Abstract
It has been demonstrated that MDM2 can differentially regulate subcellular distribution of p53 and its close structural homologue p73. In contrast to MDM2-mediated p53 nuclear export, p73 accumulates in the nucleus as aggregates that colocalize with MDM2. Distinct distribution patterns of p53 and p73 suggest the existence of unique structural elements in the two homologues that determine their MDM2-mediated relocalization in the cell. Using a series of p53/p73 chimeric proteins, we demonstrate that three regions of p53 are involved in the regulation of MDM2-mediated nuclear export. The DNA binding domain (DBD) is involved in the maintenance of a proper conformation that is required for functional activity of the nuclear export sequence (NES) of p53. The extreme C terminus of p53 harbors several lysine residues whose ubiquitination by MDM2 appears to be the initial event in p53 nuclear export, as evidenced by the impaired nucleocytoplasmic shuttling of p53 mutants bearing simultaneous substitutions of lysines 370, 372, 373, 381, 382, and 386 to arginines (6KR) or alanines (6KA). Finally, the region between the DBD and the oligomerization domain of p53, specifically lysine 305, also plays a critical role in fully revealing p53NES. We conclude that MDM2-mediated nuclear export of p53 depends on a series of ubiquitination-induced conformational changes in the p53 molecule that lead to the activation of p53NES. In addition, we demonstrate that the p53NES may be activated without necessarily disrupting the p53 tetramer.
As a result of its high turnover rate, the p53 protein has a half-life of approximately 30 to 60 min and is maintained at low levels in normal proliferating cells (2). In response to genotoxic stress or oncogenic signaling, p53 levels rapidly increase, mainly through protein stabilization (2). Recent findings suggest that p53 stabilization and accumulation can be accomplished through the inhibition of nuclear export, implicating nuclear import-export as a potential mechanism of controlling p53 stability (12). An important regulator of p53 protein level is MDM2, which possesses intrinsic E3 ligase activity and thus promotes p53 ubiquitination and subsequent degradation via proteasome-mediated proteolysis (2). In addition, MDM2 functions as a mediator of p53 nuclear export (2), and MDM2 Rev-like nuclear export sequence (NES) has been shown to be essential for this function (5). p53 has its own leucine-rich, Rev-like NES in the C terminus that has been reported to be fully capable of mediating nuclear export independently of MDM2 (21). However, the p53NES lies within the oligomerization domain (OD) of the protein. Analysis of three-dimensional structure of p53OD indicates that the NES is buried at the interface of the two dimers that form active p53 tetramer (10, 13), thus rendering it inaccessible to transport proteins. It was therefore suggested that in order to reveal the buried NES, tetrameric conformation of p53 has to be disrupted (20). Results from two recent studies indicate that the ring domain of MDM2 which contains the E3 ubiquitin ligase activity (9), rather than the NES of MDM2, is essential for p53 nuclear export, suggesting a role for ubiquitination in the regulation of p53 nuclear export (3, 6). There are multiple lysine residues in the p53 extreme C terminus (p53eCT) that have been identified as main sites of ubiquitination by MDM2 (18, 19). However, how MDM2-dependent p53 ubiquitination is translated into nuclear export is presently unclear.
p73, a new member of the p53 family, shares substantial sequence homology and a lesser degree of functional similarity with p53 (11). p73 forms a complex with MDM2 through a conserved MDM2 binding motif, and MDM2 binding results in the inhibition of p73 transcriptional activity, but not in the degradation of the p73 protein (16). In a recent study, we showed that the subcellular distribution of p53 and p73 is also differentially regulated by MDM2. In sharp contrast to p53, which undergoes nuclear export upon coexpression of MDM2, p73 accumulates in the nucleus of MDM2-expressing cells (8). In this report, we took advantage of distinct subcellular localization of these two homologues and used a domain-swapping approach to investigate structural elements that are essential for the MDM2-mediated nuclear export of p53.
MATERIALS AND METHODS
Cell culture and transfection.
H1299 cells, U2OS cells (American Type Culture Collection), and p53−/−/MDM2−/− mouse embryonic fibroblasts (MEFs) (Carl Maki, Harvard School of Public Health) were maintained in minimal essential medium. supplemented with 10% fetal bovine serum. Cells were transfected by a calcium phosphate method as previously described (7).
Preparation of whole-cell extracts and glutathione S-transferase (GST) pull- down analysis.
Cells were transfected in 60-mm-diameter plates with 8 μg of DNA and harvested at 24 h posttransfection. Cells were lysed in 100 μl of lysis buffer (10 mM Tris-HCl, pH 7.5, 1 mM EDTA, 1% Triton X-100, 150 mM NaCl, 1 mM dithiothreitol, 10% glycerol, 0.2 mM phenylmethylsulfonyl fluoride, and protease inhibitors) by incubating on ice for 30 min, and the extracts were centrifuged at 13,000 rpm for 15 min to remove cell debris. Protein concentrations were determined using a Bio-Rad protein assay (Hercules, Calif.). After addition of 5× loading buffer, the samples were incubated at 95°C for 5 min and resolved through sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). Proteins were transferred onto nitrocellulose membranes (Schleicher & Schuell) and probed with the indicated antibody. Proteins were visualized with an enhanced chemiluminescence detection system (NEN).
Oligomerization assay.
Cells were transfected with the indicated expression plasmid and lysed as described above 36 h posttransfection. Each lysate was divided into three tubes, and glutaraldehyde was added to the final concentration of 0, 0.01, or 0.1%. After incubation on ice for 15 min, equal volumes of 2× loading buffer were added and the samples were boiled and analyzed as described above.
Subcellular distribution assay.
Cells were grown on chamber slides (Nunc, Ill.) and transfected with the indicated plasmid. Cells were washed with cold phosphate-buffered saline (PBS) 36 h posttransfection and fixed with 4% paraformaldehyde (Sigma) for 30 min at 4°C. After washing with PBS, the cells were quenched by 50 mM NH4Cl at room temperature for 5 min. After washing with PBS, the cells were permeabilized with ice-cold 0.2% Triton X-100 for 5 min. The slides were washed with PBS, blocked with 0.5% bovine serum albumin in PBS for 30 min, washed with PBS, and then incubated with the indicated primary antibody at 37°C for 1 h. After washing with PBS three times, the slides were incubated with secondary antibody (Texas red-X goat anti-mouse immunoglobulin G [IgG]; Molecular Probes) and DAPI (4′-6′-diamidino-2-phenylindole) (10 μg/ml; Sigma) for 1 h. Following PBS washing, the slides were mounted with Fluoromount-G (Southern Biotechnology Associates) containing 2.5 mg of n-propyl gallate (Sigma)/ml. Specimens were examined under a fluorescence microscope (Nikon).
RESULTS
MDM2-mediated p53 and p73 subcellular distribution is determined by their intrinsic sequence elements.
