Abstract
Aims
Osteoporosis (OP) is a metabolic bone disease, characterized by a decrease in bone mineral density (BMD). However, the research of regulatory variants has been limited for BMD. In this study, we aimed to explore novel regulatory genetic variants associated with BMD.
Methods
We conducted an integrative analysis of BMD genome-wide association study (GWAS) and regulatory single nucleotide polymorphism (rSNP) annotation information. Firstly, the discovery GWAS dataset and replication GWAS dataset were integrated with rSNP annotation database to obtain BMD associated SNP regulatory elements and SNP regulatory element-target gene (E-G) pairs, respectively. Then, the common genes were further subjected to HumanNet v2 to explore the biological effects.
Results
Through discovery and replication integrative analysis for BMD GWAS and rSNP annotation database, we identified 36 common BMD-associated genes for BMD irrespective of regulatory elements, such as FAM3C (pdiscovery GWAS = 1.21 × 10-25, preplication GWAS = 1.80 × 10-12), CCDC170 (pdiscovery GWAS = 1.23 × 10-11, preplication GWAS = 3.22 × 10-9), and SOX6 (pdiscovery GWAS = 4.41 × 10-15, preplication GWAS = 6.57 × 10-14). Then, for the 36 common target genes, multiple gene ontology (GO) terms were detected for BMD such as positive regulation of cartilage development (p = 9.27 × 10-3) and positive regulation of chondrocyte differentiation (p = 9.27 × 10-3).
Conclusion
We explored the potential roles of rSNP in the genetic mechanisms of BMD and identified multiple candidate genes. Our study results support the implication of regulatory genetic variants in the development of OP.
Cite this article: Bone Joint Res 2023;12(2):147–154.
Keywords: Osteoporosis, Bone mineral density, Regulatory single nucleotide polymorphism, Genome-wide association studies, Bone mineral density (BMD), single nucleotide polymorphisms, cartilage, chondrocyte, metabolic bone disease, mesenchymal stem cell (MSC), hip, osteoblasts, RNAs
Article focus
To access the genetic mechanism of osteoporosis (OP) using the integrative analysis of genome-wide association study (GWAS) and regulatory single nucleotide polymorphism (rSNP) annotation information of bone mineral density (BMD).
We performed functional enrichment analysis of the common candidate genes associated with BMD.
Key messages
A total of 36 target regulatory genes were detected for BMD such as FAM3C, CCDC170, SOX6, and PLEKHM1.
We detected 12 BMD-associated gene ontology terms such as muscle cell differentiation, positive regulation of cartilage development, and positive regulation of chondrocyte differentiation.
Strengths and limitations
Integrating analysis of GWAS and rSNP has a boosting power for regulatory genetic variant detection.
One of the main limitations is that rSNP analysis could not take tissue or cell types into account.
Introduction
Osteoporosis (OP) is a metabolic bone disease, characterized by a decrease in the density of bone.1 Multiple risk factors contribute to the development of OP such as hormone changes, gut microbiome, the use of certain drugs, and cigarette smoking.2-4 In the USA, millions of people either already have OP or are at high risk due to low bone mass.5 OP is more common in postmenopausal women,6 which leads to heavy burden on the healthcare system and society.
Bone mineral density (BMD) is one of the major diagnostic indexes of OP. A recent study suggested that bone density had a strong genetic determination, with an estimated heritability ranging from 61% to 83%.7 More than 50 susceptibility loci have been identified for BMD or OP, such as LRP5, MEPE, SPTBN1, and DKK1.8-10 For instance, genome-wide association studies (GWASs) have led to the identification of 100 loci associated with BMD and other bone traits related to risk of fracture.11 However, only a small percentage of the heritability of BMD can be explained by the susceptibility genetic variants discovered so far,12 suggesting the existence of undiscovered causal genetic factors for BMD.
Although GWASs have achieved great success in identifying the susceptible loci of complex diseases, there are several issues and limitations associated with these genetic association studies.13,14 GWAS signals often lie in non-coding regions, which may harbour regulatory elements affecting gene expression, such as expression quantitative trait loci (eQTLs) and methylation quantitative trait loci (meQTLs).15,16 However, it is difficult to identify the precise causal SNP and causal gene for these noncoding variants.17 These causal genetic variants within non-coding regulatory regions are indistinguishable from the neighbouring markers.
Regulatory single nucleotide polymorphisms (rSNPs) refer to the SNPs that are associated with the regulation of gene expression levels through transcription factor binding, chromatin interaction, circular RNA (circRNA)-mediated post-transcriptional regulation, and so on.18,19 Hindorff et al14 have illustrated that a lot of risk SNPs could affect phenotypes in a non-coding manner, for instance impacting gene regulation. Integrating GWAS datasets with rSNPs has the potential to reveal novel susceptibility genetic variants for human complex diseases.20 A previous study has shown that a rSNP in the Cx43 promoter region plays a critical role in Tetralogy of Fallot (TOF), by impacting the transcriptional activity.21 Furthermore, Yeo et al22 reported that low-effect chronic obstructive pulmonary disease (COPD) risk SNPs were identified through enrichment of cis-regulatory SNPs in genes.
In this study, we conducted an integrative analysis of GWAS and large-scale rSNP annotation data for BMD, containing seven regulatory genetic elements. The significant loci identified by the GWAS of BMD were firstly annotated with the rSNP annotation database to obtain BMD-associated regulatory genetic elements and their target genes. To explore the functional relevance of identified candidate genes, the common candidate genes identified by both discovery and replication studies were further subjected to gene ontology (GO) functional enrichment analysis.
