Abstract
Altering the content or composition of the cell wall polymer lignin is a favored approach to valorize lignin toward biomaterial and chemical production in the biorefinery. However, modifying lignin or cellulose in transgenic plants can induce expression of defense responses and negatively affect growth. Through genetic screening for suppressors of defense gene induction in the low lignin ccr1-3 mutant of Arabidopsis thaliana, we found that loss of function of the receptor-like kinase FERONIA, although not restoring growth, affected cell wall remodeling and blocked release of elicitor-active pectic polysaccharides as a result of the ccr1-3 mutation. Loss of function of multiple wall-associated kinases prevented perception of these elicitors. The elicitors are likely heterogeneous, with tri-galacturonic acid the smallest but not necessarily the most active component. Engineering of plant cell walls will require development of ways to bypass endogenous pectin signaling pathways.
Lignin modification triggers receptor kinase-mediated cell wall remodeling.
INTRODUCTION
The polymer-rich plant cell wall would appear to be an ideal location for extracellular targeting of engineered polymeric bioproducts. For example, altering the composition of lignin, a major structural polymer in secondarily thickened plant secondary cell walls, is an approach to valorizing lignin in the biorefinery (1, 2). Lignin also contributes to the recalcitrance of lignocellulosic biomass for bioprocessing to liquid fuels (3), and engineered plants with low lignin levels have enhanced release of cell wall sugars for biofuel production (4–6). However, modifying lignin or cellulose in transgenic plants can negatively affect growth (7, 8) and/or induce expression of defense response genes in the absence of biotic or abiotic stress through activation of endogenous defense pathways (9, 10). Lignin is a complex heteropolymer of monolignol units derived from the phenylpropanoid pathway and linked through a free-radical driven process in the apoplast. The major monolignols are coniferyl alcohol, leading to guaiacyl (G) units in lignin, sinapyl alcohol, leading to syringyl (S) units, and coumaryl alcohol, leading to the less abundant hydroxyphenyl (H) units (11). Altering lignin content and/or composition can have marked effects on recalcitrance to bioprocessing (3) or, in the case of forage crops, digestibility by ruminant animals (12).
We previously showed that cell walls of Arabidopsis thaliana lines with reduced lignin content in inflorescence stems through independent down-regulation of two lignin biosynthesis enzymes, hydroxycinnamoyl coenzyme A (CoA): shikimate hydroxycinnamoyl transferase (HCT) and cinnamoyl CoA reductase (CCR), release oligosaccharide elicitors, which induce pathogenesis-related (PR) protein gene expression through the salicylic acid signaling pathway (13). The elicitors are found in cold water extracts of isolated inflorescence stem cell walls, here referred to as cell wall elicitor/extract [e.g., ccr1-3 cell wall water extract (CWE)], and are of pectic origin based on their destruction by incubation with commercial polygalacturonase (PGA) (13). Genetic and biochemical studies indicated that both in planta PR gene expression and release of elicitors from cell wall pectic polysaccharides are the result of ectopic expression of the gene encoding the ARABIDOPSIS DEHISCENCE ZONE POLYGALACTURONASE 1 (ADPG1) in the inflorescence stems of the ccr1 mutant (14). However, leaf injection experiments using water extracts from isolated cell walls of wild-type (WT) Columbia (Col-0) and ccr1 mutant lines showed that ADPG1 itself is not induced by the CWEs from the Arabidopsis lines with reduced lignin content.
There is increasing interest in the role of the cell wall in extracellular signaling (15, 16). Communication between the extracellular matrix and the inside of the cell is largely brought about through the activities of receptor-like kinases (17). These are involved in sensing a number of changes, from pathogen signals to salt stress and pH (17, 18). Lignin is believed to be associated with hemicellulosic components of the cell wall, and possibly also pectin, to provide increased structural integrity (19, 20). However, the molecular mechanisms underlying how lignin modifications are perceived in the cell wall and how plants recognize endogenously released elicitors remain poorly understood. In the case of the ccr1 mutant, total lignin levels are reduced, primarily as a result of parallel reductions in the amounts of G and S units (Fig. 1A). The simplest hypothesis is that this change results in altered cell wall integrity, which is sensed either directly via interactions of unshielded polysaccharides with receptors or indirectly via effects on physical properties such as turgor, leading to receptor-mediated cell wall remodeling to release pectic/oligosaccharide elicitors that then activate defense gene expression after perception by elicitor receptors (Fig. 1A) (14). However, the exact natures of the receptors and their ligands for initial perception and downstream response remain to be determined.
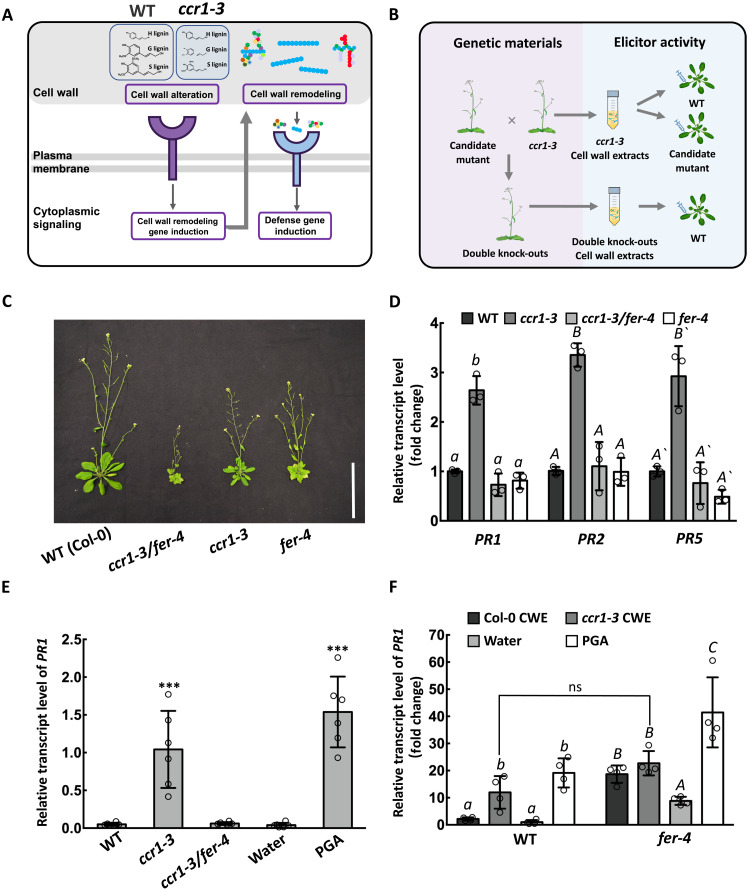
Fig. 1. FERONIA is required for elicitor release in the ccr1-3 mutant.
(A) Simplified model of the signaling hypothesis. Altered lignin is sensed by a cell wall receptor kinase to activate wall remodeling enzymes that solubilize pectin to release elicitor-active oligosaccharides that are sensed by other receptor kinases to activate defense responses. Lignin in ccr1-3 has reductions in G and S units (see Fig. 2, A and B). Colored circles represent pectin units, with galacturonic acid in blue. The three classes of pectins are rhamnogalacturonan II (left), homogalacturonan (middle), and rhamnogalacturonan I (right). (B) Strategy for identification of the two types of receptors in (A). ccr1-3 and cell wall receptor kinase mutants were crossed to screen for suppressors of ccr1-mediated PR expression in inflorescence stems. CWEs from stems were tested for the presence of elicitors by injection into WT Col-0 leaves and subsequent measurement of PR transcripts. (C) Vegetative phenotypes of Col-0 and the ccr1, fer-4, and ccr1/fer-4 mutants. Scale bar, 10 cm. (D) PR1, PR2, and PR5 transcript levels in inflorescence stems of ccr1-3, fer-4, and ccr1-3/fer-4 relative to Col-0. Transcript levels in Col-0 were set as 1. (E) PR1 transcript levels in Col-0 leaves after injection with CWEs from Col-0, ccr1-3, or ccr1-3/fer-4 (0.1 mg/ml uronic acid equivalents). Water and PGA (0.1 mg/ml) were injected as negative and positive controls, respectively. (F) PR1 transcript levels in Col-0 and fer-4 leaves after injection with CWEs from Col-0 or ccr1-3, with water and PGA as controls. Transcript level of the water control was set as 1. Bars represent means ± SD. n = 3 (D), n = 6 (E), and n = 4 (F). Hollow dots represent individual data points. Letters and asterisks indicate statistically significant differences according to Student’s t test, ***P < 0.001, in (E) and (F) and one-way ANOVA followed by Tukey’s honestly significant difference (HSD) test, α = 0.05, in (D).
RESULTS
Loss of function of FERONIA prevents PR expression and elicitor release in lignin-modified plants
To interrogate the initial sensing mechanism, we screened for suppressor mutants of defense signaling in the ccr1-3 mutant background (Fig. 1B). By searching the protein function annotation database, combined with the Arabidopsis eFP Browser gene expression database, we collected loss-of-function mutants of cell wall proteins, membrane proteins, membrane receptors, and receptor-like kinases that have been predicted to sense cell wall integrity or interact with plant cell wall oligosaccharides (21–29), and which are expressed in the inflorescence stem (table S1 and fig. S1). These selected mutants (two independent alleles where possible) were genetically crossed with the ccr1-3 mutant (30) to obtain homozygous double mutants. The transcript abundance of the PR1 gene in the inflorescence stems of the homozygous progeny was used as a genetic selection marker to screen for suppression of cell wall defense signaling associated with the ccr1-3 mutation. Subsequently, we tested whether CWEs from isolated cell walls of these mutants could induce PR gene expression following injection into Col-0 leaves, to confirm whether the mutation affected elicitor release (Fig. 1B). To determine whether the mutation affected a receptor(s) involved in recognizing the ccr1-CWEs, we injected leaves of the candidate loss-of-function mutants with ccr1-CWEs and again measured PR gene transcript levels (Fig. 1B).
