Abstract
The bundle-forming pilus (BFP) of enteropathogenic Escherichia coli (EPEC), an established virulence factor encoded on the EPEC adherence factor (EAF) plasmid, has been implicated in the formation of bacterial autoaggregates and in the localized adherence of EPEC to cultured epithelial cells. While understanding of the pathogenic mechanism of this organism is rapidly improving, a receptor ligand for BFP has not yet been identified. We now report, using both solid-phase and liposome binding assays, that BFP expression correlates with phosphatidylethanolamine (PE) binding. In a thin-layer chromatogram overlay assay, specific recognition of PE was documented for BFP-expressing strains, including E2348/69, a wild-type EPEC clinical isolate, as well as a laboratory strain, HB101, transformed with a bfp-carrying plasmid. Strains which did not express BFP did not bind PE, including a bfpA disruptional mutant of E2348/69, EAF plasmid-cured E2348/69, and HB101. E2348/69 also aggregated PE-containing liposomes but not phosphatidylcholine- or phosphatidylserine-containing liposomes, while BFP-negative strains did not produce aggregates with any tested liposomes. Purified BFP preparations bound commercial PE standards as well as a PE-containing band within lipid extracts from human epithelial cells and from E2348/69. Our results therefore indicate a specific interaction between BFP and PE and suggest that PE may serve as a BFP receptor for bacterial autoaggregation and may promote localized adherence to host cells, both of which contribute to bacterial pathogenesis.
Infection with enteropathogenic Escherichia coli (EPEC) is a major cause of severe infantile diarrhea, particularly in parts of the developing world (33). Knowledge of EPEC pathogenesis has rapidly expanded in the past 5 years, with the result that a fairly comprehensive model of attachment and infection has been developed. Based on studies with cultured epithelial cell lines, a three-stage model of EPEC attachment to host cells was developed (9). In this model, initial attachment is mediated by a type IV pilus, the bundle-forming pilus (BFP), encoded on the EPEC adherence factor (EAF) plasmid. Recent studies with pediatric intestinal biopsy samples have suggested that BFP plays a role in interbacterial aggregation, leading to the formation of complex three-dimensional colonies producing the characteristic localized adherence pattern (15). Whether BFP mediates host attachment, interbacterial aggregation, or both, it is a recognized virulence factor of EPEC, as has recently been confirmed in studies with volunteers (3). The expression of BFP requires a cluster of 14 genes, including bfpA, which encodes bundlin, the major structural BFP subunit (36, 37). A transcriptional activator encoded by the perA operon (also called bfpT), located elsewhere on the EAF plasmid, is required for bfp operon expression (20, 39). Despite intense interest in BFP, a receptor has not yet been identified.
In earlier studies, it was found that EPEC (E2348/69), like a number of other pathogens, including enterhemorrhagic Escherichia coli, Helicobacter pylori, Helicobacter mustelae, Haemophilus influenzae, Chlamydia pneumoniae, Chlamydia trachomatis, and Campylobacter upsaliensis (2, 5, 16, 29, 30, 38), bound in a specific and dose-dependent manner to phosphatidylethanolamine (PE). It was also demonstrated that a number of bfp-positive clinical isolates recognized PE, while a plasmid-cured (bfp-negative) strain, JPN15, and several bfp-negative clinical isolates did not (2).
In this study, we examined the relationship between BFP expression and PE recognition for both wild-type E2348/69 and a number of bfp-negative strains and found that specific elimination of bfp ablates PE recognition and that transformation of E. coli with the bfp gene results in the induction of PE binding. Purified BFP also binds to commercial PE standards and to PE extracted from human epithelial cells and from E2348/69.
MATERIALS AND METHODS
Materials.
Thin-layer chromatography (TLC) plates (Polygram-SilG) were purchased from Macherey-Nagel (Duren, Germany). Phospholipids (PE from E. coli, lysophosphatidylethanolamine (lysoPE), phosphatidylcholine [PC] from egg yolk, and phosphatidylserine [PS] from bovine liver), sulfogalactosylceramide (SGC), and cholesterol were obtained from Sigma Chemical Co. (St. Louis, Mo.). Ganglioside GM1 was isolated from bovine brain tissue as previously described (32), and gangliotetraosylceramide (Gg4) was prepared by desialylation of GM1 (30). Rabbit antiserum to an outer membrane preparation (anti-OMP) of HS, a human commensal E. coli strain, was kindly provided by P. Sherman, Division of Gastroenterology/Nutrition, Hospital for Sick Children, Toronto, Ontario, Canada. A polyclonal antiserum to BFP was prepared as previously described (12). Goat anti-rabbit–fluorescein isothiocyanate conjugate (GAR-FITC) and bovine serum albumin were purchased from Sigma.
Bacterial strains and growth conditions.
