Abstract
Retrotransposons and retroviruses shape genome evolution and can negatively impact genome function. Saccharomyces cerevisiae and its close relatives harbor several families of LTR-retrotransposons, the most abundant being Ty1 in several laboratory strains. The cytosolic foci that nucleate Ty1 virus-like particle (VLP) assembly are not well-understood. These foci, termed retrosomes or T-bodies, contain Ty1 Gag and likely Gag-Pol and the Ty1 mRNA destined for reverse transcription. Here, we report a novel intrinsically disordered N-terminal prion-like domain (PrLD) within Gag that is required for transposition. This domain contains amino-acid composition similar to known yeast prions and is sufficient to nucleate prionogenesis in an established cell-based prion reporter system. Deleting the Ty1 PrLD results in dramatic VLP assembly and retrotransposition defects but does not affect Gag protein level. Ty1 Gag chimeras in which the PrLD is replaced with other sequences, including yeast and mammalian prionogenic domains, display a range of retrotransposition phenotypes from wildtype to null. We examine these chimeras throughout the Ty1 replication cycle and find that some support retrosome formation, VLP assembly, and retrotransposition, including the yeast Sup35 prion and the mouse PrP prion. Our interchangeable Ty1 system provides a useful, genetically tractable in vivo platform for studying PrLDs, complete with a suite of robust and sensitive assays, and host modulators developed to study Ty1 retromobility. Our work invites study into the prevalence of PrLDs in additional mobile elements.
Keywords: retrotransposon, virus-like particle, prion-like domain, Saccharomyces cerevisiae
Introduction
Retrotransposons are pervasive across diverse eukaryotes and influence genome evolution and affect host fitness. The budding yeast Saccharomyces cerevisiae contains Ty1–5 long-terminal repeat (LTR)-retrotransposons, with Ty1 as the most abundant element in many laboratory strains (1, 2). LTR-retrotransposons are the evolutionary progenitors of retroviruses; Ty1 elements share many structural hallmarks with retroviral genomic RNA and undergo an analogous replication cycle but lack an extracellular phase. Ty1 is transcribed from LTR-to-LTR and contains two partially overlapping open reading frames: GAG and POL. Ty1 RNA serves as a template for protein synthesis and reverse transcription. Translation of Ty1 POL requires a programmed +1 frameshift near the C-terminus of GAG, resulting in a large Gag-Pol precursor (p199) (3). Like retroviral RNA, Ty1 RNA is specifically packaged into virus-like particles (VLPs) where RNA is present in a dimeric form (4–7). Proteolytic protein maturation occurs within VLPs by a protease (PR) encoded within GAG and POL. Ty1 PR cleaves the Gag-p49 precursor near the C-terminus to generate p45, the capsid protein, and Gag-Pol-p199 to form mature PR, integrase (IN), and reverse transcriptase (RT) (8, 9). Reverse transcription occurs within mature VLPs and, like HIV-1, requires a complex formed between RT and IN (10, 11). Ty1 preferentially integrates upstream of genes actively transcribed by RNA Polymerase III (Pol III) due to interactions between IN and Pol III subunits (12–14).
Ty1 Gag performs the same functions as retroviral capsid and nucleocapsid. Amino acids 159–355 encode NTD and CTD capsid folds, assembling VLPs (15), and a C-terminal domain of Gag displays nucleic acid chaperone (NAC) activity (16, 17). Sequences in the Ty1 RNA encoding the Gag protein are required for packing, dimerization, and reverse transcription (3). The N-terminal region of Gag has unknown function, and it is not known whether it is required for transposition.
While several steps of retrotransposon life cycles have been investigated, it is not wellunderstood how their RNA genomes and protein machinery associate within the cellular milieu to facilitate VLP assembly and replication. Retroviral particle assembly often occurs in subcellular domains, referred to as “viral factories” or “viral inclusions” (18, 19). The sites of Ty1 VLP assembly are cytoplasmic foci termed retrosomes, or T-bodies, which contain Ty1 RNA, Gag, Gag-Pol, and perhaps additional cellular proteins (20–22). What drives the biogenesis of retrosomes is not understood. Mounting evidence suggests liquid-liquid phase separation (LLPS) underlies many examples of membraneless compartments (23, 24). Aggregation-prone proteins that drive LLPS have overlapping properties with prions, and both are implicated in age-related disease (25–34). Spontaneous demixing in these systems is often facilitated by intrinsically disordered domains, multivalent proteins, and scaffolding around nucleic acids. Indeed, prion-like and LLPS mechanisms provide intriguing models for retroelement assembly steps. Ty1 retrosomes contain Ty1 RNA and Gag oligomers associated with the RNA. Several viruses utilize LLPS in replication and assembly, including rabies virus (35), influenza A (36), herpes simplex virus 1 (37), measles virus (38), HIV-1 (39), and SARS-CoV-2 (40). Also, the human retrotransposon LINE-1 has been reported to phase separate in vitro (41). Here, we present evidence that the Ty1 Gag protein contains a prion-like domain required for VLP assembly and transposition, raising the possibility that Ty1 Gag facilitates prion-like or phase separating behaviors within retrosomes.
Results
Bioinformatic analyses reveal a prion-like domain in Ty1 Gag.
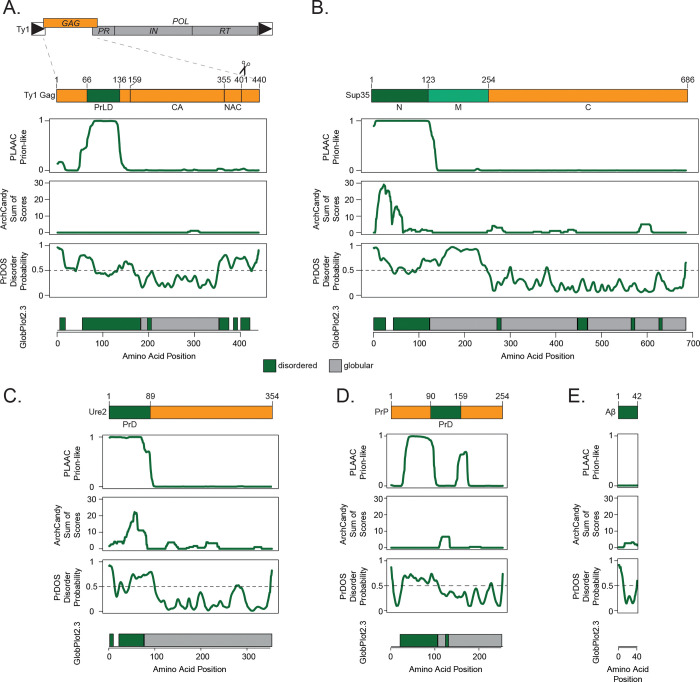
Ty1 Gag contains several protein features, including capsid and nucleic acid chaperone domains (15, 17). The N-terminal region of the protein, meanwhile, is predicted to be unstructured and does not have previously reported function. We analyzed Ty1 Gag (Fig. 1A) and Gag-Pol (Fig. S1) using several bioinformatic tools designed to predict protein disorder, amyloidogenic secondary structures, and amino acid composition similarity to known yeast prions (42–45). For comparison, we ran the well-studied yeast prions Sup35 and Ure2, the mouse prion protein PrP, and Alzheimer’s disease-associated human Aβ1–42 through the same bioinformatic analyses (Fig. 1B–E). Ty1 Gag contains a 71-amino acid domain with strikingly similar amino acid composition to yeast prions in its disordered N-terminus, comparable to Sup35 and Ure2. This Gag prion-like domain (PrLD) is predicted to be unstructured by AlphaFold (46) and no published structures of the region are available, similar to canonical prions (15, 47–50) (Fig. S2). Given the computational predictions and the requirement for Gag in forming Ty1 retrosomes, we further investigated prionogenic properties of the Gag PrLD, which we define as amino acid residues 66–136.
Fig 1.
The Ty1 retrotransposon Gag contains a prion-like domain. Schematic of the Ty1 retrotransposon gene organization, with a detailed view of domains of the Gag protein (A), yeast prion Sup35 (B), yeast prion Ure2 (C), mouse prion protein (PrP) (D), and human amyloid beta (Aβ) (E); PrLD = prion-like domain, capsid domain (CA) and nucleic acid chaperone domain (NAC) are defined in (15). Below are bioinformatic analyses of each protein aligned with the schematic above: yeast prion-like amino acid composition (PLAAC), predicted amyloidogenic regions (ArchCandy), predicted protein disorder (PrDOS), predicted disordered (green) and globular (grey) regions (GlobPlot2.3).
Prionogenic properties of the GagPrLD.
