCIS diagnosis, we retrieved primary and recurrent tumor slides and blocks for pathology review, secured a whole slide image of each sample, marked for TMA cores, and generated TMA blocks with 84 1.5-mm cores, including additional tonsil and normal breast tissue sourced from reduction mammoplasty. Serial sections (5 μm) of each TMA slide were cut onto glass slides for hematoxylin and eosin (H&E) staining, onto laser-capture slides for LCM-RNaseq (SMART-3SEQ) and cut onto gold- and tantalum-sputtered slides for MIBI-TOF imaging. H&E slides were inspected by a breast cancer pathologist to address DCIS purity and to demarcate regions of DCIS to guide MIBI imaging and laser dissection of epithelial and stromal area.
Antibody Preparation
Antibodies listed in the Key Resources Table Metal Conjugated Antibodies were conjugated to isotopic metal reporters as described previously (Keren et al., 2018; McCaffrey et al., 2020). Following conjugation, antibodies were diluted in Candor PBS Antibody Stabilization solution (Candor Bioscience). Antibodies were either stored at 4°C or lyophilized in 100 mM D-(+)-Trehalose dehydrate (Sigma Aldrich) with ultrapure distilled H2O for storage at −20°C. Prior to staining, lyophilized antibodies were reconstituted in a buffer of Tris (Thermo Fisher Scientific), sodium azide (Sigma Aldrich), ultrapure water (Thermo Fisher Scientific), and antibody stabilizer (Candor Bioscience) to a concentration of 0.05 mg/mL. Some metal-conjugated antibodies in this study were used as secondary antibodies targeting hapten groups on hapten-conjugated primary antibodies, including the pairs PDL1-Biotin and Anti-Biotin149Sm, and ER-Alexa488 and Anti-Alexa488142Nd, see Key Resources Table for antibody vendor information.
Tissue Staining
Tissues were sectioned (5 μm thick) from tissue blocks on gold- and tantalum-sputtered microscope slides. Slides were baked at 70°C overnight followed by deparaffinization and rehydration with sequential washes in xylene (3x), 100% ethanol (2x), 95% ethanol (2x), 80% ethanol (1x), 70% ethanol (1x), and ddH2O with a Leica ST4020 Linear Stainer (Leica Biosystems). Tissues next underwent antigen retrieval by submerging sides in 3-in-1 Target Retrieval Solution (pH 9, DAKO Agilent) and incubating them at 97°C for 40 min in a Lab Vision PT Module (Thermo Fisher Scientific). After cooling to room temperature, slides were washed in 1x phosphate-buffered saline (PBS) IHC Washer Buffer with Tween 20 (Cell Marque) with 0.1% (w/v) bovine serum albumin (Thermo Fisher). Next, all tissues underwent two rounds of blocking, the first to block endogenous biotin and avidin with an Avidin/Biotin Blocking Kit (Biolegend). Tissues were then washed with wash buffer and blocked for 1 h at room temperature with 1x TBS IHC Wash Buffer with Tween 20 with 3% (v/v) normal donkey serum (Sigma-Aldrich), 0.1% (v/v) cold fish skin gelatin (Sigma Aldrich), 0.1% (v/v) Triton X-100, and 0.05% (v/v) sodium azide. The first antibody cocktail was prepared in 1x TBS IHC Wash Buffer with Tween 20 with 3% (v/v) normal donkey serum (Sigma-Aldrich) and filtered through a 0.1-μm centrifugal filter (Millipore) prior to incubation with tissue overnight at 4°C in a humidity chamber. Following the overnight incubation slides were washed twice for 5 min in wash buffer. On the second day, antibody cocktail was prepared as described above and incubated with the tissues for 1 h at 4°C in a humidity chamber. Following staining, slides were washed twice for 5 min in wash buffer and fixed in a solution of 2% glutaraldehyde (Electron Microscopy Sciences) in low-barium PBS for 5 min. Slides were sequentially washed in PBS (1x), 0.1 M Tris at pH 8.5 (3x), ddH2O (2x), and then dehydrated by serially washing in 70% ethanol (1x), 80% ethanol (1x), 95% ethanol (2x), and 100% ethanol (2x). Slides were dried under vacuum prior to imaging.
MIBI-TOF Imaging
Imaging was performed using a MIBI-TOF instrument (IonPath) with a Hyperion ion source. Xe+ primary ions were used to sequentially sputter pixels for a given field of view (FOV). The following imaging parameters were used: acquisition setting: 80 kHz; field size: 500×500 μm, 1024 × 1024 pixels; dwell time: 5 ms; median gun current on tissue: 1.45 nA Xe+; ion dose: 4.23 nAmp h/mm2 for 500×500 μm FOVs. For each FOV, Mass-spec pixel data were then converted to TIFF images where the counts for each mass were taken between the ‘Start’ and ‘Stop’ values defined in Table S2.
QUANTIFICATION AND STATISTICAL ANALYSIS
Low-level Image Processing
Multiplexed images were pre-processed using an image processing pipeline previously developed in our lab specifically for multiplexed mass based images – MAUI (https://doi.org/10.1371/journal.pcbi.1008887, software publicly available at https://github.com/angelolab/MAUI). MAUI includes several steps:
Subtracting background from bare regions in the slide and gold, using empty mass channels and the gold channel as reference.
Noise removal, by filtering out very sparse and pixelated signals.
Aggregate filtering-removal of antibody aggregates recognized as very small, connected components in the image.
All user defined parameters used for this pipeline can be found in Table S2.
Single-cell Segmentation
Cell segmentation was performed on pre-processed images using deep learning based software previously developed in our lab, Mesmer (Greenwald et al., 2021b), publicly available at https://www.deepcell.org/predict. The input to Mesmer is a two-channel image containing a nuclear marker in one channel and membrane or cytoplasmic markers in the other to accurately delineate single cell nuclei. For the cell nuclei channel we combined HH3 and endogenous phosphorous (P) signal, and a combination channel of E-cadherin, PanCK, CD45, CD44, and GLUT1 was used as the membrane channel input. To more effectively capture the range of cell shapes and morphologies present in DCIS, we generated two distinct segmentation parameter sets optimized for non-epithelial and epithelial cells, that were then combined for final cell segmentation. The non-epithelial settings used a radial expansion of two pixels from the nuclear border detected by Mesmer to generate a cell object, and a stringent threshold for splitting cells (Figure S2, Stroma Parameters). The epithelial settings used a radial expansion of three pixels and a more lenient threshold for splitting cells (Figure S2, Epithelial Parameters). We combined these masks using a post-processing step that gave preference to the epithelial segmentation objects, overriding stromal-parameter-detected objects in the same area. Smaller cells identified by the stromal settings and missed in the epithelial settings were combined to the final cell segmentation mask. All segmentation masks are publicly available with the raw images on Mendeley, see resource table for DOI.
Single-cell Phenotyping and Composition
Single-cell expression of each marker was measured through total signal counts in each cell object, normalized by object area. Single-cell data were then linearly rescaled by the average cell area across the cohort, and subsequently asinh-transformed with a co-factor of 5. All mass channels were scaled to 99.9th percentile.
In order to assign each cell to a lineage and subsequent cell type, the FlowSOM clustering algorithm was used in iterative rounds with the Bioconductor “FlowSOM” package in R (v.1.16.0, Van Gassen et al., 2015). The first clustering round separated cells into 100 clusters (xdim = 10, ydim = 10), which were assigned to one of five major cell lineages based on well-established combinations of lineage marker expression, including: epithelial cells (PanCK+, ECAD+, CD45−, CK7+/−, VIM+/−), myoepithelial cells (SMA+, CD45−, PanCK+/−, ECAD+/−, CK5+/−, VIM+/−), fibroblasts (VIM+, PanCK−, ECAD−, CK7−, CD45−, SMA+/−, FAP+/−, CD36+/−), endothelial cells (CD31+, VIM+, PanCK−, ECAD−, CK7−, CD45−, SMA+/−), and immune cells (CD45+, PanCK−, ECAD−). Accurate lineage assignment was assessed by reviewing cells from each FlowSOM cluster in image overlays of lineage-defining markers. In clusters with rare, non-canonical combinations of marker expression, cluster assignments were extensively reviewed across images of various tissue types with pathologist assistance, utilizing morphometric and histological organization features in addition lineage marker expression to accurately phenotype the cells. See Figure S2E for examples of cell reassignment.
