Abstract
We previously reported a mouse monoclonal antibody (MAb), termed L2, specific for Helicobacter pylori urease strongly inhibited its enzymatic activity. Here, to gain insight into how this antibody affects urease activity, the epitope that was recognized by the antibody was determined. By screening a panel of overlapping synthetic peptides covering the entire sequence of the two subunits (UreA and UreB), we identified a stretch of UreB-derived 19 amino acid (aa) residues (UB-33; aa 321 to 339, CHHLDKSIKEDVQFADSRI) that was specifically recognized by the L2 antibody. Further sequential amino acid deletion of the 19-mer peptide from either end allowed us to determine the minimal epitope as 8 amino acid residues (F8; SIKEDVQF) for L2 reactivity. This epitope appears to lie exactly on a short sequence which formed a flap over the active site of urease, suggesting that binding of the L2 antibody sterically inhibits access of urea, the substrate of urease. Finally, immunization of rabbits with either the 19-mer peptide or the 8-mer minimal epitope resulted in generation of antiurease antibodies that were capable of inhibiting the enzymatic activity. Since urease is critical for virulence of H. pylori, antigenic peptides that induce production of antibodies to inhibit its enzymatic activity may potentially be a useful tool as a vaccine for prevention and treatment of H. pylori infection.
Helicobacter pylori, the curved bacilli discovered in the stomach of patients with gastritis and peptic ulceration by Marshall and Warren (20), has turned out to be the main cause of various gastroduodenal diseases. With the establishment of methods for identification of H. pylori infection, such as enzyme-linked immunosorbent assay (ELISA) for detecting specific antibodies in the serum (12) and the urea breath test (14), it has now been realized that more than half of the world's population have suffered from the infection (13, 22).
Nevertheless, a significant number of people infected with H. pylori do not develop gastroduodenal diseases and remain asymptomatic for a long period. The resistance may relate to the difference in H. pylori strains. Indeed, some strains have a very low virulence and fail to generate gastroduodenal disorders after infection (19). Alternatively, the prevention of disease development may correlate with host resistance controlled genetically by the immune system. In fact, Azuma et al. have reported that those who carry a particular HLA-DQA gene show resistance to H. pylori infection (1). Also, efficient induction of H. pylori-specific antibodies (6, 18) and T cells (7) may contribute to host resistance against H. pylori infection.
The H. pylori urease is a high-molecular-mass (550 kDa) multimeric enzyme composed of two distinct subunits, UreA (29.5 kDa) and UreB (66 kDa). Generally, UreA is called the small subunit and UreB is the large subunit. We have reported that purified H. pylori urease, especially UreB, is a major target for immune recognition in patients with H. pylori-induced gastroduodenal diseases among Japanese subjects (11) and H. pylori urease-specific serum immunoglobulin A (IgA) and IgG antibodies appear to reflect different stages of chronic gastritis, the surface inflammatory response and gastric atrophy, respectively (10). These findings suggest that H. pylori urease-specific humoral immune responses are associated with the progression of various gastroduodenal diseases caused by H. pylori infection, and H. pylori urease-specific antibodies may help to aggravate the gastric disorder. In contrast, urease itself seems to be an important virulence factor for H. pylori colonization (8) and elicits damage of gastric mucosa by inducing apoptosis of gastric epithelial cells expressing class II major histocompatibility complex (MHC) molecules (9). Thus, both H. pylori urease and some of its specific antibodies may potentially be unfavorable to the body.
Nonetheless, we have observed that an IgG monoclonal antibody (MAb) against H. pylori urease, termed L2, strongly inhibited its enzymatic activity, whereas urease-specific polyclonal IgG antibodies generated by immunization with purified urease protein did not induce this inhibitory effect at all (27). The former type of antibodies might neutralize the urease by inhibiting its enzymatic activity necessary for H. pylori to attach to and persist in the gastric mucosa. In addition, it has recently been reported that poor response to the urese may favor persistence of H. pylori infection, and the antiurease response might enhance clearance of bacteria (17). There might be two types of H. pylori urease-specific antibodies; one may help to induce the gastric disorder, and the other may be beneficial in preventing bacterial growth and attachment to the gastric mucosa.
In this study, using the H. pylori urease-specific murine MAb L2 having a strong capacity to neutralize the urease activity, we tried to identify the neutralizing epitope with a series of overlapping peptides covering the entire sequence of H. pylori urease to gain insight into how this antibody affects urease activity.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
H. pylori NCTC 11637 was cultured on modified Belo-Horizonte medium (pylori agar medium) (Nikken Bio Medical Lab., Kyoto, Japan) for 3 days at 37°C under a microaerophilic atmosphere (5% O2, 15% CO2, and 80% N2) by AnaeroPack Campylo (Mitsubishi Gas Chemical Co., Inc., Tokyo, Japan). A colony was inoculated into 20 ml of Brucella broth (Becton Dickinson, Cockeysville, Md.) containing 0.1% β-cyclodextrin (Wako Pure Chemical Industries, Ltd., Osaka, Japan) supplemented with 5% (vol/vol) horse serum in a 250-ml culture bottle, and cultured under the same conditions. After incubation for 24 h with shaking (100 rpm) at 37°C, 2 ml of culture was transferred to a 250-ml culture bottle containing 40 ml of fresh medium, and the cells were reincubated twice under the same conditions. One milliliter of the incubated medium containing the cells, which were mostly spiral rather than coccoid, was plated on Brucella agar (Becton Dickinson) containing 7% (vol/vol) defibrinated horse blood and cultured for an additional 3 days at 37°C in a microaerophilic atmosphere. Bacterial cells were harvested and washed twice with cold phosphate-buffered saline (PBS) at pH 7.0. Then the cells were sedimented by centrifugation (5,000 × g for 10 min at 4°C), and the cell pellet was stored at −80°C.
