Abstract
Human bocavirus (HBoV) is an important respiratory pathogen, especially in children, but it is often found in co-detection with other respiratory viruses, which makes the diagnostic approach challenging. We compared multiplex PCR and quantitative PCR for HBoV with multiplex tandem PCR (MT-PCR) in 55 cases of co-detection of HBoV and other respiratory viruses. In addition, we investigated whether there is a connection between the severity of the disease, measured by the localization of the infection, and amount of virus detected in the respiratory secretions. No statistically significant difference was found, but children with large amount of HBoV and other respiratory virus had a longer stay in hospital.
Keywords: human bocavirus, children, respiratory tract, viral load, MT-PCR
1. Introduction
Human bocavirus, a small non-enveloped DNA virus that belongs to the Parvoviridae family, was first discovered 2005 by Thomas Allander in the nasopharyngeal samples of children with acute respiratory illness by random molecular screening [1]. Later, three more viruses were found in fecal samples and named human bocavirus 2, 3, and 4, to differentiate them from the first isolated subtype, named human bocavirus 1 (HBoV1); their connection with gastrointestinal infections was investigated [2]. Human bocavirus 1 (HBoV1, further in the text defined as HBoV) is recognized as an important respiratory pathogen, especially in children, which is connected to rhinitis, acute otitis media, pneumonia, bronchiolitis, and asthma exacerbations [3,4,5,6,7]. HBoV is often found in combination with other respiratory pathogens, with rate of co-detection between 8.3% to 100% [2]. It can also be found in asymptomatic children, probably because of prolonged shedding [8] and possible persistence [9]. HBoV1 has only been found to be capable of infection in one in vitro system, which is the human airway epithelium (HAE) cultured at an air–liquid interface (HAE-ALI) made from differentiated (nondividing) epithelial cells. The other system mentioned is the HEK 293 cell line, although replication is possible after the transfection of a pIHBoV1 plasmid that holds the complete double-stranded genome. Other cell lines (HEp-2, Vero MCR-5, etc.) are not susceptible to the virus, probably due to the lack of a specific receptor, which has yet to be discovered [2,10,11]. Due to difficulties of growth in cell culture and the fact that serology is complicated with the existence of the other three types of virus, the diagnostic approach for the detection of HBoV is almost exclusively by molecular methods [2,3,12], mainly by multiplex PCR, as it is part of the standard respiratory panels. One of proposed methods is quantitative PCR, which provides more information about viral load of HBoV [3,12]. For quantitative PCR, a viral load higher than 104 copies/mL is thought to be important [13,14,15], although some studies suggest a higher viral load as a cut off for acute viral infection, more than 106 copies/mL. The advancement and wider use of new molecular techniques during the pandemic era offer new opportunities for investigating the virus. One of those methods is multiplex tandem PCR (MT-PCR) [16]. MT-PCR is a nested PCR, which employs two sequential PCR steps. Step 1 is a short multiplex pre-amplification reaction using primers homologous to all targets offered by the product. Each pair of nested primers are specific to one target. The next step contains primer pairs for every target offered by the kit and Step 2 primers are designed to be “nested inside” the Step 1 primers, which increases the sensitivity and specificity of the assay. The next step is real-time PCR for each target. MT-PCR is not a traditional quantitative method as it cannot determine the viral load in terms of copies per mL, but it can provide information on the amount of viruses by comparing them to a standard amount (spike control). It presents results in the form of intensity, a 5-star rating system for each targeted pathogen. The aim of this work is to compare MT-PCR with multiplex PCR and quantitative PCR for the detection of HBoV, with an emphasis on co-detection of HBoV with other respiratory viruses.
2. Materials and Methods
2.1. Inclusion Criteria and Sample Collection
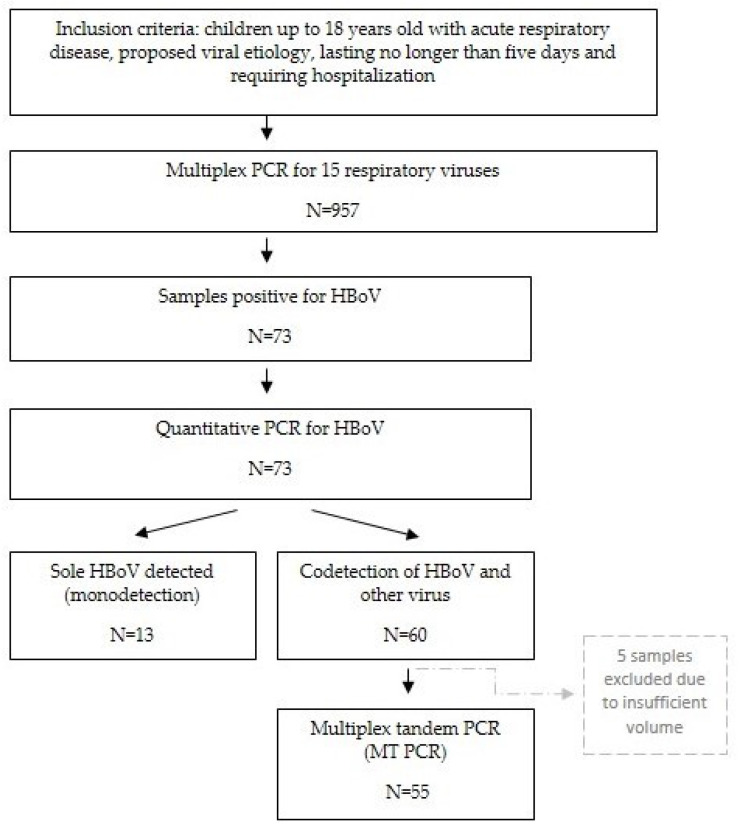
In the period from May 2017 to March 2012, we collected samples from patients aged 0 to 18 years old who were hospitalized in two Croatian hospitals, Children’s Hospital Zagreb and General Hospital Karlovac. The inclusion criteria were acute respiratory disease, proposed viral etiology, lasting no longer than five days, and requiring hospitalization.
