Abstract
Phosphatidylinositol (PI)regulating enzymes are frequently altered in cancer and have become a focus for drug development. Here, we explore the phosphatidylinositol-5-phosphate 4-kinases (PI5P4K), a family of lipid kinases that regulate pools of intracellular PI, and demonstrate that the PI5P4Kα isoform influences androgen receptor (AR) signaling, which supports prostate cancer (PCa) cell survival. The regulation of PI becomes increasingly important in the setting of metabolic stress adaptation of PCa during androgen deprivation (AD), as we show that AD influences PI abundance and enhances intracellular pools of PI-4,5-P2. We suggest that this PI5P4Kα-AR relationship is mitigated through mTORC1 dysregulation and show that PI5P4Kα colocalizes to the lysosome, the intracellular site of mTORC1 complex activation. Notably, this relationship becomes prominent in mouse prostate tissue following surgical castration. Finally, multiple PCa cell models demonstrate marked survival vulnerability following stable PI5P4Kα inhibition. These results nominate PI5P4Kα as a target to disrupt PCa metabolic adaptation to castrate resistance.
An understanding of the metabolic function of PI5P4Kα shows potential for targeting it in prostate cancer.
INTRODUCTION
In 1997, the Cantley laboratory found a kinetic function of an unexplored family of lipid kinases called phosphatidylinositol-5-phosphate 4-kinases (PI5P4Ks) (1). PI5P4K acts by phosphorylating the minor lipid phosphatidylinositol-5-phosphate (PI-5-P) at the 4 position of the inositol ring to generate phosphatidylinositol-4,5-bisphosphate (PI-4,5-P2; PIP2) (Fig. 1A). Humans and mice have three distinct genes—PIP4K2A, PIP4K2B, and PIP4K2C—encoding enzymes called PI5P4Kα, PI5P4Kβ, and PI5P4Kγ, respectively. Knockout mice have been generated for all three isoforms and are viable with normal life span and subtle phenotypes (2–4). Although mice deficient in the two most active PI5P4Ks (Pip4k2a–/–Pip4k2b–/–) develop into normal embryos, they fail to survive longer than 12 hours after birth (2). Furthermore, a strong connection of the PI5P4K isoforms has been established in cancers, as they are dysregulated in several cancer subtypes, including breast, glioblastomas, acute myeloid leukemia (AML), and sarcomas (2, 5–8). In previous work, knocking down the PI5P4Kα and PI5P4Kβ isoforms in TP53-mutant breast cancer cells inhibits cell viability, and that deletion of the enzymes in mice suppresses tumor formation with Trp53 deletion (2). Moreover, our recent studies demonstrated the requirement of both the PI5P4Kα and PI5P4Kβ isoforms for sarcoma tumor initiation and maintenance (9).
Fig. 1. PIP4K2A expression correlates with low androgen receptor signaling.
(A) Schematic comparing known kinase activity of noncanonical (left) and canonical (right) phosphoinositide kinase pathways. PI5P4K generates PI-4,5-P2 from a PI-5-P lipid instead of PI-4-P. PI5P4K functions at internal organelles of the endomembrane system as opposed to the exterior cell membrane–like canonical pathway. The importance of PI3K signaling has been established in prostate cancer (PCa), but the question remains how PI5P4K affects prostate biology and androgen receptor (AR) signaling. (B) Analysis of PIP4K2A transcript expression shows inverse correlation with AR signature score (28) on available transcript data from The Cancer Genome Atlas (PRAD-TCGA; N = 333, P = 4.9 × 10−16) and (C) Stand Up to Cancer datasets (SU2C; N = 149, P = 5.1 × 10−6). (D) RNA sequencing (RNA-seq) transcript abundance for established PCa cells lines for PIP4K2A, PIP4K2B, and PIP4K2C isoforms. (E) Highest levels of PIP4K2A transcript expression occurs in AR and KLK3 negative models (ARpos mean, 1.127; ARneg mean, 2.941; t test, P = 0.048). (F) Western blot analysis with mouse embryonic fibroblast (MEF) double knockout (negative control) and BT474 (positive control) confirms protein expression of PI5P4Kα in PCa cell lines. (G) PIP4K2A expression further characterized in a panel of human PCa organoids using RNA-seq (R2 = 0.2146, P = 4.5 × 10−2). t test values: *P < 0.05; **P < 0.01; and ***P < 0.001.
Recently, the roles of the PI5P4K isoforms in controlling cellular metabolism have been found (9, 10), and we posit that targeting PI5P4K could expose critical vulnerabilities in cancer. While most of the cellular PI-4,5-P2 is localized at the plasma membrane and generated by the canonical pathway in which PI-4-P is phosphorylated at the 5 position of the inositol ring (11, 12), intracellular PI-4,5-P2 can be synthesized by an alternative type 2 pathway in which PI5P4Ks convert PI-5-P to PI-4,5-P2 at the membranes of endosomes, lysosomes, Golgi apparatus, endoplasmic reticulum (ER), nucleus, and peroxisomes (13, 14). Notably, while PI5P4K-generated peroxisomal PI-4,5-P2 is necessary for the peroxisome-lysosome membrane contacts that facilitate cholesterol transport between organelles (15), the PI-4,5-P2 lipid has many other functions that could be important. This includes the control of the activity of membrane transport proteins that allow influx and efflux of metabolites from lysosomes, as well as roles in the localization of signaling complexes through lipid-protein binding interactions (16). While the precise spatial and temporal control of PI-4,5-P2 at intracellular membranes is important for cellular stress response, the influence of PI5P4K and its relationship with many key cancer pathways have yet to be explored.
The PI5P4Ks are lesser-known members of a network of enzymes that regulate phosphorylated phosphatidylinositol (PI). These kinases and phosphatases that control the addition or removal of phosphates on the inositol ring of PI has been established as a cornerstone of multiple biological processes and have a high frequency of alteration across most cancer types (17, 18). The most well-known class is the phosphoinositide 3-kinases (type 1 PIP kinase pathway; PI3Ks), which modulate critical cellular functions for development, immunity, and tumorigenesis. The PI3K pathway also has a major role in regulating metabolism. It conducts downstream signals from the stimulated insulin receptor through phosphorylation of AKT serine/threonine kinase to control glucose homeostasis. To date, there have been more than 700 clinical trials involving PI3K/AKT targeted molecules, with an estimated 334 of these trials listed as ongoing or currently planned (ClinicalTrials.gov). These involve a spectrum of cancer types including acute lymphocytic leukemia, AML, glioma, melanoma, colorectal, lung, breast, and prostate cancer (PCa) (19). There is tremendous progress in the development of PI3K inhibitors for the treatment of cancer; however, as with many targeted therapies, full efficacy may be stunted by dose-limiting toxicities and the acquisition of resistance. By exploring PI5P4K, we hope to understand the broader network of phosphoinositide regulators with the goal of exposing metabolic cancer vulnerabilities that support clinically used therapeutics.
The functional role of PI5P4K can vary markedly between tissue and cell types. In this study, we focused our efforts solely on PCa and, to our knowledge, are the first to characterize PI5P4K in this disease. An estimated one of seven men will develop PCa by the age of 60. While treatment strategies for localized disease can be curative, 10 to 20% of PCa will progress to advanced castrate-resistant disease [castrate-resistant PCa (CRPC)] within 5 years (20). The prostate is a specialized reproductive gland that is particularly susceptible to cancer development. Normal prostate cell growth and development are heavily dependent on androgen receptor (AR) signaling. Androgens strongly influence the metabolic state of prostate cells to favor sustained cellular growth (21). Reinforcing this growth, the PI3K pathway has the highest frequency of alteration in both primary and metastatic PCa (22). A direct connection has been established between the PI3K-AKT-mTOR (mammalian target of rapamycin) and AR signaling axis in the prostate. Pathway deregulation resulting from loss of phosphatase and tensin homolog (PTEN) is associated with the down-regulation of AR target genes (23–25). While foundational studies support the importance of phosphoinositide signaling in AR regulation, the entire picture of the intricate cross-talk between these systems remains unclear.
There are a growing number of inhibitors being tested in combination with androgen-targeted therapies in clinical trials (26, 27) but still almost nothing is known about the potential cross-talk of the greater phosphoinositide kinase network. Here, we suggest an interplay of PI5P4K in AR signaling and lay the foundation for phenotypic understanding of the role of PI5P4Kα in prostate metabolism.
RESULTS
PIP4K2A inversely correlates with AR signaling in PCa
To establish the degree and frequency of PIP4K2 in PCa, we queried two publicly available patient datasets for patterns of gene expression. PIP4K2A, PIP4K2B, and PIP4K2C transcript expression was detected in clinically localized [The Cancer Genome Atlas (TCGA)-Prostate Adenocarcinoma (PRAD), N = 498] and advanced, metastatic PCa [Stand Up 2 Cancer-Prostate Cancer Foundation (PCF) (SU2C, N = 209)] datasets. PIP4K2A and PIP4K2B demonstrate similar expression patterns, while the PIP4K2C transcript was expressed differentially from the other two isoforms (fig. S1, A and B). Previous work associated PIP4K2B with tumor protein p53 (TP53) mutant breast cancer (2). TP53 and PTEN are among the most altered genes in CRPC; however, we found that none of the PIP4K2 isoforms associate with TP53 nor PTEN mutation in CRPC (fig. S1, C and D). In addition, the genes coding for the PIP4K2 isoforms are not commonly mutated in either TCGA or SU2C datasets.
To determine whether PI5P4K relates to other know drivers of PCa, we tested whether there is a relationship between PIP4K2A and AR signaling. A previously established AR signaling score, which includes gene signature changes resulting from pathway stimulation, can be used as a metric of AR activity in tumor material (table S1) (28). AR signaling generally will increase with up-regulation of AR in early PCa development then diminish with disease progression into AR-independent CRPC. In PCa patient datasets, the highest levels of PIP4K2A correlated with lower AR signaling in both TCGA (PRAD, P = 4.9 × 10−16, N = 498) and SU2C datasets (SU2C, P = 5.1 × 10−6, N = 209) (Fig. 1, B and C). PIP4K2B and PIP4K2C isoforms showed no correlation to the AR pathway (fig. S1E). These transcript observations prompted us to optimize immunohistochemical (IHC) reagents to confirm protein-level expression in patient tissue. PI5P4Kα staining of human clinical samples revealed high cytoplasmic positivity in areas of basal cell hyperplasia and inflammation (fig. S2, A and B). Antibody specificity was further supported by comparing serial section localization of an RNA–in situ hybridization (ISH) probe for PIP4K2A (fig. S2, C and D). Together, these data show that PI5P4Kα is present in human PCa tissue and correlates with decreasing AR signaling.
Next, we profiled a panel of PCa cell lines for transcript expression of the PIP4K2 isoforms (Fig. 1D). While the transcript abundance of PIP4K2B and PIP4K2C did not vary between cell lines, PIP4K2A, the most catalytically active isoform (29, 30), shows a marked range of expression between cell lines with NCI-H660 (neuroendocrine PCa model) and VCaP (AR responsive) being the highest and lowest, respectively. An enrichment for PIP4K2A occurs in AR-negative cells that model CRPC (i.e., PC3, DU145, and NCI-H660) compared to AR-responsive cells that retain AR and KLK3 transcript expression (i.e., VCaP, LNCaP, 22RV1, and MDAPCA2B) (Fig. 1E). In addition, the PIP4K2A relative transcript levels are paralleled on the protein level (Fig. 1F). Furthermore, we queried the expression of PIP4K2 isoforms in a panel of PCa patient-derived organoids. This panel includes rare patient-derived cell models that recapitulate the disease of PCa to a high standard of in vitro culture technology (tables S2 and S3) (31–33). Here, an inverse correlation of PIP4K2A is again found in the analysis of bulk RNA sequencing (RNA-seq) data (Fig. 1G and fig. S3A). Collectively, cell lines and organoid models demonstrate that PIP4K2A expression levels are the highest in PCa models with diminished AR signaling.
