Abstract
Background: A bidirectional kidney–gut axis was described in patients with chronic kidney disease (CKD). On the one hand, gut dysbiosis could promote CKD progression, but on the other hand, studies reported specific gut microbiota alterations linked to CKD. Therefore, we aimed to systematically review the literature on gut microbiota composition in CKD patients, including those with advanced CKD stages and end-stage kidney disease (ESKD), possibilities to shift gut microbiota, and its impact on clinical outcomes. Materials and methods: We performed a literature search in MEDLINE, Embase, Scopus, and Cochrane databases to find eligible studies using pre-specified keywords. Additionally, key inclusion and exclusion criteria were pre-defined to guide the eligibility assessment. Results: We retrieved 69 eligible studies which met all inclusion criteria and were analyzed in the present systematic review. Microbiota diversity was decreased in CKD patients as compared to healthy individuals. Ruminococcus and Roseburia had good power to discriminate between CKD patients and healthy controls (AUC = 0.771 and AUC = 0.803, respectively). Roseburia abundance was consistently decreased in CKD patients, especially in those with ESKD (p < 0.001). A model based on 25 microbiota dissimilarities had an excellent predictive power for diabetic nephropathy (AUC = 0.972). Several microbiota patterns were observed in deceased ESKD patients as compared to the survivor group (increased Lactobacillus, Yersinia, and decreased Bacteroides and Phascolarctobacterium levels). Additionally, gut dysbiosis was associated with peritonitis and enhanced inflammatory activity. In addition, some studies documented a beneficial effect on gut flora composition attributed to synbiotic and probiotic therapies. Large randomized clinical trials are required to investigate the impact of different microbiota modulation strategies on gut microflora composition and subsequent clinical outcomes. Conclusions: Patients with CKD had an altered gut microbiome profile, even at early disease stages. Different abundance at genera and species levels could be used in clinical models to discriminate between healthy individuals and patients with CKD. ESKD patients with an increased mortality risk could be identified through gut microbiota analysis. Modulation therapy studies are warranted.
Keywords: chronic kidney disease, hemodialysis, end-stage kidney disease, gut microbiota, gut dysbiosis, mortality, outcomes
1. Introduction
Gut microbiota represents one of the most diverse microbiota of the human body and encompasses more than 35,000 bacterial species with 10 million genes [1]. For that reason, gut microbiota has been referred to by some authors as an additional organ and has been extensively studied in recent years [2,3,4].
Although gut microbiota varies across individuals, the most frequently encountered phyla are Firmicutes and Bacteroidetes, which constitute approximately 90% of the microbiota. Other gut microbiota phyla are represented by Actinobacteria, Fusobacteria, Proteobacteria, and Verrucomicrobia [5]. Among the Firmicutes phylum, Clostridium, Lactobacillus, Bacillus, Ruminococcus, and Enterococcus are the most frequent genera.
In addition to the local effects attributed to gut microbiota, it could also have systemic effects through secreting different active compounds, including short-chain fatty acids (SCFA) (acetate, butyrate, propionate), neurotransmitters (dopamine, serotonin, noradrenaline), bile acids, trimethylamine, cortisol, and gastrointestinal hormones (glucagon-like peptide-1, leptin, peptide YY) [4]. Therefore, gut microbiota could be regarded as a genuine endocrine organ that modulates nutrient and drug metabolism, antimicrobial protection, and immune response and ensures the integrity of the gastrointestinal tract [1,4].
In CKD patients, a bidirectional kidney–gut axis has been described [6]. The underlying cause for renal dysfunction, dietary restrictions, prolonged colonic transition time, or therapeutic intervention such as antibiotics, iron supplementation, or phosphate binders could cause dysbiosis.
Alternatively, gut dysbiosis triggers the production of detrimental metabolites such as indoxyl sulfate (IS) and p-cresyl sulfate, already associated with increased mortality and cardiovascular risk and a reduced number of valuable SCFA; the latter is implicated in energy homeostasis, maintaining the gut barrier, blood pressure control, and immune regulation. Moreover, dysbiosis induces an increase in gut permeability, which favors the translocation of bacterial species and microbial products through systemic circulation, promoting systemic inflammation and, possibly, alterations of glucose and lipid metabolism [7]. This leaky gut syndrome has also been observed in patients with inflammatory bowel disease, colorectal cancer, Parkinson’s disease, and Huntington’s disease [8,9]. An additional unfavorable effect of dysbiosis is the loss of diversity and imbalance in composition. This was related to poor survival in patients undergoing allogeneic hematopoietic-cell transplantation [10] and in patients hospitalized for chronic obstructive pulmonary disease [11].
Moreover, a heart–gut axis was described, linked to atherosclerosis and heart failure pathogenesis and development [12]. Gut dysbiosis observed in patients with chronic kidney disease (CKD) might partly explain the increased rate of cardiovascular-related deaths (almost 35% of all deaths) in this subgroup of patients [13,14]. Thus, gut dysbiosis could be regarded as a potential cardiovascular risk factor in patients with CKD in addition to other traditional risk factors. Therefore, the interplay between the gut, kidney, and heart is of great clinical importance, as gut microbiota composition could be modulated by diet, physical activity, probiotics, and prebiotics [12].
Consequently, we aimed to systematically review the literature on gut microbiota composition in CKD patients, including those with advanced CKD stages and end-stage kidney disease (ESKD), possibilities to shift gut microbiota, and its impact on clinical outcomes.
2. Materials and Methods
We conducted our systematic review in line with the updated Preferred Reporting Items for Systematic Review and Meta-analysis (PRISMA) guidelines in order to obtain reliable results [15]. The protocol of the systematic review was registered in the PROSPERO database (CRD42022369573).
2.1. Data Sources and Search Strategy
We sought to find eligible studies in MEDLINE (PubMed), Embase, Scopus, and Cochrane databases. A literature search was performed in the aforementioned databases, using various combinations between pre-specified keywords and MeSH terms or Emtree terms (respectively, for MEDLINE or Embase database): “microbiota”, “gut”, “microbiome”, “gastrointestinal”, “microflora”, “intestinal”, “composition”, “diversity”, “chronic kidney disease”, “end-stage kidney disease”, “hemodialysis”, “renal decline”, “disease severity”, “progression”, “mortality”, “major adverse cardiovascular events”, “inflammation”, “inflammatory response”, “probiotic”, “prebiotic”, and “symbiotic”. Studies were published from the inception of databases to 30 June 2022. We did not apply any language filters or restrictions in the search strategy. In addition, the ClinicalTrials.gov registry of clinical trials, Google Scholar engine, and references from cited manuscripts were screened and checked for additional eligible studies, which is compliant with PRISMA guidelines. The final search strategy for all databases and references obtained is presented in Table S1.
2.2. Eligibility Criteria and Outcomes
A multistep approach was used to assess retrieved references for eligibility. In the first step, two independent investigators evaluated the title and abstracts of articles for inclusion and exclusion criteria. In the next step, the full text of studies that met the eligibility criteria based on title and abstract was appraised.
Several inclusion criteria were pre-defined and were applied for eligibility assessment. (1) Both randomized clinical trials and observational studies were considered for inclusion. (2) Patients ≥ 18 years old with CKD of all stages were enrolled. (3) Healthy subjects or patients with early stages of CKD were included in the control group (when available). (4) Studies reporting original data on the following outcomes: (a) gut microbiota composition in CKD patients (when available, CKD patients versus healthy subjects or early CKD patients versus ESKD patients); (b) association between identified microbiota species in CKD patients and major adverse cardiovascular events (MACE), mortality, CKD severity, and disease progression; (c) the impact of prebiotics, probiotics, and symbiotics on flora composition and outcomes of CKD patients.
Early CKD patients were considered those presenting with G2 and G3 CKD stages (estimated glomerular filtration rate of 30–90 mL/min/1.73 m2). Prebiotics were defined as “non-digestible food ingredient” which enhance the growth or activity of certain beneficial gut bacteria [16]. Probiotics were defined as “live microorganisms” which exhibit beneficial health effects when they are prescribed in appropriate concentration [17]. Likewise, we defined synbiotics as a “mixture of probiotics and prebiotics” which display beneficial health effects [18].
In addition, some key exclusion criteria were established: unpublished data, studies available only in abstract, overlapping populations, case reports, meta-analyses, editorials, missing data, and inability to extract data regarding the population enrolled and outcomes investigated.
2.3. Data Collection and Synthesis
After eligibility assessment, two independent investigators extracted the following data from included studies: first author, year of publication, study design, number of patients enrolled and their age, clinical setting, reported outcome of interest, and follow-up period. We performed a qualitative synthesis of included studies to provide a better understanding of reported outcomes. Additionally, when available, data were reported as numbers, median and mean values, odds ratio (OR), and p-value.
2.4. Quality and Risk of Bias Assessment
In the case of randomized controlled clinical trials, the risk of bias was judged using the revised Cochrane risk-of-bias tool for randomized trials (RoB 2) [19]. The Newcastle–Ottawa scale (NOS) was used to guide the quality assessment of observational, non-randomized studies. NOS is a tool based on designating stars for signaling questions, which were grouped into three domains: population sampling, comparability of groups, and evaluation of outcomes of interest [20].
3. Results
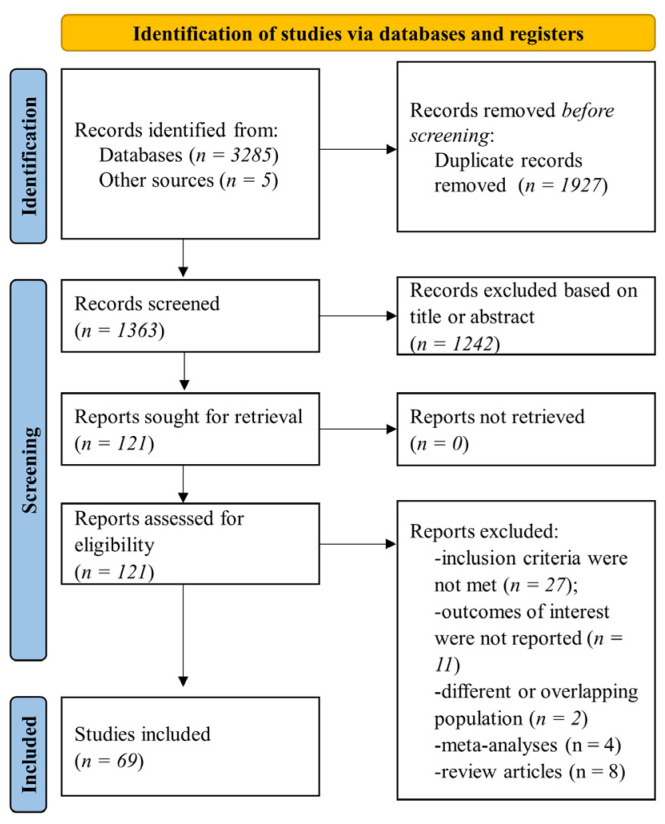
We searched the specified databases and retrieved 3290 references. Duplicate records were removed (n = 1927), leaving 1363 references for title or abstract screening. Finally, 121 records were assessed for eligibility in full-text, and 69 studies were included in the present systematic review. The flowchart of the selection process is presented in Figure 1.
Figure 1.
Flow diagram of selected studies in the present analysis.
General data of the analyzed studies, including publication year, study design, number of patients enrolled and their age, as well as gut microbiota composition in CKD patients, are reported in Table 1. The majority of included studies had an observational non-randomized design [21,22,23,24,25,26,27,28,29,30,31,32,33,34,35,36,37,38,39,40,41,42,43,44,45,46,47,48,49,50,51,52,53,54,55,56,57,58,59,60,61,62,63,64,65,66,67,68,69,70,71,72,73,74,75,76,77], while 12 studies were randomized trials [78,79,80,81,82,83,84,85,86,87,88,89]. Additionally, 21 studies investigated gut microbiota composition in ESKD patients (including patients with hemodialysis or peritoneal dialysis) [24,28,29,30,31,33,34,40,41,42,45,52,53,54,55,60,62,78,83]. Moreover, 4 studies investigated gut microbiota differences in early CKD stages as compared to healthy controls [30,51,56] or advanced-stage CKD [38].