Methods
Discovery GWAS summary dataset of BMD
The GWAS summary dataset of BMD was drawn from the GEnetic Factors for OSteoporosis (GEFOS) Consortium,23 containing a total of 52,236 individuals of European ancestry. Briefly, the GEFOS project used meta-analysis of whole genome sequencing, whole exome sequencing, and deep imputation of genotype data to identify candidate variants associated with the risk of BMD. Single variants with a minor allele frequency (MAF) > 0.5% were tested for an additive effect on femoral neck BMD (FN-BMD), lumbar spine BMD (LS-BMD), and forearm BMD (FA-BMD), adjusting for sex, age, age_squared, and weight as covariates. Detailed information of the subjects, genotyping, imputation, and quality control can be found in a previously published study.8
Replication GWAS summary dataset of BMD
The replication GWAS summary dataset of BMD was drawn from the UK Biobank database.24-26 In summary, the UK Biobank BMD GWAS dataset contains 452,264 participants. The BMD values of bones and joints were measured using dual-energy X-ray absorptiometry in this study. DNA was extracted from stored blood samples and shipped to Affymetrix Research Services Laboratory for genotyping. SNP genotyping was performed using the UK Biobank Axiom array.27 Genotypes were imputed by IMPUTE4.28 Principal component analysis (PCA) was used to account for potential population structure in both marker- and sample-based quality control procedures. Detailed information of the subjects, genotyping, imputation, and quality control can be found in two previously published studies.24,25
Annotation datasets of rSNPs
The rSNP annotation data were collected from the rSNPBase 3.1 database.29,30 There are seven types of regulatory elements, including 7,562,592 annotation terms for transcription factor binding regions (TFBRs) from the Encyclopaedia of DNA Elements (ENCODE) project,31 212,837 for chromatin interactive regions (CIRs) from the ENCODE project,31 2,794 for mature microRNA (miRNA) regions from miRbase,32 384,284 for predicted miRNA target sites from TargetScan33 and miRNAda,34-37 211,749 for long non-coding RNA (lncRNA) regions from LNCipedia,38 38,916 for topologically associated domains (TADs) from ENCODE-processed data, and 312,673 for circRNAs from CircNet.39 Except for circRNAs, target gene analyses were performed on the other six types of regulatory elements to get corresponding elements-gene (E-G) pairs.
For regulatory E-G pairs of transcriptional regulation, chromosome location was acquired from ENCODE Consortium, among which TFBR information was acquired from proceeded ChIP-seq peak data, CIR information was acquired from Chromatin Interactions by 5 C and ChIA-PET, and TAD information was acquired from proceeded Hi-C data. Ensembl recorded genes on hg19 coordinate40 was used to analyze the genome locations of these three types of regulatory elements through the potential promoter region (from 2 k upstream to 1 k downstream of transcription). For E-G pairs of non-coding RNAs, miR2Disease, miRTarBase, and lncRNA2Target databases were integrated into rSNPBase for miRNA-gene and lncRNA-gene pairs with experimentally supported target relations through RT-PCR, microarray, or RNA-seq.41-43 Moreover, TargetScan and miRnada databases were used to obtain predicted miRNA target sites,34-36 which were mapped to human genome sequence (on hg 19 coordinate). The functional association between the included regulatory elements and target genes was analyzed with genomic proximity or by using widely used reference databases as mentioned above. 31,32,34-36–38-43
Integrating analysis of GWAS and rSNP annotation information
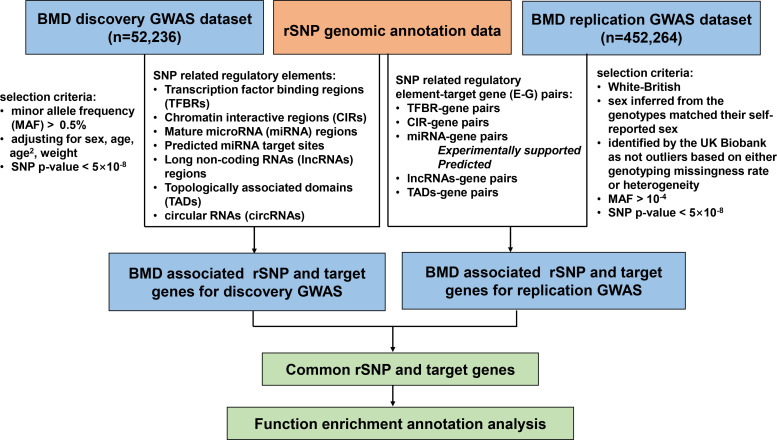
Firstly, significant SNPs were selected from the discovery GWAS dataset with p-value < 5 × 10-8, and then were annotated in rSNPBase 3.1 to obtain BMD-associated SNP regulatory elements and E-G pairs. Then, the other GWAS replicate summary dataset of BMD was used to validate the integration results with p-value < 5 × 10-8. Finally, we selected the common genes between the discovery and replication analysis results. A flowchart for this study is depicted in Figure 1.
Fig. 1.
Flowchart for integrating analysis of genome-wide association study (GWAS) and regulatory single nucleotide polymorphism (rSNP) annotation data for bone mineral density (BMD).
Functional enrichment and annotation analysis
For the BMD-associated genes overlapping in both discovery and replicate studies, GO enrichment analysis was conducted by HumanNet v2,44 a database of human gene networks.45 The network of HumanNet-XC (Functional gene network extended by Co-citation) and the network-based disease gene prediction of HumanNet were used in our study.
Statistical analysis
The significant SNPs with GWAS p-value < 5 × 10-8 were selected from both the discovery and replication GWAS summary dataset of BMD. To explore the functional relevance of identified common genes shared by both the discovery and replicate studies, GO and pathway enrichment analyses were performed by HumanNet-XC and the network-based disease gene prediction of HumanNet. The significance threshold was set as p < 0.05.
Results
BMD-associated rSNPs and their target genes
In discovery GWAS dataset study, for FA-BMD, we detected six rSNPs for TFBRs, 14 rSNPs for TADs, and 12 rSNPs for CIRs, corresponding to four, three, and six target regulatory genes, respectively. For circRNA region, 108 rSNPs were identified for FA-BMD, 16 of which have been demonstrated as eQTLs in a previous study by Guo et al29 (Supplementary Table i). For FN-BMD, we detected 20 rSNPs for TFBRs, 140 rSNPs for TADs, 53 rSNPs for CIRs, and 15 rSNPs for lncRNAs, corresponding to 19, 33, 49, and four target regulatory genes, respectively. For circRNA region, 135 rSNPs were identified for BMD, 100 of which have been demonstrated as eQTLs by Guo et al29 (Supplementary Table ii). For LS BMD, we detected 32 rSNPs for TFBRs, 268 rSNPs for TADs, 69 rSNPs for CIRs, and six rSNPs for lncRNAs, corresponding to 18, 31, 44, and four target regulatory genes, respectively. For circRNA region, 400 rSNPs were observed for BMD, 204 of which have been identified as eQTLs by Guo et al29 (Supplementary Table iii).
