Abstract
The growth inhibitory functions of p53 are controlled in unstressed cells by rapid degradation of the p53 protein. One of the principal regulators of p53 stability is MDM2, a RING finger protein that functions as an E3 ligase to ubiquitinate p53. MDM2 promotes p53 nuclear export, and in this study, we show that ubiquitination of the C terminus of p53 by MDM2 contributes to the efficient export of p53 from the nucleus to the cytoplasm. In contrast, MDM2 did not promote nuclear export of the p53-related protein, p73. p53 nuclear export was enhanced by overexpression of the export receptor CRM1, although no significant relocalization of MDM2 was seen in response to CRM1. However, nuclear export driven by CRM1 overexpression did not result in the degradation of p53, and nuclear export was not essential for p53 degradation. These results indicate that MDM2 mediated ubiquitination of p53 contributes to both nuclear export and degradation of p53 but that these activities are not absolutely dependent on each other.
The p53 tumor suppressor protein plays an important role in preventing malignant development, and p53 function is lost or compromised in most human cancers (37). One of the principal functions of p53 is to inhibit cell growth, and p53 shows strong cell cycle arrest and apoptotic activities (43). While these functions play an important role in preventing the growth of abnormal or damaged cells, p53 activity must be tightly regulated in normal tissue to allow growth and development. One of the principal regulators of p53 is the MDM2 protein (26), and loss of MDM2 results in early embryonic lethality associated with deregulated p53-mediated apoptosis (10). MDM2 expression is transcriptionally activated by p53, establishing a feedback loop in which p53 controls expression of its own negative regulator (4, 45).
MDM2 shows several functions that contribute to the inhibition of p53 activity. p53 is a transcription factor, and the activation of cell cycle arrest and apoptotic responses to p53 are dependent, at least in part, on the expression of p53 target genes (43). The p53 protein contains domains for sequence-specific DNA binding and an N-terminal transactivation domain that forms direct contacts with a number of proteins that are involved in transcriptional control (22). Since the MDM2 binding site is also within the N-terminal region of p53 (7), one of the consequences of MDM2 binding is to inhibit p53-mediated transcriptional activity by blocking the p53-transcriptional coactivator interactions (31, 33, 44). This effect may be further enhanced by MDM2-mediated inhibition of the acetylation of p53 by factors such as p300 (21, 23) and an ability of MDM2 to function directly as a transcriptional repressor (42).
Another function of MDM2 that efficiently abolishes all p53 activity is the ability of MDM2 to target p53 for degradation through the ubiquitin-dependent proteasome pathway (17, 24). Interaction between MDM2 and p53 leads to the ubiquitination and degradation of p53, and this is likely to play a key role in maintaining p53 at low levels in normal cells. Inhibition of MDM2 in response to stress leads to the rapid stabilization of the p53 protein and activation of the p53 response (2). MDM2 has been shown to function as a ubiquitin ligase for p53 (11, 19, 30), and the ability of MDM2 to directly conjugate ubiquitin onto p53 potentially allows p53 to be recognized by the proteasome for degradation. However, the role of MDM2 in regulating p53 appears to be more complex, and there is evidence that MDM2 function is also required for the efficient export of p53 from the nucleus to the cytoplasm (13). The ability of MDM2 to export from the nucleus to the cytoplasm may contribute to the export of p53 under some circumstances (36). However, recent reports have shown that the ability of MDM2 to function as a ubiquitin ligase, rather than its ability to shuttle to the cytoplasm, is critical in regulating p53 nuclear export (5, 14), although the target of MDM2 ubiquitination in regulation of nuclear export was not defined. p53 contains two nuclear export sequences (NES)—one in the N terminus and one in the C terminus of the protein (40, 49). Oligomerization of p53, which occurs through the C-terminal domain, obscures the C-terminal NES and inhibits nuclear export (40). It is therefore possible that the ubiquitination of p53 within this C-terminal region reveals the NES and allows p53 export. In support of this model, we show here that mutation of the six lysine residues in the C terminus of p53 that are targeted for ubiquitination by MDM2 results in a p53 protein that is defective for MDM2 driven nuclear export. By contrast, the p53-related protein p73 was not exported by MDM2. Nuclear export of p53 is augmented by ectopic expression of the export protein CRM1, which enhances the activity of both N-terminal and C-terminal NES in p53. However, CRM1-mediated nuclear export was not accompanied by enhanced degradation of p53, and nuclear export of p53 was not absolutely required for degradation of p53. These results indicate that ubiquitination of p53 by MDM2 can contribute to two independent responses, the nuclear export of p53 and the targeting of p53 to the proteasome.
MATERIALS AND METHODS
Plasmids and cell culture.
Plasmids encoding human wild-type p53, p53K3I, p53NES, human wild-type MDM2, human wild-type p73α, p73β, and green fluorescent protein (GFP)-CRM1 have been described previously (6, 8, 9, 40, 51). p53K3R was constructed using site-directed mutagenesis as described previously (29) to exchange lysines 370, 372, and 373 to arginine. p53K6I and p53K6R were generated using site-directed mutagenesis, changing lysines 370, 372, 373, 381, 382, and 386 to isoleucine and arginine, respectively.
p73ΔI mutants (amino acids 9 to 22 deleted) were generated by PCR in two steps. First, using cDNA3-HAp73 as a template, the DNA coding for the N terminus of the protein was amplified with 5′ phosphorylated primer pGGAGGTGGCGGTGGACTG and a vector-specific T7 primer. The C-terminal part of the p73 coding sequence was synthesized with primer pGAACCAGACAGCACCTACTTC and a vector-specific SP6 primer. The two parts were ligated, and the product was used as a template for the amplification with vector-specific primers. The final PCR product was subcloned and sequenced on both strands.
Wild-type p53 expressing human U2OS cells, p53 null human H1299, and p53/MDM2 double null mouse embryo fibroblasts (MEFs) were maintained in Dulbecco modified Eagle medium supplemented with 10% fetal calf serum at 37°C. Cells were transfected using calcium phosphate coprecipitation and harvested for protein analysis 24 h posttransfection. To monitor for equal transfection efficiency and protein loading, 1 μg of pEGFP-N1 (Clontech) was included in each transfection.
In vitro ubiquitination assay.
In vitro ubiquitin modification of p53 was conducted essentially as previously described (11). In brief, recombinant glutathione S-transferase (GST)-MDM2 expressed in Escherichia coli cells was purified on glutathione-Sepharose, mixed with in vitro-translated p53 (10,000 cpm), and incubated at 4°C for 1 h. The beads were washed three times in reaction buffer and incubated with bacterial recombinant UbcH5B (200 ng) rabbit E1 (Calbiochem) (50 ng), and His6-Ubiquitin (Calbiochem)(1 μg) in 30 μl of reaction buffer. The reaction was stopped after 60 min at 30°C by the addition of sodium dodecyl sulfate (SDS) sample buffer. Reaction products were resolved and visualized by SDS–8% polyacrylamide gel electrophoresis and autoradiography.
Immunofluorescence.