Following lineage assignment, each lineage was subclustered to identify immune cell types including B cells (CD20+, CD4+/−), CD4 T cells (CD4T; CD3+, CD4+, CD8−/low), CD8 T cells (CD8T; CD3+, CD8+, CD4−/low), monocytes (Mono; CD14+, CD11c−, CD68−, CD3−), monocyte-derived dendritic cells (MonoDCs; CD14+, CD11c+, HLADR+, CD68−, CD3−), dendritic cells (DCs; CD11c+, HLADR+, CD3−), macrophages (Macs; CD68+, HLADR+, CD14+/−), mast cells (Mast; Tryptase+), double-negative T cells (dnT; CD3+, CD4−, CD8−), and HLADR+ APC cells (APC; HLADR+, CD45+/low). CD45+-only immune cells were annotated as “immune other.” Neutrophils were rare in the dataset; they were assigned last based on the positivity threshold (> 0.25) of MPO expression in immune cells. Tumor and fibroblast cells were similarly subclustered to reveal phenotypic subsets, including luminal (ECAD+, PanCK+, CK7+), basal (ECAD+, PanCK+, CK5+), epithelial-to-mesenchymal (EMT; ECAD+/−, PanCK+, VIM+), CK5/7-low (ECAD+, PanCK+) tumor cells, and normal (VIM+, CD36+), myo− (VIM+, SMA+), resting (VIM+ only), and CAF (VIM+, FAP+) fibroblasts (Figure S2). Overall, we assigned 94% (N = 127,451 of 134,631) of cells to 16 subsets, with the remaining nucleated cells with absent or very low levels of lineage markers assigned as “other.”
Comparative manual cell assignments were performed with Cytobank software (Cytobank.com) using nuclear intensity and area gates to define single cells, and iterative gating of established markers in biaxial plots that define the major cell lineages, cell types of each lineage, and phenotypic subsets of tumor and fibroblast cells as shown in Figure S3. A table of all single cells in this study, their marker expression and their assigned lineage using both FlowSOM and manual gating is available on Mendeley Data, see Key Resources Table for DOI.
Throughout this work cellular data are presented as 1) the frequency of a cell type of its parental lineage across the entire image (e.g., luminal tumor cells as % of total tumor cells in image), 2) a cell type’s density within a particular compartment of the image (e.g., 50 fibroblasts per mm2 of stroma (see Region Masking for compartment definition)), or 3) for immune cells, the frequency of immune cell types (of total immune) calculated for both epithelial and stromal regions (e.g., % macrophages of total epithelial immune). To calculate myoepithelial cell density, the number of cells phenotyped as myoepithelium in each image is normalized by the area of the myoepithelial mask in that image.
Region Masking
Region masks were generated to define histologic regions of each FOV including the epithelium, stroma, myoepithelial (periductal) zone, and duct. We removed gold-positive areas, which marked regions of bare slide from holes in the tissue, providing an accurate measurement of tissue area. This area measurement was used to calculate cellular density in specific histologic regions (e.g., fibroblast density in the stroma) to normalize observed cell abundances by the amount of tissue sampled.
The epithelial mask was first generated though merging the ECAD and PanCK signals and applying smoothing (Gaussian blur, radius 2 px) and radial expansion (20 px) to incorporate the myoepithelial zone; the insides of ducts were filled. The stromal mask included all of the image area outside of the epithelial mask. Duct masks were generated through the erosion of the epithelial masks by 25 px. The myoepithelial mask was generated by subtracting the duct mask from the epithelial mask, leaving a ~15 μm-wide periductal ribbon following the duct edge. To calculate the area in each mask, a bare slide mask was generated from the gold (Au) channel and this area was removed from the measurement, and pixel area was converted to mm2 of tissue.
Cellular Spatial Enrichment Analyses
A spatial enrichment approach was used as previously described (Keren et al., 2018, 2019; McCaffrey et al., 2020) for enrichment or exclusion across all cell-type pairs. HH3 was excluded from the analysis. For each cell type pair of cell type X and cell type Y, the number of times the centroid of cell X was within a ~50 μm radius of cell Y was counted. A null distribution was produced by performing 100 bootstrap permutations in which the locations of cell Y were randomized. A z-score was calculated comparing the number of true co-occurrences of cell X and cell Y relative to the null distribution. Importantly, symmetry was assumed: the values of the spatial enrichment of cell X close to cell Y are the same as the values with cell Y close to cell X. For each pair of cell types, the average z-score was calculated across all DCIS FOVs.
To analyze cellular associations with the edge of the epithelium, the distances between all cell centroids to the nearest perimeter location of the epithelial mask (described above) were calculated.
Cell neighborhoods were produced by first generating a cell neighbor matrix in which each row represents an index cell and columns indicate the relative frequency of each cell phenotype within a 36-μm radius of the index cell. Next, the neighbor matrix was clustered to 10 clusters using k-means clustering, with the number of clusters being determined as the number that best separated distinct immune cell mixtures and tumor/myoepithelial spatial relationships. The neighborhood cellular profile was determined by assessing the mean prevalence of each cell phenotype within a 36-μm radius of the index cell.
Distinguishing Feature Analysis
To determine features that distinguish among normal breast tissue, DCIS, and IBC, means of all 433 features were compared between groups using the Kruskal-Wallis H test. Features with significance under p = 0.05 (Table S3) were subsequently clustered using k-means clustering into the 4 TME clusters. For paired analyses, feature means were compared between DCIS and IBC samples from the same patient (Table S3).
ECM Gene Analysis
To analyze ECM components by gene expression, an ECM gene signature (GO ECM structural constituent, GO:0030021) was downloaded from the GSEA website (www.gsea-msigdb.org) and used to compare MIBI-identified samples with the top and bottom quartiles of cancer-associated fibroblast density in the stroma. Stromal LCM-RNaseq samples were used for this analysis from a paired transcriptomic study of the RAHBT cohort (Strand et al., 2021). Raw reads were normalized with DESeq2 R package (version 1.30.0, Anders and Huber, 2010) and a paired t test was compared to the log2 ratio of group means to generate the volcano plot (Table S3).
Myoepithelial Morphology Analysis
In order to quantify myoepithelial continuity and thickness, we defined a window of myoepithelial signal quantitation. For this window, we used a topology-preserving operation and defined a curve 5 pixels out from the epithelial mask edge (see Region Masking) and a curve 30 pixels in from the epithelium mask edge; we defined those pixels between these two curves as the myoepithelium mask. We subdivided the outer curve into 5-px arc segments, and for each point on the outer edge between two segments, we found the nearest point on the inner edge, dividing the myoepithelium into a string of quadrilaterals or “wedges.” Wedges were then subdivided along the in-out (of the epithelium) axis into 10 segments. Wedges were merged when both their combined inner and outer edges had an arc length < 15 px.
We took pre-processed (background subtracted, de-noised) SMA pixels within the mesh and smoothed them with a Gaussian blur of radius of 1. We then calculated the density of SMA signal within each mesh segment as the mean pixel value of smoothed SMA within that mesh segment. This density was then binarized to create a SMA-positivity mesh using a threshold of 0.5 (density > 0.5 as positive).
