Abstract
Multi-subunit RNA Polymerases (msRNAPs) are responsible for transcription in all kingdoms of life. At the heart of these msRNAPs is an ultra-conserved active site domain, the trigger loop (TL), coordinating transcription speed and fidelity by critical conformational changes impacting multiple steps in substrate selection, catalysis, and translocation. Previous studies have observed several different types of genetic interactions between eukaryotic RNA polymerase II (Pol II) TL residues, suggesting that the TL’s function is shaped by functional interactions of residues within and around the TL. The extent of these interaction networks and how they control msRNAP function and evolution remain to be determined. Here we have dissected the Pol II TL interaction landscape by deep mutational scanning in Saccharomyces cerevisiae Pol II. Through analysis of over 15000 alleles, representing all single mutants, a rationally designed subset of double mutants, and evolutionarily observed TL haplotypes, we identify interaction networks controlling TL function. Substituting residues creates allele-specific networks and propagates epistatic effects across the Pol II active site. Furthermore, the interaction landscape further distinguishes alleles with similar growth phenotypes, suggesting increased resolution over the previously reported single mutant phenotypic landscape. Finally, co-evolutionary analyses reveal groups of co-evolving residues across Pol II converge onto the active site, where evolutionary constraints interface with pervasive epistasis. Our studies provide a powerful system to understand the plasticity of RNA polymerase mechanism and evolution, and provide the first example of pervasive epistatic landscape in a highly conserved and constrained domain within an essential enzyme.
INTRODUCTION
Transcription from cellular genomes is carried out by conserved multi-subunit RNA polymerases (msRNAPs) (1–3). Bacteria and Archaea use a single msRNAP to transcribe all genomic RNAs (4–6), while Eukaryotes have at least three msRNAPs (Pol I, II, and III) for different types of RNAs (7–10). RNA synthesis by msRNAPs occurs by iterative nucleotide addition cycles (NAC) of nucleotide selection, catalysis and polymerase translocation (11–15). msRNAPs active sites accomplish all three steps in the NAC using two conformationally flexible domains termed the bridge helix (BH) and the trigger loop (TL) (5, 12, 16–19). The multi-functional natures of the BH and TL likely underlie their striking conservation, serving as interesting models for studying the function and evolution of extremely constrained protein domains.
Nearly all catalytic cycle events are associated with the concerted conformational changes in the TL, and potentially the BH (12–14, 19–23).The BH is a straight helix in most msRNAP structures (7, 8, 12, 13, 24), but was found to be kinked in the Thermus thermophilus (Bacteria) RNAP structures(5, 25). The dynamics between the straight and kink conformations has been simulated and proposed to promote msRNAP translocation (16, 25–27). Even more importantly, the TL has been observed in various conformations that confer different functions. Among the observed conformations, a catalytic disfavoring “open” state facilitates translocation and a catalytic favoring “closed” conformation promotes catalysis (12, 13, 28). During each NAC, the TL nucleotide interaction region discriminates correct NTP over non-matched NTPs or dNTPs and initiates a TL conformational change from the open to the closed state (11, 20, 29, 30). The closure of the TL promotes phosphodiester bond formation (20, 31). Pyrophosphate release accompanies TL opening, which is proposed to support polymerase translocation to the next position downstream on the template DNA, allowing for the subsequent NAC (32–34)(Fig. 1A). Consistent with the model, mutations in the TL conferred diverse effects in every step of transcription (13, 17, 23, 35–39). For instance, mutations in the TL NIR impair interactions between TL and substrates, resulting in hypoactive catalysis and reduced elongation rate in vitro (Loss of function, LOF) (17, 23, 36, 37, 40, 41). Mutations in the TL hinge region and C-terminal portion appear to disrupt the inactive state of TL (open state) and shift the TL towards the active state (closed state), leading to hyperactive catalysis and increased elongation rate but impaired transcription fidelity (Gain of function, GOF) (17, 28, 42). TL conformational dynamics and functions are likely balanced by residue interactions within and around the TL (41, 43–46).
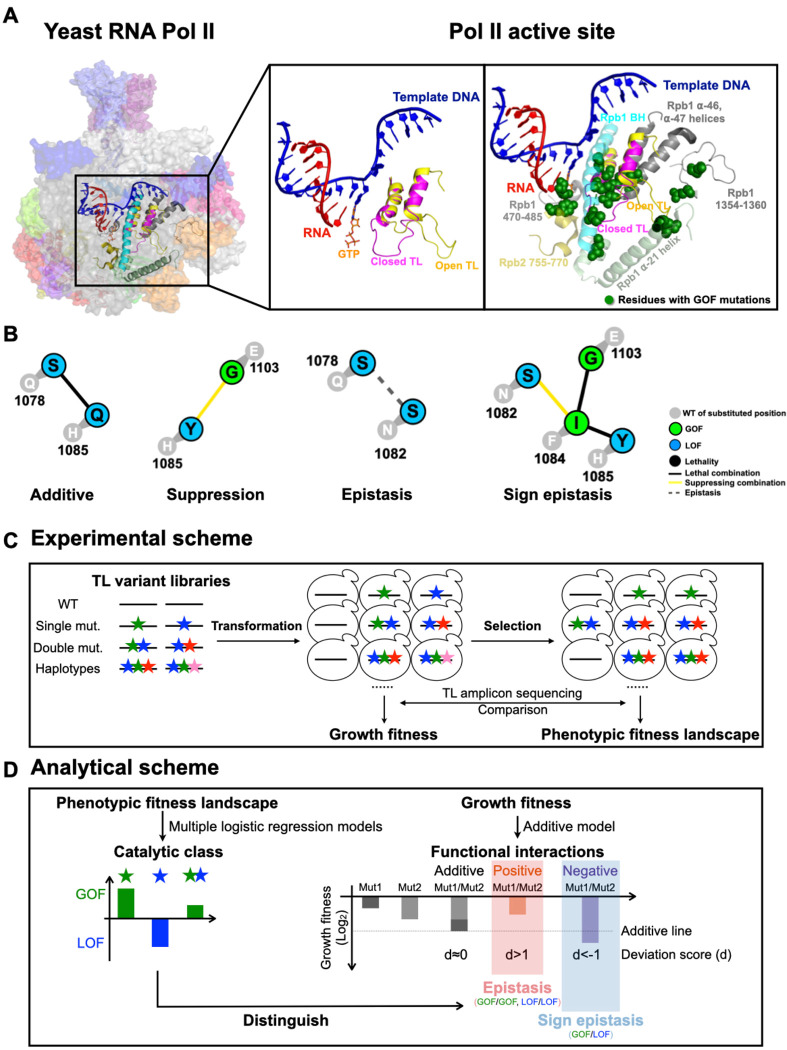
Figure 1. Schematics of the Pol II active site interaction landscape.
A. The Pol II active site is embedded in the center of a 12-subunit complex (left panel). Pol II functions are supported by distinct TL conformational states. An open TL (PDB: 5C4X)(28) and closed TL (PDB: 2E2H)(12) conformations are shown in the middle panel. GOF mutations have been identified in the TL and its proximal domains (right panel), suggesting TL mobility and function may be impacted by adjacent residues. B. Examples of inter-residue genetic interactions. WT residues are shown in grey circles with number indicating residue position in Rpb1. Mutant substitutions are shown in colored circles, with color representing mutant class. Colored lines between mutant substitutions represent types of genetic interactions. C. Overview of experimental approach. We synthesized 10 libraries of TL variants represented by colored stars. Libraries were transformed into WT or mutated yeast strains. A selection assay was subsequently performed by scraping and replating the transformants onto different media for phenotyping. DNA was extracted from yeast from all conditions, and went through TL region amplification, and Illumina sequencing. Read counts for variants on general conditions were used to determine growth fitness, while read counts on other conditions were used to determine the phenotypic fitness landscape (see Methods). D. Overview of analytical approach for determining interaction landscape. Mutant conditional growth fitnesses were calculated using allele frequencies under selective growth conditions and subjected to two logistic regression models for classification/prediction of catalytic defects. Double mutant interactions were computed using growth fitness. Classification allowed epistatic interactions to be deduced from double mutant growth fitness (see Methods).
Intramolecular interactions in the active sites of msRNAPs control catalytic activity and underpin transcriptional fidelity. The TL is embedded in the conserved active site and interacts with other domains such as the BH, α-46 and α-47 helices, which form a five helix bundle with two TL helices enclosing a hydrophobic pocket (Fig. 1A) (12, 28). Many residue interactions observed between the TL and its proximal domains are critical for proper transcription, as catalytic activity and transcription fidelity can be altered by active site mutations within the TL (described above) and domains close to the TL in many msRNAPs (examples include BH GOF T834P and LOF T834A, funnel helix α-21 GOF S713P, and Rpb2 GOF Y769F) (17, 23, 25, 26, 35–37). These mutant phenotypes suggest that TL conformational dynamics and function are finely balanced and could be sensitive to allosteric effects from proximal domains (47–50). Understanding how “connected” the TL is to the rest of the polymerase will reveal the networks that integrate its dynamics with the rest of the enzyme and pathways for how msRNAP activity and evolution might be controlled.
Physical and functional intramolecular interactions between amino acids define the protein function and evolvability (51–54). Dependence of mutant phenotypes on the identities of other amino acids (epistasis) contributes to protein evolvability by providing a physical context and an evolutionary window in which some intolerable mutations may be tolerated (55–57). Recent studies have shown that mutations can alter the protein function, allostery, and evolvability, suggesting that even conserved residues are subject to distinct epistatic constraints dependent on context (58–65). In line with this prediction, distinct phenotypes for the same conserved residues have been observed in a number of proteins, including Pol I and Pol II in yeast(63, 64, 66). For example, the yeast Pol I TL domain is incompatible when introduced into Pol II even though about 70% of residues in the two yeast TLs are identical (67). The results strongly imply even functions of ultra-conserved domains are shaped by individually evolved enzymatic contexts (higher order epistasis).
Functional interactions between residues can be revealed by genetic interactions of double mutants of interacting residues (17, 37, 68–70). Previous studies from our lab on a small subset of site-directed substitutions have identified distinct types of Pol II double mutant interactions including suppression, enhancement, epistasis, and sign-epistasis(17, 37, 69). Suppression was common between LOF and GOF mutants as expected if each mutant is individually acting in the double mutant and therefore, opposing effects on activity are balanced. Similarly, synthetic sickness and lethality were commonly observed between mutants of the same (GOF or LOF) class, consistent with the combination of mutants with partial loss of TL function having greater defects when combined. However, we have also observed lack of enhancement between mutants of similar classes (epistasis), suggesting single mutants might be functioning at the same step, and in one case, sign-epistasis, where a mutant phenotype appears dependent on the identity of a residue at another position. For example, the GOF TL substitution Rpb1 F1084I was unexpectedly lethal with the LOF TL substitution Rpb1 H1085Y (instead of the predicted mutual suppression for independently (37, 69). This was interpreted as F1084I requiring H1085 for its GOF characteristics and becoming a LOF mutant in the presence of H1085Y (Fig. 1B). How representative these interactions are, and the nature of interactions across the Pol II active site requires a more systemic analysis to fully describe and understand the networks that control Pol II activity and the requirements for each mutant phenotype.