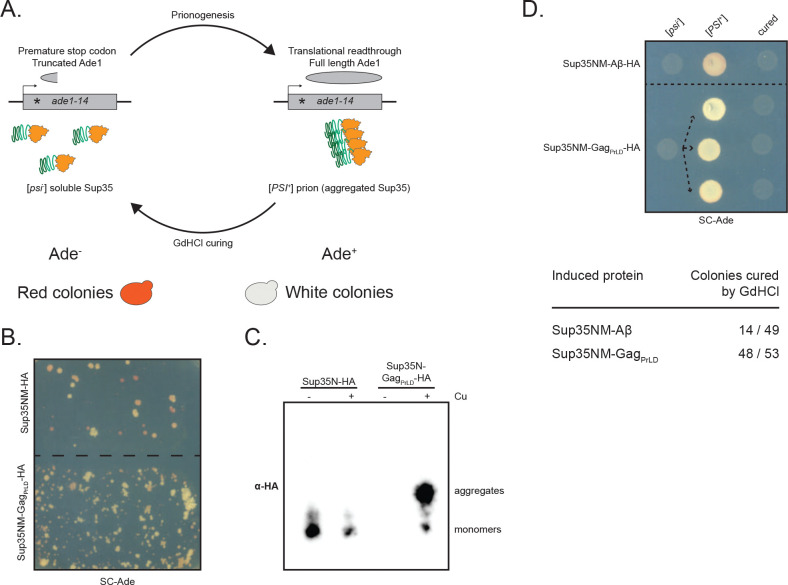
We used a well-characterized Sup35-based in vivo reporter system to assess the ability of the Gag PrLD to promote prionogenesis in a yeast strain harboring a mutant allele of the adenine biosynthesis gene, ade1–14, which contains a premature stop codon (Fig. 2A) (51–53). Soluble Sup35 functions as a translation termination factor, resulting in a truncated non-functional Ade1 protein. Yeast fails to grow on media lacking adenine and appears red due to the buildup of a metabolic intermediate. However, formation of a prion state (termed [PSI+]) aggregates Sup35 away from the ribosome, allowing for translational readthrough. This can be detected by adenine prototrophy and yeast colonies appearing white. Fusion of the PrLD of interest to the Sup35 N or NM domains promotes prion nucleation and has previously been used to study mammalian PrP and Aβ (53). Expression of Sup35NM-GagPrLD fusion under the CUP1 copper-inducible promoter stimulates prionogenesis, as detected by increased papillation on SC-Ade when compared to the reporter alone (Fig. 2B). Growth is copper responsive, however, we found that the Sup35N reporter construct displays a high background growth independent of induction (Fig. S3A–B). We next biochemically monitored prion aggregation using semi-denaturing detergent-agarose gel electrophoresis (SDD-AGE) (54). GagPrLD fusions formed large, slow-migrating, copper-inducible aggregates with both Sup35N (Fig. 2C) and Sup35NM (Fig. S2C) above reporter alone. Finally, we verified prion nucleation specifically, as opposed to colony growth due to accumulating suppressor mutations, by curing colonies of the prion after passaging cells on guanidine hydrochloride (GdHCl) (55, 56). Representative cells are shown for the naïve [psi−], induced [PSI+], and cured states for Sup35NM fusions to GagPrLD or positive control Aβ, both HA-tagged (Fig. 2D) and untagged (Fig. S2D). A large fraction of Sup35NM-GagPrLD Ade+ colonies were curable by GdHCl.
Fig 2.
GagPrLD nucleates a Sup35-based prion reporter. (A) Schematic of the prionogenesis assay using the ade1–14 allele containing a premature stop codon. Soluble Sup35 terminates translation at the premature stop codon, yielding a non-functional, truncated Ade1 (N-succinyl-5-aminoimidazole-4-carboxamide ribotide synthetase); yeast cannot grow on media lacking adenine (SC-Ade) and a red pigment develops. Sup35 aggregated into the prion state allows for translational readthrough and production of functional Ade1; yeast grow on SC-Ade and appear white. (B) Qualitative prionogenesis of Sup35NM fusions; growth on SC-Ade indicates either a suppressor mutation or [PSI+] prionogenesis. Expression of Sup35 fusions were induced with 150 μM CuSO4. A representative image of at least 3 experiments is shown. (C) SDD-AGE analysis of Sup35N-HA with and without GagPrLD fusion. Expression of Sup35 fusions were induced with 100 μM CuSO4. Monomers and high-molecular weight aggregates of chimeric proteins were detected with anti-HA antibody. A representative image of at least 3 experiments is shown. (D) Curing of Ade+ colonies by guanidine hydrochloride (GdHCl) of Sup35NM-HA chimeras. One [psi−] Sup35NM-Aβ fusion control strain is shown induced to [PSI+] and cured. Three independent inductions of a [psi−] Sup35NM-GagPrLD fusion are shown induced to [PSI+] and cured. [PSI+] yeast grow on SC-Ade while [psi−] and cured yeast do not. The table below shows the guanidine curability of Ade+ colonies induced by chimeric constructs.
The GagPrLD is required for Ty1 transposition.
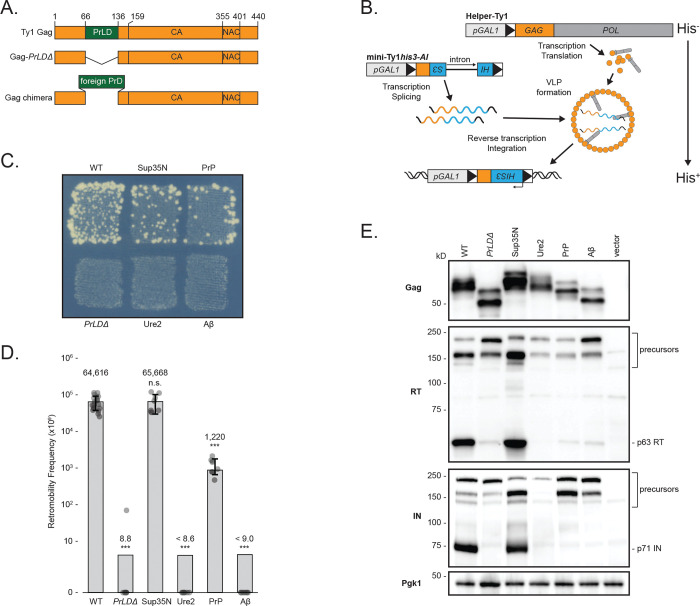
Given that the GagPrLD promotes prionogenesis of a Sup35-based reporter, we investigated its functional role in Ty1 transposition. In a Saccharomyces paradoxus strain lacking genomic Ty1 elements (57, 58), we first deleted the PrLD from Gag in a Ty1 element provided on a plasmid and tagged with the robust and sensitive his3-AI retrotranscript indicator gene (59) (Fig. 3A). This marker contains a mutant his3 gene split by an antisense artificial intron (AI) that is inserted at the 3’ untranslated region of Ty1 in the opposite transcriptional orientation. The AI is in the correct orientation to be spliced only in Ty1his3-AI RNA; cDNA reverse transcribed from this product results in a functional HIS3 allele. Insertion into the genome, either by integration or recombination, allows cells to grow on media lacking histidine. The frequency of His+ prototrophy is a direct measure of Ty1his3-AI retrotransposition or cDNA recombination, collectively known as retromobility. Deletion of the GagPrLD in a complete Ty1his3-AI element overexpressed under the GAL1 promoter completely abolished retromobility (Fig. S4A), despite retaining normal Gag protein levels (Fig. S4B). However, the PrLD region of Gag contains cis-acting RNA signals required for efficient reverse transcription (60, 61). To distinguish between a functional role in retrotransposition of the PrLD in the Gag protein versus the role of the RNA sequences that encode for the PrLD, we used a two-plasmid system to separate Ty1 RNA and protein functions (Fig. 3B). A helper-Ty1 encodes a functional mRNA, providing protein products, but lacks a 3’ LTR thus disrupting cis-acting signals required for reverse transcription. Mini-Ty1his3-AI lacks complete open-reading frames (ORFs) but contains cis-acting signals for dimerization, packaging, and reverse transcription of mini-Ty1his3-AI RNA (61, 62). Retromobility is monitored through the his3-AI reporter. In the two-plasmid assay, deletion of the GagPrLD also inhibits retromobility (Fig. 3C–D), despite producing normal levels of Gag protein (Fig. 3E), confirming a critical contribution from the PrLD in the Gag protein to retromobility.
Fig 3.
