Abstract
In principle, the generation, transmission, and dissipation of supercoiling forces are determined by the arrangement of the physical barriers defining topological boundaries and the disposition of enzymes creating (polymerases and helicases, etc.) or releasing (topoisomerases) torsional strain in DNA. These features are likely to be characteristic for individual genes. By using topoisomerase inhibitors to alter the balance between supercoiling forces in vivo, we monitored changes in the basal transcriptional activity and DNA conformation for several genes. Every gene examined displayed an individualized profile in response to inhibition of topoisomerase I or II. The expression changes elicited by camptothecin (topoisomerase I inhibitor) or adriamycin (topoisomerase II inhibitor) were not equivalent. Camptothecin generally caused transcription complexes to stall in the midst of transcription units, while provoking little response at promoters. Adriamycin, in contrast, caused dramatic changes at or near promoters and prevented transcription. The response to topoisomerase inhibition was also context dependent, differing between chromosomal or episomal c-myc promoters. In addition to being well-characterized DNA-damaging agents, topoisomerase inhibitors may evoke a biological response determined in part from transcriptional effects. The results have ramifications for the use of these drugs as antineoplastic agents.
Transcription, replication, recombination, DNA repair, and DNA compaction generate torsional stress in prokaryotic and eukaryotic chromosomes and episomes. This stress must either be accommodated by conformational changes in DNA structure (e.g., supercoils) or else dissipated. If not dissipated, high levels of torsional stress can halt RNA polymerase and deform chromosomal structure (4). Torsional stress may be dissipated by rotation of a free DNA end, i.e., chromosome termini or strand breaks. Alternatively, stresses accumulating within topological domains may be dissipated by topoisomerases. A topological domain is formed whenever both ends of an intact DNA segment are restricted from rotating relative to each other. The boundaries of these domains may be delimited by DNA loops via protein-protein interactions or tethering of DNA to an immobile matrix or scaffold. The energy required to rotate a large, free-ended DNA segment with bound proteins through a viscous medium may become so great that torsional strain accumulates within a pseudo-domain bounded at one end by a kinetic barrier (40). Topological microdomains may be nested within larger and larger macrodomains (24, 70). These domains may be short-lived or stable, depending on the nature of the particular protein-protein and protein-nucleic acid interactions creating their boundaries. A loop formed between a DNA-bound factor and a complex tracking along and around the double helix, such as RNA polymerase II, creates a mobile boundary. Little is known about the arrangement, interlinks, and transmission of torsional stress between topological domains in vivo. It is likely that the influence of DNA topology on genetic transactions may be determined by the architecture and arrangement of cis elements and factors governing the distribution of torsional stress.
Topoisomerases I and II relieve torsional strain incrementally within a domain by using controlled breakage of one or both strands, respectively; passage of DNA through the strand break; and reunion (73). Adjacent domains are not affected. The efficiency of topoisomerase is modified by domain size, binding site preference, and site accessibility. The intranuclear distribution of topoisomerase is not known. A large fraction of topoisomerase II is bound by the nuclear matrix and so is available only to local DNA sequences (13, 46). The packaging of DNA into chromatin restrains approximately one negative-supercoil on the surface of each nucleosome (51). This packaging may hinder the operation of topoisomerases and delay the relief or transmission of torsional strain (55). Inhibitors of topoisomerases I and II freeze these enzymes as protein-DNA complexes at various steps in their reaction pathways (31, 49). Topoisomerase-DNA-inhibitor complexes (cleavable complexes) are poisoned and are unable to execute a complete enzymatic cycle. Topoisomerase-DNA covalent adducts are converted into DNA strand breaks upon protein removal. The topological state of the domains encompassing these frozen complexes remains fixed; even in cleavable complexes torsional strain is not liberated until the topoisomerase subunits covalently coupled to the DNA ends dissociate, allowing the ends to rotate independently. Topoisomerase inhibitors have proven to be potent antineoplastic agents. The efficacy of these agents for cancer therapy is explained only in part by their ability to damage DNA. The response of individual genes to topoisomerase inhibitors may result directly from enzyme inhibition or may arise through secondary mechanisms.
Structural considerations dictate that no global generalizations summarize the role of DNA topology for the regulation of any given gene. The microarchitecture of matrix attachments, protein-protein-mediated loops, the arrangement of promoter sites, and the disposition of topoisomerases and nucleosomes all mold the physiological or pathological response of a transcription unit. The expression of the c-myc gene is particularly sensitive to perturbations of its normal chromosomal milieu. Translocations, regional amplifications, and viral insertions and mutations, sometimes at vast distances either 5′ or 3′ from the c-myc promoters, all deregulate c-myc transcription (28, 36, 50). Although topoisomerase inhibitors influence c-myc expression (2, 6, 16, 19, 43, 48, 56, 57, 58), it is unknown whether this results from perturbation of c-myc DNA and chromatin structure driven by changes in the localization and levels of torsional strain or whether this results from indirect effects. If c-myc transcription is sensitive to torsional strain, then changes in c-myc mRNA levels, promoter structure, and the distribution of RNA polymerase II molecules along the gene should all respond to topoisomerase inhibitors. Moreover, because every gene is subject to unique structural and physical constraints, the features controlling the expression of a particular transcription unit should be characteristically altered in response to manipulations perturbing supercoiling.
To explore the relationship between torsional strain and gene activity, the features of c-myc, c-fos, hsp70, gapdh, and rRNA transcription units were examined in response to the inhibition of topoisomerase I and/or II. A polymorphous pattern of activation or repression was observed for these genes. Redistribution of transcriptionally engaged RNA polymerases and structural changes in and around promoters were also noted. Notably, the response of the c-myc promoter was modified when carried as a stable episome. These results indicate that the interplay between torsional strain and chromatin architecture helps to define the expression profiles of many genes.
MATERIALS AND METHODS
Plasmids.
The vector pREP9/CAT (Invitrogen) encoding chloramphenicol acetyltransferase (CAT) was cut with XbaI and NotI (blunted); an XbaI-HindIII fragment containing five GAL4 DNA binding sites as well as the human c-myc promoter from the HindIII site at nucleotide 1 to SexAI (blunted) at position 2882 (accession number X00364) was inserted. This resulting plasmid, pMYC/CAT, contains the Epstein-Barr virus (EBV) DNA replication origin P1 and the EBNA1 gene and so is maintained as a circular episome.
Tissue culture.
Raji cells were grown in RPMI with 10% fetal calf serum at a density of <106/ml. Raji (5 × 106 cells) were electroporated with the pMYC/CAT (190 V and 1180 μF; BRL Cell Porator). The transfected cells were selected with 1 mg of G418 (BRL)/ml. The pool of stable transfectants, called Raji pMYC/CAT, was maintained in medium containing 0.5 mg of G418/ml.
Chemicals.
G418 (BRL) was dissolved in 100 mM HEPES (pH 7.3) at 100 mg/ml and stored at 4°C. Camptothecin (Sigma) was dissolved and diluted in dimethyl sulfoxide (DMSO) at 1, 5, or 10 mM. Adriamycin, also called doxorubicin, (Sigma) was dissolved and diluted in H2O at 1, 5, or 10 mM. Sodium butyrate (Sigma) was dissolved in H2O at 3 M. Trichostatin A (Sigma) was dissolved in DMSO at 500 ng/μl. α-Amanitin (CalBiochem) was dissolved in H2O at 1 mg/ml. All solutions except G418 were stored at –20°C. For these experiments, 1 μl of the camptothecin or trichostatin A stock was added per ml of medium. When appropriate, 0.1% DMSO was added to control samples and to the adriamycin and sodium butyrate samples to ensure comparability.
RNase protection.
A total of 3 × 107 cells in 60 ml of medium were incubated with DMSO, camptothecin, adriamycin, sodium butyrate, or trichostatin A at the concentrations indicated. After 4 h of drug treatment, cells were pelleted and resuspended in 3 ml of Trizol (BRL), and RNA was isolated according to the manufacturer's protocol (BRL). The RNA was treated with RNase-free DNase I, phenol-chloroform extracted, ethanol precipitated, washed with 75% ethanol, air dried, resuspended in 100 μl of H2O, and stored at –80°C. Then, 10 μg of total RNA was resuspended in Hybridization Buffer (PharMingen RPA Kit) and hybridized overnight with gel-purified RNA probes. After RNase and proteinase K treatments, the samples were separated on 5% acrylamide–bisacrylamide (29:1) gels with 50% urea, dried, and autoradiographed.
The gapdh template for the RNA probe was purchased from PharMingen. The pSP72/hsp70RNPA template for the hsp70 RNA probe contains the 543-bp SmaI fragment at the 3′ end of the hsp70 coding sequence. The CAT, c-myc exon 2, c-fos, and rRNA templates used to synthesize RNA probes were created by inserting gene-specific PCR fragments into the pGEM-T Easy vector (Promega). The CAT and c-myc exon 2 probes were specific for episomal or endogenous c-myc promoter-driven transcripts, respectively. For a complete list of the oligonucleotides used, see supplemental material (www-dcs.nih.gov/branches/lop /Research/genreg/levens/html). To obtain signals of comparable intensity, the RNA probes were transcribed by T7 RNA polymerase by using different ratios of radioactive and cold UTP as follows: CAT and c-fos 1 radioactive UTP out of 10 total UTPs, hsp70 and c-myc 1 radioactive UTP out of 100 total UTPs, and rRNA and gapdh 1 radioactive UTP out of 500 total UTPs.
Nuclear run-on.
A total of 4 × 107 cells in 80 ml of medium were incubated with DMSO, camptothecin, adriamycin, sodium butyrate, or trichostatin A at the indicated concentrations. These chemicals were not included in phosphate-buffered saline (PBS), Lysis Buffer, or Glycerol Buffer. Nuclear isolation and nuclear run-on were as described previously (11). Nuclei were resuspended in Glycerol Buffer (80 μl) and frozen in liquid nitrogen. For run-on experiments, 150 μCi of [α-32P]UTP (800 Ci/mmol) was included in a 200-μl reaction volume. After a 10-min incubation at 30°C, 10 μl of a 10-U/μl solution of DNase I (Roche) was added for another 10 min; then, 20 μl of the sodium dodecyl sulfate-Proteinase K Stop Solution was added, followed by incubation at 42°C for 30 min. After phenol-chloroform and ethanol precipitation, the DNase treatment was repeated. The RNA was passed over a G25 Sephadex spin column (Roche) and then hybridized in Church Buffer for 36 to 48 h at 65°C with Hybond N+ membranes (Amersham) loaded with 1 μg each of antisense DNA oligonucleotides corresponding to the genes of interest. For a complete list of the oligonucleotides used, see supplemental material (www-dcs.nih.gov/branches/lop/Research/genreg/levens/html). Exposures at –80°C of Kodak BioMax MS film with the transcreen HE are shown (Fig. 2A is 14 days; Fig. 2B is 7 days; Fig. 3A is 9 h for rRNA and 10 days for the Raji cells and the Raji pMYC/CAT cells; Fig. 3B is 1 day for rRNA, 12 days for RAJI cells, and 14 days for the Raji pMYC/CAT cells).