U2OS or H1299 cells were plated on 10-cm2 dishes (106 cells) containing 1-cm diameter glass coverslips and transfected as described. Twenty-four hours after transfection, cells on the coverslips were washed three times with phosphate-buffered saline (PBS), and then fixed in 4% paraformaldehyde for 10 min at room temperature. After fixation, cells were washed in PBS three times and then permeabilized in ice-cold PBS containing 0.2% Triton X-100 for 5 min. Cells were blocked in PBS containing 0.5% bovine serum albumin at room temperature for 30 min and then incubated overnight at 4°C with anti-p53 CM-1 (Novocastra Sciences), anti-HA (Santa Cruz), or anti-MDM2 AB1 (Oncogene Science) antibody in blocking solution respectively. Cells were washed three times with PBS and incubated for 1 h at room temperature with fluorescein isothiocyanate (FITC-conjugated rabbit anti-mouse antibody (1:100; DAKO), FITC-conjugated donkey anti-rabbit antibody (1:100; Amersham), Cy3-conjugated donkey anti-rabbit antibody (1:500; Jackson ImmunoResearch), Cy3-conjugated donkey anti-mouse antibody (1:500; Jackson ImmunoResearch), Oregon-green conjugated goat anti-mouse antibody (1:500; Molecular Probes), or Oregon-green conjugated goat anti-rabbit antibody (1:500; Molecular Probes) in blocking solution, containing 1 μg of DAPI (Sigma)/ml. Cells were washed three times with PBS and slides were mounted with PBS/glycerol mount.
Heterokaryon assays.
Heterokaryons of U2OS cells and p53/MDM2−/− MEFs were formed as described previously (40), with the following modifications. Twenty-four hours after transfections, U2OS cells were mixed with p53/MDM2−/− MEFs and reseeded onto glass coverslips. After 3 h, the cells were incubated for another 3 h in the presence of 50 μg of cycloheximide (Sigma)/ml. After 30 min in the presence of 100 μg of cycloheximide/ml, the cells were fused with 50% polyethylene glycol (PEG 3350 (Sigma) for 2 min, washed in PBS, and returned to medium containing 100 μg of cycloheximide/ml for another 1-h incubation. After paraformaldehyde fixation, immunostaining was performed as described above. Human and mouse nuclei were distinguished by DAPI (4′,6′-diamidino-2-phenylindole) staining or reactivity with a human specific Ku86 antibody (Santa Cruz).
Protein analysis.
To assess MDM2 or p53 protein levels, proteins from whole-cell extracts were separated by SDS–12% polyacrylamide gel electrophoresis and analyzed by Western blotting with anti-p53 DO-1 or 1801 (Pharmingen), anti-MDM2 AB1 and SMP14 (Oncogene Science), and anti-GFP (Clontech) antibody. To detect ubiquitinated p53, the cells were incubated in the proteasome inhibitor LLnL (N-acetyl-leucyl-leucyl-norleucinal) or MG132 for 2 h prior to harvesting. To assay for MDM2 and p53 association, 24 h after transfection, cells were washed three times in ice-cold PBS and lysed in 500 μl (per 10-cm-diameter dish) of NP-40 lysis buffer (100 mM NaCl, 100 mM Tris [pH 8.0], 1% NP-40) for 30 min at 4°C. MDM2 protein was immunoprecipitated overnight at 4°C by incubation with a mixture of anti-MDM2 SMP14 and AB1-protein A-Sepharose beads equilibrated in NP-40 lysis buffer. The immunoprecipitated protein was washed three times with NP-40 buffer, and samples were resuspended in 50 μl of 2× SDS sample buffer and incubated at room temperature for 10 min. The samples were then separated by SDS–12% polyacrylamide gel electrophoresis and analyzed by Western blotting with anti-p53 and anti-MDM2 antibodies.
RESULTS
C-terminal lysine residues in p53 are targets of ubiquitination.
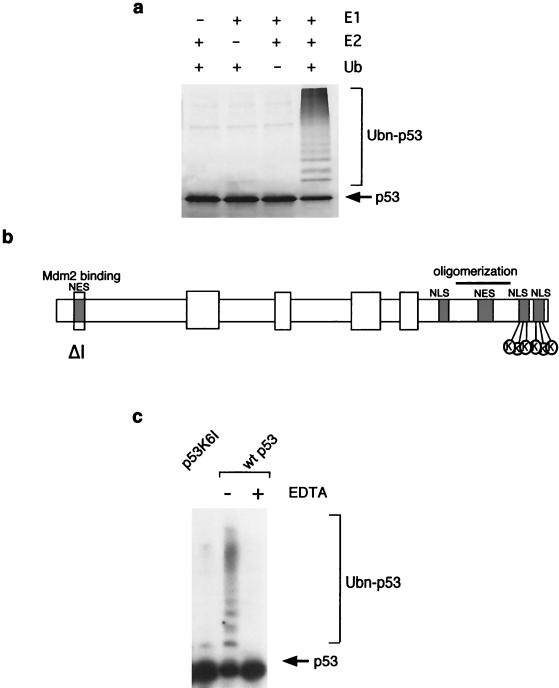
In vitro ubiquitination of p53 by MDM2 results in the accumulation of polyubiquitinated p53, with several distinct bands that may represent ubiquitination at multiple lysine residues (Fig. 1a). We showed previously that deletion of the C-terminal 23 amino acids of p53 prevents efficient degradation by MDM2 (25), and we considered that the in vitro ubiquitination pattern might represent conjugation of ubiquitin onto each of the six lysine residues within this region. We therefore generated a p53 mutant, p53K6I, carrying substitutions of lysine to isoleucine at these six residues (Fig. 1b). Ubiquitination of this mutant p53 protein by MDM2 in the in vitro assay was significantly reduced, although at least one ubiquitinated band was consistently seen, suggesting that other lysine residues in p53 could be targeted by MDM2 when the six C-terminal lysines are missing (Fig. 1c). These results are consistent with two previous publications identifying C-terminal lysine residues in p53 as targets for MDM2-mediated ubiquitination (32, 35).
FIG. 1.
In vitro ubiquitination of p53 proteins. (a) Ubiquitination of in vitro-translated p53 protein following incubation with recombinant E1, UbcH5B (E2), and ubiquitin as indicated. (b) Cartoon of the p53 protein showing the MDM2 binding region and nuclear export sequence at the N terminus and the oligomerization, nuclear localization, and nuclear export sequences at the C terminus. The positions of the six lysine residues mutated in the p53 mutants p53K6I and p53K6R, K370, 372, 373, 381, 382, and 386, are also shown. p53ΔI lacks conserved region I and has lost the ability to interact with MDM2. (c) In vitro ubiquitination of wild-type p53 or p53K6I. Ubiquitination was lost following addition of EDTA or mutation of the six C-terminal lysine residues in p53.
Subcellular localization of p53 lysine mutants.
