Abstract
Expression of the Xist gene, a key player in mammalian X inactivation, has been proposed to be controlled by the antisense Tsix transcript. Targeted deletion of the Tsix promoter encompassing the DPXas34 locus leads to nonrandom inactivation of the mutant X, but it remains unresolved whether this phenotype is caused by loss of Tsix transcription or by deletion of a crucial DNA element. In this study we determined the role of Tsix transcription in random X inactivation by using mouse embryonic stem (ES) cells as a model system. Two approaches were chosen to modulate Tsix transcription with minimal disturbance of genomic sequences. First, Tsix transcription was functionally inhibited by introducing a transcriptional stop signal into the transcribed region of Tsix. In the second approach, an inducible system for Tsix expression was created. We found that the truncation of the Tsix transcript led to complete nonrandom inactivation of the targeted X chromosome. Induction of Tsix transcription during ES cell differentiation, on the other hand, caused the targeted chromosome always to be chosen as the active chromosome. These results for the first time establish a function for antisense transcription in the regulation of X inactivation.
Dosage compensation in mammals is achieved by a unique mechanism that leads to transcriptional silencing of almost all genes present on one of the two X chromosomes in female cells, a process known as X-chromosome inactivation (X inactivation) (24). In undifferentiated cells, all X chromosomes are active and X inactivation is initiated at the onset of differentiation both in vivo and in vitro (31). The number of X chromosomes that are to be inactivated is determined relative to the ploidy of the cell, and X inactivation is initiated only when the number of X chromosomes exceeds one in a diploid nucleus. Each cell then makes the epigenetic choice to keep one X chromosome active and to inactivate all supernumerary X chromosomes (the n – 1 rule, with n the total number of X chromosomes in the cell, and n – 1 the number of X chromosomes that are being inactivated). One model suggests the presence of a blocking factor in limited quantities that can only protect one X chromosome from inactivation per diploid set of chromosomes (2). X-chromosome choice is random in the embryonic lineage of the mouse and other mammals but is influenced by a number of epigenetic and genetic factors. In metatherian mammals, such as kangaroos (8), and in the extraembryonic tissues of some eutherian mammals, including mice (43), the paternal X chromosome is imprinted and always undergoes inactivation. In addition, the choice in embryonic tissues of mice is influenced by the X-controlling element (Xce) such that X inactivation is skewed toward the chromosome possessing the weaker Xce allele (29). It has been shown that at least four alleles exist in mice: Xcea, Xceb, Xcec, and Xced, with Xcea being the weakest and Xced being the strongest allele.
X-autosomal translocations and transgenic studies in mice have defined a master switch required for X inactivation, the Xic (X-inactivation center), a multifunctional locus carrying elements for choosing and silencing (14, 20, 21, 36). One of these elements is the Xist gene, which produces a spliced and polyadenylated untranslated RNA that coats the inactive X chromosome (4, 5, 6, 28). Targeted deletions have demonstrated that Xist is necessary for both the initiation of X inactivation and the silencing of X-linked genes (26, 34), as well as for X-chromosome choice (25). Xist RNA shows a distinctive expression pattern during differentiation as analyzed by fluorescent in situ hybridization (FISH). Prior to the onset of X inactivation, Xist is expressed in an unstable form from every X chromosome in male and female cells. Upon differentiation Xist transcription is downregulated in male cells and on the future active X in female cells. On the future inactive X the Xist transcript is stabilized and accumulates along the chromosome in cis (33, 41).
Recently, a new Xic element, Tsix, has been identified as a potential negative regulator of Xist expression. Tsix is an antisense gene to Xist whose major promoter is located 15 kb downstream of the Xist gene (18). The promoter overlaps with the previously defined DXPas34 locus (18), a 2.4-kb CpG island that is hypermethylated on the active X chromosome (as opposed to other CpG islands on the active X which remain hypomethylated) (9). The primary Tsix transcript completely overlaps Xist and has been shown to cover at least 40 kb (18). Recently, Tsix has been shown to undergo complex processing including alternative splicing and polyadenylation (39). According to this study the Tsix gene contains four exons, one of which (exon 4) overlaps with the highly conserved XCR. However, the functional significance and cellular localization of the spliced transcript remain to be characterized.
Like Xist, Tsix shows a characteristic expression pattern during X inactivation. In undifferentiated ES cells, Tsix is transcribed at low levels from all X chromosomes. After differentiation, Tsix transcription is shut off in adult tissues. Interestingly, at the onset of X inactivation Tsix expression becomes confined to one of the two X's and appears to specifically mark the future active X leading to the hypothesis that Tsix may regulate Xist expression during the initiation of X inactivation (10, 18, 30).
Targeted deletions that encompass the Tsix promoter, as well as the CpG island, were shown to lead to skewed Xist expression and nonrandom inactivation of the mutated X chromosome in ES cells. Introduction of the targeted alleles into mice demonstrated the importance of this locus for imprinted X inactivation in extraembryonic tissues (17, 39). Tsix expression in early blastocysts (3.5 days post coitum) is imprinted oppositely of Xist such that it is transcribed only from the maternal allele. Expression from both alleles is observed in late blastocysts at the time when the imprint is erased and both X's are primed for random X inactivation. Paternal inheritance of the deletion has no effect, but maternal inheritance leads to ectopic expression of maternal Xist in males and biallelic expression from both X chromosomes in female extraembryonic tissues. The majority of the embryos that inherit the deletion from the mother die during development due to placental defects. Lee proposed the presence of an imprinting center within the CpG-rich domain overlapping the Tsix promoter which responds to activating maternal and repressive paternal signaling to facilitate imprinted Tsix expression (17). Analysis of the DXPas34 locus and the Tsix promoter by bisulfite sequencing revealed that methylation does not represent the imprinting mark and is also unlikely to play a role in the regulation of Tsix transcription from this site (35). However, it cannot be excluded that the imprinting is associated with a methylation-independent mechanism. Taken together, in vitro and in vivo data suggest that the Tsix locus and/or transcript influences X-chromosome choice and imprinting of the X chromosome.
A number of genes have been identified whose imprinted expression is associated with the transcription of noncoding RNAs. For example at the H19/Igf2 locus, H19 encodes an untranslated RNA whose expression is imprinted oppositely to Igf2 (32). In many instances these noncoding RNAs are transcribed in the antisense direction, as is the case for the Igf2r gene and its antisense transcript Air (46). Antisense transcripts have also been identified at the UBE3A locus (38) and the Gnas locus (23). However, no evidence for the functionality of the transcripts exists in any of these cases. In fact the only noncoding transcript that has been analyzed for its functionality, H19, has been shown to play no role in the regulation of Igf2 (16, 40).
In the case of Tsix the role of antisense transcription also still remains to be established. Published results relied on targeted deletions that not only abolished Tsix transcription but also eliminated significant portions of genomic sequence such as portions of the DXPas34 locus that might harbor crucial regulatory elements (17, 19, 39).