The characteristics of the bacterial strains used in this study are listed in Table 1. Strains 31-6-1(1), HB101(pMAR7), and E2348/69(pOG127) were generously provided by J. Kaper, University of Maryland School of Medicine, Baltimore. Bacteria were stored in 40% glycerol–5% citrate at −70°C. Prior to use, bacteria were cultured on Luria agar supplemented with the appropriate antibiotics (Table 1). For Western blot assays, bacteria were subcultured overnight on either horse blood agar or Trypticase soy agar with 5% defibrinated sheep blood (TSA blood agar) or for 4 h in Dulbecco minimum essential medium (DMEM) (to maximize BFP expression) (12). For binding assays, two suspension protocols were compared: overnight cultures from TSA blood agar suspended in phosphate-buffered saline (PBS) with 1% bovine serum albumin and overnight Luria agar cultures suspended and grown for 4 h in DMEM. Both suspensions were used immediately after preparation in the binding assays.
TABLE 1.
Bacterial strains used in this studya
| Strain | Description | Reference | EAF plasmid | bfpA genotype |
|---|---|---|---|---|
| E2348/69 | Wild-type EPEC | 27 | + | + |
| JPN15 | EAF plasmid-cured E2348/69 | 17 | − | − |
| HB101 | Nonpathogenic laboratory E. coli K-12 strain | 31 | − | − |
| 31-6-1(1) | E2348/69 mutated in bfpA; Kanr | 8 | + | − |
| HB101(pMAR7) | HB101 complemented with bfpA-containing plasmid pMAR7; Ampr | 14 | + | + |
| E2348/69(pOG127) | Derivative of E2348/69 mutated in perA; Cmr | 14 | + | + |
+, positive; −, negative.
Lipid extracts from human epithelial cells and from bacteria.
The human epithelial cell line HEp-2 (American Type Culture Collection, Manassas, Va.) was grown in minimum essential medium supplemented with 10% decomplemented fetal calf serum, 0.5% glutamine, 1% sodium bicarbonate, 2% streptomycin-penicillin, and 1% amphotericin. A lower-phase lipid extract of the cells was prepared as described previously (2, 13) and stored at −70°C. A lower-phase lipid extract was similarly prepared from a pellet of E2348/69.
TLC overlay binding assay.
A TLC overlay assay (2) was used to assess bacterial interactions with PE, PC, Gg4, GM1, and lipid extracts from HEp-2 cells and from E2348/69. The choice of the phospholipid standards and glycolipids was based on previous work in which E2348/69 binding was assayed with a large panel of glycolipids and phospholipids, with binding being seen only for PE and Gg4 (2). The effect of anti-BFP antiserum on bacterium-lipid interactions was assayed by preincubating bacterial suspensions with equivalent dilutions of either anti-BFP or rabbit nonimmune serum (30 min at 37°C) prior to overlaying on phospholipid standards immobilized on TLC plates. Bacterial binding was visualized immunologically as previously described (2).
Bacterial binding to liposomes.
Bacterial recognition of phospholipids was also determined by assessing the aggregation of bacteria with various phospholipid vesicles as previously described (2). Bacterial suspensions (100 μl of 109 CFU/ml) were incubated for 1 h at 37°C with 50 μl of vesicles containing PE, PC, PS, or cholesterol (0.20 or 0.10 mg of lipid/ml). The degree of binding was estimated by the size and number of liposome aggregates observed by differential interference contrast microscopy.
Western blot analysis of BFP expression.
Bacterial expression of BFP was analyzed by detection of the 19.5-kDa BFP structural subunit, BfpA, by Western blot analysis as previously described (8). Blots were blocked with 5% skim milk powder–50 mM Tris-buffered saline–0.05% Tween 20, reacted with rabbit anti-BFP antiserum (12), incubated with goat anti-rabbit–fluorescein isothiocyanate conjugate, and developed with hydrogen peroxide.
Isolation and purification of BFP.
BFP was purified as previously described (12).
BFP-lipid binding.
BFP was radiolabeled with iodine-125 as previously described (40) and overlaid on TLC-separated lipids. Binding was detected by autoradiography.
RESULTS
Expression of BFP correlates with PE binding.
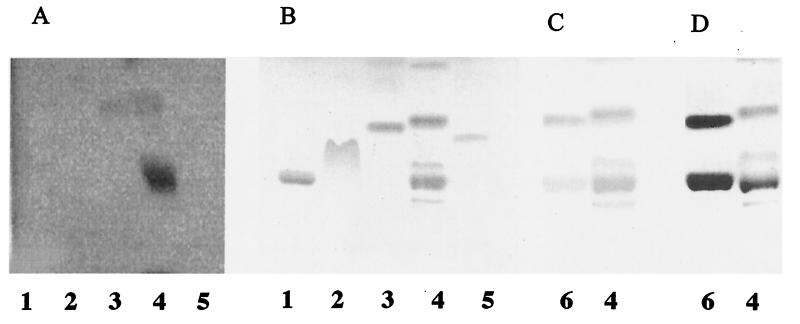
The expression of BFP was determined by Western blotting of all bacterial extracts using a polyclonal anti-BFP antiserum (Fig. 1). BFP expression was detected for all bfp-positive strains, including E2348/69, HB101(pMAR7), and E2348/69(pOG127), grown on TSA blood agar (Fig. 1) or horse blood agar (data not shown) or subcultured in DMEM (data not shown). No BFP expression was detected for HB101, EAF plasmid-cured E2348/69 (JPN15), or the bfpA disruptional mutant of E2348/69 [31-6-1(1)].