Purification of H. pylori urease.
To obtain purified H. pylori urease, the stored cell pellet was thawed, resuspended in 10% n-octyl β-d-glucopyranoside (Wako Pure Chemical Industries, Ltd.), vortexed five times for 10 s each, and then centrifuged (10,000 × g for 20 min at 4°C). dl-Dithiothreitol (DTT) (final, 0.1 mM) (Sigma Chemical Co., St. Louis, Mo.) was added to the supernatant, and the mixture was applied to a PD-10 column (Sephadex G-25) (Pharmacia Biotech AB, Uppsala, Sweden), and the elute was dialyzed against PBS containing DTT. The elute was applied to a column (Shoei Works Co., Ltd., Tokyo, Japan) with Affi-Gel Hz hydrazide gel (Bio-Rad Laboratories, Tokyo, Japan) coupled to D12 (S3) (H. pylori urease small-subunit-specific MAb) (27) overnight in a cold room. Then the column was washed with PBS containing DTT, and elution was performed with 5 M urea in 0.1 M potassium phosphate buffer (pH 6.5) containing 0.1 mM DTT. The eluate was dialyzed with the PD-10 column. The amount of protein was determined by Lowry's procedure.
Measurement of H. pylori urease enzymatic activity.
The urease was prepared from various fractions of bacteria: the urease fraction, purified by column chromatography; the distilled water-extracted fraction of intact cells; and the centrifuged supernatant (10,000 × g for 30 min at 4°C) fraction from cells disrupted by sonication. The urease fraction (25 μl) was incubated with either the IgG fraction of the ascitic fluid containing each MAb or the IgG fraction of polyclonal anti-H. pylori urease antibodies (25 μl in PBS) overnight at 4°C in 96-well microtiter plates. On the following day, 100 μl of 50 mM phosphate buffer (pH 6.8) containing 500 mM urea, 0.02% phenol red, and 0.1 mM DTT, was added to each well. The color development was monitored at 550 nm with a microplate reader (model 3550; Bio-Rad) over a 3-h incubation period at 23°C, and the rates of development were calculated from the linear portions of the curves. Urease activity was estimated with jack bean urease as a standard, the specific activity of which was 33,600 U/g, dry weight. One unit is equivalent to 1 μmol of ammonia liberated per min.
Urease inhibition (neutralizing) test using specific antibodies.
The H. pylori urease inhibition test was carried out similar to the urease assay described above. In brief, 25 μl of H. pylori urease was incubated with either 25 μl of urease-specific MAb or IgG purified from immune rabbit serum (equivalent to 0 to 25 μg of IgG protein) in 96-well microtiter plates overnight at 4°C, and then 50 μl of a 50 mM phosphate buffer (pH 6.8) containing 500 mM urea, 0.02% phenol red, and 0.1 mM DTT was added to each well. The color development was measured at 550 nm. Percent inhibition was determined by the following equation: [(activity without Ab − activity with Ab)/(activity without Ab)] × 100.
Peptide synthesis.
As shown in Fig. 1, 23 sets (UA-1 through UA-23) of 20-residue peptides (overlapping by 10 amino acids) covering residues 1 to 238 of UreA and 56 sets (UB-1 through UB-56) of 19 residue peptides (overlapping by 10 amino acids) spanning residues 1 to 569 of UreB were synthesized by the solid-phase method of Merrifield (23) as modified by Corley et al. (4). In addition, as indicated in Table 1, a series of 18 truncated peptides originating from UB-33, UB-33(10R) through UB-33(18R) and UB-33(10L) through UB-33(18L) were also synthesized to determine the minimum epitope in UB-33 by the same procedure. In brief, the tert-butyloxycarbonyl (t-Boc)-protected amino acids were obtained with a model 430 multipeptide synthesizer (Applied Biosystems, Foster City, Calif.). Each peptide was partially deblocked and cleaved from the resin in liquid anhydrous HF. All synthetic peptides were purified and isolated by reverse-phase high-pressure liquid chromatography (RP-HPLC) performed with a liquid chromatograph (Waters, Milford, Mass.). The purity and molar concentration were analyzed by RP-HPLC on a C18 column using a gradient of 0.1% trifluoroacetic acid (TFA) in water–0.1% TFA in acetonitrile and further purified on a Bio-Gel P4 gel filtration column (Bio-Rad) in 9% formic acid. Identification and concentration were confirmed by amino acid analysis.
FIG. 1.
Epitope mapping of the H. pylori urease-specific MAb (L2). A series of overlapping 20-residue peptides of H. pylori urease (UA-1 to UA-23) corresponding to the small subunit (UreA) and 19-residue peptides of H. pylori urease (UB-1 to UB-56) to the large subunit (UreB) were tested for their recognition by the L2 antibody, using the ELISA method. Note that the L2 reactivity was detected to the purified H. pylori urease (positive) and to the UB-33 peptide derived from UreB, but not to the other synthetic peptides analyzed.
