Abstract
Crop plants are vulnerable to various biotic and abiotic stresses, whereas plants tend to retain their physiological mechanisms by evolving cellular regulation. To mitigate the adverse effects of abiotic stresses, many defense mechanisms are induced in plants. One of these mechanisms is the mitogen-activated protein kinase (MAPK) cascade, a signaling pathway used in the transduction of extracellular stimuli into intercellular responses. This stress signaling pathway is activated by a series of responses involving MAPKKKs→MAPKKs→MAPKs, consisting of interacting proteins, and their functions depend on the collaboration and activation of one another by phosphorylation. These proteins are key regulators of MAPK in various crop plants under abiotic stress conditions and also related to hormonal responses. It is revealed that in response to stress signaling, MAPKs are characterized as multigenic families and elaborate the specific stimuli transformation as well as the antioxidant regulation system. This pathway is directed by the framework of proteins and stopping domains confer the related associates with unique structure and functions. Early studies of plant MAPKs focused on their functions in model plants. Based on the results of whole-genome sequencing, many MAPKs have been identified in plants, such as Arbodiposis, tomato, potato, alfalfa, poplar, rice, wheat, maize, and apple. In this review, we summarized the recent work on MAPK response to abiotic stress and the classification of MAPK cascade in crop plants. Moreover, we highlighted the modern research methodologies such as transcriptomics, proteomics, CRISPR/Cas technology, and epigenetic studies, which proposed, identified, and characterized the novel genes associated with MAPKs and their role in plants under abiotic stress conditions. In-silico-based identification of novel MAPK genes also facilitates future research on MAPK cascade identification and function in crop plants under various stress conditions.
Keywords: abiotic stresses, climate change, plant physiology, signaling pathway, stress tolerance, transcription factor
1. Introduction
One of the sustainable development goals is to end world hunger, and feeding a growing population is a significant worldwide societal concern (Raza et al., 2021; Farooq et al., 2022; Rivero et al., 2022). Despite the world’s population doubling, the long-term drop in global undernourishment has been caused by a substantial rise in food availability since 1960 (Ritchie and Roser, 2020). Nevertheless, there are currently more than 820 million hungry people around the globe (FAO et al., 2019). Only 9% of the world’s agricultural land is suitable for growing crops, while the remaining 91% is subjected to abiotic stress, which frequently occurs in combination. Abiotic stresses cause losses in agricultural productivity of more than 50% (Raza, 2021; Rivero et al., 2022). Still, due to climate change and the overuse of natural resources, their severity and adverse effects are anticipated to increase significantly, which not only reduce crop production but also cause food insecurity in the near future (Minhas et al., 2017; Farooq et al., 2022; Rivero et al., 2022).
Due to industrialization and climate change in recent decades, plants are normally exposed to various abiotic stresses such as drought, salinity, extreme temperature ranges, nutrient deficiency, high heavy metal concentrations, and osmosis stress (Haider et al., 2021; Raza et al., 2021; Raza et al., 2022a; Raza et al., 2022b; Raza et al., 2022c; Raza et al., 2022d; Raza et al., 2022e). They cause a lot of damage to plants’ physiology and also reduce growth and development that ultimately minimizes the productivity of crop plants (Anjum et al., 2017). To ensure good crop growth and optimum productivity, stress tolerance mechanisms are imperative to be studied for combating abiotic stresses in crop plants (Baer-Nawrocka and Sadowski, 2019; Raza et al., 2022a; Raza et al., 2022b; Raza et al., 2022c). Understanding these stress tolerance mechanisms will enable the generation of more climate-smart and stress-tolerant lines, which will maintain stability in the growth and productivity of agricultural productivity. For this, it is necessary to understand the genetic basis of a plant’s interaction when encountering ecological stress. Several studies considering transcriptomics, genomics, proteomics, metabolomics, and genome editing via CRISPR/Cas technology provide a roadmap toward the acclimatization mechanism in plants and crops (Wang et al., 2019; Raza et al., 2022a; Raza et al., 2022b; Raza et al., 2022c; Raza et al., 2022d; Yaqoob et al., 2023). In-depth molecular studies aid us in developing varieties and cultivars through biotechnology, genetic engineering, and other advanced breeding methods to develop plants that could adapt to different abiotic stresses in a short time (Popescu et al., 2009; Raza et al., 2022a; Raza et al., 2022b; Yaqoob et al., 2023). Post-translational modification and signal transduction are mediated by a process called phosphorylation, which changes the expression of genes through transmission of protein signals. A serine/threonine-protein kinase family called mitogen-activated protein kinase (MAPK) is one of the widely studied gene families and contributes to plant productivity under fluctuating environmental conditions (Hamel et al., 2006).
In this regard, one of the major signal transduction pathways that transduce extracellular stimuli into intracellular responses related to stress is mitogen-activated protein kinase (Mathobo et al., 2017). The first MAPK encoding gene was cloned in the 1990s; to date, many MAPK genes have been identified and isolated from different plants (Zaıdi et al., 2010), which are activated under abiotic stresses like AtMPK4 and AtMPK6 in Arabidopsis and in rice (Oryza sativa) and OsMAPK5 and OsMAPK2 under drought stress (Nadarajah and Sidek, 2010). The abscisic acid (ABA) signaling pathway regulates plant growth and development under abiotic stress conditions, such as drought or high salinity (Cutler et al., 2010). Phosphorylation of two ABA-responsive transcription factors (ABF1 and ABF4) by AtCPK4, 11, and 32 suggested the role of kinases in regulating ABA signaling through these transcription factors under stress conditions (Choi et al., 2005). Using a yeast two-hybrid test, researchers looked at the interactions between 30 members of the MPK family, 9 CPKs, 8 PP2Cs, 5 SnRKs, and 8 PP2Cs in maize’s (Zea mays) MAPK signaling pathways. Moreover, three ZmCPKs connect with three distinct ZmSnRK members, whereas four ZmCPK members positively interact with 13 different ZmMPK members in various combinations. These four ZmCPK proteins originate from three distinct maize groupings. These physical connections between ZmCPKs, ZmSnRKs, and ZmMPKs revealed that these signaling pathways might interact directly with the defense mechanism in maize and have indirect effects. The current work might contribute to a better understanding of plant signal transduction (Khalid et al., 2019). Concerning the downregulation of ZmMPK5, due to ZmCPK11 silencing, the role of ZmCPK11 upstream of ZmMPK5 has been proposed (Ding et al., 2013).
MAPK gene families consist of a vast number of genes that are classified into four different groups: A, B, C, and D. Owing to the evolutionary divergence in different plants, these groups contain a different number of MAPK genes; some of them are listed in Figure 1 and Table 1 . This MAPK signaling cascade works like a chain reaction as mitogen-activated protein kinsase kinase (MAPKK) is activated by the upstream of the mitogen-activated kinase (MAPKKK), which, in turn, activates the mitogen-activated protein kinase (MAPK) (Wang et al., 2014; Mitula et al., 2015). This full chain of MAPKs is conserved in plants, which signifies the evolutionary perspective of MAPKs. One of the most important methods that is triggered by posttranslational modification of signal transduction is called phosphorylation (Wang et al., 2019). MAPKKK phosphorylates MAPKK on the conserved serine/threonine motifs (Rodriguez et al., 2010), which finally brings about the phosphorylation of TXY (T is threonine, Y is tyrosine, and X is any amino acid) in MAPKs (Taj et al., 2010).
Figure 1.
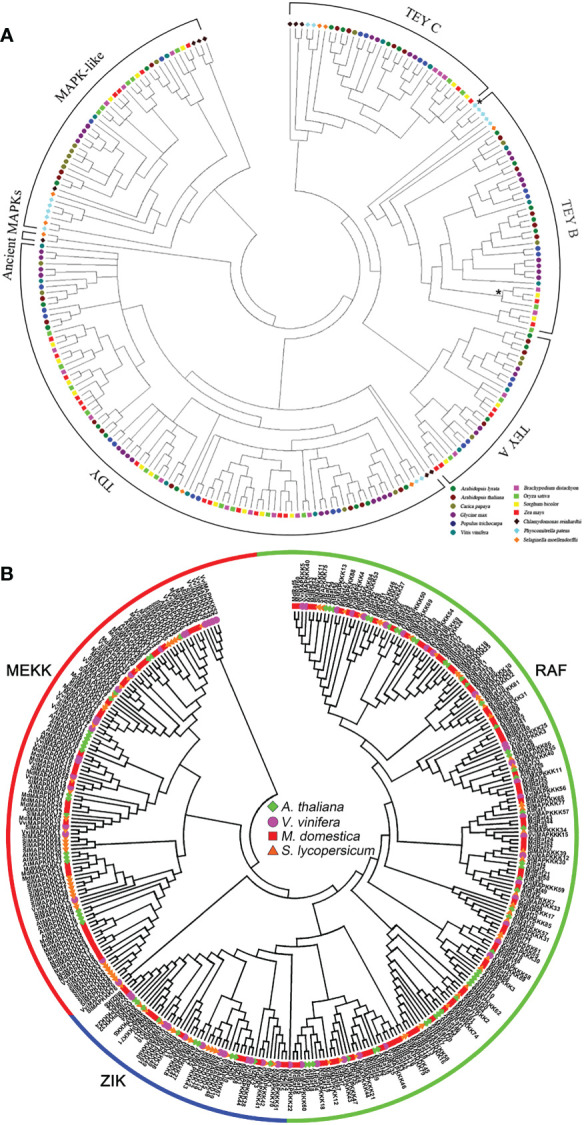
(A) Phylogenetic analysis of plant mitogen-activated protein kinases (MAPKs) and MAPK-likes in 13 plant species following Bayesian inference. Different colors indicate different species. The monocot clade and the PP sequence with the atypical MEY activation loop within the TEY-B clade are marked by asterisks. (B) Phylogenetic analysis of MAPKKKs in grapevine, apple, tomato, and Arabidopsis using the neighbor-joining (NJ) tree using MEGA-X with 1,000 bootstraps. Adapted from He et al. (2020) and Janitza et al. (2012), open-access articles distributed under the terms of the Creative Commons Attribution License (CC BY).
Table 1.
Identification of groups of MAPK gene families in different plant species.
| Plant species | Group A | Group B | Group C | Group D | Total | References |
| Arabidopsis | 3 | 5 | 4 | 8 | 20 | Cçakır and Kılıcçkaya (2015) |
| Rice | 2 | 1 | 2 | 10 | 15 | Reyna and Yang (2006) |
| Wheat | 11 | 13 | 22 | 63 | 109 | Lian et al. (2012) |
| Maize | 7 | 7 | 6 | 19 | 39 | Wang et al. (2010) |
| Cotton | 4 | 6 | 5 | 12 | 27 | Wang et al. (2016) |
| Poplar | 4 | 4 | 4 | 9 | 21 | Zhao et al. (2018) |
| Tomato | 3 | 4 | 2 | 7 | 16 | Kong et al. (2012) |
| Apple | 5 | 6 | 5 | 10 | 26 | Zhang et al. (2013) |
| Potato | 1 | 4 | 2 | 8 | 15 | Zaynab et al. (2021) |
Transcription factors and downstream kinases activate the cell after receiving extracellular signals from the activated MAPKs, which describe the variation of the cellular development factors (Zhang et al., 2018). The process of transmission and amplification of signals occurs in stepwise phosphorylation (Hamel et al., 2006). When plants are encountered by any abiotic stress or wounding, MAPK, as well as other hormones like ethylene, jasmonic acid, and salicylic acid, are activated. Studies also suggested that pathogen stimuli also cause the induction of MAPKs in various plants like alfalfa (Medicago sativa), rice (Orzya sativa), maize (Z. mays), and potato (Solanum tuberosum) (Andrasi et al., 2019). Under adverse environmental conditions like high temperature and water scarcity, MAPKs play a key role in signal transduction (Muhammad et al., 2019). Studies also have revealed that MAPK cascade is also activated under salt stress and freezing temperature (Teige et al., 2004). Shreds of evidence provide the signaling activation of MAPKs during early wounding in different plants such as Arbodiposis, apple (Malus hupehensis), poplar (Populous alba), and rice, as reported previously (Takahashi et al., 2011). Recently, many MAPK genes have been identified, which play a role under various stress conditions, but there is little knowledge on the molecular mechanisms of MAPK triggering and signaling ( Figure 1 ) (Mohanta et al., 2015). The available knowledge about the molecular characterization of the MAPK gene family under abiotic stress is summarized owing to need for more information about MAPK signaling and triggering in many crops. In this review, we will focus on the action of transcription factors, transcriptomic studies, and the molecular basis for understanding the biological, biochemical, and physiological processes of different plants under abiotic stresses. Furthermore, we summarized the classification of different MAPK gene families in response to abiotic stress in plants and molecular and cellular signaling pathways in plants for acclimatization under an adverse environment. After all, many phosphorylated MAPKs play a role in signal transmission. In short, incorporating MAPK gene families in plant breeding to produce stress-smart varieties is another aspect of research in MAPKs. This review may help to comprehend the ecological importance of MAPKs in plants to overcome the abiotic stresses for sustainable crop production and also provide new insights for breeders to incorporate MAPK gene families in plant breeding program to produce abiotic stress resistance varieties.