FIG. 1.
Western blot of bacterial extracts. Extracts from overnight cultures grown on TSA blood agar supplemented with the appropriate antibiotics (Table 1) were electrophoresed on 15% polyacrylamide gels, transferred to blots, and visualized with polyclonal BFP-specific antiserum (1/1,000 dilution). The positions of molecular mass markers are shown on the left, and that of the 19.5-kDa BFP structural subunit is shown on the right. Lane 1, E2348/69. Lane 2, JPN15. Lane 3, HB101. Lane 4, 31-6-1(1). Lane 5, E2348/69(pOG127). Lane 6, HB101(pMAR7).
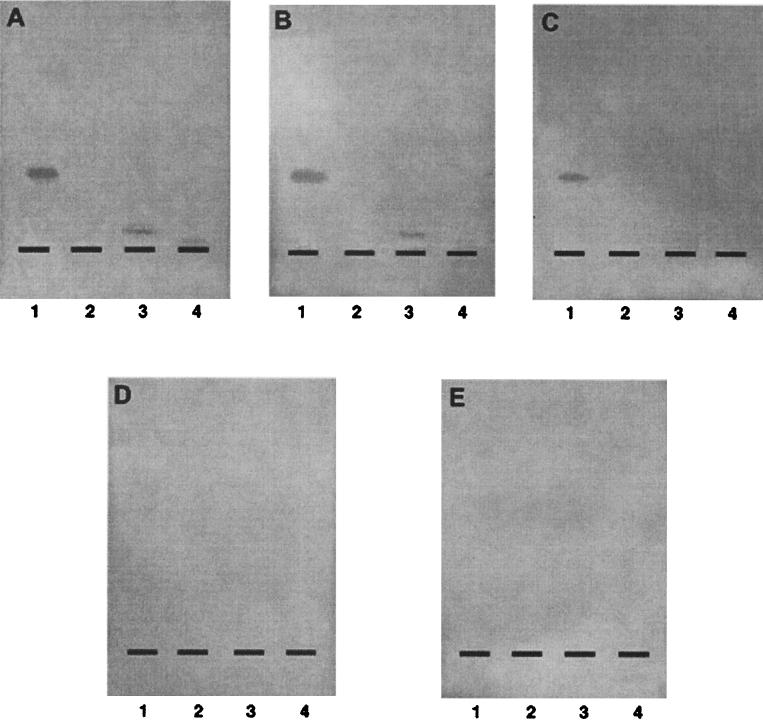
BFP expression directly correlated with PE binding, as determined by a TLC overlay assay. All BFP-expressing strains, E2348/69 (wild type), E2348/69(pOG127), and HB101(pMAR7), bound specifically to PE (Fig. 2). In contrast, all non-BFP-expressing strains, 31-6-1(1), HB101, and JPN15 (data not shown), did not bind PE. None of the strains recognized PC or GM1. Binding to Gg4, a glycolipid previously shown to bind E2348 in a TLC overlay assay (2), did not correlate with BFP expression. All BFP-expressing strains also bound to a band within the lower-phase lipid extracts from HEp2 cells and from E. coli E2348/69 (data not shown). This band comigrated with commercial PE and was stained with iodine, ninhydrin, and molybdenum blue in a manner identical to that of commercial PE. The BFP-expressing strains also recognized within the same lipid extracts a band of lower mobility which comigrated with lysoPE and was stained similarly with iodine, ninhydrin, and molybdenum blue.
FIG. 2.
TLC overlay assay with bacterial suspensions (109 CFU/ml) overlaid on TLC-separated lipids. Detection was done with anti-OMP (1/1,000 dilution). (A) E2348/69. (B) E2348/69(pOG127). (C) HB101(pMAR7). (D) 31-6-1(1). (E) HB101. Each lane contains 5 μg of lipid. Lane 1, PE from E. coli. Lane 2, PC. Lane 3, Gg4. Lane 4, GM1.
To verify the correlation between BFP expression and PE binding, bacterial suspensions were used simultaneously for the TLC overlay assay and for Western blot analysis of BFP expression. The degree of PE binding was proportional to the level of BFP expression, with E2348/69 and E2348/69 (pOG127) showing strong PE binding and high levels of BFP expression and HB101(pMAR7) showing slightly weaker PE binding and lower levels of BFP expression. It was curious that E2348/69(pOG127), a perA disruptional mutant, showed BFP expression similar to that of the wild type, but even this outcome was consistent with the PE binding phenotype.
BFP-positive strains bind to PE-containing liposomes.