PIP4K2A has a functional relationship with AR
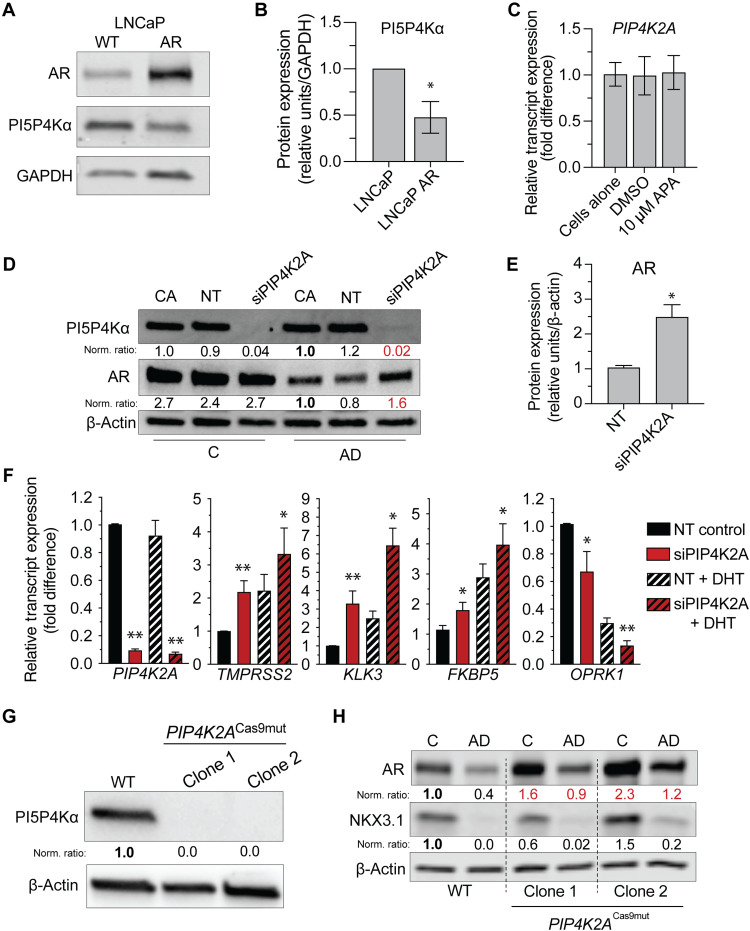
To explore the correlation between PIP4K2A and AR, we modulated PCa AR signaling in vitro. We did this using the LNCaP and LNCaP-AR models of human adenocarcinoma, as these cells retain AR pathway activity with LNCaP-AR modified to overexpress AR (34). We found LNCaP-AR to have only 0.48-fold the PI5P4Kα protein relative to wild-type LNCaP (N = 3, P = 6.0 × 10−3; Fig. 2, A and B). Compared to AR-negative CRPC models (i.e., PC3), wild-type LNCaP has relatively low PI5P4Kα that is reduced further with the overexpression of AR. Conversely, the acute inhibition of AR with apalutamide (APA) does not affect PIP4K2A transcript (Fig. 2C). This suggests that PIP4K2A is not a direct target of AR transcription factor activity, and the PI5P4Kα-AR correlation is mechanistically indirect.
Fig. 2. PIP4K2A has a functionally inverse relationship with AR in PCa cell models.
(A) Stable overexpression of AR in LNCaP model corresponds to decrease in PI5P4Kα protein by Western blot, which is quantified with (B) protein densitometry (N = 3, 0.48-fold of control, P = 6.0 × 10−3). GAPDH, glyceraldehyde-3-phosphate dehydrogenase. (C) Following a 48-hour treatment of AR inhibitor, 10 μM APA does not alter PIP4K2A transcript or (D) PI5P4Kα protein expression (CA, cells alone; NT, nontargeting control). (E) Protein densitometry of AR following siPIP4K2A knockdown in androgen depletion (AD) (n = 3, 2.49-fold of control, P = 2.9 × 10−2). (F) AR signature transcript expression is boosted following 48-hour siRNA transfection of LNCaP cells and 18-hour DHT stimulation in AD. (G) Western blot of stable PIP4K2A-targeted LNCaP clones using CRISPR-Cas9 lacking protein expression and (H) shows up-regulated AR relative to wild-type (WT) control clones and mutant clones (PIP4K2ACas9mut). Immunoblot quantification values shown following housekeeper and control normalization (relative to bolded sample). Red values highlight experimental change. t test values: *P < 0.05; **P < 0.01.
To investigate whether inhibition of PI5P4Kα functionally affects the AR activity, we further examined the outcome of its in vitro down-regulation. Small interfering RNA (siRNA) targeting of PIP4K2A (siPIP4K2A) in LNCaP cells demonstrated minimal change to AR or AR target genes in complete medium conditions; however, when cultured in androgen deprivation (AD) conditions, siPIP4K2A treatment resulted in an up-regulation of AR signature transcripts and AR protein (Fig. 2, D to F, and fig. S3, B and C). Depleted of hormones and large hydrophobic molecules, the AD medium contains charcoal-stripped (C/S) serum that lacks dihydrotestosterone (DHT) and other androgen substrate derivatives known to activate AR (35). This boost in AR transcript is comparable to levels following control DHT stimulation, and the combination of siPIP4K2A with DHT resulted in an additive boost of AR signature genes (i.e., TMPRSS2, KLK3, and FKBP5 increase; OPRK1 decrease; Fig. 2F). CRISPR-Cas9 tools and PIP4K2A-targeted small guide RNAs allowed production of LNCaP clones lacking PI5P4Kα protein (Fig. 2G). Similar to the acute siRNA treatments, PIP4K2ACas9mut LNCaP, expanded from rare single-cell clones, had up-regulated AR expression (Fig. 2H). These data provide evidence that PI5P4Kα has a functionally dynamic interplay with AR.
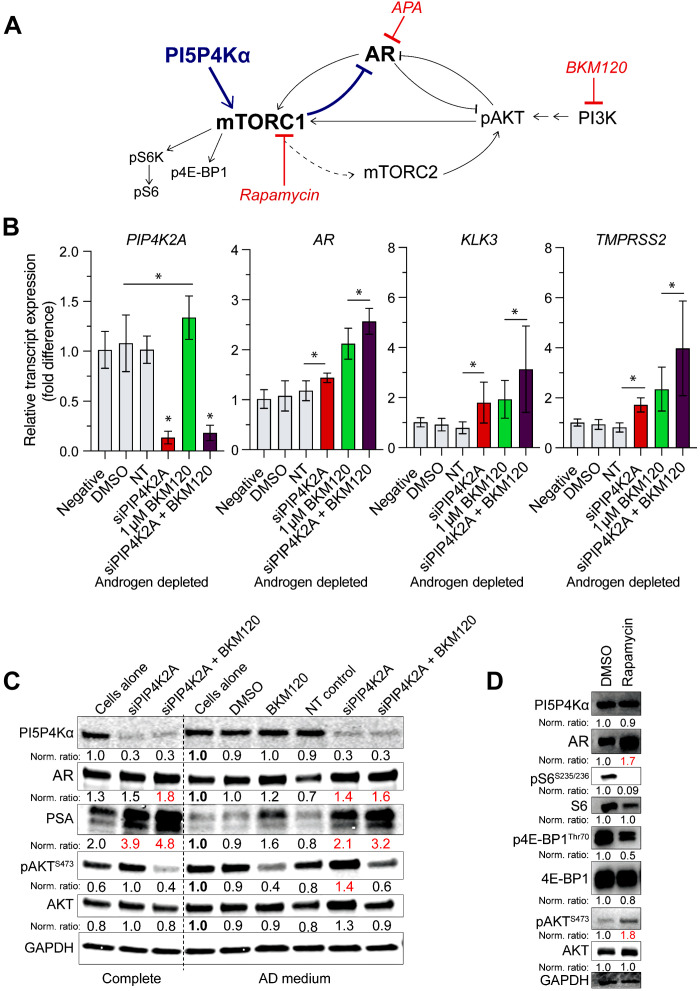
PIP4K2A-AR relationship is nonredundant to PI3K inhibition
PI5P4Kα is not the only metabolic factor with which AR dynamically interacts. Inverse relationships with AR have also been shown following the inhibition of mTOR complex 1 (mTORC1) (36, 37) as well as through targeting PI3K, a well-known PI kinase (Fig. 3A) (23, 38). Therefore, we questioned whether the boost in AR activity resulting from PIP4K2A depletion is mitigated through the PI3K-AR relationship. Buparlisib (BKM120) used at 1 μM concentration was used to compare PI3K inhibition to PIP4K2A knockdown in LNCaP cells. As expected, siPIP4K2A and BKM120 treatments as single agents caused a boost to AR and AR target genes. Unexpectedly, combination treatment of siPIP4K2A with 24 hours of BKM120 showed an additive boost in AR target genes even in AD conditions (Fig. 3B). This boost was further confirmed on protein level in both complete medium and AD conditions; however, siPIP4K2A differed from BKM120, as it produced an up-regulation of phospho-AKTS473 (Fig. 3C). Up-regulation of AR and feedback that increases phospho-AKTS473 is a phenotype of mTORC1 inhibition (36, 39). Figure 3D demonstrates this following a 24-hour treatment of LNCaP cells with 1 nM rapamycin. Down-regulation of mTORC1 is exemplified by a reduction in targets phospho-S6S235/236 and phospho–4E-BP1Thr70, whereas AR protein and phospho-AKTS473 are elevated. Levels of PI5P4Kα remain the same following rapamycin treatment (Fig. 3D). Concerning AR signaling, these findings suggest that PIP4K2A depletion is not redundant with PI3K inhibition.
Fig. 3. PI5P4Kα is nonredundant to the PI3K pathway.
(A) A relationship has been established between AR and the PI3K (type 1) pathway. Graphical summary of suggested feedback of AR when PI3K/AKT or mTORC1 is inhibited. (B) LNCaP cells in androgen deprivation (AD) were treated with siRNA targeting PIP4K2A (siPIP4K2A) for 48 hours and/or 10 μM BKM120 for 24 hours. Changes in AR transcript expression and AR signature genes (KLK3 and TMPRSS2) by quantitative real-time polymerase chain reaction (qRT-PCR) are represented. (C) Western blot analysis also compares protein-level changes following siPIP4K2A and/or 10 μM BKM120. (D) Signaling changes from siPIP4K2A treatment of LNCaP appear to phenocopy treatment with rapamycin. Phospho-target changes are represented using Western blot comparison of 1 nM rapamycin compared to dimethyl sulfoxide (DMSO) control for 24 hours. Immunoblot quantification values shown following housekeeper and control normalization (relative bolded sample). Red values highlight experimental change. t test values: *P < 0.05.
PI5P4Kα influences AR signaling through dysregulation of mTORC1
Next, we sought to understand which molecular pathways are influenced by targeting PI5P4Kα. We conducted transcriptomic sequencing of LNCaP cells following PIP4K2A knockdown in AD conditions. There were few single transcripts changed between nontargeting (NT) and siPIP4K2A based on a fold change cut of 50% (fig. S4A). However, many biological processes involving fundamental metabolism pathways were affected by PIP4K2A depletion, as exemplified by selected gene set enrichment analysis (GSEA) pathway enrichment from whole-exome RNA-seq (Fig. 4A). Notably, androgen response genes were of the highest up-regulation with PIP4K2A knockdown. Statistically significant enrichment scores were also observed for pathways involving ER stress, steroid biosynthesis, cholesterol metabolism, phospholipid metabolism, hypoxia, and glycolysis. AR and lipid pathways were further implicated in PIP4K2A regulation on a companion Western blot showing up-regulation of AR, prostate-specific antigen, and sterol regulatory element–binding protein 1 (Fig. 4B). A −2.3 normalized enrichment score (Padj = 0.012) for mTORC1 signaling stood out as a Hallmark pathway as one of the most reduced in RNA-seq (Fig. 4A and fig. S4B). Transcriptomic analysis confirmed that depletion of PIP4K2A boosts AR signature genes and dysregulates essential pathways such as lipid metabolism and mTORC1 signaling.
Fig. 4. PI5P4Kα affects mTORC1 signaling.
(A) Whole-exome transcript expression was measured with bulk RNA-seq. Statistically significant differential GSEA pathways between nontargeting siRNA control (NT) and siRNA specific for PIP4K2A (siPIP4K2A) treatments are represented. Pathways shown all with Padj significant P < 0.05. (B) Western blotting confirms changes in AR pathway targets following a knockdown in PI5P4Kα protein expression. (C) Further analysis of patient data from TCGA (PRAD-TCGA) shows a positive association with PIP4K2A (R = 0.17, P = 9.8 × 10−5) and an mTOR activation score (17). In addition, PIP4K2B (R = 0.27, P = 2 × 10−9) but not PIP4K2C (R = 0.0044, P = 0.92). (D) LNCaP cells infected with pLKO.1 lentiviral PIP4K2A shRNAs (shPIP4K2A) show reduced phospho-S6235/236 48 hours after GFP sorting. Immunoblot quantification values shown following housekeeper and control normalization (relative bolded sample). Red values highlight experimental change. (E) LNCaP cells in androgen deprivation (AD) were treated with siPIP4K2A for 48 hours. Changes of AR transcript expression and TFEB signature genes (CTSD, NEU1, HEXA, and CTSF) by qRT-PCR are represented. Data representative of three independent experiments, mean ± SEM. t test values: *P < 0.05; **P < 0.01; and ***P < 0.001.
