Abstract
The 987P fimbriae of enterotoxigenic strains of Escherichia coli bind to both glycoprotein and glycolipid receptors on the brush borders of piglet enterocytes. A mutation in lysine residue 117 of the adhesive subunit FasG [fasG(K117A)] previously shown to abrogate 987P binding to the lipid receptor sulfatide did not affect the interaction with the glycoprotein receptors. Both the fimbriae and the FasG subunits of the wild type and the fasG(K117A) mutant bound to the glycoprotein receptors, confirming that lysine 117 was not required for binding to the glycoprotein receptors. Truncated FasG molecules were used to identify domains required for glycoprotein receptor recognition. At least two segments which did not include lysine117, namely, residues 211 (glutamine) to 220 (serine) and 20 (aspartic acid) to 41 (serine), were shown to be involved in the FasG-glycoprotein receptor interactions by ligand-blotting assays. Changing isoleucine 217 or leucine 215 of FasG to alanine abolished the property of a truncated FasG fusion protein to inhibit 987P recognition of its glycoprotein receptors. Thus, the K117 residue of FasG is required only for binding to the glycolipid receptor, whereas the newly identified hydrophobic residues of the FasG subunit are required specifically for the recognition of the glycoprotein receptor. Taken together, our data indicate that different residues of the FasG adhesin are important in 987P fimbrial binding to sulfatide and glycoprotein receptors, suggesting different mechanisms of interaction.
The 987P fimbriae of porcine enterotoxigenic Escherichia coli mediate bacterial adherence to both glycoprotein and glycolipid receptors on the brush borders of porcine intestinal epithelial cells (5, 8). Both types of interaction were shown to involve the 987P minor and tip subunit FasG (14, 15). However, because isolated FasG subunits bound only to the glycoprotein receptors, it was suggested that interactions with the glycolipid receptor depends either on the quaternary structure of FasG or on the cooperative interactions of several FasG subunits on different fimbriae (14). The FasG-specific glycolipid receptor was characterized as a series of sulfatide molecules (14), whereas the FasG-specific glycoprotein receptors were identified by ligand-blotting assays as two or three distinct proteins of 32 to 35 kDa (15). FasG molecules isolated from the periplasm specifically inhibit fimbrial binding to these proteins (15), indicating that FasG is already folded appropriately in the periplasm for glycoprotein receptor recognition. Similar properties were also observed for periplasmic P and type 1 adhesins (12, 16).
We showed recently that the fimbriae of the fasG(K117A) mutant did not mediate bacterial binding to sulfatide-containing liposomes. An additional small group of fasG mutants (K17A, R116A, K118A, and R200A) bound with lower affinity to such liposomes (3). Curiously, these fimbriae bound equally well to the glycoprotein receptors and wild-type fimbriae. Here, we present new data supporting a model in which FasG utilizes distinct mechanisms of recognition involving separate domains for its interactions with two different types of receptors.
MATERIALS AND METHODS
Bacterial strains, media, and reagents.
E. coli strain JM109 (29) was used for recombinant DNA work. Strain DMS741, a malE derivative of strain MC4100 (15), was used for studies with the maltose-binding protein (MBP) fusions. Strain B834(DE3) (Novagen, Madison, Wis.) was used for 35S labeling of 987P proteins. The nonfimbriated host strain SE5000 (25) was used for all the other studies. Cultures for colony isolation or plasmid purification were grown in L media (25) supplemented with ampicillin (200 μg/ml), chloramphenicol (30 μg/ml), or tetracycline (10 μg/ml) when appropriate. Medium components were purchased from Difco (Detroit, Mich.), and unless specified, reagents were purchased from Sigma (St. Louis, Mo.). Restriction and modification enzymes were from New England BioLabs (Beverly, Mass.). Oligonucleotides were prepared with an Applied Biosystems model 380B synthesizer.
Plasmid constructs.
The plasmids used in this study are listed in Table 1. Constructs encoding the MBP fused to the NH2 (FasG1–211 or FasG1–220) or COOH (FasG212–372 or FasG220–372) end of FasG were prepared by using PCR. For this, upper primer GU151 (5′ CGCGGATCCATTAATAGTGGCGCGATGA 3′) and lower primer GL771 (5′ GCGGGATCCTCTAGGAAACTGATTCAGAA 3′) or GL809 (5′ TTTTCTGCAGCTACGATCCGCTAA 3′), or upper primer GU781 (5′ CGCGGATCCCAAGTTGCTTTA 3′) or GU820-1 (5′ GGAAGATCTATTACCCTACCTCAACGCTGTT 3′) and lower primer GL1262 (5′ GCGGGATCCTCTAACCTTTTTCCCATC 3′), were synthesized and used to perform PCR with pBKC2 DNA as a template and Expand high-fidelity DNA polymerase (Boehringer Mannheim, Indianapolis, Ind.) (initial denaturation step of 2 min at 94°C; 25 cycles of 10 s at 94°C, 30 s at 56°C, and 1 min at 72°C; terminal extension step of 10 min at 72°C), essentially as described previously (2). The resulting PCR products were purified and cloned as BamHI and/or PstI fragments into the pMAL-p2 vector (New England Biolabs) for the creation of translational fusions to the MBP and were designated pBKC-P2 (MBP-FasG1–211), pBKC-P5 (MBP-FasG212–372), pBKC-P9 (MBP-FasG1–220), and pBKC-P12 (MBP-FasG220–372). Plasmids