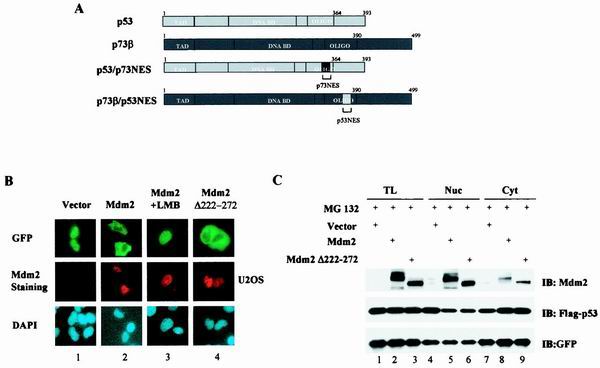
We have recently shown that while p53 undergoes nuclear export in MDM2-expressing cells, p73 accumulates in the form of nuclear aggregates that colocalize with MDM2. The inability of p73 to relocate into the cytoplasm of MDM2-expressing cells has been attributed to its functionally defective NES (8). Since p53NES has been shown to direct nuclear export of a reporter protein (20), it was of interest to determine whether introduction of p53NES could render p73 capable of nuclear export. Green fluorescent protein (GFP)-tagged plasmids expressing either p53 or p73 cDNAs with the NES exchanged (Fig. 1A) were generated to study the MDM2-mediated subcellular redistribution. GFP fusion proteins have been widely used in the study of nuclear import/export. However, it is crucial to ensure that the observed subcellular distribution of GFP-fusion proteins really results from the shuttling of the protein and is not due to protein degradation. Therefore, MG321, an inhibitor of proteasome, was included to facilitate the detection of cytoplasmically localized p53. We first tested our system by examining the MDM2-dependent relocalization of GFP-tagged wild-type p53. Consistent with its role in mediating p53 nuclear export, expression of MDM2 resulted in a redistribution of a subpopulation of GFP-p53 from the nucleus (Fig. 1B, panel 1, top) to the cytoplasm (Fig. 1B, panel 2, top). Double staining with anti-MDM2 antibody demonstrated that the cytoplasmic distribution of GFP-p53 was indeed associated with the expression of MDM2 (Fig. 1B, panel 2, middle). As expected, leptomycin B (LMB), an inhibitor of nuclear export, abolished the MDM2-mediated p53 nuclear export (Fig. 1B, panel 3). However, it is possible that protein degradation contributed to the diminished nuclear population due to incomplete inhibition of proteasome activity by the inhibitor used. To test this possibility, we generated an MDM2 deletion mutant that lacked amino acid residues 222 to 272 [MDM2 (del.222–272)]. This mutant has been shown to be unable to degrade p53 but still capable of mediating p53 nuclear export (1). As shown in Fig. 1B, panel 4, cytoplasmic localization of p53 almost identical to that observed in the wild-type MDM2-expressing cells was recorded. To further substantiate this observation, protein levels were determined by Western analysis. As shown in Fig. 1C, under the conditions used to analyze subcellular distribution, no significant degradation of the proteins was evident (lane 1 versus lane 2), even though MDM2 coexpression was associated with the translocation of the GFP-tagged proteins from the nucleus into the cytoplasm (lanes 4 and 7 versus lanes 5 and 8). A similar result was obtained when MDM2(del.222–272) was used (lanes 3, 6, and 9). Together, these results demonstrate that the observed MDM2-mediated redistribution of p53 is indeed a consequence of p53 nuclear export.
FIG. 1.
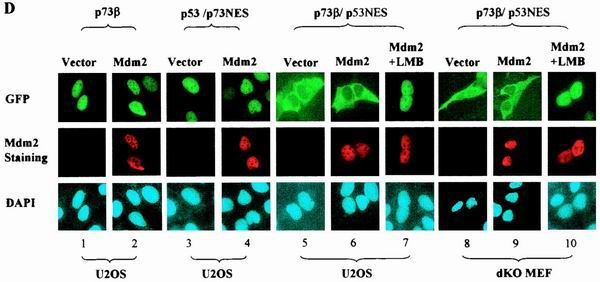
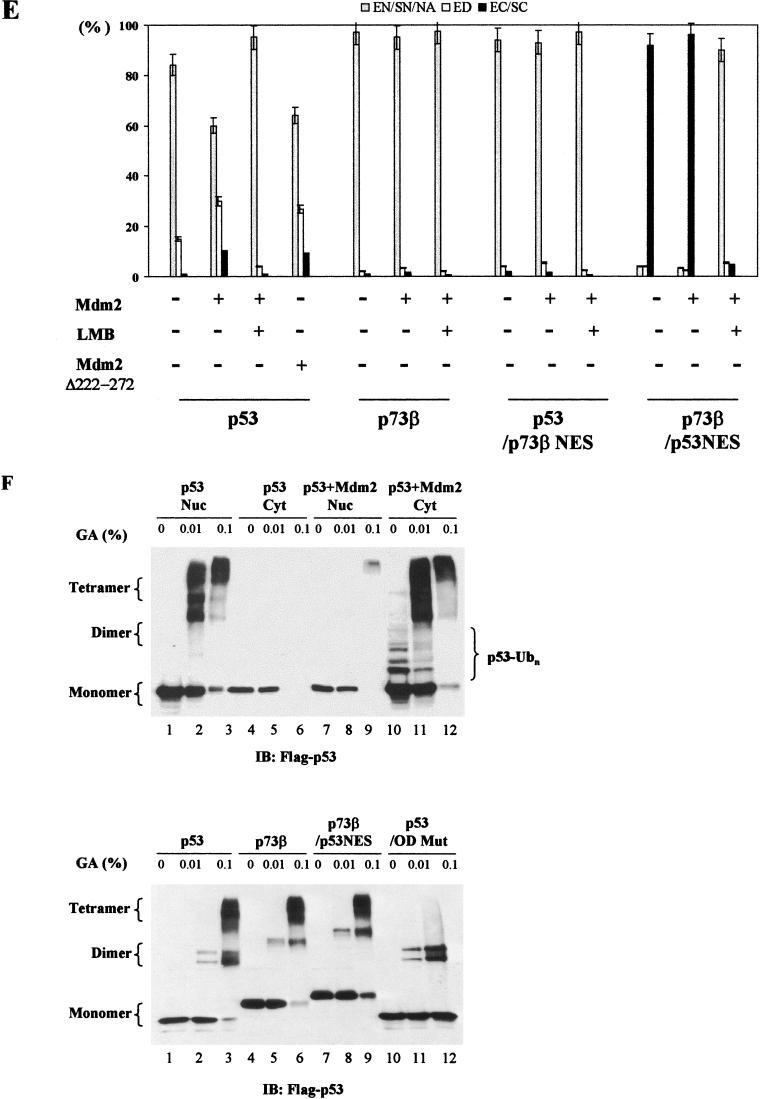
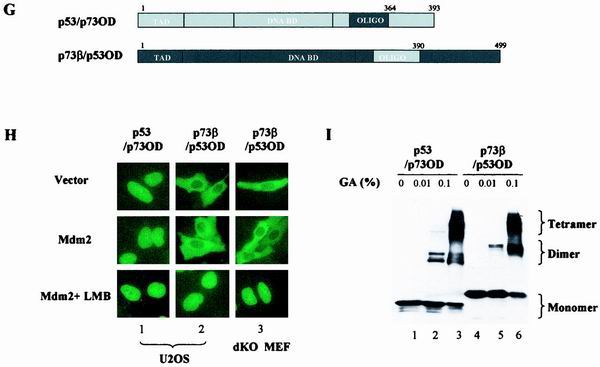
MDM2-mediated subcellular distribution of p53 and p73 is determined by their intrinsic sequence elements. (A) p73β/p53NES and p53/p73NES chimeras were generated by switching corresponding segments between p73β and p53. (B) GFP-p53 was cotransfected with a pCMV empty control vector (panel 1), pCMV-wild-type MDM2 (panels 2 and 3) or pCMV-MDM2 (del.222–272) (panel 4) into U2OS cells. MG132 (2 μM) was added to the culture to inhibit proteasome activity. MDM2-coexpressing cells were also treated with LMB (60 nM, panels 3). Cells were fixed at 36 h posttransfection, stained with anti-MDM2 antibody and DAPI, and then visualized under a fluorescent microscope. (C) Total lysates (TL) and cytoplasmic (Cyt) or nuclear (Nuc) fractions isolated from the indicated transfectants were analyzed by immunoblotting (IB) with anti-MDM2 (top) or anti-Flag (middle) antibody. MG132 (2 μM) was added to the culture to inhibit proteasome activity. An anti-GFP antibody blot was included as a transfection efficiency control (bottom). (D) GFP-tagged p53/p73NES (panels 3 and 4) or p73β/p53NES (panels 5 and 6) was cotransfected with a control vector (panels 3 or 5) or with a vector expressing MDM2 (panels 4 or 6) in U2OS cells. GFP-p73β (wt) (panels 1 and 2) was included as control. Subcellular distribution of p73β/p53NES was also analyzed in p53−/−/MDM2−/− MEFs (dKO MEF; panels 8 to 10). MDM2-coexpressing cells were also treated with LMB (60 nM) (panels 7 and 10). (E) Quantitative analysis of fluorescence data. Cells were scored as having GFP-tagged proteins distributed in the following manner: exclusive nuclear/strong nuclear/nuclear aggregates (EN/SN/EA), equally distributed in two compartments (ED), or exclusive cytoplasmic/strong cytoplasmic (EC/SC). For each condition, 200 cells from random fields were scored. Values are averages ± the standard error of the means from two separate experiments. In MDM2-coexpressing cells, the values were calculated from MDM2 and GFP double-positive cells. (F) Cytoplasmic and nuclear fractions isolated from cells expressing p53 in the presence (lanes 7 to 12) or absence (lanes 1 to 6) of MDM2 were subjected to the tetramer formation assay described in Materials and Methods. MG132 (2 μM) and cycloheximide (10 μM) were added to the culture 10 h before harvesting to block proteasome activity and protein synthesis, respectively. Proteins were resolved by SDS-PAGE, transferred onto nitrocellulose membrane, and then immunobloted with anti-Flag antibody (top). Lysates prepared from cells expressing Flag-p73β/p53NES were analyzed for tetramer formation (bottom, lanes 7 to 9). p53(wt) and p73β(wt) were included as positive controls (lanes 1 to 6). Substitution mutant (L348, 350A) of p53 was included as a negative control (lanes 10 to 12). (G) p73β/p53OD and p53/p73OD chimeras were generated by exchanging domains at the indicated positions. (H) GFP-tagged plasmids expressing the indicated cDNAs were transfected into U2OS cells (panels 1 and 2) or p53−/−/MDM2−/− MEFs (panels 3), and the subcellular distribution of the proteins was analyzed and quantitated (data not shown) as described for panels B and E. (I) Cell lysates of p73β/p53OD or p53/p73OD-expressing cells were assayed for tetramer formation as described for Fig. 1F.
