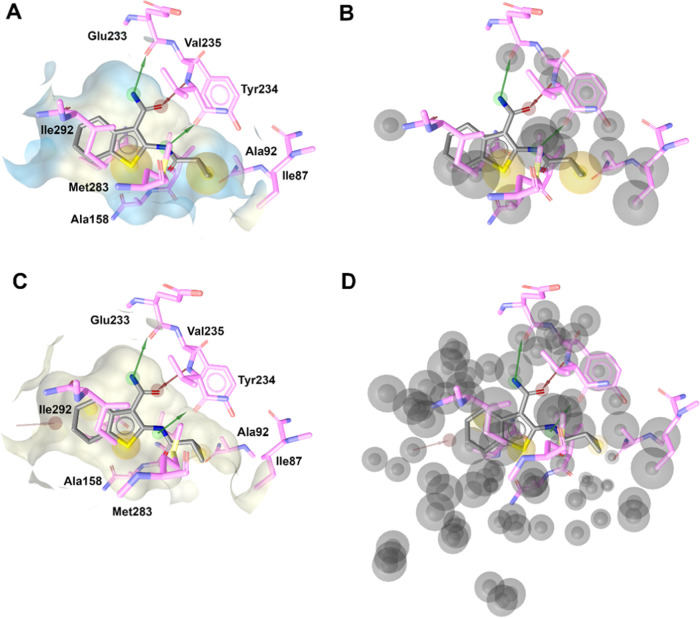
Abstract

Tuberculosis (TB) is an infectious disease caused by Mycobacterium tuberculosis (Mtb). Despite being considered curable and preventable, the increase of antibiotic resistance is becoming a serious public health problem. Mtb is a pathogen capable of surviving in macrophages, causing long-term latent infection where the mycobacterial serine/threonine protein kinase G (PknG) plays a protective role. Therefore, PknG is an important inhibitory target to prevent Mtb from entering the latency stage. In this study, we use a pharmacophore-based virtual screening and biochemical assays to identify the compound RO9021 (CHEMBL3237561) as a PknG inhibitor. In detail, 1.5 million molecules were screened using a scalable cloud-based setup, identifying 689 candidates, which were further subjected to additional screening employing molecular docking. Molecular docking spotted 62 compounds with estimated binding affinities of −7.54 kcal/mol (s.d. = 0.77 kcal/mol). Finally, 14 compounds were selected for in vitro experiments considering previously reported biological activities and commercial availability. In vitro assays of PknG activity showed that RO9021 inhibits the kinase activity similarly to AX20017, a known inhibitor. The inhibitory effect was found to be dose dependent with a relative IC50 value of 4.4 ± 1.1 μM. Molecular dynamics simulations predicted that the PknG-RO9021 complex is stable along the tested timescale. Altogether, our study indicates that RO9021 is a noteworthy drug candidate for further developing new anti-TB drugs that hold excellent reported pharmacokinetic parameters.
1. Introduction
Tuberculosis (TB) is an infectious disease caused by the Mycobacterium tuberculosis (Mtb).1,2 TB, which is considered an airborne disease, mainly affects the lungs but can also affect other sites of the human body.1,2 Despite being considered curable and preventable, TB is the second leading infectious killer after COVID-19.3 According to the latest Global Tuberculosis Report 2021,3 an estimated 9.9 million people fell ill with TB, leading to 1.3 million deaths. The latter is high due to the large drop in new people diagnosed with TB in 2020. Alarmingly, the global progress in reducing the number of people who die from TB in the last few years has been reversed by the COVID-19 pandemic. TB is present in all countries around the world. However, most TB cases (86%) were reported in WHO regions of South-East Asia, Africa, and the Western Pacific.3
Currently, anti-TB drugs have been used for decades varying the treatment according to drug sensitivity.1,2,4 In the case of a drug-sensitive disease, the drug treatment can last six months, while drug-resistant disease treatment can last up to 2 years.4 The drug resistance phenomenon arises when anti-TB drugs are used inappropriately, due to incorrect prescription by healthcare providers, poor drug quality, or premature discontinuation of treatment by patients.3 For this reason, multidrug-resistant tuberculosis (MDR-TB), TB unresponsive to at least isoniazid and rifampicin, has become a serious public health problem. The occurrence of extensively drug-resistant tuberculosis (XDR-TB) is equally problematic, implying that MDR-TB strains are resistant to fluoroquinolones and second-line injectable drugs.4 Resistance is the consequence of the fact that Mtb has great success as a pathogen by surviving within macrophages, causing long-term persistent infection (latency stage).5,6
The mycobacterial eukaryotic-like serine/threonine kinase, protein kinase G (PknG), plays a crucial role in keeping the phagosomes intact within the macrophages.7 It has been shown that PknG is a key regulator in the mycobacterial metabolism of carbon and nitrogen,8,9 but more importantly, PknG leads to the prevention of phagosome–lysosome fusion within infected macrophages.7 Assays inhibiting PknG activity using the AX20017 compound, a reference compound that targets the ATP binding site of the kinase domain, resulted in the death of internalized mycobacteria by effective lysosomal delivery.7,10 Since the interest of the scientific community has increased in the last few years, PknG of Mtb has been extensively studied from the structural point of view (Figure 1), regions such as (1) the ATP binding region, conformed by residues Glu233, Tyr234, and Val235; (2) the gatekeeper residue Met232; (3) the catalytic lysine Lys181, and (4) the Asp–Phe–Gly (DFG) motif with aspartate Asp293 are well documented and taken as reference for the Mtb kinase family. PknG is the only kinase with the Asp–Leu–Gly (DLG) motif.10−12 Therefore, PknG represents a new therapeutic target for the discovery of new anti-TB drugs. As a consequence, it is important to increase the number of hits and lead compounds that inhibit PknG activity.
Figure 1.
Structure of the PknG-AX20017 complex (ID PDB: 2PZI). (A) Ribbon representation of the PknG kinase domain. Typical secondary structure elements are indicated: DLG motif is colored black, P-loop is colored pink, activation loop is colored yellow, the catalytic loop is colored red, and helix C is colored green. The binding pocket of inhibitor AX20017 is shown in light blue. (B) Interacting residues are shown. Inhibitor AX20017 is shown in gray color. Residues Glu233 and Val235 form hydrogen bonds.
Recent studies aimed to find Mtb PknG inhibitors using target-based and cell-based approaches and suggested that synthetic chalcones, flavanones, and aminopyrimidine derivatives were promising sources of new compounds.13 The strategy of searching structural analogs of AX20017 through the exploration of its scaffold has proven to be useful, as it allowed for the discovery of the lead structural analog AX35510.14 European initiatives such as the Nested Chemical Library screening campaign containing 19000 molecules and more than 600 scaffolds represented a great coordinated effort.15 A similar effort is the discovery of compounds R406 and NU-6027;16,17 the former, as a result of the screening of a library of 80 kinase inhibitory compounds16 and the latter, with cross-reactivity against PnkD and PknG.17 From a computational point of view, 84 secondary metabolites from medicinal plants (Pelargonium reniforme and Pelargonium sidoides) were studied by molecular docking-based virtual screening achieving 10 potential inhibitors with favorable energy affinities.18 Using a similar approach, 477 flavanones from PubChem were docked against the PknG obtaining 6 potential inhibitors.19 In these last two cases, there was no verification of the potential inhibitors employing biochemical tests. Remarkably, a hybrid approach by pharmacophore-based virtual screening and in vitro studies resulted in the proposal of the validated compound NRB04248.20 Specifically, the pharmacophore screening was performed using ligand-based pharmacophore modeling and a database of approximately 55 000 molecules.20 Nonetheless, more studies with structure-based pharmacophore models are necessary to validate the approach as a valid strategy to detect new hits against PknG. Although the crystallographic structure of Mtb PknG is available,10 there is no pharmacophoric model based on the kinase structure reported in the literature.
Thus, exploration and development of new pharmacophore models for the detection of Mtb PknG inhibitors require: (i) generating structure-based pharmacophore models using the X-ray diffraction structure of PknG; (ii) validating the models theoretically using active, inactive, and decoy compounds, (iii) expanding the chemical space with the use of a more extensive chemical library, and, (iv) validating the potential inhibitors by biochemical methods.
In the present study, we set up a cloud-based high-throughput screening platform capable of using a structure-based pharmacophore model on the ChEMBL21 library, consisting of 1 578 014 molecules (Figure 2). The screening led to 689 candidates with the potential to bind to the PknG active site. The candidates from the pharmacophore screening were subjected to additional screening employing molecular docking to identify potential inhibitors. The molecular docking study led to 62 promising compounds with binding affinities of −7.54 kcal/mol (s.d. = 0.77 kcal/mol). A complementary analysis considering previously reported biological activities, price, and commercial availability led to 14 molecules. From this set, compounds with favorable binding affinities and interactions with PknG active site’s Glu233 and Val235 were selected for biochemical assessments. The PknG inhibition and the IC50 were determined by measuring kinase activity. Finally, to understand the binding modes of potential candidates and to analyze the stability of proposed binding interactions, protein–ligand complexes were subjected to molecular dynamics simulations.
Figure 2.
(A) Workflow to identify compounds from CHEMBL21 database as PknG inhibitors (B) top 14 compounds biologically evaluated for PknG inhibition.
