Abstract
Magnaporthe oryzae, the rice blast fungus, is one of the most dangerous rice pathogens, causing considerable crop losses around the world. In order to explore the rice blast-resistant sources, initially performed a large-scale screening of 277 rice accessions. In parallel with field evaluations, fifty-two rice accessions were genotyped for 25 major blast resistance genes utilizing functional/gene-based markers based on their reactivity against rice blast disease. According to the phenotypic examination, 29 (58%) and 22 (42%) entries were found to be highly resistant, 18 (36%) and 29 (57%) showed moderate resistance, and 05 (6%) and 01 (1%), respectively, were highly susceptible to leaf and neck blast. The genetic frequency of 25 major blast resistance genes ranged from 32 to 60%, with two genotypes having a maximum of 16 R-genes each. The 52 rice accessions were divided into two groups based on cluster and population structure analysis. The highly resistant and moderately resistant accessions are divided into different groups using the principal coordinate analysis. According to the analysis of molecular variance, the maximum diversity was found within the population, while the minimum diversity was found between the populations. Two markers (RM5647 and K39512), which correspond to the blast-resistant genes Pi36 and Pik, respectively, showed a significant association to the neck blast disease, whereas three markers (Pi2-i, Pita3, and k2167), which correspond to the blast-resistant genes Pi2, Pita/Pita2, and Pikm, respectively, showed a significant association to the leaf blast disease. The associated R-genes might be utilized in rice breeding programmes through marker-assisted breeding, and the identified resistant rice accessions could be used as prospective donors for the production of new resistant varieties in India and around the world.
Introduction
Rice blast disease, caused by filamentous fungus Magnaporthe oryzae (anamorph Pyricularia oryzae), remains a potential threat to global rice production [1, 2]. The blast pathogen can be found in all stages of plants growth and development, causing damage to leaves (leaf blast), nodes (nodal blast), and panicles (neck blast), as well as decreasing grain yield by up to 90% in favourable environmental conditions [3–5].
The M. oryzae has been documented all over the world and can infect more than 50 host species in the family Poaceae, including rice, wheat, pearl millet, foxtail millet, and finger millet [6–8]. Across most of the world’s rice-growing regions, including India, blast disease epidemics have occurred [9, 10]. Between 1980 and 1987, India experienced several deadly blast disease epidemics in Himachal Pradesh, Tamil Nadu, Andhra Pradesh, and Haryana [11, 12].
Chemical fungicides have been useful in controlling the disease, but they are expensive [13, 14], ineffective when disease pressure is high [15], and may contribute to pathogen resistance [16]. As a result, the most cost-effective and environmentally acceptable strategy for controlling rice blast disease is to leverage host resistance (R genes). Around 118 R genes have been discovered so far, with 35 of them being successfully cloned and characterized for leaf blast resistance [17, 18]. However, the cloned R genes that possess broad-spectrum resistance to leaf blast, have not been tested for neck blast disease [19]. Even though neck blast is the most devastating stage of the disease, there is relatively little information on the genetic processes that underpin neck blast resistance. Nevertheless, 14 QTLs [18] and a few R genes have been found for neck blast resistance, including Pi25(t) [20], Pb1 [21], Pi64 [22], Pi-jnw1 [23], and Pi68(t) [24]. A large majority of the cloned blast R genes share nucleotide-binding site (NBS) and leucine-rich repeat (LRR) domains in their protein sequences, except for a few (Pid2, pi21, and Ptr) [17, 25, 26]. According to gene-for-gene theory, these R genes are race-specific and related to the hypersensitive response (HR) [27]. The M. oryzae’s genome contains numerous repetitive DNA and retro-transposons [28], which might cause mutations in genes that mediate the pathogen’s virulence and host range [29–31], allowing the fungus to develop new deadly races. The emergence of these races results in a change in pathogenicity, posing a threat to existing blast-resistant rice cultivars [32].
By permitting the integration of the desired gene(s) in early breeding generations, marker-assisted selection (MAS) has emerged as a potent method that has advanced the rice breeding effort for blast disease resistance [33]. Many rice cultivars have been improved via MAS by pyramiding targeted R genes, resulting in the rapid release of rice varieties with durable resistance against blast disease [34]. In recent years, molecular markers have been utilized to capitalize on natural variety and pinpoint the gene of interest influencing essential features in different germplasm [35].
There is indeed a lot of genetic variation in the Indian rice germplasm collection [12, 36]. Many of these rice varieties have been reported to have resistance to biotic and abiotic stresses, including blast disease [37–39]. However, the distribution of R genes in Indian rice cultivars that confer long-term resistance to leaf and neck blast has not been adequately explored. As a result, it’s critical to comprehend R gene information in rice germplasm as well as the resistant spectrum of relevant R genes against prevailing pathogen races to use the most successful ones in the rice breeding programme to combat blast disease. The present study was carried out to explore the genetic association of 25 mapped resistance genes in 52 rice accessions, including released varieties, advanced breeding materials, and traditional rice varieties using linked/functional markers. The main goal of this study was to find an association between the leaf and neck blast R genes, which impart blast resistance to these lines, and novel blast resistance donor sources (R genes/alleles).
Materials and methods
Plant materials used in the current research
A total of 50 rice accessions were collected based on documented rice blast resistant information from the Rice Genetics laboratory, Crop Improvement Division, ICAR- Vivekananda Parvatiya Krishi Anusandhan Sansthan (VPKAS), Almora, Uttarakhand, India (Tables 1 & 2). The test material includes released varieties (04), advanced breeding materials (44), and traditional rice varieties (02). In addition, two genotypes, PB-1 and Bala, were chosen as leaf and neck blast susceptible controls, respectively (Tables 1 and 2).
Table 1. List of 52 rice accession used in this study.
| Planting materials | Genotypes |
|---|---|
| Released varieties | VL Dhan 158, VL Dhan 68, VL Dhan 221 and VL Dhan 206 |
| Advanced breeding materials | VL 8083, VL 8214, VL 8394, VL 8549, VL 8654, VL 20231, VL 20279, VL 20287, VL 20298, VL 20299, VL 20302, VL 20289, VL 31430, VL 31451, VL 31598, VL 31615, VL 31616, VL 31619, VL 31674, VL 31679, VL 31694, VL 31716, VL 31743, VL 31802, VL 31817, VL 31851, VL 31870, VL 31916, VL 31997, VL 32092, VL 32131, VL 32132, VL 32168, A-57, BL-122, BL-245, GSR-102, GSR-106, GSR-124, GSR-125, GSR-132, GSR-142, VOHP-3102 and VL 32197 |
| Traditional rice varieties | VLK 39 and Someshwar |
| Susceptible checks | PB-1 and Bala |
Table 2. List of rice genotypes along with their pedigree.
