Abstract
Background
Tumor-derived exosomes are involved in the process of tumor metastasis and angiogenesis. MicroRNAs (miRNAs) are the most widely investigated factors in exosomes. Therefore, we hope to find a new therapeutic target in bladder cancer (BLCA), which has high incidence rate and mortality.
Methods
Exosomal microRNA(miR)-93-5p expression level, downstream target molecules, and biological functions were examined with bioinformatics technology. Exosomes were extracted by sequential differential centrifugation and verified by transmission electron microscopy. The exosomal miR-93-5p on cell proliferation, invasion, and angiogenesis abilities in 5637 and T24 cells was determined by Cell Counting Kit 8 (CCK-8), colony-forming assay, Transwell assay, and vascular ring formation assay. A mouse xenograft model with intratumor injection was adopted to evaluate the correlation between BLCA-derived exosomes and tumor growth in vivo.
Results
The results revealed that exosomes play an important role in the biological progression of BLCA, with miR-93-5p being a particularly important molecule. Compared to normal cells, more malignant cells release more exosomal miR-93-5p, and tumor-derived exosomal miR-93-5p could significantly promote cell proliferation, invasion, and angiogenesis in vitro and in vivo. We identified phosphatase and tensin homolog (PTEN) as the most significant target of miR-93-5p in BLCA and human umbilical vein endothelial cells.
Conclusions
Our study successfully revealed the biological role and mechanism of BLCA-derived exosomes in tumor progression. Target at tumor exosomes and exosomal miR-93-5p may be an effective treatment in BLCA.
Keywords: Exosomes, tumor microenvironment, miR-93-5p, bladder cancer, angiogenesis
Highlight box.
Key findings
• Our study revealed that the exosomes are crucial in the progression of bladder cancer (BLCA). Among them, exosomal microRNA(miR)-93-5p was proven to be a key oncogene.
What is known and what is new?
• Previous studies have found that miR-93-5p can promote the growth and metastasis of many cancers. However, the relation of miR-93-5p to exosomes in BLCA is unclear.
• Our study revealed that exosomal miR-93-5p was a predictive index for the diagnosis of BLCA. In vitro and in vivo experiments showed that miR-93-5p can accelerate the proliferation, metastasis and angiogenesis of BLCA. Mechanically, phosphatase and tensin homolog (PTEN) may be a vital downstream effector of exosomal miR-93-5p.
What is the implication, and what should change now?
• Our study further revealed that exosomes may be an important intervention tool for the treatment of BLCA and that miR-93-5p is a potential target for diagnosis and treatment.
Introduction
Bladder cancer (BLCA) is the most diagnosed cancer of the urinary system. In recent years, the incidence and mortality rate of BLCA have increased gradually, with more than 440,000 new cases and 130,000 deaths being recorded in 2020 (1). Although the treatment of surgery combined with chemotherapy diagnostic has continuously improved for BLCA, the validity and specificity of this are limited, and most tumors will recur and clinically progress (2). More effective treatments and targets urgently need to be identified and developed to better diagnose and cure BLCA.
The tumor microenvironment is significantly affected in the occurrence and development of malignant tumors, and the regulation of the microenvironment affects tumor proliferation, metastasis, and microvascular formation (3). Exosomes are members of the extracellular vesicles, with a diameter of 30–100 nm. Exosomes are released by cancer cells and have a key role in communication in the tumor microenvironment, including promoting the tumor angiogenesis and tumor associated macrophage infiltration, etc., which could promote tumor progression (4). Study revealed that cancer cells generally produce more exosomes than normal cell. Importantly, the proportion of cancer suppressor genes and oncogenes in tumor-derived exosomes is out of balance, which promotes the growth and metastasis of tumors and inhibit the immune response of the body (5). Research showed that lots of exosomes transmit oncogenic information, like microRNA (miRNA), into targeted cancer cells to promote the progression of tumor (6,7). miRNA, which is endogenous noncoding RNA 18–22 nt in length, regulates gene expression at the posttranscriptional level by binding to the 3' untranslated region (3’UTR) region of the target gene (8). A large number of studies have shown that exosome derived miRNA regulates tumor proliferation, metastasis, and angiogenesis by silencing target genes (9,10). miR-93-5p has been noted as having a critical role in the occurrence and development of many tumors and has been associated with the clinical prognosis of patients with cancer (11-14). miR-93-5p was found to be increased in urinary exosomes of patients with BLCA and is considered to be a sensitive biomarker in BLCA (15). In our research, we clarified its effect on tumor cells and the surrounding vascular endothelial cells in relation to BLCA cell exosomes using in vitro and in vivo experiments. Exosomal miR-93-5p was found to be critically involved in tumor promotion, and the overexpression of BLCA cell exosomal miR-93-5p was positively correlated with the degree of malignancy. This may explain the increased pathological grade during cancer progression in BLCA. The presence of miR-93-5p in exosomes may be an important clinical indicator for patients with BLCA. We present the following article in accordance with the ARRIVE reporting checklist (available at https://tau.amegroups.com/article/view/10.21037/tau-22-872/rc).
Methods
Clinical specimens
A total of 25 paired BLCA and adjacent normal tissues samples were collected from patients diagnosed with BLCA at The Second Affiliated Hospital of Soochow University. All patients underwent transurethral resection or radical bladder resection, and the postoperative pathology was urothelial carcinoma. None of the patients received radiotherapy or chemotherapy. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). This study was approved by ethical committee of the Second Affiliated Hospital of Soochow University (ethical code: JD-LK-2019-104-01) and informed consent was taken from all the patients.
Cell lines and cell culture
The cell lines used in this experiment were purchased from the Cancer Institute at the Chinese Academy of Medical Sciences. Human bladder urothelial carcinoma cell lines 5637, T24, SW780, and J82 were respectively cultured in RPMI 1640 (Procell), McCoy’s 5A (Procell), L-15 (Procell), and Dulbecco’s Modified Eagle’s Medium (DMEM)/F12 (Procell) medium supplemented with 10% fetal bovine serum (FBS; Gibco, Thermo Fisher Scientific). Human umbilical vein endothelial cells (HUVECs) were also cultured in DMEM/F12 medium. T24, 5637, and J82 and were cultured in an incubator at 37 ℃ with 5% CO2, while SW780 incubation did not require extra CO2.
RNA extraction and real-time polymerase chain reaction
TRIzol was used to extract total RNA from cells and crushed tissues. Quantitative reverse transcription and real-time polymerase chain reaction (qRT-PCR) of miRNA and messenger RNA (mRNA) were performed using a reverse transcription kit (Takara Bio). U6 and GAPHD were used as internal controls for qRT-PCR quantification. The 2-ΔΔCt formula (where Ct is the cycle threshold) was used to calculate the relative expression of RNA. The primers were designed as follows in Table 1.
