Abstract
The size of the cerebral cortex increases dramatically across amniotes, from reptiles to great apes. This is primarily due to different numbers of neurons and glial cells produced during embryonic development. The evolutionary expansion of cortical neurogenesis was linked to changes in neural stem and progenitor cells, which acquired increased capacity of self‐amplification and neuron production. Evolution works via changes in the genome, and recent studies have identified a small number of new genes that emerged in the recent human and primate lineages, promoting cortical progenitor proliferation and increased neurogenesis. However, most of the mammalian genome corresponds to noncoding DNA that contains gene‐regulatory elements, and recent evidence precisely points at changes in expression levels of conserved genes as key in the evolution of cortical neurogenesis. Here, we provide an overview of basic cellular mechanisms involved in cortical neurogenesis across amniotes, and discuss recent progress on genetic mechanisms that may have changed during evolution, including gene expression regulation, leading to the expansion of the cerebral cortex.
Keywords: cerebral cortex, enhancer, ferret, intermediate progenitor, OSVZ, radial glia
1. INTRODUCTION
The human forebrain is one of the most fascinating animal organs, specialized in extremely complex computation and integration of information, which underlies high cognitive abilities and a unique capacity to modify the environment. This extraordinary computational capacity comes in part from its remarkable volume, containing billions of highly diverse neuron types and other cell classes (Herculano‐Houzel, 2009). While occasionally referred to as the pinnacle of brain evolution, the human forebrain is not the largest in absolute size, surpassed by few species like whales and elephants (Manger, 2006; Manger et al., 2013; Shoshani et al., 2006). Nevertheless, brain size clearly increased during the evolution of vertebrates and, particularly, amniotes, as observed across phylogeny from reptiles to large mammals. Differences in brain size are mostly related to the number of brain cells (neurons and glia), although significant variations in cell packing and in size of neuronal soma may also influence brain size, as seen particularly in birds, with potentially significant physiological consequences (Azevedo et al., 2009; Kverková et al., 2022; Olkowicz et al., 2016).
Neurons are generated during embryonic development by neural progenitor cells (NPCs); hence, the number of neurons in the mature cerebral cortex depends on the number, dynamics, and lineage of NPCs (Taverna et al., 2014). Studies of large‐brained mammals in the last decade have identified an unsuspected diversity of NPCs and complexity of cortical germinal zones, as compared to the small mouse, which exerted key roles in the expansion and complexification of the cerebral cortex during evolution (Dehay & Kennedy, 2007; Llinares‐Benadero & Borrell, 2019; Polioudakis et al., 2019; Taverna et al., 2014). During embryogenesis, the emergence of germinal zones and the proliferative and neurogenic activity of NPCs are under specific genetic programs. Accordingly, the evolutionary expansion and increased complexity of the cerebral cortex resulted from changes in these developmental genetic programs (Villalba et al., 2021). The search for those genetic changes during evolution has largely focused on the identification of human‐specific genes (Florio et al., 2017). Some human‐specific or primate‐specific genes with a role in the embryonic development of the cerebral cortex have been identified, but they are very few and their effects on cortical development are insufficient to explain the exponential increase in brain size during hominid evolution (Florio et al., 2018; Mora‐Bermudez et al., 2016). Most of those genes emerged from genetic duplications, followed by some degree of sequence modification (Charrier et al., 2012; Dennis & Eichler, 2016; Dougherty et al., 2017; Fiddes et al., 2018; Florio et al., 2015; Suzuki et al., 2018). Although gene duplications seem to have been favored in the recent human lineage, genes are also frequently used and reused (co‐opted) for multiple purposes in different tissues and across lifetime periods, so the duplication or loss of one gene is likely to affect many processes during development and in the adult organism, easily becoming deleterious.
An alternative mechanism for genetic evolution is to modify the regulation of gene expression. In the vast majority of amniotes, most of the genome corresponds to noncoding DNA, with a large part of it containing gene regulatory elements (promoters, enhancers, etc.). Most of the DNA in modern humans that was inherited from Neanderthals is regulatory or related to noncoding elements, and most differences between our genomes are in gene regulatory elements (Burbano et al., 2012; Maricic et al., 2013). Noncoding RNAs, including microRNAs (miRNAs), long noncoding RNAs (lncRNAs), circular RNAs (circRNAs), and pico RNAs (picRNAs), are very abundant in the genome and a major source of gene expression regulation in health and disease is playing central roles in the regulation of NPC dynamics and fate (Aprea & Calegari, 2015; Arcila et al., 2014; Hansen et al., 2013; Kosik & Nowakowski, 2018; Prieto‐Colomina et al., 2021). Endogenous retroviral sequences comprise more than 8% of the modern human genome, and while initially believed to be genomic junk, many can be important players in gene regulation, particularly related to stem cell pluripotency (Izsvák et al., 2016). Regulation of gene expression is an ultimate plasticity mechanism for gene function, and thus the evolutionary expansion and diversification of the amniote cerebral cortex may have been largely based on the modification of gene regulatory mechanisms.
2. MECHANISMS OF DEVELOPMENT IMPACTING CEREBRAL CORTEX SIZE
The size, structure, and composition of the mature cerebral cortex is the result of embryonic development. The cerebral cortex stems from the dorsal telencephalic vesicles, initially as a pseudostratified neuroepithelium composed of a monolayer of neuroepithelial cells (NECs) (Bayer & Altman, 1991; Rakic & Sidman, 1968). These cells undergo multiple rounds of self‐amplification, increasing their own abundance and the surface area of the cortical primordium. NECs have high apical–basal polarity, contacting the apical and basal surfaces of the cortical primordium with a thin process in each direction (Sidman & Rakic, 1973). The cell nucleus of NECs moves along their apical–basal extent during cell cycle, always undergoing S‐phase (DNA synthesis) at the basal side and mitosis at the apical side, a behavior known as interkinetic nuclear migration (INM) that confers the neuroepithelium its typical false appearance of multiple layering (Sauer & Walker, 1959; Takahashi et al., 1996a). Following the initial expansion of the cortical primordium, NECs transform into apical radial glia cells (aRGCs), the primary type of NPC that gives rise to all cortical excitatory neurons (Götz & Huttner, 2005; Malatesta et al., 2000; Miyata et al., 2001; Noctor et al., 2001). At early stages, aRGCs mostly undergo self‐amplificative divisions, generating few neurons. A gradual shift during development toward neurogenic divisions leads to the predominance of the latter in the last stages of development, until neurogenesis is complete (Takahashi et al., 1996b). Similar to NECs, aRGCs are anchored to the apical and basal surfaces and undergo INM (Götz & Huttner, 2005; Noctor et al., 2001; Sidman & Rakic, 1973). However, as neurons are born, they move to the basal side of the cortical primordium, and the cell bodies of aRGCs remain together at the apical side, now forming a distinct germinal zone named ventricular zone (VZ) (Boulder Committee, 1970). The generation of neurons from aRGCs may be either direct or indirect. In the direct mode, neurons are born directly from aRGC divisions at the apical surface. In the indirect mode, aRGCs generate basal progenitor cells, which coalesce on the basal border of the VZ forming a secondary germinal zone, the subventricular zone (SVZ). Basal progenitors then undergo additional rounds of cell division prior to producing neurons, thus amplifying the total number of neurons born from each initial aRGC (Fernandez et al., 2016; Kriegstein et al., 2006).
Comparison across amniote species has revealed some principles of development that have a substantial impact on cortex size. The initial expansion of the NEC cell population forming the neuroepithelium, which determines the size of the aRGC cell pool at the onset of neurogenesis, is already very different between species, in accordance with the eventual size of their mature cortex (Kriegstein et al., 2006; Uzquiano et al., 2018). The subsequent lineage from aRGCs to neurons also differs substantially (Figure 1). In lepidosaur reptiles (lizards and snakes), aRGCs in the dorsal cortex only undergo direct neurogenesis, not producing basal progenitors, and the final number of cortical neurons is relatively small, arranged in a single neuronal layer (Cárdenas et al., 2018; Martínez‐Cerdeño et al., 2016, 2006). Thus, both the proliferative and neurogenic activities of the aRGC lineage are very limited in reptiles. In birds, the equivalent to the mammalian cerebral cortex and the reptilian dorsal cortex is the dorsal pallium, and aRGCs in this region combine direct with indirect neurogenesis (Cárdenas et al., 2018; Martínez‐Cerdeño et al., 2016; Nomura et al., 2013). Remarkably, direct neurogenesis predominates in the medial region of the dorsal pallium, whereas indirect neurogenesis predominates in the lateral region, as observed in chick embryos (Cárdenas et al., 2018). Basal progenitors in the chick dorsal pallium align at the basal border of the VZ, reminiscent of a nascent SVZ, and are molecularly heterogeneous, some expressing the paired box transcription factor Pax6 (marker of mammalian aRGCs), and some expressing the t‐box transcription factor Tbr2 (marker of mouse intermediate progenitor cells [IPCs]) (Cárdenas et al., 2018) (Figure 1).
FIGURE 1.
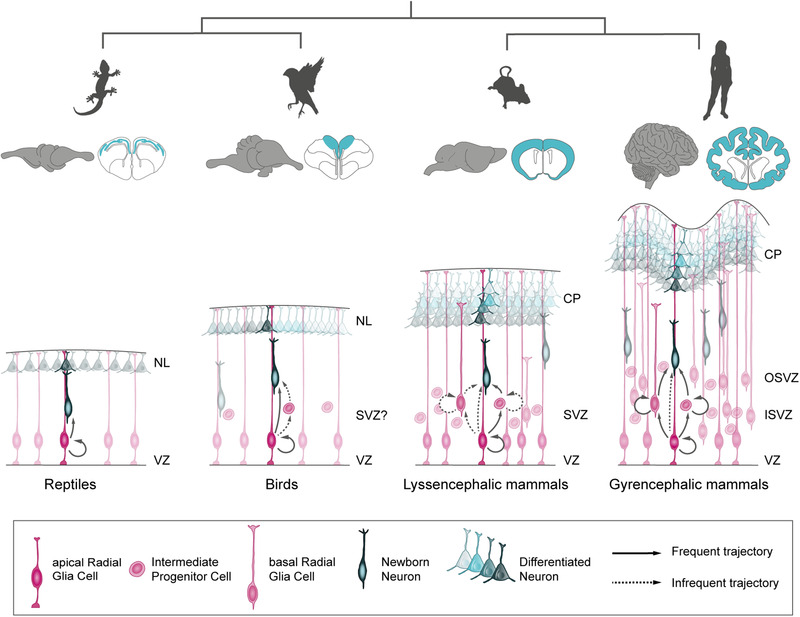
Regulation of gene expression levels across amniotes determines their predominant mode of neurogenesis, with consequences on the final size, folding, and cellular complexity of the cerebral cortex. CP, cortical plate; NL, neuronal layer; ISVZ, inner subventricular zone; OSVZ, outer subventricular zone; SVZ, subventricular zone; VZ, ventricular zone
In mammals, cortical aRGCs mostly undergo indirect neurogenesis, generating IPCs and other types of basal neurogenic progenitors (Haubensak et al., 2004; Miyata et al., 2004; Noctor et al., 2001, 2004; Taverna et al., 2014) (Figure 1). IPCs are multipolar cells with multiple short processes that express Tbr2, populate the SVZ, and usually produce neurons (Miyata et al., 2004; Noctor et al., 2004). Basal radial glia cells (bRGCs) are also basal progenitors but with remarkable similarity to aRGCs: they are cells polarized morphologically along the apico–basal axis (Florio & Huttner, 2014; Taverna et al., 2014), extend a long process pointing in the apical or basal direction (frequently reaching the pial surface), and express Pax6. However, unlike aRGCs, the cell soma of bRGCs is in the SVZ and frequently they lack an apical process anchored to the apical ventricular surface (Betizeau et al., 2013; Fietz et al., 2010; Hansen et al., 2010; Reillo et al., 2011; Shitamukai et al., 2011; Wang et al., 2011). In small mammals like mice, most basal progenitors are IPCs, with a very small proportion of bRGCs, both of which undergo one or two rounds of cell division prior to producing neurons (Kowalczyk et al., 2009; Noctor et al., 2004; Shitamukai et al., 2011; Wang et al., 2011). In mammals with large brains, like carnivores and ungulates, basal progenitors are much more proliferative and thus more abundant, forming a very thick SVZ subdivided in inner and outer compartments (ISVZ and OSVZ, respectively; Figure 1) (Reillo et al., 2011). In contrast to small rodents with a small and smooth brain, bRGCs are very abundant and self‐amplify, greatly increasing neurogenesis and driving cortex folding (Fietz et al., 2010; Pilz et al., 2013; Reillo et al., 2011). In primates and great apes with a very large and highly folded cortex, the OSVZ is thickest and basal progenitors are extremely abundant, highly proliferative, diverse, and with very plastic lineage dynamics. Altogether, this contributes to a dramatic increase in neurogenesis and cortical expansion (Figure 1) (Betizeau et al., 2013; Fietz et al., 2010; Hansen et al., 2010; LaMonica et al., 2013; Lukaszewicz et al., 2005; Reillo et al., 2011; Smart et al., 2002).
Experimental manipulation demonstrates the key importance of these distinct progenitor cell types and lineages on the abundance of neurogenesis and cortex size. Genetic enhancement of NEC self‐renewal drives an abnormal expansion of the early neuroepithelium and size of the embryonic cerebral cortex (Chenn & Walsh, 2002; Siegenthaler et al., 2009). Delay in the transition from NEC to aRGC drives increased aRGC abundance and cortex size (Hsu et al., 2015; Sahara & O'Leary, 2009). Manipulation of cell cycle proteins to force aRGC cell cycle re‐entry induces megalencephaly in mouse (Nonaka‐Kinoshita et al., 2013), whereas impairment of aRGC proliferation causes microcephaly (Feng & Walsh, 2004; Johnson et al., 2018). Increased abundance of bRGCs induces folding of the otherwise smooth cortex of mouse and marmoset monkey (Florio et al., 2015; Heide et al., 2020; Stahl et al., 2013). Enhanced or reduced bRGC proliferation in ferret causes the gain or loss of cortical folds, respectively (Masuda et al., 2015; Nonaka‐Kinoshita et al., 2013; Poluch & Juliano, 2015; Reillo et al., 2011).
All these cellular mechanisms of cortical development are under tight genetic regulation, frequently via signaling pathways that are highly conserved across phylogeny. Yet, progenitor cell lineage, neurogenesis, and cortex development unfold quite distinctly between species, raising the fundamental question of how these differences emerge. In the following sections, we address in some detail key genetic mechanisms emerged during evolution that contributed to cortex expansion and complexification.
3. THE EVOLUTION OF GENOMES PARALLELED CORTICAL EVOLUTION
The cerebral cortex of amniotes has a homologous embryonic origin (Brox et al., 2004; Dugas‐Ford & Ragsdale, 2015; Puelles et al., 2000; Striedter, 1997), such that variations in its structure and function are due to the divergence of developmental programs from a common starting point. This developmental divergence is the result of genomic changes occurred along the evolution of each lineage, which modified the pre‐existing plan in the last common ancestor (LCA) while keeping a record of its evolutionary history. Three main trajectories of cortical development evolved from the LCA of amniotes (Figure 2a). First, in the mammalian lineage there was a very significant increase in neurogenesis, concomitant with the spatial and functional specialization of neurons into the modern six‐layered neocortex (Sousa et al., 2017). Second, in the paraphyletic group of reptiles, the developmental plan remained relatively simple, forming a small and plain cortex with a single neuronal layer (Naumann & Laurent, 2017). Third, in birds a major deviation occurred in the plan of cortical development compared to their paraphyletic reptile counterparts, with the cortex becoming functionally organized into nuclei (Nomura & Izawa, 2017). Some avian groups also increased neurogenesis significantly, achieving a number of telencephalic neurons similar to small primates (Olkowicz et al., 2016).
FIGURE 2.
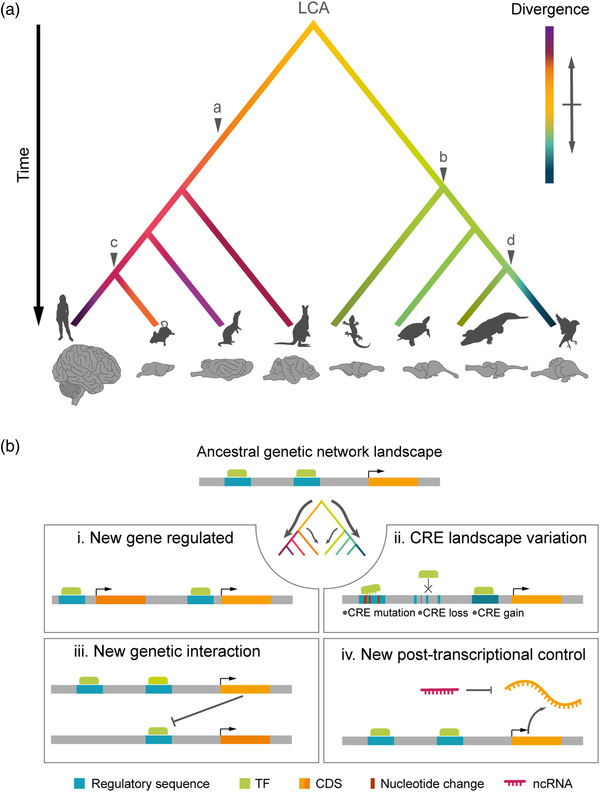
The evolution of genomes paralleled the evolution of the cerebral cortex. (a) Schematic phylogenetic tree illustrating the relationships between amniote clades (from left to right: primates, rodents, carnivores, marsupials, lepidosaurs, testudines, crocodilians, and birds) and the accumulation of genomic changes on developmental programs leading to the diversification of brain size and structure. Evolutionary time and brain are not to scale. Phenotypic divergence from the last common ancestor (LCA) is color coded. Major changes in corticogenesis are indicated: (a) mammalian six‐layered neocortex, (b) reptile single‐layered cortex, (c) secondary loss of gyrencephaly in some mammalian clades, and (d) avian dorsal pallium. Animal silhouettes are from http://phylopic.org/. (B) Schemas of typical genomic changes occurring during evolution that alter gene regulatory networks within developmental programs. (i) Integration of new genes in the network, introducing new functions. (ii) Changes in cis‐regulatory elements (CRE) that control gene expression. (iii) Emergence of new genetic interactions between elements already existing in the network. (iv) New posttranscriptional control, such as the emergence of new noncoding RNAs (ncRNAs). CDS, coding sequence; TF, transcription factor
It has been proposed that the amniote LCA had a simple cortical organization (Briscoe & Ragsdale, 2018), somewhat similar to the reptilian cortex. From this starting point, the developmental programs of the mammalian cortex and avian dorsal pallium would have been subject to greater evolutionary change than reptiles (Figure 2). This notion can be first approached by identifying the evolutionary novelty in cortex development via comparative genomics (Katju & Bergthorsson, 2013; Lynch, 2010; Scally & Durbin, 2012). Studies focused on primates, notably human, revealed that the sequence of genes related to cortical development underwent a higher rate of change in humans than rodents, including both coding (Dorus et al., 2004; Yu et al., 2006) and noncoding sequences (Haygood et al., 2010, 2007; Pollard et al., 2006; Prabhakar et al., 2006). Moreover, several studies have shown an acceleration in the generation of new genes in great apes (Bailey et al., 2002; Dennis et al., 2017; Marques et al., 2005; Marques‐Bonet et al., 2009; Nielsen et al., 2005; Sudmant et al., 2013; Zhang et al., 2010), some of them related to neurodevelopment (Fortna et al., 2004; Zhang et al., 2011). Whether similarly high rates of evolutionary change in coding and noncoding DNA also occurred in the avian and reptilian phyla remains unknown.
But did these genomic changes actually exert an effect on corticogenesis? How was this effect implemented? Any genomic novelty introduced during evolution must integrate within the pre‐existing genomic context, and act in concert with the pre‐existing core gene regulatory network (Figure 2b). This involves the control of pleiotropic effects, spatial–temporal regulation, and the conservation of functions with deep homology (Carroll, 2008, 2005; Stern, 2000). As these constraints are met, the individual genomic innovations allow for the chiseling of the developmental process in a given direction, altering the genetic network and the cellular behavior, introducing new cell types and finally giving rise to structural and physiological innovation (Figure 2) (Arendt et al., 2016; Yuste et al., 2020). Regarding corticogenesis, this relates to the diversity of neural progenitors, their proliferative capabilities, the balance between proliferation and neurogenesis, the diversity of neuron types generated, and their migration and integration into neural circuits (Florio & Huttner, 2014; Llinares‐Benadero & Borrell, 2019; Taverna et al., 2014).
