Abstract
Adherence of enterohemorrhagic Escherichia coli (EHEC) to the intestinal epithelium is critical for initiation of a bacterial infection. An in vitro infection study previously indicated that EHEC bacteria initially adhere diffusely and then proliferate to develop MC, a process that is mediated by various secreted proteins, such as EspA, EspB, EspD, Tir, and intimin, as well as other putative adherence factors. In the present study, we investigated the role of a large 93-kb plasmid (pO157) in the adherence of O157:H7 (O157Sakai) and found the toxB gene to be involved in the full adherence phenotype. A pO157-cured strain of O157Sakai (O157Cu) developed microcolonies on Caco-2 cells; however, the number of microcolonies was lower than that of O157Sakai, as were the production and secretion levels of EspA, EspB, and Tir. Introduction of a mini-pO157 plasmid (pIC37) composed of the toxB and ori regions restored full adherence capacity to O157Cu, including production and secretion of the proteins. In contrast, introduction of a pO157 mutant possessing toxB::Km into O157Cu could not restore the full adherence phenotype. Expression of truncated versions of His-tagged ToxB also promoted EspB production and/or secretion by O157Cu. These results suggest that ToxB contributes to the adherence of EHEC to epithelial cells through promotion of the production and/or secretion of type III secreted proteins.
Enterohemorrhagic Escherichia coli (EHEC) is responsible for a range of illnesses, including nonbloody diarrhea, hemorrhagic colitis, and hemolytic-uremic syndrome. The Centers for Disease Control and Prevention estimated that strains of O157:H7 cause approximately 73,000 illnesses and 60 deaths per year in the United States, and non-O157:H7 Shiga toxin-producing E. coli adds an additional 37,000 estimated cases (11, 20). The pathogenesis of EHEC (represented by O157:H7) has been shown to be associated with several characteristics, including the production of Shiga-like toxins, the ability to produce an attaching-and-effacing intestinal lesion, and the ability to induce enterohemolytic activity (2, 12, 22, 27). Among its characteristics is bacterial attachment to the intestinal epithelium, which constitutes an early step and is believed to be crucial to the development of illness. Adherence of EHEC to epithelial cells has been indicated to be mediated by various secreted proteins, such as EspA, EspB, EspD, Tir, and intimin, which are encoded by genes of the locus for enterocyte effacement (LEE) (6, 7, 17, 33, 41). We recently reported that an O157:H7 strain (O157Sakai) initially adhered diffusely to Caco-2 cells and proliferated to develop microcolonies (MC) and that mini-Tn5Km2 insertion mutants that showed a reduction in the number and size of MC were defective in an initial stage and the later stages of adherence, respectively (32). In that study, disruption of the eae gene encoding intimin in O157Sakai influenced MC formation but not initial attachment, while mutants defective in type III secretion were unable to adhere to epithelial cells. It is likely that proteins secreted via the type III system, such as EspA, and intimate adherence, which is mediated by the interaction of intimin with Tir, are involved in initial attachment and subsequent development of MC, respectively (6, 7, 32).
All O157:H7 strains, including O157Sakai, possess a large plasmid ranging from 93 to 104 kb in size (29). Although the significance of the plasmid for infection has been debated, recent studies suggest that the large plasmid (pO157) in O157:H7 is important for pathogenicity. Karch et al. reported that an O157:H7 strain (E. coli 933) produced fimbriae, which disappeared when pO157 was lost (14). Although fimbriae were not detected in other O157:H7 strains, such as 7785, when 7785 was cured of pO157, its capacity to adhere to epithelial cells was decreased (36). Fratamico et al., however, indicated that when pO157 was removed from some O157:H7 strains, their adherence was not substantially changed (9). Recently, Burland et al. and Makino et al. reported the complete nucleotide sequences of the pO157 plasmids from two independently isolated O157:H7 strains, 7785 and O157Sakai (3, 19). The entire plasmid sequences, including several putative virulence-associated genes, such as toxB, katP, espP, tagA, etcC-O, and hlyA-D, were almost identical. Recent findings may help us to understand the function of ToxB. For example, Nicholls et al. found that a clinical isolate, O111:H− EHEC, that displayed strong adherence to cultured Chinese hamster ovary cells showed a reduction in adherence capacity upon insertion of TnPhoA into an open reading frame (ORF) (efa-1) on the chromosome (23). In their report, they indicated that the predicted 365-kDa Efa-1 protein displays 28% amino acid identity with the predicted 362-kDa toxB product of pO157. Klapproth et al. recently reported that enteropathogenic E. coli (EPEC) possesses an ORF (lifA) and that the gene is associated with the ability to inhibit lymphocyte activation (15). In that study, they found that the predicted LifA protein shares 28% amino acid identity with the predicted ToxB protein and that the lifA homologue sequences were extensively distributed among various EHEC and EPEC strains, including Citrobacter rodentium (15). In this context, we wished to elucidate whether the toxB gene on pO157 is important for bacterial adherence to epithelial cells.
MATERIALS AND METHODS
Bacterial strains, media, and tissue culture.
O157:H7 strain RIMD 0509952 (referred to as O157Sakai in our previous paper [32]) was originally isolated from a patient in the biggest outbreak to occur in 1996 in Sakai City, Japan (19, 32, 40). EHEC was maintained as described previously (32). Caco-2 cells were maintained as previously reported (32).
Plasmid constructions.
Mini-pO157 plasmid pIC37 was constructed by ligating a 25-kb BamHI DNA fragment of pO157 with a Kmr-encoding gene cassette-bearing the BamHI DNA fragment of pUT mini-Tn5Km2 (5) and designated pIC37. pIC37 was maintained with 5 μg of kanamycin per ml in the host strain throughout the study. pMH900 was constructed by deleting a 10.3-kbp AatII fragment of pIC37.
Construction and detection of truncated ToxB versions tagged with six histidines.
The toxB gene and the various truncated toxB versions were constructed and tagged with six histidines as follows. pHis-ToxB(SacI) was constructed by ligating the 9.8-kb SacI-PstI fragment of pIC37 with the SacI-PstI fragment of pQE30 (QIAGEN). pHis-ToxB(HindIII) and pHis-ToxB(SphI) were constructed by deleting 5.3-kb HindIII and 5.6-kb SphI fragments from pHis- ToxB(SacI), respectively. pHis-ToxB(EcoT14I) was constructed by deleting a 1.5-kb EcoT14I fragment from pHis-ToxB(HindIII). Expression of the His-ToxB derivatives was detected by using the protocol for QIAexpress (QIAGEN) and the immunoblot method as described previously (32). Briefly, E. coli JM109 or XL-1 blue was used as a host strain. L broth (1.5 ml) containing ampicillin (50 μg/ml) was inoculated with overnight cultures (500 μl), which were then grown at 37°C for 30 min with vigorous shaking. Expressions were induced by adding isopropyl-β-d-thiogalactopyranoside (IPTG) to a final concentration of 1.5 mM. The cultures were grown for 3 h and transferred in 1-ml aliquots to microtubes. The cells and supernatants were concentrated by adding 340 μl of 24% trichloroacetic acid. The His-toxB derivatives were detected by using anti-RGSHHHH mouse immunoglobulin G antibody (QIAGEN).
