Abstract
The maturation pathway for the nickel-dependent enzyme urease utilizes the protein UreE as a metallochaperone to supply Ni(II) ions. In Helicobacter pylori urease maturation also requires HypA and HypB, accessory proteins that are commonly associated with hydrogenase maturation. Herein we report on the characterization of a protein complex formed between HypA and the UreE2 dimer. Nuclear magnetic resonance (NMR) coupled with molecular modelling show that the protein complex apo, Zn-HypA•UreE2, forms between the rigorously conserved Met-His-Glu (MHE motif) Ni-binding N-terminal sequence of HypA and the two conserved His102A and His102B located at the dimer interface of UreE2. This complex forms in the absence of Ni(II) and is supported by extensive protein contacts that include the use of the C-terminal sequences of UreE2 to form additional strands of β-sheet with the Ni-binding domain of HypA. The Ni-binding properties of apo, Zn-HypA•UreE2 and the component proteins were investigated by isothermal titration calorimetry using a global fitting strategy that included all of the relevant equilibria, and show that the Ni,Zn-HypA•UreE2 complex contains a single Ni(II)-binding site with a sub-nanomolar KD. The structural features of this novel Ni(II) site were elucidated using proteins produced with specifically deuterated amino acids, protein point mutations, and the analyses of X-ray absorption spectroscopy, hyperfine shifted NMR features, as well as molecular modeling coupled with quantum-mechanical calculations. The results show that the complex contains a six-coordinate, high-spin Ni(II) site with ligands provided by both component proteins.
Keywords: enzyme maturation, Helicobacter pylori, metallochaperone, nickel trafficking
Graphical Abstract
Graphical Abstract.
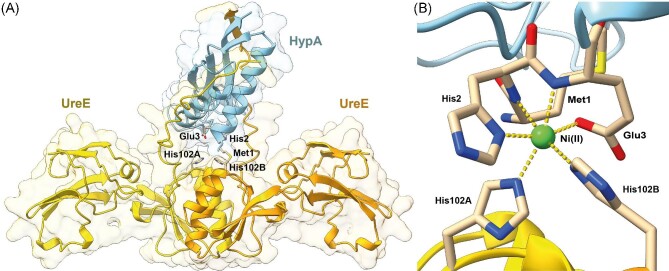
Models of the HypA•UreE2 protein complex (A) and novel Ni(II) site (B) formed at the interface.
Introduction
HypA is a monomeric metallochaperone that is generally associated with Ni(II) incorporation into the large subunit of [Ni,Fe]-hydrogenases,1–4 while the dimeric UreE2 serves this nickel transport function in bacteria that produce urease.4–9 In Helicobacter pylori (Hp), HypA serves both functions in that HypA is also required for urease Ni(II) incorporation under physiological conditions.4,10-13 Hp is a human pathogen that infects the stomach and is a major cause of ulcers, gastric cancers, and mucosa-associated lymphoid tissue lymphoma.4,14,15 Its ability to survive the acidic environment of the stomach depends on the activity of urease, a nickel-dependent enzyme that catalyzes the hydrolysis of urea to ammonia and CO2 and thereby helps Hp maintain its internal pH as well as modify the pH of its environment.16 Although [Ni,Fe]-hydrogenase is not required for acidic survival,17 it is involved in utilization of H2 as a source of energy and is associated with the virulence of the organism in terms of its ability to colonize gastric mucosa and to induce cancer.4,18,19
The activity of urease is partly controlled by Ni(II) insertion, as Hp expresses the urease protein under neutral conditions, but it remains largely apo-protein until the organism experiences acid shock, which results in rapid nickelation of the enzyme.16,20 Numerous studies have shown that mutations that lead to defective nickel trafficking to urease (HypA/B or UreE2 deletion, or interfere with the ability of HypA or UreE2 to function as a nickel metallochaperone) lead to strains of Hp that are unable to survive acidic conditions,10,12,13,21–23 as well as to inactive H2ase.17 Thus, interfering with urease nickel incorporation provides a novel antibacterial strategy for treating Hp infections, including the growing number of clarithromycin-resistant infections.24
Recent work has shown that HypA and UreE2 form a 1:1 protein complex that contains a single high-affinity nickel-binding site.25 The work described here is focused on characterizing the Ni-binding properties of the HypA•UreE2 complex and the structure of the novel Ni(II) site. Isothermal titration calorimetry (ITC) studies employing a global fitting strategy show that the affinity of this site (KD ∼ 10−10 M) is much tighter than typical for metallochaperones (KD ∼ μM)26 and is more characteristic of Ni-responsive transcriptional regulators, such as InrS (KD ∼ 10−10 M)27 or NikR (KD ∼ 10−8 M).28 This tight Ni binding, as well as the strong interactions between HypA and UreE2, allowed for the isolation and characterization of the Ni,Zn-HypA•UreE2 complex and three protein mutants.
X-ray-absorption spectroscopy (XAS) performed on isolated Ni-HypA•UreE2 reveals a six-coordinate Ni(II) center containing three His residues. This six-coordinate Ni(II) site also gives rise to well-defined hyperfine-shifted resonances in the 1H-NMR (nuclear magnetic resonance) spectrum. Detailed analysis of this NMR spectrum employing amino acid modifications of both UreE2 and HypA, as well as proteins produced with specifically deuterated amino acids, reveals a structure that involves coordination of Ni(II) by His residues from both proteins (His102A and His102B from UreE2, and His2 from HypA), provides evidence regarding the remaining three ligands, proposes a model for the structure of the HypA•UreE2 complex, and suggests a mechanism for coordination of Ni,Zn-HypA by UreE2. Density functional theory (DFT)-based computational studies corroborate the experimental results, yielding experimentally supported models for the Ni site in HypA as well as in the HypA•UreE2 complex.
Materials and methods
Protein production
Wild-type (WT) HypA and its mutant L2*HypA, where a Leu residue was inserted between the N-terminal Met1 and the His2 residue, were produced by modifying a previously reported protocol (see Supplementary Information).23 WT HypA containing perdeuterated methionine or histidine was produced with the same protocol, using a modified M9 medium (12 g/L Na2HPO4, 6 g/L KH2PO4, 1 g/L NaCl); when OD600 = 0.6 was reached, 0.16 mM of (2,3,3,4,4-D5, methyl-D3, 98%)-L-methionine (Cambridge Isotope Laboratories, MA, USA) or (2,3,3-D3, ring-D2, 98%)-L-histidine (Eurisotop, Saint Aubin, France) was added to the cellular growth medium and incubated at 25 °C for 1 h before adding 10 μM ZnSO4 and IPTG (Isopropyl β-D-1-thiogalactopyranoside) as described in the procedure provided in the Supplementary Information.
A cell-free expression protocol was applied to produce HypA containing perdeuterated aspartate or glutamate. The sequence corresponding to hypA was cloned into a pIVEX 2.3d vector; in particular, the pIVEX2.3d vector was PCR amplified using the primers 5′-gtcgactcgagcgagctcc-3′ and 5′-atgtgcatatgtatatctccttcttaaag-3′, which contain the sequences (bold characters) for XhoI and NdeI, respectively. The obtained sequence was digested with these restriction enzymes and ligated to the sequence of hypA obtained by restriction of pET22b-hypA with the same endonucleases.29 Positive clones for pIVEX 2.3d-hypA were identified by colony PCR, restriction tests and sequencing on both strands. The recombinant plasmid was purified using the Nucleobond Xtra Maxi plus kit (Macherey-Nagel). The protein was synthesized in vitro using a cell-free expression system (ISBG, Grenoble). HypA was expressed under RNAse-free conditions in dialysis mode, in a volume of 15 mL with a 1/10 ratio of reaction mixture to feeding mixture, for 16 h at 27 °C under gentle agitation. The cell-free mixture30 contained 16 μg/mL of pIVEX 2.3d-hypA, 1 mM of 19 essential protonated amino acids, 0.8 mM of each rNTPs (guanosine-, uracil-, and cytidine-5′-triphosphate), 55 mM HEPES (4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid) (pH 7.5), 68 μM folinic acid, 0.64 mM cyclic adenosine monophosphate, 3.4 mM dithiothreitol, 27.5 mM ammonium acetate, 2 mM spermidine, 80 mM creatine phosphate, 200 mM potassium acetate, 18 mM magnesium acetate, 250 μg/mL creatine kinase, 27 μg/mL T7 RNA polymerase, 0.175 μg/mL tRNA, 5 μM ZnSO4 and 400 μL/mL S30 E. coli bacterial extract. The latter was previously treated with 20 mM NaBH4, 20 mM D-malate and 20 mM amino-oxy-acetate to inhibit transaminase activity. The HypA samples containing 2H-Glu and 2H-Asp were prepared with the addition of 1 mM (2,3,3,4,4-D5, 98%)-L-glutamic acid or (2,3,3-D3, 98%)-L-aspartic acid, respectively. In both cases, the reaction mixture was clarified after incubation by centrifugation for 20 min at 15 000 ×g and 4 °C. The supernatant was diluted to 45 mL in 20 mM TrisHCl buffer at pH 7.5 containing 20 mM NaCl and 1 mM TCEP, and then loaded onto a RESSOURCE-Q 6 mL column (GE Healthcare) pre-equilibrated with 20 mM TrisHCl buffer at pH 7.2, containing 20 mM NaCl and 1 mM TCEP (tris(2-carboxyethyl)phosphine). The column was washed using a flow rate of 4 mL min−1 with the starting buffer until the baseline was stable. A linear gradient of NaCl (220 mL, from 0.02 to 0.6 M) was applied to the column and the fractions containing HypA were combined, concentrated using 3 kDa ultra-filtration units (Millipore) to a final volume of 2 mL, and loaded onto a Superdex 75 10/300GL column equilibrated with 20 mM HEPES at pH 7.2, containing 200 mM NaCl and 1 mM TCEP for a final purification step. The fractions containing the purified protein, evaluated using SDS-PAGE, were pooled together, concentrated with Amicon Ultra concentrators (Millipore, GE) with a 3 kDa cutoff, and stored at −80 °C until use.
The protein–protein complexes were prepared starting from the single apo, Zn-HypA and UreE2 or their mutants, mixing the components in equimolar amounts. Following addition of one equivalent of Ni(II), the samples were kept at room temperature for 2 h before loading the reaction mixture onto a Superdex S75 10/300 (GE Healthcare) size exclusion chromatographic column. The fractions containing the purified complexes were pooled together, concentrated with Amicon Ultra concentrators (Millipore, GE) with a 3 kDa cutoff, and stored at −80 °C until use.
The purity and integrity of UreE2 and HypA containing deuterated amino acids were tested using mass spectrometry (EMBL Proteomics Core Facility, https://www.embl.de/proteomics/proteomics_services/). The absence of Ni in the purified apo-proteins was established using inductively coupled plasma optical emission spectrometry (ICP–OES), as previously described.31
Size exclusion chromatography/multiple angle light scattering
The oligomerization properties of the single HypA and UreE2 proteins and their complexes were investigated using size exclusion chromatography (SEC) coupled with multiple angle light scattering (MALS). SEC–MALS measurements were performed using an Agilent HPLC with a Superdex 75 10/300 GL column (GE Healthcare) connected downstream to a multi-angle laser light scattering (MALS, at 690.0 nm) DAWN EOS (Wyatt Technology) photometer and to a 90° angle quasi-elastic (dynamic) light scattering (QELS) device (Wyatt Technology). The concentration of the eluted protein was determined using a refractive index detector (Optilab DSP, Wyatt). All experiments were performed at room temperature, with the system equilibrated with 20 mM HEPES pH 7.2 and 200 mM NaCl at 0.6 mL/min. Single protein samples consisted of 200 μL of 130 μM Ni,Zn-HypA, 130 μM Ni,Zn-L2*HypA, 45 μM Ni-UreE2, 45 μM Ni-H152A-UreE2 and 45 μM Ni-H102K-UreE2. The samples of protein complexes (100 μL, 85 μM) were prepared by mixing equimolar amounts of each protein prior to injection into the SEC column. All data were analysed using the ASTRA 4.90.07 software (Wyatt Technology), following the manufacturer's instructions.
Isothermal titration calorimetry
The thermodynamics of the interaction among HypA, UreE2, and Ni(II) were investigated using isothermal titration calorimetry (ITC). Titrations were performed using a high-sensitivity VP-ITC microcalorimeter (MicroCal LLC, MA, USA.) at 25 °C in 20 mM HEPES, pH 7.2, 200 mM NaCl. The reference cell was filled with MilliQ water. The solutions containing the single proteins or their mutants (20–40 μM) were loaded into the sample cell (1.4093 mL). Solutions (0.250 to 1.5 mM) of NiCl2, apo, Zn-HypA, apo, Zn-L2*HypA, Ni,Zn-HypA or Ni,Zn-L2*HypA in the same buffer were added (60 injections of 5 μL) using a computer-controlled microsyringe. Control experiments were carried out by titrating the same solutions into buffer alone, under identical conditions, verifying that the heat of dilution was negligible.
The integrated heat data obtained for each titration were fitted using a nonlinear least-squares minimization algorithm to a theoretical titration curve, using the AFFINImeter software and a global fitting approach.32 The used fitting model initially involved independent sites for simple equilibria, whereas the stoichiometric equilibria approach was used for the global fitting procedure. The ∆H (reaction enthalpy change, cal mol−1) and Ka (binding constant, M−1) were the thermodynamic fitting parameters. The parameters rM (scaling parameter for the protein concentration) and Qdil (heat of dilution, cal mol−1) were also adjusted as fitting parameters. The reaction entropy was calculated using the relationships ∆G = −RTlnKa (R = 1.9872 cal mol−1 K−1, T = 298 K) and ∆G = ∆H − T∆S. A global fitting analysis was performed for the curves representing the titrations involving three species [HypA, UreE2 and Ni(II)]. The statistical GoF (goodness of fit) parameter was used to obtain the best fit.
X-ray absorption spectroscopy
The coordination environment of Ni(II) in the HypA-UreE2 complex was monitored using X-ray absorption spectroscopy (XAS). Samples of each protein were rapidly buffer exchanged using a Zeba Spin Desalting column, 7 kDa MWCO (molecular weight cut-off, ThermoScientific) pre-equilibrated with NTP buffer (20 mM HEPES, 100 mM NaCl, 100 mM KCl, 5 mM MgCl2, 1 mM TCEP) at pH 7.2 or 6.3. Each protein was further diluted to working concentrations of 100–300 μM in NTP buffer at the target pH prior to metal additions. A nickel acetate [Ni(OAc)2] stock solution (500 mM) was prepared in distilled and deionized water, and then further diluted to 9 mM working stock in NTP buffer at pH 7.2 or 6.3. The Ni(II) concentration in each stock solution was then accurately determined by ICP–OES as previously described.23 Ni,Zn-HypA•UreE2 complexes at pH 7.2 or 6.3 were prepared by mixing equal molar equivalents of UreE2 with Ni,Zn-HypA (after excess metals were removed by Chelex treatment) at each respective pH. Complexes were prepared with each protein at target concentrations between 100 and 300 μM and equilibrated at room temperature for 30 minutes. Protein complexes were concentrated by spin filtration to approximately 100 μL and then mixed with 400 μL of NTP buffer at target pH, and then concentrated to target protein concentration of approximately 1 mM, and then mixed with 80% glycerol to a final glycerol concentration of 12%. The final flow through from spin filtration and a portion of each sample was reserved for protein and metal analyses (by ICP–OES) and found to contain nearly equal molar ratios of Ni and Zn. The sample prepared at pH 7.2 was found to contain 0.90 mM Ni and 0.85 mM Zn (Ni: Zn = 1: 0.94). The sample prepared at pH 6.3 was found to contain 0.75 mM Ni and 0.77 mM Zn (Ni: Zn = 1:1.03). The Ni,Zn-HypA•UreE2 complexes were loaded into Kapton-taped polycarbonate sample holders and flash frozen with liquid N2. Samples were stored at −80 °C prior to data collection.
XAS data on the frozen samples were collected at the Stanford Synchrotron Radiation Lightsource (SSRL) at the SLAC National Accelerator Laboratory using dedicated ring conditions (3 GeV and 450–500 mA) on beamline 9-3 with a Si (220) double crystal monochromator. The frozen samples in polycarbonate holders with Kapton windows were immobilized on aluminum prongs and cooled to ∼10 K using a liquid helium cryostat (Oxford Instruments). A 100 element Ge detector (Canberra) was used for collecting X-ray fluorescence data. To minimize scattering, a 3 μm Z-1 filter and Soller slits were installed between the detector and sample. X-ray fluorescence data on Ni K-edge of the Ni,Zn-HypA•UreE2 complex were energy calibrated by concurrently collecting spectra of a Ni metal foil in transmission mode. Extended X-ray absorption fine structure (EXAFS) data were collected to 15k above the K-edge for both metals.
