Abstract
Molecular technologies, including high-throughput sequencing, have expanded our perception of the microbial world. Unprecedented insights into the composition and function of microbial communities have generated large interest, with numerous landmark studies published in recent years relating the important roles of microbiomes and the environment—especially diet and nutrition—in human, animal, and global health. As such, food microbiomes represent an important cross-over between the environment and host. This is especially true of fermented food microbiomes, which actively introduce microbial metabolites and, to a lesser extent, live microbes into the human gut. Here, we discuss the history of fermented foods, and examine how molecular approaches have advanced research of these fermented foods over the past decade. We highlight how various molecular approaches have helped us to understand the ways in which microbes shape the qualities of these products, and we summarize the impacts of consuming fermented foods on the gut. Finally, we explore how advances in bioinformatics could be leveraged to enhance our understanding of fermented foods. This review highlights how integrated molecular approaches are changing our understanding of the microbial communities associated with food fermentation, the creation of unique food products, and their influences on the human microbiome and health.
Keywords: fermented foods, microbiome, high-throughput sequencing, metagenomics, transcriptomics, metabolomics
Multi-omic approaches are rapidly improving our knowledge of fermented foods and the application of these technologies can further enhance our control over these foods.
Introduction
Microbial exposures are critical for health, both with respect to the human microbiome and from external, environmental organisms. Food microbiomes are uniquely positioned to span both of these ecologies. This is particularly true of fermented foods, which are defined as those made through desired microbial growth and enzymatic conversions of food components (Marco et al. 2021). These foods consist of >5000 global varieties (Tamang and Kailasapathy 2010), many of which are unique to specific locations and populations. Generally, fermented foods are grouped based on the food substrate, e.g. dairy, vegetables, and meat. However, within these groupings there can be many subcategories, and corresponding differences in the associated microbiota (Leech et al. 2020). Indeed, even the microbiota of particular families of fermented foods can vary, e.g. temporally or spatially, with consequences for the underlying basic biology, product quality (Walsh et al. 2016), and induced phenotypes (van de Wouw et al. 2020).
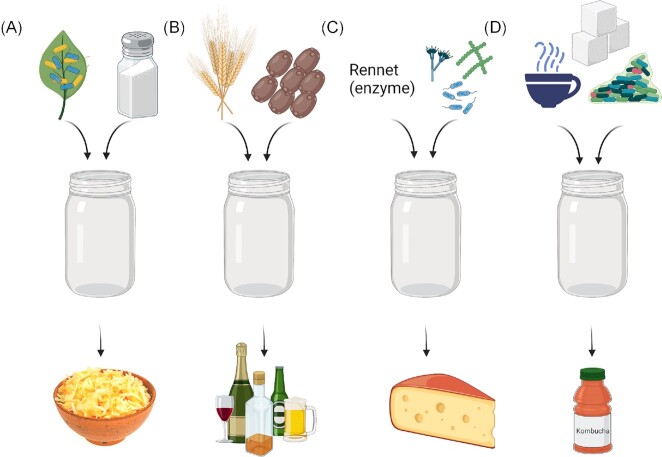
Fermentations can be driven by microorganisms that are endogenous to the raw food substrate or the production environment, i.e. spontaneous fermentations, or driven by microorganisms that are added to the food (Fig. 1). Traditionally, the latter was often achieved using ‘backslopping’, which involves adding a fraction of the previously fermented food to new food substrates to instigate a new fermentation. This is one example of a broader approach to the use of undefined cultures, which are consortia of the microorganisms that are responsible for initiating the fermentation, i.e. starters, as well as adjuncts, which do not contribute initially but become important later in the process. In the 19th century, the realization that microorganisms are responsible for fermentation led to the isolation of strains from foods (Caplice and Fitzgerald 1999). Subsequently, fermented foods have been produced in industry using these strains as defined starters, i.e. known cultures that are added to foods in specific quantities, and the optimization of these processes has been the subject of extensive research (Gibson et al. 2017, Hatti-Kaul et al. 2018).
Figure 1.
Schematic diagram of four methods of fermentation. (A) Spontaneous fermentation: For sauerkraut, cabbage is processed into small pieces. Salt is added at the ratio of 2% per weight of cabbage. This draws out moisture from the plant cells. The resulting mixture of cabbage, salt, and water is placed in a vessel and left to ferment for at least 5 days. The microbes present on the cabbage are mainly responsible for fermentation. (B) Simple starter: Many alcoholic beverages are produced using a simple starter, such as the yeast Saccharomyces cerevisiae. For beer, there are several important steps: malting, milling, mashing, lautering, boiling, fermenting, conditioning, filtering, and packaging. The simple starter, S.cerevisiae, is added to the wort after boiling, and then removed after fermentation by filtration. (C) Complex starter: For the production of certain cheese (e.g. cheddar), milk is innoculated with a multiple-strain (three to six strains) starter culture of known strains and rennet. These strains are typically lactic acid bacteria such as Lactococcus lactis subspecies cremoris and Lactococcus lactis subspecies lactis. (D) Undefined starter culture: Kombucha can be produced using a defined starter powder or using a kombucha ‘SCOBY’ (Symbiotic Colony Of Bacteria and Yeast). For the latter traditional method, a microbially produced SCOBY is added to sugary tea. Roughly 10% of the previous fermented kombucha is also added to the tea. The SCOBY and the added fermented kombucha contain an undefined community of bacteria and yeasts that ferment the tea over 7–14 days.
Over the last 20 years, molecular profiling techniques have revolutionized our understanding of the microbiota of fermented foods (Walsh et al. 2017). Genome-based characterization of individual starters and adjuncts provided valuable initial insights, while characterization of spoilage and pathogenic microorganisms provided information as to how to better control these contaminants (Garrigues et al. 2013). Marker gene and metagenomic sequencing techniques have proliferated in recent years (Fig. S1), with the latter approach in particular making it possible to study the broader microbial ecologies of these foods (De Filippis et al. 2017), facilitating greater understanding of the taxonomic composition of the microorganisms present and of their associated genetic arsenal, responsible for the associated biochemical changes in an environment. It is important to acknowledge the limits of high-throughput DNA sequencing (HTS), which include strain-level characterization of certain taxa or detection of less abundant microbes in a given food. Most recently, transcriptomics have begun to provide information on, how, and when relevant microbial genes are expressed (De Filippis et al. 2018; Fig. S1). Other molecular approaches add further layers of information regarding fermentations’ biochemical environments. These include proteomics (Carrasco-Castilla et al. 2012) for identifying important enzymes and metabolomics for profiling volatile compounds or other metabolites (Rizo et al. 2020). Studies deploying combinations of these technologies are increasing. These are critical for understanding, targeting, and developing previously unharnessed or under-utilized properties and bioactivities of fermented foods. This is particularly important in light of the current resurgence in popularity of artisanal fermented foods in Western society, largely driven by a health-conscious market. This review addresses how combining information from metagenomic, transcriptomic, and other molecular studies can be used to address important research questions of relevance to fermented foods, and indeed of broader significance to microbiology, while also improving upon the characteristics that consumers and industry seek to fully realize the potential of these foods.
High-throughput sequencing approaches
Food fermentation is the result of the biological activity of microbes present within food matrices (Marco et al. 2021). HTS enables high-quality culture-independent characterization of microbial communities, including those present in fermented foods (Leech et al. 2020). Three different HTS approaches have been used to characterize the microbiota of fermented foods: amplicon sequencing, whole metagenome shotgun sequencing, and metatranscriptomics (also known as RNA-Seq). We have compiled a comprehensive list of studies to have used HTS to characterize the microbiota of fermented foods that includes those published from 2009 until September 2019 (Table S1).
Amplicon sequencing has been the most frequently used HTS approach for the characterization of the microbiota of fermented foods (Cao et al. 2017, De Filippis et al. 2017; Fig. S1). Although it has yielded many novel insights into the microbial diversity in these foods (Kergourlay et al. 2015), it has some inherent limitations, including the absence of functional information. Additionally, in general, short-read amplicon sequencing is limited to genus-level classification, although it has been demonstrated that long-read amplicon sequencing can achieve (sub)species-level classification (Karst et al. 2021). Shotgun metagenomics yields considerably more information than amplicon sequencing, including the functional profile of the microbiome (Leech et al. 2020), but at a higher cost. Strain-level identification is also possible with shotgun metagenomics but this is often only suitable for dominant species. It can also be challenging to disentangle mixtures of strains from the same species within a sample, although improved tools are starting to make this achievable (Vicedomini et al. 2021, van Dijk et al. 2022). Notably, long-read shotgun metagenomics could facilitate improved strain-level analysis, and tools such as Strainberry have already been developed that use long reads to recover strain-resolved genomes from metagenomes (Vicedomini et al. 2021). Metatranscriptomics is more costly again, and fermented foods can be difficult to extract high-quality mRNA. However, it can be a powerful tool for examining foods as it allows the examination of gene expression.
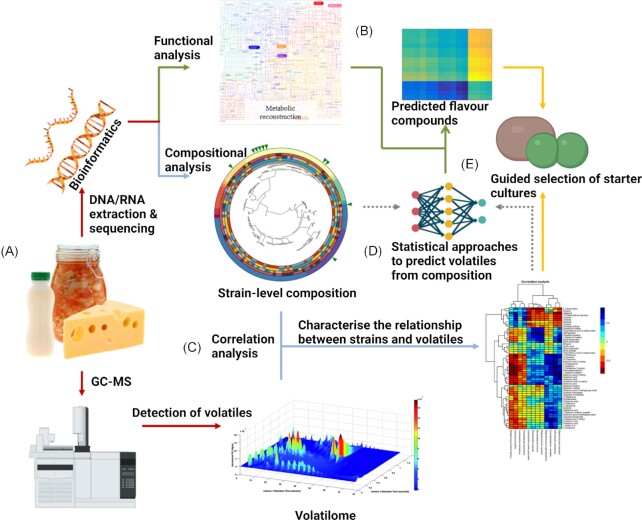
Each sequencing approach mentioned above can be used in conjunction with other omics methods, such as metabolomics or proteomics, to achieve multi-omic (Franzosa et al. 2015) analyses of fermented foods (Fig. 2). Such analyses are employed to link changes in the proportions, functional potential, or gene expression of microbes with biochemical changes that occur during food fermentations.
