Abstract
Investigation of the potential applications of artificial intelligence (AI), including machine learning (ML) and deep learning (DL) techniques, is an exponentially growing field in medicine and healthcare. These methods can be critical in providing high-quality care to patients with chronic rheumatological diseases lacking an optimal treatment, like rheumatoid arthritis (RA), which is the second most prevalent autoimmune disease. Herein, following reviewing the basic concepts of AI, we summarize the advances in its applications in RA clinical practice and research. We provide directions for future investigations in this field after reviewing the current knowledge gaps and technical and ethical challenges in applying AI. Automated models have been largely used to improve RA diagnosis since the early 2000s, and they have used a wide variety of techniques, e.g., support vector machine, random forest, and artificial neural networks. AI algorithms can facilitate screening and identification of susceptible groups, diagnosis using omics, imaging, clinical, and sensor data, patient detection within electronic health record (EHR), i.e., phenotyping, treatment response assessment, monitoring disease course, determining prognosis, novel drug discovery, and enhancing basic science research. They can also aid in risk assessment for incidence of comorbidities, e.g., cardiovascular diseases, in patients with RA. However, the proposed models may vary significantly in their performance and reliability. Despite the promising results achieved by AI models in enhancing early diagnosis and management of patients with RA, they are not fully ready to be incorporated into clinical practice. Future investigations are required to ensure development of reliable and generalizable algorithms while they carefully look for any potential source of bias or misconduct. We showed that a growing body of evidence supports the potential role of AI in revolutionizing screening, diagnosis, and management of patients with RA. However, multiple obstacles hinder clinical applications of AI models. Incorporating the machine and/or deep learning algorithms into real-world settings would be a key step in the progress of AI in medicine.
Keywords: Rheumatoid arthritis, Autoimmune diseases, Artificial intelligence, Deep learning, Diagnosis, Imaging, Machine learning, Natural language processing, Precision medicine, Treatment
Key Summary Points
| Rheumatoid arthritis (RA) is among the most common rheumatologic diseases. |
| Precision medicine with the aid of artificial intelligence (AI) is becoming more common each day. |
| Numerous machine learning and deep learning algorithms exist that could assist physicians in every step of RA care, including primary prevention, diagnosis, treatment, and rehabilitation. |
| Nonetheless, many challenges exist in the path of expanding AI-guided precision medicine, and especially its application in RA, which could and should be overcome through multi-disciplinary scientific effort. |
Introduction
Artificial intelligence (AI) is defined as "the capability of a machine to imitate intelligent human behavior" [1]. In today's world, technologies are expanding faster than ever, with capabilities one could have never thought of in the past. Machines are now able to perform tasks not only as good as humans, but even at higher qualities in many instances. AI is being used in various scientific fields, and medicine is not an exception [2]. Researchers in almost all healthcare sectors and specialties are now studying potential applications of AI, ranging from image processing in pathology [3] and radiology [4], precision medicine, and drug discovery [5] to making estimations and predictions in public health [6]. Machine learning (ML) is a branch of AI, in which the intelligence mentioned above is acquired through practice, similar to how a human learns skills. ML improved significantly in the early 2010s with the introduction of deep learning (DL) [7], which is basically combining multiple ML processes with each other [8].
Rheumatoid arthritis (RA) is the second most prevalent autoimmune disease, with an estimated global prevalence of nearly 20 million cases as of 2019 [9, 10]. The disease is characterized by destructive joint changes starting in the small joints of extremities and may continue to involve larger joints if left untreated. Rheumatoid arthritis is diagnosed clinically, and the lack of well-established diagnostic criteria [11] or a gold standard test makes the diagnosis challenging. Several classification methods have been proposed to distinguish RA from other autoimmune diseases and also stratify patients based on their disease characteristics [11]. Currently, the 2010 American College of Rheumatology/European League Against Rheumatism (ACR/EULAR) classification system is the most commonly used criteria for RA diagnosis and classification [12]. Treatment of RA aims to reduce inflammation and joint destruction. Initial therapies include non-steroidal anti-inflammatory drugs (NSAIDs) and corticosteroids, followed by disease-modifying anti-rheumatic drugs (DMARDs) [13]. Methotrexate (MTX) is the initial DMARD choice, although it may be substituted or accompanied by other treatments if indicated [13].
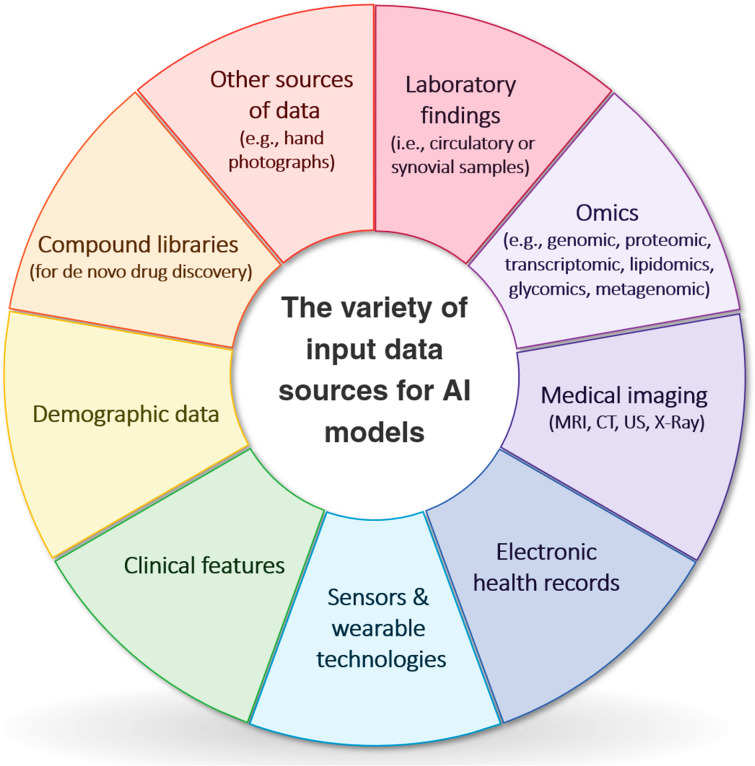
The medicine we know today is a result of experiments and, more precisely, data analysis. Therefore, utilizing the vast amount of the currently available data in the most efficient way is of great value. As evaluating all these data is virtually impossible for humans, AI helps us achieve this goal by incorporating machine-like speed and human-like comprehension. Almost all available data could be used by AI systems: laboratory findings, omics data, medical images, electronic health records (EHRs), data derived from sensors and wearable technologies, clinical features, demographic data, etc. (Fig. 1). The results obtained from these inputs could provide us with useful insights into various aspects of a disease, such as its pathophysiology and epidemiologic features. They could also assist researchers in discovering novel diagnostic methods and biomarkers, leading to quicker and more accurate diagnoses. Moreover, given the invaluable benefits of precision medicine [14], AI algorithms are able to tailor medical services and treatments for each patient according to their unique biological profile (e.g., genomics) and disease status.
Fig. 1.
The variety of input data sources for artificial intelligence (AI) models, CT computed tomography, MRI magnetic resonance imaging, US ultrasound
Given the emerging role of AI in diagnosis, monitoring, and management of autoimmune rheumatologic diseases, including RA, a thorough understanding of the achievements that have been obtained so far in the field and the existing knowledge gaps is critical to facilitate their incorporation into clinical practice and delineate the path for future studies. In this study, after reviewing the basic concepts of AI, we provide an updated comprehensive summary of the advances and applications of AI in RA clinical practice and research. Furthermore, we point out areas with a paucity of literature and challenges that have to be addressed and provide future directions for researchers on this topic.
Methods
We conducted an online search using PubMed in March 2022 using the following keywords: "rheumatoid arthritis" AND ("artificial intelligence" OR "machine learning" OR "machine intelligence" OR "computational intelligence" OR "deep learning" OR "neural network*" OR "convolutional network*" OR "Bayesian learning" OR "random forest" OR "reinforcement learning" OR "hierarchical learning" OR "computer vision"). No publication date or study type limit was applied to the search. We also searched the reference lists of the retrieved studies for identification of potentially relevant studies. Study selection was independently performed by two reviewers (SM and AN). This study was conducted in accordance with the ethical principles of the Declaration of Helsinki of 1964 and its later amendments. It is based on previously conducted studies and does not contain any new studies with human participants or animals performed by any of the authors.
Artificial Intelligence, Machine Learning, and Deep Learning
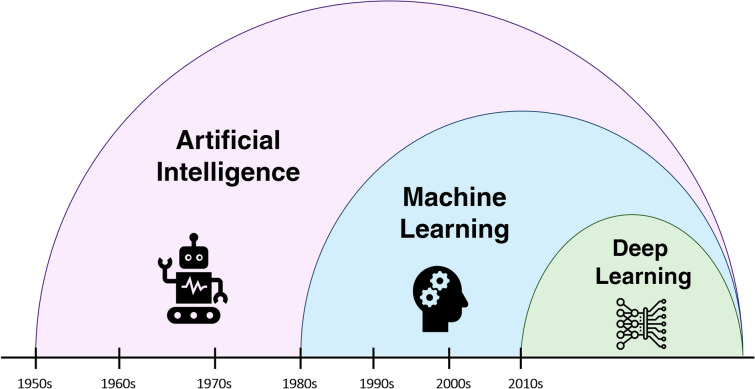
Artificial intelligence is a domain of computer sciences referring to a wide variety of interdisciplinary approaches aimed at enhancing machine capabilities. Machine learning is a subdiscipline of AI constituted of techniques for complex problem solving by automatedly learning the patterns of interaction between variables without explicit programming [15]. Compared to traditional statistical models that are hypothesis-driven and aim to identify relationships between outcomes and datapoints, ML approaches learn from the data, and their goal is to make accurate predictions with less focus on inference. Deep learning is a subset of ML identifying patterns in data using a layered structure of artificial neural networks (Fig. 2). In the past decade, due to the enhancement of computational power and availability of massive datasets, DL has been at the forefront of image analysis, genomic analysis, and drug discovery [16]. Compared to ML approaches (e.g., logistic regression, support vector machine (SVM), and random forest), DL models can perform more complex tasks; however, they require larger training data and longer training time. Moreover, DL models are able to process high-dimensionality data, such as medical images and EHRs [17]. Table 1 depicts the fundamental concepts in the most commonly used ML algorithms and neural networks.
Fig. 2.
Evolution of artificial intelligence, machine learning, and deep learning
Table 1.
Fundamental concepts in the most commonly used artificial intelligence algorithms

The process in which an ML algorithm learns to produce the desired outcome is called "training". Machine learning approaches are commonly categorized into three broad classes based on their training method, namely supervised, unsupervised, and reinforcement learning [18]. In supervised learning, models are trained to predict future values by learning patterns from known input and output data. Random forest, SVM, neural networks, and natural language processing (NLP) models are some of the most popular supervised approaches (Table 1). Natural language processing models aim to analyze text and speech by inferring the words and can be utilized in EHR analysis [19]. In contrast to supervised learning, in unsupervised learning, the goal is not assigning the correct label, but inferring underlying patterns and relationships within the input (e.g., finding clusters within the data by reducing data dimensionality) [15]. In reinforcement learning, the model learns to achieve a specific goal by interacting with its environment through trial and error, demonstration, or a hybrid approach. In healthcare, reinforcement learning is commonly used in models applied in robotic surgery [19].
Understanding the fundamental concepts of AI familiarize physicians with the potential application of AI-based models in their clinical practice and helps them detect robust models applicable in practice. Several guidelines have been developed to ensure production of reliable models. Multiple items should be considered when assessing the robustness of an algorithm, including the size of the dataset used to train the model (as more training data results in a more precise model), external validation of the model, significance of the clinical problem addressed by the model, performance of the model compared to other algorithms or clinician performance, and availability of the utilized algorithm on public repositories, which can enable independent validation of the performance and reproducibility of the model [17, 20–24].
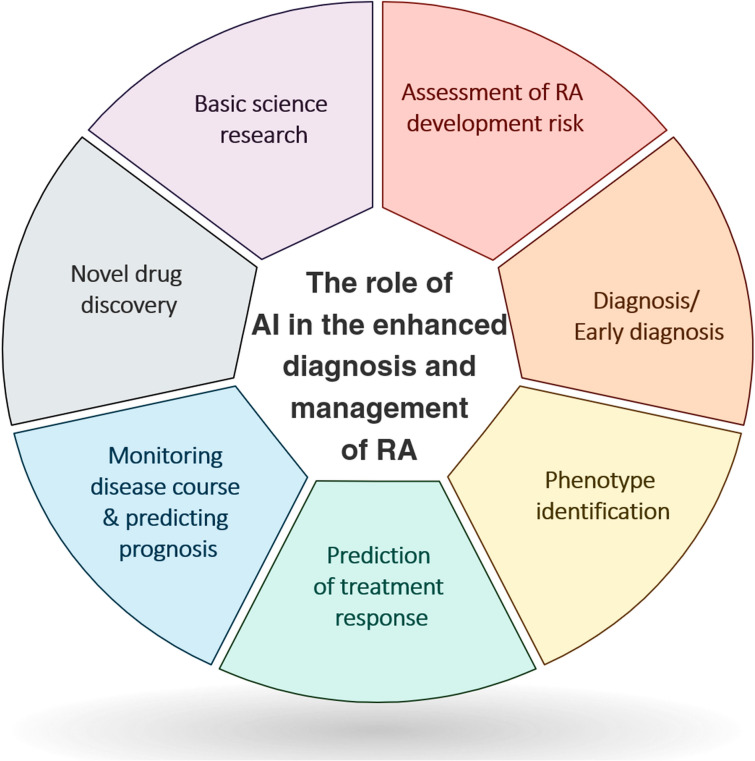
Artificial Intelligence in RA
Assessment of RA Development Risk
Currently, the most commonly used method for detecting pre-clinical RA in individuals is by measuring autoantibodies such as anti-citrullinated protein antibodies (ACPAs) or rheumatoid factor (RF), which could be present even years before the symptomatic disease [25]. However, they have a poor positive predictive value [26]. Hence, a reliable predictor of future RA development is yet to be found, and artificial intelligence could assist in this regard. O'Neil et al. [25] designed regression models with serum proteome as input to identify patients who are likely to eventually develop RA (i.e., progressors) among first-degree relatives of those with confirmed disease (i.e., at-risk population). Among ACPA-negative cases, least absolute shrinkage and selection operator (LASSO) regression recognized progressors using 17 proteins with an accuracy of 100%. However, another model for ACPA-positive individuals was less accurate (accuracy = 86.9%). Among all at-risk individuals, a third model was developed using 23 proteins as variables which demonstrated 91.2% accuracy (area under the curve (AUC) = 0.93) in the validation set in identifying progressors.
Multiple studies have attempted to identify single-nucleotide polymorphisms (SNPs) associated with RA development risk and the epistatic relationships among them. Kruppa et al. [27] used a random-jungle model and identified a 496-SNP panel closely associated with RA (AUC = 0.89). Negi and colleagues [28] also investigated SNPs and found that four SNPs were significantly associated with the disease, with maximum and minimum odds ratios (OR) being 1.42 and 0.86, respectively. One gene in which polymorphisms are associated with RA is PTPN22 [29, 30]. Briggs et al. [31] identified epistatic relations between PTPN22 and several SNPs that could augment the effect of PTPN22 on susceptibility to RA. Epistatic relationships were also probed by Gonzalez-Reico et al. [32], where they evaluated interactions between human leukocyte antigen (HLA) and non-HLA genes using Bayesian LASSO regression.
Jin et al. demonstrated that some eye diseases are associated with RA development in patients aged 50 and above [33]. In their study, cataract and other non-glaucoma eye diseases significantly increased the risk of developing RA, after adjusting for multiple other covariates (ORs = 1.33 and 1.43, respectively).
Table 2 summarizes studies incorporating ML for the assessment of RA development risk [25, 27, 28, 31–36].
Table 2.
Studies incorporating AI for the assessment of RA development risk
| First author | Model | Algorithms applied | No. of data | Type of the primary data | Validation/Test | Objective | Prominent outcomes presented |
|---|---|---|---|---|---|---|---|
| Gola et al. (2021) [34] | Supervised ML | Model-based MDR, random forest and Elastic Net |
RA = 868 Controls = 1194 |
Omics data | Nested tenfold cross-validation | Disease prediction | Model: Elastic Net, AUC = 0.86 |
| O’Neil et al. (2021) [25] | Supervised ML | LASSO regression |
Total at-risk = 127 ACPA-negative (not progressor) = 47 ACPA-positive (not progressor) = 63 Progressors = 17 |
Omics data | Whole data (for models 1 and 2), dependent test set (for model 3) | To identify RA susceptibility protein markers | Model 3 (validation, n = 34): accuracy = 91.2%, AUC = 0.931 |
| Jin et al. (2021) [33] | Supervised ML | Logistic regression, random forest |
Arthritis = 2272 No arthritis = 6151 |
Clinical and lab data | N/A | To find eye diseases that increase the risk of arthritis development |
Cataract OR = 1.331 (1.057–1.664) Glaucoma OR = 1.155 (0.703–1.805) (but not statistically significant) Other eye diseases OR = 1.428 (1.174–1.730) Gini Index for other eye diseases = 4.22 (higher than cataract and glaucoma) |
| Chin et al. (2018) [35] | Supervised and unsupervised ML | SVM + NMF |
RA = 1007 Controls = 921,192 |
EHR | Tenfold cross-validation | To identify RA risk factors | Best accuracy using 200 risk factors |
| Negi et al. (2013) [28] | Supervised ML | SVM |
Discovery: RA = 706 Controls = 761 Replication: RA = 927 Controls = 1148 |
Omics | Replication set, cross-validation | To identify SNPs associated with RA | Four SNPs were associated with RA upon replication (highest OR = 1.42, lowest OR = 0.86) |
| Kruppa et al. (2012) [27] | Supervised ML | LASSO regression, logistic regression, random jungle |
RA = 707 Controls = 738 |
Omics | Tenfold cross-validation, Dependent test set | To identify associations between SNPs and RA | Model: random jungle (496 SNPs): AUC = 0.8925 (0.8644–0.9206), sensitivity = 80.09% (74.46–84.73%), specificity = 80.48% (75.13–84.91%) |
| Liu et al. (2011) [36] | Supervised ML | Logistic regression, random forest |
NARAC1 (training): RA = 908 Controls = 1260 NARAC2 (validation): RA = 952 Controls = 1760 |
Omics data | Independent test set | To find a set of SNPs to predict RA status |
Using mean decrease in Gini: Training set: 93 out of 696 SNPs selected (error rate = 0.2, sensitivity = 83%, specificity = 75%) Validation: error rate = 0.3, sensitivity = 74%, specificity = 66% Validation cohort used as training: 88 SNPs selected (error rate = 0.28) |
| Briggs et al. (2010) [31] | Supervised ML | Logistic regression |
Extension: RA = 677 Controls = 750 Replication: RA = 947 Controls = 1756 |
Omics | Replication set | To identify epistatic relationships with the PTPN22 gene in RA susceptibility | Out of 449 SNPs found in extension stage, 7 were replicated (highest ROR = 2.42, lowest ROR = 0.51) |
| Gonzalez-Recio et al. (2009) [32] | Supervised ML | Bayesian LASSO regression |
RA = 868 Controls = 1194 |
Omics | N/A | To identify epistatic relationships between HLA and non-HLA SNPs associated with RA | Highest interaction was between rs10484560 (HLA) and rs2476601 (non-HLA) |
ACPA anti-citrullinated protein antibody, AUC area under the curve, EHR electronic health record, LASSO least absolute shrinkage and selection operator, MDR multi-factor dimensionality reduction, ML machine learning, N/A not available, NARAC North American Rheumatoid Arthritis Consortium, NMF non-negative matrix factorization, OR odds ratio, RA rheumatoid arthritis, ROR ratio of odds ratios, SNP single-nucleotide polymorphism, SVM support vector machine
Diagnosis/Early diagnosis
Early diagnosis of RA is of paramount importance as early interventions in the disease course can impede inflammatory destruction of the joints and lead to better outcomes [37].
According to the ACR/EULAR 2010 RA classification criteria, RF, ACPAs (often tested as anti-cyclic citrullinated peptide (anti-CCP) antibodies), erythrocyte sedimentation rate (ESR), and C-reactive protein (CRP) can be used as biomarkers for diagnosis of RA [38]. Nevertheless, RF and ACPA lack optimal sensitivity [39], while ESR and CRP have limited specificity. The absence of an optimal biomarker with high sensitivity and specificity necessitates the development of novel biomarker panels for early identification of RA [40]. Analysis of omics, i.e., genomics, transcriptomics, proteomics, metabolomics, lipidomics, glycomics, or metagenomic, using ML approaches enables simultaneous assessment of the association of numerous biomolecules with RA [41, 42]. Incorporating omics data into medical decision-making has several benefits. They are easily acquired from body fluids and are objectively interpreted. Furthermore, their extensiveness provides us with a vast amount of information. Of course, their limitation must also be kept in mind, such as being more complex and expensive.
