Abstract
Yersinia pestis possesses a heme-protein acquisition system (Hmu) that allows it to utilize heme and heme-protein complexes as the sole sources of iron. Analysis of the Y. pestis CO92 genomic sequence revealed a second heme-protein acquisition gene cluster that shares homology with the hemophore-dependent heme acquisition system (Has system) of Serratia marcescens. This locus consisted of the hasRyp receptor gene, the hasAyp hemophore gene, and genes encoding components of the HasAyp dedicated ABC transporter factor (hasDEyp), as well as a tonB homologue (hasByp). By using a reconstituted secretion system in Escherichia coli, we showed that HasAyp is a secreted heme-binding protein and that expression of HasAyp is iron regulated in E. coli. The use of a transcriptional reporter fusion showed that the hasRADEB promoter is Fur regulated and has increased activity at 37°C. Hemoglobin utilization via the Hasyp system was studied with both E. coli and Y. pestis, for which has and has hmu mutant strains were used. No contribution of the Has system to heme utilization was observed in either E. coli or Y. pestis under the conditions we tested. Previously it was shown that a deletion of the Hmu system had no effect on the virulence of Y. pestis in a mouse model of bubonic plague. An Hmu− Has− double mutant also retained full virulence in this model of infection. This report constitutes the first attempt to investigate the contribution of the hemophore-dependent heme acquisition system in bacterial pathogenicity.
Iron plays a central role in the metabolism of most bacteria, in which the Fe2+/Fe3+ redox pair and its wide range of electron transfer capacity are well suited to many enzymatic reactions. Despite iron abundance in nature, bacteria face an acute iron supply problem due to the extremely low concentration of soluble Fe3+ at physiological pH under aerobic conditions. In vertebrate hosts, iron deficiency is further increased by a set of iron-withholding defenses leading to the sequestration of most intracellular iron by ferritin or hemoproteins such as hemoglobin, whereas extracellular iron is complexed by transferrin, lactoferrin, hemopexin, and haptoglobin (5, 42).
To overcome this problem, gram-negative bacteria have evolved numerous energy-dependent iron-regulated systems to acquire iron from iron- or heme-containing compounds by three distinct acquisition strategies. These include (i) the release of small molecules with a high affinity for iron called “siderophores,” (ii) the direct binding of iron-containing protein compounds to outer membrane receptors, or (iii) the extracellular secretion of heme-binding proteins called “hemophores” that subsequently transport and deliver heme to their cognate outer membrane receptor. The coexistence of more than one of the above iron-scavenging systems is relatively common in bacteria (for review, see reference 42). Iron acquisition has been shown to be critical for the survival of pathogenic bacteria during the course of infection. In some instances, the first two strategies described above have been directly related to bacterial virulence (2, 14, 27). However, it is still not known to what extent the more recently characterized hemophore-dependent heme acquisition system contributes to the virulence of the bacteria for which it has been described so far: Serratia marcescens (22), Pseudomonas aeruginosa (24, 25), and Pseudomonas fluorescens (29).
Yersinia pestis, the bacterial agent of bubonic and pneumonic plague, possesses several iron acquisition systems that may be important for survival in its various iron-deficient niches. Inorganic iron can be acquired via an iron and manganese uptake system (termed Yfe) or through the secretion of a siderophore called “yersiniabactin” (6, 15, 30a). Y. pestis can also use hemin and a wide variety of host heme-containing compounds through a heme transport system (termed “Hmu”) (17) composed of at least five components: HmuR, -S, -T, -U, and -V. Under iron-depleted conditions, HmuR, the outer membrane receptor, and HmuTUV, a putative ABC-dependent transport system, are required for the use of hemin, hemin-albumin, and myoglobin, while HmuR is the only Hmu component needed to use hemoglobin and heme-hemopexin as iron sources. HmuS (HemS in Yersinia enterocolitica) may participate in removal of iron from hemin; however, conclusive demonstration of its role has been elusive (38, 39, 41, 42). Studies performed with mice showed that although both the iron/manganese and the yersiniabactin transport systems are important for Y. pestis, a deletion of the Hmu system has no effect on virulence in mice by a subcutaneous route of infection (2, 41)
During the course of the Hmu study, an hmu deletion mutant still grew under iron-depleted conditions in the presence of high concentrations of hemoglobin, leading to the hypothesis that an Hmu-independent uptake system may be involved in hemoglobin utilization (41). One possible candidate was identified in the Y. pestis CO92 genomic sequence released by the Sanger Centre as a sequence sharing homology with the hemophore-dependent heme acquisition system (Has system) of S. marcescens (22), P. aeruginosa (24, 25), and P. fluorescens (29). HasA hemophores are small extracellular proteins secreted by an ABC-dependent pathway, a process involving their carboxy-terminal secretion signal and its recognition by a tricomponent membrane ABC transporter. HasA is a heme-binding protein that delivers heme to a dedicated outer membrane receptor, HasR. Interaction between HasA and HasR induces a 100-fold increase in the capacity of the system to bind heme compounds and broadens the spectrum of potential heme sources for HasR (22, 24, 42).
In this study, we have begun characterizing the Has system of Y. pestis and investigated its biological function. We showed that HasAyp is a secreted heme-binding protein. Secretion, hemoglobin utilization, and induction conditions of the Hasyp system were studied with both Y. pestis and Escherichia coli. The effect of the deletion of the Y. pestis Has system in conjunction with a deletion of the Hmu system on the virulence of Y. pestis was investigated with a mouse model of infection. This report constitutes the first attempt to investigate the contribution of the hemophore-dependent heme acquisition system to bacterial pathogenicity.
MATERIALS AND METHODS
Bacterial strains, plasmids, and media.
The bacterial strains and plasmids used in this study are listed in Table 1. E. coli strains were cultivated in Luria broth (LB) (28). Y. pestis was grown in LB or on tryptose blood agar base (TBA). For growth under iron-restricted conditions, Y. pestis cells were grown in the defined medium PMH or PMH2 (13, 37). PMH2 is a modified version of PMH that contains 50 μM PIPES (piperazine-N, N′-bis[2-ethanesulfonic acid]) instead of HEPES (N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid). This change reduces medium acidification due to bacterial growth. PMH and PMH2 were deferrated with Chelex 100 resin (Bio-Rad) prior to filter sterilization as previously described (37). Alternatively, to reduce iron available to E. coli or Y. pestis, cells were grown in LB medium containing 0.2 mM 2,2′-dipyridyl (LBD). Hemin, hemin-agarose, and bovine hemoglobin were obtained from Sigma Chemical Co. Bovine hemoglobin agar plates were prepared as described in reference 12.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant genetic markers or features | Source or reference |
|---|---|---|
| Straina | ||
| Y. pestis | ||
| KIM5(pCD1Ap)+ | Wild type, Ampr | 13 |
| KIM6+ | Lcr− (pCDI−) | 30 |
| KIM6 | Lcr− Pgm− (Ybt− Δpgm) | 30 |
| KIM6-2030 | Lcr− Pgm− Fur− (fur::kan), Kmr | 36 |
| KIM6-2061.1 | Lcr− Pgm− HmuR− | This study |
| KIM6-2085 | Lcr− Pgm− HmuR− Has− | This study |
| KIM6-2045.1 | Lcr− Psn− | 10 |
| KIM6-2081.1+ | Lcr− Hmu− Has− | This study |
| KIM5-2045.7 | Lcr+ (pCDIAp) Ampr Psn− | This study |
| KIM5-2081.1+ | Lcr+ (pCDIAp) Ampr Hmu− Has− | This study |
| 6/69 | Wild type | 3 |
| 6/69c | Lcr− (pYV−) | This study |
| 6/69c(hasA) | Lcr− HasA− | This study |
| 6/69c (pPla−) | Lcr− pPla− | This study |
| 6/69 (pPla−) | pPla− | This study |
| 6/69 (HPI−) | Pgm− (Δpgm) | 7 |
| E. coli | ||
| C600 | F−thr leu fhuA lacY thi supE | Laboratory collection |
| TG1 | supE thi Δlac-proAB F traD36 proAB lacIqZΔM15 | Laboratory collection |
| MC4100 | F−araD139 ΔlacU169 relA1 rpsL150 thi mot flb5301 deoC7 ptsF25 rbsR | Laboratory collection |
| MC4100 fur::cat | fur | Laboratory collection |
| MC4100 hemA | hemA | R. Kadner |
| Plasmids | ||
| pACYC184 | Medium copy no., Tetr Cmr | 9 |
| pSyc150 | HasDEsm Cmr | 22 |
| pEU730 | Single-copy-no. reporter vector, promoterless lacZ, Spr | 11 |
| pCD1Ap | bla cassette inserted into ′yadA of pCD1 from Y. pestis KIM5, Ampr | 13 |
| pLG338 | Low copy number, Tetr Kmr | 40 |
| pR10K | HasRxm Kmr | 12 |
| pBGS18+ | Kmr | 35 |
| pHas98 | HasRADEByp 8.828-kb Sau3A Y. pestis KIM6 fragment inserted at BamHI site | This study |
| pHas98ΔBg12 | HasByp Deletion from hasR through hasE, 5.375-kb BglII deletion of pHas98 | This study |
| pACYCHas | HasRADEByp' Cmr, pHas98 8.5-kb EcoRI-StuI into pACYC184 EcoRI-ScaI | This study |
| pHasRA | HasRAyp+D′ Ampr, pHas98 4.3-kb HindIII-Asp718 into pUC19 HindIII-KpnI | This study |
| PHasRA2 | HasRAyp+D′, Kmr, pHasRA EcoRI-HindIII insert into pBGS18 | This study |
| pHasA | HasRAyp+ Ampr, PCR fragment into EcoRI-BamHI sites of pUC18 | This study |
| pEUHas | Single-copy-no. hasR::lacZ reporter, 383-bp hasR promoter ligated into pEU730, Spr | This study |
For Y. pestis KIM strains, + designates strains with an intact pgm locus.