TABLE 1.
Truncated and substituted synthetic peptides of UB-33
| Peptide | Residues | Sequencea | OD405b
|
||
|---|---|---|---|---|---|
| L2 | UB33-KLH rabbit IgG | F8-MAP rabbit IgG | |||
| UB-33(10L) | 321–330 | CHHLD KSIKE | − | − | − |
| UB-33(11L) | 321–331 | CHHLD KSIKE D | − | − | ± |
| UB-33(12L) | 321–332 | CHHLD KSIKE DV | − | − | +++ |
| UB-33(13L) | 321–333 | CHHLD KSIKE DVQ | − | − | +++ |
| UB-33(14L) | 321–334 | CHHLD KSIKE DVQF | +++ | +++ | +++ |
| UB-33(15L) | 321–335 | CHHLD KSIKE DVQFA | +++ | +++ | +++ |
| UB-33(16L) | 321–336 | CHHLD KSIKE DVQFA D | +++ | +++ | +++ |
| UB-33(17L) | 321–337 | CHHLD KSIKE DVQFA DS | +++ | +++ | +++ |
| UB-33(18L) | 321–338 | CHHLD KSIKE DVQFA DSR | +++ | +++ | +++ |
| UB-33 | 321–339 | CHHLD KSIKE DVQFA DSRI | +++ | +++ | +++ |
| UB-33(18R) | 322–339 | HHLD KSIKE DVQFA DSRI | +++ | +++ | +++ |
| UB-33(17R) | 323–339 | HLD KSIKE DVQFA DSRI | +++ | +++ | +++ |
| UB-33(16R) | 324–339 | LD KSIKE DVQFA DSRI | +++ | +++ | +++ |
| UB-33(15R) | 325–339 | D KSIKE DVQFA DSRI | +++ | +++ | ++ |
| UB-33(14R) | 326–339 | KSIKE DVQFA DSRI | +++ | +++ | ++ |
| UB-33(13R) | 327–339 | SIKE DVQFA DSRI | +++ | +++ | + |
| UB-33(12R) | 328–339 | IKE DVQFA DSRI | ± | +++ | − |
| UB-33(11R) | 329–339 | KE DVQFA DSRI | − | +++ | − |
| UB-33(10R) | 330–339 | E DVQFA DSRI | − | + | − |
| UB-33(327E) | CHHLD KEIKE DVQFA DSRI | +++ | +++ | ++ | |
| UB-33(329P) | CHHLD KSIPE DVQFA DSRI | − | +++ | − | |
| UB-33(332L) | CHHLD KSIKE DLQFA DSRI | +++ | +++ | ++ | |
| UB-33(333A) | CHHLD KSIKE DVAFA DSRI | +++ | +++ | +++ | |
| UB-33(336H) | CHHLD KSIKE DVQFA HSRI | +++ | +++ | ++ | |
| UB-33 (P. mirabilis) | CHHLD PSIPE DVAFA ESRI | − | +++ | − | |
| UB-33 (jack bean) | CHHLD REIPE DLAFA HSRI | − | ++ | − | |
| UB-33 (H. felis) | CHHLD KSIKE DVQFA DSRI | +++ | +++ | +++ | |
The substituted amino acid is underlined. A critical amino acid (329K) in the minimal epitope (SIKEDVQF) for neutralizing antibody L2 among UB-33 substituted peptides is indicated in bold.
OD405, optical density at 405 nm. Symbols: −, 0.0 to 0.5; ±, 0.5 to 1.0; +, 1.0 to 1.5; ++, 1.5 to 2.0; +++, >2.0.
ELISA.
Each series of nested peptides or the purified H. pylori urease antigen was diluted in 0.1 mM carbonate buffer (pH 9.6) to yield the optimal protein concentration of 10 μg/ml. A 50-μl aliquot of each peptide solution was added to wells of flat-bottomed Immulon 2 plates (Dynatech Laboratories Inc., Alexandria, Va.), incubated for 60 min at 37°C, and washed with PBS three times. After overnight blocking with 25% Block-Ace (Dainihon Seiyaku, Osaka, Japan), a 50-μl aliquot of an appropriate dilution of the anti-H. pylori urease antibody (sequentially diluted at 1:100 to 1:300) was plated for 60 min at 37°C. After washing three times with 10% Block-Ace containing 0.05% Tween 20, a 50-μl aliquot of an appropriately diluted class-specific (IgG) alkaline phosphatase-conjugated goat anti-mouse immunoglobulin (Cappel, Durham, N.C.) or alkaline phosphatase-conjugated goat anti-rabbit immunoglobulin (Cappel) was added for another 60 min at 37°C. The activity of alkaline phosphatase bound to the well was determined by measuring the hydrolysis of p-nitrophenylphosphate (Sigma Chemical Co.) to the yellow product, p-nitrophenolate, which was quantitated by absorbance at 405 nm with a microplate reader.
Rabbit immunization protocols.
