Abstract
We have used FRET-based biosensors in live cells, in a robust high-throughput screening (HTS) platform, to identify small-molecules that alter the structure and activity of the cardiac sarco/endoplasmic reticulum calcium ATPase (SERCA2a). Our primary aim is to discover drug-like small-molecule activators that improve SERCA’s function for the treatment of heart failure. We have previously demonstrated the use of an intramolecular FRET biosensor, based on human SERCA2a, by screening a small validation library using novel microplate readers that can detect the fluorescence lifetime or emission spectrum with high speed, precision, and resolution. Here we report results from a 50,000-compound screen using the same biosensor, with hit compounds functionally evaluated using Ca2+-ATPase and Ca2+-transport assays. We focused on 18 hit compounds, from which we identified eight structurally unique compounds and four compound classes as SERCA modulators, approximately half of which are activators and half are inhibitors. While both activators and inhibitors have therapeutic potential, the activators establish the basis for future testing in heart disease models and lead development, toward pharmaceutical therapy for heart failure.
Keywords: calcium ATPase, calcium transport, fluorescence, drug screening, fluorescence resonance energy transfer (FRET), sarcoplasmic reticulum (SR), fluorescence lifetime, biosensor, SERCA, SERCA activators
Introduction
Sarco/endoplasmic reticulum calcium ATPase (SERCA), integrated in the sarcoplasmic reticulum (SR, muscle cells) or endoplasmic reticulum (ER, non-muscle cells) membrane in most mammalian cells, is integral for using Ca2+-dependent hydrolysis of ATP to fuel active transport of cytosolic Ca2+ into the SR or ER. In muscle, the activity of SERCA1a (skeletal isoform) or SERCA2a (cardiac isoform) is essential for relaxation (diastole), restoring SR Ca2+ following its release via Ca2+ channels (ryanodine receptors, RyR) for muscle contraction (systole). Decreased SERCA activity and excessive RyR leak results in failure to maintain the high gradient of [Ca2+] between the cytoplasm (mM) and the SR (sub-μM) during diastole (muscle relaxation) and are associated with heart failure (HF) in human and animal models1. Decreased SERCA activity has been attributed to multiple factors, including reduced SERCA gene expression, increased post-translational modifications, and altered interaction with regulatory proteins1. Overall, decreased SERCA activity and increased Ca2+-leak lead to a pathophysiological state of the cardiac myocyte2 (HF, cardiac hypertrophy, diabetic hypertrophy), skeletal myocyte (Brody’s disease and myotonic dystrophy) and non-muscle cells (Darier’s disease, diabetes, Alzheimer’s disease)3. Altered SERCA interaction with regulatory proteins (regulins), such as phospholamban (PLB), have been linked to ventricular dysfunction and maladaptive remodeling in failing hearts4. Of the seven regulins discovered5, the dwarf open reading frame (DWORF) regulatory peptide is the only one known to activate muscle-specific SERCA activity, both by direct activation of SERCA6,7 and by competing with PLB binding8,9, and to prevent HF in a mouse model of dilated cardiomyopathy10. Current therapeutic measures include beta-blockers, angiotensin-converting enzyme (ACE) inhibitors, angiotensin-receptor blockers (ARB), and mineralocorticoid receptor antagonists. However, these do not directly target proteins responsible for dysfunctional Ca2+ cycling. Discovery of small molecules (potential drugs) that target specific proteins is needed to exert improved control measures for positive therapeutic outcomes.
In the present study, we seek primarily SERCA2a activators to alleviate heart failure11,12. However, SERCA uncouplers are also of potential interest in other indications such as in nonshivering thermogenesis, enhancing metabolism and thus reducing obesity13,14. Targeted SERCA uncouplers or inhibitors may be useful for treatment of cancer or malaria15.
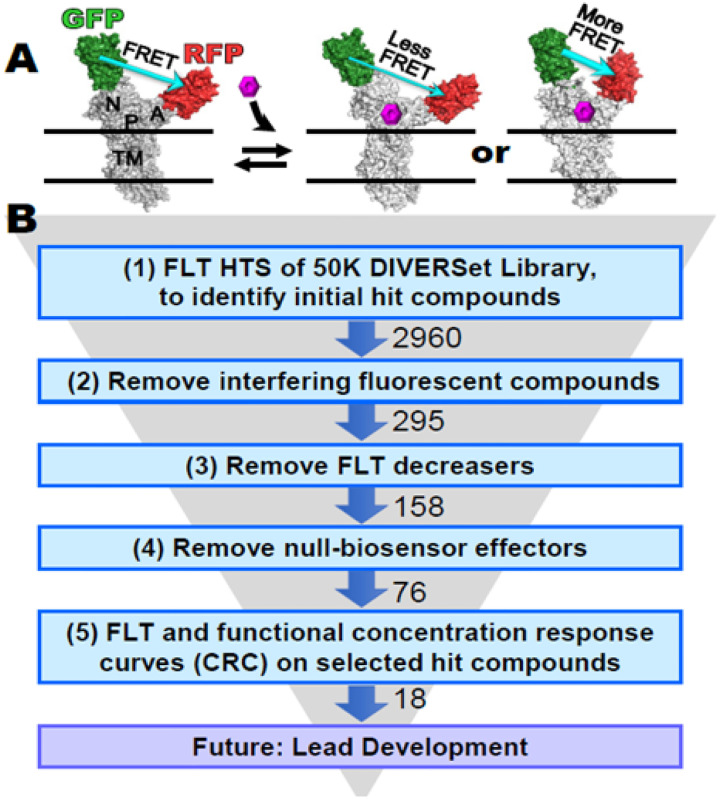
SERCA2a is a large transmembrane protein, with the phosphorylation (P) and nucleotide-binding (N) domains forming the catalytic site, coupled by the actuator (A) domain (Fig. 1A). Large (5–10 nm) relative movements of these domains are coupled to Ca2+ transport, as detected in living cells by an intramolecular FRET biosensor (Fig. 1A)16. The interaction of small molecules with SERCA can induce measurable structural changes, detectable by this biosensor, that often correlate with function, making this biosensor a powerful tool for HTS discovery of SERCA-binding compounds16,17.
Figure 1.
(A) FRET biosensor, human two-color SERCA2a (2CS), showing SERCA domains: nucleotide binding (N), phosphorylation (P), actuator (A) and transmembrane (TM). GFP is fused to N and RFP is fused to A. DFRET, measured from DFLT (fluorescence lifetime), is used to detect SERCA structural changes (B) Screening funnel describing the 5-step process undertaken in this study, involving measurements of FLT and SERCA function, with SERCA in live mammalian cells and in isolated pig cardiac SR membranes, respectively. (1) FLT changes caused by test compounds were measured using the target SERCA-specific FRET biosensor 2CS, to identify initial hit compounds. False hits were ruled out as compounds that (2) are fluorescent or affect the donor directly, (3) decrease FLT, or (4) affect FRET in a null biosensor, in which donor and acceptor are separated by a non-functional flexible peptide. (5) Concentration-dependence of FLT and SERCA function was measured to further prioritize hit compounds that could result in future lead development. Experimental details are provided in Methods.
In several previous early-stage drug discovery campaigns, we have focused on the SERCA regulator phospholamban (PLB)18,19, but we have also tested and validated FRET biosensor constructs of SERCA alone16,17,20, to detect binding of drug-like small-molecule compounds directly to SERCA. For this, we engineered a “two-color” SERCA (2CS, Fig. 1A) construct with fused eGFP and tagRFP fluorescent protein tags to the cytoplasmic N- and A-domains of SERCA, which detect relative motions of these domains during the enzymatic cycle responsible for Ca2+-transport17,20,21. Using the intramolecular FRET measurement of 2CS, stably expressed in HEK293 cells, we previously validated this biosensor using the NIH Clinical Collection (NCC, 727 compounds)17 and the LOPAC library (1280 compounds)20. Along with the fluorescent biosensors, high-throughput is enabled by a FLT-PR (fluorescence lifetime plate reader) that scans a 1536-well plate with unprecedented precision and speed, determining FRET with 0.3% CV (30 times better than conventional intensity detection) in 2.5 min18,22,23, enabling a high-precision screen of 50,000-compounds in two days. This instrument is equipped with simultaneous FLT detection at two emission wavelengths (two-channel lifetime detection)21, a function used to filter out fluorescent compounds that would be falsely identified as hit compounds. We have demonstrated the additional high throughput acquisition of fluorescence emission spectra using a spectral unmixing plate reader (SUPR), which helps detect (and rule out) interfering compounds, based on changes in the line shape in the donor-only region of the spectrum21. As in our previous projects involving other drug targets, the next logical step is to screen a 50,000-compound DIVERSet library, a diverse collection of drug-like small molecules that has yielded effective hit compounds in previous drug discovery projects22–24.
Here we apply our HTS platform using FRET lifetime measurements of two-color human cardiac SERCA (2CS) in living cells (Fig. 1B) to identify hit compounds. To validate selected hit compounds, we then acquire concentration response curves (CRCs) using both FRET and functional assays (Ca2+-ATPase activity and Ca2+-transport). We hypothesize that the combination of using improved fluorescence technology and screening a larger compound library (50K DIVERSet) will result in a larger and more diverse collection of hit compounds that more effectively regulate cardiac SERCA function, thus increasing the potential for discovering lead compounds for new heart failure therapeutics.
