Abstract
Transcription is a major regulatory mechanism for the generation of slow- and fast-twitch myofibers. We previously identified an upstream region of the slow TnI gene (slow upstream regulatory element [SURE]) and an intronic region of the fast TnI gene (fast intronic regulatory element [FIRE]) that are sufficient to direct fiber type-specific transcription in transgenic mice. Here we demonstrate that the downstream half of TnI SURE, containing E box, NFAT, MEF-2, and CACC motifs, is sufficient to confer pan-skeletal muscle-specific expression in transgenic mice. However, upstream regions of SURE and FIRE are required for slow and fast fiber type specificity, respectively. By adding back upstream SURE sequences to the pan-muscle-specific enhancer, we delineated a 15-bp region necessary for slow muscle specificity. Using this sequence in a yeast one-hybrid screen, we isolated cDNAs for general transcription factor 3 (GTF3)/muscle TFII-I repeat domain-containing protein 1 (MusTRD1). GTF3 is a multidomain nuclear protein related to initiator element-binding transcription factor TF II-I; the genes for both proteins are deleted in persons with Williams-Beuren syndrome, who often manifest muscle weakness. Gel retardation assays revealed that full-length GTF3, as well as its carboxy-terminal half, specifically bind the bicoid-like motif of SURE (GTTAATCCG). GTF3 expression is neither muscle nor fiber type specific. Its levels are highest during a period of fetal development that coincides with the emergence of specific fiber types and transiently increases in regenerating muscles damaged by bupivacaine. We further show that transcription from TnI SURE is repressed by GTF3 when overexpressed in electroporated adult soleus muscles. These results suggest a role for GTF3 as a regulator of slow TnI expression during early stages of muscle development and suggest how it could contribute to Williams-Beuren syndrome.
Skeletal muscles are composed of fast-twitch myofibers, responsible for movement and fast power generation, and slow-twitch fibers, which are important for endurance and for maintaining posture. Distinct fiber types emerge during development (7, 31, 58) and can be modified in the adult by complex interactions of intrinsic and extrinsic signals (47).
While there is a general consensus that in adult vertebrates motoneuron activity is an important stimulus regulating the maintenance and transition of fiber types, the relative contributions of intrinsic and extrinsic factors in establishing fiber type diversity during fetal development are less well understood. In vivo and in vitro studies on the emergence of fiber types with chicks support the importance of both the innervating motoneuron and myoblast cell lineage (16, 17, 48, 59). In rats, formation of primary myofibers, a major source of slow fibers, and secondary myotubes, which give rise to most of the adult fast fibers, can occur in the absence of functional innervation (15, 29, 30). Moreover, most primary fibers express slow myosin heavy chain even when innervation is prevented by injection of bungarotoxin (15). However, analysis of the expression of genes encoding fiber type-specific isoforms of contractile proteins in aneural fetal and regenerating adult muscles suggests that the manifestation of a “slow” gene expression program is more dependent on the presence of the nerve than the “fast” program (19, 63).
Transcription is a major regulatory mechanism restricting gene expression to specific fiber types (10). To identify the molecular basis of fiber type-specific gene expression during development and in response to motoneuron activity in the mature muscle (11), we chose to study the transcriptional regulation of genes encoding the troponin slow (TnIs) and fast (TnIf) isoforms. In rodents, TnI expression proceeds in two distinct stages. During fetal development, expression of TnI isoforms is initially broad and later segregates according to prospective slow and fast fiber types (72; this paper). After birth, TnI expression is upregulated in a motoneuron-dependent fashion and confined to either slow- or fast-twitch fibers (11; D. Vullhorst and A. Buonanno, unpublished observations). The developmental program is partially recapitulated in the adult when muscles are treated with myotoxins to induce degeneration, followed by satellite cell proliferation and subsequent regeneration of myofibers. Regenerating muscles initially coexpress both TnI isoforms, and as muscles are reinnervated TnI expression is again restricted to either slow or fast muscles (19). We previously isolated and characterized enhancers for both TnI genes that confer fiber type-specific transcription in transgenic mice (3, 43); a 128-bp rat slow upstream regulatory element (SURE) and a 144-bp quail fast intronic regulatory element (FIRE) (71). The downstream halves of SURE and FIRE share three conserved cis elements (Fig. 1), namely, E box, MEF-2, and CACC motifs, which bind MyoD/myogenin, MEF-2, and SP-1 transcription factors, respectively (11, 43). A fourth conserved site, the CAGG motif, is located in their 5′ halves; a factor binding this site has not been identified. We have shown in transfected myocytes and transgenic mice the functional importance of interactions between these four motifs for full enhancer activity (12, 43). In addition, we recently identified a sequence immediately upstream of the CAGG motif unique to TnI SURE that was recognized by a computer search as a bicoid-like motif (BLM) and that is necessary for enhancer function in cultured muscle cells (12). In electrophoretic mobility shift assays (EMSAs) probes harboring the BLM form a low-mobility complex with nuclear factors that are abundantly expressed in immature muscles and to a lesser extent in adult muscles and in a variety of other cell types (12). However, these studies did not directly address the role of those DNA elements for fiber type specificity of the TnI SURE.
FIG. 1.
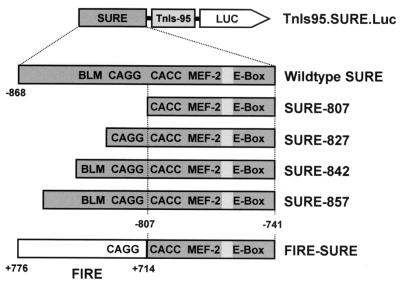
TnI SURE reporter constructs used in this study. Wild-type SURE (−868 to −741) and all its derivatives were placed upstream of the −95 TnIs basal promoter driving firefly luciferase in pGL3Basic. The shortest SURE deletion (SURE-807) terminates upstream of the CACC box and contains three of the four motifs conserved between SURE and FIRE (CACC, MEF-2, and E box). The boxed area between the MEF-2 site and E box harbors a sequence that conforms to an NFAT element. In SURE-827, 20 bp of the upstream sequence was added back to SURE-807 to obtain a truncated enhancer that includes the CAGG motif in addition to the aforementioned sites. SURE-842 and SURE-857 also contain the BLM (GTTAATCCG), which is not found in FIRE. On the opposite strand, this sequence resembles an initiator element (32, 55). The FIRE-SURE enhancer was generated by fusing the 5′ half of FIRE (from +776 to +714) to the 3′ half of SURE (from −807 to −741), resulting in a chimeric enhancer with a preserved spatial organization of all four conserved motifs.
Here we report that the DNA elements necessary for fiber type specificity of the TnI SURE reside in regions different from those that confer general skeletal muscle specificity. We analyzed transgenic mice harboring a chimeric enhancer composed of sequences from SURE and FIRE, as well as numerous deletions of sequences from TnI SURE. This analysis resulted in the identification of a 15-bp sequence in the upstream region of SURE that restricts the activity of the nonspecific downstream half to slow fibers. A sequence including the BLM and CAGG motif was used in a yeast one-hybrid screen to isolate cDNAs encoding a multidomain nuclear protein known as general transcription factor 3 (GTF3) or muscle TFII-I repeat domain-containing protein 1 (MusTRD1) (44, 61), a factor structurally homologous to transcription factor TFII-I (49, 50). Other groups have independently cloned GTF3 and named it WBSCR11 (45), GTF2IRD1 (22), and CREAM (69) in humans and BEN in mice (4). We use the designation GTF3 to emphasize its relation with TFII-I (gene designation: GTF2-I) and because, like that of TFII-I, its expression pattern in mice (4; this paper) does not support the notion that it is specific for skeletal muscle tissue, as suggested by the name MusTRD1 (44). We also report for the first time that GTF3 is highly expressed in developing and regenerating muscles, its expression thus coinciding with myofiber diversification, whereas expression in mature fibers is below the sensitivity of in situ hybridization. Furthermore, we demonstrate that forced overexpression of the GTF3 protein in vivo reduces the transcriptional activity of the TnI SURE. These findings emphasize for the first time an important function of TnI SURE in repressing expression of TnIs in prospective fast fibers during development and indicate the contribution of GTF3, acting alone or in concert with other factors, in this regulatory pathway. The possible contribution of GTF3 to the regulation of fiber type-specific transcription, which differs from that initially proposed for MusTRD1 (44), and its relevance to Williams-Beuren syndrome (WBS) are discussed.
MATERIALS AND METHODS
Generation of luciferase reporter constructs.
All constructs are based on TnIs95.Luc, which contains the −95 basal promoter of the rat TnIs gene upstream of the luciferase gene in pGL3Basic (Promega). The generation of this construct is described elsewhere (12). We have previously shown that the proximal TnIs promoter is inactive in stably transfected muscle cells and in transgenic mice (12, 43). To map regulatory sequences in TnI SURE and FIRE, different regions of these enhancers were subcloned into the TnIs95.Luc basal promoter construct (Fig. 1).
The chimeric FIRE-SURE enhancer was generated by the recombinant PCR method (27). The sequence from +776 to +714 of FIRE was inserted upstream of the CACC motif of SURE (nucleotide −807). This chimeric enhancer fragment was then inserted between the SstI and NheI sites of TnIs95.Luc; this construct is designated TnIs95.FIRE-SURE. A SURE deletion mutant lacking the region upstream of the CACC motif was generated by PCR amplification of the sequence between −807 and −741 and insertion of this fragment between the SstI and NheI sites of TnIs95.Luc; this construct was denoted TnIs95.SURE-807. A series of nested deletions in the 5′ half of SURE was generated by progressively adding more upstream sequences to TnIs95.SURE-807. The corresponding fragments were amplified using a common 3′ noncoding primer (5′-gat cgc tag cAG GGC CAC ACC TGT TTC CTG-3′) and coding primers beginning at positions −827 (SURE −827: 5′-gat cga gct cGC AGG CAT TGT CTT TCT CTG-3′), −842 (SURE −842: 5′-gat cga gct cTA CCG GAT TAA CAT AGC AGG-3′), and −857 (SURE −857: 5′-gat cga gct cAC CGA CTA TAA TAG CTA CCG-3′); SstI and NheI sites used for ligation are in lowercase. PCR fragments were purified on agarose gels and subcloned into TnIs95.Luc to obtain constructs TnIs95.SURE-827, TnIs95.SURE-842, and TnIs95.SURE-857, respectively. All constructs generated by PCR were verified by sequencing.
Generation of transgenic mouse lines.
Transgenic mice were made essentially as described previously (3, 43). All reporter constructs were digested with SstI and BamHI to separate the reporter gene transcription unit from vector sequences. The fragments were isolated on agarose gels, electroeluted, and purified on ELUTIP-D columns (Schleicher & Schuell). Transgenic mice were generated and propagated in an FVB/N background using the methods previously described (28). Putative founders and their offspring were screened by Southern blot analysis of tail DNA using a luciferase probe. All transgenic mice used to analyze tissue- and muscle-type-specific expression of luciferase activity were 6 to 8 weeks old. A variety of tissues including brain, liver, kidney, and heart as well as skeletal muscles from the body wall, intercosta, diaphragm, tongue, and hind limbs were collected and snap-frozen in liquid nitrogen for the preparation of cell extracts.