Ty1 Gag chimeras containing known PrDs produce stable Gag but have a range of transposition and proteolytic maturation phenotypes. (A) Schematic of Ty1 Gag constructs. The Ty1 Gag PrLD is intact in wildtype (WT), deleted in PrLDΔ, and replaced with known PrDs in the chimeras. (B) Schematic illustrating the two-plasmid system separating Ty1 RNA and protein functions. Helper-Ty1 encodes a functional mRNA, providing protein products, but lacks a 3’ LTR thus disrupting cis-acting signals required for reverse transcription. Mini-Ty1his3-AI lacks complete ORFs but contains cis-acting signals for dimerization, packaging, and reverse transcription of mini-Ty1his3-AI RNA. The his3-AI indicator gene detects retromobility of mini-Ty1HIS3 cDNA. (C) Qualitative retromobility of chimeric Gag constructs in the two-plasmid system. Colony growth on a medium lacking histidine indicates a retromobility event. A representative image of at least 3 replicates is shown. (D) Quantitative mobility assay of galactose-induced cells. Each bar represents the mean of at least eight independent measurements, displayed as points, and the error bar ± the standard deviation. Error bars are omitted for PrLDΔ, Ure2, and Aβ chimeras that did not transpose; one retromobility event was observed in one replicate of PrLDΔ. Adjusted retromobility frequency is indicated above the bars. For Ure2 and Aβ, frequencies are indicated as less than the calculated frequency if one retromobility event had been observed. Significance is calculated from a two-sided Student’s t-test compared with WT (n.s. not significant, ***p < 0.001. Exact p-values are provided in Supplementary Table 1). (E) Protein extracts prepared from galactose-induced cells expressing the indicated Gag constructs in the two-plasmid system were immunoblotted for the protein indicated on left. Polypeptide precursors are bracketed and mature RT and IN sizes are noted on right. Pgk1 serves as a loading control. Migration of molecular weight standards is shown alongside the immunoblots. A representative image of at least 3 replicates is shown.
Ty1 mobility of Gag chimeras containing foreign PrLDs.
To better understand the nature of the PrLD’s contribution to retromobility, we asked whether the GagPrLD sequence is uniquely capable of facilitating retromobility. Since the GagPrLD has prionogenic properties and sequence similarity to prions, we created chimeric Ty1 Gags in which the PrLD is replaced with prion domains from well-studied prions and aggregating proteins (Fig. 3A). We chose the yeast prions Sup35 and Ure2, the mouse prion protein PrP, and the Alzheimer’s disease-associated human Aβ1–42 using domains predicted computationally (Fig. 1) (52, 53, 63). Chimeric Ty1 elements on the helper-Ty1 plasmid were co-expressed with mini-Ty1his3-AI, and the level of Ty1 mobility was determined. Remarkably, substitution of the GagPrLD with the prion domain from yeast Sup35 or mouse PrP supported Ty1 retromobility in qualitative (Fig. 3C) and quantitative retromobility assays (Fig. 3D). GagSup35N retromobility is not significantly different from wildtype, whereas GagPrP is an order of magnitude lower, although still readily detectable on a qualitative plate assay. Replacing the PrLD sequence disrupts RNA signals which is reflected in the single plasmid assay, in which GagSup35N and GagPrP chimeras have dramatically reduced retromobility (Fig. S4A), despite producing similar Gag protein levels (Fig. S4B), highlighting the importance of separating protein and RNA function with the two-plasmid assay.
Retromobility measured as the frequency of His+ prototrophs formed from his3-AI tagged elements includes both new chromosomal integrations likely created via retrotransposition, and recombination of the spliced cDNA with homologous sequences present on the mini-Ty1his3-AI plasmid. To assess whether the chimeras support retrotransposition or merely recombination, we distinguished the two by monitoring histidine prototrophy after segregating the helper and mini-Ty1his3-AI plasmids (Fig. S4C). In our strain background with the wildtype two-plasmid system, 4% of retromobility events were due to recombination with either of the plasmids. The GagSup35N and GagPrP chimeras had modestly increased recombination events, although only GagSup35N reached statistical significance (p=0.024) (Fig. S4D). We conclude that the Gag chimeras support de novo retrotransposition and cDNA recombination remains a minor pathway (64, 65).
Effect of GagPrLD chimeras on Ty1 protein level and maturation.
The result that the GagPrLD can be replaced by foreign prion sequences indicates its function is not unique to the PrLD sequence and may be the same as provided in aggregation-prone proteins. However, not all the disordered domains tested in Gag chimeras supported transposition. Ty1 chimeras containing the domains from yeast Ure2 or human Aβ did not transpose (Fig. 3C–D). All the chimeric Gags were expressed at similar levels (Fig. 3E), arguing against different transposition phenotypes due to effects on protein stability from the foreign prion domains. The substituted prion domains are of various sizes, and Gag chimeras had predicted electrophoretic mobilities. Gag proteolytically matures from p49 to p45 and is subject to post-translational modifications, often resulting in multiple bands observed by western blot (3). To determine whether the Gag chimeras affected protein maturation, we assessed the relative levels of mature RT and IN by western blotting with antibodies specific to each protein. Deletion of the PrLD results in dramatically reduced mature RT and IN levels (Fig. 3E). The GagSup35N chimera transposed as well as wildtype and produced equivalent levels of mature RT and IN. The transposition-deficient chimeras, GagUre2 and GagAβ, have very reduced levels, comparable to GagPrLDΔ. Interestingly, GagPrP supports transposition, although reduced from wildtype, and has low levels of mature RT and IN. These results raise the possibility that Gag chimeras can block PR function and production of mature RT and IN that are essential for Ty1 mobility.
Ty1 GagPrLDΔ and Gag chimeras fused to GFP affect aggregation and localization.
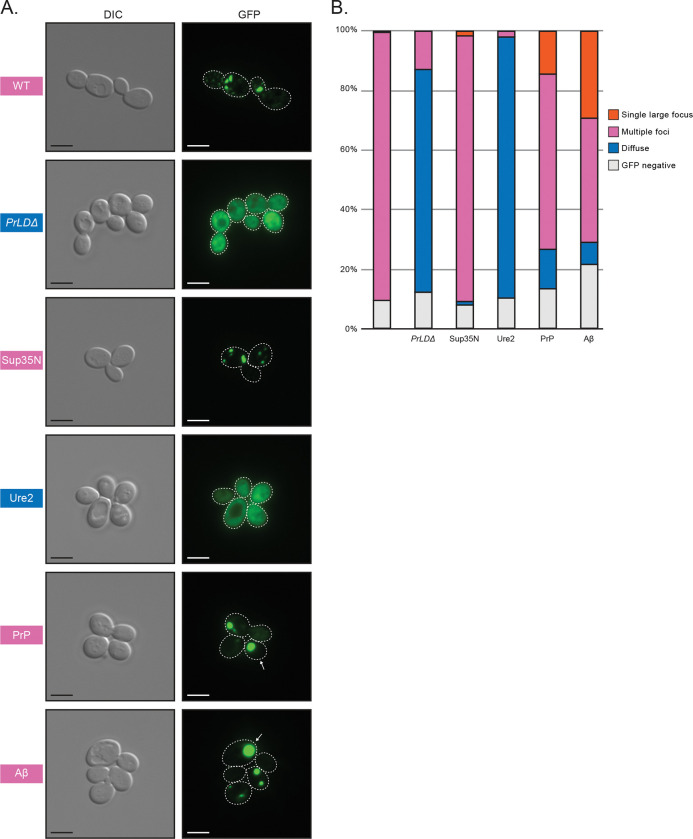
Proteolytic maturation of RT and IN via PR occurs within VLPs (66), which are believed to be assembled in retrosomes (20–22). Gag fused to green fluorescent protein (GFP) has been used as a reporter for retrosome assembly and location (67), therefore we examined formation of cytoplasmic foci of wildtype, mutant, and chimeric Gag-GFP in the Ty-less background. Wildtype Gag-GFP fusions formed discrete cytoplasmic foci, as previously reported using this construct, but deleting the PrLD resulted in diffuse localization throughout the cytoplasm (Fig. 4). We found that a 24 hr galactose induction, shorter than 48 hr-induction used above, was ideal for live-cell microscopy and GFP-detection as yeast cultures are in log-phase growth (Fig. S5). 24 hr induced GagSup35N formed similarly discrete foci patterns as wildtype Gag, whereas GagUre2 had diffuse localization similar to GagPrLDΔ. GagPrP supports transposition and predominately formed foci similar to wildtype, but also had a modest fraction of cells containing a visually distinct fluorescent morphology that appears as a single, large, very bright focus. Ty1 GagAβ does not transpose, yet formed foci and an even larger fraction of cells contained these single, large foci. Forming Gag-GFP foci correlates with a requirement for transposition but, as GagAβ shows, is not sufficient.
Fig 4.
Foci detected in cells expressing wildtype Gag, the GagPrLDΔ mutant, and Gag-PrLD chimeras fused to GFP. (A) Live-cell yeast fluorescence microscopy of strains expressing chimeric Gag-GFP after 24 hr galactose induction. Normaski (DIC) and GFP channels are shown with cell outlines added to GFP channels based on DIC images. The strain labels are colored to match the most common foci observed. White arrows indicate cells with a single large focus. Scale bars represent 5 μm. (B) Quantitation of categories of foci observed as a percentage in at least 300 cells. The multiple foci category includes cells with multiple large foci, one or more small foci, or a combination of both sizes. Cell counts are provided in Supplementary Table 2.