Figure 2.
A schematic workflow to determine the contribution of microbes to the development of flavour in fermented foods using omics approaches. (A) Fermented foods are sampled for microbiome and/or volatilome analysis (red arrows). (B) Metabolic reconstruction to predict what volatiles are produced by the microbiome (green arrows). (C) Correlation analysis is used to determine whether strains are linked to volatiles (blue arrows). (D) Statistical approaches are used to predict the volatilome of a food based on its strain-level composition (grey arrows). (E) The information obtained from at least one of these approaches is used to select starters with desirable (and predictable) properties (gold arrows).
Here, we review studies that have used high-throughput sequencing, with an emphasis on shotgun metagenomics, metatranscriptomics, or multi-omic approaches that have been used to analyse several common fermented foods. We discuss the ways in which the information gained from such analyses might be applied to enhance food qualities such as flavour. Additionally, we explore the potential for novel bioinformatics or computational biology methods to further our understanding of food fermentations. A schematic outline of these approaches is presented in Fig. 2.
Cataloguing fermented food microbiomes
A cornucopia of fermented foods have been characterized using HTS, initially by amplicon sequencing (Kim et al. 2002), followed later by shotgun metagenomics (Jung et al. 2011), propelling our understanding of these products. The majority of studies have focused on cataloging what taxa are present in fermented foods.
Like any other microbial ecosystem, HTS can reveal many taxa that cannot be found by current culture-based techniques, therefore revealing deeper insights into these communities. Taxonomic profiling of fermented foods has revealed a large degree of interchangeability between species/strains fermenting the same food/substrate, highlighting the variable nature of fermented food microbiomes. Longitudinal studies that examine the community dynamics over the course of a fermentation reveal the periods at which certain taxa dominate, and pairing with metabolomics data, the contributions of specific taxa for the progression of the fermentation can be uncovered (Walsh et al. 2016). Such approaches are vital for understanding the contribution of the overall community to the final product.
More recently, several studies have examined large numbers of fermented foods to examine broader patterns of microbial ecology. Walsh et al. (2020) examined 184 cheeses, including 77 new samples and 107 samples in publicly available databases. The study included volatile data on the 77 new samples. By examining multiple samples simultaneously, the authors uncovered that strain-level differences within the cheese microbiome had a significant impact on the resulting volatile profile. The authors also uncovered antimicrobial resistance genes, but with a low risk of transfer between microbes. This study is also a great example of the power of HTS, as the interactions between viruses and prokaryotes were examined. Analyses of CRISPR and anti-CRISPR proteins revealed the importance of phage for transferring genetic information between bacteria. Another recent study examined the microbial diversity between sourdough starter cultures from four different continents (Landis et al. 2021). Interestingly, the study did not see a geographic effect between sourdoughs from different countries. The study uncovered strong co-occurrence patterns between different microbes, highlighting the importance of a microbial-mediated community structure. The study also revealed the importance of acetic acid bacteria and their contribution to the flavour and dough-rise of sourdough bread. Finally, Leech et al. (2020) examined 58 artisanal-produced fermented foods from eight different countries. Again, no geographic signal was discovered across the 58 food microbiomes. The authors did uncover strong patterns in microbial ecology across the different substrates used, showing that dairy-based, salt-based (foods fermented in 2% saline, e.g. sauerkraut), and sugar-based (foods containing high sucrose at the beginning of fermentation, e.g. kombucha) fermented foods had different communities and functional profiles to each other. The study also revealed that dairy-based samples had lower alpha diversity than salt-based and sugar-based fermented foods. These three studies are a demonstration of the utility of HTS for looking at broader patterns across fermented foods, and how these insights can be useful for the commercial application of these ecological insights.
Applications of fermented food microbiome research
Fermented dairy products as model microbial communities
A 2014 study by Wolfe et al. pioneered the use of fermented foods as models to explore the forces that might shape microbiota. The authors reported that microorganisms from cheese rinds were easy to culture (Wolfe et al. 2014). Importantly, they were able to recreate the cheese rind microbiota in vitro. In this study, HTS revealed that moisture influenced the composition of these communities; although this finding was unsurprising, it proved that fermented foods can be used as models to assess the impact of abiotic factors on microbiota.
Subsequently, the use of cheeses as models has been extended to study several processes that might mould microbiota, including horizontal gene transfer (HGT). Bonham et al. sequenced the genomes of 165 bacteria isolated from cheese rinds, and they reported that HGT was frequent between these genomes (Bonham et al. 2017). Notably, many of the transferred genes were involved in the acquisition of nutrients from the environment, including iron, which is limiting on cheese rinds; such findings highlight the importance of the availability of nutrients on microbiota.
Cheese models have also been used to study interactions between the members of microbiota. Morin et al. grew mutants of Escherichia coli in cheese rinds to identify genes that were involved in interactions between microorganisms in this environment (Morin et al. 2018). The mutants were generated using an approach called Random Barcode Transposon-site Sequencing (RB-Tn-Seq), which measures the fitness of genes under a given condition by counting the abundance of random barcodes at transposon insertion sites. The mutants were co-cultured separately with each member of a three-species community (Hafnia alvei + Geotrichum candidum + Penicillium camemberti), and they were also grown with the entire community simultaneously. RB-Tn-Seq indicated that many of the interactions that occurred when mutants were grown as part of a pair did not occur when the mutants were grown as part of the community. In other words, the way in which strains interacted differed depending on the context in which they were grown.
Cheese models have highlighted the importance of cross-kingdom interactions in fermented foods. For example, studies have shown that the release of siderophores by fungi can influence the growth of bacteria on cheese rinds. Kastman et al. observed that the establishment of Staphylococcus equorum on cheese rinds was assisted by the mould Scopulariopsis, and RNA-Seq suggested that this might have been explained by the release of iron by the mould, which was utilized by the bacterium (Kastman et al. 2016). Similarly, Pierce et al. used RB-Tn-Seq to show that fungi could influence the growth of bacteria on cheese rinds by modulating the availability of cofactors (Pierce and Dutton 2022). Intriguingly, Zhang et al. discovered that Serratia on cheese rinds can travel along the hyphae of the fungi Mucor (Zhang et al. 2018). To determine the mechanism by which this phenomenon occurred, they employed transposon mutagenesis to generate Serratia mutants, and Whole Genome Sequencing (WGS) of Serratia mutants revealed that flagella were essential for dispersal along the hyphae. Elsewhere, Cosetta et al. used metabolomics to show that volatiles produced by fungi promoted the growth of Vibrio in cheese rinds (Cosetta et al. 2020).
Aside from cheese models, the kefir microbiome has been proposed as another model for studying microbial communities. Recently, Blasche et al. used integrated approaches to unravel the interactions between species in kefir over the course of fermentation (Blasche et al. 2021). 16S rRNA gene sequencing analysis revealed that Lactobacillus kefiranofaciens dominated in the kefir but, surprisingly, it was found that L. kefiranofaciens was unable to grow in milk by itself. Consequently, the authors postulated that L. kefiranofaciens needed to cooperate with other members of the community to propagate during fermentation. Indeed, when L. kefiranofaciens was co-cultured with Leuconostoc mesenteroides, cross-feeding between the two species was observed, wherein L. kefiranofaciens made amino acids available for L. mesenteroides, which in turn made lactate available for L. kefiranofaciens.
Flavour
Here, we discuss the use of HTS approaches to predict what volatiles are produced by microorganisms, and we examine the use of integrated and experimental approaches that combine HTS with metabolomics to understand the relationship between the microbes and volatiles. We refer to these two approaches as (a) correlative/predictive (Fig. 2B, C, and D) and (b) causative/experimental.
The correlative/predictive approach leverages the findings of research from the 20th century that established the enzymology of the development of flavours during fermentation (Smit et al. 2005). Specifically, HTS enables us to detect genes that are associated with the production of volatiles in foods. For example, analysis of the koumiss microbiome, a traditional fermented milk product consumed in parts of Central Asia, revealed that it contained genes that are important for flavour, including those associated with proteolysis (Yao et al. 2017). In dairy products, proteolysis of caseins releases amino acids that can serve as precursors to volatiles. The koumiss microbiome also contained an aminotransferase involved in the transaminase pathway that initiates the formation of volatiles (e.g. aldehydes, acids, alcohols, esters, etc.) or lyases that are involved in the production of sulphur compounds. The Cojita microbiome, a Mexican cheese, contained aldehyde dehydrogenases that convert aldehydes to acids, alcohol dehydrogenases that can convert aldehydes to alcohols, and genes involved in lipolysis, a source of volatiles (Escobar-Zepeda et al. 2016).
Metatranscriptomics analysis of an industrial Camembert-type cheese over a 77-day ripening period indicated that genes involved in the production of sulphur compounds were expressed mostly by Geotrichum candidum, while those involved in lipolysis were expressed mostly by Penicillium camemberti, which suggested that the contributions of these fungi to the flavour of the cheese were distinct (Lessard et al. 2014). Interestingly, metatranscriptomics analysis of a traditional Italian Caciocavallo Silano cheese revealed that nonstarter lactic acid bacteria (LAB) contributed to amino acid metabolism during ripening (De Filippis et al. 2016). Shotgun metagenomic analysis of the cheese rind microbiome revealed that Pseudoalteromonas species contained methionine gamma-lyase, which is involved in the production of sulphur compounds. Only Brevibacterium linens was previously described to produce this enzyme in cheese (Yeluri Jonnala et al. 2018). In a recent meta-analysis of the cheese microbiome, metagenome-assembled genomes were found belonging to species that had not been characterized before, several of which were predicted to secrete compounds that might influence the quality of cheese, including acetate (Walsh et al. 2020). Notably, in this study, Walsh et al. also integrated metagenomics with metabolomics, and it was found that strain-level variation corresponded to differences in the volatilome, the set of volatile compounds present in a sample, which is consistent with results reported elsewhere (Niccum et al. 2020). For example, two strains of B. linens showed distinct patterns of correlation with butanoic acid, wherein one strain was significantly positively correlated with the compound, whereas the second strain was significantly negatively correlated with it. Such findings highlight that species-level correlations need to be interpreted with caution.
