Abstract
Pathogenic variants in KMT5B, a lysine methyltransferase, are associated with global developmental delay, macrocephaly, autism, and congenital anomalies (OMIM# 617788). Given the relatively recent discovery of this disorder, it has not been fully characterized. Deep phenotyping of the largest (n = 43) patient cohort to date identified that hypotonia and congenital heart defects are prominent features that were previously not associated with this syndrome. Both missense variants and putative loss-of-function variants resulted in slow growth in patient-derived cell lines. KMT5B homozygous knockout mice were smaller in size than their wild-type littermates but did not have significantly smaller brains, suggesting relative macrocephaly, also noted as a prominent clinical feature. RNA sequencing of patient lymphoblasts and Kmt5b haploinsufficient mouse brains identified differentially expressed pathways associated with nervous system development and function including axon guidance signaling. Overall, we identified additional pathogenic variants and clinical features in KMT5B-related neurodevelopmental disorder and provide insights into the molecular mechanisms of the disorder using multiple model systems.
Loss of KMT5B results in decreased cellular proliferation and differential regulation of genes important for neurodevelopment.
INTRODUCTION
Histone methylases and demethylases are increasingly being found to play a role in neurodevelopment and neurodevelopmental disorders (1–4). These proteins add and remove methyl groups to histones H3 and H4 to alter transcription (5–7). Histone methylation can cause either transcriptional activation or suppression depending on which residue is methylated (4). Disruptive variants in lysine methyl transferases (KMTs) and lysine demethylases (KDMs) lead to various neurodevelopmental syndromes that share similar phenotypes including developmental delay (DD), intellectual disability (ID), and craniofacial anomalies (4, 8–11). Computational analysis (12) and large-scale sequencing studies have highlighted the importance of the lysine methyltransferase gene, KMT5B, in the pathogenesis of neurodevelopmental disorders (11, 13–16). KMT5B (OMIM: 610881), also denominated suppressor of variegation 4-20 (SUV420H1), is located on chromosome 11q13.2 and contains 13 exons (17). The encoded protein has an N domain (pre-SET domain), a catalytic SET domain, and a C domain with a Zn binding site (post-SET domain) (18). Few disruptive variants in the general population have been reported in the catalytic domain, suggesting that this region is critical for proper protein function. KMT5 family members methylate histone H4 at lysine-20 (K20), which can occur in three different states mono- (H4K20me1), di- (H4K20me2), or trimethylated (H4K20me3). Increasing methylation causes increased chromatin compaction and repression of transcription (19). Evidence suggests that different methylases are responsible for each methylation state of H4K20 (KMT5A for H4K20me1, KMT5B for H4K20me2, and KMT5C for H4K20me3) (18). An additional role for KMT5B-H4K20me2 has been proposed in the repair of DNA double-stranded breaks, through the nonhomologous end joining mechanism (18, 20).
Pathogenic variants in KMT5B cause neurodevelopmental delay with multisystemic involvement (OMIM# 617788) (11, 13, 14, 16, 18, 21). Clinical features reported include ID, autism spectrum disorder, DD, macrocephaly, febrile seizures, hypotonia, attention deficits, failure to thrive, sleep problems, tall stature, gastrointestinal issues, and characteristic facial features (11, 14, 16, 18, 21). Similar to other KMTs and KDMs, haploinsufficiency has been proposed as the most likely pathogenic mechanism for KMT5B (11); although, functional studies remain limited. The Allen BrainSpan Atlas (22) shows that peak human brain KMT5B mRNA expression occurs before 20 weeks after conception, falling after birth to a steady state (18). However, a detailed time course of KMT5B expression in model systems is lacking. Loss of KMT5B results in altered development of both the brain and cardiopulmonary system (23, 24). Evidence of KMT5B’s role in neurogenesis is available for many models [zebrafish (25, 26), amphibian (27), rodent (28), and primate (29) model systems]. These studies suggest that proper H4K20 methylation is necessary for typical neurodevelopment (18, 22), although further research is necessary to elucidate the pathways affected by pathogenic KMT5B variants in humans.
We combined deep clinical phenotyping data, in silico modeling and RNA sequencing (RNA-seq) data from the largest patient cohort to date with in vitro and in vivo studies using both mouse and zebrafish to identify KMT5B-linked pathways governing brain development (Fig. 1). We report a comprehensive clinical and functional characterization of KMT5B-related neurodevelopmental disorder including the clinical features of 43 patients. Expression data from the zebrafish and the mouse show that Kmt5b is highly and ubiquitously expressed in the developing central nervous system (CNS). In addition, RNA-seq from patient-derived lymphoblasts shows differential regulation of genes important for neurodevelopment from both patients with putative loss-of-function (pLOF) variants and with de novo missense variants. Many of these genes and pathways are shared with embryonic Kmt5b haploinsufficient mouse brains. Predicted changes in cell proliferation are further supported by primary patient fibroblast studies and by mice exhibiting decreased growth.
Fig. 1. Visual summary of experiments.
Here, we evaluate a cohort of 43 individuals with pathogenic variants in KMT5B. We perform experiments in patient-derived fibroblasts and lymphoblasts, zebrafish, and a knockout mouse model, as well as in silico analysis of the missense variants.
RESULTS
Genetic variation in KMT5B among humans
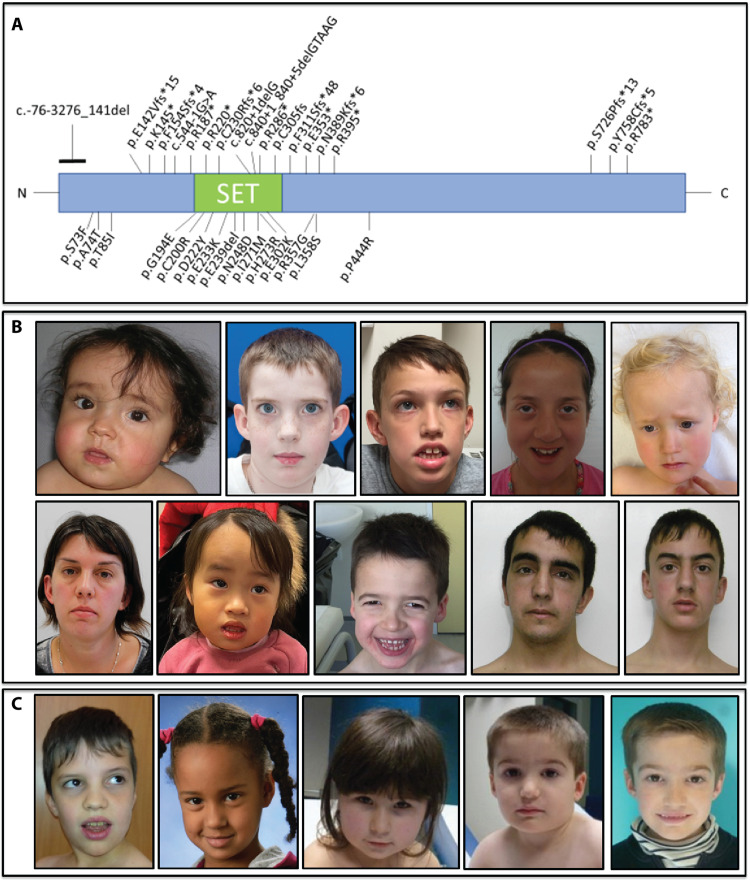
In total, we collected genetic and detailed phenotypic information from 43 individuals with KMT5B variants (Figs. 2, A to C, and 3, A and B, and table S1). All variants identified were heterozygous and, of the cases where inheritance could be tested (n = 33), 90% were confirmed as de novo. Two variants were maternally inherited [p.(Asp222Tyr) and p.(Arg395*)], and one variant was paternally inherited [p.(Pro444Arg)]. For both missense variants, the transmitting parent is also an affected patient in this cohort (patients 18 and 19 and 23 and 24; parent/child pairs). The same variant [p.(Arg220*)] was identified in two nontwin brothers (patients 37 and 38) that was not detected in either parent’s blood, suggesting germline mosaicism in one of the parents. All other patients in the study were unrelated. In our cohort, nine patients had frameshift variants, two had partial deletions, one had a deletion of a single amino acid, 11 had nonsense variants, three had splice site variants, and 16 had missense variants (Fig. 2A). Most of the missense variants were in the SET domain. Human population-level genetic data [gnomAD (Genome Aggregation Database)] show that pLOF variation in KMT5B is highly constrained (i.e., extremely rare; LOF pLI = 1.00) but that some level of missense variation may be tolerated (Z = 2.79). This is likely domain specific with less variation tolerated within the SET domain. The missense variants in our study were in regions that are highly conserved (Fig. 4A). One of the frameshift variants and two of the missense variants have been reported in gnomAD [p.(Tyr758Cysfs*5), p.(Ala74Thr) and p.(Pro444Arg)]. The p.(Tyr758Cysfs*5) and p.(Ala74Thr) variants have each been observed once in the “neuropsych” cohort in gnomAD raising the possibility of missed diagnoses. The p.(Pro444Arg) variant was seen in gnomAD twice.
Fig. 2. Location of KMT5B variants and pictures of affected patients.
In (A), we show the location of all variants aligned to NM_017635.5 in this patient cohort. Loss-of-function (pLOF) variants are shown on top, and missense variants are shown below. Photos of patients with (B) pLOF variants and (C) missense variants are shown. Some common features are long face, arched eyebrows, wide-spaced eyes with upslanting palpebral fissures, prominent ears, and mild prognathism. The final two photos are of the same patient at two different ages.
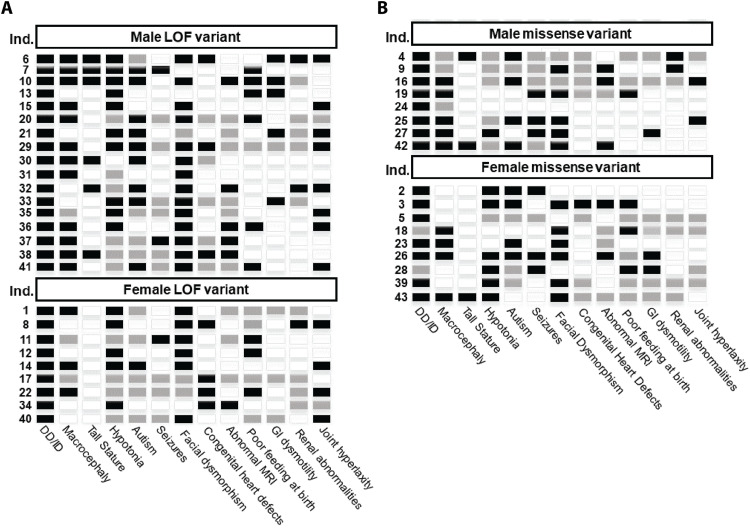
Fig. 3. KMT5B patient phenotype summary.
Primary phenotypes are shown for each patient (row) in the study. Individuals with pLOF variants are in (A), and individuals with missense variants are in (B). Black indicates that the feature is present; white indicates that the feature is absent, and gray indicates that the status of the feature is unknown or not applicable. All individuals for which information was available had DD or ID. Sixty-three percent (24 of 38) had macrocephaly, but only 21% (9 of 43) had tall stature. Sixty-two percent (18 of 29) were diagnosed with autism. Eighty-one percent (22 of 27) had hypotonia. Thirty-one percent (9 of 29) had seizures. Facial dysmorphism is one of the most common features in the phenotype, affecting 79% (31 of 39) of individuals in the cohort. Twenty-seven percent (8 of 30) had congenital heart defects including atrial or ventricular septal defects. A full clinical summary can be found for each patient in table S1.