Table 1.
Gut microbiota composition in CKD patients as reported in analyzed studies.
| Author, Year | Design | Patients, No | Age, Median/Mean ± SD | Gut Microbiota Composition |
|---|---|---|---|---|
| Barros et al., 2015 [21] | Cross-sectional study | 20 (CKD stage 3–4) | 64.4 ± 9.1 | Similar number of bands in patients with CKD vs. healthy controls. CKD: Listeria monocytogenes, Flavobacteriaceae bacterium. Healthy controls: Uncultured Lachnospiraceae bacterium, Butyrivibrio crossotus. |
| 19 (healthy participants) | 51.6 ± 6.6 | |||
| Belova et al., 2020 [78] | Randomized, parallel-group, controlled trial | 32 (HD patients who received basic therapy + symbiotic) | 57.1 ± 7.9 | HD patients displayed an increased number and diversity of Bacteroides spp., Clostridium spp., Collinsella spp., Eggerthella spp., and other bacteria. Grade I dysbiosis was observed in 37.5% of patients, grade II dysbiosis in 50.0% of patients, and grade III dysbiosis in 12.5% of patients. |
| 30 (HD patients who received basic therapy + placebo) | 54.7 ± 8.4 | |||
| Chen et al., 2021 [22] | Cross-sectional study | 96 (CKD stage 1–5) | 71 (CKD stage 3–5) | No significant difference was observed in α-diversity across different groups. Relative levels of Streptococcus, Klebsiella pneumonia, and Haemophilus Parainfluenzae increased progressively in advanced CKD. Increased levels of Fusobacterium varium and Fusobacterium mortiferum were observed in CKD stages 3–5. |
| 60 (healthy participants) | 66 | |||
| Du et al., 2021 [23] | Cross-sectional study | 43 (diabetic nephropathy stage 3–4) | 60.86 ± 5.69 | In patients with diabetic nephropathy, several genera were more abundant as compared to healthy individuals: Acidaminococcus, Lactobacillus, Megasphaera, Mitsuokella, Olsenella, Prevotella_7, and Sutterella. A model based on 25 genera discrepancies had a good prediction power for diabetic nephropathy (AUC = 0.972). |
| 37 (healthy individuals) | 61.78 ± 6.40 | |||
| Ebrahim et al., 2022 [80] | Randomized controlled trial | 59 (CKD stage 3–5) | 41.0 ± 11.6 | In CKD patients, several genera were abundant: Faecalibacterium, Prevotella, Bacteroides, Blautia, and Roseburia. |
| Gao et al., 2021 [24] | Cross-sectional study | 52 (CKD stage 3–5, including 10 patients with ESKD) | – |
Eubacterium rectale and Collinsealla genera correlated with kidney disease severity. Bifidobacterium increased with kidney disease severity, while Lactobacillus decreased. Methanobacteria was abundant in advanced CKD stages but was not present in ESKD patients. |
| Gryp et al., 2021 [25] | Cross-sectional study | 111 (CKD stage 1–5) | – |
Faecalibacterium, Bacteroides, and Roseburia were the most abundant genera in all CKD groups. Butyricicoccus (butyrate-generating properties) decreased in CKD stage 4–5 as compared to CKD stage 1–2 (p = 0.043). |
| Gryp et al., 2020 [26] | Cross-sectional study | 138 (CKD stage 1–5) and 14 controls | – |
A. muciniphila, C. dicile, Enterobacteriaceae, Lactobacillus spp., and Streptococcus spp. were increased in HD patients compared to other CKD stages. Bifidobacterium spp. and Streptococcus spp. decreased with kidney function decline, while Enterobacteriaceae and E. coli increased. |
| Guirong et al., 2018 [27] | Cross-sectional study | 16 (kidney transplant) | 42.8 ± 11.5 | Microbial richness was lowest in kidney transplant patients (Chao1 index 249.6 ± 118.7) as compared to CKD group (Chao1 index 286.4 ± 89.3) and healthy controls (Chao1 index 394.5 ± 86.8). Bacteroides and Enterobacteriaceae abundance were increased in CKD and kidney transplant patients, while Lachnospira, Ruminococcaceae, and Faecalibacterium levels were decreased. Gut microbiota profile had a good power to discriminate between CKD and healthy controls (AUC 0.921). |
| 84 (CKD) | 55.9 ± 18.2 | |||
| 53 (healthy controls) | 54.7 ± 12.8 | |||
| Hanifi et al., 2021 [28] | Cross-sectional study | 20 (CKD or ESKD) | 53.20 ± 12.03 | CKD and ESKD patients had similar abundance of different Bifidobacteriaceae species as compared to those without CKD or ESKD. |
| 20 (non-CKD/ESKD) | 59.3 ± 7.89 | |||
| He et al., 2020 [73] | Cross-sectional study | 109 (ESKD patients) | 56.8 ± 15.5 (HD patients) |
Bifidobacterium and Lactobacillus acidophilus decreased in ESKD patients as compared to healthy controls, while Escherichia coli and Enterococcus faecalis increased. Bifidobacterium and Lactobacillus acidophilus were higher in HD patients as compared to ESKD patients without RRT, while Escherichia coli and Enterococcus faecalis were decreased. |
| He et al., 2021 [29] | Cross-sectional study | 30 (HD) | 56.3 ± 13.6 | Patients with CKD (including HD) had a higher abundance of Escherichia coli and Enterococcus faecalis compared to healthy controls (p < 0.05), while Bifidobacterium and Lactobacillus acidophilus were decreased (p < 0.05). |
| 24 (non-HD) | 57.2 ± 15.1 | |||
| 30 (healthy controls) | 57.4 ± 14.9 | |||
| Hu et al., 2020 [71] | Cross-sectional study | 95 (CKD stages 1–5, non-HD) | 57.45 ± 11.68 (CKD stage 5) | Several genera (Escherichia-Shigella, Parabacteroides, Roseburia, Pyramidobacter rectale_group, Ruminococcaceae_NK4A214_group, Prevotellaceae_UCG.001, Hungatella, Intestinimonas, Pyramidobacter) discriminated between CKD stage 5 and healthy controls (AUC = 0.938, 95% CI, 0.853–1.000). Patients with CKD exhibited increased levels of Proteobacteria and decreased levels of Synergistetes as compared to healthy controls. |
| Hu et al., 2020 [74] | Case-control study | 47 (early CKD) | 43.2 ± 12.6 | α-diversity was lower in CKD versus healthy control participants. Proteobacteria and Actinobacteria were increased in CKD group vs. control group. Thirty-one species were different in CKD patients compared to healthy control (highest diagnostic power for Ruminococcus and Roseburia). Ruminococcus displayed the highest AUC for CKD prediction (0.771, 95% CI, 0.771–0.852). |
| 150 (healthy controls) | 38.5 ± 15.4 | |||
| Hu et al., 2020 [30] | Case-control study | 166 (47 non-dialysis CKD, 49 HD group, 53 PD group, and 17 healthy controls) | 57.80 ± 10.03 | α-diversity and β-diversity were lower in PD patients as compared to HD patients and control participants. Enterobacteriaceae and Enterococcaceae were highly expressed in PD patients, while Bifidobacteriaceae and Prevotellaceae were increased in the rest of the patients. |
| Iguchi et al., 2020 [31] | Cohort study | 38 (HD patients) | 66.17 ± 12.38 (sucroferric oxyhydroxide group) | Baseline phyla in HD patients: Firmicutes 67.5%; Proteobacteria 11.0%; Actinobacteria 12.2%; Bacteroides 9.2%. PD patients had lower levels of Bifidobacteriaceae and Prevotellaceae compared to other groups, while Enterobacteriaceae and Enterococcaceae were increased. |
| Jiang et al., 2016 [32] | Case-control study | 65 (CKD) | 43.45 ± 16.90 | Patients with advanced CKD and those with ESKD had significantly decreased abundance of Roseburia spp. and F. prausnitzii (respectively, p = 0.000 and p = 0.003). |
| 20 (healthy controls) | 43.05 ± 9.88 | |||
| Jiang et al., 2017 [33] | Cross-sectional study | 52 (ESKD patients) | 51.58 ± 18.33 |
E. coli, Bifidobacterium, Bacteroides fragilis group, Enterococcus spp., Clostridium coccoides group, Faecalibacterium prausnitzii, Roseburia spp., and Prevotella were decreased in ESKD patients as compared to healthy controls. Lactobacillus group levels were similar in both groups. |
| 60 (healthy controls) | 52.53 ± 13.98 | |||
| Kemp et al., 2021 [81] | Randomized, double-blind, controlled clinical trial | 10 (Resistant starch type-2 group) | 53.2 ± 12.3 |
Firmicutes phylum prevailed in HD patients. Subdoligranum, Fusicatenibacter, Prevotella, and Blautia were observed in HD patients. |
| 10 (placebo group) | 55.1 ± 11.1 | |||
| Khiabani et al., 2022 [34] | Cross-sectional | 20 (CKD/ESKD) | 53.20 ± 12.03 | Clostridium spp. abundance was similar in patients with CKD (including ESKD patients) and in healthy controls (p < 0.05). |
| 20 (healthy controls) | 59.3 ± 7.89 | |||
| Kim et al., 2020 [35] | Cross-sectional study | 103 (CKD stage 1–5) | 48.9 ± 12.2 (ESKD) |
Alistipes, Oscillibacter, Lachnospira, Veillonella, and Dialister were higher in control group as compared to patients with moderate to severe CKD. Alistipes, Oscillibacter, Lachnospira, and Veillonella were increased in mild CKD patients as compared to the moderate to severe CKD group. |
| 46 (healthy controls) | 47.0 ± 10.8 | |||
| Kumar et al., 2021 [36] | Cross-sectional study | 36 (IgAN) | 45.5 ± 13.4 | α-diversity was similar between IgAN patients and healthy controls but increased in advanced CKD stages (p = 0.025). Fusobacteria phylum was increased, while Euryarchaoeota phylum was decreased in patients with IgAN, as compared to healthy controls. |
| 12 (healthy controls) | 46.5 ± 13.5 | |||
| Lai et al., 2019 [37] | Observational, prospective study | 16 (CKD stages 3–4) | – | Patients with CKD had increased levels of Bacteroidaceae, Enterobacteriaceae, and Rickenellaceae as compared to healthy controls. |
| 16 (healthy controls) | ||||
| Lecamwasam et al., 2021 [38] | Observational, prospective study | 95 (diabetes-associated CKD stages 1–5) | 66.24 ± 10.22 (early CKD) | β-diversity and α-diversity were similar across all CKD stages. Firmicutes (the most abundant) and Bacteroidetes phyla abundance were similar in early and late CKD stages. Prevotellaceae was decreased across all CKD stages. |
| 72.68 ± 10.21 (late CKD) | ||||
| Li et al., 2019 [39] | Observational, prospective study | 50 (CKD) | 52.40 ± 13.49 | The most prevalent bacteria in CKD patients were Firmicutes (42.27%), Bacteroidetes (37.85%), Proteobacteria (16.70%), Actinobacteria (1.48%), and Verrucomicrobia (0.67%). As compared to healthy controls, patients with CKD had decreased Akkermansia (p = 0.001) and Parasutterella (p = 0.007) levels, while Lactobacillus (p < 0.001), Clostridium IV (p = 0.015), and Alloprevotella (p < 0.001) levels were higher in CKD patients. Akkermansia associated with Lactobacillus had a good predictive value for CKD (AUC 0.830). |
| 22 (healthy controls) | 50.27 ± 7.77 | |||