Molecular alterations to pathways converging on mTORCs are common across most cancer types (17). Zhang et al. (17) previously defined a score for mTOR using multicancer TCGA analysis of proteomic and transcriptomic data. In the present study, the score for mTOR pathway activation had a positive correlation with PIP4K2A expression in PCa cases (TCGA PRAD, R = 0.17, P = 9.8 × 10−5). Furthermore, PIP4K2B was also correlated (R = 0.27, P = 2 × 10−9) but with a narrow expression profile distribution. No correlation was found for PIP4K2C with mTOR pathway activation (R = 0.0044, P = 0.92; Fig. 4C). This relationship was validated using RNA-seq data from PIP4K2A knockdown in LNCaP cells (fig. S4C), as well as with Western blot measurement of mTORC1 target phospho-S6 ribosomal protein (phospho-S6235/236) following stable lentiviral infection of a short hairpin RNA (shRNA) targeting PIP4K2A (shPIP4K2A) for 72 hours (0.64-fold of control; t test, P = 0.006; Fig. 4D). Note that under conditions of nutrient stress, catabolic pathways controlled by mTORC1 are activated. For example, the loss of phosphorylation of the mTORC1 target, transcription factor EB (TFEB), allows it to translocate to the nucleus and activate transcription programs for lysosome biogenesis and stress metabolism. TFEB transcription activation is a biomarker of loss of mTORC1 anabolic activity, and we observed an up-regulation of three of four tested TFEB target genes (CTSD, NEU1, HEXA, and CTSF) following siPIP4K2A depletion in LNCaP cells in AD (Fig. 4E). Activation of TFEB via PI5P4K-mediated mTORC1 suppression is established for both genetic knockout and kinase drug inhibitor studies (10, 40). These data imply an association of PI5P4Kα with mTORC1 signaling, a key pathway used in PCa progression.
Androgen deprivation stress enriches for PI lipids
Clues into how PCa transitions to CRPC may exist in understanding the metabolic adaptations of the prostate under conditions of AD. For example, the activity of cellular lysosome, the site of mTORC1 activation, increases under conditions of in vivo castration (41). In vitro, we found that a 48-hour treatment of APA prompts the accumulation of endolysosomes in LNCaP cells by 1.33-fold (Fig. 5, A and B). We question whether PI5P4Kα influences the metabolic adaptation of PCa to AD and used a high-coverage targeted lipidomics platform to characterize changes to key signaling lipids. LNCaP cultured in AD were compared to cells grown in complete serum conditions [RPMI 1640 + 5% fetal bovine serum (FBS)] for 48 hours. A total of 273 lipid species were semiquantified and assessed for changes based on lipid (sub)class and fatty acid chain composition (fig. S5, A and B). Figure S5C summarizes 53 lipids with levels that statistically differ between culture conditions. Six of seven assessed PI lipids were increased under AD conditions with an average of 141% higher relative fold lipid abundance (t test, P = 6.04 × 10−9; Fig. 5C). Androgen deprivation and AR inhibition has previously been shown to alter PI metabolism and membrane lipid concentrations (42, 43). Next, we examined directly how down-regulating PIP4K2A affects the lipidome of PCa cells. The combined impact of siPIP4K2A in AD medium–enriched PI lipid species was higher than medium and NT controls alone (Fig. 5C). Paradoxically, inhibition of PIP4K2 isoforms is reported to increase the total cell content of the PI-4,5-P2 lipid, but this increase is suggested to be in cell membrane–localized pools (44). Disruption to metabolic function can markedly shift membrane lipid pools leading to the question of how PI dysregulation could expose survival vulnerabilities in cancer cells. Notably, PI-4,5-P2, a species of phosphorylated PI, is an essential substrate activating some compensatory pathways that allow the survival through the progression to CRPC (45, 46). With known importance, this raised the question of whether PCa depends on specific pools of PI to cope with cell stress.
Fig. 5. AR inhibition dysregulates PI signaling at the endolysosome.
(A) LysoTracker Red IF staining of LNCaP cells treated with 10 μM APA or DMSO for 48 hours. Scale bars, 10 μm. (B) Relative quantification of LysoTracker Red IF intensity in cell populations following 48 hours androgen deprivation (AD) or APA treatment. Data representative of three independent experiments. (C) LNCaP cultured in complete RPMI 1640 + 5% FBS compared to AD conditions. Total PI lipids were measured in LNCaP cells transfected with siRNA targeting PIP4K2A (siPIP4K2A) for 24 hours and then switched into AD conditions for an additional 48 hours. Relative PI lipid content was measured using lipidomic liquid chromatography–tandem mass spectrometry (LC-MS/MS) methodology. CA, cell alone in complete medium unless indicated with AD; NT, nontargeting siRNA control. Increases between CA and CA in AD (0.32-fold, P = 6.04 × 10−9) and NT and siPIP4K2A (0.15-fold, P = 3. × 10−4) are noted. (D) qRT-PCR shows suppression of AR target gene KLK3 in paired experiments where (E) intracellular pools of PI-4,5-P2 are detected with IF between LNCaP treated for 48 hours of 10 μM APA or DMSO control. Scale bars, 10 μm. (F) Image quantification of 50 cells per group showed elevated intensity of PI-4,5-P2 pool staining intensity in APA treatment. DAPI, 4′,6-diamidino-2-phenylindole. t test values: *P < 0.05; **P < 0.01; and ***P < 0.001.
Intracellular PI-4,5-P2 is enriched following AR pathway inhibition
Recently, we have shown that the PI-4,5-P2 produced by PI5P4K is important for regulating cell metabolism (9). To query whether PI-4,5-P2 is modified under conditions of AR down-regulation, we used immunofluorescent (IF) staining protocols. Most of the PI-4,5-P2 are found on the cell surface membrane, and it is a critical lipid substrate for the PI3K/AKT (type 1 PIP kinase) pathway (47). However, alterations to type 1 alone do not account for the entire interplay of PI in intracellular metabolic compartments. A method to exclusively detect intracellular pools of PI-4,5-P2 using IF staining has previously been established (9, 48). LNCaP cells treated with APA for 48 hours show strong suppression of AR target gene KLK3 (Fig. 5D). While the AR pathway is suppressed, pools of intracellular PI-4,5-P2 were found to be enriched using IF intensity (Fig. 5, E and F). PI-4,5-P2 enrichment following APA treatment also increased the number of foci per cell (fig. S5D). Intracellular PI-4,5-P2 is a key signaling molecule for pathways involved with metabolic stress, and its enrichment may be functionally important for PCa disease progression. PCa relies on the AR pathway for growth; therefore, stress compensatory pathways involving intracellular PI-4,5-P2 could support cell survival during AD.
PI5P4Kα localizes to the endolysosome and influences gland volume following in vivo androgen depletion
Targeting PI5P4Kα expression influences AR signaling in vitro, thereby prompting us to explore this relationship in vivo. We queried how AD resulting from surgical castration affects PI5P4Kα expression in mouse prostate. Eight-week-old C57BL/6J mice were castrated or subjected to a control sham procedure then harvested 10 weeks following surgery (Fig. 6A). PI5P4Kα protein expression was first confirmed in all lobes of the mouse prostate by Western blot (fig. S6A), and then, IHC and Pip4k2a RNA-ISH was used to positively detect PI5P4Kα patterns in tissue (fig. S6B). The mass of harvested urogenital (UG) tissue was markedly reduced in castrated animals as expected (fig. S6C); in addition, loss of nuclear AR staining in the prostate luminal epithelium was observed (fig. S6D). Whole castrated glands and an equal number of representative sham prostate regions were quantified for the number of high PI5P4Kα-positive stained cells. This pattern of PI5P4Kα staining is primarily observed along the luminal cell basal membrane and in the basal cell compartment (Fig. 6B). Sham tissue had 2.3% of prostate cells expressing high PI5P4Kα staining, while 11.13% of cells were positive in castrated glands (Fig. 6C). A relative enrichment of PI5P4Kα occurs in castrated mouse prostate.
Fig. 6. Lysosome localized PI5P4Kα is linked to castration adaptation of prostate tissue.
(A) C57BL/6J mice were aged to 8 weeks, surgically castrated or used for control sham surgery procedure. Tissues were harvested 10 weeks after surgery. (B) PI5P4Kα detection was optimized and characterized in normal adult mouse prostate. Examples of positive 3,3′-diaminobenzidine (DAB) staining by immunohistochemistry are shown (scale bars, 20 μm) and (C) quantified between sham and castrated tissue groups as a percentage of positive DAB cells per number of cells counted per gland (P = 5.18 × 10−9). (D) Representative images of normal mouse prostate compared to castrated tissue when stained with LAMP1 endolysosome marker with IF-IHC. Scale bars, 10 μm. (E) Quantification of Lamp1 detection in castrated versus sham surgery mouse prostate tissue (Welch’s t test, P = 0.0193). DAPI nuclear stains are used as positive cell detection marks for IF assays. (F) IF staining of normal adult mouse prostate for colocalization of LAMP1 (green) and PI5P4Kα (red). Yellow indicates areas of colocalization. Scale bars, 10 μm. See fig. S7 for Zeiss software colocalization statistics. DAPI nuclear stains are used as positive cell detection marks for IF assays. t test values: *P < 0.05; and ***P < 0.001.
Targeted suppression of PIP4K2 results in an increase of lysosome cell content potentially through dysregulation of lysosomal membrane contacts with other organelles (10, 44). In addition, lysosomes are involved in survival adaptation programs such as autophagy and mobilization of nutrients in the absence of AR pathway support (49). To determine whether lysosome regulation is altered in our model of AD, prostate tissue sections were stained for Lysosomal Associated Membrane Protein 1 (LAMP1). The cytoplasmic volume of prostate cells is atrophic compared to sham controls; however, the IF signal intensity of LAMP1 foci was elevated in the atrophic luminal cells of castrate surgery prostate glands (3.2-fold increase; Welch’s t test, P = 0.019; Fig. 6, D and E). Furthermore, we assessed whether PI5P4Kα protein expression correlates with patterns of LAMP1 positivity. LAMP1 positivity is in direct intracellular proximity with PI5P4Kα protein and has some areas of colocalization detected from IF confocal image analysis (Fig. 6F and fig. S7, A and B). Antibodies testing markers of the Golgi apparatus and ER were also tested but not found to correlate with focal PI5P4Kα expression patterns. Transmission electron microscopy images confirmed that there are concentrated lysosome numbers in mouse basal cells (fig. S7C). These data tie together PI5P4Kα with the regulation of the endolysosome and suggests a potential relationship with mTORC1, a complex highly dependent on lysosomal dynamics.
Under conditions of castration, lysosome-meditated degradation is used to catabolize cellular material of luminal prostate cells. Next, we questioned whether PI5P4Kα affects prostate tissue preservation under androgen deprivation conditions. A prostate-specific genetically engineered mouse (GEM) model was generated using a probasin (PB)–driven Cre recombinase combined with a homozygous Pip4k2a floxed (fl) allele (Pip4k2afl/fl). PB-Pip4k2afl/fl indicates allele deletion in the context of luminal prostate cell PB expression. No malignancy was observed in prostate tissues aged up to 300 and 400 days with either heterozygote or homozygote deletion of Pip4k2a (fig. S8, A to D). The only histomorphological features seen in a notable number of cases included (i) rare mitosis, (ii) rare individual cell apoptosis, and (iii) multifocal vacuolar cytoplasmic changes of prostate luminal epithelial cells. None of these features appeared to have been associated with a specific condition. An observed prostate luminal epithelial cytoplasmic vacuolation is compatible with a degenerative feature known to occur as an age-related change in laboratory mice.
Including an R26eYFP Cre reporter allowed lineage tracking of mutant prostate cells following Cre activation from endogenous hormone signaling at the onset of sexual maturity (6 to 8 weeks; Fig. 7, A and B). IF staining of castrated tissue showed expression of enhanced yellow fluorescent protein (eYFP) Cre reporter in luminal prostate cells from both eYFP-only control and PB-Pip4k2afl/fl genotypes. eYFP-expressing glands from PB-Pip4k2afl/fl prostates had a greater remaining gland epithelium area, which could explain the elevated UG mass of these tissues (Fig. 7, C and D). On average, castrated PB-Pip4k2afl/fl prostates had higher UG mass with a percentage relative to body weight of 0.32% compared to 0.26% of eYFP-only control animals (t test, P = 4.0 × 10−4; Fig. 7E). Together, these data could imply that inhibiting PI5P4Kα disrupts prostate tissue degradation under in vivo androgen deprivation conditions.
Fig. 7. PI5P4Kα affects prostate gland regression during androgen deprivation.
(A) Prostate-specific GEM model with R26eYFP reporter gene. PB-driven Cre recombinase system is used to delete Pip4k2afl/fl alleles at time of mouse sexual maturity. An eYFP fluorophore is permanently “turned on” from Cre recombinase activation. WT, wild type. (B) Graphical summary of in vivo androgen deprivation experiment using mouse castration. PB-Cre+;R26eYFP control and PB-Cre+;R26eYFP;Pip4k2afl/fl mice were aged to sexual maturity to activate Cre recombinase system, which can be detected with eYFP Cre reporter. Male mice were castrated at 8 weeks of age and harvested 10 weeks following surgery. (C) Representative images of GFP/eYFP detection with IF-IHC staining of castrated tissues. PB-expressing luminal cells show diffuse cytoplasmic GFP/eYFP staining of glandular epithelium. Scale bars, 100 μm. (D) Quantification of gland area of eYFP+ glands following surgical castration. Data are represented as median with Mann-Whitney test for significance (P = 0.0002). DAPI nuclear stains are used as positive-cell detection marks for IF assays. (E) Castrated UG mass relative to total mouse body weight shows an average of 0.26% for eYFP controls (PB-Cre+;R26eYFP; n = 9) compared to 0.32% for Pip4k2afl/fl animals (PB-Cre+;R26eYFP Pip4k2afl/fl; n = 8, one outlier, t test, P = 0.0004). t test values: **P < 0.01; and ***P < 0.001.
