ABSTRACT
Over the past decade, CRISPR/Cas-based gene editing has become a powerful tool for generating mutations in a variety of model organisms, from Escherichia coli to zebrafish, rodents and large mammals. CRISPR/Cas-based gene editing effectively generates insertions or deletions (indels), which allow for rapid gene disruption. However, a large proportion of human genetic diseases are caused by single-base-pair substitutions, which result in more subtle alterations to protein function, and which require more complex and precise editing to recreate in model systems. Precise genome editing (PGE) methods, however, typically have efficiencies of less than a tenth of those that generate less-specific indels, and so there has been a great deal of effort to improve PGE efficiency. Such optimisations include optimal guide RNA and mutation-bearing donor DNA template design, modulation of DNA repair pathways that underpin how edits result from Cas-induced cuts, and the development of Cas9 fusion proteins that introduce edits via alternative mechanisms. In this Review, we provide an overview of the recent progress in optimising PGE methods and their potential for generating models of human genetic disease.
Keywords: CRISPR/Cas, HDR, Precise genome editing, Base/prime editing, Human disease modelling
Summary: CRISPR/Cas technology can effectively disrupt gene function by generating insertions and deletions, but precise gene editing remains challenging. Here, we discuss recent advances in this field.
Introduction
Gene editing via CRISPR/Cas technology has become a key tool for researchers in many areas, including plant and agricultural biology, human disease and synthetic biology. The relative ease of its implementation in many systems, availability of bioinformatic tools (Heigwer et al., 2014; Labun et al., 2019; Liu et al., 2015), commercially available reagents, flexibility and effectiveness all combine to make CRISPR/Cas an invaluable tool for modifying gene and protein function in vitro and in vivo. The Streptococcus pyogenes Cas9 (SpCas9) endonuclease has become a workhorse for generating genetic knockouts, as well as facilitating more precise edits (Anders et al., 2014; Jinek et al., 2012). Cas9-based gene-editing approaches exploit two key features of this endonuclease: its ability to recognise and bind to specific DNA sequences based on the complementarity of a guide RNA (gRNA) (Anders et al., 2014), and its ability to introduce double-stranded breaks (DSBs) in the target DNA strand (Jinek et al., 2012) (Fig. 1). Cas9 gRNAs are composed of two small non-coding RNAs: a uniform trans-activating CRISPR RNA (tracrRNA) that is recognised by the Cas9 and a CRISPR RNA (crRNA) that contains the locus-specific sequence. Together, these form a duplex called a single-guide RNA (sgRNA), which then forms a ribonucleoprotein (RNP) complex with Cas9 (Karvelis et al., 2013). Other Cas endonucleases, such as Cas12a, only require crRNAs (Zetsche et al., 2015; Zetsche et al., 2017). To cover all types of Cas endonucleases, we use the collective term ‘gRNA’ in this Review. gRNAs require the presence of a protospacer adjacent motif (PAM) (Anders et al., 2014), a specific sequence of nucleotides without which Cas will not cut. PAM site specificity, such as NGG for SpCas9, however, limits the loci that Cas can be targeted to.
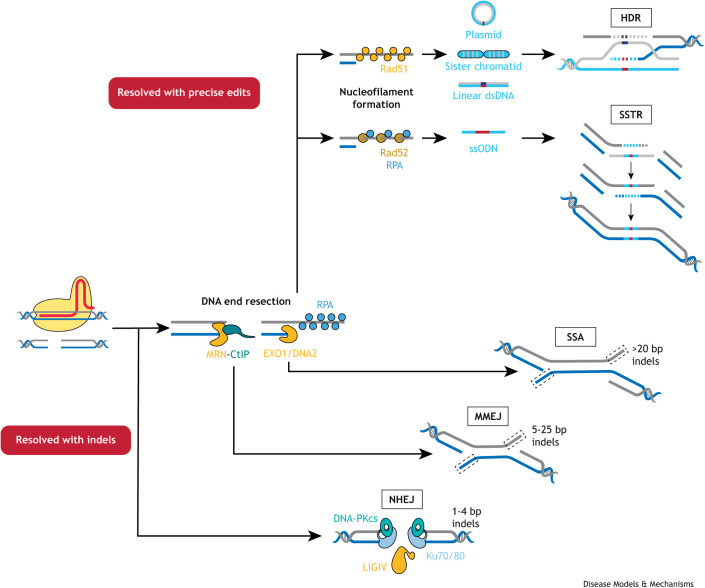
Fig. 1.
Summary of known double-stranded break (DSB) repair pathways. After initiation of a DSB, the initial repair pathway decision is between the binding of the dsDNA ends with Ku70/80 and subsequent error-prone NHEJ (bottom) or DNA end resection by the MRN complex, which is activated by CtIP (middle). Before extensive DNA end resection occurs, the micro-homologies between the exposed ssDNA ends can facilitate a second alternative error-prone repair mechanism, MMEJ. Alternatively, more extensive DNA end resection is carried out by EXO1 and DNA2, after which the exposed ssDNA is bound by RPA. This initiates one of two faithful repair mechanisms (top). When Rad51 is successfully loaded onto the ssDNA strand and a nucleofilament is therefore formed, the DSB can be resolved via HDR in the presence of a dsDNA donor. Alternatively, two other DSB resolution fates can occur: homologies within the exposed ssDNA may facilitate the third error-prone repair mechanism, SSA, or Rad52 may be loaded onto the ssDNA and can trigger the faithful repair mechanism, SSTR, if a ssODN template is present at the site of the DSB. Importantly, NHEJ, the main competitor of faithful repair mechanisms, can readily occur at the earliest stage in the DSB resolution process, with subsequent error-prone repair mechanisms still occurring either part way through and/or after extensive DNA end resection. By contrast, both known faithful repair mechanisms require extensive DNA end resection and can be subdivided into Rad51-dependent HDR and Rad51-independent SSTR. dsDNA, double-stranded DNA; HDR, homology-directed repair; indel, insertion or deletion; MMEJ, microhomology-mediated end joining; MRN, MRE11, Rad50 and NSB1; NHEJ, non-homologous end joining; SSA, single-strand annealing; ssODN, single-stranded oligodeoxynucleotide; SSTR, single-stranded template repair.
The ability of Cas endonucleases to generate DSBs triggers the recruitment of the endogenous DNA repair machinery to the break, and a specific genetic locus can be targeted via a gRNA. The most common DNA repair pathway, non-homologous end joining (NHEJ), results in the introduction of insertions or deletions (indels) due to the error-prone nature of this repair mechanism (Fig. 1). However, the initiation of DSBs also allows for the possibility of faithful homologous recombination (HR)-driven repair mechanisms, such as homology-directed repair (HDR) or single-stranded template repair (SSTR), which utilise a donor sequence to repair the DSB, allowing the engineered incorporation of specific edits into the target strand (Fig. 1). Although NHEJ is active throughout the cell cycle, HDR and SSTR are limited to the S and G2 phases, when sister chromatids would naturally be located close together to act as a repair template (Heyer et al., 2010).
Gene knockout or mutant disease models have been generated using numerous techniques, including chemical mutagenesis, such as with N-ethyl-N-nitrosourea (ENU) (de Angelis et al., 2000; Fossett et al., 1990; Hitotsumachi et al., 1985; Solnica-Krezel et al., 1994), engineered endonucleases such as transcription activator-like effector nucleases (TALENs) (Ke et al., 2016; Li et al., 2011), targeted genetic modification of mammalian embryonic stem cells to generate chimeric modified mice (Mak, 2007) or rescue of knockout models with humanised sequences containing disease-associated mutations (Switonski et al., 2015). However, these techniques can be laborious, chemical mutagenesis cannot be targeted to a specific region of the genome, and rescue with humanised mutation-bearing constructs is not always faithful to the expression level of the endogenous gene. More recently, the use of Cas endonucleases to generate indels has been optimised in many systems: efficiencies can exceed 90% in vivo in zebrafish (Kroll et al., 2021; Burger et al., 2016), with recent adaptions for high-throughput screening (Parvez et al., 2021), 85% in mammalian cell culture (Seki and Rutz, 2018), 66% in Arabidopsis thaliana (Tsutsui and Higashiyama, 2016) and 85.7% in rats (Bae et al., 2020). Indel formation, arising as a result of DSB repair via NHEJ and leading to loss-of-function gene disruption, can be used to study gene and protein function and can model many diseases (Hai et al., 2014; Kang et al., 2015; Yuan et al., 2016; Wang et al., 2019; Stenson et al., 2003). However, in many cases, diseases are caused by single-nucleotide substitutions rather than full gene disruption (Stenson et al., 2003), and the resulting subtle changes in function cannot always be fully replicated with knockout models (Hancox et al., 2019; De Gobbi et al., 2006; Ingram, 1957). Therefore, generation of targeted, specific mutations in endogenous genes is desirable.
Modelling such genetic alterations requires precise genome editing (PGE) methods. However, PGE methods that rely on HDR/SSTR to resolve Cas-induced DSBs are significantly less efficient than NHEJ-driven indel formation. PGE methods often achieve <4% efficiency (Cui et al., 2018; Prykhozhij et al., 2017 preprint; Wu et al., 2013) (Table 1), making the generation of precise human disease models, particularly in vivo (Box 1), laborious and expensive. Therefore, optimisations to PGE methods and efficiencies are urgently required. Increasing PGE efficiency is a multifaceted challenge that will likely require a combination of approaches to solve. These include efficient DNA cleavage, efficient delivery of PGE components into the target cell, the timing of editing, careful design and choice of donor DNA, the ability to effectively pass on mutations to the next generation in model organisms and the capability to subsequently screen for desired mutations. Importantly, these also require optimisation of strategies to manipulate the competitive DSB repair pathways that can either give rise to indels or PGE. Although robustly and reproducibly increasing PGE efficiency remains a challenge, there have been considerable recent advances, which we summarise in this Review. We discuss recent progress in multiple model systems and provide an overview of methods that can be employed to increase PGE efficiency.
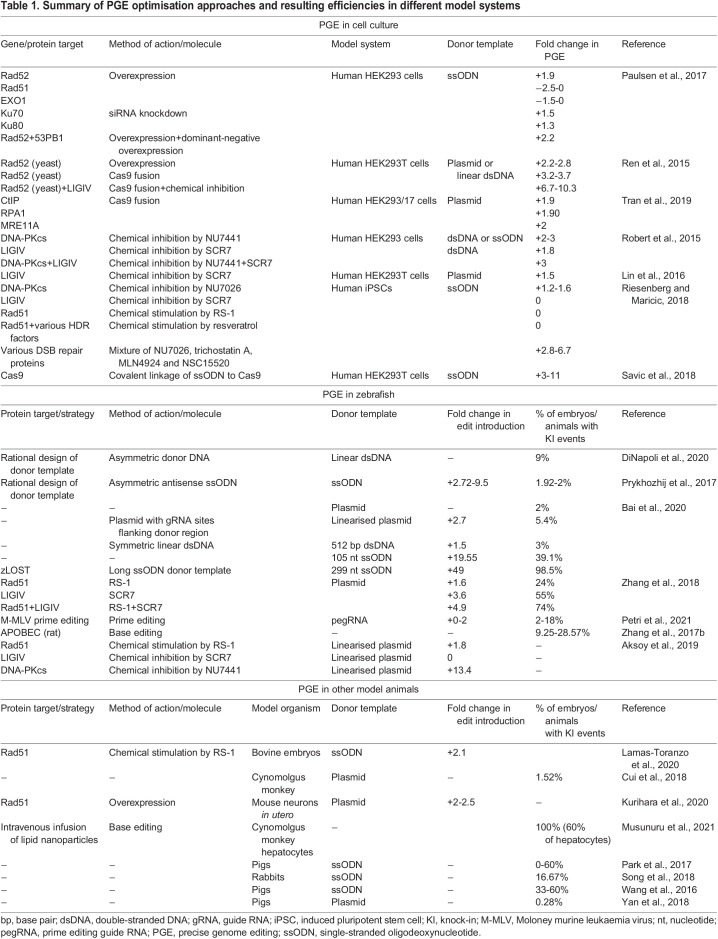
Table 1.
Summary of PGE optimisation approaches and resulting efficiencies in different model systems
Box 1. Use of precise genome editing (PGE) in vivo for the generation of disease models.
Many developments in CRISPR technology are first investigated and proven in vitro (Table 1) as these platforms allow researchers to elucidate how the genome-editing process and the resulting mutations affect protein and cell function. Translation of these methods to generate in vivo human disease models is a powerful use of this technology. However, adapting methods developed in vitro for use in vivo is challenging. In cell culture, thousands of cells can be transfected with PGE components simultaneously, whereas germ-line mutations in whole animals typically require skilled and time-consuming manual microinjection of PGE components into fertilised embryos (the F0 generation) (Cui et al., 2018; Lamas-Toranzo et al., 2020; Park et al., 2017; Zhang et al., 2018; Song et al., 2018; Tessadori et al., 2018; Aksoy et al., 2019). Furthermore, the end goal of in vivo modelling is typically a stable precisely edited strain, which also requires efficient transmission of the edits to offspring (the F1 generation).
Despite these challenges, PGE has been achieved in several model organisms, including mice (Miura et al., 2018; Wu et al., 2013), zebrafish (Hoshijima et al., 2016; Irion et al., 2014; Wierson et al., 2020; Zhang et al., 2018) rabbits (Song et al., 2016; Song et al., 2018), pigs (Song et al., 2016; Song et al., 2018) and non-human primates (Lamas-Toranzo et al., 2020; Wang et al., 2016; Yan et al., 2018). The efficiency of in vivo PGE methods, however, varies considerably between experiment, model organism and targeted loci (Table 1). For example, in vitro use of the DNA-PKcs inhibitor NU7441, which blocks non-homologous end joining and thus promotes PGE repair pathways, increased PGE efficiency to between 6.2% and 15% in HEK293 cells and induced pluripotent stem cells (Cui et al., 2018; Kang et al., 2019). When used in vivo, in zebrafish, NU7441 addition resulted in 50% somatic PGE and three of six fish produced chimeric-edited F1 progeny (Aksoy et al., 2019). Conversely, using single-stranded oligodeoxynucleotides (ssODNs) and foregoing chemical modulation for PGE in pigs resulted in between 0% and 60% of viable F0 piglets being mosaic for the desired edits and this varied between litters/experiments (Park et al., 2017). These differences in PGE efficiencies make it difficult to compare methods between studies and underscore the broad variability between organisms and protocols.