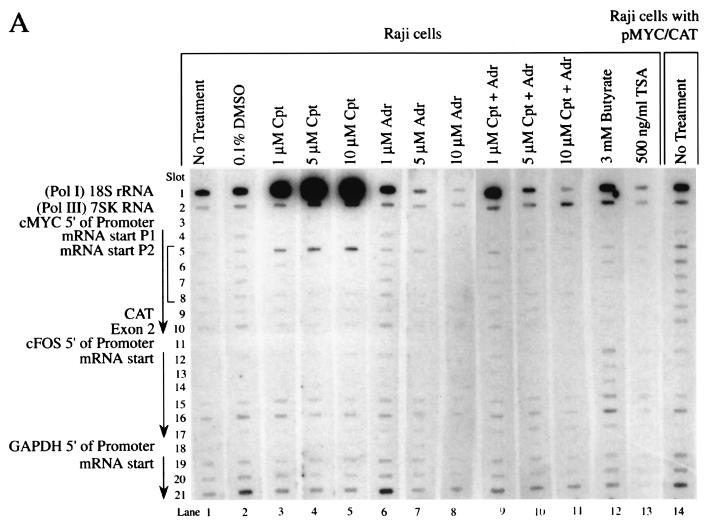
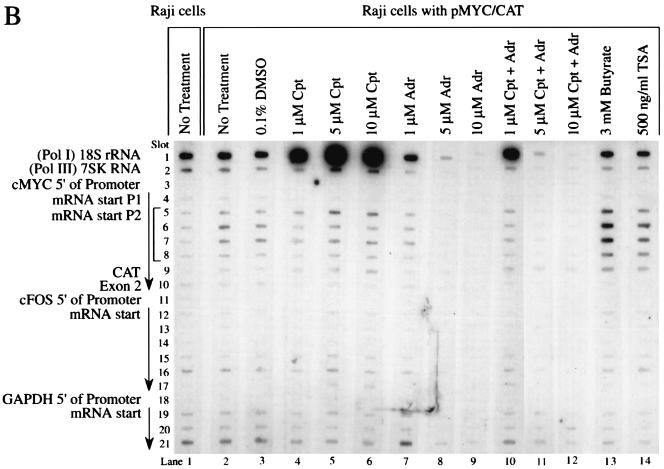
FIG. 2.
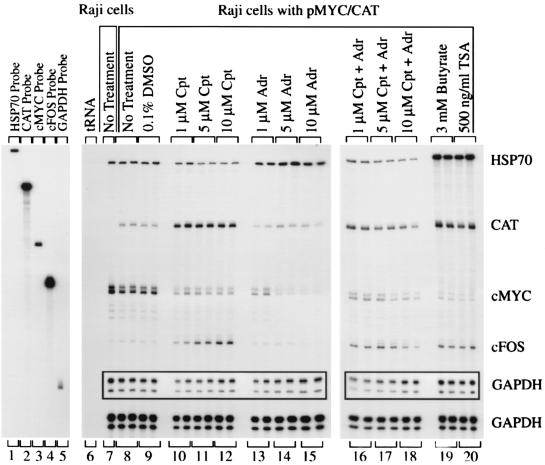
Heterogeneous response of promoter activity to topoisomerase inhibition. (A) Nuclear run-ons of Raji cells. (B) Nuclear run-ons of Raji cells with pMYC/CAT. c-myc exon 2 (slot 10) and CAT (slot 9) distinguish between the c-myc RNAs encoded by the endogenous c-myc gene and the hybrid MYC/CAT mRNA encoded by the episome pMYC/CAT. The arrow indicates the direction of transcription. Cpt, camptothecin; Adr, adriamycin; TSA, trichostatin A. The cells were incubated with inhibitors for 4 h. A 10-min run-on reaction was performed.
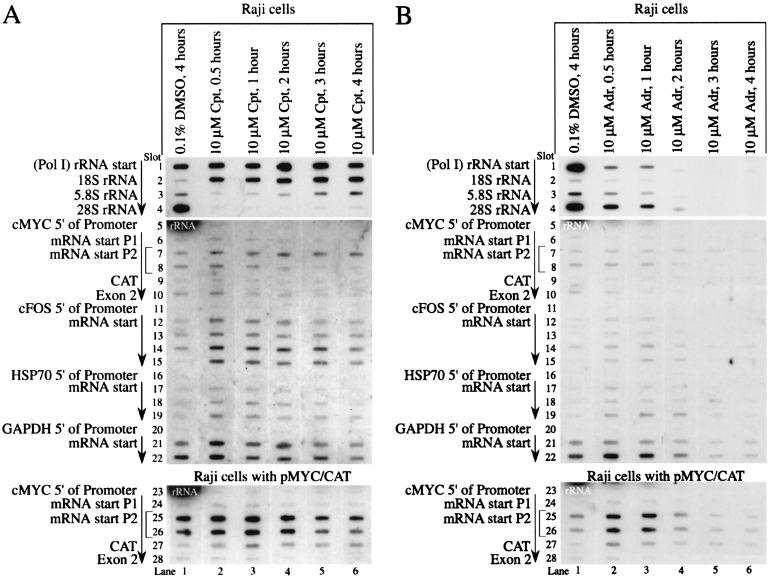
FIG. 3.
Rapid response of promoter activity to topoisomerase inhibition as shown by nuclear run-on. (A) Time course of camptothecin inhibition. In lane 1, slots 5 and 23 show the strong 28S rRNA signal spreading from the slot above. (B) Time course of adriamycin inhibition. c-myc exon 2 (slot 10) and CAT (slot 9) distinguish between the c-myc RNAs encoded by the endogenous c-myc gene and the hybrid MYC/CAT mRNA encoded by the episome pMYC/CAT. The arrow indicates the direction of transcription. Cpt, camptothecin; Adr, adriamycin. Lane 1 shows cells treated with DMSO for 4 h. Lanes 2 to 6 were incubated with either camptothecin or adriamycin for 0.5, 1, 2, 3, or 4 h before nuclei were harvested. A 10-min run-on reaction was performed.
Ligation-mediated PCR (LM-PCR).
The in vivo footprinting was done as described previously (17, 27, 38) with the following modifications. A total of 2 × 107 cells were treated with 25 mM KMnO4 for 2 min at room temperature. For the PCRs, 2 μg of KMnO4-treated template DNA was used with NEB ThermoPol Buffer, including a total of 4, 5, or 6 mM MgSO4 as needed for the different oligonucleotide sets. For a complete list of the oligonucleotides used, see supplemental material available online (www-dcs.nih.gov/branches/lop /Research/genreg/levens/html). The samples were run on a denaturing 8% acrylamide–bisacrylamide (29:1) gel with 50% urea and then compared with a DNA sequencing ladder.
Southern blot analysis.
A total of 2 × 107 cells in 40 ml of medium were incubated with the following chemicals: DMSO, camptothecin, or adriamycin for 4 h and 16 h. When cells were harvested, these chemicals were added to both the PBS (without Ca or Mg) and the Digestion Buffer (67). Cells were washed twice in 10 ml of cold PBS and then resuspended in 0.2 ml of PBS. Next, 1.8 ml of Digestion Buffer containing 0.4 mg of proteinase K/ml was added, mixed by gentle inversion, and incubated at 37°C overnight. Using large-bore pipette tips, the samples were extracted once with phenol, extracted twice with phenol-chloroform, ethanol precipitated, and resuspended in Tris-EDTA. After RNase A treatment, the samples were phenol-chloroform extracted, ethanol precipitated, and resuspended in 1 ml of H2O. DNA was digested with the restriction enzyme NheI and separated electrophoretically on a 20-by-25-cm 0.5% agarose gel in 1.5× Tris-borate-EDTA at 50 V for 48 h. The gels were transferred to Amersham Hybond N+ membranes, and the DNA was cross-linked to the membrane with a Stratagene Stratalinker.
The c-myc probe was a 1.7-kb PstI fragment containing exon 2. The CAT probe was the 3.1-kb NotI-to-BglII fragment from the pREP9/CAT plasmid (Invitrogen). The gel eluted DNA fragments were labeled with [α-32P]dATP (3,000 Ci/mmol) by using the RadPrime DNA Labeling System (BRL). The membranes were hybridized overnight at 65°C. A 2-day exposure at –80°C of Kodak BioMax MS film with the transcreen HE is shown (Fig. 9A and C). As a reference, samples were compared with the plasmid pMYC/CAT before and after topoisomerase I (BRL) treatment for 1 h at 37°C.
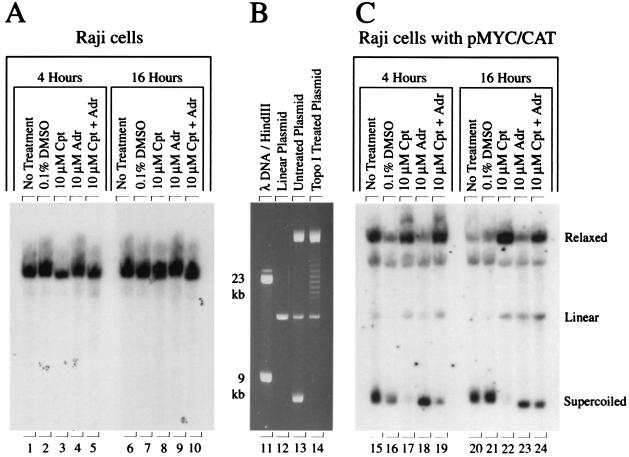
FIG. 9.
DNA damage is unlikely to account fully for the transcriptional response to topoisomerase inhibition. Southern blots showed that brief camptothecin treatment yielded relaxed, but not linear plasmid. Adriamycin yielded hypersupercoiled episomal DNA. (A) Raji cells probed for c-myc exon 2. (B) Pure preparation of pMYC/CAT and the effects of topoisomerase I treatment. (C) Raji pMYC/CAT cell line probed with CAT sequences. Cpt, camptothecin; Adr, adriamycin. The cells were incubated with inhibitors for 4 and 16 h.
RESULTS
This study was undertaken to assess the influence of DNA supercoiling on transcription in vivo. Eukaryotic topoisomerases I and II remove both positive and negative supercoils, resulting in relaxed DNA (7, 22). When these enzymes are inhibited by drugs, the superhelical density of the affected DNA is altered. To examine the transcriptional response to topoisomerase inhibition, cells were incubated with camptothecin, a topoisomerase I inhibitor, or adriamycin (doxorubicin) to inhibit topoisomerase II. The transcriptional response of specific genes to drug treatment was monitored by RNase protection and by nuclear run-on, and conformational and/or topological changes were visualized by in vivo footprinting and Southern blotting.
Two cell lines were used for these experiments. Raji cells are a B-lymphocyte Burkitt lymphoma cell line in which one c-myc gene is translocated near an immunoglobulin gene [chromosome, t(8, 14)] (20). In these cells the translocated myc allele is highly transcribed, while the wild-type copy is repressed (41). The translocation breakpoint is at position −1398 relative to the c-myc P2 promoter start site (9). The translocated c-myc allele also has scattered mutations within exon 1, intron 1, and exon 2 (53).