2. Methods
2.1. Computational Studies
2.1.1. Software
The protein structure was prepared using QuickPrep tool from MOE 2020.0901.21 Binding site prediction, drawing and editing of chemical structures as Simplified Molecular Input Line Entry Specification (SMILE), generation and evaluation of structured-based models, remote virtual screening, and creation of a virtual screening library were performed using LigandScout 4.4.3 Expert from Inte:Ligand GmbH.22 For virtual screenings, LigandScout algorithms such as Icon conformer generation and idbgen were used to create multi-conformational compound libraries.23,24 NAMD (version 2.14)25 and AMBER force fields26 were used for MD simulations. Molecular docking calculations were performed using AutoDock Vina 1.127 implemented in LigandScout 4.4.3 Expert.
2.1.2. Binding Site and Docking Validation
The X-ray diffraction structure of Mtb PknG in complex with the reference compound AX20017 (PDB ID: 2PZI, resolution 2.40 Å)10 was employed and obtained from the RCSB Protein Data Bank.28 The complex structure was prepared using the QuickPrep tool from MOE 2020.0901,21 which involves steps such as reparation of structural problems, rebuilding of the hydrogen-bond network, protonation, and energy minimization. The LigandScout’s algorithm allows the identification of ligands inside the protein–ligand complexes and automatically generates the grid box. In this work, the grid box was automatically generated using the AX20017’s PknG binding site of crystal 2PZI. AutoDock Vina 1.1 implemented in LigandScout was used to perform the docking experiments, employing a rectangular box of 17 × 12 × 14 Å3 centered at 21.557, −10.365, and −4.111 (x, y, z). The accuracy of this procedure was verified by redocking the AX20017 compound as a positive control. After that, a library of active compounds of series A15 and C29 were docked reproducing the main interactions of the tetrahydrobenzothiophene (THBT) family (hydrogen bond formation with residues Glu233 and Val235) (Table S1). Additionally, previous structural studies reported that all of the active compounds from series A and C have hydrophobic interactions with the N-terminal segment of PknG.12 The following default settings were employed: 80 of exhaustiveness, 10 maximum number of modes, and the maximum energy difference of 3 kcal/mol. Poses for AX20017 and active series A and C were prioritized based on AutoDock’s binding affinity score and LigandScout’s total number of pharmacophore feature interactions. The RMSD between the THBT scaffold from series A and C’s docking poses and AX20017’s crystallographic binding mode was computed with LigRMSD’s flexible mode.30
2.1.3. Pharmacophore Hypothesis Generation
The previously prepared structure of the PknG-AX20017 complex was used to generate the initial structure-based pharmacophore hypothesis using the structure-based module of LigandScout 4.4.3. The docked conformations of active series A and C, also called THBT derivatives, were used to generate pharmacophore models. The active compounds were selected based on an IC50 value below 5 μM. Starting from the prepared protein–ligand complexes, LigandScout can automatically generate chemical feature interactions including (a) hydrophobicity, (b) hydrogen-bond donor, and (c) hydrogen-bond acceptor. Then, the alignment module of LigandScout was used to align and merge structure-based models of AX20017 and compound SR_A6 (AX20017 as a reference point). Lastly, the initial merged model was optimized by adding exclusion volumes considering inactive data of series A and C.
2.1.4. Cloud-Based High-Throughput Screening Platform
Based on the client-server model architecture, “LigandScout Remote” module was implemented on the Amazon Web Service (AWS). Detailed information about its implementation can be found on the LigandScout website (https://docs.inteligand.com/ls-remote/) or in the respective literature.31 The virtual cluster was composed of five instances type c5.2xlarge and 40 cores.
2.1.5. Virtual Screening Using the Optimized Merged Pharmacophore Model
Chemical structures of 1 578 014 molecules from the CHEMBL21 database32,33 underwent virtual screening using the developed pharmacophore. Using the “Remote” screening module of LigandScout and an Amazon Web Service virtual cluster, the virtual screening was performed to identify potential inhibitors of PknG. The parameters included (1) scoring function: Pharmacophore fit; (2) screening mode: match all query features; (3) retrieval mode: stop after first matching conformation; (4) maximum number of omitted features: zero; and (5) check exclusion volumes: true. Virtual hits were ranked based on the pharmacophore fit score.
2.1.6. Virtual Screening Using Molecular Docking
Initial screening using the optimized pharmacophore model yielded 689 hits with the probability to interact with the PknG catalytic site. This subset of molecules was then subjected to molecular docking against the 2PZI PknG crystal to screen the most promising candidates. Molecular docking-based virtual screening was performed employing AutoDock Vina 1.1 implemented in LigandScout 4.4.3. The procedure used was the same as detailed in the docking validation section. The inclusion of Glu233 and Val235 in the search box was verified. The binding affinity score of AutoDock Vina 1.1 was applied to rank the candidates. Potential candidates were selected based on a careful inspection of (1) hydrogen bonding interactions with Glu233 or Val235, (2) characteristic hydrophobic contacts in the PknG binding pocket, and (3) surface complementarity.
2.1.7. Molecular Dynamics
The protonated complexes (pH 7) between ligands RO9021, AX20017, 6(4), and PknG, obtained from docking, were subjected to 100 ns molecular dynamics simulations (MD) using NAMD 2.14 (simulations of holo-structures).25 Additionally, a simulation of just the PknG structure (simulations of the apo-structure) was also conducted. PknG is capable of coordinating with a metal atom by residues Cys106, Cys109, Cys128, and Cys131, which are located in a rubredoxin-like domain.34,35 Rubredoxin domains usually coordinate iron, however, the metal coordinated by PknG remains unknown.35 This region is natively disordered if not coordinating a metal.34 Given the proximity of this domain to the ligands’ binding pocket, not considering a coordinating metal could lead to binding instability. Hence, a zinc atom in the coordination site was considered in the PknG structure as it already has precomputed parameters within the AMBER force field26 and there is experimental evidence that PknG can coordinate zinc.11
Each system was solvated in a TIP3P orthorhombic water box using the “tleap” module of AmberTools 21.36,37 NaCl counterions were placed to neutralize the system based on a Coulombic potential grid. After neutralization, NaCl concentration was set to 0.154 M placing its ions randomly. Box’s edges were set to be at least 15 Å of distance from the protein’s surface. Final topologies and parameters for each system were generated in the same module, using the ff19SB force field38 to model the protein and the zinc AMBER force field (ZAFF) for the coordination site.26 Ligands were parameterized with ACPYPE,39 assigning AM1-BCC charges40,41 and force constants based on the second generation of the general AMBER force field (GAFF2).42 Ions were modeled using Li/Merz parameters with the 12-6-4 Lennard–Jones-type nonbonded model.43
As the protein was previously refined and minimized (see Section 2.1.2), a minimization using only the conjugate gradient algorithm was conducted. Initially, only water atoms and ions were minimized for 5000 steps. Then, an NVT MD of the same atoms was performed for 30 ps at 298.15 K. Finally, a 5000-step minimization of the whole system was performed, until a convergence at around −672 298 kcal/mol was achieved for all of the systems (Figure S1). Subsequently, the temperature and pressure were equilibrated to 298.15 K and 1 bar, according to the procedure described elsewhere.44 Both the Langevin thermostat and Nosé–Hoover Langevin barostat were used in the NPT ensemble. Briefly, the system was heated from 50 to 298 K, increasing the temperature by 4 K every 10 ps while applying harmonic restraints to the protein backbone and the ligand with a force constant of 5 kcal/mol. Then, the restraints were reduced by 10% every 0.05 ns. Finally, a 100 ns MD of the NPT ensemble at 298.15 was performed. All simulations under the NPT ensemble were set at 1 bar. All of the processes described were performed under periodic boundary conditions, with an integration time of 2 fs/timestep and Particle Mesh Ewald (PME) grid spacing of 1.0 Å. The cut-off for nonbonded interactions was 12 Å. Water molecules were treated as rigid using the SETTLE algorithm. Other hydrogen-containing bonds were constrained using the SHAKE algorithm.
2.1.7.1. RMSD and RMSF Analysis
The root-mean-square deviation (RMSD) of each protein–ligand trajectories was computed with an in-house Tcl script using VMD and the module “bigdcd”.45 The complex coordinates from docking were used as a reference, to which all frames of the trajectory were aligned. Protein’s RMSDs were calculated using α carbons, while ligands were calculated using all non-hydrogen atoms. The effect of each ligand on protein flexibility was assessed using root-mean-square fluctuation (RMSF) measurements. For them, the last 70 ns of each trajectory were loaded with a step of 1, aligned against the complex coordinates from docking, and the RMSF computed using protein’s α carbons. RMSF measurements of the apo-simulations were also calculated for comparison.
2.1.7.2. RMSD-Based Clustering
The frames of the last 70 ns of each protein–ligand trajectory were clustered based on the ligand’s RMSD using Wordom v.0.22,46 using the algorithm proposed by Daura et. al.47,48 with a threshold of 2 Å. The centroids of each cluster were retrieved for further analysis, as they contain the representative binding modes of each ligand. RMSDs of the centroids with respect to the initial pose from docking were computed using DockRMSD.49
2.1.7.3. Interaction Analysis
MD of protein–ligand complexes was analyzed to detect and count the total hydrogen bonds, salt bridges, halogen bonds, cation-π, π-stacking, and hydrophobic interactions established by the ligands through the last 70 ns of MD. To that end, a bash script (“run_interact.sh”) freely available at https://github.com/tavolivos/Molecular-dynamics-Interaction-plot was employed. This script was successfully used in the previous work.50 The evaluation was carried out with 100 ps of resolution. The frequency of each ligand’s interactions per protein residue was expressed as a percentage of the total interactions established by the ligand in stacked bar plots. Residues with less than 2.5% of total interactions were omitted.