Deducing complex residue interaction networks on a large scale is challenging. To accomplish this for Pol II, we have previously established genetic phenotypes predictive of biochemical defects (37) and coupled this with a yeast Pol II TL deep mutational scanning system (Phenotypic landscape) (17, 69, 71). Here we develop experimental and analytical schemes to extend this system to a wide range of double and multiple mutants within the S. cerevisiae Pol II TL and between the TL and adjacent domains (Interaction landscape). By analyzing 11818 alleles including single mutants and a curated subset of double mutants, we have identified intricate intra- and inter-TL residue interactions that strongly impact TL function. Additionally, the examination of 3373 haplotypes including evolutionarily observed TL alleles and co-evolved residues revealed that TL function is heavily dependent on the msRNAP context (epistasis between TL and the rest of Pol II). These results suggest that despite being highly conserved, epistasis within msRNAPs contexts functions through derived residues and potentially reshapes functions of conserved residues. Finally, statistical coupling analyses reveals putative allosteric pathways appear to converge on the TL and may modulate active site activity upon factor binding. Our analyses indicate TL function and evolution are dominated by widespread epistasis.
RESULTS
Systematic dissection of the Pol II active site interaction landscape.
We developed an experimental and analytical framework, which we term the Pol II TL interaction landscape, to dissect residue interactions that shape Pol II TL function and evolution in S. cerevisiae. We designed and synthesized 15174 variants representing all possible Pol II TL single mutants, a subset of targeted double mutants, evolutionary haplotypes and potential intermediates in ten libraries (Supplemental Table 4). This approach follows our prior analysis of the TL phenotypic landscape (17) with modifications (see Methods and Fig. S1A). Libraries were transformed, screened under diverse conditions and phenotyped by deep sequencing (Fig. 1C, Fig. S1A). Growth phenotypes of mutants are calculated as the relative allele frequency shift from a control condition and normalized to the WT under the same conditions. Biological replicates indicate high reproducibility (Fig. S1B–C). Individual libraries were min-max normalized (72) to account for scaling differences between libraries (Fig. S2A) and same mutants present among different libraries indicate high correlation of fitness determinations in each library (Fig. S2B–C).
We defined a conceptual framework for evaluating genetic interactions among TL mutations. First, we assume that independence of mutant effects would result in log additive defects. This means that predicted double mutant fitness defects should be the combination of both single mutant defects, as is standardly assumed (57, 68, 70, 73). Deviation from log additive fitness defects represents potential genetic interactions between single mutants: either less than expected (i.e. suppression) or more than expected (i.e. synthetic sickness or lethality). Second, Pol II has two classes of active site mutants (GOF and LOF) that each confer fitness defects, and we previously observed activity additive interactions, meaning suppression between mutants of different classes (GOF+LOF) or synthetic sickness/lethality between mutants of the same class (GOF+GOF or LOF+LOF) in a set of mutants. We wished to distinguish specific epistatic interactions from activity-dependent suppression or synthetic interactions with mutant catalytic defects. For the purposes of our analysis, we defined an interaction as epistasis when we observed positive deviation in mutants of the same activity class (GOF+GOF, LOF+LOF), where we would expect synthetic sickness or lethality if mutants were functioning independently. We defined an interaction as sign epistasis for situations where we observed negative interaction for combinations between the classes (GOF+LOF), where we would expect suppression if mutants were functioning independently (Fig. S3).
Finally, the Pol II active site interaction landscape is based on accurate classification of mutant classes. We have previously demonstrated that mutant growth profiles across a select set of growth conditions are predictive of in vitro measured catalytic effects (Pol II TL phenotypic landscape). We extended this analysis by training multiple logistic regression models to predict phenotypic classes. We trained two models based on 65 mutants with measured in vitro catalytic defects and their conditional growth fitness to distinguish between GOF or LOF classes. Both models worked well in classifying GOF or LOF mutants (Fig. S4A). These two models were applied to all viable mutants (fitness score > −6.5 for control growth condition) and classified the mutants into three groups, GOF, LOF and those that did not belong to either one of the two groups (“unclassified”). To visually inspect the classification results, we applied t-SNE projection and k-means clustering for all measured mutants in all growth conditions to examine clustering relationships to predictions from multiple logistic regression models. As shown in Figure S4B, we observed separated GOF and LOF clusters consistent with logistic regression classifications. With all phenotypic data, GOF and LOF mutants were further classified into different clusters, suggesting more fine-grained separation using additional phenotypes (Fig. S4C). In summary, we developed an experimental and analytical framework to dissect Pol II active site residue-residue interaction landscape in high throughput.
Widespread epistasis in the Pol II TL interaction landscape.
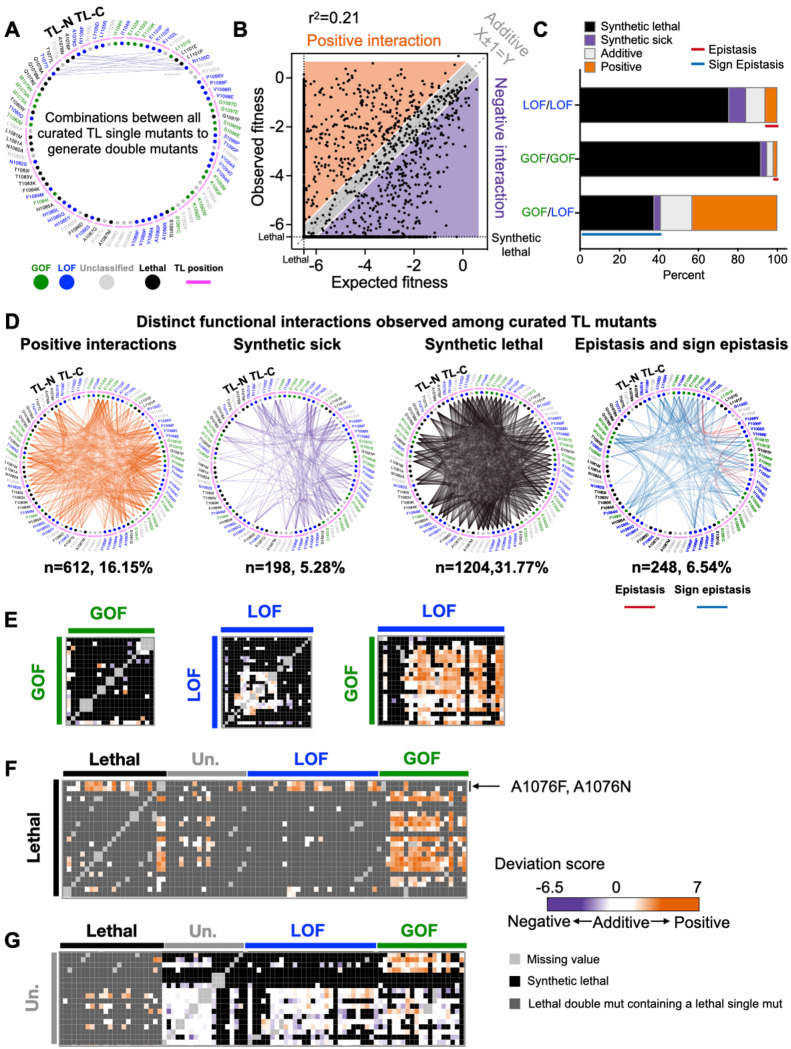
To determine the TL-internal interaction networks, we rationally selected 2–4 different substitutions for each TL residue and combined them with the selected substitutions at all other TL positions. Substitutions were chosen to represent diverse phenotypes (GOF, LOF, lethal, or unclassified mutants). This curated set of 3790 double mutants represents potential interactions between any two TL residues (Fig. 2A). We compared the observed fitness of these double mutants with expectations from the additive model, and noticed the observed double mutant fitness deviated from the predicted fitness (r2=0.21), which is much smaller than the r2 (about 0.65–0.75) reported in other studies (54, 70, 74–76) (Fig. 2B), suggesting epistasis in the ultra-conserved TL domain might be more prevalent. About half of the combinations (1776/3790) matched the additive model (observed fitness ≈ expected fitness), while the rest showed positive (observed fitness > expected fitness, n=612) or negative (observed fitness < expected fitness, n=1402) interactions (Fig. 2B). From these positive or negative interactions, we distinguished the ratio of epistasis relative to activity-additive interactions. In all GOF/LOF combinations, we observed 43% activity-additive suppression and 41% negative interactions (sign epistasis). In all GOF/GOF or LOF/LOF combinations, activity-additive synthetic sick or lethal interactions were much more common than epistasis in combinations within the same class. We observed ~2% positive (epistasis) and 95% negative (activity-additive synthetic sick or lethal) interactions in GOF/GOF combinations, and 6% positive (epistasis) and 84% negative (synthetic sick or lethal interactions) interactions in LOF/LOF combinations (Fig. 2C, Fig 2E and Fig S5). Interactions were distributed throughout the TL and covered every TL residue, supporting connectivity across the TL. Observed epistasis was concentrated within the C-terminal TL helix and adjacent regions (Fig 2D), supporting functional dependency of TL-C terminal residues and consistent with their proposed function to collaboratively stabilize the TL open state.
Figure 2. Widespread epistasis in the Pol II TL interaction landscape.
A. Design of the pairwise double mutant library. We curated 2–4 substitutions for each TL residue (in total 90 substitutions, n(GOF) = 18, n(LOF) = 30, n(Unclassified) = 19, n(Lethal) = 23), and combined them with each other to generate double mutants. 3910 double mutants representing combinations between any two TL residues were measured and 3790 of them passed the reproducibility filter. WT TL residue positions are indicated with magenta arch. Phenotype classes of single substitutions are shown as colored circles (GOF in green, LOF in blue) while unclassified mutants are in grey and lethal mutants are in black. B. An xy-plot of observed double mutant growth fitness measured in our experiment (Y-axis) and expected fitness from the addition of two constituent single mutants’ fitnesses (X-axis). N (positive) = 612. N (Negative) = 1402. N (Additive) = 1776. N (Sum)=3790. Lethal threshold (−6.5) is labeled with dotted lines on X and Y axis. The additive line where X ± 1 =Y is indicated with dashed line. Simple linear regression was performed, and the best fit equation is Y = 0.52X −2.55, r2 = 0.21, P < 0.0001. C. Percent of interactions observed from each combination group. N (LOF/LOF) = 412. N (GOF/GOF) = 156. N (GOF/LOF) = 534. Epistasis and sign epistasis are indicated with colored lines. D. Various groups of interactions are displayed in network format. E-G. The intra-TL functional interaction heatmaps of various combinations. Double mutant deviation scores are shown in the heatmap. Annotations at the top and left indicate the curated single mutants and their predicted phenotypic classes from multiple logistic regression modeling. GOF/GOF, LOF/LOF, and GOF/LOF combinations are shown in E. Combinations with lethal single substitutions are in F. Combinations with unclassified mutants are in G.
