Abstract
In the present study we sought to determine the source of heat-induced oxidative stress. We investigated the involvement of mitochondrial respiratory electron transport in post-diauxic-phase cells under conditions of lethal heat shock. Petite cells were thermosensitive, had increased nuclear mutation frequencies, and experienced elevated levels of oxidation of an intracellular probe following exposure to a temperature of 50°C. Cells with a deletion in COQ7 leading to a deficiency in coenzyme Q had a much more severe thermosensitivity phenotype for these oxidative endpoints following heat stress compared to that of petite cells. In contrast, deletion of the external NADH dehydrogenases NDE1 and NDE2, which feed electrons from NADH into the electron transport chain, abrogated the levels of heat-induced intracellular fluorescence and nuclear mutation frequency. Mitochondria isolated from COQ7-deficient cells secreted more than 30 times as much H2O2 at 42 as at 30°C, while mitochondria isolated from cells simultaneously deficient in NDE1 and NDE2 secreted no H2O2. We conclude that heat stress causes nuclear mutations via oxidative stress originating from the respiratory electron transport chains of mitochondria.
Oxygen-free radicals and other reactive oxygen species such as hydrogen peroxide are produced as byproducts of aerobic respiration and cause damage to proteins, lipids, and DNA, resulting in mutation and loss of viability (reviewed in references 2 and 14). Induction and repression of specific antioxidant proteins occur in response to the conditions leading to an increased flux of reactive oxygen species (13, 27), for example, when cells are exposed to redox cycling xenobiotics, hyperbaric oxygen, or heat stress (5).
Heat and oxygen stress responses involve the induction of an overlapping set of genes (8). In Saccharomyces cerevisiae, sublethal H2O2 exposure elicits the induction of a variety of heat shock response genes involved in thermoprotection, including the HSP104 gene (involved in the solubilization of aggregated proteins and the protection of RNA splicing) and the HSP70 genes (protein folding and refolding) as well as the antioxidant enzyme genes catalase (CTT1), thioredoxin peroxidase (TPX1 and TPX2), and cytochrome c peroxidase (CCP1) (13). Heat exposure provides cross-tolerance to later lethal H2O2 exposure (8). Deletion of protein antioxidant defenses, including catalase, cytochrome c peroxidase, superoxide dismutase, and thioredoxin peroxidase, sensitizes cells to lethal heat exposures, and anaerobic cells are greatly thermoresistant compared to their aerobic counterparts (9, 16). This evidence strongly suggests that heat stress produces an oxidative stress.
Mitochondrial electron transport consumes >90% of cellular oxygen and generates superoxide anions as minor by-products (1a, 23, 28). Mitochondria contain superoxide dismutase (SOD2), which rapidly converts the superoxide radical to neutrally charged hydrogen peroxide (22). Hydrogen peroxide can diffuse out of mitochondria, which avoids the dangerous buildup within the matrix of free radicals that could attack vulnerable mitochondrial DNA (25) or proteins containing iron sulfur centers (17). Cytochrome c peroxidase, located in the inner mitochondrial space, can detoxify H2O2 to water and oxygen, and catalase located in the yeast cytosol can do likewise (15, 30).
In the yeast S. cerevisiae, ubiquinone, or coenzyme Q, (CoQ), associates with high affinity with components of the electron transport chain to form single functional units that vary with respect to their associated ubiquinone oxidoreductases (6). Different CoQ oxidoreductases are utilized depending upon what carbon source is available; for example, upon depletion of glucose from the medium, cells enter into the post-diauxic phase of growth, in which the electron transport chain is adapted to use ethanol as the primary carbon source. The diauxic shift is also associated with the derepression of genes involved in respiration, the proliferation of mitochondria, and the oxidation of ethanol to produce NADH (10, 11). S. cerevisiae has three NADH dehydrogenase proteins associated with the inner mitochondrial membrane capable of coupling the oxidation of NADH to the reduction of CoQ (4, 18, 26). One faces the matrix space (NDI) and utilizes mitochondrial NADH; the other two NADH dehydrogenases face outward and can receive electrons from cytosolic NADH (NDE1/NHD1 and NDE2/NDH2, hereinafter referred to as NDE1 and NDE2). Growth on ethanol is unaffected by deletion of the internal NADH dehydrogenase (NDI) but is severely reduced by deletion of both external NADH dehydrogenases (NDE1 and NDE2) (18, 26). This finding suggests that the externally oriented dehydrogenases NDE1 and NDE2 are primarily involved in respiration during growth on ethanol.
In this study, nuclear mutations, oxidation of an intracellular fluorescent probe, and cellular viability were measured in petite cells ([rho−]) as well as in cells lacking CoQ and both of the externally oriented NADH dehydrogenases (NDE1 and NDE2) during the post-diauxic phase of growth. In addition, hydrogen peroxide secreted from mitochondria isolated from these strains was measured at 30 or 42°C. As mitochondrial electron transport is a large contributor to oxidative stress (24), we have investigated whether the observed oxidative effects of heat result from an augmentation of mitochondrial free-radical flux.
MATERIALS AND METHODS
Strains and plasmids.
Yeast strains used in this study are derived from JM43 MATa ura3-52 lys2-801 leu2-3,112 and JM43 coq7Δ-1::LEU2, donated kindly by Catherine F. Clarke. JDY35 was made by deletion disruption of NDE1 and NDE2 in strain JM43 using the deletion or disruption constructs detailed below. JDY43 was made by deletion or disruption of NDE1 and NDE2 in strain JM43 coq7Δ-1::LEU2 using the deletion or disruption constructs detailed below.