2. Mechanism of MAPKs in crop plants
Plants have acquired different acclimatization strategies under harsh environmental conditions over time via a number of molecular systems that consist of sensing, signaling, and expression under stress conditions by stress-responsive genes (Mahmood et al., 2020). MAPKs are one of the tools that regulate growth and development, cell division, proliferation, apoptosis, hormonal response, and other stress responses by an extremely conserved network. It consists of the three protein kinases MAPK, MAPKK, and MAPKKK. Sequential phosphorylation activates these cascades such as activation of MAPKKK phosphorylates the S/T-X (S/T is a serine/threonine and X is an arbitrary amino acid) conserved motif that is present in the activation loop of MAPKK (Rodriguez et al., 2010); then, the activation of MAPKK phosphorylates the T-X-Y (T is threonine, Y is tyrosine, and X is any amino acid) in the variant motif present in the activation loop of MAPK (Taj et al., 2010). Then, these MAPK cascades send the message in a well-designed manner to the primary genes responsive to tolerance and then to secondary genes, which induce tolerance in crops under stress conditions (Hamel et al., 2006). The MAPK cascades are also phosphorylated by the following activities such as regulation of microtubule proteins, cytoskeletal activities, and other transcription factors that help in numerous responses and phospholipases (Danquah et al., 2014). The transmission of extracellular stimuli into cells and downstream kinases is also activated by MAPK cascades (Xu et al., 2010). The main channel of signal transduction and post-transcriptional modification is carried out by phosphorylation. The process of phosphorylation is a post-translational modification process that alters the expression of downstream genes as well as diffuses and intensifies the external signals (Colcombet and Hirt, 2008).
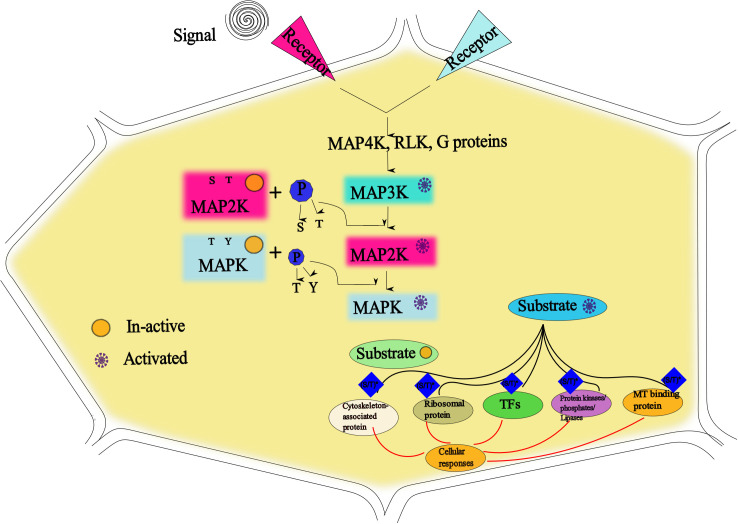
One of the wide-ranging gene families is the serine/threonine protein kinase family of MAPKs, which are protein kinases and are enzymatic in nature and mediate phosphorylation (Rodriguez et al., 2010; Xu et al., 2010). Conventionally, MAPK cascades transmit signals downstream by the activation of the stimulated receptors of the cell membrane (Wang et al., 2014). By following this mechanism, MAPKs, after activation, control the expression of many genes and proteins by the phosphorylation of transcription factors. This mechanism in plants plays a key role in cell differentiation, cell growth, development, and hormonal movement, and in response to various biotic and abiotic stresses (Komis et al., 2018). In crop species, the MAPK gene families are recognized owing to the many tolerant genes in abiotic stress response, evident from genetic studies of MAPK activity in recent years (SŠamajovaí et al., 2013). The MAPK mechanism is depicted in Figure 2 , where extracellular signals are detected by a plasma membrane and by sensors in cytoplasm, which act like a chain reaction, leading to the activation of MAPKKK and, subsequently, MAPKK and, as a result, phosphorylation of MAPK, which helps in the phosphorylation of proteins, enzymes, and post-transcriptional factors in the nucleus. These factors finally send a message to stress-tolerant responsive genes (Bigeard and Hirt, 2018; Singh et al., 2019). By understanding this novel mechanism, MAPK genes can be identified in different crop plants, which may help in maintaining plant growth under various abiotic stress conditions, but still, there is a limitation due to the activation of other intricate stress responsive mechanisms.
Figure 2.
The receptors after receiving the signals activate the specific mitogen-activated protein kinase (MAPK) proteins that are inactive before. Subsequently, MAP4K activates other MAPKs (including MAP2K and MAP3K) by phosphorylating the ST (S/T is serine/threonine) and TXY (T is threonine, Y is tyrosine, and X is any amino acid) motif in MAPKs (Rodriguez et al., 2010; Taj et al., 2010). As a result, MAPKs activate the transcription factors (TFs), enzymes, and other downstream kinases that transmit extracellular environmental signals to the cells that play a role in growth, development, stress response, and other physiological and biochemical processes (Zhang et al., 2018).
3. MAPK complexity in abiotic stress signaling interaction
Abiotic stresses, such as drought, cold, heat, salinity, and metals, are closely associated with an adverse influence on the physiology of plants (Krasensky and Jonak, 2012; Raza, 2021; Raza et al., 2021; Raza et al., 2022a; Raza et al., 2022b; Raza et al., 2022c; Raza et al., 2022d). For example, when the temperature rises, water deficiency, salinity, and osmotic pressure in the tissues of plants may be encountered (Bita and Gerats, 2013). Likewise, due to salinity and water scarcity, plants suffer from osmotic stress, and signaling molecules are activated in response to stress conditions (Rengasamy, 2006). The mechanism of expression of resistance genes in various plants like maize (Z. mays), rice (O. sativa), potato (S. tuberosum), and Arabidopsis is regulated under various abiotic stress conditions, which are activated by messenger-mediated signal transduction (Zhu, 2016; Zandalinas et al., 2019). The large numbers of stress-responsive genes like MAPK genes or MAPK cascades participate in a variety of abiotic stresses for the protection and survival of plants (Ji et al., 2017; Yanagawa et al., 2017). MAPK gene families have multiple functions in plants, such as growth and development, immune defense system, and response to biotic and abiotic stresses. Certain studies also point toward the role of MAPK cascade in the regulation of cell death and defense responses (Qiu et al., 2008; Zhou et al., 2017).
Recently, it was also reported that MAPK cascade has a dual function in plant immunity: the basal resistance is regulated positively and immunity regulation is facilitated negatively by kinase protein (Zhang et al., 2012). One case reported in Arabidopsis also provides evidence that MAPKs play a vital role in pathogen signaling (Doczi et al., 2007). Furthermore, MAPK cascade mediates Ca2+ reactive oxygen species (ROS) by signaling during early wounding (Pitzschke and Hirt, 2009). MAPK pathways regulate the synthesis of ROS, and certain genes of MAPKs are induced by ROS generation (Colcombet and Hirt, 2008). H2O2 also plays a key role in activating orthologs of MAPKs in many crops like tobacco (Nicotiana tobacum) and Arabidopsis (Xing et al., 2008). MAPK cascade in Arabidopsis regulates H2O2 metabolism (Nakagami et al., 2006), and in tobacco, it is an important component in ROS metabolism (Ning et al., 2010). In maize, H2O2 induces transcription and expression of MAPK cascade, whereas in tobacco, it protects against ROS-mediated injury under osmotic stress (Zhang et al., 2010; Kong et al., 2011). This is due to the different expression patterns of MAPK gene families under different stresses in different plant species. In maize, the MAPK network works as an early signaling response by regulating the production of ROS in plants subjected to drought stress (Gao and Xiang, 2008).
In later developmental stages, one MAPKKK gene called MAPKKK20 in Arabidopsis improves salt tolerance (Jammes et al., 2009). Similarly, water loss by transpiration in the Arabidopsis double mutant named MPK9/MPK12 is less as compared to its wild type. Moreover, it was also documented that ABA regulates the physiological response under abiotic stress (Tajdel et al., 2016). In Arabidopsis, one MAPKKK gene named MAPKKK18 shows reduced stomatal opening under normal conditions in mutant plants (Mitula et al., 2015). Furthermore, this mutant also shows ABA-induced stomatal closure, which shows that MAPKKK18 is directly interacting with ABA components, which plays a key role in signal modulation as SnRK2-6 kinase and PP2C phosphatase ABI1 (Zhang et al., 2006). An increment of ABA in plants is an indication of oxidative stress. An abnormal level of ROS causes the oxidation of free radicals such as hydrogen peroxides, which is called oxidative stress and leads to the injury of cells and tissues. The MAPK gene families reinfoirces the ABA-induced antioxidant defense system by decreasing the ROS production in many crops like maize (Shi et al., 2011).
Under physiological and biochemical conditions of abiotic stresses, the intricate role of kinase proteins by signal transduction of MAPKs still has research gaps. It is required to associate the link between MAPKs and their corresponding stress in vivo. In the future, further functional analysis on MAPK members for physiological and biochemical roles in stress management can be helpful in breeding programs for innovation and advancement of agricultural science.
4. Role of MAPKs under abiotic stresses
Plants often experience various abiotic stresses (drought, low and high temperature, salinity, osmotic, etc.), which significantly affect their growth, productivity, and nutritional quality (Yu et al., 2019; Sun et al., 2020; Raza, 2021; Raza et al., 2021; Raza et al., 2022a; Raza et al., 2022b; Raza et al., 2022c; Raza et al., 2022d). The role of MAPKs against various abiotic stresses are briefly discussed in the subsequent sections.
4.1. The role of MAPKs under drought and oxidative stress
Drought stress is a major environmental factor affecting crops’ growth and productivity, leading to significant socioeconomic damage (Sinha et al., 2011; Zhang et al., 2018; Raza et al., 2022a). Drought occurs when water uptake within the plant by root is reduced due to low moisture in the soil, afflicting root morphology growth and physiology owing to wilt conditions, and consequently reducing the annual yield in plants (Aditi et al., 2020; Ahmad et al., 2021; Feng et al., 2022; Lu et al., 2022; Raza et al., 2022a). Biochemical and transcriptional studies have elaborated the MAPK response in drought in grassy and woody plants (Huang et al., 2020). Recently, it was studied that the MAPK cascade was induced with Arbuscular mycorrhizal fungi (AMF) inoculation in apple (Malus hupehensis) seedling following increased expression level of MAPKs such as MdMAPK16-2, MdMAPK17, and MdMAPK20-1 by 36.93%, 58.14%, and 54.14%, respectively, compared to those that do not have AMF inoculation (Xu and Chua, 2012). By regulating the RNA de-capping process in Arabidopsis, MAK6 improves the tolerance to dehydration (Hua et al., 2006). In Arabidopsis, MAPK gene families are activated. The promoter RD29, which is a dehydration-responsive gene via transient expression assay, suggests that MAPK cascade plays a vital role in drought signaling (Zaheer and Akhtar, 2016). In Arabidopsis, the transcriptional regulation of 44 MAPKs has been identified, out of which some are induced by water stress such as MPK2, MPK4, MPK5, MPK12, and MAPKKK4 (Moustafa et al., 2014).
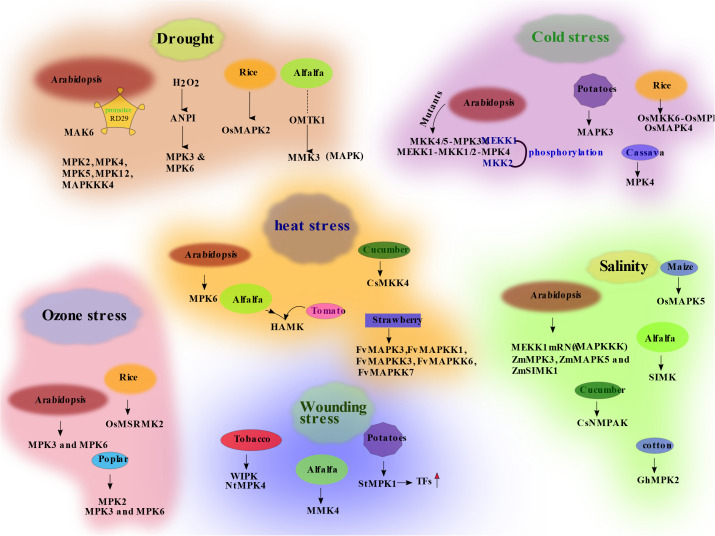
Potato (S. tuberosum L.) is known as one of the drought-sensitive crops, and serious yield loss has been threatened by drought stress (Handayani et al., 2019; Sattar et al., 2021), as well as lessened the quality of potato crops (Iftikhar et al., 2017). Studies revealed that 108 MAPK protein-coding genes have been found in potatoes (Zhu et al., 2021). More recent studies confirmed that 22 MAPK genes had been reported in the potato genome, such as StMAPK1 to StMAPK22 (Zaynab et al., 2021; Zhu et al., 2021), of which, six MAPKs have been related to abiotic stress, as well as six types of plants hormones (Boguszewska-Mankowska et al., 2020). StMAPK11 response is well studied for drought stress in potato, and it shows reasonable drought tolerance in potato when subjected under drought conditions (Zhu et al., 2021). The drought sensitivity of potato is due to the shallow root system that cannot explore the moisture from the deeper soil layers (Kumar et al., 2008). Further studies related to the characterization of MAPKs in drought stress can help improve the gene expression and physiology of potato. Many MAPKs related to drought stress tolerance are also observed in rice such as MAPKKK protein drought-hypersensitive mutant1 (DSM1), which shows reduced ROS generation under water-stressed conditions and increases the survival of plants under drought stress (Gao and Xiang, 2008). Overexpression of OsMAPK5, which is an ortholog of Arabidopsis MAPK3, also acts as a drought-responsive gene. Studies also suggested that the OsMAPK2 responds to drought stress and salt stress signaling within 15 min in rice crops, as shown in Figure 3 (Popescu et al., 2009).
Figure 3.