HTS has been particularly valuable when applied in spontaneously fermented foods. Metatranscriptomics analysis of kimchi revealed that only Leuconostoc species expressed mannitol dehydrogenase genes during fermentation, and thus were solely responsible for mannitol production (Jung et al. 2013). Additionally, genes involved in the production of the acetoin, diacetyl, and 2,3-butanediol were highly expressed in L. mesenteroides, further highlighting its importance with respect to flavour development in kimchi (Chun et al. 2017).
An extension of the predictive approaches described above is metagenome-scale metabolic modelling, a method which uses the metagenome to predict which enzymes, and ultimately metabolites, may be produced by the microbiome (Magnúsdóttir and Thiele 2018). It has been demonstrated that such an approach accurately predicted the metabolites produced by the gut microbiota in obese humans (Shoaie et al. 2015). Given the relative simplicity of fermented food microbiota compared to, e.g. the communities found in the human gut, it is plausible that metagenome-scale metabolic modelling may be applied to these communities to predict the production of flavour compounds. A challenge posed by modelling is the requirement for curation to ensure the quality of models, which, until recently, was done manually (Henry et al. 2010). However, with the release of CarveMe (Machado et al. 2018), it is now possible to automatically construct genome-scale metabolic models for the members of a community. CarveMe might be used to construct models from genomes that are recovered from metagenomes, using tools like MetaBAT (Kang et al. 2015), to predict the volatiles produced by the entire community or by different combinations of its members. Such an approach may lead to the large-scale identification of candidates as starters, but it might also inform efforts to tailor ingredients for the development of flavours of interest.
The integration of HTS with metabolomics has been employed to explore the potential contributions of microorganisms to the flavour of fermented foods. Typically, this has involved correlating the abundances of taxa with the concentrations of metabolites, and most studies to use this approach have integrated amplicon sequencing with metabolomics to examine the contribution of genera to volatiles. In a study of Chinese liquor fermentations, the approach revealed that microorganisms that originated from the surfaces of the facility in which the beverages were produced were correlated with the levels of metabolites in the products (Bokulich et al. 2014). Other studies have integrated shotgun metagenomics with metabolomics to examine the contributions of species to volatiles. For example, analysis of kefir over the course of 24-hour fermentations revealed that Lactobacillus kefiranofaciens correlated with carboxylic acids, ketones and esters. In contrast, Leuconostoc mesenteroides correlated with 2,3-butanedione and acetic acid (Walsh et al. 2016). These correlations suggested a causal relationship between the microbiota and the flavour of kefir, which was supported by experimental evidence, spiking milk kefir with both organisms separately. Sensory analysis validated these findings by showing that a kefir with a high relative abundance of L. mesenteroides had a likeable buttery flavour, whereas another kefir with a high relative abundance of L. kefiranofaciens had a less likeable but fruitier flavour. Similarly, an integrated approach was employed to characterize surface-ripened cheeses during a 30-day ripening period (Bertuzzi et al. 2018). Again, strong correlations were identified between the relative abundances of individual species and the levels of flavour compounds in the cheeses. Importantly, these correlations were supported by evidence from prior studies that had shown that these species can produce such compounds. Interestingly, Staphylococcus xylosus, which had only previously been associated with sulphur compounds in meats, was also found to correlate with sulphur compounds in these cheeses.
It is crucial to recall the adage that ‘correlation is not causation’ when interpreting findings from integrated approaches. Noecker et al. reported that in simulated microbiome–metabolome datasets, wherein the producers of each metabolite were known, correlation analysis produced false-positives in 50% of cases, and identified relationships between unrelated variables (Noecker et al. 2016). However, researchers can take factors into account to mitigate this issue when interpreting correlations, including: (a) the likelihood that a correlation is valid increases if that correlation persists across subsets of data, and (b) if a microorganism contains genes associated with the production of a metabolite. Correlations should also be validated experimentally.
As previously mentioned, strains of the same species can influence flavour differently (Pereira et al. 2017, Niccum et al. 2020, Walsh et al. 2020, Jimenez-Lorenzo et al. 2021). Strain-level characterization can be a challenge but several tools have been released that enable strain-level characterization of the microbiome (Truong et al. 2017, Olm et al. 2021, Vicedomini et al. 2021, van Dijk et al. 2022). The integration of these tools with metabolomics represents an opportunity to evolve the integrated approach discussed above (Walsh et al. 2020). Potentially, such an improvement might help in the identification of strains that produce volatiles of interest, thus facilitating: (a) the selection of starters, and (b) the construction of models that can predict the metabolome of a sample based on the strains that are present in that sample to track the progress of fermentation.
Ultimately, these approaches can only help us to predict the role of microbes in the development of flavour, and experimentation would be required to test the validity of these predictions. For example, such an experiment might involve (1) spiking a fermented food with an inoculum of a given microbe, and (2) performing sensory analysis to determine if the microbe caused a change in flavour. A Zhenjiang vinegar study is a great example of the experimental approach where shotgun metagenomics combined with pathway analysis indicated that A. pasteurianus, in addition to Lactobacilli species, had the ability to synthesize acetoin (Wang et al. 2016). Co cultures of isolated bacteria from the vinegar were found to produce more acetoin than mono-cultures in vitro and in situ when inoculated in vinegar. Also, in Chinese liquor, metatranscriptomics was used to measure the expression of genes associated with the production of two sulphur compounds, 3-(methylthio)-1-propanol and dimethyl sulphide. Saccharomyces expressed every gene necessary to produce both compounds, while Lactobacilli expressed genes involved in recycling methionine, a precursor to these compounds. Saccharomyces cerevisiae and L. buchnerii were isolated from the liquor and cultivated in monoculture or co-culture. While L. buchnerii monocultures produced neither compound, co-cultures produced significantly more of the sulphur compounds than S. cerevisiae monocultures, thus confirming a synergistic relationship between these species. Finally, as mentioned above, experimental evidence confirmed the contribution of specific kefir bacteria to the flavour profile (Walsh et al. 2016). To date, too few studies have adopted such an approach, but it is crucial that the field moves in this direction to translate our improved understanding of the fermented food microbiome into action.
Defects
As discussed above, microorganisms can produce metabolites that improve the organoleptic qualities of fermented foods. The opposite is also true: spoilage can be caused by overfermentation, which occurs when microbial metabolites (e.g. acids, alcohols) are excessively produced. Often, this can be rectified by modifying the parameters of fermentation (e.g. duration, temperature). Alternatively, spoilage can be caused by microbial contaminants. In Chinese rice wine, shotgun metagenomics provided strong evidence that Levilactobacillus brevis caused spoilage of the beverage (Hong et al. 2016). Taxonomic analysis revealed that this species was prevalent in spoiled wine, while functional analysis revealed the presence of genes involved in biotin biosynthesis and short-chain fatty acid production, which were thought to contribute to off-flavours (Hong et al. 2016). In nunu, a yoghurt-like, milk-based fermented product widely consumed in West Africa, Enterobacteriaceae genes for the biosynthesis of putrescine, which causes an unpleasant odour, were identified (Walsh et al. 2017). In fermented seafood, several studies have observed that levels of Halanaerobium corresponded with increases in spoilage metabolites, including methylamines (Lee et al. 2014, 2015).
Other defects in the qualities of fermented foods, such as pigments that discolour cheeses, are costly for producers. Shotgun metagenomics revealed that Thermus thermophilus, which is not associated with the typical cheese microbiota, has been found to be enriched in cheeses with a pink discolouration defect (Quigley et al. 2016). Carotenoid biosynthesis genes were correspondingly enriched in those cheeses. It was observed that the pinking defect was reproduced in experimental cheeses inoculated with T. thermophilus (Quigley et al. 2016). Similarly, Psychrobacter has been linked to the purpling of cheese (Kamelamela et al. 2018). Analysis of Psychrobacter genomes recovered from cheeses revealed that these bacteria contained an enzyme, which can result in the production of the pigment indigo.
Safety
While spoilage during fermentation is unpleasant and can have economic consequences, in extreme cases, specific pathogens and negative microbial biochemistry can pose a true health hazard. In addition to the insights that genomics has provided into various food pathogens, HTS can also be used to detect pathogens in fermented foods and/or associated production facilities. Indeed, some such studies have detected Enterobacteriaceae in African fermented milk products (Walsh et al. 2017, Parker et al. 2018) and, in one instance, shotgun metagenomics of nunu samples highlighted the presence of pathogenic strains, similar to strains that had caused illness (Walsh et al. 2017). It is crucial to note that the sensitivity of shotgun metagenomics for the detection of pathogens is lower than that of methods such as qPCR (Andersen et al. 2017), which has a limit of detection of 104–105 CFU/ml (Hazards et al. 2019). Instead, as currently employed, shotgun metagenomics is perhaps more suited for identifying the source of outbreaks by analysing suspected foods (Buytaers et al. 2021).
HTS has also been used to identify producers of histamine, a biogenic amine that can be harmful to some consumers, in cheese and other fermented foods (O’Sullivan et al. 2015). Also of potential concern is the presence of bacteria with antibiotic resistance genes (ARGs), especially ARGs present on mobile elements, which could in principle transfer to members of the gut microbiota after ingestion (Maeusli et al. 2020). Analysis of the resistome of cheeses revealed that ARGs were not present on the plasmids of LAB, but were detected on the plasmids of Enterobacteriaceae (Walsh et al. 2020), which are generally microbial contaminants. Thus, improving hygiene during cheese manufacture might reduce reservoirs of resistance in the food.