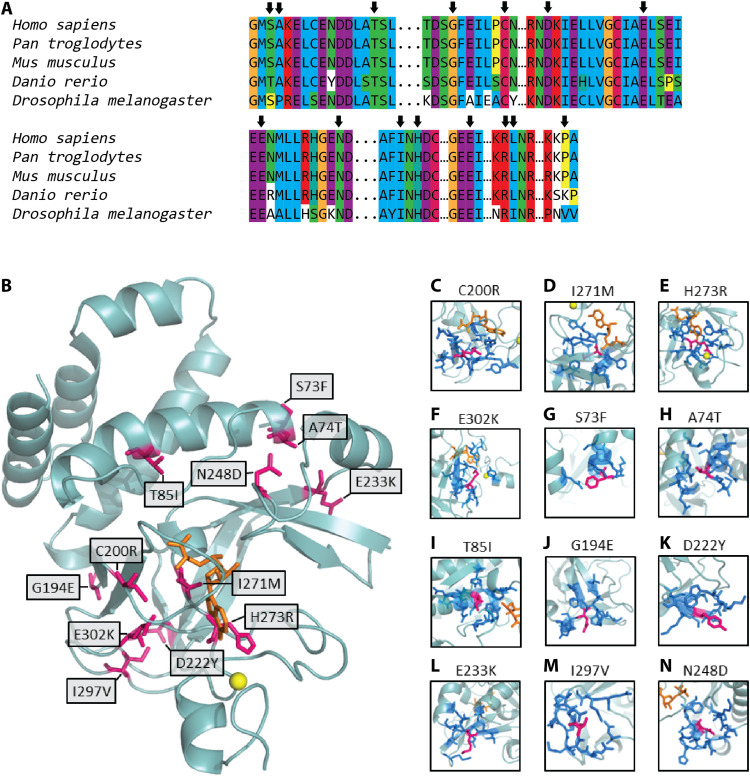
Fig. 4. Structural analyses of KMT5B variants.
In (A), we evaluated the evolutionary conservation of the residues altered by the missense variants in our cohort. Arrows indicate the locations of the missense variants. In (B), we show the location of the missense variants (pink) in the structure of the SET domain of KMT5B (light blue ribbon). The cofactor SAM is shown in orange, and the zinc atom is shown as a yellow sphere. (C to N) Local environment of each variant residue (pink). Neighbors (dark blue) are defined as those residues with at least one interatomic contact (atom-atom distance, <5 Å) with the variant residue. A homology model was built and used for each variant.
We performed in silico modeling using mCSM (mutation Cutoff Scanning Matrix) (30) and RING (Residue Interaction Network Generator) (31) to evaluate the functional impact of our 14 unique missense variants. Twelve variants mapped to the known three-dimensional (3D) structure of the SET domain of KMT5B (NP_060105.3) (32). Three variants (P444R, R357G, and L358S) could not be modeled because of a lack of adequate local 3D structure data. Visual inspection of the locations of these variants (Fig. 4B), combined with contact analyses (table S2), suggested varying functional/structural impacts. Four variants (C200R, I271M, H273R, and E302K) likely affect the binding of KMT5B to its S-adenosyl methionine (SAM) cosubstrate (Fig. 4, C to F), with which these residues share 37, 64, 161, and 3 interatomic contacts, respectively. Furthermore, zinc binding may also be affected by the H273R variant (table S2). A second group of variants (S73F, A74T, T85I, G194E, D222Y, E233K, and I297V) are located at exposed locations (Fig. 4, G to M), making few atomic contacts with other residues. In accordance with this, stability computations for these variants (table S2) indicated an unimportant destabilizing effect (|ΔΔG| < 2 in all cases). However, in a multiple sequence alignment of KMT5B orthologs (Fig. 4A), these variant amino acids appear highly conserved, particularly relative to positions outside the SET domain and its vicinity. Together with solvent accessibility calculations, it is likely that this second group of variants affect functional protein interactions. The same is also likely for N248D, which has a substantial number of interatomic contacts with its neighbors but is positioned on an external loop (Fig. 4N).
Clinical characterization of KMT5B patients
Our cohort included 26 patients with pLOF variants and phenotypic data. Representative images of some of these individuals are shown in Fig. 2B. All have been diagnosed with DD or ID, depending on their age (Fig. 3A). Autism was found in 10 of 17 (59%) of patients and seizures in 3 of 17 (18%). Facial dysmorphisms were observed in 20 of 23 patients (87%). Six of 26 patients had tall stature, and 15 of 23 had macrocephaly (23 and 65%, respectively). Hypotonia and joint hyperlaxity were observed in 15 of 16 (94%) and 12 of 24 (50%) of patients, respectively. Congenital heart defects were identified in 7 of 21 patients (33%), including atrial septal defects and patent ductus arteriosus (table S1). Brain magnetic resonance imaging (MRI) was performed on 17 of 26 of the patients with pLOF variants. Of these, six (35%) had abnormal findings including two with Chiari malformations (table S1). Previous studies did not evaluate congenital heart defects and rarely evaluated hypotonia and joint hyperlaxity. Almost all previously reported individuals had a diagnosis of autism, while only a little over half of the individuals in our cohort had autism. This characterization both expands the physical phenotype and establishes that autism is not a defining feature of the phenotype.
Seventeen patients in our cohort had a missense variant or deletion of a single amino acid. The patients with missense variants had very similar phenotypes to those with pLOF variants (Figs. 2C and 3B), although some features were milder. All patients with missense variants had DD or ID. Autism was reported in 8 of 13 (62%) of patients, and seizures were reported in 6 of 12 (50%). Only 3 of 17 patients had tall stature (18%), and 3 of 17 had short stature but not microcephaly (18%). Macrocephaly was observed in 9 of 14 of the patients with missense variants (64%). Eleven of 15 patients have some degree of unique facial characteristics (73%). Hypotonia and joint hyperlaxity were observed in 7 of 11 (64%) and 2 of 12 (17%) patients, respectively. One individual (1 of 10; 10%) had a congenital heart defect. MRI was performed on 13 of 17 of the patients with missense variants. Of these, five (38%) had abnormal findings including two with Chiari malformations (table S1).
To evaluate potential sex biases in phenotype, we considered pLOF and missense patients together (25 males and 17 females). Eighty-one percent of males had macrocephaly compared to only 44% of the females (P = 0.0357; Fisher’s exact test). Only one female had tall stature compared to 32% of the males in the cohort; however, this trend was not statistically significant (P = 0.0571). No other phenotypic feature was significantly different between the sexes (table S1).
KMT5B expression across the developing CNS
In humans, the highest levels of brain KMT5B mRNA are expressed in utero, dropping sharply by birth (18, 22). Recent studies have shown that Kmt5b haploinsufficiency in mice mimics several aspects of the KMT5B patient population (21, 33). To better understand where and when KMT5B is expressed in the developing CNS, we used zebrafish and mouse models. Human KMT5B has a single ortholog in each of these two model systems with 52% (fig. S1) and 87% (fig. S2) amino acid identity, respectively. Whole-mount in situ hybridization for zfKmt5b mRNA expression in wild-type (WT) fish identified broad expression across the embryo at the 256-cell stage (fig. S3, A and B) that became progressively restricted to the CNS at the 18-somite stage (fig. S3, C and D) through 24 hours after fertilization (fig. S3, E to G). ZfKmt5b expression became further restricted to the head at 5 days after fertilization (fig. S3H).
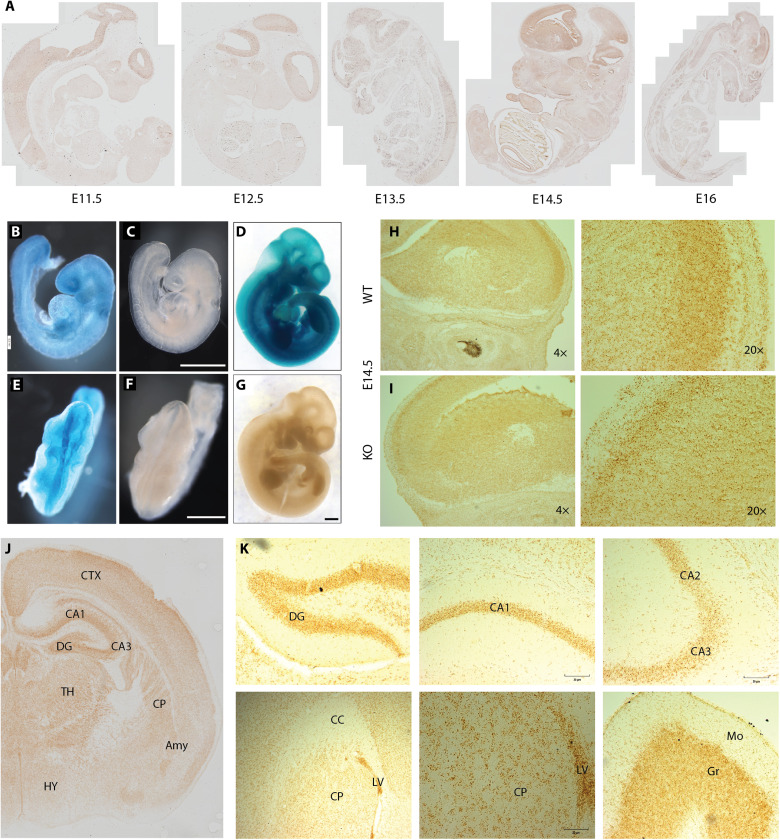
WT Kmt5b mRNA expression was also assessed using a mouse Kmt5b gene trap model (fig. S4A) from the Knockout Mouse Project (34). Using RNAscope in situ hybridization (fig. S4, B and C), we identified constitutive Kmt5b expression in all organs and tissues across embryonic day 11.5 (E11.5) 11.5 to E16 (e.g., brain, lung, liver, heart, skin, and bone; table S3) with specific enrichments in the developing CNS (Fig. 5A). Over time, CNS enrichment for Kmt5b expression focused to the brain with particularly high expression at E14.5. Embryos carrying one copy of the LacZ gene trapping cassette (Kmt5btm1a(KOMP)Wtsi/+; HET) were used to further validate KMT5B protein expression using the β-galactosidase surrogate. Basal β-galactosidase staining was identified across all tissues by E8.75 (Fig. 5, B and E) with elevated levels in the developing head, specifically the eye, first branchial arch, and the unfused neural plates (Fig. 5, C and F). The constitutive expression of β-galactosidase continued throughout E10.5 (Fig. 5, D and G) and was consistent with identified mRNA expression patterns. Closer observation of the E14.5 brain in WT (Fig. 5H) and homozygous gene trap [Kmt5b knockout (KO)] animals revealed a distinct decrease in Kmt5b expression in KO (Fig. 5I) animals in the dorsal pallium/isocortex yet no change in constitutive Kmt5b expression in other surrounding regions, suggesting reduced formation of this region in the KO brain (Fig. 5, H and I). While ubiquitous low-level Kmt5b expression in the brain continued through postnatal days 1 (P1) 1 to P2 and P10 and adult stages (P56; table S4), we noted clear enrichments in the dentate gyrus, hippocampus, and cortex at P1 to P2 (Fig. 5J). In adulthood, Kmt5b expression became more restricted to the dentate gyrus, hippocampus, and the lining of the ventricles (Fig. 5I), all known locations of adult neurogenesis. We also noted high Kmt5b expression in the granular layer of the adult cerebellum (Fig. 5I), which contains a high density of small granule cells. The expression patterns of zfKmt5b in the zebrafish and Kmt5b and β-galactosidase in the mouse supported a role for KMT5B protein function in the CNS during embryonic development.
