Abstract
Mutations in the human CSB gene cause Cockayne syndrome (CS). In addition to increased photosensitivity, CS patients suffer from severe developmental abnormalities, including growth retardation and mental retardation. Whereas a deficiency in the preferential repair of UV lesions from the transcribed strand accounts for the increased photosensitivity of CS patients, the reason for developmental defects in these individuals has remained unclear. Here we provide in vivo evidence for a role of RAD26, the counterpart of the CSB gene in Saccharomyces cerevisiae, in transcription elongation by RNA polymerase II, and in addition we show that under conditions requiring rapid synthesis of new mRNAs, growth is considerably reduced in cells lacking RAD26. These findings implicate a role for CSB in transcription elongation, and they strongly suggest that impaired transcription elongation is the underlying cause of the developmental problems in CS patients.
Cockayne syndrome (CS) in humans is characterized by severe growth retardation that has the outward appearance of cachetic dwarfism, and CS patients suffer from progressive neurologic dysfunction and mental retardation. CS individuals also exhibit mild sun sensitivity, but they do not suffer from the increased incidence of skin cancers so prevalent in xeroderma pigmentosum patients. The mean age of death in CS patients is ∼12 years (13). Mutations in two human genes, CSA and CSB, account for over 90% of CS cases (8). CS cells are impaired in their ability to perform preferential repair of DNA lesions from the transcribed strand (21), a phenomenon known as transcription-coupled repair (TCR) (11). Although the defect in preferential repair of UV lesions from the transcribed strand explains the photosensitivity of CS patients, it fails to account for the characteristic growth and neurological defects associated with CS.
RAD26 is the CSB counterpart in Saccharomyces cerevisiae, and inactivation of this gene causes a defect in the TCR of UV-damaged DNA (20). The proteins encoded by the RAD26 and CSB genes are members of the SWI2/SNF2 family of ATPases, and both proteins have DNA-dependent ATPase activities (6, 17). Interestingly, in vitro studies with the purified human CSB protein have suggested a role for CSB as an RNA polymerase II (Pol II) elongation factor (16). Here we utilize S. cerevisiae as a model to investigate the role of RAD26 in transcription elongation in vivo and to examine the possibility that the clinical features of CS patients derive from defects in transcription elongation.
Elongation factor SII enables Pol II to transcribe through intrinsic arrest sites in DNA. SII binds arrested Pol II and activates the cleavage of nascent transcript by a latent endoribonuclease intrinsic to Pol II, which eventually results in the clearance of the impediment (15). In S. cerevisiae, DST1, the gene encoding SII, is not essential for viability; however, the dst1Δ mutant exhibits enhanced sensitivity to the base analog 6-azauracil (6AU) (12), which depletes cellular levels of the RNA precursors GTP and UTP (5). Because of the decrease in nucleoside triphosphate concentrations in 6AU-treated cells, the elongation rate of Pol II is lowered and it suffers more arrest. SII releases Pol II from the arrested state and enables it to resume elongation. Yeast cells that lack SII and, additionally, that harbor a conditional mutation in RPB2, the gene encoding the second largest subunit of Pol II and which confers a 6AU-sensitive phenotype, also manifest an elongation defect in biochemical assays in vitro (14) and exhibit reduced levels of specific mRNAs following 6AU treatment (9). Thus, efficient mRNA synthesis in vivo is dependent on an optimally functioning elongation machinery.
Since CS patients are viable and the rad26Δ mutant displays no growth defects under normal conditions, inactivation of RAD26 in yeast or of CSB in humans may confer only a subtle defect in transcription. To be able to discern any such changes in the rate of transcription, we combined the rad26Δ mutation with the elongation-defective dst1Δ mutation. The underlying assumption here was that if Rad26 functions independently of SII in transcription elongation, then a more severe phenotype would result upon the simultaneous loss of both proteins. Also, we examined the mRNA levels of genes that were in a state of high transcriptional activity because any transcriptional deficiency may then become more apparent. Here we provide evidence for a role of RAD26 in the elongation phase of Pol II transcription in vivo and show that, in the absence of RAD26, growth impairment results under conditions requiring new mRNA synthesis.
