Abstract
Ribozymes are small catalytic RNA molecules that can be engineered to enzymatically cleave RNA transcripts in a sequence-specific fashion and thereby inhibit expression and function of the corresponding gene product. With their simple structures and site-specific cleavage activity, they have been exploited as potential therapeutic agents in a variety of human disorders, including hepatitis C virus (HCV) infection. We have designed a hairpin ribozyme (Rz3′X) targeting the HCV minus-strand replication intermediate at position 40 within the 3′X tail. Surprisingly, Rz3′X was found to induce ganciclovir (GCV)-resistant colonies in a bicistronic cellular reporter system with HCV internal ribosome entry site (IRES)-dependent translation of herpes simplex virus thymidine kinase (TK). Rz3′X-transduced GCV-resistant HeLa reporter cells showed substantially reduced IRES-mediated HCV core protein translation compared with control vector-transduced cells. Since these reporter systems do not contain the HCV 3′X tail sequences, the results indicate that Rz3′X probably exerted an inhibitory effect on HCV IRES activity fortuitously through another gene target. A novel technique of ribozyme cleavage-based target gene identification (cleavage-specific amplification of cDNA ends) (M. Krüger, C. Beger, P. J. Welch, J. R. Barber, and F. Wong-Staal, Nucleic Acids Res. 29:e94, 2001) revealed that human 20S proteasome α-subunit PSMA7 mRNA was a target RNA recognized and cleaved by Rz3′X. We then showed that additional ribozymes directed against PSMA7 RNA inhibited HCV IRES activity in two assay systems: GCV resistance in the HeLa IRES TK reporter cell system and a transient transfection assay performed with a bicistronic Renilla-HCV IRES-firefly luciferase reporter in Huh7 cells. In contrast, ribozymes were inactive against IRES of encephalomyocarditis virus and human rhinovirus. Additionally, proteasome inhibitor MG132 exerted a dose-dependent inhibitory effect on HCV IRES-mediated translation but not on cap-dependent translation. These data suggest a principal role for PSMA7 in regulating HCV IRES activity, a function essential for HCV replication.
Ribozymes are small catalytic RNA molecules that hybridize to complementary sequences of a particular target mRNA and that can be engineered to enzymatically cleave and destroy RNA transcripts in a sequence-specific fashion, thereby preventing expression and function of the corresponding gene product. Ribozymes have been studied in connection with a variety of diseases and human disorders as potential therapeutic molecules (for recent reviews see references 2, 9, 15, 19, and 22). Hairpin ribozymes fold into a two-dimensional hairpin structure, consisting of a small catalytic region with four helical domains (see Fig. 1A). Based on Watson-Crick base pairing, the ribozyme binding arms (helices 1 and 2) hybridize to sequences flanking the cleavage site (GUC) within the target RNA, thereby determining specificity of the recognized target sequence.
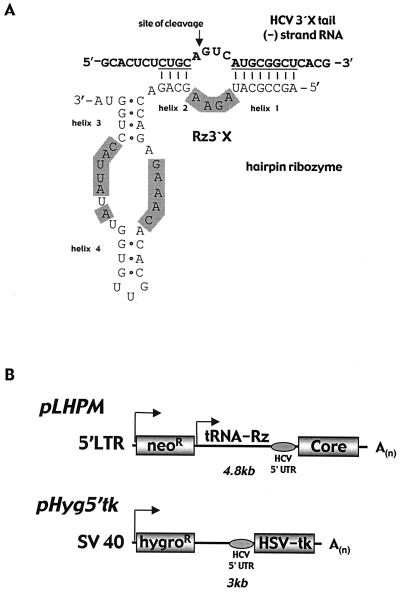
FIG. 1.
Hairpin ribozyme Rz3′X in a GCV-reporter assay with HeLa 5′tk cells stably expressing a bicistronic hygromycin-HCV IRES-HSV TK transcript. (A) Schematic representation of Rz3′X binding to the antigenomic HCV RNA within the highly conserved 3′X tail. The substrate is cleaved 5′ of the GUC triplet required for hairpin ribozyme recognition (arrow). Conserved areas of the hairpin ribozyme are shaded. (B) Vector constructs for retrovirus vector production of the ribozyme (pLHPM) (including IRES-mediated translation of HCV core protein) and for expression of a bicistronic hygromycin-HCV 5′UTR-HSV TK transcript for negative selection in cell culture (14). The ribozyme is driven independently from the retrovirus transcript by a tRNAVal promoter expression cassette. (C) GCV-resistant HeLa 5′tk reporter colonies derived from transduction with Rz3′X retrovirus particles. Colonies were enumerated 21 days after GCV application (20 μM, 4 days), and numbers are fold enrichment values compared with that for control ribozyme RzBR1-transduced reporter cells.
Recently, we introduced a novel “inverse genomics” procedure based on a retrovirus hairpin ribozyme library with randomized target recognition sequences for gene discovery in different experimental systems. These include (i) genes regulating the BRCA1 promoter (3), (ii) cellular genes mediating hepatitis C virus (HCV) internal ribosome entry site (IRES) activity (14), (iii) genes involved in anchorage-independent cell growth control (23), and (iv) genes involved in suppression of fibroblast transformation (16). Ribozymes that repeatedly conferred distinct cellular phenotypes were selected in these systems. Single ribozyme candidates were identified, and the binding sequence flanking the GUC site required for ribozyme cleavage was exploited to identify partial sequence information of the target gene responsible for the observed phenotype. For the HCV IRES project, a cellular selection scheme was developed using a reporter system based on herpes simplex virus (HSV) thymidine kinase (TK) as a negative selectable marker under translational control of the HCV IRES (14). This cellular selection system allowed the identification of ribozymes that actively inhibited HCV IRES-mediated translation of HSV TK and thereby conferred a ganciclovir (GCV)-resistance phenotype. By using the ribozyme binding sequences, potential cellular cofactors for HCV IRES were discovered (14). For these experiments, Rz3′X, a hairpin ribozyme engineered against the minus-strand HCV replication intermediate at position 40 within the terminal 98-nucleotide (nt) (3′X tail) (11, 21), as well as a ribozyme directed against human hepatitis B virus (RzBR1), served as negative controls. Surprisingly, Rz3′X repeatedly induced a strong GCV resistance phenotype upon transduction into reporter cells and reduced HCV IRES-mediated core protein translation. Here we show that Rz3′X fortuitously cleaved the cellular mRNA corresponding to 20S proteasome α-subunit PSMA7 and that additional ribozymes designed to cleave PSMA7 mRNA also inhibit HCV IRES activity but lack activity against IRES of encephalomyocarditis virus (EMCV) and human rhinovirus (HRV). Furthermore, inhibition of the 20S proteasome by proteasome inhibitor MG132 was found to exert a dose-dependent inhibitory effect on HCV IRES activity without an effect on cap-dependent translation. These data suggest a principal role for proteasome subunits in regulating essential functions of HCV replication.