| Lin et al., 2021 [72] | Prospective, cohort study | 109 (ESKD) | 68.4 ± 10.4 | In the high diversity group of patients, as well as in those with lower microbiota diversity, Bacteroidetes, Firmicutes, and Proteobacteria were the most prevalent phyla. |
| Lin et al., 2020 [41] | Case-control study | 96 (HD) | 68.1 ± 1.0 | α-diversity was decreased in patients with normal-weight obesity as compared to those with normal weight or obesity (p = 0.001). Firmicutes/Bacteroidetes ratio was similar in all weight groups. Faecalibacterium prausnitzii, Faecalibacterium, and Coprococcus were decreased in the normal-weight obesity group. |
| Lin et al., 2020 [40] | Case-control study | 88 (HD) | 68.6 ± 11.0 (protein-energy wasting) | α-diversity was decreased in HD patients with moderate protein-energy wasting as compared to those with a normal nutritional status (p = 0.018). Faecalibacterium prausnitzii was significantly lower in protein-energy wasting patients, while Akkermansia muciniphila levels were higher. |
| 68.6 ± 9.7 (normal nutritional status) | ||||
| Lin et al., 2022 [42] | Case-control study | 11 (ESKD) | 30.93 ± 4.85 | ESKD patients had a higher abundance of Escherichia coli (p < 0.001), Bacteroides fragilis (p = 0.010), Bacteroides fragilis (p = 0.010), and Bacteroides caccae (p = 0.047), as compared to healthy controls. |
| 11 (healthy controls) | 27.99 ± 2.31 | |||
| Liu et al., 2021 [43] | Case-control study | 100 (CKD) | 56.64 ± 17.25 | The Shannon index decrease was associated with CKD (p < 0.05). Actinobacteria levels were higher in CKD patients and predicted CKD prevalence (OR 1.037, 95% CI 1.007–1.068). Bacteroidetes was decreased in CKD patients and predicted CKD prevalence (OR 0.971, 95% CI, 0.951–0.991). Bifidobacterium, Enterococcus, and Streptococcus were more abundant in CKD patients. |
| 100 (healthy controls) | 60.64 ± 16.51 | |||
| Liu et al., 2020 [83] | Randomized controlled trial | 22 (HD + probiotic) | – | Desulfovibrionaceae, Veillonellaceae, and Lactobacillaceae abundance was increased in HD patients with diabetes mellitus, while Halomonadaceae and Bradyrhizobiaceae were decreased as compared to non-diabetic HD patients. |
| 23 (HD + placebo) | ||||
| Lun et al., 2018 [44] | Cross-sectional study | 49 (CKD) | 54 ± 14 | Patients with CKD had increased abundance of Bacteroidetes and Proteobacteria, while Firmicutes was lower compared to healthy controls. Ruminococcus gnavus had the best discrimination power for CKD (AUC 0.764, 95% CI, 0.656–0.873, p = 0.000). |
| 24 (healthy controls) | 56 ± 9 | |||
| Luo et al., 2021 [45] | Observational, cohort study | 73 (ESKD) | 49.71 ± 14.81 (HD) | HD and PD patients had an increased abundance of Blautia and Dorea, while Prevotella was decreased. Akkermansia, Coprococcus, Acinetobacter, Proteus, and Pseudomonas were increased in HD patients. |
| 48.95 ± 10.23 (healthy controls) | ||||
| Margiotta et al., 2020 [46] | Cross-sectional study | 64 (CKD stages 3b-4) | 80.7 ± 6.2 | α-diversity was similar in CKD patients as compared to controls. Patients with CKD had a higher abundance of Lactobacillus, Coprobacillus, Anaerotruncus, Citrobacter, and Ruminococcus torques. Patients with CKD had lower levels of saccharolytic and butyrate-producing bacteria (Prevotella spp., F. prausnitzii, and Roseburia spp.). |
| 15 (healthy controls) | 73.7 ± 7.6 | |||
| Al-Obaide et al., 2017 [47] | Cross-sectional study | 20 (T2DM and advanced CKD) | 64.4 ± 2.3 | In patients with advanced CKD and T2DM, Bifidobacterium abundance was decreased, while Clostridium, Escherichia, Enterobacter, Acinetobacter, Proteus, and Lactobacillus levels were increased as compared to healthy controls. |
| 20 (healthy controls) | 54.3 ± 3.2 | |||
| Pivari et al., 2022 [48] | Observational, cohort study | 24 (CKD) | 72 (67.5–78.8) | CKD patients had higher α-diversity as compared to healthy controls. CKD patients had decreased abundance of Bacteroides (p = 0.037), Lachnoclostridium spp. (p = 0.018), and Escherichia-Shigella (p = 0.048) compared to controls. |
| 20 (healthy controls) | 74 (68.5–78.7) | |||
| Ren et al., 2020 [49] | Observational, cohort study | 110 (CKD) | 51.75 ± 14.60 | In CKD patients, 36 genera were increased (including Klebsiella, Veillonella, and Desulfovibrio), while 16 genera were decreased (including Blautia, Roseburia, and Lachnospira). Clostridia, Verrucomicrobia, and Cyanobacteria were decreased in CKD patients compared to controls. |
| 210 (healthy controls) | 50.02 ± 4.56 | |||
| Salguero et al., 2019 [50] | Cross-sectional study | 20 (T2DM and CKD) | 62.8 ± 3.6 | Proteobacteria, Verrucomicrobia, and Fusobacteria abundance were increased in CKD patients with T2DM as compared to healthy controls (p < 0.05 for all). |
| 20 (healthy controls) | 58.5 ± 4.1 | |||
| Sato et al., 2021 [51] | Cross-sectional study | 30 (early CKD) | 68.83 ± 10.14 | CKD patients had increased abundance of Bacteroides coprocora and Bacteroides caccae, while Roseburia inulinivorans, Ruminococcus torques, and Ruminococcus lactaris were more abundant in the non-CKD group. |
| 60 (non-CKD) | 67.80 ± 11.48 | |||
| Simeoni et al., 2019 [88] | Randomized, placebo-controlled study | 14 (CKD stage 3a + probiotics) | 61.3 ± 5.2 | Lactobacillales and Bifidobacteria had decreased levels in patients with CKD stage 3a (respectively, 2.3 × 103 CFU/gr and 1.7 × 104 CFU/gr). |
| 14 (CKD stage 3a, placebo) | 58.2 ± 6.2 | |||
| Stadlbauer et al., 2017 [52] | Cross-sectional study | 30 (dialysis patients) | 61 (HD patients) | HD and PD patients had lower α-diversity index as compared to the control group (p < 0.05), but it was similar between HD and PD patients. Blautia obeum, Clostridium citroniae, and Clostridium bolteae levels were higher in HD patients compared to the control group. Clostridium citroniae and Clostridium bolteae were increased in PD patients compared to the control group. Faecalibacterium prausnizii, Roseburia intestinalis, and Clostridium nexile were decreased in HD patients compared to the control group. |
| 21 (healthy controls) | 58 | |||
| Wang et al., 2019 [53] | Cross-sectional study | 56 (CKD stages 1–4) | 47.45 ± 15.47 | Patients with CKD stage 5 had lower Enterobacter, Enterococcus, Bifidobacterium, Bacteroides, and Clostridium levels as compared to controls and patients with CKD stages 1–4 (p < 0.01 for all). Faecalibacterium and Roseburia were reduced in CKD stage 5 patients compared to CKD stages 1–4 and healthy controls (respectively, p = 0.018 and p < 0.001). |
| 72 (CKD stage 5) | 51.69 ± 14.05 | |||
| 61 (healthy controls) | 46.80 ± 10.47 | |||
| Wang et al., 2019 [54] | Observational, cohort study | 28 (ESKD group) | 43.9 ± 13.8 | α-diversity was similar between ESKD and healthy controls group. Patients with ESKD had increased levels of Prevotella, Faecalibacterium, and Fusobacterium, while Roseburia, Lachnospira, Dialister, and Bifidobacterium abundance were decreased. |
| 19 (healthy controls) | 44.1 ± 10.0 | |||
| Wang et al., 2020 [55] | Cross-sectional study | 223 (ESKD group) | – | ESKD patients displayed a higher abundance of Eggerthella lenta, Flavonifractor spp., Alistipes spp., Ruminococcus spp., and Fusobacterium spp., while Prevotella spp., Clostridium spp., Roseburia spp., Faecalibacterium prausnitzii, and Eubacterium rectale were decreased. |
| 69 (healthy controls) | ||||
| Wu et al., 2020 [56] | Cross-sectional study | 72 (CKD group) | 65.00 ± 5.94 (advanced CKD) | Bacteroides eggerthii had a good discriminatory power between early-stage CKD and healthy controls (AUC = 0.80, 95% CI, 0.67–0.93), which was higher than in the case of protein/creatinine ratio (AUC = 0.64) and serum urea (AUC = 0.72). |
| 20 (non-CKD group) | 64.00 ± 7.06 | |||
| Wu et al., 2020 [57] | Cross-sectional study | 92 (CKD group) | 66.2 ± 7.4 (advanced CKD) | CKD patients had increased abundance of Bacteroides, Blautia, Escherichia-Shigella, Collinsella, Lachnoclostridium, and Lactobacillus. Paraprevotella displayed a good discriminatory power between CKD patients and the control group (AUC = 0.78, 95% CI, 0.70–0.87). |
| 30 (control group) | 61.6 ± 8.7 | |||
| Wu et al., 2021 [58] | Cross-sectional study | 39 (CKD stages 4–5) | 56.52 ± 15.72 | CKD patients had increased abundance of Proteobacteria, Enterobacteriaceae, Enterobacteriales, Gammaproteobacteria, Lactobacillales, Escherichia_Shigella, Enterococcus, Enterococcaceae, and Lactobacillaceae. |
| 40 (healthy controls) | 56.35 ± 10.96 | |||
| Xu et al., 2017 [59] | Cross-sectional study | 32 (CKD group) | 53.34 ± 14.47 |
Enterobacteriaceae and Corynebacteriaceae were more abundant in CKD patients as compared to controls, while Ruminococcaceae levels were decreased. Enterococcus and Clostridium were increased in CKD patients, whereas Roseburia and Coprococcus were decreased. |
| 32 (healthy controls) | 55.03 ± 10.38 | |||
| Zhang et al., 2021 [60] | Cross-sectional study | 46 (ESKD group) | Stratified in groups | Ruminococcus gnavus, Ruminococcus spp., Eubacterium dolichum, Bacteroides ovatus, and Phascolarctobacterium were more abundant in ESKD patients (including immunoglobulin A nephropathy), compared to healthy controls, while Megamonas spp., Roseburia spp., and Eubacterium biforme were decreased. |
| 15 (healthy controls) | ||||
| Zhang et al., 2020 [61] | Cross-sectional study | 80 (CKD stages 3–5) | 49.50 ± 24.80 |
Megamonas, Megasphaera, Akkermansia, Lachnospira, Roseburia, and Fusobacterium were increased in healthy controls as compared to patients with CKD and nephrotic syndrome. Patients with CKD and nephrotic syndrome had increased levels of Parabacteroides. Oscillospira and Ruminococcus were more abundant in the CKD group. |
| 48 (nephrotic syndrome) | 48.47 ± 20.47 | |||
| 30 (healthy controls) | 46.50 ± 22.67 | |||
| Zheng et al., 2020 [62] | Observational, cohort study | 28 (ESKD) | 43.9 ± 13.8 | Patients with ESKD had increased levels of Holdemania, Eggerthella, and Phascolarctobacterium, while Roseburia, Bifidobacterium, and Lachnospira were decreased as compared to healthy controls. |
| 19 (healthy controls) | 44.1 ± 10.0 |
AUC = area under the curve; CKD = chronic kidney disease; ESKD = end-stage kidney disease; HD = hemodialysis; IgAN = immunoglobulin A nephropathy; PD = peritoneal dialysis; RRT = renal replacement therapy; T2DM = type 2 diabetes mellitus.