Moreover, imaging findings, e.g., evidence of synovitis, in combination with clinical data and data derived from sensors, play a critical role in diagnosis, monitoring, and management of RA. Improved data analysis using AI can facilitate early detection of the disease and more efficient use of human resources [38, 43]. Herein, we summarize the applications of ML approaches in the diagnosis of RA using omics, imaging, clinical, and sensor data.
Using omics data in the diagnosis of RA
Several studies developed panels of multiple coding or non-coding ribonucleic acid (RNAs) within the serum or plasma to establish an accurate RA diagnosis using ML approaches. In a recent study, Liu and colleagues assessed gene expression profiles of peripheral blood cells and identified 52 differentially expressed genes in patients with RA. Further protein–protein analysis identified nine hub genes with crucial roles in the development of RA, which are fundamental in immune regulation, namely CFL1, COTL1, ACTG1, PFN1, LCP1, LCK, HLA-E, FYN, and HLA-DRA. The logistic regression and random forest models showed an AUC ≥ 0.97 for the panel of these nine messenger RNAs (mRNAs) in distinguishing RA from healthy samples [44]. In one other investigation of gene expression profile, Pratt et al. showed that a 12-gene transcriptional pattern in peripheral blood cluster of differentiation (CD) 4 + T cells could predict the development of RA in patients with undifferentiated arthritis during a median follow-up of 28 months. While the autoantibody showed a higher sensitivity in the ACPA-positive patients, the newly developed expression signature had a higher sensitivity and specificity in seronegative patients. Notably, the expression of most of these genes was induced by interleukin (IL)-6-mediated STAT3 upregulation. The combination of the 12-gene risk metric with the Leiden prediction rule (AUC = 0.84) outperformed the Leiden prediction rule alone—which is a classic tool for predicting RA progression from undifferentiated arthritis—in seronegative patients (AUC = 0.78), highlighting the clinical significance of these biomarkers [45, 46]. Lastly, recently, non-coding RNAs have garnered considerable research attention as diagnostic biomarkers in RA [47]. Ormseth and colleagues used LASSO variable selection with logistic regression to develop a panel of microRNAs (miRNA) differentiating patients with RA from controls, which resulted in the selection of miR-22-3p, miR-24-3p, miR-96-5p, miR-134-5p, miR-140-3p, and miR-627-5p, all of which were upregulated in patients with RA. The miRNA panel showed an AUC of approximately 0.8 in discriminating patients with RA (seropositive or seronegative) from controls. However, the panel might be an unspecific signature in autoimmune diseases as it could not differentiate RA from systemic lupus erythematosus [48].
Multiple investigations employed proteomic approaches to discover circulating diagnostic biomarkers using mass spectrometry. In such studies, the sample sizes are commonly relatively small, whereas each sample includes a large number of input variables. This atypical data pattern makes decision tree-based algorithms suitable for analysis of the data as they can handle the disproportionate high dimensionality of the input data compared to the number of samples [49]. In such settings, Geurts and colleagues showed that the boosted decision tree outperformed other ML approaches, including SVM and k-nearest neighbors (kNN) [49]. Using this method, several patterns of protein peaks were proposed to differentiate patients with RA from controls and patients with other autoimmune diseases with high sensitivity and specificity [49–51]. The association of the positivity of the serum for the proteomic analysis and intensity of the peaks with levels of anti-CCP antibody highlights the potential role of the patterns of protein peaks in early diagnosis of RA [51]. However, the lack of absolute protein quantification or protein identification is a limitation of these studies, which needs to be addressed by detecting the protein species represented by the peaks on the spectra [50].
Several other diagnostic models have been developed using omics data derived from serum, particularly inflammatory and oxidative stress markers. Analysis of circulatory levels of 38 cytokines using an artificial neural network (ANN) resulted in a model with a sensitivity and specificity of 100% in differentiating patients with RA from controls and patients with osteoarthritis (OA). Nevertheless, the ANN is a Blackbox providing limited information for further clinical inference. Therefore, Heard and colleagues utilized a single decision tree to identify cytokines leading the program to its output. These cytokines included CD40L, transforming growth factor (TGF)-α, epidermal growth factor (EGF), interferon (IFN)-γ, eotaxin, macrophage inflammatory protein (MIP)-1β, tumor necrosis factor (TNF)-α, IL-1α, granulocyte colony-stimulating factor (G-CSF), fractalkine, growth-regulated oncogene (GRO), and vascular endothelial growth factor (VEGF) in a descending order of importance for classification of RA, OA, and controls. Of the mentioned cytokines, eotaxin, G-CSF, IL-1alpha, TGF-α, and TNF-α levels were not statistically different between the groups when analyzed using conventional statistics. This finding highlights the necessity of applying ML algorithms in addition to conventional statistical methods for development of optimal diagnostic panels [52]. 4-hydroxy 2-nonenal (HNE) is another inflammatory marker inducing inflammation in various diseases, including RA (with elevated circulatory levels in patients with RA). A recent study investigated the diagnostic value of autoantibodies against unmodified and HNE-modified peptides in detecting RA in Taiwanese women. The model identified three isotypes of anti-HNE-modified peptides discriminative between RA and controls [53].
Machine learning approaches using metabolomics and glycomics have also shown promising results in the diagnosis of RA. Ahmed and colleagues assessed the diagnostic value of damaged proteins of the joints, including oxidized, nitrated, and glycated proteins and oxidation, nitration, and glycation free adducts released in the circulation by investigating plasma, serum, and synovial samples. Their algorithm, which featured levels of ten damaged amino acids in plasma, hydroxyproline, and anti-CCP antibody status, successfully differentiated early RA from controls and patients with other arthritis. Notably, the levels of damaged amino acids were higher in patients with advanced than early stages [54]. Chocholova et al. trained ML-based diagnostic models using glycomics data with a comparable diagnostic accuracy between ANN and LASSO regression in seropositive patients. Nevertheless, ANN outperformed LASSO regression in detecting seronegative patients in their study [55].
In addition to the circulatory biomarkers, major advancements have been accomplished in diagnosis and patient stratification by assessment of synovial tissue [56]. Long et al. found a 16-gene profile expressed in the synovial samples differentiating patients with RA and OA using supervised ML approaches. This can be particularly useful in seronegative and elderly patients having an inflammatory presentation of OA [57]. Correspondingly, Yeo and colleagues found a panel of ten most informative chemokine genes discriminating patients with established RA from uninflamed controls using ML methods. As shown by their study, synovial biomarkers can assist in the early identification of patients developing RA as well. They found that mRNA levels of chemokine (C-X-C motif) ligand (CXCL)4 and CXCL7 can accurately distinguish early RA from resolving arthritis with higher levels in early RA compared to longer established RA or controls [58].
Furthermore, even within RA patients, ML algorithms can facilitate patient stratification. Orange et al. identified three patterns of synovial gene expression using a clustering algorithm, including a high inflammatory subtype with extensive infiltration of leukocytes, a low inflammatory subtype specified by enrichment in pathways mediated by TGF-β, glycoproteins, and neuronal genes, and a mixed subtype. Subsequently, they developed a model predicting the synovial subtype according to the histological features. Notably, in the high inflammatory subgroup, the severity of pain significantly correlated with the CRP levels. Therefore, they concluded that pain mechanisms might be variable in patients with different synovial subtypes. This finding can result in potential clinical application for patient treatment stratification for pain management [59].
In addition to the above-mentioned omics data, the human microbiome has recently drawn immense research attention. Dysbiosis can be associated with various diseases, including RA. Machine learning-based approaches analyzing metagenomic data are optimal for exploiting the large biological datasets created by the evolving microbiome research [60]. Wu and colleagues used a logistic regression prediction algorithm to improve multiclass classification between patients with RA, type 2 diabetes mellitus, liver cirrhosis, and controls. While no biomarker was specific to type 2 diabetes mellitus and RA, their model had a favorable diagnostic performance with an AUC near 0.95, highlighting the value of microbiome biomarkers in disease diagnostics, especially disease screening, within a large-scale population [61]. However, in a recently published meta-analysis, Volkova and colleagues found specific features in the gut microbiome distinguishing RA from healthy controls and other autoimmune diseases using random forest algorithms. They found that increased levels of Clostridiaceae Clostridium and Lachnospiraceae and reduced abundance of Erysipelotrichaceae were the most distinctive features in RA compared to other autoimmune diseases [62]. In addition to the gut microbiome, assessment of the oral microbiome using ML approaches may also provide promising diagnostic biomarkers [63].
Table 3 illustrates studies incorporating ML for diagnosis of RA using omics data [44, 45, 48–55, 57–59, 61, 62, 64, 65].
Table 3.
Studies incorporating AI for diagnosis of RA using omics data
| First author | Model | Algorithms applied | No. of data | Type of the primary data | Validation/ Test | Objective | Prominent Outcomes presented |
|---|---|---|---|---|---|---|---|
| Volkova et al. (2021) [62] | Supervised ML | Random forest, XGBoost, ridge regression, and SVM | RA = 371 |
16S rRNA sequencing or shotgun metagenomics or both |
Seven-fold-3-times cross-validation | To identify a microbial signature predictive of autoimmune diseases, including RA, MS, and IBD |
Autoimmunity (at genus level, adults): Random forest: AUC = 0.887, F1 = 0.681, XGBoost: AUC = 0.909, F1 = 0.676, SVM RFE: AUC = 0.826, F1 = 0.636, ridge regression: AUC = 0.778, F1 = 0.603, RA (at species level, adults): Random forest: AUC = 0.879, F1 = 0.664, XGBoost: AUC = 0.847, F1 = 0.650, SVM RFE: AUC = 0.845, F1 = 0.647, ridge regression: AUC = 0.795, F1 = 0.628 Most predictive features for RA: reduced concentration of Desulfovibrionaceae Bilophila, Akkermansia, and Veillonellaceae Dialister and increased levels of Lachnospiraceae Clostridium |
| Jung et al. (2021) [64] | Unsupervised ML | Naïve Bayes classifier |
RA = 152 Controls = 28 |
RNA sequencing (synovial tissue) | Tenfold cross-validation | Classifying RA patients to assess clinical features and treatment response |
Classified patients with RA into three subtypes: C1: neutrophil-enriched signature, C2: fibroblast-enriched signature, C3: prominent immune cells and proinflammatory signatures and associated with presence of ACPA and a better treatment response Key regulatory genes in each subtype were also identified |
| Xiao (2021) [65] | Supervised ML | LASSO regression, SVM, random forest, Xgboost, BPNN, and CNN |
Training: RA = 416 Controls = 318 Test: RA = 10 Controls = 13 |
mRNA expression profiling (blood samples) | Independent test set | To select the genes highly associated with RA | The algorithms were based on pre-defined key genes: BPNN: AUC = 0.99, LASSO regression: AUC = 0.91, SVM: AUC = 0.95 |
| Liu et al. (2021) [44] | Supervised ML | Logistic regression, random forest |
RA = 112 Controls = 53 |
mRNA expression profiling data (serum samples) | Fivefold cross-validation | To assess the diagnostic value of a 9 mRNAs-based panel for diagnosis of RA |
Logistic regression: AUC = 0.97 Random forest: AUC = 0.98 |
| Ormseth et al. (2020) [48] | Supervised ML | Random forest, LASSO, and logistic regression |
Discovery: RA = 167 Controls = 91 Validation: RA = 32 SLE = 12 Controls = 32 |
Plasma samples | Nested cross-validation, Independent test set | To assess the diagnostic value of a panel of miRNAs for diagnosis of RA |
Validation cohort: RA vs. controls: AUC = 0.71 (0.58–0.84), seropositive RA vs. controls: AUC = 0.73 (0.58–0.87), seronegative RA vs. controls: AUC = 0.73 (0.57–0.89) Discovery cohort: RA vs. controls: AUC = 0.79 (0.73–0.86), seropositive RA vs. controls: AUC = 0.79 (0.73–0.86) seronegative RA vs. controls: AUC = 0.84 (0.77–0.91) RA remission vs. controls: AUC = 0.85 (0.78–0.92) |
| Long et al. (2019) [57] | Supervised ML | Random forest, SVM, kNN, naïve Bayes, decision tree |
RA = 53 OA = 41 Controls = 25 |
Genome-wide transcriptional profiles from synovial tissue | Tenfold cross-validation, external test set | To assess the diagnostic value of a 16 gene biomarker panel for diagnosis of RA and differentiating RA from OA | Differentiation of RA and OA: Random forest: accuracy = 0.96, sensitivity = 1.00, specificity = 0.90, SVM: accuracy = 0.96, sensitivity = 1.00, specificity = 0.90, kNN: accuracy = 0.96, sensitivity = 0.92, specificity = 1.00, naïve Bayes: accuracy = 0.96, sensitivity = 0.92, specificity = 1.00, decision tree: accuracy = 0.91, sensitivity = 1.00, specificity = 0.80 |
| Wu et al. (2018) [61] | Supervised ML | Logistic regression, kNN, random forest, SVM, GBDT, SGD, adaptive boosting |
RA = 130 T2D = 170 Liver cirrhosis = 123 Controls = 383 |
Microbiome and phenotype information | Fivefold cross-validation | To develop a multi-class classifier for identification of type of disease using shotgun metagenome sequencing | Logistic regression: AUC = 0.96, F1-score = 0.92, kNN: F1-score = 0.86, random forest: F1-score = 0.83, SVM: F1-score = 0.91, GBDT: F1-score = 0.87, SGD: F1-score = 0.84, adaptive boosting: F1-score = 0.90 |
| Yeo et al. (2016) [58] | Supervised ML | Multivariate analysis |
Uninflamed Controls = 10 Resolving Arthritis = 9 Early RA = 17 Established RA = 12 |
Synovial mRNA expression |
N/A | To differentiate RA in different stages using synovial cytokine production profile | Established RA vs. Uninflamed: AUC = 0.996 Early RA vs. Resolving RA: AUC = 0.764 |
| Pratt et al. (2012) [45] | Supervised ML | SVM |
Training cohort: RA = 47 Controls = 64 Test cohort: Undifferentiated arthritis = 62 |
CD4 T cell transcriptome data and serum samples | Hold out validation | To identify potential biomarkers for early RA using markers expressed by peripheral blood CD4 T cells |
Sensitivity = 0.68% (0.48–0.83), specificity = 0.70 (0.60 to 0.87), PLR = 2.2 (1.2–3.8), NLR = 0.4 (0.2–0.8) Removing ACPA-positive subset: sensitivity = 0.85 (0.58–0.96), specificity = 0.75 (0.59–0.86) |
| Orange et al. (2018) [59] | Supervised & Unsupervised ML | SVM and consensus clustering |
RA = 123 OA = 6 |
RNA sequence and histology data | Leave-one-out cross-validation | To classify patients according to synovial tissue inflammation and predict the classification using histological features |
Consensus clustering: Identification of three distinct synovial subtypes SVM: Prediction of subtypes using histological data: high inflammatory vs. other: AUC = 0.88, low inflammatory vs. other: AUC = 0.71, mixed subtype vs. other: AUC = 0.59 |
| Tsai et al. (2021) [53] | Supervised ML | Decision trees, random forest, SVM |
RA = 60 OA = 35 Controls = 60 |
Levels of specific autoantibodies from serum samples | Ten-fold cross-validation | To identify RA patients using serum levels of anti-unmodified and anti-HNE-modified peptide autoantibodies |
HC vs. RA: random forest: AUC = 0.92, SVM: AUC = 0.82, decision tree: AUC = 0.86 OA vs. RA: random forest: AUC = 0.92, SVM: AUC = 0.88, decision tree: AUC = 0.84 |
| Chocholova et al. (2018) [55] | Supervised DL | ANN, L1-regularized logistic regression |
RA = 47 Controls = 53 |
Immunoassays Serum samples |
Hold-out validation, testing set | To differentiate between healthy people and seropositive/ seronegative RA patients by incorporating glycomics using serum samples |
ANN: Seropositive RA vs. non-RA (using anti-CCP and total RF combined with ELLBA-based RCA glycol profiling): AUC = 0.96, sensitivity = 80.6%, specificity = 100.0%, accuracy = 92.5%, NPV = 89.1, PPV = 100 Seronegative RA vs. non-RA (using RCA ELLBA using adsorbed protein A and serum samples): AUC = 0.86, sensitivity = 43.8%, specificity = 90.6%, accuracy = 79.7%, NPV = 84.2, PPV = 58.3 |
| Ahmed et al. (2016) [54] | Supervised ML | Random forest |
RA = 67 OA = 63 Non-RA inflammatory arthritis = 42 Controls = 53 |
Mass spectrometry (plasma and synovial fluid samples) | Fivefold cross-validation, independent test set | To identify patients with early-stage RA and OA by profiling glycated, oxidized, and nitrated proteins and amino acids in synovial fluid and plasma |
Early arthritis vs. HC: Test set validation: AUC = 0.77 (0.69–0.85), sensitivity = 0.73 (0.56–0.86), specificity = 0.72 (0.62–0.81), NPV = 0.05, PPV = 0.62, PLR = 2.6, NLR = 0.38 Early RA vs. other arthritis: Test set validation: AUC = 0.62 (0.5–0.75), sensitivity = 0.60 (0.42–0.76), specificity = 0.61 (0.46–0.72), NPV = 0.97, PPV = 0.23, PLR = 1.5, NLR = 0.67 |
| Heard et al. (2014) [52] | Supervised DL | ANN and decision tree |
RA = 100 OA = 100 Controls = 100 |
Serum inflammatory proteins (LUMINEX assays) | Hold-out validation, independent testing set | To categorize HC, patients with RA and patients with OA using a panel of inflammatory cytokines expressed in serum samples |
For RA: ANN: (both trained using all (N = 38) proteins and using only differently expressed (N = 12) proteins): specificity = 100%, sensitivity = 100% Multi-decision tree (trained using all (N = 38) proteins): specificity = 100%, sensitivity = 95% |
| Niu et al. (2010) [50] | Supervised ML | Boosted decision tree |
Training set: RA = 22 OAID = 26 Controls = 25 Test set: RA = 21 OAID = 24 Controls = 25 |
Mass spectrometry (serum samples) | Hold-out validation | To identify the serum proteomic pattern for classifying patients with RA and OAID | For RA: accuracy = 85.7%, sensitivity = 85.71%, specificity = 87.76% |
| Geurts et al. (2005) [49] | Supervised ML | Decision tree ensemble methods, kNN, SVM | RA: N = 206 (RA: N = 68, controls: N = 138), | Mass spectrometry (serum samples) | Leave-one-out cross-validation | To identify biomarkers related to a given disease from datasets obtained from mass spectrometry |
For RA: Boosted decision tree: sensitivity = 83.82%, specificity = 94.93% kNN: sensitivity = 82.35%, specificity = 82.61% SVM: sensitivity = 88.24%, specificity = 89.86% |
| de Seny et al. (2005) [51] | Supervised ML | Decision tree boosting |
RA = 34 Other inflammation group = 39 Non-inflammation group = 30 |
Mass spectrometry (serum samples) | Leave-one-out cross-validation | To identify serum protein biomarkers specific for RA |
RA versus controls: Classifying 2 spectra from one patient independently: sensitivity = 85%, specificity = 91% Combining classification of the 2 spectra sensitivity = 94%, specificity = 90% RA versus PsA: Classifying 2 spectra from one patient independently: sensitivity = 94%, specificity = 86% Combining classification of the 2 spectra sensitivity = 97%, specificity = 76% |
ACPA anti-citrullinated peptide antibodies, ANN artificial neural network, AUC area under the curve, BPNN backpropagation neural network, CCP cyclic citrullinated peptide, CNN convolutional neural network, DL deep learning, GBDT gradient boosted decision tree, IBD inflammatory bowel disease, kNN k-nearest neighbors, LASSO least absolute shrinkage and selection operator, ML machine learning, MS multiple sclerosis, NLR negative likelihood ratio, NPV negative predictive value, OA osteoarthritis, PLR positive likelihood ratio, PPV positive predictive value, PsA psoriatic arthritis, RA rheumatoid arthritis, SGD stochastic gradient descent, SLE systemic lupus erythematosus, SVM support vector machine, XGBoost gradient boosting decision tree (eXtreme Gradient Boosting)
Using imaging Data in the Diagnosis of RA
Radiological findings are critical in the diagnosis and staging of RA [66]. Conventional radiography is a commonly available and widely used modality. Multiple models have been developed to diagnose RA using inputs of hand X-ray data [67, 68], such as convolutional neural networks (CNN), with an accuracy as high as near 95% [67]. Compared with conventional radiography and computed tomography (CT), magnetic resonance imaging (MRI) and ultrasound are superior in detecting early soft tissue changes [66]. The characteristic imaging features of RA are synovitis, bone erosions, bone marrow edema, joint space narrowing, joint effusion, and subcortical cysts. Late imaging findings may include subluxation or luxation, scar formation, fibrosis, and bony ankylosis [66]. To the best of our knowledge, AI-based models have been exploited in the detection of synovitis [69–71], bone erosions [72, 73], bone marrow edema [74], and joint space narrowing [75]. However, we did not find investigations on other features, such as subcortical cysts, joint effusion, or late imaging findings.
