Abstract
Specific wiring is essential for sensory systems to precisely relay information to higher brain regions. The retina, an approachable part of the brain, is an ideal model for studying neural circuits due to its well-organized structure. In the retina, S-cone photoreceptors sense and relay short-wavelength (e.g., blue) light signals for encoding color information and other environmental cues. S-cones usually account for less than 10% of cones and are precisely connected to S-cone bipolar cells (SCBCs). This connection is ancient and highly conserved across species, indicating essential functions. How this wiring specificity is formed and maintained, however, is not understood. To unveil the molecular mechanisms underlying this highly specific connection, we sequenced the transcriptomes of thirteen-lined ground squirrel (TLGS) photoreceptors. We chose TLGS for their cone-rich retina and the absence of cones that co-express multiple opsin proteins, as compared to mice. We used a targeted SMART-seq approach to obtain high-resolution transcriptomes from S- and M-cone photoreceptors and identified a cell-adhesion molecule, Nrxn3, as a potential candidate mediating the S-cone to SCBC connection. Given the limitations of genetic manipulation in TLGS, we utilized mouse models to study the function of Nrxn3 in S-cones. In ‘true’ S-cones (S-opsin+/M-opsin−) that lack Nrxn3 expression, the number of connections with SCBCs was drastically reduced, indicating a critical role of Nrxn3 for this synapse. While neurexins are well known for their diverse roles in regulating various synapses, this study is the first to document its crucial role in mediating or maintaining a specific synapse in the central nervous system. In addition, the differentially expressed genes identified here provide a valuable resource for further investigating cone subtype-specific functions.
Introduction
The current knowledge of the formation and maintenance of accurate neuronal connections remains scarce but is critical for understanding neural development and neuroregeneration. Highly specific wiring is a fundamental requirement for sensory systems to relay precise information to higher brain areas. The neural retina is an ideal model to study the principles of synapse formation and circuit organization because of its organized, well-known structure and accessibility (Masland, 2001). For vision, two types of photoreceptors, rods and cones, receive light input in the mammalian retina. While rods are functioning in dim light conditions (e.g., moon light), cones are active under day light conditions and detect color via different cone subtypes. Most mammals are dichromats, having two cone subtypes: M- and S-cones (medium- and short-wavelength-sensitive cones, respectively). The cone subtypes express distinct opsin proteins that are tuned to specific wavelengths of light, thus allowing comparison by downstream neurons to generate color-contrast coding. S-cones expresses a short wavelength-sensitive opsin 1 (SWS1) pigment with a peak absorption wavelength (λmax) ranging from ultraviolet-sensitive (350–370 nm; e.g., murid rodents, bats) to blue-sensitive (400–457 nm; e.g., primates, cats, dogs) (Jacobs et al., 1991; Emerling et al., 2015). The S-cone pathway is ancient and highly conserved across species (Kolb, 1995). At the first synapse of this circuit, the S-cone forms a specific connection with the S-cone bipolar cell (SCBC). This bipolar cell type is highly conserved in mammals (Cohen et al., 1990a, 1990b; Famiglietti, 1981; Haverkamp et al., 2005; Li and DeVries, 2006; Mariani, 1984). Conversely, the SCBC receives its input exclusively from S-cones, which often only account for a small fraction (<10%) of total cones (Long and Fisher, 1983; Müller and Peichl, 1989; Nadal-Nicolás et al., 2020). Such specific wiring is therefore challenging but critical for conveying short wavelength signals. However, the molecular mechanism for this highly specific S-cone synapse is unknown.
To unveil the mechanisms of S-cone specific synaptic connections, we sequenced the transcriptome of thirteen-lined ground squirrel (TLGS) photoreceptors, aiming to identify differentially expressed genes (DEGs) such as those that encode cell adhesion molecules, which can distinguish cone subtypes. We used retinas from thirteen-lined ground squirrels because of their high cone abundance (~85%) and absence of mixed cones that are highly abundant in mice (Long and Fisher, 1983; Kryger et al., 1998; Applebury et al., 2000; Nadal-Nicolás et al., 2020). Previous studies used droplet-based high-throughput sequencing methods to profile mammalian retinas of multiple species (Macosko et al., 2015; Clark et al., 2019; Peng et al., 2019; Yan et al., 2020). While cone subtype clusters could be identified using these methods, DEGs for S-cone specific wiring have not been successfully identified. One possibility is that the limited sequencing depth might fail to unveil low expressing genes (Vieth et al., 2019). Thus, we used a targeted deep-sequencing approach (SMART-seq) of single photoreceptors. With this approach we identified S-cone-specific expression of Nrxn3. Using cone-subtype specific knockout mice, we confirmed that Nrxn3 is required for S-cone to S-cone bipolar cell synapse formation and/or maintenance. This result demonstrated that our approach and dataset can facilitate the discovery of molecular underpinnings of the precise photoreceptor connections in the retina. Such information can ultimately be used to combat photoreceptor degeneration and help to reintegrate transplanted or reprogramed cells to reinstate normal circuits and visual functions.