In addition, we investigated the structures formed by Gag-GFP chimeras in fixed yeast cells by thin section transmission electron microscopy (TEM) (Fig. S6) using methods similar to those used for detecting Ty1 VLPs (22). Wildtype Gag-GFP produced electron-dense structures that appear similar to VLPs but look incomplete or incorrectly assembled, lacking a circular shell with a hollow interior. GagPrLDΔ did not form any VLP-like structures detectable in micrographs. The GagSup35N strain produced tubular or filamentous structures, also not resembling proper VLPs. And strikingly, the GagAβ strain formed large densities in defined regions of the cell, instead of clusters of particles or filaments across the cytoplasm, perhaps corresponding to the single large foci seen by fluorescent microscopy. These results suggest that Gag-GFP can reveal severe assembly defects as evidenced by GagPrLDΔ but GFP may confer aberrant VLP assembly properties when wildtype or chimeric Gag-GFP fusions are produced in cells.
The Ty1 GagPrLDΔ and Gag chimeras affect VLP assembly.
To evaluate VLP assembly in the chimeras using the two-plasmid system, we examined Gag sedimentation profiles of yeast lysate run through a 7–47% continuous sucrose gradient, as previously reported (15, 58, 68). Wildtype VLPs accumulated in more dense sucrose fractions near the bottom half of the gradient, with peak fractions indicated by a bar (Fig. 5). GagPrLDΔ appears unable to assemble complete VLPs, as Gag in these mutants accumulated in less dense sucrose fractions near the top of the gradient. The transposition-competent chimeras GagSup35N and GagPrP had similar sedimentation profiles as wildtype, whereas transposition-deficient GagUre2 accumulated near the top of the gradient like GagPrLDΔ. GagAβ does not support retrotransposition, but peaked in similar fractions as wildtype, although somewhat more broadly distributed across the gradient.
Fig 5.
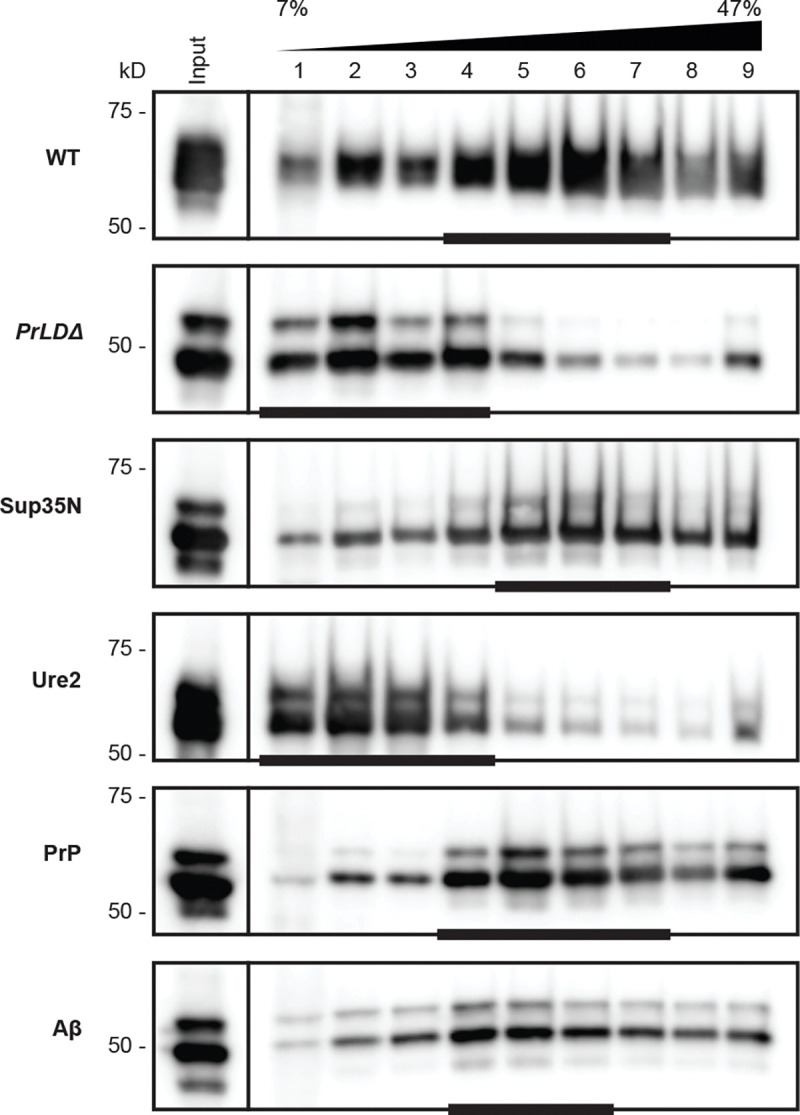
Transposition-incompetent Gag chimeras disrupt VLP assembly. Protein extracts from galactose-induced yeast cells (Input) were fractionated over a 7–47% continuous sucrose gradient and immunoblotted for Gag. Expression plasmids and molecular weight standards are noted alongside the blots. The bars at the bottom of blots denote peak Gag fractions containing more than 1/9 of the Gag signal across the gradient, as determined by densitometric analysis. A representative image of at least 3 replicates is shown.
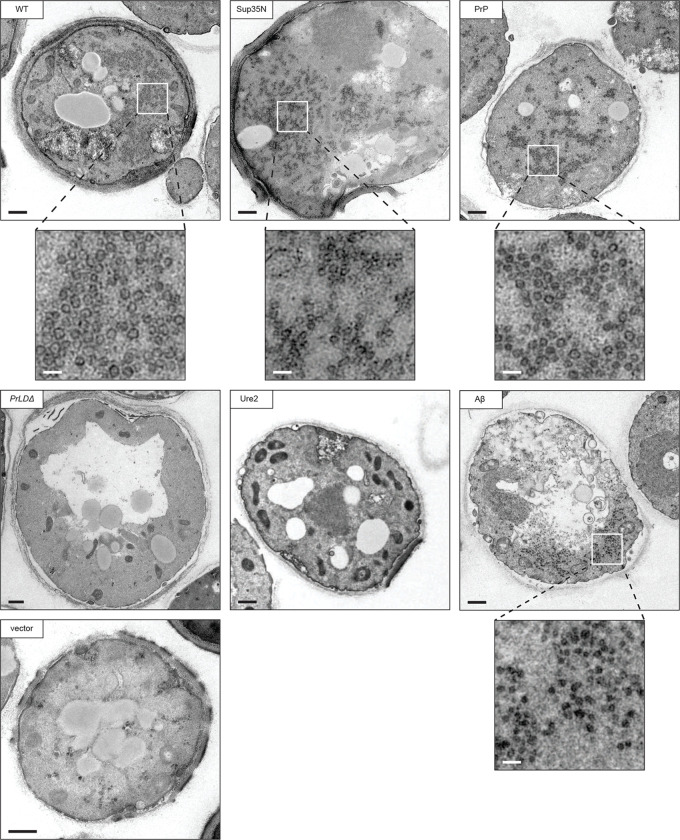
To further examine the VLPs assembled by each chimera, we visualized thin sections of fixed yeast cells by TEM. Cells overexpressing the wildtype two-plasmid Ty1 system produced large clusters of VLPs (Fig. 6). VLPs are characteristically round with an electron dense shell and their interior appears hollow in micrographs. Importantly, these particles were not observed in the parental yeast strain expressing empty vectors. Ty1 VLPs are heterogeneously sized and are approximately 30–80 nm in diameter, based on previous measurements of purified particles (69, 70). In thin section TEM, particles may be in different Z-planes when sectioned, therefore masking the diameter of a roughly spherical particle, and preventing quantitative particle size data collection from thin section TEM. With this limitation in mind, we measured particle diameters from several cells in multiple micrographs to estimate an approximate size, and found wildtype particles ranging from 40–80 nm, with a median diameter of 59 nm (Fig. S7), largely agreeing with previous reports of purified particles.
Fig 6.
Transposition competent Gag chimeras support VLP production. Thin-section TEM of galactose-induced cells expressing Gag chimeras. Representative cells are shown, those containing VLP clusters include zoomed in cutouts to highlight VLPs. The black bars represent 500 nm, the white bars represent 100 nm.
We did not observe any cells producing VLPs in the GagPrLDΔ mutant, in agreement with GagPrLDΔ-GFP imaging and sucrose sedimentation profiles. Taken together, these data lead us to conclude that the PrLD is required for Ty1 VLP assembly. The transposition-deficient GagUre2 chimera also did not assemble VLPs as monitored by thin section TEM, again agreeing with sucrose sedimentation results. The two transposition-competent chimeras, GagSup35N and GagPrP, assembled VLPs similar in size and appearance to wildtype. These chimeras also produced large numbers of particles in each cell, although consistently appearing somewhat more dispersed throughout the cell than wildtype particle clusters. Interestingly, GagAβ does not support retrotransposition, but has a similar sucrose sedimentation profile as wildtype, suggesting it may assemble particles that are defective for transposition. In thin section TEM, we observed particles in cells expressing GagAβ that are visually distinct from wildtype. The most striking difference is that these particles do not have the characteristic hollow center and instead appear electron-dense throughout. They are smaller than wildtype with a median diameter of 42 nm (Fig. S7), and, like the GagSup35N and GagPrP chimeras, are produced in large numbers of particles but are dispersed throughout the cell. Together, these results illustrate the robust and flexible nature of VLP assembly. However, our data also underscore the requirement for PrLD functionality as yeast and mammalian Gag-prionogenic chimeras form VLPs in vivo whereas the GagPrLDΔ mutant does not.