Once our assay system had been verified, the distribution of the chimeric p53/p73 proteins was analyzed. In contrast to the p53/p73NES chimera, which was confined to the nucleus of U2OS cells regardless of the expression of MDM2 (Fig. 1D, panels 3 and 4, top), p73β/p53NES was almost exclusively cytoplasmically localized even in the absence of MDM2 expression (Fig. 1D, panel 5, top). Coexpression of MDM2, as shown by MDM2 double staining (Fig. 1D, panel 6, middle row), did not significantly alter the cytoplasmic distribution of p73β/p53NES (Fig. 1D, panel 6, top).
The observed nuclear exclusion of p73/p53NES could be the result of either impaired nuclear import or hyperactive export. LMB was used to differentiate between these two possibilities. As shown in Fig. 1D (panel 7, top), incubation of cells with LMB resulted in nuclear retention of the p73/p53NES protein, indicating that the observed nuclear exclusion is indeed a consequence of hyperactive nuclear export. Quantitative analysis of immunofluorescence data is presented in Fig. 1E.
Even though the p73/p53NES chimera underwent nuclear export without coexpression of MDM2, endogenously expressed MDM2 might be responsible for this effect. p53−/−/MDM2−/− MEFs were therefore used to test this possibility. Subcellular distribution almost identical to that recorded in U2OS cells was observed: the p73/p53NES protein was predominantly cytoplasmically localized regardless of MDM2 expression (Fig. 1D, panels 8 and 9), and addition of LMB resulted in nuclear retention of the protein (Fig. 1D, panel 10). These results indicate that nuclear export of p73/p53NES is independent of MDM2.
Structural analysis of the p53 molecule shows that the NES is buried when p53 is in its tetrameric conformation (10, 13). MDM2-independent nuclear export of p73β/p53NES would suggest that this chimera has lost its ability to form a tetramer, thus rendering the NES accessible to the export proteins. We utilized a tetramer formation assay to investigate this possibility. The validity of the assay system was first verified by analyzing the cytoplasmic and nuclear fractions isolated from wild-type p53-expressing cells in the presence or absence of MDM2 expression. Cycloheximide, an inhibitor of protein synthesis, was used to ensure that cytoplasmic localization of p53 was not due to de novo protein synthesis. In addition, proteasome inhibitor was included to prevent p53 protein degradation. Consistent with the MDM2-mediated p53 nuclear export, the p53 tetramer redistributed from the nucleus into the cytoplasm upon MDM2 coexpression (Fig. 1F, top, lanes 1 to 6 versus lanes 7 to 12). Interestingly, the cytoplasmically localized p53 tetramer in the MDM2-expressing cells seemed ubiquitinated, as demonstrated by a shift of the lower typical ladder in the monomer to the higher tetramer (lanes 10 to 12). This finding implies that (i) ubiquitination may not necessarily disrupt the p53 tetramer and (ii) ubiquitinated p53 could be exported into the cytoplasm without tetramer disassociation. Under experimental conditions that allowed p53 tetramer formation (Fig. 1F, bottom, lanes 1 to 3), both wild-type p73β and p73β/p53NES were also capable of forming tetrameric complexes (lanes 4 to 6 and 7 to 9). p53 which carried a double mutation in the OD that prevented tetramer assembly (6) was included as a negative control to ensure the specificity of the assay (lanes 10 to 12). These data demonstrate that fusion of the p53NES into p73 does not affect the ability of this chimeric protein to tetramerize, thus excluding impaired tetramer formation as the cause of p73β/p53NES hyperactive nuclear export.
To further confirm that substitution of p53NES into the backbone of p73β results in a hyperactively exported protein, the entire OD that included the NES was switched between p53 and p73β (Fig. 1G). Analysis of subcellular distribution of these constructs yielded results almost identical to those obtained with the “NES only” swap chimeras. In contrast to p53/p73OD, which lost its ability to undergo nuclear export, the p73β/p53OD chimeric protein displayed mainly cytoplasmic localization regardless of MDM2 coexpression (Fig. 1H, panel 2, top and middle). Analogous to p73β/p53NES, the cytoplasmic distribution of the p73β/p53OD chimeric protein was due to hyperactive nuclear export as demonstrated by the treatment with LMB (Fig. 1H, panel 2, bottom). Results obtained with p53−/−/MDM2−/− MEFs once again ruled out the potential involvement of endogenous MDM2 in the nuclear export of this chimera (Fig. 1H, panel 3). In addition, the tetramer formation assay showed that both chimeric proteins retained their ability to form tetramers (Fig. 1I). Taken together, these results indicate that the subcellular distribution of p53 and p73 depends on their intrinsic sequence elements and that p53NES can be revealed without the disruption of p53 tetrameric complex.
Specific p53 sequence elements contribute to unmasking of p53NES.