2.2. Biological Evaluation
2.2.1. Cloning, Expression, and Purification of M. tuberculosis PknG and GarA
The DNA sequences of PknG (NCBI gene ID 886397) and GarA (NCBI gene ID 885735) were cloned into a pET-24a plasmid (KanR) using GeneScript (USA). The codon sequence of both proteins was optimized for expression in E. coli BL21(DE3) pLysS, and a His-Tag label was added to the C-terminus. Competent cell preparation and transformation were performed according to the Mix & Go E. coli transformation kit protocol (Zymo Research). Transformed cells were cultivated in Luria–Bertani (LB) agar supplemented with 30 μg/mL kanamycin (KAN). For protein expression, a single colony was inoculated in LB medium supplemented with 30 μg/mL KAN and incubated at 37 °C and 150 rpm overnight. Then an aliquot was transferred to fresh LB medium + KAN to get an optical density at 600 nm (OD600) of 0.05 and incubated at 37 °C and 150 rpm until reaching an OD600 of 0.6. Induction of protein expression was performed by adding 0.2 mM IPTG and incubation at 22 °C and 150 rpm for 16 h. Afterward, cell pellets were collected by centrifugation at 5000 rpm at 4 °C for 15 min. For PknG, the cell pellet was washed twice with Lysis P buffer (25 mM Na2HPO4, 500 mM NaCl, 25 mM Imidazole, 5% glycerol, pH 8.0), and resuspended with Lysis P buffer supplemented with 100 μg/mL lysozyme, 40 μg/mL DNase I, and complete protease inhibitor (Roche) and stored at −80 °C. Cell lysis was performed by three freeze and thaw cycles. The lysate was clarified by centrifugation at 10 000 rpm and 4 °C for 45 min. The supernatant was filtered through a 0.45-μm Millex-HV syringe filter (Sigma-Aldrich). PknG was purified using a HisTrap FF Crude column (Cytiva) equilibrated with Lysis P buffer. Protein washing and elution were performed with an imidazole gradient range of 25–300 mM in Lysis P buffer. Eluted fractions were analyzed by 15% SDS-PAGE and Coomassie blue staining. Fractions containing protein were pooled and dialyzed in a D-tube Dialyzer, MWCO 12–14 kDa (Merck, Germany) overnight at 4 °C against Dialysis P buffer (50 mM Tris–HCl, 500 mM NaCl, 5% glycerol, pH 8.0). For GarA, the used buffers were replaced with Lysis G buffer (25 mM HEPES, 500 mM NaCl, 25 mM imidazole, 10% glycerol, pH 8.0) and dialysis G buffer (25 mM Tris–HCl, 150 mM NaCl, 1 mM DTT, 5% glycerol, pH 8.0), respectively. Protein quantification was performed by the Bradford protein assay (Bio-Rad) using BSA (Sigma-Aldrich) as the standard.
2.2.2. Luminescence-Based Inhibitory Assay
Compounds were dissolved in DMSO at a 10 mM concentration. PknG phosphorylation activity was evaluated with GarA as a substrate using the luminescence-based ADP-Glo assay (Promega). The reaction used 170 nM PknG, 7 μM GarA, and 10 μM ATP in reaction buffer (25 mM HEPES, 100 mM NaCl, 5 mM MnCl2, 1 mM DTT, pH 7.4) at 37 °C for 40 min. For an initial evaluation, CHEMBL compound 10 μM each was evaluated using the reported PknG inhibitor AX20017 as a positive control.10 The phosphorylation reaction without inhibitor was used as non-inhibitor control (NIC), and a reaction without enzyme was used as background assay control (BAC), respectively. The ADP-Glo reagent was mixed with the phosphorylation reaction in a ratio of 1:1 and incubated at 25 °C for 45 min. Subsequently, the Kinase Detection reagent was added in a ratio of 1:1 and incubated at 25 °C for 45 min. Luminescence signal detection was performed in the Cytation 5 Cell Imaging Multi-Mode Reader (BioTek) at 25 °C for 1 h. Inhibition percentage (Inh%) was calculated using the following formula: Inh% = 100 × ((NIC – Inhibitor)/(NIC – BAC)). The IC50 values were determined by fitting the luminescence signal and compound concentrations, ranging from 0.1 μM to 100 μM, with an inhibitor–response model of three parameters using GraphPad Prism v9 (GraphPad Software Inc.).
2.3. Tested Compounds
Compound AX20017 was received from the MedChem Express Library (MCE; Monmouth Junction, NJ); and the CHEMBL compounds were received from Chemspace US Inc. (Chemspace; Monmouth Junction, NJ 08852). The purity of the compounds ranged from 96 to 99 percent.
3. Results and Discussion
3.1. Pharmacophore Model Generation and Virtual Screening
The crystallographic structure of the PknG-AX20017 complex is available in the Protein Data Bank (PDB code 2PZI).10 The crystal 2PZI represents a crucial starting point to study mandatory interactions to inhibit PknG (Figure 1). However, AX20017 is not the unique compound of the THBT family evaluated against PknG. A set of THBT derivatives called series A15 and C29, reported inhibitory activities against PknG of Mtb (IC50 values ranging from 0.01 to 30 μM). To explore and integrate the structure-based information and available biochemical data of PknG inhibitors, a pharmacophore model was created using structural information of the PknG-AX20017 complex and the docked active compounds. The docking parameters using AutoDock Vina were validated by redocking the 2PZI native inhibitor AX20017, achieving similar three-dimensional (3D) spatial conformation compared to the crystal structure, confirming the optimal choice of parameters. The calculated root-mean-square deviation (RMSD) for the docked AX20017 was 0.53 Å (Figure S2). Two hydrogen bonding interactions, including Glu233 and Val235, stabilized the PknG–AX20017 complex. The docked AX20017 structure reproduces the two hydrogen bonds. The structure-based pharmacophore creation was used to generate the initial merged feature model. Based on the guidelines available in LigandScout and the validated docking protocol, first, the binding poses for the twenty active derivatives were generated by docking experiments (Figure 3A). Then, twenty pharmacophores were generated for each binding mode of the THBT derivatives. After that, using the alignment perspective in LigandScout, the pharmacophore models for AX20017 and compound SR_A6 were aligned and merged. The combination of the respective merged models allows the detection of the twenty molecules. The initial model included seven features (Figure 3B): three hydrophobic, two hydrogen-bond acceptors, and two hydrogen bond donors (Figure 3).
Figure 3.
Binding modes of inhibitor AX20017 and THBT derivatives. (A) X-ray reference structure of AX20017 (in gray) (PDB ID: 2PZI) and docked conformations of twenty THBT derivatives (in cyan) used in pharmacophore modeling. Notice the alignment of common chemical features shown in the active site. RMSDs of the benzothiophene scaffold between AX20017 and THBT derivatives were in the range of 0.615 and 1.254 Å. (B) The starting merged pharmacophore that was created from specific binding modes for AX20017 and compound SR_A6. Yellow circle for hydrophobic interaction, red arrow for the hydrogen-bond acceptor, and green arrow for the hydrogen-bond donor. RMSD of the tetrahydrobenzothiophene scaffold between SR_A6 and AX20017 was 0.735 Å.
A final step was the addition of exclusion volumes. Although exclusion volumes can be added using parameters by default to the model (Figure 4A,B), they must be added manually and carefully considering information from experimentally inactive compounds. In this way, the specificity of the initial merged pharmacophore model can be considerably improved (Figure 4C). Thus, resulting in a final optimized final model (Figure 4D).
Figure 4.
Pharmacophore models created using the structured-based approach of LigandScout. (A) Pharmacophore model for PknG-AX20017 (PDB: 2PZI) using parameters by default. (B) Pharmacophore model for PknG-AX20017 with excluded volumes (gray spheres) added by default. (C) Initial pharmacophore model for PknG by the merged approach. Pharmacophore models for compounds AX20017 and SR_A6 were aligned and then merged using the alignment module of LigandScout. The merged pharmacophore is composed of three hydrophobic features, two hydrogen-bond donor features, and two hydrogen-bond acceptor features. (D) Optimized pharmacophore model for PknG with excluded volumes (gray spheres) manually added to avoid inactive molecules. Yellow circle for hydrophobic interaction, red arrow for the hydrogen-bond acceptor, and green arrow for the hydrogen-bond donor.
3.1.1. Pharmacophore Model Performance Analysis
Before a large virtual screening process, the validation of the pharmacophore model is crucial to provide reliable hits on a real-life project. To test the ability of the optimized merged pharmacophore model (Figure 4D) to maximize the number of active hits and reject inactive ones, a test set database of 1122 compounds was prepared consisting of twenty active compounds of series A and C (Table S1), twelve inactive compounds of series C, ninety-two inactive compounds reported previously for PknG,16 and nine hundred ninety-eight virtual decoys generated using the DUDE-E homepage. Therefore, performance parameters were calculated such as sensitivity (Se), specificity (Sp), % ratio of actives (RA), % yield of actives (YA), false positives (Fp), false negatives (Fn), enrichment factor (EF), and goodness of hit (GH). The performance parameters for the optimized merged model are summarized in Table 1. The pharmacophore model showed mainly Se of 1, Sp of 0.99, EF of 43.21, GH of 0.82, and RA of 64.70.