Discussion
The data presented here permit several conclusions about prionogenic domains, the functional organization of Ty1 Gag, and VLP assembly. Our results demonstrate that the Ty1 Gag protein contains a novel prion-like domain that is required for VLP assembly and retrotransposition. The GagPrLD has intrinsic prionogenic properties demonstrated by a cell-based Sup35 reporter assay, and its function in Ty1 transposition can be replaced by certain yeast and mammalian bona fide prion domains. Our findings also raise interesting unresolved questions about sequence constraints of PrLDs and how widespread PrLD functions are across retroelements. Finally, this work suggests using Ty1 as an in vivo screening platform to study intrinsically disordered domains.
Prion properties of the Ty1 GagPrLD.
We have examined prionogenic properties of the Ty1 GagPrLD using an established cell-based assay in which the newly discovered GagPrLD is fused to the N- and NM- domains of Sup35. Nonsense readthrough is measured by auxotrophic growth and colony color, aggregate formation is monitored biochemically with SDD-AGE, and curability is assessed after GdHCl treatment. It will be informative to further characterize prionogenic properties of the Ty1 GagPrLD using additional assays on various GagPrLD fusion constructs, including fused to Sup35C, and measuring binding of the amyloid-sensitive dye thioflavin-T, non-Mendelian inheritance, aggregation in SDD-AGE, and GFP localization patterns (71, 72).
RNA-contributions to GagPrLD function.
To disentangle protein-level effects of mutations in the Ty1 Gag PrLD from mutations of cis-acting RNA sequences, we separated RNA and protein function in a two-plasmid system. Retromobility is considerably lower in the two-plasmid system (Fig. 3C) than a single-plasmid expressing the intact transposon (Fig. S4A). Nonetheless, the two-plasmid system provides a wide dynamic range allowing for sensitive measurement of the impact of GagPrLD chimeras on retromobility. The GagSup35N chimera restored retromobility in the two-plasmid system but had a severe retromobility defect in the single-plasmid assay. Based on our current knowledge of functional contributions from Ty1 cis-acting RNA sequences to retrotransposition, this is likely due to disruption of the pseudoknot sequences in the RNA region that encodes for the PrLD (60, 61). It may be possible to engineer an equivalent pseudoknot sequence in the Sup35N-encoding RNA and restore retromobility in a single-plasmid GagSup35N chimera. Our chimeric proteins may provide a useful platform to interrogate RNA requirements.
Sequence requirements of the Ty1 GagPrLD.
We replaced the GagPrLD with exogenous prion domains selected based on computational predictions and the literature. We chose the entirety of Aβ1–42 and the complete N-terminal domain of Sup352–123. We introduced the highest scoring 60 amino acid stretch predicted by PLAAC, Ure217–76, which is within the established prion domain reported as the first 89 amino acids (52, 63). The infectious PrP 27–30 isoform initially isolated is roughly 142 amino acids long and spans from approximately residues 90 to 230 (73), but shorter truncations still display prion phenotypes (74–76) and PrP90–159 is sufficient to induce prionogenesis in a yeast-based assay (53). The PrP121–231 fragment is soluble, and its structure has been determined using solution NMR (49, 77). We introduced PrP90–159 as a Ty1 Gag chimera based on prior success in yeast. It will be interesting to examine other regions of PrP for function when present in Gag.
The sequence features constraining Ty1 PrLD function are not yet well-defined. Intriguingly, both the transposition-competent Gag chimeras (Sup35 and PrP) are from proteins with oligopeptide repeats associated with prionogenesis (78, 79). However, the PrP sequence introduced as a Ty1 Gag chimera in this study does not contain these repeats. Moreover, the Ty1 GagPrLD does not have equivalent repeats of 8–10 amino acids. Instead, like other reported prion domains, the GagPrLD is Q/N-rich and is depleted of charged residues. Additionally, a large number of prolines in the GagPrLD likely prevents secondary structure formation and starkly contrasts with the highly alpha-helical folding of the Gag capsid domain (15, 68). Further investigation will be required to understand the sequence parameters, such as length, amino acid composition, charge, or oligorepeats, that govern function of the GagPrLD. Transposition-deficient GagPrLD chimeras may be analyzed by reversion analysis to select mutations that restore transposition. Characterizing the revertants could reveal incompatibility with PR or other Pol proteins, rather than early VLP assembly steps.
Interaction of the GagPrLD with the host and environment.
Many host genes have been identified that activate or restrict Ty1 transposition (80–83), and examining genetic interactions with the GagPrLD may provide regulatory insights. Prion domains have been proposed to be protein-specific stress sensors that allow cells to respond to environmental conditions (84–86). Substituting the Gag PrLD may therefore change Ty1 regulation and create genetic interaction partners selective for specific Gag chimeras. For example, modulators of Sup35 prionogenesis could specifically modulate GagSup35N but not wildtype Ty1. Conversely, Gag chimeras may no longer be subject to regulation by Ty1 modulators.
Gag chimeras reveal varied deficiencies across the Ty1 life cycle.
Different GagPrLD chimeras had different phenotypes across the Ty1 life cycle. GagPrP supported retromobility, although less well than wildtype or GagSup35N. Whereas GagPrP produces VLPs that appear to have wildtype morphology by TEM (Fig. 6) and have similar sedimentation profiles to wildtype (Fig. 5), GagPrP accumulates low levels of mature RT and IN (Fig. 3E). This could indicate an incompatibility of GagPrP as a substrate for PR. Another possibility is that GagPrP VLPs inefficiently incorporate Gag-Pol or are partially defective in ways not detectable by TEM or sedimentation. The reduced retromobility of GagPrP may be explained by impaired RT and IN protein maturation. Meanwhile, GagAβ produces particles that do not support retrotransposition or Pol maturation. These particles lack the characteristic hollow center observed in TEM of wildtype VLPs (Fig. 6) and are noticeably smaller in diameter (Fig. S7). These observations highlight that VLP assembly is robust but underscores the point that simply assembling particles is not sufficient for transposition and that assembling correct VLPs is required for proteolytic maturation. It will be informative to measure packaging of the mini-Ty1 RNA into chimeric VLPs. Ultimately, cDNA synthesis requires both the mature enzymes and the RNA substrate to be present in VLPs. Our sedimentation and TEM results presented here build upon previously published sedimentation experiments (15, 58, 68), and strengthen the value of sedimentation as a proxy for VLP assembly. Nonetheless, the value of TEM is exemplified by the GagAβ chimera, which sediments similarly to wildtype but TEM reveals aberrant particle morphology.
Gag-GFP fusions have been used as a proxy for Ty1 retrosomes (67), although we have not formally tested for Ty1 RNA co-localization in our specific system. We used a previously published wildtype GFP-fusion construct that contains the mature Gag (p45) and not a full-length element. The utility of Gag-GFP is shown by the cellular mislocalization observed in Gag chimeras. However, GFP is a 26 kD protein and fusion impaired proper VLP formation (Fig. S6), perhaps interfering with Gag-Gag contacts that must be made to assemble the complete particle structure. Examining the PrLD fused to GFP alone, without the full Gag protein, or testing a Gag truncation that lacks the NAC domain, will indicate the minimal region that promotes foci formation and if RNA recruitment is required. Ty1 Gag contains a NAC and binds Ty1 RNA, but also binds diverse RNAs in vitro and cellular mRNAs associate with Ty1 VLPs (16, 17, 62, 87–89). Whether Ty1 RNA, specifically, is required to form foci or to nucleate VLP assembly, or if there is an RNA requirement at all, will require further study. It remains to be determined whether the GFP foci and VLP nucleation site is associated with any subcellular locales, as has been previously proposed at the endoplasmic reticulum (67).
The Gag-GFP foci may mature over time, as an increased percentage of GagPrLDΔ cells had foci after 48 hr of induction compared to 24 hr. These foci are proxies for retrosomes and therefore represent an early step of the Ty1 life cycle preceding VLP assembly, protein maturation, and transposition. After 48 hr of galactose induction, cultures enter stationary-phase growth and have higher levels of GFP-negative cells which often appear to have a wrinkled, potentially senescent morphology (Fig. S5). We, therefore, chose to examine Gag chimeras by fluorescent microscopy after 24 hr, but our pilot experiments at 48 hr anecdotally suggested more bright, single large foci. This observation would be consistent with a kinetic component to retrosome formation and may represent a progression from LLPS towards hydrogel formation. Whether such a gel would be an irreversible phase that is unable to dissolve and proceed with VLP formation remains to be determined.
Does the Ty1 retrosome constitute a phase-separated compartment?