MATERIALS AND METHODS
Plasmid construction.
Plasmid pHyg-5′tk (translation of HSV TK mediated by the HCV 5′-untranslated region (UTR) (HCV nt 38 to 341 of genotype 1b) and Moloney retrovirus genome-based ribozyme expression vector pLHPM were constructed as described previously (14). pLHPM contains a ribozyme expression cassette driven by the tRNAVal promoter and a neomycin phosphotransferase gene that confers resistance against G-418. Ribozymes Rz3′X (directed against the minus-strand replication intermediate at position 40 within the 3′X tail), RzBR1 (directed against human hepatitis B virus), and validation ribozymes VRz1 to -4 (target sequences: VRz1, 5′-GGATAGTCATCAACAG-3′; VRz2, 5′-GCCAAGTCAGTGCGCG-3′; VRz3, 5′-GTTCAGTCAGGTGGCA-3′; VRz4, 5′-TTGCTGTCATGAGGCG-3′) were constructed by annealing overlapping ribozyme-specific oligonucleotides and were cloned into BamHI-MluI sites of pLHPM as described previously (15, 24). Similarly, disRz3′X was constructed with a three-base change in the catalytic site of the ribozyme sequence (24). To generate plasmid pRL-5′-CFL (Renilla luciferase [RL]-HCV 5′-UTR-HCV core protein-firefly luciferase [FL]), PCR was performed with oligonucleotide primers P1 (5′-GCAAGCTTGAATTCGCCAGCCCCCTGATGGGGGCG-3′) and P3 (5′-CTGATCTCATGAAGGCTGAAGCGGGCACAGTCAG-3′) and then the PCR products were digested with EcoRI and BspHI and inserted into pGL3RL (kindly provided by Anne E. Willis, Department of Biochemistry, University of Leicester, Leicester, United Kingdom) following digestion with EcoRI and NcoI. Reporter plasmid pΔRLΔ5′-CFL lacking RL and HCV IRESs while expressing the HCV core protein and FL via a cap-dependent mechanism was generated from pRL-5′-CFL by restriction digestion using EcoRV and NruI. Plasmids pRL-EMCV-FL (EMCV IRES-dependent FL translation) and pRL-HRV-FL (HRV IRES-dependent FL translation) were also provided by Anne E. Willis. To generate plasmid pRL-HCV-FL (RL-HCV 5′UTR-FL), PCR was performed with oligonucleotide primers P1 and P2 (5′-CTGATCTCATGATGCACGGTCTACGAGACC-3′) and then the products were digested with EcoRI and BspHI and inserted into pGL3RL.
Cell culture and reagents.
HeLa cervical carcinoma cells, thymus CF2 cells, and Huh7 hepatoblastoma cells were cultured at 37°C in 5% CO2 in a humidified incubator containing Dulbecco's modified Eagle's medium supplemented with 10% heat-inactivated fetal calf serum, 1% penicillin-streptomycin, 1 mM sodium pyruvate, and nonessential amino acids (all from Gibco). GCV (Cytovene) was obtained from Syntex (Palo Alto, Calif.). MG132 (Z-Leu-Leu-Leu-CHO) was obtained from BIOMOL Research Laboratories (Plymouth Meeting, Pa.).
Retrovirus vector production, transduction, and GCV selection of HeLa 5′tk reporter cells.
Retrovirus particles were produced on CF2 cells by triple transfection (TransIT-LT1; PanVera) of pLHPM-Rz (expressing individual ribozymes) with plasmid gag-pol (expressing retrovirus helper function) and plasmid pVSV-G expressing the vesicular stomatitis virus glycoprotein (VSV-G) under conditions described previously (14). HeLa 5′tk reporter cells stably expressing the hygromycin-HCV 5′UTR-HSV TK transcript (14) were transduced with retrovirus supernatant containing Rz3′X or the control RzBR1 in the presence of 8 μg of Polybrene/ml, followed by selection with 500 μg of G-418/ml for 2 weeks. Viral titers of G-418-resistant polyclonal cell populations were on the order of 4 × 105 PFU/ml, as determined with HeLa cells. Stable ribozyme-expressing reporter cells were plated at 0.4 × 104/cm2 and exposed to 10 μM GCV for 4 days, and surviving colonies were counted under low-power light microscopy 21 days after GCV application. GCV-resistant colonies (defined as clusters of at least 25 cells) were harvested and were further expanded for immunoblot or Northern blot analyses.
Northern blot analysis.
Fifteen micrograms of total RNA extracted from cultures grown to 80% confluence (RNeasy minikit; Qiagen) was denatured in a solution containing 50% formamide, 17 mM MOPS (morpholinepropanesulfonic acid), and 2.2 M formaldehyde for 15 min at 70°C and chilled on ice. RNAs were separated by electrophoresis in a 0.8% agarose gel containing 2.2 M formaldehyde and 20 mM MOPS buffer and subsequently transferred onto a nylon membrane (Zeta-Probe; Bio-Rad, Hercules, Calif.) for 20 h in 20× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) and cross-linked by UV (Stratalinker; Stratagene). Membranes were prehybridized for 1 h at 65°C in hybridization solution (QuickHyb; Stratagene) and then incubated for 16 h in hybridization solution containing an α-32P-labeled HCV core, human PSMA7 or a glyceraldehyde-3-phosphate dehydrogenase (GAPDH) probe (4 × 108 to 6 × 108 cpm/ml). Probes were generated from the following plasmids by restriction enzyme digestion: HCV core protein, 902-bp fragment from plasmid pGEM-HCV5′C following PstI and EcoRI digestion; HSV TK, 1,342-bp fragment from plasmid pHyg-5′tk following BamHI and MluI digestion. The probe for GAPDH (nt 81 to 1042) and the probe for PSMA7 mRNA were generated by PCR. DNA probes were labeled with [α-32P]dCTP by a random-priming method (High Prime DNA-labeling kit; Boehringer Mannheim). Filters were washed twice for 15 min in 2× SSC–0.1% sodium dodecyl sulfate (SDS) and once in 0.1× SSC–0.1% SDS for 30 min and were exposed to Kodak X-OMAT AR films at −70°C with intensifying screens. Quantitation was achieved by phosphorimager analysis (Molecular Dynamics, Sunnyvale, Calif.) and computer-assisted densitometry (ImageQuant Software). Transcript levels were normalized to GAPDH mRNA, and values are expressed as percentages compared with the levels for control RzBR1, set to 100%.