The percentage of duct perimeter covered by myoepithelium was calculated by assigning an “SMA-present” variable to each wedge: “0” if no mesh segments in the wedge were positive for SMA, and “1” otherwise. Each wedge was weighted by its area relative to the myoepithelium area. The sum over all wedges of the product of the “SMA-present” variable and the weight was defined as the percent perimeter SMA positivity.
The average (non-zero) thickness of the myoepithelium for each duct was calculated by finding the weighted average “wedge thickness” for SMA-positive wedges (“SMA-present” was 1). The wedge thickness was calculated as the distance between the innermost and outermost positive mesh segments. Positive wedges were weighted by their area relative to the total area of positive wedges.
The percent myoepithelial-covered perimeter and average myoepithelial thickness metrics were weighted over meshes (ducts) in a given image by assigning a weight to each duct equal to the total area of the duct myoepithelium divided by the sum of the total areas of all myoepithelium in the image that met a minimum size filter of 7500 px.
To assess automated thickness and continuity accuracy, myoepithelial SMA continuity and thickness were quantified manually in 5 progressor and 5 non-progressor SMA images by a board-certified pathologist using ImageJ, blinded to tumor outcome. For continuity, the total periductal perimeter in each image was first quantified by manually outlining each epithelial region. Then, gaps in the myoepithelial layer along this manual outline with no discernable SMA signal where identified. The length for each of these gaps along the periductal perimeter was quantified. Lastly, gap measurements were the summed and divided by total duct perimeter. Smooth muscle thickness was calculated by taking the average of 10 representative linear measurements.
Myoepithelial Pixel Clustering Analysis
Pre-processed (background subtracted, de-noised) images were first subset for pixels within the myoepithelium mask (see Region Masking). Pixels within the myoepithelium mask were then further subset for pixels with SMA expression > 0. For all SMA+ pixels within the myoepithelium mask, a Gaussian blur was applied using a standard deviation of 1.5 for the Gaussian kernel. Pixels were normalized by their total expression such that the total expression of each pixel was equal to 1. A 99.9% normalization was applied for each marker. Pixels were clustered into 100 clusters using FlowSOM (Van Gassen et al., 2015) based on the expression of six markers: PanCK, CK5, vimentin, ECAD, CD44, and CK7. The average expression of each of the 100-px clusters was found and the z-score for each marker across the 100-px clusters was computed, with a maximum z-score of 3. Using these z-scored expression values, the 100-px clusters were hierarchically clustered using Euclidean distance into six metaclusters. SMA+ pixels that were negative for the six markers used for FlowSOM were annotated as the SMA-only metacluster, resulting in a total of seven metaclusters. These metaclusters were mapped back to the original images to generate overlay images colored by pixel metacluster.
Collagen Morphometrics
To identify collagen fibers, background-removed Col1 images were first preprocessed: Col1 pixel intensities were capped at 5, gamma transformed (1 of 2), and contrast enhanced. Images were then blurred via Gaussian with a sigma of 2. While this process enhances fidelity, it yields less clear “0-borders.” This effect was mitigated by generating a “0-region” mask and setting all values to 0 in that region. Then, highly localized contrast enhancement was applied. Since raw fiber signal intensity can vary greatly within a FOV, this step helps enhance locally recognizable—but globally dim—fiber candidates. After this process, contrast was globally enhanced via a reverse gamma transformation (2 of 2).
Collagen fiber objects were generated by watershed segmentation on the preprocessed images. An adaptive thresholding method was developed to appreciate variability in total image intensities across the large dataset. A dilated and eroded version of each preprocessed image was produced and subjected to multi-Otsu thresholding. Elevation maps for watershed were generated via the Sobel gradient of a blurred version of the preprocessed images. Once objects were extracted and segmented, length, global orientation, perimeter, and width were computed for each object. Objects that covered low-intensity regions of the image were treated as preprocessing artifacts and were not included in averaging. Average collagen fiber lengths and average collagen branch number were calculated in the entire stromal region. Collagen fiber density (#/area) and total collagen signal were also calculated in specific histological zones defined by distance from the epithelial mask. These zones comprised the periepithelial stroma region (0–20 px from the epithelial edge), mid-stroma region (20–60 px), and distal stroma region (60+ px).
Collagen fiber-fiber alignment and fiber-epithelial edge alignment were also measured. For fiber-fiber alignment, fibers were filtered for elongated shape (length > 2*width) and alignment was scored as the normalized total paired squared difference over its k nearest neighbors (k = 4 was chosen). To accommodate for the elongated shape of these objects, k-nearest neighbors were computed with the ellipsoidal membrane distance, which is the Euclidean centroid distance minus the portion of that distance that lies within the ellipse representation of the object.
To compute the myoepithelial-to-fiber (myo-fib) alignment score, the myoepithelial region was identified as the boundary of a manually annotated epithelial mask. This region was then subdivided and labeled as separate objects. The global angle of each object is then compared to the global angle of the k-nearest fiber objects, via the same metric described in the fiber-fiber method.
Prediction of Recurrence
To predict recurrence, we compared tissue procured at the time of diagnosis in two sets of patients with primary DCIS. The first set, referred to as “progressor,” consisted of 14 patients who had a new ipsilateral invasive breast event following a diagnosis of pure DCIS (median time to recurrence = 9.1 years). The second set, referred to as “non-progressor,” consisted of 44 patients with pure DCIS that did not have a new breast event following primary tumor resection (median time of follow = 11.4 years). For each patient, a vector of summary statistics was generated from MIBI data using only images derived from the original lesion. The cohort was split into training (80%) and test (20%) sets; all model optimization and predictor selection steps used only the training set. Any missing values were replaced with the set’s predictor mean. Predictors with < 12 unique values in the training set were dropped from the analysis. We removed correlated parameters because they could confound predictor importance: all predictors were ranked in importance by performing a Kolmogorov-Smirnov test between progressor and non-progressor within the training set. Greater importance was placed on predictors with lower p values, with ties broken by weighting predictors with greater effect sizes between patient groups. We quantified pairwise correlation for all predictors (Spearman method). For each group of highly correlated predictors (R > 0.85), only the highest-ranked predictor was used in the model. We varied this cutoff and found no difference in model accuracy (Figure S6G). Two-class random forest probability models (ranger package) (Wright and Ziegler, 2017) were trained to discriminate progressors versus non-progressors. Hyperparameters were tuned on the training set to minimize out-of-bag error. The optimized random forest model was evaluated on the test set and a receiver operating characteristic curve was generated for calculating the area under the curve (pROC package) (Robin et al., 2011) using the model’s assigned probability scores. Each predictor’s importance was evaluated in the model by its Gini index (Table S4). All analyses were repeated with 10 distinct random seeds for partitioning patients into training and test sets. For each seed, we additionally trained models using randomly permuted patient group labels (Figure 5C).
Myoepithelial Immunofluorescence Quantification
To identify the myoepithelial regions of interest, the SMA channel was first passed through a Gaussian filter, and had its maximum intensity capped, to mitigate intense autofluorescent signatures. Next, after being passed through a locally scaled gamma transform to enhance ridge-like features, the channel went through a Meijering ridge filter. To identify candidate myoepithelial “ridges,” the channel was thresholded and all “ridge” objects were labeled. To filter out distant candidates, their respective distances to a manually annotated mask of the epithelium were measured and gated, only classifying ridges within 80px as the myoepithelial region. The co-expression of SMA and ECAD was measured in these generated regions.