Results
FLT HTS of 50K DIVERSet library
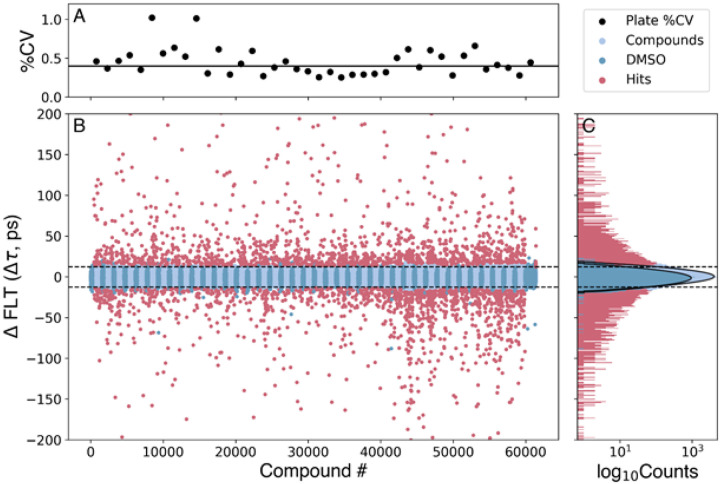
2CS expressed in live HEK-293 cells was incubated with 5 nL of compound at a final [compound] of 10μM (from a DIVERSet library of 50,000 compounds) or DMSO (as a control), preloaded on 40 assay plates for 20 min prior to being read on the FLT-PR. FLT measurements were observed to be normally distributed, with a coefficient of variation (CV) of 0.4% across all 40 plates (Fig. 2A). Plate-by-plate CV varied by < 1 % (Fig. 2A).
Figure 2.
The human cardiac 2-color SERCA2a biosensor (2CS) in HEK293 cells was used to screen the 50,000-compound DIVERSet library. (A) Screen precision was determined by computing % CV for each plate using DMSO control wells, with a median value of 0.4% across 40 plates. (B) Change in lifetime (Dt) was computed to find potential hits (red points) with a hit threshold set at a robust z-score of 3, resulting in 2960 FLT hit compounds for triage with the SUPR instrument and two-channel lifetime detection. DMSO controls (dark blue) were grouped in the plot to better illustrate plate boundaries. (C) The histogram of compounds not affecting 2CS (light blue, 1ps bin width) are normally distributed, similar to that of DMSO alone as shown by a fit of the populations to Gaussian distributions, which are shown in log scale. The horizontal lines in B, C illustrate the approximate cutoffs used, though actual cutoffs were determined on a plate-by-plate basis.
Compounds that significantly altered the structure of 2CS were determined by computing the change in lifetime (Δτ) for each compound compared to DMSO control sample (2CS plus DMSO), and the magnitude of this change was compared to the normal statistical fluctuation of the biosensor by computing the robust z-score (Methods). The distinct FLT changes induced by the potential hit compounds (Fig. 2B, red) are illustrated by the normal distributions of compounds not affecting SERCA (Fig. 2C, blue) and the control (Fig. 2C, dark blue). A hit threshold was set at a robust z-score of ± 3, resulting in 2960 FLT hit compounds (Fig. 1B, step 1). Interference from fluorescent compounds was removed (Fig. 1B, step 2) using both the similarity index (SI, detected by SUPR)17 and two-channel lifetime detection21. We also eliminated compounds that affected the lifetime detected from a donor-only (1CS) sample; 295 compounds remained.
More FLT decreasers than increasers were found to fail these tests, in alignment with previous studies18,20,21,25. FLT increasers are more advantageous for two additional reasons: (a) They offer greater reproducibility between repeats of a screen18,20,21. (b) Most previously identified SERCA modulators have been shown to be FLT increasers16,17,20. Therefore, we prioritized the 158 FLT increasers (termed “hit compounds” (Fig. 1B, step 3) for follow-up retesting and CRC evaluation.
FLT retests of select hits compounds with 2CS and null biosensor
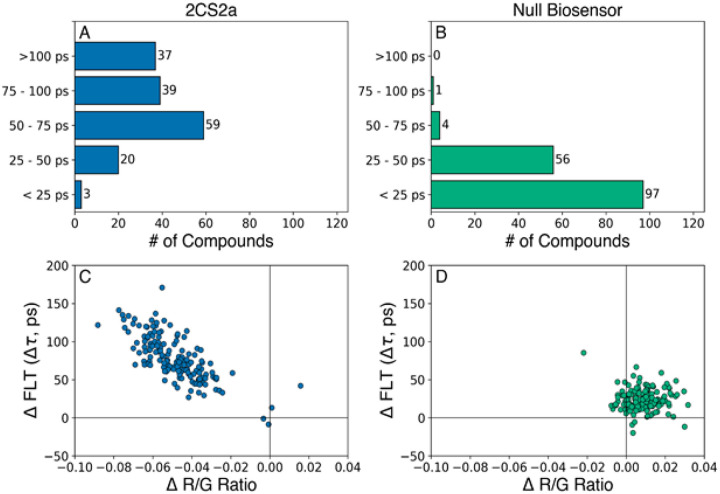
158 hit compounds were retested using 2CS (Step 4 in Fig. 1B; see Fig. 3A and C) and a null biosensor construct (Step 4 in Fig. 1B; see Fig. 3B and D), which consisted of GFP and RFP connected by a 32-residue unstructured flexible linker peptide (G32R)20. The null biosensor was used to rule out compounds that alter FLT by directly binding to the fluorescent proteins. A plot of the change in lifetime (Δτ) vs. the ΔR/G ratio (Fig. 3C and D) shows that the 2CS hits had little to no effects on the null biosensor.
Figure 3.
DIVERSet hit compound FRET retests. The most promising 158 hit compounds were selected (“cherry-picked”) and dispensed into 1536-well plates at 10 and 30 mM concentrations (n=3 wells for each drug concentration). Data is shown from the 30 mM screens for SERCA2a (A, C) and null biosensor (B, D). (A) Distribution of significant lifetime changes for the 2CS biosensor. (B) A null biosensor was counter-screened against the 158 hit compounds. Only five compounds displayed a lifetime change greater than 50 ps, indicating that our method for eliminating fluorescent compounds removes nearly all false positives. (C) and (D) Plots of the change in fluorescence lifetime vs the R/G ratio show excellent, reproducible correlation between the two techniques of lifetime (DFLT) and SUPR (DR/G) for the 2CS biosensor (C) and null biosensor (D).
Hit compounds that produced > 75 ps Δτ (76 compounds, Fig. 1B, step 4) (Fig. 3A) were targeted for further functional testing. We focused on 18 of these for the CRC testing, after the FLT data were subjected to the first four steps of the screening funnel (Fig. 1B) and compound repurchasing availability was determined. None of these compounds were in the PAINS (Pan-Assay INterference compoundS) category26, nor were they redox agents or metal chelators.
Validation of hit compounds using FLT CRC
To further evaluate the 18 hit compounds, we determined the FLT response to compound concentrations ranging 0.78–100.μM (Step 5 in Fig. 1B). All 18 hit compounds (Table 1) decreased FRET (increased FLT) of the 2CS biosensor, suggesting that the compounds induced a structural change (Fig. 1, top) in the cytosolic headpiece region of SERCA2a. Compounds 1 and 4 showed a significant decrease in FRET at the lowest concentration, but no further effect at higher concentrations. The remaining 16 compounds decreased FRET with detectable EC50 (Fig. 4–7B, Table 1).
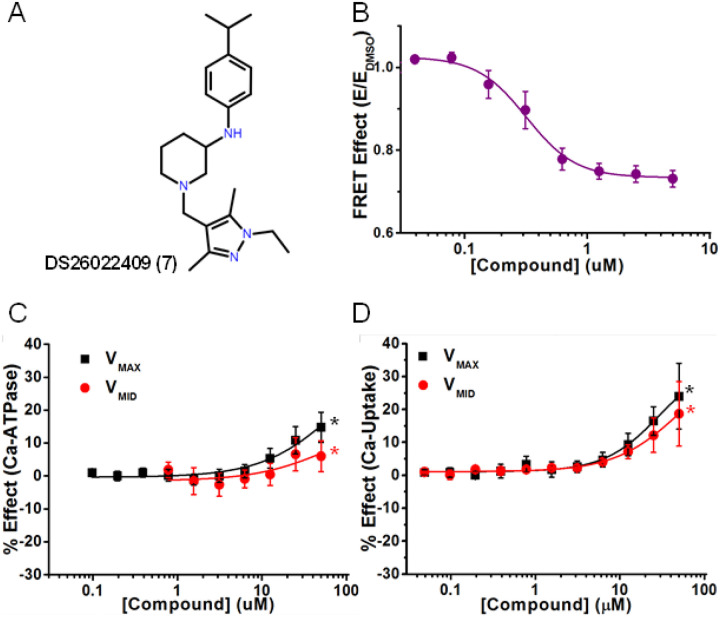
Figure 4.
A representative activator compound of both Ca2+-ATPase and Ca-transport at saturating Ca2+ (VMAX). (A) Chemical structure of Compound DS26022409 (Compound 7). (B) CRC of normalized FRET E in live HEK cells shows decreasing FRET response with increasing [compound]. (C) CRC of Ca2+-ATPase of SERCA2a in pCSR vesicles show activation at high Ca2+ (VMAX, black) and at midpoint Ca2+ (VMID, red). (D) CRC of Ca-transport shows activation at both VMAX and VMID Ca2+ (black and red, respectively). Data are presented as mean ± SEM, n = 3, *p<0.05.