Table 1. Performance Parameters for Optimized Merged Pharmacophore Model (AX20017-SR_A6).
| parameter | optimized model |
|---|---|
| total no. of molecules in the database (D) | 1122 |
| total number of actives (A) | 20 |
| total number of inactives (I) | 1102 |
| total hits (Ht) | 26 |
| active hits (Ha) | 20 |
| sensitivity | 1 |
| specificity | 0.99 |
| % yield of actives (Ha/Htx100) | 76.9% |
| % ratio of actives (Ha/Ax100) | 100% |
| enrichment factor (EF) | 43.21 |
| false positives (Ht-Ha) | 6 |
| false negatives (A-Ha) | 0 |
| goodness of hit score (GH) | 0.82 |
3.1.2. Pharmacophore-Based Virtual Screening
Using a 3D pharmacophoric pattern as a 3D probe filter, pharmacophore-based virtual screening is a computational approach where the retrieval of compounds with similar and desired properties from large libraries is the main objective. As a result, hit molecules emerge as a starting point for the drug development process. To identify potential new hits for Mtb PknG, the optimized merged pharmacophore model was used to filter 1 578 014 candidates from the CHEMBL21 database (https://www.ebi.ac.uk/chembl/) using the “remote screening” module of LigandScout. In this way, high-performance computing in the cloud through Amazon Web Service was used to search for a potential candidate. The virtual screening time was 5 h and 15 min, resulting in 689 hits and a hit rate of 0.043% (Table S2). The potential 689 hits, which met the specified pharmacophoric requirements, moved forward to the molecular docking calculations against the binding site of PnkG. According to the literature, this is the first case of pharmacophore-based virtual screening using cloud computing to search for a PknG inhibitor in a compound library of 1.5 million molecules. The calculation time and the hit rate demonstrate its feasibility and importance when searching for new inhibitors for infectious diseases.51
3.2. Molecular Docking and Candidate Selection
The hits obtained from the pharmacophore-based virtual screening were further filtered through molecular docking-based screening. The docking experiment assesses interactions of different states of compounds into the substrate-binding pocket of PknG to rank them based on the docking score (binding affinity: kcal/mol). Using the validated docking protocol, the molecular docking-based virtual screening led to 62 promising inhibitors (Table S3) selected based on a careful inspection of (1) hydrogen bonding interactions with Glu233 or Val235, (2) characteristic hydrophobic contacts in the PknG binding pocket, and (3) surface complementarity. From the latter subset, eleven candidates were selected according to (1) commercial availability, (2) price, (3) previous literature information, and (4) possible establishment of the structure–activity relationship. Thus, these eleven compounds were purchased and assayed for biological activity (Table 2).
Table 2. Selected CHEMBL Candidates Based on the Two Sets of Criteria toward Mtb PknG.
| interaction
features |
||||||
|---|---|---|---|---|---|---|
| CHEMBL ID | docking score (kcal/mol) | pharmacophore fit-score | hydrophobic | hydrogen bond acceptor | hydrogen bond donor | total number of interactions |
| CHEMBL1584623 | –8.3 | 43.5 | 7 | 1 (TYR234) | 1 (GLU233) | 9 |
| CHEMBL1530562 | –7.8 | 62.88 | 9 | 2 (VAL235) | 1 (GLU233) | 12 |
| CHEMBL1466996 | –7.8 | 42.84 | 8 | 1 (GLU233) | 9 | |
| CHEMBL3462040 | –7.7 | 43.78 | 10 | 1 (VAL235) | 1 (GLU233) | 12 |
| RO9021 | –7.7 | 43.75 | 9 | 1 (VAL235) | 2 (GLU233, VAL235) | 12 |
| CHEMBL2152572 | –7.6 | 43.93 | 8 | 1 (VAL235) | 2 (GLU233, VAL235) | 11 |
| CHEMBL1325582 | –7.3 | 43.66 | 10 | 1 (VAL235) | 2 (GLU233, VAL235) | 13 |
| CHEMBL1915540 | –7 | 51.26 | 8 | 1 (VAL235) | 1 (GLU233) | 10 |
| CHEMBL158113 | –6.8 | 51.53 | 9 | 1 (GLU233) | 10 | |
| CHEMBL158466 | –6.8 | 51.37 | 5 | 1 (GLU233) | 6 | |
| CHEMBL1884808 | –5.5 | 42.95 | 9 | 1 (GLU233) | 10 | |
| AX20017 | –7.1 | 51.82 | 8 | 1 (VAL235) | 1 (GLU233) | 10 |
Additionally, to test the criteria only based on the pharmacophore-match and pharmacophore fit-score, starting from 689 hits identified previously, three compounds with a low probability to bind the active site of PknG according to docking calculations were purchased and subjected to biological evaluation (Table 3).
Table 3. Selected CHEMBL Candidates Based on Pharmacophore-Match and Pharmacophore Fit-Score toward Mtb PknG.
| interaction
features |
|||||
|---|---|---|---|---|---|
| CHEMBL ID | pharmacophore fit-score | hydrophobic | hydrogen-bond acceptor | hydrogen-bond donor | total number of interactions |
| CHEMBL3479284 | 62.78 | 5 | 1 (VAL235) | 1 (GLU233) | 7 |
| CHEMBL1076619 | 54.76 | 7 | 1 (VAL235) | 1 (GLU233) | 9 |
| CHEMBL3458727 | 52.08 | 8 | 1 (VAL235) | 9 | |
| AX20017 | 51.82 | 8 | 1 (VAL235) | 1 (GLU233) | 10 |
Therefore, fourteen compounds in total were purchased and subjected to biological evaluation (Table S4).
3.3. Biological Evaluation of Compounds against Mycobacterial PknG
3.3.1. Performance Evaluation of the Phosphorylation Assay
Phosphorylation activity can be evaluated by measuring substrate depletion (i.e., ATP molecules) or product formation (i.e., ADP molecules).52 Luminescence-based assays have been used widely to evaluate phosphorylation activity and high-throughput compound screening.53 In the present study, the product formation assay ADP-Glo was used to evaluate the phosphorylation activity of Mtb PknG. This commercial system has been reported to evaluate potential PknG inhibitors in the past.13,16,20,54 However, to date, details of its validation and fine-tuning have been scanty or unclear. First, to optimize the phosphorylation reaction, we have evaluated the use of metal ions as PknG cofactors. The Mg2+ and Mn2+ ions have been described to be essential for PknG activity.10 At a concentration of 5 mM, the reaction buffer with MnCl2 showed a 1.5× higher luminescent signal than the reaction buffer with MgCl2. Furthermore, a signal-to-noise ratio > 2 was observed (NIC/BAC), and inhibition by 100 μM AX20017 was reported (two-fold less signal) (Figure S3A). High luminescence signals (more than 2000 RLU (a.u.) were observed in the background assay control (BAC) that included only the substrate (GarA) (Figure S3B). Second, to rule out the presence of any contaminant kinase in the protein preparations, a synthetic peptide with the phosphorylation region of GarA (SDEVTVETTSVFRADFL, corresponding to residues 14–30) was evaluated.55 Phosphorylation reaction was performed with 25 nM PknG, 10 μM ATP, and two substrate/enzyme (S/E) ratios were evaluated (1:50 and 1:100). Phosphorylation reactions were incubated for 1 and 2 h independently. A signal-to-noise ratio of 1.5 was observed for reactions with S/E ratios of 100, with a BAC reaction signal at around 1500 RLU (a.u.) (Figure S3C). These results suggested that the high background signal could be due to an unspecific ATP depletion. To confirm the observation, the stability of ATP was evaluated under the above-described reaction conditions. Buffer reaction with 10 μM ATP was incubated at 37 °C for 1 h. The luminescence signal of ATP incubated was compared to reactions without previous incubation at 37 °C (ATP, ATP—2.5 μM peptide, and ATP—7 μM GarA). In general, the results indicated a similar luminescence signal (>1200 RLU (a.u.)) in reactions without incubation independently of the presence and type of substrate. ATP incubated showed a faintly higher signal (Figure S3D). Regardless of the background signal, the specific inhibitory activity of the AX20017 molecule was observed (Figure S3).