When plants of the ccr1-3 mutant were crossed with the bups1/2, anx1/2, herk1/2, fei1/2, cap(eru)1, the1, or perk4 mutants selected as described above (table S1) and PR1 transcript levels compared in stems of homozygous progeny to those in WT and ccr1-3, the presence of the receptor mutation did not significantly reduce the PR transcript level to below that of ccr1-3 (fig. S2). The fer-4 mutant of FERONIA, a Catharanthus roseus receptor-like kinase 1 (CrRLK1)–like subfamily receptor-like kinase implicated in hormone signaling, the maintenance of cell wall integrity under high salt stress and control of plant immunity (18, 31, 32), has been well characterized (18, 33). Loss of function of FERONIA resulted in loss of PR1, PR2, and PR5 gene transcript induction in stems of the ccr1-3/fer-4 double mutant accompanied by further reduction in plant stature (Fig. 1, C and D). Furthermore, leaf injection assays with CWEs from the double mutant revealed loss of the PR gene–inducing activity compared to that of CWEs from ccr1-3 mutant plants with functional FERONIA (Fig. 1E). However, ccr1-3–derived CWEs and PGA (positive control elicitor) could still induce PR1 gene expression in fer-4 mutant leaves (Fig. 1F), indicating that FERONIA is involved in generating, but not sensing, the elicitors of PR gene expression. We also consistently observed that fer-4 leaves responded to CWE from WT Col-0 by activation of PR1, suggesting the possible loss of an inhibitory effect on recognition of components from unperturbed WT cell walls. It is also possible that the higher PR1 expression in fer-4 than in Col-0 for all leaf injection treatments including water (Fig. 1F) is associated with defective sensing of turgor pressure in fer-4 (33) or enhanced sensitivity of fer-4 to mechanical damage.
Lignin modification combined with loss of function of FERONIA has profound effects on cell wall integrity
Previous studies showed that the reduction in lignin content in the ccr1 mutant resulted in release of water-soluble pectic polysaccharides from isolated cell walls (14). To explore the role of FERONIA in altering cell wall properties in ccr1-3, we first compared the lignin content and composition in inflorescence stems of ccr1-3 and the ccr1-3/fer-4 double mutants by thioacidolysis, a method in which the lignin polymer is chemically depolymerized to release derivatives of the different monomer units, which can then be separated and quantified by gas chromatography–mass spectrometry (GC-MS). The lignin thioacidolysis monomer yield was lower in the ccr1-3/fer-4 double mutant than in ccr1-3 (Fig. 2A), and only small differences in lignin composition were observed (Fig. 2B). We interpret these results as unlikely to bear on defense signaling, since the lower lignin amount in ccr1-3/fer-4 would be predicted to result in higher PR gene expression if FERONIA were not involved in the signaling process, and alterations in lignin composition without changes in lignin amount do not appear to induce PR gene expression in Arabidopsis (13). The apparent pectin content (measured as uronic acid equivalents) in the water extract from cell walls of the ccr1-3/fer-4 double mutant was not significantly different from that of the ccr1-3 single mutant (Fig. 2C) but around threefold higher than in Col-0 or fer-4. Therefore, loss of function of FERONIA itself does not affect the overall content of water-soluble pectin-like polysaccharides in the cell walls of ccr1-3/fer-4 mutants. However, because PR genes are not induced in the ccr1-3/fer-4 double mutant, loss of function of FERONIA may qualitatively affect the release of pectic materials via cell wall remodeling enzymes. Loss of function of FERONIA strongly reduced the induction of ADPG1 resulting from the ccr1-3 mutation (Fig. 2D), and enzymatic fingerprinting of homogalacturonans present in Col-0, ccr1-3, fer-4, and ccr1-3/fer-4 CWE preparations by digestion with endopolygalacturonase (EPG) from Aspergillus aculeatus suggested that the pectins were qualitatively different (fig. S3). Pectins from ccr1-3/fer-4 and fer-4 were less sensitive to treatment with EPG. Oligosaccharides across a large size range (from 6 to 8 min) produced diagnostic fragment ions of demethylesterified oligogalacturonide (OG) upon in-source fragmentation (fig. S3). Treatment of Col, fer-4, and ccr1-3/fer-4 CWEs with EPG released more acetylated and slightly more methylesterified OGs than were released from ccr1-3 CWE (fig. S3).
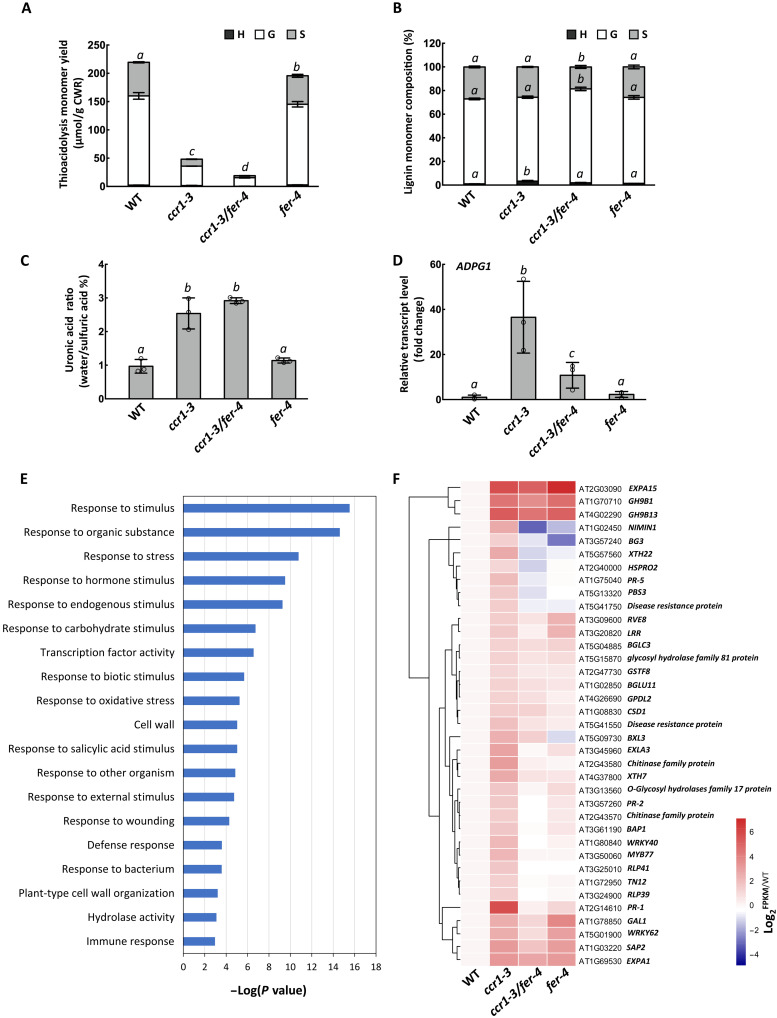
Fig. 2. Loss of function of FERONIA results in down-regulation of defense response and cell wall remodeling genes in inflorescence stems of ccr1-3.
(A) Thioacidolysis lignin monomer yield of stems of Col-0, ccr1-3, fer-4, and ccr1-3/fer-4. (B) Lignin monomer compositions of alcohol-insoluble cell wall extracts from Col-0, ccr1-3, fer-4, and ccr1-3/fer-4. H, hydroxyphenyl; G, guaiacyl; S, syringyl lignin monomer units. (C) Ratio of water-soluble to total uronic acid content of Col-0, ccr1- 3, fer-4, and ccr1-3/fer-4. (D) Levels of ADPG1 transcripts in inflorescence stems of Col-0, ccr1-3, fer-4, and ccr1-3/fer-4. Bars represent means ± SD, n = 3 (A to D), and hollow dots represent individual data points. Letters (A to D) indicate statically significant differences between treatments of each genotype according to one-way ANOVA and Tukey’s HSD test, α = 0.05. (E) Gene ontology analysis of the DEGs that are highly expressed in ccr1-3 compared with Col-0 but decreased in ccr1-3/fer-4 double mutants compared with ccr1-3. (F) Genes involved in biotic stress responses that are highly expressed in ccr1-3 compared with Col-0 but decreased in ccr1-3/fer-4 double mutants compared with ccr1-3.