Samples collected for viral detection were nasopharyngeal and pharyngeal flocked swabs from each patient, combined, and placed in viral transport medium (UTM®, Copan, Brescia, Italy), and stored in laboratory at −80 °C until tested.
The conceptual framework of the research is presented in Figure 1.
Figure 1.
Conceptual framework of the research.
2.2. Multiplex PCR for 15 Respiratory Viruses
After isolation of viral DNA and RNA from viral transport medium, which was performed using the RibospinTM vRD Kit (GeneAll Biotechnology, Seoul, Korea), multiplex RT-PCR for 15 respiratory viruses, including adenovirus (AdV), human coronavirus (HCoV) 229E/Nl63, HCoV OC43, parainfluenza virus (PIV) types 1-4, RV groups A/B/C, RSV type A and B, influenza (Flu) type A and B, human bocavirus (HBoV), human metapneumovirus (HMPV), and human enterovirus (HEV), was performed using the Seeplex® RV15 OneStep ACE Detection (Seegene Inc., Seoul, Republic of Korea). Amplification was performed with a thermal cycler GeneAmp® 9700 PCR System (Applied Biosystems, Foster City, United States). Detection of PCR products was done by microchip electrophoresis on the MCE®-202 MultiNA device (Shimadzu, Kyoto, Japan). Seeplex® RV15 OneStep ACE Detection includes two internal controls: one is PCR control, in order to check the presence of substances that may interfere with PCR amplification. The other control is called the whole process control (WPC) and includes the human RNase P target as an internal control to check the entire experimental process from nucleic acid extraction to RT-PCR.
2.3. Quantitative PCR for HBoV
Quantitative PCR for samples positive for human bocavirus was performed using the LightMix® Modular Bocavirus kit with a LightCycler 480® System (Roche Diagnostics, Rotkreuz, Switzerland). PCR primers BoF (GGAAGAGACACTGGCAGACAA), BoR (GGGTGTTCCTGATGATATGAGC), and a hydrolysis probe BoP (Cy5-CTGCGGCTCCTGCTCCTGTGAT-BHQ2) targeting the NP-1 gene of HBoV have been described previously [6,17].
2.4. Multiplex Tandem PCR
Samples in which HBoV was detected in combination with at least one other respiratory virus were further analyzed on High-Plex 24 System (AusDiagnostics, Mascot, NSW, Australia), based on the principle of Multiplex Tandem PCR method (MT-PCR) using the respiratory viruses 16 well test, which targets 16 viruses: influenza A, influenza B, RSV, rhinovirus/enterovirus, enterovirus, human bocavirus, human parainfluenza viruses 1-4, human parechovirus, human adenoviruses, human coronaviruses, and human metapneumovirus. The test consists of several controls: positive and negative control, sample adequacy and human DNA control, suitable nucleic acid control, and sample inhibition and instrument function control to ensure the whole process works well.
2.5. Clinical Data Collection
Demographic and clinical data were collected by a retrospective review of patient medical records: including duration of hospitalization, antimicrobial use record, need for oxygen supplementation, need for mechanical ventilation, routine microbiology studies, and data from chest radiographs, which were taken only if necessary, in order to avoid unnecessary X-rays.
3. Results
3.1. Multiplex PCR Results
In the period from May 2017 to March 2012, we collected 957 samples. Using multiplex PCR, human bocavirus was found in 73 samples (73/957; 7.6%), in 41 male and 32 female patients. The median age of the patients positive for HBoV was 1.36 years (the youngest child was 7 days old and the oldest was 15 years old). In 60 cases (60/73, 82%), HBoV was detected in combination with at least one other respiratory virus, and in 13 cases, sole HBoV was discovered.
3.2. Quantitative PCR Results
Quantitative PCR for HBoV samples was performed for 73 samples positive for HBoV and it was successful in 65/73 samples. Eight (8/73) samples were negative in this assay, in all of them HBoV was detected with other viruses on multiplex PCR. Viral load ranged from 9.71 to 2.14 × 107 copies/mL (median 3.91 × 104 copies/mL). In 39 (39/73) samples, a high concentration (more than 104 copies/mL) was found.