Molecular biological procedures.
Standard procedures were used for cloning and analysis of DNA, PCR, electroporation, and transformation. Enzymes used to manipulate DNA were from New England Biolabs. Oligonucleotides were from Gibco BRL or Integrated DNA Technologies, Inc. Some DNA sequencing was performed by Genome Express S.A. In some instances, sequencing reactions were performed via the dideoxynucleotide chain termination method (33) by using 35S-dATP (Amersham/USB), Sequenase version 2.0 (Amersham/USB), and 7-deaza-dGTP (Boehringer Mannheim Biochemicals). Samples were electrophoresed through a 6% polyacrylamide gel containing 8.3 M urea (Sigma) cast in Tris-borate-EDTA buffer (32). Dried gels were exposed at room temperature to Kodak Biomax MR film.
Sequence analysis.
Amino acid sequence comparisons were performed with the Clustal package program (16). Homology searches were performed with Blast 2.0 at http://www.ncbi.nlm.nih.gov/BLAST/unfinishedgenome.html. DNA sequence manipulations were done with DNAStrider 1.3 (26) or DNAMAN Version 4.21 (Lynnon Biosoft).
Construction of recombinant plasmids.
pHasA was created as follows. The hasAyp gene was amplified by PCR with oligonucleotides YpHasArbs-5 (5′- CCCGAATTCTTAATAATTAAAAGGACATTATCATGAGTA-3′) and YhasAend-3 (5′-CCCAAGCTTAGCCCTAGCGGTGGTATCAATGACTCA GCC-3′) as primers and with Y. pestis 6/69 genomic DNA as a template. The amplified DNA was digested with EcoRI and HindIII and ligated into the same sites of pUC18. For the construction of pEUHas, in which the lacZ reporter gene was placed under the control of the hasR promoter region, a 383-bp region upstream of the putative initiating methionine residue for HasR was amplified from pHas98 by using PCR primers Hasp1 (5′-GATCTTTTAAACTTAATAGACTG-3′) and Hasp2 (5′-TTATGAAACTTCATCCTTAAC-3′). The products were blunt-end ligated into the PmeI site of pEU730 (11), and the resulting plasmid was designated pEUHas. All constructs were checked by PCR with specific and vector-based primers. Then each construct was sequenced.
Construction of has mutations in Y. pestis strains.
The Sanger Centre database of Y. pestis CO92 DNA was searched for has-related sequences by using the hasRADE sequence from S. marcescens. Based on the Y. pestis sequence, PCR primers HasYP1 (5′-CCGATTACAGCATCTCCT-3′) and HasYP2 (5′-GCGCATCAGACATATAACC-3′) were designed and used to produce a probe for screening a Y. pestis KIM6+ genomic DNA library. The resultant clone, pHas98, contains an approximately 8.8-kb fragment inserted into pLG338. An ∼5.4-kb BglII fragment was removed from pHas98 to create pHas98ΔBgl2. This deletion removes all of hasA and hasD as well as portions of hasR and hasE. A 2.5-kb StuI-to-HpaI fragment was cloned from pHas98ΔBgl2 into the SmaI site of pKNG101 (18) to yield pKNGhas. The mutated has alleles were introduced into Y. pestis KIM6 strains by allelic exchange as previously described (2). The genotype of all strains was confirmed by PCR with HasYP1 and HasYP2 primers, which are missing from the mutants, as well as HasD1 (5′-GATGCGCCATACAGAGCC-3′) and HasD2 (5′-GGCATTGTCGCCATCAGG-3′) primers, which flank the deleted region.
Protein analysis.
E. coli strains harboring various plasmids, S. marcescens SM365, and Y. pestis strains were grown at 28°C (Y. pestis) or 37°C (Y. pestis, S. marcescens, and E. coli) in LB, LBD, or PMH2. Cells from overnight or exponential cultures were centrifuged for 10 min at 5,000 × g at 4°C. The supernatants were concentrated by precipitation with either 10% trichloroacetic acid (inactive supernatants that do not retain HasA heme-binding activity) or 80% ammonium sulfate (active supernatants that retain HasA heme-binding activity). Inactive concentrated cell extracts were prepared by boiling whole cells in sodium dodecyl sulfate (SDS) buffer. Proteins were analyzed by SDS-polyacrylamide gel elctrophoresis (PAGE) followed by either Coomassie blue staining or Western blot analysis. Anti-HasAyp antibodies were prepared against HasAyp protein purified on hemin-agarose, separated on SDS-polyacrylamide gel, and electroeluted. The protein was mixed with Titer Max adjuvant (Interbiotech) and injected into a rabbit to raise antibodies. The rabbit serum was used in Western blot analysis at a dilution of 1/2,000. Nondenaturating PAGE was performed as described in reference 25.
For analysis of in vitro-synthesized products, ∼5 μg of purified, circular, plasmid DNA was added to an E. coli S30 extract system (Promega Corp.) and supplemented with 35S-labeled amino acids (DuPont NEN Research Products). After a 30-min incubation at 37°C, the products were acetone precipitated, resuspended in sample buffer, and electrophoresed on a 12% polyacrylamide gel containing SDS. The gels were dried and exposed overnight to Kodak BioMax MR film.
β-Galactosidase assays.
Lysates of Y. pestis strains carrying pEUHas were prepared from cells exponentially growing in PMH2 in the presence or absence of iron through two transfers for a total of approximately 6 generations. The β-galactosidase activity, with ONPG (o-nitrophenyl-β-d-galactopyranoside) used as a substrate, was measured spectrophotometrically and expressed in Miller units (28).
HasAyp purification heme affinity chromatography.
Five hundred milliliters of E. coli C600(pSyc150, pHasA) culture grown overnight at 37°C was centrifuged for 10 min at 5,000 × g at 4°C. The supernatant was concentrated by 80% ammonium sulfate precipitation. The protein pellet was resuspended and extensively dialyzed against 10 mM Tris-HCl (pH 7.5) and then subjected to heme affinity chromatography as described in reference 25. According to the manufacturer's specifications, there is a 10- to 100-fold excess of putatively available hemin compared to the estimated number of HasA molecules used in the hemin-agarose procedure Inactive and active supernatants (for biological tests) were prepared as described in reference 25.
Role of Has in hemoglobin utilization.
Growth stimulation of an E. coli hemA mutant producing HasRsm by exogenously supplied HasA hemophores was tested as follows. The HasRsm-producing strain was mixed with 3 ml of top agar and poured onto LBD plates supplemented with 10−6 M hemoglobin. Five-millimeter-diameter wells were cut in the agar and filled with 50 μl of hemin-agarose-purified HasAyp or HasAsm protein. Growth around the wells was recorded after overnight incubation at 37°C.