Although efficient in vivo gene editing remains challenging, human diseases caused by single-nucleotide substitutions have been successfully modelled in vivo using PGE methods. Base editing has been employed in zebrafish to generate cancer models by precise mutation of tp53 and nras (Rosello et al., 2021). ssODN donor templates have been used to generate gene-edited mice that carry single-nucleotide polymorphisms implicated in human thrombosis and platelet function, which were identified from genome-wide association studies (Zhu et al., 2017). A linearised plasmid has also been used to generate knock-in pig models of Huntington's disease that exhibited germ-line transmission of the edited alleles to F1 and F2 generations (Yan et al., 2018). Such examples show the potential of Cas-based PGE for producing animal models for human genetic diseases, which can be used to better understand the function of disease-associated mutations and develop more personalised treatments.
The use of the CRISPR/Cas system for precise genome editing
The induction of DNA repair mechanisms upon Cas-induced breaks creates the opportunity to supply exogenous donor DNA (dDNA) that contains the required change of sequence. dDNA templates for HDR or SSTR are composed of homology arms flanking the desired edits. These edits can be single-base-pair changes, specific deletions or insertions ranging from a few base pairs to complete loxP sites (Yang et al., 2013), or molecular tags or fluorescent reporters (Wierson et al., 2020; Aksoy et al., 2019; Kurihara et al., 2020). When generating such precise edits, scarless integration is desired at a specific locus, driven by the specificity of the gRNA, with the flanking recipient genomic DNA sequence remaining unaltered. Such an event, regardless of method, is encapsulated by the term PGE. Different methods exist within this umbrella term, separated by their mode of action: HDR and SSTR are separate DNA repair mechanisms that can be harnessed using dDNA (Fig. 1), whereas the more recently described base editing (BE) and prime editing (PE) employ different mechanisms to alter the target DNA sequence, which we discuss in more detail below.
Researchers have used various strategies to tackle PGE optimisation and each will be discussed in this Review. Some studies have focused on altering which DSB repair pathway is favoured, either through chemical modulation (Aksoy et al., 2019; Bischoff et al., 2020; Zhang et al., 2018) or through overexpression or silencing of key genes in the HDR/SSTR or NHEJ pathways (Kurihara et al., 2020; Li et al., 2018). The search for alternative Cas endonucleases (Makarova et al., 2011) has expanded the genome-editing toolbox to include Cas12a (Cpf1) and variants of Cas9, such as Staphylococcus aureus Cas9 (SaCas9) (Kleinstiver et al., 2015a; Ran et al., 2015) and Neisseria meningitidis Cas9 (NmCas9) (Hou et al., 2013), which have differing target preferences (Chen et al., 2018; Swarts and Jinek, 2018; Zetsche et al., 2015; Nakade et al., 2017) and thus allow the targeting of new loci. Alteration (Walton et al., 2020) of the Cas9 PAM preference (Doudna and Charpentier, 2014; Mojica et al., 2009) has likewise increased the spectrum of potential genetic targets, while identification of high-fidelity Cas9 variants has increased specificity (Walton et al., 2020; Kleinstiver et al., 2016, 2015b).
The ability of Cas to act as a genetic homing mechanism has also been exploited independently of its endonuclease activity. Cas9 variants that lack the ability to introduce DSBs are termed dead Cas9 (dCas9) if they lack any endonuclease activity or Cas9 nickase (nCas9) if they only cut one DNA strand (Cong et al., 2013). dCas9 and nCas9 have been used to reduce observed off-target effects (Ran et al., 2013; Mali et al., 2013) and as a chassis onto which to fuse other enzymes, such as deaminases (Gaudelli et al., 2017; Kim et al., 2017b; Komor et al., 2016, 2017) and reverse transcriptases (Anzalone et al., 2019; Liang et al., 2022; Lin et al., 2020; Petri et al., 2021), which can induce PGE via non-HR methods, such as BE and PE. Cas9 fusions have also been used to increase PGE efficiency by assisting with the colocalisation of the dDNA or of key PGE effector proteins to the DSB site (Charpentier et al., 2018; Savic et al., 2018) (Table 1).
Different dDNA conformations have also been investigated. Researchers have compared PGE efficiencies of circular plasmid DNA, double-stranded DNA (dsDNA), single-stranded oligodeoxynucleotides (ssODNs) and long single-stranded oligodeoxynucleotides (lssODNs) (Bai et al., 2020; Miura et al., 2018; Ranawakage et al., 2021; Song and Stieger, 2017), dDNA length, strand complementarity and symmetry. These parameters have all been reported to affect HR efficiency (Okamoto et al., 2019; Richardson et al., 2016) (Table 1). In addition, researchers have developed a number of bioinformatics tools to assist with the design of dDNA and gRNAs for optimal HDR efficiencies (O'Brien et al., 2019; Prykhozhij et al., 2021).
Manipulation of key DSB repair proteins to alter repair pathway choice
Processes involved in DNA repair are a necessary bedrock on which to build an understanding of PGE optimisation. In-depth descriptions of DSB repair pathways have been provided elsewhere (Shrivastav et al., 2008; Ceccaldi et al., 2016; Hustedt and Durocher, 2017) and will not be covered in detail here. Briefly, numerous proteins are involved in the repair of DSBs (outlined in Fig. 1). Six core components of NHEJ have been identified: Ku70/80, DNA-PKcs (also known as PRKDC), Artemis, LIGIV and XRCC4. Ku70 and Ku80 form a heterodimer that binds to DNA ends and recruits DNA-PKcs. DNA-PKcs is a kinase that phosphorylates Artemis, an endonuclease that trims the DSB ends and prepares them for ligation by LIGIV. Finally, XRCC4 acts as a scaffolding protein (Andres et al., 2012) and forms a complex with LIGIV (Drouet et al., 2005) to assist with its nuclear import (Berg et al., 2011). The suppression of LIGIV activity in zebrafish (Zhang et al., 2018) and silencing of Ku70/80 in pig foetal fibroblasts (Li et al., 2018) or XRCC4 in plants (Populus trichocarpa) (Movahedi et al., 2022) all increase PGE events, indicating that downregulation of NHEJ components is a promising avenue for increasing CRISPR-mediated PGE efficiency (Fig. 2).
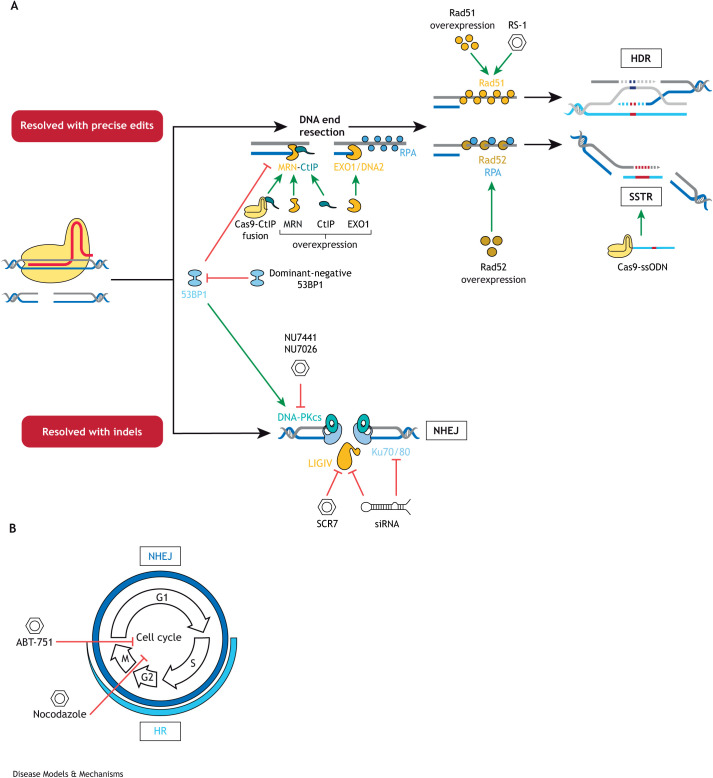
Fig. 2.
Summary of strategies to modulate DSB repair to increase precise genome editing (PGE) efficiency. (A) The upper track represents the desired route towards PGE, subdivided into HDR and SSTR. The lower route represents error-prone NHEJ. Several components of each pathway have been targeted for genetic or chemical inhibition and for upregulation, for example, via overexpression. These interventions skew endogenous DNA repair towards faithful mechanisms that allow PGE. (B) Modulating the cell cycle by arresting the cells in the G2/M phases can also increase PGE, as HDR and SSTR occur exclusively in those phases. HDR, homology-directed repair; HR, homologous recombination; MRN, MRE11, Rad50 and NSB1; NHEJ, non-homologous end joining; siRNA, small interfering RNA; ssODN, single-stranded oligodeoxynucleotide; SSTR, single-stranded template repair.
An alternative approach to increasing PGE is to upregulate components of faithful repair mechanisms. As described in Fig. 1, DNA end resection is the first step towards HDR or SSTR. As such, the end resection-initiating MRN complex, formed of MRE11, Rad50 and NSB1 and activated by CtIP (also known as RBBP8), is a potential target for upregulation (Figs 1 and 2). Indeed, overexpression of CtIP and MRE11 (Movahedi et al., 2022) and fusion of a CtIP domain to Cas9 (Charpentier et al., 2018) increase PGE events. Additionally, the nucleases EXO1 and DNA2 catalyse the more extensive DNA end resection that faithful repair mechanisms require, and overexpression of an EXO1 mimic increases SSTR in mammalian cell culture (Seigel, 2018) (Fig. 2).
Once extensive DNA end resection has occurred, the resulting exposed single-stranded DNA (ssDNA) is bound by RPA (Wobbe et al., 1987; Wold, 1997), and the repair pathways available to the cell then include HDR, SSTR and single-strand annealing (SSA) (Fig. 1). The recombinases Rad51 and Rad52 appear to be key mediators of DSB resolution at this stage, but the type of template available for repair is also critical. Rad51 facilitates HDR using a dsDNA template, whereas overexpression of Rad52 has been shown to increase PGE via SSTR with ssODN templates (Gallagher and Haber, 2021; Gallagher et al., 2020). Overexpression of Rad51 increases PGE events with double-stranded plasmid dDNA in utero in mice (Kurihara et al., 2020), in rabbit embryos (Song et al., 2016) and in human embryonic and induced pluripotent stem cells (Takayama et al., 2017) (Fig. 2). In addition, small molecules selected for their ability to stimulate Rad51 drive similar increases in PGE in rabbit (Song et al., 2016) and zebrafish (Zhang et al., 2018) embryos. However, when using ssODN donors, which require SSTR, overexpression of Rad51 appears to be detrimental to the rate of PGE (Paulsen et al., 2017). Instead, overexpression of Rad52 increases PGE events when using single-stranded dDNA (Paulsen et al., 2017) (Fig. 2).
BRCA1 and BRCA2 also play key roles in facilitating DSB repair via HDR and SSTR (Roy et al., 2012). BRCA1 likely facilitates DNA end resection by recruiting CtIP to the DSB (Yun and Hiom, 2009; Chen et al., 2008). In a separate function, BRCA1 promotes BRCA2 localisation to resected ssDNA via the intermediary protein PALB2 (Xia et al., 2006; Zhang et al., 2009). Subsequently, BRCA2 loads Rad51 onto the RPA-coated ssDNA, enabling nucleofilament formation (Thorslund et al., 2010; Liu et al., 2010). Overexpression of BRCA1 variants that display a hyper-recombination phenotype increases PGE when using plasmid donors in vitro compared to wild-type BRCA1 overexpression (Pinder et al., 2015).
The examples above demonstrate a clear potential to increase the efficiency of PGE events via the manipulation of individual components of the DNA repair pathways. This approach presents a promising avenue of inquiry towards the ultimate development of super-high-efficiency Cas-mediated PGE in both in vitro and in vivo systems (Table 1). The rational step of combining simultaneous downregulation of proteins that promote error-prone repair with upregulation of proteins that mediate faithful repair has also been explored. For example, combining overexpression of Rad52 and a dominant-negative form of 53BP1 (also known as TP53BP1), a protein that supresses DNA end resection to favour NHEJ (Chapman et al., 2013), significantly increases ssODN donor-mediated PGE events in induced pluripotent stem cells (Paulsen et al., 2017). In a similar approach, the small molecules RS-1 and SCR7, which are proposed to upregulate Rad51 and downregulate LIGIV activity, respectively, significantly increase the frequency of PGE events in zebrafish embryos (Zhang et al., 2018).
Chemical modulation of PGE
As exemplified in the study summarised above (Zhang et al., 2018), small molecules are an attractive option for the modulation of DNA repair pathways and optimisation of PGE. One benefit is that they can interact directly with proteins that already exist within the target cell, unlike small interfering RNAs (siRNAs) or morpholinos that target mRNA transcripts. This direct function facilitates more rapid effects. From a practical standpoint, it may also be easier to dissolve pre-synthesised chemicals in cell culture or embryo medium for delivery into the cells or tissue, rather than producing and delivering RNA for overexpression or knockdown of endogenous transcripts. For these reasons, researchers have investigated a range of chemicals that affect DNA repair proteins. Although promising, chemical interventions using small molecules to alter the activity of DSB repair proteins have had somewhat variable success (Table 1). For example, RS-1 and SCR7 were effective in some cases yet had small or negative effects on PGE efficiency in others (Zhang et al., 2018; Riesenberg and Maricic, 2018). The specific interplays between chemicals and the dDNA type are a potential explanation for this variability, especially when considering the Rad51-dependent nature of HDR compared to Rad52-mediated SSTR. The proposed effect of RS-1 is to stabilise the binding of Rad51 to the resected target strand (Jayathilaka et al., 2008). Studies manipulating key proteins in combination with different dDNA types can go some way to explaining the variable effects of small molecules on PGE rates. However, a more complete understanding of the precise roles of these proteins in faithful repair mechanisms is still required. For example, Lamas-Toranzo and colleagues suggested that RS-1 can stimulate other Rad51-like proteins, and this could explain how combining HDR/Rad51-promoting RS-1 with an SSTR-related ssODN donor can still increase the rate of PGE (Lamas-Toranzo et al., 2020).