| Sl. No. | Entry name | Pedigree | Sl. No. | Entry name | Pedigree |
|---|---|---|---|---|---|
| 1 | VL 8083 | VL 6394/VL 6446 | 27 | VL 31817 | Vivek Dhan 82/BL122 |
| 2 | VL 8214 | VL Dhan 81/VR539-2 | 28 | VL 31851 | VL 30424/IR78 |
| 3 | VL 8394 | VL6394/VL6446 | 29 | VL 31870 | BL 122/IR 785–36 |
| 4 | VL 8549 | VL 3861/VL 6394 | 30 | VL 31916 | VL Dhan 85/BL 245 |
| 5 | VL 8654 | RCPL 1-45/Vivek Dhan 154 | 31 | VL 31997 | Vivek Dhan 62/MAS-52 |
| 6 | VL Dhan 158 | RCPL 1-45/VL 3861 | 32 | VL 32092 | VL Dhan 85/VOHP 3102 |
| 7 | VL 20231 | VL Dhan 81/Vandana | 33 | VL 32131 | VL 10689/UPRI2005-15 |
| 8 | VL 20279 | VL 20240/Sawdhan | 34 | VL 32132 | VL 10689/UPRI2005-15 |
| 9 | VL 20287 | VHC 1462/VL 10499 | 35 | VL 32168 | VL Dhan 65/VL30919 |
| 10 | VL 20298 | Annada/C101-A51 | 36 | A-57 | - |
| 11 | VL 20299 | Annada/C101-A51 | 37 | BL-122 | - |
| 12 | VL 20302 | VL Dhan 221/ VL 30927 | 38 | BL-245 | - |
| 13 | VL 20289 | VHC 1462/VL 10499 | 39 | VL Dhan 221 | IR 2053-521-1-1-1/Ch 1039 |
| 14 | VL 31430 | Pant Dhan 6/VL 3288 | 40 | VLK 39 | China 1039/IR580-19-2-3-1 |
| 15 | VL 31451 | IR 72979/PSB RC 2 (IR 32809-26-3-3) | 41 | GSR-102 | - |
| 16 | VL 31598 | VL 3861/IR57257-34-1-2-1 | 42 | GSR-106 | - |
| 17 | VL Dhan 68 | VL 3861/SR 1818BF-4B-1-2-1-2 | 43 | GSR-124 | - |
| 18 | VL 31615 | VL 3861/SR 1818BF-4B-1-2-1-2 | 44 | GSR-125 | - |
| 19 | VL 31616 | VL 3861/SR 1818BF-4-B1-2-1-2 | 45 | GSR-132 | - |
| 20 | VL 31619 | VL 3861/SR 1818BF-4-B1-2-1-2 | 46 | GSR-142 | - |
| 21 | VL 31674 | C101-A51/O. minuta | 47 | VOHP-3102 | Local collection |
| 22 | VL 31679 | O. minuta/Vivek Dhan 82 | 48 | VL Dhan 206 | Pure line selection from Bamni (local variety) |
| 23 | VL 31694 | Vivek Dhan 82/IR57257-34 | 49 | VL 32197 | VL Dhan 81/Vandana |
| 24 | VL 31716 | O. minuta/IR57257-34 | 50 | Someshwar | Local collection |
| 25 | VL 31743 | VL 30424/IR32809 | 51 | Bala | N 22/T(N)1 |
| 26 | VL 31802 | VL 66/VL30424 | 52 | PB-1 | Pusa 167/Karnal Local |
Phenotyping of rice germplasm lines for blast disease resistance
A set of 50 rice hill germplasm collections were evaluated under the natural conditions at the rice blast hotspot area, ICAR-VPKAS, experimental farm, Hawalbagh (29o56’N, 79o40’E, and 1250m MSL), Almora, for their reactivity against leaf and neck blast. The evaluations were carried out in three replications over three years, from 2018 to 2020, during the rainy (Kharif) seasons. Sowings were done in two sets, one for leaf blast evaluations and the other for neck blast evaluations. Each rice entry (30 plants/test entry) was raised in 50 cm long rows on nursery beds with a 10 cm row spacing in a uniform blast nursery for leaf blast (UBN). One line of PB-1 (susceptible check) was sown after every 5 entries of test accessions, as well as along the boundaries, to ensure adequate disease transmission. From 25 days after sowing until the susceptibility check showed 85% of the blast disease symptom, the disease spectrum of all the test entries was recorded. A 0–9 scale devised by IRRI, Philippines [39], was used to visually record the disease reaction on each test entry.
Similarly, the other set was also tested for neck blast disease, but Bala was used as a susceptible control. The severity of the disease was graded on a 0–9 scale (IRRI, 2002), with 0 = no lesion or one or two tiny lesions on the panicles; 1 = symptom on several pedicels or secondary branches; 3 = lesions on a few primary branches or the middle part of panicle axis; 5 = moderate infection with lesions covering half of the node or the uppermost internode or the lower part of panicle axis; 7 = heavy infection, lesions abundant on the panicle base or uppermost internode or panicle axis near the base with more than 30% of filled grains; 9 = very heavy infection, around the panicle base or uppermost internode or the panicle axis near the base with less than 30% of filled grains. At physiological maturity, the disease reaction was recorded, and the affected plants were evaluated on a disease scale, Highly resistant (HR) (0–3 score), moderately resistant (MR) (4–5), and susceptible (S) (6–9) were assigned to the test entries, respectively. Whenever differences in the disease spectrum were recorded, the higher disease was taken into account.
DNA isolation and genotyping
Genomic DNA was extracted from the young leaves of 50 rice germplasm lines and two susceptible controls using the CTAB technique [40]. The quality and quantity of isolated genomic DNA were determined using a Thermo Fisher Scientific NanoDropTM 1000 Spectrophotometer. After that, the isolated DNA samples were diluted to a concentration of 25 ng/μl in nuclease-free water for PCR amplification. Molecular profiling of 52 rice lines for the presence of major blast resistance genes was carried out using 25 linked or functional molecular markers. The detailed information on blast resistance genes and their corresponding primer pairs used in this investigation is listed in Table 2. About 25 ng of template DNA, 10 pmol of each forward and reverse primers, 25 mM MgCl2, 2 mM of each dNTPs, 1X Taq buffer, 1U Taq DNA polymerase, and nuclease-free water were used in the PCR amplification. The PCR conditions were set as follows: initial denaturation at 94°C for 5 minutes was followed by 35 cycles of denaturation for 40 seconds at 94°C, primer annealing for 40 seconds at varied temperatures (Table 3), and extension for 2 minutes at 72°C were performed, followed by a final 10-minute extension at 72°C. To double-check the results, PCR amplification was done twice for each marker. The amplified PCR products were resolved in ethidium bromide-stained 3% agarose gels and the scoring were done for the PCR analysis as presence (1) or absence (0).
Table 3. Details of markers used for molecular screening of blast resistance genes in 52 rice accessions.
| Genes | Markers | Forward (5’ - 3’) | Reverse (5’ - 3’) | Type of Marker* | Annealing Temperature (°C) | References |
|---|---|---|---|---|---|---|
| Pit | tk59-1 | ATGATAACCTCATCCTCAATAAGT | GTTGGAGCTACGGTTGTTCAG | FM | 54 | [48] |
| Pid1(t) | RM262 | CATTCCGTCTCGGCTCAACT | CAGAGCAAGGTGGCTTGC | LM | 55 | [63] |
| Pish | RM6648 | GATCGATCATGGCCAGAGAG | ACAGCAGGTTGATGAGGACC | LM | 55 | [34] |
| Pb1 | RM26998 | ACGCACGCACATCCTCTTCC | CGGTTCTCCATCTGAAATCCCTAGC | LM | 55 | [21] |
| Pi33 | RM72 | CCGGCGATAAAACAATGAG | GCATCGGTCCTAACTAAGGG | LM | 55 | [64] |
| Pikhahe-1(t) | RM17496 | TAAACGGTGTGCAGCTTCTG | TATTATGGGCGGTCGCTAAC | LM | 54 | [65] |
| pi21 | pi21-79-3 | GATCCTCATCGTCGACGTCTGGC | AGGGTACGGCACCAGCTTG | InDel | 55 | [27] |
| Pi56 | CRG4-2 | CCTGTCAGTCTTTCCGAGAG | GAATCCGGTAGCTCAAGGTG | Gene-specific | 55 | [66] |
| Pi65 | SNP_3 | TGCCACCAGCCATCTTCAACAT | ACCACATCACTCATCGCCATCC | InDel | 54 | [71] |
| Pi36 | RM5647 | ACTCCGACTGCAGTTTTTGC | AACTTGGTCGTGGACAGTGC | LM | 55 | [72] |
| Pi49 | RM6094 | TGCTTGATCTGTGTTCGTCC | TAGCAGCACCAGCATGAAAG | LM | 55 | [67] |
| Pi48 | RM5364 | GTATTACGCTCGATAGCGGC | GTATCCTTTCTCGCAATCGC | LM | 55 | [68] |
| Pib | Pb28 | GACTCGGTCGACCAATTCGCC | ATCAGGCCAGGCCAGATTTG | SNP | 60 | [48] |
| Piz | Z56592 | GGACCCGCGTTTTCCACGTGTAA | AGGAATCTATTGCTAAGCATGAC | SNP | 60 | [48] |
| Piz-t | Zt56591 | TTGCTGAGCCATTGTTAAACA | ATCTCTTCATATATATGAAGGCCAC | SNP | 60 | [48] |
| Pik | K39512 | GCCACATCAATGGCTACAACGTT | CCAGAATTTACAGGCTCTGG | SNP | 60 | [48] |
| Pik-p | K3957 | ATAGTTGAATGTATGGAATGGAAT | CTGCGCCAAGCAATAAAGTC | SNP | 60 | [48] |
| Pik-h | Candidate gene marker | CATGAGTTCCATTTACTATTCCTC | ACATTGGTAGTAGTGCAATGTCA | Gene-based marker | 55 | [69] |
| Pi9 | Pi9-i | GCTGTGCTCCAAATGAGGAT | GCGATCTCACATCCTTTGCT | FNP | 54 | [52] |
| Pi2 | Pi2-i | CAGCGATGGTATGAGCACAA | CGTTCCTATACTGCCACATCG | FNP | 52 | [52] |
| Pita/Pita2 | Pita3 | AGTCGTGCGATGCGAGGACAGAAAC | GCATTCTCCAACCCTTTTGCATGCAT | SNP | 59 | [48] |
| Pi1 | RM1233 | GTGTAAATCATGGGCACGTG | AGATTGGCTCCTGAAGAAGG | SSR | 55 | [40] |
| Pi5 | 40N23R | TGTGAGGCAACAATGCCTATTGCG | CTATGAGTTCACTATGTGGAGGCT | InDel | 55 | [40] |
| Pikm | k2167 | CGTGCTGTCGCCTGAATCTG | CACGAACAAGAGTGTGTCGG | InDel | 55 | [40] |
| Pi25 | CAP1 | TGAAATGGGTGAAAGATGAG | GCCACATCATAATTCCTTGA | CAPS | 55 | [70] |
* FM, functional marker; LM, linked marker; InDel, insertion-deletion marker; FNP, functional nucleotide polymorphism; SNP, single nucleotide polymorphism; CAPS, Cleaved Amplified Polymorphism Sequences
Allele scoring and genetic diversity analysis
The presence or absence of an allele was indicated as 1 and 0, respectively, in the amplified PCR products of 25 markers, which were scored as a binary matrix. Using a binary data matrix of 25 markers, the genetic distance and similarity coefficients for 52 rice accessions were calculated. Using the Cervus 3.0 programme (Field Genetics Ltd., London, England) and POPGENE 32 software, different parameters such as the number of different alleles per locus (Na), number of effective alleles per locus (Ne), Shannon’s Information Index (I), and Expected Heterozygosity (HE) for each marker were calculated [41]. Subsequently, a heatmap of all the rice accessions was constructed using the pheatmap package with complete linkage clustering method and euclidean distance measure by R version 4.0.3 statistical software for the presence or absence of 25 markers for both leaf and neck blast.