Wildtype cells assemble discrete VLPs that can be found throughout the cell but are often observed in a particular region of the cytoplasm, and even the wildtype Gag-GFP assembled discrete structures, observed by TEM. However, the GagAβ-GFP strain produced large densities that may correspond to large foci observed by fluorescence microscopy. These assemblies would be consistent with LLPS compartments containing high concentrations of Gag-GFP that stall and cannot complete VLP assembly; however, we have not examined LLPS properties such as concentration-dependence, droplet merging, or internal mixing (23). Prion-like domains can drive formation of a gradient of assemblies, from LLPS to hydrogels and amyloid-like fibers. The Ty1 Gag chimeras may exhibit a spectrum of these morphologies. The filamentous assemblies formed by GagSup35N-GFP are potentially similar to Sup35 amyloid fibers observed in vitro, and GagAβ-GFP may form liquid droplets. Sup35, while canonically known for its ability to form amyloid fibers as a prion, has more recently been appreciated to undergo LLPS upon a decrease in cytosolic pH and can mature over time into a gel-like condensate (85, 90). Whereas wildtype Gag allows for VLP assembly to proceed and supports transposition, perhaps transiently existing in an LLPS state, chimeras may become blocked along the retrosome and VLP assembly pathway, resulting in the striking structures observed by fluorescence microscopy and TEM. Further work will be required for the rigorous characterization necessary to declare the Ty1 retrosome or other assemblies formed by Gag chimeras an LLPS compartment. Ty1 provides a promising system to unite studies of prion and LLPS pathways.
An interchangeable platform to study PrLD and LLPS domains in living cells.
The condensate-forming property, but not the prion-forming property, of Sup35 is conserved across 400 million years from S. cerevisiae to Schizosaccharomyces pombe, emphasizing the evolutionary importance of this ancient phenotype (85). Our discovery of the Ty1 PrLD raises the possibility that LLPS may, too, be widespread among retroelements. Our preliminary computational analyses of Pseudoviridae (Ty1/copia) retroelement family members reveal predicted PrLDs in not only the closely-related yeast Ty2, but also in distantly-related plants in the Oryza element Retrofit and the Arabidopsis elements Evelknievel and AtRE1. The human retrotransposon LINE-1 phase separates and retrotransposition is associated with cancer (91) and age-associated inflammation (92, 93). A condensate-hardening drug was found to block human respiratory syncytial virus replication which occurs in virus-induced inclusion bodies (94), highlighting the potential of the Ty1 platform to contribute to new anti-viral and other humanhealth therapeutics. The Ty1 Gag chimera strategy developed here may prove to be a useful platform to study prion-like and LLPS-forming domains due to the genetic tractability of yeast and the suite of robust and sensitive in vivo assays developed for Ty1.
Materials and Methods
Bioinformatic analyses.
PLAAC (http://plaac.wi.mit.edu/) (42) was used with default settings: core length of 60 and 100% S. cerevisiae background probabilities. ArchCandy (https://bioinfo.crbm.cnrs.fr/index.php?route=tools&tool=7) (43) was used with a score threshold of 0.500 and the transmembrane regions filter off; the sum of scores data is presented. PrDOS (https://prdos.hgc.jp/) (44) was used with the default 5% FDR and the disordered probability threshold set to 0.5. GlobPlot2.3 (http://globplot.embl.de/) (45) was used with default settings with Russell/Linding propensities. Bioinformatic outputs were uniformly plotted using a custom script using the base plot() and rect() functions in R version 3.5.2. Structure analysis was performed using PyMOL v1.5.0.5 with the “align” command.
Yeast strains and media.
Yeast strains with full genotypes are listed in Supplementary Table 3. Standard yeast genetic and microbiological techniques were used in this work (95). Prion nucleation experiments were performed in GT409, an S. cerevisiae strain that is [psi− pin−] and harbors the ade1–14 allele which contains a premature stop codon (kindly provided by Y. Chernoff) (53). Ty1 assays were performed in the DG3582 background, a Ty-less S. paradoxus derivative of DG1768 (57, 58). For galactose induction in liquid media, starter cultures were grown overnight at 30 °C in synthetic complete (SC) dropout media containing 2% raffinose, diluted 1:20 into media containing 2% galactose, and grown at 22 °C for 48 hours.
Plasmids and cloning.
Plasmids, primers, and gene fragments are listed in Supplementary Tables 4–6. Detailed descriptions of plasmids and cloning are provided in SI Materials and Methods.
Prion nucleation and curing.
[PSI+] induction was assayed in a [psi−] strain for chimeric plasmids under a PCUP1 promoter; yeast cells were grown at 30 °C. Yeast were grown on SC-Ura for 2 days, replica plated to SC-Ura ± 150 μM CuSO4 and grown for 2 days, then replica plated to SC-Ade and grown for approximately 10 days until imaged. Following prion nucleation, Ade+ colonies were cured of [PSI+] by guanidine hydrochloride (GdHCl). First, the induction plasmid was counter selected on FOA and single colonies were isolated. Then, Ade+/Ura− colonies were passaged as single colonies on YPD spotted with 10 or 25 μL of 5 M GdHCl until red-pigmented colonies developed.
SDD-AGE.
Semi-denaturing detergent-agarose gel electrophoresis (SDD-AGE) was adapted from published methods (53, 54). Detailed protocols are described in SI Materials and Methods.
Ty1his3-AI mobility assays.
Ty1 retromobility events were detected using the his3-AI retromobility indicator gene (59) by qualitative and quantitative assays as previously described (15, 58). Detailed protocols are described in SI Materials and Methods.
Immunoblotting.
Total yeast protein was prepared by trichloroacetic acid precipitation and immunoblotted using standard techniques (58, 96). Detailed protocols are described in SI Materials and Methods.
Yeast microscopy.
Detailed protocols for live-cell fluorescence microscopy and transmission electron microscopy preparation and imaging of yeast cells are described in SI Materials and Methods.
Sucrose gradient sedimentation.
Sucrose gradient sedimentation was performed as previously described (15). Detailed protocols are described in SI Materials and Methods.
Supplementary Material
Significance.
Retrovirus-like retrotransposons help shape the genome evolution of their hosts and replicate within cytoplasmic particles. How their building blocks associate and assemble within the cell is poorly understood. Here, we report a novel prion-like domain (PrLD) in the budding yeast retrotransposon Ty1 Gag protein that builds virus-like particles. The PrLD has similar sequence properties to prions and disordered protein domains that can drive the formation of assemblies that range from liquid to solid. We demonstrate that the Ty1 PrLD can function as a prion and that certain prion sequences can replace the PrLD and support Ty1 transposition. This interchangeable system is an effective platform to study additional disordered sequences in living cells.
Acknowledgements
This work was supported by an NIH grant to DJG (R01GM124216) and an NIH Postdoctoral Fellowship to SLB (F32GM139247). This study was also supported by the Robert P. Apkarian Integrated Electron Microscopy Core (RPAIEMC), which is subsidized by the Emory University School of Medicine and the Emory College of Arts and Sciences. Additional support was provided by the Georgia Clinical & Translational Science Alliance of the National Institute of Health under award number UL1TR000454. Some of the data reported here were collected on the JEOL JEM1400 TEM supported by the National Institutes of Health Grant S10 RR025679. We thank Joan Curcio, Katarzyna Pachulska-Wieczorek, Yury Chernoff, and Pavithra Chandramowlishwaran for providing reagents and advice, and Adam Hannon-Hatfield for valuable discussions and comments on the manuscript.
Footnotes
The authors declare no competing interests.
Data Availability
All data is presented within this article and supplementary information.