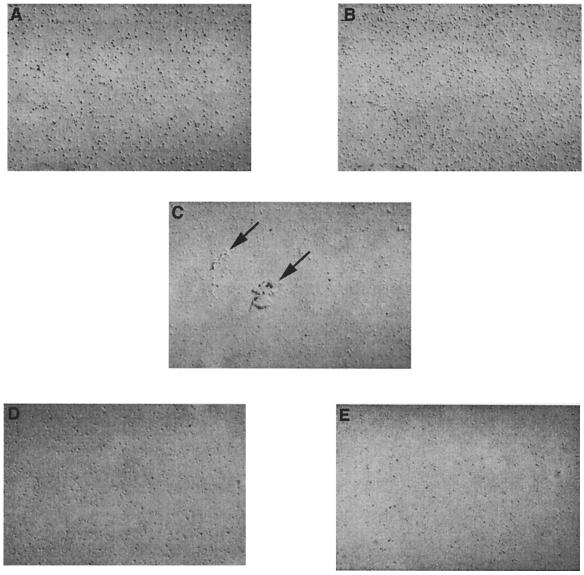
Since the TLC overlay assay detects binding to a lipid conformer which may be of questionable physiological relevance, binding was also assessed by means of bacterium-liposome aggregation, where lipid presentation may more closely model that of a membrane receptor (Fig. 3). Incubation of E2348/69 with PE-containing liposomes produced aggregates whose size and number increased with increasing PE concentration. At a concentration of 0.1 mg of PE/ml, averages of 2 aggregates per high-power field (Fig. 3C) and 25 aggregates per low-power field were detected. At a concentration of 0.2 mg of PE/ml, averages of 5 aggregates per high-power field and 80 aggregates per low-power field were detected (data not shown). In contrast, BFP-negative strains JPN15 and 31-6-1(1) showed no aggregation with PE-containing liposomes. Phospholipid recognition was specific, since liposomes containing PC, PS, or cholesterol alone did not aggregate with any of the strains (only results for E2348/69 are shown). Incubation of any of the strains with PBS also did not result in the formation of autoaggregates (data not shown).
FIG. 3.
Liposome-bacterium aggregation. Bacterial suspensions (109 CFU/ml) were incubated with liposome suspensions (0.1 mg/ml) for 1 h at 37°C. Aggregates (arrows) were examined and photographed using differential interference contrast microscopy. (A) JPN15 with PE-containing liposomes. (B) 31-6-1(1) with PE-containing liposomes. (C) E2348/69 with PE-containing liposomes. (D) E2348/69 with PC-containing liposomes. (E) E2348/69 with PS-containing liposomes.
Inhibition of binding with anti-BFP antiserum.
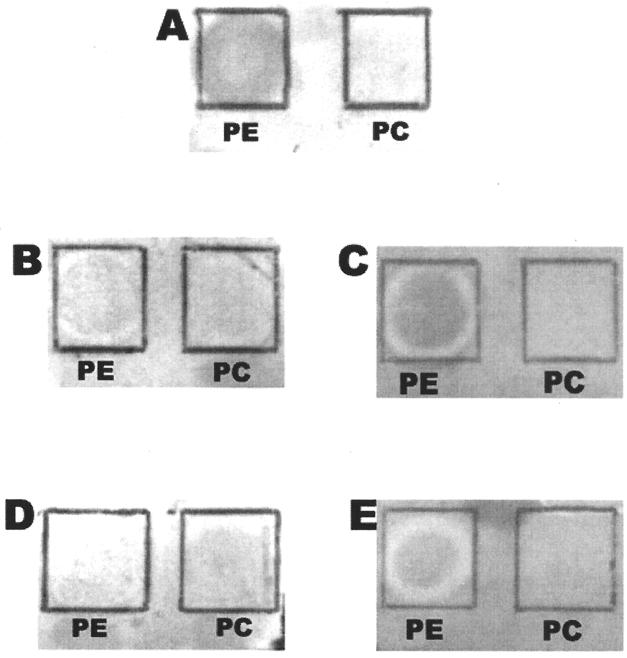
The interaction of EPEC with PE could be inhibited by preincubation with anti-BFP antiserum (Fig. 4). Binding of E2348/69 to PE was disrupted in a dose-dependent manner by preincubation with increasing concentrations of anti-BFP antiserum (dilutions of 1/50 and 1/100 are shown in Fig. 4). Preincubation with equivalent concentrations of rabbit nonimmune serum or buffer alone did not affect PE binding. Once again, binding specificity was limited to PE and not PC.
FIG. 4.
Inhibition of PE binding with anti-BFP antiserum. Suspensions of E2348/69 (109 CFU/ml) were preincubated with PBS (A), anti-BFP antiserum (1/100 dilution) (B), rabbit nonimmune serum (1/100) (C), anti-BFP antiserum (1/50) (D), and rabbit nonimmune serum (1/50) (E) and then overlaid on PE and PC immobilized on TLC plates.
Binding of BFP to PE.