Association analysis
To study the genetic relationship between blast resistance genes and the disease spectrum, we used TASSEL version 5.0 software with a general linear model (GLM) function [42]. Only the P-value was seen in 5% of the permutations for the most significant polymorphism in a region when the GLM model of TASSEL (v 5.0) software was performed with 1000 permutations of data. Using genotypic data collected with 25 molecular markers and pheatmap-based clustering with complete linkage clustering method and Euclidean distance measure, the genetic distance between the 52 rice accessions was estimated using R version 4.0.3 statistical programme.
Population structure analysis
Based on genotyping data from 25 markers, the STRUCTURE software v 2.3.4 [43] was used to evaluate the population structure of 52 rice accessions. Using the admixture and correlated allele frequencies model, each subpopulation (K) was estimated at different K values ranging from one to ten, with five runs per K value. A total of 200000 burn-in periods and 200,000 Markov chain Monte Carlo (MCMC) iterations were used in the STRUCTURE runs. Using the STRUCTURE HARVESTER software, the highest delta K (ΔK) value was estimated to determine the most likely K-value [44]. The pairwise fixation index (FST) was calculated using principal coordinate analysis (PCoA) based on a binary data matrix of 25 markers, and analysis of molecular variance (AMOVA) was performed using the GenAlEx version 6.502 software [45].
Results
Phenotyping of hill germplasm lines
Initially, the responsiveness of 277 rice accessions to rice blast disease was assessed. From these 277 accessions, we chose 52 genotypes based on their reaction to rice blast disease. i.e., resistant, moderately resistant, and susceptible (Tables 1 & 2).
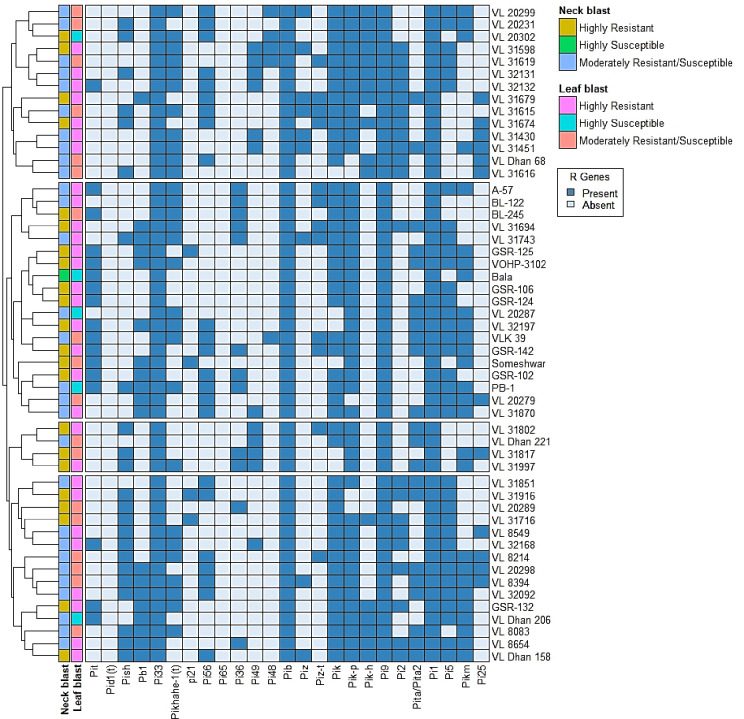
Of 52 rice genotypes, 29 (58%) and 22 (42%) rice genotypes were found to be highly resistant, 18 (36%) and 29 (57%) were moderately resistant, while 05 (6%) and 01 (1%) were highly susceptible to leaf and neck blast, respectively. Incidentally, sixteen genotypes showed high resistance to both leaf and neck blasts (Fig 1).
Fig 1. A clustered analysis based on the 25 molecular markers and Heatmap representing the summary of phenotypic and genotypic data of 52 rice genotypes analyzed in this study.
Genetic diversity of blast-resistant R genes
The present study used a set of twenty-five markers (functional/linked markers) that corresponded to the twenty-five R genes (Table 3). The gene frequency of the twenty-five blast R genes ranged from 32 to 60%, with the number of positive R-gene alleles ranging from 0 to 100%. Using a tk59-1 marker to visualize a 733 bp amplicon, the rice blast R-gene Pit was discovered in 17 rice genotypes. Pish on chromosome 1 was amplified with marker RM6648, resulting in a 207-bp band that was detected in 23 genotypes.
A 137-bp amplicon corresponding to the RM26998 marker was used to find the Pb1 gene on chromosome 11 in 19 genotypes. In 29 rice germplasm lines, the marker RM17496 was able to amplify the Pikhahe-1(t) gene with a fragment size of 84 bp. For the recessive blast-resistant gene pi21, only four genotypes were determined to be positive. The existence of the blast resistance gene Pi56 on chromosome 9 was detected using the gene-specific marker CRG4-2, which was found in 23 genotypes. Using the linked marker RM5647, the blast resistance genes Pi36 (chromosome 8) were discovered in 12 genotypes. The Pi49 gene, which is located on chromosome 11, was found in 12 genotypes after 182 bp were seen with the RM6094 marker. Using the RM5364 primer, the Pi48 gene was discovered in five genotypes.
The R genes, Piz, and Piz-t on chromosome 6 were amplified using SNP markers Z56592 and Zt56591, which revealed their presence in nine and twelve entries, respectively. Visualization of 112 bp, 148 bp, and 1500 bp amplicons corresponding to the K39512, K3957, and a gene-based marker, respectively, revealed the Pik, Pik-p, and Pik-h genes on chromosome 11. The genes Pik, Pik-p, and Pik-h were found in 46, 47, and 17 accessions, respectively. The Pi2 gene was discovered using the Pi2-i primer in twenty-one entries, resulting in positive bands. The major blast resistance gene Pita/Pita2, which was scored by visualization of an 861 bp amplicon utilizing the Pita3 marker, was found in 22 genotypes. The Pi5 gene was discovered in 35 genotypes, which was confirmed using the marker 40N23R. The Pikm gene was found in twenty-seven genotypes after PCR amplification. Pi25 was found in twelve genotypes using the CAP1 primer, which produced a 406-bp amplicon. The R genes Pi33, Pib, Pi9, and Pi1 were detected in all genotypes; however, the Pid1(t) and Pi65 genes were not discovered in any of the fifty-two genotypes examined in this study. The phenotypic and genotypic data of the 52 rice accessions studied in this investigation are summarized in Fig 1.
Cluster analysis
R-software was used to do the cluster analysis, which separated the 52 rice accessions into two primary clusters. Cluster I had 14 genotypes, seven and four of which were found to be highly resistant to leaf and neck blast, respectively, and three of which, VL31598, VL31679, and VL31674, were shown to be resistant to both leaf and neck blast. Cluster II was divided into three subgroups, the first of which contained a large number of genotypes (19), including nine genotypes resistant to both leaf and neck blast. On the other hand, two susceptible checks (Bala and PB-1) were also clustered together. Subgroup II is made up of four genotypes: VL 31802, VL 31817, VL 31997, and VL Dhan 221. Except for VL Dhan 221, all three genotypes are resistant to neck blast. Subgroup III has fifteen genotypes, eight and five genotypes showed high resistance to leaf and neck blast, respectively, and three of which were common for both leaf and neck blast resistance, including VL Dhan 158, GSR-132, and VL31916. Except for VL Dhan 206, the majority of the genotypes exhibited moderate resistance to either leaf or neck blast (Fig 1).