The types of genomic changes modifying the core gene regulatory network come in two main flavors: changes in the genetic repertoire or changes in gene regulation. Changes that alter the genetic repertoire can be achieved in several ways: alteration of gene copy number with a dosage‐dependent effect (Heide et al., 2017), generation of new genes, or mutation of already existing ones, the latter two resulting in neofunctionalization (Chen et al., 2013). New genes can quickly acquire a key role in development (Chen et al., 2013), whereas loss of genes frequently has deleterious effects, thus being less prone to evolutionary selection (Carroll, 2008; McLean et al., 2011). Changes in gene regulation modify the levels of expression as well as the temporal and spatial patterns (Chou et al., 2016; García‐moreno & Molnár, 2020). This may involve changes at various levels: from chromatin compartmentalization and epigenetic state (Acemel et al., 2017; Stillman, 2018), to the cis‐regulatory landscape and posttranscriptional modifications (Lennox et al., 2018) (Figure 1). Evolution of regulatory sequences has been proposed to be a source of continuous change for the generation of morphological variation (Wray, 2007) and may be a primary mechanism in the evolution of cortical development. In contrast, sequence variation of trans‐regulatory elements (transcription factors [TFs], transcriptional activators or repressors, chromatin‐remodeling complexes) is highly constrained due to its profound deleterious effects (Carroll, 2008). Instead, conserved TFs may act differently in different species via modification of the DNA sequence at gene regulatory elements, creating or eliminating their specific DNA binding motifs. In the next sections, we will review how these different modalities of genomic innovation have shaped corticogenesis in phylogeny.
4. EXPANSION OF THE GENE REPERTOIRE FOR CORTICAL EXPANSION
Genetic neofunctionalization may be a driving force for evolutionary change when the action of novelties incorporated is kept within specific tissues or organs (Carroll, 2008, 2005; Chen et al., 2013; Stern, 2000). In the case of cortical evolution, several studies have shown evidence about modification of the genetic arsenal involved in cortical development. For example, more than 30 gene families expanded specifically in the recent human lineage, and many of those are involved in neurodevelopment (Fortna et al., 2004; Sudmant et al., 2010; Zhang et al., 2011). Several studies have identified a series of genes emerged only in the primate and human lineage, and whose expression is enriched in the germinal layers of the developing cortex (Camp et al., 2015; Dennis et al., 2012; Fietz et al., 2012; Florio et al., 2015, 2018; Ju et al., 2016; Keeney et al., 2015; Suzuki et al., 2018). Next, we present a series of novel genetic elements that appeared in the primate and human lineage via different mechanisms and likely contributed to neocortex expansion.
4.1. Increase in gene copy number
Variation in the number of copies of a coding gene may lead to increased expression levels with phenotypic consequences (Figure 3a). In the primate and human lineage, genomic duplications have been frequent (Bailey et al., 2002; Fortna et al., 2004; Marques‐Bonet & Eichler, 2009). Some of these duplications have contributed to key changes in the program of cortical development, as is the case of TBC1D3. This hominoid oncoprotein is encoded by several paralogues on chromosome 17q12 in humans, present in only one copy in the chimpanzee genome, and altogether absent in other species (Hodzic et al., 2006; Zody et al., 2006). Expression of TBC1D3 in the developing mouse neocortex leads to increased bRGC generation and incipient cortical folding (Ju et al., 2016), supporting the notion that this gene exerts a copy number‐dependent effect on neocortex growth.
FIGURE 3.
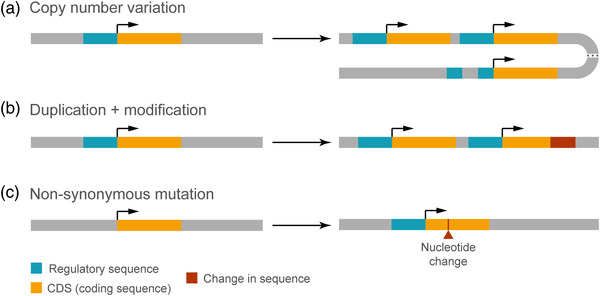
Mechanisms of genetic evolution. (a) Copy number variation caused by gene duplication, which may be complete or partial, and may go along with an original regulatory sequence. (b) Generation of a new gene by complete or partial duplication of an ancestral gene. The duplicated gene may be further subject to sequence modification. (c) Neofunctionalization of an existing gene by nonsynonymous mutation. Protein function may be altered by mutations modifying its gross structure or its functional properties, or even by nonsynonymous point mutations.
Another case is that of the NBPF (neuroblastoma breakpoint family) gene family. This is present only in placental mammals, and the number of its members is highly variable between species, being larger in primates and largest in humans (Vandepoele et al., 2005). NBPF genes contain several copies of the Olduvai (DUF1220) domain (Sikela & van Roy, 2017) and are mostly located at locus 1q21.1 (Gregory et al., 2006; Vandepoele et al., 2005). Deletions and duplications of DUF1220 domains within this locus have been related to micro‐ and macrocephaly, respectively, in addition to other severe neuropathologies (Brunetti‐Pierri et al., 2008). Moreover, the number of DUF1220 copy number is very variable in primates, with circa 270 copies in humans and 90–125 in other great apes (O'Bleness et al., 2012; Popesco et al., 2006). This suggests a relationship between DUF1220 domain abundance and brain size during primate evolution (Keeney et al., 2014). In agreement with the above notions, overexpression of DUF1220 in human neural stem cells in vitro increases their proliferation (Keeney et al., 2015). In summary, NBPF genes are likely candidates to have played an important role in the recent evolutionary expansion of the human brain.
4.2. Generation of new genes via duplication and modification
One of the main mechanisms to generate genes with a new function is via duplication of a pre‐existing gene followed by sequence modification (Figure 3b) (Bailey et al., 2002; Chen et al., 2013; Fortna et al., 2004). A few of the human‐specific genes generated by this process are specifically expressed in the germinal layers of the embryonic neocortex, and are involved in its developmental expansion. ARHGAP11B is a paradigmatic example of this scenario. This gene is expressed in aRGCs and bRGCs of the embryonic human neocortex (Fietz et al., 2012; Florio et al., 2015, 2018), and emerged from a partial duplication of ARHGAP11A followed by a C‐to‐G substitution (Antonacci et al., 2014; Riley et al., 2002). This changed a splicing site leading to the loss of 26 amino acids of the original GAP domain, substituted by a unique C‐terminal sequence of 47 amino acids (Florio et al., 2016). This modification critically changed the new protein as compared to ARHGAP11A in two ways: (1) new subcellular localization, at the mitochondrial membrane; (2) new function, modulating the mitochondrial metabolism via a Ca2+‐mediated increase of the Krebs cycle, necessarily supported by glutaminolysis (Namba et al., 2021). A key developmental consequence of the emergence of ARHGAP11B was the increase in proliferative activity of basal progenitors, with a concomitant expansion of the neocortex. This has been probed in several models including mouse (Florio et al., 2015, 2016; Xing et al., 2021), ferret (Kalebic et al., 2018), marmoset (Heide et al., 2020), and chimpanzee and human organoids (Fischer et al., 2020).
Another well‐known case of a neofunctionalized duplicated gene is NOTCH2NL. This is a human‐specific gene also expressed in NPCs that originated from a partial duplication of NOTCH2 in hominins, followed by a later modification in humans (Dougherty et al., 2017; Fiddes et al., 2018; Florio et al., 2018; Suzuki et al., 2018). The duplicated segment contains the promoter and six N‐terminal EGF‐like domains, but lacks the transmembrane and cytoplasmic domains (Fiddes et al., 2018). NOTCH2NL has four paralogues, three of them at locus 1q21.1 (NOTCH2NLA, NOTCH2NLB, and NOTCH2NLC) and a fourth NOTCH2NLR considered a pseudogene. Each of the four paralogues is very close to a gene of the abovementioned NBPF genes, which suggests their co‐evolution (Heide & Huttner, 2021). Among the main differences between NOTCH2NLB and NOTCH2NLA/C is that the former presents a 39‐amino acid N‐terminal signal peptide that the latter do not, and therefore are thought to be secreted via a noncanonical pathway. Hence, in addition to acquiring a new function, NOTCH2NL was further subspecified in multiple paralogues. Three independent studies demonstrated that NOTCH2NL promotes NPC proliferation (Fiddes et al., 2018; Florio et al., 2018; Suzuki et al., 2018). NOTCHN2L proteins have been proposed to induce trans‐activation of Notch signaling via physically interacting with Notch receptors (Fiddes et al., 2018) or via prevention of DLL1‐mediated cis‐inhibition of Notch signaling (Suzuki et al., 2018). Given that Notch signaling promotes neural stem cell self‐renewal and prevents the differentiation of cortical progenitors (Gaiano et al., 2000; Hansen et al., 2010; Imayoshi et al., 2010; Kageyama et al., 2009; Mizutani et al., 2007), these mechanisms may explain the effect of NOTCH2NL on the increased abundance of NPCs in human.
A third case is SRGAP2C. This gene arose in a similar period as NOTCH2NL from two partial duplication events from its ancestral gene, SRGAP2A, a Slit‐Robo Rho GTPase‐activating protein (Dennis et al., 2012). Although SRGAP2C contains the F‐BAR domain of SRGAP2A, it lacks its last 49 amino acids (Charrier et al., 2012). SRGAP2C is expressed in the cortical germinal layers as well as in the cortical plate of the human embryo. Its role in neurodevelopment seems not related to NPC proliferation, but to dendrite and synapse formation. Forced expression of SRGAP2C in the developing mouse neocortex leads to increased synaptic density, prolonged spine maturation, and increased speed of radial migration (Charrier et al., 2012; Fossati et al., 2016). These effects result from the antagonizing interaction of SRGAP2C on SRGAP2A, which otherwise promotes synapse maturation while limiting their density (Fossati et al., 2016). Importantly, expression of SRGAP2C in mice improves their ability to learn cortex‐dependent sensory discrimination tasks (Schmidt et al., 2021).
Finally, HYDIN2 is an example of a human‐specific gene that emerged from a large duplication event of 79 exons of HYDIN from the locus 16q22.2 into 1q21.1 (Doggett et al., 2006; Dougherty et al., 2017). HYDIN2 expression is highest in neural tissues, but no functional experiments have so far demonstrated any influence on cortical progenitor cell dynamics.
4.3. Variation of a protein function via mutations in existing genes
A third mechanism of gene neofunctionalization is via amino acid substitutions that alter protein function (Figure 3c). This is especially relevant if the gene is expressed in a tissue‐specific manner, escaping pleiotropic effects. A classic example is FOXP2, a TF highly expressed in the developing and adult human neocortex associated to language and vocal learning (Ferland et al., 2003; Feuk et al., 2006; Lai et al., 2001; MacDermot et al., 2005). Human FOXP2 differs in two amino acids from the chimpanzee counterpart, which changes its DNA‐binding motif (Enard et al., 2002; Konopka et al., 2009). Knockdown of Foxp2 in the developing mouse neocortex reduces neurogenesis, partially via blocking the transition from aRGCs to IPCs. Conversely, expression of the human FOXP2 in mouse causes an increase in basal progenitors and neurons (Tsui et al., 2013), as well as increased synaptic plasticity in the striatum (Enard et al., 2009). The effect on synapse formation is mediated by inhibition of Mef2c expression, a TF implicated in the suppression of corticostriatal synapse formation (Chen et al., 2016). This is an example of how discrete amino acid substitutions may change a gene network, with a dramatic effect on cortical development and function, such as speech development (Figure 2b). Importantly, these observations demonstrate that small modifications in protein sequence may have been key in the last steps of human cortical evolution.
Notably, comparison of the genomes of modern and ancient humans has revealed 87 proteins with single‐amino acid substitutions fixed in Homo sapiens and absent in Neanderthals and Denisovans (Prufer et al., 2014). Five of these genes are expressed in the cortical germinal layers VZ and SVZ (CASC5, KIF18A, TKTL1, SPAG5, and VCAM1), and three of them are involved in kinetochore dynamics during mitosis (CASC5, KIF18A, and SPAG5). Importantly, the orientation of the mitotic spindle and duration of the metaphase influences the fate of daughter cells during NPC division, so these gene modifications may have been relevant in the refinement of cortical development in Homo sapiens (Mora‐Bermúdez et al., 2022). Another gene involved in neurodevelopment that differs in a single‐amino acid substitution from the Neanderthal ortholog is NOVA1. This gene regulates alternative splicing of genes involved in synapse formation during nervous system development (Buckanovich & Darnell, 1997; Jensen et al., 2000; Ule et al., 2005). Replacement of the modern gene with the Neanderthal homologue in human cerebral organoids leads to changes in alternative splicing, slower development, and modification of synaptic protein interactions, of electrophysiological properties, and of excitatory signaling neural network function (Trujillo et al., 2021). Altogether, the gene changes occurred between ancient and modern humans indicate that minor adjustments in cortical development continued to occur in the very last steps of human evolution, and likely are still underway.
5. REGULATION OF GENE EXPRESSION LEVELS AND PATTERNS
The emergence of novel genes is a major driving force of phenotypic variation during evolution, but it falls short to account for the great diversity that we observe in nature. In fact, many of the major genetic regulators of cortical development are conserved along the vertebrate lineage (Fernandez et al., 1998; Puelles et al., 2000; Yamashita et al., 2018; Yun et al., 2001), but their patterns of expression vary widely across species, and even among distinct brain areas within the same species. For instance, attenuation across amniotes of Slit/Robo expression levels in the embryonic cortex is responsible for the amplification of basal progenitors and cortical expansion (Cárdenas et al., 2018). In reptiles and birds, high levels of Robo decrease Dll1 expression and promote direct neurogenesis, whereas in mammals low Robo signaling leads to high Dll1 and indirect neurogenesis, with increased generation of basal progenitors forming an SVZ. Intriguingly, Robo signaling is high in the mouse olfactory bulb and leads to a high rate of direct neurogenesis, an example of a conserved mechanism regulating the mode of neurogenesis that acts on opposite directions in adjacent regions of the developing telencephalon (Cárdenas et al., 2018). This and other examples highlighted in the following sections indicate that evolution acts on gene expression regulation at many levels, driving cortical development divergence.
Regulation of gene expression is a multilayered phenomenon that displays its full complexity in brain development, thus an ideal target for evolutionary change (Figure 4). The production of mRNA begins with the assembly of the transcriptional machinery at the promoter region, which requires binding of TFs (Zabidi & Stark, 2016). In order for TFs to bind DNA and promote transcription, DNA itself must be accessible, and this is regulated by a variety of epigenetic DNA modifications such as acetylation and methylation (Moore et al., 2013; Vastenhouw & Schier, 2013; Zhang & Reinberg, 2001). Next, transcription will depend on the expression of the specific TFs that recognize the DNA sequence of the relevant gene regulatory elements, including promoters and/or enhancers. Then, the genome inside the nucleus forms loops and other three‐dimensional conformations that approach distal enhancers and trans‐regulatory elements to the transcriptional complex, adding an extra layer of regulation (Dixon et al., 2012; Gonzalez‐Sandoval & Gasser, 2016). Finally, following transcription mRNAs are subject to multiple processes including splicing, editing, and nuclear export, prior to the starting translation at the cytoplasm. Still, once at the cytoplasm, mRNAs must be actively protected from degradation to confer sufficient stability for sustained translation. The modulation of each of these steps will result in changes of gene expression and different protein levels.
FIGURE 4.
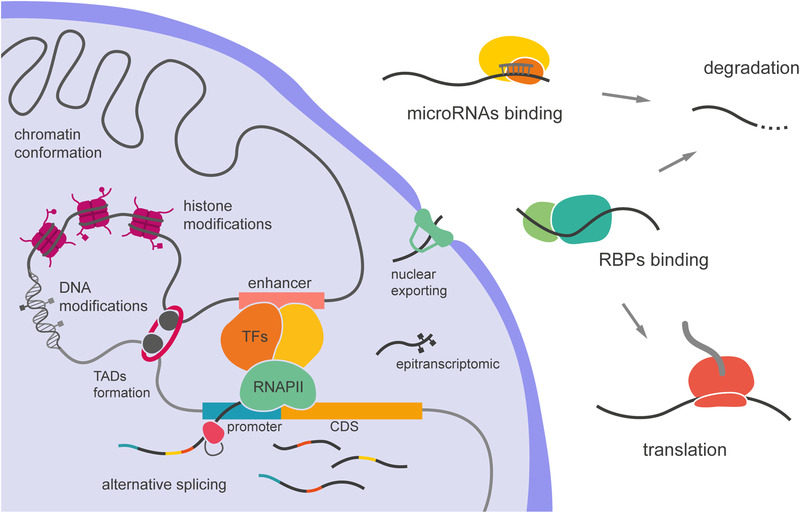
Layers of gene regulation. Pretranscriptional modalities include chromatin conformation, DNA and histone modifications, and the formation of topologically associated domains (TADs), which bring together different types of trans‐regulatory elements, affecting the binding of transcription factors (TFs). Posttranscriptional regulation involves alternative splicing, mRNA modification (epitranscriptomics), and binding to RNA binding proteins (RBPs), which control nuclear export and mRNA stability, including degradation by miRNA–RISC complex. CDS, coding sequence
5.1. TF expression
TFs are proteins that act as trans‐regulatory elements by binding on cis‐regulatory elements (CREs) of DNA (Latchman, 1997). TFs have a DNA‐binding domain, usually consisting of zinc finger domains, which binds to a specific nucleotide sequence and promotes or prevents the recruitment of RNA polymerase (Latchman, 1997; Roeder, 1996). This promotes or represses the transcription of genes proximal to the CRE, or promoter (Mitchell & Tjian, 1988; Ptashne & Gann, 1997). Several TFs are known to play key roles in different processes of cortical development, with both conserved and novel functions linked to species‐specific features (Somel et al., 2013).
TFs are critical from the very onset of corticogenesis, as they are involved in the regionalization of the telencephalon. The embryonic telencephalon has classically been divided in two distinct regions: the subpallium ventrally, which gives rise to the basal ganglia, and the pallium dorsally, which develops into the cerebral cortex. The identity of these regions is initially established early in development by extracellular signals or morphogens, including sonic hedgehog and bone morphogenic proteins, and maintained through regionally restricted cross‐regulatory interactions involving basic helix–loop–helix TFs (Wilson & Rubenstein, 2000). Ngn1 and NeuroD are two TFs expressed in the pallium and associated with the production of glutamatergic neurons, whereas Mash1 and Zash1a are expressed in the subpallium and related to GABAergic neuron production (Wullimann & Mueller, 2004). The pallium and subpallium can also be distinguished in the embryonic telencephalon of different vertebrates by the differential expression of TFs involved in glutamatergic or GABAergic neuron production, such as Tbr1 in the pallium and Dlx1/2 in the subpallium (Brox et al., 2004; Casarosa et al., 1999; Puelles et al., 2000). Nkx2.1 and Gsx1/2 contribute to the further regionalization and specification of the ventral telencephalon, the latter by repressing Pax6 expression (Chapman et al., 2018; Corbin et al., 2000; Toresson et al., 2000; Wilson & Rubenstein, 2000; Yun et al., 2001).
The dorsal pallium is further subdivided into four radial domains consisting of medial, dorsal, lateral, and ventral pallium, each giving rise to distinct regions in the adult brain (Puelles et al., 2000). The identity of these pallial regions is established by the regionally restricted expression of TFs, such as Pax6 or Emx1/Emx2. These two TFs are expressed in opposing gradients across the embryonic telencephalon, with Pax6 high in the rostrolateral pallium and low caudomedially, and Emx1/2 in the opposite direction. Loss‐of‐function of Pax6 or Emx2 in mouse leads to agenesis of the cerebral cortex, including a re‐specification of the remaining tissue to become subpallial, highlighting the fundamental importance of these two TFs in the specification and maintenance of the pallial identity (Muzio et al., 2002). Pax6 expression pattern is conserved across vertebrates, and it regulates several downstream targets in response to both cell‐intrinsic and extrinsic signals, including its own levels (Holm et al., 2007; Manuel et al., 2015; Puelles et al., 2000; Sansom et al., 2009; Yamashita et al., 2018; Ypsilanti & Rubenstein, 2016; Yun et al., 2001). Pax6 is involved in the regulation of many aspects of corticogenesis including progenitor cell proliferation, neurogenesis, differentiation, and mode of cell division (Asami et al., 2011; Estivill‐Torrus et al., 2002; Heins et al., 2002; Quinn et al., 2007; Sansom et al., 2009; Warren et al., 1999). Intriguingly, Pax6 also has species‐specific functions during pallial development. For example, sustained expression of Pax6 in progenitors of the mouse embryonic neocortex is sufficient to induce their primate‐like dynamics and an increase in basal progenitors, suggesting that sustained Pax6 expression may be linked to SVZ enlargement and neocortex expansion (Wong et al., 2015). However, in the avian brain it has divergent regulatory functions on cellular dynamics and target genes compared to the mammalian brain. For example, the role of Pax6 on progenitor cell maintenance is unique to mammals, while its role in promoting neuronal differentiation is conserved in both phyla (Yamashita et al., 2018).
