Abstract
The identification of a novel coregulator for nuclear hormone receptors, designated NRIF3, was recently reported (D. Li et al., Mol. Cell. Biol. 19:7191–7202, 1999). Unlike most known coactivators, NRIF3 exhibits a distinct receptor specificity in interacting with and potentiating the activity of only TRs and RXRs but not other examined nuclear receptors. However, the molecular basis underlying such specificity is unclear. In this report, we extended our study of NRIF3-receptor interactions. Our results suggest a bivalent interaction model, where a single NRIF3 molecule utilizes both the C-terminal LXXIL (receptor-interacting domain 1 [RID1]) and the N-terminal LXXLL (RID2) modules to cooperatively interact with TR or RXR (presumably a receptor dimer), with the spacing between RID1 and RID2 playing an important role in influencing the affinity of the interactions. During the course of these studies, we also uncovered an NRIF3-NRIF3 interaction domain. Deletion and mutagenesis analyses mapped the dimerization domain to a region in the middle of NRIF3 (residues 84 to 112), which is predicted to form a coiled-coil structure and contains a putative leucine zipper-like motif. By using Gal4 fusion constructs, we identified an autonomous transactivation domain (AD1) at the C terminus of NRIF3. Somewhat surprisingly, full-length NRIF3 fused to the DNA-binding domain of Gal4 was found to repress transcription of a Gal4 reporter. Further analyses mapped a novel repression domain (RepD1) to a small region at the N-terminal portion of NRIF3 (residues 20 to 50). The NRIF3 gene encodes at least two additional isoforms due to alternative splicing. These two isoforms contain the same RepD1 region as NRIF3. Consistent with this, Gal4 fusions of these two isoforms were also found to repress transcription. Cotransfection of NRIF3 or its two isoforms did not relieve the transrepression function mediated by their corresponding Gal4 fusion proteins, suggesting that the repression involves a mechanism(s) other than the recruitment of a titratable corepressor. Interestingly, a single amino acid residue change of a potential phosphorylation site in RepD1 (Ser28 to Ala) abolishes its transrepression function, suggesting that the coregulatory property of NRIF3 (or its isoforms) might be subjected to regulation by cellular signaling. Taken together, our results identify NRIF3 as an interesting coregulator that possesses both transactivation and transrepression domains and/or functions. Collectively, the NRIF3 family of coregulators (which includes NRIF3 and its other isoforms) may play dual roles in mediating both positive and negative regulatory effects on gene expression.
The nuclear receptor superfamily makes up a large group of transcription factors that play diverse roles in cell growth, differentiation, development, and homeostasis (35, 37, 58). The superfamily is composed of type I receptors that mediate the actions of steroid hormones and type II receptors that mediate the actions of a variety of structurally diverse ligands such as thyroid hormone, retinoids, and 1, 25-(OH)2 vitamin D3, as well as a number of orphan receptors whose ligands (if any) remain unknown (34, 35). Members of the superfamily share similar domain structures. Generally, a receptor molecule consists of a highly variable N-terminal domain (region A/B), a highly conserved central DNA-binding domain (region C), and a C-terminal ligand-binding domain (LBD; region DEF) that is diverse in sequence but exhibits some conservation in overall structure (5, 18, 58, 61). Typically, a receptor harbors two activation functions: an AF1 within the A/B region and a ligand-dependent AF2 in the LBD (3, 13, 39, 43, 56).
Ligand binding is a critical event in receptor biology, since it induces a conformational change in the receptor that bears broad functional consequences. For example, in the absence of ligand, type I receptors are associated with heat shock protein chaperones and do not bind DNA (5, 44, 45, 46). Ligand binding dissociates these receptors from the chaperones, which enables both their binding to target DNA sequences and the recruitment of coactivators that leads to transactivation (37). In contrast, type II receptors are not associated with heat shock proteins and appear to bind DNA in the absence of their ligands (11, 17, 51). As a consequence, for some of the type II receptors (such as TR and RAR), their unliganded forms can function to repress transcription through the recruitment of corepressor complexes (6, 10, 25, 43). Thus, in such cases, ligand binding promotes both the dissociation of corepressors and recruitment of coactivators, resulting in receptor-mediated transactivation (20).
Efforts to gain deeper insights into the molecular mechanisms of receptor-mediated transactivation have led to the identification of a number of putative coactivators (20, 37), such as the p160 family (1, 9, 24, 28, 32, 41, 55, 57, 59), CBP/p300 (7, 22, 28), the TRAP/DRIP complexes (16, 27, 47, 48), PGC-1 (42), p/CAF (4), NRIF3 (31), and hNRC (33). For many of these coactivators, their interaction with liganded nuclear receptors appears to involve one or more LXXLL motifs contained within their receptor interacting domains (RIDs) (20, 23, 37). The combination of molecular biological and structural studies has indicated that ligand binding induces a conformational change in the receptor LBD that repositions helix 12 which, together with helices 3, 4, and 5, forms a hydrophobic cleft that serves as the docking site for the LXXLL motif contained within the RIDs of coactivators (12, 14, 40).
Members of the p160 family are among the best-characterized examples for coactivator-receptor interactions (20, 37). Their receptor interaction domains contain three LXXLL boxes (often referred as NR boxes), which are in turn differentially utilized by different nuclear receptors (12, 36). For example, coactivation of ER requires the second LXXLL box of SRC-1/NCoA-1, while the function of TR or RAR requires both the second and the third LXXLL boxes (36). Evidence from several studies has suggested that sequences surrounding the LXXLL core are important in determining the specific utilization of different LXXLL boxes by different nuclear receptors (12, 36). This notion is reinforced by a recent study with combinatorial libraries to isolate different LXXLL-containing peptides that exhibit diverse receptor interaction patterns (8). Interestingly, although the various LXXLL boxes of a p160 family member such as SRC-1 show differential receptor preferences, the entire coactivator molecule does not appear to exhibit receptor specificity, since it generally interacts with many nuclear receptors (37).
We recently reported the cloning of a novel coactivator (NRIF3) from a yeast two-hybrid screen (31). One of the unique properties of NRIF3 is its receptor specificity. Most known coactivators identified thus far (such as the p160 family) appear to interact with many nuclear receptors (37). NRIF3, in contrast, only interacts with liganded TR and RXR but not any other examined receptors (31), raising an interesting question about the mechanism of its receptor specificity. Our previous study identified an essential RID (referred to here as RID1) at the C terminus of NRIF3 which, interestingly, contains an LXXIL module, a variant of the canonical LXXLL motif (31). Computer modeling and mutagenesis analyses have indicated that RID1 docks to the hydrophobic groove of a liganded LBD through its LXXIL motif in a way similar to that of the canonical LXXLL module utilized by other coactivators (31). However, RID1 alone does not appear to harbor the same specificity as NRIF3. For example, while NRIF3 interacts with only TR and RXR but not RAR, RID1 was found to interact with all three receptors (31). Thus, the molecular mechanism underlying the receptor specificity of NRIF3 remained unclear.
In this report, we extended our analysis of NRIF3 in a number of areas. First, we further analyzed the NRIF3-receptor interaction in order to gain additional insight into the receptor specificity of NRIF3. These results, together with our previous study, suggest a bivalent interaction model, in which a single NRIF3 molecule utilizes both the C-terminal LXXIL (RID1) and the N-terminal LXXLL (RID2) modules to cooperatively interact with the receptors (presumably a receptor dimer). The spacing between RID1 and RID2 appears to play a critical role in influencing the affinity of the interactions and thus is likely a determinant in the receptor specificity of NRIF3. Second, during the course of such analyses, we also uncovered and mapped a dimerization domain in the middle of the NRIF3 molecule, which may have functional implications in NRIF3 action(s). Third, by using Gal4 fusion constructs, we found that NRIF3 harbors both transactivation and transrepression functions. A transactivation domain (AD1) and a novel repression domain (RepD1) were mapped to residues 162 to 177 and residues 20 to 50, respectively. Fourth, we also analyzed the two alternatively spliced isoforms of NRIF3. Fluorescence microscopy showed that, like NRIF3, these two isoforms are also nucleus localized. Consistent with the fact that both isoforms contain the same RepD1 as NRIF3, Gal4 fusions of these two isoforms repress the transcription of a Gal4 reporter. Since these two isoforms do not interact with nuclear receptors because they both lack the essential RID1, our study raises the possibility that they may function as coregulators for other transcription factors. Taken together, these studies suggest that the NRIF3 gene encodes an interesting family of coregulators that, collectively, may play dual roles in mediating both positive and negative regulation of gene expression.
MATERIALS AND METHODS
Plasmids for the yeast two-hybrid assay.
All yeast plasmids expressing various LexA fusion proteins were constructed from a derivative of pEG202 (21) that contains a modified polylinker. Such plasmids include LexA-NRIF3, LexA-cTRα LBD, LexA-hRXRα LBD, and LexA-RID1. All yeast plasmids expressing various B42 fusions were derived from pJG4-5 (21). These plasmid include the following: B42-cTRα LBD(120–408), B42-cTRα LBD(120–398), B42-hRXRα LBD, B42-hRARα LBD, B42-RID1, B42-NRIF3, B42-EnS, B42-EnL, B42-NRIF3(112–177), B42-NRIF3(Δ112–161), B42-NRIF3 L9A, B42-NRIF3(Δ87–111), B42-NRIF3 L89R, B42-NRIF3 L96R, and B42-NRIF3 DM.
Domain and mutagenesis analyses.
The plasmid expressing LexA-RID1 has been described previously (as LexA-NCD) (31). LexA-NRIF3 was constructed by ligating the full-length NRIF3 fragment derived from pEx-NRIF3 (31) by NcoI and XhoI digestions into a pEG202 vector digested with the same pair of enzymes. To construct B42-RID1, synthetic oligonucleotides that encode the RID1 region of NRIF3 (residues 162 to 177) were annealed and ligated to a pJG4-5-derived vector digested with NcoI and XhoI. B42-NRIF3, B42-EnS, B42-EnL, and B42-NRIF3 L9A have been described previously (31). To construct B42-NRIF3(112–177), primers PNC1 (5′-CGC GAC GTG CCA TGG CTT TGG AGG GCA GTA GAG AGC-3′) and NFCP2 (5′-CGC GAC GTG AGA TCT CGA GCT GGT ATT TAC TGG GCA G-3′) were used to PCR amplify the cognate region of NRIF3. The PCR product was then digested with NcoI and BglII and cloned into a pJG4-5-derived vector digested with the same pair of enzymes. The resulting construct was confirmed by sequencing analysis. B42-NRIF3(Δ112–161) was constructed in two steps. First, pEx-NRIF3 was digested with BstZ17I and BglII and ligated to annealed oligonucleotides encoding residues 162 to 177 of NRIF3 to generate pEx-NRIF3(Δ112–161). pEx-NRIF3(Δ112–161) was then digested with NcoI and XhoI, and the resulting NRIF3(Δ112–161) fragment was ligated into a pJG4-5-derived vector digested with the same pair of enzymes. B42-NRIF3(Δ87–111), B42-NRIF3 L89R, B42-NRIF3 L96R, and B42-NRIF3 DM were all constructed by PCR-based methods. The primers used for these PCRs were PNC3 (5′-CGC GAC GTG GAA TTC GCT TTG GAG GGC AGT AGA GAG C-3′), PNC4 (5′-GAA GTT GGT GCT CAT GGT GAG TGC-3′), PNC5 (5′-GAC AAT GAT GAA TTC ATG ATG AGA CTA TCA AAA GTT GAG AAA TTG TCA GAA G-3′), PNC6 (5′-ATG ATG AAT TCA TGA TGT TGC TAT CAA AAG TTG AGA AAA GAT CAG AAG AAA TCA TGG AG-3′), and PNC7 (5′-ATG ATG AAT TCA TGA TGA GAC TAT CAA AAG TTG AGA AAA GAT CAG AAG AAA TCA TGG AG-3′). PNC4 was the common downstream primer used for all PCRs. The upstream primers were as follows: PNC3 for B42-NRIF3(Δ87–111), PNC5 for B42-NRIF3 L89R, PNC6 for B42-NRIF3 L96R, and PNC7 for B42-NRIF3 DM. For each of these PCRs, the resulting product was digested with EcoRI and ligated to a B42-NRIF3 vector that was digested with the same enzyme. All resulting constructs were confirmed by sequencing analysis. The suitable combinations of plasmids expressing various LexA or B42 fusion proteins were then used in a yeast two-hybrid assay to examine protein-protein interactions (31).