Table 1. Sequences of the primers used in qPCR.
| Gene | Sense primer | Anti-sense primer |
|---|---|---|
| GAPDH | AGAAGGCTGGGGCTCATTTG | AGGGGCCATCCACAGTCTTC |
| PTEN | GAGGGCCAGGTCATAAATAA | ACCATAAAATGTAAGCAAGGC |
| U6 | CGAGCACAGAATCGCTTCA | CTCGCTTCGGCAGCACATAT |
| miR-93-5p | CGCAAAGTGCTGTTCGTGC | AGTGCAGGGTCCGAGGTATT |
qPCR, real-time quantitative polymerase chain reaction; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; PTEN, phosphatase and tensin homolog.
Western blotting
Protein was extracted from T24 and 5637 cells, which were transfected by miR-93-5p-exosomes and negative controls (NC) for 36 hours (h). Protein was separated by 10% SDS-PAGE and then transferred to polyvinylidene fluoride (PVDF) membrane. After being blocked with fetal bovine serum, strips were incubated overnight in the primary antibody solution at 4 ℃. The primary antibodies were as follows: PTEN (1:1000, Abcam), TSG101 (1:500, Abcam), CD63 (1:600, Abcam). Strips with protein were the incubated with secondary horseradish peroxidase (HRP)-labeled antibody for 2 h at room temperature. An enhanced chemiluminescent kit and imaging system were used for protein quantification.
Cell proliferation assays
Cell proliferation experiment was carried out with Cell Counting Kit 8 (CCK-8). After the cells were seeded into the 6-well plate, they were successfully transfected with exosomes and then replanted in a 96-well plate at a density of 2,000 cells/well. On the first, second, third, and fourth day, 10 µL of CCK8 reagent was added into each well. Cells were incubated at 37 ℃ for 2 h, after which the absorbance was determined with an enzyme-linked immunosorbent assay (ELISA) reader at a wavelength of 450 nm.
Transwell assay
The Matrigel matrix (Corning) was dissolved at 4 ℃ and diluted in serum-free RPMI-1640 (volume ratio 1:8). Following this, 50 µL of the mixed solution above was added to the chamber, and then incubated at 4 ℃ for 2 h and 37 ℃ for 1 h to solidify the solution. After transfection with exo-miR-93-5p or negative control, the cells were resuspended in FBS-free medium, and 100,000 cells were seeded in the upper chamber of a 24-well plate while the lower chamber of the 24-well plate was filled with 20% FBS serum medium. After 24 h of incubation, a cotton swab was used to wipe the cells inside the chamber, and the cells were then fixed in methanol solution for 30 min. Before observation and photographing, 0.1% crystal violet solution was used for cell staining.
Vascular ring formation assay
Matrigel (60 µL; Corning) was added into a 96-well plate which was placed into incubator for 30 min. After being resuspended in 10% medium, cell density of HUVECs were adjusted to 100,000/mL. Following this, 10 µL of the cell suspension above was added to a 96-well plate, and tube formation of the cells was observed after 4 h. The number of nodes and length of branches were taken to represent the angiogenesis ability of cells in vitro. Four nonrepetitive fields were randomly selected and counted under a low power microscope (100×). ImageJ (National Institutes of Health) was used to count the length of branches and the number of nodes formed by the HUVECs.
Bioinformatic analysis
OncomiR (http://www.oncomir.org/) is an online resource for exploring miRNA dysregulation in cancer. We used it for analysis and downloaded the clinically significant differential genes. The ENCORI Pan-Cancer Analysis Platform (starbase.sysu.edu.cn/panCancer.php) is designed for decoding the pan-cancer networks of long noncoding RNAs (lncRNAs), miRNAs, pseudogenes, small nucleolar RNAs (snoRNAs), RNA-binding proteins (RBPs), and all protein-coding genes by analyzing their expression profiles across 32 cancer types (~10,000 RNA-sequencing (RNA-seq) and ~9900 miRNA-seq samples) integrated from The Cancer Genome Atlas (TCGA) project. We used this website to analyze the expression of miR-93-5p in cancer tissue and normal tissues as well as the correlation between miR-93-5p and downstream target genes. The target genes of miR-93-5p were predicted by miRTargetLink (https://ccb-web.cs.uni-saarland.de/mirtargetlink/network.php). Metascape (https://metascape.org/) was used for Kyoto Encyclopedia of Genes and Genomes (KEGG) and Gene Ontology (GO) analysis of the target genes of miR-93-5p. GeneMANIA (https://genemania.org) was used to analyze the functions of 18 strongly related target genes.
Isolation of exosomes
Exosomes were isolated at 4 ℃ by differential centrifugation from the cell culture medium (1,000 g for 5 min for removal of cells and 3,000 g for 5 min for removal of cell debris). After centrifugation at 10,000 g for 20 min for removal of the larger vesicles, the supernatant was centrifuged at 11,000 g for 70 min for the collection of the exosomes. Subsequently, 100 µL of phosphate-buffered saline (PBS) was used to resuspend the isolated exosomes.
Transmission electron microscopy
Exosomes were collected by differential centrifugation and fixed in 2.5% glutaraldehyde overnight at 4 ℃. Next, 5–10 µL of exosomal solution was dripped to the copper mesh and absorbed at room temperature for about 10 min, after which any excess liquid was carefully removed with filter paper. Following this, 10 µL of 2% phosphotungstic acid solution (pH=6.5) was used to stain the exosomes for 2 min at room temperature, which were then dried in a copper mesh at room temperature. Finally, the processed sample was observed with transmission electron microscopy (TEM) under 120 kV voltage.
Luciferase assay
miRTargetLink Human (ccb-web.cs.uni-saarland.de/mirtargetlink/index) was used to predict the miR-93-5p target gene and site. PTEN 3’UTR wild type and PTEN 3'UTR mutant were cloned into the psi-check-2 vector. miR-93-5p mimics or negative control and PTEN 3'UTR wild type or PTEN 3'UTR mutant were cotransfected into 5637 and HUVECs. A dual-luciferase reporter assay kit (Promega Corporation) was used to detect the binding of miR-93-5p and PTEN.