For Y. pestis strains, cells were grown at 28°C in the deferrated, defined medium, PHM2, in the absence of iron through two transfers for a total of approximately 6 generations. Third-transfer cultures were inoculated to an optical density at 620 nm (OD620) of 0.1 into PMH2 containing 50 μM ethylenediamine-N,N′-diacetic acid (EDDA) and either 0, 1.25, 2.5, 5, or 10 μM hemoglobin, assuming a molecular weight of 64,640 for hemoglobin. Growth of cells was monitored at regular intervals with a Genesys5 spectrophotometer (Spectronics Instruments, Inc.).
Dot blot HasA-binding assay.
The interaction between HasAsm or HasAyp with HasRsm was examined as follows: Cultures of MC4100 fur::cat carrying pBGS18+ or pR10K were grown exponentially in LB medium at 37°C and harvested when they reached an OD600 of 1. Cell pellets were suspended in Tris-buffered saline (TBS) buffer (50 mM Tris-HCl [pH 7.5], 150 mM NaCl) to an OD600 of 0.5. Aliquots of 50 ml were applied to nitrocellulose filters. The filters were dried for 10 min at room temperature and saturated by incubation for 1 h at 37°C in BS blocking solution (50 mM Tris-HCl [pH 7.5], 150 mM NaCl, 0.5% skim milk, 0.001% antifoam, 0.01% NaN3). The filters were then incubated for 30 min at 30°C in 1 ml of 50 mM Tris-HCl (pH 7.5) containing serial dilutions of either HasAyp or HasAsm or containing no HasA, washed three times with BS, and then incubated for 1 h at 37°C with polyclonal anti-HasAsm or anti-HasAyp antibodies diluted 1:2,000 in blocking solution. After three washes in BS, the filters were incubated with an alkaline phosphatase-conjugated goat anti-rabbit serum diluted 1:7,000 in BS. After three washes in BS, antibody binding was revealed as in other immunodetection tests (23).
Virulence testing.
Y. pestis strains lacking the low-calcium-response virulence plasmid are avirulent by any route of infection. A derivative of this plasmid, containing an ampicillin resistance gene inserted into the pseudogene yadA′/′yadA (pCD1Ap) (13), was electroporated into KIM6+, KIM6–2045.1, and KIM6–2081+, generating KIM5(pCD1Ap)+, KIM5–2045.7, and KIM5–2081.1+. All strains were constructed and tested for virulence in a BL3 facility. Bacteria were grown at 26°C in PMH2 containing 50 μM hemin and ampicillin (100 μg/ml) through two transfers for a total of approximately 7 generations. Cells were harvested at an OD620 of between 0.5 and 0.7, pelleted, and resuspended in mouse isotonic phosphate-buffered saline (PBS; 149 mM NaCl, 16 mM Na2HPO4, 4 mM NaH2PO4 [pH 7.0]). Five- to 7-week-old female NIH/Swiss Webster mice were injected subcutaneously with 0.1 ml of 10-fold serial dilutions of the bacterial suspensions. Groups of four mice were used for each bacterial dose. The number of cells injected was determined by plating serial dilutions on TBA plates containing ampicillin. Mice were monitored daily for 2 weeks. Fifty percent lethal doses (LD50s) were calculated by the method of Reed and Muench (31).
RESULTS
Identification of the Y. pestis Has system.
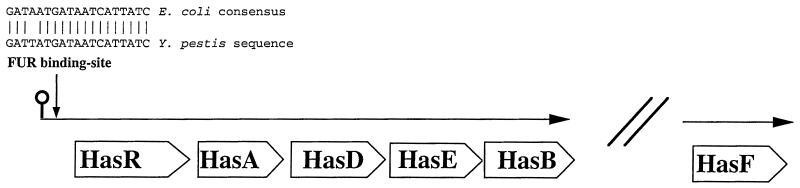
Analysis of the Y. pestis CO92 (biotype orientalis) complete genomic sequence released by the Sanger Centre (http://www.sanger.ac.uk/Projects/Y_pestis) revealed the existence of a locus sharing homology with the hemophore-dependent heme aquisition system (Has, for heme acquisition system) of S. marcescens, P. aeruginosa, and P. fluorescens. The Y. pestis has operon contains five open reading frames (ORFs) (Fig. 1) called “hasRyp,” “hasAyp,” “hasDyp,” “hasEyp,” and “hasByp ” according to the nomenclature used for the homologous genes of the S. marcescens has operon (12). The predicted sizes, protein locations, and putative functions, as well as percent identities of the deduced amino acid sequences with those found in protein databases, are shown in Table 2. A comparison of the CO92 has sequences with those from Y. pestis KIM10+ (biotype mediaevalis) (http://magpie.genome.wisc.edu/cgi-bin/Authenticate.cgi/uwgp_blast.html), shows that the deduced amino acid sequences of these five ORFs are nearly identical between these two biotypes (data not shown). The deduced amino acid sequence of HasRyp, the putative outer membrane receptor of the system, displayed a high level of identity with HasRsm, except for a short N-terminal extension missing in HasRyp (Fig. 2B). A well-conserved Fur binding site sharing 94% identity with the E. coli consensus sequence (GATAATGATAATCATTATC) was identified 340 bp upstream of the hasRyp initiation codon, suggesting that hasyp genes may be iron regulated. Further upstream of hasRyp, a gene coding for a protein 90% identical to the E. coli argino-succinate lyase ArgH was identified. The intergenic region between argH and hasR contains a stem-loop structure, suggesting that argH is not part of the hasRADEB putative operon. While this stem-loop structure may act as a transcription terminator, its localization close to the Fur binding site suggests that this structure may play a role in the Has system regulation. Finally, the deduced amino acid sequence of a protein 50.6% identical to HasFsm, the third component of the HasA transporter, was found to be located outside the main hasyp putative operon (Fig. 1). A single ORF was found in hasFyp locus, located 281 kb away from hasB. Thus, HasFyp does not seem to be part of an operon and does not contain any Fur binding site located upstream of the coding sequence.
FIG. 1.
Genetic organization of the Y. pestis Has system. Genetic organization of the 8.3-kb Y. pestis Hasyp system region identified in this study. The partial sequence comparison of the Fur binding site is shown above the diagram. The stem-loop structure located 40 bp upstream of the Fur binding site is indicated.
TABLE 2.
Properties and homologies of proteins of the Y. pestis Has system
| Protein | Mass (kDa) | Location or putative functiona | % Identity with system:
|
||
|---|---|---|---|---|---|
| Hassm | Haspa | Haspf | |||
| HasR | 93.4/89.6b | OM, HasA receptor for uptake | 48.0 | 38.0 | 44.0 |
| HasA | 22.2 | Secreted hemophore | 33.8 | 28.7 | 20.3 |
| HasD | 65.0 | IM, ATP-binding protein, HasA export | 55.0 | 56.1 | 56.9 |
| HasE | 48.9 | IM, ABC permease, HasA export | 44.2 | 43.0 | 39.1 |
| HasB | 29.3 | IM TonB-like protein | 25.3 | NAc | NA |
| HasF | 50.6 | OM, HasA export | 69.6 | 13.4 | 27.8 |
OM, outer membrane; IM, inner membrane.
Unprocessed/processed.
NA, not applicable.
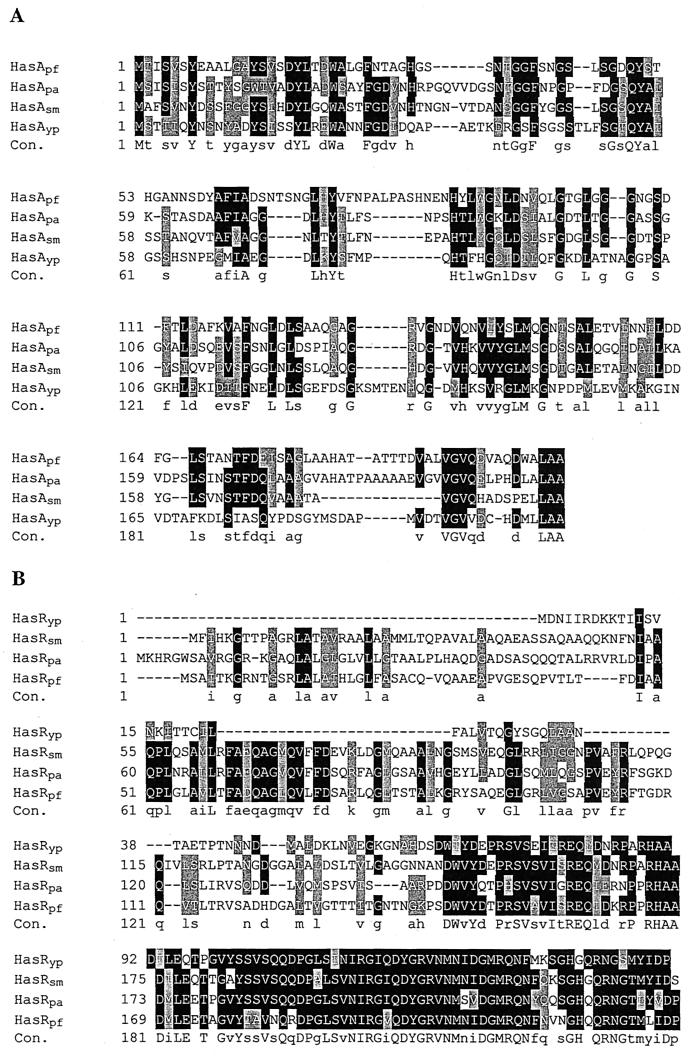
FIG. 2.