Most of the plasmids that express various Gal4 fusion proteins in mammalian cells were constructed based on a backbone vector (referred to here as Gal4/NK) that was generated by digesting an RSV-Gal4-cT3Rα expression vector (6, 43) with NcoI and KpnI to remove the cT3Rα insert. Appropriate NRIF3, EnS, and EnL inserts were generated by digesting the cognate pEx-based plasmids with NcoI and KpnI. Such inserts were then ligated to the Gal4/NK vector to generate Gal4-NRIF3, Gal4-EnS, and Gal4-EnL. To construct Gal4-NRIF3(162–177), the B42-NRIF3(162–177) vector was digested with NcoI and KpnI to liberate the cognate insert, which was then ligated to the Gal4/NK vector. Gal4-NRIF3(112–177) was constructed by a PCR-based method. Primers PNC1 (see above) and PNC2 (5′-CGC GAC GTG GGT ACC CGA GCT GGT ATT TAC TGG GCA G-3′) were used to amplify the cognate region of NRIF3. The PCR product was then digested with NcoI and KpnI and subsequently ligated to the Gal4/NK vector. The resulting Gal4-NRIF3(112–177) construct was confirmed by sequencing analysis. To construct Gal4-NRIF3(Δ87–111) and Gal4-NRIF3 DM, a slightly different Gal4 backbone vector (referred to here as Gal4/NB) was generated by digesting the Gal4-NRIF3 vector with NcoI and BglII to remove the NRIF3 insert. Inserts corresponding to NRIF3(Δ87–111) and NRIF3 DM were released from the cognate pJG4-5-based vectors by NcoI and BglII digestions and subsequently ligated to the Gal4/NB vector.
To further define the repression domain in the N-terminal region of NRIF3, plasmids expressing Gal4 fusions of various NRIF3 nested deletions were constructed by a PCR-based procedure. The PCR primers were NFCP1 (5′-CGC GAC GTG CAA TTG GCC ATG GCG CCT GTT AAA AGA TCA CTG AAG-3′), 86DP1 (5′-CGC GAC GTG AGA TCT TCA GAA TTC ATC ATT GTC TTT TGT TG-3′), 50DP1 (5′-CGC GAC GTG AGA TCT TCA AGA ACT TGT GGG AGA AGC AAA TAG-3′), 20DP1 (5′-CGC GAC GTG AGA TCT TCA AGG ATC AAA TGA ATT TTC TTC TAA C-3′), 20UP1 (5′-CGC GAC GTG CCA TGG CTC CTT CAA AAA TCA CAA GGA AG-3′), and 47UP1 (5′-CGC GAC GTG CCA TGG CTC CCA CAA GTT CTG AAG AGC AAA AG-3′). A plasmid containing the wild-type NRIF3 was used as the template. The pairing of primers was as follows: NFCP1 and 86DP1 for generating NRIF3(1–86), NFCP1 and 50DP1 for generating NRIF3(1–50), NFCP1 and 20DP1 for generating NRIF3(1–20), 20UP1 and 86DP1 for generating NRIF3(20–86), 47UP1 and 86DP1 for generating NRIF3(47–86), and 20UP1 and 50DP1 for generating NRIF3(20–50). The PCR-amplified fragments were digested with NcoI and BglII and then purified from an agarose gel. Each of the purified fragments was then ligated to the Gal4/NB vector. All constructs were confirmed by sequence analysis. Plasmids expressing various Gal4 fusion proteins were then used in transfection studies to evaluate the transactivation or transrepression functions of these proteins. A similar PCR-based procedure was used to generate the S28A mutant form of RepD1, with primers 50DP1 and S28AUP1 (5′-CGC GAC GTG CCA TGG CTC CTT CAA AAA TCA CAA GGA AGA AAG CTG TTA TAA CTT ATT CTC CAA C-3′).
The yeast two-hybrid and in vitro binding assays.
The bait and prey plasmids used for the yeast two-hybrid assay in this study have been described above. Generally, the yeast strain EGY48 harboring the LacZ reporter plasmid (pSH18-34) (21) was transformed with appropriate bait and prey plasmids. For each transformation, 8 to 10 transformants were randomly selected and analyzed on appropriate X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) plates for preliminary evaluations. Typical colonies were then selected for quantitative β-galactosidase assays as previously described (31). For in vitro binding assays, 35S-labeled wild-type cTRα and the mutant cTRα L398R were generated by in vitro transcription and translation with a reticulocyte lysate system (Promega). The glutathione S-transferase (GST) control and GST-NRIF3 were expressed in Escherichia coli and affinity purified with glutathione-agarose beads (31). The in vitro binding assay was then carried out as previously described (31), with a slightly modified binding buffer (20 mM HEPES [pH 7.8], 1 mM MgCl2, 1 mM dithiothreitol, 10% glycerol, 0.05% Triton X-100, 1 μM ZnCl2, 150 mM KCl, 0.15 mg of bovine serum albumin/ml).
Transfection studies.
The G5-tk-chloramphenicol acetyltransferase (CAT) and G5-SVB-CAT reporters used in this study have been described previously (43). Various Gal4 fusion constructs have been described above. Appropriate plasmids were transfected into HeLa cells by calcium phosphate coprecipitation, with typically 2 μg of G5-tk-CAT or 500 ng of G5-SVB-CAT. When appropriate, the Gal4 control or various Gal4 fusion vectors (400 ng to 1.2 μg) were cotransfected. After transfection, cells were incubated at 37°C for 42 h before being harvested. CAT assays were then carried out, and CAT activities were calculated as previously described (31). The experiments were repeated two to four times, with similar results. GH4C1 cells were transfected by using the Geneporter 2 reagent (Gene Therapy Systems). At 24 h before transfection, cells were set at 1.5 million per well in a six-well plate. Transfections were carried out according to the manufacturer's protocol. Typically, 400 to 750 ng of G5-tk-CAT and 250 to 500 ng of various Gal4 fusion constructs were used for each transfection. When appropriate, 2 μg of empty control vector or 2.5 μg of expression vectors for NRIF3, EnL, or EnS were cotransfected. About 48 h after transfection, cells were harvested for the determination of CAT activities. When applicable, the fold repression was calculated by comparing the CAT activity from the reporter transfected alone with that from the reporter cotransfected with the examined Gal4 fusion.
Fluorescence microscopy.
The green fluorescent protein (GFP) fusion technique was used to study the subcellular location of examined proteins. Vectors expressing GFP-EnS (29) and GFP-EnL were provided by Sanford Shattil. The GFP control and GFP-NRIF3 vectors have been described previously (31). Each vector was transfected into HeLa cells by calcium phosphate coprecipitation. After transfection, cells were incubated at 37°C for 24 h before the examination with a fluorescence microscope to determine the subcellular location of the examined protein (31).
RESULTS
NRIF3 and its isoforms.
We initially cloned NRIF3 in a yeast two-hybrid screen as a factor interacting with TR in a ligand-dependent manner (31). A search of the GenBank identified two highly related proteins, which are alternatively spliced products of the same gene (31). These two proteins were previously designated β3-endonexin short (referred to here as EnS) and long (referred to here as EnL) forms (54). EnS was cloned from a yeast two-hybrid screening with the cytoplasmic tail of β3-integrin as bait (54). EnL was then cloned as an alternatively spliced product of the same gene. However, EnL does not bind to integrin β3 (54). Interestingly, despite their extensive identity with NRIF3 (Fig. 1), our previous study indicated that EnS and EnL do not interact with nuclear receptors (31).
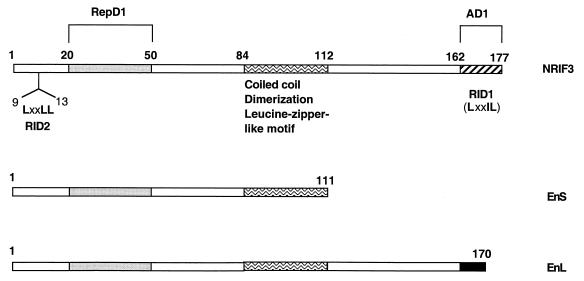
FIG. 1.
Domain organization of NRIF3 and its isoforms. NRIF3 consists of 177 amino acids. EnS consists of 111 amino acids and is 100% identical to the corresponding region of NRIF3. The first 161 amino acids of EnL are identical to those of NRIF3. EnL and NRIF3 differ in their unique C termini (9 residues in EnL, filled box; 16 residues in NRIF3, hatched box). The unique C terminus of NRIF3 (hatched box) harbors RID1, which contains an LXXIL motif. Another RID, RID2, is located at the N terminus of NRIF3 and contains the canonical LXXLL motif. A coiled-coil (dimerization) domain is mapped to the center of the NRIF3 molecule (residues 84 to 112, waved box) and is also found in EnS and EnL. This region also contains a putative leucine zipper-like motif. A transactivation domain (AD1) is mapped to the unique C terminus of NRIF3 (hatched box), a region that also harbors RID1. A transrepression domain (RepD1) is mapped in the N-terminal portion of NRIF3 (residues 20 to 50, dotted box), a region also common to EnS and EnL.
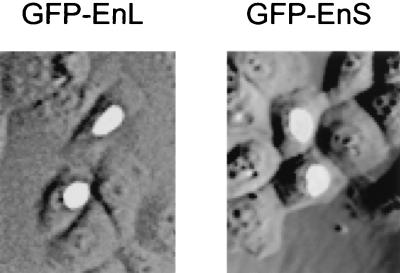
While the precise functions of EnS and EnL remain to be defined, our identification of NRIF3 as a nucleus-localized transcriptional coregulator (31) raised the possibility that they may also function in transcriptional regulation. As a first test, we examined the subcellular location of EnS and EnL to determine whether they localize to the cytoplasm, nucleus, or both compartments. HeLa cells were transfected with vectors expressing GFP alone, GFP-NRIF3, GFP-EnS, or GFP-EnL and the subcellular location of the resulting fluorescent protein was then examined. As we previously reported (31), GFP alone was distributed throughout the whole cell while GFP-NRIF3 localized exclusively to the nucleus (data not shown). Interestingly, both GFP-EnS and GFP-EnL were also found to localize to the nucleus (Fig. 2). This localization pattern is consistent with the observation that NRIF3 harbors a putative nuclear localization signal which is also present in EnS and EnL (31). The finding that both EnS and EnL are localized to the nucleus has prompted us to characterize these two isoforms, together with NRIF3, in a number of experiments (see below).
FIG. 2.
EnS and EnL are nucleus localized. HeLa cells were transfected with an expression vector for GFP-EnS or GFP-EnL. Cells were then incubated at 37°C for 24 h before being examined under a fluorescence microscope to determine the subcellular location of the fusion protein. GFP fusions of both EnS and EnL are localized in the nucleus. The control GFP alone was found to be distributed throughout the cell (data not shown). GFP-NRIF3 was used as another control, and its pattern was found to be similar to that of GFP-EnS or GFP-EnL (data not shown).
Integrity of the receptor AF2 helix is required for the NRIF3-TR interaction.