MATERIALS AND METHODS
Yeast strains.
In this study, the wild-type strain EMY73 (MATa his3-Δ1 leu2-3,-112 trp1Δ) and its isogenic derivative strains YR26.1, YR26.7, and YRP127 carrying the rad26Δ, rad26Δ dst1Δ, and dst1Δ mutations, respectively, were used. In the rad26Δ mutant, amino acids 21 to 1009 of the 1,085-amino-acid RAD26-encoded protein are deleted, and in the dst1Δ mutant, amino acids 33 to 226 of the 309-amino-acid DST1-encoded protein are deleted.
Transcription analyses.
For the examination of GAL7 and GAL10 transcription, cells were grown at 30°C in YPL (1% yeast extract, 2% peptone, 3.7% lactate) medium to saturation. The cells were diluted in the identical fresh medium to an optical density at 600 nm (OD600) of 0.5 with a final concentration of 2% galactose. Samples were removed at the time points indicated in Fig. 1 after being transferred to galactose-containing medium. Cells were pelleted and quickly frozen in crushed dry ice. Frozen cells were maintained at −80°C until RNA isolation.
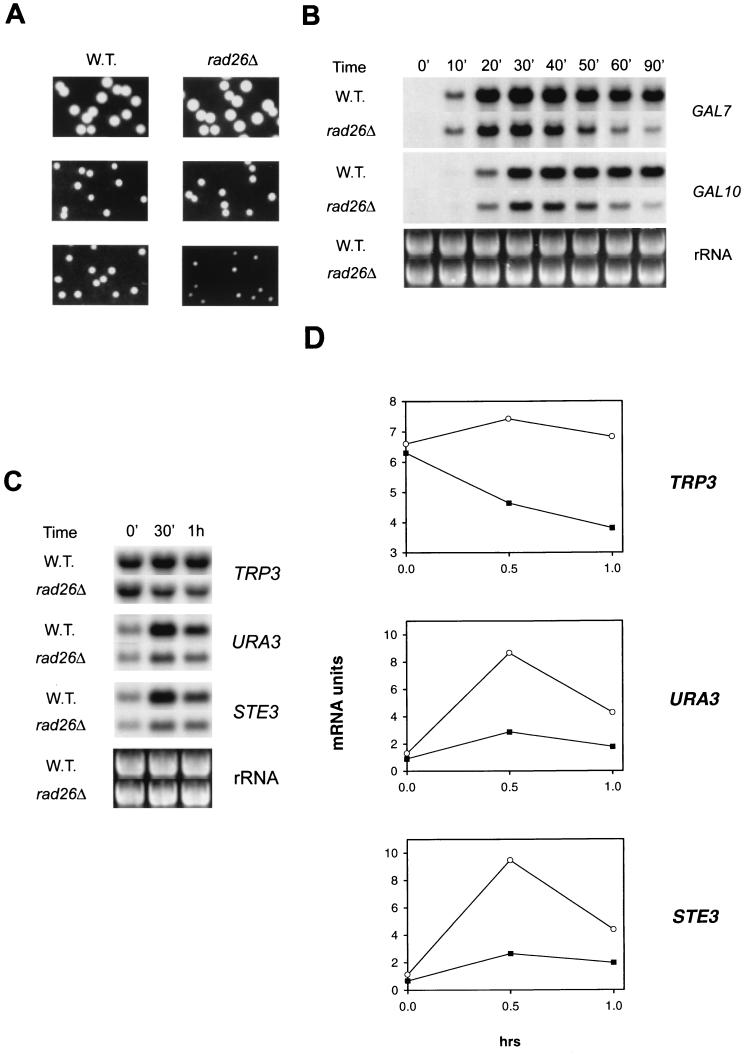
FIG. 1.