Fig. 5. Expression of Kmt5b over mouse development.
(A) RNAscope in situ hybridization using a Kmt5b probe is shown in representative samples over an embryonic developmental time course (E11.5 to E16). (B to G) Whole-mount β-galactosidase staining of HET (B to D) or WT (E to G) embryos at E8.75 (B, C, E, and F) and E10.5 (D and G). Scale bars, 0.8 mm. Matched Kmt5b RNAscope images are shown in E14.5 (H) WT and (I) KO brains at 4× and 20× resolution. Representative regions from RNAscope on postnatal brains collected at (J) P1 to P2 and (K) P56 are shown. Qualitative Kmt5b expression is compared in tables S3 and S4. DG, dentate gyrus; CA, hippocampal fields 1, 2, and 3; CC, corpus callosum; LV, lateral ventricle; CP, caudoputamen; Mo, molecular layer; Gr, granular layer; CTX, cortex; TH, thalamus; HY, hypothalamus; Amy, amygdala.
RNA-seq of KMT5B haploinsufficiency in human and mouse
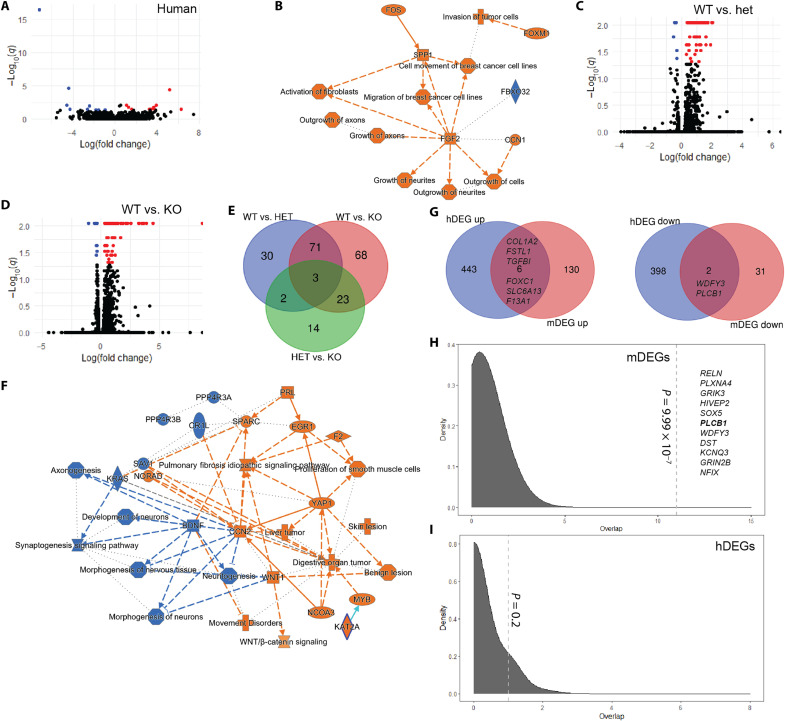
Given the chromatin modifying functions of KMT5 enzymes, we subjected our patient-derived lymphoblast cell lines and mouse resources to further gene expression experiments to identify genes and pathways whose expression patterns are regulated by KMT5B. We subjected transformed lymphoblast cell lines (35) from seven patients in the study to RNA-seq in tandem with seven age- and sex-matched controls. KMT5B is expressed in human Epstein-Barr virus–transformed lymphocytes according to GTEx with expression levels similar to most adult brain regions. Quality control metrics for this dataset are shown in fig. S5. Human differentially expressed genes (hDEGs) were defined as those that met a raw P value of P < 0.05. By this metric, 654 genes were up-regulated and 648 genes were down-regulated in cases compared to the controls. Twenty-one genes survived false discovery rate (FDR) correction (q < 0.05; Fig. 6A). See table S5 for a full list of hDEGs. Ingenuity Pathway Analysis (IPA) of all hDEGs with P < 0.05 provided an unbiased evaluation of pathway enrichment and revealed significant enrichments for axon guidance signaling (P = 1.85 × 10−9), glycoprotein 6 signaling (P = 9.63 × 10−9), and CXCR4 signaling (P = 1.61 × 10−6) pathways (Table 1). While the top disease pathway enrichment in the hDEG dataset was cancer (P = 2.06 × 10−4 to 6.78 × 10−21), there was also a strong signal for nervous system development and function pathways (P = 2.13 × 10−4 to 2.46 × 10−9; table S6), predicting significantly increased growth of neurites (P = 4.95 × 10−6), outgrowth of axons (P = 1.15 × 10−5), outgrowth of neurites (P = 1.43 × 10−5), and growth of axons (P = 2.10 × 10−4). Significant predicted upstream regulators included Paired box gene 3 (PAX3; P = 1.82 × 10−8), Proline-, glutamic acid-, and leucine-rich protein 1 (PELP1; P = 3.76 × 10−7), β-estradiol (P = 5.48 × 10−7), and transforming growth factor–β receptor 2 (TGFBR2; P = 1.37 × 10−6; table S7). A summary of all IPA findings highlighted brain-relevant phenotypes focused on changes in fibroblast growth factor 2 (FGF2) signaling (Fig. 6B) and further that cell growth and migration may be affected by KMT5B expression. Notably, several genes linked to the phenotypes in our cohort were differentially regulated in the hDEG dataset including MRAS, GNAQ, WDFY3, PLCB1, and NR2F2.
Fig. 6. RNA-seq results from human and mouse models of KMT5B haploinsufficiency.
(A, C, and D) Volcano plots illustrate the genes that are significantly up- (red) or down-regulated (blue) in the (A) human and (C and D) mouse datasets. HET, heterozygous; KO, homozygous knockout; black dots, nonsignificant genes. A functional enrichment summary is shown for the human dataset (hDEGs, P < 0.05) in (B) and for the KO mouse (mDEGs, q < 0.05) in (F). Orange, activated; blue, inhibited. Venn diagrams show (E) the number of mDEGs that overlap between the genotypes tested and (G) genes that overlapped between the hDEG and mDEG datasets. (F) Simulation testing for gene overlaps between the (H) mDEG and (I) hDEG down-regulated genes and high-risk autism genes from SFARI Gene. Dashed line, observed overlap. Gene in bold was found in both the human and mouse datasets (q < 0.05).
Table 1. Top-ranked canonical pathway enrichments.
| hDEG | mDEG (HET) | mDEG (KO) |
|---|---|---|
| Axonal guidance signaling | Hepatic fibrosis/hepatic stellate cell activation | WNT/β-catenin signaling |
| GP6 signaling pathway | Tumor microenvironment pathway | Pulmonary fibrosis idiopathic signaling pathway |
| Renin-angiotensin signaling | Apelin liver signaling pathway | Synaptogenesis signaling pathway |
| CXCR4 signaling | GP6 signaling pathway | Molecular mechanisms of cancer |
| Sperm motility | Atherosclerosis signaling | Hepatic fibrosis signaling pathway |
| P2Y purinergic receptor signaling pathway | Intrinsic prothrombin activation pathway | Axonal guidance signaling |
| Apelin endothelial signaling pathway | Wound healing signaling pathway | Apelin liver signaling pathway |
| Dopamine-DARPP32 feedback in cAMP signaling | Pulmonary fibrosis idiopathic signaling pathway | Hepatic fibrosis/hepatic stellate cell activation |
| α-Adrenergic signaling | Natural killer cell signaling | Regulation of the epithelial mesenchymal transition in development pathway |
| Synaptic long term depression | Axonal guidance signaling | Pulmonary healing signaling pathway |
| Gap junction signaling | TR/RXR activation | Osteoarthritis pathway |
| GPCR-mediated nutrient sensing in enteroendocrine cells | HOTAIR regulatory pathway | Factors promoting cardiogenesis in vertebrates |
| Natural killer cell signaling | Coronavirus replication pathway | Ovarian cancer signaling |
| Cholecystokinin/gastrin-mediated signaling | LXR/RXR activation | Mouse embryonic stem cell pluripotency |
| Role of NFAT in cardiac hypertrophy | FXR/RXR activation | BEX2 signaling pathway |
| Colorectal cancer metastasis signaling | Osteoarthritis pathway | Iron homeostasis signaling pathway |
| Thrombin signaling | Iron homeostasis signaling pathway | Intrinsic prothrombin activation pathway |
| Molecular mechanisms of cancer | NAD signaling pathway | Tumor microenvironment pathway |
| Endothelin-1 signaling | Remodeling of epithelial adherens junctions | Epithelial adherens junction signaling |
| GNRH signaling | Phagosome maturation | GP6 signaling pathway |
We have previously shown that the embryonic mouse brains of Kmt5b haploinsufficient mice show a robust down-regulation of Kmt5b expression and an increase of H4K20me1 (33). As human brain tissue was not available from our KMT5B patients, we used this mouse model to further specify genes whose expression is regulated by KMT5B in the developing brain. We performed RNA-seq on forebrain homogenates at E14.5 in Kmt5b WT, HET, and KO embryos of both sexes (n = 3 biological replicates per genotype and sex) and compared this to our hDEG data. Initial quality control analyses identified one potential litter effect driven by three samples (one KO male and two WT females from the same litter). These samples were removed, and the data were reanalyzed. Data quality was similar across all remaining samples in the mouse RNA-seq experiment (fig. S6, A and B), indicating that comparisons could be made across genotypes (fig. S6, C and D). A full list of mouse differentially expressed genes (mDEGs) that met statistical significance (raw P < 0.05) can be found in table S6. After FDR correction, we identified 161 significantly up-regulated (q > 0.05) and 35 significantly down-regulated mDEGs in either the HET (Fig. 6C) or KO (Fig. 6D) compared to the WT condition, highlighting a net increase in transcription with Kmt5b down-regulation.
Pairwise comparisons were made between the WT and HET, WT and KO, and HET and KO conditions with the hypothesis that Kmt5b may show a dose effect related to transcriptional regulation such that KO > HET > WT. Genes shared between the HET and KO conditions were considered high-confidence genes under this model. KO embryos are still present that this stage of development but die shortly after birth. Seventy-four mDEGs were shared between the WT versus HET and WT versus KO comparisons (Fig. 6E). Of these mDEGs, 68 were up-regulated and six were down-regulated in the HET/KO compared to the WT condition. However, only two genes (Kmt5b and Pisd-ps1) had a significant dose-dependent relationship, further supporting a haploinsufficient disease model. Functional enrichment analyses by IPA of the WT versus HET and WT versus KO mDEG (q < 0.05) datasets separately showed foremost that the KO state predicts significantly increased organismal death (P = 2.01 × 10−25; z = 3.229) that was not present in the HET state. Both the HET and the KO conditions predict significantly decreased morphogenesis of neurons (P = 7.52 × 10−8 to 5.58 × 10−28; KO > HET) and decreased neuritogenesis (P = 3.01 × 10−7 to 9.61 × 10−27; KO > HET; table S9). Furthermore, microtubule dynamics and organization of the cytoskeleton were both also predicted to be significantly decreased in the HET and KO conditions (table S9). We conclude that the KO condition is already not compatible with life at this stage because of amplified transcriptional responses from genes conserved with the HET condition. This may explain the presence of unique genes in the HET and KO conditions where the KO brain is actively dying and the HET brain is adjusting for survival. Unique to the HET condition, we noted increased cell migration signatures (P = 5.25 × 10−5 to 3.96 × 10−8), increased growth of vessel (P = 4.00 × 10−7), and increased adhesion of connective tissues (P = 6.35 × 10−6; table S9) driven by robust increases in extracellular matrix (ECM) proteins (e.g., collagens). A summary of all IPA findings highlighted inhibition of brain developmental processes and activation of ECM components that were amplified in the KO compared to the HET condition (Fig. 6F and table S9). Shared upstream regulators of the HET and KO conditions included Harvey rat sarcoma viral oncogene homolog (HRAS; 1.4 × 10−13 to 3.69 × 10−13), FGF2 (1.5 × 10−9 to 1.06 × 10−11), estrogen (1.25 × 10−8 to 4.4 × 10−11), and TGFB1 (3.2 × 10−6 to 8.4 × 10−8).