3.1. Early-Stage CKD
Hu et al. observed different gut microbiota compositions even in patients with early-stage CKD as compared to healthy controls [74]. Gut flora diversity was significantly decreased in early-stage CKD patients (p < 0.001) compared to those without CKD. At the genera level, Ruminococcus had a good power to discriminate between early-stage CKD patients and healthy controls (AUC = 0.771, 95% CI, 0.771–0.852), while Roseburia accurately identified healthy controls (AUC = 0.803, 95% CI, 0.804–0.864) [74]. Wu et al. reported similar discriminatory capacity for early-stage CKD patients in the case of Bacteroides eggerthii (AUC = 0.80, 95% CI, 0.67–0.93), which was higher than in the case of protein/creatinine ratio (AUC = 0.64) [56]. Studies investigating gut microbiota diversity are of particular importance, as decreased diversity could be considered a reliable marker of gut dysbiosis [90]. Consequently, even patients with early-stage CKD had gut dysbiosis (low microbiota diversity) when compared to healthy controls [74]. Thus, improving gut microbiota diversity constitutes an important target for future interventional studies involving CKD patients. Available data on Roseburia suggest a key role in the modulation of gut barrier homeostasis and inflammation [91]. Hence, Roseburia is a marker of a normal gut microbiome, and lower abundance was reported in CKD patients from early stages [74]. Moreover, Roseburia abundance decreases along with CKD progression (lower in ESKD patients compared to early-stage CKD) [53].
3.2. ESKD
Some studies reported a shift in gut microbiota profile along with CKD progression, especially in ESKD patients and those requiring renal replacement therapy (RRT). Gao et al. reported a progressively increasing abundance of Bifidobacterium in advanced CKD stages, while Lactobacillus levels decreased [24]. He et al. observed a lower abundance of both Bifidobacterium and Lactobacillus in ESKD patients compared to healthy controls. Nevertheless, Bifidobacterium and Lactobacillus were increased in hemodialysis patients compared to ESKD patients without RRT [29]. Most studies documented decreased levels of Roseburia in advanced CKD stages, including ESKD [33,52,53,54,55,60,62]. Additionally, Roseburia abundance was lower in ESKD patients as compared to those with CKD stages 1–4 and healthy controls (p < 0.001) [53]. Bifidobacterium, like other beneficial components of a normal gut microbiome, exerts modulatory effects on gut homeostasis, inflammation, and immune response [92]. Although some studies documented Bifidobacterium depletion in CKD patients (including ESKD), these results should be confirmed in larger clinical trials.
3.3. Diabetic Nephropathy
Patients with diabetic nephropathy stage 3–4 had a different microbiota profile compared to healthy individuals [23]. Du et al. developed a model based on 25 gut microbiota dissimilarities, which had an excellent predictive power for diabetic nephropathy (AUC = 0.972) [23]. Additionally, in patients with diabetes-associated CKD, Ruminococcaceae and Bacteroidaceae abundance was significantly increased, while Prevotellaceae levels were decreased. These microbiome alterations were observed across all CKD stages, highlighting early gut dysbiosis in diabetic patients, which maintains in advanced CKD stages [38]. In addition, hemodialysis patients with diabetes had an increased abundance of Desulfovibrionaceae, Veillonellaceae, and Lactobacillaceae as compared to hemodialysis patients without diabetes [83].
Outcomes of CKD patients (inflammation, renal function, disease progression, mortality, and peritonitis) linked to gut microbiota composition are displayed in Table 2. The Simpson index and the Shannon index were significantly lower in deceased hemodialysis patients, as compared to those who survived (respectively, p = 0.007 and p = 0.028). Moreover, several bacteria were increased in hemodialysis patients from the non-survivor group (Oscillospira, Achromobacter, Agrobacterium, Lactobacillus, Alloscardovia, Anoxybacillus, Devosia, Yersinia) [40]. Additionally, Luo et al. reported a lower abundance of Bacteroides and Phascolarctobacterium in ESKD patients with cardiovascular mortality compared to those who survived (p < 0.05 for both).
Table 2.
Outcomes in CKD patients related to gut microbiota.
| Study, Year | Outcomes | Results |
|---|---|---|
| Barros et al., 2015 [21] | Inflammation | VCAM-1 levels were negatively correlated with number of bands in CKD patients (r = −0.50, p = 0.03) |
| Ebrahim et al., 2022 [80] | Renal function decline | Creatinine levels were similar between intervention (β-glucan prebiotic) and control group during follow-up (14 weeks). |
| Jiang et al., 2017 [33] | Inflammation | Roseburia spp., Faecalibacterium prausnitzii, and Prevotella were negatively correlated with CRP (respectively, r = −0.452, p = 0.001; r = −0.431, p = 0.002 and r = −0.480, p = 0.000) |
| Renal function |
Roseburia spp., Faecalibacterium prausnitzii, Clostridium coccoides group, Prevotella were negatively correlated with Cystatin C levels (respectively, r = −0.414, p = 0.003; r = −0.395, p = 0.005; r = −0.400, p = 0.001 and r = −0.441, p = 0.001) Bifidobacterium was correlated with creatinine and blood urea nitrogen (r = −0.538, p = 0.000 and r = −0.495, p = 0.000, respectively) |
|
| Jiang et al., 2016 [32] | Inflammation | In CKD patients, Roseburia spp. and F. prausnitzii were negatively correlated with CRP (respectively, (r = −0.493, p = 0.00; r = -0.528, p = 0.000). |
| Disease progression | In CKD patients, Roseburia spp. and F. prausnitzii were negatively correlated with Cystatin C (r = −0.321, p = 0.006; r = −0.445, p = 0.000) and positively corelated with eGFR (respectively, r = 0.347, p = 0.002 and r = 0.416, p = 0.000). | |
| Lin et al., 2021 [72] | Mortality | The Simpson index and the Shannon index were lower in non-survivors as compared to patients who survived (respectively, p = 0.007 and p = 0.028). Non-survivors had higher levels of Oscillospira, Achromobacter, Agrobacterium, PSB_M_3, Lactobacillus, vadinCA02, Alloscardovia, Anoxybacillus, Devosia, and Yersinia. |
| Lin et al., 2020 [41] | Inflammation | The Shannon diversity index was negatively corelated with IL-6 (r = −0.253, p = 0.015) and TNFα (r = −0.260, p = 0.011). Faecalibacterium prausnitzii was negatively correlated with TNFα (r = −0.204, p = 0.047). |
| Lin et al., 2020 [40] | Inflammation | The Shannon diversity index was negatively corelated with IL-6 (r = −0.339, p = 0.001) and TNFα (r = −0.331, p = 0.002). |
| Luo et al., 2021 [45] | Mortality | ESKD patients with cardiovascular mortality had a lower proportion of Bacteroides and Phascolarctobacterium compared to survivors (p < 0.05). |
| Peritonitis | PD patients with peritonitis had decreased Dorea, Clostridium, and SMB53 proportions as compared to those without peritonitis (p < 0.05). | |
| Margiotta et al., 2020 [46] | Inflammation |
Mogibacteriaceae and Oscillospira were correlated with CRP levels. Akkermansia, Ruminococcus, and Eubacterium were negatively correlated with the neutrophil-to-lymphocyte ratio. |
| Zhou et al., 2022 [70] | Peritonitis | PD patients with Escherichia coli peritonitis had higher abundance of Bacteroidetes and Synergistetes compared to the non-peritonitis group, while Bacilli and Lactobacillus were decreased. |
| Zhu et al., 2022 [69] | Responsiveness to erythropoietin |
Neisseria, Streptococcus, Porphyromonas, Fusobacterium, Prevotella_7, Rothia, Leptotrichia, Prevotella, and Actinomyces could predict a poor response to erythropoietin in ESKD patients. Neisseria had an excellent power to discriminate between good and poor response to erythropoietin in ESKD patients (AUC 0.9535, 95% CI, 0.902–1.0, p < 0.0001). |
CKD = chronic kidney disease; CRP = C-reactive protein; ESKD = end-stage kidney disease; IL-6 = interleukin 6; PD = peritoneal dialysis; TNFα = tumor necrosis factor alpha; VCAM-1 = vascular cell adhesion molecule 1.
Peritonitis was associated with altered gut microbiota composition in peritoneal dialysis patients, as was reported by some authors. Zhou et al. observed a higher abundance of Bacteroidetes and Synergistetes in the peritonitis group as compared to patients without peritonitis, while Bacilli and Lactobacillus were decreased [70]. Additionally, Dorea and Clostridium abundance was decreased in peritoneal dialysis patients [45].
Moreover, available gut microbiota modulation interventions (including dietary interventions, probiotics, prebiotics, and synbiotics) are presented in Table 3. Concerning intestinal flora modulation, synbiotics increased Bifidobacterium levels up to 5-fold from baseline (p = 0.003) [87], which was concordant in clinical studies [84,85]. Lactobacillus levels were decreased following synbiotic therapy in one study [85], whereas in another study, Lactobacillus abundance was similar before and after the treatment [87]. Nevertheless, synbiotic therapy was linked to eGFR decrease with 3.14 mL/min/1.73 m2 (p < 0.01), requiring further research [84].
Table 3.
Studies reporting gut microbiota modulation in CKD patients.