To obtain a better understanding of the impact of loss of function of FERONIA on extractable cell wall polysaccharide composition, we performed total oligosaccharide profiling by LC-MS, comparing CWE preparations from inflorescence stems of Col-0, fer-4, ccr1-3, and ccr1-3/fer-4 before and after digestion with EPG (Fig. 3). The main peaks in the mass spectra of CWEs from Col-0 and fer-4 represented mannose-glucuronic acid-inositol-P [mass-to-charge ratio (m/z) = 597.107], presumably originating from the hydrolysis of polar heads of glycosyl inositol phosphoryl ceramide lipids (34), along with uronic acid-(hexose)2-(pentose)2 (m/z = 781.225) and uronic acid-(hexose)2-(pentose)3 (m/z = 913.267), potentially originating from rhamnogalacturonan I (Fig. 3A, upper trace). Treatment of the Col-0 CWE with EPG resulted in appearance of lower molecular weight peaks, including one corresponding to tri-galacturonic acid (GalA3) (m/z = 545.099), a product of the degradation of homoglacturonan or rhamnogalacturonan II (Fig. 3A, lower trace). Very similar profiles were observed for the CWEs from the fer-4 mutant (Fig. 3B). In contrast, CWEs from the ccr1-3 mutant showed, in addition, a number of oligosaccharide peaks that were not present in Col-0 or fer-4 extracts; these included hexose oligomers (pentamers, hexamers, and septamers), a pentose pentamer (m/z = 721.205) likely originating from xylan, and GalA3, the amount of which was increased following digestion of the CWE with EPG (Fig. 3C). This suggests that, in addition to release of pectic material, the cell walls of the ccr1-3 mutant have undergone remodeling to solubilize hemicellulosic components.
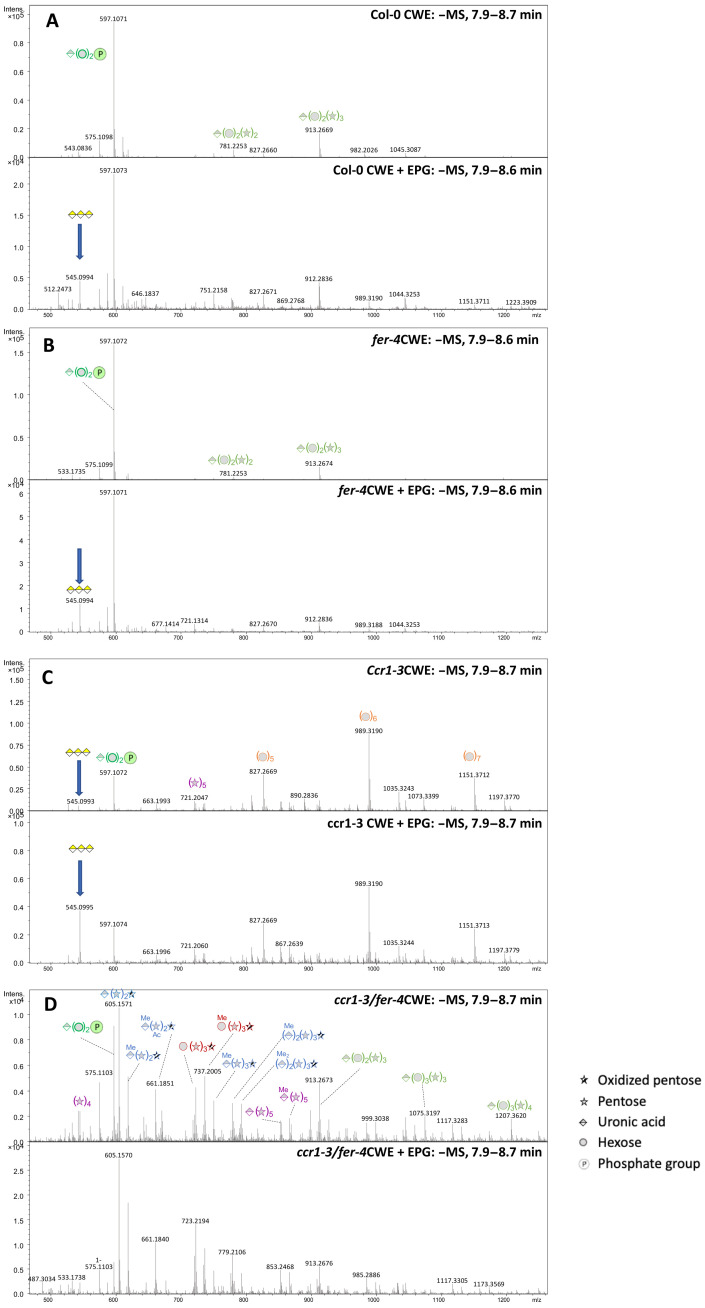
Fig. 3. Oligosaccharide profiling of water extracts from cell walls of WT and mutant Arabidopsis inflorescence stems.
Panels show mass spectra of the 7.9- to 8.7-min regions of chromatograms generated and processed as described in (57). In each set of panels, the upper panel is an undigested extract and the lower panel is the same extract after digestion with commercial PGA from A. aculeatus. Extracts were from (A) Col-0, (B) fer-4, (C) ccr1-3, and (D) ccr1-3/fer-4. Peak identities are indicated by symbols. Blue arrows indicate the peak originating from GalA3.
The largest change in the composition of the CWEs was seen in extracts from the ccr1-3 mutant in the fer-4 background (Fig. 3D). Notably, GalA3 was not observed, even after digestion with EPG, consistent with the lack of induction of ADPG1 in the ccr1-3/fer-4 stems (Fig. 2D). Even more striking, however, was the appearance of many new oligosaccharide peaks that were not present in extracts from the single ccr1-3 or fer-4 mutant stems. These included several methylated and nonmethylated uronic acid pentose oligosaccharides, some with one oxidized pentose unit (likely the result of lytic polysaccharide monooxygenase activity), and hexose-pentose oligosaccharides (compositions given in Fig. 3D). A key to the fragmentation of xylan-derived oligosaccharides and evidence for the presence/absence of GalA3 in selected cell wall extracts are given in fig. S4A.
Clearly, loss of function of FERONIA increases the solubility of hemicellulosic fractions in the ccr1-3 mutant background but prevents the solubilization of moieties that can be cleaved to yield GalA3. Considering the high amount of glucuronic acid–containing xylan oligosaccharides in the ccr1-3 and ccr1-3/fer-4 fraction and the low amount of GalA3 released by EPG, the high content of total uronic acid in water extracts of cell walls from ccr1-3 and ccr1-3/fer-4 (Fig. 2C) is more related to glucuronoxylan than pectin content.
Loss of function of FERONIA blocks the induction of cell wall remodeling enzymes in response to lignin modification
We next used RNA sequencing (RNA-seq) analysis of total RNA from inflorescence stems of WT, ccr1-3, fer-4, and ccr1-3/fer-4 to determine the extent to which loss of function of FERONIA affected the expression of other genes that are up-regulated in ccr1-3. Gene ontology (GO) analysis was performed on the 314 differentially expressed genes found (dataset S1), and these were enriched in the functions of biotic stress response and cell wall composition (Fig. 2E and fig. S5). Heat map analysis showed that, compared with ccr1-3, the transcript abundances of disease resistance and cell wall remodeling–related genes were decreased to varying degrees in ccr1-3/fer-4 (Fig. 2F). The loss of induction of various cell wall remodeling enzymes that are induced in ccr1-3 is consistent with involvement of FERONIA in processes underlying the release of cell wall elicitors. However, it should be noted that blocking of defense gene expression through loss of function of FERONIA does not restore growth in the ccr1-3 mutant background; rather, growth is further reduced beyond that seen in fer-4. This indicates that the growth reduction in ccr1-3 is not simply the result of a growth defense trade-off.