References
- 1.Kim J. M., Vanguri S., Boeke J. D., Gabriel A., Voytas D. F., Transposable elements and genome organization: a comprehensive survey of retrotransposons revealed by the complete Saccharomyces cerevisiae genome sequence. Genome Res 8, 464–78 (1998). [DOI] [PubMed] [Google Scholar]
- 2.Bleykasten-Grosshans C., Fabrizio R., Friedrich A., Schacherer J., Species-Wide Transposable Element Repertoires Retrace the Evolutionary History of the Saccharomyces cerevisiae Host. Mol Biol Evol 38, 4334–4345 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Curcio M. J., Lutz S., Lesage P., The Ty1 LTR-Retrotransposon of Budding Yeast, Saccharomyces cerevisiae. Microbiol Spectr 3, MDNA3–0053–2014 (2015). [DOI] [PubMed] [Google Scholar]
- 4.Bolton E. C., Coombes C., Eby Y., Cardell M., Boeke J. D., Identification and characterization of critical cis-acting sequences within the yeast Ty1 retrotransposon. RNA 11, 308–22 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Feng Y. X., Moore S. P., Garfinkel D. J., Rein A., The genomic RNA in Ty1 virus-like particles is dimeric. J Virol 74, 10819–21 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gumna J., Purzycka K. J., Ahn H. W., Garfinkel D. J., Pachulska-Wieczorek K., Retroviral-like determinants and functions required for dimerization of Ty1 retrotransposon RNA. RNA Biol, 1–15 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Purzycka K. J., et al. , Exploring Ty1 retrotransposon RNA structure within virus-like particles. Nucleic Acids Res 41, 463–73 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.v Merkulov G., Swiderek K. M., Brachmann C. B., Boeke J. D., A critical proteolytic cleavage site near the C terminus of the yeast retrotransposon Ty1 Gag protein. J Virol 70, 5548–56 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.v Merkulov G., Lawler J. F., Eby Y., Boeke J. D., Ty1 proteolytic cleavage sites are required for transposition: all sites are not created equal. J Virol 75, 638–44 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wilhelm M., Wilhelm F.-X., Cooperation between Reverse Transcriptase and Integrase during Reverse Transcription and Formation of the Preintegrative Complex of Ty1. Eukaryot Cell 5, 1760–1769 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dobard C. W., Briones M. S., Chow S. A., Molecular Mechanisms by Which Human Immunodeficiency Virus Type 1 Integrase Stimulates the Early Steps of Reverse Transcription. J Virol 81, 10037–10046 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bridier-Nahmias A., et al. , An RNA polymerase III subunit determines sites of retrotransposon integration. Science 348, 585–8 (2015). [DOI] [PubMed] [Google Scholar]
- 13.Asif-Laidin A., et al. , A small targeting domain in Ty1 integrase is sufficient to direct retrotransposon integration upstream of tRNA genes. EMBO J in press, 1–17 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Devine S. E., Boeke J. D., Integration of the yeast retrotransposon Ty1 is targeted to regions upstream of genes transcribed by RNA polymerase III. Genes Dev 10, 620–633 (1996). [DOI] [PubMed] [Google Scholar]
- 15.Cottee M. A., et al. , Structure of a Ty1 restriction factor reveals the molecular basis of transposition copy number control. Nat Commun 12, 5590 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cristofari G., Ficheux D., Darlix J. L., The GAG-like protein of the yeast Ty1 retrotransposon contains a nucleic acid chaperone domain analogous to retroviral nucleocapsid proteins. J Biol Chem 275, 19210–7 (2000). [DOI] [PubMed] [Google Scholar]
- 17.Nishida Y., et al. , Ty1 retrovirus-like element Gag contains overlapping restriction factor and nucleic acid chaperone functions. Nucleic Acids Res 43, 7414–31 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brocca S., Grandori R., Longhi S., Uversky V., Liquid-Liquid Phase Separation by Intrinsically Disordered Protein Regions of Viruses: Roles in Viral Life Cycle and Control of Virus-Host Interactions. Int J Mol Sci 21, 1–31 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Etibor T. A., Yamauchi Y., Amorim M. J., Liquid Biomolecular Condensates and Viral Lifecycles: Review and Perspectives. Viruses 13, 9–14 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Malagon F., Jensen T. H., The T body, a new cytoplasmic RNA granule in Saccharomyces cerevisiae. Mol Cell Biol 28, 6022–32 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Malagon F., Jensen T. H., T-body formation precedes virus-like particle maturation in S. cerevisiae. RNA Biol 8, 184–9 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Checkley M. A., Nagashima K., Lockett S. J., Nyswaner K. M., Garfinkel D. J., P-body components are required for Ty1 retrotransposition during assembly of retrotransposition-competent virus-like particles. Mol Cell Biol 30, 382–98 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Alberti S., Gladfelter A., Mittag T., Considerations and Challenges in Studying Liquid-Liquid Phase Separation and Biomolecular Condensates. Cell 176, 419–434 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Alberti S., Dormann D., Liquid–Liquid Phase Separation in Disease. Annu Rev Genet 53, 171–194 (2019). [DOI] [PubMed] [Google Scholar]
- 25.Harrison A. F., Shorter J., RNA-binding proteins with prion-like domains in health and disease. Biochemical Journal 474, 1417–1438 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Aguzzi A., Altmeyer M., Phase Separation: Linking Cellular Compartmentalization to Disease. Trends Cell Biol 26, 547–558 (2016). [DOI] [PubMed] [Google Scholar]
- 27.King O. D., Gitler A. D., Shorter J., The tip of the iceberg: RNA-binding proteins with prion-like domains in neurodegenerative disease. Brain Res 1462, 61–80 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.March Z. M., King O. D., Shorter J., Prion-like domains as epigenetic regulators, scaffolds for subcellular organization, and drivers of neurodegenerative disease. Brain Res 1647, 9–18 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Patel A., et al. , A Liquid-to-Solid Phase Transition of the ALS Protein FUS Accelerated by Disease Mutation. Cell 162, 1066–1077 (2015). [DOI] [PubMed] [Google Scholar]
- 30.Molliex A., et al. , Phase Separation by Low Complexity Domains Promotes Stress Granule Assembly and Drives Pathological Fibrillization. Cell 163, 123–133 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lin Y., Protter D. S. W., Rosen M. K., Parker R., Formation and Maturation of Phase-Separated Liquid Droplets by RNA-Binding Proteins. Mol Cell 60, 208–219 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lee M., et al. , Somatic APP gene recombination in Alzheimer’s disease and normal neurons. Nature (2018) 10.1038/s41586-018-0718-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.O’Brien R. J., Wong P. C., Amyloid precursor protein processing and Alzheimer’s disease. Annu Rev Neurosci 34, 185–204 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stefanis L., α-Synuclein in Parkinson’s disease. Cold Spring Harb Perspect Med 2, a009399 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nikolic J., et al. , Negri bodies are viral factories with properties of liquid organelles. Nat Commun 8, 1–12 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Alenquer M., et al. , Influenza A virus ribonucleoproteins form liquid organelles at endoplasmic reticulum exit sites. Nat Commun 10, 1629 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Metrick C. M., Koenigsberg A. L., Heldwein E. E., Conserved Outer Tegument Component UL11 from Herpes Simplex Virus 1 Is an Intrinsically Disordered, RNA-Binding Protein. mBio 11, 1–22 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Guseva S., et al. , Measles virus nucleo- and phosphoproteins form liquid-like phase-separated compartments that promote nucleocapsid assembly. Sci Adv 6, eaaz7095 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Monette A., et al. , Pan-retroviral Nucleocapsid-Mediated Phase Separation Regulates Genomic RNA Positioning and Trafficking. Cell Rep 31, 107520 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Savastano A., Ibáñez de Opakua A., Rankovic M., Zweckstetter M., Nucleocapsid protein of SARS-CoV-2 phase separates into RNA-rich polymerase-containing condensates. Nat Commun 11, 6041 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Newton J. C., et al. , Phase separation of the LINE-1 ORF1 protein is mediated by the N-terminus and coiled-coil domain. Biophys J 120, 2181–2191 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lancaster A. K., Nutter-Upham A., Lindquist S., King O. D., PLAAC: a web and command-line application to identify proteins with prion-like amino acid composition. Bioinformatics 30, 2501–2 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ahmed A. B., Znassi N., Château M. T., A. v. Kajava, A structure-based approach to predict predisposition to amyloidosis. Alzheimer’s and Dementia 11, 681–690 (2015). [DOI] [PubMed] [Google Scholar]
- 44.Ishida T., Kinoshita K., PrDOS: prediction of disordered protein regions from amino acid sequence. Nucleic Acids Res 35, W460–4 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Linding R., Russell R. B., Neduva V., Gibson T. J., GlobPlot: Exploring protein sequences for globularity and disorder. Nucleic Acids Res 31, 3701–8 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jumper J., et al. , Highly accurate protein structure prediction with AlphaFold. Nature 596, 583–589 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Preis A., et al. , Cryoelectron microscopic structures of eukaryotic translation termination complexes containing eRF1-eRF3 or eRF1-ABCE1. Cell Rep 8, 59–65 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bousset L., Belrhali H., Janin J., Melki R., Morera S., Structure of the globular region of the prion protein Ure2 from the yeast Saccharomyces cerevisiae. Structure 9, 39–46 (2001). [DOI] [PubMed] [Google Scholar]