Clustal W amino acid sequence alignments of HasA and the N-terminal region. For both panels, identical amino acids are shown in black boxes, while similar amino acids are shown in gray boxes. The consensus line (Con.) shows identical (uppercase) and similar (lowercase) amino acids. (A) Amino acid sequence comparison of the N-terminal regions of the Y. pestis and S. marcescens HasR hemophore-dependent receptors HasRyp and HasR sm, respectively. (B) Amino acid sequence comparison of HasA hemophores identified in Y. pestis, S. marcescens, P. aeruginosa, and P. fluorescens (HasAyp, HasAsm, HasApa, and HasApf, respectively).
HasAyp is a secreted heme-binding protein.
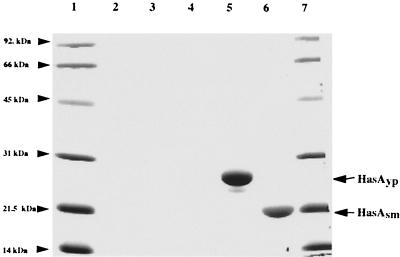
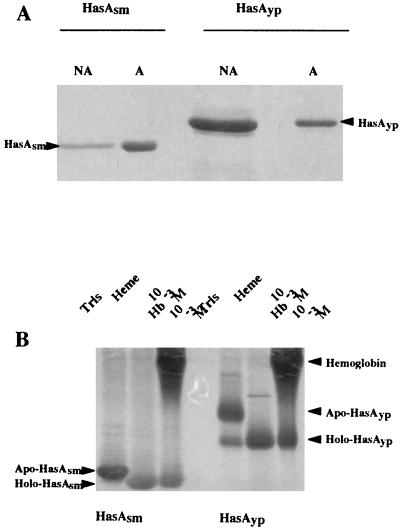
The homology between HasAsm and HasAyp (33.8% [Table 2]) suggested that, like HasAsm, HasAyp may be a secreted protein, although the differences between HasAyp and other HasA hemophores (Fig. 2A), particularly in the C-terminal region corresponding to the secretion signal, raised some questions about the capacity of HasAyp to be secreted. Previous studies showed that HasAsm is secreted in E. coli by a hybrid transporter composed of two S. marcescens inner membrane proteins, HasDsm and HasEsm, and the E. coli outer membrane protein TolC, which complements HasFsm function (22). We tested whether this transporter also secretes HasAyp. E. coli MC4100, carrying pSYC150, which expresses hasDsm and hasEsm, was cotransformed with pHasAyp, which contains hasAyp cloned with its own ribosome binding site under Plac promoter control (Table 1). Figure 3 shows that HasAyp was as efficiently secreted by the Has hybrid transporter as HasAsm.
FIG. 3.
HasAyp is secreted through the S. marcescens Has system. Samples were prepared as described in Materials and Methods and Coomassie blue stained after SDS-PAGE analysis. The equivalent of 1 ml of supernatants from cultures at an OD600 of 0.5 was loaded in each lane. Lanes: 1, molecular size markers; 2, MC4100 hemA; 3, MC4100 hemA(pSyc150); 4, MC4100 hemA(pHasAyp); 5, MC4100 hemA(pSyc150, pHasAyp); 6, MC4100 hemA(pSyc150, pHasAsm); 7, molecular size markers.
To study the biological function of HasAyp, the culture supernatant of strain E. coli MC4100(pSyc150, pHasAyp) grown in LB medium was precipitated with 80% ammonium sulfate, solubilized, and subjected to affinity chromatography on hemin-agarose. Unbound and bound proteins were analyzed by SDS-PAGE and Coomassie blue staining. A 19-kDa protein corresponding to HasAyp was retained on hemin-agarose, showing that HasAyp is a heme-binding protein (Fig. 4A). To test whether HasAyp produced in E. coli was able to acquire heme from hemoglobin, the samples were loaded in a nondenaturing polyacrylamide gel in which hemoproteins are known to migrate without dissociating from heme. In these gels, heme causes a significant shift in the migration of the bound polypeptide. As observed in previous studies (21, 25), the electrophoretic mobility of both HasAsm and HasAyp was increased in the presence of heme or hemoglobin (Fig. 4B), thus demonstrating that HasAyp can acquire heme from hemoglobin.
FIG. 4.
HasAyp is a heme-binding protein. (A) Isolation of HasAyp by heme affinity chromatography. Coomassie blue staining is shown. Concentrated supernatants were incubated with hemin-agarose as described in Materials and Methods. (Left) HasAsm, NA, E. coli MC4100(pSyc150, pHasAsm) concentrated supernatant not adsorbed (flowthrough material in excess) on hemin-agarose; A, E. coli MC4100(pSyc150, pHasAsm) concentrated supernatant adsorbed on hemin-agarose. (Right) HasAyp, NA, E. coli MC4100(pSyc150, pHasAyp) concentrated supernatant not adsorbed on hemin-agarose; A, E. coli MC4100(pSyc150, pHasAyp) concentrated supernatant adsorbed on hemin-agarose. The presence of HasAsm and HasAyp in NA lanes indicates that the capacity of the hemin-agarose columns was saturated by the supernatant concentrates used here. (B) Heme acquisition and formation of holoHasAsm and holoHasAyp after incubation of HasAsm or HasAyp with heme or hemoglobin. Four microliters (1 μg/ml) of HasAsm prepared from ammonium sulfate-concentrated supernatants of E. coli MC4100(pSyc150, pHasAsm) and 2 μl of HasAyp (2 μg/ml) prepared from ammonium sulfate-concentrated supernatant of E. coli MC4100(pSyc150, pHasAyp) were incubated with equal volumes of heme or hemoglobin (10−3 M) for 45 min at room temperature. The mixtures were loaded onto a 15% polyacrylamide gel and electrophoresed in the absence of SDS. The gel was stained with Coomassie blue. Protein bands complexed with heme (holoproteins) have a faster migration rate than uncomplexed protein (apoproteins).
Search for HasAyp in Y. pestis supernatants.
In order to investigate HasAyp production and secretion in Y. pestis, the culture supernatants and cell pellets of strains 6/69 and KIM6+ grown under iron-rich or iron-depleted conditions were harvested and probed with anti-HasAyp antibodies. Iron deprivation of Y. pestis was successfully obtained, since the iron-regulated high-molecular-weight proteins (8) were present in cell extracts (data not shown). Only a very faint band cross-reacting with the HasA antibodies was detected at ca. 15 kDa, in the cell pellet of iron-replete and iron-starved bacteria (data not shown). This band was still detected in the supernatant of Y. pestis 6/69c (has mutant), in which the chromosomal hasAyp gene was replaced by a truncated copy by reverse genetics, suggesting that it corresponds to some cross-reacting material. The effect of temperature on hasAyp expression was evaluated by growing the cells at 28 or 37°C. Neither temperature allowed the visualization of HasAyp. The influence of the presence or absence of the Y. pestis plasmids pYV and pPla and of the pgm locus (containing the high-pathogenicity island [HPI]) was also investigated by using the isogenic strains Y. pestis 6/69, 6/69c, 6/69c (pPla−), 6/69(pPla−), and 6/69(HPI−), but none of the mutants produced HasAyp. Similar negative results were obtained with Y. pestis KIM6+ (data not shown).
The reasons for the absence of HasA detection in Y. pestis may be that (i) the induction conditions used were not appropriate, (ii) HasA is unstable or rapidly degraded, (iii) its secretion rate is too low to be detected, or (iv) the Has system is not functional. Some of these hypotheses were investigated.
In Y. pestis, the hasRADEB promoter is Fur regulated and presents increased activity at 37°C.