Among mDEGs (HET or KO) with an annotated human ortholog (Ensembl release 107), six genes—COL1A2, FSTL1, TGFB1, FOXC1, SLC6A13, and F13A1—were up-regulated in both the mouse (q < 0.05) and the hDEG (P < 0.05) datasets (Fig. 6G). Similarly, the down-regulation of WDFY3 and PLCB1 were shared between the mDEG and the hDEG datasets (Fig. 6G). Axonal guidance signaling and the Glycoprotein VI Platelet (GP6) signaling pathway were both predicted in the top 20 significant canonical pathways for the hDEG (P < 0.05) and mDEG (q < 0.05; HET and KO) datasets (Table 1). Furthermore, FGF2 and CCR2 were predicted as significant upstream regulators in all three datasets (hDEG and both mDEG sets; tables S7 and S10), highlighting shared brain and ECM effects. Last, down-regulated, but not up-regulated mDEGs (q < 0.05), were enriched for known autism-associated genes by simulation testing (P = 9.99 × 10−7; Fig. 6H), suggesting that down-regulated mDEGs (specifically, Reln, Plxna4, Grik3, Hivep2, Sox5, Plcb1, Wdfy3, Dst, Kcnq3, Grin2b, and Nfix) may contribute to the phenotypes observed. Four of these genes—Grin2b, Plcb1, Reln, and Wdfy3—were validated by quantitative reverse transcription polymerase chain reaction (PCR) testing, confirming down-regulation in the HET versus WT mouse brain (fig. S7). While PLCB1 down-regulation survived FDR correction in the hDEG dataset, this gene alone was not a strong enough signal for significance in this simulation test. Both up-regulated and down-regulated hDEGs (P < 0.05) were significant for autism-associated genes by simulation testing (P = 3.9 × 10−5 to 9.99 × 10−7); however, these datasets are lower confidence and likely contained some false-positive signal.
Growth changes in primary patient cells and mouse models
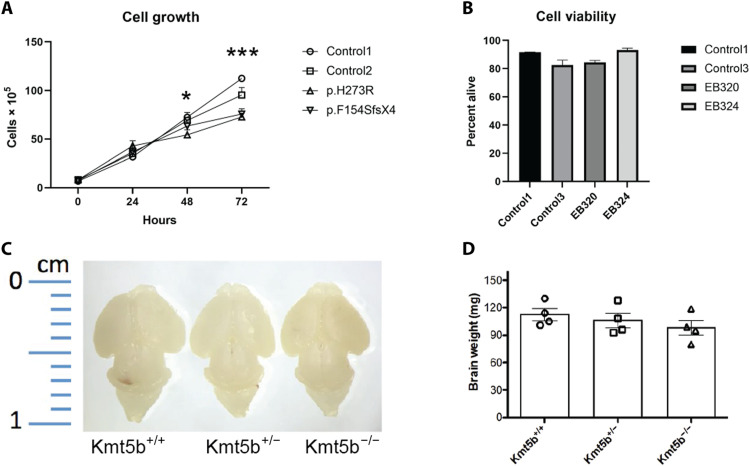
To test the RNA-seq predicted effects of KMT5B haploinsufficiency, we evaluated fibroblasts from two patients in the study with different variants [p.(H273R) and p.(F154SfsX4); patients 2 and 10, respectively]. KMT5B mutant primary cell lines grew significantly slower than two control lines (P < 0.05 at 48 hours and P < 0.005 at 72 hours; Student’s t test; Fig. 7A); this was not due to increased cell death in the mutants (Fig. 7B), supporting the hypothesis that KMT5B expression contributes to regulation of cell division (36, 37) and predicted as cancer enrichments in the hDEG analyses. We have reported persistent lower body weight and shorter body length in the Kmt5b gene trap haploinsufficient mouse model on a C57BL/6N strain background (33). We independently confirmed this growth defect using the gene trap allele on a C57BL/6JInv background (fig. S4D). Homozygous KO pups were significantly smaller than WT littermates at birth [P0; P < 0.001; one-way analysis of variance (ANOVA) with Tukey’s correction]. In both strain backgrounds, KO embryos were overtly smaller at E14. Brain size and weight in P0 mouse pups (C57BL/6JInv) also showed a dose-dependent trend that was not statistically significant (one-way ANOVA) (Fig. 7, C and D), suggesting relative macrocephaly in KO animals at birth. This relative macrocephaly was not likely due to increased brain cell proliferation, as staining for Ki67 (a marker of active proliferation), NeuN (neuron marker), and H3K20me3 in the P0 brains showed no differences (fig. S8). Nissl staining of the corpus callosum in P0 WT, HET, and KO mice suggested increased numbers of pyknotic cells in HET and KO compared to WT P0 brains (fig. S8, A and B), supporting the predicted increased organismal death signature in the mDEG KO dataset.
Fig. 7. Cell growth and death in human fibroblasts and mouse brains.
(A) Cell growth was measured for primary fibroblast lines derived from unaffected individuals and two individuals with heterozygous KMT5B variants and was repeated three times for all cell lines. Cell growth was significantly different between control lines and lines from affected individuals at 48 hours (P < 0.05) and 72 hours (P < 0.001) as determined by Student’s t test. (B) Cell viability for primary fibroblast cell lines was measured using annexin V/propidium iodide staining and evaluated using flow cytometry. There was no significant difference between the lines determined by Student’s t test. (C and D) Brains from P0 mouse pups were weighed. There was no statistically significant difference in the mouse brain weight of the different genotype determined by Student’s t test.
DISCUSSION
Through this study, we have further refined the KMT5B phenotype in humans using the largest cohort reported to date. On the basis of our experiments and the known role of KMT5B, the main conclusion of our paper is that monoallelic pathogenic variants in KMT5B perturb normal epigenetic regulation of neurological development. Similar to previous reports, all participants in this study had ID/DD, and many had unique facial characteristics and autism (fig. S9) (11, 14–16). Furthermore, our large cohort identified a high prevalence of hypotonia and congenital heart defects among this population. We have recently reported early motor reflex, strength, and muscle mass deficits in a germline Kmt5b haploinsufficient mouse model (33, 38). Kmt5b has also been linked to the maintenance of quiescent muscle satellite cell (i.e., muscle stem cell) populations in mice (39), supporting the hypothesis that KMT5B has a role in the development and/or maturation of the skeletal muscle system.
Growth abnormalities (macrocephaly and/or tall stature) were common in our cohort and have been reported in previous KMT5B patient studies (11, 14–16, 40). Eight patients in the study had both macrocephaly and tall stature, whereas 22 patients had macrocephaly without tall stature. Only one patient was reported as tall without macrocephaly. Of the two male siblings carrying the same pLOF variant in this study (patients 37 and 38), both had macrocephaly, but only one was also tall. One important caveat to height is that parental data were not collected; therefore, we cannot compare whether “tall” patients in this study are taller than would be expected on the basis of their unaffected parents. We conclude that macrocephaly, not a systemic overgrowth phenotype, is a core symptom of KMT5B haploinsufficiency. This is supported by multiple lines of evidence. First, macrocephaly appears more often in this cohort independent of tall stature, arguing against a broad overgrowth phenotype that has been previously suggested (11, 16). Primary fibroblasts isolated from KMT5B patients grow significantly slower without increased cell death compared to controls including cells from patient 10 who presented with both macrocephaly and tall stature. Kmt5b haploinsufficient mice are significantly smaller (body weight and length) than their WT littermates (33). We also identified this trend in KO animals at birth in this study. Brain weight is not proportionally decreased in these animals, suggesting relative macrocephaly. We have recently reported relative macrocephaly (i.e., increased brain weights) in male, but not female, Kmt5b HET mice at P17 on a different strain background (C57BL/6N) (38). Macrocephaly disproportionate to height is a key neurophenotype in autism being found in ~15% of males compared to ~6% of typically developing male peers (41). Much lower rates of macrocephaly are found among females with autism. Macrocephaly was more common among males than females in this study, the only phenotype that showed a significant sex bias.
While relative macrocephaly may be a shared feature of KMT5B patients and our mouse model, Kmt5b HET and KO mice have smaller bodies overall, a feature that is not found in KMT5B patients. These differences in body size effect are likely due to differences between mouse and man, perhaps even to systemic hormone effects. Head circumference correlates with brain volume but only in young children (up to the age of 6 years old (42)), and macrocephaly is often assumed to be a sign of increased brain size/volume in autism. However, we found no evidence in this study of increased cell proliferation when KMT5B expression was decreased, suggesting that KMT5B-associated macrocephaly may be due to other causes. Relative macrocephaly can have other causes, including increased cerebrospinal fluid (CSF; i.e., hydrocephalus), dilated blood vessels, thickened bone, expanded extracerebral spaces, and/or intracranial cysts (43). These etiologies cannot be distinguished without neuroimaging. Several reported imaging phenotypes in our patients—enlarged ventricles (which can be a sign of hydrocephalus), syringomyelia, subdural hygroma, and Chiari malformations—could suggest that KMT5B helps regulate CSF dynamics. Increased CSF pressured could also be causing the increased number of pyknotic cells (i.e., cell death) that we have observed in Kmt5b HET and KO mouse brains. We noted a significant up-regulation of choroid plexus markers in our mouse model in HET compared to WT brains of both sexes. The choroid plexus is composed of both epithelial and stromal cell populations and is responsible for generating CSF for the CNS (44). The most significant mDEG contributing to this enrichment was transthyretin (Ttr), which was increased in HETs fourfold among females and fivefold among males (table S6). TTR protein is secreted into the CSF by the choroid plexus and acts as a transporter for thyroid hormone thyroxine (T4). Analysis of circulating T4 levels in adult WT and HET mice of both sexes showed no significant differences (fig. S10), suggesting that increased expression of Ttr is not due to hyperthyroidism. TTR is often used as a diagnostic marker for choroid plexus tissue. In mice, the choroid plexus is established between E12.5 and E13, reaching full size by E16.5 (45). Ttr expression remains high until birth and decreases postnatally (46). Overabundance of choroid plexus tissue has been linked to increased CSF production and hydrocephalus (47).