pBKC-P9 (MBP-FasG1–220 L215A), pBKC-P9 (MBP-FasG1–220 I217A), and pBKC-P9 (MBP-FasG1–220 L215A I217A) were prepared by the same method, using template DNA from the corresponding mutants constructed as described below. To construct plasmids pBKC12, pBKC18, and pBKC19, internal segments of the fasG gene were deleted by inverse PCR, using upper primer GU820 (5′ ATGGCTAGCATTACCCTACCTCAACGCTGTT 3′) or GU283 (5′ ATGGCTAGCACTGCTGAAAGCCATA 3′) and lower primer GL197 (5′ ATGGCTAGCACCGGAAATGCTTTGATTC 3′) or GL776 (5′ ATGGCTAGCAGAGGAAACTGATT 3′), with pDMS127 as a template and Expand high-fidelity DNA polymerase (initial denaturation step of 2 min at 94°C; 10 cycles of 10 s at 94°C, 30 s at 56°C, and 3 min 30 s at 68°C; 15 cycles of 10 s at 94°C, 30 s at 56°C, and 3 min 30 s, plus 20 s per cycle, at 68°C; terminal extension step of 7 min at 68°C). pBKC20 was generated by inverse PCR essentially as described above, using pBKC18 as a template. Plasmid pBKC8 was constructed by PCR cloning, using the upper primer GU1 (5′ CGCGGATCCGATATCATACGAGT 3′) and the lower primer GL809 (5′ TTTTCTGCTACGATCCGCTAA 3′) to introduce a stop codon after residue 220. The amplified product was cloned into BamHI and PstI sites in pBluescript KS (Stratagene, La Jolla, Calif.). Plasmid pBKC10 was constructed by deleting the ScaI-ClaI fragment in the fasG gene of pDMS127. The sequences of all of the PCR constructs were confirmed by DNA sequencing.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Genotype or relevant characteristics | References |
|---|---|---|
| E. coli | ||
| SE5000 | MC4100 recA56(Fim−) | 25 |
| ES1301 mutS | lacZ53 mutS201::Tn5 thyA36 rha-5 metB1 deoC IN(rmD-rmE) | Promega Corp. |
| DMS741 | MC4100 ΔmalE | 15 |
| B834(DE3) | F ompT hsdSB(rB− mB−) gal dcm met (DE3) | Novagen |
| Plasmids | ||
| pGP1-2 | pACYC177 (T7 RNA polymerase) | 26 |
| pDMS127 | pBluescript II KS fasG | 15 |
| pBKC1 | pDMS158 fasG (deletion of BstXI fragment) | 3 |
| pBKC2 | pALTER-1 fasG | 3 |
| pMALp-HHH | pMAL-p malE::fasG (MBP-FasG1–372) | 3 |
| pBKC-P2 | pMAL-p2 malE::fasG (MBP-FasG1–211) | This study |
| pBKC-P5 | pMAL-p2 malE::fasG (MBP-FasG212–372) | This study |
| pBKC-P9 | pMAL-p2 malE::fasG (MBP-FasG1–220) | This study |
| pBKC-P12 | pMAL-p2 malE::fasG (MBP-FasG221–372) | This study |
| pBKC-P9(L215A) | pMAL-p2 malE::fasG [MBP-FasG1–220(L215A)] | This study |
| pBKC-P9(I217A) | pMAL-p2 malE::fasG [MBP-FasG1–220(I217A)] | This study |
| pBKC-P9(L215A I217A) | pMAL-p2 malE::fasG [MBP-FasG1–220(L215A I217A)] | This study |
| pBKC8 | pDMS127 (FasG1–220) | This study |
| pBKC10 | pDMS127 (FasG ΔThr42–Ser220) | This study |
| pBKC12 | pDMS127 (FasG ΔAsp20–Ser220) | This study |
| pBKC18 | pDMS127 (FasG ΔAsp20–Ser41) | This study |
| pBKC19 | pDMS127 (FasG ΔGln211–Ser220) | This study |
| pBKC20 | pDMS127 (FasG ΔAsp20–Ser41 ΔGln211–Ser220) | This study |
| pBKC-F100A | pBKC2 [FasG(F100A)] | This study |
| pBKC-W173A | pBKC2 [FasG(W173A)] | This study |
| pBKC-L215A | pBKC2 [FasG(L215A)] | This study |
| pBKC-F216A | pBKC2 [FasG(F216A)] | This study |
| pBKC-I217A | pBKC2 [FasG(I217A)] | This study |
| pBKC-L215A/I216A | pBKC2 [FasG(L215A/I217A)] | This study |
| pBKC-T222A | pBKC2 [FasG(T222A)] | This study |
| pBKC-L223A | pBKC2 [FasG(L223A)] | This study |
Site-directed mutagenesis.
The fasG gene of pBKC2 was subjected to site-directed mutagenesis using the Altered Sites II in vitro-mutagenesis system (Promega Corp.) with synthetic oligonucleotides encoding alanine or serine residues at the targeted mutated sites. The desired mutants were selected by restriction analysis, each mutagenic primer having been designed to carry a diagnostic restriction site as described previously (3). All of the mutations were confirmed by DNA sequencing, and the mutants obtained are listed in Table 1.
Complementation assays.
E. coli strain SE5000 (25) containing pBKC1, which expresses all the Fas proteins with the exception of FasG (nonfimbriated and nonadhesive phenotype), was complemented for fimbriation and adhesion with the pBKC2 derivatives containing the various mutations in fasG. Plasmid pBKC2, which contains the wild-type fasG gene, was used as a positive control, as described previously (3). Plasmid copy numbers were not significantly different and thus did not affect the interpretation of the data.
Seroagglutination.
Slide agglutinations were performed with preadsorbed rabbit anti-987P fimbrial antiserum or with an anti-FasG polyclonal antibody as described previously (2, 23, 24).
Isolation of fimbriae.
Fimbriae were isolated from wild-type clinical strain 987 (19) or strain SE5000 (pBKC1 and pBKC2 derivatives) essentially as described previously (15). Briefly, bacteria were pelleted by centrifugation, resuspended in 0.5 mM Tris-HCl (pH 7.4)–75 mM NaCl, and treated at 60°C for 30 min. After a subsequent centrifugation clearing step, ammonium sulfate was added to the supernatants to 20% final concentration to precipitate the fimbrial proteins overnight on ice. The supernatants were centrifuged at 10,000 × g for 20 min, and the pellets were resuspended in phosphate-buffered saline (PBS; 10 mM NaHPO4, 1.8 mM KH2PO4, 2.7 mM KCl, 137 mM NaCl [pH 7.5]) or Tris-buffered saline (TBS; 10 mM Tris-HCl [pH 7.4], 154 mM NaCl). Excess ammonium sulfate was removed by ultrafiltration (Centricon 30; Amicon, Beverley, Mass.).