Abundant experimental evidence suggests that MDM2-mediated ubiquitination is crucial to p53 nuclear export (3, 6). However, precise conformational changes that are induced by the ubiquitination and contribution of these structural modifications to the unmasking of p53NES remain unclear. Having established that p73β can be utilized to study the activity of p53NES (Fig. 1), we set out to search for the region of p53 that contributes to the functional regulation of p53NES. The MDM2-independent nuclear export of p73β/p53OD suggested that p73 lacks the necessary sequence element that keeps the p53NES from exposure to transport proteins in tetrameric conformation. We reasoned that if such a structural element existed in p53, fusion of this element into p73β/p53OD would restore the dependence of nuclear export on MDM2. We therefore generated additional chimeras where individual domains of p73β were replaced with the corresponding regions of p53.
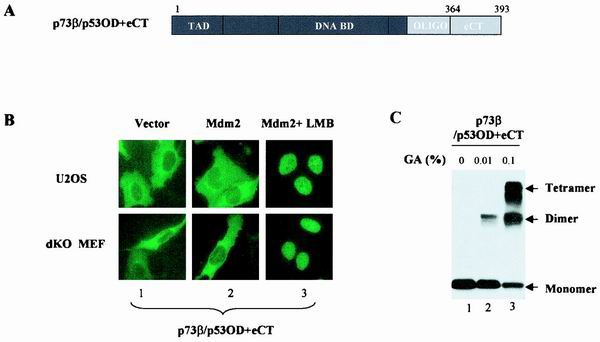
Given the fact that the p53eCT is much less conserved in p73β (9), we first fused this region into p73β/p53OD (p73β/p53OD+eCT) (Fig. 2A) to test if the extreme C terminus of p53 was able to restore the MDM2 dependence of nuclear export. GFP-tagged vector expressing p73β/p53OD+eCT was cotransfected with the empty control vector or pCMV-MDM2, and subcellular localization of this chimera was analyzed 36 h posttransfection. Surprisingly, the p53eCT failed to restore the MDM2 dependence, as evidenced by the predominantly cytoplasmic distribution of p73β/p53OD+eCT in both U2OS cells (Fig. 2B, upper panels) and p53−/−/MDM2−/− MEFs (lower panels), regardless of the MDM2 expression. LMB treatment indicated that nuclear exclusion was again attributable to the hyperactive nuclear export (Fig. 2B, panel 3). The tetramer formation ability of p73β/p53OD+eCT was apparently unaffected (Fig. 2C). These results suggest that the p53eCT is not sufficient to shield the p53NES from contacting nuclear export proteins.
FIG. 2.
Extreme C terminus of p53 is not sufficient to preserve functional latency of p53NES. (A) p73β/p53OD+eCT was generated by the swap of the p53OD+eCT into the corresponding region of p73. (B) Subcellular distribution of GFP-p73β/p53OD+eCT was analyzed in U2OS cells (top) or p53−/−/MDM2−/− MEFs (dKO MEF) (bottom) as described for Fig. 1. Exogenous MDM2-expressing cells were identified from MDM2-staining (not shown) for the analysis of chimera distribution. (C) The ability of p73β/p53OD+eCT to form tetramers was assayed as described above.
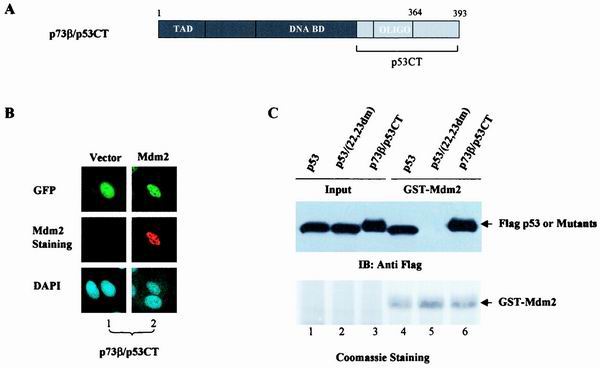
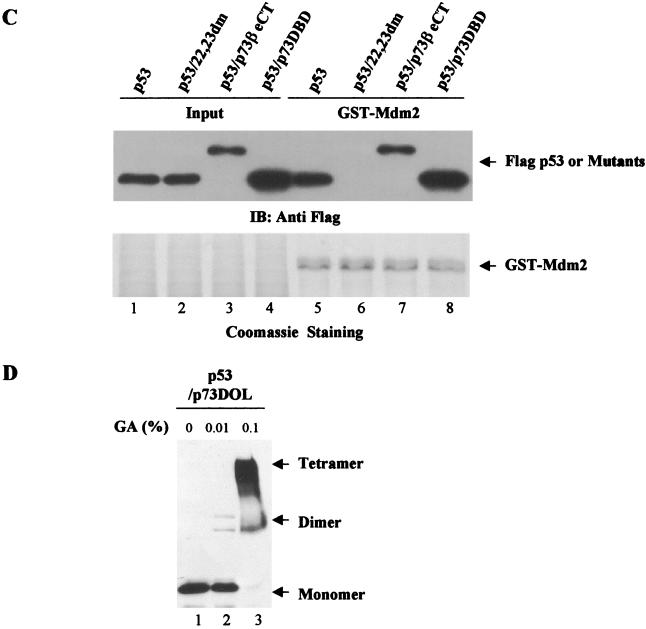
Amino acid sequence analysis showed that the region between the DBD and the OD of p53, namely, amino acids (aa) 291 to 318, is also poorly conserved between p53 and p73. We went on to examine whether this region played any role in the regulation of p53NES accessibility. To this end, the entire C terminus of p53 (aa 291 to 393) was swapped into the backbone of p73β (p73β/p53CT) (Fig. 3A) and subcellular distribution of this chimera was studied. Interestingly, p73β/p53CT protein exhibited restored nuclear distribution when the chimera was expressed alone (Fig. 3B, top), suggesting that residues 291 to 318 of p53 are necessary to keep the p53NES in its inactive form in tetrameric conformation. However, in the MDM2-expressing cells, as confirmed by anti-MDM2 antibody staining (Fig. 3B, panel 2, middle), the p73β/p53CT protein remained localized in the nucleus (Fig. 3B, panel 2 top), indicative of a resistance of this chimera to MDM2-mediated nuclear export. Since binding of MDM2 is essential for p53 nuclear export, the inability of MDM2 to direct nuclear export of the p73β/p53CT chimera could be due to its impaired binding to MDM2. Results of the GST pull-down assay, which was carried out to test this possibility, revealed that p73β/p53CT successfully bound to MDM2 (Fig. 3C, lanes 3 and 6) with an affinity comparable to that of wild-type p53 (lanes 1 and 4), while mutant p53 (22,23dm) failed to exhibit any apparent binding (Fig. 3C, lanes 2 and 5). The restored nuclear distribution of p73β/p53CT signals the importance of p53 (aa 291 to 318) to the regulation of p53NES accessibility to carrier proteins. However, the inability of this chimera to undergo MDM2-mediated nuclear export suggests the involvement of an additional sequence element.
FIG. 3.
DOL of p53 is essential for regulation of p53NES. (A) Schematic representation of the p73β/p53CT chimera. (B) GFP-p73β/p53CT was cotransfected with a control vector (panels 1) or pCMV-MDM2 (panels 2) into U2OS cells, and the subcellular distribution of the GFP-tagged proteins (top) and MDM2 (middle) was analyzed as described for Fig. 1. (C) Binding of p73β/p53CT to MDM2 was analyzed by incubating lysates from cells expressing Flag-p73β/p53CT with GST-MDM2. The adsorbates were resolved by SDS-PAGE and Western blotted with an anti-Flag antibody (top) or stained with Coomassie (bottom). Flag-p53 and p53 double mutant (p53/22.23dm) were included as positive and negative controls, respectively.