Radiolabeled BFP bound to a commercial PE standard (from E. coli) and to a band within an HEp-2 cell lower-phase lipid extract which comigrated with the PE standard and stained with iodine, ninhydrin, and molybdenum blue in a manner identical to that of commercial PE (Fig. 5). BFP also bound to an HEp-2 cell lipid extract band which had a mobility similar to that of lysoPE and which also stained with iodine, ninhydrin, and molybdenum blue in a manner similar to that of a commercial lysoPE standard (Fig. 5, lanes 4 and 6, lower band). Binding to PE and lysoPE by these strains is consistent with previous studies showing that organisms which recognize PE also recognize lysoPE (4, 5, 13). BFP did not bind to an equivalent quantity of PS, PC, or SGC.
FIG. 5.
BFP binding to selected lipids. (A) Iodine-125-labeled BFP was overlaid on TLC-separated lipids, and binding was detected by autoradiography. (B and C) Iodine staining of lipids. (D) Molybdenum blue staining of lipids. Lane 1, PC (2.5 μg). Lane 2, PS (2.5 μg). Lane 3, PE from E. coli (2.5 μg). Lane 4, HEp-2 cell lower-phase lipid extract (20 μg). Lane 5, SGC (2.5 μg). Lane 6, PE from E. coli and lysoPE (lower band) (2.5 μg each).
DISCUSSION
Our results indicate that BFP-expressing strains of EPEC specifically recognize PE in both solid-phase binding and liposome aggregation assays. In the TLC overlay assay, PE binding was directly correlated with BFP expression. All of the bfp-positive strains expressed BFP and bound PE (from E. coli and human epithelial cells), and the levels of BFP expression were similar to those for PE binding. This finding is consistent with earlier work which showed a correlation between PE binding and the bfp genotype of several clinical EPEC isolates (2). On the other hand, recognition of glycolipid Gg4, previously reported to bind a variety of pathogens, including E2348/69 (2, 5, 25, 26, 28), did not correlate with the bfp genotype. None of the strains tested recognized PC or GM1. These results confirm previous findings in which the BFP expression of several clinical EPEC isolates correlated with PE binding (2). The formation of aggregates of bacteria and PE-containing liposomes with BFP-positive E2348/69 and the absence of aggregates with the BFP-negative strains 31-6-1(1), JPN15, and HB101 not only confirm the results of the TLC assay but also indicate that the recognition of PE by BFP-expressing strains is not limited to the solid-phase presentation. The absence of interactions of E2348/69 with the PC- and PS-containing liposomes attests to the specificity of the PE interaction. Furthermore, the interaction of BFP-expressing EPEC with PE can be inhibited by preincubation of the bacteria with anti-BFP antiserum, clearly indicating the specificity of the interaction between BFP and PE.
EPEC expresses BFP as a long, rope-like structure of intertwining filaments composed of repeating subunits of bundlin (12). The multimeric nature of BFP likely contributes to its PE binding affinity. Certainly, the ability of BFP to intertwine and create a fibrous network is important to its role both in bacterial autoaggregation and in localized adherence to cultured epithelial cells (9, 12, 15, 24). The former refers to unattached EPEC aggregates in liquid cultures. The latter refers to adherent microcolonies of EPEC where BFP forms a meshlike network of interbacterial fibers which may enhance the stability of the attached microcolonies. BFP has also been shown to attach directly to human epithelial cells (12, 24), although the physiological relevance is not clear (15).
PE may serve as a receptor for both interbacterial and bacterium-host attachments. This phospholipid is present in bacterial and eukaryotic membranes, although the specific levels and presentations in these sites are not well established (34). The majority of plasma membrane PE, like PS, is localized on the cytosolic leaflet. However, studies have suggested that levels of both aminophospholipids in outer leaflets may constitute 10 to 30% of total membrane PE and PS (19). It was previously shown that the adherence of E2348/69 to epithelial cells correlates with the level of outer leaflet PE and that this adherence can be partially inhibited with anti-PE antiserum (2). All of the BFP-expressing strains as well as isolated BFP filaments recognized a PE-containing component within human epithelial cell lipid extracts. Since BFP has been reported to promote EPEC adherence to cultured epithelial cells (8), our findings suggest that host cell membrane PE may play a role in BFP-mediated localized adherence of EPEC to epithelial cells.
PE may also serve as a BFP receptor in the formation of interbacterial aggregates involved in autoaggregation and localized adherence. In our studies, BFP-expressing EPEC consistently recognized PE from E. coli in all of the binding assays, including the TLC overlay assay, the receptor-based enzyme-linked immunosorbent assay, and the liposome aggregation assay (2). In this study, we also found that isolated BFP recognized PE from E. coli. Myxococcus xanthus and Pseudomonas aeruginosa, both of which express type IV pili, show directed chemotaxis toward PE, and it has been postulated that PE serves as an autoattractant (21–23). Bacterial PE in EPEC may similarly serve as an autoattractant for BFP, thereby inducing the formation of bacterial autoaggregates that produce the patterns of localized adherence characteristic of EPEC infection.