Genetic interactions reveal further insight into the nature of previously lethal or unclassified individual mutants. First, most lethal mutants could be suppressed by at least one predicted GOF mutant (Fig. 2F, Fig. S5), suggesting that most lethal mutants likely have reduced activity (LOF) below a viable threshold, as might be predicted from greater probability of any individual mutant being a LOF than a GOF. However, two lethal mutations could be suppressed by most LOF mutations or specific other lethal mutants, but not GOF mutants, implying that their lethality resulted from being GOF (select A1076 substitutions). Second, unclassified single mutants mostly did not show widespread interactions with GOF, LOF, or lethal classes. However, a few unclassified mutants showed suppression in combination with GOF mutants, suggesting potential atypical LOF not detected by phenotypic analysis, or potential sign epistasis (Fig. 2G, Fig. S5).
Allele-specific interactions suggest unique properties of individual mutants with similar phenotypes.
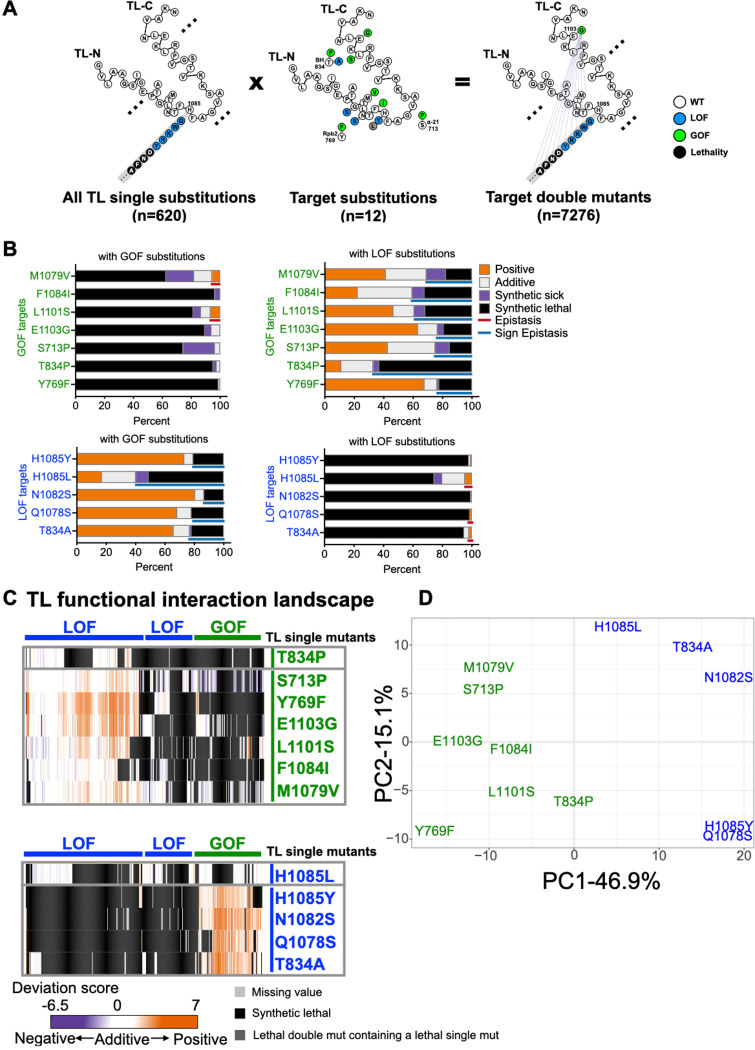
TL conformational dynamics and function are balanced by residue interactions within the TL (TL-internal interactions) and between the TL and TL-proximal domains (TL-external interactions). The properties of GOF and LOF mutants adjacent to the TL appear similar to those inside but how they behave upon TL perturbation is not known. We analyzed the scope and nature of TL-internal and TL-external interactions by exploring interaction space of 12 previously studied GOF and LOF mutants (8 within the TL and 4 outside) each combined with all possible single TL mutants (Fig. 3A). These 12 mutants function as probes for the genetic interaction space of the TL and how it might be altered in allele-specific fashion by perturbation of the “probe” mutation. TL adjacent mutants showed similar scale of widespread interactions with TL substitutions as when TL-internal mutants were used as probes (Fig. 3B, Fig. S6). For these TL adjacent substitutions, we conclude their impact on Pol II function is of similar magnitude and connection as substitutions within the TL.
Figure 3. Pol II TL interaction landscape distinguishes mutants with similar phenotypes.
A. Design of the targeted double mutant libraries. All possible substitutions at each TL residue (represented with a simplified format in the left panel) and twelve “probe” mutations (eight within the TL and four in TL-proximal domains) (middle panel) were combined with to generate 7280 double mutants (right panel). 7276 mutants passed the reproducibility filter and were used for interaction analyses. B. The percentage of functional interactions observed for each probe mutant with viable GOF or LOF TL substitutions. Epistasis and sign epistasis are labeled with colored lines. C. Pol II-TL functional interaction landscape with interactions represented by deviation scores. The upper panel shows interactions of GOF probe mutants in combination of viable GOF or LOF TL substitutions. The lower panel shows interactions of combinations with LOF probe mutants. D. Principal component analysis (PCA) of deviation scores across double mutant interactions for 12 probe mutants (see Methods).
We further compared the similarity of interaction networks for substitutions with apparently similar biochemical and phenotypic defects. These analyses were designed to detect if changes to TL function might reflect simple alteration to TL dynamics, or additional alteration to folding trajectories or conformations. In the former case, mostly additive interactions might be predicted due to TL operating in the same fashion in double mutants versus single mutants, with phenotypes deriving from differences in kinetics or distributions of existing states. In the latter case where a mutation alters TL folding trajectories or changes TL conformations, it might be predicted that individual mutants that are superficially similar will show allele-specific genetic interactions reflecting epistatic changes to TL function. A subset of probe mutants showed widespread expected activity-additive suppression between GOF/LOF mutations and activity-additive synthetic lethality between same classes of substitutions (LOF/LOF or GOF/GOF). However, allele-specific epistasis and sign epistasis were also observed and were much higher for some mutants than others (Fig. 3B–C, Fig. S6–S7). 127/620 TL substitutions showed unique interactions with specific probe mutants; for example, some lethal substitutions could only be suppressed by Y769F, a GOF TL-proximal probe mutant in Rpb2 (Fig. S8). Moreover, two TL-adjacent GOF probe mutants, Rpb1 S713P (funnel α-helix 21) and the BH allele Rpb1 T834P displayed greatly distinct interaction networks despite similarly increased activities. Rpb1 S713P exhibited widespread suppression of LOF TL substitutions (96 instances) consistent with generic enhancement of activity but preservation of TL function. In contrast, Rpb1 T834P exhibited much lower suppression ability (33 instances). In addition to much lower ability to suppress, T834P showed a much greater amount of sign epistasis than Rpb1 S713P (102 instances to 38 instances) (Fig 3C, Fig. S9A). These results are consistent with a model that perturbation to the BH structure is coupled to extensive changes to TL functional space and that T834P function as a GOF mutant requires most TL residues to be WT.
A similar distinction as above but between two internal TL GOF substitutions, Rpb1 E1103G and Rpb1 F1084I, was also apparent (Fig. S9B). Rpb1 E1103G showed widespread suppression of LOF TL substitutions (184 instances), consistent with site-directed mutagenesis studies (37) (Fig 3C, Fig. S6 and Fig S9B). These results suggest E1103G primarily may alter TL dynamics consistent with biochemical data that it promotes TL closure (23) and that it allows TL mutants primarily to maintain their effects. In contrast, Rpb1 F1084I showed more limited suppression of LOF alleles (43 instances) while showing much more widespread synthetic lethality (Fig 3C, Fig. S6 and Fig S9B). These results indicate F1084I has a much greater requirement for WT residues at many TL positions to maintain its GOF characteristics. When TL function is additionally perturbed, F1084I appears to switch from a GOF to a LOF. These results imply that individual probe mutants distinctly reshape the Pol II active site, though they might share catalytic and phenotypic defects as single mutants.
An even more striking example of this phenomenon can be observed by comparison of the interaction networks of two LOF substitutions at the exact same position, the ultra-conserved H1085 residue (Fig. S9C). This histidine contacts incoming NTP substrates (5, 12), is the target for the Pol II inhibitor α-amanitin (36), and promotes catalysis (12, 13). Initial structural data and molecular dynamics simulations were interpreted as H1085 potentially functioning as a general acid for Pol II catalysis (77–80). Our discovery that H1085L was especially well-tolerated (17), and subsequent experiments from the Landick lab (81, 82), have led to their proposal that the TL histidine functions as a positional catalyst and a similarly sized leucine supports catalysis with relatively mild effects on biochemistry and growth. If H1085Y and L substitutions are acting on a continuum of positional catalyst activity, we might predict their interaction networks would be similar and only be distinguished by magnitude of interactions, but not identity or type of interactions. In contrast to this prediction, distinct interaction patterns were observed (Fig. 3C, Fig. S6 and S9C). Most GOF mutants were able to suppress H1085Y but not H1085L. Instead, H1085L showed synthetic lethality with most GOF mutants (putative sign epistasis). For example, almost all substitutions at E1103 showed sign epistasis with H1085L but not H1085Y (Fig. S7B, S9C). Distinction between H1085L and H1085Y is evident in the PCA plot of probe mutants (Fig. 3D). The partially unique nature of each probe mutant is also evident in the PCA plot (Fig. 3D). Altogether, distinguishable interaction networks of probe mutants, despite their similarity in catalytic and growth defects, even within the same residue, suggest that each mutant has ability to propagate effects across the Pol II active site. To some extent, each Pol II mutant creates a new enzyme.
Pol II TL interaction landscape reveal functional dependency of proximal residues.