Western blot analysis.
Cells were harvested by trypsinization, pelleted, resuspended in Tris-glycine sample buffer (Novex), and boiled for 15 min at 95°C. Following quantitation (Bradford protein assay reagent; Bio-Rad) 15 μg of HCV core protein or 5 μg of β-actin was separated by gel electrophoresis under reducing conditions on Tris-glycine–14% polyacrylamide gels (Novex) and transferred to a nitrocellulose membrane (Immobilon P; Millipore) by semidry transfer (Trans-Blot semidry transfer cell; Bio-Rad) for 2 h at 14 V. The membranes were incubated with the primary antibody for 3 h using an enhanced chemiluminescence Western blotting kit (Novex). An anti-HCV core protein monoclonal antibody (6C7) was generously provided by Harry Greenberg (Stanford University, Stanford, Calif.). The PSMA7 antibody was purchased from Affiniti Research Products (Exeter, United Kingdom). Blots were exposed to film for 1 or 10 s for quantitation of β-actin and PSMA7 or 20 or 60 s for quantitation of the HCV core protein. Signals for β-actin were used to normalize core or PSMA7 protein signals. Band intensities were measured by using National Institutes of Health (NIH) Image software.
Ribozyme cleavage-based target RNA identification using C-SPACE.
The C-SPACE technique (cleavage-specific amplification of cDNA ends) is described in detail elsewhere (15a). Oligonucleotide primers SM3′X-5/6 (5′-TACGCGGGGTCATGCGG-3′), oligo(dT)-TAG (5′-GGCCACGCGTCGACTAGTACVTTTTTTTTTTTTTTTTT-3′ [V is any nucleotide except T), and TAG (5′-GGCCACGCGTCGACTAGTACT-3′) (IDT, Coralville, Iowa) were used for PCR amplification on Rz3′X-cleaved mRNA.
PSMA7-directed ribozymes and proteasome inhibitor MG132 in stable and transient IRES-dependent bicistronic reporter systems.
Huh7 cells were transiently transfected with 0.3 μg of plasmid pRL-5′-CFL (RL-HCV 5′UTR-HCV core protein-FL), pRL-HCV-FL, pRL-EMCV-FL, or pRL-HRV-FL/cm2 and 0.3 μg of ribozyme vector pLHPM/cm2 using a calcium phosphate technique (CalPhos; Clontech). Luciferase light units were measured by a dual luciferase assay (Promega) in a luminometer (Lumat LB9507; Perkin-Elmer Life Sciences) in cell lysates obtained 48 h after transfection. Signal intensities for FL and RL present in control ribozyme RzBR1-transfected cells were set to 100%. To generate stable transfectants for proteasome inhibitor experiments, HeLa cells were transfected with plasmid pRL-5′-CFL (0.3 μg/cm2) and plasmid pIREShygro (Clontech; 0.03 μg/cm2). Following selection of stable transfectants with hygromycin B (250 μg/ml) single-cell clones were obtained according to standard techniques and characterized for RL and FL expression. A particular cell clone was selected and seeded at 1.5 × 104/cm2, exposed to increasing doses of MG132 (0.5 to 20 μM) for 24 h, harvested, and measured for luciferase activities. Luciferase activities in untreated cells were set to 100%. MG132 effects in Huh7 cells (1.5 × 104/cm2) were assessed following transient transfection with pRL-5′-CFL (0.3 μg/cm2) or pΔRLΔ5′-CFL (0.3 μg/cm2) in the presence or absence of MG132 (1 μM) 6 h prior to transfection. Cells were harvested after 72 h and analyzed for luciferase activities.
RESULTS
Rz3′X retrovirus vector transduction of HeLa 5′tk reporter cells leads to enrichment in GCV-resistant colonies.
Hairpin ribozyme Rz3′X was engineered to recognize the GUC triplet at position 40 of the antigenomic strand within the 98-nt 3′X tail (Fig. 1A). Cleavage activity against HCV RNA was demonstrated by in vitro cleavages on corresponding short substrate RNAs (data not shown). HeLa 5′tk reporter cells stably expressing the bicistronic hygromycin-HCV IRES-HSV TK transcript (Fig. 1B) (14) were transduced with retrovirus vectors expressing Rz3′X, catalytically inactive Rz3′X (disRz3′X), or control ribozyme RzBR1. Cells were exposed to GCV and cultured under hygromycin selection. After 3 weeks, Rz3′X-transduced cells revealed a more-than-60-fold increase in the number of hygromycin- and GCV-resistant colonies compared with cells transduced with the vector containing catalytically inactive Rz3′X or the RzBR1 control (Fig. 1C).
Rz3′X-transduced GCV-resistant 5′tk reporter cells exhibit a reduction in HCV core protein translation.