Results
SMART-seq of manually collected TLGS photoreceptors generated high-quality genetic profiles of cone subtypes
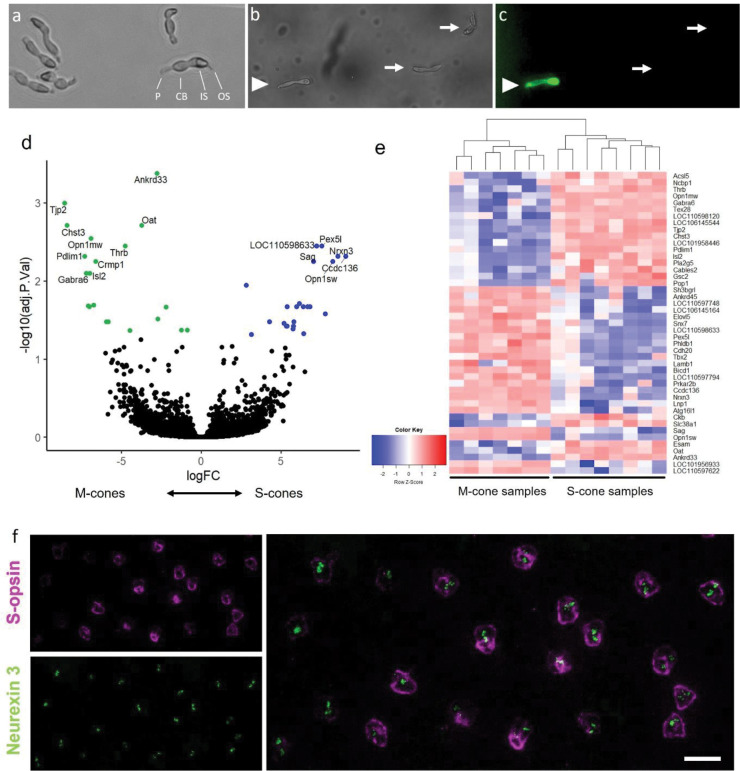
To identify molecules responsible for cone-subtype specific synaptic wiring, we turned to sequencing of cones in the TLGS retina, where cones are highly abundant and S- and M-cone types are clearly distinguished by the opsin they express. High throughput single cell RNA-seq applications are a common tool to rapidly investigate thousands of cells for gene expression, but sequencing-depth per cell remains low. Instead, we used a SMART-seq approach that puts the focus on sequencing a smaller number of isolated cells, with the advantage of receiving more reads per cell of interest, resulting in a higher gene detection rate (Vieth et al., 2019). Hence, we developed a protocol to dissociate and isolate live S- and M-cone photoreceptors (Angueyra et al., 2023). Live cells were labeled using an anti-S-opsin antibody —targeted against an extracellular domain—easily allowed us to distinguish live S- and M-cones after tissue dissociation and collect them via a micromanipulator for targeted sequencing (Figure 1 a–c).
Figure 1.
SMART-seq approach on manually collected M- and S-cone samples. a) Cones after dissociation. P: pedicle, CB: cell body, IS: inner segment, OS: outer segment. b-c) Fluorescently labeled live cones after antibody incubation under bright field (b) and excitation light (c) conditions. Labeled cone (arrowhead) was categorized as S-cone, while cells with cone morphology but without label (arrows) were categorized as M-cones. d)-e) Differentially expressed genes in S-and M-cone photoreceptors. d) Volcano plot highlighting significant genes enriched in M-cones (logFC < 0) and S-cones (logFC > 0). e) Heat map showing expression of significant differentially expressed genes in all samples. f) Validation of cone subtype-specific gene expression of Nrxn3. Flat-mounted ground squirrel retina after fluorescent in situ hybridization (FISH) with Nrxn3-probe, followed by S-opsin antibody-staining. Magenta: anti-S-opsin antibody. Green: Nrxn3 FISH probe. Scale bar: 10 μm.
A comparison of common marker genes reveals little-to-no contamination of our cone samples by rods or other cell types (supplementary table S1). General cone marker genes like Gnat2, Cnga3, Cngb3 and Grk7 were measured at similar levels in both cone subtypes as expected, whereas the opsin genes Opn1sw and Opn1mw were enriched in S-cones and M-cones, respectively. Marker genes of other cell types, like Rbpms (retinal ganglion cells), Vim (macroglia), Calb1 (horizontal cells), and Vsx2 (bipolar cells) were either not present or only detected at low levels, confirming a lack of contamination by other cell types and a high purity of the collected samples.
Differentially expressed genes in ground squirrel photoreceptors: cell-adhesion molecule Nrxn3
We detected 58 genes that showed significant differentially expression in S- or M-cones (adj. p-value < 0.1). 28 of these genes were enriched in M-cones, 30 in S-cones (Figure 1 d–e). The focus of our study lies on the identification of molecules that potentially mediate the formation or maintenance of the S-cone synapse. For this, we filtered the differentially expressed genes for cell-adhesion molecules in S-cones, resulting in the identification of S-cone-specific expression of Nrxn3 (neurexin 3).
Neurexins are classified as cell-adhesion molecules and they are involved in protein-protein interactions at synapses, where they form trans-synaptic complexes with many different protein families, e.g., neurexophilins, neuroligins, LRRTMs, latrophilins, and others (Missler et al., 1998; Hauser et al., 2022; Südhof, 2008; de Wit et al., 2009; Ko et al., 2009; Boucard et al., 2012; Südhof, 2017; Gomez et al., 2021). We confirmed the specificity via fluorescent in situ hybridization combined with antibody-based detection of S-opsin (Figure 1f). The detected probe confirms that, in the photoreceptor layer, Nrxn3 expression is restricted to S-cones. Thus, we identified Nrxn3 as a molecule that is potentially required to mediate the specific targeting of S-cones by SCBCs.