The schematic diagram of different mitogen-activated protein kinase (MAPK) signaling molecules under different stress conditions in different crops. Solid arrows show verified pathways; dashed arrows indicate assumed pathways; question marks indicate unknown cascade components.
Mostly abiotic stresses such as drought, cold, heat, and osmotic stress interrupt the metabolic equilibrium of the cell, which causes oxidative stress (Gill and Tuteja, 2010; Mittler et al., 2022; Raza et al., 2022b). The abnormal level of ROS such as free radicals and non-radicals leads to damage of specific molecules, which injured the cells or tissues in broad terms. It is called oxidative stress, which is due to the oxidation of molecules (Xing et al., 2007; Mittler et al., 2022). Antioxidants like endogenous or exogenous compounds help in the removal of ROS. Scavenger enzymes in plants like catalase decompose H2O2 and overcome oxidative stress. ABA regulates the A. thaliana CAT1, and MAPKK inhibitor PD98059 delayed CAT1 expression, which is mediated by ABA signaling (Ning et al., 2010). Under dehydration stress, the MKK1 and MPK6 mutant in A. thaliana alter their response to ABA. It was proposed from the above findings that MKKI–MPK6 controls the metabolism of H2O2 by CAT1 with the absence of ABA-mediated activation of MPK6 and MMKI (Nakagami et al., 2006). Plant defense mechanism and salicylic acid (SA) accumulation are controlled by the CAT2 expression, which is stimulated by MEKK1 and MPK4 (Colcombet and Hirt, 2008). ROS metabolism is regulated by the MEKK1–MPK4 pathways (Ning et al., 2010). Many other MPAKKKs in A. thaliana are activated by H2O2, such as ANPI, which causes the downstream activation of MPK3 and MPK6, as shown in Figure 3 (Ding et al., 2009). These findings suggest that MAPKs not only are induced by ROS but also control ROS and arbitrate oxidative stress in crops. Oxidative stress is a communal response under abiotic and biotic stress as ROS is a conjunction point to indicate the stress in plants. Recently, ROS-mediated MAPK signaling in plants has been described. In A. thaliana mutants, MAPK studies reveal the specific protein association related to ROS control (Ning et al., 2010). The significant roles of MAPK genes in different crops like alfalfa (M. sativa), rice (O. sativa), cotton (Gossypium hirsutum), maize (Z. mays), apple (Malus hupehensis), potato (S. tuberosum), cucumber (Cucumis sativus), strawberry (Fragaria vesca), mulberry (Moraceaemorus), and cassava (Manihot esculenta) under drought condition are mentioned in Table 2 . From the above discussion, it is concluded that many MAPK gene families in different crops integrate stress-related proteins and regulate stimulus response. It is necessary to identify and isolate more MAPK genes for crop improvement.
Table 2.
MAPK gene signaling responses in different plants under drought stress conditions.
| Plant species | MAPK genes | Activation | Plant stage | References |
|---|---|---|---|---|
| Arabidopsis | AtMPK2, AtMPK3, AtMPK4, AtMPK5, AtMPK12, and AtMKK | Activated at the transcriptional level Activated by promoter RD29 under dehydration |
Seedling | Chen et al. (2015) |
| Alfalfa | MsP44, MsMKK4 | Activated at the post-transcriptional level | Seedlings | Chen et al. (2015) |
| Rice | OsMPK3,4 | Activated by the regulation of protein DSM1 (drought-sensitive mutant1) | Seedling | Huang et al. (2020) |
| OsMKK6 | Transcriptionally regulated by drought | Seedling | Huang et al. (2020) | |
| OsDSM1 | Gao and Xiang (2008) | |||
| OsMSRMK2,5 and OsMAPKK1 | Activated under combined drought and osmotic stress | Seedling | Wang et al. (2010); Chen et al. (2015) | |
| Cotton | GhMAPK2, GhMAPK16 | Activated under combined drought and osmotic stress | Germination | Wang et al. (2007) |
| Maize | ZmMPK2 | Activated under reducing water and osmotic pressure | Seedling | Zhang et al. (2011) |
| Apple | MaMPK | Activated at the transcriptional level | Seedling | Sun et al. (2017) |
| Potato | StMAPK11 | Activated by induction of drought and hormone stress | Flowering | Zhu et al. (2021) |
| Cucumber | CsMAPKs | Activated by the regulation of stress-associated genes and drought stress | Flowering | Wei et al. (2014) |
| Strawberry | FvMAPK5,8 | Activated at the transcriptional level | Reproductive | Zhou et al. (2014) |
| Mulberry | MnMAPK1, MnMAPK2 | Negatively activated by drought stress | Reproductive | Yan et al. (2016) |
| Cassava | MeMAPK | Activated 20% in leaves and 70% in roots under high drought conditions | Flowering | Chinnusamy et al. (2007) |
4.2. The role of MAPKs under cold stress
One of the most critical factors that affect the growth and development of crops is cold stress or chilling temperatures like sudden frost and snow, and freezing temperatures cause serious damage to crop production and quality (Raza et al., 2021; Ma et al., 2022; Raza et al., 2022c). When plants survive under cold or freezing temperatures, it is called cold tolerance or winter hardiness (Peng et al., 2006). The physiological and metabolic status of crops is changed by the altered expression of many genes during cold acclimatization (Agarwal et al., 2010; Medina et al., 2016; Raza et al., 2021; Raza et al., 2022a; Raza et al., 2022c). In Arabidopsis, MAPK genes like MEKK1, MKK2, MPK4, and MPK6 were shown to be activated under cold stress (Teige et al., 2004). The cold-sensitive gene MKK2 does not have any effect on MPK4 or MPK6, which suggests that MKK2 is an upregulator of MPK4 and MPK6 in low temperature. This outcome shows that chilling temperature activates MEKK1, MPK3, MPK4, and MPK6 (Krasensky and Jonak, 2012). Another MAPKK gene named SbMAPKK in halophytes, such as sea bean (Salicornia brachiate), shows upregulation under cold stress (Meng and Zhang, 2013), because cold as well as salt stress induces some kinase molecules that allow plants to survive under adverse environments.
Recently, in tomato, CRISPR/Cas9-mediated SlMAPK3 mutants were used to investigate the relationship between ferulic acid (FA) and SlMAPK3 under chilling temperatures. It was shown that under low temperature, i.e., 4°C, FA content increases, which then increases the FA synthesis-related gene (SlPAL5, SlC3H, and SlCOMT) expression. However, the knockout of SlMAPK3 inhibited the content of FA and the expression of those genes compared with the control, which suggested a close relationship between SlMAPK3 and FA. Plant response to cold stress depends heavily on the CBF/DREB1 (C-repeat binding factor/dehydration resistance element binding protein 1)- and ICE1 (inducer of CBF expression1)-dependent transcriptional regulatory mechanisms. It has been demonstrated that CBFs may bind to cis elements in the COR (COLD RESPONSIVE) gene promoters, sufficiently activating the expression of COR genes and inducing resistance to cold stress. It was revealed that the FA in tomato fruit provides resistance to chilling stress by upregulating the gene expression of the repeat binding transcription factor (CBF), a transcriptional pathway, in a MAPK3-dependent manner (Shu et al., 2022). Under cold stress, MKK4/5-MPK3/6 and MEKK1-MKK1/2-MPK4, the two MAPK gene mutants, show response to external stimuli produced by cold stress (Furuya et al., 2013; Pitzschke et al., 2014). It is also documented that MEKK1-MKK2-MPK4/MPK6 pathways positively regulate the cold response and chilling temperature tolerance in Arabidopsis (Teige et al., 2004). To induce the kinase mechanism, MKK2 activity is phosphorylated by MEKK1 under cold conditions (Raina et al., 2013), which further activates MKK2 that phosphorylates MPK4 and MPK6 (Teige et al., 2004), likely adjusting downstream molecules to regulate the cellular status. These findings showed that the MMK2 pathway regulates the freezing response (Teige et al., 2004). SlMPK3 upregulates the growth of tomato under low temperature (Yu et al., 2015). In potatoes, it is reported that the MAPK3 showed a response under cold stress (Zhang et al., 2017) and chilling temperature stress (Xie et al., 2012). MAPK3 also activates the phosphorylation downstream for ACC synthase and transcriptional factor OsbHLH002/OsICE1 for chilling outbreaks (Xie et al., 2012). Under cold stress, MKK2-MPK4/MPK6 cascades are activated (Teige et al., 2004). In recent years, the MAPK genes in rice OsMKK6-OsMPK3 have been identified, which are activated in cold stress (Fu et al., 2012). cDNA library screening revealed another well-studied MAPKK gene OsMEK1 that showed interaction with OsMAP1, which is important in freezing temperature tolerance (An et al., 2012).
Under cold stress, the MPKK genes ZmMKK4 (Wu et al., 2011) and ZmMPK7, as shown in Figure 3 (Osthoff et al., 2019), showed upregulation in maize. These two genes are found in the nucleus and control the transcription factors at low temperature. Similarly, five more MAPKs have been identified, which responds positively in roots of maize under cold stress (Osthoff et al., 2019). It is described that the seedlings of cotton at low temperature (4°C) showed optimum growth against cold stress, especially in root cells because of the over-expression of the MAPK gene named GhMAPK (Zhang et al., 2011). MAPK genes in different crops like Arabidopsis, alfalfa, blue mustard (Chorispora Bungeana), cotton, maize, rice, and sea bean (Salicornia brachiate) under cold and heat stress conditions are mentioned in Table 3 . Cold stress causes severe damage in many regions of the world. Therefore, research should incorporate MAPK cascades with the help of modern technologies in breeding resistance cultivars of different crops by identifying novel MAPK gene families.
Table 3.
MAPK gene signaling responses in different plants under cold and heat stress conditions.
| Plant species | MAPK genes | Activation | References |
|---|---|---|---|
| Cold | |||
| Arabidopsis | AtMEKK1,6 | Calcium-dependent activation, to regulate vacuolar processing enzyme (VPE) | Zhang et al. (2006) |
| AtMPK1,6 | MKK2 is the upstream activator of MPK4 and MPK6 | Zhang et al. (2018) | |
| AtMAPKK, AtMKK2 | |||
| Alfalfa | MsSAMK, MsP44MKK4 | Activated by cold shock domain-containing proteins (CSDPs) | Agarwal et al. (2010) |
| Blue mustard | CbMAPK3 | Activated under low temperature | Nakagami et al. (2005) |
| Cotton | GhMAPK, GhMAPK2, 7 | Activated under 4°C | Yang et al. (2019) |
| Maize | ZmMPK3,5 | Activated under high accumulation of proline | Zhang et al. (2011) |
| ZmMPK17 | Activated under chilling conditions | Kong et al. (2011) | |
| Rice | OsMSRMK2 | Activated at 12°C | Meng and Zhang (2013) |
| OsMAPKK2, 4,6 and 10 | Activated by low temperature stress at the transcriptional level | ||
| OsMKK6, OsMPK3 | Activated by cold shock domain-containing proteins (CSDPs) | Xie et al. (2012) | |
| OsMAPK5 | Activation under chilling temperature | Xiong and Yang, 2003 | |
| Sea bean | SbMAPKK | Activated by cold shock domain-containing proteins (CSDPs) | Meng and Zhang (2013) |
| Tomato | SlMPK3 | Upregulation under low temperature | Yu et al. (2015) |
| Heat | |||
| Rice | OsMKK4,6 | Activated when temperature increased from 37°C | Wen et al. (2002); Kumar et al. (2008) |
| OsMSRMK2 | Activated at high temperature < 37°C | Agrawal et al. (2002) | |
| Alfalfa | MsHAMK | Activated by the activation of heat shock protein. | Sangwan et al. (2002) |
| Potato | StMPK1 | Activated by high temperature stress at the transcriptional level | Blanco et al. (2006) |
| Cucumber | CsMKK4 | Activated by the upregulation of heat shock transcription factors (HSFs) | Wei et al. (2014) |
| Strawberry | FvMAPKK3 | Activated by the upregulation of HSFs | Lohani et al. (2020) |
| Mulberry | MnMAPK1,5,6 | Activated at 40°C under high temperature | Yan et al. (2016) |
| Tomato | SlMPK1 | Negitively regulated under high temperature | Ding et al., 2018 |
4.3. The role of MAPKs under heat stress
Temperature is considered the most essential factor to achieve optimum metabolic processes, growth, development, and production of plants (Yin et al., 2014; Haider et al., 2022; Raza et al., 2022c). The amount of water in cells under different temperatures is a fundamental feature for cell survival, which is directly proportional to plant growth and damages the cell structure and organelles and reduces productivity (Jiang et al., 2015; Xing et al., 2015; Raza et al., 2021). High temperature also causes overproduction of ROS due to the high rate of respiration and photosynthesis, which significantly affects the growth and development of plants (Li et al., 2012). MAPKs play an important role under heat stress in plants such as aspen (Brachypodium distachyon); 60% of genes activated under high temperature by MAPKs signal transduction cascade (Suri and Dhindsa, 2008). To date, few studies about heat stress signaling of MAPKs provide evidence that only specific MAPKs are upregulated or activated under high temperature stress (Munns et al., 2020). It has been experimentally proven that MAPK3 is activated under heat stress and ultra-irradiation and helps to mitigate stress by controlling the growth of plants (Tuteja, 2007).