| Study, Year | Type of Therapy | Results |
|---|---|---|
| Abdelbary et al., 2022 [75] | Sucroferric oxyhydroxide | In hemodialysis patients, Veillonella spp. and Ruminococcus torques levels increased (p = 0.0351 for both), while Subdoligranulum decreased (p = 0.0496). |
| Belova et al., 2020 [78] | Immobilized synbiotic LB-complex L vs. placebo | In 56% of patients in the treatment group, gut microbiota recovered as compared to placebo (grade III dysbiosis was absent after therapy). CRP decreased from 6.8 ± 3.1 g/L to 5.3 g/L in the treatment group. |
| Borges et al., 2017 [79] | Probiotics | Gut microbiota profile was similar in the probiotic group (Streptococcus thermophilus, Lactobacillus acidophilus, and Bifidobacteria longum strains) and placebo group after 3 months of therapy (similar number of bands). |
| Ebrahim et al., 2022 [80] | β-glucan prebiotic | Prevotella tended to increase in the intervention group (β-glucan) as compared to the control group, while Bacteroides and Blautia tended to decrease. |
| Hu et al., 2022 [66] | Dietary intervention | HD patients from the protein-energy wasting group had lower abundance of Roseburia as compared to HD patients in the non-protein energy wasting group (p = 0.022). Escherichia abundance was increased in PD patients from the protein-energy wasting group compared to PD patients from the non-protein-energy wasting group (p = 0.022). |
| Iguchi et al., 2020 [31] | Sucroferric oxyhydroxide | In HD patients, sucroferric oxyhydroxide did not affect major phyla (p = 0.849 for Firmicutes, p = 0.776 for Proteobacteria, p = 0.517 for Actinobacteria, p = 0.728 for Bacteroides). |
| Jiang et al., 2020 [67] | Dietary intervention | Patients with CKD stage 5 who received a very low protein diet had higher levels of Escherichia, Shigella, and Klebsiella, while Blautia was decreased. |
| Kemp et al., 2021 [81] | Resistant starch type-2 | Resistant starch type-2 increased Oscillosperaceae, Roseburia, and Ruminococcus gauvreauii levels. Resistant starch type-2 decreased Ruminococcus champanellens, Dialister, and Coprococcus. |
| Kimber et al., 2020 [82] | Rifaximin | Rifaximin was linked to reduced diversity and richness of microbiota as compared to placebo. Rifaximin reduced 10 bacterial taxa from Firmicutes and Actinobacteria phyla (including Clostridium, Turicibacter, and Anaerotruncus). |
| Laffin et al., 2019 [89] | Amylose-resistant starch | Amylose-resistant starch increased levels of Faecalibacterium in ESKD patients as compared to placebo (from 0.40 ± 0.50% to 3.21 ± 4.97%, p = 0.03), while Parabacteroides, Bifidobacteria, Ruminococcus, and Prevotella levels did not change. |
| Lai et al., 2019 [37] | Low-protein diet | Low-protein diet increased Akkermansiaceae and Bacteroidaceae and decreased Christensenellaceae, Clostridiaceae, Lactobacillaceae, and Pasteurellaceae levels. |
| Low-protein diet + inulin | Low-protein diet associated with inulin therapy increased Bifidobacteriaceae levels. | |
| Inulin | Inulin decreased Enterobacteriaceae family. | |
| Liu et al., 2020 [83] | Probiotics | Probiotics increased Bacteroidaceae and Enterococcaceae abundance compared to placebo. Probiotics decreased Ruminococcaceae, Halomonadaceae, Peptostreptococcaceae, and Clostridiales Family XIII levels compared to placebo. |
| Liu et al., 2022 [76] | Iron supplementation | After oral iron supplementation, α-diversity and Firmicutes levels decreased, while Bacteroides increased. Moreover, Blautia and Coprococcus levels decreased, while Bacteroidetes increased. |
| McFarlane et al., 2021 [84] | Synbiotics vs. placebo | Synbiotic therapy increased Bifidobacterium animalis (p < 0.001) and Blautia spp. levels (p = 0.004). Synbiotics decreased Bacteroides cellulosilyticus and Ruminiclostridium spp. (p < 0.05 for both). Synbiotic therapy was linked to eGFR decrease with 3.14 mL/min/1.73 m2 (p < 0.01). |
| Miao et al., 2018 [63] | Lanthanum carbonate | In HD patients, lanthanum carbonate decreased Bacteroides and Proteobacteria but increased Actinobacteria levels. Shannon index decreased following lanthanum carbonate therapy. |
| Cruz-Mora et al., 2014 [85] | Synbiotics | In HD patients, synbiotic therapy increased Bifidobacterium abundance (p = 0.0344) but decreased Lactobacillus levels. |
| Nazzal et al., 2017 [77] | Oral vancomycin | Following vancomycin therapy, Clostridia, Roseburia, Enterococcaceae, and Bacteroidales decreased, while Veillonellaceae increased. |
| Pivari et al., 2022 [48] | Curcumin supplementation | After 6 months of dietary intervention, Escherichia-Shigella levels significantly decreased, while Lachnoclostridium and Lactobacillaceae spp. increased. |
| Rocchetti et al., 2021 [86] | Dietary intervention | The keto analogs-supplemented Mediterranean diet reduced Clostridiaceae, Methanobacteriaceae, Prevotellaceae, and Lactobacillaceae abundance, while Bacteroidaceae and Lachnospiraceae levels increased. |
| Rossi et al., 2016 [87] | Synbiotics | Compared to placebo, synbiotics were linked to a 5-fold increase in Bifidobacterium spp. (p = 0.003), while Lactobacillus spp. abundance was similar. |
| Simeoni et al., 2019 [88] | Probiotics | Compared to the placebo group, probiotics increased Lactobacillales and Bifidobacteria levels from 2.1 × 103 CFU/gr and 1.9 × 104 CFU/gr to 2.2 × 106 CFU/gr and 2.5 × 107 CFU/gr, respectively (p < 0.001 for both). Iron and ferritin levels were significantly increased after probiotic therapy (p < 0.001 for both), while CRP, total cholesterol, and triglycerides levels were decreased in patients who received probiotics (respectively, p < 0.001, p < 0.01, and p < 0.01). |
| Wu et al., 2020 [64] | Dietary intervention | CKD patients who received a low protein diet had lower levels of Lachnospiraceae and Bacteroidaceae as compared to those receiving a normal protein diet. |
| Wu et al., 2020 [65] | Phosphate binders | α-diversity and Simpson index were decreased in HD patients receiving calcium carbonate compared to the ferric citrate group (respectively, p = 0.049 and p = 0.001). Patients receiving ferric citrate had increased levels of Bacteroidetes phylum levels, while Firmicutes phylum was decreased. |
| Yacoub et al., 2017 [68] | Advanced glycation end products | PD patients who received a one-month advanced glycation end-product restriction had a lower abundance of Prevotella copri compared to those with a normal diet. |
CKD = chronic kidney disease; CRP = C-reactive protein; eGFR = estimated glomerular filtration rate; HD = hemodialysis; PD = peritoneal dialysis.
Liu et al. observed that probiotics increased Bacteroidaceae and Enterococcaceae abundance in hemodialysis patients, while Ruminococcaceae, Halomonadaceae, Peptostreptococcaceae, and Clostridiales family were decreased [83]. Additionally, probiotics significantly increased Lactobacillales and Bifidobacteria levels (p < 0.001) in another study [88].
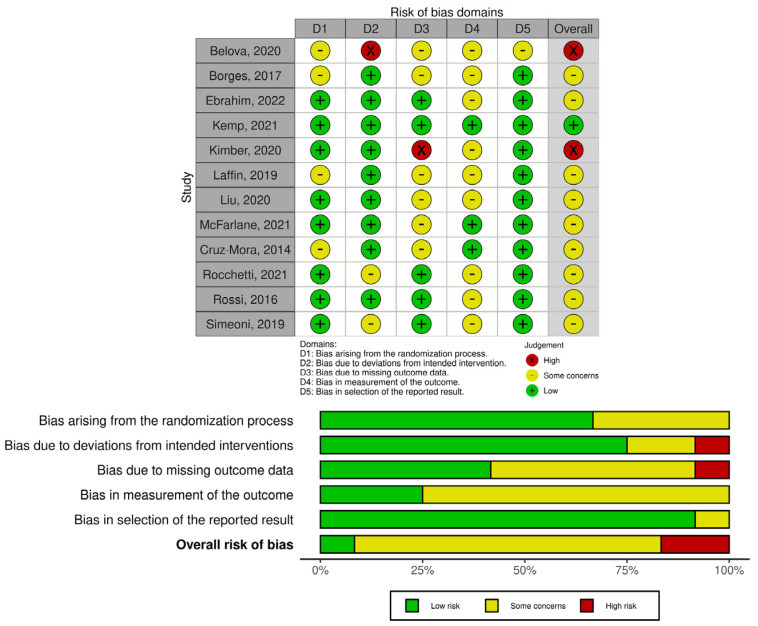
The quality of non-randomized studies was fair to good as assessed by NOS adapted for cross-sectional, case-control, and cohort studies (Tables S2–S4). The risk of bias in randomized trials was appraised using the RoB 2 tool and is displayed in Figure 2.
Figure 2.
Risk of bias of randomized trials (RoB 2). Belova et. al., 2020 [78], Borges et al., 2017 [79], Ebrahim et al., 2022 [80], Kemp et al., 2021 [81], Kimber et al., 2020 [82], Laffin et al., 2019 [89], Liu et al., 2020 [83], McFarlane et al., 2021 [84], Cruz-Mora et al., 2014 [85], Rocchetti et al., 2021 [86], Rossi et al., 2016 [87], Simeoni et al., 2019 [88].
4. Discussion
This study assessed gut microbiota alteration across a spectrum of kidney disease through a systematic review. The main findings are: (1) a different microbiota composition and a decreased gut diversity were reported from early stages to advanced CKD; (2) patients with CKD shared the depletion of anti-inflammatory butyrate-producing microbes (i.e., Roseburia, Prevotella, Bacteroides) and the enrichment of pro-inflammatory microbes (Proteobacteria, Actinobacteria); (3) there are limited data regarding the impact of dysbiosis on inflammation, mortality, or cardiovascular risk.
This review was designed to comprehensively illustrate the alteration of the gut microbiome in CKD patients. In more than half of the analyzed studies, gut diversity was significantly decreased. Similar data were reported last year by Zhao et al. [93]. Six (6/9) and four studies (4/6) that included patients with CKD and ESKD, respectively, suggested that the α-diversity of gut microbiota was significantly lower in patients than in healthy controls. Additionally, ten (10/11) studies that analyzed patients with CKD and five (5/6) studies that focused on ESKD reported a significantly altered composition of the gut microbiota in patients as compared to healthy controls [93].
At the phylum level, Fusobacteria, Verrucomicrobia, and Proteobacteria abundances were significantly higher in CKD [39,61]. At the genus and species levels, there were some substantial differences; probably, the individual variances determined by genetic and environmental factors and the etiology and the severity of CKD could explain the variation and inconsistency of the studies.
Proteobacteria, including common bacteria such as Escherichia coli, Salmonella, or Desulfovibrio, are increased in CKD [49,83]. An increased level was associated with abnormal gut barrier function, which might result in increased epithelial permeability, allowing microbial fragments and products to enter the sub-epithelial space and lamina propria, lipopolysaccharides translocation, and enhancing inflammatory response [94]. Additionally, people with an increased level of Desulfovibrio spp. had more severe renal dysfunction [94].
Concerning phylum Firmicutes, we noticed an increase in the genus Streptococcus and a lower abundance of the genus Roseburia, Faecalibacterium, and Prevotella. Roseburia produces butyrate, which could promote the proliferation of extrathymic regulatory T cells (Tregs). Tregs, as vital anti-inflammatory lymphocytes, produce interleukin-10, transforming growth factor beta, and interferon gamma. Microbial butyrate has been established to contribute to the pro- and anti-inflammatory balance by inducing Tregs differentiation [95]. Roseburia was negatively related to inflammatory status, renal function, and CKD progression [32,33].
From the phylum Verrucomicrobia, the genus Akkermansia plays an essential role in improving gut-barrier function and viscosity of the mucus. It also promotes the growth of bacteria-producing SCFAs, such as butyrate, by offering them carbon, nitrogen, and energy produced as a consequence of mucus degradation [96]. It also has anti-inflammatory properties. Unfortunately, the abundance was decreased in CKD patients [41,83] and was negatively related to inflammation [46].
Regarding the Bacteroidetes phylum, we observed an increase in the genus Bacteroides and a lower abundance of the genus Prevotella. In the general population, it was noticed that the Western diet was associated with Bacteroides and Clostridiales abundance in the gut microbiome, while rural populations with a high-fiber, low-protein diet tended to have Prevotella, which can produce SCFA. The authors from one study evaluated fecal microbiota composition differences between ESKD patients and 60 healthy controls. They found that Prevotella was enriched in the healthy group, whereas Bacteroides was prevalent in the ESKD group. Moreover, Prevotella was negatively related to inflammatory status and renal function [33]. Bacteroides was increased in most studies, whereas only two studies reported decreased levels [48,53]. Further analysis confirmed that Bacteroides was related to cardiovascular mortality in patients with dialysis. Nevertheless, the authors reported only five cardiovascular-related deaths, thus limiting the extrapolation of the results to all CKD patients. Consequently, these data should be confirmed in large clinical trials [45]. In addition, patients with peritoneal dialysis and peritonitis had a higher abundance of Bacteroides compared with the non-peritonitis group [70].
Our data are comparable to those obtained by Zhao et al.; the same abundance of Proteobacteria in CKD and ESKD was identified [93]. However, we identified a greater abundance of Bacteroides in CKD patients as compared to previous data. Zhao et al. reported enriched levels of bacteria from the Bacteroides genus in 3/11 studies involving CKD patients and in 3/9 studies on ESKD patients [93]. Furthermore, it was connected with infections and cardiovascular mortality [93].
Gut microbial dysbiosis has also been reported in diabetes mellitus. Patients with diabetic nephropathy exhibited increased levels of multiple pathogenic genera such as Acidaminococcus, Lactobacillus, Megasphaera, Clostridium, Sutterella, and Desulfovibrionaceae, while healthy controls had a high abundance of butyrate-producing bacteria [23]. The data are similar to those from a recently published systematic review [97]. Nevertheless, additional studies are warranted to investigate which specific microbes are involved in the pathophysiology of CKD linked to diabetes mellitus.
Data concerning phosphate binders on the gut microbiome in CKD patients are limited. Two studies documented a lower gut flora diversity in HD patients receiving phosphate binder [63,65]. In another study, sucroferric oxyhydroxide supplementation increased levels of Ruminococcus torques, which could influence gut barrier permeability [75,98]. Likewise, after oral iron supplementation, α-diversity and Firmicutes levels decreased, while Bacteroides increased [76]. These findings warrant further safety analysis of phosphate binders and iron supplementation in terms of gut homeostasis and microbiome composition in CKD patients.
Evidence sustaining the benefit of probiotic, prebiotic, and synbiotic supplementation in the management of CKD is mixed. Some studies insinuated that they might be useful by decreasing uremic and inflammatory toxins [99]. They could also improve oxidative stress, as well as lipid profiles in patients with CKD, which are well-known cardiovascular risk factors [100].
