Abstract
Simple Summary
Gastrointestinal stromal tumors (GISTs) are rare malignancies of the gastrointestinal tract recognized by their clinical presentation and using specific immunohistochemical staining for CD117 and DOG1. In recent years, prognoses of GISTs patients have significantly improved due to the introduction of tyrosine kinase inhibitors (TKIs) in clinical practice. KIT/PDGFRA-activating mutations are major driver alterations in the development of GISTs, leading to ligand-independent activation of KIT/PDGFRA receptors. The activated KIT/PDGFRA receptor is sensitive to the TKI imatinib. However, not all GIST patients respond to imatinib. Precise characterization of the driver mutations in GISTs—in particular, in the KIT and PDGFRA genes—and an understanding of the molecular mechanisms underlying resistance to imatinib and other TKIs should allow clinicians to select the most effective targeted drug as part of regular clinical practice. The correct choice of the TKI in the sequence of targeted agents should lead to improved survival for metastatic patients.
Abstract
Gastrointestinal stromal tumors (GISTs) are soft tissue sarcomas that mostly derive from Cajal cell precursors. They are by far the most common soft tissue sarcomas. Clinically, they present as gastrointestinal malignancies, most often with bleeding, pain, or intestinal obstruction. They are identified using characteristic immunohistochemical staining for CD117 and DOG1. Improved understanding of the molecular biology of these tumors and identification of oncogenic drivers have altered the systemic treatment of primarily disseminated disease, which is becoming increasingly complex. Gain-of-function mutations in KIT or PDGFRA genes represent the driving mutations in more than 90% of all GISTs. These patients exhibit good responses to targeted therapy with tyrosine kinase inhibitors (TKIs). Gastrointestinal stromal tumors lacking the KIT/PDGFRA mutations, however, represent distinct clinico-pathological entities with diverse molecular mechanisms of oncogenesis. In these patients, therapy with TKIs is hardly ever as effective as for KIT/PDGFRA-mutated GISTs. This review provides an outline of current diagnostics aimed at identifying clinically relevant driver alterations and a comprehensive summary of current treatments with targeted therapies for patients with GISTs in both adjuvant and metastatic settings. The role of molecular testing and the selection of the optimal targeted therapy according to the identified oncogenic driver are reviewed and some future directions are proposed.
Keywords: GIST, KIT, PDGFRA, mutations, targeted therapy
1. Introduction
Gastrointestinal stromal tumors (GISTs) are the most common mesenchymal neoplasms of the gastrointestinal tract, but they account for less than 1% of all gastrointestinal tumors. Incidence varies worldwide from 4.3/1 × 106 inhabitants per year to 21.1/1 × 106 inhabitants per year [1,2,3,4,5,6,7]. The incidence of GISTs is expected to be underestimated [8]. Most GISTs are indolent in their course and are discovered incidentally, but some are aggressive and disseminate early [8,9]. Gastrointestinal stromal tumors generally arise from pacemaker (Cajal) cells anywhere in the digestive tube—from the esophagus to the rectum—but, more recently, evidence has shown that they can also arise from telocytes or smooth muscle cells [10,11,12]. They are a heterogeneous group of diseases of different molecular subtypes, with oncogenesis mainly resulting from mutually exclusive activating mutations, most commonly in the KIT proto-oncogene (KIT) or the platelet-derived growth factor receptor alpha gene (PDGFRA) [13,14]. Around 5% of GISTs are syndromic and associated with germline mutations in genes encoding for KIT, PDGFRA, succinate dehydrogenase B/C/D (SDHB/C/D) (Carney Stratakis syndrome), and neurofibromin or with epigenetic silencing of SDHC (nonhereditary Carney triad syndrome) [15,16,17,18,19,20,21]. Patients confirmed to have GISTs with neurofibromin 1 (NF1) mutation or a deficient SDH complex should be referred to the outpatient clinic for genetic counseling.
Surgery is the only curative treatment for GISTs [22,23]. Improved understanding of the molecular biology of the disease and the identification of driver alterations and mechanisms of resistance to systemic therapies have resulted in advances in the systemic treatment of GISTs, thereby broadening the systemic therapy armamentarium. Tyrosine kinase inhibitors (TKIs) represent the standard systemic therapy and comprise various targeted drugs [22,23]. Chemotherapy is ineffective in patients with GISTs, and the time to disease progression with chemotherapy is less than 3 months [24]. Targeted therapy with the TKI of KIT/PDGFRA imatinib mesylate (imatinib) prolonged overall survival (OS) after surgery for high-risk GISTs [25]. Metastatic disease remains incurable; however, treatment with TKIs prolongs survival from 1.5 to over 5 years [22,23,26]. Despite the convincing achievements with TKI treatment, targeted therapy eventually leads to the development of drug resistance. Secondary mutations play a major role in this process, allowing for the selection of cells that are resistant to the treatment applied [27,28,29,30].
This review provides an outline of current knowledge on the molecular mechanisms of GISTs and the selection of the optimal systemic treatment according to the identified molecular mechanisms, both in limited and metastatic disease, and proposes future directions for research.
2. Molecular Classification of GISTs
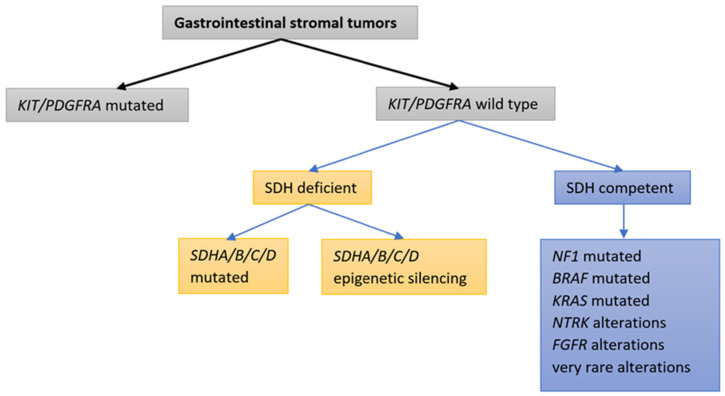
The proto-oncogene KIT encodes the KIT receptor, which is a type III receptor tyrosine kinase (RTK); it belongs to a family of RTKs that also includes PDGFRA, platelet-derived growth factor receptor beta (PDGFRB), colony-stimulating factor 1 receptor (CSF1R), and Fms-like RTK 3 (FLT3). The receptor tyrosine kinase KIT (CD 117) is normally expressed in Cajal cells in the gastrointestinal tract. It plays an important role in the development of a normal pacemaker system in the gut [10]. In 1998, Hirota and colleagues discovered that activating KIT mutations are the major mechanism of GIST oncogenesis [14]. Activating mutations in KIT lead to the formation of a permanently active protein that is a target for imatinib binding. Of equal importance—and the second most common driver mutations in GISTs—are mutations in PDGFRA, which encodes the PDGFRA receptor tyrosine kinase. The PDGFRA RTK itself is homologous to the KIT RTK and, again, an activating mutation in PDGFRA results in the formation of a permanently active RTK that is also a target for imatinib. KIT/PDGFRA-induced oncogenesis mediates the rapidly accelerated fibrosarcoma (RAF)–mitogen-activated protein kinase (MEK)–mitogen-activated protein kinases (MAPK) (RAF-MEK-MAPK) and phosphatidylinositol 3-kinase (PI3K)/protein kinase B (AKT) (PI3K-AKT) signaling pathways [10,31,32]. At present, it is believed that KIT/PDGFRA-activating mutations are mutually exclusive [28]. In recent years, it has been discovered that only 85–90% of GISTs have an activating mutation in KIT/PDGFRA, while in the remaining 10–15%, the molecular mechanism of oncogenesis has not been determined (historical wild-type (WT) GISTs). With advances in molecular diagnostic techniques—in particular, with the use of next-generation sequencing (NGS) technology—the mechanisms of oncogenesis can be more precisely defined, even in historical WT GISTs. With the increased sensitivity of the NGS method, it has been discovered that driver mutations in KIT/PDGFRA are also the most frequent in historical WT GISTs. Taking this fact into account, KIT/PDGFRA mutations represent the driving mutations in as many as 92–93% of all GISTs [22,33,34,35,36,37,38]. In 5–7.5% of all GISTs, the driving oncogene mechanism is related to a deficiency in the succinate dehydrogenase (SDH) complex [16,18,19,28,39,40,41,42,43,44,45,46]. When no KIT/PDGFRA mutations are detected and the SDH complex is competent, various very rare driver alterations have been identified: alterations in the Rat sarcoma virus (RAS) gene family, the v-Raf murine sarcoma viral oncogene homolog B1 (BRAF) gene, the NF1 gene, the neurotropic tyrosine receptor kinase 1–3 (NTRK1–3) genes, and the fibroblast growth factor receptor 1–4 (FGFR1–4) genes [18,28,33,39,40,44,47,48,49,50,51]. “True” WT GISTs for which advanced molecular techniques fail to demonstrate any driving alteration are very rare [33,34,35,36,37]. Figure 1 illustrates the principal molecular classifications of GISTs.
Figure 1.
Classifications of GISTs based on tumor genetic alterations: molecular classifications.
Different molecular mechanisms in GISTs lead to different clinical courses of the disease and, in particular, different responses to systemic imatinib treatment. Therefore, it is necessary to identify the driving alterations before initiating the first systemic therapy, both in (neo)adjuvant settings and as the first systemic therapy for metastatic disease [22,23].
3. Identifying the Molecular Mechanisms of GIST
Molecular analysis is an important factor influencing decision about systemic treatment for both limited and metastatic disease. Molecular markers have both prognostic and predictive value [52,53,54,55]. The two methods so far predominantly used to identify molecular markers are reverse transcription polymerase chain reaction (RT-PCR) and direct Sanger sequencing. Both RT-PCR and Sanger sequencing have their limitations, primarily reflected in the restricted testing for known driver changes (hot spot mutations) and in the time-consuming process of identifying driver changes in the selected gene. Even then, these methods are only successful in cases with a relatively high allele frequency for the altered gene. For Sanger sequencing, the detection limit is set at 20% of the altered deoxyribonucleic acid (DNA) in the sample. For these reasons, NGS, which has a substantially lower detection limit and allows for the identification of changes in a large number of genes in several samples simultaneously, is progressively entering routine diagnostics. Several studies have been published demonstrating that NGS outperforms RT-PCR and direct Sanger sequencing in its ability to identify molecular alterations in GISTs [34,48,49,50,51]. Next-generation sequencing also exhibits a higher positive predictive value than RT-PCR and direct Sanger sequencing [33,34]. The European Society of Medical Oncology (ESMO) guidelines for the management of patients with GISTs recommend centralization of molecular testing, although they allow either Sanger sequencing or NGS as the applied method. If no driver mutation in KIT/PDGFRA/BRAF is demonstrated, they advise immunohistochemical (IHC) staining to determine SDH B protein (SDHB) expression. In GISTs where no driver alteration in KIT/PDGFRA/BRAF is established and SDHB is expressed, they further recommend excluding driver alterations in NF1 [22]. The National Comprehensive Cancer Network (NCCN) recommendations, on the other hand, are more specific: patients in whom sequencing fails to demonstrate mutation in KIT/PDGFRA and who have a competent SDH complex, as confirmed by IHC staining, should be tested with NGS to identify any targetable driver alterations in other genes (BRAF, NTRK, FGFR) [23].
3.1. GISTs with KIT or PDGFRA Mutations
The human KIT proto-oncogene is located on chromosome 4 (q12). This gene was discovered in 1987 [56]. A nonmutated KIT encodes a type III RTK. KIT protein is a transmembrane RTK with extracellular (EC), transmembrane (TM), and intracellular (IC) domains. The EC domain is composed of five Ig-like regions; three are responsible for stem cell factor (SCF) binding and the other two for protein dimerization after SCF binding. The TM helix domain links the EC to the IC domain; the IC domain is composed of a juxtamembrane domain (JMD) and a tyrosine kinase domain (TKD). The TKD is composed of a phosphotransferase domain (PTD), an adenosine triphosphate (ATP)-binding site, and an activation loop. The JMD part controls and regulates the function of TKDs [57]. Binding of SCF causes homodimerization of the two KIT RTKs, leading to autophosphorylation of the homodimer. This releases adenosine diphosphate (ADP) and binds ATP to the active site. Further phosphorylations of parts of the IC domain of the KIT RTK follow, and only when these are complete is KIT fully active [58]. Activating mutations in KIT are the major contributor to oncogenesis in GISTs since they lead to permanent activation of KIT protein without the need for prior SCF (ligand) binding. The majority (approximately 60%) of mutations in KIT are in exon 11, including deletions (the largest number between codons 550 and 560), deletions and insertions (indels), and insertions and missense mutations, resulting in structural changes in the JMD [32,38,58]. This region has an autoinhibitory function in kinase activation, which is reduced by the mutation. Less frequently (9–10%), mutations may occur in exon 9, which encodes the EC domain of KIT (mostly tandem duplications) [32,38,58]. Mutations in KIT exon 8 (EC domain) and in exons 13 and 14 (ATP binding site on TKD), and 17 (IC domain of activation loop) are very rare events in GIST oncogenesis [32,38].
Activated KIT triggers different signaling pathways: RAS/MAP/MAPK, PI3K/AKT, phospholipase C gamma (PLC-gamma), Janus kinase (JAK)/signal transducer and activator of transcription (STAT) (JAK/STAT), and Scr kinase pathways. Which pathway will be activated depends on which tyrosine residue of the IC domain is phosphorylated [57].