Yeast one-hybrid screen.
Three tandem copies of a SURE double-stranded oligonucleotide extending from −844 to −808 were inserted in front of HIS3 and lacZ reporter genes in pHISi-1 and placZi vectors (Clontech), respectively. These 3×SURE (−844/−808)-HISi-1 and 3×SURE (−844/−808)-lacZ constructs were sequentially integrated into chromosomal DNA of yeast strain YM4271, in accordance with the procedure outlined by the manufacturer. A control screen with sequences lacking the BLM was performed in parallel with a 3×SURE (−832/−808)-HISi-1 construct. The dual-reporter-containing YM4271 was mated with pretransformed yeast strain Y187, which carried a GAL4-AD human adult skeletal muscle fusion library in yeast expression vector pACT2 (Clontech). Diploids were selected on histidine- and uracil-deficient minimal (SD) medium that contained 20 mM 3-amino-1,2,4-triazole. The his- and ura-positive clones were tested for lacZ expression using colony lifts in a membrane-based β-galactosidase assay. Library plasmid DNA from β-galactosidase-positive clones was isolated and used to transform KC8 bacterial cells. Sequences of library inserts were obtained and run against a nonredundant nucleotide database using BLAST to identify the cDNAs (1).
Generation of GTF3 expression plasmids.
All GTF3 expression constructs were subcloned into pCMV-Sport2 (Life Technologies). To generate a human GTF3 cDNA encompassing the entire open reading frame, a fragment containing 302 bp of upstream coding sequence lacking from yeast one-hybrid clone 81 (see Table 1) was generated by reverse transcription-PCR (RT-PCR) from total RNA of cultured HEK293 cells. The sequences of oligonucleotides were as follows: hGTF3(5′-F), 5′-gtc gac gcc acc ATG GCC TTG CTG GGT AAG CGC TGT-3′; hGTF3(5′-R), 5′-CCT GCT TGA GCT CTC GGA TGG CGT GGC-3′; lowercase letters represent a nonhomologous sequence, including a SalI site used for subcloning and a Kozak consensus motif for efficient translation initiation (35). The PCR product was cloned into pGemT (Promega) and verified by sequencing. The 5′ region of GTF3 was released as an 821-bp SalI-SacI fragment and inserted between the corresponding sites of clone 81. The entire GTF3 open reading frame was subsequently released from the parental plasmid, pACT2, and inserted between the SalI and XhoI sites of pCMV-Sport2. A truncated human GTF3 cDNA lacking the sequence encoding the amino terminus as well as the first and second TFII-I like repeats (amino acids [aa] 529 to 944) was generated by PCR using yeast one-hybrid clone 81 as the template. The upper-strand oligonucleotide was hGTF3Δ1 + 2, 5′-gtc gac gcc acc ATG GAT TCT GGT TAT GGG ATG GAG AGA TG-3′. The sequence for the downstream oligonucleotide (pACT2-R1, 5′-GTG AAC TTG CGG GGT TTT TCA GTA TCT ACG-3′) was located 3′ of the cDNA insertion site in library plasmid pACT2, such that its XhoI site was included in the PCR fragment. The partial GTF3 cDNA fragment was inserted between the SalI and XhoI sites of pCMVSport2. This construct was denoted hGTF3Δ1 + 2. The mouse full-length ortholog of human GTF3 was obtained from the American Type Culture Collection as an expressed sequence tag (EST) clone in pCMV-Sport2 (clone 555547; GenBank accession no. AA111609) (36). For transfection experiments, the 5′ untranslated region sequence was replaced by a Kozak consensus sequence. An N-terminal expression construct (aa 1 to 463), denoted mGTF3Δ3-6, was generated by SphI digestion and subsequent religation.
TABLE 1.
Overview of GTF3 clones obtained from the yeast one-hybrid screen
| 5′ boundarya | Length (bp) | TFII-I repeat domains | No. of clones | Clone designation(s) |
|---|---|---|---|---|
| +302 | 2,682 | 1–5 | 4 | 64, 66, 68, 81 |
| +972 | 2,012 | 2–5 | 1 | 65 |
| +1731 | 1,253 | 3 (partial), 4, 5 | 1 | 21 |
| +1757 | 1,227 | 3 (partial), 4, 5 | 1 | 29 |
| +1802 | 1,182 | 3 (partial), 4, 5 | 2 | 18, 30 |
| +1824 | 1,160 | 3 (partial), 4, 5 | 1 | 74 |
Relative to translation initiation site in GTF3 (GenBank accession no. AF118270).
EMSAs.
Full-length and partial GTF3 proteins used for (EMSAs) were generated in vitro from cDNAs subcloned into CMVSport2 (see above). Proteins were synthesized from 1 μg of plasmid DNA using 50 μl of the TNT reticulocyte-coupled transcription-translation system (Promega). Relative efficiency of translation was monitored in parallel reactions by the addition of [35S]methionine and assessed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis.
Double-stranded complementary oligonucleotides with the following sequences were used in EMSAs (mutant nucleotides are in boldface): SURE −842/−815: 5′-TAC CGG ATT AAC ATA GCA GGC ATT GTC T-3′; SURE −844/−827: 5′-GCT ACC GGA TTA ACA TAG-3′; SURE −844/−827M: 5′-GCT AGA ATT CTA ACA TAG-3′; SURE −827/−808: 5′-GCA GGC ATT GTC TTT CTC TG-3′. Probes for EMSAs were generated from double-stranded oligonucleotides labeled with [γ-32P]ATP (6,000 Ci/mmol; Amersham) and polynucleotide kinase and purified on acrylamide gels. Two microliters of in vitro-translated proteins was mixed with binding buffer (20 mM HEPES [pH 7.9], 50 mM KCl, 4 mM MgCl2, 4% Ficoll, 5% glycerol, 0.2 mM EDTA, 0.5 mM dithiothreitol), 2 μg of poly(dI-dC), and 32P-labeled probe (20,000 cpm) and incubated at room temperature for 15 min. For competition assays, 10 pmol of unlabeled competitor oligonucleotides was used along with the labeled probe. The DNA-protein complexes were resolved by electrophoresis at 4°C on a 5% polyacrylamide–2.5% glycerol gel in 0.5× Tris-borate-EDTA buffer and visualized by autoradiography.
Northern blots.
Total RNA was extracted from cells and tissues using Trizol (Life Technologies). Ten micrograms of RNA was size fractionated by electrophoresis in 1.5% agarose–2.2 M formaldehyde gels and electroblotted onto a nylon membrane (Gene Screen; NEN). A mouse multitissue membrane containing 2 μg of poly(A+) RNA was also utilized (Clontech). Blots were hybridized with a mouse GTF3 32P-labeled full-length or 5′-specific 620-bp PstI fragment derived from EST clone 555547 (see above) and washed stringently (65°C, 0.1× SSC [1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate]). Signal intensities were directly quantified using a PhosphorImager (Molecular Dynamics). As previously reported (61), we observed no cross-hybridization of the probes with TFII-I transcripts; BLAST nucleic acid sequence alignments also failed to identify areas of significant homology between GTF3 and TFII-I.
In situ hybridization histochemistry.
Hind limbs of embryonic day 14 (E14) to E18 mouse embryos, postnatal day 0 (P0) through P9 neonates, and individual muscles from adults were fixed overnight in 4% paraformaldehyde, embedded in paraffin, and sectioned at a thickness of 7 μm. For analysis of regenerating muscle, solei from 6-week-old mice were injected with 100 μl of 0.75% bupivacaine in saline. Muscles were harvested after 4 days and processed as described above. In situ hybridization was performed with 33P-radiolabeled and digoxigenin-labeled cRNA probes as previously described (66, 67). The template for the TnI slow cRNA probe was an 807-bp mouse cDNA fragment obtained by RT-PCR from adult mouse soleus muscle using primers 5′-CGA GGA GCG AGA GGC TGA GAA GG-3′ and 5′-CTT GTG AAT ACT GCT GCA GGT AGC AAC-3′ and cloned into pGEM-T (Promega). Plasmid cM113aR (34), used to generate the TnI fast cRNA probe, was a gift from K. Hastings. For analysis of GTF3 expression, cRNA probes were generated from the full-length mouse cDNA (EST 555547) or partial templates (nucleotides 186 to1571 and 2895 to the poly(A) tail). All three probes yielded the same results. An 800-bp probe for Pax7 was generated by RT-PCR from E15 mouse hind limb RNA using primers 5′-CTT CAT CAA CGG TCG ACC-3′ and 5′-GGC CTG TGT ACT GTG CTG-3′.
Transfection of muscles by DNA injection and in vivo electroporation.
Plasmid DNA for transfection experiments was extracted from Escherichia coli spheroplasts to reduce endotoxin load (65) and purified on two CsCl gradients. Ten micrograms of plasmid RSV-βGal per muscle was used as an internal control in all transfections. Plasmid DNA cocktails contained reporter and control vectors at a molar ratio of 5:5:2. To analyze the effects of GTF3 on TnI SURE activity in vivo, electroporation-mediated gene transfer was utilized to deliver DNA to mature, nonregenerating fibers essentially as described previously (38). First, 40 μl of plasmid DNA in saline was injected into the soleus muscle. Two platinum electrodes (1-mm diameter) connected to a homemade stimulator were then used to expose the muscle to a series of five trains of 1,000 bipolar 200-μs pulses at a frequency of 1 kHz. With a maximal output voltage of 25 V and 2-mm gap between electrodes, the calculated electric field strength was ∼125 V/cm. Muscles were harvested 8 to 10 days posttransfection and stored at −80°C.
Determination of luciferase reporter activity.
To quantify reporter enzyme activities in transgenic mice, tissues were collected in liquid nitrogen, pulverized, and immediately homogenized in reporter lysis buffer (Promega) supplemented with a cocktail of protease inhibitors (Roche Molecular Biochemicals). The homogenates were cleared at 12,000 × g for 10 min at 4°C and assayed in duplicate on a Berthold Lumat LB9507 luminometer. Samples from wild-type mice were used to determine background values. Luciferase values were normalized to protein concentration.
Transiently transfected muscles were homogenized in 750 μl of reporter lysis buffer (Promega) plus protease inhibitors using a PowerGen 35 homogenizer (Fisher Scientific) and processed as outlined above. Luciferase light units were normalized to β-galactosidase, which was assayed as the fluorescence of 4-methylumbelliferyl (4-MU) released from the fluorogenic substrate 4-MU-β-d-galactoside in a DyNA Quant 200 fluorometer (Hoefer Pharmacia Biotech). Muscle extracts were preincubated for 1 h at 50°C to reduce endogenous galactosidase activity.
RESULTS
The downstream region of SURE confers general muscle specificity but not fiber type specificity.