Kmt5b mRNA is highly expressed in the mouse brain at E14.5, the time point at which we collected additional transcriptomic data. Our mDEG dataset highlights a net effect of gene up-regulation with Kmt5b loss. This is consistent with other published studies (21) and with the increased H4K20me1 (permissive chromatin state) reported among mouse KMT5B HET and KO brains previously (33). Our data did not show an up-regulation of Kmt5c or Nsd2/Mmset/Whsc1 transcripts (table S6), two genes that have been proposed as an alternative H4K20 dimethylating enzymes (48), as a compensatory reaction to Kmt5b loss. We did not identify any overlaps between our mDEG dataset and that of Wang et al. (21) who used adeno-associated virus–short hairpin RNA to knock down Kmt5b expression specifically in the prefrontal cortex of young adult mice. These results suggest that where and when KMT5B is present in the developing brain may matter. A recent study identified a difference in the fraction of migrating cells during peak cortical neurogenesis when ventricular zone Kmt5b was knocked down at E13.5 via in utero electroporation (15), in agreement with our transcriptomic predicted changes in cell migration over this time period.
Collectively, the data described in this study provide many exciting avenues for future research connecting KMT5B to the developing brain. This study is the first to document the in situ expression of Kmt5b transcripts in the mouse brain over developmental time (embryonic stages through adulthood). It is also the first report of transcriptomic data from the embryonic brain in a model of germline Kmt5b loss, a period that is likely critical to the development of humans carrying disruptive KMT5B variants (18). Both our hDEG and mDEG datasets predict increases in TGF-β signaling concomitant with ECM remodeling in the mutant conditions. TGF-β is thought to be neuroprotective and is up-regulated following many types of brain damage (49). Whether increased cell stress due to loss of KMT5B/H4K20me2-mediated DNA repair contributes to the phenotypes observed in this patient population should be further investigated. TGF-β is also known to be a potent stimulator of ECM synthesis, including collagens, which were strongly up-regulated in the mDEG and hDEG datasets. Collagen I (Col1a1/Col1a2), proteoglycan link protein 1 (Hapln1), and lumican (Lum) have been shown to contribute to folding of the neocortex (50). All three of these genes were significantly up-regulated in the mDEG mutant dataset. Given the report of polymicrogyria in patient 34, changes in cortical folding may be an interesting avenue for future research. Last, TGF-β has been shown to regulate axon growth in neurons (51). Axon guidance signaling was significantly enriched in our hDEG and mDEG datasets. Given the additional roles for the ECM in axon growth and guidance (52), human cell and mouse resources should be used to further explore this process in the context of KMT5B function.
MATERIALS AND METHODS
Experimental design
The objective of this study was to determine the effects of heterozygous KMT5B mutations on development. To achieve this, we evaluated the clinical phenotype of 43 individuals with pathogenic or likely pathogenic variants in KMT5B. We performed in silico analysis on the missense variants to gain a better understanding of how they would affect protein function. We evaluated cell growth and viability from patient-derived cell lines and performed RNA-seq to determine changes in the transcriptome. In addition to the patient cell lines, we evaluated Kmt5b mRNA expression in both mice and zebrafish. We determined the timing and location of kmt5b and Kmt5b in zebrafish and mice using in situ hybridization and β-galactosidase staining. We also evaluated the effect of heterozygous and homozygous Kmt5b KO in the mice by evaluating brain size and transcriptomic changes.
Clinical cohort
Clinical exome sequencing was performed at GeneDx for the initial patient and others (patients 3, 4, 7, 11, 13, 27, and 29) or using the Autism/ID Xpanded panel, as previously published (53). GeneMatcher (54) or word of mouth was used to recruit other participants. Participants were consented into a Children’s Hospital of Philadelphia Institutional Review Board (IRB)–approved protocol or consented locally for participation (IRB no: 16-013278 approved on 17 April 2019). All KMT5B variants were aligned to NM_017635.5 using Mutalyzer (55).
In silico analyses
For multiple sequence alignment, orthologs were extracted from the UniProt database using Blast (56) with the human KMT5B (NP_060105.3) as the reference amino acid sequence. The homologous sequences retrieved were aligned using COBALT (57) with default parameters.
For each missense variant, the protein stability changes upon mutation (ΔΔG; negative values indicate destabilization) using mCSM (30), and the number of interatomic contacts of the native residue using RING (31) was calculated. For both computations, the 3D structure of the SET domain of KMT5B in complex with S-adenosyl-l-methionine [Protein Data Bank code: 3S8P (58)] was used. The structure representations were created with PyMOL (59). Structural models for the variants were obtained using MODELLER (60).
Animal models, husbandry, and embryo collection
Zebrafish (AB strain) were bred and maintained in a recirculating aquaculture system at Agency for Science, Technology and Research (A*STAR) Skin Research Labs, Singapore. All animal studies were performed in accordance with approved A*STAR Institutional Animal Care and Use Committee (IACUC) protocols #161172 and #171238. Zebrafish embryos were produced by natural mating, cultured at 28.5°C, and staged by hours after fertilization (61).
All mouse experiments were performed in animals carrying the Kmt5btm1a(KOMP)Wtsi allele [KOMP (62); fig. S4A] at two independent laboratories. Kmt5b WT, heterozygous (HET), and homozygous null (KO) progeny had genotypes of Kmt5bwt/wt, Kmt5bwt/tm1a, and Kmt5btm1a/tm1a, respectively (62). Animals at A*STAR were derived from sperm purchased from KOMP (University of California, Davis) and established on a C57BL/6JInv background through 10 generations of backcrossing. These mice were housed and bred at the Biological Resource Centre, A*STAR. Animals at Creighton University were cryorecovered at KOMP (University of California, Davis, MGI:2444557, Targeting Project: CSD28648) on a C57BL/6N background. Colony offspring were genotyped using tail biopsies with specific probes designed for Kmt5b and LacZ sequences (Transnetyx, Cordova, TN). All mouse work was approved and monitored by the Creighton University IACUC under protocol numbers 1039, 1040, and 1118. Mice were housed in a temperature-controlled vivarium maintained on a 12-hour light/12-hour dark cycle with food and water provided ad libitum. Mice were weaned at 3 to 4 weeks of age and housed in unisex groups of two to five mice in individually ventilated cages.
For timed embryo collections, stud males >8 weeks of age were housed individually. Group-housed females >8 to 12 weeks were introduced, one to two at a time, into a male cage to initiate mating. Vaginal plugs were assessed the following morning; the presence of a plug was noted as embryonic day E0.5.
In situ hybridization and β-galactosidase staining
Whole-mount in situ hybridization in zebrafish was performed using probe templates targeting zfKmt5b amplified from cDNA using primers: F-“GGATCCAATGCAGAGGCAGA” and R-“CTGCAGCCCGAACCGCGTGC.” Amplicons were subcloned into pCR-BluntII-TOPO (Life Technologies, Carlsbad, CA). The pCR-BluntII-TOPO-zfKmt5bprobe plasmids were linearized with Bam HI or Pst I. Antisense zfKmt5b probes were made by in vitro transcription using T7 polymerase. Sense probes were made by in vitro transcription using SP6 polymerase. In situ hybridizations were performed as described (25).
The expression of mouse Kmt5b transcripts across typical developmental time was assessed using RNAscope [Advanced Cell Diagnostics (ACD), Newark, CA, USA] on whole-mount embryos at E11.5, E12.5, E13.5, E14.5, and E15.5 to E16 (combined). Whole brains were also collected at P1 to P2 (combined), P10, and P56. All progeny used for RNAscope were from WT × WT or HET x HET mating. Harvested embryos (E11.5 to E16) were immersion-fixed in 4% paraformaldehyde, dehydrated through an alcohol series, embedded in paraffin, and sectioned at 10-μm thickness using a Leica Reichert-Jung 2030 microtome (Leica Microsystems, Buffalo Grove, IL, USA). For the postnatal time points (P1 to P2, P10, and P56), mice were transcardially perfused with 1× phosphate-buffered saline (PBS)/4% paraformaldehyde (PFA); brains were dissected out and further immersion-fixed in 4% PFA. Brains were cryoprotected in 30% sucrose (Thermo Fisher Scientific, Waltham, MA, USA), embedded in optimal cutting temperature (OCT) medium (VWR, Radnor, PA, USA) and cryosectioned at 30 μm using a cryostat (Leica Microsystems).
RNAscope was performed on paraffin-embedded embryos according to the manufacturer’s protocol using the RNAscope 2.5 HD reagent detection kit (ACD). Frozen cryosections were processed according to the “Sample preparation technical note for fixed frozen tissue using RNAscope 2.5 chromogenic assay” protocol obtained from the manufacturer (ACD), followed by RNAscope 2.5 HD detection. A negative control probe (#310043) and a Kmt5b-specific probe (#489621; ACD) were used for detection. The specificity of the Kmt5b probe was confirmed using consecutive sections stained with either the Kmt5b or negative control probe (fig. S4, B and C). Because of high cell density and broad Kmt5b expression across most brain regions, counterstaining was not possible on RNAscope slides. Cell density and brain regions were determined by performing hematoxylin and eosin (H&E) staining on adjacent tissue sections (fig. S4B). Briefly, H&E staining was performed (Epredia Gemini AS Automated Slide Stainer, Thermo Fisher Scientific) using standard techniques of rehydration, staining with Hematoxylin 7212 (Thermo Fisher Scientific), clarification in acetic acid, bluing, and staining in Eosin Y (Thermo Fisher Scientific), followed by dehydration. Slides were imaged on a VS120 Virtual Slide Scanner (Olympus, Tokyo, Japan) at ×20 magnification.
Whole-mount β-galactosidase staining was performed on WT and HET mouse embryos from the same litters at E8.75 and E10.5 as described in a protocol provided by the Jackson Laboratory (www.jax.org/research-and-faculty/resources/cre-repository/whole-mount-staining-protocol).
RNA-seq and gene set enrichments
RNA-seq was performed on patient-derived lymphoblasts. The lymphoblast cell lines were created by the Center for Applied Genomics and the Children’s Hospital of Philadelphia. B cells obtained from a peripheral blood draw were transformed using Epstein-Barr virus. The lymphoblast cell lines were created from three boys (3, 6, and 11 years old) and four girls (3, 5, 5, and 12 years old). Age- and sex-matched controls were identified for each cell line.
For RNA-seq of human-derived lymphoblast cell lines, total RNA was extracted by homogenizing lymphoblasts using the Maxwell kit and was performed on Promega’s robotics platform according to the manufacturer’s instructions. The method used paramagnetic particles, which provide a mobile solid phase to optimize sample capture, washing, and purification of nucleic acid. RNA purity was measured by determination of the ratio for absorbance at 260 nm versus absorbance at 280 nm (A260/A280) using a NanoDrop 8000 (Thermo Fisher Scientific). RNA integrity was also evaluated with the Agilent Tape Station by determining RNA integrity number (RIN) values using gel electrophoresis.
Preparation of samples for stranded total RNA-seq on the Illumina platform was performed following the manufacturer’s instructions. The total RNA was subjected to the first step involving the removal of ribosomal RNA (rRNA) using biotinylated, target-specific oligos combined with Ribo-Zero rRNA removal beads. The Ribo-Zero Human/Mouse/Rat kit depletes samples of cytoplasmic rRNA, and the Ribo-Zero Gold kit depletes samples of both cytoplasmic and mitochondrial rRNA. Following purification, the RNA was fragmented into small pieces using divalent cations under elevated temperature. The cleaved RNA fragments were copied into first-strand cDNA using reverse transcriptase and random primers, followed by second-strand cDNA synthesis using DNA polymerase I and ribonuclease H. These cDNA fragments then have the addition of a single “A” base and subsequent ligation of the adapter. The products were purified and enriched with PCR to create the final cDNA library. rRNA-depleted strand-specific RNA libraries were generated with the TruSeq Total Ribo-Zero rRNA Removal (Illumina, San Diego, CA, USA). Each QCed library was sequenced on an Illumina Novaseq6000 (Illumina) using V1.5 chemistry, in paired-end mode with a read length of 2 × 100 base pairs, generating approximately 40 million reads per sample.