In humans, the proto-oncogene PDGFRA is also located on chromosome 4 (q12), in the same region as KIT, and encodes a class III RTK that is structurally homologous to KIT RTK. The activation of downstream pathways is also similar to KIT; PDGFRA activation predominantly activates the RAS/RAF/MAPK and PI3K/AKT signaling pathways [10,31,32]. In GIST, mutations of PDGFRA are more infrequent than mutations of KIT [32]. The greater number (up to 15% of all GISTs) of PDGFRA mutations affect exon 18 (the activation loop of the IC domain); less frequently (up to 2% of all GISTs), they may affect exon 12 (in the JMD) and, even less frequently (1%), exon 14 (the ATP binding site TKD) [38,57]. PDGFRA exon 18 D842V mutation is the most common PDGFRA mutation (50–70% of PDGFRA mutant GISTs and about 8% of all GISTs) and results in a stable conformational structure for tyrosine kinase (TK) in its active form [59].
3.1.1. Targeted Therapy for GISTs with KIT or PDGFRA Mutations
Three years of adjuvant treatment with 400 mg/day imatinib in KIT/PDGFRA-mutated GISTs with high risk for disease recurrence prolongs recurrence-free survival (RFS) (ten-year RFS: 52.5% vs. 41.8%) and OS (ten-year OS: 79.0% vs. 65.3%) compared to 1 year of treatment alone [25,60,61]. A high estimated risk of recurrence (higher than 50%) is assessed according to the modified National Institutes of Health (NIH) consensus criteria [62,63]. The efficacy of adjuvant imatinib treatment depends on the type of KIT/PDGFRA mutation. Patients with PDGFRA mutations, KIT exon 11 duplications, insertions, and substitutions have longer RFS than patients with KIT exon 11 deletions or indel mutations. Patients with KIT exon 9 mutations (most commonly an AY duplication) have the shortest RFS. Patients with KIT exon 11 deletions or indels (especially if they affect codons 557 and/or 558) have a significantly longer RFS if treated with 3 years of imatinib compared to 1 year. This difference in RFS with regard to the duration of adjuvant imatinib treatment has not been observed in other groups (KIT exon 11 substitutions and KIT exon 9 mutations) [61]. Given the longer progression-free survival (PFS [64]) and better response to treatment in KIT exon 9-mutated metastatic patients with the 800 mg dose compared to the 400 mg/day dose, the question of an escalated dose of imatinib for KIT exon 9 mutation also arises in the context of adjuvant therapy. Results from a multi-institutional European retrospective study of patients with KIT exon 9-mutated GISTs treated with adjuvant imatinib revealed that a higher daily dose of 800 mg versus 400 mg did not improve survival outcomes (RFS: hazard ratio (HR), 1.24; mRFS: HR, 1.69; imatinib failure-free survival (IFFS): HR, 1.35; 95% CI, 0.79–2.28) [65]. However, no adjuvant prospective randomized studies with an escalated dose of imatinib in this population have been published so far [53,66,67].
Taking into consideration that a longer duration of adjuvant imatinib therapy leads to longer survival, another dilemma appears regarding further prolongation of adjuvant imatinib therapy beyond 3 years. In a phase 2 clinical trial involving imatinib-sensitive GISTs with 5 years of adjuvant imatinib therapy following resection in patients with a high risk of recurrence, the five-year RFS estimate was 90% and the five-year OS estimate was 95% [68]. Two more phase 3 clinical trials are currently underway comparing 3 versus 5 years of adjuvant imatinib and 3 versus 6 years of adjuvant imatinib for GISTs with a high risk of recurrence (NCT02413736 and NCT02260505). Currently, the standard adjuvant treatment is recommended for 3 years [22,23]. Table 1 shows the results of pivotal clinical trials employing imatinib as adjuvant therapy.
The type of KIT/PDGFRA mutation is not the only factor influencing adjuvant treatment efficacy. Other factors, such as the duration of adjuvant treatment, primary site (non-gastric primary sites being connected to poorer disease-free survival (DFS)), tumor size (larger tumor size being associated with a poorer DFS), mitotic index (high mitotic index being associated with poorer DFS), and female sex (an independent prognostic factor for a higher PFS and OS), have also been shown to contribute [52,69].
Table 1.
Pivotal clinical studies with imatinib in the adjuvant setting.
| Study First Author/Publication Information | Number of Patients, Patient Population | Clinical Phase | Intervention | Molecular Analysis | Primary Endpoint | Results |
|---|---|---|---|---|---|---|
| Dematteo et al., Lancet 2009 [70] | 713 Patients resected for ≥3 cm GIST |
3 | 1 y adjuvant imatinib 400 mg/d vs. placebo |
No | RFS (primary endpoint changed from OS to RFS) | 1 y RFS: 98% (imatinib) vs. 83% (placebo), HR 0.35. No OS benefits |
| Casali et al., JCO 2015 [71] | 908 Patients after R0-1 surgery for localized high- or intermediate-risk GISTs according to NIH criteria [72] |
3 | 2 y adjuvant imatinib 400 mg/d vs. no treatment |
No | IFFS (primary endpoint changed from OS to IFFS) | 5 y IFFS: 87.0% (imatinib) vs. 84.1% (control), p = 0.21, HR 0.79. No OS benefits |
| Joensuu et al., JAMA 2020 [25] | 397 Patients resected for high-risk GISTs according to modified NIH criteria [62] |
3 | 1 y vs. 3 y adjuvant imatinib 400 mg/d |
Yes (366/397) | RFS | 10 y RFS: 52.5% (3 y treatment) and 41.8% (1 y treatment), HR 0.66, p = 0.003 10 year OS: 79.0% (3 y treatment) and 65.3% (1 y treatment), p = 0.004, HR: 0.55. |
R0—radical surgery, R1—surgery with positive microscopic margin, OS—overall survival, RFS—recurrence-free survival, NIH—National Institutes of Health, IFFS—imatinib failure-free survival, d—day, y—year, HR—hazard ratio.
The goal of systemic treatment of metastatic or unresectable KIT/PDGFRA-mutated GISTs with imatinib is to prolong survival with good quality of life. The initial dose of imatinib is 400 mg/day, except for patients where KIT exon 9 mutation is known to be the oncogenic driver; in these patients, treatment is started upfront with an escalated dose of 800 mg/day [67]. In most cases, the dose of 400 mg daily results in up to 5% complete response, 40–68% partial response, and 14–32% stable disease, with a PFS of approximately 40 months [24,66,73,74,75,76].
However, there is a group of patients who respond worse to imatinib initiation; namely, some patients with KIT exon 11 mutations who, at the same time, bear polymorphisms in genes involved in imatinib metabolism (reducing metabolic capacity), patients with KIT exon 9 mutations, patients without KIT/PDGFRA mutations (WT GISTs), and patients with primary resistance to imatinib, especially those carrying PDGFRA D842V and KIT D816V mutations [32,77,78]. Patients with KIT exon 9 mutation achieve a PFS of only 12.6–16.7 months with 400 mg/day imatinib [53,66,67]. However, an escalated dose of 800 mg/d of imatinib has been shown to be more effective in these patients, giving an objective response rate (ORR) of 47% (vs. 21%) and a better PFS (HR = 0.57; p = 0.017) but without improving the OS [53]. Patients with “true” WT GISTs do not respond to imatinib treatment because their tumors have no target to which imatinib would bind [34,66,67]. Primary resistance to imatinib is supposed to be a landmark of GISTs bearing PDGFRA D842V or KIT D816V mutations [79,80,81]. Nevertheless, cases of partial response to imatinib in patients with the PDGFRA D842V mutation have been described [34,37,82].
3.1.2. Progression of GISTs with KIT or PDGFRA Mutations after the First-Line Targeted Therapy
Disease progression is frequently due to the occurrence of secondary mutations resulting from evolutionary selection pressure during imatinib treatment [83]. They occur in 85 to 90% of patients, with a median time to onset for secondary mutations—and, thus, disease progression—of 20–24 months [35,74,84,85]. Secondary mutations result in a modified RTK structure. As an outcome, imatinib is no longer effective, as no optimal binding sites remain for imatinib; thus, the inhibition of signaling via mutated RTK ceases. Continuous activation of RTK is re-established and the disease progresses.
In everyday clinical practice, we do not strive to confirm the exact mechanism of resistance due to the heterogeneity of mutations occurring at progression. If the progression is focal (a “nodule within a mass” up to one or a few nodule(s)/mass(es) while the rest of the disease is still responding), surgery or nonsurgical procedures (ablation, radiotherapy) may be selected [22,23]. When local ablative therapy is not feasible, second-line therapy with multitargeted TKIs is initiated.
Secondary mutations occur mainly at specific sites in KIT or PDGFRA. However, the response to second-line TKI treatment is not always the same and depends on the mechanism of resistance and the efficacy of the TKI applied. Secondary mutations, typically missense mutations, most commonly affect the coding region of the ATP binding site (exons 13 and 14 of KIT and exon 14 of PDGFRA), as well as the coding region of the TKD activation loop (exon 17 of KIT and exon 18 of PDGFRA) [30]. KIT/PDGFRA receptor tyrosine kinases with secondary mutations in regions encoding the ATP binding site are sensitive to treatment with sunitinib, ripretinib, avapritinib, and ponatinib but do not respond to treatment with regorafenib or sorafenib [86,87,88,89,90,91]. KIT/PDGFRA receptor tyrosine kinases with secondary mutations in genes encoding the activation loop are sensitive to regorafenib, ripretinib, avapritinib, sorafenib, nilotinib, and ponatinib [87,89,90,92,93,94,95].
Following disease progression with imatinib, the current guidelines recommend the use of three multitargeted TKIs—sunitinib, regorafenib, and ripretinib. There are also data in the literature on the efficacy of other multitargeted TKIs in this setting (clinical trials on the efficacy of sorafenib, dovitinib, masitinib, ponatinib, nilotinib, and pazopanib), but none of them exceeded a 6 month barrier for mPFS [22,23,96]. The prototype for multitargeted TKIs is sunitinib, a small molecule that inhibits the activity of multiple (over 80) RTKs involved in tumor growth, pathological angiogenesis, and malignant cell proliferation. Sunitinib inhibits PDGFRA and PDGFRB; vascular endothelial growth factor receptors 1, 2, and 3 (VEGFR1, VEGFR2, and VEGFR3); KIT; FLT3; CSF-1R; and glial neurotropic factor receptor (RET), among others [97]. Other multitargeted TKIs have similar but not identical spectra of activity.
The mechanisms of secondary mutations in PDGFRA are poorly understood, with well-known primary resistance to imatinib only in PDGFRA D482V (and its homologue KIT exon 17 mutation D816V) [32]. Avapritinib represents a new option for systemic therapy targeting PDGFRA D842V. In a phase 3 clinical study, it exhibited good efficacy and an objective response of 84% [90].
Other mechanisms of GIST resistance to imatinib are infrequent and poorly understood. Resistance could potentially be induced by activation of other oncogenic pathways—e.g., dysregulation of cyclin-dependent kinase inhibitor 2A (CDKN2A); loss of tumor protein 53 (TP53) function; inactivation of dystrophin; genomic alterations in chromosomes 1p, 14q, and 22q; and overexpression of KIT [47,59,98,99,100].
Table 2 shows pivotal clinical trials of targeted drugs recommended for systemic treatment of metastatic GISTs. Sunitinib exhibited moderate efficacy after progression with imatinib in a randomized phase 3 clinical trial (RCT) involving metastatic GISTs, with a significantly longer PFS (27.3 weeks vs. 6.4 weeks with placebo; HR 0.33; p < 0.0001) [86]. The efficacy of sunitinib is, however, not universal. It depends on the type of primary mutation and secondary mutation. In patients with the primary mutation in KIT exon 9, sunitinib elicits improved PFS in OS compared to those with the primary mutation in KIT exon 11 [101,102]. Furthermore, the type of secondary mutation influences PFS and OS [30,86,102]. In a phase 3 RCT, regorafenib prolonged median PFS after treatment with imatinib and sunitinib compared to placebo (4.8 months vs. 0.9 months; HR 0.27; p < 0.0001) [92]. The efficacy of regorafenib is again not universal as it depends on the type of secondary mutation [30,64]. In a phase 3 RCT, ripretinib prolonged median PFS after treatment with imatinib, sunitinib, and regorafenib compared to placebo (6.3 months vs. 1.0 month; HR 0.15; p < 0.0001). The efficacy of ripretinib is also not universal, and it depends on the type of secondary mutation [103]. In a phase 1 clinical trial, avapritinib employed for patients with unresectable GISTs and PDGFRA D842V mutations, regardless of prior systemic therapy, achieved an objective response in 88% of patients (9% complete response, 79% partial response) [90]. The efficacy of avapritinib in KIT/PDGFRA-mutated, imatinib-pretreated (PDGFRA D842V excluded) patients is only modest, with an ORR of 17%, median duration of response (mDOR) of 10.2 months, and mPFS of 3.7 months [104].
Table 2.