The second cell line, Raji pMYC/CAT, was a Raji cell line stably transfected with the plasmid pREP9/GAL45 MYC/CAT (pMYC/CAT). In this EBV-based autonomously replicating, neomycin-selectable plasmid, 2.9 kb of c-myc genomic DNA, starting 2.3 kb upstream of promoter P1, was fused with the CAT coding sequence (CAT). Similar MYC EBV-based plasmids have previously been shown to bear nucleosomes positioned identically as seen for the endogenous c-myc gene (38, 52). Raji pMYC/CAT was used to follow myc promoter function. Driven by neighboring c-myc upstream and downstream sequences, and properly assembled into chromatin, the episomal c-myc promoter was expected to recapitulate many features of c-myc regulation; the high-copy-number plasmid was expected to yield an amplified signal.
Polymorphous response of RNA synthesis to inhibition of topoisomerases I and II.
To assess the effect of topoisomerase inhibitors on mRNA abundance, the levels of the transcripts of several genes were directly measured by using RNase protection (Fig. 1). Cells were treated with camptothecin and adriamycin separately or together for 4 h. (Camptothecin inhibits topoisomerase I enzyme, and adriamycin inhibits topoisomerase II.) The influence of the histone deacetylase inhibitors butyrate and trichostatin A was also observed (35, 60). Typically, histone deacetylase inhibition leads to increased chromatin acetylation and conditions conducive for increased gene activity. Each of the mRNAs tested displayed a distinctive profile in response to topoisomerase inhibition:
FIG. 1.
The steady-state level of each mRNA displays a distinctive profile in response to inhibition of topoisomerase I or II, by monitoring RNase protection. The c-myc exon 2 versus CAT probes used in this experiment distinguish between the c-myc RNAs encoded by the endogenous c-myc gene and the hybrid MYC/CAT mRNA encoded by the episome pMYC/CAT. Cpt, camptothecin; Adr, adriamycin; TSA, trichostatin A. The cells were incubated with inhibitors for 4 h. The duplicate lanes are each from different independent experiments. A 3-day exposure made with Kodak XAR film is shown, along with a 13-h exposure for the full-length probes and the GAPDH (glyceraldehyde-3-phosphate dehydrogenase) inset.
hsp70 mRNA, which was high in all control samples (Fig. 1, lanes 7 to 9), was decreased by camptothecin (Fig. 1, lanes 10 to 12) but increased by adriamycin (Fig. 1, lanes 13 to 15), butyrate (Fig. 1, lane 19), and trichostatin A (Fig. 1, lane 20). The hsp70 mRNA half-life is 50 min (69).
The c-fos message, which was quite low in the control cells (Fig. 1, lanes 7 to 9), was strongly increased by 5 and 10 μM camptothecin (Fig. 1, lanes 11 and 12) and was raised by both butyrate (Fig. 1, lane 19) and trichostatin A (Fig. 1, lane 20). Induction of c-fos transcription by these concentrations of camptothecin has been reported previously (66). In contrast, adriamycin decreased c-fos mRNA (Fig. 1, lanes 13 to 15).
Unexpectedly, endogenous c-myc and the episomal pMYC/CAT respond differently to the drugs tested. Endogenous c-myc levels were high in the controls (Fig. 1, lanes 7 to 9) but were lowered by all of the drug treatments (camptothecin [lanes 10 to 12], adriamycin [lanes 13 to 15], butyrate [lane 19], and trichostatin A [lane 20]). In contrast, the c-myc promoter driven CAT mRNA was dramatically increased by camptothecin (Fig. 1, lanes 10 to 12), butyrate (lane 19), and trichostatin A (lane 20) but decreased by adriamycin (lanes 13 to 15). CAT mRNA levels generally paralleled c-fos. It is important to note that, although c-fos and c-myc messages both have short half-lives (54, 65), these genes responded very differently to drug treatment. Moreover, the half-life of CAT mRNA is also short (3). Because some of these RNAs increased, while others decreased during the 4-h drug treatment (long relative to the half-lives of these molecules), differential kinetics of RNA degradation do not explain these results unless these topoisomerase inhibitors differentially modify mRNA half-life.
Of all of the mRNAs analyzed, gapdh was least affected by the drug treatments. It is clear from the behavior of these genes that there is no stereotypical response of mRNA levels after topoisomerase inhibition. In contrast, butyrate and trichostatin A increased hsp70, c-fos, and MYC/CAT, as expected for histone deacetylase inhibition; only endogenous c-myc decreased after histone deacetylase inhibition, as reported previously by others (45).
The steady-state mRNA levels assayed by RNase protection indicate the net effect on RNA synthesis and degradation. If, for example, a message is increased, this may be due to an increase in synthesis or a decrease in degradation. One way to discriminate between these possibilities is to measure RNA synthesis directly with nuclear run-on assays. These assays allow a limited extension of RNA polymerases transcriptionally engaged in vivo. A battery of genes transcribed by RNA polymerase I, II, or III was analyzed with nuclear run-on assays. To assess structural changes at promoters after topoisomerase inhibition, several genes were examined by in vivo footprinting with the conformation-sensitive DNA reagent potassium permanganate.
Context-dependent response of the c-myc promoter to topoisomerase inhibition.
c-myc expression is very context dependent. In Burkitt lymphoma cells such as Raji cells, the translocated c-myc allele is deregulated, while the unrearranged allele is underexpressed. How immunoglobulin regulatory sequences project their influence over vast stretches of DNA (sometimes exceeding hundreds of kilobases) to activate the translocated allele is unknown. Disturbances of c-myc expression are also associated with far-3′-genetic irregularities in cis at the PVT locus in some tumors (62). To explore directly c-myc promoter activity in different chromosomal contexts, chromosomal or episomal transcription was compared between Raji cells and Raji pMYC/CAT by using nuclear run-on assays. After 4 h of drug treatment, nuclei were harvested, and RNA labeled in a brief run-on reaction was hybridized with a panel of oligonucleotides derived from the genes of interest. The run-on findings showed that all of the drug treatments repressed endogenous c-myc in agreement with the RNase protection results.
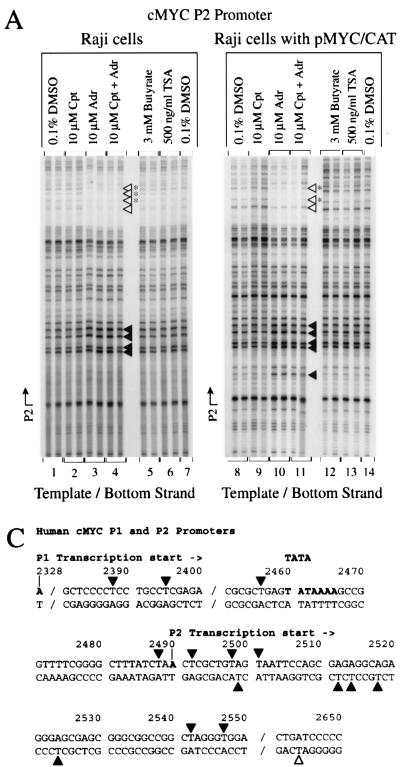
Labeled nascent transcripts from untreated Raji cells and cells treated with the vehicle DMSO hybridized similarly with antisense myc oligonucleotides extending from the P1 promoter through the P2 promoter and into exon 2 (Fig. 2A, lanes 1 and 2). Little evidence of holdback of RNA polymerase at the P2 promoter was noted. (Holdback would be indicated by strong hybridization with slot 5, with declining signals in slots 6 to 10.) The loss of RNA polymerase holdback in Burkitt lymphoma has been previously described (10). Camptothecin, even at the lowest dose, caused dramatic holdback of the RNA polymerase at the endogenous c-myc P2 promoter (Fig. 2A, lanes 3 to 5). The RNA labeled during the 10-min run-on reaction hybridized only with the first P2 sequence (slot 5), indicating that the RNA polymerase is loaded at the promoter but progresses less than 50 nucleotides. Hybridization with P1 oligonucleotides was lost (slot 4), suggesting that this promoter is vacant after camptothecin treatment. Similar results were observed with nuclei from camptothecin-treated BJAB cells (that possess only untranslocated c-myc), showing that this drug-induced holdback requires no immunoglobulin sequences (data not shown). Adriamycin (Fig. 2A, lanes 6 to 8), butyrate (lane 12), and trichostatin A (lane 13) all yielded uniformly weaker myc signals than the DMSO control. With 5 μM adriamycin, only weak holdback at P2 was noted (Fig. 2A, lane 7).
Transcription from the episome in Raji pMYC/CAT was also analyzed by using nuclear run-on (Fig. 2B). The intensity of the hybridization to sequences downstream of c-myc P2 (slots 6 and 7) was increased in Raji pMYC/CAT compared with Raji cells (Fig. 2B, lanes 2 and 3 versus lane 1) as expected, due to the increased copy number of this template. Overexpression from the episome was confirmed by comparing the relative intensities of the hybridization with CAT (slot 9) versus c-myc exon 2 (slot 10); CAT is specific for the episome whereas c-myc exon 2 is specific for the endogenous gene. The relative transcription of sequences shortly downstream of P2 (Fig. 2B, lanes 1 to 3, slots 5 to 7) was increased in Raji pMYC/CAT compared with Raji cells, a finding indicative of greater utilization of P2 from the episome, whereas P1 usage was not increased (slot 4). The intensity of the c-myc run-on signal from pMYC/CAT cells declined progressively further 3′ of the start site, suggesting that on the episome there is holdback of RNA polymerase near the c-myc P2 promoter, and thus fewer polymerases are located distally. P2 utilization with promoter proximal holdback of episomal c-myc has been noted previously (1, 34).
Although RNase protection of Raji pMYC/CAT cells showed simply that camptothecin induced CAT, the run-on transcription from pMYC/CAT in the presence of increasing camptothecin gave a more complex pattern (Fig. 2B, lanes 4 to 6). The first P2 oligonucleotide (slot 5) hybridized more strongly than the 3′ sequences, indicating more RNA polymerase holdback, but the P2 distal sequences and CAT were also more intensely transcribed, suggesting that transcription penetrated further into the gene with less holdback. These two contradictory observations are reconciled by superimposition of the transcription profiles of the episomal and endogenous c-myc promoters. As described above, camptothecin caused strong holdback of the endogenous gene at the first P2 oligonucleotide (Fig. 2A, lanes 3 to 5, slot 5). Therefore, increased holdback of the endogenous c-myc gene overlying increased downstream transcription from the plasmid pMYC/CAT reconciles these results with the RNase protection data. Adriamycin strongly repressed the episomal c-myc promoter just as it did the endogenous gene, abolishing hybridization with all probes (Fig. 2B, lanes 7 to 9). Interestingly, the run-ons obtained with both camptothecin and adriamycin most closely resembled those obtained with adriamycin alone (Fig. 2A, lanes 9 to 11, and B, lanes 10 to 12). Butyrate and trichostatin A caused upregulated transcription of all MYC/CAT sequences without altering their relative intensities (Fig. 2B, lanes 13 and 14). Adriamycin inhibition of transcription was not mitigated by butyrate (data not shown). If immobilized topoisomerase II recruited histone deacetylase to inhibit transcription (23, 71), then these drugs should have increased transcription in the presence of adriamycin; this did not occur.
Importantly, inclusion of adriamycin or camptothecin directly in the nuclear run-on reactions neither inhibited nor stimulated transcription of c-myc or other genes (data not shown); thus, the changes observed in these experiments reflect conditions set up in vivo. Run-on analysis of nuclei harvested during a time course of adriamycin drug treatment demonstrated full promoter responses in less than 2 h (Fig. 3B).