To investigate transcription from the hasRADEB promoter region, we cloned the putative promoter region of hasRADEB upstream of lacZ in the reporter plasmid pEU730 (11). This construct, termed pEUHas, was electroporated into Y. pestis strains KIM6+, KIM6, and KIM6–2030. The cells were grown in the presence or absence of iron and assayed for β-galactosidase activity. The results indicated that expression of lacZ from this promoter fragment was iron regulated, showing an approximately three- to fivefold induction when the cells were grown in the absence of iron (Table 3). Transcription of hasR::lacZ apppears to be regulated by Fur, because the levels of β-galactosidase activity were constitutively high in the fur mutant, KIM6–2030(pEUHas). We also observed a minor but reproducible difference in the β-galactosidase values obtained with KIM6(pEUHas)+ and KIM6(pEUHas), which were slightly higher in the KIM6+ strain, suggesting that a component of the 102-kb pgm locus may participate in the expression of hasRADEB. Finally, β-galactosidase activity was six- to sevenfold higher in KIM6(pEUHas)+ cells grown at 37°C than in those grown at 26°C.
TABLE 3.
β-Galactosidase activities of Y. pestis strains containing lacZ reporters
| Straina | β-Galactosidase activity (mean ± SE Miller units)a
|
−Fe/+Fe ratio | |
|---|---|---|---|
| +Fe | −Fe | ||
| KIM6(pEU730)+ | 118 ± 14 | 203 ± 71 | 1.7 |
| KIM6(pEUHas)+ | 8,264 ± 1,532 | 39,738 ± 6,182 | 4.8 |
| KIM6(pEU730) | 227 ± 49 | 285 ± 26 | 1.3 |
| KIM6(pEUHas) | 5,822 ± 1,208 | 19,023 ± 6,597 | 3.3 |
| KIM6-2030(pEU730) | 200 ± 46 | 446 ± 62 | 2.2 |
| KIM6-2030(pEUHas) | 22,152 ± 4,492 | 28,674 ± 5,235 | 1.3 |
Bacterial cells were grown in deferrated PMH2 at 37°C and harvested during mid-log-phase growth. Whole-cell lysates were assayed for β-galactosidase activity as described in reference 28.
Expression of HasAyp is iron regulated in E. coli.
To study expression from the hasRADEByp promoter region in E. coli, we monitored HasAyp secretion in E. coli MC4100(pSYC150) carrying pHasRA2 containing hasRyp and hasAyp cloned with their own putative promoter region. As revealed by Coomassie blue staining or immunodetection with anti-HasAyp antibodies, HasAyp was secreted into the supernatant of E. coli MC4100(pSYC150, pHasRA2) grown in iron-depleted but not iron-rich media (Fig. 5). These results indicate that in E. coli, hasAyp (and possibly hasRyp) is expressed from its own promoter and that expression is iron regulated.
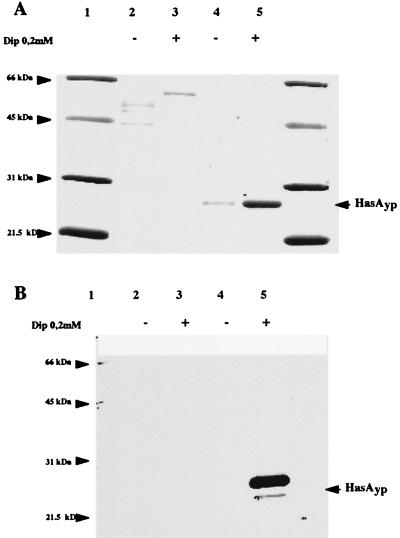
FIG. 5.
Iron regulation of HasAyp secretion in E. coli. Supernatant samples were prepared as described in Materials and Methods and subjected to SDS-PAGE analysis followed either by Coomassie blue staining (A [equivalent of 10 ml of supernatant from cultures at OD600 of 0.5 loaded in each lane]) or immunodetection with anti-HasAyp antibodies (B [equivalent of 1 ml of culture at OD600 of 0.5 loaded in each lane]). (A and B) Lanes: 1, molecular size markers; 2, MC4100(pHasRA2) in LB medium; 3, MC4100(pHasRA2) in LB medium supplemented with 0.2 mM 2,2′-dipyridyl; 4, MC4100(pSyc150, pHasRA2) in LB medium; 5, MC4100(pSyc150, pHasRA2) in LB medium supplemented with 0.2 mM 2,2′-dipyridyl.
Study of the functionality of the Y. pestis Has secretion system in E. coli
To test the functionality of the Y. pestis HasAyp transport system, the entire hasRADEB locus was cloned into pLG338 or pACYC184 to produce plasmids pHas98 and pACYCHas (Table 1). pHas98 and pACYCHas were introduced into E. coli MC4100, and the recombinant strains were grown in iron-rich (LB) or iron-depleted (LBD) medium and monitored for HasAyp secretion. No secreted HasAyp could be detected in the iron-depleted or iron-rich medium (data not shown).
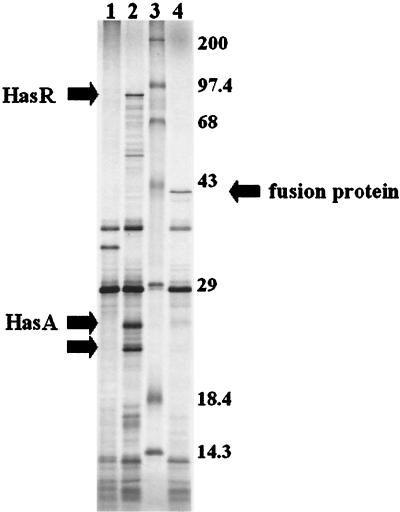
In vitro transcription or translation experiments with pHas98 showed Has-specific bands at ca. 87, 25, and 22 kDa (see arrows in Fig. 6, lane 2). The same bands were observed when a clone containing only hasRAD′ (pHasRA) was used in the transcription/translation system (data not shown), but are absent from reactions containing the vector plasmid, pLG338 (Fig. 6, lane 1). The 87-kDA band likely corresponds to HasRyp, while the 25- and 22-kDa proteins probably represent HasAyp and a degradation product generated from HasAyp. None of these bands was present in reactions containing pHas98ΔBglII (Fig. 6, lane 4). Instead, a new protein of ca. 41 kDa is seen, which is similar in size to the ca. 43-kDa fusion product predicted to occur as a result of the deletion. No bands corresponding to HasD, HasE, and HasB were convincingly observed. All three proteins contain at least one cysteine as well as several methionine residues and thus should have been labeled in the in vitro reactions. These results indicate that at least HasRyp and HasAyp are expressed from pHas98 and that the HasAyp secretion system encoded by pHas98 or pACYCHas may be expressed but not functional when reconstituted in E. coli. The biological function of the Y. pestis Has system was further investigated as described below.
FIG. 6.
Autoradiogram of plasmid-encoded proteins labeled with 35S-labeled amino acids by in vitro transcription and translation. The sizes of molecular mass standards (lane 3) are shown on the right side of the figure. The locations of HasR, HasA, and a HasA degradation product (lane 2) are indicated by the arrows on the left side of the figure. The arrow on the right points to a fusion protein (lane 4) generated by a deletion within the has operon. The plasmids used for the in vitro transcription and translation reactions were pLG338 vector (lane 1), pHas98 (lane 2), and pHas98ΔBgl2 (lane 4).
Study of the Y. pestis HasA-dependent heme acquisition system in E. coli
The biological function of HasAsm is the delivery of heme scavenged from various heme sources to the outer membrane receptor HasRsm. In S. marcescens, HasAsm is required for iron acquisition from heme and hemoglobin and was shown to interact with its outer membrane receptor, HasRsm, both in vivo and in vitro (10). To test whether HasAyp could substitute for the biological function of HasAsm, we performed an in vivo growth stimulation test with hemoglobin plates (see Materials and Methods). In this test, a lawn composed of HasRsm-expressing cells [MC4100 hemA(pR10K)] is plated on LB agar containing a low concentration of hemoglobin, and a solution of HasAsm or HasAyp is placed in a well in the agar. A productive interaction between HasAsm and HasRsm leads to bacterial growth around the HasA-containing well. As expected, HasAsm promoted bacterial growth, but this promotion effect was not observed around the well containing native HasAyp (data not shown). Consistently, in vitro dot blot binding experiments did not show any interaction between HasAyp and HasRsm (data not shown). These results suggest that HasAyp cannot substitute for the biological function of HasAsm, probably due to its lack of functional interaction with HasRsm.