Preservation
Finally, throughout history, fermentation has been used as a means of preserving foods. Fermentation continues to contribute to shelf-stability today as a means to avoid or decrease reliance on chemical preservatives and to preserve foods in regions where refrigeration is limited. During fermentation, microorganisms secrete compounds (e.g. acids, alcohols, diacetyl, bacteriocins, and others) that inhibit the growth of contaminants (including spoilers and pathogens) (Ross et al. 2002). Notably, HTS has been used to screen a variety of fermented food microbiomes for bacteriocin genes (Leech et al. 2020, Walsh et al. 2020). Approaches available for bioprospecting of bacteriocin genes in fermented food microbiomes include detection by homology to known bacteriocin gene clusters (Hammami et al. 2010), Hidden Markov models (HMM)-based tools (Morton et al. 2015, van Heel et al. 2018), or machine learning (Hamid and Friedberg 2019). The application of these can contribute to the selection of starters that produce bacteriocins targeting undesirable microorganisms.
Health, nutrition science, epidemiology, and translational applications of fermented foods
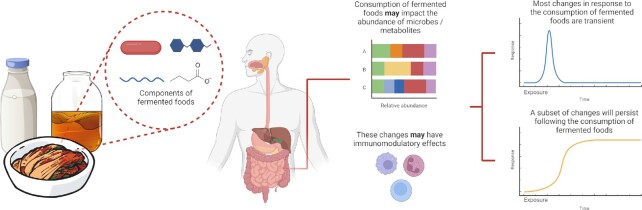
In addition to food quality considerations for consumers, fermented foods have great potential in maintaining and improving human health (Fig. 3) (Soedamah-Muthu et al. 2013, Eussen et al. 2016, Wastyk et al. 2021). This has long been of interest, but the great diversity and variability both of fermentation microbiomes and of the human microbiome have made it difficult to design and target benefits in a precision manner. Molecular techniques now allow us to engineer not just the fermentation process, but the resulting live cell and chemical effects in human hosts and populations.
Figure 3.
The effects of fermented foods on humans. Fermented foods are consumed and the components of the product (including microorganisms, metabolites, prebiotics, and proteins) could modulate the host gut microbiome, which consequently results in increased production of a metabolite. Subsequently, this metabolite could elicit an immune response and/or enter circulation. Alternatively, the components of the fermented foods themselves could cause this effect. Typically, the effect of consuming fermented foods is transient, but some effects (e.g. changes in the composition of the microbiota) may persist afterwards.
At a population scale, fermented foods, or at least those containing living microorganisms, occupy a unique position with respect to long-term human health, as one of the only practical ways to ‘chronically’ deliver beneficial live microbes concordantly with microbial chemical products outside of a specifically therapeutic context (Rezac et al. 2018). That is, while live cell therapies and fecal microbiota transplants are under intense study for treatment of acute conditions (Kelly et al. 2014, Youngster et al. 2016), they are not generally appropriate for regular, long-term use. Other types of dietary or prebiotic interventions are practical for maintenance of microbes already present in the gut (Scott et al. 2008, Cotillard et al. 2013), but they are not capable of reliably introducing new ones unless part of synbiotics. As food fermentation microbes can have beneficial effects on the gut biochemical environment during transit, but do not generally engraft stably after short exposures (Derrien and van Hylckama Vlieg 2015), adherence to regular consumption must be part of any practical ‘wellness maintenance’ applications of fermented foods.
Despite this potential, to date, there has been a shortage of large-scale investigations into the relationship between fermented food consumption and with gut microbiome modulation or, indeed, health biomarkers. It is also important to appreciate that the microbiomes and metabolomes, of specific fermented foods can differ greatly and thus the findings from studies with specific fermented foods cannot be extrapolated across to other fermented foods. Smaller cohort studies have been carried out on a range of foods, most commonly dairy, with variable results (Marco et al. 2017, Markowiak and Śliżewska 2017, Stiemsma et al. 2020). Consistent benefits have been shown for circulating lipid metabolism and corresponding cardiometabolic health in particular (Soedamah-Muthu et al. 2013, Chen et al. 2014, Lim et al. 2015, Eussen et al. 2016), while gut-local conditions such as Inflammatory Bowel Disease (IBD) (Bengmark 2007, Geier et al. 2007), Irritable Bowel Syndrome (IBS) (Laatikainen et al. 2016), or diarrhoea (Parvez et al. 2006, Nagata et al. 2016) often respond in a much more population- and study-specific manner.
Perhaps one of the most critical, least studied links between fermented foods and health outcomes is their role as part of the ‘disappearing microbiome’ (Segata 2015). During the period of time during which fermented foods became less popular in Western society, there was a corresponding increase in the application of other food preservation approaches. As a consequence, and as introduced above, the shelf life of many products across the globe has been extended considerably over the past century (Boor et al. 2017). As with other changes in early life microbial exposure—e.g. livestock (Kim et al. 2019), antibiotics (Langdon et al. 2016), breastfeeding (Bäckhed et al. 2015), and Caesarian section (Bokulich et al. 2016)—it is possible that the decreased exposure to microbes, either within fermented foods or ‘unwanted’ growth in the form of nonharmful food spoilage, may have unintended consequences on immune development over the course of a lifetime (Olivares et al. 2006). Indeed, a recent study examining the effect of consuming fermented foods found an increase in microbial diversity and an improvement of the inflammatory status of the participants who increased their daily fermented food intake (Wastyk et al. 2021). Despite this promise, a great deal of additional study is required in this area.
There are frequent misunderstandings about the expected influences of fermented foods, and associated microorganisms, on the gut microbiome. In most cases, fermentation-associated microorganisms evolved to, by definition, ferment food substrates rather than persist in the human gut (Bachmann et al. 2012). However, it has been shown that particular species of LAB found in foods are often closely related to those found in the gut (Pasolli et al. 2020). As a large bolus of live microorganisms, the presence and molecular effects of fermentation-associated microorganisms are transiently quite apparent via HTS or metabolomics (Zhang et al. 2016). Although not always the case, these transient effects can have quantifiable beneficial influences on resident gut microbiota structure, metabolism, or host immunity, inflammation or the gut brain axis (Derrien and van Hylckama Vlieg 2015, van de Wouw et al. 2020, Wastyk et al. 2021). In human subjects, live microorganisms from fermented foods can remain metabolically active in the gut, even without long-term engraftment, although the health consequences are less well understood (David et al. 2014). In limited cases, individual microbes do persist (Zhang et al. 2016, Milani et al. 2019) or transfer genetic material (Hehemann et al. 2010). However, although they may influence the dynamics of the process (Derrien and van Hylckama Vlieg 2015), in general, studies (or at least studies of faecal microbiomes) indicate that fermented food strains do not colonize (Fig. 3). In addition to the limitations of relying on faecal samples, it should also be noted that the majority of these examples derive from fermented milk products, with only extremely limited data generated to date for other fermented foods (Han et al. 2015, Abbondio et al. 2019, Jung et al. 2019).
Specific examples of the consequences of transient impacts of fermented food consumptions, and in some cases its underlying molecular mechanisms, have been studied both in human subjects and animal models. In one of the earliest HTS-based examples, while consumption of a fermented milk product did not change gut microbial composition in gnotobiotic mice, it transiently shifted carbohydrate utilization towards an upregulation of plant glycan catabolism, possibly due to contributions from Bifidobacterium animalis subsp. lactis (McNulty et al. 2011). Similar effects proved to be locally anti-inflammatory due to mechanisms as diverse as resident ecological disruption (Veiga et al. 2010), lipid metabolism (Bourrie et al. 2018), serotonergic signalling (van de Wouw et al. 2020), quorum sensing inhibition (Rooks et al. 2017), and reactive oxygen mitigation (Ballal et al. 2015) in mouse models.
At a molecular level, while breaking down complex compounds to simple molecules, the microorganisms responsible for fermentation synthesize enzymes, vitamins, essential amino acids, bioactive components, and potentially remove undesirable compounds (allergens, antinutritional factors). This leads to changes in texture and interaction between macro- and micronutriments. As a consequence, fermentation can improve the nutritional qualities and, in turn, health benefits of foods (Tamang et al. 2016, Şanlier et al. 2019). Kimchi is an example of a nutritionally enriched food with high levels of vitamins (e.g. vitamin C, β-carotene, and vitamin B), minerals, dietary fibers, and other bioactive compounds such as capsaicin, allyl compounds, gingerol, isothiocyanate, and chlorophyll. While the quantity of many of the vitamins are enhanced due to the biosynthesis of these vitamins through fermentation, other nutritional benefits are realized through increased bioavailability due to fermentation. These active compounds have, in turn, been associated with many health benefits attributed to kimchi (Park et al. 2014). The same is true for koumiss (Dhewa et al. 2015), miso (Watanabe 2013), and many other fermented foods.
In all cases, specific microbiome components are responsible for the biotransformations, but in some cases, the specific microbes and enzymatic reactions have been elucidated thanks to insights provided by metagenomics and metatransciptomics. In kimchi, metatranscriptomics revealed that genes associated with folate biosynthesis were expressed by Latilactobacillus sakei, while genes associated with riboflavin biosynthesis were expressed by Leuconostoc mesenteroides (Jung et al. 2013). In fermented mung beans, Rhizopus induces high amounts of free amino acid and γ-amino butyric acid and has been proposed to have antidiabetic and antioxidant properties (Yeap et al. 2015). In kombucha, genes involved in the biosynthesis of B-vitamins were present in Komagataeibacter rhaeticus (Arıkan et al. 2020) but as with the flavour-enhancing biochemical mechanisms above, much work remains to identify nutrition-enhancing pathways in the general case.