Nrxn3 is required for synaptic connections between S-cones and S-cone bipolar cells in mice
While sequencing of photoreceptors in the cone-dominant TLGS retina facilitated the identification of candidate genes for S-cone specific synaptic connections, the necessary genetic tools for confirming its function remain unavailable in TLGS. Thus, we proceeded with loss-of-function studies in the mouse, where such tools are readily available. Our objective was to specifically ablate expression of Nrxn3 in S-cones to assess possible changes in S-cone- to S-cone bipolar cell-connectivity. To achieve this, we crossed the cre-dependent Nrxn3 flox mouse Nrxn3tm3SudJ (Aoto et al., 2015) with our S-opsin-cre and Cpne9-venus mouse lines (Nadal-Nicolás et al., 2020). Cre recombinase is expressed under control of the Opn1sw promoter in about 12.3 % of S-opsin-expressing cones and provides cone-specific knockout of functional α- and β-neurexin 3. The Cpne9-venus line functions as S-cone bipolar cell (SCBC) reporter line because it expresses the yellow fluorescent protein venus under the control of the Cpne9-promoter which is highly specific for SCBCs (Shekhar et al., 2016). In conjunction with endogenously expressed fluorescent proteins, antibody labeling of retinas from these mice allowed the clear identification of postsynaptic SCBCs as well as three presynaptic cone subtypes: mixed cones (expressing both M- and S-opsin) as well as “true” s-cones (expressing S-opsin but lacking M-opsin; referred to here as S-cones) that either express cre (Nrxn3−/−) or had a lack thereof (control).
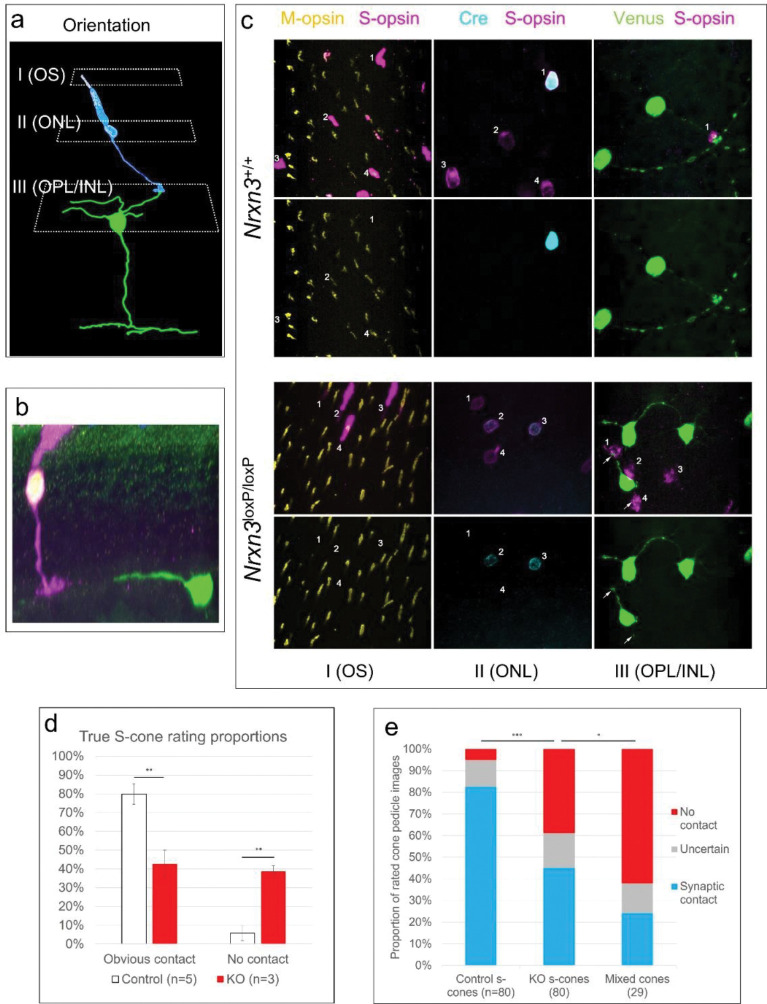
In Nrxn3 KO mice (Nrxn3loxP/loxP, cre+), Nrxn3−/− S-cones often lacked clear synaptic contact with SCBCs, whereas such connections were readily identified in control mice (Figure 2b–c). To confirm that such connections were reliably absent in Nrxn3 KO mice, we acquired images of fluorescently labeled samples from Nrxn3 KO and control mice for quantitative analysis. An image-processing based quantification of this result was confounded by heterogeneous labeling intensity and the presence of apparently passing SCBC dendrites that did not contact cone pedicles. To overcome this limitation, we adopted an analysis strategy designed to be free from bias (Hallgren, 2012): Namely, images of labeled synaptic terminals were isolated, anonymized, and scored by trained raters who were instructed to determine whether each image depicted a synaptic connection between labeled cells (see Methods for details). We reasoned that human error would apply equally to each condition under this blinded analysis strategy.
Figure 2.
S-cone to S-cone bipolar cell (SCBC) synaptic connectivity in Nrxn3 KO mice. a) To quantify synaptic connectivity, we first identified cone identity by imaging S- and M-opsin antibody signals in the outer segment (OS, plane I). Next, the cell bodies of the outer nuclear layer (ONL) were imaged to identify cre-expression in cone cells <image> (plane II). Lastly, cone pedicles and SCBC-dendrites were imaged to identify contacts (plane III). b) Example of a Nrxn3−/− S-cone that does not get contacted by a SCBC-dendrite in close proximity. Magenta: S-opsin, red: cre, green: venus (SCBC). c) The upper panel shows S-cone to SCBC connectivity in a Nrxn3 wild type control mouse. Cone 1 is a cre+ S-cone which is contacted by two SCBCs in the same frame and potentially two further SCBCs which are out of frame. Cone 2 is a mixed cone (M-opsin+, S-opsin+) and cre-. Nearby SCBCs appear to avoid this cone. The lower panel shows a Nrxn3loxp/loxp sample in which cre+ cells are lacking functional α- and β-neurexin 3. Cones 2 and 3 are both true S-cones (S+, M−) and express cre. Nearby SCBCs appear to avoid these Nrxn3−/− pedicles while pedicles from cre- cones 1 and 4 appear to receive at least one SCBC-contact (arrows). Yellow: M-opsin, magenta: S-opsin, cyan: cre, green: venus. d) Rating proportions of contacts or no contacts on a per-animal basis. e) Quantitative analysis of S-cone to SCBC contacts in control S-cones, Nrxn3−/− S-cones, and mixed cones (per-cone basis).