Production and isolation of periplasmic FasG proteins.
FasG and its truncates were specifically labeled in an in vivo T7 expression system using a mixture of [35S]methionine and [35S]cysteine (NEN Research Products, Boston, Mass.), and periplasmic fractions were prepared as described previously (15, 22). MBP-FasG fusion proteins were induced by adding isopropyl-β-d-thiogalactopyranoside (IPTG; final concentration, 0.3 mM) to DMS741 grown to log phase (A600 = 0.5) and containing the appropriate plasmids. After 2 h of induction at 37°C with IPTG, the bacteria were harvested and periplasmic fractions were isolated as described elsewhere (28). The periplasmic fractions were concentrated by ultrafiltration (Centricon 30) and equilibrated in TBS containing a cocktail of protease inhibitors (0.1 mM phenylmethylsulfonyl fluoride, 0.1 mM sodium metabisulfite, 1 mg of pepstatin per ml, 1 mg of leupeptin per ml, 2 mg of aprotinin per ml).
35S labeling of 987P fimbriae.
E. coli B834(DE3) carrying pBKC1 and pBKC2 was grown overnight. The bacteria were centrifuged, washed three times with M9 medium, and used to inoculate (1/50 dilution) freshly prepared M9 medium containing methionine (40 μg/ml) and 18 amino acids without cysteine (22). The cells were grown to log phase (A600 = 0.5) and washed three times with M9 medium. The growth medium was replaced with M9 medium containing IPTG (final concentration, 0.4 mM), methionine (10 μg/ml), and 18 amino acids without cysteine, and the cells were grown for 2 h followed by the addition of a mixture of [35S]methionine and [35S]cysteine (5 μCi/ml) and 10 min of incubation. 35S-labeled fimbriae were isolated as described above.
Preparation of BBV.
Brush border vesicles (BBV) were prepared from small-intestinal epithelial cells of 3-day-old piglets as described previously (17). The purity of the BBV was assessed by microscopy. Protein concentrations were determined (17), and the BBV were used immediately or frozen at −80°C for long-term storage.
Binding assay with BBV in solution.
To study the binding of FasG and its truncates to BBV in solution, purified BBV were washed three times, resuspended in PBS containing 0.5% bovine serum albumin and the cocktail of protease inhibitors described above, and mixed with 35S-labeled periplasmic FasG proteins (15). Isolated 35S-labeled periplasmic FasG proteins were equilibrated at room temperature and then centrifuged at 12,000 × g for 10 min to remove any precipitates. The resulting supernatants were normalized, and comparable amounts of radiolabeled FasG molecules were added to the BBV (100 μg), as determined by sodium dodecyl sulfate (SDS)–15% polyacrylamide gel electrophoresis (PAGE) fluorography, and densitometry with NIH Image software (National Institutes of Health, Bethesda, Md.) (15). After 2 h of incubation at room temperature, the BBV with bound FasG were washed three times by centrifugation (800 × g) with PBS containing a cocktail of protease inhibitors. BBV-associated FasG proteins were separated by SDS–15% PAGE, detected by fluorography, and analyzed by densitometry, as described above (15). For each experiment, the signals for total proteins were reevaluated to calculate percentages of binding by using values from the same blot.
SDS-PAGE and Western blot analysis.
Separation and analysis of fimbrial preparations or periplasmic proteins associated with BBV were undertaken by SDS-PAGE on 12% gels as described previously (15). Relative concentrations of the isolated periplasmic FasG subunits and FasG truncates, or MBP-FasG fusion protein and fusion truncates, were evaluated by fluorography or by Western blot analysis and enhanced chemiluminescence (Renaissance; NEN) using previously described specific FasG antibodies (2, 15) and densitometry.
Ligand blotting assays.
Ligand blotting assays were performed as described elsewhere (6, 15). Briefly, BBV proteins (30 μg for fimbrial binding assays or 50 μg for FasG binding assays) were separated by SDS–12% PAGE in the absence of reducing agents and electroblotted onto nitrocellulose membrane (Schleicher & Schuell, Leene, N.H.). After being blocked with 3% BSA-TBS for 3 h at room temperature, the blots were incubated for 2 h at room temperature with periplasmic 35S-labeled FasG proteins isolated from the wild type or fasG mutants and were normalized as described above. FasG binding was monitored by fluorography and densitometry (15). For each experiment, percentages of binding were determined by measuring signals for both bound and total protein on the same blot.
For the ligand blotting inhibition assay, BBV-blotted nitrocellulose membranes were blocked with 3% BSA-TBS, and membrane strips were incubated overnight at 4°C with isolated periplasmic fractions of each strain studied. For comparative purposes, only normalized quantities of the periplasmic FasG proteins and truncates were used, as determined by Western blotting, enhanced chemiluminescence, and densitometry. For several assays, different amounts of each FasG protein or truncate were studied under nonsaturated binding conditions to confirm dose-response effects. The strips were washed with TBS, and fimbrial binding was assessed with isolated 987P fimbriae using quaternary-structure-specific monoclonal antibody (MAb) E11 (23), horseradish peroxidase-conjugated goat anti-mouse antibodies (Organon Teknika Corp., Durham, N.C.), and enhanced chemiluminescence. As an alternative, 35S-labeled fimbriae were used, and binding was monitored by fluorography and densitometry.
Liquid phase binding assays with liposomes.
Sulfatide-containing liposomes were prepared and used for bacterial agglutinations as described previously (14).
RESULTS
Binding of allelic FasG subunits to the 987P glycoprotein receptors.