For the replication results, we detected 533 rSNPs for TFBRs, 4298 rSNPs for TADs, 1336 rSNPs for CIRs, one rSNP for miRNA, six rSNPs for predicted miRNA target site, and 365 rSNPs for lncRNAs, corresponding to 1,136, 2,645, 2,275, four, 12, and 380 target regulatory genes, respectively. For circRNA region, 6,446 rSNPs were identified for BMD, 90 of which have been demonstrated as eQTLs by Guo et al29 (Supplementary Table iv).
After comparing the discovery and replication study results, we identified four overlapped target regulatory genes for TFBRs, 30 overlapped target regulatory genes for TADs, and 14 overlapped target regulatory genes for CIRs, respectively (Table I). Irrespective of different regulatory genetic elements, 36 target regulatory genes were detected for BMD, such as FAM3C for TADs and CIRs (pdiscovery GWAS = 1.21 × 10-25, beta = 0.186, preplication GWAS = 1.80 × 10-12), CCDC170 for TADs (pdiscovery GWAS = 1.23 × 10-11, beta = -0.053, preplication GWAS = 3.22 × 10-9), SOX6 for TADs (pdiscovery GWAS = 4.41 × 10-15, beta = 0.080, preplication GWAS = 6.57 × 10-14), and PLEKHM1 for TADs (pdiscovery GWAS = 1.75 × 10-8, beta = 0.085, preplication GWAS = 1.56 × 10-15).
Table I.
List of common regulatory single nucleotide polymorphisms and their target regulatory genes shared by both discovery and replication studies.
| Gene | Discovery | Replication | Regulatory elements | ||||
|---|---|---|---|---|---|---|---|
| GWAS-rSNPs | EAF | Beta | pdiscovery | GWAS-rSNPs | preplication | ||
| FAM3C | rs7776725 | 0.259 | 0.186 | 1.21 × 10-25 | rs7776725 | 1.80 × 10-12 | CIRs TADs |
| PTPRZ1 | rs7776725 | 0.259 | 0.186 | 1.21 × 10-25 | rs7776725 | 1.80 × 10-12 | CIRs TADs |
| CCDC122 | rs9533094 | 0.404 | -0.083 | 2.80 × 10-20 | rs150081494 | 1.05 × 10-15 | TADs |
| DCLK1 | rs9533094 | 0.404 | -0.083 | 2.80 × 10-20 | rs191115702 | 1.52 × 10-16 | TADs |
| TPTE2P5 | rs9594738 | 0.403 | -0.082 | 3.63 × 10-20 | rs150081494 | 1.05 × 10-15 | TADs |
| ENSG00000253603 | rs7006553 | 0.385 | 0.078 | 9.99 × 10-18 | rs140103265 | 1.80 × 10-12 | CIRs |
| C11orf58 | rs16931831 | 0.195 | 0.080 | 4.41 × 10-15 | rs138547759 | 6.57 × 10-14 | TADs |
| SOX6 | rs16931831 | 0.195 | 0.080 | 4.41 × 10-15 | rs138547759 | 6.57 × 10-14 | TADs |
| ULK4 | rs369145 | 0.451 | -0.057 | 1.57 × 10-13 | rs117982632 | 2.26 × 10-9 | CIRs |
| rs10896328 | 0.238 | -0.081 | 2.09 × 10-13 | rs117982632 | 2.26 × 10-9 | TADs | |
| PPP6R3 | rs12272917 | 0.237 | -0.074 | 4.58 × 10-13 | rs17149179 | 3.14 × 10-10 | CIRs |
| rs12274114 | 0.255 | -0.072 | 1.17 × 10-12 | rs67446678 | 2.32 × 10-9 | TFBRs | |
| rs10896328 | 0.238 | -0.081 | 2.09 × 10-13 | rs72936524 | 5.15 × 10-10 | TADs | |
| CHKA | rs12272917 | 0.237 | -0.074 | 4.58 × 10-13 | rs2186937 | 6.70 × 10-10 | CIRs |
| CTTN | rs4084149 | 0.237 | -0.075 | 3.43 × 10-13 | rs374538301 | 1.16 × 10-11 | TADs |
| ELMO1 | rs1357651 | 0.355 | -0.068 | 3.75 × 10-13 | rs147344245 | 2.23 × 10-14 | TADs |
| AMPH | rs1357651 | 0.355 | -0.068 | 3.75 × 10-13 | rs147344245 | 2.23 × 10-14 | TADs |
| NDUFA3P2 | rs12272917 | 0.237 | -0.074 | 4.58 × 10-13 | rs2186937 | 6.70 × 10-10 | CIRs TADs |
| ENSG00000234985 | rs7776725 | 0.259 | 0.072 | 6.85 × 10-13 | rs7776725 | 1.80 × 10-12 | CIRs TADs |
| GEMIN6 | rs73215359 | 0.080 | 0.183 | 4.77 × 10-12 | rs114805039 | 5.80 × 10-11 | CIRs TFBRs |
| ADGB | rs7740522 | 0.505 | -0.053 | 1.23 × 10-11 | rs115947573 | 9.21 × 10-24 | TADs |
| SYNE1 | rs7740522 | 0.505 | -0.053 | 1.23 × 10-11 | rs115947573 | 9.21 × 10-24 | TADs |
| PLEKHG1 | rs7740522 | 0.505 | -0.053 | 1.23 × 10-11 | rs145135839 | 4.74 × 10-17 | TADs |
| CCDC170 | rs7740522 | 0.505 | -0.053 | 1.23 × 10-11 | rs182402542 | 3.22 × 10-9 | TADs |
| CBX5 | rs3803042 | 0.417 | -0.061 | 3.23 × 10-11 | rs57292110 | 7.11 × 10-16 | CIRs TFBRs |
| COG6 | rs9533110 | 0.496 | 0.055 | 9.83 × 10-10 | rs116613004 | 1.38 × 10-11 | TADs |
| EPSTI1 | rs9533110 | 0.496 | 0.055 | 9.83 × 10-10 | rs554647317 | 4.39 × 10-8 | TADs |
| ENSG00000254789 | rs12797380 | 0.477 | 0.047 | 2.05 × 10-9 | rs539468022 | 1.63 × 10-10 | TADs |
| PSMA1 | rs12797380 | 0.477 | 0.047 | 2.05 × 10-9 | rs539468022 | 1.63 × 10-10 | TADs |
| SUV420H1 | rs7935347 | 0.153 | -0.075 | 3.08 × 10-9 | rs183720308 | 1.03 × 10-10 | CIRs |
| RNU2-63P | rs4528686 | 0.362 | 0.071 | 6.58 × 10-9 | rs116151937 | 5.28 × 10-14 | CIRs TFBRs |
| EPDR1 | rs1376264 | 0.271 | 0.055 | 9.30 × 10-9 | rs147344245 | 2.23 × 10-14 | TADs |
| ANLN | rs1376264 | 0.271 | 0.055 | 9.30 × 10-9 | rs138651438 | 6.42 × 10-10 | TADs |
| OTOG | rs11024028 | 0.147 | 0.066 | 1.44 × 10-8 | rs372278011 | 1.02 × 10-14 | TADs |
| ENSG00000246225 | rs11024028 | 0.147 | 0.066 | 1.44 × 10-8 | rs77490678 | 3.00 × 10-16 | TADs |
| USAH1C | rs11024028 | 0.147 | 0.066 | 1.44 × 10-8 | rs138547759 | 6.57 × 10-14 | TADs |
| BDNF-AS | rs11024028 | 0.147 | 0.066 | 1.44 × 10-8 | rs187573374 | 3.33 × 10-17 | TADs |
| PLEKHM1 | rs34814687 | 0.053 | 0.085 | 1.75 × 10-8 | rs144518135 | 1.56 × 10-15 | TADs |
| CSAD | rs4759021 | 0.248 | 0.053 | 4.04 × 10-8 | rs556893559 | 8.42 × 10-12 | CIRs |
pdiscovery denotes the p-value of discovery GWAS-rSNPs; preplication denotes the p-value of replication GWAS-rSNPs.