Pivotal clinical trials of TKIs with European Medicines Agency (EMA) approval for the treatment of unresectable or metastatic GISTs.
| First Author, Publication Information | Number of Patients, Patient Population |
Clinical Phase | Intervention | Molecular Analysis Performed |
Primary Endpoint | Response Evaluation Criteria |
Results |
|---|---|---|---|---|---|---|---|
| Demetri et al., NEJM 2002 [76] | 147 Advanced GISTs |
2 | Imatinib 400 mg/d vs. imatinib 600 mg/d | No | ORR | Southwest Oncology Group criteria [105] |
ORR: 49.3% (400 mg) vs. 58.1% (600 mg) Estimated 1 y OS: 88%. |
| Verweij et al., Lancet 2004 [24] |
946 Metastatic or unresectable GISTs |
3 | Imatinib 400 mg/d vs. imatinib 800 mg/d | No | PFS | RECIST 1.0 | PFS longer in the group with 800 mg vs. 400 mg, HR 0.82, p = 0.026 1 y OS: 85% (400 mg) vs. 86% (800 mg) ORR: 50.1% (400 mg) vs. 54.3% (800 mg) |
| Blanke et al., JCO 2008 [75] | 746 Metastatic or unresectable GISTs |
3 | Imatinib 400 mg/d vs. imatinib 800 mg/d | No | PFS and OS | RECIST 1.0 | Median PFS: 18 m (400 mg) vs. 20 m (800 mg), p = 0.13 Median OS: 55 m (400 mg) vs. 51 m (800 mg), p = 0.83 No difference in RR between the two arms |
| Demetri et al., Lancet 2006 [86] | 312 Advanced GISTs resistant or intolerant to imatinib |
3 | Sunitinib 50 mg vs. placebo | No | TTF | RECIST 1.0 or WHO (WHO Handbook for Reporting Results of Cancer Treatment) |
Median TTP: 6.3 m (sunitinib) vs. 1.5 m (placebo), HR 0.33, p < 0.0001 Median PFS: 5.5 m (sunitinib) vs. 1.4 m (placebo), HR 0.33, p < 0.0001 Median OS: not reached. HR 0.49, p = 0.007 ORR: 7% (sunitinib) and 0% (placebo), p = 0.006 |
| George et al., EJC 2009 [106] | 60 Patients with unresectable GISTs resistant or intolerant to imatinib |
2 | Sunitinib 37.5 mg/d | No | DCR | RECIST 1.0 |
DCR: 53% Median PFS: 7.8 m Median OS: 24.6 m ORR: 13% 1 y survival rate: 70% |
| Demetri et al., Lancet 2013 [92] | 199 Metastatic or unresectable GISTs resistant to imatinib and sunitinib |
3 | Regorafenib 160 mg/d vs. placebo | Yes * | PFS | Modified RECIST 1.1 [92] | Median PFS 4.8 m (regorafenib) vs. 0.9 m (placebo), HR 0.27, p < 0.0001 HR OS: 0.77, p = 0.199 DCR 52.6% (regorafenib) vs. 9.1% (placebo) |
| Blay et al., Lancet Oncol 2020 [89] | 129 Advanced GISTs with resistance or intolerance to imatinib, sunitinib, and regorafenib |
3 | Ripretinib 150 mg/d + BSC vs. placebo + BSC | Yes (112/129) |
PFS | Modified RECIST 1.1 [92] | Median PFS: 6.3 m (ripretinib) vs. 1.0 m (placebo), HR: 0.15, p < 0.0001 Median OS: 15.1 m (ripretinib) vs. 6.6 m (placebo), HR 0.36 1 y estimated OS: 65.4% (ripretinib) vs. 25.9% (placebo) ORR: 9% |
| Heinrich et al. Lancet Oncol 2020 [90] | 56 Unresectable PDGFRA D842V-GISTs, regardless of previous therapy |
1 | Avapritinib 300/400 mg/d | Yes (56/56) |
ORR | Modified RECIST 1.1 [92] | ORR: 91% CBR: 98% Median DOR: 27.6 m Median PFS: 34.0 m |
*—number not provided; ORR—objective response rate, OS—overall survival, PFS—progression-free survival, RECIST—response evaluation in solid tumors, TTF—time to treatment failure, WHO—World Health Organization, DCR—disease control rate, CBR—clinical benefit rate, d—day, m—months, y—year, HR—hazard ratio.
3.2. GISTs without KIT/PDGFRA Mutations
3.2.1. GISTs without KIT/PDGFRA Mutations and with Deficient SDH Complex
The SDHA tumor suppressor gene (TSG) is located on chromosome 5 (p15.33), the SDHB TSG on chromosome 1 (p36.13), the SDHC TSG on chromosome 1 (q23.3), and the SDHD TSG on chromosome 11 (q23.1). They encode the four subunits (SDHA, SDHB, SDHC, and SDHD) of heterotetrametric enzyme SDH, the key enzyme in the Krebs cycle and respiratory chain in mitochondria.
The SDH enzyme complex catalyzes the oxidation of succinate to fumarate. Loss of function of mitochondrial SDH (due to mutations in the SDHA, SDHB, SDHC, or SDHD Miettinen et Lasotation of SDHB [41]. Succinate accumulates and inhibits the activity of dioxygenases (ten-eleven translocation methylcytosine dioxygenases (TETs) and histone lysine (K) demethylases (KDMs)). These enzymes degrade hypoxia-inducible factor 1a (HIF-1a) protein, which accumulates in the absence of dioxygenases and increases transcription of the genes it regulates: the insulin-like growth factor 1 receptor (IGF1R) and VEGFR. Furthermore, a lack of dioxygenase activity triggers hypermethylation of DNA; i.e., epigenetic silencing. Consequently, activation of IGF1R and VEGFR and/or an increase in DNA methylation lead to malignant transformation of normal interstitial Cajal cells into GISTs [21,39,107].
Deficiency in the SDH complex is a rare event in GISTs, present in up to 5–7.5% of GIST patients [16,18,19,28,39,40,41,42,43,44,45,46]. When the gene coding for any subunit is biallelically inactivated, IHC staining for SDHB is absent and shows only KIT and DOG1 [18,46]. Complex mutations in genes for subunits of the SDH complex are found in non-syndromic GISTs with a deficient SDH complex [46]. Since somatic changes are extremely rare, the absence of IHC staining for SDHB is highly likely to indicate syndromic disease caused by germline mutation [46]. Indeed, in half of patients with deficient SDH complexes, the trigger of the GIST is a germline-inactivating (“loss of function”) mutation of a gene encoding one of the SDH subunits (typically a germline SDHA mutation) in combination with a somatic mutation (“frameshift” deletion with stop codon, “missense”, “nonsense”, and “splice site” mutations) [16,46]. Germline mutations in genes coding for the SDHB/C or D subunits are associated with a rare hereditary Carney–Stratakis syndromic disease encompassing the dyad of a GIST and paraganglioma [16,17]. In the other half of the population of patients with deficient SDH complexes, the origin of the GIST is an epigenetic silencing of SDHC (post-zygotic hypermethylation of the promoter region). The specific hypermethylation pattern of the SDHC gene is associated with a rare non-hereditary Carney triad syndrome (a GIST, paraganglioma, and lung chondroma with a deficient SDH complex) [18,19,21,41,46,108].
Targeted Therapy for GISTs without KIT/PDGFRA Mutations and with a Deficient SDH Complex
Gastrointestinal stromal tumors with a deficient SDH complex do not respond to imatinib treatment but, in line with the mechanism of oncogenesis, they respond to multitargeted TKIs, which are potent antiangiogenetic agents, such as sunitinib, regorafenib, and pazopanib [23,40,43,97,109,110,111,112]. Linsitinib, the IGF1R inhibitor, is moderately effective in these patients [113]. Temozolomide (TMZ) has been shown to cause DNA damage and apoptosis in a pre-clinical study in patient-derived SDH-deficient GIST models [114]. Preliminary results from nine patients enrolled in a phase 2 clinical trial of TMZ demonstrated an ORR at 6 months of 22.2% and disease stabilization at 6 months in 22.2% [115]. A clinical phase 1 trial is presently enrolling patients with SDH-deficient GISTs to be treated with INBRX-109 (tetravalent death receptor 5 (DR5) agonist antibody) in combination with temozolomide (NCT03715933). A phase 2 clinical trial with rogaratinib is currently underway in metastatic GIST patients with a deficient SDH complex (NCT04595747).
3.2.2. GISTs without KIT/PDGFRA Mutations and with a Mutation in Neurofibromin 1 (NF1)
The NF1 gene is a huge TSG located on chromosome 17 (q11.2). It codes for the protein neurofibromin, which plays a role in the RAS/MEK/MAPK and mammalian target of rapamycin (mTOR) oncogenic pathways. Neurofibromin acts as a guanine triphosphate (GTP) hydrolase (GTPase), converting RAS-GTP to the inactive RAS guanosine diphosphate (GDP). Inactivating mutations in NF1 result in the accumulation of RAS-GTP, followed by increased RAS signaling in the RAS/MEK/MAPK signaling pathway [116]. Germline NF1 inactivation causes neurofibromatosis type 1 (NF1), a relatively common autosomal dominant genetic disorder characterized by predisposition to cancer development. Clinical studies have demonstrated that different inactivating mutations of NF1 exhibit different phenotypic variants and clinical presentations [117]. Due to the highly variable phenotype of NF1, other genes (so-called modifier genes) are likely to be involved in the pathogenesis in addition to the NF1 gene mutation [117]. Inactivating mutations of NF1 in GIST patients have been described as “missense” mutations, “nonsense” mutations, “frameshift-induced” protein truncation, “splice site” mutations, and larger deletions [28,117,118,119,120,121]. It has been confirmed that up to 7% of patients with NF1 develop GISTs. However, patients with NF1 GISTs account only for approximately 1–2.4% of all patients with GISTs [38,44,116,118,120]. Neurofibromatosis type I-associated GISTs exhibit immunohistochemical expression of KIT, DOG1, and SDHB, frequently present with loss of heterozygosity at 14q and 22q and, occasionally, with KIT mutations and/or mutations in the Notch signaling pathway [121,122,123]. Loss of 14q and 22q heterozygosity correlates with early disease presentation [121]. As already stated above, NF 1 is a negative regulator of the RAS/MEK/MAPK signaling pathway and, thereby, inactivating mutations in the NF1 gene result in increased signaling in this pathway independent of RTK KIT activation [119]. This explains the ineffectiveness of imatinib in patients with NF1 mutations and the current lack of any effective systemic treatment for NF1 GISTs.
3.2.3. GISTs without KIT/PDGFRA Mutations and with Mutations in BRAF
The human BRAF proto-oncogene is located on chromosome 7 (7q34). It encodes the BRAF protein, which is a serine threonine kinase with the function of activating the MAPK signaling pathway. Mutations in the BRAF gene are divided into three classes according to the impacts they have on the function of BRAF protein [124]. Class one involves mutations that allow BRAF to function as a constitutively active monomer. Class two involves mutations that allow the formation of a constitutively active dimer, and class three involves mutations that weaken or completely abolish the kinase activity of the BRAF protein. The greater part of the clinically relevant mutations in BRAF are mutations in exon 15, the most common of which is a substitution of valine with aspartate at codon 600 (BRAF V600E). This mutation leads to phosphorylation of the activation domain of the BRAF kinase and, consequently, its constitutive activation [125]. The BRAF V600E mutation is a driver mutation in various solid cancers, but it is a rare event in GIST tumorigenesis [126]. According to the literature, it occurs in less than 1% of adult GIST patients [125,127,128,129,130,131].
Targeted Therapy of GISTs without KIT/PDGFRA Mutations and with Mutations in BRAF
Gastrointestinal stromal tumors with BRAF V600E do not respond to treatment with imatinib and sunitinib, but a successful case of treatment with regorafenib has been described [132]. Treatment with dabrafenib (BRAF inhibitor), with or without trametinib (MEK inhibitor), has been proven successful as tumor agnostic therapy for solid cancers with a verified BRAF V600E mutation and is approved by the US Food and Drug Administration (FDA), but not by the EMA, as tumor agnostic therapy [133]. To date, one case of successful treatment of a metastatic BRAF V600E GIST with the BRAF inhibitor dabrafenib alone (without the MEK inhibitor trametinib) has been described [134]. There are no data on adjuvant treatment with BRAF/MEK inhibitors; therefore, adjuvant treatment is not recommended [22,23]. Additionally, other BRAF rearrangements (PRKAR1B-BRAF), the clinical significance of which is so far unknown, have been recently recognized [135,136].
3.2.4. GISTs without KIT/PDGFRA Mutations and with Mutations in KRAS
The human KRAS proto-oncogene is located on chromosome 12 (12p11.1–12p12.1). It codes for KRAS protein, a hydrolase that converts the nucleotide GTP to GDP and is part of the RAS super-family of GTPases. Active KRAS has the GTP molecule bound and, under physiological conditions, allows transduction of signals downstream to the RAS/MEK/MAPK and PI3K/AKT signaling pathways. Mutations in KRAS, predominantly point mutations, can be either primary or secondary in GIST evolution. Both primary and secondary mutations result in a permanently active KRAS protein and, thus, incessantly active signaling pathways [137]. Primary driver mutations of KRAS are very rare in GISTs (less than 0.5% of cases). Secondary mutations in KRAS occur as an after-effect of imatinib treatment in KIT/PDGFRA mutant GISTs. The incidence is unknown [42,138,139,140,141].
Gastrointestinal stromal tumors with a KRAS mutation do not respond to imatinib treatment. Sotorasib is a specific KRAS G12C inhibitor, the efficacy of which has been determined in a phase 1/2 clinical trial; however, it included no patients with metastatic, KRAS G12C-mutated GISTs [142].
3.2.5. GISTs without KIT/PDGFRA Mutations and with NTRK Alterations
The NTRK1 proto-oncogene is located on chromosome 1 (q23.1), the NTRK2 proto-oncogene on chromosome 9 (q21.33), and the NTRK3 proto-oncogene on chromosome 15 (q25.3) [143,144]. The proto-oncogenes NTRK1, NTRK2, and NTRK3 encode a family of receptor tropomyosin kinases—TRKA, TRKB, and TRKC—involved in neuronal development. Rearrangements of the NTRK1, NTRK2, and NTRK3 genes with different partner genes result in the formation of a constitutively active (ligand-independent) TK, which thereby leads to the development of several solid cancers. Rearrangements are rare in common cancers and common in very rare cancers [144]. NTRK rearrangements are agnostic driver alterations, and NTRK inhibitors are effective in tumors with NTRK fusions irrespective of the site of origin of metastatic disease [145,146].