Across the mice tested (3 KO, 2 control), a total of 160 cones were imaged and rated. Of the control S-cones in both wild type (WT) mice and cre− S-cones in Nrxn3 KO mice, 83% were rated as having obvious contacts with SCBCs (66 of 80), compared to 45% for cre+ S-cones in Nrxn3−/− mice (36 of 80; Figure 2e). In comparison, 24% of mixed cones were judged as making contact with SCBCs (7 of 29, but see Discussion). On a per-animal basis (Figure 2d), the proportion of cone terminals judged as clearly forming no possible synapse with SCBCs was significantly higher (p<0.01) for Nrxn3−/− S-cones in Nrxn3 KO mice (38.7 ± 3.0%, SEM; n=3) than for control S-cones from either KO or WT mice (5.7 ± 3.9%, SEM; n=5). Despite the uncertainty of the nature of these contacts as observed using light microscopy, we can conservatively estimate at least 40% of true S-cone contacts with S-cone bipolar cells were eliminated in the Nrxn3−/− KO mouse.
Discussion
Role of Nrxn3 in mammalian S-cones
Here we combined experiments to navigate around the limitations of mouse and TLGS models—likewise taking advantage of their respective benefits—ultimately establishing that Nrxn3 is required for the proper connection between S-cones and their S-cone bipolar cell postsynaptic partners.
In this study we were able to highlight differences in the genetic profiles of S- and M-cone photoreceptors. We presented candidate genes that putatively account for key differences in S- and M-cone photoreceptors, specifically for wiring. We found evidence that Nrxn3 is important for forming or maintaining the first synapse in the S-cone pathway. Previous studies have primarily characterized Nrxn3 as playing various modulatory roles. In studies of neurexins in the mammalian brain, deletion of one or more neurexin types resulted in a diverse spectrum of phenotypes, dependent on the brain area, and the pre-/postsynaptic partner combination, including the assembly of presynaptic machinery (Missler et al., 1998), the stability of postsynaptic receptors (Aoto et al., 2015), the plasticity of axonal processes, and a variety of synaptic activities in a neuronal subtype specific manner (Chen et al., 2017; Hauser et al., 2022). While neurexins are well known for their diverse splice variants, this study, to our knowledge, is the first to document a specific synapse in the central nervous system that is mediated by Nrxn3. Whether the loss of S-cone to S-cone bipolar cell synapse reflects a direct cell-cell interaction role of neurexin or an indirect outcome of synaptic function defect in Nrxn3 KO cells remains to be determined. Regardless, our findings add to the diversity of neurexin functions in the nervous system, by showing a reduction of specific synaptic connections in the outer plexiform layer of the retina.
Differential gene expression analysis of ground squirrel photoreceptors resulted in the identification of S-cone-specific expression of Nrxn3. To understand the potential function of Nrxn3 in mammalian S-cones we used a cre-dependent Nrxn3 flox mouse model combined with a transgenic Opn1sw-cre mouse and an S-cone bipolar cell (SCBC) reporter line (Aoto et al., 2015; Nadal-Nicolás et al., 2020). The cre-dependent deletion of α- and β-neurexin 3 resulted in a loss of S-cone to SCBC contacts in approximately half of cre+ true S-cones (i.e., cones that are S-opsin+ and M-opsin−), indicating an important role for Nrxn3 in this highly specific synapse. Based on previous studies (Applebury et al., 2000; Haverkamp et al., 2005; Nadal-Nicolás et al., 2020), we would not expect SCBCs to form proper contacts with mixed cones. Nevertheless, our trained readers identified 25% of such cones to potentially contact SCBCs. While this result could imply an actual number of unspecific baseline contacts between SCBCs and mixed cones, a more likely interpretation is the presence of false positive cases mis-judged by the trained readers. Notably, post-hoc examination of mixed cones rated as highly likely to make contact with SCBCs revealed that many such cones resemble true S-cones with a very weak M-cone signal (Suppl.Fig.S1). Regardless, deletion of Nrxn3 significantly lowered the proportion of S-cones rated as making proper SCBC contacts, implying that the wiring specificity was disrupted after the deletion of Nrxn3.
Our Nrxn3 loss-of-function study in S-cones focused on young adult mice with an age range of 60 – 70 days, thus we cannot judge if deletion of neurexin 3 prevents synapse formation during development or results in failure of maintenance. If they fail to make initial contacts, it is possible due either to S-cones fail to attract SCBC dendrites or they come close but fail to form synapses. A follow-up study is warranted to investigate S-cone bipolar cell dendrites during earlier timepoints of retinal development. In the light of neural plasticity, it will be imperative to investigate if the number of contacts from other types of bipolar cells is increased, and how it might affect color-contrast coding when the number of S-cone to SCBC contacts is reduced. The incomplete cre recombinase expression in our transgenic Opn1sw-cre line gave us the advantage of having cre+ Nrxn3-KO cone pedicles in close proximity to cre- cones with normal Nrxn3 expression in the same sample (Figure 2c). This was beneficial for determining morphological differences in SCBC contacts side-by-side. However, to assess how disrupting the first synapse of the S-cone pathway affects mouse behavior and global response to light, e.g., in electroretinogram (ERG) recordings, we would need a different cre-line that produces Nrxn3-loss across all S-cones and not just a fraction.