CIRs, chromatin interactive regions; EAF, effect allele frequency; GWAS, genome-wide association study; rSNP, regulatory single nucleotide polymorphism; TADs, topologically associated domains; TFBRs, transcription factor binding regions.
Gene ontology and pathway enrichment analysis
For the 36 target genes shared by both discovery and replication study, GO enrichment analysis detected 12 GO terms with p < 0.01 for BMD, such as positive regulation of cartilage development (p = 9.27 × 10-3), positive regulation of chondrocyte differentiation (p = 9.27 × 10-3), muscle cell differentiation (p = 5.31 × 10-3), and regulation of p38 mitogen-activated protein kinase (p38MAPK) cascade (p = 5.31 × 10-3) (Table II).
Table II.
Gene ontology enrichment analysis results of the common genes identified by both discovery and replication studies.
| Term ID | Term description | p-value |
|---|---|---|
| GO:0007417 | central nervous system development | 2.86 × 10-3 |
| GO:2000741 | positive regulation of mesenchymal stem cell differentiation | 5.31 × 10-3 |
| GO:1900744 | regulation of p38MAPK cascade | 5.31 × 10-3 |
| GO:0042692 | muscle cell differentiation | 5.31 × 10-3 |
| GO:0090286 | cytoskeletal anchoring at nuclear membrane | 6.63 × 10-3 |
| GO:2000726 | negative regulation of cardiac muscle cell differentiation | 6.63 × 10-3 |
| GO:0007096 | regulation of exit from mitosis | 6.63 × 10-3 |
| GO:0045987 | positive regulation of smooth muscle contraction | 6.63 × 10-3 |
| GO:0043666 | regulation of phosphoprotein phosphatase activity | 7.95 × 10-3 |
| GO:0050957 | equilibrioception | 7.95 × 10-3 |
| GO:0061036 | positive regulation of cartilage development | 9.27 × 10-3 |
| GO:0032332 | positive regulation of chondrocyte differentiation | 9.27 × 10-3 |
GO, gene ontology; MAPK, mitogen-activated protein kinase.
Discussion
Recent studies have demonstrated the important roles of regulatory genetic variants in the genetic mechanism of human complex diseases.21,22 To explore the functional relevance of regulatory genetic variants with BMD and identified novel candidate genes for OP, we conducted an integrative analysis of GWAS with rSNP annotation information. We identified multiple rSNPs and their target genes for BMD. Further functional analysis of the identified target genes supports the important roles of rSNPs in the development of OP.
One important finding of this study is FAM3C, which was detected between discovery and replication studies. FAM3C, a member of the family with sequence similarity 3 (FAM3), encodes a secreted protein with a GG domain. In a three-stage genome-wide association (GWA) meta-analysis study, FAM3C was confirmed to be associated with BMD.46 Additionally, a follow-up study in Caucasians observed that rs7776725 influenced BMD at multiple skeletal sites.47 Interestingly, the variant rs7776725 was located in the first intron of the FAM3C gene. Notably, several other SNPs identified in this gene were also associated with BMD at multiple sites,47 indicating that FAM3C may play a notable role in bone metabolism. Furthermore, the SNP rs7776725 in FAM3C was also proved to be associated with a genome-wide substantially increased risk of forearm fracture.48FAM3C was demonstrated to play a functional role in the regulation of osteoblast differentiation. In differentiating osteoblasts, knockdown of FAM3C increased alkaline phosphatase expression and activity whereas overexpression of FAM3C reduced it. Furthermore, FAM3C and TGF-β1 were found to regulate each other reciprocally.49 Furthermore, our analysis found that rs7776725 targeted FAM3C and PTPRZ1 through regulating CIRs and TADs, which belong to transcriptional regulation. Previous studies have shown that FAM3C indeed has an effect on bone metabolism-related disease through transcriptional regulation, which was consistent with our analysis.46-49
SOX6 is another important gene, which encodes transcription factor SOX-6. It has been reported that the SOX6 gene is an essential transcription factor in chondrogenesis and cartilage formation.50,51 For example, SOX6 was discovered to have differential expression during osteoblast development.52 Homozygous Sox6 mutant (Sox6-/-) mice were born with relatively mild skeletal anomalies and mesenchymal condensations, but there was no overt chondrocyte differentiation.50 Moreover, expression of chondrocyte marker genes was severely reduced in Sox6 mutant mice. Sox5, Sox6, and Sox9 are necessary for chondrogenic differentiation at different steps, the combination of which provides a new in vitro chondrogenic differentiation model.51 In addition, several studies have identified that hip BMD was associated with rs7117858, which is located downstream of the SOX6 gene.52-54 Furthermore, multiple SNPs located in the SOX6 gene were identified to be associated with hip BMD in both Caucasian and Chinese populations,55 which further highlights the importance of the role of SOX6 in influencing BMD variation. Besides, Sox9/Sox6 and Sp1 are involved in the insulin-like growth factor-I-mediated upregulation of human type II collagen gene expression in articular chondrocytes,56 which is consistent with our findings that rs16931831 and rs138547759, as regulatory SNPs, target SOX6 through TADs.