Data reduction and analyses were performed according to previously published procedures adjusted for Ni and Zn K-edge XAS data (see Supplementary Information).33 The Artemis software program34 with FEFF6 and IFEFFIT algorithm was used to generate and fit single and multiple-scattering paths for each data set as described previously (see Supplementary Information).6,23,35 To compare the different models fit to the data set, IFEFFIT utilizes three goodness of fit parameters: χ2, reduced χ2 and the R-factor (see Supplementary Information). In comparing different models, minimizing the R-factor and reduced χ2 parameter, and reasonable values of σ2 were used to determine the models that best fit the data. The R-factor will generally improve with increasing number of adjustable parameters, while reduced χ2 will go through a minimum and then increase, indicating that the model is overfitting the data. The resolution of the data (π/2Δk) was determined using Δk-values of 12.5 Å−1 and 10.5 Å−1 for Zn and Ni, yielding resolution values of 0.13 Å and 0.15 Å, respectively.
Nuclear magnetic resonance spectroscopy
The interaction of HypA, UreE2 and Ni(II) was monitored using nuclear magnetic resonance (NMR) spectroscopy. Protein samples consisted of (i) 0.5 mM 15N-labeled apo, Zn-HypA, (ii) 0.3 mM apo, Zn-HypA(15N)•UreE2(14N) complex, and (iii) the same complex added with one equivalent of Ni(II) in 20 mM HEPES buffer at pH 7.2, containing 200 mM NaCl, in 90% H2O and 10% D2O, in the absence and in the presence of TCEP. NMR spectra were obtained using a Bruker AVANCE 950 spectrometer, operating at the proton nominal frequency of 950.2 MHz (22.3 T) and equipped with a 5 mm TCI-HCN z-gradient cryo-probe; the temperature was calibrated at 298 K. Due to the high salt concentration, shaped NMR tubes (Bruker BioSpin AG) were used to improve the signal-to-noise ratio during NMR data collection. The 1H chemical shifts were referenced to 2,2-dimethyl-2-silapentane-5-sulfonic acid sodium salt (DSS), while the 15N chemical shifts were referenced indirectly to DSS, using the ratios of the gyromagnetic constants. The 1H-15N HSQC spectra were recorded using spectral widths of 13297.872 Hz (1H, 14.0 ppm) and 3852.080 Hz (15N, 40.0 ppm) with maximal evolution times of 57.7 ms (1H, 2048 points) and 16.6 ms (15N, 256 points). All NMR spectra were processed using NMRpipe.36 The original data were zero-filled (2048 × 1024 points), a cosine-squared apodization function was applied in both dimensions, and a linear prediction algorithm was applied to the indirect dimension prior to Fourier transformation. The spectra were analysed using POKY.37
1H-NMR experiments tailored for the identification of hyperfine shifted and fast relaxing signals38 were performed on an AVANCE 400 Bruker NMR spectrometer equipped with a 5 mm 1H selective probe and operating at 400.13 MHz 1H Larmor Frequency. Spectra were collected with the superWEFT pulse sequence, using 52 ms, 20 ms and 62 ms as acquisition, recovery, and inter-pulse delays, respectively. The spectral window was 156 kHz (390 ppm). The number of acquired scans ranged from 400 K to 800 K, and the experiment time was typically 16–48 h. Prior to Fourier transform, FIDs were multiplied by a cosine square weighting function followed by a 20 Hz Lorentzian line broadening. Phase and baseline correction were performed manually.
Modeling of the Ni,Zn-HypA•UreE2 complex
An initial model for the apo,Zn-HypA•UreE2 complex, devoid of any metal ion, was calculated using ColabFold,39 AlphaFold2,40 and RoseTTAFold.41 The best model was selected on the basis of predicted local-distance difference test (lDDT).42 The Ni(II)-binding site at the HypA•UreE2 interface was modelled on the protein complex achieved in the previous modelling step through a computational procedure already used to reconstruct the metal binding site in other proteins,43–45 involving the use of the loop optimization routines available in Modeller.46 The van der Waals parameters for Ni(II) were derived from the Zn(II) parameters included in the CHARMM22 force field47 implemented in the Modeller v9.18 package by applying a scale factor of 1.12 calculated on the basis of the Ni(II) ionic radius. Constraints were imposed using a Gaussian-shaped energy potential for distances, angles and dihedrals, in order to correctly position the Ni(II) ions with respect to the experimentally identified ligated residues (see Results). During the loop optimization procedure, 500 models were generated. The best model was selected on the basis of the lowest value of the DOPE score included in Modeller.48 Structural analyses were conducted using ProCheck,49 UCSF Chimera,50 and UCSF ChimeraX.51,52
Modeling of the Ni-sites in Ni,Zn-HypA and in the Ni,Zn-HypA•UreE2 complex
The starting structural model for the coordination sphere of Ni(II) in Ni,Zn-HypA•UreE2 was built using the structure of the HypA-UreE2 apo-protein complex that resulted from the previously described modelling stage. In particular, the N-terminal N atom of Met1, the amide and side chain imidazole Nδ1 atoms of His2, and the amide N and side chain carboxylate Oε1 and Oε2 atoms of Glu3 from HypA, together with the Nε2 atoms of His102A and His102B from the homodimeric UreE2, were included in the model. These selected residues were capped with a -CH3 and a -NH2 group at the N- and C-termini, respectively. A starting model of the Ni(II) site in Ni,Zn-HypA was then derived by removing the two histidine residues His102A and His102B from UreE2, and adding the HypA Met1 N-terminal N atom in the coordination sphere of the metal ion through the rotation of the protein backbone using UCSF Chimera.50 A second starting model of the Ni(II) site in Ni,Zn-HypA was generated in the same way, but with the addition of a water molecule as the sixth ligand of the Ni(II) ion instead of Glu3 Oε2. These three models were optimized using DFT computations, carried out using the program ORCA 4.0.153 and the Becke three-parameter hybrid functional combined with Lee–Yang–Parr correlation functional (B3LYP/G)54,55 as defined in the Gaussian software.56 All atoms except Ni(II) were described by the Pople-style 6–31(p, d) basis set with diffuse functions on all atoms.57 The Ni(II) ion was described with the Los Alamos effective core potentials (LANL2DZ ECP).58 TightSCF criteria for SCF [energy change <1.0 × 10−8 atomic unit (au)] and standard criteria for the geometry optimization (energy change <5 × 10−6 au, RMS gradient <1 × 10−4 au, maximum element of gradient <3 × 10−4 au, root mean square deviation (RMSD) displacement <2 × 10−3 au, maximum displacement <4 × 10−3 au) were used. Frequency computations were executed to determine the nature of the critical points. Supplementary Table SI-5 reports the Ni-ligand distances obtained for the calculated models.
Results
Protein and Ni(II) interactions by SEC–MALS
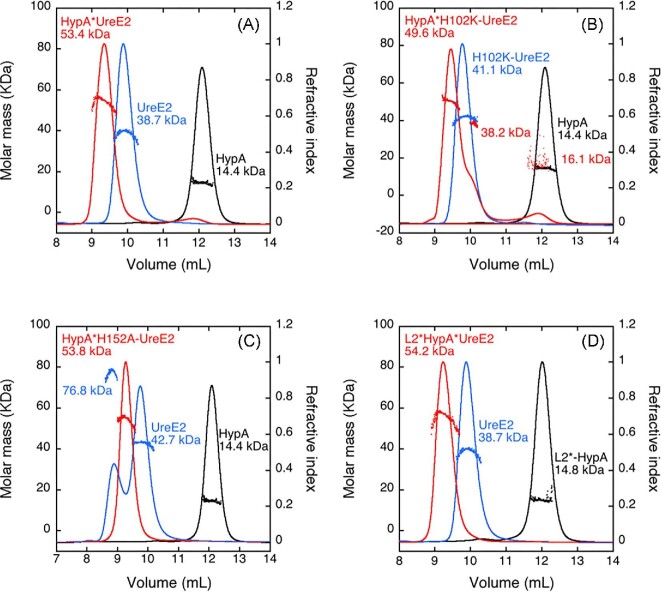
UreE2 and HypA were expressed and purified as previously reported.5,23 Mass spectrometry, performed on protein samples, confirmed that both proteins were purified as intact polypeptides, and no evidence of C-terminal protein degradation, which was reported in a previous work,23 was observed for UreE2, in the absence or in the presence of TCEP. This result is consistent with SDS-PAGE under denaturing conditions, which presented a unique band at ca. 20 kDa, corresponding to the UreE monomer. In addition to studying the native proteins, the L2*HypA mutant,23,25 as well as the H102K-UreE and H152A-UreE variants,5 in which residues known to be involved in Ni(II) binding were mutated, were also analysed to dissect the role of each metal binding residue for metal binding in the hetero-complex. The molecular mass of the Ni(II) complexes of UreE2, HypA, and of their mutants in solution was determined using a combination of SEC and MALS (Fig. 1).
Fig. 1.
Molar mass distribution plot for Ni,Zn-HypA, Ni-UreE2 and Ni,Zn-HypA•UreE2 (A) and their L2*HypA (B), H102K-UreE2 (C), and H152A-UreE2 (D) mutants as derived from size exclusion chromatography (SEC) coupled with multiple angle light scattering (SEC–MALS). The solid lines indicate the traces from the refractive index detector after elution from SEC, and the dots are the weight-average molecular weights for each slice (measured every half second) as calculated from the MALS data.
The elution profile of the single proteins is consistent with the presence of a monomer for Ni,Zn-HypA and Ni,Zn-L2*HypA (MW = 14.4 and 14.8 kDa respectively, theoretical mass of the monomer 13.2 kDa, Fig. 1A, B) and a dimer for Ni-UreE2 and Ni-H102K-UreE2 (MW = 38.7 and 42.4 kDa, theoretical mass of the monomer 19.5 kDa, Fig. 1A, C), confirming previously reported data for the WT proteins and showing that the two protein mutants possess the same oligomeric state in solution as the native proteins.5,23,25,59 In contrast, Ni-H152A-UreE2 eluted in two peaks, showing MW = 76.8 and 42.7 kDa (Fig. 1D). This indicates that, in this case, the presence of Ni(II) drives a protein tetramerization equilibrium in solution. SEC–MALS data obtained mixing equimolar amounts of Ni,Zn-HypA and UreE2 showed the formation of a peak with a lower retention volume, corresponding to MW = 53.4 kDa, compatible with the interaction of one UreE2 dimer with one HypA monomer (theoretical mass 52 kDa) (Fig. 1A). Such a Ni,Zn-HypA•UreE2 complex is formed also when Ni,Zn-HypA is present in solution together with H152A-UreE2 (Fig. 1D) or when UreE2 is co-eluted in the presence of Ni,Zn-L2*HypA (Fig. 1B), indicating that both mutants maintain the ability to interact as the native proteins, in the presence of Ni(II). On the other hand, in the case of Ni,Zn-HypA co-eluted with the H102K-UreE2 mutant, a complex chromatogram is observed (Fig. 1C) in which the prevalent species in solution corresponds to the Ni,Zn-HypA•H102K-UreE2 complex (MW = 49.6 kDa), together with two additional peaks that correspond to the separate proteins. This indicates that the complex partially dissociates during elution, likely because of a decreased affinity of the protein–protein interaction caused by the H102K mutation.
Examination of protein and Ni(II) interactions by ITC
All thermodynamic parameters obtained from ITC titrations are listed in Table 1. Ni(II) titration of either apo,Zn-HypA (Supplementary Fig. SI-1) or UreE2 (Supplementary Fig. SI-2) produced an exothermic reaction, as indicated by negative peaks following each injection of Ni(II) (Supplementary Figs. SI-1A and SI-2A).
Table 1.
Data from isothermal titration calorimetry experiments
| Equilibriuma | Complex formed | K A (×10−6) | K D (μM) | ΔH (kcal/mol) | ΔS [cal/(mol*K)] | ΔG (kcal/mol) |
|---|---|---|---|---|---|---|
| Apo,Zn-HypA + Ni (GoF = 90.2 %) | ||||||
| 1 | Ni,Zn-HypA | 1.11 ± 0.04 | 0.90 ± 0.04 | −0.83 ± 0.02 | 24.9 | −8.23 ± 0.03 |
| UreE2 + Ni (GoF = 57.4 %) | ||||||
| 2 | Ni-UreE2 | 3.4 ± 0.1 | 0.294 ± 0.009 | −11.58 ± 0.03 | −8.98 | −8.91 ± 0.03 |
| UreE2 + Apo,Zn-HypA (GoF = 69.9 %) | ||||||
| 3 | Apo,Zn-HypA•UreE2 | 1.19 ± 0.02 | 0.84 ± 0.01 | −6.33 ± 0.01 | 6.56 | −8.28 ± 0.02 |
| UreE2 + Ni,Zn-HypA (GoF = 58.8 %) | ||||||
| 1 | Ni,Zn-HypA | 1.07 ± 0.03 | 0.94 ± 0.02 | −0.83 ± 0.02 | 24.7 | −8.22 ± 0.03 |
| 2 | Ni-UreE2 | 2.30 ± 0.02 | 0.435 ± 0.004 | −11.99 ± 0.02 | −11.1 | −8.67 ± 0.01 |
| 3 | Apo,Zn-HypA•UreE2 | 1.28 ± 0.04 | 0.78 ± 0.02 | −5.98 ± 0.05 | 7.87 | −8.33 ± 0.03 |
| 4 | Ni,Zn-HypA•UreE2 | 6500 ± 400 | 0.000154 ± 0.000 009 | −12.42 ± 0.02 | 3.22 | −13.38 ± 0.03 |
| 5 | Ni,Ni,Zn-HypA•UreE2 | 7.7 ± 0.6 | 0.13 ± 0.01 | −3.31 ± 0.03 | −9.40 | −9.38 ± 0.08 |
| H102K-UreE2 + Ni,Zn-HypA (GoF = 68.4 %) | ||||||
| 1 | Ni,Zn-HypA | 2.23 ± 0.09 | 0.45 ± 0.02 | −0.787 ± 0.002 | 26.4 | −8.66 ± 0.04 |
| 4 | Ni,Zn-HypA•H102K-UreE2 | 16.9 ± 0.9 | 0.060 ± 0.003 | −4.686 ± 0.003 | 17.3 | −9.86 ± 0.05 |
| H152A-UreE2 + Ni (GoF = 47.9 %) | ||||||
| 2 | Ni-H152A-UreE2 | 59 ± 5 | 0.017 ± 0.009 | −5.88 ± 0.02 | 15.8 | −10.59 ± 0.08 |
| 6 | Ni-H152A-UreE4 | 1.1 ± 0.2 | 0.909 ± 0.009 | −0.80 ± 0.02 | 25.1 | −8.2 ± 0.2 |
| H152A-UreE2 + Ni,Zn-HypA (GoF = 58.4 %) | ||||||
| 1 | Ni,Zn-HypAb | 1.11 | 0.90 | −1.55 ± 0.09 | 22.5 | −8.2 |
| 2 | Ni-H152A-UreE2b | 59 | 0.017 | −6.01 ± 0.07 | 15.4 | −10.59 |
| 4 | Ni,Zn-HypA•H152A-UreE2 | 16.0 ± 0.3 | 0.063 ± 0.001 | −2.69 ± 0.01 | 23.9 | −9.82 ± 0.02 |
| 6 | Ni-H152A-UreE4 | 1.61 ± 0.04 | 0.62 ± 0.01 | −0.61 ± 0.08 | 26.3 | −8.46 ± 0.02 |
| UreE2 + Ni,Zn-L2*HypA (GoF = 45.5 %) | ||||||
| 1 | Ni,Zn-L2*HypA | 0.0014 ± 0.0001 | 710 ± 50 | −66 ± 3 | −206 | −4.289 ± 0.007 |
| 2 | Ni-UreE2 | 3.6 ± 0.1 | 0.278 ± 0.008 | −11.98 ± 0.07 | −8.21 | −8.94 ± 0.03 |
| 3 | Apo,Zn-L2*HypA•UreE2 | 0.59 ± 0.03 | 1.69 ± 0.08 | −14.06 ± 0.03 | −20.8 | −7.87 ± 0.05 |
| 4 | Ni,Zn-L2*HypA•UreE2 | 19 ± 2 | 0.053 ± 0.006 | −78 ± 3 | −228 | −9.9 ± 0.1 |
aThe first column refers to the equilibria indicated in Eqns. 1–8.
bThe values for KA (and KD) were fixed for this fit, as derived from the analogous constants obtained for the single proteins.