Pax6 regulates neuronal differentiation, mostly affecting upper layer neurons (Tuoc et al., 2009), via the positive regulation of proneural genes, such as Ngn2 or Tbr2 (Heins et al., 2002; Holm et al., 2007; Sansom et al., 2009; Scardigli et al., 2003). Trb2 is a member of the T‐box family of TFs and is directly related to the amplification of IPCs in mammals. Since IPCs expand the neuronal output, Tbr2 is directly related to cortical expansion (Sessa et al., 2008). Loss of Tbr2 function in the developing ferret cortex results in reduced folding (Toda et al., 2016), and its deletion in mouse results in a great reduction of IPC‐dependent neurogenesis (Mihalas et al., 2016), confirming its role in basal progenitor amplification. Cell lineage and single‐cell transcriptomic studies in mouse show that most cortical glutamatergic neurons transit through intermediate states of Tbr2 expression (Mihalas & Hevner, 2018; Mihalas et al., 2016), suggesting that Tbr2 regulates the transition from RGCs to IPCs by gradually turning off the RGC genetic program in favor of neurogenesis. Mitotic Tbr2+ cells have been reported in the developing pallium of many vertebrates including sharks, lizards, turtles, chicken, and doves (Cárdenas et al., 2018; Clinton et al., 2014; Docampo‐Seara et al., 2018; Martínez‐Cerdeño et al., 2016), raising the possibility that neuronal output amplification through Tbr2+ progenitors may be an ancient mechanism in vertebrates.
Another TF seemingly involved in the generation of basal progenitors in the mammalian cerebral cortex is Hopx. This is a member of the homeodomain‐containing protein family that lacks the ability to bind DNA, as other members of its family, and is expressed by bRGCs in the developing human cortex (Pollen et al., 2015; Thomsen et al., 2015). Hopx is also expressed in the developing ferret and mouse cerebral cortex, where it is sufficient to promote bRGC expansion without altering aRGC dynamics (Vaid et al., 2018). However, the cell types expressing Hopx in the developing mammalian cerebral cortex, and its role in cortical development, remain unclear.
5.2. Gene regulatory elements
CREs are regions of noncoding DNA that contain binding motifs for several TFs, which upon binding may regulate the transcription of nearby genes, either enhancing or silencing their expression (Figure 4). CREs thus contribute to the specificity and dynamics of gene expression across cell types and species (Nord & West, 2020). In the mammalian genome, enhancers are the most common type of CRE, and a great source of phenotypic variation across species. Human accelerated regions (HARs) are genomic regions well conserved in nonhuman primates but with a high rate of mutation in humans (Hubisz & Pollard, 2014). Around 50% of HARs (genomic sequences quickly evolved in the recent human lineage) have been predicted to function as regulatory enhancers (Capra et al., 2013; Girskis et al., 2021; Uebbing et al., 2021). Many HARs are in the proximity of genes important in brain development, including GLI2, GLI3, and TBR1, suggesting a role in proliferation and differentiation during cortical development (Won et al., 2019).
HARs have been associated with cortical expansion during evolution. Human and chimpanzee HAR Enhancer 5 (HARE5) show different enhancer activity, as evidenced in mouse transgenic models via its activity promoting Fzd8 expression. Importantly, expression of the human HARE5 leads to accelerated cell cycle and increased brain size in mouse (Boyd et al., 2015). HARs have been recently shown to extensively rewire the expression pattern of PPP1R17 in the developing primate cortex, where it is expressed in progenitor cells in OSVZ and ISVZ. This gene shortens the cell cycle, promotes the transient amplification of progenitor cells, and delays neuronal differentiation, suggesting a central role in primate corticogenesis of this HAR‐regulated gene (Girskis et al., 2021). PPP1R17 is not expressed in ferret or mouse NPCs, further highlighting the key role of regulatory mechanisms in cortical evolution.
Accelerated regions are not only present in humans, as nearly 5000 noncoding accelerated regions have been identified within the therian lineage, with several of them proven to have gained transcriptional enhancer function compared to nonmammalian orthologous regions (Holloway et al., 2016). Many of these accelerated regions are located near developmental TFs, suggesting they may influence the expression of already existing genes. It is important to point out that small changes in regulatory elements have been demonstrated to drive major evolutionary shifts, such as loss of limbs in snakes (Kvon et al., 2016; Sagai et al., 2005, 2004), suggesting that variation in the sequence of regulatory elements is a major force in evolution.
5.3. Chromatin and TADs
The accessibility of the transcriptional machinery to DNA also regulates gene expression. DNA is organized around a histone scaffold where the basic unit is the nucleosome, composed of a section of DNA filament wrapped around a core of eight histone proteins (McGinty & Tan, 2015). The position of nucleosomes along DNA affects the accessibility of the RNA polymerase. More importantly, histones are characterized by amino‐terminal tails that are susceptible to posttranslational modifications, involved in activation or repression of transcription (Stillman, 2018). A recent study has uncovered the relevance of H3 acetylation in cortical development (Kerimoglu et al., 2021). Human basal progenitors show a higher level of this modification compared to mouse cells, leading to an increased expression of Trnp1 and promoting progenitor cell proliferation. Nucleobases can also be modified. The most studied example is the addition of methyl groups onto cytosines, usually in CG pairs (otherwise called CH methylation). This may reduce gene expression when present on a promoter, and thus be relevant in cortical evolution (Lister et al., 2013; Moore et al., 2013; Price et al., 2019). Recent studies show that the human prefrontal cortex is enriched in hypomethylated sites compared to the chimpanzee, with those more accessible sites containing binding motifs for specific TFs such as FOXP1 (Jeong et al., 2021). Given that CH methylation is more common in the human genome compared to other primates, the transcriptional inhibition caused by this change may be important for the refined distinction of neuronal cell types.
Modifying the chromatin structure and organization is a powerful way to regulate the transcriptional landscape of cells, with multiple examples in the developing nervous system. During corticogenesis, aRGCs shift from neurogenesis at embryonic stages to gliogenesis postnatally, and this is paralleled by a shift in chromatin conformation, from being open and widely accessible to much more compact (Kishi et al., 2012). This phenomenon is not limited to a few protein‐coding regions involved in neural differentiation, but it involves a change in chromatin dynamics across the whole genome mediated by HMGA proteins, which bind DNA directly and promote an accessible configuration. Once chromatin is open and accessible, gene expression is further regulated by the specific topological conformation of DNA at a large scale, as revealed by chromatin conformation capture techniques (Belton et al., 2012; Bonev et al., 2017). Open chromatin forms loops that bring into proximity distantly located gene regulatory elements, favoring new enhancer–promoter interactions and thus dramatically increasing the possibilities for gene expression regulation (Figure 4). These topologically associated domains (TADs) formed by chromatin looping are dynamic and subject to change. For example, during neural differentiation the long‐range interactions between transcriptionally active chromatin domains (type‐A) decrease, increasing between type‐B domains. The boundaries of these interaction domains are largely conserved across cell types, but then the chromatin binding sites may be modified by insulator proteins such as CTCF or cohesins, causing a shift in the interactions. This is the case of Sox2, its expression driven by an Embryonic Stem (ES)‐specific enhancer at early embryonic stages, and then by an NPC‐specific enhancer at late stages (Bonev et al., 2017).
The conformational organization of the genome in TADs seems to be conserved across bilaterian animals (Acemel et al., 2017), so they may have been relevant as a gene‐regulatory mechanism during evolution. For example, the organization in TADs may have favored that evolutionarily new CREs be functionally relevant on multiple promoters within a given TAD. Similarly, changes in TAD boundaries during evolution may have brought into proximity genomic regions that were previously existing but distant, as might be the case with the evolution of the Hox cluster in chordates. These events have been studied in the context of cortical development in human, macaque, and mouse (Luo et al., 2021). This study demonstrates the existence of multiple human‐specific TADs, whose boundaries are enriched in evolutionarily young transposable elements, and have reduced binding to CTCF compared to conserved elements. Moreover, human TADs seem to be more cell type specific compared to those shared with macaque and mouse, supporting the idea of evolutionary acquired ways to fine tune neurodevelopment in human (Luo et al., 2021). Analysis of TADs and their related loops revealed the emergence of new enhancer interactions mediated by anchors that contain HARs. Comparison of different species also shows that genes present in human and macaque, but not mouse, share loops that are enriched in the VZ, a key germinal layer in cortical development (Luo et al., 2021).
5.4. Posttranscriptional mechanisms
Differences in mRNA levels alone cannot explain the full variability observed in protein abundance (Grassi et al., 2019; Vogel & Marcotte, 2012), supporting the relevance of posttranscriptional mechanisms. Posttranscriptional regulation is a complex process involving many players that influence each other, both in the nucleus and cytoplasm (Figure 4). The first mechanism from a chronological perspective is alternative splicing, linked with the transcriptional process itself. The spliceosome complex assembles while the RNA‐polymerase is still ongoing, and it regulates the splicing process per se and the choice of specific exons. This choice is also influenced by the structure of chromatin (Naftelberg et al., 2015). Alternative splicing may have consequences on the posttranslational modifications and on protein–protein interactions, for example, through the insertion of microexons (Ule & Blencowe, 2019). Alternative splicing sites are present in the whole vertebrate lineage, but they are more common in primates and more conserved in brain (Barbosa‐Morais et al., 2012; Merkin et al., 2012). In particular, its importance has emerged in neural development, where paralogs of splicing regulator proteins are associated with cell identity changes. The switch from PTBP1 to PTBP2 is essential for cell identity, since the former promotes expression of progenitor‐specific splicing variants and inhibits the expression of the latter, which then is upregulated during neuronal differentiation (Su et al., 2018; Vuong et al., 2016).
Similar to DNA, mRNA is susceptible to covalent modifications of its nucleotides, known as epitranscriptomics (Fu et al., 2014; Yoon et al., 2018) (Figure 4). The importance of these changes has not been deeply explored yet, but the most common modification, methylation on adenosine N6, has effects on nuclear exporting, microRNA‐mediated decay, and, in some cases, on the splicing process (Noack & Calegari, 2018). Furthermore, this specific modification is important for NPC cycle and stability of specific neuronal genes (Yoon et al., 2017). The dynamic patterning of this specific modification is related to cell identity specification, as shown in the developing brain where it increases along neuronal differentiation (Meyer et al., 2012). Second, mRNAs can bind multiple proteins RNA binding proteins (RBPs) that play roles in the regulation of splicing, polyadenylation, nuclear exporting, cytoplasmic localization, and stabilization (Hsu et al., 2019; Lennox et al., 2018; Salamon & Rasin, 2022; Sena et al., 2021) (Figure 4). One mechanism of action of RBPs is to shuttle transcripts into RNA granules containing repressors of translation (Tian et al., 2020). The subcellular localization of mRNA is particularly important in highly polarized cells and asymmetric processes, like RGCs, where perturbation of the apical complex containing the RBP Stau2 causes premature cortical neuron differentiation (Amadei et al., 2015; Hoye & Silver, 2021; Vessey et al., 2012). Epitranscriptomics influences other gene modulation mechanisms, such as chromatin modifications. For instance, the RBP protein RBM15 controls the level of certain transcripts through RNA methylation, and its direct targets include the chromatin remodeling subunit BAF155, which regulates basal progenitor formation during corticogenesis (Narayanan et al., 2018; Xie et al., 2019).
Control of mRNA stability is another mechanism to regulate the accessibility of mRNAs to the translation machinery, and thus their expression into protein. The role of miRNAs in this process is well documented. MiRNAs are single‐strand short noncoding RNAs that guide the RISC complex to bind specific mRNAs through the recognition of a complementary sequence, usually at the 3′‐UTR. This binding leads to inhibition of translation or degradation of the target mRNA (Prieto‐Colomina et al., 2021). The canonical pathway of miRNA biosynthesis relies on the sequential processing of an immature miRNA, first by Drosha in the nucleus and then by Dicer in the cytoplasm (Miyoshi et al., 2010; Prieto‐Colomina et al., 2021). This process can be modulated itself by posttranscriptional mechanisms, such as RBP interaction (Michlewski & Cáceres, 2019). Multiple studies have explored the role of miRNAs in the development of the nervous system using conditional Dicer knockout mice (Sun & Shi, 2015). In the developing dorsal telencephalon, loss of Dicer (and hence canonical miRNAs) after the onset of neurogenesis leads to the formation of a smaller cortex and impaired neuronal differentiation (De Pietri Tonelli et al., 2008). More recent analyses using very early Cre‐driver mouse lines (E7.5) show that Dicer‐dependent miRNAs are critical already for the homeostatic expansion of the primordial telencephalic neuroepithelium (Fernández et al., 2020). Other studies have explored the role of specific miRNAs such as miR‐7, which controls the production of intermediate progenitors and their apoptosis (Pollock et al., 2014), and miR‐9, which negatively regulates progenitor proliferation and promotes neural differentiation (Zhao et al., 2009).
MiRNAs are important also in brain evolution, via the differential expression of conserved miRNAs or via the emergence of new ones, as is the case of a eutherian‐specific cluster containing miRNAs involved in the acquisition of neuronal identity (Diaz et al., 2020; Martins et al., 2021). The proliferation of NPCs and the emergence of features key in cortical expansion and gyrification, such as the expansion of the OSVZ, are affected by the evolution of posttranscriptional regulation by miRNAs. For example, miR‐137 and miR‐122 are highly expressed in the OSVZ of the developing cerebral cortex of ferret, macaque, and human, three gyrencephalic species distantly related (Tomasello et al., 2022). However, in the embryonic mouse cortex, which does not have an OSVZ, these two miRNAs are not expressed. Ectopic expression of miR‐137 in mouse causes an increase in progenitor proliferation and abundance in the SVZ. MiR‐122 instead acts on neuronal maturation, favoring the identity of superficial cortical layers at the cost of deep layers. A recent study further unravels that miRNAs were also important in recent events of mammalian brain evolution, such as the secondary loss of cortex size and folding typical of small rodents (Chinnappa et al., 2022). miR‐3607 is expressed in cortical germinal zones of gyrencephalic but not lissencephalic species. When overexpressed in the early embryonic mouse cortex, miR3607 blocks APC expression, thus disinhibiting Wnt/beta‐catenin signaling, resulting in the amplification of NPCs. Blockade of endogenous miR‐3607 in the ferret cortex causes a reduction in proliferation and a loss of progenitor polarity accompanied by an increase of APC, consistent with it regulating the Wnt pathway (Chinnappa et al., 2022). These results support that species‐specific patterns of expression of conserved miRNAs contributed to brain evolution. Moreover, evolutionary changes in miRNA action may have also occurred by changes in their target sequences at the 3′‐UTR of target genes. Other classes of noncoding RNAs may have been also important in brain evolution, such as circRNAs and lncRNAs. These ncRNAs regulate miRNA activity by acting as sponges: they contain repeated sequences complementary to specific miRNAs that act as scavengers, blocking their availability to target coding genes (Hansen et al., 2013). Noncoding RNAs may interact in an evolutionary context to modulate otherwise conserved signaling pathways. LncND is a lncRNA with a primate‐specific region that presents multiple miRNA‐responsive elements. LncND is expressed in human NPCs but not later on during neuronal differentiation, and via binding miR‐143‐3p increases the levels of its target genes, including the highly relevant Notch receptors (Rani et al., 2016).
6. TEMPORAL–SPATIAL REGULATION OF GENE EXPRESSION AND FUNCTION
The development of an extremely complex structure like the cerebral cortex requires a tight regulation of gene expression both in space and time. The former is key to pattern the formation of the different brain structures. The latter is important to regulate the process of progenitor cell amplification followed by neurogenesis (Figure 5). A shorter cell cycle is related to a higher proliferative potential of progenitor cells (Caviness Jr. et al., 1995; Dehay & Kennedy, 2007; Salomoni & Calegari, 2010), as recently shown also in newly identified progenitor cell types in reptiles, rodents, carnivores, and primates (Betizeau et al., 2013; Nomura et al., 2013; Pilz et al., 2013; Reillo & Borrell, 2012). However, developmental time and cell cycle length can be dramatically different between species, where, for example, cortical progenitor cells in primates have a longer cell cycle than in rodents at equivalent stages of brain development (Dehay et al., 1993; Kornack & Rakic, 1998; Takahashi et al., 1995). This is contrary to intuition, as cortical progenitor cells in primates are more amplificative than rodents, and thus one would expect shorter cell cycles in the former. However, gestational time and duration of the neurogenic period are also very different among species, and longer neurogenic periods allow for more cell cycles. Indeed, lengthening the neurogenic period alone is sufficient to increase neurogenesis, as shown for upper‐layer neurons (Stepien et al., 2020). Comparative studies performed in cerebral organoids have also identified key differences in developing timing between human and other primates. Apical progenitors in human organoids have a significantly longer metaphase than those in chimpanzee (Mora‐Bermudez et al., 2016), and have a longer proliferative period compared to macaque (Otani et al., 2016). Similarly, NECs transit to RGCs later in development in human than in gorilla organoids (Benito‐Kwiecinski et al., 2021). Altogether, this points to a delayed maturation in the human brain when compared to other primates (Mora‐Bermúdez et al., 2022), which is further supported by differences in chromatin accessibility. At equivalent developmental stages, chimpanzee cells show increased expression of genes related to neuron maturation compared to human cells, enriched in genes linked to RG proliferation (Kanton et al., 2019).
FIGURE 5.
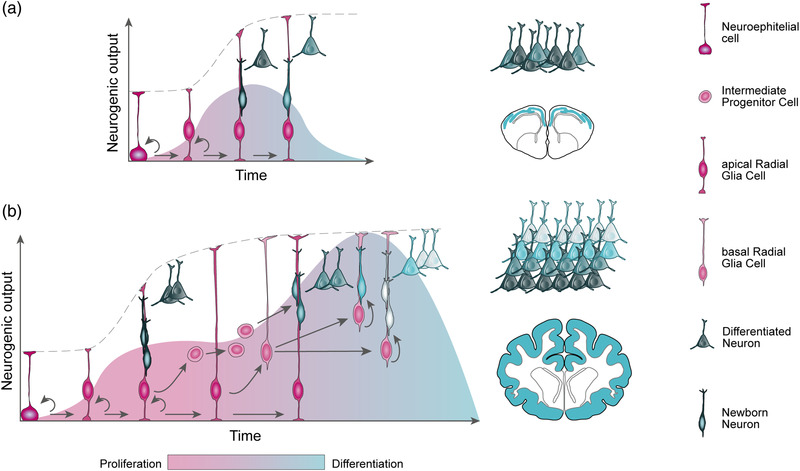
Temporal progression of cortical neurogenesis. (a) Temporal progression of neurogenesis in the reptilian dorsal pallium. At the onset of neurogenesis, self‐amplifying neuroepithelial cells transit to apical radial glia cells, which then undergo long cell cycles to directly produce neurons, rapidly depleting the progenitor pool during the short neurogenic period. As a result, a low number and diversity of neurons are produced, arranged in a thin cortex. (b) Temporal progression of neurogenesis in the mammalian dorsal pallium. Apical radial glia cells self‐amplify prior to begin producing deep‐layer neurons (dark gray). A long neurogenic period allows for the unfolding of genetic programs that drive the emergence of basal progenitors, such as intermediate progenitor cells and basal radial glia cells. Basal progenitors amplify the output and diversity of neurons produced, especially those destined to upper layers, producing an expanded six‐layered neocortex. Developmental time is not to scale.