Additionally, we detected several novel candidate genes for BMD, such as PLEKHM1, PTPRZ1, CCDC122, and DCLK1. PLEKHM1 encodes pleckstrin homology domain-containing family M member 1, which is suggested to be involved in vesicular transport in osteoclasts.57PLEKHM1 gene mutation results in osteopetrosis both in humans and rats, which is characterized by diminished bone resorption by osteoclasts. Mechanistically, loss of PLEKHM1 abrogates the peripheral distribution of lysosomes and bone resorption in osteoclasts through regulating lysosome positioning and secretion through RAB7.58 Rptpzeta, which is encoded by PTPRZ1, has been proved to play a physiological role in bone remodelling, and was thus identified as the first protein tyrosine phosphatase (PTP) regulating bone formation in vivo.59CCDC122 was identified as a susceptibility gene for leprosy in 3,614 individuals, involving two family-based and three independent case-control samples.60 Moreover, a study for exploring the relationship of testicular atrophy to bone metabolism in 31 male leprosy patients and 31 healthy control men was conducted and identified that BMD of the forearm significantly correlated with free testosterone levels (r = 0.689, p < 0.001), which indicates that low BMD may be due to testicular atrophy in leprosy patients.61 In addition, DCLK1 is upregulated in osteoblast-prone clones compared with non-differentiating clones through transcriptome sequencing and confirmed by real-time quantitative PCR.62 rs7776725 targeting PTPRZ1, rs34814687 and rs144518135 targeting PLEKHM1, rs9533094 targeting CCDC122 and DCLK1, rs150081494 targeting CCDC122, and rs191115702 targeting DCLK1 are all regulated through TADs. Further biological studies are warranted to explore the potential roles of PLEKHM1, PTPRZ1, CCDC122, and DCLK1 in the development of BMD.
We identified 12 GO terms enriched in the overlapped candidate genes identified by discovery and replication studies, such as positive regulation of mesenchymal stem cell (MSC) differentiation GO term (GO:2000741) and positive regulation of cartilage development (GO:0061036). Bone-forming osteoblasts are derived from MSCs. In a study of transcriptional profiling of human femoral MSCs in OP, some genetic changes in MSCs were found to be involved in the pathophysiology of OP.63 Research of mouse pathological models and patients with postmenopausal osteoporosis (PMOP) indicated that MEG3 regulates the expression of miR-133a-3p, and inhibits the osteogenic differentiation of bone marrow mesenchymal stem cell (BMSC)-induced PMOP.64 It has also been reported that cell-based arthroplasty therapy via the use of MSCs may become one of the strategies for OP treatment.65
Another interesting GO term is positive regulation of cartilage development. There is a growing amount of research focus on the relationship between systemic BMD and cartilage properties. For example, Zhu et al66 have demonstrated that low BMD, particularly at the hip, was positively associated with knee cartilage defects. In addition, it has been reported that longitudinal BMD loss is related to progressive cartilage loss in knees with osteoarthritis.67 Furthermore, a cross-sectional study demonstrated that systemic BMD is positively associated with knee cartilage volume in healthy, asymptomatic adult females.68
To the best of our knowledge, this study represents one of the larger studies to explore the roles of regulatory genetic variants in the genetic mechanism of BMD. The main advantage of this study is that rSNPBase 3.1 extends the scope of SNP-related annotations, the regulatory elements to SNP-related regulatory element target gene pairs, therefore it supports SNP-based gene regulatory network analysis. In addition, we further used the replication analysis to validate the discovery results, enhancing the reliability and persuasiveness of our study. However, there are several limitations in our study. First, although data used in our study were from a large sample and we performed a replication analysis, further studies need to be conducted using larger sample sizes, different genetic populations, and different gene variations. Additionally, the biological effect of target genes in different tissue or cell types was not considered in this functional annotation information of rSNP. Our rSNP analysis could not take tissue or cell types into account; therefore, the roles of different tissue and cell types should be warranted in further studies. Moreover, in this analysis, integrative analysis and functional analysis were performed, however some other regulatory elements are not included in the rSNPBase 3.1 database, for instance m6A.69 Finally, this study focused on rSNP, which occupies a small part of the whole genome.
In conclusion, we identified SNP-related regulatory elements and regulatory element-target gene pairs associated with BMD by integrating rSNPBase 3.1 with both discovery and replication GWAS summary datasets of BMD. We anticipate that the findings of this investigation will bring new insights into the aetiology and treatment of OP. Further research is required to substantiate and elucidate the underlying mechanisms of the identified genes implicated in the development of OP.
Author contributions
Y. Jia: Data curation, Methodology, Writing – original draft.
X. Qi: Data curation, Methodology.
M. Ma: Data curation, Writing – original draft.
S. Cheng: Methodology.
B. Cheng: Methodology.
C. Liang: Methodology.
X. Guo: Writing – review & editing.
F. Zhang: Funding acquisition, Writing – review & editing.
Funding statement
The authors disclose receipt of the following financial or material support for the research, authorship, and/or publication of this article: funding from the National Natural Scientific Foundation of China (82103959, 81673112), the Fundamental Research Funds for the Central Universities (xzy012020082), as well as the Key projects of international cooperation among governments in scientific and technological innovation (2016YFE0119100).
ICMJE COI statement
The authors declare no conflict of interest.