3.3. Comparison between Multiplex PCR and MT-PCR
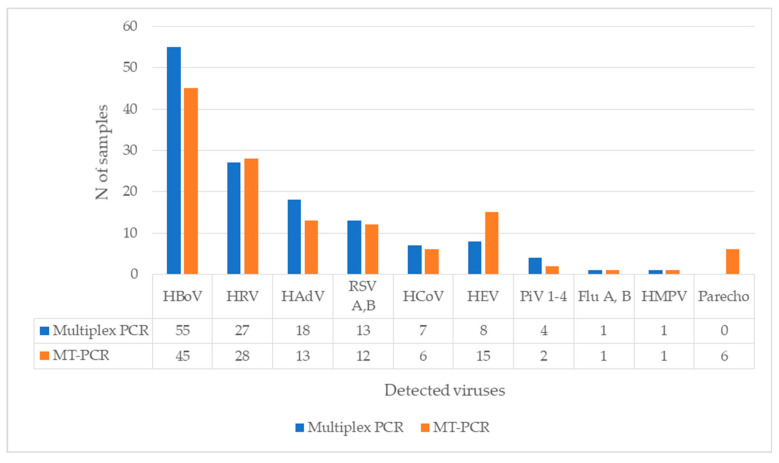
From 60 samples with co-detection, 55 were suitable for further analysis with the MT-PCR method. Of the 55 positive samples for HBoV and other respiratory virus on multiplex PCR, HBoV was present in 45 (45/55) samples on MT-PCR. The most common virus found in co-detection with both methods was human rhinovirus, followed by human adenovirus and RSV. A comparison of results for each virus are presented in Figure 2. Note that multiplex PCR does not consist of primers for parechovirus, while MT-PCR reported enterovirus (EV) in 15 samples, all of them were also positive on the rhinovirus assay (RV/EV).
Figure 2.
Codetection of HBoV with other respiratory viruses, a comparison of multiplex PCR and MT-PCR (N of samples = 55).
HRV—human rhinovirus, AdV—human adenovirus, RSV A B—Respiratory syncytial virus type A and B, HCoV—seasonal human coronavirus, HEV—human enterovirus, PIV 1-4—parainfluenza virus 1-4, HMPV—human metapneumovirus, Flu A B—influenza type A and B, parecho-parechovirus.
In four samples, sole HBoV was detected. Results of the multiplex PCR and quantitative HBoV PCR for those samples are shown in Table 1.
Table 1.
Samples in which only HBoV was detected on MT-PCR with the results of two other tests for those samples.
| Multiplex PCR | HBoV Quantitative PCR (Number of Copies/mL) | MT-PCR | MT-PCR HBoV Intensity |
|---|---|---|---|
| HBoV, RSV B | 9.70 × 106 | HBoV | 5 |
| HBoV, PiV 4, AdV | 8.40 × 105 | HBoV | 5 |
| HBoV, PiV 4 | 2.03 × 102 | HBoV | 1 |
| HBoV, HRV | 5.86 × 102 | HBoV | 2 |
In 10 samples, HBoV was not detected (one sample was negative for all tested viruses). All of them had very low concentration of HBoV or were negative on quantitative PCR, as it is shown in Table 2.
Table 2.
Comparison of results between three methods for samples negative for HBoV on MT-PCR (N = 10).
| Multiplex PCR | HBoV Quantitative PCR (Number of Copies/mL) | MT-PCR | MT-PCR Sample Adequacy |
|---|---|---|---|
| HBoV, HRV | 6.29 × 103 | RV/EV * | 3 |
| HBoV, HRV, RSV A | 37.1 | RSV | 4 |
| HBoV, HRV, HEV, HCoV | 9.71 | HCoV, RV/EV, EV * | 4 |
| HBoV, HRV | 0 | RV/EV * | 3 |
| HBoV, HEV, AdV | 0 | RV/EV, EV * | 5 |
| HBoV, HRV | 0 | RV/EV * | 3 |
| HBoV, HRV, AdV, PiV 1, PiV 4 |
0 | PiV 4 | 4 |
| HBoV, RSV A, HCoV | 0 | RSV, RV/EV * | 4 |
| HBoV, HRV, AdV | 0 | AdV | 4 |
| HBoV, AdV | 0 | 0 | 3 |
* Note that MT-PCR has an assay for human rhinovirus (RV/EV), which is designed to detect human rhinoviruses groups A to C and human enterovirus groups A to D, and an assay for human enterovirus (EV), which is designed to detect enterovirus groups A to D. If both RV/EV and EV assays are positive, the sample is enterovirus. If only RV/EV assay is positive, the sample is rhinovirus.
3.4. Comparison of Quantitative PCR for HBoV and MT PCR
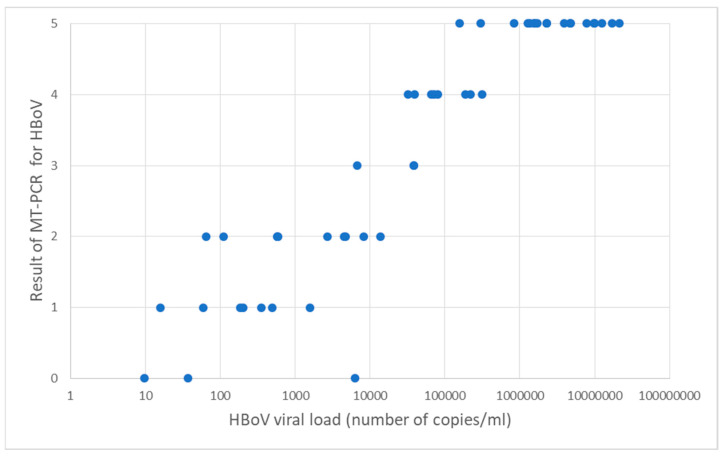
A high HBoV concentration (more than 104 copies/mL) was found in 28/55 samples. Although MT-PCR does not express absolute quantitative concentrations of viruses in a sample, it can provide additional information how much of the targeted gene is present in the sample by 5-star-based representation marked as intensity. We compared results of HBoV quantitative PCR with results on MT-PCR. Of 28 samples with a high concentration on quantitative PCR (more than 104 copies/mL), 25 samples were marked with 4–5 stars on MT-PCR. A comparison of the results are presented in Figure 3.