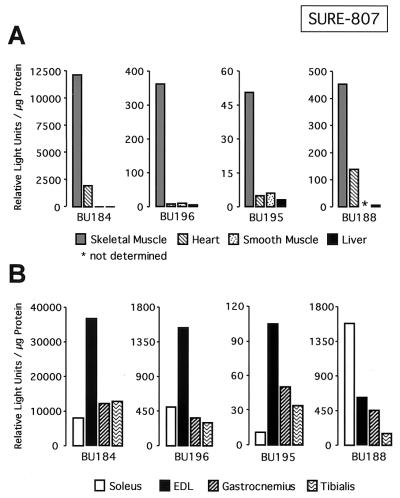
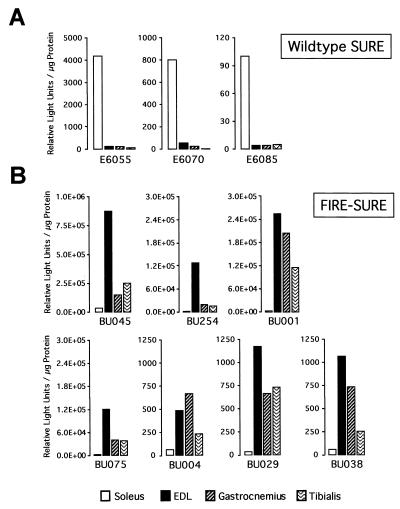
We recently demonstrated that multiple DNA motifs in the TnI SURE are important for the activity of this enhancer (12). To identify the cis elements that confer the fiber type specificity of the TnIs gene, we generated transgenic mice with a luciferase reporter construct driven by the 3′ half of SURE (−807 to −741) and the 95-bp TnIs basal promoter (Fig. 1). The −95 promoter alone was previously shown to be inactive in stably transfected myocytes and transgenic mice (3, 43). As shown in Fig. 2A, mice that harbor the TnIs95.SURE-807 construct expressed the luciferase reporter specifically in skeletal muscles. Only transgenic lines BU184 and BU188 exhibited significant luciferase activity in the heart, where the TnIs gene is expressed during embryonic and perinatal development (53, 72). Very low luciferase activities were observed in the stomach, suggesting that SURE-807 is not active in smooth muscle and in nonmuscle tissues such as liver and kidney (not shown). More important, however, is the fact that the fiber type specificity of this truncated TnI SURE construct was lost or diminished in all four transgenic founders that were analyzed (Fig. 2B). In lines BU184, BU195, and BU196 luciferase reporter levels were higher in the fast-twitch extensor digitorum longus (EDL) than in the slow-twitch soleus muscle. In these transgenic lines luciferase levels in the gastrocnemius and tibialis, two other crural muscles that are rich in fast-twitch fibers, were similar to those measured in the soleus. Only one founder (BU188) showed a moderate preference for expression in slow muscles. These results differed dramatically from those obtained with transgenes harboring the full-length SURE enhancer (founders E6055, E6070, and E6085), where luciferase reporter levels were 25- to 50-fold higher in the soleus muscle than in either the EDL, gastrocnemius, or tibialis muscles (Fig. 3A). Thus, removal of the 5′ half of SURE, which leaves the E box as well as the MEF-2 and CACC motifs intact, results in an active element that confers general muscle specificity but not fiber type-specific transcription. This finding is in accordance with previous studies on other muscle promoters showing that DNA elements which bind MyoD, MEF-2, and Sp1 and which are present in the 3′ half of SURE are sufficient to confer skeletal muscle specificity (see Discussion).
FIG. 2.
The 3′ half of TnI SURE confines reporter expression to skeletal muscle but does not confer fiber type specificity. (A) Luciferase activities in various tissues of transgenic mice harboring the SURE-807 construct. Whole-cell extracts of muscle and nonmuscle tissues from four 6-week-old founder mice were prepared and assayed for luciferase activity as described in Materials and Methods. Data for skeletal muscle are for the gastrocnemius. Heart and stomach were used to assess reporter activity in other muscle types. Liver was included as a nonmuscle tissue. Enzyme activities are normalized for protein concentration. (B) Luciferase activity mediated by TnIs95.SURE-807 in slow and fast muscles. Extracts from the slow soleus, and the fast EDL, gastrocnemius, and tibialis muscles were used to determine the fiber type specificity of SURE-807. Cell extracts were prepared and assayed as described above.
FIG. 3.
The specificity of TnI SURE can be redirected toward fast fibers by swapping 5′ sequences of SURE and FIRE. (A) Slow muscle specificity of wild-type SURE. Normalized luciferase values for slow and fast muscles are shown for three transgenic lines expressing the wild-type TnIs95.SURE construct. Samples were prepared and assayed as described for Fig. 2. (B) Fast muscle specificity of the chimeric FIRE-SURE enhancer. Founders of seven transgenic lines expressing the chimeric TnIs95.FIRE-SURE construct were analyzed for reporter activity.
The upstream halves of the TnI SURE and FIRE are necessary to confer fiber type specificity.
The sequences in the upstream halves of the TnI SURE and FIRE differ markedly, except for the CAGG motif. This and the lack of fiber type specificity of the SURE-807 construct suggested that sequences residing in the 5′ half of the TnI enhancers might restrict expression to slow or fast fibers. Taking advantage of the conserved spatial arrangement of the E box, MEF-2, CACC, and CAGG motifs in SURE and FIRE, we generated a chimeric fast/slow TnI enhancer. The TnIs95.FIRE-SURE construct (Fig. 1) containing the upstream region of FIRE (+776 to +714) was fused to the downstream region of SURE (−807 to −741) and used to generate transgenic mice for the analysis of fiber type-specific expression. As shown in Fig. 3B, all seven founders expressing luciferase were selective for fast-twitch muscles. The extent of fast-muscle expression is evident from comparing the ratios of reporter activity between EDL and soleus muscles in FIRE-SURE transgenic mice (ranging between 125 and 8), in mice lacking the upstream region of SURE (SURE-807; 10 to 0.38), and in mice harboring the wild type enhancer (WT-SURE; 0.03 to 0.07). Taken together, these results indicate that the 5′ regions of SURE and FIRE confer fiber type specificity, at least in part, by repressing transcription in fast- and slow-twitch muscles, respectively.
Expression of the TnIs gene is repressed in presumptive fast muscles during development.
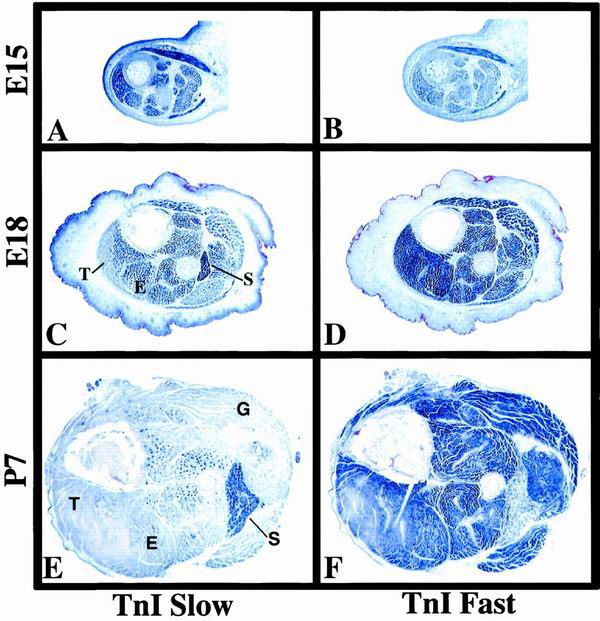
The results observed with the SURE-807 transgenes were initially unexpected. TnIs transcript levels are already low in the EDL by the time of birth, suggesting that confinement of its expression to presumptive slow fibers may begin earlier during fetal stages of muscle development (D. Vullhorst, unpublished observations). These findings prompted us to analyze fetal and perinatal expression pattern of TnI genes in the mouse by in situ hybridization. As shown in Fig. 4A and B, high levels of TnIs transcripts and low levels of TnIf transcripts are detected at E15 in all crural hind limb muscles, consistent with their expression in primary fibers. As development proceeds, TnIs expression increases in the soleus but decreases in prospective fast-twitch muscles (Fig. 4C). Decrease of TnIs mRNA levels in future fast muscles begins in peripheral regions of superficial muscles, such as the tibialis. The general pattern of TnI expression is essentially established by P7 (Fig. 4E and F). These results, in conjunction with the observed loss of fiber specificity in SURE-807 transgenic mice, suggest that the upstream half of SURE functions to repress TnIs transcription during fetal development in presumptive fast-twitch muscles.
FIG. 4.
Expression of slow and fast TnI genes during development. Cross sections of crural muscles from mouse embryos and neonates were probed for the expression of both TnI isoforms by in situ hybridization as described in Materials and Methods. (A and B) TnIs and TnIf transcripts are detected in all muscle fibers at E15. (C and D) By E18, TnIs expression has increased selectively in prospective slow fibers, most notably in the soleus muscle, while beginning to decrease in superficial areas of peripheral fast muscles such as the tibialis. Up to this stage, TnIf transcripts continue to be abundant in all muscles (E and F). By P7, expression of TnIs and TnIf is largely confined to slow and fast muscle, respectively. S, soleus; E, EDL; G, gastrocnemius; T, tibialis.
Mapping sequences in the TnI SURE conferring fiber type specificity.
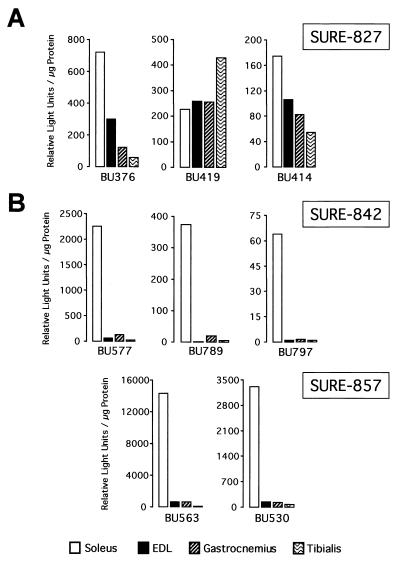
With the purpose of defining the minimal sequence requirement for slow fiber specificity conferred by the 5′ half of SURE, three luciferase reporter constructs were generated by progressively adding upstream sequences back to the TnIs95.SURE-807 construct. These constructs were denoted TnIs95.SURE-827, TnIs95.SURE-842, and TnIs95.SURE-857 (Fig. 1A). As shown in Fig. 5A, the SURE-827 transgenes exhibit only small differences in reporter expression between the slow soleus and the fast EDL, gastrocnemius, and tibialis muscles. For founders BU414 and BU376 luciferase was merely 1.6- to 2.4-fold higher in the soleus than in the EDL; for BU419 levels were approximately equal. This demonstrates that the addition of 20 bp of DNA, including the CAGG motif, is not sufficient to impart specificity to the SURE-807 construct. In stark contrast, mice harboring constructs with 35 or 50 bp of the SURE upstream sequence (SURE-842 and SURE-857) exhibited the same degree of specificity as WT-SURE (compare Fig. 5B and 4A). Luciferase reporter activities of founders BU577, BU789, and BU797 (SURE-842) and founders BU530 and BU563 (SURE-857) were on average about 50-fold higher in the soleus muscle than in the EDL, gastrocnemius, and tibialis muscles. Interestingly, the DNA stretch between −842 and −827 harbors BLM (GTTAATCCG), which we had shown previously to be critical for SURE function in stably transfected myocytes and which binds to nuclear factors from numerous cell types of mesodermal and endodermal origin (12). Thus, our results demonstrate that DNA elements necessary for slow fiber type-specific transcription reside between positions −842 and −827 of the TnI SURE.