Wall-associated kinases act redundantly in elicitor reception and signaling
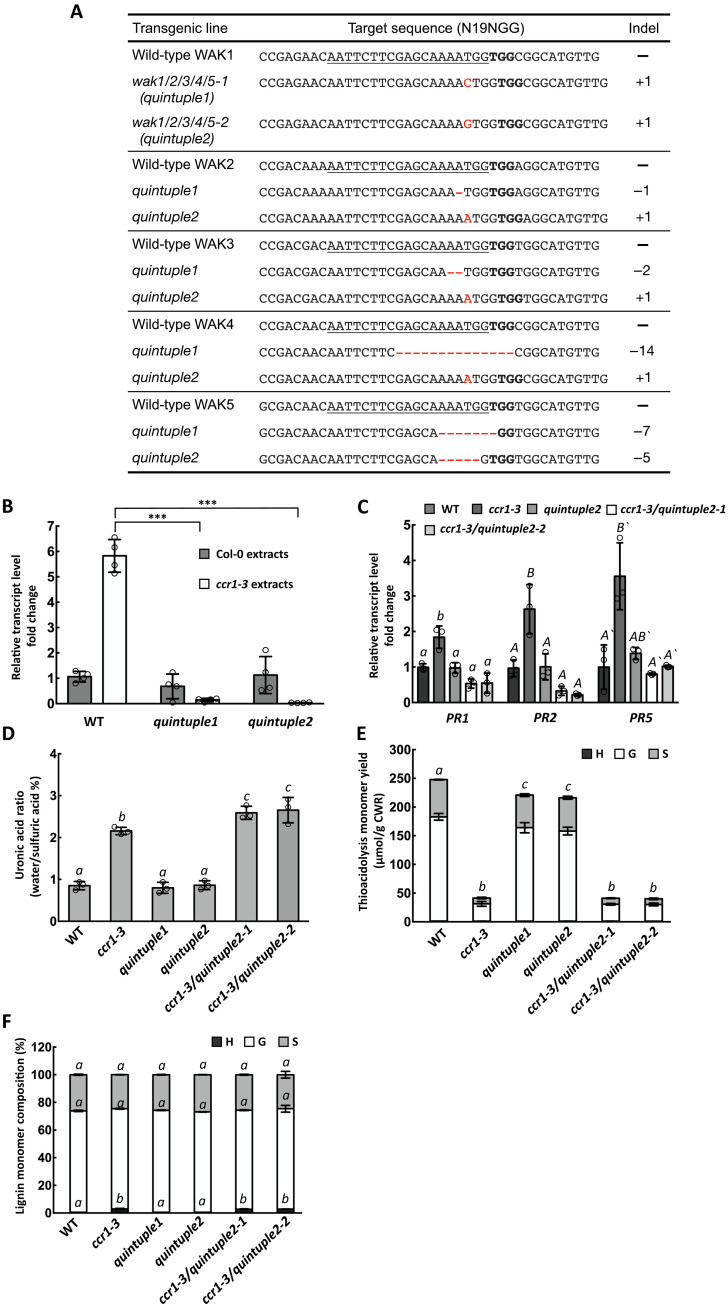
Because the fer-4 mutant remains sensitive to ccr1-CWE or PGA treatment (Fig. 1F), other receptors are necessary to perceive the elicitor signal. As previous studies suggest the strong likelihood that the elicitors are released from pectin, we focused on receptors that have been reported to interact with pectin or OGs. Wall-associated kinase (WAK) family proteins have been shown to interact with pectin and transmit defense and developmental signals in plant cells (35–37). There are five WAK family genes in Arabidopsis that are located next to each other on chromosome 1. WAK1 and WAK2 have the highest transcript expression levels in stems, followed by WAK3. WAK5 shows the lowest transcript expression level among all the five WAK family genes (fig. S6A). We therefore crossed ccr1-3 with the wak1, wak2, wak3, and wak4 single mutants. PR1 and PR2 transcript levels remained high in stems of the homozygous double mutants (fig. S6B), and ccr1-derived CWEs could still induce PR genes in wak1 and wak2 single mutant leaves (fig. S6C). Considering the potential redundancy of WAK family proteins, we next generated the wak1/2 double mutant by CRISPR-Cas9 gene editing (fig. S7A). However, ccr1-CWEs could still induce PR gene expression in the wak1/2 double mutant background (fig. S7B). Last, we designed a single-guide RNA (sgRNA) to target all five WAK genes and obtained two lines of wak1/2/3/4/5 quintuple mutants (Fig. 4A). The quintuple mutant appeared to grow normally (fig. S8). In the leaf injection assay, ccr1-CWEs could not induce PR1, PR2, or PR5 expression in wak quintuple mutant leaves (Fig. 4B). To test whether WAKs are responsible for perceiving ccr1-CWEs in vivo, we crossed the ccr1-3 single mutant with each of the two wak quintuple mutant lines to generate ccr1/wak1/2/3/4/5 sextuple mutants. Similar to the situation with ccr1-3/fer-4, loss of function of WAKs did not restore growth in the ccr1-3/wak1/2/3/4/5 sextuple mutant (fig. S8). PR1, PR2, and PR5 transcript levels in the sextuple mutants were similar to or lower than those in the WT (Fig. 4C). The elevated uronic acid–containing polysaccharide content of cell wall extracts of ccr1-3 was maintained in the ccr1/wak1/2/3/4/5 sextuple mutants (Fig. 4D), and there was no significant difference in lignin monomer composition or total lignin thioacidolysis yield (H + G + S) in the inflorescence stems (Fig. 4, E and F), suggesting that loss of function of all five WAK genes does not noticeably alter the cell wall properties of ccr1-3. These results suggest that WAKs are likely involved redundantly in perceiving ccr1-CWEs. Another possibility, based on the negative results with wak1, wak2, wak3, and wak4 single mutants (fig. S6B), is that WAK5 is the elicitor receptor, although this appears unlikely in view of its very low expression level in inflorescence stems.
Fig. 4. WAK is required for perceiving the water-soluble elicitors released from cell walls of the ccr1-3 mutant.
(A) Gene-editing mutations generated in the five WAK genes in the wak1/2/3/4/5 (wakQ) quintuple mutant. (B) PR1 transcript levels in Col-0 and wakQ mutants after treatment with CWEs from Col-0 or ccr1-3 (0.1 mg/ml uronic acid equivalents). The transcript level of WT treated by Col-0 extracts was set as 1. (C) PR1, PR2, and PR5 transcript levels in inflorescence stems of Col-0, ccr1-3, wakQ, and ccr1-3/wakQ hextuple mutant. The transcript level of each gene in Col-0 was set as 1. (D) Relative ratio of water-soluble to total uronic acid content of Col-0, ccr1-3, wakQ, and ccr1-3/wakQ. (E) Thioacidolysis lignin monomer yield of inflorescence stems of Col-0, ccr1-3, wakQ, and ccr1-3/wakQ. (F) Lignin monomer compositions of alcohol-insoluble cell wall extracts from Col-0, ccr1-3, wakQ, and ccr1-3/wakQ. H, hydroxyphenyl; G, guaiacyl; S, syringyl. Bars represent means ± SD. n = 4 (B) and n = 3 (C to F). Hollow dots represent individual data points. Letters and asterisks indicate statistically significant differences among samples according to Student’s t test, ***P < 0.001, in (B) and one-way ANOVA followed by Tukey’s HSD test, α = 0.05, in (C) to (F).
ADPG1 processes pectins as ligands for WAKs
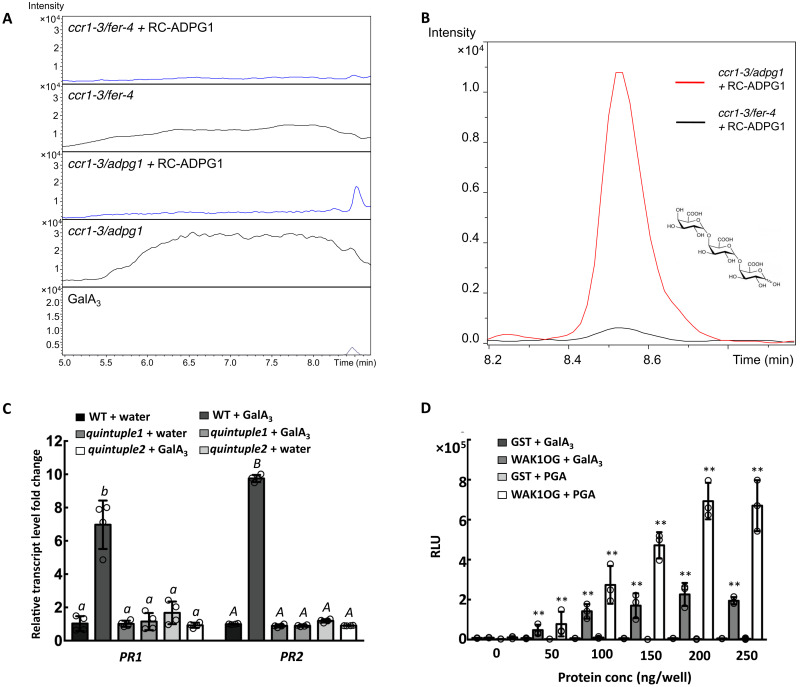
ADPG1 activity is required for the release of elicitors from the remodeled cell wall matrix, and no elicitor activity is observed in cell wall extracts from the ccr1-3/adpg1 double mutant (14). However, elicitor activity is recovered after incubating ccr1-3/adpg1 CWEs with recombinant ADPG1 protein (14). We therefore used a high-resolution oligosaccharide profiling liquid chromatography–mass spectrometry (LC-MS) platform to profile oligosaccharides in the water-soluble extracts from cell walls of ccr1-3/adpg1 and ccr1-3/fer-4 before and after digestion with ADPG1 to attempt to identify elicitor-active molecules and address their binding to WAK.
Extracts from ccr1-3/adpg1 contained a complex mixture of oligosaccharides across a large size range that produce diagnostic OG fragments upon in-source fragmentation (Fig. 5A). Treatment of ccr1-3/adpg1 extracts with recombinant ADPG1 resulted in the loss of most of the higher molecular weight oligosaccharides and generated a large peak around the region of GalA3 (Fig. 5A). Extracts from ccr1-3/fer-4 also had a broad range of oligosaccharides (Fig. 5A). However, treating the ccr1-3/fer-4 extracts with recombinant ADPG1, although shifting the molecular weight profile to smaller oligosaccharides, did not result in the release of a predominance of oligomers around the size of GalA3 (Fig. 5A).
Fig. 5. Pectins released from cell walls of the ccr1 mutant are cleaved by ADPG1 to release GalA3, which binds to the WAK OG-binding domain and induces PR proteins.
(A) Extracted ion chromatograms of OGs from DP3 to DP9 in CWEs from ccr1-3/fer-4 and ccr1-3/adpg1 with and without digestion with recombinant ADPG1. (B) Selected ion monitoring for GalA3 (at m/z = 545) of LC-MS traces of oligosaccharides from CWEs from ccr1-3/fer-4 (black) and ccr1-3/adpg1 (red). CWEs were preincubated with recombinant ADPG1. (C) PR1 and PR2 transcript levels in leaves of Col-0 or wak1/2/3/4/5 quintuple mutant after injection of water (control) or GalA3 (0.05 mg/ml). (D) ELISA of the binding of GalA3 (20 μg per well) and polygalacturonic acid (20 μg per well, positive ligand control) to the WAK1 OG-binding domain, expressed in E. coli as a GST fusion protein (fig. S9B). GST serves as a negative binding protein control. Bars represent means ± SD. n = 3. Hollow dots represent individual data points in (C) and (D). Letters and asterisks indicate statistically significant differences among samples according to one-way ANOVA followed by Tukey’s HSD test, α = 0.05, in (C) and Student’s t test, ***P < 0.001, in (D).