A synthetic epitope peptide, UB-33 (CHHLDKSIKEDVQFADSRI), from H. pylori urease was synthesized by solid-phase peptide synthesis as described above. Then, keyhole limpet hemocyanin (KLH) was conjugated to the N-terminal cystein of UB-33 by the Maleimido method (16) using meta-maleimidobenzyl N-hydroxysuccinimide ester (MBS) as a coupling reagent. Preimmune sera (negative control) were obtained from three rabbits, which were then immunized subcutaneously (s.c.) with 1 mg of UB-33-KLH dissolved in PBS emulsified 1:1 in complete Freund's adjuvant (CFA) in a total volume of 1 ml per animal. The rabbits were bled once (prebooster), and then they were given boosters in the same manner, except that CFA was changed to incomplete Freund's adjuvant (IFA), at 2-week intervals. The immune rabbits were bled every 7 days after the booster immunization, and each serum sample was tested for H. pylori urease-specific antibody activity by ELISA. Similarly, another three sets of rabbits (each set composed of three rabbits) were immunized with either 1 mg of purified H. pylori urease, KLH-conjugated minimum epitope (F8; SIKEDVQF) (F8-KLH), or F8-based multiple antigenic peptides (MAP) (29) composed of eight residues of F8 (F8-MAP) in a unit. In this unit, first, two F8 residues were conjugated to the C terminus with lysine (K) having two basic amines (NH2) as binders to make F8-K-F8, and then two sets of F8-K-F8 were further conjugated with a similar procedure using lysine as a binder to make a four-F8 unit. Finally, an octabranching F8-MAP consisting of a core matrix made up of three levels of lysine and eight N-terminals for anchoring F8 peptides.
All animal experiments were performed according to the guideline of the NIH Guide for the Care and Use of Laboratory Animals and approved by the Review Board of Nippon Medical School.
RESULTS
Mapping of neutralization epitope identified with H. pylori urease-specific MAb L2.
We have previously reported that we happened to obtain a murine hybridoma specific for H. pylori urease, termed L2, that could significantly inhibit the urease activity (27), which we have confirmed again in the present study (Fig. 2, indicated as open circles). Using a series of overlapping peptides covering the entire sequence of H. pylori urease, we tried to identify the neutralizing epitope by ELISA. The L2 reactivity to each peptide fragment derived from UreA and UreB is shown in Fig. 1. Among 79 overlapping peptides tested, only 1 showed a significant response to L2. The epitopic peptide was identified in the large subunit of H. pylori urease, UreB, as a UB-33 (aa 321 to 339; CHHLDKSIKEDVQFADSRI [28a]) that showed a predominant response, and the titer was almost equal to that against the whole purified H. pylori urease (positive control).
FIG. 2.
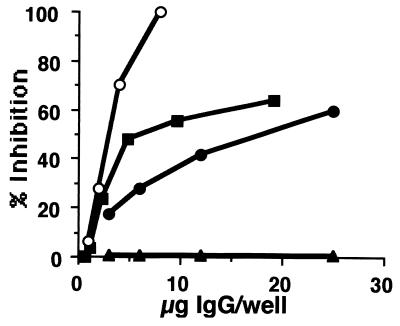
Inhibition of H. pylori urease enzymatic activity by specific antibodies. The distilled water-extracted urease fraction from NCTC 11637 and various concentrations of antibodies (0 to 25 μg of IgG protein/well) purified by staphylococcal protein A binding affinity column: anti-H. pylori urease-specific mouse MAb (L2) (○), anti-F8-MAP-specific rabbit IgG Abs (▪), anti-UB-33-specific rabbit IgG Abs (●), or anti-H. pylori urease-specific rabbit IgG Abs (▴) were preincubated overnight at 4°C on 96-well microplates, and phosphate buffer containing urea, phenol red, and DTT was added. Optical density at 550 nm was measured and percent inhibition was calculated as described in the text.
Induction of linear epitope (UB-33)-specific antibody by immunization with UB-33-KLH.
Then we tried to confirm whether the UB-33 (linear epitope)-specific antibody could actually inhibit (neutralize) H. pylori urease activity. We made a carrier protein (KLH)-conjugated UB-33 (UB-33-KLH) and immunized three rabbits with that UB-33-KLH together with CFA to obtain UB-33-specific antibody. The mixture of sera from three rabbits was purified into IgG with an affinity column, and its specific response was measured by ELISA. The obtained rabbit IgG antibody was specific for the linear peptide UB-33 and does not cross-react with the neighboring peptides UB-28 to UB-35 (data not shown), suggesting that the UB-33-derived antibody may respond to its central portion.
Capacity of UB-33-specific rabbit IgG antibody to inhibit (neutralize) H. pylori urease activity.
Based on the inhibition assay described in the Materials and Methods section, we examined whether the obtained UB-33-specific rabbit IgG antibody had the capacity to inhibit H. pylori urease activity. As demonstrated in Fig. 2, in comparison with purified IgG from the mixed sera of three rabbits immunized with purified H. pylori urease, the rabbit UB-33-specific IgG antibody showed a significant inhibition of H. pylori urease activity. As also shown in Fig. 2, less than 10 μg of IgG/well of L2 MAb induced complete inhibition of the H. pylori urease activity, whereas around 25 μg of IgG/well of UB-33-specific rabbit antibody showed 60% inhibition, a value obtained with approximately 3 μg of IgG/well of L2. Thus, the inhibitory capacity of L2 appears to be 8 to 9 times greater than that of UB-33-specific rabbit IgG antibody.
Comparison of minimum antigenic epitope between L2 and UB-33-specific rabbit IgG antibody.