3.3.2. Screening of Selected Candidates and Determination of IC50 against Kinase Activity
The selected candidates were screened as inhibitors of the kinase activity of PknG. Fourteen candidates were evaluated with the luminescence-based assay to determine inhibitory activity using the ADP-Glo assay kit. The compounds CHEMBL1584623, CHEMBL1530562, CHEMBL1466996, CHEMBL3462040, RO9021 (CHEMBL3237561), CHEMBL2152572, CHEMBL1325582, CHEMBL1915540, CHEMBL158113, CHEMBL158466, CHEMBL1884808, CHEMBL3479284, CHEMBL1076619, and CHEMBL3458727 showed inhibitory percentage (INH%) values of 41, 42, 32, 32, 61, 30, 31, 39, 34, 43, 39, 48, 40, and 33% respectively against Mtb PknG at 10 μM (Figure 5A). In the same assay, the control AX20017 showed an INH% value of 56%. Only AX20017 and RO9021 showed INH% values of more than 50% at a concentration of 10 μM. Dose–response assays demonstrated that only AX20017 and RO9021 showed specific inhibitory activity against PknG (Figure 5B). A substantial reduction of the luminescent signal was observed for these molecules with a negative dependence at an increasing inhibitor concentration (Figure 5B). AX20017 exhibited, under our experimental conditions, a relative IC50 of 0.2 ± 0.04 μM (Figure 5C) while compound RO9021 presented a relative IC50 of 4.4 ± 1.1 μM (Figure 5D). The present results are in agreement with other screening attempts for detecting PnkG inhibitors,16 where 50% of INH% was considered the cut-off point for successful dose–response tests.
Figure 5.
Screening of selected candidates against kinase activity. (A) Inhibitory percentage values reported for AX20017 and selected candidates from the CHEMBL database. Initial screenings were performed with 10 μM compounds. The dashed line represents the maximum inhibitory percentage reached by AX20017 under our experimental conditions (B) Dose–response assay. The luminescence signal was normalized by subtracting the BAC signal and then calculating a ratio against the NIC reaction. (C) Dose–response assay for AX20017. The luminescence signal was plotted against the log compound concentration. Compounds were tested ranging from 100 nM to 100 μM. (D) As in (C), for RO9021.
In that sense, the present work has identified RO9021 (6-[(1R,2S)-2-amino-cyclohexylamino]-4-(5,6-dimethyl-pyridin-2-ylamino)-pyridazine-3-carboxylic) as a new hit for TB. However, it has been reported that this compound is also an ATP-competitive inhibitor of the human enzyme spleen tyrosine kinase (SYK), developed to treat rheumatoid arthritis and multiple sclerosis.56,57 Hence, selectivity improvement by chemical modifications to achieve interactions with amino acids unique to Mtb PknG is required. RO9021’s excellent pharmacokinetic parameters make it a promising template to introduce these modifications. For instance, its oral bioavailability56 is of particular interest as it is a key feature to take into account when choosing scaffolds to design new anti-tuberculosis drugs for low-income countries. The discovery of such an adequate starting point and the fact that proposed PknG inhibitors are often other kinase inhibitors,15−17 support the strategy and results applied and obtained in this study.
3.4. Proposed Mode of Binding of Active Compound RO9021
3.4.1. Molecular Docking
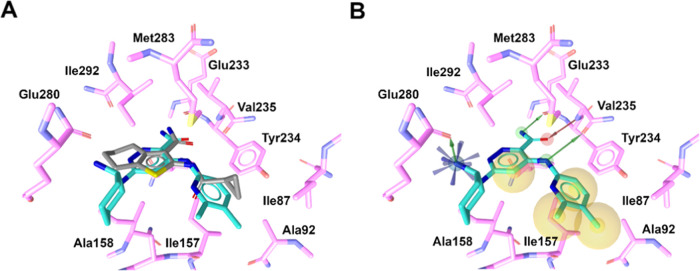
To propose the ligand–receptor binding mechanism, a molecular docking simulation was performed. The predicted binding mode for the active compound RO9021 with an INH% value of 61% and IC50 of 4.4 μM in the kinase assay is shown in Figure 6. RO9021 was predicted to bind inside the substrate-binding pocket in a similar 3D arrangement to AX20017, reproducing a crucial pattern of hydrogen bond formation (Figure 6A). In this pose, it had a binding energy of −7.7 kcal/mol, higher than the −7.1 kcal/mol of the reference ligand (AX20017). The RO9021 compound was hydrogen-bonded to the hinge region residues Glu233 and Val235 due to the amide group of the ligand. Additionally, two hydrogen bonds with the CO backbone of V235 and Glu280 were also observed (Figure 6B). Except for the interaction with Glu280, all of the interactions are in agreement with the hydrogen-bond donor and hydrogen-bond acceptor features of the optimized pharmacophore proposed. Hydrophobic interactions were detected with residues Ile87, Ala92, Ile157, Ala158, Ile165, Val179, Tyr234, Met283, and Ile292.
Figure 6.
Potential binding mode of compound RO9021. (A) X-ray reference structure of AX20017 (in gray) (PDB: 2PZI) and docked conformation of RO9021 (cyan). (B) Main interactions in the PknG binding site for hit compound RO9021. Yellow circle for hydrophobic interaction, red arrow for the hydrogen-bond acceptor, blue star for the positive ionizable area, and green arrow for the hydrogen-bond donor.
3.4.2. Molecular Dynamics Simulations
3.4.2.1. RMSD Measurements
MD is a valuable tool for assessing the stability of RO9021 within the targeted binding pocket, as well as to evaluate drug-induced conformational changes of PknG that could contribute to inhibition. In this work, 100 ns of MD were conducted for the complex between PknG and RO9021 obtained from molecular docking. The same was done for PknG complexes with AX20017 and a previously reported inactive compound, 6(4).13 Additionally, an apo-PknG was simulated for the same amount of time. Constant temperature, density, and potential energy in 100 ns was verified before the analysis (Figure S4).
For each protein–ligand trajectory, the protein and ligand RMSDs were computed. The RMSD of AX20017 remained practically constant during the 100 ns of MD, with an average value of 1.34 Å (s.d. = 0.36 Å) (Figure 7A). This value plus two standard deviations were used as a threshold to determine the stability of the docking binding mode. We considered that the docking binding mode of a ligand was less stable than AX20017’s if its RMSDs persistently fluctuated around a value above this threshold. As expected, the RMSD of 6(4) oscillated above the threshold after the first 20 ns of MD, which reflects its binding instability (Figure 7B). Notably, RMSD of RO9021 always fluctuated below the threshold, which indicates promising stability of the binding mode obtained from molecular docking (Figure 7C).
Figure 7.
RMSDs of RO9021 as compared to AX20017 during 100 ns of MD. (A) Time courses of MD simulation of PknG apo (gray) or AX20017-bound (pink). (B, C) as A for 6(4) and RO9021, respectively. Pink horizontal line shows the ligand thresholds based on the PknG-AX20017 complex simulation (RMSD average + 2 s.d.). (D) Representative binding mode of the MD (centroids) obtained for AX20017 RMSD-based clustering compared to the initial docked state, RMSD obtained 0.908 Å. (E) as (D) for compound 6(4), RMSD range between 1.968 and 5.610 Å (F) as (D) for RO9021, RMSD obtained 1.676 Å. In (D)–(F). Protein atoms are represented by transparent cartoons. Ligands are depicted as sticks and their atoms are colored by the CPK convention. The initial docked state is colored gray (D), orange (E), and cyan (F). The unique representative binding modes of AX20017 and RO9021 are colored in green RO9021. For clarity, only 3 of the 10 representative binding modes of 6(4) are shown, for a complete list see Table S4. The binding states are colored using a grayscale where lighter grays indicate that the centroid corresponds to a time frame further along the simulation.
3.4.2.2. RMSD-Based Clustering
A ligand RMSD-based clustering was performed over the last 70 ns per protein–ligand complex. The centroids of each cluster were retrieved as the representative ligand binding modes. AX20017 formed only 1 cluster (and hence, 1 representative structure) (Table S5). In addition, the representative binding mode was similar to that obtained from molecular docking (Figure 7D), which is confirmed by an RMSD of 0.908 Å. In contrast, compound 6(4) formed 10 clusters (Table S5). On average, its representative binding modes differed in 3.792 Å (s.d. = 1.176 Å) with the initial docked pose. Notably, the results of the RMSD-based clustering of RO9021 were like that of AX20017’s (Table S5 and Figure 7F). It formed a single cluster with a representative binding mode similar to the initial docked pose, as RMSD between both was 1.676 Å.
The agreement of AX20017’s docking and MD binding modes suggests that a very similar conformation is stably adopted by this ligand in reality, which also indicates inhibitory potential. This is confirmed, as AX20017 is an active compound against PknG and docking and MD binding modes converge with the crystallographically determined pose. Hence, the present methodology seems to be able to predict active compounds and their real binding poses. Moreover, the divergence between 6(4)’s MD multiple binding modes and its initial docked pose suggest instability and correlates with its inactivity. Thus, the methodology followed here also seems capable of predicting binding instability and, therefore, rejecting inert molecules. In that sense, convergence between RO9021’s MD and docking binding modes suggests that a similar and stable conformation is expected in reality. This indicates a promising potential as a PknG inhibitor.
3.4.2.3. RMSF Analysis
Protein RMSF calculations were performed to assess the effect of the two stable compounds, AX20017 and RO9021, on the protein conformational landscape. RMSFs were computed along the last 70 ns for each protein–ligand complex to avoid measuring effects caused by the ligand initial stabilization. AX20017 destabilized residues 127–134 and restrained residues 150–159 and 241–246 (Figure 8A). RO9021 caused the same effects on similar regions of the protein (Figure 8B). The fact that RO9021 shared these effects with AX20017, an extensively tested active molecule, reaffirms its promising inhibitory capacity. Furthermore, the sharing of these effects between two active molecules might indicate that they are important for understanding the structural inhibitory mechanism.
Figure 8.