Construction of a toxB mutant by insertion of a Kmr cassette gene.
Bacteria were cultured at 30°C for all constructions. A set of oligonucleotides, toxB3 (5′-CCGGGATCCATCCTGAAACAAAACGAAACG-3′) and toxB4 (5′-CGGAATTCGATGCAAAATTGTATGCTCTAG-3′), was used to amplify DNA fragments of 3,033 bp corresponding to the internal region of the toxB gene from plasmid pIC37 by PCR as described previously (32). The corresponding nucleotides contained restriction sites for the endonucleases BamHI and EcoRI (shown in bold in the oligonucleotide sequences). These enzymes were used to clone an amplification product into pBluescript II KS(+) (30). The resulting 3,017-bp BamHI-XbaI fragment, containing the toxB gene, at the ScaI site of which the blunted PstI fragment, containing the Kmr-encoding gene of pUK4K (34) was introduced, was subcloned into temperature- and sucrose-sensitive suicide vector pCACTUS (31), and the plasmid was introduced into O157Sakai by electroporation. The transformants were grown on L agar plates supplemented with kanamycin at 5 μg/ml and 5% sucrose without NaCl at 42°C. The resulting sucrose- and kanamycin-resistant colonies were tested for chloramphenicol sensitivity, which is indicative of loss of the suicide vector. The chloramphenicol-sensitive colonies thus selected were confirmed to contain an insertion of the Kmr-encoding gene in the toxB gene, as determined by restriction enzyme digestion of the PCR-amplified segment.
Adherence assay.
The adherence of O157 derivatives to Caco-2 cells was evaluated as previously described (32). Briefly, bacteria were grown in Dulbecco modified Eagle medium (DMEM) containing 0.45% glycerol for 2 h at 37°C (optical density at 600 nm, ∼1.8) and inoculated at a multiplicity of infection of 50:1 onto semiconfluent Caco-2 cell monolayers grown on coverslips in a 24-well plastic plate. Bacteria and cells were incubated for 1.5 h, washed five times with 1 ml of sterile phosphate-buffered saline, and overlaid with 1 ml of fresh DMEM containing 0.45% glycerol for an additional 2.5 h. The monolayers were fixed and stained with Giemsa's solution for microscopic evaluation. Bacterial clusters on Caco-2 cells consisting of eight or more bacteria were considered MC. All MC were counted by a technical assistant who did not know which strain was shown on the slide. The numbers of MC that developed on Caco-2 cells monolayers were adjusted by CFU measured by plating the inoculated bacteria on agar plates.
Secretion and expression of type III secreted proteins.
The secretion and expression of EspA, EspB, and Tir were analyzed as described previously (32), but in the case of O157Cu cells with pHis-ToxB(SacI) derivatives, bacteria were grown in DMEM-glycerol with 1.5 mM IPTG.
Fluorescence actin staining.
The ability of O157Sakai derivatives to induce intimate attachment was assessed by using the fluorescence actin staining test (16).
Northern blotting.
Cells harboring pIC37 were grown in 10 ml of DMEM containing 0.45% glycerol at 37°C. At an optical density at 600 nm of 0.27, the cells were harvested and total cellular RNA was prepared. The total RNA (5 or 15 μg) was resolved by 1.0% agarose-gel electrophoresis in the presence of formamide and blotted onto a Hybond-N+ membrane (Amersham) as previously described (28). The 563-bp EcoRV-SacII fragment containing the internal region of the espB structural gene and the 395-bp SspI fragment containing the entire ler structural gene were used as probes for the espB and ler mRNAs, respectively. The membrane was hybridized to the DNA probe labeled with the BrightStar Psoralen-Biotin Nonisotopic Labeling Kit (Ambion) and washed, and the signals were visualized with a BrightStar BioDetect Nonisotopic Detection Kit (Ambion).
RESULTS
Construction of pO157-cured strains and the adherence phenotype.
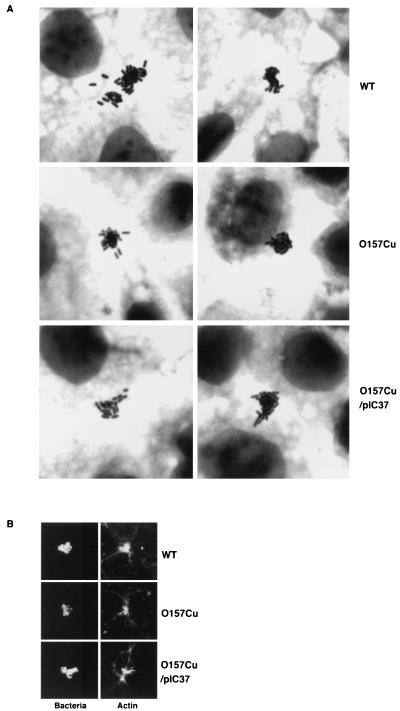
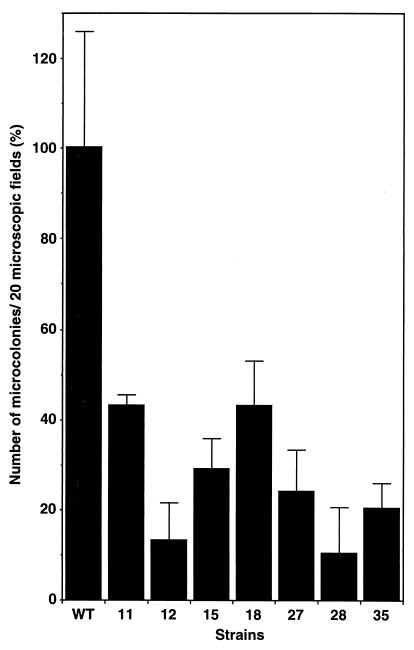
To eliminate pO157 from O157Sakai, mini-R plasmid pKP2368 encoding chloramphenicol (CM) resistance, which has the same incompatibility as pO157 (T. Miki et al., unpublished data), was introduced into O157Sakai by transformation. Of 41 transformants that were taken from the same electroporation and purified on LB agar containing CM at 25 μg/ml, 7 lost pO157, as confirmed by agarose gel electrophoresis. These seven Cmr transformants were then cured of pKP2368 by subculturing in LB broth without CM, since pKP2368 in E. coli K-12 had been shown to be less stable when grown in Luria-Bertani (LB) broth without CM (Miki et al., unpublished data). Seven independent isolates that displayed Cms were confirmed to be cured of pKP2368, but not pOSAK, which was the other small plasmid in O157Sakai (19), by agarose gel electrophoresis, and the mutants were tested for the capacity to adhere to Caco-2 cells. Caco-2 cells were infected with each of the mutants for 1.5 h, the nonadherent bacteria were washed out, and the infected cells were further incubated in fresh DMEM for another 2.5 h to allow the adherent bacteria to develop MC visible by Giemsa staining. The MC were then enumerated directly under a microscope, and the number of MC per 20 microfields was corrected by the CFU measured by placing bacteria on agar plates (see Materials and Methods). In the assay, the plasmid-cured strains developed MC on Caco-2 cells and induced actin condensation underneath the MC (Fig. 1A and B); however, the number of MC was lower than for the parental strain (Fig. 2). The results suggested that the presence of pO157 in O157 affects the initial adherence but not the intimate attachment involved in the later stage.