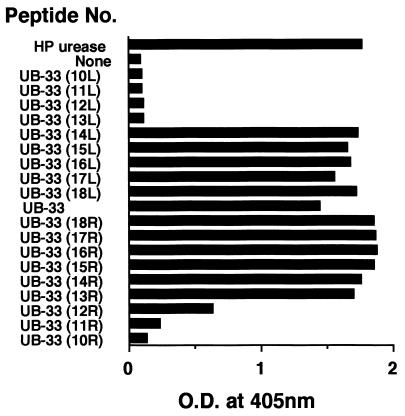
We tried to compare the minimum antigenic epitope(s) for both L2 and UB-33-specific rabbit IgG antibody using a series of truncated peptides (Table 1). As shown in Fig. 3 as well as in Table 1, since both UB-33(12R) (IKEDVQFADSRI) and UB-33(13L) (CHHLDKSIKEDVQ) significantly reduced the L2 response, the minimum antigenic epitope for L2 was identified as an 8-mer peptide, SIKEDVQF (UB-33-F8) within UB-33 (CHHLDKSIKEDVQFADSRI, shown in italics). In contrast, because the UB-33-specific rabbit polyclonal IgG did respond to both UB-33(12R) and UB-33(11R) (KEDVQFADSRI) but weakly to UB-33(10R) (EDVQFADSRI), the minimum antigenic epitope among the rabbit polyclonal antibodies was a 5- or 6-mer peptide, (K)EDVQF. However, these polyclonal antibodies also seem to contain UB-33-F8-specific MAb to some extent that might elicit L2-like inhibition shown in Fig. 2. To examine whether the 8-mer peptide can induce neutralizing antibody production, we made a KLH-conjugated L2 epitope (F8-KLH; SIKEDVQF-KLH) and immunized rabbits with it. However, although we performed the rabbit immunization protocol several times, we failed to obtain an epitope-specific antibody response at all (data not shown). This may be because the minimal L2 epitope (UB-33-F8) is far less immunogenic than UB-33 and B-cell responses were directed predominantly to the carrier protein KLH.
FIG. 3.
Identification of minimal antigenic epitope for L2 using a series of truncated peptides shown in Table 1. A series of 18 truncated peptides originated from UB-33 were synthesized, and the L2-specific antibody titer was measured by ELISA. The minimal epitope of L2 in UB-33 was determined to be an 8-mer peptide (UB-33-F8: SIKEDVQF).
Induction of minimal epitope-specific antibody by F8-MAP peptide.
To induce antibodies specific for minimal epitope (UB-33-F8), we then tried to immunize rabbits with F8-based MAP composed of 8 residues of F8 (F8-MAP) in a unit using lysine (K) as binders (see Materials and Methods). Because the generated rabbit IgG showed clear UB-33 specificity, we tried to examine the capacity to inhibit H. pylori urease activity. As also shown in Fig. 2, rabbit IgG fraction purified from F8-MAP-immunized rabbits inhibited the urease activity better than UB-33-KLH-derived IgG. Thus, we tried to identify the minimal epitope of that antibody using similar sets of truncated peptides. As demonstrated in Table 1, the minimal epitope of F8-MAP-elicited antibodies was determined to be SIKEDV. However, Table 1 also indicated that the F8-MAP-induced polyclonal rabbit IgG antibodies also seem to contain F8 (SIKEDVQF)-specific MAbs that might elicit L2-like inhibition shown in Fig. 2.
Cross-reactivity to ureases of other species and peptide corresponding to UB-33 of each urease.
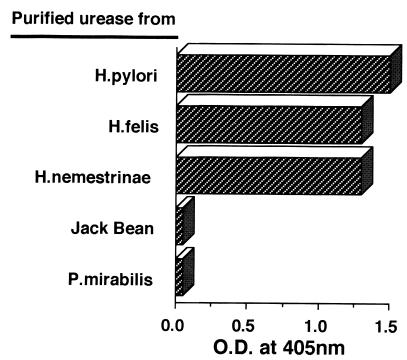
We further studied whether UB-33-specific antibody might cross-react with the ureases of other species using L2. As shown in Fig. 4, although L2 did cross-react with purified ureases of the Helicobacter species H. pylori, H. felis, and H. nemestrinae, the MAb did not cross-react with ureases of Proteus mirabillis or jack bean. Thus, we synthesized peptides from the ureases corresponding to the UB-33 portion to examine the cross-reactivity using three distinct antibodies, such as L2, UB-33-specific IgG, and F8-MAP IgG. As indicated in Table 1, although L2 and F8-MAP IgG did not cross-react, the urease-derived peptides from jack bean and P. mirabilis, UB-33-specific IgG did respond. Therefore, we synthesized a series of peptides with a single amino acid substitution in the epitope of UB-33 corresponding to the jack bean urease (327E, 329P, 332L, 333A, and 336H) to determine the critical amino acid(s) for the response. The results clearly demonstrated that both L2- and UB-33-specific antibodies cross-reacted with 327E, 332L, 333A, and 336H, while L2 and F8-MAP antibodies did not respond to 329P at all, though the UB-33-specific antibody did cross-react with that substituted peptide. These results strongly indicated the importance of 329K for F8-specific recognition, and 329K is not a critical amino acid for the recognition of UB-33-KLH-induced antibodies.
FIG. 4.
To examine whether H. pylori urease-specific MAb L2 has the capacity to cross-react with purified ureases from other species in addition to the H. pylori urease, purified ureases of H. felis, H. nemestrinae, Proteus mirabilis, and jack bean were obtained by the same purification method and used as antigenic molecules for ELISA. As demonstrated in the figure, L2 did cross-react with purified ureases of H. pylori, H. felis, and H. nemestrinae, whereas the MAb did not cross-react with ureases of P. mirabilis and jack bean.