FIG. 5.
Addition of 35 bp of upstream sequence to SURE-807 restores slow fiber-restricted enhancer activity. Slow and fast muscles from founders of three independent transgenic lines harboring TnIs95.SURE-827 (A) and three lines of TnIs95.SURE-842 as well as two lines of TnIs95.SURE-857 (B) expressing the luciferase reporter were analyzed for reporter activity. SURE-827 includes the CAGG motif but not the BLM, whereas SURE-842 and SURE-857 contain both motifs. All transgenic lines selectively expressed luciferase in skeletal muscle.
Isolation of a nuclear factor binding the TnI SURE upstream element.
A yeast one-hybrid screen was performed using three tandem copies of the sequence between −844 and −808 of TnI SURE as “bait” and an adult skeletal muscle cDNA library fused to the GAL-4 activation domain (see Materials and Methods). Clones were selected for growth in the absence of both histidine and uracil and screened for β-galactosidase expression. A total of 2.7 × 107 transformants yielded 88 His+ Ura+ clones, and of these 30 were also positive for β-galactosidase activity. Sixteen of these clones were successfully transferred to E. coli and sequenced. Analysis of their cDNA inserts revealed that 10 out of these 16 clones contained partial cDNA sequences coding for GTF3 (Table 1) (49). A similar screen performed with TnI SURE sequences between −832 and −808 did not yield any GTF3 clones (data not shown), indicating that the area between −844 and −832 is necessary for binding. Most clones examined (clones 30, 29, 21, 18, and 74) contained the short form of exon 19, and one (clone 81) contained the long form (61). All 10 GTF3 partial cDNAs shared sequences encoding the carboxy-terminal half of the protein but differed in length at the 5′ ends (Table 1). The longest clones were missing 302 bp of sequence encoding the amino terminus, whereas the shortest clone (clone 74) initiated within the sequence encoding the third TFII-I-like repeat. These results suggested that the amino-terminal half of GTF3 is not required for DNA binding (see below).
GTF3 binds to a DNA motif in the upstream region of the TnI SURE that is similar to the binding site for TFII-I.
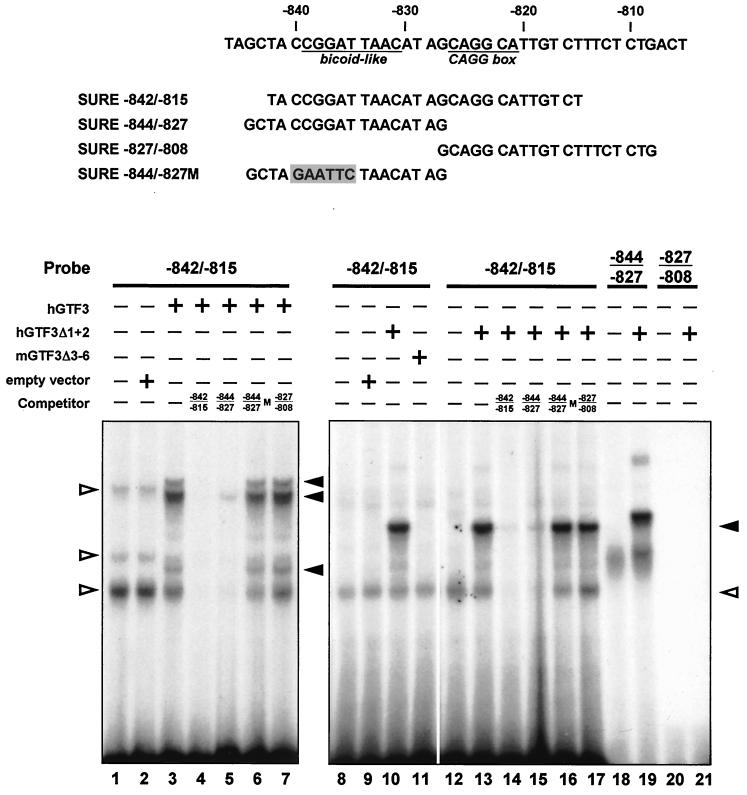
A full-length human GTF3 cDNA was constructed and subcloned into a mammalian expression vector for in vitro translation, and its capacity to specifically bind SURE sequences was assessed by EMSAs (Fig. 6, lanes 1 to 7). The lysate programmed with GTF3 but not with the empty vector showed selective binding of the 32P-labeled oligonucleotide probe containing the sequence between −842 and −815 of SURE (lanes 1 to 3). The observation that all GTF3 clones selected by the yeast one-hybrid screen share sequences encoding the C-terminal half of GTF3 suggested that this region is sufficient for DNA binding (Table 1). Expression vectors for GTF3 encoding either its N- or C-terminal half were translated in vitro and used in competitive EMSAs to assess the DNA binding activity of both regions of the protein (lanes 8 to 11). Lysates containing the C-terminal half of GTF3 (aa 529 to 959; hGTF3Δ1 + 2), which corresponds to the shortest cDNA clone obtained from the yeast one-hybrid screen (clone 74), specifically bound the −842/−815 oligonucleotide probe (lane 10). In contrast, in our experimental conditions the N-terminal half of GTF3 (aa 1 to 463; mGTF3Δ3-6) failed to form a visible complex (lane 11). These results confirmed that the C-terminal half of GTF3 harbors a DNA binding domain.
FIG. 6.
GTF3 binds specifically to the bicoid-like sequence of the TnI SURE. The ability of in vitro-translated GTF3 to interact with the TnI SURE was assessed by EMSA. Sequences of oligonucleotides used in this experiment are shown at the top. Oligonucleotides spanning the region between −842 and −815 (lanes 1 to 17), −844 and −827 (lanes 18 and 19), and −827 and −808 (lanes 20 and 21) were used as probes. Full-length and partial GTF3 proteins were translated from expression plasmids containing cDNAs for hGTF3, hGTF3Δ1 + 2 (C-terminal half; lanes 10, 13 to 17, 19, and 21), mGTF3Δ3–6 (N-terminal half; lane 11) and added to the reactions where indicated. Reticulocyte lysates containing no plasmid or the parental expression vector served as negative controls. For competition experiments, an excess of unlabeled oligonucleotide was added. Competitor oligonucleotides were −842/−815 (self; lanes 4 and 14), −844/−827 (upstream half; lanes 5 and 15), −827/−808 (downstream half; lanes 7 and 17), and −844/−827 M, which harbors a mutation (shaded) in the sequence corresponding to the BLM (lanes 6 and 16). Filled arrowheads, specific binding of probes to translated proteins; open arrowheads, bands that are due to nonspecific interactions between the probe and components of the reticulocyte lysate.
The −842/−815 probe includes BLM and the CAGG site of the TnI SURE. By using shorter probes that harbor only one of these motifs in competitive EMSAs, we delineated the sequences bound by the full-length GTF3 (lanes 4 to 7) and the C-terminal half of the protein (lanes 12 to 21). The BLM-containing oligonucleotide SURE −844/−827 effectively competes with SURE −842/−815 for GTF3 binding (lanes 5 and 15), whereas the mutated oligonucleotide missing this motif failed to compete (lanes 6, 16). Moreover, SURE oligonucleotides −827/−808 and −832/−815 (not shown) harboring the CAGG site but not the BLM failed to compete for GTF3 binding (lanes 7 and 17). We also found that the C-terminal half of GTF3 is sufficient to bind the oligonucleotide probe harboring only the BLM (SURE −844/−827) and failed to shift the CAGG-containing probe (SURE −827/−808), confirming that GTF3 binds the TnI SURE via the BLM (lanes 18 to 21). Interestingly, the sequence of this motif on the complementary strand (GTTAATCCG) conforms to the consensus site for the initiator element (Inr: YYANWYY) (55) that is a binding site for TFII-I (50), indicating that GTF3 and TFII-I may recognize similar DNA motifs.
Analysis of GTF3 expression in developing and adult muscles using Northern blot and in situ hybridization.
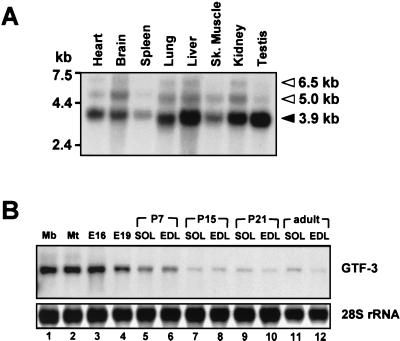
Northern blots were used to assess the expression pattern of GTF3 in various adult mouse tissues and during muscle development. Using a full-length and a 5′-specific probe that lacked sequences for the TFII-I repeated domains (not shown), the main 3.9-kb transcript was detected in all tissues analyzed, which include those containing cardiac and skeletal muscles as well as liver, brain, and kidney (Fig. 7A). The wide distribution of GTF3 expression that we observe in the mouse is consistent with previous studies (4). Weakly hybridizing bands corresponding to transcripts of approximately 6.5 and 5 kb were also detected in most tissues. These bands are likely to represent either transcripts not fully processed or alternatively spliced forms of GTF3 as previously reported (22). The signals are unlikely to stem from cross-hybridizations with TFII-I due to the limited sequence homology between both transcripts and because a 5′-specific probe that lacked sequences for conserved TFII-I repeat domains yielded indistinguishable hybridization patterns.
FIG. 7.
GTF3 expression in adult tissues and during muscle development. (A) Expression pattern of GTF3 in different mouse tissues. A commercial poly(A) blot containing 2 μg of mRNA from various adult mouse tissues per lane (mouse MTN; Clontech) was probed with a 32P-labeled full-length mouse GTF3 cDNA. The positions of RNA molecular weight markers are shown on the left. Filled arrowhead, position and size of the major GTF3 transcript; open arrowheads, additional bands that possibly represent incompletely processed or alternative GTF3 transcripts. Sk. muscle, skeletal muscle. (B) GTF3 expression is downregulated during muscle development. Total RNA from cultured myogenic cells and rat tissues (10 μg) was separated on a 1.5% formaldehyde-agarose gel and probed as described above. 28S rRNA hybridization signals are shown as a loading control. Total RNA from C2C12 myoblasts (lane 1) and myotubes (differentiated for 72 h; lane 2) were used to assess GTF3 expression during myogenic differentiation in vitro. RNA from whole hind limbs of E16 and E19 (excluding skin) rat embryos was used to approximate GTF3 expression during the fetal stage of appendicular muscle development (lanes 3 and 4). RNAs from postnatal stages of muscle development (from P7 to the adult; lanes 5 to 12) were used to determine GTF3 expression in maturating slow soleus (SOL) and fast EDL muscles.