SMART-seq of thirteen-lined ground squirrel photoreceptors
We used the SMART-seq approach to investigate differential gene expression in ground squirrel cone subtypes. Compared to high-throughput droplet-based sequencing methods (Vieth et al., 2019), this approach is cost-effective and allows for targeting healthy, intact, subtype-specific photoreceptors for deeper sequencing. Thirteen-lined ground squirrels were advantageous for our cone-focused study for three main reasons: first, they have a cone-dominant retina. Second, their S- and M-cones are clearly distinguishable since there is no co-expression of M- and S-opsin in the same cell. Third, squirrel neurons are more resilient to damage and provide higher quality material as they showed constantly high cell viability of > 92% after dissociation (determined with propidium iodide staining; data not shown). We speculate this could be due to the cell-protective mechanisms present in this hibernating animal model, which has the genetic program available to withstand stressful conditions like cold temperatures (von der Ohe et al., 2006; Ou et al., 2018). The combination of above-mentioned features enabled us to generate high-quality low-contamination genetic profiles for S- and M-cones and a higher number (58) of differentially expressed genes (DEGs), compared to those described in high-throughput droplet-based sequencing publications (Peng et al., 2019; Yan et al., 2020). While droplet-based sequencing method is ideal for classifying cell types, this single cell picking based SMART-seq method can facilitate the detection of lower expression genes, such as Nrxn3, which has not been detected as a DEG in previous works.
Thus, our dataset provides a valuable roadmap to explore the functions of these DEGs and their cone subtype specific physiology. For example, we identified a higher expression of rod arrestin (Sag) in S-cones but not in M-cones while cone arrestin (Arr3) expression has the opposite pattern (Fig.1e–d). This may underly the difference in the kinetics of S- and M-cone light response, especially the decay phase to which the arrestin molecules contribute (Angueyra and Rieke, 2013). Conversely, the gene Tjp2, specific to M-cones, encodes a gap junction scaffold protein that may be responsible for the M-cone specific cell membrane localization of Cx36 (Li and DeVries, 2004). This localization ensures electrical coupling only occurs between M-cones. Thus, our results highlight the advantage of SMART-seq when investigating a small number of specialized cells, such as S-cones, which account for less than 10% of photoreceptors in many mammalian species (Long and Fisher, 1983; Müller and Peichl, 1989; Nadal-Nicolás et al., 2020). This strategy will also be valuable for targeting postsynaptic SCBCs to uncover the molecular mechanisms involved in synaptic formation and maintenance. This knowledge will aid in developing cell replacement therapies for the retina.
Methods
SMART-seq of ground squirrel photoreceptors
Eyes were enucleated from euthanized ground squirrels by sharp dissection following preparation of the retina and removal of the retinal pigment epithelium. A ~10 mm2 piece of retina was incubated in digestion buffer (5 U/mL papain, 0.667 mg/mL L-cysteine, 1 mM EDTA in Hank’s balanced salt solution (HBSS)) for 20 min at 37 °C. The sample was washed two times with enriched Hibernate-A media (Hibernate-A medium, 2% B27, 1 % streptavidin/penicillin, 0.25 % Glutamax, 0.25 % L-glutamine, 10 mM NaCl) followed by antibody incubation (anti-S-opsin) for 45 min (4 °C). After two washes, the retina was incubated with Donkey-anti-goat antibody for 45 min at 4 °C. The tissue was triturated 10 times using a 1000 μL pipette tip to dissociate cells. The single cell suspension was filtered through a cell strainer (35 μm) and centrifuged to remove debris (2000 × g, 3 min, 4 °C). The pellet was carefully resuspended in 500 μL enriched Hibernate-A and placed on an inverted microscope (Evos cell imaging system). S-opsin-positive cells (S-cones) and negative cells with cone morphology (M-cones) were separately collected using a micromanipulator (Eppendorf TransferMan 4r) and transferred into 8 μL lysis buffer (Clontech SMARTer Ultra Low Input RNA Kit for Sequencing - v3). For each sample, 16–20 single cells were pooled. Samples were flash frozen in dry ice and temporarily stored at −80 °C until used for RNA extraction and library preparation. RNA-extraction of single cell samples and generation of double stranded cDNA was performed using the Clontech SMARTer Ultra Low Input RNA Kit. For the construction of the library, the Low Input Library Prep Kit v2 (Clontech) was used. cDNA quality was analyzed with the Agilent 2100 BioAnalyzer and concentrations were measured with the Qubit dsDNA HS assay. Up to eight samples with different Illumina barcodes were pooled in one lane of a flow cell (Illumina HiSeq 2500; 50 bp read length, single-end mode). All kits were used as instructed by the manufacturers.
Data analysis
Analysis was performed as described (Law et al., 2018). Briefly, we used RefSeq transcriptome and annotation GCF_000236235 to estimate abundance of ground squirrel transcripts using the Kallisto package (Bray et al., 2016) on the NIH high performance computer Biowulf cluster. Resulting output was imported into R (R Core Team, 2022) using the packages rhdf5 (Fischer et al., 2022) and tximport (Soneson et al., 2015) and then normalized and analyzed for differential gene expression using R packages EdgeR (Robinson et al., 2010) and limma (Ritchie et al., 2015).