The expression of hasAyp and hasRyp from plasmids pHasRA and pHasRA2, carrying hasRyp and hasAyp, raised the possibility that HasRyp could reach the outer membrane and is able to interact with hemophores available at the bacterial cell surface. To test this, we performed either in vivo growth stimulation tests with hemoglobin or in vitro dot blot binding experiments with cells potentially expressing HasRyp [MC4100 hemA(pHasRA)]. We did not see any growth stimulation or dot blot interaction with either HasAyp or HasAsm (data not shown). These results suggest that if HasRyp is correctly expressed in pHasRA and interacts with Y. pestis or S. marcescens HasA hemophores, this interaction does not lead to heme uptake under the conditions tested with E. coli.
We then investigated whether such a functional interaction could be detected with the whole Y. pestis Has system reconstituted in E. coli. To determine if the Y. pestis heme acquisition system allows cells to use hemoglobin as a source of iron, we introduced pHas98 or pACYCHas (encoding hasRADEB) and pHasRA (encoding hasRA) into an E. coli hemA mutant strain (Table 1). This mutant grows aerobically if supplemented with δ-aminolevulinic acid, but not with exogenously supplied hemoglobin. Growth was tested on iron-rich or iron-depleted agar plates in the presence of hemoglobin at concentrations ranging from 10−4 to 10−7 M. No growth was observed under any of the conditions tested, while a control strain carrying the Hassm system grew at these various hemoglobin concentrations (data not shown). These results indicate that the Y. pestis Has-dependent heme acquisition system is not functional in E. coli and are consistent with the lack of HasAyp secretion from pHas98- and pACYCHas-harboring cells.
Growth studies of has and has hmu mutant strains of Y. pestis
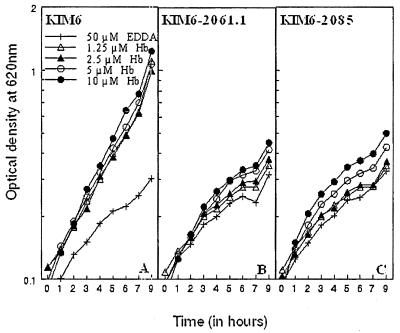
In Y. pestis, the Hmu system is the primary system that enables the bacteria to use heme and heme-containing compounds as a source of iron (17, 41). However, hmu mutants can still grow in the presence of high concentrations of hemoglobin, suggesting that there may be a second, lower-affinity system that the bacteria can use to obtain iron from hemoglobin. To determine if the Has system is responsible for this activity in hmu mutant strains, we examined the growth of KIM6 (Ybt− Has+ Hmu+), KIM6–2061.1 (Ybt− Has+ Hmu−), and KIM6–2085 (Ybt− Has− Hmu−) at 28°C in a defined medium containing 50 μM EDDA and different concentrations of hemoglobin (Fig. 7). Similar results were obtained with cells grown at 37°C. KIM6 readily grew in this medium with as little as 1.25 μM hemoglobin (Fig. 7A). However, the growth of KIM6–2061.1 (Fig. 7B) and KIM6–2085 (Fig. 7C) was not significantly stimulated by any tested concentration of hemoglobin. Furthermore, the growth of the hmu mutant was not significantly different from that of the hmu has mutant strain with any of the tested concentrations of hemoglobin. These results suggest that the Has system does not allow the bacteria to use hemoglobin as an iron source under the conditions tested in this study.
FIG. 7.
Growth of Y. pestis KIM6 (has+ hmu+ ), KIM6–2061.1 (has+ hmu mutant), and KIM6–2085 (has hmu double mutant) cells at 28°C in deferrated PMH2 with hemoglobin as the sole iron source. Iron-depleted cells were transferred to PMH2 containing 50 μM EDDA alone (+) or with hemoglobin at concentrations of 1.25 (▵), 2.5 (▴), 5 (○), or 10 (●) μM. These graphs depict one of two separate experiments with similar results.
Virulence testing of Y. pestis has mutants.
Previously, we had shown that a Y. pestis strain (KIM5–2060.21+) with a mutation in the hemin utilization system (Hmu) was fully virulent in mice by a subcutaneous route of infection (41). To determine if the Y. pestis Has system contributes to virulence in mammals, we constructed a has hmu double mutant, KIM6–2081.1+. The virulence plasmid with an ampicillin gene cassette inserted into the yadA′/′yadA pseudogene (pCD1Ap) was electroporated into KIM6–2081.1+, generating KIM5–2081.1+. Y. pestis strains lacking the virulence plasmid are avirulent by any route of infection. Groups of four mice were infected subcutaneously with 10-fold dilutions of bacterial cells grown at 26°C. The LD50s are listed in Table 4. The has hmu mutant was fully virulent by this route of infection, with an LD50 identical to that of the wild-type strain [KIM5(pCD1Ap)+]. In contrast, a strain bearing a mutation in the receptor (Psn) for the siderophore yersiniabactin is completely avirulent. This result suggests that the heme uptake systems Hmu and Has do not play a role in the virulence of Y. pestis for mice, at least by a subcutaneous route of infection.
TABLE 4.
Virulence testing of Y. pestis derivatives in mice infected subcutaneously
| Strain | Relevant characteristics | LD50 (CFU) |
|---|---|---|
| KIM5(pCDIAp)+ | Psn+ Hmu+ Has+ Lcr+ | <4.6 |
| KIM5-2081.1+ | Psn+ Hmu− Has− Lcr+ | <4.2 |
| KIM5-2045.7 | Psn− Hmu+ Has+ Lcr+ | >5.4 × 106 |
DISCUSSION
As a flea-mammal parasite, Y. pestis experiences a wide range of environmental conditions in which the ability to scavenge iron and heme contributes to its colonization potential and virulence. Accordingly, many inorganic iron or heme uptake systems in Y. pestis were characterized (Perry et al., submitted). The heme acquisition systems identified in Y. enterolitica (HemRSTUV) and Y. pestis (HmuRSTUV) (17, 38, 39, 41) involve an outer membrane TonB-dependent receptor and a periplasmic binding protein transport apparatus. The availability of Y. pestis genome sequences allowed the identification of a potential second category of heme uptake system relying on type I secretion of an extracellular heme-binding protein hemophore called “HasA” (24, 41; Perry et al., submitted). Here we characterized the Has system of Y. pestis and studied its function in E. coli and Y. pestis.
The comparison of the four Has systems identified so far in S. marcescens, P. aeruginosa, P. fluorescens, and Y. pestis showed that the genetic organization of the Y. pestis Has system is closely related to the homologous system of S. marcescens. It encodes the outer membrane receptor (HasR) and the two inner membrane components (HasD and HasE) of the ABC transporter of the extracellular hemophore HasA. As in the S. marcescens has operon, a gene showing similarities in its deduced amino acid sequence to tonB, hasB, is located downstream of hasE. HasB was shown to be a TonB-like protein specifically dedicated to the HasA-dependent heme acquisition system (29a). A similar function may be associated with the Y. pestis protein HasB. A hasB equivalent was not found associated with the two other Has systems identified so far in P. aeruginosa and P. fluorescens.
Comparisons between Y. pestis HasAyp and previously characterized hemophores revealed that they display relatively low levels of identity (33, 28, and 20% identity with HasAsm, HasApa, and HasApf, respectively). Amino acid sequence comparisons showed that HasAyp contains a 15-amino-acid insertion near the C terminus compared to HasAsm (Fig. 2). This insertion was also found in P. aeruginosa and P. fluorescens HasA hemophores.
We showed here, by using a reconstituted secretion system in E. coli, that HasAyp exhibits secretion and heme-binding properties very similar to those of HasAsm. However, in vivo and in vitro experiments indicated that HasAyp does not interact with HasRsm. This lack of interaction between heterologous pairs of hemophores and the HasR receptor was previously investigated in the case of HasApa and HasRsm, and it was shown that the ability of HasApf to interact with HasRsm depends on the removal of the C-terminal insertion. The additional stretch of 15 amino acids inserted at the C terminus of HasApf and HasApa was proposed to be responsible for the observed inhibition of heme delivery of HasApf or HasApa to HasRsm (1, 24). While it may also be the case for HasAyp (M. S. Rossi and J. M. Ghigo, unpublished results), these data further support the hypothesis that HasR-HasA couples constitute bipartite receptor entities that closely interact. These interactions may well be highly species specific, preventing eventual detrimental cross talk between hemophore-dependent systems expressed by two different bacterial species. Further characterization of functional HasR receptors may help clarify this point.