The Illumina DRAGEN Bio-IT Platform (v3.6.3) was used to generate FASTQ files for each human sample by first demultiplexing the raw sequencing data. DRAGEN was then used to align these files to the Homo sapiens (GRCh37.75) reference genome from the Broad Institute with a Smith-Waterman alignment scoring algorithm (63). Gene expression quantification was also calculated on this platform with use of the corresponding gene transfer format (GTF) file. Bioconductor Rsubread package (v4.1.0) was used to generate gene counts and mapping metrics against the GTF file. Differential expression was performed using the Bioconductor DESeq2 package (v1.28.1) (64). Comparison groups of interest during this step were described as all samples, sex-specific, and age-specific. Results were filtered by a log fold change > 1.5 or < 1.5 and P < 0.05 before performing pathway analysis.
For RNA-seq of mouse, E14.5 embryos were collected from HET × HET crosses, and brains were dissected out in cold 1× PBS and flash-frozen in liquid nitrogen. Tail samples from each embryo were collected for genotyping and sex determination (Transnetyx). RNA was extracted using the RNeasy Mini Kit including deoxyribonuclease I treatment (QIAGEN, Hilden, Germany) to eliminate genomic DNA. Samples were quantified using a Qubit 3.0 Fluorometer and RNA BR Assay Kit (Invitrogen, Waltham, MA, USA). RNA library preparation and sequencing were performed at BGI (Cambridge, MA, USA) using a paired-end 2 × 150 cycle protocol. Three biological replicates were run for each sex (n = 2) and genotype (n = 3). All samples had a RIN of 10 before sequencing. Sample data were analyzed at Creighton University using the Tuxedo tools suite (65). Briefly, samples were mapped to the mm10 mouse reference genome (https://support.illumina.com/sequencing/sequencing_software/igenome.html; downloaded 7 September 2018) using TopHat v2.1.0 (Bowtie2 v2.2.6), followed by transcript assembly using cufflinks v2.2.1. Transcript differences were assessed using cuffdiff on merged transcriptome replicates by genotype. Data visualization and mining were performed using cummeRbund for R (v3.5). Gene orthologs (human and mouse) were identified using the Ensembl release 106 Biomart tool.
Gene set enrichment analyses were performed using QIAGEN Ingenuity Pathway Analysis (IPA; v01-20-04) where noted. We also compared our up- and down-regulated mDEG and hDEG gene lists to annotated autism risk genes from the SFARI Gene Database (downloaded 31 March 2021; n = 1003) that were performed using R v 4.1.1. For comparison, 106 random sets of genes of the same relative sizes of the test and SFARI dataset were sampled. Density plots were used to show the number of genes from each of the simulated random sets that are coexpressed. These data were used to calculate the probability of the overlaps that we identified as being random. The following categories were included: SFARI gene category “syndromic genes,” SFARI gene category 1 “high confidence gene,” and SFARI gene category 2 “strong candidate gene.” A full description of SFARI categories can be found at https://gene.sfari.org/about-gene-scoring/.
Cell proliferation and viability assays
Two patient-derived fibroblast lines and two control fibroblast lines were evaluated for cellular proliferation. The cell lines were established using skin biopsies from a 10-year-old female and a 7-year-old male. The skin samples were cut into small pieces and placed in a six-well dish to adhere to the dish for 30 min. Medium was then added to the plate, and the fibroblasts migrated out of the skin samples over the next 2 weeks. The fibroblasts were trypsinized and transferred to a new plate, establishing the fibroblast lines.
For the proliferation and viability assays, the fibroblasts were plated at 3 × 104 cells per well and then were manually counted at baseline, 24 hours, 48 hours, and 72 hours. Three biological replicates were performed for each line, and each biological replicate had three technical replicates.
Two patient-derived fibroblast lines and two control fibroblast lines were evaluated for cellular viability. The cells were grown until they were ~80% confluent. Then, they were stained for annexin V/propidium iodide and evaluated using flow cytometry on an Accuri C6 (BD Biosciences). Four biological replicates with two technical replicates each were analyzed.
Histology and immunocytochemistry
Mouse brains from P0 pups were extracted and further fixed in 4% PFA overnight at 4°C and washed three times with PBS. Coronal sections of 50 μm were obtained in a vibratome and stored at 4°C until staining. For Nissl staining, brain sections were mounted on frosted slides and left to dry overnight. Staining with cresyl violet (Millipore, C5042) was followed by dehydration in an alcohol gradient, xylene clearance, and coverslipping. Stained brain sections were examined under a microscope.
For immunofluorescence in brain sections, sections were permeabilized/blocked with blocking solution (5% fetal bovine serum plus 0.5% Triton X-100 in PBS) for 2 hour at room temperature. Sections were incubated with primary antibodies [H4K20me3 (1:500; Millipore, AB5700175), Ki-67 (1:1000; Abcam, ab15580), and NeuN (1:2000; Millipore, MAB377)] in blocking solution overnight at 4°C. Sections were washed three times with 0.1% Triton X-100 in PBS, incubated with secondary antibodies [Alexa Fluor 488–, Alexa Fluor 555–, Alexa Fluor 633–labeled goat anti-mouse, goat anti-rabbit, or goat anti-rat immunoglobulin G (H+L); 1:500; Thermo Fisher Scientific] and 4′,6-diamidino-2-phenylindole (1:100; Thermo Fisher Scientific, D3571) in blocking solution at room temperature for 2 hours, then washed three times with 0.1% Triton X-100 in PBS, coverslipped using ProLong Gold, and dried overnight. Imaging was performed using a Leica confocal microscope. For z-stack images, 5-μm z-stack confocal images were acquired at 1-μm intervals. Image processing was performed using ImageJ Software (National Institutes of Health, USA).
Statistical analysis
Statistical analyses have been described in the above Materials and Methods sections. To compare the clinical characteristics in males and females, we performed Fisher’s exact tests. For the RNA-seq results, we used Bioconductor DESeq2 to compare expression levels in the human samples and cuffdiff to compare transcript levels in the mouse samples. Raw P values were FDR-corrected for both datasets. When comparing the growth of the cell lines, we used Student’s t tests. The size of the mouse brains were compared using a one-way ANOVA with Tukey’s correction.
Acknowledgments
The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.. This research was partially conducted using the Histology Core and Integrated Biomedical Imaging Facility (IBIF) at Creighton University, Omaha, NE. The IBIF facility is supported by the Creighton University School of Medicine and grants GM103427 and GM139762 from NIGMS. The facility was constructed with support from grants from the National Center for Research Resources (RR016469) and the NIGMS (GM103427).
Funding: This work was supported by LB692 Nebraska Tobacco Settlement Biomedical Research Development Program (to H.A.F.S.); The Simons Foundation Autism Research Initiative–Bridge to Independence Award SFARI 381192 (to H.A.F.S.); The A*STAR, Singapore, IAF-PP Program H17/01/a0/004 (to C.Y.L.); The Wong Boon Hock Society research program Yong Loo Lin School of Medicine (to Z.X.C.); NIH training grant 2T32GM008638-25 (L.B.); The Intramural Research Program of the National Human Genome Research Institute (to W.G.); The National Center for Advancing Translational Sciences of the NIH award number TL1TR001880 (to S.E.S.); The Eunice Kennedy Shriver National Institute of Child Health and Human Development award number HD009003-01 (to S.E.S.); Institute for Translational Medicine and Therapeutics of the Perelman School of Medicine at the University of Pennsylvania (to S.E.S.); and Swiss National Science Foundation (SNSF) grant 320020_179547 and funds from the University of Zurich Research Priority Programs (URPP) AdaBD: Adaptive Brain Circuits in Developments (to A.Rau.). F.J.K. was funded by the Deutsche Forschungsgemeinschaft grant number FOR 2488. In silico modeling was supported by the Spanish Ministerio de Ciencia e Innovación grant number PID2019-111217RB-I00 (to X.d.l.C.). This study used data from the DDD study. The DDD study presents independent research commissioned by the Health Innovation Challenge Fund (grant number HICF-1009-003). This study makes use of DECIPHER (www.deciphergenomics.org), which is funded by Wellcome (grant number 223718/Z/21/Z). See Nature PMID: 25533962 or www.ddduk.org/access.html for full acknowledgement.
Author contributions: Conceptualization: R.N.W., H.A.F.S., S.E.S., and E.J.B. Data collection: R.N.W., B.R., J.Ha., J.Hu., C.J.W., P.L., M.F.-M., A.D.-R., M.Ma., and J.S. Data analysis: R.N.W., J.A.S., H.A.F.S., S.E.S., L.B., C.V., P.L., M.C.S., M.F.-M., N.A., D.L., and R.P.D.S. In silico modeling: X.d.l.C. and N.P. Simulation testing: J.A.S. Clinical and molecular evaluation of affected individuals: C.Du., H.J., F.D, M.Mu., C.A., N.L., M.An., D.B., J.C., A.v.H., I.S.-S., P.C., A.H., J.M.G.J., M. Au, A.V., L.F., W.S., Ro.M., J.P., D.V., W.G., C.T., E.M., N.H., Re.M., J.M., A.A., D.Do., B.K., P.A., S.M.-M., I.M., B.Ca., B.Co., A.Re., C.Z., C.B., A.S., M.F.S., Aj.K., F.E., C.De., M.N., B.C., Y.v.I., M.W., M.v.S., J.Y.C., D.Dr., D.P., S.Wo., S.We., E.-J.K., J.K., D.H., L.P., H.T., K.R., A.-S.D.-P., R.P.d.l.F., J.S.d.P., P.J., K.S., A.Rau., D.M., R.G., J.L., F.T.M.-T., C.P., B.D., L.R., S.M., A.M.C.-G., T.Y.T., C.M., F.R., S.G., T.G.D., L.H., A. Ra., I.P., F.J.K., Al.K., Ø.L.B., L.I., Z.X.C., C.Y.L., S.E.S., L.B.H., J.J., R.P., R.E.S., S.B., V.F., Y.D., I.P.d.B., J.M.L.R., and E.J.B. Functional evaluation of variants: L.B., S.E.S., E.J.B., X.d.l.C., and N.P. R.N.W., H.A.F.S., S.E.S., E.J.B., and L.B. wrote the paper with contributions from all authors.
Competing interests: L.B.H., J.J., R.P., and R.E.S. are employees of GeneDx LLC. A.M.C.-G. is a part of the European Reference Network on Rare Congenital Malformations and Rare Intellectual Disability (ERN ITHACA). All other authors declare that they have no competing interests.
Data and materials availability: The RNA-seq data has been deposited at GEO (accession GSE184953). All other data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. The fibroblast cell lines can be provided by CHOP pending scientific review and a completed material transfer agreement. Requests for the fibroblast cell lines should be submitted to E.B.