Figure 3.
Comparison of 5-star based representation on MT-PCR (intensity) and quantitative HBoV PCR: x axis represents concentration of HBoV by quantitative PCR and y axis number of stars on MT-PCR test.
3.5. Characterization of Co-Detections and Comparison of the Amount of HBoV and Other Viruses
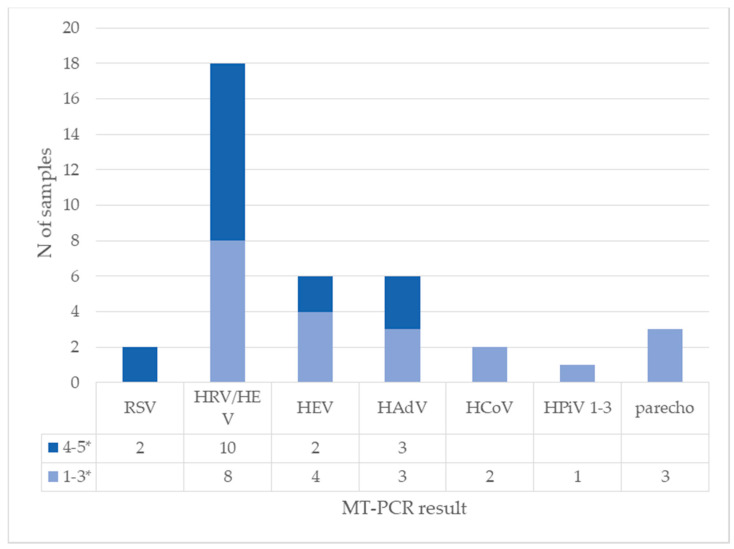
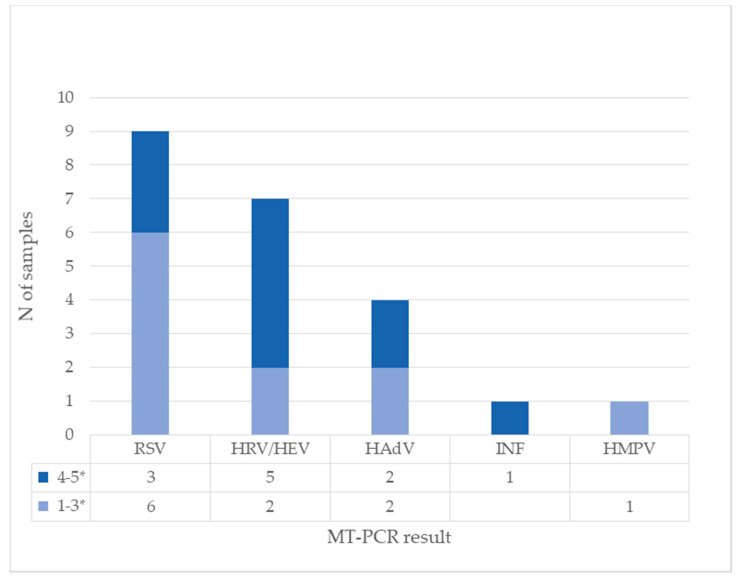
Next, the quantitative nature of MT-PCR potentially allows us to compare relative quantity of viruses in cases of co-infection. We analyzed the amount of other viruses in samples with HBoV to see if there is a difference between samples with high and low HBoV concentrations. In 28 samples with a high concentration of HBoV on quantitative PCR (more than 104 copies/mL), four viruses were present in a high amount (marked with 4–5 stars on MT-PCR): RSV, human rhinovirus, human enterovirus, and human adenovirus. Other viruses were detected in small concentrations (marked 1–3 stars on MT-PCR). The results are presented in Figure 4 and Figure 5.
Figure 4.
Detection of other viruses on MT-PCR in samples with high concentration of HBoV on quantitative PCR (N = 28). Results of MT-PCR are expressed by 5-star representation marked as intensity (1–3* and 4–5*) In some samples, more than one other virus is present.
Figure 5.
Detection of other viruses on MT-PCR in samples with low concentration of HBoV on quantitative PCR (N = 27). Results of MT-PCR are expressed by a 5-star-based representation marked as intensity (1–3* and 4–5*). In some samples, more than one other virus is present.
3.6. Clinical Relevance of Different Amounts of HBoV in Cases of Co-Detection
Clinically, of 55 analyzed patients with co-infection, 23 had infections in the upper respiratory tract (URTI) and 32 had a lower respiratory tract infection (LTRI). The average duration of hospitalization was 6.5 days. Eight patients needed oxygen supplementation and 33 of them received antibiotics. Additional microbiological investigation (nasopharyngeal and throat swab, urine sample, blood culture and serology for M. pneumoniae and C. pneumoniae) for each child was undertaken upon clinical indication. In one child, S. pneumoniae was grown in blood culture, one child had H. influenzae in the nasopharyngeal swab, and one child had S. pyogenes group A in a throat swab. The other children had no bacteriological isolates and received antimicrobial therapy empirically. Radiologic imaging of the lung was indicated in 36/55 patients and in 23 patients’ pathologic findings were found. The average body temperature was 37.2 °C. The median CRP was 28.15 mg/L and median leukocyte count was 7.08 × 109/L.