Depletion of PI5P4Kα exposes survival vulnerability in models of PCa
PI5P4Ks are promising targets for cancer therapeutic development in recent preclinical reports (8). Therefore, we first questioned whether vulnerabilities result from genetic Pip4k2a deletion. Noncancer organoids were isolated from mouse prostate tissue using the cell sorting for the R26eYFP reporter (Fig. 8A). Compared to wild-type controls, cell cultures generated from PB-Pip4k2afl/fl prostate tissue had distinct vacuolized morphology in two-dimensional (2D) organoid culture (Fig. 8B), a phenotype associated with metabolic stress, lipid accumulation, and lysosome dysregulation (50, 51). Transcript expression confirmed down-regulation of Pip4k2a with a subsequent increase in AR expression (Fig. 8C). Isolation of cells from PB-Pip4k2afl/fl mouse prostates did not demonstrate major differences in cell proliferation compared to controls (Pip4k2awt); however, when metabolically challenged with a glycolysis inhibitor, 2-deoxy-d-glucose (2DG), organoid cells had a greater reduction in proliferation (Fig. 8D). The role of Pip4k2a was further explored in a mouse model of PCa. Pip4k2a was up-regulated in aggressive PB-Cre+;Trp53fl/fl;Ptenfl/fl prostate tumors and was inversely correlated with the AR gene expression signature (Fig. 8, E and F).
Fig. 8. Pip4k2a is elevated in GEM cancer models, and deletion reduces prostate cell metabolic stress survival.
(A) Graphical representation of PB-Cre+;R26eYFP;Pip4k2afl/fl prostate luminal cells with and without Cre recombinase activation and eYFP reporter expression in PB-expressing cells. (B) Light phase microscopy image of PB-Pip4k2afl/fl organoids cells in 2D culture demonstrates vacuole morphologic features. Relative to eYFP+ control cells. Scale bars, 400 μm. (C) qRT-PCR gene expression shows that Pip4k2a fl/fl cells have less Pip4k2a transcript (0.23-fold, P = 1.32 ×10−6) and have an increase in Ar (1.83-fold, P = 0.002). (D) PB-Pip4k2afl/fl cells were treated with 1, 2, 5, or 10 μM of 2DG or H2O control for 72 hours. Differences in cell confluency are represented compared to eYFP+ only control cells from three independent experiments. (E to F) Transcript expression was measured from mouse anterior prostate from animals with normal prostates, genotypes, and histology indicating early progression of prostatic intraepithelial neoplasia (PIN) and tumors from PB-Cre+; Ptenfl/fl; Trp53fl/fl prostates. Ar signature genes, Nkx3.1 (0.01 tumor, P = 2.55 ×10−8) and Fkbp5 (7.2-fold in PIN, P = 0.01; 5.4-fold in tumor, P = 6.19 × 10−5) increase with early tumor development but are lost in advanced-stage disease. Pip4k2a is elevated in prostate tumors (32-fold in tumor, P = 2.52 ×10−7). t test values: *P < 0.05; **P < 0.01; and ***P < 0.001.
Next, we asked whether acute PI5P4Kα depletion affects the proliferation of human PCa models representative of different stages of disease. Lentiviral-mediated shRNA depletion of PIP4K2A enhances and sustains a knockdown beyond transient siRNA tools. A pLKO.1-GFP-shRNA lentiviral construct paired with green fluorescent protein (GFP) selection demonstrates that sustained PIP4K2A depletion prevents the viable proliferation of LNCaP, C4-2, and 22RV1 cells while reducing the doubling time of DU145 (control, 44.93 hours; shPIP4K2A, 48.80 hours; Fig. 9, A to D). In addition to cell lines, the PM154 organoid model of advanced PCa also demonstrated strong growth reduction following lentiviral delivery of shPIP4K2A (doubling time control, 62.45 hours; shPIP4K2A, 95.56 hours; Fig. 9E)
Fig. 9. Acute depletion of PIP4K2A exposes survival vulnerability in models of PCa.
Human PCa cell lines are impaired in proliferation when infected with stable PIP4K2A-targeted shRNA (shPIP4K2A) construct relative to control pLKO.1 lentivirus. PI5P4Kα knockdown confirmed by Western blots and cell proliferation changes measured by changes in confluence over time shown for (A) LNCaP, (B) C4-2, (C) 22RV1, and (D) DU145, which represents a model of castrate-resistant PCa (CRPC). (E) Changes in fold growth over time shown for the 3D Neuroendocrine PCa (NEPC) organoid line PM154. t test values: *P < 0.05 . Data are represented as means ± SD. (F) Graphical representation of the proposed mechanism. PI5P4Kα is an important regulator of metabolic organelles from the endolysosome system, which regulates the nutrient sensing and metabolic stress pathway driven by mTORC1. In PCa, AR and mTORC1 are established to have a dynamic interplay that is critical in prostate cell growth and survival. (i and ii) We show that the depletion of PIP4K2 increases PCa cell stress and results in the up-regulation of AR and suggest that this is mitigated via the down-regulation of mTORC1. (iii) Patient data suggests an increase in PIP4K2A in AR-independent PCa cases (CRPC). PI5P4Kα may become critical in maintaining CRPC growth by promoting mTORC1 and thereby exposes a vulnerability in the cancer metabolism.
Lastly, down-regulation of PIP4K2A in LNCaP cells enriched the proportion of G1 cycle–arrested cells (fig. S9, A and B). PIP4K2ACas9mut LNCaP cells demonstrate reduced proliferative ability following low-dose treatment of 0.5 μM BKM120 (fig. S9C), as well as when exposed to 5 and 10 μM APA (fig. S9D). Together, targeting PIP4K2A exposes a vulnerability in PCa through the dysregulation of key metabolic pathways.
DISCUSSION
The treatment of advanced PCa requires more than a mutational profile; clinicians need actionable interventions founded on functional prostate studies. We propose targeting a previously unexplored lipid kinase for PCa, PI5P4Kα. Here, we are the first to question how PI5P4K is expressed and functions in prostate luminal biology. We show that (i) PIP4K2A expression is enhanced in PCa with AR signaling reflecting CRPC, (ii) PI5P4Kα localizes to the lysosome where its influence on mTORC1 signaling may affect AR, and (iii) inhibiting PI5P4Kα exposes PCa cells to a metabolic vulnerability that may enhance efficacy of targeted therapeutics. This line of evidence prompts a model where, under AR high conditions, PI5P4Kα supports basal mTORC1 signaling and becomes more important as PCa cells metabolically adapt to androgen independence. CRPC relies on up-regulating mTORC1 for growth and survival through therapeutic stress; therefore, it is beneficial to increase the expression of enzymes like PI5P4Kα to orchestrate pathway dynamics (Fig. 9F).
We sought to examine whether PI5P4Kα inhibition affected AR and found an inverse relationship that we propose is mitigated via mTORC1. Previous literature strongly supports the involvement of the PI5P4K isoforms in promoting metabolic stress survival through metazoan mTORC pathways. A single PIP4K2 ortholog exists in the model organisms Caenorhabditis elegans, Drosophila, and zebrafish (Danio rerio), whereas mice and humans share three isoforms (α, β, and γ) (2, 3, 10, 52, 53). Gupta et al. (52) demonstrate that PIP4K2 mutant Drosophila have reduced mTORC1 signaling and growth that can be rescued with overexpression of the mTORC1 activator, Rheb. C. elegans lacking PPK-2 have a decreased life span and impaired lipid droplet recycling, a phenotype linked to dysfunctional lysosomal autophagy (10). Similarly, in mouse models, genetic targeting of Pip4k2a/b also altered lysosome-autophagosome fusion, resulting in mTORC1 suppression, liver lipid droplet accumulation, increased lysosome content, and defeats in lipid metabolism (9, 10). Using human cells, targeting PI5P4Kα/β inhibits mTORC1 phospho-target expression and exposes metabolic vulnerability to sarcoma and breast cancer cell survival under conditions of nutrient stress (9, 54). These observations are not restricted to the α and β isoforms, as PI5P4Kγ is reported to maintain a regulatory loop with mTORC1 to control activation in conditions of low nutrients (55). While less is known about the role of PI5P4K specifically in cancer biology, one hypothesis is that transformed cells could hijack PI5P4K to reprogram mTORC1 to maintain activation of cancer growth. In PCa, this may be particularly advantageous as AR-sensitive adenocarcinoma adapts to a “fuel switch,” as it lacks lipogenesis pathway stimulation following AD.
In this study, we associate elevated PIP4K2A expression with AR-independent PCa (i.e., CRPC). Targeting PI5P4K could offer a strategy to control mTORC1, as advanced PCa is highly reliant on the activation of mTORC1 for growth and survival. For example, the gene for the leucine transporter, LAT3, was first characterized because of its high overexpression in PCa (56). Leucine is an essential activator of the mTORC1 complex, and PCa up-regulates amino acid transporters with the purpose of meeting demands for supporting tumor growth (57). Furthermore, a reciprocal relationship has been previously established between AR and mTOR activity. Multiple studies report sub-baseline mTOR activation from nutrient stress or rapamycin-increased AR protein levels (36, 58). The precise mechanism coordinating this feedback is up for debate and could involve transcriptional regulation of mTOR repressors TSC Complex Subunit 1 and 2 (TSC1/2) by AR (58), deactivation of the tumor suppressor phosphatase PH Domain And Leucine Rich Repeat Protein Phosphatase 1 (PHLPP1) (59), reactivation of AKT via mTORC2 (36), or reactivation of insulin receptor substrate 1. Therefore, it is expected that most mTOR inhibitors as monotherapies have not succeeded past phase 2 clinical trials in PCa. This is thought to be due to a high degree of reciprocal feedback loops involving PI3K and AR that may function to develop drug resistance (23). It is presently accepted that therapeutic strategies that involve the inhibition of multiple arms of the PI3K-mTOR-AR network may offer the greatest promise in minimizing the risk of recurrence.
To our knowledge, this study is the first to directly show the colocalization of PI5P4Kα at the lysosome in tissue. The role of PI5P4K is especially context dependent; specifically, it varies between different cell types and the status of nutrient availability. The added complexity is layered onto the mechanistic details of the PI5P4K mode of action if also considering kinase-dependent or independent functions (44). One limitation of this study is that we do not directly address whether scaffolding functions of PI5P4K are involved in its relationship with AR signaling. While not presently in the scope of our investigation, further work comparing kinase inhibitors versus PI5P4K protein degraders will be valuable in demonstrating exact functions that PI5P4K relies on to support cancer metabolism.
mTORC1 is considered the “master regulator” of cell metabolism, and it is necessary for this protein complex to be localized to the surface membranes of the intracellular organelles, the lysosome, and endosome (60, 61). Endomembrane PIP lipids have major implications in mTORC1 regulation. Class II PI3K ß (PI3KC2ß) synthesizes PI-3,4-P2 at the late endosome, which can repress mTORC1 activity through recruitment of inhibitory binding proteins to Raptor (62). Recently, PI-3,5-P2 and PI-4,5-P2 were shown to be required for mTORC1 regulation at the lysosome surface through TSC translocation and complex binding (16). Although a very minor fraction of phospholipids, the PIPs embedded into lipid membranes within and surrounding the cell regulate a large number of molecules that have a variety of functions [reviewed in (63)]. There is still limited knowledge regarding the full spectrum of signaling cascades that are orchestrated by PIP lipids in PCa. Goto et al. (64) nominate PIP lipids as potential PCa biomarkers after conducting the first spatial high-resolution matrix-assisted laser desorption/ionization imaging mass spectrometry analysis of benign versus cancer patient tissue. Recently, shifts in PIP abundance and acyl chain saturation have also been associated with PTEN mutant PCa (45). A recurring limitation to most published studies involving PIPs in cancer, however, is the technical challenge in deciphering individual PI classes (PI, PIP1, PIP2, and PIP3) and regioisomers (PI-4,5-P2 versus PI-3,4-P2).
More than 150 kinase-targeted drugs (mostly protein kinases) are currently in cancer clinical trials, with about 37 having received U.S. Food and Drug Administration approval since the 1980s (65). In May 2019, alpelisib, a PI3K inhibitor, was the first of its kind to be approved for breast cancer. Kinases are key signaling molecules that offer effective drug targets in a pharmaceutical environment already adapted for their development. Interest in the PI5P4Ks is growing, as seen with the publication of a pan isoform covalent inhibitor (THZ-P1-2) (40), a dual PI5P4Kα/β inhibitor (54) and earlier single-isoform targeted compounds (66, 67).