Outcomes of small-molecule treatments could potentially also be affected by the chosen model system or Cas variant. For example, Cas12 produces staggered DNA ends, whereas Cas9 can generate blunt and staggered ends (Riesenberg and Maricic, 2018; Shou et al., 2018). This may alter how DSB repair proteins and the chemicals that modulate them take effect. However, the reasons why small molecules may not reproduce the same results in all model organisms, cell types or with different dDNA remain unclear. Typically, studies in which panels of compounds have been assessed for their effects on PGE identified at least one chemical that increased PGE efficiency, but different studies often identified different beneficial chemicals (Aksoy et al., 2019; Zhang et al., 2018; Riesenberg and Maricic, 2018). These discrepancies could be caused by the aforementioned differences in dDNA and cell type or by other minor protocol differences, which add additional variables and make direct comparisons difficult. Despite the lack of consensus, these studies do suggest that small molecules can effectively increase PGE, even if the optimal choice must be carefully selected based on the model system and dDNA. The range of chemicals, and their effects on PGE, are broad. We have summarised these in Table 1 and they have been covered in other recent reviews (Bischoff et al., 2020; Yeh et al., 2019).
Cell cycle arrest and optimisation of DSB resolution timing
One of the reasons why PGE may be less efficient than NHEJ-mediated indels is the limited cell cycle window in which faithful DNA repair can occur. Therefore, researchers have sought to synchronise the cell cycle phase for timed delivery of Cas proteins and donors to increase PGE rates (Fig. 2B). The microtubule polymerisation inhibitors nocodazole and ABT-751 have been used to arrest human embryonic cell lines, induced pluripotent stem cells, neural progenitor cells and HEK293T cells in the S/G2 phases of the cell cycle, resulting in a significant increase in both HDR and NHEJ events (Yang et al., 2016; Lin et al., 2014). As an alternative approach, researchers have generated a Cas9-Geminin fusion. The presence of Geminin targets this fusion for ubiquitin-mediated degradation in the M and G1 phases of the cell cycle. Expressing this fusion in HEK293T cells limits Cas9, and therefore DSB formation, to the HDR/SSTR-active cell cycle stages, leading to increases in PGE rates (Gutschner et al., 2016).
These techniques, however, may be difficult to translate into in vivo model generation, where fertilised embryos may be harvested at different time points. In practice, many embryos need to be microinjected in sequence. Holding embryo development at the one-cell stage or in the cell cycle phases that permit HDR/SSTR, therefore, is desirable. This is especially important in organisms with rapid post-fertilisation cell cycles, such as zebrafish (Kimmel et al., 1995). A simple protocol modification to incubate zebrafish embryos on ice before microinjection of the gene-editing components showed a non-significant increase in PGE in ice-cooled compared to room temperature embryos. However, when the addition of the small molecules NU7441 and RS-1 was combined with ice incubation, this resulted in a significant 1.5- to 2-fold increase in PGE over the use of the small molecules alone (Aksoy et al., 2019). This suggests that, in zebrafish embryos at least, a straightforward protocol adaptation to slow the cell cycle, when combined with alteration of DNA repair mechanisms via small molecules, can produce significant improvements to PGE efficiency.
The importance of gRNA design
gRNAs enable the ‘homing mechanism’ of Cas endonucleases and are thus a core part of the CRISPR system that have also been targeted for optimisation. However, some gRNAs direct Cas to the target site more efficiently than others, inducing DSBs more frequently. The reasons for this variable efficiency are incompletely understood, but sequence preferences that allow the complex to locate the target loci are thought to be crucial (Moreb and Lynch, 2021). In addition, poor gRNA specificity can lead to off-target effects (Wong et al., 2015; Manghwar et al., 2020; Schaefer et al., 2017; Varshney et al., 2015; Akcakaya et al., 2018). Furthermore, the requirement of a precise PAM site means that the positioning options of a gRNA and therefore the Cas-induced break are finite (Sternberg et al., 2014). To assist optimal gRNA design, researchers have designed a multitude of computational tools based on large datasets of on-target and off-target efficiencies (Heigwer et al., 2014; Montague et al., 2014; Moreno-Mateos et al., 2015) that provide metrics by which users can judge candidate gRNA sequences. The on-target score predicts the rate at which a given gRNA should induce cutting at the target locus, and the off-target score provides the predicted Cas endonuclease activity at unintended loci. These design tools have been comprehensively described and compared elsewhere (Chuai et al., 2017). It is clear, however, that the in silico-designed gRNAs still require in vivo and in vitro validation, and the efficacy of design tools across different cell types has been questioned (Chuai et al., 2017). In zebrafish, it has been found that two gRNA design tools underestimated the indel generation rate of gRNAs by ∼20% (Uribe-Salazar et al., 2022). In our own experience in zebrafish using Cas9 RNPs, even gRNAs with predicted low on-target scores (<40) can achieve cutting of >80% (as judged by indel rates in the absence of dDNA). Many tools, however, are based on algorithms trained on gRNA screens in mouse and human cells (Doench et al., 2014), and so may be more accurate in mammalian systems. The development of species-specific gRNA design tools, especially with in-built functionality for PGE, could be a valuable contribution to the field.
Although off-target edits do not directly affect PGE efficiency, they have the potential to reduce the validity of the modified cell or organism through unwanted indels in other genes (Schaefer et al., 2017). When attempting to achieve PGE, the distance from the DSB to the intended mutation site is also important. Cas9 induces a cut 3 bp upstream of the PAM recognition site (Chen et al., 2014), and mutation site distances of >15 bp from this cut result in suboptimal PGE efficiency, with an increase in distance having a more pronounced negative impact (O'Brien et al., 2019; Schubert et al., 2021; Wang et al., 2016). However, these studies also showed that the cutting efficiency of the gRNA still has a greater magnitude of effect on PGE efficiency than distance between cut and mutation site. Experimentally, gRNAs with higher cutting efficiencies can outperform gRNAs with lesser cutting efficiencies that cut closer to the edit site (Schubert et al., 2021). When choosing gRNAs, it is advisable to aim for those with a PAM site as close as possible to the site of the desired edit, with a high on-target score and low off-target score to avoid unwanted mutations.
Rational design of donor sequences and limitations
As PGE mediated by HDR and SSTR requires a donor template, several studies have compared different dDNA designs and their effects on PGE efficiency, again often reaching different conclusions (Zhang et al., 2018; Bai et al., 2020). One group investigated whether ssODNs with either symmetrical or asymmetrical arms of homology (with relation to the length of the sequence either side of the cut site) could improve PGE efficiency. Indeed, they achieved HDR rates of up to 60% in HEK293 cells by using asymmetric ssODNs with complementarity to the non-target DNA strand (which is not bound by the gRNA) (Richardson et al., 2016). The use of ssODNs with non-target DNA strand specificity has also been validated by other researchers (Janssen et al., 2019). Multiple groups have investigated the length of dDNA homology arms, often reaching different conclusions (Table 1). According to these studies, the optimal length of ssODN homology arms varies between 30 bp and 60 bp (Okamoto et al., 2019; Wang et al., 2016; Schubert et al., 2021). However, the inability to generate ssODNs longer than 200 bp has previously been a limiting factor, both to the length of homology arms and for the length of the exogenous insert, e.g. limiting the ability to insert fluorophore tags at a specific site. Subsequent research has allowed the development of lssODNs that are up to ∼2.0 kb in length (Miura et al., 2018), enabling the design of 300 bp homology arms and the insertion of up to 200 bp of exogenous sequence (Bai et al., 2020; Ranawakage et al., 2021).
An alternative approach to lssODNs is to deliver donor templates as plasmids, which are linearised in the cell by Cas9 to produce dsDNA donors in situ (Wierson et al., 2020; Zhang et al., 2017a). This approach has achieved high PGE efficiencies in Drosophila melanogaster (Kanca et al., 2019) and up to 77% PGE in zebrafish (Hisano et al., 2015). However, there is disagreement over the optimal length of homology arms in the linearised dDNA, with one study suggesting very short, 24-48 bp homology arms for PGE in zebrafish and in porcine and human cell lines (Wierson et al., 2020), and other studies suggesting relatively long homology arms of 600 bp as being the most efficient (Zhang et al., 2017a).
The introduction of silent mutations within dDNA sequences should also be considered. Several studies have identified that a silent ‘blocking’ mutation within the PAM site of the linearised dDNA, which should prevent re-cutting of the edited locus by residual Cas, increases PGE rates (Okamoto et al., 2019; Paquet et al., 2016; Schubert et al., 2021). To aid subsequent screening, silent mutations that add or remove restriction enzyme sites (Paulsen et al., 2017; Schubert et al., 2021) can identify edited loci without the need to resort to sequencing.
Aside from specific dDNA design, dDNA availability is also a key aspect of optimising PGE. Because dDNA is physically required at the site of the DSB to act as an HDR template, increasing dDNA concentration is a logical approach to maximise its availability. However, high concentrations of ssODNs (Kagita et al., 2021; Nakanishi et al., 2015) and dsDNA can be cytotoxic (Luecke et al., 2017; Nguyen et al., 2020). An alternative approach is to fuse dDNA directly to the Cas endonuclease (Savic et al., 2018) or the gRNA (Lee et al., 2017), thereby optimising the spatial and temporal colocalisation of the template to the DSB site and increasing PGE rates, which has been effective in cell culture.
Other considerations to optimise PGE via modulation of DSB resolution
As we discussed above, manipulation of key proteins within the DSB repair pathways, either via chemical modulation, overexpression or knockdown, can have robust and reproducible success in increasing PGE rates. In theory, there is no limit to the number of DNA repair proteins that could be simultaneously targeted for upregulation or downregulation. Indeed, combinations of up to seven chemicals to alter DSB repair protein function in cell culture have been investigated (Riesenberg and Maricic, 2018). The resulting degree of change in PGE rates depended on cell type, and whether Cas9, nCas9 or Cas12a was used, with the DNA-PKcs inhibitor NU7026 providing the bulk of PGE improvement when using standard Cas9. However, in other cases, the use of only two agents, one downregulating NHEJ and one upregulating HDR/SSTR, have successfully increased PGE rates in cell culture (Paulsen et al., 2017) and in zebrafish (Zhang et al., 2018). It remains to be seen whether the increases in PGE rates that can be achieved by directly altering DSB repair protein function have an upper limit.
Overexpression of key faithful repair components or knockdown of NHEJ proteins aim to increase the abundance of proteins interacting with a DSB leading to PGE. It is also important to consider the rates of NHEJ alongside those of PGE. In vivo, where genome editing affects a whole organism, the resulting animals often carry both NHEJ and PGE events (Zhang et al., 2018; Cui et al., 2018). As such, it can be useful to compare rates of PGE relative to indel formation as each animal will likely carry both types of genetic edit. The absolute degree of somatic PGE is also relevant for its germline transmission (Aksoy et al., 2019), which is important when the goal is to generate a stable line (see Box 1). The use of nCas9 variants and other methods that avoid DSBs (Anzalone et al., 2019; Gaudelli et al., 2017) sidestep the possibility of indel formation via NHEJ, thereby effectively increasing PGE rates.
The physical properties of the genomic landscape targeted for PGE are also worth considering. In HEK293 cells, targeting physically restrained heterochromatin rather than open euchromatin improved the PGE-to-NHEJ ratio (Janssen et al., 2019). In this study, HDR rates using dsDNA donors varied little between heterochromatin and euchromatin, whereas using ssODNs increased absolute PGE rates at euchromatin over heterochromatin. However, in both experiments, the authors observed higher rates of NHEJ when euchromatin was targeted, offsetting any gains in PGE. The use of multiple histone deacetylase inhibitors, which promote an open chromatin state, has also resulted in PGE and NHEJ increases in induced pluripotent stem cells (Zhang et al., 2021). The researchers noted, however, that some of this improvement was due to increased Cas9 and gRNA expression from their plasmid. However, at least in zebrafish, Cas9 reportedly lacks a preference for binding to heterochromatin or euchromatin (Moreno-Mateos et al., 2015). Gene locus- and cell-type-dependent variation in PGE efficiency has been noted in many studies (Bosch et al., 2020; Miyaoka et al., 2016; Remy et al., 2017; Petri et al., 2021), which may be due to chromatin structure or gRNA design limitations for a specific locus, although the factors controlling this variation are incompletely understood and warrant further investigation.
The best approach for optimising CRISPR-based PGE varies by model system and by the kind of edits that are desired. A consensus on optimal approaches has yet to be reached in the field, but the recent advancements described here provide some general advice. For small insertions and substitutions, the use of ssODNs complementary to the non-target strand, with chemical or protein modulation, may yield good PGE rates, whereas PGE of larger inserts may require the use of plasmid or lssODN donors, each with their own complement of chemical or protein modulation. When choosing targets for PGE, it may also be worth considering targeting multiple genes of interest, as some may be more amenable to editing.