Here we determined the role of Tsix transcription in random X inactivation by using embryonic stem (ES) cells as a model system for random X inactivation (19, 21, 34, 37). Two approaches were chosen to modulate Tsix transcription with minimal perturbation of genomic sequences. In the first approach, Tsix transcription was functionally inhibited by introducing a transcriptional stop signal into the transcribed region of Tsix. In the second approach, we created an inducible system for Tsix expression. We found that the truncation of the Tsix transcript recapitulates the phenotype of a targeted Tsix deletion, leading to complete nonrandom inactivation of the targeted X chromosome. Induction of Tsix transcription during ES cell differentiation, on the other hand, caused the targeted chromosome always to be chosen as the active chromosome. These results for the first time establish a function for antisense transcription in the regulation of X inactivation.
MATERIALS AND METHODS
Plasmid construction.
The 5-kb SacII-SalI fragment of fragment 2 (cosmid MB4-14A, accession number U41395) cloned into pBluescript (Stratagene), pSacSal, was used for all targeting constructs. This fragment contains a unique HindIII site located 10,959 bp away from the end of the Xist cDNA (4) (GenBank accession number L04961) as an insertion site.
For the stop construct, a 0.65- to 0.1-kb XhoI-HindIII fragment from pPGKneotpAlox2 (42) containing a triple poly(A) signal (tpA) flanked by a loxP site was ligated into the SalI-HindIII-digested vector pSAβgeo (42) containing a splice acceptor (SA). The SAtpA1lox cassette from this vector was then released by XhoI digestion and ligated into a XhoI-digested pSacSal derivative lacking a XhoI site in the polylinker and harboring a XhoI adapter at the unique HindIII site, yielding vector pSS-SAtpA1lox. In order to create a unique restriction site for linearization of the finished construct, an SfiI adapter was ligated into the unique SacII site of this vector. A HygTK selection cassette flanked by two loxP sites was isolated from vector pBS246CMVHygTK (a gift from Chris Wilson and Peggy Lee) lacking the SfiI site 5′ of the selection cassette by NotI-SfiI digestion and inserted in the correct orientation into a unique SmaI site of pSS-SAtpA1lox by blunt end ligation, yielding plasmid pSS-Stop. For electroporation, pSS-Stop was linearized by SfiI digestion.
For the inducible construct, a ClaI adapter was introduced into the unique SalI site of pSacSal, yielding pSSC. This step facilitated linearization of the final construct. A 0.8-kb XhoI-HindIII fragment from pTETOP-H/X (45) was cloned into the unique SfiI site pBS246CMVHygTK-SfiI by blunt-end ligation creating pTetopHygTK. The NotI-XbaI fragment containing the tetracycline-responsive promoter (tetop) and the HygTK selectable marker flanked by loxP sites was then inserted into the unique HindIII site of pSSC by blunt-end ligation, yielding pIndTsix. For electroporation, pIndTsix, was linearized with ClaI.
Cell culture and generation of transgenic ES cells.
Undifferentiated ES cells were grown on mouse embryonic fibroblasts in Dulbecco modified Eagle medium (Gibco), 15% fetal calf serum (HyClone), and 1,000 U of leukemia-inhibiting factor (LIF)/ml. Differentiation of ES cells was induced in medium containing 10% fetal calf serum without LIF and with 40 ng of all-trans-retinoic acid (Sigma)/ml. Doxycycline (Sigma) was added to the medium at a final concentration of 1 μg/ml.
Transgenic ES cells were generated by transfecting cells with 30 μg of linearized plasmid by electroporation by using a Bio-Rad Genepulser set at 25 μF and 400 V. After selection with 140 μg of hygromycin B (Roche)/ml for 6 to 9 days, the colonies were screened by Southern analysis.
For both the stop and the inducible construct, KpnI-digested genomic DNA was probed with the 1-kb probe. A 4.8-kb fragment identified the targeted allele (wild type at 16 kb). To check the integrity of the recombination event of the stop construct, blots were stripped in 0.04 M NaOH and reprobed with the 3′ internal probe. Fifty percent of the male and 6% of the female cells were targeted correctly. The integrity of the recombination event for the inducible construct was confirmed on SmaI-digested DNA with the 3′ internal probe. The correct targeting event was detected in 30% of the male cells and 3% of the female cells.
Cre-mediated recombination was achieved by electroporating targeted cells with 25 μg of supercoiled pOG231 and selecting for the absence of the TK gene in medium containing 2 μM ganciclovir. Resistant clones were analyzed by Southern blotting. For the stop construct, EcoRI-digested DNA was probed with the 5′ internal probe. The wild-type allele yielded a 1.7-kb band, whereas the 1 lox and the 2 lox (stop) alleles (that is, alleles containing one and two loxP sites, respectively) were detected by 1.1- and 1.8-kb bands, respectively. For the inducible construct, XbaI-digested DNA was probed with the 5′ internal probe. The loop-out was confirmed by a 4.4-kb band, compared to the wild type, and the targeted band which gave 3.8- and 7.7-kb bands, respectively.
For this study, two independent clones were chosen for each construct, which behaved similarly in all experiments.
Southern probes.
The 1-kb probe is a 986-bp PCR product specific for a region 2,207 to 3,193 bp downstream of the insertion site (relative to the direction of the Xist gene) and was created by using the primers indF (5′-AAGGCAGGGATTTTAGCGAT-3′) and IndR (5′-TGCAGCCATTCTTTTCTGTG-3′). This probe was specific for a region outside the right arm of the targeting vector. The 3′ internal probe is an 876-bp PCR product specific for a region 1,291 to 2,167 bp downstream of the insertion site (right arm of both constructs) obtained by using primers HS3intF (5′-AATCCGCATCAAAACCAAAG-3′) and HS3intR (5′-GCAGAGCAGAGGTGATGAAA-3′). The 5′ internal probe is a 183-bp PCR product specific for a region 743 to 926 bp upstream of the insertion site (left arm of the construct) created by using primers 5s and 5as (see below).
RT-PCR and primers.
RNA was prepared by using RNAzol B reagent (Teltest) and treated with RNase-free DNase I (Gibco) according to the manufacturer's instructions. Strand-specific reverse transcription-PCR (RT-PCR) was carried out essentially as described previously (18) by using 1 μg of total RNA per reaction. Control reactions that were carried out in the absence of reverse transcriptase were included in all cases. The following primers are described relative to the first bp of Xist cDNA: 1s (positions 272 to 294), 1as (positions 755 to 775), 2s (positions 10162 to 10183), 2sE7 (positions 11011 to 11030), and 2as (positions 11279 to 11308). The following primers are located relative to the end of Xist cDNA: 3s (positions 1131 to 1150), 3as (positions 1553 to 1572), 4s (positions 5118 to 5757), 4as (positions 6142 to 6161), 5s (positions 10033 to 10056), 5as (positions 10197 to 10216), 6s (positions 12250 to 12270), and 6as (positions 12714 to 12734). Primers 2s and 2as were used to amplify a product specific for the spliced Xist transcript (referred to as primer pair 2a). Primers 2sE7 and 2as were used to amplify the Tsix transcript.