Our previous study indicated that a unique C-terminal domain in NRIF3 (referred to here as RID1; see Fig. 1) is essential for its interaction with liganded TR and RXR (31). The importance of RID1 is highlighted by the fact that EnS and EnL, the two alternatively spliced isoforms of NRIF3 that lack the RID1 (Fig. 1), do not interact with TR or RXR (31). RID1 contains an LXXIL motif (Fig. 1), a variant of the canonical LXXLL motif. On the basis of a combination of computer modeling and subsequent experimental analysis, we proposed a model where RID1 docks to the hydrophobic groove of the liganded TR or RXR LBD via its LXXIL motif in a fashion similar to that of the canonical LXXLL motif (31). In support of this model, we found that LexA-RID1 can directly interact with the liganded LBDs, and such interaction is completely abolished when the conserved hydrophobic residues of the LXXIL core are mutated (31).
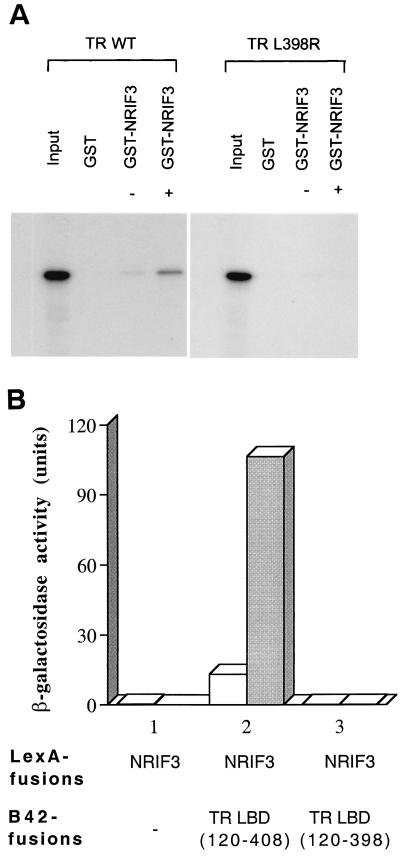
Since the integrity of helix 12 (often referred to also as the AF2 helix) is essential for the proper formation of the hydrophobic cleft on the liganded LBD (14) to which the LXXIL motif is predicted to bind (31), we further tested this model by examining the interaction of NRIF3 with two TRα mutants. One mutant, L398R, contains a single point mutation (Leu398 to Arg398) in helix 12 which abolishes its ability to transactivate but does not appear to affect ligand binding (53). An in vitro GST pull-down assay was carried out with purified GST-NRIF3 and in vitro-translated 35S-labeled L398R TR in the presence or absence of T3. As shown in Fig. 3A, while wild-type TR exhibited a ligand-dependent interaction with GST-NRIF3, such interaction was completely abolished for L398R TR. In another study, we examined the interaction of NRIF3 with TR(120–398) in a yeast two-hybrid assay. TR(120–398) harbors a deletion in the LBD that removes the last 10 amino acids of helix 12 and is consequently defective in ligand-mediated transactivation (18, 19, 53). As shown in Fig. 3B, NRIF3 interacted with wild-type TR LBD(120–408) in a ligand-dependent manner. However, no interaction was detected when the mutant TR LBD(120–398) was used in the assay. Taken together, the in vitro binding and yeast two-hybrid assays suggest that helix 12 plays an essential role in NRIF3 interaction with liganded TR LBD, as predicted from our computer modeling.
FIG. 3.
Integrity of helix 12 (the AF2 helix) is essential for the NRIF3-TR interaction. (A) 35S-labeled wild-type TR (WT) or the mutant TR (L398R) was generated by in vitro translation. The labeled receptors were then examined for binding to purified GST control or GST-NRIF3 in the presence (+) or absence (–) of T3 as described in Materials and Methods. (B) Yeast two-hybrid assay. LexA-NRIF3 was examined for interaction with the control B42 alone, B42-TR LBD(120–408), or B42-TR LBD(120–398) that deletes 10 amino acid residues from helix 12. β-Galactosidase activities were determined in the presence (shaded columns) or absence (open columns) of T3.
Interactions of the NRIF3 RID1 with receptor LBDs in yeast are fusion partner dependent.
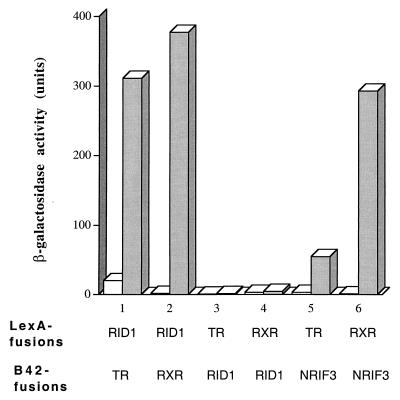
Yeast two-hybrid assays were used to further characterize NRIF3-receptor interactions and to map an essential receptor interacting surface of NRIF3 (RID1) to its C terminus (31) (Fig. 1). As shown in Fig. 4, LexA-RID1 interacts with the LBDs of TR and RXR (fused to B42) in a ligand-dependent manner, while LexA alone exhibits no such interactions (data not shown). However, in a reciprocal experiment with LexA-TR (or RXR) LBD as the bait and B42-RID1 as the prey, we did not observe any interactions with or without ligand (Fig. 4). As a positive control, LexA-TR LBD or LexA-RXR LBD was found to interact with full-length B42-NRIF3 in a ligand-dependent manner (Fig. 4). Therefore, the interactions of RID1 with receptor LBDs appear to depend on the identity of its fusion partner, with the LexA-RID1 proficient and B42-RID1 deficient in the interaction.
FIG. 4.
Interactions of RID1 with liganded LBDs in yeast are fusion partner dependent. The indicated baits LexA-RID1, LexA-TR LBD, and LexA-RXR LBD were examined for interactions with the indicated preys (as B42 fusions) in a yeast two-hybrid assay as described in Materials and Methods. β-Galactosidase activities were determined in the presence (shaded columns) or absence (open columns) of cognate ligands: T3 for TR, 9-cis RA for RXR.
NRIF3-NRIF3 interaction.
A plausible explanation for the difference in the abilities of LexA-RID1 and B42-RID1 to interact with the receptor LBDs is that two copies of RID1 are required for efficient association with the liganded LBDs. However, we cannot exclude the possibility that the B42-RID1 fusion protein might be unstable or fold incorrectly. LexA-RID1 can potentially provide two copies of RID1 for interaction since the LexA DNA-binding domain (DBD) binds to its cognate operator sequences as a dimer (30, 38). In contrast, the B42 activation domain is not known to dimerize and therefore the B42-RID1 monomer would be expected to be monovalent for RID1. Thus, the functional differences found between LexA-RID1 and B42-RID1 raise the possibility that bivalency might be required for a productive interaction between the RID(s) of NRIF3 and the liganded receptor LBDs.
To begin to explore this, we examined whether NRIF3 can form a dimer in a yeast two-hybrid assay, since dimerization of NRIF3 could conceivably provide two copies of RID1. As negative controls, coexpression of LexA-NRIF3 and B42 alone (Fig. 5), or LexA alone and B42-NRIF3 (data not shown), did not activate the LacZ reporter in yeast. However, coexpression of LexA-NRIF3 and B42-NRIF3 resulted in a strong activation of the LacZ reporter (Fig. 5), suggesting the formation of an NRIF3 dimer.
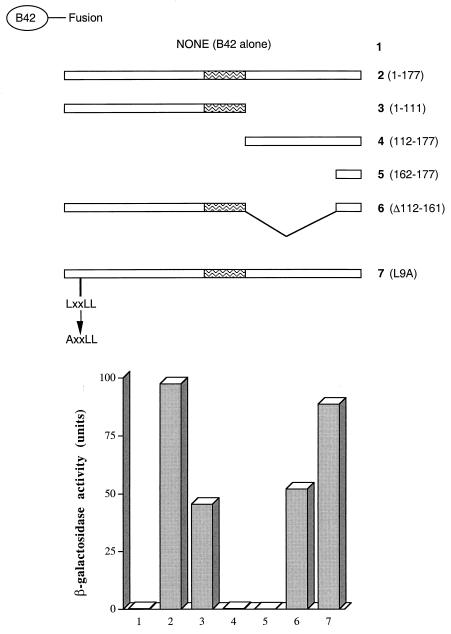
FIG. 5.
Characterization of the NRIF3-NRIF3 interaction. LexA-NRIF3 was studied in a yeast two-hybrid assay for interactions with various B42 fusions as depicted in the figure. The region of the coiled-coil domain is indicated as a waved box.
To further characterize which domain in NRIF3 is responsible for the dimerization, the N (residues 1 to 111)- and C (residues 112 to 177)-terminal portions of NRIF3 were separately fused to B42 and tested for interactions with LexA-NRIF3. The NRIF3(1–111) used in this assay is equivalent to one of the alternatively spliced isoforms, EnS (Fig. 1). As shown in Fig. 5, while NRIF3(1–111) largely retained the interaction with NRIF3, NRIF3(112–177) was completely deficient in the interaction. Not surprisingly, the very C terminus of NRIF3 (residues 162 to 177) that harbors the RID1 also failed to exhibit any interaction (Fig. 5). NRIF3(Δ112–161), which harbors an internal deletion that removes amino acid residues 112 to 161, exhibited a level of interaction similar to that of NRIF3(1–111) (Fig. 5). Taken together, these results suggest that the dimerization domain is contained within amino acid residues 1 to 111 of NRIF3. Interestingly, the L9A mutant of NRIF3, which contains a point mutation (Leu9 to Ala9) in the first leucine residue of the putative LXXLL motif (31), showed a level of interaction similar to that of wild-type NRIF3 (Fig. 5), suggesting that the LXXLL motif is not involved in the NRIF3-NRIF3 interaction.
A central coiled-coil domain is essential for the NRIF3-NRIF3 interaction.
We further defined the NRIF3 dimerization surface in order to generate an appropriate mutant(s) to examine whether dimerization of NRIF3 plays a role in its interaction with TR or RXR. Structural analysis of NRIF3 by computer modeling (52) revealed the presence of a putative coiled-coil domain in the middle of the NRIF3 molecule that spans amino acid residues 84 to 112 (Fig. 1 and 6A). Close inspection of this putative coiled-coil domain identified a leucine zipper-like motif (Fig. 6A). Interestingly, virtually the entire portion of this putative coiled-coil domain is contained within the region of NRIF3 found to be involved in its dimerization (Fig. 5).
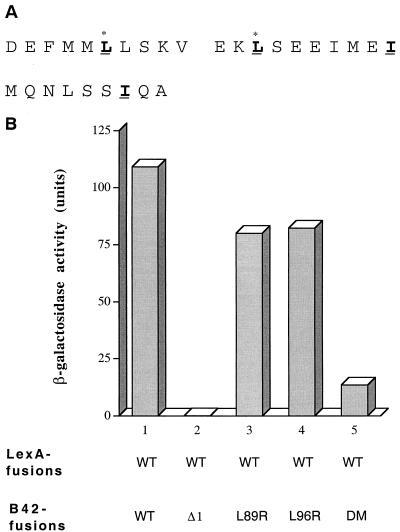
FIG. 6.
The central coiled-coil domain is essential for the NRIF3-NRIF3 interaction. (A) Amino acid sequence of the coiled-coil domain of NRIF3 (residues 84 to 112). The putative leucine zipper-like structure is shown in boldface and underlined, with the occurrence of Leu, Leu, Ile, and Ile at every seventh position between residues 89 and 110. Leu89 and Leu96 are marked with asterisks. (B) LexA-NRIF3 (WT) was examined in a yeast two-hybrid assay for interactions with the following preys (as B42 fusions), respectively: wild-type NRIF3 (WT), mutant NRIF3 with an internal deletion of the coiled-coil domain (Δ1), and mutant NRIF3s L89R, L96R, and DM.