A recent study concludes that differences in the timing of gene expression may account for some of the main differences of cortical development between marsupials and eutherians. Transcriptomic comparisons at early stages of corticogenesis reveal more mature gene networks in marsupials compared to mouse, which reverts at later stages, suggesting that neuronal maturation programs are triggered sooner and more prolonged in marsupials (Kozulin et al., 2022). While eutherians have a corpus callosum formed by axonal projections of cortical neurons specified by the TF Satb2, marsupials lack this feature and instead project via the anterior commissure. The differential timing in neuronal maturation programs between marsupials and eutherians may account for these differences, as a substantial delay in Satb2 expression in mouse seems to impact the fate of projection neurons (Kozulin et al., 2022; Paolino et al., 2020; Suárez et al., 2018). This may be related to a new regulation by miR‐541 (Martins et al., 2021).
A long neurogenic period may be important for the unfolding of refined genetic programs giving rise to new progenitor cell types and a greater diversity of neurons (Figure 5). For example, the embryonic cortex of gyrencephalic species contains a large diversity of basal progenitor cell types, especially in OSVZ and ISVZ, including a variety of bRGC morphotypes (Betizeau et al., 2013; Fietz et al., 2010; Hansen et al., 2010; Reillo et al., 2011). Ferret studies show that bRGCs destined to the OSVZ are massively produced during a restricted time window of embryonic development, and this is dynamically regulated by the expression levels of Cdh1 and Trnp1 (Martínez‐Martínez et al., 2016). The coincident downregulation of both genes is necessary to generate this burst of bRGCs, which then seed the OSVZ for subsequent self‐amplification, establishing a self‐sustained lineage. Before and after this brief period, high levels of either of these genes restrict the formation of bRGC. Experimental increase of expression during the critical period is also sufficient to impair bRGC production, indicating that the dynamic temporal regulation of these genes is key for the emergence of the OSVZ (Martínez‐Martínez et al., 2016; Stahl et al., 2013). Similarly, in macaque monkeys cortical neurogenesis is interrupted by an interim period of progenitor cell amplification, necessary prior to proceeding with the massive neurogenesis for supragranular layers (II and III) at late stages (Betizeau et al., 2013). The temporal control of gene expression is here also key to switch from neurogenesis to amplification, and back to neurogenesis.
Differences in gene expression regulation occur not only at the temporal and spatial levels, but also at the cellular level. In the mammalian cerebral cortex, the developmental transition from aRGCs to IPCs, and then to neurons, follows a stereotyped sequence of TF expression: Pax6, Tbr2, and Tbr1 (Elsen et al., 2018; Englund et al., 2005). Expression of these TFs is mutually exclusive in the smooth cortex of mouse and rat, but not in the large and folded cortices of ferret and macaque. In these species, Pax6 and Tbr2 are extensively co‐expressed in their highly proliferative basal progenitor cells (which in mouse only express Tbr2), suggesting that sustained Pax6 expression favors progenitor self‐renewal and/or proliferation (Betizeau et al., 2013; Reillo et al., 2011). Accordingly, artificially sustained expression of Pax6 in the embryonic mouse cortex leads to enlargement of the SVZ and increased abundance of basal progenitors, which exhibit primate‐like behavior with increased cell cycle re‐entry and production of upper layer neurons, as mentioned above (Wong et al., 2015).
Lineage tracing studies have analyzed the transcriptional identities of successive generations of cortical aRGCs and their progeny, identifying a set of evolutionary conserved genes that drive temporal aRGC progression, such as PRC2 (Telley et al., 2019). Temporally dependent molecular states are epigenetically transmitted to daughter neurons, thus regulating neuron identity in a temporal manner (Telley et al., 2019). This temporal regulation seems to become progressively restricted toward neuronal identity, as highlighted by experiments of heterochronic transplantation of aRGCs and IPCs from late into early embryos. While aRGCs show some temporal plasticity and can re‐enter previous molecular states, IPCs remain similarly committed (Oberst et al., 2019). In contrast to the temporal transitions of neurogenesis in the mammalian cortex, birds seem to have adopted a spatial transition, where different modes of neurogenesis and different types of dorsal pallial neurons are generated in different territories (Cárdenas et al., 2018; Suzuki et al., 2012). In this context, extrinsic factors seem to restrict the multipotency of NPCs in different areas of the avian pallium, resulting in divergent cytoarchitectural structures (Suzuki et al., 2012; Yamashita et al., 2018).
7. CONCLUDING REMARKS
The mammalian cerebral cortex is the ultimate achievement of brain evolution, following a dramatic increase in size and cellular complexity. Cortex size and complexity are the result of embryonic development, where a variety of progenitor cell types linked by defined lineage relationships proliferate and produce the full repertoire of neuron types. The evolutionary expansion of the cerebral cortex involved an increased proliferation of neural stem and progenitor cells, combined with the emergence of new types of neurogenic progenitors with greater amplificative potential. These phenotypic changes were based on genomic changes occurred during evolution, including the emergence of new genes and, most notably, the differential regulation of expression of conserved genes and signaling pathways, including changes in time and space. While some of these evolutionary changes have begun to be identified, in parallel to our increased understanding of the molecular regulation of cortical development, much remains uncharted. The advent of new analytical tools such as single‐cell omics and methods for genomic manipulation such as CRISPR‐mediated genome editing have open an entirely new world of possibilities for studying unconventional animal species with critically strategic phylogenetic value. Improved understanding of genomic and epigenomic mechanisms that regulate cortical development, and how these emerged and were differentially exploited during evolution, will help us unravel the natural history of human cognition.
CONFLICT OF INTEREST
The authors declare no conflict of interest.
ACKNOWLEDGMENTS
We thank people in our lab and in Scientific Program 1 at IN for helpful discussions. We apologize that, due to space limitations, many primary and historical publications could not be cited.
Espinós, A. , Fernández‐Ortuño, E. , Negri, E. , & Borrell, V. (2022). Evolution of genetic mechanisms regulating cortical neurogenesis. Developmental Neurobiology, 82, 428–453. 10.1002/dneu.22891
Alexandre Espinós, Eduardo Fernández‐Ortuño, and Enrico Negri contributed equally to this work and listed alphabetically.
REFERENCES
- Acemel, R. D. , Maeso, I. , & Gómez‐Skarmeta, J. L. (2017). Topologically associated domains: A successful scaffold for the evolution of gene regulation in animals. Wiley Interdisciplinary Reviews: Developmental Biology, 6, 1–19. 10.1002/wdev.265 [DOI] [PubMed] [Google Scholar]
- Amadei, G. , Zander, M. A. , Yang, G. , Dumelie, J. G. , Vessey, J. P. , Lipshitz, H. D. , Smibert, C. A. , Kaplan, D. R. , & Miller, F. D. (2015). A smaug2‐based translational repression complex determines the balance between precursor maintenance versus differentiation during mammalian neurogenesis. Journal of Neuroscience, 35, 15666–15681. 10.1523/JNEUROSCI.2172-15.2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antonacci, F. , Dennis, M. Y. , Huddleston, J. , Sudmant, P. H. , Steinberg, K. M. , Rosenfeld, J. A. , Miroballo, M. , Graves, T. A. , Vives, L. , Malig, M. , Denman, L. , Raja, A. , Stuart, A. , Tang, J. , Munson, B. , Shaffer, L. G. , Amemiya, C. T. , Wilson, R. K. , & Eichler, E. E. (2014). Palindromic GOLGA8 core duplicons promote chromosome 15q13.3 microdeletion and evolutionary instability. Nature Genetics, 46, 1293–1302. 10.1038/ng.3120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aprea, J. , & Calegari, F. (2015). Long non‐coding RNAs in corticogenesis: Deciphering the non‐coding code of the brain. EMBO Journal, 34, 2865–2884. 10.15252/embj.201592655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arcila, M. L. , Betizeau, M. , Cambronne, X. A. , Guzman, E. , Doerflinger, N. , Bouhallier, F. , Zhou, H. , Wu, B. , Rani, N. , Bassett, D. S. , Borello, U. , Huissoud, C. , Goodman, R. H. , Dehay, C. , & Kosik, K. S. (2014). Novel primate miRNAs coevolved with ancient target genes in germinal zone‐specific expression patterns. Neuron, 81, 1255–1262. 10.1016/j.neuron.2014.01.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arendt, D. , Musser, J. M. , Baker, C. V. H. , Bergman, A. , Cepko, C. , Erwin, D. H. , Pavlicev, M. , Schlosser, G. , Widder, S. , Laubichler, M. D. , & Wagner, G. P. (2016). The origin and evolution of cell types. Nature Reviews Genetics, 17, 744–757. 10.1038/nrg.2016.127 [DOI] [PubMed] [Google Scholar]
- Asami, M. , Pilz, G. A. , Ninkovic, J. , Godinho, L. , Schroeder, T. , Huttner, W. B. , & Götz, M. (2011). The role of Pax6 in regulating the orientation and mode of cell division of progenitors in the mouse cerebral cortex. Development, 138, 5067–5078. 10.1242/dev.074591 [DOI] [PubMed] [Google Scholar]
- Azevedo, F. A. C. , Carvalho, L. R. B. , Grinberg, L. T. , Farfel, J. M. , Ferretti, R. E. L. , Leite, R. E. P. , Filho, W. J. , Lent, R. , & Herculano‐Houzel, S. (2009). Equal numbers of neuronal and nonneuronal cells make the human brain an isometrically scaled‐up primate brain. Journal of Comparative Neurology, 513, 532–541. 10.1002/cne.21974 [DOI] [PubMed] [Google Scholar]
- Bailey, J. A. , Gu, Z. , Clark, R. A. , Reinert, K. , Samonte, R. V. , Schwartz, S. , Adams, M. D. , Myers, E. W. , Li, P. W. , & Eichler, E. E. (2002). Recent segmental duplications in the human genome. Science, 297, 1003–1007. 10.1126/science.1072047 [DOI] [PubMed] [Google Scholar]
- Barbosa‐Morais, N. L. , Irimia, M. , Pan, Q. , Xiong, H. Y. , Gueroussov, S. , Lee, L. J. , Slobodeniuc, V. , Kutter, C. , Watt, S. , Çolak, R. , Kim, T. H. , Misquitta‐Ali, C. M. , Wilson, M. D. , Kim, P. M. , Odom, D. T. , Frey, B. J. , & Blencowe, B. J. (2012). The evolutionary landscape of alternative splicing in vertebrate species. Science, 338, 1587–1593. 10.1126/science.1230612 [DOI] [PubMed] [Google Scholar]
- Bayer, S. A. , & Altman, J. (1991). Neocortical development. Raven Press. [Google Scholar]
- Belton, J. M. , McCord, R. P. , Gibcus, J. H. , Naumova, N. , Zhan, Y. , & Dekker, J. (2012). Hi‐C: A comprehensive technique to capture the conformation of genomes. Methods, 58, 268–276. 10.1016/j.ymeth.2012.05.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benito‐Kwiecinski, S. , Giandomenico, S. L. , Sutcliffe, M. , Riis, E. S. , Freire‐Pritchett, P. , Kelava, I. , Wunderlich, S. , Martin, U. , Wray, G. A. , McDole, K. , & Lancaster, M. A. (2021). An early cell shape transition drives evolutionary expansion of the human forebrain. Cell, 184, 2084.e19–2102.e19. 10.1016/j.cell.2021.02.050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betizeau, M. , Cortay, V. , Patti, D. , Pfister, S. , Gautier, E. , Bellemin‐Ménard, A. , Afanassieff, M. , Huissoud, C. , Douglas, R. J. , Kennedy, H. , & Dehay, C. (2013). Precursor diversity and complexity of lineage relationships in the outer subventricular zone of the primate. Neuron, 80, 442–457. 10.1016/j.neuron.2013.09.032 [DOI] [PubMed] [Google Scholar]
- Bonev, B. , Mendelson Cohen, N. , Szabo, Q. , Fritsch, L. , Papadopoulos, G. L. , Lubling, Y. , Xu, X. , Lv, X. , Hugnot, J. P. , Tanay, A. , & Cavalli, G. (2017). Multiscale 3D genome rewiring during mouse neural development. Cell, 171, 572.e24–572.e24. 10.1016/j.cell.2017.09.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boulder Committee . (1970). Embryonic vertebrate central nervous system: Revised terminology. Anatomical Record, 166, 257–261. [DOI] [PubMed] [Google Scholar]
- Boyd, J. L. , Skove, S. L. , Rouanet, J. P. , Pilaz, L. J. , Bepler, T. , Gordân, R. , Wray, G. A. , & Silver, D. L. (2015). Human‐chimpanzee differences in a FZD8 enhancer alter cell‐cycle dynamics in the developing neocortex. Current Biology, 25, 772–779. 10.1016/j.cub.2015.01.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briscoe, S. D. , & Ragsdale, C. W. (2018). Homology, neocortex, and the evolution of developmental mechanisms. Science, 362, 190–193. 10.1126/science.aau3711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brox, A. , Puelles, L. , Ferreiro, B. , & Medina, L. (2004). Expression of the genes Emx1, Tbr1, and Eomes (Tbr2) in the telencephalon of Xenopus laevis confirms the existence of a ventral pallial division in all tetrapods. Journal of Comparative Neurology, 474, 562–577. 10.1002/cne.20152 [DOI] [PubMed] [Google Scholar]
- Brunetti‐Pierri, N. , Berg, J. S. , Scaglia, F. , Belmont, J. , Bacino, C. A. , Sahoo, T. , Lalani, S. R. , Graham, B. , Lee, B. , Shinawi, M. , Shen, J. , Kang, S.‐H. L. , Pursley, A. , Lotze, T. , Kennedy, G. , Lansky‐Shafer, S. , Weaver, C. , Roeder, E. R. , Grebe, T. A. , … Patel, A. (2008). Recurrent reciprocal 1q21.1 deletions and duplications associated with microcephaly or macrocephaly and developmental and behavioral abnormalities. Nature Genetics, 40, 1466–1471. 10.1038/ng.279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckanovich, R. J. , & Darnell, R. B. (1997). The neuronal RNA binding protein Nova‐1 recognizes specific RNA targets in vitro and in vivo. Molecular and Cellular Biology, 17, 3194–3201. 10.1128/mcb.17.6.3194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burbano, H. A. , Green, R. E. , Maricic, T. , Lalueza‐Fox, C. , de la Rasilla, M. , Rosas, A. , Kelso, J. , Pollard, K. S. , Lachmann, M. , & Pääbo, S. (2012). Analysis of human accelerated DNA regions using archaic hominin genomes. PLoS ONE, 7, 1–8. 10.1371/journal.pone.0032877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camp, J. G. , Badsha, F. , Florio, M. , Kanton, S. , Gerber, T. , Wilsch‐Bräuninger, M. , Lewitus, E. , Sykes, A. , Hevers, W. , Lancaster, M. , Knoblich, J. A. , Lachmann, R. , Pääbo, S. , Huttner, W. B. , Treutlein, B. , Wilsch‐Brauninger, M. , Lewitus, E. , Sykes, A. , Hevers, W. , … Treutlein, B. (2015). Human cerebral organoids recapitulate gene expression programs of fetal neocortex development. Proceedings of the National Academy of Sciences of the United States of America, 112, 15672–15677. 10.1073/pnas.1520760112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capra, J. A. , Erwin, G. D. , McKinsey, G. , Rubenstein, J. L. , & Pollard, K. S. (2013). Many human accelerated regions are developmental enhancers. Philosophical Transactions of the Royal Society B: Biological Sciences, 368, 20130025. 10.1098/rstb.2013.0025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cárdenas, A. , Villalba, A. , de Juan Romero, C. , Picó, E. , Kyrousi, C. , Tzika, A. C. , Tessier‐Lavigne, M. , Ma, L. , Drukker, M. , Cappello, S. , & Borrell, V. (2018). Evolution of cortical neurogenesis in amniotes controlled by Robo signaling levels. Cell, 174, 590.e21–606.e21. 10.1016/j.cell.2018.06.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll, S. B. (2008). Evo‐devo and an expanding evolutionary synthesis: A genetic theory of morphological evolution. Cell, 134, 25–36. 10.1016/j.cell.2008.06.030 [DOI] [PubMed] [Google Scholar]
- Carroll, S. B. (2005). Evolution at two levels: On genes and form. PLoS Biology, 3, 1159–1166. 10.1371/journal.pbio.0030245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casarosa, S. , Fode, C. , & Guillemot, F. (1999). Mash1 regulates neurogenesis in the ventral telencephalon. Development, 126, 525–534. [DOI] [PubMed] [Google Scholar]
- Caviness, V. S., Jr. , Takahashi, T. , & Nowakowski, R. S. (1995). Numbers, time and neocortical neuronogenesis: A general developmental and evolutionary model. Trends in Neuroscience, 18, 379–383. [DOI] [PubMed] [Google Scholar]
- Chapman, H. , Riesenberg, A. , Ehrman, L. A. , Kohli, V. , Nardini, D. , Nakafuku, M. , Campbell, K. , & Waclaw, R. R. (2018). Gsx transcription factors control neuronal versus glial specification in ventricular zone progenitors of the mouse lateral ganglionic eminence. Developmental Biology, 442, 115–126. 10.1016/J.YDBIO.2018.07.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charrier, C. , Joshi, K. , Coutinho‐Budd, J. , Kim, J. E. , Lambert, N. , De Marchena, J. , Jin, W. L. , Vanderhaeghen, P. , Ghosh, A. , Sassa, T. , & Polleux, F. (2012). Inhibition of SRGAP2 function by its human‐specific paralogs induces neoteny during spine maturation. Cell, 149, 923–935. 10.1016/j.cell.2012.03.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, S. , Krinsky, B. H. , & Long, M. (2013). New genes as drivers of phenotypic evolution. Nature Reviews Genetics, 14, 645–660. 10.1038/nrg3521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, Y. C. , Kuo, H. Y. , Bornschein, U. , Takahashi, H. , Chen, S. Y. , Lu, K. M. , Yang, H. Y. , Chen, G. M. , Lin, J. R. , Lee, Y. H. , Chou, Y. C. , Cheng, S. J. , Chien, C. T. , Enard, W. , Hevers, W. , Pääbo, S. , Graybiel, A. M. , & Liu, F. C. (2016). Foxp2 controls synaptic wiring of corticostriatal circuits and vocal communication by opposing Mef2c. Nature Neuroscience, 19, 1513–1522. 10.1038/nn.4380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chenn, A. , & Walsh, C. A. (2002). Regulation of cerebral cortical size by control of cell cycle exit in neural precursors. Science, 297, 365–369. [DOI] [PubMed] [Google Scholar]