A fit of the integrated heat data (Supplementary Figs. SI-1B and SI-2B) using a single set of independent binding sites (Eqns. 1, 2) indicated the interaction of one Ni(II) ion per apo,Zn-HypA monomer (KD = 0.90 μM) and one Ni(II) ion per UreE2 dimer (KD = 0.29 μM). In both cases, the enthalpic contribution is favorable, while the entropic contribution is positive for apo,Zn-HypA binding and negative for UreE2 binding. These results largely confirm the previously obtained thermodynamic parameters for Ni(II) titrations of apo,Zn-HypA25 (KD = 0.97 μM) while they contrast with those obtained by Sun et al. on the same protein prepared using a different protocol59 (KD = 14 μM). In the case of UreE2, the value obtained for the dissociation constant agrees with those obtained by Bellucci et al.5 (KD = 0.15 μM) and by Yang et al.59 (KD = 0.53 μM) while it differs by one order of magnitude with the value reported by Hu et al.25 (KD = 0.067 μM), a discrepancy possibly caused by the slow protein degradation reported in the latter study.
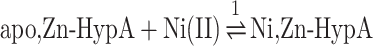 |
(1) |
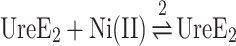 |
(2) |
Interaction of UreE2 and apo,Zn-HypA in the absence of Ni(II) also occurs with an exothermic reaction (Supplementary Fig. SI-3A) whose thermodynamic parameters were obtained from the fit of the integrated heat to Eqn. 3 (Supplementary Fig. SI-3B, Table 1). The dissociation constant of this apo,Zn-HypA•UreE2 complex (KD = 0.84 μM) confirmed the values previously obtained by Hu et al.23,25 (KD = 0.90 μM) and Yang et al.59 (KD = 1.2 μM).
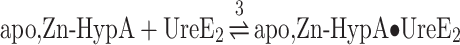 |
(3) |
Titration of UreE2 with Ni,Zn-HypA produced an exothermic reaction (Fig. 2A) and a multipartite binding isotherm (Fig. 2B) similar to the one previously reported.25 In prior work, these data were interpreted using an independent sites approach, suggesting the occurrence of two binding events with different affinities: one
Fig. 2.
Ni,Zn-HypA titration of UreE2. (A) Heat response for injections of 0.45 mM Ni,Zn-HypA into 30 μM UreE2. (B) Integrated heat data of the titration as a function of Ni,Zn-HypA/UreE2 molar ratio. The continuous line represents the best fit (GoF = 58.8%) obtained with a model involving a global fit according to Eqn. 4. (C) Concentration distribution of the different species occurring during the titration described in (B), calculated using the thermodynamic parameters obtained from the fit and the binding model described by Eqn. 4.
in the sub-to-low nanomole range and another in the sub-to-low micromole range.25 This approach led to the conclusion that the apo,Zn-HypA•UreE2 complex contains a new high-affinity Ni(II)-binding site that does not exist for the two separate proteins. However, the complexity of the system, involving several equilibria between the three species present in solution (i.e. Ni(II), apo,Zn-HypA and UreE2), did not allow the multipartite binding isotherm to be attributed solely to Ni(II) binding.25
In this work, the data were re-analysed by considering “all” the equilibria among the diverse species in solution, using the “global fitting approach” available in the AFFINImeter software.32 Using the model builder tool of AFFINImeter, a binding scheme was constructed, consisting of five equilibria (Eqn. 4): (1) Ni(II) binding to apo,Zn-HypA, (2) Ni(II) binding to UreE2, (3) UreE2 interaction with apo,Zn-HypA, (4) UreE2 binding to Ni,Zn-HypA, (5) additional Ni(II) binding to the ternary Ni,Zn-HypA•UreE2 complex. The latter reaction makes a small contribution to the thermodynamics but was necessary to obtain a good fit of the data. Most notably, this binding scheme was used to simultaneously fit the binding isotherms representing the single equilibria illustrated in Eqns. 1–3 (Supplementary Figs. SI-1, SI-2, and SI-3), as well as the isotherm resulting from the titration of UreE2 with Ni,Zn-HypA, resulting in the formation of the ternary complex (Fig. 2B).
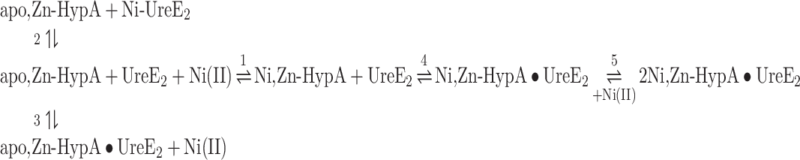 |
(4) |
The fit derived from these equilibria reproduced the integrated data well (Fig. 2B and Table 1). The dissociation constants of the binary complexes derived using this global fit are consistent with those obtained by fitting the single binding isotherms (Table 1), and additionally indicate the formation of two binding sites for Ni(II) in the protein complex, one with affinity in the sub-nanomole range (KD = 0.15 nM), which was not present in the separate proteins, and one with affinity in the sub-micromole range (KD = 0.13 μM). A plot of species distribution as a function of the UreE2/apo,Zn-HypA molar ratio is reported in Fig. 2C.
To dissect the Ni(II) and protein binding events, and to investigate the roles of Ni(II) binding residues for the formation of the Ni(II) ternary complex, titrations were replicated with forms of UreE2 and HypA in which known Ni(II) binding residues (His102 and His152 for UreE25 and the N-terminus of HypA23) were mutated to obtain the H102K and H152A UreE2 as well as the L2*HypA (an insertion of Leu after the N-terminal Met residue) mutants. As previously reported,5 Ni(II) titration of H102K-UreE2 did not produce any heat of binding (Supplementary Fig. SI-4A), indicating of the absence of reaction under the experimental conditions. Similarly, apo,Zn-HypA titration of H102K-UreE2 provided a very low reaction heat that did not significantly change with increasing molar ratio (Supplementary Fig. SI-4B), indicating that mutation of His102 has a detrimental effect on the protein–protein interaction when Ni(II) is not present in solution.
In contrast, Ni,Zn-HypA titration of the H102K-UreE2 mutant produced an exothermic event (Fig. 3A), indicating that the presence of the Ni(II) ion in the system restores interactions between the two proteins. A global fit of the data, obtained using the AFFINImeter software as described earlier, was performed using a modified scheme (Eqn. 5) that considers two binding equilibria: (1) the apo,Zn-HypA interaction with Ni(II) and (4) the formation of the ternary complex with H102K-UreE2, while it does not include the single equilibria for Ni(II) binding to H102K-UreE2 or for apo,Zn-HypA binding to H102K-UreE2, neither of which produced significant heats of binding (Supplementary Fig. SI-4). The global fitting procedure combined the isotherms for the interaction between HypA and Ni(II) (Supplementary Fig. SI-1) and that for the formation of the ternary complex, resulting in the fit shown in Fig. 3B.
Fig. 3.
Ni,Zn-HypA titration of H102K-UreE2. (A) Heat response for injections of 0.45 mM Ni,Zn-HypA into 30 μM H102K-UreE2. (B) Integrated heat data of the titration as a function of Ni,Zn-HypA/H102K-UreE2 molar ratio. The continuous line represents the best fit (GoF = 68.4%) obtained with a model involving a global fit according to Eqn. 5. (C) Concentration distribution of the different species occurring during the titration described in (B), calculated using the thermodynamic parameters obtained from the fit and the binding model described by Eqn. 5.
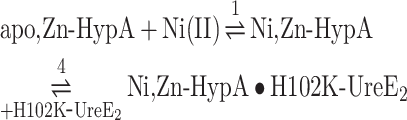 |
(5) |
The dissociation constant of the Ni,Zn-HypA complex thus derived is consistent with that obtained by fitting the single binding isotherm (Table 1). This fit additionally provides a value for the dissociation constant of the ternary complex (KD = 60 nM), which indicates that the H102K mutation decreases the affinity of the ternary complex for Ni(II) by ∼400 times.
Similar experiments were conducted for the H152A-UreE2 mutant. Titration of this protein with Ni(II) provides an exothermic binding (Supplementary Fig. SI-5A) as previously reported.5 A fit using a single site binding model provided the thermodynamic parameters KD = 640 ± 20 nM, ΔHH152A-Ni = −6.93 ± 0.03 kcal mol−1, ΔSH152A-Ni = +5.09 cal mol−1 K−1, confirming previous values.5 As this mutant was proven to undergo protein tetramerization upon Ni(II) binding using SEC–MALS experiments (vide supra), the fitting was performed including the dimer of dimer formation in the model (Eqn. 6).
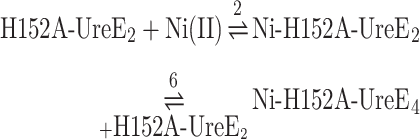 |
(6) |
A fit performed using this approach (Supplementary Fig. SI-5B) determined that Ni(II) binding to the H152A-UreE2 mutant occurs (Table 1) with a dissociation constant KD = 17 nM, while protein dimerization features a dissociation constant one order of magnitude lower affinity (KD = 0.909 μM). Substitution of His152 with an alanine residue in the H152A-UreE2 mutant strongly decreases the protein affinity for apo,Zn-HypA (Supplementary Fig. SI-6): indeed, injections of apo,Zn-HypA into a H152A-UreE2 solution generates a moderate thermal response that is observed as negative peaks following each injection. This effect does not decrease with the progress of the titration, indicating either a low interaction affinity (not sufficient to produce a binding isotherm) or a non-specific effect. The titration of the H152A-UreE2 with Ni,Zn-HypA, on the other hand, revealed the restoration of protein–protein interactions, occurring with an exothermic reaction (Fig. 4A). A fit of the integrated heat data (Fig. 4B) was carried out using a model that considers all equilibria present in this solution (Eqn. 7): (1) Ni(II) binding to apo,Zn-HypA, (2) Ni(II) binding to H152A-UreE2, (4) Ni(II) binding to apo,Zn-HypA•H152A-UreE2 complex, and (6) formation of the Ni-H152A-UreE4 complex.
Fig. 4.
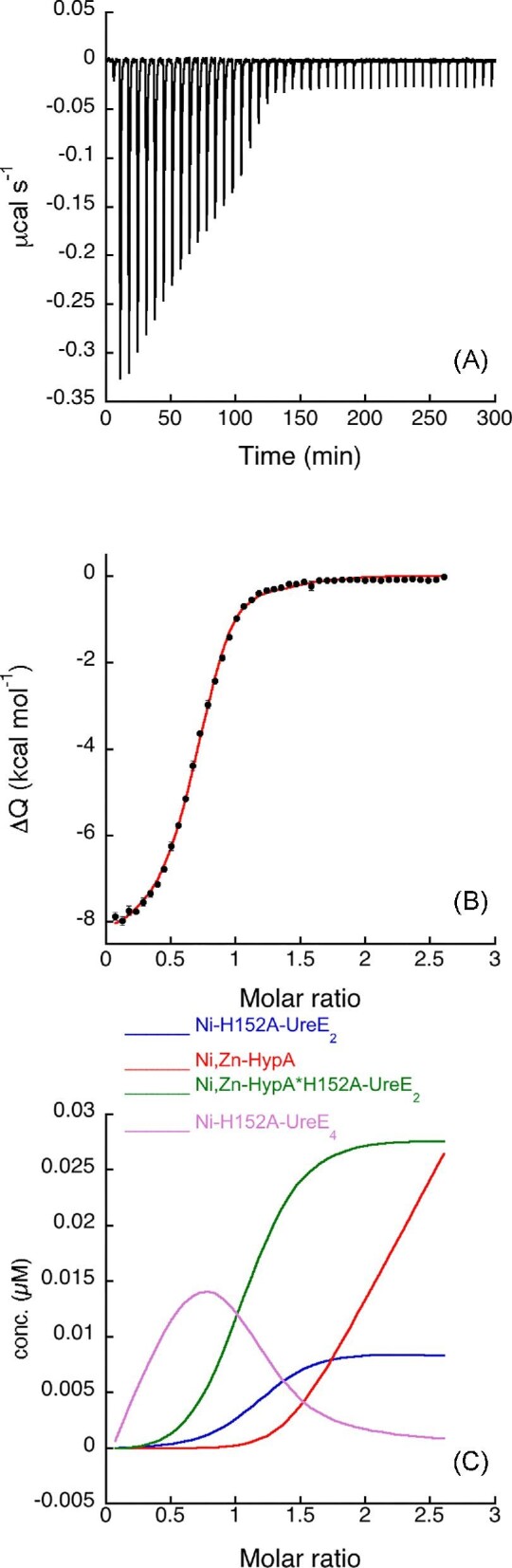
Ni,Zn-HypA titration of H152A-UreE2. (A) Heat response for injections of 0.45 mM Ni,Zn-HypA into 30 μM H152A-UreE2. (B) Integrated heat data of the titration as a function of Ni,Zn-HypA/H152A-UreE2 molar ratio. The continuous line represents the best fit (GoF = 58.4%) obtained with a model involving a global fit according to Eqn. 7. (C) Concentration distribution of the different species occurring during the titration described in B), calculated using the thermodynamic parameters obtained from the fit and the binding model described in Eqn. 7.
A good fit could be obtained only by fixing the affinity constants for the apo,Zn-HypA and of H152A-UreE2 interactions with Ni(II) to the values provided by the single isotherms (Table 1). Although this procedure provides an interpolation curve (Fig. 4B) that is consistent with the experimental data, this suggests that some other event might occur during the titration, such as conformational change or protein aggregation, that was not considered. The dissociation constants obtained from the fit are KD = 63 nM and KD = 0.62 μM for the Ni,Zn-HypA•H152A-UreE2 and for the Ni-H152A-UreE4 complexes, respectively. These values indicate that the affinity of the high-affinity binding site for Ni(II) decreases by two-orders of magnitude in the mutant ternary complex Ni,Zn-HypA•H152A-UreE2 relative to the WT, as also observed for the analogous complex in the case of the H102K-UreE2 mutant, the latter featuring similar affinity for Ni(II) as the H152A-UreE2 mutant (KD = 60 nM, Table 1). The value for the Ni-UreE4 dissociation constant is similar (0.62 vs. 0.91 μM) to that obtained by directly titrating the H152A-UreE2 mutant with Ni(II), indicating a good consistency of the analysis. The concentration distribution of the different species upon the increasing HypA/H152A-UreE2 molar ratio is shown in Fig. 3C.
 |
(7) |
Titration of apo, Zn-L2*HypA with Ni(II) shows a small endothermic effect indicative of the absence of metal binding or of binding with low affinity (Supplementary Fig. SI-7A), analogous with previous observations.23 Addition of Ni(II) to the syringe solution containing apo, Zn-L2*HypA in the titration of UreE2 restores protein–protein interactions. The exothermic peaks at the beginning of the titration are initially negative, then progressively shift to positive, indicating an ensemble of exothermic and endothermic effects (Fig. 5A).
Fig. 5.
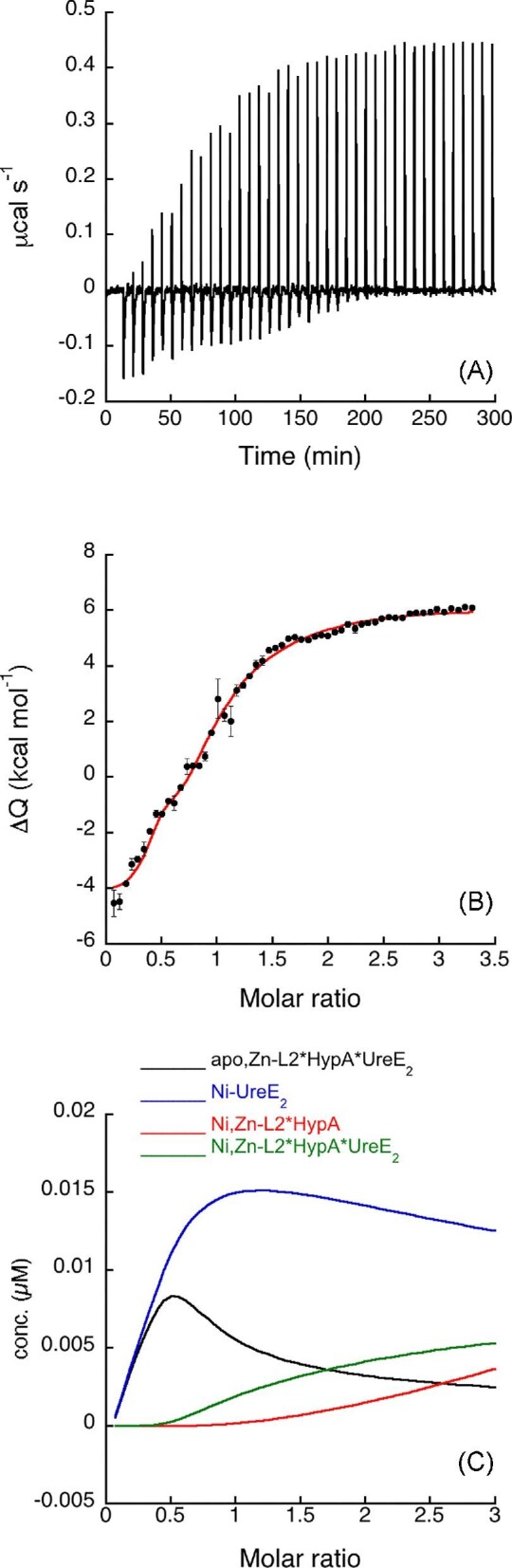
Ni,Zn-L2*HypA titration of UreE2. (A) Heat response for injections of 0.412 mM apo, Zn-L2*HypA into 27.5 μM UreE2. (B) Integrated heat data of the titration as a function of apo, Zn-L2*HypA/UreE2 molar ratio. The continuous line represents the best fit (GoF = 45.5%) obtained with a model involving a global fit according to Eqn. 8. (C) Concentration distribution of the different species occurring during the titration described in (B), calculated using the thermodynamic parameters obtained from the fit and the binding model described by Eqn. 8.