Protein RMSF and ligand interaction mapping from PknG-AX20017 and PknG-RO9021 complexes. (A, B) Protein RMSFs of protein–ligand complexes (holo-PknG) are given (pink lines). Protein RMSF of the apo-PknG simulation is also shown in each plot (black line) to facilitate comparison. Blue arrows indicate differences between apo- and holo-structures present in both, PknG-AX20017 and PknG-RO9021 complexes. (C, D) The frequency of different types of interactions per protein residue is expressed as a percentage of the ligand’s total interactions and shown in stacked bar plots. Residues with less than 2.5% of the ligand’s total interactions were omitted. The panels show analysis for complexes with (A/C) AX20017 and (B/D) RO9021.
3.4.2.4. Interaction Analysis
Interaction mapping is a crucial step to evaluate the structural basis and biological significance of ligand binding and inhibition to a protein target. In addition, analysis of the nature of the most frequent interactions formed by the ligand could give a notion about its stability within the binding pocket. In this research, identification and counting of different types of ligand interactions formed during the last 70 ns of MD were performed. AX20017’s more frequent interactions were with Cys106, Glu233, and Val235 (Figure 8C). Notably, all of the contacts with these residues were hydrogen bonds. Hydrogen bonds with Glu233 and Val235 were the same as those identified in the optimized pharmacophore model created for the complex with this ligand (Figure 4D). The most frequent interactions of RO9021 were with Glu233, Val235, and Glu280 (Figure 8D). Exactly as AX20017, interactions with Glu233 and Val235 were exclusively hydrogen bonds, the same as the ones identified in molecular docking (Figure 6B). This confirms that RO9021 is able to persistently reproduce the hydrogen-bond features of the optimized pharmacophore proposed for the complex with AX20017. The stability of both hydrogen bonds for AX20017 and the fact that RO9021 replicates it further validate their selection as features for the pharmacophore model.
4. Conclusions
PknG leads to the blocking of phagosome–lysosome fusion within the infected macrophages and therefore represents a promising target for new anti-TB drugs, especially for persistent mycobacteria. Consequently, it is important to increase the number of hits and lead compounds that inhibit PknG activity. In this study, the conduction of a structural pharmacophore-based virtual screening integrated with molecular docking and biochemical assays led to the identification of compound RO9021 as a promising PknG inhibitor. To our knowledge, this is the first structure-based pharmacophore model that used cloud computing for Mtb PknG, and its design and theoretical validation are described. A hit rate of 0.043% against 1.5 M molecules supports its high specificity during theoretical validation. Biochemical assays showed an INH% value of 60.6% for compound RO9021 with a relative IC50 value of 4.4 ± 1.1 μM. Both values are shown to be promising due to the kinase activity similarity to AX20017 and can be optimized in future medicinal chemistry projects. Molecular dynamics simulations showed the binding mode and stability of the predicted PknG-RO9021 complex. Reference AX20017 and compound RO9021 were stable within the PknG binding pocket supporting in vitro experiments. Further biochemical studies of mycobacterial growth with infected macrophage cells are required to establish a better understanding of the RO9021 potential. RO9021 is an excellent starting point to develop new anti-TB drugs due to its excellent reported pharmacokinetic parameters, especially oral bioavailability. Future medicinal chemistry projects should focus on improving its selectivity against the PknG of Mtb.
Acknowledgments
This work was funded by the Peruvian Consejo Nacional de Ciencia, Tecnología e Innovación Tecnológica and Fondo Nacional de Desarrollo Científico, Tecnológico y de Innovación Tecnológica to M.Q. during the call for funding “Proyecto Investigación Básica 2019-01” [FONDECYT, 406-2019]. Funding for seed idea and proof of concept was provided by the Universidad Peruana de Ciencias Aplicadas (C-19-2019 and C-014-2021-2) to M.Q. This work was supported by grants from the Peruvian Fondo Nacional de Desarrollo Científico, Tecnológico y de Innovación Tecnológica (Grants 154-2017-FONDECYT and 036-2019-FONDECYT-BM-INC.INV) to P.M. We are very thankful to Victor Zegarra for seeding the project and for exploratory experiments setting up protocols for PknG and GarA expression and purification. We especially thank Hewlett-Packard Company (HP) for the computer donation of a PC workstation used in this study, allowing us to perform molecular modeling simulations.
Glossary
Abbreviations
- AWS
Amazon web service
- EF
enrichment factor
- XDR-TB
extensively drug-resistant tuberculosis
- Fn
false negatives
- Fp
false positives
- GH
goodness of hit
- INH%
inhibitory percentage
- MD
molecular dynamics simulations
- MDR-TB
multidrug-resistant tuberculosis
- Mtb
Mycobacterium tuberculosis
- RA
ratio of actives
- RMSD
root-mean-square deviation
- Se
sensitivity
- Sp
specificity
- SMILE
simplified molecular input line entry specification
- SYK
spleen tyrosine kinase
- TB
tuberculosis
- THBT
tetrahydrobenzothiophene
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsomega.2c02093.
5000-step whole system minimizations of protein–ligand systems (Figure S1); molecular docking protocol validation (Figure S2); evaluation of the phosphorylation reaction conditions for PknG activity evaluation (Figure S3); active compounds from series A and C against Mtb PknG (Table S1); selected CHEMBL candidates based on molecular pharmacophore-based virtual screening (Table S2); selected CHEMBL candidates based on molecular docking-based virtual screening (Table S3); final CHEMBL candidates purchased and subjected to biological evaluation (SMILE representation) (Table S4); and results of the RMSD-based clustering (Table S5) (PDF)
Author Contributions
M.Q. and P.M. conceived the project. A.A.-S., R.A., G.J.-A., and M.Q. performed experiments. All authors analyzed the data. M.Q., M.Z., and P.M. wrote the manuscript with input of all authors.
The authors declare no competing financial interest.
Notes
In silico and experimental data discussed in this paper are shared in the Supporting Information. LigandScout 4.4.3 Expert software used for binding site prediction; drawing and editing of chemical structures as SMILES; pharmacophore modeling; and virtual screening campaigns is available via an academic license for users and can be downloaded at http://www.inteligand.com/. Molecular Operating Environment (MOE) MOE 2020.0901 software used for the protein preparation, which involves reparation of structural problems, the rebuilding of the hydrogen-bond network, protonation, and energy minimization is available via an academic license for users and can be downloaded at https://www.chemcomp.com/Products.htm. The NAMD 2.14 software used for MD can be downloaded free of charge at https://www.ks.uiuc.edu/Research/namd/. The AmberTools 21 software used to solvate each system can be downloaded free of charge at https://ambermd.org/GetAmber.php#ambertools. The Wordom 0.22 software used to cluster MD can be downloaded free of charge at https://sourceforge.net/projects/wordom/files/0.22/. VMD used for molecular visualization is available free of charge at https://www.ks.uiuc.edu/Research/vmd/.
Supplementary Material
References
- Pai M.; Behr M. A.; Dowdy D.; Dheda K.; Divangahi M.; Boehme C. C.; Ginsberg A.; Swaminathan S.; Spigelman M.; Getahun H.; Menzies D.; Raviglione M. Tuberculosis. Nat. Rev. Dis. Primers 2016, 2, 16076. 10.1038/nrdp.2016.76. [DOI] [PubMed] [Google Scholar]
- Furin J.; Cox H.; Pai M. Tuberculosis. Lancet 2019, 393, 1642–1656. 10.1016/S0140-6736(19)30308-3. [DOI] [PubMed] [Google Scholar]
- Global tuberculosis report; World Health Organization: Geneva, 2021; p 30.