Furthermore, we investigated if there was a difference between the localization of infection in the respiratory tract (URTI and LRTI) in relation to relative amount of viruses in sample. For that purpose, we divided patients in four groups:
-
A-
Samples with a high concentration of HBoV (more than 104 copies/mL in quantitative PCR) and a high score of other viruses (intensity 4–5 stars on MT-PCR);
-
B-
Samples with a high concentration of HBoV (more than 104 copies/mL in quantitative PCR of and a low score of other viruses (intensity 1–3 stars on MT-PCR);
-
C-
Low concentration of HBoV (less than 104 copies/mL in quantitative PCR) or negative and high score of other viruses (intensity 4–5 stars on MT-PCR);
-
D-
Low concentration of HBoV (less than 104 copies/mL in quantitative PCR) or negative and low score of other viruses (intensity 1–3 stars on MT-PCR).
The results are presented in Table 3. There was no statistically significant difference between groups on the chi-square test (p = 0.53).
Table 3.
Patients divided in four groups according to amount of HBoV and other respiratory viruses, and localization of infection in the respiratory tract (UTRI—upper respiratory tract infection, LRTI—lower respiratory tract infection).
| Group | Quantitative PCR Concentration of HBoV | MT-PCR Intensity of Other Viruses | URTI (N of Patients) |
LRTI (N of Patients) |
|---|---|---|---|---|
| A | high (more than 104 copies/mL) |
4–5 stars | 6 | 8 |
| B | high (more than 104 copies/mL) |
1–3 stars | 8 | 6 |
| C | low or negative (less than 104 copies/mL) |
4–5 stars | 5 | 9 |
| D | low or negative (less than 104 copies/mL) |
1–3 stars | 4 | 9 |
| Total | 23 | 32 |
4. Discussion
The last three years of the pandemic has pointed viral respiratory infections as a focus of medical and social interest, thus, understanding their etiology is of outmost importance. Since its first discovery 2005, HBoV is recognized as an important respiratory pathogen in the child population. However, diagnostic limitations and lack of animal models are still leaving questions about clinical significance and pathogenesis of this infection open. Our study revealed a high prevalence of HBoV (7.6%) among children hospitalized for acute respiratory infection during a four-year period, which points to it as the fifth most frequently detected virus in respiratory tract samples after RV, RSV, AdV, and PiV [18]. The results are in accordance with other prevalence studies. Guido et al. [2] estimated a global HBoV prevalence of 6.3 and 5.9% in respiratory and gastrointestinal infections, respectively, and a large systematic analysis from 2022 found that the HBoV prevalence in Europe varied from 2.0 to 45.69% with a pooled estimate of 9.57% [19]. Additionally, a high occurrence of co-detection with other respiratory viruses was present in our study (82.2%), which is also a well-documented characteristic of HBoV. In a review that included 311 studies from 50 countries all over the world performed between 2005 and 2016, HBoV-positivity ranged from 8.3% to 100%, with total co-infection estimates in the 193 studies covered of 52.4% [2]. It is probably result of prolonged shedding from the nasopharynx, found both in immunocompetent and immunocompromised children [2,20,21], and possibly persistence [2,22]. The diagnosis of this virus and the interpretation of its test results, often in conjunction with other respiratory viruses, is controversial. It is clear that relying solely on qualitative PCR is not sufficient for a definitive diagnosis due to a high rate of co-detection with other respiratory viruses. Other proposed methods are quantitative PCR, mRNA detection [3,23,24], and serology, which are complicated with the existence of the three other types of viruses, HBoV 2-4, causing immunological cross-reactivity and a phenomena called original antigenic sin [25]. Quantitative PCR suggests that a viral load above 104 copies/mL is significant; however, some research indicates that for acute viral infections, a cut off of over 106 copies/mL is more appropriate [3,16,26]. A study that compared performances between mRNA detection and quantitative PCR showed that quantitative PCR has sensitivity of 100%, specificity of 93–99%, and positive predictive values (PPV) of 56–87%. When serology was used, the standard sensitivity of quantitative PCR was 81%, specificity was 92%, and the PPV was 87% [24].
In the era of pandemics, we are witnessing to broader application of molecular methods and the wider use of automated and semi-automated systems, which opens doors for easier diagnostic approach to this virus.
In this work, we compared classic quantitative PCR for HBoV with MT-PCR, which is one of those methods.