BE and PE: the next generation
Employing Cas nucleases to form DSBs followed by endogenous DNA repair is only one approach to achieve PGE. Newer techniques that rely on fusing Cas to effector enzymes, chiefly reverse transcriptase and cytidine deaminase (Fig. 3), effectively achieve PGE in cell culture and in vivo in zebrafish and mice (Anzalone et al., 2019; Böck et al., 2022; Gaudelli et al., 2017; Kim et al., 2017a; Koblan et al., 2018; Komor et al., 2016; Liu et al., 2020; Petri et al., 2021; Rosello et al., 2022; Sasaguri et al., 2018; Walton et al., 2020; Xu et al., 2020; Zhang et al., 2017b) (summarised in Table 2). Typically, these approaches utilise nCas9 variants that cause single-stranded DNA nicks, avoiding the bulk of deleterious NHEJ events that can result from DSB repair (Fig. 3).
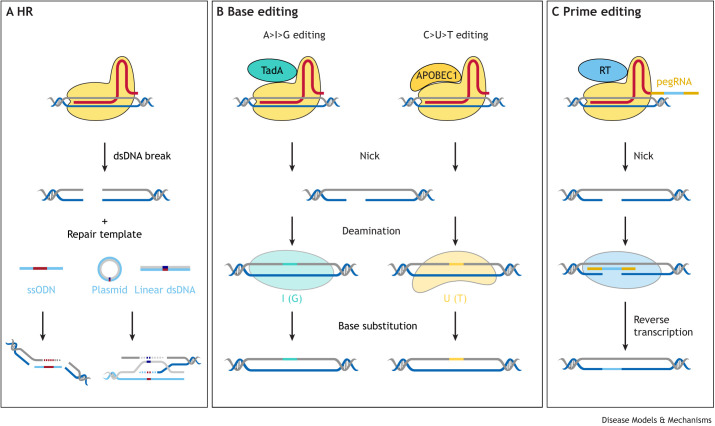
Fig. 3.
Comparison of next-generation PGE methods. (A) HR-based PGE, which encompasses HDR and SSTR, requires a Cas nuclease that induces a DSB in the endogenous genomic DNA, an exogenous donor DNA template that includes the desired edit, and resolution by an endogenous DNA repair pathway. (B) Base editing typically uses Cas9 nickase (nCas9), which only cuts one DNA strand (nick), fused to a deaminase. The deaminase converts adenosine to guanine or cytosine to thymine. No exogenous repair template is required. (C) Prime editing uses nCas9 fused to a reverse transcriptase and an extended gRNA (pegRNA) that acts as a repair template for the reverse transcriptase. A cut on only one DNA strand is necessary. A, adenosine; C, cytosine; dsDNA, double-stranded DNA; G, guanine; HR, homologous recombination; I, inosine; pegRNA, prime editing guide RNA; RT, reverse transcriptase; ssODN, single-stranded oligodeoxynucleotide; T, thymine; U, uracil.
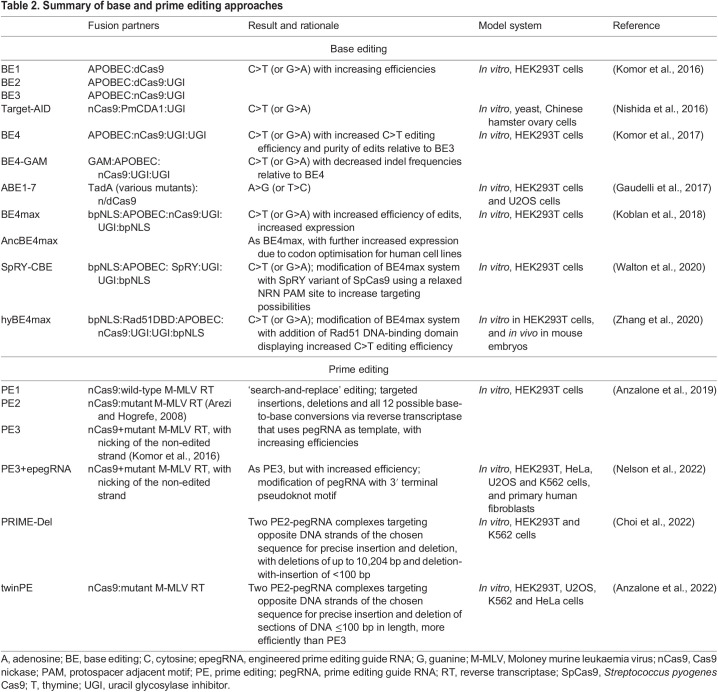
Table 2.
Summary of base and prime editing approaches
BE was the first to step away from manipulating endogenous DSB repair pathways. The original iteration of this system used dCas9 fused to the deaminase APOBEC1 (Harris et al., 2002; Petersen-Mahrt et al., 2002), which can ultimately convert any cytidine residues within a five-nucleotide PAM-upstream activity window to thymine (Komor et al., 2016). The more recently developed adenosine base editors are constructed with nCas9 fused to an evolved transfer RNA adenosine deaminase, which expands the BE toolbox for A>G conversion (Gaudelli et al., 2017). Researchers have continued to improve this system, increasing the base conversion edit efficiency, narrowing the deaminase activity window to increase targeting specificity, and adding PAM-less Cas9 nucleases to increase the targeting scope of the system (Gaudelli et al., 2017; Koblan et al., 2018; Komor et al., 2016; Walton et al., 2020) (Table 2). These improvements, culminating in the development of BE4, achieve a maximum editing rate of almost 70% base conversion in human cell culture (Komor et al., 2017), but these rates vary depending on the application and organism (Table 2). They range from low efficiencies of 1.3% in wheat (Zong et al., 2017) and 9-28% in zebrafish (Zhang et al., 2017b) to high efficiencies of 96% in yeast (Nishida et al., 2016), with varied levels of efficiency reported for mammalian in vitro (∼30-70%) and in vivo (44-57%) systems (Komor et al., 2017; Kim et al., 2017a). Recent attempts to apply BE to zebrafish have yielded better results, with some experiments generating up to 50% of larvae showing high rates of BE homozygosity, and up to 100% bearing some degree of BE (Rosello et al., 2022; Liang et al., 2022). Importantly, concurrent indel formation, which can be an issue in DSB pathway-based PGE methods (Zhang et al., 2018; Cui et al., 2018), is typically very low in most applications of BE and ranges between 2% and 10% (Anzalone et al., 2019; Zhang et al., 2017b), giving the technique an advantage.
The purpose of using BE is often to replicate diseases caused by single-base changes in model organisms, as has been done so successfully in zebrafish (Rosello et al., 2021), mice (Sasaguri et al., 2018) and Drosophila (Marr and Potter, 2021). However, the somewhat indiscriminate activity window of the deaminase is a key drawback, as it introduces a downstream screening issue, which reduces the potentially higher efficiency of the technology due to fewer indels. If a desired edit is a single-base conversion within an area that contains multiple target residues, then it is likely that BE will generate unwanted mutations. Another limitation of BE is its inability to introduce larger targeted insertions or multiple different base pair changes simultaneously. BE does have advantages over methods that rely on DSBs, but it is not yet a silver bullet for the targeted generation of single-base changes, although continued improvements may ameliorate these issues.
PE (Anzalone et al., 2019) is a still more recent advancement and again uses a nCas9 protein as a targeting chassis for an enzyme. However, in place of a deaminase, nCas9 is fused with a reverse transcriptase, such as the Moloney murine leukaemia virus (M-MLV) reverse transcriptase and bound to an extended gRNA that acts as a reverse transcription template, referred to as prime editing gRNA (pegRNA). PE has wider applicability than BE, as it can insert up to 44 bp of exogenous sequence and delete up to 80 bp (Lin et al., 2020; Anzalone et al., 2019), and it can generate any combination of base substitutions within those boundaries (Liu et al., 2020; Xu et al., 2020). Initial efforts to construct, optimise and apply PE in HEK293T cells resulted in the generation of three prime editor versions, PE1-3 (Table 2). PE3 achieved an average point mutation efficiency of 36±8.7%, with an average concurrent indel formation of 8.6±2.0% (Anzalone et al., 2019), and is more efficient than previous PE versions at generating larger precise deletions of varied sizes, from 5 bp to 80 bp, displaying editing efficiencies of 52-78% with average indel formation rates of 11±4.8% (Anzalone et al., 2019).
PE has been successfully applied to in vivo models, although, as is often the case with PGE, in vivo efficiency is generally much lower than that in cell culture. In zebrafish, PE3 did not always generate more efficient PGE than its predecessor PE2, although altering microinjection and embryo incubation temperature improved overall PE efficiency (Petri et al., 2021). The authors achieved average somatic PGE rates of up to 3.33% and 6.53% for two separate loci. From 14 edited F0 fish, only one passed PGEs onto the F1 generation at a rate of 8.3% (Petri et al., 2021). A separate study of PE in mice also identified discrepancies between the in vivo efficiencies of PE2 and PE3 compared to their efficiencies in mammalian cell culture (Aida et al., 2020 preprint). Here, PGE occurred in 44-75% of blastocysts, but the frequency of those PGEs accounted for only 1.1-18.5% of total embryo genomic DNA. The degree of editing within an embryo is an important factor, and one which complicates the process of generating PGEs in vivo. Higher rates of mosaicism (differences in the genetic sequence harboured by different cells within an organism) cause complications when trying to establish future generations that are genetically homogenous carriers of the desired edit. In addition, low rates of PGE within a single organism mean that few of the organism's offspring will likely contain the desired edit. Both outcomes cause more downstream work for researchers. An ideal technique will achieve a high number of individual embryos that bear any degree of editing, and each of those embryos should have a high percentage of their cells that bear the PGE. In another study, adeno-associated virus-mediated PE of newborn mice achieved G>C conversion with an efficiency of 14.4±6.6% in primary hepatocytes isolated at 4 weeks post injection, and the authors did not detect any indels (Böck et al., 2022). A preprint by Aida et al. indicates that PE in mouse embryos resulted in 10.5-13% PGE compared to a rate of 24-36.95% PGE when using Cas9 with ssODN donors. However, they also observed a much higher rate of PE-mediated indel formation than initially reported for mammalian cell culture (Aida et al., 2020 preprint). In Drosophila, PE2 has also been successfully implemented to generate efficient germline transmission of specific edits, with 36% transmission of PGEs to progeny (Bosch et al., 2021).
PE is clearly a promising step forward in PGE; but, as is often the case with new technologies, there are hurdles that need to be overcome before it can become a routine and robust tool for use in varied model organisms. The generally low indel formation and versatility of precise deletion, insertion and substitution in one platform are advantages that certainly make the continued development of PE worthwhile. In summary, BE is effective, especially when off-target mutations in the activity window are acceptable or if the targeted base is the only C or A residue in the activity window. PE can also be used to target larger indels. It is currently unclear whether previously tested chemical or protein modulation can further increase BE or PE efficiencies.
Conclusions
Using CRISPR systems to generate gene knockouts via NHEJ remains easier than PGE. However, the refinement and improvement of methods to leverage endogenous HDR and SSTR, and the development of new techniques such as BE and PE, are paving the way for routine PGE. These advances can be effectively applied to disease modelling but still require careful selection and design of the PGE components, optimisation for a specific locus or model system, and the screening of large numbers of animals or cells to isolate those that carry the desired edits. Comprehensive comparisons of technical modifications and their effect on PGE efficiency across loci and model systems would aid a methodological consensus benefitting multiple fields of research.
With continued improvement of PGE methods, however, the future of engineering precise human disease models looks bright. Increased reliability and reduction of labour time facilitates the high-throughput or more specialised use of PGE. For example, the introduction of patient-specific mutations into animal models to generate patient ‘avatars’ would allow for pre-testing of drugs and treatments. This strategy is already being used in zebrafish and Drosophila for cancer patients (Bangi et al., 2021; Fazio et al., 2020). Additionally, genome-wide association studies are identifying thousands of disease-related single-nucleotide polymorphisms (Visscher et al., 2012; Yengo et al., 2022) and novel genetic pathways from gene networks (Mäkinen et al., 2014), which ultimately require validation in vivo. Large-scale PGE screens could be carried out to investigate these, potentially in combination, to model complex traits and assess the phenotypic impact of multiple genetic variants. Precise mutations and tagging of endogenous genes also allow for the study of how misfunctioning proteins interact.
Generating disease models to study the subtle effects of genes bearing deleterious mutations is only one application of Cas-based PGE, however. There is also huge potential for PGE to treat human disease. Recently, the CRISPR system has been used to knock out genes repressing foetal haemoglobin production to treat sickle cell anaemia in somatic cells, and additional clinical trials are underway (Kan and Doudna, 2022). Additionally, multiple clinical trials of CRISPR-edited T cells for cancer immunotherapy are underway (Lu et al., 2020; Stadtmauer et al., 2020; Kan and Doudna, 2022). Although the transition from disease modelling to treatment raises further efficacy and ethical implications, with efficient delivery, off-target screening and safety protocols all requiring careful implementation, this future application of PGE represents an exciting new use for this technology.
Footnotes
Funding
This work was supported by the Biotechnology and Biological Sciences Research Council-funded South West Biosciences Doctoral Training Partnership [training grant reference BB/T008741/1].
Contributor Information
Robert N. Kelsh, Email: r.n.kelsh@bath.ac.uk.
Rebecca J. Richardson, Email: rebecca.richardson@bristol.ac.uk.