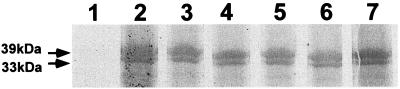
The adhesive property of the 987P fimbrial subunit FasG was originally identified by direct binding to porcine BBV and by inhibition of 987P binding to the glycoprotein receptor using either binding assays with BBV in solution or ligand-blotting assays (15). Using the latter assay, the FasG protein was also found to bind directly to the receptors, although detection of this interaction was less sensitive (Fig. 1, lane 2) than fimbrial binding (3). A weaker signal was expected, because of the absence of labeled major subunits amplifying the detection signals when whole fimbriae are used. The fimbriae of five previously characterized site-directed fasG mutants (residue replacements K17A, R116A, K117A, K118A, and R200A), which were defective for binding to the glycolipid receptor sulfatide, had been found to bind normally to the glycoprotein receptors by ligand-blotting assays (3). Confirming that the mutated FasG proteins themselves were responsible for this interaction, direct binding assays showed that all five mutated FasG proteins bound to the glycoproteins on ligand blots (Fig. 1). Moreover, no significant differences between the binding of wild-type FasG and that of the mutated molecules could be detected by densitometry after the ratios of band signal versus background signal for each lane were compared. This result further supported the possibility that the mechanisms of FasG binding to the glycolipid and glycoprotein receptors are different.
FIG. 1.
Direct binding of wild-type and mutated FasG fimbrial subunits to blotted BBV proteins. Each membrane strip was incubated with 35S-labeled periplasmic proteins isolated from a different strain. FasG binding was visualized by fluorogaphy. Periplasmic proteins were isolated from the following strains: control strain, E. coli SE5000(pGP1-2) (lane 1), SE5000(pGP1-2, pDMS127) expressing wild-type FasG (lane 2), SE5000(pGP1-2, pBKC-R17A) expressing FasG(R17A) (lane 3), SE5000(pGP1-2, pBKC-R116A) expressing FasG(R116A) (lane 4), SE5000(pGP1-2, pBKC-K117A) expressing FasG(K117A) (lane 5), SE5000(pGP1-2, pBKC-K118A) expressing FasG(K118A) (lane 6), and SE5000(pGP1-2, pBKC-R200A) expressing FasG(R200A) (lane 7).
Inhibition of 987P binding with FasG fragments fused to the MBP.
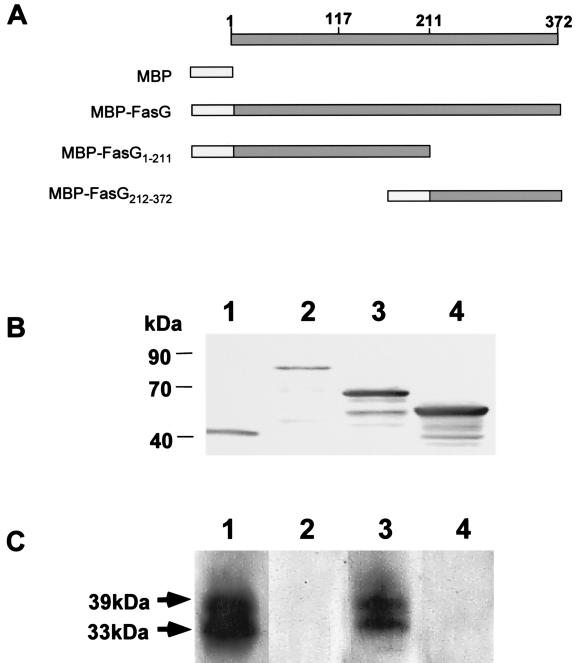
FasG subunits were previously shown to inhibit 987P binding to glycoprotein receptors (17), demonstrating that these interactions relate not only to monomeric FasG but also to the relevant in vivo fimbria-associated FasG. Here, we used this property to dissect the glycoprotein binding domain(s) of FasG. Assuming that this binding domain consists of a limited number of continuous segments of FasG, we first determined whether a major binding segment would be detectable in the NH2 or COOH half of FasG. To prepare sufficient amounts of FasG proteins as inhibitors of fimbrial binding, we increased the solubility of FasG and of its truncates in the periplasm by fusing them to MBP. As shown in Fig. 2, both MBP-FasG1–372 (full-length FasG) and MBP-FasG212–372 completely inhibited 987P binding, whereas inhibition by MBP-FasG1–211 was significantly weaker (25% inhibition). This result indicated that the binding property of FasG involved mainly amino acids found between residues 212 and 372.
FIG. 2.
Ligand-blotting inhibition assay. (A) Constructs of FasG and truncated FasG proteins fused to the MBP of E. coli. The residue numbers relate to mature FasG. (B) Western blot analysis of periplasmic fractions of bacteria expressing MBP or MBP-FasG fusion products detected with anti-MBP antibodies. Lane 1, MBP; lane 2, MBP-FasG; lane 3, MBP-FasG1–211; lane 4, MBP-FasG212–372. (C) Fimbrial ligand-blotting assays using MBP or MBP-FasG fusion proteins as inhibitors. Each membrane strip with SDS-PAGE-separated BBV proteins was incubated with the periplasmic fraction. The periplasmic fractions used were isolated from bacteria expressing the following proteins: MBP (lane 1), MBP-FasG (lane 2), MBP-FasG1–210 (lane 3), and MBP-FasG211–372 (lane 4).
Glycoprotein receptor-binding domain of FasG.