Establishment of tumor xenografts in mice
Female nude mice (BALB/c-nu, 6–8 weeks, 18–20 g) were purchased from the Model Animal Center of Soochow University, and they were fed in a specific-pathogen-free animal facility. According to the minimum number requirements of previous animal experiments, the number of animals = the number of groups + 10. The mice were randomly divided into 2 groups of 6, with only 1 researcher (FY) being aware of the group assignment of the mice. Following this, 5637 cells were subcutaneously injected into mice (3×106 cells in 0.2 mL of PBS per mouse; 6 mice per group). A total of 60 µg exosomes per mouse was injected intratumourally every 3 days (at this maximum dose, mice and subcutaneous tumors can grew well), and the tumor volume of the mice was measured every 6 days until the termination of the experiment. Mice were killed 24 days after the injection for removal of the xenografted tumors, and the volumes and weights of the tumors were recorded. Tumor volume was calculated with the following formula: 0.5× length × width2. Immunohistochemical staining in tumor tissue with CD31 was used to compare the density of the tumor vascularity. A protocol was prepared before the study without registration. Animal experiments were approved by ethical committee of the Second Affiliated Hospital of Soochow University, in compliance with institutional guidelines for the care and use of animals.
Immunohistochemistry
Immunohistochemical staining was performed at the Department of Pathology in The Second Affiliated Hospital of Soochow University. The tumors were fixed in 4% paraformaldehyde, embedded in paraffin, sectioned, and then stained with 1,2-dibromoethane–conjugated anti-CD31 antibody (Abcam). The fluorescence was measured from at least 3 sections.
Cellular internalization of exosomes
Exosomes from 1.5×106 cells were suspended in 100 µL of PBS with 1 mL of mixed PKH67 (in diluent C; Sigma-Aldrich), resuspended in 10% exosome-depleted FBS serum-free medium, and then added to the HUVECs at 60% confluence. To stain the nuclei, 4',6-diamidino-2-phenylindole (DAPI, Sigma-Aldrich) was added for 10 min, and the stained cells were observed with a fluorescence microscope.
Statistical analysis
The associations of the expression of miR-93-5p with clinical data were evaluated by the chi-squared test or the Mann-Whitney test. Subsequent experimental results are presented as the average of at least 3 experiments with standard error. Data are described with median values ± standard error of the mean (SEM) and analyzed with using Student t-test for 2-group comparisons. A P value <0.05 was considered to be statistically significant. Statistical analyses and graphical presentations were conducted with GraphPad Prism 7.0 (GraphPad Software).
Results
Exosomal miR-93-5p was associated with the clinical progression of BLCA
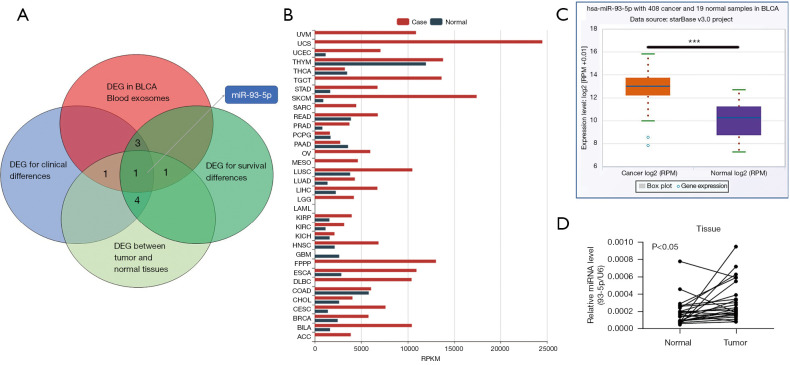
We obtained miRNAs from the oncomiR database with high relevance for survival and clinical staging. Among them, the expression of 4 miRNAs in normal tissues and tumor tissues was significantly different. A study by Armstrong et al. (16) has shown that among these miRNAs, miR-93-5p is expressed significantly differently BLCA exosomes, which may be an important factor in the exertion of BLCA exosomes (Figure 1A). Notably, high miR-93-5p expression was found in most tumors in TCGA database (Figure 1B). Our results confirmed data from TCGA that showed the expression of miR-93-5p to be significantly higher in cancer tissues (Figure 1C). The relative miR-93-5p expression of 25 paired BLCA and normal bladder tissues was detected by qRT-PCR. The expression of miR-93-5p in BLCA tissue was significantly higher than that in matched normal tissue (Figure 1D). Furthermore, the correlation analysis between the expression of miR-93-5p and clinical data showed that miR-93-5p was significantly related to the tumor stage and lymph node metastasis of the patients (Table 2).
Figure 1.
miR-93-5p had significant clinical relevance and differential expression in BLCA. (A) The online website based on the TCGA database selected 144 survival-related DEG, 155 clinical stage-related genes, and 25 genes with the most significant expression differences. These genes intersected with the 25 genes that were most significantly elevated in exosomes in patients with BLCA. (B) The expression of miR-93-5p in different cancer and normal tissues in TCGA database. (C) The expression of miR-93-5p in BLCA and paracancerous tissues in TCGA database. (D) The expression of miR-93-5p in tumor and paracancerous tissues in 25 paired samples collected by our team. ***, P<0.001. TCGA, The Cancer Genome Atlas; BLCA, bladder cancer; DEG, differentially expressed genes; RPKM, reads per kilobase per million mapped reads; RPM, reads per million mapped reads.
Table 2. Relationship between miR-93-5p expression and clinicopathologic factors in 25 patients.
| Parameters | N | Relative miR-93-5p expression | P value |
|---|---|---|---|
| Age (years) | 0.641 | ||
| <60 | 10 | 0.0003481±7.043e-005 | |
| ≥60 | 15 | 0.0003042±5.936e-005 | |
| Gender | 0.849 | ||
| Male | 23 | 0.0003243±4.822e-005 | |
| Female | 2 | 0.0002919±9.9e-005 | |
| Grade | 0.146 | ||
| Low | 7 | 0.0002166±6.561e-005 | |
| High | 18 | 0.0003626±5.452e-005 | |
| Pathologic T stage | 0.008* | ||
| pT1-T2 | 18 | 0.0002506±3.963e-005 | |
| pT3-T4 | 7 | 0.0005046±9.666e-005 | |
| Pathologic N stage | 0.0311* | ||
| N0 | 19 | 0.0002686±4.157e-005 | |
| N1 | 6 | 0.00049±1.131 e-006 | |
Data are described with median ± standard error of the mean. *, P<0.05.
Exosomes in high-grade tumors cell carried more miR-93-5p
Exosomes were isolated by ultracentrifugation from HUVECs and different BLCA cell media (Figure 2). We observed round-like extracellular vesicles through TEM (Figure 2A). Particle size analysis showed that the diameter of the extracellular vesicles was about 100 nm (Figure 2B), and exosomes were verified by protein markers TSG101 and CD63 (Figure 2C). The relative expression of miR-93-5p in exosomes was then quantified by qRT-PCR. Results showed that the relative expression of exosomal miR-93-5p was higher in J82 and T24 (BLCA cell lines) than in low-grade cell lines (Figure 2D). Interestingly, the HUVEC exosomes barely contained miR-93-5p. This suggested that exosomal miR-93-5p may play a critical role in tumor development.