Acknowledgements
We thank the GEnetic Factors for OSteoporosis (GEFOS) Consortium and UK Biobank team for their work in collecting, processing, and disseminating these data for analysis.
Open access funding
The authors report that they received open access funding for their manuscript from the National Natural Scientific Foundation of China.
Supplementary material
Tables showing lists of the top 50 significant single nucleotide polymorphisms (SNPs) identified by integrating forearm, femoral neck, and lumbar spine bone mineral density from discovery genome-wide association study and regulatory SNP annotation data, as well as a list of the top 50 significant single nucleotide polymorphisms from replication genome-wide association study dataset (analyzed by rSNPBase analysis).
© 2023 Author(s) et al. This is an open-access article distributed under the terms of the Creative Commons Attribution Non-Commercial No Derivatives (CC BY-NC-ND 4.0) licence, which permits the copying and redistribution of the work only, and provided the original author and source are credited. See https://creativecommons.org/licenses/by-nc-nd/4.0/
Contributor Information
Yumeng Jia, Email: jiayumemg@mail.xjtu.edu.cn.
Xin Qi, Email: xinqi0@stu.xjtu.edu.cn.
Mei Ma, Email: captainooo@stu.xjtu.edu.cn.
Shiqiang Cheng, Email: chengsq0701@stu.xjtu.edu.cn.
Bolun Cheng, Email: cblbs1@stu.xjtu.edu.cn.
Chujun Liang, Email: liangchujun2018@stu.xjtu.edu.cn.
Xiong Guo, Email: guox@mail.xjtu.edu.cn.
Feng Zhang, Email: fzhxjtu@mail.xjtu.edu.cn.
References
- 1. Ammann P, Rizzoli R. Bone strength and its determinants. Osteoporos Int. 2003;14 Suppl 3:S13–8. doi: 10.1007/s00198-002-1345-4. [DOI] [PubMed] [Google Scholar]
- 2. Lane NE. Epidemiology, etiology, and diagnosis of osteoporosis. Am J Obstet Gynecol. 2006;194(2 Suppl):S3–11. doi: 10.1016/j.ajog.2005.08.047. [DOI] [PubMed] [Google Scholar]
- 3. Li S, Mao Y, Zhou F, Yang H, Shi Q, Meng B. Gut microbiome and osteoporosis: a review. Bone Joint Res. 2020;9(8):524–530. doi: 10.1302/2046-3758.98.BJR-2020-0089.R1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Li J, Ho WTP, Liu C, et al. The role of gut microbiota in bone homeostasis. Bone Joint Res. 2021;10(1):51–59. doi: 10.1302/2046-3758.101.BJR-2020-0273.R1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Qaseem A, Forciea MA, McLean RM, et al. Treatment of low bone density or osteoporosis to prevent fractures in men and women: A clinical practice guideline update from the American College of Physicians. Ann Intern Med. 2017;166(11):818–839. doi: 10.7326/M15-1361. [DOI] [PubMed] [Google Scholar]
- 6. Ji M-X, Yu Q. Primary osteoporosis in postmenopausal women. Chronic Dis Transl Med. 2015;1(1):9–13. doi: 10.1016/j.cdtm.2015.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Piroska M, Tarnoki DL, Szabo H, et al. Strong genetic effects on bone mineral density in multiple locations with two different techniques: Results from a cross-sectional twin study. Medicina (Kaunas) 2021;57(3):57. doi: 10.3390/medicina57030248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Zheng H-F, Forgetta V, Hsu Y-H, et al. Whole-genome sequencing identifies EN1 as a determinant of bone density and fracture. Nature. 2015;526(7571):112–117. doi: 10.1038/nature14878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Mo XB, Lu X, Zhang YH, Zhang ZL, Deng FY, Lei SF. Gene-based association analysis identified novel genes associated with bone mineral density. PLoS One. 2015;10(3):e0121811. doi: 10.1371/journal.pone.0121811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Urano T, Inoue S. Genetics of osteoporosis. Biochem Biophys Res Commun. 2014;452(2):287–293. doi: 10.1016/j.bbrc.2014.07.141. [DOI] [PubMed] [Google Scholar]
- 11. Sabik OL, Farber CR. Using GWAS to identify novel therapeutic targets for osteoporosis. Transl Res. 2017;181:15–26. doi: 10.1016/j.trsl.2016.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Boudin E, Fijalkowski I, Hendrickx G, Van Hul W. Genetic control of bone mass. Mol Cell Endocrinol. 2016;432:3–13. doi: 10.1016/j.mce.2015.12.021. [DOI] [PubMed] [Google Scholar]
- 13. Debette S. How to interpret a genome-wide association study (GWAS)? Sang Thrombose Vaisseaux. 2012;24(5):240–247. [Google Scholar]
- 14. Hindorff LA, Sethupathy P, Junkins HA, et al. Potential etiologic and functional implications of genome-wide association loci for human diseases and traits. Proc Natl Acad Sci U S A. 2009;106(23):9362–9367. doi: 10.1073/pnas.0903103106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Tak YG, Farnham PJ. Making sense of GWAS: using epigenomics and genome engineering to understand the functional relevance of SNPs in non-coding regions of the human genome. Epigenetics Chromatin. 2015;8:57. doi: 10.1186/s13072-015-0050-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Grubert F, Zaugg JB, Kasowski M, et al. Genetic control of chromatin states in humans involves local and distal chromosomal interactions. Cell. 2015;162(5):1051–1065. doi: 10.1016/j.cell.2015.07.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Soldner F, Stelzer Y, Shivalila CS, et al. Parkinson-associated risk variant in distal enhancer of α-synuclein modulates target gene expression. Nature. 2016;533(7601):95–99. doi: 10.1038/nature17939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Molineris I, Schiavone D, Rosa F, Matullo G, Poli V, Provero P. Identification of functional cis-regulatory polymorphisms in the human genome. Hum Mutat. 2013;34(5):735–742. doi: 10.1002/humu.22299. [DOI] [PubMed] [Google Scholar]
- 19. Riva A. Large-scale computational identification of regulatory SNPs with rSNP-MAPPER. BMC Genomics. 2012;13 Suppl 4(Suppl 4):S7. doi: 10.1186/1471-2164-13-S4-S7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Macintyre G, Bailey J, Haviv I, Kowalczyk A. is-rSNP: a novel technique for in silico regulatory SNP detection. Bioinformatics. 2010;26(18):i524–30. doi: 10.1093/bioinformatics/btq378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Gu R, Xu J, Lin Y, et al. The role of histone modification and a regulatory single-nucleotide polymorphism (rs2071166) in the Cx43 promoter in patients with TOF. Sci Rep. 2017;7(1):10435. doi: 10.1038/s41598-017-10756-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Yeo J, Morales DA, Chen T, et al. RNAseq analysis of bronchial epithelial cells to identify COPD-associated genes and SNPs. BMC Pulm Med. 2018;18(1):42. doi: 10.1186/s12890-018-0603-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.No authors listed GEnetic Factors for OSteoporosis Consortium (GEFOS): Home. gefos.org. 2023. [18 January 2023]. http://www.gefos.org/ date last. accessed.