Transcription of GAL7, GAL10, and PHO5 genes in wild-type (W.T.) and rad26Δ, dst1Δ, and rad26Δ dst1Δ mutant strains. Total RNAs from cells grown in YPL medium containing galactose were subjected to Northern analysis. (A) Transcript levels of GAL7 (top left panel), GAL10 (middle left panel), and PHO5 (top right panel) genes. The ethidium bromide-stained gel shown in the bottom right panel indicates the levels of RNAs loaded for PHO5, and that which is shown in the bottom left panel indicates the levels of RNAs loaded for the GAL genes. mRNA levels were examined at the indicated times after cells were transferred to galactose-containing medium or to low-phosphate medium. (B) Quantitation of GAL7, GAL10, and PHO5 mRNA levels. mRNA units at each time point are relative to the highest mRNA level in the wild-type strain. Symbols: ●, wild type; ■, rad26Δ; ▵, dst1Δ; □, rad26Δ dst1Δ.
To examine GAL gene transcription in the presence of 6AU, cells were grown to log phase at 30°C in synthetic complete (SC) medium containing 3% glycerol–2% lactate and lacking uracil. Cells were harvested by centrifugation and resuspended in the identical fresh medium with 100 μg of 6AU/ml. Following incubation in 6AU for 2 h at 30°C, galactose was added to reach a final concentration of 2%. Samples were removed at the time points indicated in Fig. 3B after the addition of galactose. Cells were pelleted and quickly frozen in crushed dry ice. Frozen cells were maintained at −80°C until RNA isolation.
FIG. 3.
Inhibition of growth and transcription in 6AU-treated rad26Δ cells. W. T., wild type. (A) Growth of wild-type and rad26Δ mutant cells in the absence or presence of 6AU. Wild-type and rad26Δ mutant cells were grown on YPD plates in the absence of 6AU (top panels), on SC medium lacking uracil and in the absence of 6AU (middle panels), or on SC medium lacking uracil but containing 50 μg of 6AU/ml (bottom panels). (B) Transcription of GAL7 and GAL10 genes in the presence of 6AU. Cells grown to log phase at 30°C in SC medium containing 3% glycerol and 2% lactate and lacking uracil were harvested and resuspended in the identical fresh medium with 100 μg of 6AU/ml. After incubation for 2 h at 30°C, galactose was added to reach a final concentration of 2%. Samples were removed at the indicated time points after the addition of galactose, and GAL7 (top) and GAL10 (middle) mRNA levels were examined. The ethidium bromide-stained gel shown in the bottom panel indicates the levels of RNAs loaded. The time points indicate the period in minutes after the addition of galactose to the medium. (C) mRNA levels of TRP3, URA3, and STE3 in wild-type and rad26Δ mutant cells treated with 6AU. Cells grown to log phase at 30°C in SC medium lacking uracil were diluted in identical fresh medium to an OD600 of 0.5, and 6AU was added to reach a final concentration of 75 μg/ml. Samples were removed at the indicated time points after the addition of 6AU. The ethidium bromide-stained gel shown in the bottom panel indicates the levels of RNAs loaded. (D) Quantitation of TRP3, URA3, and STE3 mRNAs in the presence of 6AU. mRNA units at each time point are relative to the highest mRNA level in the wild-type strain. Symbols: ○, wild type; ■, rad26Δ mutant.
For the examination of transcription of the PHO5 gene, YP (1% yeast extract, 2% peptone) medium containing a high or low concentration of phosphate (Pi) was prepared as described previously (7). Cells were grown to log phase at 30°C in YP–high-Pi medium. Cells were harvested by centrifugation, washed twice with distilled water, and diluted in YP–low-Pi medium to an OD600 of 0.5. Samples were removed at the indicated time after being transferred to YP–low-Pi medium. Cells were pelleted and quickly frozen in crushed dry ice. Frozen cells were maintained at −80°C until RNA isolation.
Total RNA was isolated using the hot phenol method (1a) and fractionated by electrophoresis on 1.4% agarose–6% formaldehyde gels. RNA was transferred to Hybond nylon membranes (Amersham). Each DNA probe was 32P labeled by the Multiprime DNA-labeling system (Amersham). Hybridization was performed at 42°C in 40% formamide–5% dextran sulfate–1% sodium dodecyl sulfate (SDS)–5× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate)–5× Denhardt's solution–1 M KPO4 containing 100 μg of denatured herring sperm DNA/ml. The blots were washed twice with 2× SSC–0.1% SDS for 5 and 10 min at room temperature, once with 0.5× SSC–0.1% SDS for 30 min at 50°C, and once with 0.1× SSC–0.1% SDS for 15 min at 50°C. Quantitation of mRNA levels was performed in a PhosphorImager using ImageQuant software.