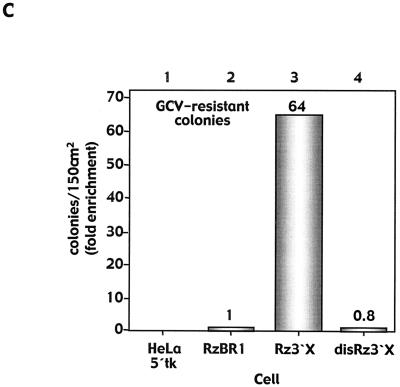
To demonstrate a ribozyme-mediated effect on HCV IRES activity, cell lysates of GCV-resistant reporter cells were analyzed for the HCV core protein by immunoblotting. In these reporter cells, the HCV core protein (21 kDa) was translated under the control of the HCV 5′-UTR (nt 342 to 874) as part of the retrovirus ribozyme vector transcript (Fig. 1B). Levels of the core protein were reduced by 50% in Rz3′X-transduced GCV-resistant colonies compared with levels in control ribozyme-transduced cells (Fig. 2A and B). RNA analysis for these cells revealed a slight increase in core transcript levels (Fig. 2A), while titers of retrovirus ribozyme vectors were in a comparable range (4 × 105 to 4.5 × 105 PFU/ml, defined by G-418 resistance). However, to eliminate differences in expression of the retrovirus transcript (harboring the 5′-UTR core sequence) between cells transduced with Rz3′X vector and those transduced with control vector RzBR1, the ratio of core protein to the corresponding RNA transcript was used as an indicator of HCV IRES translational activity. HCV IRES activity showed a 60% reduction in GCV-resistant cells derived from transduction with Rz3′X compared with controls (Fig. 2C). These data strongly suggest that the reduced levels of core protein were caused by interference of the ribozyme with IRES-mediated core protein translation.
FIG. 2.
Protein and RNA analyses of GCV-resistant HeLa 5′tk reporter cells. (A) The HCV core protein was determined by immunoblotting with cell lysates, and Northern blot analysis was performed on total RNA extracted from GCV-resistant reporter cells. Parental cells expressing bicistronic vector 5′tk (lane 1) lack the HCV core protein and core RNA expression derived from transduction with the retrovirus ribozyme construct. (B) Bands were quantified by densitometry using NIH Image software or phosphorimager analysis (Molecular Dynamics), and signals were normalized against β-actin (for protein) or GAPDH (for RNA). Results from triplicate experiments are expressed as percentages of control and are presented as means ± standard errors of the means. (C) Relative HCV IRES activity expressed as the ratio of core protein translation relative to the amount of corresponding transcript detected in GCV-resistant cells.
C-SPACE for ribozyme cleavage-based target RNA identification.
Since the GCV reporter system does not contain RNA sequences corresponding to the HCV 3′X tail in either the sense or antisense orientation, we hypothesized that the GCV resistance phenotype and the reduction in core protein expression by Rz3′X resulted from cleavage of a cellular mRNA involved in HCV IRES-mediated translation. We therefore attempted to clone the target gene(s) responsible for the observed phenotypic changes. Generally, hairpin ribozymes recognize a particular target RNA by Watson-Crick base pairing of their substrate binding nucleotides. Using the 16 nt derived from the binding sequence of Rz3′X (8-nt helix 1 and 4-nt helix 2, flanking the GUC triplet) to search various databases, we failed to identify perfect or nearly perfect matches to potential target genes in nonredundant or expressed sequence tag databases for Rz3′X (data not shown). Nor did we obtain candidate cDNAs by PCR amplification strategies such as 5′- or 3′-rapid amplification of cDNA ends or by phage library screening with oligonucleotide primers derived from a ribozyme binding sequence (data not shown). Therefore, we developed a method for target gene identification based on ribozyme cleavage activity, termed C-SPACE (described in detail previously [15a]). C-SPACE can easily be applied to identify potential target molecule(s) for any particular hairpin ribozyme. We performed C-SPACE on Rz3′X-cleaved HeLa mRNA and identified a 290-nt product, which was absent in control ribozyme-cleaved mRNA (data not shown). Cloning and sequencing this band a revealed partial sequence of the human 20S proteasome α-subunit PSMA7 mRNA (GenBank accession no. NM 002792) (7).
Validation of human 20S proteasome α-subunit PSMA7 mRNA targeted by Rz3′X as a gene involved in HCV IRES-mediated translation.
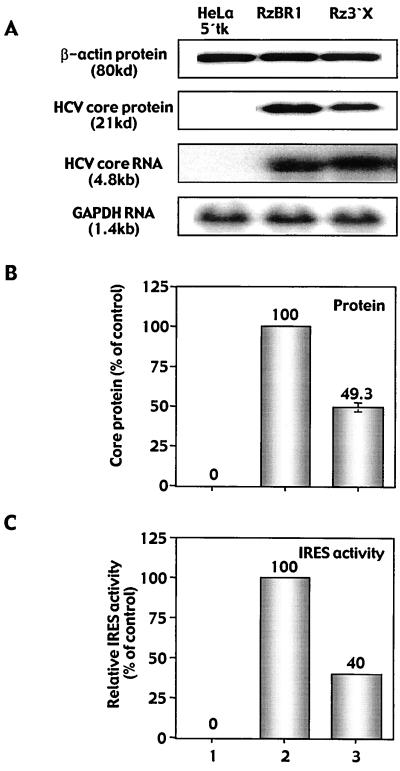
The putative binding site for Rz3′X at the GUC cleavage site (position 652) within the 20S proteasome α-subunit PSMA7 mRNA revealed a mismatch at two positions in helix 1 of the binding arms. Additionally, a G-U base pair in helix 2 was noted at the terminal 3′ position of the Rz3′X binding sequence of PSMA7 mRNA (Fig. 3A). However, the BamHI restriction enzyme recognition sequence (derived from cloning individual ribozymes into retrovirus vector pLHPM) present in the ribozyme sequence adjacent to helix 1 matches the target at four positions, thereby stabilizing the ribozyme-target interaction. Overall, Rz3′X binding arms match PSMA7 mRNA at 14 of 18 nt.
FIG. 3.