Figure 2.
miR-93-5p as an important transmitter in exosomes secreted by BLCA cells. (A) Scanning electron microscopy images of exosomes isolated from T24 cells; scale bars: 100 nm (B) The diameter of extracellular vesicles in Figure A as determined by particle size analysis. (C) Western blotting images of exosome markers TSG101 and CD63 in T24 cells and T24 exosomes (exo). (D) The expression of miR-93-5p in exosomes secreted by BLCA cells and HUVECs. (E) The expression of miR-93-5p in exosomes secreted by T24 cells with transfection of mimics. Exosomes transfected with miR-93-5p (exo-93-5p) expressed more miR-93-5p than exosomes transfected with negative control (exo-NC). (F) Exosomes from T24 cells fused with HUVECs. DAPI and PKH67 were used to stain HUVECs and exosomes, respectively, and exosomes were found to enter HUVECs at 6 h; scale bars: 25 µm. ***, P<0.001. BLCA, bladder cancer; HUVEC, human umbilical vein endothelial cells; DAPI, 4',6-diamidino-2-phenylindole.
miR-93-5p transferred by exosomes promoted the proliferation, migration, and invasion of BLCA cells
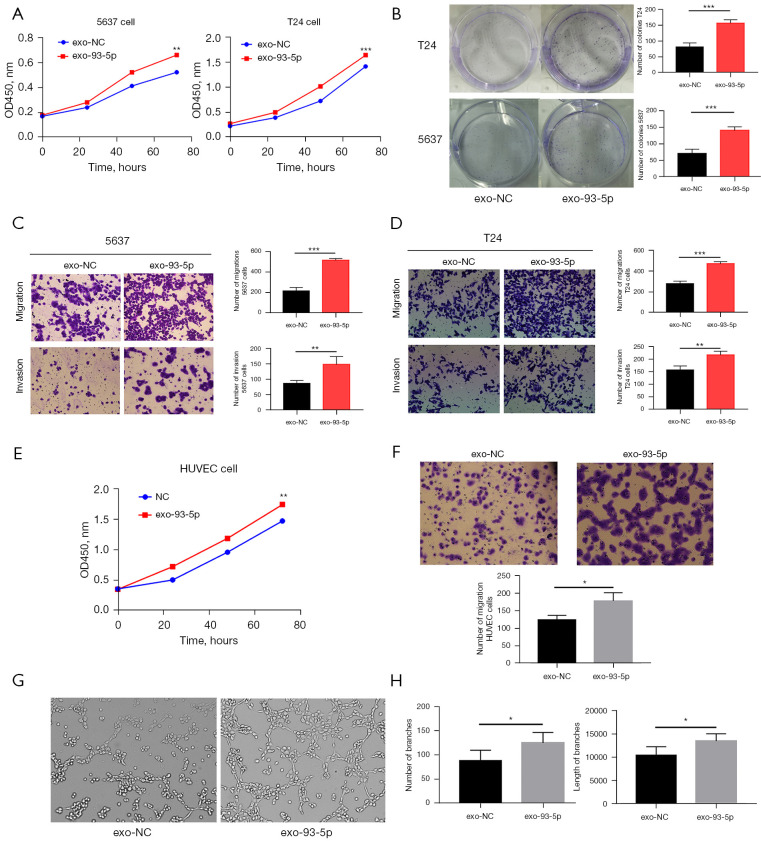
To study the role of exosomal miR-93-5p in BLCA cells, we used mimics of miR-93-5p and (NC) to transfect T24 cells. The T24 cell exosomes showed higher expression of miR-93-5p after transfection with miR-93-5p mimics compared with the control group (Figure 2E). We cultured processed exosomes with 5637 and T24 cells to determine the role of exosomes in cell proliferation and invasion. After 24-h coculture with different processed exosomes, we performed CCK-8 assay, Transwell migration, and invasion assay to detect the proliferation and metastatic abilities of 5637 and T24 cells.
CCK8 assay showed that the proliferation rate of 5637 and T24 cells cocultured with miR-93-5p-exosomes was significantly accelerated compared with that of the control group (Figure 3A). Colony-forming assay revealed the promotive proliferation function of exosomal miR-93-5p in BLCA cells (Figure 3B), while Transwell assay revealed a higher migration and invasion rate of the cancer cells (both of T24 and 5637) as well as an overexpression of miR-93-5p-exosomes as compared to the control group (Figure 3C,3D).
Figure 3.
The role of BLCA exosomes in cancer cells and angiogenesis. T24 cells, 5637 cells, and HUVECs were incubated with the exosomes above (Figure 2E), exosomes transfected with miR-93-5p (exo-93-5p) and exosomes transfected with negative control (exo-NC), and the migration, invasion, and proliferation of the HUVECs were assessed at 24 h. (A) T24 cells with exosomal miR-93-5p promoted the proliferation of BLCA cells. (B) The proliferation rate of T24 and 5637 cells measured by colony-forming assays. (C) Exosomal miR-93-5p promoted the cell migration and invasion in 5637 cells (magnification: 100×). (D) Exosomal miR-93-5p promoted the cell migration and invasion of T24 cells (magnification: 100×). (E) T24 cells with exosomal miR-93-5p promoted the proliferation of HUVECs. (F) Transwell assay detected the migration of exosomal miR-93-5p in HUVECs (magnification: 100×). (G) Exosomal miR-93-5p derived from T24 cells promoted the angiogenesis of HUVECs in vitro (magnification: 100×). (H) Quantitative analysis of angiogenesis data by ImageJ software. *, P<0.05, **, P<0.01, and ***, P<0.001. 0.1% crystal violet solution was used for cell staining in B-D and F. BLCA, bladder cancer; HUVEC, human umbilical vein endothelial cells.
The results above indicated that BLCA exosomes can transmit information between different tumor cells, with miR-93-5p playing a critical role in this process as a cancer promoting factor.