Response of purified H. pylori urease immune sera to epitope UB-33.
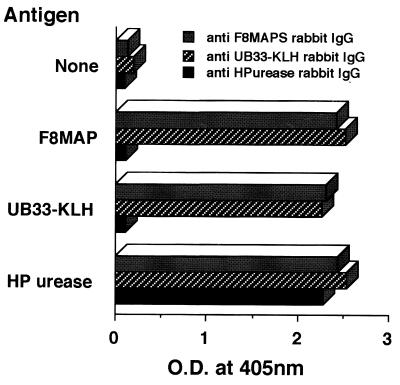
Finally, we tried to confirm whether purified H. pylori urease immune sera could respond to the neutralization epitope UB-33. The result indicates that the mixed IgG antibodies from the sera of three rabbits immunized with purified H. pylori urease did not respond to UB-33 at all, though it showed the strong response to purified H. pylori urease at almost the same level as UB-33-KLH- or F8-MAP-immunized rabbit IgG antibodies (Fig. 5). In addition, as far as we have investigated, we could not find any specific linear epitope recognized by that immune serum (data not shown). Thus, when we use purified urease as an immunogen, we might obtain antibodies mostly specific for conformational structure of the H. pylori urease.
FIG. 5.
Antibodies from purified H. pylori urease immune sera did not respond to linear UB-33 and F8 peptides. Diluted antigens (F8-MAP, UB-33, or H. pylori [HP]urease) were coated onto 96-well ELISA plates overnight with blocking, and then 50 μl of each sample (diluted 1:20) (anti-F8-MAP rabbit IgG, anti-UB-33-KLH rabbit IgG, anti-H. pylori urease rabbit IgG) was added. After washing, either alkaline phosphatase-conjugated goat anti-mouse immunoglobulin or goat anti-rabbit immunoglobulin was added, and optical density (O.D.) was measured at 405 nm by ELISA.
DISCUSSION
In this paper, we demonstrated that there are two types of IgG antibodies against H. pylori urease. One is conformational structure specific and the other is linear epitope (UB-33) specific. Our present observation indicates that dominant production of the former antibodies that have no effect on H. pylori urease activity when the whole purified urease was used as the immunogen. In contrast, when the linear epitope was used as the immunogen, we could obtain a good amount of the latter epitope-specific antibodies, which inhibit the enzymatic activity of H. pylori urease.
Indeed, Mooney and collegues demonstrated that inhibitors for the urease abrogated H. pylori persistence in the gastric mucosa and prevent infection (25). To date, there are several reports on H. pylori vaccination using H. pylori urease (24) or its B subunit as immunogens (5). However, the present study in an animal model suggests that the whole H. pylori urease seems to generate insufficient immunity that might not abolish the enzymatic activity. If H. pylori urease is an actual target for making an effective vaccine, it is better to elicit antibodies that abrogate urease activity. Therefore, we would like to recommend that the epitope UB-33, in particular the minimal 8-mer peptide SIKEDVQF, be included to make a vaccine that prevents H. pylori infection as well as persistence.
In the present study, we synthesized a series of overlapping peptides covering the entire sequence of H. pylori urease and tried to identify the epitope of a neutralizing antibody specific for the urease and found the linear epitope UB-33 (residues 321 to 339; CHHLDKSIKEDVQFADSRI). As reported by Regenmortel et al., in general most MAbs for the intact protein tend to be directed against the conformational discontinuous epitopes, and few antibodies are generated against the linear epitope(s) (30). Our results are consistent with their observation in that only one MAb (L2) recognizing a linear epitope could be generated among plenty of MAbs, as we have shown previously (27), that did not recognize any synthesized peptides (data not shown). There are reports that the MAbs can recognize a short linear epitope (21, 28) and the linear epitope-specific antibodies have the capacity to neutralize the antigens. In addition, Bittle et al. reported that neutralizing antibody induced by synthetic peptide responds much more strongly to the antigen particles containing the liner epitope (2). Thus, one might speculate that the antibodies against the linear epitope seem to have greater ability to neutralize the antigens than the antibodies against discontinuous determinants.
It is interesting that the 8-mer epitope, SIKEDVQF, is also involved in the region of H2-H4 flap among the large subunit from Klebsiella aerogenes urease (UreC) (26), which corresponds to UreB in H. pylori (3). Based on the recent report of Jabri et al. on determining actual structure for UreC (15), the H2-H4 flap region appears to be highly related to the active site of urease. Moreover, Ha et al. just reported the crystal structure of H. pylori urease and that its characteristic flap structure contributed to the active site (14a). The flap structure is composed of amino acid residues 313 to 346 of the UreB subunit, forming a helix-turn-helix motif and showing unique flap motion. As expected, the 8-mer epitope, SIKEDVQF (aa 327 to 334 of UreB), is involved in the central core region of the turn-helix of this flap structure and formed the active site of H. pylori urease. Thus, L2 may fit to the flap region and consequently inhibit enzymatic activity. The amino acid residues corresponding to the flap were highly conserved among ureases from H. pylori, Klebsiella aerogenes, and Bacillus pasteurii except for a few amino acids in the turn-helix (14a). It will be interesting to examine the inhibitory activity of L2 against the ureases of K. aerogenes and B. pasteurii.