Allele specific detection of Xist and Tsix
The NcoI polymorphism in Xist exon 1 has been described previously (19). First-strand synthesis with primers specific for Xist (1as) or Tsix (1s), followed by PCR and detection, was as described previously (19). PCR was carried out for 28 cycles.
The PstI polymorphism is located in Xist exon 7. First-strand synthesis with primers specific for Xist (2as) or Tsix (2sE7) was carried out at 50°C. cDNA was amplified by using Xist specific primer pair 2a or Tsix specific primer pair 2b for 30 cycles (linear range). PCR products were purified through QIAquick columns (Qiagen) and digested with PstI for 3 h. Fragments specific for the 129 and cast alleles were resolved on 2% agarose gels and detected with ethidium bromide.
FISH.
Cells were harvested by trypsinization and allowed to adhere on poly-l-lysine-coated microscope slides (Sigma) for 2 min before fixation in 4% formaldehyde–5% acetic acid–saline for 18 min. FISH was performed as previously described (44) with the following modification. After initial washes in 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) at 37°C, unhybridized RNA was digested with 25 μg of RNase A/ml in 0.5 M NaCl–10 mM Tris (pH 7.5)–0.1% Tween 20 under a coverslip for 45 min at 37°C.
Strand-specific probes were generated by riboprobe synthesis (Promega) by using T3 or T7 polymerase in the presence of biotin-16-UTP (Roche). Antibody detection included three amplification steps with mouse anti-biotin antibodies (Roche), followed by treatment with rhodamine-conjugated donkey anti-mouse and goat anti-horse antibodies (Jackson Laboratories).
We used pBgl5K containing a 5-kb Xist cDNA fragment from exons 4 to 7 in pBluescript (Stratagene) as a template for Tsix- and Xist-specific probes. A template for a probe specific for Tsix exon 4, pTsixE7, was generated by cloning a bp −1615 to +1883 BamHI fragment (relative to the start of Xist cDNA) into pBluescript (Stratagene).
RESULTS
Generation of ES cells expressing a truncated Tsix allele.
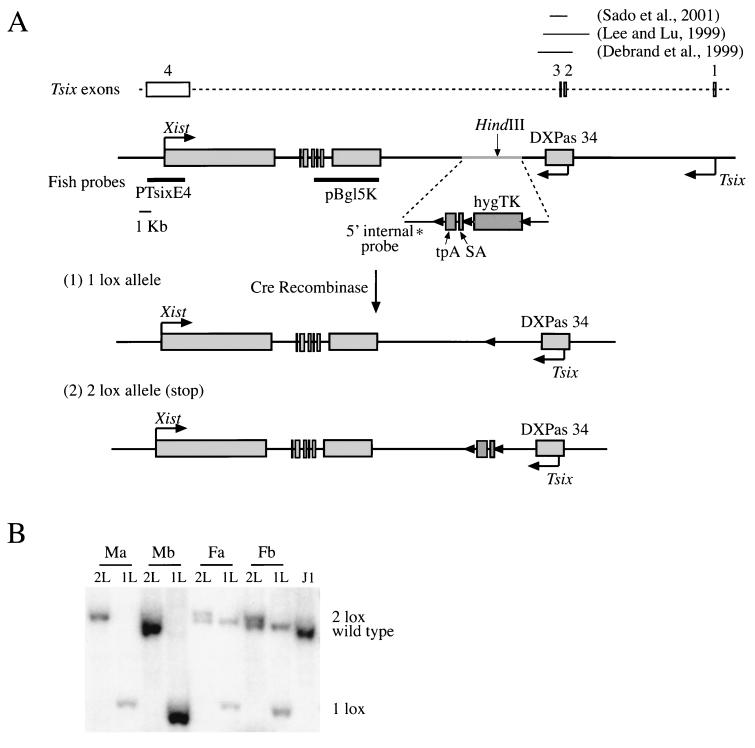
In order to assess the functionality of the Tsix transcript, we created a truncated Tsix allele by introducing a transcriptional stop signal (triple polyadenylation site, i.e., tpA) (42) into a HindIII site 4 kb downstream of the major Tsix transcriptional start site (Fig. 1A). The presence of a strong splice acceptor (i.e., SA) (11) ensured trapping of any transcripts originating from all upstream promoters. In addition, the construct contained a HygroTK selectable marker and three loxP sites. After introduction of the construct into male and female ES cells by homologous recombination, the loxP sites facilitated the generation of two alleles after transient transfection with Cre-recombinase and selection for the loss of the TK gene as follows. (i) A 2 lox allele (stop allele) was created by deleting the selectable marker that was transcribed from the cytomegalovirus promoter. (ii) A 1 lox allele was generated by removal of both the selectable marker and the tpA/SA element. The 1 lox allele served as a control to ensure that effects observed in the presence of the stop allele did not simply arise from physical disturbance of the locus.
FIG. 1.
(A) Scheme for the generation of the truncated Tsix (2 lox) and the control 1 lox alleles (see the text for details). The Tsix intron/exon structure, the two putative promoters, and regions deleted in previous studies (10, 19, 39) are indicated. (B) Southern analysis of targeted male and female cell lines after Cre-mediated recombination. DNA was digested with EcoRI and probed with the 5′ internal probe (indicated in panel A) in order to detect the wild-type (1.7-kb), 1 lox (1.1-kb), and 2 lox (1.8-kb) alleles.
We targeted the X chromosome in the male ES cell line J1 (22) and the 129 allele in the female cell line F1-2.1 (33) that carries a 129 and a Mus castaneus X chromosome. Two independent clones from each cell line were analyzed, and subclones that carried either the 1 lox (male Ma1L Mb1L and female Fa1L Fb1L) or the 2 lox (stop) allele (male Ma2L Mb2L and female Fa2L Fb2L) were generated by transient transfection with Cre-recombinase (Fig. 1B).