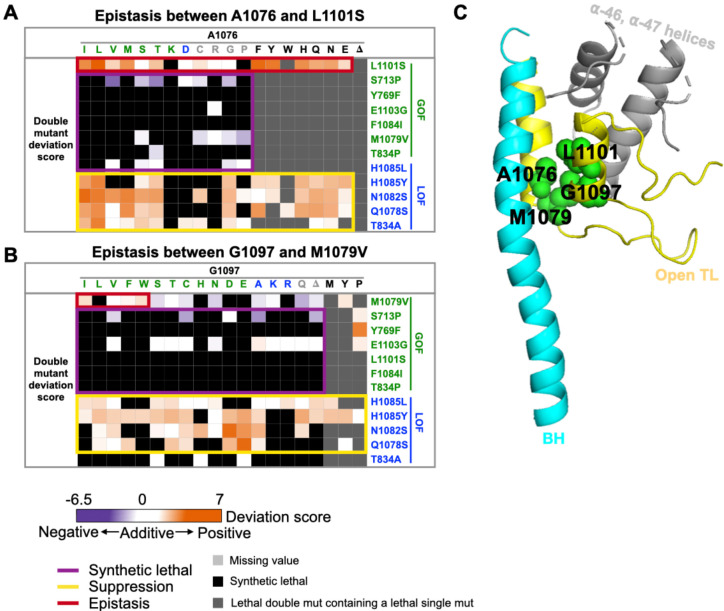
Several allele-specific epistatic interactions were also observed. Some of the strongest epistatic interactions were between A1076 substitutions and L1101S, which differed from all other GOF probe mutants (Fig. 4A), suggesting tight coupling between A1076 and L1101 for Pol II function. These two hydrophobic residues, together with other hydrophobic residues in TL proximal helices, form a five-helix bundle in the Pol II active site likely stabilizing the open TL conformation (Fig. 4C). Consistent with this, another pair adjacent residues, M1079 and G1097, also showed allele-specific epistasis (Fig. 4B).
Figure 4. Pol II TL interaction landscape reveal functional dependency of proximal residues.
A-B. Specific epistatic interactions observed between hydrophobic residues A1076 and L1101 (A), and M1079 and G1097 are shown as heatmaps (B). The x-axis of both heatmaps are 20 substitutions ordered by predicted phenotypic classes, and the color of substitution represents the phenotypic class of the substitution. GOF substitution is in green, LOF is in blue, unclassified is in gray, and lethal (fitness < −6.5) is in black. C. The epistatic interactions we identified between A1076 and L1101, together with M1079 and G1097 are shown on the five-helix bundle of Pol II active site (PDB:5C4X)(28).
The epistasis we identified in combinations within the same class (GOF/GOF or LOF/LOF) might also be sign epistasis (GOF suppressing GOF or LOF suppressing LOF due to a switch in residue class). We distinguished regular epistasis (lack of additivity) from sign epistasis suppression by checking conditional phenotypes predictive of biochemical defects. We reasoned that epistatic interactions would exhibit double mutant conditional phenotypes similar to single mutants while sign epistasis suppression would also exhibit suppression of conditional phenotypes. Therefore, we examined double mutants with our logistic regression models for determining phenotypic class. The majority of double mutants within each class showing positive epistasis (GOF/GOF or LOF/LOF) maintained single mutant classification. 6/10 GOF/GOF doubles showing positive epistasis were classified as GOF while 30/38 LOF/LOF doubles were classified as LOF, suggesting classic epistasis (Fig. S10A). In three cases of GOF/GOF combinations, all between L1101S and A1076 substitutions, the resulting double mutants were unclassified, consistent with nearly WT behavior. Here, each constituent single mutant conferred a GOF phenotype, but the double mutants show mutual suppression. This suggests tight coupling between 1101 and 1076 (see Discussion).
We also observed allele-specific interactions for predicted lethal mutants. Our threshold for lethality is likely higher than that in actuality, and very slow growing mutants may fall below our lethal threshold while still having enough data on conditional fitness assessment for logistic regression to predict mutant class. For 21 ultra sick/lethal TL substitutions predicted as GOF themselves, we observed suppression when combined with other GOF mutants (Fig. S10B–C). Lethal substitutions of A1076 could be suppressed by LOF probe mutants and the GOF probe L1101S, consistent with specific combinations between 1076 and 1101 showing sign-epistasis suppression or allele-specific mutual suppression. F1084R is a predicted lethal GOF but can be suppressed specifically by GOF probe Y769F. F1084 and Y769 are close to each other when the TL is in the closed, substrate bound state. Additionally, 5 ultra-sick/lethal substitutions predicted as LOF could be suppressed by a LOF allele (Fig. S10B). As an example, S1091G could be suppressed by almost all curated GOF mutants, yet it was also specifically suppressed by the LOF V1094D (Fig. S10C). S1091G and V1094D appear to compensate for each other in a allele-specific fashion. We suggest that these are the types of interactions that will allow the TL and adjacent residues to evolve and differentiate while maintaining essential functions.
We note that strong epistasis is much more prevalent in the Pol II system than in other proteins where it has been quantified (54, 74–76, 83) (Fig. S10D). We attribute this difference to the much higher rate of suppressive interactions due to Pol II mutants having opposing effects on catalysis.
TL evolution is shaped by contextual epistasis.
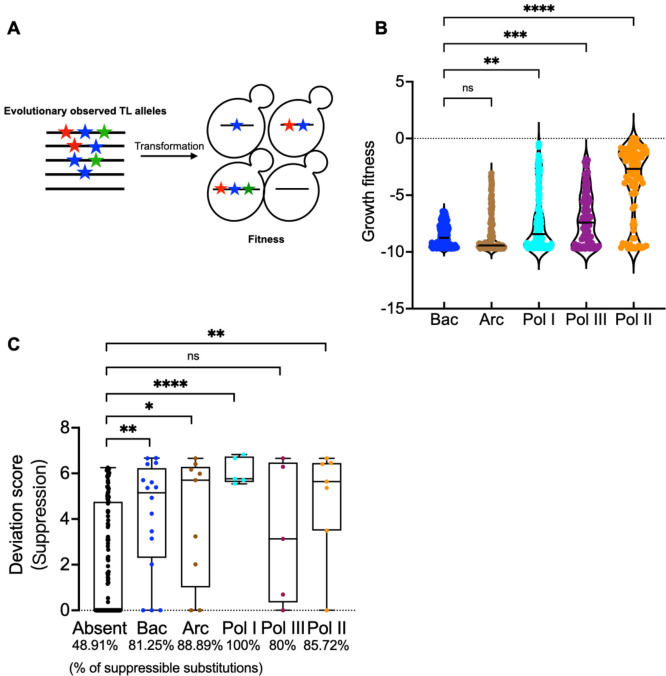
We previously found that identical mutations in a residue conserved between the Pol I and Pol II TLs yielded different biochemical phenotypes (67, 84). Furthermore, the yeast Pol I TL was incompatible within the yeast Pol II enzyme, implying that TL function is shaped by the enzymatic context (67, 84). To determine the generality and scope of TL-Pol II incompatibility, we designed a library containing evolutionary TL variants from bacterial, archaeal, and eukaryotic msRNAPs and determined their compatibility in the yeast Pol II context (Fig. 5A). TL alleles of eukaryotic Pols were more compatible than those from Archaea and Bacteria, and Pol II alleles were the most compatible (Fig. 5B, Fig. S11A), consistent with evolutionary distance. The total number of TL substitutions in haplotypes were slightly negatively correlated with growth fitness in the Pol II background for Archaeal, Pol I, II and III TLs (Fig. S11C), though not for Bacterial TLs, likely because the bacterial TLs were almost entirely incompatible in the Pol II context (Fig. S11C). Conservation of TL sequence and function was high enough that some archaeal sequences could provide viability to yeast Pol II, yet at the same time a number of Pol II TLs from other species were defective if not lethal. These results suggest widespread coevolution of TL sequence outside of ultra-conserved positions shapes TL function (see Discussion).
Figure 5. Contextual epistasis shapes TL evolution.
A. Schematic for the TL evolutionary haplotypes library. We selected 662 TL haplotypes representing TL alleles from bacterial, archaeal and the three conserved eukaryotic msRNAPs. These TL alleles were transformed into yeast and were phenotyped under selective conditions. B. Fitness of evolutionarily observed TL haplotypes in the yeast Pol II background. The Pol II WT TL fitness (0) is labeled as dotted line. Kruskal-Wallis test was performed for comparison and significant levels (P < 0.05) were labeled. C. A comparison of the maximum deviation score of each TL lethal single substitution that is present in any evolutionary TL haplotypes from bacterial, archaeal or eukaryotic Pols versus those that have not been observed in any species. The evolutionary TL haplotypes were from multiple sequence alignments (MSA). 9 substitutions were found in an MSA of 542 archaeal TL sequences that are lethal when present in yeast as a single substitution. 17 were found in an MSA of 1403 bacterial TLs, 5 were found in 749 Pol I TLs, 7 were found in 499 Pol II TLs, and 5 were found in 539 Pol III TLs. Evolutionarily observed lethal substitutions were compared to those unobserved in our TL MSA. The percentage of in total suppressible lethal single mutants for each group is labeled at the bottom of the plot. Statistical comparison was done with the Mann-Whitney test and the significant levels (P < 0.05) are shown in the figure.
We reasoned that evolutionarily observed lethal substitutions might be closer to functional than non-evolutionarily observed and would therefore be more likely to be suppressible by Pol II GOF alleles. To compare suppressibility between evolutionarily observed and unobserved substitutions lethal to Pol II, we extracted the highest positive deviation scores among all double mutants containing each lethal substitution. Maximum deviation scores for Pol II lethal substitutions present in TLs of existing msRNAPs were higher than for lethal substitutions that were absent, indicating the Pol II lethal mutants present in existing msRNAPs on average maintain a greater functionality and/or are suppressible by single changes (Fig. 5C, Fig. S11B). The TL has been estimated as providing 500–1000 fold enhancement on catalytic activity (85–87), while we estimate only ~10-fold effects are tolerated for yeast viability (36). We conclude that lethal mutants observed as functional residues in other species are more likely to be close to the viability threshold as might result from a series of small steps to allow them to function.
TL residues co-evolve with the rest of Rpb1 through diverse pathways.