Myoepithelial Features LDA
All myoepithelial features (Table S5) were standardized (mean subtracted and divided by the standard deviation). DCIS (primary and recurring) samples were defined as training data while normal samples were defined as the test set. We then used a dimensionality reduction technique based on Linear Discriminant Analysis (LDA) (Tsai et al., 2020) on the DCIS-only training set in order to capture the main differences in myoepithelial character between progressors and non-progressors. This supervised method finds the optimal linear combination of a subset of features that maximizes the separation between pre-labeled classes. By combining the myoepithelial features with a progressor/non-progressor label, we separated the DCIS patients in a one-dimensional LDA-generated space (LD1 coordinate) with respect to their progression status. LD1 is therefore the optimized linear combination of the myoepithelial- and SMA-related features for separating progressors from non-progressors (Table S5). We then calculated LD1 values for our test data—the normal samples based on the trained model (Table S5). The code for this LDA-based method was provided by (Tsai et al., 2020) and was made available on GitHub at https://github.com/davidrglass. p values for comparing LD1 distributions between sample types were calculated with the Kruskal-Wallis H test using the MATLAB function kruskalwallis.
Feature Ontology Enrichment Analysis
Taking into account DCIS samples only, we calculated the correlation of features in Table S1 with LD1. In this calculation we excluded the 21 features used to define LD1 in the LDA analysis described above. We then sorted the features by correlation with LD1, creating a ranked list of features (Table S5). Features were also annotated based on belonging to one (or none) of the following functional modules or pathways: Desmoplasia and ECM remodeling (terms: CAFs, MMP9 expression, collagen deposition and fibers), Immune: immunoregulation (immune cells + PD1/PDL1/IDO1/COX2), Lipid metabolism (CD36), Lymphoid: growth/proliferation (CD4T, CD8T, B cell, dnT cell + Ki67/pS6), Myeloid: growth/proliferation (Macs, Mono, MonoDC, DC, APC + Ki67/pS6), Immune density in stroma (immune cell + stroma density), Stroma: growth/proliferation (Fibroblast or endothelium + Ki67/pS6), Tumor: ER/AR/HER2 expression (tumor + ER/AR/HER2), Tumor: immunoregulation (tumor + PDL1/IDO1/COX2), Tumor: growth/proliferation (tumor + Ki67/pS6), and Hypoxia and Glycolysis (HIF1a + GLUT1) (see Table S5 for specific ontology features). This ranked list of features combined with their annotations into pathways was used to perform gene set enrichment analysis (GSEA) using the R package FGSEA (Korotkevich et al., 2021). This procedure identified functionally related groups of features that were enriched either among the features highly correlated with LD1 or significantly anti-correlated with LD1 (Table S5).
Software used for data analysis
Image processing was conducted with MATLAB 2016a and MATLAB 2019b. Data visualization and plots were generated in R with ggplot and pheatmap packages, in GraphPad Prism, and in Python using the scikitimage, matplotlib, and seaborn packages. Representative images were processed in Adobe Photoshop CS6. Schematic visualizations were produced with Biorender. R packages used for GSEA were AnnotationDbi (1.52.0) and org.Hs.eg.db, (3.12.0), clusterProfiler (3.19.0), msigdbr (7.2.1), for C2 curated datasets. Python packages used for spatial enrichment analysis and collagen morphometrics were sckikit-image, pandas, numpy, xarray, scipy, statsmodels.
Statistical analysis
All statistical analyses were performed using GraphPad Prism (9.1.0), MATLAB (2016b), or R (1.2.5033). Grouped data are presented with individual sample points throughout, and where not applicable, data are presented as mean and standard deviation. For determining significance, grouped data were first tested for normality with the D’Agostino & Pearson omnibus normality test. Normally distributed data were compared between two groups with the two-tailed Student’s t test. Non-normal data were compared between two groups using the Mann–Whitney test. Multiple groups were compared using the Kruskal-Wallis test. Statistical tests that were performed on multiple features were corrected for multiple comparisons using the Benjamini-Hochberg FDR Procedure and the corrected Q-values were then used for subsequent feature selection.
Supplementary Material
Figure S1. Single marker staining controls, related to Figure 1
Representative images of MIBI conjugate staining for all immune markers, with immune control tissues (tonsil, lymph node, and placenta).
Figure S2. Single-cell segmentation and annotation strategy, related to Figure 2
(A) Workflow for Deepcell-based segmentation of single cells from multiplexed images. Workflow shows (1) the input data to model training, (2) the model output data of nuclear segmentation, and (3) the multiple sets of parameters used in this study to optimally segment and expand nuclei to identify the diverse cell populations in DCIS.
(B) Schematic of steps involved in single-cell phenotyping, including marker normalization (left), cell clustering into major cellular lineages (middle), and clustering within lineages into cell types (right).
(C) The major cell subset divisions in each iterative round of phenotype clustering are shown. Cells are first subdivided into cellular lineage, then lineages are further clustered to identify cell types (immune) or phenotypic subsets (tumor, fibroblast).
(D) Heatmap of the 100 clusters from the round1 lineage clustering. Clusters are annotated by color based on their cell compartment (epithelial: “EPI,” teal; stroma: brown; other: black), as well as their determined final lineage (EPI, green; myoepithelial (“MYOEP”) blue; fibroblast (“FIBRO”) red); endothelial (“ENDO”) brown; immune, gold; other, black.
(E) Examples of image-based interrogation of cell clusters expressing non-canonical combinations of markers, including a SMA+/CK7+ myoepithelial cluster (Cluster 57, top) and a PanCK+/VIM+/CK7-low tumor cluster (12, bottom).
(F) Heatmap of marker expression in immune lineage cell type clustering, with assigned cell type phenotype to right.
(G) Heatmap of epithelial marker expression in epithelial lineage cell type clustering.
(H) Heatmap of fibroblast marker expression in epithelial lineage cell type clustering.
Figure S3. Robustness analysis of cell annotations, related to Figure 2
(A) Biaxial manual gating scheme to assign the cell types, denoted by the colored text: Myoepithelium “MYOEP,” Basal Tumor “BASAL,” Luminal Tumor “LUMINAL,” EMT Tumor “EMT,” CK5/7-low Tumor “CK5/7-low,” Endothelial “ENDO,” Fibroblasts “FIBRO,” CAFs “CAF,” Normal Fibroblasts “NORM.FIBRO,” Resting Fibroblasts “REST.FIBRO,” MyoFibroblasts “MYOFIBRO,” B Cells “BCELL,” Mast cells “MAST,” Neutrophils “NEUT,” CD8T cells “CD8T,” CD4 T cells “CD4T,” Macrophages “MACS,” Antigen Presenting Cells “APC,” Monocytes “MONO’, Dendritic Cells “DC,” MonoDCs “MONODC,” double negative T cells “dnT,” Other Immune Cells “IMM. OTHER.”
(B) Bar plot comparing cell type abundance across the entire cohort by FlowSOM assignment (blue) and manual gating assignment (orange).
(C) Scatterplot comparing cell type abundances between FlowSOM (X) and manual gating (Y) with a linear regression with accompanying R2 and slope values. Residuals for the regression are displayed below.
(D) Scatterplot comparing the significance of tissue distinguishing features (listed in Figure 3D, Table S3) after calculating features with FlowSOM cell annotation (X) versus manual gating cell annotation (Y), residuals for the regression are displayed to the right.
Figure S4. Tumor cell state profiling, related to Figure 2
(A) Representative MIBI image overlays showing an ER+HER2− tumor (left) and ER-HER2+ (right), scale bars = 100 μm.
(B) Criteria used to define tumors as ER, AR, HER2, or Ki67 positive, and HER2-intense.
(C) Area plots showing the frequency of receptor expression states in tumor cells (top), and immune cell type composition (bottom) in all DCIS, IBC, and normal patient samples profiled in this study. Tissue and PAM50 subtype are denoted by color in the top row.
Figure S5. Tumor microenvironment spatial and structural analyses, related to Figure 3
(A) Representative MIBI image overlay of a pure DCIS tumor with major immune cell type markers. Zoomed inset (left) and arrow highlighting intraductal immune phenotypes. Right inset, masked stromal and duct regions where immune cell density is measured. All scale bars = 100 μm.
(B) Heatmap of z-score-normalized cell-type frequency for each cellular neighborhood (CN).