Immunohistochemistry and image acquisition
Dissected mouse retinae were incubated in PBS with 4 % PFA for 30 min at room temperature. After three washes with PBS for 15 min, the tissue was incubated with primary antibody in blocking solution (PBS with 4 % normal donkey serum, 0.01 % sodium azide, 0.5 % Triton X-100) for 5 days at 4 °C. After washing 5 times (15 min) with PBST (PBS + 0.5 % Triton X-100), the tissue was incubated with secondary antibody (1:500) in blocking solution for 2 days at 4 °C. The samples were washed 5 times with PBST before mounting on a microscope slide. Retinal whole-mounts were imaged with a 60x objective on a Nikon A1R confocal microscope controlled by Nikon NIS-Elements software.
Mouse strains
The transgenic Opn1sw-cre mouse was generated by the NEI Genetic Engineering Core by BAC insertion. Strain generation was identical to that of our previously described Opn1sw-venus mouse (Ishibashi et al., 2022), but with the cre recombinase sequence instead of venus. Cre expression is under control of the Opn1sw-promoter. To quantify the proportion of cre-expressing S-opsin+ cones, we imaged retinal flat-mounts from three Opn1sw-cre mice after staining with S-opsin- and cre-antibody and corresponding fluorescent secondary antibodies. On average, 12.3% of S-opsin+ cells also expressed cre. The S-cone bipolar cell Cpne9-venus mouse was previously described in Nadal-Nicolás et al. (2020). The Nrxn3tm3SudJ mouse was generated by the Südhof lab (Aoto et al., 2015) and obtained from The Jackson Laboratory (strain #014157), and backcrossed over multiple generations to C57BL/6J mice (strain #000664). Mice in this study were both female and male, 60–70 days old.
All experiments and animal care were conducted in accordance with protocols approved by the Animal Care and Use Committee of the National Institutes of Health.
Imaging rating and statistical analysis
From each mouse (5 total: 3 Nrxn3−/− and 2 wild types (WT)), at least 20 S-opsin+ cones were imaged, and each cone was categorized as a “true” S-cone vs. a mixed cone (using the presence vs. absence of M-opsin labeling in the cone outer segment), and S-cones were further divided into Nrxn3 KO vs. control (according to the cre antibody signal in the cone nucleus). From each mouse, at least 18 S-cones were identified. From the acquired z-stack images, sections of the image surrounding the synaptic terminal were extracted that contained only the S-opsin and venus labels, thus isolating context-free images of the S-cone terminal and S-cone bipolar cell dendrites. Cone categorization was confirmed independently by two researchers.
These images were assigned randomized numbers to anonymize the cone categorization and were then shared with volunteer raters (2 participants; see also ref (Hallgren, 2012)) who were first trained on the task—without having been informed of the expected outcome of this analysis—and then asked to score each terminal from 1 (“clearly no contact”) to 5 (“a clear synaptic connection”). Raters were experienced with using ImageJ to view 3D multichannel confocal z-stacks for judging each terminal. Completed ratings were returned to a third expert rater, who noted the rankings for each anonymized image. Final ratings for each image were taken as the average of the two raters’ scores, or in the case of major disagreements (defined as score differences of 2 points or more), the expert rater assigned a replacement score to the still-anonymized images (21 of 195 images). For each mouse, the proportions of S-cone images scored higher than 3.5 were computed for KO vs. control S-cones and compared. KO S-cones were defined as M-opsin−, cre+ cones in Nrxn3loxP/loxP mice, and control S-cones were defined as either M-opsin−, cre− cones in Nrxn3loxP/loxP mice, or M-opsin− cones in wild-type mice regardless of cre labeling. Observed proportions of these highly-rated S-cone images were compared using a one-tailed t-test using Microsoft Excel, where significance was denoted as follows: *: p<0.05, **: p<0.01.
Supplementary Material
Table 1.
Antibodies used in this study. IF: immunofluorescence.
| Antibody | Source | Catalog number | Dilution |
|---|---|---|---|
| Anti-GFP (chicken) | Aves labs | GFP-1020 | IF: 1:100 |
| Anti-S-opsin (goat) | Santa Cruz | sc-14363 | IF: 1:200; Picking: 1:50 |
| Anti-Cre (guinea pig) | Synaptic Systems | 257 004 | IF: 1:250 |
| Anti-M/L-opsin (rabbit) | Millipore | AB5405 | IF: 1:200 |
| Anti-chicken Alexa 488 | Jackson ImmunoResearch | 703-545-155 | IF: 1:500 |
| Anti-goat Alexa 647 | Jackson ImmunoResearch | 705-606-147 | IF: 1:500 |
| Anti-goat Alexa 488 | ThermoFisher | A-11055 | Picking: 1:50 |
| Anti-guinea pig Cy3 | Jackson ImmunoResearch | 706-165-148 | IF: 1:500 |
| Anti-rabbit DyLight 405 | Jackson ImmunoResearch | 711-475-152 | IF: 1:500 |
References