FIG. 1.
Adherence phenotypes of a pO157-cured mutant (O157Cu) and O157Cu/pIC37 (toxB gene). The representative slides were chosen by the first author, who did not know the identity of the bacteria being examined by the second. The other authors checked that the selection of slides was reasonable. Bacteria grown in DMEM-glycerol for 2 h were used to infect Caco-2 monolayers. Infected monolayers were incubated for 1.5 h and washed five times with phosphate-buffered saline. After another 2.5 h of incubation, the monolayers were again washed three times and fixed with methanol and stained with Giemsa solution to visualize the adherent bacterial colonies (A) or fixed with 1% paraformaldehyde and shown as fluorescent views after treatment with anti-O157 lipopolysaccharide rabbit antibody, followed by anti-rabbit goat antibody conjugated with fluorescein isothiocyanate (Bacteria), or as fluorescent views of actin stained by rhodamine-phalloidin (Actin) (B). Strains are indicated at the right of the photos. WT, wild type.
FIG. 2.
Adherence efficiency of pO157-cured mutants (O157Cu). Seven clones of O157Cu were identified as 11, 12, 15, 18, 27, 28, and 35. The number of MC on Caco-2 cells infected by bacteria and visualized as described in Fig. 1 was scored as the sum of 20 microscopic fields. Independent experiments were done at least 33 times with clone 11, 18 times with clone 12, 21 times with clone 15, 24 times with clone 18, 27 times with clone 27, 30 times with clone 28, 27 times with clone 35, and 18 times with clone 38. The data shown are the means and standard deviations of three representative experiments. The experiments were performed on separate days; therefore, each value was normalized to that of the wild type (WT; 100%).
The toxB region in pO157 affects EHEC adherence to host cells.
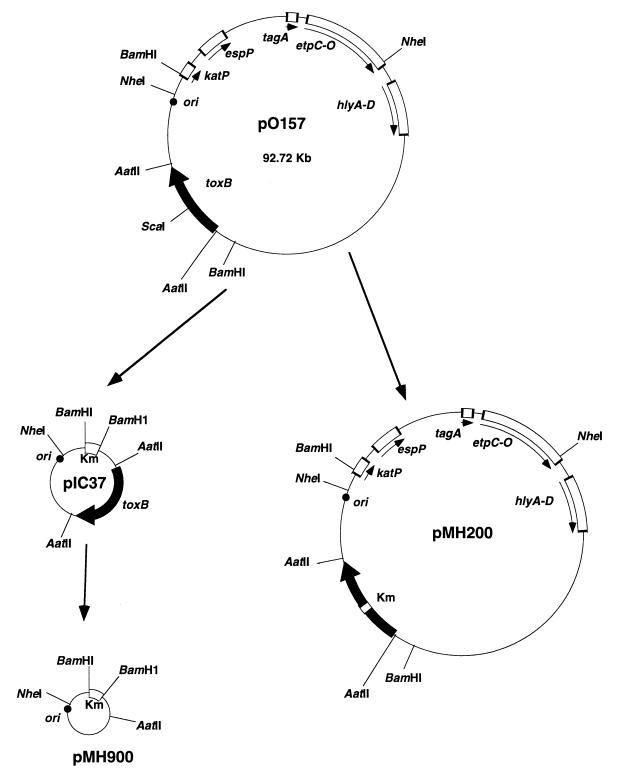
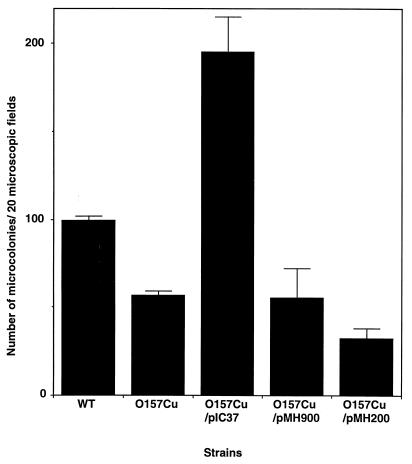
We attempted to tag the large plasmid by inserting a kanamycin resistance-encoding gene and reintroduce it into the cured strain, but the insertion was not successful. We next attempted direct cloning of the toxB gene by using a conventional vector such as pWSK29 (a pSC101-derived low-copy-number vector) (39) or pUC119 (a pBR322-deroved high-copy-number vector) (37) but were unsuccessful. We therefore constructed a pO157-derived miniplasmid by ligating the 25-kb BamHI segment with a Kmr cassette since the 25-kb segment contained the toxB gene and its replication origin (ori) (Fig. 3). The resulting mini-pO157 plasmid, named pIC37, was then introduced into one of the pO157-cured derivatives (O157Cu), and O157Cu carrying pIC37 was investigated for the ability to adhere to Caco-2 cells. As shown in Fig. 4, the adherence capacity of O157Cu carrying pIC37 was increased remarkably (197%) compared to that of parental strain O157Sakai (100%). In contrast, when a pIC37 derivative with toxB deleted (pMH900) was introduced into O157Cu, the bacteria showed a low (56%) adherence capacity compared with that of the wild type (100%) (Fig. 4). To further confirm the contribution of toxB to bacterial adherence, we inserted the Kmr-encoding gene cassette into the gene on pO157 (pMH200). The resultant mutant strain had a reduced adherence capacity compared to that of parental wild-type strain O157Sakai (data not shown). pMH200 was also introduced into O157Cu, and the adherence capacity of that strain was decreased (32%) compared with that of O157 (100%), indicating that the toxB gene is involved in promoting bacterial adherence.
FIG. 3.
Physical maps of pO157 derivatives. The names of the genes that have been implicated in EHEC pathogenesis and localized to the large plasmid derived from other EHEC O157: H7 strains (19) are inside the circles. Black arrows and squares indicate the toxB gene. White squares indicate the coding regions of the other genes, which are on the strands indicated by the arrows. Km, kanamycin resistance-encoding gene.
FIG. 4.
Complementation of the adherence deficiency of the O157Cu mutant by the toxB gene. The number of MC on Caco-2 cells infected by O157Cu clone 11 derivatives (Fig. 2) was scored as described in the legend to Fig. 2. The data shown are the means and standard deviations of three representative experiments. Adherence assays of the wild type (WT), O157Cu, and O157Cu/pIC37 were repeated 24 times. Essentially the same results were obtained with O157Cu clone 28.
The toxB region of pO157 affects the production and secretion of type III translocated proteins.