The findings shown here offer important information for making a vaccine that neutralizes H. pylori urease in preventing both H. pylori infection as well as its persistence in the stomach.
ACKNOWLEDGMENTS
We thank Kazushi Ohhashi, Masao Katsumata, and Toshiki Nishizawa for assistance with this work and Masahiko Sugita for critical reading of the manuscript.
This work was supported in part by grants from the Ministry of Education, Science, Sport, and Culture, from the Ministry of Health and Labor and Welfare, and CREST, JST, Japan.
REFERENCES
- 1.Azuma T, Konishi J, Tanaka Y, Hirai M, Ito S, Kato T, Kohli Y. Contribution of HLA-DQA gene to host's response against Helicobacter pylori. Lancet. 1994;343:542–543. doi: 10.1016/s0140-6736(94)91496-6. [DOI] [PubMed] [Google Scholar]
- 2.Bittle J L, Houghten R A, Alexander H, Shinnick T M, Sutcliffe J G, Lerner R A, Rowlands D J, Brown F. Protection against foot-and-mouth disease by immunization with a chemically synthesized peptide predicted from the viral nucleotide sequence. Nature. 1982;298:30–33. doi: 10.1038/298030a0. [DOI] [PubMed] [Google Scholar]
- 3.Clayton C L, Pallen M J, Kleanthous H, Wren B W, Tabaqchali S. Nucleotide sequence of two genes from Helicobacter pylori encoding for urease subunits. Nucleic Acids Res. 1990;18:362. doi: 10.1093/nar/18.2.362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Corley L, Sachs D H, Anfinsen C B. Rapid solid-phase synthesis of bradykinin. Biochem Biophys Res Commun. 1972;47:1353–1359. doi: 10.1016/0006-291x(72)90221-5. [DOI] [PubMed] [Google Scholar]
- 5.Corthesy-Theulaz I, Porta N, Glauser M, Saraga E, Vaney A C, Haas R, Kraehenbuhl J P, Blum A L, Michetti P. Oral immunization with Helicobacter pylori urease B subunit as a treatment against Helicobacter infection in mice. Gastroenterology. 1995;109:115–121. doi: 10.1016/0016-5085(95)90275-9. [DOI] [PubMed] [Google Scholar]
- 6.Czinn S J, Nedrud J G. Oral immunization against Helicobacter pylori. Infect Immun. 1991;59:2359–2363. doi: 10.1128/iai.59.7.2359-2363.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.D'Elios M M, Manghetti M, Almerigogna F, Amedei A, Costa F, Burroni D, Baldari C T, Romagnani S, Telford J L, Del Prete G. Different cytokine profile and antigen-specificity repertoire in Helicobacter pylori-specific T cell clones from the antrum of chronic gastritis patients with or without peptic ulcer. Eur J Immunol. 1997;27:1751–1755. doi: 10.1002/eji.1830270723. [DOI] [PubMed] [Google Scholar]
- 8.Eaton K A, Brooks C L, Morgan D R, Krakowka S. Essential role of urease in pathogenesis of gastritis induced by Helicobacter pylori in gnotobiotic piglets. Infect Immun. 1991;59:2470–2475. doi: 10.1128/iai.59.7.2470-2475.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fan X, Gunasena H, Cheng Z, Espejo R, Crowe S E, Ernst P B, Reyes V E. Helicobacter pylori urease binds to class II MHC on gastric epithelial cells and induces their apoptosis. J Immunol. 2000;165:1918–1924. doi: 10.4049/jimmunol.165.4.1918. [DOI] [PubMed] [Google Scholar]
- 10.Futagami S, Takahashi H, Norose Y, Kobayashi M. Systemic and local immune responses against Helicobacter pylori urease in patients with chronic gastritis: distinct IgA and IgG productive sites. Gut. 1998;43:168–175. doi: 10.1136/gut.43.2.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Futagami S, Takahashi H, Norose Y, Nagata K, Kobayashi M, Nomura T. Analysis of immune response to Helicobacter pylori; identification of the protein recognized by anti-Helicobacter pylori antibodies from sera of patients with gastroduodenal diseases. Jpn J Gastroenterol. 1994;91:2202–2213. [PubMed] [Google Scholar]
- 12.Goodwin C S, Blincow E, Peterson G, Sanderson C, Cheng W, Marshall B, Warren J R, McCulloch R. Enzyme-linked immunosorbent assay for Campylobacter pyloridis: correlation with presence of C. pyloridis in the gastric mucosa. J Infect Dis. 1987;155:488–494. doi: 10.1093/infdis/155.3.488. [DOI] [PubMed] [Google Scholar]
- 13.Graham D Y, Adam E, Reddy G T, Agarwal J P, Agarwal R, Evans D J, Jr, Malaty H M, Evans D G. Seroepidemiology of Helicobacter pylori infection in India. Comparison of developing and developed countries. Dig Dis Sci. 1991;36:1084–1088. doi: 10.1007/BF01297451. [DOI] [PubMed] [Google Scholar]
- 14.Graham D Y, Klein P D, Evans D J, Jr, Evans D G, Alpert L C, Opekun A R, Boutton T W. Campylobacter pylori detected noninvasively by the 13C-urea breath test. Lancet. 1987;i:1174–1177. doi: 10.1016/s0140-6736(87)92145-3. [DOI] [PubMed] [Google Scholar]