To test the role of the putative coiled-coil domain in NRIF3 dimerization, we constructed an internal deletion mutant of NRIF3 (referred as NRIF3Δ1) that essentially removes this entire region (residues 87 to 111). As shown in Fig. 6B, NRIF3Δ1 was found to be completely deficient in interaction with wild-type NRIF3, indicating that the coiled-coil domain is indeed indispensable for dimerization. To test the role of the leucine zipper-like motif, we constructed mutants of NRIF3 (L89R and L96R) which changed the first (Leu89) or second (Leu96) leucine residue of the motif into arginine. The mutant DM (double mutant) contains mutations of both residues (L89R and L96R). As shown in Fig. 6B, dimerization of NRIF3 is not affected by the single L89R or L96R change. However, interaction with NRIF3 is severely reduced when both leucine residues are mutated. Taken together, these results suggest that the NRIF3-NRIF3 interaction is mediated by the central coiled-coil domain, probably through a leucine zipper-like structure.
Dimerization of NRIF3 is not required for its interaction with TR or RXR.
If the NRIF3-receptor association involves a bivalent interaction, one possible model is that dimerization of NRIF3 facilitates the interaction of two copies of RID1 with the receptor LBDs. To test this possibility, we examined the three NRIF3 mutants—L89R, L96R, and DM—for their interactions with TR and RXR. As shown in Fig. 7, L89R and L96R, which are still capable of dimerization, remain proficient in interacting with TR and RXR LBDs in a ligand-dependent manner. In addition, the DM mutant that is deficient in NRIF3-NRIF3 dimerization exhibited interactions with liganded LBDs of TR and RXR which were equal to or greater than those of wild-type NRIF3 (Fig. 7), suggesting that NRIF3 dimerization is not required for NRIF3-receptor interactions.
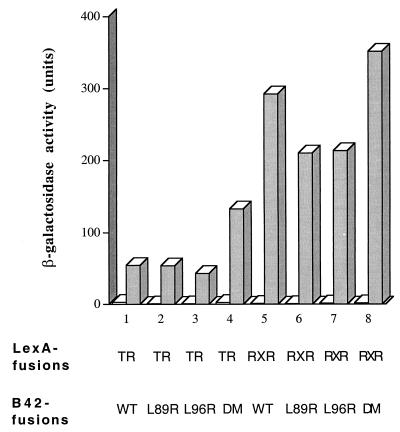
FIG. 7.
Dimerization of NRIF3 is not required for receptor interactions. The baits LexA-TR LBD and LexA-RXR LBD were examined in a yeast two-hybrid assay for interactions with the following preys (as B42 fusions), respectively: wild-type NRIF3 (WT), NRIF3 L89R, NRIF3 L96R, and the double mutant (DM). β-Galactosidase activities were determined in the presence (shaded columns) or absence (open columns) of cognate ligands: T3 for TR, 9-cis RA for RXR.
Spacing between RID1 and RID2 influences NRIF3-receptor interactions.
Since dimerization of NRIF3 appears to be dispensable for TR or RXR interactions, it seems unlikely that such interactions would involve two copies of RID1 contributed by a NRIF3 dimer. We previously reported that the N-terminal LXXLL motif is required for optimum NRIF3-receptor interactions, since a mutation in one of the core leucine residues reduces NRIF3 interactions with TR or RXR (31). Thus, an alternative possibility is that NRIF3-receptor interactions involve and require both the C-terminal RID1 and the N-terminal LXXLL motif (referred to here as RID2). This model is consistent with our findings that (i) RID1 is essential and RID2 is important for optimum NRIF3-receptor interactions (31); (ii) a monovalent version of RID1(B42-RID1; see Fig. 4), or EnS and EnL molecules that contain only RID2 (31), fail to interact with receptors; and (iii) dimerization of NRIF3 is dispensable for receptor interactions.
This model is further supported by an experiment shown in Fig. 8. While the B42 fusion of full-length NRIF3 interacted with LexA-TR LBD in a ligand-dependent manner, the interaction was completely abolished when either the N (residues 1 to 111)- or C (residues 112 to 177)-terminal portion of NRIF3 is fused with B42. Similar results were obtained when the LexA-RXR LBD was used as bait (Fig. 8, columns 6 to 8).
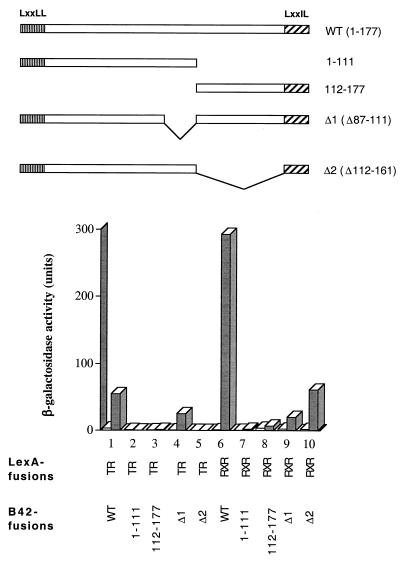
FIG. 8.
NRIF3-receptor interaction requires regions of both RID1 and RID2 and is influenced by the spacing between RID1 and RID2. The baits LexA-TR LBD and LexA-RXR LBD were examined in a yeast two-hybrid assay for interactions with the following preys (as B42 fusions), respectively: full-length NRIF3 (WT), the N-terminal portion of NRIF3(1–111) that contains RID2, the C-terminal portion of NRIF3(112–177) that contains RID1, and mutant NRIF3s that delete residues 87 to 111 (Δ1), or residues 112 to 161 (Δ2). β-Galactosidase activities were determined in the presence (shaded columns) or absence (open columns) of cognate ligands: T3 for TR, 9-cis RA for RXR.
Since an optimum NRIF3-receptor interaction appears to require both RID1 and RID2, the receptor specificity of NRIF3 might arise from the interplay between these two RIDs. To explore this, we examined two internal deletion mutants of NRIF3 for receptor interactions. NRIF3(Δ112–161) contains a deletion of 50 amino acids from the end of the coiled-coil domain to the beginning of RID1, while NRIF3(Δ87–111) deletes 25 amino acids comprising essentially the entire coiled-coil domain. It should be noted that both mutants retain RID1 and RID2. As shown in Fig. 8 (columns 1, 4, and 5 and columns 6, 9, and 10), interactions with liganded TR or RXR LBDs were reduced for both mutants, compared with wild-type NRIF3. This result suggests that proper spacing between RID1 and RID2 is important for receptor interactions. Moreover, the two deletions were found to have somewhat different effects on interactions with TR and RXR. Specifically, the deletion of residues 87 to 111 resulted in a relatively modest 2-fold decrease in the interaction with liganded TR LBD, while the interaction with liganded RXR LBD was reduced by about 15-fold (Fig. 8, columns 1 and 4 and columns 6 and 9). In contrast, the deletion of residues 112 to 161 affects the interaction with TR much more than that with RXR (Fig. 8). NRIF3(Δ112–161) showed a reduced (∼5-fold) but nevertheless significant interaction with the liganded RXR LBD (Fig. 8, columns 6 and 10), whereas its interaction with the liganded TR LBD was completely abolished (Fig. 8, columns 1 and 5). Therefore, the two deletions not only reduce the overall affinity for the receptor LBDs but also influence the specificity profile of the resulting mutant NRIF3s. Thus, while wild-type NRIF3 selectively interacts with liganded TR and RXR, it interacts more efficiently with RXR than with TR (Fig. 8, columns 1 and 6). The preference for RXR is eliminated in NRIF3(Δ87–111), which exhibits similar levels of interaction with TR and RXR (Fig. 8, columns 4 and 9). In contrast, the other internal deletion mutant, NRIF3(Δ112–161), is specific for RXR and does not interact with TR (Fig. 8, columns 5 and 10). Taken together, these results suggest that the spacing between RID1 and RID2 plays a critical role in determining the interaction (or lack of interaction) with different receptor LBDs and thus likely contributes to the receptor specificity of NRIF3.
The C terminus of NRIF3 contains an autonomous transactivation domain.
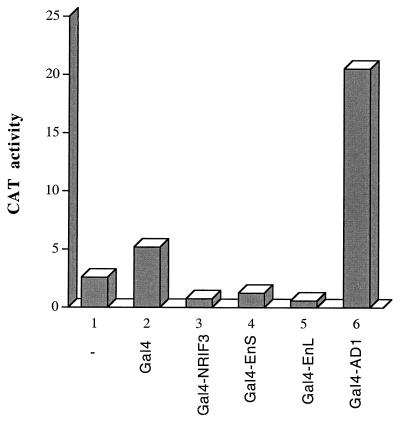
To gain further insights into NRIF3-mediated coactivation, cDNAs encoding full-length NRIF3, its derived fragments, or its related isoforms were fused in frame with the Gal4 DBD. The resulting Gal4 fusion constructs were transfected into HeLa cells with G5-tk-CAT, a reporter under the control of the basal tk promoter linked to five copies of a Gal4 response sequence. As shown in Fig. 9, G5-tk-CAT exhibited a relatively low basal activity, and cotransfection of the Gal4 DBD resulted in a modest activation (∼2-fold) that is commonly observed with this reporter. In contrast, cotransfection of Gal4-NRIF3(162–177) (also referred to as Gal4-AD1) resulted in a 10-fold activation (Fig. 9), suggesting that this region (the very C terminus) of NRIF3 harbors an autonomous transactivation domain (AD1). However, all other Gal4 fusions, including that of full-length NRIF3(1–177), NRIF3(1–111)/EnS, and the alternatively spliced isoform EnL, failed to activate G5-tk-CAT.
FIG. 9.
The C-terminal region of NRIF3 contains an autonomous transactivation domain (AD1). HeLa cells were transfected with the G5-tk-CAT reporter either alone or together with one of the following constructs: Gal4, Gal4-NRIF3 (full-length, 1 to 177), Gal4-EnS (equivalent to residues 1 to 111 of NRIF3, see Fig. 1), Gal4-EnL (full length; Fig. 1), and Gal4-AD1 (residues 162 to 177 of NRIF3, see Fig. 1). Cells were harvested 42 h after transfection, and CAT activities were then determined as described in Materials and Methods.
Since AD1 overlaps RID1, the possibility exists that an endogenous receptor (e.g., an RXR) might tether to Gal4-AD1, which in turn is responsible for its observed transactivation function. We believe this is unlikely, since we have shown previously that the RID1 has little binding to receptor LBDs in the absence of cognate ligands (31), and the experiments described in Fig. 9 were carried out by using cells cultured with ligand-depleted serum. Nevertheless, we tested this by examining the effect of endogenous RXRs. Cells transfected with G5-tk-CAT and Gal4-AD1 (or the Gal4 control) were treated with or without 9-cis RA, and the resulting reporter activities were compared. If transactivation by Gal4-AD1 is a result of its association with an endogenous RXR, then 9-cis RA would be expected to affect Gal4-AD1 activity since ligand would promote such an association. However, we found that 9-cis RA had no effect on Gal4-AD1-mediated transactivation (data not shown). In another experiment, G5-tk-CAT and Gal4-AD1 (or the Gal4 control) were cotransfected with TR in the presence or absence of T3. TR was also found to have no effect on Gal4-AD1-mediated transactivation with or without T3 (data not shown). Taken together, these results suggest that the activation function of AD1 is independent of receptor binding.
The N-terminal portion of NRIF3 harbors a transrepression function.