Determination of growth.
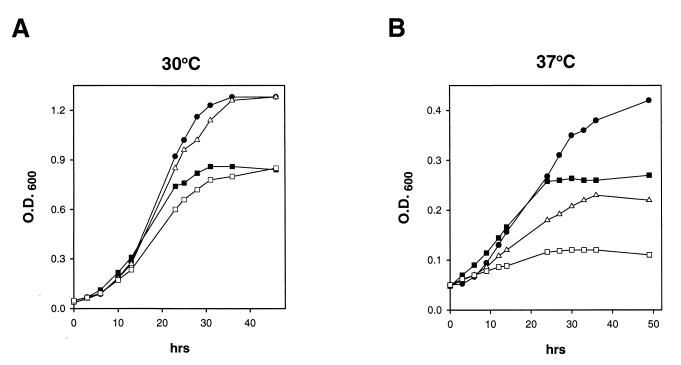
Cells were grown on YPD (1% yeast extract, 2% peptone, 2% dextrose) plates and diluted in YPL medium to an OD600 of 0.05. The cultures were then split, and half of the culture was left at 30°C while the other half was shifted to 37°C. Cell density (OD600) was determined at the time points indicated in Fig. 2.
FIG. 2.
Growth of wild-type (●) and rad26Δ (■), dst1Δ (▵), and rad26Δ dst1Δ (□) mutant strains. Cells were grown on YPD plates and diluted in YPL medium to an OD600 of 0.05. Half of the culture was left at 30°C (A), the other half was shifted to 37°C (B), and cell density (OD600) was determined at the indicated time points.
RESULTS
Effect of rad26Δ mutation on inducible synthesis of mRNAs.
To investigate the role of RAD26 in transcription, we examined the inducible synthesis of GAL7 and GAL10 mRNAs and of PHO5 mRNA in the wild-type and rad26Δ, dst1Δ, and rad26Δ dst1Δ mutant strains. Transcription of these GAL genes, which is induced upon the addition of galactose, was lowered in the rad26Δ mutant (Fig. 1). Transcription was also affected in the dst1Δ strain but to a lesser degree than in the rad26Δ strain. The most severe reduction in transcription occurred when the rad26Δ mutation was combined with the dst1Δ mutation. Transcription of the PHO5 gene, which is induced when phosphate is limiting in the medium, was also reduced in the rad26Δ strain. For PHO5, transcription impairment was greater in the dst1Δ strain than in the rad26Δ strain, and for this gene also, mRNA levels became more severely depressed in the rad26Δ dst1Δ double mutant strain than in the rad26Δ or dst1Δ single mutant strain (Fig. 1). For example, the levels of PHO5 transcripts at 5 h were reduced to 85, 70, and 34% in the rad26Δ, dst1Δ, and rad26Δ dst1Δ strains, respectively, compared to that in the wild-type strain, indicating that a synergistic decline in PHO5 transcription occurs in the double mutant (Fig. 1B). In summary, for all three genes examined, deletion of RAD26 reduces the levels of their encoded mRNAs and a more severe reduction in transcription occurs in the absence of both SII and Rad26 proteins.
Synergistic enhancement of growth defects in the rad26Δ dst1Δ double mutant.