Next, we analyzed GTF3 transcript levels in cultured myogenic cells and rat skeletal muscle during different stages of development (Fig. 7B). Strong expression was observed in undifferentiated and differentiated C2C12 myocytes, as well as in E16 and E19 hind limbs. GTF3 transcript levels decrease as development proceeds, most significantly between P7 and P15, and remain low after the second postnatal week. We did not detect a pronounced preference for either slow or fast muscles at any stage of postnatal development. Transcript levels were maximally 1.4-fold higher in adult soleus than in EDL, indicating that GTF3 expression is not fiber type specific. In contrast, cultured myoblasts and myotubes have 8- to 10-fold-higher GTF3 transcript levels than the adult soleus, suggesting that GTF3 may have a more important role during development than in the mature muscle.
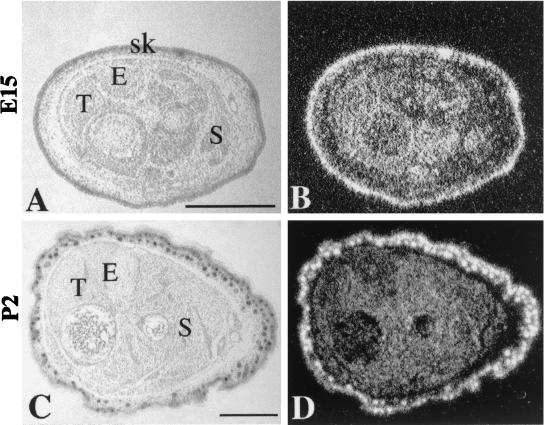
In situ hybridization was used to determine the cellular distribution of GTF3 transcripts during fetal muscle development and during regeneration of adult muscle. A 33P-labeled full-length probe synthesized from the mouse GTF3 cDNA (EST clone 555547) was used to hybridize transverse sections of hind limb muscles from E15 through adult. Indistinguishable results were obtained with 5′- and 3′-specific probes (see Materials and Methods). At E15, prominent hybridization is observed in the forming muscle masses, as well as in skin and bones (Fig. 8A and B). The hybridization intensity is greatly reduced in perinatal muscles (Fig. 8C and D). In adult myofibers GTF3 transcripts levels are very low or not significantly higher than background when analyzed by in situ hybridization (Fig. 9B). In agreement with Northern blot data on postnatal muscle (see above), in situ analysis of GTF3 expression during fetal muscle development suggests that it follows neither a muscle- nor a fiber type-specific pattern; thus, part of the signals observed on Northern blots using adult muscle tissue may originate from nonmyogenic cell types.
FIG. 8.
Expression of GTF3 in developing hind limb muscles. (B and D) Dark-field images showing in situ hybridization of GTF3; (A and C) hematoxylin images of the same sections. (A and B) At E15, transcripts are abundant in all muscle fibers, as well as in cells of bone and skin. (C and D) By P2, GTF3 expression has decreased to low levels in muscle fibers, while remaining high in skin. S, soleus; E, EDL; T, tibialis; sk, skin. Bar, 0.6 mm.
FIG. 9.
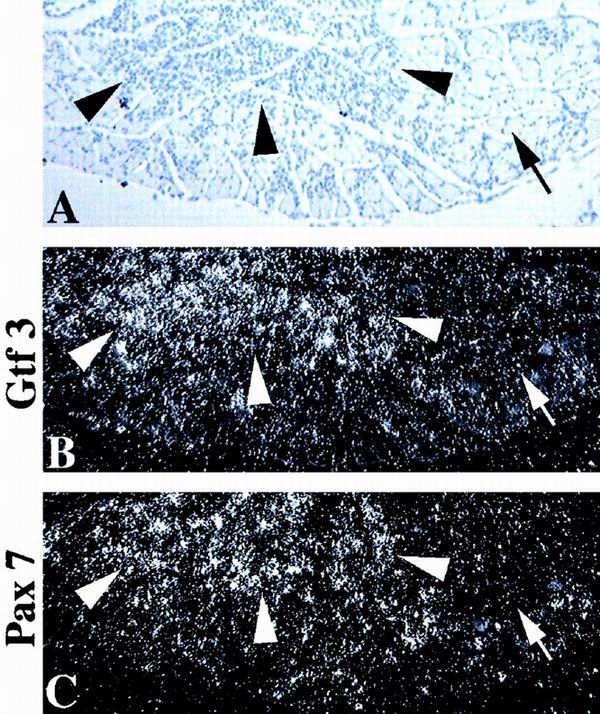
GTF3 and Pax7 are upregulated in regenerating muscle. (A) Hematoxylin image of a transverse section of an adult mouse soleus muscle 4 days after injection of bupivacaine. Regenerating (arrowheads) and nonregenerating areas (arrows) are indicated. (B and C) Dark-field images of strictly consecutive sections, hybridized with probes for GTF3 and Pax7. (B) GTF3 is strongly expressed in regenerating areas (arrowheads) but not in normal areas (arrows). (C) Pax7 hybridization shows the expansion of the satellite cell population in regenerating areas compared to nonregenerating areas.
Muscle regeneration recapitulates many aspects of early myogenesis including myoblast proliferation, formation of syncytial myotubes, and the reestablishment of fiber type specificity. Because of the strong expression of GTF3 in undifferentiated C2C12 myoblasts and in nascent myofibers, we speculated that its gene might be reactivated in proliferating satellite cells. We therefore induced degeneration in adult soleus muscles by injection of the myotoxin bupivacaine and analyzed tissues 4 days later. As shown in Fig. 9B, regenerating areas of the muscle are strongly positive for GTF3, whereas nondegenerated areas are essentially devoid of GTF3 hybridization signals. Consecutive sections were hybridized with a probe specific for Pax7, a marker for mouse muscle satellite cells (54), to assess the extent to which GTF3 is confined to this cell type in regenerating muscles (Fig. 9C). Pax7 hybridization was observed to be more blotchy and confined than GTF3 signals, suggesting that GTF3 expression in regenerating fibers is not restricted to satellite cells and is consistent with its lack of cell type specificity. The results from Northern blot and in situ hybridization experiments indicate that GTF3 expression is highest during a period of muscle differentiation that coincides with fiber type diversification.
GTF3 represses activity from the TnI SURE.
The functional role of GTF3 in TnIs transcription was examined in adult rat muscles by coexpressing GTF3 and the TnIs95.SURE luciferase reporter in soleus muscles transfected by electroporation in vivo. Electroporation promotes DNA uptake by adult, nonregenerating muscle fibers (38), hence enabling us to study the effect of GTF3 overexpression in fibers that have very low endogenous GTF3 transcript levels (see Fig. 9). The contralateral soleus of each rat, transfected with the TnIs95.SURE luciferase reporter and the empty expression vector, served as the negative control. A cotransfected RSV-βGal expression vector was used to normalize for transfection efficiency. As shown in Fig. 10, muscles transfected with the GTF3 construct expressed normalized luciferase levels that were 3- to 10-fold lower than their contralateral controls (nonnormalized luciferase activities were 5- to 38-fold lower). The levels of β-galactosidase reporter used for normalization were one- to fivefold lower in experimental muscle than in control muscle, indicating that, although GTF3 significantly repressed transcription from the TnI SURE, a nonspecific component partially contributed to the overall loss of reporter activity. Based on the importance of sequences containing the BLM for fiber type specificity of the TnI SURE, the early expression of GTF3 in skeletal muscle, and the inhibition of SURE activity by GTF3, we propose that GTF3 contributes to myofiber diversification during early development.
FIG. 10.
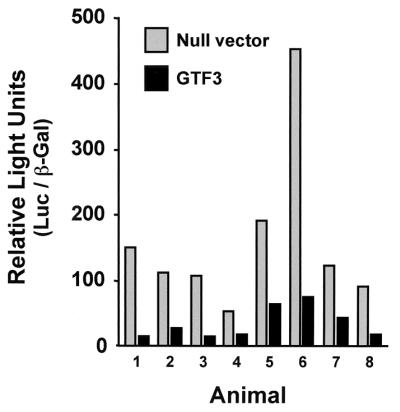
GTF3 represses TnI SURE enhancer activity in transiently transfected rat soleus muscle. Muscles were transfected with equimolar amounts of TnIs95.SURE and mGTF3 and harvested after 9 days for analysis of reporter activity. In each animal, the contralateral soleus was cotransfected with the reporter plasmid and the empty parental expression vector. Luciferase values were normalized for β-galactosidase expression using RSV-βGal. Each pair of bars represents values obtained from a single animal. Black bars, experimental muscles; gray bars, control muscles.
DISCUSSION
The transcriptional programs that are responsible for generating skeletal muscle diversity and that confine the expression of contractile proteins and metabolic enzymes to specific fiber types are not understood. As an important step toward unraveling these programs, we have identified cis- and trans-acting factors that are important for fiber type-specific transcription of the slow and fast TnI isoforms. We have shown that a TnIs enhancer construct containing the downstream half of the SURE is sufficient for general skeletal muscle expression in transgenic mice and that an additional region of 15 bp in the upstream half of SURE is necessary to confer fiber type specificity. This region harbors a BLM that binds the nuclear factor GTF3. Interestingly, this sequence is unique to the slow enhancer, whereas other cis elements are conserved between SURE and FIRE. Northern blot and in situ hybridization analyses demonstrate that GTF3 is expressed during myogenesis and in regenerating muscles, paralleling the emergence of distinct fiber types. Overexpression of the GTF3 protein in electroporated adult muscles represses transcription from the TnI SURE. Below, we propose how GTF3 may contribute to fiber type specificity and its possible relevance to WBS.
DNA elements and factors conferring general skeletal muscle-specific transcription.
In this study we were able to dissociate the TnI SURE regulatory sequences necessary for general muscle specificity from those required for transcription in slow-twitch muscles. The deletion analysis of the TnI SURE shows that its downstream half, harboring the E box, MEF-2, and CACC motifs, which, respectively, bind MyoD/myogenin, MEF-2, and Sp1, is sufficient to confer pan-muscle specificity in transgenic mice (SURE-807 and SURE-827). These cis elements are also important for the regulation of the TnIs gene during differentiation of cultured myocytes (11). We have previously shown the TnI SURE and FIRE E boxes to be functionally equivalent and not to confer muscle type specificity (12); the involvement of MEF-2 and CACC motifs in fiber type-specific transcription was unknown. Interestingly, one or more of these motifs are important for enhancer function of other slow and fast muscle genes, such as the MLC2v, MLC1/3f, and aldolase genes (20, 37, 51, 64). In almost all cases, the mutation of any one of these sites results in the reduction or abolishment of overall enhancer activity but not in the loss of fiber type specificity. Consistent with the role of E box, MEF-2, and CACC motifs in general muscle specificity is the observation that these cis elements are frequently found in enhancers for muscle genes not expressed in a fiber type-specific fashion, such as the α-actin (6, 52), muscle creatine kinase (2), MRF-4 (42), and myogenin (9, 13, 70) genes. These findings corroborate the idea that the interactions of myogenic basic helix-loop-helix factors with MEF-2 (40) and possibly Sp1 are sufficient to confer muscle-specific transcription.