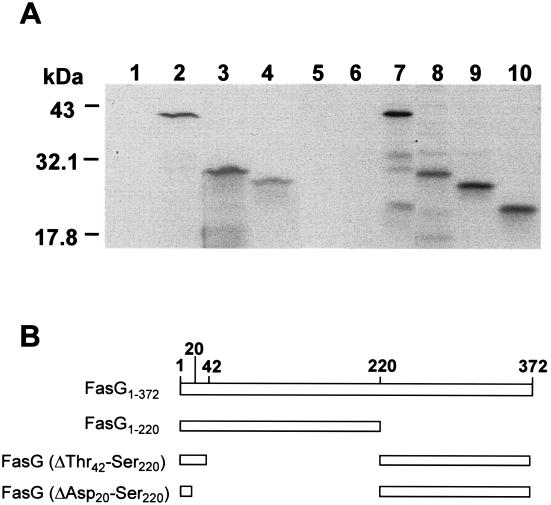
In contrast to 987P-associated FasG, FasG monomers do not interact with the sulfatide receptor (14). Thus, piglet BBV, which offer optimal receptor display in membranes, can be used to selectively study the FasG-glycoprotein receptor interaction. To further map the glycoprotein binding domain of FasG, we took advantage of the property of FasG monomers of directly binding to the glycoprotein receptors of BBV membranes (15). Full-length and truncated FasG proteins were specifically radiolabeled using an in vivo T7 expression system, and binding of periplasmic FasG or FasG truncates to BBV was analyzed by SDS-PAGE and fluorography. As shown in Fig. 3, the FasG1–220 truncate bound as well as the full-length protein (100% binding), whereas less-truncated FasG(ΔThr42–Ser220) protein bound to the BBV (approximately 50% binding). Binding by FasG(ΔAsp20–Ser220) was not significant (<10%). These results indicated that the FasG domain for glycoprotein receptor recognition localizes between residues 20 and 220 of FasG. Taken together with the results of the binding inhibition assay, the direct-binding data also suggested that binding can be attributed independently to amino acids of at least two continuous segments of FasG comprising residues 20 to 41 and 212 to 220.
FIG. 3.
Direct binding of FasG and its truncates to BBV. (A) Comparable amounts of all isolated 35S-labeled full-length or truncated FasG proteins were mixed with BBV. After 2 h of incubation, the BBV with bound FasG were washed by centrifugation, as described in Materials and Methods. BBV-associated FasG protein or truncates were separated by SDS–15% PAGE and detected by fluorography. Periplasmic FasG proteins were from the following strains: control strain, E. coli SE5000(pGP1-2) expressing no FasG (lanes 1 and 6), SE5000(pGP1-2, pDMS127) expressing full-length FasG (lanes 2 and 7), SE5000(pGP1-2, pBKC8) expressing FasG1–220 (lanes 3 and 8), SE5000(pGP1-2, pBKC10) expressing FasG(ΔThr42–Ser220) (lanes 4 and 9), and SE5000(pGP1-2, pBKC12) expressing FasG(ΔAsp20–Ser220) (lanes 5 and 10). Lanes 1 to 5, periplasmic FasG proteins which bound to the BBV; lanes 6 to 10, periplasmic FasG proteins before addition to the BBV. (B) DNA segments of the fasG gene included in each construct. The numbers correspond to the amino acid residues of mature FasG.
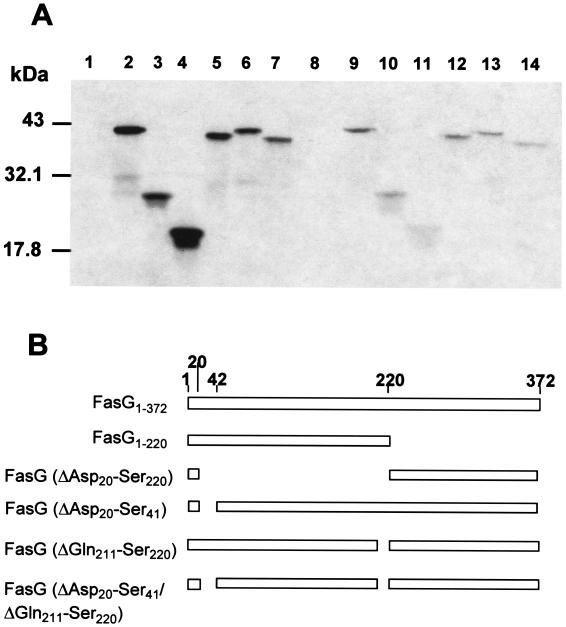
In addition, to confirm that the binding of FasG was mediated by FasG1–220 while the binding properties of FasG221–372 were not significant (<10%) (Fig. 4A, lanes 9 to 11), the products of three new fasG constructs were evaluated for their binding properties (Fig. 4A, lanes 12 to 14). FasG proteins expressed from plasmid pBKC18 or pBKC19 have residues 20 to 41 or 211 to 220, respectively, internally deleted, and FasG expressed from plasmid pBKC20 has both short segments internally deleted. FasG(Δ20–41 Δ211–220) bound the least to the receptor, followed by FasG(Δ211–220) and FasG(Δ20–41). Whether the weak binding signal of FasG(Δ20–41 Δ211–220) is due to additional minor effects of amino acid residues in the FasG segment 42 to 210 on the FasG-receptor interaction remains to be investigated.
FIG. 4.
Direct binding of internally deleted FasG proteins to BBV. (A) Comparable amounts of isolated 35S-labeled full-length, truncated, or internally deleted FasG proteins were mixed with BBV. After 2 h of incubation, the BBV with bound FasG were washed by centrifugation, as described in Materials and Methods. BBV-associated FasG proteins were separated by SDS–15% PAGE and detected by fluorography. The periplasmic FasG proteins were from the following strains: control strain, E. coli SE5000(pGP1-2) expressing no FasG (lanes 1 and 8), SE5000(pGP1-2, pDMS127) expressing full-length FasG (lanes 2 and 9), SE5000(pGP1-2, pBKC8) expressing FasG1–220 (lanes 3 and 10), SE5000(pGP1-2, pBKC12) expressing FasG(ΔAsp20–Ser220) (lanes 4 and 11), SE5000(pGP1-2, pBKC18) expressing FasG(ΔAsp20–Ser41) (lanes 5 and 12), SE5000(pGP1-2, pBKC19) expressing FasG(ΔGln211–Ser220) (lanes 6 and 13), and SE5000(pGP1-2, pBKC20) expressing FasG(ΔAsp20–Ser41 ΔGln211–Ser220) (lanes 7 and 14). Lanes 1 to 7, periplasmic FasG proteins before addition to the BBV; lanes 8 to 14, periplasmic FasG proteins which bound to the BBV. (B) DNA segments of the fasG gene included in each construct. The numbers correspond to the amino acid residues of mature FasG.