- Seung K. J.; Keshavjee S.; Rich M. L. Multidrug-Resistant Tuberculosis and Extensively Drug-Resistant Tuberculosis. Cold Spring Harb. Perspect. Med. 2015, 5, a017863. 10.1101/cshperspect.a017863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gengenbacher M.; Kaufmann S. H. E. Mycobacterium tuberculosis: success through dormancy. FEMS Microbiol. Rev. 2012, 36, 514–532. 10.1111/j.1574-6976.2012.00331.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Upadhyay S.; Mittal E.; Philips J. A. Tuberculosis and the art of macrophage manipulation. Pathog. Dis. 2018, 76, fty037 10.1093/femspd/fty037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walburger A.; Koul A.; Ferrari G.; Nguyen L.; Prescianotto-Baschong C.; Huygen K.; Klebl B.; Thompson C.; Bacher G.; Pieters J. Protein kinase G from pathogenic mycobacteria promotes survival within macrophages. Science 2004, 304, 1800. 10.1126/science.1099384. [DOI] [PubMed] [Google Scholar]
- Cowley S.; Ko M.; Pick N.; Chow R.; Downing K. J.; Gordhan B. G.; Betts J. C.; Mizrahi V.; Smith D. A.; Stokes R. W.; Av-Gay Y. The Mycobacterium tuberculosis protein serine/threonine kinase PknG is linked to cellular glutamate/glutamine levels and is important for growth in vivo. Mol. Microbiol. 2004, 52, 1691–1702. 10.1111/j.1365-2958.2004.04085.x. [DOI] [PubMed] [Google Scholar]
- Ventura M.; Rieck B.; Boldrin F.; Degiacomi G.; Bellinzoni M.; Barilone N.; Alzaidi F.; Alzari P. M.; Manganelli R.; O’Hare H. M. GarA is an essential regulator of metabolism in Mycobacterium tuberculosis. Mol. Microbiol. 2013, 90, 356–366. [DOI] [PubMed] [Google Scholar]
- Scherr N.; Honnappa S.; Kunz G.; Mueller P.; Jayachandran R.; Winkler F.; Pieters J.; Steinmetz M. O. Structural basis for the specific inhibition of protein kinase G, a virulence factor of Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. U.S.A. 2007, 104, 12151–12156. 10.1073/pnas.0702842104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisa M.-N.; Gil M.; André-Leroux G.; Barilone N.; Durán R.; Biondi Ricardo M.; Alzari Pedro M. Molecular Basis of the Activity and the Regulation of the Eukaryotic-like S/T Protein Kinase PknG from Mycobacterium tuberculosis. Structure 2015, 23, 1039–1048. 10.1016/j.str.2015.04.001. [DOI] [PubMed] [Google Scholar]
- Caballero J.; Morales-Bayuelo A.; Navarro-Retamal C. Mycobacterium tuberculosis serine/threonine protein kinases: structural information for the design of their specific ATP-competitive inhibitors. J. Comput. Aided Mol. Des. 2018, 32, 1315–1336. 10.1007/s10822-018-0173-3. [DOI] [PubMed] [Google Scholar]
- Anand N.; Singh P.; Sharma A.; Tiwari S.; Singh V.; Singh D. K.; Srivastava K. K.; Singh B. N.; Tripathi R. P. Synthesis and evaluation of small libraries of triazolylmethoxy chalcones, flavanones and 2-aminopyrimidines as inhibitors of mycobacterial FAS-II and PknG. Biorg. Med. Chem. 2012, 20, 5150–5163. 10.1016/j.bmc.2012.07.009. [DOI] [PubMed] [Google Scholar]
- Székely R.; Wáczek F.; Szabadkai I.; Németh G.; Hegymegi-Barakonyi B.; Erős D.; Szokol B.; Pató J.; Hafenbradl D.; Satchell J.; Saint-Joanis B.; Cole S. T.; Őrfi L.; Klebl B. M.; Kéri G. A novel drug discovery concept for tuberculosis: Inhibition of bacterial and host cell signalling. Immunol. Lett. 2008, 116, 225–231. 10.1016/j.imlet.2007.12.005. [DOI] [PubMed] [Google Scholar]
- Sipos A.; Pató J.; Székely R.; Hartkoorn R. C.; Kékesi L.; Őrfi L.; Szántai-Kis C.; Mikušová K.; Svetlíková Z.; Korduláková J.; Nagaraja V.; Godbole A. A.; Bush N.; Collin F.; Maxwell A.; Cole S. T.; Kéri G. Lead selection and characterization of antitubercular compounds using the Nested Chemical Library. Tuberculosis 2015, 95, S200–S206. 10.1016/j.tube.2015.02.028. [DOI] [PubMed] [Google Scholar]
- Kanehiro Y.; Tomioka H.; Pieters J.; Tatano Y.; Kim H.; Iizasa H.; Yoshiyama H. Identification of Novel Mycobacterial Inhibitors Against Mycobacterial Protein Kinase G. Front. Microbiol. 2018, 9, 1517 10.3389/fmicb.2018.01517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kidwai S.; Bouzeyen R.; Chakraborti S.; Khare N.; Das S.; Priya Gosain T.; Behura A.; Meena C. L.; Dhiman R.; Essafi M.; Bajaj A.; Saini D. K.; Srinivasan N.; Mahajan D.; Singh R. NU-6027 Inhibits Growth of Mycobacterium tuberculosis by Targeting Protein Kinase D and Protein Kinase G. Antimicrob. Agents Chemother. 2019, 63, e00996-19 10.1128/AAC.00996-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qasaymeh R. M.; Rotondo D.; Oosthuizen C. B.; Lall N.; Seidel V. Predictive Binding Affinity of Plant-Derived Natural Products Towards the Protein Kinase G Enzyme of Mycobacterium tuberculosis (MtPknG). Plants 2019, 8, 477 10.3390/plants8110477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swain S. P.; Gupta S.; Das N.; Franca T. C. C.; Goncalves A. dS.; Ramalho T. C.; Subrahmanya S.; Narsaria U.; Deb D.; Mishra N. Flavanones: A potential natural inhibitor of the ATP binding site of PknG of Mycobacterium tuberculosis. J. Biomol. Struct. Dyn. 2021, 1–15. 10.1080/07391102.2021.1965913. [DOI] [PubMed] [Google Scholar]
- Singh N.; Tiwari S.; Srivastava K. K.; Siddiqi M. I. Identification of Novel Inhibitors of Mycobacterium tuberculosis PknG Using Pharmacophore Based Virtual Screening, Docking, Molecular Dynamics Simulation, and Their Biological Evaluation. J. Chem. Inf. Model. 2015, 55, 1120–1129. 10.1021/acs.jcim.5b00150. [DOI] [PubMed] [Google Scholar]
- Molecular Operating Environment (MOE), 2020.0901; Chemical Computing Group ULC: 1010 Sherbooke St. West, Suite #910, Montreal, QC, Canada, H3A 2R7, 2022.
- Wolber G.; Langer T. LigandScout: 3-D Pharmacophores Derived from Protein-Bound Ligands and Their Use as Virtual Screening Filters. J. Chem. Inf. Model. 2005, 45, 160–169. 10.1021/ci049885e. [DOI] [PubMed] [Google Scholar]
- Friedrich N. O.; de Bruyn Kops C.; Flachsenberg F.; Sommer K.; Rarey M.; Kirchmair J. Benchmarking Commercial Conformer Ensemble Generators. J. Chem. Inf. Model. 2017, 57, 2719–2728. 10.1021/acs.jcim.7b00505. [DOI] [PubMed] [Google Scholar]
- Poli G.; Seidel T.; Langer T. Conformational Sampling of Small Molecules With iCon: Performance Assessment in Comparison With OMEGA. Front. Chem. 2018, 6, 229 10.3389/fchem.2018.00229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips J. C.; Hardy D. J.; Maia J. D. C.; Stone J. E.; Ribeiro J. V.; Bernardi R. C.; Buch R.; Fiorin G.; Hénin J.; Jiang W.; McGreevy R.; Melo M. C. R.; Radak B. K.; Skeel R. D.; Singharoy A.; Wang Y.; Roux B.; Aksimentiev A.; Luthey-Schulten Z.; Kalé L. V.; Schulten K.; Chipot C.; Tajkhorshid E. Scalable molecular dynamics on CPU and GPU architectures with NAMD. J. Chem. Phys. 2020, 153, 044130 10.1063/5.0014475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters M. B.; Yang Y.; Wang B.; Füsti-Molnár L.; Weaver M. N.; Merz K. M. Structural Survey of Zinc-Containing Proteins and Development of the Zinc AMBER Force Field (ZAFF). J. Chem. Theory Comput. 2010, 6, 2935–2947. 10.1021/ct1002626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trott O.; Olson A. J. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010, 31, 455–461. 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berman H. M.; Westbrook J.; Feng Z.; Gilliland G.; Bhat T. N.; Weissig H.; Shindyalov I. N.; Bourne P. E. The Protein Data Bank. Nucleic Acids Res. 2000, 28, 235–242. 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- János Pató G. K.; Örfi L.; Wáczek F.; Horváth Z.; Banhegyi P.; Szabadkai I.; Marosfalvi J.; Hegymegi-barakonyi B.; Székélyhidi Z.; Greff Z.; Choidas A.; Bacher G.; Missio A.; Koul A.. Novel Therapeutic Targets for the Treatment of Mycobacterial Infections and Compounds Useful Therefor. U.S. Patent US20090298842A12009.