Machine learning-based algorithms, both supervised and unsupervised, have been developed to detect and quantify synovitis using MRI images [71, 76]. Computer-aided diagnostic approaches have been highly consistent with manual synovitis quantifications in dynamic-contrast enhanced (DCE) MRI, while they can significantly reduce the time spent by the observer reading the image [76, 77]. We did not find any DL-based study assessing synovitis on wrist MRI. Moreover, few studies were designed to classify and quantify synovitis using ultrasound images [70, 78, 79]. In a recent investigation, Wu and colleagues developed a DL-based model assessing the severity of RA by classifying synovial proliferation captured by ultrasound [78].
Several studies used images obtained from different modalities to create models detecting and grading bone lesions. Most studies utilized hand X-ray images to identify erosions [73, 80]. A recent study showed that severity scores acquired from a DL-based model analyzing hand X-ray images could be comparable to the scoring of a human assessor [81]. Artificial intelligence-based models also performed well in detecting joint space narrowing in RA on plain X-rays [75, 80]. However, conventional radiography may underestimate number and size of erosions because of their projectional character [72]. Therefore, utilizing CT images for automatic detection and quantification of bone erosions can facilitate a more accurate assessment of disease activity [72, 82]. Moreover, clustering methods have been useful in detecting and quantifying bone marrow edema, a prominent feature in RA, on wrist MRI [74].
Other than conventional radiography, CT, ultrasound, and MRI, molecular imaging can also play a key role in diagnosis and management of patients with RA [83]. Nevertheless, we did not find any AI-based investigation of enhancing or analyzing molecular imaging data in RA. In addition to the radiologic modalities, reliable diagnostic models have been developed using hand photographs [84] or a combination of thermal and RGB hand images, demographic data, and hand gripping force [85]. Notably, given the accessibility of acquiring the required data, such algorithms can be used as screening tools for RA [85].
Table 4 provides a summary of the ML and DL studies that used imaging data as input to diagnose patients with RA.
Table 4.
Studies incorporating AI for diagnosis of RA using imaging data
| First author | Model | Algorithms applied | No. of data | Type of the primary data | Validation/ Test | Objective | Prominent Outcomes presented | Comparison with conventional methods if performed |
|---|---|---|---|---|---|---|---|---|
| Wu et al. (2022) [78] | Supervised DL | DenseNet | RA = 1337 (L0 = 313, L1 = 657, L2 = 178, L3 = 189) | Ultrasound images (the wrist, proximal interphalangeal, and the MCP | Holdout test set | To classify synovial proliferation in ultrasound images of patients with RA |
Synovial proliferation (SP)-no versus SP-yes (grade L0 versus grades L1 and L2 and L3 in OESS): AUC = 0.886 (95% CI 0.836, 0.936), accuracy = 82.1%, sensitivity = 70.0%, specificity = 94.3% Healthy versus Diseased (grades L0 and L1 versus grades L2 and L3): AUC = 0.916 (95% CI 0.883, 0.952), accuracy = 80.4%, sensitivity = 90.8%, specificity = 70.0% |
N/A |
| Alarcon-Paredes et al. (2021) [85] | Supervised ML | A collection of classifiers, including random forest, and wrapper feature selection method |
Training: RA = 100 Controls = 100 Test: RA = 18 Controls = 20 |
Thermal and RGB images recording gripping force + demographic data |
Tenfold cross-validation, independent validation set | To develop an algorithm for diagnosis of RA using easy-to-acquire variables |
RGB images, age, and grip force: random forest accuracy = 0.945, sensitivity = 0.941, specificity = 0.95, AUC = 0.962 Thermal images, age, and grip force: random forest: accuracy = 0.90, sensitivity = 0.888, specificity = 0.912, AUC = 0.954 |
N/A |
| Mate et al. (2021) [67] | Supervised ML | CNN, SVM, ANN |
RA = 160 Controls = 130 |
Hand X-ray | Part of data as test set | To diagnose RA using hand X-ray | Using CNN: accuracy = 94.46%, sensitivity = 0.95, specificity = 0.82 | N/A |
| Ureten et al. (2020) [68] | Supervised DL | CNN |
Testing set: RA = 25 Controls = 20 |
Hand X-ray | Part of data as test set | To diagnose RA using hand X-ray | Inflammatory arthritis: accuracy = 73.33%, sensitivity = 0.6818, specificity = 0.7826, precision = 0.75, error rate = 0.0167 | N/A |
| Reed et al. (2020) [84] | Supervised ML | SVM, random forest, logistic regression, Google's TensorFlow (TF) Inception v3 model for the photographic algorithm |
RA = 117 OA = 56 PsA = 38 OA/RA = 45 OA/PsA = 17 Gout = 7 |
Hand photograph, a 9-part questionnaire, and clinical data | Leave-one-out cross-validation | To establish a diagnosis of hand arthritis using several types of input data |
Differentiating inflammatory arthritis from OA: logistic regression: accuracy = 0.975, PPV = 0.982, sensitivity = 0.986, specificity = 0.937, SVM: accuracy = 0.971, PPV = 0.973%, sensitivity = 0.991%, specificity = 0.905% Differentiating inflammatory arthritis from OA with inclusion of RF, CCP, ESR and CRP results: logistic regression: accuracy = 0.971, PPV = 0.977%, sensitivity = 0.986%, specificity = 0.921%, Differentiating RA from other arthritis: SVM: accuracy = 0.911, PPV = 0.911%, sensitivity = 0.938%, specificity = 0.873%, random forest: accuracy = 0.911, PPV = 0.926%, sensitivity = 0.920%, specificity = 0.898% |
N/A |
| Hirano et al. (2019) [80] | Supervised DL | CNN |
RA = 108 Radiographs = 216 (training = 186, test = 30) |
Hand X-ray | Part of data as validation and test sets | To assess radiographic finger joint destruction in RA |
For joint space narrowing: accuracy = 49.3–65.4% For erosion: accuracy = 70.6–74.1% |
The correlation coefficient between scores by the model and clinicians per image: for joint space narrowing: 0.72–0.88 and for erosion: 0.54–0.75 |
| Rohrbach et al. (2019) [81] | Supervised DL | CNN | Images = 102,265 | Hands and feet X-ray | Hold-out test dataset | Bone erosion scoring | Global accuracy for scoring eroded joints in the test set = 65.8% | Yes, the agreement between the CNN's predictions and the human scores was comparable with the agreement between different human scorers |
| Aizenberg et al. (2018) [74] | Supervised ML | atlas-based segmentation, fuzzy C-means clustering |
Training = 56 Validation = 485 |
Wrist MRI (T1-Gd scans) | Leave-one-out cross-validation | Automatic quantification of bone marrow edema in early arthritis |
Accuracy of atlas-based segmentation compared to manual segmentation: Lowest recall in pisiform (mean ± SD) = 0.58 ± 0.09 Highest recall in capitate (mean ± SD) = 0.82 ± 0.03 |
Yes, correlation with visual BME scores: r = 0.83, p < 0.dee |
| Murakami et al. (2017) [73] | Supervised DL | MSGVF Snakes algorithm and DCNN classifier |
Training: RA = 90 Controls = 39 Test: RA = 30 |
Hand X-ray | Threefold cross-validation, Independent testing dataset | identification of bone erosions | True-positive rate (sensitivity) = 80.5%, False-positive rate = 0.84% | N/A |
| Czaplicka et al. (2015) [76] | Supervised ML | Automatic segmentation | RA = 32 | Pre-and post-contrast wrist MRI | N/A | To determine inflamed synovial membrane volume | Following segmentation of wrist bones and automatic quantification of volume of synovitis: Correlation between the total RAMRIS score and the total volume of synovitis (automated segmentation): rs = 0.87, which is as same as manual segmentation | Yes: Manual versus automated segmentation: Pearson’s coefficient of correlation = 0.82, rs = 0.70) |
| Töpfer et al. (2014) [72] | Supervised DL | 3D segmentation | N = 18 | HR-pQCT of the second to fourth metacarpophalangeal joints | N/A | Quantification of bone erosions |
for erosions with volumes > 10 mm3: Intraoperator precision error = 3.02%/0.92 mm3, Interoperator precision error = 5.99%/1.53 mm3 for smaller erosions: Intraoperator precision error = 6.11%/0.32 mm3, Interoperator precision error = 8.27%/0.35 mm3 Intraoperator and interoperator precision error for erosions segmented fully automatically < manually edited erosions |
Yes, The correlation between manual measurements and segmentation volumes: r = 0.61 |
| Boesen et al. (2012) [77] | N/A | DYNAMIKA software | N = 54 | DCE MRI of the wrist | N/A | To assess the correlation of DCE MRI analyzed by a computer-aided approach and the scores of the RAMRIS system | Computed aided analysis of DCE MRI correlated with RAMRIS synovitis and BME with a shorter performance time for the observer | The time the observer spent was compared between the computer-aided approach and RAMRIS synovitis and BME |
| Langs et al. (2008) [75] | Supervised ML | Automated segmentation using LLM and ASM |
Set A = 40 radiographs Set B = 17 radiographs |
Hand X-ray | Cross-validation | To measure joint space widths and detect erosions on the bone contour |
Joint space widths measurement: coefficient of variation = 2–7% for repeated measurements AUC for erosion detection = 0.89 |
Yes, joint space widths and erosions, detected by a radiologist was the standard of reference |
| Tripoliti et al. (2007) [71] | Unsupervised ML | Fuzzy C-means algorithm | N = 25 patients (Both in baseline and 1-year follow-up = 17 comprising 504 images [300 (baseline) and 204 (follow-up)] | Contrast-enhanced T1-weighted MRI | N/A | Segmentation and quantification of inflammatory tissue of the hand | Performance in identifying regions compared with physicians: sensitivity = 97.7%, PPV = 83.35% | Yes |
| Scheel et al. (2002) [69] | Supervised ML | Neural network |
RA = 22 (72 joints) Controls = 8 (64 joints) |
Laser imaging data | N/A | To assess proximal finger joint inflammation using laser-based imaging technique | Accuracy = 83%, sensitivity = 80%, specificity = 89% (in detecting inflammatory changes) | N/A |
ANN artificial neural network, ASM active shape models, AUC area under the curve, BME bone marrow edema, CCP cyclic citrullinated peptide, CNN convolutional neural network, CRP C-reactive protein, CT computed tomography, DCE dynamic contrast-enhanced, DL deep learning, ESR erythrocyte sedimentation rate, HR-pQCT high-resolution peripheral quantitative CT, LLM local linear mapping, ML machine learning, MRI magnetic resonance imaging, MSGVF multiscale gradient vector flow, OA osteoarthritis, PPV positive predictive value, PsA psoriatic arthritis, RA rheumatoid arthritis, RAMRIS rheumatoid arthritis MRI scoring system, RF rheumatoid factor, SVM support vector machine
Using Clinical and Sensor Data for Diagnosis of RA
Several models have been developed for the diagnosis of RA using clinical data (Table 5) [86–88]. Singh and colleagues showed that a fuzzy inference system could have an acceptable diagnostic performance when fed with data on clinical symptoms [87]. In a novel approach, Fukae et al. converted clinical information to two-dimensional array images and used CNN (AlexNet) to distinguish patients with RA. The results of their algorithm showed a favorable agreement with the diagnosis made by three rheumatologists [88].
Table 5.
Studies incorporating AI for diagnosis of RA using clinical or sensors data
| First author | Model | Algorithms applied | No. of data | Type of the primary data | Validation/ Test | Objective | Prominent Outcomes presented |
|---|---|---|---|---|---|---|---|
| Fukae et al. (2020) [88] | Supervised ML | CNN (AlexNet and Resnet-18) |
Training: RA = 225 Controls = 785 Test: RA = 10 Controls = 40 |
Clinical data converted to two-dimensional array images | Independent testing dataset | To diagnose RA using clinical data converted to two-dimensional array images |
AlexNet: accuracy = 98%, precision = 91%, recall = 100% |
| Sharon et al. (2019) [90] | Unsupervised ML | kNN and random forest |
Dataset 1 = 40 Dataset 2 = 310 |
Microscopic features of lymphocytes captured by electronic image sensor | Hold-out and tenfold cross-validation | To classify RA patients using microscopic images of lymphocytes |
Tenfold cross-validation method: Random Subspace classifier: random forest: precision = 97.6, recall = 97.5, F-measure = 97.55, AUC = 100.0, accuracy rate = 97.5, kNN: precision = 97.6, recall = 97.5, F-measure = 97.55, AUC = 99.7, accuracy rate = 97.5; bagging classifier: random forest: precision = 89.8, recall = 90.0, F-measure = 89.9, AUC = 97.7, accuracy rate = 90.0, kNN: precision = 91.1, recall = 90.0, F-measure = 90.54, AUC = 94.7, accuracy rate = 90.0 Hold-out method: Random subspace classifier: random forest: precision = 83.3, recall = 75.0, F-measure = 78.93, AUC = 100, accuracy rate = 75.0, kNN: precision = 68.8, recall = 66.7, F-measure = 67.73, AUC = 87.5 accuracy rate = 66.67; bagging classifier: random forest: precision = 86.6, recall = 87.0, F-measure = 86.8, AUC = 97.1, accuracy rate = 86.96, kNN: precision = 81.0, recall = 71.4, F-measure = 75.9, AUC = 95.8, accuracy rate = 71.43 |
| Bardhan et al. (2019) [91] | Unsupervised ML | K-means, fuzzy C-means, Otsu, single and multi-seeded region growing, SVM |
RA = 60 Controls = 50 |
Knee joint thermograms | Three-fold cross-validation | To identify arthritis and RA using knee thermograms | RA classification rate obtained with accuracy-based feature selection = 73% |
| Singh et al. (2012) [87] | Supervised ML | Fuzzy inference system | N = 150 | Clinical symptoms | N/A | N/A | The performance of the diagnostic system for arthritis was acceptable |
| Wyns et al. (2004) [86] | Supervised & Unsupervised DL | Kohonen neural network (includes self-organizing maps) |
RA = 51 SpA = 43 Other = 26 No definite diagnosis = 40 |
Clinical data | Hold-out test set | Prediction of diagnosis in patients with early arthritis | Accuracy = 62.3%, 65.3% (without undetermined samples) |
AUC area under the curve, CNN convolutional neural network, DL deep learning, kNN k-nearest neighbors, ML machine learning, RA rheumatoid arthritis, SVM support vector machine
Sensor data, which are rich datasets for disease diagnosis and monitoring, are acquired using technologies such as wearable devices, thermography sensors, and image sensors [89–91]. In a recent study, ML algorithms using features extracted from lymphocyte images generated by an electronic image sensor were highly accurate for RA classification, with accuracy rates as high as 97.5%. Notably, electronic image sensors convert optical images into electronic data [90]. Furthermore, thermograms are noninvasive methods used to assess joint inflammation in RA [92]. Bardhan et al. developed a two-stage classification algorithm correctly labeling nearly three-fourths of the knee thermograph scans (stage one was detection of arthritis-affected knees, and stage two was detection of knees affected by RA) [91].
Phenotype identification using EHRs
In the context of EHRs, "phenotype" is a clinical condition or characteristic that can be obtained via an automated method from EHR system or clinical data repository using a specific group of data elements and logical expressions. Electronic health records contain a comprehensive pool of data, which can be widely used in clinical and translational research. Nevertheless, due to the large amount of data, the manual review and extraction can be extremely time-consuming and inefficient. Both rule-based and ML (supervised or unsupervised) models have been used to identify disease status using EHRs. Phenotype identification algorithms usually combine various sources of information, e.g., billing codes, laboratory data, medication exposures, and NLP, to make accurate predictions [93, 94].
Several models have been developed to identify patients with RA efficiently from EHRs using NLP and ML (Table 6) [95–109]. Support vector machine is one of the most commonly used algorithms for phenotype identification. In 2010, Carrol and colleagues developed an SVM model with a favorable performance (AUC > 0.90) in predicting RA disease status using naïve and refined data (i.e., naïve data curated to only include RA-related items). Notably, the SVM model had higher patient identification precision than a deterministic model [108]. Importantly, given the changes in EHR systems, addition of novel DMARDs, and updates of the ICD codes, the validity of such phenotype identification algorithms should be routinely investigated with contemporary data. A recent assessment of the performance of Carrol et al.'s model using 2017 data showed that even though the diagnostic codes and medications have changed from 2010, the model still performed robustly and outperformed rule-based algorithms. Nevertheless, updating the model using ICD-10 codes resulted in a slight improvement in the sensitivity of the model [100]. In a recent study, Maarseveen et al. found that between naïve Bayes, SVM, gradient boosting, random forest, decision tree, neural networks, and a random classifier, SVM outperformed others in disease identification using EHR [99]. They showed that the performance of the proposed model was similar to a manual chart review using the 1987 and 2010 RA classification criteria [110].
Table 6.
Studies incorporating AI for phenotyping RA using EHR
| First author | Model | Algorithms applied | No. of data | Type of the primary data | Validation/ Test | Objective | Prominent Outcomes presented |
|---|---|---|---|---|---|---|---|
| Cai et al. (2021) [95] | Supervised ML | Random forest, logistic regression | N = 4001 | EHR | Independent testing dataset | Efficient identification of eligible patients for clinical trial recruitment |
At the tertiary hospital: sensitivity = 98%, PPV = 21.8% At the community hospital: sensitivity = 98%, PPV = 24.3% The model resulted in reduction of ineligible patients from chart review by 40.5% at the tertiary care center and by 57.0% at the community hospital |
| Fernández-Gutiérrez et al. (2021) [96] | Supervised ML | Decision trees | N = 9657 (RA = 1484) | EHR | Tenfold cross-validation, independent testing dataset | To identify patients with a condition from EHR | Accuracy = 86.19, sensitivity = 72.2, specificity = 92.64, PPV = 81.92, NPV = 87.83 |
| Ferte et al. (2021) [97] | Unsupervised ML | SAFE algorithm, random forest, logistic regression |
Training = 9102 Test = 2359 |
EHR | Cross validation | Extending PheNorm [102] by combining diagnosis codes and medical concepts | For RA: AUC = 0.943 (0.940–0.945), AUPRC = 0.754 (0.744–0.763) |
| Maarseveen et al. (2021) [98] | Supervised ML | SVM |
Training = 2000 Test = 1000 |
EHR | Independent testing dataset | Extending PheNorm [102] by combining diagnosis codes and medical concepts | sensitivity = 0.85, specificity = 0.99, PPV = 0.86, NPV = 0.99 |
| Maarseveen et al. (2020) [99] | Supervised ML | SVM, gradient boosting, random forest, decision tree, neural networks, and a random classifier | N = 30,000 | EHR | Tenfold cross-validation | To identify patients with RA from EHR | SVM: F1 score = 0.81, PPV = 0.94, NPV = 0.97, sensitivity = 0.71, specificity = 1.00 |
| Huang et al. (2020) [100] | Supervised ML | SVM | EMR | Independent validation dataset | To evaluate the performance of a phenotyping algorithm trained by a previous version of diagnostic codes and effect of updating diagnostic codes |
In all patients with RA: Previous model: AUC = 0.93, PPV = 91%, NPV = 0.87, specificity = 0.95, sensitivity = 0.76 Updated version: AUC = 0.94, PPV = 91%, NPV = 0.88, specificity = 0.95, sensitivity = 0.77 |
|
| Ning et al. (2019) [101] | Supervised & Unsupervised ML | SEmantics-Driven Feature Extraction (SEDFE), adaptive elastic-net penalized logistic regression, PheNorm | Training = 100, 150, 200, 250, and 300 | EHR | Manual validation | To develop a feature extraction model independent from the EHR and classify rheumatoid arthritis, CAD, CD, UC, and pediatric PAH |
For RA: Supervised (with SEDFE and 300 labels): AUC = 0.940 PheNorm (with SEDFE): AUC = 0.944 |
| Yu et al. (2018) [102] | Supervised & Unsupervised ML | PheNorm, adaptive elastic-net penalized logistic regression, XPRESS algorithms, Anchor algorithms |
Training = 100, 150, 200, 250, and 300 For XPRESS: N = 750 (except for CAD), N = 741 (for CAD) |
EHR | Manual validation, fivefold cross-validation | To classify rheumatoid arthritis, CAD, CD, and UC using unlabeled data |
For RA: PheNormvote (with SAFE): AUC = 0.937 Supervised (with SAFE and 300 labels): AUC = 0.935 XPRESS algorithms: AUC = 0.896 Anchor algorithms: AUC = 0.890 |
| Gronsbell et al. (2019) [103] | Supervised & Unsupervised ML | Unsupervised clustering, followed by regularized regression on | N = 435 | EMR | Independent validation dataset | To identify disease status and predict the most informative features using unlabeled data | AUC = 0.928 |
| Gronsbell et al. (2018) [104] | Semi-supervised ML | Semi-supervised approach | N = 44,014 (Labeled = 500, Unlabeled = 43,514) | EMR | Tenfold cross-validation | To develop a semi-supervised phenotyping algorithm | For RA: AUC = 94.93 |
| Zhou et al. (2016) [105] | Supervised ML | Random forest and C5.0 decision tree |
Two data sets: N= 5208 and N = 475,580 |
EHR | Two independent testing datasets | To accurately and rapidly identify the most informative predictors for classification of RA in primary care EHR in a cost-effective manner |
Using the Cardiff-Cellma population with a prevalence of 27% for RA: PPV = 85.6%, specificity = 94.6%, sensitivity = 86.2% and overall accuracy = 92.29% Using the primary care population: in the worst-case scenario: PPV = 30.9%, specificity = 99%, sensitivity 83% = in the best-case scenario: PPV = 91.3%, specificity = 99.9%, and sensitivity 94% |
| Lin et al. (2015) [106] | Supervised ML | NLP and classification rules |
Case = 600 Controls = 430 |
EMR | Tenfold cross-validation, independent test set | To identify RA patients with methotrexate-induced liver toxicity | F1-score = 0.847, Precision = 0.8, Recall = 0.899 |
| Chen et al. (2013) [107] | Supervised ML | Active learning and SVM |
RA = 185 Controls = 191 |
EHR | Five-fold cross-validation | Phenotype identification using active learning to reduce the number of required annotated samples | AUC > 0.95 |
| Carrol et al. (2011) [108] | Supervised ML | SVM | N = 376 | EHR | Ten-fold cross-validation | To predict RA disease status | Naïve dataset: Precision = 93.3 ± 0.5, Recall = 79.7 ± 5.2, F measure = 85.1 ± 3.7, AUC = 94.2 ± 1.3; Refined dataset: Precision = 93.3 ± 0.5, Recall = 85.8 ± 5.7, F measure = 88.6 ± 4.0, AUC = 96.6 ± 1.1 |
| Liao et al. (2010) [109] | Supervised ML | Penalized logistic regression with adaptive LASSO procedure |
Training: RA = 96 Controls = 404 Validation: NN = 400 |
EMR | Threefold cross-validation, hold-out test set | To classify RA and non-RA cases |
Complete algorithm: PPV = 94% (95% CI 91–96%) sensitivity = 63% (95%CI 51–75%) |
AUC area under the curve, CAD coronary artery disease, CD Crohn's disease, EHR electronic health record, EMR electronic medical record, LASSO least absolute shrinkage and selection operator, ML machine learning, NLP natural language processing, NPV negative predictive value, PAH pulmonary arterial hypertension, PPV positive predictive value, RA rheumatoid arthritis, SVM support vector machine, UC ulcerative colitis
Several other supervised ML models have been developed for phenotype identification. Zhou and colleagues applied random forests algorithm and proposed a model identifying the most informative predictors of RA status using a large pool of data from patients in primary and secondary care settings, with an overall accuracy of 92.3%, which was comparable with methods derived from expert clinical opinion [105].