These data suggest that loss of function of FERONIA blocks the release/availability of higher molecular weight pectic material (a substrate for ADPG1) that can be further processed by ADPG1 to yield smaller oligomers such as GalA3. To explore this further, we used selected ion monitoring at m/z = 545 to examine production of GalA3 after digestion of CWEs from ccr1-3/fer-4 and ccr1-3/adpg1. GalA3 was efficiently released from ccr1-3/adpg1 CWE by ADPG1, but only a small peak of GalA3 was observed in ADPG1-treated ccr1-3/fer-4 CWE (Fig. 5B). Parallel analysis of ADPG1 digestion of WT Col-0 CWE revealed generation of some GalA3, but less than from ccr1/adpg1 (fig. S9). Together, these results, combined with the MS profiling in Fig. 3 (C and D), confirm that altered lignin content results in FERONIA-dependent solubilization of a specific fraction of pectin that is further processed by ADPG1 to lower molecular weight products.
GalA3 is an elicitor
We next used the leaf injection assay to verify whether GalA3 itself has elicitor activity and is perceived by WAK receptors. GalA3 could induce PR1 and PR2 gene expression in WT leaves but not in wak quintuple mutant leaves (Fig. 5C). To test whether GalA3 could interact with WAK protein, we expressed the WAK1 OG-binding domain (38) (fig. S10A) as a glutathione S-transferase (GST) fusion in Escherichia coli and purified the recombinant protein, confirming the band by protein gel blot analysis (fig. S10B). Enzyme-linked immunosorbent assay (ELISA) assays showed that GalA3 can interact with the OG-binding domain of WAK1 (Fig. 5D). Last, we used the immobilized OG-binding domain of WAK1 in pull-down assays of CWEs from Col-0 and ccr1-3 (fig. S10C). To simplify the oligosaccharide profiling, the bound polysaccharides were first digested by EPG before LC-MS analysis. The results indicated that more pectic material with degree of polymerization (DP) between DP3 to DP20 was bound from the ccr1-3–derived CWE than from Col-0 and that GalA3 was the major product.
DISCUSSION
Together, our results suggest that FERONIA is essential for controlling the processing of cell walls for release of defense signals in response to lignin modification and that GalA3 is one resultant signal that can induce PR gene expression by binding with the WAK extracellular domain. However, pectic fractions of various sizes are known to be elicitor active (35), and the complexity of the present oligosaccharide profiles makes it hard to ascribe elicitor activity to any single moiety, especially as complex pectins with DPs from 3 to 20 are revealed in pull-down assays from ccr1-3 CWEs with the WAK OG-binding domain. Furthermore, treatment of ccr1-3 CWE with recombinant EPG from A. aculeatus destroys elicitor activity, whereas similar treatment with the endogenous EPG ADPG1 does not (14). This indicates that GalA3 cannot be the only elicitor but may be an epitope found on (and maybe a marker for) larger, and possibly more bioactive, pectic oligosaccharides released in a FERONIA-dependent process.
We do not suggest that GalA3 or oligosaccharides with related epitopes are necessarily the ligands in all WAK-mediated responses. Furthermore, OG signaling through WAKs appears quite complex. For example, it has been shown that PR1 is not induced in Arabidopsis by OGs up to 3 hours after application, whereas a FAD-linked oxidase that is a marker for OG-induced responses is induced (39). Induction of reactive oxygen species (ROS; an early response to OG signaling) is blocked in an Arabidopsis wak quintuple mutant that retains a potentially functional WAK4 extracellular domain, but induction of the FAD-linked oxidase is not suppressed (40). These results suggest that WAKs 1 to 5 may not be the only OG receptors in plants. Last, the previously described wak quintuple mutant is also defective in ROS induction in response to chitin and bacterial flagellin, suggesting a role beyond OG recognition (40). However, the requirement for ADPG1 for elicitor release and the loss of elicitor activity on digestion of ccr1-CWE preparations with commercial EPG (14) place the OG binding of WAKs in the signaling pathway between lignin modification and PR gene expression.
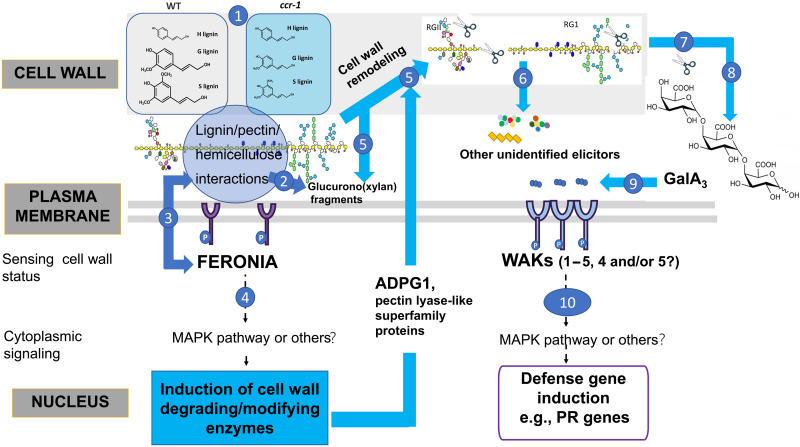
A conservative model based on our data in the present paper and in (14), and explained further in the model in Fig. 6, proposes that the reduced lignin content in secondary cell walls of the ccr1-3 mutant affects interactions with pectin and other wall polysaccharides and that these perturbations are sensed by FERONIA, leading to activated expression of a number of cell wall remodeling enzymes, including ADPG1 and pectin lyase superfamily members as shown by RNA-seq analysis (14). We suggest that the latter enzymes release pectic material (primary cell wall remodeling) that harbors latent elicitors, which require ADPG1 for their release in planta (secondary cell wall remodeling). It is likely that these molecules, which include GalA3, are heterogeneous but function through binding to WAK receptors.
Fig. 6. Model showing the involvement of FERONIA, ADPG1, GalA3, and WAK in the activation of defense responses following lignin modification.
The model is based on the data in the present paper and in (14). (1) Loss of function of CCR1 results in reduced lignin levels that disrupt lignin/pectin/hemicellulose interactions. (2) The altered wall structure in ccr1 results in release of wall-derived oligosaccharides such as (glucurono)xylan fragments. (3) FERONIA senses pectins or cell wall turgor to transduce the “status” of the cell wall. (4) By sensing the changes to the cell wall resulting from reduced lignin content, FERONIA activates a cytoplasmic signaling pathway that leads to transcriptional activation of a set of cell wall–degrading/modifying enzymes, including pectin lyases and ADPG1. (5 to 7) The cell wall–modifying enzymes partially degrade pectins to directly or indirectly release a range of oligosaccharides; these may come from pectin itself via the action of ADPG1 or pectin lyases or may include other wall components including (glucurono)xylans that are more accessible because of the reduced lignification of the secondary wall. (8) One or more pectic poly-/oligosaccharides are further cleaved by ADPG1 to release smaller OGs including GalA3. (9) GalA3 and related OGs are recognized by WAK receptor kinases. (10) WAKs transduce signals for the transcriptional activation of defense response genes such as PR1.
FERONIA has previously been implicated in binding to pectin and sensing wall softening, leading to the maintenance of cell wall integrity under high salt conditions that is disrupted in the fer-4 mutant (18). Our results now provide direct evidence for alterations in pectin composition and extractability directed by FERONIA in response to an endogenous, cell wall–localized signal, leading to release of elicitors that are perceived by WAK receptors. Many questions remain, however. These include understanding the relationship between FERONIA’s ability to sense lignin-mediated chemical and/or mechanical perturbations in the wall through its complex with pectin (18, 41) and its ability to coordinate expression of pectin-modifying enzymes to regulate cell wall integrity, determining the full spectrum of latent and active elicitor molecules generated as a result of lignin modifications, and understanding the exact roles of the redundant WAKs in PR gene induction in response to lignin modification. It is also possible that other receptors are involved in recognition of the perturbed cell wall status of ccr1-3. The the1-4 mutant we chose to analyze is a hypermorphic gain-of-function allele that encodes a truncated protein, lacking the kinase domain, and could trigger stress responses through the interaction with other membrane receptor kinases (42).
The signaling events immediately downstream of the FERONIA and WAK receptors will presumably involve kinase cascades, which require further study. Last, it will be important to determine the cellular localization of the triggering, signaling, and response events. Lignin is a feature of secondarily thickened cell walls, so the signaling is likely initiated there. ADPG1 was first associated with anther dehiscence, a process involving separation of lignified cell layers (43), so it is likely that the cell wall remodeling also takes place in the secondary walls of the vascular system. We cannot, however, rule out the generation of OG signals originating from remodeling of primary cell walls in response to initial changes in secondary cell walls. Answers to all these questions will facilitate balancing engineering polymers in plant cell walls in a way that does not overactivate or compromise plant defense.
MATERIALS AND METHODS
Plant materials and growth conditions
WT and mutant A. thaliana lines used in this study were obtained from the Arabidopsis Biological Resource Center (https://abrc.osu.edu). Detailed information is listed in table S1. Seeds were stratified on 1/2 strength Murashige and Skoog (MS) agar medium containing 1.5% sucrose at 4°C for 3 days and then transferred to a growth chamber set at 22°C under a 16-hour light/8-hour dark photoperiod (light intensity, 110 μmol m−2 s−1, supplied by both incandescent and fluorescent lights) for 10 days. The small seedlings were then transferred to soil (Metro-Mix 360, SUN GRO with controlled fertilizer: 20-10-20; 20% N, 10% P2O5, and 20% K2O at 200 ppm/gallon), and growth continued at 22°C under the same photoperiod. For crossing experiments, ccr1-3 was used as male parent, and the other mutants were used as female parents. All mutants were in the A. thaliana Columbia ecotype background. Primer pairs for genotyping mutant alleles are listed in table S2. Analyses of cell wall composition, extractability, and transcript levels were performed on inflorescence stems of 8-week-old plants. The siliques, leaves, and apical parts of the stem were removed, leaving the basal lignified parts for preparation of cell wall extracts.