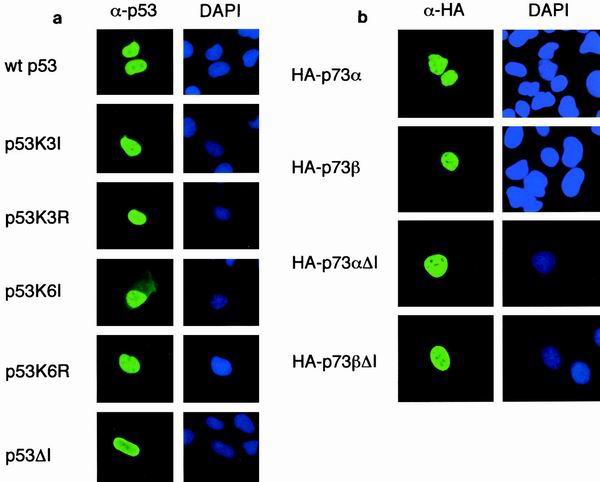
The C terminus of p53 has been shown to contain three nuclear localization signals, two of which contain lysine residues that are targets for ubiquitination (Fig. 1b). We have previously shown that deletion of the C-terminal 23 amino acids of p53 containing all six lysines and two nuclear localization signals gave rise to a protein localized to the nucleus, like wild-type p53 (25). However, analysis of the subcellular localization of p53K6I, in which the six ubiquitinated lysines were substituted for isoleucine, revealed both cytoplasmic and nuclear localization which was also evident following mutation of only the last three lysines (381, 382, and 386) to isoleucine (Fig. 2a). A similar cytoplasmic localization of mutants carrying alanine substitutions at these lysine residues has been attributed to enhanced export (15, 32). In light of these observations, we constructed another p53 mutant in which the lysine residues were mutated to arginine, to mimic the nuclear localization signals, and confirmed previously published results that this mutant localizes to the nucleus like the wild-type protein and the p53ΔI protein lacking the MDM2 binding site (Fig. 2a) (35). Similarly, the p53-related proteins p73α and p73β and p73 mutants that lacked the MDM2 binding site (p73αΔI and p73βΔI) were localized to the nucleus in transfected cells (Fig. 2b).
FIG. 2.
(a) Subcellular localization of transfected wild-type p53, p53K3I, p53K3R, p53K6I, p53K6R, and p53ΔI proteins in U2OS cells. Costaining with DAPI shows localization of the nucleus. Identical results were obtained with H1299 cells (data not shown). (b) Subcellular localization of transfected p73α, p73β, p73αΔI, and p73βΔI proteins in U2OS cells. The p73 proteins were HA tagged and detected with an anti-HA antibody. Costaining with DAPI shows localization of the nucleus.
Ubiquitination and degradation of p53 lysine mutant in cells.
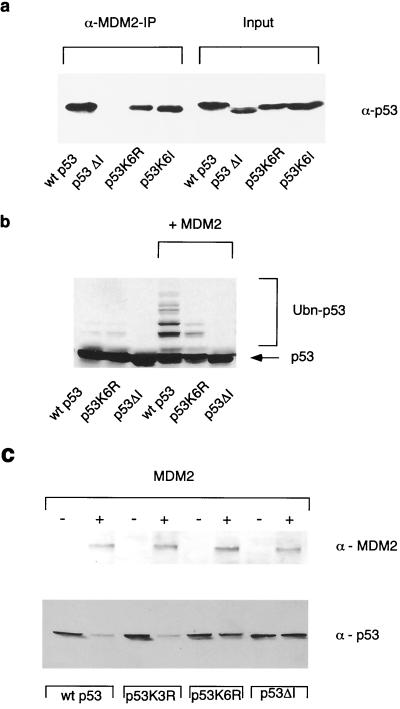
Our studies and previous reports suggested that the loss of the six lysines at the C terminus of p53 would not affect MDM2 binding but would render the p53 protein resistant to degradation. To confirm these predictions in our cell systems, we examined the interaction of each p53 protein with MDM2 by coimmunoprecipitation from transfected cells (Fig. 3a). These studies confirmed that while the deletion of the MDM2 binding region in p53ΔI prevented interaction with MDM2, mutation of the six lysine residues in p53K6R did not significantly affect binding to MDM2. We also examined the accumulation of ubiquitinated p53 proteins expressed in U2OS cells following the inhibition of degradation through the proteasome (Fig. 3b). In agreement with the in vitro assay, deletion of the MDM2 binding site abolished ubiquitination of p53, while mutation of the six C-terminal lysine residues significantly decreased ubiquitination. We did note, however, that following overexpression of exogenous ubiquitin the p53K6R mutant showed clear evidence of ubiquitination (data not shown), supporting the presence of at least one other ubiquitination site in p53. p53 mutants that were not efficiently ubiquitinated by MDM2, either because of an inability to bind MDM2 (p53ΔI) or mutation of the C-terminal lysine residues (p53K6R), were also resistant to MDM2-mediated degradation (Fig. 3c), although at higher MDM2 levels some degradation of p53K6R was observed, as previously reported for a p53 mutant lacking this C-terminal region (25). These results are again consistent with the retention of some ubiquitination for the p53K6R mutant in cells.
FIG. 3.
(a) Association of p53, p53ΔI, and p53K6R with MDM2 following cotransfection into U2OS cells. MDM2 protein was immunoprecipitated from whole-cell lysates, and bound p53 was detected by Western blotting (α-MDM2-IP). Levels of expression of each p53 protein was determined by Western blotting using the whole-cell lysate (input). (b) Ubiquitination of p53, p53K6R, and p53ΔI following transfection with or without MDM2 in U2OS cells. Cells were treated with 50 μM LLnL 2 h prior to harvesting. Ubiquitination of the p53 proteins was detected by Western blotting. (c) Degradation of p53, p53ΔI, p53K3R, and p53K6R following cotransfection of p53-null Saos-2 cells with MDM2 as indicated. The levels of p53 and MDM2 proteins were assessed by Western blotting 24 h after transfection. Identical results were obtained in U2OS cells (data not shown).
Effect of MDM2 on subcellular localization of p53.
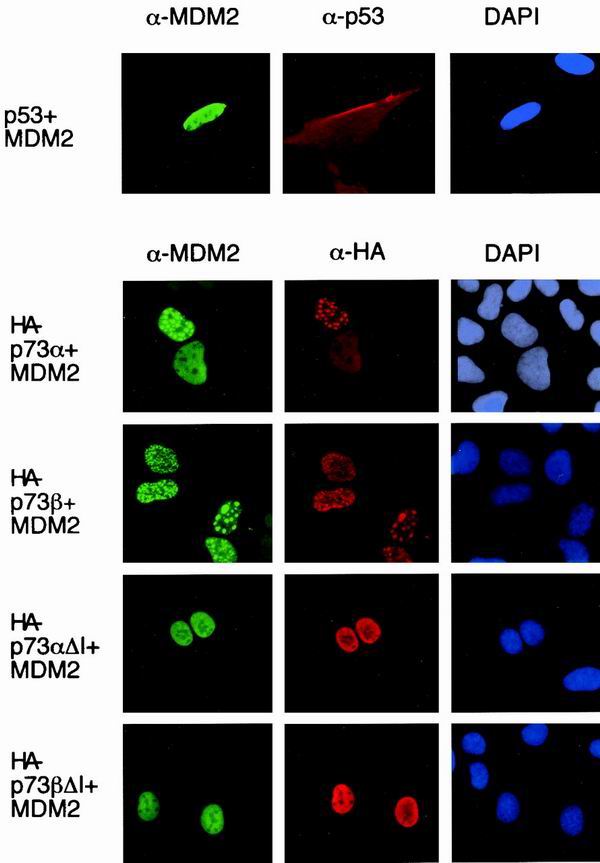
Previous studies have shown that the export of p53 from the nucleus to the cytoplasm depends on nuclear export sequences within the C terminus of p53, but that efficient nuclear export depends on the ubiquitin ligase activity of MDM2 (5, 14, 40). Consistent with these studies, we found that coexpression of MDM2 with wild-type p53 enhanced nuclear export of p53, increasing the proportion of cells in which cytoplasmic p53 was detected (Fig. 4). By contrast, nuclear export of p73α and p73β was not enhanced by MDM2 (Fig. 4). Instead, coexpression of MDM2 resulted in the localization of both p73 and MDM2 to nuclear aggregates in some cells, while other cells retained p73 and MDM2 in the nucleoplasm, as recently reported (16). Relocalization to the nuclear aggregates appeared to correlate with high levels of MDM2 expression and was dependent on the ability of p73 to interact with MDM2, since p73 proteins mutated in the MDM2 binding site (p73αΔI and p73βΔI) did not show this relocalization (Fig. 4). Although the significance of the nuclear localization of p73 and MDM2 is not clear, these results show some specificity in the ability of MDM2 to promote nuclear export.