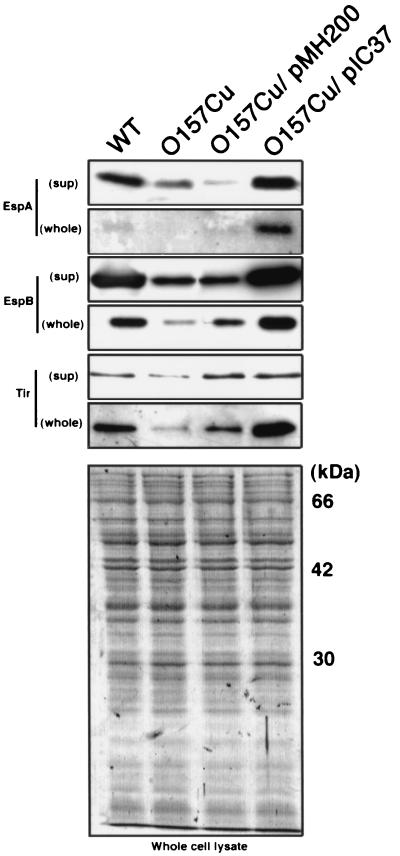
Since the secretion of EspA, EspB, and Tir from EHEC is crucial for the adherence of EHEC to epithelial cells, the production and secretion of the secreted proteins in O157 (wild type), O157Cu (pO157 free), O157Cu carrying pIC37 (toxB +), and O157Cu carrying pMH200 (toxB −) were compared by immunoblotting with anti-EspA, anti-EspB, and anti-Tir antibodies. The growth rates of the strains were essentially the same, although O157Cu carrying pIC37 grew slightly slower than O157Cu (data not shown). As shown in Fig. 5, production of EspA, EspB, and Tir was lower in O157Cu or O157Cu carrying pMH200 than in O157 or O157Cu carrying pIC37. Consistent with these results, the level of EspA and EspB secreted into the medium by O157Cu or O157Cu carrying pMH200 was lower than that secreted by O157 or O157Cu carrying pIC37. The same was also true of O157/pMH900, whose level of protein secretion into the medium was lower than that of O157 or O157Cu carrying pIC37 (data not shown). These results suggest that toxB expression in O157 affects the production and secretion of type III secreted proteins.
FIG. 5.
Levels of secretion and expression of EspA, EspB, and Tir by O157Cu clone 11. Trichloroacetic acid-precipitated culture supernatants (sup) and bacterial cell lysates (whole) derived from equal amounts of wild-type (WT) O157Sakai (lane 1), O157Cu (lane 2), O157Cu/pMH200 (lane 3), and O157/pIC37 (lane 4) were resolved by sodium dodecyl sulfate–12% polyacrylamide gel electrophoresis and stained with Coomassie brilliant blue (bottom photo) or transferred to a nitrocellulose membrane and probed with a polyclonal rabbit antiserum specific to each protein (indicated at the left). Each antibody was purified by using the peptide itself as an antigen, and no other bands reacted with it, demonstrating antibody specificity. Furthermore, no band reacted with each antibody in the culture supernatant of type III-defective mutants (32). Essentially the same results were obtained in O157Cu clone 28.
ToxB may affect the production of type III secreted proteins at the posttranscriptional level.
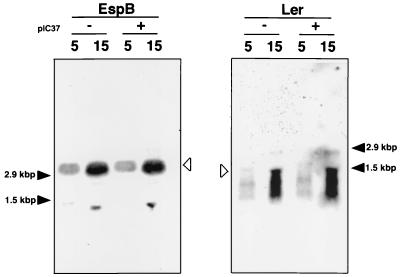
To further investigate the role of toxB in the increase in the production and secretion of EspA, EspB, and Tir in O157, the level of espB mRNA expressed in O157Cu with or without pIC37 was investigated by Northern blotting with a DNA probe specific for espB. Figure 6 shows that the levels of espB mRNA in O157Cu carrying pIC37 (toxB+) and O157Cu were similar. The same was true of O157Cu carrying pMH900 (toxB−), in which the level of espB mRNA was similar to that in O157 (data not shown). Since the espADB operon and other operons encoding the type III secretion system in LEE has been indicated to be positively regulated at the transcriptional level by Ler (LEE-encoded regulator) in EPEC and EHEC (8, 10, 21, 24), the level of ler mRNA in O157Cu with or without pIC37 was examined by Northern blotting with a DNA probe specific for ler. As shown in Fig. 6, the levels of ler mRNA in the two strains were similar.
FIG. 6.
Analysis of espB and ler mRNAs generated from O157Cu with or without pIC37 by Northern blotting. Total RNA (5 or 15 μg) prepared from each strain was subjected to Northern blot analysis. Open arrowheads indicate the mRNA investigated.
The N-terminal half of ToxB may be important for production and secretion of type III secreted proteins.
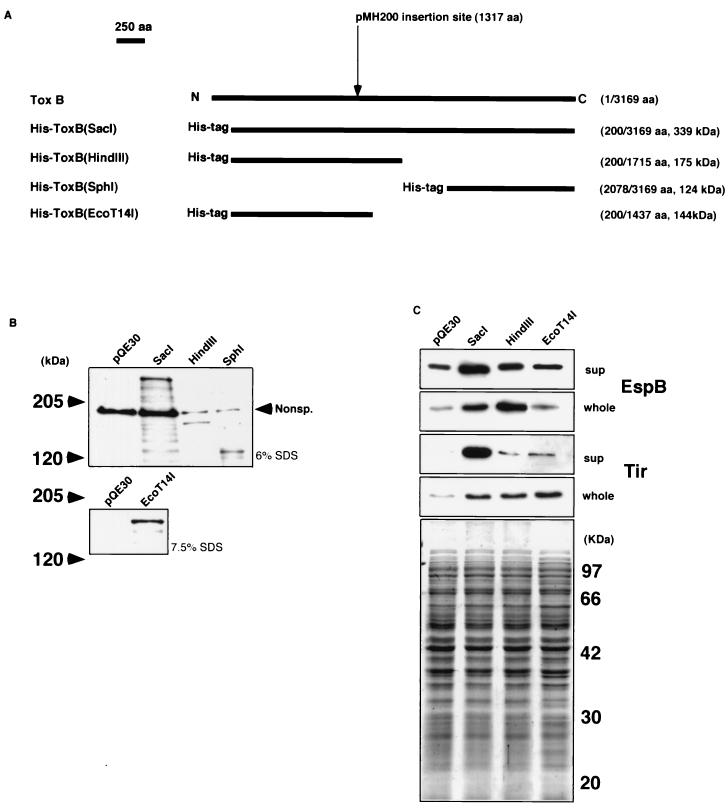
The deduced ToxB sequence was predicted to be a slightly hydrophilic 362-kDa protein and shared amino acid sequences with other putative adherence factors, such as Efa-1 of O111:H− (see introduction). However, since the predicted 362-kDa protein expressed by the toxB gene on pO157, including the cognate genes in EPEC and other EHEC strains, has yet to be reported, we undertook to detect ToxB protein by immunoblotting. We prepared two synthetic peptides corresponding to the 23 amino acids starting from Val-74 in the N terminus and the 21 amino acids starting from Glu-3087 in the C terminus and raised antibodies by injecting rabbits. Although both of the antibodies reacted with their own synthetic peptides in immunoblots, neither reacted with a putative 362-kDa toxB product, as examined by using a whole bacterial cell lysate of O157 or O157Cu carrying pIC37. Therefore, the toxB gene and various truncated toxB versions were constructed and tagged with six histidines by cloning in pQE30, a histidine-tagged vector. The resulting plasmids, each encoding a His-tagged ToxB derivative and referred to as pHis-ToxB(SacI), pHis-ToxB(HindIII), and pHis-ToxB(SphI) (Fig. 7A), were examined for His-tagged products by immunoblotting with an anti-His antibody in JM109 or XL1-blue. As shown in Fig. 7B, pHis-ToxB(SacI), pHis-ToxB(HindIII), and pHis-ToxB(SphI) produced the expected 339-kDa, 175-kDa, and 124-kDa proteins, respectively. When pHis-ToxB(SacI) was introduced into O157Cu, the level of EspB production, including secretion into the medium, was increased (Fig. 7C). When pHis-ToxB(HindIII) was introduced into O157Cu, the level of EspB production was increased (Fig. 7C). The O157Cu/pHis-ToxB(SphI) strain, meanwhile, showed reduced growth and EspB production, including secretion into the medium (data not shown). To narrow down further the ToxB region involved in the promotion of EspB production, we constructed a C-terminally truncated version of pHis-ToxB(HindIII); plasmid pHis-ToxB(EcoT14I) encoded a His-ToxB(HindIII) version lacking 278 C-terminal amino acids (Fig. 7A). However, when pHis-ToxB(EcoT14I) was introduced into O157Cu, EspB production and secretion were not increased, as examined by immunoblotting with the anti-EspB antibody (Fig. 7C).