The genotypic data from the 25 markers was used to calculate genetic diversity measures including the number of distinct alleles per locus (Na), the number of effective alleles per locus (Ne), Shannon’s Information Index (I), and Expected Heterozygosity (HE). A total of 44 alleles were generated from 25 loci or markers (Table 4). The average number of alleles per locus (Na) was 1.76, with a range of 1 to 2. The number of effective alleles per locus (Ne) ranged from 1 to 1.99, with an average of 1.49. Shannon’s Information Index (I) ranged from 0 to 0.692 (Pikm), with an average of 0.42. The Expected Heterozygosity (HE) ranged from 0 (Pid1(t), Pi33, Pi65, Pib, Pi9(Pi9-i), and Pi1) to 0.499 (Pikm) with an average of 0.285.
Table 4. Analysis of the number of alleles, Shannon’s Information Index, observed and expected Heterozygosity.
| Locus | Na | Ne | I | He |
|---|---|---|---|---|
| Pit | 2.000 | 1.786 | 0.632 | 0.440 |
| Pid1(t) | 1.000 | 1.000 | 0.000 | 0.000 |
| Pish | 2.000 | 1.974 | 0.686 | 0.493 |
| Pb1 | 2.000 | 1.865 | 0.656 | 0.464 |
| Pi33 | 1.000 | 1.000 | 0.000 | 0.000 |
| Pikhahe-1(t) | 2.000 | 1.974 | 0.686 | 0.493 |
| pi21 | 2.000 | 1.166 | 0.271 | 0.142 |
| Pi56 | 2.000 | 1.974 | 0.686 | 0.493 |
| Pi65 | 1.000 | 1.000 | 0.000 | 0.000 |
| Pi36 | 2.000 | 1.550 | 0.540 | 0.355 |
| Pi49 | 2.000 | 1.550 | 0.540 | 0.355 |
| Pi48 | 2.000 | 1.210 | 0.317 | 0.174 |
| Pib | 1.000 | 1.000 | 0.000 | 0.000 |
| Piz | 2.000 | 1.401 | 0.461 | 0.286 |
| Piz-t | 2.000 | 1.550 | 0.540 | 0.355 |
| Pik | 2.000 | 1.257 | 0.358 | 0.204 |
| Pik-p | 2.000 | 1.210 | 0.317 | 0.174 |
| Pik-h | 2.000 | 1.786 | 0.632 | 0.440 |
| Pi9 (Pi9-i) | 1.000 | 1.000 | 0.000 | 0.000 |
| Pi2 (Pi2-i) | 2.000 | 1.929 | 0.675 | 0.482 |
| Pita (Pita3) | 2.000 | 1.954 | 0.681 | 0.488 |
| Pi1 | 1.000 | 1.000 | 0.000 | 0.000 |
| Pi5 | 2.000 | 1.786 | 0.632 | 0.440 |
| Pikm | 2.000 | 1.997 | 0.692 | 0.499 |
| Pi25 | 2.000 | 1.550 | 0.540 | 0.355 |
Association analysis
The genetic association of markers with leaf and neck blast disease was examined using the general linear model (GLM) function to see if there was any evidence of a significant link between gene-specific markers and the disease reaction. Only two markers (RM5647 and K39512), which correspond to the blast-resistant genes Pi36 and Pik, respectively, showed a significant association with the neck blast disease, while only three markers (Pi2-i, Pita3, and k2167), which correspond to the blast-resistant genes Pi2, Pita/Pita2, and Pikm, respectively, showed a significant association with the leaf blast disease (Table 5). For leaf blast, the associated markers showed a phenotypic variance of 7.2% to 12.2%. The marker k2167, which is linked to the Pikm gene, was shown to have the maximum phenotypic variance. The markers K39512 and RM5647, corresponding to the blast-resistant genes Pik and Pi36, respectively, showed a phenotypic variance of 4.7 and 5.2% for neck blast. The remaining twenty markers, on the other hand, showed no significant association with blast disease (p≤0.1).
Table 5. Genetic association of blast resistant genes with rice neck and leaf blast disease in 52 genotypes.
| Marker | Neck blast | Leaf blast | ||
|---|---|---|---|---|
| P value | marker_R2 | P value | marker_R2 | |
| Pit | 0.31011 | 0.0206 | 0.94482 | 9.68E-05 |
| Pid1(t) | NaN | 0 | NaN | 0 |
| Pish | 0.97709 | 1.67E-05 | 0.76334 | 0.00183 |
| Pb1 | 0.7654 | 0.0018 | 0.97428 | 2.10E-05 |
| Pi33 | NaN | 0 | NaN | 0 |
| Pikhahe-1(t) | 0.26063 | 0.02524 | 0.77591 | 0.00164 |
| pi21 | 0.23418 | 0.02818 | 0.98122 | 1.12E-05 |
| Pi56 | 0.85031 | 7.19E-04 | 0.40692 | 0.0138 |
| Pi65 | NaN | 0 | NaN | 0 |
| Pi36 | 0.1001 | 0.05253 * | 0.46899 | 0.01054 |
| Pi49 | 0.74584 | 0.00212 | 0.16333 | 0.03849 |
| Pi48 | 0.66804 | 0.00371 | 0.18689 | 0.03458 |
| Pib | NaN | 0 | NaN | 0 |
| Piz | 0.62943 | 0.00469 | 0.31383 | 0.02028 |
| Piz-t | 0.93256 | 1.45E-04 | 0.16943 | 0.03742 |
| Pik | 0.10002 | 0.0479 * | 0.1949 | 0.03337 |
| Pik-p | 0.84113 | 8.11E-04 | 0.46584 | 0.01069 |
| Pik-h | 0.78264 | 0.00154 | 0.90339 | 2.98E-04 |
| Pi9 (Pi9-i) | NaN | 0 | NaN | 0 |
| Pi2 (Pi2-i) | 0.43981 | 0.01198 | 0.01374 | 0.1154 ** |
| Pita (Pita3) | 0.62468 | 0.00482 | 0.05363 | 0.07247 ** |
| Pi1 | NaN | 0 | NaN | 0 |
| Pi5 | 0.77264 | 0.00168 | 0.33685 | 0.01846 |
| Pikm | 0.1383 | 0.04341 | 0.01114 | 0.12202 ** |
| Pi25 | 0.64389 | 0.00431 | 0.6185 | 0.005 |
* & ** Significant at P value <0.1 and <0.05 respectively
Population structure analysis
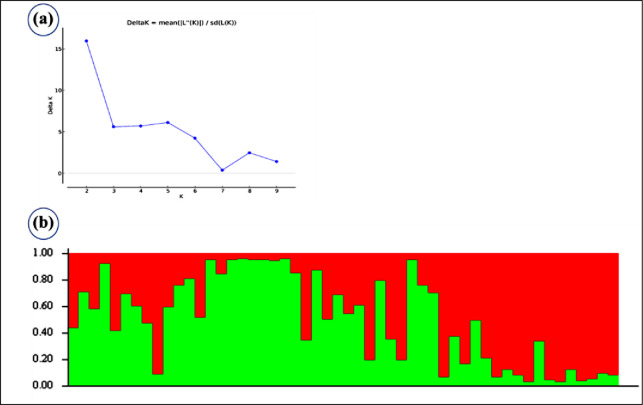
Using STRUCTURE software, all 52 rice genotypes were examined for population structure estimation for leaf and neck blast disease based on 25 markers. The Adhoc Measure K peak plateau was discovered to be K = 2 (Fig 2), indicating that the complete 52 rice genotypes were divided into two subgroups (SG1 and SG2).
Fig 2.
Population structure analysis of 52 rice genotypes (a) The maximum of ad hoc measure ΔK was observed to be K = 3 (b) Estimated population structure graph separated the whole population into two subgroups.
All populations were divided into two major subgroups with eight admixture levels based on an ancestry threshold of >60% (Table 6). SG1 was made up of the most genotypes identified to be highly resistant to neck blast. The majority of genotypes identified to be highly resistant to leaf blast, on the other hand, were concentrated in SG2. Genotypes with moderate resistance to both leaf and neck blast were clustered together in SG2, while genotypes with high susceptibility to both leaf and neck blast were grouped together in SG1.