Not only can ML models facilitate disease status prediction, but they also could aid in stratification of patients. For instance, Lin et al. developed a classification algorithm to predict cases with MTX-induced liver toxicity. They found that incorporating temporality, i.e., the temporal relation between the presence of liver toxicity events and receiving MTX, can improve the performance of the model [106].
In a novel approach, Cai et al. developed a supervised model to facilitate participant selection for clinical trials by providing an alternative solution for the costly and time-consuming process of eligibility screening and chart review. They combined random forest and logistic LASSO regression to produce a model identifying potentially eligible patients from EHRs for an RA clinical trial. Compared with two rule-based systems, the AI algorithm had a better positive predictive value than one and a better sensitivity than the other; therefore, creating a balance between including and excluding too many patients for manual review [95].
Requirement of a large number of labeled data for training the supervised models is a major challenge in their application for phenotype identification. The quantity of needed annotated samples can be reduced by using semi-supervised and unsupervised models [101, 102]. Semi-supervised models usually use a small-sized labeled dataset and also a large-sized unlabeled dataset to classify data. Few semi-supervised models have been created for phenotype identification using EHRs. Gronsbell and colleagues developed a semi-supervised model that was validated with real data from patients with RA and multiple sclerosis (MS) with a performance comparable to the supervised methods [104]. Moreover, Chen et al. combined SVM and active learning, a form of semi-supervised learning method, and developed a model that outperformed passive learning and reduced the number of the required annotated samples by approximately two-thirds [107]. PheNorm is an unsupervised phenotyping algorithm that has been validated using four phenotypes, namely coronary artery disease, RA, Crohn's disease, and ulcerative colitis, with an accuracy comparable to that of supervised models [102]. Lastly, Gronsbell et al. developed a two-step model, with the first step being an unsupervised clustering method followed by a regularized regression as the second step using unlabeled observations to identify the most informative features from text fields available in the entire EHR. Their model showed a favorable performance (AUC = 0.93) with improved efficiency by reducing the number of labels required [103].
Importantly, the potential of EHRs can be further unraveled by enhancing the performance of the models through developing more complex networks incorporating DL and ANN [111, 112]. Algorithms with high performance can ultimately supersede ICD billing codes, which have the limitation of considerable error rates due to inconsistent terminology [113].
Predicting Treatment Response
Methotrexate is generally the initial DMARD choice for RA. If MTX fails to suppress the disease (which is the case in half of MTX monotherapy patients [114]), the treatment is stepped-up, and other anti-inflammatory drugs are administered, which are usually more expensive [115]. However, treatment failure still persists in some patients on second- or third-line medications, which can only be overcome by trial and error. Hence, a precision medicine treatment approach (also known as personalized or individualized medicine) based on each patient's biological profile could reduce treatment irresponsiveness and its consequences for both the patient and the healthcare system. The data used for choosing the proper treatment plan for a patient could range from simple variables, such as sex and age, to complex data, such as proteomics and transcriptomics.
Patients' demographic and clinical information are generally easily accessible. Such availability of vast amounts of input can result in accurate precision medicine algorithms. Machine learning algorithms have been shown to be able to predict response to MTX with AUCs as high as 0.84 using demographic and clinical data, such as past medical history and laboratory measures [116, 117]. Patients who do not respond to initial treatment should be stepped-up to more powerful medications. Morid et al. [118] evaluated multiple supervised and semi-supervised ML techniques to find the most accurate one to forecast a need for treatment step-up within 1 year among 120,237 patients. One-class SVM showed the best performance with a sensitivity and specificity of 89% and 83%, respectively. Despite the step-up therapy and trying several regimens, response failure persists in some patients (i.e., difficult-to-treat patients) [119]. An extreme gradient boosting algorithm [119] was able to identify these patients with a comparatively high accuracy (AUC = 0.73, sensitivity = 79%, specificity = 50%).
Omics are valuable input sources for predicting treatment response and vary greatly between patients due to different genetic materials and disease molecular basis. Artacho et al. created a random forest model that could identify MTX responders using gut microbiome data with an AUC of 0.84 [114]. When only patients with high (≥ 80%) or low (≤ 20%) chances of response were taken into account, the AUC of the algorithm increased to 0.94. The algorithm did not select pharmacogenetic predictors when provided as input, demonstrating a close relationship between gut microbiota and treatment response [114]. In another study, Plant et al. [120] incorporated transcriptomics and were able to predict MTX response among patients in early treatment stages with an AUC of 0.78. Not all studies yielded such favorable results, and AUCs for predicting MTX response reached as low as 0.61 [115].
Utilizing omics data seems more beneficial in predicting response to second- or third-line biological DMARDs (bDMARDs) than MTX [121, 122]. For instance, an SVM algorithm recognized patients responding to infliximab with an AUC of 0.92 using genomics data [122]. Some studies fed clinical data (e.g., lab results and disease activity measurements) in addition to omics, to their algorithms [123–125] and produced treatment response prediction AUCs as high as 0.83 [126], although the results were fairly heterogeneous.
Imaging data can also be employed in models predicting response to treatment. Kato et al. [127] developed a scoring system based on severity of synovitis, tenosynovitis, and enthesitis on ultrasound images in patients with RA and spondyloarthritis, assessing treatment response. An unsupervised random forest, in addition to uniform manifold approximation and a projection algorithm, was implemented, which divided patients into two clusters with significantly different responses to treatment as measured by the American College of Rheumatology 20, 50, and 70 (ACR20/50/70) criteria.
However, several shortcomings need to be acknowledged in studies applying AI to predict response to treatment. The variety of evaluation methods in determining treatment response makes the comparison of the results between different studies difficult and inaccurate. The EULAR criteria [128] was the most commonly used measure of response, which takes disease activity scores, ESR, and patient's global assessment into account (several variations exist). However, some studies used other definitions for treatment responsiveness, such as the continuation of MTX administration [117] and dose adjustments [129]. Furthermore, most studies are performed on MTX, and few have evaluated treatment outcomes using other RA treatments, especially non-biological DMARDs. Identifying patients for whom non-biological DMARDs are safe and effective substitutes using AI algorithms can be immensely helpful considering the higher cost of bDMARDs and their unavailability to many patients [130].
Table 7 lists studies incorporating ML for predicting treatment response in RA [114–127, 129, 131–134].
Table 7.
Studies incorporating AI for assessment of treatment response in RA
| First author | Model | Algorithms applied | No. of data | Type of the primary data | Validation/Test | Objective | Prominent outcomes presented |
|---|---|---|---|---|---|---|---|
| Lim et al. (2022) [131] | Supervised ML | Neural networks, SVM, logistic regression, elastic nets, random forest, boosted trees |
Training = 279 Test = 70 |
Demographic, clinical, lab, and omics data | Five-fold cross-validation, hold-out test set |
To predict response to MTX Criteria: DAS28 |
100 features (95 genetic), Model: boosted trees, AUC = 0.828, sensitivity = 0.6875, specificity = 0.8684 |
| Amin Shipa et al. (2021) [116] | Supervised ML | Logistic regression, LASSO logistic regression, SVM, naïve bayes, random forest, bagging, decision single tree, gradient boosting |
Training = 655 (358 responders) Validation = 225 (130 responders) |
Demographic, clinical, and lab data | Independent test set |
To predict response to MTX Criteria: DAS28-ESR ≤ 3.2 at 6 months |
Model: SVM, accuracy = 86%, AUC = 0.84 |
| Artacho et al. (2021) [114] | Supervised ML | Random forest |
Training = 26 (10 responders) Validation 1 = 21 Validation 2 (RA patients not on MTX) = 20 |
Gut microbiome | Test set |
To predict response to MTX in patients with new-onset RA Criteria: 1.8 DAS28 improvement by month 4 with no additional biologic drug |
AUC = 0.84, True negative rate = 83.3%, True positive rate = 78% AUC = 0.94 (for patients with high (≥ 80%) or low (≤ 20%) chance of responding) |
| Gosselt et al. (2021) [125] | Supervised ML | Logistic regression, LASSO regression, random forest, extreme gradient boosting |
Training = 249 (125 responders) Test = 106 (53 responders) |
Demographic, clinical, and genotype data | Tenfold cross-validation, hold-out test set |
To predict response to MTX Criteria: DAS28 ≤ 3.2 at 3 months |
Model: logistic regression, AUC = 0.77 (0.68–0.86), sensitivity = 81% specificity = 60%, accuracy = 71%, PPV = 67%, NPV = 76% |
| Jung et al. (2021) [64] | Unsupervised ML | Naïve Bayes | N = 152 | Omics data | N/A | To predict treatment response based on synovial tissue subtype |
Classification yielded 3 groups: C1: 17.6% response to infliximab C2: 40% response to triple DMARDs and 29.4% response to infliximab C3: 77.8% response to triple DMARDs and 63.6% response to infliximab |
| Kato et al. (2021) [127] | Unsupervised ML | Random forest + uniform manifold approximation and projection |
N = 38 [RA (26) and spondyloarthritis (12)] ACR20 = 26 ACR50 = 21 ACR70 = 17 |
Ultrasound imaging | N/A |
To predict response to MTX at 12 weeks Criteria: ACR20/50/70% |
Significantly more ACR20, ACR50, and ACR70 in cluster group 1 (p = 0.007, 0.034, and 0.016, respectively) |
| Koo et al. (2021) [132] | Supervised ML | LASSO linear regression, ridge linear regression, SVM, random forest, extreme gradient boosting |
N = 1397 (564 responders) TNF inhibitors = 793 (252 responders) Non-TNF Inhibitors = 504 (312 responders) Adalimumab = 289 (91 responders) Etanercept = 220 (75 responders) Golimumab = 122 (41 responders) Infliximab = 162 (45 responders) Abatacept = 194 (62 responders) Tocilizumab = 410 (250 responders) |
Demographic, clinical, and lab data | Five-fold cross-validation, hold-out test set |
To predict response to bDMARDs Criteria: DAS28-ESR ≤ 2.6 |
All bDMARDs → Model: Ridge AUC = 0.619, accuracy = 61.5%, sensitivity = 29.6%, specificity = 83.1% TNF inhibitors → Model: Ridge AUC = 0.655, accuracy = 70.0%, sensitivity = 21.3%, specificity = 92.6% Non-TNF inhibitors → Model: Ridge AUC = 0.607, accuracy = 57.8%, sensitivity = 64.5%, specificity = 51.7% Adalimumab → Model: Ridge AUC = 0.688, accuracy = 69.8%, sensitivity = 29.6%, specificity = 88.1% Etanercept → Model: Ridge, random forest AUC = 0.656, accuracy = 67.2%, 66.2%, sensitivity = 36.4%, 0%, specificity = 83.7%, 100% Golimumab→ Model: Ridge AUC = 0.694, accuracy = 63.9%, sensitivity = 41.7%, specificity = 79.2% Infliximab→ Model: Ridge AUC = 0.626, accuracy = 70.8%, sensitivity = 23.1%, specificity = 88.6% Abatacept→ Model: Ridge AUC = 0.679, accuracy = 68.4%, sensitivity = 30.6%, specificity = 84.6% Tocilizumab → Model: SVM AUC = 0.556, accuracy = 61.0%, sensitivity = 80.0%, specificity = 22.9% |
| Luque-Tevar et al. (2021) [126] | Supervised ML | LASSO regression, ridge regression |
Training = 74 (responders = 52) Validation = 25 (responders = 14) |
Clinical, lab, and omics data | Independent test set |
To predict response to TNF inhibitors Criteria: EULAR |
Model: LASSO regression (mixed clinical and molecular parameters), AUC = 0.83 |
| Maciejewski et al. (2021) [115] | Supervised ML | Linear regression, random forest, SVM with kernel, LASSO/ridge regression | N = 100 (responders = 50) | Omics data | Five-fold cross-validation |
To predict response to MTX Criteria: EULAR |
Model: LASSO/ridge regression, AUC = 0.61 ± 0.02 |
| Messelink et al. (2021) [119] | Supervised ML | Extreme gradient boosting | N = 116 (28 D2T) | Demographic, clinical, and lab data | tenfold cross-validation |
To identify difficult to treat patients Criteria: EULAR |
AUC = 0.73 (0.71–0.75), sensitivity = 79%, specificity = 50% |
| Morid et al. (2021) [118] | Supervised and semi-supervised ML | Naïve Bayes, SVM, extreme gradient boosting, kNN, random forest, logistic regression, one-class SVM, support vector mapping convergence, POSC4.5, nearest neighbor description, naïve bayes positive class | N = 120,237 (17,602 step-up) | Demographic and clinical data | Validation set, hold-out test set | To predict treatment step-up necessity within one year | Model: one-class SVM, PPV = 51%, NPV = 97%, sensitivity = 89%, specificity = 83% |
| Tao et al. (2021) [121] | Supervised ML | Random forest | N = 38 (responders = 20) | Omics data | Five-fold cross-validation, Independent test set |
To predict response to adalimumab Criteria: EULAR |
Using the PBMC RNA model: accuracy = 85.9% |
| N = 42 (responders = 19) |
To predict response to etanercept Criteria: EULAR |
Using the PBMC DNA model: accuracy = 88% | |||||
| Westerlind et al. (2021) [117] | Supervised ML | LASSO regression, elastic net regularization, SVM with linear kernel, extreme gradient boosting | Training = 4927, validation = 548 | Demographic, clinical, and past medical data | Five-fold cross-validation, hold-out test set |
To predict response to MTX within one year Criteria: continuation of MTX administration |
Models: LASSO and elastic net, AUC = 0.67 (0.62–0.71/0.72) using covariate sets A, B, and C, AUC = 0.66 (0.62–0.71) using covariate set D |
| Yoosuf et al. (2021) [124] | Supervised ML | Logistic regression | N = 28 (baseline data) | Demographic, clinical, lab, and response data | Independent test set |
To identify differentially expressed genes between anti-TNF responders and non-responders Criteria: EULAR |
7 genes after leave on out analysis: 2 with no errors, 5 with one error |
| Mixed LASSO and ridge linear regression, random forest, SVM with radial basis function kernel | N = 25 (responders = 17) | Lab, clinical, and omics data | Five-fold cross-validation |
To predict response to anti-TNF treatment Criteria: EULAR |
Model: linear model using transcriptomic data, AUC = 0.81 (SEM = 0.17) | ||
| Gomez et al. (2020) [133] | Supervised ML | SVM, random forest | N = 58 (responders = 36) | Blood lipid mediator profiles | Independent test set |
To predict response to DMARDs Criteria: EULAR DAS28-ESR |
Model: random forest, AUC = 0.8, accuracy = 83%, sensitivity = 83%, specificity = 59% |
| Guan et al. (2019) [123] | Supervised ML | Gaussian process regression |
Training: N = 1892 (responders = 1456) Test: N = 680 (responders = 442) |
Demographic, clinical, and omics data | Two-fold cross-validation, independent test set |
To predict anti-TNF response Criteria: DAS28 change and EULAR |
DAS28 change correlation = 0.393 Response prediction AUC = 0.615 |
| Kim et al. (2019) [122] | Supervised ML | Naïve-Bayes, decision Trees, kNN, SVM | N = 62 (responders = 18) | Pathway and omics data | Tenfold cross-validation |
To predict response to infliximab Criteria: EULAR |
Data: genomics data, Model: SVM, AUC = 0.92 |
| Plant et al. (2019) [120] | Supervised ML | Ridge logistic regression, random forest, network-based approach | N = 85 (responders = 42) | Omics data | Tenfold nested cross-validation |
To predict response to MTX Criteria: EULAR |
Model: network-based models, AUC = 0.78 (SEM = 0.06), Balanced accuracy = 0.68 (SEM = 0.06) |
| Miyoshi et al. (2016) [134] | Supervised ML | Multi-layer perceptron |
N = 179 (responders = 138) Training = 141 Test = 38 |
Clinical and lab data | Independent test set |
To predict response to infliximab Criteria: EULAR |
AUC = 0.75, accuracy = 92.1%, sensitivity = 96.7%, specificity = 75% |
| Van looy et al. (2006) [129] | Supervised and unsupervised ML | Self-organizing maps, multi-layer perceptron, SVM, linear discrimination analysis, logistic regression | RA = 511 | Clinical and lab data | Cross-validation, Hold-out test set |
To predict response to infliximab Criteria: dose increment at week 22 |
Using 4 features: Model: logistic regression, AUC = 0.878, sensitivity at 0.95 specificity = 0.385, specificity at 0.95 sensitivity = 0.575 |
(b)DMARD (biological) disease-modifying anti-rheumatic drug, ACR American college of rheumatology, AUC area under the curve, DAS28 disease activity score in 28 joints, DNA deoxy-ribonucleic acid, ESR erythrocyte sedimentation rate, EULAR European alliance of associations for rheumatology (also, European league against rheumatism), kNN k-nearest neighbor, LASSO least absolute shrinkage and selection operator, ML machine learning, MTX methotrexate, N/A not available, NPV negative predictive value, PBMC peripheral blood mononuclear cell, PPV positive predictive value, RA rheumatoid arthritis, RNA ribonucleic acid, SEM standard error of the mean, SVM support vector machine, TNF tumor necrosis factor
Monitoring Disease Course and Predicting Prognosis
Measuring disease activity is crucial in choosing the optimal treatment plan, determining response to therapy, and prognosis. Moreover, predicting disease severity early on could assist in timely administration of the most suitable medications. Disease activity score in 28 joints (DAS28) is one of the most utilized severity measures of RA [135, 136]. This index could be calculated based on various inflammatory markers, including ESR or CRP [137]. An adaptive deep neural network [137] was able to outperform non-DL methods in predicting DAS28-ESR from demographical and clinical data with an AUC of 0.73 (categorical prediction) and mean standard error of 0.9 (numerical prediction). However, the attempt by Rychkov et al. [138] to predict DAS28 using omics data yielded unsatisfactory results, and their novel RA score showed only a weak (r = 0.33) correlation with DAS28. The clinical disease activity index (CDAI) [139] is another scoring system that only uses clinical data and can be calculated more rapidly than DAS28. Norgeot et al. developed a model using neural networks with a remarkable AUC of 0.91 in predicting disease activity according to the CDAI [140].