- Chinnappa, K. , Cárdenas, A. , Prieto‐Colomina, A. , Villalba, A. , Márquez‐Galera, Á. , Soler, R. , Nomura, Y. , Llorens, E. , Tomasello, U. , López‐Atalaya, J. P. , & Borrell, V. (2022). Secondary loss of miR‐3607 reduced cortical progenitor amplification during rodent evolution. Science Advances, 8, eabj4010. 10.1126/sciadv.abj4010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou, S. J. , Wang, C. , Sintupisut, N. , Niou, Z. X. , Lin, C. H. , Li, K. C. , & Yeang, C. H. (2016). Analysis of spatial‐temporal gene expression patterns reveals dynamics and regionalization in developing mouse brain. Science Reports, 6, 1–13. 10.1038/srep19274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clinton, B. K. , Cunningham, C. L. , Kriegstein, A. R. , Noctor, S. C. , & Martinez‐Cerdeno, V. (2014). Radial glia in the proliferative ventricular zone of the embryonic and adult turtle, Trachemys scripta elegans . Neurogenesis, 1, e970905. 10.4161/23262125.2014.970905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corbin, J. G. , Gaiano, N. , Machold, R. P. , Langston, A. , & Fishell, G. (2000). The Gsh2 homeodomain gene controls multiple aspects of telencephalic development. Development, 127, 5007–5020. [DOI] [PubMed] [Google Scholar]
- De Pietri Tonelli, D. , Pulvers, J. N. , Haffner, C. , Murchison, E. P. , Hannon, G. J. , & Huttner, W. B. (2008). miRNAs are essential for survival and differentiation of newborn neurons but not for expansion of neural progenitors during early neurogenesis in the mouse embryonic neocortex. Development, 135, 3911–3921. 10.1242/dev.025080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dehay, C. , Giroud, P. , Berland, M. , Smart, I. , & Kennedy, H. (1993). Modulation of the cell cycle contributes to the parcellation of the primate visual cortex. Nature, 366, 464–466. [DOI] [PubMed] [Google Scholar]
- Dehay, C. , & Kennedy, H. (2007). Cell‐cycle control and cortical development. Nature Reviews Neuroscience, 8, 438–450. [DOI] [PubMed] [Google Scholar]
- Dennis, M. Y. , & Eichler, E. E. (2016). Human adaptation and evolution by segmental duplication. Current Opinion in Genetics & Development, 41, 44–52. 10.1016/j.gde.2016.08.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennis, M. Y. , Harshman, L. , Nelson, B. J. , Penn, O. , Cantsilieris, S. , Huddleston, J. , Antonacci, F. , Penewit, K. , Denman, L. , Raja, A. , Baker, C. , Mark, K. , Malig, M. , Janke, N. , Espinoza, C. , Stessman, H. A. F. , Nuttle, X. , Hoekzema, K. , Lindsay‐Graves, T. A. , … Eichler, E. E. (2017). The evolution and population diversity of human‐specific segmental duplications. Nature Ecology & Evolution, 1, 69. 10.1038/s41559-016-0069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennis, M. Y. , Nuttle, X. , Sudmant, P. H. , Antonacci, F. , Graves, T. A. , Nefedov, M. , Rosenfeld, J. A. , Sajjadian, S. , Malig, M. , Kotkiewicz, H. , Curry, C. J. , Shafer, S. , Shaffer, L. G. , de Jong, P. J. , Wilson, R. K. , & Eichler, E. E. (2012). Evolution of human‐specific neural SRGAP2 genes by incomplete segmental duplication. Cell, 149, 912–922. 10.1016/j.cell.2012.03.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz, J. L. , Siththanandan, V. B. , Lu, V. , Gonzalez‐Nava, N. , Pasquina, L. , MacDonald, J. L. , Woodworth, M. B. , Ozkan, A. , Nair, R. , He, Z. , Sahni, V. , Sarnow, P. , Palmer, T. D. , Macklis, J. D. , & Tharin, S. (2020). An evolutionarily acquired microRNA shapes development of mammalian cortical projections. Proceedings of the National Academy of Sciences of the United States of America, 117, 29113–29122. 10.1073/pnas.2006700117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon, J. R. , Selvaraj, S. , Yue, F. , Kim, A. , Li, Y. , Shen, Y. , Hu, M. , Liu, J. S. , & Ren, B. (2012). Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature, 485, 376–380. 10.1038/nature11082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Docampo‐Seara, A. , Lagadec, R. , Mazan, S. , Rodríguez, M. A. , Quintana‐Urzainqui, I. , Candal, E. , Rodriguez, M. A. , Quintana‐Urzainqui, I. , & Candal, E. (2018). Study of pallial neurogenesis in shark embryos and the evolutionary origin of the subventricular zone. Brain Structure and Function, 223, 3593–3612. 10.1007/s00429-018-1705-2 [DOI] [PubMed] [Google Scholar]
- Doggett, N. A. , Xie, G. , Meincke, L. J. , Sutherland, R. D. , Mundt, M. O. , Berbari, N. S. , Davy, B. E. , Robinson, M. L. , Rudd, M. K. , Weber, J. L. , Stallings, R. L. , & Han, C. (2006). A 360‐kb interchromosomal duplication of the human HYDIN locus. Genomics, 88, 762–771. 10.1016/j.ygeno.2006.07.012 [DOI] [PubMed] [Google Scholar]
- Dorus, S. , Vallender, E. J. , Evans, P. D. , Anderson, J. R. , Gilbert, S. L. , Mahowald, M. , Wyckoff, G. J. , Malcom, C. M. , & Lahn, B. T. (2004). Accelerated evolution of nervous system genes in the origin of Homo sapiens. Cell, 119, 1027–1040. [DOI] [PubMed] [Google Scholar]
- Dougherty, M. L. , Nuttle, X. , Penn, O. , Nelson, B. J. , Huddleston, J. , Baker, C. , Harshman, L. , Duyzend, M. H. , Ventura, M. , Antonacci, F. , Sandstrom, R. , Dennis, M. Y. , & Eichler, E. E. (2017). The birth of a human‐specific neural gene by incomplete duplication and gene fusion. Genome Biology, 18, 49. 10.1186/s13059-017-1163-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dugas‐Ford, J. , & Ragsdale, C. W. (2015). Levels of homology and the problem of neocortex. Annual Review of Neuroscience, 38, 351–368. 10.1146/annurev-neuro-071714-033911 [DOI] [PubMed] [Google Scholar]
- Elsen, G. E. , Bedogni, F. , Hodge, R. D. , Bammler, T. K. , MacDonald, J. W. , Lindtner, S. , Rubenstein, J. L. R. , & Hevner, R. F. (2018). The epigenetic factor landscape of developing neocortex is regulated by transcription factors Pax6→ Tbr2→ Tbr1. Frontiers in Neuroscience, 12, 571. 10.3389/fnins.2018.00571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enard, W. , Gehre, S. , Hammerschmidt, K. , Hölter, S. M. , Blass, T. , Somel, M. , Brückner, M. K. , Schreiweis, C. , Winter, C. , Sohr, R. , Becker, L. , Wiebe, V. , Nickel, B. , Giger, T. , Müller, U. , Groszer, M. , Adler, T. , Aguilar, A. , Bolle, I. , … Pääbo, S. (2009). A humanized version of Foxp2 affects cortico‐basal ganglia circuits in mice. Cell, 137, 961–971. 10.1016/j.cell.2009.03.041 [DOI] [PubMed] [Google Scholar]
- Enard, W. , Przeworsky Simone, E. M. , Lai, C. S. L. , Wiebe, V. , Kitano, T. , Monaco, A. P. , Paabo, S. , Przeworski, M. , Fisher, S. E. , Lai, C. S. L. , Wiebe, V. , Kitano, T. , Monaco, A. P. , & Paabo, S. (2002). Molecular evolution of FOXP2, a gene involved in speech and language. Nature, 418, 869–872. 10.1038/nature01025 [DOI] [PubMed] [Google Scholar]
- Englund, C. , Fink, A. , Lau, C. , Pham, D. , Daza, R. A. M. , Bulfone, A. , Kowalczyk, T. , & Hevner, R. F. (2005). Pax6, Tbr2, and Tbr1 are expressed sequentially by radial glia, intermediate progenitor cells, and postmitotic neurons in developing neocortex. Journal of Neuroscience, 25, 247–251. 10.1523/JNEUROSCI.2899-04.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estivill‐Torrus, G. , Pearson, H. , van Heyningen, V. , Price, D. J. , & Rashbass, P. (2002). Pax6 is required to regulate the cell cycle and the rate of progression from symmetrical to asymmetrical division in mammalian cortical progenitors. Development, 129, 455–466. [DOI] [PubMed] [Google Scholar]
- Feng, Y. , & Walsh, C. A. (2004). Mitotic spindle regulation by Nde1 controls cerebral cortical size. Neuron, 44, 279–293. 10.1016/j.neuron.2004.09.023 [DOI] [PubMed] [Google Scholar]
- Ferland, R. J. , Cherry, T. J. , Preware, P. O. , Morrisey, E. E. , & Walsh, C. A. (2003). Characterization of Foxp2 and Foxp1 mRNA and protein in the developing and mature brain. Journal of Comparative Neurology, 460, 266–279. 10.1002/cne.10654 [DOI] [PubMed] [Google Scholar]
- Fernandez, A. S. , Pieau, C. , Repérant, J. , Boncinelli, E. , & Wassef, M. (1998). Expression of the Emx‐1 and Dlx‐1 homeobox genes define three molecularly distinct domains in the telencephalon of mouse, chick, turtle and frog embryos: Implications for the evolution of telencephalic subdivisions in amniotes. Development, 125, 2099–2111. 10.1242/dev.125.11.2099 [DOI] [PubMed] [Google Scholar]
- Fernandez, V. , Llinares‐Benadero, C. , Borrell, V. , Fernández, V. , Llinares‐Benadero, C. , & Borrell, V. (2016). Cerebral cortex expansion and folding: What have we learned? EMBO Journal, 35, 1021–1044. 10.15252/embj.201593701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernández, V. , Martínez‐Martínez, M. Á. , Prieto‐Colomina, A. , Cárdenas, A. , Soler, R. , Dori, M. , Tomasello, U. , Nomura, Y. , López‐Atalaya, J. P. , Calegari, F. , & Borrell, V. (2020). Repression of Irs2 by let‐7 miRNAs is essential for homeostasis of the telencephalic neuroepithelium. EMBO Journal, 39, e105479. 10.15252/embj.2020105479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feuk, L. , Kalervo, A. , Lipsanen‐Nyman, M. , Skaug, J. , Nakabayashi, K. , Finucane, B. , Hartung, D. , Innes, M. , Kerem, B. , Nowaczyk, M. J. , Rivlin, J. , Roberts, W. , Senman, L. , Summers, A. , Szatmari, P. , Wong, V. , Vincent, J. B. , Zeesman, S. , Osborne, L. R. , … Hannula‐Jouppi, K. (2006). Absence of a paternally inherited FOXP2 gene in developmental verbal dyspraxia. American Journal of Human Genetics, 79, 965–972. 10.1086/508902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiddes, I. T. , Lodewijk, G. A. , Mooring, M. , Bosworth, C. M. , Ewing, A. D. , Mantalas, G. L. , Novak, A. M. , van den Bout, A. , Bishara, A. , Rosenkrantz, J. L. , Lorig‐Roach, R. , Field, A. R. , Haeussler, M. , Russo, L. , Bhaduri, A. , Nowakowski, T. J. , Pollen, A. A. , Dougherty, M. L. , Nuttle, X. , … Haussler, D. (2018). Human‐specific NOTCH2NL genes affect notch signaling and cortical neurogenesis. Cell, 173, 1356.e22–1369.e22. 10.1016/j.cell.2018.03.051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fietz, S. A. , Kelava, I. , Vogt, J. , Wilsch‐Brauninger, M. , Stenzel, D. , Fish, J. L. , Corbeil, D. , Riehn, A. , Distler, W. , Nitsch, R. , Huttner, W. B. , Wilsch‐Bräuninger, M. , Stenzel, D. , Fish, J. L. , Corbeil, D. , Riehn, A. , Distler, W. , Nitsch, R. , & Huttner, W. B. (2010). OSVZ progenitors of human and ferret neocortex are epithelial‐like and expand by integrin signaling. Nature Neuroscience, 13, 690–699. 10.1038/nn.2553 [DOI] [PubMed] [Google Scholar]
- Fietz, S. A. , Lachmann, R. , Brandl, H. , Kircher, M. , Samusik, N. , Schröder, R. , Lakshmanaperumal, N. , Henry, I. , Vogt, J. , Riehn, A. , Distler, W. , Nitsch, R. , Enard, W. , Pääbo, S. , Huttner, W. B. , Schroder, R. , Lakshmanaperumal, N. , Henry, I. , Vogt, J. , … Huttner, W. B. (2012). Transcriptomes of germinal zones of human and mouse fetal neocortex suggest a role of extracellular matrix in progenitor self‐renewal. Proceedings of the National Academy of Sciences of the United States of America, 109, 11836–11841. 10.1073/pnas.1209647109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer, J. , Peters, J. , Namba, T. , Huttner, W. B. , & Heide, M. (2020). Human and chimpanzee cerebral organoids reveal that human‐specific ARHGAP11B is necessary and sufficient for a human‐type basal progenitor abundance. bioRxiv, 10.1101/2020.10.01.322792 [DOI] [Google Scholar]
- Florio, M. , Albert, M. , Taverna, E. , Namba, T. , Brandl, H. , Lewitus, E. , Haffner, C. , Sykes, A. , Wong, F. K. , Peters, J. , Guhr, E. , Klemroth, S. , Prufer, K. , Kelso, J. , Naumann, R. , Nusslein, I. , Dahl, A. , Lachmann, R. , Paabo, S. , … Huttner, W. B. (2015). Human‐specific gene ARHGAP11B promotes basal progenitor amplification and neocortex expansion. Science, 347, 1465–1470. 10.1126/science.aaa1975 [DOI] [PubMed] [Google Scholar]
- Florio, M. , Borrell, V. , & Huttner, W. B. (2017). Human‐specific genomic signatures of neocortical expansion. Current Opinion in Neurobiology, 42, 33–44. 10.1016/j.conb.2016.11.004 [DOI] [PubMed] [Google Scholar]
- Florio, M. , Heide, M. , Pinson, A. , Brandl, H. , Albert, M. , Winkler, S. , Wimberger, P. , Huttner, W. B. , & Hiller, M. (2018). Evolution and cell‐type specificity of human‐specific genes preferentially expressed in progenitors of fetal neocortex. Elife, 7, e32332. 10.7554/eLife.32332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Florio, M. , & Huttner, W. B. (2014). Neural progenitors, neurogenesis and the evolution of the neocortex. Development, 141, 2182–2194. 10.1242/dev.090571 [DOI] [PubMed] [Google Scholar]
- Florio, M. , Namba, T. , Paabo, S. , Hiller, M. , & Huttner, W. B. (2016). A single splice site mutation in human‐specific ARHGAP11B causes basal progenitor amplification. Science Advances, 2, e1601941. 10.1126/sciadv.1601941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fortna, A. , Kim, Y. , MacLaren, E. , Marshall, K. , Hahn, G. , Meltesen, L. , Brenton, M. , Hink, R. , Burgers, S. , Hernandez‐Boussard, T. , Karimpour‐Fard, A. , Glueck, D. , McGavran, L. , Berry, R. , Pollack, J. , & Sikela, J. M. (2004). Lineage‐specific gene duplication and loss in human and great ape evolution. PLoS Biology, 2, 937–954. 10.1371/journal.pbio.0020207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fossati, M. , Pizzarelli, R. , Schmidt, E. R. , Kupferman, J. V. , Stroebel, D. , Polleux, F. , & Charrier, C. (2016). SRGAP2 and its human‐specific paralog co‐regulate the development of excitatory and inhibitory synapses. Neuron, 91, 356–369. 10.1016/j.neuron.2016.06.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu, Y. , Dominissini, D. , Rechavi, G. , & He, C. (2014). Gene expression regulation mediated through reversible m 6 A RNA methylation. Nature Reviews Genetics, 15, 293–306. 10.1038/nrg3724 [DOI] [PubMed] [Google Scholar]
- Gaiano, N. , Nye, J. S. , & Fishell, G. (2000). Radial glial identity is promoted by Notch1 signaling in the murine forebrain. Neuron, 26, 395–404. 10.1016/S0896-6273(00)81172-1 [DOI] [PubMed] [Google Scholar]
- García‐moreno, F. , & Molnár, Z. (2020). Variations of telencephalic development that paved the way for neocortical evolution. Progress in Neurobiology, 194, 101865. 10.1016/j.pneurobio.2020.101865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girskis, K. M. , Stergachis, A. B. , DeGennaro, E. M. , Doan, R. N. , Qian, X. , Johnson, M. B. , Wang, P. P. , Sejourne, G. M. , Nagy, M. A. , Pollina, E. A. , Sousa, A. M. M. , Shin, T. , Kenny, C. J. , Scotellaro, J. L. , Debo, B. M. , Gonzalez, D. M. , Rento, L. M. , Yeh, R. C. , Song, J. H. T. , … Walsh, C. A. (2021). Rewiring of human neurodevelopmental gene regulatory programs by human accelerated regions. Neuron, 109, 3239.e7–3251.e7. 10.1016/j.neuron.2021.08.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez‐Sandoval, A. , & Gasser, S. M. (2016). On TADs and LADs: Spatial control over gene expression. Trends in Genetics, 32, 485–495. 10.1016/j.tig.2016.05.004 [DOI] [PubMed] [Google Scholar]
- Götz, M. , & Huttner, W. B. (2005). The cell biology of neurogenesis. Nature Reviews Molecular Cell Biology, 6, 777–788. 10.1038/nrm1739 [DOI] [PubMed] [Google Scholar]
- Grassi, D. A. , Brattås, P. L. , Valdés, J. G. , Rezeli, M. , Jönsson, M. E. , Nolbrant, S. , Parmar, M. , Marko‐Varga, G. , & Jakobsson, J. (2019). Post‐transcriptional mechanisms distinguish human and chimp forebrain progenitor cells. bioRxiv, 10.1101/582197 [DOI] [Google Scholar]
- Gregory, S. G. , Barlow, K. F. , McLay, K. E. , Kaul, R. , Swarbreck, D. , Dunham, A. , Scott, C. E. , Howe, K. L. , Woodfine, K. , Spencer, C. C. A. , Jones, M. C. , Gillson, C. , Searle, S. , Zhou, Y. , Kokocinski, F. , McDonald, L. , Evans, R. , Phillips, K. , Atkinson, A. , … Bentley, D. R. (2006). The DNA sequence and biological annotation of human chromosome 1. Nature, 441, 315–321. 10.1038/nature04727 [DOI] [PubMed] [Google Scholar]
- Hansen, T. B. , Jensen, T. I. , Clausen, B. H. , Bramsen, J. B. , Finsen, B. , Damgaard, C. K. , & Kjems, J. (2013). Natural RNA circles function as efficient microRNA sponges. Nature, 495, 384–388. 10.1038/nature11993 [DOI] [PubMed] [Google Scholar]
- Hansen, D. V. , Lui, J. H. , Parker, P. R. L. L. , & Kriegstein, A. R. (2010). Neurogenic radial glia in the outer subventricular zone of human neocortex. Nature, 464, 554–561. 10.1038/nature08845 [DOI] [PubMed] [Google Scholar]
- Haubensak, W. , Attardo, A. , Denk, W. , & Huttner, W. B. (2004). Neurons arise in the basal neuroepithelium of the early mammalian telencephalon: A major site of neurogenesis. Proceedings of the National Academy of Sciences of the United States of America, 101, 3196–3201. 10.1073/pnas.0308600100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haygood, R. , Babbitt, C. C. , Fedrigo, O. , & Wray, G. A. (2010). Contrasts between adaptive coding and noncoding changes during human evolution. Proceedings of the National Academy of Sciences of the United States of America, 107, 7853–7857. 10.1073/pnas.0911249107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haygood, R. , Fedrigo, O. , Hanson, B. , Yokoyama, K. D. , & Wray, G. A. (2007). Promoter regions of many neural‐ and nutrition‐related genes have experienced positive selection during human evolution. Nature Genetics, 39, 1140–1144. 10.1038/ng2104 [DOI] [PubMed] [Google Scholar]
- Heide, M. , Haffner, C. , Murayama, A. , Kurotaki, Y. , Shinohara, H. , Okano, H. , Sasaki, E. , & Huttner, W. B. (2020). Human‐specific ARHGAP11B increases size and folding of primate neocortex in the fetal marmoset. Science, 369, 546–550. 10.1126/science.abb2401 [DOI] [PubMed] [Google Scholar]
- Heide, M. , & Huttner, W. B. (2021). Human‐specific genes, cortical progenitor cells, and microcephaly. Cells, 10, 1209. 10.3390/CELLS10051209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heide, M. , Long, K. R. , & Huttner, W. B. (2017). Novel gene function and regulation in neocortex expansion. Current Opinion in Cell Biology, 49, 22–30. 10.1016/j.ceb.2017.11.008 [DOI] [PubMed] [Google Scholar]
- Heins, N. , Malatesta, P. , Cecconi, F. , Nakafuku, M. , Tucker, K. L. , Hack, M. A. , Chapouton, P. , Barde, Y. A. , & Goötz, M. (2002). Glial cells generate neurons: The role of the transcription factor Pax6. Nature Neuroscience, 5, 308–315. 10.1038/nn828 [DOI] [PubMed] [Google Scholar]
- Herculano‐Houzel, S. (2009). The human brain in numbers: A linearly scaled‐up primate brain. Frontiers in Human Neuroscience, 3, 31. 10.3389/neuro.09.031.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodzic, D. , Kong, C. , Wainszelbaum, M. J. , Charron, A. J. , Su, X. , & Stahl, P. D. (2006). TBC1D3, a hominoid oncoprotein, is encoded by a cluster of paralogues located on chromosome 17q12. Genomics, 88, 731–736. 10.1016/j.ygeno.2006.05.009 [DOI] [PubMed] [Google Scholar]