(C) CN map of the spatial localization of distinct CNs, denoted by color as in (B). Insets: Color overlays for lymphocyte-enriched (green dotted line, top) or tumor-interface (red dotted line, bottom) CNs. Scale bar = 100 μm.
(D) Images of SMA signal in normal breast and DCIS with a projected measurement lattice to quantify myoepithelial SMA signal continuity and thickness. Zoomed inset (left) shows myoepithelial SMA signal with nuclear signal (Nuc) and ductal cytokeratin expression (CK); the right inset shows this SMA signal in its binarized form (white) for continuity and thickness measurement.
(E) Scatterplot of the automated SMA thickness measurement from the method in (D) compared to SMA thickness measurements made in ImageJ by a blinded pathologist.
(F) Scatterplot of the automated SMA continuity measurement compared to SMA continuity measurements made in ImageJ by a blinded pathologist.
(G) Workflow showing the measurement of collagen signal density and collagen fiber morphometrics in three stromal regions (periepithelial, mid stroma, distal stroma). Fiber orientation was measured compared to other fibers as well as the epithelial edge.
(H) Area plot of the distribution of each feature class in all features measured.
(I) Heatmap of the distinguishing feature prevalence in normal breast, DCIS, and recurrent IBC samples from the TME4: DCIS Low cluster, with all features annotated to the left.
Figure S6. Interrogation of fibroblast differentiation and classifier results, related to Figures 4 and 5
(A) Cell phenotype maps of normal breast tissue, DCIS, and IBC samples showing the distribution of normal fibroblast and CAF states in the stroma, as well as two epithelial states. Insets (left) highlight areas with representative fibroblast makeup with MIBI marker overlays of the same region with fibroblast and epithelial markers shown to the right of the same region. Scale bars = 100 μm.
(B) Boxplot of the quantification of collagen signal in the periepithelial zone of normal breast tissue, DCIS, and IBC samples; p value from Kruskal-Wallis H test.
(C) Boxplot of the quantitation of collagen fiber density in the stroma of normal breast tissue, DCIS, and IBC samples; p value from Kruskal-Wallis H test.
(D) Boxplot of the quantification of collagen fiber branching in normal breast tissue, DCIS, and IBC samples; p value from Kruskal-Wallis H test.
(E) Stacked bar plot of the frequency of mastectomy, radiation therapy, and tamoxifen therapy in the progressor (P) and non-progressor (NP) outcome groups in the training data for the recurrence model.
(F) Distribution of mastectomy, radiation, and tamoxifen therapy is shown by color in the model-predicted progressors (orange) and non-progressors (green), with the random forest prediction probability shown for each patient. P values comparing the treated frequency of total between groups is displayed, Wilcoxon signed-rank test.
(G) Column plot of the model’s AUC after modifying the correlation cutoff for feature inclusion.
(H) Stacked column plot of the distribution of spatial versus non-spatial features for all features used in model training (“All”), and those determined to be the 20 most important features by Gini importance test (“Top 20 Gini”).
(I) Column plot of accumulative Gini importance of features that involve APC cells, dnT cells, or mast cells.
Figure S7. Characterizing myoepithelial phenotype, related to Figure 6
(A) Workflow schematic for pixel-based clustering of myoepithelial phenotype.
(B) Heatmap of mean marker expression in the seven myoepithelial expression clusters, with a bar plot (left) of cluster abundance out of total identified myoepithelium in the cohort.
(C) Pseudo-colored image illustrating the spatial distribution of myoepithelial pixel clusters defined in (B) for a DCIS patient tumor. Scale bars = 50 μm.
(D) Representative immunofluorescent image overlay of DAPI, SMA, and ECAD with zoomed inset of ducts (left) and the myoepithelial objects (right) used to quantify SMA and ECAD coexpression.
(E) Scatterplot of the quantified myoepithelial ECAD-SMA pixel coexpression by MIBI versus the coexpression quantified in the same patient samples by immunofluorescence.
(F) Boxplot comparing the frequency of EAD+ myoepithelium between progressor (P) and non-progressor (NP) tumors.
Highlights.
A spatial atlas of breast cancer progression using MIBI-TOF and tissue transcriptomics
Coordinated changes in the tumor microenvironment (TME) track invasive transition of DCIS
DCIS TME structure is predictive of invasive relapse within 10 years of diagnosis
Recurrence risk is heavily influenced by myoepithelial phenotype and morphology
ACKNOWLEDGMENTS
This publication is part of the HTAN (Human Tumor Atlas Network) Consortium paper package. A list of HTAN members is available at humantumoratlas.org/htan-authors/. The authors thank the HTAN Consortium for intellectual and collaborative support of this work. We thank Pauline Chu and the Stanford Human Histology Core for providing technical assistance. T.R. was supported by American Cancer Society Postdoctoral Fellowship 133099-PF-19–002-01-CCE and Stanford Immunology Training Grant 5 T32 AI07290–33. D.R.G. was supported by the Bio-X Stanford Interdisciplinary Graduate Fellowship. C.C.L. was supported by the Stanford Graduate Fellowship. E.S.H. is funded by RFA-CA-17–035 (NIH), 1505–30497 (PCORI), and BCRF 19–074 (BCRF); E.S.H. and C.M. are funded by DOD BC132057 and R01 CA185138–01. R.B.W. was supported by R01CA193694 and U2C CA233254. M.A. was supported by 1-DP5-OD019822. S.C.B. and M.A. were jointly supported by 1R01AG056287, 1R01AG057915, 1U24CA224309, the Bill and Melinda Gates Foundation, and a Translational Research Award from the Stanford Cancer Institute. Graphical abstract was created with Biorender.com.
Footnotes
DECLARATION OF INTERESTS
M.A. and S.C.B. are inventors on patent US20150287578A1. M.A. and S.C.B. are board members and shareholders in IonPath Inc. T.R. and E.F.M. have previously consulted for IonPath Inc.
SUPPLEMENTAL INFORMATION
Supplemental information can be found online at https://doi.org/10.1016/j.cell.2021.12.023.