Another family of well-characterized nutrition-enhancing biotransformations includes the enhancement of antioxidant potential in fermented foods. Surprisingly, due to this process, some cheeses can have antioxidant levels close to that of vegetables or fruit juices (Fardet and Rock 2018). In kombucha, changes in the microbiota were found to correspond with increases in the levels of antioxidants, which may have arisen from microorganisms degrading polyphenols in the tea (Chakravorty et al. 2016). Furthermore, in tempeh, polyphenol, and isoflavone contents were significantly enhanced due to Rhizopus oligosporus activities (Kuligowski et al. 2017). Although it is not clear in each case what caused these enhancements, correlations within the data set point towards deglucosidation as a mechanism in some strains, but not in others. Information on the genetic profile and gene expression of these strains via metagenomics and metatranscriptomics can supplement such studies to better understand the strain-level differences and mechanisms for food enhancement.
Fermentation also provides a natural means to reduce undesirable compounds. Lactose (of concern to lactose-intolerant individuals) is commonly reduced to free glucose, galactose, and/or lactate; antinutrients such as phytate and tannins are degraded to release and enhance bioavailability of minerals such as iron, zinc, and calcium (Blandino et al. 2003, Poutanen et al. 2009); protease inhibitors and lectins are reduced in favour of protein absorption (Nkhata et al. 2018); beta-galactosides responsible for gut discomfort (e.g. stachyose and raffinose) are hydrolysed (Mukherjee et al. 2016); and toxic substrates present in raw materials can be eliminated. For the latter, examples include reductions in aflatoxin B1 levels in Ogi (a Nigerian fermented sorghum porridge) (Dada and Muller 1983), and removal of cyanide from cassava fermented with S. cerevisiae (Iyayi and Losel 2000). While the reduction of aflatoxins in fermented foods is mainly attributed to cellular binding (both viable and heat killed) and growth inhibition of aflatoxin-producing Aspergillus spp., other toxicity-mitigating mechanisms are emerging. For instance, Huang et al. (2017) showed that L. plantarum may reduce toxicity by altering gene expression in the liver of the consumer.
Gaps in our knowledge of fermented food microbiomes
Some of our most basic gaps regarding fermented food microbiomes have been dictated by the assays used to study them to date. The majority of studies have used amplicon sequencing, i.e. 16S rRNA or ITS sequencing to determine the composition of the bacteriome and mycobiome, respectively. The adoption of shotgun metagenomics has been slow relative to other microbial community types, and among studies that have adopted the approach, the greatest focus has remained on taxonomic profiling of the bacteriome, with corresponding approaches for the mycobiome being limited by the lower numbers of food-relevant eukaryotic genomes in reference databases and/or the higher complexity of eukaryotic genomes (Table S1, Fig. S1).
The application of shotgun metagenomics to study viromes also remains a relatively emerging area, despite the importance of phage in fermented foods (O’Sullivan et al. 2015). In the dairy industry, for example, phage infection remains the biggest cause of fermentation failure and leads to significant economic loss (Samson and Moineau 2013). However, phages can also be used as antimicrobial agents in food production, and thus be used to prevent contamination (Fernández et al. 2017). As in most microbial communities, fermented food viromes are technically challenging to study. They are best accessed through the enrichment of viral particles, but protocols for the extraction of viral nucleic acids from fermented foods are not yet standardized. Chloroform-based preparations adapted from other environments have been effective for cheeses (Dugat-Bony et al. 2020), while Muhammed et al. (2017) optimized ultracentrifugation for the purification of viral particles from dairies. Subsequent in silico analysis of viral sequences is notoriously challenging due to their great diversity, and under-representation in reference databases, limiting their analytical tractability even in the best case (Garmaeva et al. 2019, Wang 2020). Therefore, going forward it is imperative to expand reference databases, which might be achieved by sequencing isolates (Lagier et al. 2018) or recovering genomes from metagenomes in silico (Chen et al. 2020). Long-read sequencing has emerged as a promising technology that could be used for the characterization of the virome. Notably, using long-read metagenome assembly, Somerville et al. (2019) were able to recover phage genomes from natural whey cultures used in the production of Swiss Gruyère cheese. Since long-read sequencers generate reads longer than many phage genomes, long-read sequencing can also be used to recover complete phage genomes from samples without the requirement for assembly (Beaulaurier et al. 2020).
Shotgun metagenomics is further hampered by the prevalence of genes with unknown functions in reference databases. Over the past decade, the number of available genomes has exploded, while the annotation of these genomes has stagnated (Salzberg 2019). If we do not know the function of genes in the fermented food microbiome, shotgun metagenomics by itself cannot explain the roles of microorganisms in fermentation, completely. Improving reference databases will be important to address this issue. This might be achieved by annotating the genomes of isolates from fermented foods, e.g. CRISPR interference (Wang et al. 2018) or Tn-Seq (Van Opijnen et al. 2009). Additionally, the utilization of multi-omics (including metabolomics and metaproteomics) in parallel might prove useful for inferring the function of members of fermented food microbiomes. One approach would be to replicate communities in vitro and investigate what happens to the metabolome or metaproteome when a specific strain is added to or removed from the community. The implementation of such approaches has the potential to maximize our understanding of the biology of fermented foods microbiomes. It should be noted that high-throughput metaproteomics is still somewhat challenging and, in the context of fermented foods, is further complicated by the frequently high levels of background proteins present in the food substrate. If such technical hurdles can be overcome, the rewards have the potential to be particularly great.
How can we influence fermented food microbiomes?
Once the field is able to fill gaps relating to the composition of a broader variety of fermented food microbial systems and how they function, the logical progression is to influence and modify those functions towards desirable, beneficial outcomes. This is especially the case for products using ‘undefined starters’, which contain complex mixtures of microorganisms that can vary with time. In fermented foods, variability in the microbiota equates to variability in quality and, potentially, health benefits. The challenge for industry is to meet this demand while replicating the products authentically. We propose two avenues that producers might take to achieve this, i.e. controlling abiotic factors to shape the microbiota or using defined microbiota.
The first strategy can be used to forcibly shape or select for desirable properties even when beginning from a nominally undefined community. As mentioned above, Wolfe et al. (2014) demonstrated that moisture influenced the convergence of cheese rind microbiota, and temperature can act as a similar, simple selective pressure. Indeed, De Filippis et al. (2016) showed that an increase in temperature accelerated the rate at which cheeses ripened by inducing an increase in the expression of genes involved in the development of flavour. Controlling the availability of nutrients can also affect fermented food microbiota. Analysis of kefir revealed that Lactobacillus kefiranofaciens decreased during fermentation. Interestingly, L. kefiranofaciens lacked genes involved in the biosynthesis of tyrosine, and its decrease coincided with a reduction in the concentrations of tyrosine during fermentation (Walsh et al. 2016). Overall, these studies suggest that HTS can inform efforts to optimize fermentations in ways that favour the growth of microorganisms with the phenotype of interest, by modifying abiotic factors.
The second option, i.e. the use of defined starters, is already standard industrial practice, since it ensures the consistency of products. This sometimes comes at the cost of desirable properties lost due to the limitations of these controlled communities. Defined microbiota can be considered as a possible extension of this approach whereby strains identified as being key components of the undefined starters, on the basis of HTS-related insights into properties relating to flavour, tractability, and health benefits, could be targeted for isolation and combined. With respect to production, especially at scale, it will also be important to choose combinations of strains that will grow well together. In limited cases, it has already been demonstrated that the cheese rind microbiota can be replicated in vitro (Wolfe 2018), and therefore it is plausible that this can be done with engineered communities for other fermented food microbiota.
Understanding the (purported) benefits of fermented foods
Owing to the great variety of fermented foods, health benefit claims across different regulators and countries are a challenge. Beyond this, we also suffer from gaps in our basic biological knowledge of the potential health benefits of many fermented foods. These health benefits may derive from many sources, likely in combination: microorganisms in these foods, the metabolites they produce, or their impact on the raw food substrate on the host directly or indirectly through the resident human gut microbiome. Untangling the specific effects of microorganisms and metabolites/food bioactives can be facilitated through parallel investigations to determine the health benefits of foods from which the microorganisms have been removed (e.g. through centrifugation, filtration, or pasteurization) or the microbial cultures when not grown in the food substrate. These experiments, along with microbial genetic modifications and whole organism knock-ins/knock-outs from fermentation communities, can readily be performed in animal models, as performed by Bourrie et al. (2021), and in some cases in human subjects, given the wide range of strains already approved for food use. Multi-omic approaches to analyse host responses to fermented foods can also play a major role in elucidating health benefits. While population-scale observational studies are useful for investigating potential health benefits (Taylor et al. 2020), ultimately, there is a need for truly randomized, blinded, placebo-controlled trials using fermented foods, which have been largely absent to date. It will be impossible to standardize precision health or wellness maintenance based on fermented foods, however, without a much more substantial understanding of the underlying biochemical and microbial mechanisms.
Conclusion
The ability to probe the composition and functionalities of the microbial populations present in fermented foods using the integrated approaches discussed above affords us an unprecedented opportunity to optimize various attributes of these foods. It is important that future studies validate observed correlations experimentally so that these insights can be translated. Importantly, integrated approaches have also helped to establish fermented foods as models for studying the interaction between microorganism within microbiomes, and we expect that technological improvements, such as enhancements relating to long-read sequencing, will help us to e.g. explore the role of phage in these microbiomes. Finally, integrated approaches are beginning to provide insights into impact of fermented foods on our health, although more studies are needed to support some of the claims attributed to these products. In conclusion, while integrated approaches have become central to the field of microbiology as a whole, they have, and are likely to continue to be, particularly transformative with respect to the microbiomes of fermented foods.
Supplementary Material
Acknowledgements
The authors thank Patrick Veiga for helpful discussion.
Contributor Information
Aaron M Walsh, Teagasc Food Research Centre, Moorepark, Fermoy, Cork and APC Microbiome Ireland, P61 C996, Ireland; Broad Institute of MIT and Harvard, Cambridge, MA 02142, USA; Department of Biostatistics, Harvard T.H. Chan School of Public Health, Boston, MA 02115, USA.
John Leech, Teagasc Food Research Centre, Moorepark, Fermoy, Cork and APC Microbiome Ireland, P61 C996, Ireland.