RNase protection and nuclear run-on experiments indicated that different functional states of the c-myc promoter, directly affecting transcriptional levels, exist in the chromosome versus the epsiome and in response to topoisomerase I versus topoisomerase II inhibition.
Context-dependent KMnO4 sensitivity of the c-myc promoter to topoisomerase inhibition.
Topoisomerase I and II inhibitors modify transcription of the endogenous and episomal c-myc genes. What structural alterations of the template DNA occur concomitant with topoisomerase inhibition? In vivo footprinting was performed with KMnO4 to see whether the changes in transcription due to drug treatments are reflected in the conformational state of promoter DNA sequences. KMnO4 is most reactive with pyrimidines in single-stranded or otherwise conformationally distorted DNA (17, 27, 38). Concomitant alterations or adjustments of protein contacts with melted or strained DNA would further modify the chemical reactivity of promoter and regulatory DNA. After treatment with permanganate, cellular DNA was extracted, and the phosphodiester backbone at the modified bases was cleaved with piperidine; ligation-mediated PCR was then performed with gene specific primers to amplify and display the sites of altered KMnO4 reactivity.
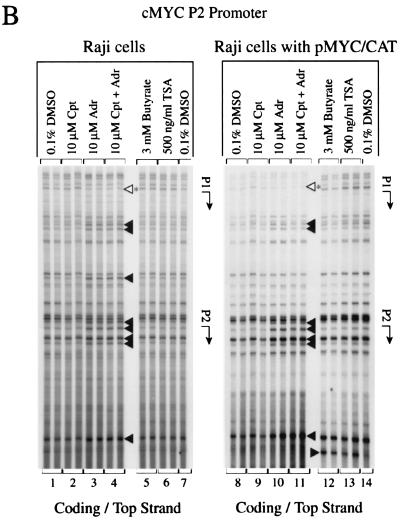
The endogenous c-myc promoter region demonstrated dramatic changes at the P2 promoter in response to topoisomerase II inhibition with adriamycin. Enhanced reactivity at multiple specific bases on both strands extended 87 bases, from −34 upstream of the P2 promoter transcription start site to +53 downstream (positions 2456 to 2542, with the mRNA start at position 2490 [accession number X00364]) (Fig. 4A and B, lanes 3). Sequences further 3′ displayed generalized hyporeactivity, presumably reflecting the depletion of elongation complexes and consequently fewer transcription bubbles to generate permanganate targets. Decreased sensitivity to permanganate was also seen at the c-myc P1 promoter start site in response to adriamycin. Similar adriamycin sensitivity was observed on both the endogenous and episomal c-myc sequences within the P2 promoter proximal region (Fig. 4A and B, lanes 3 and 10). These data indicate that adriamycin freezes a hyper-open complex at c-myc P2 transcription start site but depresses open DNA downstream. The promoter-frozen complexes are not activated during nuclear run-on experiments and so must either be inactivated or else have encountered an insurmountable barrier.
FIG. 4.
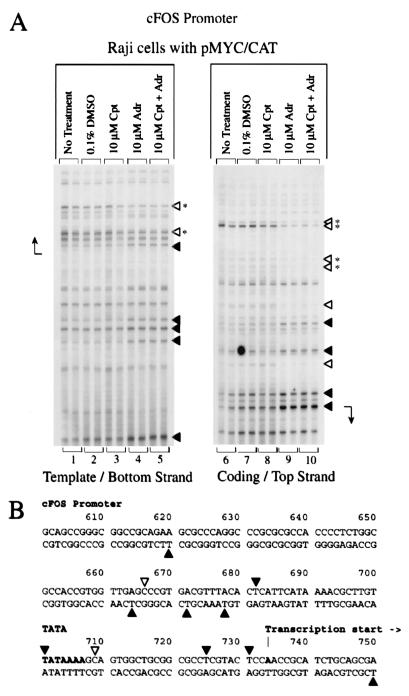
Response of c-myc promoter structure to topoisomerase inhibition. In vivo potassium permanganate footprint. (A) Bottom DNA strand. (B) Top DNA strand. Cpt, camptothecin; Adr, adriamycin; TSA, trichostatin A. The cells were incubated with inhibitors for 4 h. The arrow indicates the transcription start site. Bases made partially hyposensitive by camptothecin are present but are not annotated (panel B, lane 2). (C) Nucleotides with altered intensities in the footprint are identified. Symbols: ◂, hypersensitivity; ◃, hyposensitivity. The asterisk indicates reactivity at bases that were not exactly mapped due to DNA compression in this region of the gel. The duplicate lanes are each from different independent experiments.
Camptothecin, in contrast to adriamycin, elicited only subtle changes in permanganate sensitivity from the chromosomal c-myc in Raji. Reactivity at several bases from positions 2450 to 2501 surrounding the P2 start site was slightly reduced after drug treatment (Fig. 4B, lane 2). Opposite to the endogenous gene, camptothecin enhanced the KMnO4 sensitivity of downstream-transcribed episomal template, indicating more elongating complexes (Fig. 4A, lane 9), a finding consistent with the increase of CAT mRNA (Fig. 1, lanes 10 to 12). Butyrate (Fig. 4B, lane 12) and trichostatin A (lane 13) augmented the permanganate reactivity of residue +58 relative to residue +53 on the nontemplate strand (positions 2547 and 2542, respectively) and increased the overall reactivity farther downstream on the template strand (Fig. 4A, lanes 12 and 13); this pattern often correlates with high-output states of c-myc. Thus, unlike the endogenous gene, histone deacetylase inhibition correlated with increased output and openness of episomal c-myc DNA. The footprints obtained upon combination of camptothecin and adriamycin most closely resembled those obtained with the latter alone (Fig. 4A and B, compare lanes 2, 3, and 4). Footprint changes after topoisomerase I or II inhibition were not simply a secondary result of transcription inhibition, since treatment with α-amanitin, which freezes transcription complexes in place, generated no specific response to KMnO4 at promoters (data not shown).
c-myc upstream CT element becomes conformationally stressed after topoisomerase II inhibition.
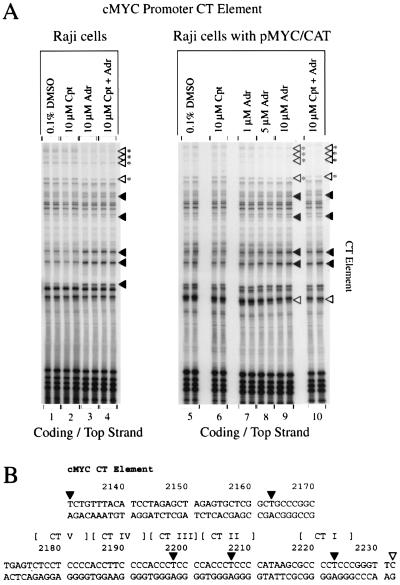
When stressed by torsional strain, particular DNA segments adopt altered DNA conformation or structure. FUSE and CT are two such elements upstream of active c-myc genes that are peculiarly sensitive to KMnO4 (38). If topoisomerase inhibition perturbs the degree or distribution of torsional strain, then altered reactivity of FUSE and CT should follow. The permanganate footprint of the CT element, located 300 bp 5′ of the MYC P2 start site, was dramatically altered after adriamycin treatment. The endogenous c-myc gene seen in the Raji footprint (Fig. 5A, lane 3) shows several dramatically darker DNA bands corresponding to the T nucleotides of the three CT repeats closest to the promoter; these same repeats are the preferred targets for hnRNP K binding in vitro. The two most upstream elements exhibited sharply reduced reactivity; these sites are the preferred binding sites for the transcription factor Sp1. Competition between hnRNP K and Sp1 for binding in this region has previously been reported (38). Raji pMYC/CAT cells (Fig. 5A, lanes 7 to 9) showed only minor changes in this same region, due either to reduced reactivity in all plasmids or to heterogeneous CT reactivity between plasmids; the number of active episomes at any instant is unknown. (By comparing the chromosomal and episomal exposure times, it was estimated that these cells have at least 20 copies of the episome [Fig. 5].) The increased reactivity of the CT element after adriamycin treatment might indicate a greater propensity toward melting, perhaps due to the failure of topoisomerase II to remove accumulated strain at the promoter and at the CT element 250 to 300 bp upstream of the promoter.
FIG. 5.
Topoisomerase II inhibition provokes conformational changes at the CT-element of the endogenus c-myc gene. (A) In vivo potassium permanganate footprint. Cpt, camptothecin; Adr, adriamycin. The cells were incubated with inhibitors for 4 h. (B) Nucleotides with altered intensities in the footprint are identified. Symbols are as defined for Fig. 4. The duplicate lanes are each from different independent experiments.
In addition the AT-rich FUSE element, found 1,700 bp 5′ of the MYC P2 start site, was examined in the Raji pMYC/CAT cell line (data not shown). Here adriamycin treatment showed subtle changes in the footprint. Several DNA bands with decreased sensitivity to permanganate were seen within the FUSE element, indicating a more closed, double-stranded character. The FUSE element in the parental Raji cell line was not examined because in the expressed c-myc allele the FUSE element is translocated away from the myc coding sequence. The decreased reactivity of the FUSE element after adriamycin treatment might be due to repression of the episomal MYC/CAT gene, since a closed FUSE sequence is associated with a repressed c-myc gene (38). Camptothecin failed to modify the permanganate footprints of both the FUSE and the CT upstream elements, indicating that topoisomerase I was not active in these topological domains (Fig. 5A, lanes 2 and 6, and data not shown).
Adriamycin alters the structure of the c-fos and hsp70 promoters.
c-fos transcription was also examined by using nuclear run-on assays. Promoter proximal segments of c-fos were undertranscribed relative to sequences that are more distal (Fig. 2A, lanes 1 and 2, and Fig. 2B, lanes 1 to 3, slots 12 to 14 versus slots 15 and 16), a finding consistent with the transcriptional pause reported in murine c-fos intron 1 (44). In fact, the signals arising from the 5′-most sequences of the transcript were difficult to discern above the background. Adriamycin induced a biphasic response of the c-fos promoter. Within the first hour of treatment, transcription was increased, most noticeably at the 5′ end (Fig. 3B, lanes 2 and 3). Subsequently, transcription from the entire gene was shut down (Fig. 3B, lanes 4 to 6). Transient augmentation of c-fos transcription by camptothecin was noted (Fig. 3A, lanes 2 to 6). Butyrate and trichostatin A slightly elevated transcription at the c-fos start site (Fig. 2A, lane 12, and Fig. 2B, lanes 13 and 14).