FIG. 7.
(A) Schematic maps of the truncated ToxB versions tagged with six histidines. The names of the products are indicated on the left of the maps. The numbers correspond to the positions of the amino acids (aa) in wild-type ToxB, and the predicted molecular masses are indicated on the right of the maps. The insertion site of the kanamycin resistance-encoding gene in pMH200 is indicated above the maps. (B) Expression of truncated ToxB tagged with six histidines. Trichloroacetic acid-precipitated cultures derived from equal amounts of JM109/pQE30, JM109/pHis-toxB(SacI), XL1-blue/pHis-toxB(HindIII), and XL1-blue/pHis-toxB(SphI) (upper photo) and JM109/pHis-toxB(EcoT14I) (lower photo) were resolved by sodium dodecyl sulfate (SDS)–6% (indicated on the right of the upper photo) and –7.5% (indicated on the right of the lower photo) polyacrylamide gel electrophoresis, transferred to a nitrocellulose membrane, and probed with anti-RGSHHHH mouse immunoglobulin G antibody. Nonsp., a band cross-reacting nonspecifically with the serum. (C) Levels of secretion and expression of EspB and Tir by O157Cu cells with pHis-toxB(SacI), pHis-toxB(HindIII), and pHis-toxB(EcoT14I). Trichloroacetic acid-precipitated culture supernatants (sup) and bacterial cell lysates (whole) derived from the same amount of O157Cu with either pHis-toxB(SacI) derivatives or pQE30 (used as a control) were resolved by sodium dodecyl sulfate–12% polyacrylamide gel electrophoresis and stained with Coomassie brilliant blue (bottom photo) or transferred to a nitrocellulose membrane and probed with polyclonal rabbit antiserum specific to each protein indicated at the right.
DISCUSSION
In this study, we investigated the significance of pO157 for the pathogenicity of EHEC and found that the toxB gene on pO157 is important for full expression of adherence in that it affects the production and secretion of EspA, EspB, and Tir, which are required for bacterial attachment. Our conclusion was based on the following results: (i) removal of pO157 from O157Sakai reduced adherence capacity; (ii) introduction of mini-pO157 (pIC37) encoding the toxB region into O157Cu (O157 cured of pO157), but not pO157 derivatives lacking toxB, such as pMH200 (pO157 toxB::Km) or pMH900 (a pIC37 derivative without the toxB region), increased adherence capacity; and (iii) O157Cu carrying pIC37, but not pMH200, stimulated the production and/or secretion of EspA, EspB, and Tir.
We have recently proposed that O157Sakai may adhere to epithelial cells through a two-step process; the first step is diffuse adherence, and the second is a more intimate attachment and the development of MC on epithelial cells (32). We also indicated that type III secreted proteins, such as EspA, or additional putative adherence factors are involved in the initial adherence and that the intimin-Tir interaction is required for subsequent development of MC but that the development rate appears to depend on the bacterial growth rate rather than on the level of intimin and/or Tir expression (32). This idea was agreed with by Ebel et al., who suggested that EspA is required at the initial stage of bacterial adherence to the host cells in O26 Shiga toxin-producing E. coli (7). According to the model, the loss of pO157 from O157 mostly affects the initial adherence stage, since O157Cu (pO157-cured derivative) could develop MC on Caco-2 cells, with actin condensation occurring beneath the attached bacteria. The apparent size of O157Cu MC, including the accumulation of F-actin, as observed by Giemsa staining of infected Caco-2 cells at 4 h postinfection, was similar to that of the wild type MC (Fig. 1). Nevertheless, as shown in Fig. 5, the level of Tir secretion into the culture medium by O157Cu was less than that by the wild type, implying that ToxB also, if only in part, involves the later stage of bacterial adherence. Although the exact reason why O157Cu can still form MC remains to be elucidated, there is a possible explanation. For example, the smaller amount of Tir secreted may still be sufficient for bacteria to allow a specific interaction with intimin during bacterial infection of Caco-2 cell monolayers. This idea is supported by previous findings obtained with a less adherent O157 mutant isolated by random mini-Tn5Km2 insertion mutagenesis (32). In that study, the mutant named H7-C4 was still able to form MC sufficiently on Caco-2 cells at 4 h after infection, even though the levels of Tir secreted into the medium were low compared with those secreted by the wild type.
Interestingly, the capacity of O157Cu carrying pIC37 (toxB+) to adhere to Caco-2 cells was remarkably high compared with that of the wild type. This increase caused by pIC37 did not appear to be due to an increase in the plasmid copy number, since the copy number of pIC37 in O157Cu was similar to that of pO157 in O157Sakai, as estimated by agarose gel electrophoresis (data not shown). Thus, although no direct evidence has been obtained, toxB expression from pIC37 may be increased in O157Cu.
To account for the positive effect of the presence of ToxB in EHEC on the production and secretion of secreted proteins, the levels of ler mRNA and espB mRNA in O157Cu in the presence or absence of pIC37 were investigated by using Northern blotting with a DNA probe specific for the ler gene or espB mRNA. The results indicated that pO157 in O157Sakai had no appreciable effect on the level of ler mRNA or espB mRNA. To confirm this, a pler::cat fusion plasmid was constructed and introduced into O157Cu with or without pIC37 to monitor the effect of the plasmid on promoter activity. The chloramphenicol acetyltransferase activities in O157Cu carrying pIC37/pler::cat and in O157Cu/pler::cat were similar (H. Abe et al., unpublished data). On the basis of the results described above, we assume that ToxB somehow affects the production of type III secreted proteins at the posttranscriptional level and that the increased level of EspB secretion in O157Cu/pIC37, meanwhile, may depend on the production level rather than the activity of the type III secretion system itself, which is regulated by Ler.