Secondly, the heat shock protein HSP70 plays a vital role in response to heat stress signaling through the HSP gene by the activation of the MAPK named HAMK in tobacco (Wei et al., 2014). In cucumber (C. sativus), an MAPK gene named CsMKK4 is activated after 8 h of heat treatment. Most of the MAPK genes showed activation under heat stress except CsMPK3 and CsMPK7 (Wei et al., 2014). The upregulated transcript level of FvMAPK3, FvMAPKK1, FvMAPKK3, FvMAPKK6, and FvMAPKK7 in strawberry (F. vesca) has been observed under high temperature (Zhou et al., 2014). In the same way, under heat stress, the gene named FvMAPKK3 is activated (Zhou et al., 2014). In mulberry (morus) under heat stress, eight MAPK genes have been identified, which showed upregulation (MnMAPK1, MnMAPK5, MnMAPK6, and MnMAPK9) and downregulation (MnMAPK2, MnMAPK3, MnMAPK8, and MnMAPK10) (Yan et al., 2016). Likewise, under high temperature, the downward growth regulation of the tomato gene named SlMPK1 is shown (Ding et al., 2018). The above studies indicated that under heat stress conditions, MAPK cascade is regulated in plants. MAPK gene families related to heat resistance is negligible; due to climate change, this stress will be a severe threat in coming years. This stress can be overcome by identifying desirable MAPK gene families in commercial crops.
4.4. The role of MAPKs under salinity and osmotic stress
In plants, there are three major salt tolerance mechanisms, namely, osmotic pressure tolerance, ionic balance management, and decreasing the Na+ and Cl− cytoplasmic concentrations (Mohamed et al., 2022; Raza et al., 2022d). Salinity stress is a very serious threat in many regions of the world; more than 50% of irrigated soils all over the world face reduction in the productivity of major agricultural crops (Shi et al., 2010). Under salt stress, cell membrane and protein disruption occurs due to the overproduction of ROS in plants, which is one of serious problems for crops grown under salt conditions (Nakagami et al., 2005; Raza et al., 2022d).
It was also reported that MAPK proteins are activated when plants are subjected to salt stress; this leads to speeding up the expression level of V-H+-ATPase, which increases the tolerance to salinity stress (Krasensky and Jonak, 2012). In Arabidopsis, a MAPKK gene named MEKK1 mRNA accumulated in response to high salt stress. The protein–protein interaction between MMK2/MEK1 and MPK4 MAPKs, MMK21 and MKK2/MEK1 MAPKKs, and MPK4 and MEKK1 was shown by yeast two-hybrid analysis (Teige et al., 2004). It was also documented that salt stress signal transmission occurred at two MAPK cascades such as MPK4 MAPK cascade with genes named MEKK1-MEK1/MKK2-MPK4 and MPK6 and a second MAPK cascade with a gene named p44MAPK involved in salt tolerance in Arabidopsis, as shown in Table 4 . MEKK1 involve as more upstream of MKK2 and more downstream MAPKs MPK4 and MPK6 under salinity stress (Teige et al., 2004). MAKs such as MPK6, MPK4, and MKP1 play a negative role under salinity stress conditions (Faried et al., 2017). A salt stress-induced MAPK named 46kDa SIMK showed response under salt stress in alfalfa (Krasensky and Jonak, 2012). In vivo as well as in vitro tolerance to salt stress produced by upstream kinase named SIMKK intermingles with SIMK by yeast two-hybrid (Krasensky and Jonak, 2012).
Table 4.
MAPK gene signaling responses in different plants under salinity stress.
| Plant species | MAPK genes | Activation | Concentration | References |
|---|---|---|---|---|
| Arabidopsis | AtMEKK1,2 | Activation by accumulation of sodium chloride | High salt accumulation | Teige et al. (2004); Moustafa et al. (2008) |
| AtMPK1,3 | Activation of RD29A and RD29B gene promoters | Excess of salts | Yu et al. (2010) | |
| AtMPK4,6 | Activated by phosphatidic acid (PA) | Acid accumulation | Moustafa et al. (2008) | |
| Maize | ZmMPK3,5 | Activated under very high salt concentration | High amount of salts | Wang et al. (2010) |
| ZmSIMK1 | Activated by high salinity | Higher salt accumulation | Gu et al. (2010) | |
| ZmMKK4 | Activated by NaCl accumulation | Excess amount of NaCl | Kong et al. (2011) | |
| Rice |
OsMSRMK2, MAPKK4, 6 |
Activated in response to high salt and cytotoxicity. | NaCl accumulation | Agrawal et al. (2002) |
| OsMAPK5, | Activated by salt accumulation | Very high concentration | Xiong and Yang (2003) | |
| OsMPK3, 4, 5 | Activated by accumulation of arsenic stress and salts | High arsenic and salt concentration | Rao et al. (2010) | |
| OsMKK6 | Activated in hypersensitivity of other stresses | High salinity | ||
| OsEDR1 | Activated by high salt accumulation | Excess salt amount | Kim et al. (2003) | |
| OsMAPK4 | Activated under salt accumulation | NaCl excess amount | Rao et al. (2010) | |
| Cotton |
GhMAPK, GhMPK2 |
Activated by salt stress and osmotic adjustment | High amount of salts | Wang et al. (2007); Zhang et al. (2011) |
| Blue mustard | CbMAPK3 | Activated under salt accumulation | High salt accumulation | Zhang et al. (2006) |
| Sea bean | SbMAPKK | Activated under salt accumulation | Water reduction and salt accumulation | Agarwal et al. (2010) |
| Cucumber | CsNMAPK | Activated under salt accumulation | Salt accumulation | Xu et al. (2010) |
| Tobacco | NtSIPK | Activated by salt stress and osmotic adjustment | NaCl accumulation | Mikolajczyk et al. (2000) |
| Potato | StMAPK3 | Activated by salt stress and osmotic adjustment | PEG, menthol, and NaCl treatment | Zhu et al. (2020) |
| Osmotic stress | ||||
| Arabidopsis | AtMPK1, 4, 6, and 20 | Activated under high salt accumulation and improved the osmosis | Hyper-osmolarity | Droillard et al. (2004); Moustafa et al. (2008) |
| AtMKK7,9 | Activated under higher NaCl concentration | Sodium chloride excess | Moustafa et al. (2008) | |
| AtMPK9, 17,18 | Activated under higher salt accumulation | Hyper-osmolarity | Moustafa et al. (2014) | |
| Maize | ZmMPK7 | Activated under salt accumulation and molecular imbalance | Hyper-osmolarity | Zong et al. (2009) |
| Tobacco | NtSIPK | Activated by salicylic acid under high salt concentration | Higher osmotic stress | Mikolajczyk et al. (2000) |
| Alfalfa | MsSIMK | Upstream activation by SIMKK | Higher osmolarity | Jonak et al. (2004) |
| Potato | StMAPK3 | Activated by NaCl (40 mM and 80 mM) | Salt accumulation | Zhu et al. (2020) |
Crops respond to salt stress through different processes like elimination and appropriation of ions (Na+ and Cl−) into vacuole to lessen cytotoxicity; research has shown that osmotic stress is also regulated by MAPKs (Kim et al., 2003). Protective proteins like late embryogenesis abundant (LEA) and chaperones, a heat shocking protein also speed up to defense the negativity of these toxic ions. MAPKs also respond by conveying signals for osmotic stress to specific effectors and play a key role to survive in the cell under high salt concentration. It is already reported that MAPK cascade is activated under salt and osmotic stress at both transcriptional and protein levels (Iftikhar et al., 2017). In A. thaliana, genes like AtMEKK1, AtMKK2, and AtMPK4, as shown in Figure 3 , have stress tolerance under salinity conditions. MMK gene named as SIMKK an upward regulator of SIMK, a MAPK gene response upregulation in alfalfa under salinity as well as osmotic stress (Xu et al., 2010). In addition, salicylic acid-induced protein kinase (SIPK) regulates osmotic stress in tobacco in a very short time (5 to 10 min) (Ghorbel et al., 2019; Habiba et al., 2021); SIPK is an Arabidopsis MPK6 homolog in tobacco.
In another study, it was also shown that plant growth recovered in the overexpression lines of StMAPK3 in potato, and the lethal effects of osmotic stress and salinity were reduced by the MAPK gene family (Zhu et al., 2020). The increased content of many oxidative markers like NaCl, H2O2, polyethylene glycol (PEG), and menthol is weakened by StMAPK3 overexpression in potato (Zhu et al., 2020). Similarly, the opposite effect is shown by catalase (CAT), peroxidase (POD), superoxide dismutase (SOD), and proline content by StMAPK3. In Arabidopsis, interchangeable names MPK4 and MPK6 have been identified, whereas salinity signal responding genes like MEKK1 and MKK2 have also been recognized (Teige et al., 2004). Salt tolerance is shown by the stress marker gene MKK2 in transgenic plants. Gene promoters called RD29A and RD29B were activated by the expression of MKK and MPK, respectively (Zaheer and Akhtar, 2016), which shows that the MAPKs regulate upward under salinity and osmotic stress. Many MPKs (MPK9, MPK10, MPK11, MPK17, and MPK18), MKKs (MKK7 and MKK9), and MEKKs (MEKK3, MEKK5, MEKK6, and MEKK7) have been screened for salt and osmotic stress (Moustafa et al., 2008; Chen et al., 2015). In maize, the MAPK named ZmSIMK1 was identified as having high salt tolerance by the overexpression of Arabidopsis transgenic plants. In the same way, in maize, ZmMPK17 also showed high osmotic stress tolerance by the activation of Arabidopsis MPK17 (Yang et al., 2019), which provides an idea that further description and practical study of MPK17 and its ortholog can provide better achievement in abiotic stress tolerance gene identification and screening.
It is also described that, in rice, MAPKs control salinity and osmotic stress; OsMPK4, OsMPK3, OsMSRMK2, OsEDR1, OsEDR1, OsMAPK5, and OsMAPK4 have been classsified as salinity tolerance genes (Xie et al., 2012). Salt tolerance and osmotic pressure are also affected by the activation of OsMPK5 and OsMPK4 in transgenic maize line (Xiong and Yang, 2003). Over-expressing GhMPK2 shows the osmotic stress tolerance in transgenic cotton (G. hirsutum) (Yang et al., 2019). Similarly, cucumber roots and transgenic tobacco seeds showed overexpression of MAPK genes called CsNMAPK and CsNMPAK, which showed a germination rate higher than wild type (Gomi et al., 2005), which proves that CsNMPAK performs better under salt stress condition in the early stages. Salinity is associated with osmotic stress; from the above studies, it is strongly depicted that MAPK genes that are activated under salinity conditions are also upregulated under osmotic stress conditions. The MAPK genes in different crops like Arabidposis, maize, rice, cotton, blue mustard, alfalfa, rice, tobacco, potato, sea bean, cucumber, tobacco, and potato under salinity stress and Arabidposis, maize, tobacco, alfalfa, and potato under osmotic stress are mentioned in Table 4 .
The above discussion revealed that many MAPK genes related to salinity and osmotic stress had been identified, and highly expressed under saline and osmotic stress conditions. By identifying more MAPK genes in crops, crop yield and productivity can be improved, and saline soil can be brought under cultivation.
4.5. The role of MAPKs under heavy metal stress
Heavy metal ions play a vital role in the development and growth as well as in the physiological processes of plants (Lee and Ellis, 2007; Raza et al., 2022e). However, at higher concentrations, they can have lethal effects on plant physiology. With the increase in concentration of heavy metals in the soil, the cellular response of the plants is activated (Raza et al., 2022e). A novel type of MAPK gene called OsMSRMK2 in rice is activated when plants have high levels of cadmium, mercury, and copper ions (Yang et al., 2019). Studies have also confirmed the activation of MAPK genes in rice (O. sativa) with the high level of cadmium ions (Haider et al., 2021).
Many MAPK genes like SIMK, MMK2, MMK3, and SAMK in alfalfa (M. sativa) seedling are activated when they are exposed to copper and cadmium ions stress (Gupta et al., 2009). Studies have also confirmed that the activation of SIMKK is only under copper ion stress but not under cadmium ion stress, as shown in Table 5 . It was documented that this MAPK activity is specific in response as specific treatment indicates the specific signals’ transduction (Yeh et al., 2004). Involvement of MAPK genes called OsMPK3, OsMPK4, and OsMKK4 mediated heavy metal stress tolerance in rice seedlings as shown in Figure 3 (Gupta et al., 2009). In maize (Z. mays), under heavy metal stress, the activation of MAPK genes has also been confirmed (Lumbreras et al., 2010). These findings show the role of MAPKs in signaling activation under various heavy metal stresses.
Table 5.
MAPK gene signaling responses in different plants under heavy metal stress conditions.
| Plant species | MAPK genes | Stress causing metals | Methods | Reference |
|---|---|---|---|---|
| Alfalfa | MsMMK2, MsMMK3, MsSAMK | Cadmium and copper | Complex activation pattern of MAPKs: SIMK, MMK2, MMK3, and SAMK | Jonak et al. (2004) |
| Rice | OsMSRMK2, OsMPK3,4, and OsMKK4 | Mercury, copper, cadmium, and arsenic | Overexpression of functions of an OsMSRMK2 and MBP kinase | Agrawal et al. (2002) |
| OsHMA3 | Cadmium | Overexpression of a functional allele of OsHMA3 | Ueno et al. (2010); Shao et al. (2018) | |
| OsLCT1 | Cadmium | Knockdown of OsLCT1 | Ueno et al. (2010) | |
| OsNRAMP5 | Cadmium | CRISPR/Cas9-mediated editing of OsNRAMP5 | Uraguchi et al. (2011) | |
| OsHMA3 | Cadmium | Articulation and tissue restrictions of OsHMA3 | Shao et al. (2018) | |
| Maize | ZmMPK3 | Cadmium | Signaling activation of ZmMPK3 | Wang et al. (2010) |
From the above discussion, except rice, maize, and alfalfa, there are no other crops in which MAPK gene families have been identified. In the near future, heavy metal stress will severely threaten crops due to climate change and heavy industrialization. The researcher’s job is to verify the functional roles of MAPK genes and other mechanisms to develop heavy metal resistance cultivars.