The dramatic shutoff of c-fos due to adriamycin treatment was explored further by using KMnO4-LM-PCR in vivo footprinting. Dramatic alterations in the promoter conformation were caused by topoisomerase II inhibition. Increased reactivity within the immediate vicinity of the start site was prominent, while just 3′ of the start site on the bottom strand reactivity was diminished (Fig. 6A, lanes 4 and 9). (The DNA region shown in the c-fos footprint Fig. 6 maps to slots 11 and 12 of the run-on transcription reactions shown in Fig. 2 and 3.) Farther downstream, the same intron 1 segments holding paused RNA polymerases detected with nuclear run-on (Fig. 2, slots 15 and 16, and Fig. 3, slots 13 and 14), showed bases with altered reactivity (both increased and diminished), perhaps indicating that the drug treatment thwarted downstream transit of complexes (data not shown). Consistent with this interpretation, the DNA from adriamycin-treated cells became exclusively hyporeactive distal to the downstream pause region (data not shown). As with c-myc, topoisomerase II inhibition freezes complexes at the c-fos promoter and prevents RNA synthesis either by inactivating the transcription machinery or by imposing an insurmountable barrier such as accumulated torsional strain or drug-frozen topoisomerase II bound to the DNA template. It is also likely that unrelieved torsional strain perturbs the reactivity of downstream segments.
FIG. 6.
Topoisomerase II inhibition alters the structure of the c-fos promoter. (A) In vivo potassium permanganate footprint. Cpt, camptothecin; Adr, adriamycin. The cells were incubated with inhibitors for 4 h. (B) Nucleotides with altered intensities in the footprint are identified. Symbols are as defined for Fig. 4. The duplicate lanes are each from different independent experiments.
Also like c-myc, camptothecin evoked no clear changes in the pattern of KMnO4 reactivity for c-fos (Fig. 6A, lanes 3 and 8). Camptothecin perturbs c-fos transcription by elevating basal expression (as detected with RNase protection [Fig. 1, lanes 10 to 12] and with run-on assays within 3 h of drug treatment [Fig. 3A, lanes 2 to 5]), while delaying and dampening induced expression (66). These effects occur without disturbing promoter-DNA conformation. Therefore, it is likely that the primary effect of this drug is exerted at the level of elongation throughout the body of the gene.
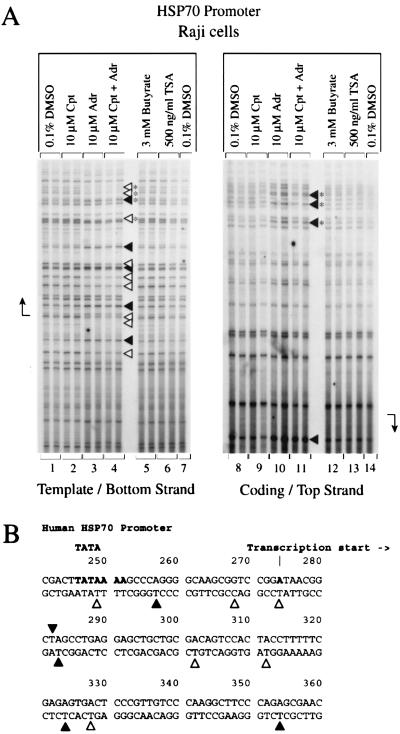
Basal (non-heat shocked) hsp70 RNA levels were depressed by camptothecin (Fig. 1, lanes 10 to 12) but increased by adriamycin (Fig. 1, lanes 13 to 15) and HDAC inhibitors (Fig. 1, lanes 19 and 20). hsp70 nuclear run-on transcription increased transiently within the first hour of camptothecin or adriamycin treatment and then declined by 4 h (Fig. 3, lanes 2 to 6). Adriamycin developed an alternating hyphenated pattern of augmented and diminished permanganate reactivity throughout the region of the paused polymerase (Fig. 7A, lane 3). While on the opposite DNA strand, a single downstream residue (+9) (position 282, accession number M11717) became intensely reactive after adriamycin treatment (Fig. 7A, lane 10). Thus, as with c-myc and c-fos, adriamycin seemed to cause increased hsp70 promoter occupancy after a transient increase in promoter activity.
FIG. 7.
Topoisomerase II inhibition alters the structure of the hsp70 promoter. (A) In vivo potassium permanganate footprint. An alternating pattern of increased and decreased intensity near the hsp70 transcription start site is seen. Cpt, camptothecin; Adr, adriamycin; TSA, trichostatin A. The cells were incubated with inhibitors for 4 h. (B) Nucleotides with altered intensities in the footprint are identified. Symbols are as in Fig. 4. The duplicate lanes are each from different independent experiments.
gapdh is often employed as a normalization standard. Considering the relative stability of gapdh mRNA levels (Fig. 1) and the modest response of gapdh nuclear run-on activity (Fig. 2 and 3) in the face of assorted pharmacological challenges, it may be likely that several molecular devices cooperate to enforce homeostasis on this gene.
Topoisomerase I inhibition forces downstream stalling in rRNA transcription units.
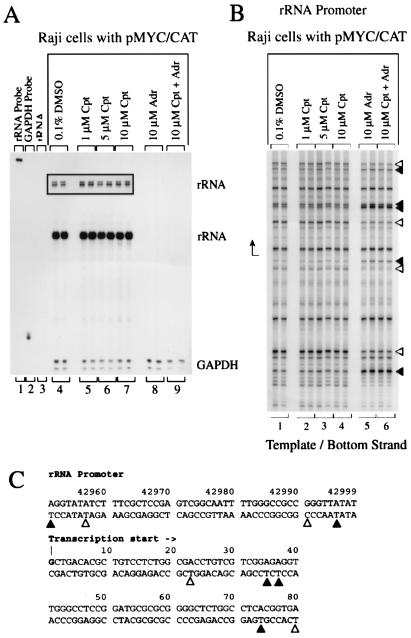
Humans have 200 to 300 copies of the 43-kb ribosomal DNA repeat unit, each transcribed from a single promoter. Each 13-kb primary transcript consists of a 3.6-kb 5′ leader sequence, followed by a 1.9-kb 18S gene, a 1-kb spacer, a 0.15-kb 5.8S gene, a 1.1-kb spacer, and a 5-kb 28S gene; the transcript encoding segments are separated by a 30-kb spacer (accession number U13369). The long half-life and large pool of ribosomes and rRNA precursors buffer rRNA levels from rapid fluctuation. Therefore, failure of camptothecin to depress rRNA levels as measured with RNase protection (by using a probe at the rRNA transcription start) was not surprising (Fig. 8A, lanes 5 to 7), despite the well-established requirement of proper topoisomerase function for rRNA transcription (12, 75). More perplexing was the dramatic increase of the rRNA nuclear run-on signal seen in response to camptothecin (Fig. 2A, lanes 3 to 5, and Fig. 2B, lanes 4 to 6). Nascent rRNA synthesized by nuclear run-on was detected by hybridization with sequences at the proximal segment of the 18S rRNA, ca. 4 kb downstream of the start site. The conformation of the rRNA promoter before and after camptothecin treatment was assessed by using LM-PCR after permanganate oxidation of intact cells. Camptothecin induced no significant changes of the rRNA promoter, suggesting that topoisomerase I inhibition provoked no alteration of transcription complexes at most of the rRNA promoters (Fig. 8B, lanes 2 to 4). The most straightforward explanation of these data requires the stalling of elongation complexes within the proximal one-third of the rRNA transcription unit after topoisomerase I inhibition. To test this possibility, nuclear run-on assays were performed by using nuclei from cells treated with camptothecin for various times; the nascent rRNA transcripts elongated in vitro were hybridized with a battery of oligonucleotide sequences derived from segments throughout the rRNA transcription unit. Indeed, the predicted holdback was observed in the proximal one third of the transcribed region (Fig. 3A, lanes 2 to 6). Similar over-representation of proximal rRNA gene sequences in nuclear run-on assays has been noted previously (75). Initiated RNA polymerase I fails to penetrate very far into the body of the gene, perhaps due to accumulated torsional strain.
FIG. 8.
Adriamycin, but not camptothecin, depresses rRNA promoter activity and alters promoter structure. (A) RNase Protection with a probe for the rRNA transcription start site. A 13-h Kodak XAR film exposure and a 2-h exposure (inset) are shown. (B) In vivo potassium permanganate footprint. Cpt, camptothecin; Adr, adriamycin. The cells were incubated with inhibitors for 4 h. Symbols are as in Fig. 4. (C) Nucleotides with altered intensities in the footprint are identified. The duplicate lanes are each from different independent experiments.
Adriamycin depressed rRNA levels in RNase protection studies (Fig. 8A, lane 8) and diminished rRNA synthesis as detected by nuclear run-on (Fig. 2A, lanes 6 to 8, Fig. 2B, lanes 7 to 9, and Fig. 3B, lanes 2 to 6). In contrast to topoisomerase I inhibition, this topoisomerase II inhibitor augmented KMnO4 reactivity at some nucleotides at the promoter while depressing oxidation at others, implying considerable reorganization of the template at the transcription start site (Fig. 8B, lane 5).
Transcription by RNA polymerase III of 7SK RNA in Raji cells was fully resistant to camptothecin and partly resistant to adriamycin as measured by nuclear run-on (Fig. 2A, lanes 3 to 8, and Fig. 2B, lanes 4 to 9).
Effect of topoisomerase I and II inhibition on the state of c-myc sequences in vivo.
To assess more directly the influence of adriamycin and camptothecin on DNA topology in vivo, the episome from drug-treated Raji pMYC/CAT was recovered and analyzed for linear, relaxed and/or nicked (relaxed/nicked), and supercoiled forms by Southern blotting. Plasmid pMYC/CAT extracted from bacteria provided the reference for this analysis. As expected, the plasmid linearized with NotI migrated as a single band in the middle of the gel (Fig. 9B, lane 12). The untreated plasmid revealed three bands: the fastest-migrating band represented supercoiled plasmid; the band of intermediate mobility was the linearized plasmid, while the slowest species was the relaxed/nicked plasmid (Fig. 9B, lane 13). Figure 9C shows the DNA recovered from the Raji pMYC/CAT cell line electrophoretically separated, blotted to a membrane and probed with the plasmid-specific CAT sequence. DNA from untreated or DMSO-only treated cells displayed a mixture of supercoiled and relaxed/nicked episomal DNA with minimal linearization (Fig. 9C, lanes 15 and 16). Four hours of camptothecin treatment yielded the expected conversion from supercoiled to nicked plasmid (Fig. 9C, lane 17). Linear forms accrued only slowly because of the low probability that two closely spaced single-stranded nicks occurred on opposite DNA strands. In contrast to camptothecin, adriamycin had the opposite effect. Adriamycin treatment yielded more supercoiled and less relaxed episomal DNA (Fig. 9C, lane 18). These data suggest that under the conditions used in this study, adriamycin inhibited topoisomerase II prior to formation of the protein-DNA adduct cleavable complexes. (DNA fragmentation passes through a maximum and then declines as the concentration of adriamycin is increased [15, 64].) Such inhibition would result in hypersupercoiled episomal DNA in contrast to the linearized DNA expected from poisoned topoisomerase II-DNA complexes. The minimal linearization promoted by adriamycin on the endogenous c-myc in Raji (Fig. 9A, lane 4) or on the episome recovered from Raji pMYC/CAT (Fig. 9C, lane 18) indicated that the transcriptional holdback and promoter remodeling occurring in response to this drug did not result from local DNA damage or repair complexes. Moreover, the rapidity of these changes might suggest direct involvement of topoisomerase II in maintaining a transcriptionally conducive chromatin environment.