In studies that used the TnI SURE as a model for genes regulated by neural activity, it was recently proposed that a calcineurin-dependent transcriptional pathway controls skeletal muscle fiber types in response to slow, tonic motoneuron activity (14, 68). The authors proposed that activation of the calcium/calmodulin-dependent phosphatase calcineurin by neural depolarization results in the binding of NFAT and MEF-2 to DNA elements present in the downstream half of the SURE, which in turn leads to SURE activation specifically in slow muscles. This proposal is difficult to reconcile with our previous studies showing that mutation of the NFAT site in the TnI SURE alters neither the activity nor the specificity of the enhancer (12). Moreover, here we provide evidence that constructs harboring the E-box, NFAT, MEF-2, and CACC motifs of SURE are expressed in both fast and slow muscles (SURE-807 and SURE-827) and that cis elements in the 5′ half of SURE and FIRE are necessary to restrict expression to slow and fast fibers, respectively. Our results are in agreement with experiments by O'Mahoney and colleagues using direct DNA injection into adult muscles to demonstrate that the downstream half of the human TnIs enhancer (highly homologous to SURE) is expressed similarly in soleus and EDL muscles (44). It is also important to emphasize that numerous studies have demonstrated that separate enhancers are often required to faithfully recapitulate diverse aspects of fiber type-specific gene regulation, as in the case of the β-myosin heavy chain (39) and TnIf (25) regulatory elements. Thus, we believe that, even though the TnIs gene is regulated by patterned motoneuron activity in the adult (8, 11), it is premature to conclude that the TnI SURE mediates these effects. A role for calcineurin in the regulation of other fiber type-specific genes, namely, the promoters for the slow myosin light chain and fast sarcoplasmic reticulum calcium ATPase, has also been investigated (60). This study directly measured calcineurin activity and NFAT binding in slow and fast muscles and the effects of calcineurin overexpression on the function of promoters for both genes. Based on their results the authors concluded that neither calcineurin nor NFAT appears to have dominant roles in the induction or maintenance of slow or fast fiber types in the adult. It is apparent from these conflicting findings that additional studies will be necessary to clarify a possible role for NFAT and MEF-2 as mediators of the calcineurin pathway in the specific regulation of genes in slow-twitch muscles.
DNA regulatory sequences and factors involved in fiber type-specific transcription.
The results presented here demonstrate that the upstream regions of the TnI SURE (−842 to −808) and FIRE (+776 to +714) are necessary to confer fiber type specificity. The deletion analysis performed with transgenic mice showed that removal of this sequence from the 5′ half of SURE results in similar transcription levels in fast- and slow-twitch muscles, suggesting a repressive function for this area. Within this region there are two elements that we have previously studied, the CAGG motif and the BLM (11, 43); the possibility that other elements in this region contribute to fiber type specificity cannot be excluded. Mutation of the CAGG motif in the context of the full-length enhancer abolished SURE activity in all muscles (12), thus precluding interpretation of its role in fiber type specificity. Here we showed that this cis element is not sufficient to confer slow muscle specificity to the TnI SURE, because the CAGG motif is present in the SURE-827 construct that is active in both fiber types. The core of the CAGG motif (GCAGGCA) is similar to the MEF-3 site (TCAGGTT[A/T]C) initially described for the cardiac troponin C enhancer and also found in the myogenin and fast pM aldolase promoters (26, 46, 56, 57). Spitz et al. (56, 57) found that the MEF-3 site in the aldolase pM promoter is necessary, but not sufficient, to direct transcription in a subset of mouse fast-twitch muscles. The Six/sine oculis family of homeodomain proteins binds the MEF-3 site in the aldolase promoter (56); nonetheless, the contribution of these factors to fiber type specificity remains to be established.
A smaller deletion in the upstream half of TnIs SURE (from −842 to −827) that only removes the BLM and that maintains the CAGG motif also results in pan-muscle-specific transcription. Two lines of evidence indicate that GTF3 interacts with SURE via the BLM included in this region. First, the yeast one-hybrid screen and gel retardation assays demonstrate an affinity of GTF3 for the BLM. Second, forced expression of GTF3 protein in adult fibers represses transcription from the TnI SURE, thus revealing a functional importance of GTF3 for SURE activation.
Using an upstream sequence element of the human TnI slow enhancer (USE B1) corresponding to the same region of SURE, O'Mahoney et al. recently cloned MusTRD1, the human ortholog of mouse GTF3 (44). Based on their findings and/or interpretations, the authors reached conclusions that differ from those drawn in this study. First, based on a Northern blot experiment it was suggested that MusTRD1 is predominantly expressed in adult skeletal and cardiac muscle. We have demonstrated that GTF3 is expressed in a variety of tissues and is weakly expressed in, and not restricted to, adult skeletal muscle of rodents. In addition, by in situ hybridization we found that GTF3 expression is relatively strong during early muscle differentiation but is dramatically reduced in adult myofibers. Our results are consistent with those of Bayarsaihan and Ruddle (4), who also found broad expression of GTF3 in mouse tissues, and with those of Tassabehji et al. (62), who found that GTF3 transcripts are abundant in human fetal tissues and that GTF3 levels decline during development. Thus, we believe that skeletal muscle specificity is not a defining feature of GTF3 and that the discrepancy observed for GTF3 tissue distribution could reflect a simple species difference. As we discuss below, the issue of when during development GTF3 interacts with SURE can greatly influence the interpretation of how this factor may contribute to the fiber specificity of TnIs transcription. The second discrepancy concerns the location of the DNA binding site in GTF3. O'Mahoney and colleagues proposed that a basic region in MusTRD1, located between the first and second TFII-I repeat, is involved in DNA-protein interaction (44). We have found both by inspection of GTF3 clones obtained from the yeast one-hybrid screen (Table 1) and by EMSAs using an N-terminally truncated GTF3 protein (hGTF3Δ1 + 2) that this basic region is not required for DNA binding. Moreover, we have directly compared the binding of hGTF3Δ1 + 2 with that of mGTF3Δ3-6, a truncated protein that mimics the protein used by O'Mahoney et al., and showed that in our experimental conditions the amino-terminal half of GTF3 has no detectable affinity for the BLM. In agreement with our findings, Bayarsaihan and Ruddle (4) showed that N-terminally truncated BEN/GTF3 bound to the EFG site of the Hoxc8 early enhancer (homologous to BLM) in a yeast one-hybrid screen and on EMSAs. In conclusion, although discrepancies concerning the expression profile and DNA binding domains of GTF3 exist, there is a general consensus that GTF3 binds the BLM. However, knowing when and in which muscle types GTF3 is expressed during development is important for understanding how this factor contributes to the regulation of the TnIs gene.
How may GTF3 contribute to fiber type-specific transcription of the TnIs gene during development?
Both intrinsic and extrinsic factors are thought to contribute to the establishment of different fiber types during development. Motoneuron innervation and myoblast lineage are important for the expression of specific fiber type properties in chicks (16, 17, 48, 59). The TnIs gene is initially expressed in all myofibers of hind limb muscles during embryonic development. As muscles mature, the gene is selectively repressed in prospective fast-twitch fibers but continues to be expressed at high levels in myofibers of the slow soleus muscle (Fig. 4). Shortly after birth expression of the TnI genes is upregulated by nerve-dependent mechanisms; distinct frequencies and patterns of motoneuron activity regulate TnI gene levels in the adult (8, 11).
Our deletion analysis of transgenic mice indicates that removal of the 15-bp region containing the BLM motif results in a loss of fiber type specificity. These results indicate that this region may contribute to the repression of TnIs expression in prospective fast myofibers during prenatal development; it also could serve to augment transcription in slow muscles. Interestingly, the developmental loss of TnIs expression in prospective fast fibers coincides with a period of strong GTF3 expression and thus could result from GTF3 binding to the BLM of SURE. In addition, we observed an increase of GTF3 expression in regenerating myofibers, which, like developing muscles, transiently coexpress slow and fast isoforms of contractile proteins before they are restricted to specific fiber types (19). Last, the cotransfection experiments with electroporated adult muscles show that forced expression of GTF3 represses transcription from the TnI SURE. Taken together, our analyses of cis- and trans-acting regulatory factors strongly suggest that the interactions of GTF3 with the BLM contribute to the fiber type-specific expression of TnIs during early muscle development.
Various mechanisms can be invoked for the contribution of GTF3 to the establishment of fiber types, even though its expression is not confined to prospective slow or fast myofibers during development. (i) Studies on the structurally related multidomain protein TFII-I demonstrate that it interacts with numerous other transcription factors, such as USF-1, SRF, Phox1, and NF-κB (24, 33, 41, 49), presumably via its six helix-loop-helix-like domains. Greuneberg et al. (24) reported that TFII-I promotes the formation of higher-order complexes between SRF and Phox1 on the fos promoter and proposed that it coordinates the linkage of activator complexes with the general transcription machinery. Based on the structural similarities between GTF3 and TFII-I, it is conceivable that GTF3 interacts with different combination of transcription factors in slow and fast muscles. We have previously demonstrated that interactions of factors binding to the conserved cis elements of the TnI SURE, such as the E box and MEF-2 site, are critical for transcription in the context of native chromatin (11, 12, 43). Based on the observed cooperation between the distinct SURE elements and the structural similarities between GTF3 and TFII-I, we believe that GTF3 may regulate SURE function by stabilizing or coordinating the interaction of multiple transcription factors and establishing a link to the basal transcription machinery. (ii) Currently, we cannot eliminate the possibility that other factors, in addition to GTF3, could bind the region between −842 and −808. Mutually exclusive interactions for overlapping cis elements located in this region between GTF3 and other trans-acting factors specific to slow and fast fibers could occur to regulate muscle specificity. (iii) Posttranscriptional modifications could differentially regulate GTF3 function or its interactions with mediator proteins in slow and fast myofibers. (iv) GTF3 splice variants could be differentially expressed to regulate transcription in slow and fast muscles. An additional GTF3 isoform generated by alternate splicing has been reported, but its distribution in muscle is unknown (22). Although the yeast one-hybrid screen using a quadriceps muscle (predominantly fast muscle) library yielded mostly one type of transcript, it is feasible that other forms of GTF3 that function as activators or repressors could be expressed in different muscles. Studies to determine which of these regulatory mechanisms are utilized to regulate slow-specific transcription are in progress.
An intriguing question is how can these early developmental events contribute to the fiber type specificity in adult muscles? One possibility is that distinct transcriptional complexes (enhanceosomes) or chromatin modifications could serve to differentially imprint TnI genes in slow and fast myocytes to influence their response to epigenetic signals elicited by motoneuron innervation later in postnatal development. For example, cultured chick myoblasts retain their slow or fast phenotypes when transplanted back into the limbs of embryos, indicating their commitment to specific fates (16). Transgenic mice harboring the MLC1/3fast enhancer also retain their transcriptional activity and positional information in culture by a mechanism that involves differential DNA methylation (18, 23). It is therefore conceivable that GTF3 not only participates in setting up fiber type-specific transcription from the TnI SURE but also contributes to establishing a molecular memory of fiber fate that persists into adulthood through chromatin remodeling. Because the levels of GTF3 protein in adult myofibers could not be determined in these studies, it is also feasible that lower levels GTF3 would be sufficient to maintain fiber type differences in the adult. Transgenic and knockout mice will be instrumental to resolve the involvement of GTF3 in the establishment and maintenance of slow fiber-specific gene expression.