T24. cells exosomal miR-93-5p- promoted the cell proliferation, migration, and angiogenesis of HUVECs
In this study, we stained exosomes and HUVECs with PKH67 and DAPI respectively and then cocultured them. Result showed that the exosomes released by T24 cells entered the HUVECs after 6 h of coculture. This observation suggests that HUVECs can ingest exosomes secreted by T24 cells (Figure 2F). Following this, HUVECs were co-cultured with NC and miR-93-5p exosomes. Cell proliferation, invasion, and angiogenesis abilities was determined by Cell Counting Kit 8 (CCK-8), colony-forming assay, Transwell assay, and vascular ring formation assay. Coculture with miR-93-5p exosomes exhibited increased HUVEC proliferation and migration (Figure 3E,3F). Furthermore, vascular ring formation assay showed an increased length and number of branches in HUVECs cultured with miR-93-5p-exosomes as compared with the control group (Figure 3G,3H). These results suggested that miR-93-5p-exosomes could facilitate the cell proliferation, migration. and angiogenesis of HUVECs in vitro.
Bioinformation analysis of the target genes of miR-93-5p
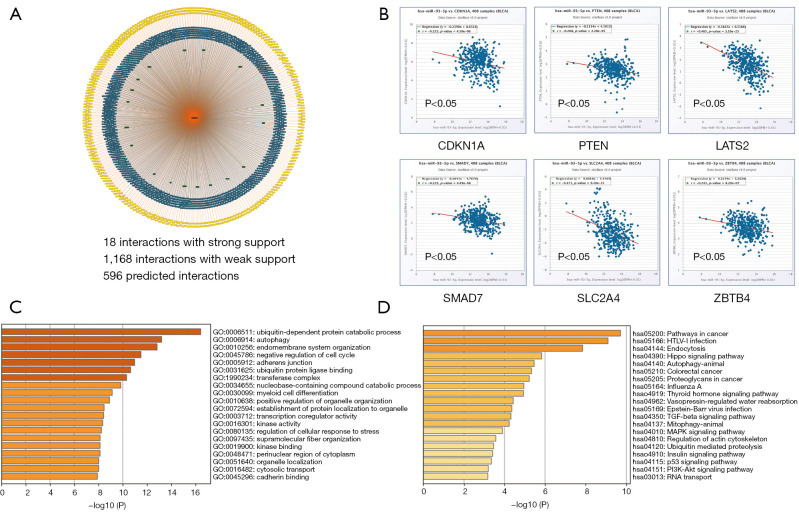
The target genes of miR-93-5p were predicted through miRTargetLink Human. Result showed that 18 genes were strongly interacted with miR-93-5p, 1168 genes were weakly interacted with miR-93-5p, and 596 predicted genes may be interacted with miR-93-5p (Figure 4A). The 18 strongly related genes were TP53INP1, CDKN1A, E2F1, MAPK9, VEGFA, ITGB8, KAT2B, TUSC2, PTEN, PURA, LATS2, TGFBR2, SLC2A4, ATG16L1, DAB2, SMAD7, ZBTB4, and IL8. Further correlation analysis showed that there were 6 genes negatively correlated with the expression of miR-93-5p in BLCA (Figure 4B). GO and KEGG enrichment analysis of candidate target genes performed by Metascape indicated 18 strongly related genes and 1,168 weakly related genes.
Figure 4.
Target genes predicted by bioinformatics. (A) The target genes of miR-93-5p were predicted by miRTargetLink. (B) Co-expression analysis for miR-93-5p and the predicted gene. (C) GO analysis of the 1,000 predicted genes. (D) 1,000 predicted genes were analyzed in KEGG. KEGG, Kyoto Encyclopedia of Genes and Genomes; GO, Gene Ontology.
GO analysis showed that candidate genes were related to 162 GO classifications. The molecular functions involved ubiquitin protein ligase activity and binding, transcription coregulator activity, protein kinase activity, and cadherin binding, among others. The cell components involved adherent junction, transferase complex, and the perinuclear region of cytoplasm. The biological processes involved ubiquitin-dependent protein catabolic process, autophagy, endomembrane system organization, and negative regulation of cell cycle, among others (Figure 4C). KEGG pathway analysis showed that miR-93-5p target genes were significantly involved in the pathways in cancer, Hippo signaling pathway, TGF-β pathway, MAPK signaling pathway, and P53 and PI3K-Akt signaling pathway, among others (Figure 4D).
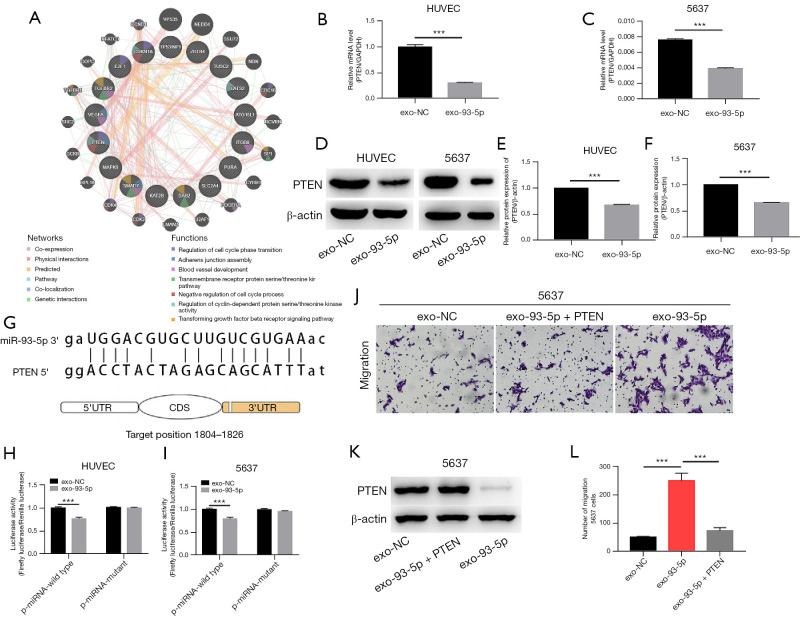
We used GeneMANIA to analyze the function of 18 strongly related target genes, and the results showed that several candidate genes are closely related to cell cycle, adhesion junction, and angiogenesis. PTEN, as an inhibitor of PI3K-Akt signaling pathway, was mainly involved in these biological functions (Figure 5).
Figure 5.
Exosomal miR-93-5p targeted PTEN directly in HUVECs and BLCA cells. (A) Analysis of the function of 18 strongly related genes in GeneMANIA. (B) The expression of PTEN in HUVECs was determined by qRT-PCR, after transfected with the exosomes above (Figure 2E), exosomes transfected with miR-93-5p (exo-93-5p) and exosomes transfected with negative control (exo-NC). (C) The expression of PTEN in 5637 cells was determined by qRT-PCR. (D) Western blotting analyzed the PTEN in each group. (E, F) Quantitative analysis of PTEN protein expression with ImageJ software. (G) The predicted miR-93-5p-binding sites in the 3'UTR of PTEN mRNA. The miRNA that potentially interacts with PTEN was calculated with miRTargetLink. (H, I) Relative levels of luciferase in 5637 cells and HUVECs cotransfected with firefly luciferase reporter plasmid containing either wild-type (WT) or mutant PTEN 3'UTR and exo-93-5p. (J-L) Transwell assay in 5637 cells that were cotransfected with a PTEN-overexpression plasmid and exo-93-5p (Figure magnification: 100×). 0.1% crystal violet solution was used for cell staining in (J). ***, P<0.001. PTEN, phosphatase and tensin homolog; BLCA, bladder cancer; HUVEC, human umbilical vein endothelial cells.