5. Conclusion
There is growing evidence that the MAPK cascade is the hub of a sophisticated network structure that transduces signals related to plant stress tolerance. The MAPK cascade gradually amplifies and conveys stress signals to downstream response components through phosphorylation and dephosphorylation, leading to various stress responses. Furthermore, a better understanding of the MAPK cascades’ process should make it easier to create new methods for enhancing plants’ ability to withstand stress. Numerous genetic engineering methods are available to increase abiotic stress tolerance in crops. As key players in signal transduction and regulators of gene transcription, MAPK cascades have already been used to improve abiotic stress tolerance, as discussed in this review. Molecular biology analysis of the MAPK cascade’s components and its function is essential for enhancing crop improvement. Research on the function of MAPK genes or the mechanism by which the MAPK cascade regulates plant stress resistance is still limited, despite many studies in plants that have demonstrated the involvement of MAPK cascades in numerous biological processes in response to abiotic and biotic stresses. In addition, different stress stimuli can activate the same MAPK cascade genes. The above discussion concludes that the MAPK cascades and the molecular mechanisms of plant stress resistance have great significance for elucidating the entire stress tolerance signal transduction pathway in plants.
6. Future perspective
Abiotic stress in crops helps to expand the tolerance of crops under different abiotic stresses by various methods of genetic manipulation. A number of studies recounted that by exploiting MAPK pathways in crops like Arabidopsis and many other crops like potato, maize, rice, and poplar, stress tolerance has improved. In the near future, investigation should include detecting the MAPK substrate, and by using advanced breeding methods and molecular methodologies, the development of new lines would be able to withstand harsh environments to meet the food requirements of the increasing population. Interacting protein is the main factor that needs to identify and quantify for producing multiple environmental stress, also with their mode of action, which is like that (MAPKKK-MAPKK-MAPK) chain and accumulated into one functional ‘MAPK transgenic circuits’ which could be inserted into target sequence or cell through genetic engineering and biotechnology to develop a tolerance species for specific function having specific kinase protein.
It is important to study the function of MAPKs related to abiotic stress in crops, and previous studies have confirmed their role in crops related to abiotic as well as biotic stress. In the future, there is a need to identify more MAPK gene families in crops related to environmental stresses and also identify their functional analysis through advanced methodologies like transcriptomics, proteomics, metabolomics, bioinformatics, CRISPR/Cas technology, and DNA/RNA sequencing to encourage analysis of a regulation network that controls abiotic stress response. In addition, identification and functional analysis of MAPKs can be further processed by mutation, gene silencing, and microRNA techniques to produce MAPK mutants and genetically engineered gene families, which transform into crops for tolerance to biotic stresses.
Author contributions
YM, HS, and XZ conceived the idea. YM, XZ, NZ, Nu-A, AR, and FH contributed to writing and literature search. YM and Nu-A contributed to organizing tables and figures. All authors contributed to the article and approved the submitted version.
Funding Statement
The research program was sponsored by the National Natural Science Foundation of China (No. 31960444), the Gansu Provincial Key Laboratory of Aridland Crop Science, Gansu Agricultural University (No. GSCS-2019-Z03), and the Gansu Major Science and Technology Project (No. GZGG-2021-6).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
- Aditi G., Andrés R.-M., Ana I.-C. (2020). The physiology of plant responses to drought. Science 368, 266–269. doi: 10.1126/science.aaz7614 [DOI] [PubMed] [Google Scholar]
- Agarwal P.-K., Gupta K., Jha B. (2010). Molecular characterization of the salicornia brachiata SbMAPKK gene and its expression by abiotic stress. Mol. Biol. Rep. 37, 981–986. doi: 10.1007/s11033-009-9774-1 [DOI] [PubMed] [Google Scholar]
- Agrawal G.-K., Rakwal R., Iwahashi H. (2002). Isolation of novel rice (Oryza sativa l.) multiple stress responsive MAP kinase gene, OsMSRMK2, whose mRNA accumulates rapidly in response to environmental cues. Biochem. Biophys. Res. Commun. 294, 1009–1016. doi: 10.1016/S0006-291X(02)00571-5 [DOI] [PubMed] [Google Scholar]
- Ahmad A., Aslam Z., Naz M., Hussain S., Javed T., Aslam S., et al. (2021). Exogenous salicylic acid-induced drought stress tolerance in wheat (Triticum aestivum l.) grown under hydroponic culture. PloS One 16 (12), e0260556. doi: 10.1371/journal.pone.0260556 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- An D., Yang J., Zhang P. (2012). Transcriptome profiling of low temperature-treated cassava apical shoots showed dynamic responses of tropical plant to cold stress. BMC Genom 13, 64. doi: 10.1186/1471-2164-13-64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrasi N., Rigo G., Zsigmond L., Perez-Salamo I., Papdi C., Klement E., et al. (2019). The mitogen-activated protein kinase 4-phosphorylated heat shock factor A4A regulates responses to combined salt and heat stresses. J. Exp. Bot. 70, 4903–4918. doi: 10.1093/jxb/erz217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anjum S.-A., Ashraf U., Tanveer M., Khan I., Hussain S., Shahzad B., et al. (2017). Drought induced changes in growth, osmolyte accumulation and antioxidant metabolism of three maize hybrids. Front. Plant Sci. 8, 69. doi: 10.3389/fpls.2017.00069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baer-Nawrocka A., Sadowski A. (2019). Food security and food self-sufficiency around the world: A typology of countries. PloS One 14. doi: 10.1371/journal.pone.0213448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bigeard J., Hirt H. (2018). Nuclear signaling of plant MAPKs. Front. Plant Sci. 9, 469. doi: 10.3389/fpls.2018.00469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bita C.-E., Gerats T. (2013). Plant tolerance to high temperature in a changing environment: Scientific fundamentals and production of heat stress-tolerant crops. Front. Plant Sci. 4, 273. doi: 10.3389/fpls.2013.00273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanco F.-A., Zanetti M.-E., Casalongue C.-A., Daleo G.-R. (2006). Molecular characterization of a potato MAP kinase transcriptionally regulated by multiple environmental stresses. Plant Physiol. Biochem. PPB/Societe Francaise physiologie vegetale 44, 315–322. doi: 10.1016/j.plaphy.2006.05.005 [DOI] [PubMed] [Google Scholar]
- Boguszewska-Mankowska D., Zarzynska K., Nosalewicz A. (2020). Drought differentially affects root system size and architecture of potato cultivars with differing drought tolerance. Am. J. Potato Res. 97, 54–62. doi: 10.1007/s12230-019-09755-2 [DOI] [Google Scholar]
- Cçakır B., Kılıcçkaya O. (2015). Mitogen-activated protein kinase cascades in Vitis vinifera. Front. Plant Sci. 6. doi: 10.3389/fpls.2015.00556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X., Wang J., Zhu M., Jia H., Liu D., Hao L., et al. (2015). A cotton raf-like MAP3K gene, GhMAP3K40, mediates reduced tolerance to biotic and abiotic stress in nicotiana benthamiana by negatively regulating growth and development. Plant Sci. 240, 10–24. doi: 10.1016/j.plantsci.2015.08.012 [DOI] [PubMed] [Google Scholar]
- Chinnusamy V., Zhu J., Zhu J.-K. (2007). Cold stress regulation of gene expression in plants. Trends Plant Sci. 12, 444–451. doi: 10.1016/j.tplants.2007.07.002 [DOI] [PubMed] [Google Scholar]
- Choi H.-I., Park H.-J., Park J.-H., Kim S., Im M.-Y., Seo H.-H., et al. (2005). Arabidopsis calcium-dependent protein kinase AtCPK32 interacts with ABF4, a transcriptional regulator of abscisic acid-responsive gene expression, and modulates its activity. Plant Physiol. 139, 1750–1761. doi: 10.1104/pp.105.069757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colcombet J., Hirt H. (2008). Arabidopsis MAPKs: A complex signaling network involved in multiple biological processes. Biochem. J. 413, 217–226. doi: 10.1042/BJ20080625 [DOI] [PubMed] [Google Scholar]
- Cutler S.-R., Rodriguez P.-L., Finkelstein R.-R., Abrams S.-R. (2010). Abscisic acid: Emergence of a core signaling network. Annu. Rev. Plant Biol. 61, 651–679. doi: 10.1146/annurev-arplant-042809-112122 [DOI] [PubMed] [Google Scholar]
- Danquah A., de-Zelicourt A., Colcombet J., Hirt H. (2014). The role of ABA and MAPK signaling pathways in plant abiotic stress responses. Biotechnol. Adv. 32, 40–52. doi: 10.1016/j.biotechadv.2013.09.006 [DOI] [PubMed] [Google Scholar]
- Ding Y., Cao J., Ni L., Zhu Y., Zhang A., Tan M., et al. (2013). ZmCPK11 is involved in abscisic acid-induced antioxidant defence and functions upstream of ZmMPK5 in abscisic acid signalling in maize. J. Exp. Bot. 64, 871–884. doi: 10.1093/jxb/ers366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding H., He J., Wu Y., Wu X., Ge C., Wang Y., et al. (2018). The tomato mitogen-activated protein kinase SlMPK1 is as a negative regulator of the hightemperature stress response. Plant Physiol. 177, 633–651. doi: 10.1104/pp.18.00067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding H.-D., Zhang X.-H., Xu S.-C., Sun L.-L., Jiang M.-Y., Zhang A.-Y. (2009). Induction of protection against paraquat-induced oxidative damage by abscisic acid in maize leaves is mediated through mitogen-activated protein kinase. J. Integr. Plant Bio l51, 961–972. doi: 10.1111/j.1744-7909.2009.00868.x [DOI] [PubMed] [Google Scholar]
- Doczi R., Brader G., Pettko-Szandtner A., Rajh I., Djamei A., Pitzschke A. (2007). The arabidopsis mitogen-activated protein kinase kinase MKK3 is upstream of group c mitogen-activated protein kinases and participates in pathogen signaling. Plant Cell 19, 3266–3279. doi: 10.1105/tpc.106.050039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Droillard M.-J., Boudsocq M., Barbier-Brygoo H., Lauriere C. (2004). Involvement of MPK4 in osmotic stress response pathways in cell suspensions and plantlets of arabidopsis thaliana: Activation by hypoosmolarity and negative role in hyperosmolarity tolerance. FEBS Lett. 574, 42–48. doi: 10.1016/j.febslet.2004.08.001 [DOI] [PubMed] [Google Scholar]
- FAO. IFAD. UNICEF. WFP. WHO (2019). The state of food security and nutrition in the world 2019. safeguarding against economic slowdowns and downturns (tech. rep.) (Rome: FAO; ). [Google Scholar]
- Faried H.-N., Ayyub C.-M., Amjad M., Ahmed R., Wattoo F.-M., Butt M., et al. (2017). Salicylic acid confers salt tolerance in potato plants by improving water relations, gaseous exchange, antioxidant activities and osmoregulation. J. Sci. Food Agric. 97, 1868–1875. doi: 10.1002/jsfa.7989 [DOI] [PubMed] [Google Scholar]
- Farooq M. S., Uzair M., Raza A., Habib M., Xu Y., Yousuf M., et al. (2022). Uncovering the research gaps to alleviate the negative impacts of climate change on food security: A review. Front. Plant Sci. 13, 927535. doi: 10.3389/fpls.2022.927535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng W., Liu Y., Cao Y., Zhao Y., Zhang H., Sun F., et al. (2022). Maize ZmBES1/BZR1-3 and-9 transcription factors negatively regulate drought tolerance in transgenic arabidopsis. Int. J. Mol. Sci. 23 (11), 6025. doi: 10.3390/ijms23116025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu S.-F., Chou W.-C., Huang D.-D., Huang H.-J. (2012). Transcriptional regulation of a rice mitogen. Physiol. J. 43, 958–963. doi: 10.1093/pcp/pcf111 [DOI] [PubMed] [Google Scholar]
- Furuya T., Matsuoka D., Nanmorim T. (2013). Phosphorylation of arabidopsis thaliana MEKK1 via Ca2+signaling as a part of the cold stress response. J. Plant Res. 126, 833–840. doi: 10.1007/s10265-013-0576-0 [DOI] [PubMed] [Google Scholar]