REFERENCES
- Afghahi A, Forgó E, Mitani AA, Desai M, Varma S, Seto T, Rigdon J, Jensen KC, Troxell ML, Gomez SL, et al. (2015). Chromosomal copy number alterations for associations of ductal carcinoma in situ with invasive breast cancer. Breast Cancer Res. 17, 108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aguiar FN, Cirqueira CS, Bacchi CE, and Carvalho FM (2015). Morphologic, molecular and microenvironment factors associated with stromal invasion in breast ductal carcinoma in situ: Role of myoepithelial cells. Breast Dis. 35, 249–252. [DOI] [PubMed] [Google Scholar]
- Casasent AK, Schalck A, Gao R, Sei E, Long A, Pangburn W, Casasent T, Meric-Bernstam F, Edgerton ME, and Navin NE (2018). Multiclonal Invasion in Breast Tumors Identified by Topographic Single Cell Sequencing. Cell 172, 205–217.e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders S, and Huber W (2010). Differential expression analysis for sequence count data. Genome Biol. 11, R106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aponte-López A, Fuentes-Pananá EM, Cortes-Muñoz D, and MuñozCruz S (2018). Mast Cell, the Neglected Member of the Tumor Microenvironment: Role in Breast Cancer. J. Immunol. Res 10.1155/2018/2584243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barsky SH, and Karlin NJ (2005). Myoepithelial cells: autocrine and paracrine suppressors of breast cancer progression. J. Mammary Gland Biol. Neoplasia 10, 249–260. [DOI] [PubMed] [Google Scholar]
- Barth PJ, Moll R, and Ramaswamy A (2005). Stromal remodeling and SPARC (secreted protein acid rich in cysteine) expression in invasive ductal carcinomas of the breast. Virchows Arch. 446, 532–536. [DOI] [PubMed] [Google Scholar]
- Bartova M, Ondrias F, Muy-Kheng T, Kastner M, Singer C.h., and Pohlodek K (2014). COX-2, p16 and Ki67 expression in DCIS, microinvasive and early invasive breast carcinoma with extensive intraductal component. Bratisl. Lek Listy 115, 445–451. [DOI] [PubMed] [Google Scholar]
- Betsill WL Jr., Rosen PP, Lieberman PH, and Robbins GF (1978). Intraductal carcinoma. Long-term follow-up after treatment by biopsy alone. JAMA 239, 1863–1867. [DOI] [PubMed] [Google Scholar]
- Buerger H, Otterbach F, Simon R, Poremba C, Diallo R, Decker T, Riethdorf L, Brinkschmidt C, Dockhorn-Dworniczak B, and Boecker W (1999). Comparative genomic hybridization of ductal carcinoma in situ of the breast-evidence of multiple genetic pathways. J. Pathol. 187, 396–402. [DOI] [PubMed] [Google Scholar]
- Cancer Genome Atlas Network (2012). Comprehensive molecular portraits of human breast tumours. Nature 490, 61–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conklin MW, Eickhoff JC, Riching KM, Pehlke CA, Eliceiri KW, Provenzano PP, Friedl A, and Keely PJ (2011). Aligned collagen is a prognostic signature for survival in human breast carcinoma. Am. J. Pathol. 178, 1221–1232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtis C, Shah SP, Chin S-F, Turashvili G, Rueda OM, Dunning MJ, Speed D, Lynch AG, Samarajiwa S, Yuan Y, et al. ; METABRIC Group (2012). The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature 486, 346–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding L, Su Y, Fassl A, Hinohara K, Qiu X, Harper NW, Huh SJ, Bloushtain-Qimron N, Jovanović B, Ekram M, et al. (2019). Perturbed myoepithelial cell differentiation in BRCA mutation carriers and in ductal carcinoma in situ. Nat. Commun. 10, 4182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erbas B, Provenzano E, Armes J, and Gertig D (2006). The natural history of ductal carcinoma in situ of the breast: a review. Breast Cancer Res. Treat. 97, 135–144. [DOI] [PubMed] [Google Scholar]
- Esbona K, Yi Y, Saha S, Yu M, Van Doorn RR, Conklin MW, Graham DS, Wisinski KB, Ponik SM, Eliceiri KW, et al. (2018). The Presence of Cyclooxygenase 2, Tumor-Associated Macrophages, and Collagen Alignment as Prognostic Markers for Invasive Breast Carcinoma Patients. Am. J. Pathol. 188, 559–573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eusebi V, Feudale E, Foschini MP, Micheli A, Conti A, Riva C, Di Palma S, and Rilke F (1994). Long-term follow-up of in situ carcinoma of the breast. Semin. Diagn. Pathol. 11, 223–235. [PubMed] [Google Scholar]
- Foley JW, Zhu C, Jolivet P, Zhu SX, Lu P, Meaney MJ, and West RB (2019). Gene expression profiling of single cells from archival tissue with laser-capture microdissection and Smart-3SEQ. Genome Res. 29, 1816–1825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman G, Levi-Galibov O, David E, Bornstein C, Giladi A, Dadiani M, Mayo A, Halperin C, Pevsner-Fischer M, Lavon H, et al. (2020). Cancer-associated fibroblast compositions change with breast-cancer progression linking S100A4 and PDPN ratios with clinical outcome. bioRxiv. 10.1101/2020.01.12.903039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii H, Szumel R, Marsh C, Zhou W, and Gabrielson E (1996). Genetic progression, histological grade, and allelic loss in ductal carcinoma in situ of the breast. Cancer Res. 56, 5260–5265. [PubMed] [Google Scholar]
- Gil Del Alcazar CR, Huh SJ, Ekram MB, Trinh A, Liu LL, Beca F, Zi X, Kwak M, Bergholtz H, Su Y, et al. (2017). Immune Escape in Breast Cancer During In Situ to Invasive Carcinoma Transition. Cancer Discov. 7, 1098–1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenwald NF, Miller G, Moen E, Kong A, Kagel A, Fullaway CC, McIntosh BJ, Leow K, Schwartz MS, Dougherty T, et al. (2021a). Whole-cell segmentation of tissue images with human-level performance using large-scale data annotation and deep learning. bioRxiv. 10.1101/2021.03.01.431313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenwald NF, Miller G, Moen E, Kong A, Kagel A, Dougherty T, Fullaway CC, McIntosh BJ, Leow KX, Schwartz MS, et al. (2021b). Whole-cell segmentation of tissue images with human-level performance using large-scale data annotation and deep learning. Nat. Biotechnol. 10.1038/s41587-021-01094-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibrahim AM, Moss MA, Gray Z, Rojo MD, Burke CM, Schwertfeger KL, Dos Santos CO, and Machado HL (2020). Diverse Macrophage Populations Contribute to the Inflammatory Microenvironment in Premalignant Lesions During Localized Invasion. Front. Oncol. 10, 569985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones JL, Shaw JA, Pringle JH, and Walker RA (2003). Primary breast myoepithelial cells exert an invasion-suppressor effect on breast cancer cells via paracrine down-regulation of MMP expression in fibroblasts and tumour cells. J. Pathol. 201, 562–572. [DOI] [PubMed] [Google Scholar]
- Keren L, Bosse M, Marquez D, Angoshtari R, Jain S, Varma S, Yang S-R, Kurian A, Van Valen D, West R, et al. (2018). A Structured Tumor-Immune Microenvironment in Triple Negative Breast Cancer Revealed by Multiplexed Ion Beam Imaging. Cell 174, 1373–1387.e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keren L, Bosse M, Thompson S, Risom T, Vijayaragavan K, McCaffrey E, Marquez D, Angoshtari R, Greenwald NF, Fienberg H, et al. (2019). MIBI-TOF: A multi-modal multiplexed imaging platform for tissue pathology. Sci. Adv. 5, eaax5851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SY, Jung S-H, Kim MS, Baek I-P, Lee SH, Kim T-M, Chung Y-J, and Lee SH (2015). Genomic differences between pure ductal carcinoma in situ and synchronous ductal carcinoma in situ with invasive breast cancer. Oncotarget 6, 7597–7607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korotkevich G, Sukhov V, Budin N, Shpak B, Artyomov MN, and Sergushichev A (2021). Fast gene set enrichment analysis. bioRxiv. 10.1101/060012. [DOI] [Google Scholar]
- Malanchi I, Santamaria-Martínez A, Susanto E, Peng H, Lehr H-A, Delaloye J-F, and Huelsken J (2011). Interactions between cancer stem cells and their niche govern metastatic colonization. Nature 481, 85–89. [DOI] [PubMed] [Google Scholar]
- McCaffrey EF, Donato M, Keren L, Chen Z, Fitzpatrick M, Jojic V, Delmastro A, Greenwald NF, Baranski A, Graf W, et al. (2020). Multiplexed imaging of human tuberculosis granulomas uncovers immunoregulatory features conserved across tissue and blood. bioRxiv. 10.1101/2020.06.08.140426. [DOI] [Google Scholar]