Exosomes transported miR-93-5p into 5637 cells and HUVECs and degraded PTEN directly
To explore the regulatory effect of miR-93-5p on PTEN in BLCA, we confirmed the silencing effect of miR-93-5p on PTEN gene at the mRNA level (Figure 5B,5C). Then, we verified the direct regulatory effect of miR-93-5p on PTEN with a luciferase reporter assay. Based on the prediction of the miR-93-5p binding sites on PTEN by miRTargetLink, we mutated the PTEN binding site in the base sequence and then performed a luciferase assay. We then cotransfected the mutated PTEN reporter and miR-93-5p into HUVECs and 5637 cells. The results showed that the luciferase activity was significantly inhibited when miR-93-5p mimics were cotransfected with the PTEN wild type, but the functionality was lost when PTEN binding site was mutated (Figure 5G-5I). Furthermore, the PTEN protein was significantly downregulated in 5637 cells and HUVECs after miR-93-5p-exosome administration for 36 h (Figure 5D-5F), and the downregulation of PTEN could be partially reversed by artificial PTEN overexpression (Figure 5J-5L).
Exosome-mediated delivery of miR-93-5p induced angiogenesis and tumor growth in vivo
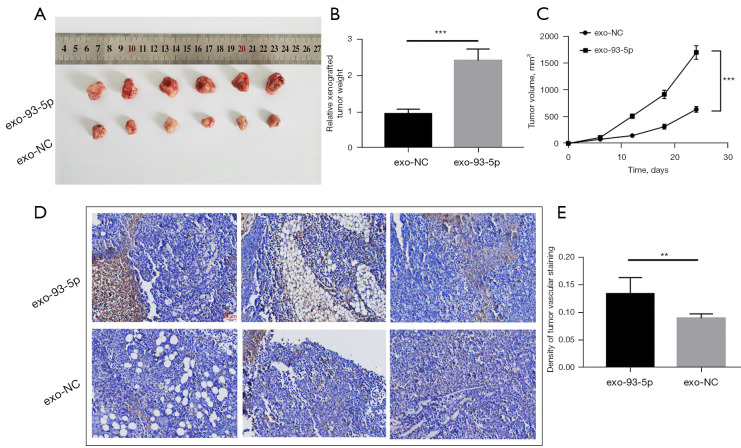
We tested whether cancer-derived exosomes enriched in miR-93-5p can function as angiogenesis and tumorigenesis promoters by performing experiments in vivo. Consequently, 5637 cells were subcutaneously injected into BALB/c mice to establish the experimental group and control group. We then stimulated tumor growth through intratumoral injection of the corresponding exosomes mentioned above, and 24 days later, the tumors were harvested by surgical removal (Figure 6A). We regularly measured the volume of tumor in mice. Tumors stimulated by exo-93-5p presented with a high rate of tumor growth (Figure 6B,6C). From a histological perspective, cancer angiogenesis was detected by immunohistochemistry using CD31. The expression intensity of CD31 was used to measure the angiogenesis rate in cancer tissue. Our experiments revealed that compared with angiogenesis intensity in the control group, the angiogenesis intensity in the exo-93-5p group was strengthened as indicated by the enhanced expression of CD31 (Figure 6D,6E).
Figure 6.
In vivo verification for T24 exosome-derived miR-93-5p promotion of angiogenesis and tumor growth. (A) Tumor tissues excised from tumor-implanted mice (n=6), which were received intra-tumoral injection of the exosomes above (Figure 1F) respectively, exosomes transfected with miR-93-5p (exo-93-5p) and exosomes transfected with negative control (exo-NC). (B, C) Quantitative analysis of xenografted tumor weight (B) and volume (C) (n=6). (D, E) Immunohistochemical analysis (D) of the paraffin-embedded tumor tissues with a CD31 antibody (n=6; CD31 is widely used to demonstrate the existence of endothelial tissue and to assess tumor angiogenesis) and the corresponding quantification (E); scale bars: 50 µm. **, P<0.01, and ***, P<0.001.
Discussion
Although surgery combined with radiotherapy and chemotherapy can control most cases of BLCA, effective treatment strategy or targets are still lacking for locally advanced and metastatic tumors (17). Abundant clinical data show that tumor metastasis is closely related to the prognosis of patients (18), and patients with metastatic BLCA have a shorter survival and poor prognosis. Cancer invasion ability and its influence on the surrounding angiogenesis are closely related to clinical metastasis (19-21). Understanding the mechanisms of BLCA metastasis and angiogenesis may lead to the identification of meaningful intervention targets and provide new strategies for guiding the prognosis and treatment of BLCA.
Cancer-derived exosomes play a vital role in the tumor microenvironment. They act as carriers that deliver factors to tumor cell spaces and surrounding structures, which may promote the generation of the tumor microenvironment. Exosomes are involved in many aspects of tumor progression, such as tumor metastasis, recurrence (22), drug resistance (23), and immune response (24). A growing body of evidence shows that exosomes can be used as predictors of tumor prognosis and targets for therapeutic intervention in numerous cancers (25).
Exosomes carry biologically active molecules to perform diverse functions. Among them, miRNA is the most studied molecule. Once entering cells through exosomes, exosomal miRNA can inhibit the transcription of target genes, thereby regulating the proliferation and metastasis of tumor cells. Studies show that miR-93-5p acts as a accelerator in the progression of many tumors, such as esophageal squamous cancer (26) and small cell lung cancer (27), and it can be considered a hallmark to guide treatment and prognosis. Yang et al. showed that miR-93-5p is correlated with tumor progression and immune regulation in BRCA and that it directly targets PD-L1 (programmed cell death 1 ligand 1) and Cyclin D1 to execute its functions (28). However, miR-93-5p may play an opposing role in other tumors. Wu et al. showed that miR-93-5p was downregulated in glioma and could suppress the proliferation and metastasis of glioma cells by targeting MMP2 (matrix metallopeptidase 2) (29). In another study, Xiang et al. demonstrated miR-93-5p inhibits the Epithelial-mesenchymal transition (EMT) of breast cancer cells through suppressing the expression of MKL-1 (Megakaryoblastic leukemia 1) and STAT3 (activator of transcription 3) (30). According to these studies, the expression levels and functions of miR-93-5p may vary in different tumors and may control signal network nodes to exert its function.