Site-directed mutations of the major glycoprotein receptor-binding segment of FasG.
MBP-FasG212–372 was a stronger inhibitor of binding than MBP-FasG1–211 (Fig. 2), and FasG221–372 was essentially not involved in binding (Fig. 3 and 4), suggesting that major residues involved in the glycoprotein binding interactions are located on a FasG212–220 segment. This FasG segment contains a high ratio of hydrophobic amino acid residues, as previously observed for the binding domains of adhesins of other fimbriae (9, 11). Early attempts to use synthetic peptides for mapping the FasG binding domains failed, as the peptides were poorly soluble. As an alternative approach, some of the hydrophobic residues in the most relevant 212- to-220 segment of FasG were targeted by site-directed mutagenesis. FasG residues W173 (control), L215, F216, I217, L223, and S218 (control) were all replaced with alanine. With the exception of mutant W173A, all of the mutants produced normal amounts of exported FasG and 987P fimbriae. Moreover, these mutants did not show any reduced binding to sulfatide-containing liposomes. The results of ligand-blotting assays with isolated fimbriae of each mutant suggested that binding for three mutated fimbriae [with FasG(L215A), FasG(F216A), or FasG(I217A)] was somewhat reduced, albeit weakly (data not shown). A double mutant expressing FasG(L215A I217A) did not produce fimbriae and thus could not be tested. Since our data suggest that the binding of 987P fimbriae to the glycoprotein receptors involves at least two independent segments of the primary structure of FasG, it is plausible that direct fimbrial binding, as detected by ligand blotting, may not be significantly affected by a single mutation in one segment. A comparison of the ligand-blotting assay with the ligand-blotting inhibition assay indicates that the latter method is more sensitive. For example, direct FasG binding was reduced by only approximately 50% in the absence of residues 212 to 220 (Fig. 3 and 4), whereas a FasG truncate containing only the residues involved in the binding of segment 212 to 220 completely inhibited fimbrial binding (Fig. 2). Thus, to determine whether hydrophobic residues of segment 212 to 220 were involved in its binding property, new constructs expressing mutated MBP-FasG truncates were prepared [pBKC-P9, pBKC-P9(L215A), pBKC-P9(I217A), and pBKC-P9(L215A I217A)]. As shown in Fig. 5, using three dilutions of normalized concentrations of periplasmic MBP-FasG fusion truncates, fimbrial binding to the glycoprotein receptors was fully inhibited by MBP-FasG1–220, whereas MBP-FasG220–372 did not inhibit fimbrial binding. This result confirmed our previous finding that the COOH-terminal segment FasG220–372 does not include residues required for glycoprotein receptor recognition. Most interestingly, all three mutated fusion truncates showed reduced inhibition. Using the smallest amount of inhibitors, none of the mutated fusion truncates inhibited 987P binding (Fig. 5, lanes 13 to 15), whereas the wild-type truncate remained fully inhibitory (Fig. 5, lane 11). A dose-response effect was detected by using higher concentrations of inhibitors (Fig. 5, lanes 1 to 10), allowing all three mutated fusion truncates to partially inhibit 987P binding and showing that the highest loss of inhibition was detectable with the double mutant (Fig. 5, lane 5). These results confirmed the importance of hydrophobic residues such as L215 and I217 in segment 212 to 220 for the FasG binding interactions with the glycoprotein receptors.
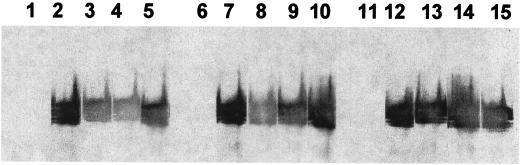
FIG. 5.
Ligand-blotting inhibition assay. Periplasmic MBP-FasG fusion proteins were isolated from the following strains: SE5000(pGP1-2, pBKC-P9) expressing MBP-FasG1–220 (lanes 1, 6, and 11), SE5000(pGP1-2, pBKC12) expressing MBP-FasG221–372 (lanes 2, 7, and 12), SE5000[pGP1-2, pBKC-P9(L215A)] expressing MBP-FasG1–220 (L215A) (lanes 3, 8, and 13), SE5000[pGP1-2, pBKC-P9(I217A)] expressing MBP-FasG1–220 (I217A) (lanes 4, 9, and 14), and SE5000[pGP1-2, pBKC-P9(L215A I217A)] expressing MBP-FasG1–220 (L215AI217A) (lanes 5, 10, and 15). Three dilutions (lanes 1 to 5, undiluted; lanes 6 to 10, diluted 1/2; lanes 11 to 15, diluted 1/4) of normalized concentrations of MBP-FasG fusion proteins from periplasmic fractions were used to inhibit fimbrial binding to glycoprotein receptors on the membrane.
DISCUSSION
In addition to intestinal sulfatide receptors for the 987P fimbriae (14), previous studies using brush borders from piglet enterocytes had identified 32- and 35-kDa glycoprotein receptors for the same fimbriae (7, 15). All the site-directed fasG mutants showing altered sulfatide-binding properties still adhered to the glycoprotein receptors (14), raising the possibility of separate mechanisms of binding for the two types of receptors. This was confirmed by the results of the present study. At least two relatively short segments of FasG (residues 20 to 41 and 212 to 220), which do not contain any of the lysine or arginine residues previously shown to be required for sulfatide binding (16), interacted with the glycoprotein receptors (Fig. 3 and 4). Most interestingly, these segments interacted independently with the receptors, since each of them inhibited binding. However, only one of the fragments (FasG212–220) fully inhibited binding by itself (Fig. 2 and 5). This result suggested that this fragment has the strongest affinity for the receptor and that it is adjacent to the other binding segment in the tertiary structure of FasG, enabling it to sterically interfere with the binding of whole fimbriae. In contrast, segment FasG20–41 has a weaker affinity for the receptor (Fig. 2B, lane 3, FasG1–211) and interferes less efficiently with the binding of whole fimbriae, whose avidity for the receptor is mainly determined by FasG212–220. These data are consistent with a conformational binding domain whose specificity for the glycoprotein receptor is determined by residues located in at least two separate segments of FasG. It remains possible that additional residues between these two segments participate in this binding domain, albeit less significantly.