Supplementary Materials
This PDF file includes:
Supplementary Text
Figs. S1 to S10
Legends for tables S1, S5 to S10
Tables S2 to S4
Other Supplementary Material for this : manuscript includes the following:
Tables S1, S5 to S10
REFERENCES AND NOTES
- 1.M. Podobinska, I. Szablowska-Gadomska, J. Augustyniak, I. Sandvig, A. Sandvig, L. Buzanska, Epigenetic modulation of stem cells in neurodevelopment: The role of methylation and acetylation. Front. Cell. Neurosci. 11, 23 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Y. J. Loke, A. J. Hannan, J. M. Craig, The role of epigenetic change in autism spectrum disorders. Front. Neurol. 6, 107 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.S. Rangasamy, S. R. D'Mello, V. Narayanan, Epigenetics, autism spectrum, and neurodevelopmental disorders. Neurotherapeutics 10, 742–756 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.M. S. Fallah, D. Szarics, C. M. Robson, J. H. Eubanks, Impaired regulation of histone methylation and acetylation underlies specific neurodevelopmental disorders. Front. Genet. 11, 613098 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Z. Wu, J. Connolly, K. K. Biggar, Beyond histones—The expanding roles of protein lysine methylation. FEBS J. 284, 2732–2744 (2017). [DOI] [PubMed] [Google Scholar]
- 6.C. Martin, Y. Zhang, The diverse functions of histone lysine methylation. Nat. Rev. Mol. Cell Biol. 6, 838–849 (2005). [DOI] [PubMed] [Google Scholar]
- 7.T. Lilja, N. Heldring, O. Hermanson, Like a rolling histone: Epigenetic regulation of neural stem cells and brain development by factors controlling histone acetylation and methylation. Biochim. Biophys. Acta 1830, 2354–2360 (2013). [DOI] [PubMed] [Google Scholar]
- 8.W. D. Jones, D. Dafou, M. McEntagart, W. J. Woollard, F. V. Elmslie, M. Holder-Espinasse, M. Irving, A. K. Saggar, S. Smithson, R. C. Trembath, C. Deshpande, M. A. Simpson, De novo mutations in MLL cause Wiedemann-Steiner syndrome. Am. J. Hum. Genet. 91, 358–364 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.L. Micale, B. Augello, C. Maffeo, A. Selicorni, F. Zucchetti, C. Fusco, P. De Nittis, M. T. Pellico, B. Mandriani, R. Fischetto, L. Boccone, M. Silengo, E. Biamino, C. Perria, S. Sotgiu, G. Serra, E. Lapi, M. Neri, A. Ferlini, M. L. Cavaliere, P. Chiurazzi, M. D. Monica, G. Scarano, F. Faravelli, P. Ferrari, L. Mazzanti, A. Pilotta, M. G. Patricelli, M. F. Bedeschi, F. Benedicenti, P. Prontera, B. Toschi, L. Salviati, D. Melis, E. Di Battista, A. Vancini, L. Garavelli, L. Zelante, G. Merla, Molecular analysis, pathogenic mechanisms, and readthrough therapy on a large cohort of Kabuki syndrome patients. Hum. Mutat. 35, 841–850 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.N. Kurotaki, K. Imaizumi, N. Harada, M. Masuno, T. Kondoh, T. Nagai, H. Ohashi, K. Naritomi, M. Tsukahara, Y. Makita, T. Sugimoto, T. Sonoda, T. Hasegawa, Y. Chinen, H. A. Tomita Ha, A. Kinoshita, T. Mizuguchi, K.-i. Yoshiura, T. Ohta, T. Kishino, Y. Fukushima, N. Niikawa, N. Matsumoto, Haploinsufficiency of NSD1 causes Sotos syndrome. Nat. Genet. 30, 365–366 (2002). [DOI] [PubMed] [Google Scholar]
- 11.V. Faundes, W. G. Newman, L. Bernardini, N. Canham, J. Clayton-Smith, B. Dallapiccola, S. J. Davies, M. K. Demos, A. Goldman, H. Gill, R. Horton, B. Kerr, D. Kumar, A. Lehman, S. McKee, J. Morton, M. J. Parker, J. Rankin, L. Robertson, I. K. Temple; Clinical Assessment of the Utility of Sequencing and Evaluation as a Service (CAUSES) Study; Deciphering Developmental Disorders (DDD) Study, S. Banka, Histone lysine methylases and demethylases in the landscape of human developmental disorders. Am. J. Hum. Genet. 102, 175–187 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Y. Jiang, Y. Han, S. Petrovski, K. Owzar, D. B. Goldstein, A. S. Allen, Incorporating functional information in tests of excess de novo mutational load. Am. J. Hum. Genet. 97, 272–283 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Deciphering Developmental Disorders Study , Prevalence and architecture of de novo mutations in developmental disorders. Nature 542, 433–438 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.H. A. Stessman, B. Xiong, B. P. Coe, T. Wang, K. Hoekzema, M. Fenckova, M. Kvarnung, J. Gerdts, S. Trinh, N. Cosemans, L. Vives, J. Lin, T. N. Turner, G. Santen, C. Ruivenkamp, M. Kriek, A. van Haeringen, E. Aten, K. Friend, J. Liebelt, C. Barnett, E. Haan, M. Shaw, J. Gecz, B. M. Anderlid, A. Nordgren, A. Lindstrand, C. Schwartz, R. F. Kooy, G. Vandeweyer, C. Helsmoortel, C. Romano, A. Alberti, M. Vinci, E. Avola, S. Giusto, E. Courchesne, T. Pramparo, K. Pierce, S. Nalabolu, D. G. Amaral, I. E. Scheffer, M. B. Delatycki, P. J. Lockhart, F. Hormozdiari, B. Harich, A. Castells-Nobau, K. Xia, H. Peeters, M. Nordenskjold, A. Schenck, R. A. Bernier, E. E. Eichler, Targeted sequencing identifies 91 neurodevelopmental-disorder risk genes with autism and developmental-disability biases. Nat. Genet. 49, 515–526 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.G. Chen, L. Han, S. Tan, X. Jia, H. Wu, Y. Quan, Q. Zhang, B. Yu, Z. Hu, K. Xia, H. Guo, Loss-of-function of KMT5B leads to neurodevelopmental disorder and impairs neuronal development and neurogenesis. J. Genet. Genomics 49, 881–890 (2022). [DOI] [PubMed] [Google Scholar]
- 16.A. Eliyahu, O. Barel, L. Greenbaum, G. Zaks Hoffer, Y. Goldberg, A. Raas-Rothschild, A. Singer, I. Bar-Joseph, V. Kunik, E. Javasky, O. Staretz-Chacham, N. Pode-Shakked, L. Bazak, N. Ruhrman-Shahar, E. Pras, M. Frydman, M. Shohat, B. Pode-Shakked, Refining the phenotypic spectrum of KMT5B-associated developmental delay. Front. Pediatr. 10, 844845 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.R. C. Twells, M. L. Metzker, S. D. Brown, R. Cox, C. Garey, H. Hammond, P. J. Hey, E. Levy, Y. Nakagawa, M. S. Philips, J. A. Todd, J. F. Hess, The sequence and gene characterization of a 400-kb candidate region for IDDM4 on chromosome 11q13. Genomics 72, 231–242 (2001). [DOI] [PubMed] [Google Scholar]
- 18.R. N. Wickramasekara, H. A. F. Stessman, Histone 4 lysine 20 methylation: A case for neurodevelopmental disease. Biology 8, 11 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.G. Schotta, M. Lachner, K. Sarma, A. Ebert, R. Sengupta, G. Reuter, D. Reinberg, T. Jenuwein, A silencing pathway to induce H3-K9 and H4-K20 trimethylation at constitutive heterochromatin. Genes Dev. 18, 1251–1262 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.M. Zimmermann, T. de Lange, 53BP1: Pro choice in DNA repair. Trends Cell Biol. 24, 108–117 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Z. J. Wang, B. Rein, P. Zhong, J. Williams, Q. Cao, F. Yang, F. Zhang, K. Ma, Z. Yan, Autism risk gene KMT5B deficiency in prefrontal cortex induces synaptic dysfunction and social deficits via alterations of DNA repair and gene transcription. Neuropsychopharmacology 46, 1617–1626 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.J. A. Miller, S. L. Ding, S. M. Sunkin, K. A. Smith, L. Ng, A. Szafer, A. Ebbert, Z. L. Riley, J. J. Royall, K. Aiona, J. M. Arnold, C. Bennet, D. Bertagnolli, K. Brouner, S. Butler, S. Caldejon, A. Carey, C. Cuhaciyan, R. A. Dalley, N. Dee, T. A. Dolbeare, B. A. Facer, D. Feng, T. P. Fliss, G. Gee, J. Goldy, L. Gourley, B. W. Gregor, G. Gu, R. E. Howard, J. M. Jochim, C. L. Kuan, C. Lau, C. K. Lee, F. Lee, T. A. Lemon, P. Lesnar, B. McMurray, N. Mastan, N. Mosqueda, T. Naluai-Cecchini, N. K. Ngo, J. Nyhus, A. Oldre, E. Olson, J. Parente, P. D. Parker, S. E. Parry, A. Stevens, M. Pletikos, M. Reding, K. Roll, D. Sandman, M. Sarreal, S. Shapouri, N. V. Shapovalova, E. H. Shen, N. Sjoquist, C. R. Slaughterbeck, M. Smith, A. J. Sodt, D. Williams, L. Zollei, B. Fischl, M. B. Gerstein, D. H. Geschwind, I. A. Glass, M. J. Hawrylycz, R. F. Hevner, H. Huang, A. R. Jones, J. A. Knowles, P. Levitt, J. W. Phillips, N. Sestan, P. Wohnoutka, C. Dang, A. Bernard, J. G. Hohmann, E. S. Lein, Transcriptional landscape of the prenatal human brain. Nature 508, 199–206 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.B. Paulsen, S. Velasco, A. J. Kedaigle, M. Pigoni, G. Quadrato, A. J. Deo, X. Adiconis, A. Uzquiano, R. Sartore, S. M. Yang, S. K. Simmons, P. Symvoulidis, K. Kim, K. Tsafou, A. Podury, C. Abbate, A. Tucewicz, S. N. Smith, A. Albanese, L. Barrett, N. E. Sanjana, X. Shi, K. Chung, K. Lage, E. S. Boyden, A. Regev, J. Z. Levin, P. Arlotta, Autism genes converge on asynchronous development of shared neuron classes. Nature 602, 268–273 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.H. Qi, H. Liu, S. S. Pullamsetti, S. Gunther, C. Kuenne, A. Atzberger, N. Sommer, S. Hadzic, A. Gunther, N. Weissmann, Y. Zhou, X. Yuan, T. Braun, Epigenetic regulation by Suv4-20h1 in cardiopulmonary progenitor cells is required to prevent pulmonary hypertension and chronic obstructive pulmonary disease. Circulation 144, 1042–1058 (2021). [DOI] [PubMed] [Google Scholar]
- 25.B. Thisse, V. Heyer, A. Lux, V. Alunni, A. Degrave, I. Seiliez, J. Kirchner, J. P. Parkhill, C. Thisse, Spatial and temporal expression of the zebrafish genome by large-scale in situ hybridization screening. Methods Cell Biol. 77, 505–519 (2004). [DOI] [PubMed] [Google Scholar]