- Holloway, A. K. , Bruneau, B. G. , Sukonnik, T. , Rubenstein, J. L. , & Pollard, K. S. (2016). Accelerated evolution of enhancer hotspots in the mammal ancestor. Molecular Biology and Evolution, 33, 1008–1018. 10.1093/molbev/msv344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holm, P. C. , Mader, M. T. , Haubst, N. , Wizenmann, A. , Sigvardsson, M. , & Götz, M. (2007). Loss‐ and gain‐of‐function analyses reveal targets of Pax6 in the developing mouse telencephalon. Molecular and Cellular Neuroscience, 34, 99–119. 10.1016/j.mcn.2006.10.008 [DOI] [PubMed] [Google Scholar]
- Hoye, M. L. , & Silver, D. L. (2021). Decoding mixed messages in the developing cortex: Translational regulation of neural progenitor fate. Current Opinion in Neurobiology, 66, 93–102. 10.1016/j.conb.2020.10.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu, L. C. L. , Nam, S. , Cui, Y. , Chang, C. P. , Wang, C. F. , Kuo, H. C. , Touboul, J. D. , & Chou, S. J. (2015). Lhx2 regulates the timing of β‐catenin‐dependent cortical neurogenesis. Proceedings of the National Academy of Sciences of the United States of America, 112, 12199–12204. 10.1073/pnas.1507145112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu, P. J. , Shi, H. , Zhu, A. C. , Lu, Z. , Miller, N. , Edens, B. M. , Ma, Y. C. , & He, C. (2019). The RNA‐binding protein FMRP facilitates the nuclear export of N6‐methyladenosine–containing mRNAs. Journal of Biological Chemistry, 294, 19889–19895. 10.1074/jbc.AC119.010078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubisz, M. J. , & Pollard, K. S. (2014). Exploring the genesis and functions of Human Accelerated Regions sheds light on their role in human evolution. Current Opinion in Genetics & Development, 29, 15–21. 10.1016/j.gde.2014.07.005 [DOI] [PubMed] [Google Scholar]
- Imayoshi, I. , Sakamoto, M. , Yamaguchi, M. , Mori, K. , & Kageyama, R. (2010). Essential roles of Notch signaling in maintenance of neural stem cells in developing and adult brains. Journal of Neuroscience, 30, 3489–3498. 10.1523/JNEUROSCI.4987-09.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izsvák, Z. , Wang, J. , Singh, M. , Mager, D. L. , & Hurst, L. D. (2016). Pluripotency and the endogenous retrovirus HERVH: Conflict or serendipity? Bioessays, 38, 109–117. 10.1002/BIES.201500096 [DOI] [PubMed] [Google Scholar]
- Jensen, K. B. , Dredge, B. K. , Stefani, G. , Zhong, R. , Buckanovich, R. J. , Okano, H. J. , Yang, Y. Y. L. , & Darnell, R. B. (2000). Nova‐1 regulates neuron‐specific alternative splicing and is essential for neuronal viability. Neuron, 25, 359–371. 10.1016/S0896-6273(00)80900-9 [DOI] [PubMed] [Google Scholar]
- Jeong, H. , Mendizabal, I. , Berto, S. , Chatterjee, P. , Layman, T. , Usui, N. , Toriumi, K. , Douglas, C. , Singh, D. , Huh, I. , Preuss, T. M. , Konopka, G. , & Yi, S. V. (2021). Evolution of DNA methylation in the human brain. Nature Communication, 12, 2021. 10.1038/s41467-021-21917-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson, M. B. , Sun, X. , Kodani, A. , Borges‐Monroy, R. , Girskis, K. M. , Ryu, S. C. , Wang, P. P. , Patel, K. , Gonzalez, D. M. , Woo, Y. M. , Yan, Z. , Liang, B. , Smith, R. S. , Chatterjee, M. , Coman, D. , Papademetris, X. , Staib, L. H. , Hyder, F. , Mandeville, J. B. , … Bae, B.‐I. I. (2018). Aspm knockout ferret reveals an evolutionary mechanism governing cerebral cortical size. Nature, 556, 370–375. 10.1038/s41586-018-0035-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ju, X.‐C. C. , Hou, Q.‐Q. Q. , Sheng, A.‐L. L. S. , Wu, K.‐Y. Y. , Zhou, Y. , Jin, Y. , Wen, T. , Yang, Z. , Wang, X. , & Luo, Z.‐G. G. (2016). The hominoid‐specific gene TBC1D3 promotes generation of basal neural progenitors and induces cortical folding in mice. Elife, 5, e18197. 10.7554/eLife.18197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kageyama, R. , Ohtsuka, T. , Shimojo, H. , & Imayoshi, I. (2009). Dynamic regulation of Notch signaling in neural progenitor cells. Current Opinion in Cell Biology, 21, 733–740. 10.1016/j.ceb.2009.08.009 [DOI] [PubMed] [Google Scholar]
- Kalebic, N. , Gilardi, C. , Albert, M. , Namba, T. , Long, K. R. , Kostic, M. , Langen, B. , & Huttner, W. B. (2018). Human‐specific ARHGAP11B induces hallmarks of neocortical expansion in developing ferret neocortex. Elife, 7, e41241. 10.7554/eLife.41241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanton, S. , Boyle, M. J. , He, Z. , Santel, M. , Weigert, A. , Sanchís‐Calleja, F. , Guijarro, P. , Sidow, L. , Fleck, J. S. , Han, D. , Qian, Z. , Heide, M. , Huttner, W. B. , Khaitovich, P. , Pääbo, S. , Treutlein, B. , & Camp, J. G. (2019). Organoid single‐cell genomic atlas uncovers human‐specific features of brain development. Nature, 574, 418–422. 10.1038/s41586-019-1654-9 [DOI] [PubMed] [Google Scholar]
- Katju, V. , & Bergthorsson, U. (2013). Copy‐number changes in evolution: Rates, fitness effects and adaptive significance. Frontiers in Genetics, 4, 1–12. 10.3389/fgene.2013.00273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keeney, J. G. , Davis, J. M. , Siegenthaler, J. A. , Post, M. D. , Nielsen, B. S. , Hopkins, W. D. , & Sikela, J. M. (2015). DUF1220 protein domains drive proliferation in human neural stem cells and are associated with increased cortical volume in anthropoid primates. Brain Structure and Function, 220, 3053–3060. 10.1007/s00429-014-0814-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keeney, J. G. , Dumas, L. , & Sikela, J. M. (2014). The case for DUF1220 domain dosage as a primary contributor to anthropoid brain expansion. Frontiers in Human Neuroscience, 8, 427. 10.3389/fnhum.2014.00427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerimoglu, C. , Pham, L. , Tonchev, A. B. , Sakib, M. S. , Xie, Y. , Sokpor, G. , Ulmke, P. A. , Kaurani, L. , Abbas, E. , Nguyen, H. , Rosenbusch, J. , Michurina, A. , Capece, V. , Angelova, M. , Maricic, N. , Brand‐Saberi, B. , Esgleas, M. , Albert, M. , Minkov, R. , … Tuoc, T. (2021). H3 acetylation selectively promotes basal progenitor proliferation and neocortex expansion. Science Advances, 7, 1–18. 10.1126/sciadv.abc6792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kishi, Y. , Fujii, Y. , Hirabayashi, Y. , & Gotoh, Y. (2012). HMGA regulates the global chromatin state and neurogenic potential in neocortical precursor cells. Nature Neuroscience, 15, 1127–1133. 10.1038/nn.3165 [DOI] [PubMed] [Google Scholar]
- Konopka, G. , Bomar, J. M. , Winden, K. , Coppola, G. , Jonsson, Z. O. , Gao, F. , Peng, S. , Preuss, T. M. , Wohlschlegel, J. A. , & Geschwind, D. H. (2009). Human‐specific transcriptional regulation of CNS development genes by FOXP2. Nature, 462, 213–217. 10.1038/nature08549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornack, D. R. , & Rakic, P. (1998). Changes in cell‐cycle kinetics during the development and evolution of primate neocortex. Proceedings of the National Academy of Sciences of the United States of America, 95, 1242–1246. 10.1073/pnas.95.3.1242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosik, K. S. , & Nowakowski, T. (2018). Evolution of new miRNAs and cerebro‐cortical development. Annual Review of Neuroscience, 41, 119–137. 10.1146/annurev-neuro-080317-061822 [DOI] [PubMed] [Google Scholar]
- Kowalczyk, T. , Pontious, A. , Englund, C. , Daza, R. A. M. , Bedogni, F. , Hodge, R. , Attardo, A. , Bell, C. , Huttner, W. B. , & Hevner, R. F. (2009). Intermediate neuronal progenitors (basal progenitors) produce pyramidal–projection neurons for all layers of cerebral cortex. Cerebral Cortex, 19, 2439–2450. 10.1093/cercor/bhn260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozulin, P. , Suárez, R. , Zhao, Q. Y. , Paolino, A. , Richards, L. J. , & Fenlon, L. R. (2022). Divergent evolution of developmental timing in the neocortex revealed by marsupial and eutherian transcriptomes. Development, 149, dev200212. 10.1242/DEV.200212 [DOI] [PubMed] [Google Scholar]
- Kriegstein, A. , Noctor, S. , & Martinez‐Cerdeno, V. (2006). Patterns of neural stem and progenitor cell division may underlie evolutionary cortical expansion. Nature Reviews Neuroscience, 7, 883–890. [DOI] [PubMed] [Google Scholar]
- Kverková, K. , Marhounová, L. , Polonyiová, A. , Kocourek, M. , Zhang, Y. , Olkowicz, S. , Straková, B. , Pavelková, Z. , Vodička, R. , Frynta, D. , & Němec, P. (2022). The evolution of brain neuron numbers in amniotes. Proceedings of the National Academy of Sciences of the United States of America, 119, e2121624119. 10.1073/PNAS.2121624119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kvon, E. Z. , Kamneva, O. K. , Melo, U. S. , Barozzi, I. , Osterwalder, M. , Mannion, B. J. , Tissières, V. , Pickle, C. S. , Plajzer‐Frick, I. , Lee, E. A. , Kato, M. , Garvin, T. H. , Akiyama, J. A. , Afzal, V. , Lopez‐Rios, J. , Rubin, E. M. , Dickel, D. E. , Pennacchio, L. A. , & Visel, A. (2016). Progressive loss of function in a limb enhancer during snake evolution. Cell, 167, 633.e11–642.e11. 10.1016/j.cell.2016.09.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai, C. S. , Fisher, S. E. , Hurst, J. A. , Vargha‐Khadem, F. , & Monaco, A. P. (2001). A forkhead‐domain gene is mutated in a severe speech and language disorder. Nature, 413, 519–523. 10.1038/35097076 [DOI] [PubMed] [Google Scholar]
- LaMonica, B. E. , Lui, J. H. , Hansen, D. V. , & Kriegstein, A. R. (2013). Mitotic spindle orientation predicts outer radial glial cell generation in human neocortex. Nature Communication, 4, 1665. 10.1038/ncomms2647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latchman, D. S. (1997). Transcription factors: An overview. International Journal of Biochemistry & Cell Biology, 29, 1305–1312. 10.1016/S1357-2725(97)00085-X [DOI] [PubMed] [Google Scholar]
- Lennox, A. L. , Mao, H. , & Silver, D. L. (2018). RNA on the brain: Emerging layers of post‐transcriptional regulation in cerebral cortex development. Wiley Interdisciplinary Reviews: Developmental Biology, 7, 1–18. 10.1002/wdev.290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lister, R. , Mukamel, E. A. , Nery, J. R. , Urich, M. , Puddifoot, C. A. , Johnson, N. D. , Lucero, J. , Huang, Y. , Dwork, A. J. , Schultz, E. D. , Yu, M. , Tonti‐filippini, J. , Heyn, H. , Hu, S. , Wu, J. C. , Rao, A. , Esteller, M. , He, C. , Haghighi, F. G. , … Ecker, J. R. (2013). Global epigenomic reconfiguration during mammalian brain development. Science, 341, 1237905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llinares‐Benadero, C. , & Borrell, V. (2019). Deconstructing cortical folding: Genetic, cellular and mechanical determinants. Nature Reviews Neuroscience, 20, 161–176. 10.1038/s41583-018-0112-2 [DOI] [PubMed] [Google Scholar]
- Lukaszewicz, A. , Savatier, P. , Cortay, V. , Giroud, P. , Huissoud, C. , Berland, M. , Kennedy, H. , & Dehay, C. (2005). G1 phase regulation, area‐specific cell cycle control, and cytoarchitectonics in the primate cortex. Neuron, 47, 353–364. 10.1016/j.neuron.2005.06.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo, X. , Liu, Y. , Dang, D. , Hu, T. , Hou, Y. , Meng, X. , Zhang, F. , Li, T. , Wang, C. , Li, M. , Wu, H. , Shen, Q. , Hu, Y. , Zeng, X. , He, X. , Yan, L. , Zhang, S. , Li, C. , & Su, B. (2021). 3D Genome of macaque fetal brain reveals evolutionary innovations during primate corticogenesis. Cell, 184, 723.e21–740.e21. 10.1016/j.cell.2021.01.001 [DOI] [PubMed] [Google Scholar]
- Lynch, M. (2010). Evolution of the mutation rate. Trends in Genetics, 26, 345–352. 10.1016/j.tig.2010.05.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacDermot, K. D. , Bonora, E. , Sykes, N. , Coupe, A. M. , Lai, C. S. L. , Vernes, S. C. , Vargha‐Khadem, F. , McKenzie, F. , Smith, R. L. , Monaco, A. P. , & Fisher, S. E. (2005). Identification of FOXP2 truncation as a novel cause of developmental speech and language deficits. American Journal of Human Genetics, 76, 1074–1080. 10.1086/430841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malatesta, P. , Hartfuss, E. , Gotz, M. , & Götz, M. (2000). Isolation of radial glial cells by fluorescent‐activated cell sorting reveals a neuronal lineage. Development, 127, 5253–5263. [DOI] [PubMed] [Google Scholar]
- Manger, P. R. (2006). An examination of cetacean brain structure with a novel hypothesis correlating thermogenesis to the evolution of a big brain. Biological Reviews of the Cambridge Philosophical Society, 81, 293–338. 10.1017/S1464793106007019 [DOI] [PubMed] [Google Scholar]
- Manger, P. R. , Spocter, M. A. , & Patzke, N. (2013). The evolutions of large brain size in mammals: The “Over‐700‐gram club quartet”. Brain, Behavior and Evolution, 82, 68–78. 10.1159/000352056 [DOI] [PubMed] [Google Scholar]
- Manuel, M. N. , Mi, D. , Mason, J. O. , & Price, D. J. (2015). Regulation of cerebral cortical neurogenesis by the Pax6 transcription factor. Frontiers in Cellular Neuroscience, 9, 70. 10.3389/fncel.2015.00070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maricic, T. , Gunther, V. , Georgiev, O. , Gehre, S. , Curlin, M. , Schreiweis, C. , Naumann, R. , Burbano, H. A. , Meyer, M. , Lalueza‐Fox, C. , de la Rasilla, M. , Rosas, A. , Gajovic, S. , Kelso, J. , Enard, W. , Schaffner, W. , & Paabo, S. (2013). A recent evolutionary change affects a regulatory element in the human FOXP2 gene. Molecular Biology and Evolution, 30, 844–852. 10.1093/molbev/mss271 [DOI] [PubMed] [Google Scholar]
- Marques‐Bonet, T. , & Eichler, E. E. (2009). The evolution of human segmental duplications and the core duplicon hypothesis. Cold Spring Harbor Symposia on Quantitative Biology, 74, 355–362. 10.1101/sqb.2009.74.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marques‐Bonet, T. , Kidd, J. M. , Ventura, M. , Graves, T. A. , Cheng, Z. , Hillier, L. W. , Jiang, Z. , Baker, C. , Malfavon‐Borja, R. , Fulton, L. A. , Alkan, C. , Aksay, G. , Girirajan, S. , Siswara, P. , Chen, L. , Cardone, M. F. , Navarro, A. , Mardis, E. R. , Wilson, R. K. , & Eichler, E. E. (2009). A burst of segmental duplications in the genome of the African great ape ancestor. Nature, 457, 877–881. 10.1038/nature07744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marques, A. C. , Dupanloup, I. , Vinckenbosch, N. , Reymond, A. , & Kaessmann, H. (2005). Emergence of young human genes after a burst of retroposition in primates. PLoS Biology, 3, 1970–1979. 10.1371/journal.pbio.0030357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez‐Cerdeño, V. , Cunningham, C. L. , Camacho, J. , Keiter, J. A. , Ariza, J. , Lovern, M. , & Noctor, S. C. (2016). Evolutionary origin of Tbr2‐expressing precursor cells and the subventricular zone in the developing cortex. Journal of Comparative Neurology, 524, 433–447. 10.1002/cne.23879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez‐Cerdeño, V. , Noctor, S. C. , & Kriegstein, A. R. (2006). The role of intermediate progenitor cells in the evolutionary expansion of the cerebral cortex. Cerebral Cortex, 16(1), i152–i161. [DOI] [PubMed] [Google Scholar]
- Martínez‐Martínez, M. Á. , De Juan Romero, C. , Fernández, V. , Cárdenas, A. , Götz, M. , & Borrell, V. (2016). A restricted period for formation of outer subventricular zone defined by Cdh1 and Trnp1 levels. Nature Communication, 7, 1–15. 10.1038/ncomms11812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martins, M. , Galfrè, S. , Terrigno, M. , Pandolfini, L. , Appolloni, I. , Dunville, K. , Marranci, A. , Rizzo, M. , Mercatanti, A. , Poliseno, L. , Morandin, F. , Pietrosanto, M. , Helmer‐Citterich, M. , Malatesta, P. , Vignali, R. , & Cremisi, F. (2021). A eutherian‐specific microRNA controls the translation of Satb2 in a model of cortical differentiation. Stem Cell Reports, 16, 1496–1509. 10.1016/j.stemcr.2021.04.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masuda, K. , Toda, T. , Shinmyo, Y. , Ebisu, H. , Hoshiba, Y. , Wakimoto, M. , Ichikawa, Y. , & Kawasaki, H. (2015). Pathophysiological analyses of cortical malformation using gyrencephalic mammals. Science Reports, 5, 15370. 10.1038/srep15370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGinty, R. K. , & Tan, S. (2015). Nucleosome structure and function. Chemical Reviews, 115, 2255–2273. 10.1021/cr500373h [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLean, C. Y. , Reno, P. L. , Pollen, A. A. , Bassan, A. I. , Capellini, T. D. , Guenther, C. , Indjeian, V. B. , Lim, X. , Menke, D. B. , Schaar, B. T. , Wenger, A. M. , Bejerano, G. , & Kingsley, D. M. (2011). Human‐specific loss of regulatory DNA and the evolution of human‐specific traits. Nature, 471, 216–219. 10.1038/nature09774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merkin, J. , Russell, C. , Chen, P. , & Burge, C. B. (2012). Evolutionary dynamics of gene and isoform regulation in mammalian tissues. Science, 338, 1593–1599. 10.1126/science.1228186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer, K. D. , Saletore, Y. , Zumbo, P. , Elemento, O. , Mason, C. E. , & Jaffrey, S. R. (2012). Comprehensive analysis of mRNA methylation reveals enrichment in 3′ UTRs and near stop codons. Cell, 149, 1635–1646. 10.1016/j.cell.2012.05.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michlewski, G. , & Cáceres, J. F. (2019). Post‐transcriptional control of miRNA biogenesis. RNA, 25, 1–16. 10.1261/rna.068692.118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mihalas, A. B. , Elsen, G. E. , Bedogni, F. , Daza, R. A. M. M. , Ramos‐Laguna, K. A. , Arnold, S. J. , & Hevner, R. F. (2016). Intermediate progenitor cohorts differentially generate cortical layers and require Tbr2 for timely acquisition of neuronal subtype identity. Cell Reports, 16, 92–105. 10.1016/j.celrep.2016.05.072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mihalas, A. B. , & Hevner, R. F. (2018). Clonal analysis reveals laminar fate multipotency and daughter cell apoptosis of mouse cortical intermediate progenitors. Development, 145, dev164335. 10.1242/dev.164335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell, P. J. , & Tjian, R. (1988). Transcriptional regulation in mammalian cells by sequence‐specific DNA binding proteins. Science, 245, 371–378. [DOI] [PubMed] [Google Scholar]