- 24. Canela-Xandri O, Rawlik K, Tenesa A. An atlas of genetic associations in UK Biobank. Nat Genet. 2018;50(11):1593–1599. doi: 10.1038/s41588-018-0248-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Bycroft C, Freeman C, Petkova D, et al. The UK Biobank resource with deep phenotyping and genomic data. Nature. 2018;562(7726):203–209. doi: 10.1038/s41586-018-0579-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.No authors listed GeneATLAS. The University of Edinburgh. 2023. [18 January 2023]. http://geneatlas.roslin.ed.ac.uk/ date last. accessed.
- 27. Wain LV, Shrine N, Miller S, et al. Novel insights into the genetics of smoking behaviour, lung function, and chronic obstructive pulmonary disease (UK BiLEVE): a genetic association study in UK Biobank. Lancet Respir Med. 2015;3(10):769–781. doi: 10.1016/S2213-2600(15)00283-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.No authors listed Software - jmarchini. jmarchini. 2023. [18 January 2023]. https://jmarchini.org/software/ date last. accessed.
- 29. Guo L, Wang J. rSNPBase 3.0: an updated database of SNP-related regulatory elements, element-gene pairs and SNP-based gene regulatory networks. Nucleic Acids Res. 2018;46(D1):D1111–D1116. doi: 10.1093/nar/gkx1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.No authors listed rSNPBase 3.1. Bioinformatics Lab, Institute of Psychology, Chinese Academy of Sciences. 2017. [18 January 2023]. http://rsnp3.psych.ac.cn/index.do date last. accessed.
- 31. Sloan CA, Chan ET, Davidson JM, et al. ENCODE data at the ENCODE portal. Nucleic Acids Res. 2016;44(D1):D726–32. doi: 10.1093/nar/gkv1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Griffiths-Jones S. miRBase: the microRNA sequence database. Methods Mol Biol. 2006;342:129–138. doi: 10.1385/1-59745-123-1:129. [DOI] [PubMed] [Google Scholar]
- 33.No authors listed TargetScan. Whitehead Institute for Biomedical Research. 2023. [18 January 2023]. https://www.targetscan.org/vert_80/ date last. accessed.
- 34. Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120(1):15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 35. Friedman RC, Farh K-H, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009;19(1):92–105. doi: 10.1101/gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Betel D, Wilson M, Gabow A, Marks DS, Sander C. The microRNA.org resource: targets and expression. Nucleic Acids Res. 2008;36(Database issue):D149–D153. doi: 10.1093/nar/gkm995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Bai X, Huang Y, Zhang K, et al. CircNf1-mediated CXCL12 expression in the spinal cord contributes to morphine analgesic tolerance. Brain Behav Immun. 2023;107:140–151. doi: 10.1016/j.bbi.2022.09.018. [DOI] [PubMed] [Google Scholar]
- 38. Volders PJ, Verheggen K, Menschaert G, et al. An update on LNCipedia: a database for annotated human lncRNA sequences. Nucleic Acids Res. 2015;43(8):4363–4364. doi: 10.1093/nar/gkv295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Liu Y-C, Li J-R, Sun C-H, et al. CircNet: a database of circular RNAs derived from transcriptome sequencing data. Nucleic Acids Res. 2016;44(D1):D209–15. doi: 10.1093/nar/gkv940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Aken BL, Ayling S, Barrell D, et al. The Ensembl gene annotation system. Database (Oxford) 2016;2016:baw093. doi: 10.1093/database/baw093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Jiang Q, Wang Y, Hao Y, et al. miR2Disease: a manually curated database for microRNA deregulation in human disease. Nucleic Acids Res. 2009;37(Database issue):D98–104. doi: 10.1093/nar/gkn714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Chou C-H, Chang N-W, Shrestha S, et al. miRTarBase 2016: updates to the experimentally validated miRNA-target interactions database. Nucleic Acids Res. 2016;44(D1):D239–47. doi: 10.1093/nar/gkv1258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Jiang Q, Wang J, Wu X, et al. LncRNA2Target: a database for differentially expressed genes after lncRNA knockdown or overexpression. Nucleic Acids Res. 2015;43(Database issue):D193–D196. doi: 10.1093/nar/gku1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.No authors listed HumanNet Search. inetbio.org. 2023. [18 January 2023]. http://www.inetbio.org/humannet/ date last. accessed.