References
- Aida, T., Wilde, J. J., Yang, L., Hou, Y., Li, M., Xu, D., Lin, J., Qi, P., Lu, Z. and Feng, G. (2020). Prime editing primarily induces undesired outcomes in mice. bioRxiv 2020.08.06.239723. 10.1101/2020.08.06.239723 [DOI] [Google Scholar]
- Akcakaya, P., Bobbin, M. L., Guo, J. A., Malagon-Lopez, J., Clement, K., Garcia, S. P., Fellows, M. D., Porritt, M. J., Firth, M. A., Carreras, A.et al. (2018). In vivo CRISPR editing with no detectable genome-wide off-target mutations. Nature 561, 416-419. 10.1038/s41586-018-0500-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aksoy, Y. A., Nguyen, D. T., Chow, S., Chung, R. S., Guillemin, G. J., Cole, N. J. and Hesselson, D. (2019). Chemical reprogramming enhances homology-directed genome editing in zebrafish embryos. Commun. Biol. 2, 198. 10.1038/s42003-019-0444-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andres, S. N., Vergnes, A., Ristic, D., Wyman, C., Modesti, M. and Junop, M. (2012). A human XRCC4-XLF complex bridges DNA. Nucleic Acids Res. 40, 1868-1878. 10.1093/nar/gks022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders, C., Niewoehner, O., Duerst, A. and Jinek, M. (2014). Structural basis of PAM-dependent target DNA recognition by the Cas9 endonuclease. Nature 513, 569-573. 10.1038/nature13579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anzalone, A. V., Randolph, P. B., Davis, J. R., Sousa, A. A., Koblan, L. W., Levy, J. M., Chen, P. J., Wilson, C., Newby, G. A., Raguram, A.et al. (2019). Search-and-replace genome editing without double-strand breaks or donor DNA. Nature 576, 149-157. 10.1038/s41586-019-1711-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anzalone, A. V., Gao, X. D., Podracky, C. J., Nelson, A. T., Koblan, L. W., Raguram, A., Levy, J. M., Mercer, J. A. M. and Liu, D. R. (2022). Programmable deletion, replacement, integration and inversion of large DNA sequences with twin prime editing. Nat. Biotechnol. 40, 731-740. 10.1038/s41587-021-01133-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arezi, B. and Hogrefe, H. (2008). Novel mutations in Moloney Murine Leukemia Virus reverse transcriptase increase thermostability through tighter binding to template-primer. Nucleic Acids Res. 37, 473-481. 10.1093/nar/gkn952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bae, H. S., Jin, Y.-K., Ham, S., Kim, H. K., Shin, H., Cho, G.-B., Lee, K. J., Lee, H., Kim, K.-M., Koo, O.-J.et al. (2020). CRISPR/Cas9-mediated knockout of Mct8 reveals a functional involvement of Mct8 in testis and sperm development in a rat. Scientific Reports 10, 11148. 10.1038/s41598-020-67594-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai, H., Liu, L., An, K., Lu, X., Harrison, M., Zhao, Y., Yan, R., Lu, Z., Li, S., Lin, S.et al. (2020). CRISPR/Cas9-mediated precise genome modification by a long ssDNA template in zebrafish. BMC Genomics 21, 67. 10.1186/s12864-020-6493-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bangi, E., Smibert, P., Uzilov, A. V., Teague, A. G., Gopinath, S., Antipin, Y., Chen, R., Hecht, C., Gruszczynski, N., Yon, W. J.et al. (2021). A Drosophila platform identifies a novel, personalized therapy for a patient with adenoid cystic carcinoma. iScience 24, 102212. 10.1016/j.isci.2021.102212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg, E., Christensen, M. O., Dalla Rosa, I., Wannagat, E., Jänicke, R. U., Rösner, L. M., Dirks, W. G., Boege, F. and Mielke, C. (2011). XRCC4 controls nuclear import and distribution of Ligase IV and exchanges faster at damaged DNA in complex with Ligase IV. DNA Repair 10, 1232-1242. 10.1016/j.dnarep.2011.09.012 [DOI] [PubMed] [Google Scholar]
- Bischoff, N., Wimberger, S., Maresca, M. and Brakebusch, C. (2020). Improving precise CRISPR genome editing by small molecules: is there a magic potion? Cells 9, 1318. 10.3390/cells9051318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Böck, D., Rothgangl, T., Villiger, L., Schmidheini, L., Matsushita, M., Mathis, N., Ioannidi, E., Rimann, N., Grisch-Chan, H. M., Kreutzer, S.et al. (2022). In vivo prime editing of a metabolic liver disease in mice. Sci. Transl. Med. 14, eabl9238. 10.1126/scitranslmed.abl9238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosch, J. A., Colbeth, R., Zirin, J. and Perrimon, N. (2020). Gene knock-ins in drosophila using homology-independent insertion of universal donor plasmids. Genetics 214, 75-89. 10.1534/genetics.119.302819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosch, J. A., Birchak, G. and Perrimon, N. (2021). Precise genome engineering in. Drosophila using prime editing. Proc. Natl Acad. Sci. USA 118, e2021996118. 10.1073/pnas.2021996118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burger, A., Lindsay, H., Felker, A., Hess, C., Anders, C., Chiavacci, E., Zaugg, J., Weber, L. M., Catena, R., Jinek, M.et al. (2016). Maximizing mutagenesis with solubilized CRISPR-Cas9 ribonucleoprotein complexes. Development 143, 2025-2037. [DOI] [PubMed] [Google Scholar]
- Ceccaldi, R., Rondinelli, B. and D'andrea, A. D. (2016). Repair pathway choices and consequences at the double-strand break. Trends in Cell Biol. 26, 52-64. 10.1016/j.tcb.2015.07.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman, J. R., Barral, P., Vannier, J. B., Borel, V., Steger, M., Tomas-Loba, A., Sartori, A. A., Adams, I. R., Batista, F. D. and Boulton, S. J. (2013). RIF1 is essential for 53BP1-dependent nonhomologous end joining and suppression of DNA double-strand break resection. Mol Cell 49, 858-871. 10.1016/j.molcel.2013.01.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charpentier, M., Khedher, A. H. Y., Menoret, S., Brion, A., Lamribet, K., Dardillac, E., Boix, C., Perrouault, L., Tesson, L., Geny, S.et al. (2018). CtIP fusion to Cas9 enhances transgene integration by homology-dependent repair. Nat. Commun. 9, 1133. 10.1038/s41467-018-03475-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, L., Nievera, C. J., Lee, A. Y. and Wu, X. (2008). Cell cycle-dependent complex formation of BRCA1.CtIP.MRN is important for DNA double-strand break repair. J. Biol. Chem. 283, 7713-7720. 10.1074/jbc.M710245200 [DOI] [PubMed] [Google Scholar]
- Chen, H., Choi, J. and Bailey, S. (2014). Cut site selection by the two nuclease domains of the Cas9 RNA-guided endonuclease. J. Biol. Chem. 289, 13284-13294. 10.1074/jbc.M113.539726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, J. S., Ma, E., Harrington, L. B., Costa, M. D., Tian, X., Palefsky, J. M. and Doudna, J. A. (2018). CRISPR-Cas12a target binding unleashes indiscriminate single-stranded DNase activity. Science 360, 436-439. 10.1126/science.aar6245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi, J., Chen, W., Suiter, C. C., Lee, C., Chardon, F. M., Yang, W., Leith, A., Daza, R. M., Martin, B. and Shendure, J. (2022). Precise genomic deletions using paired prime editing. Nat. Biotechnol. 40, 218-226. 10.1038/s41587-021-01025-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuai, G.-H., Wang, Q.-L. and Liu, Q. (2017). In silico meets in vivo: towards computational CRISPR-based sgRNA design. Trends Biotechnol. 35, 12-21. 10.1016/j.tibtech.2016.06.008 [DOI] [PubMed] [Google Scholar]
- Cong, L., Ran, F. A., Cox, D., Lin, S., Barretto, R., Habib, N., Hsu, P. D., Wu, X., Jiang, W., Marraffini, L. A.et al. (2013). Multiplex Genome Engineering Using CRISPR/Cas Systems. Science 339, 819-823. 10.1126/science.1231143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui, Y., Niu, Y., Zhou, J., Chen, Y., Cheng, Y., Li, S., Ai, Z., Chu, C., Wang, H., Zheng, B.et al. (2018). Generation of a precise Oct4-hrGFP knockin cynomolgus monkey model via CRISPR/Cas9-assisted homologous recombination. Cell Res. 28, 383-386. 10.1038/cr.2018.10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Angelis, M. H., Flaswinkel, H., Fuchs, H., Rathkolb, B., Soewarto, D., Marschall, S., Heffner, S., Pargent, W., Wuensch, K., Jung, M.et al. (2000). Genome-wide, large-scale production of mutant mice by ENU mutagenesis. Nat. Genet. 25, 444-447. 10.1038/78146 [DOI] [PubMed] [Google Scholar]
- De Gobbi, M., Viprakasit, V., Hughes, J. R., Fisher, C., Buckle, V. J., Ayyub, H., Gibbons, R. J., Vernimmen, D., Yoshinaga, Y., De Jong, P.et al. (2006). A Regulatory SNP causes a human genetic disease by creating a new transcriptional promoter. Science 312, 1215-1217. 10.1126/science.1126431 [DOI] [PubMed] [Google Scholar]
- Dinapoli, S. E., Martinez-Mcfaline, R., Gribbin, C. K., Wrighton, P. J., Balgobin, C. A., Nelson, I., Leonard, A., Maskin, C. R., Shwartz, A., Quenzer, E. D.et al. (2020). Synthetic CRISPR/Cas9 reagents facilitate genome editing and homology directed repair. Nucleic Acids Res. 48, e38. 10.1093/nar/gkaa085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doench, J. G., Hartenian, E., Graham, D. B., Tothova, Z., Hegde, M., Smith, I., Sullender, M., Ebert, B. L., Xavier, R. J. and Root, D. E. (2014). Rational design of highly active sgRNAs for CRISPR-Cas9–mediated gene inactivation. Nat. Biotechnol. 32, 1262-1267. 10.1038/nbt.3026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doudna, J. A. and Charpentier, E. (2014). The new frontier of genome engineering with CRISPR-Cas9. Science 346, 1258096. 10.1126/science.1258096 [DOI] [PubMed] [Google Scholar]
- Drouet, J., Delteil, C., Lefrançois, J., Concannon, P., Salles, B. and Calsou, P. (2005). DNA-dependent protein kinase and XRCC4-DNA Ligase IV mobilization in the cell in response to DNA double strand breaks*. J. Biol. Chem. 280, 7060-7069. 10.1074/jbc.M410746200 [DOI] [PubMed] [Google Scholar]
- Fazio, M., Ablain, J., Chuan, Y., Langenau, D. M. and Zon, L. I. (2020). Zebrafish patient avatars in cancer biology and precision cancer therapy. Nat. Rev. Cancer 20, 263-273. 10.1038/s41568-020-0252-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fossett, N. G., Arbour-Reily, P., Kilroy, G., Mcdaniel, M., Mahmoud, J., Tucker, A. B., Chang, S. H. and Lee, W. R. (1990). Analysis of ENU-induced mutations at the Adh locus in Drosophila melanogaster. Mutat. Res. 231, 73-85. 10.1016/0027-5107(90)90178-7 [DOI] [PubMed] [Google Scholar]
- Gallagher, D. N. and Haber, J. E. (2021). Single-strand template repair: key insights to increase the efficiency of gene editing. Curr. Genet. 67, 747-753. 10.1007/s00294-021-01186-z [DOI] [PubMed] [Google Scholar]
- Gallagher, D. N., Pham, N., Tsai, A. M., Janto, A. N., Choi, J., Ira, G. and Haber, J. E. (2020). A Rad51-independent pathway promotes single-strand template repair in gene editing. PLoS Genet 16, e1008689. 10.1371/journal.pgen.1008689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaudelli, N. M., Komor, A. C., Rees, H. A., Packer, M. S., Badran, A. H., Bryson, D. I. and Liu, D. R. (2017). Programmable base editing of A•T to G•C in genomic DNA without DNA cleavage. Nature 551, 464-471. 10.1038/nature24644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutschner, T., Haemmerle, M., Genovese, G., Draetta, G. F. and Chin, L. (2016). Post-translational regulation of Cas9 during G1 enhances homology-directed repair. Cell Reports 14, 1555-1566. 10.1016/j.celrep.2016.01.019 [DOI] [PubMed] [Google Scholar]
- Hai, T., Teng, F., Guo, R., Li, W. and Zhou, Q. (2014). One-step generation of knockout pigs by zygote injection of CRISPR/Cas system. Cell Res. 24, 372-375. 10.1038/cr.2014.11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hancox, J. C., Whittaker, D. G., Zhang, H. and Stuart, A. G. (2019). Learning from studying very rare cardiac conditions: the example of short QT syndrome. J. Congenital Cardiol. 3, 3. 10.1186/s40949-019-0024-7 [DOI] [Google Scholar]
- Harris, R. S., Petersen-Mahrt, S. K. and Neuberger, M. S. (2002). RNA editing enzyme APOBEC1 and some of its homologs can act as DNA mutators. Mol. Cell 10, 1247-1253. 10.1016/S1097-2765(02)00742-6 [DOI] [PubMed] [Google Scholar]
- Heigwer, F., Kerr, G. and Boutros, M. (2014). E-CRISP: fast CRISPR target site identification. Nat. Method. 11, 122-123. 10.1038/nmeth.2812 [DOI] [PubMed] [Google Scholar]