- 14a.Ha N-C, Oh S T, Sung J Y, Cha K A, Lee M H, Oh B-H. Supramolecular assembly and acid resistance of Helicobacter pylori urease. Nat Struct Biol. 2001;8:505–509. doi: 10.1038/88563. [DOI] [PubMed] [Google Scholar]
- 15.Jabri E, Carr M B, Hausinger R P, Karplus P A. The crystal structure of urease from Klebsiella aerogenes. Science. 1995;268:998–1004. [PubMed] [Google Scholar]
- 16.Kitagawa T, Aikawa T. Enzyme coupled immunoassay of insulin using a novel coupling reagent. J Biochem (Tokyo) 1976;79:233–236. doi: 10.1093/oxfordjournals.jbchem.a131053. [DOI] [PubMed] [Google Scholar]
- 17.Leal-Herrera Y, Torres J, Perez-Perez G, Gomez A, Monath T, Tapia-Conyer R, Munoz O. Serologic IgG response to urease in Helicobacter pylori-infected persons from Mexico. Am J Trop Med Hyg. 1999;60:587–592. doi: 10.4269/ajtmh.1999.60.587. [DOI] [PubMed] [Google Scholar]
- 18.Lee C K, Weltzin R, Thomas W D, Jr, Kleanthous H, Ermak T H, Soman G, Hill J E, Ackerman S K, Monath T P. Oral immunization with recombinant Helicobacter pylori urease induces secretory IgA antibodies and protects mice from challenge with Helicobacter felis. J Infect Dis. 1995;172:161–172. doi: 10.1093/infdis/172.1.161. [DOI] [PubMed] [Google Scholar]
- 19.Marchetti M, Arico B, Burroni D, Figura N, Rappuoli R, Ghiara P. Development of a mouse model of Helicobacter pylori infection that mimics human disease. Science. 1995;267:1655–1658. doi: 10.1126/science.7886456. [DOI] [PubMed] [Google Scholar]
- 20.Marshall B J, Warren J R. Unidentified curved bacilli in the stomach of patients with gastritis and peptic ulceration. Lancet. 1984;i:1311–1315. doi: 10.1016/s0140-6736(84)91816-6. [DOI] [PubMed] [Google Scholar]
- 21.Matsushita S, Robert-Guroff M, Rusche J, Koito A, Hattori T, Hoshino H, Javaherian K, Takatsuki K, Putney S. Characterization of a human immunodeficiency virus neutralizing monoclonal antibody and mapping of the neutralizing epitope. J Virol. 1988;62:2107–2114. doi: 10.1128/jvi.62.6.2107-2114.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Megraud F, Brassens-Rabbe M P, Denis F, Belbouri A, Hoa D Q. Seroepidemiology of Campylobacter pylori infection in various populations. J Clin Microbiol. 1989;27:1870–1873. doi: 10.1128/jcm.27.8.1870-1873.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Merrifield R B. Automated synthesis of peptides. Science. 1965;150:178–185. doi: 10.1126/science.150.3693.178. [DOI] [PubMed] [Google Scholar]
- 24.Michetti P, Corthesy-Theulaz I, Davin C, Haas R, Vaney A C, Heitz M, Bille J, Kraehenbuhl J P, Saraga E, Blum A L. Immunization of BALB/c mice against Helicobacter felis infection with Helicobacter pylori urease. Gastroenterology. 1994;107:1002–1011. doi: 10.1016/0016-5085(94)90224-0. [DOI] [PubMed] [Google Scholar]
- 25.Mooney C, Munster D J, Bagshaw P F, Allardyce R A. Helicobacter pylori acid resistance. Lancet. 1990;335:1232. doi: 10.1016/0140-6736(90)92764-9. [DOI] [PubMed] [Google Scholar]
- 26.Mulrooney S B, Hausinger R P. Sequence of the Klebsiella aerogenes urease genes and evidence for accessory proteins facilitating nickel incorporation. J Bacteriol. 1990;172:5837–5843. doi: 10.1128/jb.172.10.5837-5843.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nagata K, Mizuta T, Tonokatu Y, Fukuda Y, Okamura H, Hayashi T, Shimoyama T, Tamura T. Monoclonal antibodies against the native urease of Helicobacter pylori: synergistic inhibition of urease activity by monoclonal antibody combinations. Infect Immun. 1992;60:4826–4831. doi: 10.1128/iai.60.11.4826-4831.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Roivainen M, Narvanen A, Korkolainen M, Huhtala M L, Hovi T. Antigenic regions of poliovirus type 3/Sabin capsid proteins recognized by human sera in the peptide scanning technique. Virology. 1991;180:99–107. doi: 10.1016/0042-6822(91)90013-2. [DOI] [PubMed] [Google Scholar]
- 28a.Takahashi, H. November 1999. Japanese patent 3007037.
- 29.Tam J P. Synthetic peptide vaccine design: synthesis and properties of a high-density multiple antigenic peptide system. Proc Natl Acad Sci USA. 1988;85:5409–5413. doi: 10.1073/pnas.85.15.5409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Westhof E, Altschuh D, Moras D, Bloomer A C, Mondragon A, Klug A, Van Regenmortel M H. Correlation between segmental mobility and the location of antigenic determinants in proteins. Nature. 1984;311:123–126. doi: 10.1038/311123a0. [DOI] [PubMed] [Google Scholar]