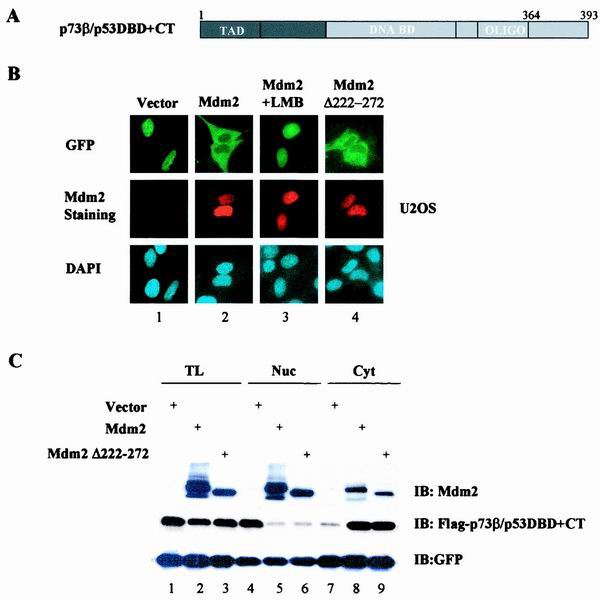
To search for this additional sequence element, we utilized p53 sequences further towards the N terminus, including the DNA-binding domain (DBD), to generate p73β/p53DBD+CT (Fig. 4A). Strikingly, the addition of the DBD completely restored this chimera's ability to undergo MDM2-dependent nuclear export. While mainly localized in the nuclei in vector-expressing cells (Fig. 4B, panel 1), the GFP-p73β/p53DBD+CT proteins in the MDM2-expressing cells exhibited predominantly cytoplasmic distribution that was abolished by LMB (Fig. 4B, panels 2 and 3). A similar cytoplasmic distribution of the chimera was observed in the MDM2(del.222–272)-expressing cells, excluding the involvement of p53 degradation (Fig. 4B, panel 4). Despite considerable efforts, we were unable to further define the minimal region within DBD that is required for MDM2-mediated nuclear export since any swapping within the DBD resulted in chimeras that were no longer responsive to MDM2 (not shown). Protein levels were also measured by Western analysis to ensure that nuclear export, rather than protein degradation, was responsible for the observed nuclear exclusion of the chimeric protein (Fig. 4C). In fact, we demonstrated earlier that p73β/p53DBD+CT is inherently resistant to MDM2-mediated degradation (7). These observations suggest that the intact p53DBD is essential for MDM2-dependent nuclear export.
FIG. 4.
Role of p53 DBD in MDM2-mediated nuclear export. (A) Schematic presentation of p73β/p53DBD+CT. (B) GFP-p73β/p53DBD+CT was cotransfected into U2OS cells with either an empty vector (panel 1), pCMV-MDM2 (wt) (panels 2 and 3), or pCMV-MDM2(del.222–272), and the subcellular distribution of the chimeras was analyzed as described for Fig. 1. (C) Total lysates (lanes 1 to 3), nuclear fraction (lanes 4 to 6), or cytoplasmic fraction (lanes 7 to 9) isolated from the transfectants were immunoblotted with anti-MDM2 (top), anti-Flag (middle), or anti-GFP (bottom) antibody as described for Fig. 1C.
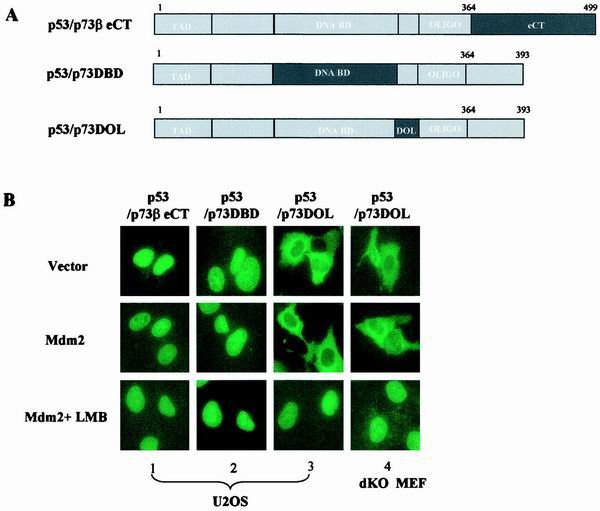
To gain additional evidence for the role of the p53OD, C terminus, and DBD in the regulation of p53 subcellular distribution, we employed yet another strategy and systematically replaced each of these three domains in p53 with the corresponding sequence of p73β (Fig. 5A). We then analyzed MDM2-mediated nuclear export of these chimeras. Substitution of the p53eCT resulted in the protein with nuclear distribution that was no longer exported upon coexpression of MDM2 (Fig. 5B, panel 1), further implicating p53eCT in the MDM2-mediated nucleocytoplasmic shuttling of p53. An almost identical result was obtained with the p53/p73DBD chimera that had mainly nuclear distribution even in the cells expressing MDM2 (Fig. 5B, panel 2). The latter observation mirrored the result described for Fig. 3, where the p53DBD was shown to be essential for the MDM2-mediated nuclear export. Both p53/p73βeCT and p53/p73DBD retained their ability to bind to MDM2 (Fig. 5C), thus eliminating defective MDM2 binding as a probable cause of impaired nuclear export. Interestingly, replacement of p53 amino acid residues 291 to 318 (DBD and OD linker region [DOL]) with the corresponding region of p73 abolished the MDM2 dependence: the chimeric protein was mainly cytoplasmically localized even in the absence of MDM2 expression (Fig. 5B, panel 3, top). The cytoplasmic distribution of this chimera was due to the hyperactive nuclear export (Fig. 5B, panel 3, bottom), which was independent of MDM2 as demonstrated by the results obtained with p53−/−/MDM2−/− MEFs (Fig. 5B, panel 4). The tetramerization ability of p53/p73DOL was similar to that of wild-type p53 (Fig. 5D). Taken together, these data demonstrate that while residues 291 to 318 of p53 appear to be required for the maintenance of p53NES in its inactive form, the DBD and the eCT of p53 seem to be essential for the MDM2-mediated activation of the p53NES.
FIG. 5.
DBD, eCT, and DOL of p53 contribute to regulation of p53NES. (A) Chimeras were generated by the swap of each segment of p73β into the backbone of p53 at the indicated position. (B) Subcellular distribution of chimeric proteins was analyzed as described for Fig. 1 in U2OS cells (panels 1 to 3). Distribution of p53/p73DOL was also analyzed in p53−/−/MDM2−/− MEFs (dKO MEF; panel 4). (C) Binding of p53/p73DBD or p53/p73βeCT to MDM2 was examined as described for Fig. 3. (D) Tetramer formation of the p53/p73DOL protein was assayed as described for Fig. 1.
Potential mechanisms of p53NES regulation.
Having demonstrated the critical role of three regions of p53 in the regulation of MDM2-mediated activation of p53NES, we proceeded to obtain mechanistic insights into the contribution of these p53 domains to the regulation of p53NES. As mentioned previously, the ring domain of MDM2, which contains the E3 ligase activity, has been shown to be essential for MDM2-mediated p53 nuclear export. It is therefore plausible that the ubiquitination of p53 lysine residues residing within DBD, eCT, or DOL may induce conformational changes that will result in the unmasking of p53NES.