Table 6. Population structure group of 52 genotypes based on inferred ancestry values.
| Genotypes | Inferred Ancestry | Structure group | |
|---|---|---|---|
| Q1 | Q2 | ||
| VL 8083 | 0.560 | 0.44 | AD |
| VL 8214 | 0.291 | 0.709 | SG2 |
| VL 8394 | 0.419 | 0.581 | AD |
| VL 8549 | 0.076 | 0.924 | SG2 |
| VL 8654 | 0.582 | 0.418 | AD |
| VL Dhan 158 | 0.302 | 0.698 | SG2 |
| VL 20231 | 0.398 | 0.602 | SG2 |
| VL 20279 | 0.527 | 0.473 | AD |
| VL 20287 | 0.913 | 0.087 | SG1 |
| VL 20298 | 0.403 | 0.601 | SG2 |
| VL 20299 | 0.237 | 0.763 | SG2 |
| VL 20302 | 0.186 | 0.814 | SG2 |
| VL 20289 | 0.484 | 0.516 | AD |
| VL 31430 | 0.047 | 0.953 | SG2 |
| VL 31451 | 0.151 | 0.849 | SG2 |
| VL 31598 | 0.045 | 0.955 | SG2 |
| VL Dhan 68 | 0.039 | 0.961 | SG2 |
| VL 31615 | 0.048 | 0.952 | SG2 |
| VL 31616 | 0.05 | 0.95 | SG2 |
| VL 31619 | 0.055 | 0.945 | SG2 |
| VL 31674 | 0.04 | 0.96 | SG2 |
| VL 31679 | 0.145 | 0.855 | SG2 |
| VL 31694 | 0.657 | 0.343 | SG1 |
| VL 31716 | 0.123 | 0.877 | SG2 |
| VL 31743 | 0.495 | 0.505 | AD |
| VL 31802 | 0.313 | 0.687 | SG2 |
| VL 31817 | 0.453 | 0.547 | AD |
| VL 31851 | 0.388 | 0.612 | SG2 |
| VL 31870 | 0.806 | 0.194 | SG1 |
| VL 31916 | 0.201 | 0.799 | SG2 |
| VL 31997 | 0.647 | 0.353 | SG1 |
| VL 32092 | 0.807 | 0.193 | SG1 |
| VL 32131 | 0.044 | 0.956 | SG2 |
| VL 32132 | 0.24 | 0.76 | SG2 |
| VL 32168 | 0.296 | 0.704 | SG2 |
| A-57 | 0.933 | 0.067 | SG1 |
| BL-122 | 0.626 | 0.374 | SG1 |
| BL-245 | 0.832 | 0.168 | SG1 |
| VL Dhan 221 | 0.503 | 0.497 | AD |
| VLK 39 | 0.79 | 0.21 | SG1 |
| GSR-102 | 0.933 | 0.067 | SG1 |
| GSR-106 | 0.875 | 0.125 | SG1 |
| GSR-124 | 0.915 | 0.085 | SG1 |
| GSR-125 | 0.968 | 0.032 | SG1 |
| GSR-132 | 0.658 | 0.342 | SG1 |
| GSR-142 | 0.954 | 0.046 | SG1 |
| VOHP-3102 | 0.965 | 0.035 | SG1 |
| VL Dhan 206 | 0.878 | 0.122 | SG1 |
| VL 32197 | 0.964 | 0.036 | SG1 |
| Someshwar | 0.944 | 0.056 | SG1 |
| Bala | 0.904 | 0.096 | SG1 |
| PB-1 | 0.92 | 0.08 | SG1 |
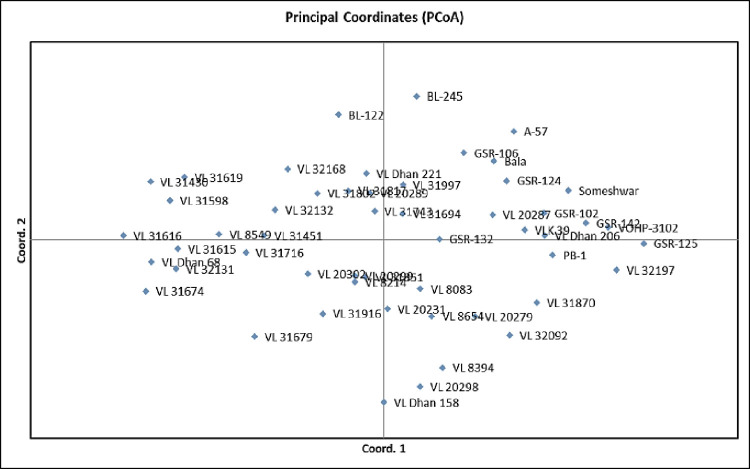
PCoA analysis has been carried out to establish the genetic relationship among the rice genotypes. PCoA analysis revealed that the first two axes explained 17.18% and 12.29% of the total variance (Table 7 and Fig 3). In PCoA, leaf blast-resistant genotypes were largely distributed among 1st and 2nd quadrants; on the other hand, most genotypes showed neck blast-resistant were concentrated in the 2nd quadrant. The genotypes found moderately resistant to both leaf and neck blast resistance were mostly distributed among the 1st, 3rd, and 4th quadrants, whereas susceptible genotypes were concentrated in the 2nd quadrant.
Table 7. Percentage of variation explained by the first 3 axes using blast resistance gene in PCoA.
| Axis | 1 | 2 | 3 |
|---|---|---|---|
| Variation of the individual axis (%) | 17.18 | 12.29 | 9.21 |
| Cumulative variation (%) | 17.18 | 29.47 | 38.67 |
Fig 3. PCoA of 25 molecular markers linked to blast resistance in 52 rice genotypes.
AMOVA analysis
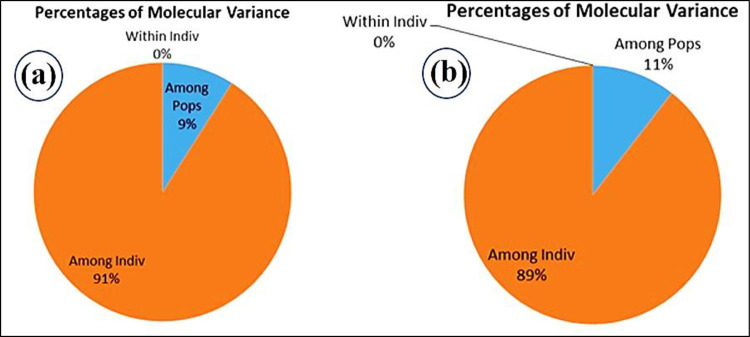
The genetic variations within and between the populations were assessed using AMOVA analysis. The leaf blast score was used to separate 52 rice genotypes into three populations: 29 (HR), 18 (MR), and 05 (S). Similarly, based on neck blast score, 22(HR), 29(MR), and 01(S) were separated. Furthermore, the maximum variance (91%) and (89%) was found within the population, while the least (9%) and (11%), respectively, were found between the populations for leaf and neck blast scores (Fig 4).
Fig 4.
AMOVA analysis based on populations separated with leaf blast scores (a) and neck blast scores (b).
Discussion
Rice genetic diversity has been reduced as a result of large-scale cultivation of high-yielding rice varieties, which have replaced landraces and traditional cultivars, limiting varietal improvement possibilities with existing resources [12, 46]. As a result of the widespread cultivation of genetically similar cultivars across a large area, the pathogen population is subjected to selection pressure, causing it to establish new races. Rice production has become a global threat as a result of the emergence of these new harmful races. However, the problem can be avoided by finding possible donors for unique functional genes or alleles that will help to overcome the disease and ensure future rice harvests [12, 37]. The present experiment investigated the genetic diversity of released varieties, advanced breeding materials, and traditional rice varieties for blast resistance genes using 25 molecular markers.
In this study, we used functional/gene-based molecular markers to genotype fifty-two rice hill germplasm collections for 25 major blast-resistant genes, in addition to field evaluations. We examined 52 rice accessions for leaf blast disease resistance in the uniform blast nursery and found that 29 (58%) and 22 (42%) genotypes were highly resistant to leaf and neck blast disease, respectively. Surprisingly, 16 accessions were found to be common for both leaf and neck blast resistance among the highly resistant rice accessions. With one released variety, VL Dhan 158, the vast majority of these accessions are advanced breeding materials.
Identification of the individual resistance based on phenotype is typically challenging because it is heavily influenced by developmental stage and environmental factors. However, using a linked marker associated with the R genes is the easiest and most reliable way for identifying individual/multiple gene(s) [47, 48]. The frequency of R-gene positive alleles ranged from 0% to 100%, with the genetic frequency of 25 major blast resistance genes ranging from 32% to 60%. The most positive alleles for the fifteen resistance genes are found in only two accessions (VL 8394 and VL Dhan 158) [49–51]. Our findings are similar to those of Yadav et al. [40] and Susan et al. [47], who reported gene frequencies ranging from 0% to 100% in 80 rice varieties released by National Rice Research Institute (NRRI), Cuttack, 9.4% to 100% in 32 Chinese rice germplasm, and 6% to 27% in 288 Indian landraces, respectively. The R-genes Pib, Pi9, Pi1, and Pi33 appeared to be present in all rice accessions. Our findings match those of Yadav et al. [40], who discovered the Pib gene in all eighty rice accessions studied. Similarly, the Pi9 gene was discovered in 51 Indian landraces [40] and 40 Chinese rice varieties [52]. However, just a few studies have documented the Pi9 gene’s rare prevalence [53, 40]. This could be owing to the Pi9 gene’s origin in the wild species O. minuta and its subsequent introduction into Indica rice [53]. The Pi1 gene was detected in 39 landraces with a frequency of 46.98%, according to Ingole et al. [50]. The presence of the Pi33 gene was discovered in 77 accessions in another investigation [5].