There is a growing list of proteins which show a binding specificity for PE. These include a 19-amino-acid cyclic peptide, Ro09-0198, from Streptoverticillium griseoverticillatum (7); a 23-kDa protein from rat sperm plasma membrane (18); and a 21-kDa protein from bovine brain cytosol (35). Ro09-0198, whose interaction with PE has been well characterized (6, 10, 11), is an antibiotic peptide which induces the lysis of erythrocytes and fibroblasts and increases the permeability of PE-containing liposomes (6, 7). It is also able to arrest cytokinesis by ligating outer leaflet PE on the cytoplasmic bridge of dividing cells. It is possible that BFP shares some or all of these capabilities. We have shown that enterohemorrhagic E. coli and EPEC both induce apoptosis in human epithelial cell lines, an event which leads to an increase in the level of outer leaflet PE on the cells and also enhances bacterial binding (1). Furthermore, the level of PE on the outer leaflet correlates with the level of bacterial binding, whether by natural variation of outer leaflet PE across cell types or by time-dependent induction of apoptosis. These findings were confirmed by studies in which the addition of exogenous PE augmented outer leaflet PE levels and enhanced bacterial binding.
The correlation between bacterial recognition of PE and the induction of apoptosis suggests that PE binding may be involved in the initiation of apoptotic signaling. Consequently, we examined the ability of bfp-positive and bfp-negative strains to induce apoptosis and found that BFP-expressing EPEC strains induced significantly higher levels of apoptosis than non-BFP-expressing strains (unpublished observations). Furthermore, HB101(pMAR7) was also able to induce apoptosis at the same level as wild-type E2348/69. These results, taken together with the current findings, suggest that BFP interaction with host cell membrane PE plays a role in bacterial induction of apoptosis. However, further studies are necessary to characterize the involvement of membrane PE-BFP binding in these events.
The identification of PE as a receptor candidate, albeit a host receptor, a bacterial ligand, or both, for the virulence factor BFP of EPEC provides the basis for future studies that will enable a more detailed analysis of the pathogenic mechanism and that may permit new therapeutic opportunities for the treatment of EPEC-induced diarrheal disease. Furthermore, the identification of a PE binding adhesin in EPEC may indicate the presence of as-yet-unidentified BFP analogues in other PE binding pathogens, particularly enterohemorrhagic E. coli, another attaching and effacing gastrointestinal pathogen that also forms clusters of bacteria on cultured cells.
ACKNOWLEDGMENTS
This work was supported by grants from the Crohn's and Colitis Foundation of Canada (to D. E. Barnett Foster) and the Medical Research Council of Canada (MT1 3073) (to C. A. Lingwood).
We are grateful for the excellent technical support provided by Ying Wu, Beth Boyd, and Anita Nutikka.
REFERENCES
- 1.Barnett Foster D, Abul-Milh M, Huesca M, Lingwood C A. Enterohemorrhagic Escherichia coli induces apoptosis which augments bacterial binding and phosphatidylethanolamine exposure on the plasma membrane outer leaflet. Infect Immun. 2000;68:3108–3115. doi: 10.1128/iai.68.6.3108-3115.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barnett Foster D E, Philpott D, Abul-Milh M, Huesca M, Sherman P M, Lingwood C A. Phosphatidylethanolamine recognition mediates enteropathogenic and enterohemorrhagic Escherichia coli host cell attachment. Microb Pathog. 1999;27:289–301. doi: 10.1006/mpat.1999.0305. [DOI] [PubMed] [Google Scholar]
- 3.Bieber D, Ramer S W, Wu C-Y, Murray W J, Tabe T, Fernadez R, Schoolnik G K. Type IV Pili, transient bacterial aggregates and virulence of enteropathogenic Escherichia coli. Science. 1998;280:2114–2118. doi: 10.1126/science.280.5372.2114. [DOI] [PubMed] [Google Scholar]
- 4.Bitzan M M, Gold B D, Philpot D J, Huesca M, Sherman P M, Karch H, Lissner R, Lingwood C A, Karmali M A. Inhibition of Helicobacter pylori and Helicobacter mustelae binding to lipid receptors by bovine colostrum. J Infect Dis. 1998;177:955–961. doi: 10.1086/515256. [DOI] [PubMed] [Google Scholar]
- 5.Busse J, Hartmann E, Lingwood C A. Receptor affinity purification of a lipid-binding adhesin from Haemophilus influenzae. J Infect Dis. 1996;175:77–83. doi: 10.1093/infdis/175.1.77. [DOI] [PubMed] [Google Scholar]