- Miyata, T. , Kawaguchi, A. , Okano, H. , & Ogawa, M. (2001). Asymmetric inheritance of radial glial fibers by cortical neurons. Neuron, 31, 727–741. 10.1016/S0896-6273(01)00420-2 [DOI] [PubMed] [Google Scholar]
- Miyata, T. , Kawaguchi, A. , Saito, K. , Kawano, M. , Muto, T. , & Ogawa, M. (2004). Asymmetric production of surface‐dividing and non‐surface‐dividing cortical progenitor cells. Development, 131, 3133–3145. [DOI] [PubMed] [Google Scholar]
- Miyoshi, K. , Miyoshi, T. , & Siomi, H. (2010). Many ways to generate microRNA‐like small RNAs: Non‐canonical pathways for microRNA production. Molecular Genetics and Genomics, 284, 95–103. 10.1007/s00438-010-0556-1 [DOI] [PubMed] [Google Scholar]
- Mizutani, K. I. , Yoon, K. , Dang, L. , Tokunaga, A. , & Gaiano, N. (2007). Differential Notch signalling distinguishes neural stem cells from intermediate progenitors. Nature, 449, 351–355. 10.1038/nature06090 [DOI] [PubMed] [Google Scholar]
- Moore, L. D. , Le, T. , & Fan, G. (2013). DNA methylation and its basic function. Neuropsychopharmacology, 38, 23–38. 10.1038/npp.2012.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mora‐Bermudez, F. , Badsha, F. , Kanton, S. , Camp, J. G. , Vernot, B. , Kohler, K. , Voigt, B. , Okita, K. , Maricic, T. , He, Z. , Lachmann, R. , Paabo, S. , Treutlein, B. , & Huttner, W. B. (2016). Differences and similarities between human and chimpanzee neural progenitors during cerebral cortex development. Elife, 5, e18683. 10.7554/eLife.18683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mora‐Bermúdez, F. , Taverna, E. , & Huttner, W. B. (2021). From stem and progenitor cells to neurons in the developing neocortex: Key differences among hominids. FEBS Journal, 289, 1524–1535. 10.1111/febs.15793 [DOI] [PubMed] [Google Scholar]
- Muzio, L. , DiBenedetto, B. , Stoykova, A. , Boncinelli, E. , Gruss, P. , & Mallamaci, A. (2002). Emx2 and Pax6 control regionalization of the pre‐neuronogenic cortical primordium. Cerebral Cortex, 12, 129–139. [DOI] [PubMed] [Google Scholar]
- Naftelberg, S. , Schor, I. E. , Ast, G. , & Kornblihtt, A. R. (2015). Regulation of alternative splicing through coupling with transcription and chromatin structure. Annual Review of Biochemistry, 84, 165–198. 10.1146/annurev-biochem-060614-034242 [DOI] [PubMed] [Google Scholar]
- Namba, T. , Nardelli, J. , Gressens, P. , & Huttner, W. B. (2021). Metabolic regulation of neocortical expansion in development and evolution. Neuron, 109, 408–419. 10.1016/j.neuron.2020.11.014 [DOI] [PubMed] [Google Scholar]
- Narayanan, R. , Pham, L. , Kerimoglu, C. , Watanabe, T. , Castro Hernandez, R. , Sokpor, G. , Ulmke, P. A. , Kiszka, K. A. , Tonchev, A. B. , Rosenbusch, J. , Seong, R. H. , Teichmann, U. , Frahm, J. , Fischer, A. , Bonn, S. , Stoykova, A. , Staiger, J. F. , & Tuoc, T. (2018). Chromatin remodeling BAF155 subunit regulates the genesis of basal progenitors in developing cortex. iScience, 4, 109–126. 10.1016/j.isci.2018.05.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naumann, R. K. , & Laurent, G. (2016). Function and evolution of the reptilian cerebral cortex. In Kaas J. H. & Striedter G. F. (Eds.), Evolution of nervous systems (2nd ed., pp. 491–518). Elsevier. 10.1016/B978-0-12-804042-3.00022-1 [DOI] [Google Scholar]
- Nielsen, R. , Bustamante, C. , Clark, A. G. , Glanowski, S. , Sackton, T. B. , Hubisz, M. J. , Fledel‐Alon, A. , Tanenbaum, D. M. , Civello, D. , White, T. J. , Sninsky, J. J. , Adams, M. D. , & Cargill, M. (2005). A scan for positively selected genes in the genomes of humans and chimpanzees. PLoS Biology, 3, 976–985. 10.1371/journal.pbio.0030170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noack, F. , & Calegari, F. (2018). Epitranscriptomics: A new regulatory mechanism of brain development and function. Frontiers in Neuroscience, 12, 1–9. 10.3389/fnins.2018.00085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noctor, S. C. , Flint, A. C. , Weissman, T. A. , Dammerman, R. S. , & Kriegstein, A. R. (2001). Neurons derived from radial glial cells establish radial units in neocortex. Nature, 409, 714–720. [DOI] [PubMed] [Google Scholar]
- Noctor, S. C. , Martínez‐Cerdeño, V. , Ivic, L. , Kriegstein, A. R. , Martinez‐Cerdeno, V. , Ivic, L. , & Kriegstein, A. R. (2004). Cortical neurons arise in symmetric and asymmetric division zones and migrate through specific phases. Nature Neuroscience, 7, 136–144. 10.1038/nn1172 [DOI] [PubMed] [Google Scholar]
- Nomura, T. , Gotoh, H. , & Ono, K. (2013). Changes in the regulation of cortical neurogenesis contribute to encephalization during amniote brain evolution. Nature Communication, 4, 2206. 10.1038/ncomms3206 [DOI] [PubMed] [Google Scholar]
- Nomura, T. , & Izawa, E. I. (2017). Avian brains: Insights from development, behaviors and evolution. Development Growth and Differentiation, 59, 244–257. 10.1111/DGD.12362 [DOI] [PubMed] [Google Scholar]
- Nonaka‐Kinoshita, M. , Reillo, I. , Artegiani, B. , Martínez‐Martínez, M. A. , Nelson, M. , Borrell, V. V. , & Calegari, F. (2013). Regulation of cerebral cortex size and folding by expansion of basal progenitors. EMBO Journal, 32, 1817–1828. 10.1038/emboj.2013.96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nord, A. S. , & West, A. E. (2020). Neurobiological functions of transcriptional enhancers. Nature Neuroscience, 23, 5–14. 10.1038/s41593-019-0538-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Bleness, M. S. , Dickens, C. M. , Dumas, L. J. , Kehrer‐Sawatzki, H. , Wyckoff, G. J. , & Sikela, J. M. (2012). Evolutionary history and genome organization of DUF1220 protein domains. G3: Genes, Genomes, Genetics, 2, 977–986. 10.1534/G3.112.003061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oberst, P. , Fièvre, S. , Baumann, N. , Concetti, C. , Bartolini, G. , & Jabaudon, D. (2019). Temporal plasticity of apical progenitors in the developing mouse neocortex. Nature, 573, 370–374. 10.1038/s41586-019-1515-6 [DOI] [PubMed] [Google Scholar]
- Olkowicz, S. , Kocourek, M. , Lucan, R. K. , Portes, M. , Fitch, W. T. , Herculano‐Houzel, S. , & Nemec, P. (2016). Birds have primate‐like numbers of neurons in the forebrain. Proceedings of the National Academy of Sciences of the United States of America, 113, 7255–7260. 10.1073/pnas.1517131113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otani, T. , Marchetto, M. C. , Gage, F. H. , Simons, B. D. , & Livesey, F. J. (2016). 2D and 3D stem cell models of primate cortical development identify species‐specific differences in progenitor behavior contributing to brain size. Cell Stem Cell, 18, 467–480. 10.1016/j.stem.2016.03.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paolino, A. , Fenlon, L. R. , Kozulin, P. , Haines, E. , Lim, J. W. C. , Richards, L. J. , & Suárez, R. (2020). Differential timing of a conserved transcriptional network underlies divergent cortical projection routes across mammalian brain evolution. Proceedings of the National Academy of Sciences of the United States of America, 117, 10554–10564. 10.1073/pnas.1922422117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilz, G.‐A. A. , Shitamukai, A. , Reillo, I. , Pacary, E. , Schwausch, J. , Stahl, R. , Ninkovic, J. , Snippert, H. J. , Clevers, H. , Godinho, L. , Guillemot, F. , Borrell, V. , Matsuzaki, F. , & Götz, M. (2013). Amplification of progenitors in the mammalian telencephalon includes a new radial glial cell type. Nature Communication, 4, 2125. 10.1038/ncomms3125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polioudakis, D. , de la Torre‐Ubieta, L. , Langerman, J. , Elkins, A. G. , Shi, X. , Stein, J. L. , Vuong, C. K. , Nichterwitz, S. , Gevorgian, M. , Opland, C. K. , Lu, D. , Connell, W. , Ruzzo, E. K. , Lowe, J. K. , Hadzic, T. , Hinz, F. I. , Sabri, S. , Lowry, W. E. , Gerstein, M. B. , … Geschwind, D. H. (2019). A single‐cell transcriptomic atlas of human neocortical development during mid‐gestation. Neuron, 103, 785.e8–801.e8. 10.1016/j.neuron.2019.06.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard, K. S. , Salama, S. R. , Lambert, N. , Lambot, M. A. , Coppens, S. , Pedersen, J. S. , Katzman, S. , King, B. , Onodera, C. , Siepel, A. , Kern, A. D. , Dehay, C. , Igel, H. , Ares, Jr., M. , Vanderhaeghen, P. , & Haussler, D. (2006). An RNA gene expressed during cortical development evolved rapidly in humans. Nature, 443, 167–172. 10.1038/nature05113 [DOI] [PubMed] [Google Scholar]
- Pollen, A. A. , Nowakowski, T. J. , Chen, J. , Retallack, H. , Sandoval‐Espinosa, C. , Nicholas, C. R. , Shuga, J. , Liu, S. J. J. , Oldham, M. C. , Diaz, A. , Lim, D. A. , Leyrat, A. A. , West, J. A. , & Kriegstein, A. R. (2015). Molecular identity of human outer radial glia during cortical development. Cell, 163, 55–67. 10.1016/j.cell.2015.09.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollock, A. , Bian, S. , Zhang, C. , Chen, Z. , & Sun, T. (2014). Growth of the developing cerebral cortex is controlled by microRNA‐7 through the p53 pathway. Cell Reports, 7, 1184–1196. 10.1016/j.celrep.2014.04.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poluch, S. , & Juliano, S. L. (2015). Fine‐tuning of neurogenesis is essential for the evolutionary expansion of the cerebral cortex. Cerebral Cortex, 25, 346–364. 10.1093/cercor/bht232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popesco, M. C. , MacLaren, E. J. , Hopkins, J. , Dumas, L. , Cox, M. , Meltesen, L. , McGavran, L. , Wyckoff, G. J. , & Sikela, J. M. (2006). Human lineage–specific amplification, selection, and neuronal expression of DUF1220 domains. Science, 313, 1304–1307. 10.1126/SCIENCE.1127980 [DOI] [PubMed] [Google Scholar]
- Prabhakar, S. , Noonan, J. P. , Pääbo, S. , & Rubin, E. M. (2006). Accelerated evolution of conserved noncoding sequences in humans. Science, 314, 786. 10.1126/science.1130738 [DOI] [PubMed] [Google Scholar]
- Price, A. J. , Collado‐Torres, L. , Ivanov, N. A. , Xia, W. , Burke, E. E. , Shin, J. H. , Tao, R. , Ma, L. , Jia, Y. , Hyde, T. M. , Kleinman, J. E. , Weinberger, D. R. , & Jaffe, A. E. (2019). Divergent neuronal DNA methylation patterns across human cortical development reveal critical periods and a unique role of CpH methylation. Genome Biology, 20, 1–20. 10.1186/s13059-019-1805-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prieto‐Colomina, A. , Fernández, V. , Chinnappa, K. , & Borrell, V. (2021). MiRNAs in early brain development and pediatric cancer: At the intersection between healthy and diseased embryonic development. Bioessays, 43, 1–20. 10.1002/bies.202100073 [DOI] [PubMed] [Google Scholar]
- Prufer, K. , Racimo, F. , Patterson, N. , Jay, F. , Sankararaman, S. , Sawyer, S. , Heinze, A. , Renaud, G. , Sudmant, P. H. , De Filippo, C. , Li, H. , Mallick, S. , Dannemann, M. , Fu, Q. , Kircher, M. , Kuhlwilm, M. , Lachmann, M. , Meyer, M. , Ongyerth, M. , … Pääbo, S. (2014). The complete genome sequence of a Neanderthal from the Altai Mountains. Nature, 505, 43–49. 10.1038/nature12886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ptashne, M. , & Gann, A. (1997). Transcriptional activation by recruitment. Nature, 386, 569–577. 10.1038/386569a0 [DOI] [PubMed] [Google Scholar]
- Puelles, L. , Kuwana, E. , Puelles, E. , Bulfone, A. , Shimamura, K. , Keleher, J. , Smiga, S. , & Rubenstein, J. L. R. (2000). Pallial and subpallial derivatives in the embryonic chick and mouse telencephalon, traced by the expression of the genes Dlx‐2, Emx‐1, Nkx‐2.1, Pax‐6, and Tbr‐1. Journal of Comparative Neurology, 424, 409–438. 10.1002/1096-9861(20000828)424:3<409::AID‐CNE3>3.0.CO;2‐7 [DOI] [PubMed] [Google Scholar]
- Quinn, J. C. , Molinek, M. , Martynoga, B. S. , Zaki, P. A. , Faedo, A. , Bulfone, A. , Hevner, R. F. , West, J. D. , & Price, D. J. (2007). Pax6 controls cerebral cortical cell number by regulating exit from the cell cycle and specifies cortical cell identity by a cell autonomous mechanism. Developmental Biology, 302, 50–65. 10.1016/j.ydbio.2006.08.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rakic, P. , & Sidman, R. L. (1968). Autoradiographic study of supravital DNA synthesis in fetal human brain. Journal of Neuropathology and Experimental Neurology, 27, 139–140. [PubMed] [Google Scholar]
- Rani, N. , Nowakowski, T. J. , Zhou, H. , Godshalk, S. E. , Lisi, V. , Kriegstein, A. R. , & Kosik, K. S. (2016). A primate lncRNA mediates notch signaling during neuronal development by sequestering miRNA. Neuron, 90, 1174–1188. 10.1016/j.neuron.2016.05.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reillo, I. , & Borrell, V. V. (2012). Germinal zones in the developing cerebral cortex of ferret: Ontogeny, cell cycle kinetics, and diversity of progenitors. Cerebral Cortex, 22, 2039–2054. 10.1093/cercor/bhr284 [DOI] [PubMed] [Google Scholar]
- Reillo, I. , De Juan Romero, C. , García‐Cabezas, M. Á. , & Borrell, V. (2011). A role for intermediate radial glia in the tangential expansion of the mammalian cerebral cortex. Cerebral Cortex, 21, 1674–1694. 10.1093/cercor/bhq238 [DOI] [PubMed] [Google Scholar]
- Riley, B. , Williamson, M. , Collier, D. , Wilkie, H. , & Makoff, A. (2002). A 3‐Mb map of a large segmental duplication overlapping the alpha7‐nicotinic acetylcholine receptor gene (CHRNA7) at human 15q13‐q14. Genomics, 79, 197–209. 10.1006/geno.2002.6694 [DOI] [PubMed] [Google Scholar]
- Roeder, R. G. (1996). The role of general initiation factors in transcription by RNA polymerase II. Trends in Biochemical Sciences, 21, 327–335. 10.1016/0968-0004(96)10050-5 [DOI] [PubMed] [Google Scholar]
- Sagai, T. , Hosoya, M. , Mizushina, Y. , Tamura, M. , & Shiroishi, T. (2005). Elimination of a long‐range cis‐regulatory module causes complete loss of limb‐specific Shh expression and truncation of the mouse limb. Development, 132, 797–803. 10.1242/dev.01613 [DOI] [PubMed] [Google Scholar]
- Sagai, T. , Masuya, H. , Tamura, M. , Shimizu, K. , Yada, Y. , Wakana, S. , Gondo, Y. , Noda, T. , & Shiroishi, T. (2004). Phylogenetic conservation of a limb‐specific, cis‐acting regulator of Sonic hedgehog (Shh). Mammalian Genome, 15, 23–34. 10.1007/s00335-033-2317-5 [DOI] [PubMed] [Google Scholar]
- Sahara, S. , & O'Leary, D. D. M. M. (2009). Fgf10 regulates transition period of cortical stem cell differentiation to radial glia controlling generation of neurons and basal progenitors. Neuron, 63, 48–62. 10.1016/j.neuron.2009.06.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salamon, I. , & Rasin, M. R. (2022). Evolution of the neocortex through RNA‐binding proteins and post‐transcriptional regulation. Frontiers in Neuroscience, 15, 1–31. 10.3389/fnins.2021.803107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salomoni, P. , & Calegari, F. (2010). Cell cycle control of mammalian neural stem cells: Putting a speed limit on G1. Trends in Cell Biology, 20, 233–243. 10.1016/j.tcb.2010.01.006 [DOI] [PubMed] [Google Scholar]
- Sansom, S. N. , Griffiths, D. S. , Faedo, A. , Kleinjan, D. J. , Ruan, Y. , Smith, J. , van Heyningen, V. , Rubenstein, J. L. , & Livesey, F. J. (2009). The level of the transcription factor Pax6 is essential for controlling the balance between neural stem cell self‐renewal and neurogenesis. PLoS Genetics, 5, e1000511. 10.1371/journal.pgen.1000511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauer, M. E. , & Walker, B. E. (1959). Radioautographic study of interkinetic nuclear migration in the neural tube. Proceedings of the Society for Experimental Biology and Medicine, 101, 557–560. [DOI] [PubMed] [Google Scholar]
- Scally, A. , & Durbin, R. (2012). Revising the human mutation rate: Implications for understanding human evolution. Nature Reviews Genetics, 13, 745–753. [DOI] [PubMed] [Google Scholar]
- Scardigli, R. , Bäumer, N. , Gruss, P. , Guillemot, F. , & Le Roux, I. (2003). Direct and concentration‐dependent regulation of the proneural gene Neurogenin2 by Pax6. Development, 130, 3269–3281. 10.1242/dev.00539 [DOI] [PubMed] [Google Scholar]
- Schmidt, E. R. E. , Zhao, H. T. , Park, J. M. , Dipoppa, M. , Monsalve‐Mercado, M. M. , Dahan, J. B. , Rodgers, C. C. , Lejeune, A. , Hillman, E. M. C. , Miller, K. D. , Bruno, R. M. , & Polleux, F. (2021). A human‐specific modifier of cortical connectivity and circuit function. Nature, 599, 640–644. 10.1038/S41586-021-04039-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sena, R. M. , Twiss, J. L. , Gardiner, A. S. , Dell'orco, M. , Linsenbardt, D. N. , & Perrone‐Bizzozero, N. I. (2021). The RNA‐binding protein HuD regulates alternative splicing and alternative polyadenylation in the mouse neocortex. Molecules, 26, 2836. 10.3390/molecules26102836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sessa, A. , Mao, C. A. , Hadjantonakis, A. K. , Klein, W. H. , & Broccoli, V. (2008). Tbr2 directs conversion of radial glia into basal precursors and guides neuronal amplification by indirect neurogenesis in the developing neocortex. Neuron, 60, 56–69. 10.1016/j.neuron.2008.09.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shitamukai, A. , Konno, D. , & Matsuzaki, F. (2011). Oblique radial glial divisions in the developing mouse neocortex induce self‐renewing progenitors outside the germinal zone that resemble primate outer subventricular zone progenitors. Journal of Neuroscience, 31, 3683–3695. 10.1523/JNEUROSCI.4773-10.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoshani, J. , Kupsky, W. J. , & Marchant, G. H. (2006). Elephant brain. Part I: Gross morphology, functions, comparative anatomy, and evolution. Brain Research Bulletin, 70, 124–157. 10.1016/j.brainresbull.2006.03.016 [DOI] [PubMed] [Google Scholar]
- Sidman, R. L. , & Rakic, P. (1973). Neuronal migration, with special reference to developing human brain: A review. Brain Research, 62, 1–35. 10.1016/0006-8993(73)90617-3 [DOI] [PubMed] [Google Scholar]