- 49.Riek R., et al. , NMR structure of the mouse prion protein domain PrP(121–231). Nature 382, 180–2 (1996). [DOI] [PubMed] [Google Scholar]
- 50.Crescenzi O., et al. , Solution structure of the Alzheimer amyloid beta-peptide (1–42) in an apolar microenvironment. Similarity with a virus fusion domain. Eur J Biochem 269, 5642–8 (2002). [DOI] [PubMed] [Google Scholar]
- 51.Chernoff Y. O., Lindquist S. L., Ono B., Inge-Vechtomov S. G., Liebman S. W., Role of the chaperone protein Hsp104 in propagation of the yeast prion-like factor [psi+]. Science 268, 880–4 (1995). [DOI] [PubMed] [Google Scholar]
- 52.Liebman S. W., Chernoff Y. O., Prions in yeast. Genetics 191, 1041–72 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chandramowlishwaran P., et al. , Mammalian amyloidogenic proteins promote prion nucleation in yeast. Journal of Biological Chemistry 293, 3436–3450 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Halfmann R., Lindquist S., Screening for amyloid aggregation by Semi-Denaturing Detergent-Agarose Gel Electrophoresis. J Vis Exp, 11–13 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tuite M. F., Mundy C. R., Cox B. S., Agents that cause a high frequency of genetic change from [psi+] to [psi−] in Saccharomyces cerevisiae. Genetics 98, 691–711 (1981). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ferreira P. C., Ness F., Edwards S. R., Cox B. S., Tuite M. F., The elimination of the yeast [PSI+] prion by guanidine hydrochloride is the result of Hsp104 inactivation. Mol Microbiol 40, 1357–1369 (2001). [DOI] [PubMed] [Google Scholar]
- 57.Chen J., et al. , Genome Assembly of the Ty1-Less Saccharomyces paradoxus Strain DG1768. Microbiol Resour Announc 11, e0086821 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Saha A., et al. , A trans-dominant form of Gag restricts Ty1 retrotransposition and mediates copy number control. J Virol 89, 3922–38 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Curcio M. J., Garfinkel D. J., Single-step selection for Ty1 element retrotransposition. Proc Natl Acad Sci U S A 88, 936–40 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Huang Q., et al. , Retrotransposon Ty1 RNA contains a 5’-terminal long-range pseudoknot required for efficient reverse transcription. Rna 19, 320–322 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gamache E. R., et al. , Structure-function model for kissing loop interactions that initiate dimerization of ty1 RNA. Viruses 9, 1–23 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Xu H., Boeke J. D., Localization of sequences required in cis for yeast Ty1 element transposition near the long terminal repeats: analysis of mini-Ty1 elements. Mol Cell Biol 10, 2695–702 (1990). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Baxa U., et al. , Characterization of beta-sheet structure in Ure2p1–89 yeast prion fibrils by solid-state nuclear magnetic resonance. Biochemistry 46, 13149–62 (2007). [DOI] [PubMed] [Google Scholar]
- 64.Sharon G., Burkett T. J., Garfinkel D. J., Efficient homologous recombination of Ty1 element cDNA when integration is blocked. Mol Cell Biol 14, 6540–51 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Melamed C., Nevo Y., Kupiec M., Involvement of cDNA in homologous recombination between Ty elements in Saccharomyces cerevisiae. Mol Cell Biol 12, 1613–20 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Adams S. E., et al. , The functions and relationships of Ty-VLP proteins in yeast reflect those of mammalian retroviral proteins. Cell 49, 111–9 (1987). [DOI] [PubMed] [Google Scholar]
- 67.Doh J. H., Lutz S., Curcio M. J., Co-translational localization of an LTR-retrotransposon RNA to the endoplasmic reticulum nucleates virus-like particle assembly sites. PLoS Genet 10, e1004219 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Tucker J. M., Larango M. E., Wachsmuth L. P., Kannan N., Garfinkel D. J., The Ty1 Retrotransposon Restriction Factor p22 Targets Gag. PLoS Genet 11, e1005571 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.AL-Khayat H. A., et al. , Yeast Ty retrotransposons assemble into virus-like particles whose T-numbers depend on the C-terminal length of the capsid protein. J Mol Biol 292, 65–73 (1999). [DOI] [PubMed] [Google Scholar]
- 70.Burns N. R., et al. , Symmetry, flexibility and permeability in the structure of yeast retrotransposon virus-like particles. EMBO J 11, 1155–64 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sondheimer N., Lindquist S., Rnq1: an epigenetic modifier of protein function in yeast. Mol Cell 5, 163–72 (2000). [DOI] [PubMed] [Google Scholar]
- 72.Alberti S., Halfmann R., King O., Kapila A., Lindquist S., A systematic survey identifies prions and illuminates sequence features of prionogenic proteins. Cell 137, 146–58 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Prusiner S. B., Prions. Proc Natl Acad Sci U S A 95, 13363–83 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lorenz H., Windl O., Kretzschmar H. A., Cellular phenotyping of secretory and nuclear prion proteins associated with inherited prion diseases. J Biol Chem 277, 8508–16 (2002). [DOI] [PubMed] [Google Scholar]
- 75.Kitamoto T., Iizuka R., Tateishi J., An amber mutation of prion protein in Gerstmann-Sträussler syndrome with mutant PrP plaques. Biochem Biophys Res Commun 192, 525–31 (1993). [DOI] [PubMed] [Google Scholar]
- 76.Ghetti B., et al. , Prion protein amyloidosis. Brain Pathol 6, 127–45 (1996). [DOI] [PubMed] [Google Scholar]
- 77.Hornemann S., Glockshuber R., Autonomous and reversible folding of a soluble amino-terminally truncated segment of the mouse prion protein. J Mol Biol 261, 614–9 (1996). [DOI] [PubMed] [Google Scholar]
- 78.Liu J. J., Lindquist S., Oligopeptide-repeat expansions modulate “protein-only” inheritance in yeast. Nature 400, 573–6 (1999). [DOI] [PubMed] [Google Scholar]
- 79.Parham S. N., Resende C. G., Tuite M. F., Oligopeptide repeats in the yeast protein Sup35p stabilize intermolecular prion interactions. EMBO J 20, 2111–9 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Scholes D. T., Banerjee M., Bowen B., Curcio M. J., Multiple regulators of Ty1 transposition in Saccharomyces cerevisiae have conserved roles in genome maintenance. Genetics 159, 1449–65 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Dakshinamurthy A., Nyswaner K. M., Farabaugh P. J., Garfinkel D. J., BUD22 affects Ty1 retrotransposition and ribosome biogenesis in Saccharomyces cerevisiae. Genetics 185, 1193–205 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Nyswaner K. M., Checkley M. A., Yi M., Stephens R. M., Garfinkel D. J., Chromatin-associated genes protect the yeast genome from Ty1 insertional mutagenesis. Genetics 178, 197–214 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Risler J. K., Kenny A. E., Palumbo R. J., Gamache E. R., Curcio M. J., Host co-factors of the retrovirus-like transposon Ty1. Mob DNA 3, 12 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Franzmann T. M., Alberti S., Protein Phase Separation as a Stress Survival Strategy. Cold Spring Harb Perspect Biol 11, a034058 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Franzmann T. M., et al. , Phase separation of a yeast prion protein promotes cellular fitness. Science 359 (2018). [DOI] [PubMed] [Google Scholar]
- 86.Chernoff Y. O., Stress and prions: lessons from the yeast model. FEBS Lett 581, 3695–701 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Gumna J., Romanowska A. A., Garfinkel D. J., Wieczorek K. P., RNA Binding Properties of the Ty1 LTR - Retrotransposon Gag Protein. Int J Mol Sci, 1–16 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Maxwell P. H., Curcio M. J., Retrosequence formation restructures the yeast genome. Genes Dev 21, 3308–18 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Maxwell P. H., et al. , Ty1 mobilizes subtelomeric Y’ elements in telomerase-negative Saccharomyces cerevisiae survivors. Mol Cell Biol 24, 9887–98 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Lyke D. R., Dorweiler J. E., Manogaran A. L., The Three Faces of Sup35. Yeast, 0–3 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Lee E., et al. , Landscape of somatic retrotransposition in human cancers. Science 337, 967–71 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.de Cecco M., et al. , L1 drives IFN in senescent cells and promotes age-associated inflammation. Nature 566, 73–78 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Simon M., et al. , LINE1 Derepression in Aged Wild-Type and SIRT6-Deficient Mice Drives Inflammation. Cell Metab 29, 871–885.e5 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Risso-Ballester J., et al. , A condensate-hardening drug blocks RSV replication in vivo. Nature 595, 596–599 (2021). [DOI] [PubMed] [Google Scholar]
- 95.Guthrie C., Fink G., Guide to yeast genetics and molecular biology. Methods Enzymol 194, 1–863 (1991). [PubMed] [Google Scholar]
- 96.Ohashi A., Gibson J., Gregor I., Schatz G., Import of proteins into mitochondria. The precursor of cytochrome c1 is processed in two steps, one of them heme-dependent. J Biol Chem 257, 13042–7 (1982). [PubMed] [Google Scholar]
- 97.Brachmann C. B., et al. , Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast 14, 115–32 (1998). [DOI] [PubMed] [Google Scholar]
- 98.Faul F., Erdfelder E., Buchner A., Lang A.-G., Statistical power analyses using G*Power 3.1: tests for correlation and regression analyses. Behav Res Methods 41, 1149–60 (2009). [DOI] [PubMed] [Google Scholar]
- 99.Bastin P., Bagherzadeh A., Matthews K. R., Gull K., A novel epitope tag system to study protein targeting and organelle biogenesis in Trypanosoma brucei. Mol Biochem Parasitol 77, 235–239 (1996). [DOI] [PubMed] [Google Scholar]
- 100.Schindelin J., et al. , Fiji: an open-source platform for biological-image analysis. Nat Methods 9, 676–82 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data is presented within this article and supplementary information.