- Moen E, Bannon D, Kudo T, Graf W, Covert M, and Van Valen D (2019). Deep learning for cellular image analysis. Nat. Methods 16, 1233–1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newburger DE, Kashef-Haghighi D, Weng Z, Salari R, Sweeney RT, Brunner AL, Zhu SX, Guo X, Varma S, Troxell ML, et al. (2013). Genome evolution during progression to breast cancer. Genome Res. 23, 1097–1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page DL, Dupont WD, Rogers LW, and Landenberger M (1982). Intraductal carcinoma of the breast: follow-up after biopsy only. Cancer 49, 751–758. [DOI] [PubMed] [Google Scholar]
- Pelon F, Bourachot B, Kieffer Y, Magagna I, Mermet-Meillon F, Bonnet I, Costa A, Givel A-M, Attieh Y, Barbazan J, et al. (2020). Cancer-associated fibroblast heterogeneity in axillary lymph nodes drives metastases in breast cancer through complementary mechanisms. Nat. Commun. 11, 404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez AA, Balabram D, Rocha RM, da Silva Souza Á, and Gobbi H (2015). Co-Expression of p16, Ki67 and COX-2 Is Associated with Basal Phenotype in High-Grade Ductal Carcinoma In Situ of the Breast. J. Histochem. Cytochem. 63, 408–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rakovitch E, Nofech-Mozes S, Hanna W, Narod S, Thiruchelvam D, Saskin R, Spayne J, Taylor C, and Paszat L (2012). HER2/neu and Ki-67 expression predict non-invasive recurrence following breast-conserving therapy for ductal carcinoma in situ. Br. J. Cancer 106, 1160–1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robin X, Turck N, Hainard A, Tiberti N, Lisacek F, Sanchez J-C, and Müller M (2011). pROC: an open-source package for R and S+ to analyze and compare ROC curves. BMC Bioinformatics 12, 77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryser MD, Weaver DL, Zhao F, Worni M, Grimm LJ, Gulati R, Etzioni R, Hyslop T, Lee SJ, and Hwang ES (2019). Cancer Outcomes in DCIS Patients Without Locoregional Treatment. J. Natl. Cancer Inst. 111, 952–960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shani O, Vorobyov T, Monteran L, Lavie D, Cohen N, Raz Y, Tsarfaty G, Avivi C, Barshack I, and Erez N (2020). Fibroblast-Derived IL33 Facilitates Breast Cancer Metastasis by Modifying the Immune Microenvironment and Driving Type 2 Immunity. Cancer Res. 80, 5317–5329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sirka OK, Shamir ER, and Ewald AJ (2018). Myoepithelial cells are a dynamic barrier to epithelial dissemination. J. Cell Biol. 217, 3368–3381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sprague BL, Vacek PM, Mulrow SE, Evans MF, Trentham-Dietz A, Herschorn SD, James TA, Surachaicharn N, Keikhosravi A, Eliceiri KW, et al. (2021). Collagen Organization in Relation to Ductal Carcinoma In Situ Pathology and Outcomes. Cancer Epidemiol. Biomarkers Prev. 30, 80–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strand SH, Rivero-Gutiérrez B, Houlahan KE, Seoane JA, King L, Risom T, Simpson LA, Vennam S, Khan A, Cisneros L, et al. (2021). DCIS genomic signatures define biology and correlate with clinical outcome: a Human Tumor Atlas Network (HTAN) analysis of TBCRC 038 and RAHBT co-horts. bioRxiv. 10.1101/2021.06.16.448585. [DOI] [Google Scholar]
- Tsai AG, Glass DR, Juntilla M, Hartmann FJ, Oak JS, Fernandez-Pol S, Ohgami RS, and Bendall SC (2020). Multiplexed single-cell morphometry for hematopathology diagnostics. Nat. Med. 26, 408–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Gassen S, Callebaut B, Van Helden MJ, Lambrecht BN, Demeester P, Dhaene T, and Saeys Y (2015). FlowSOM: Using self-organizing maps for visualization and interpretation of cytometry data. Cytometry A 87, 636–645. [DOI] [PubMed] [Google Scholar]
- Van Valen DA, Kudo T, Lane KM, Macklin DN, Quach NT, DeFelice MM, Maayan I, Tanouchi Y, Ashley EA, and Covert MW (2016). Deep Learning Automates the Quantitative Analysis of Individual Cells in Live-Cell Imaging Experiments. PLoS Comput. Biol. 12, e1005177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright MN, and Ziegler A (2017). ranger: A Fast Implementation of Random Forests for High Dimensional Data in C++ and R. J. Stat. Softw. 10.18637/jss.v077.i01. [DOI] [Google Scholar]
- Yang M, Li Z, Ren M, Li S, Zhang L, Zhang X, and Liu F (2018). Stromal Infiltration of Tumor-Associated Macrophages Conferring Poor Prognosis of Patients with Basal-Like Breast Carcinoma. J. Cancer 9, 2308–2316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Wang X-H, Zhao Y-X, Chen C, Xu X-Y, Sun Q, Wu H-Y, Chen M, Sang J-F, Su L, et al. (2018). Cancer-Associated Fibroblasts Correlate with Tumor-Associated Macrophages Infiltration and Lymphatic Metastasis in Triple Negative Breast Cancer Patients. J. Cancer 9, 4635–4641. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Single marker staining controls, related to Figure 1
Representative images of MIBI conjugate staining for all immune markers, with immune control tissues (tonsil, lymph node, and placenta).
Figure S2. Single-cell segmentation and annotation strategy, related to Figure 2
(A) Workflow for Deepcell-based segmentation of single cells from multiplexed images. Workflow shows (1) the input data to model training, (2) the model output data of nuclear segmentation, and (3) the multiple sets of parameters used in this study to optimally segment and expand nuclei to identify the diverse cell populations in DCIS.
(B) Schematic of steps involved in single-cell phenotyping, including marker normalization (left), cell clustering into major cellular lineages (middle), and clustering within lineages into cell types (right).
(C) The major cell subset divisions in each iterative round of phenotype clustering are shown. Cells are first subdivided into cellular lineage, then lineages are further clustered to identify cell types (immune) or phenotypic subsets (tumor, fibroblast).
(D) Heatmap of the 100 clusters from the round1 lineage clustering. Clusters are annotated by color based on their cell compartment (epithelial: “EPI,” teal; stroma: brown; other: black), as well as their determined final lineage (EPI, green; myoepithelial (“MYOEP”) blue; fibroblast (“FIBRO”) red); endothelial (“ENDO”) brown; immune, gold; other, black.
(E) Examples of image-based interrogation of cell clusters expressing non-canonical combinations of markers, including a SMA+/CK7+ myoepithelial cluster (Cluster 57, top) and a PanCK+/VIM+/CK7-low tumor cluster (12, bottom).
(F) Heatmap of marker expression in immune lineage cell type clustering, with assigned cell type phenotype to right.
(G) Heatmap of epithelial marker expression in epithelial lineage cell type clustering.
(H) Heatmap of fibroblast marker expression in epithelial lineage cell type clustering.
Figure S3. Robustness analysis of cell annotations, related to Figure 2
(A) Biaxial manual gating scheme to assign the cell types, denoted by the colored text: Myoepithelium “MYOEP,” Basal Tumor “BASAL,” Luminal Tumor “LUMINAL,” EMT Tumor “EMT,” CK5/7-low Tumor “CK5/7-low,” Endothelial “ENDO,” Fibroblasts “FIBRO,” CAFs “CAF,” Normal Fibroblasts “NORM.FIBRO,” Resting Fibroblasts “REST.FIBRO,” MyoFibroblasts “MYOFIBRO,” B Cells “BCELL,” Mast cells “MAST,” Neutrophils “NEUT,” CD8T cells “CD8T,” CD4 T cells “CD4T,” Macrophages “MACS,” Antigen Presenting Cells “APC,” Monocytes “MONO’, Dendritic Cells “DC,” MonoDCs “MONODC,” double negative T cells “dnT,” Other Immune Cells “IMM. OTHER.”
(B) Bar plot comparing cell type abundance across the entire cohort by FlowSOM assignment (blue) and manual gating assignment (orange).
(C) Scatterplot comparing cell type abundances between FlowSOM (X) and manual gating (Y) with a linear regression with accompanying R2 and slope values. Residuals for the regression are displayed below.
(D) Scatterplot comparing the significance of tissue distinguishing features (listed in Figure 3D, Table S3) after calculating features with FlowSOM cell annotation (X) versus manual gating cell annotation (Y), residuals for the regression are displayed to the right.