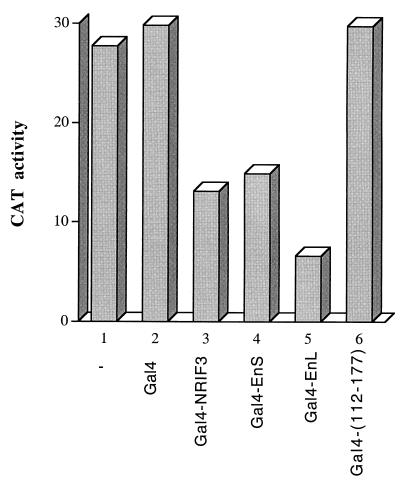
Although NRIF3 contains AD1, we found that full-length NRIF3 fused to Gal4 did not activate G5-tk-CAT (Fig. 9). In fact, the reporter activity appears to be lowered in the presence of Gal4-NRIF3, suggesting possible repression by this fusion protein (Fig. 9, columns 1 to 3). Since the basal activity of G5-tk-CAT is quite low in our transfected HeLa cells (Fig. 9), we used the G5-SVB-CAT reporter, which exhibits higher basal activity (Fig. 10), to further evaluate the potential repression function of Gal4-NRIF3. G5-SVB-CAT was transfected into HeLa cells alone, with the Gal4 DBD control, or with one of the Gal4 DBD fusions depicted in Fig. 10. As shown in Fig. 10, while Gal4 DBD alone had little effect on the reporter activity, Gal4-NRIF3 (full length, 1 to 177) significantly repressed the reporter activity. A Gal4 fusion of the N-terminal portion of NRIF3 (residues 1 to 111, which is also equivalent to EnS) exhibited a similar extent of repression (Fig. 10). In contrast, a Gal4 fusion with the C-terminal portion of NRIF3 (residues 112 to 177) failed to repress the reporter activity. These results suggest that the N-terminal portion of NRIF3 (residues 1 to 111) harbors a transrepression function. Thus, NRIF3 appears to harbor both transactivation and transrepression functions. In the context of a Gal4 DBD fusion with full-length NRIF3 [Gal4-NRIF3(1–177)], the transrepression function appears to be dominant. We also examined the Gal4 DBD fusion of EnL, an alternatively spliced isoform of NRIF3, and found it also mediated repression (Fig. 10).
FIG. 10.
The N-terminal portion of NRIF3 harbors a transrepression function. HeLa cells were transfected with the G5-SVB-CAT reporter either alone or together with one of the following constructs: Gal4, Gal4-NRIF3 (full length, 1 to 177), Gal4-EnS (equivalent to residues 1 to 111 of NRIF3; Fig. 1), Gal4-EnL (full length; Fig. 1), and Gal4-(112–177) (residues 112 to 177 of NRIF3). Cells were harvested 42 h after transfection, and CAT activities were then determined as described in Materials and Methods.
The repression function of NRIF3 does not require its coiled-coil domain.
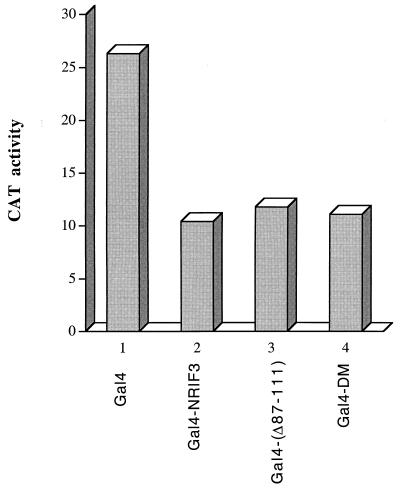
The repression function of NRIF3 is localized to its N-terminal region which also contains the coiled-coil domain essential for mediating dimerization (Fig. 1). Since the coiled-coil domain and its leucine zipper-like motif are also candidate structures for mediating protein-protein interactions, we tested whether the coiled-coil domain may function as a surface for a docking factor(s) responsible for repression. To this end, we constructed Gal4 fusions of two mutant forms of NRIF3: Gal4-NRIF3(Δ87–111), which deletes essentially the entire coiled-coil domain, and Gal4-NRIF3 DM, which contains mutations in the first and second leucine residues of the putative leucine zipper-like motif. These two fusions were then evaluated for the ability to repress G5-SVB-CAT in transfected HeLa cells. As shown in Fig. 11, both mutants exhibited levels of repression similar to that of wild-type Gal4-NRIF3, indicating that the coiled-coil domain is not required for the repression function of NRIF3 and that the repression function of NRIF3 appears to reside in amino acid residues 1 to 86, which are also common to EnS and EnL.
FIG. 11.
The coiled-coil domain is not required for the transrepression function of NRIF3. HeLa cells were transfected with the G5-SVB-CAT reporter, together with one of the following constructs: Gal4, Gal4-NRIF3 (wild type), Gal4-(Δ87–111) (a mutant NRIF3 that deletes the coiled-coil domain), and Gal4-DM (a mutant NRIF3 containing double mutations L89R and L96R). Cells were harvested 42 h after transfection, and CAT activities were then determined as described in Materials and Methods.
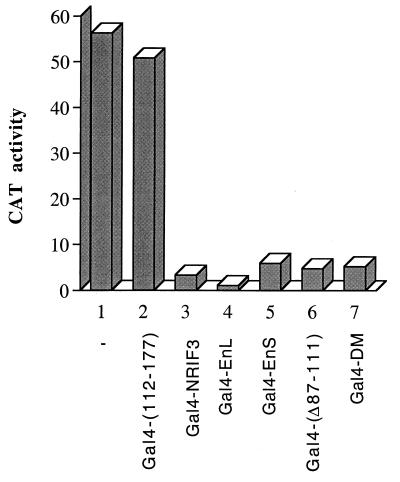
Gal4 fusions of NRIF3 and its isoforms function as potent repressors in GH4C1 cells.
Our study of Gal4 fusions of NRIF3 and its isoforms was first carried out with transfected HeLa cells. While these experiments reproducibly showed that Gal4-NRIF3, Gal4-EnS, and Gal4-EnL function as repressors (Fig. 10), the extent of repression is nevertheless moderate (about two- to threefold). To test whether different cell types might affect the transcriptional activity of Gal4-NRIF3 (or its isoforms), we performed similar studies with another cell line, GH4C1, which has been used extensively to study nuclear receptor actions (50). Since a simian virus 40 promoter-controlled reporter exhibits minimal activity in GH4C1 cells, we used the G5-tk-CAT reporter, which showed high basal activity under our transfection conditions (Fig. 12). As shown in Fig. 12, Gal4-NRIF3, Gal4-EnS, and Gal4-EnL all function as potent repressors in GH4C1 cells. The extent of repression is between 10- and 20-fold, compared to the 2- to 3-fold repression observed in HeLa cells. We also examined several NRIF3 derivatives. Gal4-(112–177) did not repress in HeLa cells (Fig. 10, column 6) and also showed no repression in GH4C1 cells (Fig. 12, column 2). The coiled-coil domain deletion mutant Gal4-(Δ87–111) and the leucine zipper mutant Gal4-DM exhibited a repression similar to that of Gal4-NRIF3 in HeLa cells (Fig. 11), and both were found to function as potent repressors in GH4C1 cells as well. Thus, the overall repression pattern of Gal4 fusions of NRIF3 or its isoforms or its derivatives is conserved between HeLa and GH4C1 cells, despite the fact that the extent of repression is greater in GH4C1 cells.
FIG. 12.
Gal4 fusions of NRIF3, EnL, and EnS function as potent repressors in GH4C1 cells. Constructs expressing Gal4-NRIF3, Gal4-EnL, or Gal4-EnS were transfected into GH4C1 cells, together with the G5-tk-CAT reporter, to evaluate the potential repression function by the Gal4 fusion proteins. Gal4-(112–177) (which shows no repression and was included as a control), Gal4-(Δ87–111 (a mutant NRIF3 that deletes the coiled-coil domain), and Gal4-DM (a mutant NRIF3 containing double mutations L89R and L96R) were used to examine the potential role of the coiled-coil domain.
One mechanism by which transcription factors or cofactors mediate repression is through the recruitment of additional corepressors. For example, some nuclear receptors repress transcription in the absence of their ligands (2, 11). The finding that overexpression of an unliganded receptor in trans leads to relief of repression provided the first line of evidence suggesting that a limiting cofactor (corepressor) is involved in receptor-mediated repression (6). To test whether such a mechanism might also be involved in the repression mediated by Gal4-NRIF3, we examined its ability to repress the G5-tk-CAT reporter in GH4C1 cells when cotransfected with wild-type NRIF3. Cotransfection of NRIF3 did not reverse Gal4-NRIF3-mediated repression (data not shown). Similarly, cotransfection of EnS and EnL did not reverse the repression mediated by Gal4-EnS and Gal4-EnL, respectively (data not shown). Thus, repression by Gal4 fusions of NRIF3, EnS, and EnL may involve a mechanism(s) other than the recruitment of a titratable corepressor. One possibility is that the repression is mediated through a direct contact of these proteins with a member of the basal transcription machinery. We tested two such factors, TBP and TFIIB, in transfected GH4C1 cells. Neither factor was found to significantly influence repression mediated by Gal4-NRIF3 (or EnS or EnL) (data not shown).
A novel repression domain maps to residues 20 to 50 of NRIF3.
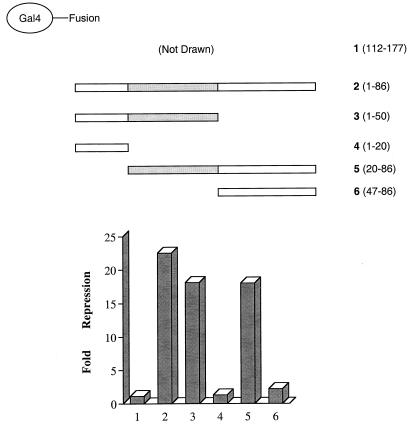
Our transfection studies indicated that the major transrepression function of NRIF3 is contained in its N terminus (residues 1 to 111), since Gal4-EnS (which is equivalent to the region including residues 1 to 111 of NRIF3 [Fig. 1]) showed an extent of repression similar to that of full-length Gal4-NRIF3 in both HeLa and GH4C1 cells (Fig. 10 and 12). Moreover, deletion of the bulk of the coiled-coil domain (residues 87 to 111) appeared to have little effect on repression (Fig. 11 and 12). From these results, we inferred that the repression function of NRIF3 is probably contained within residues 1 to 86. To confirm this, we constructed Gal4-NRIF3(1–86) and found that it indeed mediates potent repression in GH4C1 cells (Fig. 13). To further define the domain(s) responsible for repression, a series of Gal4 fusions that contain various deletions of NRIF3 were constructed, including Gal4-(1–20), Gal4-(1–50), Gal4-(20–86), and Gal4-(47–86) (see Fig. 13 for schematic drawings of these constructs). These Gal4 fusions were designed based on a secondary structure analysis of NRIF3 so that the junction points of various deletions are in the predicted nonstructured regions of NRIF3 in order to minimize the potential conformational disruption to the resulting deletion molecules. As shown in Fig. 13, while Gal4-(1–50) and Gal4-(20–86) were found to be potent repressors in GH4C1 cells, repression is largely abolished for Gal4-(1–20) and Gal4-(47–86), suggesting that the region spanning residues 20 to 50 is the essential domain (termed RepD1) required for repression. RepD1 shares no homology with known domains in the database. Secondary structure analysis suggests that RepD1 is composed of two short β-strands separated by an unstructured linker.
FIG. 13.
An essential repression domain (RepD1) is mapped to residues 20 to 50 of NRIF3. Constructs expressing Gal4 fusions of various regions of NRIF3 as illustrated were transfected into GH4C1 cells, together with the G5-tk-CAT reporter to evaluate repression. Gal4-(112–177), which shows no repression, was included as a control (construct 1 [not drawn]). The fold repression was calculated as described in Materials and Methods. The mapped RepD1 region spanning residues 20 to 50 is shown as a dotted box.
A single amino acid substitution (Ser28 to Ala) in RepD1 abolishes its transrepression function.