In conclusion, this is the first report to link PI5P4Kα to the AR pathway and its regulation in PCa. Evidence reported here opens the question of whether PI5P4Kα is vital for maintaining mTORC1 activation under stress conditions, specifically at the endolysosome, which relies heavily on PI to mitigate organelle dynamics. We propose that the influence of PI5P4Kα on lysosomal mTORC1 is responsible for stress activation of AR and exposes survival vulnerability in cancer. Herein prompts the rationale for the therapeutic development of PI5P4K-targeted inhibitors for the multimodal treatment of PCa.
MATERIALS AND METHODS
AR signaling score and mTOR score analysis of patient datasets
AR signaling score was calculated as previously described, namely, by calculating the Pearson’s correlation coefficient between the log2-transformed fragments per kilobase of exon per million mapped fragments (FPKM) values of a set of 30 AR-regulated genes and a reference vector (tables S1 and S2) (68). The mTOR signaling score was calculated as previously described (17). Briefly, a t test was performed between the log2-transformed FPKM values of genes up-regulated on mTOR pathway activation, against those down-regulated on mTOR pathway activation. The absolute value of the resulting t statistic was then used as the “mTOR score” for each sample.
Cell models and culture conditions
LNCaP [male; American Type Culture Collection (ATCC), RRID: CVCL_1379], C4-2 (male; ATCC, RRID: CRL-3314), VCaP (male; ATCC, RRID: CRL-2876), PC3 (male; ATCC, RRID: CRL-3470), DU145 (male; ATCC, RRID: CVCL_0105), 22RV1, and RWPE (male; ATCC, RRID: CVCL_3791) cells were acquired from the ATCC. LNCaP-AR cells were a gift from Charles L. Sawyers and Ping Mu (Memorial Sloan Kettering Cancer Center) (34). LNCaP were grown in RPMI 1640 (Gibco, A1049101) supplemented with 5% FBS (Gibco, 10270106) and 1% penicillin-streptomycin (Gibco, 11548876) on poly-l-lysine–coated plates. VCaP, PC3, DU145, and 22RV1 were maintained in Dulbecco’s modified Eagle’s medium (Gibco, 31966021), supplemented with 10% FBS and 1% penicillin-streptomycin. RWPE were maintained in keratinocyte serum-free medium (Gibco, 17005075) supplemented with bovine pituitary extract and human recombinant EGF (included) and 1% penicillin-streptomycin (Gibco, 11548876). Androgen deprivation (AD) was modeled in vitro by supplementing RPMI 1640, lacking phenol red (Fisher Scientific, 11-835-055) with 5% C/S serum (Thermo Scientific, A3382101) and penicillin-streptomycin. All cell lines were grown at 37°C with 5% CO2, regularly tested for mycoplasma and authenticated using short tandem repeat (STR) profiling.
GSEA and RNA-seq
Cell line transcript data were acquired from the Cancer Cell Line Encyclopedia (69) database, and organoid cell models were sequenced as reported (table S3) (33, 70). Bulk RNA-seq was conducted on RNA extracted from phosphate-buffered saline (PBS)–washed cell pellets using a QIAGEN RNeasy kit. RNA was deoxyribonuclease-treated (Ambion, AM1906) and quality-assessed using Bioanalyzer and Qubit. Sequencing libraries were prepared using 500 ng of RNA and Illumina TruSeq Stranded mRNA with IDT for Illumina TruSeq UD RNA indexes. FASTQ read files were analyzed as previously described (70) using STARv2.3.0e (71), RSEQtools (72), and GENCODE v19 (www.gencodegenes.org/human/release_19.html). GSEA analysis was conducted as previously described (9).
Chemical inhibitors and cell proliferation assays
A summary of key reagent product information is listed in table S4.
siRNA targeted knockdown
Cell lines were plated on poly-l-lysine–coated six-well plates and then transfected with pooled siRNA targeting PIP4K2A (siPIP4K2A) or NT control oligos (ON-TARGETplus, Dharmacon) using recommended conditions of the Lipofectamine RNAiMAX Transfection Reagent (Thermo Fisher Scientific, 13778150). The next day, we switched to C/S medium and light RPMI 1640 + 5% C/S FBS for 48 hours for AD conditions.
Inhibitor treatment
Cells were treated with 5 or 10 μM of APA (Selleck Chemicals, S2840), 1 nM rapamycin (Selleck Chemicals, S1039), or 10 μM BKM120 (Selleck Chemicals, S2247) with dimethyl sulfoxide (DMSO) solvent control in culture medium in either six-well or 96-well plate formats. Inhibitor(s) efficacy was confirmed with quantitative real-time polymerase chain reaction (qRT-PCR) or Western blot of known downstream targets. Glycolysis inhibition was done using 2DG (Sigma-Aldrich, D8375) in sterile H2O at indicated dilutions. DHT (Sigma-Aldrich, 10300) treatments were done for 18 hours in AD medium, which is made up of 5% C/S serum. Ethanol was used for DHT solvent control.
Cell proliferation
Cell models were seeded on poly-l-lysine–coated 96-well plates at 5000 cells per well between three and six replicates per condition per experiment repeat. Cell confluence was determined using the IncuCyte S3 instrument and the IncuCyte S3 2018B software (Essen Bioscience, Germany). Changes in proliferation over time are estimated on the basis of the digital area mask coverage of field of view from microscope images captured at ×20 magnification.
Lentiviral hairpin systems
Plasmids used were previously generated for stable shRNA knockdown of PIP4K2A (pLKO.1-GFP-shPIP4K2A and control pLKO.1-GFP) (10) and doxycycline-induced knockdown of PIP4K2A (Ez-tet-Puro-Scramble, Ind5145, and Ind6009) (9). Lentivirus was produced as previously reported using transfection of human embryonic kidney 293T cells. The target cell population was infected with 1:4 volume virus:medium ratio in medium containing polybrene (0.8 μg/ml), then centrifuged at 1000g for 30 min, and left to recover for 48 hours in a cell culture incubator. Selection was conducted using fluorescence-activated cell sorting (FACS) for GFP+ cells.
Western blot protein detection
Protein lysates were extracted from PBS-washed cell pellets using radioimmunoprecipitation assay with protease and phosphatase inhibitors. Total protein concentration was measured using the Pierce BCA Protein Assay Kit (Thermo Fisher Scientific). Between 20 and 50 μg of protein was used for standard SDS–polyacrylamide gel electrophoresis procedures with Mini-Protean TGX gels (4 to 15%; Bio-Rad, 456-1084). Membrane transfer to nitrocellulose was done using an iBlot2 quick transfer system (Thermo Fisher Scientific, IB23001). Blocking and secondary antibody incubations were done using 5% milk in Tris-buffered saline with 0.1% Tween® 20 detergent (TBST), and primary antibodies were probed with optimized conditions in 5% BSA unless stated otherwise (table S5). A minimum of three 5-min wash steps in TBST (1% tween) were used between incubations and before visualization. Final detection with chemiluminescence was done with the Luminata Forte substrate (Thermo Fisher Scientific, WBLUF0100), and multiple image exposures were acquired with the FUSION FX7 EDGE Imaging System (Witec AG). Band densitometry was measured using ImageJ version 1.53a and normalized to housekeepers.
qRT-PCR transcript detection
RNA was extracted from cell pellets using the QIAGEN RNeasy Kit (QIAGEN, 74106). cDNA was synthesized using the FIREScript RT cDNA Synthesis Kit (Solis BioDyne, 06-15-00200); then, qRT-PCR was conducted on a ViiA 7 system (Applied Biosystems) with the HOT FIREPol EvaGreen qPCR master mix (Solis BioDyne, 08-24-00020). Gene cycle thresholds were calculated using triplicate replicates and normalized to housekeeping controls. See table S6 for primer list and sequences.
LysoTracker flow cytometry
LNCaP (3 × 105) were treated with 10 μM of APA or DMSO for 48 hours. Cells were incubated with 50 nM LysoTracker Deep Red (Invitrogen), freshly prepared in RPMI 1640 + 5% FBS, for 1 hour in the dark at 37°C. At least 10 000 live cells were captured using ImageStream-X MKII flow cytometry and analyzed by the IDEAS version 6.2.
Lipidomics analysis
Lipid extraction
Cell metabolism was quenched by the addition of cold 80% methanol (MeOH) following cell scraping. This methanolic solution of collected cells was evaporated, and lipids were extracted by the addition of butanol:MeOH (1:1) through mechanic homogenization (2 × 20 s in Precellys tissue homogenizer). Protein quantification was performed on the remaining cell pellets, and the total protein content was used for sample amount normalization.
Targeted analysis of lipids
Analysis on extract lysates was performed by hydrophilic interaction liquid chromatography coupled to tandem mass spectrometry (MS/MS) in both positive and negative ionization modes using a Q-TRAP 6500+ liquid chromatography–MS/MS (LC-MS/MS) system (Sciex Technologies) (73, 74).
Quality control
Pooled quality control (QC) samples were analyzed periodically (every five to six samples) throughout the overall analytical run to assess the data quality, correct the signal intensity drift (if any), and remove the peaks with poor reproducibility [coefficient of variation (CV) > 30%]. In addition, a series of diluted QCs were prepared by dilution with butanol:MeOH: 100, 50, 25, 12.5, and 6.25% QC.
Data processing
Raw LC-MS/MS data were processed using the MultiQuant Software (version 3.0.3, Sciex Technologies). Relative quantification of metabolites was based on extracted ion chromatogram areas for the monitored multiple reaction monitoring (MRM) transitions. Signal intensity drift correction on peak area data was done within the LOWESS/Spline normalization program 8 (R software), followed by noise filtering [CV (QC features) > 30%] and visual inspection of linear response (75).
Intracellular PI-4,5-P2 detection by IF
PI-4,5-P2 detection in intracellular membranes was done using IF protocols as previously described (9, 48). Briefly, cells were plated and drug-treated on poly-l-lysine–coated coverslips in six-well dishes. At a final time point, cells were fixed in 2% paraformaldehyde (PFA) and then permeabilized with 20 mM digitonin. Samples were blocked in 5% normal goat serum in buffer A with 50 mM NH4Cl before being incubated in primary anti–PI-4,5-P2 (1:100; Abcam, ab11039) overnight. Samples were washed three times and then incubated with secondary antibodies, followed by postfixation with 2% PFA in PBS. DAPI (4′,6-diamidino-2-phenylindole) was added to the wash buffer during the final wash steps. Samples slides were imaged with 60× oil immersion on Zeiss LSM 710 confocal microscope. Images were quantified using the ImageJ Spot Localization ComDet plug-in. A minimum of 50 cells were quantified per condition per replicate.
GEM models
All animal studies were approved by the Cantonal Veterinary Ethical Committee, Switzerland (license BE 41/2018). Animals were housed in ventilated cages with unrestricted access to presterilized food and fresh water. A maximum of five animals were maintained per cage on Aspen bedding. Ambient temperature was 20° ± 2°C, kept at a constant humidity of 50 ± 10%, and on a 12-hour automatic light-dark cycle. Pip4k2atm1.2Lca animals (MGI:5568930), referred to as Pip4k2afl/fl, were a gift of the group of L. Cantley. The generation of Pip4k2afl/fl (2, 10), Probasin-Cre (PB-Cre) (76), and Rosa26eYFP (77) mice has been previously described. These strains were then crossbred to generate the PB-Cre+/−; R26eYFP+/+; Pip4k2afl/fl mice used in the mouse adult prostate and castration study. Breeding schemes limited inclusion of the PB-Cre+ allele exclusively through male mice to avoid off-target effects. Animals were genotyped using the Transnetyx real-time PCR platform.
Castration surgery
Surgical castration or sham surgery was performed on 8-week-old C57BL/6JRj mice. Standard procedures are previously described (78). Mice are preemptively dosed with buprenorphine, and the surgical area was shaved following confirmation of suitable anesthetic plane using isoflurane and a 37°C heat pad. Marcaine (0.25 to 0.5%) solution was used for local anesthetic, and the surgical site was prepared with alternating 70% isopropyl alcohol and povidone-iodine scrub. A midline incision was made in the scrotum, and testicles were removed with cauterization. The incision site was closed using one sterilized wound clip (Autoclip), which was removed between 10 and 14 days postoperatively. Animals were monitored for the duration of the experiment, and analgesic treatments were applied for 3 days following the procedure. UG tissue mass, including prostate, bladder, and seminal vesicle was recorded, while prostate tissue was collected 10 weeks following castration or sham procedure.