Changing specific hydrophobic residues of segment 212 to 220 (L215 and/or I217) to alanine dramatically affected the binding-inhibitory property of this segment, pointing to the importance of hydrophobic interactions for glycoprotein receptor recognition. Whether these interactions occur between FasG and the receptor or in FasG to maintain a cognate binding domain for the receptor remains to be determined. Since at least one other binding segment largely compensated for the effect of these mutations when intact 987P fimbrial binding was evaluated, additional residues are likely essential and sufficient to create a productive surface for interaction with the glycoprotein receptors. Additional residues in the two segments will have to be mutated to determine whether glycoprotein-specific binding can be disrupted without affecting fimbrial biogenesis.
Recent X-ray studies of the type 1 and Pap fimbrial adhesins revealed similar conformational properties (4, 10). Both adhesins, FimH and PapG, are folded into two domains, an N-terminal lectin domain and a C-terminal pilin domain. The lectin domain of FimH consists of an 11-stranded elongated β barrel with a jelly roll-like topology. Similarly, the receptor-binding domain of PapG is mostly a β-sheet structure composed of two regions. One region forms a β barrel resembling the mannose-binding domain of FimH (10), and the other region consists of a central antiparallel β sheet flanked by two double-stranded β sheets and one α helix (10). Although the similarities at the primary structure level between FimH or PapG and FasG are low (14 and 13%, respectively), secondary-structure predictions suggest that FasG consists mainly of β strands. A protein sequence alignment (Clustal; DNASTAR, Madison, Wis.) (not shown) suggests that the two continuous segments of FasG found in this study to be involved in glycoprotein receptor recognition (segments 20 to 41 and 212 to 220) align with three flanking parallel β strands of FimH (strands 3, 4a, and 11) and overlap with some of the residues lining the carbohydrate-binding pocket in FimH (4). This is in agreement with the interpretation of our data suggesting that the two FasG segments involved in glycoprotein receptor recognition participate in one conformational domain. Interestingly, the positively charged residue patch of FasG shown previously to be required for sulfatide binding (K116, K117, and R118) does not align with the FimH receptor-binding domain (4, 20, 21, 27) but with a portion of the PapG receptor binding domain (10). Although the structural significance of the sequence alignments has to be confirmed by biophysical methods of structure analysis, the comparisons obtained are consistent with a model proposing that FasG harbors two independent and spatially separated surfaces of interaction for its two types of porcine intestinal receptors.
In conclusion, our data strongly suggest that the 987P fimbrial adhesin FasG harbors two distinct functional domains for its two types of glycoconjugate receptors. Whether these binding domains are utilized simultaneously or sequentially in the intestines of pigs is not known. Nevertheless, based on the two-step model of microbial binding to host cell surfaces (13), it is possible that sulfatide accessibility to 987P on enterocytes is optimized only after glycoprotein receptors are targeted for bacterial binding. Low-affinity binding may then be increased by lateral diffusion and reorganization of sulfatide in the membrane to anchor the bacteria by multivalent interactions (1, 13). Our current investigations are aimed at determining the biological relevance of each set of interactions by in vivo experimentation. A major goal is to design model therapeutic and prophylactic approaches, based on ligand or carbohydrate receptor analogues (18), for interfering with pathogen colonization.
ACKNOWLEDGMENTS
We thank W. M. Armstead for the piglets and R. N. Harty for critically reading the manuscript.
This work was supported by U.S. Department of Agriculture Grant 1999-02327.
REFERENCES
- 1.Ben-Tal N, Honig B, Peitzsch R M, Denisov G, McLaughlin S. Binding of small basic peptides to membranes containing acidic lipids: theoretical models and experimental results. Biophys J. 1996;71:561–575. doi: 10.1016/S0006-3495(96)79280-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cao J, Khan A S, Bayer M E, Schifferli D M. Ordered translocation of 987P fimbrial subunits through the outer membrane of Escherichia coli. J Bacteriol. 1995;177:3704–3713. doi: 10.1128/jb.177.13.3704-3713.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Choi B-K, Schifferli D M. Lysine residue 117 of the FasG adhesin of enterotoxigenic Escherichia coli is essential for binding of 987P fimbriae to sulfatide. Infect Immun. 1999;67:5755–5761. doi: 10.1128/iai.67.11.5755-5761.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Choudhury D, Thompson A, Stojanoff V, Langermann S, Pinkner J, Hultgren S J, Knight S D. X-ray structure of the FimC-FimH chaperone-adhesin complex from uropathogenic Escherichia coli. Science. 1999;285:1061–1066. doi: 10.1126/science.285.5430.1061. [DOI] [PubMed] [Google Scholar]