FIG. 4.
Subcellular localization of p53, p73α, p73β p73αΔI, and p73βΔI proteins in U2OS cells following cotransfection with MDM2. Costaining with DAPI shows localization of the nucleus.
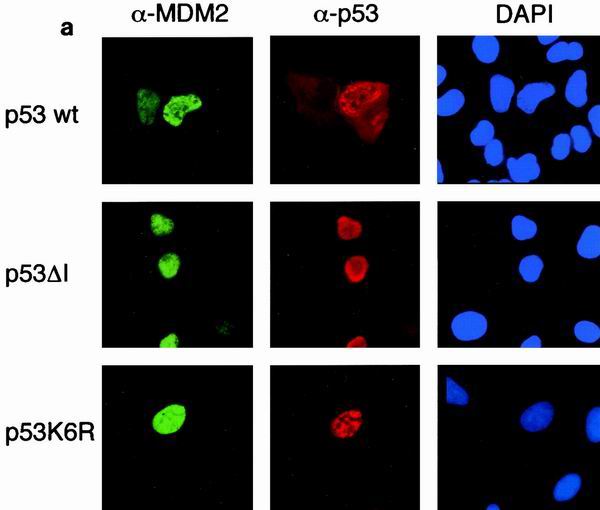
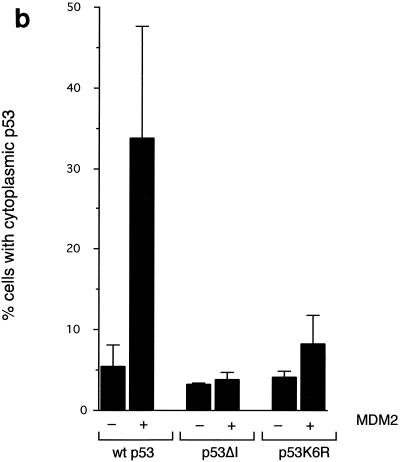
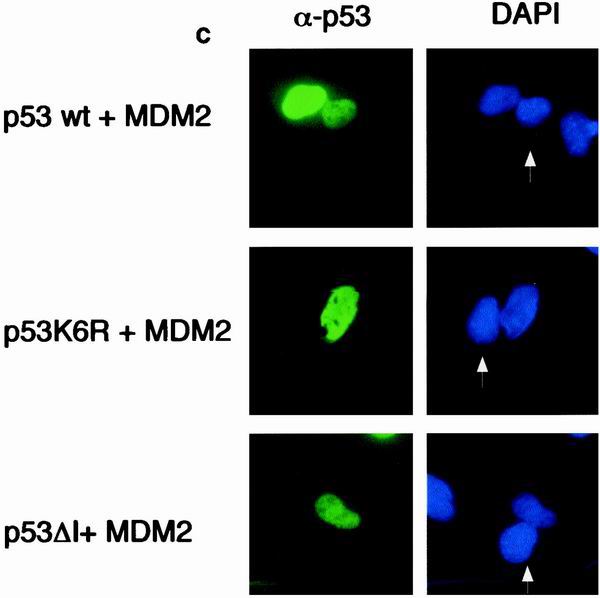
Although p73 can bind MDM2, this does not target p73 for degradation (3, 34, 48). To investigate whether nuclear export of p53 is related to its ubiquitination, we examined the subcellular localization of the p53 mutants which localized to the nucleus in the absence of cotransfected MDM2 (Fig. 2) in response to MDM2 (Fig. 5). The ability of MDM2 to promote nuclear export of p53 was dependent on the ability to bind p53, since a p53 mutant that cannot bind MDM2 (p53ΔI) remained in the nucleus following MDM2 expression (Fig. 5a). The same staining patterns were seen using GFP-tagged p53 proteins (Fig. 5b). Interestingly, nuclear export of the p53K6R mutant by MDM2 was also significantly reduced (Fig. 5a and b), suggesting that ubiquitination of p53 on these residues is necessary for efficient nuclear export. To confirm that nuclear localization of the mutant p53 proteins is the results of a failure to export from the nucleus, as opposed enhanced nuclear import, we carried out heterokaryon assays (Fig. 5c). Although the transport of wild-type p53 between the nuclei of transfected human U2OS cells and p53/MDM2−/− mouse embryo fibroblasts was seen in the presence of MDM2, both the p53K6R mutant and p53ΔI proteins were restricted to the human nucleus in which they were expressed. These results confirm that MDM2 expression cannot enhance nuclear export of these p53 mutants and are consistent with those described in the accompanying paper (15).
FIG. 5.
(a) Subcellular localization of p53, p53ΔI, and p53K6R in the presence of cotransfected MDM2. Costaining with DAPI shows localization of the nucleus. Identical results were obtained with H1299 cells (data not shown). (b) Graph summarizing three independent experiments with U2OS cells, showing the proportion of cells expressing cytoplasmic p53 following transfection with GFP-p53, GFP-p53ΔI, GFP-p53K6R, and MDM2 as indicated. (c) Heterokaryon assays between U2OS cells transfected with p53, p53K6R, or p53ΔI and p53/MDM2 −/− MEFs. After cell fusion, p53 protein was detected in human and mouse nuclei (white arrow).