Petite strains were generated by overnight growth of [rho+] cells in liquid YPAD (10 g of yeast extract per liter, 20 g of peptone per liter, 48 mg of adenine per ml, 2% glucose) containing 10 μM ethidium bromide. Following incubation, cultures were streaked onto solid YPAD and replica plated onto medium containing 3% glycerol as the sole carbon source. [rho−] cells were classed according to their inability to grow on the nonfermentable carbon source. This treatment resulted in >90% petite mutant formation.
Strains with deletions or disruptions in both genes NDE1 and NDE2 were generated from plasmids pJD55 and pJD59, respectively. When both genes are inactivated, the mitochondria are completely unable to utilize external (cytoplasmic) NADH (18, 26).
The NDE1 deletion construct was made from a 2.2-kb PCR fragment containing the NDE1 gene, which was amplified using the primers NDE1UP (3′-CGGGAATTCCGGCAATGAGAAGAGTTTTGG-5′) and NDE1LOW (5′-CGCGGATCCGCATAAAAAAGGGACAAGGC-3′). BamHI and EcoRI digestion of the PCR fragments facilitated cloning into the BamHI and EcoRI sites of pUC19 to create pJD53. Digestion of pJD53 with the BbsI restriction enzyme releases a 1.1-kb fragment from within the open reading frame. Fill-in reaction with the Klenow fragment enzyme and insertion of a BglII linker created pJD54. Insertion of a 1.1-kb BamHI fragment containing the URA3 gene into the BglII site of pJD54 created pJD55. Digestion of pJD55 with BamHI/EcoRI releases the deletion or the disruption construct.
The NDE2 deletion construct was made from a 1.6-kb PCR fragment containing the NDE2 gene, which was amplified using the primers NDE2UP (5′-CGCGGATCCGAGTGAAATAATAGAGCCCG-3′) and NDE2LOW (5′-CCGGAATTCAGTATTCGCCTTCCTGATGTGC-3′). BamHI and EcoRI digestion of the PCR fragments facilitated cloning into the BamHI and EcoRI sites of pUC19 to create pJD57. Digestion of pJD57 with EcoRV/StuI released a 1.2-kb fragment from within the open reading frame. Insertion of a BglII linker created pJD58. The BamHI/BglII fragment of pNKY51 contains the hisG-URA3-hisG cassette (1). Insertion of the hisG-URA3-hisG BamHI/BglII fragment into the BglII site of pJD58 created pJD59. Digestion of pJD59 with EcoRI and BamHI released the deletion or the disruption construct. The URA3 marker can be recovered in mutants generated with the hisG cassette by selection on 5-fluoroorotic acid.
Oxidative fluorescence.
Intracellular fluorescence was measured using the oxidant-sensitive probe 2′,7′-dichlorofluoroscin diacetate (DCFH-DA). Uptake of this dye, followed by the removal of the acetate groups, results in the formation of the nonfluorescing substrate DCFH. Oxidation of DCF produces 2′,7′-dichlorofluoroscein, which absorbs at a wavelength of 504 nm and emits at a wavelength of 524 nm. Five milliliters of YPAD cultures were grown for 48 h and washed in distilled water. Cells were resuspended at 107 cells/ml in 5 ml of the original YPAD saved from the cultures. Cells were incubated with shaking at 30°C in 50 μM DCFH-DA for 1 h. Following loading, cells were washed three times in distilled water and resuspended in 100 μl of phosphate-buffered saline (pH 7.4) (PBS); Heating at 50°C was performed for various time periods, followed by incubation on ice. Cells were diluted with 1 ml of PBS, 25 μl of chloroform, and 50 μl of 0.1% sodium dodecyl sulfate; all of the ingredients were added and mixed thoroughly before incubation on ice for 15 min. Following lysis, cells and debris were removed by centrifugation for 5 min at high speed in a microcentrifuge. One milliliter of the supernatant was added to 1 ml of PBS, and fluorescence was measured at an excitation wavelength of 504 nm and an emission wavelength of 524 nm (Hitachi Spectrofluorimeter F-2000) at 25°C. Anaerobic experiments were performed as described above, except that the aerobically grown cells were introduced into the anaerobic glove box 1 h prior to the addition of DCFH-DA. All washing and heat stress steps were performed within the anaerobic chamber using preequilibrated anaerobic solutions.
Mutation frequency.
Forward mutations were measured at the CAN1 locus. Any mutation at the CAN1 locus which results in the disruption of the function of the arginine permease leads to the development of resistance to the drug canavanine.
Lethal heat assays were performed on cells grown in YPAD for 48 h. Cells were plated onto solid synthetic complete (SC) medium lacking arginine (SC−Arg) and onto SC−Arg with 60 mg of l-canavinine per liter added. Forward mutation frequencies were measured by counting the number of canavanine-resistant colonies per viable cell.
Microtiter mutation rate assay.
This assay is a measure of spontaneous mutation rates in cells during the log phase of growth and has the advantage of avoiding the “jackpot effect.” Mutation rates were performed using a modification of the Von Borstel 1,000-well assay (29). Cultures were grown overnight in 5 ml of liquid YPAD, washed in water, and diluted to 2.8 × 103 cells/ml in 40 ml of SC−Arg liquid medium. Each well of a 96-well microtiter plate was filled with 350 μl of the original culture (1,000 cells/well). A 100-μl aliquot was plated onto solid YPAD medium to determine the exact number of initial inocula per well. The cultures on the microtiter plates were grown at 30°C for six generations to populations of approximately 7 × 104 cells per well before the addition of l-canavanine to a final concentration of 60 mg/liter for each well. The microtiter plates were sealed with Parafilm to prevent evaporation and incubated at 30°C for 6 days. Wells containing colonies were scored. Wells containing no colonies were counted with a Coulter Counter to determine the final concentrations of cells per well. The mutation rate was calculated according to the method of Von Borstel (29) and expressed as the number of mutation events per cell per generation, where the number of generations corresponds to the number of new cells generated during the assay.