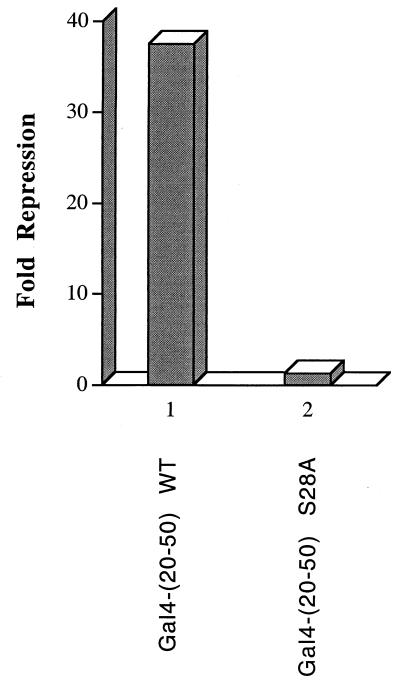
The nested deletion experiment shown in Fig. 13 suggested that the transrepression domain of NRIF3 is located within residues 20 to 50. To confirm that this region can indeed mediate repression, we constructed Gal4-NRIF3(20–50) and examined its effect on the G5-tk-CAT reporter in transfected GH4C1 cells. As shown in Fig. 14, Gal4-NRIF3(20–50) was found to mediate potent repression in GH4C1 cells. Thus, the RepD1 region (residues 20 to 50) is both necessary and sufficient for mediating repression. As discussed earlier, RepD1 shares no homology with known domains in the database. Moreover, our cotransfection study argues against a corepressor recruitment model for NRIF3 family-mediated repression. To shed some more light on RepD1 function, we performed a motif search with an online bioinformatics tool (62) by using the full-length NRIF3 as a query. The search predicted a potential phosphorylation site (Ser28) in NRIF3 which, interestingly, is located within the RepD1 region. To test the potential role of Ser28 in RepD1 function, we constructed Gal4-NRIF3(20–50, S28A), which contains a single amino acid substitution (Ser28 to Ala) in RepD1. Remarkably, while wild-type RepD1 elicits potent repression (more than 35-fold in this experiment), the ability to repress is virtually abolished for the mutant RepD1 (about 1.3-fold [Fig. 14]). This result suggests that phosphorylation at Ser28 is essential for RepD1 function in vivo and raises the interesting possibility that cellular signaling may influence the regulatory property of the NRIF3 family through the regulation of Ser28 modification.
FIG. 14.
A single amino acid substitution (Ser28 to Ala) abolishes RepD1-mediated repression. Gal4 fusions of the wild-type RepD1 [Gal4-(20–50) WT] or the mutant RepD1 [Gal4-(20–50) S28A] were examined for the ability to repress the G5-tk-CAT reporter in transfected GH4C1 cells. The fold repression was calculated as described in Materials and Methods.
DISCUSSION
Domain structure of the NRIF3 gene family.
A summary of the domain organization of NRIF3 and its isoforms, EnS and EnL, is shown in Fig. 1. NRIF3 harbors two RIDs (RID1 and RID2; Fig. 1). The C terminus of NRIF3 harbors an autonomous transactivation domain (AD1), which is moderately active when fused to the heterologous Gal4 DBD (Fig. 9). A novel repression domain (RepD1) is mapped to the N-terminal part of NRIF3 (residues 20 to 50) (Fig. 1, 13, and 14). In addition, the central region of NRIF3 contains a coiled-coil (dimerization) domain and a putative leucine zipper-like structure (Fig. 1 and 6). The gene that encodes NRIF3 also expresses two other alternatively spliced isoforms, EnS and EnL (Fig. 1). EnS is 100% identical to the corresponding portions (residues 1 to 111) of NRIF3 and EnL (Fig. 1). EnL and NRIF3 share 100% identity in their first 161 amino acid residues but differ in their small C-terminal portions (Fig. 1). EnS and EnL do not interact with nuclear receptors, while the unique C-terminal RID1 (which contains an LXXIL motif) of NRIF3 allows for a high-affinity ligand-dependent interaction with the TRs and RXRs (31). Thus, EnS and EnL lack the RID1 and AD1 region but contain the RepD1 and the coiled-coil (dimerization) domain (Fig. 1). Fluorescence microscopy of GFP fusions indicates that, like NRIF3 (31), EnS and EnL localize to the cell nucleus (Fig. 2). Our finding that EnS localizes only to the cell nucleus is somewhat different from a report by Kashiwagi et al. (29), which showed that the GFP fusion of EnS was found in both the cytoplasm and the nucleus. This discrepancy may arise from the differences in the cell lines used for transfection and/or the expression levels of the fusion proteins. Nevertheless, we have consistently observed the nuclear localization of EnS and EnL under the described experimental conditions.
Receptor specificity of NRIF3.
One of the unique features of NRIF3 is its receptor specificity. While most known coactivators exhibit interactions with a broad array of receptors, NRIF3 interacts only with liganded TRs and RXRs and not with other examined nuclear receptors (31). Our previous study mapped an essential RID (i.e., RID1) to the unique C terminus of NRIF3 (31; also Fig. 1). Interestingly, RID1 contains an LXXIL motif, a variant of the canonical LXXLL motif. Computer simulation suggested that RID1 docks to the hydrophobic cleft of liganded receptor LBDs via its LXXIL motif in a similar fashion as to the canonical LXXLL motif utilized by the p160 family members (31). This conclusion is supported by several lines of experimental evidence. First, RID1 is essential for receptor interactions (31). Second, LexA-RID1 interacts with the TR and RXR LBDs in a ligand-dependent fashion (31; also Fig. 4), and such interactions are abolished by mutations in the core leucine residues of the LXXIL motif (31). Third, the integrity of helix 12 (the AF2 helix), a critical part in the formation of the hydrophobic cleft in the liganded LBDs, is essential for the NRIF3-receptor interaction (Fig. 3).
Although full-length NRIF3 physically and functionally interacts with TR and RXR but not with other nuclear receptors, LexA-RID1 does not appear to have the same specificity (31). Studies of the p160 family have suggested that residues flanking an LXXLL core play roles in specific interactions of a coactivator with different nuclear receptors (12, 36). While a similar mechanism may operate with the LXXIL motif of RID1, it nevertheless cannot account for the receptor specificity of NRIF3, as RID1 itself does not appear to be receptor specific (31). An interesting finding was that LexA-RID1 efficiently interacts with liganded LBDs, whereas the B42 fusion of RID1 fails to exhibit such interactions (Fig. 4). Although other potential explanations exist, this observation led us to consider the possibility that a stable NRIF3-receptor interaction might require the participation of two copies of RIDs. One model is that two copies of RID1 are provided through an NRIF3 dimer. In exploring this possibility, we mapped and characterized a dimerization surface in the central region of NRIF3 (Fig. 5 and 6). However, a subsequent study indicated that dimerization of NRIF3 is not required for receptor interactions (Fig. 7).
The results of Fig. 7 and our previous studies suggest an alternative model where a high-affinity NRIF3-receptor interaction involves and requires both RID1 and RID2. This model is supported by several experimental findings. First, while RID1 is essential for receptor interactions, RID2 allows for an increase in the association of NRIF3 with TR or RXR (31). Second, monovalent versions of RID1 [such as the B42-RID1 fusion or the B42-NRIF3(112–177) fusion] fail to interact with receptor LBDs (Fig. 4 and 8). Third, dimerization of NRIF3 (a potentially alternative form of bivalency) is dispensable for receptor interactions (Fig. 7).
Further studies suggest that proper spacing between RID1 and RID2 is important for productive NRIF3-receptor interactions, since the two internal deletions of NRIF3 (which remove residues 87 to 111 and residues 112 to 161, respectively) result in reduced interactions with liganded TR or RXR LBDs (Fig. 8). Interestingly, these deletions were found to change the specificity profile of the resulting mutant NRIF3s, due to their differential effects with respect to TR and RXR LBDs. Wild-type NRIF3 selectively interacts with TR and RXR but shows a preference for RXR (Fig. 8). This preference is lost with NRIF3(Δ87–111), which now exhibits similar levels of interactions with TR and RXR. In contrast, NRIF3(Δ112–161) no longer interacts with TR but retains a significant interaction with RXR and thus is “specific” for RXR (Fig. 8). These results suggest that the spacing between RID1 and RID2 plays a critical role in determining the interaction (or lack of) with receptor LBDs and thus influences the receptor specificity of NRIF3.
The finding that the expression of monovalent forms of RID1 or RID2 is not sufficient for receptor interactions suggests that a productive association of NRIF3 with liganded TR or RXR involves the cooperative effect of both RID1 and RID2. Interestingly, the crystal structure of liganded PPARγ complexed with a region of SRC-1 supports the notion that a receptor dimer is simultaneously contacted by two LXXLL boxes in the coactivator (40). Moreover, studies with SRC-1/NCoA-1 and TRAP220 have also suggested that proper spacing between the LXXLL boxes contained within the coactivator is important for coactivator-receptor interactions (36, 49). These results, along with our findings, support a model for the receptor specificity of NRIF3 where a single NRIF3 molecule employs both RID1 and RID2 to simultaneously and cooperatively contact the LBDs of certain receptor dimers. The ability or inability to make cooperative contacts would be determined by (i) the spatial orientation of the two hydrophobic clefts on receptor LBDs (determined by the conformation of the receptor dimer) and (ii) the spatial alignment of the two RIDs (determined by the conformation of NRIF3 and/or the spacing between RID1 and RID2). In this model, the specificity of a NRIF3-receptor interaction results from the proper spatial alignment of RID1, RID2, and hydrophobic clefts of the receptor LBDs, which is presumably only satisfied for nuclear hormone receptors in the case of liganded TR and RXR.
Repression mediated by the NRIF3 family is cell type dependent.
Although we initially observed the transrepression function of Gal4 fusions of the NRIF3 family in transfected HeLa cells, the relative magnitude of repression is only moderate (Fig. 10). In contrast, these Gal4 fusion proteins were found to be potent repressors in GH4C1 cells (Fig. 12). The precise mechanism underlying such an apparent cell type specificity in repression by the NRIF3 family is unclear. A previous study with the orphan nuclear receptor Rev-erb showed that it can elicit different levels of repression in different cell types depending on the presence or absence of the corepressor N-CoR (63). Thus, one potential explanation for the cell type specificity in repression by the NRIF3 family would be that they mediate repression through the recruitment of an additional corepressor(s), whose abundance or availability or activity may vary in HeLa and GH4C1 cells. Although we cannot exclude this possibility, our finding that transrepression by Gal4 fusions of the NRIF3 family is not relieved by cotransfection of the corresponding wild-type proteins argues against this model.
An alternative possibility is that the repression by the NRIF3 family is mediated through the direct contact with a basal promoter factor or a member of the basal machinery. For example, basal factors TBP and TFIIB have been suggested to be direct targets for certain transcriptional repressors or corepressors (15, 60). We tested TBP and TFIIB in cotransfection experiments and found that they showed little effect on NRIF3 family-mediated repression (data not shown). Thus, TFIIB and TBP are unlikely to be the target of NRIF3 family-mediated repression. By using nested deletion analysis, we mapped an essential transrepression domain (termed RepD1) of NRIF3 to a short region spanning residues 20 to 50. RepD1 shares no homology with known domains in the database. A computer-based secondary structure analysis suggests that RepD1 is composed of two short β-strands flanked by unstructured linker regions. Since RepD1 is relatively small in size, a future two-hybrid screen with RepD1 as the bait may be a useful approach in identifying targets of NRIF3 family-mediated repression.
Interestingly, a motif search with a Web-based bioinformatics tool predicted a potential phosphorylation site (Ser28) in RepD1. Substitution of Ser28 to Ala was found to abolish the repression function of RepD1 (Fig. 14), suggesting that phosphorylation at Ser28 is essential for RepD1 function in vivo. This result also raises the possibility that cellular signaling may regulate the function of NRIF3 and its isoform through modulating RepD1 function by events such as phosphorylation or dephosphorylation and thus may account for (some of) the cell type specificity of NRIF3 family-mediated repression. Future studies are needed to verify the suggested Ser28 phosphorylation in vivo, as well as to define the responsible kinase(s) and cellular pathway(s) if applicable.
Potential dual regulatory roles of NRIF3 and its isoforms.