Although the rad26Δ mutation has no discernible effect on growth in rich medium (YPD or SC medium), the requirement of RAD26 for efficient transcription suggested that growth impairment might become more apparent under conditions requiring rapid synthesis of new mRNAs. To investigate such a possibility, the wild-type and rad26Δ, dst1Δ, and rad26Δ dst1Δ mutant strains were transferred from glucose to lactate medium and grown at 30 and 37°C. At 30°C, growth was not significantly affected by the dst1Δ mutation but the rad26Δ strain grew at a lower rate than did the wild-type or the dst1Δ strain, and a slight further decrease in growth was noted in the rad26Δ dst1Δ strain (Fig. 2A). The growth defects, however, became much more striking at 37°C, presumably because under these conditions, in addition to the change in the carbon source, cells have to adapt to the exigencies of high temperature. At 37°C, growth was retarded in both the rad26Δ and dst1Δ strains but the dst1Δ mutation had a more pronounced debilitating effect on growth than did the rad26Δ mutation and the rad26Δ dst1Δ strain barely grew (Fig. 2B). A comparison of OD600s at 50 h shows that cell density was reduced to 65, 45, and 22% in the rad26Δ, dst1Δ, and rad26Δ dst1Δ strains, respectively, compared to that in the wild-type strain (Fig. 2B). Thus, a synergistic decline in cell density occurs in the rad26Δ dst1Δ double mutant strain. The extreme growth defect of the rad26Δ dst1Δ double mutant most likely stems from the more severe transcriptional defect in the absence of both the Rad26 and SII proteins.
Reduced mRNA levels in 6AU-treated rad26Δ mutant cells.
Since nucleotide depletion occurring in the presence of 6AU affects the elongation efficiency of Pol II, sensitivity to 6AU has been exploited as a means for identifying elongation factors. Thus, mutations in SII or in the RPB2 subunit of Pol II, defective in transcription elongation, yield a 6AU-sensitive phenotype (9). In the presence of 6AU, growth is impaired in the rad26Δ strain (Fig. 3A). Also, the transcriptional defect of the rad26Δ strain was further enhanced upon 6AU treatment, as the levels of GAL7 and GAL10 mRNAs suffered more drastic reductions in 6AU-treated rad26Δ cells (Fig. 3B) than in untreated cells (Fig. 1). In Table 1, we show the levels of GAL7 and GAL10 mRNAs in 6AU-treated and untreated rad26Δ cells relative to those in the wild-type strain. While transcription is affected in untreated rad26Δ cells, the transcriptional defect becomes more pronounced in 6AU-treated rad26Δ cells. For example, in the 60-min samples, GAL7 and GAL10 mRNA levels in untreated rad26Δ cells were about 70% of the levels in the wild-type strain, whereas in 6AU-treated rad26Δ cells, these mRNA levels were reduced to about 30% of the levels in wild-type cells.
TABLE 1.
Levels of GAL7 and GAL10 mRNAs in the rad26Δ strain
| Time (min)a | mRNA levelb
|
|||
|---|---|---|---|---|
|
GAL7
|
GAL10
|
|||
| −6AU | +6AU | −6AU | +6AU | |
| 0 | ||||
| 10 | 86 | 87 | ||
| 20 | 68 | 64 | 71 | 80 |
| 30 | 68 | 64 | 56 | 67 |
| 40 | 67 | 53 | 53 | 51 |
| 50 | 73 | 45 | 60 | 44 |
| 60 | 74 | 30 | 66 | 28 |
| 90 | 11 | 13 | ||
Time point of sample removal.
mRNA levels of GAL7 and GAL10 in the rad26Δ strain are given relative to the mRNA level in the wild type times 100. They were measured in the presence (+) or absence (−) of 6AU.