IHC detection
Tissue sections were processed using standard histology procedures, fixed with 4% formaldehyde overnight, and paraffin-embedded, and sections were cut at 2.5 μm. Antibody staining was optimized for PI5P4Kα (Proteintech, 1246-1-AP) with 30 min tris buffer antigen retrieval with 1:200 dilution antibody and with 30-min citrate buffer antigen retrieval. Conditions for additional antibodies are listed in table S5. Slide staining was done using the BOND RX autostainer (Leica Biosystems) as previously described (75, 79)
IF tissue staining
Adult C57BL/6JRj mouse prostates were harvested and paraffin-embedded. Tissue serial sections were deparaffinized in xylene and rehydrated using decreasing concentrations of ethanol (100 to 95 to 70% to H2O). Heat-induced epitope retrieval was used at a setting of 98°C for 35 min in tris-EDTA buffer. Slides were left to return to room temperature, then washed in Tris-buffered saline (TBS), blocked in 5% normal goat serum at room temperature, and probed with primary antibodies overnight at 4°C in an immunostain moisture chamber. Protein targets were detected using 1:200 PI5P4Kα polyclonal antibody (Proteintech, 1249-1-AP) and 1:200 LAMP1 monoclonal antibody (Abcam; Ab24871) with 1:300 goat anti-rabbit Cy3 (Jackson ImmunoResearch, 111-165-003) or donkey anti-rabbit A488 (Jackson ImmunoResearch, 712-545-150) secondary antibody. DAPI (100 ng/ml) was used as nuclear counterstain, and ProLong Gold (Thermo Fisher Scientific; P36930) was used for mounting. Images were acquired with 60× oil immersion on Zeiss LSM 710 confocal microscope.
Digital tissue quantification
Whole-slide images were acquired on a Pannoramic 250 (3DHistech) slide scanner, and images were exported from CaseView software (version 2.5). File codes were used to blind the investigator from the experimental group during data acquisition and annotation. Sample group blinding was also implemented for pathology evaluation of tissue samples. Images were analyzed using QuPath v0.2 (80) and as previously described (81).
Primary mouse organoid models
Mouse organoids were isolated as previously described (82). Adult prostates were harvested, dissociated overnight, and FACS-sorted for eYFP+ cell population from mouse lines containing activated R26eYFP Cre recombinase reporter. Organoids were maintained in 3D Matrigel culture and transferred to collagen-coated 96-well plates for 2D proliferation assays (as described above).
Statistics and data availability
Data are expressed as means ± SD. Statistical analyses for all data, including microscopy quantification, qRT-PCR data, metabolomics, and lipidomics, were done using Student’s two-tailed t test or analysis of variance (ANOVA) as indicated using GraphPad Prism. Statistical significance is indicated in the figure legends [*P < 0.05; **P < 0.01; ***P < 0.001; and not significant (n.s.), P > 0.05] unless specified otherwise. The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request. RNA-seq, lipidomics, and Western blot source data are available in data S1 to S3.
Acknowledgments
We are especially appreciative to all the help from our entire team and collaborators.
Funding: This work was funded by the Swiss National Science Foundation (SNF#31003A_175609 and SNF#310030_207635), an EU Commission Marie Skłodowska-Curie Individual Fellowship (PCAPIP), and The Johanna Dürmüller-Bol Foundation. Work in the B.M.E. laboratory was supported by NCI (R01 CA237536, CBC under contract 1053 no.75N91019D00024, task order no. 75N91020F00003) and ACS (RSG-20-064-01-TBE and TLC-21-156-01). L.C. was supported by the National Cancer Institute (R35 CA197588). The metabolomics platform thanks SNF for the financial support: R’Equip grant no. 183377. Additional support was provided by the Englander Institute for Precision Medicine (EIPM).
Author contributions: J.T.: Conceptualization, data curation, formal analysis, funding acquisition, investigation, methodology, project administration, resources, supervision, validation, visualization, and writing the original draft. M.R.: Data curation and methodology. F.C.M.: Data curation. M.Le.: Data curation and methodology. J.G.: Computational analysis. L.K.: Data curation and validation. A.R.: Methodology. G.K.A.: Methodology. S.d.B.: Data curation and methodology. M.Lu.: Conceptualization. H.G.-A.: Data curation and computational analysis. J.I.: Data curation and computational analysis. S.P.: Computational analysis and supervision. L.C.: Conceptualization. B.M.E.: Conceptualization, methodology, supervision, and manuscript writing. M.A.R.: Conceptualization, methodology, supervision, manuscript writing, and funding acquisition.
Competing interests: L.C. is a founder and scientific advisory board member of Agios Pharmaceuticals, Faeth Therapeutics, Petra Pharma Corporation, Larkspur Therapeutics, and Volastra Pharmaceuticals and a scientific advisory board member for Scorpion Therapeutics. All other authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials.
Supplementary Materials
This PDF file includes:
Figs. S1 to S9
Tables S1 to S6
Other Supplementary Material for this : manuscript includes the following:
Data S1 to S3
REFERENCES AND NOTES
- 1.L. E. Rameh, K. F. Tolias, B. C. Duckworth, L. C. Cantley, A new pathway for synthesis of phosphatidylinositol-4,5-bisphosphate. Nature 390, 192–196 (1997). [DOI] [PubMed] [Google Scholar]
- 2.B. M. Emerling, J. B. Hurov, G. Poulogiannis, K. S. Tsukazawa, R. Choo-Wing, G. M. Wulf, E. L. Bell, H. S. Shim, K. A. Lamia, L. E. Rameh, G. Bellinger, A. T. Sasaki, J. M. Asara, X. Yuan, A. Bullock, G. M. Denicola, J. Song, V. Brown, S. Signoretti, L. C. Cantley, Depletion of a putatively druggable class of phosphatidylinositol kinases inhibits growth of p53-null tumors. Cell 155, 844–857 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.K. A. Lamia, O. D. Peroni, Y. Kim, L. E. Rameh, B. B. Kahn, L. C. Cantley, Increased insulin sensitivity and reduced adiposity in phosphatidylinositol 5-phosphate 4-kinase β−/− mice. Society. 24, 5080–5087 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.H. Shim, C. Wu, S. Ramsamooj, K. N. Bosch, Z. Chen, B. M. Emerling, Deletion of the gene Pip4k2c, a novel phosphatidylinositol kinase, results in hyperactivation of the immune system. Proc. Natl. Acad. Sci. U.S.A. 113, 1–10 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.J. G. Jude, G. J. Spencer, X. Huang, T. D. D. Somerville, D. R. Jones, N. Divecha, T. C. P. Somervaille, A targeted knockdown screen of genes coding for phosphoinositide modulators identifies PIP4K2A as required for acute myeloid leukemia cell proliferation and survival. Oncogene 34, 1–10 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.K. Lima, D. M. Ribeiro, P. de Melo Campos, F. F. Costa, F. Traina, S. T. Olalla Saad, M. de Fátima Sonati, J. A. Machado-Neto, Differential profile of PIP4K2A expression in hematological malignancies. Blood Cells Mol. Dis. 55, 228–235 (2015). [DOI] [PubMed] [Google Scholar]
- 7.Y. J. Shin, J. K. Sa, Y. Lee, D. Kim, N. Chang, H. J. Cho, M. Son, M. Y. T. Oh, K. Shin, J.-K. Lee, J. Park, Y. K. Jo, M. Kim, P. J. Paddison, V. Tergaonkar, J. Lee, D.-H. Nam, PIP4K2A as a negative regulator of PI3K in PTEN-deficient glioblastoma. J. Exp. Med. 216, 1120–1134 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.G. K. Arora, L. Palamiuc, B. M. Emerling, Expanding role of PI5P4Ks in cancer: A promising druggable target. FEBS Lett. 596, 3–16 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.A. Ravi, L. Palamiuc, R. M. Loughran, J. Triscott, G. K. Arora, A. Kumar, V. Tieu, C. Pauli, M. Reist, R. J. Lew, S. L. Houlihan, C. Fellmann, C. Metallo, M. A. Rubin, B. M. Emerling, PI5P4Ks drive metabolic homeostasis through peroxisome-mitochondria interplay. Dev. Cell , 1661–1676.e10 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.M. R. Lundquist, M. D. Goncalves, R. M. Loughran, E. Possik, T. Vijayaraghavan, A. Yang, C. Pauli, A. Ravi, A. Verma, Z. Yang, J. L. Johnson, J. C. Y. Wong, Y. Ma, K. S. K. Hwang, D. Weinkove, N. Divecha, J. M. Asara, O. Elemento, M. A. Rubin, A. C. Kimmelman, A. Pause, L. C. Cantley, B. M. Emerling, Phosphatidylinositol-5-phosphate 4-kinases regulate cellular lipid metabolism by facilitating autophagy. Mol. Cell 70, 531–544.e9 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Y. Rong, M. Liu, L. Ma, W. Du, H. Zhang, Y. Tian, Z. Cao, Y. Li, H. Ren, C. Zhang, L. Li, S. Chen, J. Xi, L. Yu, Clathrin and phosphatidylinositol-4,5-bisphosphate regulate autophagic lysosome reformation. Nat. Cell Biol. 14, 924–934 (2012). [DOI] [PubMed] [Google Scholar]
- 12.Y. Sun, A. C. Hedman, X. Tan, N. J. Schill, R. A. Anderson, Endosomal type Iγ PIP 5-kinase controls EGF receptor lysosomal sorting. Dev. Cell 25, 144–155 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.D. A. Sarkes, L. E. Rameh, A novel HPLC-based approach makes possible the spatial characterization of cellular PtdIns5P and other phosphoinositides. Biochem. J. 428, 375–384 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.J. Viaud, F. Boal, H. Tronchère, F. Gaits-Iacovoni, B. Payrastre, Phosphatidylinositol 5-phosphate: A nuclear stress lipid and a tuner of membranes and cytoskeleton dynamics. BioEssays 36, 260–272 (2014). [DOI] [PubMed] [Google Scholar]
- 15.A. Hu, X.-T. Zhao, H. Tu, T. Xiao, T. Fu, Y. Wang, Y. Liu, X.-J. Shi, J. Luo, B.-L. Song, PIP4K2A regulates intracellular cholesterol transport through modulating PI(4,5)P 2 homeostasis. J. Lipid Res. 59, 507–514 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.K. Fitzian, A. Brückner, L. Brohée, R. Zech, C. Antoni, S. Kiontke, R. Gasper, A. L. Linard Matos, S. Beel, S. Wilhelm, V. Gerke, C. Ungermann, M. Nellist, S. Raunser, C. Demetriades, A. Oeckinghaus, D. Kümmel, TSC1 binding to lysosomal PIPs is required for TSC complex translocation and mTORC1 regulation. Mol. Cell 81, 1–18 (2021). [DOI] [PubMed] [Google Scholar]