Predicting risk of needing treatment step up to tocilizumab in patients who do not respond to initial therapy is another example of applications of AI in monitoring disease course in RA. A logistic regression model [141] showed that higher age and remission CDAI were the most important risk and protective factors for tocilizumab monotherapy, respectively (OR = 1.04 and 0.17, respectively) when excluding other treatments as variables. For any tocilizumab use (either monotherapy or in combination), the highest and lowest ORs belonged to the number of comorbidities (OR = 1.16) and remission CDAI (OR = 0.20) (excluding other treatments as factors).
Rheumatoid arthritis is associated with a wide range of comorbidities, particularly cardiovascular, atherosclerotic, musculoskeletal, and neurological diseases [142–144]. Preventing these complications requires timely identification of patients at risk. Carotid ultrasound is a non-invasive and efficient modality to assess atherosclerotic plaques. ML and DL algorithms enable enhanced cardiovascular risk stratification in patients with RA by analyzing these images [145]. Machine learning algorithms developed by Wei et al. using demographic, clinical, and laboratory data as input performed satisfactorily in predicting the incidence of coronary heart disease (CHD) in patients with RA (AUC = 0.79, accuracy = 76%). Their logistic regression model outperformed conventional cardiovascular disease (CVD) risk score, i.e., Framingham Risk Score [146]. However, another investigation found a statistically comparable AUC for predicting stroke using a complex logistic regression model fed with laboratory data compared to the Framingham Risk Model [147]. Remarkably, in a recent investigation, ML classifiers outperformed the classical cardiovascular disease risk score when they were fed with cardiovascular risk factors, including conventional risk factors, laboratory-based blood biomarkers, and ultrasound images [148].
Musculoskeletal complications are one of the other major comorbidities in patients with RA. Risk factors for bone loss in patients with RA were identified by Hu et al. [149] using conventional logistic regression, LASSO regression, and random forest methods. The highest and lowest OR belonged to age for femoral neck bone loss (OR = 1.17) and TNF inhibitor use in the past year for lumbar spine bone loss (OR = 0.27). Other affecting factors included body mass index (BMI) and serum vitamin D levels.
Wearable and portable devices can play a substantial role in monitoring disease activity as well. Many of the devices used in today's medicine have become portable, such as pulse oximeters and cardiac Holter monitors. Newer wearable devices can measure a wide variety of indicators and have the capacity to be programmed to produce the most helpful outputs. The most common use of wearable sensors is probably tracking physical activity [150], which in recent years has been finding its way into medicine [151, 152]. Patients with RA may experience flares throughout their disease course, which will most likely hinder their physical activity due to the acute inflammation [153, 154]. Furthermore, flares are associated with disease progression and worse outcomes [155], even in those with low disease activity [156]. Hence, keeping an accurate track of flares could greatly improve patient care. Gossec et al. [157] developed a naïve Bayes model that utilized physical activity input from a watch to detect flares (as reported by the patients themselves). Their algorithm showed 95.7% sensitivity and 96.7% specificity for detecting flares, suggesting wearable sensors as potentially reliable devices for monitoring flares.
Table 8 summarizes studies implementing ML and DL for monitoring disease course and predicting prognosis [79, 137, 138, 140, 141, 146, 147, 149, 157–166].
Table 8.
Studies incorporating AI for assessment of disease course and prognosis in RA
| First author | Model | Algorithms applied | No. of data | Type of the primary data | Validation/Test | Objective | Prominent Outcomes presented |
|---|---|---|---|---|---|---|---|
| Hu et al. (2021) [149] | Supervised ML | Conventional logistic regression, LASSO regression, random forest | RA = 340 (osteopenia = 88, osteoporosis = 186) | Demographic, clinical, and lab data | N/A | To identify risk factors for osteopenia and osteoporosis in patients with RA |
Lumbar spine: Model: conventional logistic regression, Predictors: age (OR = 1.1), BMI (OR = 0.88), serum 25(OH)D3 level (OR = 0.99), TNF inhibitor usage in the past year (OR = 0.27) Femoral neck: Model: LASSO regression, Predictors: age (OR = 1.17), BMI (OR = 0.85), rheumatoid factor concentration (OR = 1, p > 0.05) Total hip: Model: conventional logistic and LASSO regressions, Predictors: age (OR = 1.11), BMI (OR = 0.84), TNF inhibitor usage in the past year (OR = 0.37) |
| Hur et al. (2021) [158] | Supervised ML | Generalized linear model |
Training = 64 Test = 12 |
Omics data | Cross-validation, Independent test set | To predict disease activity Criteria: DAS28-CRP |
Mean absolute error = 0.97 ± 0.47 (using feature selection and 51 metabolites) Spearman’s p = 0.69 |
| Kalweit et al. (2021) [137] | Supervised ML, DL | Adaptive deep neural network (AdaptiveNet), random forest, logistic regression, SVM | 28,601 visits | Demographic, clinical, and lab data | Five-fold cross-validation |
To predict disease activity Criteria: DAS28-ESR |
Model: Adaptive deep neural network, Categorical prediction AUC = 0.728 ± 0.01, Numerical prediction MSE = 0.90 ± 0.05 |
| Rychkov et al. (2021) [138] | Supervised ML | Logistic regression, elastic net, random forest | N = 411 | Omics data | N/A |
To predict disease activity Criteria: DAS28 |
Calculated RA score from 13 gene markers RA score correlation with DAS28 = 0.33 (0.24–0.41) |
| Vodencarevic et al. (2021) [159] | Supervised ML | Logistic regression, kNN, naïve Bayes, random forest, a stacking classifier | N = 135 visits (31 flares) | Demographic, clinical, and lab data | Two-layer cross-validation | To predict flare in patients in remission (based on DAS28-ESR) | Model: stacking classifier, AUC = 0.808 ± 0.090, sensitivity = 0.78 ± 0.11, specificity = 0.86 ± 11, accuracy = 0.81 ± 0.08 |
| Solomon et al. (2021) [141] | Supervised ML | Logistic regression | N = 7300 (287 tocilizumab monotherapy) | Demographic, clinical, and lab data | N/A | To predict the use of tocilizumab monotherapy |
ORs for variables: Age (baseline + follow-up) = 1.04 (1.00–1.08) Disease duration (baseline + follow-up) = 1.04 (1.00–1.07) Positive serological status (baseline) = 0.73 (0.57–0.93) Conventional synthetic DMARD use (baseline) = 0.48 (0.37–0.63) Tocilizumab combination therapy (baseline + follow-up) = 327 (80–1343) TNF inhibitor therapy (baseline + follow-up) = 0.21 (0.09–0.49) No DMARD use (baseline + follow-up) = 0.09 (0.03–0.33) Moderate CDAI (compared to severe, baseline + follow-up) = 0.43 (0.23–0.78) Low CDAI (compared to severe, baseline + follow-up) = 0.39 (0.21–0.74) Remission CDAI (compared to severe, baseline + follow-up) = 0.17 (0.07–0.42) |
| To predict the use of tocilizumab |
ORs for variables: Age (baseline) = 0.99 (0.98–0.99) Age (baseline + follow-up) = 1.05 (1.02–1.08) Disease duration (baseline + follow-up) = 1.06 (1.03–1.08) Positive serological status (baseline) = 0.66 (0.56–0.78) Positive serological status (baseline + follow-up) = 0.11 (0.04–0.29) Conventional synthetic DMARD use (baseline) = 0.71 (0.59–0.86) Conventional synthetic DMARD use (baseline + follow-up) = 0.18 (0.12–0.27) TNF inhibitor therapy (baseline + follow-up) = 0.48 (0.26–0.90) Non-TNF-inhibitor, Non-tocilizumab biological DMARD use (baseline + follow-up) = 4.59 (2.91–7.25) Only conventional synthetic DMARD (baseline + follow-up) = 5.83 (3.73–9.13) No DMARD use (baseline + follow-up) = 0.40 (0.18–0.88) Moderate CDAI (compared to severe, baseline + follow-up) = 0.65 (0.46–0.91) Low CDAI (compared to severe, baseline + follow-up) = 0.48 (0.33–0.70) Remission CDAI (compared to severe, baseline + follow-up) = 0.20 (0.11–0.37) Number of comorbidities (baseline + follow-up) = 1.16 (1.11–1.22) |
||||||
| Xin et al. (2021) [147] | Supervised ML | Logistic regression, SVM, random forest, extreme gradient boosting, gradient boosting decision tree, kNN |
Training = 1354 (218 with stroke) Test = 518 (95 with stroke) |
Lab data | Five-fold cross-validation, Hold-out test set | To predict stroke in RA patients | Adjusted (complex) model: logistic regression, AUC = 0.784 (0.750–0.818), F1-score = 0.630 |
| Bonakdari et al. (2020) [160] | Supervised DL | GS-GMDH | N = 17,505 (total hip replacement = 465, total knee replacement = 650) | Time-series data | Hold-out test set | To predict the incidence of total hip and total knee replacements in RA patients |
Total hip replacement: R = 0.98, Scatter index = 0.09, Mean absolute relative error = 0.10, Mean prediction error = 0.000 ± 0.484 Total knee replacement: R = 0.78, Scatter index = 0.11, Mean absolute relative error = 0.09, Mean prediction error = 0.000 ± 1.182 |
| Christensen et al. (2020) [161] | Supervised DL | Cascaded CNN |
Training = 1678 Test = 322 |
Ultrasound images | Validation and hold-out test sets |
To grade disease activity based on ultrasound images Criteria: EULAR-OMERACT synovitis scoring system |
Four-class accuracy = 83.9% Binary classification: accuracy = 89.9%, AUC = 0.96, sensitivity = 90.5%, specificity = 89.3% |
| Lotsch et al. (2020) [162] | Supervised ML | Classification and regression trees, kNN, SVM, multilayer perceptron, naïve Bayes | N = 288 | Demographic, clinical, and lab data | Three-fold cross-validation | To identify pain-related phenotype patients | Model: kNN, accuracy = 71.5%, F1-score = 61.5, sensitivity = 63.2%, specificity = 80.9%, Positive predictive value = 61.5%, Negative predictive value = 82.9% |
| Petrackova et al. (2020) [163] | Supervised ML | Neural network |
Training = 57 Test = 10 |
Omics data | tenfold cross-validation, Independent test set |
To identify patients with active disease Criteria: DAS28 |
Two sets of genes discriminated between patients with active and inactive disease |
| Wei et al. (2020) [146] | Supervised ML | Logistic regression, SVM, random forest, extreme gradient boosting, gradient boosting decision tree, kNN |
Training = 1012 (294 with coronary heart disease) Test = 274 (70 with coronary heart disease) |
Demographic, clinical, and lab data | tenfold cross-validation | To predict coronary heart disease in RA patients | Adjusted model: logistic regression, AUC = 0.79, accuracy = 76%, F1-score = 0.68, Recall (sensitivity) = 68%, Precision (positive predictive value) = 70%, Balance error = 0.32 |
| Andersen et al. (2019) [79] | Supervised DL | CNN |
N = 40 patients Training images = 1342 Validation images = 176 Test images = 176 (44 of each grade) |
Ultrasound images | Validation and hold-out test sets |
To grade disease activity based on ultrasound images Criteria: EULAR-OMERACT synovitis scoring system |
Four-class accuracy = 75% Binary classification: accuracy = 86.9%, sensitivity = 87.5%, specificity = 86.4% |
| Feldman et al. (2019) [164] | Supervised ML | LASSO regression | N = 300 (154 high disease activity) | Demographic, clinical, and lab data | tenfold cross-validation |
To estimate disease activity Criteria: DAS28-CRP |
Continuous DAS28-CRP estimation max R2 = 0.18 Binary disease activity classification: AUC = 0.77, sensitivity = 83.1% (65.9–76.4), specificity = 58.9% (50.5–67.0), PPV = 68.1% (60.9–74.7), NPV = 76.8% (67.9–84.2) |
| Gossec et al. (2019) [157] | Supervised ML | Naïve Bayes |
N = 155 (1339 weekly flare assessments, ~ 13.5 million activity points) RA = 82 (axial spondyloarthritis = 73) |
Physical activity data | Hold-out test sets | To predict flares |
Sensitivity = 95.7% (94.4–97.0), specificity = 96.7% (96.0–97.3), PPV = 89% (88–91) NPV = 99% (98–100), accuracy = 96.4% |
| Norgeot et al. (2019) [140] | Supervised DL | Neural networks |
Primary cohort = 578 (116 test cohort) Secondary cohort = 242 (117 test cohort) |
Demographic, clinical, and lab data | Hold-out and mixed test sets | To predict disease activity Criteria: CDAI | Primary cohort AUC = 0.91 (0.86–0.96) |
| Joo et al. (2017) [165] | Supervised ML | SVM (for selecting the SNPs), Logistic regression |
Training = 374 Test = 399 |
Clinical and omics data | Tenfold cross-validation, Independent test set |
To predict radiographic progression Criteria: Sharp/Van der Heijde modified score |
Average accuracy = 61.43% |
| Lezcano-Valverde et al. (2017) [166] | Supervised ML | Random survival forest |
Training = 1461 Test = 280 |
Demographic, clinical, and lab data | Independent test set | To predict mortality in RA patients | Prediction error = 0.233 |
AUC area under the curve, BMI body mass index, CDAI clinical disease activity index, CNN convolutional neural network, CRP C-reactive protein, DAS28 disease activity score in 28 joints, DL deep learning, DMARD disease-modifying anti-rheumatic drug, ESR erythrocyte sedimentation rate, EULAR European alliance of associations for rheumatology (also, European league against rheumatism), GS-GMDH generalized structure group method of data handling, kNN k-nearest neighbor, LASSO least absolute shrinkage and selection operator, ML machine learning, MSE: mean squared error, N/A not available, NPV negative predictive value, OMERACT outcomes measures in rheumatology, OR odds ratio, PPV positive predictive value, RA rheumatoid arthritis, SNP single-nucleotide polymorphism, SVM support vector machine, TNF tumor necrosis factor
Drug Discovery
Rheumatic diseases are generally chronic in nature and require long-term treatment. Hence, developing novel drugs that are well tolerated and effective is of utmost importance. Drug discovery is an expensive process [167]; thus, it is necessary to make the involved procedures as efficient as possible. Many pharmaceutical projects fail due to incorrect target selection [168], which is an inevitable consequence of hypothesis-driven testing. Zhao and colleagues [169] addressed this issue by creating ML models that proposed potential treatments by inspecting expression profiles of patients being treated with a drug already proven to be effective and presenting targets that, if targeted, result in similar expression profiles. Their results for finding candidate targets for RA using random forest and gradient boosting machine algorithms showed significant concordance with an external database listing potential. Such investigations shift research flow from assumption-based and hypothesis-derived studies to studies based on known and proven data, which was not possible until recently due to challenges in handling the colossal amount of available information.
Basic Science Research
Similar to many other rheumatic diseases, not all aspects of the pathways involved in RA pathogenesis are known (133), mainly due to the complexity and extensiveness of involving factors. Machine learning algorithms are specifically designed to handle such conditions. For instance, two recent studies [170, 171] have pointed toward the possible role of gut microbiota in RA pathogenesis. Devaprasad and colleagues [172] acquired the immunome of 316 samples with immune-mediated inflammatory diseases, which were used to identify disease-related genes and cells of 12 inflammatory conditions, including RA. Their non-negative matrix factorization algorithm identified two main clusters of patients with different sets of cells and genes, further shedding light on immunological pathways involved in RA pathophysiology.
Discussion
This comprehensive updated study reviewed published investigations incorporating AI, including ML and DL related to RA, the second most prevalent autoimmune disease. Artificial intelligence models are used to assess RA development risk, diagnose RA using omics, imaging, clinical, and sensor data, detect RA patients within EHR, predict treatment response, monitor disease course, determine prognosis, discover novel drugs, and enhance basic science research (Fig. 3). We showed that a growing body of evidence supports the potential role of AI in revolutionizing screening, diagnosis, and management of patients with RA. However, the proposed models may vary significantly in their performance and reliability. Notably, since every decision made in the healthcare setting may have dire and irreversible consequences, considering the limitations of AI and the challenges of its implementation in healthcare is immensely important.
Fig. 3.
The role of artificial intelligence (AI) in enhanced diagnosis and management of rheumatoid arthritis (RA)
In 2020, Stafford and colleagues systematically reviewed the available literature on AI applications in autoimmune diseases [113]. After MS, the RA had the highest number of manuscripts dedicated to itself (41 and 32, respectively), followed by inflammatory bowel syndrome (30) and type 1 diabetes (17). Although less in number, RA studies investigated more types of outcomes than MS, utilized more data sources and AI methods, and had a higher median sample size (338 versus 99). In fact, RA had the widest range of input data sources among all autoimmune diseases, indicating the vast potential of AI application in the field. Furthermore, AI-based precision medicine approaches could especially be effective in RA due to the diversity in treatment options and disease phenotypes.
Challenges and Limitations of Implementing AI
Multiple technical challenges hinder applying AI models in patient care. The need for large and accurately labeled data is a major issue in training supervised models. Importantly, small training datasets can result in over-fitted models. Creating large and high-quality open-access databases can aid in tackling this challenge. The presence of such datasets also facilitates performance comparison between different models. The variability of test datasets in various studies does not usually allow for making accurate comparisons [173, 174]. The osteoarthritis initiative study is an example of such datasets, which has been used to test and train dozens of AI models to improve diagnosis and prediction of pain progression and outcome in osteoarthritis [175–177].
Moreover, the clinical applicability of AI models cannot necessarily be represented by the accuracy of the model. In many cases, the accuracy measures reported in a scientific paper may represent the performance of the model in a small dataset from a specific population instead of providing generalizable results to other populations [178]. The variation between the input datasets is a limiting factor in the clinical implementation of AI models [179]. Datasets obtained from different healthcare environments may vary in data acquisition method, coding, and patient population. As a result, the model might perform differently when applied to datasets different from the training input. External validation can show the effect of input data variation on the performance of the model. However, in most of the studies included in this review (approximately 70%), validation using an independent external dataset was not performed.
The AI models are technically prone to several other challenges as well. These models use any signal that helps them achieve the highest performance. However, these signals may include unknown confounders, incorporation of which in the model may damage the generalizability of the model. For instance, a model designed to detect hip fractures used confounding features, including the scanner model and "priority" marks on scans, to classify the input data [180]. Moreover, data manipulation (adversarial attack) can have damaging effects on the performance of the AI model. Adversarial examples are inputs with small changes made to fool the model intentionally [181, 182].
The retrospective study design in most investigations in this field can also limit the real-world application of AI models. While historically labeled data are the most commonly used resources for training and testing AI models, the true additional value of AI algorithms in the diagnosis and management of patients can be best captured by trials with a prospective design. Nevertheless, only a few prospective studies have been conducted on the real-world applications of AI in the medical field [183], and research related to RA is not an exemption. As an example of prospective trials, a multi-center randomized controlled trial was performed to compare the accuracy of an AI algorithm with senior consultants in diagnosing childhood cataracts and choosing optimal treatment options [184].
In addition to the mentioned challenges, in many cases, particularly for neural networks, it is very difficult to convey the intuitive notions driving the conclusion of the model. These models that are too complicated for a straightforward interpretation of the factors involved in the decision making are also referred to as the "black box". The opaque rationale behind decisions made by the model can cause ethical and social challenges. Such models may fail in engendering user trust as transparency is a fundamental factor in gaining credence. Additionally, not understanding the rationale behind the decisions and the potential sources of error may increase the chances of inaccuracy in the decisions made by the model, especially in new datasets obtained in a different setting. Notably, given that healthcare is a high-stakes field, it is critical to minimize the margin of error as much as possible [185, 186].
Algorithmic bias is another ethical challenge raised by the use of AI. In 2019, Panch et al. defined algorithmic bias as when the application of an AI model aggravates existing inequities in society, such as racial and sexual discrimination [187]. For instance, a recent paper showed that one of the commonly used algorithms in healthcare is racially biased, considering the same risk score for White patients and Black patients while the Black patients are considerably sicker. They found that the underlying cause of this bias is that the algorithm predicts healthcare costs instead of disease severity. Due to the discrimination in access to care, as less money is spent on the care of Black patients compared to White patients, the model generates biased results [188]. In another example, under-representation of skin cancer images from patients with darker skin can result in less accurate results for patients of color as the model has not been trained on a sufficient number of observations representing these populations [173, 189].
The intention behind the development of AI algorithms should also be acknowledged as one of the potential ethical challenges of implementing AI in healthcare. Given the growing importance of quality measures, private-sector developers may be inclined to create algorithms suggesting clinical decisions that improve quality metrics without necessarily enhancing quality of care [190]. An example of this action has been observed in the car industry, where software was used to reduce emissions [191]. Additionally, AI algorithms might be designed in a way profiting their developers or buyers by suggesting certain drugs, tests, or devices to increase profit, while the clinicians using the algorithm may not be aware of such biases [190].