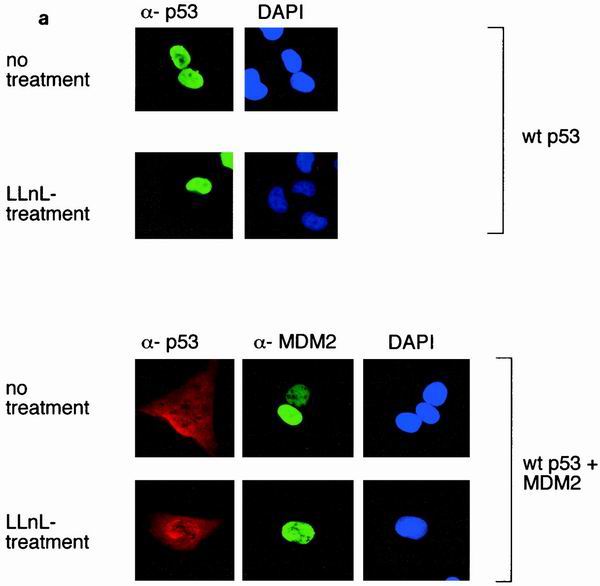
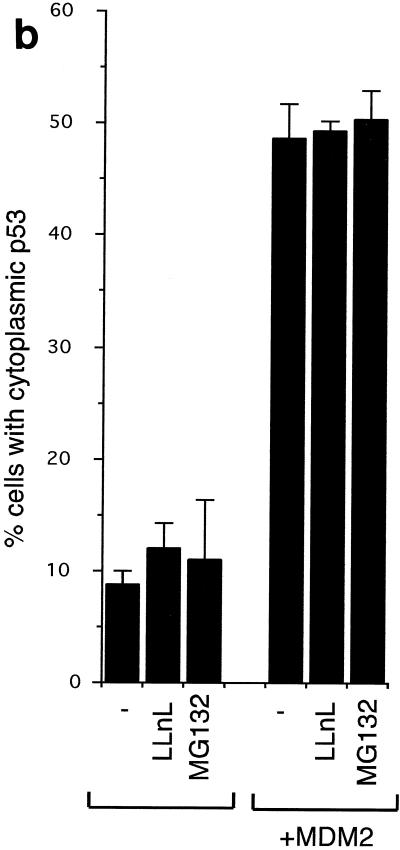
The accessibility of the nuclear export signal in the C terminus of p53 to the export machinery has been shown to be regulated by the oligomerization status of the p53 protein (40), which may be affected by protein concentration. It is possible that p53 at a low concentration exists mostly in the monomeric form, exposing the NES and allowing export to the cytoplasm, while increased concentration of p53 leads to oligomerization, concealing the NES and therefore resulting in nuclear sequestration. Since both the p53ΔI and p53K6R mutants are not degraded by MDM2 and may therefore be expressed at higher levels than wild-type p53, we considered the possibility that failure of these mutants to export was related simply to an increased proportion of oligomerized p53 in the nucleus. To address this point, we examined the ability of MDM2 to influence nuclear export in the presence of proteasome inhibitors, which prevent p53 degradation and allow expression of comparable levels of wild-type and mutant p53 proteins. MDM2 clearly retained the ability to enhance the export of wild-type p53 following treatment of cells with either LLnL or MG132 for 1.5 (Fig. 6), 3, or 4 h (data not shown), indicating that increased protein expression, which might affect the oligomerization status of p53, is not sufficient to sequester p53 in the nucleus.
FIG. 6.
(a) Subcellular localization of p53 and MDM2 in U2OS cells with or without treatment with LLnL (50 μM LLnL for 1.5 h). (b) Graph showing the proportion of cells expressing cytoplasmic p53 following transfection of GFP-p53 with or without MDM2 in the presence of LLnL (50 μM) or MG132 (1 μM) as indicated.
CRM1 enhances nuclear export of p53.
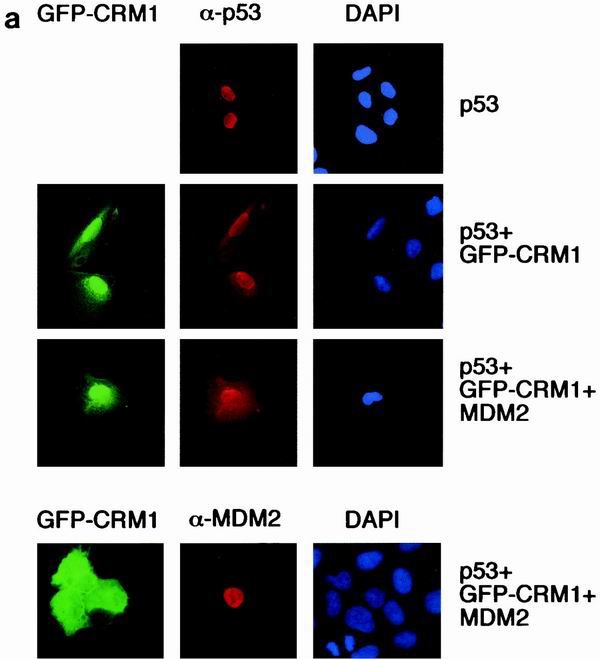
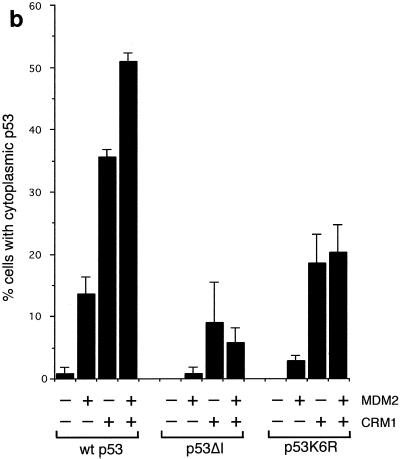
The export of p53 from the nucleus has been shown to be inhibited by treatment with leptomycin-B, suggesting that export is mediated through a CRM1-dependent mechanism (40). Coexpression of exogenous CRM1 efficiently enhanced export of p53 (Fig. 7a), which was further increased following transfection of MDM2 (Fig. 7b). Interestingly, although reduced compared to wild-type p53, the p53K6R mutant showed a significant increase in nuclear export following CRM1 expression, probably reflecting enhanced export through the N-terminal NES. As predicted, the addition of MDM2 did not further enhance the export of these p53 mutants, even in the presence of additional CRM1 (Fig. 7b). These results support previous studies showing MDM2 independent export of p53 through both N- and C-terminal NES (40, 49).
FIG. 7.
(a) Subcellular localization of p53, MDM2, and GFP-CRM1 following transfection into U2OS cells. Costaining with DAPI shows localization of the nucleus. (b) Graph showing the proportion of cells expressing cytoplasmic p53 following transfection of wild-type p53, p53ΔI, or p53K6R, with GFP-CRM1 and MDM2 in U2OS cells. Note the reduced sensitivity of detection of cytoplasmic localization of untagged p53 compared to GFP-p53 shown in Fig. 5b and 6b.
Unlike p53, MDM2 nuclear export was not significantly enhanced by expression of CRM1 in these studies (Fig. 7a). Although MDM2 has also been shown to contain a nuclear export signal (36), this observation suggests that the export of MDM2 is independent of the export of p53 and regulated by a different mechanism.
Nuclear export and degradation of p53 are not dependent on each other.