Targeted Therapy of GISTs without KIT/PDGFRA Mutations and with NTRK Alterations
The ETV6-NTRK3 and LMNA-NTRK1 rearrangements in GISTs have already been described [147,148,149,150]. Gastrointestinal stromal tumors with NTRK rearrangements do not respond to treatment with imatinib or sunitinib but do respond to treatment with NTRK inhibitors (larotrectinib and entrectinib). A pooled analysis of three phase 1 and 2 clinical trials on the efficacy and safety of larotrectinib enrolled four patients with metastatic GISTs with NTRK rearrangements. All four patients treated with larotrectinib had an objective response [151]. Furthermore, three patients with metastatic GISTs and ETV6-NTRK3 rearrangement treated with larotrectinib have been reported, all of whom responded to treatment, one with a complete response [152]. In another integrated analysis of three phase 1 and 2 clinical trials on the efficacy and safety of entrectinib, one patient with a metastatic GIST was enrolled, but individual efficacy was not reported [150].
3.2.6. GISTs without KIT/PDGFRA Mutations and with FGFR Alterations
Fibroblast growth factor receptor 1 (FGFR1) proto-oncogene is located on chromosome 8 (p11.23), FGFR2 on chromosome 10 (q26.13), FGFR3 on chromosome 4 (p16.3), and FGFR4 on chromosome 5 (q35.2). They encode the FGFR family of proteins, which are transmembrane receptors with an IC tyrosine kinase domain [153]. Their activation depends on the conformation (homo- or hetero-dimerization) and consequent (auto- or trans-) phosphorylation of the kinase domain. Activated FGFR is involved in the RAS/MEK/MAPK and PI3K/AKT signaling pathways, among others [153,154,155]. Constitutive activation and overexpression of FGFR may be due to rearrangements or mutations in FGFR1–4 [47,154]. Primary FGFR driver alterations are very rare events in GIST evolution but have been described as a mechanism of resistance to imatinib in addition to secondary KIT/PDGFRA mutations [47,98].
Targeted Therapy for GISTs without KIT/PDGFRA Mutations and with FGFR Alterations
Multitargeted TKIs that have been found to be active in cases of FGFR driver alterations are the standard systemic treatment for metastatic GISTs (regorafenib), while other multitargeted TKIs are currently in clinical trials in this setting: dovitinib, masitinib, ponatinib, lenvatinib, pazopanib, and nintedanib [47,92]. Selective FGFR inhibitors (erdafitinib, infigratinib, pemigatinib, and futibatinib) are available but have so far not been investigated in relation to GISTs [156].
3.2.7. GISTs without KIT/PDGFRA Mutations and with Very Rare Mutations of Unknown Clinical Significance
Since the introduction of NGS into routine clinical practice, several other rare alterations in various genes have been described in relation to GISTs (PIK3CA, MAX, MEN1, ARID1A, ARID1B, ATR CBL, LTK, MEN1, PARK2, SUFU, and ZNF217) [28,51,99,157]. Most of them are accompanying (passenger) alterations that may affect the clinical course of the disease. However, as the number of cases described so far is limited, there are no data on the therapeutic implications of these changes.
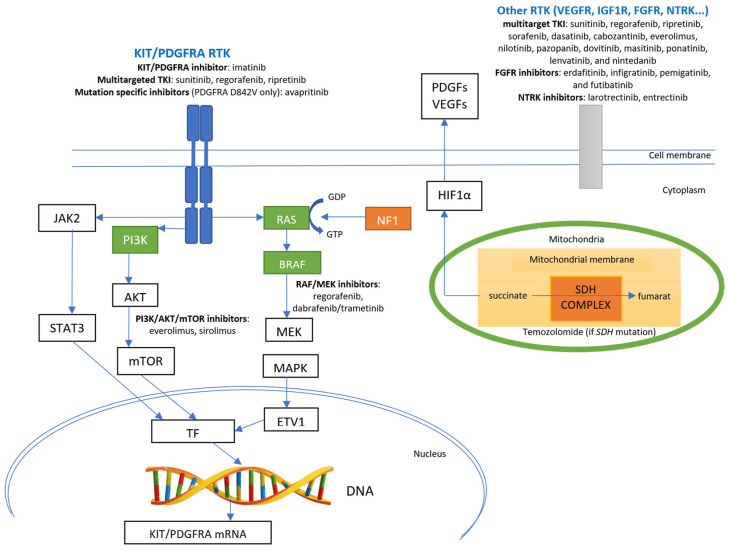
Figure 2 presents the principal molecular pathways for GISTs with the corresponding sites of action of different targeted drugs.
Figure 2.
Principal molecular pathways for GISTs and sites of action of specific pathways’ inhibitors. Green boxes indicate kinases with gain-of-function mutations (RAS, BRAF, and PI3K genes); orange boxes indicate proteins with loss-of-function (mutations in the NF1 gene and loss of function of the SDH complex—either loss of function mutations in genes coding for SDH subunits or epigenetic silencing of the SDHC gene). TF—transcription factor; ETV1—ETS variant transcription factor 1; RTK—receptor tyrosine kinase; TKI—tyrosine kinase inhibitor; DNA—deoxyribonucleic acid.
3.3. Future Directions
Currently, there are several interventional prospective clinical trials active for GISTs, either in (neo)adjuvant or metastatic settings [158]. Novel drugs or “old” drugs in combination with novel ones are being tested and the results are eagerly awaited. However, a comprehensive review of the novel targeted therapies in development is beyond the scope of this article.
On the other hand, growing research interest has been focused on so-called liquid biopsies. In patients with advanced malignancy, circulating tumor DNA (ctDNA) assays could be utilized to identify clinically relevant mutations for directing targeted therapy. However, the limitations of these assays should be considered carefully [159]. In metastatic GISTs, ctDNA has shown promising applicative value in detecting KIT primary and secondary mutations, particularly after progression with imatinib, and in assessing tumor dynamics with serial monitoring [160,161]. Additionally, NGS-based sequencing of ctDNA has demonstrated the potential to foretell the clinical benefit of sunitinib or ripretinib as second-line therapies in patients with advanced KIT-mutated GISTs [162].
4. Conclusions
Gastrointestinal stromal tumors are rare diseases that vary in their clinical and molecular characteristics. Decision regarding their systemic treatment should be governed by molecular predictive markers (driver alterations). Due to the rarity of the disease and the complexity of treatment necessitated by the variety of molecular characteristics, these patients should be managed in centers that offer the possibility of genuinely comprehensive treatment. The introduction of wide-ranging genomic profiling has improved knowledge about the molecular features of GISTs and opened new perspectives for systemic treatment. Last but not least, increased enrollment of patients in clinical trials is essential, as this leads to improved survival.
Author Contributions
Conceptualization, M.U., B.J.N. and S.N.; writing—original draft preparation, M.U., B.J.N. and S.N.; writing—review and editing, M.U., B.J.N. and S.N.; supervision, S.N. and B.J.N.; project administration, M.U. and S.N. All authors have read and agreed to the published version of the manuscript.
Conflicts of Interest
The authors declare no conflict of interest.
Funding Statement
This research received no external funding.
Footnotes
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content.
References
- 1.Wang Z.H., Liang X.B., Wang Y., Ma G.L., Qu Y.Q., Tian X.W. Epidemiology survey of gastrointestinal stromal tumor in Shanxi Province in 2011. Zhonghua Yi Xue Za Zhi. 2013;93:2541–2544. [PubMed] [Google Scholar]
- 2.Chan K.H., Chan C.W., Chow W.H., Kwan W.K., Kong C.K., Mak K.F., Leung M.Y., Lau L.K. Gastrointestinal stromal tumors in a cohort of Chinese patients in Hong Kong. World J. Gastroenterol. 2006;12:2223–2228. doi: 10.3748/wjg.v12.i14.2223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lv M., Wu C., Zheng Y., Zhao N. Incidence and survival analysis of gastrointestinal stromal tumors in shanghai: A population-based study from 2001 to 2010. Gastroenterol. Res. Pract. 2014;2014:834136. doi: 10.1155/2014/834136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cho M.Y., Sohn J.H., Kim J.M., Kim K.M., Park Y.S., Kim W.H., Jung J.S., Jung E.S., Jin S.Y., Kang D.Y., et al. Current trends in the epidemiological and pathological characteristics of gastrointestinal stromal tumors in Korea, 2003–2004. J. Korean Med. Sci. 2010;25:853–862. doi: 10.3346/jkms.2010.25.6.853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chiang N.J., Chen L.T., Tsai C.R., Chang J.S. The epidemiology of gastrointestinal stromal tumors in Taiwan, 1998–2008: A nation-wide cancer registry-based study. BMC Cancer. 2014;14:102. doi: 10.1186/1471-2407-14-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Steigen S.E., Eide T.J. Trends in incidence and survival of mesenchymal neoplasm of the digestive tract within a defined population of northern Norway. Apmis. 2006;114:192–200. doi: 10.1111/j.1600-0463.2006.apm_261.x. [DOI] [PubMed] [Google Scholar]
- 7.Søreide K., Sandvik O.M., Søreide J.A., Giljaca V., Jureckova A., Bulusu V.R. Global epidemiology of gastrointestinal stromal tumours (GIST): A systematic review of population-based cohort studies. Cancer Epidemiol. 2016;40:39–46. doi: 10.1016/j.canep.2015.10.031. [DOI] [PubMed] [Google Scholar]
- 8.Agaimy A., Wünsch P.H., Hofstaedter F., Blaszyk H., Rümmele P., Gaumann A., Dietmaier W., Hartmann A. Minute gastric sclerosing stromal tumors (GIST tumorlets) are common in adults and frequently show c-KIT mutations. Am. J. Surg. Pathol. 2007;31:113–120. doi: 10.1097/01.pas.0000213307.05811.f0. [DOI] [PubMed] [Google Scholar]
- 9.Kawanowa K., Sakuma Y., Sakurai S., Hishima T., Iwasaki Y., Saito K., Hosoya Y., Nakajima T., Funata N. High incidence of microscopic gastrointestinal stromal tumors in the stomach. Hum. Pathol. 2006;37:1527–1535. doi: 10.1016/j.humpath.2006.07.002. [DOI] [PubMed] [Google Scholar]
- 10.Corless C.L., Barnett C.M., Heinrich M.C. Gastrointestinal stromal tumours: Origin and molecular oncology. Nat. Rev. Cancer. 2011;11:865–878. doi: 10.1038/nrc3143. [DOI] [PubMed] [Google Scholar]
- 11.Ricci R., Giustiniani M.C., Gessi M., Lanza P., Castri F., Biondi A., Persiani R., Vecchio F.M., Risio M. Telocytes are the physiological counterpart of inflammatory fibroid polyps and PDGFRA-mutant GISTs. J. Cell. Mol. Med. 2018;22:4856–4862. doi: 10.1111/jcmm.13748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kondo J., Huh W.J., Franklin J.L., Heinrich M.C., Rubin B.P., Coffey R.J. A smooth muscle-derived, Braf-driven mouse model of gastrointestinal stromal tumor (GIST): Evidence for an alternative GIST cell-of-origin. J. Pathol. 2020;252:441–450. doi: 10.1002/path.5552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.De Pinieux G., Karanian M., Le Loarer F., Le Guellec S., Chabaud S., Terrier P., Bouvier C., Batistella M., Neuville A., Robin Y.M., et al. Nationwide incidence of sarcomas and connective tissue tumors of intermediate malignancy over four years using an expert pathology review network. PLoS ONE. 2021;16:e0246958. doi: 10.1371/journal.pone.0246958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hirota S., Isozaki K., Moriyama Y., Hashimoto K., Nishida T., Ishiguro S., Kawano K., Hanada M., Kurata A., Takeda M., et al. Gain-of-function mutations of c-kit in human gastrointestinal stromal tumors. Science. 1998;279:577–580. doi: 10.1126/science.279.5350.577. [DOI] [PubMed] [Google Scholar]
- 15.Ricci R. Syndromic gastrointestinal stromal tumors. Hered. Cancer Clin. Pract. 2016;14:15. doi: 10.1186/s13053-016-0055-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Miettinen M., Killian J.K., Wang Z.F., Lasota J., Lau C., Jones L., Walker R., Pineda M., Zhu Y.J., Kim S.Y., et al. Immunohistochemical loss of succinate dehydrogenase subunit A (SDHA) in gastrointestinal stromal tumors (GISTs) signals SDHA germline mutation. Am. J. Surg. Pathol. 2013;37:234–240. doi: 10.1097/PAS.0b013e3182671178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stratakis C.A., Carney J.A. The triad of paragangliomas, gastric stromal tumours and pulmonary chondromas (Carney triad), and the dyad of paragangliomas and gastric stromal sarcomas (Carney-Stratakis syndrome): Molecular genetics and clinical implications. J. Intern. Med. 2009;266:43–52. doi: 10.1111/j.1365-2796.2009.02110.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Belinsky M.G., Rink L., von Mehren M. Succinate dehydrogenase deficiency in pediatric and adult gastrointestinal stromal tumors. Front. Oncol. 2013;3:117. doi: 10.3389/fonc.2013.00117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Italiano A., Chen C.-L., Sung Y.-S., Singer S., DeMatteo R.P., LaQuaglia M.P., Besmer P., Socci N., Antonescu C.R. SDHA loss of function mutations in a subset of young adult wild-type gastrointestinal stromal tumors. BMC Cancer. 2012;12:408. doi: 10.1186/1471-2407-12-408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Janeway K.A., Liegl B., Harlow A., Le C., Perez-Atayde A., Kozakewich H., Corless C.L., Heinrich M.C., Fletcher J.A. Pediatric KIT wild-type and platelet-derived growth factor receptor alpha-wild-type gastrointestinal stromal tumors share KIT activation but not mechanisms of genetic progression with adult gastrointestinal stromal tumors. Cancer Res. 2007;67:9084–9088. doi: 10.1158/0008-5472.CAN-07-1938. [DOI] [PubMed] [Google Scholar]
- 21.Pantaleo M.A., Astolfi A., Urbini M., Nannini M., Paterini P., Indio V., Saponara M., Formica S., Ceccarelli C., Casadio R., et al. Analysis of all subunits, SDHA, SDHB, SDHC, SDHD, of the succinate dehydrogenase complex in KIT/PDGFRA wild-type GIST. Eur. J. Hum. Genet. 2014;22:32–39. doi: 10.1038/ejhg.2013.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Casali P.G., Blay J.Y., Abecassis N., Bajpai J., Bauer S., Biagini R., Bielack S., Bonvalot S., Boukovinas I., Bovee J.V.M.G., et al. Gastrointestinal stromal tumours: ESMO-EURACAN-GENTURIS Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann. Oncol. 2022;33:20–33. doi: 10.1016/j.annonc.2021.09.005. [DOI] [PubMed] [Google Scholar]
- 23.Network N.C.C. NCCN Guidelines. [(accessed on 9 December 2022)]. Available online: https://www.nccn.org/guidelines/guidelines-detail?category=1&id=1507.