5. Conclusions
In conclusion, patients with CKD had an altered gut microbiome profile, even at early disease stages, as was documented consistently in clinical studies. Moreover, studies reported a shift in gut microbiota composition along with CKD progression, especially in ESKD patients and those requiring RRT. Different abundance at genera and species levels could be used in clinical models to discriminate between healthy individuals and patients with CKD (including those with diabetic nephropathy), with excellent predictive power. In addition to traditional risk factors, ESKD patients with an increased mortality risk could also be identified through gut microbiota analysis. In addition, studies established gut microbiome patterns linked to enhanced inflammatory activity and to a higher risk of peritonitis in patients receiving peritoneal dialysis. Nevertheless, clinical studies with larger sample sizes are required to confirm the association between altered gut microbiota composition and adverse outcomes in CKD patients, including all-cause and cardiovascular mortality. Due to data inconsistency, randomized clinical trials are needed to analyze the effect of different microbiota modulation therapies on gut bacterial composition and adverse end-points in CKD patients.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/jcm12051948/s1.
Author Contributions
Conceptualization, L.V., A.B., A.C. (Andreea Covic), A.C. (Adrian Covic); methodology, A.B., C.B.; investigation, A.B., C.B., G.G.B., I.N., C.P., S.H.; resources, L.V., A.C. (Andreea Covic); data curation, A.C. (Adrian Covic), L.V.; writing—original draft preparation, A.B., C.B., L.V., A.C. (Andreea Covic); writing—review and editing, A.B., L.V., A.C. (Adrian Covic); visualization, A.B., C.B.; supervision, A.C. (Adrian Covic), A.B.; project administration, A.C. (Adrian Covic), I.N., A.B. All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The data presented in this study are available on request from the corresponding author.
Conflicts of Interest
The authors declare no conflict of interest.
Funding Statement
This research received no external funding.
Footnotes
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content.
References
- 1.Jandhyala S.M., Talukdar R., Subramanyam C., Vuyyuru H., Sasikala M., Nageshwar Reddy D. Role of the normal gut microbiota. World J. Gastroenterol. 2015;21:8787–8803. doi: 10.3748/wjg.v21.i29.8787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.O’Hara A.M., Shanahan F. The gut flora as a forgotten organ. EMBO Rep. 2006;7:688–693. doi: 10.1038/sj.embor.7400731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baquero F., Nombela C. The microbiome as a human organ. Clin. Microbiol. Infect. 2012;18((Suppl. 4)):2–4. doi: 10.1111/j.1469-0691.2012.03916.x. [DOI] [PubMed] [Google Scholar]
- 4.Clarke G., Stilling R.M., Kennedy P.J., Stanton C., Cryan J.F., Dinan T.G. Minireview: Gut microbiota: The neglected endocrine organ. Mol. Endocrinol. 2014;28:1221–1238. doi: 10.1210/me.2014-1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rinninella E., Raoul P., Cintoni M., Franceschi F., Miggiano G.A.D., Gasbarrini A., Mele M.C. What is the Healthy Gut Microbiota Composition? A Changing Ecosystem across Age, Environment, Diet, and Diseases. Microorganisms. 2019;7:14. doi: 10.3390/microorganisms7010014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Al Khodor S., Shatat I.F. Gut microbiome and kidney disease: A bidirectional relationship. Pediatr. Nephrol. 2017;32:921–931. doi: 10.1007/s00467-016-3392-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Masenga S.K., Hamooya B., Hangoma J., Hayumbu V., Ertuglu L.A., Ishimwe J., Rahman S., Saleem M., Laffer C.L., Elijovich F., et al. Recent advances in modulation of cardiovascular diseases by the gut microbiota. J. Hum. Hypertens. 2022;36:952–959. doi: 10.1038/s41371-022-00698-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Twardowska A., Makaro A., Binienda A., Fichna J., Salaga M. Preventing Bacterial Translocation in Patients with Leaky Gut Syndrome: Nutrition and Pharmacological Treatment Options. Int. J. Mol. Sci. 2022;23:3204. doi: 10.3390/ijms23063204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.de Waal G.M., de Villiers W.J.S., Pretorius E. The Link Between Bacterial Inflammagens, Leaky Gut Syndrome and Colorectal Cancer. Curr. Med. Chem. 2021;28:8534–8548. doi: 10.2174/0929867328666210219142737. [DOI] [PubMed] [Google Scholar]
- 10.Schwabkey Z.I., Jenq R.R. Microbiome Anomalies in Allogeneic Hematopoietic Cell Transplantation. Annu. Rev. Med. 2020;71:137–148. doi: 10.1146/annurev-med-052918-122440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ananya F.N., Ahammed M.R., Fahem M.M., Kafle S., Viswanathan M., Desai D., Akku R., Khan F., Hernandez T.E., Bala S.K., et al. Association of Intestinal Microbial Dysbiosis With Chronic Obstructive Pulmonary Disease. Cureus. 2021;13:e19343. doi: 10.7759/cureus.19343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Forkosh E., Ilan Y. The heart-gut axis: New target for atherosclerosis and congestive heart failure therapy. Open Heart. 2019;6:e000993. doi: 10.1136/openhrt-2018-000993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Navaneethan S.D., Schold J.D., Arrigain S., Jolly S.E., Nally J.V., Jr. Cause-Specific Deaths in Non-Dialysis-Dependent CKD. J. Am. Soc. Nephrol. 2015;26:2512–2520. doi: 10.1681/ASN.2014101034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Witkowski M., Weeks T.L., Hazen S.L. Gut Microbiota and Cardiovascular Disease. Circ. Res. 2020;127:553–570. doi: 10.1161/CIRCRESAHA.120.316242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Page M.J., McKenzie J.E., Bossuyt P.M., Boutron I., Hoffmann T.C., Mulrow C.D., Shamseer L., Tetzlaff J.M., Akl E.A., Brennan S.E., et al. The PRISMA 2020 statement: An updated guideline for reporting systematic reviews. BMJ. 2021;372:n71. doi: 10.1136/bmj.n71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Davani-Davari D., Negahdaripour M., Karimzadeh I., Seifan M., Mohkam M., Masoumi S.J., Berenjian A., Ghasemi Y. Prebiotics: Definition, Types, Sources, Mechanisms, and Clinical Applications. Foods. 2019;8:92. doi: 10.3390/foods8030092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hill C., Guarner F., Reid G., Gibson G.R., Merenstein D.J., Pot B., Morelli L., Canani R.B., Flint H.J., Salminen S., et al. The International Scientific Association for Probiotics and Prebiotics consensus statement on the scope and appropriate use of the term probiotic. Nat. Rev. Gastroenterol. Hepatol. 2014;11:506–514. doi: 10.1038/nrgastro.2014.66. [DOI] [PubMed] [Google Scholar]
- 18.Swanson K.S., Gibson G.R., Hutkins R., Reimer R.A., Reid G., Verbeke K., Scott K.P., Holscher H.D., Azad M.B., Delzenne N.M., et al. The International Scientific Association for Probiotics and Prebiotics (ISAPP) consensus statement on the definition and scope of synbiotics. Nat. Rev. Gastroenterol. Hepatol. 2020;17:687–701. doi: 10.1038/s41575-020-0344-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sterne J.A.C., Savović J., Page M.J., Elbers R.G., Blencowe N.S., Boutron I., Cates C.J., Cheng H.-Y., Corbett M.S., Eldridge S.M., et al. RoB 2: A revised tool for assessing risk of bias in randomised trials. BMJ. 2019;366:l4898. doi: 10.1136/bmj.l4898. [DOI] [PubMed] [Google Scholar]
- 20.Stang A. Critical evaluation of the Newcastle-Ottawa scale for the assessment of the quality of nonrandomized studies in meta-analyses. Eur. J. Epidemiol. 2010;25:603–605. doi: 10.1007/s10654-010-9491-z. [DOI] [PubMed] [Google Scholar]
- 21.Barros A.F., Borges N.A., Ferreira D.C., Carmo F.L., Rosado A.S., Fouque D., Mafra D. Is there interaction between gut microbial profile and cardiovascular risk in chronic kidney disease patients? Future Microbiol. 2015;10:517–526. doi: 10.2217/fmb.14.140. [DOI] [PubMed] [Google Scholar]
- 22.Chen T.-H., Liu C.-W., Ho Y.-H., Huang C.-K., Hung C.-S., Smith B., Lin J.-C. Gut Microbiota Composition and Its Metabolites in Different Stages of Chronic Kidney Disease. J. Clin. Med. 2021;10:3881. doi: 10.3390/jcm10173881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Du X., Liu J., Xue Y., Kong X., Lv C., Li Z., Huang Y., Wang B. Alteration of gut microbial profile in patients with diabetic nephropathy. Endocrine. 2021;73:71–84. doi: 10.1007/s12020-021-02721-1. [DOI] [PubMed] [Google Scholar]
- 24.Gao B., Jose A., Alonzo-Palma N., Malik T., Shankaranarayanan D., Regunathan-Shenk R., Raj D.S. Butyrate producing microbiota are reduced in chronic kidney diseases. Sci. Rep. 2021;11:23530. doi: 10.1038/s41598-021-02865-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gryp T., Faust K., Van Biesen W., Huys G.R.B., Verbeke F., Speeckaert M., Raes J., Vaneechoutte M., Joossens M., Glorieux G. Gut Microbiome Profiling Uncovers a Lower Abundance of Butyricicoccus in Advanced Stages of Chronic Kidney Disease. J. Pers. Med. 2021;11:1118. doi: 10.3390/jpm11111118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gryp T., Huys G.R.B., Joossens M., Van Biesen W., Glorieux G., Vaneechoutte M. Isolation and Quantification of Uremic Toxin Precursor-Generating Gut Bacteria in Chronic Kidney Disease Patients. Int. J. Mol. Sci. 2020;21:1986. doi: 10.3390/ijms21061986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Guirong Y.E., Minjie Z., Lixin Y.U., Junsheng Y.E., Lin Y., Lisha S. [Gut microbiota in renal transplant recipients, patients with chronic kidney disease and healthy subjects] Nan Fang Yi Ke Xue Xue Bao. 2018;38:1401–1408. doi: 10.12122/j.issn.1673-4254.2018.12.01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hanifi G.R., Samadi Kafil H., Tayebi Khosroshahi H., Shapouri R., Asgharzadeh M. Bifidobacteriaceae Family Diversity in Gut Microbiota of Patients with Renal Failure. Arch. Razi Inst. 2021;76:521–528. doi: 10.22092/ari.2020.352271.1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.He H., Xie Y. Alteration of Intestinal Microflora in Uremia Patients With or Without Blood Purification. Niger. J. Clin. Pract. 2021;24:1133–1137. doi: 10.4103/njcp.njcp_484_19. [DOI] [PubMed] [Google Scholar]
- 30.Hu J., Zhong X., Yan J., Zhou D., Qin D., Xiao X., Zheng Y., Liu Y. High-throughput sequencing analysis of intestinal flora changes in ESRD and CKD patients. BMC Nephrol. 2020;21:12. doi: 10.1186/s12882-019-1668-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Iguchi A., Yamamoto S., Oda A., Tanaka K., Kazama J.J., Saeki T., Yamazaki H., Ishioka K., Suzutani T., Narita I. Effect of sucroferric oxyhydroxide on gastrointestinal microbiome and uremic toxins in patients with chronic kidney disease undergoing hemodialysis. Clin. Exp. Nephrol. 2020;24:725–733. doi: 10.1007/s10157-020-01892-x. [DOI] [PubMed] [Google Scholar]
- 32.Jiang S., Xie S., Lv D., Zhang Y., Deng J., Zeng L., Chen Y. A reduction in the butyrate producing species Roseburia spp. and Faecalibacterium prausnitzii is associated with chronic kidney disease progression. Antonie Leeuwenhoek. 2016;109:1389–1396. doi: 10.1007/s10482-016-0737-y. [DOI] [PubMed] [Google Scholar]