Elicitation assays
The elicitor activity of CWEs and pectic compounds was determined with a leaf injection assay as previously described in (14). The assays used 0.1 mg uronic acid equivalents/ml CWEs, and treated leaves were harvested after 24 hours. For testing elicitor activity of pectic compounds, 0.05 mg uronic acid equivalents/ml elicitors were used, and treated leaves were harvested after 6 hours for RNA extraction.
RNA-seq analysis
RNA was extracted from the main inflorescence stems of 2-month-old WT Col-0, ccr1-3, fer-4, and ccr1-3/fer-4 double mutant plants. Total RNA was isolated with TRIzol reagent (Invitrogen, USA) and treated with the TURBO DNA-free Kit (Ambion) to remove genomic DNA contaminants. RNA was cleaned using an RNA cleanup kit (Qiagen). Purified RNA was quantified using a NanoDrop ND-100 spectrophotometer (NanoDrop Technologies) and evaluated for purity with an Agilent 4200 TapeStation system (Agilent Technologies). Three biological replicates for each sample were used for RNA-seq. Libraries were constructed using a TruSeq stranded mRNAseq prep kit following the standard protocols and ran on an Illumina Nextseq 500 platform [2 × 75 base pairs (bp)].
For each sample, RNA-seq raw reads were trimmed and filtered using in-house perl scrips as described previously (44). Clean reads were mapped to the A. thaliana genome sequence and the reference-annotated genes (TAIR10) using Bowtie v2.3.2.0 (45) and TopHat v2.1.1 (46) with default parameters. The gene expression levels for each sample were estimated by Cufflinks v2.2.1 (47) based on the FPKM (fragments per kilobase of exon model per million mapped fragments) method. Significantly differentially expressed genes were analyzed by Cuffdiff in the cufflinks package (48) using twofold change and multiple test P value of <0.05. The GO analysis was performed on differentially expressed genes by AgriGO (49, 50).
Quantitative real-time polymerase chain reaction
Total RNA was extracted as described above. The first-strand complementary DNA (cDNA) was synthesized using the High-Capacity cDNA Reverse Transcription Kit (Thermo Fisher Scientific). The 10× diluted cDNA samples were used as templates, and quantitative real-time polymerase chain reaction (qRT-PCR) was performed with a QuantStudio 6 Flex system (Applied Biosystems) using PowerUp SYBR Green Master Mix (Thermo Fisher Scientific). qRT-PCR analysis was performed as described previously (51). Gene-specific primers are listed in table S2.
Determination of lignin and pectin
Mature inflorescence stems were harvested from 2-month-old A. thaliana WT Col-0, and mutant plants of ccr1-3, fer-4, ccr1-3/fer-4, wak1/2/3/4/5, and wak1/2/3/4/5/ccr1-3. The alcohol-insoluble cell wall residues (AIRs) were prepared as previously described (52). The lignin monomer content and composition were determined by an optimized thioacidolysis method as described in (53). Extraction of water-soluble elicitors from AIRs was performed as described previously (14) with the following minor modifications; after 24 hours of incubation of AIR with water at room temperature, the extracts were filtered through a 0.22-μm filter (Millipore). After that, the filtered extracts were either lyophilized for further storage or directly used for elicitation assay. The pectin in AIRs and pectic material released by cold-water extraction were determined by the m-hydroxydiphenyl method in the form of uronic acid (54). For each measurement, three independent biological replicates were analyzed.
CRISPR-Cas9 vector construction and generation of transgenic lines
To make the wak1/2 double mutant, sgRNA was designed to target the third exons of WAK1 and WAK2 at the same time. To generate the wak1/2/3/4/5 quintuple mutant, the sgRNA was designed to target the third exons of all the five WAKs. The sgRNAs were designed using the software Geneious R7 and cloned into the pCambia1300-35S-Cas9 plasmid after the AtU6 promoter. Transformation of A. thaliana was performed by the floral dip method (55). Transgenic plants were isolated on 1/2 MS medium containing hygromycin (50 μg/ml), and positive seedlings were transferred to soil. The homozygous double or quintuple mutants in the T4 generation of transgenic plants were used for further analysis. Sanger sequencing was used for genotyping of the mutant alleles. DNA was isolated from 3-week-old A. thaliana leaves and was amplified using a forward PCR primer 500 bp upstream and a reverse primer 400 bp downstream of the sgRNA target site. The homozygous and biallele mutation results were analyzed using the method previously described in (56). All primers are listed in table S2.
Digestion of CWEs by ADPG1
The expression and purification of recombinant ADPG1 protein were performed as described in (14). The CWEs of WT and mutant A. thaliana plants were prepared as described above and lyophilized. One-milligram batches of lyophilized CWEs were weighed and incubated with 1.5 μg of recombinant ADPG1 protein in 500 μl of 20 mM sodium acetate buffer (pH 5.0) at 37°C for 1.5 hours. An amount of 1.5 units of the commercial EPG M2 from A. aculeatus (Megazyme) was incubated with CWEs in parallel with recombinant ADPG1 protein as the positive control. After incubation, an equal volume of absolute ethanol was added to stop reactions. After centrifuging at 5000g for 10 min, the supernatants were collected and either dried in a speed vacuum concentrator to prepare samples for oligosaccharide profiling or lyophilized for use in elicitation assays.
Oligosaccharide profiling and analysis
The oligosaccharides present in CWEs or released upon digestion were separated and analyzed according to (57). The chromatographic separation was performed on an ACQUITY UPLC Protein BEH SEC Column (125A, 1.7 μm, 4.6 mm × 300 mm; Waters Corporation, Milford, MA, USA) coupled with BEH SEC Guard Column (125A, 1.7 μm, 4.6 mm × 30 mm).
Expression of recombinant WAK1 OG-binding domain in E. coli
A 651-bp cDNA sequence encoding amino acids 67 to 254 containing the galacturonan binding subdomain of the extracellular domain of A. thaliana WAK1 (58) was amplified and fused with an X-press tag peptide (DLYDDDDK) at the N terminus. To construct this recombinant protein, a 24-bp DNA sequence encoding the X-press peptide was synthesized into the PCR primer and fused with the 651-bp WAK1 sequencing in frame. The construct was cloned into pGEX-6p-1 vector to provide a GST tag at the N terminus of the expressed protein. The resulting plasmid was transformed into E. coli strain Origami 2(DE3) pLysS cells (Novagen). Recombinant protein expression was induced with isopropyl-β-d-thiogalactopyranoside (IPTG) (0.5 mM) at 28°C for 5 hours. After induction, soluble recombinant protein was extracted and purified using Glutathione Sepharose 4B (Cytiva) according to the manufacturer’s protocol. Protein purification was verified by SDS–polyacrylamide gel electrophoresis (PAGE) and anti–X-press tag Western blot, and protein concentrations were determined using the Bradford Protein assay (Bio-Rad). After verification, purified protein was desalted and concentrated using Amicon Ultra4 centrifugal filters (Sigma-Aldrich) against corresponding buffer used in the binding tests.
Enzyme-linked immunosorbent assays
ELISA was performed as described (35) with modifications. The Maxisorp microplates were pretreated with polylysine-HBr (50 μg/ml in H20, 50 μl per well) for 1 hour at 25°C. The polylysine-HBr was then discarded, and the wells were washed five times with 50 μl per well of tris-NaCl buffer (20 mM tris-HCl and 150 mM NaCl, pH 8.2). CaCl2 was not included in the buffer to restrict the interaction of recombinant WAK1 OG-binding domain protein with long-chain pectin homogalacturonans. The microplates were then coated with 20 μg of PGA or GalA3 dissolved in tris-NaCl buffer overnight at 4°C. After coating, the wells were washed five times with 50 μl per well of tris-NaCl buffer. The wells were then blocked with 50 μl per well of 3% skimmed milk for 2 hours at 25°C, and the wash step was repeated three times after removing the blocking buffer. The recombinant WAK1 OG-binding domain protein (100 ng/μl in tris-NaCl buffer with 10 mM dithiothreitol) was added to the wells in increments, and the GST tag protein was used as the negative control. The wells were washed and incubated with 1:1000 diluted anti-Xpress primary antibody (Invitrogen) in tris-NaCl buffer containing 1% skimmed milk for 1 hour at 25°C. The primary antibody was removed, and the wells were washed 20 times. Then, 1:10,000 diluted horseradish peroxidase anti-mouse secondary antibody in tris-NaCl buffer containing 1% skimmed milk was added and the wells were shaken for 1 hour at 25°C. The secondary antibody was removed, and the wash step was repeated 20 times. Last, working solution (50 μl per well) of SuperSignal ELISA Femto maximum sensitivity substrate (Thermo Fisher Scientific) was added. The liquid in the wells was mixed for 1 min using a microplate mixer, and then a Synergy Mx luminometer (BioTeK) was used to measure relative light units at 425 nm for between 1 and 5 min.