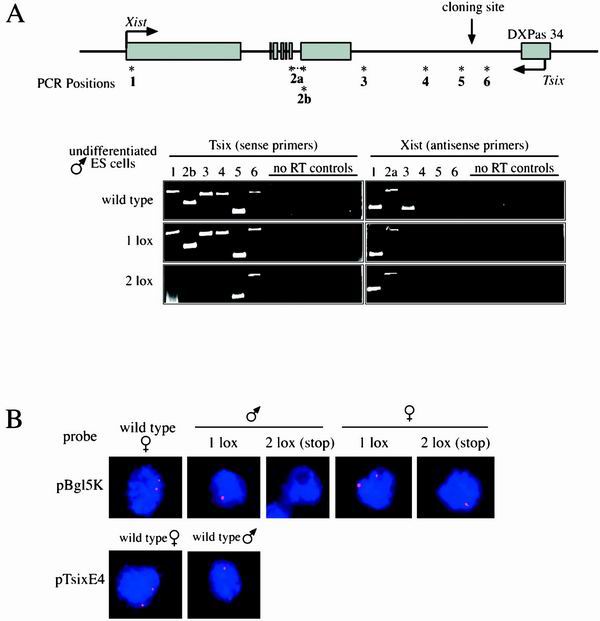
We determined whether the presence of the 2 lox (stop) allele led to the expression of a truncated Tsix transcript. This was particularly important because transcriptional readthrough has been seen even in the presence of an ectopic stop signal (27). Total RNA was prepared from undifferentiated male ES cells that were wild type or carried either the 1 lox or the 2 lox (stop) allele, and strand-specific RT-PCR (30 cycles) was carried out at six sites across the Xist/Tsix locus (Fig. 2A). In wild-type cells, as well as in the 1 lox cell line, the Tsix transcript was detectable with all tested primers (Fig. 2A, left panel), indicating that the presence of the 1 loxP site did not inhibit Tsix transcription. In contrast, in the 2 lox (stop) cell line Tsix RNA could be detected only with primers 5 and 6, which encompass the insertion site. No Tsix signal was detected at the other four positions tested. Primer 6 corresponded to a region upstream of the insertion site, whereas primer 5 was located 927 nucleotides downstream of it, indicating a transcriptional readthrough of at least 1 kb. These results were confirmed by RNA FISH by using a strand-specific probe for a Tsix region downstream of the stop signal (probe pBgl5K, Fig. 1A): male and female 1 lox cells showed a wild-type pattern of Tsix expression, whereas no Tsix signal was detectable in male 2 lox (stop) cells, and only one signal was detectable in female 2 lox (stop) cells (Fig. 2B).
FIG. 2.
Presence of the 2 lox (stop) allele leads to expression of a truncated Tsix transcript, while leaving Xist expression unaffected. The 1 lox allele has no effect on Tsix transcription. (A) Strand-specific RT-PCR for Tsix and Xist was performed on wild-type and targeted undifferentiated male ES cells. The location of primer pairs is indicated as “PCR Positions.” Primer pair 2a spans intron 6 and was used to amplify a product specific for the spliced Xist transcript. Primer pair 2b is located in exon 7 and was used to amplify the Tsix transcript. “No RT controls” refers to reactions carried out in the absence of reverse transcriptase. (B) Strand-specific RNA FISH for Tsix in undifferentiated ES cells. Male and female wild-type and targeted ES cells were probed with riboprobes specific for either the Tsix region overlapping Xist exons 4 to 7 (probe pBgl5K) or Tsix exon 4 (probe pTsixE4). The location of both probes is indicated in Fig. 1A.
Xist transcription was detectable in all three cell lines with primers 1 and 2 by RT-PCR (Fig. 2A, right panel). The signal obtained with primer 3 was variable, which may have resulted from sporadic transcriptional readthrough since this primer corresponded to an area 1 kb downstream of the end of the published Xist cDNA sequence (4) (GenBank accession number L04961).
We conclude that the presence of the 2 lox (stop) allele led to the transcription of a truncated Tsix allele without affecting Xist expression in undifferentiated cells. This effect was reversed by Cre-mediated deletion of the stop cassette.
Tsix transcription is a negative regulator of Xist expression.
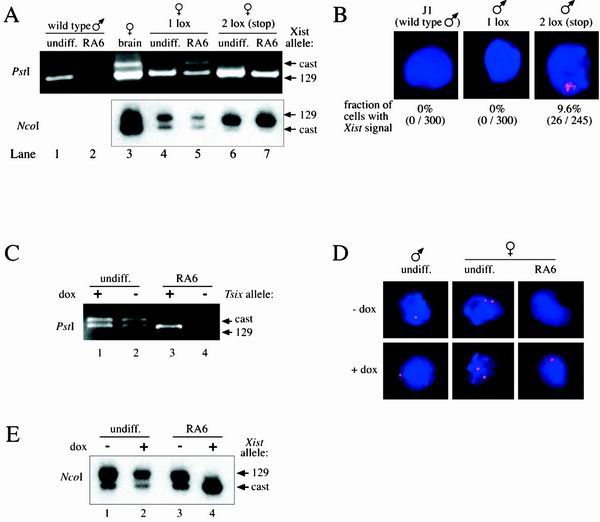
Recent work has shown that deletion of the major Tsix transcriptional start site, including portions of the surrounding CpG island, leads to primary nonrandom inactivation of the mutant X in differentiated female ES cells, as well as in the epiblast lineages of the embryo proper (17, 19, 39). However, it remained unresolved whether Tsix action was mediated by a DNA element that had been deleted in the mutant locus or whether the observed phenotype was caused by loss of the antisense transcript and/or transcription through the region. We reasoned that, if the phenotype was caused by loss of Tsix transcription rather than by deletion of a crucial DNA element, then the 2 lox (stop) allele would lead to skewed X inactivation in differentiated female ES cells. In order to assess this hypothesis, DNA and RNA were isolated from undifferentiated ES cells, as well as from ES cells that were induced to differentiate with all-trans-retinoic acid in the absence of LIF (15) for 6 days. The presence of two X chromosomes of different parental origin in the female cell lines as confirmed by Southern analysis provided a way to analyze allele-specific expression of Xist by strand-specific RT-PCR. Two restriction fragment length polymorphisms in exon 1 (NcoI polymorphism) (19) and exon 7 (PstI polymorphism), respectively, were analyzed (see Fig. 4A). In wild-type male cells, Xist was expressed in undifferentiated cells and undetectable in differentiated male cells (see Fig. 4A, compare lanes 1 and 2), in agreement with Xist expression being silenced in male cells upon differentiation. Control female brain cells showed expression from both Xist alleles (see Fig. 4A, lane 3), because either X chromosome in these cells can be stochastically chosen for X inactivation. However, Xist expression from the 129 allele carrying the lower-strength Xce (Xcea) relative to that of M. castaneus (Xcec) (3) was more abundant. A similar pattern of Xist expression was observed in differentiated 1 lox cells (see Fig. 4A, lane 5) and differentiated wild-type ES cells (data not shown), indicating that random X inactivation was not affected by the presence of the loxP site. In contrast, cells heterozygous for the 2 lox (stop) allele showed complete skewing of Xist expression toward the targeted 129 allele which does not express Tsix (see Fig. 4A, lane 7).
FIG. 4.