Cell viability.
Following heat stress at 50°C, cells were incubated on ice prior to dilution and plating onto solid SC medium. Unheated controls were also plated to enable survival percentages to be determined.
Measurement of released hydrogen peroxide from isolated mitochondria.
Isolation of crude mitochondria was performed as described previously (12). Hydrogen peroxide released from mitochondria was measured using the fluorescent horseradish peroxidase (HRP) substrate A6550, as described previously (21). Briefly, 2.5 mg of mitochondria per ml was suspended in 1.2 M sorbitol–20 mM potassium phosphate–50 μM NADH (pH 7.4) (Sigma, St. Louis, Mo.) to a final volume of 85 μl and incubated at 30 or 42°C for 60 min. Following the incubation, HRP and A6550 (Molecular Probes, Eugene, Oreg.) were added to final concentrations of 1 U/ml and 100 μM, respectively. Catalase, when added, was at 1,000 U/ml. ADP, when added, was at 1 mM. The HRP reaction was allowed to proceed for 15 min at 37°C. Fluorescence was measured using an excitation wavelength of 570 nm and an emission wavelength of 645 nm with a fluorescence 96-well plate reader (Packard). No change in fluorescence over background values was observed in the absence of either NADH or mitochondria (data not shown). The baseline fluorescence obtained at the zero time point was subtracted from the fluorescence at the 60-min time point for each sample. The result, expressed in picomolar units of H2O2, was calculated from the standard curve. The results and standard curve data are the means ± standard deviations (SD) calculated from the results of four experiments.
RESULTS
Heat stress increases the reactive oxygen flux in S. cerevisiae. As mitochondrial electron transport is known to be a major source of superoxide anion generation, we decided to investigate the involvement of mitochondrial electron transport in the generation of heat-induced reactive oxygen during respiratory growth in post-diauxic-phase cultures.
Petite and CoQ-deficient cells are thermosensitive.
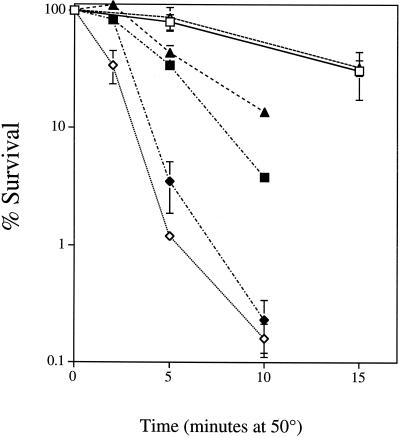
We determined the heat sensitivity of isogenic petite strains and of strains deficient in ubiquinone biosynthesis and the mitochondrial NADH dehydrogenases NDE1 and NDE2. Cultures were inoculated at 104 cells/ml and grown for 48 h in liquid YPAD medium for all experiments. Cells deficient in ubiquinone (coq7Δ) exhibit an even more severe thermosensitivity phenotype than petite cells (Fig. 1). Cells with deletions in the NDE1 and NDE2 external NADH dehydrogenases have wild-type [rho+] viability profiles. The thermosensitivity phenotype for both petite and ubiquinone-deficient cells is partially alleviated by the deletion of NDE1 and NDE2 in these backgrounds.
FIG. 1.
Viability of respiratory mutants after heat stress. Viability was calculated as described in Materials and Methods. Cells were grown for 48 h in YPAD before heat stress. □, JM43 [rho+]; ■, JM43 [rho−]; ◊, JM43 coq7Δ; ▵, JM43 nde1 nde2; ⧫, JM43 nde1 nde2 coq7Δ; ▴, JM43 nde1 nde2 [rho−]. Each point represents the mean ± SD of the results from at least three experiments.
Petite and CoQ-deficient cells have elevated heat-induced nuclear mutations.
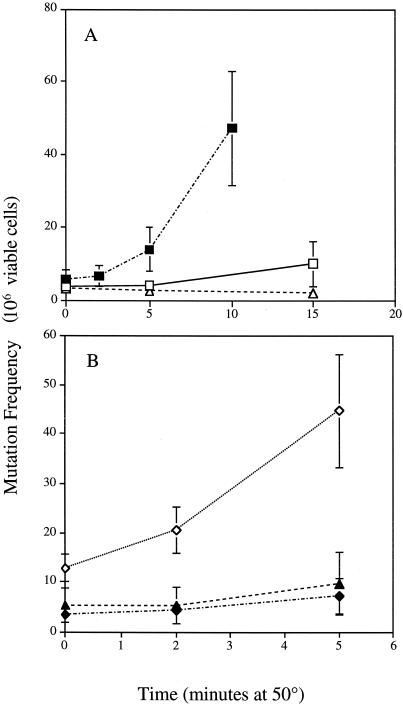
The generation of oxidants during heat stress may lead to oxidative DNA damage. Forward mutation frequencies were measured at the nuclear CAN1 locus in petite, [rho+], JM43 coq7Δ, and JDY35 (nde1 nde2) cells following a 50°C heat stress (Fig. 2). CoQ-deficient coq7Δ cells display a mild but significant mutator phenotype during normal growth conditions (Table 1), which was abolished in the triple mutant JM43 coq7Δ nde1 nde2. The heat-induced-mutation frequency was calculated by subtraction of the spontaneous unheated-control frequency from the treatment frequency and is an indicator of damage developed during heat exposure. When heated, the coq7Δ mutant showed dramatic increases in mutation frequencies at the nuclear CAN1 locus (Fig. 2B). Mutation frequencies at the nuclear CAN1 locus were likewise increased in [rho−] cells by heat stress exposure compared to those of wild-type [rho+] controls (Fig. 2A). After 5 min at 50°C, cells deficient in CoQ displayed approximately 32 induced mutation events at the CAN1 locus per 106 viable cells (P < 0.01). For [rho−] cells there were approximately 8 induced mutation events per viable cell (P < 0.05), while for wild-type [rho+] cells there was around 0.4 induced event per 106 viable cells. Mutation frequencies could not be measured at longer heat exposures due to the thermosensitivity of the coq7Δ and petite strains; however, even after 15 min at 50°C, the wild-type [rho+] cells displayed only approximately 6.4 induced mutation events per 106 viable cells (Fig. 2A).