- 17.Y. Zhang, P. K.-S. Ng, M. Kucherlapati, F. Chen, Y. Liu, Y. H. Tsang, G. de Velasco, K. J. Jeong, R. Akbani, A. Hadjipanayis, A. Pantazi, C. A. Bristow, E. Lee, H. S. Mahadeshwar, J. Tang, J. Zhang, L. Yang, S. Seth, S. Lee, X. Ren, X. Song, H. Sun, J. Seidman, L. J. Luquette, R. Xi, L. Chin, A. Protopopov, T. F. Westbrook, C. S. Shelley, T. K. Choueiri, M. Ittmann, C. van Waes, J. N. Weinstein, H. Liang, E. P. Henske, A. K. Godwin, P. J. Park, R. Kucherlapati, K. L. Scott, G. B. Mills, D. J. Kwiatkowski, C. J. Creighton, A pan-cancer proteogenomic atlas of PI3K/AKT/mTOR pathway alterations. Cancer Cell 31, 820–832.e3 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.D. Robinson, E. M. Van Allen, Y.-M. Wu, N. Schultz, R. J. Lonigro, J.-M. Mosquera, B. Montgomery, M.-E. Taplin, C. C. Pritchard, G. Attard, H. Beltran, W. Abida, R. K. Bradley, J. Vinson, X. Cao, P. Vats, L. P. Kunju, M. Hussain, F. Y. Feng, S. A. Tomlins, K. A. Cooney, D. C. Smith, C. Brennan, J. Siddiqui, R. Mehra, Y. Chen, D. E. Rathkopf, M. J. Morris, S. B. Solomon, J. C. Durack, V. E. Reuter, A. Gopalan, J. Gao, M. Loda, R. T. Lis, M. Bowden, S. P. Balk, G. Gaviola, C. Sougnez, M. Gupta, E. Y. Yu, E. A. Mostaghel, H. H. Cheng, H. Mulcahy, L. D. True, S. R. Plymate, H. Dvinge, R. Ferraldeschi, P. Flohr, S. Miranda, Z. Zafeiriou, N. Tunariu, J. Mateo, R. Perez-Lopez, F. Demichelis, B. D. Robinson, M. Schiffman, D. M. Nanus, S. T. Tagawa, A. Sigaras, K. W. Eng, O. Elemento, A. Sboner, E. I. Heath, H. I. Scher, K. J. Pienta, P. Kantoff, J. S. de Bono, M. A. Rubin, P. S. Nelson, L. A. Garraway, C. L. Sawyers, A. M. Chinnaiyan, Integrative clinical genomics of advanced prostate cancer. Cell 161, 1215–1228 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.P. Castel, E. Toska, J. A. Engelman, M. Scaltriti, The present and future of PI3K inhibitors for cancer therapy. Nat. Cancer 2, 587–597 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.M. Kirby, C. Hirst, E. D. Crawford, Characterising the castration-resistant prostate cancer population: A systematic review. Int. J. Clin. Pract. 65, 1180–1192 (2011). [DOI] [PubMed] [Google Scholar]
- 21.E. Audet-Walsh, T. Yee, S. McGuirk, M. Vernier, C. Ouellet, J. St-Pierre, V. Giguere, Androgen-dependent repression of ERRγ reprograms metabolism in prostate cancer. Cancer Res. 77, 378–389 (2016). [DOI] [PubMed] [Google Scholar]
- 22.J. Armenia, S. A. M. Wankowicz, D. Liu, J. Gao, R. Kundra, E. Reznik, W. K. Chatila, D. Chakravarty, G. C. Han, I. Coleman, B. Montgomery, C. Pritchard, C. Morrissey, C. E. Barbieri, H. Beltran, A. Sboner, Z. Zafeiriou, S. Miranda, C. M. Bielski, A. V. Penson, C. Tolonen, F. W. Huang, D. Robinson, Y. M. Wu, R. Lonigro, L. A. Garraway, F. Demichelis, P. W. Kantoff, M.-E. Taplin, W. Abida, B. S. Taylor, H. I. Scher, P. S. Nelson, J. S. de Bono, M. A. Rubin, C. L. Sawyers, A. M. Chinnaiyan, N. Schultz, E. M. Van Allen, The long tail of oncogenic drivers in prostate cancer. Nat. Genet. 50, 645–651 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.B. S. Carver, C. Chapinski, J. Wongvipat, H. Hieronymus, Y. Chen, S. Chandarlapaty, V. K. Arora, C. Le, J. Koutcher, H. Scher, P. T. Scardino, N. Rosen, C. L. Sawyers, Reciprocal feedback regulation of PI3K and androgen receptor signaling in PTEN-deficient prostate cancer. Cancer Cell 19, 575–586 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.D. J. Mulholland, L. M. Tran, Y. Li, H. Cai, A. Morim, S. Wang, S. Plaisier, I. P. Garraway, J. Huang, T. G. Graeber, H. Wu, Cell autonomous role of PTEN in regulating castration-resistant prostate cancer growth. Cancer Cell. 19, 792–804 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.S. Wang, J. Gao, Q. Lei, N. Rozengurt, C. Pritchard, J. Jiao, G. V. Thomas, G. Li, P. Roy-Burman, P. S. Nelson, X. Liu, H. Wu, Prostate-specific deletion of the murine Pten tumor suppressor gene leads to metastatic prostate cancer. Cancer Cell 4, 209–221 (2003). [DOI] [PubMed] [Google Scholar]
- 26.J. S. De Bono, U. De Giorgi, D. N. Rodrigues, C. Massard, S. Bracarda, A. Font, J. A. A. Arija, K. C. Shih, G. D. Radavoi, N. Xu, W. Y. Chan, H. Ma, S. Gendreau, R. Riisnaes, P. H. Patel, D. J. Maslyar, V. Jinga, Randomized phase II study evaluating AKT blockade with ipatasertib, in combination with abiraterone, in patients with metastatic prostate cancer with and without PTEN loss. Clin. Cancer Res. 25, 928–936 (2019). [DOI] [PubMed] [Google Scholar]
- 27.C. Sweeney, S. Bracarda, C. N. Sternberg, K. N. Chi, D. Olmos, S. Sandhu, C. Massard, N. Matsubara, B. Alekseev, F. Parnis, V. Atduev, G. L. Buchschacher, R. Gafanov, L. Corrales, M. Borre, D. Stroyakovskiy, G. V. Alves, E. Bournakis, J. Puente, M. L. Harle-Yge, J. Gallo, G. Chen, J. Hanover, M. J. Wongchenko, J. Garcia, J. S. de Bono, Ipatasertib plus abiraterone and prednisolone in metastatic castration-resistant prostate cancer (IPATential150): a multicentre, randomised, double-blind, phase 3 trial. Lancet 398, 131–142 (2021). [DOI] [PubMed] [Google Scholar]
- 28.H. Hieronymus, J. Lamb, K. N. Ross, X. P. Peng, C. Clement, A. Rodina, M. Nieto, J. Du, K. Stegmaier, S. M. Raj, K. N. Maloney, J. Clardy, W. C. Hahn, G. Chiosis, T. R. Golub, Gene expression signature-based chemical genomic prediction identifies a novel class of HSP90 pathway modulators. Cancer Cell 10, 321–330 (2006). [DOI] [PubMed] [Google Scholar]
- 29.J. H. Clarke, R. F. Irvine, Evolutionarily conserved structural changes in phosphatidylinositol 5-phosphate 4-kinase (PI5P4K) isoforms are responsible for differences in enzyme activity and localization. Biochem. J. 454, 49–57 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Y. Bultsma, W. J. Keune, N. Nivecha, PIP4Kβ interacts with and modulates nuclear localization of the high-activity PtdIns5P-4-kinase isoform PIP4Kα. Biochem. J. 430, 223–235 (2010). [DOI] [PubMed] [Google Scholar]
- 31.L. Puca, R. Bareja, D. Prandi, R. Shaw, M. Benelli, W. R. Karthaus, J. Hess, M. Sigouros, A. Donoghue, M. Kossai, D. Gao, J. Cyrta, V. Sailer, A. Vosoughi, C. Pauli, Y. Churakova, C. Cheung, L. D. Deonarine, T. J. Mcnary, R. Rosati, S. T. Tagawa, D. M. Nanus, J. M. Mosquera, C. L. Sawyers, Y. Chen, G. Inghirami, R. A. Rao, C. Grandori, O. Elemento, A. Sboner, F. Demichelis, M. A. Rubin, H. Beltran, Patient derived organoids to model rare prostate cancer phenotypes. Nat. Commun. 9, 1–10 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.D. Gao, I. Vela, A. Sboner, P. J. Iaquinta, W. R. Karthaus, A. Gopalan, C. Dowling, J. N. Wanjala, E. A. Undvall, V. K. Arora, J. Wongvipat, M. Kossai, S. Ramazanoglu, L. P. Barboza, W. Di, Z. Cao, Q. F. Zhang, I. Sirota, L. Ran, T. Y. MacDonald, H. Beltran, J.-M. Mosquera, K. A. Touijer, P. T. Scardino, V. P. Laudone, K. R. Curtis, D. E. Rathkopf, M. J. Morris, D. C. Danila, S. F. Slovin, S. B. Solomon, J. A. Eastham, P. Chi, B. Carver, M. A. Rubin, H. I. Scher, H. Clevers, C. L. Sawyers, Y. Chen, Organoid cultures derived from patients with advanced prostate cancer. Cell 159, 176–187 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.F. Tang, D. Xu, S. Wang, C. K. Wong, A. Martinez-Fundichely, C. J. Lee, S. Cohen, J. Park, C. E. Hill, K. Eng, R. Bareja, T. Han, E. M. Liu, A. Palladino, W. Di, D. Gao, W. Abida, S. Beg, L. Puca, M. Meneses, E. de Stanchina, M. F. Berger, A. Gopalan, L. E. Dow, J. M. Mosquera, H. Beltran, C. N. Sternberg, P. Chi, H. I. Scher, A. Sboner, Y. Chen, E. Khurana, Chromatin profiles classify castration-resistant prostate cancers suggesting therapeutic targets. Science 376, eabe1505 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.C. Tran, S. Ouk, N. J. Clegg, Y. Chen, P. A. Watson, V. Arora, J. Wongvipat, P. M. Smith-jones, D. Yoo, A. Kwon, T. Wasielewska, D. Welsbie, C. D. Chen, C. S. Higano, T. M. Beer, D. T. Hung, H. I. Scher, M. E. Jung, C. L. Sawyers, Development of a second-generation antiandrogen for treatment of advanced prostate cancer. Science 826, 787–790 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.C. A. Rothermund, V. K. Gopalakrishnan, J. K. Vishwanatha, Androgen signaling and post-transcriptional downregulation of Bcl-2 in androgen-unresponsive prostate cancer. Prostate Cancer Prostatic Dis. 7, 158–164 (2004). [DOI] [PubMed] [Google Scholar]
- 36.Y. Wang, M. Mikhailova, S. Bose, C. X. Pan, R. W. D. V. White, P. M. Ghosh, Regulation of androgen receptor transcriptional activity by rapamycin in prostate cancer cell proliferation and survival. Oncogene. 27, 7106–7117 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.L. Wu, D. C. Birle, I. F. Tannock, Effects of the mammalian target of rapamycin inhibitor CCI-779 used alone or with chemotherapy on human prostate cancer cells and xenografts. Cancer Res. 65, 2825–2831 (2005). [DOI] [PubMed] [Google Scholar]
- 38.S. H. Lee, D. Johnson, R. Luong, Z. Sun, Crosstalking between androgen and PI3K/AKT signaling pathways in prostate cancer cells. J. Biol. Chem. 290, 2759–2768 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.K. E. O’Reilly, mTOR inhibition induces upstream receptor tyrosine kinase signaling and activates Akt. Cancer Res. 66, 1500–1508 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.S. C. Sivakumaren, H. Shim, T. Zhang, F. M. Ferguson, M. R. Lundquist, C. M. Browne, H. S. Seo, M. N. Paddock, T. D. Manz, B. Jiang, M. F. Hao, P. Krishnan, D. G. Wang, T. J. Yang, N. P. Kwiatkowski, S. B. Ficarro, J. M. Cunningham, J. A. Marto, S. Dhe-Paganon, L. C. Cantley, N. S. Gray, Targeting the PI5P4K lipid kinase family in cancer using covalent inhibitors. Cell Chem. Biol. 27, 525–537.e6 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.J. E. Paris, D. Brandes, E. B. Anton, S. Barnard, Effect of castration on histochemistry and biochemistry of acid hydrolases in rat prostate gland12. J. Natl. Cancer Inst. 49, 1685–1696 (1972). [DOI] [PubMed] [Google Scholar]
- 42.K. D. Tousignant, A. Rockstroh, B. L. J. Poad, A. Talebi, R. S. E. Young, A. Taherian Fard, R. Gupta, T. Zang, C. Wang, M. L. Lehman, J. V. Swinnen, S. J. Blanksby, C. C. Nelson, M. C. Sadowski, Therapy-induced lipid uptake and remodeling underpin ferroptosis hypersensitivity in prostate cancer. Cancer Metab. 8, 1–21 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.G. Wilding, E. P. Gelmann, C. E. Freter, Phosphoinositide metabolism in human prostate cancer cells in vitro. Prostate 16, 15–27 (1990). [DOI] [PubMed] [Google Scholar]
- 44.D. G. Wang, M. N. Paddock, M. R. Lundquist, J. Y. Sun, O. Mashadova, S. Amadiume, T. W. Bumpus, C. Hodakoski, B. D. Hopkins, M. Fine, A. Hill, T. J. Yang, J. M. Baskin, L. E. Dow, L. C. Cantley, PIP4Ks suppress insulin signaling through a catalytic-independent mechanism. Cell Rep. 27, 1991–2001.e5 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.A. Koizumi, S. Narita, H. Nakanishi, M. Ishikawa, S. Eguchi, H. Kimura, S. Takasuga, M. Huang, T. Inoue, J. Sasaki, T. Yoshioka, T. Habuchi, T. Sasaki, Increased fatty acyl saturation of phosphatidylinositol phosphates in prostate cancer progression. Sci. Rep. 9, 1–8 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.D. Raucher, T. Stauffer, W. Chen, K. Shen, S. Guo, J. D. York, M. P. Sheetz, T. Meyer, Phosphatidylinositol 4,5-bisphosphate functions as a second messenger that regulates cytoskeleton-plasma membrane adhesion. Cell 100, 221–228 (2000). [DOI] [PubMed] [Google Scholar]
- 47.R. L. Doughman, A. J. Firestone, R. A. Anderson, Phosphatidylinositol phosphate kinases put PI4,5P2 in its place. J. Membr. Biol. 194, 77–89 (2003). [DOI] [PubMed] [Google Scholar]
- 48.G. R. V. Hammond, G. Schiavo, R. F. Irvine, Immunocytochemical techniques reveal multiple, distinct cellular pools of PtdIns4P and PtdIns(4,5)P2. Biochem. J. 422, 23–35 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.F. Lamoureux, C. Thomas, C. Crafter, M. Kumano, F. Zhang, B. R. Davies, M. E. Gleave, A. Zoubeidi, Blocked autophagy using lysosomotropic agents sensitizes resistant prostate tumor cells to the novel Akt inhibitor AZD5363. Clin. Cancer Res. 19, 833–844 (2013). [DOI] [PubMed] [Google Scholar]