Although MT-PCR does not provide an exact number of copies (viral load), as does classic quantitative PCR, it is naturally a quantitative method [27] and expresses results as intensity with a 5-star-based representation for every targeted pathogen. We compared those results for human bocavirus with results of quantitative PCR and high congruence was present, 25/28 samples with a high concentration of HBoV on quantitative PCR (more than 104 copies/mL) had 4–5 stars on the MT-PCR intensity report for this virus. Ten samples positive for HBoV on multiplex PCR were negative on the MT-PCR test, but for seven of them, quantitative PCR was also negative, and two samples had very low number of copies of HBoV, at less than 50 copies. MT-PCR has endogenous control of extraction and amplification, named Sample Adequacy Control, which targets a human reference gene as an indicator of suitability of nucleic acid extract and it was appropriate for all 10 negative samples. Sample inhibition control, named SPIKE, was also positive, so the negative MT-PCR result could not be explained with extraction or amplification failure. The low level of viral DNA in the sample may have resulted from degradation over time due to extended freezing. Next, the majority of practitioners still approach respiratory infections assuming single pathogen etiology. However, sensitive molecular methods have discovered that multiple pathogens can be detected, even in cases of influenza or respiratory syncytial virus infection [28], but the influence of such co-detection on severity of respiratory diseases is still unclear [29,30]. In order to highlight cases of co-detection in which HBoV is present in combination with other viruses, we divided the patients in four groups to see is there a difference in clinical presentation between them. It would be expected that patients with high amount of human bocavirus and high intensity of other respiratory virus had a more severe clinical presentation, as some studies suggest [14,31,32], and more often develop infection of the lower respiratory system (LRTI). Moreover, Moesker et al. [33] reported a higher viral load in HBoV-positive patients admitted to an intensive care unit. Contrarily, other studies are questioning such findings, including one multicentric study which compared viral load in the upper respiratory tract of children with pneumonia and age-matched community controls and the results were equivocal [34]. In our study, we did not find a significant difference in the localization of infection in the respiratory tract between the groups of patients divided according to the amount of detected viruses in nasopharyngeal/pharyngeal samples. However, we did find that children from group A (high amount of both HBoV and other virus) had a slightly longer average stay in hospital, 6.64 days, respectively, compared to the other three groups, where the average hospitalization days were 5.38, 4.25, and 5.75 days for group B, C, and D, repsectively. The average stay in hospital for children with mono-infection with HBoV was 5.75 days.
The study has a few limitations, including the relatively small sample size that hinders a detailed analysis of the clinical differences between groups. Further research, ideally with multiple centers involved, is needed to better understand the clinical outcomes of HBoV co-infections with other viruses.
Subsequently, a complete analysis of all samples with all three methods was not possible. Specifically, five samples detected with co-detection could not undergo further analysis using MT-PCR due to insufficient sample volume. As a result, the final analysis was based on 55 specimens.
Next, we eliminated the possibility of bacterial co-infections at the time of admission based on clinical evaluation, CRP, and white blood cell count, and the available microbiological information. However, empiric antibiotic therapy was started in 33 cases during the stay in the hospital, and bacterial superinfection could not be proven or ruled out by subsequent sampling of these children.
In conclusion, diagnostic advances, including nucleic acid amplification platforms, have greatly improved the detection of respiratory viral pathogens, but the high sensitivity of such molecular methods has raised question about meaning of co-detection of multiple pathogens. One of the major challenges nowadays is distinguishing true infection from asymptomatic carriage [35,36]. In addition, it is difficult to assess the direct impact of HBoV on respiratory tract infections due to its frequent co-detection with other respiratory viruses. A comparison of MT-PCR with quantitative PCR showed that MT-PCR could be powerful tool for distinguishing the true nature of HBoV detection on the basis of amount of virus compared to co-detected pathogens.
Acknowledgments
Authors wish to thank Matea Kvaternik Celjak and Suzana Ćesić for their technical assistance.
Author Contributions
Conceptualization M.M. and S.L.-S.; methodology M.M. and S.L.-S.; analysis M.M. and S.L.-S.; investigation M.M., S.L.-S. and I.I.-J.; resources, I.I.-J., S.L.-S. and J.V.; data curation M.M., S.L.-S.; writing—original draft preparation, M.M. and S.L.-S.; writing—review and editing S.L.-S. and J.V.; supervision S.L.-S. and J.V.; funding acquisition S.L.-S. and J.V. All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement
The study was conducted according to the guidelines of the Declaration of Helsinki and approved by the ethics committee of the Dr Andrija Štampar Teaching Institute of Public Health (protocol code 500-04/15-01/01; date of approval 23 May 2016).
Informed Consent Statement
Written informed consent was obtained from all participants (childrens’parents or their legal guardians).
Data Availability Statement
The data presented in this study are available on request from the corresponding author. The data are not publicly available due to privacy consideration.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the designof the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Funding Statement
This research was funded by the Croatian Science Foundation under the project titled “New and neglected respiratory viruses in vulnerable groups of patients” (grant no. IP-2016-06-7556 to S.L.J.-S.)
Footnotes
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content.