FIG. 2.
Heat-induced mutation frequency of respiratory mutants. The mutation frequency at the CAN1 locus was measured as described in Materials and Methods. Cells were grown for 48 h in YPAD before heat stress and plating onto solid agar plates containing canavanine. (A) □, JM43 [rho+]; ■, JM43 [rho−]; ▵, JM43 nde1 nde2. (B) ◊, JM43 coq7Δ; ⧫, JM43 nde1 nde2 coq7Δ; ▴, JM43 nde1 nde2 [rho−]. Each point represents the mean ± SD of the results from at least three experiments.
TABLE 1.
Mutation rates of strains used in this study
| Strain | Mutation rate (10−7)a |
|---|---|
| JM43 [rho+] | 4.2 |
| JM43 [rho−] | 4.1 |
| JM43 coq7Δ | 7.7∗ |
| JM43 nde1 nde2 | 4.9 |
| JM43 coq7Δ nde1 nde2 | 3.7 |
Mutation rates were calculated from the results of three independent 96-well experiments. ∗, P < 0.05 by the two-tailed t test for comparison of means of mutants versus those of the wild type [rho+].
In both the petite and the coq7Δ strain, the increases in heat-induced mutations were completely dependent on the function of the NDE1 and NDE2 genes, since they were absent in both the coq7Δ nde1 nde2 and the petite nde1 nde2 mutant (Fig. 2B).
The amount of heat-induced oxidation of an intracellular-oxidant-sensitive probe is greater in petite and CoQ-deficient cells than in cells deficient in the external NADH dehydrogenases.
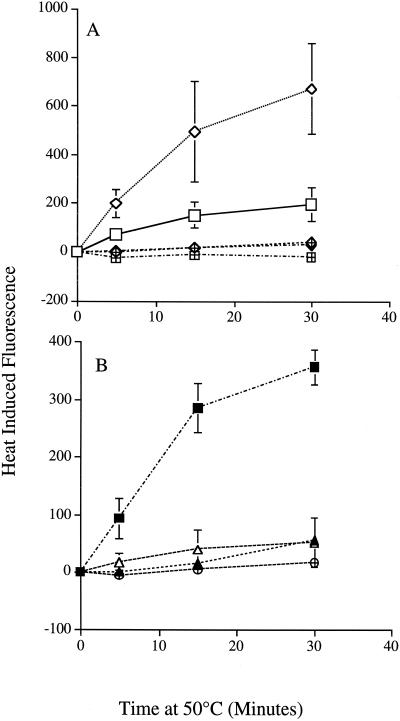
We measured the oxidation of DCFH-DA, an intracellular-oxidant-sensitive probe, following exposure of isogenic petite strains and strains deficient in ubiquinone biosynthesis and the mitochondrial NADH dehydrogenases NDE1 and NDE2 to 50°C heat stress. The ubiquinone-deficient strain showed the highest increase in oxidative fluorescence at all heat exposures, reaching 670 relative fluorescent units (RFU) after 30 min at 50°C (Fig. 3A). Cells deficient in the external dehydrogenase genes NDE1 and NDE2 displayed less intracellular oxidation than wild-type [rho+] cells, having approximately 50 RFU after 30 min at 50°C compared to wild-type levels of approximately 200 RFU after 30 min (Fig. 3B). A triple-deletion mutant lacking NDE1, NDE2, and COQ7Δ reduced the intracellular oxidation of 2′,7′-dichlorofluoroscein to the levels observed for anaerobic cells (Fig. 3A). Likewise for the petite nde1 nde2 strain, the heat-induced oxidative stress was reduced to [rho+] nde1 nde2 strain levels, indicating that the heat-induced increase in oxidative stress in the petite strain was dependent on the function of the external NADH dehydrogenases (Fig. 3B).
FIG. 3.
Heat-induced intracellular oxidation. Intracellular oxidation of DCFH fluorescent dye was measured in cells that were grown for 48 h in YPAD and subjected to heat stress for various times as described in Materials and Methods. (A) □, JM43 [rho+]; ◊, JM43 coq7Δ;  , JM43 coq7Δ −O2; ⧫, JM43 nde1 nde2 coq7Δ; ⊞, JM43 [rho+] −O2. (B) ■, JM43 [rho−]; ⊕, JM43 [rho−] −O2; ▵, JM43 nde1 nde2; ▴, JM43 nde1 nde2 [rho−]. Each point represents the mean ± SD of the results from at least three experiments.