- Heyer, W. D., Ehmsen, K. T. and Liu, J. (2010). Regulation of homologous recombination in eukaryotes. Annu. Rev. Genet. 44, 113-139. 10.1146/annurev-genet-051710-150955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hisano, Y., Sakuma, T., Nakade, S., Ohga, R., Ota, S., Okamoto, H., Yamamoto, T. and Kawahara, A. (2015). Precise in-frame integration of exogenous DNA mediated by CRISPR/Cas9 system in zebrafish. Sci. Rep. 5, 8841. 10.1038/srep08841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hitotsumachi, S., Carpenter, D. A. and Russell, W. L. (1985). Dose-repetition increases the mutagenic effectiveness of N-ethyl-N-nitrosourea in mouse spermatogonia. Proc. Natl. Acad. Sci. USA 82, 6619-6621. 10.1073/pnas.82.19.6619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoshijima, K., Jurynec, M. J. and Grunwald, D. J. (2016). Precise editing of the zebrafish genome made simple and efficient. Dev. Cell 36, 654-667. 10.1016/j.devcel.2016.02.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou, Z., Zhang, Y., Propson, N. E., Howden, S. E., Chu, L.-F., Sontheimer, E. J. and Thomson, J. A. (2013). Efficient genome engineering in human pluripotent stem cells using Cas9 from Neisseria meningitidis. Proc. Natl Acad. Sci. USA 110, 15644-15649. 10.1073/pnas.1313587110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hustedt, N. and Durocher, D. (2017). The control of DNA repair by the cell cycle. Nat. Cell Biol. 19, 1-9. 10.1038/ncb3452 [DOI] [PubMed] [Google Scholar]
- Ingram, V. M. (1957). Gene mutations in human haemoglobin: the chemical difference between normal and sickle cell haemoglobin. Nature 180, 326-328. 10.1038/180326a0 [DOI] [PubMed] [Google Scholar]
- Irion, U., Krauss, J. and Nüsslein-Volhard, C. (2014). Precise and efficient genome editing in zebrafish using the CRISPR/Cas9 system. Development 141, 4827-4830. 10.1242/dev.115584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janssen, J. M., Chen, X., Liu, J. and Gonçalves, M. A. F. V. (2019). The chromatin structure of CRISPR-Cas9 target DNA controls the balance between mutagenic and homology-directed gene-editing events. Mol. Ther. Nucleic Acids 16, 141-154. 10.1016/j.omtn.2019.02.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jayathilaka, K., Sheridan, S. D., Bold, T. D., Bochenska, K., Logan, H. L., Weichselbaum, R. R., Bishop, D. K. and Connell, P. P. (2008). A chemical compound that stimulates the human homologous recombination protein RAD51. Proc. Natl. Acad. Sci. USA 105, 15848-15853. 10.1073/pnas.0808046105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinek, M., Chylinski, K., Fonfara, I., Hauer, M., Doudna, J. A. and Charpentier, E. (2012). A programmable dual-RNA–Guided DNA endonuclease in adaptive bacterial immunity. Science 337, 816-821. 10.1126/science.1225829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagita, A., Lung, M. S. Y., Xu, H., Kita, Y., Sasakawa, N., Iguchi, T., Ono, M., Wang, X. H., Gee, P. and Hotta, A. (2021). Efficient ssODN-mediated targeting by avoiding cellular inhibitory RNAs through precomplexed CRISPR-Cas9/sgRNA ribonucleoprotein. Stem Cell Rep. 16, 985-996. 10.1016/j.stemcr.2021.02.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kan, M. J. and Doudna, J. A. (2022). Treatment of genetic diseases With CRISPR genome editing. JAMA 328, 980-981. 10.1001/jama.2022.13468 [DOI] [PubMed] [Google Scholar]
- Kanca, O., Zirin, J., Garcia-Marques, J., Knight, S. M., Yang-Zhou, D., Amador, G., Chung, H., Zuo, Z., Ma, L., He, Y.et al. (2019). An efficient CRISPR-based strategy to insert small and large fragments of DNA using short homology arms. eLife 8, e51539. 10.7554/eLife.51539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang, Y., Zheng, B., Shen, B., Chen, Y., Wang, L., Wang, J., Niu, Y., Cui, Y., Zhou, J., Wang, H.et al. (2015). CRISPR/Cas9-mediated Dax1 knockout in the monkey recapitulates human AHC-HH. Hum. Mol. Genet. 24, 7255-7264. 10.1093/hmg/ddv425 [DOI] [PubMed] [Google Scholar]
- Kang, Y., Chu, C., Wang, F. and Niu, Y. (2019). CRISPR/Cas9-mediated genome editing in nonhuman primates. Dis. Model. Mech. 12, dmm039982. 10.1242/dmm.039982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karvelis, T., Gasiunas, G., Miksys, A., Barrangou, R., Horvath, P. and Siksnys, V. (2013). crRNA and tracrRNA guide Cas9-mediated DNA interference in Streptococcus thermophilus. RNA Biol. 10, 841-851. 10.4161/rna.24203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ke, Q., Li, W., Lai, X., Chen, H., Huang, L., Kang, Z., Li, K., Ren, J., Lin, X., Zheng, H.et al. (2016). TALEN-based generation of a cynomolgus monkey disease model for human microcephaly. Cell Res. 26, 1048-1061. 10.1038/cr.2016.93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, K., Ryu, S.-M., Kim, S.-T., Baek, G., Kim, D., Lim, K., Chung, E., Kim, S. and Kim, J.-S. (2017a). Highly efficient RNA-guided base editing in mouse embryos. Nat. Biotechnol. 35, 435-437. 10.1038/nbt.3816 [DOI] [PubMed] [Google Scholar]
- Kim, Y. B., Komor, A. C., Levy, J. M., Packer, M. S., Zhao, K. T. and Liu, D. R. (2017b). Increasing the genome-targeting scope and precision of base editing with engineered Cas9-cytidine deaminase fusions. Nat. Biotechnol. 35, 371-376. 10.1038/nbt.3803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimmel, C. B., Ballard, W. W., Kimmel, S. R., Ullmann, B. and Schilling, T. F. (1995). Stages of embryonic development of the zebrafish. Dev. Dyn. 203, 253-310. 10.1002/aja.1002030302 [DOI] [PubMed] [Google Scholar]
- Kleinstiver, B. P., Prew, M. S., Tsai, S. Q., Nguyen, N. T., Topkar, V. V., Zheng, Z. and Joung, J. K. (2015a). Broadening the targeting range of Staphylococcus aureus CRISPR-Cas9 by modifying PAM recognition. Nat. Biotechnol. 33, 1293-1298. 10.1038/nbt.3404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleinstiver, B. P., Prew, M. S., Tsai, S. Q., Topkar, V. V., Nguyen, N. T., Zheng, Z., Gonzales, A. P. W., Li, Z., Peterson, R. T., Yeh, J.-R. J.et al. (2015b). Engineered CRISPR-Cas9 nucleases with altered PAM specificities. Nature 523, 481-485. 10.1038/nature14592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleinstiver, B. P., Pattanayak, V., Prew, M. S., Tsai, S. Q., Nguyen, N. T., Zheng, Z. and Joung, J. K. (2016). High-fidelity CRISPR–Cas9 nucleases with no detectable genome-wide off-target effects. Nature 529, 490-495. 10.1038/nature16526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koblan, L. W., Doman, J. L., Wilson, C., Levy, J. M., Tay, T., Newby, G. A., Maianti, J. P., Raguram, A. and Liu, D. R. (2018). Improving cytidine and adenine base editors by expression optimization and ancestral reconstruction. Nat. Biotechnol. 36, 843-846. 10.1038/nbt.4172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komor, A. C., Kim, Y. B., Packer, M. S., Zuris, J. A. and Liu, D. R. (2016). Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature 533, 420-424. 10.1038/nature17946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komor, A. C., Zhao, K. T., Packer, M. S., Gaudelli, N. M., Waterbury, A. L., Koblan, L. W., Kim, Y. B., Badran, A. H. and Liu, D. R. (2017). Improved base excision repair inhibition and bacteriophage Mu Gam protein yields C:G-to-T:A base editors with higher efficiency and product purity. Sci. Adv. 3, eaao4774. 10.1126/sciadv.aao4774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroll, F., Powell, G. T., Ghosh, M., Gestri, G., Antinucci, P., Hearn, T. J., Tunbak, H., Lim, S., Dennis, H. W., Fernandez, J. M.et al. (2021). A simple and effective F0 knockout method for rapid screening of behaviour and other complex phenotypes. eLife 10, e59683. 10.7554/eLife.59683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurihara, T., Kouyama-Suzuki, E., Satoga, M., Li, X., Badawi, M., Thiha, B. D. N., Yanagawa, T., Uemura, T., Mori, T. and Tabuchi, K. (2020). DNA repair protein RAD51 enhances the CRISPR/Cas9-mediated knock-in efficiency in brain neurons. Biochem. Biophys. Res. Commun. 524, 621-628. 10.1016/j.bbrc.2020.01.132 [DOI] [PubMed] [Google Scholar]
- Labun, K., Montague, T. G., Krause, M., Torres Cleuren, Y. N., Tjeldnes, H. and Valen, E. (2019). CHOPCHOP v3: expanding the CRISPR web toolbox beyond genome editing. Nucleic Acids Res. 47, W171-W174. 10.1093/nar/gkz365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamas-Toranzo, I., Martínez-Moro, A., O′Callaghan, E., Millán-Blanca, G., Sánchez, J. M., Lonergan, P. and Bermejo-Álvarez, P. (2020). RS-1 enhances CRISPR-mediated targeted knock-in in bovine embryos. Mol. Reprod. Dev. 87, 542-549. 10.1002/mrd.23341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, K., Mackley, V. A., Rao, A., Chong, A. T., Dewitt, M. A., Corn, J. E. and Murthy, N. (2017). Synthetically modified guide RNA and donor DNA are a versatile platform for CRISPR-Cas9 engineering. eLife 6, e25312. 10.7554/eLife.25312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, G., Liu, D., Zhang, X., Quan, R., Zhong, C., Mo, J., Huang, Y., Wang, H., Ruan, X., Xu, Z.et al. (2018). Suppressing Ku70/Ku80 expression elevates homology-directed repair efficiency in primary fibroblasts. Int. J. Biochem. Cell Biol. 99, 154-160. 10.1016/j.biocel.2018.04.011 [DOI] [PubMed] [Google Scholar]
- Li, T., Huang, S., Jiang, W. Z., Wright, D., Spalding, M. H., Weeks, D. P. and Yang, B. (2011). TAL nucleases (TALNs): hybrid proteins composed of TAL effectors and FokI DNA-cleavage domain. Nucleic Acids Res. 39, 359-372. 10.1093/nar/gkq704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang, F., Zhang, Y., Li, L., Yang, Y., Fei, J.-F., Liu, Y. and Qin, W. (2022). SpG and SpRY variants expand the CRISPR toolbox for genome editing in zebrafish. Nat. Commun. 13, 3421. 10.1038/s41467-022-31034-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, S., Staahl, B. T., Alla, R. K. and Doudna, J. A. (2014). Enhanced homology-directed human genome engineering by controlled timing of CRISPR/Cas9 delivery. eLife 3, e04766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, C., Li, H., Hao, M., Xiong, D., Luo, Y., Huang, C., Yuan, Q., Zhang, J. and Xia, N. (2016). Increasing the Efficiency of CRISPR/Cas9-mediated Precise Genome Editing of HSV-1 Virus in Human Cells. Sci. Rep. 6, 34531. 10.1038/srep34531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, Q., Zong, Y., Xue, C., Wang, S., Jin, S., Zhu, Z., Wang, Y., Anzalone, A. V., Raguram, A., Doman, J. L.et al. (2020). Prime genome editing in rice and wheat. Nat. Biotechnol. 38, 582-585. 10.1038/s41587-020-0455-x [DOI] [PubMed] [Google Scholar]
- Liu, J., Doty, T., Gibson, B. and Heyer, W.-D. (2010). Human BRCA2 protein promotes RAD51 filament formation on RPA-covered single-stranded DNA. Nat. Struct. Mol. Biol. 17, 1260-1262. 10.1038/nsmb.1904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, H., Wei, Z., Dominguez, A., Li, Y., Wang, X. and Qi, L. S. (2015). CRISPR-ERA: a comprehensive design tool for CRISPR-mediated gene editing, repression and activation. Bioinformatics 31, 3676-3678. 10.1093/bioinformatics/btv423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, Y., Li, X., He, S., Huang, S., Li, C., Chen, Y., Liu, Z., Huang, X. and Wang, X. (2020). Efficient generation of mouse models with the prime editing system. Cell Discovery 6, 27. 10.1038/s41421-020-0165-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu, Y., Xue, J., Deng, T., Zhou, X., Yu, K., Deng, L., Huang, M., Yi, X., Liang, M., Wang, Y.et al. (2020). Safety and feasibility of CRISPR-edited T cells in patients with refractory non-small-cell lung cancer. Nat. Med. 26, 732-740. 10.1038/s41591-020-0840-5 [DOI] [PubMed] [Google Scholar]
- Luecke, S., Holleufer, A., Christensen, M. H., Jønsson, K. L., Boni, G. A., Sørensen, L. K., Johannsen, M., Jakobsen, M. R., Hartmann, R. and Paludan, S. R. (2017). cGAS is activated by DNA in a length-dependent manner. EMBO Rep. 18, 1707-1715. 10.15252/embr.201744017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mak, T. W. (2007). Gene targeting in embryonic stem cells scores a knockout in stockholm. Cell 131, 1027-1031. 10.1016/j.cell.2007.11.033 [DOI] [PubMed] [Google Scholar]