Following 6AU treatment, steady-state levels of mRNAs, such as those made by genes involved in amino acid biosynthesis, declined in the elongation-defective dst1Δ rpb2-10 double mutant, while the wild-type cells maintained near-normal levels of these mRNAs (9). In wild-type yeast cells treated with 6AU for up to 1 h, steady-state levels of TRP3 mRNA also remained unchanged, whereas in the similarly treated rad26Δ cells, these mRNA levels were reduced to approximately 50% of wild-type levels (Fig. 3C and D). 6AU reduces intracellular GTP and UTP levels by inhibiting the enzymes IMP dehydrogenase and orotidylate decarboxylase, respectively. Upon 6AU treatment, wild-type yeast cells induce transcription of PUR5, a gene that encodes IMP dehydrogenase. The capacity to induce PUR5 transcription, however, is greatly reduced in yeast cells deficient in transcription elongation machinery, such as those harboring the dst1Δ mutation or the rpb2-10 mutation (18). These and other results have suggested that yeast cells respond to nucleotide depletion by transcriptional induction of genes involved in nucleotide biosynthesis and that this induction is somehow coupled to efficient elongation (18). The levels of mRNAs made by the URA3 gene, which encodes orotidylate decarboxylase, also increase in wild-type cells treated with 6AU for 30 min; by contrast, only a slight increase was evident in rad26Δ mutant cells treated similarly (Fig. 3C and D). Also, the mRNA levels of the STE3 gene increased almost 10-fold in wild-type cells treated with 6AU for 30 min, wherease there was little increase in rad26Δ mutant cells (Fig. 3C and D).
DISCUSSION
Here we show that transcription of the three inducible genes examined, GAL7, GAL10, and PHO5, is reduced in the absence of Rad26 or SII protein and that this transcriptional defect becomes more severe in cells lacking both of these proteins. Moreover, the rad26Δ mutation confers sensitivity to 6AU, a sensitive indicator of transcription elongation defect, and compared to the wild-type strain, GAL7 and GAL10 mRNA levels were more severely depressed in 6AU-treated rad26Δ cells than in untreated cells. The steady-state levels of TRP3, URA3, and STE3 mRNAs were also much lower in 6AU-treated rad26Δ cells than in similarly treated wild-type cells. Additionally, under conditions requiring new mRNA synthesis, growth is affected in cells lacking Rad26 or SII and a further inhibition of growth occurs in the absence of both Rad26 and SII. Together, these observations implicate a role for RAD26 in transcription elongation and suggest that Rad26 and SII contribute independently to this process.
Because of the lack of isogenicity of CSB-deficient cell lines, studies such as those reported here would be difficult to conduct with humans, since any effect on transcription may be due to differences in the genetic backgrounds of different cell lines. Also, in view of the fact that inactivation of CSB is not lethal, we expect the effect of the CSB protein on transcription elongation to be subtle and possibly limited to particular genes. Therefore, it is not surprising that studies of transcription in CSB-deficient cells have yielded conflicting results. Thus, while in one study, Pol II transcription was reported to be lower in some CSB-deficient cell lines than it was in normal cells (2), in another study, there was no difference in the levels of transcription supported by extracts of normal, CSA, or CSB cells when undamaged DNA was used as the template (4). In another study, microinjection of antibodies against the CSA or CSB proteins also had no effect on the overall level of transcription in unirradiated cultured fibroblasts (19).
In addition to increased photosensitivity, individuals suffering from CS exhibit developmental defects such as severe growth retardation, mental retardation, neurodysmyelination, and skeletal and retinal abnormalities (13). The high degree of conservation of Rad26 and CSB proteins strongly suggests that the two proteins act similarly in S. cerevisiae and humans, respectively. Thus, from our observations with RAD26 in S. cerevisiae, we deduce a role for CSB in Pol II-dependent transcription elongation and suggest that the various developmental defects in CSB-deficient individuals accrue from defects in transcription. In humans, p53 levels rise in response to transcription inhibition and cell death is associated with prolonged induction of p53 (1). p53-dependent apoptosis resulting from deficiencies in transcription elongation would further contribute to developmental defects in CS.
Recently, based upon the findings that the TCR of oxidative lesions 8-oxoguanine and thymine glycol is defective in CS cells, failure to repair oxidative damage from the transcribed strand was suggested to be the basis of developmental defects in CS (3, 10). Our results showing the involvement of the RAD26 gene in Pol II transcription elongation in the absence of any exogenous DNA damage and the finding that, under conditions requiring new mRNA synthesis, growth impairment occurs in the absence of RAD26 imply, however, that developmental defects in CS patients arise from defects in transcription elongation and not from faulty DNA repair.