Tsix transcription is a negative regulator of Xist expression. (A) Allele-specific RT-PCR for Xist expression was performed on control female brain cells, undifferentiated ES cells (undiff.), and ES cells that were induced for differentiated with all-trans-retinoic acid for 6 days (RA6). PstI indicates a polymorphism in Xist exon 7 that was detected after PstI digestion of a PCR product amplified with primer pair 2a (28 cycles). First-strand synthesis was performed with primer 2as. NcoI indicates a polymorphism in Xist exon 1 that has been described previously (19). Amplification was carried out with the primer pair 1s/as (28 cycles) after first-strand synthesis with primer 1as. No bands were detected in control reactions performed in the absence of reverse transcriptase (not shown). The polymorphic bands are indicated: “129” corresponds to the targeted 129 derived X chromosome, whereas “cast” corresponds to the nontargeted M. castaneus X. (B) Wild-type and targeted male ES cells were induced to differentiate with all-trans-retinoic acid for 2 days. Strand-specific RNA FISH for Xist was performed with a riboprobe specific for Xist exons 4 to 7 (probe pBgl5K, Fig. 1A). The proportion of cells expressing ectopic Xist signals is indicated below each panel. (C) Allele-specific expression of Tsix in the presence (+dox) and absence (−dox) of the inducing agent doxycycline in female ES cells. Cells were either undifferentiated (undiff.) or were differentiated for 6 days in all-trans-retinoic acid (RA6). To detect the PstI polymorphism, strand-specific RT-PCR for Tsix was carried out with primer 2sE7 for first-strand synthesis, followed by amplification with primer pair 2b (30 cycles) and digestion of the PCR product with PstI. “129” corresponds to the Tsix allele derived from the 129 X chromosome that harbors the inducible promoter; “cast” refers to the nontargeted M. castaneus-derived X. (D) Tsix signals in induced cells are indistinguishable from the signals in uninduced and wild-type cells. Cells were either undifferentiated (undiff.) or differentiated with all-trans-retinoic acid for 6 days (RA6) and grown in the presence (+dox) or absence (−dox) of doxycycline. Strand-specific RNA FISH was performed with a riboprobe specific for the Tsix region overlapping Xist exons 4 to 7 (probe pBgl5K, Fig. 1A). (E) Allele-specific expression of Xist in female ES cells. The X chromosome that constitutively expresses Tsix during differentiation is always chosen to remain active. Xist expression was skewed almost completely toward the nontargeted M. castaneus allele (cast) when Tsix was constitutively expressed from the 129 allele (129) (compare lanes 3 [−dox] and 4 [+dox]). In undifferentiated cells induction of Tsix expression had no effect on Xist expression (compare lanes 1 [−dox] and 2 [+dox]). Strand-specific RT-PCR was performed as described above.
The presence of the 2 lox (stop) allele also resulted in increased Xist expression from the targeted allele in undifferentiated cells (see Fig. 4A, lane 6), similar to what had been observed in cells heterozygous for the Tsix promoter deletion (19). In contrast, in undifferentiated 1 lox cells as in wild-type ES cells, Xist was expressed from both alleles (Fig. 4A, lane 4, and data not shown). To examine whether the skewing of Xist expression observed in undifferentiated ES cells was caused by a high proportion of cells in the population that had entered differentiation, Xist expression was examined by RNA FISH by using a strand-specific Xist probe. A Xist cloud characteristically seen only in differentiated cells was detectable only in 6.5% of 2 lox (stop) cells (n = 184) and 8.6% of 1 lox cells (n = 209), suggesting that this effect was specific for undifferentiated cells. It cannot be excluded, however, that the PCR assays were biased toward the more abundant Xist allele. Taken together, these results indicate that loss of Tsix transcription led to skewed Xist expression, and the X chromosome lacking Tsix transcription was always chosen for inactivation.
To assess the effect of loss of Tsix transcription on Xist expression in the absence of a second X chromosome, male cells were differentiated for 2 or 3 days in all-trans-retinoic acid in the absence of LIF, and Xist expression was analyzed by strand-specific RNA FISH (Fig. 4B). In wild-type and 1 lox cells Xist accumulation was never detected (n > 300). In contrast, ectopic high-level Xist expression was found in 9.6% (n = 271) of 2 lox (stop) cells on day 2 of differentiation and in 9.5% (n = 232) on day 3.
These results indicate that Tsix transcription acts as a negative regulator of Xist expression, resulting in skewed Xist expression toward the chromosome lacking Tsix transcription in female cells and stochastic upregulation of Xist expression in male cells upon differentiation.
Generation of an inducible Tsix allele.
To generate an experimental system that allows regulated expression of Tsix RNA in response to doxycycline addition to the medium, we inserted a tetracycline-responsive promoter (12) into the HindIII site analogous to the insertion site of the stop construct (tetop, Fig. 3A). In addition, the construct contained a HygroTK selectable marker flanked by loxP sites. By using homologous recombination, the construct was introduced into a male (J1) ES cell line and into the 129 Xist allele of a female (F1-2.1) ES cell line carrying the nls-rtTA cDNA encoding a doxycycline-controlled transcriptional activator at the ubiquitously expressed ROSA 26 locus (45). Targeted male and female cell lines were transfected with Cre-recombinase and selected for loss of the TK gene. Male lines M.ind1a and M.ind1b and female lines F.ind1a and F.ind1b containing the inducible promoter were identified by Southern blotting (Fig. 3B).
FIG. 3.
(A) Strategy for the generation of an inducible Tsix allele (see the text for details). (B) Southern analysis of targeted male and female cell lines after Cre-mediated recombination. XbaI-digested DNA was probed with the 5′ internal probe (indicated in Fig. 1A). The wild-type allele gave a 3.8-kb band compared to the 1 lox (loop-out, 4.4-kb) and 2 lox (no loop-out, 7.7-kb) alleles.
Tsix expression was assessed in cells that were grown in the presence or absence of doxycyline. Strand-specific RT-PCR and RNA FISH analysis revealed that Tsix was expressed at similar levels from both X chromosomes in uninduced and induced female cells prior to differentiation (Fig. 4C, lanes 1 and 2; Fig. 4D, middle panel). Doxycycline treatment of undifferentiated male cells did not result in an increased Tsix signal, as detected by strand-specific RNA FISH (Fig. 4D, left panel), suggesting that it might be impossible to induce Tsix expression above the physiological level in undifferentiated cells. Alternatively, it is possible that elevated Tsix levels were not detectable due to the instability of the transcript. In contrast, when cells were differentiated in the presence, but not in the absence, of doxycycline for 6 days, Tsix was expressed from the targeted 129 allele at a level that was comparable to that in undifferentiated cells (Fig. 4C, lanes 3 and 4; Fig. 4D, right panel). These results indicate that the presence of doxycycline does not lead to detectable overexpression of Tsix in undifferentiated cell. However, when cells were differentiated in the presence of doxycyline, Tsix expression was clearly induced.