References
- 1.Allander T., Tammi M.T., Eriksson M., Bjerkner A., Tiveljung-Lindell A., Andersson B. Cloning of a human parvovirus by molecular screening of respiratory tract samples. Proc. Natl. Acad. Sci. USA. 2005;102:12891–12896. doi: 10.1073/pnas.0504666102. Erratum in Proc. Natl. Acad. Sci. USA 2005, 102, 15712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Guido M., Tumolo M.R., Verri T., Romano A., Serio F., De Giorgi M., De Donno A., Bagordo F., Zizza A. Human bocavirus: Current knowledge and future challenges. World J. Gastroenterol. 2016;22:8684–8697. doi: 10.3748/wjg.v22.i39.8684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Christensen A., Kesti O., Elenius V., Eskola A.L., Døllner H., Altunbulakli C., Akdis C.A., Söderlund-Venermo M., Jartti T. Human bocaviruses and paediatric infections. Lancet Child Adolesc. Health. 2019;3:418–426. doi: 10.1016/S2352-4642(19)30057-4. [DOI] [PubMed] [Google Scholar]
- 4.Petrarca L., Nenna R., Frassanito A., Pierangeli A., Di Mattia G., Scagnolari C., Midulla F. Human bocavirus in children hospitalized for acute respiratory tract infection in Rome. World J. Pediatr. 2020;16:293–298. doi: 10.1007/s12519-019-00324-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Meriluoto M., Hedman L., Tanner L., Simell V., Mäkinen M., Simell S., Mykkänen J., Korpelainen J., Ruuskanen O., Ilonen J., et al. Association of humanbocavirus 1 infection with respiratory disease in childhoodfollow-up study, Finland. Emerg. Infect. Dis. 2012;18:264–271. doi: 10.3201/eid1802.111293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Allander T., Jartti T., Gupta S., Niesters H.G., Lehtinen P., Osterback R., Vuorinen T., Waris M., Bjerkner A., Tiveljung-Lindell A., et al. Human bocavirus and acute wheezing in children. Clin. Infect. Dis. 2007;44:904–910. doi: 10.1086/512196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nokso-Koivisto J., Pyles R.B., Miller A.L., Jennings K., Loeffelholz M., Chonmaitree T. Role of Human Bocavirus in Upper Respiratory Tract Infections and Acute Otitis Media. J. Pediatr. Infect. Dis. Soc. 2014;3:98–103. doi: 10.1093/jpids/pit061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Martin E.T., Fairchok M.P., Kuypers J., Magaret A., Zerr D.M., Wald A., Englund J.A. Frequent and prolonged shedding of bocavirus in young children attending daycare. J. Infect. Dis. 2010;201:1625–1632. doi: 10.1086/652405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wagner J.C., Pyles R.B., Miller A.L., Nokso-Koivisto J., Loeffelholz M.J., Chonmaitree T. Determining Persistence of Bocavirus DNA in the Respiratory Tract of Children by Pyrosequencing. Pediatr. Infect. Dis. J. 2016;35:471–476. doi: 10.1097/INF.0000000000001058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang Z., Shen W., Cheng F., Deng X., Engelhardt J.F., Yan Z., Qiu J. Parvovirus Expresses a Small Noncoding RNA That Plays an Essential Role in Virus Replication. J. Virol. 2017;91:e02375-16. doi: 10.1128/JVI.02375-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jartti T., Hedman K., Jartti L., Ruuskanen O., Allander T., Söderlund-Venermo M. Human bocavirus-the first 5 years. Rev. Med. Virol. 2012;22:46–64. doi: 10.1002/rmv.720. [DOI] [PubMed] [Google Scholar]
- 12.Kantola K., Sadeghi M., Antikainen J., Kirveskari J., Delwart E., Hedman K., Söderlund-Venermo M. Real-time quantitative PCR detection of four human bocaviruses. J. Clin. Microbiol. 2010;48:4044–4050. doi: 10.1128/JCM.00686-10. Erratum in J. Clin. Microbiol. 2011, 49, 4029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kantola K., Hedman L., Tanner L., Simell V., Mäkinen M., Partanen J., Sadeghi M., Veijola R., Knip M., Ilonen J., et al. B-Cell Responses to Human Bocaviruses 1-4: New Insights from a Childhood Follow-Up Study. PLoS ONE. 2015;10:e0139096. doi: 10.1371/journal.pone.0139096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jiang W., Yin F., Zhou W., Yan Y., Ji W. Clinical significance of different virus load of human bocavirus in patients with lower respiratory tract infection. Sci. Rep. 2016;6:20246. doi: 10.1038/srep20246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Söderlund-Venermo M., Lahtinen A., Jartti T., Hedman L., Kemppainen K., Lehtinen P., Allander T., Ruuskanen O., Hedman K. Clinical assessment and improved diagnosis of bocavirus-induced wheezing in children, Finland. Emerg. Infect. Dis. 2009;15:1423–1430. doi: 10.3201/eid1509.090204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stanley K.K., Szewczuk E. Multiplexed tandem PCR: Gene profiling from small amounts of RNA using SYBR Green detection. Nucleic Acids Res. 2005;33:e180. doi: 10.1093/nar/gni182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Koskenvuo M., Möttönen M., Waris M., Allander T., Salmi T.T., Ruuskanen O. Human bocavirus in children with acute lymphoblastic leukemia. Eur. J. Pediatr. 2008;167:1011–1015. doi: 10.1007/s00431-007-0631-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ljubin-Sternak S., Slović A., Mijač M., Jurković M., Forčić D., Ivković-Jureković I., Tot T., Vraneš J. Prevalence and Molecular Characterization of Human Bocavirus Detected in Croatian Children with Respiratory Infection. Viruses. 2021;13:1728. doi: 10.3390/v13091728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Polo D., Lema A., Gándara E., Romalde J.L. Prevalence of human bocavirus infections in Europe. A systematic review and meta-analysis. Transbound. Emerg. Dis. 2022;69:2451–2461. doi: 10.1111/tbed.14233. [DOI] [PubMed] [Google Scholar]