Implications of GTF3 function in WBS.
WBS is a rare, sporadic disorder resulting from the loss of approximately 20 contiguous genes in a microdeletion on chromosome band 7q11.23, which include GTF2I (encodes TFII-I) and GTF2IRD1 (encodes GTF3), which reside in the telomeric end of the deleted segment (21). Individuals with WBS have distinctive physical, cognitive, and behavior abnormalities, which may include pulmonary and supravalvular aortic stenosis, transient neonatal hypercalcemia, growth retardation, impaired cognitive skills, and clinical and morphological evidence of myopathies (5, 21). Persons with microdeletions that spare the telomeric end of the critical region do not manifest most of the abnormalities associated with WBS (62), suggesting that haploinsufficiency of GTF2I and GTF2IRD1 may contribute to these deficiencies. Alterations in the expression of proteins that regulate the contractile or metabolic properties of skeletal muscles could account either directly or indirectly for the myopathies associated with WBS. We have found that TFII-I and GTF3 are expressed at high levels in developing musculature, brain, and other tissues (I. Karavanova, unpublished data), raising the possibility that reduced levels of these factors during early development could result in the later downregulation or misexpression of target genes. This observation has important implications for studies on the etiology of WBS.
ACKNOWLEDGMENTS
We are grateful to Daniel Abebe for expert technical assistance with transgenic animals.
D.V. was supported by a postdoctoral fellowship from the Deutsche Forschungsgemeinschaft.
REFERENCES
- 1.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 2.Amacher S L, Buskin J N, Hauschka S D. Multiple regulatory elements contribute differentially to muscle creatine kinase enhancer activity in skeletal and cardiac muscle. Mol Cell Biol. 1993;13:2753–2764. doi: 10.1128/mcb.13.5.2753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Banerjee-Basu S, Buonanno A. cis-acting sequences of the rat troponin I slow gene confer tissue- and development-specific transcription in cultured muscle cells as well as fiber type specificity in transgenic mice. Mol Cell Biol. 1993;13:7019–7028. doi: 10.1128/mcb.13.11.7019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bayarsaihan D, Ruddle F H. Isolation and characterization of BEN, a member of the TFII-I family of DNA-binding proteins containing distinct helix-loop-helix domains. Proc Natl Acad Sci USA. 2000;97:7342–7347. doi: 10.1073/pnas.97.13.7342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bellugi U, Lichtenberger L, Jones W, Lai Z, St. George M. I. The neurocognitive profile of Williams syndrome: a complex pattern of strengths and weaknesses. J Cogn Neurosci. 2000;12:7–29. doi: 10.1162/089892900561959. [DOI] [PubMed] [Google Scholar]
- 6.Biesiada E, Hamamori Y, Kedes L, Sartorelli V. Myogenic basic helix-loop-helix proteins and Sp1 interact as components of a multiprotein transcriptional complex required for activity of the human cardiac alpha-actin promoter. Mol Cell Biol. 1999;19:2577–2584. doi: 10.1128/mcb.19.4.2577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Buckingham M. Making muscle in mammals. Trends Genet. 1992;8:144–148. doi: 10.1016/0168-9525(92)90373-C. [DOI] [PubMed] [Google Scholar]
- 8.Buonanno, A., J. Cheng, P. Venepally, J. Weis, and S. Calvo. Activity-dependent regulation of muscle genes: repressive and stimulatory effects of innervation. Acta Physiol. Scand. 163:S17–S26. [DOI] [PubMed]
- 9.Buonanno A, Edmondson D G, Hayes W P. Upstream sequences of the myogenin gene convey responsiveness to skeletal muscle denervation in transgenic mice. Nucleic Acids Res. 1993;21:5684–5693. doi: 10.1093/nar/21.24.5684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Buonanno A, Rosenthal N. Molecular control of muscle diversity and plasticity. Dev Genet. 1996;19:95–107. doi: 10.1002/(SICI)1520-6408(1996)19:2<95::AID-DVG1>3.0.CO;2-V. [DOI] [PubMed] [Google Scholar]
- 11.Calvo S, Stauffer J, Nakayama M, Buonanno A. Transcriptional control of muscle plasticity: differential regulation of troponin I genes by electrical activity. Dev Genet. 1996;19:169–181. doi: 10.1002/(SICI)1520-6408(1996)19:2<169::AID-DVG9>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- 12.Calvo S, Venepally P, Cheng J, Buonanno A. Fiber-type-specific transcription of the troponin I slow gene is regulated by multiple elements. Mol Cell Biol. 1999;19:515–525. doi: 10.1128/mcb.19.1.515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cheng T C, Wallace M C, Merlie J P, Olson E N. Separable regulatory elements governing myogenin transcription in mouse embryogenesis. Science. 1993;261:215–218. doi: 10.1126/science.8392225. [DOI] [PubMed] [Google Scholar]
- 14.Chin E R, Olson E N, Richardson J A, Yang Q, Humphries C, Shelton J M, Wu H, Zhu W, Bassel-Duby R, Williams R S. A calcineurin-dependent transcriptional pathway controls skeletal muscle fiber type. Genes Dev. 1998;12:2499–2509. doi: 10.1101/gad.12.16.2499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Condon K, Silberstein L, Blau H M, Thompson W J. Differentiation of fiber types in aneural musculature of the prenatal rat hindlimb. Dev Biol. 1990;138:275–295. doi: 10.1016/0012-1606(90)90197-q. [DOI] [PubMed] [Google Scholar]
- 16.DiMario J X, Fernyak S E, Stockdale F E. Myoblasts transferred to the limbs of embryos are committed to specific fibre fates. Nature. 1993;362:165–167. doi: 10.1038/362165a0. [DOI] [PubMed] [Google Scholar]
- 17.DiMario J X, Stockdale F E. Both myoblast lineage and innervation determine fiber type and are required for expression of the slow myosin heavy chain 2 gene. Dev Biol. 1997;188:167–180. doi: 10.1006/dbio.1997.8619. [DOI] [PubMed] [Google Scholar]
- 18.Donoghue M J, Patton B L, Sanes J R, Merlie J P. An axial gradient of transgene methylation in murine skeletal muscle: genomic imprint of rostrocaudal position. Development. 1992;116:1101–1112. doi: 10.1242/dev.116.4.1101. [DOI] [PubMed] [Google Scholar]
- 19.Esser K, Gunning P, Hardeman E. Nerve-dependent and -independent patterns of mRNA expression in regenerating skeletal muscle. Dev Biol. 1993;159:173–183. doi: 10.1006/dbio.1993.1231. [DOI] [PubMed] [Google Scholar]
- 20.Esser K, Nelson T, Lupa-Kimball V, Blough E. The CACC box and myocyte enhancer factor-2 sites within the myosin light chain 2 slow promoter cooperate in regulating nerve-specific transcription in skeletal muscle. J Biol Chem. 1999;274:12095–12102. doi: 10.1074/jbc.274.17.12095. [DOI] [PubMed] [Google Scholar]
- 21.Francke U. Williams-Beuren syndrome: genes and mechanisms. Hum Mol Genet. 1999;8:1947–1954. doi: 10.1093/hmg/8.10.1947. [DOI] [PubMed] [Google Scholar]
- 22.Franke Y, Peoples R J, Francke U. Identification of GTF2IRD1, a putative transcription factor within the Williams-Beuren syndrome deletion at 7q11.23. Cytogenet Cell Genet. 1999;86:296–304. doi: 10.1159/000015322. [DOI] [PubMed] [Google Scholar]
- 23.Grieshammer U, McGrew M J, Rosenthal N. Role of methylation in maintenance of positionally restricted transgene expression in developing muscle. Development. 1995;121:2245–2253. doi: 10.1242/dev.121.7.2245. [DOI] [PubMed] [Google Scholar]
- 24.Grueneberg D A, Henry R W, Brauer A, Novina C D, Cheriyath V, Roy A L, Gilman M. A multifunctional DNA-binding protein that promotes the formation of serum response factor/homeodomain complexes: identity to TFII-I. Genes Dev. 1997;11:2482–2493. doi: 10.1101/gad.11.19.2482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hallauer P L, Bradshaw H L, Hastings K E. Complex fiber-type-specific expression of fast skeletal muscle troponin I gene constructs in transgenic mice. Development. 1993;119:691–701. doi: 10.1242/dev.119.3.691. [DOI] [PubMed] [Google Scholar]
- 26.Hidaka K, Yamamoto I, Arai Y, Mukai T. The MEF-3 motif is required for MEF-2-mediated skeletal muscle-specific induction of the rat aldolase A gene. Mol Cell Biol. 1993;13:6469–6478. doi: 10.1128/mcb.13.10.6469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Higuchi R. Recombinant PCR. In: Innis M A, Gelfand D H, Sninsky J J, White T J, editors. PCR protocols. A guide to methods and applications. San Diego, Calif: Academic Press; 1990. pp. 177–183. [Google Scholar]
- 28.Hogan B. Manipulating the mouse embryo: a laboratory manual. 2nd ed. Plainview, N.Y: Cold Spring Harbor Laboratory Press; 1994. [Google Scholar]
- 29.Hughes D S, Ontell M. Morphometric analysis of the developing, murine aneural soleus muscle. Dev Dyn. 1992;193:175–184. doi: 10.1002/aja.1001930209. [DOI] [PubMed] [Google Scholar]
- 30.Hughes D S, Schade R R, Ontell M. Ablation of the fetal mouse spinal cord: the effect on soleus muscle cytoarchitecture. Dev Dyn. 1992;193:164–174. doi: 10.1002/aja.1001930208. [DOI] [PubMed] [Google Scholar]
- 31.Hughes S M, Salinas P C. Control of muscle fibre and motoneuron diversification. Curr Opin Neurobiol. 1999;9:54–64. doi: 10.1016/s0959-4388(99)80007-5. [DOI] [PubMed] [Google Scholar]
- 32.Javahery R, Khachi A, Lo K, Zenzie-Gregory B, Smale S T. DNA sequence requirements for transcriptional initiator activity in mammalian cells. Mol Cell Biol. 1994;14:116–127. doi: 10.1128/mcb.14.1.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kim D W, Cheriyath V, Roy A L, Cochran B H. TFII-I enhances activation of the c-fos promoter through interactions with upstream elements. Mol Cell Biol. 1998;18:3310–3320. doi: 10.1128/mcb.18.6.3310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Koppe R I, Hallauer P L, Karpati G, Hastings K E. cDNA clone and expression analysis of rodent fast and slow skeletal muscle troponin I mRNAs. J Biol Chem. 1989;264:14327–14333. [PubMed] [Google Scholar]