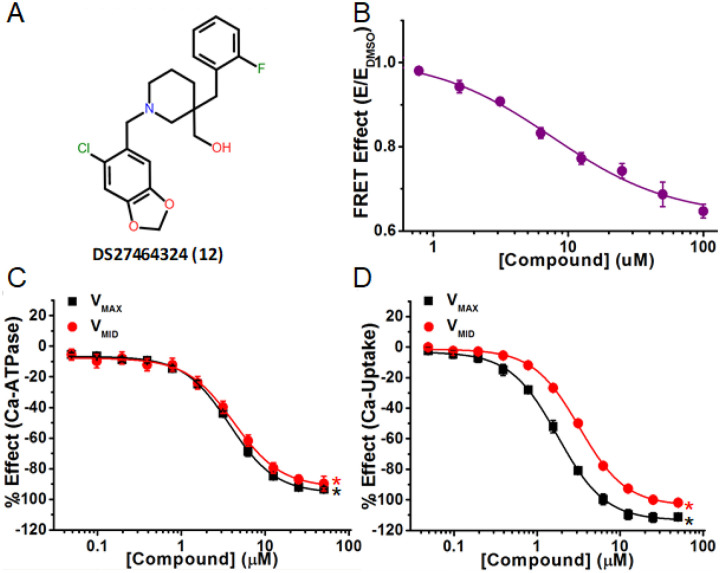
Figure 7.
A representative inhibitor compound that severely decreases FRET and inhibits both Ca-ATPase and Ca-Transport activity. (A) Chemical structure of Compound DS27464324 (Compound 12). (B) CRC of normalized FRET E shows decreased FRET response in 2CS biosensor. (C) CRC shows inhibition of Ca-ATPase in pCSR under both VMAX (black) and VMID (red) [Ca]. (D) CRC shows inhibition of Ca-transport in pCSR under VMAX (black) and VMID (red) [Ca]. Data is presented as mean ± SEM, n = 3, *p<0.05.
Effects of hit compounds on SERCA2a activity using Ca2+-ATPase and Ca2+-transport assays
To assess the impacts of FRET hit compounds on SERCA2a function, we used an absorbance-based enzyme-coupled NADH-linked Ca2+-dependent ATPase assay (referred to below as ATPase) and a fluorescence-based Ca2+-transport assay (referred to below as transport), using pig cardiac SR vesicles enriched for SERCA2a18 (Step 5 in Fig. 1B). Ca2+-ATPase and Ca2+-transport activities were measured at VMAX (saturating, pCa 5.4) and VMID (subsaturating, midpoint, pCa 6.2) [Ca2+], revealing activators (Compounds 1–9) and inhibitors (Compounds 10–18) (Table 1 and Supplementary Fig. S1), identified on the basis of potency (1/EC50) and efficacy (amplitude of the effect, Δ) (Table 1). Activators were compounds that increased Ca2+-ATPase and/or Ca2+-transport activities (at one or both [Ca2+]) (Table 1, Fig. 4–6). We grouped the activators in two subcategories based on the functional effects at the two Ca2+ concentrations: (1) activates both Ca2+-ATPase and Ca2+-transport (Compounds 2, 4, 7, 8, and 9), and (2) activates Ca2+-ATPase with divergent effects on Ca2+-transport (Compounds 1, 3, 5, and 6). We define “divergent” to indicate that the compound induces opposing effects at two different [Ca] (an increase at one [Ca] and a decrease at the other) in one assay. These effects indicate induced changes in the coupling ratio (CR), which is optimally two Ca2+ ions transported per molecule of ATP hydrolyzed27–30. This change was determined from the ratio of VMAX (Ca2+-transport) to VMAX (Ca2+-ATPase) (Table 1), as discussed below in SERCA Activators.
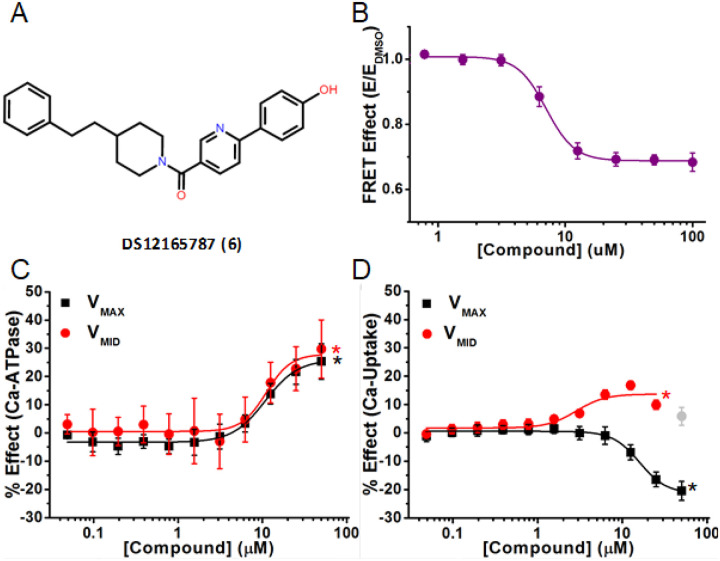
Figure 6.
A representative activator compound that decreases FRET, increases Ca2+-ATPase activity, and has divergent effects on Ca2+-transport activity. (A) Chemical structure of Compound DS12165787 (Compound 6). (B) Concentration response curve (CRC) of normalized FRET E shows decreased FRET with a half maximal effect (EC50) at 7.1 ± 0.2 mM. (C) CRC shows Ca2+-ATPase activation of SERCA2a in pig CSR vesicles at VMAX (black, pCa 5.4) and VM!D (red, pCa 6.2) [Ca2+] (D) CRC of Ca2+-transport of SERCA2a in pig CSR vesicles, showing inhibition for VMAX (black) and activation for VMID (red). Grey data point was omitted from fitting. D, C10, and EC50 are reported in Table 1 for (C) and (D). Data is presented as mean ± SEM, n = 3, *p<0.05.
We identified inhibitors that induced strong (≥ 68%), moderate (34 to 67%), and mild (≤ 33%) inhibition of SERCA function. Therefore, we define four subcategories of compound inhibition: 1) strong inhibition of Ca2+-ATPase and Ca2+-transport (Compounds 11, 12, 13), 2) moderate inhibition of Ca2+-ATPase and strong inhibition of Ca2+-transport (Compounds 15 and 17), 3) mild inhibition of Ca2+-ATPase and moderate-to-strong inhibition of Ca2+-transport (Compounds 14 and 16), and 4) mild inhibition on Ca2+-ATPase and mild-to-moderate inhibition on Ca2+-transport (Compounds 10 and 18) (Table 1, Fig. 7, and discussed below under SERCA Inhibitors).
Classification of compounds by physicochemical characteristics
The 18 hit compounds were subjected to cheminformatic analysis, to determine whether any of the compounds shared a common chemical scaffold. Compounds with a Tanimoto coefficient and maximum common substructure (MCS)31 scores above 0.4 were binned as clusters, while those with scores below 0.4 were classified as singletons. The analysis yielded diverse scaffolds31,32 of the hit compounds (Supplementary Fig. S1 and Table S1).
Four clusters of compounds with multiple examples (A-D in Table 1) were found, and the remaining eight were unique compounds (singletons) (E-L in Table 1 and Supplementary Fig. S1 and Table S1). The three compounds in cluster A (Compounds 1, 2, and 3 in Table 1) have a common 5-(aryloxymethyl)oxazole-3-carboxamide)33, while those in cluster B (Compounds 4 and 5) share a N-heteroaryl-N-alkylpiperazine. Cluster C (Compounds 9 and 10) is defined by an amide linkage and Cluster D (Compounds 11, 12, and 13) by a piperidine scaffold. Clusters E-L (Compounds 6, 7, 8, 14, 15, 16, 17, and 18) contain a single compound (singleton) with no common scaffold with any other hit compound in this study. All of the hit compounds have physicochemical properties34, conducive of favorable drug disposition in vivo, including a low molecular weight (< 500), low cLogP (calculated partition coefficient for lipophilicity) values (< 5), low non-H rotatable bonds that describe the molecular flexibility (< 10), low degree of possible hydrogen bond formation (total number of hydrogen bond acceptors and donors should be less than 8), and low total polar surface area (tPSA < 140 Å) (Supplementary Table S1).
Next we describe in more detail the nine activators (Fig. 4–6) that are grouped into two subcategories, and the nine inhibitors (Fig. 7) that are grouped into three classes and four subcategories.
SERCA Activators
In the first category of activators, Compounds 2, 4, 7, 8, and 9 activated both Ca2+-ATPase and Ca2+-transport. Compound 7 (Fig. 4A) (singleton F) decreased FRET of 2CS in live cells so that EC50 = 0.3 μM, indicating stabilization of the open conformation of SERCA, and accelerated Ca2+-ATPase to induce ΔVmax = 14% and ΔVMID = 7% (Fig. 4C and Table 1). Compound 7 induced the highest increase in Ca2+-transport of all the compounds at both VMAX (24%) and VMID (19%), (Fig. 4D), which was greater than that of Ca2+-ATPase activity (Fig. 4D). The CR increased to 0.74 compared to control (0.66, Table 1). Saturation of CRC was not reached at the highest [compound] measured, so the functional EC50 was not determined, therefore we determined C10, the compound concentration that increases function by 10%. At VMAX and VMID, C10 was 25 μM for Ca2+-ATPase, 14 μM and 21 μM for Ca2+-transport (Table 1). This compound will be placed at high priority for future optimization by medicinal chemistry and testing in animal models.