- 20.Blessing K., Neske F., Herre U., Kreth H.W., Weissbrich B. Prolonged detection of human bocavirus DNA in nasopharyngeal aspirates of children with respiratory tract disease. Pediatr. Infect. Dis. J. 2009;28:1018–1019. doi: 10.1097/INF.0b013e3181a854ae. [DOI] [PubMed] [Google Scholar]
- 21.Martin E.T., Kuypers J., McRoberts J.P., Englund J.A., Zerr D.M. Human Bocavirus 1 Primary Infection and Shedding in Infants. J. Infect. Dis. 2015;212:516–524. doi: 10.1093/infdis/jiv044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xu M., Perdomo M.F., Mattola S., Pyöriä L., Toppinen M., Qiu J., Vihinen-Ranta M., Hedman K., Nokso-Koivisto J., Aaltonen L.M., et al. Persistence of Human Bocavirus 1 in Tonsillar Germinal Centers and Antibody-Dependent Enhancement of Infection. MBio. 2021;12:e03132-20. doi: 10.1128/mBio.03132-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xu M., Arku B., Jartti T., Koskinen J., Peltola V., Hedman K., Söderlund-Venermo M. Comparative Diagnosis of Human Bocavirus 1 Respiratory Infection with Messenger RNA Reverse-Transcription Polymerase Chain Reaction (PCR), DNA Quantitative PCR, and Serology. J. Infect. Dis. 2017;215:1551–1557. doi: 10.1093/infdis/jix169. [DOI] [PubMed] [Google Scholar]
- 24.Schlaberg R., Ampofo K., Tardif K.D., Stockmann C., Simmon K.E., Hymas W., Flygare S., Kennedy B., Blaschke A., Eilbeck K., et al. Human Bocavirus Capsid Messenger RNA Detection in Children with Pneumonia. J. Infect. Dis. 2017;216:688–696. doi: 10.1093/infdis/jix352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Proença-Modena J.L., Gagliardi T.B., Paula F.E., Iwamoto M.A., Criado M.F., Camara A.A., Acrani G.O., Cintra O.A., Cervi M.C., Arruda L.K., et al. Detection of human bocavirus mRNA in respiratory secretions correlates with high viral load and concurrent diarrhea. PLoS ONE. 2011;6:e21083. doi: 10.1371/journal.pone.0021083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Christensen A., Nordbø S.A., Krokstad S., Rognlien A.G., Døllner H. Human bocavirus in children: Mono-detection, high viral load and viraemia are associated with respiratory tract infection. J. Clin. Virol. 2010;49:158–162. doi: 10.1016/j.jcv.2010.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Szewczuk E., Thapa K., Anninos T., McPhie K., Higgins G., Dwyer D.E., Stanley K.K., Iredell J.R. Rapid semi-automated quantitative multiplex tandem PCR (MT-PCR) assays for the differential diagnosis of influenza-like illness. BMC Infect. Dis. 2010;10:113. doi: 10.1186/1471-2334-10-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Esper F.P., Spahlinger T., Zhou L. Rate and influence of respiratory virus co-infection on pandemic (H1N1) influenza disease. J. Infect. 2011;63:260–266. doi: 10.1016/j.jinf.2011.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Goka E.A., Vallely P.J., Mutton K.J., Klapper P.E. Single and multiple respiratory virus infections and severity of respiratory disease: A systematic review. Paediatr. Respir Rev. 2014;15:363–370. doi: 10.1016/j.prrv.2013.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mazur N.I., Bont L., Cohen A.L., Cohen C., von Gottberg A., Groome M.J., Hellferscee O., Klipstein-Grobusch K., Mekgoe O., Naby F., et al. South African Severe Acute Respiratory Illness (SARI) Surveillance Group. Severity of Respiratory Syncytial Virus Lower Respiratory Tract Infection with Viral Coinfection in HIV-Uninfected Children. Clin. Infect. Dis. 2017;64:443–450. doi: 10.1093/cid/ciw756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhao B., Yu X., Wang C., Teng Z., Wang C., Shen J., Gao Y., Zhu Z., Wang J., Yuan Z., et al. High human bocavirus viral load is associated with disease severity in children under five years of age. PLoS ONE. 2013;8:e62318. doi: 10.1371/journal.pone.0062318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Deng Y., Gu X., Zhao X., Luo J., Luo Z., Wang L., Fu Z., Yang X., Liu E. High viral load of human bocavirus correlates with duration of wheezing in children with severe lower respiratory tract infection. PLoS ONE. 2012;7:e34353. doi: 10.1371/journal.pone.0034353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Moesker F.M., van Kampen J.J., van der Eijk A.A., van Rossum A.M., de Hoog M., Schutten M., Smits S.L., Bodewes R., Osterhaus A.D., Fraaij P.L. Human bocavirus infection as a cause of severe acute respiratory tract infection in children. Clin. Microbiol. Infect. 2015;21:964.e1–964.e8. doi: 10.1016/j.cmi.2015.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Feikin D.R., Fu W., Park D.E., Shi Q., Higdon M.M., Baggett H.C., Brooks W.A., Deloria Knoll M., Hammitt L.L., Howie S.R.C., et al. Is Higher Viral Load in the Upper Respiratory Tract Associated With Severe Pneumonia? Findings From the PERCH Study. Clin. Infect. Dis. 2017;64((Suppl. S3)):S337–S346. doi: 10.1093/cid/cix148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Walter J.M., Wunderink R.G. Severe Respiratory Viral Infections: New Evidence and Changing Paradigms. Infect. Dis. Clin. N. Am. 2017;31:455–474. doi: 10.1016/j.idc.2017.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Self W.H., Williams D.J., Zhu Y., Ampofo K., Pavia A.T., Chappell J.D., Hymas W.C., Stockmann C., Bramley A.M., Schneider E., et al. Respiratory Viral Detection in Children and Adults: Comparing Asymptomatic Controls and Patients with Community-Acquired Pneumonia. J. Infect. Dis. 2016;213:584–591. doi: 10.1093/infdis/jiv323. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data presented in this study are available on request from the corresponding author. The data are not publicly available due to privacy consideration.