DISCUSSION
The experiments reported here reveal a protean and pleomorphic response of transcription to topoisomerase inhibition. Increased or decreased torsional strain may alter transcription and may antagonize or synergize with chromatin modification and remodeling. Nucleosomes with acetylated histones restrain and stabilize negative supercoils less tightly than when unmodified (39, 42). Chromatin remodeling machines generate and are influenced by torsional strain (14, 21). Thus, there is considerable potential for mechanical linkage and cross-regulation of all of these processes by using DNA as a force-bearing cable. The pattern of protein-tethering and DNA-looping coupled with the distinctive properties of individual genes are likely to determine the effect of topoisomerases and their inhibition on gene activity. RNA levels for particular genes may rise or fall in response to camptothecin and adriamycin treatment. How might topoisomerase inhibition differently influence the expression of diverse genes? Several nonexclusive possibilities include: (i) disturbance of the distribution and transmission of torsional strain; (ii) direct protein-protein interaction of topoisomerases with transcription factors, the basal transcription apparatus or chromatin modifying or remodeling machinery altering the expression of specific genes; (iii) immobilized topoisomerase-inhibitor-DNA complexes forming a roadblock hindering the movement of transcription elongation complexes; and (iv) topoisomerase inhibitors as DNA-damaging agents indirectly modifying gene expression pursuant to the activation of the particular signal transduction pathways regulating DNA repair.
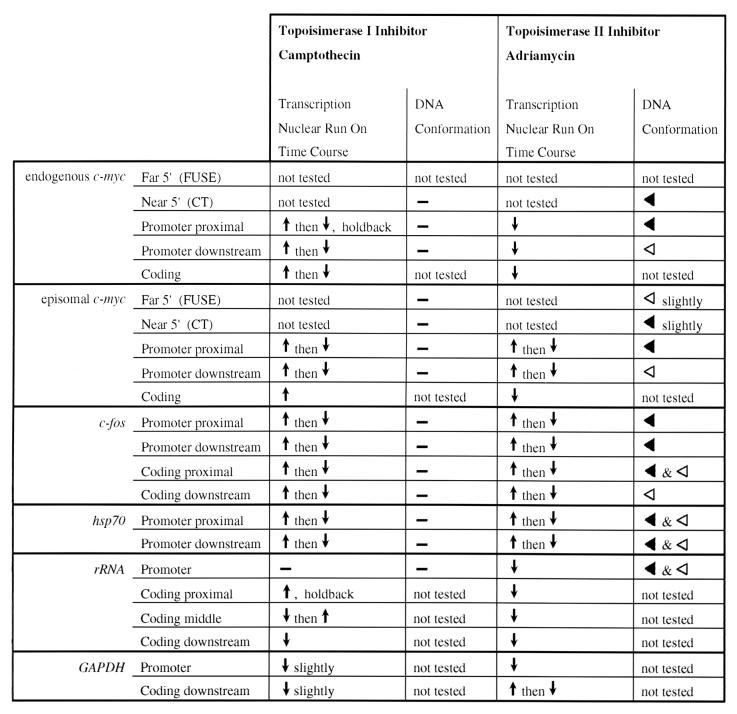
When constrained by physical barriers, transcriptionally generated torsional strain diffusing behind and in front of a moving transcription complex accumulates within topological domains unless dissipated by topoisomerases I and/or II (32). So the consequences of camptothecin and/or adriamycin treatment on gene expression should relate to the rate of transcription and the specific architecture of the affected targets. The results described here clearly show that the changes produced by these two drugs on mRNA levels, nuclear run-on rates, and on the conformation of promoters and upstream regulatory sequences are not equivalent (Fig. 10).
FIG. 10.
Summary of response to camptothecin or adriamycin treatment. Symbols: ↑, increased transcription; ↓, decreased transcription. A ↑ then ↓ combination indicates that in the run-on time course experiment, transcription was induced at the early time point but then repressed by the late time point. The dash (—) reflects no change in transcription. The black triangle (◂) indicates hypersensitivity, while the open triangle (◃) indicates hyposensitivity in the in vivo potassium permanganate footprint. The combination “◂ and ◃” indicates a mixed hypersensitivity and hyposensitivity in the in vivo potassium permanganate footprint.
Camptothecin produces few if any detectable changes in promoter architecture despite activating the transcription of some genes while repressing others. Therefore, topoisomerase I more likely influences RNA synthesis away from the promoter, probably at the level of elongation. Topoisomerase I has been implicated in the penetration of elongation complexes into the body of a gene; c-fos, Drosphila hsp70, dhfr, and rRNA transcription are all accompanied by recruitment of topoisomerase I to downstream sequences (25, 33, 66, 75). Failure of topoisomerase I to remove the positive supercoils accruing in front of RNA polymerase may lead to transcription arrest. Superhelical densities of ca. 0.1 generate sufficient torque to oppose RNA polymerase translocation (74). Transcription within a large topological domain may proceed for a considerable distance before arresting forces are achieved; in the absence of any release of torsional strain, elongation through 10% of the transcription unit would be required. Therefore, camptothecin inhibition would be predicted to stall polymerases away from the promoter, within the body of the gene, just as occurred with rRNA transcription. This explanation conceivably may explain the opposite response of the native and the episomal c-myc promoters to camptothecin. If a topological boundary were imposed on the native gene within exon I or at the 5′ end of intron I, but not on the episome (which lacks these sequences), then minimal transcription of the cellular gene would generate sufficient force to hold back transcription; without this barrier, episomal transcription would progress further. Indeed, low levels of positive torsion might even destabilize the wrapping of negative supercoils on nucleosomes to facilitate elongation transiently.
Adriamycin, in contrast, caused clear changes at all promoters analyzed. In addition, in the case of c-myc, topoisomerase II inhibition increased the permanganate sensitivity of the upstream CT element (Raji and Raji pMYC/CAT), in a region likely to experience strong negative supercoiling forces. In the case of c-fos dramatic changes were present downstream of the transcription start site, at the region of strongest run-on basal c-fos transcription. Thus, topoisomerase II inhibition alters DNA conformation most dramatically at and near promoters, at sites likely to experience strong unwinding stresses. Southern blot analysis, in fact, visualizes the accumulation of negative supercoils within the episomal DNA. Conformational changes at elements such as CT or FUSE may change the spectrum of bound factors at these elements, thus altering gene activity.
Thus, topoisomerase II operates predominantly within a domain embracing the promoter and nearby regulatory sequences. Simultaneous inhibition of topoisomerase I and II neither abrogated nor exacerbated permanganate reactivity, nor did it modify the nuclear run-on pattern, suggesting that each enzyme operates in an architecturally separate topological domain during transcription. If topoisomerase I and II enzymes have the same biological activity, then incubating cells with both topoisomerase I and II inhibitors together should give an additive or synergistic effect, rather than the adriamycin dominance seen in the run-ons and footprints or the compromise between the two drugs seen with RNase protection.
Direct interaction of topoisomerases with the transcription factors, the transcription apparatus or chromatin modification and/or remodeling machinery.
How topoisomerases are recruited to genes is unknown. Topoisomerase II is a component of nuclear matrix (13, 46). Topoisomerase I is a promoter-specific repressor of basal transcription operating through TATA binding protein in vitro (37). Activators abolish this repression. This repression does not demand the camptothecin-sensitive topoisomerase I catalytic activity, indicating that topoisomerase I most likely plays a structural role at promoters. The drug, nevertheless, may interfere with the architecture of preinitiation complexes as topoisomerase I enhances assembly of the transcription apparatus at some promoters (63). Furthermore, topoisomerase activity might facilitate subsequent stages of transcription, such as via interaction with RNA polymerase II (61). Overlying any general influence on basal transcription is the pattern of recruitment of topoisomerase via DNA-bound, sequence-specific transcription and chromatin modifying and/or remodeling factors. A variety of factors reported to bind with topoisomerases I or II, including p53, jun, CREB, HTLV1-Tax, simian virus 40 T antigen, histone deacetylase, Drosophila CHRAC, and pRb, are recruited to particular genes depending upon the particular assortment of cis elements at a given promoter (5, 18, 23, 26, 47, 49, 68, 71, 72). Interaction of these factors with topoisomerases may augment or diminish topoisomerase activity. Reciprocally, masking of transcription factor effector domains by interaction with topoisomerases, although unreported, would seem possible.
In this scheme, if topoisomerases served solely as architectural components of transcription complexes, then topoisomerase inhibitors might not alter transcription. Alternatively, topoisomerase inhibitors might lock the enzyme onto the DNA, providing a stable platform for recruiting factors. For example, increased recruitment of histone deacetylase by topoisomerase II frozen on the DNA could repress gene activity. (The opposite actions of the deacetylase inhibitors butyrate and trichostatin A on episomal versus chromosomal c-myc promoters argue against this alternative.) Topoisomerase enzymatic activity might help dissipate the strain generated by the wrapping of DNA in topological microdomains created by looping of chromatin bound factors during transcription complex assembly or during chromatin remodeling (14, 21, 24, 70); topoisomerase inhibitors might then lessen the activity of the transcription complexes dependent on enzyme activity for assembly. These conditions impose no a priori requirement at any particular gene for topoisomerases I or II to be recruited upstream or downstream.
Drug-immobilized topoisomerases in principle might impose a downstream impediment to the passage of elongation complexes. However, such a mechanism does not provide a general explanation for drug effects because (i) drug-induced upregulation of transcription is not explained and (ii) the camptothecin-induced holdback at the promoter of the endogenous c-myc gene and loss of nascent RNAs hybridizing with downstream sequences indicates that distal elongating complexes cleared the gene (drug removal during the run-on assay should have allowed measurable downstream activity).
Although topoisomerase inhibitors damage DNA, several factors argue that such damage is not directly responsible for the transcriptional effects observed here. (i) In the case of adriamycin, under the chosen conditions minimal damage of either the endogenous c-myc gene or episomal sequences occurred (15, 64). (ii) Drug treatments were quite brief; after such brief treatment with topoisomerase inhibitors, cleavable complex formation is fully reversible upon drug removal (29, 31). Cleavable complexes are converted into a site of DNA damage after dissociation of the topoiosmerases frozen for a protracted period at the strand breaks. During the brief interval of drug treatment, most cells would not transit S phase, and so replication forks would convert cleavable complexes into nicks or breaks (30) in only a minority of cells. (iii) Raji and Raji pMYC/CAT express only mutant p53 and so would not mount an effective apoptotic response (8). (iv) Microarray classification of the global gene response of 60 cell lines tested with 118 compounds revealed that topoisomerase I inhibitors (including campthothecin) cluster together; likewise, various topoisomerase II inhibitors (including adriamycin) all cluster together (59). These two clusters together form a supercluster distinct from DNA-damaging agents. The data indicate that these drugs, including camptothecin and adriamycin, target topoisomerase I or II to produce the observed biological response by mechanisms distinct from or in addition to secondary DNA damage.