- Makarova, K. S., Aravind, L., Wolf, Y. I. and Koonin, E. V. (2011). Unification of Cas protein families and a simple scenario for the origin and evolution of CRISPR-Cas systems. Biol. Direct 6, 38. 10.1186/1745-6150-6-38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mäkinen, V.-P., Civelek, M., Meng, Q., Zhang, B., Zhu, J., Levian, C., Huan, T., Segrè, A. V., Ghosh, S. and Vivar, J. (2014). Integrative genomics reveals novel molecular pathways and gene networks for coronary artery disease. PLoS Genet. 10, e1004502. 10.1371/journal.pgen.1004502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mali, P., Aach, J., Stranges, P. B., Esvelt, K. M., Moosburner, M., Kosuri, S., Yang, L. and Church, G. M. (2013). CAS9 transcriptional activators for target specificity screening and paired nickases for cooperative genome engineering. Nat. Biotechnol. 31, 833-838. 10.1038/nbt.2675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manghwar, H., Li, B., Ding, X., Hussain, A., Lindsey, K., Zhang, X. and Jin, S. (2020). CRISPR/Cas Systems in genome editing: methodologies and tools for sgRNA design, off-target evaluation, and strategies to mitigate off-target effects. Adv. Sci. 7, 1902312. 10.1002/advs.201902312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marr, E. and Potter, C. J. (2021). Base editing of somatic cells using CRISPR–Cas9 in Drosophila. CRISPR J. 4, 836-845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miura, H., Quadros, R. M., Gurumurthy, C. B. and Ohtsuka, M. (2018). Easi-CRISPR for creating knock-in and conditional knockout mouse models using long ssDNA donors. Nat. Protocols 13, 195-215. 10.1038/nprot.2017.153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyaoka, Y., Berman, J. R., Cooper, S. B., Mayerl, S. J., Chan, A. H., Zhang, B., Karlin-Neumann, G. A. and Conklin, B. R. (2016). Systematic quantification of HDR and NHEJ reveals effects of locus, nuclease, and cell type on genome-editing. Sci. Rep. 6, 23549. 10.1038/srep23549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mojica, F. J. M., Díez-Villaseñor, C., García-Martínez, J. and Almendros, C. (2009). Short motif sequences determine the targets of the prokaryotic CRISPR defence system. Microbiology 155, 733-740. 10.1099/mic.0.023960-0 [DOI] [PubMed] [Google Scholar]
- Montague, T. G., Cruz, J. M., Gagnon, J. A., Church, G. M. and Valen, E. (2014). CHOPCHOP: a CRISPR/Cas9 and TALEN web tool for genome editing. Nucleic Acids Res. 42, W401-W407. 10.1093/nar/gku410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreb, E. A. and Lynch, M. D. (2021). Genome dependent Cas9/gRNA search time underlies sequence dependent gRNA activity. Nat. Commun. 12, 5034. 10.1038/s41467-021-25339-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno-Mateos, M. A., Vejnar, C. E., Beaudoin, J.-D., Fernandez, J. P., Mis, E. K., Khokha, M. K. and Giraldez, A. J. (2015). CRISPRscan: designing highly efficient sgRNAs for CRISPR-Cas9 targeting in vivo. Nat. Method. 12, 982-988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Movahedi, A., Wei, H., Zhou, X., Fountain, J. C., Chen, Z.-H., Mu, Z., Sun, W., Zhang, J., Li, D., Guo, B.et al. (2022). Precise exogenous insertion and sequence replacements in poplar by simultaneous HDR overexpression and NHEJ suppression using CRISPR-Cas9. Hortic. Res. 9, uhac154. 10.1093/hr/uhac154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musunuru, K., Chadwick, A. C., Mizoguchi, T., Garcia, S. P., Denizio, J. E., Reiss, C. W., Wang, K., Iyer, S., Dutta, C., Clendaniel, V.et al. (2021). In vivo CRISPR base editing of PCSK9 durably lowers cholesterol in primates. Nature 593, 429-434. 10.1038/s41586-021-03534-y [DOI] [PubMed] [Google Scholar]
- Nakade, S., Yamamoto, T. and Sakuma, T. (2017). Cas9, Cpf1 and C2c1/2/3—What's next? Bioengineered 8, 265-273. 10.1080/21655979.2017.1282018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakanishi, T., Kato, Y., Matsuura, T. and Watanabe, H. (2015). TALEN-mediated homologous recombination in Daphnia magna. Sci. Rep. 5, 18312. 10.1038/srep18312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson, J. W., Randolph, P. B., Shen, S. P., Everette, K. A., Chen, P. J., Anzalone, A. V., An, M., Newby, G. A., Chen, J. C., Hsu, A.et al. (2022). Engineered pegRNAs improve prime editing efficiency. Nat. Biotechnol. 40, 402-410. 10.1038/s41587-021-01039-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen, D. N., Roth, T. L., Li, P. J., Chen, P. A., Apathy, R., Mamedov, M. R., Vo, L. T., Tobin, V. R., Goodman, D., Shifrut, E.et al. (2020). Polymer-stabilized Cas9 nanoparticles and modified repair templates increase genome editing efficiency. Nat. Biotechnol. 38, 44-49. 10.1038/s41587-019-0325-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishida, K., Arazoe, T., Yachie, N., Banno, S., Kakimoto, M., Tabata, M., Mochizuki, M., Miyabe, A., Araki, M., Hara, K. Y.et al. (2016). Targeted nucleotide editing using hybrid prokaryotic and vertebrate adaptive immune systems. Science 353, aaf8729. 10.1126/science.aaf8729 [DOI] [PubMed] [Google Scholar]
- O'brien, A. R., Wilson, L. O. W., Burgio, G. and Bauer, D. C. (2019). Unlocking HDR-mediated nucleotide editing by identifying high-efficiency target sites using machine learning. Sci. Rep. 9, 2788. 10.1038/s41598-019-39142-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamoto, S., Amaishi, Y., Maki, I., Enoki, T. and Mineno, J. (2019). Highly efficient genome editing for single-base substitutions using optimized ssODNs with Cas9-RNPs. Sci. Rep. 9, 4811. 10.1038/s41598-019-41121-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paquet, D., Kwart, D., Chen, A., Sproul, A., Jacob, S., Teo, S., Olsen, K. M., Gregg, A., Noggle, S. and Tessier-Lavigne, M. (2016). Efficient introduction of specific homozygous and heterozygous mutations using CRISPR/Cas9. Nature 533, 125-129. 10.1038/nature17664 [DOI] [PubMed] [Google Scholar]
- Park, K.-E., Powell, A., Sandmaier, S. E. S., Kim, C.-M., Mileham, A., Donovan, D. M. and Telugu, B. P. (2017). Targeted gene knock-in by CRISPR/Cas ribonucleoproteins in porcine zygotes. Sci. Rep. 7, 42458. 10.1038/srep42458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parvez, S., Herdman, C., Beerens, M., Chakraborti, K., Harmer, Z. P., Yeh, J.-R. J., Macrae, C. A., Yost, H. J. and Peterson, R. T. (2021). MIC-Drop: A platform for large-scale in vivo CRISPR screens. Science 373, 1146-1151. 10.1126/science.abi8870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulsen, B. S., Mandal, P. K., Frock, R. L., Boyraz, B., Yadav, R., Upadhyayula, S., Gutierrez-Martinez, P., Ebina, W., Fasth, A., Kirchhausen, T.et al. (2017). Ectopic expression of RAD52 and dn53BP1 improves homology-directed repair during CRISPR-Cas9 genome editing. Nat. Biomed. Eng. 1, 878-888. 10.1038/s41551-017-0145-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen-Mahrt, S. K., Harris, R. S. and Neuberger, M. S. (2002). AID mutates E. coli suggesting a DNA deamination mechanism for antibody diversification. Nature 418, 99-104. 10.1038/nature00862 [DOI] [PubMed] [Google Scholar]
- Petri, K., Zhang, W., Ma, J., Schmidts, A., Lee, H., Horng, J. E., Kim, D. Y., Kurt, I. C., Clement, K., Hsu, J. Y.et al. (2021). CRISPR prime editing with ribonucleoprotein complexes in zebrafish and primary human cells. Nat. Biotechnol. 40, 189-193. 10.1038/s41587-021-00901-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinder, J., Salsman, J. and Dellaire, G. (2015). Nuclear domain ‘knock-in’ screen for the evaluation and identification of small molecule enhancers of CRISPR-based genome editing. Nucleic Acids Res. 43, 9379-9392. 10.1093/nar/gkv993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prykhozhij, S. V., Rajan, V., Ban, K. and Berman, J. N. (2021). CRISPR knock-in designer: automatic oligonucleotide design software to introduce point mutations by gene editing methods. Re:GEN Open 1, 53-67. 10.1089/regen.2021.0025 [DOI] [Google Scholar]
- Prykhozhij, S. V., Fuller, C., Steele, S. L., Veinotte, C. J., Razaghi, B., Robitaille, J., Mcmaster, C., Shlien, A., Malkin, D. and Berman, J. N. (2017). Successful optimization of CRISPR/Cas9-mediated defined point mutation knock-in using allele-specific PCR assays in zebrafish. bioRxiv 194936. 10.1101/194936 [DOI] [Google Scholar]
- Ran, F. A., Cong, L., Yan, W. X., Scott, D. A., Gootenberg, J. S., Kriz, A. J., Zetsche, B., Shalem, O., Wu, X., Makarova, K. S.et al. (2015). In vivo genome editing using Staphylococcus aureus Cas9. Nature 520, 186-191. 10.1038/nature14299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ran, F. A., Hsu, P. D., Lin, C.-Y., Gootenberg, J. S., Konermann, S., Trevino, A. E., Scott, D. A., Inoue, A., Matoba, S., Zhang, Y.et al. (2013). Double nicking by RNA-guided CRISPR Cas9 for enhanced genome editing specificity. Cell 154, 1380-1389. 10.1016/j.cell.2013.08.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranawakage, D. C., Okada, K., Sugio, K., Kawaguchi, Y., Kuninobu-Bonkohara, Y., Takada, T. and Kamachi, Y. (2021). Efficient CRISPR-Cas9-mediated knock-in of composite tags in zebrafish using long ssDNA as a donor. Front. Cell Dev. Biol. 8, 598634. 10.3389/fcell.2020.598634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remy, S., Chenouard, V., Tesson, L., Usal, C., Ménoret, S., Brusselle, L., Heslan, J.-M., Nguyen, T. H., Bellien, J., Merot, J.et al. (2017). Generation of gene-edited rats by delivery of CRISPR/Cas9 protein and donor DNA into intact zygotes using electroporation. Sci. Rep. 7, 16554. 10.1038/s41598-017-16328-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren, C., Xu, K., Liu, Z., Shen, J., Han, F., Chen, Z. and Zhang, Z. (2015). Dual-reporter surrogate systems for efficient enrichment of genetically modified cells. Cell. Mol. Life Sci. 72, 2763-2772. 10.1007/s00018-015-1874-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson, C. D., Ray, G. J., Dewitt, M. A., Curie, G. L. and Corn, J. E. (2016). Enhancing homology-directed genome editing by catalytically active and inactive CRISPR-Cas9 using asymmetric donor DNA. Nat. Biotechnol. 34, 339-344. 10.1038/nbt.3481 [DOI] [PubMed] [Google Scholar]
- Riesenberg, S. and Maricic, T. (2018). Targeting repair pathways with small molecules increases precise genome editing in pluripotent stem cells. Nat. Commun. 9, 2164. 10.1038/s41467-018-04609-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robert, F., Barbeau, M., Éthier, S., Dostie, J. and Pelletier, J. (2015). Pharmacological inhibition of DNA-PK stimulates Cas9-mediated genome editing. Genome Med 7, 93. 10.1186/s13073-015-0215-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosello, M., Vougny, J., Czarny, F., Mione, M. C., Concordet, J.-P., Albadri, S. and Del Bene, F. (2021). Precise base editing for the in vivo study of developmental signaling and human pathologies in zebrafish. eLife 10, e65552. 10.7554/eLife.65552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosello, M., Serafini, M., Mignani, L., Finazzi, D., Giovannangeli, C., Mione, M. C., Concordet, J.-P. and Del Bene, F. (2022). Disease modeling by efficient genome editing using a near PAM-less base editor in vivo. Nat. Commun. 13, 3435. 10.1038/s41467-022-31172-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy, R., Chun, J. and Powell, S. N. (2012). BRCA1 and BRCA2: different roles in a common pathway of genome protection. Nat. Rev. Cancer 12, 68-78. 10.1038/nrc3181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaguri, H., Nagata, K., Sekiguchi, M., Fujioka, R., Matsuba, Y., Hashimoto, S., Sato, K., Kurup, D., Yokota, T. and Saido, T. C. (2018). Introduction of pathogenic mutations into the mouse Psen1 gene by Base Editor and Target-AID. Nat. Commun. 9, 2892. 10.1038/s41467-018-05262-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savic, N., Ringnalda, F. C., Lindsay, H., Berk, C., Bargsten, K., Li, Y., Neri, D., Robinson, M. D., Ciaudo, C., Hall, J.et al. (2018). Covalent linkage of the DNA repair template to the CRISPR-Cas9 nuclease enhances homology-directed repair. Elife 7, e33761. 10.7554/eLife.33761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaefer, K. A., Wu, W. H., Colgan, D. F., Tsang, S. H., Bassuk, A. G. and Mahajan, V. B. (2017). Unexpected mutations after CRISPR-Cas9 editing in vivo. Nat Methods 14, 547-548. 10.1038/nmeth.4293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schubert, M. S., Thommandru, B., Woodley, J., Turk, R., Yan, S., Kurgan, G., Mcneill, M. S. and Rettig, G. R. (2021). Optimized design parameters for CRISPR Cas9 and Cas12a homology-directed repair. Sci. Rep. 11, 19482. 10.1038/s41598-021-98965-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seigel, K. E. (2018). Enhancing the Efficiency of CRISPR/Cas9 Precise Gene Editing for Cystic Fibrosis Gene Therapy. M.Sc., University of Toronto (Canada).