Induction of Tsix transcription prevents the inactivation of an X chromosome in cis.
We have shown that the X chromosome lacking Tsix transcription was always chosen for X inactivation. If the role of Tsix transcription was to block the accumulation of Xist, then the overexpression of Tsix should protect the X chromosome in cis from inactivation. To test this hypothesis, we compared Xist expression patterns from cells that were grown in the presence or absence of doxycycline. In both induced and uninduced undifferentiated cells, Xist was expressed from both alleles (Fig. 4E, lanes 1 and 2). This result was in agreement with the finding that Tsix overexpression from the 129 allele was not detectable in undifferentiated cells (Fig. 4C and D).
In contrast, biased X inactivation was observed when cells were differentiated in the presence of doxycycline for 6 days (Fig. 4E, compare lanes 3 and 4). Ectopic Tsix transcription from the induced 129 allele led to almost exclusive upregulation of Xist expression from the noninduced cast allele (>90% as quantified by phosphorimaging). Background expression from the 129 allele was probably attributable to cells that had initiated differentiation and undergone random X inactivation before the addition of doxycycline. In agreement with this, an Xist cloud was detectable in 6 to 10% of cells before the induction of differentiation by strand-specific RNA FISH. The presence of both X chromosomes in these cells was confirmed by Southern blotting (data not shown). These results demonstrate that an X chromosome that ectopically expresses Tsix RNA is always chosen to remain active.
DISCUSSION
In this study, we generated two Tsix alleles in order to assess the role of Tsix transcription in the regulation of X-chromosome choice during X inactivation. We used, as a model system, ES cells which are known to undergo random X inactivation when induced to differentiate. In the first approach, a transcriptional stop signal was introduced into the transcribed region of Tsix flanked by a strong splice acceptor. The splice acceptor ensured trapping of all transcripts arising from upstream promoters, including the major promoter initially identified by Lee et al. (18) and the minor promoter recently characterized by Sado et al. (39). In the second approach, we created a doxycycline-inducible allele that enabled us to ectopically activate Tsix expression in a regulated manner. The objective was to functionally modify Tsix expression, while leaving potentially important regulatory DNA elements intact. This approach differs from previously published studies in which targeted deletions of Tsix not only abolished Tsix transcription but, in addition, deleted major parts of the DXPas34 locus (10, 19, 39), a CpG island that has been shown to be differentially methylated on the active and inactive X chromosome (9).
The results described here clearly establish for the first time the importance of active transcription for Tsix function. Loss of Tsix transcription leads to nonrandom inactivation of the X chromosome, with the mutant X being always inactivated (Fig. 4A), whereas constitutive expression of Tsix causes the mutant X chromosome to remain active (Fig. 4E). This unequivocally demonstrates that Tsix transcription through the Xist locus is crucial for preventing X-chromosome inactivation and confirms the role of Tsix as a negative regulator of Xist.
We found, in agreement with this, that the loss of Tsix transcription led to ectopic Xist expression in a subset of differentiating male cells (Fig. 4B), an effect that was specific for differentiating cells since Xist accumulation is detectable only in ca. 1% (n = 367) of undifferentiated cells. These results differ from previous studies that found ectopic Xist expression to be restricted to cells of extraembryonic tissues (17, 19, 39). In these tissues X inactivation is normally imprinted such that expression from the maternal Xist allele is silenced. Deletion of the Tsix promoter/DXPas34 locus erased this imprint, leading to ectopic Xist expression from the maternal allele in addition to upregulation of Xist expression from the paternal allele. However, it is possible that ectopic Xist upregulation still occurred in the epiblast lineage of these embryos. Cells that undergo aberrant dosage compensation are not viable and are progressively lost from the population. Since our findings suggest that only a small fraction of cells of the epiblast lineage are affected, the loss of these cells may not have an obvious effect on the overall cell population. Interestingly, Sado et al. (39) did observe Xist accumulation in a subset of mutant male ES cells grown under nondifferentiating conditions, a finding consistent with these cells representing a subpopulation that had already entered differentiation and initiated stochastic upregulation of Xist expression.
In addition, the phenotype described here differs from that of a 65-kb knockout that included the Tsix promoter (7). This knockout also resulted in nonrandom X inactivation of the deleted X in female ES cells. However, in cells carrying only one intact X chromosome (X0), ectopic Xist upregulation was observed in virtually all of the cells. In contrast, we see ectopic expression only in a subset of cells. An explanation for this difference may be provided by a model proposed by Lee and Lu (19). This model suggests that Xist expression is regulated by a complex interplay of positive and negative factors. A blocking factor, present in limited amount, may mark one X chromosome per diploid set of autosomes as the future active X. Our work indicates that this blocking factor most likely acts by regulating Tsix transcription and may perhaps consist of a complex of autosomal and X-linked factors that bind to the Tsix promoter. In addition, Lee and Lu suggest the presence of competence factors that are required to initiate Xist upregulation on the future inactive X once the active X is chosen. These competence factors are thought to be expressed only in cells that need to undergo dosage compensation, i.e., in cells that carry more than one X per diploid set of autosomes. Therefore, loss of Tsix transcription as the target of the blocking factor would not be sufficient to induce upregulation of Xist expression in male cells. However, the observation that stochastic ectopic Xist expression is still observed in some cells may indicate that competence factors are not strictly limited to cells that carry more than one X chromosome. Male cells might simply be less competent to upregulate Xist expression in the absence of the blocking factor because they are not poised for X inactivation and might therefore provide a less-permissive environment for Xist upregulation and stabilization.
Recent findings suggest that Tsix consists of four exons and is subject to complex alternative splicing (39). We found that the localization of these spliced forms most likely overlaps with the localization of the unspliced transcript, as indicated by FISH with a strand-specific probe specific for the common exon 4 (probe pTsixE4; Fig. 2B). In our study, both the transcriptional stop signal and the inducible promoter were inserted downstream of exons 1 through 3, leaving exon 4 as the only exon whose expression was modified. In view of our results two types of models for the mechanism of Tsix action can be proposed. (i) Tsix function could be mediated by a functional RNA. According to this model, exon 4 would be the only exon that is necessary and sufficient for Tsix function. Tsix RNA may interact directly with Xist DNA or local chromatin to interfere with Xist transcription. Alternatively, the formation of an Xist/Tsix RNA duplex may trigger RNA degradation or mask functional domains in Xist RNA. Interestingly, exon 4 overlaps with the Xist transcription unit and the XCR, a conserved region that is crucial for Xist function (1). (ii) It is possible that transcriptional activity of the locus per se, rather than the RNA itself, is important for function so that the processed form of Tsix would play only a minor role in the regulation of Xist expression. Antisense transcription might, for example, be involved in regulating the chromatin domain around the Xist locus. This mechanism has been shown to be crucial for the expression of the human β-globin gene (13), where intergenic transcription was found to be required for the remodeling of chromatin structure to allow expression of this locus in a developmentally regulated manner. Similarly, Tsix transcription may directly modulate the chromatin structure through the Xist to allow access of developmentally regulated factors. Alternatively, transcriptional interference between Tsix and Xist might inhibit efficient transcription of the Xist gene, thereby preventing Xist accumulation.