- Gao L., Xiang C.-B. (2008). The genetic locus At1g73660 encodes a putative MAPKKK and negatively regulates salt tolerance in arabidopsis. Plant Mol. Bio l67, 125–134. doi: 10.1007/s11103-008-9306-8 [DOI] [PubMed] [Google Scholar]
- Ghorbel M., Zaidi I., Ebel C., Hanin M. (2019). Differential regulation of the durum wheat MAPK phosphatase 1 by calmodulin, bivalent cations and possibly mitogen activated protein kinase. Plant Physiol. Biochem. 135, 242–252. doi: 10.1016/j.plaphy.2018.12.016 [DOI] [PubMed] [Google Scholar]
- Gill S.-S., Tuteja N. (2010). Reactive oxygen species and antioxidant machinery in abiotic stress tolerance in crop plants. Plant Physiol. Biochem. 48 (12), 909–930. doi: 10.1016/j.plaphy.2010.08.016 [DOI] [PubMed] [Google Scholar]
- Gomi K., Ogawa D., Katou S., Kamada H., Nakajima N., Saji H. (2005). A mitogen-activated protein kinase NtMPK4 activated by SIPKK is required for jasmonic acid signaling and involved in ozone tolerance via stomatal movement in tobacco. Plant Cell Physiol. 46, 1902–1914. doi: 10.1093/pcp/pci211 [DOI] [PubMed] [Google Scholar]
- Gu L., Liu Y., Zong X., Liu L., Li D.-P., Li D.-Q. (2010). Overexpression of maize mitogen-activated protein kinase gene, ZmSIMK1 in arabidopsis increases tolerance to salt stress. Mol. Biol. Rep. 37, 4067–4073. doi: 10.1007/s11033-010-0066-6 [DOI] [PubMed] [Google Scholar]
- Gupta M., Sharma P., Sarin N.-B., Sinha A.-K. (2009). Differential response of arsenic stress in two varieties of Brassica juncea l. Chemosphere 74, 1201–1208. doi: 10.1016/j.chemosphere.2008.11.023 [DOI] [PubMed] [Google Scholar]
- Habiba, Xu J., Gad A. G., Luo Y., Fan C., Uddin J. B. G., et al. (2021). Five OsS40 family members are identified as senescence-related genes in rice by reverse genetics approach. Front. Plant Sci. 12, 1865. doi: 10.3389/fpls.2021.701529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haider F.-U., Liqun C., Coulter J.-A., Cheema S.-A., Wu J., Zhang R., et al. (2021). Cadmium toxicity in plants: Impacts and remediation strategies. Ecotoxicol Environ. Saf. 211, 111887. doi: 10.1016/j.ecoenv.2020.111887 [DOI] [PubMed] [Google Scholar]
- Haider S., Raza A., Iqbal J., Shaukat M., Mahmood T. (2022). Analyzing the regulatory role of heat shock transcription factors in plant heat stress tolerance: A brief appraisal. Mol. Biol. Rep. 49, 5771–5785. doi: 10.1007/s11033-022-07190-x [DOI] [PubMed] [Google Scholar]
- Hamel L.-P., Nicole M.-C., Sritubtim S., Morency M.-J., Ellis M., Ehlting J., et al. (2006). Ancient signals: comparative genomics of plant MAPK and MAPKK gene families. Trends Plant Sci. 11, 192–198. doi: 10.1016/j.tplants.2006.02.007 [DOI] [PubMed] [Google Scholar]
- Handayani T., Gilani S.-A., Watanabe K.-N. (2019). Climatic changes and potatoes: How can we cope with the abiotic stresses? Breed Sci. 69, 545–563. doi: 10.1270/jsbbs.19070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He X., Wang C., Wang H., Li L., Wang C. (2020). The function of MAPK cascades in response to various stresses in horticultural plants. Front. Plant Sci. 11, 952. doi: 10.3389/fpls.2020.00952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua Z.-M., Yang X., Fromm M.-E. (2006). Activation of the NaCl and drought-induced RD29A and RD29B promoters by constitutively active arabidopsis MAPKK or MAPK proteins. Plant Cell Environ. 29, 1761–1770. doi: 10.1111/j.1365-3040.2006.01552.x [DOI] [PubMed] [Google Scholar]
- Huang D., Ma M.-N., Wang Q., Zhang M.-X., Jing G.-Q., Li C. (2020). Arbuscular mycorrhizal fungi enhanced drought resistance in apple by regulating genes in the MAPK pathway. Plant Physiol. Biochem. 149, 245–255. doi: 10.1016/j.plaphy.2020.02.020 [DOI] [PubMed] [Google Scholar]
- Iftikhar H., Naveed N., Virk N., Bhatti M.-F., Song F. (2017). In silica analysis reveals widespread presence of three gene families, MAPK, MAPKK and MAPKKK, of the MAPK cascade from crop plants of solanaceae in comparison to the distantly related syntenic species from rubiaceae; coffee. Peer J. 5, e3255. doi: 10.7717/peerj.3255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jammes F., Song C., Shin D., Munemasa S., Takeda K., Gu D., et al. (2009). MAP kinases MPK9 and MPK12 are preferentially expressed in guard cells and positively regulate ROS-mediated ABA signaling. Proc. Natl. Acad. Sci. U.S.A. 106, 20520–20525. doi: 10.1073/pnas.0907205106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janitza P., Ullrich K. K., Quint M. (2012). Toward a comprehensive phylogenetic reconstruction of the evolutionary history of mitogen-activated protein kinases in the plant kingdom. Front. Plant Sci. 3. doi: 10.3389/FPLS.2012.00271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji T., Li S., Huang M., Di Q., Wang X., Wei M. (2017). Overexpression of cucumber phospholipase d alpha gene (CsPLDalpha) in tobacco enhanced salinity stress tolerance by regulating na(+)-k(+) balance and lipid peroxidation. Front. Plant Sci. 8, 499. doi: 10.3389/fpls.2017.00499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang M., Wen F., Cao J., Li P., She J., Chu Z. (2015). Genome-wide exploration of the molecular evolution and regulatory network of mitogenactivated protein kinase cascades upon multiple stresses in brachypodium distachyon. BMC Genomics 16, 228. doi: 10.1186/s12864-015-1452-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonak C., Nakagami H., Hirt H. (2004). Heavy metal stress. activation of distinct mitogen-activated protein kinase pathways by copper and cadmium. Plant Physiol. 136, 3276–3283. doi: 10.1104/pp.104.045724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khalid M.-H.-B., Raza M.-A., Yu H.-Q., Khan I., Sun F.-A., Feng L.-Y., et al. (2019). Expression, subcellular localization, and interactions of CPK family genes in maize. Int. J. Mol. Sci. 20 (24), 6173. doi: 10.3390/ijms20246173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J.-A., Agrawal G.-K., Rakwal R., Han K., Kim K.-N., Yun C.-H., et al. (2003). Molecular cloning and mRNA expression analysis of a novel rice (Oryza sativa l.) MAPK kinase kinase, OsEDR1, an ortholog of arabidopsis AtEDR1, reveal its role in defense/stress signaling pathways and development. Biochem. Biophys. Res. Commun. 300, 868–876. doi: 10.1016/S0006-291X(02)02944-3 [DOI] [PubMed] [Google Scholar]
- Komis G., SŠamajovaí-OvecŠka M., sSŠamaj J. (2018). Cell and developmental biology of plant mitogen-activated protein kinases. Annu. Rev. Plant Bio l69, 237–265. doi: 10.1146/annurev-arplant-042817-040314 [DOI] [PubMed] [Google Scholar]
- Kong X., Pan J., Zhang M., Xing X., Zhou Y., Liu Y. (2011). ZmMKK4, a novel group c mitogen-activated protein kinase kinase in maize (Zea mays), confers salt and cold tolerance in transgenic arabidopsis. Plant Cell Environ. 34, 1291–1303. doi: 10.1111/j.1365-3040.2011.02329.x [DOI] [PubMed] [Google Scholar]
- Kong F., Wang J., Cheng L., Liu S., Wu J., Peng Z., et al. (2012). Genome-wide analysis of the mitogen-activated protein kinase gene family in Solanum lycopersicum . Gene 499, 108–120. doi: 10.1016/j.gene.2012.01.048 [DOI] [PubMed] [Google Scholar]
- Krasensky J., Jonak C. (2012). Drought, salt, and temperature stress-induced metabolic rearrangements and regulatory networks. J. Exp. Bot. 63, 1593–1608. doi: 10.1093/jxb/err460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar K., Rao K.-P., Sharma P., Sinha A.-K. (2008). Differential regulation of rice mitogen activated protein kinase kinase (MKK) by abiotic stress. Plant Physiol. Biochem.(PPB/Societe Francaise physiologie vegetale) 46, 891–897. doi: 10.1016/j.plaphy.2008.05.014 [DOI] [PubMed] [Google Scholar]
- Lee J.-S., Ellis B.-E. (2007). Arabidopsis MAPK phosphatase 2 (MKP2) positively regulates oxidative stress tolerance and inactivates the MPK3 and MPK6 MAPKs. J. Biol. Chem. 282, 250–259. doi: 10.1074/jbc.M701888200 [DOI] [PubMed] [Google Scholar]
- Li Z., Yue H., Xing D. (2012). MAP kinase 6-mediated activation of vacuole processing enzyme modulates heat shock-induced programmed cell death in arabidopsis. New Phytol. 195, 85–96. doi: 10.1111/j.1469-8137.2012.04131.x [DOI] [PubMed] [Google Scholar]
- Lian W.-W., Tang Y.-M., Gao S.-Q., Zhang Z., Zaho X., Chang P., et al. (2012). Phylogenetic analysis and expression patterns of the MAPK gene family in wheat (L.). J. Integr. Agric. 11 (8), 1227–1235. doi: 10.1016/S2095-3119(12)60119-1 [DOI] [Google Scholar]
- Lohani N., Singh M. B., Bhalla P. L. (2020). High temperature susceptibility of sexual reproduction in crop plants. J. Exp. Bot. 71, 555–568. doi: 10.1093/jxb/erz426 [DOI] [PubMed] [Google Scholar]
- Lu F., Li W., Peng Y., Cao Y., Qu J., Sun F., et al. (2022). ZmPP2C26 alternative splicing variants negatively regulate drought tolerance in maize. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.851531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lumbreras V., Vilela B., Irar S., Solé M., Capellades M., Valls M. (2010). MAPK phosphatase MKP2 mediates disease responses in arabidopsis and functionally interacts with MPK3 and MPK6. Plant J. 63, 1017–1030. doi: 10.1111/j.1365-313X.2010.04297.x [DOI] [PubMed] [Google Scholar]
- Ma B., Nian L., Ain N., Liu X., Yang Y., Zhu X., et al. (2022). Genome-wide identification and expression profiling of the SRS gene family in melilotus albus reveals functions in various stress conditions. Plants 11 (22), 3101. doi: 10.3390/plants11223101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahmood T., Khalid S., Abdullah M., Ahmed Z., Shah N.-K.-M., Ghafoor A., et al. (2020). Insights into drought stress signaling in plants and the molecular genetic basis of cotton drought tolerance. Cells 9, 105. doi: 10.3390/cells9010105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathobo R., Marais D., Steyn J.-M. (2017). The effect of drought stress on yield, leaf gaseous exchange and chlorophyll fluorescence of dry beans (Phaseolus vulgaris l.). Agric. Water Manage. 180, 118–125. doi: 10.1016/j.agwat.2016.11.005 [DOI] [Google Scholar]
- Medina J., Catala R., Salinas J. (2016). The CBFs: Three arabidopsis transcription factors to cold acclimate. Plant Sci. 180, 3–11. doi: 10.1016/j.plantsci.2010.06.019 [DOI] [PubMed] [Google Scholar]
- Meng X., Zhang S. (2013). MAPK cascades in plant disease resistance signaling. Annu. Rev. Phytopatho l51, 245–266. doi: 10.1146/annurev-phyto-082712-102314 [DOI] [PubMed] [Google Scholar]
- Mikolajczyk M., Awotunde O.-S., Muszynska G., Klessig D.-F., Dobrowolska G. (2000). Osmotic stress induces rapid activation of a salicylic acid-induced protein kinase and a homolog of protein kinase ASK1 in tobacco cells. Plant Cell. 12, 165–178. doi: 10.2307/3871037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minhas P.-S., Rane J., Pasala R.-K. (2017). “Abiotic stresses in agriculture: An overview,” in Abiotic stress management for resilient agriculture (Singapore: Springer; ). doi: 10.1007/978-981-10-5744-1_1 [DOI] [Google Scholar]
- Mittler R., Zandalinas S. I., Fichman Y., Van Breusegem F. (2022). Reactive oxygen species signalling in plant stress responses. Nat. Rev. Mol. Cell Biol. 23 (10), 663–679. doi: 10.1038/s41580-022-00499-2 [DOI] [PubMed] [Google Scholar]
- Mitula F., Tajdel M., Ciesla A., Kasprowicz-Maluski A., Kulik A., Babula-Skowronska D., et al. (2015). Arabidopsis ABA-activated kinase MAPKKK18 is regulated by protein phosphatase 2C ABI1 and the ubiquitin-proteasome pathway. Plant Cell Physiol. 56, 2351–2367. doi: 10.1093/pcp/pcv146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohamed I.-A.-A., Shalby N., El-Badri M.-A., Batool M., Wang C., Wang Z., et al. (2022). RNA-Seq analysis revealed key genes associated with salt tolerance in rapeseed germination through carbohydrate metabolism, hormone, and MAPK signaling pathways. Ind. Crops Products 176, 114262. doi: 10.1016/j.indcrop.2021.114262 [DOI] [Google Scholar]