Figure S4. Tumor cell state profiling, related to Figure 2
(A) Representative MIBI image overlays showing an ER+HER2− tumor (left) and ER-HER2+ (right), scale bars = 100 μm.
(B) Criteria used to define tumors as ER, AR, HER2, or Ki67 positive, and HER2-intense.
(C) Area plots showing the frequency of receptor expression states in tumor cells (top), and immune cell type composition (bottom) in all DCIS, IBC, and normal patient samples profiled in this study. Tissue and PAM50 subtype are denoted by color in the top row.
Figure S5. Tumor microenvironment spatial and structural analyses, related to Figure 3
(A) Representative MIBI image overlay of a pure DCIS tumor with major immune cell type markers. Zoomed inset (left) and arrow highlighting intraductal immune phenotypes. Right inset, masked stromal and duct regions where immune cell density is measured. All scale bars = 100 μm.
(B) Heatmap of z-score-normalized cell-type frequency for each cellular neighborhood (CN).
(C) CN map of the spatial localization of distinct CNs, denoted by color as in (B). Insets: Color overlays for lymphocyte-enriched (green dotted line, top) or tumor-interface (red dotted line, bottom) CNs. Scale bar = 100 μm.
(D) Images of SMA signal in normal breast and DCIS with a projected measurement lattice to quantify myoepithelial SMA signal continuity and thickness. Zoomed inset (left) shows myoepithelial SMA signal with nuclear signal (Nuc) and ductal cytokeratin expression (CK); the right inset shows this SMA signal in its binarized form (white) for continuity and thickness measurement.
(E) Scatterplot of the automated SMA thickness measurement from the method in (D) compared to SMA thickness measurements made in ImageJ by a blinded pathologist.
(F) Scatterplot of the automated SMA continuity measurement compared to SMA continuity measurements made in ImageJ by a blinded pathologist.
(G) Workflow showing the measurement of collagen signal density and collagen fiber morphometrics in three stromal regions (periepithelial, mid stroma, distal stroma). Fiber orientation was measured compared to other fibers as well as the epithelial edge.
(H) Area plot of the distribution of each feature class in all features measured.
(I) Heatmap of the distinguishing feature prevalence in normal breast, DCIS, and recurrent IBC samples from the TME4: DCIS Low cluster, with all features annotated to the left.
Figure S6. Interrogation of fibroblast differentiation and classifier results, related to Figures 4 and 5
(A) Cell phenotype maps of normal breast tissue, DCIS, and IBC samples showing the distribution of normal fibroblast and CAF states in the stroma, as well as two epithelial states. Insets (left) highlight areas with representative fibroblast makeup with MIBI marker overlays of the same region with fibroblast and epithelial markers shown to the right of the same region. Scale bars = 100 μm.
(B) Boxplot of the quantification of collagen signal in the periepithelial zone of normal breast tissue, DCIS, and IBC samples; p value from Kruskal-Wallis H test.
(C) Boxplot of the quantitation of collagen fiber density in the stroma of normal breast tissue, DCIS, and IBC samples; p value from Kruskal-Wallis H test.
(D) Boxplot of the quantification of collagen fiber branching in normal breast tissue, DCIS, and IBC samples; p value from Kruskal-Wallis H test.
(E) Stacked bar plot of the frequency of mastectomy, radiation therapy, and tamoxifen therapy in the progressor (P) and non-progressor (NP) outcome groups in the training data for the recurrence model.
(F) Distribution of mastectomy, radiation, and tamoxifen therapy is shown by color in the model-predicted progressors (orange) and non-progressors (green), with the random forest prediction probability shown for each patient. P values comparing the treated frequency of total between groups is displayed, Wilcoxon signed-rank test.
(G) Column plot of the model’s AUC after modifying the correlation cutoff for feature inclusion.
(H) Stacked column plot of the distribution of spatial versus non-spatial features for all features used in model training (“All”), and those determined to be the 20 most important features by Gini importance test (“Top 20 Gini”).
(I) Column plot of accumulative Gini importance of features that involve APC cells, dnT cells, or mast cells.
Figure S7. Characterizing myoepithelial phenotype, related to Figure 6
(A) Workflow schematic for pixel-based clustering of myoepithelial phenotype.
(B) Heatmap of mean marker expression in the seven myoepithelial expression clusters, with a bar plot (left) of cluster abundance out of total identified myoepithelium in the cohort.
(C) Pseudo-colored image illustrating the spatial distribution of myoepithelial pixel clusters defined in (B) for a DCIS patient tumor. Scale bars = 50 μm.
(D) Representative immunofluorescent image overlay of DAPI, SMA, and ECAD with zoomed inset of ducts (left) and the myoepithelial objects (right) used to quantify SMA and ECAD coexpression.
(E) Scatterplot of the quantified myoepithelial ECAD-SMA pixel coexpression by MIBI versus the coexpression quantified in the same patient samples by immunofluorescence.
(F) Boxplot comparing the frequency of EAD+ myoepithelium between progressor (P) and non-progressor (NP) tumors.
Data Availability Statement
All single channel images and area masks are present as single Tiffs in this public Mendeley Data repository. The DOI is listed in the key resources table. Accession numbers are listed in the key resources table.
All original code has been deposited at Mendeley and is publicly available as of the date of publication. DOIs are listed in the key resources table.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
KEY RESOURCES TABLE.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
|
| ||
| Antibodies | ||
|
| ||
| A full list of antibodies is provided in Table S2 | N/A | N/A |
|
| ||
| Biological samples | ||
|
| ||
| The Resource of Archival Breast Tissue (RAHBT) cohort TMAs were compiled at Washington University in St. Louis, all patient information is included in Table S1 | N/A | N/A |
|
| ||
| Chemicals, peptides, and recombinant proteins | ||
|
| ||
| TBS IHC Wash Buffer with Tween 20 | Cell Marque | Cat#935B-09 |
| PBS IHC Wash Buffer with Tween 20 | Cell Marque | Cat#934B-09 |
| Target Retrieval Solution, pH 9, (3:1) | Agilent (Dako) | Cat#S2375 |
| Avidin/Biotin Blocking Kit | Biolegend | Cat#927301 |
| Gelatin (cold water fish skin) | Sigma-Aldrich | Cat#G7765–250 |
| Xylene Histological grade | Sigma-Aldrich | Cat#534056–500 |
| Glutaraldehyde 8% Aqueous Solution EM Grade | EMS | Cat#16020 |
| Normal Donkey serum | Sigma-Aldrich | Cat#D9663–10ML |
| Bovine Albumin (BSA) | Fisher | Cat#BP1600–100 |
| Centrifugal filters (0.1 μm) | Millipore | Cat#UFC30VV00 |
|
| ||
| Critical commercial assays | ||
|
| ||
| MIBItag Conjugation Kit | IONpath | Cat#600XXX |
| ImmPRESS UNIVERSAL (Anti-Mouse/Anti-Rabbit) IgG KIT (HRP) | Vector Laboratories | Cat#MP-7500–15 |
| ImmPACT DAB (For HRP Substrate) | Vector Laboratories | Cat#SK-4105 |
|
| ||
| Deposited data | ||
|
| ||
| All image data, single cell data, and tissue feature data, is located in a public Mendeley data repository: https://data.mendeley.com/datasets/d87vg86zd8 | Mendeley Data | https://data.mendeley.com/datasets/d87vg86zd8 |
|
| ||
| Software and algorithms | ||
|
| ||
| Data analysis was done using MATLAB 2016 | Mathworks | N/A |
| Data analysis was done using R 3.6.1 | R | N/A |
| Data analysis was done using Python | Python | N/A |
| Analysis code for MATLAB, R, and Python is available at the Mendeley data and code repository: https://data.mendeley.com/datasets/d87vg86zd8 | Mendeley Data | https://data.mendeley.com/datasets/d87vg86zd8 |