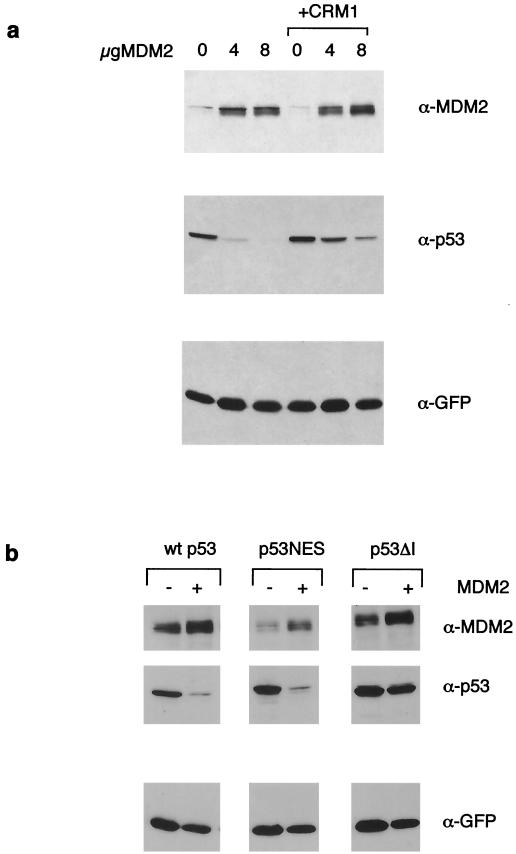
The ability of CRM1 to enhance the export of p53 independently of MDM2 allowed us to examine whether export of p53 is sufficient to allow degradation. Despite the efficient nuclear export of wild-type p53 in the presence of CRM1, we were unable to detect enhanced degradation following CRM1 expression. Indeed, enhanced export by overexpression of CRM1 slightly reduced the sensitivity of p53 to MDM2-mediated degradation (Fig. 8a). These results indicate that MDM2 ubiquitination of p53 contributes to both export and degradation of the p53 protein, and that these may be separable activities. In the light of these observations, and our previous studies showing that proteasome inhibition leads to the accumulation of p53 in the nucleus and not in the cytoplasm (25), we reassessed the importance of nuclear export of p53 for p53 degradation. In the cell systems under study here, a p53 protein mutated in the C-terminal NES (40) was efficiently degraded by MDM2 (Fig. 8b). However, as shown previously (5, 14), MDM2 did not enhance the nuclear export of this mutant (data not shown), despite the retention of the N-terminal NES. These results suggest that nuclear export and degradation of p53 are not necessarily interdependent and may be regulated separately.
FIG. 8.
(a) Degradation of transfected wild-type p53 following cotransfection of U2OS cells with increasing amounts of MDM2 in the presence or absence of 6 μg of transfected CRM1, as indicated. The levels of p53 and MDM2 proteins were assessed by Western blotting 24 h after transfection. Equal expression of cotransfected GFP was used to control for transfection efficiency. (b) Degradation of transfected p53, p53NES, or p53ΔI following cotransfection of U2OS cells with MDM2. The levels of p53 and MDM2 protein were assessed by Western blotting 24 h after transfection. Cotransfected GFP expression was monitored as a control for transfection efficiency.
DISCUSSION
Nuclear-cytoplasmic shuttling of p53 depends on nuclear export signals in the N and C termini of the p53 protein (5, 14, 40, 49), and MDM2 can enhance nuclear export through the C-terminal signal. The C-terminal NES is concealed following the tetramerization of p53, but revealed and accessible to the nuclear export machinery in monomers or dimers of p53 (40). Since nuclear localization is necessary for p53 to function to suppress cell growth (38), export of p53 to the cytoplasm represents an efficient mechanism to negatively regulate p53 activity. Indeed, the cytoplasmic localization of p53 in some tumors is associated with hyperactive nuclear export (40).
Our data indicate that ubiquitination at the C terminus of p53 contributes to the nuclear export of the p53 protein. Since MDM2-dependent export of p53 occurs with equal efficiency even when degradation is blocked using proteasome inhibitors, it seems unlikely that our results reflect differences in wild-type and mutant p53 protein concentrations, leading to altered oligomerization states that might reveal the nuclear export sequence in p53. Our results support a model in which ubiquitination of the C terminus of p53, in a region close to both the nuclear export and oligomerization sequences (Fig. 1b), reveals the nuclear export signal and allows interaction of p53 with the CRM1-dependent nuclear export machinery.
Unlike the effect on p53, MDM2 failed to drive nuclear export of p73, and at higher expression levels, some relocalization of both p73 and MDM2 to nuclear aggregates was observed, as recently reported (16). Interestingly, although p73 is also targeted for proteasome-mediated degradation (3), this is not mediated by MDM2, and inhibition of nuclear export using leptomycin B (LMB), which results in the stabilization of p53, fails to stabilize p73 (39). Taken together, these results suggest that the mechanisms regulating p73 stability are different from those controlling p53, with no apparent role for MDM2 or nuclear export. The significance of the MDM2-dependent relocalization of p73 to nuclear aggregates remains to be elucidated.
Inhibition of p53 nuclear export by LMB suggested a role for the export receptor CRM1, and we have shown that ectopic expression of CRM1 can strongly enhance nuclear export of p53. In contrast, the export of MDM2 is not enhanced by the ectopic expression of CRM1, supporting the previous observation that the export of p53 can occur independently of nuclear export of MDM2 (5, 14).
MDM2 has been shown to ubiquitinate p53 in the nucleus (47), and our data suggest that nuclear export is not essential for degradation, which is likely to be carried out, to some extent, by nuclear proteasomes. These results are consistent with another recent report showing degradation of p53 by MDM2 in the nucleus (46) and suggest that the stabilization of p53 by treatment with LMB (12, 28) may reflect the consequences of blocking nuclear export of proteins other than p53. However, others have shown that p53 proteins with mutations in the C-terminal NES are resistant to MDM2-mediated degradation (5, 14), indicating that although not essential, cytoplasmic factors can contribute to the efficiency of degradation of p53. Nevertheless, our results indicate that the ability to MDM2 to ubiquitinate p53 can contribute to both degradation and nuclear export, but that these two responses are not absolutely dependent on each other. Efficient nuclear export of p53 following expression of exogenous CRM1 alone does not enhance p53 degradation, and inhibition of nuclear export by mutation within the C-terminal NES in p53 does not abolish MDM2 mediated degradation. These results raise the possibility that nuclear export and degradation of p53 are regulated independently and that enhancement of one response may not lead to an increase in the other. This may be reflected in some tumors, where enhanced export of p53 is associated with cytoplasmic accumulation, rather than enhanced degradation (40).
The observation that nuclear export and proteasome targeting of p53 following ubiquitination by MDM2 are separable also leads to the possibility that additional steps may be necessary for one or other response. A recent report suggests that when using purified components, MDM2 may only direct multiple monoubiquitination of p53 (27). In order for a protein to be targeted to the proteasome, it must be polyubiquitinated with chains of at least four ubiquitin residues (41), and it is clear that polyubiquitination of p53 by MDM2 can be detected in assays that contain components isolated from cells or reticulocyte lysate (11, 19, 20, 30). These results suggest that additional cellular factors may be required for polyubiquitination of p53 in response to MDM2. Although not sufficient for degradation, the ability of MDM2 to monoubiquitinate p53 could regulate p53 in other ways (18) and may be enough to allow nuclear export of p53. How the balance between mono- and polyubiquitiation of p53 is regulated remains to be determined, but it is interesting to consider that the additional factors required for polyubiquitination and degradation of p53 may also be recruited by MDM2. In support of this suggestion, several recent studies have described MDM2 mutants that retain ubiquitin ligase activity but fail to target the degradation of p53 (1, 50).