Curtis Huttenhower, Broad Institute of MIT and Harvard, Cambridge, MA 02142, USA; Department of Biostatistics, Harvard T.H. Chan School of Public Health, Boston, MA 02115, USA.
Hue Delhomme-Nguyen, Danone Nutricia Research, Centre Daniel Carasso, Palaiseau 91120, France.
Fiona Crispie, Teagasc Food Research Centre, Moorepark, Fermoy, Cork and APC Microbiome Ireland, P61 C996, Ireland.
Christian Chervaux, Danone Nutricia Research, Centre Daniel Carasso, Palaiseau 91120, France.
Paul D Cotter, Teagasc Food Research Centre, Moorepark, Fermoy, Cork and APC Microbiome Ireland, P61 C996, Ireland.
Author contributions
All authors contributed towards writing, reviewing, and editing.
Competing interests
H.D.-N. and C.C. are employees of Danone, a producer of fermented foods.
Funding
Research in the Cotter laboratory is conducted with the financial support of the MASTER project, an Innovation Action funded by the European Commission under the Horizon 2020 Programme under grant number 818368, by Science Foundation Ireland (SFI) under grant numbers SFI/12/RC/2273P1 and SFI/12/RC/2273P2 (APC Microbiome Ireland), by SFI and the Department of Agriculture, Food and Marine under grant 16/RC/3835 (VistaMilk), by the Enterprise Ireland Technology Centre, Food for Health Ireland, and by a number of commercial companies, including Danone.
References
- Abbondio M, Palomba A, Tanca Aet al. Fecal metaproteomic analysis reveals unique changes of the gut microbiome functions after consumption of sourdough Carasau bread. Front Microbiol. 2019;10:1733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen SC, Kiil K, Harder CBet al. Towards diagnostic metagenomics of Campylobacter in fecal samples. BMC Microbiol. 2017;17:1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arıkan M, Mitchell AL, Finn RDet al. Microbial composition of Kombucha determined using amplicon sequencing and shotgun metagenomics. J Food Sci. 2020;85:455–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachmann H, Starrenburg MJ, Molenaar Det al. Microbial domestication signatures of Lactococcus lactis can be reproduced by experimental evolution. Genome Res. 2012;22:115–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bäckhed F, Roswall J, Peng Yet al. Dynamics and stabilization of the human gut microbiome during the first year of life. Cell Host Microbe. 2015;17:690–703. [DOI] [PubMed] [Google Scholar]
- Ballal SA, Veiga P, Fenn Ket al. Host lysozyme-mediated lysis of Lactococcus lactis facilitates delivery of colitis-attenuating superoxide dismutase to inflamed colons. Proc Natl Acad Sci USA. 2015;112:7803–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaulaurier J, Luo E, Eppley JMet al. Assembly-free single-molecule sequencing recovers complete virus genomes from natural microbial communities. Genome Res. 2020;30:437–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bengmark S. Bioecological control of inflammatory bowel disease. Clin Nutr. 2007;26:169–81. [DOI] [PubMed] [Google Scholar]
- Bertuzzi AS, Walsh AM, Sheehan Jet al. Omics-based insights into flavor development and microbial succession within surface-ripened cheese. mSystems. 2018;3:e00211–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blandino A, Al-Aseeri M, Pandiella Set al. Cereal-based fermented foods and beverages. Food Res Int. 2003;36:527–43. [Google Scholar]
- Blasche S, Kim Y, Mars RAet al. Metabolic cooperation and spatiotemporal niche partitioning in a kefir microbial community. Nat Microbiol. 2021;6:196–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bokulich NA, Chung J, Battaglia Tet al. Antibiotics, birth mode, and diet shape microbiome maturation during early life. Sci Transl Med. 2016;8:343ra382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bokulich NA, Ohta M, Lee Met al. Indigenous bacteria and fungi drive traditional kimoto sake fermentations. Appl Environ Microbiol. 2014;80:5522–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonham KS, Wolfe BE, Dutton RJ. Extensive horizontal gene transfer in cheese-associated bacteria. Elife. 2017;6:e22144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boor KJ, Wiedmann M, Murphy Set al. A 100-year review: microbiology and safety of milk handling. J Dairy Sci. 2017;100:9933–51. [DOI] [PubMed] [Google Scholar]
- Bourrie BC, Cotter PD, Willing BP. Traditional kefir reduces weight gain and improves plasma and liver lipid profiles more successfully than a commercial equivalent in a mouse model of obesity. J Funct Foods. 2018;46:29–37. [Google Scholar]
- Bourrie BC, Ju T, Fouhse JMet al. Kefir microbial composition is a deciding factor in the physiological impact of kefir in a mouse model of obesity. Br J Nutr. 2021;125:129–38. [DOI] [PubMed] [Google Scholar]
- Buytaers FE, Saltykova A, Mattheus Wet al. Application of a strain-level shotgun metagenomics approach on food samples: resolution of the source of a Salmonella food-borne outbreak. Microb Genom. 2021;7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Y, Fanning S, Proos Set al. A review on the applications of next generation sequencing technologies as applied to food-related microbiome studies. Front Microbiol. 2017;8:1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caplice E, Fitzgerald GF. Food fermentations: role of microorganisms in food production and preservation. Int J Food Microbiol. 1999;50:131–49. [DOI] [PubMed] [Google Scholar]
- Carrasco-Castilla J, Hernández-Álvarez AJ, Jiménez-Martínez Cet al. Use of proteomics and peptidomics methods in food bioactive peptide science and engineering. Food Eng Rev. 2012;4:224–43. [Google Scholar]
- Chakravorty S, Bhattacharya S, Chatzinotas Aet al. Kombucha tea fermentation: microbial and biochemical dynamics. Int J Food Microbiol. 2016;220:63–72. [DOI] [PubMed] [Google Scholar]
- Chen L-X, Anantharaman K, Shaiber Aet al. Accurate and complete genomes from metagenomes. Genome Res. 2020;30:315–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M, Sun Q, Giovannucci Eet al. Dairy consumption and risk of type 2 diabetes: 3 cohorts of US adults and an updated meta-analysis. BMC Med. 2014;12:215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chun BH, Kim KH, Jeon HHet al. Pan-genomic and transcriptomic analyses of Leuconostoc mesenteroides provide insights into its genomic and metabolic features and roles in kimchi fermentation. Sci Rep. 2017;7:1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosetta CM, Kfoury N, Robbat Aet al. Fungal volatiles mediate cheese rind microbiome assembly. Environ Microbiol. 2020;22:4745–60. [DOI] [PubMed] [Google Scholar]
- Cotillard A, Kennedy SP, Kong LCet al. Dietary intervention impact on gut microbial gene richness. Nature. 2013;500:585–8. [DOI] [PubMed] [Google Scholar]
- Dada L, Muller H. The fate of aflatoxin B1 in the production of ogi, a Nigerian fermented sorghum porridge. J Cereal Sci. 1983;1:63–70. [Google Scholar]
- David LA, Maurice CF, Carmody RNet al. Diet rapidly and reproducibly alters the human gut microbiome. Nature. 2014;505:559–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Filippis F, Genovese A, Ferranti Pet al. Metatranscriptomics reveals temperature-driven functional changes in microbiome impacting cheese maturation rate. Sci Rep. 2016;6:21871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Filippis F, Parente E, Ercolini D. Metagenomics insights into food fermentations. Microb Biotechnol. 2017;10:91–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Filippis F, Troise AD, Vitaglione Pet al. Different temperatures select distinctive acetic acid bacteria species and promotes organic acids production during Kombucha tea fermentation. Food Microbiol. 2018;73:11–6. [DOI] [PubMed] [Google Scholar]
- Derrien M, van Hylckama Vlieg JE. Fate, activity, and impact of ingested bacteria within the human gut microbiota. Trends Microbiol. 2015;23:354–66. [DOI] [PubMed] [Google Scholar]
- Dhewa T, Mishra V, Kumar Net al. Koumiss: the nutritional and therapeutic values. In: Puniya AK (ed.), Fermented Milk and Dairy Products. CRC Press, 2015, 483–94. [Google Scholar]
- Dugat-Bony E, Lossouarn J, De Paepe Met al. Viral metagenomic analysis of the cheese surface: a comparative study of rapid procedures for extracting viral particles. Food Microbiol. 2020;85:103278. [DOI] [PubMed] [Google Scholar]
- Escobar-Zepeda A, Sanchez-Flores A, Baruch MQ. Metagenomic analysis of a Mexican ripened cheese reveals a unique complex microbiota. Food Microbiol. 2016;57:116–27. [DOI] [PubMed] [Google Scholar]
- Eussen SJ, van Dongen MC, Wijckmans Net al. Consumption of dairy foods in relation to impaired glucose metabolism and type 2 diabetes mellitus: the Maastricht Study. Br J Nutr. 2016;115:1453–61. [DOI] [PubMed] [Google Scholar]
- Fardet A, Rock E. In vitro and in vivo antioxidant potential of milks, yoghurts, fermented milks and cheeses: a narrative review of evidence. Nutr Res Rev. 2018;31:52–70. [DOI] [PubMed] [Google Scholar]
- Fernández L, Escobedo S, Gutiérrez Det al. Bacteriophages in the dairy environment: from enemies to allies. Antibiotics. 2017;6:27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franzosa EA, Hsu T, Sirota-Madi Aet al. Sequencing and beyond: integrating molecular ‘omics’ for microbial community profiling. Nat Rev Microbiol. 2015;13:360–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garmaeva S, Sinha T, Kurilshikov Aet al. Studying the gut virome in the metagenomic era: challenges and perspectives. BMC Biol. 2019;17:1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrigues C, Johansen E, Crittenden R. Pangenomics–an avenue to improved industrial starter cultures and probiotics. Curr Opin Biotechnol. 2013;24:187–91. [DOI] [PubMed] [Google Scholar]
- Geier MS, Butler RN, Howarth GS. Inflammatory bowel disease: current insights into pathogenesis and new therapeutic options; probiotics, prebiotics and synbiotics. Int J Food Microbiol. 2007;115:1–11. [DOI] [PubMed] [Google Scholar]
- Gibson B, Geertman J, Hittinger Cet al. New yeasts—new brews: modern approaches to brewing yeast design and development. FEMS Yeast Res. 2017;17. [DOI] [PubMed] [Google Scholar]