A fit of the integrated heat data (Fig. 5B) could be obtained using a global fitting approach that contained all equilibria described in Eqn. 8, which included Ni(II) binding to either (1) apo, Zn-L2*HypA or (2) UreE2, as well as protein–protein interaction (3) in the absence or (4) in the presence of Ni(II). The dissociation constants obtained from the fit indicate that Ni(II) binding to the L2*HypA mutant occurs with a three-orders of magnitude lower affinity as compared to the WT protein. Moreover, the interaction of the two proteins in the absence of Ni(II) occurs with similar affinity for the WT and the L2*HypA variant, as previously observed.23 On the other hand, the stability of the ternary complex formed by Ni(II), UreE2 and L2*HypA is ∼350 times lower as compared to that involving the WT HypA.
 |
(8) |
The concentration distribution of the different species that occurs upon increasing the L2*HypA/UreE2 molar ratio, shown in Fig. 5C, indicates that the amount of the ternary complex in solution remains minor, while the major species that forms is Ni-UreE2, consistent with the much lower affinity of L2*HypA for Ni(II) as compared to UreE2.
Ni(II)–protein interactions examined by XAS
Ni K-edge XAS was used to probe the structure of the high-affinity Ni(II) site in the Ni,Zn-HypA•UreE2 complex at pH 7.2 and pH 6.3. X-ray absorption near edge structure (XANES) analysis can provide information regarding the coordination number/geometry and oxidation state of the metal-complexes.60 Ni(II) complexes show features associated with high-energy electronic transitions in the pre-edge XANES region of XAS spectra.60 These transitions involve the promotion of a Ni 1s electron to either the 3d manifold (1s → 3d), which occurs near 8331 eV, or to a 4pz orbital (1s → 4pz), which occurs near 8336 eV.60
XANES analysis on the nickel site of Ni,Zn-HypA•UreE2 complex at pH 7.2 show a small 1s → 3d feature at 8331.6 eV (Fig. 6) with a peak area of 0.035(2) eV2. This peak area coupled with the absence of any feature near 8336 eV is consistent with a six-coordinate distorted octahedral geometry.60 Similarly, the XANES of the Ni(II) site in the Ni,Zn-HypA•UreE2 complex at pH 6.3 also shows a small 1s → 3d feature at 8331.6 eV (Supplementary Fig. SI-8) with a peak area of 0.036(2) eV2, and is also consistent with a six-coordinate nickel site.
Fig. 6.
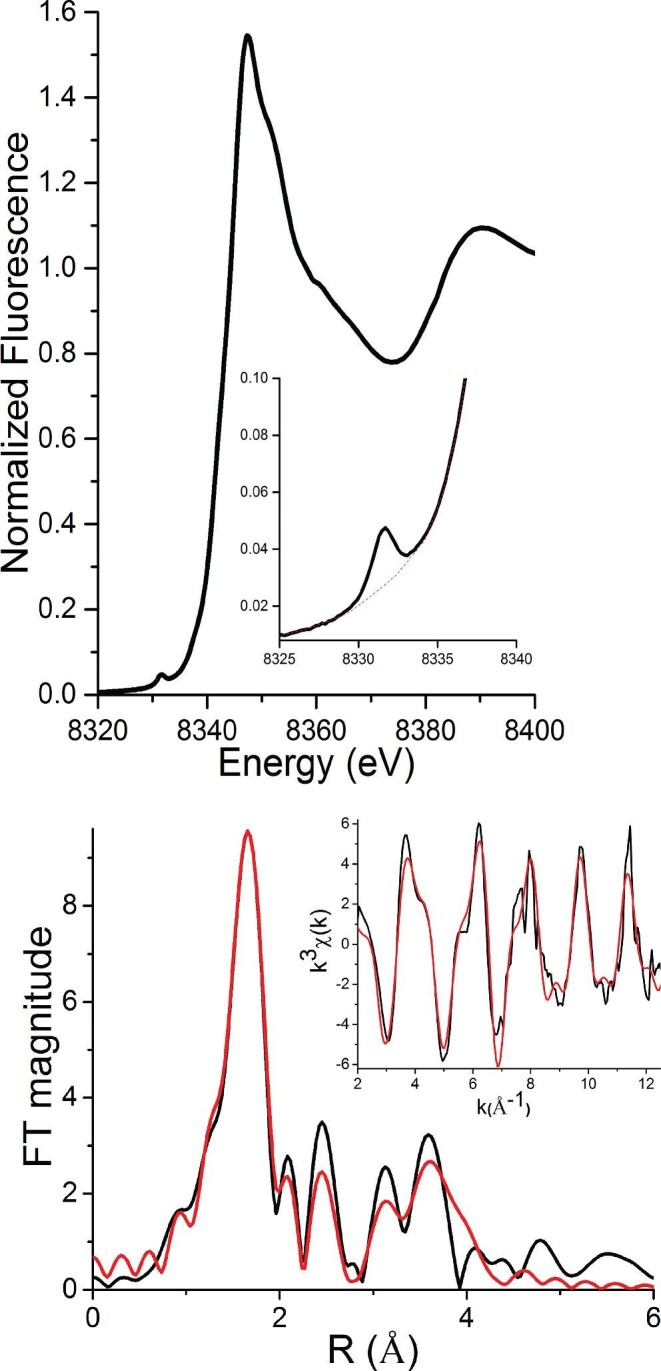
Ni K-edge XAS of the Ni,Zn-HypA•UreE2 complex at pH 7.2. Top: XANES spectrum Inset: Pre-edge feature associated with a 1s → 3d electronic transition (The baseline for area calculation is shown by the red dotted line.) Bottom: The Fourier-transformed (FT window = 2.0–12.5 Å−1) EXAFS spectrum (black line) and best fit from Table 2 (red line). Inset: Unfiltered k3-weighted EXAFS spectrum and calculated best fit model.
Analysis of the EXAFS region of the XAS spectra of metal-complexes provides information on the identity of ligand-donor atoms (Z ± 1) around the metal center, the M-L distances (±0.02Å), as well as a second measure of coordination number (± ∼20%).61 The Ni K-edge EXAFS spectrum of the Ni(II) site in the Ni,Zn-HypA•UreE2 complex is shown in Fig. 6, with the fits leading to the best fit model of the nickel site structure summarized in Table 2. Single-scattering analysis of the EXAFS data of this complex indicates a nickel site composed of N/O-donor atoms in a six-coordinate geometry (Supplementary Table SI-1), which is also consistent with the coordination number/geometry determined from the XANES analysis (vide supra). Systematic splitting of the single shell of six-coordinate N/O atoms into two different shells corresponding to N/O atoms at two different distances did not improve the fit (Supplementary Table SI-1).
Table 2.
Selected EXAFS fits for the Ni-site in the Ni,Zn-HypA•UreE2 complex at pH 7.2a
| Shell | r (Å) | σ2 (×10−3 Å−2) | ΔE0 | R-factor | Red. χ2 |
|---|---|---|---|---|---|
| 6 N/O | 2.08 (1) | 4 (0) | 1 (2) | 17.2 | 214.18 |
| 5 N/O 1 Im0o |
2.08 (1) 2.09 (1) |
6(1) neg |
0 (1) | 10.2 | 144.00 |
| 4 N/O 2 Im0o |
2.07 (1) 2.09 (1) |
5 (2) 2 (2) |
1 (1) | 8.1 | 114.11 |
| 3 N/O 3 Im0o |
2.08 (1) 2.09 (2) |
2 (1) 6 (2) |
1 (1) | 6.6 | 93.18 |
| 2 N/O 4 Im0o |
2.08 (1) 2.08 (2) |
0 (1) 8 (1) |
0 (1) | 5.4 | 76.39 |
| 1 N/O 5 Im0o |
2.08 (1) 2.08 (1) |
neg 8 (1) |
0 (1) | 5.3 | 76.04 |
| 3 N/O 3 Im5o |
2.06 (1) 2.10 (2) |
2 (1) 5 (2) |
0 (1) | 6.5 | 91.57 |
| 3 N/O 3 Im10o |
2.04 (1) 2.10 (1) |
4 (1) 2 (0) |
0 (1) | 5.3 | 74.76 |
|
3 N/O 1 Im0o 2 Im10o |
2.03 (1)
2.07 (1) 2.09 (1) |
7 (2)
1 (2) 1 (1) |
2 (1) | 3.6 | 59.44 |
| 2 N/O 2 Im0o 2 Im10o |
2.03 (2) 2.09 (3) 2.09 (1) |
3 (2) 7 (3) 0 (1) |
0(1) | 3.4 | 55.9 |
aThe best fit is shown in bold.
neg = a negative value; parameters shown in red are unacceptable.
Errors indicated by numbers in parentheses represent uncertainties estimated by Artemis and are the changes in variables required to generate an increase in χ2 of the value of reduced χ2.
Multiple-scattering pathways were then added to account for the features in the FT-EXAFS spectra arising from second and third coordination sphere scattering atoms of coordinated His imidazole ligands using a rigid five-membered ring with a single adjustable Ni–N distance, as previously described.6,23,35 Addition of multiple-scattering pathways from single shells of one to five His imidazole ligands with α = 0o, 5o or 10o, along with one shell of one to five N/O donors and totaling six ligands were examined.
Fits that included three or four imidazole ligands resulted in lower R-factors and reduced χ2 values and had acceptable Debye–Waller factors (σ2). However, none produced a fit with R < 5%, a criterion for a good model. Splitting the single shell of N/O scatterers in models with three to four His ligands resulted in modest improvements in R-factor but produced negative values of σ2 for N/O shells (Supplementary Table SI-1). In contrast, splitting the shell of imidazoles into shells with different α angles dramatically improved the fits and produced fits with acceptable R-factors (<5%) and Debye–Waller values. The fit containing three N/O donors and three His ligands produced the best fit: three His imidazole ligands (one with a Ni–N distance of 2.07 Å and α = 0° and two with Ni–N = 2.09 Å and α = 10°) and three other N/O-donor ligands with an ave. Ni–N/O distance of 2.03 Å. The progression leading to this fit is shown in Table 2. Similar attempts to split the single shell of four imidazoles into two or three different shells resulted in an alternative fit that has a slightly better R-factor but features a less acceptable Debye–Waller factor. (Table 2). Attempts to add additional second-coordination sphere C-scattering atoms, as might be ordered by backbone amide N-coordination and the formation of five-membered chelate rings, led to poorer fits (R-factor).
The Ni K-edge XAS spectrum of the Ni(II) site in the Ni,Zn-HypA•UreE2 complex at pH 6.3 (Supplementary Fig. SI-8) is nearly superimposable with that obtained at pH 7.2 (Supplementary Fig. SI-9), indicating that the structure of the site is not greatly altered by this pH change. The analysis of the EXAFS for the Ni-site in the Ni,Zn-HypA•UreE2 complex at pH 6.3 (Supplementary Fig. SI-8) was performed as described for the pH 7.2 sample (vide supra) and also reveals six N/O-donor ligands (Supplementary Table SI-2) and leads to a best fit model featuring three N/O scatterers and three imidazole ligands at similar distances. However, unlike the fits to the spectra obtained at pH 7.2, splitting the shell of three imidazoles did not improve the fit and resulted in negative Debye–Waller factors (Supplementary Table SI-2). The best model featured three N/O scatterers at 2.04 Å and a single shell of three imidazole ligands with average Ni–N bond distance of 2.13 Å at an α-angle 10o, and R = 4.9%.
Zn K-edge XAS was used to characterize the Zn(II)-site of HypA in the Ni,Zn-HypA•UreE2 complex at pH 7.2 (Supplementary Fig. SI-10; Supplementary Table SI-3) and 6.3 (Supplementary Fig. SI-11; Supplementary Table SI-4). The EXAFS analysis on the Zn-site is consistent with four S-donor ligands with average Zn-S distances of 2.32(2) Å at both pH 7.2 and 6.3.
Protein and Ni(II) interactions by NMR spectroscopy
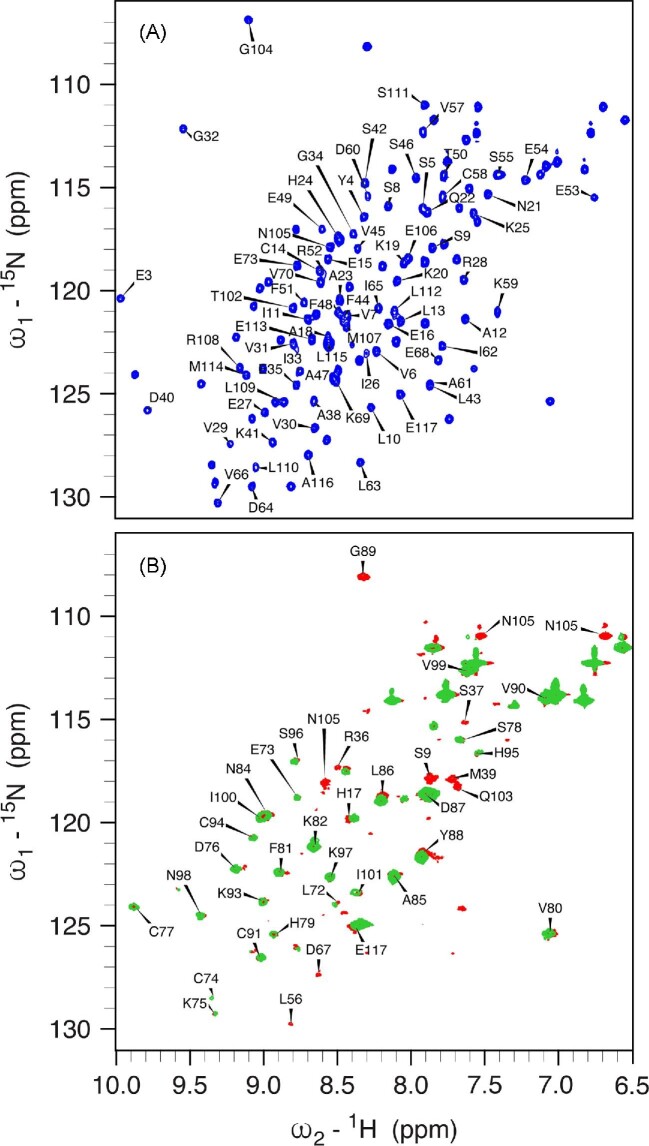
The 950 MHz 1H,15N HSQC spectrum of apo,Zn-HypA (Fig. 7A), reporting the previously published assignment,62 is not perturbed by the presence of 1 mM TCEP, so that all the following discussion refers to spectra obtained in its absence. In this spectrum, the signals of Met1 and His2 are not visible: the N-terminal NH3+ group of Met1 disappears because of fast exchange with the water signal, while the NH signal of His2 might undergo conformational exchange phenomena that broaden its line width beyond detection.
Fig. 7.
950 MHz 1H,15N HSQC spectra of apo,Zn-HypA (A, blue) and of the purified apo,Zn-HypA•UreE2 complex (B, red). In panel A, the labels refer to the signals of residues of HypA that disappear in the spectrum of the complex with UreE2, while in panel B the labels indicate the residues of HypA that are unperturbed or only slightly shifted upon interaction with UreE2. In panel B, the spectrum of the Ni,Zn-HypA•UreE2 complex is also shown in green.
The spectrum of the apo,Zn-HypA•UreE2 complex, containing 15N-labeled HypA and unlabeled UreE2 and pre-purified by SEC, is shown in Fig. 7B and reveals three main types of modifications as compared to the spectrum of isolated apo,Zn-HypA: some signals are not perturbed, some are only slightly shifted, and others are completely erased. Mapping the three types of signals on the NMR structure of apo,Zn-HypA62 allowed us to conclude that the Zn-binding domain of HypA (residues 72–103), whose signals are not affected upon complex formation, is not involved in the interaction of HypA with UreE2, while essentially all of the Ni-binding domain (residues 1–68 and 107–117) features either a small recognizable shift or a large change in the H-N peak frequency. These perturbations are not localized on a distinct surface patch of the Ni-binding domain, but are generally spread throughout the domain, suggesting that significant structural perturbations are generated upon complex formation, which, even though likely involving a specific region, are allosterically transmitted to the whole domain.