- 24.Verweij J., Casali P.G., Zalcberg J., LeCesne A., Reichardt P., Blay J.Y., Issels R., van Oosterom A., Hogendoorn P.C., Van Glabbeke M., et al. Progression-free survival in gastrointestinal stromal tumours with high-dose imatinib: Randomised trial. Lancet. 2004;364:1127–1134. doi: 10.1016/S0140-6736(04)17098-0. [DOI] [PubMed] [Google Scholar]
- 25.Joensuu H., Eriksson M., Sundby Hall K., Reichardt A., Hermes B., Schütte J., Cameron S., Hohenberger P., Jost P.J., Al-Batran S.E., et al. Survival Outcomes Associated with 3 Years vs. 1 Year of Adjuvant Imatinib for Patients with High-Risk Gastrointestinal Stromal Tumors: An Analysis of a Randomized Clinical Trial after 10-Year Follow-up. JAMA Oncol. 2020;6:1241–1246. doi: 10.1001/jamaoncol.2020.2091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Demetri G.D., Benjamin R.S., Blanke C.D., Blay J.Y., Casali P., Choi H., Corless C.L., Debiec-Rychter M., DeMatteo R.P., Ettinger D.S., et al. NCCN Task Force report: Management of patients with gastrointestinal stromal tumor (GIST)—Update of the NCCN clinical practice guidelines. J. Natl. Compr. Canc. Netw. 2007;5((Suppl. 2)):S1-29. doi: 10.6004/jnccn.2007.2002. quiz S30. [DOI] [PubMed] [Google Scholar]
- 27.Pierotti M.A., Tamborini E., Negri T., Pricl S., Pilotti S. Targeted therapy in GIST: In silico modeling for prediction of resistance. Nat. Rev. Clin. Oncol. 2011;8:161–170. doi: 10.1038/nrclinonc.2011.3. [DOI] [PubMed] [Google Scholar]
- 28.Brčić I., Argyropoulos A., Liegl-Atzwanger B. Update on Molecular Genetics of Gastrointestinal Stromal Tumors. Diagnostics. 2021;11:194. doi: 10.3390/diagnostics11020194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hu X., Wang Z., Su P., Zhang Q., Kou Y. Advances in the research of the mechanism of secondary resistance to imatinib in gastrointestinal stromal tumors. Front. Oncol. 2022;12:933248. doi: 10.3389/fonc.2022.933248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Serrano C., Mariño-Enríquez A., Tao D.L., Ketzer J., Eilers G., Zhu M., Yu C., Mannan A.M., Rubin B.P., Demetri G.D., et al. Complementary activity of tyrosine kinase inhibitors against secondary kit mutations in imatinib-resistant gastrointestinal stromal tumours. Br. J. Cancer. 2019;120:612–620. doi: 10.1038/s41416-019-0389-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Heinrich M.C., Corless C.L., Duensing A., McGreevey L., Chen C.J., Joseph N., Singer S., Griffith D.J., Haley A., Town A., et al. PDGFRA activating mutations in gastrointestinal stromal tumors. Science. 2003;299:708–710. doi: 10.1126/science.1079666. [DOI] [PubMed] [Google Scholar]
- 32.Klug L.R., Khosroyani H.M., Kent J.D., Heinrich M.C. New treatment strategies for advanced-stage gastrointestinal stromal tumours. Nat. Rev. Clin. Oncol. 2022;19:328–341. doi: 10.1038/s41571-022-00606-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Astolfi A., Indio V., Nannini M., Saponara M., Schipani A., De Leo A., Altimari A., Vincenzi B., Comandini D., Grignani G., et al. Targeted Deep Sequencing Uncovers Cryptic KIT Mutations in KIT/PDGFRA/SDH/RAS-P Wild-Type GIST. Front. Oncol. 2020;10:504. doi: 10.3389/fonc.2020.00504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Unk M., Bombač A., Jezeršek Novaković B., Stegel V., Šetrajčič Dragoš V., Blatnik O., Klančar G., Novaković S. Correlation of treatment outcome in sanger/RT-qPCR. Oncol. Rep. 2022;48:167. doi: 10.3892/or.2022.8382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Heinrich M.C., Rankin C., Blanke C.D., Demetri G.D., Borden E.C., Ryan C.W., von Mehren M., Blackstein M.E., Priebat D.A., Tap W.D., et al. Correlation of Long-term Results of Imatinib in Advanced Gastrointestinal Stromal Tumors with Next-Generation Sequencing Results: Analysis of Phase 3 SWOG Intergroup Trial S0033. JAMA Oncol. 2017;3:944–952. doi: 10.1001/jamaoncol.2016.6728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vanden Bempt I., Vander Borght S., Sciot R., Spans L., Claerhout S., Brems H., Lehnert S., Dehaspe L., Fransis S., Neuville B., et al. Comprehensive targeted next-generation sequencing approach in the molecular diagnosis of gastrointestinal stromal tumor. Genes Chromosomes Cancer. 2021;60:239–249. doi: 10.1002/gcc.22923. [DOI] [PubMed] [Google Scholar]
- 37.Bombac A., Zakotnik B., Bucic M., Setrajcic Dragos V., Gazic B., Stegel V., Klancar G., Novakovic S. Mutational spectrum and classification of novel mutations in patients with metastatic gastrointestinal stromal tumours. Int. J. Oncol. 2020;56:1468–1478. doi: 10.3892/ijo.2020.5028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Blay J.Y., Kang Y.K., Nishida T., von Mehren M. Gastrointestinal stromal tumours. Nat. Rev. Dis. Primers. 2021;7:22. doi: 10.1038/s41572-021-00254-5. [DOI] [PubMed] [Google Scholar]
- 39.Boikos S.A., Stratakis C.A. The genetic landscape of gastrointestinal stromal tumor lacking KIT and PDGFRA mutations. Endocrine. 2014;47:401–408. doi: 10.1007/s12020-014-0346-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Boikos S.A., Pappo A.S., Killian J.K., LaQuaglia M.P., Weldon C.B., George S., Trent J.C., von Mehren M., Wright J.A., Schiffman J.D., et al. Molecular Subtypes of KIT/PDGFRA Wild-Type Gastrointestinal Stromal Tumors: A Report from the National Institutes of Health Gastrointestinal Stromal Tumor Clinic. JAMA Oncol. 2016;2:922–928. doi: 10.1001/jamaoncol.2016.0256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Janeway K.A., Kim S.Y., Lodish M., Nosé V., Rustin P., Gaal J., Dahia P.L., Liegl B., Ball E.R., Raygada M., et al. Defects in succinate dehydrogenase in gastrointestinal stromal tumors lacking KIT and PDGFRA mutations. Proc. Natl. Acad. Sci. USA. 2011;108:314–318. doi: 10.1073/pnas.1009199108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lasota J., Xi L., Coates T., Dennis R., Evbuomwan M.O., Wang Z.F., Raffeld M., Miettinen M. No KRAS mutations found in gastrointestinal stromal tumors (GISTs): Molecular genetic study of 514 cases. Mod. Pathol. 2013;26:1488–1491. doi: 10.1038/modpathol.2013.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liu W., Zeng X., Wu X., He J., Gao J., Shuai X., Wang G., Zhang P., Tao K. Clinicopathologic study of succinate-dehydrogenase-deficient gastrointestinal stromal tumors: A single-institutional experience in China. Medicine. 2017;96:e7668. doi: 10.1097/MD.0000000000007668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mathias-Machado M.C., de Jesus V.H.F., de Carvalho Oliveira L.J., Neumann M., Peixoto R.D. Current Molecular Profile of Gastrointestinal Stromal Tumors and Systemic Therapeutic Implications. Cancers. 2022;14:5330. doi: 10.3390/cancers14215330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mei L., Smith S.C., Faber A.C., Trent J., Grossman S.R., Stratakis C.A., Boikos S.A. Gastrointestinal Stromal Tumors: The GIST of Precision Medicine. Trends Cancer. 2018;4:74–91. doi: 10.1016/j.trecan.2017.11.006. [DOI] [PubMed] [Google Scholar]
- 46.Miettinen M., Lasota J. Succinate dehydrogenase deficient gastrointestinal stromal tumors (GISTs)—A review. Int. J. Biochem. Cell Biol. 2014;53:514–519. doi: 10.1016/j.biocel.2014.05.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Astolfi A., Pantaleo M.A., Indio V., Urbini M., Nannini M. The Emerging Role of the FGF/FGFR Pathway in Gastrointestinal Stromal Tumor. Int. J. Mol. Sci. 2020;21:3313. doi: 10.3390/ijms21093313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nannini M., Biasco G., Astolfi A., Pantaleo M.A. An overview on molecular biology of KIT/PDGFRA wild type (WT) gastrointestinal stromal tumours (GIST) J. Med. Genet. 2013;50:653–661. doi: 10.1136/jmedgenet-2013-101695. [DOI] [PubMed] [Google Scholar]
- 49.Nannini M., Urbini M., Astolfi A., Biasco G., Pantaleo M.A. The progressive fragmentation of the KIT/PDGFRA wild-type (WT) gastrointestinal stromal tumors (GIST) J. Transl. Med. 2017;15:113. doi: 10.1186/s12967-017-1212-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pantaleo M.A., Nannini M., Corless C.L., Heinrich M.C. Quadruple wild-type (WT) GIST: Defining the subset of GIST that lacks abnormalities of KIT, PDGFRA, SDH, or RAS signaling pathways. Cancer Med. 2015;4:101–103. doi: 10.1002/cam4.325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pantaleo M.A., Urbini M., Indio V., Ravegnini G., Nannini M., De Luca M., Tarantino G., Angelini S., Gronchi A., Vincenzi B., et al. Genome-Wide Analysis Identifies MEN1 and MAX Mutations and a Neuroendocrine-Like Molecular Heterogeneity in Quadruple WT GIST. Mol. Cancer Res. 2017;15:553–562. doi: 10.1158/1541-7786.MCR-16-0376. [DOI] [PubMed] [Google Scholar]
- 52.Wozniak A., Rutkowski P., Schöffski P., Ray-Coquard I., Hostein I., Schildhaus H.U., Le Cesne A., Bylina E., Limon J., Blay J.Y., et al. Tumor genotype is an independent prognostic factor in primary gastrointestinal stromal tumors of gastric origin: A european multicenter analysis based on ConticaGIST. Clin. Cancer Res. 2014;20:6105–6116. doi: 10.1158/1078-0432.CCR-14-1677. [DOI] [PubMed] [Google Scholar]
- 53.(MetaGIST) G.S.T.M.-A.G. Comparison of two doses of imatinib for the treatment of unresectable or metastatic gastrointestinal stromal tumors: A meta-analysis of 1640 patients. J. Clin. Oncol. 2010;28:1247–1253. doi: 10.1200/JCO.2009.24.2099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Szucs Z., Thway K., Fisher C., Bulusu R., Constantinidou A., Benson C., van der Graaf W.T., Jones R.L. Molecular subtypes of gastrointestinal stromal tumors and their prognostic and therapeutic implications. Future Oncol. 2017;13:93–107. doi: 10.2217/fon-2016-0192. [DOI] [PubMed] [Google Scholar]
- 55.Zhang H., Liu Q. Prognostic Indicators for Gastrointestinal Stromal Tumors: A Review. Transl. Oncol. 2020;13:100812. doi: 10.1016/j.tranon.2020.100812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yarden Y., Kuang W.J., Yang-Feng T., Coussens L., Munemitsu S., Dull T.J., Chen E., Schlessinger J., Francke U., Ullrich A. Human proto-oncogene c-kit: A new cell surface receptor tyrosine kinase for an unidentified ligand. EMBO J. 1987;6:3341–3351. doi: 10.1002/j.1460-2075.1987.tb02655.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sheikh E., Tran T., Vranic S., Levy A., Bonfil R.D. Role and significance of c-KIT receptor tyrosine kinase in cancer: A review. Bosn. J. Basic Med. Sci. 2022;22:683–698. doi: 10.17305/bjbms.2021.7399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lennartsson J., Rönnstrand L. Stem cell factor receptor/c-Kit: From basic science to clinical implications. Physiol. Rev. 2012;92:1619–1649. doi: 10.1152/physrev.00046.2011. [DOI] [PubMed] [Google Scholar]
- 59.Kelly C.M., Gutierrez Sainz L., Chi P. The management of metastatic GIST: Current standard and investigational therapeutics. J. Hematol. Oncol. 2021;14:2. doi: 10.1186/s13045-020-01026-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Joensuu H., Eriksson M., Sundby Hall K., Hartmann J.T., Pink D., Schütte J., Ramadori G., Hohenberger P., Duyster J., Al-Batran S.E., et al. One vs three years of adjuvant imatinib for operable gastrointestinal stromal tumor: A randomized trial. JAMA. 2012;307:1265–1272. doi: 10.1001/jama.2012.347. [DOI] [PubMed] [Google Scholar]
- 61.Joensuu H., Wardelmann E., Sihto H., Eriksson M., Sundby Hall K., Reichardt A., Hartmann J.T., Pink D., Cameron S., Hohenberger P., et al. Effect of KIT and PDGFRA Mutations on Survival in Patients with Gastrointestinal Stromal Tumors Treated with Adjuvant Imatinib: An Exploratory Analysis of a Randomized Clinical Trial. JAMA Oncol. 2017;3:602–609. doi: 10.1001/jamaoncol.2016.5751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Joensuu H. Risk stratification of patients diagnosed with gastrointestinal stromal tumor. Hum. Pathol. 2008;39:1411–1419. doi: 10.1016/j.humpath.2008.06.025. [DOI] [PubMed] [Google Scholar]
- 63.Miettinen M., Lasota J. Gastrointestinal stromal tumors: Pathology and prognosis at different sites. Semin. Diagn. Pathol. 2006;23:70–83. doi: 10.1053/j.semdp.2006.09.001. [DOI] [PubMed] [Google Scholar]