Ribozyme-mediated identification of human PSMA7 proteasome α-subunit mRNA and confirmation of functional relevance for HCV IRES-mediated translation. (A) Rz3′X binds to its target site (position 652) within the human PSMA7 proteasome α-subunit mRNA. Partial sequence information from the gene was obtained by using the sequence flanking the GUC site as the oligonucleotide primer binding sites in a C-SPACE amplification (Krüger et al., submitted). The entire cDNA of human PSMA7 mRNA was cloned by additional 5′- and 3′-rapid amplification of cDNA ends using cDNA derived from HeLa 5′tk cells and confirmed by standard sequencing techniques (GenBank accession no. NM 002792) (7). The BamHI recognition sequence (underlined) derived from ribozyme cloning into retrovirus vector pLHPM stabilizes the ribozyme-target interaction. (B) Four validation ribozymes (VRz1 to -4) were engineered against unique GUC sites within the human PSMA7 mRNA. The binding arms of VRz4 are designed to perfectly match the Rz3′X GUC site at position 652. (C) Analysis of GCV-resistant HeLa 5′tk reporter cells derived from stable transduction with retrovirus ribozyme vector particles. Colonies were enumerated 21 days after GCV application (20 μM, 4 days), and numbers are fold enrichment values compared with that for control ribozyme RzBR1-transduced reporter cells. GCV-resistant colonies were analyzed by immunoblotting and Northern blotting. Bands were quantified by densitometry using NIH Image software or by phosphorimager analysis (Molecular Dynamics), and signals were normalized against β-actin (for protein) or GAPDH (for RNA). Results from triplicate experiments are expressed as percentages of control and are presented as means ± standard errors of the means.
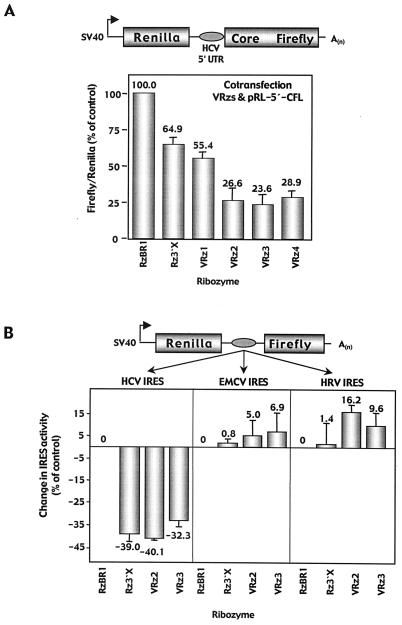
To confirm the relevance of PSMA7 for HCV IRES-mediated translation, we designed four “validation” ribozymes (VRz1 to -4) directed against unique GUC sites within PSMA7 mRNA (Fig. 3B). The binding arms of VRz4 are designed to exactly match the Rz3′X GUC site at position 652 of the gene. Each ribozyme was cloned into the retrovirus expression vector (pLHPM) and HeLa 5′tk cells were stably transduced with retrovirus VRz vectors. The number of GCV-resistant colonies, enumerated 21 days after exposure to GCV, was increased 18- to 40-fold for VRz-transduced cells compared with control RzBR1-transduced cells (Fig. 3C, upper left). In contrast, the catalytically inactive version of Rz3′X (disRz3′X) did not generate enough GCV-resistant colonies for them to be distinguishable from background (data not shown). These data confirmed the ability of the gene-specific VRzs to confer GCV resistance in a selection system based on HCV IRES-mediated HSV TK expression. Next we assessed the effect of VRzs on IRES-mediated core protein translation. Cell lysates from GCV-resistant reporter cells were analyzed for core protein by immunoblotting. Core protein expression normalized to β-actin revealed a 57 to 86% reduction in core protein for Rz3′X and the VRzs, while RNA levels for this transcript were slightly elevated (range 101 to 131%) compared with controls (data not shown). Again, the ratio of core protein translated from the corresponding RNA transcript indicated a 67 to 87% reduction of HCV IRES activity (Fig. 3C, upper right). Next, PSMA7 RNA and protein expression was analyzed in ribozyme-transduced GCV-resistant reporter cells. Northern analysis indicated a 25 to 40% reduction in PSMA7 RNA in cells derived from transduction with the VRzs compared with controls (Fig. 3C, lower right). When cell lysates were analyzed for protein expression, a 33 to 84% reduction in PSMA7 levels was observed for VRzs compared with controls (Fig. 3C, lower left). Interestingly, VRz4, exactly matching the GUC triplet recognized by Rz3′X, revealed the highest degree of PSMA7 protein inhibition. These data demonstrate a marked reduction in HCV IRES activity by PSMA7-specific ribozymes, thereby establishing that PSMA7 mRNA is a cellular target for Rz3′X as identified by in vitro C-SPACE.
Rz3′X and PSMA7-directed ribozymes reduce HCV IRES-mediated translation in transiently transfected human hepatocellular carcinoma cells (Huh7).
Since HCV mainly replicates in liver cells, we subsequently performed transient transfection experiments with Huh7 cells. Rz3′X as well as PSMA7-directed ribozymes VRz1 to -4 and control RzBR1 were cotransfected into Huh7 cells with bicistronic reporter construct RL-5′-CFL, which directs cap-dependent translation of the RL gene and cap-independent HCV IRES-mediated translation of HCV core protein and FL genes (Fig. 4A). RL, as the upstream translation product, serves as an internal control to account for differences in transfection efficiency. Core protein and FL translated from the downstream cistron are posttranslationally processed as two separate proteins, with core protein levels (measured by immunoblotting) correlating with FL activities (data not shown). Rz3′X and PSMA7-directed ribozymes substantially reduced HCV IRES activity, as indicated by a 55 to 76% reduction in FL as the IRES-dependent downstream translation product (Fig. 4A). In repeated experiments, the absolute signal intensities for RL remained unchanged after transfection of the different ribozymes, indicating that cap-dependent translation was not affected by the expression of ribozymes targeting PSMA7 RNA (data not shown).
FIG. 4.
Transient transfection of Huh7 cells with Rz3′X or PSMA7-directed ribozymes and bicistronic reporter plasmids expressing FL dependent on HCV, EMCV, or HRV IRES. (A) PSMA7-directed ribozymes VRz1 to -4, Rz3′X, and control RzBR1 were cotransfected into Huh7 cells with a bicistronic reporter construct (RL-5′-CFL), allowing for cap-dependent translation of the RL gene, followed by the HCV 5′UTR for IRES-mediated translation of the HCV core protein and FL gene. Luciferase activities were measured by dual-luciferase assay, and the FL/RL ratio detected in RzBR1-transfected cells was set to 100%. (B) Rz3′X, PSMA7-directed ribozymes VRz2 and -3, and control RzBR1 were cotransfected into Huh7 cells with bicistronic reporter construct RL-HCV-FL (HCV 5′ UTR, allowing for IRES-mediated translation of the FL gene), RL-EMCV-FL, or RL-HRV-FL. Changes in IRES activities (FL/RL ratios) from triplicate experiments are expressed relative to the activity of the RzBR1 control and are presented as means ± standard errors of the means.