Although the mechanism by which ToxB enhances the production of EspA, EspB, and Tir is still vague, certain characteristics of the protein in bacteria could be predicted from the deduced toxB sequence. For example, ToxB has no typical signal sequence at the N terminus for secretion from the bacterial cytoplasm. Based on the methods of Kyte and Doolittle (18), ToxB is predicted to be slightly hydrophilic throughout the length of its polypeptide, suggesting that ToxB is a cytoplasmic protein and serves to promote the production of type III translocated proteins. In this regard, recent studies have indicated that type III translocated proteins in bacteria such as Yersinia, Shigella, and Salmonella spp. and EPEC require chaperons for stability in the cytoplasm or targeting to the type III secretion machinery (1, 4). The possibility that ToxB itself serves as a chaperon is less likely, since the size of ToxB (362 kDa) seems to be extraordinarily large compared with those of known chaperons. We have therefore checked the possibility that another ORF is present in the ToxB region of O157 by searching DNA fragments of 1,000 bp by BLASTX (42; http: //www.blast.genome.ad.jp/), but there is no potential ORF encoding a protein homologous to a known chaperon in the frame other than the toxB gene or in the complementary strand of the DNA. Although the toxB gene is predicated to encode ToxB, some other internal translated proteins or processing versions smaller than the full length of the ToxB protein might be expressed and the small ToxB version(s) might have some ability to enhance the production of EspA, EspB, and Tir. As another example, it is still possible that ToxB exists in association with the inner membrane in EHEC, since the ToxB sequence possesses a putative transmembrane region located at positions 1961 to 1980, as shown by a previous study (3). If that is the case, it might be possible that ToxB somehow affects type III secretion activity by being involved in the sensing of some extracellular signals required for the stimulation of secretion, as suggested previously (38, 41). This idea might be supported by the finding that His-ToxB(SacI), but not His-ToxB(HindIII) without the transmembrane region, induces the secretion of EspB (Fig. 7). Alternatively, ToxB expression may also influence gross surface properties, although there was no alteration in the lipopolysaccharide properties of O157Sakai derivatives, as examined by an agglutination assay with antiserum specific for O157. In addition, no surface appendixes on O157Sakai, O157Cu, and O157Cu carrying pIC37 were observed by electron microscopy of parental O157Sakai, O157Cu, and O157Cu carrying pIC37. In any case, it is important to elucidate the mechanisms underlying the enhancement of the production and/or secretion of type III secreted proteins by the toxB product(s).
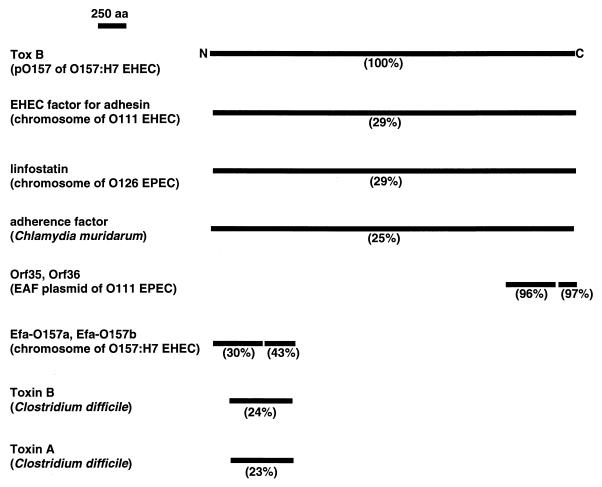
The deduced ToxB sequence shares amino acid similarity with a variety of proteins, including some putative adherence factors, although the extent of the similarity or the size of the corresponding ToxB sequence varies, depending on the counterpart proteins (Fig. 8). For example, efa-1, which is associated with the ability to adhere to Chinese hamster ovary cells, was found on the chromosome of a clinical isolate of O111:H− EHEC strain E45035 (23) and the predicted 365-kDa Efa-1 protein showed 29% identity to ToxB. It has recently been suggested that the lifA gene in EPEC encodes a factor that is 99% identical to Efa-1 and inhibits lymphocyte functions such as interleukins 2 and 4 and gamma interferon (15). Genome sequences of a Chlamydia muridarum strain (synonym: Chlamydia trachomatis MoPn) revealed that the strain contained three copies of a novel gene homologous to toxB from O157:H7 (26). The orf35 and orf36 genes were recently identified on an adherence factor plasmid of EPEC strain O111 (35). The C termini of ToxB contained two contiguous sequences that shared 96 and 97% identity with the amino acids of Orf35 and Orf36, respectively. We recently identified a putative adherence-associated gene, named efa-1 (renamed efa-O157a in this study), in O157Sakai chromosomal DNA by random insertion mutagenesis using mini-Tn5Km2 (13, 25, 32). The downstream DNA sequence possessed another cognate gene, named efa-O157b. The deduced efa-O157a and efa-O157b products were 433- and 275-amino-acid peptides, respectively, that shared 30 and 43% identity with the two N-terminal sequences in ToxB, respectively. Surprisingly, the Efa-O157a and Efa-O157b sequences shared complete identity with the two N-terminal contiguous sequences in Efa-1 of O111:H−. The N termini of ToxB in O157Sakai were found to contain 707 and 708 amino acids and exhibit up to 23 and 24% identity to Clostridium difficile toxins A and B, respectively, when analyzed by BLAST (42; http://www.blast.genome.ad.jp/). Therefore, these data led us to speculate that ToxB possesses several distinctive, if not separable, domains each associated with either full production of type III secreted proteins (this study), adherence to epithelial cells (23), or inhibition of lymphocyte activation (15).
FIG. 8.
Schematic analysis of the amino acid (aa) sequence deduced from toxB. Regional amino acid similarity was based on high BLAST search scores and is shown in parentheses under each solid bar. Product names (and the names of the organisms containing the genes) are indicated on the left of the bars. A scale bar is in the upper left corner.
Under the circumstances, we investigated the region of ToxB essential for the full production of type III secreted proteins in O157Sakai by constructing various histidine-tagged ToxB derivatives. In the construction, His was tagged at amino acid 200 from the N terminus of the ToxB peptide by using pQE30 due to the feasibility of this construction. Estimation of EspB production and secretion in O157Cu expressing the histidine-tagged ToxB derivatives suggested that the N-terminal ToxB sequence encompassing amino acids 200 through to 1715 would be important in affecting EspB production (Fig. 7). Importantly, the region indicated was consistent with the results obtained with pMH200, which failed to fully produce EspB in O157Cu, since the Kmr cassette was placed in the middle of toxB, at a position corresponding to residue 1317 in ToxB peptides (Fig. 3 and 7). This finding further supports the idea that ToxB is required for the full production and/or secretion of type III secreted proteins by O157.
In summary, our results provide evidence supporting the notion that pO157, conserved in EHEC strain O157:H7, is important for the expression of full pathogenicity since ToxB encoded by pO157 can contribute to full expression of the type III secreted proteins required for bacterial adherence to host cells.
ACKNOWLEDGMENTS
This research was supported by the Research for the Future Program of the Japan Society for the Promotion of Science and by grant 11770135 from the Ministry of Education, Science and Culture of the Japanese Government.
We thank Chizu Sasako and Asaomi Kuwae for technical assistance and Kumar Rajakumar for providing suicide vector pCACTUS. We gratefully acknowledge Naresh Verma and Toru Tobe for helpful discussions.