- 64.Yeh C.N., Chen M.H., Chen Y.Y., Yang C.Y., Yen C.C., Tzen C.Y., Chen L.T., Chen J.S. A phase II trial of regorafenib in patients with metastatic and/or a unresectable gastrointestinal stromal tumor harboring secondary mutations of exon 17. Oncotarget. 2017;8:44121–44130. doi: 10.18632/oncotarget.17310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Vincenzi B., Napolitano A., Fiocco M., Mir O., Rutkowski P., Blay J.Y., Reichardt P., Joensuu H., Fumagalli E., Gennatas S., et al. Adjuvant Imatinib in Patients with GIST Harboring Exon 9 KIT Mutations: Results from a Multi-institutional European Retrospective Study. Clin. Cancer Res. 2022;28:1672–1679. doi: 10.1158/1078-0432.CCR-21-1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Heinrich M.C., Owzar K., Corless C.L., Hollis D., Borden E.C., Fletcher C.D., Ryan C.W., von Mehren M., Blanke C.D., Rankin C., et al. Correlation of kinase genotype and clinical outcome in the North American Intergroup Phase III Trial of imatinib mesylate for treatment of advanced gastrointestinal stromal tumor: CALGB 150105 Study by Cancer and Leukemia Group B and Southwest Oncology Group. J. Clin. Oncol. 2008;26:5360–5367. doi: 10.1200/JCO.2008.17.4284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Debiec-Rychter M., Sciot R., Le Cesne A., Schlemmer M., Hohenberger P., van Oosterom A.T., Blay J.Y., Leyvraz S., Stul M., Casali P.G., et al. KIT mutations and dose selection for imatinib in patients with advanced gastrointestinal stromal tumours. Eur. J. Cancer. 2006;42:1093–1103. doi: 10.1016/j.ejca.2006.01.030. [DOI] [PubMed] [Google Scholar]
- 68.Raut C.P., Espat N.J., Maki R.G., Araujo D.M., Trent J., Williams T.F., Purkayastha D.D., DeMatteo R.P. Efficacy and Tolerability of 5-Year Adjuvant Imatinib Treatment for Patients with Resected Intermediate- or High-Risk Primary Gastrointestinal Stromal Tumor: The PERSIST-5 Clinical Trial. JAMA Oncol. 2018;4:e184060. doi: 10.1001/jamaoncol.2018.4060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Patrikidou A., Domont J., Chabaud S., Ray-Coquard I., Coindre J.M., Bui-Nguyen B., Adenis A., Rios M., Bertucci F., Duffaud F., et al. Long-term outcome of molecular subgroups of GIST patients treated with standard-dose imatinib in the BFR14 trial of the French Sarcoma Group. Eur. J. Cancer. 2016;52:173–180. doi: 10.1016/j.ejca.2015.10.069. [DOI] [PubMed] [Google Scholar]
- 70.Dematteo R.P., Ballman K.V., Antonescu C.R., Maki R.G., Pisters P.W., Demetri G.D., Blackstein M.E., Blanke C.D., von Mehren M., Brennan M.F., et al. Adjuvant imatinib mesylate after resection of localised, primary gastrointestinal stromal tumour: A randomised, double-blind, placebo-controlled trial. Lancet. 2009;373:1097–1104. doi: 10.1016/S0140-6736(09)60500-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Casali P.G., Le Cesne A., Poveda Velasco A., Kotasek D., Rutkowski P., Hohenberger P., Fumagalli E., Judson I.R., Italiano A., Gelderblom H., et al. Time to Definitive Failure to the First Tyrosine Kinase Inhibitor in Localized GI Stromal Tumors Treated with Imatinib as an Adjuvant: A European Organisation for Research and Treatment of Cancer Soft Tissue and Bone Sarcoma Group Intergroup Randomized Trial in Collaboration with the Australasian Gastro-Intestinal Trials Group, UNICANCER, French Sarcoma Group, Italian Sarcoma Group, and Spanish Group for Research on Sarcomas. J. Clin. Oncol. 2015;33:4276–4283. doi: 10.1200/jco.2015.62.4304. [DOI] [PubMed] [Google Scholar]
- 72.Fletcher C.D., Berman J.J., Corless C., Gorstein F., Lasota J., Longley B.J., Miettinen M., O’Leary T.J., Remotti H., Rubin B.P., et al. Diagnosis of gastrointestinal stromal tumors: A consensus approach. Hum. Pathol. 2002;33:459–465. doi: 10.1053/hupa.2002.123545. [DOI] [PubMed] [Google Scholar]
- 73.Heinrich M.C., Corless C.L., Demetri G.D., Blanke C.D., von Mehren M., Joensuu H., McGreevey L.S., Chen C.J., Van den Abbeele A.D., Druker B.J., et al. Kinase mutations and imatinib response in patients with metastatic gastrointestinal stromal tumor. J. Clin. Oncol. 2003;21:4342–4349. doi: 10.1200/JCO.2003.04.190. [DOI] [PubMed] [Google Scholar]
- 74.Blanke C.D., Demetri G.D., von Mehren M., Heinrich M.C., Eisenberg B., Fletcher J.A., Corless C.L., Fletcher C.D., Roberts P.J., Heinz D., et al. Long-term results from a randomized phase II trial of standard- versus higher-dose imatinib mesylate for patients with unresectable or metastatic gastrointestinal stromal tumors expressing KIT. J. Clin. Oncol. 2008;26:620–625. doi: 10.1200/JCO.2007.13.4403. [DOI] [PubMed] [Google Scholar]
- 75.Blanke C.D., Rankin C., Demetri G.D., Ryan C.W., von Mehren M., Benjamin R.S., Raymond A.K., Bramwell V.H., Baker L.H., Maki R.G., et al. Phase III randomized, intergroup trial assessing imatinib mesylate at two dose levels in patients with unresectable or metastatic gastrointestinal stromal tumors expressing the kit receptor tyrosine kinase: S0033. J. Clin. Oncol. 2008;26:626–632. doi: 10.1200/JCO.2007.13.4452. [DOI] [PubMed] [Google Scholar]
- 76.Demetri G.D., von Mehren M., Blanke C.D., Van den Abbeele A.D., Eisenberg B., Roberts P.J., Heinrich M.C., Tuveson D.A., Singer S., Janicek M., et al. Efficacy and safety of imatinib mesylate in advanced gastrointestinal stromal tumors. N. Engl. J. Med. 2002;347:472–480. doi: 10.1056/NEJMoa020461. [DOI] [PubMed] [Google Scholar]
- 77.Grunewald S., Klug L.R., Mühlenberg T., Lategahn J., Falkenhorst J., Town A., Ehrt C., Wardelmann E., Hartmann W., Schildhaus H.U., et al. Resistance to Avapritinib in PDGFRA-Driven GIST Is Caused by Secondary Mutations in the PDGFRA Kinase Domain. Cancer Discov. 2021;11:108–125. doi: 10.1158/2159-8290.CD-20-0487. [DOI] [PubMed] [Google Scholar]
- 78.Ravegnini G., Urbini M., Simeon V., Genovese C., Astolfi A., Nannini M., Gatto L., Saponara M., Ianni M., Indio V., et al. An exploratory study by DMET array identifies a germline signature associated with imatinib response in gastrointestinal stromal tumor. Pharm. J. 2019;19:390–400. doi: 10.1038/s41397-018-0050-4. [DOI] [PubMed] [Google Scholar]
- 79.Corless C.L., Schroeder A., Griffith D., Town A., McGreevey L., Harrell P., Shiraga S., Bainbridge T., Morich J., Heinrich M.C. PDGFRA mutations in gastrointestinal stromal tumors: Frequency, spectrum and in vitro sensitivity to imatinib. J. Clin. Oncol. 2005;23:5357–5364. doi: 10.1200/JCO.2005.14.068. [DOI] [PubMed] [Google Scholar]
- 80.Cassier P.A., Fumagalli E., Rutkowski P., Schöffski P., Van Glabbeke M., Debiec-Rychter M., Emile J.F., Duffaud F., Martin-Broto J., Landi B., et al. Outcome of patients with platelet-derived growth factor receptor alpha-mutated gastrointestinal stromal tumors in the tyrosine kinase inhibitor era. Clin. Cancer Res. 2012;18:4458–4464. doi: 10.1158/1078-0432.CCR-11-3025. [DOI] [PubMed] [Google Scholar]
- 81.Yoo C., Ryu M.H., Jo J., Park I., Ryoo B.Y., Kang Y.K. Efficacy of Imatinib in Patients with Platelet-Derived Growth Factor Receptor Alpha-Mutated Gastrointestinal Stromal Tumors. Cancer Res. Treat. 2016;48:546–552. doi: 10.4143/crt.2015.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Farag S., Somaiah N., Choi H., Heeres B., Wang W.L., van Boven H., Nederlof P., Benjamin R., van der Graaf W., Grunhagen D., et al. Clinical characteristics and treatment outcome in a large multicentre observational cohort of PDGFRA exon 18 mutated gastrointestinal stromal tumour patients. Eur. J. Cancer. 2017;76:76–83. doi: 10.1016/j.ejca.2017.02.007. [DOI] [PubMed] [Google Scholar]
- 83.Smith C.I.E. Enigmas in tumor resistance to kinase inhibitors and calculation of the drug resistance index for cancer (DRIC) Semin. Cancer Biol. 2017;45:36–49. doi: 10.1016/j.semcancer.2016.11.008. [DOI] [PubMed] [Google Scholar]
- 84.Casali P.G., Zalcberg J., Le Cesne A., Reichardt P., Blay J.Y., Lindner L.H., Judson I.R., Schöffski P., Leyvraz S., Italiano A., et al. Ten-Year Progression-Free and Overall Survival in Patients with Unresectable or Metastatic GI Stromal Tumors: Long-Term Analysis of the European Organisation for Research and Treatment of Cancer, Italian Sarcoma Group, and Australasian Gastrointestinal Trials Group Intergroup Phase III Randomized Trial on Imatinib at Two Dose Levels. J. Clin. Oncol. 2017;35:1713–1720. doi: 10.1200/JCO.2016.71.0228. [DOI] [PubMed] [Google Scholar]
- 85.Heinrich M.C., Corless C.L., Blanke C.D., Demetri G.D., Joensuu H., Roberts P.J., Eisenberg B.L., von Mehren M., Fletcher C.D., Sandau K., et al. Molecular correlates of imatinib resistance in gastrointestinal stromal tumors. J. Clin. Oncol. 2006;24:4764–4774. doi: 10.1200/JCO.2006.06.2265. [DOI] [PubMed] [Google Scholar]
- 86.Demetri G.D., van Oosterom A.T., Garrett C.R., Blackstein M.E., Shah M.H., Verweij J., McArthur G., Judson I.R., Heinrich M.C., Morgan J.A., et al. Efficacy and safety of sunitinib in patients with advanced gastrointestinal stromal tumour after failure of imatinib: A randomised controlled trial. Lancet. 2006;368:1329–1338. doi: 10.1016/S0140-6736(06)69446-4. [DOI] [PubMed] [Google Scholar]
- 87.Heinrich M.C., vonMehren M., Demetri G.D., Fletcher J.A., Sun J., Hodgson J.G., Rivera V.M., Turner C.D., George S. A phase 2 study of ponatinib in patients (pts) with advanced gastrointestinal stromal tumors (GIST) after failure of tyrosine kinase inhibitor (TKI) therapy: Initial report. J. Clin. Oncol. 2014;32:10506. doi: 10.1200/jco.2014.32.15_suppl.10506. [DOI] [Google Scholar]
- 88.Lostes-Bardaji M.J., García-Illescas D., Valverde C., Serrano C. Ripretinib in gastrointestinal stromal tumor: The long-awaited step forward. Ther. Adv. Med. Oncol. 2021;13:1758835920986498. doi: 10.1177/1758835920986498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Blay J.Y., Serrano C., Heinrich M.C., Zalcberg J., Bauer S., Gelderblom H., Schöffski P., Jones R.L., Attia S., D’Amato G., et al. Ripretinib in patients with advanced gastrointestinal stromal tumours (INVICTUS): A double-blind, randomised, placebo-controlled, phase 3 trial. Lancet Oncol. 2020;21:923–934. doi: 10.1016/S1470-2045(20)30168-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Heinrich M.C., Jones R.L., von Mehren M., Schöffski P., Serrano C., Kang Y.K., Cassier P.A., Mir O., Eskens F., Tap W.D., et al. Avapritinib in advanced PDGFRA D842V-mutant gastrointestinal stromal tumour (NAVIGATOR): A multicentre, open-label, phase 1 trial. Lancet Oncol. 2020;21:935–946. doi: 10.1016/S1470-2045(20)30269-2. [DOI] [PubMed] [Google Scholar]
- 91.Kang Y.K., George S., Jones R.L., Rutkowski P., Shen L., Mir O., Patel S., Zhou Y., von Mehren M., Hohenberger P., et al. Avapritinib Versus Regorafenib in Locally Advanced Unresectable or Metastatic GI Stromal Tumor: A Randomized, Open-Label Phase III Study. J. Clin. Oncol. 2021;39:3128–3139. doi: 10.1200/JCO.21.00217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Demetri G.D., Reichardt P., Kang Y.K., Blay J.Y., Rutkowski P., Gelderblom H., Hohenberger P., Leahy M., von Mehren M., Joensuu H., et al. Efficacy and safety of regorafenib for advanced gastrointestinal stromal tumours after failure of imatinib and sunitinib (GRID): An international, multicentre, randomised, placebo-controlled, phase 3 trial. Lancet. 2013;381:295–302. doi: 10.1016/S0140-6736(12)61857-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Heinrich M.C., Marino-Enriquez A., Presnell A., Donsky R.S., Griffith D.J., McKinley A., Patterson J., Taguchi T., Liang C.W., Fletcher J.A. Sorafenib inhibits many kinase mutations associated with drug-resistant gastrointestinal stromal tumors. Mol. Cancer Ther. 2012;11:1770–1780. doi: 10.1158/1535-7163.MCT-12-0223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Reichardt P., Blay J.Y., Gelderblom H., Schlemmer M., Demetri G.D., Bui-Nguyen B., McArthur G.A., Yazji S., Hsu Y., Galetic I., et al. Phase III study of nilotinib versus best supportive care with or without a TKI in patients with gastrointestinal stromal tumors resistant to or intolerant of imatinib and sunitinib. Ann. Oncol. 2012;23:1680–1687. doi: 10.1093/annonc/mdr598. [DOI] [PubMed] [Google Scholar]
- 95.Gardino A.K., Evans E.K., Kim J.L., Brooijmans N., Hodous B.L., Wolf B., Lengauer C. Targeting kinases with precision. Mol. Cell. Oncol. 2018;5:e1435183. doi: 10.1080/23723556.2018.1435183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Serrano C., George S., Valverde C., Olivares D., García-Valverde A., Suárez C., Morales-Barrera R., Carles J. Novel Insights into the Treatment of Imatinib-Resistant Gastrointestinal Stromal Tumors. Target. Oncol. 2017;12:277–288. doi: 10.1007/s11523-017-0490-9. [DOI] [PubMed] [Google Scholar]