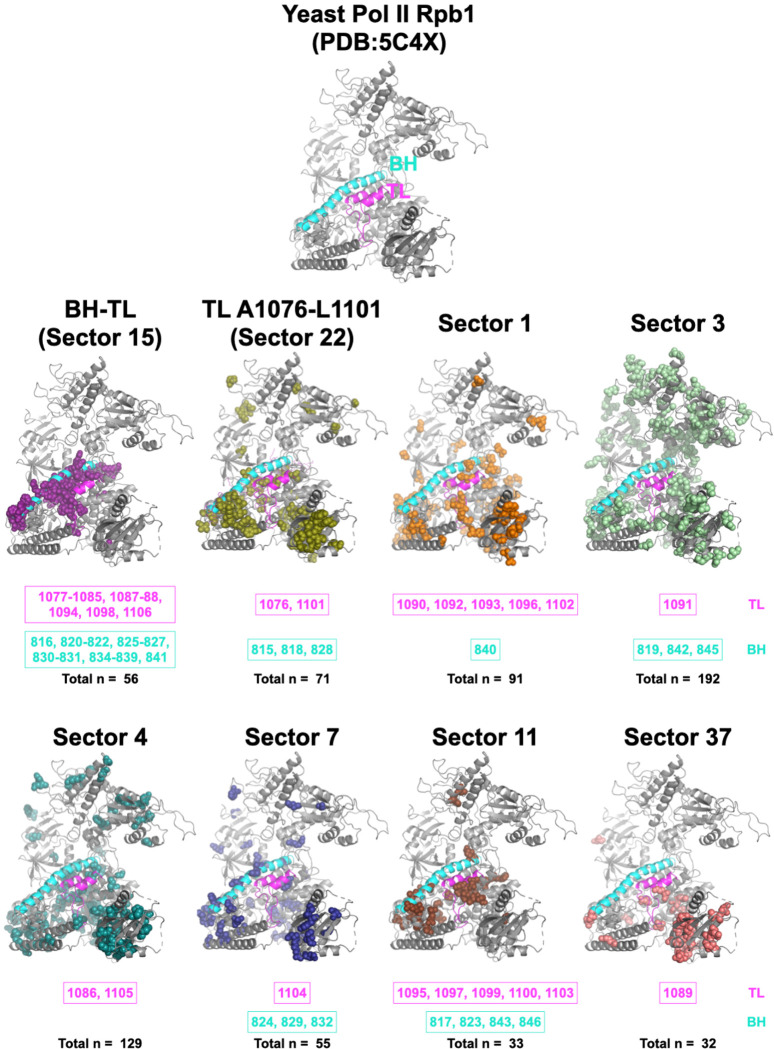
Our analyses suggest that even a highly conserved domain such as the Pol II TL can be sensitive to identity of adjacent residues and that changing networks of interactions shape the Pol II active site across evolution. We employed statistical coupling analysis (SCA) to identify if there are any coevolving residue networks in Rpb1 and ask about pathways that might co-evolve the TL. SCA “Sector” analysis is especially useful for identify subgroups of coupled residues that might form allosteric communication networks (88–90). We extracted 410 yeast Pol II Rpb1 sequences from the recently published msRNAP large subunit multiple sequence alignment (MSA) from the Landick lab (82) and performed SCA (see Methods)(89). We identified 40 coevolving sectors (Fig. S12), and every single TL residue was found within one of the eight sectors that form generally continuous network of interactions within Rpb1(Fig. 6). TL residues within the TL NIR were coupled with most BH residues and the alanine-glycine linker (Rpb1 1087–1088). These residues are highly conserved (17), indicating this sector is driven by conservation primarily. Six of eight Rpb1 sectors containing TL residues also contained at least one BH residue, supporting functional coupling between these two domains. Coupling is not limited to residues that are close to the active site. Distal residues can potentially modulate TL function through allosteric interactions. For example, the greatest distance between a TL residue and another Rpb1 residue in the same sector is ~ 55 Å. Interestingly, the residue pair 1076–1101, for which we observed extensive epistasis, are the sole TL residues within a very large cluster containing >150 residues across Rpb1. Our epistasis studies indicate multiple allele-specific interactions between 1076 and 1101 of exactly the type that might appear as evolutionary coupling between specific substitutions at these positions. The hydrophobic TL pocket is an attractive linchpin for potential communication to the TL from throughout Pol II, and multiple sectors converge on this domain.
Figure 6. Ultra-conserved TL co-evolves with Pol II residues through diverse pathways.
The eight coevolution sectors containing any TL residues that were identified from statistical coupling analysis are shown on the yeast Pol II Rpb1 structure (PDB:5C4X)(28). The TL is labeled with magenta and the BH is labeled with cyan. The TL and BH residues in each sector are labeled at the bottom of each sector. The total number of residues within each sector is also shown. The details of the statistical coupling analysis are in Methods.
DISCUSSION
How individual mutants alter a protein’s function is not necessarily straightforward at the mechanistic level. Amino acid substitutions both remove functionality of the WT residue but replace that functionality with something different. By altering the local environment within a protein or potentially propagating effects to distant locations through allosteric changes, each substitution potentially can be quite different. These differences may not be apparent as phenotypic outputs and phenotypic assays may not have granularity to distinguish different biophysical behaviors if they result in similar outputs. For Pol II mutants, even high-resolution phenotypic analyses, such as gene expression profiling or genetic interaction profiling between Pol II mutants and deletions in other yeast genes (49), suggest that LOF and GOF mutants represent a continuum of defects that match enzymatic activity in vitro. Therefore, these profiles also appear dependent on the output of Pol II activity defects and can’t distinguish potential differences in underlying mechanism.
Through systematic detection of genetic interactions within the Pol II active site, we have identified functional relationships between amino acids across the TL and between TL substitutions and others. In the absence of double mutant epistasis analyses it would not be possible to differentiate similar alleles from one another. L1101S and E1103G, for example, are two GOF alleles very close to each other in Pol II structure and confer similar phenotypic landscapes across various growth conditions. Here, we find that their distinct interactions support that substitutions at 1101 and 1103 target distinct residue networks (Fig. 3C, Fig. 4A, and Fig. S6–S7). 1101 functions in the five-helix bundle hydrophobic pocket while 1103 interacts and co-evolves with a number of TL external residues that together support interactions that promote the open TL conformation (Fig. 4C, Fig. 6). We also observed connections between TL C-terminal residues that suggest a limit to how disruptions to structure there can alter Pol II activity (Fig. 2D, Fig. 6). Helix-disrupting LOF proline substitutions in at least two TL positions showed epistasis with multiple substitutions in the back of the TL (1094–1098), suggesting that their functions require TL C-terminal helix structure and in the absence of that structure (proline disruption) effects are no longer additive.
The strongest epistatic interactions observed were between two pairs of hydrophobic residues, A1076 and L1101, and M1079 and G1097 (Fig. 4). Each of these contributes to the structure of a hydrophobic pocket that bundles two TL proximal helices with the BH and two others in a five-helix bundle. Supporting the dependence of these residues on each other for maintaining function, identity at these positions over evolution also shows coupling. Interestingly, these A1076 and L1101 were coupled uniquely out of TL residues with a great number of other positions in Rpb1 (Fig. 6).
Elongation factors bind Pol II and alter its activity, but the mechanisms by which they do so are not known (91, 92). We observed a high level of genetic interactions between residues outside the TL and residues within it, including allele-specific reshaping of TL mutant space upon single substitution outside the TL (Fig. 3). The fact that minor mutational changes outside the TL can apparently functionally perturb the TL would be consistent with the idea that minor alterations to Pol II structure upon elongation factor binding could easily propagate into the active site via the TL or the BH. As an example, human Rtf1 has been observed to project a domain into the Pol II structure adjacent to the BH (in yeast, this region is occupied instead by Rpb2 (93)). These contacts have been proposed to alter Pol II activity. We would propose that the paths for such alteration activity would follow the coupling sectors we have observed by SCA.
How different individual substitutions are under the surface is critical for understanding plasticity in protein mechanisms and how they might be altered by evolutionary change. A key open question in nucleic acid polymerase mechanisms is the paths for protons in the reaction (for example, deprotonation of the synthesized strand 3′-OH and protonation of pyrophosphate leaving group, for example) (e.g.(77, 78, 80, 82, 94–96)). For msRNAPs, the association with incoming NTP by a nearly universally conserved histidine led to the proposal that this residue might donate a proton during the reaction (12, 79, 94). Some substitutions at this position can provide minimal essential function (e.g. tyrosine, arginine), while others are only moderately defective (glutamine). Surprisingly, we found that H1085L was very-well tolerated for growth (17) and the Landick lab has proposed this substitution supports catalysis through positional but not chemical effects (81, 82). Our studies here were quite surprising in that they indicated that L1085 Pol II has unique behavior when perturbed by all possible TL substitutions and is entirely distinct from H1085Y (where we have direct observations of all possible intra-TL doubles) or H1085A or H1085Q (curated doubles) (Fig. S9C, Fig. S13). These residue specific behaviors suggest that each substitution may have different properties, and compatibility with function may not necessarily represent similar function under the surface.
Evolutionary change over time can alter protein function but it can also alter protein functional plasticity. Recent work from the Thornton lab elegantly demonstrates that phenotypes of substitutions to residues conserved over hundreds of millions of years can change over evolutionary time and can do so unpredictably and transiently during evolution (59). msRNAPs have structures and functions conserved over billions of years, and deep within their active sites is a mobile domain, the TL, that has large functional constraints on its sequence. The TL sequence must be able to fold into multiple states and maintain recognition of the same substrates across evolutionary space and is shows high identity even between distantly related species. Here we show that the TL, and likely the entire Pol II active site, exhibits a great amount of plasticity through non-conserved positions that are essential for compatibility of the TL and surrounding domains. Our results illustrating widespread epistasis and allele-specific effects of single and double mutants predict that comparative analyses among Pol I, II, and III will reveal widespread and enzyme-specific mechanisms due to higher order epistasis shaping function of conserved residues.
METHODS
Design and Synthesis of TL mutant libraries.
We updated and extended the fitness dataset of Qiu et al (17). Using a similar methodology, but with adjusted conditions and a second-generation mutant library strategy, in order to generate a complete Pol II TL mutation-phenotype map and examine genetic interactions. Mutants were constructed by synthesis with Agilent and screened for phenotypes previously established as informative for Pol II mutant biochemical defects. Programmed oligonucleotide library pools included all 620 single TL residue substitutions and deletions for Rpb1 amino acids 1076–1106 (Library 1), 3914 pairwise double substitutions (Library 2), 4800 targeted double substitutions (Library 6), and 3373 multiple substitutions (Library 3–5), along with the WT S. cerevisiae Pol II TL allele at a level of ~15% of the total variants, enabling precise quantification (see Supplemental Table 4). Each synthesized region contained a mutated or WT Pol II TL sequence and two flanking regions at the 5′ and 3′’ ends of the TL-encoding sequence. These flanking regions also contained designed “PCR handle” (20bp) sequences, allowing distinct subsets of oligos to be amplified from synthesized pools using selected primers for PCR, and additional flanking WT Pol II sequences allow for further extension of homology arms by PCR “sewing” (Details are in Supplemental Method 2 and 3).
Introduction of Libraries into yeast and phenotyping.