- Angueyra J.M., Kunze V.P., Patak L.K., Kim H., Kindt K., Li W., 2023. Transcription factors underlying photoreceptor diversity. eLife 12, e81579. 10.7554/eLife.81579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angueyra J.M., Rieke F., 2013. Origin and effect of phototransduction noise in primate cone photoreceptors. Nat. Neurosci. 16, 1692–1700. 10.1038/nn.3534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aoto J., Földy C., Ilcus S.M.C., Tabuchi K., Südhof T.C., 2015. Distinct circuit-dependent functions of presynaptic neurexin-3 at GABAergic and glutamatergic synapses. Nat Neurosci 18, 997–1007. 10.1038/nn.4037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Applebury M.L., Antoch M.P., Baxter L.C., Chun L.L., Falk J.D., Farhangfar F., Kage K., Krzystolik M.G., Lyass L.A., Robbins J.T., 2000. The murine cone photoreceptor: a single cone type expresses both S and M opsins with retinal spatial patterning. Neuron 27, 513–523. 10.1016/s0896-6273(00)00062-3 [DOI] [PubMed] [Google Scholar]
- Boucard A.A., Ko J., Südhof T.C., 2012. High Affinity Neurexin Binding to Cell Adhesion G-protein-coupled Receptor CIRL1/Latrophilin-1 Produces an Intercellular Adhesion Complex*. Journal of Biological Chemistry 287, 9399–9413. 10.1074/jbc.M111.318659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bray N.L., Pimentel H., Melsted P., Pachter L., 2016. Near-optimal probabilistic RNA-seq quantification. Nat Biotechnol 34, 525–527. 10.1038/nbt.3519 [DOI] [PubMed] [Google Scholar]
- Chen L.Y., Jiang M., Zhang B., Gokce O., Südhof T.C., 2017. Conditional Deletion of All Neurexins Defines Diversity of Essential Synaptic Organizer Functions for Neurexins. Neuron 94, 611–625.e4. 10.1016/j.neuron.2017.04.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark B.S., Stein-O’Brien G.L., Shiau F., Cannon G.H., Davis-Marcisak E., Sherman T., Santiago C.P., Hoang T.V., Rajaii F., James-Esposito R.E., Gronostajski R.M., Fertig E.J., Goff L.A., Blackshaw S., 2019. Single-Cell RNA-Seq Analysis of Retinal Development Identifies NFI Factors as Regulating Mitotic Exit and Late-Born Cell Specification. Neuron 102, 1111–1126.e5. 10.1016/j.neuron.2019.04.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen E., Sterling P., Boycott B.B., 1990a. Convergence and divergence of cones onto bipolar cells in the central area of cat retina. Philosophical Transactions of the Royal Society of London. Series B: Biological Sciences 330, 323–328. 10.1098/rstb.1990.0202 [DOI] [PubMed] [Google Scholar]
- Cohen E., Sterling P., Boycott B.B., 1990b. Demonstration of cell types among cone bipolar neurons of cat retina. Philosophical Transactions of the Royal Society of London. Series B: Biological Sciences 330, 305–321. 10.1098/rstb.1990.0201 [DOI] [PubMed] [Google Scholar]
- de Wit J., Sylwestrak E., O’Sullivan M.L., Otto S., Tiglio K., Savas J.N., Yates J.R., Comoletti D., Taylor P., Ghosh A., 2009. LRRTM2 Interacts with Neurexin1 and Regulates Excitatory Synapse Formation. Neuron 64, 799–806. 10.1016/j.neuron.2009.12.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emerling C.A., Huynh H.T., Nguyen M.A., Meredith R.W., Springer M.S., 2015. Spectral shifts of mammalian ultraviolet-sensitive pigments (short wavelength-sensitive opsin 1) are associated with eye length and photic niche evolution. Proceedings of the Royal Society B: Biological Sciences 282, 20151817. 10.1098/rspb.2015.1817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Famiglietti E.V., 1981. Functional architecture of cone bipolar cells in mammalian retina. Vision Research 21, 1559–1563. 10.1016/0042-6989(81)90032-8 [DOI] [PubMed] [Google Scholar]
- Fischer B., Smith M., Pau G., Morgan M., van Twisk D., 2022. rhdf5: R Interface to HDF5. R package. [Google Scholar]
- Gomez A.M., Traunmüller L., Scheiffele P., 2021. Neurexins: molecular codes for shaping neuronal synapses. Nat Rev Neurosci 22, 137–151. 10.1038/s41583-020-00415-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallgren K.A., 2012. Computing Inter-Rater Reliability for Observational Data: An Overview and Tutorial. Tutorials in quantitative methods for psychology 8, 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauser D., Behr K., Konno K., Schreiner D., Schmidt A., Watanabe M., Bischofberger J., Scheiffele P., 2022. Targeted proteoform mapping uncovers specific Neurexin-3 variants required for dendritic inhibition. Neuron S0896627322003622. 10.1016/j.neuron.2022.04.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haverkamp S., Wässle H., Duebel J., Kuner T., Augustine G.J., Feng G., Euler T., 2005. The primordial, blue-cone color system of the mouse retina. J. Neurosci. 25, 5438–5445. 10.1523/JNEUROSCI.1117-05.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishibashi M., Keung J., Morgans C.W., Aicher S.A., Carroll J.R., Singer J.H., Jia L., Li W., Fahrenfort I., Ribelayga C.P., Massey S.C., 2022. Analysis of rod/cone gap junctions from the reconstruction of mouse photoreceptor terminals. eLife 11, e73039. 10.7554/eLife.73039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs G.H., Neitz J., Deegan J.F., 1991. Retinal receptors in rodents maximally sensitive to ultraviolet light. Nature 353, 655–656. 10.1038/353655a0 [DOI] [PubMed] [Google Scholar]
- Ko J., Fuccillo M.V., Malenka R.C., Südhof T.C., 2009. LRRTM2 Functions as a Neurexin Ligand in Promoting Excitatory Synapse Formation. Neuron 64, 791–798. 10.1016/j.neuron.2009.12.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolb H., 1995. S-Cone Pathways, in: Kolb H., Fernandez E., Nelson R. (Eds.), Webvision: The Organization of the Retina and Visual System. University of Utah Health Sciences Center, Salt Lake City (UT). [PubMed] [Google Scholar]