- Siegenthaler, J. A. , Ashique, A. M. , Zarbalis, K. , Patterson, K. P. , Hecht, J. H. , Kane, M. A. , Folias, A. E. , Choe, Y. , May, S. R. , Kume, T. , Napoli, J. L. , Peterson, A. S. , & Pleasure, S. J. (2009). Retinoic acid from the meninges regulates cortical neuron generation. Cell, 139, 597–609. 10.1016/j.cell.2009.10.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikela, J. M. , & van Roy, F. (2017). A proposal to change the name of the NBPF/DUF1220 domain to the Olduvai domain. F1000Research, 6, 2185. 10.12688/f1000research.13586.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smart, I. H. M. , Dehay, C. , Giroud, P. , Berland, M. , & Kennedy, H. (2002). Unique morphological features of the proliferative zones and postmitotic compartments of the neural epithelium giving rise to striate and extrastriate cortex in the monkey. Cerebral Cortex, 12, 37–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somel, M. , Liu, X. , & Khaitovich, P. (2013). Human brain evolution: Transcripts, metabolites and their regulators. Nature Reviews Neuroscience, 14, 112–127. 10.1038/nrn3372 [DOI] [PubMed] [Google Scholar]
- Sousa, A. M. M. M. , Meyer, K. A. , Santpere, G. , Gulden, F. O. , & Sestan, N. (2017). Evolution of the human nervous system function, structure, and development. Cell, 170, 226–247. 10.1016/j.cell.2017.06.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stahl, R. , Walcher, T. , De Juan Romero, C. , Pilz, G. A. , Cappello, S. , Irmler, M. , Sanz‐Aquela, J. M. , Beckers, J. , Blum, R. , Borrell, V. , Götz, M. , De Juan Romero, C. , Pilz, G. A. , Cappello, S. , Irmler, M. , Sanz‐Aquela, J. M. , Beckers, J. , Blum, R. , Borrell, V. , & Götz, M. (2013). Trnp1 regulates expansion and folding of the mammalian cerebral cortex by control of radial glial fate. Cell, 153, 535–549. 10.1016/j.cell.2013.03.027 [DOI] [PubMed] [Google Scholar]
- Stepien, B. K. , Naumann, R. , Holtz, A. , Helppi, J. , Huttner, W. B. , & Vaid, S. (2020). Lengthening neurogenic period during neocortical development causes a hallmark of neocortex expansion. Current Biology, 30, 4227.e5–4237.e5. 10.1016/j.cub.2020.08.046 [DOI] [PubMed] [Google Scholar]
- Stern, D. L. (2000). Perspective: Evolutionary developmental biology and the problem of variation. Evolution, 54(4), 1079–1091. 10.1111/j.0014-3820.2000.tb00544.x [DOI] [PubMed] [Google Scholar]
- Stillman, B. (2018). Histone modifications: Insights into their influence on gene expression. Cell, 175, 6–9. 10.1016/j.cell.2018.08.032 [DOI] [PubMed] [Google Scholar]
- Striedter, G. F. (1997). The telencephalon of tetrapods in evolution. Brain, Behavior and Evolution, 49, 179–194. 10.1159/000112991 [DOI] [PubMed] [Google Scholar]
- Su, C. H. , Dhananjaya, D. , & Tarn, W. Y. (2018). Alternative splicing in neurogenesis and brain development. Frontiers in Molecular Biosciences, 5, 1–9. 10.3389/fmolb.2018.00012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suárez, R. , Paolino, A. , Fenlon, L. R. , Morcom, L. R. , Kozulin, P. , Kurniawan, N. D. , & Richards, L. J. (2018). A pan‐mammalian map of interhemispheric brain connections predates the evolution of the corpus callosum. Proceedings of the National Academy of Sciences of the United States of America, 115, 9622–9627. 10.1073/pnas.1808262115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudmant, P. H. , Huddleston, J. , Catacchio, C. R. , Malig, M. , Hillier, L. W. , Baker, C. , Mohajeri, K. , Kondova, I. , Bontrop, R. E. , Persengiev, S. , Antonacci, F. , Ventura, M. , Prado‐Martinez, J. , Great Ape Genome Project , Marques‐Bonet, T. , & Eichler, E. E.. (2013). Evolution and diversity of copy number variation in the great ape lineage. Genome Research, 23, 1373–1382. 10.1101/gr.158543.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudmant, P. H. , Kitzman, J. O. , Antonacci, F. , Alkan, C. , Malig, M. , Tsalenko, A. , Sampas, N. , Bruhn, L. , Shendure, J. , Genomes, P. , & Eichler, E. E. (2010). Diversity of human copy number variation and multicopy genes. Science, 330, 641–646. 10.1126/science.1197005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun, E. , & Shi, Y. (2015). MicroRNAs: Small molecules with big roles in neurodevelopment and diseases. Experimental Neurology, 268, 46–53. 10.1016/j.expneurol.2014.08.005 [DOI] [PubMed] [Google Scholar]
- Suzuki, I. K. , Gacquer, D. , Van Heurck, R. , Kumar, D. , Wojno, M. , Bilheu, A. , Herpoel, A. , Lambert, N. , Cheron, J. , Polleux, F. , Detours, V. , & Vanderhaeghen, P. (2018). Human‐specific NOTCH2NL genes expand cortical neurogenesis through delta/notch regulation. Cell, 173, 1370.e16–1384.e16. 10.1016/j.cell.2018.03.067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki, I. K. , Kawasaki, T. , Gojobori, T. , & Hirata, T. (2012). The temporal sequence of the mammalian neocortical neurogenetic program drives mediolateral pattern in the chick pallium. Developmental Cell, 22, 863–870. 10.1016/j.devcel.2012.01.004 [DOI] [PubMed] [Google Scholar]
- Takahashi, T. , Nowakowski, R. S. , & Caviness, V. S., Jr. (1996a). Interkinetic and migratory behavior of a cohort of neocortical neurons arising in the early embryonic murine cerebral wall. Journal of Neuroscience, 16, 5762–5776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi, T. , Nowakowski, R. S. , & Caviness, V. S., Jr. (1996b). The leaving or Q fraction of the murine cerebral proliferative epithelium: A general model of neocortical neuronogenesis. Journal of Neuroscience, 16, 6183–6196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi, T. , Nowakowski, R. S. , & Caviness, V. S., Jr. (1995). Early ontogeny of the secondary proliferative population of the embryonic murine cerebral wall. Journal of Neuroscience, 15, 6058–6068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taverna, E. , Götz, M. , & Huttner, W. B. (2014). The cell biology of neurogenesis: Toward an understanding of the development and evolution of the neocortex. Annual Review of Cell and Developmental Biology, 30, 465–502. 10.1146/annurev-cellbio-101011-155801 [DOI] [PubMed] [Google Scholar]
- Telley, L. , Agirman, G. , Prados, J. , Amberg, N. , Fièvre, S. , Oberst, P. , Bartolini, G. , Vitali, I. , Cadilhac, C. , Hippenmeyer, S. , Nguyen, L. , Dayer, A. , & Jabaudon, D. (2019). Temporal patterning of apical progenitors and their daughter neurons in the developing neocortex. Science, 364, eaav2522. 10.1126/science.aav2522 [DOI] [PubMed] [Google Scholar]
- Thomsen, E. R. , Mich, J. K. , Yao, Z. , Hodge, R. D. , Doyle, A. M. , Jang, S. , Shehata, S. I. , Nelson, A. M. , Shapovalova, N. V. , Levi, B. P. , & Ramanathan, S. (2015). Fixed single‐cell transcriptomic characterization of human radial glial diversity. Nature Methods, 13, 87–93. 10.1038/nmeth.3629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian, S. , Curnutte, H. A. , & Trcek, T. (2020). RNA granules: A view from the RNA perspective. Molecules, 25, 3130. 10.3390/molecules25143130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toda, T. , Shinmyo, Y. , Dinh Duong, T. A. , Masuda, K. , & Kawasaki, H. (2016). An essential role of SVZ progenitors in cortical folding in gyrencephalic mammals. Science Reports, 6, 29578. 10.1038/srep29578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomasello, U. , Klingler, E. , Niquille, M. , Mule, N. , Santinha, A. J. , de Vevey, L. , Prados, J. , Platt, R. J. , Borrell, V. , Jabaudon, D. , & Dayer, A. (2022). miR‐137 and miR‐122, two outer subventricular zone non‐coding RNAs, regulate basal progenitor expansion and neuronal differentiation. Cell Reports, 38, 110381. 10.1016/J.CELREP.2022.110381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toresson, H. , Potter, S. S. , & Campbell, K. (2000). Genetic control of dorsal‐ventral identity in the telencephalon: Opposing roles for Pax6 and Gsh2. Development, 127, 4361–4371. 10.1242/DEV.127.20.4361 [DOI] [PubMed] [Google Scholar]
- Trujillo, C. A. , Rice, E. S. , Schaefer, N. K. , Chaim, I. A. , Wheeler, E. C. , Madrigal, A. A. , Buchanan, J. , Preissl, S. , Wang, A. , Negraes, P. D. , Szeto, R. A. , Herai, R. H. , Huseynov, A. , Ferraz, M. S. A. , Borges, F. S. , Kihara, A. H. , Byrne, A. , Marin, M. , Vollmers, C. , … Muotri, A. R. (2021). Reintroduction of the archaic variant of NOVA1 in cortical organoids alters neurodevelopment. Science, 371, eaax2537. 10.1126/SCIENCE.AAX2537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsui, D. , Vessey, J. P. , Tomita, H. , Kaplan, D. R. , & Miller, F. D. (2013). FoxP2 regulates neurogenesis during embryonic cortical development. Journal of Neuroscience, 33, 244–258. 10.1523/JNEUROSCI.1665-12.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuoc, T. C. , Radyushkin, K. , Tonchev, A. B. , Piñon, M. C. , Ashery‐Padan, R. , Molnár, Z. , Davidoff, M. S. , & Stoykova, A. (2009). Selective cortical layering abnormalities and behavioral deficits in cortex‐specific Pax6 knock‐out mice. Journal of Neuroscience, 29, 8335–8349. 10.1523/JNEUROSCI.5669-08.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uebbing, S. , Gockley, J. , Reilly, S. K. , Kocher, A. A. , Geller, E. , Gandotra, N. , Scharfe, C. , Cotney, J. , & Noonan, J. P. (2021). Massively parallel discovery of human‐specific substitutions that alter enhancer activity. Proceedings of the National Academy of Sciences of the United States of America, 118, 1–11. 10.1073/pnas.2007049118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ule, J. , & Blencowe, B. J. (2019). Alternative splicing regulatory networks: Functions, mechanisms, and evolution. Molecular Cell, 76, 329–345. 10.1016/j.molcel.2019.09.017 [DOI] [PubMed] [Google Scholar]
- Ule, J. , Ule, A. , Spencer, J. , Williams, A. , Hu, J. S. , Cline, M. , Wang, H. , Clark, T. , Fraser, C. , Ruggiu, M. , Zeeberg, B. R. , Kane, D. , Weinstein, J. N. , Blume, J. , & Darnell, R. B. (2005). Nova regulates brain‐specific splicing to shape the synapse. Nature Genetics, 37, 844–852. 10.1038/ng1610 [DOI] [PubMed] [Google Scholar]
- Uzquiano, A. , Gladwyn‐Ng, I. , Nguyen, L. , Reiner, O. , Götz, M. , Matsuzaki, F. , Francis, F. , Götz, M. , Matsuzaki, F. , & Francis, F. (2018). Cortical progenitor biology: Key features mediating proliferation versus differentiation. Journal of Neurochemistry, 146, 500–525. 10.1111/jnc.14338 [DOI] [PubMed] [Google Scholar]
- Vaid, S. , Camp, J. G. , Hersemann, L. , Eugster Oegema, C. , Heninger, A. K. , Winkler, S. , Brandl, H. , Sarov, M. , Treutlein, B. , Huttner, W. B. , & Namba, T. (2018). A novel population of Hopx‐dependent basal radial glial cells in the developing mouse neocortex. Development, 145, dev169276. 10.1242/dev.169276 [DOI] [PubMed] [Google Scholar]
- Vandepoele, K. , Van Roy, N. , Staes, K. , Speleman, F. , & van Roy, F. (2005). A novel gene family NBPF: Intricate structure generated by gene duplications during primate evolution. Molecular Biology and Evolution, 22, 2265–2274. 10.1093/MOLBEV/MSI222 [DOI] [PubMed] [Google Scholar]
- Vastenhouw, N. L. , & Schier, A. F. (2013). Bivalent histone modifications in early embryogenesis. Current Opinion in Cell Biology, 24, 374–386. 10.1016/j.ceb.2012.03.009.Bivalent [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vessey, J. P. , Amadei, G. , Burns, S. E. , Kiebler, M. A. , Kaplan, D. R. , & Miller, F. D. (2012). An asymmetrically localized staufen2‐dependent RNA complex regulates maintenance of mammalian neural stem cells. Cell Stem Cell, 11, 517–528. 10.1016/j.stem.2012.06.010 [DOI] [PubMed] [Google Scholar]
- Villalba, A. , Götz, M. , & Borrell, V. (2021). The regulation of cortical neurogenesis. Current Topics in Developmental Biology, 142, 1–66. 10.1016/bs.ctdb.2020.10.003 [DOI] [PubMed] [Google Scholar]
- Vogel, C. , & Marcotte, E. M. (2012). Insights into the regulation of protein abundance from proteomic and transcriptomic analyses. Nature Reviews Genetics, 13, 227–232. 10.1038/nrg3185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vuong, C. K. , Black, D. L. , & Zheng, S. (2016). The neurogenetics of alternative splicing. Nature Reviews Neuroscience, 17, 265–281. 10.1038/nrn.2016.27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, X. , Tsai, J.‐W. W. , LaMonica, B. , & Kriegstein, A. R. (2011). A new subtype of progenitor cell in the mouse embryonic neocortex. Nature Neuroscience, 14, 555–561. 10.1038/nn.2807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren, N. , Caric, D. , Pratt, T. , Clausen, J. A. , Asavaritikrai, P. , Mason, J. O. , Hill, R. E. , & Price, D. J. (1999). The transcription factor, Pax6, is required for cell proliferation and differentiation in the developing cerebral cortex. Cerebral Cortex, 9, 627–635. [DOI] [PubMed] [Google Scholar]
- Wilson, S. W. , & Rubenstein, J. L. R. (2000). Induction and dorsoventral patterning of the telencephalon. Neuron, 28, 641–651. 10.1016/S0896-6273(00)00171-9 [DOI] [PubMed] [Google Scholar]
- Won, H. , Huang, J. , Opland, C. K. , Hartl, C. L. , & Geschwind, D. H. (2019). Human evolved regulatory elements modulate genes involved in cortical expansion and neurodevelopmental disease susceptibility. Nature Communication, 10, 1–11. 10.1038/s41467-019-10248-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong, F. K. , Fei, J.‐F. F. , Mora‐Bermudez, F. , Taverna, E. , Haffner, C. , Fu, J. , Anastassiadis, K. , Stewart, A. F. , Huttner, W. B. , Mora‐Bermúdez, F. , Taverna, E. , Haffner, C. , Fu, J. , Anastassiadis, K. , Stewart, A. F. , & Huttner, W. B. (2015). Sustained Pax6 expression generates primate‐like basal radial glia in developing mouse neocortex. PLoS Biology, 13, e1002217. 10.1371/journal.pbio.1002217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wray, G. A. (2007). The evolutionary significance of cis‐regulatory mutations. Nature Reviews Genetics, 8, 206–216. 10.1038/nrg2063 [DOI] [PubMed] [Google Scholar]
- Wullimann, M. F. , & Mueller, T. (2004). Teleostean and mammalian forebrains contrasted: Evidence from genes to behavior. Journal of Comparative Neurology, 475, 143–162. 10.1002/cne.20183 [DOI] [PubMed] [Google Scholar]
- Xie, Y. , Castro‐Hernández, R. , Sokpor, G. , Pham, L. , Narayanan, R. , Rosenbusch, J. , Staiger, J. F. , & Tuoc, T. (2019). RBM15 modulates the function of chromatin remodeling factor BAF155 through RNA methylation in developing cortex. Molecular Neurobiology, 56, 7305–7320. 10.1007/s12035-019-1595-1 [DOI] [PubMed] [Google Scholar]
- Xing, L. , Kubik‐Zahorodna, A. , Namba, T. , Pinson, A. , Florio, M. , Prochazka, J. , Sarov, M. , Sedlacek, R. , & Huttner, W. B. (2021). Expression of human‐specific ARHGAP11B in mice leads to neocortex expansion and increased memory flexibility. EMBO Journal, 40, e107093. 10.15252/embj.2020107093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita, W. , Takahashi, M. , Kikkawa, T. , Gotoh, H. , Osumi, N. , Ono, K. , & Nomura, T. (2018). Conserved and divergent functions of Pax6 underlie species‐specific neurogenic patterns in the developing amniote brain. Development, 145, dev159764. 10.1242/dev.159764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon, K.‐J. J. , Vissers, C. , Ming, G. l. , & Song, H. (2018). Epigenetics and epitranscriptomics in temporal patterning of cortical neural progenitor competence. Journal of Cell Biology, 217, 1901–1914. 10.1083/jcb.201802117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon, K. J. , Ringeling, F. R. , Vissers, C. , Jacob, F. , Pokrass, M. , Jimenez‐Cyrus, D. , Su, Y. , Kim, N. S. , Zhu, Y. , Zheng, L. , Kim, S. , Wang, X. , Dore, L. C. , Jin, P. , Regot, S. , Zhuang, X. , Canzar, S. , He, C. , Ming, G. l. , … Song, H. (2017). Temporal control of mammalian cortical neurogenesis by m6A methylation. Cell, 171, 877.e17–889.e17. 10.1016/j.cell.2017.09.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ypsilanti, A. R. , & Rubenstein, J. L. R. (2016). Transcriptional and epigenetic mechanisms of early cortical development: An examination of how Pax6 coordinates cortical development. Journal of Comparative Neurology, 524(3), 609–629. 10.1002/cne.23866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, X. J. , Zheng, H. K. , Wang, J. , Wang, W. , & Su, B. (2006). Detecting lineage‐specific adaptive evolution of brain‐expressed genes in human using rhesus macaque as outgroup. Genomics, 88, 745–751. 10.1016/j.ygeno.2006.05.008 [DOI] [PubMed] [Google Scholar]
- Yun, K. , Potter, S. , & Rubenstein, J. L. (2001). Gsh2 and Pax6 play complementary roles in dorsoventral patterning of the mammalian telencephalon. Development, 128, 193–205. [DOI] [PubMed] [Google Scholar]
- Yuste, R. , Hawrylycz, M. , Aalling, N. , Aguilar‐Valles, A. , Arendt, D. , Arnedillo, R. A. , Ascoli, G. A. , Bielza, C. , Bokharaie, V. , Bergmann, T. B. , Bystron, I. , Capogna, M. , Chang, Y. , Clemens, A. , de Kock, C. P. J. J. , DeFelipe, J. , Santos, D. , SE, D. , K, F. , … Lein, E. (2020). A community‐based transcriptomics classification and nomenclature of neocortical cell types. Nature Neuroscience, 23, 1456–1468. 10.1038/s41593-020-0685-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zabidi, M. A. , & Stark, A. (2016). Regulatory enhancer–core‐promoter communication via transcription factors and cofactors. Trends in Genetics, 32, 801–814. 10.1016/j.tig.2016.10.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Y. , & Reinberg, D. (2001). Transcription regulation by histone methylation: Interplay between different covalent modifications of the core histone tails. Genes & Development, 15, 2343–2360. 10.1101/gad.927301 [DOI] [PubMed] [Google Scholar]
- Zhang, Y. E. , Landback, P. , Vibranovski, M. D. , & Long, M. (2011). Accelerated recruitment of new brain development genes into the human genome. PLoS Biology, 9, e1001179. 10.1371/journal.pbio.1001179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Y. E. , Vibranovski, M. D. , Landback, P. , Marais, G. A. B. , & Long, M. (2010). Chromosomal redistribution of male‐biased genes in mammalian evolution with two bursts of gene gain on the X chromosome. PLoS Biology, 8, e1000494. 10.1371/journal.pbio.1000494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, C. , Sun, G. , Li, S. , & Shi, Y. (2009). A feedback regulatory loop involving microRNA‐9 and nuclear receptor TLX in neural stem cell fate determination. Nature Structural & Molecular Biology, 16, 365–371. 10.1038/nsmb.1576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zody, M. C. , Garber, M. , Adams, D. J. , Sharpe, T. , Harrow, J. , Lupski, J. R. , Nicholson, C. , Searle, S. M. , Wilming, L. , Young, S. K. , Abouelleil, A. , Allen, N. R. , Bi, W. , Bloom, T. , Borowsky, M. L. , Bugalter, B. E. , Butler, J. , Chang, J. L. , Chen, C. K. , … Nusbaum, C. (2006). DNA sequence of human chromosome 17 and analysis of rearrangement in the human lineage. Nature, 440, 1045–1049. 10.1038/nature04689 [DOI] [PMC free article] [PubMed] [Google Scholar]