Synthesized mutant pools were transformed into yeast (CKY283) along with an RPB1-encoding plasmid where the TL-encoding sequence was replaced with an MluI restriction site for linearization as described in Qiu et al (17). This strategy allows construction of rpb1 mutant libraries by gap repair between library fragments and the linearized vector. Briefly, the synthesized oligo pools were amplified by limited cycles of emulsion PCR to limit template switching. Extension of flanking homology arms of ~200 bp were added by PCR sewing. Amplified TL sequences with extended flanking regions were co-transformed with linearized pRS315-derived CEN LEU2 plasmid (pCK892) into CKY283, allowing gap repair via homologous flanking regions. To detect potential residue-residue interactions between the TL and TL-proximal domains including the Rpb1 Bridge Helix (BH), Funnel Helix alpha-21 and Rpb2, the Pol II TL single mutant pool (Library 1, 620 mutant alleles and 111 WT alleles) was co-transformed individually with gapped plasmids encoding an additional rpb1 allele (Rpb1 BH T834P, T834A, or Funnel Helix alpha-21 S713P) into CKY283 respectively, or with the gapped WT RPB1 plasmid into a strain with the genomic mutation, rpb2 Y769F. These co-transformations created double mutants between the TL and TL-proximal mutants. The WT allele in single mutant pool represented the single probe mutant due to substitutions outside the TL on the plasmid or in the strain background. To distinguish between a fully WT TL and a WT TL representing the TL of a mutant allele elsewhere, a WT Pol II TL allele with a silent mutant at T1083 (WT codon ACC was replaced with ACT) was co-transformed with plasmid containing gapped WT RPB1 in a WT strain in parallel. 15% of the transformants with silent mutation were mixed with transformants of double mutants. The silent mutation allowed us to distinguish the WT and the single mutants. Each transformation was done in three biological replicates. After transformation, Leu+ colonies were collected from SC-Leu plates by scraping into sterile water and replated on SC-Leu+5FOA to select for cells having lost the RPB1 URA3 plasmid. 5-FOA-resistant colonies were scraped into sterile water from SC-Leu+5FOA and replated on SC-Leu, SC-Leu + 20mg/ml MPA (Fisher Scientific), SC-Leu + 15 mM Mn (Sigma), YPRaf, YPRafGal, SC-Lys, and SC-Leu + 3% Formamide (JT Baker) for phenotyping. Details of cell numbers plated on each plate and screening time of each plate are in Supplemental Table 3. Details of high efficiency transformation protocol is in supplemental method 1.
Generation of libraries for quantification by amplicon sequencing.
Genomic DNA of each screened library was extracted using the Yeastar genomic DNA kit according to manufacturer’s instructions (Zymo Research). To ensure adequate DNA for sequencing, the TL regions of all libraries were amplified with PCR cycles that were verified to be in the linear range by qPCR to minimize disturbance of allele distributions, and under emulsion PCR conditions (EURx Micellula DNA Emulsion & Purification (ePCR) PCR kit) to limit template switching. Details are in Supplemental Method 2 and 3. To multiplex samples, we employed a dual indexing strategy wherein 10 initial barcodes for differentiating 10 mutant libraries were added during the initial amplification using 10 pairs of custom primers. In a second amplification, 28 primers containing 28 NEB indices were used to add a second index for distinguishing conditions and replicates (NEBNext Multiplex Oligos for Illumina) (see Supplemental Table 2). As a result, a sample-specific barcodes were present for each set of variants. The indexed, pooled samples were sequenced by single end sequencing on an Illumina Next-Seq (150nt reads). On average, over 11 million reads were obtained for individual samples with high reproducibility from two rounds of sequencing. Raw sequencing data has been deposited on the NCBI SRA (Sequence Read Archive) database under BioProject PRJNA948661. Processed mutants counts and fitnesses are available through GitHub (https://github.com/Kaplan-Lab-Pitt/TLs_Screening.git).
Data cleaning and fitness calculation and normalization.
Reads of mutants were sorted into appropriate libraries and conditions by detecting particular indices after sequencing. Read counts were estimated by a codon-based alignment algorithm to distinguish reads that exactly matched designated codons of mutants (97). To clean the data, mutant reads with coefficients of variation greater than 0.5 in the control condition (SC-Leu) were excluded from the analysis. The mutant read count was increased by 1 to calculate the allele frequency under different conditions. To measure and compare the phenotypes of all mutants, mutant phenotypic score (fitness) was calculated by allele frequency change of a mutant under selective conditions relative to the unselective condition comparing to the frequency change of WT. The formula for calculating fitness is shown below.
We applied min-max normalization to bring the median growth fitness of mutants measured at ten libraries to the same level for direct comparison (formula is shown below). In each library, we divided mutants into several groups based on their allele counts on the control condition. Mutants with read count differences of less than 10 are present in one group. The WT growth fitness was set as the maximum value and the minimum fitness in each group was the minimum. Min-max normalization was used to equalize the growth fitness into the same range between various groups inside each library. Additionally, we utilized min-max normalization to level the mutant fitness across all ten libraries with WT fitness as Max and minimal fitness in each library as the minimum. As a result, mutant growth fitness was scaled to one range and could be used to determine genetic interactions.
Determination of functional interactions.
The genetic interactions between single substitutions were determined by comparing the multiple-substitution mutant normalized median fitness to the log additive of the single substitution normalized median fitness. The simplified formula is as follows:
−1 < Deviation score < 1, the interaction among the constituent single mutants is additive and mutants are acting independently.
Deviation score ≥ 1, the interaction is non-additive and is positive, including suppression and epistatic interactions.
Deviation score ≤ −1. the interaction is non-additive and is negative, including synthetic sick, synthetic lethal, and sign epistasis interactions.
Any mutation with fitness smaller than the lethal threshold (−6.50) was classified as an ultra-sick/lethal mutant and its fitness was normalized to −6.50 for calculation of the deviation score. Synthetic sickness and synthetic lethality were distinguished by whether a double mutant is viable or lethal (fitness is greater than or equals to the lethal threshold −6.5) when two constituent mutations are viable. Synthetic lethality can be further classified into two types. First, additive-synthetic lethality was determined when the expected double mutant fitness calculated by additive model was lethal (expected fitness = −6.5) and the observed double mutant fitness was also lethal (fitness = −6.5) (in this case the deviation score = 0). Second, the beyond-additive synthetic lethality was determined when the expected double mutant was viable (expected fitness > −6.5) while the observed double mutant fitness was lethal (fitness = −6.5) (in this case the deviation score < 0). To separate these two situations in our figures, we labeled additive synthetic lethality as black and beyond-additive synthetic lethality as purple.
Details of formulas are in Supplemental Method 4. The codes for calculating deviation scores and generating figures are available in GitHub (https://github.com/Kaplan-Lab-Pitt/TLs_Screening.git).
Mutant classification using two multiple logistic regression models.
We trained two multiple logistic regression models to distinguish GOF and LOF mutants using the phenotypic fitness on SC-Leu+MPA, SC-Lys, and YPRafGal conditions of 65 single mutants, including 25 previously identified GOF mutants, 33 LOF mutants, one WT, and six that were not GOF or LOF mutants. Intercept, main effects, and two-way interactions were involved in defining both models. 0.75 was used as the cutoff threshold for both the GOF and LOF models.
Model for predicting the probability of a mutant being a GOF:
Model for predicting the probability of a mutant being LOF:
Both models showed accuracy, with the area under ROC close to one (Fig. S3A). The details are provided in Supplemental Table 5.
Principal component analysis (PCA).
Deviation scores of curated and probe double mutants were analyzed in PCA. The scripts using R language v4.0.3 (https://www.R-project.org/) with R packages tidyverse v1.3.1 (https://www.tidyverse.org), prompt (stats v3.6.2 (https://www.rdocumentation.org/packages/stats/versions/3.6.2/topics/prcomp)), ggplot2 v3.3.3 (https://ggplot2.tidyverse.org), dplyr v1.0.6 (https://dplyr.tidyverse.org), and missMDA v1.18 (https://dplyr.tidyverse.org), are available in GitHub (https://github.com/Kaplan-Lab-Pitt/TLs_Screening.git).
t-SNE projection.
Allele frequencies for all mutants in nine conditions with three replicates were analyzed by t-SNE (Perplexity = 50) or k-means (clusters =20). Thirteen clusters with ultra-sick to lethal mutants as majority were eliminated. The remaining mutants were analyzed again with t-SNE (Perplexity = 100) and k-means (cluster =10). The scripts utilizing R language v4.0.3 (https://www.R-project.org/), along with R packages Rtsne v0.15 (https://github.com/jkrijthe/Rtsne), ggplot2 v3.3.3 (https://ggplot2.tidyverse.org), k-means (stats v3.6.2 (https://www.rdocumentation.org/packages/stats/versions/3.6.2/topics/kmeans)), are available through GitHub (https://github.com/Kaplan-Lab-Pitt/TLs_Screening.git).
Statistical coupling analysis.
A published multiple sequence alignment (MSA) containing 5787 eukaryotic homologous sequences of yeast Rpb1 was used in the statistical coupling analysis (82). 1464 sequences were retained after sequence identity reducing to 90% with T-coffee package v12.00.7fb08c2 (98) through conda v4.6.14. Pol I, II, and III sequences were separated based on an ML tree constructed with FastTree 2 (99) and 410 Pol II Rpb1 homologous sequences were re-aligned with T-coffee, and the newly generated MSA was used for statistical coupling analysis with the python-based package pySCA v6.1 (89). The scripts were adapted from https://github.com/ranganathanlab/pySCA and are available via GitHub (https://github.com/Kaplan-Lab-Pitt/TLs_Screening.git).
Supplementary Material
ACKNOWLEDGMENTS
We thank Dr. Anne-Ruxandra Carvunis (U. Pittsburgh) and Dr. Steve Lockless (Texas A&M) for discussions and advice. We thank Zhizhen Wang and Muyao Lin from the Pitt Statistical Consulting Center for their advice on checking the reproducibility of our data. We acknowledge funding from NIH R01GM097260 for initiation of this project and NIH R35GM144116 for this work. This research was supported in part by the University of Pittsburgh Center for Research Computing, RRID:SCR_022735, through the resources provided. Specifically, this work used the HTC cluster, which is supported by NIH award number S10OD028483.