Future Directions
Our study shed light on eight recommendations for future investigations. Notably, these directions can be used in studies related to other autoimmune musculoskeletal disorders as well. (1) Adherence to guidelines ensuring good conduct is critical in AI studies. The Checklist for Artificial Intelligence in Medical Imaging (CLAIM) [22] and the guideline released by the National Health Service (NHS) for "good practice for digital and data-driven health technologies" [192] are examples of such recommendations. (2) Open communication of the complete source codes is indispensable for verifying the reproducibility of the results by testing them on external datasets. Nevertheless, among studies reviewed in this paper, only a few provided open-access codes [97–99, 102, 103, 133, 140, 157, 158]. (3) It is vital that AI studies conduct external validation as it is a key component in assessing performance of a model in the real-world setting. However, among studies included in this review, almost half of the studies did not have an independent external dataset to validate the model. (4) As an AI model can be only as good as the data used to train it, future investigations need to ensure using high-quality data in large quantities. This can be achieved by creating large-scale multimodal datasets containing data on demographic, clinical, laboratory, genomic, imaging, and lifestyle features of the patients. (5) Future studies require consideration of the potential risk of algorithm bias during model development, and they should include sufficient data points representing minorities to reduce the risk of bias. (6) AI algorithms can be further used to assess extra-articular involvement, such as skin and ocular manifestations, in patients with RA. (7) Furthermore, currently, most investigations have compared the performance of AI algorithms with human experts. However, evaluating the performance of the collaboration of AI algorithms and human experts versus human experts alone would provide more realistic and applicable results [174]. (8) Lastly, real-world, and wide application of AI algorithms would heavily rely on design of prospective trials, ideally multi-center and randomized, assessing the performance of these models. Of note, our study paved the way for future reviews focusing on applications of AI in other high-burden autoimmune and inflammatory rheumatological and musculoskeletal diseases, such as MS and systemic lupus erythematosus.
Conclusions
Artificial intelligence (AI) can facilitate screening, diagnosis, monitoring, risk assessment, prognosis determination, achieving optimal treatment outcome, and de novo drug discovery for patients with rheumatoid arthritis, as well as broadening the knowledge of the disease pathophysiology by enhancing basic science research. Incorporating these machine and/or deep learning algorithms into real-world settings would be a key step in the progress of AI in medicine. Future investigations are required to ensure development of reliable and generalizable algorithms while they carefully look for any potential source of bias or misconduct.
Acknowledgements
Funding
No funding or sponsorship was received for this study or publication of this article.
Authorship
All named authors meet the International Committee of Medical Journal Editors (ICMJE) criteria for authorship for this article, take responsibility for the integrity of the work as a whole, and have given their approval for this version to be published.
Authors' Contributions
SM: Conceptualization, Investigation, Writing—Original Draft, Writing—Review & Editing, Visualization, AN: Conceptualization, Investigation, Writing—Original Draft, Writing—Review & Editing, Visualization, NR: Conceptualization, Writing—Review & Editing, Supervision, All authors approved the submitted version.
Disclosures
Sara Momtazmanesh, Ali Nowroozi, and Nima Rezaei have nothing to disclose.
Compliance with Ethics Guidelines
This study was conducted in accordance with the ethical principles of the Declaration of Helsinki of 1964 and its later amendments. Ethics committee approval was not required for this review article as it is based on previously conducted studies and does not contain any new studies with human participants or animals performed by any of the authors.
Data Availability
Data sharing is not applicable to this article as no datasets were generated or analyzed during the current study.
Footnotes
Sara Momtazmanesh and Ali Nowroozi have contributed equally to this work and share first authorship.
References
- 1.Artificial intelligence. https://www.merriam-webster.com/dictionary/artificial%20intelligence. Accessed 15 Feb 2022.
- 2.Hamet P, Tremblay J. Artificial intelligence in medicine. Metabolism. 2017;69S:S36–S40. doi: 10.1016/j.metabol.2017.01.011. [DOI] [PubMed] [Google Scholar]
- 3.Niazi MKK, Parwani AV, Gurcan MN. Digital pathology and artificial intelligence. Lancet Oncol. 2019;20:e253–e261. doi: 10.1016/S1470-2045(19)30154-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hosny A, Parmar C, Quackenbush J, Schwartz LH, Aerts H. Artificial intelligence in radiology. Nat Rev Cancer. 2018;18:500–510. doi: 10.1038/s41568-018-0016-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vamathevan J, Clark D, Czodrowski P, Dunham I, Ferran E, Lee G, Li B, Madabhushi A, Shah P, Spitzer M, Zhao S. Applications of machine learning in drug discovery and development. Nat Rev Drug Discov. 2019;18:463–477. doi: 10.1038/s41573-019-0024-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Benke K, Benke G. Artificial intelligence and Big Data in public health. Int J Environ Res Public Health. 2018;15:2796. doi: 10.3390/ijerph15122796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu X, Faes L, Kale AU, Wagner SK, Fu DJ, Bruynseels A, Mahendiran T, Moraes G, Shamdas M, Kern C, et al. A comparison of deep learning performance against health-care professionals in detecting diseases from medical imaging: a systematic review and meta-analysis. Lancet Digit Health. 2019;1:e271–e297. doi: 10.1016/S2589-7500(19)30123-2. [DOI] [PubMed] [Google Scholar]
- 8.Cao C, Liu F, Tan H, Song D, Shu W, Li W, Zhou Y, Bo X, Xie Z. Deep learning and its applications in biomedicine. Genom Proteom Bioinform. 2018;16:17–32. doi: 10.1016/j.gpb.2017.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.GBD Results Tool. http://ghdx.healthdata.org/gbd-results-tool. Accessed 15 Feb 2022.
- 10.Cooper GS, Stroehla BC. The epidemiology of autoimmune diseases. Autoimmun Rev. 2003;2:119–125. doi: 10.1016/S1568-9972(03)00006-5. [DOI] [PubMed] [Google Scholar]
- 11.van der Woude D, van der Helm-van Mil AHM. Update on the epidemiology, risk factors, and disease outcomes of rheumatoid arthritis. Best Pract Res Clin Rheumatol. 2018;32:174–187. doi: 10.1016/j.berh.2018.10.005. [DOI] [PubMed] [Google Scholar]
- 12.Aletaha D, Neogi T, Silman AJ, Funovits J, Felson DT, Bingham CO, 3rd, Birnbaum NS, Burmester GR, Bykerk VP, Cohen MD, et al. 2010 Rheumatoid arthritis classification criteria: an American College of Rheumatology/European League Against Rheumatism collaborative initiative. Arthritis Rheum. 2010;62:2569–2581. doi: 10.1002/art.27584. [DOI] [PubMed] [Google Scholar]
- 13.Bullock J, Rizvi SAA, Saleh AM, Ahmed SS, Do DP, Ansari RA, Ahmed J. Rheumatoid arthritis: a brief overview of the treatment. Med Princ Pract. 2018;27:501–507. doi: 10.1159/000493390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mathur S, Sutton J. Personalized medicine could transform healthcare. Biomed Rep. 2017;7:3–5. doi: 10.3892/br.2017.922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yu K-H, Beam AL, Kohane IS. Artificial intelligence in healthcare. Nat Biomed Eng. 2018;2:719–731. doi: 10.1038/s41551-018-0305-z. [DOI] [PubMed] [Google Scholar]
- 16.LeCun Y, Bengio Y, Hinton G. Deep learning. Nature. 2015;521:436–444. doi: 10.1038/nature14539. [DOI] [PubMed] [Google Scholar]
- 17.Meskó B, Görög M. A short guide for medical professionals in the era of artificial intelligence. npj Digit Med. 2020;3:126. doi: 10.1038/s41746-020-00333-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Iglesias LL, Bellón PS, del Barrio AP, Fernández-Miranda PM, González DR, Vega JA, Mandly AAG, Blanco JAP. A primer on deep learning and convolutional neural networks for clinicians. Insights Imaging. 2021;12:117. doi: 10.1186/s13244-021-01052-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Esteva A, Robicquet A, Ramsundar B, Kuleshov V, DePristo M, Chou K, Cui C, Corrado G, Thrun S, Dean J. A guide to deep learning in healthcare. Nat Med. 2019;25:24–29. doi: 10.1038/s41591-018-0316-z. [DOI] [PubMed] [Google Scholar]
- 20.Bluemke DA, Moy L, Bredella MA, Ertl-Wagner BB, Fowler KJ, Goh VJ, Halpern EF, Hess CP, Schiebler ML, Weiss CR. Assessing radiology research on artificial intelligence: a brief guide for authors, reviewers, and readers-from the radiology editorial board. Radiology. 2020;294:487–489. doi: 10.1148/radiol.2019192515. [DOI] [PubMed] [Google Scholar]
- 21.Liu Y, Chen PC, Krause J, Peng L. How to read articles that use machine learning: users' guides to the medical literature. JAMA. 2019;322:1806–1816. doi: 10.1001/jama.2019.16489. [DOI] [PubMed] [Google Scholar]
- 22.Mongan J, Moy L, Kahn CE. Checklist for artificial intelligence in medical imaging (CLAIM): a guide for authors and reviewers. Radiol Artif Intell. 2020;2:e200029. doi: 10.1148/ryai.2020200029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kohane IS, Aronow BJ, Avillach P, Beaulieu-Jones BK, Bellazzi R, Bradford RL, Brat GA, Cannataro M, Cimino JJ, Garcia-Barrio N, et al. What every reader should know about studies using electronic health record data but may be afraid to ask. J Med Internet Res. 2021;23:e22219. doi: 10.2196/22219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Scott I, Carter S, Coiera E. Clinician checklist for assessing suitability of machine learning applications in healthcare. BMJ Health Care Inf. 2021;28:e100251. doi: 10.1136/bmjhci-2020-100251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.O'Neil LJ, Spicer V, Smolik I, Meng X, Goel RR, Anaparti V, Wilkins J, El-Gabalawy HS. Association of a serum protein signature with rheumatoid arthritis development. Arthritis Rheumatol. 2021;73:78–88. doi: 10.1002/art.41483. [DOI] [PubMed] [Google Scholar]
- 26.Tanner S, Dufault B, Smolik I, Meng X, Anaparti V, Hitchon C, Robinson DB, Robinson W, Sokolove J, Lahey L, et al. A prospective study of the development of inflammatory arthritis in the family members of Indigenous North American people with rheumatoid arthritis. Arthritis Rheumatol. 2019;71:1494–1503. doi: 10.1002/art.40880. [DOI] [PubMed] [Google Scholar]
- 27.Kruppa J, Ziegler A, Konig IR. Risk estimation and risk prediction using machine-learning methods. Hum Genet. 2012;131:1639–1654. doi: 10.1007/s00439-012-1194-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Negi S, Juyal G, Senapati S, Prasad P, Gupta A, Singh S, Kashyap S, Kumar A, Kumar U, Gupta R, et al. A genome-wide association study reveals ARL15, a novel non-HLA susceptibility gene for rheumatoid arthritis in North Indians. Arthritis Rheum. 2013;65:3026–3035. doi: 10.1002/art.38110. [DOI] [PubMed] [Google Scholar]
- 29.Abbasifard M, Imani D, Bagheri-Hosseinabadi Z. PTPN22 gene polymorphism and susceptibility to rheumatoid arthritis (RA): Updated systematic review and meta-analysis. J Gene Med. 2020;22:e3204. doi: 10.1002/jgm.3204. [DOI] [PubMed] [Google Scholar]
- 30.Begovich AB, Carlton VE, Honigberg LA, Schrodi SJ, Chokkalingam AP, Alexander HC, Ardlie KG, Huang Q, Smith AM, Spoerke JM, et al. A missense single-nucleotide polymorphism in a gene encoding a protein tyrosine phosphatase (PTPN22) is associated with rheumatoid arthritis. Am J Hum Genet. 2004;75:330–337. doi: 10.1086/422827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Briggs FB, Ramsay PP, Madden E, Norris JM, Holers VM, Mikuls TR, Sokka T, Seldin MF, Gregersen PK, Criswell LA, Barcellos LF. Supervised machine learning and logistic regression identifies novel epistatic risk factors with PTPN22 for rheumatoid arthritis. Genes Immun. 2010;11:199–208. doi: 10.1038/gene.2009.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.González-Recio O, de Maturana EL, Vega AT, Engelman CD, Broman KW. Detecting single-nucleotide polymorphism by single-nucleotide polymorphism interactions in rheumatoid arthritis using a two-step approach with machine learning and a Bayesian threshold least absolute shrinkage and selection operator (LASSO) model. BMC Proc. 2009;3(Suppl 7):S63. doi: 10.1186/1753-6561-3-S7-S63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jin W, Yao Q, Liu Z, Cao W, Zhang Y, Che Z, Peng H. Do eye diseases increase the risk of arthritis in the elderly population? Aging (Albany NY) 2021;13:15580–15594. doi: 10.18632/aging.203122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gola D, Konig IR. Empowering individual trait prediction using interactions for precision medicine. BMC Bioinform. 2021;22:74. doi: 10.1186/s12859-021-04011-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chin CY, Hsieh SY, Tseng VS. eDRAM: Effective early disease risk assessment with matrix factorization on a large-scale medical database: a case study on rheumatoid arthritis. PLoS ONE. 2018;13:e0207579. doi: 10.1371/journal.pone.0207579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu C, Ackerman HH, Carulli JP. A genome-wide screen of gene-gene interactions for rheumatoid arthritis susceptibility. Hum Genet. 2011;129:473–485. doi: 10.1007/s00439-010-0943-z. [DOI] [PubMed] [Google Scholar]
- 37.van der Linden MP, le Cessie S, Raza K, van der Woude D, Knevel R, Huizinga TW, van der Helm-van Mil AH. Long-term impact of delay in assessment of patients with early arthritis. Arthritis Rheum. 2010;62:3537–3546. doi: 10.1002/art.27692. [DOI] [PubMed] [Google Scholar]
- 38.Kay J, Upchurch KS. ACR/EULAR 2010 rheumatoid arthritis classification criteria. Rheumatology (Oxford) 2012;51(Suppl 6):vi5–9. doi: 10.1093/rheumatology/kes279. [DOI] [PubMed] [Google Scholar]
- 39.Pecani A, Alessandri C, Spinelli FR, Priori R, Riccieri V, Di Franco M, Ceccarelli F, Colasanti T, Pendolino M, Mancini R, et al. Prevalence, sensitivity and specificity of antibodies against carbamylated proteins in a monocentric cohort of patients with rheumatoid arthritis and other autoimmune rheumatic diseases. Arthritis Res Ther. 2016;18:276. doi: 10.1186/s13075-016-1173-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Savvateeva E, Smoldovskaya O, Feyzkhanova G, Rubina A. Multiple biomarker approach for the diagnosis and therapy of rheumatoid arthritis. Crit Rev Clin Lab Sci. 2021;58:17–28. doi: 10.1080/10408363.2020.1775545. [DOI] [PubMed] [Google Scholar]
- 41.Song X, Lin Q. Genomics, transcriptomics and proteomics to elucidate the pathogenesis of rheumatoid arthritis. Rheumatol Int. 2017;37:1257–1265. doi: 10.1007/s00296-017-3732-3. [DOI] [PubMed] [Google Scholar]
- 42.Lin E, Lane H-Y. Machine learning and systems genomics approaches for multi-omics data. Biomark Res. 2017;5:2. doi: 10.1186/s40364-017-0082-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tins BJ, Butler R. Imaging in rheumatology: reconciling radiology and rheumatology. Insights Imaging. 2013;4:799–810. doi: 10.1007/s13244-013-0293-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liu J, Chen N. A 9 mRNAs-based diagnostic signature for rheumatoid arthritis by integrating bioinformatic analysis and machine-learning. J Orthop Surg Res. 2021;16:44. doi: 10.1186/s13018-020-02180-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pratt AG, Swan DC, Richardson S, Wilson G, Hilkens CM, Young DA, Isaacs JD. A CD4 T cell gene signature for early rheumatoid arthritis implicates interleukin 6-mediated STAT3 signalling, particularly in anti-citrullinated peptide antibody-negative disease. Ann Rheum Dis. 2012;71:1374–1381. doi: 10.1136/annrheumdis-2011-200968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.van der Helm-van Mil AH, Detert J, le Cessie S, Filer A, Bastian H, Burmester GR, Huizinga TW, Raza K. Validation of a prediction rule for disease outcome in patients with recent-onset undifferentiated arthritis: moving toward individualized treatment decision-making. Arthritis Rheum. 2008;58:2241–2247. doi: 10.1002/art.23681. [DOI] [PubMed] [Google Scholar]
- 47.Wang J, Yan S, Yang J, Lu H, Xu D, Wang Z. Non-coding RNAs in rheumatoid arthritis: from bench to bedside. Front Immunol. 2019;10:3129. doi: 10.3389/fimmu.2019.03129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ormseth MJ, Solus JF, Sheng Q, Ye F, Wu Q, Guo Y, Oeser AM, Allen RM, Vickers KC, Stein CM. Development and validation of a MicroRNA panel to differentiate between patients with rheumatoid arthritis or systemic lupus erythematosus and controls. J Rheumatol. 2020;47:188–196. doi: 10.3899/jrheum.181029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Geurts P, Fillet M, de Seny D, Meuwis MA, Malaise M, Merville MP, Wehenkel L. Proteomic mass spectra classification using decision-tree based ensemble methods. Bioinformatics. 2005;21:3138–3145. doi: 10.1093/bioinformatics/bti494. [DOI] [PubMed] [Google Scholar]
- 50.Niu Q, Huang Z, Shi Y, Wang L, Pan X, Hu C. Specific serum protein biomarkers of rheumatoid arthritis detected by MALDI-TOF-MS combined with magnetic beads. Int Immunol. 2010;22:611–618. doi: 10.1093/intimm/dxq043. [DOI] [PubMed] [Google Scholar]
- 51.de Seny D, Fillet M, Meuwis MA, Geurts P, Lutteri L, Ribbens C, Bours V, Wehenkel L, Piette J, Malaise M, Merville MP. Discovery of new rheumatoid arthritis biomarkers using the surface-enhanced laser desorption/ionization time-of-flight mass spectrometry ProteinChip approach. Arthritis Rheum. 2005;52:3801–3812. doi: 10.1002/art.21607. [DOI] [PubMed] [Google Scholar]
- 52.Heard BJ, Rosvold JM, Fritzler MJ, El-Gabalawy H, Wiley JP, Krawetz RJ. A computational method to differentiate normal individuals, osteoarthritis and rheumatoid arthritis patients using serum biomarkers. J R Soc Interface. 2014;11:20140428. doi: 10.1098/rsif.2014.0428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tsai KL, Chang CC, Chang YS, Lu YY, Tsai IJ, Chen JH, Lin SH, Tai CC, Lin YF, Chang HW, et al. Isotypes of autoantibodies against novel differential 4-hydroxy-2-nonenal-modified peptide adducts in serum is associated with rheumatoid arthritis in Taiwanese women. BMC Med Inform Decis Mak. 2021;21:49. doi: 10.1186/s12911-020-01380-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ahmed U, Anwar A, Savage RS, Thornalley PJ, Rabbani N. Protein oxidation, nitration and glycation biomarkers for early-stage diagnosis of osteoarthritis of the knee and typing and progression of arthritic disease. Arthritis Res Ther. 2016;18:250. doi: 10.1186/s13075-016-1154-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chocholova E, Bertok T, Jane E, Lorencova L, Holazova A, Belicka L, Belicky S, Mislovicova D, Vikartovska A, Imrich R, et al. Glycomics meets artificial intelligence—potential of glycan analysis for identification of seropositive and seronegative rheumatoid arthritis patients revealed. Clin Chim Acta. 2018;481:49–55. doi: 10.1016/j.cca.2018.02.031. [DOI] [PubMed] [Google Scholar]
- 56.Orr C, Vieira-Sousa E, Boyle DL, Buch MH, Buckley CD, Cañete JD, Catrina AI, Choy EHS, Emery P, Fearon U, et al. Synovial tissue research: a state-of-the-art review. Nat Rev Rheumatol. 2017;13:463–475. doi: 10.1038/nrrheum.2017.115. [DOI] [PubMed] [Google Scholar]