- 26.X. J. Sun, P. F. Xu, T. Zhou, M. Hu, C. T. Fu, Y. Zhang, Y. Jin, Y. Chen, S. J. Chen, Q. H. Huang, T. X. Liu, Z. Chen, Genome-wide survey and developmental expression mapping of zebrafish SET domain-containing genes. PLOS ONE 3, e1499 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.D. Nicetto, M. Hahn, J. Jung, T. D. Schneider, T. Straub, R. David, G. Schotta, R. A. Rupp, Suv4-20h histone methyltransferases promote neuroectodermal differentiation by silencing the pluripotency-associated Oct-25 gene. PLOS Genet. 9, e1003188 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.G. Schotta, R. Sengupta, S. Kubicek, S. Malin, M. Kauer, E. Callen, A. Celeste, M. Pagani, S. Opravil, I. A. De La Rosa-Velazquez, A. Espejo, M. T. Bedford, A. Nussenzweig, M. Busslinger, T. Jenuwein, A chromatin-wide transition to H4K20 monomethylation impairs genome integrity and programmed DNA rearrangements in the mouse. Genes Dev. 22, 2048–2061 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.C. T. Rhodes, R. S. Sandstrom, S.-W. A. Huang, Y. Wang, G. Schotta, M. S. Berger, C.-H. A. Lin, Cross-species analyses unravel the complexity of H3K27me3 and H4K20me3 in the context of neural stem progenitor cells. Neuroepigenetics 6, 10–25 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.D. E. V. Pires, D. B. Ascher, T. L. Blundell, mCSM: Predicting the effects of mutations in proteins using graph-based signatures. Bioinformatics 30, 335–342 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.D. Piovesan, G. Minervini, S. C. E. Tosatto, The RING 2.0 web server for high quality residue interaction networks. Nucleic Acids Res. 44, W367–W374 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.J. Gureasko, O. Kuchment, D. L. Makino, H. Sondermann, D. Bar-Sagi, J. Kuriyan, Role of the histone domain in the autoinhibition and activation of the Ras activator Son of Sevenless. Proc. Natl. Acad. Sci. U.S.A. 107, 3430–3435 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.R. N. Wickramasekara, B. Robertson, J. Hulen, J. Hallgren, H. A. F. Stessman, Differential effects by sex with Kmt5b loss. Autism Res. 14, 1554–1571 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.C. P. Austin, J. F. Battey, A. Bradley, M. Bucan, M. Capecchi, F. S. Collins, W. F. Dove, G. Duyk, S. Dymecki, J. T. Eppig, F. B. Grieder, N. Heintz, G. Hicks, T. R. Insel, A. Joyner, B. H. Koller, K. C. Lloyd, T. Magnuson, M. W. Moore, A. Nagy, J. D. Pollock, A. D. Roses, A. T. Sands, B. Seed, W. C. Skarnes, J. Snoddy, P. Soriano, D. J. Stewart, F. Stewart, B. Stillman, H. Varmus, L. Varticovski, I. M. Verma, T. F. Vogt, H. von Melchner, J. Witkowski, R. P. Woychik, W. Wurst, G. D. Yancopoulos, S. G. Young, B. Zambrowicz, The knockout mouse project. Nat. Genet. 36, 921–924 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.J. Hui-Yuen, S. McAllister, S. Koganti, E. Hill, S. Bhaduri-McIntosh, Establishment of Epstein-Barr virus growth-transformed lymphoblastoid cell lines. J. Vis. Exp. 57, 3321 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.D. B. Beck, A. Burton, H. Oda, C. Ziegler-Birling, M.-E. Torres-Padilla, D. Reinberg, The role of PR-Set7 in replication licensing depends on Suv4-20h. Genes Dev. 26, 2580–2589 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Y. Wu, Y. Wang, M. Liu, M. Nie, Y. Wang, Y. Deng, B. Yao, T. Gui, X. Li, L. Ma, C. Guo, C. Ma, J. Ju, Q. Zhao, Suv4-20h1 promotes G1 to S phase transition by downregulating p21(WAF1/CIP1) expression in chronic myeloid leukemia K562 cells. Oncol. Lett. 15, 6123–6130 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.J. Hulen, D. Kenny, R. Black, J. Hallgren, K. G. Hammond, E. C. Bredahl, R. N. Wickramasekara, P. W. Abel, H. A. F. Stessman, KMT5B is required for early motor development. Front. Genet. 13, 901228 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.V. Boonsanay, T. Zhang, A. Georgieva, S. Kostin, H. Qi, X. Yuan, Y. Zhou, T. Braun, Regulation of skeletal muscle stem cell quiescence by Suv4-20h1-dependent facultative heterochromatin formation. Cell Stem Cell 18, 229–242 (2016). [DOI] [PubMed] [Google Scholar]
- 40.J. Trinh, K. K. Kandaswamy, M. Werber, M. E. R. Weiss, G. Oprea, S. Kishore, K. Lohmann, A. Rolfs, Novel pathogenic variants and multiple molecular diagnoses in neurodevelopmental disorders. J Neurodev. Disord. 11, 11 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.D. G. Amaral, D. Li, L. Libero, M. Solomon, J. Van de Water, A. Mastergeorge, L. Naigles, S. Rogers, C. Wu Nordahl, In pursuit of neurophenotypes: The consequences of having autism and a big brain. Autism Res. 10, 711–722 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.H. H. Bartholomeusz, E. Courchesne, C. M. Karns, Relationship between head circumference and brain volume in healthy normal toddlers, children, and adults. Neuropediatrics 33, 239–241 (2002). [DOI] [PubMed] [Google Scholar]
- 43.S. Jones, D. Samanta, Macrocephaly, in StatPearls (StatPearls Publishing, 2022). [PubMed] [Google Scholar]
- 44.M. P. Lun, E. S. Monuki, M. K. Lehtinen, Development and functions of the choroid plexus-cerebrospinal fluid system. Nat. Rev. Neurosci. 16, 445–457 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.V. S. Chen, J. P. Morrison, M. F. Southwell, J. F. Foley, B. Bolon, S. A. Elmore, Histology atlas of the developing prenatal and postnatal mouse central nervous system, with emphasis on prenatal days E7.5 to E18.5. Toxicol. Pathol. 45, 705–744 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.W. P. Fung, T. Thomas, P. W. Dickson, A. R. Aldred, J. Milland, M. Dziadek, B. Power, P. Hudson, G. Schreiber, Structure and expression of the rat transthyretin (prealbumin) gene. J. Biol. Chem. 263, 480–488 (1988). [PubMed] [Google Scholar]
- 47.H. M. Eisenberg, J. G. McComb, A. V. Lorenzo, Cerebrospinal fluid overproduction and hydrocephalus associated with choroid plexus papilloma. J. Neurosurg. 40, 381–385 (1974). [DOI] [PubMed] [Google Scholar]
- 48.H. Pei, L. Zhang, K. Luo, Y. Qin, M. Chesi, F. Fei, P. L. Bergsagel, L. Wang, Z. You, Z. Lou, MMSET regulates histone H4K20 methylation and 53BP1 accumulation at DNA damage sites. Nature 470, 124–128 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.D. Vivien, C. Ali, Transforming growth factor-β signalling in brain disorders. Cytokine Growth Factor Rev. 17, 121–128 (2006). [DOI] [PubMed] [Google Scholar]
- 50.K. R. Long, B. Newland, M. Florio, N. Kalebic, B. Langen, A. Kolterer, P. Wimberger, W. B. Huttner, Extracellular matrix components HAPLN1, lumican, and collagen I cause hyaluronic acid-dependent folding of the developing human neocortex. Neuron 99, 702–719.e6 (2018). [DOI] [PubMed] [Google Scholar]
- 51.J. J. Yi, A. P. Barnes, R. Hand, F. Polleux, M. D. Ehlers, TGF-β signaling specifies axons during brain development. Cell 142, 144–157 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.J. P. Myers, M. Santiago-Medina, T. M. Gomez, Regulation of axonal outgrowth and pathfinding by integrin-ECM interactions. Dev. Neurobiol. 71, 901–923 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.M. J. G. Sacoto, I. A. Tchasovnikarova, E. Torti, C. Forster, E. H. Andrew, I. Anselm, K. W. Baranano, L. C. Briere, J. S. Cohen, W. J. Craigen, C. Cytrynbaum, N. Ekhilevitch, M. J. Elrick, A. Fatemi, J. L. Fraser, R. C. Gallagher, A. Guerin, D. Haynes, F. A. High, C. N. Inglese, C. Kiss, M. K. Koenig, J. Krier, K. Lindstrom, M. Marble, H. Meddaugh, E. S. Moran, C. F. Morel, W. Mu, E. A. Muller 2nd, J. Nance, M. R. Natowicz, A. L. Numis, B. Ostrem, J. Pappas, C. E. Stafstrom, H. Streff, D. A. Sweetser, M. Szybowska; Undiagnosed Diseases Network, M. A. Walker, W. Wang, K. Weiss, R. Weksberg, P. G. Wheeler, G. Yoon, R. E. Kingston, J. Juusola, De novo variants in the ATPase module of MORC2 cause a neurodevelopmental disorder with growth retardation and variable craniofacial dysmorphism. Am. J. Hum. Genet. 107, 352–363 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.N. Sobreira, F. Schiettecatte, C. Boehm, D. Valle, A. Hamosh, New tools for Mendelian disease gene identification: PhenoDB variant analysis module; and GeneMatcher, a web-based tool for linking investigators with an interest in the same gene. Hum. Mutat. 36, 425–431 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.M. Lefter, J. K. Vis, M. Vermaat, J. T. den Dunnen, P. E. M. Taschner, J. F. J. Laros, Mutalyzer 2: Next generation HGVS nomenclature checker. Bioinformatics 37, 2811–2817 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.S. F. Altschul, T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, D. J. Lipman, Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 25, 3389–3402 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.J. S. Papadopoulos, R. Agarwala, COBALT: Constraint-based alignment tool for multiple protein sequences. Bioinformatics 23, 1073–1079 (2007). [DOI] [PubMed] [Google Scholar]
- 58.H. Wu, A. Siarheyeva, H. Zeng, R. Lam, A. Dong, X. H. Wu, Y. Li, M. Schapira, M. Vedadi, J. Min, Crystal structures of the human histone H4K20 methyltransferases SUV420H1 and SUV420H2. FEBS Lett. 587, 3859–3868 (2013). [DOI] [PubMed] [Google Scholar]
- 59.XSchrödinge LLC.
- 60.B. Webb, A. Sali, Comparative protein structure modeling using MODELLER. Curr. Protoc. Bioinformatics 54, 5.6.1–5.6.37 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.C. B. Kimmel, W. W. Ballard, S. R. Kimmel, B. Ullmann, T. F. Schilling, Stages of embryonic development of the zebrafish. Dev. Dyn. 203, 253–310 (1995). [DOI] [PubMed] [Google Scholar]
- 62.R. H. Friedel, C. Seisenberger, C. Kaloff, W. Wurst, EUCOMM—The European conditional mouse mutagenesis program. Brief. Funct. Genomic. Proteomic. 6, 180–185 (2007). [DOI] [PubMed] [Google Scholar]
- 63.T. F. Smith, M. S. Waterman, Identification of common molecular subsequences. J. Mol. Biol. 147, 195–197 (1981). [DOI] [PubMed] [Google Scholar]
- 64.M. I. Love, W. Huber, S. Anders, Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.C. Trapnell, A. Roberts, L. Goff, G. Pertea, D. Kim, D. R. Kelley, H. Pimentel, S. L. Salzberg, J. L. Rinn, L. Pachter, Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 7, 562–578 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Text
Figs. S1 to S10
Legends for tables S1, S5 to S10
Tables S2 to S4
Tables S1, S5 to S10