Research found that the expression level of miR-31-5p, miR-93-5p, and miR-191-5p was significantly higher than other miRNAs in the urine of patients with BLCA, and they can be used as a noninvasive detection method for these patients (31). Our study found that the expression of miR-93-5p was significantly related to the T and N staging of BLCA. Lin et al. reported that miR-93-5p could facilitate proliferation, migration, and invasion in BLCA cells by targeting BTG2 (15); however, their study did not examine the role of exosomes in the potential diagnosis and treatment of BLCA. Previous experiments also lacked the corroboration of experiments in vivo, which is an indispensable prerequisite for clinical application. Our team successfully constructed exosomes with a high expression of miR-93-5p from T24 cells. More importantly, we demonstrated that exosomes secreted by T24 cells can communicate between tumor cells and vascular endothelial cells. Subsequent, in vitro and in vivo experiments showed that exosomes expressing high miR-93-5p could promote the proliferation, invasion, and angiogenesis of BLCA
PTEN is a cancer-related transcription factor that negatively regulates the proliferation and invasion of BLCA cells via PI3K/Akt/mTOR pathway (32). Downregulation of PTEN is associated with poor prognosis, chemoresistance, and the progression of cancer cells (33). Notably, antitumor drugs, such as kaempferol and sorafenib, inhibit BLCA cell proliferation and invasion through upregulating PTEN (34,35). We predicted 18 downstream target genes by bioinformatics. The expression levels of 6 genes among them were negatively correlated with miR-93-5p. Gene functional analysis revealed that PTEN and SMAD7 may play important roles in patients with BLCA. Further mRNA quantitative analysis showed that after overexpression of miR-93-5p in BLCA, the expression of SMAD7 did not change significantly, while the expression of PTEN was significantly downregulated. Based on a literature review, we came to speculate that PTEN is an important target of miR-93-5p in BLCA, and this was further confirmed by the luciferase reporter assay and the Western blotting assay.
In brief, our team found that miR-93-5p is highly expressed in tumor tissues and is closely related to the pathological stage of patients. The expression of miR-93-5p was higher in exosomes from malignant cells, while the exosomes derived from HUVECs had an extremely low expression of miR-93-5p. Furthermore, exosomal miR-93-5p can was found to promote the proliferation, migration, and invasion of BLCA cells and facilitate angiogenesis in vitro. Further in vivo experiments yielded similar results. Moreover, PTEN was found to be a vital downstream target of miR-93-5p in BLCA cells. However, the role of exosomal miR-93-5p in the tumor microenvironment and its specific mechanisms still need to be clarified by further research.
Conclusions
Our study successfully revealed the biological role and mechanism of BLCA-derived exosomes in tumor progression. Target at tumor exosomes and exosomal miR-93-5p may be an effective treatment in BLCA.
Supplementary
The article’s supplementary files as
Acknowledgments
Funding: This study was supported in part by funding from the Youth Excellence Projects of Suzhou Health (No. GSWS2020031) and pre-research fund of the Second Affiliated Hospital of Soochow University (No.SDFEYQN2014).
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). This study was approved by ethical committee of the Second Affiliated Hospital of Soochow University (ethical code: JD-LK-2019-104-01), and informed consent was taken from all the patients. Animal experiments were approved by ethical committee of the Second Affiliated Hospital of Soochow University, in compliance with institutional guidelines for the care and use of animals.
Reporting Checklist: The authors have completed the ARRIVE reporting checklist. Available at https://tau.amegroups.com/article/view/10.21037/tau-22-872/rc
Data Sharing Statement: Available at https://tau.amegroups.com/article/view/10.21037/tau-22-872/dss
Peer Review File: Available at https://tau.amegroups.com/article/view/10.21037/tau-22-872/prf
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://tau.amegroups.com/article/view/10.21037/tau-22-872/coif). The authors have no conflicts of interest to declare.
(English Language Editor: J. Gray)
References
- 1.Sung H, Ferlay J, Siegel RL, et al. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin 2021;71:209-49. 10.3322/caac.21660 [DOI] [PubMed] [Google Scholar]
- 2.Fonteyne V, Ost P, Bellmunt J, et al. Curative Treatment for Muscle Invasive Bladder Cancer in Elderly Patients: A Systematic Review. Eur Urol 2018;73:40-50. 10.1016/j.eururo.2017.03.019 [DOI] [PubMed] [Google Scholar]
- 3.Talib WH, Alsayed AR, Farhan F, et al. Resveratrol and Tumor Microenvironment: Mechanistic Basis and Therapeutic Targets. Molecules 2020;25:4282. 10.3390/molecules25184282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mao Y, Wang Y, Dong L, et al. Hypoxic exosomes facilitate angiogenesis and metastasis in esophageal squamous cell carcinoma through altering the phenotype and transcriptome of endothelial cells. J Exp Clin Cancer Res 2019;38:389. 10.1186/s13046-019-1384-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hu JL, Wang W, Lan XL, et al. CAFs secreted exosomes promote metastasis and chemotherapy resistance by enhancing cell stemness and epithelial-mesenchymal transition in colorectal cancer. Mol Cancer 2019;18:91. 10.1186/s12943-019-1019-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang L, Yu D. Exosomes in cancer development, metastasis, and immunity. Biochim Biophys Acta Rev Cancer 2019;1871:455-68. 10.1016/j.bbcan.2019.04.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Han M, Hu J, Lu P, et al. Exosome-transmitted miR-567 reverses trastuzumab resistance by inhibiting ATG5 in breast cancer. Cell Death Dis 2020;11:43. 10.1038/s41419-020-2250-5 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 8.Ni L, Xu J, Zhao F, et al. MiR-221-3p-mediated downregulation of MDM2 reverses the paclitaxel resistance of non-small cell lung cancer in vitro and in vivo. Eur J Pharmacol 2021;899:174054. 10.1016/j.ejphar.2021.174054 [DOI] [PubMed] [Google Scholar]
- 9.Liu MX, Liao J, Xie M, et al. miR-93-5p Transferred by Exosomes Promotes the Proliferation of Esophageal Cancer Cells via Intercellular Communication by Targeting PTEN. Biomed Environ Sci BES. 2018;31:171-85. [DOI] [PubMed] [Google Scholar]