WAK1 pull-down assays
Fifty milliliters of E. coli cells containing recombinant WAK1 OG-binding domain protein (around 125 μg of GST fusion proteins) was collected and broken down by sonication in 15 ml of 20 mM tris and 150 mM NaCl buffer (pH 8.0). Then, the cell lysate was incubated with 1.5 ml of balanced Glutathione Sepharose 4B (Cytiva) for 2 hours at 4°C. The beads were then washed three times with 10 ml of tris-NaCl buffer. Next, recombinant WAK1 OG-binding domain–GST beads were divided into three equal portions. The portions were incubated overnight at 4°C with either 1 ml of Col-0 CWE (1 mg/ml), 1 ml of ccr1-3 CWE (1 mg/ml) dissolved in tris-NaCl buffer, or 1 ml of tris-NaCl buffer alone. The concentration of CWE was calculated as the total weight of freeze-dried powder/volume. As a control, 0.5 ml of balanced GST beads was incubated with 1 ml of ccr1-3 CWE (1 mg/ml). After binding, the beads were washed five times with 2 ml of water. The beads were then treated with 1 ml of 70% ethanol for 2 hours at room temperature. The supernatant was collected and dried in a speed vacuum concentrator, and the dried samples were then used in the analysis of bound oligosaccharide fragments.
Statistical analysis
In general, data are represented by means ± SD of biological triplicates. Statistical significance was assessed using either Student’s t test or one-way analysis of variance (ANOVA).
Acknowledgments
We thank Y. Mao, Shanghai Center for Plant Stress Biology (PSC), for providing the pCambia1300-Cas9 Vector; T. Kwon, Genomic Center of University of North Texas, for library construction and RNA sequencing; and B. De Meester and W. Boerjan, Center for Plant Systems Biology, Vlaams Instituut voor Biotechnologie, 9052 Ghent, Belgium, for providing the Arabidopsis ccr1-3 mutant.
Funding: This work was funded by the University of North Texas. We acknowledge the support of IJPB’s Plant Observatory technological platforms. The IJPB benefits from the support of Saclay Plant Sciences-SPS (ANR-17-EUR-0007).
Author contributions: R.A.D. and C.L. conceived the study. C.L., H.Y., and A.V. performed experiments. C.L., H.Y., A.V., X.R., and R.A.D. analyzed data. R.A.D. and C.L. wrote the paper.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: RNA-seq data are deposited in the GenBank GEO under the accession number GSE210734. All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials.
Supplementary Materials
This PDF file includes:
Figs. S1 to S10
Tables S1 and S2
Legend for dataset S1
Other Supplementary Material for this : manuscript includes the following:
Dataset S1
REFERENCES AND NOTES
- 1.A. J. Ragauskas, G. T. Beckham, M. J. Biddy, R. Chandra, F. Chen, M. F. Davis, B. H. Davison, R. A. Dixon, P. Gilna, M. Keller, P. Langan, A. K. Naskar, J. N. Saddler, T. J. Tschaplinski, G. A. Tuskan, C. E. Wyman, Lignin valorization: Improving lignin processing in the biorefinery. Science 344, 1246843 (2014). [DOI] [PubMed] [Google Scholar]
- 2.S. Sesupathy, G. M. Morales, L. Gao, H. Wang, B. Yang, J. Jiang, J. Sun, D. Zhu, Lignin valorization: Status, challenges and opportunities. Bioresour. Technol. 347, 126696 (2022). [DOI] [PubMed] [Google Scholar]
- 3.F. Chen, R. A. Dixon, Lignin modification improves fermentable sugar yields for biofuel production. Nat. Biotechnol. 25, 759–761 (2007). [DOI] [PubMed] [Google Scholar]
- 4.X. Li, J.-K. Weng, C. Chapple, Improvement of biomass through lignin modification. Plant J. 54, 569–581 (2008). [DOI] [PubMed] [Google Scholar]
- 5.A. Chanoca, L. de Vries, W. Boerjan, Lignin engineering in forest trees. Front. Plant Sci. 10, 912 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.C. Halpin, Lignin engineering to improve saccharification and digestibility in grasses. Curr. Opin. Biotechnol. 56, 223–229 (2019). [DOI] [PubMed] [Google Scholar]
- 7.F. Muro-Villanueva, X. Mao, C. Chapple, Linking phenylpropanoid metabolism, lignin deposition, and plant growth inhibition. Curr. Opin. Biotechnol. 56, 202–208 (2019). [DOI] [PubMed] [Google Scholar]
- 8.C. Hernández-Blanco, D. X. Feng, J. Hu, A. Sánchez-Vallet, L. Deslandes, F. Llorente, M. Berrocal-Lobo, H. Keller, X. Barlet, C. Sánchez-Rodríguez, L. K. Anderson, S. Somerville, Y. Marco, A. Molina, Impairment of cellulose synthases required for Arabidopsis secondary cell wall formation enhances disease resistance. Plant Cell 19, 890–903 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.C. M. Ha, D. Fine, A. Bahtia, X. Rao, M. Z. Martin, N. Engle, D. J. Wherritt, T. J. Tschaplinski, L. W. Sumner, R. A. Dixon, Ectopic defense gene expression is associated with growth defects in Medicago truncatula lignin pathway mutants. Plant Physiol. 181, 63–84 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.C. M. Ha, X. Rao, G. Saxena, R. A. Dixon, Growth-defense trade-offs and yield loss in plants with engineered cell walls. New Phytol. 231, 60–74 (2021). [DOI] [PubMed] [Google Scholar]
- 11.N. G. Lewis, E. Yamamoto, Lignin: Occurrence, biogenesis and biodegradation. Annu. Rev. Plant Biol. 41, 455–496 (1990). [DOI] [PubMed] [Google Scholar]
- 12.J. Barros-Rios, S. Temple, R. A. Dixon, Development and commercialization of reduced lignin alfalfa. Curr. Opin. Biotechnol. 56, 48–54 (2018). [DOI] [PubMed] [Google Scholar]
- 13.L. Gallego-Giraldo, S. Pose-Albacete, S. Pattathil, A. G. Peralta, M. Hahn, B. G. Ayre, J. Sunuwar, J. Hernandez, M. Patel, J. Shah, X. Rao, J. P. Knox, R. A. Dixon, Elicitors and defense gene induction in plants with altered lignin compositions. New Phytol. 219, 1235–1251 (2018). [DOI] [PubMed] [Google Scholar]
- 14.L. Gallego-Giraldo, C. Liu, S. Pose-Albacete, S. Pattathil, A. G. Peralta, J. Young, J. Westphaling, M. Hahn, X. Rao, B. de Meester, J. P. Knox, W. Boerjan, R. A. Dixon, ARABIDOPSIS DEHISCENCE ZONE POLYGALACTURONASE 1 (ADPG1) releases latent defense signals in stems with reduced lignin content. Proc. Natl. Acad. Sci. U.S.A. 117, 3281–3290 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Q. Zhao, R. A. Dixon, Altering the cell wall and its impact on plant disease: From forage to bioenergy. Annu. Rev. Phytopathol. 52, 62–91 (2014). [DOI] [PubMed] [Google Scholar]
- 16.S. Wolf, Cell wall signaling in plant development and defense. Annu. Rev. Plant Biol. 73, 323–353 (2022). [DOI] [PubMed] [Google Scholar]
- 17.S. Wolf, Plant cell wall signalling and receptor-like kinases. Biochem. J. 474, 471–492 (2017). [DOI] [PubMed] [Google Scholar]
- 18.W. Feng, D. Kita, A. Peaucelle, H. N. Cartwright, V. Doan, Q. Duan, M. C. Liu, J. Maman, L. Steinhorst, I. Schmitz-Thom, R. Yvon, J. Kudla, H. M. Wu, A. Y. Cheung, J. R. Dinneny, The FERONIA receptor kinase maintains cell-wall integrity during salt stress through Ca2+ signaling. Curr. Biol. 28, 666–675.e5 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.X. Kang, A. Kirui, M. C. Dickwella Widanage, F. Mentink-Vigier, D. J. Cosgrove, T. Wang, Lignin-polysaccharide interactions in plant secondary cell walls revealed by solid-state NMR. Nat. Commun. 10, 347 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.D. Tarasov, M. Leitch, P. Fatehi, Lignin–carbohydrate complexes: Properties, applications, analyses, and methods of extraction: A review. Biotechnol. Biofuels 11, 269 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.B. D. Kohorn, M. Kobayashi, S. Johansen, J. Riese, L. F. Huang, K. Koch, S. Fu, A. Dotson, N. Byers, An Arabidopsis cell wall-associated kinase required for invertase activity and cell growth. Plant J. 46, 307–316 (2006). [DOI] [PubMed] [Google Scholar]
- 22.H. Guo, H. Ye, L. Li, Y. Yin, A family of receptor-like kinases are regulated by BES1 and involved in plant growth in Arabidopsis thaliana. Plant Signal. Behav. 4, 784–786 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.H. Guo, L. Li, H. Ye, X. Yu, A. Algreen, Y. Yin, Three related receptor-like kinases are required for optimal cell elongation in Arabidopsis thaliana. Proc. Natl. Acad. Sci. U.S.A. 106, 7648–7653 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.S.-L. Xu, A. Rahman, T. I. Baskin, J. J. Kieber, Two leucine-rich repeat receptor kinases mediate signaling, linking cell wall biosynthesis and ACC synthase in Arabidopsis. Plant Cell 20, 3065–3079 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.L. Bai, G. Zhang, Y. Zhou, Z. Zhang, W. Wang, Y. Du, Z. Wu, C.-P. Song, Plasma membrane-associated proline-rich extensin-like receptor kinase 4, a novel regulator of Ca2+ signalling, is required for abscisic acid responses in Arabidopsis thaliana. Plant J. 60, 314–327 (2009). [DOI] [PubMed] [Google Scholar]