- 57.Long NP, Park S, Anh NH, Min JE, Yoon SJ, Kim HM, Nghi TD, Lim DK, Park JH, Lim J, Kwon SW. Efficacy of integrating a novel 16-gene biomarker panel and intelligence classifiers for differential diagnosis of rheumatoid arthritis and osteoarthritis. J Clin Med. 2019;8:50. doi: 10.3390/jcm8010050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yeo L, Adlard N, Biehl M, Juarez M, Smallie T, Snow M, Buckley CD, Raza K, Filer A, Scheel-Toellner D. Expression of chemokines CXCL4 and CXCL7 by synovial macrophages defines an early stage of rheumatoid arthritis. Ann Rheum Dis. 2016;75:763–771. doi: 10.1136/annrheumdis-2014-206921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Orange DE, Agius P, DiCarlo EF, Robine N, Geiger H, Szymonifka J, McNamara M, Cummings R, Andersen KM, Mirza S, et al. Identification of three rheumatoid arthritis disease subtypes by machine learning integration of synovial histologic features and RNA sequencing data. Arthritis Rheumatol. 2018;70:690–701. doi: 10.1002/art.40428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Marcos-Zambrano LJ, Karaduzovic-Hadziabdic K, Loncar Turukalo T, Przymus P, Trajkovik V, Aasmets O, Berland M, Gruca A, Hasic J, Hron K, et al. Applications of machine learning in human microbiome studies: a review on feature selection, biomarker identification, disease prediction and treatment. Front Microbiol. 2021;12:634511. doi: 10.3389/fmicb.2021.634511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wu H, Cai L, Li D, Wang X, Zhao S, Zou F, Zhou K. Metagenomics biomarkers selected for prediction of three different diseases in Chinese population. Biomed Res Int. 2018;2018:2936257. doi: 10.1155/2018/2936257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Volkova A, Ruggles KV. Predictive metagenomic analysis of autoimmune disease identifies robust autoimmunity and disease specific microbial signatures. Front Microbiol. 2021;12:621310. doi: 10.3389/fmicb.2021.621310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bellando-Randone S, Russo E, Venerito V, Matucci-Cerinic M, Iannone F, Tangaro S, Amedei A. Exploring the oral microbiome in rheumatic diseases, state of art and future prospective in personalized medicine with an AI approach. J Pers Med. 2021;11:625. doi: 10.3390/jpm11070625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Jung SM, Park KS, Kim KJ. Deep phenotyping of synovial molecular signatures by integrative systems analysis in rheumatoid arthritis. Rheumatology (Oxford) 2021;60:3420–3431. doi: 10.1093/rheumatology/keaa751. [DOI] [PubMed] [Google Scholar]
- 65.Xiao J, Wang R, Cai X, Ye Z. Coupling of co-expression network analysis and machine learning validation unearthed potential key genes involved in rheumatoid arthritis. Front Genet. 2021;12:604714. doi: 10.3389/fgene.2021.604714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sommer OJ, Kladosek A, Weiler V, Czembirek H, Boeck M, Stiskal M. Rheumatoid arthritis: a practical guide to state-of-the-art imaging, image interpretation, and clinical implications. Radiographics. 2005;25:381–398. doi: 10.1148/rg.252045111. [DOI] [PubMed] [Google Scholar]
- 67.Mate GS, Kureshi AK, Singh BK. An efficient CNN for hand X-ray classification of rheumatoid arthritis. J Healthc Eng. 2021;2021:6712785. doi: 10.1155/2021/6712785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ureten K, Erbay H, Maras HH. Detection of rheumatoid arthritis from hand radiographs using a convolutional neural network. Clin Rheumatol. 2020;39:969–974. doi: 10.1007/s10067-019-04487-4. [DOI] [PubMed] [Google Scholar]
- 69.Scheel AK, Krause A, Rheinbaben IM, Metzger G, Rost H, Tresp V, Mayer P, Reuss-Borst M, Müller GA. Assessment of proximal finger joint inflammation in patients with rheumatoid arthritis, using a novel laser-based imaging technique. Arthritis Rheum. 2002;46:1177–1184. doi: 10.1002/art.10226. [DOI] [PubMed] [Google Scholar]
- 70.Cupek R, Ziębiński A. Automated assessment of joint synovitis activity from medical ultrasound and power doppler examinations using image processing and machine learning methods. Reumatologia. 2016;54:239–242. doi: 10.5114/reum.2016.63664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Tripoliti EE, Fotiadis DI, Argyropoulou M. Automated segmentation and quantification of inflammatory tissue of the hand in rheumatoid arthritis patients using magnetic resonance imaging data. Artif Intell Med. 2007;40:65–85. doi: 10.1016/j.artmed.2007.02.003. [DOI] [PubMed] [Google Scholar]
- 72.Topfer D, Finzel S, Museyko O, Schett G, Engelke K. Segmentation and quantification of bone erosions in high-resolution peripheral quantitative computed tomography datasets of the metacarpophalangeal joints of patients with rheumatoid arthritis. Rheumatology (Oxford) 2014;53:65–71. doi: 10.1093/rheumatology/ket259. [DOI] [PubMed] [Google Scholar]
- 73.Murakami S, Hatano K, Tan J, Kim H, Aoki T. Automatic identification of bone erosions in rheumatoid arthritis from hand radiographs based on deep convolutional neural network. Multimed Tools Appl. 2018;77:10921–10937. doi: 10.1007/s11042-017-5449-4. [DOI] [Google Scholar]
- 74.Aizenberg E, Roex EAH, Nieuwenhuis WP, Mangnus L, van der Helm-van Mil AHM, Reijnierse M, Bloem JL, Lelieveldt BPF, Stoel BC. Automatic quantification of bone marrow edema on MRI of the wrist in patients with early arthritis: a feasibility study. Magn Reson Med. 2018;79:1127–1134. doi: 10.1002/mrm.26712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Langs G, Peloschek P, Bischof H, Kainberger F. Automatic quantification of joint space narrowing and erosions in rheumatoid arthritis. IEEE Trans Med Imaging. 2009;28:151–164. doi: 10.1109/TMI.2008.2004401. [DOI] [PubMed] [Google Scholar]
- 76.Czaplicka K, Wojciechowski W, Włodarczyk J, Urbanik A, Tabor Z. Automated assessment of synovitis in 0.2T magnetic resonance images of the wrist. Comput Biol Med. 2015;67:116–125. doi: 10.1016/j.compbiomed.2015.10.009. [DOI] [PubMed] [Google Scholar]
- 77.Boesen M, Kubassova O, Bouert R, Axelsen MB, Ostergaard M, Cimmino MA, Danneskiold-Samsoe B, Horslev-Petersen K, Bliddal H. Correlation between computer-aided dynamic gadolinium-enhanced MRI assessment of inflammation and semi-quantitative synovitis and bone marrow oedema scores of the wrist in patients with rheumatoid arthritis–a cohort study. Rheumatology (Oxford) 2012;51:134–143. doi: 10.1093/rheumatology/ker220. [DOI] [PubMed] [Google Scholar]
- 78.Wu M, Wu H, Wu L, Cui C, Shi S, Xu J, Liu Y, Dong F. A deep learning classification of metacarpophalangeal joints synovial proliferation in rheumatoid arthritis by ultrasound images. J Clin Ultrasound. 2022;50:296–301. doi: 10.1002/jcu.23143. [DOI] [PubMed] [Google Scholar]
- 79.Andersen JKH, Pedersen JS, Laursen MS, Holtz K, Grauslund J, Savarimuthu TR, Just SA. Neural networks for automatic scoring of arthritis disease activity on ultrasound images. RMD Open. 2019;5:e000891. doi: 10.1136/rmdopen-2018-000891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Hirano T, Nishide M, Nonaka N, Seita J, Ebina K, Sakurada K, Kumanogoh A. Development and validation of a deep-learning model for scoring of radiographic finger joint destruction in rheumatoid arthritis. Rheumatol Adv Pract. 2019;3:rkz047. doi: 10.1093/rap/rkz047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Rohrbach J, Reinhard T, Sick B, Dürr O. Bone erosion scoring for rheumatoid arthritis with deep convolutional neural networks. Comput Electr Eng. 2019;78:472–481. doi: 10.1016/j.compeleceng.2019.08.003. [DOI] [Google Scholar]
- 82.Jintao R, Arash Moaddel H, Ellen MH, Kresten KK, Rasmus KJ, François L. Automatic detection and localization of bone erosion in hand HR-pQCT. In: ProcSPIE. vol 10950. Medical Imaging 2019: Computer-Aided Diagnosis, SPIE; 2019. p. 1095022.
- 83.Put S, Westhovens R, Lahoutte T, Matthys P. Molecular imaging of rheumatoid arthritis: emerging markers, tools, and techniques. Arthritis Res Ther. 2014;16:208. doi: 10.1186/ar4542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Reed M, Le Souef T, Rampono E. A pilot study of a machine-learning tool to assist in the diagnosis of hand arthritis. Intern Med J. 2022;52(6):959–967. doi: 10.1111/imj.15173. [DOI] [PubMed] [Google Scholar]
- 85.Alarcon-Paredes A, Guzman-Guzman IP, Hernandez-Rosales DE, Navarro-Zarza JE, Cantillo-Negrete J, Cuevas-Valencia RE, Alonso GA. Computer-aided diagnosis based on hand thermal, RGB images, and grip force using artificial intelligence as screening tool for rheumatoid arthritis in women. Med Biol Eng Comput. 2021;59:287–300. doi: 10.1007/s11517-020-02294-7. [DOI] [PubMed] [Google Scholar]
- 86.Wyns B, Sette S, Boullart L, Baeten D, Hoffman IE, De Keyser F. Prediction of diagnosis in patients with early arthritis using a combined Kohonen mapping and instance-based evaluation criterion. Artif Intell Med. 2004;31:45–55. doi: 10.1016/j.artmed.2004.01.002. [DOI] [PubMed] [Google Scholar]
- 87.Singh S, Kumar A, Panneerselvam K, Vennila JJ. Diagnosis of arthritis through fuzzy inference system. J Med Syst. 2012;36:1459–1468. doi: 10.1007/s10916-010-9606-9. [DOI] [PubMed] [Google Scholar]
- 88.Fukae J, Isobe M, Hattori T, Fujieda Y, Kono M, Abe N, Kitano A, Narita A, Henmi M, Sakamoto F, et al. Convolutional neural network for classification of two-dimensional array images generated from clinical information may support diagnosis of rheumatoid arthritis. Sci Rep. 2020;10:5648. doi: 10.1038/s41598-020-62634-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Snekhalatha U, Anburajan M, Sowmiya V, Venkatraman B, Menaka M. Automated hand thermal image segmentation and feature extraction in the evaluation of rheumatoid arthritis. Proc Inst Mech Eng H. 2015;229:319–331. doi: 10.1177/0954411915580809. [DOI] [PubMed] [Google Scholar]
- 90.Sharon H, Elamvazuthi I, Lu CK, Parasuraman S, Natarajan E: Development of Rheumatoid Arthritis Classification from Electronic Image Sensor Using Ensemble Method. Sensors (Basel) 2019, 20. [DOI] [PMC free article] [PubMed]
- 91.Bardhan S, Bhowmik MK. 2-Stage classification of knee joint thermograms for rheumatoid arthritis prediction in subclinical inflammation. Australas Phys Eng Sci Med. 2019;42:259–277. doi: 10.1007/s13246-019-00726-9. [DOI] [PubMed] [Google Scholar]
- 92.Pauk J, Wasilewska A, Ihnatouski M. Infrared thermography sensor for disease activity detection in rheumatoid arthritis patients. Sensors (Basel) 2019;19:3444. doi: 10.3390/s19163444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Shivade C, Raghavan P, Fosler-Lussier E, Embi PJ, Elhadad N, Johnson SB, Lai AM. A review of approaches to identifying patient phenotype cohorts using electronic health records. J Am Med Inform Assoc. 2014;21:221–230. doi: 10.1136/amiajnl-2013-001935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Banda JM, Seneviratne M, Hernandez-Boussard T, Shah NH. Advances in electronic phenotyping: from rule-based definitions to machine learning models. Annu Rev Biomed Data Sci. 2018;1:53–68. doi: 10.1146/annurev-biodatasci-080917-013315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Cai T, Cai F, Dahal KP, Cremone G, Lam E, Golnik C, Seyok T, Hong C, Cai T, Liao KP. Improving the efficiency of clinical trial recruitment using an ensemble machine learning to assist with eligibility screening. ACR Open Rheumatol. 2021. [DOI] [PMC free article] [PubMed]
- 96.Fernandez-Gutierrez F, Kennedy JI, Cooksey R, Atkinson M, Choy E, Brophy S, Huo L, Zhou SM. Mining primary care electronic health records for automatic disease phenotyping: a transparent machine learning framework. Diagnostics (Basel) 2021;11:1908. doi: 10.3390/diagnostics11101908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Ferte T, Cossin S, Schaeverbeke T, Barnetche T, Jouhet V, Hejblum BP. Automatic phenotyping of electronical health record: PheVis algorithm. J Biomed Inform. 2021;117:103746. doi: 10.1016/j.jbi.2021.103746. [DOI] [PubMed] [Google Scholar]
- 98.Maarseveen TD, Maurits MP, Niemantsverdriet E, van der Helm-van Mil AHM, Huizinga TWJ, Knevel R. Handwork vs. machine: a comparison of rheumatoid arthritis patient populations as identified from EHR free-text by diagnosis extraction through machine-learning or traditional criteria-based chart review. Arthritis Res Ther. 2021;23:174. doi: 10.1186/s13075-021-02553-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Maarseveen TD, Meinderink T, Reinders MJT, Knitza J, Huizinga TWJ, Kleyer A, Simon D, van den Akker EB, Knevel R. Machine learning electronic health record identification of patients with rheumatoid arthritis: algorithm pipeline development and validation study. JMIR Med Inf. 2020;8:e23930. doi: 10.2196/23930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Huang S, Huang J, Cai T, Dahal KP, Cagan A, He Z, Stratton J, Gorelik I, Hong C, Cai T, Liao KP. Impact of ICD10 and secular changes on electronic medical record rheumatoid arthritis algorithms. Rheumatology (Oxford) 2020;59:3759–3766. doi: 10.1093/rheumatology/keaa198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Ning W, Chan S, Beam A, Yu M, Geva A, Liao K, Mullen M, Mandl KD, Kohane I, Cai T, Yu S. Feature extraction for phenotyping from semantic and knowledge resources. J Biomed Inf. 2019;91:103122. doi: 10.1016/j.jbi.2019.103122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Yu S, Ma Y, Gronsbell J, Cai T, Ananthakrishnan AN, Gainer VS, Churchill SE, Szolovits P, Murphy SN, Kohane IS, et al. Enabling phenotypic Big Data with PheNorm. J Am Med Inf Assoc. 2018;25:54–60. doi: 10.1093/jamia/ocx111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Gronsbell J, Minnier J, Yu S, Liao K, Cai T. Automated feature selection of predictors in electronic medical records data. Biometrics. 2019;75:268–277. doi: 10.1111/biom.12987. [DOI] [PubMed] [Google Scholar]
- 104.Gronsbell JL, Cai T. Semi-supervised approaches to efficient evaluation of model prediction performance. J R Stat Soc Ser B (Statistical Methodology) 2018;80:579–594. doi: 10.1111/rssb.12264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Zhou SM, Fernandez-Gutierrez F, Kennedy J, Cooksey R, Atkinson M, Denaxas S, Siebert S, Dixon WG, O'Neill TW, Choy E, et al. Defining disease phenotypes in primary care electronic health records by a machine learning approach: a case study in identifying rheumatoid arthritis. PLoS ONE. 2016;11:e0154515. doi: 10.1371/journal.pone.0154515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Lin C, Karlson EW, Dligach D, Ramirez MP, Miller TA, Mo H, Braggs NS, Cagan A, Gainer V, Denny JC, Savova GK. Automatic identification of methotrexate-induced liver toxicity in patients with rheumatoid arthritis from the electronic medical record. J Am Med Inform Assoc. 2015;22:e151–161. doi: 10.1136/amiajnl-2014-002642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Chen Y, Carroll RJ, Hinz ER, Shah A, Eyler AE, Denny JC, Xu H. Applying active learning to high-throughput phenotyping algorithms for electronic health records data. J Am Med Inform Assoc. 2013;20:e253–259. doi: 10.1136/amiajnl-2013-001945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Carroll RJ, Eyler AE, Denny JC. Naïve electronic health record phenotype identification for rheumatoid arthritis. AMIA Annu Symp Proc. 2011;2011:189–196. [PMC free article] [PubMed] [Google Scholar]
- 109.Liao KP, Cai T, Gainer V, Goryachev S, Zeng-treitler Q, Raychaudhuri S, Szolovits P, Churchill S, Murphy S, Kohane I, et al. Electronic medical records for discovery research in rheumatoid arthritis. Arthritis Care Res (Hoboken) 2010;62:1120–1127. doi: 10.1002/acr.20184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Blaiss MS, Hammerby E, Robinson S, Kennedy-Martin T, Buchs S. The burden of allergic rhinitis and allergic rhinoconjunctivitis on adolescents: a literature review. Ann Allergy Asthma Immunol. 2018;121:43–52.e43. doi: 10.1016/j.anai.2018.03.028. [DOI] [PubMed] [Google Scholar]
- 111.Yang Z, Dehmer M, Yli-Harja O, Emmert-Streib F. Combining deep learning with token selection for patient phenotyping from electronic health records. Sci Rep. 2020;10:1432. doi: 10.1038/s41598-020-58178-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Gehrmann S, Dernoncourt F, Li Y, Carlson ET, Wu JT, Welt J, Foote J, Jr, Moseley ET, Grant DW, Tyler PD, Celi LA. Comparing deep learning and concept extraction-based methods for patient phenotyping from clinical narratives. PLoS ONE. 2018;13:e0192360. doi: 10.1371/journal.pone.0192360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Stafford IS, Kellermann M, Mossotto E, Beattie RM, MacArthur BD, Ennis S. A systematic review of the applications of artificial intelligence and machine learning in autoimmune diseases. npj Digit Med. 2020;3:30. doi: 10.1038/s41746-020-0229-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Artacho A, Isaac S, Nayak R, Flor-Duro A, Alexander M, Koo I, Manasson J, Smith PB, Rosenthal P, Homsi Y, et al. The pretreatment gut microbiome is associated with lack of response to methotrexate in new-onset rheumatoid arthritis. Arthritis Rheumatol. 2021;73:931–942. doi: 10.1002/art.41622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Maciejewski M, Sands C, Nair N, Ling S, Verstappen S, Hyrich K, Barton A, Ziemek D, Lewis MR, Plant D. Prediction of response of methotrexate in patients with rheumatoid arthritis using serum lipidomics. Sci Rep. 2021;11:7266. doi: 10.1038/s41598-021-86729-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Amin Shipa MR, Yeoh SA, Embleton-Thirsk A, Mukerjee D, Ehrenstein MR. The synergistic efficacy of hydroxychloroquine with methotrexate is accompanied by increased erythrocyte mean corpuscular volume. Rheumatology (Oxford). 2022;61(2):787–793. doi: 10.1093/rheumatology/keab403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Westerlind H, Maciejewski M, Frisell T, Jelinsky SA, Ziemek D, Askling J. What is the persistence to methotrexate in rheumatoid arthritis, and does machine learning outperform hypothesis-based approaches to its prediction? ACR Open Rheumatol. 2021;3:457–463. doi: 10.1002/acr2.11266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Morid MA, Lau M, Del Fiol G. Predictive analytics for step-up therapy: supervised or semi-supervised learning? J Biomed Inform. 2021;119:103842. doi: 10.1016/j.jbi.2021.103842. [DOI] [PubMed] [Google Scholar]
- 119.Messelink MA, Roodenrijs NMT, van Es B, Hulsbergen-Veelken CAR, Jong S, Overmars LM, Reteig LC, Tan SC, Tauber T, van Laar JM, et al. Identification and prediction of difficult-to-treat rheumatoid arthritis patients in structured and unstructured routine care data: results from a hackathon. Arthritis Res Ther. 2021;23:184. doi: 10.1186/s13075-021-02560-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Plant D, Maciejewski M, Smith S, Nair N, Hyrich K, Ziemek D, Barton A, Verstappen S, Maximising Therapeutic Utility in Rheumatoid Arthritis Consortium tRSG Profiling of gene expression biomarkers as a classifier of methotrexate nonresponse in patients with rheumatoid arthritis. Arthritis Rheumatol. 2019;71:678–684. doi: 10.1002/art.40810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Tao W, Concepcion AN, Vianen M, Marijnissen ACA, Lafeber F, Radstake T, Pandit A. Multiomics and machine learning accurately predict clinical response to adalimumab and etanercept therapy in patients with rheumatoid arthritis. Arthritis Rheumatol. 2021;73:212–222. doi: 10.1002/art.41516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Kim KJ, Kim M, Adamopoulos IE, Tagkopoulos I. Compendium of synovial signatures identifies pathologic characteristics for predicting treatment response in rheumatoid arthritis patients. Clin Immunol. 2019;202:1–10. doi: 10.1016/j.clim.2019.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Guan Y, Zhang H, Quang D, Wang Z, Parker SCJ, Pappas DA, Kremer JM, Zhu F. Machine learning to predict anti-tumor necrosis factor drug responses of rheumatoid arthritis patients by integrating clinical and genetic markers. Arthritis Rheumatol. 2019;71:1987–1996. doi: 10.1002/art.41056. [DOI] [PubMed] [Google Scholar]