- 45. Hwang S, Kim CY, Yang S, et al. HumanNet v2: human gene networks for disease research. Nucleic Acids Res. 2019;47(D1):D573–D580. doi: 10.1093/nar/gky1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Zhang L, Choi HJ, Estrada K, et al. Multistage genome-wide association meta-analyses identified two new loci for bone mineral density. Hum Mol Genet. 2014;23(7):1923–1933. doi: 10.1093/hmg/ddt575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Zhang L-S, Hu H-G, Liu Y-J, et al. A follow-up association study of two genetic variants for bone mineral density variation in Caucasians. Osteoporos Int. 2012;23(7):1867–1875. doi: 10.1007/s00198-011-1863-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Zheng H-F, Tobias JH, Duncan E, et al. WNT16 influences bone mineral density, cortical bone thickness, bone strength, and osteoporotic fracture risk. PLOS Genet. 2012;8(7):e1002745. doi: 10.1371/journal.pgen.1002745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Bendre A, Büki KG, Määttä JA. Fam3c modulates osteogenic differentiation by down-regulating Runx2. Differentiation. 2017;93:50–57. doi: 10.1016/j.diff.2016.11.005. [DOI] [PubMed] [Google Scholar]
- 50. Dy P, Smits P, Silvester A, et al. Synovial joint morphogenesis requires the chondrogenic action of Sox5 and Sox6 in growth plate and articular cartilage. Dev Biol. 2010;341(2):346–359. doi: 10.1016/j.ydbio.2010.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Ikeda T, Kawaguchi H, Kamekura S, et al. Distinct roles of Sox5, Sox6, and Sox9 in different stages of chondrogenic differentiation. J Bone Miner Metab. 2005;23(5):337–340. doi: 10.1007/s00774-005-0610-y. [DOI] [PubMed] [Google Scholar]
- 52. Hsu Y-H, Zillikens MC, Wilson SG, et al. An integration of genome-wide association study and gene expression profiling to prioritize the discovery of novel susceptibility Loci for osteoporosis-related traits. PLoS Genet. 2010;6(6):e1000977. doi: 10.1371/journal.pgen.1000977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Rivadeneira F, Styrkársdottir U, Estrada K, et al. Twenty bone-mineral-density loci identified by large-scale meta-analysis of genome-wide association studies. Nat Genet. 2009;41(11):1199–1206. doi: 10.1038/ng.446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Styrkarsdottir U, Halldorsson BV, Gudbjartsson DF, et al. European bone mineral density loci are also associated with BMD in East-Asian populations. PLoS One. 2010;5(10):e13217. doi: 10.1371/journal.pone.0013217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Yang T-L, Guo Y, Liu Y-J, et al. Genetic variants in the SOX6 gene are associated with bone mineral density in both Caucasian and Chinese populations. Osteoporos Int. 2012;23(2):781–787. doi: 10.1007/s00198-011-1626-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Renard E, Porée B, Chadjichristos C, et al. Sox9/Sox6 and Sp1 are involved in the insulin-like growth factor-I-mediated upregulation of human type II collagen gene expression in articular chondrocytes. J Mol Med (Berl) 2012;90(6):649–666. doi: 10.1007/s00109-011-0842-3. [DOI] [PubMed] [Google Scholar]
- 57. Van Wesenbeeck L, Odgren PR, Coxon FP, et al. Involvement of PLEKHM1 in osteoclastic vesicular transport and osteopetrosis in incisors absent rats and humans. J Clin Invest. 2007;117(4):919–930. doi: 10.1172/JCI30328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Fujiwara T, Ye S, Castro-Gomes T, et al. PLEKHM1/DEF8/RAB7 complex regulates lysosome positioning and bone homeostasis. JCI Insight. 2016;1(17):e86330. doi: 10.1172/jci.insight.86330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Schinke T, Gebauer M, Schilling AF, et al. The protein tyrosine phosphatase Rptpzeta is expressed in differentiated osteoblasts and affects bone formation in mice. Bone. 2008;42(3):524–534. doi: 10.1016/j.bone.2007.11.009. [DOI] [PubMed] [Google Scholar]
- 60. Sales-Marques C, Salomão H, Fava VM, et al. NOD2 and CCDC122-LACC1 genes are associated with leprosy susceptibility in Brazilians. Hum Genet. 2014;133(12):1525–1532. doi: 10.1007/s00439-014-1502-9. [DOI] [PubMed] [Google Scholar]
- 61. Ishikawa S, Tanaka H, Mizushima M, Hashizume H, Ishida Y, Inoue H. Osteoporosis due to testicular atrophy in male leprosy patients. Acta Med Okayama. 1997;51(5):279–283. doi: 10.18926/AMO/30786. [DOI] [PubMed] [Google Scholar]
- 62. Kim Y-H, Cho K-A, Lee H-J, et al. Identification of WNT16 as a predictable biomarker for accelerated osteogenic differentiation of tonsil-derived mesenchymal stem cells in vitro. Stem Cells Int. 2019;2019:8503148. doi: 10.1155/2019/8503148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Choi YJ, Song I, Jin Y, et al. Transcriptional profiling of human femoral mesenchymal stem cells in osteoporosis and its association with adipogenesis. Gene. 2017;632:7–15. doi: 10.1016/j.gene.2017.08.015. [DOI] [PubMed] [Google Scholar]
- 64. Wang Q, Li Y, Zhang Y, et al. LncRNA MEG3 inhibited osteogenic differentiation of bone marrow mesenchymal stem cells from postmenopausal osteoporosis by targeting miR-133a-3p. Biomed Pharmacother. 2017;89:1178–1186. doi: 10.1016/j.biopha.2017.02.090. [DOI] [PubMed] [Google Scholar]
- 65. Phetfong J, Sanvoranart T, Nartprayut K, et al. Osteoporosis: the current status of mesenchymal stem cell-based therapy. Cell Mol Biol Lett. 2016;21:12. doi: 10.1186/s11658-016-0013-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Zhu Q, Xu J, Wang K, et al. Associations between systemic bone mineral density, knee cartilage defects and bone marrow lesions in patients with knee osteoarthritis. Int J Rheum Dis. 2018;21(6):1202–1210. doi: 10.1111/1756-185X.13148. [DOI] [PubMed] [Google Scholar]
- 67. Lee JY, Harvey WF, Price LL, Paulus JK, Dawson-Hughes B, McAlindon TE. Relationship of bone mineral density to progression of knee osteoarthritis. Arthritis Rheum. 2013;65(6):1541–1546. doi: 10.1002/art.37926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Brennan SL, Pasco JA, Cicuttini FM, et al. Bone mineral density is cross sectionally associated with cartilage volume in healthy, asymptomatic adult females: Geelong Osteoporosis Study. Bone. 2011;49(4):839–844. doi: 10.1016/j.bone.2011.06.015. [DOI] [PubMed] [Google Scholar]
- 69. Harper JE, Miceli SM, Roberts RJ, Manley JL. Sequence specificity of the human mRNA N6-adenosine methylase in vitro. Nucleic Acids Res. 1990;18(19):5735–5741. doi: 10.1093/nar/18.19.5735. [DOI] [PMC free article] [PubMed] [Google Scholar]