, JM43 coq7Δ −O2; ⧫, JM43 nde1 nde2 coq7Δ; ⊞, JM43 [rho+] −O2. (B) ■, JM43 [rho−]; ⊕, JM43 [rho−] −O2; ▵, JM43 nde1 nde2; ▴, JM43 nde1 nde2 [rho−]. Each point represents the mean ± SD of the results from at least three experiments.
Intracellular oxidation during heat stress is oxygen dependent.
Intracellular oxidation of 2′,7′-dichlorofluoroscein was completely abolished in wild-type cells and was reduced from 670 to approximately 41 RFU in coq7Δ cells shifted to anaerobic conditions (<1 ppm O2) 1 h prior to heat stress. Oxygenated respiratory-competent wild-type cells showed a dose-dependent increase in fluorescence from 0 to approximately 200 RFU after 30 min at 50°C. Respiratory-incompetent [rho−] cells likewise showed increased intracellular oxidation of the fluorescent probe compared to that of wild-type cells to approximately 350 RFU. Intracellular oxidation of the molecular probe in [rho−] cells was reduced to 17 RFU in the absence of oxygen (Fig. 3B).
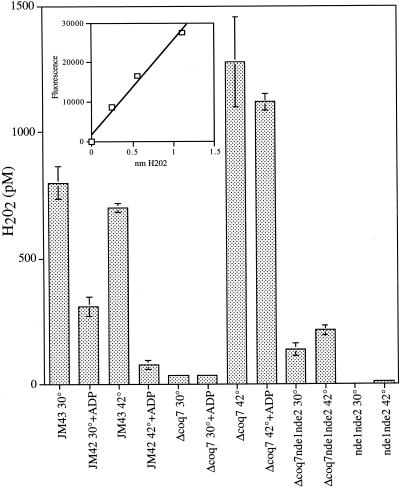
Release of H2O2 from mitochondria is increased in coq7 cells during heating.
Undamaged mitochondria incubated with NADH in the absence of ADP will enter state IV respiration, in which the electron transport carriers become fully reduced. These conditions are optimal for univalent reduction of oxygen by mitochondrial electron carriers and for production of superoxide anions. Superoxide rapidly dismutates to H2O2, the substrate for HRP-catalyzed A6550 fluorescence. In every case, the addition of catalase either abolished development of fluorescence or reduced it to <10% (data not shown), indicating that the reaction was specific for H2O2. In the presence of ADP, CoQ-deficient mitochondria evolved approximately 6.5 times less H2O2 than wild-type mitochondria at 30°C and over 13 times more H2O2 at 42°C. Under ADP-limiting type IV conditions, we observed that the mitochondria isolated from CoQ-deficient cells evolved approximately 20 times less H2O2 at 30°C than wild-type mitochondria and over 1.5 times more H2O2 at 42°C. This represents a >30-fold increase in H2O2 production in mitochondria derived from coq7-deficient cells at the elevated temperature. Mitochondria derived from cells deficient in the NADH dehydrogenases NDE1 and NDE2 showed no measurable release of H2O2, and the mitochondria derived from cells with the triple deletion coq7 nde1 nde2 showed only a 1.5-fold increase of H2O2 production at 42°C over the amounts produced at 30°C.
The majority of H2O2 produced from the respiration of NADH during heat stress is, therefore, derived from the electrons that flow through the external NADH dehydrogenases to CoQ.
DISCUSSION
Heat stress is associated with the development of an oxidative stress, the source of which was previously unknown. Here we report not only that the production of reactive oxygen species following heat stress is modulated by the deletion of components of the mitochondrial electron transport chain but also that mitochondrion-derived oxidative stress results in an increased mutation frequency in nuclear DNA.
During normal aerobic respiration, tetravalent reduction of oxygen is the terminal reducing step of mitochondrial electron transport, with minimal univalent reduction of oxygen occurring (11). A greater flux of superoxide anions is produced from mitochondria under conditions where electron flow is disrupted by specific inhibitors or ADP becomes limiting (state IV respiration). In these situations, electron carriers upstream of the blockage remain reduced, increasing the rate of single electron transfers to oxygen (28).
The deletion of COQ7 in yeast results in the production of cells with no measurable CoQ (19). These cells were greatly thermosensitized compared to wild-type cells, displayed elevated levels of intracellular oxidation, and had higher mutation frequencies at the nuclear CAN1 locus following heat stress exposure. The lack of ubiquinone in these cells rules out the ubisemiquinone radical as the single electron donor to oxygen during heat stress. CoQ resides within the hydrophobic interior of the inner mitochondrion membrane, where it associates with and receives electrons from several CoQ oxidoreductases that are imbedded in the inner membrane, including the internal and external NADH dehydrogenases, succinate dehydrogenase, and glycerol-3-phosphate dehydrogenase (reviewed in reference 11). CoQ receives electrons stepwise from these sources and funnels them through complex III, delivering them to cytochrome c. By removing CoQ from the electron transport chain, all upstream carriers, including the CoQ oxidoreductases, would be unable to deliver the electrons they receive and would eventually become fully reduced.
Each CoQ oxidoreductase (internal NADH dehydrogenase, succinate dehydrogenase, and glycerol-3-phosphate dehydrogenase) contains flavin adenine dinucleotide as a cofactor, and succinate dehydrogenase has an additional iron sulfur protein as a redox factor. Flavosemiquinones can form and react with oxygen to produce superoxide anions (3), and flavin adenine dinucleotide autoxidation from NADH dehydrogenase in Escherichia coli has been reported to be a primary source of superoxide anion formation, which also increases in ubiquinone mutants in a temperature-dependent manner (20). Perhaps thermal denaturation of flavin-containing redox proteins might enhance the rate of reaction of reduced flavoproteins with oxygen to form superoxide anions. In this model, more superoxide anions would be produced during heat stress conditions when a higher ratio of reduced redox centers exists. Wild-type cells with intact electron transport would be expected to have much lower ratios of reduced electron carriers than cells with blocked electron transport.