- 10.Wang QZ, Zhao ZL, Liu C, et al. Exosome-derived miR-196b-5p facilitates intercellular interaction in infantile hemangioma via down-regulating CDKN1B. Ann Transl Med 2021;9:394. 10.21037/atm-20-6456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liao W, He J, Disoma C, et al. Hsa_circ_0107593 Suppresses the Progression of Cervical Cancer via Sponging hsa-miR-20a-5p/93-5p/106b-5p. Front Oncol 2021;10:590627. 10.3389/fonc.2020.590627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shi X, Liu TT, Yu XN, et al. microRNA-93-5p promotes hepatocellular carcinoma progression via a microRNA-93-5p/MAP3K2/c-Jun positive feedback circuit. Oncogene 2020;39:5768-81. 10.1038/s41388-020-01401-0 [DOI] [PubMed] [Google Scholar]
- 13.Santangelo A, Rossato M, Lombardi G, et al. A molecular signature associated with prolonged survival in glioblastoma patients treated with regorafenib. Neuro Oncol 2021;23:264-76. 10.1093/neuonc/noaa156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen X, Liu J, Zhang Q, et al. Exosome-mediated transfer of miR-93-5p from cancer-associated fibroblasts confer radioresistance in colorectal cancer cells by downregulating FOXA1 and upregulating TGFB3. J Exp Clin Cancer Res 2020;39:65. 10.1186/s13046-019-1507-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lin H, Shi X, Li H, et al. Urinary Exosomal miRNAs as biomarkers of bladder Cancer and experimental verification of mechanism of miR-93-5p in bladder Cancer. BMC Cancer 2021;21:1293. 10.1186/s12885-021-08926-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Armstrong DA, Green BB, Seigne JD, et al. MicroRNA molecular profiling from matched tumor and bio-fluids in bladder cancer. Mol Cancer 2015;14:194. 10.1186/s12943-015-0466-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ghandour R, Singla N, Lotan Y. Treatment Options and Outcomes in Nonmetastatic Muscle Invasive Bladder Cancer. Trends Cancer 2019;5:426-39. 10.1016/j.trecan.2019.05.011 [DOI] [PubMed] [Google Scholar]
- 18.Jin X, Demere Z, Nair K, et al. A metastasis map of human cancer cell lines. Nature 2020;588:331-6. 10.1038/s41586-020-2969-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zeng Z, Li Y, Pan Y, et al. Cancer-derived exosomal miR-25-3p promotes pre-metastatic niche formation by inducing vascular permeability and angiogenesis. Nat Commun 2018;9:5395. 10.1038/s41467-018-07810-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Niu W, Luo Y, Zhou Y, et al. BRD7 suppresses invasion and metastasis in breast cancer by negatively regulating YB1-induced epithelial-mesenchymal transition. J Exp Clin Cancer Res 2020;39:30. 10.1186/s13046-019-1493-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wu Y, Tan X, Liu P, et al. ITGA6 and RPSA synergistically promote pancreatic cancer invasion and metastasis via PI3K and MAPK signaling pathways. Exp Cell Res 2019;379:30-47. 10.1016/j.yexcr.2019.03.022 [DOI] [PubMed] [Google Scholar]
- 22.Ge Y, Mu W, Ba Q, et al. Hepatocellular carcinoma-derived exosomes in organotropic metastasis, recurrence and early diagnosis application. Cancer Lett 2020;477:41-8. 10.1016/j.canlet.2020.02.003 [DOI] [PubMed] [Google Scholar]
- 23.Jena BC, Mandal M. The emerging roles of exosomes in anti-cancer drug resistance and tumor progression: An insight towards tumor-microenvironment interaction. Biochim Biophys Acta Rev Cancer 2021;1875:188488. 10.1016/j.bbcan.2020.188488 [DOI] [PubMed] [Google Scholar]
- 24.Yin T, Xin H, Yu J, et al. The role of exosomes in tumour immunity under radiotherapy: eliciting abscopal effects? Biomark Res 2021;9:22. 10.1186/s40364-021-00277-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yu W, Hurley J, Roberts D, et al. Exosome-based liquid biopsies in cancer: opportunities and challenges. Ann Oncol 2021;32:466-77. 10.1016/j.annonc.2021.01.074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Su X, Xue C, Xie C, et al. lncRNA-LET Regulates Glycolysis and Glutamine Decomposition of Esophageal Squamous Cell Carcinoma Through miR-93-5p/miR-106b-5p/SOCS4. Front Oncol 2022;12:897751. 10.3389/fonc.2022.897751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu W, Liang F, Yang G, et al. LncRNA LINC01116 sponges miR-93-5p to promote cell invasion and migration in small cell lung cancer. BMC Pulm Med 2021;21:50. 10.1186/s12890-020-01369-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yang M, Xiao R, Wang X, et al. MiR-93-5p regulates tumorigenesis and tumor immunity by targeting PD-L1/CCND1 in breast cancer. Ann Transl Med 2022;10:203. 10.21037/atm-22-97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wu H, Liu L, Zhu JM. MiR-93-5p inhibited proliferation and metastasis of glioma cells by targeting MMP2. Eur Rev Med Pharmacol Sci 2019;23:9517-24. [DOI] [PubMed] [Google Scholar]
- 30.Xiang Y, Liao XH, Yu CX, et al. MiR-93-5p inhibits the EMT of breast cancer cells via targeting MKL-1 and STAT3. Exp Cell Res 2017;357:135-44. 10.1016/j.yexcr.2017.05.007 [DOI] [PubMed] [Google Scholar]
- 31.Juracek J, Peltanova B, Dolezel J, et al. Genome-wide identification of urinary cell-free microRNAs for non-invasive detection of bladder cancer. J CELL MOL MED 2018;22(3):2033-8. 10.1111/jcmm.13487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ashrafizadeh M, Zarrabi A, Samarghandian S, et al. PTEN: What we know of the function and regulation of this onco-suppressor factor in bladder cancer? Eur J Pharmacol 2020;881:173226. 10.1016/j.ejphar.2020.173226 [DOI] [PubMed] [Google Scholar]
- 33.Kashiwagi E, Inoue S, Mizushima T, et al. Prostaglandin receptors induce urothelial tumourigenesis as well as bladder cancer progression and cisplatin resistance presumably via modulating PTEN expression. Br J Cancer 2018;118:213-23. 10.1038/bjc.2017.393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cao JY, Yin HS, Li HS, et al. Interleukin-27 augments the inhibitory effects of sorafenib on bladder cancer cells. Braz J Med Biol Res 2017;50:e6207. 10.1590/1414-431x20176207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xie F, Su M, Qiu W, et al. Kaempferol promotes apoptosis in human bladder cancer cells by inducing the tumor suppressor, PTEN. Int J Mol Sci 2013;14:21215-26. 10.3390/ijms141121215 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The article’s supplementary files as