ACKNOWLEDGMENTS
We are grateful to Barbara Felber for GFP-CRM1 and Arnie Levine for the MDM2 expression construct, Gerry Melino for the p73 constructs, and Allan Weissman for advice and reagents for the in vitro ubiquitination assay. We also thank Geoff Wahl, David Lane, Zhi-Min Yuan, and members of the Vousden lab for advice and discussions.
REFERENCES
- 1.Argentini M, Barboule N, Wasylyk B. The contribution of the acidic domain of MDM2 to p53 and MDM2 stability. Oncogene. 2001;20:1267–1275. doi: 10.1038/sj.onc.1204241. [DOI] [PubMed] [Google Scholar]
- 2.Ashcroft M, Vousden K H. Regulation of p53 stability. Oncogene. 1999;18:7637–7643. doi: 10.1038/sj.onc.1203012. [DOI] [PubMed] [Google Scholar]
- 3.Balint E, Bates S, Vousden K H. Mdm2 binds p73α without targeting degradation. Oncogene. 1999;18:3923–3929. doi: 10.1038/sj.onc.1202781. [DOI] [PubMed] [Google Scholar]
- 4.Barak Y, Juven T, Haffner R, Oren M. Mdm-2 expression is induced by wild type p53 activity. EMBO J. 1993;12:461–468. doi: 10.1002/j.1460-2075.1993.tb05678.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boyd S D, Tsai K Y, Jacks T. An intact HDM2 RING-finger domain is required for nuclear exclusion of p53. Nat Cell Biol. 2000;2:563–568. doi: 10.1038/35023500. [DOI] [PubMed] [Google Scholar]
- 6.Chen J, Wu X, Lin J, Levine A J. mdm-2 inhibits the G1 arrest and apoptosis functions of the p53 tumor suppressor protein. Mol Cell Biol. 1996;16:2445–2452. doi: 10.1128/mcb.16.5.2445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen J D, Marechal V, Levine A J. Mapping of the p53 and mdm-2 interaction domains. Mol Cell Biol. 1993;13:4107–4114. doi: 10.1128/mcb.13.7.4107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Crook T, Ludwig R L, Marston N J, Willkomm D, Vousden K H. Sensitivity of p53 lysine mutants to ubiquitin-directed degradation targeted by human papillomavirus E6. Virology. 1996;217:285–292. doi: 10.1006/viro.1996.0115. [DOI] [PubMed] [Google Scholar]
- 9.De Laurenzi V, Costanzo A, Barcaroli D, Terrinoni M, Falco M, Annicchiarico-Petruzzelli M, Levrero M, Melino G. Two new p73 splice variants, gamma and delta, with different transcriptional activity. J Exp Med. 1998;188:1763–1768. doi: 10.1084/jem.188.9.1763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.de Rozieres S, Maya R, Oren M, Lozano G. The loss of mdm2induces p53 mediated apoptosis. Oncogene. 2000;19:1691–1697. doi: 10.1038/sj.onc.1203468. [DOI] [PubMed] [Google Scholar]
- 11.Fang S, Jensen J P, Ludwig R L, Vousden K H, Weissman A M. Ubiquitin protein ligase activity of Mdm2: differential RING finger requirements for ubiquitination and proteasomal targeting of Mdm2 and p53. J Biol Chem. 2000;275:8945–8951. doi: 10.1074/jbc.275.12.8945. [DOI] [PubMed] [Google Scholar]
- 12.Freedman D A, Levine A J. Nuclear export is required for degradation of endogenous p53 by MDM2 and human papillomavirus E6. Mol Cell Biol. 1998;18:7288–7293. doi: 10.1128/mcb.18.12.7288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Freedman D A, Wu L, Levine A J. Functions of the MDM2 oncoprotein. Cell Mol Life Sci. 1999;55:96–107. doi: 10.1007/s000180050273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Geyer R K, Yu Z K, Maki C G. The MDM2 RING-finger domain is required to promote p53 nuclear export. Nat Cell Biol. 2000;2:569–573. doi: 10.1038/35023507. [DOI] [PubMed] [Google Scholar]
- 15.Gu J, Wiederschain D, Yuan Z-M. Identification of p53 sequence elements that are required for MDM2-mediated nuclear export. Mol Cell Biol. 2001;21:8533–8546. doi: 10.1128/MCB.21.24.8533-8546.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gu J J, Nie L H, Kawai H, Yuan Z M. Subcellular distribution of p53 and p73 are differentially regulated by MDM2. Cancer Res. 2001;61:6703–6707. [PubMed] [Google Scholar]
- 17.Haupt Y, Maya R, Kazaz A, Oren M. Mdm2 promotes the rapid degradation of p53. Nature. 1997;387:296–299. doi: 10.1038/387296a0. [DOI] [PubMed] [Google Scholar]
- 18.Hicke L. Protein regulation by monoubiquitination. Nat Rev Mol Cell Biol. 2001;2:195–201. doi: 10.1038/35056583. [DOI] [PubMed] [Google Scholar]
- 19.Honda R, Tanaka H, Yasuda H. Oncoprotein MDM2 is a ubiquitin ligase E3 for tumor suppressor p53. FEBS Lett. 1997;420:25–27. doi: 10.1016/s0014-5793(97)01480-4. [DOI] [PubMed] [Google Scholar]
- 20.Honda R, Yasuda H. Association of p19ARFwith Mdm2 inhibits ubiquitin ligase activity of Mdm2 for tumor suppressor p53. EMBO J. 1999;18:22–27. doi: 10.1093/emboj/18.1.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ito A, Lai C H, Zhao X, Saito S, Hamilton M H, Appella E, Yao T-P. p300/CBP-mediated p53 acetylation is commonly induced by p53-activating agents and inhibited by MDM2. EMBO J. 2001;20:1331–1340. doi: 10.1093/emboj/20.6.1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ko L J, Prives C. p53: puzzle and paradigm. Genes Dev. 1996;10:1054–1072. doi: 10.1101/gad.10.9.1054. [DOI] [PubMed] [Google Scholar]
- 23.Kobet E, Zeng X, Zhu Y, Keller D, Lu H. MDM2 inhibits p300-mediated p53 acetylation and activation by forming a ternary complex with the two proteins. Proc Natl Acad Sci USA. 2000;97:12547–12552. doi: 10.1073/pnas.97.23.12547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kubbutat M H G, Jones S N, Vousden K H. Regulation of p53 stability by Mdm2. Nature. 1997;387:299–303. doi: 10.1038/387299a0. [DOI] [PubMed] [Google Scholar]
- 25.Kubbutat M H G, Ludwig R L, Ashcroft M, Vousden K H. Regulation of Mdm2 directed degradation by the C-terminus of p53. Mol Cell Biol. 1998;18:5690–5698. doi: 10.1128/mcb.18.10.5690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kubbutat M H G, Vousden K H. Keeping an old friend under control: regulation of p53 stability. Mol Med Today. 1998;4:250–256. doi: 10.1016/s1357-4310(98)01260-x. [DOI] [PubMed] [Google Scholar]