NRIF3 represents an unusual example of a coregulator that harbors both a transactivation and a transrepression domain. Although AD1 coresides in the same region with RID1 (Fig. 1), transactivation by AD1 appears to be independent of receptor binding. We previously showed that, in transfected HeLa cells, NRIF3 enhances wild-type TR- or RXR-mediated transactivation of reporters controlled by their cognate hormone response elements. Thus, it is intriguing that NRIF3 also contains a transrepression domain (RepD1) (Fig. 1, 13, and 14). In the context of Gal4-NRIF3, RepD1 appears to be dominant over AD1, since Gal4-NRIF3 was found to repress transcription in both HeLa and GH4C1 cells.
A precedent of a coregulator with such a domain organization is NSD1, a 284-kDa protein that interacts with nuclear receptors and contains separate activation and repression domains (26). Although the role of NSD1 in nuclear receptor function remains to be defined, the possibility was raised that NSD1 may act as a bifunctional transcriptional intermediary factor (26). The functional significance of using both AD1 and RepD1 in a single NRIF3 molecule is currently unclear. One possibility is that such a domain organization provides regulatory flexibility, which would enable NRIF3 to differentially coactivate (e.g., with liganded TR or RXR) or corepress (e.g., with other transcription factors), depending on the nature of the NRIF3-interacting factor(s) and/or specific promoter or cellular context. In this regard, phosphorylation or dephosphorylation may serve as a molecular mechanism to modulate or even switch the regulatory property of NRIF3. Since NRIF3 enhances TR- or RXR-mediated transactivation, it is likely that when DNA-bound wild-type TR or RXR associates with NRIF3, the activation function of NRIF3 dominates over its repression activity. Nevertheless, it remains to be determined whether and how AD1 and/or RepD1 might contribute to NRIF3 coactivation of nuclear receptor activity. Future study of NRIF3-associated factors may provide clues to the functional mechanisms of AD1 and RepD1, as well as facilitate more-detailed understanding of NRIF3-mediated coregulation.
During the course of studying the receptor specificity of NRIF3, we identified a dimerization domain in NRIF3 (Fig. 1), which is predicted to form a coiled-coil structure by computer analysis and contains a putative leucine zipper-like motif (Fig. 1 and 6A). Interestingly, mutations in the leucine zipper-like motif diminish the NRIF3-NRIF3 interaction but do not affect the NRIF3-receptor interaction (Fig. 6 and 7) or NRIF3 enhancement of TR-mediated transactivation in HeLa cells (data not shown). In addition, the coiled-coil domain is not required for the repression activity found in Gal4-NRIF3 (Fig. 11 and 12). Preliminary yeast two-hybrid screens for factors interacting with NRIF3 identified a novel leucine zipper protein with homology to several transcription factors (Li et al., unpublished data). Thus, an intriguing possibility is that NRIF3 may also function as a coregulator for other transcription factors (in addition to TR and RXR), with its coiled-coil domain serving as an interacting surface for such factors.
Since the RID1 region of NRIF3 is missing in EnS and EnL, these two isoforms lack the AD1 activity found in RID1 (Fig. 1). Nevertheless, they share with NRIF3 the coiled-coil (dimerization) domain, as well as the transrepression domain RepD1 (Fig. 1). Consequently, EnS and EnL are capable of interacting with NRIF3 in a yeast two-hybrid assay (Fig. 6 and data not shown) and Gal4 fusions of these two proteins mediate repression (Fig. 10 and 12). These properties of EnS and EnL are consistent with their potential role(s) as transcriptional coregulators (or, more likely, corepressors). Taken together, our results support the notion that NRIF3, EnS, and EnL constitute a new family of coregulators that, collectively, may play dual roles in mediating both positive and negative regulation of gene expression.
ACKNOWLEDGMENTS
We thank Sanford Shattil for plasmids expressing GFP-EnL and GFP-EnS, Richard Heyman for providing the retinoids, Gordon Fishell for help with fluorescence microscopy, and Fred Stanley for advice on graphic preparations.
This research was supported by NIH grant DK16636 (to H.H.S.) and NRSA postdoctoral fellowship award DK09581 (to D.L.). H.H.S is a member of the NYUMC Cancer Center (CA16087).
REFERENCES
- 1.Anzick S L, Kononen J, Walker R L, Azorsa D O, Tanner M M, Guan X Y, Sauter G, Kallioniemi O P, Trent J M, Meltzer P S. AIB1, a steroid receptor coactivator amplified in breast and ovarian cancer. Science. 1997;277:965–968. doi: 10.1126/science.277.5328.965. [DOI] [PubMed] [Google Scholar]
- 2.Baniahmad A, Kohne C A, Renkawitz R. A transferable silencing domain is present in the thyroid hormone receptor, in the v-erbA oncogene product and in the retinoic acid receptor. EMBO J. 1992;11:1015–1023. doi: 10.1002/j.1460-2075.1992.tb05140.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barettino D, Ruiz M D M V, Stunnenberg H G. Characterization of the ligand-dependent transactivation domain of thyroid hormone receptor. EMBO J. 1994;13:3039–3049. doi: 10.1002/j.1460-2075.1994.tb06603.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blanco J C G, Minucci S, Lu J, Yang X J, Walker K K, Chen H, Evans R M, Nakatani V, Ozato K. The histone acetylase PCAF is a nuclear receptor coactivator. Genes Dev. 1998;12:1638–1651. doi: 10.1101/gad.12.11.1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carson-Jurica M A, Schrader W T, O'Malley B W. Steroid receptor family: structure and functions. Endocrine Rev. 1990;11:201–218. doi: 10.1210/edrv-11-2-201. [DOI] [PubMed] [Google Scholar]
- 6.Casanova J, Helmer E, Selmi-Ruby S, Qi J-S, Au-Fliegner M, Desai-Yajnik V, Koudinova N, Yarm F, Raaka B M, Samuels H H. Functional evidence for ligand-dependent dissociation of thyroid hormone and retinoic acid receptors from an inhibitory cellular factor. Mol Cell Biol. 1994;14:5756–5765. doi: 10.1128/mcb.14.9.5756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chakravarti D, LaMorte V J, Nelson M C, Nakajima T, Schulman I G, Juguilon H, Montminy M, Evans R M. Role of CBP/P300 in nuclear receptor signalling. Nature. 1996;383:99–103. doi: 10.1038/383099a0. [DOI] [PubMed] [Google Scholar]
- 8.Chang C Y, Norris J D, Gron H, Paige L A, Hamilton P T, Kenan D J, Fowlkes D, McDonnell D P. Dissection of the LXXLL nuclear receptor-coactivator interaction motif by using combinatorial peptide libraries: discovery of peptide antagonists of estrogen receptors α and β. Mol Cell Biol. 1999;19:8226–8239. doi: 10.1128/mcb.19.12.8226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen H, Lin R J, Schiltz R L, Chakravarti D, Nash A, Nagy L, Privalsky M L, Nakatani Y, Evans R M. Nuclear receptor coactivator ACTR is a novel histone acetyltransferase and forms a multimeric activation complex with P/CAF and CBP/p300. Cell. 1997;90:569–580. doi: 10.1016/s0092-8674(00)80516-4. [DOI] [PubMed] [Google Scholar]
- 10.Chen J D, Evans R M. A transcriptional co-repressor that interacts with nuclear hormone receptors. Nature. 1995;377:454–457. doi: 10.1038/377454a0. [DOI] [PubMed] [Google Scholar]
- 11.Damm K, Thompson C C, Evans R M. Protein encoded by v-erbA functions as a thyroid hormone receptor antagonist. Nature. 1989;339:593–597. doi: 10.1038/339593a0. [DOI] [PubMed] [Google Scholar]
- 12.Darimont B D, Wagner R L, Apriletti J W, Stallcup M R, Kushner P J, Baxter D, Fletterick R J, Yamamoto K R. Structure and specificity of nuclear receptor-coactivator interactions. Genes Dev. 1998;12:3343–3356. doi: 10.1101/gad.12.21.3343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Durand B, Saunders M, Gausdon C, Roy B, Losson R, Chambon P. Activation function 2 (AF-2) of retinoic acid receptor and 9-cis retinoic acid receptor: presence of a conserved autonomous constitutive activating domain and influence of the nature of the response element on AF-2 activity. EMBO. 1994;13:5370–5382. doi: 10.1002/j.1460-2075.1994.tb06872.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Feng W, Ribeiro R C, Wagner R L, Nguyen H, Apriletti J W, Fletterick R J, Baxter J D, Kushner P J, West B L. Hormone-dependent coactivator binding to a hydrophobic cleft on nuclear receptors. Science. 1998;280:1747–1749. doi: 10.1126/science.280.5370.1747. [DOI] [PubMed] [Google Scholar]
- 15.Fondell J D, Brunel F, Hisatake K, Roeder R G. Unliganded thyroid hormone receptor alpha can target TATA-binding protein for transcriptional repression. Mol Cell Biol. 1996;16:281–287. doi: 10.1128/mcb.16.1.281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fondell J D, Ge H, Roeder R G. Ligand induction of a transcriptionally active thyroid hormone receptor coactivator complex. Proc Natl Acad Sci USA. 1996;93:8329–8333. doi: 10.1073/pnas.93.16.8329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Forman B M, Casanova J, Raaka B M, Ghysdael J, Samuels H H. Half-site spacing and orientation determines whether thyroid hormone and retinoic acid receptors and related factors bind to DNA response elements as monomers, homodimers, or heterodimers. Mol Endocrinol. 1992;6:429–442. doi: 10.1210/mend.6.3.1316541. [DOI] [PubMed] [Google Scholar]
- 18.Forman B M, Samuels H H. Interactions among a subfamily of nuclear hormone receptors: the regulatory zipper model. Mol Endocrinol. 1990;4:1293–1301. doi: 10.1210/mend-4-9-1293. [DOI] [PubMed] [Google Scholar]
- 19.Forman B M, Yang C-R, Au M, Casanova J, Ghysdael J, Samuels H H. A domain containing leucine zipper like motifs mediates novel in vivo interactions between the thyroid hormone and retinoic acid receptors. Mol Endocrinol. 1989;3:1610–1626. doi: 10.1210/mend-3-10-1610. [DOI] [PubMed] [Google Scholar]
- 20.Glass C K, Rosenfeld M G. The coregulator exchange in transcriptional functions of nuclear receptors. Genes Dev. 2000;14:121–141. [PubMed] [Google Scholar]
- 21.Gyuris J, Golemis E, Chertkov H, Brent R. Cdi1, a human G1 and S phase protein phosphatase that associates with Cdk2. Cell. 1993;75:791–803. doi: 10.1016/0092-8674(93)90498-f. [DOI] [PubMed] [Google Scholar]
- 22.Hanstein B, Eckner R, DiRenzo J, Halachmi S, Liu H, Searcy B, Kurokawa R, Brown M. p300 is a component of an estrogen receptor coactivator complex. Proc Natl Acad Sci USA. 1996;93:11540–11545. doi: 10.1073/pnas.93.21.11540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Heery D M, Kalkhoven E, Hoare S, Parker M G. A signature motif in transcriptional co-activators mediates binding to nuclear receptors. Nature. 1997;387:733–736. doi: 10.1038/42750. [DOI] [PubMed] [Google Scholar]