Rz3′X and PSMA7-directed ribozymes reduce HCV IRES activity independently of HCV core protein but lack activity on the EMCV IRES or HRV IRES in transiently transfected Huh7 cells.
A bicistronic reporter (RL-HCV-FL) similar to the RL-5′-CFL vector but lacking the HCV core protein coding sequence was cotransfected with Rz3′X, PSMA7-directed ribozymes VRz2 and -3, or control RzBR1. Following transfection into Huh7 cells, Rz3′X, VRz2, and VRz3 reduced HCV IRES activity by 32 to 40% compared with control RzBR1 (Fig. 4B). This confirms the HCV IRES inhibitory effect in a transient system independently of the HCV core protein and indicates that reduction of core protein levels associated with expression of active ribozymes in the stably selected GCV-resistant cells (Fig. 2B) was not caused by decreased core protein stability. To further characterize the IRES inhibitory effect of Rz3′X and the PSMA7 RNA validation ribozymes compared with that for the control ribozyme, we used additional bicistronic reporter plasmids for transient transfection assays with Huh7 cells. Since we were mainly interested in viral IRESs, we chose to investigate the effect of the active ribozymes on translation derived from IRES of EMCV or HRV in place of the HCV IRES. Upon transient transfection in Huh7 cells, Rz3′X and the PSMA7-directed ribozymes did not exhibit inhibitory effects on translation derived from EMCV or HRV IRES (Fig. 4B). In these repeated experiments, the absolute values of RL activities did not differ significantly between the cells transfected with active ribozymes and controls (data not shown), indicating that the ribozyme-mediated effect is not caused by inhibition of general protein stability.
Proteasome inhibitor MG132 reduces HCV IRES activity.
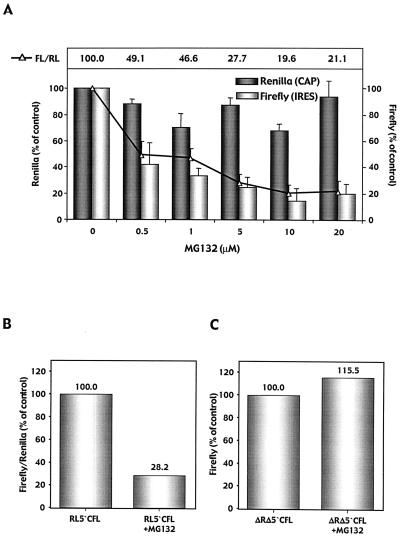
Next, we investigated whether the observed inhibition of IRES activity can also be achieved by a peptide inhibitor of the 20S proteasome complex. MG132, a potent and selective reversible inhibitor of the 20S proteasome, was administered to HeLa cells stably expressing the simian virus 40-driven bicistronic reporter transcript (RL-5′-CFL). FL activities were decreased 58 to 81% upon MG132 application (Fig. 5A). In contrast, cap-dependent translation of RL from this stably expressed bicistronic reporter remained largely unchanged. These data indicate a dose-dependent inhibitory effect of proteasome inhibitor MG132 on HCV IRES activity.
FIG. 5.
Effect of proteasome inhibitor MG132 on HCV IRES activity. (A) HeLa cells stably expressing the RL-5′-CFL reporter construct were analyzed for luciferase activities following exposure to increasing doses of MG132. Signals for RL and FL as well as the FL/RL ratio (relative HCV IRES activity) measured in untreated cells were set to 100%. Results from triplicate experiments are expressed as percentages of control and are presented as means ± standard errors of the means. (B) Huh7 cells were transfected with plasmid pRL-5′-CFL in the presence or absence of MG132 (1 μM). (C) A reporter plasmid lacking RL and the HCV IRES sequence while expressing HCV core protein and FL via a cap-dependent mechanism (pΔRLΔ5′-CFL) was transfected into Huh7 cells in the presence or absence of MG132.
We also investigated the inhibitory effect of MG132 in a transient transfection assay with Huh7 cells using the same bicistronic luciferase reporter construct (RL-5′-CFL) (1 μM MG132 starting 6 h prior to transfection). Luciferase activities measured 48 h after transfection revealed a 72% decrease in HCV IRES activity (indicated by the FL/RL ratio) in MG132 treated cells compared with that in controls (Fig. 5B). To demonstrate that there is an IRES-specific decrease in FL activity rather than interference of the drug with intracellular FL protein degradation, we repeated these experiments with a reporter plasmid expressing HCV core protein and FL via a cap-dependent mechanism (pΔRLΔ5′-CFL). Here, we did not observe significant changes in luciferase activities in cells treated with the proteasome inhibitor versus controls (Fig. 5C). This indicated a preferential, dose-dependent effect of proteasome inhibitor MG132 on HCV IRES activity and underlines the functional relevance of the proteasome complex in HCV IRES-mediated translation.
DISCUSSION
Here, we provide evidence for an involvement of a proteasome subunit with HCV translation by demonstrating that inhibition of HCV IRES activity by the ribozyme Rz3′X, originally engineered to target the 3′X sequence of the HCV minus strand, was mediated by targeting the proteasome subunit PSMA7 RNA. Rz3′X was found to induce GCV-resistant colonies in a bicistronic HCV IRES-HSV TK reporter system and reduced HCV IRES-mediated core protein translation, indicating an inhibitory effect of the ribozyme on HCV IRES activity. Using C-SPACE, a ribozyme cleavage-based technique for target gene identification, we discovered human 20S proteasome α-subunit PSMA7 mRNA as a target RNA cleaved by Rz3′X. Additional ribozymes directed against PSMA7 RNA also substantially inhibited HCV IRES activity in a stable HeLa IRES-TK reporter system as well as in an independent transient Huh7 IRES-luciferase reporter assay. Rz3′X and PSMA7-directed ribozymes reduced HCV IRES activity independently of HCV core protein and did not significantly influence EMCV IRES or HRV IRES activities in transiently transfected Huh7 cells. These data indicate that the reduction in core protein associated with expression of active ribozymes in the stably selected GCV-resistant cells was not caused by decreased core protein stability and that the IRES-inhibitory effect is mediated by factors other than the core protein. The data obtained from different reporter systems and in particular the lack of activity on other viral IRESs suggest a preferential effect of the ribozymes on HCV IRES and indicate that the ribozymes do not generally decrease overall translation or protein stability.