- 97.National Center for Biotechnology Information . Hazardous Substances Data Bank. National Library of Medicine; Bethesda, MD, USA: 2022. [Google Scholar]
- 98.Javidi-Sharifi N., Traer E., Martinez J., Gupta A., Taguchi T., Dunlap J., Heinrich M.C., Corless C.L., Rubin B.P., Druker B.J., et al. Crosstalk between KIT and FGFR3 Promotes Gastrointestinal Stromal Tumor Cell Growth and Drug Resistance. Cancer Res. 2015;75:880–891. doi: 10.1158/0008-5472.CAN-14-0573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Lasota J., Felisiak-Golabek A., Wasag B., Kowalik A., Zięba S., Chłopek M., Wang Z.F., Coates T., Kopczynski J., Gozdz S., et al. Frequency and clinicopathologic profile of PIK3CA mutant GISTs: Molecular genetic study of 529 cases. Mod. Pathol. 2016;29:275–282. doi: 10.1038/modpathol.2015.160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Li K., Cheng H., Li Z., Pang Y., Jia X., Xie F., Hu G., Cai Q., Wang Y. Genetic progression in gastrointestinal stromal tumors: Mechanisms and molecular interventions. Oncotarget. 2017;8:60589–60604. doi: 10.18632/oncotarget.16014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Reichardt P., Demetri G.D., Gelderblom H., Rutkowski P., Im S.A., Gupta S., Kang Y.K., Schöffski P., Schuette J., Soulières D., et al. Correlation of KIT and PDGFRA mutational status with clinical benefit in patients with gastrointestinal stromal tumor treated with sunitinib in a worldwide treatment-use trial. BMC Cancer. 2016;16:22. doi: 10.1186/s12885-016-2051-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Heinrich M.C., Maki R.G., Corless C.L., Antonescu C.R., Harlow A., Griffith D., Town A., McKinley A., Ou W.B., Fletcher J.A., et al. Primary and secondary kinase genotypes correlate with the biological and clinical activity of sunitinib in imatinib-resistant gastrointestinal stromal tumor. J. Clin. Oncol. 2008;26:5352–5359. doi: 10.1200/JCO.2007.15.7461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Janku F., Abdul Razak A.R., Chi P., Heinrich M.C., von Mehren M., Jones R.L., Ganjoo K., Trent J., Gelderblom H., Somaiah N., et al. Switch Control Inhibition of KIT and PDGFRA in Patients with Advanced Gastrointestinal Stromal Tumor: A Phase I Study of Ripretinib. J. Clin. Oncol. 2020;38:3294–3303. doi: 10.1200/JCO.20.00522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.George S., Jones R.L., Bauer S., Kang Y.K., Schöffski P., Eskens F., Mir O., Cassier P.A., Serrano C., Tap W.D., et al. Avapritinib in Patients with Advanced Gastrointestinal Stromal Tumors Following at Least Three Prior Lines of Therapy. Oncologist. 2021;26:e639–e649. doi: 10.1002/onco.13674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Green S., Weiss G.R. Southwest Oncology Group standard response criteria, endpoint definitions and toxicity criteria. Investig. New Drugs. 1992;10:239–253. doi: 10.1007/BF00944177. [DOI] [PubMed] [Google Scholar]
- 106.George S., Blay J.Y., Casali P.G., Le Cesne A., Stephenson P., Deprimo S.E., Harmon C.S., Law C.N., Morgan J.A., Ray-Coquard I., et al. Clinical evaluation of continuous daily dosing of sunitinib malate in patients with advanced gastrointestinal stromal tumour after imatinib failure. Eur. J. Cancer. 2009;45:1959–1968. doi: 10.1016/j.ejca.2009.02.011. [DOI] [PubMed] [Google Scholar]
- 107.Nannini M., Astolfi A., Paterini P., Urbini M., Santini D., Catena F., Indio V., Casadio R., Pinna A.D., Biasco G., et al. Expression of IGF-1 receptor in KIT/PDGF receptor-α wild-type gastrointestinal stromal tumors with succinate dehydrogenase complex dysfunction. Future Oncol. 2013;9:121–126. doi: 10.2217/fon.12.170. [DOI] [PubMed] [Google Scholar]
- 108.Oudijk L., Gaal J., Korpershoek E., van Nederveen F.H., Kelly L., Schiavon G., Verweij J., Mathijssen R.H.J., den Bakker M.A., Oldenburg R.A., et al. SDHA mutations in adult and pediatric wild-type gastrointestinal stromal tumors. Mod. Pathol. 2013;26:456–463. doi: 10.1038/modpathol.2012.186. [DOI] [PubMed] [Google Scholar]
- 109.Rutkowski P., Magnan H., Chou A.J., Benson C. Treatment of gastrointestinal stromal tumours in paediatric and young adult patients with sunitinib: A multicentre case series. BMC Cancer. 2017;17:717. doi: 10.1186/s12885-017-3727-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Andrzejewska M., Czarny J., Derwich K. Latest Advances in the Management of Pediatric Gastrointestinal Stromal Tumors. Cancers. 2022;14:4989. doi: 10.3390/cancers14204989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Janeway K.A., Albritton K.H., Van Den Abbeele A.D., D’Amato G.Z., Pedrazzoli P., Siena S., Picus J., Butrynski J.E., Schlemmer M., Heinrich M.C., et al. Sunitinib treatment in pediatric patients with advanced GIST following failure of imatinib. Pediatr. Blood Cancer. 2009;52:767–771. doi: 10.1002/pbc.21909. [DOI] [PubMed] [Google Scholar]
- 112.Ganjoo K.N., Villalobos V.M., Kamaya A., Fisher G.A., Butrynski J.E., Morgan J.A., Wagner A.J., D’Adamo D., McMillan A., Demetri G.D., et al. A multicenter phase II study of pazopanib in patients with advanced gastrointestinal stromal tumors (GIST) following failure of at least imatinib and sunitinib. Ann. Oncol. 2014;25:236–240. doi: 10.1093/annonc/mdt484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Von Mehren M., George S., Heinrich M.C., Schuetze S.M., Yap J.T., Yu J.Q., Abbott A., Litwin S., Crowley J., Belinsky M., et al. Linsitinib (OSI-906) for the Treatment of Adult and Pediatric Wild-Type Gastrointestinal Stromal Tumors, a SARC Phase II Study. Clin. Cancer Res. 2020;26:1837–1845. doi: 10.1158/1078-0432.CCR-19-1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Yebra M., Bhargava S., Kumar A., Burgoyne A.M., Tang C.M., Yoon H., Banerjee S., Aguilera J., Cordes T., Sheth V., et al. Establishment of Patient-Derived Succinate Dehydrogenase-Deficient Gastrointestinal Stromal Tumor Models for Predicting Therapeutic Response. Clin. Cancer Res. 2022;28:187–200. doi: 10.1158/1078-0432.CCR-21-2092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Burgoyne A., Heinrich M.C., von Mehren M., Trent J.C., Messer E., Messer K., Metallo C., Sicklick J.K. An open-label, phase 2 efficacy study of temozolomide in advanced succinate dehydrogenase-mutant/deficient gastrointestinal stromal tumor; Proceedings of the CTOS 2022 Annual Meeting; Vancouver, BC, Canada. 16–19 November 2022. [Google Scholar]
- 116.Bergoug M., Doudeau M., Godin F., Mosrin C., Vallée B., Bénédetti H. Neurofibromin Structure, Functions and Regulation. Cells. 2020;9:2365. doi: 10.3390/cells9112365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Wang W., Wei C.-J., Cui X.-W., Li Y.-H., Gu Y.-H., Gu B., Li Q.-F., Wang Z.-C. Impacts of NF1 Gene Mutations and Genetic Modifiers in Neurofibromatosis Type 1. Front. Neurol. 2021;12:704639. doi: 10.3389/fneur.2021.704639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Gasparotto D., Rossi S., Polano M., Tamborini E., Lorenzetto E., Sbaraglia M., Mondello A., Massani M., Lamon S., Bracci R., et al. Quadruple-Negative GIST Is a Sentinel for Unrecognized Neurofibromatosis Type 1 Syndrome. Clin. Cancer Res. 2017;23:273–282. doi: 10.1158/1078-0432.CCR-16-0152. [DOI] [PubMed] [Google Scholar]
- 119.Maertens O., Prenen H., Debiec-Rychter M., Wozniak A., Sciot R., Pauwels P., De Wever I., Vermeesch J.R., de Raedt T., De Paepe A., et al. Molecular pathogenesis of multiple gastrointestinal stromal tumors in NF1 patients. Hum. Mol. Genet. 2006;15:1015–1023. doi: 10.1093/hmg/ddl016. [DOI] [PubMed] [Google Scholar]
- 120.Miettinen M., Fetsch J.F., Sobin L.H., Lasota J. Gastrointestinal stromal tumors in patients with neurofibromatosis 1: A clinicopathologic and molecular genetic study of 45 cases. Am. J. Surg. Pathol. 2006;30:90–96. doi: 10.1097/01.pas.0000176433.81079.bd. [DOI] [PubMed] [Google Scholar]
- 121.Yamamoto H., Tobo T., Nakamori M., Imamura M., Kojima A., Oda Y., Nakamura N., Takahira T., Yao T., Tsuneyoshi M. Neurofibromatosis type 1-related gastrointestinal stromal tumors: A special reference to loss of heterozygosity at 14q and 22q. J. Cancer Res. Clin. Oncol. 2009;135:791–798. doi: 10.1007/s00432-008-0514-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Burgoyne A.M., De Siena M., Alkhuziem M., Tang C.M., Medina B., Fanta P.T., Belinsky M.G., von Mehren M., Thorson J.A., Madlensky L., et al. Duodenal-Jejunal Flexure GI Stromal Tumor Frequently Heralds Somatic. JCO Precis. Oncol. 2017;2017:PO.17.00014. doi: 10.1200/PO.17.00014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Mussi C., Schildhaus H.U., Gronchi A., Wardelmann E., Hohenberger P. Therapeutic consequences from molecular biology for gastrointestinal stromal tumor patients affected by neurofibromatosis type 1. Clin. Cancer Res. 2008;14:4550–4555. doi: 10.1158/1078-0432.CCR-08-0086. [DOI] [PubMed] [Google Scholar]
- 124.Śmiech M., Leszczyński P., Kono H., Wardell C., Taniguchi H. Emerging BRAF Mutations in Cancer Progression and Their Possible Effects on Transcriptional Networks. Genes. 2020;11:1342. doi: 10.3390/genes11111342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Hostein I., Faur N., Primois C., Boury F., Denard J., Emile J.F., Bringuier P.P., Scoazec J.Y., Coindre J.M. BRAF mutation status in gastrointestinal stromal tumors. Am. J. Clin. Pathol. 2010;133:141–148. doi: 10.1309/AJCPPCKGA2QGBJ1R. [DOI] [PubMed] [Google Scholar]
- 126.Cohn A.L., Day B.M., Abhyankar S., McKenna E., Riehl T., Puzanov I. BRAF(V600) mutations in solid tumors, other than metastatic melanoma and papillary thyroid cancer, or multiple myeloma: A screening study. Onco Targets Ther. 2017;10:965–971. doi: 10.2147/OTT.S120440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Huss S., Pasternack H., Ihle M.A., Merkelbach-Bruse S., Heitkötter B., Hartmann W., Trautmann M., Gevensleben H., Büttner R., Schildhaus H.U., et al. Clinicopathological and molecular features of a large cohort of gastrointestinal stromal tumors (GISTs) and review of the literature: BRAF mutations in KIT/PDGFRA wild-type GISTs are rare events. Hum. Pathol. 2017;62:206–214. doi: 10.1016/j.humpath.2017.01.005. [DOI] [PubMed] [Google Scholar]
- 128.Agaimy A., Terracciano L.M., Dirnhofer S., Tornillo L., Foerster A., Hartmann A., Bihl M.P. V600E BRAF mutations are alternative early molecular events in a subset of KIT/PDGFRA wild-type gastrointestinal stromal tumours. J. Clin. Pathol. 2009;62:613–616. doi: 10.1136/jcp.2009.064550. [DOI] [PubMed] [Google Scholar]
- 129.Daniels M., Lurkin I., Pauli R., Erbstösser E., Hildebrandt U., Hellwig K., Zschille U., Lüders P., Krüger G., Knolle J., et al. Spectrum of KIT/PDGFRA/BRAF mutations and Phosphatidylinositol-3-Kinase pathway gene alterations in gastrointestinal stromal tumors (GIST) Cancer Lett. 2011;312:43–54. doi: 10.1016/j.canlet.2011.07.029. [DOI] [PubMed] [Google Scholar]
- 130.Jašek K., Váňová B., Grendár M., Štanclová A., Szépe P., Hornáková A., Holubeková V., Plank L., Lasabová Z. BRAF mutations in KIT/PDGFRA positive gastrointestinal stromal tumours (GISTs): Is their frequency underestimated? Pathol. Res. Pract. 2020;216:153171. doi: 10.1016/j.prp.2020.153171. [DOI] [PubMed] [Google Scholar]
- 131.Agaram N.P., Wong G.C., Guo T., Maki R.G., Singer S., Dematteo R.P., Besmer P., Antonescu C.R. Novel V600E BRAF mutations in imatinib-naive and imatinib-resistant gastrointestinal stromal tumors. Genes Chromosomes Cancer. 2008;47:853–859. doi: 10.1002/gcc.20589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Nannini M., Valerio D.S., Gruppioni E., Altimari A., Chiusole B., Saponara M., Pantaleo M.A., Brunello A. Complete radiological response to first-line regorafenib in a patient with abdominal relapse of BRAF V600E mutated GIST. Therap. Adv. Gastroenterol. 2020;13:1756284820927305. doi: 10.1177/1756284820927305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.U.S. Food and Drug Administration Dabrafenib–Trametinib Combination Approved for Solid Tumors with BRAF Mutations. [(accessed on 9 December 2022)]; Available online: https://www.cancer.gov/newsevents/cancer-currents-blog/2022/fda-dabrafenib-trametinib-braf-solidtumors.