- 50.W. J. Lee, M. H. Chien, J. M. Chow, J. L. Chang, Y. C. Wen, Y. W. Lin, C. W. Cheng, G. M. Lai, M. Hsiao, L. M. Lee, Nonautophagic cytoplasmic vacuolation death induction in human PC-3M prostate cancer by curcumin through reactive oxygen species -mediated endoplasmic reticulum stress. Sci Rep. 5, 1–14 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.H. Hino, N. Iriyama, H. Kokuba, H. Kazama, S. Moriya, N. Takano, M. Hiramoto, S. Aizawa, K. Miyazawa, Abemaciclib induces atypical cell death in cancer cells characterized by formation of cytoplasmic vacuoles derived from lysosomes. Cancer Sci. 111, 2132–2145 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.A. Gupta, S. Toscano, D. Trivedi, D. R. Jones, S. Mathre, J. H. Clarke, N. Divecha, P. Raghu, Phosphatidylinositol 5-phosphate 4-kinase (PIP4K) regulates TOR signaling and cell growth during Drosophila development. Proc. Natl. Acad. Sci. U.S.A. 110, 5963–5968 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.D. Elouarrat, Y. U. Van Der Velden, D. R. Jones, W. H. Moolenaar, N. Divecha, A. P. G. Haramis, Role of phosphatidylinositol 5-phosphate 4-kinase α in zebrafish development. Int. J. Biochem. Cell Biol. 45, 1293–1301 (2013). [DOI] [PubMed] [Google Scholar]
- 54.S. Chen, C. C. Tjin, X. Gao, Y. Xue, H. Jiao, R. Zhang, M. Wu, Z. He, J. Ellman, Y. Ha, Pharmacological inhibition of PI5P4Kα/β disrupts cell energy metabolism and selectively kills p53-null tumor cells. Proc. Natl. Acad. Sci. U.S.A. 118, e2002486118 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.A. M. Mackey, D. A. Sarkes, I. Bettencourt, J. M. Asara, L. E. Rameh, PIP4kγ is a substrate for mTORC1 that maintains basal mTORC1 signaling during starvation. Sci. Signal. 7, ra104 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.K. A. Cole, R. F. Chuaqui, K. Katz, S. Pack, Z. Zhuang, C. E. Cole, J. C. Lyne, W. M. Linehan, L. A. Liotta, M. R. Emmert-Buck, cDNA sequencing and analysis of POV1 (PB39): A novel gene up-regulated in prostate cancer. Genomics 51, 282–287 (1998). [DOI] [PubMed] [Google Scholar]
- 57.Q. Wang, C. G. Bailey, C. Ng, J. Tiffen, A. Thoeng, V. Minhas, M. L. Lehman, S. C. Hendy, G. Buchanan, C. C. Nelson, J. E. J. Rasko, J. Holst, Androgen receptor and nutrient signaling pathways coordinate the demand for increased amino acid transport during prostate cancer progression. Cancer Res. 71, 7525–7536 (2011). [DOI] [PubMed] [Google Scholar]
- 58.Y. Wu, R. R. Chhipa, J. Cheng, H. Zhang, J. L. Mohler, C. Ip, Androgen receptor-mTOR crosstalk is regulated by testosterone availability: Implication for prostate cancer cell survival. Anticancer Res. 30, 3895–3901 (2010). [PMC free article] [PubMed] [Google Scholar]
- 59.M. Chen, C. P. Pratt, M. E. Zeeman, N. Schultz, S. Barry, A. O. Neill, M. Castillo-martin, D. G. Nowak, A. Naguib, M. Grace, J. Murn, N. Navin, G. S. Atwal, C. Sander, L. William, C. Cordon-cardo, A. C. Newton, B. S. Carver, C. Lloyd, Identification of PHLPP1 as a tumor suppressor reveals the role of feedback activation in PTEN-mutant prostate cancer progression. Cancer Cell 20, 173–186 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Y. Sancak, T. R. Peterson, Y. D. Shaul, R. A. Lindquist, C. C. Thoreen, L. Bar-Peled, D. M. Sabatini, The Rag GTPases bind raptor and mediate amino acid signaling to mTORC1. Science 320, 1496–1501 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Y. Sancak, L. Bar-Peled, R. Zoncu, A. L. Markhard, S. Nada, D. M. Sabatini, Ragulator-rag complex targets mTORC1 to the lysosomal surface and is necessary for its activation by amino acids. Cell 141, 290–303 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.A. L. Marat, A. Wallroth, W. T. Lo, R. Müller, G. D. Norata, M. Falasca, C. Schultz, V. Haucke, mTORC1 activity repression by late endosomal phosphatidylinositol 3,4-bisphosphate. Science 356, 968–972 (2017). [DOI] [PubMed] [Google Scholar]
- 63.S. Kolay, U. Basu, R. Padinjat, Control of diverse subcellular processes by a single multi-functional lipidphosphatidylinositol 4,5-bisphosphate [PI(4,5)P2]. Biochem. J. 473, 1681–1692 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.T. Goto, N. Terada, T. Inoue, K. Nakayama, Y. Okada, T. Yoshikawa, Y. Miyazaki, M. Uegaki, S. Sumiyoshi, T. Kobayashi, T. Kamba, K. Yoshimura, O. Ogawa, The expression profile of phosphatidylinositol in high spatial resolution imaging mass spectrometry as a potential biomarker for prostate cancer. PLOS ONE 9, 1–9 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.K. S. Bhullar, N. O. Lagarón, E. M. McGowan, I. Parmar, A. Jha, B. P. Hubbard, H. P. V. Rupasinghe, Kinase-targeted cancer therapies: Progress, challenges and future directions. Mol. Cancer 17, 1–20 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.M. D. Voss, W. Czechtizky, Z. Li, C. Rudolph, S. Petry, H. Brummerhop, T. Langer, A. Schiffer, H. L. Schaefer, Discovery and pharmacological characterization of a novel small molecule inhibitor of phosphatidylinositol-5-phosphate 4-kinase, type II, β. Biochem. Biophys. Res. Commun. 449, 327–331 (2014). [DOI] [PubMed] [Google Scholar]
- 67.J. H. Clarke, M. Giudici, J. E. Burke, R. L. Williams, D. J. Maloney, J. Marugan, R. F. Irvine, The function of phosphatidylinositol 5-phosphate 4-kinase γ (PI5P4Kγ) explored using a specific inhibitor that targets the PI5P-binding site. Biochem. J. 466, 359–367 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.H. Beltran, D. Prandi, J. M. Mosquera, M. Benelli, L. Puca, J. Cyrta, C. Marotz, E. Giannopoulou, B. V. S. K. Chakravarthi, S. Varambally, S. A. Tomlins, D. M. Nanus, S. T. Tagawa, E. M. Van Allen, O. Elemento, A. Sboner, L. A. Garraway, M. A. Rubin, F. Demichelis, Divergent clonal evolution of castration-resistant neuroendocrine prostate cancer. Nat. Med. 22, 298–305 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.J. Barretina, G. Caponigro, N. Stransky, K. Venkatesan, A. A. Margolin, S. Kim, C. J. Wilson, J. Lehár, G. V. Kryukov, D. Sonkin, A. Reddy, M. Liu, L. Murray, M. F. Berger, J. E. Monahan, P. Morais, J. Meltzer, A. Korejwa, J. Jané-Valbuena, F. A. Mapa, J. Thibault, E. Bric-Furlong, P. Raman, A. Shipway, I. H. Engels, J. Cheng, G. K. Yu, J. Yu, P. Aspesi, M. De Silva, K. Jagtap, M. D. Jones, L. Wang, C. Hatton, E. Palescandolo, S. Gupta, S. Mahan, C. Sougnez, R. C. Onofrio, T. Liefeld, L. MacConaill, W. Winckler, M. Reich, N. Li, J. P. Mesirov, S. B. Gabriel, G. Getz, K. Ardlie, V. Chan, V. E. Myer, B. L. Weber, J. Porter, M. Warmuth, P. Finan, J. L. Harris, M. Meyerson, T. R. Golub, M. P. Morrissey, W. R. Sellers, R. Schlegel, L. A. Garraway, The cancer cell line encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature 483, 603–607 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.J. Cyrta, A. Augspach, M. R. de Filippo, D. Prandi, P. Thienger, M. Benelli, V. Cooley, R. Bareja, D. Wilkes, S. S. Chae, P. Cavaliere, N. Dephoure, A. C. Uldry, S. B. Lagache, L. Roma, S. Cohen, M. Jaquet, L. P. Brandt, M. Alshalalfa, L. Puca, A. Sboner, F. Feng, S. Wang, H. Beltran, T. Lotan, M. Spahn, M. K.-d. Julio, Y. Chen, K. V. Ballman, F. Demichelis, S. Piscuoglio, M. A. Rubin, Role of specialized composition of SWI/SNF complexes in prostate cancer lineage plasticity. Nat. Commun. 11, 5549 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.A. Dobin, C. A. Davis, F. Schlesinger, J. Drenkow, C. Zaleski, S. Jha, P. Batut, M. Chaisson, T. R. Gingeras, STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.L. Habegger, A. Sboner, T. A. Gianoulis, J. Rozowsky, A. Agarwal, M. Snyder, M. Gerstein, RSEQtools: A modular framework to analyze RNA-Seq data using compact, anonymized data summaries. Bioinformatics 27, 281–283 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.J. Medina, V. van der Velpen, T. Teav, Y. Guitton, H. Gallart-Ayala, J. Ivanisevic, Single-step extraction coupled with targeted hilic-ms/ms approach for comprehensive analysis of human plasma lipidome and polar metabolome. Metabolites 10, 1–17 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.W. B. Dunn, D. Broadhurst, P. Begley, E. Zelena, S. Francis-McIntyre, N. Anderson, M. Brown, J. D. Knowles, A. Halsall, J. N. Haselden, A. W. Nicholls, I. D. Wilson, D. B. Kell, R. Goodacre; The Human Serum Metabolome (HUSERMET) Consortium , Procedures for large-scale metabolic profiling of serum and plasma using gas chromatography and liquid chromatography coupled to mass spectrometry. Nat. Protoc. 6, 1060–1083 (2011). [DOI] [PubMed] [Google Scholar]
- 75.H. Tsugawa, M. Arita, M. Kanazawa, A. Ogiwara, T. Bamba, E. Fukusaki, MRMPROBS: A data assessment and metabolite identification tool for large-scale multiple reaction monitoring based widely targeted metabolomics. Anal Chem. 85, 5191–5199 (2013). [DOI] [PubMed] [Google Scholar]
- 76.X. Wu, J. Wu, J. Huang, W. Powell, J. Zhang, R. Matusik, F. Sangiorgi, R. Maxson, H. Sucov, P. Roy-Burman, Generation of a prostate epithelial cell-specific Cre transgenic mouse model for tissu.pdf. Mech. Dev. 101, 61–69 (2001). [DOI] [PubMed] [Google Scholar]
- 77.S. Srinivas, T. Watanabe, C. S. Lin, C. M. William, Y. Tanabe, T. M. Jessell, F. Costantini, Cre reporter strains produced by targeted insertion of EYFP and ECFP into the ROSA26 locus. BMC Dev. Biol. 1, 1–8 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.K. C. Valkenburg, S. R. Amend, K. J. Pienta, Murine prostate micro-dissection and surgical castration. J. Vis. Exp. , e53984 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.S. Berezowska, J. A. Galván, "Immunohistochemical Detection of the Autophagy Markers LC3 and p62/SQSTM1 in Formalin-Fixed and Paraffin-Embedded Tissue BT-Histochemistry of Single Molecules: Methods and Protocols" in, C. Pellicciari, M. Biggiogera, Eds. (Springer New York, 2017), pp. 189–194.
- 80.P. Bankhead, M. B. Loughrey, J. A. Fernández, Y. Dombrowski, D. G. McArt, P. D. Dunne, S. McQuaid, R. T. Gray, L. J. Murray, H. G. Coleman, J. A. James, M. Salto-Tellez, P. W. Hamilton, QuPath: Open source software for digital pathology image analysis. Sci Rep. 7, 1–7 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.R. Mevel, I. Steiner, S. Mason, L. C. A. Galbraith, R. Patel, M. Z. H. Fadlullah, I. Ahmad, H. Y. Leung, P. Oliveira, K. Blyth, E. Baena, G. Lacaud, RUNX1 marks a luminal castration-resistant lineage established at the onset of prostate development. eLife 9, e60225 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.J. Drost, W. R. Karthaus, D. Gao, E. Driehuis, C. L. Sawyers, Y. Chen, H. Clevers, Organoid culture systems for prostate epithelial and cancer tissue. Nat. Protoc. 11, 347–358 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S9
Tables S1 to S6
Data S1 to S3