- 27.Lai Z, Ferry K V, Diamond M A, Wee K E, Kim Y B, Ma J, Yamg T, Benfield P A, Copland R A, Auger K R. Human mdm2 mediates multiple mono-ubiquitination of p53 by a mechanism requiring enzyme isomerization. J Biol Chem. 2001;276:31357–31367. doi: 10.1074/jbc.M011517200. [DOI] [PubMed] [Google Scholar]
- 28.Lain S, Midgley C, Sparks A, Lane E B, Lane D P. An inhibitor of nuclear export activates the p53 response and induces the localization of HDM2 and p53 to U1A-positive nuclear bodies associated with the PODS. Exp Cell Res. 1999;248:457–472. doi: 10.1006/excr.1999.4433. [DOI] [PubMed] [Google Scholar]
- 29.Lohrum M A E, Ashcroft M, Kubbutat M H G, Vousden K H. Identification of a cryptic nucleolar localization signal in MDM2. Nat Cell Biol. 2000;2:179–181. doi: 10.1038/35004057. [DOI] [PubMed] [Google Scholar]
- 30.Midgley C A, Desterro J M, Saville M K, Howard S, Sparks A, Hay R T, Lane D P. An N-terminal p14ARF peptide blocks Mdm2-dependent ubiquitination in vitro and can activate p53 in vivo. Oncogene. 2000;19:2312–2323. doi: 10.1038/sj.onc.1203593. [DOI] [PubMed] [Google Scholar]
- 31.Momand J, Zambetti G P, George D L, Levine A J. The mdm-2 oncogene product forms a complex with the p53 protein and inhibits p53-mediated transactivation. Cell. 1992;69:1237–1245. doi: 10.1016/0092-8674(92)90644-r. [DOI] [PubMed] [Google Scholar]
- 32.Nakamura S, Roth J A, Muckopadhay T. Multiple lysine mutations in the C-terminal domain of p53 interfere with MDM2-dependent protein degradation and ubiquitination. Mol Cell Biol. 2000;20:9391–9398. doi: 10.1128/mcb.20.24.9391-9398.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Oliner J D, Pietenpol J A, Thiagalingam S, Gyuris J, Kinzler K W, Vogelstein B. Oncoprotein MDM2 conceals the activation domain of tumour suppressor p53. Nature. 1993;362:857–860. doi: 10.1038/362857a0. [DOI] [PubMed] [Google Scholar]
- 34.Ongkeko W M, Wang X Q, Siu W Y, Lau A W S, Yamashita K, Harris A L, Cox L S, Poon R Y C. MDM2 and MDMX bind and stabilize the p53-related protein p73. Curr Biol. 1999;9:829–832. doi: 10.1016/s0960-9822(99)80367-4. [DOI] [PubMed] [Google Scholar]
- 35.Rodriguez M, Desterro J M P, Lain S, Lane D P, Hay R T. Multiple C-terminal lysine residues target p53 for ubiquitin-proteasome-mediated degradation. Mol Cell Biol. 2000;20:8458–8467. doi: 10.1128/mcb.20.22.8458-8467.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Roth J, Dobbelstein M, Freedman D A, Shenk T, Levine A J. Nucleo-cytoplasmic shuttling of the hdm2 oncoprotein regulates the levels of the p53 protein via a pathway used by the human immunodeficiency virus rev protein. EMBO J. 1998;17:554–564. doi: 10.1093/emboj/17.2.554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ryan K M, Phillips A C, Vousden K H. Regulation and function of the p53 tumor suppressor protein. Curr Biol. 2001;13:332–337. doi: 10.1016/s0955-0674(00)00216-7. [DOI] [PubMed] [Google Scholar]
- 38.Shaulsky G, Goldfinger N, Tosky M S, Levine A, Rotter V. Nuclear localization is essential for the activity of p53 protein. Oncogene. 1991;6:2055–2065. [PubMed] [Google Scholar]
- 39.Smart P, Lane E B, Lane D P, Midgley C, Vojtesek B, Laín S. Effects on normal fibroblasts and neuroblastoma cells of the activation of the p53 response by the nuclear export inhibitor leptomycin B. Oncogene. 1999;18:7378–7386. doi: 10.1038/sj.onc.1203260. [DOI] [PubMed] [Google Scholar]
- 40.Stommel J M, Marchenko N D, Jimenez G S, Moll U M, Hope T J, Wahl G M. A leucine-rich nuclear export signal in the p53 tetramerization domain: regulation of subcellular localization and p53 activity by NES masking. EMBO J. 1999;18:1660–1672. doi: 10.1093/emboj/18.6.1660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Thrower J S, Hoffman L, Rechsteiner M, Pickart C M. Recognition of the ployubiquitin proteolytic signal. EMBO J. 2000;19:94–102. doi: 10.1093/emboj/19.1.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Thut C J, Goodrich J A, Tjian R. Repression of p53-mediated transcription by MDM2: a dual mechanism. Genes Dev. 1997;11:1974–1986. doi: 10.1101/gad.11.15.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vousden K H. p53: death star. Cell. 2000;101:691–694. doi: 10.1016/s0092-8674(00)00171-9. [DOI] [PubMed] [Google Scholar]
- 44.Wadgaonkar R, Collins T. Murine double minute (MDM2) blocks p53-coactivator interaction, a new mechanisms for inhibition of p53-dependent gene expression. J Biol Chem. 1999;274:13760–13767. doi: 10.1074/jbc.274.20.13760. [DOI] [PubMed] [Google Scholar]
- 45.Wu X W, Bayle J H, Olson D, Levine A J. The p53 mdm-2 autoregulatory feedback loop. Genes Dev. 1993;7:1126–1132. doi: 10.1101/gad.7.7a.1126. [DOI] [PubMed] [Google Scholar]
- 46.Xirodimas D P, Stephen C W, Lane D P. Co-compartmentalisation of p53 and Mdm2 is a major determinant for Mdm2-mediated degradation of p53. Exp Cell Res. 2001;270:66–77. doi: 10.1006/excr.2001.5314. [DOI] [PubMed] [Google Scholar]
- 47.Yu Z K, Geyer R K, Maki C G. MDM2-dependent ubiquitination of nuclear and cytoplasmic p53. Oncogene. 2000;19:5892–5897. doi: 10.1038/sj.onc.1203980. [DOI] [PubMed] [Google Scholar]
- 48.Zeng X, Chen L, Jost C A, Maya R, Keller D, Wang X, Kaelin W G J, Oren M, Chen J, Lu H. MDM2 suppresses p73 function without promoting p73 degradation. Mol Cell Biol. 1999;19:3257–3266. doi: 10.1128/mcb.19.5.3257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang Y, Xiong Y. A p53 amino-terminal nuclear export signal inhibited by DNA damage-induced phosphorylation. Science. 2001;292:1910–1915. doi: 10.1126/science.1058637. [DOI] [PubMed] [Google Scholar]
- 50.Zhu Q, Y J, Wani G, Wanit M A, Wani A A. Mdm2 mutant defective in binding p300 promotes ubiquitination but not degradation of p53. J Biol Chem. 2001;276:29695–29701. doi: 10.1074/jbc.M102634200. [DOI] [PubMed] [Google Scholar]
- 51.Zolotukhin A S, Felber B K. Nucleoporins nup98 and nup214 participate in nuclear export of human immunodeficiency virus type 1 Rev. J Virol. 1999;73:120–127. doi: 10.1128/jvi.73.1.120-127.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]