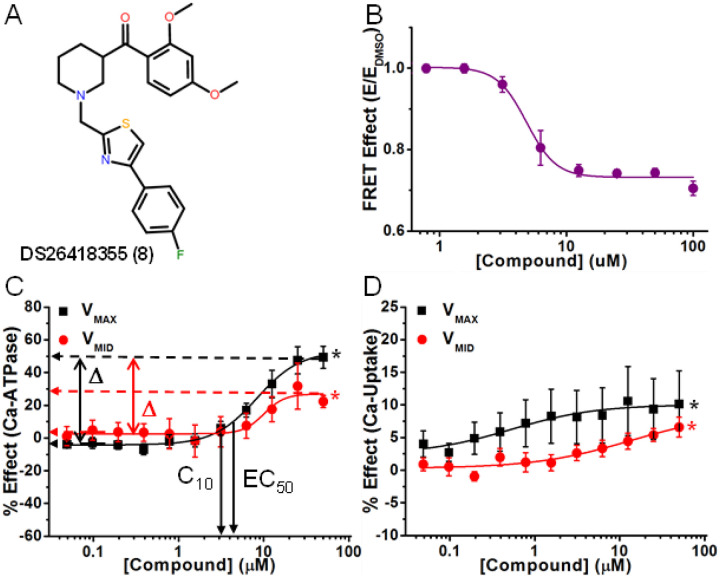
Compound 8 (singleton G) (Fig. 5A) decreased FRET (EC50 = 4.9 μM, Table 1) and increased Ca2+-ATPase at both VMAX (49%, the largest increase observed in the screen) and VMID (31 %) (Table 1 and Fig. 5C). For Ca2+-transport, effects (Δ values) were lower (10% for VMAX and 7% for VMID, Fig. 5D), decreasing CR to 0.4 (Table 1). EC50 values for Ca2+-ATPase were not significantly different at VMAX and VMID, and were ~ 2x greater than the values observed by FRET. C10 was 4.3 ^M (VMAX) and 7.8 ^M (VMID), indicating significant ATPase activation at low dosage. C10 for Ca-transport was 25 μM at VMAX, and was not determined at VMID.
Figure 5.
A representative activator compound that decreases FRET and uncouples Ca2+-ATPase from Ca2+-transport activity. (A) Chemical structure of Compound DS26418355 (Compound 8). (B) CRC of normalized FRET E in 2CS biosensor in live HEK cells shows decreasing FRET response with increasing [compound]. (C) CRC shows Ca2+-ATPase activation in pCSR vesicles under both VMAX (black) and VMID (red) Ca2+. (D) Activation was less for Ca2+-transport of SERCA2a in pCSR vesicles. Mean ± SEM, n = 3, *p<0.05.
Compound 2 (cluster A) induced similar Ca2+-ATPase activation (16% at VMAX, 7% at VMID) as observed with Compound 7, but induced smaller increases on Ca2+-transport functions (Table 1), decreasing CR to 0.53. Compounds 4 (cluster B) and 9 (cluster C) also showed similar effects as Compound 7, increasing both activities. Compound 4 increased Ca2+-ATPase at both VMAX and VMID by ~ 10% and increased Ca2+-transport at both VMAX (0.5%) and VMID (11%). Compound 9 increased both VMAX (21 %) and VMID (10%) for Ca2+-ATPase, with smaller increases in Ca2+-transport (3–5%). Compounds 4 and 9 decreased CR to similar extents (0.51) (Table 1).
In the second category of activators, Compounds 1 and 3 (cluster A), 5 (cluster B), and 6 (singleton E) activated Ca2+-ATPase at both VMAX and VMID, but induced divergent effects on Ca2+-transport. Compound 6 (Fig. 6A) decreased FRET with EC50 = 7.1 μM (Fig. 6B, Table 1), while moderately activating Ca2+-ATPase activity by 25% at VMAX and by 30% at VMID (Fig. 6C), with EC50 = 11 μM for both VMAX and VMID. It induced divergent effects during Ca2+-transport, inhibiting VMAX by 21% and activating VMID by 17% (Table 1 and Fig. 6D). C10 was similar at the two Ca2+ concentrations (9.7 μM and 8.7 μM), but was significantly different for Ca2+-transport (15 μM at VMAX, 4 μM at VMID). CR was decreased to 0.48 by Compound 6.
Compound 5 activated Ca2+-ATPase moderately at VMAX (25%) and VMID (35%). Ca2+-transport was inhibited slightly at VMAX (2%), but activated at VMID (20%) (Table 1). Compounds 1, and 3 induced low activating effects at VMID for Ca2+-transport (2% and 6%), but they inhibited Ca2+-transport at VMAX (−10% and − 20%) (Table 1). Compounds 1 and 5 induced similar decreases in the CR at VMAX (0.0.49 and 0.47), while Compound 3 induced a slightly smaller CR of 0.39 (Table 1). These effects are similar to that of unphosphorylated phospholamban (PLB) in cardiac SR35.
SERCA Inhibitors
Compounds 10–18 all decreased Ca2+-ATPase and Ca2+-transport activities at both VMAX and VMID. Compared with FRET EC50 of the activators (0.3–7 μM), most of the inhibitors (Compounds 10–18) showed weaker affinity, with FRET EC50 values in the range of 5–32 μM, but the maximum functional effects (efficacies) of the inhibitors tended to be greater (Table 1).
Compound 12 (cluster D) strongly inhibited both the Ca2+-ATPase and Ca2+-transport activities (Fig. 7C and D) to levels similar to the well-known SERCA inhibitor, thapsigargin, although thapsigargin acts with much greater affinity (EC50 ≈ 7.5 nM18) than compound 12 (EC50 = 3.8 μM) (Table 1).
Compounds 11, 12, and 13 (cluster D) showed similar inhibition of both SERCA2a functions: Ca2+-ATPase was inhibited by 61 %, 93%, and 81 % at VMAX; by 59%, 90%, and 72% at VMID. Ca2+-transport was completely inhibited. Compared with Ca2+-ATPase, Ca2+-transport inhibition at VMID required slightly higher compound concentration as shown by the shift to the right of the red curve (Fig. 7D).
Compounds 15 (singleton I) and 17 (singleton K) induced moderate inhibition of both activities, decreasing VMAX and VMID by ~ 50% for Ca2+-ATPase and slightly more for Ca2+-transport (70–85%). C10 values for Compound 15 were ~ 1 μM or less, slightly higher for Compound 17 (5–6%) (Table 1) for Ca2+-ATPase, and similar for Ca2+-uptake at both VMAX and VMID ranging from (0.9–1.7%).
Compounds 14 (singleton H) and 16 (singleton J) induced mild inhibition of Ca2+-ATPase, but a considerably larger effect of moderate-to-strong inhibition of the Ca2+-transport. Ca2+-ATPase decreased by 13% and 8% at VMAX, 26% and 16% at VMID. Ca2+-transport was inhibited by 83% and 66% at VMAX, 62% and 48% at VMID. C10 values ranged from 0.5 to 7μM, for Ca2+-ATPase at VMAX and VMID.
Compounds 10 (cluster C) and 18 (singleton L) induced mild inhibition of Ca2+-ATPase and moderate inhibition of Ca2+-transport. At VMAX, Ca2+-ATPase was inhibited by 31% and 24%, respectively; while at VMID it was inhibited by 24 and 16%, respectively. At VMAX Ca-transport was inhibited by 57% and 41 %, and at VMID by 34% and 16%. C10 values were 1 μM and 7 μM for Ca2+-ATPase, and 4.1 μM and 22.7 μM for Ca2+-transport.
Discussion
We identified new compounds based on an increase in donor FLT, within a human cardiac 2CS biosensor expressed in live mammalian cells, indicating a decrease in FRET, implying that the actuator (A) and nucleotide-binding domains (N) of SERCAC2a moved further apart. We confirmed that these compounds affect SERCA activity using Ca2+-ATPase and Ca2+-transport assays with SERCA2a in native SR preparations, where we further categorized them as activators or inhibitors. We identified two subcategories of activators, whereby the compound either (1) activates both Ca2+-ATPase and Ca2+-transport activities (Compounds 2, 4, 7, 8, and 9) (Figs. 4 and 5) and (2) activates Ca2+-ATPase with divergent effects on Ca-transport (Compounds 1, 3, 5, and 6) (Fig. 6). We identified four subcategories of inhibitors based on the extent of Ca2+ATPase and Ca2+-transport decrease for cardiac SERCA2a (Table 1 and Fig. 7).
In general, the FRET EC50 values were smaller (indicating higher potencies) for activators (Compounds 1–9; 0.3–7μM) than for inhibitors (Compounds 10–18; (3–32μM) (Table 1). The potencies observed by FRET and function are not precisely correlated, probably because the assays were performed on different types of samples (live cells vs purified proteins), measuring different properties (structure vs function).