- 33.Jiang S., Xie S., Lv D., Wang P., He H., Zhang T., Zhou Y., Lin Q., Zhou H., Jiang J., et al. Alteration of the gut microbiota in Chinese population with chronic kidney disease. Sci. Rep. 2017;7:2870. doi: 10.1038/s41598-017-02989-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Khiabani S.A., Haghighat S., Khosroshahi H.T., Asgharzadeh M., Kafil H.S. Clostridium species diversity in gut microbiota of patients with renal failure. Microb. Pathog. 2022;169:105667. doi: 10.1016/j.micpath.2022.105667. [DOI] [PubMed] [Google Scholar]
- 35.Kim J.E., Kim H.E., Park J.I., Cho H., Kwak M.J., Kim B.Y., Yang S.H., Lee J.P., Kim D.K., Joo K.W., et al. The Association between Gut Microbiota and Uremia of Chronic Kidney Disease. Microorganisms. 2020;8:907. doi: 10.3390/microorganisms8060907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sugurmar A.N.K., Mohd R., Shah S.A., Neoh H.M., Cader R.A. Gut microbiota in Immunoglobulin A Nephropathy: A Malaysian Perspective. BMC Nephrol. 2021;22:145. doi: 10.1186/s12882-021-02315-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lai S., Molfino A., Testorio M., Perrotta A.M., Currado A., Pintus G., Pietrucci D., Unida V., La Rocca D., Biocca S., et al. Effect of Low-Protein Diet and Inulin on Microbiota and Clinical Parameters in Patients with Chronic Kidney Disease. Nutrients. 2019;11:3006. doi: 10.3390/nu11123006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lecamwasam A., Nelson T.M., Rivera L., Ekinci E.I., Saffery R., Dwyer K.M. Gut Microbiome Composition Remains Stable in Individuals with Diabetes-Related Early to Late Stage Chronic Kidney Disease. Biomedicines. 2020;9:19. doi: 10.3390/biomedicines9010019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li F., Wang M., Wang J., Li R., Zhang Y. Alterations to the Gut Microbiota and Their Correlation With Inflammatory Factors in Chronic Kidney Disease. Front. Cell. Infect. Microbiol. 2019;9:206. doi: 10.3389/fcimb.2019.00206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lin T.Y., Hung S.C. Association of subjective global assessment of nutritional status with gut microbiota in hemodialysis patients: A case-control study. Nephrol. Dial. Transplant. 2021;36:1104–1111. doi: 10.1093/ndt/gfaa019. [DOI] [PubMed] [Google Scholar]
- 41.Lin T.Y., Wu P.H., Lin Y.T., Hung S.C. Characterization of Gut Microbiota Composition in Hemodialysis Patients With Normal Weight Obesity. J. Clin. Endocrinol. Metab. 2020;105:2006–2014. doi: 10.1210/clinem/dgaa166. [DOI] [PubMed] [Google Scholar]
- 42.Lin X., Liang W., Li L., Xiong Q., He S., Zhao J., Guo X., Xiang S., Zhang P., Wang H., et al. The Accumulation of Gut Microbiome-derived Indoxyl Sulfate and P-Cresyl Sulfate in Patients With End-stage Renal Disease. J. Ren. Nutr. 2022;32:578–586. doi: 10.1053/j.jrn.2021.09.007. [DOI] [PubMed] [Google Scholar]
- 43.Liu F., Xu X., Chao L., Chen K., Shao A., Sun D., Hong Y., Hu R., Jiang P., Zhang N., et al. Alteration of the Gut Microbiome in Chronic Kidney Disease Patients and Its Association With Serum Free Immunoglobulin Light Chains. Front. Immunol. 2021;12:609700. doi: 10.3389/fimmu.2021.609700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lun H., Yang W., Zhao S., Jiang M., Xu M., Liu F., Wang Y. Altered gut microbiota and microbial biomarkers associated with chronic kidney disease. MicrobiologyOpen. 2019;8:e00678. doi: 10.1002/mbo3.678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Luo D., Zhao W., Lin Z., Wu J., Lin H., Li Y., Song J., Zhang J., Peng H. The Effects of Hemodialysis and Peritoneal Dialysis on the Gut Microbiota of End-Stage Renal Disease Patients, and the Relationship Between Gut Microbiota and Patient Prognoses. Front. Cell. Infect. Microbiol. 2021;11:579386. doi: 10.3389/fcimb.2021.579386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Margiotta E., Miragoli F., Callegari M.L., Vettoretti S., Caldiroli L., Meneghini M., Zanoni F., Messa P. Gut microbiota composition and frailty in elderly patients with Chronic Kidney Disease. PLoS ONE. 2020;15:e0228530. doi: 10.1371/journal.pone.0228530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Al-Obaide M.A.I., Singh R., Datta P., Rewers-Felkins K.A., Salguero M.V., Al-Obaidi I., Kottapalli K.R., Vasylyeva T.L. Gut Microbiota-Dependent Trimethylamine-N-oxide and Serum Biomarkers in Patients with T2DM and Advanced CKD. J. Clin. Med. 2017;6:86. doi: 10.3390/jcm6090086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pivari F., Mingione A., Piazzini G., Ceccarani C., Ottaviano E., Brasacchio C., Dei Cas M., Vischi M., Cozzolino M.G., Fogagnolo P., et al. Curcumin Supplementation (Meriva(®)) Modulates Inflammation, Lipid Peroxidation and Gut Microbiota Composition in Chronic Kidney Disease. Nutrients. 2022;14:231. doi: 10.3390/nu14010231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ren Z., Fan Y., Li A., Shen Q., Wu J., Ren L., Lu H., Ding S., Ren H., Liu C., et al. Alterations of the Human Gut Microbiome in Chronic Kidney Disease. Adv. Sci. 2020;7:2001936. doi: 10.1002/advs.202001936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Salguero M.V., Al-Obaide M.A.I., Singh R., Siepmann T., Vasylyeva T.L. Dysbiosis of Gram-negative gut microbiota and the associated serum lipopolysaccharide exacerbates inflammation in type 2 diabetic patients with chronic kidney disease. Exp. Ther. Med. 2019;18:3461–3469. doi: 10.3892/etm.2019.7943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sato N., Kakuta M., Hasegawa T., Yamaguchi R., Uchino E., Murashita K., Nakaji S., Imoto S., Yanagita M., Okuno Y. Metagenomic profiling of gut microbiome in early chronic kidney disease. Nephrol. Dial. Transplant. 2021;36:1675–1684. doi: 10.1093/ndt/gfaa122. [DOI] [PubMed] [Google Scholar]
- 52.Stadlbauer V., Horvath A., Ribitsch W., Schmerböck B., Schilcher G., Lemesch S., Stiegler P., Rosenkranz A.R., Fickert P., Leber B. Structural and functional differences in gut microbiome composition in patients undergoing haemodialysis or peritoneal dialysis. Sci. Rep. 2017;7:15601. doi: 10.1038/s41598-017-15650-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang S., Lv D., Jiang S., Jiang J., Liang M., Hou F., Chen Y. Quantitative reduction in short-chain fatty acids, especially butyrate, contributes to the progression of chronic kidney disease. Clin. Sci. 2019;133:1857–1870. doi: 10.1042/CS20190171. [DOI] [PubMed] [Google Scholar]
- 54.Wang Y.F., Zheng L.J., Liu Y., Ye Y.B., Luo S., Lu G.M., Gong D., Zhang L.J. The gut microbiota-inflammation-brain axis in end-stage renal disease: Perspectives from default mode network. Theranostics. 2019;9:8171–8181. doi: 10.7150/thno.35387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wang X., Yang S., Li S., Zhao L., Hao Y., Qin J., Zhang L., Zhang C., Bian W., Zuo L., et al. Aberrant gut microbiota alters host metabolome and impacts renal failure in humans and rodents. Gut. 2020;69:2131–2142. doi: 10.1136/gutjnl-2019-319766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wu I.W., Gao S.S., Chou H.C., Yang H.Y., Chang L.C., Kuo Y.L., Dinh M.C.V., Chung W.H., Yang C.W., Lai H.C., et al. Integrative metagenomic and metabolomic analyses reveal severity-specific signatures of gut microbiota in chronic kidney disease. Theranostics. 2020;10:5398–5411. doi: 10.7150/thno.41725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wu I.W., Lin C.Y., Chang L.C., Lee C.C., Chiu C.Y., Hsu H.J., Sun C.Y., Chen Y.C., Kuo Y.L., Yang C.W., et al. Gut Microbiota as Diagnostic Tools for Mirroring Disease Progression and Circulating Nephrotoxin Levels in Chronic Kidney Disease: Discovery and Validation Study. Int. J. Biol. Sci. 2020;16:420–434. doi: 10.7150/ijbs.37421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wu R., Ruan X.L., Ruan D.D., Zhang J.H., Wang H.L., Zeng Q.Z., Lu T., Gan Y.M., Luo J.W., Wu J.B. Differences in gut microbiota structure in patients with stages 4-5 chronic kidney disease. Am. J. Transl. Res. 2021;13:10056–10074. [PMC free article] [PubMed] [Google Scholar]
- 59.Xu K.Y., Xia G.H., Lu J.Q., Chen M.X., Zhen X., Wang S., You C., Nie J., Zhou H.W., Yin J. Impaired renal function and dysbiosis of gut microbiota contribute to increased trimethylamine-N-oxide in chronic kidney disease patients. Sci. Rep. 2017;7:1445. doi: 10.1038/s41598-017-01387-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhang P., Fang J., Li G., Zhang L., Lai X., Xu L., Liu L., Xiong Y., Li L., Zhang T., et al. Sex Differences in Fecal Microbiota Correlation With Physiological and Biochemical Indices Associated With End-Stage Renal Disease Caused by Immunoglobulin a Nephropathy or Diabetes. Front. Microbiol. 2021;12:752393. doi: 10.3389/fmicb.2021.752393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhang J., Luo D., Lin Z., Zhou W., Rao J., Li Y., Wu J., Peng H., Lou T. Dysbiosis of gut microbiota in adult idiopathic membranous nephropathy with nephrotic syndrome. Microb. Pathog. 2020;147:104359. doi: 10.1016/j.micpath.2020.104359. [DOI] [PubMed] [Google Scholar]
- 62.Zheng L.J., Lin L., Zhong J., Zhang Z., Ye Y.B., Zhang X.Y., Wang Y.F., Zhang H., Liu Y., Lu G.M., et al. Gut dysbiosis-influence on amygdala-based functional activity in patients with end stage renal disease: A preliminary study. Brain Imaging Behav. 2020;14:2731–2744. doi: 10.1007/s11682-019-00223-3. [DOI] [PubMed] [Google Scholar]
- 63.Miao Y.Y., Xu C.M., Xia M., Zhu H.Q., Chen Y.Q. Relationship between Gut Microbiota and Phosphorus Metabolism in Hemodialysis Patients: A Preliminary Exploration. Chin. Med. J. 2018;131:2792–2799. doi: 10.4103/0366-6999.246059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wu I.W., Lee C.C., Hsu H.J., Sun C.Y., Chen Y.C., Yang K.J., Yang C.W., Chung W.H., Lai H.C., Chang L.C., et al. Compositional and Functional Adaptations of Intestinal Microbiota and Related Metabolites in CKD Patients Receiving Dietary Protein Restriction. Nutrients. 2020;12:2799. doi: 10.3390/nu12092799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wu P.H., Liu P.Y., Chiu Y.W., Hung W.C., Lin Y.T., Lin T.Y., Hung S.C., Delicano R.A., Kuo M.C., Wu C.Y. Comparative Gut Microbiome Differences between Ferric Citrate and Calcium Carbonate Phosphate Binders in Patients with End-Stage Kidney Disease. Microorganisms. 2020;8:2040. doi: 10.3390/microorganisms8122040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hu J., Zhong X., Liu Y., Yan J., Zhou D., Qin D., Xiao X., Zheng Y., Wen L., Tan R., et al. Correlation between intestinal flora disruption and protein–energy wasting in patients with end-stage renal disease. BMC Nephrol. 2022;23:130. doi: 10.1186/s12882-022-02762-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Jiang S., Wang B., Sha T., Li X. Changes in the Intestinal Microbiota in Patients with Stage 5 Chronic Kidney Disease on a Low-Protein Diet and the Effects of Human to Rat Fecal Microbiota Transplantation. Med. Sci. Monit. 2020;26:e921557. doi: 10.12659/MSM.921557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yacoub R., Nugent M., Cai W., Nadkarni G.N., Chaves L.D., Abyad S., Honan A.M., Thomas S.A., Zheng W., Valiyaparambil S.A., et al. Advanced glycation end products dietary restriction effects on bacterial gut microbiota in peritoneal dialysis patients; a randomized open label controlled trial. PLoS ONE. 2017;12:e0184789. doi: 10.1371/journal.pone.0184789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zhu Y., Tang Y., He H., Hu P., Sun W., Jin M., Wang L., Xu X. Gut Microbiota Correlates With Clinical Responsiveness to Erythropoietin in Hemodialysis Patients With Anemia. Front. Cell. Infect. Microbiol. 2022;12:919352. doi: 10.3389/fcimb.2022.919352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zhou J., Yang C., Lei W., Yang Z., Chen J., Lin H., Li Q., Yuan W. Exploration of the correlation between intestinal flora and Escherichia coli peritoneal dialysis-related peritonitis. BMC Nephrol. 2022;23:76. doi: 10.1186/s12882-022-02704-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hu X., Ouyang S., Xie Y., Gong Z., Du J. Characterizing the gut microbiota in patients with chronic kidney disease. Postgrad. Med. 2020;132:495–505. doi: 10.1080/00325481.2020.1744335. [DOI] [PubMed] [Google Scholar]