REFERENCES
- 1.Cramer P., Multisubunit RNA polymerases. Curr Opin Struct Biol 12, 89–97 (2002). [DOI] [PubMed] [Google Scholar]
- 2.Werner F., Grohmann D., Evolution of multisubunit RNA polymerases in the three domains of life. Nat Rev Microbiol 9, 85–98 (2011). [DOI] [PubMed] [Google Scholar]
- 3.Allison L. A., Moyle M., Shales M., Ingles C. J., Extensive homology among the largest subunits of eukaryotic and prokaryotic RNA polymerases. Cell 42, 599–610 (1985). [DOI] [PubMed] [Google Scholar]
- 4.Zhang G., Campbell E. A., Minakhin L., Richter C., Severinov K., Darst S. A., Crystal Structure of Thermus aquaticus Core RNA Polymerase at 3.3 Å Resolution. Cell 98, 811–824 (1999). [DOI] [PubMed] [Google Scholar]
- 5.Vassylyev D. G., Sekine S., Laptenko O., Lee J., Vassylyeva M. N., Borukhov S., Yokoyama S., Crystal structure of a bacterial RNA polymerase holoenzyme at 2.6 A resolution. Nature 417, 712–719 (2002). [DOI] [PubMed] [Google Scholar]
- 6.Hirata A., Klein B. J., Murakami K. S., The X-ray crystal structure of RNA polymerase from Archaea. Nature 451, 851–854 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gnatt A. L., Cramer P., Fu J., Bushnell D. A., Kornberg R. D., Structural basis of transcription: an RNA polymerase II elongation complex at 3.3 A resolution. Science 292, 1876–1882 (2001). [DOI] [PubMed] [Google Scholar]
- 8.Cramer P., Bushnell D. A., Kornberg R. D., Structural basis of transcription: RNA polymerase II at 2.8 angstrom resolution. Science 292, 1863–1876 (2001). [DOI] [PubMed] [Google Scholar]
- 9.Fernandez-Tornero C., Moreno-Morcillo M., Rashid U. J., Taylor N. M., Ruiz F. M., Gruene T., Legrand P., Steuerwald U., Muller C. W., Crystal structure of the 14-subunit RNA polymerase I. Nature 502, 644–649 (2013). [DOI] [PubMed] [Google Scholar]
- 10.Hoffmann N. A., Jakobi A. J., Moreno-Morcillo M., Glatt S., Kosinski J., Hagen W. J., Sachse C., Muller C. W., Molecular structures of unbound and transcribing RNA polymerase III. Nature 528, 231–236 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Malinen A. M., Turtola M., Parthiban M., Vainonen L., Johnson M. S., Belogurov G. A., Active site opening and closure control translocation of multisubunit RNA polymerase. Nucleic Acids Res 40, 7442–7451 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang D., Bushnell D. A., Westover K. D., Kaplan C. D., Kornberg R. D., Structural basis of transcription: role of the trigger loop in substrate specificity and catalysis. Cell 127, 941–954 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kaplan C. D., Basic mechanisms of RNA polymerase II activity and alteration of gene expression in Saccharomyces cerevisiae. Biochimica Et Biophysica Acta-Gene Regulatory Mechanisms 1829, 39–54 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dangkulwanich M., Ishibashi T., Liu S., Kireeva M. L., Lubkowska L., Kashlev M., Bustamante C. J., Complete dissection of transcription elongation reveals slow translocation of RNA polymerase II in a linear ratchet mechanism. Elife 2, e00971 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bar-Nahum G., Epshtein V., Ruckenstein A. E., Rafikov R., Mustaev A., Nudler E., A ratchet mechanism of transcription elongation and its control. Cell 120, 183–193 (2005). [DOI] [PubMed] [Google Scholar]
- 16.Weinzierl R. O., The nucleotide addition cycle of RNA polymerase is controlled by two molecular hinges in the Bridge Helix domain. BMC Biol 8, 134 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Qiu C., Erinne O. C., Dave J. M., Cui P., Jin H., Muthukrishnan N., Tang L. K., Babu S. G., Lam K. C., Vandeventer P. J., Strohner R., Van den Brulle J., Sze S. H., Kaplan C. D., High-Resolution Phenotypic Landscape of the RNA Polymerase II Trigger Loop. PLoS Genet 12, e1006321 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Da L. T., Pardo-Avila F., Xu L., Silva D. A., Zhang L., Gao X., Wang D., Huang X., Bridge helix bending promotes RNA polymerase II backtracking through a critical and conserved threonine residue. Nat Commun 7, 11244 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mazumder A., Lin M., Kapanidis A. N., Ebright R. H., Closing and opening of the RNA polymerase trigger loop. Proc Natl Acad Sci U S A 117, 15642–15649 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang B., Predeus A. V., Burton Z. F., Feig M., Energetic and structural details of the trigger-loop closing transition in RNA polymerase II. Biophys J 105, 767–775 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Larson M. H., Zhou J., Kaplan C. D., Palangat M., Kornberg R. D., Landick R., Block S. M., Trigger loop dynamics mediate the balance between the transcriptional fidelity and speed of RNA polymerase II. Proc Natl Acad Sci U S A 109, 6555–6560 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fouqueau T., Zeller M. E., Cheung A. C., Cramer P., Thomm M., The RNA polymerase trigger loop functions in all three phases of the transcription cycle. Nucleic Acids Res 41, 7048–7059 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kireeva M. L., Nedialkov Y. A., Cremona G. H., Purtov Y. A., Lubkowska L., Malagon F., Burton Z. F., Strathern J. N., Kashlev M., Transient reversal of RNA polymerase II active site closing controls fidelity of transcription elongation. Mol Cell 30, 557–566 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu X., Bushnell D. A., Kornberg R. D., RNA polymerase II transcription: structure and mechanism. Biochim Biophys Acta 1829, 2–8 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kaplan C. D., Kornberg R. D., A bridge to transcription by RNA polymerase. J Biol 7, 39 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tan L., Wiesler S., Trzaska D., Carney H. C., Weinzierl R. O., Bridge helix and trigger loop perturbations generate superactive RNA polymerases. J Biol 7, 40 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Silva D. A., Weiss D. R., Pardo Avila F., Da L. T., Levitt M., Wang D., Huang X., Millisecond dynamics of RNA polymerase II translocation at atomic resolution. Proc Natl Acad Sci U S A 111, 7665–7670 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Barnes C. O., Calero M., Malik I., Graham B. W., Spahr H., Lin G., Cohen A. E., Brown I. S., Zhang Q., Pullara F., Trakselis M. A., Kaplan C. D., Calero G., Crystal Structure of a Transcribing RNA Polymerase II Complex Reveals a Complete Transcription Bubble. Mol Cell 59, 258–269 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fong N., Kim H., Zhou Y., Ji X., Qiu J., Saldi T., Diener K., Jones K., Fu X. D., Bentley D. L., Pre-mRNA splicing is facilitated by an optimal RNA polymerase II elongation rate. Genes Dev 28, 2663–2676 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xu L., Butler K. V., Chong J., Wengel J., Kool E. T., Wang D., Dissecting the chemical interactions and substrate structural signatures governing RNA polymerase II trigger loop closure by synthetic nucleic acid analogues. Nucleic Acids Res 42, 5863–5870 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vassylyev D. G., Vassylyeva M. N., Zhang J., Palangat M., Artsimovitch I., Landick R., Structural basis for substrate loading in bacterial RNA polymerase. Nature 448, 163–168 (2007). [DOI] [PubMed] [Google Scholar]
- 32.Da L. T., Wang D., Huang X., Dynamics of pyrophosphate ion release and its coupled trigger loop motion from closed to open state in RNA polymerase II. J Am Chem Soc 134, 2399–2406 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu B., Zuo Y., Steitz T. A., Structures of E. coli sigmaS-transcription initiation complexes provide new insights into polymerase mechanism. Proc Natl Acad Sci U S A 113, 4051–4056 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Seibold S. A., Singh B. N., Zhang C., Kireeva M., Domecq C., Bouchard A., Nazione A. M., Feig M., Cukier R. I., Coulombe B., Kashlev M., Hampsey M., Burton Z. F., Conformational coupling, bridge helix dynamics and active site dehydration in catalysis by RNA polymerase. Biochim Biophys Acta 1799, 575–587 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Malagon F., Kireeva M. L., Shafer B. K., Lubkowska L., Kashlev M., Strathern J. N., Mutations in the Saccharomyces cerevisiae RPB1 gene conferring hypersensitivity to 6-azauracil. Genetics 172, 2201–2209 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kaplan C. D., Larsson K. M., Kornberg R. D., The RNA polymerase II trigger loop functions in substrate selection and is directly targeted by alpha-amanitin. Mol Cell 30, 547–556 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kaplan C. D., Jin H., Zhang I. L., Belyanin A., Dissection of Pol II trigger loop function and Pol II activity-dependent control of start site selection in vivo. PLoS Genet 8, e1002627 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Larsona Matthew H., Zhoub Jing,1Kaplanc Craig D.,1, Palangatd Murali,2, Kornberge Roger D., Landickd Robert, and Blocka Steven M.,b,f,3, Trigger loop dynamics mediate the balance between the transcriptional fidelity and speed of RNA polymerase II. (2012). [DOI] [PMC free article] [PubMed]
- 39.Kireeva M. L., Opron K., Seibold S. A., Domecq C., Cukier R. I., Coulombe B., Kashlev M., Burton Z. F., Molecular dynamics and mutational analysis of the catalytic and translocation cycle of RNA polymerase. BMC Biophys 5, 11 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Windgassen T. A., Mooney R. A., Nayak D., Palangat M., Zhang J., Landick R., Trigger-helix folding pathway and SI3 mediate catalysis and hairpin-stabilized pausing by Escherichia coli RNA polymerase. Nucleic Acids Res 42, 12707–12721 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nayak D., Voss M., Windgassen T., Mooney R. A., Landick R., Cys-pair reporters detect a constrained trigger loop in a paused RNA polymerase. Mol Cell 50, 882–893 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cheung A. C., Cramer P., Structural basis of RNA polymerase II backtracking, arrest and reactivation. Nature 471, 249–253 (2011). [DOI] [PubMed] [Google Scholar]
- 43.Kettenberger H., Armache K. J., Cramer P., Complete RNA polymerase II elongation complex structure and its interactions with NTP and TFIIS. Mol Cell 16, 955–965 (2004). [DOI] [PubMed] [Google Scholar]
- 44.Lennon C. W., Ross W., Martin-Tumasz S., Toulokhonov I., Vrentas C. E., Rutherford S. T., Lee J. H., Butcher S. E., Gourse R. L., Direct interactions between the coiled-coil tip of DksA and the trigger loop of RNA polymerase mediate transcriptional regulation. Genes Dev 26, 2634–2646 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sekine S., Murayama Y., Svetlov V., Nudler E., Yokoyama S., The ratcheted and ratchetable structural states of RNA polymerase underlie multiple transcriptional functions. Mol Cell 57, 408–421 (2015). [DOI] [PubMed] [Google Scholar]
- 46.Hein P. P., Kolb K. E., Windgassen T., Bellecourt M. J., Darst S. A., Mooney R. A., Landick R., RNA polymerase pausing and nascent-RNA structure formation are linked through clamp-domain movement. Nat Struct Mol Biol 21, 794–802 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Taatjes A. C. S. a. D. J., Structure and mechanism of the RNA polymerase II transcription machinery. (2020). [DOI] [PMC free article] [PubMed]