Functional CRC assays showed that inhibitors tended to induce larger changes (indicating higher efficacies) than activators, in both Ca2+-ATPase activity and Ca2+-uptake. Also, most inhibitors induce a larger change in Ca2+-uptake than in Ca2+-ATPase, decreasing the coupling ratio. In general, the C10 and EC50 values were smaller, indicating greater potency, for inhibitors than for activators.
Effects of most activators were to reduce the CR, as they induced larger changes in the Ca2+-ATPase than in corresponding the Ca2+-uptake assay. The most notable exception is Compound 7, which activates Ca2+-transport even more than it activates Ca2+-ATPase, increasing CR. This compound will be a high priority as a lead compound for future efforts in medicinal chemistry and assays of physiological function. Ten compounds were binned into four clusters (A-D), while eight compounds were classified as singleton (E-L) (Table 1). Many compounds showed similar functional traits, suggesting that there are ligand-sensing sites in SERCA2a that recognize a range of scaffolds, or that the ligand-binding sites are close to each other, providing potentially powerful tools in the design of future compounds36,37,38. Compounds 2 (cluster A), 4 (cluster B), 7 (singleton F), 8 (singleton G), and 9 (cluster C) induced similar effects of moderate activation on VMAX in the Ca2+-ATPase assay with smaller activating effects on Ca2+-transport. Compounds in activator clusters A (Compounds 1 and 3) and B (Compound 5) along with Compound 6 from singleton E, showed similar functional effects: moderate activation of VMAX, with smaller activation of Ca2+-transport (decreased coupling) at both [Ca2+]. The compounds in clusters D (11, 12, and 13), C (10), and H-L (14–18) of inhibitors also induced similar functional effects.
There was little or no overlap in the hit compounds identified in our previous FRET HTS screen of the same (DIVERSet) library with another biosensor for tumor necrosis factor receptor 1 (TNFR122. There was 81% overlap in the fluorescent compounds detected in these two HTS screens, indicating that our FRET HTS screening methodologies independently and successfully removed the fluorescently interfering compounds21,24. In another HTS study of the DIVERSet library, using a SERCA functional (Ca2+-ATPase) assay in the primary screen, we discovered several activators, several of which showed isoform specificity for either SERCA1a or SERCA2a28. However, there was negligible overlap between hit compounds identified in that ATPase HTS study and in the current study that used FRET in the primary screen. This observation highlights the value of complementary HTS assays for the same target.
SERCA activators are needed when cardiac relaxation is impaired, as in early-onset diastolic dysfunction that precedes systolic impairment in HF1, diabetic cardiomyopathy39, Alzheimer’s disease40, or Duchenne muscular dystrophy (DMD)41. The stimulation of SERCA2a activity, as a novel therapeutic measure to relieve cardiac dysfunction in heart failure without arrhythmogenic effects, is a promising strategy to be used in combination with other first-line therapeutic agents such as β-blockers and ACE inhibitors42. Until recently, only a few compounds were known to stimulate SERCA: CDN1163 (stimulates Ca2+ transport)43,19, CP-154526 (increases the apparent Ca2+ affinity of SERC2a)44, Ro 41–0960 (increases SERCA maximal activity in high Ca2+)44, and istaroxime (stimulates SERCA activity)45. However, our recent HTS using Ca2+-ATPase activity as the target HTS assay identified ~ 19 new activators of SERCA28, and we identified nine activator compounds in the present study. A SERCA activator from our previous work shows promise as a therapeutic target for Alzheimer’s disease, as it rescued memory function in a mouse model for the disease40 as well as for DMD as it was shown to ameliorate dystrophic phenotypes in dystrophin-deficient mdx mice41. Of all these SERCA2a activators only istaroxime, a known Na+/K+ transporting ATPase inhibitor as well as an inotropic/lusitropic agent acting to enhance SERCA2a activity, has been in phase IIb clinical trials for treatment of heart failure45,46. However, because of its unsuitability for human usage (poor gastrointestinal absorption, high clearance rate, and extensive metabolic transformation)46, istaroxime derivatives were designed from QSAR studies and a new promising class of SERCA2a activators has been identified47,48,49.
Compounds 1, 3, 5, and 6 induced small effects on the VMAX of Ca2+-ATPase (~ 10–25% increase) and induced a negative effect on the VMAX of Ca2+-transport (Fig. 6C and D), thus decreasing the CR, which is likely to increase heat output13,52,54. These effects are similar to that of SLN on SERCA1a (skeletal muscle), where SLN induces no observable effect on the VMAX of Ca-ATPase but reduces the VMAX of Ca2+-transport (SERCA1a uncoupling), thus reducing CR50. This results in futile cycling of SERCA1a and higher usage of ATP resulting in increased non-shivering thermogenesis (NST)50. Another contributor to NST is Ca2+ leak from SR to sarcoplasm through RyR channels, stimulating SERCA to re-sequester Ca2+ into the SR, thus using more ATP and generating heat51. This increase in energy expenditure in muscle has been suggested as a potential therapeutic strategy for weight loss50,52. Thus, the SERCA uncouplers in this study may serve as the basis for further drug development targeting weight loss.
Over the past several decades (~ 60 years), research on the potential for small-molecule SERCA inhibitors as oncology therapeutics has yielded hundreds of SERCA inhibitors with varying potencies and efficacies15. Similarly, our discovery of new SERCA inhibitors with a range of potencies and efficacies is likely to be advantageous for treatment of non-cardiac applications15,53.
In the present study, the 2CS biosensor has been used to identify novel small-molecule effectors of SERCA that have diverse chemical scaffolds for binding to SERCA, resulting in an array of hit compounds that are activators and inhibitors. Most importantly, we discovered a potential lead compound (Compound 7) that activates Ca2+-uptake more than the Ca2+-ATPase, increasing the CR, so this will be a high riority for future efforts in medicinal chemistry and assays of physiological function. The enabling technology included three novel plate-readers, the FLT instrument used in the primary screen, and two spectral instruments that were used to remove interference of fluorescent compounds, allowing us to focus on valid SERCA activators and inhibitors. In the future, hits from the present study will be evaluated in more functional detail, including studies on multiple SERCA isoforms and on intact muscles and animals. Medicinal chemistry will be applied to elucidate structure-activity relationships, with the goal of designing analogs with greater potency and specificity22,54. We will also expand our approach to much larger compound libraries, since our primary screening technology is capable of evaluating thousands of compounds per hour.
Methods
Molecular biology
A two-color intramolecular human SERCA2a (2CS) biosensor, based on human cardiac SERCA2a fused to green fluorescent protein (eGFP) and red fluorescent protein (tagRFP) was developed to detect structural changes that are related to the functional changes of SERCA20. Briefly, tagRFP was genetically fused to the N-terminus of SERCA and eGFP was inserted as an intrasequence tag before residue 509 in the nucleotide-binding domain (N-domain)55,56. A donor-only (1CS) biosensor was created in a similar manner as the 2CS biosensor with the exception of the construct containing only eGFP The fluorescent proteins fused to SERCA in 2CS and 1CS do not significantly affect SERCA activity, in membranes purified from HEK cells18,20. A null biosensor construct consisting of eGFP and tagRFP connected by a 32-residue unstructured flexible linker peptide (G32R) was created as described previously18,20. All constructs were cloned into expression vectors containing the genes for antibiotic resistance to G418, puromycin, or blasticidin.
Cell culture
Stable cell lines were generated using either HEK293 (ATCC, Manassas, VA) or HEK293–6E (National Research Council, Canada) cells20. Briefly, cells were transiently transfected with 2CS, 1CS, or G32R null biosensor plasmids using Lipofecatime 3000 or 293fectin (Thermo Fisher Scientific). Flow cytometry was used to select and enrich for the population of cells expressing respective biosensors. Stable HEK293 cell lines were maintained in phenol red-free DMEM media (Gibco, Waltham, MA) supplemented with 2 mM GlutaMAX (Gibco, Waltham, MA), 10% fetal bovine serum (FBS) (Atlanta Biologicals, Lawrenceville, GA), 1 IU/mL penicillin/streptomycin (Gibco, Waltham, MA), and 250μg/mL G418 (Fisher Scientific). Stable HEK293–6E cell lines were maintained in F17 media (Sigma Aldrich) supplemented with Kolliphor p188 (Sigma Aldrich, St. Louis, MO), 200 nM/mL GlutaMAX, and either 1 μg/mL puromycin (Invitrogen, Carlsbad, CA) or 2μg/mL blasticidin (Goldbio) as a selection antibiotic. All cell lines were grown at 37°C with 5% CO2.
Compound handling
A DIVERSet 50,000 compound library was purchased from ChemBridge Corporation (San Diego, CA) at a 10 mM stock concentration for each compound. All compounds met the high quality standard of 100% identification by NMR and/or LC-MS and have a minimum purity of 85% and their identity verified using LC-MS/ELSD as confirmed by the ChemBridge Corporation. For the FRET HTS initial screens, the compound library was reformatted into 384 well Echo compatible plates using the Biomek FX (Beckman Coulter, Miami, FL) and 5 nL of either compound (columns 3–22 and 27–46) or DMSO (columns 1–2, 23–26, and 47–48) was dispensed into 1536 well black polystyrene assay plates (Greiner, Kremsmunste, Austria) using an Echo 550 liquid dispenser (Beckman Coulter) to yield a final assay screening concentration of 10μM. The low autofluorescence and low interwell cross-talk of these plates made them advantageous for FRET measurements. Plates were heat sealed with a PlateLoc Thermal Microplate Sealer (Agilent, Santa Clara, CA) and stored at −20°C prior to use. The same methods were applied for subsequent FRET retesting of the reproducible hit compounds identified in the pilot screen, except that the [compound] was tested at 10μM and 30 μM in triplicate.