We showed that HasAyp expressed from pHasAyp can be secreted through a heterologous transporter in complementation experiments with the S. marcescens Has transporter. However, the introduction in E. coli of the whole Y. pestis has region led neither to HasA secretion nor to the reconstitution of a functional heme acquisition system. In particular, secretion studies did not provide evidence that the Y. pestis transporter components HasDyp and HasEyp are functional in E. coli. However, the lack of in vivo function in E. coli cannot be explained only by a HasAyp secretion defect, since a heme-deficient strain expressing HasRyp and HasAyp was unable to acquire heme, even at a high hemoglobin concentration at which hasR-alone-dependent heme acquisition should be observed. This suggests either that HasR is not properly expressed at the cell surface or, less trivially, that other Has-specific components are missing in these heterologous experiments. These components may be required either for the proper function of the receptor or for further heme internalization.
Previous studies demonstrated that Has systems are iron regulated, in good agreement with the identification of Fur binding sites (or “Fur boxes”) in these systems (21, 25, 29). Fur boxes are found in the promoter regions of iron-regulated genes and constitute fixation sites for the DNA-binding repressor Fur, which tightly regulates (represses) these genes when associated with ferrous iron as a corepressor. In Y. pestis, such a potential Fur binding site, albeit unusually distant upstream of the start codon for hasR, was also identified. Moreover, the use of a reporter plasmid where the hasRADEB putative promoter region was placed upstream of a promoterless lacZ gene showed that the Y. pestis has operon is active and iron regulated through a Fur-dependent control mechanism. We therefore tested and probed the supernatants of various Y. pestis 6/69 and KIM6 derivatives with anti-HasAyp antibodies by using cells grown in iron-chelated synthetic and rich media at 28 and 37°C. None of these conditions allowed the detection of an extracellular HasAyp. Although we provided evidence that the has operon is at least partially expressed in E. coli and that the has promoter is active and iron inducible when introduced in Y. pestis, we cannot formally rule out the possibility that the observed lack of function comes from the absence of expression of some of the components of the Y. pestis Has system. Such situations in which genes potentially involved in Yersinia environmental fitness are nonfunctional in Y. pestis have been reported (34). However, neither a frameshift mutation nor an obvious deletion was seen in the genes of the hasRADEB Y. pestis putative operon.
Previous studies suggested that, in addition to the Hmu system, there may be a second, lower-affinity system that Y. pestis can use to obtain iron from hemoglobin. However, the growth study presented here suggests that the Has system does not allow the bacteria to use hemoglobin as an iron source. The results obtained with the ΔhmuP′RSTUV mutant differ from those of earlier studies in which a graded response to increasing concentrations of hemoglobin was observed (41). Previous experiments used Y. pestis strains that produced the siderophore yersiniabactin (Ybt). Thus, the previous results with the ΔhmuP′RSTUV mutant were probably due to Ybt-mediated acquisition of iron from either hemoglobin degradation products or the EDDA-Fe complex, and not to the existence of a functional low-affinity heme transport system.
The recognition that an intact functional yersiniabactin system is a prerequisite for mouse virulence in the early stages of infection emphasized the fact that iron acquisition is critical for the progression of Y. pestis infections (30a). Our study, with a has hmu double mutant, constitutes the first investigation of the contribution of a hemophore-dependent system to bacterial pathogenicity. Our results with subcutaneous infection in mice suggest that the Has heme uptake system, as shown for the Hmu system, does not play a role in Y. pestis virulence in the mouse model of bubonic plague. Further experiments will be required to determine whether other routes of infection or infection in other animal hosts rely on Hmu and/or Has heme acquisition systems.
Taken together, our results suggest that a plausible reason for the absence of a HasA-related phenotype in Y. pestis may be that induction of the Has system was not appropriate in our experimental model and under the conditions used. While our data indicate that iron starvation participates in the regulation of the Has system, other factors may have to be considered. Comparison between HasR homologues revealed that HasRyp lacks an N-terminal extension present in HasRsm and other hemophore-dependent receptors (Fig. 2A) (data not shown). In E. coli, the amino terminus of the FecA outer membrane receptor is involved in regulating the Fec iron-dicitrate acquisition system (4, 19). Thus, regulation of the Hasyp system may be different from those of other Has systems and could require other factors. We noted the presence of strong stem-loop structures in the hasRADEB promoter region. Moreover, we showed that this promoter displays an increased hasRADEB promoter activity at 37°C compared to 26°C. This suggests that expression of the Has system may be temperature responsive, as shown to be the case for several bacterial virulence genes (20).
This study suggests that two distinct heme acquisition systems may coexist in Y. pestis and in other Yersinia species, supporting the hypothesis that hemophore-dependent heme acquisition systems are widespread in members of the family Enterobacteriaceae. While some of these systems have now been characterized at the structural and molecular level, their role in environmental survival and growth of gram-negative bacteria still awaits conclusive experimental demonstration.
ACKNOWLEDGMENTS
M.-S. Rossi was supported by the Conselho Nacional de Pesquisa (CNPq) grant no. 201031/97–3, Brazil. Work on this study by R.D.P. and J.D.F. was supported by Public Health Service grant AI-25098. J.-M.G., S.L., and E.C. were supported by the Institut Pasteur, Paris, France.
We are grateful to Jan Thompson and Steve Zink for initial work on the Has system in Y. pestis KIM6+ and to Cécile Wandersman for helpful discussions and critical reading of the manuscript.
REFERENCES
- 1.Akatsuka H, Binet R, Kawai E, Wandersman C, Omori K. Lipase secretion by bacterial hybrid ATP-binding cassette exporters: molecular recognition of the LipBCD, PrtDEF, and HasDEF exporters. J Bacteriol. 1997;179:4754–4760. doi: 10.1128/jb.179.15.4754-4760.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bearden S W, Perry R D. The Yfe system of Yersinia pestis transports iron and manganese and is required for full virulence of plague. Mol Microbiol. 1999;32:403–414. doi: 10.1046/j.1365-2958.1999.01360.x. [DOI] [PubMed] [Google Scholar]
- 3.Bonacorsi S P, Scavizzi M R, Guiyoule A, Amouroux J H, Carniel E. Assessment of a fluoroquinolone, three β-lactams, two aminoglycosides, and a cycline in treatment of murine Yersinia pestis infection. Antimicrob Agents Chemother. 1994;38:481–486. doi: 10.1128/aac.38.3.481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Braun V. Surface signaling: novel transcription initiation mechanism starting from the cell surface. Arch Microbiol. 1997;167:325–331. doi: 10.1007/s002030050451. [DOI] [PubMed] [Google Scholar]
- 5.Braun V, Hantke K, Koster W. Bacterial iron transport: mechanisms, genetics, and regulation. Met Ions Biol Syst. 1998;35:67–145. [PubMed] [Google Scholar]
- 6.Carniel E. The Yersinia high-pathogenicity island. Int Microbiol. 1999;2:161–167. [PubMed] [Google Scholar]
- 7.Carniel E, Guiyoule A, Mercereau-Puijalon O, Mollaret H H. Chromosomal marker for the ‘high pathogenicity’ phenotype in Yersinia. Contrib Microbiol Immunol. 1991;12:192–197. [PubMed] [Google Scholar]