REFERENCES
- 1.Bennett J C, Hughes C. From flagellum assembly to virulence: the extended family of type III export chaperones. Trends Microbiol. 2000;8:202–204. doi: 10.1016/s0966-842x(00)01751-0. [DOI] [PubMed] [Google Scholar]
- 2.Boyce T G, Swerdlow D L, Griffin P M. Current concepts: Escherichia coli O157:H7 and the hemolytic-uremic syndrome. N Engl J Med. 1995;333:364–368. doi: 10.1056/NEJM199508103330608. [DOI] [PubMed] [Google Scholar]
- 3.Burland V, Shao Y, Perna N T, Plunkett G, Sofia H J, Blattner F R. The complete DNA sequence and analysis of the large virulence plasmid of Escherichia coli O157:H7. Nucleic Acids Res. 1998;26:4196–4204. doi: 10.1093/nar/26.18.4196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cornelis G R. Type III secretion: a bacterial device for close combat with cells of their eukaryotic host. Philos Trans R Soc Lond B Biol Sci. 2000;355:681–693. doi: 10.1098/rstb.2000.0608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.de Lorenzo V, Herrero M, Jakubzik U, Timmis K N. Mini-Tn5 transposon derivatives for insertion mutagenesis, promoter probing, and chromosomal insertion of cloned DNA in gram-negative eubacteria. J Bacteriol. 1990;172:6568–6572. doi: 10.1128/jb.172.11.6568-6572.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.DeVinney R, Stein M, Reinscheid D, Abe A, Ruschkowski S, Finley B B. Enterohemorrhagic Escherichia coli O157: H7 produces Tir, which is translocated to the host cell membrane but is not tyrosine phosphorylated. Infect Immun. 1999;67:2389–2398. doi: 10.1128/iai.67.5.2389-2398.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ebel F, Podzadel T, Rohde M, Kresse A U, Krämer S, Deibel C, Guzmán C A, Chakraborty T. Initial binding of Shiga toxin-producing Escherichia coli to host cells and subsequent induction of actin rearrangement depend on filamentous EspA-containing surface appendages. Mol Microbiol. 1998;30:147–161. doi: 10.1046/j.1365-2958.1998.01046.x. [DOI] [PubMed] [Google Scholar]
- 8.Elliott S J, Sperandio V, Girón J A, Shin S, Mellies J L, Wainwright L, Hutcheson S W, McDaniel T K, Kaper J B. The locus of enterocyte effacement (LEE)-encoded regulator controls expression of both LEE- and non-LEE-encoded virulence factors in enteropathogenic and enterohemorrhagic Escherichia coli. Infect Immun. 2000;68:6115–6126. doi: 10.1128/iai.68.11.6115-6126.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fratamico P M, Bhaduri S, Buchanan R L. Studies on Escherichia coli serotype O157:H7 strains containing a 60-MDa plasmid and on 60-Mda plasmid-cured derivatives. J Med Microbiol. 1993;39:371–381. doi: 10.1099/00222615-39-5-371. [DOI] [PubMed] [Google Scholar]
- 10.Friedberg D, Umanski T, Fang Y, Rosenshine I. Hierarchy in the expression of the locus of enterocyte effacement genes of enteropathogenic Escherichia coli. Mol Microbiol. 1999;34:941–952. doi: 10.1046/j.1365-2958.1999.01655.x. [DOI] [PubMed] [Google Scholar]
- 11.Gansheroff L J, O'Brien A D. Escherichia coli O157:H7 in beef cattle presented for slaughter in the U.S.: higher prevalence rates than previously estimated. Proc Natl Acad Sci USA. 2000;97:2959–2961. doi: 10.1073/pnas.97.7.2959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Griffin P M, Tauxe R V. The epidemiology of infections caused by Escherichia coli O157:H7: other enterohemorrhagic E. coli and the associated hemolytic uremic syndrome. Epidemiol Rev. 1991;13:60–97. doi: 10.1093/oxfordjournals.epirev.a036079. [DOI] [PubMed] [Google Scholar]
- 13.Hayashi T, Makino K, Ohnishi M, Kurokawa K, Ishii K, Yokoyama K, Han C-G, Ohtsubo E, Nakayama K, Murata T, Tanaka M, Tobe T, Iida T, Takami H, Honda T, Sasakawa C, Ogasawara N, Yasunaga T, Kuhara S, Shiba T, Hattori M, Shinagawa H. Complete genome sequence of enterohemorrhagic Escherichia coli O157:H7 and genomic comparison with a laboratory strain K-12. DNA Res. 2001;8:11–22. doi: 10.1093/dnares/8.1.11. [DOI] [PubMed] [Google Scholar]
- 14.Karch H, Heesemann J, Laufs R, O'Brien A D, Tacket C O, Levine M M. A plasmid of enterohemorrhagic Escherichia coli O157:H7 is required for expression of a new fimbrial antigen and for adhesion to epithelial cells. Infect Immun. 1987;55:455–461. doi: 10.1128/iai.55.2.455-461.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Klapproth J M, Scaletsky I C, McNamara B P, Lai L C, Malstrom C, James S P, Donnenberg M S. A large toxin from pathogenic Escherichia coli strains that inhibits lymphocyte activation. Infect Immun. 2000;68:2148–2155. doi: 10.1128/iai.68.4.2148-2155.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Knutton S, Baldwin T, Williams P H, McNeish A S. Actin accumulation at sites of bacterial adhesion to tissue culture cells: basis of a new diagnostic test for enteropathogenic and enterohemorrhagic Escherichia coli. Infect Immun. 1989;57:1290–1298. doi: 10.1128/iai.57.4.1290-1298.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kresse A U, Rohde M, Guzmán C A. The EspD protein of enterohemorrhagic Escherichia coli is required for the formation of bacterial surface appendages and is incorporated in the cytoplasmic membrane of target cells. Infect Immun. 1999;67:4834–4842. doi: 10.1128/iai.67.9.4834-4842.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kyte J, Doolittle R F. A simple method for displaying the hydropathic character of a protein. J Mol Biol. 1982;157:105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- 19.Makino K, Ishii K, Yasunaga T, Hattori M, Yokoyama K, Yutsudo C H, Kubota Y, Yamaichi Y, Iida T, Yamamoto K, Honda T, Han C G, Ohtsubo E, Kasamatsu M, Hayashi T, Kuhara S, Shinagawa H. Complete nucleotide sequences of 93-kb and 3.3-kb plasmids of an enterohemorrhagic Escherichia coli O157:H7 derived from Sakai outbreak. DNA Res. 1998;28:1–9. doi: 10.1093/dnares/5.1.1. [DOI] [PubMed] [Google Scholar]
- 20.Mead P S, Slutsker L, Dietz V, McCaig L F, Bresee J S, Shapiro C, Griffin P M, Tauxe R V. Food-related illness and death in the United States. Emerg Infect Dis. 1999;5:607–625. doi: 10.3201/eid0505.990502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mellies J L, Elliott S J, Sperandio V, Donnenberg M S, Kaper J B. The Per regulon of enteropathogenic Escherichia coli : identification of a regulatory cascade and a novel transcriptional activator, the locus of enterocyte effacement (LEE)-encoded regulator (Ler) Mol Microbiol. 1999;33:296–306. doi: 10.1046/j.1365-2958.1999.01473.x. [DOI] [PubMed] [Google Scholar]