- 124.Yoosuf N, Maciejewski M, Ziemek D, Jelinsky SA, Folkersen L, Muller M, Sahlstrom P, Vivar N, Catrina A, Berg L, et al. Early Prediction of clinical response to anti-TNF treatment using multi-omics and machine learning in rheumatoid arthritis. Rheumatology (Oxford). 2022;61(4):1680–1689. doi: 10.1093/rheumatology/keab521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Gosselt HR, Verhoeven MMA, Bulatovic-Calasan M, Welsing PM, de Rotte M, Hazes JMW, Lafeber F, Hoogendoorn M, de Jonge R. Complex machine-learning algorithms and multivariable logistic regression on par in the prediction of insufficient clinical response to methotrexate in rheumatoid arthritis. J Pers Med. 2021;11:44. doi: 10.3390/jpm11010044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Luque-Tevar M, Perez-Sanchez C, Patino-Trives AM, Barbarroja N, Arias de la Rosa I, Abalos-Aguilera MC, Marin-Sanz JA, Ruiz-Vilchez D, Ortega-Castro R, Font P, et al. Integrative clinical, molecular, and computational analysis identify novel biomarkers and differential profiles of anti-TNF response in rheumatoid arthritis. Front Immunol. 2021;12:631662. doi: 10.3389/fimmu.2021.631662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Kato M, Ikeda K, Sugiyama T, Tanaka S, Iida K, Suga K, Nishimura N, Mimura N, Kasuya T, Kumagai T, et al. Associations of ultrasound-based inflammation patterns with peripheral innate lymphoid cell populations, serum cytokines/chemokines, and treatment response to methotrexate in rheumatoid arthritis and spondyloarthritis. PLoS ONE. 2021;16:e0252116. doi: 10.1371/journal.pone.0252116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Fransen J, van Riel PL. The Disease Activity Score and the EULAR response criteria. Clin Exp Rheumatol. 2005;23:S93–99. [PubMed] [Google Scholar]
- 129.Looy SV, Cruyssen BV, Meeus J, Wyns B, Westhovens R, Durez P, Bosch FVd, Vastesaeger N, Geldhof A, Boullart L, Keyser FD. Prediction of dose escalation for rheumatoid arthritis patients under infliximab treatment. Eng Appl Artif Intell. 2006;19:819–828. doi: 10.1016/j.engappai.2006.05.001. [DOI] [Google Scholar]
- 130.Parida JR, Misra DP, Wakhlu A, Agarwal V. Is non-biological treatment of rheumatoid arthritis as good as biologics? World J Orthop. 2015;6:278–283. doi: 10.5312/wjo.v6.i2.278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Lim AJW, Lim LJ, Ooi BNS, Koh ET, Tan JWL, Group TRS, Chong SS, Khor CC, Tucker-Kellogg L, Leong KP, Lee CG. Functional coding haplotypes and machine-learning feature elimination identifies predictors of methotrexate response in rheumatoid arthritis patients. EBioMedicine. 2022;75:103800. doi: 10.1016/j.ebiom.2021.103800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Koo BS, Eun S, Shin K, Yoon H, Hong C, Kim DH, Hong S, Kim YG, Lee CK, Yoo B, Oh JS. Machine learning model for identifying important clinical features for predicting remission in patients with rheumatoid arthritis treated with biologics. Arthritis Res Ther. 2021;23:178. doi: 10.1186/s13075-021-02567-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Gomez EA, Colas RA, Souza PR, Hands R, Lewis MJ, Bessant C, Pitzalis C, Dalli J. Blood pro-resolving mediators are linked with synovial pathology and are predictive of DMARD responsiveness in rheumatoid arthritis. Nat Commun. 2020;11:5420. doi: 10.1038/s41467-020-19176-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Miyoshi F, Honne K, Minota S, Okada M, Ogawa N, Mimura T. A novel method predicting clinical response using only background clinical data in RA patients before treatment with infliximab. Mod Rheumatol. 2016;26:813–816. doi: 10.3109/14397595.2016.1168536. [DOI] [PubMed] [Google Scholar]
- 135.Prevoo ML. van 't Hof MA, Kuper HH, van Leeuwen MA, van de Putte LB, van Riel PL: Modified disease activity scores that include twenty-eight-joint counts. Development and validation in a prospective longitudinal study of patients with rheumatoid arthritis. Arthritis Rheum. 1995;38:44–48. doi: 10.1002/art.1780380107. [DOI] [PubMed] [Google Scholar]
- 136.Anderson J, Caplan L, Yazdany J, Robbins ML, Neogi T, Michaud K, Saag KG, O'Dell JR, Kazi S. Rheumatoid arthritis disease activity measures: American College of Rheumatology recommendations for use in clinical practice. Arthritis Care Res (Hoboken) 2012;64:640–647. doi: 10.1002/acr.21649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Kalweit M, Walker UA, Finckh A, Muller R, Kalweit G, Scherer A, Boedecker J, Hugle T. Personalized prediction of disease activity in patients with rheumatoid arthritis using an adaptive deep neural network. PLoS ONE. 2021;16:e0252289. doi: 10.1371/journal.pone.0252289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Rychkov D, Neely J, Oskotsky T, Yu S, Perlmutter N, Nititham J, Carvidi A, Krueger M, Gross A, Criswell LA, et al. Cross-tissue transcriptomic analysis leveraging machine learning approaches identifies new biomarkers for rheumatoid arthritis. Front Immunol. 2021;12:638066. doi: 10.3389/fimmu.2021.638066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Aletaha D, Smolen J. The Simplified Disease Activity Index (SDAI) and the Clinical Disease Activity Index (CDAI): a review of their usefulness and validity in rheumatoid arthritis. Clin Exp Rheumatol. 2005;23:S100–108. [PubMed] [Google Scholar]
- 140.Norgeot B, Glicksberg BS, Trupin L, Lituiev D, Gianfrancesco M, Oskotsky B, Schmajuk G, Yazdany J, Butte AJ. Assessment of a deep learning model based on electronic health record data to forecast clinical outcomes in patients with rheumatoid arthritis. JAMA Netw Open. 2019;2:e190606. doi: 10.1001/jamanetworkopen.2019.0606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Solomon DH, Xu C, Collins J, Kim SC, Losina E, Yau V, Johansson FD. The sequence of disease-modifying anti-rheumatic drugs: pathways to and predictors of tocilizumab monotherapy. Arthritis Res Ther. 2021;23:26. doi: 10.1186/s13075-020-02408-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Chauhan K, Jandu JS, Goyal A, Bansal P, Al-Dhahir MA. Rheumatoid arthritis. Treasure Island: StatPearls; 2022. [Google Scholar]
- 143.Kim JW, Suh CH. Systemic Manifestations and Complications in Patients with Rheumatoid Arthritis. J Clin Med. 2020;9:2008. doi: 10.3390/jcm9062008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Dougados M, Soubrier M, Antunez A, Balint P, Balsa A, Buch MH, Casado G, Detert J, El-Zorkany B, Emery P, et al. Prevalence of comorbidities in rheumatoid arthritis and evaluation of their monitoring: results of an international, cross-sectional study (COMORA) Ann Rheum Dis. 2014;73:62–68. doi: 10.1136/annrheumdis-2013-204223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Khanna NN, Jamthikar AD, Gupta D, Piga M, Saba L, Carcassi C, Giannopoulos AA, Nicolaides A, Laird JR, Suri HS, et al. Rheumatoid arthritis: atherosclerosis imaging and cardiovascular risk assessment using machine and deep learning-based tissue characterization. Curr Atheroscler Rep. 2019;21:7. doi: 10.1007/s11883-019-0766-x. [DOI] [PubMed] [Google Scholar]
- 146.Wei T, Yang B, Liu H, Xin F, Fu L. Development and validation of a nomogram to predict coronary heart disease in patients with rheumatoid arthritis in northern China. Aging (Albany NY) 2020;12:3190–3204. doi: 10.18632/aging.102823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Xin F, Fu L, Yang B, Liu H, Wei T, Zou C, Bai B. Development and validation of a nomogram for predicting stroke risk in rheumatoid arthritis patients. Aging (Albany NY) 2021;13:15061–15077. doi: 10.18632/aging.203071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Konstantonis G, Singh KV, Sfikakis PP, Jamthikar AD, Kitas GD, Gupta SK, Saba L, Verrou K, Khanna NN, Ruzsa Z, et al. Cardiovascular disease detection using machine learning and carotid/femoral arterial imaging frameworks in rheumatoid arthritis patients. Rheumatol Int. 2022;42:215–239. doi: 10.1007/s00296-021-05062-4. [DOI] [PubMed] [Google Scholar]
- 149.Hu Z, Zhang L, Lin Z, Zhao C, Xu S, Lin H, Zhang J, Li W, Chu Y. Prevalence and risk factors for bone loss in rheumatoid arthritis patients from South China: modeled by three methods. BMC Musculoskelet Disord. 2021;22:534. doi: 10.1186/s12891-021-04403-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Smuck M, Odonkor CA, Wilt JK, Schmidt N, Swiernik MA. The emerging clinical role of wearables: factors for successful implementation in healthcare. NPJ Digit Med. 2021;4:45. doi: 10.1038/s41746-021-00418-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Ravalli S, Roggio F, Lauretta G, Di Rosa M, D'Amico AG, D'Agata V, Maugeri G, Musumeci G. Exploiting real-world data to monitor physical activity in patients with osteoarthritis: the opportunity of digital epidemiology. Heliyon. 2022;8:e08991. doi: 10.1016/j.heliyon.2022.e08991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Teixeira E, Fonseca H, Diniz-Sousa F, Veras L, Boppre G, Oliveira J, Pinto D, Alves AJ, Barbosa A, Mendes R, Marques-Aleixo I. Wearable devices for physical activity and healthcare monitoring in elderly people: a critical review. Geriatrics (Basel) 2021;6:38. doi: 10.3390/geriatrics6020038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Hernandez-Hernandez V, Ferraz-Amaro I, Diaz-Gonzalez F. Influence of disease activity on the physical activity of rheumatoid arthritis patients. Rheumatology (Oxford) 2014;53:722–731. doi: 10.1093/rheumatology/ket422. [DOI] [PubMed] [Google Scholar]
- 154.Brophy S, Cooksey R, Davies H, Dennis MS, Zhou SM, Siebert S. The effect of physical activity and motivation on function in ankylosing spondylitis: a cohort study. Semin Arthritis Rheum. 2013;42:619–626. doi: 10.1016/j.semarthrit.2012.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Markusse IM, Dirven L, Gerards AH, van Groenendael JH, Ronday HK, Kerstens PJ, Lems WF, Huizinga TW, Allaart CF. Disease flares in rheumatoid arthritis are associated with joint damage progression and disability: 10-year results from the BeSt study. Arthritis Res Ther. 2015;17:232. doi: 10.1186/s13075-015-0730-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Bechman K, Tweehuysen L, Garrood T, Scott DL, Cope AP, Galloway JB, Ma MHY. Flares in rheumatoid arthritis patients with low disease activity: predictability and association with worse clinical outcomes. J Rheumatol. 2018;45:1515–1521. doi: 10.3899/jrheum.171375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Gossec L, Guyard F, Leroy D, Lafargue T, Seiler M, Jacquemin C, Molto A, Sellam J, Foltz V, Gandjbakhch F, et al. Detection of flares by decrease in physical activity, collected using wearable activity trackers in rheumatoid arthritis or axial spondyloarthritis: an application of machine learning analyses in rheumatology. Arthritis Care Res (Hoboken) 2019;71:1336–1343. doi: 10.1002/acr.23768. [DOI] [PubMed] [Google Scholar]
- 158.Hur B, Gupta VK, Huang H, Wright KA, Warrington KJ, Taneja V, Davis JM, 3rd, Sung J. Plasma metabolomic profiling in patients with rheumatoid arthritis identifies biochemical features predictive of quantitative disease activity. Arthritis Res Ther. 2021;23:164. doi: 10.1186/s13075-021-02537-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Vodencarevic A, Tascilar K, Hartmann F, Reiser M, Hueber AJ, Haschka J, Bayat S, Meinderink T, Knitza J, Mendez L, et al. Advanced machine learning for predicting individual risk of flares in rheumatoid arthritis patients tapering biologic drugs. Arthritis Res Ther. 2021;23:67. doi: 10.1186/s13075-021-02439-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Bonakdari H, Pelletier JP, Martel-Pelletier J. A reliable time-series method for predicting arthritic disease outcomes: New step from regression toward a nonlinear artificial intelligence method. Comput Methods Programs Biomed. 2020;189:105315. doi: 10.1016/j.cmpb.2020.105315. [DOI] [PubMed] [Google Scholar]
- 161.Christensen ABH, Just SA, Andersen JKH, Savarimuthu TR. Applying cascaded convolutional neural network design further enhances automatic scoring of arthritis disease activity on ultrasound images from rheumatoid arthritis patients. Ann Rheum Dis. 2020;79:1189–1193. doi: 10.1136/annrheumdis-2019-216636. [DOI] [PubMed] [Google Scholar]
- 162.Lotsch J, Alfredsson L, Lampa J. Machine-learning-based knowledge discovery in rheumatoid arthritis-related registry data to identify predictors of persistent pain. Pain. 2020;161:114–126. doi: 10.1097/j.pain.0000000000001693. [DOI] [PubMed] [Google Scholar]
- 163.Petrackova A, Horak P, Radvansky M, Fillerova R, Smotkova Kraiczova V, Kudelka M, Mrazek F, Skacelova M, Smrzova A, Kriegova E. Revealed heterogeneity in rheumatoid arthritis based on multivariate innate signature analysis. Clin Exp Rheumatol. 2020;38:289–298. doi: 10.55563/clinexprheumatol/qb2ha3. [DOI] [PubMed] [Google Scholar]
- 164.Feldman CH, Yoshida K, Xu C, Frits ML, Shadick NA, Weinblatt ME, Connolly SE, Alemao E, Solomon DH. Supplementing claims data with electronic medical records to improve estimation and classification of rheumatoid arthritis disease activity: a machine learning approach. ACR Open Rheumatol. 2019;1:552–559. doi: 10.1002/acr2.11068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Joo YB, Kim Y, Park Y, Kim K, Ryu JA, Lee S, Bang SY, Lee HS, Yi GS, Bae SC. Biological function integrated prediction of severe radiographic progression in rheumatoid arthritis: a nested case control study. Arthritis Res Ther. 2017;19:244. doi: 10.1186/s13075-017-1414-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Lezcano-Valverde JM, Salazar F, León L, Toledano E, Jover JA, Fernandez-Gutierrez B, Soudah E, González-Álvaro I, Abasolo L, Rodriguez-Rodriguez L. Development and validation of a multivariate predictive model for rheumatoid arthritis mortality using a machine learning approach. Sci Rep. 2017;7:10189. doi: 10.1038/s41598-017-10558-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.DiMasi JA, Grabowski HG, Hansen RW. Innovation in the pharmaceutical industry: new estimates of R&D costs. J Health Econ. 2016;47:20–33. doi: 10.1016/j.jhealeco.2016.01.012. [DOI] [PubMed] [Google Scholar]
- 168.Shih HP, Zhang X, Aronov AM. Drug discovery effectiveness from the standpoint of therapeutic mechanisms and indications. Nat Rev Drug Discov. 2018;17:19–33. doi: 10.1038/nrd.2017.194. [DOI] [PubMed] [Google Scholar]
- 169.Zhao K, Shi Y, So HC. Prediction of drug targets for specific diseases leveraging gene perturbation data: a machine learning approach. Pharmaceutics. 2022;14:234. doi: 10.3390/pharmaceutics14020234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Forbes JD, Chen CY, Knox NC, Marrie RA, El-Gabalawy H, de Kievit T, Alfa M, Bernstein CN, Van Domselaar G. A comparative study of the gut microbiota in immune-mediated inflammatory diseases-does a common dysbiosis exist? Microbiome. 2018;6:221. doi: 10.1186/s40168-018-0603-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Kishikawa T, Maeda Y, Nii T, Motooka D, Matsumoto Y, Matsushita M, Matsuoka H, Yoshimura M, Kawada S, Teshigawara S, et al. Metagenome-wide association study of gut microbiome revealed novel aetiology of rheumatoid arthritis in the Japanese population. Ann Rheum Dis. 2020;79:103–111. doi: 10.1136/annrheumdis-2019-215743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Devaprasad A, Radstake T, Pandit A. Integration of immunome with disease-gene network reveals common cellular mechanisms between IMIDs and drug repurposing strategies. Front Immunol. 2021;12:669400. doi: 10.3389/fimmu.2021.669400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Kelly CJ, Karthikesalingam A, Suleyman M, Corrado G, King D. Key challenges for delivering clinical impact with artificial intelligence. BMC Med. 2019;17:195. doi: 10.1186/s12916-019-1426-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Rajpurkar P, Chen E, Banerjee O, Topol EJ. AI in health and medicine. Nat Med. 2022;28:31–38. doi: 10.1038/s41591-021-01614-0. [DOI] [PubMed] [Google Scholar]
- 175.Eckstein F, Wirth W, Nevitt MC. Recent advances in osteoarthritis imaging–the osteoarthritis initiative. Nat Rev Rheumatol. 2012;8:622–630. doi: 10.1038/nrrheum.2012.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Guan B, Liu F, Mizaian AH, Demehri S, Samsonov A, Guermazi A, Kijowski R. Deep learning approach to predict pain progression in knee osteoarthritis. Skeletal Radiol. 2022;51(2):363–373. doi: 10.1007/s00256-021-03773-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Leung K, Zhang B, Tan J, Shen Y, Geras KJ, Babb JS, Cho K, Chang G, Deniz CM. Prediction of total knee replacement and diagnosis of osteoarthritis by using deep learning on knee radiographs: data from the osteoarthritis initiative. Radiology. 2020;296:584–593. doi: 10.1148/radiol.2020192091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Keane PA, Topol EJ. With an eye to AI and autonomous diagnosis. npj Digit Med. 2018;1:40. doi: 10.1038/s41746-018-0048-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Obermeyer Z, Emanuel EJ. Predicting the future—Big Data, machine learning, and clinical medicine. N Engl J Med. 2016;375:1216–1219. doi: 10.1056/NEJMp1606181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Badgeley MA, Zech JR, Oakden-Rayner L, Glicksberg BS, Liu M, Gale W, McConnell MV, Percha B, Snyder TM, Dudley JT. Deep learning predicts hip fracture using confounding patient and healthcare variables. NPJ Digit Med. 2019;2:31. doi: 10.1038/s41746-019-0105-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Finlayson SG, Bowers JD, Ito J, Zittrain JL, Beam AL, Kohane IS. Adversarial attacks on medical machine learning. Science. 2019;363:1287–1289. doi: 10.1126/science.aaw4399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Hirano H, Minagi A, Takemoto K. Universal adversarial attacks on deep neural networks for medical image classification. BMC Med Imaging. 2021;21:9. doi: 10.1186/s12880-020-00530-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Nagendran M, Chen Y, Lovejoy CA, Gordon AC, Komorowski M, Harvey H, Topol EJ, Ioannidis JPA, Collins GS, Maruthappu M. Artificial intelligence versus clinicians: systematic review of design, reporting standards, and claims of deep learning studies. BMJ. 2020;368:m689. doi: 10.1136/bmj.m689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Lin H, Li R, Liu Z, Chen J, Yang Y, Chen H, Lin Z, Lai W, Long E, Wu X, et al. Diagnostic efficacy and therapeutic decision-making capacity of an artificial intelligence platform for childhood cataracts in eye clinics: a multicentre randomized controlled trial. EClinicalMedicine. 2019;9:52–59. doi: 10.1016/j.eclinm.2019.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.The Lancet Respiratory M Opening the black box of machine learning. Lancet Respir Med. 2018;6:801. doi: 10.1016/S2213-2600(18)30425-9. [DOI] [PubMed] [Google Scholar]
- 186.Price WN. Big Data and black-box medical algorithms. Sci Transl Med. 2018;10(471):eaao5333. doi: 10.1126/scitranslmed.aao5333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Panch T, Mattie H, Atun R. Artificial intelligence and algorithmic bias: implications for health systems. J Glob Health. 2019;9:010318. doi: 10.7189/jogh.09.020318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Obermeyer Z, Powers B, Vogeli C, Mullainathan S. Dissecting racial bias in an algorithm used to manage the health of populations. Science. 2019;366:447–453. doi: 10.1126/science.aax2342. [DOI] [PubMed] [Google Scholar]
- 189.Wen D, Khan SM, Ji XuA, Ibrahim H, Smith L, Caballero J, Zepeda L, de Blas PC, Denniston AK, Liu X, Matin RN. Characteristics of publicly available skin cancer image datasets: a systematic review. The Lancet Digit Health. 2022;4:e64–e74. doi: 10.1016/S2589-7500(21)00252-1. [DOI] [PubMed] [Google Scholar]
- 190.Char DS, Shah NH, Magnus D. Implementing machine learning in health care—addressing ethical challenges. N Engl J Med. 2018;378:981–983. doi: 10.1056/NEJMp1714229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Barrett SRH, Speth RL, Eastham SD, Dedoussi IC, Ashok A, Malina R, Keith DW. Impact of the Volkswagen emissions control defeat device on US public health. Environ Res Lett. 2015;10:114005. doi: 10.1088/1748-9326/10/11/114005. [DOI] [Google Scholar]
- 192.A guide to good practice for digital and data-driven health technologies https://www.gov.uk/government/publications/code-of-conduct-for-data-driven-health-and-care-technology/initial-code-of-conduct-for-data-driven-health-and-care-technology. Accessed 16 Mar 2022.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data sharing is not applicable to this article as no datasets were generated or analyzed during the current study.