- Mohanta T.-K., Arora P.-K., Mohanta N., Parida P., Bae H. (2015). Identification of new members of the MAPK gene family in plants shows diverse conserved domains and novel activation loop variants. BMC Genomics 16 (1), 1–20. doi: 10.1186/s12864-015-1244-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moustafa K., Abu-Qamar S., Jarrar M., Al-Rajab J.-A., Tremouillaux-Guiller J. (2014). MAPK cascades and major abiotic stresses. Plant Cell Rep. 33, 1217–1225. doi: 10.1007/s00299-014-1629-0 [DOI] [PubMed] [Google Scholar]
- Moustafa K., Lefebvre-De Vos D., Leprince A.-S., Savoure´e A., Laurie`re C. (2008). Analysis of the arabidopsis mitogen-activated protein kinase families: Organ specificity and transcriptional regulation upon water stresses. Sch Res. Exch. p, 12. doi: 10.3814/2008/143656 [DOI] [Google Scholar]
- Muhammad T., Zhang J., Ma Y., Li Y., Zhang F., Zhang Y., et al. (2019). Overexpression of a mitogen-activated protein kinase SiMAPK3 positively regulates tomato tolerance to cadmium and drought stress. Molecules 24, 556. doi: 10.3390/molecules24030556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munns R., Passioura J.-B., Colmer T.-D., Byrt C.-S. (2020). Osmotic adjustment and energy limitations to plant growth in saline soil. New Phytol. 225, 1091–1096. doi: 10.1111/nph.15862 [DOI] [PubMed] [Google Scholar]
- Nadarajah K., Sidek H.-M. (2010). The green MAPKs. Asian J. @ Plant Sci. 9, 1–10. doi: 10.3923/ajps.2010.1.10 [DOI] [Google Scholar]
- Nakagami H., Pitzschke A., Hirt H. (2005). Emerging MAP kinase pathways in plant stress signaling. Trends Plant Sci. 10, 339–346. doi: 10.1016/j.tplants.2005.05.009 [DOI] [PubMed] [Google Scholar]
- Nakagami H., Soukupova H., Schikora A., Zarsky V., Hirt H. (2006). ). a mitogen-activated protein kinase kinase kinase mediates reactive oxygen species homeostasis in Arabidopsis . J. Biol. Chem. 281, 38697–38704. doi: 10.1074/jbc.M605293200 [DOI] [PubMed] [Google Scholar]
- Ning J., Li X., Hicks L.-M., Xiong L. (2010). A raf-like MAPKKK gene DSM1 mediates drought resistance through reactive oxygen species scavenging in rice. Plant Physiol. 152, 876–890. doi: 10.1104/pp.109.149856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osthoff A., Rose P.-D.-D., Baldauf J.-A., Piepho H.-P., Hochholdinger F. (2019). Transcriptomic reprogramming of barley seminal roots by combined water deficit and salt stress. BMC Genomics 20, 325. doi: 10.1186/s12864-019-5634-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng L.-X., Gu L.-K., Zheng C.-C., Li D.-Q., Shu H.-R. (2006). Expression of MaMAPK gene in seedlings of malus l. under water stress. Acta Biochim. Biophys. Sin. 38, 281–286. doi: 10.1093/abbs/38.4.281 [DOI] [PubMed] [Google Scholar]
- Pitzschke A., Datta S., Persak H. (2014). Salt stress in arabidopsis: Lipid transfer protein AZI1 and its control by mitogen-activated protein kinase MPK3. Mol. Plant 7, 722–738. doi: 10.1093/mp/sst157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitzschke A., Hirt H. (2009). Disentangling the complexity of mitogen-activated protein kinases and reactive oxygen species signaling. Plant Physiol. 149, 606–615. doi: 10.1104/pp.108.131557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popescu S.-C., Popescu G.-V., Bachan S., Zhang Z., Gerstein M., Snyder M., et al. (2009). MAPK target networks in arabidopsis thaliana revealed using functional protein microarrays. Genes Dev. 23, 80–92. doi: 10.1101/gad.1740009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu J.-L., Zhou L., Yun B.-W., Nielsen H.-B., Fiil B.-K., Petersen K. (2008). Arabidopsis mitogen activated protein kinase kinases MKK1 and MKK2 have overlapping functions in defense signaling mediated by MEKK1 MPK4, and MKS1. Plant Physiol. 148, 212–222. doi: 10.1104/pp.108.120006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raina S.-K., Wankhede D.-P., Sinha A.-K. (2013). Catharanthus roseus mitogen-activated protein kinase 3 confers UV and heat tolerance to saccharomyces cerevisiae. Plant Signal Behav. J. 8, e22716. doi: 10.4161/psb.22716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao K.-P., Vani G., Kumar K., Wankhede D.-P., Mishra M., Gupta M., et al. (2010). Arsenic stress activates MAP kinase in rice roots and leaves. Arch. Biochem. Biophys. 506, 73–82. doi: 10.1016/j.abb.2010.11.006 [DOI] [PubMed] [Google Scholar]
- Raza A. (2021). Eco-physiological and biochemical responses of rapeseed (Brassica napus l.) to abiotic stresses: Consequences and mitigation strategies. J. Plant Growth Regul. 40 (4), 1368–1388. doi: 10.1007/s00344-020-10231-z [DOI] [Google Scholar]
- Raza A., Charagh S., García-Caparrós P., Rahman M. A., Ogwugwa V. H., Saeed F., et al. (2022. c). Melatonin-mediated temperature stress tolerance in plants. GM Crops Food 13 (1), 196–217. doi: 10.1080/21645698.2022.2106111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raza A., Mubarik M. S., Sharif R., Habib M., Jabeen W., Zhang C., et al. (2022. a). Developing drought-smart, ready-to-grow future crops. Plant Genome 10, e20279. doi: 10.1002/tpg2.20279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raza A., Salehi H., Rahman M. A., Zahid Z., Madadkar Haghjou M., Najafi-Kakavand S., et al. (2022. b). Plant hormones and neurotransmitter interactions mediate antioxidant defenses under induced oxidative stress in plants. Front. Plant Sci. 13, 961872. doi: 10.3389/fpls.2022.961872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raza A., Tabassum J., Fakhar A. Z., Sharif R., Chen H., Zhang C., et al. (2022. d). Smart reprograming of plants against salinity stress using modern biotechnological tools. Crit. Rev. Biotechnol. 12, 1–28. doi: 10.1080/07388551.2022.2093695 [DOI] [PubMed] [Google Scholar]
- Raza A., Tabassum J., Kudapa H., Varshney R. K. (2021). Can omics deliver temperature resilient ready-to-grow crops? Crit. Rev. Biotechnol. 41 (8), 1209–1232. doi: 10.1080/07388551.2021.1898332 [DOI] [PubMed] [Google Scholar]
- Raza A., Tabassum J., Zahid Z., Charagh S., Bashir S., Barmukh R., et al. (2022. e). Advances in “Omics” approaches for improving toxic Metals/Metalloids tolerance in plants. Front. Plant Sci. 12, 794373. doi: 10.3389/fpls.2021.794373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rengasamy P. (2006). World salinization with emphasis on Australia. J. Exp. Bot. 57, 1017–1023. doi: 10.1093/jxb/erj108 [DOI] [PubMed] [Google Scholar]
- Reyna N.-S., Yang Y. (2006). Molecular analysis of the rice MAP kinase gene family in relation to Magnaporthe grisea infection. Mol. Plant-Microbe Interact. 19 (5), 530–540. doi: 10.1094/MPMI-19-0530 [DOI] [PubMed] [Google Scholar]
- Ritchie H., Roser M. (2020). Environmental impacts of food production. Our World Data. [Google Scholar]
- Rivero R. M., Mittler R., Blumwald E., Zandalinas S. I. (2022). Developing climate-resilient crops: Improving plant tolerance to stress combination. Plant J. 109 (2), 373–389. doi: 10.1111/tpj.15483 [DOI] [PubMed] [Google Scholar]
- Rodriguez M.-C., Petersen M., Mundy J. (2010). Mitogen-activated protein kinase signaling in plants. Annu. Rev. Plant Biol. 61, 621–649. doi: 10.1146/annurev-arplant-042809-112252 [DOI] [PubMed] [Google Scholar]
- SŠamajovaí O., Plıíhal O., Al-Yousif M., Hirt H., SŠamaj J. (2013). Improvement of stress tolerance in plants by genetic manipulation of mitogen-activated protein kinases. Biotechnol. Adv. 31, 118–128. doi: 10.1016/j.biotechadv.2011.12.002 [DOI] [PubMed] [Google Scholar]
- Sangwan V., Orvar B.-L., Beyerly J., Hirt H., Dhindsa R.-S. (2002). Opposite changes in membrane fluidity mimic cold and heat stress activation of distinct plant MAP kinase pathways. Plant J. Cell Mol. Biol. 31, 629–638. doi: 10.1046/j.1365-313X.2002.01384.x [DOI] [PubMed] [Google Scholar]
- Sattar F., Hamooh B., Wellman G., Ali A., Shah S., Anwar Y., et al. (2021). Growth and biochemical responses of potato cultivars under In vitro lithium chloride and mannitol simulated salinity and drought stress. Plants 10, 924. doi: 10.3390/plants10050924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao J.-F., Xia J., Yamaji N., Shen R.-F., Ma J.-F. (2018). Effective reduction of cadmium accumulation in rice grain by expressing OsHMA3 under the control of theOsHMA2 promoter. J. Exp. Bot. 69 (10), 2743e–22752. doi: 10.1093/jxb/ery107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi J., An H.-L., Zhang L., Gao Z., Guo X.-Q. (2010). GhMPK7, a novel multiple stress-responsive cotton group c MAPK gene, has a role in broad spectrum disease resistance and plant development. Plant Mol. Biol. 74, 1–17. doi: 10.1007/s11103-010-9661-0 [DOI] [PubMed] [Google Scholar]
- Shi J., Zhang L., An H., Wu C., Guo X. (2011). GhMPK16, a novel stress responsive group d MAPK gene from cotton, is involved in disease resistance and drought sensitivity. BMC Mol. Biol. 12, S22. doi: 10.1186/1471-2199-12-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shu P., Li Y., Li Z., Sheng J., Shen L. (2022). SlMAPK3 enhances tolerance to salt stress in tomato plants by scavenging ROS accumulation and up-regulating the expression of ethylene signaling related genes. Environ. Exp. Bot. 193, 104698. doi: 10.1016/j.envexpbot.2021.104698 [DOI] [Google Scholar]
- Singh P., Ara H., Tayyeba S., Pandey C., Sinha A.-K. (2019). Development of efficient protocol for rice transformation overexpressing MAP kinase and their effect on root phenotypic traits. Protoplasma [J] 256, 997–1011. doi: 10.1007/s00709-019-01359-1 [DOI] [PubMed] [Google Scholar]
- Sinha K.-A., Jaggi M., Raghuram B., Tuteja N. (2011). Mitogen-activated protein kinase signaling in plants under abiotic stress. Plant Signaling Behav. 6, 2. doi: 10.4161/psb.6.2.14701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun M., Xu Y., Huang J., Jiang Z., Shu H., Wang H., et al. (2017). Global identification, classification, and expression analysis of MAPKKK genes: Functional characterization of MdRaf5 reveals evolution and drought-responsive profile in apple. Sci. Rep. 7, 13511. doi: 10.1038/s41598-017-13627-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun F., Yu H., Qu J., Cao Y., Ding L., Feng W., et al. (2020). Maize ZmBES1/BZR1-5 decreases ABA sensitivity and confers tolerance to osmotic stress in transgenic arabidopsis. Int. J. Mol. Sci. 21 (3), 996. doi: 10.3390/ijms21030996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suri S.-S., Dhindsa R.-S. (2008). A heat-activated MAP kinase (HAMK) as a mediator of heat shock response in tobacco cells. Plant Cell Environ. 3, 218–226. doi: 10.1111/j.1365-3040.2007.01754.x [DOI] [PubMed] [Google Scholar]
- Taj G., Agarwal P., Grant M., Kumar A. (2010). MAPK machinery in plants: recognition and response to different stresses through multiple signal transduction pathways. Plant Signal Behav. 5, 1370–1378. doi: 10.4161/psb.5.11.13020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tajdel M., Mituła F., Ludwików A. (2016). Regulation of arabidopsis MAPKKK18 by ABI1 and SnRK2, components of the ABA signaling pathway. Plant Signal. Behav. 11, e39277. doi: 10.1080/15592324.2016.1139277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi F., Mizoguchi T., Yoshida R., Ichimura K., Shinozaki K. (2011). Calmodul independent activation of MAP kinase for ROS homeostasis in arabidopsis. Mol. Cell 41, 649–660. doi: 10.1016/j.molcel.2011.02.029 [DOI] [PubMed] [Google Scholar]
- Teige M., Scheikl E., Eulgem T., Doczi R., Ichimura K., Shinozaki K., et al. (2004). The MKK2 pathway mediates cold and salt stress signaling in arabidopsis. Mol. Cell 15, 141–152. doi: 10.1016/j.molcel.2004.06.023 [DOI] [PubMed] [Google Scholar]
- Tuteja N. (2007). Mechanisms of high salinity tolerance in plants. Methods Enzymo l428, 419–438. doi: 10.1016/S0076-6879(07)28024-3 [DOI] [PubMed] [Google Scholar]
- Ueno D., Yamaji N., Kono I., Huang C.-F., Ando T., Yano M., et al. (2010). Gene limiting cadmium accumulation in rice. Proc. Natl. Acad. Sci. Unit States Am. 107 (38), 16500e–116505. doi: 10.1073/pnas.1005396107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uraguchi S., Kamiya T., Sakamoto T., Kasai K., Sato Y., Nagamura Y., et al. (2011). Low-affinity cation transporter (OsLCT1) regulates cadmium transport into rice grains. Proc. Natl. Acad. Sci. Unit States Am. 108 (52), 20959e–220964. doi: 10.1073/pnas.1116531109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Ding H., Zhang A., Ma F., Cao J., Jiang M. (2010). A novel mitogen-activated protein kinase gene in maize (Zea mays), ZmMPK3, is involved in response to diverse environmental cues. J. Integr. Plant Biol. 52 (5), 442–452. doi: 10.1111/j.1744-7909.2010.00906.x [DOI] [PubMed] [Google Scholar]