In addition to the implications for mechanisms for transcriptional regulation, these results have important implications for the utilization of topoisomerase inhibitors as antineoplastic agents. When acting as topoisomerase poisons, topoisomerase inhibitors damage DNA. This damage, in the face of normal homeostatic mechanisms, leads to cell cycle arrest or apoptosis via standard pathways. Acting as topoisomerase inhibitors, these drugs produce a protean genetic response, modifying the expression of many genes. Chromosomal c-myc expression is particularly susceptible to downregulation by topoisomerase inhibition. Combinations of drugs acting in concert to up- or downregulate special targets, such as c-myc, may allow the manipulation of genetic programs to enhance or retard the performance and selectivity of a pharmacologic regimen. Modification of the expression profiles of various genes would be expected to alter cell growth and proliferation and hence drug sensitivity. For example, camptothecin kills cells in S phase; so if c-myc shutoff in some cells prevents entry into S phase, then these cells may be less susceptible to killing by this drug. Utilization of microarray data may help to identify and classify transcriptional targets responsive to topoisomerase inhibitors, alone or in combination with other agents. One caveat to this approach is the relative insensitivity of microarray analysis to rapid changes in the regulation of stable and/or abundant transcripts. Dramatic effects at the level of nuclear run-on or DNA conformation may precede by many hours correlative changes at the RNA level. Elucidation of the mechanisms generating the regulatory diversity conferred by topoisomerase inhibitors will emphasize the importance of the topological architecture of genes for physiological expression and afford opportunities for pharmacological intervention to mediate the recruitment and activity of the machinery controlling transcription and chromatin through protein-protein interactions.
ACKNOWLEDGMENTS
We thank L. Liotta, S. Mackem, and Y. Pommier for helpful comments, critical review, and fruitful discussions.
REFERENCES
- 1.Albert T, Mautner J, Funk J O, Hortnagel K, Pullner A, Eick D. Nucleosomal structures of c-myc promoters with transcriptionally engaged RNA polymerase II. Mol Cell Biol. 1997;17:4363–4371. doi: 10.1128/mcb.17.8.4363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aller P, Rius C, Mata F, Zorrilla A, Cabanas C, Bellon T, Bernabeu C. Camptothecin induces differentiation and stimulates the expression of differentiation-related genes in U-937 human promonocytic leukemia cells. Cancer Res. 1992;52:1245–1251. [PubMed] [Google Scholar]
- 3.Amara F M, Entwistle J, Kuschak T I, Turley E A, Wright J A. Transforming growth factor-β1 stimulates multiple protein interactions at a unique cis-element in the 3′-untranslated region of the hyaluronan receptor RHAMM mRNA. J Biol Chem. 1996;271:15279–15284. doi: 10.1074/jbc.271.25.15279. [DOI] [PubMed] [Google Scholar]
- 4.Bates A D, Maxwell A. DNA Topology. Oxford, England: IRL Press; 1993. [Google Scholar]
- 5.Bhat U G, Raychaudhuri P, Beck W T. Functional interaction between human topoisomerase IIα and retinoblastoma protein. Proc Natl Acad Sci USA. 1999;96:7859–7864. doi: 10.1073/pnas.96.14.7859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bunch R T, Povirk L F, Orr M S, Randolph J K, Fornari F A, Gewirtz D A. Influence of amsacrine (m-AMSA) on bulk and gene-specific DNA damage and c-myc expression in MCF-7 breast tumor cells. Biochem Pharmacol. 1994;47:317–329. doi: 10.1016/0006-2952(94)90023-x. [DOI] [PubMed] [Google Scholar]
- 7.Champoux J J. Mechanistic aspects of type-I topoisomerases. In: Cozzarelli N R, Wang J C, editors. DNA topology and its biological effects. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1990. pp. 217–242. [Google Scholar]
- 8.Duthu A, Debuire B, Romano J, Ehrhart J C, Fiscella M, May E, Apella E, May P. p53 mutations in Raji cells: characterization and localization relative to other Burkitt's lymphomas. Oncogene. 1992;7:2161–2167. [PubMed] [Google Scholar]
- 9.Dyson P J, Rabbitts T H. Chromatin structure around the c-myc gene in Burkitts lymphomas with upstream and downstream translocation points. Proc Natl Acad Sci USA. 1985;82:1984–1988. doi: 10.1073/pnas.82.7.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Eick D, Bornkamm G W. Expression of normal and translocated c-myc alleles in Burkitt's lymphoma cells: evidence for different regulation. EMBO J. 1989;8:1965–1972. doi: 10.1002/j.1460-2075.1989.tb03602.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Eick D, Polack A, Kofler E, Bornkamm G W. The block of elongation in c-myc exon 1 is abolished in Burkitt's lymphoma cell lines with variant translocation. Oncogene. 1988;3:397–403. [PubMed] [Google Scholar]
- 12.Farabegoli F, Govoni M, Novello F. Effects of camptothecin, an inhibitor of DNA topoisomerase I, on ribosomal gene structure and function in TG cells. Biol Cell. 1992;74:281–286. doi: 10.1016/0248-4900(92)90039-4. [DOI] [PubMed] [Google Scholar]
- 13.Freeman L A, Garrard W T. DNA supercoiling in chromatin structure and gene expression. Crit Rev Eukaryot Gene Expr. 1992;2:165–209. [PubMed] [Google Scholar]
- 14.Gavin I, Horn P J, Peterson C L. SWI/SNF chromatin remodeling requires changes in DNA topology. Mol Cell. 2001;7:97–104. doi: 10.1016/s1097-2765(01)00158-7. [DOI] [PubMed] [Google Scholar]
- 15.Gewirtz D A. A critical evaluation of the mechanisms of action proposed for the antitumor effects of the anthracycline antibiotics adriamycin and daunorubicin. Biochem Pharmacol. 1999;57:727–741. doi: 10.1016/s0006-2952(98)00307-4. [DOI] [PubMed] [Google Scholar]
- 16.Gewirtz D A, Randolph J K, Chawla J, Orr M S, Fornari F A. Induction of DNA damage, inhibition of DNA synthesis and suppression of c-myc expression by the anthracycline analog, idarubicin (4-demethoxy-daunorubicin) in the MCF-7 breast tumor cell line. Cancer Chemother Pharmacol. 1998;41:361–369. doi: 10.1007/s002800050752. [DOI] [PubMed] [Google Scholar]
- 17.Giardina C, Perez-Riba M, Lis J T. Promoter melting and TFIID complexes on Drosophila genes in vivo. Genes Dev. 1992;6:2190–2200. doi: 10.1101/gad.6.11.2190. [DOI] [PubMed] [Google Scholar]
- 18.Gobert C, Skladanowski A, Larsen A K. The interaction between p53 and DNA topoisomerase I is regulated differently in cells with wild-type and mutant p53. Proc Natl Acad Sci USA. 1999;96:10355–10360. doi: 10.1073/pnas.96.18.10355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gromova I I, Thomsen B, Razin S V. Different topoisomerase II antitumor drugs direct similar specific long-range fragmentation of an amplified c-MYC gene locus in living cells and in high-salt-extracted nuclei. Proc Natl Acad Sci USA. 1995;92:102–106. doi: 10.1073/pnas.92.1.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hamlyn P H, Rabbitts T H. Translocation joins c-myc and immunoglobulin gamma 1 genes in a Burkitt lymphoma revealing a third exon in the c-myc oncogene. Nature. 1983;304:135–139. doi: 10.1038/304135a0. [DOI] [PubMed] [Google Scholar]
- 21.Havas K, Flaus A, Phelan M, Kingston R, Wade P A, Lilley D M J, Owen-Hughes T. Generation of superhelical torsion by ATP-dependent chromatin remodeling activities. Cell. 2000;103:1133–1142. doi: 10.1016/s0092-8674(00)00215-4. [DOI] [PubMed] [Google Scholar]
- 22.Hsieh T-S. Mechanistic aspects of type-II topoiosmerase. In: Cozzarelli N R, Wang J C, editors. DNA topology and its biological effects. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1990. pp. 243–263. [Google Scholar]
- 23.Johnson C A, Padget K, Austin C A, Turner B M. Deacetylase activity associates with topoisomerase II and is necessary for etoposide-induced apoptosis. J Biol Chem. 2001;276:4539–4542. doi: 10.1074/jbc.C000824200. [DOI] [PubMed] [Google Scholar]
- 24.Jupe E R, Sinden R R, Cartwright I L. Stably maintained microdomain of localized unrestrained supercoiling at a Drosophila heat shock gene locus. EMBO J. 1993;12:1067–1075. doi: 10.1002/j.1460-2075.1993.tb05748.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kroeger P E, Rowe T C. Analysis of topoisomerase I and II cleavage sites on the Drosophila Actin and Hsp70 heat shock genes. Biochem. 1992;31:2492–2501. doi: 10.1021/bi00124a008. [DOI] [PubMed] [Google Scholar]
- 26.Kroll D J, Sullivan D M, Gutierrez-Hartmann A, Hoeffler J P. Modification of DNA topoisomerase II activity via direct interactions with the cyclic adenosine-3′,5′-monophosphate response element-binding protein and related transcription factors. Mol Endocrinol. 1993;7:305–318. doi: 10.1210/mend.7.3.8387155. [DOI] [PubMed] [Google Scholar]
- 27.Krumm A, Meulia T, Brunvand M, Groudine M. The block to transcriptional elongation within the human c-myc gene is determined in the promoter-proximal region. Genes Dev. 1992;6:2201–2213. doi: 10.1101/gad.6.11.2201. [DOI] [PubMed] [Google Scholar]
- 28.Lavenu A, Pournin S, Babinet C, Morello D. The cis-acting elements known to regulate c-myc expression ex vivo are not sufficient for correct transcription in vivo. Oncogene. 1994;9:527–536. [PubMed] [Google Scholar]
- 29.Li T-K, Chen A Y, Yu C, Mao Y, Wang H, Liu L F. Activation of topoisomerase II-mediated excision of chromosomal DNA loops during oxidative stress. Genes Dev. 1999;13:1553–1560. doi: 10.1101/gad.13.12.1553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li T-K, Liu L F. Tumor cell death induced by topoisomerase-targeting drugs. Annu Rev Pharmacol Toxicol. 2001;41:53–77. doi: 10.1146/annurev.pharmtox.41.1.53. [DOI] [PubMed] [Google Scholar]
- 31.Liu L F. Anticancer drugs that convert DNA topoisomerases into DNA-damaging agents. In: Cozzarelli N R, Wang J C, editors. DNA topology and its biological effects. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1990. pp. 371–389. [Google Scholar]
- 32.Liu L F, Wang J C. Supercoiling of the DNA template during transcription. Proc Natl Acad Sci USA. 1987;84:7024–7027. doi: 10.1073/pnas.84.20.7024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ljungman M, Hanawalt P C. The anti-cancer drug camptothecin inhibits elongation but stimulates initiation of RNA polymerase II transcription. Carcinogen. 1996;17:31–35. doi: 10.1093/carcin/17.1.31. [DOI] [PubMed] [Google Scholar]