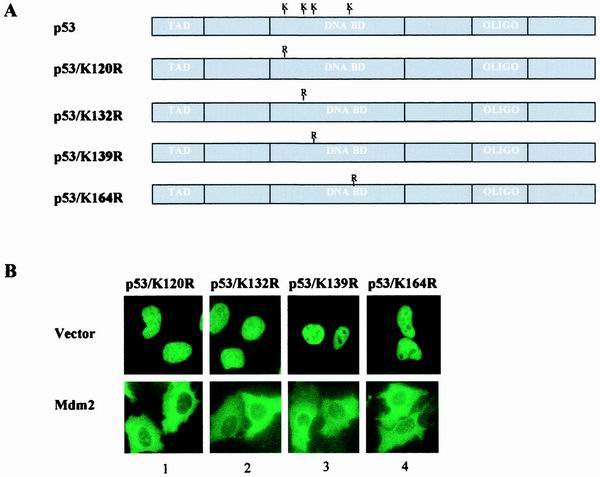
We first focused on the DBD of p53. At least three possibilities exist that could explain the importance of p53DBD in the regulation of the NES: (i) an additional NES harbored in the DBD contributes to p53 nuclear export; (ii) lysine residues within the DBD of p53 can be targeted by MDM2 for ubiquitination and thereby contribute to the activation of p53NES; and (iii) the DBD of p53 is critical to the maintenance of overall three-dimensional conformation that regulates p53NES. Examination of the subcellular distribution of the entire DBD, or smaller regions of the DBD, that were fused into the GFP vector did not support the presence of an additional NES in the DBD of p53 (data not shown). Analysis of the MDM2-mediated subcellular redistribution of p53 mutants where lysines in the DBD were replaced with arginines (Fig. 6A) revealed no apparent difference between the mutants and wild-type p53 (Fig. 6B), thus ruling out the possibility that the ubiquitination of these lysine residues contributes to the activation of p53NES. Together, these results suggest that the DBD of p53 likely contributes to the maintenance of an overall molecular conformation that is necessary for the functional regulation of p53NES.
FIG. 6.
Lysine residues in DBD of p53 are dispensable for MDM2-mediated p53 nuclear export. (A) Schematic representation of p53 mutant bearing substitutions of lysine 120, 132, 139, or 164 to arginine. (B) The indicated p53 mutant was cotransfected with a control vector (top) or MDM2 (bottom), and the subcellular distribution of the mutant was analyzed and quantitated as described for Fig. 1B and E.
We next directed our attention to the six C-terminal lysine residues of p53 since recently obtained evidence indicates that these lysines are main sites of MDM2-dependent ubiquitination (18, 19). Situated next to the OD where p53NES resides, the ubiquitination of these residues could induce conformational changes that would lead to the exposure of p53NES. If it is the case, then the observed MDM2-independent nuclear export of p73β/p53OD might be explained by the lack of these C-terminal lysine residues in p73β. In addition, the p53 chimera lacking its extreme C terminus, p53/p73βeCT, has shown resistance to the MDM2-mediated nuclear export (Fig. 4B), which would also implicate C-terminal lysines in p53 nuclear export.
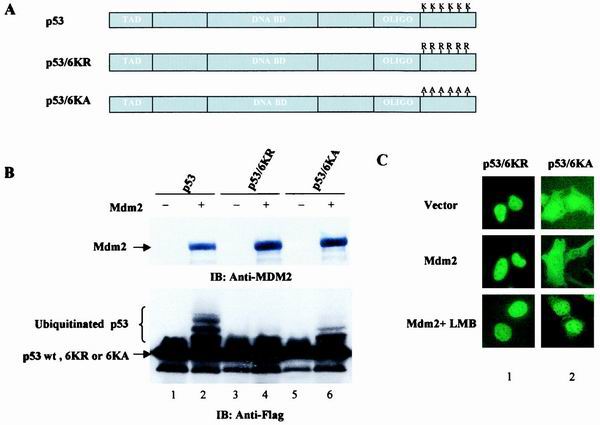
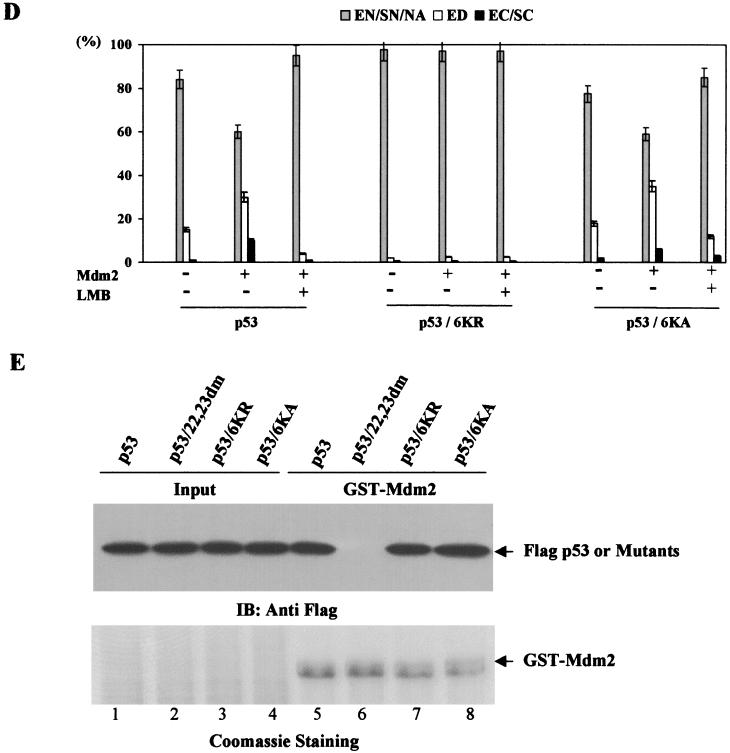
To assess the role of the C-terminal lysine residues in the regulation of p53NES, we prepared p53 mutants bearing simultaneous substitutions of lysines 370, 372, 373, 381, 382, and 386 to arginines (6KR) or to alanines (6KA) (Fig. 7A). Since these C-terminal lysine residues have been shown to undergo ubiquitination by MDM2 (14, 15), both 6KR and 6KA mutants of p53 exhibited significantly reduced ubiquitination in comparison to wild-type p53 (Fig. 7B). Nonetheless, the 6KA mutant retained a partial ability to be ubiquitinated by MDM2 (Fig. 7B, lane 4 versus lane 6). The latter observation has been reported previously (18) and indicates that lysine residues other than those in the C terminus can also be ubiquitinated. Subcellular distribution analysis revealed that the K-to-R substitution completely abolished the ability of this p53 mutant to be nuclear exported, as demonstrated by the exclusive nuclear staining of the protein even in the MDM2-expressing cells (Fig. 7C, panel 1, middle). This is consistent with the results described in the accompanying paper by Lohrum et al. (15). Interestingly, the 6KA mutant of p53, when transfected alone, exhibited a slight increase in the cytoplasmic localization (Fig. 7C, panel 2, top) when compared with wild-type p53 (Fig. 1B, panel 1), and this increased cytoplasmic staining was completely abolished by the treatment with LMB. Moreover, coexpression of MDM2 resulted in a further increase of the cytoplasmically localized 6KA mutant (Fig. 7C, panel 2, middle), suggesting that this p53 mutant retained partial ability to undergo MDM2-mediated nuclear export. Quantitative data from these experiments are presented in Fig. 7D. Consistent with previous reports (18, 19), both 6KR and 6KA mutants of p53 were able to bind to MDM2 (Fig. 7E).
FIG. 7.
Six C-terminal p53 lysine residues are essential for MDM2-mediated nuclear export. (A) Schematic representation of p53 mutants bearing substitutions of lysines 370, 372, 373, 381, 382, and 386 to arginines or alanines. (B) Flag-tagged p53wt (lanes 1 and 2), p53/6KR (lanes 3 and 4), or p53/6KA (lanes 5 and 6) were cotransfected with an empty vector (lanes 1, 3, and 5) or pCMV-MDM2 (lanes 2, 4, and 6). Cells were harvested 36 h posttransfection and subjected to Western analysis with anti-MDM2 (top) or anti-Flag (bottom) antibody. (C) Subcellular distribution of GFP-p53/6KR or 6KA was analyzed as described for Fig. 1. (D) Quantitative analysis of fluorescence data as described for Fig. 1. (E) p53/6KR or 6KA protein binding to MDM2 was assessed as described in Materials and Methods.