Addition of one equivalent of Ni(II) to the preformed apo,Zn-HypA•UreE2 complex (Fig. 7B) has the effect of making the amide NH signals of Ser9, Met39, and Glu103 as well as the side chain -NH2 signals of Asn105, disappear. These residues are located well within 10 Å of the N-terminal Ni-binding site on HypA, so that this effect can be attributed to the paramagnetism of the Ni(II) ion. The concomitant disappearance of Gly89, located on a loop in the Zn-binding domain and far from the Ni-binding site, could be caused by some undefined long distance allosteric modification, although mutation of Gly89 to Ala did not affect the delivery of Ni to urease, nor was it found to affect acidic viability in Hp.62
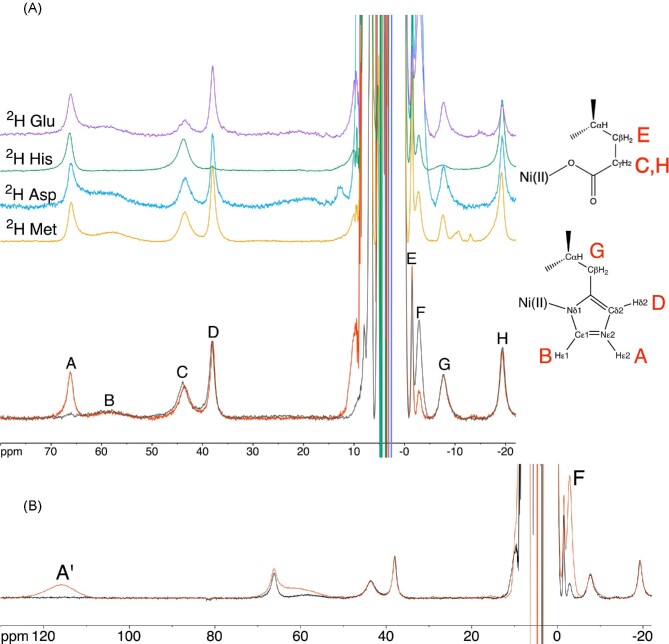
The interaction between Ni,Zn-HypA and UreE2 was then investigated using 1H NMR spectra tailored for the observation of signals of residues bound to the Ni(II) center and affected by its paramagnetism. The spectrum of Ni,Zn-HypA (Fig. 8A) contains eight hyperfine-shifted signals (A–H) in the range from +80 to −20 ppm, arising primarily from contact and pseudo-contact shifts involving a single high spin (S = 1) Ni(II) center coordinated to the N-terminal Met1 N atom, the amide N atoms of His2 and Glu3, and to an imidazole N atom of His2, with additional ligands filling the octahedral coordination environment of the metal ion coming from the side chain carboxylate of Glu3 and possibly a water molecule.23,62 These signals have previously been tentatively assigned based on their chemical shift, linewidths, longitudinal relaxation times, temperature dependence, field dependence, solvent-exchange phenomena, and mono-dimensional NOE (Nuclear Overhauser Effect) experiments.62
Fig. 8.
1H NMR spectra (400 MHz) of (A) Ni,Zn-HypA in H2O (red) and D2O (gray), together with the 1H NMR spectra of the corresponding selectively deuterated forms (2H Met in orange, 2H Asp in blue, 2H His in green and 2H Glu in purple). The assignment of signals belonging to His2 is shown. Panel B shows the 1H NMR spectrum of Ni,Zn-HypA in the presence (red) and absence (black) of excess Ni.
In order to resolve some ambiguous assignments, we recorded the spectra of samples in which the putative Ni-binding amino acids had been selectively deuterated. No change in the paramagnetic 1H NMR spectra of Ni,Zn-HypA containing deuterated methionine or aspartate residues could be observed (Fig. 8A), thus excluding Asp40 from the coordination sphere of Ni(II), in contrast to previous hypotheses,62,63 as well as excluding any contribution of Met1 nuclei to the spectrum. On the other hand, the spectrum of HypA containing selectively deuterated histidines (Fig. 8A) shows that signals B, D, and G are essentially obliterated, indicating unequivocally that these signals belong to non-exchangeable protons of His2; concomitantly, the D2O-exchangeable signal A is assigned to either Hδ1 or Hε2 of the side chain imidazole of the Ni-bound His2. Considering the relatively larger linewidth of signal B as compared to that of signal D, and the fact that signal D features a significant NOE with signal A,62 it can be concluded unambiguously that His2 must be bound to Ni(II) through its imidazole Nδ1 atom (Fig. 8A). In this way, signal A is assigned to Hε2, signal B to the ortho-like Hε1, broadened by dipolar interactions because of its proximity to Ni(II), and signal D to the meta-like Hδ2, which experiences a smaller dipolar broadening because of its relatively larger distance from Ni(II). This assignment can be extended to signal G: the previously described lack of NOEs between signal G and any other signal assigned to His262 indicates that it belongs to either Hα or a Hβ of the side chain of His2.
The intensities of signals C, E, and H show a significantly decreased intensity in the NMR spectrum of HypA containing deuterated glutamate residues (Fig. 8A), indicating that they belong to the Ni-bound Glu3. This assignment is consistent with the observation of significant NOEs among signals C, E, and H in the NMR spectrum of WT Ni,Zn-HypA,62 confirming that they belong to the same residue. The fact that they are not completely abolished is interpreted as an indication that the cell-free protocol used to produce this deuterated sample did not fully inhibit the transaminase activity, yielding only a partially, and not fully, deuterated sample as confirmed by mass spectrometry analysis (Supplementary Fig. SI-19). This is evidenced by the coexistence of several species, distributed broadly between two main peaks (13 242 and 13 268 Da) corresponding to partially deuterated samples (−35 and −9 Da as compared to the theoretical mass for the sample with fully deuterated glutamate residues, that is 13 277 Da), suggesting that there are multiple species partially deuterated. The relatively higher NOE intensities between signals C and H as compared to the NOEs involving signal E62 suggests that signals C and H belong to the two Hγ geminal methylene proton pairs of the Glu3 side chain, while the much sharper and less shifted signal E can be safely assigned to a vicinal Hβ of Glu3, respectively. The different linewidth of the signals of the two geminal protons could be explained by a conformation of the side chain of Glu3 that brings one of the two protons closer to the Ni(II) center with respect to the other proton. These experiments showed also that signal F is not affected by deuteration of residues located in the coordination environment of Ni(II) in the Met-His-Glu (MHE) N-terminal motif of HypA. Considering that the intensity of signal F positively correlates with a broad signal A′ found at 115.6 ppm, belonging to a non-exchangeable proton and observable only when an excess of Ni(II) is added to the protein (Fig. 8B), it can be concluded that signals A′ and F are related to protons belonging to Cys14 and Cys58, found closely spaced in the Ni-binding domain. These residues are potentially able to bind an additional Ni(II) ion using their thiolate side chains. Indeed, Hβ signals of Ni(II)-bound cysteines are typically broad and observed above 100 ppm64–66 consistent with signal A′, while the sharper and upfield-shifted signal F might be assigned to a cysteine Hα proton.
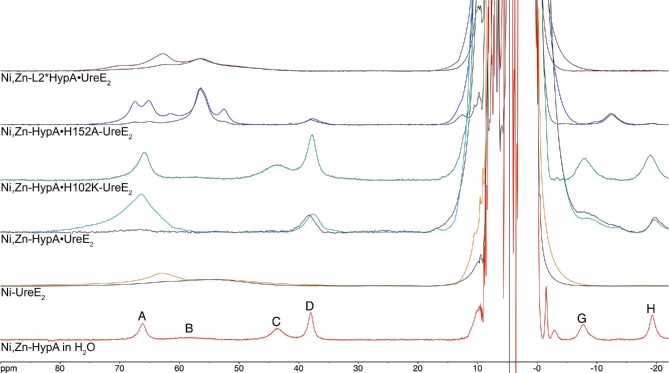
The 1H NMR spectra of Ni,Zn-HypA, Ni-UreE2, the Ni, Zn-HypA•UreE2 protein complex, and of the complexes formed between Ni,Zn-HypA and H102K-UreE2 or H152A-UreE2, or between Ni-L2*HypA and UreE2 (Fig. 9) were essential to investigate the interaction of Ni,Zn-HypA with UreE2. In contrast to the well-defined hyperfine-shifted 1H-NMR spectrum observed for Ni,Zn-HypA (Fig. 9), the Ni-UreE2 dimer reveals only a broad solvent-exchangeable envelope centered near 63 ppm and a very broad overlapping envelope of non-exchangeable resonances centered near 55 ppm (Fig. 9). These signals have been assigned to the exchangeable H-Nδ1 and non-exchangeable Hδ2 and Hε1 resonances of the two Ni-bound His102A and His102B imidazole rings.67 At variance with Ni,Zn-HypA, the absence of another relatively sharp non-exchangeable resonance is consistent with the coordination of Ni(II) by the imidazole Nε atom, in agreement with crystallography and XAS results.6
Fig. 9.
1H NMR spectra (400 MHz) of Ni,Zn-HypA in H2O (red), Ni-UreE2 (orange), Ni,Zn-HypA•UreE2 (cyan), Ni,Zn-HypA•H102K-UreE2 (green), Ni,Zn-HypA•H152A-UreE2 (blue), and Ni,Zn-L2*HypA•UreE2 (brown). The corresponding spectra of the same proteins in D2O are shown in dark gray.
The spectrum of the Ni,Zn-HypA•UreE2 WT protein complex (Fig. 9) exhibits a broad peak centered at ca. 67 ppm consisting of exchangeable histidine imidazole H-Nδ1 or H-Nε2, in addition to underlying non-exchangeable resonances (Hδ2 or Hε1). The intensity of the exchangeable resonances indicates coordination of the Ni(II) center in the protein complex by several His imidazole side chains, and is consistent with the EXAFS analysis that estimates the presence of three-four imidazole ligands (vide supra).
In addition, two non-exchangeable resonances are observed at 38 and −19 ppm, which are absent in the spectrum of Ni-UreE2 and correspond well to Ni,Zn-HypA signals D and H, assigned to His2 Hδ2 (signal D) and to a Hγ of Glu3 (signal H) in Ni,Zn-HypA. The concomitant absence of a signal corresponding to resonance C in Ni,Zn-HypA, assigned to the geminal Hγ partner of signal H, poses the question of the substantial larger broadening that affects signal C in the complex so that it is no longer visible, while signal H is slightly sharper than in the H102K-UreE2 mutant complex above (from 970 to 900 Hz). This could be attributed to a modification of the metal-to-proton distances of the two Glu3 Hγ protons, which in turn could be caused by a variation of the Chi3 dihedral angle of the side chain of Glu3 (i.e. the dihedral angle Ni-Cδ-Cγ-Hγ, see Discussion).
The hyperfine shifted 1H-NMR spectrum of Ni,Zn-HypA•H102K-UreE2 mutant (Fig. 9) is very similar to that obtained for the Ni,Zn-HypA protein, indicating that the coordination of the Ni(II) ion in HypA is not modified upon formation of the mutant complex, and that the presence of the pairs of His102A, B in the UreE2 dimer is crucial for the formation of the Ni-binding site in the WT complex. The linewidths of the resonances observed in the Ni,Zn-HypA•H102K-UreE2 complex are however significantly larger than in HypA alone (Fig. 9). The mere increase of the rotational correlation time of the larger protein complex (from 8 to 30 ns) is not sufficient to justify the ca. two-fold increase of the observed linewidths (see the quantitative treatment of the paramagnetic effects on relaxation in the Supplementary Information), while the data can be reasonably fitted by considering an electronic correlation time of 8 × 10−11 s, slightly larger than that in Ni,Zn-HypA complex (5 × 10−11 s). Such an increase of the electronic relaxation rate could be explained with a slight change in the coordination geometry of Ni(II) without significantly affecting the chemical shifts because the hyperfine constants are not modified.
The hyperfine-shifted 1H-NMR spectrum obtained for the Ni,Zn-HypA•H152A-UreE2 complex does not closely resemble the spectra obtained from either the HypA or UreE2 Ni(II) complexes (Fig. 9). At least four solvent exchangeable imidazole HN resonances are observed for a protein complex that has only three of the original imidazole residues (a pair of His102 residues from UreE2 and the His2 residue from HypA). Based on the species distribution obtained by the analysis of the ITC data (Fig. 4C), we suggest that these signals might arise from a mixture of Ni,Zn-HypA•H152A-UreE2, Ni-H152A-UreE2 and Ni-H152A-UreE4, possibly coordinated to a mixture of Nε and Nδ imidazole atoms, with some residual Ni,Zn-HypA.
The insertion variant L2*HypA was also used to form a complex with UreE2 and examined by NMR (Fig. 9). The fact that L2*HypA forms a Ni,Zn-L2*HypA•UreE2 protein complex is confirmed by SEC–MALS, and that the protein complex binds Ni(II) is established by ITC (Table 1 and in Hu et al.25). However, the hyperfine spectrum obtained for this complex contains none of the non-exchangeable proton resonances associated with HypA, and the remaining broad resonances resemble those of the Ni(II) complex of UreE2. This result indicates that Ni(II) is bound solely to UreE2 in the complex formed between L2*HypA, and is consistent with the large decrease in Ni(II) binding affinity in the L2*HypA variant (vide supra).
Computation of the structural model of the Ni,Zn-HypA•UreE2 complex
Based on all the available experimental evidence, a viable structural model of the Ni,Zn-HypA•UreE2 protein complex was calculated. The structural modelling procedure consisted of two steps: an initial model of the apo,Zn-HypA•UreE2 protein complex was generated by using a procedure that takes advantage of two state-of-the-art approaches, namely AlphaFold240 and RoseTTAFold41; these algorithms yielded excellent results in the most recent CASP14 experiment68 and were also applied to model proteins involved in the import/export of Ni(II) ions through cellular membranes.69 Subsequently, a model of the Ni-binding site located at the interface between HypA and UreE2 was constructed through a data-driven procedure already used in the case of other metalloproteins involved in the activation of urease43 or nickel binding transcriptional regulators.44,45
The best model structure of the apo,Zn-HypA•UreE2 complex is reported in Fig. 10A (see Supplementary Figs. SI-12–15 for more details on the generated models). Both HypA and UreE2 are well-modelled and show structures nearly identical to the available experimental NMR and X-ray crystal structures, respectively (see Supplementary Fig. SI-16 for a comparison). The main differences are the presence, in the UreE2 model, of the C-terminal regions that were not observed in the crystal structure because of disorder in the solid state, and a slightly different orientation of the HypA Zn-binding domain caused by the flexible unstructured linker region between the Zn- and the Ni-binding domains. In this model, HypA interacts with the UreE2 dimer in two regions: (i) the N-terminal α-helix that is in close contact with the Ni(II) binding region of UreE2, specifically with the His102A and His102B from both UreE monomers, and (ii) the β-sheet located in the Ni-binding domain, which is found to interact with one of the C-terminal tails of UreE2 and results in an extended β-sheet. Such an extended intermolecular β-sheet is also present in all the other apo,Zn-HypA•UreE2 model structures (see Supplementary Figs. SI-12–15), even though the involved UreE2 C-terminus is different in each model (in one case, both C-terminals of UreE2 are engaged in the interaction with HypA). This result suggests an unanticipated role for the UreE2 C-terminal regions in stabilizing the protein–protein complex. In agreement with the 1H,15N HSQC NMR spectrum discussed earlier (Fig. 7), the HypA Zn-domain does not appear to be involved in the formation of the complex with the UreE2 dimer. Interestingly, the calculation of the model structure of the apo,Zn-HypA•UreE2 complex without including the C-terminal regions of UreE2 (residues 152–170) results in a completely different structure that is inconsistent with the experimental NMR observations. This model features larger portions of HypA, including the Zn-domain, involved in the protein–protein interaction, and the N-terminal region of HypA no longer poised to build a Ni(II)-binding site in correspondence to the His102A and His102B residues on UreE2 (Supplementary Fig. SI-17), highlighting the importance of the elongated C-termini.
Fig. 10.
(A) Ribbon diagram and molecular surface of the apo,Zn-HypA•UreE2 complex model structure. HypA is in light blue, while UreE2 monomers are in yellow and orange. Residues involved in Ni(II) binding are in sticks, colored according to the atom type. (B) Data-based model of the Ni(II)-binding site located at the interface between HypA and UreE2. The Ni(II) ion is shown as a green sphere, while the coordination bonds are indicated with yellow dashed lines.