During the post-diauxic phase of growth, cells utilize the ethanol produced during the fermentation of glucose (31). Ethanol is preferentially oxidized to acetaldehyde by cytosolic alcohol dehydrogenase, forming NADH in the process. As the mitochondrial NADH and cytosolic pools are separate, ethanol oxidation results in a rise in the cytosolic NADH/NAD+ ratio (31). The excess NADH is oxidized by the mitochondrial respiratory chain via NADH dehydrogenases located on the inner mitochondrial membrane. Any disruption of electron flow, such as would occur in a petite or coq7 cell, would be expected to result in a backlog of electrons along the NADH-utilizing electron transport pathway.
Petite cells generated from ethidium bromide treatment contain large deletion mutations in the mitochondrial DNA, resulting in the loss of components of the mitochondrial protein synthesis machinery or in the direct loss of cytochrome b and cytochrome c oxidase subunits (7). The bulk of mitochondrion genes are nuclearly encoded and regulated, becoming derepressed following the exhaustion of glucose by fermentation. As such, the petite phenotype can be viewed as a dysfunctional state in which the cell induces genes for nonfermentable carbon source metabolism but in which electron transport and respiration are unable to occur due to the absence of key mitochondrion-encoded elements, specifically cytochrome b and cytochrome c oxidase. NADH generated by the oxidation of ethanol to acetaldehyde will still feed into the defunct electron transport chain through the internal and external mitochondrial NADH dehydrogenases, but a logjam of electrons will ensue, as the electrons are not transferred through the missing links. The downstream components, including the CoQ oxidoreductases, will become fully reduced and capable of leaking electrons to oxygen. One expects that removing the source of electrons would alleviate the oxidative stress. Deletion of the external dehydrogenases did indeed reduce the heat-induced mutation frequencies in [rho−] and coq7Δ cells, the heat-induced oxidation of the intracellular fluorescent probe, and the production of H2O2 from mitochondria. These findings suggest that the oxidative stress generated in these cells during heat stress derived from electrons donated by the external NADH dehydrogenases during growth in the post-diauxic phase.
The deletion of both external mitochondrial NADH dehydrogenase genes (NDE1 and NDE2) reduced the intracellular oxidation of the 2′,7′-dichlorofluoroscein fluorescent probe compared to that in the [rho+] wild type. Double nde1 nde2 deletion mutations had no effect on viability after a heat stress exposure in the wild type [rho+] but did ameliorate slightly the thermosensitivity phenotype of the coq7Δ and petite strains. In addition, double nde1 nde2 mutations in the [rho−] and coq7Δ backgrounds completely abolished the heat-induced mutations at the nuclear CAN1 gene and the intracellular oxidation of the fluorescent probe (Fig. 2 and 3). These results suggest a role for the external NADH dehydrogenases as a conduit for the electrons that eventually lead to oxidative stress. Direct measurement of the H2O2 released from mitochondria isolated from coq7 showed that elevated temperatures result in a large increase in H2O2 release from ubiquinone-deficient mitochondria (Fig. 4). The effect of nde1 and nde2 deletions both alone and in conjunction with a coq7 deletion was to greatly reduce the production of H2O2.
FIG. 4.
Heat-induced hydrogen peroxide leakage from mitochondria. Hydrogen peroxide was measured in mitochondria isolated from JM43, JM43 coq7Δ, JM43 nde1 nde2, and the triple mutant JM43 nde1 nde2 coq7 at 30 and 42°C in the presence and absence of ADP as described in Materials and Methods. Each bar represents the mean ± SD of the results of four experiments.
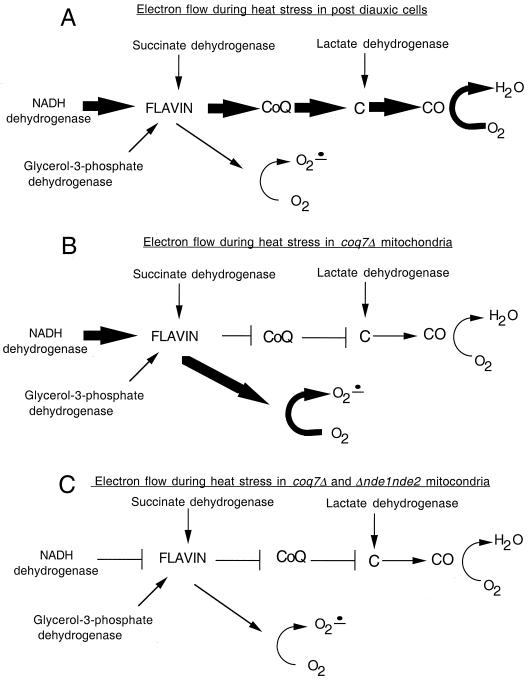
Interestingly, the levels of H2O2 released from ubiquinone-deficient mitochondria at 30°C were substantially reduced compared to those of wild-type mitochondria, suggesting that, at nonstressful temperatures, CoQ itself appears to generate the majority of free radicals, possibly through the radical intermediate ubisemiquinone (23). When heat is applied, however, the situation reverses dramatically and CoQ-deficient mitochondria generate significant amounts of H2O2. As CoQ is absent, one can infer that the molecule responsible for reducing the supply of oxygen to the superoxide anion during heat stress is not the ubisemiquinone radical but some other reductant. The flavin cofactor from the NADH dehydrogenase enzymes themselves or another unidentified electron carrier located in the transport chain between those enzymes and CoQ may be the culprit responsible for the heat-induced production of H2O2. We propose a model describing electron flow during heat stress in Fig. 5. In this model, electrons become diverted to molecular oxygen during heat stress as flavoproteins denature, exposing or releasing the reduced flavin cofactor. Conditions that favor an increased ratio of reduced flavin electron carriers, such as mitochondria deficient in CoQ and [rho−] mitochondria, result in a greater superoxide anion flux.