- Hamid M-N, Friedberg I. Identifying antimicrobial peptides using word embedding with deep recurrent neural networks. Bioinformatics. 2019;35:2009–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammami R, Zouhir A, Le Lay Cet al. BACTIBASE second release: a database and tool platform for bacteriocin characterization. BMC Microbiol. 2010;10:1–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han K, Bose S, Wang Jhet al. Contrasting effects of fresh and fermented kimchi consumption on gut microbiota composition and gene expression related to metabolic syndrome in obese Korean women. Mol Nutr Food Res. 2015;59:1004–8. [DOI] [PubMed] [Google Scholar]
- Hatti-Kaul R, Chen L, Dishisha Tet al. Lactic acid bacteria: from starter cultures to producers of chemicals. FEMS Microbiol Lett. 2018;365:fny213. [DOI] [PubMed] [Google Scholar]
- Hazards EPoB, Koutsoumanis K, Allende Aet al. Whole genome sequencing and metagenomics for outbreak investigation, source attribution and risk assessment of food-borne microorganisms. EFSA J. 2019;17:e05898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hehemann J-H, Correc G, Barbeyron Tet al. Transfer of carbohydrate-active enzymes from marine bacteria to Japanese gut microbiota. Nature. 2010;464:908–12. [DOI] [PubMed] [Google Scholar]
- Henry CS, Dejongh M, Best AAet al. High-throughput generation, optimization and analysis of genome-scale metabolic models. Nat Biotechnol. 2010;28:977–82. [DOI] [PubMed] [Google Scholar]
- Hong X, Chen J, Liu Let al. Metagenomic sequencing reveals the relationship between microbiota composition and quality of Chinese Rice Wine. Sci Rep. 2016;6:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang L, Duan C, Zhao Yet al. Reduction of aflatoxin B1 toxicity by Lactobacillus plantarum C88: a potential probiotic strain isolated from Chinese traditional fermented food “tofu”. PLoS One. 2017;12:e0170109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyayi EA, Losel DM. Cyanide detoxifition in cassava by-products by fungal solid state fermentation. J Food Technol Afr. 2000;5:48–51. [Google Scholar]
- Jimenez-Lorenzo R, Bloem A, Farines Vet al. How to modulate the formation of negative volatile sulfur compounds during wine fermentation?. FEMS Yeast Res. 2021;21:foab038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung Y, Kim I, Mannaa Met al. Effect of Kombucha on gut-microbiota in mouse having non-alcoholic fatty liver disease. Food Sci Biotechnol. 2019;28:261–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung JY, Lee SH, Jin HMet al. Metatranscriptomic analysis of lactic acid bacterial gene expression during kimchi fermentation. Int J Food Microbiol. 2013;163:171–9. [DOI] [PubMed] [Google Scholar]
- Jung JY, Lee SH, Kim JMet al. Metagenomic analysis of kimchi, the Korean traditional fermented food. Appl Environ Microbiol. 2011;77:2264–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamelamela N, Zalesne M, Morimoto Jet al. Indigo- and indirubin-producing strains of Proteus and Psychrobacter are associated with purple rind defect in a surface-ripened cheese. Food Microbiol. 2018;76:543–52. [DOI] [PubMed] [Google Scholar]
- Kang DD, Froula J, Egan Ret al. MetaBAT, an efficient tool for accurately reconstructing single genomes from complex microbial communities. PeerJ. 2015;3:e1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karst SM, Ziels RM, Kirkegaard RHet al. High-accuracy long-read amplicon sequences using unique molecular identifiers with Nanopore or PacBio sequencing. Nat Methods. 2021;18:165–9. [DOI] [PubMed] [Google Scholar]
- Kastman EK, Kamelamela N, Norville JWet al. Biotic interactions shape the ecological distributions of Staphylococcus species. MBio. 2016;7:e01157–01116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly CR, Ihunnah C, Fischer Met al. Fecal microbiota transplant for treatment of Clostridium difficile infection in immunocompromised patients. Am J Gastroenterol. 2014;109:1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kergourlay G, Taminiau B, Daube Get al. Metagenomic insights into the dynamics of microbial communities in food. Int J Food Microbiol. 2015;213:31–9. [DOI] [PubMed] [Google Scholar]
- Kim T-W, Lee J-Y, Jung S-Het al. Identification and distribution of predominant tactic acid bacteria in Kimchi, a Korean traditional fermented food. J Microbiol Biotechnol. 2002;12:635–42. [Google Scholar]
- Kim H, Sitarik AR, Woodcroft Ket al. Birth mode, breastfeeding, pet exposure, and antibiotic use: associations with the gut microbiome and sensitization in children. Curr Allergy Asthma Rep. 2019;19:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuligowski M, Pawłowska K, Jasińska-Kuligowska Iet al. Isoflavone composition, polyphenols content and antioxidative activity of soybean seeds during tempeh fermentation. CyTA J Food. 2017;15:27–33. [Google Scholar]
- Laatikainen R, Koskenpato J, Hongisto SMet al. Randomised clinical trial: low-FODMAP rye bread vs. regular rye bread to relieve the symptoms of irritable bowel syndrome. Aliment Pharmacol Ther. 2016;44:460–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagier J-C, Dubourg G, Million Met al. Culturing the human microbiota and culturomics. Nat Rev Microbiol. 2018;16:540–50. [DOI] [PubMed] [Google Scholar]
- Landis EA, Oliverio AM, McKenney EAet al. The diversity and function of sourdough starter microbiomes. Elife. 2021;10:e61644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langdon A, Crook N, Dantas G. The effects of antibiotics on the microbiome throughout development and alternative approaches for therapeutic modulation. Genome Med. 2016;8:1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SH, Jung JY, Jeon CO. Effects of temperature on microbial succession and metabolite change during saeu-jeot fermentation. Food Microbiol. 2014;38:16–25. [DOI] [PubMed] [Google Scholar]
- Lee SH, Jung JY, Jeon CO. Bacterial community dynamics and metabolite changes in myeolchi-aekjeot, a Korean traditional fermented fish sauce, during fermentation. Int J Food Microbiol. 2015;203:15–22. [DOI] [PubMed] [Google Scholar]
- Leech J, Cabrera-Rubio R, Walsh AMet al. Fermented-food metagenomics reveals substrate-associated differences in taxonomy and health-associated and antibiotic resistance determinants. mSystems. 2020;5:e00522–00520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lessard M-H, Viel C, Boyle Bet al. Metatranscriptome analysis of fungal strains Penicillium camemberti and Geotrichum candidum reveal cheese matrix breakdown and potential development of sensory properties of ripened Camembert-type cheese. BMC Genom. 2014;15:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim J-H, Jung E-S, Choi E-Ket al. Supplementation with Aspergillus oryzae-fermented kochujang lowers serum cholesterol in subjects with hyperlipidemia. Clin Nutr. 2015;34:383–7. [DOI] [PubMed] [Google Scholar]
- Machado D, Tramontano M, Andrejev Set al. Fast automated reconstruction of genome-scale metabolic models for microbial species and communities. Nucleic Acids Res. 2018;46:7542–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNulty NP, Yatsunenko T, Hsiao Aet al. The impact of a consortium of fermented milk strains on the gut microbiome of gnotobiotic mice and monozygotic twins. Sci Transl Med. 2011;3:106ra106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeusli M, Lee B, Miller Set al. Horizontal gene transfer of antibiotic resistance from Acinetobacter baylyi to Escherichia coli on lettuce and subsequent antibiotic resistance transmission to the gut microbiome. mSphere. 2020;5:e00329–00320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magnúsdóttir S, Thiele I. Modeling metabolism of the human gut microbiome. Curr Opin Biotechnol. 2018;51:90–6. [DOI] [PubMed] [Google Scholar]
- Marco ML, Heeney D, Binda Set al. Health benefits of fermented foods: microbiota and beyond. Curr Opin Biotechnol. 2017;44:94–102. [DOI] [PubMed] [Google Scholar]
- Marco ML, Sanders ME, Gänzle Met al. The International Scientific Association for Probiotics and Prebiotics (ISAPP) consensus statement on fermented foods. Nat Rev Gastroenterol Hepatol. 2021;18:196–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markowiak P, Śliżewska K. Effects of probiotics, prebiotics, and synbiotics on human health. Nutrients. 2017;9:1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milani C, Duranti S, Napoli Set al. Colonization of the human gut by bovine bacteria present in Parmesan cheese. Nat Commun. 2019;10:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morin M, Pierce EC, Dutton RJ. Changes in the genetic requirements for microbial interactions with increasing community complexity. Elife. 2018;7:e37072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morton JT, Freed SD, Lee SWet al. A large scale prediction of bacteriocin gene blocks suggests a wide functional spectrum for bacteriocins. BMC Bioinf. 2015;16:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muhammed MK, Kot W, Neve Het al. Metagenomic analysis of dairy bacteriophages: extraction method and pilot study on whey samples derived from using undefined and defined mesophilic starter cultures. Appl Environ Microbiol. 2017;83:e00888–00817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee R, Chakraborty R, Dutta A. Role of fermentation in improving nutritional quality of soybean meal—a review. Asian-Australas J Anim Sci. 2016;29:1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagata S, Asahara T, Wang Cet al. The effectiveness of Lactobacillus beverages in controlling infections among the residents of an aged care facility: a randomized placebo-controlled double-blind trial. Ann Nutr Metab. 2016;68:51–9. [DOI] [PubMed] [Google Scholar]
- Niccum BA, Kastman EK, Kfoury Net al. Strain-level diversity impacts cheese rind microbiome assembly and function. mSystems. 2020;5:e00149–00120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nkhata SG, Ayua E, Kamau EHet al. Fermentation and germination improve nutritional value of cereals and legumes through activation of endogenous enzymes. Food Sci Nutr. 2018;6:2446–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noecker C, Eng A, Srinivasan Set al. Metabolic model-based integration of microbiome taxonomic and metabolomic profiles elucidates mechanistic links between ecological and metabolic variation. mSystems. 2016;1:e00013–00015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Sullivan DJ, Fallico V, O’Sullivan Oet al. High-throughput DNA sequencing to survey bacterial histidine and tyrosine decarboxylases in raw milk cheeses. BMC Microbiol. 2015;15:266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olivares M, Díaz-Ropero MP, Gómez Net al. Dietary deprivation of fermented foods causes a fall in innate immune response. Lactic acid bacteria can counteract the immunological effect of this deprivation. J Dairy Res. 2006;73:492–8. [DOI] [PubMed] [Google Scholar]