- 24.Hong H, Kohli K, Garabedian M, Stallcup M R. GRIP1, a transcriptional coactivator for the AF-2 transactivation domain of steroid, thyroid, retinoid, and vitamin D receptors. Mol Cell Biol. 1997;17:2735–2744. doi: 10.1128/mcb.17.5.2735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Horlein A J, Naar A M, Heinzel T, Torchia J, Gloss B, Kurokawa R, Ryan A, Kamil Y, Soderstrom M, Glass C K, Rosenfeld M G. Ligand-independent repression by the thyroid hormone receptor mediated by a nuclear receptor co-repressor. Nature. 1995;377:397–404. doi: 10.1038/377397a0. [DOI] [PubMed] [Google Scholar]
- 26.Huang N, vom Baur E, Garnier J M, Lerouge T, Vonesch J L, Lutz Y, Chambon P, Losson R. Two distinct nuclear receptor interaction domains in NSD1, a novel SET protein that exhibits characteristics of both corepressors and coactivators. EMBO J. 1998;17:3398–3412. doi: 10.1093/emboj/17.12.3398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ito M, Yuan C X, Malik S, Gu W, Fondell J D, Yamamura S, Fu Z Y, Zhang X, Qin J, Roeder R G. Identity between TRAP and SMCC complexes indicates novel pathways for the function of nuclear receptors and diverse mammalian activators. Mol Cell. 1999;3:361–370. doi: 10.1016/s1097-2765(00)80463-3. [DOI] [PubMed] [Google Scholar]
- 28.Kamei Y, Xu L, Heinzel T, Torchia J, Kurokawa R, Gloss B, Lin S-C, Heyman R A, Rose D W, Glass C K, Rosenfeld M G. A CBP integrator complex mediates transcriptional activation and AP-1 inhibition by nuclear receptors. Cell. 1996;85:403–414. doi: 10.1016/s0092-8674(00)81118-6. [DOI] [PubMed] [Google Scholar]
- 29.Kashiwagi H, Schwartz M A, Eigenthaler M, Davis K A, Ginsberg M H, Shattil S J. Affinity modulation of platelet integrin αIIbβ3 by β3-endonexin, a selective binding partner of the β3 integrin cytoplasmic tail. J Cell Biol. 1997;137:1433–1443. doi: 10.1083/jcb.137.6.1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kim B, Little J W. Dimerization of a specific DNA-binding protein on the DNA. Science. 1992;255:203–206. doi: 10.1126/science.1553548. [DOI] [PubMed] [Google Scholar]
- 31.Li D, Desai-Yajnik V, Lo E, Schapira M, Abagyan R, Samuels H H. NRIF3 is a novel coactivator mediating functional specificity of nuclear hormone receptors. Mol Cell Biol. 1999;19:7191–7202. doi: 10.1128/mcb.19.10.7191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li H, Gomes P J, Chen J D. RAC3, a steroid/nuclear receptor-associated coactivator that is related to SRC-1 and TIF2. Proc Natl Acad Sci USA. 1997;94:8479–8484. doi: 10.1073/pnas.94.16.8479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mahajan M A, Samuels H H. A new family of nuclear receptor coregulators that integrate nuclear receptor signaling through CREB-binding protein. Mol Cell Biol. 2000;20:5048–5063. doi: 10.1128/mcb.20.14.5048-5063.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mangelsdorf D J, Evans R M. The RXR heterodimers and orphan receptors. Cell. 1995;83:841–850. doi: 10.1016/0092-8674(95)90200-7. [DOI] [PubMed] [Google Scholar]
- 35.Mangelsdorf D J, Thummel C, Beato M, Herrlich P, Schutz G, Umesono K, Blumberg B, Kastner P, Mark M, Chambon P, Evans R M. The nuclear receptor superfamily: the second decade. Cell. 1995;83:835–839. doi: 10.1016/0092-8674(95)90199-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McInerney E M, Rose D W, Flynn S E, Westin S, Mullen T M, Krones A, Inostroza J, Torchia J, Nolte R T, Assa-Munt N, Milburn M V, Glass C K, Rosenfeld M G. Determinants of coactivator LXXLL motif specificity in nuclear receptor transcriptional activation. Genes Dev. 1998;12:3357–3368. doi: 10.1101/gad.12.21.3357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McKenna N J, Lanz R B, O'Malley B W. Nuclear receptor coregulators: cellular and molecular biology. Endocrinol Rev. 1999;20:321–344. doi: 10.1210/edrv.20.3.0366. [DOI] [PubMed] [Google Scholar]
- 38.Mohana-Borges R, Pacheco A B, Sousa F J, Foguel D, Almeida D F, Silva J L. LexA repressor forms stable dimers in solution. The role of specific DNA in tightening protein-protein interactions. J Biol Chem. 2000;275:4708–4712. doi: 10.1074/jbc.275.7.4708. [DOI] [PubMed] [Google Scholar]
- 39.Nagpal S, Friant S, Nakshatri H, Chambon P. RARs and RXRs: evidence for two autonomous transactivation functions (AF-1 and AF-2) and heterodimerization in vivo. EMBO J. 1993;12:2349–2360. doi: 10.1002/j.1460-2075.1993.tb05889.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nolte R T, Wisely G B, Westin S, Cobb J E, Lambert M H, Kurokawa R, Rosenfeld M G, Willson T M, Glass C K, Milburn M V. Ligand binding and co-activator assembly of the peroxisome proliferator-activated receptor-γ. Nature. 1998;395:137–143. doi: 10.1038/25931. [DOI] [PubMed] [Google Scholar]
- 41.Onate S A, Tsai S Y, Tsai M-J, O'Malley B W. Sequence and characterization of a coactivator of the steroid hormone receptor superfamily. Science. 1995;270:1354–1357. doi: 10.1126/science.270.5240.1354. [DOI] [PubMed] [Google Scholar]
- 42.Puigserver P, Wu Z, Park C W, Graves R, Wright M, Spiegelman B M. A cold-inducible coactivator of nuclear receptors linked to adaptive thermogenesis. Cell. 1998;92:829–839. doi: 10.1016/s0092-8674(00)81410-5. [DOI] [PubMed] [Google Scholar]
- 43.Qi J-S, Desai-Yajnik V, Greene M E, Raaka B M, Samuels H H. The ligand binding domains of the thyroid hormone/retinoid receptor gene subfamily function in vivo to mediate heterodimerization, gene silencing, and transactivation. Mol Cell Biol. 1995;15:1817–1825. doi: 10.1128/mcb.15.3.1817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Raaka B M, Finnerty M, Samuels H H. The glucocorticoid antagonist 17α-methyltestosterone binds to the 10 S glucocorticoid receptor and blocks agonist-mediated dissociation of the 10 S oligomer to the 4 S deoxyribonucleic acid-binding subunit. Mol Endocrinol. 1989;3:332–341. doi: 10.1210/mend-3-2-332. [DOI] [PubMed] [Google Scholar]
- 45.Raaka B M, Finnerty M, Sun E, Samuels H H. Effects of molybdate on steroid receptors in intact GH1 cells. Evidence for dissociation of an intracellular 10 S receptor oligomer prior to nuclear accumulation. J Biol Chem. 1985;260:14009–14015. [PubMed] [Google Scholar]
- 46.Raaka B M, Samuels H H. The glucocorticoid receptor in GH1 cells. Evidence from dense amino acid labeling and whole-cell studies for an equilibrium model explaining the influence of hormone on the intracellular distribution of receptor. J Biol Chem. 1983;258:417–425. [PubMed] [Google Scholar]
- 47.Rachez C, Lemon B D, Suldan Z, Bromleigh V, Gamble M, Naar A M, Erdjument-Bromage H, Tempst P, Freedman L P. Ligand-dependent transcription activation by nuclear receptors requires the DRIP complex. Nature. 1999;398:824–828. doi: 10.1038/19783. [DOI] [PubMed] [Google Scholar]
- 48.Rachez C, Suldan Z, Ward J, Chang C P, Burakov D, Erdjument-Bromage H, Tempst P, Freedman L P. A novel protein complex that interacts with the vitamin D3 receptor in a ligand-dependent manner and enhances VDR transactivation in a cell-free system. Genes Dev. 1998;12:1787–1800. doi: 10.1101/gad.12.12.1787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ren Y, Behre E, Ren Z, Zhang J, Wang Q, Fondell J D. Specific structural motifs determine TRAP220 interactions with nuclear hormone receptors. Mol Cell Biol. 2000;20:5433–5446. doi: 10.1128/mcb.20.15.5433-5446.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Samuels H H, Forman B M, Horowitz Z D, Ye Z-S. Regulation of gene expression by thyroid hormone. J Clin Investig. 1988;81:957–967. doi: 10.1172/JCI113449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Samuels H H, Perlman A J, Raaka B M, Stanley F. Organization of the thyroid hormone receptor in chromatin. Recent Prog Horm Res. 1982;38:557–599. doi: 10.1016/b978-0-12-571138-8.50018-4. [DOI] [PubMed] [Google Scholar]
- 52.Schultz J, Copley R R, Doerks T, Ponting C P, Bork P. SMART: a web-based tool for the study of genetically mobile domains. Nucleic Acids Res. 2000;28:231–234. doi: 10.1093/nar/28.1.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Selmi-Ruby S, Casanova J, Malhotra S, Roussett B, Raaka B M, Samuels H H. Role of the conserved C-terminal region of thyroid hormone receptor-α in ligand-dependent transcriptional activation. Mol Cell Endocrinol. 1998;138:105–114. doi: 10.1016/s0303-7207(98)00016-1. [DOI] [PubMed] [Google Scholar]
- 54.Shattil S J, O'Toole T, Eigenthaler M, Thon V, Williams M, Babior B M, Ginsberg M H. β3-Endonexin, a novel polypeptide that interacts specifically with the cytoplasmic tail of the integrin β3 subunit. J Cell Biol. 1995;131:807–816. doi: 10.1083/jcb.131.3.807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Takeshita A, Cardona G R, Koibuchi N, Suen C S, Chin W W. TRAM-1, A novel 160-kDa thyroid hormone receptor activator molecule, exhibits distinct properties from steroid receptor coactivator-1. J Biol Chem. 1997;272:27629–27634. doi: 10.1074/jbc.272.44.27629. [DOI] [PubMed] [Google Scholar]
- 56.Tora L, White J, Brou C, Tasset D, Webster N, Scheer E, Chambon P. The human estrogen receptor has two independent nonacidic transcriptional activation functions. Cell. 1989;59:477–487. doi: 10.1016/0092-8674(89)90031-7. [DOI] [PubMed] [Google Scholar]
- 57.Torchia J, Rose D W, Inostroza J, Kamei Y, Westin S, Glass C K, Rosenfeld M G. The transcriptional co-activator p/CIP binds CBP and mediates nuclear-receptor function. Nature. 1997;387:677–684. doi: 10.1038/42652. [DOI] [PubMed] [Google Scholar]
- 58.Tsai M J, O'Malley B W. Molecular mechanisms of action of steroid/thyroid receptor superfamily members. Annu Rev Biochem. 1994;63:451–486. doi: 10.1146/annurev.bi.63.070194.002315. [DOI] [PubMed] [Google Scholar]
- 59.Voegel J J, Heine M J S, Zechel C, Chambon P, Gronemeyer H. TIF2, a 160 kDa transcriptional mediator for the ligand-dependent activation function AF-2 of nuclear receptors. EMBO J. 1996;15:3667–3675. [PMC free article] [PubMed] [Google Scholar]
- 60.Wong C W, Privalsky M L. Transcriptional repression by the SMRT-mSin3 corepressor: multiple interactions, multiple mechanisms, and a potential role for TFIIB. Mol Cell Biol. 1998;18:5500–5510. doi: 10.1128/mcb.18.9.5500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wurtz J M, Bourguet W, Renaud J P, Vivat V, Chambon P, Moras D, Gronemeyer H. A canonical structure for the ligand-binding domain of nuclear receptors. Nat Struct Biol. 1996;3:87–94. doi: 10.1038/nsb0196-87. [DOI] [PubMed] [Google Scholar]
- 62.Yaffe M B, Leparc G G, Lai J, Obata T, Volinia S, Cantley L C. A motif-based profile scanning approach for genome-wide prediction of signaling pathways. Nat Biotechnol. 2001;19:348–353. doi: 10.1038/86737. [DOI] [PubMed] [Google Scholar]
- 63.Zhang J, Guenther M G, Carthew R W, Lazar M A. Proteasomal regulation of nuclear receptor corepressor-mediated repression. Genes Dev. 1998;12:1775–1780. doi: 10.1101/gad.12.12.1775. [DOI] [PMC free article] [PubMed] [Google Scholar]