The genes Pit, Pish, and Pikhahe-1(t) were found in 17, 23, and 29 accessions, respectively [12, 40]. They are also found in the majority of accessions, according to earlier studies [12, 40]. In twenty-three accessions, the Pi56 gene was found. Although it has previously been detected in 27 landraces from northeastern India [40] and 26 NRRI Cuttack, released varieties, the gene Pi5 was found in 35 accessions [40]. Nine and twelve accessions, respectively, have the R genes Piz and Piz-t. However, there was no significant correlation was found between these two R genes and observed phenotypes. Similarly, they show partial resistance to the genotypes examined by Yadav et al. [40] and Susan et al. [47]. The Pi2 and Pita/Pita2 genes were detected in the majority of the rice accessions with high resistance to leaf blast [53]; however, a few genotypes without either of these genes were also resistant to leaf blast and may contain other unique R-genes/alleles. Except for VL 20287, which tested positive for the Pita/Pita2 gene, the genotypes that rated highly vulnerable to leaf blast did not include either of these two genes [53]. A blast resistance (BR) gene Pik-l was delineated in the region ~168.05 kb of the telomeric end of long of chromosome 11 of Japonica rice cv. Nipponbare. Based on its genomic position and distinct resistance spectra and also compared to previously identified Pik alleles, the new BR gene Pik-l was inferred to be a new allele of Pik locus [54]. Others earlier findings found that genes Pi2 and Pita/Pita2 express an NBS-LRR type R protein which are responsible to increase the resistant ability rice against leaf blast disease across a broad spectrum of pathogenic races [55,56].
In 19 accessions, the panicle blast resistance gene Pb1 was found. Only 9 accessions were found to have high resistance to neck blast, while the other ten showed moderate resistance. The Pb1 gene is a quantitative resistance gene that confers broad-spectrum resistance to all races. Despite having the Pb1 gene, 10 accessions were found to have only moderate resistance to neck blast. This could be owing to the involvement of at least four QTLs in neck blast resistance, three of which, Chr7, Chr9, and Chr11, have a negative impact on Pb1-mediated resistance, while Chr8, on the other hand, has a positive impact. These four QTLs are expected to influence the Pb1-mediated resistance either individually or in combination with others [57]. As of today, a few R genes, Pi25, Pb1, Pi64, Pi-jnw1, and Pi68(t) [20–24] and QTLs like, qNBL-9, qNBL-10, qNBL-5 [58], qNB11-1, qNB11-3, qNB1-1, qNB1-2, qNB1-3 [59], qPbh11-1 and qPbh7-1, [60] were found to confer resistance to neck blast. Among them, Pi64, and Pi68(t) were identified for the leaf as well as neck blast resistance. The pi21 gene was discovered in just four accessions. Surprisingly, all four accessions had high resistance to neck blast, while only the two genotypes had high and moderate resistance to leaf blast disease. The pi21 gene is a quantitative resistance gene for rice blast disease that offers broad-spectrum resistance [61].
The distance-based clustering was evaluated using genotype data, which divided the 52 germplasm into two primary groupings. Cluster I genotypes was moderately resistant to leaf and neck blast, whereas Cluster II genotypes are highly resistant to both leaf and neck blast. Similarly, the population structure analysis separated the 52 rice accessions into two subpopulations (SG1 and SG2), each with eight admixtures.
The leaf and neck blast-resistant genotypes are found in the first and second quadrants of the PCoA analysis, whereas moderately resistant genotypes were found in the first, third, and fourth quadrants. Previous research has also divided resistant and susceptible germplasm into distinct categories [40, 47]. A statistical approach for estimating molecular variance in a single species is the analysis of molecular variance (AMOVA). The AMOVA analysis revealed that there is the highest diversity within the population and minimal diversity between populations.
As a result of association mapping investigations, several genes influencing significant features have been uncovered, and it is now being utilized to deconstruct the genetic basis of many new qualities [62]. Two markers related to blast resistant genes Pi36 and Pik were found to be strongly associated with neck blast resistance, whereas three markers related to blast resistant genes Pi2, Pita/Pita2, and Pikm were found to be significantly associated with leaf blast resistance. Previous research on association mapping and blast disease resistance has shown its effectiveness in identifying markers associated with QTLs and/or resistance genes giving blast resistance [12, 33, 40]. The identified resistant rice accessions could be used as donors in future breeding projects because they come from a variety of genetic origins. These resistant accessions might then be studied for the existence of novel functional genes/alleles, allowing them to be exploited in rice improvement programs tailored to the needs of agricultural systems.
Conclusions
The identification of resistant germplasm for both leaf and neck blast will be facilitated by phenotyping along with the molecular characterization of blast resistance genes. Our current research on leaf and neck blast screening provided significant germplasm for breeders to employ as parent material for blast resistance transfer, particularly neck blast resistance, in the production of resistant breeding lines. Further identified resistant lines could be a valuable resource for blast resistance gene mapping, particularly in the case of neck blast disease.
Acknowledgments
The authors are grateful to the ICAR-Vivekananda Parvatiya Krishi Anusandhan Sansthan, Almora, Uttarakhand, India and the Taif University, Taif, Saudi Arabia for providing all facilities and support during conducting the study.
Data Availability
All relevant data are within the paper.
Funding Statement
This research was funded by ICAR-Vivekananda Parvatiya Krishi Anusandhan Sansthan, Almora, Uttarakhand, India. This research was also partially funded by the Taif University Researchers for funding this research with Supporting Project number (TURSP-2020/39), Taif University, Taif, Saudi Arabia. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Pennisi E. Armed and dangerous. Science 2010, 327, 804–805. doi: 10.1126/science.327.5967.804 [DOI] [PubMed] [Google Scholar]
- 2.Fernandez J.1; Orth K Rise of a Cereal Killer: The Biology of Magnaporthe oryzae Biotrophic Growth. Trends Microbiol. 2018, 26, 582–597. doi: 10.1016/j.tim.2017.12.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Le M.T.; Arie T.; Teraoka T. Population dynamics and pathogenic races of rice blast fungus, Magnaporthe oryzae in the Mekong Delta in Vietnam. J. Gen. Plant Pathol. 2010, 76, 177–182. [Google Scholar]
- 4.He X.; Liu X.; Wang L.; Lin F.; Cheng Y.; Chen Z.; et al. Identification of the novel recessive gene pi55(t) conferring resistance to Magnaporthe oryzae. Sci. China Life Sci. 2012, 55, 141–149. doi: 10.1007/s11427-012-4282-2 [DOI] [PubMed] [Google Scholar]
- 5.Singh A.K.; Singh P.K.; Arya M.; Singh N.K.; Singh U.S. Molecular screening of blast resistance genes in rice using SSR markers. Plant Pathol. J. 2015, 31, 12–24. doi: 10.5423/PPJ.OA.06.2014.0054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ou S.H. Rice Diseases, 2nd ed.; Commonwealth Agricultural Bureaux International: Wallingford, UK, 1985; p. 380. [Google Scholar]