- Olm MR, Crits-Christoph A, Bouma-Gregson Ket al. inStrain profiles population microdiversity from metagenomic data and sensitively detects shared microbial strains. Nat Biotechnol. 2021;39:727–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park K-Y, Jeong J-K, Lee Y-Eet al. Health benefits of kimchi (Korean fermented vegetables) as a probiotic food. J Med Food. 2014;17:6–20. [DOI] [PubMed] [Google Scholar]
- Parker M, Zobrist S, Donahue Cet al. Naturally fermented milk from northern Senegal: bacterial community composition and probiotic enrichment with Lactobacillus rhamnosus. Front Microbiol. 2018;9:2218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parvez S, Malik KA, Ah Kang Set al. Probiotics and their fermented food products are beneficial for health. J Appl Microbiol. 2006;100:1171–85. [DOI] [PubMed] [Google Scholar]
- Pasolli E, De Filippis F, Mauriello IEet al. Large-scale genome-wide analysis links lactic acid bacteria from food with the gut microbiome. Nat Commun. 2020;11:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira GV, Alvarez JP, Neto DPdCet al. Great intraspecies diversity of Pichia kudriavzevii in cocoa fermentation highlights the importance of yeast strain selection for flavor modulation of cocoa beans. LWT. 2017;84:290–7. [Google Scholar]
- Pierce EC, Dutton RJ. Putting microbial interactions back into community contexts. Curr Opin Microbiol. 2022;65:56–63. [DOI] [PubMed] [Google Scholar]
- Poutanen K, Flander L, Katina K. Sourdough and cereal fermentation in a nutritional perspective. Food Microbiol. 2009;26:693–9. [DOI] [PubMed] [Google Scholar]
- Quigley L, O'Sullivan DJ, Daly Det al. Thermus and the pink discoloration defect in cheese. mSystems. 2016;1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rezac S, Kok CR, Heermann Met al. Fermented foods as a dietary source of live organisms. Front Microbiol. 2018;9:1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizo J, Guillén D, Farrés Aet al. Omics in traditional vegetable fermented foods and beverages. Crit Rev Food Sci Nutr. 2020;60:791–809. [DOI] [PubMed] [Google Scholar]
- Rooks MG, Veiga P, Reeves AZet al. QseC inhibition as an antivirulence approach for colitis-associated bacteria. Proc Natl Acad Sci. 2017;114:142–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross RP, Morgan S, Hill C. Preservation and fermentation: past, present and future. Int J Food Microbiol. 2002;79:3–16. [DOI] [PubMed] [Google Scholar]
- Salzberg SL. Next-generation genome annotation: we still struggle to get it right. BioMed Central. 2019;20:1–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samson JE, Moineau S. Bacteriophages in food fermentations: new frontiers in a continuous arms race. Annu Rev Food Sci Technol. 2013;4:347–68. [DOI] [PubMed] [Google Scholar]
- Şanlier N, Gökcen BB, Sezgin AC. Health benefits of fermented foods. Crit Rev Food Sci Nutr. 2019;59:506–27. [DOI] [PubMed] [Google Scholar]
- Scott KP, Duncan SH, Flint HJ. Dietary fibre and the gut microbiota. Nutr Bull. 2008;33:201–11. [Google Scholar]
- Segata N. Gut microbiome: westernization and the disappearance of intestinal diversity. Curr Biol. 2015;25:R611–3. [DOI] [PubMed] [Google Scholar]
- Shoaie S, Ghaffari P, Kovatcheva-Datchary Pet al. Quantifying diet-induced metabolic changes of the human gut microbiome. Cell Metab. 2015;22:320–31. [DOI] [PubMed] [Google Scholar]
- Smit G, Smit BA, Engels WJ. Flavour formation by lactic acid bacteria and biochemical flavour profiling of cheese products. FEMS Microbiol Rev. 2005;29:591–610. [DOI] [PubMed] [Google Scholar]
- Soedamah-Muthu SS, Masset G, Verberne Let al. Consumption of dairy products and associations with incident diabetes, CHD and mortality in the Whitehall II study. Br J Nutr. 2013;109:718–26. [DOI] [PubMed] [Google Scholar]
- Somerville V, Lutz S, Schmid Met al. Long-read based de novo assembly of low-complexity metagenome samples results in finished genomes and reveals insights into strain diversity and an active phage system. BMC Microbiol. 2019;19:1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stiemsma LT, Nakamura RE, Nguyen JGet al. Does consumption of fermented foods modify the human gut microbiota?. J Nutr. 2020;150:1680–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamang JP, Kailasapathy K. Fermented Foods and Beverages of the World. Boca Raton: CRC press, 2010. [Google Scholar]
- Tamang JP, Shin D-H, Jung S-Jet al. Functional properties of microorganisms in fermented foods. Front Microbiol. 2016;7:578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor BC, Lejzerowicz F, Poirel Met al. Consumption of fermented foods is associated with systematic differences in the gut microbiome and metabolome. mSystems. 2020;5:e00901–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truong DT, Tett A, Pasolli Eet al. Microbial strain-level population structure and genetic diversity from metagenomes. Genome Res. 2017;27:626–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van de Wouw M, Walsh AM, Crispie Fet al. Distinct actions of the fermented beverage kefir on host behaviour, immunity and microbiome gut-brain modules in the mouse. Microbiome. 2020;8:1–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Dijk LR, Walker BJ, Straub TJet al. StrainGE: a toolkit to track and characterize low-abundance strains in complex microbial communities. Genome Biol. 2022;23:1–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Heel AJ, de Jong A, Song Cet al. BAGEL4: a user-friendly web server to thoroughly mine RiPPs and bacteriocins. Nucleic Acids Res. 2018;46:W278–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Opijnen T, Bodi KL, Camilli A. Tn-seq: high-throughput parallel sequencing for fitness and genetic interaction studies in microorganisms. Nat Methods. 2009;6:767–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veiga P, Gallini CA, Beal Cet al. Bifidobacterium animalis subsp. lactis fermented milk product reduces inflammation by altering a niche for colitogenic microbes. Proc Natl Acad Sci. 2010;107:18132–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vicedomini R, Quince C, Darling AEet al. Strainberry: automated strain separation in low-complexity metagenomes using long reads. Nat Commun. 2021;12:1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh AM, Crispie F, Claesson MJet al. Translating omics to food microbiology. Annu Rev Food Sci Technol. 2017;8:113–34. [DOI] [PubMed] [Google Scholar]
- Walsh AM, Macori G, Kilcawley KNet al. Meta-analysis of cheese microbiomes highlights contributions to multiple aspects of quality. Nat Food. 2020;1:500–10. [DOI] [PubMed] [Google Scholar]
- Walsh AM, Crispie F, Daari Ket al. Strain-level metagenomic analysis of the fermented dairy beverage nunu highlights potential food safety risks. Appl Environ Microbiol. 2017;83:01144–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh AM, Crispie F, Kilcawley Ket al. Microbial succession and flavor production in the fermented dairy beverage kefir. mSystems. 2016;1:e00052–00016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D. 5 challenges in understanding the role of the virome in health and disease. PLoS Pathog. 2020;16:e1008318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T, Guan C, Guo Jet al. Pooled CRISPR interference screening enables genome-scale functional genomics study in bacteria with superior performance. Nat Commun. 2018;9:1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z-M, Lu Z-M, Shi J-Set al. Exploring flavour-producing core microbiota in multispecies solid-state fermentation of traditional Chinese vinegar. Sci Rep. 2016;6:26818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wastyk HC, Fragiadakis GK, Perelman Det al. Gut-microbiota-targeted diets modulate human immune status. Cell. 2021;184:4137–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe H. Beneficial biological effects of miso with reference to radiation injury, cancer and hypertension. J Toxicol Pathol. 2013;26:91–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfe BE. Using cultivated microbial communities to dissect microbiome assembly: challenges, limitations, and the path ahead. mSystems. 2018;3:e00161–00117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfe BE, Button JE, Santarelli Met al. Cheese rind communities provide tractable systems for in situ and in vitro studies of microbial diversity. Cell. 2014;158:422–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao G, Yu J, Hou Qet al. A perspective study of koumiss microbiome by metagenomics analysis based on single-cell amplification technique. Front Microbiol. 2017;8:165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeap SK, Beh BK, Ho WYet al. In vivo antioxidant and hypolipidemic effects of fermented mung bean on hypercholesterolemic mice. Evid Based Complement Altern Med. 2015;2015:1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeluri Jonnala B, McSweeney PL, Sheehan JJet al. Sequencing of the cheese microbiome and its relevance to industry. Front Microbiol. 2018;9:1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youngster I, Mahabamunuge J, Systrom HKet al. Oral, frozen fecal microbiota transplant (FMT) capsules for recurrent Clostridium difficile infection. BMC Med. 2016;14:1–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Derrien M, Levenez Fet al. Ecological robustness of the gut microbiota in response to ingestion of transient food-borne microbes. ISME J. 2016;10:2235–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Kastman EK, Guasto JSet al. Fungal networks shape dynamics of bacterial dispersal and community assembly in cheese rind microbiomes. Nat Commun. 2018;9:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.