- 34.Madisen L, Krumm A, Hebbes T R, Groudine M. The immunoglobulin heavy chain locus control region increases histone acetylation along linked c-myc genes. Mol Cell Biol. 1998;18:6281–6292. doi: 10.1128/mcb.18.11.6281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Marks P A, Richon V M, Rifkind R A. Histone deacetylase inhibitors: inducers of differentiation or apoptosis of transformed cells. J Natl Cancer Inst. 2000;92:1210–1216. doi: 10.1093/jnci/92.15.1210. [DOI] [PubMed] [Google Scholar]
- 36.Mautner J, Behrends U, Hortnagel K, Brielmeier M, Hammerschmidt W, Strobl L, Bornkamm G W, Polack A. c-myc expression is activated by the immunoglobulin κ-enhancers from a distance of at least 30 kb but not by elements located within 50 kb of the unaltered c-myc locus in vivo. Oncogene. 1996;12:1299–1307. [PubMed] [Google Scholar]
- 37.Merino A, Madden K R, Lane W S, Champoux J J, Reinberg D. DNA topoisomerase I is involved in both repression and activation of transcription. Nature. 1993;365:227–232. doi: 10.1038/365227a0. [DOI] [PubMed] [Google Scholar]
- 38.Michelotti G A, Michelotti E F, Pullner A, Duncan R C, Eick D, Levens D. Multiple single-stranded cis elements are associated with activated chromatin of the human c-myc gene in vivo. Mol Cell Biol. 1996;16:2656–2669. doi: 10.1128/mcb.16.6.2656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Morales V, Richard-Foy H. Role of histone N-terminal tails and their acetylation in nucleosome dynamics. Mol Cell Biol. 2000;20:7230–7237. doi: 10.1128/mcb.20.19.7230-7237.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nelson P. Transport of torsional stress in DNA. Proc Natl Acad Sci USA. 1999;96:14342–14347. doi: 10.1073/pnas.96.25.14342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nishikura K, Erikson J, ar-Rushdi A, Huebner K, Croce C M. The translocated c-myc oncogene of Raji Burkitt lymphoma cells is not expressed in human lymphoblastoid cells. Proc Natl Acad Sci USA. 1985;82:2900–2904. doi: 10.1073/pnas.82.9.2900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Norton V G, Marvin K W, Yau P, Bradbury E M. Nucleosome linking number change controlled by acetylation of histones H3 and H4. J Biol Chem. 1990;265:19848–19852. [PubMed] [Google Scholar]
- 43.Orr M S, Fornari F A, Randolph J K, Gewirtz D A. Transcriptional down-regulation of c-myc expression in the MCF-7 breast tumor cell line by the topoisomerase II inhibitor, VM-26. Biochim Biophys Acta. 1995;1262:139–145. doi: 10.1016/0167-4781(95)00064-n. [DOI] [PubMed] [Google Scholar]
- 44.Plet A, Eick D, Blanchard J M. Elongation and premature termination of transcripts initiated from c-fos and c-myc promoters show dissimilar patterns. Oncogene. 1995;10:319–328. [PubMed] [Google Scholar]
- 45.Polack A, Eick D, Koch E, Bornkamm G W. Truncation does not abrogate transcriptional downregulation of the c-myc gene by sodium butyrate in Burkitt's lymphoma cells. EMBO J. 1987;6:2959–2964. doi: 10.1002/j.1460-2075.1987.tb02601.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Poljak L, Kas E. Resolving the role of topoisomerase II in chromatin structure and function. Trends Cell Biol. 1995;5:348–354. doi: 10.1016/s0962-8924(00)89068-6. [DOI] [PubMed] [Google Scholar]
- 47.Pommier Y, Kohlhagen G, Wu C, Simmons D T. Mammalian DNA topoisomerase I activity and poisoning by camptothecin are inhibited by simian virus 40 large T antigen. Biochemistry. 1998;37:3818–3823. doi: 10.1021/bi972067d. [DOI] [PubMed] [Google Scholar]
- 48.Pommier Y, Orr A, Kohn K W, Riou J-F. Differential effects of amsacrine and epipodophyllotoxins on topoisomerase II cleavage in the human c-myc protooncogene. Cancer Res. 1992;52:3125–3130. [PubMed] [Google Scholar]
- 49.Pommier Y, Pourquier P, Fan Y, Strumberg D. Mechanism of action of eukaryotic DNA topoisomerase I and drugs targeted to the enzyme. Biochim Biophys Acta. 1998;1400:83–105. doi: 10.1016/s0167-4781(98)00129-8. [DOI] [PubMed] [Google Scholar]
- 50.Potter M, Marcu K B. The c-myc story: where we've been, where we seem to be going. Curr Top Microbiol Immunol. 1997;224:1–17. doi: 10.1007/978-3-642-60801-8_1. [DOI] [PubMed] [Google Scholar]
- 51.Prunell A. A topological approach to nucleosome structure and dynamics: the linking number paradox and other issues. Biophys J. 1998;74:2531–2544. doi: 10.1016/S0006-3495(98)77961-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pullner A, Mautner J, Albert T, Eick D. Nucleosomal structure of active and inactive c-myc genes. J Biol Chem. 1996;271:31452–31457. doi: 10.1074/jbc.271.49.31452. [DOI] [PubMed] [Google Scholar]
- 53.Rabbitts T H, Forster A, Baer R, Hamlyn P H. Transcription enhancer identified near the human C mu immunoglobulin heavy chain gene is unavailable to the translocated c-myc gene in a Burkitt lymphoma. Nature. 1983;306:806–809. doi: 10.1038/306806a0. [DOI] [PubMed] [Google Scholar]
- 54.Rahmsdorf H J, Schonthal A, Angel P, Litfin M, Ruther U, Herrlich P. Posttranscriptional regulation of c-fos mRNA expression. Nucleic Acids Res. 1987;15:1643–1659. doi: 10.1093/nar/15.4.1643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ramsperger U, Stahl H. Unwinding of chromatin by the SV40 large T antigen DNA helicase. EMBO J. 1995;14:3215–3225. doi: 10.1002/j.1460-2075.1995.tb07324.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Riou J-F, Gabillot M, Riou G. Analysis of topoisomerase II-mediated DNA cleavage of the c-myc gene during HL60 differentiation. FEBS Lett. 1993;334:369–372. doi: 10.1016/0014-5793(93)80714-6. [DOI] [PubMed] [Google Scholar]
- 57.Riou J-F, Lefevre D, Riou G. Stimulation of the topoisomerase II induced DNA cleavage sites in the c-myc protooncogene by antitumor drugs is associated with gene expression. Biochemistry. 1989;28:1904–1910. doi: 10.1021/bi00449a022. [DOI] [PubMed] [Google Scholar]
- 58.Rius C, Zorrilla A R, Cabanas C, Mata F, Bernabeu C, Aller P. Differentiation of human promonocytic leukemia U-937 cells with DNA topoisomerase II inhibitors: induction of vimentin gene expression. Mol Pharmacol. 1991;39:442–448. [PubMed] [Google Scholar]
- 59.Scherf U, Ross D T, Waltham M, Smith L H, Lee J K, Tanabe L, Kohn K W, Reinhold W C, Myers T G, Andrews D T, Scudiero D A, Eisen M B, Sausville E A, Pommier Y, Botstein D, Brown P O, Weinstein J N. A gene expression database for the molecular pharmacology of cancer. Nat Genet. 2000;24:236–244. doi: 10.1038/73439. [DOI] [PubMed] [Google Scholar]
- 60.Schlake T, Klehr-Wirth D, Yoshida M, Beppu T, Bode J. Gene expression within a chromatin domain: the role of core histone hyperacetylation. Biochemistry. 1994;33:4197–4206. doi: 10.1021/bi00180a012. [DOI] [PubMed] [Google Scholar]
- 61.Shaiu W-L, Hsieh T-S. Targeting to transcriptionally active loci by the hydrophilic N-terminal domain of Drosophila DNA topoiosmerase I. Mol Cell Biol. 1998;18:4358–4367. doi: 10.1128/mcb.18.7.4358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Shtivelman E, Bishop J M. Effects of translocations on transcription from PVT. Mol Cell Biol. 1990;10:1835–1839. doi: 10.1128/mcb.10.4.1835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Shykind B M, Kim J, Stewart L, Champoux J J, Sharp P A. Topoisomerase I enhances TFIID-TFIIA complex assembly during activation of transcription. Genes Dev. 1997;11:397–407. doi: 10.1101/gad.11.3.397. [DOI] [PubMed] [Google Scholar]
- 64.Smith P J, Rackstraw C, Cotter F. DNA fragmentation as a consequence of cell cycle traverse in doxorubicin- and idarubicin-treated human lymphoma cells. Ann Hematol. 1994;69:S7–S11. doi: 10.1007/BF01757348. [DOI] [PubMed] [Google Scholar]
- 65.Spencer C A, Groudine M. Control of c-myc regulation in normal and neoplastic cells. Adv Cancer Res. 1991;56:1–48. doi: 10.1016/s0065-230x(08)60476-5. [DOI] [PubMed] [Google Scholar]
- 66.Stewart A F, Herrera R E, Nordheim A. Rapid induction of c-fos transcription reveals quantitative linkage of RNA polymerase II and DNA topoisomerase I enzyme activities. Cell. 1990;60:141–149. doi: 10.1016/0092-8674(90)90724-s. [DOI] [PubMed] [Google Scholar]
- 67.Strauss W M. Preparation of genomic DNA from mammalian tissue. In: Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Protocols in molecular biology. Current protocols in molecular biology. Vol. 1. New York, N.Y: John Wiley & Sons, Inc.; 1994. pp. 2.2.1–2.2.3. [Google Scholar]
- 68.Suzuki T, Uchida-Toita M, Andoh T, Yoshida M. HTLV-1 Tax oncoprotein binds to DNA topoisomerase I and inhibits its catalytic activity. Virology. 2000;270:291–298. doi: 10.1006/viro.2000.0266. [DOI] [PubMed] [Google Scholar]
- 69.Theodorakis N G, Morimoto R I. Posttranscriptional regulation of hsp70 expression in human cells: effects of heat shock, inhibition of protein synthesis, and adenovirus infection on translation and mRNA stability. Mol Cell Biol. 1987;7:4357–4368. doi: 10.1128/mcb.7.12.4357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Travers A, Muskhelishvili G. DNA microloops and microdomains: a general mechanism for transcription activation by torsional transmission. J Mol Biol. 1998;279:1027–1043. doi: 10.1006/jmbi.1998.1834. [DOI] [PubMed] [Google Scholar]
- 71.Tsai S-C, Valkov N, Yang W-M, Gump J, Sullivan D, Seto E. Histone deacetylase interacts directly with DNA topoisomerase II. Nat Genet. 2000;26:349–353. doi: 10.1038/81671. [DOI] [PubMed] [Google Scholar]
- 72.Varga-Weisz P D, Wilm M, Bonte E, Dumas K, Mann M, Becker P B. Chromatin-remodelling factor CHRAC contains the ATPases ISWI and topoisomerase II. Nature. 1997;388:598–602. doi: 10.1038/41587. [DOI] [PubMed] [Google Scholar]
- 73.Wang J C. DNA topoisomerases. Annu Rev Biochem. 1996;65:635–692. doi: 10.1146/annurev.bi.65.070196.003223. [DOI] [PubMed] [Google Scholar]
- 74.Yin H, Wang M D, Svoboda K, Landick R, Block S M, Gelles J. Transcription against an applied force. Science. 1995;270:1653–1657. doi: 10.1126/science.270.5242.1653. [DOI] [PubMed] [Google Scholar]
- 75.Zhang H, Wang J C, Liu L F. Involvement of DNA topoisomerase I in transcription of human ribosomal RNA genes. Proc Natl Acad Sci USA. 1988;85:1060–1064. doi: 10.1073/pnas.85.4.1060. [DOI] [PMC free article] [PubMed] [Google Scholar]