- 6.Choung S Y, Kobayashi T, Inoue J, Takemoto K, Ishitsuka H, Inoue K. Hemolytic activity of a cyclic peptode Ro09–0198 isolated from Streptoverticullium. Biochim Biophys Acta. 1988;940:171–179. doi: 10.1016/0005-2736(88)90192-7. [DOI] [PubMed] [Google Scholar]
- 7.Choung S Y, Kobayashi T, Takemoto K, Ishitsuka H, Inoue K. Interaction of a cyclic peptide, Ro09–198, with phosphatidylethanolamine in liposomal membranes. Biochim Biophys Acta. 1988;940:180–187. doi: 10.1016/0005-2736(88)90193-9. [DOI] [PubMed] [Google Scholar]
- 8.Donnenberg M S, Giron J A, Nataro J P, Kaper J B. A plasmid-encoded type IV fimbrial gene of enteropathogenic Escherichia coli associated with localized adherence. Mol Microbiol. 1992;6:3427–3437. doi: 10.1111/j.1365-2958.1992.tb02210.x. [DOI] [PubMed] [Google Scholar]
- 9.Donnenberg M S, Kaper J B, Finlay B B. Interactions between enteropathogenic Escherichia coli and host epithelial cells. Trends Microbiol. 1997;5:109–114. doi: 10.1016/S0966-842X(97)01000-7. [DOI] [PubMed] [Google Scholar]
- 10.Emoto K, Toyama-Sorimachi N, Karasuyama H, Inoue K, Umeda M. Exposure of phosphatidylethanolamine on the surface of apoptotic cells. Exp Cell Res. 1997;232:430–434. doi: 10.1006/excr.1997.3521. [DOI] [PubMed] [Google Scholar]
- 11.Emoto K, Umeda M. An essential role for a membrane lipid in cytokinesis: regulation of contractile ring disassembly by redistribution of phosphatidylethanolamine. J Cell Biol. 2000;149:1215–1224. doi: 10.1083/jcb.149.6.1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Giron J A, Ho A S, Schoolnik G K. An inducible bundle-forming pilus of enteropathogenic Escherichia coli. Science. 1991;254:710–713. doi: 10.1126/science.1683004. [DOI] [PubMed] [Google Scholar]
- 13.Gold B, Huesca M, Sherman P, Lingwood C. Helicobacter mustelae and Helicobacter pylori bind to common lipid receptors in vitro. Infect Immun. 1993;61:2632–2638. doi: 10.1128/iai.61.6.2632-2638.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gomez-Duarte O G, Kaper J B. A plasmid-encoded regulatory region activates chromosomal eaeA expression in enteropathogenic Escherichia coli. Infect Immun. 1995;63:1767–1776. doi: 10.1128/iai.63.5.1767-1776.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hicks S, Frankel G, Kaper J B, Dougan G, Phillips A D. Role of intimin and bundle-forming pili in enteropathogenic Escherichia coli adhesion to pediatric intestinal tissue in vitro. Infect Immun. 1998;66:1570–1578. doi: 10.1128/iai.66.4.1570-1578.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huesca M, Borgia S, Hoffman P, Lingwood C A. Acidic pH changes receptor binding of Helicobacter pylori: a binary adhesion model in which surface heat shock (stress) proteins mediate sulfatide recognition in gastric colonization. Infect Immun. 1996;64:2643–2648. doi: 10.1128/iai.64.7.2643-2648.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jerse A E, Yu J, Tall B D, Kaper J B. A genetic locus of enteropathogenic Escherichia coli necessary for the production of attaching and effacing lesions on tissue culture cells. Proc Natl Acad Sci USA. 1990;87:7839–7843. doi: 10.1073/pnas.87.20.7839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jones R, Hall L. A 23 kDa protein from rat sperm plasma membranes shows a sequence similarity and phospholipid binding properties to a bovine brain cytosolic protein. Biochim Biophys Acta. 1991;1080:78–82. doi: 10.1016/0167-4838(91)90114-f. [DOI] [PubMed] [Google Scholar]
- 19.Kamp D, Haest C W M. Evidence for a role of the multidrug resistance protein (MRP) in the outward translocation of NBD-phospholipids in the erythrocyte membrane. Biochim Biophys Acta. 1998;1372:91–101. doi: 10.1016/s0005-2736(98)00049-2. [DOI] [PubMed] [Google Scholar]
- 20.Kaper J, Gomez-Duarte O G. The per regulator of enteropathogenic Escherichia coli. Mol Microbiol. 1997;23:179–181. doi: 10.1046/j.1365-2958.1997.1841559.x. [DOI] [PubMed] [Google Scholar]
- 21.Kearns D B, Robinson J, Shimkets L J. Pseudomonas aeruginosa exhibits directed twitching motility up phosphatidylethanolamine gradients. J Bacteriol. 2001;183:763–767. doi: 10.1128/JB.183.2.763-767.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kearns D B, Shimkets L J. Chemotaxis in a gliding bacterium. Proc Natl Acad Sci USA. 1998;95:11957–11962. doi: 10.1073/pnas.95.20.11957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kearns D B, Shimkets L J. Directed movement and surface-borne motility in Myococcus xanthus and Pseudomonas aeruginosa. Methods Enzymol. 2001;336:94–102. doi: 10.1016/s0076-6879(01)36582-5. [DOI] [PubMed] [Google Scholar]