FRET CRC assay plates (0.78–100 μM compound range) with at least ten different compound concentrations were made by adding the appropriate volume of compound or DMSO into black 384 well plates (Greiner Bio-One) using a Mosquito HV (SPTLabTech, United Kingdom). Subsequent ATPase and Ca2+-transport CRC assay plates (0–50 μM compound range) with repurchased compounds were made in a similar manner using with the Echo 550 (Beckman Coulter) using either 384 well transparent plates (Greiner Bio-One) or black-walled plates with transparent bottoms (Greiner Bio-One), respectively.
HTS sample preparation and FRET measurements
On each day of screening, cells were harvested, washed three times with PBS, and centrifuged at 300g for 5min. Cells were filtered using a 70μm cell strainer and diluted to 1–2 × 106 cells/mL. Cell concentration and viability were assessed using the Cell countess (Invitrogen) and trypan blue assay. During assays, cells were constantly and gently stirred using a magnetic stir bar at room temperature, keeping the cells in suspension and evenly distributed to avoid clumping. Cells were dispensed at 5μL or 50μL per well into assay plates (dispensed into 40 assay plates, each containing 1536 wells) pre-plated with either compound or DMSO using a Multdrop Combi liquid dispenser (Thermo Fisher Scientific, Pittsburg, PA) and sealed until needed. Because the kinetics of membrane permeability, diffusion, and/or binding of the compound to live cells may be compound-dependent, we tested two incubation times, 20 min and 120 min, for the FLT CRC. FRET EC50 values determined from both incubations were similar, but the 120 min incubation yielded a more reproducible and sigmoidal curve. Plates containing eight-point concentration curves of three tool compounds (known SERCA effectors) were also included on the plates as positive controls for the HTS FRET assay. The FRET HTS screen was performed over two days with a custom HTS fluorescence lifetime plate reader (FLT-PR) and spectral plate reader (SUPR) provided by Photonic Pharma LLC (Minneapolis, MN)18.
The same methods were applied for subsequent FRET retesting of the reproducible hit compounds identified in the pilot screen, except that the compound tested at 10μM and 30 μM [compound]. 158 hit compounds were picked from the library master plates and reloaded onto new assay plates for retesting with 2CS and a null biosensor. Then 18 hit compounds were selected and purchased from ChemBridge Corporation to determine CRC from FRET, ATPase, and Ca-transport assays using at least ten different concentrations by repeatedly scanning the 1536-well plates.
FRET HTS instrumentation and data analysis
A detailed description of the high-throughput fluorescence lifetime plate reader (FLT-PR) and spectral unmixing plate reader (SUPR), manufactured by Fluorescence Innovations Inc and provided by Photonic Pharma, LLC was described previously18,21. Briefly, for lifetime measurement with the FLT-PR, the observed donor-fluorescence waveform, F (t) was fit by a convolution of the measured instrument response function (IRF) and a single-exponential decay to obtain the lifetime (τ) of the donor fluorophore57 in the absence (τD) and presence (τDA) of the acceptor as described in Eq. (1):
| 1 |
FRET efficiency (E) was determined as the fractional decrease of donor FLT in the absence and in the presence of acceptor as in Eq. (2):
| 2 |
E was determined in the presence and absence of compound and normalized relative to E of the DMSO control. For spectral detection with the SUPR, the observed fluorescence emission spectrum F(λ) was fit by least-squares minimization of a linear combination of component spectra for donor (D), acceptor (A), cellular autofluorescence (C) and water Raman (W) as described previously17.
HTS data analysis
FLT-PR data was used as the primary metric for flagging potential hit compounds. After fitting waveforms with a single exponential decay to quantify donor lifetime, the change in fluorescence lifetime (Δ FLT) was computed by performing a moving average subtraction in the order the plate was scanned with a window size of half a plate row (24 columns). The reasons for this are twofold: 1) plate gradients are often observed due to heating of the digitizer during acquisition and 2) performing ΔFLT computations with DMSO controls alone can sometimes result in artifacts as a half of the DMSO wells are on the edge of plates, which occasionally exhibit artifacts due to processes needed for the preparation of the drug library being tested. As most compounds are likely to be non-hits, and therefore DMSO like, computation of a moving average is an effective alternative to solving both gradient issues and edge-effect distortion of the primary metric for hit selection, Δτ. As hit compounds from FLT-PR were to be further triaged with a secondary technique (using the spectral plate reader), a generous cutoff was set at a robust z-score of 3 on a plate-by-plate basis. The robust z-score was used, where the median (M) and median absolute deviation (MAD) are used in place of the mean and standard deviation (Eq. 3), to best capture the most hits, as the standard z-score is more subject to strong outliers (compounds that fall outside of the defined upper and lower limits)18.
| 3 |
To remove “false positive” fluorescent compounds, the similarity index (SI17 was computed by comparing a region (500–540nm) of the donor only spectrum (l(a)) for each well to that of the plate-wide average DMSO spectrum (l(b)) in the same wavelength band as described in Eq. 421. Compounds that exceeded an SI robust z-score of 5 (corresponding to an SI of 2×10−4) were deemed likely fluorescent compounds and removed from consideration.
| 4 |
Spectral (SUPR) data was processed similarly to FLT-PR data, with the ΔR/G ratio being computed by applying the same moving average filter on the initial measurement of the ratio of the acceptor amplitude over the donor amplitude as found by fitting basis sets of the component spectra through least squares minimization. The hit threshold was also set using a robust z-score of 3. While the FLT-PR data and SUPR data showed a robust correlation, the FLT-PR data exhibited some strong outliers, presumably due to compounds directly modifying the donor lifetime. To eliminate these likely interfering compounds, correlation was enforced by eliminating compounds that exceed a robust z-score of 3 from the median value of the ratio of ΔFLT over the ΔR/G ratio metric. Additional interfering compounds were removed using two-channel lifetime detection18.
Cardiac SR preparation
Cardiac SR vesicles were isolated from fresh porcine left ventricular tissue using differential centrifugation of the homogenized tissue as previously described58. The SR vesicles were flash-frozen and stored at −80°C until needed.
Enzymatic SERCA activity assays of FRET hit compounds
Functional assays were performed using porcine cardiac SR vesicles16. An enzyme-coupled, NADH-linked ATPase assay was used to measure SERCA ATPase activity in 384-well microplates. Each well contained 50 mM MOPS (pH 7.0), 100 mM KCl, 1 mM EGTA, 0.2 mM NADH, 1 mM phosphoenol pyruvate, 10 IU/mL of pyruvate kinase, 10 IU/mL of lactate dehydrogenase, 7 μM of the calcium ionophore A23187 (Sigma), and CaCl2 was added to set free [Ca2+] to three different concentrations59. The Ca2+-ATPase were measured at VMAX (saturating, pCa 5.4), VM!D (subsaturating, midpoint, pCa 6.2), and basal (non-activating, pCa 8.0) [Ca2+]. 10 μg/mL of SR vesicle, calcium, compound (0.048 to 50 μM), and assay mix were incubated for 20 min at room temperature before measurement of functional assays with each of the 18 hit compounds, because a shorter incubation time than the FRET live-cell assays achieved optimal responses. The assay was started upon the addition of MgATP at a final concentration of 5 mM (total volume to 80 μL), and absorbance was read in a SpectraMax Plus microplate spectrophotometer from Molecular Devices (Sunnyvale, CA) at 340nm.
Ca2+-transport assays of FRET hit compounds
Ca2+-transport assays were performed with similar porcine SR samples as in the Ca2+-ATPase assays described above. The compound effect on the Ca2+-transport activity of SERCA2a was determined using an oxalate-supported assay in which the change in fluorescence in a Ca-sensitive dye, Fluor-4, was determined as previously described18. A buffered solution containing 50 mM MOPS (pH 7.0), 100 mM KCl, 30 mg/mL sucrose, 1 mM EGTA, 10 mM potassium oxalate, 2 μM Fluo-4, 30 μg/mL porcine cardiac SR vesicles, CaCl2 calculated to reach the free [Ca2+] (pCa 8.0, 6.2, and 5.4), and compound (0.048 to 50μM) was dispensed into 384-well black walled, transparent bottomed plates (Greiner Bio-One) containing the tested small molecule and incubated at 22°C for 20 minutes while covered and protected from light. To start the reaction, MgATP was added to a final concentration of 5 mM, and the decrease in 485-nm excited fluorescence of Fluo-4 was monitored at 520 nm for 15 min using a FLIPR Tetra (Molecular Devices, San Jose, CA).
Data analysis of FRET CRC assays of hit compounds
FRET efficiency (E) (Eq. 2) was determined as the fractional decrease of donor (1CS) lifetimes (τD) in the presence of acceptor (2CS) fluorophore (τDA) due to FRET as described in Eq. 1 and normalized to DMSO controls.