- 26.A. Boisson-Dernier, S. Roy, K. Kritsas, M. A. Grobei, M. Jaciubek, J. I. Schroeder, U. Grossniklaus, Disruption of the pollen-expressed FERONIA homologs ANXUR1 and ANXUR2 triggers pollen tube discharge. Development 136, 3279–3288 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Q. Duan, M. J. Liu, D. Kita, S. S. Jordan, F. J. Yeh, R. Yvon, H. Carpenter, A. N. Federico, L. E. Garcia-Valencia, S. J. Eyles, C. S. Wang, H. M. Wu, A. Y. Cheung, FERONIA controls pectin- and nitric oxide-mediated male-female interaction. Nature 579, 561–566 (2020). [DOI] [PubMed] [Google Scholar]
- 28.Q. Duan, D. Kita, C. Li, A. Y. Cheung, H.-M. Wu, FERONIA receptor-like kinase regulates RHO GTPase signaling of root hair development. Proc. Natl. Acad. Sci. U.S.A. 107, 17821–17826 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.S. Schoenaers, D. Balcerowicz, A. Costa, K. Vissenberg, The kinase ERULUS controls pollen tube targeting and growth in Arabidopsis thaliana. Front. Plant Sci. 8, 1942 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.M. Mir Derikvand, J. B. Sierra, K. Ruel, B. Pollet, C.-T. Do, J. Thévenin, D. Buffard, L. Jouanin, C. Lapierre, Redirection of the phenylpropanoid pathway to feruloyl malate in Arabidopsis mutants deficient for cinnamoyl-CoA reductase 1. Planta 227, 943–956 (2008). [DOI] [PubMed] [Google Scholar]
- 31.H. Liao, R. Tang, X. Zhang, S. Luan, F. Yu, FERONIA receptor kinase at the crossroads of hormone signaling and stress responses. Plant Cell Physiol. 58, 1143–1150 (2017). [DOI] [PubMed] [Google Scholar]
- 32.H. Guo, T. M. Nolan, G. Song, S. Liu, Z. Xie, J. Chen, P. S. Schnable, J. W. Walley, Y. Yin, FERONIA receptor kinase contributes to plant immunity by suppressing jasmonic acid signaling in Arabidopsis thaliana. Curr. Biol. 28, 3316–3324.e6 (2018). [DOI] [PubMed] [Google Scholar]
- 33.A. Malivert, Ö. Erguvan, A. Chevallier, A. Dehem, R. Friaud, M. Liu, M. Martin, T. Peyraud, O. Hamant, S. Verger, FERONIA and microtubules independently contribute to mechanical integrity in the Arabidopsis shoot. PLOS Biol. 19, e3001454 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.A. Voxeur, J. Sechet, S. Vernhettes, Inositolphosphate glycans and a fucosylated xyloglucan oligosaccharide are accumulated upon Arabidopsis thaliana/ Botrytis cinerea infection. bioRxiv 2021.06.25.449971 [Preprint]. 9 July 2021. 10.1101/2021.06.25.449971. [DOI]
- 35.A. Decreux, J. Messiaen, Wall-associated kinase WAK1 interacts with cell wall pectins in a calcium-induced conformation. Plant Cell Physiol. 46, 268–278 (2005). [DOI] [PubMed] [Google Scholar]
- 36.V. Kanneganti, A. K. Gupta, Wall associated kinases from plants - An overview. Physiol. Mol. Biol. Plants 14, 109–118 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.B. D. Kohorn, S. L. Kohorn, The cell wall-associated kinases, WAKs, as pectin receptors. Front. Plant Sci. 3, 88 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.A. Decreux, A. Thomas, B. Spies, R. Brasseur, P. Cutsem, J. Messiaen, In vitro characterization of the homogalacturonan-binding domain of the wall-associated kinase WAK1 using site-directed mutagenesis. Phytochemistry 67, 1068–1079 (2006). [DOI] [PubMed] [Google Scholar]
- 39.C. Denoux, R. Galletti, N. Mammarella, S. Gopalan, D. Werck, G. De Lorenzo, S. Ferrari, F. M. Ausubel, J. Dewdney, Activation of defense response pathways by OGs and Flg22 elicitors in Arabidopsis seedlings. Mol. Plant 1, 423–445 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.B. D. Kohorn, B. E. Greed, G. Mouille, S. Verger, S. L. Kohorn, Effects of Arabidopsis wall associated kinase mutations on ESMERALDA1 and elicitor induced ROS. PLOS ONE 16, e0251922 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.W. Tang, W. Lin, X. Zhou, J. Guo, X. Dang, B. Li, D. Lin, Z. Yang, Mechano-transduction via the pectin-FERONIA complex activates ROP6 GTPase signaling in Arabidopsis pavement cell morphogenesis. Curr. Biol. 32, 508–517.e3 (2022). [DOI] [PubMed] [Google Scholar]
- 42.D. Merz, J. Richter, M. Gonneau, C. Sanchez-Rodriguez, T. Eder, R. Sormani, M. Martin, K. Hématy, H. Höfte, M. T. Hauser, T-DNA alleles of the receptor kinase THESEUS1 with opposing effects on cell wall integrity signaling. J. Exp. Bot. 68, 4583–4593 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.M. Ogawa, P. Kay, S. Wilson, S. M. Swain, ARABIDOPSIS DEHISCENCE ZONE POLYGALACTURONASE1 (ADPG1), ADPG2, and QUARTET2 are polygalacturonases required for cell separation during reproductive development in Arabidopsis. Plant Cell 21, 216–233 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.X. Rao, N. Krom, Y. Tang, T. Widiez, D. Havkin-Frenkel, F. C. Belanger, R. A. Dixon, F. Chen, A deep transcriptomic analysis of pod development in the vanilla orchid (Vanilla planifolia). BMC Genomics 15, 964 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.B. Langmead, S. L. Salzberg, Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.D. Kim, G. Pertea, C. Trapnell, H. Pimentel, R. Kelley, S. L. Salzberg, TopHat2: Accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 14, R36 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.C. Trapnell, B. A. Williams, G. Pertea, A. Mortazavi, G. Kwan, M. J. van Baren, S. L. Salzberg, B. J. Wold, L. Pachter, Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 28, 511–515 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.C. Trapnell, D. G. Hendrickson, M. Sauvageau, L. Goff, J. L. Rinn, L. Pachter, Differential analysis of gene regulation at transcript resolution with RNA-seq. Nat. Biotechnol. 31, 46–53 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.T. Tian, Y. Liu, H. Yan, Q. You, X. Yi, Z. du, W. Xu, Z. Su, agriGO v2. 0: A GO analysis toolkit for the agricultural community, 2017 update. Nucleic Acids Res. 45, W122–W129 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Z. Du, X. Zhou, Y. Ling, Z. Zhang, Z. Su, agriGO: A GO analysis toolkit for the agricultural community. Nucleic Acids Res. 38, W64–W70 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.C. Liu, H. Yu, X. Rao, L. Li, R. A. Dixon, Abscisic acid regulates secondary cell-wall formation and lignin deposition in Arabidopsis thaliana through phosphorylation of NST1. Proc. Natl. Acad. Sci. U.S.A. 118, e2010911118 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.L. Gallego-Giraldo, Y. Jikumaru, Y. Kamiya, Y. Tang, R. A. Dixon, Selective lignin downregulation leads to constitutive defense response expression in alfalfa (Medicago sativa L.). New Phytol. 190, 627–639 (2011). [DOI] [PubMed] [Google Scholar]
- 53.F. Chen, C. Zhuo, X. Xiao, T. H. Pendergast, K. M. Devos, A rapid thioacidolysis method for biomass lignin composition and tricin analysis. Biotechnol. Biofuels 14, 18 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.L. D. Melton, B. G. Smith, Determination of the uronic acid content of plant cell walls using a colorimetric assay. Curr. Protoc. Food Anal. Chem. , E3.3.1–E3.3.4 (2001). [Google Scholar]
- 55.S. J. Clough, A. F. Bent, Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743 (1998). [DOI] [PubMed] [Google Scholar]
- 56.H. Yu, C. Liu, R. A. Dixon, A gene-editing/complementation strategy for tissue-specific lignin reduction while preserving biomass yield. Biotechnol. Biofuels 14, 175 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.A. Voxeur, O. Habrylo, S. Guénin, F. Miart, M. C. Soulié, C. Rihouey, C. Pau-Roblot, J. M. Domon, L. Gutierrez, J. Pelloux, G. Mouille, M. Fagard, H. Höfte, S. Vernhettes, Oligogalacturonide production upon Arabidopsis thaliana–Botrytis cinerea interaction. Proc. Natl. Acad. Sci. U.S.A. 116, 19743–19752 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.A. Marchler-Bauer, Y. Bo, L. Han, J. He, C. J. Lanczycki, S. Lu, F. Chitsaz, M. K. Derbyshire, R. C. Geer, N. R. Gonzales, M. Gwadz, D. I. Hurwitz, F. Lu, G. H. Marchler, J. S. Song, N. Thanki, Z. Wang, R. A. Yamashita, D. Zhang, C. Zheng, L. Y. Geer, S. H. Bryant, CDD/SPARCLE: Functional classification of proteins via subfamily domain architectures. Nucleic Acids Res. 45, D200–D203 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S10
Tables S1 and S2
Legend for dataset S1
Dataset S1