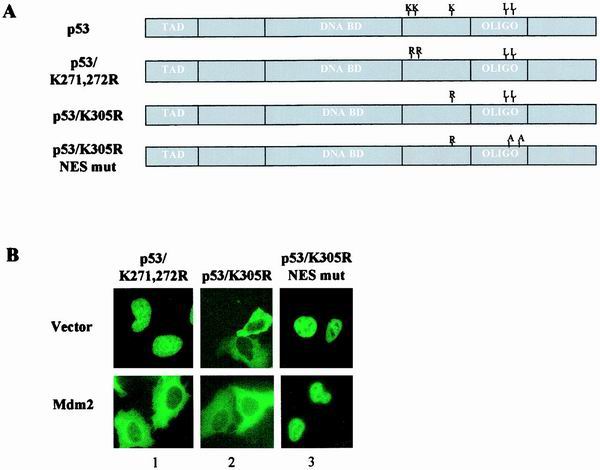
Lastly, the DOL of p53 was examined. Since this region was shown to be essential for preservation of p53NES in its inactive state, we focused on the possibility that MDM2 targets lysine residues in this region for ubiquitination, which in turn induces an additional conformational change required for fully revealing p53NES. p53 mutants bearing lysine-to-arginine substitutions as shown (Fig. 8A) were generated to test the potential involvement of DOL ubiquitination in p53 nuclear export. Analysis of MDM2-dependent subcellular distribution revealed that lysines 291 and 292 are dispensable for p53 nuclear export, as demonstrated by the fully preserved ability of the K-to-R mutant to undergo nuclear export in the MDM2-expressing cells (Fig. 8B, panel 1). In contrast, the 305(K-R) mutant of p53 exhibited predominantly cytoplasmic distribution even in the absence of MDM2 coexpression (Fig. 8B, panel 2), an observation consistent with the result reported earlier (14). The cytoplasmic distribution of this mutant, however, has been previously interpreted as a result of impaired nuclear import (14). An additional mutation that disabled the p53NES was introduced into the 305(K-R) mutant to determine whether impaired nuclear import or hyperactive nuclear export was responsible for cytoplasmic localization. As shown in Fig. 8B, panel 3, the double mutant of p53 was exclusively confined to the nucleus, indicative of a retained nuclear import function in the 305(K-R) mutant of p53. Together, these results demonstrate that lysine 305 plays a critical role in the control of p53NES.
FIG. 8.
Lysine 305 is pivotal in control of p53NES. (A) Schematic representation of p53 mutant bearing substitutions of lysines 271 and 272 or 305 to arginines or K305R plus NES mutation (L348,350A). (B) Subcellular distribution of the mutants was analyzed as described above.
DISCUSSION
It has been shown that nuclear import/export of the p53 protein is controlled by a fast, energy-dependent pathway (17). Similarly to other nuclear proteins, p53 contains both nuclear localization sequences (NLSs) and an NES. The NLSs and NES are recognized by special transporter proteins, karyopherin alpha (importin alpha), and exportin 1 (CRM1), respectively (21). Binding of the corresponding transporter to the NLSs or NES is essential to protein shuttling between the cytoplasm and the nucleus. The NES of p53, however, lies within the OD and is buried at the interface of the two dimers that form the active tetramer (10, 13). It was therefore suggested that p53 tetramer has to be disassembled into either dimer or monomer form in order to reveal the NES (20). MDM2 has been shown to function as a mediator of p53 nuclear export, and the MDM2 ring domain, which harbors the E3 ubiquitin ligase activity, is essential for this function. Thus, ubiquitination of p53 by MDM2 could be one of the required components of the nuclear export process.
In this study, we utilized a domain swap approach between p53 and its close structural homologue p73 to investigate relative contributions of various p53 domains to nuclear export by MDM2. Substantial sequence homology to p53 notwithstanding, p73 does not undergo nuclear export in the presence of MDM2 but rather colocalizes with MDM2 in the form of nuclear aggregates. Different patterns of subcellular distribution of p53 and p73 imply that a unique p53 sequence element(s) might control the MDM2-mediated nuclear export. We show here that three regions of p53, including the DBD, C terminus, and the residues from 291 to 318 of p53, appear to be involved in the regulation of the MDM2-mediated nuclear export. While the precise role of residues 291 to 318 remains to be determined, we demonstrate that the DBD of p53 is likely to be involved in the maintenance of protein conformation essential to the proper functioning of the NES, and the extreme C terminus of p53, specifically lysine residues found in that region, are necessary for the unmasking of the p53NES.
Consistent with recent reports showing that p53 C-terminal lysine residues are major sites of ubiquitination by MDM2 (18, 19), the replacement of the p53eCT with the corresponding region of p73 that lacks lysine residues resulted in a significantly reduced level of ubiquitination. Failure of the p53/p73eCT chimera to undergo MDM2-dependent nuclear export provided evidence of a potential association between the ubiquitination of p53eCT and nuclear export. These findings were further confirmed when the inefficient ubiquitination of the 6KR p53 mutant strongly correlated with its inability to undergo nuclear export. The latter observation establishes a direct link between ubiquitination of the C-terminal lysine residues and nuclear export of p53, a finding similar to that described in the accompanying paper by Lohrum et al. (15). It is plausible that ubiquitination of the C-terminal lysines could induce conformational changes that result in the exposure of the buried p53NES and permit p53 to undergo nuclear export.
However, the MDM2-independent nuclear export of p73/p53OD+eCT chimera suggests the existence of sequence elements other than the extreme p53 C terminus that are important in regulating the p53NES. Indeed, the DBD and the residues from 291 to 318 of p53 are also required for the MDM2-mediated p53 nuclear export. Since we demonstrate herein that p53DBD does not harbor any additional NES and that abrogation of ubiquitination in this region does not diminish MDM2-mediated p53 nuclear export, it is plausible that the DBD of p53 contributes to the maintenance of the overall conformation that is required for the functional activation of p53NES. The DBD of p53 features unique loops and loop-sheet-helix conformation, as revealed by the crystallographic study (4). While the crystal structure of p73DBD has not yet been solved, the finding that p73 differentially regulates cellular p53 target genes (22) suggests distinct conformational folding of p53 and p73 DBD. The resistance of the p53/p73DBD to MDM2-mediated nuclear export is consistent with this notion.
The importance of the DOL of p53 in the regulation of p53NES is evident from our study, although its precise role remains to be determined. The results obtained from the lysine-to-arginine substitution mutant identifies lysine 305 as a key residue in the control of p53NES unmasking. Although, due to a technical difficulty, we were unable to provide direct evidence of MDM2 ubiquitination of this residue, the profound change from a nuclear to cytoplasmic distribution associated with replacing lysine 305 with arginine suggests that a subtle modification of this region of p53 may be sufficient to induce conformational change and subsequently fully reveal the NES.
We therefore envision a model of stepwise ubiquitination of p53 by MDM2 which results in the unmasking of buried p53NES. In this model, MDM2 first ubiquitinates the C-terminal lysine residues, an event that renders additional lysines from outside of the C terminus susceptible to ubiquitination. Results obtained using the 6KA p53 mutant support this model. The K-to-A substitution neutralized the net positive charge on the p53 molecule, which likely caused conformational change that allowed other lysine residues in p53 to undergo ubiquitination by MDM2. Accordingly, the 6KA mutant of p53 remained ubiquitinated, albeit with a reduced efficiency, and exhibited a partial response to the MDM2-mediated nuclear export. In contrast, the net charge of the 6KR p53 mutant was not significantly affected and it maintained its conformation, thus not becoming responsive to MDM2. Observations obtained with the 6KA p53 mutant underscore the importance of ubiquitination of non-C-terminal lysines to the activation of the p53NES and nuclear export. Together, our results suggest a cascade of conformational changes in the p53 molecule in response to the MDM2-mediated stepwise ubiquitination that eventually lead to the exposure of p53NES and p53 export into the cytoplasm.