In the modelling of the Ni-binding site in the Ni,Zn-HypA•UreE2 complex, the metal ion was restrained to binding the Nε atoms of His102A and His102B from both UreE2 monomers, in agreement with the crystal structure of the Ni-bound UreE26, in addition to the backbone N atoms of His2 and Glu3, the imidazole Nδ of His2 and the carboxylate Oε1 atom of Glu3 on the HypA side, with distances taken from the XAS data analysis (Table 3), while no restraints were added to constrain a specific coordination geometry around the metal ion. In Fig. 10B the result of the metal site modelling procedure is shown: the Ni(II) is coordinated to the six ligands, spontaneously assuming a slightly distorted octahedral coordination geometry (root mean square deviation with respect to an ideal coordination geometry = 0.336 Å). The Ni(II) ion is bound to the HypA His2 imidazole ring Nδ atom, and to the Nε atoms of the imidazoles of the UreE2 His102 residues, consistent with the hyperfine-shifted NMR results (vide supra). The remaining ligands are provided by the His2 and Glu3 amide N-donors, and the Glu3 sidechain carboxylate. The Ni-bound His imidazole ligands are in a fac configuration with the His2 imidazole ring trans to the side chain of HypA Glu3, which in turn is cis with respect to the amide N atoms of HypA His2 and Glu3. The analysis of the structure did not reveal any distortion from the ideal geometries for the residues involved in the metal binding. With respect to the model structure of the apo,Zn-HypA•UreE2 complex previously calculated, a slight unfolding of the HypA N-terminal helix is observed, possibly to allow the formation of the Ni(II)-binding site. Indeed, the flexibility of the N-terminal helix had already been described by atomistic molecular dynamics calculations carried out on apo,Zn-HypA.62
Table 3.
Distances, angles, and dihedral constraints used in the modelling of Ni-HypA•UreE2. All constraints in the form mean ± 1 standard deviation
| Constrained atoms | Distance (Å) | |
|---|---|---|
| Ni(II)-His2(HypA,N) | 2.00 ± 0.05 | |
| Ni(II)-His2(HypA, Nδ) | 2.10 ± 0.05 | |
| Ni(II)-Glu3(HypA,N) | 2.00 ± 0.05 | |
| Ni(II)-Glu3(HypA,Oε1) | 2.00 ± 0.05 | |
| Ni(II)-His102(UreE,Nε) | 2.10 ± 0.05 | |
| Bonded atoms | Constrained atoms | Angle (degrees) |
| Ni(II)-His2(HypA,N) | Ni(II)-His2(N)-Met1(C) | 120 ± 10 |
| Ni(II)-His2(N)-His2(Cα) | 120 ± 10 | |
| Ni(II)-His2(HypA, Nδ) | Ni(II)-His2(Nδ)-His2(Cγ) | 120 ± 10 |
| Ni(II)-His2(Nδ)-His2(Cε) | 120 ± 10 | |
| Ni(II)-Glu3(HypA,N) | Ni(II)-Glu3(N)-His2(C) | 120 ± 10 |
| Ni(II)-Glu3(N)-Glu3(Cα) | 120 ± 10 | |
| Ni(II)-Glu3(HypA,Oε1) | Ni(II)-Glu3(Oε1)-Glu3(Cδ) | 109 ± 5 |
| Ni(II)-His102(UreE,Nε) | Ni(II)-His(Nε)-His(Cδ) | 120 ± 10 |
| Ni(II)-His(Nε)-His(Cε) | 120 ± 10 | |
| Bonded atoms | Constrained atoms | Dihedral (degrees) |
| Ni(II)-His2(HypA,N) | Ni(II)-His2(N)-Met1(C)-Met1(O) | 180 ± 10 |
| Ni(II)-His2(HypA, Nδ) | Ni(II)-His(Nδ)-His(Cε)-His(Nε) | 180 ± 10 |
| Ni(II)-His(Nδ)-His(Cγ)-His(Cδ) | 180 ± 10 | |
| Ni(II)-Glu3(HypA,N) | Ni(II)-Glu3(N)-His2(C)-His2(O) | 180 ± 10 |
| Ni(II)-His102(UreE,Nε) | Ni(II)-His(Nε)-His(Cε)-His(Nδ) | 180 ± 10 |
| Ni(II)−His(Nε)-His(Cδ)-His(Cγ) | 180 ± 10 | |
An alternative model of the Ni(II)-binding site in the Ni,Zn-HypA•UreE2 complex, generated by including the N-terminal nitrogen atom from HypA Met1 as a nickel ligand and excluding the backbone N atoms from HypA Glu3, resulted in steric clashes involving several atoms of Glu3 (see Supplementary Fig. SI-18 in the Supplementary Information). The latter clashes can be removed only through the denaturation of the initial ten HypA residues, a result that is not supported by experimental evidence or molecular dynamics simulations.62
Computation of the Ni-site in the Ni,Zn-HypA•UreE2 complex and in Ni,Zn-HypA
On the basis of the previously achieved model of the Ni, Zn-HypA•UreE2 complex, derived on the basis of restrained molecular mechanics calculations, a computational model of the Ni-site was derived and geometry optimized by using DFT computations. The geometry of the optimized model (Fig. 11A) does not change significantly from the more approximate model obtained through homology modelling, confirming the reliability of the present prediction. The Ni(II) coordination geometry is octahedral with only small distortions (RMSD from an ideal octahedral geometry = 0.150 Å).
Fig. 11.
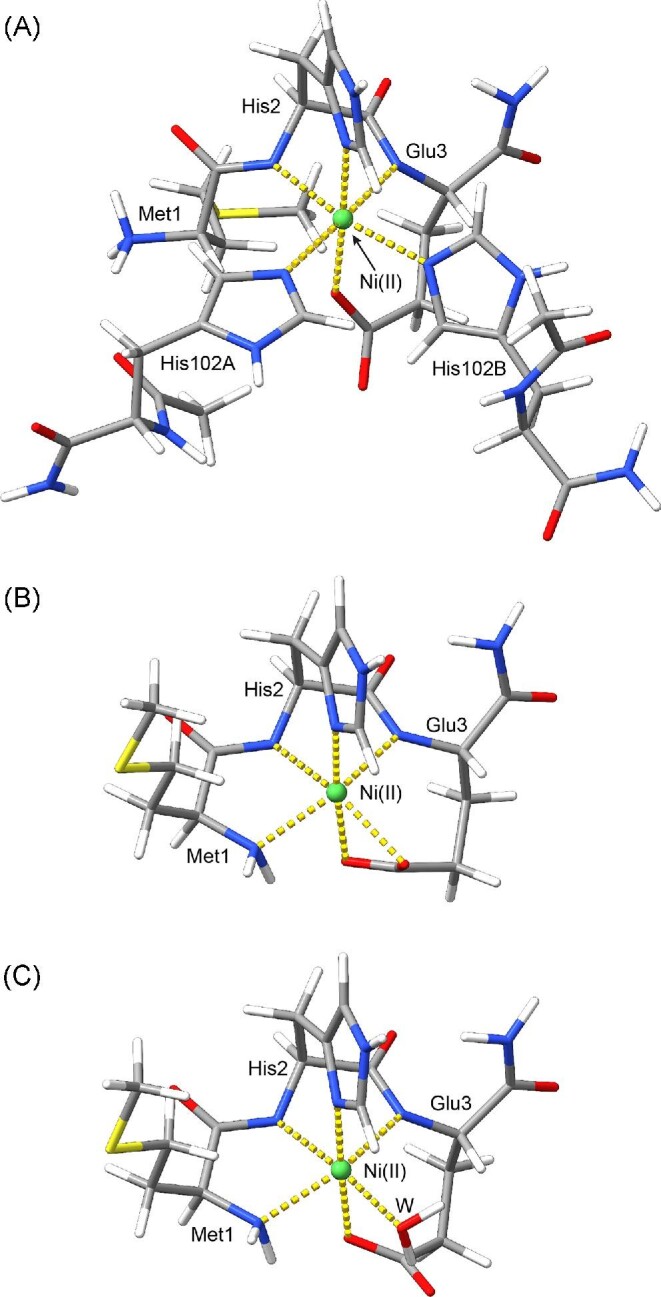
B3LYP/G 6–31(p, d) with diffuse functions on all atoms optimized geometries of the Ni-site in the Ni,Zn-HypA•UreE2 complex (A) and in Ni,Zn-HypA (B and C). Atoms are colored according to atom type and coordination bonds are reported using dashed yellow lines.
From this model of the Ni-site in the Ni,Zn-HypA•UreE2 complex, two tentative model structures of the nickel-binding site of HypA were generated. In the first model, the two histidine residues from the UreE dimer were removed and the conformation of HypA Met1 was changed manually in order to bring the N-terminal nitrogen in the vicinity of the Ni(II) ion. The optimized structure of such a model (Fig. 11B) resulted in a quite distorted octahedral Ni(II) coordination geometry (RMSD = 0.341 Å) where the Glu3 carboxylate is bound in a bidentate fashion to the Ni(II) ion.
Finally, we considered an alternative model for the Ni(II) site in HypA, which included the presence of a water molecule as Ni(II) ion sixth ligand. The result of the geometry optimization of this HypA Ni-site model are reported in Fig. 11C. In this model, the Ni(II) coordination geometry is less distorted than in the model involving a bidentate Glu3 carboxylate group (RMSD = 0.196 Å).
These models were then used to explain some features of the paramagnetically shifted NMR signals. The spectrum of Ni-HypA (Figs. 8 and 9) features signals C and H, assigned to a geminal Hγ pair of Glu3, displaying different linewidths, suggesting two different nickel-proton distances. In the DFT-optimized model of holo-HypA containing a bidentate mode for Ni(II) coordination by Glu3 these distances are 4.2 and 4.4 Å, while in the model featuring a bound solvent molecule, the distances are 4.6 and 4.7 Å. A quantitative analysis of the paramagnetic transverse relaxation (see Supplementary Information and Supplementary Fig. SI-21) suggests that a slight variation of the Chi3 dihedral angle for Glu3 with respect to the model calculated in vacuo is sufficient to bring the γCH2 protons at distances from Ni(II) compatible with the observed linewidths. Similarly, the linewidths of signals C and H in the Ni,Zn-HypA-(H102K)UreE2 mutant complex (Fig. 9), in which Ni(II) is retained by HypA because of the mutation of the nickel-binding residues on UreE2, a similar treatment suggests again that a minor rotation of Chi3, together with an minor increase of the electronic relaxation time, allows the reproduction of the experimental linewidths. Finally, in the NMR spectrum of Ni,Zn-HypA-UreE2 (Fig. 9), signal C disappears while signal H is still visible and actually decreases its linewidth. Assuming 3 kHz as a threshold limit for signal detection, using the same quantitative treatment of the nuclear relaxation as mentioned earlier, these observations support a rotation of the Chi3 angle by ca. 40° with respect to the model calculated in vacuo. In all these cases, the difference of this geometric parameter between the theoretical model and the experimental data can be justified by the presence of the protein scaffold surrounding the Ni(II)-binding site.
Discussion
A unique feature of nickel trafficking in Hp is the use of the metallochaperone HypA to mediate Ni incorporation in two distinct enzymes, urease and hydrogenase.10,11 In general, metallochaperones are able to transfer the specific metals required for enzyme active sites via formation of specific protein–protein complexes.4 In the case of [Ni,Fe]-hydrogenase, a complex formed between HypA and the apo-large subunit of the hydrogenase heterodimer appears to be involved in Ni incorporation3. For urease, the analogous transfer is mediated by UreE2.8,70 The requirement of a second metallochaperone in the delivery of Ni to urease suggests the involvement of additional protein complexes, as well as perhaps implying a higher degree of control over the process. Indeed, an interaction between HypA and UreE2 has been shown to be required for maturation of urease12 and a protein–protein complex was previously described as consisting of one HypA molecule and one UreE2 dimer.25 This complex was found to bind Ni(II) many orders of magnitude tighter than either the HypA or UreE2 proteins, implying the existence of a unique Ni(II)-binding site formed in the protein complex.25 The focus of this work is to more fully characterize and elaborate the structure of the HypA•UreE2 complex and the novel high-affinity Ni(II) site in the protein complex and elucidate mechanistic details of Ni binding.
The protein–protein interactions and the Ni-binding affinities of the complexes formed between WT- and L2*-HypA proteins and UreE2 protein and His102/152 UreE2 variants were assessed by a combination of SEC–MALS and ITC experiments that were interpreted using a global fitting model that takes into account five equilibria (Eqn. 4). This method allowed each equilibrium involved in processes of forming Ni,Zn-HypA•UreE2 to be examined and the effects of protein modifications on individual equilibria to be assessed. The results shown in Table 1 allow several observations to be made:
The SEC–MALS data show that the protein complexes form regardless of mutations in the Ni(II)-binding site (Fig. 1), although the affinity of the proteins (Table 1) is weakened in the Ni site variants (e.g. apo, Zn-L2*HypA•UreE2 (KD = 1.7 μM) vs. apo,Zn-HypA•UreE2 (KD = 0.8 μM). Therefore, the Ni ligands are not critical for protein complex formation.
Apo, Zn-HypA•UreE2 contains a novel high-affinity Ni-binding site with a KD of about 0.15 nM that is not present in either component protein. The sub-nanomole KD indicates that the apo,Zn-HypA•UreE2 protein complex binds Ni much tighter than either of the component proteins (KD = 0.9 and 0.3 mM for HypA and UreE2, respectively). The values of KD obtained for HypA and UreE2 are characteristic of metallochaperones in general, which typically feature values of KD in the μM range.26 The sub-nanomole value KD observed for Ni,Zn-HypA•UreE2 is more characteristic of Ni-responsive transcriptional regulators, such as InrS (KD ∼ 10−10 M)27 and suggests a different function for the protein complex.
Mutations involving metal-binding ligands in the component proteins (the HypA N-terminal amine in L2*, His102 and His152 in UreE2) significantly weaken the Ni affinity of the novel Ni-binding site in the protein complex (Table 1). This result shows that ligands from both component proteins are involved in forming the high-affinity binding site in apo,Zn-HypA•UreE2 and indicates that the high-affinity site forms at a protein–protein interface.
The robust formation of the Ni,Zn-HypA•UreE2 allowed for isolation and characterization of Ni,Zn-HypA•UreE2. HypA is a small, elongated protein that contains an intrinsic Zn-binding site associated with two CXXC sequences near the C-terminus and a Ni-binding site located at the N-terminus.62 The Zn-binding domain is composed of a β-structure and is connected to the larger Ni-binding domain by a flexible linker.62 The Ni-binding domain is composed of two α-helices and a three-stranded parallel/antiparallel β-sheet.62 1H,15N HSQC spectra at 950 MHz of labeled HypA protein in the apo,Zn-HypA•UreE2 complex (Fig. 7) show that the Zn binding domain is not involved in forming the protein complex. In contrast, many perturbations are found in the Ni-binding domain. This result is consistent with analogous experiments carried out using a form of HypA mutated at the N-terminus and containing a Gly-Ser extension,59 demonstrating that the protein–protein interaction does not depend greatly on the N-terminus of the HypA polypeptide. This observation is also consistent with the studies of the L2*-HypA variant (involving an insertion of Leu between Met1 and His2) that greatly weakens Ni(II) binding to HypA (by a factor of > 700, Table 1) and the HypA•UreE2 complex (by a factor of ∼400),23 but affects the protein–protein interaction to a much smaller extent (Table 1 and reference25). A potential explanation for the global structural changes in the Ni-binding domain of HypA in apo,Zn-HypA•UreE2 observed by NMR, as well as for the insensitivity of the protein–protein interaction to the HypA N-terminal sequence, was obtained from computational modelling of the apo,Zn-HypA•UreE2 complex with AlphaFold240 and RoseTTAFold.41 The resulting models show that the N-terminal Ni-binding site of HypA contacts the dimer interface of UreE2 near to the known Ni-binding ligands—a pair of His102 residues—and that the rest of the protein is locked in the embrace of the UreE2 C-terminal sequence, which forms additional strands of β-sheet with the HypA Ni-binding domain three-stranded β-sheet (Fig. 10A).
Structural characterizations of the metal sites were performed on the isolated Ni,Zn-HypA•UreE2 protein complex by XAS. These studies revealed that the Zn site in Ni,Zn-HypA•UreE2 is a Zn(SCys)4 site that is unperturbed from that found in HypA protein alone,62 in agreement with the NMR results and computational models that show that the Zn-binding domain is not involved in the formation of the complex (vide supra). The interfacial Ni site in Ni,Zn-HypA•UreE2 was shown to be a six-coordinate distorted octahedral site composed of three or four His imidazole ligands and totaling six O, N-donor ligands (Table 2 and Fig. 6). Given the odd number of His ligands available in the metal-binding sites of the two component proteins, the data are most consistent with three His imidazole ligands, His2 from HypA and a pair of His imidazole ligands from the UreE2 dimer that are assigned to a pair of His102 ligands on the basis of the ITC and NMR results (vide infra).