FIG. 5.
Model for electron pathways during heat exposure in post-diauxic-phase respiration. The sizes of the arrows relate to the relative electron flow. CoQ, coenzyme Q; C, cytochrome c; CO, cytochrome c oxidase; O2 , superoxide anion. (A) Electrons flow to the flavin cofactors of the dehydrogenases which are coupled to CoQ. During normal post-diauxic-phase growth, abundant ethanol is oxidized and contributes to the majority of the electron flow to CoQ. (B) If CoQ is absent from the chain, electrons become diverted to oxygen to form superoxide anion. (C) Removal of the external dehydrogenases alleviates the electron flow to oxygen.
, superoxide anion. (A) Electrons flow to the flavin cofactors of the dehydrogenases which are coupled to CoQ. During normal post-diauxic-phase growth, abundant ethanol is oxidized and contributes to the majority of the electron flow to CoQ. (B) If CoQ is absent from the chain, electrons become diverted to oxygen to form superoxide anion. (C) Removal of the external dehydrogenases alleviates the electron flow to oxygen.
We have demonstrated that mitochondrion-derived oxidants were able to cause mutations in the nuclear DNA in yeast, an observation that has important implications for carcinogenesis, should a similar process occur in mammalian cells. Many degenerative diseases associated with aging are proposed to be caused by the development of a greater mitochondrial oxidative burden over time, which may be exacerbated during conditions of heat stress. Periods of elevated temperature exposure for humans are common, with chronic exposures occurring during fever and heat stroke and acute and localized exposures occurring following burns and ingestion of hot beverages and during hyperthermic cancer therapy. Understanding the molecular details of heat-induced oxidative stress will extend our knowledge of molecular events following these common heat stress exposures.
ACKNOWLEDGMENTS
This work was supported by KO2 award ES00299 from the National Institute of Environmental Health Sciences, NIH, to R.H.S. J.F.D. was supported by NIH training grants 5-T32CA09078 and 2-T32ES07155.
REFERENCES
- 1.Alani E, Cao L, Kleckner N. A method for gene disruption that allows repeated use of URA3 selection in the construction of multiply disrupted yeast strains. Genetics. 1987;116:541–545. doi: 10.1534/genetics.112.541.test. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1a.Ames B N, Shigenaga M K, Hagen T M. Mitochondrial decay in aging. Biochim Biophys Acta. 1995;1271:165–170. doi: 10.1016/0925-4439(95)00024-x. [DOI] [PubMed] [Google Scholar]
- 2.Ames B N, Shigenaga M K, Hagen T M. Oxidants, antioxidants, and the degenerative diseases of aging. Proc Natl Acad Sci USA. 1993;90:7915–7922. doi: 10.1073/pnas.90.17.7915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Anderson R F. Energetics of the one-electron reduction steps of riboflavin, FMN and FAD to their fully reduced forms. Biochim Biophys Acta. 1983;722:158–162. doi: 10.1016/0005-2728(83)90169-x. [DOI] [PubMed] [Google Scholar]
- 4.Bakker B M, Overkamp K M, van Maris A J, Kotter P, Luttik M A, van Dijken J P, Pronk J T. Stoichiometry and compartmentation of NADH metabolism in Saccharomyces cerevisiae. FEMS Microbiol Rev. 2001;25:15–37. doi: 10.1111/j.1574-6976.2001.tb00570.x. [DOI] [PubMed] [Google Scholar]
- 5.Benov L, Fridovich I. Superoxide dismutase protects against aerobic heat shock in Escherichia coli. J Bacteriol. 1995;177:3344–3346. doi: 10.1128/jb.177.11.3344-3346.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boumans H, Grivell L A, Berden J A. The respiratory chain in yeast behaves as a single functional unit. J Biol Chem. 1998;273:4872–4877. doi: 10.1074/jbc.273.9.4872. [DOI] [PubMed] [Google Scholar]
- 7.Broach J R, Pringle J R, Jones E W. The molecular and cellular biology of the yeast Saccharomyces. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1991. [Google Scholar]
- 8.Collinson L P, Dawes I W. Inducibility of the response of yeast cells to peroxide stress. J Gen Microbiol. 1992;138:329–335. doi: 10.1099/00221287-138-2-329. [DOI] [PubMed] [Google Scholar]
- 9.Davidson J F, Whyte B, Bissinger P H, Schiestl R H. Oxidative stress is involved in heat-induced cell death in Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1996;93:5116–5121. doi: 10.1073/pnas.93.10.5116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.DeRisi J L, Iyer V R, Brown P O. Exploring the metabolic and genetic control of gene expression on a genomic scale. Science. 1997;278:680–686. doi: 10.1126/science.278.5338.680. [DOI] [PubMed] [Google Scholar]