- 72.Lin T.-Y., Wu P.-H., Lin Y.-T., Hung S.-C. Gut dysbiosis and mortality in hemodialysis patients. NPJ Biofilms Microbiomes. 2021;7:20. doi: 10.1038/s41522-021-00191-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.He H., Xie Y. Effect of Different Hemodialysis Methods on Microbiota in Uremic Patients. BioMed Res. Int. 2020;2020:6739762. doi: 10.1155/2020/6739762. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 74.Hu Q., Wu K., Pan W., Zeng Y., Hu K., Chen D., Huang X., Zhang Q. Intestinal flora alterations in patients with early chronic kidney disease: A case-control study among the Han population in southwestern China. J. Int. Med. Res. 2020;48:300060520926033. doi: 10.1177/0300060520926033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Abdelbary M.M.H., Kuppe C., Michael S.S.-Y., Krüger T., Floege J., Conrads G. Impact of sucroferric oxyhydroxide on the oral and intestinal microbiome in hemodialysis patients. Sci. Rep. 2022;12:9614. doi: 10.1038/s41598-022-13552-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Liu H., Wu W., Luo Y. Oral and intravenous iron treatment alter the gut microbiome differentially in dialysis patients. Int. Urol. Nephrol. 2022;55:759–767. doi: 10.1007/s11255-022-03377-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Nazzal L., Roberts J., Singh P., Jhawar S., Matalon A., Gao Z., Holzman R., Liebes L., Blaser M.J., Lowenstein J. Microbiome perturbation by oral vancomycin reduces plasma concentration of two gut-derived uremic solutes, indoxyl sulfate and p-cresyl sulfate, in end-stage renal disease. Nephrol. Dial. Transplant. 2017;32:1809–1817. doi: 10.1093/ndt/gfx029. [DOI] [PubMed] [Google Scholar]
- 78.Belova I.V., Khrulev A.E., Tochilina A.G., Khruleva N.S., Lobanova N.A., Zhirnov V.A., Molodtsova S.B., Lobanov V.N., Solovieva I.V. Colon Microbiocenosis and Its Correction in Patients Receiving Programmed Hemodialysis. Sovrem. Tekhnologii V Meditsine. 2021;12:62–68. doi: 10.17691/stm2020.12.5.07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Borges N.A., Carmo F.L., Stockler-Pinto M.B., de Brito J.S., Dolenga C.J., Ferreira D.C., Nakao L.S., Rosado A., Fouque D., Mafra D. Probiotic Supplementation in Chronic Kidney Disease: A Double-blind, Randomized, Placebo-controlled Trial. J. Ren. Nutr. 2018;28:28–36. doi: 10.1053/j.jrn.2017.06.010. [DOI] [PubMed] [Google Scholar]
- 80.Ebrahim Z., Proost S., Tito R.Y., Raes J., Glorieux G., Moosa M.R., Blaauw R. The Effect of ß-Glucan Prebiotic on Kidney Function, Uremic Toxins and Gut Microbiome in Stage 3 to 5 Chronic Kidney Disease (CKD) Predialysis Participants: A Randomized Controlled Trial. Nutrients. 2022;14:805. doi: 10.3390/nu14040805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kemp J.A., Regis de Paiva B., Fragoso Dos Santos H., Emiliano de Jesus H., Craven H., U Z.I., Alvarenga Borges N., P G.S., Mafra D. The Impact of Enriched Resistant Starch Type-2 Cookies on the Gut Microbiome in Hemodialysis Patients: A Randomized Controlled Trial. Mol. Nutr. Food Res. 2021;65:e2100374. doi: 10.1002/mnfr.202100374. [DOI] [PubMed] [Google Scholar]
- 82.Kimber C., Zhang S., Johnson C., West R.E., 3rd, Prokopienko A.J., Mahnken J.D., Yu A.S., Hoofnagle A.N., Ir D., Robertson C.E., et al. Randomized, Placebo-Controlled Trial of Rifaximin Therapy for Lowering Gut-Derived Cardiovascular Toxins and Inflammation in CKD. Kidney360. 2020;1:1206–1216. doi: 10.34067/KID.0003942020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Liu S., Liu H., Chen L., Liang S.S., Shi K., Meng W., Xue J., He Q., Jiang H. Effect of probiotics on the intestinal microbiota of hemodialysis patients: A randomized trial. Eur. J. Nutr. 2020;59:3755–3766. doi: 10.1007/s00394-020-02207-2. [DOI] [PubMed] [Google Scholar]
- 84.McFarlane C., Krishnasamy R., Stanton T., Savill E., Snelson M., Mihala G., Kelly J.T., Morrison M., Johnson D.W., Campbell K.L. Synbiotics Easing Renal Failure by Improving Gut Microbiology II (SYNERGY II): A Feasibility Randomized Controlled Trial. Nutrients. 2021;13:4481. doi: 10.3390/nu13124481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Cruz-Mora J., Martínez-Hernández N.E., Martín del Campo-López F., Viramontes-Hörner D., Vizmanos-Lamotte B., Muñoz-Valle J.F., García-García G., Parra-Rojas I., Castro-Alarcón N. Effects of a symbiotic on gut microbiota in Mexican patients with end-stage renal disease. J. Ren. Nutr. 2014;24:330–335. doi: 10.1053/j.jrn.2014.05.006. [DOI] [PubMed] [Google Scholar]
- 86.Rocchetti M.T., Di Iorio B.R., Vacca M., Cosola C., Marzocco S., di Bari I., Calabrese F.M., Ciarcia R., De Angelis M., Gesualdo L. Ketoanalogs’ Effects on Intestinal Microbiota Modulation and Uremic Toxins Serum Levels in Chronic Kidney Disease (Medika2 Study) J. Clin. Med. 2021;10:840. doi: 10.3390/jcm10040840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Rossi M., Johnson D.W., Morrison M., Pascoe E.M., Coombes J.S., Forbes J.M., Szeto C.C., McWhinney B.C., Ungerer J.P., Campbell K.L. Synbiotics Easing Renal Failure by Improving Gut Microbiology (SYNERGY): A Randomized Trial. Clin. J. Am. Soc. Nephrol. 2016;11:223–231. doi: 10.2215/CJN.05240515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Simeoni M., Citraro M.L., Cerantonio A., Deodato F., Provenzano M., Cianfrone P., Capria M., Corrado S., Libri E., Comi A., et al. An open-label, randomized, placebo-controlled study on the effectiveness of a novel probiotics administration protocol (ProbiotiCKD) in patients with mild renal insufficiency (stage 3a of CKD) Eur. J. Nutr. 2019;58:2145–2156. doi: 10.1007/s00394-018-1785-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Laffin M.R., Tayebi Khosroshahi H., Park H., Laffin L.J., Madsen K., Kafil H.S., Abedi B., Shiralizadeh S., Vaziri N.D. Amylose resistant starch (HAM-RS2) supplementation increases the proportion of Faecalibacterium bacteria in end-stage renal disease patients: Microbial analysis from a randomized placebo-controlled trial. Hemodial. International. Int. Symp. Home Hemodial. 2019;23:343–347. doi: 10.1111/hdi.12753. [DOI] [PubMed] [Google Scholar]
- 90.Mosca A., Leclerc M., Hugot J.P. Gut Microbiota Diversity and Human Diseases: Should We Reintroduce Key Predators in Our Ecosystem? Front. Microbiol. 2016;7:455. doi: 10.3389/fmicb.2016.00455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Nie K., Ma K., Luo W., Shen Z., Yang Z., Xiao M., Tong T., Yang Y., Wang X. Roseburia intestinalis: A Beneficial Gut Organism From the Discoveries in Genus and Species. Front. Cell. Infect. Microbiol. 2021;11:757718. doi: 10.3389/fcimb.2021.757718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Alessandri G., Ossiprandi M.C., MacSharry J., van Sinderen D., Ventura M. Bifidobacterial Dialogue With Its Human Host and Consequent Modulation of the Immune System. Front. Immunol. 2019;10:2348. doi: 10.3389/fimmu.2019.02348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Zhao J., Ning X., Liu B., Dong R., Bai M., Sun S. Specific alterations in gut microbiota in patients with chronic kidney disease: An updated systematic review. Ren. Fail. 2021;43:102–112. doi: 10.1080/0886022X.2020.1864404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Onal E.M., Afsar B., Covic A., Vaziri N.D., Kanbay M. Gut microbiota and inflammation in chronic kidney disease and their roles in the development of cardiovascular disease. Hypertens. Res. 2019;42:123–140. doi: 10.1038/s41440-018-0144-z. [DOI] [PubMed] [Google Scholar]
- 95.Hegazy A.N., West N.R., Stubbington M.J.T., Wendt E., Suijker K.I.M., Datsi A., This S., Danne C., Campion S., Duncan S.H., et al. Circulating and Tissue-Resident CD4(+) T Cells With Reactivity to Intestinal Microbiota Are Abundant in Healthy Individuals and Function Is Altered During Inflammation. Gastroenterology. 2017;153:1320–1337.e16. doi: 10.1053/j.gastro.2017.07.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Hänninen A., Toivonen R., Pöysti S., Belzer C., Plovier H., Ouwerkerk J.P., Emani R., Cani P.D., De Vos W.M. Akkermansia muciniphila induces gut microbiota remodelling and controls islet autoimmunity in NOD mice. Gut. 2018;67:1445–1453. doi: 10.1136/gutjnl-2017-314508. [DOI] [PubMed] [Google Scholar]
- 97.Han S., Chen M., Cheng P., Zhang Z., Lu Y., Xu Y., Wang Y. A systematic review and meta-analysis of gut microbiota in diabetic kidney disease: Comparisons with diabetes mellitus, non-diabetic kidney disease, and healthy individuals. Front. Endocrinol. 2022;13:1018093. doi: 10.3389/fendo.2022.1018093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Deaver J.A., Eum S.Y., Toborek M. Circadian Disruption Changes Gut Microbiome Taxa and Functional Gene Composition. Front. Microbiol. 2018;9:737. doi: 10.3389/fmicb.2018.00737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Ramezani A., Massy Z.A., Meijers B., Evenepoel P., Vanholder R., Raj D.S. Role of the Gut Microbiome in Uremia: A Potential Therapeutic Target. Am. J. Kidney Dis. 2016;67:483–498. doi: 10.1053/j.ajkd.2015.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Zheng H.J., Guo J., Wang Q., Wang L., Wang Y., Zhang F., Huang W.J., Zhang W., Liu W.J., Wang Y. Probiotics, prebiotics, and synbiotics for the improvement of metabolic profiles in patients with chronic kidney disease: A systematic review and meta-analysis of randomized controlled trials. Crit. Rev. Food Sci. Nutr. 2021;61:577–598. doi: 10.1080/10408398.2020.1740645. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data presented in this study are available on request from the corresponding author.