It will be interesting to see by what mechanism Tsix transcription regulates Xist expression. In addition, upstream factors that control Tsix expression and therefore constitute the potential blocking factor still remain to be identified
ACKNOWLEDGMENTS
We thank Nicki Watson for help with microscopy; Györgyi Csankovszki, Joost Gribnau, Caroline Beard, and Ted Rasmussen for discussions and critical reading of the manuscript.
This work was conducted at the W. M. Keck Foundation biological imaging facility at the Whitehead Institute and was supported by NIH grants CA44339 and CA87869 to R.J.
REFERENCES
- 1.Allaman-Pillet N, Djemai A, Bonny C, Schorderet D F. The 5′ repeat elements of the mouse Xist gene inhibit the transcription of X-linked genes. Gene Expr. 2000;9:93–101. doi: 10.3727/000000001783992632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Avner P, Heard E. X-chromosome inactivation: counting, choice and initiation. Nat Rev Genet. 2001;2:59–67. doi: 10.1038/35047580. [DOI] [PubMed] [Google Scholar]
- 3.Avner P, Prissette M, Arnaud D, Courtier B, Cecchi C, Heard E. Molecular correlates of the murine Xce locus. Genet Res. 1998;72:217–224. doi: 10.1017/s0016672398003516. [DOI] [PubMed] [Google Scholar]
- 4.Brockdorff N, Ashworth A, Kay G F, McCabe V M, Norris D P, Cooper P J, Swift S, Rastan S. The product of the mouse Xist gene is a 15 kb inactive X-specific transcript containing no conserved ORF and located in the nucleus. Cell. 1992;71:515–526. doi: 10.1016/0092-8674(92)90519-i. [DOI] [PubMed] [Google Scholar]
- 5.Brown C J, Hendrich B D, Rupert J L, Lafreniere R G, Xing Y, Lawrence J, Willard H F. The human XIST gene: analysis of a 17-kb inactive X-specific RNA that contains conserved repeats and is highly localized within the nucleus. Cell. 1992;71:527–542. doi: 10.1016/0092-8674(92)90520-m. [DOI] [PubMed] [Google Scholar]
- 6.Clemson C M, McNeil J A, Willard H F, Lawrence J B. XIST RNA paints the inactive X chromosome at interphase: evidence for a novel RNA involved in nuclear/chromosome structure. J Cell Biol. 1996;132:259–275. doi: 10.1083/jcb.132.3.259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Clerc P, Avner P. Role of the region 3′ to Xist exon 6 in the counting process of X-chromosome inactivation. Nat Genet. 1998;19:249–253. doi: 10.1038/924. [DOI] [PubMed] [Google Scholar]
- 8.Cooper D W, VandeBerg J L, Sharman G B, Poole W E. Phosphoglycerate kinase polymorphism in kangaroos provides further evidence for paternal X inactivation. Nat New Biol. 1971;230:155–157. doi: 10.1038/newbio230155a0. [DOI] [PubMed] [Google Scholar]
- 9.Courtier B, Heard E, Avner P. Xce haplotypes show modified methylation in a region of the active X chromosome lying 3′ to Xist. Proc Natl Acad Sci USA. 1995;92:3531–3535. doi: 10.1073/pnas.92.8.3531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Debrand E, Chureau C, Arnaud D, Avner P, Heard E. Functional analysis of the DXPas34 locus, a 3′ regulator of Xist expression. Mol Cell Biol. 1999;19:8513–8525. doi: 10.1128/mcb.19.12.8513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Friedrich G, Soriano P. Promoter traps in embryonic stem cells: a genetic screen to identify and mutate developmental genes in mice. Genes Dev. 1991;5:1513–1523. doi: 10.1101/gad.5.9.1513. [DOI] [PubMed] [Google Scholar]
- 12.Gossen M, Freundlieb S, Bender G, Muller G, Hillen W, Bujard H. Transcriptional activation by tetracyclines in mammalian cells. Science. 1995;268:1766–1769. doi: 10.1126/science.7792603. [DOI] [PubMed] [Google Scholar]
- 13.Gribnau J, Diderich K, Pruzina S, Calzolari R, Fraser P. Intergenic transcription and developmental remodeling of chromatin subdomains in the human β-globin locus. Mol Cell. 2000;5:377–386. doi: 10.1016/s1097-2765(00)80432-3. [DOI] [PubMed] [Google Scholar]
- 14.Herzing L B, Romer J T, Horn J M, Ashworth A. Xist has properties of the X-chromosome inactivation centre. Nature. 1997;386:272–275. doi: 10.1038/386272a0. [DOI] [PubMed] [Google Scholar]
- 15.Hogan B, Beddington R, Costantini F, Lacy F. Manipulating the mouse embryo: a laboratory manual. Cold Spring Habor, N.Y: Cold Spring Habor Laboratory Press; 1994. [Google Scholar]
- 16.Jones B K, Levorse J M, Tilghman S M. Igf2 imprinting does not require its own DNA methylation or H19 RNA. Genes Dev. 1998;12:2200–2207. doi: 10.1101/gad.12.14.2200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee J T. Disruption of imprinted X inactivation by parent-of-origin effects at Tsix. Cell. 2000;103:17–27. doi: 10.1016/s0092-8674(00)00101-x. [DOI] [PubMed] [Google Scholar]
- 18.Lee J T, Davidow L S, Warshawsky D. Tsix, a gene antisense to Xist at the X-inactivation centre. Nat Genet. 1999;21:400–404. doi: 10.1038/7734. [DOI] [PubMed] [Google Scholar]
- 19.Lee J T, Lu N. Targeted mutagenesis of Tsix leads to nonrandom X inactivation. Cell. 1999;99:47–57. doi: 10.1016/s0092-8674(00)80061-6. [DOI] [PubMed] [Google Scholar]
- 20.Lee J T, Lu N, Han Y. Genetic analysis of the mouse X inactivation center defines an 80-kb multifunction domain. Proc Natl Acad Sci USA. 1999;96:3836–3841. doi: 10.1073/pnas.96.7.3836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee J T, Strauss W M, Dausman J A, Jaenisch R. A 450-kb transgene displays properties of the mammalian X-inactivation center. Cell. 1996;86:83–94. doi: 10.1016/s0092-8674(00)80079-3. [DOI] [PubMed] [Google Scholar]