- 134.Falchook G.S., Trent J.C., Heinrich M.C., Beadling C., Patterson J., Bastida C.C., Blackman S.C., Kurzrock R. BRAF mutant gastrointestinal stromal tumor: First report of regression with BRAF inhibitor dabrafenib (GSK2118436) and whole exomic sequencing for analysis of acquired resistance. Oncotarget. 2013;4:310–315. doi: 10.18632/oncotarget.864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Charo L.M., Burgoyne A.M., Fanta P.T., Patel H., Chmielecki J., Sicklick J.K., McHale M.T. A Novel PRKAR1B-BRAF Fusion in Gastrointestinal Stromal Tumor Guides Adjuvant Treatment Decision-Making during Pregnancy. J. Natl. Compr. Canc. Netw. 2018;16:238–242. doi: 10.6004/jnccn.2017.7039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Torrence D., Xie Z., Zhang L., Chi P., Antonescu C.R. Gastrointestinal stromal tumors with BRAF gene fusions. A report of two cases showing low or absent KIT expression resulting in diagnostic pitfalls. Genes Chromosomes Cancer. 2021;60:789–795. doi: 10.1002/gcc.22991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Huang L., Guo Z., Wang F., Fu L. KRAS mutation: From undruggable to druggable in cancer. Signal Transduct. Target. Ther. 2021;6:386. doi: 10.1038/s41392-021-00780-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Ricci R., Dei Tos A.P., Rindi G. On the prevalence of KRAS mutations in GISTs. Virchows Arch. 2013;463:847. doi: 10.1007/s00428-013-1496-z. [DOI] [PubMed] [Google Scholar]
- 139.Hechtman J.F., Zehir A., Mitchell T., Borsu L., Singer S., Tap W., Oultache A., Ladanyi M., Nafa K. Novel oncogene and tumor suppressor mutations in KIT and PDGFRA wild type gastrointestinal stromal tumors revealed by next generation sequencing. Genes Chromosomes Cancer. 2015;54:177–184. doi: 10.1002/gcc.22230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Miranda C., Nucifora M., Molinari F., Conca E., Anania M.C., Bordoni A., Saletti P., Mazzucchelli L., Pilotti S., Pierotti M.A., et al. KRAS and BRAF mutations predict primary resistance to imatinib in gastrointestinal stromal tumors. Clin. Cancer Res. 2012;18:1769–1776. doi: 10.1158/1078-0432.CCR-11-2230. [DOI] [PubMed] [Google Scholar]
- 141.Antonescu C.R., Romeo S., Zhang L., Nafa K., Hornick J.L., Nielsen G.P., Mino-Kenudson M., Huang H.-Y., Mosquera J.-M., Dei Tos P.A., et al. Dedifferentiation in gastrointestinal stromal tumor to an anaplastic KIT-negative phenotype: A diagnostic pitfall: Morphologic and molecular characterization of 8 cases occurring either de novo or after imatinib therapy. Am. J. Surg. Pathol. 2013;37:385–392. doi: 10.1097/PAS.0b013e31826c1761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Hong D.S., Fakih M.G., Strickler J.H., Desai J., Durm G.A., Shapiro G.I., Falchook G.S., Price T.J., Sacher A., Denlinger C.S., et al. KRASG12C Inhibition with Sotorasib in Advanced Solid Tumors. N. Engl. J. Med. 2020;383:1207–1217. doi: 10.1056/NEJMoa1917239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Amatu A., Sartore-Bianchi A., Siena S. NTRK gene fusions as novel targets of cancer therapy across multiple tumour types. ESMO Open. 2016;1:e000023. doi: 10.1136/esmoopen-2015-000023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Manea C.A., Badiu D.C., PLoScaru I.C., Zgura A., Bacinschi X., Smarandache C.G., Serban D., Popescu C.G., Grigorean V.T., Botnarciuc V. A review of NTRK fusions in cancer. Ann. Med. Surg. 2022;79:103893. doi: 10.1016/j.amsu.2022.103893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.U.S. Food and Drug Administration FDA Approves an Oncology Drug that Targets a Key Genetic Driver of Cancer, rather than a Specific Type of Tumor. [(accessed on 3 January 2023)]; Available online: https://www.fda.gov/news-events/press-announcements/fda-approves-oncology-drug-targets-key-genetic-driver-cancer-rather-specific-type-tumor.
- 146.U.S. Food and Drug Administration FDA Approves Entrectinib NTRK Solid Tumors and ROS1 NSCLC. [(accessed on 31 December 2022)]; Available online: https://www.fda.gov/drugs/resources-information-approved-drugs/fda-approves-entrectinib-ntrk-solid-tumors-and-ros-1-nsclc.
- 147.Brenca M., Rossi S., Polano M., Gasparotto D., Zanatta L., Racanelli D., Valori L., Lamon S., Dei Tos A.P., Maestro R. Transcriptome sequencing identifies ETV6-NTRK3 as a gene fusion involved in GIST. J. Pathol. 2016;238:543–549. doi: 10.1002/path.4677. [DOI] [PubMed] [Google Scholar]
- 148.Shi E., Chmielecki J., Tang C.M., Wang K., Heinrich M.C., Kang G., Corless C.L., Hong D., Fero K.E., Murphy J.D., et al. FGFR1 and NTRK3 actionable alterations in “Wild-Type” gastrointestinal stromal tumors. J. Transl. Med. 2016;14:339. doi: 10.1186/s12967-016-1075-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.D’Alpino Peixoto R., Medeiros B.A., Cronemberger E.H. Resected High-Risk Rectal GIST Harboring NTRK1 Fusion: A Case Report and Review of the Literature. J. Gastrointest. Cancer. 2021;52:316–319. doi: 10.1007/s12029-020-00423-x. [DOI] [PubMed] [Google Scholar]
- 150.Doebele R.C., Drilon A., Paz-Ares L., Siena S., Shaw A.T., Farago A.F., Blakely C.M., Seto T., Cho B.C., Tosi D., et al. Entrectinib in patients with advanced or metastatic NTRK fusion-positive solid tumours: Integrated analysis of three phase 1–2 trials. Lancet Oncol. 2020;21:271–282. doi: 10.1016/S1470-2045(19)30691-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Hong D.S., DuBois S.G., Kummar S., Farago A.F., Albert C.M., Rohrberg K.S., van Tilburg C.M., Nagasubramanian R., Berlin J.D., Federman N., et al. Larotrectinib in patients with TRK fusion-positive solid tumours: A pooled analysis of three phase 1/2 clinical trials. Lancet Oncol. 2020;21:531–540. doi: 10.1016/S1470-2045(19)30856-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Nathenson M., Demetri G., Lassen U., Hong D., Boni V., Deeken J., Dowlati A., Cox M., Ku N., Cruickshank S., et al. O-020—Activity of larotrectinib in patients with TRK fusion GI malignancies. Ann. Oncol. 2018;29:v107. doi: 10.1093/annonc/mdy149.019. [DOI] [Google Scholar]
- 153.Krook M.A., Reeser J.W., Ernst G., Barker H., Wilberding M., Li G., Chen H.-Z., Roychowdhury S. Fibroblast growth factor receptors in cancer: Genetic alterations, diagnostics, therapeutic targets and mechanisms of resistance. Br. J. Cancer. 2021;124:880–892. doi: 10.1038/s41416-020-01157-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Ornitz D.M., Itoh N. The Fibroblast Growth Factor signaling pathway. Wiley Interdiscip Rev Dev Biol. 2015;4:215–266. doi: 10.1002/wdev.176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Katoh M., Nakagama H. FGF receptors: Cancer biology and therapeutics. Med. Res. Rev. 2014;34:280–300. doi: 10.1002/med.21288. [DOI] [PubMed] [Google Scholar]
- 156.Ellinghaus P., Neureiter D., Nogai H., Stintzing S., Ocker M. Patient Selection Approaches in FGFR Inhibitor Trials-Many Paths to the Same End? Cells. 2022;11:3180. doi: 10.3390/cells11193180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Schaefer I.M., Mariño-Enríquez A., Fletcher J.A. What is New in Gastrointestinal Stromal Tumor? Adv. Anat. Pathol. 2017;24:259–267. doi: 10.1097/PAP.0000000000000158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.U.S. National Library of Medicine ClinicalTrials.gov. [(accessed on 17 February 2023)];2023 Available online: https://clinicaltrials.gov/
- 159.Pascual J., Attard G., Bidard F.C., Curigliano G., De Mattos-Arruda L., Diehn M., Italiano A., Lindberg J., Merker J.D., Montagut C., et al. ESMO recommendations on the use of circulating tumour DNA assays for patients with cancer: A report from the ESMO Precision Medicine Working Group. Ann. Oncol. 2022;33:750–768. doi: 10.1016/j.annonc.2022.05.520. [DOI] [PubMed] [Google Scholar]
- 160.Serrano C., Vivancos A., López-Pousa A., Matito J., Mancuso F.M., Valverde C., Quiroga S., Landolfi S., Castro S., Dopazo C., et al. Clinical value of next generation sequencing of plasma cell-free DNA in gastrointestinal stromal tumors. BMC Cancer. 2020;20:99. doi: 10.1186/s12885-020-6597-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Ravegnini G., Sammarini G., Serrano C., Nannini M., Pantaleo M.A., Hrelia P., Angelini S. Clinical relevance of circulating molecules in cancer: Focus on gastrointestinal stromal tumors. Ther. Adv. Med. Oncol. 2019;11:1758835919831902. doi: 10.1177/1758835919831902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Bauer S., Jones R.L., George S., Gelderblom H., Schöffski P., von Mehren M., Zalcberg J.R., Kang Y.-K., Abdul Razak A.R., Trent J.C., et al. Mutational heterogeneity of imatinib resistance and efficacy of ripretinib vs sunitinib in patients with gastrointestinal stromal tumor: ctDNA analysis from INTRIGUE. J. Clin. Oncol. 2023;41:397784. doi: 10.1200/JCO.2023.41.36_suppl.397784. [DOI] [Google Scholar]