- Seki, A. and Rutz, S. (2018). Optimized RNP transfection for highly efficient CRISPR/Cas9-mediated gene knockout in primary T cells. J. Exp. Med. 215, 985-997. 10.1084/jem.20171626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shou, J., Li, J., Liu, Y. and Wu, Q. (2018). Precise and predictable CRISPR chromosomal rearrangements reveal principles of Cas9-mediated nucleotide insertion. Mol. Cell 71, 498-509.e4. 10.1016/j.molcel.2018.06.021 [DOI] [PubMed] [Google Scholar]
- Shrivastav, M., De Haro, L. P. and Nickoloff, J. A. (2008). Regulation of DNA double-strand break repair pathway choice. Cell Res. 18, 134-147. 10.1038/cr.2007.111 [DOI] [PubMed] [Google Scholar]
- Solnica-Krezel, L., Schier, A. F. and Driever, W. (1994). Efficient recovery of ENU-induced mutations from the zebrafish germline. Genetics 136, 1401-1420. 10.1093/genetics/136.4.1401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song, F. and Stieger, K. (2017). Optimizing the DNA donor template for homology-directed repair of double-strand breaks. Mol. Ther. Nucleic Acids 7, 53-60. 10.1016/j.omtn.2017.02.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song, J., Yang, D., Xu, J., Zhu, T., Chen, Y. E. and Zhang, J. (2016). RS-1 enhances CRISPR/Cas9- and TALEN-mediated knock-in efficiency. Nat. Commun. 7, 10548. 10.1038/ncomms10548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song, Y., Zhang, Y., Chen, M., Deng, J., Sui, T., Lai, L. and Li, Z. (2018). Functional validation of the albinism-associated tyrosinase T373K SNP by CRISPR/Cas9-mediated homology-directed repair (HDR) in rabbits. EBioMedicine 36, 517-525. 10.1016/j.ebiom.2018.09.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadtmauer, E. A., Fraietta, J. A., Davis, M. M., Cohen, A. D., Weber, K. L., Lancaster, E., Mangan, P. A., Kulikovskaya, I., Gupta, M., Chen, F.et al. (2020). CRISPR-engineered T cells in patients with refractory cancer. Science 367, eaba7365. 10.1126/science.aba7365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenson, P. D., Ball, E. V., Mort, M., Phillips, A. D., Shiel, J. A., Thomas, N. S. T., Abeysinghe, S., Krawczak, M. and Cooper, D. N. (2003). Human gene mutation database (HGMD®): 2003 update. Hum. Mutat. 21, 577-581. 10.1002/humu.10212 [DOI] [PubMed] [Google Scholar]
- Sternberg, S. H., Redding, S., Jinek, M., Greene, E. C. and Doudna, J. A. (2014). DNA interrogation by the CRISPR RNA-guided endonuclease Cas9. Nature 507, 62-67. 10.1038/nature13011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swarts, D. C. and Jinek, M. (2018). Cas9 versus Cas12a/Cpf1: Structure–function comparisons and implications for genome editing. WIREs RNA 9, e1481. [DOI] [PubMed] [Google Scholar]
- Switonski, P. M., Szlachcic, W. J., Krzyzosiak, W. J. and Figiel, M. (2015). A new humanized ataxin-3 knock-in mouse model combines the genetic features, pathogenesis of neurons and glia and late disease onset of SCA3/MJD. Neurobiol. Dis. 73, 174-188. 10.1016/j.nbd.2014.09.020 [DOI] [PubMed] [Google Scholar]
- Takayama, K., Igai, K., Hagihara, Y., Hashimoto, R., Hanawa, M., Sakuma, T., Tachibana, M., Sakurai, F., Yamamoto, T. and Mizuguchi, H. (2017). Highly efficient biallelic genome editing of human ES/iPS cells using a CRISPR/Cas9 or TALEN system. Nucleic Acids Res. 45, 5198-5207. 10.1093/nar/gkx130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tessadori, F., Roessler, H. I., Savelberg, S. M. C., Chocron, S., Kamel, S. M., Duran, K. J., Van Haelst, M. M., Van Haaften, G. and Bakkers, J. (2018). Effective CRISPR/Cas9-based nucleotide editing in zebrafish to model human genetic cardiovascular disorders. Dis. Model. Mech. 11, dmm035469. 10.1242/dmm.035469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorslund, T., Mcilwraith, M. J., Compton, S. A., Lekomtsev, S., Petronczki, M., Griffith, J. D. and West, S. C. (2010). The breast cancer tumor suppressor BRCA2 promotes the specific targeting of RAD51 to single-stranded DNA. Nat. Struct. Mol. Biol. 17, 1263-1265. 10.1038/nsmb.1905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran, N.-T., Bashir, S., Li, X., Rossius, J., Chu, V. T., Rajewsky, K. and Kühn, R. (2019). Enhancement of precise gene editing by the association of Cas9 with homologous recombination factors. Front. Genet. 10, 365. 10.3389/fgene.2019.00365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsutsui, H. and Higashiyama, T. (2016). pKAMA-ITACHI vectors for highly efficient CRISPR/Cas9-mediated gene knockout in arabidopsis thaliana. Plant Cell Physiol. 58, 46-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uribe-Salazar, J. M., Kaya, G., Sekar, A., Weyenberg, K., Ingamells, C. and Dennis, M. Y. (2022). Evaluation of CRISPR gene-editing tools in zebrafish. BMC Genomics 23, 12. 10.1186/s12864-021-08238-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varshney, G. K., Pei, W., Lafave, M. C., Idol, J., Xu, L., Gallardo, V., Carrington, B., Bishop, K., Jones, M. and Li, M. (2015). High-throughput gene targeting and phenotyping in zebrafish using CRISPR/Cas9. Genome Res. 25, 1030-1042. 10.1101/gr.186379.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visscher, P. M., Brown, M. A., Mccarthy, M. I. and Yang, J. (2012). Five years of GWAS discovery. Am. J. Hum. Genet. 90, 7-24. 10.1016/j.ajhg.2011.11.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walton, R. T., Christie, K. A., Whittaker, M. N. and Kleinstiver, B. P. (2020). Unconstrained genome targeting with near-PAMless engineered CRISPR-Cas9 variants. Science 368, 290-296. 10.1126/science.aba8853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, K., Tang, X., Liu, Y., Xie, Z., Zou, X., Li, M., Yuan, H., Ouyang, H., Jiao, H. and Pang, D. (2016). Efficient generation of orthologous point mutations in pigs via CRISPR-assisted ssODN-mediated homology-directed repair. Mol. Ther. Nucleic Acids 5, e396. 10.1038/mtna.2016.101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, R., Zhang, J.-Y., Lu, K.-H., Lu, S.-S. and Zhu, X.-X. (2019). Efficient generation of GHR knockout Bama minipig fibroblast cells using CRISPR/Cas9-mediated gene editing. In Vitro Cell. Dev. Biol. Anim. 55, 784-792. 10.1007/s11626-019-00397-6 [DOI] [PubMed] [Google Scholar]
- Wierson, W. A., Welker, J. M., Almeida, M. P., Mann, C. M., Webster, D. A., Torrie, M. E., Weiss, T. J., Kambakam, S., Vollbrecht, M. K., Lan, M.et al. (2020). Efficient targeted integration directed by short homology in zebrafish and mammalian cells. eLife 9, e53968. 10.7554/eLife.53968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wobbe, C. R., Weissbach, L., Borowiec, J. A., Dean, F. B., Murakami, Y., Bullock, P. and Hurwitz, J. (1987). Replication of simian virus 40 origin-containing DNA in vitro with purified proteins. Proc. Natl. Acad. Sci. USA 84, 1834-1838. 10.1073/pnas.84.7.1834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wold, M. S. (1997). REPLICATION PROTEIN A: a heterotrimeric, single-stranded DNA-binding protein required for eukaryotic DNA metabolism. Annu. Rev. Biochem. 66, 61-92. 10.1146/annurev.biochem.66.1.61 [DOI] [PubMed] [Google Scholar]
- Wong, N., Liu, W. and Wang, X. (2015). WU-CRISPR: characteristics of functional guide RNAs for the CRISPR/Cas9 system. Genome Biol. 16, 218. 10.1186/s13059-015-0784-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, Y., Liang, D., Wang, Y., Bai, M., Tang, W., Bao, S., Yan, Z., Li, D. and Li, J. (2013). Correction of a genetic disease in mouse via use of CRISPR-Cas9. Cell Stem Cell 13, 659-662. 10.1016/j.stem.2013.10.016 [DOI] [PubMed] [Google Scholar]
- Xia, B., Sheng, Q., Nakanishi, K., Ohashi, A., Wu, J., Christ, N., Liu, X., Jasin, M., Couch, F. J. and Livingston, D. M. (2006). Control of BRCA2 cellular and clinical functions by a nuclear partner, PALB2. Mol. Cell 22, 719-729. 10.1016/j.molcel.2006.05.022 [DOI] [PubMed] [Google Scholar]
- Xu, W., Zhang, C., Yang, Y., Zhao, S., Kang, G., He, X., Song, J. and Yang, J. (2020). Versatile nucleotides substitution in plant using an improved prime editing system. Mol. Plant 13, 675-678. 10.1016/j.molp.2020.03.012 [DOI] [PubMed] [Google Scholar]
- Yan, S., Tu, Z., Liu, Z., Fan, N., Yang, H., Yang, S., Yang, W., Zhao, Y., Ouyang, Z., Lai, C.et al. (2018). A huntingtin knockin pig model recapitulates features of selective neurodegeneration in Huntington's disease. Cell 173, 989-1002.e13. 10.1016/j.cell.2018.03.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, H., Wang, H., Shivalila, C. S., Cheng, A. W., Shi, L. and Jaenisch, R. (2013). One-step generation of mice carrying reporter and conditional alleles by CRISPR/Cas-mediated genome engineering. Cell 154, 1370-1379. 10.1016/j.cell.2013.08.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, D., Scavuzzo, M. A., Chmielowiec, J., Sharp, R., Bajic, A. and Borowiak, M. (2016). Enrichment of G2/M cell cycle phase in human pluripotent stem cells enhances HDR-mediated gene repair with customizable endonucleases. Sci. Rep. 6, 21264. 10.1038/srep21264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh, C. D., Richardson, C. D. and Corn, J. E. (2019). Advances in genome editing through control of DNA repair pathways. Nat. Cell Biol. 21, 1468-1478. 10.1038/s41556-019-0425-z [DOI] [PubMed] [Google Scholar]
- Yengo, L., Vedantam, S., Marouli, E., Sidorenko, J., Bartell, E., Sakaue, S., Graff, M., Eliasen, A. U., Jiang, Y., Raghavan, S.et al. (2022). A saturated map of common genetic variants associated with human height. Nature 610, 704-712. 10.1038/s41586-022-05275-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan, L., Sui, T., Chen, M., Deng, J., Huang, Y., Zeng, J., Lv, Q., Song, Y., Li, Z. and Lai, L. (2016). CRISPR/Cas9-mediated GJA8 knockout in rabbits recapitulates human congenital cataracts. Sci. Rep. 6, 22024. 10.1038/srep22024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yun, M. H. and Hiom, K. (2009). CtIP-BRCA1 modulates the choice of DNA double-strand-break repair pathway throughout the cell cycle. Nature 459, 460-463. 10.1038/nature07955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zetsche, B., Gootenberg, J. S., Abudayyeh, O. O., Slaymaker, I. M., Makarova, K. S., Essletzbichler, P., Volz, S. E., Joung, J., Van Der Oost, J., Regev, A.et al. (2015). Cpf1 is a single RNA-guided endonuclease of a class 2 CRISPR-Cas system. Cell 163, 759-771. 10.1016/j.cell.2015.09.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zetsche, B., Heidenreich, M., Mohanraju, P., Fedorova, I., Kneppers, J., Degennaro, E. M., Winblad, N., Choudhury, S. R., Abudayyeh, O. O., Gootenberg, J. S.et al. (2017). Multiplex gene editing by CRISPR–Cpf1 using a single crRNA array. Nat. Biotechnol. 35, 31-34. 10.1038/nbt.3737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, F., Fan, Q., Ren, K. and Andreassen, P. R. (2009). PALB2 functionally connects the breast cancer susceptibility proteins BRCA1 and BRCA2. Mol. Cancer Res. 7, 1110-1118. 10.1158/1541-7786.MCR-09-0123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, J.-P., Li, X.-L., Li, G.-H., Chen, W., Arakaki, C., Botimer, G. D., Baylink, D., Zhang, L., Wen, W., Fu, Y.-W.et al. (2017a). Efficient precise knockin with a double cut HDR donor after CRISPR/Cas9-mediated double-stranded DNA cleavage. Genome Biol. 18, 35. 10.1186/s13059-017-1164-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Y., Qin, W., Lu, X., Xu, J., Huang, H., Bai, H., Li, S. and Lin, S. (2017b). Programmable base editing of zebrafish genome using a modified CRISPR-Cas9 system. Nat. Commun. 8, 118. 10.1038/s41467-017-00175-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Y., Zhang, Z. and Ge, W. (2018). An efficient platform for generating somatic point mutations with germline transmission in the zebrafish by CRISPR/Cas9-mediated gene editing. J. Biol. Chem. 293, 6611-6622. 10.1074/jbc.RA117.001080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, X., Chen, L., Zhu, B., Wang, L., Chen, C., Hong, M., Huang, Y., Li, H., Han, H., Cai, B.et al. (2020). Increasing the efficiency and targeting range of cytidine base editors through fusion of a single-stranded DNA-binding protein domain. Nat. Cell Biol. 22, 740-750. 10.1038/s41556-020-0518-8 [DOI] [PubMed] [Google Scholar]
- Zhang, J.-P., Yang, Z.-X., Zhang, F., Fu, Y.-W., Dai, X.-Y., Wen, W., Zhang, B., Choi, H., Chen, W., Brown, M.et al. (2021). HDAC inhibitors improve CRISPR-mediated HDR editing efficiency in iPSCs. Sci. China Life Sci. 64, 1449-1462. 10.1007/s11427-020-1855-4 [DOI] [PubMed] [Google Scholar]
- Zhu, Q. M., Ko, K. A., Ture, S., Mastrangelo, M. A., Chen, M.-H., Johnson, A. D., O'donnell, C. J., Morrell, C. N., Miano, J. M. and Lowenstein, C. J. (2017). Novel thrombotic function of a human SNP in STXBP5 revealed by CRISPR/Cas9 gene editing in mice. Arterioscler. Thromb. Vasc. Biol. 37, 264-270. 10.1161/ATVBAHA.116.308614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zong, Y., Wang, Y., Li, C., Zhang, R., Chen, K., Ran, Y., Qiu, J.-L., Wang, D. and Gao, C. (2017). Precise base editing in rice, wheat and maize with a Cas9-cytidine deaminase fusion. Nat. Biotechnol. 35, 438-440. 10.1038/nbt.3811 [DOI] [PubMed] [Google Scholar]