Interestingly, we observed that a 30% decrease in PSMA7 RNA was sufficient to induce a 70 to 80% reduction in HCV IRES translation. Similarly, Rz3′X induces a high number of GCV-resistant colonies, while only moderately decreasing PSMA7 RNA and protein expression in the stable selection system presented here. In contrast, VRz4 exactly matching the Rz3′X target site within PSMA7 RNA revealed less GCV-resistant colonies, while a stronger reduction in PSMA7 RNA and protein was noted. Additionally, in the transient luciferase reporter system, VRz4 caused a higher inhibition of IRES activity than Rz3′X. This is in concordance with our previous observations that a biologically relevant change of function was often achieved by a moderate knock-down of target RNA expression by hairpin ribozymes (3, 14). Our results also suggest that, depending on the target molecule, a more active ribozyme with perfectly matching binding arms can exhibit cellular toxicity in stable selection systems, whereas this effect is less important for transient assay systems.
The ubiquitin proteasome pathway is a highly conserved intracellular pathway for the degradation of proteins. Many of the short-lived regulatory proteins that govern cell division, growth, activation, signaling, and transcription are substrates degraded by the proteasome (1). The 26S proteasome is a multisubunit protease complex that catalyzes the final step of intracellular protein degradation. It consists of a cylindrical 20S catalytic chamber, a barrel-shaped structure, shown by electron microscopy to comprise four rings, each containing seven subunits, which is capped on both ends by a 19S regulatory complex. All 14 20S proteasomal subunit sequences may be classified into two groups, α and β, each group showing distinct structural and functional roles (13). Ubiquinated proteins are recognized and unfolded by the regulatory complex and threaded through the small pores at the ends of the catalytic chamber, where they are degraded by different protease activities. The resulting peptides exit from the cylinder, upon which the ubiquitin chains from the degraded protein are recycled. A variety of proteins are processed via the ubiquitin-proteasome pathway, such as proteins involved in cell proliferation or cell cycle control, transcriptional regulators, cytosolic proteins, membrane proteins, and major histocompatibility complex class I (MHC-I) antigen processing (17). Viral proteins synthesized in virus-infected cells are (partially) degraded by the proteasome (5). The resulting peptides are bound to MHC-I molecules and presented on the cell surface to initiate antiviral immune defenses. Recent data suggested an additional antiviral function of the proteasome at the cellular level by degradation of incoming human immunodeficiency virus proteins (20). In addition, the 20S proteasome associates with a number of viral RNAs which are cleaved by proteasome-associated endonuclease activity (6, 10) and it is very likely that additional interactions of proteasome subunits with viral RNA or protein exist.
A dose-dependent inhibitory effect on HCV IRES-mediated translation was demonstrated for short-term application of MG132 in transient and stable HCV IRES-dependent luciferase reporter assays prior to the onset of cellular toxicity. However, when we applied MG132 or lactacystin to HeLa 5′tk reporter cells (1, 10, or 20 μM for 6 h) prior to GCV selection, all cells pretreated with the proteasome inhibitors died during selection with GCV (data not shown). The dose-dependent toxic effects observed upon the application of proteasome inhibitors were not observed when Rz3′X or anti-PSMA7 ribozymes were applied to inhibit proteasome function. This suggests distinct mechanisms of ribozyme-mediated inhibition of PSMA7 RNA and peptide inhibition by proteasome inhibitors.
Since the knowledge about the exact functions of the proteasome subunits and the interaction between proteasomes and viral gene products is still limited, additional studies are required to evaluate the effects of ribozyme-mediated cleavage of proteasomal subunits on modulation of proteasome function and viral antigen processing and presentation as well as viral replication in host cells. Does the proteasome influence HCV IRES translation directly or by influencing degradation of specific cellular (or viral) translation factors mediating this effect? Is this reduction in IRES activity specific for HCV IRES and not applicable to other cellular IRESs? Are the effects reproducible for subgenomic replicons (4, 18) or the potential full-length HCV replication system?
Rz3′X might confer synergistic antiviral effects on HCV by (i) cleavage of the highly conserved antigenomic 3′X sequence of HCV 3′-UTR and (ii) a reduction in IRES activity mediated by inhibition of PSMA7 RNA. Since the HCV 3′-terminal sequence is essential for productive replication in vivo (12) and significantly enhances translation of HCV RNA from the HCV IRES when supplied in cis (8), a ribozyme targeting this crucial HCV sequence might substantially inhibit translation of viral gene products and viral replication. We are currently investigating the antiviral effects of Rz3′X in cell culture systems of HCV replication.
Finally, the development of specific, nontoxic proteasome inhibitors, such as ribozymes, antisense oligonucleotides, and small molecules that inhibit active sites of proteasome subunits, might prove useful for investigating the role of the ubiquitin-proteasome pathway in diverse cellular processes, such as inflammatory events, cellular immune surveillance, tumorigenesis, and chronic infectious diseases.
ACKNOWLEDGMENTS
We are grateful to Leigh Pierce, Armando Ayala, Kathrin Baumert, and Katja Langer for excellent technical assistance.
C.B. was supported by the Deutsche Forschungsgemeinschaft (Be1980/1-1). M.K. was supported by Hochschulinterne Leistungsförderung from the Medizinische Hochschule Hannover.