- Kryger Z., Galli-Resta L., Jacobs G.H., Reese B.E., 1998. The topography of rod and cone photoreceptors in the retina of the ground squirrel. Vis Neurosci 15, 685–691. 10.1017/s0952523898154081 [DOI] [PubMed] [Google Scholar]
- Law C.W., Alhamdoosh M., Su S., Dong X., Tian L., Smyth G.K., Ritchie M.E., 2018. RNA-seq analysis is easy as 1–2-3 with limma, Glimma and edgeR. F1000Res 5, ISCB Comm J-1408. 10.12688/f1000research.9005.3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., DeVries S.H., 2006. Bipolar cell pathways for color and luminance vision in a dichromatic mammalian retina. Nat Neurosci 9, 669–675. 10.1038/nn1686 [DOI] [PubMed] [Google Scholar]
- Li W., DeVries S.H., 2004. Separate blue and green cone networks in the mammalian retina. Nat. Neurosci. 7, 751–756. 10.1038/nn1275 [DOI] [PubMed] [Google Scholar]
- Long K.O., Fisher S.K., 1983. The distributions of photoreceptors and ganglion cells in the California ground squirrel, Spermophilus beecheyi. Journal of Comparative Neurology 221, 329–340. 10.1002/cne.902210308 [DOI] [PubMed] [Google Scholar]
- Macosko E.Z., Basu A., Satija R., Nemesh J., Shekhar K., Goldman M., Tirosh I., Bialas A.R., Kamitaki N., Martersteck E.M., Trombetta J.J., Weitz D.A., Sanes J.R., Shalek A.K., Regev A., McCarroll S.A., 2015. Highly Parallel Genome-wide Expression Profiling of Individual Cells Using Nanoliter Droplets. Cell 161, 1202–1214. 10.1016/j.cell.2015.05.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mariani A.P., 1984. Bipolar cells in monkey retina selective for the cones likely to be blue-sensitive. Nature 308, 184–186. 10.1038/308184a0 [DOI] [PubMed] [Google Scholar]
- Masland R.H., 2001. The fundamental plan of the retina. Nat Neurosci 4, 877–886. 10.1038/nn0901-877 [DOI] [PubMed] [Google Scholar]
- Missler M., Hammer R.E., Südhof T.C., 1998. Neurexophilin Binding to α-Neurexins: A SINGLE LNS DOMAIN FUNCTIONS AS AN INDEPENDENTLY FOLDING LIGAND-BINDING UNIT*. Journal of Biological Chemistry 273, 34716–34723. 10.1074/jbc.273.52.34716 [DOI] [PubMed] [Google Scholar]
- Müller B., Peichl L., 1989. Topography of cones and rods in the tree shrew retina. J Comp Neurol 282, 581–594. 10.1002/cne.902820409 [DOI] [PubMed] [Google Scholar]
- Nadal-Nicolás F.M., Kunze V.P., Ball J.M., Peng B.T., Krishnan A., Zhou G., Dong L., Li W., 2020. True S-cones are concentrated in the ventral mouse retina and wired for color detection in the upper visual field. Elife 9, e56840. 10.7554/eLife.56840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ou J., Ball J.M., Luan Y., Zhao T., Miyagishima K.J., Xu Y., Zhou H., Chen J., Merriman D.K., Xie Z., Mallon B.S., Li W., 2018. iPSCs from a Hibernator Provide a Platform for Studying Cold Adaptation and Its Potential Medical Applications. Cell 173, 851–863.e16. 10.1016/j.cell.2018.03.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng Y.-R., Shekhar K., Yan W., Herrmann D., Sappington A., Bryman G.S., van Zyl T., Do M.Tri.H., Regev A., Sanes J.R., 2019. Molecular Classification and Comparative Taxonomics of Foveal and Peripheral Cells in Primate Retina. Cell 176, 1222–1237.e22. 10.1016/j.cell.2019.01.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team, 2022. R: A language and environment for statistical computing.
- Ritchie M.E., Phipson B., Wu D., Hu Y., Law C.W., Shi W., Smyth G.K., 2015. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Research 43, e47. 10.1093/nar/gkv007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson M.D., McCarthy D.J., Smyth G.K., 2010. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140. 10.1093/bioinformatics/btp616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shekhar K., Lapan S.W., Whitney I.E., Tran N.M., Macosko E.Z., Kowalczyk M., Adiconis X., Levin J.Z., Nemesh J., Goldman M., McCarroll S.A., Cepko C.L., Regev A., Sanes J.R., 2016. Comprehensive Classification of Retinal Bipolar Neurons by Single-Cell Transcriptomics. Cell 166, 1308–1323.e30. 10.1016/j.cell.2016.07.054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soneson C., Love M.I., Robinson M.D., 2015. Differential analyses for RNA-seq: transcript-level estimates improve gene-level inferences. F1000Res 4, 1521. 10.12688/f1000research.7563.2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Südhof T.C., 2017. Synaptic Neurexin Complexes: A Molecular Code for the Logic of Neural Circuits. Cell 171, 745–769. 10.1016/j.cell.2017.10.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Südhof T.C., 2008. Neuroligins and neurexins link synaptic function to cognitive disease. Nature 455, 903–911. 10.1038/nature07456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vieth B., Parekh S., Ziegenhain C., Enard W., Hellmann I., 2019. A systematic evaluation of single cell RNA-seq analysis pipelines. Nature Communications 10, 4667. 10.1038/s41467-019-12266-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- von der Ohe C.G., Darian-Smith C., Garner C.C., Heller H.C., 2006. Ubiquitous and temperature-dependent neural plasticity in hibernators. J Neurosci 26, 10590–10598. 10.1523/JNEUROSCI.2874-06.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan W., Peng Y.-R., van Zyl T., Regev A., Shekhar K., Juric D., Sanes J.R., 2020. Cell Atlas of The Human Fovea and Peripheral Retina. Sci Rep 10, 9802. 10.1038/s41598-020-66092-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.