- 22.Li E, Bestor T H, Jaenisch R. Targeted mutation of the DNA methyltransferase gene results in embryonic lethality. Cell. 1992;69:915–926. doi: 10.1016/0092-8674(92)90611-f. [DOI] [PubMed] [Google Scholar]
- 23.Li T, Vu T H, Zeng Z L, Nguyen B T, Hayward B E, Bonthron D T, Hu J F, Hoffman A R. Tissue-specific expression of antisense and sense transcripts at the imprinted Gnas locus. Genomics. 2000;69:295–304. doi: 10.1006/geno.2000.6337. [DOI] [PubMed] [Google Scholar]
- 24.Lyon M F. Gene action in the X-chromosome of the mouse (Mus musculus L.) Nature. 1961;190:372–373. doi: 10.1038/190372a0. [DOI] [PubMed] [Google Scholar]
- 25.Marahrens Y, Loring J, Jaenisch R. Role of the Xist gene in X chromosome choosing. Cell. 1998;92:657–664. doi: 10.1016/s0092-8674(00)81133-2. [DOI] [PubMed] [Google Scholar]
- 26.Marahrens Y, Panning B, Dausman J, Strauss W, Jaenisch R. Xist-deficient mice are defective in dosage compensation but not spermatogenesis. Genes Dev. 1997;11:156–166. doi: 10.1101/gad.11.2.156. [DOI] [PubMed] [Google Scholar]
- 27.Maxwell I H, Harrison G S, Wood W M, Maxwell F. A DNA cassette containing a trimerized SV40 polyadenylation signal which efficiently blocks spurious plasmid-initiated transcription. BioTechniques. 1989;7:276–280. [PubMed] [Google Scholar]
- 28.Memili E, Hong Y K, Kim D H, Ontiveros S D, Strauss W M. Murine Xist RNA isoforms are different at their 3′ ends: a role for differential polyadenylation. Gene. 2001;266:131–137. doi: 10.1016/s0378-1119(01)00353-5. [DOI] [PubMed] [Google Scholar]
- 29.Migeon B R. X-chromosome inactivation: molecular mechanisms and genetic consequences. Trends Genet. 1994;10:230–235. doi: 10.1016/0168-9525(94)90169-4. [DOI] [PubMed] [Google Scholar]
- 30.Mise N, Goto Y, Nakajima N, Takagi N. Molecular cloning of antisense transcripts of the mouse Xist gene. Biochem Biophys Res Commun. 1999;258:537–541. doi: 10.1006/bbrc.1999.0681. [DOI] [PubMed] [Google Scholar]
- 31.Monk M, Harper M I. Sequential X chromosome inactivation coupled with cellular differentiation in early mouse embryos. Nature. 1979;281:311–313. doi: 10.1038/281311a0. [DOI] [PubMed] [Google Scholar]
- 32.Moore T, Constancia M, Zubair M, Bailleul B, Feil R, Sasaki H, Reik W. Multiple imprinted sense and antisense transcripts, differential methylation and tandem repeats in a putative imprinting control region upstream of mouse Igf2. Proc Natl Acad Sci USA. 1997;94:12509–12514. doi: 10.1073/pnas.94.23.12509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Panning B, Dausman J, Jaenisch R. X chromosome inactivation is mediated by Xist RNA stabilization. Cell. 1997;90:907–916. doi: 10.1016/s0092-8674(00)80355-4. [DOI] [PubMed] [Google Scholar]
- 34.Penny G D, Kay G F, Sheardown S A, Rastan S, Brockdorff N. Requirement for Xist in X chromosome inactivation. Nature. 1996;379:131–137. doi: 10.1038/379131a0. [DOI] [PubMed] [Google Scholar]
- 35.Prissette M, El-Maarri O, Arnaud D, Walter J, Avner P. Methylation profiles of DXPas34 during the onset of X-inactivation. Hum Mol Genet. 2001;10:31–38. doi: 10.1093/hmg/10.1.31. [DOI] [PubMed] [Google Scholar]
- 36.Rastan S, Brown S D. The search for the mouse X-chromosome inactivation centre. Genet Res. 1990;56:99–106. doi: 10.1017/s0016672300035163. [DOI] [PubMed] [Google Scholar]
- 37.Rastan S, Robertson E J. X-chromosome deletions in embryo-derived (EK) cell lines associated with lack of X-chromosome inactivation. J Embryol Exp Morphol. 1985;90:379–388. [PubMed] [Google Scholar]
- 38.Rougeulle C, Cardoso C, Fontes M, Colleaux L, Lalande M. An imprinted antisense RNA overlaps UBE3A and a second maternally expressed transcript. Nat Genet. 1998;19:15–16. doi: 10.1038/ng0598-15. [DOI] [PubMed] [Google Scholar]
- 39.Sado T, Wang Z, Sasaki H, Li E. Regulation of imprinted X-chromosome inactivation in mice by Tsix. Development. 2001;128:1275–1286. doi: 10.1242/dev.128.8.1275. [DOI] [PubMed] [Google Scholar]
- 40.Schmidt J V, Levorse J M, Tilghman S M. Enhancer competition between H19 and Igf2 does not mediate their imprinting. Proc Natl Acad Sci USA. 1999;96:9733–9738. doi: 10.1073/pnas.96.17.9733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sheardown S A, Duthie S M, Johnston C M, Newall A E, Formstone E J, Arkell R M, Nesterova T B, Alghisi G C, Rastan S, Brockdorff N. Stabilization of Xist RNA mediates initiation of X chromosome inactivation. Cell. 1997;91:99–107. doi: 10.1016/s0092-8674(01)80012-x. [DOI] [PubMed] [Google Scholar]
- 42.Soriano P. Generalized lacZ expression with the ROSA26 Cre reporter strain. Nat Genet. 1999;21:70–71. doi: 10.1038/5007. [DOI] [PubMed] [Google Scholar]
- 43.Takagi N, Sasaki M. Preferential inactivation of the paternally derived X chromosome in the extraembryonic membranes of the mouse. Nature. 1975;256:640–642. doi: 10.1038/256640a0. [DOI] [PubMed] [Google Scholar]
- 44.Wijgerde M, Grosveld F, Fraser P. Transcription complex stability and chromatin dynamics in vivo. Nature. 1995;377:209–213. doi: 10.1038/377209a0. [DOI] [PubMed] [Google Scholar]
- 45.Wutz A, Jaenisch R. A shift from reversible to irreversible X inactivation is triggered during ES cell differentiation. Mol Cell. 2000;5:695–705. doi: 10.1016/s1097-2765(00)80248-8. [DOI] [PubMed] [Google Scholar]
- 46.Wutz A, Smrzka O W, Schweifer N, Schellander K, Wagner E F, Barlow D P. Imprinted expression of the Igf2r gene depends on an intronic CpG island. Nature. 1997;389:745–749. doi: 10.1038/39631. [DOI] [PubMed] [Google Scholar]