- 35.Kozak M. An analysis of 5′-noncoding sequences from 699 vertebrate messenger RNAs. Nucleic Acids Res. 1987;15:8125–8148. doi: 10.1093/nar/15.20.8125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lennon G, Auffray C, Polymeropoulos M, Soares M B. The I.M.A.G.E. consortium: an integrated molecular analysis of genomes and their expression. Genomics. 1996;33:151–152. doi: 10.1006/geno.1996.0177. [DOI] [PubMed] [Google Scholar]
- 37.Lupa-Kimball V A, Esser K A. Use of DNA injection for identification of slow nerve-dependent regions of the MLC2s gene. Am J Physiol. 1998;274:C229–C235. doi: 10.1152/ajpcell.1998.274.1.C229. [DOI] [PubMed] [Google Scholar]
- 38.Mathiesen I. Electropermeabilization of skeletal muscle enhances gene transfer in vivo. Gene Ther. 1999;6:508–514. doi: 10.1038/sj.gt.3300847. [DOI] [PubMed] [Google Scholar]
- 39.McCarthy J J, Vyas D R, Tsika G L, Tsika R W. Segregated regulatory elements direct beta-myosin heavy chain expression in response to altered muscle activity. J Biol Chem. 1999;274:14270–14279. doi: 10.1074/jbc.274.20.14270. [DOI] [PubMed] [Google Scholar]
- 40.Molkentin J D, Olson E N. Combinatorial control of muscle development by basic helix-loop-helix and MADS-box transcription factors. Proc Natl Acad Sci USA. 1996;93:9366–9373. doi: 10.1073/pnas.93.18.9366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Montano M A, Kripke K, Norina C D, Achacoso P, Herzenberg L A, Roy A L, Nolan G P. NF-kappa B homodimer binding within the HIV-1 initiator region and interactions with TFII-I. Proc Natl Acad Sci USA. 1996;93:12376–12381. doi: 10.1073/pnas.93.22.12376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Naidu P S, Ludolph D C, To R Q, Hinterberger T J, Konieczny S F. Myogenin and MEF2 function synergistically to activate the MRF4 promoter during myogenesis. Mol Cell Biol. 1995;15:2707–2718. doi: 10.1128/mcb.15.5.2707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nakayama M, Stauffer J, Cheng J, Banerjee-Basu S, Wawrousek E, Buonanno A. Common core sequences are found in skeletal muscle slow- and fast-fiber-type-specific regulatory elements. Mol Cell Biol. 1996;16:2408–2417. doi: 10.1128/mcb.16.5.2408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.O'Mahoney J V, Guven K L, Lin J, Joya J E, Robinson C S, Wade R P, Hardeman E C. Identification of a novel slow-muscle-fiber enhancer binding protein, MusTRD1. Mol Cell Biol. 1998;18:6641–6652. doi: 10.1128/mcb.18.11.6641. . (Erratum, 20:5361, 2000.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Osborne L R, Campbell T, Daradich A, Scherer S W, Tsui L C. Identification of a putative transcription factor gene (WBSCR11) that is commonly deleted in Williams-Beuren syndrome. Genomics. 1999;57:279–284. doi: 10.1006/geno.1999.5784. [DOI] [PubMed] [Google Scholar]
- 46.Parmacek M S, Ip H S, Jung F, Shen T, Martin J F, Vora A J, Olson E N, Leiden J M. A novel myogenic regulatory circuit controls slow/cardiac troponin C gene transcription in skeletal muscle. Mol Cell Biol. 1994;14:1870–1885. doi: 10.1128/mcb.14.3.1870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pette D, Staron R. Mammalian skeletal muscle fiber type transitions. Int Rev Cytol. 1997;170:143–223. doi: 10.1016/s0074-7696(08)61622-8. [DOI] [PubMed] [Google Scholar]
- 48.Rafuse V F, Milner L D, Landmesser L T. Selective innervation of fast and slow muscle regions during early chick neuromuscular development. J Neurosci. 1996;16:6864–6877. doi: 10.1523/JNEUROSCI.16-21-06864.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Roy A L, Du H, Gregor P D, Novina C D, Martinez E, Roeder R G. Cloning of an inr- and E-box-binding protein, TFII-I, that interacts physically and functionally with USF1. EMBO J. 1997;16:7091–7104. doi: 10.1093/emboj/16.23.7091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Roy A L, Malik S, Meisterernst M, Roeder R G. An alternative pathway for transcription initiation involving TFII-I. Nature. 1993;365:355–359. doi: 10.1038/365355a0. [DOI] [PubMed] [Google Scholar]
- 51.Salminen M, Lopez S, Maire P, Kahn A, Daegelen D. Fast-muscle-specific DNA-protein interactions occurring in vivo at the human aldolase A M promoter are necessary for correct promoter activity in transgenic mice. Mol Cell Biol. 1996;16:76–85. doi: 10.1128/mcb.16.1.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sartorelli V, Webster K A, Kedes L. Muscle-specific expression of the cardiac alpha-actin gene requires MyoD1, CArG-box binding factor, and Sp1. Genes Dev. 1990;4:1811–1822. doi: 10.1101/gad.4.10.1811. [DOI] [PubMed] [Google Scholar]
- 53.Schiaffino S, Gorza S, Ausoni S. Troponin isoform switching in the developing heart and its functional consequences. Trends Cardiovasc Med. 1993;3:12–17. doi: 10.1016/1050-1738(93)90022-X. [DOI] [PubMed] [Google Scholar]
- 54.Seale P, Sabourin L A, Girgis-Gabardo A, Mansouri A, Gruss P, Rudnicki M A. Pax7 is required for the specification of myogenic satellite cells. Cell. 2000;102:777–786. doi: 10.1016/s0092-8674(00)00066-0. [DOI] [PubMed] [Google Scholar]
- 55.Smale S T, Baltimore D. The “initiator” as a transcription control element. Cell. 1989;57:103–113. doi: 10.1016/0092-8674(89)90176-1. [DOI] [PubMed] [Google Scholar]
- 56.Spitz F, Demignon J, Porteu A, Kahn A, Concordet J P, Daegelen D, Maire P. Expression of myogenin during embryogenesis is controlled by Six/sine oculis homeoproteins through a conserved MEF3 binding site. Proc Natl Acad Sci USA. 1998;95:14220–14225. doi: 10.1073/pnas.95.24.14220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Spitz F, Salminen M, Demignon J, Kahn A, Daegelen D, Maire P. A combination of MEF3 and NFI proteins activates transcription in a subset of fast-twitch muscles. Mol Cell Biol. 1997;17:656–666. doi: 10.1128/mcb.17.2.656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Stockdale F E. Mechanisms of formation of muscle fiber types. Cell Struct Funct. 1997;22:37–43. doi: 10.1247/csf.22.37. [DOI] [PubMed] [Google Scholar]
- 59.Stockdale F E. Myogenic cell lineages. Dev Biol. 1992;154:284–298. doi: 10.1016/0012-1606(92)90068-r. [DOI] [PubMed] [Google Scholar]
- 60.Swoap S J, Hunter R B, Stevenson E J, Felton H M, Kansagra N V, Lang J M, Esser K A, Kandarian S C. The calcineurin-NFAT pathway and muscle fiber-type gene expression. Am J Physiol. 2000;279:C915–C924. doi: 10.1152/ajpcell.2000.279.4.C915. [DOI] [PubMed] [Google Scholar]
- 61.Tassabehji M, Carette M, Wilmot C, Donnai D, Read A P, Metcalfe K. A transcription factor involved in skeletal muscle gene expression is deleted in patients with Williams syndrome. Eur J Hum Genet. 1999;7:737–747. doi: 10.1038/sj.ejhg.5200396. [DOI] [PubMed] [Google Scholar]
- 62.Tassabehji M, Metcalfe K, Karmiloff-Smith A, Carette M J, Grant J, Dennis N, Reardon W, Splitt M, Read A P, Donnai D. Williams syndrome: use of chromosomal microdeletions as a tool to dissect cognitive and physical phenotypes. Am J Hum Genet. 1999;64:118–125. doi: 10.1086/302214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Washabaugh C H, Ontell M P, Shan Z, Hoffman E P, Ontell M. Role of the nerve in determining fetal skeletal muscle phenotype. Dev Dyn. 1998;211:177–190. doi: 10.1002/(SICI)1097-0177(199802)211:2<177::AID-AJA6>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- 64.Wentworth B M, Donoghue M, Engert J C, Berglund E B, Rosenthal N. Paired MyoD-binding sites regulate myosin light chain gene expression. Proc Natl Acad Sci USA. 1991;88:1242–1246. doi: 10.1073/pnas.88.4.1242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wicks I P, Howell M L, Hancock T, Kohsaka H, Olee T, Carson D A. Bacterial lipopolysaccharide copurifies with plasmid DNA: implications for animal models and human gene therapy. Hum Gene Ther. 1995;6:317–323. doi: 10.1089/hum.1995.6.3-317. [DOI] [PubMed] [Google Scholar]
- 66.Wilkinson D G, Green J. In situ hybridization and the three-dimensional reconstruction of serial sections. In: Copp A J, Cokroft D L, editors. Postimplantation mammalian embryos. London, United Kingdom: Oxford University Press; 1990. pp. 155–171. [Google Scholar]
- 67.Wilkinson D G, Nieto M A. Detection of messenger RNA by in-situ hybridization to tissue sections and whole mounts. Methods Enzymol. 1993;225:361–373. doi: 10.1016/0076-6879(93)25025-w. [DOI] [PubMed] [Google Scholar]
- 68.Wu H, Naya F J, McKinsey T A, Mercer B, Shelton J M, Chin E R, Simard A R, Michel R N, Bassel-Duby R, Olson E N, Williams R S. MEF2 responds to multiple calcium-regulated signals in the control of skeletal muscle fiber type. EMBO J. 2000;19:1963–1973. doi: 10.1093/emboj/19.9.1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Yan X, Zhao X, Qian M, Guo N, Gong X, Zhu X. Characterization and gene structure of a novel retinoblastoma-protein-associated protein similar to the transcription regulator TFII-I. Biochem J. 2000;345:749–757. [PMC free article] [PubMed] [Google Scholar]
- 70.Yee S P, Rigby P W. The regulation of myogenin gene expression during the embryonic development of the mouse. Genes Dev. 1993;7:1277–1289. doi: 10.1101/gad.7.7a.1277. [DOI] [PubMed] [Google Scholar]
- 71.Yutzey K E, Kline R L, Konieczny S F. An internal regulatory element controls troponin I gene expression. Mol Cell Biol. 1989;9:1397–1405. doi: 10.1128/mcb.9.4.1397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zhu L, Lyons G E, Juhasz O, Joya J E, Hardeman E C, Wade R. Developmental regulation of troponin I isoform genes in striated muscles of transgenic mice. Dev Biol. 1995;169:487–503. doi: 10.1006/dbio.1995.1163. [DOI] [PubMed] [Google Scholar]