- Wang C., Guo H., He X., Zhang S., Wang J., Wang L. (2019). Scaffold protein GhMORG1 enhances the resistance of cotton to fusarium oxysporum by facilitating the MKK6-MPK4 cascade. Plant Biotechnol. J. 18, 421–1433. doi: 10.1111/pbi.13307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G., Lovato A., Polverari A., Wang M., Liang Y.-H., Ma Y.-C. (2014). Genome-wide identification and analysis of mitogen activated protein kinase kinase kinase gene family in grapevine (Vitis vinifera). BMC Plant Biol. 14, 219. doi: 10.1186/s12870-014-0219-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C., Lu W., He X., Wang F., Zhou Y., Guo X., et al. (2016). The cotton Mitogen-Activated Protein Kinase Kinase Functions in drought tolerance by regulating stomatal responses and root growth. Plant Cell Physiol. 57, 8, 1629–1642. doi: 10.1093/pcp/pcw090 [DOI] [PubMed] [Google Scholar]
- Wang M., Zhang Y., Wang J., Wu X., Guo X. (2007). A novel MAP kinase gene in cotton (Gossypium hirsutum l.), GhMAPK, is involved in response to diverse environmental stresses. J. Biochem. Mol. Bio l40, 325–332. doi: 10.5483/BMBRep.2007.40.3.325 [DOI] [PubMed] [Google Scholar]
- Wei C., Liu X., Long D., Guo Q., Fang Y., Bian C. (2014). Molecular cloning and expression analysis of mulberry MAPK gene family. Plant Physiol. Biochem. 77, 108–116. doi: 10.1016/j.plaphy.2014.02.002 [DOI] [PubMed] [Google Scholar]
- Wen J.-Q., Oono K., Imai R. (2002). Two novel mitogen-activated protein signaling components, OsMEK1 and OsMAP1, are involved in a moderate low-temperature signaling pathway in rice. Plant Physiol. .129, 1880–1891. doi: 10.1104/pp.006072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu T., Kong X.-P., Zong X.-J., Li D.-P., Li D.-Q. (2011). Expression analysis of five maize MAP kinase genes in response to various abiotic stresses and signal molecules. Mol. Biol. Rep. 38, 3967–3975. doi: 10.1007/s11033-010-0514-3 [DOI] [PubMed] [Google Scholar]
- Xie G., Kato H., Imai R. (2012). Biochemical identification of the OsMKK6-OsMPK3 signaling pathway for chilling stress tolerance in rice. Biochem. J. 443, 95–102. doi: 10.1042/BJ20111792 [DOI] [PubMed] [Google Scholar]
- Xing Y., Chen W.-H., Jia W., Zhang J. (2015). Mitogen-activated protein kinase kinase 5 (MKK5)-mediated signaling cascade regulates expression of iron superoxide dismutase gene in arabidopsis under salinity stress. J. Exp. Bot. 66, 5971–5981. doi: 10.1093/jxb/erv305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing Y., Jia W., Zhang J. (2007). AtMEK1 mediates stress induced gene expression of CAT1 catalase by triggering H2O2 production in arabidopsis. J. Exp. Bot. 58, 2969–2981. doi: 10.1093/jxb/erm144 [DOI] [PubMed] [Google Scholar]
- Xing Y., Jia W., Zhang J. (2008). AtMKK1 mediates ABA-induced CAT1 expression and H2O2 production via AtMPK6-coupled signaling in Arabidopsis . Plant J. 54, 440–451. doi: 10.1111/j.1365-313X.2008.03433.x [DOI] [PubMed] [Google Scholar]
- Xiong L., Yang Y. (2003). Disease resistance and abiotic stress tolerance in rice are inversely modulated by an abscisic acid-inducible mitogen-activated protein kinase. Plant Cell 15, 745–759. doi: 10.1105/tpc.008714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J., Chua N.-H. (2012). Dehydration stress activates arabidopsis MPK6 to signal DCP1 phosphorylation. EMBO J. 31, 1975–1984. doi: 10.1038/emboj.2012.56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H., Li K., Yang F., Shi Q., Wang X. (2010). Overexpression of CsNMAPK in tobacco enhanced seed germination under salt and osmotic stresses. Mol. Biol. Rep. 37, 3157–3163. doi: 10.1007/s11033-009-9895-6 [DOI] [PubMed] [Google Scholar]
- Yan Y., Wang L., Ding Z., Tie W., Ding X., Zeng C. (2016). Genome-wide identification and expression analysis of the mitogen-activated protein kinase gene family in cassava. Front. Plant Sci. 7, 1294. doi: 10.3389/fpls.2016.01294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanagawa Y., Yoda H., Osaki K., Amano Y., Aono M., Seo S. (2017). Mitogen-activated protein kinase 4-like carrying an MEY motif instead of a TXY motif is involved in ozone tolerance and regulation of stomatal closure in tobacco. J. Exp. Bot. 67, 3471–3479. doi: 10.1093/jxb/erw173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang C., Zhang Y., Huang C. (2019). Reduction in cadmium accumulation in japonica rice grains by CRISPR/Cas9-mediated editing of OsNRAMP5. J. Integr. Agric. 18 (3), 688–697. doi: 10.1016/S2095-3119(18)61904-5 [DOI] [Google Scholar]
- Yang J.-W., Zhu X., Li S.-G., Tang X., Zhang N., Si H.-J. (2019). A novel potato microRNA stu-miR856 regulates mitogen-activated protein kinase genes contributing to drought tolerance. Biol. plantarum 63, 618–626. doi: 10.32615/bp.2019.067 [DOI] [Google Scholar]
- Yaqoob H., Tariq A., Bhat B. A., Bhat K. A., Nehvi I. B., Raza A., et al. (2023). Integrating genomics and genome editing for orphan crop improvement: A bridge between orphan crops and modern agriculture system. GM Crops Food 14 (1), 1–20. doi: 10.1080/21645698.2022.2146952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh C.-M., Hsiao L.-J., Huang H.-J. (2004). Cadmium activates a mitogen-activated protein kinase gene and MBP kinases in rice. Plant Cell Physiol. l45, 1306–1312. doi: 10.1093/pcp/pch135 [DOI] [PubMed] [Google Scholar]
- Yin Y.-X., Guo W.-L., Zhang Y.-L., Ji J.-J., Xiao H.-J., Yan F. (2014). Cloning and characterization of a pepper aquaporin, CaAQP, which reduces chilling stress in transgenic tobacco plants. Plant Cell Tissue Organ Cult 118, 431–444. doi: 10.1007/s11240-014-0495-3 [DOI] [Google Scholar]
- Yu L., Nie J., Cao C., Jin Y., Yan M., Wang F., et al. (2010). Phosphatidic acid mediates salt stress response by regulation of MPK6 in Arabidopsis thaliana. New Phytol. 188, 762–773. doi: 10.1111/j.1469-8137.2010.03422.x [DOI] [PubMed] [Google Scholar]
- Yu H. Q., Sun F. A., Feng W. Q., Lu F. Z., Li W. C., Fu F. L. (2019). The BES1/BZR1 transcription factors regulate growth, development and stress resistance in plants. Hereditas 41 (3), 206–214. doi: 10.16288/j.yczz.18-253 [DOI] [PubMed] [Google Scholar]
- Yu L., Yan J., Yang Y., Zhu W. (2015). Overexpression of tomato mitogenactivated protein kinase SlMPK3 in tobacco increases tolerance to low temperature stress. Plant Cell Tissue Organ Cult. 121, 21–34. doi: 10.1007/s11240-014-0675-1 [DOI] [Google Scholar]
- Zaheer K., Akhtar M.-H. (2016). Potato production, usage, and nutrition–a review. Crit. Rev. Food Sci. Nutr. 56, 711–721. doi: 10.1080/10408398.2012.724479 [DOI] [PubMed] [Google Scholar]
- Zaıdi I., Ebel C., Touzri M., Herzog E., Evrard J.-L., Schmit A.-C., et al. (2010). TMKP1 is a novel wheat stress responsive MAP kinase phosphatase localized in the nucleus. Plant Mol. Biol. 73, 325–338. doi: 10.1007/s11103-010-9617-4 [DOI] [PubMed] [Google Scholar]
- Zandalinas S.-I., Fritschi F.-B., Mittler R. (2019). Signal transduction networks during stress combination. J. Exp. Bot. 71, 1734–1741. doi: 10.1093/jxb/erz486 [DOI] [PubMed] [Google Scholar]
- Zaynab M., Hussain A., Sharif Y., Fatima M., Sajid M., Rehman N., et al. (2021). Mitogen-activated protein kinase expression profiling revealed its role in regulating stress responses in potato (Solanum tuberosum). Plants 10, 1371. doi: 10.3390/plants10071371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang A., Jiang M., Zhang J., Tan M., Hu X. (2006). Mitogen-activated protein kinase is involved in abscisic acid-induced antioxidant defense and acts downstream of reactive oxygen species production in leaves of maize plants. Plant Physiol. 141, 475–487. doi: 10.1104/pp.105.075416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z., Li J., Li F., Liu H., Yang W., Chong K., et al. (2017). OsMAPK3 phosphorylates OsbHLH002/OsICE1 and inhibits its ubiquitination to activate OsTPP1 and enhances rice chilling tolerance. Dev. Cell J. 43, 731–743. doi: 10.1016/j.devcel.2017.11.016 [DOI] [PubMed] [Google Scholar]
- Zhang H., Li Y., Zhu J.-K. (2018). Developing naturally stress-resistant crops for a sustainable agriculture. Nat. Plants 4, 989–996. doi: 10.1038/s41477-018-0309-4 [DOI] [PubMed] [Google Scholar]
- Zhang T., Liu Y., Xue L., Xu S., Chen T., Yang T. (2006). Molecular cloning and characterization of a novel MAP kinase gene in chorispora bungeana. Plant Physiol. Biochem. 44, 78–84. doi: 10.1016/j.plaphy.2006.01.001 [DOI] [PubMed] [Google Scholar]
- Zhang M., Su J., Zhang Y., Xu J., Zhang S. (2018). Conveying endogenous and exogenous signals: MAPK cascades in plant growth and defense. Curr. Opin. Plant Biol. 45, 1–10. doi: 10.1016/j.pbi.2018.04.012 [DOI] [PubMed] [Google Scholar]
- Zhang Z., Wu Y., Gao M., Zhang J., Kong Q., Liu Y. (2012). Disruption of PAMP-induced MAP kinase cascade by a pseudomonas syringae effector activates plant immunity mediated by the NB-LRR protein SUMM2. Cell Host Microbe 11, 253–263. doi: 10.1016/j.chom.2012.01.015 [DOI] [PubMed] [Google Scholar]
- Zhang L., Xi D., Li S., Gao Z., Zhao S., Shi J., et al. (2011). A cotton group c MAP kinase gene, GhMPK2, positively regulates salt and drought tolerance in tobacco. Plant Mol. Biol. 77, 17–31. doi: 10.1007/s11103-011-9788-7 [DOI] [PubMed] [Google Scholar]
- Zhang S., Xu R., Luo X., Jiang Z., Shu H. (2013). Genome-wide identification and expression analysis of MAPK and MAPKK gene family in malus domestica. Gene 531, 377–387. doi: 10.1016/j.gene.2013.07.107 [DOI] [PubMed] [Google Scholar]
- Zhang A., Zhang J., Ye N., Cao J., Tan M., Zhang J. (2010). ZmMPK5 is required for the NADPH oxidase-mediated self-propagation of apoplastic H2O2 in brassino steroid induced antioxidant defense in leaves of maize. J. Exp. Bot. 61, 4399–4411. doi: 10.1093/jxb/erq243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y., Wang D., Zhang Y., Niu Y., Zong X., Ma Y., et al. (2018). Cloning and characterization of a mitogen-activated protein kinase gene 84KMPK14 in hybrid poplar (Populus alba × P. glandulosa cv. “84K”). Am. J. Plant Sci. 9, 2567–2579. doi: 10.4236/ajps.2018.913186 [DOI] [Google Scholar]
- Zhou H., Ren S., Han Y., Zhang Q., Qin L., Xing Y. (2017). Identification and analysis of mitogen-activated protein kinase (MAPK) cascades in Fragaria vesca . Int. J. Mol. Sci. 18, 1766. doi: 10.3390/ijms18081766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J., Xia X.-J., Zhou Y.-H., Shi K., Chen Z., Yu J.-Q. (2014). RBOH1- dependent H2O2 production and subsequent activation of MPK1/2 play an important role in acclimation-induced cross-tolerance in tomato. J. Exp. Bot. 65, 595–607. doi: 10.1093/jxb/ert404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J.-K. (2016). Abiotic stress signaling and responses in plants. Cell 167, 313–324. doi: 10.1016/j.cell.2016.08.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu X., Zhang N., Liu X., Wang X., Li S., Yang J., et al. (2020). StMAPK3 controls oxidase activity, photosynthesis and stomatal aperture under salinity and osmosis stress in potato. Plant Phy Biochem. 156, 157–167. doi: 10.1016/j.plaphy.2020.09.012 [DOI] [PubMed] [Google Scholar]
- Zhu X., Zhang N., Liu X., Yang J., Hong X., Wang F., et al. (2021). Mitogen activated protein kinase 11 (MAPK11) maintains growth and photosynthesis of potato plant under drought condition. Plant Cell Rep. 40, 491–506. doi: 10.1007/s00299-020-02645-6 [DOI] [PubMed] [Google Scholar]
- Zong X.-J., Li D.-P., Gu L.-K., Li D.-Q., Liu L.-X., Hu X.-L. (2009). Abscisic acid and hydrogen peroxide induce a novel maize group c MAP kinase gene, ZmMPK7, which is responsible for the removal of reactive oxygen species. Planta 229, 485–495. doi: 10.1007/s00425-008-0848-4 [DOI] [PubMed] [Google Scholar]