Although the in vivo evidence for a role of CSB in the elongation step of Pol II transcription is lacking, purified CSB protein stimulates the rate of elongation by Pol II on oligo(dC)-tailed DNA templates in the absence of additional transcription factors (16). These in vitro biochemical studies with the CSB protein support in vivo studies with the RAD26 gene in S. cerevisiae and they suggest a direct role for Rad26 and CSB proteins in transcriptional elongation by Pol II. The DNA-dependent ATPase activity of these proteins (6, 17) may promote passage of Pol II through a variety of transcriptional impediments, including intrinsic arrest sites in DNA.
Loss of CSB in humans induces metaphase fragility of four loci, RNU1, PSU1, RNU2, and RN5S, that are highly transcriptionally active (22). RNU1 and RNU2 contain tandemly repeated U1 and U2 snRNA genes, respectively, PSU1 contains U1 pseudogenes, and RN5S contains tandemly repeated 5S rRNA genes. U1 and U2 snRNAs are transcribed by Pol II, and 5S rRNA is transcribed by Pol III. Transcription of highly structured RNAs, like U1 and U2 snRNA and 5S rRNA, might require the activity of elongation factors like CSB. In the absence of CSB, the presence of stalled RNA polymerases on DNA may block chromatin condensation, causing localized chromosome fragility at metaphase (22). Since the absence of CSB causes fragility of genes transcribed by Pol II and Pol III, CSB may function as an elongation factor for Pol III as well.
ACKNOWLEDGMENT
This work was supported by National Institutes of Health grant CA35035.
REFERENCES
- 1.Andrea L, Wasylyk B. Transcription abnormalities potentiate apoptosis of normal human fibroblasts. Mol Med. 1997;3:852–863. [PMC free article] [PubMed] [Google Scholar]
- 1a.Ausubel F M, editor. Current protocols in molecular biology. 2, unit 13.12. New York, N.Y: Wiley; 1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Balajee A S, May A, Dianov G L, Friedberg E C, Bohr V A. Reduced RNA polymerase II transcription in intact and permeabilized Cockayne syndrome group B cells. Proc Natl Acad Sci USA. 1997;94:4306–4311. doi: 10.1073/pnas.94.9.4306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cooper P K, Nouspikel T, Clarkson S G, Leadon S A. Defective transcription-coupled repair of oxidative base damage in Cockayne syndrome patients from XP group G. Science. 1997;275:990–993. doi: 10.1126/science.275.5302.990. [DOI] [PubMed] [Google Scholar]
- 4.Dianov G L, Houle J-F, Iyer N, Bohr V A, Friedberg E C. Reduced RNA polymerase II transcription in extracts of Cockayne syndrome and xeroderma pigmentosum/Cockayne syndrome cells. Nucleic Acids Res. 1997;25:3636–3642. doi: 10.1093/nar/25.18.3636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Exinger F, Lacroute F. 6-Azauracil inhibition of GTP biosynthesis in Saccharomyces cerevisiae. Curr Genet. 1992;22:9–11. doi: 10.1007/BF00351735. [DOI] [PubMed] [Google Scholar]
- 6.Guzder S N, Habraken Y, Sung P, Prakash L, Prakash S. RAD26, the yeast homolog of human Cockayne's syndrome group B gene, encodes a DNA dependent ATPase. J Biol Chem. 1996;271:18314–18317. doi: 10.1074/jbc.271.31.18314. [DOI] [PubMed] [Google Scholar]
- 7.Han M, Kim U-J, Kayne P, Grunstein M. Depletion of histone H4 and nucleosomes activates the PHO5 gene in Saccharomyces cerevisiae. EMBO J. 1988;7:2221–2228. doi: 10.1002/j.1460-2075.1988.tb03061.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hanawalt P C. The bases for Cockayne syndrome. Nature. 2000;405:415–416. doi: 10.1038/35013197. [DOI] [PubMed] [Google Scholar]