- 5.Dean E A. Comparison of receptors for 987P pili of enterotoxigenic Escherichia coli in the small intestines of neonatal and older pigs. Infect Immun. 1990;58:4030–4035. doi: 10.1128/iai.58.12.4030-4035.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dean E A, Isaacson R E. Purification and characterization of a receptor for the 987P pilus of Escherichia coli. Infect Immun. 1985;47:345–348. doi: 10.1128/iai.47.1.98-105.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dean E A, Whipp S C, Moon H W. Age-specific colonization of porcine intestinal epithelium by 987P-piliated enterotoxigenic Escherichia coli. Infect Immun. 1989;57:82–87. doi: 10.1128/iai.57.1.82-87.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dean-Nystrom E A, Samuel J E. Age-related resistance to 987P fimbria-mediated colonization correlates with specific glycolipid receptors in intestinal mucus in swine. Infect Immun. 1994;62:4789–4794. doi: 10.1128/iai.62.11.4789-4794.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Di Martino P, Girardeau J P, Der Vartanian M, Joly B, Darfeuille-Michaud A. The central variable V2 region of the CS31A major subunit is involved in the receptor-binding domain. Infect Immun. 1997;65:609–616. doi: 10.1128/iai.65.2.609-616.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dodson K W, Pinkner J S, Rose T, Magnusson G, Hultgren S J, Waksman G. Structural basis of the interaction of the pyelonephritic E. coli adhesin to its human kidney receptor. Cell. 2001;105:733–743. doi: 10.1016/s0092-8674(01)00388-9. [DOI] [PubMed] [Google Scholar]
- 11.Jacobs A A C, Venema J, Leeven J, van Pelt-Heerschap H, de Graaf F K. Inhibition of adhesive activity of K88 fibrillae by peptides derived from the K88 adhesin. J Bacteriol. 1987;169:735–741. doi: 10.1128/jb.169.2.735-741.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jones C, Pinkner J, Nicholes A, Slonim L, Abraham S, Hultgren S. FimC is a periplasmic PapD-like chaperone that directs assembly of type 1 pili in bacteria. Proc Natl Acad Sci USA. 1993;90:8397–8401. doi: 10.1073/pnas.90.18.8397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Karlsson K-A, Ångström J, Bergström J, Lanne B. Microbial interaction with animal cell surface carbohydrates. APMIS. 1992;100:71–83. [PubMed] [Google Scholar]
- 14.Khan A S, Johnston N H, Goldfine H, Schifferli D M. Porcine 987P glycolipid receptors on intestinal brush borders and their cognate bacterial ligands. Infect Immun. 1996;64:3688–3693. doi: 10.1128/iai.64.9.3688-3693.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Khan A S, Schifferli D M. A minor 987P protein different from the structural fimbrial subunit is the adhesin. Infect Immun. 1994;62:4233–4243. doi: 10.1128/iai.62.10.4233-4243.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lindberg F, Tennent J M, Hultgren S J, Lund B, Normark S. PapD, a periplasmic transport protein in P-pilus biogenesis. J Bacteriol. 1989;171:6052–6058. doi: 10.1128/jb.171.11.6052-6058.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Markwell M A, Haas S M, Bieber L L, Tolbert N E. A modification of the Lowry procedure to simplify protein determination in membrane and lipoprotein samples. Anal Biochem. 1978;87:206–210. doi: 10.1016/0003-2697(78)90586-9. [DOI] [PubMed] [Google Scholar]
- 18.Mouricout M, Petit J M, Carias J E, Julien R. Glycoprotein glycans that inhibit adhesion of Escherichia coli mediated by K99 fimbriae: treatment of experimental colibacillosis. Infect Immun. 1990;58:98–106. doi: 10.1128/iai.58.1.98-106.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nagy B, Moon H W, Isaacson R E. Colonization of porcine intestine by enterotoxigenic Escherichia coli: selection of piliated forms in vivo, adhesion of piliated forms to epithelial cells in vitro, and incidence of a pilus antigen among porcine enteropathogenic E. coli. Infect Immun. 1977;16:344–352. doi: 10.1128/iai.16.1.344-352.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sauer F G, Barnhart M, Choudhury D, Knight S D, Waksman G, Hultgren S J. Chaperone-assisted pilus assembly and bacterial attachment Curr. Opin Struct Biol. 2000;10:548–556. doi: 10.1016/s0959-440x(00)00129-9. [DOI] [PubMed] [Google Scholar]
- 21.Schembri M A, Sokurenko E V, Klemm P. Functional flexibility of the FimH adhesin: insights from a random mutant library. Infect Immun. 2000;68:2638–2646. doi: 10.1128/iai.68.5.2638-2646.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schifferli D M. Use of TnphoA and T7 RNA polymerase as tools to study fimbrial proteins. Methods Enzymol. 1995;252:242–258. doi: 10.1016/s0076-6879(95)53023-1. [DOI] [PubMed] [Google Scholar]
- 23.Schifferli D M, Abraham S N, Beachey E H. Use of monoclonal antibodies to probe subunit-and polymer-specific epitopes of 987P fimbriae of Escherichia coli. Infect Immun. 1987;55:923–930. doi: 10.1128/iai.55.4.923-930.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schifferli D M, Beachey E H, Taylor R K. Genetic analysis of 987P adhesion and fimbriation of Escherichia coli: the fas genes link both phenotypes. J Bacteriol. 1991;173:1230–1240. doi: 10.1128/jb.173.3.1230-1240.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Silhavy T J, Berman M L, Enquist L W. Experiments with gene fusion. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1984. [Google Scholar]
- 26.Tabor S, Richardson C C. A bacteriophage T7 RNA polymerase/promoter system for controlled exclusive expression of specific genes. Proc Natl Acad Sci USA. 1985;82:1074–1078. doi: 10.1073/pnas.82.4.1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Thankavel K, Madison B, Ikeda T, Malaviya R, Shah A H, Arumugam P M, Abraham S N. Localization of a domain in the FimH adhesin of Escherichia coli type 1 fimbriae capable of receptor recognition and use of a domain-specific antibody to confer protection against experimental urinary tract infection. J Clin Investig. 1997;100:1123–1136. doi: 10.1172/JCI119623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Witholt B, Boekhout M, Brock M, Kingma J, van Heerikhuizen H, de Leij L. An efficient and reproducible procedure for the formation of spheroplasts from variously grown Escherichia coli. Anal Biochem. 1976;74:160–170. doi: 10.1016/0003-2697(76)90320-1. [DOI] [PubMed] [Google Scholar]
- 29.Yanisch-Perron C, Vieira J, Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33:103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]