- 7.Choi J.; Park S.Y.; Kim B.R.; Roh J.H.; Oh I.S.; Han S.S.; et al. Comparative Analysis of Pathogenicity and Phylogenetic Relationship in Magnaporthe grisea Species Complex. PLoS ONE, 2013, 8, 1–8. doi: 10.1371/journal.pone.0057196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jeevan B.; Rajashekara H.; Mishra K.K.; Subbanna A.R.N.S.; Singh A.K.; Sharma D. Nayaka S C, Hosahatti R., Prakash G, Satyavathi C T, Sharma R Nayaka S C, Hosahatti R, Prakash G, Satyavathi C T, Sharma R.Finger millet blast disease: Potential threat to global nutrition security. In Blast Disease of Cereal Crops. Fungal Biology. Springer, Cham. 2021, pp 51–57. [Google Scholar]
- 9.Yashaswini C.; Reddy N.P.; Pushpavati B.; Rao S.C.; Madhav S.M. Prevalence of Rice blast (Magnaporthe oryzae) incidence in South India. Bulletin of Environment, Pharmacology and Life Sci. 2017, 6, 370–373. [Google Scholar]
- 10.Ariya-anandech K.; Chaipanya C.; Teerasan W.; Kate-Ngam S.; Jantasuriyarat C. Detection and allele identification of rice blast resistance gene, Pik, in Thai rice germplasm. Agric. Nat. Resour. 2018, 52, 525–535. [Google Scholar]
- 11.Sharma T.R.; Rai A.K.; Gupta S.K.; Vijayan J.; Devanna B.N.; Ray S. Rice blast management through host–plant resistance: retrospect and prospects. Agric. Res. 2012, 1:37–52. [Google Scholar]
- 12.Yadav M.K.; Aravindan S.; Ngangkham U.; Raghu S.; Prabhukarthikeyan S.R.; Keerthana U.; et al. Blast resistance in Indian rice landraces: Genetic dissection by gene-specific markers. PLoS ONE, 2019, 14(1), 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Panda G.; Sahu C.; Yadav M.K.; Aravindan S.; Umakanta N.; Raghu S.; et al. Morphological and molecular characterization of Magnaporthe oryzae from Chhattisgarh. ORYZA-An International Journal on Rice. 2017, 54, 330–336. [Google Scholar]
- 14.Sahu C.; Yadav M.K.; Panda G.; Aravindan S.; Umakanta N.; Raghu S.; et al. Morphological and molecular characterization of Magnaporthe oryzae causing rice blast disease in Odisha. ORYZA-An International Journal on Rice. 2018, 55, 467–472. [Google Scholar]
- 15.Jeevan B.; Gogoi R.; Sharma D.; Manjunatha C.; Rajashekara H.; Ram D.; et al. Genetic analysis of maydis leaf blight resistance in subtropical maize (Zea mays L.) germplasm. J. Genet. 2020, 99, 1–9. [PubMed] [Google Scholar]
- 16.Yamaguchi I. Overview on the chemical control of rice blast disease. Kluwer Academic Publishers. 2004, pp 1–13. [Google Scholar]
- 17.Wang G.L.; Valent B. Durable resistance to rice blast. Science. 2017, 355, 906–907. doi: 10.1126/science.aam9517 [DOI] [PubMed] [Google Scholar]
- 18.Kalia S.; Rathour R. Current status on mapping of genes for resistance to leaf- and neck-blast disease in rice. 3 Biotech, 2019, 9, 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ning X.; Yunyu W.; Aihong L. Strategy for use of rice blast resistance genes in rice molecular breeding, Rice Sci. 2020, 27, 263–277 [Google Scholar]
- 20.Zhuang J.Y.; Ma W.B.; Wu J.L.; Chai R.Y.; Lu J.; Fan Y.Y.; et al. Mapping of leaf and neck blast resistance genes with resistance gene analog, RAPD and RFLP in rice. Euphytica, 2002, 128, 363–370. [Google Scholar]
- 21.Hayashi N.; Inoue H.; Kato T.; Funao T.; Shirota M.; Shimizu T.; et al. Durable panicle blast-resistance gene Pb1 encodes an atypical CC-NBS-LRR protein and was generated by acquiring a promoter through local genome duplication. Plant J. 2010, 64, 498–510. [DOI] [PubMed] [Google Scholar]
- 22.Ma J.; Lei C.; Xu X.; Hao K.; Wang J.; Cheng Z.; et al. Pi64, encoding a novel CC-NBS-LRR protein, confers resistance to leaf and neck blast in rice. Mol. Plant Microbe Interact. 2015, 28, 558–568. [DOI] [PubMed] [Google Scholar]
- 23.Wang R.; Fang W.; Guan C.; He W.; Bao Y.; Zhang H. Characterization and fine mapping of a blast resistant gene Pi-jnw1 from the japonica rice landrace Jiangnanwan. PLoS One, 2016, 11, 1–12. doi: 10.1371/journal.pone.0169417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Devi S.J.S.R.; Singh K.; Umakanth B.; Vishalakshi B.; Rao K.V.S.; Suneel B.; et al. Identification and characterization of a Large Effect QTL from Oryza glumaepatula revealed Pi68(t) as putative candidate gene for rice blast resistance. Rice, 2020, 13, 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Skamnioti P.; Gurr S.J. Against the grain: safeguarding rice from rice blast disease. Trends Biotechnol. 2009, 27:141–150. doi: 10.1016/j.tibtech.2008.12.002 [DOI] [PubMed] [Google Scholar]
- 26.Liu J.; Wang X.; Mitchell T.; Hu Y.; Liu X.; Dai L.; et al. Recent progress and understanding of the molecular mechanisms of the rice- Magnaporthe oryzae interaction. Mol. Plant Pathol. 2010, 11:419–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fukuoka S.; Saka N.; Koga H.; Ono K.; Shimizu T.; Ebana K.; et al. Loss of function of a proline containing protein confers durable disease resistance in rice. Science, 2009, 325, 998–1001. doi: 10.1126/science.1175550 [DOI] [PubMed] [Google Scholar]
- 28.Dean R.A.; Talbot N.J.; Ebbole D.J.; Farman M.L.; Mitchell T.K.; Orbach M.J.; et al. The genome sequence of the rice blast fungus Magnaporthe grisea. Nature. 2005, 434, 980–986. [DOI] [PubMed] [Google Scholar]
- 29.Kang S.; Lebrun M.H.; Farrall L.; Valent B. Gain of virulence caused by insertion of a Pot3 transposon in a Magnaporthe grisea avirulence gene. Mol. Plant Microbe Interact. 2001, 14, 671–674. [DOI] [PubMed] [Google Scholar]
- 30.Farman M.L.; Eto Y.; Nakao T.; Tosa Y.; Nakayashiki H.; Mayama S.; et al. Analysis of the structure of the AVR1-CO39 avirulence locus in virulent rice-infecting isolates of Magnaporthe grisea. Mol. Plant Microbe Interact, 2002, 15, 6–16. [DOI] [PubMed] [Google Scholar]
- 31.Bohnert H.; Fudal I.; Dioh W.; Tharreau D.; Notteghem J.; Lebrun M. A putative polyketide synthase/peptide synthetase from Magnaporthe grisea signals pathogen attack to resistant rice. Plant Cell, 2004, 16, 2499–2513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hittalmani S.; Parco A.; Mew T.V.; Zeigler R.S.; Huang N. Fine mapping and DNA marker-assisted pyramiding of the three major genes for blast resistance in rice. Theor. Appl. Genet. 2000, 100, 1121–1128. [Google Scholar]
- 33.Wang C.; Yang Y.; Yuan X.; Xu Q.; Feng Y.; Yu H.; et al. Genome–wide association study of blast resistance in indica rice. BMC Plant Biol. 2014, 14, 1–11. doi: 10.1186/s12870-014-0311-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Koide Y.; Kawasaki A.; Telebanco-Yanoria M.J.; Hairmansis A.; Nguyet N.T.; Bigirimana J.; et al. Development of pyramided lines with two resistance genes, Pish and Pib, for blast disease (Magnaporthe oryzae B. Couch) in rice (Oryza sativa L.). Plant Breed., 2010, 129, 670–675. [Google Scholar]
- 35.Zhang P.; Liu X.; Tong H.; Lu Y.; Li J. Association mapping for important agronomic traits in core collection of rice (Oryza sativa L.) with SSR markers. PLoS One. 2014, 9, 1–16. doi: 10.1371/journal.pone.0111508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kumbhar S.D.; Kulwal P.L.; Patil J.V.; Sarawate C.D.; Gaikwad A.P.; Jadhav A.S. Genetic diversity and population structure in landraces and improved rice varieties from India. Rice Sci. 2015, 22, 99–107. [Google Scholar]
- 37.Vasudevan K.; Vera Cruz C.M.; Gruissem W.; Bhullar N.K. Large-scale germplasm screening for identification of novel rice blast resistance sources. Front. Plant Sci. 2014, 5, 1–9. doi: 10.3389/fpls.2014.00505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Singh N.; Choudhury D.R.; Tiwari G.; Singh A.K.; Kumar S.; Srinivasan K.; et al. Singh, R. Genetic diversity trend in Indian rice varieties: an analysis using SSR markers. BMC Genet. 2016, 17, 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.International Rice Research Institute (IRRI). Standard Evaluation System for Rice (SES). 2002. Available online: http://www.knowledgebank.irri.org/images/docs/rice‐standard‐evaluation‐system.pdf