- 11.de Vries S, Marres C A. The mitochondrial respiratory chain of yeast. Structure and biosynthesis and the role in cellular metabolism. Biochim Biophys Acta. 1987;895:205–239. doi: 10.1016/s0304-4173(87)80003-4. [DOI] [PubMed] [Google Scholar]
- 12.Glick B S, Pon L A. Isolation of highly purified mitochondria from Saccharomyces cerevisiae. Methods Enzymol. 1995;260:213–223. doi: 10.1016/0076-6879(95)60139-2. [DOI] [PubMed] [Google Scholar]
- 13.Godon C, Lagniel G, Lee J, Buhler J M, Kieffer S, Perrot M, Boucherie H, Toledano M B, Labarre J. The H2O2 stimulon in Saccharomyces cerevisiae. J Biol Chem. 1998;273:22480–22489. doi: 10.1074/jbc.273.35.22480. [DOI] [PubMed] [Google Scholar]
- 14.Halliwell B, Gutteridge J M. Oxygen toxicity, oxygen radicals, transition metals and disease. Biochem J. 1984;219:1–14. doi: 10.1042/bj2190001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kaput J, Goltz S, Blobel G. Nucleotide sequence of the yeast nuclear gene for cytochrome c peroxidase precursor. Functional implications of the pre sequence for protein transport into mitochondria. J Biol Chem. 1982;257:15054–15058. [PubMed] [Google Scholar]
- 16.Lee S M, Park J W. Thermosensitive phenotype of yeast mutant lacking thioredoxin peroxidase. Arch Biochem Biophys. 1998;359:99–106. doi: 10.1006/abbi.1998.0896. [DOI] [PubMed] [Google Scholar]
- 17.Longo V D, Liou L L, Valentine J S, Gralla E B. Mitochondrial superoxide decreases yeast survival in stationary phase. Arch Biochem Biophys. 1999;365:131–142. doi: 10.1006/abbi.1999.1158. [DOI] [PubMed] [Google Scholar]
- 18.Luttik M A, Overkamp K M, Kotter P, de Vries S, van Dijken J P, Pronk J T. The Saccharomyces cerevisiae NDE1 and NDE2 genes encode separate mitochondrial NADH dehydrogenases catalyzing the oxidation of cytosolic NADH. J Biol Chem. 1998;273:24529–24534. doi: 10.1074/jbc.273.38.24529. [DOI] [PubMed] [Google Scholar]
- 19.Marbois B N, Clarke C F. The COQ7 gene encodes a protein in Saccharomyces cerevisiae necessary for ubiquinone biosynthesis. J Biol Chem. 1996;271:2995–3004. doi: 10.1074/jbc.271.6.2995. [DOI] [PubMed] [Google Scholar]
- 20.Messner K R, Imlay J A. The identification of primary sites of superoxide and hydrogen peroxide formation in the aerobic respiratory chain and sulfite reductase complex of Escherichia coli. J Biol Chem. 1999;274:10119–10128. doi: 10.1074/jbc.274.15.10119. [DOI] [PubMed] [Google Scholar]
- 21.Mohanty J G, Jaffe J S, Schulman E S, Raible D G. A highly sensitive fluorescent micro-assay of H2O2 release from activated human leukocytes using a dihydroxyphenoxazine derivative. J Immunol Methods. 1997;202:133–141. doi: 10.1016/s0022-1759(96)00244-x. [DOI] [PubMed] [Google Scholar]
- 22.Moradas-Ferreira P, Costa V, Piper P, Mager W. The molecular defences against reactive oxygen species in yeast. Mol Microbiol. 1996;19:651–658. doi: 10.1046/j.1365-2958.1996.403940.x. [DOI] [PubMed] [Google Scholar]
- 23.Nohl H. Generation of superoxide radicals as byproduct of cellular respiration. Ann Biol Clin. 1994;52:199–204. [PubMed] [Google Scholar]
- 24.Shigenaga M K, Hagen T M, Ames B N. Oxidative damage and mitochondrial decay in aging. Proc Natl Acad Sci USA. 1994;91:10771–10778. doi: 10.1073/pnas.91.23.10771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Skulachev V P. Membrane-linked systems preventing superoxide formation. Biosci Rep. 1997;17:347–366. doi: 10.1023/a:1027344914565. [DOI] [PubMed] [Google Scholar]
- 26.Small W C, McAlister-Henn L. Identification of a cytosolically directed NADH dehydrogenase in mitochondria of Saccharomyces cerevisiae. J Bacteriol. 1998;180:4051–4055. doi: 10.1128/jb.180.16.4051-4055.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stephen D W, Rivers S L, Jamieson D J. The role of the YAP1 and YAP2 genes in the regulation of the adaptive oxidative stress responses of Saccharomyces cerevisiae. Mol Microbiol. 1995;16:415–423. doi: 10.1111/j.1365-2958.1995.tb02407.x. [DOI] [PubMed] [Google Scholar]
- 28.Turrens J F. Superoxide production by the mitochondrial respiratory chain. Biosci Rep. 1997;17:3–8. doi: 10.1023/a:1027374931887. [DOI] [PubMed] [Google Scholar]
- 29.Von Borstel R C. Measuring spontaneous mutation rates in yeast. Methods Cell Biol. 1978;20:1–24. doi: 10.1016/s0091-679x(08)62005-1. [DOI] [PubMed] [Google Scholar]
- 30.Wieser R, Adam G, Wagner A, Schuller C, Marchler G, Ruis H, Krawiec Z, Bilinski T. Heat shock factor-independent heat control of transcription of the CTT1 gene encoding the cytosolic catalase T of Saccharomyces cerevisiae. J Biol Chem. 1991;266:12406–12411. [PubMed] [Google Scholar]
- 31.Wills C. Regulation of sugar and ethanol metabolism in Saccharomyces cerevisiae. Crit Rev Biochem Mol Biol. 1990;25:245–80. doi: 10.3109/10409239009090611. [DOI] [PubMed] [Google Scholar]