- Velázquez-Libera J. L.; Durán-Verdugo F.; Valdés-Jiménez A.; Núñez-Vivanco G.; Caballero J. LigRMSD: a web server for automatic structure matching and RMSD calculations among identical and similar compounds in protein-ligand docking. Bioinformatics 2020, 36, 2912–2914. 10.1093/bioinformatics/btaa018. [DOI] [PubMed] [Google Scholar]
- Kainrad T.; Hunold S.; Seidel T.; Langer T. LigandScout Remote: A New User-Friendly Interface for HPC and Cloud Resources. J. Chem. Inf. Model. 2019, 59, 31–37. 10.1021/acs.jcim.8b00716. [DOI] [PubMed] [Google Scholar]
- Davies M.; Nowotka M.; Papadatos G.; Dedman N.; Gaulton A.; Atkinson F.; Bellis L.; Overington J. P. ChEMBL web services: streamlining access to drug discovery data and utilities. Nucleic Acids Res. 2015, 43, W612–W620. 10.1093/nar/gkv352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendez D.; Gaulton A.; Bento A. P.; Chambers J.; De Veij M.; Félix E.; Magariños M. P.; Mosquera J. F.; Mutowo P.; Nowotka M.; Gordillo-Marañón M.; Hunter F.; Junco L.; Mugumbate G.; Rodriguez-Lopez M.; Atkinson F.; Bosc N.; Radoux C. J.; Segura-Cabrera A.; Hersey A.; Leach A. R. ChEMBL: towards direct deposition of bioassay data. Nucleic Acids Res. 2019, 47, D930–D940. 10.1093/nar/gky1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittwer M.; Dames S. A. Expression and purification of the natively disordered and redox sensitive metal binding regions of Mycobacterium tuberculosis protein kinase G. Protein Expr. Purif. 2015, 111, 68–74. 10.1016/j.pep.2015.03.015. [DOI] [PubMed] [Google Scholar]
- Wittwer M.; Luo Q.; Kaila V. R. I.; Dames S. A. Oxidative Unfolding of the Rubredoxin Domain and the Natively Disordered N-terminal Region Regulate the Catalytic Activity of Mycobacterium tuberculosis Protein Kinase G. J. Biol. Chem. 2016, 291, 27062–27072. 10.1074/jbc.M116.747089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorgensen W. L.; Chandrasekhar J.; Madura J. D.; Impey R. W.; Klein M. L. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 1983, 79, 926–935. 10.1063/1.445869. [DOI] [Google Scholar]
- Case D. A.; Aktulga H. M.; Belfon K.; Ben-Shalom I. Y.; Brozell S. R.; Cerutti D. S.; Cheatham T. E. III; Cisneros G. A.; Cruzeiro V. W. D.; Darden T. A.; Duke R. E.; Giambasu G.; Gilson M. K.; Gohlke H.; Goetz A. W.; Harris R.; Izadi S.; Izmailov S. A.; Jin C.; Kasavajhala K.; Kaymak M. C.; King E.; Kovalenko A.; Kurtzman T.; Lee T. S.; LeGrand S.; Li P.; Lin C.; Liu J.; Luchko T.; Luo R.; Machado M.; Man V.; Manathunga M.; Merz K. M.; Miao Y.; Mikhailovskii O.; Monard G.; Nguyen H.; O’Hearn K. A.; Onufriev A.; Pan F.; Pantano S.; Qi R.; Rahnamoun A.; Roe D. R.; Roitberg A.; Sagui C.; Schott-Verdugo S.; Shen J.; Simmerling C. L.; Skrynnikov N. R.; Smith J.; Swails J.; Walker R. C.; Wang J.; Wei H.; Wolf R. M.; Wu X.; Xue Y.; York D. M.; Zhao S.; Kollman P. A.. Amber; University of California: San Francisco, 2021.
- Tian C.; Kasavajhala K.; Belfon K. A. A.; Raguette L.; Huang H.; Migues A. N.; Bickel J.; Wang Y.; Pincay J.; Wu Q.; Simmerling C. ff19SB: Amino-Acid-Specific Protein Backbone Parameters Trained against Quantum Mechanics Energy Surfaces in Solution. J. Chem. Theory Comput. 2020, 16, 528–552. 10.1021/acs.jctc.9b00591. [DOI] [PubMed] [Google Scholar]
- Sousa da Silva A. W.; Vranken W. F. ACPYPE - AnteChamber PYthon Parser interfacE. BMC Res. Notes 2012, 5, 367 10.1186/1756-0500-5-367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakalian A.; Bush B. L.; Jack D. B.; Bayly C. I. Fast, efficient generation of high-quality atomic charges. AM1-BCC model: I. Method. J. Comput. Chem. 2000, 21, 132–146. . [DOI] [PubMed] [Google Scholar]
- Jakalian A.; Jack D. B.; Bayly C. I. Fast, efficient generation of high-quality atomic charges. AM1-BCC model: II. Parameterization and validation. J. Comput. Chem. 2002, 23, 1623–1641. 10.1002/jcc.10128. [DOI] [PubMed] [Google Scholar]
- Wang J.; Wolf R. M.; Caldwell J. W.; Kollman P. A.; Case D. A. Development and testing of a general amber force field. J. Comput. Chem. 2004, 25, 1157–1174. 10.1002/jcc.20035. [DOI] [PubMed] [Google Scholar]
- Li P.; Song L. F.; Merz K. M. Parameterization of Highly Charged Metal Ions Using the 12-6-4 LJ-Type Nonbonded Model in Explicit Water. J. Phys. Chem. B 2015, 119, 883–895. 10.1021/jp505875v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadden J. A.; Perilla J. R., Molecular Dynamics Simulations of Protein–Drug Complexes: A Computational Protocol for Investigating the Interactions of Small-Molecule Therapeutics with Biological Targets and Biosensors. In Computational Drug Discovery and Design; Gore M.; Jagtap U. B., Eds. Springer New York: New York, NY, 2018; pp 245–270. [DOI] [PubMed] [Google Scholar]
- Humphrey W.; Dalke A.; Schulten K. VMD: Visual molecular dynamics. J. Mol. Graph. 1996, 14, 33–38. 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- Seeber M.; Felline A.; Raimondi F.; Muff S.; Friedman R.; Rao F.; Caflisch A.; Fanelli F. Wordom: A user-friendly program for the analysis of molecular structures, trajectories, and free energy surfaces. J. Comput. Chem. 2011, 32, 1183–1194. 10.1002/jcc.21688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daura X.; van Gunsteren W. F.; Jaun B.; Mark A.19 E.; Gademann K.; Seebach D. Peptide Folding: When Simulation Meets 20 Experiment. Angew. Chem., Int. Ed. 1999, 38, 236–240. . [DOI] [Google Scholar]
- González-Alemán R.; Hernández-Castillo D.; Caballero J.; Montero-Cabrera L. A. Quality threshold clustering of molecular dynamics: a word of caution. Journal of Chemical Information and Modeling. J. Chem. Inf. Model. 2020, 60, 467–472. 10.1021/acs.jcim.9b00558. [DOI] [PubMed] [Google Scholar]
- Bell E. W.; Zhang Y. DockRMSD: an Open-Source Tool for Atom Mapping and RMSD calculation of Symmetric Molecules through Graph Isomorphism. J. Cheminf. 2019, 11, 1–9. 10.1186/s13321-019-0362-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiménez-Avalos G.; Vargas-Ruiz A. P.; Delgado-Pease N. E.; Olivos-Ramirez G. E.; Sheen P.; Fernández-Díaz M.; Quiliano M.; Zimic M.; et al. Comprehensive virtual screening of 4.8 k flavonoids reveals novel insights into allosteric inhibition of SARS-CoV-2 MPRO. Sci. Rep. 2021, 11, 15452 10.1038/s41598-021-94951-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu T.; Cao S.; Su P.-C.; Patel R.; Shah D.; Chokshi H. B.; Szukala R.; Johnson M. E.; Hevener K. E. Hit Identification and Optimization in Virtual Screening: Practical Recommendations Based on a Critical Literature Analysis. J. Med. Chem. 2013, 56, 6560–6572. 10.1021/jm301916b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanega C.; Shen M.; Mott B. T.; Thomas C. J.; MacArthur R.; Inglese J.; Auld D. S. Comparison of bioluminescent kinase assays using substrate depletion and product formation. Assay Drug Dev. Technol. 2009, 7, 606–614. 10.1089/adt.2009.0230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanghera J.; Li R.; Yan J. Comparison of the Luminescent ADP-Glo Assay to a Standard Radiometric Assay for Measurement of Protein Kinase Activity. Assay Drug Dev. Technol. 2009, 7, 615–622. 10.1089/adt.2009.0237. [DOI] [PubMed] [Google Scholar]
- Chen D.; Ma S.; He L.; Yuan P.; She Z.; Lu Y. Sclerotiorin inhibits protein kinase G from Mycobacterium tuberculosis and impairs mycobacterial growth in macrophages. Tuberculosis 2017, 103, 37–43. 10.1016/j.tube.2017.01.001. [DOI] [PubMed] [Google Scholar]
- Wagner T.; André-Leroux G.; Hindie V.; Barilone N.; Lisa M.-N.; Hoos S.; Raynal B.; Vulliez-Le Normand B.; O’Hare Helen M.; Bellinzoni M.; Alzari Pedro M. Structural insights into the functional versatility of an FHA domain protein in mycobacterial signaling. Sci. Signaling 2019, 12, eaav9504 10.1126/scisignal.aav9504. [DOI] [PubMed] [Google Scholar]
- Liao C.; Hsu J.; Kim Y.; Hu D.-Q.; Xu D.; Zhang J.; Pashine A.; Menke J.; Whittard T.; Romero N.; Truitt T.; Slade M.; Lukacs C.; Hermann J.; Zhou M.; Lucas M.; Narula S.; DeMartino J.; Tan S.-L. Selective inhibition of spleen tyrosine kinase (SYK) with a novel orally bioavailable small molecule inhibitor, RO9021, impinges on various innate and adaptive immune responses: implications for SYK inhibitors in autoimmune disease therapy. Arthritis Res. Ther. 2013, 15, R146. 10.1186/ar4329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucas M. C.; Bhagirath N.; Chiao E.; Goldstein D. M.; Hermann J. C.; Hsu P.-Y.; Kirchner S.; Kennedy-Smith J. J.; Kuglstatter A.; Lukacs C.; Menke J.; Niu L.; Padilla F.; Peng Y.; Polonchuk L.; Railkar A.; Slade M.; Soth M.; Xu D.; Yadava P.; Yee C.; Zhou M.; Liao C. Using Ovality to Predict Nonmutagenic, Orally Efficacious Pyridazine Amides as Cell Specific Spleen Tyrosine Kinase Inhibitors. J. Med. Chem. 2014, 57, 2683–2691. 10.1021/jm401982j. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.