- 9.Lennon J C, III, Wind M, Saunders L, Hock M B, Reines D. Mutations in RNA polymerase II and elongation factor SII severely reduce mRNA levels in Saccharomyces cerevisiae. Mol Cell Biol. 1998;18:5771–5779. doi: 10.1128/mcb.18.10.5771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Le Page F, Kwoh E E, Avrutskaya A, Gentil A, Leadon S A, Sarasin A, Cooper P K. Transcription-coupled repair of 8-oxoguanine: requirement for XPG, TFIIH, and CSB and implications for Cockayne syndrome. Cell. 2000;101:159–171. doi: 10.1016/s0092-8674(00)80827-2. [DOI] [PubMed] [Google Scholar]
- 11.Mellon I, Spivak G, Hanawalt P C. Selective removal of transcription-blocking DNA damage from the transcribed strand of the mammalian DHFR gene. Cell. 1987;51:241–249. doi: 10.1016/0092-8674(87)90151-6. [DOI] [PubMed] [Google Scholar]
- 12.Nakanishi T, Shimoaraiso M, Kubo T, Natori S. Structure-function relationship of yeast S-II in terms of stimulation of RNA polymerase II, arrest relief, and suppression of 6-azauracil sensitivity. J Biol Chem. 1995;270:8991–8995. doi: 10.1074/jbc.270.15.8991. [DOI] [PubMed] [Google Scholar]
- 13.Nance M A, Berry S A. Cockayne syndrome: review of 140 cases. Am J Med Gen. 1992;42:68–84. doi: 10.1002/ajmg.1320420115. [DOI] [PubMed] [Google Scholar]
- 14.Powell W, Reines D. Mutations in the second largest subunit of RNA polymerase II cause 6-azauracil sensitivity in yeast and increased transcriptional arrest in vitro. J Biol Chem. 1996;271:6866–6873. doi: 10.1074/jbc.271.12.6866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Reines D. Nascent RNA cleavage by transcription elongation complexes. In: Conaway R C, Conaway J W, editors. Transcription: mechanisms and regulation. New York, N.Y: Raven Press, Ltd.; 1994. pp. 263–278. [Google Scholar]
- 16.Selby C P, Sancar A. Cockayne syndrome group B protein enhances elongation by RNA polymerase II. Proc Natl Acad Sci USA. 1997;94:11205–11209. doi: 10.1073/pnas.94.21.11205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Selby C P, Sancar A. Human transcription-repair coupling factor CSB/ERCC6 is a DNA-stimulated ATPase but is not a helicase and does not disrupt the ternary transcription complex of stalled RNA polymerase II. J Biol Chem. 1997;272:1885–1890. doi: 10.1074/jbc.272.3.1885. [DOI] [PubMed] [Google Scholar]
- 18.Shaw R J, Reines D. Saccharomyces cerevisiae transcription elongation mutants are defective in PUR5 induction in response to nucleotide depletion. Mol Cell Biol. 2000;20:7427–7437. doi: 10.1128/mcb.20.20.7427-7437.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.van Gool A J, Citterio E, Rademakers S, van Os R, Vermeulen W, Constantinou A, Egly J-M, Bootsma D, Hoeijmakers J H J. The Cockayne syndrome B protein, involved in transcription-coupled DNA repair, resides in an RNA polymerase II-containing complex. EMBO J. 1997;16:5955–5965. doi: 10.1093/emboj/16.19.5955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.van Gool A J, Verhage R, Swagemakers S M A, van de Putte P, Brouwer J, Troelstra C, Bootsma D, Hoeijmakers J H J. RAD26, the functional S. cerevisiae homolog of the Cockayne syndrome B gene ERCC6. EMBO J. 1994;13:5361–5369. doi: 10.1002/j.1460-2075.1994.tb06871.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.van Hoffen A, Natarajan A T, Mayne L V, van Zeeland A A, Mullenders L H F, Venema J. Deficient repair of the transcribed strand of active genes in Cockayne's syndrome cells. Nucleic Acids Res. 1993;21:5890–5895. doi: 10.1093/nar/21.25.5890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yu A, Fan H-Y, Liao D, Bailey A D, Weiner A M. Activation of p53 or loss of the Cockayne syndrome group B repair protein causes metaphase fragility of human U1, U2, and 5S genes. Mol Cell. 2000;5:801–810. doi: 10.1016/s1097-2765(00)80320-2. [DOI] [PubMed] [Google Scholar]