- 40.Yadav M.K.; Aravindan S.; Umakanta N.; Shubudhi H.N.; Bag M.K.; Adak T.; et al. Use of molecular markers in identification and characterization of resistance to rice blast in India. PLoS One, 2017, 12, 1–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yeh F.C. Population genetic analysis of co-dominant and dominant marker and quantitative traits. Belg. J. Bot. 1997, 130:129–157. [Google Scholar]
- 42.Bradbury P.J.; Zhang Z.; Kroon D.E.; Casstevens T.M.; Ramdoss Y.; Bucker E.S. TASSEL: software for association mapping of complex traits in diverse samples. Bioinformatics, 2007, 23, 2633–2635. doi: 10.1093/bioinformatics/btm308 [DOI] [PubMed] [Google Scholar]
- 43.Pritchard J.K.; Stephens M.; Donnelly P. Inference of population structure using multilocus genotype data. Genetics, 2000, 155, 945–959. doi: 10.1093/genetics/155.2.945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Earl D.A. STRUCTURE HARVESTER: a website and program for visualizing STRUCTURE output and implementing the Evanno method. Conservation Genetics Resources, 2012, 4, 359–361. [Google Scholar]
- 45.Peakall R.O.; Smouse P.E. GENALEX 6.5: genetic analysis in EXCEL. Population genetic software for teaching and research. Bioinformatics, 2012, 28, 2537–2539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tanksley S.D.; Mc Couch S.R. Seed banks and molecular maps: unlocking genetic potential from the wild. Science, 1997, 277, 1063–1066. doi: 10.1126/science.277.5329.1063 [DOI] [PubMed] [Google Scholar]
- 47.Susan A.; Yadav M.K.; Kar S.; Aravindan S.; Ngangkham U.; Raghu S.; et al. Molecular identification of blast resistance genes in rice landraces from northeastern India. Plant Pathol. 2019, 68, 537–546. [Google Scholar]
- 48.Hayashi K.; Yoshida H.; Ashikawa I. Development of PCR-based allele-specific and InDel marker sets for nine rice blast resistance genes. Theor. Appl. Genet. 2006, 113, 251–260. doi: 10.1007/s00122-006-0290-6 [DOI] [PubMed] [Google Scholar]
- 49.Jia Y.; Wang Z.; Fjellstrom R.G.; Moldenhauer K.A.K.; Azam M.D.A.; Correll J.; et al. Rice Pi–ta gene confers resistance to the major pathotypes of the rice blast fungus in the United States. Phytopathology, 2004, 94, 296–301. [DOI] [PubMed] [Google Scholar]
- 50.Ingole K.D.; Prashanthi S.K.; Krishnaraj P.U. Mining for major blast resistance genes in rice landraces of Karnataka. Indian J Genet Plant Breed. 2014, 74, 378–83. [Google Scholar]
- 51.Yan L.; Yan B.Y.; Peng Y.L.; Ji Z.J.; Zeng Y.X.; Wu H.L.; et al. Molecular screening of blast resistance genes in rice germplasms resistant to Magnaporthe oryzae. Rice Sci. 2017, 24, 41–47. [Google Scholar]
- 52.Yang Y.; Zhang H.; Xuan N.; Chen G.; Liu X.; Yao F.; et al. Identification of blast resistance genes in 358 rice germplasms (Oryza sativa L.) using functional molecular markers. Eur. J. Plant Pathol. 2017, 148, 567–576. [Google Scholar]
- 53.Imam J.; Alam S.; Mandal N.P.; Variar M.; Shukla P. Molecular screening for identification of blast resistance genes in North East and Eastern Indian rice germplasm (Oryza sativa L.) with PCR based makers. Euphytica, 2014, 196, 199–211. [Google Scholar]
- 54.Singh W.; Kapila R.; Sharma T.; Rathour R. Genetic and physical mapping of a new allele of Pik locus from japonica rice ‘Lijiangxintuanheigu’. Euphytica, 2015, 889–901. [Google Scholar]
- 55.Deng Y.; Zhu X.; Shen Y.; He Z. Genetic characterization and fine mapping of the blast resistance locus Pigm(t) tightly linked to Pi2 and Pi9 in a broad-spectrum resistant Chinese variety. Theor. Appl. Genet. 2006, 113, 705–713. doi: 10.1007/s00122-006-0338-7 [DOI] [PubMed] [Google Scholar]
- 56.Meng X.; Xiao G.; Telebanco-Yanoria M.J.; Siazon P.M.; Padilla J.; Opulencia R.; et al. The broad-spectrum rice blast resistance (R) gene Pita2 encodes a novel R protein unique from Pita. Rice, 2020, 13, 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Inoue H.; Nakamura M.; Mizubayashi T; Takahashi A.; Sugano S; Fukuoka S; et al. Panicle blast 1 (Pb1) resistance is dependent on at least four QTLs in the rice genome. Rice, 2017, 10, 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hittalmani S.; Srinivasachary B.P.; Shashidhar H.E. Identifying major genes and QTLs for field resistance to neck blast in rice. In: Khush GS et al. (eds) Advances in rice genetics. Rice Genetics Collection, Los Banos, 2003, pp 248–250. [Google Scholar]
- 59.Noenplab A.; Vanavichit A.; Toojinda T.; Sirithunyad P.; Tragoonrung S.; Sriprakhon S.; et al. QTL mapping for leaf and neck blast resistance in Khao Dawk Mali 105 and Jao Hom Nin recombinant inbred lines. Sci. Asia, 2006, 32, 133–142. [Google Scholar]
- 60.Fang N.; Wang R.; He W.; Yin C.; Guan C.; Chen H.; et al. QTL mapping of panicle blast resistance in japonica landrace Heikezijing and its application in rice breeding. Mol. Breed. 2016, 36, 171–179. [Google Scholar]
- 61.Angeles-Shim R.B.; Reyes V.P.; del Valle M.M.; Lapis R.S.; Shim J.; Sunohara H.; et al. Marker-assisted introgression of quantitative resistance gene pi21 confers broad spectrum resistance to rice blast. Rice Sci. 2020, 27, 113–123. [Google Scholar]
- 62.Hall D.; Tegström C.; Ingvarsson Pär K. Using association mapping to dissect the genetic basis of complex traits in plants. Brief. Funct. Genom. 2010, 9, 157–165. doi: 10.1093/bfgp/elp048 [DOI] [PubMed] [Google Scholar]
- 63.Chen X.W.; Li S.G.; Xu J.C.; Zhai W.X.; Ling Z.Z. Ma, B.T.; Wang, Y.P.; et al. Identification of two blast resistance genes in a rice variety, Digu. J Phytopathol, 2004, 152, 77–85. [Google Scholar]
- 64.Berruyer R.; Adreit H.; Milazzo J.; Gaillard S.; Berger A.; Dioh W.; et al. Identification and fine mapping of Pi33, the rice resistance gene corresponding to the Magnaporthe grisea avirulence gene ACE1. Theor. Appl. Genet. 2003, 107, 1139–1147. [DOI] [PubMed] [Google Scholar]
- 65.Xu X.; Chen H.; Fujimura T.; Kawasaki S. Fine mapping of a strong QTL of field resistance against rice blast, Pikahei– 1(t), from upland rice Kahei, utilizing a novel resistance evaluation system in the greenhouse. Theor. Appl. Genet. 2008, 117, 997–1008. doi: 10.1007/s00122-008-0839-7 [DOI] [PubMed] [Google Scholar]
- 66.Liu Y.; Liu B.; Zhu X.; Yang J.; Bordeos A.; Wang G.; et al. Fine–mapping and molecular marker development for Pi56(t), a NBS–LRR gene conferring broad–spectrum resistance to Magnaporthe oryzae in rice. Theor. Appl. Genet. 2013, 126, 985–998. doi: 10.1007/s00122-012-2031-3 [DOI] [PubMed] [Google Scholar]
- 67.Sun P.; Liu J.; Wang Y.; Jiang N.; Wang S.; Dai Y.; et al. Molecular mapping of the blast resistance gene Pi49 in the durably resistant rice cultivar Mowanggu. Euphytica. 2013, 192, 45–54. [Google Scholar]
- 68.Huang H.; Huang L.; Feng G.; Wang S.; Wang Y.; Liu J.; et al. Molecular mapping of the new blast resistance genes Pi47 and Pi48 in the durably resistant local rice cultivar Xiangzi–3150. Phytopathology, 2011, 101, 620–626. [DOI] [PubMed] [Google Scholar]
- 69.Sharma T.R.; Madhav M.S.; Singh B.K.; Shanker P.; Jana T.K.; Dalal V.; et al. High-resolution mapping, cloning and molecular characterization of the Pi-kh gene of rice, which confers resistance to Magnaporthe grisea. Mol. Genet. Genom. 2005, 274, 569–578. [DOI] [PubMed] [Google Scholar]
- 70.Wang Y.; Wang D.; Deng X.; Liu J.; Sun P.; Liu Y.; et al. Molecular mapping of the blast resistance genes Pi2–1 and Pi51(t) in the durably resistant rice ‘Tianjingyeshengdao’. Phytopathology, 2012, 102, 779–786. [DOI] [PubMed] [Google Scholar]
- 71.Zheng W.; Wang Y.; Wang L.; Ma Z.; Zhao J.; Wang P.; et al. Genetic mapping and molecular marker development for Pi65(t), a novel broad–spectrum resistance gene to rice blast using next-generation sequencing. Theor. Appl. Genet. 2016, 129 1035–1044. doi: 10.1007/s00122-016-2681-7 [DOI] [PubMed] [Google Scholar]
- 72.Liu Q.X.; Wang L.; Chen S.; Lin F.; Pan Q.H. Genetic and physical mapping of Pi36(t), a novel rice blast resistance gene located on rice chromosome 8. Mol. Genet. Genom. 2005, 274, 394–401. doi: 10.1007/s00438-005-0032-5 [DOI] [PubMed] [Google Scholar]