REFERENCES
- 1.Adams J, Palombella V J, Elliott P J. Proteasome inhibition: a new strategy in cancer treatment. Investig New Drugs. 2000;18:109–121. doi: 10.1023/a:1006321828515. [DOI] [PubMed] [Google Scholar]
- 2.Beger C, Krüger M, Wong-Staal F. Ribozymes in cancer gene therapy. In: Lattime E C, Gerson S L, editors. Gene therapy of cancer. San Diego, Calif: Academic Press; 1998. pp. 139–152. [Google Scholar]
- 3.Beger C, Pierce L N, Krüger M, Marcusson E G, Robbins J M, Welcsh P, Welch P J, Welte K, King M C, Barber J R, Wong-Staal F. Identification of Id4 as a regulator of BRCA1 expression by using a ribozyme-library-based inverse genomics approach. Proc Natl Acad Sci USA. 2001;98:130–135. doi: 10.1073/pnas.98.1.130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blight K J, Kolykhalov A A, Rice C M. Efficient initiation of HCV RNA replication in cell culture. Science. 2000;290:1972–1974. doi: 10.1126/science.290.5498.1972. [DOI] [PubMed] [Google Scholar]
- 5.Goldberg A L, Rock K L. Proteolysis, proteasomes and antigen presentation. Nature. 1992;357:375–379. doi: 10.1038/357375a0. [DOI] [PubMed] [Google Scholar]
- 6.Goldberg A L, Stein R, Adams J. New insights into proteasome function: from archaebacteria to drug development. Chem Biol. 1995;2:503–508. doi: 10.1016/1074-5521(95)90182-5. [DOI] [PubMed] [Google Scholar]
- 7.Huang J, Kwong J, Sun E C, Liang T J. Proteasome complex as a potential cellular target of hepatitis B virus X protein. J Virol. 1996;70:5582–5591. doi: 10.1128/jvi.70.8.5582-5591.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ito T, Tahara S M, Lai M M. The 3′-untranslated region of hepatitis C virus RNA enhances translation from an internal ribosomal entry site. J Virol. 1998;72:8789–8796. doi: 10.1128/jvi.72.11.8789-8796.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.James H A, Gibson I. The therapeutic potential of ribozymes. Blood. 1998;91:371–382. [PubMed] [Google Scholar]
- 10.Jarrousse A S, Gautier K, Apcher S, Badaoui S, Boissonnet G, Dadet M H, Henry L, Bureau J P, Schmid H P, Petit F. Relationships between proteasomes and viral gene products. Mol Biol Rep. 1999;26:113–117. doi: 10.1023/a:1006982023524. [DOI] [PubMed] [Google Scholar]
- 11.Kolykhalov A A, Feinstone S M, Rice C M. Identification of a highly conserved sequence element at the 3′ terminus of hepatitis C virus genome RNA. J Virol. 1996;70:3363–3371. doi: 10.1128/jvi.70.6.3363-3371.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kolykhalov A A, Mihalik K, Feinstone S M, Rice C M. Hepatitis C virus-encoded enzymatic activities and conserved RNA elements in the 3′ nontranslated region are essential for virus replication in vivo. J Virol. 2000;74:2046–2051. doi: 10.1128/jvi.74.4.2046-2051.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kopp F, Hendil K B, Dahlmann B, Kristensen P, Sobek A, Uerkvitz W. Subunit arrangement in the human 20S proteasome. Proc Natl Acad Sci USA. 1997;94:2939–2944. doi: 10.1073/pnas.94.7.2939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Krüger M, Beger C, Li Q X, Welch P J, Tritz R, Leavitt M, Barber J R, Wong-Staal F. Identification of eIF2Bgamma and eIF2gamma as cofactors of hepatitis C virus internal ribosome entry site-mediated translation using a functional genomics approach. Proc Natl Acad Sci USA. 2000;97:8566–8571. doi: 10.1073/pnas.97.15.8566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Krüger M, Beger C, Wong-Staal F. Use of ribozymes to inhibit gene expression. Methods Enzymol. 1999;306:207–225. doi: 10.1016/s0076-6879(99)06014-0. [DOI] [PubMed] [Google Scholar]
- 15a.Krüger M, Beger C, Welch P J, Barber J R, Wong-Staal F. C-SPACE (cleavage-specific amplification of cDNA ends): a novel method of ribozyme-mediated gene identification. Nucleic Acids Res. 2001;29:e94. doi: 10.1093/nar/29.19.e94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li Q X, Robbins J M, Welch P J, Wong-Staal F, Barber J R. A novel functional genomics approach identifies mTERT as a suppressor of fibroblast transformation. Nucleic Acids Res. 2000;28:2605–2612. doi: 10.1093/nar/28.13.2605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lin K I, Baraban J M, Ratan R R. Inhibition versus induction of apoptosis by proteasome inhibitors depends on concentration. Cell Death Differ. 1998;5:577–583. doi: 10.1038/sj.cdd.4400384. [DOI] [PubMed] [Google Scholar]
- 18.Lohmann V, Korner F, Koch J, Herian U, Theilmann L, Bartenschlager R. Replication of subgenomic hepatitis C virus RNAs in a hepatoma cell line. Science. 1999;285:110–113. doi: 10.1126/science.285.5424.110. [DOI] [PubMed] [Google Scholar]
- 19.Muotri A R, da Veiga Pereira L, dos Reis Vasques L, Menck C F. Ribozymes and the anti-gene therapy: how a catalytic RNA can be used to inhibit gene function. Gene. 1999;237:303–310. doi: 10.1016/s0378-1119(99)00334-0. [DOI] [PubMed] [Google Scholar]
- 20.Schwartz O, Marechal V, Friguet B, Arenzana-Seisdedos F, Heard J M. Antiviral activity of the proteasome on incoming human immunodeficiency virus type 1. J Virol. 1998;72:3845–3850. doi: 10.1128/jvi.72.5.3845-3850.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tanaka T, Kato N, Cho M J, Sugiyama K, Shimotohno K. Structure of the 3′ terminus of the hepatitis C virus genome. J Virol. 1996;70:3307–3312. doi: 10.1128/jvi.70.5.3307-3312.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Welch P J, Barber J R, Wong-Staal F. Expression of ribozymes in gene transfer systems to modulate target RNA levels. Curr Opin Biotechnol. 1998;9:486–496. doi: 10.1016/s0958-1669(98)80034-7. [DOI] [PubMed] [Google Scholar]
- 23.Welch P J, Marcusson E G, Li Q X, Beger C, Krüger M, Zhou C, Leavitt M, Wong-Staal F, Barber J R. Identification and validation of a gene involved in anchorage-independent cell growth control using a library of randomized hairpin ribozymes. Genomics. 2000;66:274–283. doi: 10.1006/geno.2000.6230. [DOI] [PubMed] [Google Scholar]
- 24.Welch P J, Tritz R, Yei S, Barber J, Yu M. Intracellular application of hairpin ribozyme genes against hepatitis B virus. Gene Ther. 1997;4:736–743. doi: 10.1038/sj.gt.3300441. [DOI] [PubMed] [Google Scholar]