Data analysis of Ca2+-ATPase and Ca2+-transport CRC assays
SERCA2a ATPase (or Ca-transport) activity at pCa 8.0 was subtracted from pCa 5.4 and pCa 6.2 values. The % effect ATPase (or Ca-transport transport) activity was normalized to the DMSO only (or 2CS in the absence of compound), and then were plotted against [Ca], and the curves were fitted using the Hill function, where V is the initial ATPase rate (or fluorescence rate), VMAX is the ATPase (or Ca2+-transport) at saturating [Ca2+], and EC50 or pKCa, or VM!D is the apparent Ca2+ dissociation constant as described previously ATPase (or Ca2+-transport) at (Midpoint Ca2+)44. These parameters and the [Ca2+] at 10% (C10) above or below baseline pCa (8.0) are reported in Table 1.
Cheminformatic analysis of hit compounds
An online interactive program was used to perform cheminformatics analysis60 to determine whether the hit compounds had structural similarity by identifying common chemical scaffolds (core structural feature) using binning, multidimensional scaling (MDS), and compound similarity methods where the Tanimoto coefficient31 and maximum common substructure31 values were used to determine clustering (Supplementary Table S1). The physicochemical properties (for e.g. Lipinski Rule of 5) and bioactivity properties of the compounds were also used in the clustering analysis34. A cluster contained two or more compounds with similarity score > 0.4, while a unique compound with a similarity score < 0.4 was referred to as a singleton.
Statistical analysis
Analysis of two-group comparisons was done by a two-tailed unpaired Student’s t-test (*p < 0.05) using the data analysis program Microsoft Excel (Santa Rosa, CA). Data are presented as mean ± SEM calculated from a minimum of three separate experiments (n = 3).
Acknowledgments
Prachi Bawaskar, Ben Grant, Simon J. Gruber, Evan W. Kleinboehl, Ang Li, Ji Li, Kurt Peterson, Seth L. Robia, and Tory M. Schaaf contributed to the conception of this project. Jesse E. McCaffrey, Bengt Svensson, Sarah Blakely Anderson, and J. Michael Autry provided helpful discussions. Marzena Brinkmann provided helpful advice on the chemical scaffold nomenclature. Fluorescence microscopy was performed at the UMN Imaging Center, flow cytometry at the UMN Lillehei Heart Institute, compound dispensing at the UMN Institute of Therapeutic Drug Discovery and Development, and spectroscopy at the UMN Biophysical Technology Center. This work was supported by NIH grants R01HL139065 (to DDT and RLC) and R37AG26160 (to DDT).
Footnotes
Competing interests
DDT and RLC hold equity in, and serve as executive officers for Photonic Pharma LLC, which had no role in this study except for providing some instrumentation. OR is the sole proprietor of Editing Science LLC, which had no role in this study. This relationship has been reviewed and managed by the University of Minnesota. SLY, ART, LNR, RTR, PAB, and CCA declare no conflicts of interest in regard to this manuscript.
Supplementary Information
Available online
Table 1
Table 1 is available in Supplementary Files section.
Supplementary Files
Contributor Information
Osha Roopnarine, University of Minnesota.
Samantha L. Yuen, University of Minnesota
Andrew R. Thompson, University of Minnesota
Lauren N. Roelike, University of Minnesota
Robyn T. Rebbeck, University of Minnesota
Phillip A. Bidwell, University of Minnesota
Courtney C. Aldrich, University of Minnesota
Razvan L. Cornea, University of Minnesota
David D. Thomas, University of Minnesota
Data availability
All the data discussed are presented within the article and Supplementary Information and are available from the corresponding authors (OR and DDT) on reasonable request.
References
- 1.Bers D. M., Eisner D. A. & Valdivia H. H. Sarcoplasmic reticulum Ca2+ and heart failure: roles of diastolic leak and Ca2+ transport. Circ Res 93, 487–490, doi: 10.1161/01.RES.0000091871.54907.6B (2003). [DOI] [PubMed] [Google Scholar]
- 2.Bers D. M. Cardiac sarcoplasmic reticulum calcium leak: basis and roles in cardiac dysfunction. Annu Rev Physiol 76, 107–127, doi: 10.1146/annurev-physiol-020911-153308 (2014). [DOI] [PubMed] [Google Scholar]
- 3.Viskupicova J. & Rezbarikova P Natural Polyphenols as SERCA Activators: Role in the Endoplasmic Reticulum Stress-Related Diseases. Molecules 27, doi: 10.3390/molecules27165095 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bers D. M. & Despa S. Cardiac myocytes Ca2+ and Na+ regulation in normal and failing hearts. J Pharmacol Sci 100, 315–322 (2006). [DOI] [PubMed] [Google Scholar]
- 5.Rathod N. et al. Nothing Regular about the Regulins: Distinct Functional Properties of SERCA Transmembrane Peptide Regulatory Subunits. Int J Mol Sci 22, doi: 10.3390/ijms22168891 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li A. et al. The transmembrane peptide DWORF activates SERCA2a via dual mechanisms. J Biol Chem 296, 100412, doi: 10.1016/j.jbc.2021.100412 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fisher M. E. et al. Dwarf open reading frame (DWORF) is a direct activator of the sarcoplasmic reticulum calcium pump SERCA. Elife 10, doi: 10.7554/eLife.65545 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rustad M. D., Roopnarine O., Cornea R. L. & Thomas D. D. Interaction of DWORF with SERCA and PLB as determined by EPR spectroscopy. Biochem Biophys Res Commun 645, 97–102, doi: 10.1016/j.bbrc.2023.01.041 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nelson B. R. et al. A peptide encoded by a transcript annotated as long noncoding RNA enhances SERCA activity in muscle. Science 351, 271–275, doi: 10.1126/science.aad4076 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Makarewich C. A. et al. The DWORF micropeptide enhances contractility and prevents heart failure in a mouse model of dilated cardiomyopathy. Elife 7, doi: 10.7554/eLife.38319 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hadri L. & Hajjar R. J. Calcium cycling proteins and their association with heart failure. Clin Pharmacol Ther 90, 620–624, doi: 10.1038/clpt.2011.161 clpt2011161 [pii] (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sato D., Uchinoumi H. & Bers D. M. Increasing SERCA function promotes initiation of calcium sparks and breakup of calcium waves. J Physiol 599, 3267–3278, doi: 10.1113/JP281579 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guarnieri A. R., Benson T. W. & Tranter M. Calcium cycling as a mediator of thermogenic metabolism in adipose tissue. Mol Pharmacol, doi: 10.1124/molpharm.121.000465 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nowack J., Giroud S., Arnold W. & Ruf T. Muscle Non-shivering Thermogenesis and Its Role in the Evolution of Endothermy. FrontPhysiol 8, 889, doi: 10.3389/fphys.2017.00889 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Michelangeli F. & East J. M. A diversity of SERCA Ca2+ pump inhibitors. Biochem Soc Trans 39, 789–797, doi: 10.1042/BST0390789 (2011). [DOI] [PubMed] [Google Scholar]
- 16.Gruber S. J. et al. Discovery of enzyme modulators via high-throughput time-resolved FRET in living cells. J BiomolScreen 19, 215–222, doi: 10.1177/108705711351074019/2/215 [pii] (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schaaf T. M. et al. High-Throughput Spectral and Lifetime-Based FRET Screening in Living Cells to Identify Small-Molecule Effectors of SERCA. SLAS Discov 22, 262–273, doi: 10.1177/1087057116680151 (2017b). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schaaf T. M. et al. Live-Cell Cardiac-Specific High-Throughput Screening Platform for Drug-Like Molecules that Enhance Ca(2+) Transport. Cells 9, doi: 10.3390/cells9051170 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cornea R. L. et al. High-throughput FRET assay yields allosteric SERCA activators. J Biomol Screen 18, 97–107, doi: 10.1177/1087057112456878 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schaaf T. M. et al. Red-Shifted FRET Biosensors for High-Throughput Fluorescence Lifetime Screening. Biosensors (Basel) 8, doi: 10.3390/bios8040099 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schaaf T. M., Peterson K. C., Grant B. D., Thomas D. D. & Gillispie G. D. Spectral Unmixing Plate Reader: High-Throughput, High-Precision FRET Assays in Living Cells. SLAS Discov 22, 250–261, doi: 10.1177/1087057116679637 (2017a). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lo C. H. et al. Noncompetitive inhibitors of TNFR1 probe conformational activation states. Sci Signal 12, doi: 10.1126/scisignal.aav5637 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lo C. H., Schaaf T. M., Thomas D. D. & Sachs J. N. Fluorescence-Based TNFR1 Biosensor for Monitoring Receptor Structural and Conformational Dynamics and Discovery of Small Molecule Modulators. Methods Mol Biol 2248, 121–137, doi: 10.1007/978-1-0716-1130-2_9 (2021). [DOI] [PubMed] [Google Scholar]
- 24.Lo C. H. et al. Discovery of Small Molecule Inhibitors of Huntingtin Exon 1 Aggregation by FRET-Based High-Throughput Screening in Living Cells. ACS Chem Neurosci 11, 2286–2295, doi: 10.1021/acschemneuro.0c00226 (2020). [DOI] [PubMed] [Google Scholar]