- 24.Knutton S, Shaw R K, Anantha R P, Donnenberg M S, Zorgani A A. The type IV bundle forming pilus of enteropathogenic Escherichia coli undergoes dramatic alterations in structure associated with bacterial adherence, aggregation and dispersal. Mol Microbiol. 1999;33:499–509. doi: 10.1046/j.1365-2958.1999.01495.x. [DOI] [PubMed] [Google Scholar]
- 25.Krivan H C, Roberts D D, Ginsburg V. Many pulmonary pathogenic bacteria bind specifically to the carbohydrate sequence GalNAcβ1–4 Gal found in some glycolipids. Proc Natl Acad Sci USA. 1988;85:6157–6161. doi: 10.1073/pnas.85.16.6157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee K K, Yu L, Macdonald D L, Paranchych W, Hodges R S, Irvin R T. Anti-adhesin antibodies that recognize a receptor-binding motif (adhesintope) inhibit pilus/fimbrial-mediated adherence of Pseudomonas aeruginosa and Candida albicans to asialo-GM1 receptors and human buccal epithelial cell surface receptors. Can J Microbiol. 1996;42:479–486. doi: 10.1139/m96-065. [DOI] [PubMed] [Google Scholar]
- 27.Levine M M, Berquist J, Nalen D R, Waterman D H, Hornich R B, Young C R, Sotman S. Escherichia coli strains that cause diarrhea but do not produce heat-labile or heat-stable enterotoxins and are non-invasive. Lancet. 1978;i:1119–1122. doi: 10.1016/s0140-6736(78)90299-4. [DOI] [PubMed] [Google Scholar]
- 28.Lingwood C A. Glycolipids as receptors. Adv Lipid Res. 1991;1:39–55. [Google Scholar]
- 29.Lingwood C A. H. pylori adhesins and receptors. In: Goodwin S, Worsley B, editors. Helicobacter pylori: biology and clinical practice. Boca Raton, Fla: CRC Press, Inc.; 1993. pp. 209–222. [Google Scholar]
- 30.Lingwood C A, Huesca M, Kuksis A. The glycerolipid receptor for Helicobacter pylori (and exoenzyme S) is phosphatidylethanolamine. Infect Immun. 1992;60:2470–2474. doi: 10.1128/iai.60.6.2470-2474.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Louie M, DeAzavedo J, Clarke R, Brunton J. Serotype distribution and sequence heterogeneity of eae gene in verotoxin-producing Escherichia coli. Epidemiol Infect. 1994;112:449–461. doi: 10.1017/s0950268800051153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Momoi T, Ando S, Magai Y. High resolution preparative column chromatographic system for gangliosides using DEAE-Sephadex and a new porus silica, Iatrobeads. Biochim Biophys Acta. 1976;441:488–497. [PubMed] [Google Scholar]
- 33.Nataro J P, Kaper J B. Diarrheagenic Escherichia coli. Clin Microbiol Rev. 1998;11:141–201. doi: 10.1128/cmr.11.1.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Op den Kamp J A F. Lipid asymmetry in membranes. Annu Rev Biochem. 1979;48:47–71. doi: 10.1146/annurev.bi.48.070179.000403. [DOI] [PubMed] [Google Scholar]
- 35.Schoentgen F, Saccaccio F, Jolles J, Bernier I, Jolles P. Complete amino acid sequence of a basic 21 kDa protein from bovine brain cytosol. Eur J Biochem. 1987;166:333–338. doi: 10.1111/j.1432-1033.1987.tb13519.x. [DOI] [PubMed] [Google Scholar]
- 36.Sohel I, Puente J L, Ramer S W, Bieber D, Wu C Y, Schoolnik G K. Enteropathogenic Escherichia coli: identification of a gene cluster coding for bundle-forming pilus morphogenesis. J Bacteriol. 1996;178:2613–2618. doi: 10.1128/jb.178.9.2613-2628.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stone K D, Zhang H-Z, Carlson L K, Donnenberg M S. A cluster of fourteen genes from enteropathogenic Escherichia coli is sufficient for the biogenesis of the type IV pilus. Mol Microbiol. 1996;20:325. doi: 10.1111/j.1365-2958.1996.tb02620.x. [DOI] [PubMed] [Google Scholar]
- 38.Sylvester F A, Philpott D, Gold B, Lastovica A, Forstner J F. Adherence to lipids and intestinal mucin by a recently recognized human pathogen, Campylobacter upsaliensis. Infect Immun. 1996;64:4060–4066. doi: 10.1128/iai.64.10.4060-4066.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tobe T, Schoolnik G K, Sohel I, Bustamante V H, Puente J L. Cloning and characterization of bfp TVW, genes required for the transcriptional activation of bfpA of enteropathogenic Escherichia coli. Mol Microbiol. 1996;21:963–975. doi: 10.1046/j.1365-2958.1996.531415.x. [DOI] [PubMed] [Google Scholar]
- 40.Tyrrell G J, Ramotar K, Toye B, Boyd B, Lingwood C A, Brunton J L. Alteration of the carbohydrate binding specificity of verotoxins from Galα1–4Gal to GalNAcβ1–3Galα1–4Gal and vice versa by site-directed mutagenesis of the binding subunit. Proc Natl Acad Sci USA. 1992;89:524–528. doi: 10.1073/pnas.89.2.524. [DOI] [PMC free article] [PubMed] [Google Scholar]