- 22.Nataro J P, Kaper J B. Diarrheagenic Escherichia coli. Clin Microbiol Rev. 1998;11:142–201. doi: 10.1128/cmr.11.1.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nicholls L, Grant T H, Robins-Browne R M. Identification of a novel genetic locus that is required for in vitro adhesion of a clinical isolate of enterohaemorrhagic Escherichia coli to epithelial cells. Mol Microbiol. 2000;35:275–288. doi: 10.1046/j.1365-2958.2000.01690.x. [DOI] [PubMed] [Google Scholar]
- 24.Perna N T, Mayhew G F, Pósfai G, Elliott S, Donnenberg M S, Kaper J B, Blattner F R. Molecular evolution of a pathogenicity island from enterohemorrhagic Escherichia coli O157:H7. Infect Immun. 1998;66:3810–3817. doi: 10.1128/iai.66.8.3810-3817.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Perna N T, Plunkett III G, Burland V, Mau B, Glasner J D, Rose D J, Mayhew G F, Evans P S, Gregor J, Kirkpatrick H A, Pósfai G, Hackett J, Klink S, Boutin A, Shao Y, Miller I, Grotbeck E J, Davis N W, Lim A, Dimalanta E, Potamousis K, Apodaca J, Anantharaman T S, Lin J, Yen G, Schwartz D C, Welch R A, Blattner F R. Genome sequence of enterohemorrhagic Escherichia coli O157:H7. Nature. 2001;409:529–533. doi: 10.1038/35054089. [DOI] [PubMed] [Google Scholar]
- 26.Read T D, Brunham R C, Shen C, Gill S R, Heidelberg J F, White O, Hickey E K, Peterson J, Utterback T, Berry K, Bass S, Linher K, Weidman J, Khouri H, Craven B, Bowman C, Dodson R, Gwinn M, Nelson W, DeBoy R, Kolonay J, McClarty G, Salzberg S L, Eisen J, Fraser C M. Genome sequences of Chlamydia trachomatis MoPn and Chlamydia pneumoniae AR39. Nucleic Acids Res. 2000;28:1397–1406. doi: 10.1093/nar/28.6.1397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Riley L W, Remis R S, Helgerson S D, McGee H B, Wells J G, Davis B R, Hebert R J, Olcott E S, Johnson L M, Hargrett N T, Blake P A, Cohen M L. Hemorrhagic colitis associated with a rare Escherichia coli serotype. N Engl J Med. 1983;308:681–685. doi: 10.1056/NEJM198303243081203. [DOI] [PubMed] [Google Scholar]
- 28.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 29.Schmidt H, Kernbach C, Karch H. Analysis of the EHEC hly operon and its location in the physical map of the large plasmid of enterohaemorrhagic Escherichia coli O157:H7. Microbiology. 1996;142:907–914. doi: 10.1099/00221287-142-4-907. [DOI] [PubMed] [Google Scholar]
- 30.Short J M, Fernandez J M, Sorge J A, Huse W D. Lambda ZAP: a bacteriophage lambda expression vector with in vivo excision properties. Nucleic Acids Res. 1988;16:7583–7600. doi: 10.1093/nar/16.15.7583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tamano K, Aizawa S, Katayama E, Nonaka T, Imajoh-Ohmi S, Kuwae A, Nagai S, Sasakawa C. Supramolecular structure of the Shigella type III secretion machinery: the needle part is changeable in length and essential for delivery of effectors. EMBO J. 2000;19:3876–3887. doi: 10.1093/emboj/19.15.3876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tatsuno I, Kimura H, Okutani A, Kanamaru K, Abe H, Nagai S, Makino K, Shinagawa H, Yoshida M, Sato K, Nakamoto J, Tobe T, Sasakawa C. Isolation and characterization of mini-Tn5Km2 insertion mutants of enterohemorrhagic Escherichia coli O157:H7 deficient in adherence to Caco-2 cells. Infect Immun. 2000;68:5943–5952. doi: 10.1128/iai.68.10.5943-5952.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Taylor K A, O'Connell C B, Luther P W, Donnenberg M S. The EspB protein of enteropathogenic Escherichia coli is targeted to the cytoplasm of infected HeLa cells. Infect Immun. 1998;66:5501–5507. doi: 10.1128/iai.66.11.5501-5507.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Taylor L A, Rose R E. A correction in the nucleotide sequence of the Tn903 kanamycin resistance determinant in pUC4K. Nucleic Acids Res. 1988;16:358. doi: 10.1093/nar/16.1.358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tobe T, Hayashi T, Han C G, Schoolnik G K, Ohtsubo E, Sasakawa C. Complete DNA sequence and structural analysis of the enteropathogenic Escherichia coli adherence factor plasmid. Infect Immun. 1999;67:5455–5462. doi: 10.1128/iai.67.10.5455-5462.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Toth I, Cohen M L, Rumschlag H S, Riley L W, White E H, Carr J H, Bond W W, Wachsmuth I K. Influence of the 60-megadalton plasmid on adherence of Escherichia coli O157:H7 and genetic derivatives. Infect Immun. 1990;58:1223–1231. doi: 10.1128/iai.58.5.1223-1231.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vieira J, Messing J. Production of single-stranded plasmid DNA. Methods Enzymol. 1987;153:3–11. doi: 10.1016/0076-6879(87)53044-0. [DOI] [PubMed] [Google Scholar]
- 38.Wachter C, Beinke C, Mattes M, Schmidt M A. Insertion of EspD into epithelial target cell membranes by infecting enteropathogenic Escherichia coli. Mol Microbiol. 1999;31:1695–1707. doi: 10.1046/j.1365-2958.1999.01303.x. [DOI] [PubMed] [Google Scholar]
- 39.Wang R F, Kushner S R. Construction of versatile low-copy-number vectors for cloning, sequencing and gene expression in Escherichia coli. Gene. 1991;100:195–199. [PubMed] [Google Scholar]
- 40.Watanabe H, Wada A, Inagaki Y, Ito K, Tamura K. Outbreaks of enterohaemorrhagic Escherichia coli O157:H7 infection by two different genotype strains in Japan 1996. Lancet. 1996;348:831–832. doi: 10.1016/s0140-6736(05)65257-9. [DOI] [PubMed] [Google Scholar]
- 41.Wolff C, Nisan I, Hanski E, Frankel G, Rosenshine I. Protein translocation into host epithelial cells by infecting enteropathogenic Escherichia coli. Mol Microbiol. 1998;28:143–155. doi: 10.1046/j.1365-2958.1998.00782.x. [DOI] [PubMed] [Google Scholar]
- 42.Zhang Z, Schäffer A A, Miller W, Madden T L, Lipman D J, Koonin E V, Altschul S F. Protein sequence similarity searches using patterns as seeds. Nucleic Acids Res. 1998;26:3986–3990. doi: 10.1093/nar/26.17.3986. [DOI] [PMC free article] [PubMed] [Google Scholar]