- 25.Rebbeck R. T. et al. Novel drug discovery platform for spinocerebellar ataxia, using fluorescence technology targeting beta-III-spectrin. J Biol Chem 296, 100215, doi: 10.1074/jbc.RA120.015417 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Baell J. B. & Holloway G. A. New substructure filters for removal of pan assay interference compounds (PAINS) from screening libraries and for their exclusion in bioassays. J Med Chem 53, 2719–2740, doi: 10.1021/jm901137j (2010). [DOI] [PubMed] [Google Scholar]
- 27.Autry J. M. et al. Sarcoplasmic Reticulum from Horse Gluteal Muscle Is Poised for Enhanced Calcium Transport. Vet Sci 8, doi: 10.3390/vetsci8120289 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bidwell P. A. et al. A Large-Scale High-Throughput Screen for Modulators of SERCA Activity. Biomolecules 12, doi: 10.3390/biom12121789 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Racker E. & Eytan E. A coupling factor from sarcoplasmic reticulum required for the translocation of Ca2+ ions in a reconstituted Ca2+ATPase pump. J Biol Chem 250, 7533–7534 (1975). [PubMed] [Google Scholar]
- 30.Yu X. & Inesi G. Variable stoichiometric efficiency of Ca2+ and Sr2+ transport by the sarcoplasmic reticulum ATPase. J Biol Chem 270, 4361–4367, doi: 10.1074/jbc.270.9.4361 (1995). [DOI] [PubMed] [Google Scholar]
- 31.Willett P The Calculation of Molecular Structural Similarity: Principles and Practice. Mol Inform 33, 403–413, doi: 10.1002/minf.201400024 (2014). [DOI] [PubMed] [Google Scholar]
- 32.Vitaku E., Smith D. T. & Njardarson J. T. Analysis of the structural diversity, substitution patterns, and frequency of nitrogen heterocycles among U.S. FDA approved pharmaceuticals. J Med Chem 57, 10257–10274, doi: 10.1021/jm501100b (2014). [DOI] [PubMed] [Google Scholar]
- 33.Shinde S. R. et al. A systematic appraisal on catalytic synthesis of 1,3-oxazole derivatives: A mechanistic review on metal dependent synthesis. Synthetic Communications 52, 1–36, doi: 10.1080/00397911.2021.1989596 (2022). [DOI] [Google Scholar]
- 34.Lipinski C. A., Lombardo F., Dominy B. W. & Feeney P J. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv Drug DelivRev 46, 3–26, doi: 10.1016/s0169-409x(00)00129-0 (2001). [DOI] [PubMed] [Google Scholar]
- 35.Kranias E. G. & Hajjar R. J. Modulation of cardiac contractility by the phospholamban/SERCA2a regulatome. Circ Res s 110, 1646–1660, doi: 10.1161/CIRCRESAHA.111.259754 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Balamurugan R., Dekker F. J. & Waldmann H. Design of compound libraries based on natural product scaffolds and protein structure similarity clustering (PSSC). Mol Biosyst 1, 36–45, doi: 10.1039/b503623b (2005). [DOI] [PubMed] [Google Scholar]
- 37.Koch M. A. et al. Charting biologically relevant chemical space: a structural classification of natural products (SCONP). Proc Natl Acad Sci U S A 102, 17272–17277, doi: 10.1073/pnas.0503647102 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ma B., Shatsky M., Wolfson H. J. & Nussinov R. Multiple diverse ligands binding at a single protein site: a matter of pre-existing populations. Protein Sci 11, 184–197, doi: 10.1110/ps.21302 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lebeche D., Davidoff A. J. & Hajjar R. J. Interplay between impaired calcium regulation and insulin signaling abnormalities in diabetic cardiomyopathy. Nat Clin Pract Cardiovasc Med 5, 715–724, doi: 10.1038/ncpcardio1347 (2008). [DOI] [PubMed] [Google Scholar]
- 40.Krajnak K. & Dahl R. A new target for Alzheimer’s disease: A small molecule SERCA activator is neuroprotective in vitro and improves memory and cognition in APP/PS1 mice. Bioorg Med Chem Let 28, 1591–1594, doi: 10.1016/j.bmcl.2018.03.052 (2018). [DOI] [PubMed] [Google Scholar]
- 41.Nogami K. et al. Pharmacological activation of SERCA ameliorates dystrophic phenotypes in dystrophin-deficient mdx mice. Hum Mol Genet 30, 1006–1019, doi: 10.1093/hmg/ddab100 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shareef M. A., Anwer L. A. & Poizat C. Cardiac SERCA2A/B: therapeutic targets for heart failure. Eur J Pharmacol 724, 1–8, doi: 10.1016/j.ejphar.2013.12.018 (2014). [DOI] [PubMed] [Google Scholar]
- 43.Sordi G., Goti A., Young H. S., Palchetti I. & Tadini-Buoninsegni F. Stimulation of Ca(2+) -ATPase Transport Activity by a Small-Molecule Drug. ChemMedChem 16, 3293–3299, doi: 10.1002/cmdc.202100350 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stroik D. R. et al. Targeting protein-protein interactions for therapeutic discovery via FRET-based high-throughput screening in living cells. Sci Rep 8, 12560, doi: 10.1038/s41598-018-29685-z (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rocchetti M. et al. Modulation of sarcoplasmic reticulum function by PST2744 [istaroxime; (E,Z)-3-((2-aminoethoxy)imino) androstane-6,17-dione hydrochloride)] in a pressure-overload heart failure model. J Pharmacol Exp Ther 326, 957–965, doi: 10.1124/jpet.108.138701 (2008). [DOI] [PubMed] [Google Scholar]
- 46.Gheorghiade M., Ambrosy A. P, Ferrandi M. & Ferrari P Combining SERCA2a activation and Na-K ATPase inhibition: a promising new approach to managing acute heart failure syndromes with low cardiac output. Discov Med 12, 141–151 (2011). [PubMed] [Google Scholar]
- 47.Luraghi A. et al. Highly Selective SERCA2a Activators: Preclinical Development of a Congeneric Group of First-in-Class Drug Leads against Heart Failure. J Med Chem 65, 7324–7333, doi: 10.1021/acs.jmedchem.2c00347 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Arici M. et al. Istaroxime Metabolite PST3093 Selectively Stimulates SERCA2a and Reverses Disease-Induced Changes in Cardiac Function. J Pharmacol Exp Ther 384, 231–244, doi: 10.1124/jpet.122.001335 (2023). [DOI] [PubMed] [Google Scholar]
- 49.Avvisato R., Jankauskas S. S. & Santulli G. Istaroxime and Beyond: New Therapeutic Strategies to Specifically Activate SERCA and Treat Heart Failure. J Pharmacol Exp Ther 384, 227–230, doi: 10.1124/jpet.122.001446 (2023). [DOI] [PubMed] [Google Scholar]
- 50.Bal N. C. & Periasamy M. Uncoupling of sarcoendoplasmic reticulum calcium ATPase pump activity by sarcolipin as the basis for muscle non-shivering thermogenesis. Philos Trans R Soc Lond B Biol Sci 375, 20190135, doi: 10.1098/rstb.2019.0135 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Meizoso-Huesca A., Pearce L., Barclay C. J. & Launikonis B. S. Ca(2+) leak through ryanodine receptor 1 regulates thermogenesis in resting skeletal muscle. Proc Natl Acad Sci U S A 119, doi: 10.1073/pnas.2119203119 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Maurya S. K. & Periasamy M. Sarcolipin is a novel regulator of muscle metabolism and obesity. Pharmacol Res 102, 270–275, doi: 10.1016/j.phrs.2015.10.020 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pagliaro L., Marchesini M. & Roti G. Targeting oncogenic Notch signaling with SERCA inhibitors. J Hematol Oncol 14, 8, doi: 10.1186/s13045-020-01015-9 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nikolaienko R. et al. New N-aryl-N-alkyl-thiophene-2-carboxamide compound enhances intracellular Ca(2+) dynamics by increasing SERCA2a Ca(2+) pumping. Biophys J, doi: 10.1016/j.bpj.2022.12.002 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hou Z. et al. 2-Color calcium pump reveals closure of the cytoplasmic headpiece with calcium binding. PLoS One 7, e40369, doi: 10.1371/journal.pone.0040369P0NE-D-12-07470 [pii] (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pallikkuth S. et al. Phosphorylated phospholamban stabilizes a compact conformation of the cardiac calcium-ATPase. Biophys J 105, 1812–1821, doi: 10.1016/j.bpj.2013.08.045S0006-3495(13)01017-5 [pii] (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Talbot C. B. et al. Correction Approach for Delta Function Convolution Model Fitting of Fluorescence Decay Data in the Case of a Monoexponential Reference Fluorophore. J Fluoresc 25, 1169–1182, doi: 10.1007/s10895-015-1583-4 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Fruen B. R., Bardy J. M., Byrem T. M., Strasburg G. M. & Louis C. F. Differential Ca(2+) sensitivity of skeletal and cardiac muscle ryanodine receptors in the presence of calmodulin. Am J Physiol Cell Physiol 279, C724–733, doi: 10.1152/ajpcell.2000.279.3.C724 (2000). [DOI] [PubMed] [Google Scholar]
- 59.Mueller B., Karim C. B., Negrashov I. V., Kutchai H. & Thomas D. D. Direct detection of phospholamban and sarcoplasmic reticulum Ca-ATPase interaction in membranes using fluorescence resonance energy transfer. Biochemistry 43, 8754–8765 (2004). [DOI] [PubMed] [Google Scholar]
- 60.Backman T. W., Cao Y. & Girke T. ChemMine tools: an online service for analyzing and clustering small molecules. Nucleic Acids Res 39, W486–491, doi: 10.1093/nar/gkr320 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All the data discussed are presented within the article and Supplementary Information and are available from the corresponding authors (OR and DDT) on reasonable request.