Finally, it has been proposed that the p53 tetramer has to be disassociated into a dimer or monomer to reveal the buried p53NES (20). Results obtained with the p73/p53NES, p73/p53OD, and p53/p73 DOL chimeric proteins, however, clearly show that it is not necessary to disrupt the tetrameric conformation of p53 in order to reveal the buried NES. Our data suggest that MDM2-mediated sequential ubiquitination of p53 is sufficient for the unmasking of NES in a p53 tetramer.
ACKNOWLEDGMENTS
We thank Carl Maki (Harvard School of Public Health) for p53−/−/MDM2−/− MEFs. We are grateful to Minoru Yoshida, Tokyo University, Tokyo, Japan, for generously providing leptomycin B.
This work was supported by an NIH research grant (RO1 CA85679-01) and the Milton Fund of Harvard Medical School.
REFERENCES
- 1.Argentini M, Barboule N, Wasylyk B. The contribution of the acidic domain of MDM2 to p53 and MDM2 stability. Oncogene. 2001;20:1267–1275. doi: 10.1038/sj.onc.1204241. [DOI] [PubMed] [Google Scholar]
- 2.Ashcroft M, Vousden K H. Regulation of p53 stability. Oncogene. 1999;18:7637–7643. doi: 10.1038/sj.onc.1203012. [DOI] [PubMed] [Google Scholar]
- 3.Boyd S D, Tsai K Y, Jacks T. An intact HDM2 RING-finger domain is required for nuclear exclusion of p53. Nat Cell Biol. 2000;2:563–568. doi: 10.1038/35023500. [DOI] [PubMed] [Google Scholar]
- 4.Cho Y, Gorina S, Jeffrey P D, Pavletich N P. Crystal structure of a p53 tumor suppressor-DNA complex: understanding tumorigenic mutations. Science. 1994;265:346–355. doi: 10.1126/science.8023157. [DOI] [PubMed] [Google Scholar]
- 5.Freedman D A, Levine A J. Nuclear export is required for degradation of endogenous p53 by MDM2 and human papillomavirus E6. Mol Cell Biol. 1998;18:7288–7293. doi: 10.1128/mcb.18.12.7288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Geyer R K, Yu Z K, Maki C G. The MDM2 RING-finger domain is required to promote p53 nuclear export. Nat Cell Biol. 2000;2:569–573. doi: 10.1038/35023507. [DOI] [PubMed] [Google Scholar]
- 7.Gu J, Chen D, Rosenblum J, Rubin R M, Yuan Z M. Identification of a sequence element from p53 that signals for Mdm2-targeted degradation. Mol Cell Biol. 2000;20:1243–1253. doi: 10.1128/mcb.20.4.1243-1253.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gu J, Nie L H, Kawai H, Yuan Z M. Subcellular distribution of p53 and p73 are differentially regulated by MDM2. Cancer Res. 2001;61:6703–6707. [PubMed] [Google Scholar]
- 9.Honda R, Tanaka H, Yasuda H. Oncoprotein MDM2 is a ubiquitin ligase E3 for tumor suppressor p53. FEBS Lett. 1997;420:25–27. doi: 10.1016/s0014-5793(97)01480-4. [DOI] [PubMed] [Google Scholar]
- 10.Jeffrey P D, Gorina S, Pavletich N P. Crystal structure of the tetramerization domain of the p53 tumor suppressor at 1.7 angstroms. Science. 1995;267:1498–1502. doi: 10.1126/science.7878469. [DOI] [PubMed] [Google Scholar]
- 11.Kaghad M, Bonnet H, Yang A, Creancier L, Biscan J C, Valent A, Minty A, Chalon P, Lelias J M, Dumont X, Ferrara P, McKeon F, Caput D. Monoallelically expressed gene related to p53 at 1p36, a region frequently deleted in neuroblastoma and other human cancers. Cell. 1997;90:809–819. doi: 10.1016/s0092-8674(00)80540-1. [DOI] [PubMed] [Google Scholar]
- 12.Lain S, Midgley C, Sparks A, Lane E B, Lane D P. An inhibitor of nuclear export activates the p53 response and induces the localization of HDM2 and p53 to U1A-positive nuclear bodies associated with the PODs. Exp Cell Res. 1999;248:457–472. doi: 10.1006/excr.1999.4433. [DOI] [PubMed] [Google Scholar]
- 13.Lee W, Harvey T S, Yin Y, Yau P, Litchfield D, Arrowsmith C H. Solution structure of the tetrameric minimum transforming domain of p53. Nat Struct Biol. 1994;1:877–890. doi: 10.1038/nsb1294-877. [DOI] [PubMed] [Google Scholar]
- 14.Liang S H, Clarke M F. The nuclear import of p53 is determined by the presence of a basic domain and its relative position to the nuclear localization signal. Oncogene. 1999;18:2163–2166. doi: 10.1038/sj.onc.1202350. [DOI] [PubMed] [Google Scholar]
- 15.Lohrum M A, Woods D, Ludwig R L, Vousden K H. C-terminal ubiquitination of p53 contributes to nuclear export. Mol Cell Biol. 2001;21:8521–8532. doi: 10.1128/MCB.21.24.8521-8532.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lohrum M A, Vousden K H. Regulation and function of the p53-related proteins: same family, different rules. Trends Cell Biol. 2000;10:197–202. doi: 10.1016/s0962-8924(00)01736-0. [DOI] [PubMed] [Google Scholar]
- 17.Middeler G, Zerf K, Jenovai S, Thulig A, Tschodrich-Rotter M, Kubitscheck U, Peters R. The tumor suppressor p53 is subject to both nuclear import and export, and both are fast, energy-dependent and lectin-inhibited. Oncogene. 1997;14:1407–17. doi: 10.1038/sj.onc.1200949. [DOI] [PubMed] [Google Scholar]
- 18.Nakamura S, Roth J A, Mukhopadhyay T. Multiple lysine mutations in the C-terminal domain of p53 interfere with MDM2-dependent protein degradation and ubiquitination. Mol Cell Biol. 2000;20:9391–9398. doi: 10.1128/mcb.20.24.9391-9398.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rodriguez M S, Desterro J M, Lain S, Lane D P, Hay R T. Multiple C-terminal lysine residues target p53 for ubiquitin-proteasome-mediated degradation. Mol Cell Biol. 2000;20:8458–8467. doi: 10.1128/mcb.20.22.8458-8467.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stommel J M, Marchenko N D, Jimenez G S, Moll U M, Hope T J, Wahl G M. A leucine-rich nuclear export signal in the p53 tetramerization domain: regulation of subcellular localization and p53 activity by NES masking. EMBO J. 1999;18:1660–1672. doi: 10.1093/emboj/18.6.1660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Weis K. Importins and exportins: how to get in and out of the nucleus. Trends Biochem Sci. 1998;23:185–189. doi: 10.1016/s0968-0004(98)01204-3. [DOI] [PubMed] [Google Scholar]
- 22.Zhu J, Jiang J, Zhou W, Chen X. The potential tumor suppressor p73 differentially regulates cellular p53 target genes. Cancer Res. 1998;58:5061–5065. [PubMed] [Google Scholar]