- 48.Leng X. Y., Iyanov M., Kindgren P., Malik I., Thieffry A., Brodersen P., Sandelin A., Kaplan C. D., Marquardt S., Organismal benefits of transcription speed control at gene boundaries. Embo Reports 21, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Braberg H., Jin H., Moehle E. A., Chan Y. A., Wang S., Shales M., Benschop J. J., Morris J. H., Qiu C., Hu F., Tang L. K., Fraser J. S., Holstege F. C., Hieter P., Guthrie C., Kaplan C. D., Krogan N. J., From structure to systems: high-resolution, quantitative genetic analysis of RNA polymerase II. Cell 154, 775–788 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kaster B. C., Knippa K. C., Kaplan C. D., Peterson D. O., RNA Polymerase II Trigger Loop Mobility: INDIRECT EFFECTS OF Rpb9. J Biol Chem 291, 14883–14895 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tesileanu T., Colwell L. J., Leibler S., Protein sectors: statistical coupling analysis versus conservation. PLoS Comput Biol 11, e1004091 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Phillips P. C., The language of gene interaction. Genetics 149, 1167–1171 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Breen M. S., Kemena C., Vlasov P. K., Notredame C., Kondrashov F. A., Epistasis as the primary factor in molecular evolution. Nature 490, 535–538 (2012). [DOI] [PubMed] [Google Scholar]
- 54.Starr T. N., Thornton J. W., Epistasis in protein evolution. Protein Sci 25, 1204–1218 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ortlund E. A., Bridgham J. T., Redinbo M. R., Thornton J. W., Crystal structure of an ancient protein: evolution by conformational epistasis. Science 317, 1544–1548 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Karageorgi M., Groen S. C., Sumbul F., Pelaez J. N., Verster K. I., Aguilar J. M., Hastings A. P., Bernstein S. L., Matsunaga T., Astourian M., Guerra G., Rico F., Dobler S., Agrawal A. A., Whiteman N. K., Genome editing retraces the evolution of toxin resistance in the monarch butterfly. Nature 574, 409–412 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Phillips P. C., Epistasis--the essential role of gene interactions in the structure and evolution of genetic systems. Nat Rev Genet 9, 855–867 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Faure A. J., Domingo J., Schmiedel J. M., Hidalgo-Carcedo C., Diss G., Lehner B., Mapping the energetic and allosteric landscapes of protein binding domains. Nature 604, 175–183 (2022). [DOI] [PubMed] [Google Scholar]
- 59.Park Y., Metzger B. P. H., Thornton J. W., Epistatic drift causes gradual decay of predictability in protein evolution. Science 376, 823–830 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ding D., Green A. G., Wang B., Lite T. V., Weinstein E. N., Marks D. S., Laub M. T., Co-evolution of interacting proteins through non-contacting and non-specific mutations. Nat Ecol Evol 6, 590–603 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kondrashov A. S., Sunyaev S., Kondrashov F. A., Dobzhansky-Muller incompatibilities in protein evolution. Proc Natl Acad Sci U S A 99, 14878–14883 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lunzer M., Golding G. B., Dean A. M., Pervasive cryptic epistasis in molecular evolution. PLoS Genet 6, e1001162 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Natarajan C., Inoguchi N., Weber R. E., Fago A., Moriyama H., Storz J. F., Epistasis among adaptive mutations in deer mouse hemoglobin. Science 340, 1324–1327 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Doud M. B., Ashenberg O., Bloom J. D., Site-Specific Amino Acid Preferences Are Mostly Conserved in Two Closely Related Protein Homologs. Mol Biol Evol 32, 2944–2960 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Starr T. N., Greaney A. J., Hannon W. W., Loes A. N., Hauser K., Dillen J. R., Ferri E., Farrell A. G., Dadonaite B., McCallum M., Matreyek K. A., Corti D., Veesler D., Snell G., Bloom J. D., Shifting mutational constraints in the SARS-CoV-2 receptor-binding domain during viral evolution. Science 377, 420–424 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Haddox H. K., Dingens A. S., Hilton S. K., Overbaugh J., Bloom J. D., Mapping mutational effects along the evolutionary landscape of HIV envelope. Elife 7, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Viktorovskaya O. V., Engel K. L., French S. L., Cui P., Vandeventer P. J., Pavlovic E. M., Beyer A. L., Kaplan C. D., Schneider D. A., Divergent contributions of conserved active site residues to transcription by eukaryotic RNA polymerases I and II. Cell Rep 4, 974–984 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Mani R., St Onge R. P., J. L. t. Hartman, G. Giaever, F. P. Roth, Defining genetic interaction. Proc Natl Acad Sci U S A 105, 3461–3466 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Qiu C., Kaplan C. D., Functional assays for transcription mechanisms in high-throughput. Methods 159–160, 115–123 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lin X., Liu Y., Liu S., Zhu X., Wu L., Zhu Y., Zhao D., Xu X., Chemparathy A., Wang H., Cao Y., Nakamura M., Noordermeer J. N., La Russa M., Wong W. H., Zhao K., Qi L. S., Nested epistasis enhancer networks for robust genome regulation. Science 377, 1077–1085 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Fowler D. M., Fields S., Deep mutational scanning: a new style of protein science. Nat Methods 11, 801–807 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sergey Ioffe C. S., Batch Normalization: Accelerating Deep Network Training by Reducing Internal Covariate Shift. arXiv:1502.03167v3, (2015). [Google Scholar]
- 73.Hill W. G., Goddard M. E., Visscher P. M., Data and theory point to mainly additive genetic variance for complex traits. PLoS Genet 4, e1000008 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Melamed D., Young D. L., Gamble C. E., Miller C. R., Fields S., Deep mutational scanning of an RRM domain of the Saccharomyces cerevisiae poly(A)-binding protein. RNA 19, 1537–1551 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Araya C. L., Fowler D. M., Chen W., Muniez I., Kelly J. W., Fields S., A fundamental protein property, thermodynamic stability, revealed solely from large-scale measurements of protein function. Proc Natl Acad Sci U S A 109, 16858–16863 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Fowler D. M., Araya C. L., Fleishman S. J., Kellogg E. H., Stephany J. J., Baker D., Fields S., High-resolution mapping of protein sequence-function relationships. Nat Methods 7, 741–746 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Carvalho A. T., Fernandes P. A., Ramos M. J., The Catalytic Mechanism of RNA Polymerase II. J Chem Theory Comput 7, 1177–1188 (2011). [DOI] [PubMed] [Google Scholar]
- 78.Huang X., Wang D., Weiss D. R., Bushnell D. A., Kornberg R. D., Levitt M., RNA polymerase II trigger loop residues stabilize and position the incoming nucleotide triphosphate in transcription. Proc Natl Acad Sci U S A 107, 15745–15750 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Castro C., Smidansky E. D., Arnold J. J., Maksimchuk K. R., Moustafa I., Uchida A., Gotte M., Konigsberg W., Cameron C. E., Nucleic acid polymerases use a general acid for nucleotidyl transfer. Nat Struct Mol Biol 16, 212–218 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Unarta I. C., Goonetilleke E. C., Wang D., Huang X., Nucleotide addition and cleavage by RNA polymerase II: Coordination of two catalytic reactions using a single active site. J Biol Chem 299, 102844 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Mishanina T. V., Palo M. Z., Nayak D., Mooney R. A., Landick R., Trigger loop of RNA polymerase is a positional, not acid-base, catalyst for both transcription and proofreading. Proc Natl Acad Sci U S A 114, E5103–E5112 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Palo M. Z., Zhu J., Mishanina T. V., Landick R., Conserved Trigger Loop Histidine of RNA Polymerase II Functions as a Positional Catalyst Primarily through Steric Effects. Biochemistry 60, 3323–3336 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Harms M. J., Thornton J. W., Analyzing protein structure and function using ancestral gene reconstruction. Curr Opin Struct Biol 20, 360–366 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Scull C. E., Ingram Z. M., Lucius A. L., Schneider D. A., A Novel Assay for RNA Polymerase I Transcription Elongation Sheds Light on the Evolutionary Divergence of Eukaryotic RNA Polymerases. Biochemistry 58, 2116–2124 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Toulokhonov I., Zhang J., Palangat M., Landick R., A central role of the RNA polymerase trigger loop in active-site rearrangement during transcriptional pausing. Mol Cell 27, 406–419 (2007). [DOI] [PubMed] [Google Scholar]
- 86.Yuzenkova Y., Bochkareva A., Tadigotla V. R., Roghanian M., Zorov S., Severinov K., Zenkin N., Stepwise mechanism for transcription fidelity. BMC Biol 8, 54 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Wang W., Walmacq C., Chong J., Kashlev M., Wang D., Structural basis of transcriptional stalling and bypass of abasic DNA lesion by RNA polymerase II. Proc Natl Acad Sci U S A 115, E2538–E2545 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Halabi N., Rivoire O., Leibler S., Ranganathan R., Protein sectors: evolutionary units of three-dimensional structure. Cell 138, 774–786 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Rivoire O., Reynolds K. A., Ranganathan R., Evolution-Based Functional Decomposition of Proteins. PLoS Comput Biol 12, e1004817 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Salinas V. H., Ranganathan R., Coevolution-based inference of amino acid interactions underlying protein function. Elife 7, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Cramer P., Organization and regulation of gene transcription. Nature 573, 45–54 (2019). [DOI] [PubMed] [Google Scholar]
- 92.Schier A. C., Taatjes D. J., Structure and mechanism of the RNA polymerase II transcription machinery. Genes Dev 34, 465–488 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Vos S. M., Farnung L., Linden A., Urlaub H., Cramer P., Structure of complete Pol II-DSIF-PAF-SPT6 transcription complex reveals RTF1 allosteric activation. Nat Struct Mol Biol 27, 668–677 (2020). [DOI] [PubMed] [Google Scholar]
- 94.Castro C., Smidansky E., Maksimchuk K. R., Arnold J. J., Korneeva V. S., Gotte M., Konigsberg W., Cameron C. E., Two proton transfers in the transition state for nucleotidyl transfer catalyzed by RNA- and DNA-dependent RNA and DNA polymerases. Proc Natl Acad Sci U S A 104, 4267–4272 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Gregory M. T., Gao Y., Cui Q., Yang W., Multiple deprotonation paths of the nucleophile 3’-OH in the DNA synthesis reaction. Proc Natl Acad Sci U S A 118, (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Belogurov G. A., Artsimovitch I., The Mechanisms of Substrate Selection, Catalysis, and Translocation by the Elongating RNA Polymerase. J Mol Biol 431, 3975–4006 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Sing-Hoi Sze C. D. K., Codon-Based Sequence Alignment for Mutation Analysis by High-Throughput Sequencing. 2018 IEEE 8th International Conference on Computational Advances in Bio and Medical Sciences (ICCABS), (2018). [Google Scholar]
- 98.Notredame C., Higgins D. G., Heringa J., T-Coffee: A novel method for fast and accurate multiple sequence alignment. J Mol Biol 302, 205–217 (2000). [DOI] [PubMed] [Google Scholar]
- 99.Price M. N., Dehal P. S., Arkin A. P., FastTree 2--approximately maximum-likelihood trees for large alignments. Plos One 5, e9490 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.