Further structural details regarding the Ni site in Ni,Zn-HypA•UreE2 were obtained by NMR. The HypA protein exhibits eight hyperfine-shifted signals (A–H) in the range from +80 to −20 ppm (Fig. 8). These signals had previously been assigned to amino acid residues based on their chemical shift, linewidths, longitudinal relaxation times, temperature dependence, field dependence, solvent-exchange phenomena and NOEs, that left some assignments ambiguous, particularly those that might be due to Met1, Glu3, or Asp40 α- or methylene-protons.62 The spectrum of Ni,Zn-HypA was re-examined using HypA samples that contained selectively deuterated amino acids. This confirmed the assignments of signals A, B, D and G to His2 protons, and allowed the unambiguous assignment of signals C, E, and H to Glu3 (Fig. 8A). No signals were found to be associated with Met1 or with Asp40. This is not surprising for Met1 bound by the N-terminal amine,23 considering that the paramagnetism of the bound Ni(II) is expected to obliterate the signals of both the terminal amine protons as well as the Hα proton of Met1, broadened beyond detection because of decreased relaxation time; moreover, this also indicates that Asp40 is not a ligand for Ni(II). The consideration of Asp40 as a ligand derives from its inclusion as a carboxylate O-donor in the Ni site based on NMR studies of a HypA complex modified with an N-terminal Gly-Ser extension.63 This extension of the N-terminus in HypA induces the formation of an artefactual planar and diamagnetic Ni(II) site, instead of a now well-established pseudo-octahedral metal-binding site in the WT protein, which causes the Ni(II) ion to be paramagnetic.23,62 Thus, the involvement of Asp40, while consistent with the disappearance of NMR signals near Asp40,62 is likely an artifact of the structural alteration of the Ni site in the N-terminally modified HypA protein. The combination of XAS structural information and this NMR data lead to a structure for the Ni complex in HypA that involves ligation by the N-terminal amine of Met1, the imidazole Nδ and amide N of His2, the side-chain carboxylate and amide N of Glu3, and one O/N-donor lacking any hyperfine-shifted 1H-NMR resonances that is consistent with a water-derived ligand or the presence of a bidentate Glu3 carboxylate.
The hyperfine-shifted 1H-NMR of the Ni,Zn-HypA•UreE2 complex (Fig. 9) reveals several resonances that are analogous to those assigned to His imidazole ligands in HypA and UreE2, including signal D. The observation of signal D confirms that the HypA His2 ligand is present in the high-affinity Ni-binding site of the complex and that its coordination mode is unaltered from the HypA site. The increase in the intensity of the envelope of resonances corresponding to solvent-exchangeable protons assigned to N-H protons of His imidazole ligands relative to signal D is consistent with the presence of three His imidazole ligands found in the EXAFS analysis (vide supra). The identity of the two additional His imidazole ligands in Ni,Zn-HypA•UreE2 was confirmed by the hyperfine-shifted NMR spectrum of the Ni,Zn-HypA•H102K-UreE2 variant, which exhibits resonances only from HypA ligands.
Notably absent from the spectrum of HypA•H102K-UreE2 is signal C, assigned to a methylene proton in the Glu3 side chain. At first glance, the absence of signal C might suggest that the Glu3 carboxylate has been lost as a Ni ligand in the protein complex. This would be consistent with the substitution of this ligand and the putative water-derived ligand by the pair of UreE2 His102 residues and the retention of all other HypA ligands, including the N-terminal amine, which cannot be detected by NMR nor unambiguously assigned by EXAFS analysis. However, signal H is still present in the hyperfine-shifted NMR spectrum of Ni,Zn-HypA•UreE2 and has been assigned to a methylene proton on Glu3 that is geminal to the proton giving rise to signal C on the basis of an NOE measurement.62 This fact argues for retention of the Glu3 carboxylate in the HypA•UreE2 complex. The disappearance of signal C can be explained as arising from a reorientation of the methylene protons (as might accompany a bidentate to monodentate conversion) such that the proton giving rise to signal C is much closer to the Ni center in the HypA•UreE2 complex than in HypA, leading to dipolar line broadening to the extent that it is no longer detected.
The inclusion of the Glu3 carboxylate in the Ni,Zn-HypA•UreE2 complex requires that some other ligand is lost in addition to substitution of water (or change in carboxylate coordination). This ligand must lack any observable hyperfine-shifted 1H-NMR signal. Based on the structure of the Ni(II) site in the HypA protein, three possibilities exist: the Met1 amine, the His2 amide N atom, or the Glu3 amide N-atom. The lack of perturbation of any resonances associated with His2 in Ni,Zn-HypA•UreE2 relative to the HypA complex argues for retention of the His2 amide N-donor, which enforces the Nδ coordination of the His2 imidazole. The fact that Ni-binding affinities are greatly reduced and the structures of the Ni sites are altered in both the L2*-HypA complex and in Ni, Zn-L2*-HypA•UreE2 argues for retention of the Met1 amine in Ni,Zn-HypA•UreE2. In the case of HypA, the L2* insertion decreases Ni binding affinity (Table 1), prevents nickelation of urease in vivo, and causes the structure to become five-coordinate and adopt a low-spin (diamagnetic) electronic configuration.23 In Ni, Zn-L2*-HypA•UreE2, the affinity of the Ni site is also decreased, and the hyperfine-shifted NMR resembles that of Ni-UreE2, consistent with the loss of coordination of HypA ligands and the complete transfer of Ni(II) from HypA to UreE2 in the protein complex. However, inclusion of the N-terminal amine in Ni,Zn-HypA•UreE2 and dissociation of the Glu3 amide N atom is not compatible with the modelling results, given the attendant constraints (vide supra), which support only the dissociation of the N-terminal amine in Ni,Zn-HypA•UreE2.
The details of how of Ni,Zn-HypA•UreE2 functions in the maturation of Hp urease remain incomplete. In other organisms, the presence of UreE2 is sufficient to provide Ni(II) to the urease maturation pathway by delivering Ni to UreG (a soluble GTPase involved in the urease maturation process) via the heterodimeric Ure(GE)2, forming a UreG dimer with an interfacial Ni(II) site.5,8,71 Ni-UreG2 then completes nickelation of apo-urease via a ternary Ure(DFG)2 protein complex.72,73 In Hp, while UreE2 is a requirement for urease maturation, the process also requires HypA under physiological conditions.4 NMR-based results show that no complex is formed between Ni,Zn-HypA•UreE2 and UreG (Supplementary Fig. SI-20), suggesting that UreE2 can interact with either HypA or UreG, but not both simultaneously, presumably because both complexes involve the same UreE2 interface that comprises the conserved His102A, B on the homodimer surface. HypA is known to acquire Ni(II) from HypB, which in turn acquires the metal ion from a complex formed by the ABC-importer NiuBDE and SlyD.4,74 It is possible that Ni(II) acquired by this specific uptake pathway is unavailable to UreE2 until HypA acquires Ni(II) from HypB and performs a handoff to deliver Ni(II) to UreE2. However, the very tight binding of nickel in the HypA•UreE2 complex would require a specific mechanism to either transfer Ni(II) to UreE2 and release HypA, or somehow decrease the affinity of the complex so that Ni(II) is available to the urease maturation pathway. It is possible that Ni,Zn-HypA•UreE2 plays a role in inhibiting nickelation of the large amount of apo-urease present under neutral conditions until the other urease chaperones are ready for correct Ni(II) incorporation. In addition, the HypA•UreE2 complex might also provide a targeted pool of Ni(II) for rapid urease activation under acid stress, when all the urease apparatus is ready for Ni(II) delivery.
Conclusions
The results presented here characterize a protein complex formed between HypA and UreE2 that contains a single high-affinity (sub-nanomolar KD) Ni(II)-binding site and presumably represents the key interaction between UreE2 and HypA that is required for maturation of urease in Hp. This novel Ni(II) site is located at the intersection of the UreE2 dimer interface and the rigorously conserved MHE N-terminus of HypA, and is supported by extensive interactions between the component proteins in the absence of Ni(II). The Ni(II) complex is hexa-coordinated and therefore high spin (S = 1), and draws on ligands from both component proteins. Direct evidence for the inclusion of the HypA His2 imidazole, Glu3 carboxylate and a pair of His102 imidazole ligands from UreE2 was obtained from XAS and hyperfine-shifted NMR studies. The addition of the two His ligands from UreE2 requires substitution of two ligands from the HypA complex. Biochemical studies are most consistent with retention of the N-terminal amine of Met1, suggesting the loss of the Glu3 amide N-donor.23,25 However, molecular modelling indicates that the N-donors from both His2 and Glu3 are retained, and that the Met1 N-terminal amine present in Ni,Zn-HypA dissociates in forming the Ni,Zn-HypA•UreE2 complex.
Supplementary Material
Acknowledgements
Fabio Calogiuri and Massimo Lucci are duly acknowledged for their support in collecting all NMR spectra at CERM—Centre for Magnetic Resonance—University of Firenze, Italy.
Contributor Information
Barbara Zambelli, Laboratory of Bioinorganic Chemistry, Department of Pharmacy and Biotechnology, University of Bologna, Bologna, Italy.
Priyanka Basak, Department of Chemistry and Program in Molecular and Cellular Biology, University of Massachusetts, Amherst, MA, USA.
Heidi Hu, Department of Chemistry and Program in Molecular and Cellular Biology, University of Massachusetts, Amherst, MA, USA.
Mario Piccioli, Centre for Magnetic Resonance, Department of Chemistry, University of Florence, Florence Italy.
Francesco Musiani, Laboratory of Bioinorganic Chemistry, Department of Pharmacy and Biotechnology, University of Bologna, Bologna, Italy.
Valquiria Broll, Laboratory of Bioinorganic Chemistry, Department of Pharmacy and Biotechnology, University of Bologna, Bologna, Italy.
Lionel Imbert, Univ. Grenoble Alpes, CNRS, CEA, Institut de Biologie Structurale (IBS), Grenoble, France.
Jerome Boisbouvier, Univ. Grenoble Alpes, CNRS, CEA, Institut de Biologie Structurale (IBS), Grenoble, France.
Michael J Maroney, Department of Chemistry and Program in Molecular and Cellular Biology, University of Massachusetts, Amherst, MA, USA.
Stefano Ciurli, Laboratory of Bioinorganic Chemistry, Department of Pharmacy and Biotechnology, University of Bologna, Bologna, Italy; Centre for Magnetic Resonance, Department of Chemistry, University of Florence, Florence Italy.
Authors contributions
Protein production was performed by BZ, PB, HH, and VB. Production of HypA containing deuterated Asp or Glu residues was performed by LI and JB. MALS experiments were performed and analyzed by BS and VB. ITC experiments were performed and analyzed by BZ and VB. XAS data collection and analysis was performed by PB. NMR experiments were performed and analyzed by MP. Computation and theoretical studies were performed by FM. Conceptualization was done by MM and SC, who also participated in the final interpretations of the data. All authors contributed to the writing and revisions of the manuscript.
Funding
The authors acknowledge support for this work from the NIH R01-069696 (M.J.M.). Use of the Stanford Synchrotron Radiation Lightsource, SLAC National Accelerator Laboratory, is supported by the U.S. Department of Energy, Office of Science, Office of Basic Energy Sciences under Contract No. DE-AC02-76SF00515. The SSRL Structural Molecular Biology Program is supported by the DOE Office of Biological and Environmental Research, and by the National Institutes of Health, National Institute of General Medical Sciences (P30GM133894). The contents of this publication are solely the responsibility of the authors and do not necessarily represent the official views of NIGMS or NIH. This work benefited from access to the Cell-Free platform of the Grenoble Instruct-ERIC center (ISBG; UMS 3518 CNRS-CEA-UGA-EMBL), an Instruct-ERIC center, within the Grenoble Partnership for Structural Biology (PSB), supported by FRISBI (ANR-10-INBS-0005-02) and GRAL, financed within the University Grenoble Alpes graduate school (Ecoles Universitaires de Recherche) CBH-EUR-GS (ANR-17-EURE-0003). Financial support was provided by Instruct-ERIC (PID: 13114). VB was supported by a grant from Consorzio Interuniversitario di Risonanze Magnetiche di Metallo-Proteine (CIRMMP). SC, BZ and FM acknowledge partial support by the University of Bologna. MJM was partially supported by the Institute of the Advanced Studies of the University of Bologna through a visiting fellowship.
Conflict of interest
The authors report no conflicts of interest.
Data availability
Data and models are available upon request to the authors.