- 8.Carniel E, Mazigh D, Mollaret H H. Expression of iron-regulated proteins in Yersinia species and their relation to virulence. Infect Immun. 1987;55:277–280. doi: 10.1128/iai.55.1.277-280.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chang A C Y, Cohen S N. Construction and characterization of amplifiable multicopy DNA cloning vehicles derived from the P15A cryptic miniplasmid. J Bacteriol. 1978;134:1141–1156. doi: 10.1128/jb.134.3.1141-1156.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fetherston J D, Lillard J W, Jr, Perry R D. Analysis of the pesticin receptor from Yersinia pestis: role in iron-deficient growth and possible regulation by its siderophore. J Bacteriol. 1995;177:1824–1833. doi: 10.1128/jb.177.7.1824-1833.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Froehlich B, Husmann L, Caron J, Scott J R. Regulation of rns, a positive regulatory factor for pili of enterotoxigenic Escherichia coli. J Bacteriol. 1994;176:5385–5392. doi: 10.1128/jb.176.17.5385-5392.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ghigo J-M, Letoffe S, Wandersman C. A new type of hemophore-dependent heme acquisition system of Serratia marcescens reconstituted in Escherichia coli. J Bacteriol. 1997;179:3572–3579. doi: 10.1128/jb.179.11.3572-3579.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gong S, Bearden S W, Geoffroy V A, Fetherston J D, Perry R D. Characterization of the Yersinia pestis Yfu ABC inorganic iron transport system. Infect Immun. 2001;69:2829–2837. doi: 10.1128/IAI.67.5.2829-2837.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Griffiths E. The iron-uptake systems of pathogenic bacteria. In: Bullen J J, Griffiths E, editors. Iron and infection: molecular, physiological, and clinical aspects. New York, N.Y: John Wiley & Sons, Inc.; 1987. pp. 69–137. [Google Scholar]
- 15.Heesemann J, et al. Virulence of Yersinia enterocolitica is closely associated with siderophore production, expression of an iron-repressible outer membrane polypeptide of 65,000 Da and pesticin sensitivity. Mol Microbiol. 1993;8:397–408. doi: 10.1111/j.1365-2958.1993.tb01583.x. [DOI] [PubMed] [Google Scholar]
- 16.Higgins D G, Thompson J D, Gibson T J. Using CLUSTAL for multiple sequence alignments. Methods Enzymol. 1996;266:383–402. doi: 10.1016/s0076-6879(96)66024-8. [DOI] [PubMed] [Google Scholar]
- 17.Hornung J M, Jones H A, Perry R D. The hmu locus of Yersinia pestis is essential for utilization of free haemin and haem-protein complexes as iron sources. Mol Microbiol. 1996;20:725–739. doi: 10.1111/j.1365-2958.1996.tb02512.x. [DOI] [PubMed] [Google Scholar]
- 18.Kaniga K, Delor I, Cornelis G R. A wide-host-range suicide vector for improving reverse genetics in gram-negative bacteria: inactivation of the blaA gene of Yersinia enterocolitica. Gene. 1991;109:137–141. doi: 10.1016/0378-1119(91)90599-7. [DOI] [PubMed] [Google Scholar]
- 19.Kim I, Stiefel A, Plantor S, Angerer A, Braun V. Transcription induction of the ferric citrate transport genes via the N-terminus of the FecA outer membrane protein, the Ton system and the electrochemical potential of the cytoplasmic membrane. Mol Microbiol. 1997;23:333–344. doi: 10.1046/j.1365-2958.1997.2401593.x. [DOI] [PubMed] [Google Scholar]
- 20.Konkel M E, Tilly K. Temperature-regulated expression of bacterial virulence genes. Microbes Infect. 2000;2:157–166. doi: 10.1016/s1286-4579(00)00272-0. [DOI] [PubMed] [Google Scholar]
- 21.Letoffe S, Ghigo J M, Wandersman C. Iron acquisition from heme and hemoglobin by a Serratia marcescens extracellular protein. Proc Natl Acad Sci USA. 1994;91:9876–9880. doi: 10.1073/pnas.91.21.9876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Létoffé S, Ghigo J M, Wandersman C. Secretion of the Serratia marcescens HasA protein by an ABC transporter. J Bacteriol. 1994;176:5372–5377. doi: 10.1128/jb.176.17.5372-5377.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Letoffe S, Nato F, Goldberg M E, Wandersman C. Interactions of HasA, a bacterial haemophore, with haemoglobin and with its outer membrane receptor HasR. Mol Microbiol. 1999;33:546–555. doi: 10.1046/j.1365-2958.1999.01499.x. [DOI] [PubMed] [Google Scholar]
- 24.Létoffé S, Omori K, Wandersman C. Functional characterization of the HasAPF hemophore and its truncated and chimeric variants: determination of a region involved in binding to the hemophore receptor. J Bacteriol. 2000;182:4401–4405. doi: 10.1128/jb.182.16.4401-4405.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Letoffe S, Redeker V, Wandersman C. Isolation and characterization of an extracellular haem-binding protein from Pseudomonas aeruginosa that shares function and sequence similarities with the Serratia marcescens HasA haemophore. Mol Microbiol. 1998;28:1223–1234. doi: 10.1046/j.1365-2958.1998.00885.x. [DOI] [PubMed] [Google Scholar]
- 26.Marck C. 'DNA Strider': a 'C' program for the fast analysis of DNA and protein sequences on the Apple Macintosh family of computers. Nucleic Acids Res. 1988;16:1829–1836. doi: 10.1093/nar/16.5.1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mietzner T A, Morse S A. The role of iron-binding proteins in the survival of pathogenic bacteria. Annu Rev Nutr. 1994;14:471–493. doi: 10.1146/annurev.nu.14.070194.002351. [DOI] [PubMed] [Google Scholar]
- 28.Miller J H. A short course in bacterial genetics: a laboratory manual and handbook for Escherichia coli and related bacteria. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1992. [Google Scholar]
- 29.Ochsner U A, Johnson Z, Vasil M L. Genetics and regulation of two distinct haem-uptake systems, phu and has, in Pseudomonas aeruginosa. Microbiology. 2000;146:185–198. doi: 10.1099/00221287-146-1-185. [DOI] [PubMed] [Google Scholar]
- 29a.Paquelin, A., J. M. Ghigo, S. Bertin, and C. Wandersma. Characterisation of HasB, a Serratia marcescens TonB-like protein specifically involved in the hemophore-dependent heme acquisition system. Mol. Microbiol., in press. [DOI] [PubMed]
- 30.Perry R D, Pendrak M L, Schuetze P. Identification and cloning of a hemin storage locus involved in the pigmentation phenotype of Yersinia pestis. J Bacteriol. 1990;172:5929–5937. doi: 10.1128/jb.172.10.5929-5937.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30a.Perry R D, Bearden S W, Fetherston J D. Iron and heme acquisition storage systems in Yersinia pestis. Recent Res Dev Microbiol. 2001;5:13–27. [Google Scholar]
- 31.Reed L J, Muench H. A simple method of estimating fifty percent endpoints. Am J Hyg. 1938;27:493–497. [Google Scholar]
- 32.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 33.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sebbane F, Devalckenaere A, Foulon J, Carniel E, Simonet M. Silencing and reactivation of urease in Yersinia pestis is determined by one G residue at a specific position in the ureD gene. Infect Immun. 2001;69:170–176. doi: 10.1128/IAI.69.1.170-176.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Spratt B G, Hedge P J, te Heesen S, Edelman A, Broome-Smith J K. Kanamycin-resistant vectors that are analogues of plasmids pUC8, pUC9, pEMBL8 and pEMBL9. Gene. 1986;41:337–342. doi: 10.1016/0378-1119(86)90117-4. [DOI] [PubMed] [Google Scholar]
- 36.Staggs T M, Fetherston J D, Perry R D. Pleiotropic effects of a Yersinia pestis fur mutation. J Bacteriol. 1994;176:7614–7624. doi: 10.1128/jb.176.24.7614-7624.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Staggs T M, Perry R D. Identification and cloning of a fur regulatory gene in Yersinia pestis. J Bacteriol. 1991;173:417–425. doi: 10.1128/jb.173.2.417-425.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stojiljkovic I, Baumler A J, Hantke K. Fur regulon in gram-negative bacteria. Identification and characterization of new iron-regulated Escherichia coli genes by a fur titration assay. J Mol Biol. 1994;236:531–545. doi: 10.1006/jmbi.1994.1163. [DOI] [PubMed] [Google Scholar]
- 39.Stojiljkovic I, Hantke K. Hemin uptake system of Yersinia enterocolitica: similarities with other TonB-dependent systems in gram-negative bacteria. EMBO J. 1992;11:4359–4367. doi: 10.1002/j.1460-2075.1992.tb05535.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stoker N G, Fairweather N F, Spratt B G. Versatile low-copy-number plasmid vectors for cloning in Escherichia coli. Gene. 1982;18:335–341. doi: 10.1016/0378-1119(82)90172-x. [DOI] [PubMed] [Google Scholar]
- 41.Thompson J M, Jones H A, Perry R D. Molecular characterization of the hemin uptake locus (hmu) from Yersinia pestis and analysis of hmu mutants for hemin and hemoprotein utilization. Infect Immun. 1999;67:3879–3892. doi: 10.1128/iai.67.8.3879-3892.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wandersman C, Stojiljkovic I. Bacterial heme sources: the role of heme, hemoprotein receptors and hemophores. Curr Opin Microbiol. 2000;3:215–220. doi: 10.1016/s1369-5274(00)00078-3. [DOI] [PubMed] [Google Scholar]