References
- 1. Peters J. W., Schut G. J., Boyd E. S., Mulder D. W., Shepard E. M., Broderick J. B., King P. W., Adams M. W. W., [FeFe]- and [NiFe]-hydrogenase diversity, mechanism, and maturation, BBA-Mol Cell Res. 2015, 1853 (12), 1350–1369. [DOI] [PubMed] [Google Scholar]
- 2. Lacasse M. J., Zamble D B., [NiFe]-hydrogenase maturation, Biochemistry 2016, 55 (12), 1689–1701. [DOI] [PubMed] [Google Scholar]
- 3. Miki K., Atomi H., Watanabe S., Structural insight into [NiFe] hydrogenase maturation by transient complexes between Hyp proteins, Acc. Chem. Res. 2020, 53 (4), 875–886. [DOI] [PubMed] [Google Scholar]
- 4. Maroney M. J., Ciurli S., Nickel as a virulence factor in the Class I bacterial carcinogen, Helicobacter pylori, Semin. Cancer Biol. 2021, 76, 143–155. [DOI] [PubMed] [Google Scholar]
- 5. Bellucci M., Zambelli B., Musiani F., Turano P., Ciurli S., Helicobacter pylori UreE, a urease accessory protein: specific Ni(II)- and Zn(II)-binding properties and interaction with its cognate UreG, Biochem. J. 2009, 422 (1), 91–100. [DOI] [PubMed] [Google Scholar]
- 6. Banaszak K., Martin-Diaconescu V., Bellucci M., Zambelli B., Rypniewski W., Maroney M. J., Ciurli S., Crystallographic and X-ray absorption spectroscopic characterization of Helicobacter pylori UreE bound to Ni2+ and Zn2+ reveals a role for the disordered C-terminal arm in metal trafficking, Biochem. J. 2012, 441 (3), 1017–1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Merloni A., Dobrovolska O., Zambelli B., Agostini F., Bazzani M., Musiani F., Ciurli S., Molecular landscape of the interaction between the urease accessory proteins UreE and UreG, Biochim. Biophys. Acta 2014, 1844, 1662–1674. [DOI] [PubMed] [Google Scholar]
- 8. Nim Y. S., Wong K.-B., The maturation pathway of nickel urease, Inorganics 2019, 7 (7), 85 [Google Scholar]
- 9. Alfano M., Cavazza C., Structure, function, and biosynthesis of nickel-dependent enzymes, Protein Sci. 2020, 1–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Olson J. W., Mehta N. S., Maier R J., Requirement of nickel metabolism proteins HypA and HypB for full activity of both hydrogenase and urease in Helicobacter pylori, Mol. Microbiol. 2001, 39 (1), 176–182. [DOI] [PubMed] [Google Scholar]
- 11. Mehta N., Olson J. W., Maier R J., Characterization of Helicobacter pylori nickel metabolism accessory proteins needed for maturation of both urease and hydrogenase, J. Bacteriol. 2003, 185 (3), 726–734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Benoit S. L., Mehta N., Weinberg M. V., Maier C., Maier R J., Interaction between the Helicobacter pylori accessory proteins HypA and UreE is needed for urease maturation, Microbiology 2007, 153 (Pt 5), 1474–1482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Benoit S. L., Zbell A. L., Maier R J., Nickel enzyme maturation in Helicobacter hepaticus: roles of accessory proteins in hydrogenase and urease activities, Microbiology 2007, 153 (Pt 11), 3748–3756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Testerman T. L., Morris J., Beyond the stomach: an updated view of Helicobacter pylori pathogenesis, diagnosis, and treatment, World J. Gastroenterol. 2014, 20 (36), 12781–12808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Zamani M., Ebrahimtabar F., Zamani V., Miller W. H., Alizadeh-Navaei R., Shokri-Shirvani J., Derakhshan M H., Systematic review with meta-analysis: the worldwide prevalence of Helicobacter pylori infection, Aliment Pharm Therap 2018, 47 (7), 868–876. [DOI] [PubMed] [Google Scholar]
- 16. Sachs G., Weeks D. L., Wen Y., Marcus E. A., Scott D. R., Melchers K., Acid acclimation by Helicobacter pylori, Physiology 2005, 20, 429–438. [DOI] [PubMed] [Google Scholar]
- 17. Blum F. C., Hu H. Q., Servetas S. L., Benoit S. L., Maier R. J., Maroney M. J., Merrell D S., Structure-function analyses of metal-binding sites of HypA reveal residues important for hydrogenase maturation in Helicobacter pylori, PLoS One 2017, 12 (8), e0183260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Wang G., Romero-Gallo J., Benoit S. L., Piazuelo M. B., Dominguez R. L., Morgan D. R., Peek R. M. Jr., Maier R. J.. Hydrogen metabolism in Helicobacter pylori plays a role in gastric carcinogenesis through facilitating CagA translocation, mBio 2016, 7 (4), e01022–e01016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Benoit S. L., Maier R. J., Sawers R. G., Greening C., Molecular hydrogen metabolism: a widespread trait of pathogenic bacteria and protists, Microbiol Mol Biol R 2020, 84 (1), e00092–e00019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hu L. T., Mobley H L., Purification and N-terminal analysis of urease from Helicobacter pylori, Infect. Immun. 1990, 58 (4), 992–998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Voland P., Weeks D. L., Marcus E. A., Prinz C., Sachs G., Scott D., Interactions among the seven Helicobacter pylori proteins encoded by the urease gene cluster, Am. J. Physiol. Gastrointest. Liver Physiol. 2003, 284 (1), G96–G106. [DOI] [PubMed] [Google Scholar]
- 22. Johnson R. C., Hu H. Q., Merrell D. S., Maroney M J., Dynamic HypA zinc site is essential for acid viability and proper urease maturation in Helicobacter pylori, Metallomics 2015, 7 (4), 674–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hu H. Q., Johnson R. C., Merrell D. S., Maroney M J., Nickel ligation of the N-terminal amine of HypA is required for urease maturation in Helicobacter pylori, Biochemistry 2017, 56 (8), 1105–1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Savoldi A., Carrara E., Graham D. Y., Conti M., Tacconelli E., Prevalence of antibiotic resistance in Helicobacter pylori: a systematic review and meta-analysis in World Health Organization regions, Gastroenterology 2018, 155 (5), 1372–1382.e17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hu H. Q., Huang H. T., Maroney M J., The Helicobacter pylori HypA•UreE2 complex contains a novel high-affinity Ni(II)-binding site, Biochemistry 2018, 57, 2932–2942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Rosenzweig A C., Metallochaperones: bind and deliver, Chem. Biol. 2002, 9 (6), 673–677. [DOI] [PubMed] [Google Scholar]
- 27. Musiani F., Zambelli B., Bazzani M., Mazzei L., Ciurli S., Nickel-responsive transcriptional regulators, Metallomics 2015, 7, 1305–1318. [DOI] [PubMed] [Google Scholar]
- 28. Zambelli B., Bellucci M., Danielli A., Scarlato V., Ciurli S., The Ni2+ binding properties of Helicobacter pylori NikR, Chem. Commun. (Camb.) 2007(35), 3649–3651. [DOI] [PubMed] [Google Scholar]
- 29. Herbst R. W., Perovic I., Martin-Diaconescu V., O'Brien K., Chivers P. T., Pochapsky S. S., Pochapsky T. C., Maroney M. J., Communication between the zinc and nickel sites in dimeric HypA: metal recognition and pH sensing, J. Am. Chem. Soc. 2010, 132 (30), 10338–10351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Imbert L., Lenoir-Capello R., Crublet E., Vallet A., Awad R., Ayala I., Juillan-Binard C., Mayerhofer H., Kerfah R., Gans P., Miclet E., Boisbouvier J., In vitro production of perdeuterated proteins in H2O for biomolecular NMR Studies, Methods Mol. Biol. 2021, 2199, 127–149. [DOI] [PubMed] [Google Scholar]
- 31. Stola M., Musiani F., Mangani S., Turano P., Safarov N., Zambelli B., Ciurli S., The nickel site of Bacillus pasteurii UreE, a urease metallo-chaperone, as revealed by metal-binding studies and X-ray absorption spectroscopy, Biochemistry 2006, 45 (20), 6495–6509. [DOI] [PubMed] [Google Scholar]
- 32. Muñoz E., Piñeiro A., AFFINImeter Software: from its beginnings to future trends-A Literature review, J Appl Bioanal 2018, 4 (4), 124–139. [Google Scholar]
- 33. Higgins K. A., Hu H. Q., Chivers P. T., Maroney M J., Effects of select histidine to cysteine mutations on transcriptional regulation by Escherichia coli RcnR, Biochemistry 2013, 52 (1), 84–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Ravel B., Newville M. A, ARTEMIS, HEPHAESTUS: data analysis for X-ray absorption spectroscopy using IFEFFIT, J. Synchrotron Radiat. 2005, 12 (Pt 4), 537–541. [DOI] [PubMed] [Google Scholar]
- 35. Martin-Diaconescu V., Maroney M J., Nickel Bioinorganic Systems, Comprehensive Inorganic Chemistry II: Elsevier, 2013, 295–322. [Google Scholar]
- 36. Delaglio F., Grzesiek S., Vuister G. W., Zhu G., Pfeifer J., Bax A., NMRPipe: a multidimensional spectral processing system based on UNIX pipes, J. Biomol. NMR 1995, 6 (3), 277–293. [DOI] [PubMed] [Google Scholar]
- 37. Lee W., Rahimi M., Lee Y., Chiu A., POKY: a software suite for multidimensional NMR and 3D structure calculation of biomolecules, Bioinformatics 2021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Piccioli M., Turano P., Transient iron coordination sites in proteins: Exploiting the dual nature of paramagnetic NMR, Coord. Chem. Rev. 2015, 284, 313–328. [Google Scholar]
- 39. Mirdita M., Schütze K., Moriwaki Y., Heo L., Ovchinnikov S., Steinegger M., ColabFold - Making protein folding accessible to all, bioRxiv 2022:2021.08.15.456425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Senior A. W., Evans R., Jumper J., Kirkpatrick J., Sifre L., Green T., Qin C., Zidek A., Nelson A. W. R., Bridgland A., Penedones H., Petersen S., Simonyan K., Crossan S., Kohli P., Jones D. T., Silver D., Kavukcuoglu K., Hassabis D., Improved protein structure prediction using potentials from deep learning, Nature 2020, 577 (7792), 706–710. [DOI] [PubMed] [Google Scholar]
- 41. Baek M., DiMaio F., Anishchenko I., Dauparas J., Ovchinnikov S., Lee G. R., Wang J., Cong Q., Kinch L. N., Schaeffer R. D., Millan C., Park H., Adams C., Glassman C. R., DeGiovanni A., Pereira J. H., Rodrigues A. V., van Dijk A. A., Ebrecht A. C., Opperman D. J., Sagmeister T., Buhlheller C., Pavkov-Keller T., Rathinaswamy M. K., Dalwadi U., Yip C. K., Burke J. E., Garcia K. C., Grishin N. V., Adams P. D., Read R. J., Baker D., Accurate prediction of protein structures and interactions using a three-track neural network, Science 2021, 373 (6557), 871–876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Mariani V., Biasini M., Barbato A., Schwede T., lDDT: a local superposition-free score for comparing protein structures and models using distance difference tests, Bioinformatics 2013, 29 (21), 2722–2728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Martin-Diaconescu V., Bellucci M., Musiani F., Ciurli S., Maroney M J., Unraveling the Helicobacter pylori UreG zinc binding site using X-ray absorption spectroscopy (XAS) and structural modeling, J. Biol. Inorg. Chem. 2012, 17 (3), 353–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Carr C. E., Musiani F., Huang H. -. T., Chivers P. T., Ciurli S., Maroney M J., Glutamate ligation in the Ni(II)- and Co(II)-responsive Escherichia coli transcriptional regulator, RcnR, Inorg. Chem. 2017, 56 (11), 6459–6476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Barchi E., Musiani F., Molecular modelling of the Ni(II)-responsive Synechocystis PCC 6803 transcriptional regulator InrS in the metal bound form, Inorganics 2019, 7 (6), 76 [Google Scholar]
- 46. Martí-Renom M. A., Stuart A. C., Fiser A., Sánchez R., Melo F., Sali A., Comparative protein structure modeling of genes and genomes, Ann. Rev. Biophys. Biomol. Struct. 2000, 29 (1), 291–325. [DOI] [PubMed] [Google Scholar]
- 47. MacKerell A., Bashford D., Bellot M., Dunbrack R., Evanseck J., Field M., All-atom empirical potential for molecular modeling and dynamics studies of proteins using the CHARMM22 force field, J. Phys. Chem. B 1998, 102, 3586–3616. [DOI] [PubMed] [Google Scholar]
- 48. Shen M-y, Sali A., Statistical potential for assessment and prediction of protein structures, Protein Sci. 2006, 15 (11), 2507–2524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Laskowski R. A., MacArthur M. W., Moss D. S., Thornton J M., PROCHECK: a program to check the stereochemical quality of protein structures, J Appl Crystallograph 1993, 26, 283–291. [Google Scholar]
- 50. Pettersen E. F., Goddard T. D., Huang C. C., Couch G. S., Greenblatt D. M., Meng E. C., Ferrin T E., UCSF Chimera - A visualization system for exploratory research and analysis, J. Comput. Chem. 2004, 25, 1605–1612. [DOI] [PubMed] [Google Scholar]
- 51. Goddard T. D., Huang C. C., Meng E. C., Pettersen E. F., Couch G. S., Morris J. H., Ferrin T E., UCSF ChimeraX: Meeting modern challenges in visualization and analysis, Protein Sci. 2018, 27 (1), 14–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Pettersen E. F., Goddard T. D., Huang C. C., Meng E. C., Couch G. S., Croll T. I., Morris J. H., Ferrin T. E., UCSF ChimeraX: Structure visualization for researchers, educators, and developers, Protein Sci. 2021, 30 (1), 70–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Neese F., The ORCA program system, Wiley Interdiscip Rev: Comput Mol Sci 2012, 2 (1), 73–78. [Google Scholar]
- 54. Becke A D., A new mixing of Hartree—Fock and local density-functional theories, J. Chem. Phys. 1993, 98 (2), 1372–1377. [Google Scholar]
- 55. Lee C., Yang W., Parr R G., Development of the Colle-Salvetti correlation-energy formula into a functional of the electron density, Phys. Rev. B 1988, 37 (2), 785–789. [DOI] [PubMed] [Google Scholar]
- 56. Frisch M. J., Trucks G. W., Schlegel H. B., Scuseria G. E., Robb M. A., Cheeseman J. R., Scalmani G., Barone V., Mennucci B., Petersson G. A., Nakatsuji H., Caricato M., Li X., Hratchian H. P., Izmaylov A. F., Bloino J., Zheng G., Sonnenberg J. L., Hada M., Ehara M., Toyota K., Fukuda R., Hasegawa J., Ishida M., Nakajima T., Honda Y., Kitao O., Nakai H., Vreven T., Montgomery J. A. Jr., Peralta J. E., Ogliaro F., Bearpark M. J., Heyd J., Brothers E. N., Kudin K. N., Staroverov V. N., Kobayashi R., Normand J., Raghavachari K., Rendell A. P., Burant J. C., Iyengar S. S., Tomasi J., Cossi M., Rega N., Millam N. J., Klene M., Knox J. E., Cross J. B., Bakken V., Adamo C., Jaramillo J., Gomperts R., Stratmann R. E., Yazyev O., Austin A. J., Cammi R., Pomelli C., Ochterski J. W., Martin R. L., Morokuma K., Zakrzewski V. G., Voth G. A., Salvador P., Dannenberg J. J., Dapprich S., Daniels A. D., Farkas Ö., Foresman J. B., Ortiz J. V., Cioslowski J., Fox D. J.. Gaussian 09. Wallingford, CT, USA: Gaussian, Inc., 2009 [Google Scholar]
- 57. Frisch M. J., Pople J A., Self-consistent molecular orbital methods 25. Supplementary functions for Gaussian basis sets, J. Chem. Phys. 1984, 80, 3265 [Google Scholar]
- 58. Hay P. J., Wadt W R., Ab initio effective core potentials for molecular calculations. Potentials for K to Au including the outermost core orbitals, J. Chem. Phys. 1985, 82 (1), 299–310. [Google Scholar]
- 59. Yang X., Li H., Cheng T., Xia W., Lai Y. T., Sun H., Nickel translocation between metallochaperones HypA and UreE in Helicobacter pylori, Metallomics 2014, 6 (9), 1731–1736. [DOI] [PubMed] [Google Scholar]
- 60. Colpas G. J., Maroney M. J., Bagyinka C., Kumar M., Willis W. S., Suib S. L., Baidya N., Mascharak P K., X-ray spectroscopic studies of nickel complexes, with application to the structure of nickel sites in hydrogenases, Inorg. Chem. 1991, 30, 920–928. [Google Scholar]
- 61. Scott R A., X-Ray Absorption Spectroscopy, In: Que L Jr.s (ed). Physical Methods in Inorganic and Bioinorganic Chemistry. Menlo Park (CA, USA): University Science Press, 2000 [Google Scholar]
- 62. Spronk C., Zerko S., Gorka M., Kozminski W., Bardiaux B., Zambelli B., Musiani F., Piccioli M., Basak P., Blum F. C., Johnson R. C., Hu H., Merrell D. S., Maroney M., Ciurli S., Structure and dynamics of Helicobacter pylori nickel-chaperone HypA: an integrated approach using NMR spectroscopy, functional assays and computational tools, J. Biol. Inorg. Chem. 2018, 23 (8), 1309–1330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Xia W., Li H., Sze K.-.H., Sun H., Structure of a nickel chaperone, HypA, from Helicobacter pylori reveals two distinct metal binding sites, J. Am. Chem. Soc. 2009, 131 (29), 10031–10040. [DOI] [PubMed] [Google Scholar]
- 64. Salgado J., Kalverda A. P., Diederix R. E. M., Canters G. W., Moratal J.-.M., Lawler A. T., Dennison C., Paramagnetic NMR investigations of Co(II) and Ni(II) amicyanin, J. Biol. Inorg. Chem. 1999, 4, 457–467. [DOI] [PubMed] [Google Scholar]
- 65. Banci L., Piccioli M., Cobalt(II)- and Nickel(II)-SubstitutedProteins, eMarRes 2007. [Google Scholar]
- 66. Goodfellow B. J., Duarte I. C., Macedo A. L., Volkman B. F., Nunes S. G., Moura I., Markley J. L., Moura J J., An NMR structural study of nickel-substituted rubredoxin, J. Biol. Inorg. Chem. 2010, 15 (3), 409–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Ciurli S., Safarov N., Miletti S., Dikiy A., Christensen S. K., Kornetzky K., Bryant D. A., Vandenberghe I., Devreese B., Samyn B., Remaut H., Van Beeumen J., Molecular characterization of Bacillus pasteurii UreE, a metal-binding chaperone for the assembly of the urease active site, J. Biol. Inorg. Chem. 2002, 7 (6), 623–631. [DOI] [PubMed] [Google Scholar]
- 68. Pereira J., Simpkin A. J., Hartmann M. D., Rigden D. J., Keegan R. M., Lupas A N., High-accuracy protein structure prediction in CASP14, Proteins 2021, 89 (12), 1687–1699. [DOI] [PubMed] [Google Scholar]
- 69. Camporesi G., Minzoni A., Morasso L., Ciurli S., Musiani F., Nickel import and export in the human pathogen Helicobacter pylori, perspectives from molecular modelling, Metallomics 2021, 13 (12). [DOI] [PubMed] [Google Scholar]
- 70. Mazzei L., Musiani F., Urease C S.. Zamble D, Rowińska-Żyrek M, Kozłowski Hs (eds). The Biological Chemistry of Nickel. London, UK: Royal Society of Chemistry, 2017, 60–97. [Google Scholar]
- 71. Yang X., Li H., Lai T. P., Sun H., UreE-UreG complex facilitates nickel transfer and preactivates GTPase of UreG in Helicobacter pylori, J. Biol. Chem. 2015, 290, 12474–12485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Soriano A., Hausinger R P., GTP-dependent activation of urease apoprotein in complex with the UreD, UreF, and UreG accessory proteins, Proc Natl Acad Sci, USA 1999, 96 (20), 11140–11144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Fong Y. H., Wong H. C., Yuen M. H., Lau P. H., Chen Y. W., Wong K B., Structure of UreG/UreF/UreH complex reveals how urease accessory proteins facilitate maturation of Helicobacter pylori urease, PLoS Biol. 2013, 11 (10), e1001678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Denic M., Turlin E., Michel V., Fischer F., Khorasani-Motlagh M., Zamble D., Vinella D., de Reuse H., A novel mode of control of nickel uptake by a multifunctional metallochaperone, PLoS Pathog. 2021, 17 (1), e1009193. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data and models are available upon request to the authors.